Significance
Many small molecule drugs are identified by phenotypic screening without any knowledge of their associated protein target(s). Identifying these protein targets remains a critical step in the drug discovery pipeline and oftentimes represents a formidable challenge. Traditional approaches use small molecule probes containing crosslinkers that can be stoichiometrically activated by ultraviolet light. However, most of the highly reactive crosslinker reacts with water, leading to low target labeling. In this report, we describe a new class of small molecule probe that activates a spatially separated crosslinker in a catalytic manner, leading to multiple labeling events and, consequently, signal amplification. This platform operates through a photophysical process called Dexter energy transfer between a photocatalyst and diazirine in the presence of blue light.
Keywords: target identification, proteomics, photocatalysis
Abstract
Over half of new therapeutic approaches fail in clinical trials due to a lack of target validation. As such, the development of new methods to improve and accelerate the identification of cellular targets, broadly known as target ID, remains a fundamental goal in drug discovery. While advances in sequencing and mass spectrometry technologies have revolutionized drug target ID in recent decades, the corresponding chemical-based approaches have not changed in over 50 y. Consigned to outdated stoichiometric activation modes, modern target ID campaigns are regularly confounded by poor signal-to-noise resulting from limited receptor occupancy and low crosslinking yields, especially when targeting low abundance membrane proteins or multiple protein target engagement. Here, we describe a broadly general platform for photocatalytic small molecule target ID, which is founded upon the catalytic amplification of target-tag crosslinking through the continuous generation of high-energy carbene intermediates via visible light-mediated Dexter energy transfer. By decoupling the reactive warhead tag from the small molecule ligand, catalytic signal amplification results in unprecedented levels of target enrichment, enabling the quantitative target and off target ID of several drugs including (+)-JQ1, paclitaxel (Taxol), dasatinib (Sprycel), as well as two G-protein-coupled receptors—ADORA2A and GPR40.
During the last decade, over 50% of drugs in phase II and III clinical trials have failed due to a lack of efficacy stemming from incomplete target validation (1). Hence, the identification of biological targets and understanding of their interactions at the molecular level (target ID) is essential for the successful design of new therapeutic candidates and reducing clinical attrition (2, 3). In recent years, however, the intrinsic challenges associated with fully characterizing drug targets has manifested in an industry-wide bottleneck within the developmental pipeline (4, 5), which has profoundly curtailed the impact of new therapeutics on society. It is clear, therefore, that the development of new methods to elucidate molecular targets has the potential to significantly increase the success of therapeutic target selections, which should ultimately lead to a reduction in patient morbidity (Fig. 1A) (2, 6, 7).
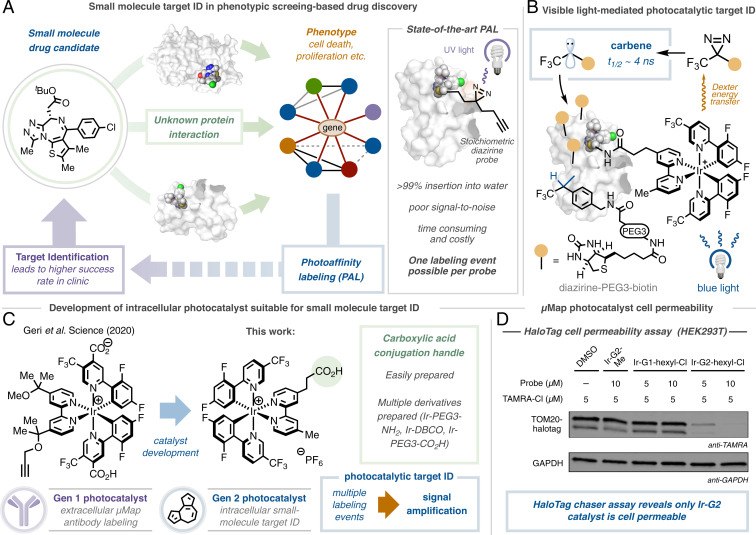
Fig. 1.
Photoaffinity labeling comprises a critical component of small molecule target ID. (A) Target ID campaigns are critical for the development of successful drugs, although they often rely on challenging photoaffinity labeling campaigns that employ the stoichiometric activation of diazirine small-molecule conjugates with UV light. (B) Our approach separates the warhead from the small molecule probe, instead employing the photocatalytic activation of diazirines using visible light, giving rise to significant signal enhancement. (C) Development of cell-penetrating, generation 2 photocatalyst suitable for small-molecule conjugation and target ID. (D) Cell permeability of Ir-photocatalysts determined by HaloTag chaser assay; photocatalyst PEG-hexyl chloride conjugate and TAMRA hexyl chloride incubated with HEK293T cells expressing TOM20-HaloTag. Western blot analysis and immunostaining TOM20 with anti-TAMRA reveals off-compete only in the presence of Ir-G2 catalyst.
Over the last two decades, technological advancements in the fields of mass spectrometry (8), chemical genetics (9), and bioinformatics (10) have transformed drug target identification leading to improvements in our understanding of biological pathways and cellular signaling (3, 11). However, while this information has provided a more focused route to the complex process of drug discovery, there remains a demand for target identification technologies for proteins without a well-described mechanism-of-action (12). To address this need, affinity-based approaches (13), and photoaffinity labeling (PAL) in particular, have now become routinely used tools in drug discovery (Fig. 1A) (14). PAL works by the incorporation of a stoichiometric photoactivatable group, such as a diazirine, and an affinity handle, such as biotin, precisely into the small-molecule architecture (15). Following ultraviolet (UV)-activation and affinity-based enrichment, immunoblotting and proteomic analysis can be used to gather information regarding the identity of the target protein (16).
