Abstract
In the last five years, the prevalence of monkeypox has been increasing both in the regions considered endemic for the disease (West and Central Africa) and worldwide. Indeed, in July 2022, the World Health Organization declared the ongoing global outbreak of monkeypox a public health emergency of international concern. The disease is caused by monkeypox virus (MPXV), a member of the Orthopoxvirus genus, which also includes variola virus (the causative agent of smallpox) and vaccinia virus (used in the smallpox eradication campaign). Here, we review aspects of MPXV genetic diversity and epidemiology, with an emphasis on its genome structure, host range, and relationship with other orthopoxviruses. We also summarize the most recent findings deriving from the sequencing of outbreak MPXV genomes, and we discuss the apparent changing of MPXV evolutionary trajectory, which is characterized by the accumulation of point mutations rather than by gene gains/losses. Whereas the availability of a vaccine, the relatively mild presentation of the disease, and its relatively low transmissibility speak in favor of an efficient control of the global outbreak, the wide host range of MPXV raises concerns about the possible establishment of novel reservoirs. We also call for the deployment of field surveys and genomic surveillance programs to identify and control the MPXV reservoirs in West and Central Africa.
Keywords: Monkeypox, Zoonotic transmission, Genome features, Geographic structure, Genetic diversity
1. Introduction
At the beginning of May 2022, an unexpected occurrence of monkeypox infections was reported from UK health officers. Soon afterwards subjects affected by the disease were identified in Portugal, Spain, Belgium, and several other European and non-European countries (Kraemer et al., 2022). Since May 7, 2022, and as of this writing (27 September 2022), more than 61,000 cases have been reported in 105 countries, making the ongoing monkeypox outbreak the largest ever recorded (https://www.who.int/emergencies/situation-reports). The fast rising number of cases is not the only unusual feature of the epidemic. Indeed, until the beginning of May 2022, monkeypox was considered to be a zoonotic disease occasionally transmitted in West and Central Africa, although exported cases had already been reported (see below). Monkeypox spread across the globe seems to herald a new twist in the epidemiology of the disease and, on July 23, 2022, the WHO declared the global monkeypox outbreak a Public Health Emergency of International Concern (PHEIC).
The disease is caused by monkeypox virus (MPXV), a member of the Orthopoxvirus genus (family Poxviridae, order Chitovirales, class Pokkesviricetes), which also includes variola virus (VARV, the causative agent of smallpox). Monkeypox has a similar but less severe disease presentation than smallpox (Simpson et al., 2020). Clinically, three distinct phases - incubation, prodrome, and rash - can be distinguished (McCollum and Damon, 2014). The incubation phase is relatively long, ranging from 7 to 14 days. The prodrome phase typically includes inguinal lymphoadenopathy, fatigue and headache (McCollum and Damon, 2014). As for the rash, lesions begin as macules and progress to papules, vesicles, pustules, and scabs; pruritus and pain may be present. Usually 10-to-150 lesions are observed, which can persist for up to four weeks. Differently from the lesions of smallpox, monkeypox lesions are similar in size and typically present at the same stage (McCollum and Damon, 2014). The lesions contain infectious virus that can be transmitted through direct contact. Secondary complications of infection include, gastroenteritis, sepsis, pneumonia, encephalitis, and bacterial skin infections (Ogoina et al., 2020). In the endemic regions, human case fatality rates (CFRs) range from 3.6% to 10.6% (Bunge et al., 2022). Disease presentation and CFRs are usually worse in patients with a diagnosis of HIV infection, and are influenced by viral genetic determinants (see below) (Bunge et al., 2022,Yinka-Ogunleye et al., 2019).
For many years, monkeypox has been a neglected disease virtually confined to Africa. In the wake of the COVID-19 pandemic, the worldwide spread of MPXV has raised concern that the virus might turn into a new global threat. While, for many reasons, this is unlikely to be the case, a better understanding of MPXV biology will be instrumental to control viral spread in both affluent and developing countries. Here, we review aspects of MPXV genetic diversity and epidemiology with an emphasis on its evolutionary history and relationship with other poxviruses.
2. Monkeypox history and epidemiology
Monkeypox was first recognized as a human disease in 1970, when a 9-month-old child in the Democratic Republic of the Congo (DRC, at the time known as Zaire) was suspected of having smallpox (Ladnyj et al., 1972). Laboratory investigations revealed that the causative agent was not VARV, but a virus previously isolated during an outbreak of pustular disease in captive macaques in Denmark (Magnus et al., 1959). Hence, the designation of monkeypox. Since 1970, human cases of monkeypox have been reported from several countries in West and Central Africa (Bunge et al., 2022). In these endemic regions, the number of cases has been fluctuating over decades (Bunge et al., 2022), but in 2017–2019, Nigeria and the DRC suffered the largest ever reported outbreaks (Yinka-Ogunleye et al., 2019,Beer and Rao, 2019).
MPXV is a zoonotic virus and, in the endemic regions, most human cases occur in rural areas and derive from independent introductions from animal reservoirs (most likely rodents, see below), although human-to-human transmission was also documented (Yinka-Ogunleye et al., 2019). The epidemiology of the disease suggests that the viral reservoir is associated with the African rainforest, although the 2017 Nigerian outbreak also included cases in savanna regions and in urban areas (Yinka-Ogunleye et al., 2019). Notably, in West Africa, the surge in human cases that started in 2017 was paralleled by an increased incidence in primate communities (Patrono et al., 2020). Specifically, Patrono and co-workers surveyed populations of human-habituated western chimpanzees in the Ivory Coast and reported multiple MPXV outbreaks starting in 2017, after decades of little viral activity. The hunting habits of the chimpanzees did not change prior to the outbreaks and rodent consumption did not increase. The authors thus suggested that the raising of cases in human and primate communities was related to an epizootic in the reservoir or to changes in the demography and distribution of this latter (Patrono et al., 2020).
The first exported monkeypox cases were registered in 2003 in the US. The US outbreak was initiated by rodents imported from Ghana as exotic pets and eventually resulted in 47 confirmed human cases (Reynolds et al., 2006). Human-to human transmission was limited and most transmissions occurred through contact with animals. In September 2018–2019, there were three unrelated importations of monkeypox to the UK, by persons traveling from Nigeria. One nosocomial human-to-human transmission and a small household cluster were reported (Simpson et al., 2020,Adler et al., 2022). Two additional infected travelers reached Singapore and Israel, with no secondary transmissions (Simpson et al., 2020,Reynolds et al., 2006,Mauldin et al., 2022,Ng et al., 2019). In 2021, two imported travel-related cases were also identified in Maryland and Texas. Again, no secondary transmission was documented (Rao et al., 2022,Costello et al., 2022). Thus, before May 2022, the epidemiology of monkeypox outside the endemic regions was characterized by infrequent, travel-related cases that either did not result in onward transmissions or caused a sporadic and self-extinguishing number of cases. These features, however, do not apply to the expanding worldwide outbreak, as most cases have no epidemiological links to Africa and human-to-human transmission is thought to be driving viral spread (Adalja and Inglesby, 2022). Indeed, patients are mostly young males, who are not vaccinated against smallpox and, in several cases, report a previous story of sexually-transmitted infections. This raises the possibility of community spread, with sexual contact being suspected to contribute to transmission. The clinical presentation also tends to differ from previous outbreaks in that the prodromal symptoms are not always present and anogenital lesions and/or rashes are often observed, whereas the face and extremities are usually not affected. Also the rash phase is often characterized by fewer than 10 lesions (Thornhill et al., 2022,Bragazzi et al., 2022).
3. Genetic features, classification, and tropism
3.1. The poxviridae family
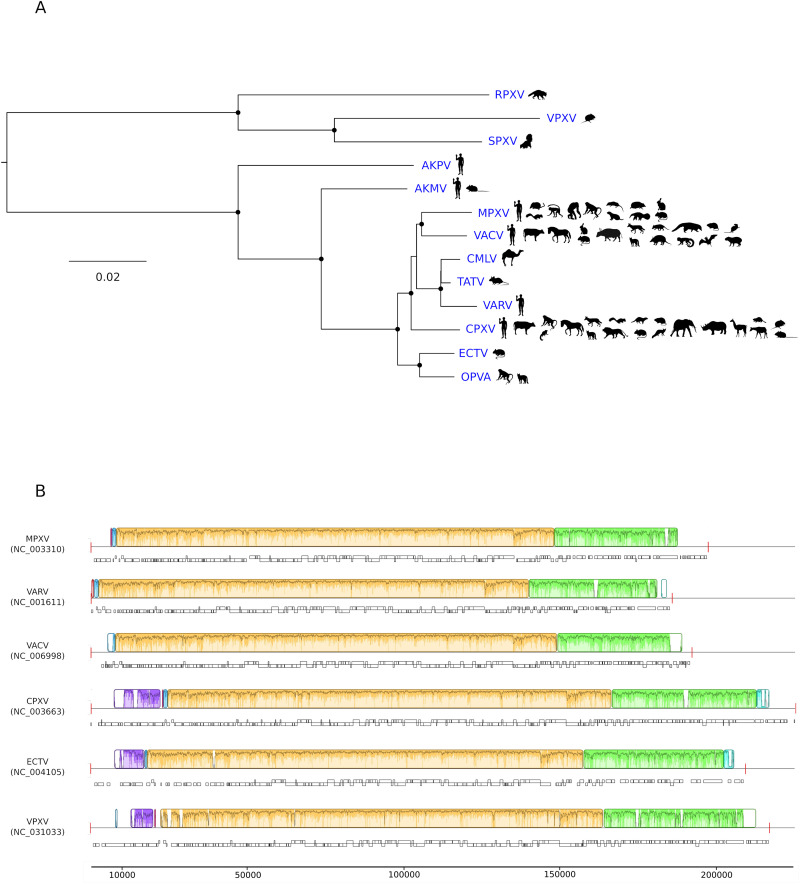
The Poxviridae family comprises many diverse large, enveloped viruses. The poxvirus genome consists of a linear, double-stranded DNA (dsDNA) molecule. Poxviruses infect a wide range of hosts, including insects, birds, reptiles, and mammals (Gyuranecz et al., 2013,Sarker et al., 2019,Alonso et al., 2020,Lefkowitz et al., 2006,Moss, 2013). The Poxviridae family is thus divided into two subfamilies, Chordopoxvirinae and Entomopoxvirinae, for viruses found in vertebrates and invertebrates, respectively. Chordopoxviruses are further classified into 18 genera (https://talk.ictvonline.org/ictv-reports/ictv_9th_report/dsdna-viruses-2011/w/dsdna_viruses/74/poxviridae). Among these, the Orthopoxvirus genus includes MPXV and several viruses of great medical relevance, such as VARV, vaccinia virus (VACV, which was used in the smallpox eradication campaign), and cowpox virus (CPXV), another zoonotic pathogen. Additional orthopoxviruses infect diverse mammals, including camels, voles, rodents, skunks, and raccoons (Fig. 1A). Whereas some of these viruses have been known for decades, others have been identified more recently in humans and other animals. For instance, Akhmeta (AKMV) and Alaska (AKPV) viruses have been sequenced in the last 15 years from people living in Georgia and Alaska (Gigante et al., 2019,Gao et al., 2018). Orthopoxvirus Abatino (OPVA) was instead isolated in Italy during an outbreak in captive macaques in 2015 (Cardeti et al., 2017) and from a case of fatal infection in a cat in 2017 (Lanave et al., 2018). To date, 12 orthopoxvirus species have been officially recognized by the ICTV (https://talk.ictvonline.org/ictv-reports/ictv_9th_report/dsdna-viruses-2011/w/dsdna_viruses/74/poxviridae). Phylogenetically, these viruses form two major clades corresponding to species sampled in the Old World and in the New World. Alaska virus, still not formally classified, forms a distinct clade (Gigante et al., 2019) (Fig. 1A).
