Abstract
Background: Lung adenocarcinoma (LUAD) is the most common subtype of non-small-cell lung cancer (NSCLC). The aim of our study was to determine prognostic risk factors and establish a novel nomogram for lung adenocarcinoma patients. Methods: This retrospective cohort study is based on the Surveillance, Epidemiology, and End Results (SEER) database and the Chinese multicenter lung cancer database. We selected 22,368 eligible LUAD patients diagnosed between 2010 and 2015 from the SEER database and screened them based on the inclusion and exclusion criteria. Subsequently, the patients were randomly divided into the training cohort (n = 15,657) and the testing cohort (n = 6711), with a ratio of 7:3. Meanwhile, 736 eligible LUAD patients from the Chinese multicenter lung cancer database diagnosed between 2011 and 2021 were considered as the validation cohort. Results: We established a nomogram based on each independent prognostic factor analysis for 1-, 3-, and 5-year overall survival (OS) . For the training cohort, the area under the curves (AUCs) for predicting the 1-, 3-, and 5-year OS were 0.806, 0.856, and 0.886. For the testing cohort, AUCs for predicting the 1-, 3-, and 5-year OS were 0.804, 0.849, and 0.873. For the validation cohort, AUCs for predicting the 1-, 3-, and 5-year OS were 0.86, 0.874, and 0.861. The calibration curves were observed to be closer to the ideal 45° dotted line with regard to 1-, 3-, and 5-year OS in the training cohort, the testing cohort, and the validation cohort. The decision curve analysis (DCA) plots indicated that the established nomogram had greater net benefits in comparison with the Tumor-Node-Metastasis (TNM) staging system for predicting 1-, 3-, and 5-year OS of lung adenocarcinoma patients. The Kaplan–Meier curves indicated that patients’ survival in the low-risk group was better than that in the high-risk group (P < .001). Conclusion: The nomogram performed very well with excellent predictive ability in both the US population and the Chinese population.
Keywords: LUAD, prognosis, SEER, nomogram, overall survival
Introduction
Lung cancer is the most common cause of cancer-related death worldwide. Approximately 236,740 new patients and 130,180 deaths occurred in the United States in 2022.1 Non-small-cell lung cancer (NSCLC), containing adenocarcinoma, squamous cell carcinoma (SCC), small cell lung carcinoma, and large cell carcinoma, accounting for 85% of primary lung cancer patients. Among them, lung adenocarcinoma (LUAD) is the most common subtype.2 LUAD is generally peripherally located and related to the surface alveolar epithelium or bronchial mucosa.3 As the most prevalent pathological type of NSCLC, LUAD generally derives from the acinar cells of the lung periphery.4 Although there has been a great development in the therapy of LUAD, the survival prognosis in LUAD cases remains unsatisfactory due to the lack of early diagnosis and efficient personalized treatment.5
Accumulating evidence indicates that several demographic, clinical, and pathological variables are considered to be associated with the survival prognosis of lung cancer.6–9 A study by Guo et al has found that age, gender, histological grade, T stage, N stage, and treatment were proved to be independent prognostic factors for overall survival (OS) in elderly locally advanced NSCLC (LA-NSCLC) patients. In addition, elderly LA-NSCLC patients who underwent surgery were reported to have the best prognosis, which was superior to chemoradiotherapy, radiotherapy alone, and chemotherapy alone.6 Moreover, Ke et al studied clear cell adenocarcinoma of the lung (CCAL) patients, and found that age, metastasis, and surgery were associated with overall prognosis. It is worth noting that surgery was significantly associated with a decreased risk of death (hazard ratio [HR] = 0.44, 95% confidence interval [CI] 0.33-0.59).7 Similarly, Zheng et al have discovered that age, sex, TNM stage, and surgery were independent predictors of OS for lung squamous cell cancer (LSCC) patients.8 Zhang et al identified that small cell lung cancer (SCLC) and LUAD exhibited higher risks of bone metastasis than other histological subtypes.9 Precise prediction of the survival prognosis of patients with LUAD is of quite important for clinicians in decision making. At present, there is still no accurate conclusion on the prognostic factors of LUAD patients, and few large multiple clinical centers studies have estimated them in order to make a comprehensive and precise prediction for the survival rate of LUAD patients. So far, the TNM staging system is a device used by clinicians to predict survival prognosis and construct treatment plan.10 However, the prognostic evaluation based on the TNM staging system could not meet the requirements for precise prognosis prediction of LUAD patients. Therefore, investigating the prognostic predictive model is an urgent desire to assist the clinician to predict the survival prognosis of LUAD patients and provide personalized treatment.
The nomogram, based on the regression coefficients of each variable, combines many important prognostic correlates to more accurately predict individual patient survival rates.11 A satisfactory accuracy and reliability nomogram would be instrumental in identifying high-risk and making appropriate clinical decisions for LUAD patients. Nomogram has been performed in the prognosis prediction of various cancers.12,13 Liao et al established a nomogram for the prediction of OS in patients with stage II and III NSCLC.14 Zhong et al developed a prognostic nomogram to predict the death rate for extensive-stage SCLC.15 In addition, the population-based Surveillance, Epidemiology, and End Results (SEER) database could be used to solve this problem on a larger number of patients with long follow-up. To the best of our knowledge, a majority of nomograms predicting the survival probability of LUAD patients have been validated in the same race or single center, which would decrease the feasibility and universality of the models. There were some differences in case characteristics between the US and Chinese populations, which might be related to prevalence, economic status, religious beliefs, and eligibility for health insurance. Moreover, it has been reported that even in China, there are different epidemiological differences between the north and the south.16
Therefore, the aim of our study was to establish and validate a prognostic nomogram to predict OS for LUAD patients using the SEER database and to additionally validate the nomogram using a multicenter Chinese cohort for wider clinical application. This comprehensive nomogram could assist clinicians in making decisions regarding appropriate personalized treatment.
