Abstract
Background
Cervical cancer is a preventable disease, but it is a major public health problem despite having a good prognosis when diagnosed early. Although the Pap smear has led to huge drops in rates of cervical cancer and death from the disease, it has some limitations, making new approaches necessary for early diagnosis and biomarkers discovery. MiRNAs have been considered a new class of non-invasive biomarkers and may have great clinical value for screening early-stage cervical intraepithelial neoplasia. Well-designed studies have emerged as a necessary strategy for the identification of miRNAs that could be used safely and reliably for a differential diagnosis. This review aims to provide an up-to-date perspective on the assessment of circulating miRNA expression from precursor lesions to cervical cancer, identifying circulating miRNAs or specific miRNA signatures that can be used as potential biomarkers of different stages of cervical carcinogenesis.
Methods
A systematic review was performed and searches were conducted in the PubMed, LILACS, and Scopus electronic databases.
Results
Most studies involved Chinese ethnic women and searched for circulating miRNAs in serum samples. Thirty three microRNAs were evaluated in the eligible studies and 17 (miR-196a, miR-16-2, miR-497, miR-1290, miR-425-5p, hsa-miR- 92a, miR-1266, miR-9, miR-192, miR-205, miR-21, miR-152, miR-15b, miR-34a, miR-218, miR-199a-5p and miR-155-5p) showed up-regulation in women with precursor lesion and cervical cancer and 16 microRNAs showed decreased expression in these same groups of women compared to healthy controls (miR-195, miR-2861, miR-145, miR-214, miR-34a, miR-200a, let-7d-3p, miR-30d-5p, miR-638, miR-203a-3p, miR-1914-5p, miR-521, miR-125b, miR-370, miR-218 and miR-100).
Conclusion
Therefore, defining promising circulating miRNAs or specific miRNA signatures of biological fluid samples can be useful for the screening, diagnosis, prognosis and clinical monitoring of women undergoing cervical carcinogenesis, but greater standardization of studies seems to be necessary for greater consolidation of information.
Keywords: Circulating microRNA, microRNAs, Uterine cervical neoplasms, Cervical intraepithelial neoplasia, Biomarkers
Introduction
Cervical cancer (CC) is one of the most important public health problems worldwide, due to its high rates of morbidity and mortality [1], especially in less developed countries. Every year, approximately 570,000 women are diagnosed with CC in the world with an estimated 311,000 deaths, being considered the fourth most common type of cancer among women in the world, affecting relatively young women, between 30 to 50 years old [2]. Its development is often slow and it may take five to 15 years for its total progression from a precursor lesion, also called cervical intraepithelial neoplasia (CIN). When associated cellular alterations are not identified and/or treated early, they can develop into a localized tumor, called carcinoma in situ. This, in turn, can progress to invasive carcinoma, the most severe form, which can also become metastatic [3]. The staging of CC is determined by its dissemination and, according to the International Federation of Gynecology and Obstetrics (FIGO), as it increases, the estimated five-year survival for women affected by this neoplasm decreases.
Human Papillomavirus (HPV) infection, present in approximately 90% of cases [3], is considered the main risk factor for the development of CC. Although necessary, HPV infection alone is not enough to develop a cancerous disease and behavioral factors, both host and the virus-related, are also correlated [4]. According to their oncogenic potential, HPV types are classified as high or low oncogenic risk viruses [5] whose risk is directly related to the behavior of the viral genome in the host cell nucleus [6] [7].
Although Pap smear is the most used preventive strategy, this test also has its limitations [8], therefore, a better understanding of molecular mechanisms and genetic alterations associated with the pathogenesis of CC would be helpful to control this disease. In this context, non-invasive, specific, and sensitive biomarkers for early detection of CC are needed to reduce its morbidity and mortality worldwide [9].
Circulating microRNAs are considered a new class of these biomarkers due to their stability and presence in almost all body fluids [8]. They are also described as having great clinical value for screening early-stage different types of cancer, including CC. Well-designed studies, which aim to identify new biomarkers related to cervical carcinogenesis, emerged as a necessary strategy for the identification of miRNAs that could be used safely and reliably for a differential diagnosis. Different studies evaluating the expression of circulating microRNAs in CC show considerable divergence in the methodology used and also in the results obtained. Taking into account the great diversity and amount of information, as well as the divergences observed in these studies, the development of a systematic review on the gene expression of circulating microRNAs in cervical carcinogenesis would be important for the systematization and compilation of information available in the literature. Therefore, this review aimed to identify circulating miRNAs or specific miRNA signatures that could be used as potential biomarkers of different stages of cervical carcinogenesis, as well as provide an updated perspective on the evaluation of circulating miRNA expression from CC precursor lesions.
Material and methods
As a study designed as a systematic literature review, the recommendations of the Preferred Reporting Items for Systematic Reviews and Meta-Analysis - PRISMA checklist were adopted [10]. The protocol for this review was submitted for registration in the International Prospective Register of Systematic Reviews (PROSPERO), under registration CRD42021259736. Permission from the ethics committee or institutional review board is not required for conducting a systematic review and meta-analysis.
Study inclusion criteria
Inclusion criteria were: original studies; articles that evaluated the expression of one or more circulating microRNAs (serum, plasma or urine); published in English, from January 2011 to April 2022.
Study exclusion criteria
The articles were excluded for the reasons described in Fig. 1: articles that evaluated the expression of microRNAs only in tissue samples and cervical cells; studies unrelated to microRNA expression, cervical cancer or diagnosis; all those that did not assess the expression of circulating microRNAs; studies not performed in humans; research not carried out in three different sample groups (healthy control, cervical intraepithelial neoplasia and cervical cancer); studies that have evaluated the expression of microRNAs using qualitative or non-quantitative methodologies; conference abstracts; comments; literature reviews; letters to the editor; editorials; meta-analyses; clinical case reports; theses, dissertations, book chapters; studies with duplication in the databases and retracted articles.
Fig. 1.
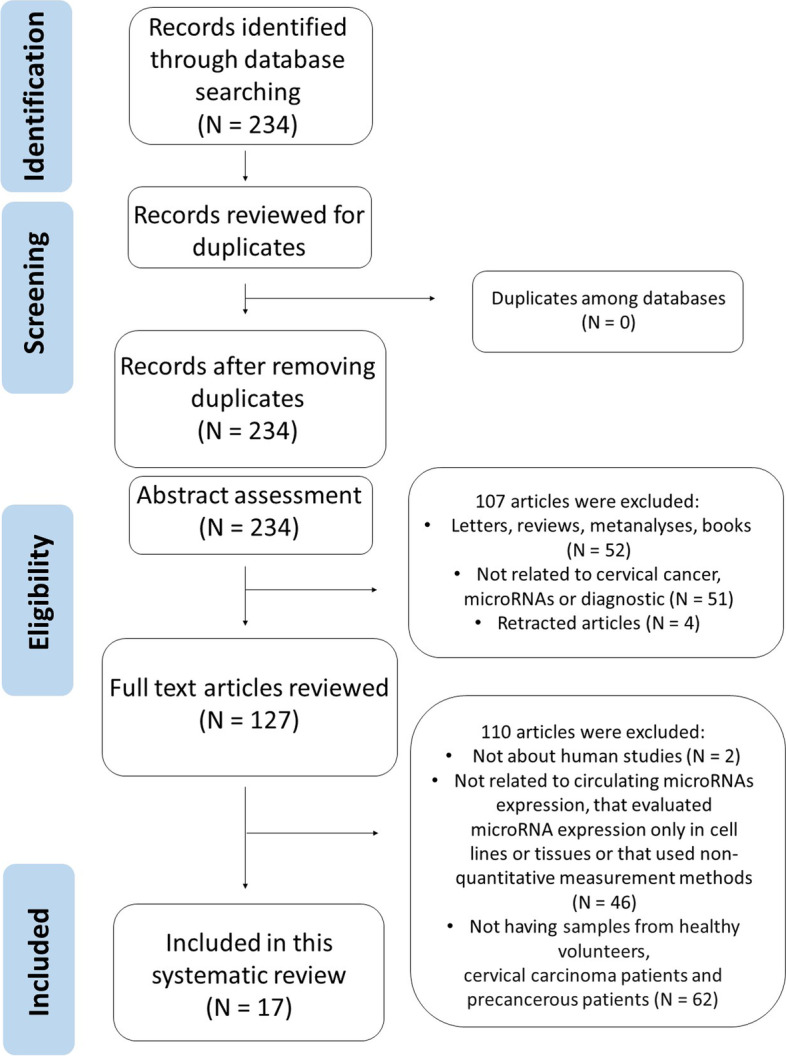
PRISMA flow chart
Searching and screening of literature
Pubmed (through pubmed.gov), LILACS (Latin American and Caribbean Health Sciences Literature – through https://lilacs.bvsalud.org/), and Scopus (through scopus.periodicos.capes.gov.br) databases were searched until April 2022. The search descriptors (terms) were as follows: cervical cancer OR uterine cervical neoplasms OR cervical intraepithelial neoplasia OR uterine cervical dysplasia AND circulating microRNAs OR plasma microRNAs OR serum microRNAs OR blood microRNAs OR cell-free microRNAs OR exosome microRNAs OR extracellular vesicles microRNAs.
