Abstract
African cichlids (subfamily: Pseudocrenilabrinae) are among the most diverse vertebrates, and their propensity for repeated rapid radiation has made them a celebrated model system in evolutionary research. Nonetheless, despite numerous studies, phylogenetic uncertainty persists, and riverine lineages remain comparatively underrepresented in higher-level phylogenetic studies. Heterogeneous gene histories resulting from incomplete lineage sorting (ILS) and hybridization are likely sources of uncertainty, especially during episodes of rapid speciation. We investigate the relationships of Pseudocrenilabrinae and its close relatives while accounting for multiple sources of genetic discordance using species tree and hybrid network analyses with hundreds of single-copy exons. We improve sequence recovery for distant relatives, thereby extending the taxonomic reach of our probes, with a hybrid reference guided/de novo assembly approach. Our analyses provide robust hypotheses for most higher-level relationships and reveal widespread gene heterogeneity, including in riverine taxa. ILS and past hybridization are identified as the sources of genetic discordance in different lineages. Sampling of various Blenniiformes (formerly Ovalentaria) adds strong phylogenomic support for convict blennies (Pholidichthyidae) as sister to Cichlidae and points to other potentially useful protein-coding markers across the order. A reliable phylogeny with representatives from diverse environments will support ongoing taxonomic and comparative evolutionary research in the cichlid model system. [African cichlids; Blenniiformes; Gene tree heterogeneity; Hybrid assembly; Phylogenetic network; Pseudocrenilabrinae; Species tree.]
A reliable phylogeny is a necessary foundation for investigating evolutionary processes. Despite substantial efforts, however, recalcitrant nodes and uncertainty persist for some groups, including diverse clades with rapid radiations considered model systems in evolutionary research (Murphy et al. 2007; Jarvis et al. 2014; Hughes et al. 2018; Takahashi and Sota 2016). African cichlids (Blenniiformes sensuGhezelayagh et al. 2021: Cichlidae: Pseudocrenilabrinae) are a celebrated model system for studies of speciation, adaptation, and rapid radiation (Seehausen 2006). They are the largest cichlid subfamily with 1700 species (Turner 2007; Seehausen 2015; Froese and Pauly 2021) inhabiting rivers and lakes across Africa and include five species in the Jordan Valley and coastal basins of southern Iran (Fig. 1). Some lineages underwent rapid radiations in east African Great Lakes (Kocher 2004; Seehausen 2006), various small lakes around the continent (Roberts and Kullander 1994; Schliewen et al. 1994; Joyce et al. 2005; Seehausen 2006; Ford et al. 2016), and the lower Congo river basin (Schwarzer et al. 2011; Stiassny and Alter 2021), but still others have remained relatively species poor. Such striking disparity among lineages has spurred research into diversification along an array of dimensions (e.g., phenotypic, molecular, and environmental) in an effort to reveal intrinsic and extrinsic factors that influence speciation and adaptation in cichlids (Hulsey and García de León 2005; Seehausen 2006; Wagner et al. 2012; López-Fernández et al. 2013; Brawand et al. 2014; Astudillo-Clavijo et al. 2015; Arbour and López-Fernández 2016; Ivory et al. 2016; Burress et al. 2017; Meier et al. 2017; Salzburger 2018; Ronco et al. 2021). Despite the significance of African cichlids in evolutionary research, uncertainty persists at several deep nodes and riverine lineages remain underrepresented in most higher-level phylogenetic hypotheses. A reliable phylogeny with broad sampling across tribes from diverse environments is crucial for furthering macroevolutionary research in the cichlid model system.
species (Turner 2007; Seehausen 2015; Froese and Pauly 2021) inhabiting rivers and lakes across Africa and include five species in the Jordan Valley and coastal basins of southern Iran (Fig. 1). Some lineages underwent rapid radiations in east African Great Lakes (Kocher 2004; Seehausen 2006), various small lakes around the continent (Roberts and Kullander 1994; Schliewen et al. 1994; Joyce et al. 2005; Seehausen 2006; Ford et al. 2016), and the lower Congo river basin (Schwarzer et al. 2011; Stiassny and Alter 2021), but still others have remained relatively species poor. Such striking disparity among lineages has spurred research into diversification along an array of dimensions (e.g., phenotypic, molecular, and environmental) in an effort to reveal intrinsic and extrinsic factors that influence speciation and adaptation in cichlids (Hulsey and García de León 2005; Seehausen 2006; Wagner et al. 2012; López-Fernández et al. 2013; Brawand et al. 2014; Astudillo-Clavijo et al. 2015; Arbour and López-Fernández 2016; Ivory et al. 2016; Burress et al. 2017; Meier et al. 2017; Salzburger 2018; Ronco et al. 2021). Despite the significance of African cichlids in evolutionary research, uncertainty persists at several deep nodes and riverine lineages remain underrepresented in most higher-level phylogenetic hypotheses. A reliable phylogeny with broad sampling across tribes from diverse environments is crucial for furthering macroevolutionary research in the cichlid model system.
Figure 1.
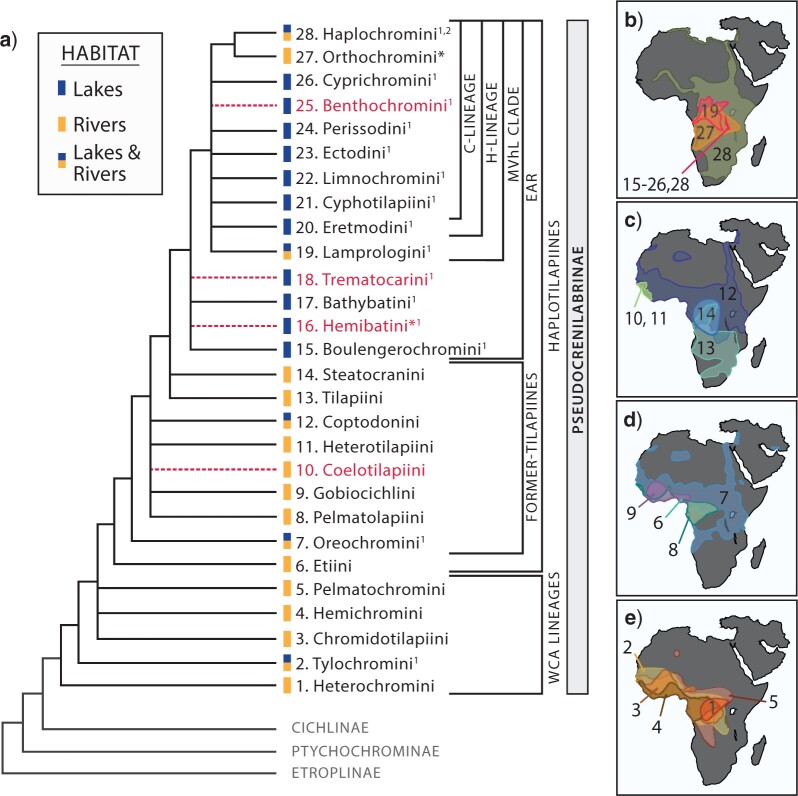
(Color online) Cichlidae and Pseudocrenilabrinae relationships prior to this work (a) and distribution of Pseudocrenilabrinae tribes (b–e). a) Pseudocrenilabrinae relationships are based on combined evidence from existing molecular phylogenies. Bifurcating branches indicate robust relationships. Polytomies show conflicting or poorly supported nodes. Dashed (red) branches are tribes that were not sampled in this study. Square brackets to the right of the phylogeny denote commonly recovered tribal assemblages or groupings discussed throughout the text: WCA, West/Central African Lineages; EAR, East African Radiations Clade; MVhL, Malawi, Victoria, H-Lineage and Lamprologini Clade; H-Lineage, Haplochromini and remaining Tanganyikan tribes; C-Lineage, H-Lineage minus Eretmodini.  Tribes with Tanganyikan endemics.
Tribes with Tanganyikan endemics.  The Malawi and Victoria system radiations are nested within Haplochromini. *Tribes with disputed taxonomic status. Rectangles next to tribe names indicate tribes with riverine and/or lacustrine lineages. Numbering of tribes matches numbers in the distribution maps (b–e). b) EAR tribes 15–28. c) Former-tilapiine tribes 10–14. d) Etiini and former-tilapiine tribes 7–9. e) WCA tribes 1–5. See Figure S1 in Appendix S1 of the Supplementary material available on Dryad for individual tribal distribution maps.
The Malawi and Victoria system radiations are nested within Haplochromini. *Tribes with disputed taxonomic status. Rectangles next to tribe names indicate tribes with riverine and/or lacustrine lineages. Numbering of tribes matches numbers in the distribution maps (b–e). b) EAR tribes 15–28. c) Former-tilapiine tribes 10–14. d) Etiini and former-tilapiine tribes 7–9. e) WCA tribes 1–5. See Figure S1 in Appendix S1 of the Supplementary material available on Dryad for individual tribal distribution maps.
Monophyly of Pseudocrenilabrinae is strongly supported by molecular data (Farias et al. 1999, 2000; Sparks and Smith 2004; Keck and Hulsey 2014), and its internal relationships have been explored to various degrees with morphological, multilocus, and genomic data (see Appendix S1 of the Supplementary material available on Dryad at http://dx.doi.org/10.5061/dryad.d7wm37q26 for review of previous phylogenetic findings). Currently, 26–28 tribes are formally or informally recognized and some form part of consistently recovered tribal assemblages (Fig. 1a). Relationships within the East African Radiations (EAR) clade, which contain most lake taxa, have been assessed most thoroughly (e.g., Dunz and Schliewen 2013; Weiss et al. 2015; Takahashi and Sota 2016; Ronco et al. 2020). By contrast, few detailed hypotheses are available for the various early diverging and largely riverine lineages. Representation of the oldest West/Central African tribes (henceforth WCA lineages) and early haplotilapiines (Schliewen and Stiassny 2003) formerly grouped into the catch-all Tilapiini (sensu Poll 1986 minus Boulengerochromini, Takahashi 2003; Dunz and Schliewen 2013; henceforth former-tilapiines) has generally been limited to a handful of species with the aim of addressing family-level (Streelman and Karl 1997; Streelman et al. 1998; Sparks and Smith 2004; Friedman et al. 2013), Neotropical (Farias et al. 1999, 2000; Smith et al. 2008; Ilves et al. 2018), or EAR relationships (Sültmann et al. 1995; Mayer et al. 1998; Salzburger et al. 2005; Genner et al. 2007; Schedel et al. 2019, Ronco et al. 2021). Studies with a larger sampling of WCA and former-tilapiine taxa are usually focused on shallower intrageneric (Schwarzer et al. 2011; Ford et al. 2019) or intratribal (Martin et al. 2015; Schwarzer et al. 2015) relationships. Consequently, densely sampled hypotheses of higher-level Pseudocrenilabrinae relationships are currently represented by primarily multilocus phylogenies (but see Irisarri et al. 2018), most of which have limited riverine representation and reveal variously conflicting topologies (Schwarzer et al. 2009; Dunz and Schliewen 2013; Matschiner et al. 2017; Schedel et al. 2019).
The growing availability of large molecular data sets is revealing genetic discordance, the presence of conflicting evolutionary histories amongst loci and between loci and the underlying species tree, as an important contributor to persistent phylogenetic uncertainty (Maddison 1997; Edwards 2009; Jarvis et al. 2014; Hughes et al. 2018; Irisarri et al. 2018; Salzburger 2018). Incomplete lineage sorting (ILS) and hybridization are foremost sources of gene heterogeneity, especially in rapidly radiating clades like cichlids (Takahashi et al. 2001; Koblmüller et al. 2010; Martin et al. 2015; Meier et al. 2017; Malinsky et al. 2018; McGee et al. 2020) and various others (Whitefield and Lockhard 2007; Litsios and Salamin 2014; Suh et al. 2015; Grant and Grant 2019; Cai et al. 2021; Morales-Briones et al. 2021). Irrespective of its source, elevated gene heterogeneity can positively mislead (Felsenstein 1978) phylogenetic inference when unaccounted for (Kubatko and Degnan 2007; Liu et al. 2015; Roch and Steel 2015; Mirarab et al. 2016).
Several high-throughput sequencing methods can now yield hundreds to thousands of loci (Miller et al. 2007; Faircloth et al. 2012; Lemmon et al. 2012) comprising more representative samples of alternative genealogies than small multilocus data sets. Targeted sequence capture is particularly appealing as it allows for the curation of data sets to meet specific challenges of the focal group. Target capture with taxon-specific probes provides added control over locus identity, capture rates, the proportion of informative sites, and capture of paralogs (Ilves and López-Fernández 2014; Chau et al. 2018; Hughes et al. 2018; Jiang et al. 2019).
Complementary phylogenetic methods that account for the diversity of sampled gene histories are also on the rise, fueled by a recognition of traditional concatenation’s vulnerability to systematic error in the presence of gene heterogeneity (Degnan and Rosenberg 2009; Roch and Warnow 2015). Coalescent species tree methods are most commonly applied as these tend to outperform concatenation with rising gene heterogeneity (Kubatko and Degnan 2007; Davidson et al. 2015; Liu et al. 2015; Roch and Steel 2015; Mirarab et al. 2016). Species tree methods assume ILS to be the prime source of discordance and do not explicitly account for other process, such as hybridization (Solís-Lemus et al. 2016, but see Davidson et al. 2015). Approaches that model gene flow in the presence of ILS are also available, but their application to phylogenomic or taxonomically rich data sets has been more limited due to high computational demands and restricted scalability (Kubatko 2009; Meng and Kubatko 2009; Yu et al. 2012, 2014; Solís-Lemus and Ané 2016).
Together, the accessibility of large curated molecular data sets, advances in phylogenetic methods that conform to theoretical models of speciation, and a growing awareness of conditions affecting the performance of traditional and emerging methods make us well poised to tackle some of the most stubborn phylogenetic challenges. Here, we investigate relationships of the African cichlid subfamily, Pseudocrenilabrinae, and its close relatives in light of ILS and hybridization using hundreds of cichlid-specific single-copy exons (Ilves and López-Fernández 2014). We sample various Blenniiformes (formerly Ovalentaria) families for a phylogenomic test of the sister relationship between Cichlidae and the phenotypically divergent marine convict blenny, Pholidichthys (Wainwright et al. 2012; Betancur-R et al. 2013; Near et al. 2013; Ghezelayagh et al. 2021). We further assess the taxonomic reach of our probes and implement a hybrid guided/de novo assembly approach to improve data recovery in distant relatives. This study provides the most densely sampled phylogenomic hypothesis of higher-level Pseudocrenilabrinae relationships and is among the first to apply both coalescent and hybrid network approaches across nearly every tribe and both lake and river taxa.
