Abstract
Summary
Numerous R packages have been developed for bioinformatics analysis in the last decade and dependencies among packages have become critical issues to consider. In this work, we proposed a new metric named dependency heaviness that measures the number of dependencies that a parent uniquely brings to a package and we proposed possible solutions for reducing the complexity of dependencies by optimizing the use of heavy parents. We implemented the metric in a new R package pkgndep which provides an intuitive way for dependency heaviness analysis. Based on pkgndep, we additionally performed a global analysis of dependency heaviness on CRAN and Bioconductor ecosystems and we revealed top packages that have significant contributions of high dependency heaviness to their child packages.
Availability and implementation
The package pkgndep and documentations are freely available from the Comprehensive R Archive Network https://cran.r-project.org/package=pkgndep. The dependency heaviness analysis for all 22 076 CRAN and Bioconductor packages retrieved on June 8, 2022 are available at https://pkgndep.github.io/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Reliable and robust software is essential for data analysis in bioinformatics research. In the last decade, R has rapidly become a major programming language for developing software for biological data analysis, including data processing and visualization, statistical modeling, interactive web application development and reproducible report generation. It is applied in broad research fields of genomics, transcriptomics, epigenomics, proteomics, metabolomics and more. Reusable and extensible code is normally organized in forms of R packages, which are distributed on public repositories such as the Comprehensive R Archive Network (CRAN) and Bioconductor. Publications describing R packages have also been increasing. By June 8, 2022, using search term ‘(R package [Title]) OR Bioconductor [Title/Abstract]’ on PubMed resulted in 2605 publications. Dependencies always exist in packages where a package imports functionalities from other packages. As the numbers of R packages increase, dependencies among packages become even more complicated. By June 8, 2022, there are 194 351 direct dependency relations among in total 18 638 and 3438 packages from CRAN and Bioconductor, respectively.
In the ecosystems of CRAN and Bioconductor, package dependencies are represented as a directed graph. Research on the dependency graph helps to reveal interesting patterns of package relations, such as top packages that play leading roles or package modules for specific analysis topics (Mora-Cantallops et al., 2020). There are also R packages for dependency graph visualizations, such as deepdep, pkgnet and pkggraph. They support visualizing subgraphs induced from single packages.
Denote a package as P in the dependency graph, then the total packages located upstream of P compose its dependencies. If P depends on a large number of upstream packages, there will be the following risks: (i) Users have to install a lot of additional packages when installing P, which would bring the risk that installation failure of any upstream package stops the installation of P. (ii) The number of packages loaded into the R session after loading P will be huge, which increases the difficulty to reproduce a completely identical working environment on other computers. (iii) Dependencies of P will spread to all its child packages. (iv) On the platforms for continuous integration such as GitHub Action or Travis CI, automatic validation of P could easily fail due to the failures of its upstream packages. P inherits all its dependencies from its parent packages. Thus, identifying the parents that contribute high dependencies to P, that is, identifying P’s heavy parents, helps to reduce the complexity of P’s own dependencies.
In this work, we proposed a new metric named dependency heaviness that measures the number of additional dependency packages that a parent brings to its child package and are mutually exclusive to the dependencies imported by all other parents. We implemented the heaviness metric in an R package named pkgndep. Additionally, pkgndep provides an intuitive way for visualizing package dependencies. Based on pkgndep, we also performed a preliminary analysis of dependency heaviness on the CRAN and Bioconductor ecosystems and we revealed top packages that contribute significantly high heaviness on their child packages.
2 Materials and methods
Every R package has a DESCRIPTION file for its metadata. In it, its dependency packages are listed in the fields of ‘Depends’, ‘Imports’, ‘LinkingTo’, ‘Suggests’ and ‘Enhances’. Denote a package as P, packages listed in ‘Depends’, ‘Imports’ and ‘LinkingTo’ are mandatory to be installed when installing P. Functions, S4 methods or S4 classes defined in packages in ‘Depends’ and ‘Imports’ are imported to the namespace of P according to the rules defined in P’s NAMESPACE file. Packages listed in ‘Suggests’ and ‘Enhances’ are not mandatory to be installed. They are optionally used in P, such as in vignettes, in examples, or in functions that are only required when they are used. We kindly refer readers to the official R manual ‘Writing R Extensions’ for more details.
R package dependencies can be modeled as a directed graph. For the ease of discussion, the following dependency categories for P are defined. (i) Strong parent packages: the packages listed in ‘Depends’, ‘Imports’ and ‘LinkingTo’. For simplicity, in the text, we always refer to them as parent packages. (ii) Weak parent packages: the packages listed in ‘Suggests’ and ‘Enhances’. (iii) Strong dependency packages: the total packages by recursively looking for their parent packages. Note all strong dependency packages are mandatory to be installed for P and a failure of any of them stops installation of P. In the text, we always refer to them as dependency packages. (iv) All dependency packages: the total packages by recursively looking for their parent packages, but at the level of P, its weak parents are also included. It simulates a situation in which full functionality of P is required, or in other words, when all its weak parents are changed to strong parents, the number of strong dependency packages it requires. (v) Child packages: the packages whose parent packages include P. These are the packages on which P has a direct contribution on dependencies.