Fig. 1.
Orthopoxvirus genetic diversity. (A) Phylogenetic tree of 12 recognized orthopoxvirus species (Akhmeta virus, AKMV; camelpox virus, CMLV; cowpox virus, CPXV; ectromelia virus, ECTV; MPXV; orthopoxvirus Abatino, OPVA; raccoonpox virus, RPXV; skunkpox virus, SPXV; taterapox virus, GBLV, also known as TATV; VACV; VARV; volepox virus, VPXV) plus Alaska virus (AKPV). The tree is mid-point rooted and was constructed with IQTREE (Trifinopoulos et al., 2016), using a concatenated alignment of 49 genes shared by all poxviruses (Upton et al., 2003). Black dots indicate internal nodes with bootstrap support >0.8. Animal silhouettes were obtained from the Phylopic website (http://www.phylopic.org/) and are representative of the host ranges of different viruses (Silva et al., 2020). (B) Whole-genome alignment of MPXV, VARV, VACV, CPXV, and ECTV obtained with progressive Mauve (v.2.3.1) (Darling et al., 2004,Darling et al., 2010). Each genome is laid out in a horizontal track, with annotated coding regions denoted as boxes. Vertical red bars represent genome boundaries. A similarity plot generated by progressive Mauve is shown above each genome, with colors indicating regions that align to part of any another genome and thus represent locally collinear blocks. A similarity profile is also plotted within blocks, with height proportional to the level of conservation in that region. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Orthopoxvirus genomes and host range
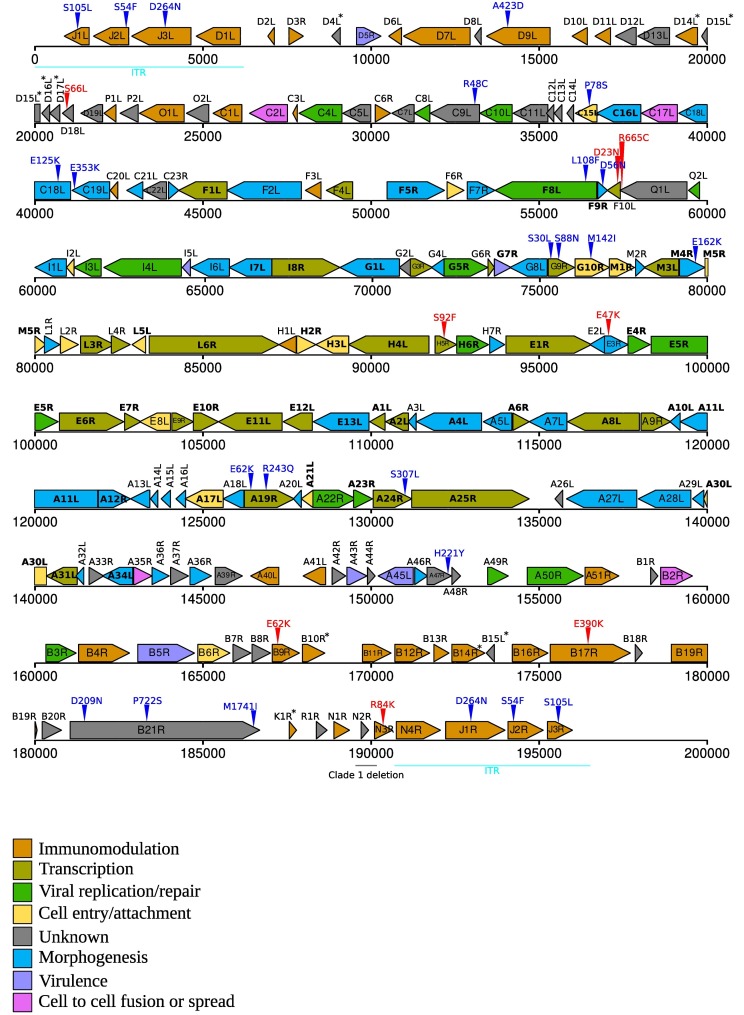
Orthopoxviruses have large and complex genomes ranging in length from ∼170 to ∼230 kb and encoding approximately 200 proteins (Hendrickson et al., 2010). Comparative genomic analyses indicated that orthopoxvirus genomes typically display a central collinear region, where the majority of core genes are located, and dynamic flanking regions, which differ in gene content among species and even among strains of the same species (Fig. 1B). Most likely, recombination, an active process in poxviruses, significantly contributes to genome plasticity (Fenner and Comben, 1958; Bedson and Dumbell, 1964; Moyer et al., 1980; Pickup et al., 1984; Evans et al., 1988; Esposito et al., 2006; Lin and Evans, 2010; Qin et al., 2011; Paszkowski et al., 2016; Senkevich et al., 2021; Evans, 2022; Molteni et al., 2022). Core genes are often conserved well beyond orthopoxviruses and some of them are present in all poxvirus genomes (Upton et al., 2003) (Fig. 1B, Fig. 2 ). The encoded proteins are often essential for virus replication and participate in processes such as virion morphogenesis, replication, and transcription (Fig. 2). Conversely, non-core genes, usually referred to as “accessory”, mainly encode proteins involved in host-virus interactions (Senkevich et al., 2021).
Fig. 2.
Structure of the MPXV genome. A schematic representation of the MPXV genome is reported based on NCBI annotation and gene positions of the reference strain (NC_003310, clade I). Arrowheads indicate gene orientation. Genes are colored based on the function/category they belong to (see legends for details). Forty-nine genes shared among all poxviruses are shown in bold and asterisks denote genes that are deleted/truncated in clade IIa/IIb genomes (see also Table 1). Vertical triangles indicate nonsynonymous single nucleotide variants that are fixed (blue) or polymorphic (red) in B.1 genomes. ITR: inverted terminal repeats. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Recent analyses indicated that the evolution of chordopoxvirus genomes was characterized by major waves of gene acquisition from host genomes. The capture of these genes was followed by extensive gene duplications, resulting in several families of paralogous genes (Senkevich et al., 2021). Nine genes were also gained during the evolution of orthopoxviruses, but many others were lost due to mutation (Senkevich et al., 2021). In fact, the evolution of orthopoxvirus genomes was dominated by gene losses that occurred independently over multiple lineages, so that it is estimated that no extant virus contains all accessory genes that were present in the orthopoxvirus ancestor (Senkevich et al., 2021).
This process of reductive evolution is intimately connected with the evolution of host range (Hendrickson et al., 2010,Senkevich et al., 2021,Hatcher et al., 2014). Indeed, orthopoxviruses with a restricted host range tend to have fewer genes than those with a broad host range. The most extreme example is VARV, which only infected humans and has the smallest genome among orthopoxviruses. On the opposite side, CPXV, with a broad host range (which includes humans), has a genome of approximately 225 Kb, with variations depending on the isolate (Fig. 1B).
The mechanism underlying reductive evolution is that adaptation to a specific host is accompanied by the mutation or deletion of genes that facilitate infection of multiple hosts(Senkevich et al., 2021). Because, unlike many other viruses, cell entry is not a determinant of host range in poxviruses (most mammalian cells are susceptible to infection), lost genes are usually involved in the subversion of the host antiviral responses. Thus, some of the best studied poxvirus host range genes target protein kinase R (PKR), the zinc-finger antiviral protein (ZAP), and the SAMD9/SAMD9L pathway (Cao et al., 2020,Meng et al., 2018,Park et al., 2021,Peng et al., 2020,Zhang et al., 2019).
3.3. MPXV genome and host range
Although far from complete, knowledge about the host range of MPXV in natural settings suggests that the main reservoir is represented by squirrels (see below) (Doty et al., 2017,Tiee et al., 2018). However, the virus has been detected in a number of small mammals (Doty et al., 2017,Tiee et al., 2018,Silva et al., 2020,Bernard and Anderson, 2006). Old World non-human primates are also susceptible to MPXV and experimental or accidental infection (e.g., in captive animals) has indicated that the virus has a wide host range, which extends to non-placental mammals such as opossums (Silva et al., 2020). These observations place MPXV among orthopoxviruses with a broad host range (Fig. 1A).
The MPXV genome of the reference strain (NC_003310, Zaire-96-I-16, clade I) is ∼197 kb long with 191 annotated protein products (Fig. 2). Like in all poxviruses, the linear MPXV genome is flanked by inverted terminal repeats (ITRs) (Fig. 2). Forty-nine genes shared among all poxviruses occupy the central portion of the genome. A number of genes encoding potential immunomodulators are located in the regions flanking core genes (Fig. 2). It should however be noted that interference with the host immune system can also be contributed by proteins that play other, unrelated function in poxvirus biology. For instance, the products of the VACV orthologs of E9R and E10R (D9 and D10 in VACV nomenclature) remove caps from mRNAs but also inhibit the RNAse L/PKR pathway (Liu et al., 2015). In VACV, the same pathway is also modulated by mutations in the A24R gene, which encodes the catalytic subunit of the viral RNA polymerase (A25R in MPXV) (Cone et al., 2017,Brennan et al., 2015). In this respect, it is worth noting that the function of several MPXV genes is simply inferred from experiments performed with VACV, the most extensively studied poxvirus. Whereas the two viruses most likely share most core processes, even subtle changes in proteins involved in interaction with the host might have significant effects on virulence or other phenotypes.
3.4. MPXV cell/tissue tropism
Because poxvirus cell entry is mediated by ubiquitous surface elements, the cell-type and tissue tropism is determined by events that occur after the virus has gained access to the cell and initiated the replication cycle (McFadden, 2005). A full understanding of MPXV tissue/cell tropism in humans is still missing, and most information derive from studies in animal models or with other orthopox viruses. MPXV infection is known to start from the respiratory tract or the skin (Lum et al., 2022). In a macaque model of areosol exposure, the virus can readily infect the oral and respiratory tract mucosae, with airway epithelial cells as the main targets of primary infection (Lum et al., 2022,Zaucha et al., 2001). Lymphoid tissues such as tonsils, mediastinal and mandibular lymph nodes are also infected early after exposure and the lymphatic system contributes to sistemic viral spread. Within draining lyph nodes, MPXV infection is thought to mainly involve macrophages and dendritic cells (Lum et al., 2022,Zaucha et al., 2001). In the macaque model, lesions are mainly detected in the thymus, spleen, skin, oral mucosa, gastrointestinal tract, and reproductive system (Zaucha et al., 2001). A similar tropism was observed in other animals (Falendysz et al., 2017,Osorio et al., 2009), including rope squirrels, which represent a likely natural reservoir of MPXV (see below). Intranasal or intradermal inoculation of these animals causes lesions to occur mainly in the skin, oral cavity, and lungs (Falendysz et al., 2017).
MPXV infection can also occur through the skin. In this case, infection is thought to be initially mediated by keratinocytes and fibroblasts, although skin-resident immune cells may also be infected (Lum et al., 2022). It is however still unclear whether migratory antigen-presenting cell contribute to virus dissemination through the lymphatic system or if the virus gets direct access to lymphatic vessels.