Materials and Methods
Taxon Selection
We sampled 24 of 26–28 Pseudocrenilabrinae tribes (Fig. 1). Nearly, every tribe containing riverine lineages was sampled. A number of endemic species flocks, beyond the African Great Lakes, were represented: lower Congo River (Nanochromis, Steatocranus, Teleogramma, and Lamprologus), Cameroonian crater lakes Barombi Mbo (Oreochromini) and Bermin (Coptodon), soda lakes Natron and Magadi (Alcolapia), Lake Fwa (Haplochromini), and Pleistocene Lake palaeo-Magadikgadi (serranochromines). Only a handful of Malawi and Victoria lake species was sampled since they represent closely related clades nested well within Haplochromini (Friedman et al. 2013; Meyer et al. 2015; Matschiner et al. 2017; McGee et al. 2020). Quotations denote taxa awaiting formal description and nonmonophyletic genera that exclude the type species. We herein recognize Chromidotilapiini and Pelmatochromini as tribes, given existing support for the monophyly of these currently informally designated clades (Schwarzer et al. 2011, 2015; Dunz and Schliewen 2013). Tribal assemblages are referred to using the suffix “ines” (e.g., haplotilapiines).
Outside Pseudocrenilabrinae, we sampled representatives of the three other cichlid subfamilies and 11 closely related Blenniiformes families, inclusive of the proposed sister to Cichlidae and Pholidichthyidae (Wainwright et al. 2012). Our final data set had 178 terminals, with 165 cichlids (Pseudocrenilabrinae: 150, Cichlinae: 3, Etroplinae: 5, Ptychochrominae: 7) and 13 noncichlid blenniiforms. Tables S1–S5 of the Supplementary material available on Dryad contain taxonomic and locus information.
DNA Sequencing and Processing: From Raw Reads to Multiple Sequence Alignments
Genomic DNA was extracted from preserved tissues using the DNeasy Blood and Tissue kit (Qiagen, Venlo, the Netherlands). Library preparation and high-throughput paired-end sequencing (Illumina HiSeq) were performed at the University of Toronto Donnelly Sequencing Centre (http://dsc.utoronto.ca/dsc/index.html). A set of probes designed based on a Nile tilapia (Oreochromis niloticus) genome (BioMart Ensembl Genes 69 database; Flicek et al. 2013) was used for targeted sequence capture of 923 single-copy exons ranging in length from 750 to 2000 bp (Ilves and López-Fernández 2014).
Reads were filtered and trimmed in prinseq 0.20.3 lite (Schmieder and Edwards 2011) and assembled into target exons with one of three approaches, depending on the amount of recoverable data (Fig. S2 of the Supplementary material available on Dryad). First, reads were mapped to tilapia reference sequences and assembled in Bowtie2 2.2.0 (Langmead and Salzberg 2012) with the –very-sensitive-local preset. Consensus sequences with a minimum read depth of 10 were generated using SAMtools 0.1.19 (Li et al. 2009) and Biopython’s SeqIO.convert (Cock et al. 2009).
Bowtie2 assembly yielded more data for African and Neotropical cichlids (Fig. S2, Tables S3 and S4 of the Supplementary material available on Dryad), which shared a common ancestor with tilapia more recently than Indo-Malagasy Etroplinae and Ptychochrominae subfamilies and noncichlid blenniiforms (Fig. 1a). Mutations accumulated since divergence may have reduced probe efficacy and interfered with read mapping to tilapia references during Bowtie2 assembly. We therefore applied two modified assembly protocols aimed at increasing bp yield for these more distant taxa.
A hybrid reference guided/de novo assembly approach was used for seven Etroplinae and Ptychochrominae with the lowest rates of data recovery. Hybrid assembly consisted of three stages (see Appendices S2 and S3 and Fig. S3 of the Supplementary material available on Dryad for details and scripts). (i) An initial set of consensus exons was obtained through guided assembly with tilapia references using BWA-MEM 0.7.12 (Li and Durbin 2009; Li 2013), with more lenient mapping parameters than Bowtie2 and a minimum read depth of six. (ii) A second set of consensus exons with a minimum depth of six was assembled de novo in Velvet 1.0.17 (Zebrino and Birney 2008; Zebrino 2009; Crawford 2010). (iii) Guided and de novo assembled exons were aligned in MUSCLE v3.8.31 (Edgar 2004), from which a final set of consensus exons was extracted. In this way, sections missing from guided-assembled exons were complemented by novel regions obtained through de novo assembly. Visual inspection of alignments in Geneious 7.0.6 (http://www.geneious.com, Kearse et al. 2012) confirmed that sections spanned by both guided and de novo assemblies were identical, providing reassurance that exclusively de novo regions were accurately recovered.
For the remaining five Indo-Malagasy cichlids and 13 Blenniiformes outgroups, we applied a reduced-sensitivity assembly approach, equivalent to the first stage of hybrid assembly. High-throughput sequencing has proven especially difficult for Pholidichthys, with our (Table S4 of the Supplementary material available on Dryad) and previous (Eytan et al. 2015; pers. obs.) attempts resulting in reduced sequence recovery relative to other taxa in the same data set. In this study, we maximized informative sites by combining sequences from three individuals into a single set of consensus exons for Pholidichthys (Tables S1 and S4 of the Supplementary material available on Dryad). We ensured that exons from different individuals were correctly aligned for consensus generation by ascertaining that sections recovered across individuals were identical.
We identified coding and noncoding regions and codon positions in our targets by aligning them to annotated tilapia mRNA retrieved from GenBank (NCBI Oreochromis niloticus Annotation Release 103; Accession No. GCF_001858045.1; Clark et al. 2016) using a custom script (Appendix S4 of the Supplementary material available on Dryad; Altschul et al. 1990; Maglott et al. 2005; R Core Team 2015). Knowledge of coding regions was used to assist with alignment editing and delineation of partition schemes for PartitionFinder analyses.
Assembled cichlid and outgroup sequences were aligned using MUSCLE. Alignments were visually inspected and manually edited to ensure open reading frame in Geneious, based on our determination of coding regions. A limited amount of missing taxonomic and character data was permitted since some missing data are unlikely to be problematic in large data sets (Jiang et al. 2014; Xi et al. 2016; Nute et al. 2018). Final alignments contained at least 70 of taxa (124 species) and sequences spanning 30
of taxa (124 species) and sequences spanning 30 or more of the corresponding tilapia reference. Exon alignments were also concatenated into a supermatrix.
or more of the corresponding tilapia reference. Exon alignments were also concatenated into a supermatrix.
PartitionFinder v1.1.1 (Lanfear et al. 2012) was run on individual exons partitioned by coding and noncoding regions and codon position, and on the supermatrix partitioned by exon and coding and noncoding regions. A single data block was identified as the best partitioning scheme for individual exons and for the supermatrix. The best fitting models were GTR G for the supermatrix, and either GTR
G for the supermatrix, and either GTR G or GTR
G or GTR G
G I for individual exons (Table S2 of the Supplementary material available on Dryad).
I for individual exons (Table S2 of the Supplementary material available on Dryad).
Phylogenetic Inference: Concatenation, Species, and Network Analyses
Maximum likelihood (ML) concatenation analysis was performed with the supermatrix in CIPRES Science Gateway (Miller et al. 2010). We used RAxML-HPC v.8 on XSEDE (8.2.10) (Stamatakis 2014) with GTR G and 1000 rapid bootstrap replicates.
G and 1000 rapid bootstrap replicates.
A species tree approach was used to infer relationships in light of gene heterogeneity under ILS. ML gene trees were estimated for 588 exons (available on GenBank, BioProject PRJNA853788) in raxmlHPC-MPI-SSE3 v8.2.8 with GTR G and 1000 rapid bootstrap replicates. A species tree was inferred from gene trees in ASTRAL-II v.4.10.8 (Mirarab and Warnow 2015) with 700 bootstrap replicates. Local posterior probabilities, alternative quartet support (ASTRAL), and gene concordance factors (IQ-TREE; Minh et al. 2020a,b) were also calculated for the ASTRAL tree to assess topological robustness under various support metrics and to quantify gene heterogeneity. Node-specific gene tree conflict was visualized with pie charts obtained using ape 5.0’s “nodelabels” function (Paradis and Schliep 2019) with “pie” equal to ASTRAL’s q1–q3 values.
G and 1000 rapid bootstrap replicates. A species tree was inferred from gene trees in ASTRAL-II v.4.10.8 (Mirarab and Warnow 2015) with 700 bootstrap replicates. Local posterior probabilities, alternative quartet support (ASTRAL), and gene concordance factors (IQ-TREE; Minh et al. 2020a,b) were also calculated for the ASTRAL tree to assess topological robustness under various support metrics and to quantify gene heterogeneity. Node-specific gene tree conflict was visualized with pie charts obtained using ape 5.0’s “nodelabels” function (Paradis and Schliep 2019) with “pie” equal to ASTRAL’s q1–q3 values.
Poorly or incorrectly resolved gene trees can mislead species trees (Gatesy and Springer 2014; Mirarab and Warnow 2015). We assessed locus information content by quantifying parsimony informative sites and summarizing node support at various gene tree depths (first 25 , 50
, 50 , 75
, 75 , and 100
, and 100 of total depth) and for different locus lengths. We also investigated the possible impacts of gene tree error, including model misspecification, by inferring three additional ASTRAL trees based on alternative sets of gene trees. One set contained gene trees with nodes supported by less than 10
of total depth) and for different locus lengths. We also investigated the possible impacts of gene tree error, including model misspecification, by inferring three additional ASTRAL trees based on alternative sets of gene trees. One set contained gene trees with nodes supported by less than 10 bootstrap collapsed (Mirarab and Warnow 2015; Zhang et al. 2018). Another set re-estimated a subset of gene trees (Table S2 of the Supplementary material available on Dryad) with the GTR
bootstrap collapsed (Mirarab and Warnow 2015; Zhang et al. 2018). Another set re-estimated a subset of gene trees (Table S2 of the Supplementary material available on Dryad) with the GTR G
G I model, as suggested by PartitionFinder (see Appendix S2). For the third set, we reassessed the fit of over 200 substitution models not implemented in RAxML and re-estimated gene trees in IQ-TREE 1.6.12 (Nguyen et al. 2015). We also investigated base compositional heterogeneity, a common source of model misspecification (Foster 2004), with
I model, as suggested by PartitionFinder (see Appendix S2). For the third set, we reassessed the fit of over 200 substitution models not implemented in RAxML and re-estimated gene trees in IQ-TREE 1.6.12 (Nguyen et al. 2015). We also investigated base compositional heterogeneity, a common source of model misspecification (Foster 2004), with  test in IQ-TREE. Alternative ASTRAL species trees were compared with the weighted Robinson–Foulds distance (Robinson and Foulds 1981) in phanghorn 2.8.1 (Schliep 2011).
test in IQ-TREE. Alternative ASTRAL species trees were compared with the weighted Robinson–Foulds distance (Robinson and Foulds 1981) in phanghorn 2.8.1 (Schliep 2011).
Finally, we tested for hybridization and its impacts on the resolution of Pseudocrenilabrinae relationships using PhyloNetworks’ SNaQ phylogenetic network analyses (Bezanson et al. 2017; Solís-Lemus et al. 2017). Due to SNaQ’s computational demands, we focused on eight taxonomic subsets to assess hybridization at various phylogenetic scales. For each subset, ASTRAL species and ML gene trees were pruned to a selection of representative species, and only gene trees containing all representatives were incorporated. Sub1 sampled across tribes to test for deep hybridization. Sub2 and Sub 3 investigated hybridization for WCA lineages. Sub4 considered hybridization amongst former-tilapiines. Sub5 investigated hybridization within Barombi Mbo, Natron/Magadi, and Bermin lake flocks, and between these and their close riverine relatives. Sub6 assessed deep hybridization between former-tilapiine and EAR taxa. Sub7 focused on hybridization among EAR tribes. Sub8 looked for hybridization within Lamprologini and between it and its close relatives. SNaQ analyses were performed with the ASTRAL starting topology and incremental increases of  for 20 runs. Analyses were considered complete when tree scores stabilized within two pseudolikelihood points. Support for hybrid networks was assessed with 100 bootstrap replicates.
for 20 runs. Analyses were considered complete when tree scores stabilized within two pseudolikelihood points. Support for hybrid networks was assessed with 100 bootstrap replicates.
Concatenated, species, hybrid network, and gene tree topologies were compared by computing multivariate “tree spaces” in R’s treespace 1.1.4.1 package (Jombart et al. 2017). Topological variability was quantified with tip–tip path difference (Steel and Penny 1993) and Kendall–Colijn (Kendall and Colijn 2016) metrics. Appendix S2 of the Supplementary material available on Dryad contains details for all methods used in this study.
Results
Target Capture and Gene Tree Resolution
Alternative assembly contributed more data for Indo-Malagasy cichlids and outgroup taxa relative to initial Bowtie2 assemblies ( ; Fig. S4 of the Supplementary material available on Dryad). The span of target sequences increased by an average of 16.8
; Fig. S4 of the Supplementary material available on Dryad). The span of target sequences increased by an average of 16.8  22.7
22.7 SD, and 1,624,734 bp and 1087 exons were added across taxa. Hybrid guided/de novo assembly showed the largest improvements, adding more characters and exons per taxon than the reduced-sensitivity approach (Fig. S4 of the Supplementary material available on Dryad). Ultimately, we obtained 588 exon alignments between 291 and 1938 bp (mean
SD, and 1,624,734 bp and 1087 exons were added across taxa. Hybrid guided/de novo assembly showed the largest improvements, adding more characters and exons per taxon than the reduced-sensitivity approach (Fig. S4 of the Supplementary material available on Dryad). Ultimately, we obtained 588 exon alignments between 291 and 1938 bp (mean  ), for a final aligned supermatrix length of 610,990 bp. Alignments contained 124–178 (mean
), for a final aligned supermatrix length of 610,990 bp. Alignments contained 124–178 (mean  ) taxa and sequences spanning an average of 89
) taxa and sequences spanning an average of 89  9.7
9.7 of their reference.
of their reference.