While these methods have been empowering for a number of protein classes (17–19), the high failure rates associated with such outdated cross-linking technologies often outweigh any potential benefits; diminished receptor occupancy, poor cross-linking yields, low protein abundance, problematic downstream processing, UV-based protein degradation, low cell permeability, and intrinsic background signal are but a few of the challenges that have to be overcome (6, 16, 20–22). The use of diazirine-based probes in particular has been challenging in this context as >99% of the carbenes generated upon UV irradiation react with water and not the target (23). These spent probes serve to further block the binding of unreacted molecules, further hampering labeling efficiency. As a result, costly and time-consuming structural optimization campaigns, nonnative cell lines, and bespoke analytic techniques are often required to overcome these shortfalls.
Indeed, the inherent difficulties associated with PAL have inspired the development of several elegant methods that hinge upon the use of stoichiometric activated electrophiles (13, 24–27), single-electron transfer events (28), or specific oxidizable residues (29) to identify target proteins. However, many of these technologies remain limited to a single labeling event per drug molecule. We therefore reasoned that the development of a catalytic target ID technology that separates the drug molecule from the reactive warhead could overcome these challenges through multiple label-ling events leading to signal amplification (Fig. 1B).
We recently disclosed an antibody-based proximity labeling platform for cell surface microenvironment elucidation, termed µMap (30). This method relies upon the activation of diazirine molecules in close proximity to a set of photocatalysts appended to an antibody via Dexter energy transfer. Inspired by this unique activation mode, we questioned whether such a tactic could be leveraged for small molecule target ID through the incorporation of an iridium photocatalyst onto a bioactive small molecule: following visible light (blue LEDs) excitation of the photocatalyst to its triplet excited state (T1), short range energy transfer to a proximal diazirine (1 nm) leads to carbene formation, following N2 extrusion, and regeneration of the ground state iridium photocatalyst. The highly reactive carbene covalently labels the surrounding residues with minimal diffusion (<4 nm), which can be identified via downstream processing. However, at the outset of the investigation, we were cognizant of several challenges inherent in developing such a technology, such as catalyst cell permeability and biocompatibility, ease of chemical manipulation, retention of biological activity, and labeling efficiency (given each antibody contained an average of six to eight photocatalysts). However, we reasoned that by “switching on” catalysis through visible light activation, labeling could be controlled both spatially and temporally, bypassing intrinsic reactivity problems and enabling the identification of novel targets across numerous drug discovery programs.
We began by investigating cell permeability: employing a HaloTag-based chaser assay off-competing a TAMRA dye in HEK293T cells, we identified that our previous catalyst design (Gen 1) was impermeable by virtue of its neutral net charge and two carboxylic acid residues (Fig. 1C). Through screening different photocatalyst structures, we realized that Ir-catalysts containing both the dFCF3-phenyl pyridine moiety and 4,4-dialkyl bpy ligand were crucial in achieving the necessary triplet energy (30). Pleasingly, by removing the carboxylic acid groups, the cationic photocatalyst (Gen 2) was rendered cell-permeable (Fig. 1D). With this in mind, we evaluated conjugation handles based around the 4,4-dMebpy ligand, opting for a distal carboxylic acid to enable facile amide coupling. Importantly, our G2-iridium catalyst can be accessed on gram-scale and be readily conjugated to a range of linkers and complex small molecules (vide infra).
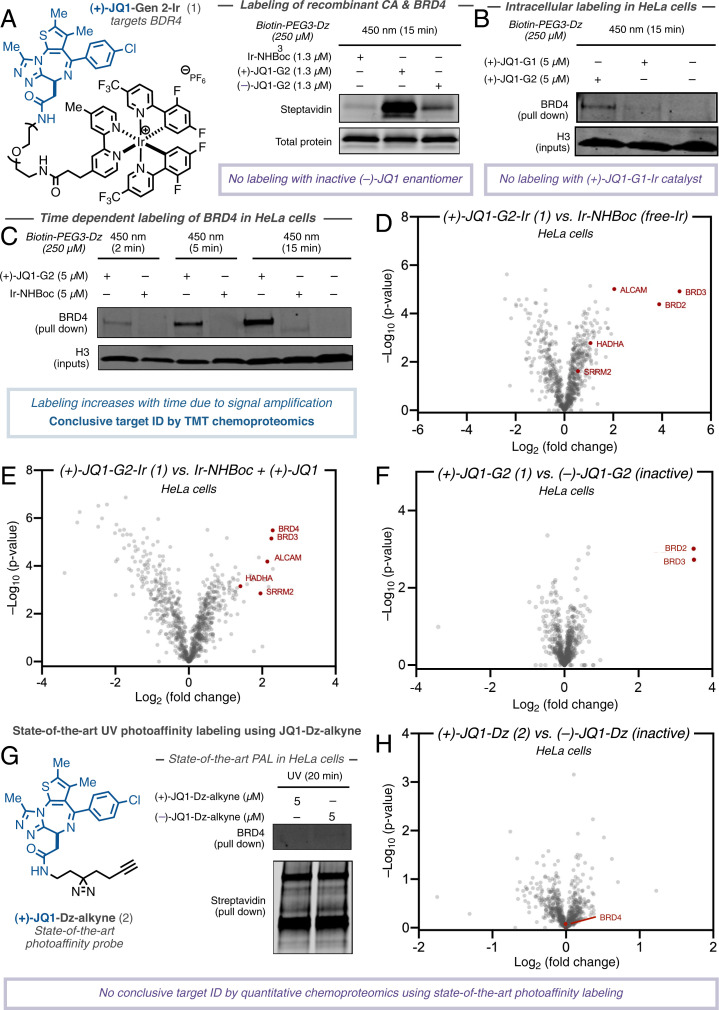
Confident in our ability to access almost any Ir-drug conjugate, we initiated our target ID campaign with the validated epigenetic tool compound (+)-JQ1 (31). A potent inhibitor of the BET family of bromodomain proteins (BRD2/3/4), several JQ1 structural analogs are in clinical trials for a variety of cancers including NUT midline carcinoma (32). We prepared the corresponding (+)-JQ1-G2 conjugate (1) (Fig. 2) and validated target engagement in vitro with recombinant BRD4 in a competition assay vs. bovine carbonic anhydrase (CA). An equimolar amount of CA and BRD4 was treated with (+)-JQ1-G2 probe (1) and an excess of diazirine-PEG3-biotin prior to irradiation at 450 nm. Labeling intensity was measured by Western blotting with a streptavidin stain. Pleasingly, these preliminary experiments revealed a 20-fold increase in labeling for BRD4 vs. CA compared to the unconjugated (free) photocatalyst (Fig. 2A). Importantly, the (–)-JQ1-G2 conjugate, which is known to not bind BRD4 (31), showed significantly reduced labeling, demonstrating that labeling is as a result of a ligand/protein binding event (Fig. 2A). In addition, we were able to confirm this through microscale thermophoresis (MST), where the addition of the Ir-catalyst made only a minor impact on the binding constant (SI Appendix, Fig. S1).