Materials and Methods
Data Source and Patients
The reporting of this study conforms to STROBE guidelines.17 This retrospective cohort study is based on the SEER database and the Chinese multicenter lung cancer database. Data on patients with the first primary LUAD histologically diagnosed between 2010 and 2015 were extracted from the SEER database. The SEER program is supported by the National Cancer Institute, which collects information of cancer patients in 18 registries, covering approximately 30% of the total US population.9 The SEER data (Incidence – SEER Research Data, 9 registries, November 2017 Sub 1975-2018) were obtained via SEER*Stat version 8.3.9. The diagnosis of LUAD was defined by the ICD-O-3/WHO2008 (International Classification of Diseases for Oncology, third edition), and histopathological types were defined using ICD-O-3 His/Behave, malignant.18 The study consisted of LUAD patients with the following ICD-O-3/WHO2008, morphology codes: 8140/3, 8141/3, 8144/3, 8211/3, 8250/3, 8251/3, 8255/3, 8260/3, 8290/3, 8310/3, 8323/3, 8333/3, 8401/3, 8441/3, 8480/3, 8481/3, 8570/3, 8574/3, and 8576/3; and the site codes: C34.0, C34.1, C34.2, C34.3, C34.8, and C34.9. According to the inclusion criteria, we initially selected 43,741 patients with LUAD in the SEER database. And then, the further identification of LUAD patients based on the exclusion criteria were as follows: (1) type of reporting source: autopsy only or death certificate only; (2) age at diagnosis less than 18 years; (3) patients with multiple primary tumors; (4) patients with any unknown epidemiology and clinical information; Eventually, we identified 22,368 eligible LUAD patients for this study (Figure 1A). The patients were randomly divided into the training cohort (n = 15,657) and the testing cohort (n = 6711), with a ratio of 7:3. Patients in the training cohort were retrospectively analyzed to construct the nomogram, which was internally validated by the testing cohort. To estimate the generalizability of the novel nomogram, we performed separate external validation using 736 eligible LUAD patients from the Chinese multicenter lung cancer database histologically diagnosed between 2011 and 2021. Inclusion and exclusion criteria were the same as the SEER cohort (Figure 1B). In brief, the Chinese multicenter lung cancer database is one of the largest nationwide cancer databases that collects lung cancer patient information from 6 areas in China. These 6 areas are distributed across northern and southern China, including Beijing, Hebei Province, Hubei Province, Gansu Province, Henan Province, and Jiangsu Province. The database provides many types of information, including health status, sociodemographic characteristics, lifestyles, and clinicopathological data of lung cancer patients.
Figure 1.
The flowchart of the patient selection in the present study from (A) the Surveillance, Epidemiology, and End Results (SEER) database and (B) the Chinese multicenter lung cancer database.
Study Variables
In the study, demographic, clinical, and pathological variables, including age, sex, tumor laterality, TNM stage (American Joint Committee on Cancer [AJCC], seventh ed.), bone metastasis, brain metastasis, liver metastasis, surgery, follow-up time, and survival outcomes were abstracted from the SEER database. Age was categorized as <59 years, 59 to 67 years, 67 to 75 years, and ≥75 years according to the quantile. Tumor laterality includes left, right, and bilateral. For the validation cohort, the same characteristics of LUAD patients were collected from the Chinese multicenter lung cancer database.
Study Outcome
The outcome of our study was OS, which was defined as the interval between the first pathological diagnosis and the date of death from any cause or the last follow-up.
Statistical Analysis
Categorical data were presented as quantity and percentage (N %), and differences were analyzed by the Pearson chi-square test. Univariate and multivariate Cox regression models were constructed to assess the independent prognostic factors of OS. OS was determined by the Kaplan–Meier method, and differences between different groups were compared using a log-rank test. The nomogram was established and validated based on the multivariate Cox regression model. Concordance index (C-index) and calibration curve are used to assess the predictive performance of nomogram. The C-index value ranges from 0.50 to 1.00 and shows a positive correlation with the predictive accuracy of the nomogram. It illustrates the model accompanied by perfect performance when the value is 1.00. And when the calibration curve is considered a perfectly calibrated model, the predicted value will fall on the diagonal 45° in the figure. Furthermore, receiver operating characteristic (ROC) curves, time-dependent ROC curves, and decision curve analysis (DCA) were performed to evaluate the predictive performance of the established nomogram.19,20 The bootstrapping technique was performed for validation of the established nomogram based on 1000 resamples. To determine whether the nomogram could successfully distinguish 2 different risk groups (low and high), each patient's prediction point was derived according to the nomogram, and the patients were categorized into the high-risk and low-risk groups based on the median values of the risk point.21 All statistical analyses were carried out using the R software version 4.0.4 (http://www.R-project.org). In all analyses, a two-sided P value <.05 was deemed statistically significant.
Results
Demographic, Clinical, and Pathological Characteristics of the Study Cohorts
According to the inclusion and exclusion criteria, a total of 23,104 LUAD patients from the SEER database and the Chinese multicenter database were included in this study. All eligible cases from the SEER database (n = 22,368) were randomly divided into the training cohort (n = 15,657) and the testing cohort (n = 6711). All eligible cases from the Chinese multicenter database were treated as the validation cohort. Table 1 showed the detailed demographic, clinical, and pathological characteristics of the patients. Among these patients, 5321 (23.03) were <59, 5696 (24.65) were 59 to 67, 5751 (24.89) were 67 to 75, 6336 (27.42) were ≥75. For sex, the number of female and male patients was 12,197 (52.79) and 10,907 (47.21), respectively. For tumor laterality, a total of 9106 (39.41) patients had tumors on the left, 13,720 (59.38) patients with tumors on the right, and 278 (1.20) patients with tumors on the bilateral. For T stage, the number of T1, T2, T3, and T4 stage tumors was 6049 (26.18), 7240 (31.34), 4650 (20.13), and 5165 (22.36), respectively. For N stage, the number of N0, N1, N2, N3, and N4 stage tumors was 9699 (41.98), 1991 (8.62), 7760 (33.59), and 3654 (15.82), respectively. For M stage, the number of M0 and M1 stage tumors was 11,130 (48.17) and 11,974 (51.83), respectively. For patients with distant organ metastasis, bone metastasis occurred in 5169 (22.37), brain metastases occurred in 3804 (16.46), and liver metastases occurred in 1870 (8.09). For surgery, surgical treatment was performed in 6745 (29.19) patients.