Loci informativeness increased with tree depth and locus length (Fig. 2b,c). IQ-TREE identified different best-fit models than PartitionFinder (Table S2 of the Supplementary material available on Dryad). Nevertheless, compositional heterogeneity tests showed that nearly no sequence violated the assumption of homogeneous base composition for GTR (mean proportion passed  ; Table S2 of the Supplementary material available on Dryad), and therefore, our application of this highly parameterized model is not necessarily inappropriate. Indeed, the fixed application of GTR has been shown to yield accurate topologies even when it is not the best-fit model (Abadi et al. 2019). Moreover, comparisons of ASTRAL trees generated with different gene tree sets revealed highly congruent topologies (wRF: 0.021–0.058, Table S6 of the Supplementary material available on Dryad), especially along backbone relationships. Disagreements were restricted to a few shallow, poorly supported nodes, like Crater lake (Coptodon and Bermin) and Congolese (Lamprologini) species flocks whose resolution may require additional taxa and faster-evolving loci (Fig. S5 of the Supplementary material available on Dryad).
; Table S2 of the Supplementary material available on Dryad), and therefore, our application of this highly parameterized model is not necessarily inappropriate. Indeed, the fixed application of GTR has been shown to yield accurate topologies even when it is not the best-fit model (Abadi et al. 2019). Moreover, comparisons of ASTRAL trees generated with different gene tree sets revealed highly congruent topologies (wRF: 0.021–0.058, Table S6 of the Supplementary material available on Dryad), especially along backbone relationships. Disagreements were restricted to a few shallow, poorly supported nodes, like Crater lake (Coptodon and Bermin) and Congolese (Lamprologini) species flocks whose resolution may require additional taxa and faster-evolving loci (Fig. S5 of the Supplementary material available on Dryad).
Figure 2.
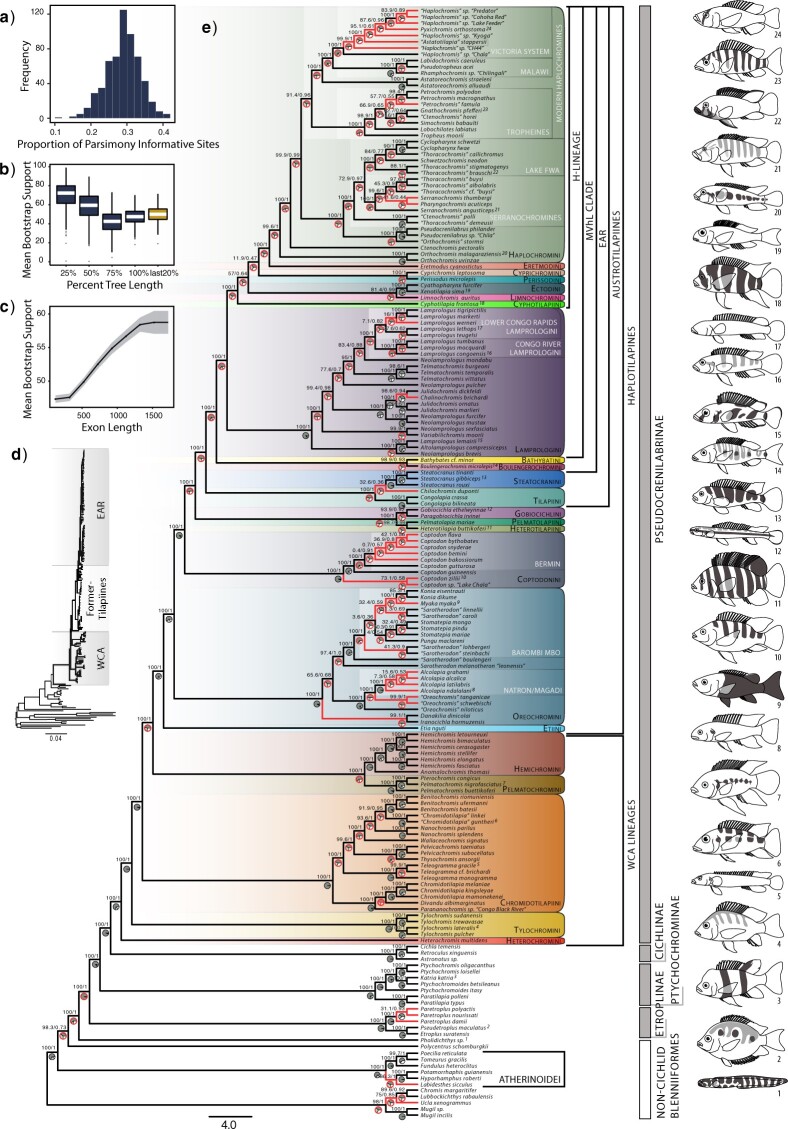
(Color online) Proportion of parsimony informative sites (a), gene tree resolution (b,c), and phylogenies (d,e) for Pseudocrenilabrinae and close relatives. b) Bootstrap support at various gene tree depths. The first four (blue) boxes correspond to increasing distances from the root, while the last (yellow) box corresponds to the shallowest 20 of tree depth. c) Bootstrap support and 95
of tree depth. c) Bootstrap support and 95 confidence interval (shaded area) for gene trees inferred from loci of various lengths. d) Concatenated RAxML tree showing branch lengths; see Figure S6 of the Supplementary material available on Dryad for full annotations. e) ASTRAL-II species tree. Numbers above branches are bootstrap and local posterior probability support. Pie charts show gene tree support for the main (dark gray), and first (light gray) and second (white) alternative quartet topologies (i.e., ASTRAL’s q1–q3 support values). Pies encircled with a thick (red) line indicate nodes that also had low gene concordance, below 50. Figure S8 of the Supplementary material available on Dryad contains full concordance factor annotations. Branches that differ between the species and concatenated phylogenies are lighter (red). Shaded (colored online) boxes with round edges and black names denote tribes, while nested boxes with square edges and white names indicate subclades discussed in the text. Square brackets to the right show tribal assemblages and noncichlid groupings. Gray and white vertical bars furthest to the right denote cichlid subfamilies and noncichlid Blenniiformes. Superscript numbers next to some species names indicate illustrated species to the right of the phylogeny. Illustrations by VAC.
confidence interval (shaded area) for gene trees inferred from loci of various lengths. d) Concatenated RAxML tree showing branch lengths; see Figure S6 of the Supplementary material available on Dryad for full annotations. e) ASTRAL-II species tree. Numbers above branches are bootstrap and local posterior probability support. Pie charts show gene tree support for the main (dark gray), and first (light gray) and second (white) alternative quartet topologies (i.e., ASTRAL’s q1–q3 support values). Pies encircled with a thick (red) line indicate nodes that also had low gene concordance, below 50. Figure S8 of the Supplementary material available on Dryad contains full concordance factor annotations. Branches that differ between the species and concatenated phylogenies are lighter (red). Shaded (colored online) boxes with round edges and black names denote tribes, while nested boxes with square edges and white names indicate subclades discussed in the text. Square brackets to the right show tribal assemblages and noncichlid groupings. Gray and white vertical bars furthest to the right denote cichlid subfamilies and noncichlid Blenniiformes. Superscript numbers next to some species names indicate illustrated species to the right of the phylogeny. Illustrations by VAC.
Phylogenetic Relationships and Gene Tree Heterogeneity
Targeted exon capture provided generally well-resolved and consistent relationships across concatenated (Fig. 2d, Fig. S6 of the Supplementary material available on Dryad, mean bootstrap  ), species (Fig. 2e, Fig. S5 and Table S6 of the Supplementary material available on Dryad) and phylogenetic network (Fig. 3) trees, despite widespread genetic discordance (Figs. 2e and 4, Figs. S7 and S8 of the Supplementary material available on Dryad). Quartet and gene concordance scores largely agreed in their identification of genetically inconsistent nodes (Fig. 2e, Fig. S8 of the Supplementary material available on Dryad). Disagreements between concatenated and species trees were limited to mainly shallow relationships (Figs. 2e and 4). Hybrid networks indicated a few more topological differences (Fig. 4b–i), some of which coincided with lineages possibly involved in hybridization events. Tree files are available in Appendices S5–S15 of the Supplementary material available on Dryad.
), species (Fig. 2e, Fig. S5 and Table S6 of the Supplementary material available on Dryad) and phylogenetic network (Fig. 3) trees, despite widespread genetic discordance (Figs. 2e and 4, Figs. S7 and S8 of the Supplementary material available on Dryad). Quartet and gene concordance scores largely agreed in their identification of genetically inconsistent nodes (Fig. 2e, Fig. S8 of the Supplementary material available on Dryad). Disagreements between concatenated and species trees were limited to mainly shallow relationships (Figs. 2e and 4). Hybrid networks indicated a few more topological differences (Fig. 4b–i), some of which coincided with lineages possibly involved in hybridization events. Tree files are available in Appendices S5–S15 of the Supplementary material available on Dryad.
Figure 3.
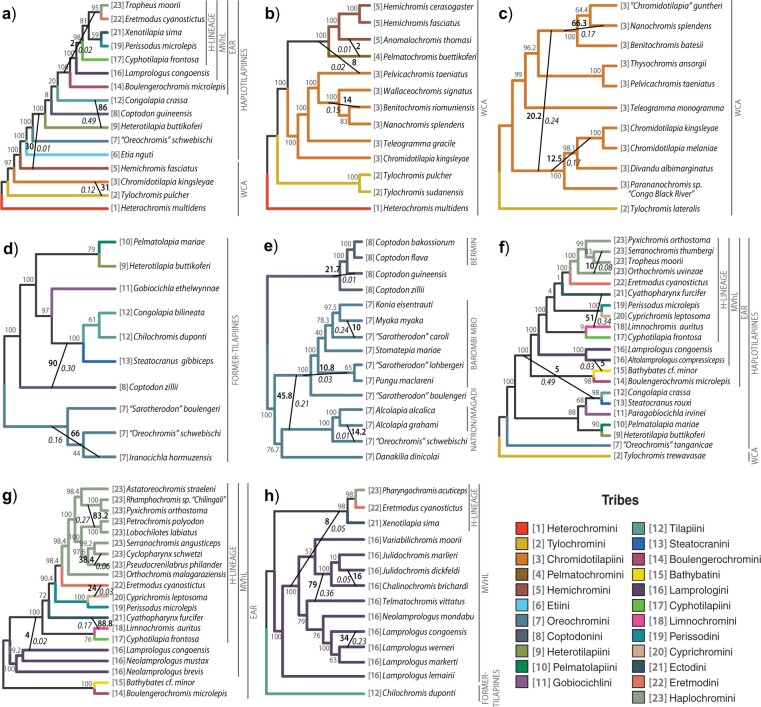
(Color online) Best phylogenetic networks estimated with SNaQ for eight taxonomic subsets. a) Sub1: Pseudocrenilabrinae tribes; b) Sub2: WCA lineages; c) Sub3: Chromidotilapiini; d) Sub4: former-tilapiine tribes; e) Sub5: Barombi Mbo, Bermin and Natron/Magadi flocks, and riverine relatives; f) Sub6: EAR, former-tilapiine and other Tanganyikan lineages; g) Sub7: EAR clade; h) Sub8: Lamprologini and close relatives. Tribes are differentiated by numbers in square brackets adjacent to species names and also by color in the online version of this figure. Vertical lines to the right of phylogenies denote multitribal groupings (a–d and f–h) or species flocks (e). Gray numbers around nodes show bootstrap support for species relationships. Diagonal black lines connecting lineages show hybrid networks. Black and italicized numbers indicate bootstrap support and inheritance probabilities (i.e.,  proportion of genes transferred during hybridization) of estimated networks.
proportion of genes transferred during hybridization) of estimated networks.
Figure 4.
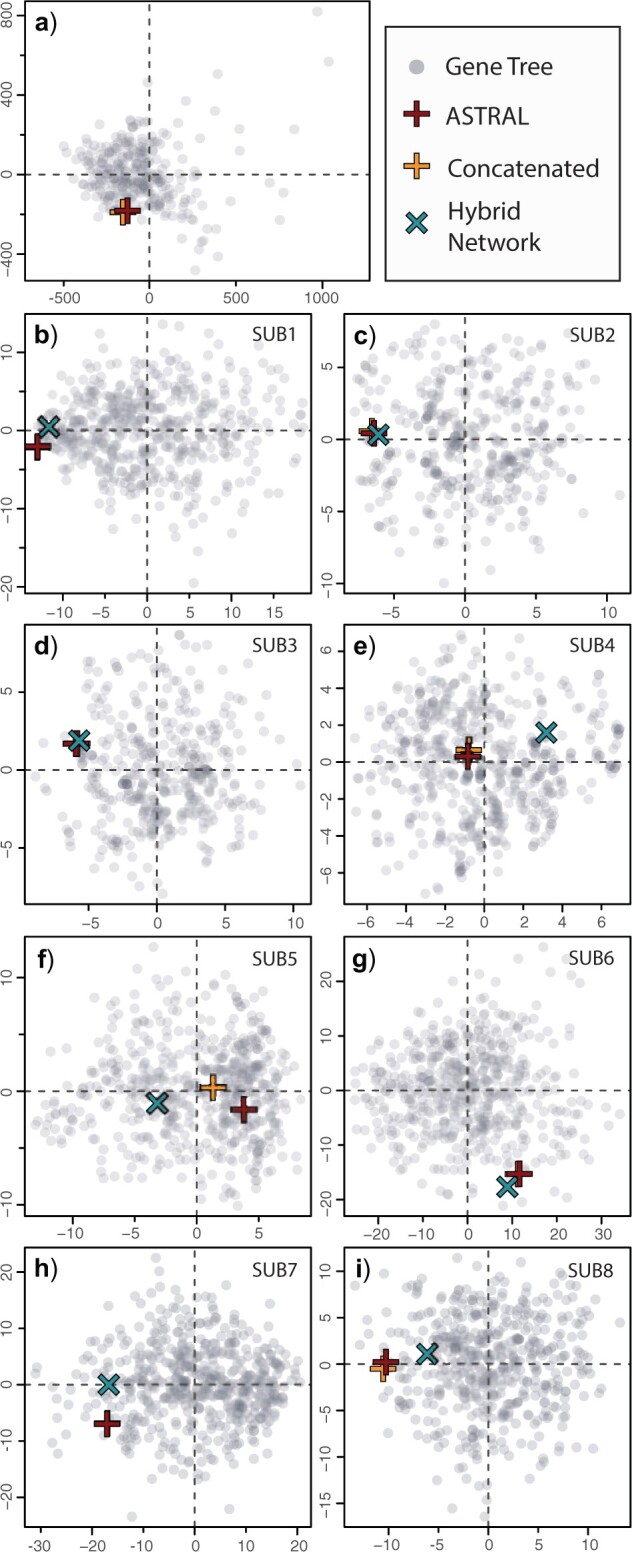
(Color online) Tree spaces comparing Pseudocrenilabrinae hypotheses across concatenated, species, hybrid network, and gene trees based on the tip–tip path difference metric. a) Comparison of full phylogeny across concatenated, species, and gene trees. b–i) Comparison of concatenated, species, hybrid network, and gene trees, restricted to taxa in hybrid network subsets. Tree spaces inferred with the Kendal–Colijn metric showed comparable similarity patterns across concatenated, species, and hybrid network trees and are thus included in Fig. S7 of Appendix S2 of the Supplementary material available on Dryad.
