Abstract
Eosinophilic esophagitis (EoE) is an emerging chronic inflammatory disease of the oesophagus and is clinically characterized by upper gastrointestinal (GI) symptoms including dysphagia and esophageal food impaction. Histopathologic manifestations, which include intraepithelial eosinophilic inflammation and alterations of the esophageal squamous epithelium, such as basal zone hyperplasia (BZH) and dilated intercellular spaces (DIS), are thought to contribute to esophageal dysfunction and disease symptoms. Corroborative clinical and discovery science‐based studies have established that EoE is characterized by an underlying allergic inflammatory response, in part, related to the IL‐13/CCL26/eosinophil axis driving dysregulation of several key epithelial barrier and proliferative regulatory genes including kallikrein (KLK) serine proteases, calpain 14 (CAPN14) and anoctamin 1 (ANO1). The contribution of these inflammatory and proliferative processes to the clinical and histological manifestations of disease are not fully elucidated. Herein, we discuss the immune molecules and cells that are thought to underlie the clinical and pathologic manifestations of EoE and the emerging therapeutics targeting these processes for the treatment of EoE.
Keywords: eosinophilic esophagitis, cytokines, mast cell, eosinophils, biologics
Key messages.
Eosinophilic esophagitis (EoE) is a chronic inflammatory disease of the oesophagus characterized epithelial remodelling that drives esophageal dysfunction and disease symptoms.
Dietary food antigens stimulate a pro‐type 2 inflammatory response in the esophageal mucosa driving dysregulation of esophageal epithelial barrier and proliferative regulatory genes and epithelial remodelling.
Biologics targeting key aspects of Type‐2 immune signals (IL‐4, IL‐5, IL‐5Rα and IL‐13) are emerging treatment modalities for the treatment of EoE.
1. INTRODUCTION
Eosinophilic esophagitis (EoE) is an emerging chronic inflammatory disease of the oesophagus characterized by upper gastrointestinal (GI) symptoms including dysphagia and esophageal food impaction. 1 , 2 The histopathological manifestations involve intraepithelial infiltration of eosinophils (≥15 Eos/HPF) and remodelling of the esophageal epithelium including basal zone hyperplasia (BZH) and dilated intercellular spaces (DIS), which can lead to strictures and narrow‐caliber oesophagus. 3 , 4
Eosinophilic esophagitis is a complex disease characterized by heterogeneous clinical presentation (age of onset, symptomology, varying clinical manifestations and comorbidities, natural history, and responsiveness to therapy). 5 Despite challenges in disease diagnosis and management, there is corroborative clinical and experimental evidence to suggest that EoE is driven by an underlying CD4+ T helper type 2 (Th2) allergic inflammatory response to dietary food allergens in the esophageal mucosa. 6 , 7 , 8 , 9 , 10 Esophageal epithelial derived signals (e.g. thymic stromal lymphopoietin (TSLP) and interleukin (IL)‐33) are thought to induce Type‐2 allergic cytokines, including IL‐5 and IL‐13, which stimulate the recruitment and activation of the allergic effector cells, eosinophils and mast cells. Eosinophil and mast cell‐derived mediators stimulate dysregulation of epithelial barrier regulatory and proliferative response genes within the esophageal epithelial compartment leading to esophageal epithelial remodelling and fibrosis. Compelling evidence supporting a role for allergic inflammatory cells (CD4+ Th2‐type cells, eosinophils, and mast cells) and cytokines in EoE has led to using biologics that target these inflammatory cells and mediators as potential treatments for EoE. Herein, we summarize the current understanding of processes that underlie the esophageal inflammation and remodelling in EoE and discuss the mechanisms of action, specific indications, benefits and side‐effects of biological therapies for EoE.
2. PREVALENCE AND INCIDENCE
Over the last three decades, EoE has evolved from a rare, case‐reportable diagnosis to an increasingly common encounter in a multitude of settings across healthcare systems throughout the world. 11 Current estimates of EoE incidence in Europe and in the United States range from 1.30 to 12.8 cases per 100,000. 12 Incidence rates have been increasing in paediatric and adult EoE, with incidence levels reported to have increased 131‐fold in the Netherlands (1996–2010), 20‐fold in Denmark (1997–2012) and 5.1‐fold in Calgary, Canada (2004–2008). 13 , 14 , 15 The prevalence of EoE is significantly higher in male patients than in female patients, in the adult population (18–65 years of age) and in Whites than in Asians and African Americans. 16 The male predominance persists across demographic groups, regardless of age, geographic region, socio‐economic status or race. 17 , 18 , 19 While there are no observed gender differences in EoE severity, the clinical and histologic presentation of EoE can differ between the sexes in both paediatric and adult EoE. 20 , 21 For example, paediatric male patients are more likely to present with food impaction and feeding refusal, whereas female patients report more abdominal pain. 17
3. RISK FACTORS OF EoE
3.1. Genetic
The demonstration of increased risk of EoE in first‐degree relatives (10‐ to 64‐fold) compared with that of the general population has led to the concept of a genetic contribution to the disease. 22 , 23 This is further supported by the observed increased risk of EoE in second‐degree relatives and first cousins who likely did not share a common environment. 24 However, genetic data indicate the EoE inheritance pattern is not clearly autosomal dominant or recessive or X‐linked suggesting, a complex inheritance pattern. 23 Candidate and GWAS studies have identified 31 independent risk loci across the genome, 25 with four loci consistently demonstrating genome‐wide significance (5q22 [TSLP/WDR36], 2p23 [CAPN14], 11q13 [LRRC32/C11orf30] and 12q13 [STAT6]). The majority of the EoE genetic risk variants are positioned within either intergenic (36.7%) or within intronic genomic sequences (42.4%), with only 2.2% variants associated with amino acid substitutions. 25 These non‐coding sequences include gene promoters, introns and genomic regulatory elements suggesting that these variants are likely to contribute to altered gene expression and function through transcriptional or epigenetic‐dependent mechanisms. 25
3.2. Environmental
While these studies suggest a genetic component, analyses of EoE concordance in a twins EoE cohort also suggest a greater environmental contribution to EoE risk. 23 This is best exemplified by the lack of significant difference in EoE concordance rates between monozygotic twins (~40%) and dizygotic twins (~30%). 23 Notably, genetics was found to contribute 14.5% and common environment 81.0% to the variation in EoE heritability. Consistent with this, antibiotic use in infancy, caesarean delivery, preterm birth, season of birth, birthweight, and lack of breastfeeding have all been identified as factors that affect EoE risk. 23 , 26 Future studies in larger patient cohorts are required to delineate the contribution of genetic versus environmental risk components to the predisposition to the development of EoE.
4. EoE PHENOTYPES AND ENDOTYPES
Eosinophilic esophagitis is becoming increasingly recognized as a heterogeneous disease with different phenotypes. 27 , 28 Patients with EoE present at different ages, and with varying clinical, endoscopic, and histopathological manifestations, comorbidities, natural history, and are responsive to different treatments. 16 , 29 , 30 , 31 , 32 For example, adults with EoE often experience symptoms of dysphagia, esophageal food impaction and upon endoscopic evaluation present with fibrostenotic phenotype with typical rings and strictures. 4 , 33 , 34 , 35 , 36 In contrast, paediatric EoE presentation includes emesis, abdominal pain, gastroesophageal reflux, feeding difficulties and failure to thrive 16 , 37 with an endoscopic phenotype characterized by mucosal findings of longitudinal furrows and exudates. 34 , 35 , 36 , 38 , 39 , 40 Initial reports suggested that patients with EoE despite the disease heterogeneity possessed similar molecular signatures 41 , 42 suggesting a common disease entity. However, recent reports in both adult and paediatric EoE suggest gender‐related differences in the molecular signature and that these differences in transcriptional profiles may contribute to observed differences in clinical presentation EoE between male patients and female patients. 43 , 44
Clinical therapy trials have identified two distinct EoE treatment phenotypes (PPI‐R EoE and PPI‐UR EoE) based upon differential responsiveness to PPI. 45 , 46 PPI‐R EoE and PPI‐UR EoE are clinically, endoscopically, and histologically indistinguishable at diagnosis. 45 , 47 , 48 , 49 , 50 , 51 , 52 Furthermore, atopy status and molecular signature are also largely similar between the two phenotypes. 5 , 53 PPI therapy in PPI‐R EoE individuals induces clinicopathological remission with reports of 20%–80% of patients achieving symptom improvement 47 , 54 , 55 , 56 with ~68% of patients achieving complete symptom‐free remission with high‐dose PPI. 57 Notably, there is not always concordance in clinical and histological response to PPI therapy with histologic remission achieved in only 33%–61% of individuals. 56 However, individuals who achieve complete histologic remission on PPI therapy often have improvement to complete symptom elimination. 45 , 47 , 58 Recognition and characterization of the different EoE phenotypes will be critical in assisting clinicians with clinical care and likely to and favourable clinical outcome.
Allergic diseases including asthma and EoE are diseases of heterogeneous phenotypes with clinical differences including clinical symptoms, atopy history, responsiveness to therapy and anthropometrics are not predictive of treatment outcomes. 31 , 59 , 60 To better understand disease heterogeneity and assist with management and treatment decisions, efforts have been made to stratify allergic diseases according to pathophysiologic mechanisms termed endotypes. These efforts have been greatly assisted by the advancement in molecular analysis and permitted classification of EoE patients based on molecular endotypes. For example, Shoda et al. recently performed a cross‐sectional study across 10 CEGIR (Consortium of Eosinophilic Gastrointestinal Disorders) centres and analysed the association between histologic and endoscopic features of paediatric and adult EoE with the 96 gene eosinophilic esophagitis diagnostic panel molecular signature. 5 The investigators identified 3 distinct EoE endotypes‐ EoEe1, EoEe2 and EoEe3. EoEe1 was associated with an endoscopically normal appearing oesophagus with mild histological changes and molecular signature that was distinguished by high expression of epithelial differentiation genes and low expression of inflammatory and remodelling genes. EoEe2 was characterized by high degree of endoscopic and histological severity for the inflammatory component, high expression of inflammatory cytokine genes and refractoriness to steroids and EoEe3 was associated with adult‐onset, fibrostenotic phenotype and a low expression of epithelial differentiation genes. 5 The identification of EoE endotypes will provide a framework for precision medicine in future therapeutic prevention strategies for specific EoE populations. For example, the EoE endotype 2 which was characterized by type‐2 immune responses and evidence of refractoriness to steroids may be more amendable to specific anti‐type‐2 immune biologic therapy such as anti‐IL4Ra or anti‐TSLP. 5
5. PATHOPHYSIOLOGY OF EoE
Histopathologic manifestations of EoE include intraepithelial eosinophilic inflammation and alterations of the esophageal squamous epithelium including BZH and DIS which are thought to contribute to esophageal dysfunction and disease symptoms. Recent studies have revealed an important role for Type‐2 immune‐induced expression and function of the ion transport proteins anoctamin 1 (ANO1) and sodium hydrogen exchange member 3 (NHE3) in the regulation of the esophageal epithelial proliferative response and DIS formation in EoE. 61 , 62 Despite these advancements, the mechanism by which these ion transport proteins mediate BZH and DIS remains largely unclear and there is a paucity of data describing the interaction of these processes that drive esophageal epithelial remodelling in EoE.
