Abstract
Screening of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) among symptomatic and asymptomatic patients offers unique opportunities for curtailing the transmission of novel coronavirus disease 2019, commonly known as COVID-19. Molecular diagnostic techniques, namely reverse transcription loop-mediated isothermal amplification (RT-LAMP), reverse transcription-polymerase chain reaction (RT-PCR), and immunoassays, have been frequently used to identify COVID-19 infection. Although these techniques are robust and accurate, mass testing of potentially infected individuals has shown difficulty due to the resources, manpower, and costs it entails. Moreover, as these techniques are typically used to test symptomatic patients, healthcare systems have failed to screen asymptomatic patients, whereas the spread of COVID-19 by these asymptomatic individuals has turned into a crucial problem. Besides, respiratory infections or cardiovascular conditions generally demonstrate changes in physiological parameters, namely body temperature, blood pressure, and breathing rate, which signifies the onset of diseases. Such vitals monitoring systems have shown promising results employing artificial intelligence (AI). Therefore, the potential use of wearable devices for monitoring asymptomatic COVID-19 individuals has recently been explored. This work summarizes the efforts that have been made in the domains from laboratory-based testing to asymptomatic patient monitoring via wearable systems.
Keywords: Asymptomatic, COVID-19, Screening, Wearable systems, Machine learning
1. Introduction
Since the first detection, the novel coronavirus disease 2019, otherwise known as COVID-19 has posed a dire threat worldwide, infecting over 500 million people so far, with 6.19 million causalities, up until April 17, 2022 [1]. Healthcare facilities have been overwhelmed by the sudden rise of COVID-19 patients, who need proper isolations to prevent the virus from spreading to healthy people. However, due to the high air-born transmissibility of COVID-19, along with its asymptomatic viral shedding, the healthcare systems have faced unprecedented challenges. Nevertheless, healthcare professionals have tried their best to manage the crises with limited facilities. Overall, providing quarantine and essential healthcare facilities to a large number of infected individuals has become one of the major challenges. Hence, extensive research has been carried out to tackle the challenges, especially with the emergence of new variants.
COVID-19 is currently detected using the reverse transcription-polymerase chain reaction (RT-PCR), which can detect the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pathogen in patients 3 days after infection [2]. However, RT-PCR-based detection is challenged by its limited capacity in that it depends upon a laboratory to test the samples [3]. Besides, RT-PCR tests are rarely able to identify pre-symptomatic patients and have difficulties identifying early-stage patients due to the sensitivity challenges of the ribonucleic acid (RNA) copy number [4]. Another reliable detection method is reverse transcription loop-mediated isothermal amplification (RT-LAMP). Yan et al. developed an RT-LAMP assay that targeted the S gene and ORF1a gene with a 100% detection sensitivity and specificity for 130 clinical samples in just about 30 min [5]. However, processing the samples requires costly reagents and laboratory equipment. On the other hand, clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (CAS) systems can detect the pathogen within 50 min [6]; however, the reduced sensitivity of this assay has been a major hindrance in the mass-scale application. The other detection methods, such as immunoassay and biosensor-based identification, have also faced severe issues due to low throughput, inaccessibility, and limited scope in analyzing large populations at once [7,8].
Due to the infectious nature, early identification of COVID-19 infection can help to reduce transmission and provide the patients with the right treatment to avoid further complications. Thus, inexpensive, faster, and more accessible mass screening methods are urgently required to identify COVID-19 patients in real-time to prevent the transmission of the virus. An intelligent identification and monitoring system using wearable devices for earlier identification of asymptomatic COVID-19 infection is considered a top-tier solution for containing the transmission. Numerous well-developed commercial wearable devices are now available, namely the Apple Watch [9], Fitbit [10], Zephyr BioHarness [11], WHOOP Strap [12], BioButton [13], Garmin [14], and smart ring [15]. Most of these have the functionality to measure heart rhythm rate, sleeping hours, steps, temperature, pure body temperature, and arterial oxygen saturation [16]. Therefore, using these devices to design an intelligent monitoring and detection system can be a cost-effective and faster solution for screening asymptomatic COVID-19 patients.
The primary objective of this work is to provide a comprehensive overview of different COVID-19 detection systems and their effectiveness in different scenarios. In this review, we start with a history of SARS-CoV-2 and then discuss contemporary molecular biology detection techniques used for screening symptomatic COVID-19 patients, focusing on their analysis strategies as well as their pros and cons. We also reviewed currently available intelligent detection and monitoring systems for early detection of asymptomatic COVID-19 patients using real-time vital data captured by wearable devices. This work is aimed to assist the researchers in exploring different COVID-19 screening methods and gaining a better understanding of the prospects and challenges as well.
2. Origin of SARS-CoV-2
The first case of SARS-CoV-2 was reported in Wuhan, Hubei Province, China, in December 2019 [17], and it took a few weeks for SARS-CoV-2 to reach most countries of the world due to travel and community interactions [18]. Initially, a sudden outbreak of pneumonia in Wuhan was documented in the World Health Organization (WHO) China office in December 2019 [19]. The cause of the disease was an unknown and unidentified pathogen among 44 patients, 11 of whom were in severe condition on January 5, 2020. Five days later, on 10 January, the WHO announced the first guideline for the new coronavirus [20,21].
The novel strain of the coronavirus was named SARS-CoV-2 in February 2020 based on phylogenetic test criteria [22]. The sequencing of the genome of this virus showed that the SARS-CoV-2 has approximately 30,000 bases [23,24] and belongs to Nidovirales in the family Coronaviridae. Furthermore, based on phylogenetic clustering, coronaviruses have been classified into alpha, beta, gamma, and delta-coronaviruses [25]. It was established that alpha and beta-coronaviruses mainly infect mammals, including humans, while gamma and delta-coronaviruses infect birds (Fig. 1 ) [26]. Moreover, the family of coronaviruses, such as Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) can cross the species barrier and lead to serious human diseases [26]. Hence, it has been considered the most likely scenario for SARS-CoV-2 as well.
Fig. 1.
Transmission of coronavirus by crossing the species barrier [26].
The rapid spread of COVID-19 has affected people all over the world, with the number of people infected by the novel coronavirus increasing daily. After the genomic sequencing of SARS-CoV-2 [27], the WHO declared the disease as “the first 21st-century pandemic” on March 11, 2020 due to its rapid global infection level [17,18]. On 13 January, Thailand reported the first case outside China [28], and by the end of the month, the WHO had reported 7818 global in 18 countries, with 170 casualties in China [18]. In less than six months from the first detection, SARS-CoV-2 went from being an unknown pathogen to becoming prevalent in practically all countries worldwide. Some of these countries suffered a second wave, and many countries, including India, Russia, Brazil, and several African nations are still on their third wave [[29], [30], [31]]. On February 11, 2020, WHO named the etiologic agent of the disease SARS-CoV-2 [32], which is in the same family of coronaviruses as was reported for MERS in 2012 and the earlier strain of SARS in 2003. The number of causalities from COVID-19 appeared much higher than with earlier coronaviruses due to its fast spread and pathogenicity [29]. The primary reason for the large-scale prevalence of SARS-CoV-2 transmission is due to the transmission of active viral particles from asymptomatic patients, who act as a reservoir for the dispersal of this disease on an exponential scale [33].
3. Laboratory-based detection methods
The detection of symptomatic COVID-19 patients was carried out with different molecular biology techniques but the use of RT-PCR remained on top priority in clinics and labs since the emergence of this disease. Other laboratory-based SARS-CoV-2 detection approaches were useful too for their high accuracy in the detection of COVID-19 patients, however, based on the available resources and testing time, all other methods except RT-PCR have been hardly used for mass testing.
3.1. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Reverse transcription-polymerase chain reaction (RT-PCR) is the most popular molecular-based technique to detect SARS-CoV-2 RNA in samples from patients with compatible signs and symptoms (dry cough, fatigue, fever, lymphopenia, pneumonia, myalgia, dyspnea, sneezing, and chills) [34]. This technique can detect a few copies of RNA from clinical and/or environmental samples as it is highly sensitive and has been proven to be effective for pathogen detection [35]. Due to better sensitivity and accuracy, RT-PCR is a well-established assay for detecting SARS-CoV-2. Different testing laboratories designed and developed this test, and there have been several modifications (Table 1 ) [36], many of which have been approved by the WHO for the laboratory testing of samples from suspected COVID-19 patients.
Table 1.
RT-qPCR assays commonly used for COVID-19 diagnosis.
| Institute | Target | Primer/Probe# | Reference |
|---|---|---|---|
| Charite | E | E_Sarbeco_F | Corman et al. (2020) |
| E_Sarbeco_R | |||
| E_Sarbeco_P1 | |||
| RdRp | RdRp_SARSr-F | ||
| RdRp_SARSr-R | |||
| RdRp_SARSr-P1 | |||
| RdRp_SARSr-P2 | |||
| HKU | N | HKU-N-F | Chu et al. (2020) |
| HKU-N-R | |||
| HKU-N-P | |||
| Nsp14 | HKU-ORF1-F | ||
| HKU-ORF1-R | |||
| HKU-ORF1-P | |||
| China CDC | N | CCDC-N-F | China CDC (2020) |
| CCDC-N-R | |||
| CCDC-N-P | |||
| nsp10 | CCDC-ORF1-F | ||
| CCDC-ORF1-R | |||
| CCDC-ORF1-P | |||
| US CDC | N | 2019-nCoV_N1–F | CDC, 2020 |
| 2019-nCoV_N1-R | |||
| 2019-nCoV_N1–P | |||
| N | 2019-nCoV_N2–F | ||
| 2019-nCoV_N2-R | |||
| 2019-nCoV_N2–P | |||
| N | 2019-nCoV_N3–F | ||
| 2019-nCoV_N3-R | |||
| 2019-nCoV_N3–P | |||
| Human RNase P | RP-F | ||
| RP-R | |||
| RP-P |
#: Primer/Probe sequences not included. Adapted from Vogels et al. [36].
RT-PCR became the benchmark for the diagnosis of COVID-19 patients, enabling researchers to identify COVID-19-positive patients who had been recently discharged from hospitals [37]. However, there have been controversies about the chemicals, procedures, and analytical techniques used in RT-PCR assays that have given rise to doubts over the accuracy and reliability of using RT-PCR to test for SARS-CoV-2. RT-PCR assay shows a cycle threshold amplification curve value which manifests that RNA is amplified (detectable indicator) and this is referred to as the quantitative cycle (Ct) [34,35]. A Ct value ranging between 25 and 28 is considered suitable and a Ct value greater than 32 is not considered accurate in terms of the nonspecific amplification of the target RNA. A clinical sample that has a relatively lower Ct value shows a higher RNA/DNA copy number and it is thus considered positive, demonstrating that there is an inverse correlation between the Ct value and the DNA/RNA copy number in the template of a given sample. The Ct value of the RT-PCR reaction varies considerably based on the type of specimens, quality of the sample, the protocol followed by a technician, and the thermal cycler brand or model itself [36].
It is worth explaining here that RT-PCR-based testing fails to detect RNA from the collected samples from patients that have well-known COVID-19 symptoms. Sometimes, a person can be SARS-CoV-2 positive by RT-PCR-based test albeit he/she does not depict any COVID-19 symptoms. This means RT-PCR results can be false positive or false negative [38]. In such circumstances, two consecutive negative test results are required to declare that person is not infected with the virus. The accuracy and reliability of the results depend upon several factors and especially on the expertise of the lab technician and the quality of reagents (RT-PCR kits) used in the reaction. As far as the quality of different RT-PCR is concerned, a study was carried out by Puck et al. to validate the usefulness of reagents of different company RT-PCR assay kits. The team evaluated the analytical performance of qRT-PCR kits purchased from seven different companies, including KH Medical, BGI, Seegene, R-Biopharm AG, Altona Diagnostics, CerTest Biotec, and Primer Design, to demonstrate their effectiveness [39]. They showed that all the kits used in this study are reliable enough to be used by experienced diagnostic laboratories for the daily diagnosis of COVID-19 patients.