For noncichlid blenniiforms, we recovered two monophyletic clades: Atherinoidei (sensuGhezelayagh et al. 2021, formerly Atherinomorpha) and an unnamed clade with the remaining families (Fig. 2e). The Pholidichthyidae Cichlidae sister relationship was well supported, and this clade was in turn sister to the South American leaf-fishes (Polycentridae).
Cichlidae sister relationship was well supported, and this clade was in turn sister to the South American leaf-fishes (Polycentridae).
As expected, Cichlidae and its four subfamilies were monophyletic across analyses. Etroplinae and Ptychochrominae diverged prior to the African Pseudocrenilabrinae and Neotropical Cichlinae split. Pseudocrenilabrinae tribes and their interrelationships were for the most part well supported. A few challenging relationships persisted particularly within the EAR and recent rapid radiations (Fig. 2d).
Early divergence of Heterochromini was followed by successive branching of four other WCA tribes. Gene heterogeneity was elevated at some WCA nodes, including a sister relationship between Pelmatochromini Hemichromini and the haplotilapiines. Hybridization was inferred among (Tylochromini–Chromidotilapiini, Hemichromini–Pelmatochromini; Fig. 3a,b) and within (Chromidotilapiini; Fig. 3c) WCA tribes with limited genetic exchange (
Hemichromini and the haplotilapiines. Hybridization was inferred among (Tylochromini–Chromidotilapiini, Hemichromini–Pelmatochromini; Fig. 3a,b) and within (Chromidotilapiini; Fig. 3c) WCA tribes with limited genetic exchange ( ), and also between WCA Hemichromini and the haplotilapiines (Fig. 3a).
), and also between WCA Hemichromini and the haplotilapiines (Fig. 3a).
The large haplotilapiine clade encompassed the remaining Pseudocrenilabrinae lineages. In contrast to WCA, haplotilapiines were characterized by widespread gene heterogeneity. This included not only clades known for their rapid diversification but also deeper nodes defining intertribal relationships. While concatenated and species trees agreed on nearly all intertribal relationships, phylogenetic networks inferred slightly different arrangements for some former-tilapiine and EAR tribes, with variably supported hybridization events.
Etiini was the sister to all other haplotilapiines, followed by Oreochromini, which contains the mouthbrooding former-tilapiines. Oreochromini formed two large clades. One contained the west/central African “Sarotherodon” and monophyletic Barombi Mbo flock. The other comprised “Oreochromis” and the soda lakes Alcolapia radiation. Barombi Mbo and Alcolapia groups had short and genetically inconsistent branches. A clade consisting of Danakilia, from the Danakil Depression in Ethiopia/Eritrea, and Iranocichla, from southern Iran, was inconsistently placed as sister to all other oreochromines (Fig. 2e and Fig. S8b of the Supplementary material available on Dryad) or Alcolapia “Oreochromis” (Figs. 3d,e and Figs. S6 and S8a,c of the Supplementarymaterial available on Dryad). Hybridization was detected within the species flocks and for Iranocichla (Fig. 3d,e).
“Oreochromis” (Figs. 3d,e and Figs. S6 and S8a,c of the Supplementarymaterial available on Dryad). Hybridization was detected within the species flocks and for Iranocichla (Fig. 3d,e).
The remaining substrate spawning former-tilapiines diversified into several tribes near the base of the EAR. The early branching of Coptodonini and Heterotilapiini Pelmatolapiini were well supported, but Gobiocichlini varied in its position as sister to Heterotilapiini
Pelmatolapiini were well supported, but Gobiocichlini varied in its position as sister to Heterotilapiini Pelmatolapiini (species and concatenated trees) or Tilapiini
Pelmatolapiini (species and concatenated trees) or Tilapiini Steatocranini (phylogenetic networks; Fig. 3d,f). Gene heterogeneity was widespread among former-tilapiines. Hybridization with high genetic exchange (
Steatocranini (phylogenetic networks; Fig. 3d,f). Gene heterogeneity was widespread among former-tilapiines. Hybridization with high genetic exchange ( ) was detected between a Tilapiini-containing lineage and other substrate spawning former-tilapiines (Fig. 3a,d).
) was detected between a Tilapiini-containing lineage and other substrate spawning former-tilapiines (Fig. 3a,d).
Tilapiini Steatocranini was sister to the EAR in concatenated and species trees, but its placement varied in phylogenetic networks. One subset concurred with a Tilapiini
Steatocranini was sister to the EAR in concatenated and species trees, but its placement varied in phylogenetic networks. One subset concurred with a Tilapiini EAR clade (Fig. 3a), but another grouped Tilapiini
EAR clade (Fig. 3a), but another grouped Tilapiini Steatocranini with former-tilapiines and inferred hybridization between Tilapiini
Steatocranini with former-tilapiines and inferred hybridization between Tilapiini Steatocranini and an EAR ancestor (Fig. 3f).
Steatocranini and an EAR ancestor (Fig. 3f).
The widely supported EAR clade comprised the endemic Tanganyikan tribes plus Lamprologini and Haplochromini. Bathybatini Boulengerochromini was sister to the remaining EAR lineages, which together formed the MVhL clade (Takahashi et al. 2001) named for its inclusion of Malawi, Victoria, Nishida’s (1991) H-lineage (Fig. 1a) and Lamprologini taxa. A Lamprologini
Boulengerochromini was sister to the remaining EAR lineages, which together formed the MVhL clade (Takahashi et al. 2001) named for its inclusion of Malawi, Victoria, Nishida’s (1991) H-lineage (Fig. 1a) and Lamprologini taxa. A Lamprologini H-Lineage sister relationship was robust.
H-Lineage sister relationship was robust.
Internal lamprologine relationships were mostly well resolved, despite short branches and high gene heterogeneity. Riverine Lamprologus endemic to the Congo basin formed a monophyletic grouping nested among Tanganyikan lineages. Several hybrid events were detected, of which the most strongly supported involved Chalinochromis Julidochromis and Telmatochromis (Fig. 3h). Lamprologines may have also hybridized beyond the tribe, with the ancestor of the H-lineage (Fig. 3g,h) or Bathybatini (Fig. 3f).
Julidochromis and Telmatochromis (Fig. 3h). Lamprologines may have also hybridized beyond the tribe, with the ancestor of the H-lineage (Fig. 3g,h) or Bathybatini (Fig. 3f).
The H-lineage was unambiguously supported. Concatenated and species trees produced identical topologies, but some highly heterogeneous nodes received equivocal support or differed from network analyses (Fig. 3a,f,g). Cyprichromini Perissodini and Eretmodini
Perissodini and Eretmodini Haplochromini were consistently inferred. Ectodini
Haplochromini were consistently inferred. Ectodini Limnochromini was supported by concatenated and species trees, but phylogenetic networks grouped Limnochromini
Limnochromini was supported by concatenated and species trees, but phylogenetic networks grouped Limnochromini Cyphotilapiini and connected Ectodini to Limnochromini only through hybridization (
Cyphotilapiini and connected Ectodini to Limnochromini only through hybridization ( 0.17–0.34; Fig. 3f,g). Additional hybrid events involving the H-lineage were inferred between the EAR ancestors and more nested Eretmodini
0.17–0.34; Fig. 3f,g). Additional hybrid events involving the H-lineage were inferred between the EAR ancestors and more nested Eretmodini Haplochromini clades (Fig. 3a), Cyprichromini and Eretmodini (Fig. 3g), and the H-Lineage ancestor and Lamprologini (Fig. 3g,h).
Haplochromini clades (Fig. 3a), Cyprichromini and Eretmodini (Fig. 3g), and the H-Lineage ancestor and Lamprologini (Fig. 3g,h).
The earliest Haplochromini divergences gave rise to Malagarasi River Orthochromis and Ctenochromis from southern Tanzanian streams. Concatenated and species trees placed the remaining lineages into two clades. One included the Congolese Pseudocrenilabrus Orthochromis as sister to a grouping of Congo/Zambezi serranochromines and Lake Fwa radiation. The other clade included Tanganyikan tropheines as sister to a group of riverine haplochromines and the Malawi and Victoria lake radiations. Phylogenetic networks placed tropheines
Orthochromis as sister to a grouping of Congo/Zambezi serranochromines and Lake Fwa radiation. The other clade included Tanganyikan tropheines as sister to a group of riverine haplochromines and the Malawi and Victoria lake radiations. Phylogenetic networks placed tropheines serranochromines instead as sister to the Malawi/Victoria radiations (Fig. 3f,g). Despite our limited sampling of haplochromines, we identified possible hybridization between serranochromines and other Congolese taxa (Fig. 3f,g) and between tropheines and a Malawi/Victoria clade ancestor (Fig. 3g).
serranochromines instead as sister to the Malawi/Victoria radiations (Fig. 3f,g). Despite our limited sampling of haplochromines, we identified possible hybridization between serranochromines and other Congolese taxa (Fig. 3f,g) and between tropheines and a Malawi/Victoria clade ancestor (Fig. 3g).
Discussion
We applied a suite of approaches that draw on emerging high-throughput, bioinformatics, and phylogenomic tools to tackle the phylogenetic challenges of Pseudocrenilabrinae, a large clade with multiple radiations and rampant gene tree conflict. Hundreds of carefully curated loci and phylogenomic methods that account for ILS, hybridization, and gene tree error ensured that relationships were informed by a comprehensive sampling of the diverse genetic histories and biological processes underlying cichlid evolutionary history, as well as possible methodological bias.
Phylogenetic Relationships
We infer strong and generally consistent phylogenetic hypotheses for Pseudocrenilabrinae and its close relatives, especially at higher taxonomic levels. Support for homogeneous base composition and highly consistent species trees based on alternative input gene trees indicate that while gene tree error is inevitable (Mirarab and Warnow 2015), it is unlikely to have considerably misled phylogenetic reconstruction. Major taxonomic contributions of this work include the resolution of: (i) WCA intertribal relationships, (ii) the WCA sister to haplotilapiines, (iii) former-tilapiine relationships, (iv) relationships near the root of the EAR clade, and (v) placement of several challenging MVhL tribes. See Appendix S1 of the Supplementary material available on Dryad for a detailed taxonomic review.
WCA tribes diverged successively, with Hemichromini Pelmatochromini supported as sister to the pan-African haplotilapiines. Previous, mostly multilocus, hypotheses for this part of the tree are either based on the incomplete sampling of WCA tribes or provide equivocal support for similar or conflicting relationships (Schwarzer et al. 2009; Dunz and Schliewen 2013; Matschiner et al. 2017; Irisarri et al. 2018). Haplotilapiines are frequently split into three clades: Oreochromini, west/central “boreotilapiines” (substrate spawning former-tilapiines less Tilapiini and Steatocranini), and central/east “austrotilapiines” (Tilapiini
Pelmatochromini supported as sister to the pan-African haplotilapiines. Previous, mostly multilocus, hypotheses for this part of the tree are either based on the incomplete sampling of WCA tribes or provide equivocal support for similar or conflicting relationships (Schwarzer et al. 2009; Dunz and Schliewen 2013; Matschiner et al. 2017; Irisarri et al. 2018). Haplotilapiines are frequently split into three clades: Oreochromini, west/central “boreotilapiines” (substrate spawning former-tilapiines less Tilapiini and Steatocranini), and central/east “austrotilapiines” (Tilapiini Steatocranini
Steatocranini EAR) (Schwarzer et al. 2009; Dunz and Schliewen 2013; Matschiner et al. 2017). We fail to recover “boreotilapiines,” but our grouping of the notably intractable Tilapiini
EAR) (Schwarzer et al. 2009; Dunz and Schliewen 2013; Matschiner et al. 2017). We fail to recover “boreotilapiines,” but our grouping of the notably intractable Tilapiini Steatocranini (Schwarzer et al. 2009; Dunz and Schliewen 2013; Irisarri et al. 2018; Schedel et al. 2019) with the EAR clade in all but one network analysis (Fig. 3f) substantiates a more nested austrotilapiine assemblage. Within the EAR, we support a monophyletic Bathybatini
Steatocranini (Schwarzer et al. 2009; Dunz and Schliewen 2013; Irisarri et al. 2018; Schedel et al. 2019) with the EAR clade in all but one network analysis (Fig. 3f) substantiates a more nested austrotilapiine assemblage. Within the EAR, we support a monophyletic Bathybatini Boulengerochromini as sister to the MVhL clade, a finding consistent with some multilocus (e.g., Takahashi et al. 2001; Salzburger et al. 2002; Clabaut et al. 2005; Friedman et al. 2013) and most phylogenomic (Meyer et al. 2015; Takahashi and Sota 2016; Matschiner et al. 2017; Irisarri et al. 2018; Ronco et al. 2021) studies.
Boulengerochromini as sister to the MVhL clade, a finding consistent with some multilocus (e.g., Takahashi et al. 2001; Salzburger et al. 2002; Clabaut et al. 2005; Friedman et al. 2013) and most phylogenomic (Meyer et al. 2015; Takahashi and Sota 2016; Matschiner et al. 2017; Irisarri et al. 2018; Ronco et al. 2021) studies.