Table 1.
Baseline Demographic, Clinical, and Pathological Characteristics of the Training, Testing, and Validation Cohorts.
| Characteristics | Total cohort | Training cohort | Testing cohort | Validation cohort |
|---|---|---|---|---|
| (n = 23,104) | (n = 15,657) | (n = 6711) | (n = 736) | |
| Age (years) | ||||
| <59 | 5321 (23.03) | 3511 (22.42) | 1507 (22.46) | 303 (41.17) |
| 59-67 | 5696 (24.65) | 3844 (24.55) | 1599 (23.83) | 253 (34.38) |
| 67-75 | 5751 (24.89) | 3900 (24.91) | 1708 (25.45) | 143 (19.43) |
| ≥75 | 6336 (27.42) | 4402 (28.12) | 1897 (28.27) | 37 (5.03) |
| Sex | ||||
| Female | 12,197 (52.79) | 8329 (53.20) | 3501 (52.17) | 367 (49.86) |
| Male | 10,907 (47.21) | 7328 (46.80) | 3210 (47.83) | 369 (50.14) |
| Tumor laterality | ||||
| Left | 9106 (39.41) | 6113 (39.04) | 2714 (40.44) | 279 (37.91) |
| Right | 13,720 (59.38) | 9384 (59.93) | 3922 (58.44) | 414 (56.25) |
| Bilateral | 278 (1.20) | 160 (1.02) | 75 (1.12) | 43 (5.84) |
| T stage | ||||
| T1 | 6049 (26.18) | 3989 (25.48) | 1763 (26.27) | 297 (40.35) |
| T2 | 7240 (31.34) | 4910 (31.36) | 2084 (31.05) | 246 (33.42) |
| T3 | 4650 (20.13) | 3196 (20.41) | 1377 (20.52) | 77 (10.46) |
| T4 | 5165 (22.36) | 3562 (22.75) | 1487 (22.16) | 116 (15.76) |
| N stage | ||||
| N0 | 9699 (41.98) | 6492 (41.46) | 2853 (42.51) | 354 (48.10) |
| N1 | 1991 (8.62) | 1366 (8.72) | 552 (8.23) | 73 (9.92) |
| N2 | 7760 (33.59) | 5329 (34.04) | 2246 (33.47) | 185 (25.14) |
| N3 | 3654 (15.82) | 2470 (15.78) | 1060 (15.79) | 124 (16.85) |
| M stage | ||||
| M0 | 11,130 (48.17) | 7458 (47.63) | 3196 (47.62) | 476 (64.67) |
| M1 | 11,974 (51.83) | 8199 (52.37) | 3515 (52.38) | 260 (35.33) |
| Bone metastasis | ||||
| No | 17,935 (77.63) | 12,116 (77.38) | 5213 (77.68) | 606 (82.34) |
| Yes | 5169 (22.37) | 3541 (22.62) | 1498 (22.32) | 130 (17.66) |
| Brain metastasis | ||||
| No | 19,300 (83.54) | 13,016 (83.13) | 5614 (83.65) | 670 (91.03) |
| Yes | 3804 (16.46) | 2641 (16.87) | 1097 (16.35) | 66 (8.97) |
| Liver metastasis | ||||
| No | 21,234 (91.91) | 14,375 (91.81) | 6171 (91.95) | 688 (93.48) |
| Yes | 1870 (8.09) | 1282 (8.19) | 540 (8.05) | 48 (6.52) |
| Surgery metastasis | ||||
| No | 16,359 (70.81) | 11,300 (72.17) | 4833 (72.02) | 226 (30.71) |
| Yes | 6745 (29.19) | 4357 (27.83) | 1878 (27.98) | 510 (69.29) |
Identification of Prognostic Factors in the Training Cohort
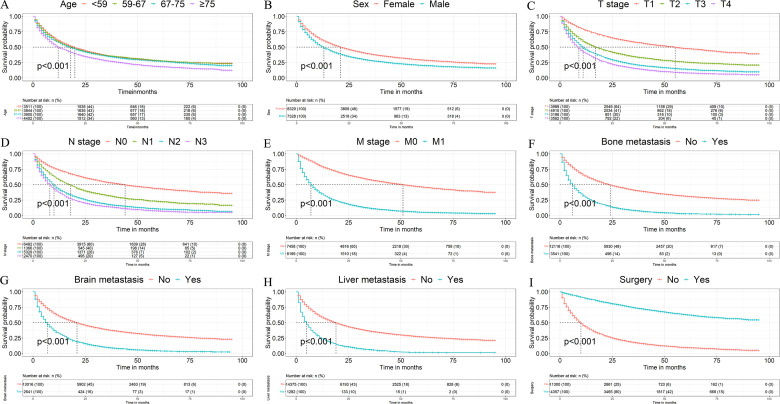
Univariate and multivariate Cox regression showed that many factors were related to the OS (Table 2). Univariate Cox regression showed that age (compared with <59 years), sex (compared with female), tumor laterality (compared with left), T stage (compared with T1), N stage (compared with N1), M stage (compared with M1), bone metastasis (compared with none-met), brain metastasis (compared with none-met), liver metastasis (compared with none-met), and surgery (compared with none-surgery) were significantly associated with OS. To control confounding factors, multivariate Cox analysis was performed, and the results showed that age, sex, T stage, N stage, M stage, bone metastasis, brain metastasis, and liver metastasis were independent risk factors for the OS, and surgery was an independent favorable factor OS. In order to more vividly demonstrate the relationship between independent prognostic factors and survival time, patients were analyzed and survival curves were drawn using the Kaplan–Meier method (Figure 2). Kaplan–Meier analysis illustrated that patients who had older age, male sex, later T, N, and M stage, metastasis to the bone, brain, and liver, patients without surgery was associated with worse prognosis.