3.5. MPXV genetic diversity
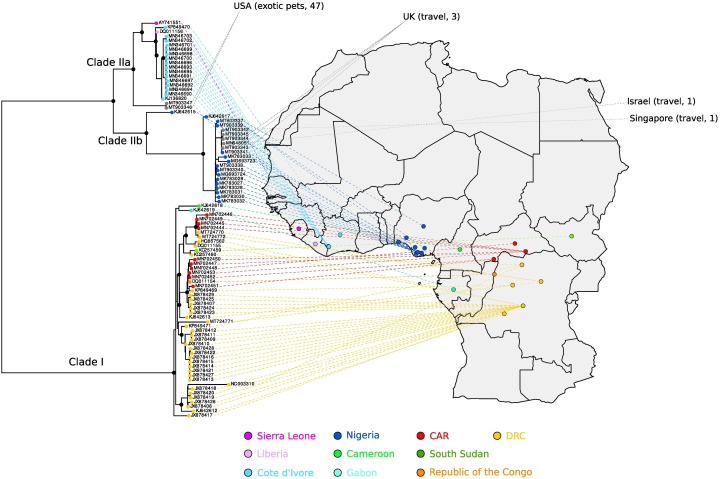
In the endemic regions, the genetic diversity of MPXV is divided into two major clades, which were originally named after the regions where the virus is mainly transmitted: West Africa and the Congo Basin (Berthet et al., 2021,Likos et al., 2005). The West African clade is further divided into two sub-clades broadly corresponding to viruses sampled in Nigeria and West of Nigeria (i.e., in Liberia, Sierra Leone, and the Ivory Coast) (Forni et al., 2022) (Fig. 3 ). Recently, though, it was proposed that, also because of the growing international attention to the current outbreak, a non-stigmatizing and non-discriminatory nomenclature for MPXV clades should be adopted. In particular, Happi and coworkers suggested that clades should be named with numbers in order of detection: clade I (mainly transmitted in the Congo Basin), clade IIa (prevailing West of Nigeria), and clade IIb (mainly Nigerian viruses) (Fig. 3) (Happi et al., 2022). The same authors also indicated that the virus which is spreading worldwide, as well as some recent Nigerian samples, have different features and should be considered as a distinct entity referred to as hMPXV1 (Happi et al., 2022). We thus adopt such nomenclature hereafter.
Fig. 3.
MPXV phylogenetic relationships in the endemic regions. A phylogenetic tree of 89 MPXV genomes from the endemic regions was generated using IQTREE (Trifinopoulos et al., 2016) and virus sampling location was placed on a geographic map using country (indicated by colors) or, when possible, sub-region information. Identical genomes were excluded. The most internal nodes with bootstrap support >0.8 are denoted with dots. The gray color denotes exported human cases (before 2021) with country of destination shown at the end of the dashed lines. The route of exposure and the total number of cases (index and secondary) are reported (Simpson et al., 2020,Reynolds et al., 2006,Mauldin et al., 2022,Ng et al., 2019). Clades are labeled based on to the nomenclature proposed by Happi (Happi et al., 2022). DRC: Democratic Republic of the Congo; CAR: Central African Republic.
Besides their names and geographic distribution, the two major clades (I and II), which show ∼0.5% genomic sequence difference, are associated with distinct disease presentation. In fact, retrospective surveys of human outbreaks and experimental infection in cynomolgus monkeys indicated that clade I MPXV is more virulent and has a higher CFR than clades IIa/IIb (Bunge et al., 2022,Likos et al., 2005,Chen et al., 2005). Understanding the genetic differences between the two clades could thus provide extremely relevant information on the virulence determinants of monkeypox.
Early studies based on the pattern of gene inactivation in the reference clade I genome (NC_003310, Zaire-96-I-16) and in the clade IIa SL-70 strain (AY741551, isolated in Sierra Leone in 1970) suggested some possible virulence determinants and indicated D14L as the most likely candidate (Chen et al., 2005). D14L encodes a complement inhibitor also referred to as MOPICE (monkeypox inhibitor of complement enzymes) which is deleted in clade IIa/IIb viruses (Fig. 2). However, using recombinant MPXV, Estep and co-workers showed that deletion of D14L from the Zaire-96 strain renders the virus more rather than less virulent, at least in a macaque model (Estep et al., 2011). Conversely, deletion of D14L was shown to decrease MPXV virulence in prairie dogs, although insertion of the gene in a clade IIa virus was not associated with changes in disease presentation (Hudson et al., 2012). Thus, these results do not support a relevant role of MOPICE as a virulence determinant, although the protein might engage complement components with different efficiency in distinct species and thus result in variable effects depending on the host.
With the aim to describe differences in gene content between viruses in the two major MPXV clades, we analyzed 20 annotated genomes from clade I and the same number from clade IIa/IIb. We used annotation and orthology analysis to identify genes that are encoded by most genomes in one clade but not in the other (Table 1 , Fig. 2). In addition to D14L, genes encoding potential immunomodulators are present in clade I genomes but not in clades IIa/IIb. These include genes for a putative regulator of apoptosis (B10R), an interleukin 1 binding protein (B14R), and a tumor necrosis factor (TNF) receptor-like protein (K1R) (Table 1). Among these, B14R, already pinpointed by Chen and co-workers (Chen et al., 2005), is particularly interesting because deletion of its ortholog in VACV (B15R) determines increased illness and mortality in mice. Loss of B15R also induces a febrile response, which is prevented by the expression of the interleukin 1 binding protein (Alcamí and Smith, 1996,Alcamí and Smith, 1992). The VARV ortholog (B16R) is also inactivated in most viral genomes, both modern and ancient (Mühlemann et al., 2020). It was thus suggested that its loss was associated with the characteristic high severity of smallpox (Alcamí, 2020). It is thus surprising that, in MPXV, the gene is inactivated in the less virulent clade. Clearly, as the MOPICE example reported above also indicates, multiple genetic factors most likely interact to determine MPXV virulence, with possible differences among hosts. For instance, clade I viruses also encode three short kelch-like proteins, which are deleted in clade IIa/IIb viruses (Table 1, Fig. 2). Such proteins play a role in different processes and, in other poxviruses, were associated with host range and virulence, cell adhesion, evasion of the innate immune system, and interaction with the ubiquitin pathway (Balinsky et al., 2007,Kochneva et al., 2005,Shchelkunov, 2010,Wang et al., 2014). Independent deletion of two kelch-like proteins in mouse models leads to increased lesion size, although the phenotype also depends on the mode of infection (i.e., intradermal or intranasal) (Beard et al., 2006,Pires de Miranda et al., 2003). Conversely, deletion of the F12L gene in VACV, also encoding a kelch-like protein, results in lower virulence (Froggatt et al., 2007). In vitro and in vivo studies will thus be necessary to assess the role of the three kelch-like proteins encoded by clade I MPXV, which are however very short, thus raising doubts about their functional potential. Along the same lines, the tumor necrosis factor receptor-like molecule encoded by clade I viruses is a truncated copy of the protein expressed by some VACV and VARV strains, where it acts as a virulence factor (Alvarez-de Miranda et al., 2021). In particular, the MXPV protein only covers the first cystein-rich domain and a portion of the second, making its ability to bind TNF uncertain.
Table 1.
Differences in gene content between the two major MPXV clades.
| Name | Positiona | Clade IIa/IIbb | Clade Ic | Function/features | Reference |
|---|---|---|---|---|---|
| D4L | 8830–9081 | 20 fragmentedd | 20 intactd | Belongs to the poxvirus C4/C10 family of immunomodulators | (Ember et al., 2012) |
| D14L | 19,060–19,710 | – | 20 intact | Complement inhibitor and virulence factor | (Chen et al., 2005) |
| D15L | 19,834–20,151 | – | 20 intact | Kelch-like protein | |
| D16L | 20,205–20,438 | – | 20 intact | Kelch-like protein | |
| D17L | 20,440–20,736 | – | 20 intact | Kelch-like protein | |
| MPXV_LIB1970_184_057 | 49,967–50,368 (DQ011156) | 17 intact, 3 fragmented | 20 fragmented | Ortholog in VACV is an inhibitor of cGAS | (Yang et al., 2021) |
| B10R | 167,957–168,622 | 15 intact, 2 fragmented | 20 intact | Possible regulator of apoptosis | (Barry et al., 1997) |
| B14R | 172,403–173,383 | 20 fragmented or truncatedd | 15 intact, 5 truncated | Interleukin 1 binding protein | (Alcamí and Smith, 1992) |
| B15L | 173,429–173,665 | 20 fragmented or truncated | 20 intact | Unknown | |
| K1R | 187,567–187,779 | 20 fragmented or truncated | 20 intact | Tumor necrosis factor receptor-like | (Alvarez-de Miranda et al., 2021) |
If not specified, the position refers to the reference genome (NC_003310).
Gene status in 20 clade IIa/IIb viruses (AY741551, DQ011156, KP849470, MT903346, MT903347, KJ642615, MT903341, MT903338, MT903342, KJ642617, MN648051, MT903337,MT903339, MT903340,MT903343, MT903344, MT903345,MG693723, MG693724, MG693725).
Gene status in 20 clade I viruses (KC257459, NC_003310, DQ011154, DQ011155,HQ857562, JX878407, JX878416, JX878429, KC257460, KJ642612,KJ642613, KJ642618, KJ642619,KP849469, KP849471, MN702446, MN702449, MN702451, MN702452, MN702453).
Intact: the predicted protein is full-length; fragmented: the predicted protein is less than 50% in length than the intact protein; truncated: the predicted protein is not full length but it is more than 50% in length compared to the intact protein.
Concerning clade IIa/IIb, we detected one gene that is present in these viruses but not in those from clade I (Table 1). The predicted encoded protein is an N-terminal and C-terminal truncated version of the E5 protein of VACV and VARV (Yang et al., 2021). E5 is inactivated in some ancient VARV strains but it is encoded by most modern genomes (Mühlemann et al., 2020). Its function is still largely unknown, although preliminary evidence suggests that the VACV protein acts as an inhibitor of the cytoplasmic DNA sensor cGAS and that it represents a virulence factor in mice (Yang et al., 2021). However, because the functional domains of the protein were not characterized, it is impossible to determine whether the short version of the protein encoded by MPXV clade IIa/IIb viruses performs the same functions as the full-length VACV ortholog.
Clearly, MPXV clades do not only differ in terms of gene content, but also display single nucleotide substitutions and small indels in coding and non-coding regions. Understanding the role of such changes for viral phenotypes is clearly challenging. Extended sampling and collection of related metadata in the endemic regions may provide valuable information. Most likely, though, such data will need to be integrated with results from in vivo and in vitro reverse genetic experiments.
Finally, it should be noted that differences also exist among viruses in the same clade and some of these might be associated with relevant features of MPXV lineages. For instance, clade I viruses carrying a 628 bp deletion were previously identified during a survey of primary and secondary human infections in the DRC (Kugelman et al., 2014). The deletion removes the N2R gene (with unknown function) and the 5’region of N3R, which encodes the orthopoxvirus major histocompatibility complex class I–like protein (OMCP). OMPC functions as an antagonist of the NKG2D activating receptor, possibly representing a strategy to avoid NK-cell-mediated killing (Campbell et al., 2007). Based on the clinical metadata, it was suggested that the deletion is associated with increased human-to-human transmission. Clearly, additional data and continuous genomic surveillance will be necessary to asses whether this is the case.
4. Monkeypox laboratory diagnosis and MPXV genome sequencing
The clinical presentation of monkeypox may resemble that of other infectious diseases or conditions, including chickenpox (VZV, varicella zoster virus), herpes simplex virus (HSV) infection, measles, bacterial skin infections, scabies, primary or secondary syphilis or other orthopoxvirus infections. Thus, accurate and rapid molecular diagnosis for confirmation of MPXV infection is critical for timely control and tracking of transmissions.
Monkeypox diagnostic tools include both serological analysis and polymerase chain reaction (PCR) tests. Serological examination is limited by cross-reactivity with different orthopoxviruses, as antigen/antibody presence might be the result of previous exposure to other orthopoxviruses or to the smallpox vaccine. Although it does not lead to a definitive diagnosis of MPXV infection, serological analysis is very useful for epidemiological purposes, especially for monitoring orthopoxvirus epidemics in non-endemic areas (Mileto et al., 2022).
