Abstract
Background
Classical swine fever virus (CSFV), the causative agent of classical swine fever (CFS), is a highly contagious disease that poses a serious threat to Chinese pig populations.
Objectives
Many provinces of China, such as Shandong, Henan, Hebei, Heilongjiang, and Liaoning provinces, have reported epidemics of CSFV, while the references to the epidemic of CSFV in Yunnan province are rare. This study examined the epidemic characteristics of the CSFV in Yunnan province.
Methods
In this study, 326 tissue samples were collected from different regions in Yunnan province from 2015 to 2021. A reverse transcription-polymerase chain reaction (RT-PCR), sequences analysis, and phylogenetic analysis were performed for the pathogenic detection and analysis of these 326 clinical specimens.
Results
Approximately 3.37% (11/326) of specimens tested positive for the CSFV by RT-PCR, which is lower than that of other regions of China. Sequence analysis of the partial E2 sequences of eleven CSFV strains showed that they shared 89.0–100.0% nucleotide (nt) and 95.0–100.0% amino acid (aa) homology, respectively. Phylogenetic analysis showed that these novel isolates belonged to the subgenotypes 2.1c and 2.1d, with subgenotype 2.1c being predominant.
Conclusions
The CSFV was sporadic in China’s Yunnan province from 2015 to 2021. Both 2.1c and 2.1d subgenotypes were found in this region, but 2.1c was dominant.
Keywords: Classical swine fever virus, sequence analysis, subgenotype 2.1c, Yunnan province
INTRODUCTION
The classical swine fever virus (CSFV), belonging to the genus Pestivirus in the family Flaviviridae, is a single positive-stranded, enveloped RNA virus that comprises a genome of approximately 12.3 kb [1]. Classical swine fever (CSF) caused by the CSFV is a highly contagious and fatal disease in pigs. The disease presents acute, subacute, chronic, and asymmetric characteristics, depending mainly on the viral virulence and host factors [2]. An outbreak of CSF causes high morbidity and mortality rates and significantly affects the development of the pig industry. Therefore, the occurrence of CSF in the pig population must be reported to the World Organization for Animal Health (OIE) [3].
The genome of the CSFV encodes only one open reading frame, which can be further hydrolyzed into four structural proteins (C, Erns, E1, and E2) and seven non-structural proteins (P7, Npro, NS2, NS3, NS4A, SN4B, NS5A, and NS5B) following co- and post-translational processing [4]. Of the genome of the CSFV, the E2 gene displaying high genetic diversity is considered a standardized molecular marker to investigate the genetic characteristics of the CSFV [5,6].
In recent years, CSFV strains circulating globally have been divided into three genotypes (1-3) and eleven subgenotypes (1.1-1.4, 2.1-2.3, and 3.1-3.4) [7]. The subgenotype 2.1 strains are the main causative agents responsible for outbreaks of CSF in pig populations [8], and they display higher genetic variation (2.1a-2.1j) than the other subgenotypes [9]. More importantly, the prevalence of subgenotype 2.1d strains has been monitored in many C-strain-vaccinated pig farms. Further investigations showed that the C-strain vaccine does not fully protect against the 2.1d strain [4].
CSF is widespread in different regions of the world, and outbreaks have been observed in Japan recently [10,11]. In China, the CSF epidemic remains, despite the wide application of lapinized-attenuated vaccine (C-strain) in pig populations [4,12]. To provide an update on the epidemic status of CSFV in Yunnan province of China, 326 tissue specimens from suspected CSF pigs were collected from this region from 2015 to 2021 to investigate this issue. The genetic characteristics of the E2 sequences from the new CSFV strains obtained were analyzed.
MATERIALS AND METHODS
Necropsy procedures
All pigs were sacrificed and dissected in strict accordance with the relative standards [13]. Briefly, the pigs were stunned by electric shock and bled to death. The necropsy steps can be summarized as follows: observing the appearance of the corpse; opening the abdominal cavity and removing and examining the abdominal organs; opening the chest cavity and examining the chest organs. These procedures were performed by a veterinarian at different farms.
Cells and reagents
Porcine testis cells (ST) were cultured in Eagle’s Minimum Essential Medium (MEM) supplemented with 5% newborn bovine serum (NBCS) free of BVDV-specific antibodies, streptomycin (100 μg/mL), and penicillin (100 IU/mL) at 37°C in a humidified 5% CO2 incubator. The virulent CSFV Shimen, rescued C-strains, and monoclonal antibody (mAb) against the E2 protein of CSFV were kept in the authors’ laboratory.