Fig. 2.
Development of photocatalytic target ID platform for interactome mapping of (+)-JQ1 in HeLa cells. Structure of JQ1-based photocatalyst conjugate and state-of-the-art PAL probe. (A) Labeling of recombinant BRD4 protein vs. spectator protein carbonic anhydrase using free iridium-, (+)-JQ1-, and (–)-JQ1-probe by immunostaining with streptavidin. (B) Comparing permeability of G1- and G2-based (+)-JQ1 probes following irradiation in HeLa cells, streptavidin bead enrichment, and immunostaining with anti-BRD4. (C) BRD4 labeling increases over time (2-min, 5-min, and 15-min irradiation) through photocatalytic signal amplification using (+)-JQ1-G2 probe in HeLa cells following streptavidin bead enrichment and immunostaining with anti-BRD4. (D) TMT-based quantitative chemoproteomic analysis of JQ1-labeling in HeLa cells comparing intracellular labeling by (+)-JQ1-G2 catalysts and unconjugated iridium catalyst (control) reveals BRD proteins as highly enriched in addition to known JQ1 off-targets. (E) TMT-based quantitative chemoproteomic analysis of JQ1-labeling in HeLa cells comparing intracellular labeling by (+)-JQ1-G2 catalysts and unconjugated iridium catalyst + (+)-JQ1 (control). (F) TMT-based quantitative chemoproteomic analysis of JQ1-labeling in HeLa cells comparing intracellular labeling between active (+)-JQ1-G2 and inactive (–)-JQ1-G2 catalysts reveals only (+)-isomer labels BRD proteins. (G) State-of-the-art PAL employing active (+)-JQ1- and inactive (–)-JQ1-Dz-alkyne probes reveals no selective enrichment of BRD4 by Western blotting despite broad biotinylation visible by immunostaining with streptavidin. (H) TMT-based quantitative chemoproteomic analysis in HeLa cells comparing state-of-the-art PAL employing active (+)-JQ1- and inactive (–)-JQ1-Dz-alkyne probes reveals no enrichment of BRD proteins.
Based on these results, we sought to apply this method to live cells. We treated HeLa cells with 5 µM (+)-JQ1-Gen 2 (1) for 3 h before the addition of 250 µM Dz-PEG3-Biotin and subsequent 15-min irradiation (450 nm). Following lysis and streptavidin-bead enrichment, Western blot analysis with anti-BRD4 showed a clear labeling of the target protein compared to dimethyl sulfoxide (DMSO) control (SI Appendix, Fig. S2). In line with previous findings, the corresponding (+)-JQ1-Gen 1 catalyst, while demonstrating similar in vitro labeling capability, showed no such enrichment of the target protein in cells (Fig. 2B. Consistent with our hypothesis, the intensity of labeling was found to be linearly related to irradiation time (see SI Appendix), demonstrating the photocatalytic signal amplification and temporal control offered by the photocatalytic platform (Fig. 2C). This was also observable by confocal microscopy, wherein the degree of biotinylation increased significantly over time (SI Appendix, Fig. S3). Encouraged by our Western blot validation data, we moved to tandem mass tag (TMT)-based quantitative chemoproteomics in order to more completely assess the interactome of (+)-JQ1; quantitative proteomic analysis represents the ‘gold standard’ within target ID, providing the highest levels accuracy and lower promiscuity (22). To our delight, by comparing the labeling by (+)-JQ1-Gen 2 (1) vs. unconjugated (free) photocatalyst in HeLa cells, we observed several BRD proteins as the most enriched, although their precise identities remain difficult to ascertain due to structural homology (Fig. 2D). We also identified two previously annotated (+)-JQ1 off-targets, HADHA (33) and SRRM2 (34). ALCAM (CD166), a transmembrane glycoprotein, was also identified as being significantly enriched, but currently has no reported interaction with (+)-JQ1. CD166 exerts a procarcinogenic role via the inhibition of transcription factors along the FOXO/AKT axis and is considered a novel therapeutic target for liver cancer (35). Interestingly, BET inhibition by (+)-JQ1 has been shown to up-regulate expression of FOXO1, although the mechanism remains unclear (36). In order to evaluate whether protein enrichment was as a direct result of labeling or up-regulation by virtue of the presence of (+)-JQ1, we repeated the experiment with an equivalent of (+)-JQ1 in the free iridium control (Fig. 2E). Upon chemoproteomic analysis, we found that CD166 was similarly enriched, indicating that it may be a putative off-target binder of (+)-JQ1, although further biological validation is required. We further compared the interactomes of the enantiomers of JQ1-G2 and found the active (+) enantiomer, 1, delivered BRD2/3/4 as top hits, and while CD166 was detected, it was not enriched, indicating that binding may not be affected by the stereogenic center (Fig. 2F). In contrast to these data, the same analysis using classical UV-based PAL employing (+)-JQ-Dz-alkyne (2) (37), in our hands, did not lead to enrichment of BRD proteins by Western blot (Fig. 2G) or chemoproteomic analysis (Fig. 2H).