6. ESOPHAGEAL EPITHELIAL CELLS
The normal human esophageal epithelium consists of non‐keratinized stratified squamous epithelium. The esophageal epithelium consists of the several layers including the stratum germinativum (basal layer) and stratum spinosum (suprabasal/prickle cell layer) and the stratum corneum (superficial/surface layer). The basal layer consists of proliferative cells and is not more than 3 cell layers thick or more than 15% of the total epithelial thickness. The suprabasal layer consists of transitional basal cells and the superficial layer consists of the simple and stratified squamous epithelial cells. In EoE, the esophageal epithelium and sub‐epithelium undergo extensive remodelling manifested as BZH, DIS, 3 , 63 , 64 fibrosis, angiogenesis and smooth muscle hyperplasia. 65 , 66 This epithelial and subepithelial remodelling is thought to contribute to endoscopic (esophageal rings and stricture formation) and clinical (dysphagia and food impaction) manifestations typical of EoE. 67 , 68 , 69 , 70
6.1. Basal zone hyperplasia
Esophageal epithelial BZ expansion in patients with EoE is defined as a basal cell layer exceeding 15% of total epithelial thickness. The underlying pathways that induce esophageal epithelial BZH are poorly understood. Recently, we identified involvement of the calcium‐activated chloride channel anoctamin 1 (ANO1) in the regulation of chloride transport and proliferation in esophageal epithelial cells. 61 We demonstrated that biopsies from patients with EoE had increased mRNA expression of ANO1 and the level of ANO1 expression correlates with BZH. 61 Immunofluorescence analyses localized ANO1 to esophageal basal cells suggesting a relationship between ANO1 expression and esophageal epithelial proliferation. In vitro studies revealed that ANO1 played an important role in esophageal epithelial chloride transport and that loss of ANO1‐dependent chloride transport reduced esophageal epithelial proliferation. 61 ANO1 was required for phosphorylation of cyclin‐dependent kinase 2 (p‐CDK2) and transition through G1/S phase cell cycle check point to permit esophageal epithelial proliferation. Interestingly, a key requirement of G1/S phase and G2/M phase transition and cellular proliferation is increased cell volume, which is regulated in part by ion channel regulation of the membrane potential (Vmem). 71 , 72 , 73 , 74 , 75 , 76 , 77 The increased Vmem across the plasma membrane osmotically drives water influx from the extracellular to intracellular space leading to cell swelling. 78 The functional relevance of BZH and ANO1 expression and relationship to human EoE is unclear. Shoda et al. identified an EoE endotype (EoE Endotype 3) 55 that was associated with a fibrostenotic (rings, narrowing, strictures) phenotype and the highest degree of endoscopic and histological severity. Molecular analyses revealed that ANO1 was one of two discriminatory genes (ANO1 and UPK1A) to provide 98% PPV of EoE endotype 3 suggesting a link between ANO1 and the fibrostenotic and histologic phenotype. 5 We speculate that ANO1's contribution to functional outcomes in EoE such as fibrostenosis is not through directly driving esophageal dysfunction but rather indirectly through chronicity of inflammation and subsequent development of secondary associated histopathologic manifestations such as BZH and DIS formation.
6.2. Dilated intracellular spaces
Dilated intercellular spaces have been consistently described in the esophageal epithelium of both adult and paediatric patients with gastroesophageal reflux disease (GERD) and EoE. 68 , 70 , 79 , 80 , 81 Histologically, DIS is significantly more intense in EoE than GERD. 82 In EoE, the presence and magnitude of the DIS correlate with esophageal eosinophil numbers and disease severity. 69 Furthermore, decreased DIS is associated with symptom improvement with steroid therapy or elimination diet in EoE suggesting that DIS contributes to clinical signs and symptoms of disease. 70 , 83 , 84 How DIS drives the clinical manifestations of EoE and the molecular basis underlying DIS formation are not fully elucidated. One proposed mechanism involves solute carrier family 9, subfamily A, member 3 (SLC9A3)‐dependent acid extrusion by esophageal epithelial cells and acidification of the intercellular spaces. 85 The acidification of the intercellular spaces promotes the formation of an electrochemical gradient and chloride (Cl−) diffusion, creating an osmotic force for water flux and intercellular space dilation. 86 , 87 SLC9A3 encodes sodium hydrogen exchange member 3 (NHE3) and is specifically up‐regulated in biopsy specimens from patients with EoE. Notably, the level of SLC9A3/NHE3 mRNA expression correlates with eosinophil count and the level of DIS. 62 In an in vitro model of EoE, IL‐13 stimulation of esophageal epithelial cells results in increased expression of SLC9A3/NHE3 and DIS formation. 62 Pharmacological antagonism of NHE3 activity reduced DIS formation. 62 In EoE, there is significant esophageal epithelial basal zone (BZ) expansion, and the basal cell layer can exceed 15% of the total epithelial thickness. 88 We speculate that the esophageal proliferative response and thickening of the epithelial basal zone leads to diminished capacity of the intercellular acid‐protective mechanisms and leading to DIS. Consistent with the concept of esophageal epithelial intercellular acid as a primary driver for DIS in EoE, luminal acid has been shown to drive acidification of the intercellular spaces and DIS in non‐erosive reflux disease. 86 , 87 , 89 , 90 To further support this concept, a recent study reported a strong positive correlation between BZH and DIS (r 2 ≥ .67) in both proximal and distal biopsy samples from paediatric patients with EoE. 69 Interestingly, the increased esophageal intercellular acid in non‐erosive reflux disease is thought to activate afferent neurons (nociceptors) within the esophageal epithelium leading to the development of heartburn. 90 Although less common, EoE symptoms can include heartburn. 4
7. IMMUNOLOGICAL PROCESSES
Corroborative clinical and experimental studies indicate that an underlying allergic sensitization to dietary food antigens and development of a CD4+ Th2 and ILC2 inflammatory response in the esophageal mucosa drive the eosinophilic inflammation and esophageal remodelling in EoE 3 , 63 , 91 , 92 (Figure 1). Dietary modification (i.e. complete or targeted food antigen avoidance) and swallowed glucocorticoids alleviate much of the disease pathology, 93 , 94 suggesting a food‐induced CD4+ Type‐2 allergic inflammatory response. 6 , 95 , 96 , 97 , 98 , 99 Consistent with this, animal‐based studies have revealed important roles for CD4+ Th2 cells, pro‐allergic cytokines [IL‐5 and IL‐13] in the histopathologic manifestations of disease. 9 , 100 , 101 These cytokines are thought to mobilize eosinophils and promote eosinophil survival, activation and degranulation and also dysregulate the expression of several key epithelial barrier regulatory genes driving the esophageal remodelling and clinical symptoms (Figure 1). 102 , 103 , 104
FIGURE 1.
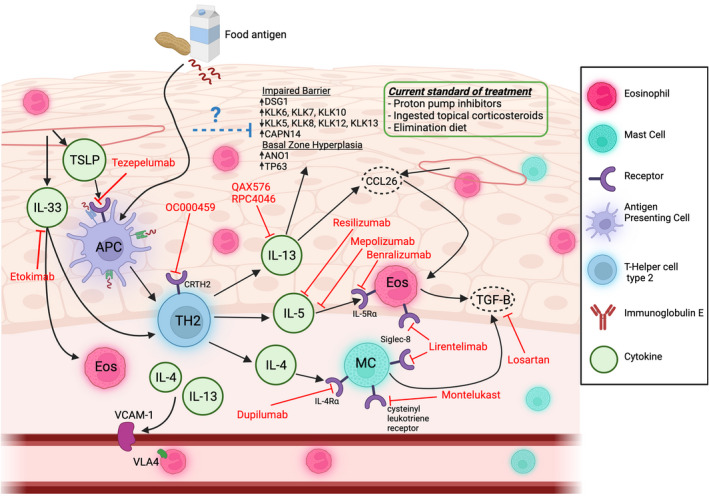
Pathophysiology and clinical management of eosinophilic esophagitis (EoE). EoE is a chronic inflammatory disease of the oesophagus driven by food allergen exposure which triggers esophageal eosinophilia and esophageal epithelial remodelling. Esophageal epithelial derived cytokines TSLP and IL‐33 stimulate antigen presenting cells (APCs) presentation of food antigens to CD4+ T helper type 2 (TH2) cells and secretion of the cytokines IL‐4, IL‐5 and IL‐13. IL‐4 is responsible for driving mast‐cell (MC) and vascular endothelial adhesion responses, IL‐5 is responsible for eosinophil maturation, activation and survival, and IL‐13 is responsible for inducing pro‐inflammatory and pro‐adhesion pathways and inducing expression of pro‐proliferation, pro‐inflammatory, and barrier regulatory genes within the esophageal epithelial compartment which contribute to the “EoE Transcriptome.” Current standards to manage EoE symptoms include proton‐pump inhibitor (PPI) therapy, food elimination diet, and ingested topical corticosteroids. Several therapeutics and biologics, including monoclonal antibodies and small molecule inhibitors are under investigation which target various aspects of EoE pathophysiology. Created with BioRender.com
7.1. The eosinophil, eotaxin subfamily and IL‐5
The eosinophil is a pleiotropic granulocytic leukocyte that is histologically characterized by a bilobed nucleus and cytoplasmic granules and arises from CD34+ progenitor cells within the bone marrow. 105 The eosinophil possesses homeostatic functions including tissue development, thymic T cell selection and innate host defense; however eosinophils also contribute to the initiation and propagation of inflammatory responses including parasitic helminth, bacterial and viral infections, tissue injury, tissue immunity and allergic diseases. 106 , 107 , 108 , 109 Under homeostatic conditions, the eosinophil is found in all portions of the gastrointestinal (GI) tract apart from the oesophagus. 110 The most characteristic histopathologic feature of EoE is esophageal intraepithelial eosinophilia (≥15 eosinophils/HPF). Eosinophil numbers can be significant with micro‐abscesses and esophageal eosinophilia correlates with histopathologic features (including DIS and BZH) and disease severity. 111 , 112 In a registry of EoE patients, O'shea et al compared patients with high‐grade esophageal eosinophilia (>350 eosinophils per hpf) to patients with low‐grade eosinophilia (15–24 eosinophils per hpf) and found statistically significant differences in histologic severity, endoscopic severity and gene expression but not symptoms. 113
The mechanisms by which eosinophils drive histopathologic features of EoE are not fully elucidated. Eosinophils possess granules that consist of cytotoxic proteins including eosinophil peroxidase (EPX), major basic protein (MBP), eosinophil cationic protein (ECP) and eosinophil‐derived neurotoxin (EDN). 114 Electron microscopy analysis of EoE esophageal biopsy specimens revealed evidence of esophageal eosinophil piecemeal degranulation or cytolysis. 115 Consistent with this finding, esophageal biopsy specimens from patients with EoE demonstrated increased EPX, MBP and EDN extracellular deposition. 116 , 117 Notably, the presence of these proteins correlated with histologic features including intercellular oedema, BZH, lamina propria elongation and lamina propria fibrosis suggesting a pathogenic role for eosinophils in disease. 116 , 118 Using an antigen‐induced model of EoE, investigators demonstrated that depletion of eosinophils using neutralizing monoclonal antibody (anti–Siglec F antibody) led to decreased esophageal eosinophilia, angiogenesis, BZH and fibronectin deposition. 119 Similarly, mice deficient in eosinophils have a decrease in allergen‐induced esophageal BZH and esophageal lamina propria thickness and do not develop esophageal strictures. 120 , 121 , 122 Notably, these mice still had evidence of esophageal motility dysfunction, suggesting the presence of eosinophil‐independent processes in histopathological manifestations of disease. 122
Eosinophil development, maturation and survival is largely regulated by the cytokine IL‐5. Patients with active EoE have increased levels of IL‐5 as compared to inactive EoE patients and healthy controls. 8 Overexpression of IL‐5 in the esophageal squamous epithelia of mice that received oxazolone (OXA) sensitization and topical challenge of the oesophagus leads to increased level of esophageal eosinophilia. Conversely, neutralization of IL‐5 reduces esophageal eosinophil numbers in an allergen‐induced EoE model 9 supporting a role for IL‐5 in maintenance of esophageal eosinophilia in EoE. Eosinophil trafficking is primarily regulated by the chemokine receptor CCR3 and eotaxin subfamily of chemokines (eotaxin‐1/CCL11; eotaxin‐2/CCL24 and eotaxin‐3/CCL26). 123 , 124 CCR3 is predominantly expressed on eosinophils and CCR3 gene deletion impairs eosinophil recruitment in models of allergic inflammation including allergen‐induced esophageal eosinophilic inflammation. 123 , 125 , 126 Transcriptional analysis of biopsy specimens from patients with EoE revealed significant up‐regulation of CCL26 mRNA compared with healthy individuals. 125 Furthermore, CCL26 mRNA expression was shown to directly correlate with esophageal eosinophilia in EoE. 127 Notably, a single‐nucleotide polymorphism (SNP) in the CCL26 gene has been associated with EoE disease susceptibility. 125 Collectively, these clinical and mouse‐based analyses suggest that IL‐5 regulates eosinophils maturation and survival in the oesophagus, whereas CCL26/eotaxin‐3 likely regulates eosinophil recruitment into the oesophagus in EoE.