The H-lineage presents some of the most stubborn challenges of the Pseudocrenilabrinae backbone. While studies based on a small number of loci propose various, often poorly supported, topologies, phylogenomic efforts are converging on a few robust relationships (Takahashi and Sota 2016; Irisarri et al. 2018, Ronco et al. 2021). Common phylogenomic groupings (also recovered here) are Ectodini Limnochromini, Perissodini
Limnochromini, Perissodini Cyprichromini, and Eretmodini
Cyprichromini, and Eretmodini Haplochromini. Eretmodini
Haplochromini. Eretmodini Haplochromini corroborates an H-Lineage (Nishida 1991) over the alternative C-Lineage (Clabaut et al. 2005; Fig. 1a), which instead groups Eretmodini
Haplochromini corroborates an H-Lineage (Nishida 1991) over the alternative C-Lineage (Clabaut et al. 2005; Fig. 1a), which instead groups Eretmodini Lamprologini.
Lamprologini.
Notable findings at shallower nodes include the first phylogenomic evidence for Danakilia Iranocichla within Oreochromini (Trewavas 1983), a Heterotilapiini
Iranocichla within Oreochromini (Trewavas 1983), a Heterotilapiini Pelmatolapiini sister relationship, and monophyly of nested radiations beyond the Great Lakes. Combined evidence for monophyly (here, Stiassny et al. 1992; Schliewen et al. 1994; Roberts and Kullander 1994; Joyce et al. 2005; Ford et al. 2015, 2016; Martin et al. 2015; Musilova et al. 2019), short branches (Fig. S6 of the Supplementary material available on Dryad), and gene tree conflict (Figs. 2e and 4, Fig. S8 of the Supplementary material available on Dryad) for sampled flocks are consistent with repeated episodes of rapid speciation in diverse habitats.
Pelmatolapiini sister relationship, and monophyly of nested radiations beyond the Great Lakes. Combined evidence for monophyly (here, Stiassny et al. 1992; Schliewen et al. 1994; Roberts and Kullander 1994; Joyce et al. 2005; Ford et al. 2015, 2016; Martin et al. 2015; Musilova et al. 2019), short branches (Fig. S6 of the Supplementary material available on Dryad), and gene tree conflict (Figs. 2e and 4, Fig. S8 of the Supplementary material available on Dryad) for sampled flocks are consistent with repeated episodes of rapid speciation in diverse habitats.
Beyond Pseudocrenilabrinae, we corroborate the Pholidichthyidae Cichlidae sister relationship (Wainwright et al. 2012) with amongst (Ghezelayagh et al. 2021) the largest phylogenomic data sets. The Cichlidae
Cichlidae sister relationship (Wainwright et al. 2012) with amongst (Ghezelayagh et al. 2021) the largest phylogenomic data sets. The Cichlidae Pholidicthyidae
Pholidicthyidae Polycentridae grouping further sheds light on the biogeographic origin of cichlids (Friedman et al. 2013; Matschiner et al. 2017; Ghezelayagh et al. 2021). This grouping suggests freshwater origins for the clade with a secondary marine transition in Pholidichthyidae. Going forward, our documentation of genetic discordance at interfamilial nodes indicates that phylogenomic tools accounting for gene heterogeneity may help improve resolution within Blenniiformes (Hughes et al. 2018; Ghezelayagh et al. 2021).
Polycentridae grouping further sheds light on the biogeographic origin of cichlids (Friedman et al. 2013; Matschiner et al. 2017; Ghezelayagh et al. 2021). This grouping suggests freshwater origins for the clade with a secondary marine transition in Pholidichthyidae. Going forward, our documentation of genetic discordance at interfamilial nodes indicates that phylogenomic tools accounting for gene heterogeneity may help improve resolution within Blenniiformes (Hughes et al. 2018; Ghezelayagh et al. 2021).
ILS and Hybridization
Complementary coalescent and network analyses support ILS and hybridization as likely sources of gene heterogeneity in African cichlids. Whereas hybridization is implicated in specific instances (Fig. 3), ILS appears to be a prevailing source of genetic incongruence throughout. Gene tree error is also expected to have contributed to genetic discordance.
Like authors before us, we document short branches (Fig. 2d, Fig. S6 of the Supplementary material available on Dryad) and gene heterogeneity (Figs. 2e and 4, Fig. S8 of the Supplementary material available on Dryad), consistent with rapid speciation and ILS, across the EAR and within smaller-scale lake radiations. Sampling of riverine taxa shows that these patterns are far from a unique property of lacustrine or recent taxa. Similar patterns within Congo species flocks, among chromidotilapiines, between WCA lineages and haplotilapiines, and among former-tilapiine tribes, indicate possible rapid diversification for recent riverine radiations and very early in Pseudocrenilabrinae history as well.
Phylogenetic networks document hybridization among taxa at various tree depths and from diverse environments, some of which have presented persistent phylogenetic challenges. Disagreements regarding an H- versus C-Lineage and its internal relationships are likely influenced by hybridization. Most studies in favor of a C-Lineage are based on 50–100 mitochondrial markers (Kocher et al. 1995; Salzburger et al. 2002; Clabaut et al. 2005; Day et al. 2008; Schwarzer et al. 2009; Muschick et al. 2012; Dunz and Schliewen 2013; Weiss et al. 2015; Matschiner et al. 2017; but see Friedman et al. 2013), while the H-Lineage is supported by mainly nuclear data (here, Nishida 1991; Meyer et al. 2015; Takahashi and Sota 2016; Irisarri et al. 2018; Schedel et al. 2019; Ronco et al. 2021). We contribute additional phylogenomic evidence for gene flow within the H-Lineage and between it and early EAR and lamprologine taxa (Fig. 3a,f–h; Weiss et al. 2015; Meyer et al. 2017). Ectodini–Limnochromini hybridization (Fig. 3f,g) in particular seems to explain the grouping of these tribes in some analyses (Fig. 2e) and conflicting placements for Ectodini and Cyphotilapiini in prior work (Weiss et al. 2015; Takahashi and Sota 2016; Meyer et al. 2017; Irisarri et al. 2018; Schedel et al. 2019; Ronco et al. 2021).
mitochondrial markers (Kocher et al. 1995; Salzburger et al. 2002; Clabaut et al. 2005; Day et al. 2008; Schwarzer et al. 2009; Muschick et al. 2012; Dunz and Schliewen 2013; Weiss et al. 2015; Matschiner et al. 2017; but see Friedman et al. 2013), while the H-Lineage is supported by mainly nuclear data (here, Nishida 1991; Meyer et al. 2015; Takahashi and Sota 2016; Irisarri et al. 2018; Schedel et al. 2019; Ronco et al. 2021). We contribute additional phylogenomic evidence for gene flow within the H-Lineage and between it and early EAR and lamprologine taxa (Fig. 3a,f–h; Weiss et al. 2015; Meyer et al. 2017). Ectodini–Limnochromini hybridization (Fig. 3f,g) in particular seems to explain the grouping of these tribes in some analyses (Fig. 2e) and conflicting placements for Ectodini and Cyphotilapiini in prior work (Weiss et al. 2015; Takahashi and Sota 2016; Meyer et al. 2017; Irisarri et al. 2018; Schedel et al. 2019; Ronco et al. 2021).
Hybridization among riverine and between river and lake lineages is also implicated. Tilapiini Steatocranini appear to be disposed to hybridization with other former-tilapiine and EAR taxa (Fig. 3a,d,f; Loh et al. 2013; Dunz and Schliewen 2013; Weiss et al. 2015; Irisarri et al. 2018). Such repeated and taxonomically far-reaching hybrid events may underlie disagreements regarding a Tilapiini
Steatocranini appear to be disposed to hybridization with other former-tilapiine and EAR taxa (Fig. 3a,d,f; Loh et al. 2013; Dunz and Schliewen 2013; Weiss et al. 2015; Irisarri et al. 2018). Such repeated and taxonomically far-reaching hybrid events may underlie disagreements regarding a Tilapiini Steatocranini grouping and its relationship to the EAR (here, Schwarzer et al. 2009; Dunz and Schliewen 2013; Weiss et al. 2015; Irisarri et al. 2018; Schedel et al. 2019). Gene flow among WCA lineages (Fig. 3a–c) is a novel finding consistent with previous suspicions. Schwarzer et al. (2015) proposed hybridization as a source of cytonuclear discordance in Chromidotilapiini, which could have been aided by past hydrological connections between West Africa and the lower Congo. Inconsistent placements of Iranocichla
Steatocranini grouping and its relationship to the EAR (here, Schwarzer et al. 2009; Dunz and Schliewen 2013; Weiss et al. 2015; Irisarri et al. 2018; Schedel et al. 2019). Gene flow among WCA lineages (Fig. 3a–c) is a novel finding consistent with previous suspicions. Schwarzer et al. (2015) proposed hybridization as a source of cytonuclear discordance in Chromidotilapiini, which could have been aided by past hydrological connections between West Africa and the lower Congo. Inconsistent placements of Iranocichla Danakilia (Fig. 2e; Schwarzer et al. 2009, Wagner et al. 2012; Matschiner et al. 2017; Schedel et al. 2019) may also be due to hybridization with other oreochromines (Fig. 3d,e). Finally, networks connecting lake flocks to riverine relatives (Fig. 3e–g) highlight a prospective role for river taxa as conduits of genetic material that could have supported nested lake radiations (Schwarzer et al. 2012a,b; Loh et al. 2013; Martin et al. 2015; Weiss et al. 2015; Ford et al. 2016).
Danakilia (Fig. 2e; Schwarzer et al. 2009, Wagner et al. 2012; Matschiner et al. 2017; Schedel et al. 2019) may also be due to hybridization with other oreochromines (Fig. 3d,e). Finally, networks connecting lake flocks to riverine relatives (Fig. 3e–g) highlight a prospective role for river taxa as conduits of genetic material that could have supported nested lake radiations (Schwarzer et al. 2012a,b; Loh et al. 2013; Martin et al. 2015; Weiss et al. 2015; Ford et al. 2016).
Together, our findings corroborate mounting evidence for ILS and hybridization in the EAR clade (Takahashi et al. 2001; Salzburger et al. 2002; Koblmüller et al. 2007, 2010; Weiss et al. 2015; Meyer et al. 2017; Meier et al. 2017; Irisarri et al. 2018), small lacustrine species flocks (Schliewen and Klee 2004; Zaccara et al. 2014; Ford et al. 2015; Martin et al. 2015; Richards et al. 2018), and some riverine taxa (Schwarzer et al. 2011, Schwarzer et al. 2012a,b; Loh et al. 2013; Alter et al. 2017) and also point to other lineages whose diversification may have been marked by these processes. The prevalence of these processes and resulting gene heterogeneity underscores the importance of incorporating diverse genetic histories alongside approaches that consider multiple sources of discordance in phylogenies for taxa with complex evolutionary histories, like cichlids.
Biogeographic and Macroevolutionary Implications
Sequential early divergence of WCA lineages supports West/Central African rivers as the place of origin for Pseudocrenilabrinae (Stiassny 1987; Mayer et al. 1998). Several WCA and former-tilapiine tribes diversified in situ until austrotilapiines dispersed and radiated further east. Most age estimates place the origin of austrotilapiines somewhere between 25 and 35 Ma (Schwarzer et al. 2009; McMahan et al. 2013; Schedel et al. 2019; Matschiner 2019; Matschiner et al. 2020), which coincides with early rifting in East Africa (Ring 2014). Slightly older (Murray et al. 2001a; Schwarzer et al. 2009; Schedel et al. 2019) and substantially younger estimates (Friedman et al. 2013; Irisarri et al. 2018) still place the austrotilapiine split well within a time range that would see their earliest descendants impacted by rifting. As rift branches migrated across the continent, rivers changed course, drainages reversed, basin connectivity was altered, and lakes formed and desiccated (Stankiewicz and de Wit 2006; Danley et al. 2012). This was a dynamic time for aquatic ecosystems that may have afforded ancestral populations opportunities to colonize and diversify in new habitats further east (Murray et al. 2001a,b; Joyce et al. 2005; Ford et al. 2016).
Lake Tanganyika was most likely colonized by a Congo basin ancestor, based on its nested position among Congolese endemics. Later-diverging taxa recolonized surrounding rivers (e.g., Lamprologus and Orthochromis) and other eastern lakes. A close relationship between Lake Victoria and “Astatotilapia” suggest that the Malagarasi or Ruzizi rivers may have served as the portal through which the Victoria system was colonized (Danley et al. 2012). In addition to seeding lakes, changing river networks likely also facilitated the transfer of genetic diversity throughout the continent by allowing for hybridization among newly interacting communities (Schwarzer et al. 2012a,b; Loh et al. 2013; Martin et al. 2015; Weiss et al. 2015; Svardal et al. 2020).
Lake colonization resulted in repeated adaptive radiations that have been linked to both extrinsic factors related to ecological opportunity (e.g., depth and energy) and intrinsic attributes that enhance standing diversity or expedite speciation (e.g., proclivity for hybridization and sexual selection) (Deutsch 1997; Seehausen 2006; Wagner et al. 2012; Brawand et al. 2014; Weiss et al. 2015; Ford et al. 2016; Ivory et al. 2016; Meier et al. 2017; McGee et al. 2020). But, lakes are not the only habitats in which cichlids thrive. Although not as prolific, the Congo River, particularly its lower stretches, is another fecund environment for cichlids (Markert et al. 2010; Schwarzer et al. 2011; Alter et al. 2017; Stiassny and Alter 2021). Geographic and ecological drivers of riverine cichlid diversification are being investigated to various degrees in the Neotropics (Piálek et al. 2012, 2019; Poll 1986; López-Fernández et al. 2013; Astudillo-Clavijo et al. 2015; Arbour and López-Fernández 2016; Burress et al. 2017); much less is known about diversification in African rivers (Markert et al. 2010; Schwarzer et al. 2011; Alter et al. 2017). Improved resolution of relationships will support comparative phylogenetic work in classic and understudied groups and enable comparisons of lineages and environments.