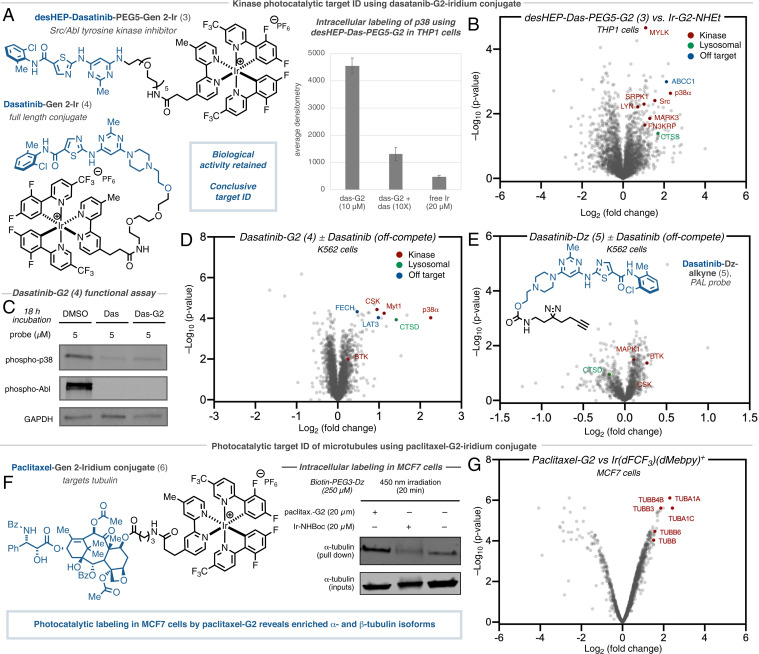
The dual Src/Abl tyrosine kinase inhibitor dasatinib (Sprycel) displays significant antileukemic effects against various imatinib-resistant mutants (38). However, despite well-documented BCR/ABL inhibition, its precise downstream cellular MOA remains to be fully understood. While the dasatinib interactome has been previously characterized (17), most methods have been performed with recombinant protein or in cell-lysate; live cell data are typically restricted to kinase-based assays that measure downstream phosphorylation or residence at engineered kinase constructs, which can be challenging to deconvolute and fail to identify nonkinase-based off targets (39–42).
As previous studies have demonstrated difficulties in maintaining potency and cell permeability using dasatinib-derived probes (17), we started by synthesizing three truncated (deshydrox-yethylpiperazinyl)-dasatinib iridium conjugates using our cell-permeable Ir-G2 catalyst with varying PEG linker lengths (n = 3–5) (3) (Fig. 3, Top). Gratifyingly, upon subjection of the desHEP-dasatinib-G2 conjugates (3) (5 µM) to our standard photocatalytic labeling protocol, all of the conjugates revealed enrichment of p38 (MAP kinase) by Western blot analysis compared to off-compete (4× dasatinib) controls in THP1 cells (SI Appendix, Fig. S4). As the corresponding PEG5-G2 conjugate showed the greatest enrichment (3.5× enrichment vs. off-compete and 9.5× enrichment vs. free-Ir) (Fig. 3A), we undertook label-free proteomic analysis of these reactions, revealing significant enrichment of p38 (SI Appendix, Fig. S5), which has been shown to play a critical role in its antileukemic properties (43), as well as several other established kinase interactors including Src and Lyn (Fig. 3B) (44). Furthermore, we identified multidrug resistance transporter ABCC1 among the most enriched proteins—an important off target; understanding the interaction between drug molecules and efflux transporters is an important consideration in many drug discovery efforts (45). Lysosomal sequestration of dasatinib (46), due to its lipophilic and weakly basic properties, was evident by the presence of cathepsin S (CTSS) among the most enriched proteins. Encouraged by these initial results, we turned our attention to the underexplored full dasatinib-PEG3-G2 catalyst (4), which retains the 2-hydroxyethylpiperazine tail. Importantly, we found a similar kinase inhibition profile against p38, in addition to Abl, by evaluation of downstream phosphorylation in Ph+ K562 cells, compared to the parent drug, again highlighting the compatibility of the iridium photocatalyst toward maintaining biological function and cell permeability (Fig. 3C and SI Appendix, Fig. S6). Gratifyingly, subjection of our photocatalytic labeling to TMT-based chemoproteomics revealed extensive enrichment of p38 as well as Myt1 and CSK kinases, both well-established binders of dasatinib (Fig. 3D) (42). Moreover, known kinase off-target ferrochelatase (FECH) (47) was also significantly enriched, alongside large amino acid transporter (LAT3) (42). Similarly, lysosomal protein cathepsin D (CTSD) was among the most enriched proteins. Notably, in our hands, state-of-the-art photoaffinity labeling, employing dasatinib-diazirine-alkyne (5), revealed only trace enrichment of CSK and the kinases BTK and MAPK1 (BTK was found to be similarly enriched by photocatalytic target ID) (Fig. 3E).
Fig. 3.