3.2. Reverse Transcriptase-Loop Mediated Amplification (RT-LAMP)
The RT-PCR reaction-based detection of SARS-CoV-2 is comparatively accurate and highly sensitive, but the prolonged testing time (∼2 h) means that there is a need for a similar analytical platform for the mass-scale monitoring of COVID-19-positive and negative cases [40]. It could hereby be replaced by reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) reaction testing, which can amplify RNA/DNA in a very short time [41,42]. Owing to its robustness and accuracy, this technique is largely used for the detection of pathogens, especially viruses, bacteria, or malaria [[43], [44], [45]]. The difference between the RT-LAMP reaction and RT-PCR is that it occurs at a uniform temperature and requires 4–6 primers, giving it high accuracy and improved efficacy [42]. The reaction is usually performed at 60–65 °C by adding four primers [41] and through the subsequent addition of two-loop primers, the reaction time can be further reduced to half [46]. The use of WarmStart RTx in the RT-LAMP reaction allows reverse transcription in a single step-reaction [47]. Since RNA-CoV‐2 is a 30 kb [48] long reverse transcription virus, using a single reverse transcription (RT) and LAMP reaction tremendously reduces the reaction time. In addition, the reaction can be performed using unpurified RNA, which also contributes to the fast processing of samples and the detection of the virus [47].
Due to the high accuracy and fast turnout, the RT-LAMP reaction has been used by many researchers for testing the clinical samples from suspected individuals. For instance, Huang et al. detected SARs-CoV-2 using RT-LAMP by considering that it requires a constant temperature (65 °C) and can be completed in only 20 min. The team found that RT-LAMP can detect 80 copies of viral RNA and the results can be visualized by a simple color-change confirming viral RNA amplification in the reaction [47]. In addition, Thi et al. developed an RT-LAMP assay without using a prior RNA isolation step from clinical samples and assay results showed high specificity (99.50%); however, very reduced sensitivity (86% for Ct < 30) compared to RT-PCR [49]. Jiang et al. also recommended the use of RT-LAMP by performing a rapid RT-LAMP reaction, concluding that this assay could considerably increase laboratory testing capacities by processing twice the clinical samples compared to qRT-PCR. Thus, this reaction can potentially replace RT-PCR in critical situations, especially when sample quantity is very high and there is a need to reduce the cost of the assay as it does not require a costly thermocycler [50].
3.3. CRISPR-Cas system
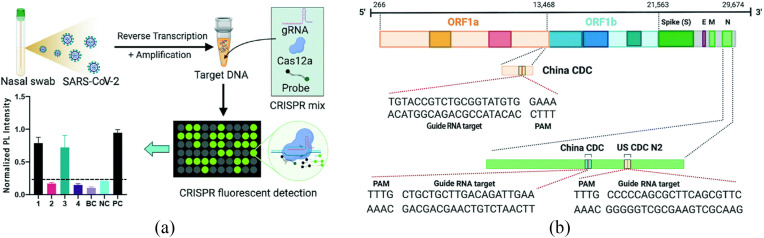
Clustered regularly interspaced short palindromic repeats (CRISPR) is a promising technology for gene editing and is used for trimming, cutting, replacing, or adding DNA sequences. Due to their functionality, CRISPR/Cas proteins are also termed “molecular scissors” [51]. In recent years, CRISPR and its associated proteins, especially Cas12a and Cas13 have been applied for diagnostics purposes for their ability to recognize specific nucleic acid sequences. The proteins used in CRISPR are combined with fluorophore quencher DNA samples which generate a signal amplification once Cas13 and Cas12a bind to the target sequences during the process, guided by RNA (GRNA) sequences. For instance, in one study, Cas12a or Cas13 was used to predict the sequence of COVID-19, and then the reporter molecule was cleaved to confirm that the samples were contaminated with the virus [52]. CRISPR technology is used for the detection of SARS-CoV-2 RNA when this technology is combined with other approaches such as recombinase polymerase amplification (RPA), RT-LAMP, and RT-PCR reaction. During the reaction, the Cas12 protein cuts the DNA into two strands. Furthermore, Cas12a along with crRNA binds with the target sequences and cleaves the DNA into single-sanded DNA (ssDNA) [53]. For instance, Huang et al. used the RT-PCR/CRISPR-Cas12a fluorescence diagnostics method which completed the detection of viral RNA in three steps, namely RNA extraction, target amplification, and fluorescent sensing – termed the CRISPR-FDS method (Fig. 2 ) [54]. This method was found immensely useful in terms of its sensitivity as it can detect five copies of RNA and can be completed within 50 min. Chiu et al. used another highly sensitive, robust, and mobile CRISPR-Cas12 technique for detecting SARS CoV-2. The reaction includes the RT-LAMP amplification of the target SARS-CoV-2 RNA within 20–30 min for the samples collected in the form of respiratory swabs. The reaction involved cleavage of target sequences by Cas12 initially for 10 min and then RT is performed at 62 °C with the use of the human RNase-P gene, N genes, and E gene [55].
Fig. 2.
CRISPR-based diagnostic system using fluorescence for the detection of SARS-CoV-2. (a) Schematic chart of a CRISPR-FDS assay to detect SARS-CoV-2 RNA; (b) SARS-CoV-2 genomic map of COVID-19 CRISPR-FDS targeted sequences [54].
One of the uniqueness of CRISPR-based RNA diagnostic techniques is that this platform has solved the sensitivity challenges faced by other molecular biology techniques. For example, CRISPR/Cas 13a platform developed by Zhang et al. displayed one million-fold improved sensitivity resulting in solving the issues of detection of Zika virus (ZIKV) or dengue virus from very low quality at a level of 1 pc per microliter [56]. Zhang et al. used SHERLOCK and the CRISPR-Cas13 method for detecting the RNA extracted from SARS-CoV-2 and found that this method can detect 10–100 copies of RNA per microliter. Furthermore, it was established that this approach is easy to use, highly sensitive, and robust, as it needs 1 h to complete [57].
3.4. Immunoassays for Point of Care testing (POCT)
POCT refers to the antigen and antibody-based detection of SARS-CoV-2 [58,59]. The SARS-CoV-2 antigen is present when a person is infected, and antigen-based detection is less sensitive compared to immunoassay techniques [60]. On the other hand, the antibody-based detection of SARS- CoV-2 relies on the immune response and is a much better method for detecting infection [61]. However, like other molecular biology techniques, it is also susceptible to false negatives. The screening of antibodies mainly involves the detection of Immunoglobulin M (IgM) and Immunoglobulin G (IgG). Once SARS-CoV-2 enters the body, it multiplies over one week; after a lapse of 1–3 weeks or more [62], the immune system responds to the viral antigens and starts generating adequate amounts of IgG antibodies. The IgG concentration in the serum is much higher than that of IgM after a SARS-CoV-2 infection [63]. Studies have proved that immunoassays are very effective methods that rely on antibody production and serve as alternative options for detecting SARS-CoV-2 at different phases of viral infection [64].
Based on various identification methodologies, immunoassays are categorized into chemiluminescence (enzyme-associated immunosorbent assay), immunosorbent enzyme assay (ELISA), and colloidal gold/immunofluorescence (lateral immunoassay, LFA). Lateral immunoassays can be performed with a very short turnaround of 10–15 min, making them time and cost-efficient. On the other hand, the chemiluminescence and ELISA techniques are more specific and sensitive, although they are time-consuming as these require complex protocols for detecting SARS-CoV-2 [65,66].
3.4.1. Lateral flow-based tests
Benjamin D. Grant et al. used a half-strip lateral flow assay method that was developed for POCT SARS-CoV-2 antigen detection [67] with a detection limit of 0.65 ng mL−1 recombinant antigen. The pH, antigen concentration, and various other lateral flow immunoassay strip parameters were optimized by Wen et al., producing specific and stable detection results for SARS-CoV-2 IgG and IgM antibodies [68]. In comparison to the RT-PCR results, the identification of IgM was 100% and 93.3% specific. Examining clinical samples, Zhenhua Chen et al. used the fluorescent reporter of lanthanide-doped polystyrene nanoparticles (LNPs) and detected anti-SARs-CoV-2 IgG. The results were consistent with those of RT-PCR [69]. Fig. 3 illustrates the assays of the abovementioned studies.
Fig. 3.
Lateral flow assays. (a) Structure and detection process of the LFIA strip [68]. (c) Design of developed assay by Chen et al. [69].
LFA has been widely used to detect COVID-19 as it uses low-cost equipment and has fewer constraints in terms of environmental requirements. It can be carried in a small package and used for on-spot testing, which expands its usability. However, any contaminant can sabotage the results since the samples are tested without any intermediate processing. It is, therefore, necessary that reagent formulations are adjusted to strengthen the anti-interference abilities to optimize the processing of band materials so that the filtering ability of the interfering substances can be improved.
3.4.2. The enzyme-linked immunosorbent assay (ELISA)-based tests
The ELISA microfluidic sandwich system for SARS-CoV-2 antibodies was proposed by Siddhartha Tripathi and Ami Agrawal, who separated plasma from blood using a T-shaped microchannel on a microfluidic chip [70]. SARS-CoV-2 ELISA plasma was used to detect the virus. The testing involved the isolation of about 10 μL of plasma from whole human blood within approximately 3 min. Meanwhile, Xudong Fan's group developed an ELISA microfluidic method to measure SARS-CoV-2 quantitatively using serum IgG and antigen-S viral protein. In this study, a 12-channel capillary sensor array was used with the ELISA reactor, and the microfluidic chip was operated using an automated system [71,72]. The detection limits were tested with humanized chimeric SARS-CoV-2 antibodies, with 2 ng mL−1 a limit of detection (LOD).
A capillary flow assay (MCFA) platform was developed by Sthitodhi Ghosh et al. using an MCFA chip, an optical sensor, and a smartphone. This platform enables ELISA detection based on chemiluminescence. The system has an on-chip capillary pump and the sample is driven amidst the channels and detection areas by capillary force [73]. The ELISA method can identify the SARS-CoV-2 pathogen quantitatively and shows better performance compared to the LFA procedure. Moreover, it has lower requirements for detection environment than the detection methods using nucleic acid. Due to the complex operating steps, however, many researchers have managed to achieve only parts of ELISA's operation or chemiluminescence using microfluidic methods. Furthermore, few have achieved POCT with ELISA or chemiluminescence as a fully automated process.
3.5. Biosensor-based detection
Besides the above methods, rapid and sensitive techniques for identifying SARS-CoV-2 as an alternative to POCT systems are also required [58]. Recently, the potential of miniature biosensors as analytic platforms has been demonstrated based on their unique characteristics including sensitivity, reliability, and rapid diagnoses [74]. A biosensor can determine the presence of an intruder object and can provide feedback upon detection, generally, in form of an optical or electrical signal. The main three components of a biosensor are a bioreceptor, a transducer, and a signal amplifier or analyzer. The continued advancements in the field of nanoscience have improved sample-to-response time for devices with a greater signal-to-noise ratio. Biosensor-based pathogen detection techniques have shown promise for COVID-19 detection and can be a viable alternative to the current sensors in wearable devices [75].