Collection and pre-treatment of specimens
From March 2015 to April 2021, 326 porcine tissue (tonsils and lymph nodes) samples from 73 pig farms with suspected CSFV infections were collected from different regions of Yunnan province, China (Table 1). The specimen homogenates in sterile phosphate-buffered saline underwent three freeze and thaw cycles. The supernatants were collected after being centrifuged with 10,000×g for 10 min at 4°C and stored at −80°C.
Table 1. Detection of the CSFV of the collected samples from different periods in Yunnan province.
| Year | Total samples | CSFV-positive samples | CSFV-positive rate | Viral isolation |
|---|---|---|---|---|
| 2015 | 47 | 1 | 2.13% | - |
| 2016 | 63 | 2 | 3.17% | 1 |
| 2017 | 71 | 1 | 1.41% | 1 |
| 2018 | 57 | 4 | 7.01% | 1 |
| 2019 | 36 | 1 | 2.78% | - |
| 2020 | 22 | 1 | 4.55% | 1 |
| 2021 | 30 | 1 | 3.33% | - |
| Total | 326 | 11 | 3.27% | 4 |
CSFV, classical swine fever virus.
Detection of CSFV in clinical specimens
The viral RNA genome from the supernatants was extracted using commercial kits (TaKaRa, Japan) according to the manufacturer’s protocols. The reverse transcription-polymerase chain reaction (RT-PCR) assay was conducted to detect the presence of CSFV, targeting the NS5B gene using one pair of specific primers [14]. A one-step RT-PCR reaction was performed in the following steps: 50°C for 30 min, 94°C for two minutes; 35 cycles of 94°C for 30 sec, 56°C for 30 sec, 72°C for 40 sec; followed by 7 min at 72°C. An expected DNA band of 449 bp from the PCR products in 1% agarose gel electrophoresis was observed, indicating that the sample was CSFV positive.
Virus isolation and identification of the presence of CSFV in the ST cells
The supernatants of eleven representative CSFV-positive samples were filtered through 0.22-μm filters (Merck Millipore, Cork, Ireland), inoculated on monolayer ST cells, and cultured for 72 h. Finally, the presence of infectious viruses was confirmed by RT-PCR and indirect immunofluorescence assay (IFA).
The CSFV-positive cells were proliferated further with continuous passages. At the tenth passage, the viral titers were determined by IFA using a monoclonal antibody (mAb) against the E2 protein of CSFV, as described previously [15]. In this research, FITC labeled E2 protein was purchased from MEDIAN Diagnostics Company in Korea. FITC labeled goat anti-mouse Ig G (secondary antibody) was purchased from Beijing Boaolong Immunology Technology Co., LTD. The FITC labeled sheep anti-pig secondary antibody was purchased from SBA Company.
Sequence and analysis of the partial E2 genes
RT-PCR assays were performed to amplify the partial E2 genes of eleven CSFV strains obtained in this study with the primers for E2-F: 5-GTAAATATGTGTGTGTTAGACCAGA-3′ and E2-R: 5-GTGTGGGTAATTGAGTTCCCTATCA-3′. After amplification, the purified PCR products were cloned into the pMD19-T vector (TakaRa, Japan). The positive plasmids containing the targeted sequence were confirmed by enzyme digestion identification and sent for sequencing. The nucleotide sequences of these novel CSFV strains were deposited in the GenBank database (Table 2).
Table 2. Detailed information on the eleven CSFV isolates collected in this study.