Next the heaviness from a strong parent is defined. If package A is a parent of P, the heaviness of A on P denoted as h is calculated as h = n1 − n2, where n1 is the number of strong dependencies of P and n2 is the number of strong dependencies of P after changing A from a strong parent to a weak parent, that is, moving A to ‘Suggests’ of P. From the aspect of the dependency graph where only the strong dependency relations are included, n1 is the number of upstream packages of P and n2 is the number of upstream packages after removing the connection between A and P. So, the heaviness measures the number of additionally required strong dependencies that A brings to P and these dependencies are not imported by any other parent.
When A is a weak parent of P, n2 is defined as the number of strong dependencies of P after changing A to a strong parent of P, that is, moving A to ‘Imports’ of P. In this scenario, the heaviness of the weak parent is calculated as h = n2 − n1. Similarly, from the aspect of the dependency graph, n2 is the number of upstream packages after adding a new connection between A and P.
Note for some (A, P) pairs, it is possible that n1 = n2 which results in h = 0, that is, moving parent A to ‘Suggests’ of P won’t reduce the number of P’s strong dependencies. It mostly occurs when a second parent, denoted as B, also depends on A (directly or indirectly). Thus, only moving A to ‘Suggests’ of P won’t affect that A is still imported as a strong dependency by B. For example, the package GenomicRanges is a parent of the package bsseq where GenomicRanges is a general-purpose package for dealing with genomic intervals. bsseq has a second parent package BSgenome that also depends on GenomicRanges. Thus, moving GenomicRanges to ‘Suggests’ of bsseq won’t change the number of strong dependencies for bsseq because BSgenome still imports it. As a result, GenomicRanges contribute zero heaviness on bsseq as its parent.
3 Results
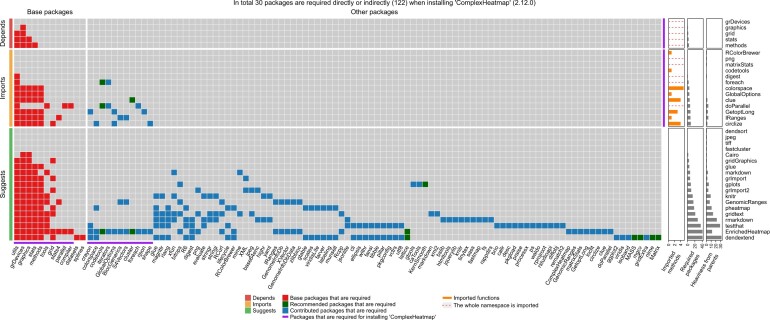
The result of a dependency heaviness analysis is represented as a customized heatmap. Figure 1 demonstrates the dependency heatmap for the package ComplexHeatmap. ComplexHeatmap’s strong and weak parents are rows split by their dependency relations, that is, ‘Depends’, ‘Imports’ or ‘Suggests’. The dependency packages imported by each parent are columns and they are split into two groups: base packages and other packages. The red, dark green and blue cells correspond to dependency packages that each parent requires. The total of all strong dependencies required for ComplexHeatmap are marked by purple lines on both rows and columns. On the right side of the dependency heatmap, there are three additional bar plot annotations. The first bar plot annotation illustrates the numbers of functions or classes imported from parents, which are parsed from ComplexHeatmap’s NAMESPACE file. In general, the bar plot can distinguish the following different scenarios of imports from a parent to a child package: (i) The whole namespace is imported. (ii) A limited number of functions are imported. (iii) A limited number of S4 methods/classes is imported (e.g. in package biovizBase). (iv) The whole namespace excluding a limited number of functions is imported (e.g. in package dplyr). (v) No function is imported but the parent still has an ‘Imports’ relation to the child package (e.g. in package ggplot2). The second bar plot annotation illustrates the number of required packages for each strong or weak parent, which is simply the number of hits on the corresponding row. The third bar plot annotation illustrates the heaviness of each strong or weak parent package on ComplexHeatmap, which is the number of required packages uniquely imported by the parent while not by all other strong parents.
Fig. 1.
Dependency heaviness analysis of the package ComplexHeatmap. The interpretation of the plot can be found in the main text
As illustrated in Figure 1, ComplexHeatmap has 30 strong dependency packages, while if including dependencies from all weak parents as well, the number will dramatically increase to 122 (more than four times more). Since we are also the developers of ComplexHeatmap, during the development, we moved some heavy parents, which only provide enhanced functionalities for ComplexHeatmap but are not expected to be frequently used, to ‘Suggests’, such as the package dendextend (contributing a heaviness of 32) which is only used for coloring dendrogram branches, and the package gridtext (contributing a heaviness of 14) which is only used for customizing text formats in heatmaps. These two packages are only required when the corresponding functionalities are used.