Conversely, detection of MPXV DNA from clinical specimens, as well as veterinary samples or MPXV-infected cell cultures, is based primarily on nucleid acid amplification testing, using real-time polymerase chain reaction (quantitative PCR, qPCR) or PCR, alone or in combination with sequencing. Given the specificity, sensitivity, low-cost, and rapidity of qPCR analysis, this approach is highly recommended for routine diagnosis (http://www.who.int/health-topics/monkeypox, https://www.cdc.gov/poxvirus/monkeypox/lab-personnel/index.html). Several validated qPCR protocols were developed for the detection of orthopoxviruses, MPXV and/or to discriminate MPXV clade I from clade IIa/IIb (WHO, https://www.cdc.gov/poxvirus/monkeypox/lab-personnel/index.html) (Li et al., 2010,Orba et al., 2015,Li et al., 2006). In addition, several commercial qPCR assays are now available for the qualitative detection of MPXV nucleic acid.
Swabs of lesion exudate and/or surface or crust sample represent the best specimens for MPXV DNA detection, although viral DNA can also be detected in blood, seminal fluid and urine (Mileto et al., 2022,Antinori et al., 2022,Peiró-Mestres et al., 2022). In the latter samples, the quantity of viral DNA is reduced compared to that present in the vesicular samples and depends on the viremic phase. Therefore an analysis carried out exclusively on these samples is at risk of false negative results.
The PAHO/WHO suggests two protocols for orthopoxvirus and MPXV detection (https://www.paho.org/en/documents/laboratory-guidelines-detection-and-diagnosis-monkeypox-virus-infection). The first consists of a qPCR using the orthopoxvirus test; positive samples are then subjected to MPXV-specific qPCR. In the second protocol, the samples are directly subjected to MPXV-specific PCR, followed by differentiation of clade I and clade IIa/IIb using primers/probes specific for the D14L and J2R genes, respectively. As mentioned above, D14L (C3L in VACV nomenclature) is deleted in clade IIa/IIb viruses, hence the specificity of the assay for clade I (Li et al., 2010). In the case of J2R, different primer/probe sets detect either all MPXV clades or clade IIa/IIb only (Li et al., 2010).
Finally, whole-genome sequencing, using high-throughput sequencing (HTS), represents a gold standard for the characterization of MPXV. This type of technology is more expensive than qPCR and requires a much longer data analysis time, making it unsuitable for a diagnostic routine. However, virus genome data are critical for evidence-based epidemiological interventions and outbreak genome surveillance. In general, whole genome sequencing can provide information on transmission chains, viral geographic spread, and microevolutionary processes. As detailed below, HTS efforts early during the recent MPXV epidemic allowed the tracing of the viral lineages and the detection of ongoing evolution in hMPXV1 genomes (Isidro et al., 2022,Gigante et al., 2022).
5. Ecology and geographic distribution of MPXV
Whether genetic differences between the two major clades represent MPXV adaption to new environments or hosts is presently unknown. Recent data indicated that the two clades separated ∼560–860 years ago, a time frame that spans from the Medieval Warm Period to the Little Ice Age (Forni et al., 2022). This time frame was characterized by hydrological changes that affected the extension of the tropical rainforest, which expanded during high rainfall periods and contracted when the climate was drier (Ngomanda et al., 2007). It was thus suggested that such environmental changes determined the movement and geographic patterning of the MPXV reservoir(s) (Forni et al., 2022). Based on the level of genetic drift in distinct populations, a possible origin of MPXV in West Africa was proposed (Forni et al., 2022). Migration of the reservoir into the Congo Basin might have resulted in the separation of the two major clades, with rainforest extension and distribution playing a major role in this process. In fact, despite major gaps in our understanding of MPXV prevalence in wild mammals, the epidemiology of human cases suggests that at least some of the reservoir species dwell in the rainforest.
Although wild non-human primates were found to be infected with MPXV, these animals are not thought to contribute significantly to MPXV circulation (Patrono et al., 2020,Radonić et al., 2014). Conversely, based on field surveys and analysis of museum specimens, rope squirrels (Funisciurus spp) are considered potential natural reservoirs for MPXV (Doty et al., 2017,Tiee et al., 2018). Indeed, the first isolation of the virus from a wild animal was from a diseased rope squirrel in the DRC (Khodakevich et al., 1986) (Table 2 ). Also, analysis of more than 1000 museum specimens from Central Africa revealed MPXV genomic DNA in specimens from different species of African rope squirrels (F. anerythrus, F. carruthersi, F. congicus, F. lemniscatus, and F. pyrropus), the highest prevalence being observed for F. anerythrus (Thomas' rope squirrel) and F. congicus (Congo rope squirrel)(Tiee et al., 2018). However, both in Central and in West Africa, additional small mammals were found to be potential carriers of MPXV. In the DRC, MPXV DNA was found in rodents (Lunda rope squirrels, target rats, giant pouched rats) and naked-tail shrews (Mariën et al., 2021) (Table 2). In West Africa, evidence of orthopoxvirus infection was obtained in giant pouched rats, African dormice, sun squirrels, and ground squirrels (Reynolds et al., 2010) (Table 2). It is thus possible, and even likely, that MPXV circulates in a wide range of wild mammals, although several serological studies could not provide definitive demonstration of MPXV infection because of potential cross-reactivity with other orthopoxviruses (Table 2).
Table 2.
List of mammals showing potential evidence of natural infection with MPXV.
| Common name(s) | Scientific name(s) | Location/provenance | Evidence | Reference |
|---|---|---|---|---|
| Thomas' rope squirrel | Funisciurus anerythrus | DRC | Virus isolation | (Khodakevich et al., 1986) |
| Sooty mangabey | Cercocebus atys | Ivory Coast | Virus isolation | (Radonić et al., 2014) |
| African rope squirrels | Funisciurus anerythrus, Funisciurus congicus, Funisciurus carruthersi, Funisciurus lemniscatus, and Funisciurus pyrropus | Museum specimens, Central Africa | RT-PCR/high-resolution melting/Sanger sequencing | (Tiee et al., 2018) |
| Thomas' rope squirrel | Funisciurus anerythrus | DRC | PCR assays/sequencing | (Mariën et al., 2021) |
| Lunda rope squirrel | Funisciurus bayonii | DRC | PCR assays/sequencing | (Mariën et al., 2021) |
| Target rat | Stochomys longicaudatus | DRC | PCR assays/sequencing | (Mariën et al., 2021) |
| Giant pouched rat | Cricetomys spp | DRC | PCR assays/sequencing | (Mariën et al., 2021) |
| Naked-tail shrews | Crocidura littoralis | DRC | PCR assays/sequencing | (Mariën et al., 2021) |
| African dormouse | Graphiurus spp | Ghana | RT-PCR, serology | (Reynolds et al., 2010) |
| Giant pouched rat | Cricetomys spp | Ghana | RT-PCR, serology | (Reynolds et al., 2010) |
| Ground squirrel | Xerus spp | Ghana | RT-PCR | (Reynolds et al., 2010) |
| Western chimpanzee | Pan troglodytes verus | Ivory Coast | RT-PCR/sequencing | (Patrono et al., 2020) |
| African rope squirrels | Funisciurus spp | Ghana | Serology | (Reynolds et al., 2010) |
| Sun squirrel | Heliosciurus spp | Ghana | Serology | (Reynolds et al., 2010) |
| African rope squirrels | Funisciurus spp | DRC | Serology | (Doty et al., 2017,Nolen et al., 2015) |
| Lorrain dormouse | Graphiurus lorraineus | DRC | Serology | (Doty et al., 2017) |
| Emin's pouched rat | Cricetomys emini | DRC | Serology | (Doty et al., 2017) |
| Sun squirrel | Heliosciurus spp | DRC | Serology | (Doty et al., 2017) |
| Common rufous-nosed rat | Oenomys hypoxanthus | DRC | Serology | (Doty et al., 2017) |
| Elephant shrew | Petrodromus tetradactylus | DRC | Serology | (Doty et al., 2017) |
With respect to the identity of the MPXV reservoir, it is interesting to note that experimental infection of African rope squirrels with a clade I strain causes significant pathology and mortality, and the infected animals shed large quantities of virus (Falendysz et al., 2017). In comparison, Gambian pouched rats are susceptible to MPXV but seem to suffer less severe disease and lower mortality when infected with the same viral strain (Falendysz et al., 2015). These animals are frequently hunted and eaten (Guagliardo et al., 2020), making them an attractive candidate as a source of human infections.
Both rope squirrels and Gambian pouched rats are distributed in the forest and savanna areas of Central and West Africa (https://www.iucnredlist.org/species/1). Conversely, other possible MPXV reservoirs such as Congo rope squirrels, Lunda rope squirrels, and naked-tail shrews have a more restricted distribution confined to Central Africa (https://www.iucnredlist.org/). It is thus possible that in the regions where the two major MPXV clades are transmitted different reservoirs sustain viral circulation and spread. Thus, geographic barriers might not primarily underlie the separation of clades I and IIa/IIb, which may instead derive from diverse ecological opportunities of infection and adaptation to different natural hosts (Forni et al., 2022).
6. hMPXV1 and the global outbreak
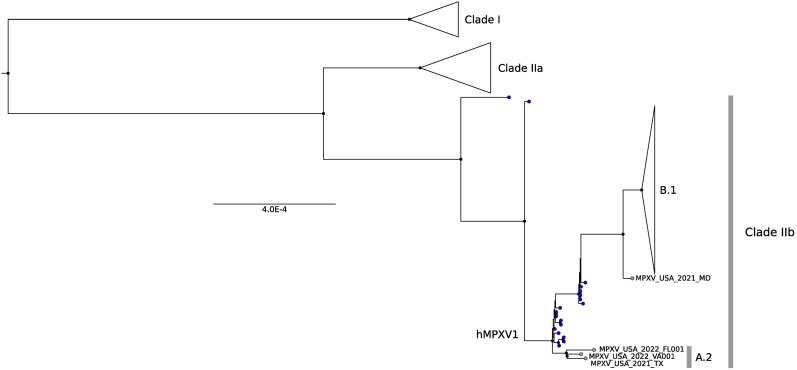
Based on the COVID-19 pandemic experience, a global genome sequencing effort was soon implemented to understand the origin and spread of the hMPXV1 outbreak. Early results showed that all hMPXV1 genomes sampled in 2022 cluster together, thus suggesting a single origin and the same transmission chain (Isidro et al., 2022,Gigante et al., 2022). Phylogenetically, those genomes belong to clade IIb (Isidro et al., 2022,Gigante et al., 2022). However, their genomic and epidemiological features are quite different compared to the hMPXV1 Nigerian strains, placing them in a separated sub-clade (B.1) (Fig. 4 ). Specifically, the B.1 genomes differ from the hMPXV1 viruses sampled before 2021 by at least 46 single nucleotide substitutions, 24 of them being missense mutations (Isidro et al., 2022,Gigante et al., 2022) (Fig. 2). This rate of evolution is much faster than expected for orthopoxviruses (Firth et al., 2010) and may represent an indication of viral adaptation to the human host. Moreover, Isidro and co-workers detected ongoing evolution among the 2022 strains, with 15 substitutions (8 missense) that differentiate the genomes they analyzed (Fig. 2) (Isidro et al., 2022). Non-synonymous mutations do not appear to be preferentially located in specific gene categories and tend to be scattered throughout the genome (Fig. 2). More interestingly, the overwhelming majority of these mutations and the ones that characterize the 2022 outbreak involve GA > AA or TC > TT replacements, suggesting the presence of some specific molecular mechanism possibly related to the action of host apolipoprotein B mRNA editing catalytic polypeptide-like 3 (APOBEC3) proteins (Isidro et al., 2022,Gigante et al., 2022). APOBEC3 proteins have an important role in the host immune system as antiviral effectors. They act by deamination of cytidine nucleotides to uridines in single-stranded DNA substrates by preferentially targeting specific nucleotide motifs (e.g. 5′-TC-3′) (Harris and Dudley, 2015).