3.5.1. Plasmonic biosensors
Plasmonic biosensors are significant and crucial POCT tools based on Localized Surface Plasmon Resonance (LSPR) [76]. In combination with the surface functionalization procedures, plasmonic nanostructures can enable fast and real-time reagent identification even in ultra-low concentrations. In the field of materials science, developments have now enabled nanomaterials with tunable plasmonic properties and sensitivities to be precisely controlled [77]. Sophisticated nano-fabrication methodologies have opened opportunities for engineering nanometric arrays on different substrates as plasmonic detecting equipment [78]. Moitra et al. presented a gold-nanoparticles (AuNPs) colorimetric test that was tailored to caps of Thiol-modified oligonucleotides (ASOs). This allows the N gene (nucleocapsid phosphoprotein) to be specifically and quickly (within 10 min) detected in samples infected by SARS-CoV-2 by spectral resonance changes [79]. Interestingly, RNaseH was also integrated by the authors to split the RNA-DNA hybrid strand into a noticeable naked-eye test that plays a significant role in bordering among AuNPs. Qiu et al. used the combinational mechanisms of plasmonic photothermal and LSPR sensing through functionalization with complementary DNA receptors [80]. Based on earlier investigations, Murugan et al. presented a plasmonic biosensor technique based on fiber-optic absorbance for using directly on saliva to provide a 1-stage wash-free monitoring platform for SARS-CoV-2 [81]. The authors considered two different types of biosensors as a conceptual idea to identify the SARS-CoV-2 N protein within 15 min, which can meet the current urgent demand for quick and low-cost diagnosis. Funari et al. devised an LSPR-based microfluidic chip to identify SARS-CoV-2 antibodies [82]. During the antibody-antigen attachment, the highest resonant wavelength shift from the gold nano spikes is encountered. With the limit of detection of around 0.08 ng mL−1, the result can be obtained within 30 min (per 0.5 pM), making it easier, cheaper, and faster to carry out a quantitative SARS-CoV-2 diagnosis.
3.5.2. Electromechanical biosensors
Due to the ease of miniaturization, low cost, and simplicity, electromechanical biosensors have recently attracted attention. Generally, a customized electrode is used in such sensors to act as the receptor or transducer for precise and real-time monitoring of the target. With the presence of the analyte of interest, a signal is generated in the sensing electrode [83]. Tripathy et al. proposed an electrodeposited AuNP-based electrical biosensor for detecting COVID-19 [84].
Researchers have recently also investigated alternative electrochemical solutions that further enhance the detection capacity and pave the way for reductions in operation or overall costs. Md. Ali and others created a 3D reduced graphene oxide electrode using an advanced 3D printing method and integrated it as an electrochemical sensor with a microflow device [85]. The viral antigens used to detect sensitive antibodies specific to SARS-CoV-2 through such 3D electrodes have a detection limit as low as 2.8-10−15 M. To achieve a miniature electrochemical sensor, Fabiani et al. applied magnet beads to a black carbon electrode [86]. The external magnetic field could offer benefits thanks to these magnetic beads - removing the wash step while preserving superior characteristics such as valid and accurate detection and promoting the pre-concentration process, etc. Two other studies have recently also introduced an electrochemical sensor substratum because of the benefits of low cost and ease of handling and disposal. Using printed electrode patterns on paper, the electric chemicals device proposed by Yakoh et al. can identify the antibodies of SARS-CoV-2 in 30 min [87]. The paper-based sensor is a potential POCT platform with acceptable sensitivity and specificity and particularly has a uniquely disposable, portable, and cost-effective nature. Moreover, Alafeef et al. proposed a device for nucleic acid testing by immobilizing the samples on a paper-based platform [88]. By meticulously designing the essential sensing materials, namely gold nanoparticles, the sensor improves sensitivity and output signals by 5 min. The sensor can also identify the targets within 585.4 μL−1 to 5854 μL−1 copies per μL range with an initial infected validation (copies per μL)−1, revealing its capacity to prove the progression of SARS-CoV-2 infection. A convenient data transmission system to facilitate fast results for end-users and at-home diagnostics is another key point for this detection mode. The smartphone-based detection of SARS-CoV-2 RNA was shown to be feasible by Zhao et al. [89]. Such a “plug-and-play” technique would provide end-users with a portable channel to easily evaluate the test results. Moreover, a recent study used mass graphene electrodes and combined a wireless module with the electrochemistry platform to rapidly identify COVID-19 [90]. This system is called SARS-CoV-2 RapidPlex and its low-cost detection along with high sensitivity makes it a potential home-testing system.
4. Non-laboratory-based detection methods
The understanding of the exact epidemiology of the disease and its subsequent impact on humans is realized through the current SARS-CoV-2 outbreak [91]. To gain complete control over this disease, it is crucial to achieve the early detection of viral infection [92]. The third wave of the COVID-19 pandemic has brought some unprecedented challenges hindering the timely data collection and dissemination, thereby hampering the efforts of public health planners and clinical managerial teams. Data from asymptomatic COVID-19 patients can be used for subclinical presentations and the planning of immediate steps before the symptoms manifest. It will also help in the accurate estimation of disease occurrence and the management of risk mitigation strategies, quarantine timing, and testing resources. To aid the screening of COVID-19 patients, the following asymptomatic detection methods have been used globally.
4.1. Smartwatch and wearable device-based COVID-19 detection
During the COVID-19 pandemic, hospital systems around the globe faced severe limitations in laboratory testing materials. It was very hard to cope with the laboratory testing of the enormous patient volume in the most populated areas during the outbreak of the first two waves of COVID-19. Moreover, some tests are highly sensitive and take a significant amount of time to process. In this scenario, the consumer-wearable devices, namely smartwatches and fitness trackers provided the perfect opportunity to monitor the baseline health parameters and detect any significant deviation to alert about a possible infection [16,[93], [94], [95]]. Among such wearable devices, smartwatches and activity trackers have become very common for daily use in developed and developing countries, which can characterize the resting heart rate (RHR) [96] and sleep [97] baselines of each user and thus, can monitor any fluctuations in these parameters [98].
Mishra et al. designed an elaborated study on a cohort of around 5300 volunteers [99]. They were registered in their study via a mobile app named MyPHD, which was developed by the team as well. Among this large cohort, there were several types of commercially available smartwatches (Fitbit, Apple Watch, Garmin, and others) users. The app synced health data, namely heart rate, steps count, and sleep duration, from their smartwatch and the users were prompted to provide any information about their symptoms, symptom onset date, diagnosis date, along with demographic information. Among the users, Fitbit constituted the highest percentage, and hence, they led their analysis based on 32 COVID-19 positive Fitbit users and found a correlation with the health data fluctuations with COVID-19 infection. They also evaluated the possibility of using such a system for possible COVID-19 infection even before the symptom onset. They also used the data to develop a real-time alerting system based on their statistical algorithm. In their presymptomatic COVID-19 detection experiment, 63% of the COVID-19 positive cases were detected using their algorithm before the onset of symptoms. Although the method is in the early stages, their findings suggest that continuous health monitoring using wearable devices on a large scale might be utilized for real-time COVID-19 detection, often in the pre-symptomatic stage [99]. Fig. 4 a illustrates one of the detection methods by the team employing the relation between heart rate and step count of the users.
Fig. 4.
Different physiological changes due to COVID-19 infection. (a) Deviation in the ratio of heart rate and steps before the onset of the COVID-19 symptoms (marked by the red dotted vertical line). The violet dotted vertical line denotes the COVID-19 diagnosis date and the red marker and star on the graph indicate the anomalous data and the first instance of the anomaly, respectively [99]. (b) Correlation of different symptoms with COVID-19 infection among 3811 untested, 54 COVID-19-positive, and 279 COVID-19-negative subjects. The asterisk denotes significant differences in symptoms (p-value < 0.05) [102].
Another work from Bogu et al. used a long short-term memory (LSTM)-based autoencoder to improve the anomalous RHR detection [100]. Their model detected 23 out of 25 COVID-19 infected subjects using smartwatch data where 14 of them were detected even before the symptom onset. On the same smartwatch dataset, Abir et al. employed LSTM-based variational autoencoder (VAE) architecture for anomalous RHR detection that improved the detection up to 25 out of 25 COVID-19 infected subjects [101]. Moreover, their model detected the possible COVID-19 infection in the presymptomatic phase for 20 out of 25 subjects.
Although these studies presented enormous promise for continuous monitoring, they are yet to be ready for real-world deployment. Mishra et al. collected smartwatch data, from a large cohort; however, only 25 COVID-19 subjects’ data were available for the studies of Bogu et al. and Abir et al. [[99], [100], [101]]. Hence, more collaborative efforts among different research groups and smartwatch manufacturers are required in this regard to collect global data from different demography that will make such models effective in the real-world scenario.
Body temperature can be another important physical parameter to monitor for possible COVID-19 infection. In a study, Richardson et al. showed that an elevated body temperature above 37.8 °C was recorded among 12% of COVID-19 positive individuals and 31% of hospitalized patients [103]. Chung et al. proposed a wrist-worn wearable device named HEARThermo, which monitored body surface temperature along with the heart rate. In most cases, the body surface temperature measured at terminal organs is not equal to the actual body temperature [104]. Hence, evaluating these two physical parameters, if the data seemed anomalous, the device would notify the user to take further thermal measurements using the thermometer. The device was given to 63 college students, 75 suspected patients, and 149 medical professionals. All these subjects’ data showed good test-retest reliability. Such a result suggests that the continuous monitoring of body temperature and heart rate via a wrist-worn device can be a viable option for point-of-care and early detection of SARS-CoV-2 infection.
Radin et al. explored the prospects of wearable devices for detecting influenza-like diseases at a mass level [105]. Quer et al. created a prospective app-based research platform, called DETECT (Digital Engagement and Tracking for Early Control and Treatment), wherein individuals could share their sensor data, self-reported symptoms, diagnoses, and electronic health records to improve the ability to identify and track individual- and population-level viral illnesses, including COVID-19 [102]. 30,529 participants were enrolled between 25 March and June 7, 2020, of whom 3811 reported symptoms. In this study, COVID-19 positive subjects were diagnosed via laboratory testing to ensure their infection. Fig. 4b depicts the correlations of different symptoms with the SARS-CoV-2 infection among positively tested, negatively tested, and untested subjects. According to Fig. 4b, fever chills or swelling, fatigue, breathing difficulty, decrease in smell, cough, and body ache are shown significantly higher in COVID-19 positive subjects compared to others. The combined evaluation of the self-reported symptom survey and wearable device data showed an area under the curve (AUC) of 0.80 with the interquartile range (IQR) of 0.73–0.86) for COVID-19 infection among the symptomatic patients. Moreover, the combination of both data showed particularly better detection (p-value < 0.01) than the symptom-based model where AUC was 0.71 with IQR of 0.63–0.79.
In another study within the quarantine facilities of Hong Kong, Wong et al. showed the usability of wearable biosensors for early screening of COVID-19 by monitoring several health parameters [106]. They proposed a randomized controlled trial on asymptomatic individuals ranging from 200 to 1000 who were in close contact with COVID-19 positive individuals. The volunteers were randomly divided into two groups for standard strategy and remote monitoring (intervention group) during the two-week quarantine. Both groups had symptoms and fever monitoring protocols; however, the latter group had biosensors on the arms for continuous monitoring of health parameters, namely blood pressure, respiration rate, activities, oxygen saturation, heart rate, and skin temperature. A mobile app named Biovitals Sentinel was used to sync the sensor data into a cloud database, where it was processed by an analytics engine. The result of this study reiterated that using wearable biosensors to continuously monitor multi-dimensional parameters and using machine learning-based analysis gives a holistic picture of a person's physiological changes due to COVID-19 infection. Moreover, the continuous monitoring system showed earlier infection detection compared to intermittent testing only.