Intracellular photocatalytic target ID and interactome mapping of dasatinib and paclitaxel. (A) Enrichment of p38 by Western blot for labeling using desHEP-dasatinib-PEG5-G2 labeling in THP1 cells. (B) Label-free proteomic analysis in THP1 cells comparing intracellular labeling by desHEP-dasatinib-PEG5-G2 catalyst vs. Ir-G2-NHEt reveals enrichment of several kinases (red), as well as lysosomal proteins (green) and off-targets (blue). (C) Kinase activity assays reveals dasatinib-G2 retains inhibition activity against Abl and p38, as well as general tyrosine phosphorylation, in K562 cells. (D) TMT-based quantitative chemoproteomic analysis in K562 cells comparing intracellular labeling by dasatinib-G2 catalyst vs. dasatinib-G2 + dasatinib (off-compete control) reveals enrichment of several kinases (red), as well as lysosomal proteins (green) and established off-targets (blue). (E) TMT-based quantitative chemoproteomic analysis in K562 cells comparing intracellular labeling by dasatinib-Dz-alkyne (PAL probe) vs. off-compete control does not reveal enrichment of kinases suitable for conclusive target ID. (F) Initial Western blot studies for paclitaxel-G2 labeling in MCF7 cells following irradiation and streptavidin bead enrichment reveals significant enrichment of a-tubulin by immunostaining compared to unconjugated iridium and DMSO controls. (G) TMT-based quantitative chemoproteomic analysis in MCF7 cells comparing intracellular labeling by paclitaxel–G2 catalyst and unconjugated iridium catalyst (control) reveals enrichment of several tubulin isoforms.
The anti-cancer properties of the natural product paclitaxel (Taxol) have been proposed to be derived from binding to microtubules, leading to stabilization and mitotic arrest; however, the full extent of its mechanism remains unclear (48). Based on its widespread use and intriguing mechanism, we prepared the corresponding paclitaxel-Gen 2-iridium conjugate (6) (Fig. 3, Bottom) and assessed its cellular activity. Through a series of cell proliferation assays, we found that our paclitaxel-G2 conjugate displayed similar anti-proliferative properties as the native compound, suggesting that the pendent Ir-catalyst did not disrupt the native function of paclitaxel (SI Appendix, Fig. S7). Encouraged by this, we proceeded to study the efficiency of labeling in the breast cancer cell line MCF7. Following our standard photocatalytic labeling protocol with 20 µM paclitaxel-G2 conjugate (6) for 3 h, Western blot analysis with anti-α-tubulin showed clear labeling of the target protein com-pared both the free iridium and DMSO controls (Fig. 3F). Subjection to our photocatalytic labeling protocol and TMT-based chemoproteomics revealed extensive labeling of tubulin isotypes αIa, βIII, βIVb, and αIc (Fig. 3G), which is in good agreement with previous photoaffinity labeling studies on extracted tubulin (49).
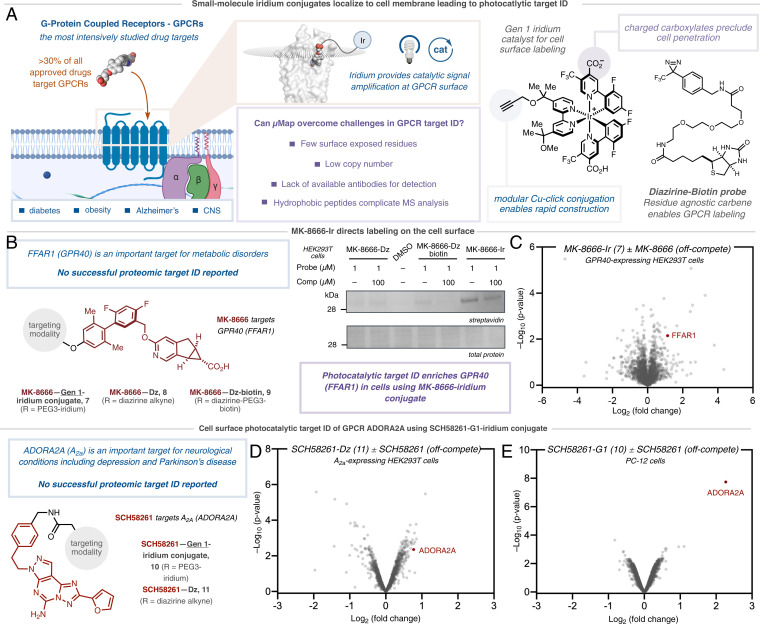
Having established the efficacy of photocatalytic target ID for intracellular proteins, we turned our attention to the cell surface. The exceedingly low abundance, lack of exposed residues, and aggregation-prone hydrophobic domains oftentimes confounds the detection and manipulation of membrane proteins, rendering target ID unfeasible (20, 50, 51). These challenges are exacerbated when combined with the high background labeling, poor sensitivity, and low cross-linking yields systemic in PAL campaigns. Given that over 30% of approved drugs target membrane-based G protein-coupled receptors (GPCRs) (52), we felt that our photocatalytic target ID platform was ideally placed to tackle these challenges by virtue of our catalytic signal amplification. To investigate this, we prepared three conjugates derived from the G protein-coupled receptor 40 (GPR40)-targeting small molecule probe MK-8666 (53) (Fig. 4A). GPR40 functions as a receptor for long-chain free fatty acids, which enhances insulin secretion via IP3 generation and PKC activation. It has therefore become an important therapeutic target for the treatment of type 2 diabetes (54). In order to compare the efficacy of photocatalytic target ID to classical UV-based PAL, we synthesized a photocatalyst conjugate based on the known small molecule binder MK-8666 (7, SI Appendix, Fig. S8), based on the more hydrophilic G1 catalyst, MK-8666-PEG2-azide diazirine-alkyne tethered probe (8) and diazirine-PEG3-biotin probe (9). Following incubation of the probes (1 µM) with GPR40-expressing HEK293T cells and labeling, Western blot visualization and streptavidin staining showed no visible GPR40 enrichment using classical PAL probes (8 and 9). However, we identified the presence of a band corresponding to the weight of GPR40 in the experiment containing the MK-8666-Ir conjugate (7) (Fig. 4B); no bands were observed when using the unconjugated iridium catalyst (SI Appendix, Fig. S9). Encouraged by these results, we questioned whether the photocatalytic labeling would translate to a successful target identification via label free proteomics. Gratifyingly, analysis of our photocatalytic-labeling method revealed considerable enrichment for GPR40 (FFAR1) when using MK-8666-Ir (7) versus off-competing with an excess of the parent MK-8666 ligand (Fig. 4C and SI Appendix, Fig. S10).