| Herd | Isolate | Place | Time | Pig group | Immunization | GenBank accession no. |
|---|---|---|---|---|---|---|
| 1 | YNWS-2021 | Wenshan, Yunnan | 2021.3 | Nursery pig | Yes | OK169300 |
| 2 | YNDL-2020* | DaLi, Yunnan | 2020.11 | Nursery pigs | Yes | OK169301 |
| 3 | YNKM-2019 | Kunming, Yunnan | 2019.9 | Weaned piglet | Yes | OK169302 |
| 4 | YNLP-2018 | Luoping, Yunnan | 2018.7 | Nursery pigs | Yes | MW392291 |
| 5 | YNMZ-2018* | Mengzi, Yunnan | 2018.6 | Weaned piglet | Yes | MW392292 |
| 6 | YNHP-2018 | Huaping, Yunnan | 2018.7 | Fattening pig | Yes | MW392289 |
| 7 | YNDL-2018 | Dali, Yunnan | 2018.3 | Nursery pigs | Yes | MW392288 |
| 8 | YNYS-2017* | Yongsheng, Yunnan | 2017.6 | Weaned piglet | Yes | MW392294 |
| 9 | YNKD-2016* | Kedu, Yunnan | 2016.8 | Nursery pigs | Yes | MW392290 |
| 10 | YNQJ-2016 | Qujing, Yunnan | 2016.5 | Weaned piglet | Yes | OK169299 |
| 11 | YNSM-2015 | Songming, Yunnan | 2015.8 | Weaned piglet | Yes | MW392293 |
*Represent the strain we isolated successfully.
The E2 nucleotide sequences of the eleven CSFV isolates obtained, and their corresponding reference strains from other regions/countries in the GenBank database were aligned using DNA Star version 7.0 software to analyze the genetic characteristics. A phylogenetic tree based on the E2 nucleotide sequences was constructed using the neighbor-joining (NJ) method with 1000 bootstrapping in MEGA 7.0 software.
Ethics approval and consent to participate
All experiments related to animals in this research have been approved by Yunnan Tropical and Subtropical Animal Virus Diseases Laboratory, Yunnan Animal Science & Veterinary Institute, Yunnan, China.
RESULTS
Pathological changes
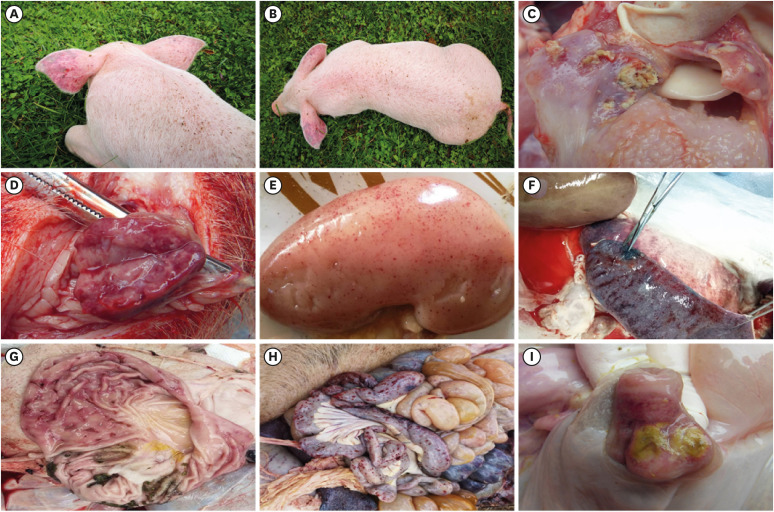
In this study, obvious pathological lesions of CSFV infection were observed from a necropsy, such as an ecchymosis on the skin (Fig. 1A and B), tonsil ulcers (Fig. 1C), petechiae of the lymph nodes (Fig. 1D), petechiated hemorrhage on the kidneys (Fig. 1E), infarction of the spleen (Fig. 1F), necrosis of the stomach and intestine (Fig. 1G and H) and button-shaped ulcers in the ileocecal valves (Fig. 1I).
Fig. 1. Typical clinical and pathological signs were observed in CSFV-infected pigs, such as bleeding points on the skin (A, B), tonsil ulcers (C), welling and hemorrhage of lymph nodes (D), hemorrhagic spots on the kidneys (E), infarction of spleen (F), necrosis of intestine (G, H) and button-shaped ulcers in the ileocecal valves (I).
CSFV, classical swine fever virus.
CSFV detection
PR-PCR was performed to confirm the infection of the CSFV. Table 1 lists the detailed detection results from 2015 to 2021. The total number of samples was 326; 11 were CSFA-positive, accounting for 3.27%. Importantly, four strains of CSFV were isolated successfully. Fig. 2 was the agarose gel electrophoresis analysis of PCR products of partial NS5B gene sequence of CSFV strains, and positive bands with the size of 449 bp were observed.