7.2. Esophageal epithelial cells
Transcriptomics analysis has revealed altered gene expression in esophageal epithelial cells from EoE individuals. 128 , 129 Esophageal epithelial cells from EoE individuals are enriched for genes involved in molecular functions including structural molecule activity (SPRR1 and 2 family members, Keratin family members), Serine‐type peptidase activity (KLK6‐8, KLK10‐12), Serine‐type endopeptidase inhibitor activity (SERPINB2‐5, 7 and SPINK5) and IL1 receptor binding (IL33, IL36B, IL36A, IL36RN, IL1A, IL1RN). 128 , 129 These molecular functions are involved in biological processes such as epidermis development, keratinocyte differentiation, epithelial cell development, desmosome organization and establishment of skin barrier which is consistent with the observed esophageal epithelial proliferative response, BZH and impaired barrier function. 128 , 129
SPRR proteins are 6‐18 kDa proteins encoded within the epidermal differentiation complex region (EDC). SPRR1 and SPRR2 proteins are predominantly expressed in the skin, oral mucosa and oesophagus and are thought to cross‐link EDC proteins including loricrin and involucrin. EoE has been associated with decreased expression of epithelial barrier function genes including SPRR proteins as well as desmoglein 1 (DSG1) and structural epithelial genes including filaggrin (FLG), involucrin (IVL) which likely contributes to loss of esophageal barrier integrity. SPRR1a and SPRR2a have recently been shown to possess potent bactericidal activity which suggests a possible role for these proteins in the regulation of esophageal host defense. 130
Kallikreins (KLKs) are a subgroup of serine proteases that possess trypsin‐ or chymotrypsin‐like activity. 131 KLK6, KLK7 and KLK10 are significantly increased, whereas KLK5, KLK8, KLK12 and KLK13 are down‐regulated in EoE. 102 KLK5 is thought to regulate squamous epithelial barrier properties through processing of barrier proteins including DSG1. 132 , 133 , 134 KLK5 is also known to stimulate pro‐inflammatory signals via cleavage and activation of protease‐activated receptor 2 (PAR2) and activate TSLP production. 135 Recent studies indicate that KLK5 is a direct target of SPINK7. 136 The two most highly expressed SPINK genes in the human oesophagus are SPINK5 and SPINK7. In EoE, both SPINK5 (1.9‐fold) and SPINK7 (16‐fold) are significantly down‐regulated as compared with control individuals. 137 Mechanistic analyses have revealed that reduced expression of SPINK7 in esophageal epithelial cells leads to increased barrier dysfunction, loss of cellular differentiation and increased pro‐allergic signals including production of TSLP and and urokinase plasminogen‐type activator (uPA). TSLP is a key cytokine involved in the generation of CD4+ Th2 responses and genetic variants have been linked with increased EoE risk. 25 , 138 , 139 UPA is a serine protease that has been shown to promote uPA receptor‐dependent eosinophil activation. 137 The altered expression of an array of structural proteins, proteases and protease inhibitors within the esophageal epithelial layer during EoE suggests a complex interaction between regulatory and counter‐regulatory processes likely in effort to promote esophageal epithelial proliferation and differentiation and sustain barrier integrity.
7.3. T cells and Th2 cytokines: IL‐4 and IL‐13
Clinical studies have demonstrated increased frequency of CD4+ Th2 cells in the peripheral blood and esophageal biopsy samples from EoE individuals. 127 , 140 , 141 scRNAseq analyses of esophageal biopsy samples from EoE patients revealed a prominent tissue resident CD4+ T cell population that expressed IL‐4, IL‐5 and IL‐13. 141 Notably, the percentage of these CD4+ Th2 cells correlated with esophageal tissue eosinophilia. 141 IL‐4 mRNA expression is increased in EoE patients and levels have been shown to be decreased in patients with EoE following glucocorticoid therapy or dietary elimination therapy suggesting a role for IL‐4 in disease. 8 , 142 IL‐4 has been shown to regulate multiple aspects of eosinophil trafficking and function including eotaxin subfamily and adhesion molecule expression. 143 , 144 IL‐13 appears to be the dominant Type‐2 cytokine involved in orchestrating the eosinophil dominant inflammatory response and the histologic manifestations of EoE. Overexpression of IL‐13 in mice is sufficient to promote esophageal eotaxin subfamily expression, esophageal eosinophilia, epithelial hyperplasia, angiogenesis and fibrosis. 10 , 145 Furthermore, the genes differentially expressed by primary esophageal epithelial cells following IL‐13 stimulation significantly overlap with the EoE transcriptome (22%, p < .05) and transcriptional changes observed in the oesophagus of mice engineered to overexpress IL‐13. 10 Furthermore, IL‐13 stimulation of esophageal epithelial cells in the absence of eosinophils is sufficient to promote esophageal remodelling including BZH and DIS formation. 62
7.4. Mast cells
Mucosal biopsies from patients with EoE also demonstrate increased frequency of mast cells and evidence of degranulation. 146 , 147 Not only do EoE patients have a greater number of mast cells in the oesophagus, but also have alterations in mast cell gene expression such as up‐regulation of carboxypeptidase A3 (CPA3) and tryptase, but not chymase. 146 , 148 Intriguingly, increased esophageal mast cell density has been observed in EoE patients with persistent symptoms and endoscopic abnormalities in the absence of an esophageal eosinophilia supporting a key role for mast cells in EoE pathogenesis. 149 A recent study examining esophagectomy specimens from patients with EoE identified tryptase‐positive mast cells infiltrating all layers of the oesophagus including mucosa, muscularis mucosa, submucosa, muscularis propria and adventitia. 65 Employing mouse models of EoE, investigators have demonstrated that mast cells increase concurrently with eosinophils in the oesophagus in response to allergen challenge. 150 Notably, the mast cell infiltration into the muscular layers of the oesophagus was associated with muscular hyperplasia and hypertrophy and this phenotype was diminished in mast cell deficient mice. 150 Mast cells are also thought to dysregulate esophageal muscle contractility and relaxation responses. 120 Tryptase‐positive mast cells within the muscularis mucosae of patients with EoE have increased expression of TGF‐β1. 65 TGFβ1 stimulation of human esophageal smooth muscle cells promotes smooth muscle contraction. 65 Collectively, these studies suggest that mast cells through release of cytokines such as TGFβ1 and autocoid mediators may stimulate esophageal smooth muscle hyperplasia and exert procontractile effects on esophageal smooth muscle in EoE. 151 The presence of mast cells in EoE may define a specific subtype that lacks significant esophageal eosinophilia and is prone to extra‐intestinal symptoms such as dysphagia. 149
7.5. Immunoglobulin
Eosinophilic esophagitis is often associated with atopic comorbidities including food allergy, asthma, allergic rhinitis and atopic dermatitis suggesting IgE involvement. 20 , 152 EoE individuals often have food‐ or environmental‐specific or elevated total IgE. 142 , 152 , 153 , 154 Furthermore, there is evidence of local immunoglobulin class switching and IgE production in the esophageal mucosa of paediatric EoE patients 155 which had led investigators to propose that eosinophilic esophagitis is a IgE‐mediated disease. 153 , 156 However, the contribution of IgE to the pathogenesis of EoE is inconclusive. Treatment of paediatric and adult EoE patients with the anti‐IgE humanized monoclonal antibody (omalizumab) resulted in only a 33% improvement in histologic and clinical outcomes despite significant reduction in tissue IgE levels. 157 Furthermore, a prospective randomized double‐blind placebo‐controlled trial of adults with EoE administered omalizumab (n = 16) or placebo (n = 14) every 2 to 4 weeks for 16 weeks revealed that while omalizumab induced a significant decrease in serum IgE, no reduction in EoE symptoms or eosinophil histologic involvement was observed. 158 Collectively, these studies suggest that specific IgE may be associated with EoE; however, it is not causative in the pathogenesis of EoE.
There is increasing evidence of a role for IgG4 in EoE. Adult EoE individuals have increased total and food‐specific serum IgG4 compared with controls. 158 , 159 Furthermore, levels of IgG4 in esophageal tissue from EoE patients are 45‐fold higher than that observed in controls and evidence of IgG4 extracellular deposits has been observed in the esophageal wall of EoE individuals. 158 Similarly, increased serum food‐specific IgG4 and increased IgG4 + plasma cells have been observed in paediatric EoE and tissue IgG4 levels positively correlate with peak eosinophil count and histologic involvement, in particularly BZH. 160 , 161 , 162 , 163 Wright et al. demonstrated that 50% of peanut‐allergic individuals that underwent peanut oral immunotherapy developed a new esophageal eosinophilia that was consistent with the pathologic criteria for EoE. Interestingly, all these individuals demonstrated marked esophageal tissue deposition of IgG4. 164 Recent studies also support a pathogenic role for IgG4 in EoE. Treatment of EoE patients with the topical steroid budesonide (1 mg BID) led to a reduction in both serum and esophageal IgG4 levels and this was associated with reduced EoE symptoms. 165 Furthermore, EoE individuals who responded to six‐food elimination diet consisting of removal of common dietary allergens were shown to have reduced total IgG4 levels and esophageal food‐specific IgG4. 166 Further studies are required to delineate whether or not IgG4 plays a pathogenic role in EoE or is simply reflective of chronic antigen exposure (e.g. OIT) and activation of memory CD4 Th2 response.
