ABSTRACT.
Sierra Leone relies heavily on histidine-rich protein 2–based diagnostics for malaria because of the high transmission of Plasmodium falciparum. During the 2015 recombinant vesicular stomatitis virus (VSV)−Zaire Ebola virus envelope glycoprotein (GP) vaccine trial, 77 participants with asymptomatic Plasmodium infection were enrolled, with all but four having P. falciparum malaria. Of the 73 participants with P. falciparum malaria, one infection (1 of 73, 1.4%; 95% CI, 0.03–7.4) showed P. falciparum with a pfhrp3 single deletion, and two P. falciparum infections (2 of 73, 2.7%; 95% CI, 0.03–9.6) showed pfhrp2/pfhrp3 dual deletions. This study shows evidence of pfhrp2- and pfhrp3-deleted parasites in Freetown, Sierra Leone. Additional studies for more precise estimates of prevalence are warranted.
INTRODUCTION
In 2021, the WHO estimated that the entire population of Sierra Leone, a total of 7,976,985 individuals, was at high risk for malaria infection.1 During the 2014 West African Ebola virus outbreak, several phase 2/3 studies of the recombinant vesicular stomatitis virus–Zaire Ebola virus envelope glycoprotein were carried out as part of the Sierra Leone Trial to Introduce a Vaccine against Ebola (STRIVE).2,3 Because Plasmodium falciparum is highly endemic in Sierra Leone, the implications of asymptomatic malaria infection on vaccine immunogenicity were examined among STRIVE participants,2 as active malaria infection has been found to limit the immunogenicity of certain vaccines.4,5
In Sierra Leone, P. falciparum is responsible for more than 99% of malaria cases, although Plasmodium ovale and Plasmodium malariae infections have also been identified.6 Malaria rapid diagnostic tests (RDTs) used in Sierra Leone are typically based only on the histidine-rich protein 2 (HRP2) antigen,7 a protein found exclusively in P. falciparum that is highly expressed during the erythrocytic stage of infection.8 Because of the presence of multiple repeat regions in HRP2 and the closely related HRP3 protein, multiple anti-HRP2 diagnostic antibodies can bind to these proteins and, as a result, lead to an amplified assay signal that makes HRP2-based RDTs a highly sensitive tool for P. falciparum diagnosis.8
Although HRP2-based RDTs have aided in the rapid diagnosis of P. falciparum malaria as a result of their low cost, high sensitivity, and minimal training required, the recent emergence of pfhrp2 and pfhrp3 (pfhrp2/3) gene deletions threatens the utility of HRP2-based RDTs.8,9 Deletions of the pfhrp2/3 genes can cause false-negative RDT results as a result of the antigens not being expressed, preventing the appropriate treatment of infected individuals and allowing for continued circulation of these strains in the community.8,9 To ensure the continued reliability of HRP2-based RDTs, the WHO established guidelines for monitoring pfhrp2 deletions, and set a 5% local prevalence of pfhrp2 deletions as the cutoff point for a change in testing strategy, recommending that affected regions switch from HRP2-based RDTs to combination antigen RDTs or microscopy.9 In this study, the assessment of P. falciparum pfhrp2/3 gene deletions was carried out using blood samples provided by STRIVE participants at enrollment.
METHODS
As described elsewhere, baseline enrollment for this arm of STRIVE occurred at Connaught Hospital in Freetown, Sierra Leone, from June through September 2015.2,3 The study protocol was approved by the Sierra Leone Ethics and Scientific Review Committee, CDC Institutional Review Board (0900f3eb81c52483), Pharmacy Board of Sierra Leone, and U.S. Food and Drug Administration. All participants provided written informed consent for the use of samples for malaria assays.
Potential participants with oral temperature greater than 38°C or any acute illness symptoms were excluded from enrollment. For post hoc laboratory identification of Plasmodium infection, total DNA was extracted from blood clots as described elsewhere.2 Photon-induced electron transfer–polymerase chain reaction (PET-PCR) for both Plasmodium genus and species were performed in duplicate,10,11 and genotyping for the pfmsp1, pfmsp2, pfhrp2, and pfhrp3 genes was performed in duplicate with previously described reactions.12,13 Detection of the HRP2 antigen was accomplished through a bead-based immunoassay, with reagents and conditions described by Rogier et al.14 A total of 73 individuals were found to be positive for P. falciparum by PET-PCR out of an initial cohort of 521 participants.