Table 2.
Univariable and Multivariable Cox Regression Analyses Overall Survival (OS) of LUAD Patients in the Training Cohort.
| Characteristics | Univariable | Multivariable | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age (years) | ||||||
| <59 | Reference | Reference | ||||
| 59-67 | 1.01 | 0.96-1.07 | .606 | 1.15 | 1.09-1.22 | <.001 |
| 67-75 | 1.07 | 1.01-1.13 | .017 | 1.35 | 1.27-1.42 | <.001 |
| ≥75 | 1.34 | 1.27-1.41 | <.001 | 1.73 | 1.64-1.83 | <.001 |
| Sex | ||||||
| Female | Reference | Reference | ||||
| Male | 1.34 | 1.29-1.39 | <.001 | 1.29 | 1.25-1.34 | <.001 |
| Tumor laterality | ||||||
| Left | Reference | Reference | ||||
| Right | 0.99 | 0.95-1.03 | .528 | 1.01 | 0.97-1.05 | .731 |
| Bilateral | 1.99 | 1.69-2.35 | <.001 | 1.13 | 0.96-1.34 | .145 |
| T stage | ||||||
| T1 | Reference | Reference | ||||
| T2 | 1.93 | 1.82-2.04 | <.001 | 1.45 | 1.37-1.54 | <.001 |
| T3 | 2.74 | 2.59-2.91 | <.001 | 1.49 | 1.40-1.59 | <.001 |
| T4 | 3.39 | 3.20-3.59 | <.001 | 1.53 | 1.44-1.62 | <.001 |
| N stage | ||||||
| N0 | Reference | Reference | ||||
| N1 | 1.80 | 1.68-1.93 | <.001 | 1.36 | 1.27-1.46 | <.001 |
| N2 | 2.72 | 2.60-2.85 | <.001 | 1.39 | 1.33-1.46 | <.001 |
| N3 | 3.20 | 3.03-3.38 | <.001 | 1.39 | 1.31-1.47 | <.001 |
| M stage | ||||||
| M0 | Reference | Reference | ||||
| M1 | 4.10 | 3.93-4.27 | <.001 | 1.81 | 1.72-1.91 | <.001 |
| Bone metastasis | ||||||
| No | Reference | Reference | ||||
| Yes | 2.83 | 2.71-2.95 | <.001 | 1.22 | 1.16-1.27 | <.001 |
| Brain metastasis | ||||||
| No | Reference | Reference | ||||
| Yes | 2.38 | 2.27-2.49 | <.001 | 1.21 | 1.15-1.27 | <.001 |
| Liver metastasis | ||||||
| No | Reference | Reference | ||||
| Yes | 2.80 | 2.63-2.97 | <.001 | 1.35 | 1.27-1.43 | <.001 |
| Surgery | ||||||
| No | Reference | Reference | ||||
| Yes | 0.18 | 0.17-0.19 | <.001 | 0.36 | 0.34-0.38 | <.001 |
Abbreviations: CI, confidence interval; HR, hazard ratio; LUAD, lung adenocarcinoma.
Figure 2.
Kaplan–Meier survival curves of overall survival (OS) based on (A) age, (B) sex, (C) T stage, (D) N stage, (E) M stage, (F) bone metastasis, (G) brain metastasis, (H) liver metastasis, (I) and surgery.
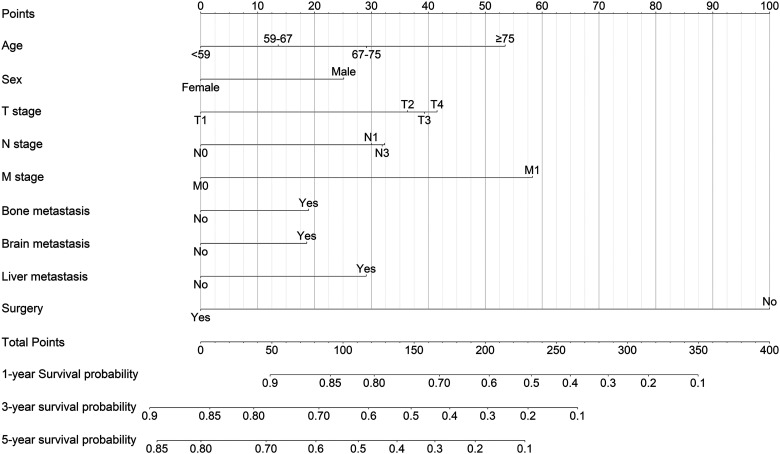
Development of the Prognosis Nomogram
We established a nomogram based on each independent prognostic factor analysis for 1-, 3-, and 5-year OS (Figure 3). Each prognostic factor can calculate a corresponding point by drawing a line straight upward to the point axis. Total point can be calculated by adding up the point of each prognostic factor, which can find a position on the total point axis. Next, the predicted probability of 1-, 3-, and 5-year OS can be obtained by drawing a line straight downw from the total point axis to the corresponding survival axis.
Figure 3.
Nomogram predicted 1-, 3-, and 5-year overall survival (OS) in lung adenocarcinoma (LUAD) patients. The factors of age, sex, T stage, N stage, M stage, bone metastasis, brain metastasis, liver metastasis, and surgery were included in the model.