7.6. TGF‐β
Surgical resection specimens have revealed that the esophageal eosinophilic inflammation in EoE can extend throughout all layers of the oesophagus beyond the squamous epithelium. Notably, the subepithelial eosinophilic inflitrate is often associated with subepithelial fibrosis and endoscopic findings including esophageal furrowing or ridging and esophageal dysfunction (e.g. dysphagia and food impaction) which can progress to a fibrostenotic disease phenotype. 4 , 118 , 167 , 168 , 169 Notably, the profibrotic cytokine TGF‐β and downstream signalling molecules (phosphorylated SMAD2/3) has been found to be increased in esophageal biopsies from patients with EoE. 66 TGF‐β promotes quiescent fibroblast to myofibroblast transdifferentiation and up‐regulation of expression of extracellular matrix (ECM) proteins including Type I collagen. 170 TGFβ is secreted by all cell types within the oesophagus, and fibroblasts when stimulated by TGF‐β secrete extracellular matrix components such as collagens (COL1A1) and fibronectin. 171 TGF‐β is also though to contribute smooth muscle dysfunction, collagen deposition, epithelial remodelling, barrier dysfunction and angiogenesis in EoE. 172
Genetic studies have previously linked a single‐nucleotide polymorphism (SNP) in the promoter region of the TGF‐β1 gene (−509) with increased risk of asthma when exposed to traffic‐related emissions. 173 Analyses of the TGFβ1 promoter C‐509 genotypes CC, CT, and TT in EoE subjects revealed of 155 EoE subjects, 52% patients possessed the ‐509CT genotype, 35% ‐509CC genotype and 13% ‐509TT genotype. Notably, −509TT subjects possessed more esophageal TGFβ1+ cells and mast cells and higher esophageal epithelial remodelling scores than −509 CC and CT subjects. It is postulated that the TT genotype at −509 of the TGFβ1 gene leads to increased TGFβ1 gene transcription either through loss of AP1 suppressive signal or YY‐1 positive transcriptional signal. 174 Consistent with this, fibroblasts with the TT genotype possessed significantly elevated baseline levels of TGFβ1 mRNA expression as well as TGFβ1 target genes including collagen 1a1 and MMP2 compared with CC genotype fibroblasts. Functional assays have revealed that the TT genotype is also associated with increased TGFβ1‐induced E‐cadherin localization and epithelial barrier function suggesting that genotype at the TGFβ1 promoter SNP −509 is also associated with altered fibroblast function. 175 Recent in vitro studies suggest that TGFβ1 induction of collagen 1a1 in esophageal fibroblasts involves Thrombopsondin‐1. 176 The frequency of ‐509TT genotype has not been shown to be significantly different between EoE and control populations indicating that this polymorphism is not likely a genetic risk factor for EoE but rather a disease modifying allele gene. 177
8. DISEASE MONITORING AND EMERGING THERAPEUTICS
With the rising prevalence and incidence of EoE, and as diagnosis requires the identification of esophageal eosinophilia, an emerging challenge in management is disease monitoring. 178 , 179 As symptoms of EoE are not consistently reliable in regard to correlation with disease activity, disease monitoring in EoE commonly requires repeated endoscopy with biopsies. 180 The development of minimally invasive monitoring tests for disease activity in EoE is an area of active investigation. Non‐invasive tests such as the esophageal string test 181 and Cytosponge 182 are currently being evaluated. Furthermore, multiple biomarkers including plasma EDN, CCL26 and eosinophil progenitor levels are also under investigation for disease monitoring in EoE. 183 , 184
The focus of EoE treatment remains alleviation of symptoms and the prevention of complications of fibrostenotic remodelling. The current mainstays of effective treatment for EoE include proton‐pump inhibitors (PPI), various elimination diets and topical corticosteroids such as swallowed fluticasone inhaler and oral viscous budesonide. 48 , 185 , 186 , 187 , 188 Dietary and pharmacologic therapies are effective in inducing and maintaining disease remission, reducing the risk of esophageal food impactions, and improving quality of life. 48 , 185 , 186 , 187 , 188 Recently, several corticosteroid formulations including fluticasone orodispersible tablet, budesonide oral solution and budesonide orodispersible tablet have been developed to specifically treat EoE. 133 , 189 Some of these oesophagus‐targeted formulations of topical steroids have demonstrated excellent response rates inducing clinical and histologic remission and improvement of QoL in patients with EoE. 133 , 189 , 190 However, several patients do not respond to these standard therapies, histologic relapse in EoE is not uncommon and long‐term efficacy remains unclear. 191
Recently, there has been increasing focus on the utilization of biologic agents focussed on targeting specific aspects of eosinophil biology (IL‐4, IL‐5, IL‐5Rα, IL‐13 and siglec‐8) for the treatment of EoE 188 (Figure 1). There are currently three IL‐5 directed agents (Mepolizumab, Reslizumab and Benralizumab) originally approved for treatment of asthma that have been examined for EoE treatment. 192 , 193 , 194 Mepolizumab, a humanized anti‐IL‐5 monoclonal antibody which inhibits IL‐5 binding to its receptor 193 was shown to significantly reduce esophageal eosinophilia in EoE patients, however, failed to show a reduction in symptoms. 195 , 196 , 197 Reslizumab a fully humanized IgG4 antibody with high affinity and specificity for IL‐5 198 also demonstrated a reduction in esophageal eosinophilia but failed to show a consistent symptomatic response. 199 , 200 Benralizumab, a fully humanized, afucosylated anti‐IL‐5 receptor α antibody is currently in Phase 3 trial for EoE. 201
Sialic acid binding immunoglobulin‐like lectin (Siglec) 8 is a surface receptor expressed on mature eosinophils, mast cells and basophils. 202 Cross‐linking of Siglec‐8 has been demonstrated to induce eosinophil apoptosis and thus it has been proposed as a therapeutic target for eosinophilic disorders. 202 , 203 Lirentelimab is a humanized anti‐Siglec 8 antibody and has been shown to elicit antibody‐dependent cell‐mediated cytotoxicity against human eosinophils and inhibit mast cell activity. 204 Lirentelimab has recently been evaluated in a clinical trial with patients with either eosinophilic gastritis (EoG) or duodenitis (EoD). 205 In this phase 2 multi‐center clinical trial, lirentelimab reduced gastrointestinal eosinophil levels and disease symptoms. 205 Notably, a subset of patients who had concomitant EoE was noted to have decreased esophageal eosinophilia after treatment. 205
Biologics targeting the IL‐4/IL‐13 signalling pathway have also been examined in the context of EoE and individuals with esophageal eosinophilia. 206 Treatment of adult patients with PPI resistant esophageal eosinophilia with QAX576, a fully human monoclonal antibody against IL‐13, reduced esophageal eosinophil counts and improved expression of esophageal transcripts involved in EoE. 206 However, significant improvement of clinical symptoms was not observed, and the study failed to meet its primary end‐point. 206 RPC4046, a recombinant humanized monoclonal antibody against IL‐13, was examined in a multicenter double‐blind placebo‐controlled trial of adults with EoE. 207 In a phase 2 16‐week short‐course treatment study, RPC4046 demonstrated statistically significant changes in histologic and endoscopic outcomes; however, limited improvement in symptomatic outcomes was observed. 207 Patients who completed the 16‐week, double‐blind, induction portion of the phase 2 study of RPC4046 (180 mg or 360 mg/wk) that enrolled into the 52‐week open‐label, long‐term extension (LTE) study receiving open‐label RPC4046 360 mg/week demonstrated sustained endoscopic, histologic and clinical improvement. 208
Dupilumab is a fully human IgG4 monoclonal antibody that binds the IL4Rα chain, and is approved the treatment of multiple allergic conditions including atopic dermatitis, asthma, and chronic rhinosinusitis with nasal polyps. 209 Through its ability to bind to the IL‐4Rα receptor subunit that is shared by both type 1 and type 2 IL‐4 receptor, it can antagonize both IL‐4 and IL‐13 signalling. 210 In a phase 2 multi‐center study, adults with active EoE who received weekly subcutaneous injections of dupilumab (300 mg/dose) (loading dose, 600 mg on Day 1) demonstrated reduced peak esophageal eosinophil count, decreased histologic and endoscopic severity scores, and increased esophageal distensibility. 211 Preliminary data from a phase 3, randomized, 3‐Part study to investigate the efficacy and safety of dupilumab in adult and adolescent patients with EoE shows that weekly dupilumab promotes similar improvements in histologic, symptomatic and endoscopic measures. 212 These findings suggest that concurrent antagonism of IL‐4 and IL‐13 may be a viable treatment option for EoE.
Anti‐IL‐33 therapies have been investigated in murine models of atopic dermatitis as well as asthma and demonstrated anti‐inflammatory effects. 213 , 214 Data in humans are limited, but Etokimab (an IgG1 anti‐IL‐33 monoclonal antibody) has been examined in both atopic dermatitis and peanut allergy in humans. 215 , 216 Initial results from these human trials suggest that antagonism of IL‐33 results in several anti‐inflammatory effects which may be clinically significant. Anti‐IL33 therapy has yet to be examined in EoE but has the potential to become a viable therapeutic in this context. Given the prominent role TSLP in inducing Th2 responses, blockade of TSLP has been investigated for treatment of various allergic diseases. 217 Tezepelumab is a human monoclonal antibody that binds to TSLP and prevents its interaction with the TSLP receptor complex and has shown efficacy in the treatment of severe asthma, 218 but limited effect in atopic dermatitis. 219 Tezepelumab has not been studied in patients with EoE.
9. CONCLUSION
Our understanding of EoE has rapidly evolved over the last two decades. There have been significant advancements in defining the complex interplay between the immune system, epithelial barrier and environmental exposures that lead to the development of EoE. EoE is now recognized as a Th2‐mediated, food‐antigen‐driven disease that is characterized by impaired barrier function. The current challenges facing EoE include the development of novel therapeutics including biologic therapies and non‐invasive testing for diagnosis and disease monitoring and defining distinct endotypes and phenotypes to tailor specific therapies for a given patient. The solution to such challenges requires further inquiry into the mechanisms of disease.
AUTHOR CONTRIBUTIONS
KD, MS and SPH drafted the manuscript: IG, JWC, CM and SPH reviewed and revised the manuscript. All authors approved the final version of the manuscript.
FUNDING INFORMATION
This work was supported by NIH DK073553, AI112626, AI138177, AI140133 (S. P. H.) and Food Allergy Research & Education, M‐FARA and Mary H. Weiser Food Allergy Center (S.P.H).
CONFLICT OF INTEREST
MC: Consultant: Regeneron, Allakos, Adare/Ellodi, Shire/Takeda, AstraZeneca, Sanofi, Bristol Myers Squibb, Phathom. Research funding: Regeneron, Allakos, Shire/Takeda, AstraZeneca, Adare/Ellodi, Danone; SPH Research funding: Regeneron.
Khokhar D, Marella S, Idelman G, Chang JW, Chehade M, Hogan SP. Eosinophilic esophagitis: Immune mechanisms and therapeutic targets. Clin Exp Allergy. 2022;52:1142‐1156. doi: 10.1111/cea.14196
Dilawar Khokhar and Sahiti Marella contributed equally to this study.
Contributor Information
Dilawar Khokhar, Email: dkhokhar@uw.edu.
Simon P. Hogan, Email: sihogan@umich.edu.