Fig. 4.
Global MPXV phylogenetic distribution. Phylogenetic tree of 79 representative MPXV/hMPXV1 strains sampled worldwide. A conserved genomic region (Kugelman et al., 2014) was aligned using MAFFT and the tree was generated using IQTREE (Trifinopoulos et al., 2016). Clades are labeled based on the nomenclature proposed by Happi and colleagues (Happi et al., 2022). Black dots indicate internal nodes with bootstrap support >0.8, blue dots indicate clade IIb strains sampled before 2021. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Humans encode 7 APOBEC3 proteins (APOBEC3A to 3H), which differ in motif preference and tissue expression (Harris and Dudley, 2015). Their action has been extensively studied in HIV and other RNA and DNA viruses, many of which have developed strategies to evade APOBEC3-mediated restriction (Harris and Dudley, 2015). For instance, HIV-1 encodes the Vif (viral infectivity factor) protein which targets APOBEC3G for degradation (Harris and Dudley, 2015). Poxviruses are not known to encode APOBEC3-counteracting products and VACV is not restricted by APOBEC3G, 3F or 3H in vitro (Kremer et al., 2006). Dedicated experiments will thus be required to study the effect of APOBEC3s on MPXV/hMPXV1 replication in human cells.
Interestingly, Gigante and co-workers also analyzed monkeypox genomes from the 2021 cases in Maryland and Texas, and revealed a more complex epidemiology than previously suspected. In fact, the 2021 Maryland genome shared several genomic substitutions with the B.1 clade (Fig. 4), although differing from the latter by 13 mutations (Gigante et al., 2022). Conversely, the 2021 Texas sequence did not cluster with B.1 genomes, although still belonging within the hMPXV1 diversity (Fig. 4). Notably, two 2022 US cases were related to the Texas genome, although, based on the differences among each other, they were not directly linked (Gigante et al., 2022). Thus, all these three genomes were assigned to a specific hMPXV1 clade: A.2 (Fig. 4). Altogether, these results suggest different introductions of the virus in the US. An important finding related to these sequences is that they share the same substitution patter found in the predominant B.1 genomes and ascribed to APOBEC3 action. Indeed, the same pattern was also found in hMPXV1 samples collected between 2017 and 2019 in Nigeria, but not in clade IIa or clade I samples (Gigante et al., 2022). These results suggest that the recent evolution of MPXV was characterized by changes in the virus biology (e.g. occurrence of adaptive mutations) or in its interaction with host proteins.
Finally, a recent preprint (Jones et al., 2022) analyzed several B.1 genomes from Germany and, after confirming the pattern described above, highlighted the presence of a genomic translocation involving the ITRs. In particular, the authors found a duplication event of a gene (D2L) located in the 5′ region of the genome that generates a copy of this gene in the 3′ region, leading to the complete or partial disruption of other 4 genes (Jones et al., 2022). This event was found in one German strain only, but analysis of older African samples identified two other rearrangements in the same regions, although with slightly different mechanisms, both in samples from clade IIa/IIb and in samples from clade I (Jones et al., 2022). Notably, two of the genes affected by the deletion in the German case are N2R and N3R, also deleted in the DRC samples mentioned above (Kugelman et al., 2014). The authors thus suggested that these events could represent an adaptation of the virus that facilitate human-to-human transmission (Jones et al., 2022).
7. Conclusions and perspectives
Epidemiological and genetic data suggest that the ongoing hMPXV1 outbreak is the result of events that occurred before 2022, most likely starting in 2017. The increased incidence of monkeypox in both West and Central Africa in the past five years has been considered to be at least partially caused by discontinuation of smallpox vaccination. However, the observation that MPXV prevalence also increased in primate communities starting in 2017 suggests that waning immunity against orthopoxviruses is not the sole determinant of monkeypox outbreak intensification (Yinka-Ogunleye et al., 2019,Patrono et al., 2020,Rimoin et al., 2010,Sklenovská and Van Ranst, 2018). In this respect, it is worth noting that, in the same period, Nigeria experienced a surge in human cases of Lassa fever, which is caused by another zoonotic virus transmitted by rodents (although unrelated from poxviruses) (Siddle et al., 2018). It is thus possible that climatic or ecological changes resulted in multiple epizootics in rodent populations with a consequent increase in the transmission to humans and non-human primates. Alternatively, widespread encroachment on wild habitats might have favored human-animal interactions eventually resulting in ample opportunities for spillovers. However, the pattern of substitution observed both in B.1 and in earlier samples of hMPXV1 also suggests that some unexplained shift in the virus biology occurred. Different hypotheses have been put forward, including cryptic transmission in human populations, circulation in a novel reservoir, and superspreader events (Isidro et al., 2022). Whereas all these are valid possibilities, the most arcane aspect remains the change in the substitution spectrum, which is suggestive of APOBEC3 action. No evidence of APOBEC3-mediated deamination is evident before 2017 or in other MPXV clades, and poxvirsuses are not considered targets of APOBEC3. Thus, it remains to be determined whether any APOBEC3 protein is indeed responsible for the observed substitution pattern and whether this has any relation to the increased human-to-human spread of hMPXV1. In this respect, it might be worth noting that, since 2018, an increase of monkeypox cases was also observed in Central Africa, where a different viral lineage, with a distinct evolutionary history and no evidence of APOBEC3-mediated changes, is transmitted (Beer and Rao, 2019). Additional analyses and experiments will be necessary to clarify these open questions, which however highlight how large dsDNA viruses, which are usually considered slow-evolving, can adopt new evolutionary strategies. An interesting corollary of this observation is that VACV was shown to adapt to the host innate immune system in vitro by gene copy number amplification and contraction, a model referred to as “genomic accordion” (Elde et al., 2012). In general, gene gains/losses seem to have played a relevant role in poxvirus evolution and in determining host range. This is not the case of the ongoing hMPXV1 outbreak as, with the exclusion of the German case mentioned above, virus evolution seems to occur through point mutations.
A major problem in the interpretation of the mutation pattern observed in hMPXV1 sequences is that the large MPXV genome is poorly studied and the function of many proteins remains largely uncharacterized. Indeed, we even have little clues about the virulence determinants of the two major clades, which have been transmitting for decades in the endemic regions. International efforts will be required to fill these knowledge gaps and to develop and widely deploy strategies to diagnose, prevent and treat monkeypox. Large scale field studies and genomic surveillance should also be undertaken to clarify the nature and distribution of the MPXV reservoirs in West and Central Africa. In this respect, we should add that the wide host range of MPXV is of concern, especially as the virus has now expanded in novel areas characterized by diverse ecological conditions and, thus, by a variety of naive potential hosts. As already noted elsewhere, American ground squirrels are susceptible to MPXV infection, and possibly many other mammals are (Tesh et al., 2004,Xiang and White, 2022). It is thus imperative that the hMPXV1 outbreak is rapidly controlled to prevent establishment of novel MPXV reservoirs that might sustain viral spread and promote its accelerated evolution.
Credit author statement
| Conceptualization | MS, DF, RC |
| Data curation | MS, DF, RC, CM, MC |
| Writing - original draft | MS, RC, DF, MC |
| Writing - review & editing | MS, DF, RC, CM, MC |
| Visualization | MS, DF, CM |
| Supervision | MS, MC |
| Funding acquisition | MS |
Funding
This work was supported by the Italian Ministry of Health (“Ricerca Corrente 2022” to MS).
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
Given her role as Editor-in-Chief, Dr. Manuela Sironi had no involvement in the peer-review of this article and has no access to information regarding its peer-review. Full responsibility for the editorial process for this article was delegated to the Editor-in-Chief Professor Fernando Gonzalez-Candelas.
Data availability
No data was used for the research described in the article.
References
- Adalja A., Inglesby T. A novel international monkeypox outbreak. Ann. Intern. Med. 2022;75:1175–1176. doi: 10.7326/M22-1581. [DOI] [PubMed] [Google Scholar]
- Adler H., Gould S., Hine P., Snell L.B., Wong W., Houlihan C.F., Osborne J.C., Rampling T., Beadsworth M.B., Duncan C.J., Dunning J., Fletcher T.E., Hunter E.R., Jacobs M., Khoo S.H., Newsholme W., Porter D., Porter R.J., Ratcliffe L., Schmid M.L., Semple M.G., Tunbridge A.J., Wingfield T., Price N.M., NHS England High Consequence Infectious Diseases (Airborne) Network Clinical features and management of human monkeypox: a retrospective observational study in the UK. Lancet Infect. Dis. 2022;22:1153–1162. doi: 10.1016/S1473-3099(22)00228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcamí A. Was smallpox a widespread mild disease? Science. 2020;369:376–377. doi: 10.1126/science.abd1214. [DOI] [PubMed] [Google Scholar]
- Alcamí A., Smith G.L. A soluble receptor for interleukin-1 beta encoded by vaccinia virus: a novel mechanism of virus modulation of the host response to infection. Cell. 1992;71:153–167. doi: 10.1016/0092-8674(92)90274-g. [DOI] [PubMed] [Google Scholar]
- Alcamí A., Smith G.L. A mechanism for the inhibition of fever by a virus. Proc. Natl. Acad. Sci. U. S. A. 1996;93:11029–11034. doi: 10.1073/pnas.93.20.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso R.C., Moura P.P., Caldeira D.F., Mendes M.H., Pinto M.H., Cargnelutti J.F., Flores E.F., de Sant’Ana F.J. Poxviruses diagnosed in cattle from Distrito Federal, Brazil (2015–2018) Transbound. Emerg. Dis. 2020;67:1563–1573. doi: 10.1111/tbed.13490. [DOI] [PubMed] [Google Scholar]