The abovementioned studies have explored the potential to extract subtle physiological changes with COVID-19 infection using wearables. This new avenue of using smartwatches and wearables to track underlying health conditions is particularly valuable for infectious diseases like COVID-19. Moreover, some studies have shown promising results towards detecting the infection even before any physical symptoms have added multifold utility to such systems, namely continuous monitoring and contact-tracing.
4.2. Radiography-based COVID-19 detection
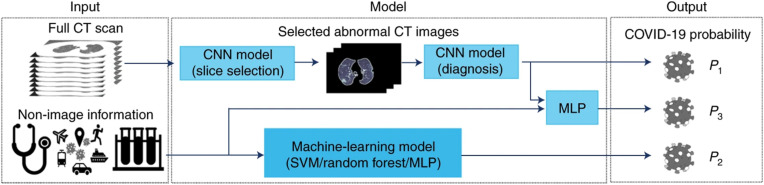
RT-PCR, the primary diagnostic tool for detecting COVID-19, has a high false alarm rate due to the damage through the viral mutations and the contamination of samples [107]. Hence, multiple groups have suggested the radiology based approach as a secondary diagnostic tool for COVID-19 [108,109]. Moreover, since the outbreak of COVID-19, a significant number of studies have shown the prospect of AI to detect the COVID-19 infection progression using chest X-rays and CT images with high accuracy. Mei et al. proposed an AI-based model to detect the COVID-19 infection using clinical history and chest CT scans [110]. The clinical history involved prior diseases, blood test results, symptoms, demographic information, exposure to COVID-19 infected individuals, and travel history. They also collected the CT scan images with the consent of the subjects. They developed a convolutional neural network (CNN) to extract and learn the normal and infection features from CT scans. Afterward, the extracted features along with the clinical data were classified into normal and infected classes using three different classifiers - multilayer perceptron (MLP), random forest, and support vector machine (SVM). Among the three classical machine learning classifiers, MLP showed the best performance and they proposed it in their final pipeline. Their final pipeline is illustrated in Fig. 5 .
Fig. 5.
AI-based COVID-19 detection pipeline using a combination of radiology and symptom data [110].
They used RT-PCR test results as the ground truth for the study among 905 patients, of which 46.3% were positively tested. In a test set of 279 patients, the AI system had a sensitivity score equal to a senior thoracic radiologist and demonstrated an AUC of 0.92. Moreover, the AI-based system labeled 68% of the patients as COVID-19 positive, which was confirmed by the RT-PCR; however, the radiologist misclassified three patients due to the relatively early stage of infection and disease progression. Thus, an AI-based system showed potential for early COVID-19 detection based on a combined evaluation of clinical and radiology data.
Likewise, Bai et al. established and evaluated an AI-based framework for distinguishing COVID-19 infection from pneumonia induced by other pathogens through chest CT [112]. The study included chest CT images of 521 patients from 10 hospitals along with their RT-PCR results. Moreover, 665 pneumonia patients' chest CTs were collected from three medical centers ranging between 2017 and 2019. A deep neural network, EfficientNet B4, was used to distinguish the COVID-19 infection from other pneumonia infections. The lung portion was segmented before the input to the model. The study involved 1186 patients' data, resulting in 132,583 CT slices and separated in a 7:2:1 ratio as train-validation-test sets. Afterward, they performed separate testing in another hospital. Six radiologists participated in the study to identify COVID-19 infection in the CT images. The AI-based model resulted in 96% accuracy, 95% sensitivity, 96% specificity, and an AUC of 0.95. This study shows the potential of an AI-assisted system for differentiating COVID-19-induced infection from other forms of pneumonia. Moreover, Qiblawey et al. proposed another AI-based pipeline for COVID-19 recognition and severity grading from the CT volumes of 1110 subjects (Fig. 6 ) [111]. The presented system detected COVID-19 detection performance with 98.72% specificity and 99.64% sensitivity. The system also classified four levels of infection severity, namely mild, moderate, severe, and critical, with 98.3%, 71.2%, 77.8%, and 100% sensitivity, respectively.
Fig. 6.
AI-based COVID-19 detection pipeline using CT images using multiple encoder-decoder networks [111].
Ozturk et al. used an altered version of DarkNet on 114 COVID-19 chest X-rays for two types of classification schemes - binary classification between normal and COVID-19 infected subjects and multi-class classification among normal, COVID-19 infected, and non-COVID-19 infected pneumonia patients. The modified model achieved a sensitivity of 90.65% and 85.35% for the binary and multi-class schemes, respectively [113]. Apostolopoulos et al. presented their study on the chest X-rays of 224 COVID-19 patients using MobileNetV2 as the classifier, which achieved a sensitivity of 98.7% [114]. Wang et al. proposed a CNN-based architecture named COVID-Net, which was specifically tuned for COVID-19 identification. The study was carried out on 358 COVID-19 chest X-rays and the model showed an accuracy of 91% [115].
A common problem in AI-based image classification is the scarcity of reliable data. To circumvent this problem, AI-based data generation techniques, specifically, Generative Adversarial Networks (GANs) have recently shown tremendous development in almost all domains. These networks produce realistic data from small samples to create a comparatively large dataset. In this regard, Waheed et al. presented an Auxiliary Classifier Generative Adversarial Network (ACGAN) network to augment synthetic images from publicly available COVID-19 chest X-rays [116]. Chowdhury et al. experimented with several CNN-based pre-trained networks, namely CheXNet, DenseNet201, MobileNetV2, ResNet101, ResNet18, and SqueezeNet on a dataset of 423 chest X-ray images for binary and multi-class classifications. Among the models, DenseNet201 demonstrated the best sensitivity of 99.7% and 97.9% in binary and multi-class classifications, respectively [117]. Yamac et al. used a combination of CheXNet and Convolution Support Estimation Network (CSEN) for classifying bacterial pneumonia, viral pneumonia, or normal [118]. In the combined network, the CheXNet was the feature extractor and CSEN was the classifier. The combined network was used on QaTa-COV19, a benchmark dataset of 462 COVID-19 chest X-ray images, which resulted in a sensitivity of 98%. Fan et al. proposed Multi-Kernel-Size Spatial-Channel Attention Network, an attention-based COVID-19 detection system [119]. It was used on 1000 chest X-ray images from an equal number of COVID-19 and non-COVID-19 patients, which reached a sensitivity of 98.1% and specificity of 98.3%. Degerli et al. experimented on several CNN-based encoder-decoder networks to get a COVID-19 infection map [120]. They compiled 2951 chest X-ray images with ground truth annotation. Among the encoder-decoder networks, the highest F1 score was 85.81% for segmenting the infection locations. Anas et al. also presented an AI-based pipeline for detecting and quantifying the infection progression of COVID-19 based on chest X-ray images [121]. The input images were passed through two parallel CNN-based encoder-decoder networks for generating two binary segmentation masks (Fig. 7 ). The generated infection masks, superimposed on the chest X-rays were used to evaluate COVID-19 infection progression (the system is deployed at qatacov.live).
Fig. 7.
Schematic representation of the COVID-19 detection and infection quantification method [121].
Among these two radiography-based techniques, despite the superior performance of CT, this method is costly and time consuming compared to chest X-ray. Moreover, it is not often available in the remote places. On the other hand, X-ray imaging is not only cheaper and faster, but also operate on a smaller amount of radiation compared to CT [122].
These AI-enabled radiography-based detection methods detect the SARS-CoV-2 infection directly from the lungs area. Despite having the potential of using chest X-ray and computed tomography (CT) for identifying the infected regions, these two systems may fail to detect COVID-19 at an early stage because of the limited infection progression [123]. Hence, this method is more suitable to determine the infection progression over the diagnosis of the infection itself. Moreover, for a highly infectious disease like COVID-19, fast detection of infection presence and its severity identification is of high priority. However, in such a scenario, fast diagnosis by expert radiologists is sometimes not possible due to the high volume of patients and lower number of experts, which increases the utility of such AI-assisted diagnosis.
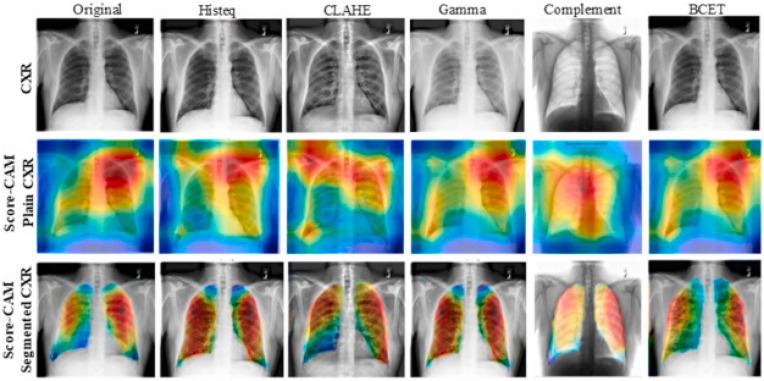
Machine learning model interpretability has become a major concern as ML algorithms are getting popularity in computer-aided diagnosis. To achieve the trust of the clinicians, it is important to know that the deep learning models are not just a black box rather the model performance should be explainable and the model should learn from the relevant areas from the X-ray and CT scan images. Different variants of class activation maps (CAM) were used in recent studies to show the interpretability of deep learning algorithms. Rahman et al. [124] using Score-CAM-based heat maps showed that the Gamma-enhanced segmented lung can detect COVID-19 reliably compared to the full Chest X-ray (CXR) image as the model can learn from irrelevant areas in full CXR images while for segmented lung X-ray images, the model can learn only from the relevant areas as shown in Fig. 8 .
Fig. 8.
Score-CAM visualization for properly classified COVID-19 X-ray images using the different enhancement techniques: CXR (top row), Score-CAM heat map on original CXR (middle row), and segmented lungs CXR (bottom row). Note: Gamma correction technique was the best performing image pre-processing technique reported in Ref. [124].
Similarly, Wang et al. [125] showed Grad-CAM-based heat maps for mild, moderate, severe, and critical patients to show the lung areas where the model is taking the decision in severity grading. Fig. 9 clearly shows that the proposed model mainly focused on the lesion regions of CT images to make decisions regarding moderate, severe, or critical patients.
Fig. 9.
Grad-CAM heat-map for Network visualization for the model interpretation [125].
4.3. COVID-19 detection using cough and breathing sounds
Recent works have shown how the respiratory sounds (e.g., coughs, breathing and voice) of patients who tested positive for COVID-19 in hospitals differ from the sounds of healthy people. Huang et al. used lung auscultation data recorded by digital stethoscope for the diagnosis of COVID-19 [126]. The coughing sound was collected from 48 COVID-19 infected subjects as well as from other individuals with pathological coughs. An ensemble of CNN models was used to differentiate COVID-19 from the other type of coughs. Moreover, to determine the health condition of the hospitalized COVID-19 infected subjects, their speech recordings were examined by automated systems to determine their infection status. This demonstrates that it is possible to identify an infected individual based on their respiratory signals, such as breathing and cough sounds [127].