Target Capture
Target enrichment with taxon-specific probes allows researchers to obtain hundreds to thousands of loci chosen to meet known challenges of the focal group. However, mutation accumulation can affect data acquisition in distant relatives of the focal group (Chau et al. 2018). Indeed, effectiveness of Ilves and López-Fernández’s (2014) tilapia-based probes decreases beyond African and Neotropical cichlids (Fig. S2 of the Supplementary material available on Dryad). Nevertheless, data loss may be ameliorated to some extent with bioinformatic modifications. Our hybrid approach proved capable of contributing novel sites through de novo assembly that are otherwise too divergent to map to a reference genome during guided assembly (Fig. S2 of the Supplementary material available on Dryad). Thus, our hybrid approach provides a tool for simultaneously reaping the benefits of a carefully curated data set for the focal group and placing it in a broader phylogenetic context with reduced missing data.
Given the advantages of protein-coding loci for phylogenetic inference, there is also a keen interest in identifying exon markers that are informative at broad phylogenetic scales (Li et al. 2013; Jiang et al. 2019). We identified close to 250 loci that are consistently retrieved across Blenniiformes (Table S5 of the Supplementary material available on Dryad) and may be suitable for investigating fish relationships beyond cichlids.
Conclusions
African cichlids have long captivated researchers with their species richness and ecomorphological diversity. Advances in molecular techniques are quickly building on the knowledge acquired from decades of meticulous morphological observation, and together this work is pulling back the curtain on a complex evolutionary history involving repeated radiations and hybridization that has played out over a dynamic continent. While genetic exchange and rapid diversification may have been important to the success of African cichlids, these processes also complicate phylogenetic inference. Thoroughly sampled phylogenetic hypotheses that account for gene heterogeneity from multiple sources will contribute to improved classification schemes and provide the necessary foundations for comparative research into drivers of cichlid diversity.
Acknowledgments
For curatorial assistance and laboratory support, we thank Mary Burridge, Margaret Zur, and Erling Holm (ROM); Tom Vigliotta, Radford Arrindell, Chloe Lewis, and Barbara Brown (AMNH), and Fabrizia Ronco and Adrian Indermaur (University of Basel). Samples not available at the ROM, AMNH, or University of Basel were provided by Oliver Lucanus (Belowwater.com). We gratefully acknowledge the field participation and support of Victor Mamonekenei and Armel Ibala (Université Marien Ngouabi, Republic of Congo); Raoul Monsembula Iyaba, José Justin Mbimbi Mayi Munene, Tobit Liyandja and Myriam Modima Yoko (Université de Kinshasa, Democratic Republic of Congo); Lawrence Makasa and Taylor Banda (Department of Fisheries, Mpulungu, Zambia); Arnold Roger Bitja-Nyom (University of Douala, Cameroon); Antoine Pariselle; Cyrille Dening; and Ngando Ephesians Jiku. For granting us permission to do research in their locations and their assistance with collection and exportation permitting, we thank the Ministère de la Recherché Scientifique et de l’Innovation Technique, Institut National de Recherche EnSciences Exactes et Naturelles (IRSEN) and Groupe d’Etude et de Recherche sur la Diversité Biologique (GERBID) (Republic of Congo); Ministère de l’Agriculture et du Développement Rural, Sécretariat Général de l’Agriculture, Pêche et Elevage, Direction des Pêches and the Université de Kinshasa, Cabinet du Recteur (Democratic Republic of Congo); local authorities of the Bemé and Barombi villages (Cameroon); and the Ministry of Scientific Research and Innovation (Cameroon).
Contributor Information
Viviana Astudillo-Clavijo, Department of Ecology and Evolutionary Biology, University of Toronto, 25 Willcocks St., Toronto, ON M5S 3B2, Canada; Department of Natural History, Royal Ontario Museum, 100 Queens Park, Toronto, ON M5S 2C6, Canada; Department of Ecology and Evolutionary Biology and Museum of Zoology, University of Michigan, Biological Science Building, 1105 N University Ave., Ann Arbor, MI 48109, USA.
Melanie L J Stiassny, Department of Ichthyology, American Museum of Natural History, 200 Central Park West, New York, NY 10024-5102, USA.
Katriina L Ilves, Department of Zoology, Canadian Museum of Nature, 240 McLeod St., Ottawa, ON K2P 2R1, Canada.
Zuzana Musilova, Department of Zoology, Charles University in Prague, Vinicna 7, Prague, Central Bohemia, CZ-128 44, Czech Republic.
Walter Salzburger, Department of Environmental Sciences, Zoological Institute, University of Basel, Vesalgasse 1, CH-4051 Basel, Basel-Stadt, Switzerland.
Hernán López-Fernández, Department of Ecology and Evolutionary Biology, University of Toronto, 25 Willcocks St., Toronto, ON M5S 3B2, Canada; Department of Natural History, Royal Ontario Museum, 100 Queens Park, Toronto, ON M5S 2C6, Canada; Department of Ecology and Evolutionary Biology and Museum of Zoology, University of Michigan, Biological Science Building, 1105 N University Ave., Ann Arbor, MI 48109, USA.
Supplementary material
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.d7wm37q26. DNA sequences are also available from GenBank, BioProject PRJNA853788.
Funding
This work was supported by the Royal Ontario Museum [ROM], the University of Michigan and Natural Sciences and Engineering Research Council of Canada [NSERC] Discovery Grants [371212-2009 and 2014-05374 to H.L.F.]; NSERC Postgraduate/Alexander Graham Bell Canada Graduate Scholarship [3-475436-2015 to V.A.C.]; ROM Rebanks Postdoctoral Fellowship and Canadian Museum of Nature Research Activity Grant to K.L.I.; (American Museum of Natural History [AMNH]) Axelrod Research Curatorship and US National Science Foundation [NSF] [DEB 1655694 to M.L.J.S.]; Czech Science Foundation [16-09784Y] and Swiss National Science Foundation [SNSF] [166550 to Z.M.], European Research Council [ERC] (CICHLID X) and SNSF [156405 and 176039 to W.S.].
X) and SNSF [156405 and 176039 to W.S.].
References
- Abadi S., Azouri D., Pupko T., Mayrose I.. 2019. Model selection may not be a mandatory step for phylogeny reconstruction. Nat. Commun. 10:934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter S., Munshi-South J., Stiassny M.L.J.. 2017. Genomewide SNP data reveal cryptic phylogeographic structure and microallopatric divergence in a rapids-adapted clade of cichlids from the Congo River. Mol. Ecol. 26:1401–1419. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Arbour J.H., López-Fernández H.. 2016. Continental cichlid radiations: functional diversity reveals the role of changing ecological opportunity in the Neotropics. Proc. Biol. Sci. 283:20160556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astudillo-Clavijo V., Arbour J.H., López-Fernández H.. 2015. Selection towards different adaptive optima drove the early diversification of locomotor phenotypes in the radiation of Neotropical geophagine cichlids. BMC Evol. Biol. 15:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur-R R., Broughton R.E., Wiley E.O., Carpenter K., López J.A., Li C., Holcroft N.I., Arcila D., Sanciangco M., Cureton li J.C., Zhang F., Buser T., Campbell M.A., Ballesteros J.A., Roa-Varon A., Willis S., Borden W.C., Rowley T., Reneau P.C., Hough D.J., Lu G., Grande T., Arratia G., Ortí G.. 2013. The tree of life and a new classification of bony fishes. PLoS Curr. 18:ecurrents.tol.53ba26640df0ccaee75bb165c8c26288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur-R R., Wiley E.O., Arratia G., Acero A., Bailly N., Miya M., Lecointre G., Orti G.. 2017. Phylogenetic classification of bony fishes. BMC Evol. Biol. 17:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanson J., Edelman A., Karpinski S., Shah V.B.. 2017. Julia: a fresh approach to numerical computing. SIAM Review 59:65–98. [Google Scholar]
- Brawand D., Wagner C.E., Li Y.I., Malinsky M., Keller I., Fan S., Simakov O., Ng A.Y., Lim Z.W., Bezault E., Turner-Maier J., Johnson J., Alcazar R., Noh H.J., Russell P., Aken B., Alföldi J., Amemiya C., Azzouzi N., Baroiller J.F., Barloy-Hubler R., Berlin A., Bloomquist R., Carleton K.L., Conte M.A., D’Cotta H., Eshel O., Gaffney L., Galibert F., Gante H.F., Gnerre S., Greuter L., Guyon R., Haddad N.S., Haerty W., Harris R.M., Hofmann H.A., Hourlier R., Hulata G., Jaffe D.B., Lara M., Lee A.P., MacCallum I., Mwaiko S., Nikaido M., Nishihara H., Ozouf-Costaz C., Penman D.J., Przybylski D., Rakotomanga M., Renn S.C.P., Ribeiro F.J., Ron M., Salzburger W., Sanchez-Pulido L., Santos M.E., Searle S., Sharpe T., Swofford R., Tan F.J., Williams L., Young S., Yin S., Okada N., Kocher T.D., Miska E.A., Lander E.S., Venkatesh B., Fernald R.D., Meyer A., Ponting C.P., Streelman J.T., Lindblad-Toh K,Seehausen O., Di Palma F.. 2014. The genomic substrate for adaptive radiation in African cichlid fish. Nature 513:375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burress E.D., Piálek, L., Casciotta J.R., Almirón A., Tan M., Armbruster J.W., Říçan O.. 2017. Island- and lake-like parallel adaptive radiations replicated in rivers. Proc. R. Soc. B 285:20171762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Xi Z., Lemmon E.M., Lemmon A.R., Mast A., Buddenhagen C.E.. 2021. The perfect storm: gene tree estimation error, incomplete lineage sorting, and ancient gene flow explain the most recalcitrant ancient angiosperm clade, Malpighiales. Syst. Biol. 70:491–507. [DOI] [PubMed] [Google Scholar]
- Chau J.H., Rahfeldt W.A., Olmstead R.G.. 2018. Comparison of taxon-specific versus general locus sets for targeted sequence capture in plant phylogenomics. Appl. Plant Sci. 6:e1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clabaut C., Salzburger W., Meyer A.. 2005. Comparative phylogenetic analyses of the adaptive radiation of Lake Tanganyika cichlid fish: nuclear sequences are less homoplasious but also less informative than mitochondrial DNA. J. Mol. Evol. 61:666–681. [DOI] [PubMed] [Google Scholar]
- Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W.. 2016. GenBank. Nucleic Acids Res. 44:D67–D72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock P.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., de Hoon M.J.L.. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford J. 2010. AssemblyAssembler1.3. GitHub repository: https://github.com/dzerbino/velvet/tree/master/contrib/AssemblyAssembler1.3. [Google Scholar]
- Danley P.D., Husemann M., Ding B., DiPietro L.M., Beverly E.J., Peppe D.J.. 2012. The impact of the geologic history and paleoclimate on the diversification of East African Cichlids. Int. J. Evol. Biol. 2012:574851–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R., Vachaspati P., Mirarab S., Warnow T.. 2015. Phylogenomic species tree estimation in the presence of incomplete lineage sorting and horizontal gene transfer. BMC Genomics 16:S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day J.J., Cotton J.A., Barraclough T.G.. 2008. Tempo and mode of diversification of Lake Tanganyika cichlids. PLoS One 3: doi: 10.1371/journal.pone.0001730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan J.H., Rosenberg N.A.. 2009. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24:332–340. [DOI] [PubMed] [Google Scholar]
- Deutsch J.C. 1997. Colour diversification in Malawi cichlids: evidence for adaptation, reinforcement or sexual selection? Biol. J. Linn. Soc. 62:1–14. [Google Scholar]
- Dunz A.R., Schliewen U.K.. 2013. Molecular phylogeny and revised classification of the haplotilapiine cichlid fishes. Mol. Phylogenet. Evol. 68:64–80. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S.V. 2009. Is a new and general theory of molecular systematics emerging? Evolution 63:1–19. [DOI] [PubMed] [Google Scholar]
- Eytan R.I., Evans B.R., Dornburg A., Lemmon A.R., Lemmon E.M., Wainwright P.C., Near T.J.. 2015. Are 100 enough? Inferring acanthomorph teleost phylogeny using Anchored Hybrid Enrichment. BMC Evol. Biol. 15:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias I.P., Ortí G., Meyer A.. 2000. Total evidence: molecules, morphology, and the phylogenetics of cichlid fishes. J. Exp. Zool. B 288:76–92. [PubMed] [Google Scholar]
- Farias I.P., Ortí G., Sampaio I., Schneider H., Meyer A.. 1999. Mitochondrial DNA phylogeny of the family Cichlidae: monophyly and fast molecular evolution of the Neotropical assemblage. J. Mol. Evol. 48:703–711. [DOI] [PubMed] [Google Scholar]
- Faircloth B.C., McCormack J.E., Crawford N.G., Harvey M.G., Brumfield R.T., Glenn T.C.. 2012. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst. Biol. 61:717–726. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1978. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 27:401–410. [Google Scholar]
- Flicek P., Ahmed I., Amode M.R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S., Fitzgerald S., Gil L., García-Girón C., Gordon L., Hourlier T., Hunt S., Juettemann T., Kähäri A.K., Keenan S., Komorowska M., Kulesha E., Longden I., Maurel T., McLaren W.M., Muffato M., Nag R., Overduin B., Pignatelli M., Pritchard B., Pritchard E., Riat H.S., Ritchie G.R., Ruffier M., Schuster M., Sheppard D., Sobral D., Taylor K., Thormann A., Trevanion S., White S., Wilder S.P., Aken B.L., Birney E., Cunningham F., Dunham I., Harrow J., Herrero J., Hubbard T.J., Johnson N., Kinsella R., Parker A., Spudich G., Yates A., Zadissa A., Searle S.M.. 2013. Ensembl 2013. Nucleic Acids Res. 41:D48–D55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P.G. 2004. Modeling compositional heterogeneity. Syst. Biol. 53:485–495. [DOI] [PubMed] [Google Scholar]