Fig. 4.
Extracellular photocatalytic target ID of GPCRs GPR40 and ADORA2A. (A) Small molecule iridium conjugate of MK-8666-Ir is prepared by Cu-click reaction and localized to the target protein GPR40 (FFAR1) via a ligand binding event on the cell surface of GPR40-expressing HEK293T cells. Irradiation and energy transfer to the diazirine leads to selective labeling of the target protein (PDB: 5TZR). (B) Comparative analysis of labeling of GPR40 through classical UV-based PAL using MK-8666-diazirne and MK-8666-diazirine-biotin probes vs. MK-8666-diazirine-G1 Iridium conjugate. Analysis by Western blot visualization and staining with streptavidin shows no labeling of the target protein following streptavidin-bead based enrichment. (C) Label-free proteomic analysis of MK-8666-Ir photocatalytic µMap reveals enrichment of the target protein compared to off-compete with the unmodified ligand. (D) TMT-based quantitative chemoproteomic analysis in A2a-expressing HEK293T cells comparing extracellular labeling by SCH58261-Dz-alkyne vs. SCH58261-Dz-alkyne + SCH58261 (off-compete control) reveals inconclusive target ID of ADORA2A. (E) TMT-based quantitative chemoproteomic analysis in PC-12 cells comparing extracellular labeling by SCH58261-G1 catalyst vs. SCH58261-G1 + SCH58261 (off-compete control) reveals significant enrichment of ADORA2A.
Finally, we chose to target the adenosine receptor A2a (ADORA2A) as a second exemplar membrane target. This GPCR has become an important potential target for immunotherapy (55), as well as psychiatric and degenerative disorders (56), but critically, has never been identified through live cell chemoproteomics (57, 58). Using a reported ligand for ADORA2A that binds from the extracellular face, SCH58261 (59), we prepared both a tethered diazirine-conjugate, SCH58261-Dz (11), and SCH58261-G1-Ir (10); the low cell permeability affording a higher effective concentration of photocatalyst probe on the cell surface. Photocatalytic labeling applied to A2a-expressing HEK293T cells, followed by Western blot visualization, revealed a stark difference in labeling between the SCH58261-G1-Ir (10) and the corresponding off-compete controls (SI Appendix, Fig. S11 A and B). TMT-based chemoproteomic analysis of these reactions confirmed our initial result, with our photocatalytic-labeling method using SCH58261-G1-Ir (10) showing a 10-fold enrichment for ADORA2A with respect to off-competing with the parent SCH58261 ligand, and >20-fold enrichment vs. free-Ir photocatalyst (SI Appendix, Fig. S12 A and B). In contrast, PAL using SCH58261-Dz (11), showed poor enrichment of ADORA2A by quantitative chemoproteomics (Fig. 4D), in line with Western blot data (SI Appendix, Fig. S11A). Based on the degree of enrichment in A2a-expressing HEK293T cells, we were keen to ascertain how the photocatalytic target ID platform performed at native levels of membrane protein concentration, wherein classical PAL remains extremely challenging. Remarkably, photocatalytic labeling using SCH58261-G1-Ir (10) in PC-12 cells, which have previously been validated to natively express A2a (60), revealed similarly high levels of enrichment for the target protein ADORA2A—highlighting the signal amplification conferred by photocatalysis (Fig. 4E).
In conclusion, we describe a general platform for photocatalytic target ID that utilizes cell-penetrating iridium-conjugated small molecules, which can bind protein targets, to locally activate proximal diazirines via Dexter energy transfer. The catalytic signal amplification conferred by photocatalytic target ID has allowed for the identification of multiple protein targets and off targets across multiple drug classes and cellular compartments where established PAL have not been successful. The unequivocal target ID provided by our quantitative proteomic analysis far out strips typical efforts in the field, providing a simple and visual measure of target engagement. As such, we envision that photocatalytic target ID will find immediate use in providing a deeper biological understanding of efficacy target networks, quickly revealing off-target pharmacology, and ultimately driving pharmacotherapy forward against novel targets within drug discovery programs in both academic and industrial settings.