Imran et al. proposed a mobile app called AI4COVID-19, which audio-records 3 s of coughing and then automatically analyzes the recordings to detect COVID-19 within 2 min using transfer learning [128]. The pipeline consisted of two stages: cough detection and collection, and COVID-19 diagnosis. In the cough detection engine, a user records 3 s of good quality cough sound, and the Mel spectrogram image of the wave is analyzed with a CNN. After the cough is detected, the system passes to the COVID-19 diagnosis to decide the result. It consists of three AI approaches, namely the classical machine learning multi-class classifier (CML-MC), deep transfer learning multi-class classifier (DTL-MC), and the deep transfer learning binary-class classifier. Some key limitations of the current AI4COVID-19 app are 1) limited training data, 2) limited data to generalize the model, and 3) the cough features of COVID-19 may overlap with those of other diseases. In another study by Pal and Sankarasubbu, the authors investigated deep neural networks (DNNs) on a dataset containing 150 patients’ data of a total of 328 instances of coughing sounds, who were afflicted with asthma, bronchitis, COVID-19, or were healthy [129]. In the study, the trained DNN could distinguish the COVID-19 coughs from others with an accuracy of 96.83%. These studies confirm that COVID-19 coughs have a unique pattern. Bagad et al. also found using a Resnet18 network that the model could identify COVID-19 cough sound with an 0.72 AUC score using COVID-19 confirmed coughing sounds from 3621 individuals recorded over the phone [130]. Laguarta, Hueto, and Subirana had an AUC of 0.97 and a sensitivity of 98.5% with a pre-trained ResNet50 network for distinguishing COVID-19 cough sounds from the other types of cough using the samples collected from 4256 subjects and evaluated on 1064 subjects [131].
Brown et al. collected both cough and breathing sounds, then investigated how such data can aid with COVID-19 diagnosis [132]. They provided handcrafted features for cough and breathing sounds, such as period, tempo, onset, duration, root mean square (RMS) of energy, zero-crossing, roll-off frequency, spectral centroid, Mel-frequency cepstrum (MFCC), and delta MFCC. Combined with deep transfer learning, VGGish, which is a convolution network designed to extract audio features automatically, achieved an 80% accuracy on average with 2-class classification problems using the cough and breathing data. A Cambridge University team shared a dataset of cough and breathing sound samples for 582 healthy individuals and 141 individuals with COVID-19 infection [133]. Among the COVID-19 infected subjects, 87 were asymptomatic while 54 were symptomatic (had a dry or wet cough). A web/android mobile application was developed to collect a cough and breathing dataset and screen the patients for COVID-19 from the comfort of their own homes. The collected dataset includes data from 245 healthy, 78 asymptomatic, and 18 symptomatic COVID-19 patients. Users can simply download a mobile application or use the application from any web browser without installation and enter their symptoms, record audio clips of their cough and breathing sounds, and upload the data anonymously. Chowdhury et al. developed two different screening pipelines based on the symptoms reported by the users, namely asymptomatic and symptomatic. Nine state-of-the-art deep learning algorithms were evaluated on the spectrogram generated from the breathing and cough sounds individually and in combination with the sounds of the Cambridge and collected dataset. The authors achieved an accuracy of 95.38% for symptomatic and 98.5% for asymptomatic COVID-19 patient detection using cough sounds. However, the detection accuracies for symptomatic and asymptomatic patients using breath sounds were 90.33% and 75.6%, respectively. A real-time deep learning-based backend server was deployed to convert the cough and breathing sounds to spectrograms and to apply the best-performing machine learning model to identify the COVID-19 patients. The result was then reported back to the test user on the web/mobile application (the system is deployed at https://qu-mlg.com/projects/qu-cough-scope).
Although the AI-assisted COVID-19 infection detection methods based on cough sound show good performance, they are far from perfect as they were not verified for a larger population. Moreover, as a passive detection technique, these systems rely on the symptoms (cough), not the pathogen itself which is not the case for laboratory-based testing methods. Hence, they cannot be used as a standalone COVID-19 detection system; however, they can provide a free-of-cost screening method during an infectious disease outbreak like COVID-19. Based on the result, medical professionals can prescribe further screening. The utility of such a method is particularly promising during resource constraint scenarios.
4.4. COVID-19 symptom tracker
The SARS-CoV-2 has shown novel behavior and infection patterns in human hosts compared to its predecessors from the same coronavirus family, namely MERS and SARS. Due to its novel nature and erratic symptoms, it took scientists several months to get a symptoms pattern for COVID-19.
A group of scientists from diverse disciplines, united by the Coronavirus Pandemic Epidemiology (COPE) Consortium, developed a COVID-19 symptom tracker to help in detecting the disease and collecting diverse data on numerous patient populations [134]. Moreover, Zoe Global Ltd., a nutritional science company, collaborated with academic researchers and launched a mobile app named COVID Symptom Study (initially named COVID Symptom Tracker) in March 2020 [135]. This app aimed to aid researchers to get an insight into the symptoms and designing protocols based on that. Moreover, the app was designed for surveying data from both the general population and the healthcare workers. Such work information, demographics, location, and prior health conditions are added to the database upon registration. Moreover, each user was notified about the research use of their data complying with the Health Insurance Portability and Accountability Act (HIPAA) and General Data Protection Regulation (GDPR). The users were prompted daily to enter their health conditions and possible symptom. From the initially reported data, 75% of the app users were female, and the age group varied from 18 to 90 years old, with an average of 41. In the UK, reported symptoms of the first 1.6 million users, the infected individuals experienced a wide range of symptoms [134], which helped the researchers and doctors get a clear picture of the symptom types and progression from this crowdsourced data [136].
Zens et al. also formulated a mobile application to evaluate the symptoms pattern of COVID-19 infection. The app was also a self-reporting tool and was downloaded by 22,327 individuals between April 8 and May 15, 2020. Similar to the app by Zoe Global Ltd., this app also recorded demographic information, namely the age, gender, and postal code of each user, along with relevant diseases and health conditions. Users got daily reminders to provide an update about any symptom. In case of the onset of symptoms or COVID-19 testing, they were requested to provide details regarding their symptoms and diagnostics. This anonymous study included only participants over 18 years old. From the self-reported data, the research team could derive a predictive value of a certain symptom among certain demography, which opened the possibility of developing effective screening systems. In a resource constraint scenario, the patients with symptoms with higher predictive values can be tested with higher priority, which provides a cost-effective solution. Based on their findings from the study, the loss of taste and smell is the prime indication of possible COVID-19 infection, and people with diabetes are in a high-risk faction [137].
Soriano et al. designed an application called the Hospital Epidemics Tracker (HEpiTracker) to specifically identify and track symptoms in healthcare workers. For data collection purposes, healthcare workers from nine hospitals across five different regions of Spain participated in the study and used the HEpiTracker mobile application. They updated their body temperature daily along with any symptoms related to COVID-19. Moreover, any diagnostic details, namely PCR, and serological test results were recorded in the app as well. The study showed a promising result of such a symptom tracking system for monitoring COVID-19 among hospital workers [138].
5. Challenges and prospects of identifying asymptomatic COVID-19 patients and other diseases
Nowadays, wearable monitoring technology is very accessible, and it offers good accuracy with a wide range of continuous (24/7) vital information monitoring. Smart wearable devices like smartwatches [9], and fitness wearables [[10], [11], [12], [13]] are widely adopted to monitor different vital parameters, namely oxygen saturation, blood pressure, body temperature, heart rate, body mass index, sleep hours, and steps [139]. These parameters are highly correlated with the symptoms of asymptomatic COVID-19 patients, which can be a promising way to identify and monitor these patients [134,140]. In the literature, several studies have been explored based on their usability in the continuous health monitoring and detection of SARS-CoV-2 infection based on cough sounds [128,141,142], body temperature [143], and heart rate [99]. However, studies have shown that asymptomatic COVID-19 patients might not exhibit abnormal vital signs, such as a fever or an abnormal heart rate. Thus, identifying COVID-19 patients based on only one variable can be misleading. Therefore, multiple variables need to be considered for the detection of a disease like COVID-19 using such devices. However, the mass deployment using wearable devices has been a major challenge in identifying COVID-19 patients using their vital information, particularly due to the lack of information regarding the usage of such wearable solutions among the local population. If these wearable devices were already used by a large proportion of the population, it would be much easier to deploy such a solution to keep track of COVID-19 patients.
Another major challenge for remote monitoring and identification systems using wearable devices is the processing of the data. Offline analysis of the collected data from a large number of patients is time-consuming and requires specialized expertise. For the real-time analysis of the data, the use of machine learning models can offer a vital and timely solution. Moreover, different wearable systems will make the process quite challenging as the data need to be integrated due to format and protocol variations. Mishra et al. performed an elaborate study on the detection of SARS-CoV-2 infection in patients, before the appearance of symptoms, through smart wearable health data [99]. 5262 participants were enrolled for continuous smartwatch sensor data monitoring, in particular heart rate and physical activity recording, using an in-house app named MyPHD. 32 Fitbit watch users eventually turned out to have a COVID-19 infection and 81% of them had visible variations in two proposed parameters, namely heart rate at rest and the ratio of heart rate and step count. Through a simple cumulative statistical anomaly detection model, it was estimated that almost 63% of COVID-19 patients could have been identified before they showed any symptoms. However, no machine learning model was utilized to detect asymptomatic COVID-19 patients, which would have increased the robustness of the model. Quer et al. performed a study on a much larger population of 30,529 participants using a smartphone application that, in addition to self-reported symptom data, monitors sensor data such as heart rate, sleep, and activity [102]. Among 3811 people with symptoms, 54 individuals were consequently found to be COVID-19 positive. The interesting outcome of a 9% increase in the AUC in identifying COVID-19 when combining sensor data with symptoms instead of symptoms alone further supports the potential of wearable health monitoring. Smarr et al. [143] further integrated temperature sensors and heart rate-related features on 50 COVID-19 patients and revealed that 38 of them could have been identified much earlier than when they started to show symptoms using these biomarkers. A similar study by Radin et al. [105] expanded the applicability of such physiological and activity data monitoring from wearables to influenza-like diseases. Bogu et al. [100] designed a wearable system to collect patients' heart rate data during rest. They used a deep learning model to identify asymptomatic COVID-19 patients. The variation in the visceral nervous system response, symbolized by heart rate variability (HRV), relates to the heart rate. This research demonstrated that COVID-19 patients can be identified before the clinical identification of infection. Hirten et al. [144] used information collected from wearable devices (Apple Watch) for the identification and prediction of COVID-19. This research work used several statistical models to identify and predict COVID-19 and its associated symptoms. It was reported that a notable change was observed in HRV for COVID-19 patients before the RT-PCR test was carried out. They found a significant deviation (p-value = 0.006) in the mean amplitude of the standard deviation of the interbeat interval of normal sinus beats (SDNN) circadian pattern among subjects with (1.23 ms, 95% CI: 1.94–3.11) and without (5.30 ms, 95% CI: 4.97–5.65) COVID-19 infection.
A crowdsourcing-based epidemiologic study often does not represent the whole population, which makes the result non-generalized. This is also the case for smartphone-based applications for COVID-19 detection. Nevertheless, such an approach can be deployed rapidly in a large population during a novel disease outbreak. With the progression of the research as well as continued use by a large number of participants, such systems can overcome the distribution issue [134].