Data extraction
Eligible studies were independently and separately reviewed by two reviewers (N.P.G.N. and T.B.G.). Disagreements were resolved by discussions and mutual consensus. The necessary information and data related to the study design were extracted from each of the included studies as follows: authorship, year of publication, country of origin, type of study, subject’s ethnicity, number of sample groups, control group, CIN group, and cancer group data, in addition, miRNAs expression test methods and other variables described in Tables 1, 2 and 3. All information was tabulated by the reviewers in a spreadsheet (Microsoft Excel). Disagreements were resolved through discussion until an agreement was reached among reviewers.
Table 1.
Characteristics of the samples in the included studies
| CONTROL GROUP (C) | CIN GROUP (CIN) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author | Year | Country | N | Mean Age (y) |
HPV tested |
HPV detection method | HPV infection |
HPV type |
N | Mean Age (y) |
HPV tested |
HPV detection method |
| Liu; Xin; Ma [11] | 2015 | China | 50 | N/I | No | N/A | N/A | N/A |
T: 86 CIN I: 19 CIN II: 25 CIN III: 42 |
N/I | N/I | N/I |
| Zhang et al [8] | 2015 | China | 193 | N/I | N/I | N/I | N/I | N/I | 186 | N/I | N/I | N/I |
| Nagamitsu et al [12] | 2016 | Japan | 31 | 39 | N/I | N/I | N/I | N/I |
T: 55 CIN I: 15 CIN II: 16 CIN III: 24 |
CIN I: 34 CIN II: 35 CIN III: 34 |
N/I | N/I |
| Sun et al [13] | 2016 | China | 32 | N/I | N/I | N/I | N/I | N/I | 13 | N/I | N/I | N/I |
| Kong et al [14] | 2017 | China | 40 | 39 | Yes | Liquid-Based Cytology | N/I | N/I |
T: 68 CIN I: 21 CIN II: 23 CIN III: 24 |
39 | Yes | Liquid-Based Cytology |
| Wei; Wen-Ming; Jun-Bo [15] | 2017 | China | 120 | N/I | N/I | N/I | N/I | N/I | 120 | N/I | N/I | N/I |
| Wang et al [16] | 2018 | China | 50 | N/I | N/I | N/I | N/I | N/I |
HSIL: 50 LSIL: 50 |
N/I | N/I | N/I |
| Farzanehpour et al [17] | 2019 | Iran | 36 | 36 | Yes | PCR | Yes |
16 66 68 |
18 | 47 | Yes | PCR |
| Wang et al [18] | 2019 | China | 42 | 40.94 | Yes | N/I | No | N/A |
T: 144 CIN I: 19 CIN II: 54 CIN III: 71 |
CIN I: 41.93 CIN II: 42.37 CIN III: 40.77 |
N/I | N/I |
| Yang; Zhang [19] | 2019 | China | 50 | 43 | Yes | N/I | No | N/A |
T: 50 CIN I: 15 CIN II: 15 CIN III: 20 |
CIN I: 38.0 CIN II: 35.3 CIN III: 36.2 |
Yes | N/I |
| Zheng et al [20] | 2019 | China | 50 | 50 | N/I | N/I | N/I | N/I |
T: 90 CIN I: 34 CIN II-III: 56 |
50 | N/I | N/I |
| Zheng et al [21] | 2020 | China | 41 | 42.71 | N/I | N/I | N/I | N/I | 40 | 43.88 | N/I | N/I |
| Qiu et al [22] | 2020 | China | 90 |
> = 50: 41 < 50: 49 |
N/I | N/I | N/I | N/I | 45 |
> = 50: 20 < 50: 25 |
N/I | N/I |
| Aftab et al [23] | 2021 | Índia | 50 | N/I | Yes | PCR | Yes | 16: 2 |
LSIL: 17 HSIL: 33 |
N/I | Yes | PCR |
| Hoelzle et al [24] | 2021 | Brasil | 12 | 27,5 | Yes | PCR | No | N/A |
LSIL: 14 HSIL: 11 |
LSIL: 32 HSIL: 39 |
Yes | PCR |
| Ocadiz-Delgado et al [25] | 2021 | México | 23 | 38 | Yes | PCR | No | N/A | 17 | 34 | Yes | PCR |
| Yamanaka et al [26] | 2021 | Japão | 34 | 42 | N/I | N/I | N/I | N/I | 64 |
CIN I: 34 CIN II: 35 CIN III: 34 |
N/I | N/I |
| CIN GROUP (CIN) | CERVICAL CANCER GROUP (CC) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Author | HPV infection |
HPV type |
N | Mean Age (y) |
Tumor stage | Metast | HPV tested |
HPV detection method |
HPV infection |
HPV type |
Specim. |
| Liu; Xin; Ma [11] | N/I | N/I | 105 |
<= 50: 64 > 50: 41 |
FIGO I-II: 74 III-IV: 31 |
Yes: 37 No: 68 |
Yes | N/I |
Yes: 92 No: 13 |
N/I | Serum |
| Zhang et al [8] | N/I | N/I | 184 | N/I | N/I | N/I | N/I | N/I | N/I | N/I | Serum |
| Nagamitsu et al [12] | N/I | N/I | 45 | 49 |
Ia: 7 Ib: 16 IIa: 7 IIb: 10 III: 3 IV: 2 |
N/I | N/I | N/I | N/I | N/I | Serum |
| Sun et al [13] | N/I | N/I | 40 |
<= 60: 22 > 60: 18 |
Stage I: 12 Stage II, III and IV: 28 |
Yes: 29 No: 11 |
Yes | N/I |
Yes: 28 No: 12 |
N/I | Serum |
| Kong et al [14] | Yes (all) | N/I | 18 | 39 |
FIGO Ia - IIa |
N/I | Yes |
Liquid- Based Cytology |
Yes (all) | N/I | Serum |
| Wei; Wen-Ming; Jun-Bo [15] | N/I | N/I | 120 |
< 50: 59 > = 50: 61 |
FIGO I-II: 77 III: 43 |
Yes: 47 No: 73 |
Yes | N/I |
Yes: 81 No: 39 |
N/I | Plasma |
| Wang et al [16] | N/I | N/I | 50 | N/I | N/I | N/I | N/I | N/I | N/I | N/I | Serum |
| Farzanehpour et al [17] | Yes |
16 53 |
18 | 61 | N/I | N/I | Yes | PCR | Yes | 16 | Serum |
| Wang et al [18] | N/I | N/I | 15 | 49.73 | N/I | N/I | N/I | N/I | N/I | N/I | Plasma |
| Yang; Zhang [19] | No | N/A | 50 | 48 |
Stage I: 15 Stage II: 15 Stages III – IV: 20 |
N/I | Yes | N/I | No | N/A | Total Blood |
| Zheng et al [20] | N/I | N/I | 63 | 50 | N/I | N/I | N/I | N/I | N/I | N/I | Exosomes |
| Zheng et al [21] | N/I | N/I | 40 | 50.39 |
FIGO I B1– I B2: 26 II A1– II B2: 12 N/I: 2 |
Yes: 9 No: 25 N/I: 6 |
N/I | N/I | N/I | N/I | Serum |
| Qiu et al [22] | N/I | N/I | 112 |
> = 50: 53 < 50: 59 |
I: 90 IIA:22 |
Yes: 20 No: 92 |
N/I | N/I | N/I | N/I | Serum |
| Aftab et al [23] | Yes |
16 LSIL: 4 HSIL: 24 |
50 | N/I |
I: 10 II: 17 III: 18 IV: 5 |
N/I | Yes | PCR | Yes |
HG 16: 37 CS 16: 40 18: 2 |
Urine |
| Hoelzle et al [24] | Yes |
LSIL 33: 2 Outros: 3 HSIL 16: 3 31: 1 Outros: 3 |
13 | 47 | N/I | N/I | Yes | PCR | Yes |
16: 4 18: 2 33: 2 Outros: 4 |
Plasma |
| Ocadiz-Delgado et al [25] | Yes | 16 | 23 | 50 | N/I | N/I | Yes | PCR | Yes | 16 | Serum |
| Yamanaka et al [26] | N/I | N/I | 46 | 49 |
FIGO IA: 4 IB: 21 IIA: 4 IIB: 10 III: 3 IV: 4 |
Yes: 19 No: 16 N/I: 11 |
N/I | N/I | N/I | N/I | Serum |
HPV Human Papillomavirus, N/I Not Informed, N/A Not appliable, CIN Cervical intraepithelial neoplasia, FIGO International Federation of Gynecology and Obstetrics, HSIL High grade squamous intraepithelial lesion, LSIL Low-grade squamous intraepithelial lesion, PCR Polymerase Chain Reaction, T Total, HG Histopathological grading, CS Clinical stage.