RESULTS
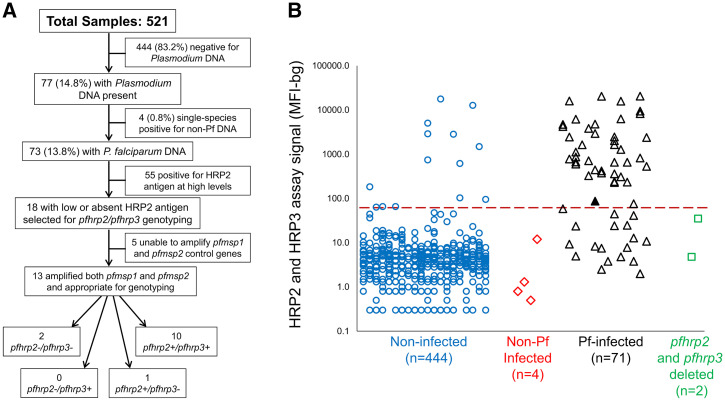
A total of 521 persons provided blood samples that were assessed for malaria infection. Of the 521 persons enrolled, 444 (83.2%) showed no evidence of Plasmodium infection by PET and nested PCRs (Figure 1A). Of the 77 blood samples positive for Plasmodium DNA, 73 (94.8%) were positive for P. falciparum DNA and four (5.2%) were single-species infections with a non-falciparum Plasmodium DNA (one P. ovale, and three P. malariae). One blood sample with a P. falciparum infection was also found to harbor a low-density P. malariae infection.
Figure 1.
Laboratory assay workflow, reporting of pfhrp2 and pfhrp3 deletions, and relationship of histidine-rich protein 2 (HRP2) antigen levels to Plasmodium falciparum genotype. (A) Flow diagram for all blood samples included in this study with terminal boxes displaying pfhrp2 and pfhrp3 genotypes of selected samples. (B) Level of HRP2 and HRP3 antigen in blood samples for persons polymerase chain reaction negative for Plasmodium DNA (n = 444, blue circles), mono-infected with non-falciparum Plasmodium (n = 4, red diamonds), infected with wild-type P. falciparum (n = 71, black triangles), or infected with P. falciparum with a pfhrp2 deletion (n = 2, green squares). Red hashed line indicates threshold for assay signal indicating positivity for HRP2. Among the P. falciparum–infected participant samples, a single sample was determined be a pfhrp3 single deletion (pfhrp2+/pfhrp3–), and is indicated as the single shaded black triangle near the threshold line. Pf = Plasmodium falciparum.
Blood samples without detectable Plasmodium DNA occasionally had HRP2 antigen present (14 of 444, 3.2%) (Figure 1B). For blood samples with P. falciparum DNA, most (61 of 73, 83.6%) were positive for the HRP2 antigen, with generally high levels of this antigen in the blood. For investigating potential deletions of the pfhrp2/3 genes, the 12 HRP2-negative samples from P. falciparum–infected persons underwent genotyping. Six additional samples were also selected from P. falciparum infections that were positive for the HRP2 antigen, but at very low levels, bringing the total number of samples for genotyping to 18. The majority of these 18 samples (13 of 18, 72.2%) had sufficient Plasmodium DNA to amplify successfully the pfmsp1 and pfmsp2 single-copy genes to qualify for appropriate reporting of pfhrp2/3 genotypes. Of the 13 samples genotyped successfully, 10 (76.9%) were found to be pfhrp2/3 wild-type parasites with successful amplification of both genes. One P. falciparum isolate was found to be a single deletion of the pfhrp3 gene (1 of 73, 1.4%; 95% CI, 0.03–7.4), and two isolates showed a pfhrp2/3 double-deletion genotype (2 of 73, 2.7%; 95% CI, 0.03–9.6).