- Alvarez-de Miranda F.J., Alonso-Sánchez I., Alcamí A., Hernaez B. TNF decoy receptors encoded by poxviruses. Pathogens. 2021;10:1065. doi: 10.3390/pathogens10081065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinori A., Mazzotta V., Vita S., Carletti F., Tacconi D., Lapini L.E., D'Abramo A., Cicalini S., Lapa D., Pittalis S., Puro V., Rivano Capparuccia M., Giombini E., Gruber C.E.M., Garbuglia A.R., Marani A., Vairo F., Girardi E., Vaia F., Nicastri E., INMI Monkeypox Group Epidemiological, clinical and virological characteristics of four cases of monkeypox support transmission through sexual contact, Italy, May 2022. Euro Surveill. 2022;27:2200421. doi: 10.2807/1560-7917.ES.2022.27.22.2200421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balinsky C.A., Delhon G., Afonso C.L., Risatti G.R., Borca M.V., French R.A., Tulman E.R., Geary S.J., Rock D.L. Sheeppox virus kelch-like gene SPPV-019 affects virus virulence. J. Virol. 2007;81:11392–11401. doi: 10.1128/JVI.01093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry M., Hnatiuk S., Mossman K., Lee S.F., Boshkov L., McFadden G. The myxoma virus M-T4 gene encodes a novel RDEL-containing protein that is retained within the endoplasmic reticulum and is important for the productive infection of lymphocytes. Virology. 1997;239:360–377. doi: 10.1006/viro.1997.8894. [DOI] [PubMed] [Google Scholar]
- Beard P.M., Froggatt G.C., Smith G.L. Vaccinia virus kelch protein A55 is a 64 kDa intracellular factor that affects virus-induced cytopathic effect and the outcome of infection in a murine intradermal model. J. Gen. Virol. 2006;87:1521–1529. doi: 10.1099/vir.0.81854-0. [DOI] [PubMed] [Google Scholar]
- Bedson H.S., Dumbell K.R. Hybrids derived from the viruses of variola major and cowpox. J. Hyg. (Lond) 1964;62:147–158. doi: 10.1017/s0022172400039887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer E.M., Rao V.B. A systematic review of the epidemiology of human monkeypox outbreaks and implications for outbreak strategy. PLoS Negl. Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard S.M., Anderson S.A. Qualitative assessment of risk for monkeypox associated with domestic trade in certain animal species, United States. Emerg. Infect. Dis. 2006;12:1827–1833. doi: 10.3201/eid1212.060454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet N., Descorps-Declère S., Besombes C., Curaudeau M., Nkili Meyong A.A., Selekon B., Labouba I., Gonofio E.C., Ouilibona R.S., Simo Tchetgna H.D., Feher M., Fontanet A., Kazanji M., Manuguerra J.C., Hassanin A., Gessain A., Nakoune E. Genomic history of human monkey pox infections in the Central African Republic between 2001 and 2018. Sci. Rep. 2021;11:13085. doi: 10.1038/s41598-021-92315-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragazzi N.L., Kong J.D., Mahroum N., Tsigalou C., Khamisy-Farah R., Converti M., Wu J. Epidemiological trends and clinical features of the ongoing monkeypox epidemic: a preliminary pooled data analysis and literature review. J. Med. Virol. 2022 doi: 10.1002/jmv.27931. (In press) [DOI] [PubMed] [Google Scholar]
- Brennan G., Kitzman J.O., Shendure J., Geballe A.P. Experimental evolution identifies vaccinia virus mutations in A24R and A35R that antagonize the protein kinase R pathway and accompany collapse of an extragenic gene amplification. J. Virol. 2015;89:9986–9997. doi: 10.1128/JVI.01233-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge E.M., Hoet B., Chen L., Lienert F., Weidenthaler H., Baer L.R., Steffen R. The changing epidemiology of human monkeypox-A potential threat? A systematic review. PLoS Negl. Trop. Dis. 2022;16 doi: 10.1371/journal.pntd.0010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J.A., Trossman D.S., Yokoyama W.M., Carayannopoulos L.N. Zoonotic orthopoxviruses encode a high-affinity antagonist of NKG2D. J. Exp. Med. 2007;204:1311–1317. doi: 10.1084/jem.20062026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Varga J., Deschambault Y. Poxvirus encoded eIF2α homolog, K3 family proteins, is a key determinant of poxvirus host species specificity. Virology. 2020;541:101–112. doi: 10.1016/j.virol.2019.12.008. [DOI] [PubMed] [Google Scholar]
- Cardeti G., Gruber C.E.M., Eleni C., Carletti F., Castilletti C., Manna G., Rosone F., Giombini E., Selleri M., Lapa D., Puro V., Di Caro A., Lorenzetti R., Scicluna M.T., Grifoni G., Rizzoli A., Tagliapietra V., De Marco L., Capobianchi M.R., Autorino G.L. Fatal outbreak in tonkean macaques caused by possibly novel orthopoxvirus, Italy, January 2015 (1) Emerg. Infect. Dis. 2017;23:1941–1949. doi: 10.3201/eid2312.162098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Li G., Liszewski M.K., Atkinson J.P., Jahrling P.B., Feng Z., Schriewer J., Buck C., Wang C., Lefkowitz E.J., Esposito J.J., Harms T., Damon I.K., Roper R.L., Upton C., Buller R.M. Virulence differences between monkeypox virus isolates from West Africa and the Congo basin. Virology. 2005;340:46–63. doi: 10.1016/j.virol.2005.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone K.R., Kronenberg Z.N., Yandell M., Elde N.C. Emergence of a viral RNA polymerase variant during gene copy number amplification promotes rapid evolution of vaccinia virus. J. Virol. 2017;91 doi: 10.1128/JVI.01428-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello V., Sowash M., Gaur A., Cardis M., Pasieka H., Wortmann G., Ramdeen S. Imported monkeypox from international traveler, Maryland, USA, 2021. Emerg. Infect. Dis. 2022;28:1002–1005. doi: 10.3201/eid2805.220292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A.C., Mau B., Blattner F.R., Perna N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A.E., Mau B., Perna N.T. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doty J.B., Malekani J.M., Kalemba L.N., Stanley W.T., Monroe B.P., Nakazawa Y.U., Mauldin M.R., Bakambana T.L., Liyandja Dja Liyandja T., Braden Z.H., Wallace R.M., Malekani D.V., McCollum A.M., Gallardo-Romero N., Kondas A., Peterson A.T., Osorio J.E., Rocke T.E., Karem K.L., Emerson G.L., Carroll D.S. Assessing monkeypox virus prevalence in small mammals at the human-animal interface in the democratic Republic of the Congo. Viruses. 2017;9:283. doi: 10.3390/v9100283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde N.C., Child S.J., Eickbush M.T., Kitzman J.O., Rogers K.S., Shendure J., Geballe A.P., Malik H.S. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell. 2012;150:831–841. doi: 10.1016/j.cell.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ember S.W.J., Ren H., Ferguson B.J., Smith G.L. Vaccinia virus protein C4 inhibits NF-κB activation and promotes virus virulence. J. Gen. Virol. 2012;93:2098–2108. doi: 10.1099/vir.0.045070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito J.J., Sammons S.A., Frace A.M., Osborne J.D., Olsen-Rasmussen M., Zhang M., Govil D., Damon I.K., Kline R., Laker M., Li Y., Smith G.L., Meyer H., Leduc J.W., Wohlhueter R.M. Genome sequence diversity and clues to the evolution of variola (smallpox) virus. Science. 2006;313:807–812. doi: 10.1126/science.1125134. [DOI] [PubMed] [Google Scholar]
- Estep R.D., Messaoudi I., O'Connor M.A., Li H., Sprague J., Barron A., Engelmann F., Yen B., Powers M.F., Jones J.M., Robinson B.A., Orzechowska B.U., Manoharan M., Legasse A., Planer S., Wilk J., Axthelm M.K., Wong S.W. Deletion of the monkeypox virus inhibitor of complement enzymes locus impacts the adaptive immune response to monkeypox virus in a nonhuman primate model of infection. J. Virol. 2011;85:9527–9542. doi: 10.1128/JVI.00199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D.H. Poxvirus recombination. Pathogens. 2022;11:896. doi: 10.3390/pathogens11080896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D.H., Stuart D., McFadden G. High levels of genetic recombination among cotransfected plasmid DNAs in poxvirus-infected mammalian cells. J. Virol. 1988;62:367–375. doi: 10.1128/jvi.62.2.367-375.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falendysz E.A., Lopera J.G., Lorenzsonn F., Salzer J.S., Hutson C.L., Doty J., Gallardo-Romero N., Carroll D.S., Osorio J.E., Rocke T.E. Further assessment of monkeypox virus infection in Gambian Pouched Rats (Cricetomys gambianus) using in vivo bioluminescent imaging. PLoS Negl. Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0004130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falendysz E.A., Lopera J.G., Doty J.B., Nakazawa Y., Crill C., Lorenzsonn F., Kalemba L.N., Ronderos M.D., Mejia A., Malekani J.M., Karem K., Carroll D.S., Osorio J.E., Rocke T.E. Characterization of monkeypox virus infection in African rope squirrels (Funisciurus sp.) PLoS Negl. Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner F., Comben B.M. Genetic studies with mammalian poxviruses. I. Demonstration of recombination between two strains of vaccina virus. Virology. 1958;5:530–548. doi: 10.1016/0042-6822(58)90043-6. [DOI] [PubMed] [Google Scholar]
- Firth C., Kitchen A., Shapiro B., Suchard M.A., Holmes E.C., Rambaut A. Using time-structured data to estimate evolutionary rates of double-stranded DNA viruses. Mol. Biol. Evol. 2010;27:2038–2051. doi: 10.1093/molbev/msq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Molteni C., Cagliani R., Sironi M. Geographic structuring and divergence time frame of monkeypox virus in the endemic region. J. Infect. Dis. 2022 doi: 10.1093/infdis/jiac298. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froggatt G.C., Smith G.L., Beard P.M. Vaccinia virus gene F3L encodes an intracellular protein that affects the innate immune response. J. Gen. Virol. 2007;88:1917–1921. doi: 10.1099/vir.0.82815-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Gigante C., Khmaladze E., Liu P., Tang S., Wilkins K., Zhao K., Davidson W., Nakazawa Y., Maghlakelidze G., Geleishvili M., Kokhreidze M., Carroll D.S., Emerson G., Li Y. Genome sequences of akhmeta virus, an early divergent old world orthopoxvirus. Viruses. 2018;10:252. doi: 10.3390/v10050252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigante C.M., Gao J., Tang S., McCollum A.M., Wilkins K., Reynolds M.G., Davidson W., McLaughlin J., Olson V.A., Li Y. Genome of alaskapox virus, a novel orthopoxvirus isolated from Alaska. Viruses. 2019;11:708. doi: 10.3390/v11080708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigante C.M., Korber B., Seabolt M.H., Wilkins K., Davidson W., Rao A.K., Zhao H., Hughes C.M., Minhaj F., Waltenburg M.A., Theiler J., Smole S., Gallagher G.R., Blythe D., Myers R., Schulte J., Stringer J., Lee P., Mendoza R.M., Griffin-Thomas L.A., Crain J., Murray J., Atkinson A., Gonzalez A.H., Nash J., Batra D., Damon I., McQuiston J., Hutson C.L., McCollum A.M., Li Y. Multiple lineages of Monkeypox virus detected in the United States, 2021–2022. bioRxiv. 2022 doi: 10.1126/science.add4153. 2022.06.10.495526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guagliardo S.A.J., Monroe B., Moundjoa C., Athanase A., Okpu G., Burgado J., Townsend M.B., Satheshkumar P.S., Epperson S., Doty J.B., Reynolds M.G., Dibongue E., Etoundi G.A., Mathieu E., McCollum A.M. Asymptomatic orthopoxvirus circulation in humans in the wake of a monkeypox outbreak among chimpanzees in cameroon. Am. J. Trop. Med. Hyg. 2020;102:206–212. doi: 10.4269/ajtmh.19-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyuranecz M., Foster J.T., Dán Á., Ip H.S., Egstad K.F., Parker P.G., Higashiguchi J.M., Skinner M.A., Höfle U., Kreizinger Z., Dorrestein G.M., Solt S., Sós E., Kim Y.J., Uhart M., Pereda A., González-Hein G., Hidalgo H., Blanco J.M., Erdélyi K. Worldwide phylogenetic relationship of avian poxviruses. J. Virol. 