Table 2.
MicroRNA candidate selection methods are found in included articles
| Author | Year | Type of Global MicroRNA Expression profiling | If yes, were samples pooled? | If samples were not pooled, how many samples per group were analyzed in large-scale analysis? | Were candidate microRNAs selected by analysis of public microRNAs datasets? | Were candidate microRNAs selected by literature review? |
|---|---|---|---|---|---|---|
| Liu; Xin; Ma | 2015 | N/A | N/A | N/A | No | Yes |
| Zhang et al | 2015 | MicroRNA PCR panel | No |
C: 5 CC: 4 |
No | No |
| Nagamitsu et al | 2016 | Microarray | No |
C: 5 CC: 5 |
No | No |
| Sun et al | 2016 | N/A | N/A | N/A | No | Yes |
| Kong et al | 2017 | N/A | N/A | N/A | No | Yes |
| Wei; Wen-Ming; Jun-Bo | 2017 | N/A | N/A | N/A | No | Yes |
| Wang et al | 2018 | N/A | N/A | N/A | No | Yes |
| Farzanehpour et al | 2019 | N/A | N/A | N/A | No | Yes |
| Wang et al | 2019 | N/A | N/A | N/A | No | Yes |
| Yang; Zhang | 2019 | N/A | N/A | N/A | No | Yes |
| Zheng et al | 2019 | Sequencing | No |
C: 23 CIN I: 5 CIN II-III: 59 SCC: 21 ACC: 13 |
No | No |
| Zheng et al | 2020 | N/A | N/A | N/A | No | Yes |
| Qiu et al [22] | 2020 | N/A | N/A | N/A | Yes | No |
| Aftab et al [23] | 2021 | N/A | N/A | N/A | Yes | No |
| Hoelzle et al [24] | 2021 | N/A | N/A | N/A | Yes | No |
| Ocadiz-Delgado et al [25] | 2021 | N/A | N/A | N/A | No | Yes |
| Yamanaka et al [26] | 2021 | N/A | N/A | N/A | No | Yes |
N/A (Not applicable), PCR (Polymerase Chain Reaction), C (Control Group), CC (Cervical Cancer Group), CIN (Cervical Intraepithelial Neoplasia group), SCC (Squamous Cell carcinoma group), ACC (Adenocarcinoma group).
Table 3.
Characteristics of the 12 studies included in our review
| Author | Year | Detection Method | Normalize. | Method for expression level calculation | Differentially expressed microRNAs between HPV+ e HPV - samples | Differentially expressed microRNAs between CIN or CC and C | Up or down regulation description only |
AUC | SEN | SPE | Main results description |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Liu; Xin; Ma [11] | 2015 | qPCR | RNU6B | 2−ΔΔCt | MiR-196a expression levels are not correlated to HPV infection | miR-196a | Yes | N/A | N/A | N/A | CC Patients showed a significantly higher level of serum miR-196a than CIN patients p < 0.05) and controls (p < 0.01). CIN patients expressed higher MiR-196a levels than C volunteers (p < 0.05). 67% of CIN III patients showed significantly higher expression of miR-196 than CIN II(32%) and CIN I (21%) patients. CC Patients with higher miR-196a serum levels had a poorer overall survival rate. |
| Zhang et al. [8] | 2015 | qPCR | cel-miR-67 | 2−ΔΔCt | N/I |
miR-497 miR-16-2 miR-195 miR-2861 |
No |
4 miRs: CC vs. C: 0.849 CC vs. NIC: 0.829 NIC vs. C: 0,734 |
4 miRs: CC vs. C: 73,1% CC vs. NIC: 71,4% NIC vs. C: 62,6% |
4 miRs: CC vs. C: 88,4% CC vs. NIC: 67,2% NIC vs. C: 88,9% |
MIR-195 expression level was significantly decreased, while miR-16-2* and miR-497 were significantly increased in CC patients, compared to CIN and control groups. miR-2861 expression levels are significantly decreased in CIN and CC patients compared to the control group. |
| Nagamitsu et al. [12] | 2016 | qPCR | miR-16 | 2−ΔΔCt | N/I | miR-1290 | No | CC vs. C: 0.7957 | CC vs. C: 90.3% | CC vs. C: 62.2% | MiR-1290 expression level was significantly elevated in CC patients compared to CIN and control patients. MiR-1290 increased expression was also related to the CC stage. |
| Sun et al. [13] | 2016 | qPCR | Geometric mean of miR-16 miR-223 and miR-31 | 2−ΔΔCt | N/I | miR-425-5p | Yes | N/A | N/A | N/A | MiR-425-5p expression levels were significantly the higher in CC group compared to CIN (p< 0.0001) or control groups (p< 0.0001). Elevated levels of miR-425-5p were significantly correlated with TNM stage (p = 0.0003) and lymph node metastasis (p = 0.0037). High levels of serum miR-425have expression that has been correlated with poor overall survival. |
| Kong et al. [14] | 2017 | qPCR | Cel-39-3p | 2−ΔΔCt | MiR-92a expression levels are not correlated to HPV infection | hsa-mir-92a | No | 0,83 | 69.6% | 80.4% | Serum miR-92a levels were significantly higher in CIN and CC patients than in healthy subjects (p< 0.01). MiR-92a expression was also significantly higher in patients with advanced-stage CC than in those with early-stage CC. |
|
Wei; Wen-Ming; Jun-Bo [15] |
2017 | qPCR | RNU6B | 2−ΔΔCt | Lower plasmatic miR-145 expression levels significantly associated to HPV infection (p 0,016) | miR-145 | No |
CC vs. C: 0,848 CC vs. CIN: 0,828 |
CC vs. C: 81,7% CC vs. CIN: 91,7% |
CC vs. C: 63,3% CC vs. CIN: 54,2% |
MiR-145 plasma levels were significantly lower in CC patients than in CIN patients and healthy controls (p < 0.01). CC patients who achiea ved complete response to radiotherapy had higher plasma miR-145 levels than incomplete responders (p 0.005). |
| Wang et al. [16] | 2018 | qPCR | U6 | 2−ΔΔCt | N/I | miR-1266 | Yes | N/A | N/A | N/A | Serum miR-126upregulatedulated in CC patients also correlates with metastatic progression. Serum MiR-1266 expression was significantly higher in patients with LSIL, HSIL and CC compared to healthy controls (p< 0.05). CC patients with high miR-1266 expression had lower overall survival rates than patients with low miR-1266 expression. |
| Farzanehpour et al. [17] | 2019 | qPCR | U6 | 2−ΔΔCt | N/I |
miR-9 miR-192 miR-205 |
No |
CC vs. C miR-9: 0,99 miR-192: miR-205: 0,96 CIN vs. C miR-9: 0,90 miR-192: 0,98 miR-205: 0,75 CC vs. CIN miR-9: 0,85 miR-192: 0,82 miR-205: 0,75 |
CC vs. C miR-9: 100% miR-192: 100% miR-205: 88,2% CIN vs. C miR-9: 77,8% miR-192: 83,3% miR-205: 66,7% CC vs. CIN miR-9: 52,9% miR-192: 58,8% miR-205: 35,3% |