DISCUSSION
Of the 73 STRIVE participants with asymptomatic P. falciparum infection at enrollment, genotyping PCR assays of these samples revealed only two parasite isolates (2 of 73, 2.7%; 95% CI, 0.03–9.6) to have dual pfhrp2/3 deletions. This low level of pfhrp2/3 deletions among P. falciparum parasites in Freetown, Sierra Leone in 2015 suggests the vast majority of these parasites retain the ability to express the HRP2 and HRP3 antigens necessary for a positive HRP2 RDT result. In addition, samples from 14 individuals that were confirmed to be negative for Plasmodium DNA by PCR had detectable HRP2 signals. The presence of HRP2 antigen in DNA-negative samples could be a result of lingering HRP2 from previous P. falciparum infections, because HRP2 can persist in the circulation for months after successful treatment and can even contribute to false-positive RDT results.8,15 In addition, DNA degradation or other poor-quality DNA could cause a false-negative PCR result.
In 2010, Gerstl et al.16 assessed RDT failures in a hyperendemic P. falciparum region of Sierra Leone and reported issues with false-positive results, but did not observe any false-negative tests for either the HRP2-based or the pan-Plasmodium lactate dehydrogenase/P. falciparum HRP2 combination tests assessed when results were compared with microscopy. Similarly, assessment of false-negative P. falciparum RDTs in Monrovia, Liberia, showed no evidence of false-negative RDTs when RDT performance was evaluated by quantitative PCR.17 Combined with the results from our study showing the presence of pfhrp2 gene deletion in Freetown, this work highlights the need for additional studies to understand the pfhrp2/3 deletion status of parasites throughout Sierra Leone.
As described previously by Mahon et al.,2 assessment of malaria infections among STRIVE participants was limited to those with asymptomatic presentation, and therefore these results would not necessarily be representative of symptomatic infections from treatment-seeking persons from this region. Furthermore, this study did not enroll children, who typically experience infections with greater parasite densities.2,18 The results presented here assume all participants had P. falciparum mono-infection, but the presence of pfhrp2 and/or pfhrp3 deleted P. falciparum could potentially be masked in a multiclonal infection if one of the isolates was wild-type, although this is less likely in asymptomatic malaria infections.19,20
As of 2016, Sierra Leone has been using the SD BIOLINE Malaria Antigen P.f. (HRP-II) RDT (Abbott Laboratories, Chicago, IL) as the approved RDT for diagnosis of malaria.21 In 2021, the U.S. President’s Malaria Initiative has estimated the number of RDTs needed for diagnosis of the at-risk population of Sierra Leone will increase from approximately 4.7 million to 5.4 million between 2021 to 2023.22
In our study, pfhrp2/3 deletions were observed among P. falciparum isolates obtained in 2015 from Freetown, Sierra Leone, but these deletions appear to be at very low levels. Because it is possible that deletions in these genes may have increased in Sierra Leone since 2015, future surveys or surveillance efforts to monitor pfhrp2/3 deletions and obtain precise prevalence estimates are required.