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
REFERENCES
- 1. Furuta GT, Katzka DA. Eosinophilic esophagitis. N Engl J Med. 2015;373(17):1640‐1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O'Shea KM, Aceves SS, Dellon ES, et al. Pathophysiology of eosinophilic esophagitis. Gastroenterology. 2018;154(2):333‐345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Collins MH. Histopathologic features of eosinophilic esophagitis. Gastrointest Endosc Clin N Am. 2008;18(1):59‐71; viii‐ix. [DOI] [PubMed] [Google Scholar]
- 4. Dellon ES, Kim HP, Sperry SL, Rybnicek DA, Woosley JT, Shaheen NJ. A phenotypic analysis shows that eosinophilic esophagitis is a progressive fibrostenotic disease. Gastrointest Endosc. 2014;79(4):577‐585.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shoda T, Wen T, Aceves SS, et al. Eosinophilic oesophagitis endotype classification by molecular, clinical, and histopathological analyses: a cross‐sectional study. Lancet Gastroenterol Hepatol. 2018;3(7):477‐488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blanchard C, Mingler MK, Vicario M, et al. IL‐13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120(6):1292‐1300. [DOI] [PubMed] [Google Scholar]
- 7. Blanchard C, Stucke EM, Burwinkel K, et al. Coordinate interaction between IL‐13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 2010;184(7):4033‐4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blanchard C, Stucke EM, Rodriguez‐Jimenez B, et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. 2011;127(1):208‐217, 217.e1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mishra A, Rothenberg ME. Intratracheal IL‐13 induces eosinophilic esophagitis by an IL‐5, eotaxin‐1, and STAT6‐dependent mechanism. Gastroenterology. 2003;125(5):1419‐1427. [DOI] [PubMed] [Google Scholar]
- 10. Zuo L, Fulkerson PC, Finkelman FD, et al. IL‐13 induces esophageal remodeling and gene expression by an eosinophil‐independent IL‐13Rα2‐inhibited pathway. J Immunol. 2010;185(1):660‐669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Attwood S, Sabri S. Historical aspects of eosinophilic esophagitis: from case reports to clinical trials. Dig Dis. 2014;32(1–2):34‐39. [DOI] [PubMed] [Google Scholar]
- 12. Dellon ES, Hirano I. Epidemiology and natural history of eosinophilic esophagitis. Gastroenterology. 2018;154(2):319‐332.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Rhijn BD, Verheij J, Smout AJ, Bredenoord AJ. Rapidly increasing incidence of eosinophilic esophagitis in a large cohort. Neurogastroenterol Motil. 2013;25(1):47‐52.e5. [DOI] [PubMed] [Google Scholar]
- 14. Dellon ES, Erichsen R, Baron JA, et al. The increasing incidence and prevalence of eosinophilic oesophagitis outpaces changes in endoscopic and biopsy practice: national population‐based estimates from Denmark. Aliment Pharmacol Ther. 2015;41(7):662‐670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Syed AA, Andrews CN, Shaffer E, Urbanski SJ, Beck P, Storr M. The rising incidence of eosinophilic oesophagitis is associated with increasing biopsy rates: a population‐based study. Aliment Pharmacol Ther. 2012;36(10):950‐958. [DOI] [PubMed] [Google Scholar]
- 16. Shaheen NJ, Mukkada V, Eichinger CS, Schofield H, Todorova L, Falk GW. Natural history of eosinophilic esophagitis: a systematic review of epidemiology and disease course. Dis Esophagus. 2018;31(8):doy015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moawad FJ, Cheng E, Schoepfer A, et al. Eosinophilic esophagitis: current perspectives from diagnosis to management. Ann N Y Acad Sci. 2016;1380(1):204‐217. [DOI] [PubMed] [Google Scholar]
- 18. Sperry SL, Woosley JT, Shaheen NJ, Dellon ES. Influence of race and gender on the presentation of eosinophilic esophagitis. Am J Gastroenterol. 2012;107(2):215‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Franciosi JP, Liacouras CA. Eosinophilic esophagitis. Immunol Allergy Clin N Am. 2009;29(1):19‐27, viii. [DOI] [PubMed] [Google Scholar]
- 20. Chehade M, Jones SM, Pesek RD, et al. Phenotypic characterization of eosinophilic esophagitis in a large multicenter patient population from the consortium for food allergy research. J Allergy Clin Immunol Pract. 2018;6(5):1534‐1544.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moawad FJ, Dellon ES, Achem SR, et al. Effects of race and sex on features of eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2016;14(1):23‐30. [DOI] [PubMed] [Google Scholar]
- 22. Kottyan LC, Parameswaran S, Weirauch MT, Rothenberg ME, Martin LJ. The genetic etiology of eosinophilic esophagitis. J Allergy Clin Immunol. 2020;145(1):9‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alexander ES, Martin LJ, Collins MH, et al. Twin and family studies reveal strong environmental and weaker genetic cues explaining heritability of eosinophilic esophagitis. J Allergy Clin Immunol. 2014;134(5):1084‐1092.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Allen‐Brady K, Firszt R, Fang JC, Wong J, Smith KR, Peterson KA. Population‐based familial aggregation of eosinophilic esophagitis suggests a genetic contribution. J Allergy Clin Immunol. 2017;140(4):1138‐1143. [DOI] [PubMed] [Google Scholar]
- 25. Kottyan LC, Davis BP, Sherrill JD, et al. Genome‐wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014;46(8):895‐900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jensen ET, Kuhl JT, Martin LJ, Rothenberg ME, Dellon ES. Prenatal, intrapartum, and postnatal factors are associated with pediatric eosinophilic esophagitis. J Allergy Clin Immunol. 2018;141(1):214‐222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ruffner MA, Cianferoni A. Phenotypes and endotypes in eosinophilic esophagitis. Ann Allergy Asthma Immunol. 2020;124(3):233‐239. [DOI] [PubMed] [Google Scholar]
- 28. Collins CA, Palmquist J, Proudfoot JA, et al. Evaluation of long‐term course in children with eosinophilic esophagitis reveals distinct histologic patterns and clinical characteristics. J Allergy Clin Immunol. 2019;144(4):1050‐1057.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ruffner MA, Kennedy K, Cianferoni A. Pathophysiology of eosinophilic esophagitis: recent advances and their clinical implications. Expert Rev Clin Immunol. 2019;15(1):83‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Atkins D, Furuta GT, Liacouras CA, Spergel JM. Eosinophilic esophagitis phenotypes: Ready for prime time? Pediatr Allergy Immunol. 2017;28(4):312‐319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ferguson AE, Mukkada VA, Fulkerson PC. Pediatric Eosinophilic Esophagitis Endotypes: Are We Closer to Predicting Treatment Response? Clin Rev Allergy Immunol. 2018;55(1):43‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wong S, Ellison S, Haj Ali S, et al. Characteristics and progression of childhood‐onset and adult‐onset eosinophilic esophagitis. J Gastroenterol Hepatol. 2021;37(1):69‐74. [DOI] [PubMed] [Google Scholar]
- 33. Miehlke S. Clinical features of eosinophilic esophagitis in children and adults. Best Pract Res Clin Gastroenterol. 2015;29(5):739‐748. [DOI] [PubMed] [Google Scholar]
- 34. Liacouras CA, Furuta GT, Hirano I, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128(1):3‐20.e6; quiz 21‐22. [DOI] [PubMed] [Google Scholar]
- 35. Straumann A, Aceves SS, Blanchard C, et al. Pediatric and adult eosinophilic esophagitis: similarities and differences. Allergy. 2012;67(4):477‐490. [DOI] [PubMed] [Google Scholar]
- 36. Lucendo AJ, Sanchez‐Cazalilla M. Adult versus pediatric eosinophilic esophagitis: important differences and similarities for the clinician to understand. Expert Rev Clin Immunol. 2012;8(8):733‐745. [DOI] [PubMed] [Google Scholar]
- 37. Spergel JM, Brown‐Whitehorn TF, Beausoleil JL, et al. 14 years of eosinophilic esophagitis: clinical features and prognosis. J Pediatr Gastroenterol Nutr. 2009;48(1):30‐36. [DOI] [PubMed] [Google Scholar]
- 38. Dellon ES. Diagnosis and management of eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2012;10(10):1066‐1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim HP, Vance RB, Shaheen NJ, Dellon ES. The prevalence and diagnostic utility of endoscopic features of eosinophilic esophagitis: a meta‐analysis. Clin Gastroenterol Hepatology. 2012;10(9):988‐996.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Visaggi P, Savarino E, Sciume G, et al. Eosinophilic esophagitis: clinical, endoscopic, histologic and therapeutic differences and similarities between children and adults. Ther Adv Gastroenterol. 2021;14:1756284820980860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wen T, Stucke EM, Grotjan TM, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology. 2013;145(6):1289‐1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shoda T, Matsuda A, Nomura I, et al. Eosinophilic esophagitis versus proton pump inhibitor‐responsive esophageal eosinophilia: transcriptome analysis. J Allergy Clin Immunol. 2017;139(6):2010‐2013.e4. [DOI] [PubMed] [Google Scholar]
- 43. Gonsalves N, Berdnikovs S, Schroeder H, Zalewski A, Bryce PJ. Gender‐specific differences in the molecular signatures of adult Eosinophilic Oesophagitis. Clin Exp Allergy. 2017;47(7):969‐971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Erwin EA, Jaramillo LM, Smith B, et al. Sex differences in blood transcriptional profiles and clinical phenotypes in pediatric patients with eosinophilic esophagitis. J Allergy Clin Immunol Pract. 2021;9(9):3350‐3358.e8. [DOI] [PubMed] [Google Scholar]
- 45. Sayej WN, Patel R, Baker RD, Tron E, Baker SS. Treatment with high‐dose proton pump inhibitors helps distinguish eosinophilic esophagitis from noneosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2009;49(4):393‐399. [DOI] [PubMed] [Google Scholar]
- 46. Dohil R, Newbury RO, Aceves S. Transient PPI responsive esophageal eosinophilia may be a clinical sub‐phenotype of pediatric eosinophilic esophagitis. Dig Dis Sci. 2012;57(5):1413‐1419. [DOI] [PubMed] [Google Scholar]
- 47. Molina‐Infante J, Ferrando‐Lamana L, Ripoll C, et al. Esophageal eosinophilic infiltration responds to proton pump inhibition in most adults. Clin Gastroenterol Hepatol. 2011;9(2):110‐117. [DOI] [PubMed] [Google Scholar]
- 48. Dellon ES, Gonsalves N, Hirano I, Furuta GT, Liacouras CA, Katzka DA. ACG clinical guideline: Evidenced based approach to the diagnosis and management of esophageal eosinophilia and eosinophilic esophagitis (EoE). Am J Gastroenterol. 2013;108(5):679‐692; quiz 693. [DOI] [PubMed] [Google Scholar]
- 49. Francis DL, Foxx‐Orenstein A, Arora AS, et al. Results of ambulatory pH monitoring do not reliably predict response to therapy in patients with eosinophilic oesophagitis. Aliment Pharmacol Ther. 2012;35(2):300‐307. [DOI] [PubMed] [Google Scholar]
- 50. Moawad FJ. Letter: distinguishing PPI‐responsive oesophageal eosinophilia from eosinophilic esophagitis ‐ still a long way to go; authors' reply. Aliment Pharmacol Ther. 2014;39(10):1249‐1250. [DOI] [PubMed] [Google Scholar]
- 51. Warners MJ, van Rhijn BD, Curvers WL, Smout AJ, Bredenoord AJ. PPI‐responsive esophageal eosinophilia cannot be distinguished from eosinophilic esophagitis by endoscopic signs. Eur J Gastroenterol Hepatol. 2015;27(5):506‐511. [DOI] [PubMed] [Google Scholar]
- 52. Moawad FJ, Schoepfer AM, Safroneeva E, et al. Eosinophilic oesophagitis and proton pump inhibitor‐responsive oesophageal eosinophilia have similar clinical, endoscopic and histological findings. Aliment Pharmacol Ther. 2014;39(6):603‐608. [DOI] [PubMed] [Google Scholar]
- 53. Wen T, Dellon ES, Moawad FJ, Furuta GT, Aceves SS, Rothenberg ME. Transcriptome analysis of proton pump inhibitor‐responsive esophageal eosinophilia reveals PPI‐reversible allergic inflammation. J Allergy Clin Immunol. 2015;135(1):187‐197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Molina‐Infante J, Hernandez‐Alonso M, Vinagre‐Rodriguez G, Martin‐Noguerol E. Proton pump inhibitors therapy for esophageal eosinophilia: simply following consensus guidelines. J Gastroenterol. 2011;46(5):712‐713; author reply 714‐715. [DOI] [PubMed] [Google Scholar]
- 55. Navarro P, Laserna‐Mendieta EJ, Guagnozzi D, et al. Proton pump inhibitor therapy reverses endoscopic features of fibrosis in eosinophilic esophagitis. Dig Liver Dis. 2021;53(11):1479‐1485. [DOI] [PubMed] [Google Scholar]