CC vs. C miR-9: 94,4% miR-192: 94,4% miR-205: 88,9% CIN vs. C miR-9: 94,4% miR-192: 94,4% miR-205: 88,9% CC vs. CIN miR-9: 94,4% miR-192: 83,3% miR-205: 94,4% |
Increased expression levels of miR-9, miR-192 and miR-205 were observed in the CC group compared to normal samples (p< 0.0001). MiR-9, miR-192, and miR-205 expression levels were significantly increased in the CIN group compared to the control group. MiR-205 expression showed to be increased in the CC group compared to the CIN group (p< 0.0001). |
| Wang et al. [18] | 2019 | qPCR | U6 | 2−ΔΔCt | N/I |
miR-21 miR-214 miR-34a miR-200a |
No |
CIN I- vs. CIN II+ miR-21: 0,613 miR-34a: 0,508 miR-200a: 0,615 miR-214: 0,505 |
N/I | N/I |
MiRNA-21 plasma expression was increased in patients with cervical lesions (including CIN I, CIN II, CI, II,I and CC patients). MiRNA-21,−34a, and -200a expression levels were decreased in patients with severe cervical lesions (CIN I, CIN II, CIN, III and CC). Differences between these miRNAs expression levels were significant when comparing controls and CIN II e III patients(p< 0.05). |
| Yang; Zhang [19] | 2019 | qPCR | U6 | 2−ΔΔCt | N/I | miR-152 | No |
CIN vs. C: 0.831 CC vs. C: 0.935 |
N/I | N/I | MiR-152 expression levels were increased in CIN patients compared to control and higher in CC patients. Furthermore, miR-152 expression level in peripheral blood was higher in patients with high-grade CIN compared to those with low-grade CIN. MiR-152 expression levels also increased with the CC stage and were associated with serum SCC-Ag levels. A miR-152 expression reduction was observed after treatment. |
| Zheng et al. [20] | 2019 | Digital PCR |
miR-128-3p miR-129-5p miR-320 let-7i-5p |
R_score | N/I |
let-7d-3p miR-30d-5p |
No |
let-7d-3p: 0.822 miR-30d-5p: 0.79 Combined: 0.828 |
N/I | N/I | Let-7d-3p and miR-30d-5p expression levels were significantly decreased in CIN II + group when compared to CIN I – group. |
| Zheng et al. [21] | 2020 | qPCR | cel-miR-39 | 2−ΔΔCt | N/I |
miR-638 miR-203a-3p miR-1914-5p miR-521 |
No |
CC vs. CIN miR-638: 0,681 miR-203a-3p: 0,651 miR-1914-5p: 0,717 miR-521: 0,716 CC vs. C miR-638: 0,734 miR-203a-3p: 0,660 miR-1914-5p: 0,626 miR-521: 0,742 |
CC vs. CIN miR-638: 85% miR-203a-3p: 42,5% miR-1914-5p: 82,5% miR-521: 62,5% CC vs. C miR-638: 80% miR-203a-3p: 75% miR-1914-5p: 85% miR-521: 80% |
CC vs. CIN miR-638: 46,15% miR-203a-3p: 82,05% miR-1914-5p: 61,54% miR-521: 76,92% CC vs. C miR-638: 60,98% miR-203a-3p: 53,66% miR-1914-5p: 43,90% miR-521: 65,85% |
Lower expression levels of miR-638, miR-203a-3p, miR-1914-5p and and MiR-521 were observed in the group compared to CIN. Furthermore, miR-638, miR-203a-3,p and miR-521 expression levels were lower in CC patients compared to healthy controls. |
| Qiu et al [22] | 2020 | qPCR | cel-miR-39 | 2−ΔΔCt | N/I |
miR-21 miR-125b miR-370 |
No |
CC vs. C miR-21: 0,783 miR-125b: 0,642 miR-370: 0,822 CC vs. CIN miR-21: 0,689 miR-125b: 0,735 miR-370: 0,821 Combined CC vs. C: 0,912 CC vs. CIN: 0,897 |
N/I | N/I | Higher miR-21 serum expression was observed in CC than in CIN and healthy women. The expression of miR-125b and miR-370 was lower in CC women compared to healthy and CIN patients. Combined expression of the three miRNAs performed well to identify early-stage CC compared to CIN or healthy controls. High levels of miR-21 and low levels of miR-125b and miR-370 were associated with metastasis and recurrences. |
| Aftab et al [23] | 2021 | qPCR | snRNA U6 | 2−ΔΔCt |
Lower miRNA-145-5p expression in samples LSIL/HSIL HPV 16 + and CC HPV 16+ samples. |
miR-21-5p miR-199a-5p miR-155-5p miR-34a-5p miR-145-5p miR-218-5p |
No |
miR-21-5p: 0,97 miR-199a-5p: 0,831 miR-155-5p: 0,719 miR-34a-5p: 0,888 miR-145-5p: 0,575 miR-218-5p: 0,609 Combined: 0,942 miR-21-5p miR-199a-5p miR-155-5p Combined: 0,969 miR-34a-5p miR-145-5p miR-218-5p Combined: 1 miR-21-5p miR-199a-5p miR-155-5p miR-34a-5p miR-145-5p miR-218-5p |
miR-21-5p: 88 miR-199a-5p: 86.7 miR-155-5p: 93.8 miR-34a-5p: 89.1 miR-145-5p: 67.7 miR-218-5p: 83.3 Combined: 100 miR-21-5p miR-199a-5p miR-155-5p Combined: 100 miR-34a-5p miR-145-5p miR-218-5p Combined: 100 miR-21-5p miR-199a-5p miR-155-5p miR-34a-5p miR-145-5p miR-218-5p |
miR-21-5p: 98 miR-199a-5p: 91.7 miR-155-5p: 88.3 miR-34a-5p: 91.7 miR-145-5p: 60 miR-218-5p: 72.7 Combined: 78.9 miR-21-5p miR-199a-5p miR-155-5p Combined: 92.8 miR-34a-5p miR-145-5p miR-218-5p Combined: 100 miR-21-5p miR-199a-5p miR-155-5p miR-34a-5p miR-145-5p miR-218-5p |
Significant up-regulation of miR-21-5p, miR-199a-5p and miR-155-5p and down-regulation of miR-145-5p, miR-34a-5p and miR-218-5p was observed in women with LSIL/HSIL and CC when compared to healthy controls. |
| Hoelzle et al [24] | 2021 | qPCR | miR-23a-3p | 2−ΔΔCt | Higher plasmatic miR-let-7a expression levels awithociated with HPV infection |
miR-21 miR-let-7a miR-214 |
Yes | N/I | N/I | N/I | No significant differences were found between groups. |
| Ocadiz-Delgado et al [25] | 2021 | qPCR | Beta2mHu | 2−ΔΔCt | N/I |
miR-15b miR-34a miR-218 |
No |
LSIL miR-34a: 0,9952 miR-15b: 0,6765 miR-218: 0,9872 CC miR-34a: 0,9505 miR-15b: 0,8173 miR-218: 0,8421 |
N/I | N/I | MiR-15b expression was higher in LSIL group and even higher in CC patients compared to healthy controls. MiR-34a and miR-218 expression increased in LSIL and decreased in CC, compared to control group. |
| Yamanaka et al [26] | 2021 | qPCR | miR-16 | 2−ΔΔCt | N/I | miR-100 | No | 0,879 | 91,2% | 80,4% | MiR-100 serum expression was high in healthy women, intermediate in women with CIN and low in CC women. |
HPV Human Papillomavirus, C Control Group, CIN Cervical Intraepithelial Neoplasia Group, CC Cervical Cancer Group, AUC Area Under Curve, SEN Sensibility, SPE Specificity, qPCR Quantitative Real-Time PCR, N/I Not Informed, N/A Not appliable.