Validation of the Prognosis Nomogram
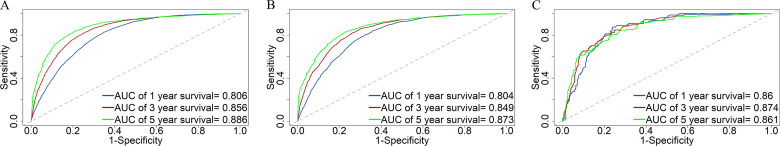
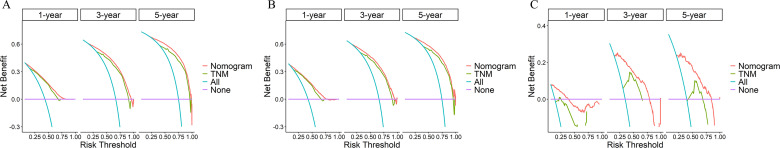
The C-index was 0.745 (95% CI, 0.741-0.749), 0.741 (95% CI, 0.733-0.749), and 0.809 (95% CI, 0.785-0.833) in the training cohort, the testing cohort, and the validation cohort, respectively. For the training cohort, the area under the curves (AUCs) for predicting the 1-, 3-, and 5-year OS were 0.806, 0.856, and 0.886. For the testing cohort, the AUCs for predicting the 1-, 3-, and 5-year OS were 0.804, 0.849, and 0.873. For the validation cohort, the AUCs for predicting the 1-, 3-, and 5-year OS were 0.86, 0.874, and 0.861. The results revealed the excellent discrimination ability of the nomogram (Figure 4). The time-dependent ROC curve is essential for further evaluating the accuracy of the prediction model. For the training cohort, the AUCs of the time-dependent ROC analysis for predicting the 1-, 3-, and 5-year OS were 0.809, 0.864, and 0.893. For the testing cohort, the AUCs of the time-dependent ROC analysis for predicting the 1-, 3-, and 5-year OS were 0.808, 0.856, and 0.881. For the validation cohort, the AUCs of the time-dependent ROC analysis for predicting the 1-, 3-, and 5-year OS were 0.852, 0.869, and 0.757. The time-dependent ROC curve indicated that nomogram has stable predictive performance in OS for each time period (Figure 5). The calibration curves are listed in Figure 6. Calibration curves exhibited a good consistency between the model prediction and the actual observation for 1-, 3-, and 5-year OS in the training cohort, the testing cohort, and the validation cohort. The DCA was carried out in the training cohort, the testing cohort, and the validation cohort. DCA plots indicated that the established nomogram had greater net benefits in comparison with the TNM staging system for predicting 1-, 3-, and 5-year OS of patients (Figure 7), indicating that it has a good clinical utility in predicting the OS for patients.
Figure 4.
Receiver operating characteristic (ROC) curves of the nomogram for predicting survival prognosis at the 1-, 3-, and 5-year points in (A) the training cohort, (B) the testing cohort, (C) and the validation cohort.
Figure 5.
Time-dependent receiver operating characteristic (ROC) curves of the nomogram in the training cohort, the testing cohort, and the validation cohort.
Figure 6.
Calibration curves of the nomogram for predicting survival prognosis at the 1-, 3-, and 5-year points in (A) the training cohort, (B) the training cohort, (C) and the validation cohort.
Figure 7.
Decision curve analysis (DCA) of the nomogram and TNM staging system for predicting survival prognosis at the 1-, 3-, and 5-year points in (A) the training cohort, (B) the training cohort, and (C) the validation cohort.
Risk Stratification
According to the total points of each patient, we also established a risk stratification system generated by nomogram. All patients in the training cohort, the testing cohort, and the validation cohort were divided into high-risk and low-risk groups according to the median value. The Kaplan–Meier curve was performed to draw the OS curves for the high-risk group and low-risk group in the training cohort, the testing cohort, and the validation cohort. The Kaplan–Meier curves indicated that patients’ survival in the low-risk group was better than that in the high-risk group (P < .001) (Figure 8).
Figure 8.
Kaplan–Meier curves of overall survival (OS) stratified by risk classification system in the (A) training cohort, (B) the testing cohort, and (C) the validation cohort.
Discussion
In recent years, some nomograms including a variety of clinical characteristics have been utilized to predict the prognosis of various types of lung cancer.22–27 Jia et al developed a new prognostic nomogram to predict survival probability for NSCLC cases who underwent surgery and explored the potential influencing factors.22 Besides, Dai et al constructed a nomogram for the prognostic prediction of lung cancer patients below the age of 45 years in both OSl and cancer-specific survival, assisting the clinicians to choose personalized treatment.23 To the best of our knowledge, our study is the first to establish a nomogram for predicting the survival prognosis of LUAD patients based on a large population-based cohort from the SEER database and a multicenter cohort from China. More importantly, the nomogram revealed excellent predictive ability in both the US population and the Chinese population.
In previous works, there existed some deficiencies such as the number of patients in the external validation cohort was small, and the external validation of model was only performed with single-center data. In contrast, our study was significantly different from other retrospective single-center designed studies. The population in our nomogram was derived from the SEER cohort and the Chinese cohort, covering multiple-center registered, multiracial, and large populations, which would greatly improve the universality of this visualized model. In our study, age, sex, T stage, N stage, M stage, bone metastasis, brain metastasis, liver metastasis, and surgery were confirmed by univariate and multivariate Cox regression analyses as independent significant risk factors for predicting OS, which were in line with the results in the previous clinical reports.22–27 Subsequently, the abovementioned 9 prognostic factors were added to a nomogram to estimate the risk probability of OS for LUAD patients. In our study, older patients were reported to exhibit a higher risk and a shorter survival prognosis. Older patients had poor survival outcomes than younger patients might due to degenerative changes in multiple organ function and increased incidence of comorbid illnesses.28 These results were consistent with the findings in other types of lung cancer, such as SCLC and SCC, suggesting that older patients have a worse survival prognosis.24,25 Wang et al have found that increased age was associated with poor OS of SCLC patients.24 At present, it is generally agreed that male patients can be seen as a risk factor for NSCLC patients. However, many research results showed that survival outcomes for female patients were obviously better than those for male patients.26,27 In our study, the male patients had poor survival than the female patients, which was in agreement with the research of Lin et al and Yu et al.29,30 NSCLC is generally considered as a hormone-independent cancer. Whether gender potentially played a central role in survival prognosis in LUAD patients requires further analysis.31 NSCLC is characterized by severe metastatic features, and bone metastasis is the most common location, followed by lung metastasis, brain metastasis, and liver metastasis.32,33 NSCLC patients with distant metastasis are closely related to significantly reduced OS.34 A previous study reported that LUAD occupied about 50% of all the lung cancer patients with bone metastasis.35 LUAD individuals also displayed a 2.86-fold higher risk of brain metastases occur compared with other types of NSCLC individuals.36 Therefore, distant metastasis is vital for the survival prognosis of LUAD. In our study, similar results were proved by the statistical analysis. Bone metastasis, brain metastasis, and liver metastasis display an unsatisfactory prognosis and short-term survival for LUAD patients. Surgery is the main treatment for the majority of lung cancer.14,37–39 In our study, surgery was also showed to be a significant prognostic predictor for LUAD in that patients with surgery could significantly improve OS. Therefore, for patients without surgical contraindications, surgery treatment should be considered. Although surgery generates the better prognosis for LUAD, the selection of therapeutic scheme may be made based on various factors, such as the TNM stage and the patient's clinical characteristics. The optimal therapeutic scheme for the individual should be identified after the comprehensive assessment. Our study showed that tumor laterality was not associated with OS of LUAD through the multivariate Cox stepwise regression analysis, which is consistent with previous studies.7,24,27