Likewise, AI-based systems also have limitations as well. Firstly, there can be subtle discrepancies among the CT images induced by different ethnicity, country, and even the CT instrument. Such discrepancies in the dataset will sabotage a study aiming for generalized detection. Secondly, the baseline data contains dissimilarities in COVID-19 and other pneumonia patients. The reason behind such dissimilarities is attributed to the data collection as well as the disease types. In the case of COVID-19, initially, most datasets were collected in China from varied age and gender groups. On the other hand, for other pneumonia patients, the majority of the data were collected in the United States and constituted the older population. Such differences in demography might cause bias in the model as well. Thirdly, as the results of such AI-based models have relied on radiologists, it can add certain human bias. The reason behind such bias is mostly due to the experiment design, where the same radiologist reviews the same image with and without the AI-based segmentation mask. However, this bias can be averted by adopting a prospective experiment. Fourthly, the CT images of COVID-19 infection contain subtle differences from the other pneumonia infection mostly during the early stage. Due to the lack of the number of such CT images at that stage, the models relied on CT images for a wide range of COVID-19 progression. Such a dataset with diverse disease progression is hard to be useful for particularly detecting COVID-19 early despite representing the real-life testing scenarios. However, with the progression of the pandemic, such scarcity of data will eventually be resolved. Lastly, a notable number of other pneumonia cases after COVID-19 breakout did not go through Respiratory Pathogen Panel (RPP) testing, which leaves a suspicion about whether it progressed to COVID-19 or not. For this reason, several studies adopted a second review scheme by another radiologist. However, selection bias can be induced in such a system. These factors can make AI-based systems unusable in a real-world scenario. However, with the recent collaboration of researchers with larger and more diverse cohorts along with more meticulous experiment design, AI-based systems have the potential to get more reliable [112].
It is clear from the previous studies that despite the negative RT-PCR test, chest X-ray images and computed tomography (CT) contributed to the detection of SARS-CoV-2 infection and measuring the extent of the infection progression in the suspected patients [110]. Therefore, multi-modal screening techniques were shown to improve the screening accuracy and helped to reduce the risk of spreading this contagious disease. Therefore, an intelligent detection model based on robust machine learning algorithms can be a promising way to accurately identify asymptomatic COVID-19 patients using vital parameters from wearable devices. Thus, introducing smart monitoring devices for early COVID-19 detection can help public health officials to provide more coherent follow-ups of the patients.
6. Conclusions
Screening of SARS-CoV-2 is currently performed with molecular cell-based POCT techniques, which are useful for performing testing of symptomatic patients at the individual level and with acceptable accuracy, precision, and robustness. Among these methods, while the RT-PCR testing is more reliable, it suffers from significant challenges in terms of giving false positive and false negative outcomes. The other molecular-based testing methods, including RT-LAMP, CRISPER/CAS system, immunoassays, and biosensors, have their benefits in terms of accuracy and reliability, but these also have limitations when mass-scale testing is required, such as during seasonally driven disease spikes. Therefore, developing such strategies which enable locating and identifying COVID-2 infected patients at a mass scale in an asymptomatic state can open a new venue for the management of patient health. Recent approaches for monitoring asymptomatic COVID-19 patients using smartwatches and mobile applications have shown promising results; hence, there is still a need for the development of AI-based systems for the mass-scale surveillance of suspected COVID-19 patients. This will eventually help public health officials to identify infected patients in the early stages to keep them in isolation and provide the necessary treatments. The technique will help to manage and monitor the health of COVID-19 patients, both asymptomatic and symptomatic, which will help to manage the available facilities systematically. The method will also help to track and manage the COVID-19 patients’ health after their recovery by continuously monitoring their health parameters, and it will also be able to mitigate any post-recovery complications.
Funding
This work was supported by the Qatar National Research Grant: UREP28-144-3-046. The statements made herein are solely the responsibility of the authors.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.WHO coronavirus (COVID-19) dashboard. https://covid19.who.int
- 2.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323:2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 3.Yang S., Rothman R.E. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 2004;4:337–348. doi: 10.1016/S1473-3099(04)01044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Z., Harrich D., Li Z., Hu D., Li D. The unique features of SARS-CoV-2 transmission: comparison with SARS-CoV, MERS-CoV and 2009 H1N1 pandemic influenza virus. Rev. Med. Virol. 2021;31 doi: 10.1002/rmv.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong D., Dai W., Gong J., Li G., Liu N., Wu W., Pan J., Chen C., Jiao Y., Deng H. Rapid detection of SARS-CoV-2 with CRISPR-Cas12a. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naresh V., Lee N. A review on biosensors and recent development of nanostructured materials-enabled biosensors. Sensors. 2021;21:1109. doi: 10.3390/s21041109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darwish I.A. Immunoassay methods and their applications in pharmaceutical analysis: basic methodology and recent advances. Int. J. Biomed. Sci.: IJBS. 2006;2:217. [PMC free article] [PubMed] [Google Scholar]
- 9.https://www.apple.com/shop/buy-watch/apple-watch Apple Inc., Apple Watch, Apple. (n.d.)
- 10.Fitbit Fitbit official site for activity trackers and more. https://www.fitbit.com/global/uk/home
- 11.ZephyrTM performance systems | performance monitoring technology. https://www.zephyranywhere.com/
- 12.D.I. Labs, WHOOP | Your Personal Digital Fitness and Health Coach, WHOOP. (n.d.). https://www.whoop.com/(accessed April 19, 2022).
- 13.BioIntelliSense https://biointellisense.com/
- 14.Garmin Smartwatches with fitness and health tracking | Garmin. https://www.garmin.com/en-US/c/sports-fitness/activity-fitness-trackers/
- 15.Best Smart Rings: Put a Ring on it in 2022. Wareable; 2022. https://www.wareable.com/fashion/best-smart-rings-1340 [Google Scholar]
- 16.Seshadri D.R., Davies E.V., Harlow E.R., Hsu J.J., Knighton S.C., Walker T.A., Voos J.E., Drummond C.K. Wearable sensors for COVID-19: a call to action to harness our digital infrastructure for remote patient monitoring and virtual assessments. Front. Digit. Health. 2020:8. doi: 10.3389/fdgth.2020.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coronavirus Disease (COVID-19) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports n.d.)
- 19.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Archived: WHO Timeline - COVID-19. https://www.who.int/news/item/27-04-2020-who-timeline---covid-19 (n.d.)
- 21.Coronavirus Disease COVID-19) - events as they happen. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen n.d.)
- 22.International Committee on Taxonomy of Viruses Executive Committee The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks. Nat. Microbiol. 2020;5:668. doi: 10.1038/s41564-020-0709-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Britannica, the Editors of Encyclopaedia, coronavirus | definition, features, & examples | Britannica, Encyclopedia Britannica. https://www.britannica.com/science/coronavirus-virus-group (n.d.)
- 24.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lefkowitz E.J., Dempsey D.M., Hendrickson R.C., Orton R.J., Siddell S.G., Smith D.B. Virus taxonomy: the database of the international committee on taxonomy of viruses (ICTV) Nucleic Acids Res. 2018;46:D708–D717. doi: 10.1093/nar/gkx932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machhi J., Herskovitz J., Senan A.M., Dutta D., Nath B., Oleynikov M.D., Blomberg W.R., Meigs D.D., Hasan M., Patel M. The natural history, pathobiology, and clinical manifestations of SARS-CoV-2 infections. J. Neuroimmune Pharmacol. 2020;15:359–386. doi: 10.1007/s11481-020-09944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novel Coronavirus – Thailand https://www.who.int/emergencies/disease-outbreak-news/item/2020-DON234 (n.d.)
- 29.Dong Y., Mo X., Hu Y., Qi X., Jiang F., Jiang Z., Tong S. Epidemiology of COVID-19 among children in China. Pediatrics. 2020;145 doi: 10.1542/peds.2020-0702. [DOI] [PubMed] [Google Scholar]
- 30.Wu J.T., Leung K., Lam T.T., Ni M.Y., Wong C.K., Peiris J., Leung G.M. Nowcasting epidemics of novel pathogens: lessons from COVID-19. Nat. Med. 2021;27:388–395. doi: 10.1038/s41591-021-01278-w. [DOI] [PubMed] [Google Scholar]
- 31.Xu S., Li Y. Beware of the second wave of COVID-19. Lancet. 2020;395:1321–1322. doi: 10.1016/S0140-6736(20)30845-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gandhi M., Yokoe D.S., Havlir D.V. 2020. Asymptomatic Transmission, the Achilles' Heel of Current Strategies to Control Covid-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. M inimum I nformation for Publication of Q uantitative Real-Time PCR E xperiments; 2009. The MIQE Guidelines. [DOI] [PubMed] [Google Scholar]
- 36.Vogels C.B., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Catherine Muenker M., Moore A.J. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lan L., Xu D., Ye G., Xia C., Wang S., Li Y., Xu H. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 2020;323:1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharfstein J.M., Becker S.J., Mello M.M. Diagnostic testing for the novel coronavirus. JAMA. 2020;323:1437–1438. doi: 10.1001/jama.2020.3864. [DOI] [PubMed] [Google Scholar]