Quality assessment
To assess the risk of bias and applicability of the studies, the Quality Assessment of Diagnostic Accuracy Studies - QUADAS-2 tool was used. This scoring system combines four key domains: patient selection; index test; reference standard and flow and time. Each domain is assessed for risk of bias, such as high risk, unclear or low risk, and the first three domains are also assessed for applicability [27].
Results
Literature search results
Figure 1 shows that 234 articles were found in the first literature search according to the screening strategy. With no duplicates to remove, the 234 articles were submitted for the title and abstract assessment and 107 articles were excluded, because they had been published in the form of letters, reviews, editorials, case reports, expert opinions, protocols, conferences or meeting abstracts, comments or meta-analyses or were either related to cervical cancer, microRNAs or diagnostic or were retracted articles. The remaining 127 articles were submitted to full-text review, of which 110 articles were excluded because were not realized in human samples because they did not deal with circulating microRNAs, they evaluated the expression of microRNAs only in cell lines or tissues because they did not use quantitative measurement or not having samples from healthy volunteers, precancerous and cervical carcinoma patients. Seventeen studies met the criteria for the final analysis and were enrolled in this systematic review. These studies were finally assessed as described in the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) guidelines for systematic review [10]. Tables 1, 2 and 3 show the characteristics of these studies.
Characteristics of included studies
As shown in Table 1, of the 17 included articles, 11 were from China, 2 from Japan, 1 from Iran, 1 from India, 1 from Brazil and 1 from Mexico. First, we focused our analysis on the characterization of patients and samples, due to its important implications for the molecular diagnosis approach. The correct characterization of healthy donors, patients with cervical intraepithelial neoplasia and cervical cancer patients are necessary and correct information about clinical implications related to cervical cancer is imperative. The control group (C) included healthy women and a mean sample size of 56 patients (ranging from 12 to 193). The mean age was 39.81 years (ranging from 27.5 to 50 years), but six studies did not report the age of included donors [11] [8] [13] [15] [16] [23]. Still, in relation to the control group, seven studies reported that samples were tested for HPV infection [14] [17] [18] [19] [23] [24] [25], and one study declared that the samples were not tested [11] and other studies did not report any information on the assessment of HPV infection. Among the studies in which samples were tested for HPV infection, one reported using the liquid-cytology method [14], and four reports of HPV infection were evaluated by PCR [17] [23] [24] [25], identifying the presence of HPV types 16, 66 and 68. Two studies reported that HPV infection was not detected in women in the control group [18] [19], without mentioning the method used for HPV detection. A total of 1171 women were included in the cervical intraepithelial neoplasia (CIN) group in all included studies, with an average sample size of 69 patients (ranging from 13 to 186), but only 9 studies discriminated the number of affected women according to stage lesion. The mean age among studies that reported this information was 38.2 years (ranging from 32 to 50). Six studies reported that CIN samples were tested for HPV infection [14] [17] [19] [23] [24] [25] and the others did not report any information about HPV infection in this patient group. Kong et al. [14] reported HPV infection in all patients included in the CIN group and the same was reported by Farzanehpour et al. [17], who also reported that HPV types 16 and 53 were identified. Aftab et al. [23] and Ocadiz-Delgado et al. [25] reported that HPV type 16 was identified and Hoelzle et al. [24] reported the identification of HPV types 16, 31, and 33, among others. Yang and Zhang reported that none of the patients included in the CIN group had HPV infection [19]. The cervical cancer group (CC) included an average of 58 patients (ranging from 13 to 184) and the mean age among the studies that reported this information, was 49.31 years old (ranging from 39 to 61). Ten studies reported the tumor stage according to the International Federation of Gynecology and Obstetrics (FIGO) and 161 women were diagnosed with metastatic cervical cancer while 285 were not, but 11 studies did not report any information related to the presence of metastasis. Regarding the histological type, most studies diagnosed squamous cell carcinoma as the most prevalent: LIU; XIN; MA (carcinoma: 84, others: 21) [11], NAGAMITSU et al., (carcinoma: 38, adenocarcinoma: 06) [12], WEI; WEN-MING; JUN-BO (squamous cell carcinoma: 101, adenocarcinoma: 19) [15], YANG; ZHANG (epithelial cell carcinoma: 39, adenocarcinoma: 11) [19], ZHENG et al.(epithelial cell carcinoma) [21], QIU et al. (carcinoma: 107, adenocarcinoma: 05) [22], YAMANAKA et al. (carcinoma: 37, adenocarcinoma: 09) [26]. Squamous cell carcinoma was the prevalent type in all CC women in Aftab et al. [23] and Ocadiz-Delgado et al. [25] studies. Wang et al. [16] reported having found both histological types but did not discriminate the prevalence. Only Sun et al. [13] found a higher number of adenocarcinoma-type cases in relation to squamous cell carcinoma.
Nine studies reported that CC samples were tested for HPV infection and Kong et al. [14] found that all the women included had HPV infection, as well as patients in the study by Farzanehpour et al. [17] and Ocadiz-Delgado et al. [25] who also identified HPV type 16. In their study, Liu, Xin and Ma [11] reported that 92 patients had HPV infection and 13 did not. Sun et al. [13] found HPV-positive patients, while 12 had negative results. Wei, Wen, Mingand Jun-Bo [15] reported that 81 women had HPV infection and 39 did not. Yang and Zang reported that all patients included in the CC group did not have this type of infection [19].
Serum samples were the specimens evaluated in 11 articles, 3 studies analyzed plasma samples, 1 analyzed total blood, 1 evaluated microRNA expression in exosomes, and 1 analyzed urine. Three of the 17 included studies employed some type of high-throughput analysis for microRNA candidate selection, of which 1 used sequencing [20], 1 performed microarray analysis [12] and 1 employed some type of microRNA panel PCR [8]. Regarding the type of samples evaluated in these high-throughput analyses, all articles used non-pooled samples, 11 target miRNAs were selected from literature and 3 target microRNAs were selected by database analysis (Table 2).
Quantitative Real-time PCR was used to evaluate microRNAs expression in almost all studies, U6 and RNU6B microRNAs were the most used endogenous reference microRNAs for normalization purposes, but other microRNAs such as MiR-16, MiR-223, cel-miR-39 and cel-miR-67 were also found in the included studies, reinforcing the importance of to evaluate the choice of a normalization method to minimize quantization errors and technical variability in experiments (Table 3). Five studies reported assessment of microRNAs expression comparing patients with and without HPV infection. Liu, Xin, and Ma [11] verified that the expression levels of MiR-196a were not correlated with HPV infection and the same was observed by Kong et al. [14] for MiR-92a. Wei, Wen-Ming and Jun-Bo [15] found that low plasma miR-145 level was significantly associated with HPV infection. Aftab et al. [23] also observed reduced expression of miRNA-145-5p in LSIL/HSIL HPV 16+ and CC HPV 16+ samples. Hoelzle et al. [24] found high plasmatic levels miR-let-7a in HPV+ samples. The expression of 33 microRNAs was evaluated in the eligible studies included in this review and 17 microRNAs (miR-196a, miR-16-2, miR-497, miR-1290, miR − 425-5p, hsa-miR-92a, miR-1266, miR-9, miR-192, miR-205, miR-21, miR-152, miR-15b, miR-34a, miR-218, miR-199a-5p and miR-155-5p) were found to show expression differential in cervical carcinogenesis. Only miR-21 was differentially expressed in more than one study [18] [22] [23].