- 56. Molina‐Infante J, Katzka DA, Gisbert JP. Review article: proton pump inhibitor therapy for suspected eosinophilic oesophagitis. Aliment Pharmacol Ther. 2013;37(12):1157‐1164. [DOI] [PubMed] [Google Scholar]
- 57. Frandsen LT, Westmark S, Melgaard D, Krarup AL. Effectiveness of PPI treatment and guideline adherence in 236 patients with eosinophilic oesophagitis‐Results from the population‐based DanEoE cohort shows a low complication rate. United European Gastroenterol J. 2021;9(8):910‐918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fujiwara Y, Sugawa T, Tanaka F, et al. A multicenter study on the prevalence of eosinophilic esophagitis and PPI‐responsive esophageal eosinophilic infiltration. Intern Med. 2012;51(23):3235‐3239. [DOI] [PubMed] [Google Scholar]
- 59. Kuruvilla ME, Lee FE, Lee GB. Understanding asthma phenotypes, endotypes, and mechanisms of disease. Clin Rev Allergy Immunol. 2019;56(2):219‐233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Agache I, Akdis CA. Precision medicine and phenotypes, endotypes, genotypes, regiotypes, and theratypes of allergic diseases. J Clin Invest. 2019;129(4):1493‐1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vanoni S, Zeng C, Marella S, et al. Identification of anoctamin 1 (ANO1) as a key driver of esophageal epithelial proliferation in eosinophilic esophagitis. J Allergy Clin Immunol. 2020;145(1):239‐254.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zeng C, Vanoni S, Wu D, et al. Solute carrier family 9, subfamily A, member 3 (SLC9A3)/sodium‐hydrogen exchanger member 3 (NHE3) dysregulation and dilated intercellular spaces in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2018;142(6):1843‐1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Collins MH. Histopathology of eosinophilic esophagitis. Dig Dis. 2014;32(1–2):68‐73. [DOI] [PubMed] [Google Scholar]
- 64. Collins MH, Blanchard C, Abonia JP, et al. Clinical, pathologic, and molecular characterization of familial eosinophilic esophagitis compared with sporadic cases. Clin Gastroenterol Hepatol. 2008;6(6):621‐629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Aceves SS, Chen D, Newbury RO, Dohil R, Bastian JF, Broide DH. Mast cells infiltrate the esophageal smooth muscle in patients with eosinophilic esophagitis, express TGF‐beta1, and increase esophageal smooth muscle contraction. J Allergy Clin Immunol. 2010;126(6):1198‐1204.e4. [DOI] [PubMed] [Google Scholar]
- 66. Aceves SS, Newbury RO, Dohil R, Bastian JF, Broide DH. Esophageal remodeling in pediatric eosinophilic esophagitis. J Allergy Clin Immunol. 2007;119(1):206‐212. [DOI] [PubMed] [Google Scholar]
- 67. Kwiatek MA, Hirano I, Kahrilas PJ, Rothe J, Luger D, Pandolfino JE. Mechanical properties of the esophagus in eosinophilic esophagitis. Gastroenterology. 2011;140(1):82‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Katzka DA, Tadi R, Smyrk TC, et al. Effects of topical steroids on tight junction proteins and spongiosis in esophageal epithelia of patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2014;12(11):1824‐1829.e1. [DOI] [PubMed] [Google Scholar]
- 69. Collins MH, Martin LJ, Alexander ES, et al. Newly developed and validated eosinophilic esophagitis histology scoring system and evidence that it outperforms peak eosinophil count for disease diagnosis and monitoring. Dis Esophagus. 2017;30(3):1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ravelli A, Villanacci V, Cadei M, Fuoti M, Gennati G, Salemme M. Dilated intercellular spaces in eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2014;59(5):589‐593. [DOI] [PubMed] [Google Scholar]
- 71. MacFarlane SN, Sontheimer H. Modulation of Kv1.5 currents by Src tyrosine phosphorylation: potential role in the differentiation of astrocytes. J Neurosci. 2000;20(14):5245‐5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. MacFarlane SN, Sontheimer H. Changes in ion channel expression accompany cell cycle progression of spinal cord astrocytes. Glia. 2000;30(1):39‐48. [DOI] [PubMed] [Google Scholar]
- 73. Habela CW, Olsen ML, Sontheimer H. ClC3 is a critical regulator of the cell cycle in normal and malignant glial cells. J Neurosci. 2008;28(37):9205‐9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ouadid‐Ahidouch H, Le Bourhis X, Roudbaraki M, Toillon RA, Delcourt P, Prevarskaya N. Changes in the K+ current‐density of MCF‐7 cells during progression through the cell cycle: possible involvement of a h‐ether.a‐gogo K+ channel. Recept Channels. 2001;7(5):345‐356. [PubMed] [Google Scholar]
- 75. Ouadid‐Ahidouch H, Roudbaraki M, Ahidouch A, Delcourt P, Prevarskaya N. Cell‐cycle‐dependent expression of the large Ca2+−activated K+ channels in breast cancer cells. Biochem Biophys Res Commun. 2004;316(1):244‐251. [DOI] [PubMed] [Google Scholar]
- 76. Ouadid‐Ahidouch H, Roudbaraki M, Delcourt P, Ahidouch A, Joury N, Prevarskaya N. Functional and molecular identification of intermediate‐conductance Ca(2+)‐activated K(+) channels in breast cancer cells: association with cell cycle progression. Am J Physiol Cell Physiol. 2004;287(1):C125‐C134. [DOI] [PubMed] [Google Scholar]
- 77. Woodfork KA, Wonderlin WF, Peterson VA, Strobl JS. Inhibition of ATP‐sensitive potassium channels causes reversible cell‐cycle arrest of human breast cancer cells in tissue culture. J Cell Physiol. 1995;162(2):163‐171. [DOI] [PubMed] [Google Scholar]
- 78. Blackiston DJ, McLaughlin KA, Levin M. Bioelectric controls of cell proliferation: ion channels, membrane voltage and the cell cycle. Cell cycle. 2009;8(21):3527‐3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ravelli AM, Villanacci V, Ruzzenenti N, et al. Dilated intercellular spaces: a major morphological feature of esophagitis. J Pediatr Gastroenterol Nutr. 2006;42(5):510‐515. [DOI] [PubMed] [Google Scholar]
- 80. Vaezi MF, Slaughter JC, Smith BS, et al. Dilated intercellular space in chronic laryngitis and gastro‐oesophageal reflux disease: at baseline and post‐lansoprazole therapy. Aliment Pharmacol Ther. 2010;32(7):916‐924. [DOI] [PubMed] [Google Scholar]
- 81. van Malenstein H, Farre R, Sifrim D. Esophageal dilated intercellular spaces (DIS) and nonerosive reflux disease. Am J Gastroenterol. 2008;103(4):1021‐1028. [DOI] [PubMed] [Google Scholar]
- 82. Mueller S, Neureiter D, Aigner T, Stolte M. Comparison of histological parameters for the diagnosis of eosinophilic oesophagitis versus gastro‐oesophageal reflux disease on oesophageal biopsy material. Histopathology. 2008;53(6):676‐684. [DOI] [PubMed] [Google Scholar]
- 83. Mueller S, Aigner T, Neureiter D, Stolte M. Eosinophil infiltration and degranulation in oesophageal mucosa from adult patients with eosinophilic oesophagitis: a retrospective and comparative study on pathological biopsy. J Clin Pathol. 2006;59(11):1175‐1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Katzka DA, Ravi K, Geno DM, et al. Endoscopic mucosal impedance measurements correlate with eosinophilia and dilation of intercellular spaces in patients with eosinophilic esophagitis. Clin Gastroenterol Hepatology. 2015;13(7):1242‐1248.e1. [DOI] [PubMed] [Google Scholar]
- 85. Swietach P, Vaughan‐Jones RD, Harris AL, Hulikova A. The chemistry, physiology and pathology of pH in cancer. Philos Trans R Soc Lond Ser B Biol Sci. 2014;369(1638):20130099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tobey NA, Gambling TM, Vanegas XC, Carson JL, Orlando RC. Physicochemical basis for dilated intercellular spaces in non‐erosive acid‐damaged rabbit esophageal epithelium. Dis Esophagus. 2008;21(8):757‐764. [DOI] [PubMed] [Google Scholar]
- 87. Orlando LA, Orlando RC. Dilated intercellular spaces as a marker of GERD. Curr Gastroenterol Rep. 2009;11(3):190‐194. [DOI] [PubMed] [Google Scholar]
- 88. Noffsinger AE. Update on esophagitis: controversial and underdiagnosed causes. Arch Pathol Lab Med. 2009;133(7):1087‐1095. [DOI] [PubMed] [Google Scholar]
- 89. Orlando RC. Current understanding of the mechanisms of gastro‐oesophageal reflux disease. Drugs. 2006;66(Suppl 1):1‐5; discussion 29‐33. [DOI] [PubMed] [Google Scholar]
- 90. Barlow WJ, Orlando RC. The pathogenesis of heartburn in nonerosive reflux disease: a unifying hypothesis. Gastroenterology. 2005;128(3):771‐778. [DOI] [PubMed] [Google Scholar]
- 91. Akei HS, Mishra A, Blanchard C, Rothenberg ME. Epicutaneous antigen exposure primes for experimental eosinophilic esophagitis in mice. Gastroenterology. 2005;129(3):985‐994. [DOI] [PubMed] [Google Scholar]
- 92. Doherty TA, Baum R, Newbury RO, et al. Group 2 innate lymphocytes (ILC2) are enriched in active eosinophilic esophagitis. J Allergy Clin Immunol. 2015;136(3):792‐794.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Abu‐Sultaneh SM, Durst P, Maynard V, Elitsur Y. Fluticasone and food allergen elimination reverse sub‐epithelial fibrosis in children with eosinophilic esophagitis. Dig Dis Sci. 2011;56(1):97‐102. [DOI] [PubMed] [Google Scholar]
- 94. Straumann A, Conus S, Degen L, et al. Budesonide is effective in adolescent and adult patients with active eosinophilic esophagitis. Gastroenterology. 2010;139(5):1526‐1537, 1537.e1. [DOI] [PubMed] [Google Scholar]
- 95. Arora AS, Perrault J, Smyrk TC. Topical corticosteroid treatment of dysphagia due to eosinophilic esophagitis in adults. Mayo Clin Proc. 2003;78(7):830‐835. [DOI] [PubMed] [Google Scholar]
- 96. Faubion WA Jr, Perrault J, Burgart LJ, Zein NN, Clawson M, Freese DK. Treatment of eosinophilic esophagitis with inhaled corticosteroids. J Pediatr Gastroenterol Nutr. 1998;27(1):90‐93. [DOI] [PubMed] [Google Scholar]
- 97. Kelly KJ, Lazenby AJ, Rowe PC, Yardley JH, Perman JA, Sampson HA. Eosinophilic esophagitis attributed to gastroesophageal reflux: improvement with an amino acid‐based formula. Gastroenterology. 1995;109(5):1503‐1512. [DOI] [PubMed] [Google Scholar]
- 98. Liacouras CA, Ruchelli E. Eosinophilic esophagitis. Curr Opin Pediatr. 2004;16(5):560‐566. [DOI] [PubMed] [Google Scholar]
- 99. Liacouras CA, Wenner WJ, Brown K, Ruchelli E. Primary eosinophilic esophagitis in children: successful treatment with oral corticosteroids. J Pediatr Gastroenterol Nutr. 1998;26(4):380‐385. [DOI] [PubMed] [Google Scholar]
- 100. Mishra A, Hogan SP, Brandt EB, Rothenberg ME. IL‐5 promotes eosinophil trafficking to the esophagus. J Immunol. 2002;168(5):2464‐2469. [DOI] [PubMed] [Google Scholar]
- 101. Mishra A, Hogan SP, Brandt EB, Rothenberg ME. An etiological role for aeroallergens and eosinophils in experimental esophagitis. J Clin Invest. 2001;107:83‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. D'Mello RJ, Caldwell JM, Azouz NP, et al. LRRC31 is induced by IL‐13 and regulates kallikrein expression and barrier function in the esophageal epithelium. Mucosal Immunol. 2016;9(3):744‐756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Davis BP, Stucke EM, Khorki ME, et al. Eosinophilic esophagitis‐linked calpain 14 is an IL‐13‐induced protease that mediates esophageal epithelial barrier impairment. JCI Insight. 2016;1(4):e86355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sherrill JD, Kc K, Wu D, et al. Desmoglein‐1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. 2014;7(3):718‐729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Fulkerson PC, Rothenberg ME. Eosinophil development, disease involvement, and therapeutic suppression. Adv Immunol. 2018;138:1‐34. [DOI] [PubMed] [Google Scholar]
- 106. Gleich GJ, Loegering DA. Immunobiology of eosinophils. Annu Rev Immunol. 1984;2(429):429‐459. [DOI] [PubMed] [Google Scholar]
- 107. Weller PF. Eosinophils: structure and functions. Curr Opin Immunol. 1994;6(1):85‐90. [DOI] [PubMed] [Google Scholar]
- 108. Rothenberg ME. Eosinophilia. N Engl J Med. 1998;338(22):1592‐1600. [DOI] [PubMed] [Google Scholar]
- 109. Hogan SP, Rosenberg HF, Moqbel R, et al. Eosinophils: biological properties and role in health and disease. Clin Exp Allergy. 2008;38(5):709‐750. [DOI] [PubMed] [Google Scholar]
- 110. Rothenberg ME, Mishra A, Brandt EB, Hogan SP. Gastrointestinal eosinophils in health and disease. Adv Immunol. 2001;78:291‐328. [DOI] [PubMed] [Google Scholar]