The dependency heatmap gives hints for reducing dependency complexity of a package. Generally speaking, if a parent has a high heaviness compared with other parents, an optimization might be considered, especially if only a few functions are imported from that parent. In Supplementary File S1 which contains dependency heaviness analysis for the package mapStats, an extremely heavy parent Hmisc can be observed where Hmisc has a heaviness of 49 which is almost 60% of the total number of required packages of mapStats. If moving Hmisc to ‘Suggests’ of mapStats, the number of strong dependencies can be reduced from 83 to 34. The first bar plot annotation shows there is only one function imported from Hmisc. A deep inspection into the source code of mapStats reveals that a function capitalize() from Hmisc is only imported to mapStats. capitalize() is a simple function that only capitalizes the first letter of a string. The 49 additional dependencies imported from Hmisc can be avoided by simply reimplementing a function capitalize() by the developer’s own.
In the package pkgndep, the function pkgndep() performs dependency heaviness analysis for a specific package. Later the function plot() generates the dependency heatmap and the function dependency_report() generates an HTML report of the complete dependency analysis.
We performed heaviness analysis for all 18 638 CRAN packages and 3438 Bioconductor packages retrieved on June 8, 2022 (Supplementary File S2, also available at https://pkgndep.github.io/). We systematically analyzed the heaviness of packages on their child packages. For a package denoted as P, the mean heaviness over those children for which P is a strong parent was calculated. This metric also measures the expected number of additional dependencies brought to a package if P is added as a new strong parent. On top of the list of packages ordered by their heaviness on children (Supplementary File S3), we found there are some popular R packages for bioinformatics analysis that have very high heaviness on their child packages, such as Seurat (85, 38 children), minfi (62, 38 children), WGCNA (52, 33 children) and Gviz (43, 37 children) where the first value in the parentheses is the average heaviness on their children. Thus, packages depending on these heavy parents should be aware of the risks from the huge dependencies they bring in.
4 Discussion and conclusion
Dependency heaviness analysis provides hints for reducing the complexity of package dependencies, but how to optimize the dependency depends on the specific use of parent packages in the corresponding package. As has been demonstrated in the example of mapStats, the heavy parent can be avoided by implementing a function with the same functionality as the imported one, but this scenario is not common. Common cases are when heavy parents are less used in analyses by users. This is typical for some bioinformatics packages which provide both core analysis and secondary analysis. Parent packages for secondary analysis might be arranged as weak parents (i.e. to be put in ‘Suggests’) if they are heavy and not expected to be frequently accessed by users (see an example in Supplementary File S4). Another solution is to separate large packages into several smaller packages where each one only focuses on a specific task, for example, the package remotes separated from the package devtools only focuses on installing packages from remote repositories, where remotes only has 4 strong dependencies, but devtools has 76. Nevertheless, there are still scenarios when reduction of heavy parents could not be performed: (i) A heavy parent provides core functionality to its child package and (ii) S4 methods or S4 classes are imported from a parent package.
The heaviness measures the number of additional dependency packages that a parent uniquely imports. However, there are scenarios when multiple parents import similar sets of dependencies, which results in heaviness for individual parents being very small. In Supplementary File S5 of the dependency analysis for the package DESeq2, its two parent packages geneplotter and genefilter import 51 and 53 dependencies, respectively, among which 50 are the same. Due to the high overlap, the heaviness of geneplotter and genefilter on DESeq2 is only 1 and 2. However, if considering the two parents together, that is, by moving both to ‘Suggests’ of DESeq2, 23 dependency packages can be reduced. pkgndep also proposes a metric named co-heaviness (by the function co_heaviness()) which measures the number of dependencies simultaneously imported by two parents (Supplementary File S5). Nevertheless, the co-effects of multiple parents can always be easily observed from the dependency heatmap.
Current studies on package dependencies rank packages’ influences by the numbers of their child packages, that is, through degree centralities in the dependency graph analysis (Mora-Cantallops et al., 2020). However, a package with more child packages does not necessarily mean it contributes more dependencies. For example, package Rcpp has 2795 child packages but it only contributes an average heaviness of 0.6 to them.
R packages may also have system dependencies, for example, on C/C++ libraries, which can involve another layer of complex dependency relations. pkgndep only considers the dependency relations on the R package level.
In this study, we proposed a new metric that helps to find parents contributing heavy dependencies to their child packages. We believe pkgndep will be a useful tool for R package developers to properly handle the dependency complexity of their packages and to build more robust and reliable packages.
Funding
This work was supported by the National Center for Tumor Diseases (NCT) Molecular Precision Oncology Program.
Conflict of Interest: none declared.
Supplementary Material
Contributor Information
Zuguang Gu, Molecular Precision Oncology Program, National Center for Tumor Diseases (NCT).
Daniel Hübschmann, Molecular Precision Oncology Program, National Center for Tumor Diseases (NCT); Heidelberg Institute of Stem Cell Technology and Experimental Medicine (HI-STEM); German Cancer Consortium (DKTK); Department of Pediatric Immunology, Hematology and Oncology, University Hospital Heidelberg, 69120 Heidelberg, Germany.
Data Availability
The data underlying this article are available in the article and in its online supplementary material.
Reference
- Mora-Cantallops M. et al. (2020) A complex network analysis of the comprehensive R archive network (CRAN) package ecosystem. J. Syst. Softw., 170, 110744. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.