Fig. 2. Agarose gel electrophoresis analysis of PCR products of partial NS5B gene sequence of CSFV strains. “+” and “–” were regarded as positive and negative controls during PCR amplification, respectively. 1–4 represents the samples from CSFV-infected pigs.
Phylogenetic tree based on partial E2
The genetic characteristics of CSFV strains in Yunnan province were examined by amplifying and sequencing the partial E2 gene sequences of eleven novel CSFV strains in this study, which were submitted to the GenBank database. Table 2 lists the detailed relative information of the eleven CSFV isolates collected. In addition, the neighbor-joining phylogenetic tree based on the E2 gene nucleotide sequences was constructed. As shown in Fig. 3, only one CSFV strain (YN-DL/2020) belonged to subgenotype 2.1d. The remaining ten (YNWS-2021, YNKM-2019, YNLP-2018, YNMZ-2018, YNHP-2018, YNDL-2018, YNYS-2017, YNKD-2016, YNQJ-2016, and YNSM-2015) prevalent in Yunnan province, obtained in this study, were clustered into subgenotype 2.1c.
Fig. 3. Phylogenetic tree based on the partial E2 gene sequence of the CSFV strains obtained in this study and available in the GenBank database generated by the neighbor-joining method in MEGA 7.0 software. The black and red diamonds represented CSFV strains obtained here and vaccine strains, respectively.
*Represents the strain isolated successfully.
Proliferation characteristics of the CSFV isolated
The proliferation characteristics of these CSFV strains from Yunnan province were investigated by inoculating the monolayer ST cells with the supernatants of the CSFV-positive specimens. Finally, four CSFV strains (YNDL-2020, YNKD-2016, YNMZ-2018, and YNYS-2017) were isolated successfully. The unique fluorescence signal of the CSFV was visualized using the mouse monoclonal antibodies against the E2 protein of CSFV in an IFA assay (Fig. 4). In addition, the cell-adapted viruses after 15 passages were obtained with infectious titers of 106.75, 106.375, 107.875, and 105.75 TCID50/mL, respectively.
Fig. 4. Indirect immunofluorescent assay for the detection of the CSFV in ST cells using anti-CSFV E2 primary antibodies. Scale bar = 100 μm.
CSFV, classical swine fever virus.
DISCUSSION
A CSFV infection causes severe clinical symptoms in pigs, including hyperpyrexia, vomiting, constipation or diarrhea, and cyanosis or bleeding points on the skin. Indeed, noticeable pathological lesions (Fig. 1) were observed in the necropsy. In this study, only eleven samples were confirmed to be CSFV-positive by RT-PCR (Fig. 2, Table 1), yielding an average prevalence of 3.37% (11/326), which is lower than those in Shandong province (11.1%, 188/4,866) [16], and other regions of China, including Henan, Hebei, Heilongjiang, and Liaoning provinces (6.0%, 21/350) [17]. Therefore, these results suggest that despite the low epidemic tendency of CFSV in Yunnan province, this pathogen is widespread in China. This phenomenon can be explained. First, different provinces have different epidemic prevention and control policies; the comparative lower positive rate in Yunnan province might be related to the better prevention rules and vaccination. More importantly, Yunnan is a mountainous province covered by extensive forest areas providing an excellent natural barrier in Yunnan province, which is beneficial for the epidemic prevention. Many farms in Yunnan province are located at some places with poor transportation systems and the farms are sparsely distributed.
The CSFV strains prevalent in China mainly belong to genotype 2, which could be divided further into three subgenotypes, namely 2.1, 2.2, and 2.3. Subgenotypes 2.2 and 2.3 are less prevalent in China than subgenotype 2.1 [2], which can be divided further into 2.1a, 2.1b, 2.1c, and 2.1d. In particular, the prevalence of subgenotypes 2.1 strains in different regions of China presents high diversity [4,6,9]. For example, subgenotype 2.1d of the CSFV is mainly dominant in the Shandong province of China [18,19,20], while CSFV strains circulating in Guangdong province of China belong to subgenotype 2.1c [6]. This study examined the genetic characteristics of the CSFV strains. There were two subgenotypes (2.1c and 2.1d) of CSFV prevalent in Yunnan province, China, with subgenotype 2.1c being predominant (Fig. 3, Table 2), which is similar to Guangdong province.