The AUC value was calculated in some studies with the aim of evaluating the diagnostic potential of microRNAs in cervical carcinogenesis. When comparing the CC group the controls, the AUC values were - miR-1290: 0.7957 [12]; miR-145: 0.848 [15] and miR-152: 0.935 [19]. The AUC value of 0.849 was obtained for miR-497, miR-16-2*, miR-195 and miR-2861 when combined to form a panel [8]. The AUC value for MiR-21, miR-125b and miR-370 was 0,912 [22]. Elevated AUC values were also observed for miR-9: 0.99; miR-192: 1 and miR-205: 0.96 [17], as well as for miR-638: 0.734; miR-203a-3p: 0.660; miR-1914-5p: 0.626 and miR-521: 0.742 [21] and miR-21: 0,783; miR-125b: 0,642 and miR-370: 0,822 [22]. When comparing patients in the CC and CIN groups, the following AUC values were obtained: miR-145: 0.828 [15]; miR-152: 0.831 [19]; miR-497, miR-16-2*, miR-195 and miR-2861 combined to form a panel: 0.829 [8]; miR-9: 0.85, miR-192: 0.82 and miR-205: 0.75 [17]; miR-638: 0.681, miR-203a-3p: 0.651, miR-1914-5p: 0.717 and miR-521: 0.716 [21]; miR-21: 0,689; miR-125b: 0,735 and miR-370: 0,821 [22]. The AUC value of 0.897 was obtained for miR-21, miR-125b e miR-370 when combined [22]. Comparison of CIN and healthy group resulted in AUC values: 0.831 for miR-152 [19]; 0.734 for combination of miR-497, miR-16-2*, miR-195 and miR-2861 [8]; 0.90, 0.98 and 0.75 for miR-9, miR-192 and miR-205, respectively [17]. The AUC value found was 0.83 for hsa-miR-92a [14], 0,879 for miR-100 [22], 0.822 and 0.79 for let-7d-3p and miR-30d-5p and 0.828 for the combination of both [20]. The AUC value was also calculated to differentiate between CC women who responded completely and incompletely to radiotherapy and a value of 0.801 was obtained for MiR-145 [15]. Comparisons between CIN- group (healthy control and CIN I cases) and CIN+ group (CIN II, CIN III and cervical cancer) were also performed, resulting in the following AUC values: miR -21: 0.613, miR-214: 0.505, miR-34a: 0.508 and miR-200a: 0.615 [18]. Ocadiz-Delgado found that AUC values were calculated for samples of patients with LSIL and the values found were: miR-34a: 0,9952, miR-15b: 0,6765 and miR-218: 0,9872, and for samples of women CC: miR-34a: 0,9505, miR-15b: 0,8173 and miR-218: 0,8421 [25]. Individual AUC values were found for miR-21-5p: 0,97, miR-199a-5p: 0,831, miR-155-5p: 0,719, miR-34a-5p: 0,888, miR-145-5p: 0,575, miR-218-5p: 0,609 [23].
In summary, 12 of 17 included studies described microRNAs with diagnostic potential for precursor lesion and cervical cancer with diagnostic performance ≥0.70. Among all 33 microRNAs evaluated, 25 showed diagnostic performance ≥0.70 (miR-497, miR-16-2, miR-195 and miR-2861 when combined to form a panel; miR-1290; miR-145; miR-9, miR-192 and miR-205; miR-152; miR-638, miR-1914-5p and miR-521; hsa-miR-92a; let-7d-3p and miR-30d-5p; miR-21, miR-125b and miR-370; miR-15b, miR-34a and miR-218; miR-100; miR-199a-5p and miR-155-5p).
Some microRNAs showed excellent diagnostic performance. The AUC values for miR-9, miR-192, and miR-205 were 0.99, 1, and 0.96 (when comparing the group of women with cervical cancer and healthy controls), and 0.90 and 0.98 (when comparing the group of CIN patients and the healthy controls) [17]. Excellent results were also obtained for miR-152 which showed an AUC value of 0.935 [19]. When combined to compare CC women and controls, miR-21, miR-125b and miR-370 showed an AUC value of 0,912 [22]. In LSIL women, AUC values of 0,9952 (miR-34a) and 0,9872 (miR-218) were found and in CC women an AUC value of 0,9505 for miR-34a was obtained [25]. Excellent performance was also observed for miR-21-5p (AUC: 0,97) and for combined miR-21-5p, miR-199a-5p and miR-155-5p (AUC: 0,942), and miR-34a-5p, miR-145-5p and miR-218-5p (AUC: 0,969) [23].
Among 8 studies that reported sensitivity, 5 exceeded 70% when comparing the CC and C groups. Among the 33 evaluated microRNAs, 19 showed sensitivity ≥70% (miR-497, miR-16-2, miR-195 and miR-2861 when combined to form a panel; miR-1290; miR-145; miR-9, miR-192 and miR-205; miR-638, miR-203a-3p, miR-1914-5p and miR-521; miR-100; miR-21-5p, miR-199a-5p, miR-155-5p, miR-34a-5p and miR-218-5p). When comparing the CIN and C groups, eight microRNAs showed sensitivity ≥80% (miR-1290, miR-145, miR-9, miR-192, miR-205, miR-638, miR-1914-5p and miR-521) and of these, miR-9 and miR-192 showed 100% sensitivity.
Eight studies reported specificity and, among them, 2 exceeded 80% when comparing the CC and C groups. Among the 33 evaluated microRNAs, 15 showed specificity ≥80% (miR-497, miR-16-2, miR-195 and miR-2861 when combined to form a panel; miR-9, miR-192, miR-205 and miR-92a; miR-100; miR-21-5p, miR-199a-5p, miR-155-5p and miR-34a-5p, when combined, miR-145-5p and miR-218-5p). MiR-9, miR-192 and miR-21-5p also had the highest specificity values (94.4 and 98%). Table 3 also shows the main results obtained for each one of the 17 eligible studies.
Thirty three differentially expressed microRNAs are shown in Table 4 that also shows that 17 microRNAs (miR-196a, miR-16-2, miR-497, miR −1290, miR-425-5p, hsa-miR-92a, miR-1266, miR-9, miR-192, miR-205, miR-21, miR-152, miR-15b, miR-34a, miR-218, miR-199a-5p and miR-155-5p) were up-regulated in CIN and CC patients compared to healthy controls and 16 microRNAs (miR-195, miR-2861, miR-145, miR-214, miR-34a, miR-200a, let- 7d-3p, miR-30d-5p, miR-638, miR-203a-3p, miR-1914-5p, miR-521, miR-125b, miR-370, miR-218 and miR-100) were downregulated in these same groups.
Table 4.