- 39.van Kasteren P.B., van Der Veer B., van den Brink S., Wijsman L., de Jonge J., van den Brandt A., Molenkamp R., Reusken C.B., Meijer A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Augustine R., Hasan A., Das S., Ahmed R., Mori Y., Notomi T., Kevadiya B.D., Thakor A.S. Loop-mediated isothermal amplification (LAMP): a rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology. 2020;9:182. doi: 10.3390/biology9080182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomita N., Mori Y., Kanda H., Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 43.Law J.W.-F., Ab Mutalib N.-S., Chan K.-G., Lee L.-H. Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front. Microbiol. 2015;5:770. doi: 10.3389/fmicb.2014.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reboud J., Xu G., Garrett A., Adriko M., Yang Z., Tukahebwa E.M., Rowell C., Cooper J.M. Paper-based microfluidics for DNA diagnostics of malaria in low resource underserved rural communities. Proc. Natl. Acad. Sci. USA. 2019;116:4834–4842. doi: 10.1073/pnas.1812296116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carter J.G., Iturbe L.O., Duprey J.-L.H., Carter I.R., Southern C.D., Rana M., Whalley C.M., Bosworth A., Beggs A.D., Hicks M.R. Ultrarapid detection of SARS-CoV-2 RNA using a reverse transcription–free exponential amplification reaction, RTF-EXPAR. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2100347118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 47.Huang W.E., Lim B., Hsu C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M. RT‐LAMP for rapid diagnosis of coronavirus SARS‐CoV‐2. Microb. Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dao Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.P., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang M., Pan W., Arasthfer A., Fang W., Ling L., Fang H., Daneshnia F., Yu J., Liao W., Pei H. Development and validation of a rapid, single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19. Front. Cell. Infect. Microbiol. 2020;10:331. doi: 10.3389/fcimb.2020.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 52.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.East-Seletsky A., O'Connell M.R., Knight S.C., Burstein D., Cate J.H., Tjian R., Doudna J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., Fusco D., Drouin A., Yin X., Hu T. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164 doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar R., Nagpal S., Kaushik S., Mendiratta S. COVID-19 diagnostic approaches: different roads to the same destination. VirusDis. 2020;31:97–105. doi: 10.1007/s13337-020-00599-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clinical decision support tool and rapid point-of-care platform for determining disease severity in patients with COVID-19 - lab on a Chip. (RSC Publishing) DOI:10.1039/D0LC00373E, (n.d.). https://pubs.rsc.org/en/content/articlehtml/2020/lc/d0lc00373e. [DOI] [PMC free article] [PubMed]
- 60.S. Lambert-Niclot, A. Cuffel, S. Le Pape, C. Vauloup-Fellous, L. Morand-Joubert, A.-M. Roque-Afonso, J. Le Goff, C. Delaugerre, On behalf of the AP-HP/Universities/INSERMCOVID-19 Research Collaboration, Evaluation of a Rapid Diagnostic Assay for Detection of SARS-CoV-2 Antigen in Nasopharyngeal Swabs, J. Clin. Microbiol.. 58 (n.d.) e00977-20. 10.1128/JCM.00977-20. [DOI] [PMC free article] [PubMed]
- 61.Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., Wang D.-Q., Hu Y., Ren J.-H., Tang N., Xu Y.-Y., Yu L.-H., Mo Z., Gong F., Zhang X.-L., Tian W.-G., Hu L., Zhang X.-X., Xiang J.-L., Du H.-X., Liu H.-W., Lang C.-H., Luo X.-H., Wu S.-B., Cui X.-P., Zhou Z., Zhu M.-M., Wang J., Xue C.-J., Li X.-F., Wang L., Li Z.-J., Wang K., Niu C.-C., Yang Q.-J., Tang X.-J., Zhang Y., Liu X.-M., Li J.-J., Zhang D.-C., Zhang F., Liu P., Yuan J., Li Q., Hu J.-L., Chen J., Huang A.-L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 62.D'Cruz R.J., Currier A.W., Sampson V.B. Laboratory testing methods for novel severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.00468. https://www.frontiersin.org/article/10.3389/fcell.2020.00468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ni L., Ye F., Cheng M.-L., Feng Y., Deng Y.-Q., Zhao H., Wei P., Ge J., Gou M., Li X., Sun L., Cao T., Wang P., Zhou C., Zhang R., Liang P., Guo H., Wang X., Qin C.-F., Chen F., Dong C. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977. doi: 10.1016/j.immuni.2020.04.023. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kilic T., Weissleder R., Lee H. Molecular and immunological diagnostic tests of COVID-19: current status and challenges. iScience. 2020;23 doi: 10.1016/j.isci.2020.101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coste A.T., Jaton K., Papadimitriou-Olivgeris M., Greub G., Croxatto A. Comparison of SARS-CoV-2 serological tests with different antigen targets. J. Clin. Virol. 2021;134 doi: 10.1016/j.jcv.2020.104690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Montesinos I., Gruson D., Kabamba B., Dahma H., Van den Wijngaert S., Reza S., Carbone V., Vandenberg O., Gulbis B., Wolff F., Rodriguez-Villalobos H. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grant B.D., Anderson C.E., Williford J.R., Alonzo L.F., Glukhova V.A., Boyle D.S., Weigl B.H., Nichols K.P. SARS-CoV-2 coronavirus nucleocapsid antigen-detecting half-strip lateral flow assay toward the development of point of care tests using commercially available reagents. Anal. Chem. 2020;92:11305–11309. doi: 10.1021/acs.analchem.0c01975. [DOI] [PubMed] [Google Scholar]
- 68.Wen T., Huang C., Shi F.-J., Zeng X.-Y., Lu T., Ding S.-N., Jiao Y.-J. Development of a lateral flow immunoassay strip for rapid detection of IgG antibody against SARS-CoV-2 virus. Analyst. 2020;145:5345–5352. doi: 10.1039/D0AN00629G. [DOI] [PubMed] [Google Scholar]
- 69.Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Bian L., Li P., Yu L., Wu Y., Lin G. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 70.Tripathi S., Agrawal A. Blood plasma microfluidic device: aiming for the detection of COVID-19 antibodies using an on-chip ELISA platform. Trans Indian Natl. Acad. Eng. 2020;5:217–220. doi: 10.1007/s41403-020-00123-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tan X., Krel M., Dolgov E., Park S., Li X., Wu W., Sun Y.-L., Zhang J., Khaing Oo M.K., Perlin D.S., Fan X. Rapid and quantitative detection of SARS-CoV-2 specific IgG for convalescent serum evaluation. Biosens. Bioelectron. 2020;169 doi: 10.1016/j.bios.2020.112572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan X., Broses L.J., Zhou M., Day K.C., Liu W., Li Z., Weizer A.Z., Munson K.A., Oo M.K.K., Day M.L., Fan X. Multiparameter urine analysis for quantitative bladder cancer surveillance of orthotopic xenografted mice. Lab Chip. 2020;20:634–646. doi: 10.1039/C9LC01006H. [DOI] [PubMed] [Google Scholar]
- 73.Ghosh S., Aggarwal K., V.T U., Nguyen T., Han J., Ahn C.H. A new microchannel capillary flow assay (MCFA) platform with lyophilized chemiluminescence reagents for a smartphone-based POCT detecting malaria. Microsyst. Nanoeng. 2020;6:1–18. doi: 10.1038/s41378-019-0108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morales-Narváez E., Dincer C. The impact of biosensing in a pandemic outbreak: COVID-19. Biosens. Bioelectron. 2020;163 doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Biosensors for managing the COVID-19 cytokine storm: challenges ahead | ACS sensors. https://pubs.acs.org/doi/abs/10.1021/acssensors.0c00979 (n.d.) [DOI] [PubMed]
- 76.Mejía-Salazar J.R., Oliveira O.N. Plasmonic Biosensing. Chem. Rev. 2018;118:10617–10625. doi: 10.1021/acs.chemrev.8b00359. [DOI] [PubMed] [Google Scholar]
- 77.Mujawar M.A., Gohel H., Bhardwaj S.K., Srinivasan S., Hickman N., Kaushik A. Nano-enabled biosensing systems for intelligent healthcare: towards COVID-19 management. Mater. Today Chem. 2020;17 doi: 10.1016/j.mtchem.2020.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weiss C., Carriere M., Fusco L., Capua I., Regla-Nava J.A., Pasquali M., Scott J.A., Vitale F., Unal M.A., Mattevi C., Bedognetti D., Merkoçi A., Tasciotti E., Yilmazer A., Gogotsi Y., Stellacci F., Delogu L.G. Toward nanotechnology-enabled approaches against the COVID-19 pandemic. ACS Nano. 2020;14:6383–6406. doi: 10.1021/acsnano.0c03697. [DOI] [PubMed] [Google Scholar]
- 79.Moitra P., Alafeef M., Dighe K., Frieman M.B., Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 81.Murugan D., Bhatia H., Sai V.V.R., Satija J. P-FAB: A fiber-optic biosensor device for rapid detection of COVID-19. Trans Indian Natl. Acad. Eng. 2020;5:211–215. doi: 10.1007/s41403-020-00122-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Funari R., Chu K.-Y., Shen A.Q. Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip. Biosens. Bioelectron. 2020;169 doi: 10.1016/j.bios.2020.112578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Eguilaz M.R., Cumba L.R., Forster R.J. Electrochemical detection of viruses and antibodies: a mini review. Electrochem. Commun. 2020;116 doi: 10.1016/j.elecom.2020.106762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tripathy S., Singh S.G. Label-free electrochemical detection of DNA hybridization: a method for COVID-19 diagnosis. Trans Indian Natl. Acad. Eng. 2020;5:205–209. doi: 10.1007/s41403-020-00103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ali Md.A., Hu C., Jahan S., Yuan B., Saleh M.S., Ju E., Gao S.-J., Panat R. Sensing of COVID-19 antibodies in seconds via aerosol jet nanoprinted reduced-graphene-oxide-coated 3D electrodes. Adv. Mater. 2021;33 doi: 10.1002/adma.202006647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fabiani L., Saroglia M., Galatà G., De Santis R., Fillo S., Luca V., Faggioni G., D'Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen. Biosens. Bioelectron. 2021;176 doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alafeef M., Dighe K., Moitra P., Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020;14:17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao H., Liu F., Xie W., Zhou T.-C., OuYang J., Jin L., Li H., Zhao C.-Y., Zhang L., Wei J., Zhang Y.-P., Li C.-P. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensor. Actuator. B Chem. 2021;327 doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Torrente-Rodríguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H.B., Gao W. SARS-CoV-2 RapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter. 2020;3:1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lipsitch M., Swerdlow D.L., Finelli L. Defining the epidemiology of covid-19 — studies needed. N. Engl. J. Med. 2020;382:1194–1196. doi: 10.1056/NEJMp2002125. [DOI] [PubMed] [Google Scholar]
- 92.FitzGerald G.A. Misguided drug advice for COVID-19. Science. 2020;367:1434. doi: 10.1126/science.abb8034. 1434. [DOI] [PubMed] [Google Scholar]
- 93.Marinsek N., Shapiro A., Clay I., Bradshaw B., Ramirez E., Min J., Trister A., Wang Y., Althoff T., Foschini L. 2020. Measuring COVID-19 and Influenza in the Real World via Person-Generated Health Data; p. 2020. 05.28.20115964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dunn J., Runge R., Snyder M. Wearables and the medical revolution. Pers. Med. 2018;15:429–448. doi: 10.2217/pme-2018-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kellogg R.A., Dunn J., Snyder M.P. Personal omics for precision health. Circ. Res. 2018;122:1169–1171. doi: 10.1161/CIRCRESAHA.117.310909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Quer G., Gouda P., Galarnyk M., Topol E.J., Steinhubl S.R. Inter- and intraindividual variability in daily resting heart rate and its associations with age, sex, sleep, BMI, and time of year: retrospective, longitudinal cohort study of 92,457 adults. PLoS One. 2020;15 doi: 10.1371/journal.pone.0227709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jaiswal S.J., Quer G., Galarnyk M., Steinhubl S.R., Topol E.J., Owens R.L. Association of sleep duration and variability with body mass index: sleep measurements in a large US population of wearable sensor users. JAMA Intern. Med. 2020;180:1694–1696. doi: 10.1001/jamainternmed.2020.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhu T., Watkinson P., Clifton D.A. Smartwatch data help detect COVID-19. Nat. Biomed. Eng. 2020;4:1125–1127. doi: 10.1038/s41551-020-00659-9. [DOI] [PubMed] [Google Scholar]
- 99.Mishra T., Wang M., Metwally A.A., Bogu G.K., Brooks A.W., Bahmani A., Alavi A., Celli A., Higgs E., Dagan-Rosenfeld O. others, Pre-symptomatic detection of COVID-19 from smartwatch data. Nat. Biomed. Eng. 2020;4:1208–1220. doi: 10.1038/s41551-020-00640-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bogu G.K., Snyder M.P. Deep learning-based detection of COVID-19 using wearables data. medRxiv. 2021 [Google Scholar]