2013;87:4938–4951. doi: 10.1128/JVI.03183-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happi C., Adetifa I., Mbala P., Njouom R., Nakoune E., Happi A., Ndodo N., Ayansola O., Mboowa G., Bedford T., Neher R.A., Roemer C., Hodcroft E., Tegally H., O'Toole Á., Rambaut A., Pybus O., Kraemer M.U.G., Wilkinson E., Isidro J., Borges V., Pinto M., Gomes J.P., Freitas L., Resende P.C., Lee R.T.C., Maurer-Stroh S., Baxter C., Lessells R., Ogwell A.E., Kebede Y., Tessema S.K., de Oliveira T. Urgent need for a non-discriminatory and non-stigmatizing nomenclature for monkeypox virus. PLoS Biol. 2022;20 doi: 10.1371/journal.pbio.3001769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R.S., Dudley J.P. APOBECs and virus restriction. Virology. 2015;479-480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatcher E.L., Hendrickson R.C., Lefkowitz E.J. Identification of nucleotide-level changes impacting gene content and genome evolution in orthopoxviruses. J. Virol. 2014;88:13651–13668. doi: 10.1128/JVI.02015-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson R.C., Wang C., Hatcher E.L., Lefkowitz E.J. Orthopoxvirus genome evolution: the role of gene loss. Viruses. 2010;2:1933–1967. doi: 10.3390/v2091933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson P.N., Self J., Weiss S., Braden Z., Xiao Y., Girgis N.M., Emerson G., Hughes C., Sammons S.A., Isaacs S.N., Damon I.K., Olson V.A. Elucidating the role of the complement control protein in monkeypox pathogenicity. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidro J., Borges V., Pinto M., Sobral D., Santos J.D., Nunes A., Mixão V., Ferreira R., Santos D., Duarte S., Vieira L., Borrego M.J., Núncio S., de Carvalho I.L., Pelerito A., Cordeiro R., Gomes J.P. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat. Med. 2022;28:1569–1572. doi: 10.1038/s41591-022-01907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.C., Schneider J., Mühlemann B., Veith T., Beheim-Schwarzbach J., Tesch J., Schmidt M.L., Walper F., Bleicker T., Isner C., Pfäfflin F., Werner R.N., Corman V.M., Drosten C. Genetic variability, including gene duplication and deletion, in early sequences from the 2022 European monkeypox outbreak. bioRxiv. 2022 2022.07.23.501239. [Google Scholar]
- Khodakevich L., Jezek Z., Kinzanzka K. Isolation of monkeypox virus from wild squirrel infected in nature. Lancet. 1986;1:98–99. doi: 10.1016/S0140-6736(86)90748-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochneva G., Kolosova I., Maksyutova T., Ryabchikova E., Shchelkunov S. Effects of deletions of kelch-like genes on cowpox virus biological properties. Arch. Virol. 2005;150:1857–1870. doi: 10.1007/s00705-005-0530-0. [DOI] [PubMed] [Google Scholar]
- Kraemer M.U.G., Tegally H., Pigott D.M., Dasgupta A., Sheldon J., Wilkinson E., Schultheiss M., Han A., Oglia M., Marks S., Kanner J., O'Brien K., Dandamudi S., Rader B., Sewalk K., Bento A.I., Scarpino S.V., de Oliveira T., Bogoch I.I., Katz R., Brownstein J.S. Tracking the 2022 monkeypox outbreak with epidemiological data in real-time. Lancet Infect. Dis. 2022;22:941–942. doi: 10.1016/S1473-3099(22)00359-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer M., Suezer Y., Martinez-Fernandez Y., Münk C., Sutter G., Schnierle B.S. Vaccinia virus replication is not affected by APOBEC3 family members. Virol. J. 2006;3:86. doi: 10.1186/1743-422X-3-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelman J.R., Johnston S.C., Mulembakani P.M., Kisalu N., Lee M.S., Koroleva G., McCarthy S.E., Gestole M.C., Wolfe N.D., Fair J.N., Schneider B.S., Wright L.L., Huggins J., Whitehouse C.A., Wemakoy E.O., Muyembe-Tamfum J.J., Hensley L.E., Palacios G.F., Rimoin A.W. Genomic variability of monkeypox virus among humans, Democratic Republic of the Congo. Emerg. Infect. Dis. 2014;20:232–239. doi: 10.3201/eid2002.130118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladnyj I.D., Ziegler P., Kima E. A human infection caused by monkeypox virus in Basankusu Territory, Democratic Republic of the Congo. Bull. World Health Organ. 1972;46:593–597. [PMC free article] [PubMed] [Google Scholar]
- Lanave G., Dowgier G., Decaro N., Albanese F., Brogi E., Parisi A., Losurdo M., Lavazza A., Martella V., Buonavoglia C., Elia G. Novel orthopoxvirus and lethal disease in cat. Italy. Emerg. Infect. Dis. 2018;24:1665–1673. doi: 10.3201/eid2409.171283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz E., Wang C., Upton C. Poxviruses: past, present and future. Virus Res. 2006;117:105–118. doi: 10.1016/j.virusres.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Li Y., Olson V.A., Laue T., Laker M.T., Damon I.K. Detection of monkeypox virus with real-time PCR assays. J. Clin. Virol. 2006;36:194–203. doi: 10.1016/j.jcv.2006.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhao H., Wilkins K., Hughes C., Damon I.K. Real-time PCR assays for the specific detection of monkeypox virus West African and Congo Basin strain DNA. J. Virol. Methods. 2010;169:223–227. doi: 10.1016/j.jviromet.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likos A.M., Sammons S.A., Olson V.A., Frace A.M., Li Y., Olsen-Rasmussen M., Davidson W., Galloway R., Khristova M.L., Reynolds M.G., Zhao H., Carroll D.S., Curns A., Formenty P., Esposito J.J., Regnery R.L., Damon I.K. A tale of two clades: monkeypox viruses. J. Gen. Virol. 2005;86:2661–2672. doi: 10.1099/vir.0.81215-0. [DOI] [PubMed] [Google Scholar]
- Lin Y.C., Evans D.H. Vaccinia virus particles mix inefficiently, and in a way that would restrict viral recombination, in coinfected cells. J. Virol. 2010;84:2432–2443. doi: 10.1128/JVI.01998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.W., Katsafanas G.C., Liu R., Wyatt L.S., Moss B. Poxvirus decapping enzymes enhance virulence by preventing the accumulation of dsRNA and the induction of innate antiviral responses. Cell Host Microbe. 2015;17:320–331. doi: 10.1016/j.chom.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum F.M., Torres-Ruesta A., Tay M.Z., Lin R.T.P., Lye D.C., Rénia L., Ng L.F.P. Monkeypox: disease epidemiology, host immunity and clinical interventions. Nat. Rev. Immunol. 2022:1–17. doi: 10.1038/s41577-022-00775-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus P.V., Andersen E.K., Petersen K.B., Birch-Andersen A. A pox-like disease in cynomolgus monkeys. Acta Pathologica Microbiologica Scandinavica. 1959;46:156–176. [Google Scholar]
- Mariën J., Laudisoit A., Patrono L., Baelo P., van Vredendaal R., Musaba P., Gembu G., Mande C., Ngoy S., Mussaw M. 2021. Monkeypox viruses circulate in distantly-related small mammal species in the Democratic Republic of the Congo. https://www.researchsquare.com/article/rs-414280/v1. [Google Scholar]
- Mauldin M.R., McCollum A.M., Nakazawa Y.J., Mandra A., Whitehouse E.R., Davidson W., Zhao H., Gao J., Li Y., Doty J., Yinka-Ogunleye A., Akinpelu A., Aruna O., Naidoo D., Lewandowski K., Afrough B., Graham V., Aarons E., Hewson R., Vipond R., Dunning J., Chand M., Brown C., Cohen-Gihon I., Erez N., Shifman O., Israeli O., Sharon M., Schwartz E., Beth-Din A., Zvi A., Mak T.M., Ng Y.K., Cui L., Lin R.T.P., Olson V.A., Brooks T., Paran N., Ihekweazu C., Reynolds M.G. Exportation of monkeypox virus from the African continent. J. Infect. Dis. 2022;225:1367–1376. doi: 10.1093/infdis/jiaa559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum A.M., Damon I.K. Human monkeypox. Clin. Infect. Dis. 2014;58:260–267. doi: 10.1093/cid/cit703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden G. Poxvirus tropism. Nat. Rev. Microbiol. 2005;3:201–213. doi: 10.1038/nrmicro1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Zhang F., Yan B., Si C., Honda H., Nagamachi A., Sun L.Z., Xiang Y. A paralogous pair of mammalian host restriction factors form a critical host barrier against poxvirus infection. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileto D., Riva A., Cutrera M., Moschese D., Mancon A., Meroni L., Giacomelli A., Bestetti G., Rizzardini G., Gismondo M.R., Antinori S. New challenges in human monkeypox outside Africa: a review and case report from Italy. Travel Med. Infect. Dis. 2022;49 doi: 10.1016/j.tmaid.2022.102386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molteni C., Forni D., Cagliani R., Clerici M., Sironi M. Genetic ancestry and population structure of vaccinia virus. NPJ Vaccines. 2022;7:92. doi: 10.1038/s41541-022-00519-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B. Poxvirus DNA replication. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer R.W., Graves R.L., Rothe C.T. The white pock (mu) mutants of rabbit poxvirus. III. Terminal DNA sequence duplication and transposition in rabbit poxvirus. Cell. 1980;22:545–553. doi: 10.1016/0092-8674(80)90364-5. [DOI] [PubMed] [Google Scholar]
- Mühlemann B., Vinner L., Margaryan A., Wilhelmson H., de la Fuente Castro C., Allentoft M.E., de Barros Damgaard P., Hansen A.J., Holtsmark Nielsen S., Strand L.M., Bill J., Buzhilova A., Pushkina T., Falys C., Khartanovich V., Moiseyev V., Jørkov M.L.S., Østergaard Sørensen P., Magnusson Y., Gustin I., Schroeder H., Sutter G., Smith G.L., Drosten C., Fouchier R.A.M., Smith D.J., Willerslev E., Jones T.C., Sikora M. Diverse variola virus (smallpox) strains were widespread in northern Europe in the Viking Age. Science. 2020;369:eaaw8977. doi: 10.1126/science.aaw8977. [DOI] [PubMed] [Google Scholar]
- Ng O.T., Lee V., Marimuthu K., Vasoo S., Chan G., Lin R.T.P., Leo Y.S. A case of imported Monkeypox in Singapore. Lancet Infect. Dis. 2019;19:1166. doi: 10.1016/S1473-3099(19)30537-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngomanda Alfred, Jolly Dominique, Bentaleb Ilhem, Chepstow-Lusty Alex, Maley Jean, Fontugne Michel, Oslisly Richard, Rabenkogo Nicaise, et al. Lowland rainforest response to hydrological changes during the last 1500 years in Gabon, Western Equatorial Africa. Quat. Res. 2007;67:411–425. [Google Scholar]
- Nolen L.D., Osadebe L., Katomba J., Likofata J., Mukadi D., Monroe B., Doty J., Kalemba L., Malekani J., Kabamba J., Bomponda P.L., Lokota J.I., Balilo M.P., Likafi T., Lushima R.S., Tamfum J.J., Okitolonda E.W., McCollum A.M., Reynolds M.G. Introduction of monkeypox into a community and household: risk factors and zoonotic reservoirs in the democratic Republic of the Congo. Am. J. Trop. Med. Hyg. 2015;93:410–415. doi: 10.4269/ajtmh.15-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogoina D., Iroezindu M., James H.I., Oladokun R., Yinka-Ogunleye A., Wakama P., Otike-Odibi B., Usman L.M., Obazee E., Aruna O., Ihekweazu C. Clinical course and outcome of human monkeypox in Nigeria. Clin. Infect. Dis. 2020;71:e210–e214. doi: 10.1093/cid/ciaa143. [DOI] [PubMed] [Google Scholar]