Up and downregulated microRNAs expressed in patients with CIN and CC compared to healthy controls reported in the included studies
| Upregulated miRNAs* | Downregulated miRNAs* |
|---|---|
| miR-196a [11] | miR-195 e miR-2861 [8] |
| miR-16-2* e miR-497 [8] | miR-145 [15] |
| miR-1290 [12] | miRNA-214, −34a e -200a [18] |
| miR-425-5p [13] | let-7d-3p e miR-30d-5p [20] |
| miR-92a [14] | miR-638, miR-203a-3p, miR-1914-5p e miR-521 [21] |
| miR-1266 [16] | miR-125b and miR-370 [22] |
| miR-9, miR-192 e miR-205 [17] | miR-34a and miR-218 [25] |
| miRNA-21 [18] | miR-100 [26] |
| miR-152 [19] | miR-34a-5p, miR-145-5p and miR-218-5p [23] |
| miR-21 [22] | |
| miR-15b, miR-34a and miR-218 [25] | |
| miR-21-5p, miR-199a-5p and miR-155-5p [23] |
* When comparing CIN and CC patients to healthy controls
Methodological quality assessment
QUADAS-2 was carried out for the 17 included studies for quality assessment. Quality assessment of included studies revealed a generally good quality. However, potential risk of bias was detected in our review and high-risk bias was found in eleven studies in the index test domain. The risk of bias and applicability concerns graph for included studies were presented in Fig. 2.
Fig. 2.
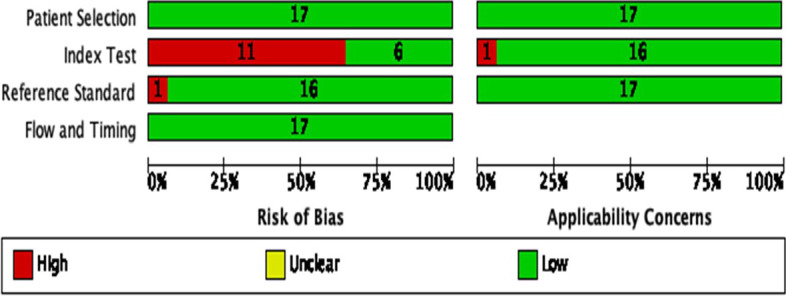
Quality assessment of studies using the Quality Assessment of Diagnostic Accuracy Studies 2 score system
Discussion
Despite having one of the highest potentials for prevention and cure among all neoplasms [28], cervical cancer is still considered a public health problem due to its high morbidity and mortality rates, affecting relatively young women, in the age group of 30 to 50 years old [2]. This type of cancer also has a good prognosis when diagnosed early and, although the Pap smear test is the most used preventive strategy for cervical cancer screening, this test also has its limitations [8], making new approaches necessary to achieve prevention, early diagnosis, treatment strategies, and prognosis to control this disease.
Due to their stability, circulating microRNAs, small non-coding RNAs that can be found freely in body fluids such as serum, plasma and urine, have been considered a new class of minimally invasive biomarkers and may have a great clinical value for early screening of cervical intraepithelial neoplasia stage. Studies aimed at the identification of new biomarkers related to cervical carcinogenesis emerged as a necessary strategy for the determination of microRNAs that could be used safely and reliably for a differential diagnosis.
In this review, we provided an updated perspective on the assessment of circulating miRNA expression from cervical cancer precursor lesions, identifying circulating miRNAs or specific miRNA signatures that can be used as potential biomarkers of different stages of cervical carcinogenesis. We identified 17 different studies, most involving women Chinese ethics, searching for circulating microRNAs differentially expressed in serum samples and using the literature review as a method for selecting microRNA candidates. However, the heterogeneity of the study was large. The sample size was found to be satisfactory (minimum of 50 patients per group) in the majority of included studies, but most of them failed to provide essential information about the characterization of CC, CIN and control evaluated samples. The description of the participant characteristics, such as age, type of lesion, tumor stage and presence of metastasis, whether the patients are undergoing chemotherapy are important to define whether the study findings can be generalized to show their possible limitations. The presence of HPV infection, for example, can also potentially alter the expression of circulating microRNAs [29] [30], which have been shown to play an important role in many physiological and pathological processes such as viral infections and oncogenesis. In this context, all samples included should must be tested for HPV infection in order to avoid the effect of this viral infection on microRNA expression, but only 6 of the eligible studies tested women from all samples group for HPV infection and some studies included women with and without HPV infection, which cause measurement errors.
Methodological differences in the same execution of experiments applied to the identification of differentially expressed circulating microRNAs were also found, potentially influencing the screening of circulating microRNAs for NIC and CC detection. The expression MicroRNAs and the final quantitative results, for example, can be greatly altered using different normalization methods [31] [32]. The absence of a universal quantitative evaluation for the expression of microRNAs and the consensus on the ideal endogenous reference microRNA to be used for the normalization of microRNA expression data from patients with NIC, CC and other types of cancer is another factor. Even though most used endogenous reference genes (RNU6B, cel-miR-39, U6 snRNA, and miR-16) were found in the included studies, other unusual microRNAs, such as MiR-128-3p, MiR-129-5p, let7i-5p, MiR-223 and MiR-31, were also found, reinforcing the importance of the development of a rigorous normalization strategy that avoids measurement errors.
The expression of 33 microRNAs was evaluated in the eligible studies included in this review and 17 of these microRNAs (miR-196a, miR-16-2, miR-497, miR-1290, miR-425-5p, hsa-miR- 92a, miR-1266, miR-9, miR-192, miR-205, miR-21, miR-152, miR-15b, miR-34a, miR-218, miR-199a-5p and miR-155-5p) were shown to be upregulated in women with precursor lesion and cervical cancer [8, 11–14, 16–19, 22, 25] and 16 microRNAs showed decreased expression in these same groups of women compared to healthy controls (miR-195, miR-2861, miR-145, miR-214, miR-34a, miR-200a, let-7d-3p, miR-30d-5p, miR-638, miR-203a-3p, miR-1914-5p, miR-521, miR-125b, miR-370, miR-218 and miR-100) [8, 15, 18, 20–23, 25, 26].
Only miR-21 was found to be differentially expressed in more than one study [18, 22, 23]. MiR-9 and miR-192 had the highest value of AUC, sensibility and specificity [17]. Therefore, defining promising circulating miRNAs or specific miRNA signatures from biological fluid samples can be useful for screening, diagnosis, prognosis and clinical follow-up of women undergoing cervical carcinogenesis, but greater standardization of studies seems to be necessary for greater consolidation of information to be obtained.
Conclusion
In conclusion, this systematic review suggests that circulating microRNAs have great potential to be used in the diagnosis of CIN and CC, but future studies should consider a more rigorous standardization sample characterization especially in evaluating HPV infection. Differential expression of miR-21 and elevated AUC values, sensitivity and specificity observed for miR-9 and miR-192 suggest that these are promising candidates, although further studies with larger samples and women of different ethnical groups are necessary. Furthermore, these studies should also consider the combination of microRNA biomarkers already evaluated for CIN and CC detection, in order to enhance diagnostic power.
Acknowledgments
This work was supported by UESC (Universidade Estadual de Santa Cruz) and UFSB (Universidade Federal do Sul da Bahia), Brazil.
Abbreviations
- AUC
Area Under Curve
- CC
Cervical cancer
- CIN
Cervical Intraepithelial Neoplasia
- FIGO
International Federation of Gynecology and Obstetrics
- HPV
Human Papillomavirus
- HSIL
High grade squamous intraepithelial lesion
- LSIL
Low-grade squamous intraepithelial lesion
- N/A
Not applicable
- N/I
Not Informed
- PCR
Polymerase Chain Reaction
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analysis
- PROSPERO
International Prospective Register of Systematic Reviews
- qPCR
Quantitative Real-Time PCR
- QUADAS-2
Quality Assessment of Diagnostic Accuracy Studies
- SEN
Sensibility
- SPE
Specificity
Authors’ contributions
NPGR, TBG and CMK extracted the data from a systematic review and drafted the manuscript. GFB performed a quality assessment analysis. CMK, GFB and LCGCL revised the article. All authors finished reading and approving the final manuscript of this study.
Funding
Not applicable.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Viii V. Cancer incidence in five continents. IARC Sci Publ. 1976;15:1–583. [PubMed] [Google Scholar]
- 2.Shah S, Senapati S, Klacsmann F, Miller D, Johnson J, Chang H-C, et al. Current Technologies and Recent Developments for Screening of HPV-Associated Cervical and Oropharyngeal Cancers. Cancers (Basel) 2016;8(9):85. doi: 10.3390/cancers8090085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Da SM. Falando Sobre Câncer Do Colo Do Útero. Ministério da Saúde. 2002;67.