- 111. Pentiuk S, Putnam PE, Collins MH, Rothenberg ME. Dissociation between symptoms and histological severity in pediatric eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2009;48(2):152‐160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Cavalli E, Brusaferro A, Pieri ES, et al. Eosinophilic esophagitis in children: doubts and future perspectives. J Transl Med. 2019;17(1):262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. O'Shea KM, Rochman M, Shoda T, Zimmermann N, Caldwell J, Rothenberg ME. Eosinophilic esophagitis with extremely high esophageal eosinophil counts. J Allergy Clin Immunol. 2020;147(1):409‐412.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006;24:147‐174. [DOI] [PubMed] [Google Scholar]
- 115. Saffari H, Hoffman LH, Peterson KA, et al. Electron microscopy elucidates eosinophil degranulation patterns in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2014;133:1728‐1734.e1. [DOI] [PubMed] [Google Scholar]
- 116. Protheroe C, Woodruff SA, de Petris G, et al. A novel histologic scoring system to evaluate mucosal biopsies from patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2009;7(7):749‐755.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kephart GM, Alexander JA, Arora AS, et al. Marked deposition of eosinophil‐derived neurotoxin in adult patients with eosinophilic esophagitis. Am J Gastroenterol. 2010;105(2):298‐307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Chehade M, Sampson HA, Morotti RA, Magid MS. Esophageal subepithelial fibrosis in children with eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2007;45(3):319‐328. [DOI] [PubMed] [Google Scholar]
- 119. Rubinstein E, Cho JY, Rosenthal P, et al. Siglec‐F inhibition reduces esophageal eosinophilia and angiogenesis in a mouse model of eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2011;53(4):409‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Mavi P, Rajavelu P, Rayapudi M, Paul RJ, Mishra A. Esophageal functional impairments in experimental eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol. 2012;302(11):G1347‐G1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Mishra A, Wang M, Pemmaraju VR, et al. Esophageal remodeling develops as a consequence of tissue specific IL‐5‐induced eosinophilia. Gastroenterology. 2008;134(1):204‐214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Davis BP. Pathophysiology of eosinophilic esophagitis. Clin Rev Allergy Immunol. 2018;55(1):19‐42. [DOI] [PubMed] [Google Scholar]
- 123. Fulkerson P, Fischetti CA, McBride ML, Hassmann LM, Hogan SP, Rothenberg ME. A central regulatory role for eosinophils and the eotaxin/CCR3 axis in chronic experimental allergic airway inflammation. Proc Natl Acad Sci U S A. 2006;103:16418‐16423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Hogan S, Mishra E, Brandt E, Foster P, Rothenberg M. A critical role for eotaxin in experimental oral antigen‐indcued eosinophilic gastrointestinal allergy. Proc Natl Acad Sci U S A. 2000;97:6681‐6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Blanchard C, Wang N, Stringer KF, et al. Eotaxin‐3 and a uniquely conserved gene‐expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116(2):536‐547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Fulkerson PC, Fischetti CA, Rothenberg ME. Eosinophils and CCR3 regulate interleukin‐13 transgene‐induced pulmonary remodeling. Am J Pathol. 2006;169(6):2117‐2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Bullock JZ, Villanueva JM, Blanchard C, et al. Interplay of adaptive th2 immunity with eotaxin‐3/c‐C chemokine receptor 3 in eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2007;45(1):22‐31. [DOI] [PubMed] [Google Scholar]
- 128. Rochman M, Azouz NP, Rothenberg ME. Epithelial origin of eosinophilic esophagitis. J Allergy Clin Immunol. 2018;142(1):10‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Rochman M, Travers J, Miracle CE, et al. Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2017;140(3):738‐749.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zhang C, Hu Z, Lone AG, et al. Small proline‐rich proteins (SPRRs) are epidermally produced antimicrobial proteins that defend the cutaneous barrier by direct bacterial membrane disruption. elife. 2022;11:e76729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kalinska M, Meyer‐Hoffert U, Kantyka T, Potempa J. Kallikreins – the melting pot of activity and function. Biochimie. 2016;122:270‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Furio L, Hovnanian A. Netherton syndrome: defective kallikrein inhibition in the skin leads to skin inflammation and allergy. Biol Chem. 2014;395(9):945‐958. [DOI] [PubMed] [Google Scholar]
- 133. Nennstiel S, Schlag C. Treatment of eosinophlic esophagitis with swallowed topical corticosteroids. World J Gastroenterol. 2020;26(36):5395‐5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Jiang R, Shi Z, Johnson JJ, Liu Y, Stack MS. Kallikrein‐5 promotes cleavage of desmoglein‐1 and loss of cell‐cell cohesion in oral squamous cell carcinoma. J Biol Chem. 2011;286(11):9127‐9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Briot A, Deraison C, Lacroix M, et al. Kallikrein 5 induces atopic dermatitis‐like lesions through PAR2‐mediated thymic stromal lymphopoietin expression in Netherton syndrome. J Exp Med. 2009;206(5):1135‐1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Azouz NP, Klingler AM, Pathre P, et al. Functional role of kallikrein 5 and proteinase‐activated receptor 2 in eosinophilic esophagitis. Sci Transl Med. 2020;12(545):eaaz7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Azouz NP, Ynga‐Durand MA, Caldwell JM, et al. The antiprotease SPINK7 serves as an inhibitory checkpoint for esophageal epithelial inflammatory responses. Sci Transl Med. 2018;10(444):eaap9736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Rothenberg ME, Spergel JM, Sherrill JD, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42(4):289‐291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Sleiman PM, Wang ML, Cianferoni A, et al. GWAS identifies four novel eosinophilic esophagitis loci. Nat Commun. 2014;5:5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Cianferoni A, Ruffner MA, Guzek R, et al. Elevated expression of activated T(H)2 cells and milk‐specific T(H)2 cells in milk‐induced eosinophilic esophagitis. Ann Allergy Asthma Immunol. 2018;120(2):177‐183.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Wen T, Aronow BJ, Rochman Y, et al. Single‐cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis. J Clin Invest. 2019;129(5):2014‐2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Straumann A, Bauer M, Fischer B, Blaser K, Simon HU. Idiopathic eosinophilic esophagitis is associated with a TH2‐type allergic inflammatory response. J Allergy Clin Immunol. 2001;108(6):954‐961. [DOI] [PubMed] [Google Scholar]
- 143. Heller NM, Gwinn WM, Donnelly RP, Constant SL, Keegan AD. IL‐4 engagement of the type I IL‐4 receptor complex enhances mouse eosinophil migration to eotaxin‐1 in vitro. PLoS One. 2012;7(6):e39673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Schleimer RP, Sterbinsky SA, Kaiser J, et al. IL‐4 induces adherence of human eosinophils and basophils but not neutrophils to endothelium. Association with expression of VCAM‐1. J Immunol. 1992;148(4):1086‐1092. [PubMed] [Google Scholar]
- 145. Dunn JLM, Shoda T, Caldwell JM, et al. Esophageal type 2 cytokine expression heterogeneity in eosinophilic esophagitis in a multisite cohort. J Allergy Clin Immunol. 2020;145(6):1629‐1640.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Abonia JP, Blanchard C, Butz BB, et al. Involvement of mast cells in eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126(1):140‐149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Strasser DS, Seger S, Bussmann C, et al. Eosinophilic oesophagitis: relevance of mast cell infiltration. Histopathology. 2018;73(3):454‐463. [DOI] [PubMed] [Google Scholar]
- 148. Hsu Blatman KS, Gonsalves N, Hirano I, Bryce PJ. Expression of mast cell‐associated genes is upregulated in adult eosinophilic esophagitis and responds to steroid or dietary therapy. J Allergy Clin Immunol. 2011;127(5):1307‐1308.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Bolton SM, Kagalwalla AF, Arva NC, et al. Mast cell infiltration is associated with persistent symptoms and endoscopic abnormalities despite resolution of eosinophilia in pediatric eosinophilic esophagitis. Am J Gastroenterol. 2020;115(2):224‐233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Niranjan R, Mavi P, Rayapudi M, Dynda S, Mishra A. Pathogenic role of mast cells in experimental eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol. 2013;304(12):G1087‐G1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Nelson M, Zhang X, Pan Z, Spechler SJ, Souza RF. Mast cell effects on esophageal smooth muscle and their potential role in eosinophilic esophagitis and achalasia. Am J Physiol Gastrointest Liver Physiol. 2021;320(3):G319‐G327. [DOI] [PubMed] [Google Scholar]
- 152. Assa'ad AH, Putnam PE, Collins MH, et al. Pediatric patients with eosinophilic esophagitis: an 8‐year follow‐up. J Allergy Clin Immunol. 2007;119(3):731‐738. [DOI] [PubMed] [Google Scholar]
- 153. Simon D, Cianferoni A, Spergel JM, et al. Eosinophilic esophagitis is characterized by a non‐IgE‐mediated food hypersensitivity. Allergy. 2016;71(5):611‐620. [DOI] [PubMed] [Google Scholar]
- 154. Simon D, Straumann A, Dahinden C, Simon HU. Frequent sensitization to Candida albicans and profilins in adult eosinophilic esophagitis. Allergy. 2013;68(7):945‐948. [DOI] [PubMed] [Google Scholar]
- 155. Vicario M, Blanchard C, Stringer KF, et al. Local B cells and IgE production in the oesophageal mucosa in eosinophilic oesophagitis. Gut. 2010;59(1):12‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Blanchard C, Simon D, Schoepfer A, Straumann A, Simon HU. Eosinophilic esophagitis: unclear roles of IgE and eosinophils. J Intern Med. 2017;281(5):448‐457. [DOI] [PubMed] [Google Scholar]
- 157. Loizou D, Enav B, Komlodi‐Pasztor E, et al. A pilot study of omalizumab in eosinophilic esophagitis. PLoS One. 2015;10(3):e0113483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Clayton F, Fang JC, Gleich GJ, et al. Eosinophilic esophagitis in adults is associated with IgG4 and not mediated by IgE. Gastroenterology. 2014;147(3):602‐609. [DOI] [PubMed] [Google Scholar]
- 159. Wright BL, Kulis M, Guo R, et al. Food‐specific IgG(4) is associated with eosinophilic esophagitis. J Allergy Clin Immunol. 2016;138(4):1190‐1192.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Schuyler AJ, Wilson JM, Tripathi A, et al. Specific IgG(4) antibodies to cow's milk proteins in pediatric patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2018;142(1):139‐148.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Pope AE, Stanzione N, Naini BV, et al. Esophageal IgG4: clinical, endoscopic, and histologic correlations in eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2019;68(5):689‐694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Mohammad N, Avinashi V, Chan E, Vallance BA, Portales‐Casamar E, Bush JW. Pediatric eosinophilic esophagitis is associated with increased lamina propria immunoglobulin G4‐positive plasma cells. J Pediatr Gastroenterol Nutr. 2018;67(2):204‐209. [DOI] [PubMed] [Google Scholar]
- 163. Rosenberg CE, Mingler MK, Caldwell JM, et al. Esophageal IgG4 levels correlate with histopathologic and transcriptomic features in eosinophilic esophagitis. Allergy. 2018;73(9):1892‐1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Wright BL, Rank MA, Shim KS, et al. Esophageal IgG4 and eosinophilic inflammation correlate in subjects undergoing peanut oral immunotherapy. J Allergy Clin Immunol. 2018;141(2 Supplement):AB404. [Google Scholar]
- 165. Weidlich S, Nennstiel S, Jesinghaus M, et al. IgG4 is elevated in eosinophilic esophagitis but not in gastroesophageal reflux disease patients. J Clin Gastroenterol. 2020;54(1):43‐49. [DOI] [PubMed] [Google Scholar]
- 166. Wright BL, Kulis M, Guo R, et al. Food‐specific IgG4 is associated with eosinophilic esophagitis. J Allergy Clin Immunol. 2016;138(4):1190‐1192.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Aceves SS, Newbury RO, Chen D, et al. Resolution of remodeling in eosinophilic esophagitis correlates with epithelial response to topical corticosteroids. Allergy. 2010;65(1):109‐116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Aceves SS, Newbury RO, Dohil MA, Bastian JF, Dohil R. A symptom scoring tool for identifying pediatric patients with eosinophilic esophagitis and correlating symptoms with inflammation. Ann Allergy Asthma Immunol. 2009;103(5):401‐406. [DOI] [PubMed] [Google Scholar]