With the development of biomedical technologies in recent years, several studies have developed nomogram based on a combination of molecular biomarkers to predict the survival probability for cancers.40–47 Xin et al have developed a model with microRNA expression profile for prognosis prediction in patients with LUAD. The C-indexes were 0.68 and 0.72 in the training and validating cohorts, respectively.40 Moreover, a nomogram integrated clinical variables and the metabolic gene was established by He et al might be useful for personalized risk predictions, achieving a C-index of 0.702.41 An immune-related gene-based prognostic nomogram for LUAD has been created and ideal results have been achieved. The highest AUC score was 0.717.42 One study established nomogram based on immune-infiltrating Treg-related genes for LUAD individuals, and the AUC values were found to be good (3-year AUC: 0.733; 5-year AUC: 0.777).43 The performance of the nomogram may also have discrimination ability and calibration accuracy, so ROC curves and calibration curves should be performed using the training, testing, and validation cohorts. In our study, a SEER cohort was used for the training and internal validation of the established nomogram, and the Chinese population was used for external validation to further assess the accuracy, applicability, and credibility of the nomogram in a broad population of patients with LUAD. Our study found that the C-index was 0.745 (95% CI, 0.741-0.749), 0.741 (95% CI, 0.733-0.749), and 0.809 (95% CI, 0.785-0.833) in the training cohort, the testing cohort, and the validation cohort, respectively. For the training cohort, the AUCs for predicting the 1-, 3-, and 5-year OS were 0.806, 0.856, and 0.886. For the testing cohort, the AUCs for predicting the 1-, 3-, and 5-year OS were 0.804, 0.849, and 0.873. For the validation cohort, the AUCs for predicting the 1-, 3-, and 5-year OS were 0.86, 0.874, and 0.861. Compared to the abovementioned prognostic nomogram indexes for LUAD, the present research demonstrated that the nomogram also had good predictive accuracy and reliability, which were consistent with previous studies.40–43 The AUCs of the training cohort, the testing cohort, and the validation cohort based on the nomogram were higher than those based on the molecular biomarker models. These results indicate that our nomogram has greater potential for accurately predicting prognosis compared to abovementioned prognostic nomograms. We also performed the time-dependent ROC curve to evaluate the discrimination ability of the nomogram. The time-dependent ROC curve of the nomogram includes all 9 risk factors. The time-dependent ROC curves of the nomogram showed that they have good predictive performance in the training cohort, the testing cohort, and the validation cohort during the follow-up time. Furthermore, our study showed excellent agreement between predicted and actual probability due to the points are close to the 45° diagonal line. What's more, our nomogram contained all the risk factors that could be easily available from clinical and pathological data, which is helpful in the clinical applicability of individual patients.
At present, the TNM staging system which identified by the laboratory and pathological results. Since the popularization of the TNM staging system in the 1970s, main revisions have been made many times.10 In our established nomogram, it is obvious that the TNM stage significantly affected the total point used for predicting the survival prognosis of LUAD patients. These results were in agreement with the previous study, which found that sex, age, TNM stage, and surgery were significant risk factors for OS in NSCLC patients.14 It is known that the TNM staging system plays a critical role in predicting survival prognosis and affecting treatment selection in NSCLC patients. However, it ignores a variety of clinical variables, such as age, sex, and biomarkers. For several tumors, the better predictive performance of nomograms than the widely performed TNM staging system has been identified.48,49 To better evaluate the net benefit of our predictive nomogram across a range of threshold risks to help clinical decision making, we demonstrated that the novel developed nomogram predicted OS with much more clinical applicability than the TNM staging system using DCA. The risk classification system regarding to this nomogram can efficiently divided LUAD patients in the training, the testing, and the validation cohorts into 2 risk groups. LUAD patients in the low-risk group had a better OS compared with those in the high-risk group. The use of this nomogram to distinguish between risk groups may help in clinical decision making for patients.
Our study had several advantages. First, to the best of our knowledge, we developed the first prognostic nomogram that can evaluate the significant risk factors related to survival prognostic in LUAD patients based on a large population-based cohort from the SEER database and a multicenter cohort from China. Second, we divided LUAD individuals into a training cohort, a testing cohort, and a validation cohort, which made our predictive nomograms reliability. Third, both internal and external validation cohorts confirmed the excellent applicability of this nomogram, indicating that the nomogram is suitable for LUAD patients of different races. Finally, all significant demographic, clinical, and pathological variables in our visualized nomograms were available and common in clinical practice. Therefore, our models were useful for clinicians to predict the survival prognosis of LUAD patients and make decisions regarding appropriate treatment strategies. Limitations have to be acknowledged in our study. First, the SEER database lacks some critical variables linked to prognosis, such as chemotherapy, molecular factors, socioeconomic status, alcohol consumption, smoking, and other lifestyle factors. A more comprehensive model considering all potential risk factors might be expected to have better prognostic performance. Second, our report is a retrospective study, and LUAD patients with incomplete variables were excluded from the analysis, which may lead to selection biases. Third, for better external validation, large samples from international multicenter cohorts are desired.