- 4.Wang SS, Hildesheim A. Chapter 5: viral and host factors in human papillomavirus persistence and progression. JNCI Monogr [Internet]. 2003;(31):35–40. Available from: https://academic.oup.com/jncimono/article-lookup/doi/10.1093/oxfordjournals.jncimonographs.a003480. 10.1093/oxfordjournals.jncimonographs.a003480. [DOI] [PubMed]
- 5.de Villiers E-M, Fauquet C, Broker TR, Bernard H-U, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324(1):17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 6.Muñoz N, Castellsagué X, de González AB, Gissmann L. Chapter 1: HPV in the etiology of human cancer. Vaccine. 2006;24:S1–10. doi: 10.1016/j.vaccine.2006.05.115. [DOI] [PubMed] [Google Scholar]
- 7.ME SCHEURER, TORTOLERO-LUNA G, ADLER-STORTHZ K. Human papillomavirus infection: biology, epidemiology, and prevention. Int J Gynecol Cancer [Internet] 2005;15(5):727–746. doi: 10.1111/j.1525-1438.2005.00246.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Zhang D, Wang F, Xu D, Guo Y, Cui W. Serum miRNAs panel (miR-16-2*, miR-195, miR-2861, miR-497) as novel non-invasive biomarkers for detection of cervical cancer. Sci Rep. 2015;5(August):17942. doi: 10.1038/srep17942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.JIA W, WU Y, ZHANG Q, GAO G, ZHANG C, XIANG Y. Expression profile of circulating microRNAs as a promising fingerprint for cervical cancer diagnosis and monitoring. Mol Clin Oncol. 2015;3(4):851–858. doi: 10.3892/mco.2015.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu P, Xin F, Ma CF. Clinical significance of serum miR-196a in cervical intraepithelial neoplasia and cervical cancer. Genet Mol Res. 2015;14(4):17995–18002. doi: 10.4238/2015.December.22.25. [DOI] [PubMed] [Google Scholar]
- 12.NAGAMITSU Y, NISHI H, SASAKI T, TAKAESU Y, TERAUCHI F, ISAKA K. Profiling analysis of circulating microRNA expression in cervical cancer. Mol Clin Oncol. 2016;5(1):189–194. doi: 10.3892/mco.2016.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun L, Jiang R, Li J, Wang B, Ma C, Lv Y, et al. MicoRNA-425-5p is a potential prognostic biomarker for cervical cancer. Ann Clin Biochem. 2017;54(1):127–133. doi: 10.1177/0004563216649377. [DOI] [PubMed] [Google Scholar]
- 14.Kong Q, Tang Z, Xiang F, Jiang J, Yue H, Wu R, et al. Diagnostic value of serum hsa-MIR-92a in patients with cervical cancer. Clin Lab. 2017;63(2):335–340. doi: 10.7754/Clin.Lab.2016.160610. [DOI] [PubMed] [Google Scholar]
- 15.Wei H, Wen-Ming C, Jun-Bo J. Plasma miR-145 as a novel biomarker for the diagnosis and radiosensitivity prediction of human cervical cancer. J Int Med Res. 2017;45(3):1054–1060. doi: 10.1177/0300060517709614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Liu Y, Wang X, Li J, Wei J, Wang Y, et al. MiR-1266 promotes cell proliferation, migrati,on and invasion in cervical cancer by targeting DAB2IP. Biochim Biophys Acta Mol basis Dis. 2018;1864(12):3623–3630. doi: 10.1016/j.bbadis.2018.09.028. [DOI] [PubMed] [Google Scholar]
- 17.Farzanehpour M, Mozhgani SH, Jalilvand S, Faghihloo E, Akhavan S, Salimi V, et al. Serum and tissue miRNAs: potential biomarkers for the diagnosis of cervical cancer. Virol J. 2019;16(1):1–9. doi: 10.1186/s12985-019-1220-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Zhang D, Chen Q, Hong Y. Plasma expression of miRNA-21, − 214, −34a, and -200a in patients with persistent HPV infection and cervical lesions. BMC Cancer. 2019;19(1):1–7. doi: 10.1186/s12885-018-5219-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang D, Zhang Q. Mir-152 may function as an early diagnostic and prognostic biomarker in patients with cervical intraepithelial neoplasia and patients with cervical cancer. Oncol Lett. 2019;17(6):5693–5698. doi: 10.3892/ol.2019.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng M, Hou L, Ma Y, Zhou L, Wang F, Cheng B, et al. Exosomal let-7d-3p and miR-30d-5p as diagnostic biomarkers for non-invasive screening of cervical cancer and its precursors. Mol Cancer. 2019;18(1):1–8. doi: 10.1186/s12943-019-0999-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng S, Li R, Liang J, Wen Z, Huang X, Du X, et al. Serum miR-638 combined with squamous cell carcinoma-related antigen as potential screening biomarkers for cervical squamous cell carcinoma. Genet Test Mol Biomarkers. 2020;24(4):188–194. doi: 10.1089/gtmb.2019.0147. [DOI] [PubMed] [Google Scholar]
- 22.Qiu H, Liang D, Liu L, Xiang Q, Yi Z, Ji Y. A novel circulating MiRNA-based signature for the diagnosis and prognosis prediction of early-stage cervical Cancer. Technol Cancer Res Treat. 2020;1(19):153303382097066. doi: 10.1177/1533033820970667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aftab M, Poojary SS, Seshan V, Kumar S, Agarwal P, Tandon S, et al. Urine miRNA signature as a potential non-invasive diagnostic and prognostic biomarker in cervical cancer. Sci Rep. 2021;11(1):10323. doi: 10.1038/s41598-021-89388-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoelzle CR, Arnoult S, Borém CRM, Ottone M, de Magalhães KCSF, da Silva IL, et al. MicroRNA levels in cervical Cancer samples and relationship with lesion grade and HPV infection. MicroRNA. 2021;10(2):139–145. doi: 10.2174/2211536610666210604123534. [DOI] [PubMed] [Google Scholar]
- 25.Ocadiz-Delgado R, Lizcano-Meneses S, Trejo-Vazquez J, Conde-Perezprina J, Garrido-Palmas F, Alvarez-Rios E, et al. Circulating miR-15b, miR-34a and miR-218 as promising novel early low-invasive biomarkers of cervical carcinogenesis. APMIS [Internet] 2021;129(2):70–79. doi: 10.1111/apm.13093. [DOI] [PubMed] [Google Scholar]
- 26.Yamanaka Z, Sasaki T, Yamanaka A, Kato K, Nishi H. Circulating and tissue miR-100 acts as a potential diagnostic biomarker for cervical cancer. Cancer Biomarkers [Internet] 2021;32(4):551–558. doi: 10.3233/CBM-201021. [DOI] [PubMed] [Google Scholar]
- 27.Whiting PF. QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann Intern Med. 2011;155(8):529. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 28.Ferlay J, Shin H-R, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 29.Pardini B, De Maria D, Francavilla A, Di Gaetano C, Ronco G, Naccarati A. MicroRNAs as markers of progression in cervical cancer: a systematic review. BMC Cancer [Internet] 2018;18(1):696. doi: 10.1186/s12885-018-4590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davidson-Moncada J, Papavasiliou FN, Tam W. MicroRNAs of the immune system: roles in inflammation and cancer. Ann N Y Acad Sci [Internet] 2010;1183:183–194. doi: 10.1111/j.1749-6632.2009.05121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods [Internet] 2010;50(4):298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prado MSJG, de Goes TC, de Jesus ML, Mendonça LSO, Nascimento JS, Kaneto CM. Identification of miR-328-3p as an endogenous reference gene for the normalization of miRNA expression data from patients with diabetic retinopathy. Sci Rep. 2019;9(1):1–8. doi: 10.1038/s41598-018-37186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