- 169. Rajan J, Newbury RO, Anilkumar A, Dohil R, Broide DH, Aceves SS. Long‐term assessment of esophageal remodeling in patients with pediatric eosinophilic esophagitis treated with topical corticosteroids. J Allergy Clin Immunol. 2016;137(1):147‐156.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Frangogiannis N. Transforming growth factor‐β in tissue fibrosis. J Exp Med. 2020;217(3):e20190103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Rieder F, Nonevski I, Ma J, et al. T‐helper 2 cytokines, transforming growth factor beta1, and eosinophil products induce fibrogenesis and alter muscle motility in patients with eosinophilic esophagitis. Gastroenterology. 2014;146(5):1266‐1277.e1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Armbruster‐Lee J, Cavender CP, Lieberman JA, Samarasinghe AE. Understanding fibrosis in eosinophilic esophagitis: are we there yet? J Leukoc Biol. 2018;104(1):31‐40. [DOI] [PubMed] [Google Scholar]
- 173. Salam MT, Gauderman WJ, McConnell R, Lin PC, Gilliland FD. Transforming growth factor‐ 1 C‐509T polymorphism, oxidant stress, and early‐onset childhood asthma. Am J Respir Crit Care Med. 2007;176(12):1192‐1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Silverman ES, Palmer LJ, Subramaniam V, et al. Transforming growth factor‐beta1 promoter polymorphism C‐509T is associated with asthma. Am J Respir Crit Care Med. 2004;169(2):214‐219. [DOI] [PubMed] [Google Scholar]
- 175. Duong LD, Rawson R, Bezryadina A, et al. TGFβ1 single‐nucleotide polymorphism C‐509T alters mucosal cell function in pediatric eosinophilic esophagitis. Mucosal Immunol. 2020;13(1):110‐117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Hsieh LY, Chiang AWT, Duong LD, et al. A unique esophageal extracellular matrix proteome alters normal fibroblast function in severe eosinophilic esophagitis. J Allergy Clin Immunol. 2021;148(2):486‐494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Rawson R, Anilkumar A, Newbury RO, et al. The TGFβ1 promoter SNP C‐509T and food sensitization promote esophageal remodeling in pediatric eosinophilic esophagitis. PLoS One. 2015;10(12):e0144651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Godwin B, Wilkins B, Muir AB. EoE disease monitoring: where we are and where we are going. Ann Allergy Asthma Immunol. 2020;124(3):240‐247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Lucendo AJ, Molina‐Infante J, Arias A, et al. Guidelines on eosinophilic esophagitis: evidence‐based statements and recommendations for diagnosis and management in children and adults. United European Gastroenterol J. 2017;5(3):335‐358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Safroneeva E, Straumann A, Coslovsky M, et al. Symptoms have modest accuracy in detecting endoscopic and histologic remission in adults with eosinophilic esophagitis. Gastroenterology. 2016;150(3):581‐590.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Ackerman SJ, Kagalwalla AF, Hirano I, et al. One‐hour esophageal string test: a nonendoscopic minimally invasive test that accurately detects disease activity in eosinophilic esophagitis. Am J Gastroenterol. 2019;114(10):1614‐1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Katzka DA, Smyrk TC, Alexander JA, et al. Accuracy and safety of the cytosponge for assessing histologic activity in eosinophilic esophagitis: a two‐center study. Am J Gastroenterol. 2017;112(10):1538‐1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Konikoff MR, Blanchard C, Kirby C, et al. Potential of blood eosinophils, eosinophil‐derived neurotoxin, and eotaxin‐3 as biomarkers of eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2006;4(11):1328‐1336. [DOI] [PubMed] [Google Scholar]
- 184. Henderson A, Magier A, Schwartz JT, et al. Monitoring eosinophilic esophagitis disease activity with blood eosinophil progenitor levels. J Pediatr Gastroenterol Nutr. 2020;70(4):482‐488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Hirano I, Chan ES, Rank MA, et al. AGA institute and the joint task force on allergy‐immunology practice parameters clinical guidelines for the management of eosinophilic esophagitis. Ann Allergy Asthma Immunol. 2020;124(5):416‐423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Arias A, Gonzalez‐Cervera J, Tenias JM, Lucendo AJ. Efficacy of dietary interventions for inducing histologic remission in patients with eosinophilic esophagitis: a systematic review and meta‐analysis. Gastroenterology. 2014;146(7):1639‐1648. [DOI] [PubMed] [Google Scholar]
- 187. Dellon ES, Liacouras CA, Molina‐Infante J, et al. Updated International Consensus diagnostic criteria for eosinophilic esophagitis: proceedings of the AGREE conference. Gastroenterology. 2018;155(4):1022‐1033.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Chehade M, Aceves SS. Treatment of eosinophilic esophagitis: diet or medication? J Allergy Clin Immunol Pract. 2021;9(9):3249‐3256. [DOI] [PubMed] [Google Scholar]
- 189. Miehlke S, Lucendo AJ, Straumann A, Jan Bredenoord A, Attwood S. Orodispersible budesonide tablets for the treatment of eosinophilic esophagitis: a review of the latest evidence. Ther Adv Gastroenterol. 2020;13:1756284820927282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Straumann A, Lucendo AJ, Miehlke S, et al. Budesonide orodispersible tablets maintain remission in a randomized, placebo‐controlled trial of patients with eosinophilic esophagitis. Gastroenterology. 2020;159(5):1672‐1685.e5. [DOI] [PubMed] [Google Scholar]
- 191. Greuter T, Godat A, Ringel A, et al. Effectiveness and safety of high‐ vs low‐dose swallowed topical steroids for maintenance treatment of eosinophilic esophagitis: a multicenter observational study. Clin Gastroenterol Hepatol. 2021;19(12):2514‐2523.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Bagnasco D, Caminati M, Ferrando M, et al. Anti‐IL‐5 and IL‐5Ra: efficacy and safety of new therapeutic strategies in severe uncontrolled asthma. Biomed Res Int. 2018;2018:5698212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Corren J. Inhibition of interleukin‐5 for the treatment of eosinophilic diseases. Discov Med. 2012;13(71):305‐312. [PubMed] [Google Scholar]
- 194. de Rooij WE, Dellon ES, Parker CE, et al. Pharmacotherapies for the treatment of eosinophilic esophagitis: state of the art review. Drugs. 2019;79(13):1419‐1434. [DOI] [PubMed] [Google Scholar]
- 195. Assa'ad AH, Gupta SK, Collins MH, et al. An antibody against IL‐5 reduces numbers of esophageal intraepithelial eosinophils in children with eosinophilic esophagitis. Gastroenterology. 2011;141(5):1593‐1604. [DOI] [PubMed] [Google Scholar]
- 196. Stein ML, Collins MH, Villanueva JM, et al. Anti‐IL‐5 (mepolizumab) therapy for eosinophilic esophagitis. J Allergy Clin Immunol. 2006;118(6):1312‐1319. [DOI] [PubMed] [Google Scholar]
- 197. Straumann A, Conus S, Grzonka P, et al. Anti‐interleukin‐5 antibody treatment (mepolizumab) in active eosinophilic oesophagitis: a randomised, placebo‐controlled, double‐blind trial. Gut. 2010;59(1):21‐30. [DOI] [PubMed] [Google Scholar]
- 198. Castro M, Mathur S, Hargreave F, et al. Reslizumab for poorly controlled, eosinophilic asthma: a randomized, placebo‐controlled study. Am J Respir Crit Care Med. 2011;184(10):1125‐1132. [DOI] [PubMed] [Google Scholar]
- 199. Spergel JM, Rothenberg ME, Collins MH, et al. Reslizumab in children and adolescents with eosinophilic esophagitis: results of a double‐blind, randomized, placebo‐controlled trial. J Allergy Clin Immunol. 2012;129(2):456‐463, 463.e1‐3. [DOI] [PubMed] [Google Scholar]
- 200. Markowitz JE, Jobe L, Miller M, Frost C, Laney Z, Eke R. Safety and efficacy of reslizumab for children and adolescents with eosinophilic esophagitis treated for 9 years. J Pediatr Gastroenterol Nutr. 2018;66(6):893‐897. [DOI] [PubMed] [Google Scholar]
- 201. FitzGerald JM, Bleecker ER, Nair P, et al. Benralizumab, an anti‐interleukin‐5 receptor alpha monoclonal antibody, as add‐on treatment for patients with severe, uncontrolled, eosinophilic asthma (CALIMA): a randomised, double‐blind, placebo‐controlled phase 3 trial. Lancet. 2016;388(10056):2128‐2141. [DOI] [PubMed] [Google Scholar]
- 202. Nutku E, Aizawa H, Hudson SA, Bochner BS. Ligation of Siglec‐8: a selective mechanism for induction of human eosinophil apoptosis. Blood. 2003;101(12):5014‐5020. [DOI] [PubMed] [Google Scholar]
- 203. Legrand F, Cao Y, Wechsler JB, et al. Sialic acid‐binding immunoglobulin‐like lectin (Siglec) 8 in patients with eosinophilic disorders: receptor expression and targeting using chimeric antibodies. J Allergy Clin Immunol. 2019;143(6):2227‐2237.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Youngblood BA, Leung J, Falahati R, et al. Discovery, function, and therapeutic targeting of Siglec‐8. Cell. 2020;10(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Dellon ES, Peterson KA, Murray JA, et al. Anti‐Siglec‐8 antibody for eosinophilic gastritis and duodenitis. N Engl J Med. 2020;383(17):1624‐1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Rothenberg ME, Wen T, Greenberg A, et al. Intravenous anti‐IL‐13 mAb QAX576 for the treatment of eosinophilic esophagitis. J Allergy Clin Immunol. 2015;135(2):500‐507. [DOI] [PubMed] [Google Scholar]
- 207. Hirano I, Collins MH, Assouline‐Dayan Y, et al. RPC4046, a monoclonal antibody against IL13, reduces histologic and endoscopic activity in patients with eosinophilic esophagitis. Gastroenterology. 2019;156(3):592‐603.e10. [DOI] [PubMed] [Google Scholar]
- 208. Dellon ES, Collins MH, Rothenberg ME, et al. Long‐term efficacy and tolerability of RPC4046 in an open‐label extension trial of patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2021;19(3):473‐483.e17. [DOI] [PubMed] [Google Scholar]
- 209. Sastre J, Davila I. Dupilumab: a new paradigm for the treatment of allergic diseases. J Investig Allergol Clin Immunol. 2018;28(3):139‐150. [DOI] [PubMed] [Google Scholar]
- 210. Junttila IS. Tuning the cytokine responses: an update on interleukin (IL)‐4 and IL‐13 receptor complexes. Front Immunol. 2018;9:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Hirano I, Dellon ES, Hamilton JD, et al. Efficacy of dupilumab in a phase 2 randomized trial of adults with active eosinophilic esophagitis. Gastroenterology. 2020;158(1):111‐122.e10. [DOI] [PubMed] [Google Scholar]
- 212. Dellon ES, Rothenberg ME, Collins MH, et al. A phase 3, randomized, 3‐part study to investigate the efficacy and safety of dupilumab in adult and adolescent patients with eosinophilic esophagitis: results from part A. American College of Gastroenterology 2020 Annual Meeting; October 23‐28th, 2020; Virtual.
- 213. Peng G, Mu Z, Cui L, et al. Anti‐IL‐33 antibody has a therapeutic effect in an atopic dermatitis murine model induced by 2, 4‐dinitrochlorobenzene. Inflammation. 2018;41(1):154‐163. [DOI] [PubMed] [Google Scholar]
- 214. Holgado A, Braun H, Van Nuffel E, et al. IL‐33trap is a novel IL‐33‐neutralizing biologic that inhibits allergic airway inflammation. J Allergy Clin Immunol. 2019;144(1):204‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Chen YL, Gutowska‐Owsiak D, Hardman CS, et al. Proof‐of‐concept clinical trial of etokimab shows a key role for IL‐33 in atopic dermatitis pathogenesis. Sci Transl Med. 2019;11(515):eaax2945. [DOI] [PubMed] [Google Scholar]
- 216. Chinthrajah S, Cao S, Liu C, et al. Phase 2a randomized, placebo‐controlled study of anti‐IL‐33 in peanut allergy. JCI Insight. 2019;4(22):e131347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Nakajima S, Kabata H, Kabashima K, Asano K. Anti‐TSLP antibodies: targeting a master regulator of type 2 immune responses. Allergol Int. 2020;69(2):197‐203. [DOI] [PubMed] [Google Scholar]
- 218. Marone G, Spadaro G, Braile M, et al. Tezepelumab: a novel biological therapy for the treatment of severe uncontrolled asthma. Expert Opin Investig Drugs. 2019;28(11):931‐940. [DOI] [PubMed] [Google Scholar]
- 219. Simpson EL, Parnes JR, She D, et al. Tezepelumab, an anti‐thymic stromal lymphopoietin monoclonal antibody, in the treatment of moderate to severe atopic dermatitis: a randomized phase 2a clinical trial. J Am Acad Dermatol. 2019;80(4):1013‐1021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.