- Orba Y., Sasaki M., Yamaguchi H., Ishii A., Thomas Y., Ogawa H., Hang'ombe B.M., Mweene A.S., Morikawa S., Saijo M., Sawa H. Orthopoxvirus infection among wildlife in Zambia. J. Gen. Virol. 2015;96:390–394. doi: 10.1099/vir.0.070219-0. [DOI] [PubMed] [Google Scholar]
- Osorio J.E., Iams K.P., Meteyer C.U., Rocke T.E. Comparison of monkeypox viruses pathogenesis in mice by in vivo imaging. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Peng C., Rahman M.J., Haller S.L., Tazi L., Brennan G., Rothenburg S. Orthopoxvirus K3 orthologs show virus- and host-specific inhibition of the antiviral protein kinase PKR. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski P., Noyce R.S., Evans D.H. Live-cell imaging of vaccinia virus recombination. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrono L.V., Pléh K., Samuni L., Ulrich M., Röthemeier C., Sachse A., Muschter S., Nitsche A., Couacy-Hymann E., Boesch C., Wittig R.M., Calvignac-Spencer S., Leendertz F.H. Monkeypox virus emergence in wild chimpanzees reveals distinct clinical outcomes and viral diversity. Nat. Microbiol. 2020;5:955–965. doi: 10.1038/s41564-020-0706-0. [DOI] [PubMed] [Google Scholar]
- Peiró-Mestres A., Fuertes I., Camprubí-Ferrer D., Marcos M.Á., Vilella A., Navarro M., Rodriguez-Elena L., Riera J., Català A., Martínez M.J., Blanco J.L., Hospital Clinic de Barcelona Monkeypox Study Group Frequent detection of monkeypox virus DNA in saliva, semen, and other clinical samples from 12 patients, Barcelona, Spain, May to June 2022. Euro Surveill. 2022;27:2200503. doi: 10.2807/1560-7917.ES.2022.27.28.2200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C., Wyatt L.S., Glushakow-Smith S.G., Lal-Nag M., Weisberg A.S., Moss B. Zinc-finger antiviral protein (ZAP) is a restriction factor for replication of modified vaccinia virus Ankara (MVA) in human cells. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickup D.J., Ink B.S., Parsons B.L., Hu W., Joklik W.K. Spontaneous deletions and duplications of sequences in the genome of cowpox virus. Proc. Natl. Acad. Sci. U. S. A. 1984;81:6817–6821. doi: 10.1073/pnas.81.21.6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires de Miranda M., Reading P.C., Tscharke D.C., Murphy B.J., Smith G.L. The vaccinia virus kelch-like protein C2L affects calcium-independent adhesion to the extracellular matrix and inflammation in a murine intradermal model. J. Gen. Virol. 2003;84:2459–2471. doi: 10.1099/vir.0.19292-0. [DOI] [PubMed] [Google Scholar]
- Qin L., Upton C., Hazes B., Evans D.H. Genomic analysis of the vaccinia virus strain variants found in Dryvax vaccine. J. Virol. 2011;85:13049–13060. doi: 10.1128/JVI.05779-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radonić A., Metzger S., Dabrowski P.W., Couacy-Hymann E., Schuenadel L., Kurth A., Mätz-Rensing K., Boesch C., Leendertz F.H., Nitsche A. Fatal monkeypox in wild-living sooty mangabey, Côte d'Ivoire, 2012. Emerg. Infect. Dis. 2014;20:1009–1011. doi: 10.3201/eid2006.131329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A.K., Schulte J., Chen T.H., Hughes C.M., Davidson W., Neff J.M., Markarian M., Delea K.C., Wada S., Liddell A., Alexander S., Sunshine B., Huang P., Honza H.T., Rey A., Monroe B., Doty J., Christensen B., Delaney L., Massey J., Waltenburg M., Schrodt C.A., Kuhar D., Satheshkumar P.S., Kondas A., Li Y., Wilkins K., Sage K.M., Yu Y., Yu P., Feldpausch A., McQuiston J., Damon I.K., McCollum A.M., July 2021 Monkeypox Response Team Monkeypox in a traveler returning from Nigeria - Dallas, Texas, July 2021. MMWR Morb. Mortal. Wkly Rep. 2022;71:509–516. doi: 10.15585/mmwr.mm7114a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds M.G., Yorita K.L., Kuehnert M.J., Davidson W.B., Huhn G.D., Holman R.C., Damon I.K. Clinical manifestations of human monkeypox influenced by route of infection. J. Infect. Dis. 2006;194:773–780. doi: 10.1086/505880. [DOI] [PubMed] [Google Scholar]
- Reynolds M.G., Carroll D.S., Olson V.A., Hughes C., Galley J., Likos A., Montgomery J.M., Suu-Ire R., Kwasi M.O., Jeffrey Root J., Braden Z., Abel J., Clemmons C., Regnery R., Karem K., Damon I.K. A silent enzootic of an orthopoxvirus in Ghana, West Africa: evidence for multi-species involvement in the absence of widespread human disease. Am. J. Trop. Med. Hyg. 2010;82:746–754. doi: 10.4269/ajtmh.2010.09-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoin A.W., Mulembakani P.M., Johnston S.C., Lloyd Smith J.O., Kisalu N.K., Kinkela T.L., Blumberg S., Thomassen H.A., Pike B.L., Fair J.N., Wolfe N.D., Shongo R.L., Graham B.S., Formenty P., Okitolonda E., Hensley L.E., Meyer H., Wright L.L., Muyembe J.J. Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc. Natl. Acad. Sci. U. S. A. 2010;107:16262–16267. doi: 10.1073/pnas.1005769107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker S., Isberg S.R., Moran J.L., Araujo R.D., Elliott N., Melville L., Beddoe T., Helbig K.J. Crocodilepox virus evolutionary genomics supports observed poxvirus infection dynamics on saltwater crocodile (Crocodylus porosus) Viruses. 2019;11:1116. doi: 10.3390/v11121116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkevich T.G., Yutin N., Wolf Y.I., Koonin E.V., Moss B. Ancient gene capture and recent gene loss shape the evolution of orthopoxvirus-host interaction genes. mBio. 2021;12 doi: 10.1128/mBio.01495-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchelkunov S.N. Interaction of orthopoxviruses with the cellular ubiquitin-ligase system. Virus Genes. 2010;41:309–318. doi: 10.1007/s11262-010-0519-y. [DOI] [PubMed] [Google Scholar]
- Siddle K.J., Eromon P., Barnes K.G., Mehta S., Oguzie J.U., Odia I., Schaffner S.F., Winnicki S.M., Shah R.R., Qu J., Wohl S., Brehio P., Iruolagbe C., Aiyepada J., Uyigue E., Akhilomen P., Okonofua G., Ye S., Kayode T., Ajogbasile F., Uwanibe J., Gaye A., Momoh M., Chak B., Kotliar D., Carter A., Gladden-Young A., Freije C.A., Omoregie O., Osiemi B., Muoebonam E.B., Airende M., Enigbe R., Ebo B., Nosamiefan I., Oluniyi P., Nekoui M., Ogbaini-Emovon E., Garry R.F., Andersen K.G., Park D.J., Yozwiak N.L., Akpede G., Ihekweazu C., Tomori O., Okogbenin S., Folarin O.A., Okokhere P.O., MacInnis B.L., Sabeti P.C., Happi C.T. Genomic analysis of lassa virus during an increase in cases in Nigeria in 2018. N. Engl. J. Med. 2018;379:1745–1753. doi: 10.1056/NEJMoa1804498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva N.I.O., de Oliveira J.S., Kroon E.G., Trindade G.S., Drumond B.P. Here, there, and everywhere: the wide host range and geographic distribution of zoonotic orthopoxviruses. Viruses. 2020;13:43. doi: 10.3390/v13010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson K., Heymann D., Brown C.S., Edmunds W.J., Elsgaard J., Fine P., Hochrein H., Hoff N.A., Green A., Ihekweazu C., Jones T.C., Lule S., Maclennan J., McCollum A., Mühlemann B., Nightingale E., Ogoina D., Ogunleye A., Petersen B., Powell J., Quantick O., Rimoin A.W., Ulaeato D., Wapling A. Human monkeypox - After 40 years, an unintended consequence of smallpox eradication. Vaccine. 2020;38:5077–5081. doi: 10.1016/j.vaccine.2020.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenovská N., Van Ranst M. Emergence of Monkeypox as the most important orthopoxvirus infection in humans. Front. Public Health. 2018;6:241. doi: 10.3389/fpubh.2018.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesh R.B., Watts D.M., Sbrana E., Siirin M., Popov V.L., Xiao S.Y. Experimental infection of ground squirrels (Spermophilus tridecemlineatus) with monkeypox virus. Emerg. Infect. Dis. 2004;10:1563–1567. doi: 10.3201/eid1009.040310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornhill J.P., Barkati S., Walmsley S., Rockstroh J., Antinori A., Harrison L.B., Palich R., Nori A., Reeves I., Habibi M.S., Apea V., Boesecke C., Vandekerckhove L., Yakubovsky M., Sendagorta E., Blanco J.L., Florence E., Moschese D., Maltez F.M., Goorhuis A., Pourcher V., Migaud P., Noe S., Pintado C., Maggi F., Hansen A.E., Hoffmann C., Lezama J.I., Mussini C., Cattelan A., Makofane K., Tan D., Nozza S., Nemeth J., Klein M.B., Orkin C.M., SHARE-net Clinical Group Monkeypox virus infection in humans across 16 Countries - April-June 2022. N. Engl. J. Med. 2022;387:679–691. doi: 10.1056/NEJMoa2207323. [DOI] [PubMed] [Google Scholar]
- Tiee M.S., Harrigan R.J., Thomassen H.A., Smith T.B. Ghosts of infections past: using archival samples to understand a century of monkeypox virus prevalence among host communities across space and time. R. Soc. Open Sci. 2018;5 doi: 10.1098/rsos.171089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J., Nguyen L.T., von Haeseler A., Minh B.Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton C., Slack S., Hunter A.L., Ehlers A., Roper R.L. Poxvirus orthologous clusters: toward defining the minimum essential poxvirus genome. J. Virol. 2003;77:7590–7600. doi: 10.1128/JVI.77.13.7590-7600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Burles K., Couturier B., Randall C.M., Shisler J., Barry M. Ectromelia virus encodes a BTB/kelch protein, EVM150, that inhibits NF-κB signaling. J. Virol. 2014;88:4853–4865. doi: 10.1128/JVI.02923-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., White A. Monkeypox virus emerges from the shadow of its more infamous cousin: family biology matters. Emerg. Microbes Infect. 2022;11:1768–1777. doi: 10.1080/22221751.2022.2095309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Wang Y., Dai P., Li T., Zierhut C., Tan A., Zhang T., Pan H., Li Z., Ordureau A., Xiang J.Z., Hendrickson R.C., Funabiki H., Chen Z., Deng L. Vaccinia E5 is a major inhibitor of the DNA sensor cGAS. bioRxiv. 2021 doi: 10.1038/s41467-023-38514-5. 2021.10.25.465197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yinka-Ogunleye A., Aruna O., Dalhat M., Ogoina D., McCollum A., Disu Y., Mamadu I., Akinpelu A., Ahmad A., Burga J., Ndoreraho A., Nkunzimana E., Manneh L., Mohammed A., Adeoye O., Tom-Aba D., Silenou B., Ipadeola O., Saleh M., Adeyemo A., Nwadiutor I., Aworabhi N., Uke P., John D., Wakama P., Reynolds M., Mauldin M.R., Doty J., Wilkins K., Musa J., Khalakdina A., Adedeji A., Mba N., Ojo O., Krause G., Ihekweazu C., CDC Monkeypox Outbreak Team Outbreak of human monkeypox in Nigeria in 2017-18: a clinical and epidemiological report. Lancet Infect. Dis. 2019;19:872–879. doi: 10.1016/S1473-3099(19)30294-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaucha G.M., Jahrling P.B., Geisbert T.W., Swearengen J.R., Hensley L. The pathology of experimental aerosolized monkeypox virus infection in cynomolgus monkeys (Macaca fascicularis) Lab. Investig. 2001;81:1581–1600. doi: 10.1038/labinvest.3780373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Meng X., Townsend M.B., Satheshkumar P.S., Xiang Y. Identification of CP77 as the Third Orthopoxvirus SAMD9 and SAMD9L inhibitor with unique specificity for a rodent SAMD9L. J. Virol. 2019;93 doi: 10.1128/JVI.00225-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.