The lengths of the amplified E2 gene sequences of the CSFV strains obtained in this study were 1343 bp, encoding 435 amino acids. Further genetic analyses showed that the eleven strains displayed identities of 89.0–100.0% and 95.0–100.0% at the nt and aa levels, respectively. In addition, the E2 nucleotide sequences of eleven CSFV strains obtained in this study were compared with six reference strains, including Shimen (1.1), 96TD (2.1a), HNLY (2.1c), SDMZ2 (2.1d), LAL-290E (2.2), and Novaka (2.3). The results showed that the eleven new strains shared 90.0–98.7% and 89.6–99.0% identity with subgenotypes 2.1c and 2.1d, respectively. Moreover, they exhibited 85.1–93.0% sequence identity with other subgenotypes 2 and 3 isolates, including 2.1a, 2.1b, 2.2, and 2.3. On the other hand, these new strains displayed lower sequence identity with subgenotype 1.1 (Shimen), 82.2–83.0%, suggesting that all CSFV strains had different genetic relationships with vaccine strains.
The proliferation characteristics of these CSFV strains from Yunnan province were investigated. Four CSFV strains were isolated and were specially recognized by the IFA assay (Fig. 4), demonstrating that the CSFV collected can proliferate well in the monolayer ST cells. Intriguingly, the viral titer of the YNMZ-2018 strain was ten times higher than these of the other strains. SA sequence comparison showed that the YNMZ-2018 strain contained a series of unique amino acids substitution in the E2 protein, such as the T to I change at the position of 49. Moreover, K71R, D97G, K174N, N192S, and T197S were also identified in this novel strain. Nevertheless, whether these amino acid substitutions would influence the proliferation characteristics of CSFV strain in vitro needs further investigation.
Historically, owing to the extensive application of effective vaccines against CSF in Chinese pig populations, this infectious disease appears to have been eradicated in some regions of China. Moreover, prevention has been neglected because of the occurrence or prevalence of other infectious diseases, such as porcine reproductive and respiratory syndrome, porcine circovirus disease, and porcine epidemic diarrhea. In the current research, a low prevalence of CSFV (3.37%, 11/218) in pigs was confirmed in Yunnan province of China, even though these pigs had been vaccinated against CSF. This suggests that this infectious disease should not be neglected, and corresponding measures for the prevention of CSF need to be conducted. Further genetic analyses showed that all CSFV strains obtained here belonged to subgenotypes 2.1c and 2.1d, with 2.1c being the dominant subgenotypes in China’s Yunnan province.
Footnotes
Funding: This research was funded by National Key Research and Development Program of China (No. 2017YFD0501800) and Major Specialized Projects of Yunnan Science and Technology (No. 202102AE090007).
Conflict of Interest: The authors declare no conflicts of interest.
- Conceptualization: Yang S, Zhang Y.
- Data curation: Chai J.
- Formal analysis: Song C.
- Funding acquisition: Shu X.
- Investigation: Yao J.
- Methodology: Su L.
- Project administration: Wang Q.
- Resources: Gao L.
- Software: Xie J.
- Supervision: Zhang Y.
- Validation: He Y.
- Visualization: Yang S.
- Writing - original draft: Yao J, Su L.
- Writing - review & editing: Yao J, Yang S, Zhang Y.