Conclusion
In conclusion, the first novel nomogram was established for predicting the OS of LUAD patients with reliability and high performance. The nomogram performed very well with excellent predictive ability in both the US population and the Chinese population. The clinical application of our nomogram was superior to the TNM staging system. These findings could be helpful in assisting clinicians to assess the OS of LUAD patients in order to conduct individualized treatment for LUAD patients.
Acknowledgments
The authors would like to thank all staff and participants of the Surveillance, Epidemiology, and End Results (SEER) program and the Chinese multicenter lung cancer database.
Abbreviations
- AUCAL
area under the curve
- AJCCAL
American Joint Committee on Cancer
- CIAL
confidence interval
- C-indexAL
concordance index
- CCALAL
clear cell adenocarcinoma of the lung
- DCAAL
decision curve analysis
- HRAL
hazard ratio
- LSCCAL
lung squamous cell
- LUADAL
lung adenocarcinoma
- LA-NSCLCAL
locally advanced NSCLC
- NSCLCAL
non-small-cell lung cancer
- OSAL
overall survival
- ROCAL
receiver operating characteristic curves
- SCCAL
squamous cell
- SCLCAL
small cell lung cancer
- SEERAL
SurveillanceAL, EpidemiologyAL, and End Results.
Footnotes
Author’s Contribution: YC conceived of and designed the study; TZ, SXZ, HYP, XNF, YSZ, JL, and KWH participated in data collection; ZQW, RJC, DNX, YJC, and XG performed literature search; ZQW, XYY, and CX created the figures and tables; ZQW, YJL, and RXW analyzed the data; HC, SBL, and FH supervised the research; ZQW wrote the manuscript; and FH critically reviewed the manuscript.
Data Availability Statement: The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Key R&D Program of China, Strategic Collaborative Innovation Team, Shanghai Health Care Commission Clinical Research Program, and Shanghai Three-year Action Plan for Public Health (grant numbers 2018YFC1705100, SSMU-ZLCX20180601, 20214Y0205, and GWV-10.1-XK18).
Ethical Approval and Consent to Participate: The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Participant consent was not necessary as this study involved the use of a previously published de-identified database according to the SEER database. We signed the data use agreement in accordance with the requirement of using the SEER database. Therefore, we could download data from the SEER database. This retrospective study of the Chinese cohort was approved by the ethics committee of the Fourth Hospital of Hebei Medical University (No. 2019075) and Dongfang Hospital Beijing University of Chinese Medicine (No. JDF-IRB-2019034101). The patient data of the Chinese cohort have been de-identified for this research.
ORCID iD: Yong Cai https://orcid.org/0000-0002-5706-7906
References
- 1.Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7‐33. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299‐311. [DOI] [PubMed] [Google Scholar]
- 3.Socinski MA, Obasaju C, Gandara D, et al. Clinicopathologic features of advanced squamous NSCLC. J Thorac Oncol. 2016;11(9):1411‐1422. [DOI] [PubMed] [Google Scholar]
- 4.Senosain MF, Massion PP. Intratumor heterogeneity in early lung adenocarcinoma. Front Oncol. 2020;10:349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolly SO, Collins DC, Sundar R, et al. Advances in the development of molecularly targeted agents in non-small-cell lung cancer. Drugs. 2017;77(8):813‐827. [DOI] [PubMed] [Google Scholar]
- 6.Guo M, Li B, Yu Y, et al. Delineating the pattern of treatment for elderly locally advanced NSCLC and predicting outcomes by a validated model: a SEER based analysis. Cancer Med. 2019;8(5):2587‐2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ke SJ, Wang P, Xu B. Clear cell adenocarcinoma of the lung: a population-based study. Cancer Manag Res. 2019;11:1003‐1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng R, Gu X, Wang M, et al. Nomograms to predict survival in patients with lung squamous cell cancer: a population-based study. J Nippon Med Sch. 2019;86(6):336‐344. [DOI] [PubMed] [Google Scholar]
- 9.Zhang C, Mao M, Guo X, et al. Nomogram based on homogeneous and heterogeneous associated factors for predicting bone metastases in patients with different histological types of lung cancer. BMC Cancer. 2019;19(1):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim W, Ridge CA, Nicholson AG, et al. The 8th lung cancer TNM classification and clinical staging system: review of the changes and clinical implications. Quant Imaging Med Surg. 2018;8(7):709‐718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang G, Wu Y, Zhang J, et al. Nomograms for predicting long-term overall survival and disease-specific survival of patients with clear cell renal cell carcinoma. Onco Targets Ther. 2018;11:5535‐5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu C, Zhang Y. Development and validation of prognostic nomogram for young patients with gastric cancer. Ann Transl Med. 2019;7(22):641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan X, Yang W, Chen Y, et al. Nomogram for predicting the overall survival of patients with inflammatory breast cancer: a SEER-based study. Breast. 2019;47:56‐61. [DOI] [PubMed] [Google Scholar]
- 14.Liao Y, Wang X, Zhong P, et al. A nomogram for the prediction of overall survival in patients with stage II and III non-small cell lung cancer using a population-based study. Oncol Lett. 2019;18(6):5905‐5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong J, Zheng Q, An T, et al. Nomogram to predict cause-specific mortality in extensive-stage small cell lung cancer: a competing risk analysis. Thorac Cancer. 2019;10(9):1788‐1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu F, Wang L, Zhou C. Lung cancer in China: current and prospect. Curr Opin Oncol. 2021;33(1):40‐46. [DOI] [PubMed] [Google Scholar]
- 17.von Elm E, Altman DG, Egger M, et al. The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med. 2007;147:573‐577. [DOI] [PubMed] [Google Scholar]
- 18.Bi G, Lu T, Yao G, et al. The prognostic value of lymph node ratio in patients with N2 stage lung squamous cell carcinoma: a nomogram and heat map approach. Cancer Manag Res. 2019;11:9427‐9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337‐344. [DOI] [PubMed] [Google Scholar]