REFERENCES
- 1. World Health Organization , 2021. World Malaria Report 2021. Geneva, Switzerland, WHO. [Google Scholar]
- 2. Mahon BE. et al. , 2021. Baseline asymptomatic malaria infection and immunogenicity of rVSVDeltaG-ZEBOV-GP vaccine: The Sierra Leone Trial to Introduce a Vaccine Against Ebola (STRIVE). J Infect Dis 224: 1907–1915. [Google Scholar]
- 3. Samai M. et al. , 2018. The Sierra Leone Trial to Introduce a Vaccine Against Ebola: an evaluation of rVSVG-ZEBOV-GP vaccine tolerability and safety during the West Africa Ebola outbreak. J Infect Dis 217: S6–S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Williamson WA, Greenwood BM, 1978. Impairment of the immune response to vaccination after acute malaria. Lancet 1: 1328–1329. [DOI] [PubMed] [Google Scholar]
- 5. Zimmermann P, Curtis N, 2019. Factors that influence the immune response to vaccination. Clin Microbiol Rev 32: 1–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. World Health Organization Global Malaria Programme , 2020. World Malaria Report 2020. Geneva, Switzerland, WHO. [Google Scholar]
- 7. World Health Organziation , 2018. WHO Malaria Country Profile: Sierra Leone 2018. Geneva, Switzerland: WHO. [Google Scholar]
- 8. Poti KE, Sullivan DJ, Dondorp AM, Woodrow CJ, 2020. HRP2: transforming malaria diagnosis, but with caveats. Trends Parasitol 36: 112–126. [DOI] [PubMed] [Google Scholar]
- 9. World Health Organization , 2019. Response Plan to pfhrp2 Gene Deletions. Geneva, Switzerland: WHO. [Google Scholar]
- 10. Snounou G, 1996. Detection and identification of the four malaria parasite species infecting humans by PCR amplification. Methods Mol Biol 50: 263–291. [DOI] [PubMed] [Google Scholar]
- 11. Lucchi NW, Narayanan J, Karell MA, Xayavong M, Kariuki S, DaSilva AJ, Hill V, Udhayakumar V, 2013. Molecular diagnosis of malaria by photo-induced electron transfer fluorogenic primers: PET-PCR. PLoS One 8: e56677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCaffery JN, Nace D, Herman C, Singh B, Sompwe EM, Nkoli PM, Ngoyi DM, Kahunu GM, Halsey ES, Rogier E, 2021. Plasmodium falciparum pfhrp2 and pfhrp3 gene deletions among patients in the DRC enrolled from 2017 to 2018. Sci Rep 11: 22979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jones S, Subramaniam G, Plucinski MM, Patel D, Padilla J, Aidoo M, Talundzic E, 2020. One-step PCR: a novel protocol for determination of pfhrp2 deletion status in Plasmodium falciparum. PLoS One 15: e0236369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rogier E. et al. , 2017. Bead-based immunoassay allows sub-picogram detection of histidine-rich protein 2 from Plasmodium falciparum and estimates reliability of malaria rapid diagnostic tests. PLoS One 12: e0172139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tjitra E, Suprianto S, McBroom J, Currie BJ, Anstey NM, 2001. Persistent ICT malaria P.f/P.v panmalarial and HRP2 antigen reactivity after treatment of Plasmodium falciparum malaria is associated with gametocytemia and results in false-positive diagnoses of Plasmodium vivax in convalescence. J Clin Microbiol 39: 1025–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gerstl S, Dunkley S, Mukhtar A, De Smet M, Baker S, Maikere J, 2010. Assessment of two malaria rapid diagnostic tests in children under five years of age, with follow-up of false-positive pLDH test results, in a hyperendemic falciparum malaria area, Sierra Leone. Malar J 9: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. King M. et al. , 2021. No evidence of false-negative Plasmodium falciparum rapid diagnostic results in Monrovia, Liberia. Malar J 20: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. White NJ, Pukrittayakamee S, Hien TT, Faiz MA, Mokuolu OA, Dondorp AM, 2014. Malaria. Lancet 383: 723–735. [DOI] [PubMed] [Google Scholar]
- 19. Plucinski MM. et al. , 2018. Screening for pfhrp2/3-deleted Plasmodium falciparum, non-falciparum, and low-density malaria infections by a multiplex antigen assay. J Infect Dis 219: 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bharti PK, Chandel HS, Ahmad A, Krishna S, Udhayakumar V, Singh N, 2016. Prevalence of pfhrp2 and/or pfhrp3 gene deletion in Plasmodium falciparum population in eight highly endemic states in India. PLoS One 11: e0157949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. National Malaria Control Programme Sierra Leone , 2016. The 2016 Sierra Leone Malaria Indicator Survey: The Demographic and Health Surveys Program. Washington, DC: USAID. [Google Scholar]
- 22. U.S. President's Malaria Initiative , 2022. Sierra Leone Malaria Operational Plan FY 2022: U.S. President’s Malaria Initiative. Atlanta, GA: U.S. President's Malaria Initiative.