References
- 1.Becher P, Avalos Ramirez R, Orlich M, Cedillo Rosales S, König M, Schweizer M, et al. Genetic and antigenic characterization of novel pestivirus genotypes: implications for classification. Virology. 2003;311(1):96–104. doi: 10.1016/s0042-6822(03)00192-2. [DOI] [PubMed] [Google Scholar]
- 2.Zhou B. Classical swine fever in China-an update minireview. Front Vet Sci. 2019;6:187. doi: 10.3389/fvets.2019.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji W, Guo Z, Ding NZ, He CQ. Studying classical swine fever virus: making the best of a bad virus. Virus Res. 2015;197:35–47. doi: 10.1016/j.virusres.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Luo Y, Ji S, Liu Y, Lei JL, Xia SL, Wang Y, et al. Isolation and characterization of a moderately virulent classical swine fever virus emerging in China. Transbound Emerg Dis. 2017;64(6):1848–1857. doi: 10.1111/tbed.12581. [DOI] [PubMed] [Google Scholar]
- 5.Luo TR, Liao SH, Wu XS, Feng L, Yuan ZX, Li H, et al. Phylogenetic analysis of the E2 gene of classical swine fever virus from the Guangxi Province of southern China. Virus Genes. 2011;42(3):347–354. doi: 10.1007/s11262-011-0578-8. [DOI] [PubMed] [Google Scholar]
- 6.Xing C, Lu Z, Jiang J, Huang L, Xu J, He D, et al. Sub-subgenotype 2.1c isolates of classical swine fever virus are dominant in Guangdong province of China, 2018. Infect Genet Evol. 2019;68:212–217. doi: 10.1016/j.meegid.2018.12.029. [DOI] [PubMed] [Google Scholar]
- 7.An DJ, Lim SI, Choe S, Kim KS, Cha RM, Cho IS, et al. Evolutionary dynamics of classical swine fever virus in South Korea: 1987–2017. Vet Microbiol. 2018;225:79–88. doi: 10.1016/j.vetmic.2018.09.020. [DOI] [PubMed] [Google Scholar]
- 8.Luo Y, Li S, Sun Y, Qiu HJ. Classical swine fever in China: a minireview. Vet Microbiol. 2014;172(1-2):1–6. doi: 10.1016/j.vetmic.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Gong W, Wu J, Lu Z, Zhang L, Qin S, Chen F, et al. Genetic diversity of subgenotype 2.1 isolates of classical swine fever virus. Infect Genet Evol. 2016;41:218–226. doi: 10.1016/j.meegid.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Brown VR, Bevins SN. A review of classical swine fever virus and routes of introduction into the United States and the potential for virus establishment. Front Vet Sci. 2018;5:31. doi: 10.3389/fvets.2018.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishi T, Kameyama KI, Kato T, Fukai K. Genome sequence of a classical swine fever virus of subgenotype 2.1, isolated from a pig in Japan in 2018. Microbiol Resour Announc. 2019;8(3):e01362-18. doi: 10.1128/MRA.01362-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leng C, Zhang H, Kan Y, Yao L, Li M, Zhai H, et al. Characterisation of newly emerged isolates of classical swine fever virus in China, 2014–2015. J Vet Res (Pulawy) 2017;61(1):1–9. doi: 10.1515/jvetres-2017-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrews JJ, Van Alstine WG, Schwartz KJ. A basic approach to food animal necropsy. Vet Clin North Am Food Anim Pract. 1986;2(1):1–29. doi: 10.1016/s0749-0720(15)31284-6. [DOI] [PubMed] [Google Scholar]
- 14.Paton DJ, McGoldrick A, Greiser-Wilke I, Parchariyanon S, Song JY, Liou PP, et al. Genetic typing of classical swine fever virus. Vet Microbiol. 2000;73(2-3):137–157. doi: 10.1016/s0378-1135(00)00141-3. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Xu J, Sun Y, Li S, Li N, Yang S, et al. Identification of a linear epitope on the capsid protein of classical swine fever virus. Virus Res. 2011;156(1-2):134–140. doi: 10.1016/j.virusres.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S, Zeng H, Yu J, Chen Z, Zhang Y, Wang X, et al. Epidemiological investigation and analysis of PRRSV, PRV and CSFV diseases in Shandong province from 2015 to 2017. China J Vet Sci. 2019;39:1064–1069. [Google Scholar]
- 17.Zhang H, Leng C, Tian Z, Liu C, Chen J, Bai Y, et al. Complete genomic characteristics and pathogenic analysis of the newly emerged classical swine fever virus in China. BMC Vet Res. 2018;14(1):204. doi: 10.1186/s12917-018-1504-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng L, Zhang H, Peng J, Liu C, Wang Q, Li Z, et al. Characterization and complete genome sequence of two classical swine fever viruses belonging to 2.1d new subgenotype. China J Prev Vet Med. 2016;38(1):1–5. [Google Scholar]
- 19.Li Y, Ma Z, Liu Z, Wang H, Meng F, Cao L, et al. Comprehensive diagnosis and analysis of virus E2 gene in typical cases of classical swine fever in a swine farm. China J Cell Biol. 2019;41(3):461–467. [Google Scholar]
- 20.Wang H, Li C, Meng F, Liu Z, Ma Z, Jiao Q, et al. Comprehensive diagnosis of swine fever cases and phylogenetic analysis on the isolated strain. China Anim Health Inspection. 2019;36(1):65–79. [Google Scholar]