Supplementary Material
Acknowledgments
The authors thank Saw Kyin and Henry H. Shwe at the Princeton Proteomics Facility. The authors thank Brande Thomas-Fowlkes and Xiaoping Zhang (MRL, Merck & Co., Inc., Kenilworth, NJ), for providing GPR40-HEK cells and running IP-1 assay, respectively. HEK-hA2aR cell line was received as a gift from Jeremy Presland (MRL, Merck & Co., Inc., Boston, MA). We acknowledge the use of Princeton’s Imaging and Analysis Centre, which is partially supported by the Princeton Centre for Complex Materials, a NSF/Materials Research Science and Engineering Centres program (DMR-1420541). We also acknowledge V.G. Vendavasi and the use of Princeton’s Biophysics Core Facility. We thank Antony Burton for assistance in performing confocal microscopy. We thank T.W. Muir and members of the Muir Laboratory for their advice and analytical support. Research reported in this publication was provided by the NIH National Institute of General Medical Sciences (NIGMS), the NIH (R35GM134897-01), the Princeton Catalysis Initiative, and gifts from Merck & Co., Inc. A.D.T. would like to thank the European Union’s Horizon 2020 research and innovation program under Marie Sklodowska-Curie Grant Agreement 891458. B.X.L. acknowledges Princeton University, E. Taylor, and the Taylor family for the Edward C. Taylor Fellowship for funding support. J.B.G. acknowledges the NIH for a postdoctoral fellowship (F32-GM133133-01). J.V.O. acknowledges the NSF Graduate Research Fellowship Program (DGE-1656466). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the NSF. This manuscript was deposited at a preprint to bioRxiv: https://doi.org/10.1101/2021.08.02.454797.
Footnotes
Reviewers: D.D., California Institute of Technology; and D.L., Harvard University.
Competing interest statement: A provisional US patent has been filed by A.D.T., C.P.S., and D.W.C.M. based in part on this work (62/982,366; 63/076,658). International Application PCT/US2021/019959. D.W.C.M. declares an ownership interest, and A.D.T. and C.P.S. declare an affiliation interest, in the company Dexterity Pharma LLC, which has commercialized materials used in this work. D.W.C.M. declares an ownership interest in Penn PhD, which has commercialized materials used in this work.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2208077119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Arrowsmith J., Miller P., Trial watch: Phase II and phase III attrition rates 2011-2012. Nat. Rev. Drug Discov. 12, 569 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Chan J. N. Y., Nislow C., Emili A., Recent advances and method development for drug target identification. Trends Pharmacol. Sci. 31, 82–88 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Schenone M., Dančík V., Wagner B. K., Clemons P. A., Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 9, 232–240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hart C. P., Finding the target after screening the phenotype. Drug Discov. Today 10, 513–519 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Williams M., Target validation. Curr. Opin. Pharmacol. 3, 571–577 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Ziegler S., Pries V., Hedberg C., Waldmann H., Target identification for small bioactive molecules: Finding the needle in the haystack. Angew. Chem. Int. Ed. Engl. 52, 2744–2792 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Morgan P., et al. , Impact of a five-dimensional framework on R&D productivity at AstraZeneca. Nat. Rev. Drug Discov. 17, 167–181 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Schirle M., Bantscheff M., Kuster B., Mass spectrometry-based proteomics in preclinical drug discovery. Chem. Biol. 19, 72–84 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Wagner B. K., Clemons P. A., Connecting synthetic chemistry decisions to cell and genome biology using small-molecule phenotypic profiling. Curr. Opin. Chem. Biol. 13, 539–548 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butcher E. C., Berg E. L., Kunkel E. J., Systems biology in drug discovery. Nat. Biotechnol. 22, 1253–1259 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Schürmann M., Janning P., Ziegler S., Waldmann H., Small-molecule target engagement in cells. Cell Chem. Biol. 23, 435–441 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Simon G. M., Niphakis M. J., Cravatt B. F., Determining target engagement in living systems. Nat. Chem. Biol. 9, 200–205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawatani M., Osada H., Affinity-based target identification for bioactive small molecules. MedChemComm 5, 277–287 (2014). [Google Scholar]
- 14.Hill J. R., Robertson A. A. B., Fishing for drug targets: A focus on diazirine photoaffinity probe synthesis. J. Med. Chem. 61, 6945–6963 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Ge S.-S., et al. , Current advances of carbene-mediated photoaffinity labeling in medicinal chemistry. RSC Adv. 8, 29428–29454 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith E., Collins I., Photoaffinity labeling in target- and binding-site identification. Future Med. Chem. 7, 159–183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi H., Zhang C.-J., Chen G. Y. J., Yao S. Q., Cell-based proteome profiling of potential dasatinib targets by use of affinity-based probes. J. Am. Chem. Soc. 134, 3001–3014 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Zuhl A. M., et al. , Chemoproteomic profiling reveals that cathepsin D off-target activity drives ocular toxicity of β-secretase inhibitors. Nat. Commun. 7, 13042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito T., et al. , Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Wright M. H., Sieber S. A., Chemical proteomics approaches for identifying the cellular targets of natural products. Nat. Prod. Rep. 33, 681–708 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murale D. P., Hong S. C., Haque M. M., Lee J.-S., Photo-affinity labeling (PAL) in chemical proteomics: A handy tool to investigate protein-protein interactions (PPIs). Proteome Sci. 15, 14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X., et al. , Target identification of natural medicine with chemical proteomics approach: Probe synthesis, target fishing and protein identification. Signal Transduct. Target. Ther. 5, 72 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J., Koh M., Koo J. Y., Lee S., Park S. B., Investigation of specific binding proteins to photoaffinity linkers for efficient deconvolution of target protein. ACS Chem. Biol. 11, 44–52 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Hayashi T., Hamachi I., Traceless affinity labeling of endogenous proteins for functional analysis in living cells. Acc. Chem. Res. 45, 1460–1469 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Yamaura K., Kiyonaka S., Numata T., Inoue R., Hamachi I., Discovery of allosteric modulators for GABAA receptors by ligand-directed chemistry. Nat. Chem. Biol. 12, 822–830 