- 20.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26(6):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranstam J, Cook JA. Kaplan-Meier curve. Br J Surg. 2017;104(4):442. [DOI] [PubMed] [Google Scholar]
- 22.Jia B, Zheng Q, Wang J, et al. A nomogram model to predict death rate among non-small cell lung cancer (NSCLC) patients with surgery in surveillance, epidemiology, and end results (SEER) database. BMC Cancer. 2020;20(1):666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai L, Wang W, Liu Q, et al. Development and validation of prognostic nomogram for lung cancer patients below the age of 45 years. Bosn J Basic Med Sci. 2021;21(3):352‐363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Pang Z, Chen X, et al. Development and validation of a prognostic model of resectable small-cell lung cancer: a large population-based cohort study and external validation. J Transl Med. 2020;18(1):237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Gao C, Du Q, et al. A prognostic model for elderly patients with squamous non-small cell lung cancer: a population-based study. J Transl Med. 2020;18(1):436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song CK, Guo ZX, Shen XY, et al. Prognostic factors analysis and nomogram construction of dual primary lung cancer: a population study. Biomed Res Int. 2020;2020:7206591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong S, Liang J, Zhai W, et al. Development and validation of an individualized nomogram for predicting overall survival in patients with typical lung carcinoid tumors. Am J Clin Oncol. 2020;43(9):607‐614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aarts MJ, Aerts JG, van den Borne BE, et al. Comorbidity in patients with small-cell lung cancer: trends and prognostic impact. Clin Lung Cancer. 2015;16(4):282‐291. [DOI] [PubMed] [Google Scholar]
- 29.Lin G, Qi K, Liu B, et al. A nomogram prognostic model for large cell lung cancer: analysis from the surveillance, epidemiology and end results database. Transl Lung Cancer Res. 2021;10(2):622‐635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu X, Gao S, Xue Q, et al. Development of a nomogram for predicting the operative mortality of patients who underwent pneumonectomy for lung cancer: a population-based analysis. Transl Lung Cancer Res. 2021;10(1):381‐391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mok TSK, Wu YL, Kudaba I, et al. KEYNOTE-042 Investigators. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. 2019;393(10183):1819‐1830. [DOI] [PubMed] [Google Scholar]
- 32.Riihimäki M, Hemminki A, Fallah M, et al. Metastatic sites and survival in lung cancer. Lung Cancer. 2014;86(1):78‐84. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Zhu H, Sun L, et al. Prognostic value of site-specific metastases in lung cancer: a population based study. J Cancer. 2019;10(14):3079‐3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashour Badawy A, Khedr G, Omar A, et al. Site of metastases as prognostic factors in unselected population of stage IV non-small cell lung cancer. Asian Pac J Cancer Prev. 2018;19(7):1907‐1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugiura H, Yamada K, Sugiura T, et al. Predictors of survival in patients with bone metastasis of lung cancer. Clin Orthop Relat Res. 2008;466(3):729‐736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang F, Zheng W, Ying L, et al. A nomogram to predict brain metastases of resected non-small cell lung cancer patients. Ann Surg Oncol. 2016;23(9):3033‐3039. [DOI] [PubMed] [Google Scholar]
- 37.Shi Y, Chen W, Li C, et al. Clinicopathological characteristics and prediction of cancer-specific survival in large cell lung cancer: a population-based study. J Thorac Dis. 2020;12(5):2261‐2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao Y, Yin G, Fan X. The positive lymph node ratio predicts survival in T1-4N1-3M0 non-small cell lung cancer: a nomogram using the SEER database. Front Oncol. 2020;10:1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdel-Rahman O. Risk of cardiac death among cancer survivors in the United States: a SEER database analysis. Expert Rev Anticancer Ther. 2017;17(9):873‐878. [DOI] [PubMed] [Google Scholar]
- 40.Xin G, Cao X, Zhao W, et al. MicroRNA expression profile and TNM staging system predict survival in patients with lung adenocarcinoma. Math Biosci Eng. 2020;17(6):8074‐8083. [DOI] [PubMed] [Google Scholar]
- 41.He L, Chen J, Xu F, et al. Prognostic implication of a metabolism-associated gene signature in lung adenocarcinoma. Mol Ther Oncolytics. 2020;19:265‐277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang T, Hao L, Cui R, et al. Identification of an immune prognostic 11-gene signature for lung adenocarcinoma. PeerJ. 2021;9:e10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, Xiao Z, Gong J, et al. A prognostic nomogram for lung adenocarcinoma based on immune-infiltrating Treg-related genes: from bench to bedside. Transl Lung Cancer Res. 2021;10(1):167‐182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abdel Ghafar MT, Soliman NA. Metadherin (AEG-1/MTDH/LYRIC) expression: significance in malignancy and crucial role in colorectal cancer. Adv Clin Chem. 2022;106:235‐280. [DOI] [PubMed] [Google Scholar]
- 45.Abdel Ghafar MT, El-Rashidy MA, Gharib F, et al. Impact of XRCC1 genetic variants on its tissue expression and breast cancer risk: a case-control study. Environ Mol Mutagen. 2021;62(7):399‐408. [DOI] [PubMed] [Google Scholar]
- 46.Abdel Ghafar MT, Elkhouly RA, Elnaggar MH, et al. Utility of serum neuropilin-1 and angiopoietin-2 as markers of hepatocellular carcinoma. J Investig Med. 2021;69(6):1222‐1229. [DOI] [PubMed] [Google Scholar]
- 47.Habib EM, Nosiar NA, Eid MA, et al. MiR-150 expression in chronic myeloid leukemia: relation to imatinib response. Lab Med. 2022;53(1):58‐64. [DOI] [PubMed] [Google Scholar]
- 48.Cheng B, Wang C, Zou B, et al. A nomogram to predict outcomes of lung cancer patients after pneumonectomy based on 47 indicators. Cancer Med. 2020;9(4):1430‐1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng XQ, Huang JF, Lin JL, et al. Incidence, prognostic factors, and a nomogram of lung cancer with bone metastasis at initial diagnosis: a population-based study. Transl Lung Cancer Res. 2019;8(4):367‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]