- Ford A.G.P., Bullen T.R., Pang L., Genner M.J., Bills R., Flouri T., Ngatunga B.P., Rüber L., Schliewen U.K., Seehausen O., Shechonge A., Stiassny M.L.J., Turner G.F., Day J.J.. 2019. Molecular phylogeny of Oreochromis (Cichlidae: Oreochromini) reveals mito-nuclear discordance and multiple colonisation of adverse aquatic environments. Mol. Phylogenet. Evol. 136:215–226. [DOI] [PubMed] [Google Scholar]
- Ford A.G.P., Dasmahapatra K.K., Rüber L., Gharbi K., Cezard T., Day J.J.. 2015. High levels of interspecific gene flow in an endemic cichlid fish adaptive radiation from an extreme lake environment. Mol. Ecol. 24:3421–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford A.G.P., Rüber L., Newton J., Dasmahapatra K.K., Balarin J.D., Brunn K., Day J.J.. 2016. Niche divergence facilitated by fine-scale ecological partitioning in a recent cichlid fish adaptive radiation. Evolution 70:2718–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman M., Keck B.P., Dornburg A., Eytan R.I., Martin C.H., Hulsey C.D., Wainwright P.C., Near T.J.. 2013. Molecular and fossil evidence place the origin of cichlid fishes long after Gondwanan rifting. Proc. R. Soc. B. 280:20131733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froese R., Pauly D.. 2021. Fishbase. www.fishbase.org [Google Scholar]
- Gatesy J., Springer M.S.. 2014. Phylogenetic analysis at deep timescales: unreliable gene trees, bypassed hidden support, and the coalescence/concatalescence conundrum. Mol. Phylogenet. Evol. 80:231–266. [DOI] [PubMed] [Google Scholar]
- Genner M.J., Seehausen O., Lunt D.H., Joyce D.A., Shaw P.W., Carvalho R., Turner G.F.. 2007. Age of cichlids: new dates for ancient lake fish radiations. Mol. Biol. Evol. 24:1269–1282. [DOI] [PubMed] [Google Scholar]
- Ghezelayagh A., Harrington R.C., Burress E.D., Campbell M.A., Buckner J.C., Chakrabarty P., Glass J.R., McCraney W.T., Unmack P.J., Thacker C.E., Alfaro M.E., Friedman S.T., Ludt W.B., Cowman P.F., Friedman M., Price S.A., Dornburg A., Faircloth B.C., Wainwright P.C., Near T.J.. 2021. Prolonged morphological expansion of spiny-rayed fishes following the end-Cretaceous. bioRxiv 2021.07.12.452083. doi: 10.1101/2021.07.12.452083. [DOI] [PubMed] [Google Scholar]
- Grant P.P.R., Grant R.. 2019. Hybridization increases population variation during adapting radiation. Proc. Natl. Acad. Sci. USA 116:23216–23224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes L.C., Ortí G., Huang Y., Sun Y., Baldwin C.C., Thompson A.W., Arcila D., Betancur-R R., Li C., Becker L., Bellora N., Zhao X., Li X., Wang M., Fang C., Xie B., Zhou Z., Huang H., Chen S., Venkatesh B., Shi Q.. 2018. Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc. Natl. Acad. Sci. USA 115:6249–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsey C.D., García de León F.. 2005. Cichlid jaw mechanics: linking morphology to feeding specialization. Funct. Ecol. 19:487–494. [Google Scholar]
- Ilves K.L., López-Fernández H.. 2014. A targeted next-generation sequencing toolkit for exon-based cichlid phylogenomics. Mol. Ecol. Resour. 14:802–811. [DOI] [PubMed] [Google Scholar]
- Ilves K.L., Torti D., López-Fernández H.. 2018. Exon-based phylogenomics strengthens the phylogeny of Neotropical cichlids and identifies remaining conflicting clades (Cichliformes: Cichlidae: Cichlinae). Mol. Phylogenet. Evol. 118:232–243. [DOI] [PubMed] [Google Scholar]
- Irisarri I., Singh P., Koblmüller S., Torres-Dowdall J., Henning F., Franchini P., Fischer C., Lemmon A.R., Lemmon E.M., Thallinger G.G., Sturmbauer C., Meyer A.. 2018. Phylogenomics uncovers early hybridization and adaptive loci shaping the radiation of Lake Tanganyika cichlid fishes. Nat. Commun. 9:3159–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivory S.J., Blome M.W., King J.W., McGlue M.M., Cole J.E., Cohen A.S.. 2016. Environmental change explains cichlid adaptive radiation at Lake Malawi over the past 1.2 million year. Proc. Natl. Acad. Sci. USA 113:11895–11900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis E.D., Mirarab S., Aberer A.J., Li B., Houde P., Li C., Ho S.Y.W., Faircloth B.C., Nabholz B., Howard J.T., Suh A., Weber C.C., da Fonseca R.R., Li J., Zhang F., Li H., Zhou L., Narula N., Liu L., Ganapathy G., Boussau B., Bayzid M.S., Zavidovych V., Subramanian S., Gabaldón T., Capella-Gutiérrez S., Huerta-Cepas J., Rekepalli B., Munch K., Schierup M., Lindow B., Warren W.C., Ray D., Green R.E., Bruford M.W, Zhan X., Dixon A., Li S., Li N., Huang Y., Derryberry E.P, Bertelsen M.F., Sheldon F.H., Brumfield R.T., Mello C.V., Lovell P.V., Wirthlin M., Schneider M.P.C., Prosdocimi F., Samaniego J.A., Velazquez A.M.V., Alfaro-Núñez A., Campos P.F., Petersen B., Sicheritz-Ponten T., Pas A., Bailey T., Scofield P., Bunce M., Lambert D.M., Zhou Q., Perelman P., Driskell A.C., Shapiro B., Xiong Z., Zeng Y., Liu S., Li Z., Liu B., Wu K., Xiao J., Yinq X., Zheng Q., Zhang Y., Yang H., Wang J., Smeds L., Rheindt F.E., Braun M., Fjeldsa J., Orlando L., Barker F.K., Jønsson K., Johnson W., Koepfli K.P., O’Brien S., Haussler D., Ryder O.A., Rahbek C., Willerslev E., Graves G.R., Glenn T.C., McCormack J., Burt D., Ellegren H., Alström P., Edwards S.V., Stamatakis A., Mindell D.P., Cracraft J., Braun E.L., Warnow T., Jun W., Gilbert M.T.P., Zhang G.. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346:1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Chen S.Y., Wang H., Li D.Z., Wiens J.J.. 2014. Should genes with missing data be excluded from phylogenetic analyses? Mol. Phylogenet. Evol. 80:308–318. [DOI] [PubMed] [Google Scholar]
- Jiang J., Yuan H., Zheng X., Wang Q., Kuang T., Li J., Liu J., Song S., Wang S., Cheng F., Li H., Huang J., Li C.. 2019. Gene markers for exon capture and phylogenomics in ray-finned fishes. Ecol. Evol. 9:3973–3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T., Kendall M., Almagro-Garcia J., Colijn C.. 2017. treespace: statistical exploration of landscapes of phylogenetic trees. Mol. Ecol. Resour. 17:1385–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce D.A., Lunt D.H., Bills R., Turner G.F., Katongo C., Duftner N., Sturmbauer C., Seehausen O.. 2005. An extant cichlid fish radiation emerged in an extinct Pleistocene Lake. Nature 435:90–95. [DOI] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A.. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck B.P., Hulsey C.D.. 2014. Continental monophyly of cichlid fishes and the phylogenetic position of Heterochromis multidens. Mol. Phylogenet. Evol. 73:53–59. [DOI] [PubMed] [Google Scholar]
- Kendall M., Colijn C.. 2016. Mapping phylogenetic trees to reveal distinct patterns of evolution. Mol. Biol. Evol. 33:2735–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblmüller S., Duftner N., Sefc K.M., Aibara M., Stipacek M., Blanc M., Egger B., Sturmbauer C.. 2007. Reticulate phylogeny of gastropod-shell-breeding cichlids from Lake Tanganyika – the result of repeated introgressive hybridization. BMC Evol. Biol. 7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblmüller S., Egger B., Sturmbauer C., Sefc K.M.. 2010. Rapid radiation, ancient incomplete lineage sorting and ancient hybridization in the endemic Lake Tanganyika cichlid tribe Tropheini. Mol. Phylogenet. Evol. 55:318–334. [DOI] [PubMed] [Google Scholar]
- Kocher T., Conroy J.A., McKaye K.R., Stauffer J.R., Lockwood S.F.. 1995. Evolution of NADH Dehydrogenase Subunit 2 in East African cichlid fish. Mol. Phylogenet. Evol. 4:420–432. [DOI] [PubMed] [Google Scholar]
- Kocher T. 2004. Adaptive evolution and explosive speciation: the cichlid fish model. Nat. Rev. Genet. 5:288–298. [DOI] [PubMed] [Google Scholar]
- Kubatko L.S. 2009. Identifying hybridization events in the presence of coalescence via model selection. Syst. Biol. 58:478–488. [DOI] [PubMed] [Google Scholar]
- Kubatko L.S., Degnan J.H.. 2007. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst. Biol. 56:17–24. [DOI] [PubMed] [Google Scholar]
- Kubatko L.S., Carstens B.C., Knowles L.L.. 2009. STEM: species tree estimation using maximum likelihood for gene trees under coalescence. Bioinformatics 25:971–973. [DOI] [PubMed] [Google Scholar]
- Lanfear R., Calcott B., Ho S.Y.W., Guindon S.. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29:1695–1701. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.. 2012. Fast gapped-read alignment with Bowtie2. Nat. Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon A.R., Emme S.A., Lemmon E.M.. 2012. Anchored hybrid enrichment for massively high-throughput phylogenomics. Syst. Biol. 61:727–744. [DOI] [PubMed] [Google Scholar]
- Li C., Hofreiter M., Straube N., Corrigan S., Naylor G.J.P.. 2013. Capturing protein-coding genes across highly divergent species. Biotechniques 54:321–326. [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997v2 doi: 10.48550/arXiv.1303.3997. [Google Scholar]
- Li H., Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsios G., Salamin N.. 2014. Hybridisation and diversification in the adaptive radiation of clownfishes. BMC Evol. Biol. 14:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wu S., Yu L.. 2015. Coalescent methods for estimating species trees from phylogenomic data. J. Syst. Evol. 53:380–390. [Google Scholar]
- Loh Y.-H.E., Bezault E., Muenzel F.M., Roberts R.B., Swofford R., Barluenga M., Kidd C.E., Howe A.E., Di Palma F., Linbald-Toh K., Hey J., Seehausen O., Salzburger W., Kocher T.D., Streelman J.T.. 2013. Origins of shared genetic variation in African cichlids. Mol. Biol. Evol. 30:906–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Fernández H., Arbour J.H., Winemiller K.O., Honeycutt R.L.. 2013. Testing for ancient adaptive radiations in Neotropical cichlid fishes. Evolution 67:1321–1337. [DOI] [PubMed] [Google Scholar]
- Maddison W.P. 1997. Gene trees in species trees. Syst. Biol. 46:523–536. [Google Scholar]
- Maglott D., Ostell J., Pruitt K.D., Tatusova T.. 2005. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 33:D52–D57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky M., Svardal H., Tyers A.M., Miska E.A., Genner M.J., Turner G.F., Durbin R.. 2018. Whole-genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nat. Ecol. Evol. 2:1940–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markert J.A., Schelly R.C., Stiassny M.L.J.. 2010. Genetic isolation and morphological divergence mediated by high-energy rapids in two cichlid genera from the lower Congo rapids. BMC Evol. Biol. 10:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C.H., Cutler J.S., Friel J.P., Dening Touokong C., Coop G., Wainwright P.C.. 2015. Complex histories of repeated gene flow in Cameroon crater lake cichlids cast doubt on one of the clearest examples of sympatric speciation. Evolution 69:1406–1422. [DOI] [PubMed] [Google Scholar]
- Matschiner M. 2019. Gondwanan vicariance or trans-Atlantic dispersal of cichlid fishes. A review of the molecular evidence. Hydrobiologia 832:9–37. [Google Scholar]
- Matschiner M., Böhne A., Ronco F., Salzburger W.. 2020. The genomic timeline of cichlid fish diversification across continents. Nat. Commun. 11:5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschiner M., Musilová Z., Barth J.M.I., Starostová Z., Salzburger W., Steel M., Bouckaert R.. 2017. Bayesian phylogenetic estimation of clade ages supports Trans-Atlantic dispersal of cichlid fishes. Syst. Biol. 66:3–22. [DOI] [PubMed] [Google Scholar]
- Mayer W.E., Tichy H., Klein J.. 1998. Phylogeny of African cichlid fishes as revealed by molecular markers. Heredity 80:702–714. [DOI] [PubMed] [Google Scholar]
- McGee M.D., Borstein S.R., Meier J.I., Marques D.A., Mwaiko S., Taabu A., Kishe M.A., O’Meara B., Bruggmann R., Excoffier L., Seehausen O.. 2020. The ecological and genomic basis of explosive adaptive radiation. Nature 586:75–59. [DOI] [PubMed] [Google Scholar]
- McMahan C.D., Chakrabarty P., Sparks J.S., Smith W.L., Davis M.P.. 2013. Temporal patterns of diversification across global cichlid biodiversity (Acanthomorpha: Cichlidae). PLoS One 8:e71162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B.Q., Hahn M.W., Lanfear R.. 2020a. New methods to calculate concordance factors for phylogenomic datasets. Mol. Biol. Evol. 37:2727–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R.. 2020b. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37:1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier J.I., Marques D.A., Mwaiko S., Wagner C.E., Excoffier L., Seehausen O.. 2017. Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 8:14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng C., Kubatko L.S.. 2009. Detecting hybrid speciation in the presence of incomplete lineage sorting using gene tree incongruence: a model. Theor. Popul. Biol. 75:35–45. [DOI] [PubMed] [Google Scholar]
- Meyer B.S., Matschiner M., Salzburger W.. 2015. A tribal level phylogeny of Lake Tanganyika fishes based on a genomic multi-marker approach. Mol. Phylogenet. Evol. 83:56–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B.S., Matschiner M., Salzburger W.. 2017. Disentangling incomplete lineage sorting and introgression to refine species-tree estimates for Lake Tanganyika cichlid fishes. Syst. Biol. 66:531–550. [DOI] [PubMed] [Google Scholar]
- Miller M.R., Dunham J.P., Amores A., Cresko W.A., Johnson E.A.. 2007. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated NDA (RAD) markers. Genome Res. 17:240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.A., Pferiffer W., Schwartz T.. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA. p. 1–8. [Google Scholar]