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Fujishima S. H., Yasui R., Miki T., Ojida A., Hamachi I., Ligand-directed acyl imidazole chemistry for labeling of membrane-bound proteins on live cells. J. Am. Chem. Soc. 134, 3961–3964 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Herner A., et al. , 2-Aryl-5-carboxytetrazole as a new photoaffinity label for drug target identification. J. Am. Chem. Soc. 138, 14609–14615 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato S., Nakamura H., Ligand-directed selective protein modification based on local single-electron-transfer catalysis. Angew. Chem. Int. Ed. Engl. 52, 8681–8684 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Sato S., Tsushima M., Nakamura H., Target-protein-selective inactivation and labelling using an oxidative catalyst. Org. Biomol. Chem. 16, 6168–6179 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Geri J. B., et al. , Microenvironment mapping via Dexter energy transfer on immune cells. Science 367, 1091–1097 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filippakopoulos P., et al. , Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shorstova T., Foulkes W. D., Witcher M., Achieving clinical success with BET inhibitors as anti-cancer agents. Br. J. Cancer 124, 1478–1490 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurzawa N., et al. , A computational method for detection of ligand-binding proteins from dose range thermal proteome profiles. Nat. Commun. 11, 5783 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyler D. S., et al. , Click chemistry enables preclinical evaluation of targeted epigenetic therapies. Science 356, 1397–1401 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu W., et al. , CD166 plays a pro-carcinogenic role in liver cancer cells via inhibition of FOXO proteins through AKT. Oncol. Rep. 32, 677–683 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Tan Y., et al. , Inhibition of BRD4 suppresses tumor growth in prostate cancer via the enhancement of FOXO1 expression. Int. J. Oncol. 53, 2503–2517 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z., et al. , “Minimalist” cyclopropene-containing photo-cross-linkers suitable for live-cell imaging and affinity-based protein labeling. J. Am. Chem. Soc. 136, 9990–9998 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Shah N. P., et al. , Overriding imatinib resistance with a novel ABL kinase inhibitor. Science 305, 399–401 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Karaman M. W., et al. , A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 26, 127–132 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Hantschel O., et al. , The Btk tyrosine kinase is a major target of the Bcr-Abl inhibitor dasatinib. Proc. Natl. Acad. Sci. U.S.A. 104, 13283–13288 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasta J. D., et al. , Quantitative, wide-spectrum kinase profiling in live cells for assessing the effect of cellular ATP on target engagement. Cell Chem. Biol. 25, 206–214.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedman Ohana R., et al. , Streamlined target deconvolution approach utilizing a single photoreactive chloroalkane capture tag. ACS Chem. Biol. 16, 404–413 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Dumka D., et al. , Activation of the p38 Map kinase pathway is essential for the antileukemic effects of dasatinib. Leuk. Lymphoma 50, 2017–2029 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams N. K., Lucet I. S., Klinken S. P., Ingley E., Rossjohn J., Crystal structures of the Lyn protein tyrosine kinase domain in its Apo- and inhibitor-bound state. J. Biol. Chem. 284, 284–291 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Gottesman M. M., Fojo T., Bates S. E., Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat. Rev. Cancer 2, 48–58 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Ruzickova E., Skoupa N., Dolezel P., Smith D. A., Mlejnek P., The lysosomal sequestration of tyrosine kinase inhibitors and drug resistance. Biomolecules 9, 675 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klaeger S., et al. , Chemical proteomics reveals ferrochelatase as a common off-target of kinase inhibitors. ACS Chem. Biol. 11, 1245–1254 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Wang T.-H., Wang H.-S., Soong Y.-K., Paclitaxel-induced cell death: Where the cell cycle and apoptosis come together. Cancer 88, 2619–2628 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Yang C.-P. H., Yap E.-H., Xiao H., Fiser A., Horwitz S. B., 2-(m-Azidobenzoyl)taxol binds differentially to distinct β-tubulin isotypes. Proc. Natl. Acad. Sci. U.S.A. 113, 11294–11299 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Comess K. M., et al. , Emerging approaches for the identification of protein targets of small molecules – A practitioners’ perspective. J. Med. Chem. 61, 8504–8535 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Helbig A. O., Heck A. J. R., Slijper M., Exploring the membrane proteome—Challenges and analytical strategies. J. Proteomics 73, 868–878 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Sriram K., Insel P. A., G protein-coupled receptors as targets for approved drugs: How many targets and how many drugs? Mol. Pharmacol. 93, 251–258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hyde A. M., et al. , Synthesis of the GPR40 partial agonist MK-8666 through a kinetically controlled dynamic enzymatic ketone reduction. Org. Lett. 18, 5888–5891 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Defossa E., Wagner M., Recent developments in the discovery of FFA1 receptor agonists as novel oral treatment for type 2 diabetes mellitus. Bioorg. Med. Chem. Lett. 24, 2991–3000 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Cekic C., Linden J., Adenosine A2A receptors intrinsically regulate CD8+ T cells in the tumor microenvironment. Cancer Res. 74, 7239–7249 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cunha R. A., Ferré S., Vaugeois J.-M., Chen J.-F., Potential therapeutic interest of adenosine A2A receptors in psychiatric disorders. Curr. Pharm. Des. 14, 1512–1524 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muranaka H., Momose T., Handa C., Ozawa T., Photoaffinity labeling of the human A2A adenosine receptor and cross-link position analysis by mass spectrometry. ACS Med. Chem. Lett. 8, 660–665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang X., et al. , An affinity-based probe for the human adenosine A2A receptor. J. Med. Chem. 61, 7892–7901 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zocchi C., et al. , The non-xanthine heterocyclic compound SCH 58261 is a new potent and selective A2a adenosine receptor antagonist. J. Pharmacol. Exp. Ther. 276, 398–404 (1996). [PubMed] [Google Scholar]
- 60.Kudlacek O., et al. , The human D2 dopamine receptor synergizes with the A2A adenosine receptor to stimulate adenylyl cyclase in PC12 cells. Neuropsychopharmacology 28, 1317–1327 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.