- 101.Abir F.F., Alyafei K., Chowdhury M.E.H., Khandaker A., Ahmed R., Hossain M.S., Mahmud S., Rahman A., Abbas T.O., Zughaier S.M., Naji K.K. PCovNet: a presymptomatic COVID-19 detection framework using deep learning model using wearables data. Comput. Biol. Med. 2022 doi: 10.1016/j.compbiomed.2022.105682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Quer G., Radin J.M., Gadaleta M., Baca-Motes K., Ariniello L., Ramos E., Kheterpal V., Topol E.J., Steinhubl S.R. Wearable sensor data and self-reported symptoms for COVID-19 detection. Nat. Med. 2021;27:73–77. doi: 10.1038/s41591-020-1123-x. [DOI] [PubMed] [Google Scholar]
- 103.Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W. And the northwell COVID-19 research Consortium, presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York city area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chung Y.-T., Yeh C.-Y., Shu Y.-C., Chuang K.-T., Chen C.-C., Kao H.-Y., Ko W.-C., Chen P.-L., Ko N.-Y. Continuous temperature monitoring by a wearable device for early detection of febrile events in the SARS-CoV-2 outbreak in Taiwan. J. Microbiol. Immunol. Infect. 2020;53:503–504. doi: 10.1016/j.jmii.2020.04.005. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Radin J.M., Wineinger N.E., Topol E.J., Steinhubl S.R. Harnessing wearable device data to improve state-level real-time surveillance of influenza-like illness in the USA: a population-based study. Lancet Digit. Health. 2020;2 doi: 10.1016/S2589-7500(19)30222-5. e85–e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wong C.K., Ho D.T.Y., Tam A.R., Zhou M., Lau Y.M., Tang M.O.Y., Tong R.C.F., Rajput K.S., Chen G., Chan S.C., Siu C.W., Hung I.F.N. Artificial intelligence mobile health platform for early detection of COVID-19 in quarantine subjects using a wearable biosensor: protocol for a randomised controlled trial. BMJ Open. 2020;10 doi: 10.1136/bmjopen-2020-038555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev. Mol. Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296:E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296:E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mei X., Lee H.-C., Diao K., Huang M., Lin B., Liu C., Xie Z., Ma Y., Robson P.M., Chung M., Bernheim A., Mani V., Calcagno C., Li K., Li S., Shan H., Lv J., Zhao T., Xia J., Long Q., Steinberger S., Jacobi A., Deyer T., Luksza M., Liu F., Little B.P., Fayad Z.A., Yang Y. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020;26:1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qiblawey Y., Tahir A., Chowdhury M.E., Khandakar A., Kiranyaz S., Rahman T., Ibtehaz N., Mahmud S., Maadeed S.A., Musharavati F. others, Detection and severity classification of COVID-19 in CT images using deep learning. Diagnostics. 2021;11:893. doi: 10.3390/diagnostics11050893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bai H.X., Wang R., Xiong Z., Hsieh B., Chang K., Halsey K., Tran T.M.L., Choi J.W., Wang D.-C., Shi L.-B., Mei J., Jiang X.-L., Pan I., Zeng Q.-H., Hu P.-F., Li Y.-H., Fu F.-X., Huang R.Y., Sebro R., Yu Q.-Z., Atalay M.K., Liao W.-H. Artificial intelligence augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other origin at chest CT. Radiology. 2020;296:E156–E165. doi: 10.1148/radiol.2020201491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang L., Lin Z.Q., Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020;10:19549. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Waheed A., Goyal M., Gupta D., Khanna A., Al-Turjman F., Pinheiro P.R. CovidGAN: data augmentation using auxiliary classifier GAN for improved covid-19 detection. IEEE Access. 2020;8:91916–91923. doi: 10.1109/ACCESS.2020.2994762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chowdhury M.E.H., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Emadi N.A., Reaz M.B.I., Islam M.T. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- 118.Yamaç M., Ahishali M., Degerli A., Kiranyaz S., Chowdhury M.E., Gabbouj M. Convolutional sparse support Estimator-based COVID-19 recognition from X-ray images. IEEE Transact. Neural Networks Learn. Syst. 2021;32:1810–1820. doi: 10.1109/TNNLS.2021.3070467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fan Y., Liu J., Yao R., Yuan X. COVID-19 detection from X-ray images using multi-kernel-size Spatial-Channel attention network. Pattern Recogn. 2021;119 doi: 10.1016/j.patcog.2021.108055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Degerli A., Ahishali M., Yamac M., Kiranyaz S., Chowdhury M.E.H., Hameed K., Hamid T., Mazhar R., Gabbouj M. COVID-19 infection map generation and detection from chest X-ray images. Health Inf. Sci. Syst. 2021;9:15. doi: 10.1007/s13755-021-00146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tahir A.M., Chowdhury M.E.H., Khandakar A., Rahman T., Qiblawey Y., Khurshid U., Kiranyaz S., Ibtehaz N., Rahman M.S., Al-Maadeed S., Mahmud S., Ezeddin M., Hameed K., Hamid T. COVID-19 infection localization and severity grading from chest X-ray images. Comput. Biol. Med. 2021;139 doi: 10.1016/j.compbiomed.2021.105002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brenner D.J., Hall E.J. Computed tomography — an increasing source of radiation exposure. N. Engl. J. Med. 2007;357:2277–2284. doi: 10.1056/NEJMra072149. [DOI] [PubMed] [Google Scholar]
- 123.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z.A., Jacobi A., Li K., Li S., Shan H. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rahman T., Khandakar A., Qiblawey Y., Tahir A., Kiranyaz S., Abul Kashem S.B., Islam M.T., Al Maadeed S., Zughaier S.M., Khan M.S., Chowdhury M.E.H. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang J., Liu C., Li J., Yuan C., Zhang L., Jin C., Xu J., Wang Y., Wen Y., Lu H., Li B., Chen C., Li X., Shen D., Qian D., Wang J. iCOVID: interpretable deep learning framework for early recovery-time prediction of COVID-19 patients. Npj Digit. Med. 2021;4:1–13. doi: 10.1038/s41746-021-00496-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huang Y., Meng S., Zhang Y., Wu S., Zhang Y., Zhang Y., Ye Y., Wei Q., Zhao N., Jiang J., Ji X., Zhou C., Zheng C., Zhang W., Xie L., Hu Y., He J., Chen J., Wang W., Cao L., Xu W., Lei Y., Jiang Z., Hu W., Qin W., Wang W., He Y., Xiao H., Zheng X., Hu Y., Pan W., Zhang C., Cai J. 2020. The Respiratory Sound Features of COVID-19 Patients Fill Gaps between Clinical Data and Screening Methods; p. 2020. 04.07.20051060. [DOI] [Google Scholar]
- 127.Han J., Qian K., Song M., Yang Z., Ren Z., Liu S., Liu J., Zheng H., Ji W., Koike T., Li X., Zhang Z., Yamamoto Y., Schuller B.W. 2020. An Early Study on Intelligent Analysis of Speech under COVID-19: Severity, Sleep Quality, Fatigue, and Anxiety.http://arxiv.org/abs/2005.00096 ArXiv:2005.00096 [Cs, Eess] [Google Scholar]
- 128.Imran A., Posokhova I., Qureshi H.N., Masood U., Riaz M.S., Ali K., John C.N., Hussain M.I., Nabeel M. AI4COVID-19: AI enabled preliminary diagnosis for COVID-19 from cough samples via an app. Inform. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pal A., Sankarasubbu M. Proceedings of the 36th Annual ACM Symposium on Applied Computing. Association for Computing Machinery; New York, NY, USA: 2021. Pay attention to the cough: early diagnosis of COVID-19 using interpretable symptoms embeddings with cough sound signal processing; pp. 620–628. [DOI] [Google Scholar]
- 130.Bagad P., Dalmia A., Doshi J., Nagrani A., Bhamare P., Mahale A., Rane S., Agarwal N., Panicker R. 2020. Cough against COVID: Evidence of COVID-19 Signature in Cough Sounds.http://arxiv.org/abs/2009.08790 ArXiv:2009.08790 [Cs, Eess] [Google Scholar]
- 131.Laguarta J., Hueto F., Subirana B. COVID-19 artificial intelligence diagnosis using only cough recordings. IEEE Open J. Eng. Med. Biol. 2020;1:275–281. doi: 10.1109/OJEMB.2020.3026928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brown C., Chauhan J., Grammenos A., Han J., Hasthanasombat A., Spathis D., Xia T., Cicuta P., Mascolo C. Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining. 2020. Exploring automatic diagnosis of COVID-19 from crowdsourced respiratory sound data; pp. 3474–3484. [DOI] [Google Scholar]
- 133.Xia T., Spathis D., Brown C., Ch J., Grammenos A., Han J., Hasthanasombat A., Bondareva E., Dang T., Floto A., Cicuta P., Mascolo C. vol. 1. 2021. COVID-19 sounds: a large-scale Audio dataset for digital respiratory screening.https://datasets-benchmarks-proceedings.neurips.cc/paper/2021/hash/e2c0be24560d78c5e599c2a9c9d0bbd2-Abstract-round2.html (Proceedings of the Neural Information Processing Systems Track on Datasets and Benchmarks). [Google Scholar]
- 134.Drew D.A., Nguyen L.H., Steves C.J., Menni C., Freydin M., Varsavsky T., Sudre C.H., Cardoso M.J., Ourselin S., Wolf J., Spector T.D., Chan A.T., COPE CONSORTIUM Rapid implementation of mobile technology for real-time epidemiology of COVID-19. Science. 2020;368:1362–1367. doi: 10.1126/science.abc0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.ZOE COVID Study https://covid.joinzoe.com
- 136.Kraemer M.U.G., Scarpino S.V., Marivate V., Gutierrez B., Xu B., Lee G., Hawkins J.B., Rivers C., Pigott D.M., Katz R., Brownstein J.S. Data curation during a pandemic and lessons learned from COVID-19. Nat. Comput. Sci. 2021;1:9–10. doi: 10.1038/s43588-020-00015-6. [DOI] [PubMed] [Google Scholar]
- 137.Zens M., Brammertz A., Herpich J., Südkamp N., Hinterseer M. App-based tracking of self-reported COVID-19 symptoms: analysis of questionnaire data. J. Med. Internet Res. 2020;22 doi: 10.2196/21956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Soriano J.B., Fernández E., de Astorza Á., de Llano L.A.P., Fernández-Villar A., Carnicer-Pont D., Alcázar-Navarrete B., García A., Morales A., Lobo M., Maroto M., Ferreras E., Soriano C., Rio-Bermudez C.D., Vega-Piris L., Basagaña X., Muncunill J., Cosio B.G., Lumbreras S., Catalina C., Alzaga J.M., Quilón D.G., Valdivia C.A., de Lara C., Ancochea J. Hospital epidemics tracker (HEpiTracker): description and pilot study of a mobile app to track COVID-19 in hospital workers. JMIR Publ. Health Surveill. 2020;6 doi: 10.2196/21653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Witt D.R., Kellogg R.A., Snyder M.P., Dunn J. Windows into human health through wearables data analytics. Curr. Opin. Biomed. Eng. 2019;9:28–46. doi: 10.1016/j.cobme.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chamola V., Hassija V., Gupta V., Guizani M. A comprehensive review of the COVID-19 pandemic and the role of IoT, drones, AI, Blockchain, and 5G in managing its impact. IEEE Access. 2020;8:90225–90265. doi: 10.1109/ACCESS.2020.2992341. [DOI] [Google Scholar]
- 141.Vijayakumar D.S., Sneha M. Low cost Covid-19 preliminary diagnosis utilizing cough samples and keenly intellective deep learning approaches. Alex. Eng. J. 2021;60:549–557. doi: 10.1016/j.aej.2020.09.032. [DOI] [Google Scholar]
- 142.Soliński M., Łepek M., Kołtowski Ł. Automatic cough detection based on airflow signals for portable spirometry system. Inform. Med. Unlocked. 2020;18 doi: 10.1016/j.imu.2020.100313. [DOI] [Google Scholar]
- 143.Smarr B.L., Aschbacher K., Fisher S.M., Chowdhary A., Dilchert S., Puldon K., Rao A., Hecht F.M., Mason A.E. Feasibility of continuous fever monitoring using wearable devices. Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-78355-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hirten R.P., Danieletto M., Tomalin L., Choi K.H., Zweig M., Golden E., Kaur S., Helmus D., Biello A., Pyzik R., Nabeel I., Charney A., Glicksberg B., Levin M., Reich D., Charney D., Bottinger E.P., Keefer L., Suarez-Farinas M., Nadkarni G.N., Fayad Z.A. 2020. Longitudinal Physiological Data from a Wearable Device Identifies SARS-CoV-2 Infection and Symptoms and Predicts COVID-19 Diagnosis; p. 2020. 11.06.20226803. [DOI] [PMC free article] [PubMed] [Google Scholar]