- Mirarab S., Warnow T.. 2015. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31:i44–i52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirarab S., Bayzid M.S., Warnow T.. 2016. Evaluating summary methods for multilocus species tree estimation in the presence of incomplete lineage sorting. Syst. Biol. 65:366–380. [DOI] [PubMed] [Google Scholar]
- Morales-Briones D.F., Kadereit G., Tefarikis D.T., Moore M.J., Smith S.A., Brockington S.F., Timoneda A., Yim W.C., Cushman J.C., Yang Y.. 2021. Disentangling sources of gene tree discordance in phylogenomic data sets: testing ancient hybridizations in Amaranthaceae s.l. Syst. Biol. 70:219–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W.J., Pringle T.H., Crider T.A., Springer M.S., Miller W.. 2007. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 17:413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A.M. 2001a. The fossil record and biogeography of the Cichlidae (Actinopterygii: Labroidei). Biol. J. Linn. Soc. 74:517–532. [Google Scholar]
- Murray A.M. 2001b. Eocene cichlid fishes from Tanzania, East Africa. J. Vertebr. Paleontol. 20:651–664. [Google Scholar]
- Muschick M., Indermaur A., Salzburger W.. 2012. Convergent evolution within an adaptive radiation of cichlid fishes. Curr. Biol. 22:2362–2368. [DOI] [PubMed] [Google Scholar]
- Musilova Z., Indermaur A., Bitja-Nyom A.R., Omelchenko D., Kłodawska M., Albergati L., Remišová K., Salzburger W.. 2019. Evolution of the visual sensory system in cichlid fishes from crater lake Barombi Mbo in Cameroon. Mol. Ecol. 28:5010–5031. [DOI] [PubMed] [Google Scholar]
- Near T.J., Dornburg A., Eytana R.I., Keck B.P., Smith W.L., Kuhn K.L., Moore J.A., Price S.A., Burbrink F.T., Friedman M., Wainwright P.C.. 2013. Phylogeny and tempo of diversification in the superradiation of spiny-rayed fishes. Proc. Natl. Acad. Sci. USA 110:12738–12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida M. 1991. Lake Tanganyika as an evolutionary reservoir of old lineages of East African cichfid fishes: inferences from allozyme data. Experiencia 47:974–979. [Google Scholar]
- Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q.. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nute M., Chou J., Molloy E.K., Warnow T.. 2018. The performance of coalescent-based species tree estimation methods under models of missing data. BMC Genomics 19:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Schliep K.. 2019. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35:526–528. [DOI] [PubMed] [Google Scholar]
- Piálek L., Burress E., Dragová K., Almirón A., Casciotta J., Říçan O.. 2019. Phylogenomics of pike cichlids. (Cichlidae: Crenicichla) of the C. manderlburgeri species complex: rapid ecological speciation in the Iguazú River basin. Hydrobiologia 832:355–375. [Google Scholar]
- Piálek L., Říçan O., Casciotta J., Almirón A., Zrzavý.. 2012. Multilocus phylogeny of Crenicichla (Teleostei: Cichlidae), with biogeography of the C. lacustris group: species flocks as a model for sympatric speciation in rivers. Mol. Phylogenet. Evol. 62:46–61. [DOI] [PubMed] [Google Scholar]
- Poll M. 1986. Classification des Cichlidae du lac Tanganika. Tribus, genres et espéces. Acad. R. Belgium Mem. Classe Sci. 45:1–163. [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. http://www.R-project.org/. [Google Scholar]
- Richards E.J., Poelstra J.W., Martin C.H.. 2018. Don’t throw out the sympatric speciation with the crater lake water: fine-scale investigation of introgression provides equivocal support for causal role of secondary gene flow in one of the clearest examples of sympatric speciation. Evol. Lett. 2:524–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring U. 2014. The East African Rift System. Aust. J. Earth. Sci. 107:132–146. [Google Scholar]
- Roberts T., Kullander S.O.. 1994. Endemic cichlid fishes of the Fwa River, Zaire: Systematics and ecology. Ichthyol. Explor. Fres. 2:97–154. [Google Scholar]
- Robinson D.R., Foulds L.R.. 1981. Comparison of phylogenetic trees. Math. Biosci. 53:131–147. [Google Scholar]
- Roch S., Steel M.. 2015. Likelihood-based tree reconstruction on a concatenation of aligned sequence data sets can be statistically inconsistent. Theor. Popul. Biol. 100C:56–62. [DOI] [PubMed] [Google Scholar]
- Roch S., Warnow T.. 2015. On the robustness to gene tree estimation error (or lack thereof) of coalescent-based species tree methods. Syst. Biol. 64:663–676. [DOI] [PubMed] [Google Scholar]
- Ronco F., Matschiner M., Böhne A., Boila A., Büscher H.H., El Taher A., Indermaur A., Malinsky M., Ricci V., Kahem A., Jentoft S., Salzburger W.. 2021. Drivers and dynamics of a massive adaptive radiation in cichlid fishes. Nature 589:76–81. [DOI] [PubMed] [Google Scholar]
- Salzburger W. 2018. Understanding explosive diversification through cichlid fish genomics. Nat. Rev. Genet. 19:705–717. [DOI] [PubMed] [Google Scholar]
- Salzburger W., Mack T., Verheyen E., Meyer A.. 2005. Out of Tanganyika: Genesis, explosive speciation, key-innovations and phylogeography of the haplochromine cichlid fishes. BMC Evol. Biol. 5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W., Meyer A., Maric S., Verheyen E., Sturmbauer C.. 2002. Phylogeny of the Lake Tanganyika cichlid species flock and its relationship to the central and East African haplochromine cichlid fish faunas. Syst. Biol. 5:113–135. [DOI] [PubMed] [Google Scholar]
- Schedel F.D.B., Musilova Z., Schliewen U.K.. 2019. East African cichlid lineages (Teleostei: Cichlidae) might be older than their ancient host lakes: new divergence estimates for the east African cichlid radiation. BMC Evol. Biol. 19:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep K. 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27:592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliewen U.K., Klee B.. 2004. Reticulate sympatric speciation in Cameroonian crater lake cichlids. Front. Zool. 41:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliewen U.K., Stiassny M.L.J.. 2003. Etia nguti, a new genus and species of cichlid fish from the River Mamfue, Upper Cross River basin, Cameroon, Central Africa. Ichthyol. Explor. Fres. 14:61–72. [Google Scholar]
- Schliewen U.K., Tautz D., Pääbo S.. 1994. Sympatric speciation suggested by monophyly of crater lake cichlids. Nature 368:629–632. [DOI] [PubMed] [Google Scholar]
- Schmieder R., Edwards R.. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer J., Lamboj A., Langen K., Misof B., Schliewen U.K.. 2015. Phylogeny and age of chromidotilapiini cichlids (Teleostei: Cichlidae). Hydrobiologia 748:185–199. [Google Scholar]
- Schwarzer J., Misof B., Ifuta S.N., Schliewen U.K.. 2011. Time and origin of cichlid colonization of the lower Congo rapids. PLoS One 6:e22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer J., Misof B., Schliewen U.K.. 2012a. Speciation within genomic networks: a case study based on Steatocranus cichlids of the lower Congo rapids. J. Evol. Biol. 25:138–148. [DOI] [PubMed] [Google Scholar]
- Schwarzer J., Misof B., Tautz D., Schliewen U.K.. 2009. The root of the East African cichlid radiations. BMC Evol. Biol. 9:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer J., Swartz E.R., Vreven E., Snoeks S.M., Cotterill F.P.D., Misof B., Schliewen U.K.. 2012b. Repeated trans-watershed hybridization among haplochromine cichlids (Cichlidae) was triggered by Neogene landscape evolution. Proc. R. Soc. 279:4389–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O. 2006. African cichlid fish: a model system in adaptive radiation research. Proc. R. Soc. B 273:1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O. 2015. Process and pattern in cichlid radiations – inferences for understanding unusually high rates of evolutionary diversification. New Phytol. 207:304–312. [DOI] [PubMed] [Google Scholar]
- Smith W.L., Chakrabarty P., Sparks J.S.. 2008. Phylogeny, taxonomy, and evolution of Neotropical cichlids (Teleostei: Cichlidae: Cichlinae). Cladistics 24:625–641. [Google Scholar]
- Solís-Lemus C., Ané C.. 2016. Inferring phylogenetic networks with maximum pseudolikelihood under incomplete lineage sorting. PLoS Genet. 12:e1005896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solís-Lemus C., Bastide P., Ané C.. 2017. Phylonetworks: a package for phylogenetic networks. Mol. Biol. Evol. 34:3292–3298. [DOI] [PubMed] [Google Scholar]
- Solís-Lemus C., Yang M., Ané C.. 2016. Inconsistency of species tree methods under gene flow. Syst. Biol. 65:843–851. [DOI] [PubMed] [Google Scholar]
- Sparks J.S., Smith W.L.. 2004. Phylogeny and biogeography of cichlid fishes (Teleostei: Perciformes: Cichlidae). Cladistics 20:501:517. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz J., de Wit M.J.. 2006. A proposed drainage evolution model for Central Africa—did the Congo flow east? J. Afr. Earth Sci. 44:75–84. [Google Scholar]
- Steel M.A., Penny D.. 1993. Distribution of tree comparison metrics – some new results. Syst Biol 42:126–141. [Google Scholar]
- Stiassny M.L.J. 1987. Cichlid familial interrelationships and the placement of the neotropical genus Cichla (Perciformes, Labroidei). J. Nat. Hist. 21:1311–1331. [Google Scholar]
- Stiassny M.L.J., Alter S.E.. 2021. Life in the fast lane: diversity, ecology, and speciation of cichlids in the lower Congo River. In: Abate M.E., Noakes D.L.G., editors. The behavior, ecology and evolution of cichlid fishes. Dordrecht, The Netherlands: Springer Publishing. p. 107–133. [Google Scholar]
- Stiassny M.L.J., Schliewen U.K., Dominey W.J.. 1992. A new species flock of cichlid fishes from Lake Bermin, Cameroon with a description of eight new species of Tilapia (Labroidei: Cichlidae). Ichthyol. Explor. Fres. 3:311–346. [Google Scholar]
- Streelman J.T., Karl S.A.. 1997. Reconstructing labroid evolution using single-copy nuclear DNA. Proc. R Soc. Lond. B 264:1011–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streelman J.T., Zardoya R., Meyer A., Karl S.A.. 1998. Multilocus phylogeny of cichlid fishes (Pisces: Perciformes): evolutionary comparison of microsatellite and single-copy nuclear loci. Mol. Biol. Evol. 15:798–808. [DOI] [PubMed] [Google Scholar]
- Suh A., Smeds L., Ellegren H.. 2015. The dynamics of incomplete lineage sorting across the ancient adaptive radiations of Neoavian birds. PLoS Biol. 13:e1002224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sültmann H., Mayer W.E., Figueroa F., Tichy H., Klein J.. 1995. Phylogenetic analysis of cichlid fishes using nuclear DNA markers. Mol. Biol. Evol. 12:1033–1047. [DOI] [PubMed] [Google Scholar]
- Svardal H., Quah F.X., Malinsky M., Ngatunga B.P., Miska E.A., Salzburger W., Genner M.J., Turner G.F., Durbin R.. 2020. Ancestral hybridization facilitated species diversification in the Lake Malawi cichlid fish adaptive radiation. Mol. Biol. Evol. 37: 1100–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Sota T.. 2016. A robust phylogeny among major lineages of the East African cichlids. Mol. Phylogenet. Evol. 100:234–242. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Terai Y., Nishida M., Okada N.. 2001. Phylogenetic relationships and incomplete lineage sorting among cichlid fishes in Lake Tanganyika as revealed by analyses of the insertion retroposons. Mol. Biol. Evol. 18:2057–2066. [DOI] [PubMed] [Google Scholar]
- Takahashi T. 2003. Systematics of Tanganyika cichlid fishes (Teleostei: Perciformes). Ichthyol. Res. 50:367–382. [Google Scholar]
- Trewavas E. 1983. Tilapiine fishes of the genera Sarotherodon, Oreochromis and Danakilia. British Museum (Natural History): London. [Google Scholar]
- Turner G.F. 2007. Adaptive radiation of cichlid fish. Curr. Biol. 19:R827–R831. [DOI] [PubMed] [Google Scholar]
- Wagner C.E., Harmon L.J., Seehausen O.. 2012. Ecological opportunity and sexual selection together predict adaptive radiation. Nature 487:366–370. [DOI] [PubMed] [Google Scholar]
- Wainwright P.C., Smith W.L., Price S.A., Tang K.L., Sparks J.S., Ferry L.A., Kuhn K.L., Eytan R.I., Near T.J.. 2012. The evolution of pharyngognathy: a phylogenetic and functional appraisal of the pharyngeal jaw key innovation in labroid fishes and beyond. Syst. Biol. 61:1001–1027. [DOI] [PubMed] [Google Scholar]
- Weiss J.D., Cotterill F.P.D., Schliewen U.K.. 2015. Lake Tanganyika- a ‘melting pot’ of ancient and young cichlid lineages (Teleostei: Cichlidae)? PLoS One 10:e0125043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitefield J.B., Lockhard P.J.. 2007. Deciphering ancient rapid radiations. Trends Ecol. Evol. 22:258–265. [DOI] [PubMed] [Google Scholar]
- Xi Z., Liu L., Davis C.C.. 2016. The impact of missing data on species tree estimation. Mol. Biol. Evol. 33:838–860. [DOI] [PubMed] [Google Scholar]
- Yu Y., Degnan L., Nakhleh L.. 2012. The probability of a gene tree topology within a phylogenetic network with applications to hybridization detection. PLoS Genet. 8:e1002660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Dong J., Liu K.J., Nakhleh L.. 2014. Maximum likelihood inference of reticulate evolutionary histories. Proc. Natl. Acad. Sci. USA 111:16448–16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S., Crosa G., Vanetti I., Binelli G., Harper D.M., Mavuti K.M., Balarin J.D., Britton J.R.. 2014. Genetic and morphological analyses indicate high population mixing in the endangered cichlid Alcolapia flock of East Africa. Conserv. Genet. 15:429–440. [Google Scholar]
- Zebrino D.R., Birney E.. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebrino D.R. 2009. extractContigReads. GitHub repository: https://github.com/dzerbino/velvet/blob/master/contrib/extractContigReads/extractContigReads.pl. [Google Scholar]
- Zhang C., Rabiee M., Sayyari E., Mirarab S.. 2018. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19:153. [DOI] [PMC free article] [PubMed] [Google Scholar]