Abstract
Background: Nasopharyngeal carcinomas (NPCs) are malignant tumors originating from the lining epithelium of the nasopharynx. Fusion genes have been confirmed to play important roles in the occurrence and development of various malignant tumors, but the role of fusion genes in NPC is poorly understood. We aimed to explore new fusion genes that promote the occurrence and development of NPC. Methods: RNA-seq was used to search for interchromosomal translocations in 18 NPC tissues. Polymerase chain reaction (PCR) and Sanger sequencing were applied to verify the presence of BCL6-SPECC1L (BS); quantitative PCR (qPCR) and Western blotting were used to measure the expression level of BCL-6 in NPC cells; MTT and in vivo tumorigenesis assays were applied to evaluate the cell proliferation ability; immunofluorescence assays were used to determine the cellular localization of BCL6 and BS; and a luciferase reporter assay was performed to evaluate the ability of BCL6 and BS to inhibit transcription. Results: BS was present in 5.34% (11/206) of primary NPC biopsies and 2.13% (1/47) of head and neck cancer biopsies. The expression of BCL6 was downregulated in NPC, and silencing of endogenous BCL6 promoted NPC cell proliferation in vitro. Overexpression of BCL6 but not BS inhibited the growth of NPC cells in vivo and in vitro. Mechanistically, BCL6 localized in the nucleus can inhibit the G1/S transition to suppress the growth of NPC cells. However, after the fusion of BCL6 and SPECC1L, the product cannot localize to the nucleus, and the transcriptional inhibitory function of BCL6 is abolished, eventually abolishing its tumor suppressor effect and leading to the development of NPC. Conclusion: BS is a novel fusion gene in NPC that may play an important role in the occurrence and development of this cancer. The clinical significance of the BS fusion gene needs further elucidation.
Keywords: fusion gene, BCL6, SPECC1L, nasopharyngeal carcinoma, RNA-seq
Introduction
Nasopharyngeal carcinoma (NPC) is a malignant tumor originating from the epithelium of the nasopharynx, with the highest incidence in southeastern China, Southeast Asia, and North Africa.1 Although great progress has been made in the treatment of NPC as the radiotherapy techniques have improved, approximately 10% of patients experience relapse after treatment. Therefore, it is very important to elucidate the molecular mechanism of NPC and to identify early diagnostic markers and therapeutic targets for this cancer.
In 2011, Weinberg proposed that genomic instability and mutation are hallmarks of cancer that can lead to chromosomal rearrangement and gene fusion.2,3 Over the past decades, many fusion genes have been identified in leukemia and lymphoma, and it has recently been found that fusion genes are widely present in solid tumors and can serve as diagnostic and therapeutic markers. For example, the TMPRSS2-ERG fusion gene was identified in 33% of prostate cancer specimens4; FGFR3-TACC3 was observed in 3.1% of glioblastoma specimens5; EML4-ALK was detected in 6.7% of NSCLC specimens6; and NAB2-STAT6 was found in 51% of solitary fibrous tumor specimens.7 However, reports on fusion genes in NPC are limited. In 2013, the fusion genes UBR5-ZNF423 and RARS-MAD1L1 were identified in the NPC cell line C666 by Chung GT et al and were found to greatly promote the malignant behaviors of NPC cells in vivo and in vitro.8,9 In 2014, researchers identified the recurrent FGFR3-TACC3 fusion gene in NPC tissues through transcriptome sequencing and polymerase chain reaction (PCR). FGFR3-TACC3 promotes the proliferation of NPC cells by continuously activating fibroblast growth factor kinase, and this effect can be significantly inhibited by the fibroblast growth factor inhibitor PD173074.10 Therefore, fusion gene may be highly important in the occurrence and development of NPC, a possibility worthy of further exploration.
BCL6 is a transcriptional suppressor reported by most studies to function as an oncogene in several cancer types, such as lymphoma and breast cancer.11 Many mutations in BCL6 occur in diffuse large-cell lymphoma, all of which are activating mutations.12 BCL6 was found to activate a malignant transformation process by forming a transcriptional repression complex with BCOR,13 SMRT,14 and NCOR15 to inhibit various tumor suppressor genes, such as TP5316 and TNFRs. In B-cell lymphoma, various fusion gene forms of BCL6, such as fusions with the chr14q32 and chr22q11 loci, were also found. The fusion forms mainly altered the promoter of BCL6,17 thus constitutively activating BCL6 expression.
However, a few studies have reported that BCL6 could be used as a tumor suppressor gene. For example, in medulloblastoma, BCL6 was found to inhibit the expression of the Gli1 and Gli2 transcription factors in the SHH pathway at the transcriptional level by recruiting the transcription inhibitor BCOR and deacetylase SIRT1 and forming a complex, thus inhibiting the occurrence and development of neuroblastoma.18
In this study, we found that BCL6 functions as a tumor suppressor gene rather than an oncogene in NPC and discovered a novel fusion gene, BS, which causes loss of the DNA-binding domain in BCL6, impairing its tumor suppressor function and thus leading to the nasopharyngeal epithelial carcinogenesis. Our research confirmed that BS may play an important role in the occurrence and development of NPC.
Materials and Methods
Reagents
The reagents used were as follows: mouse monoclonal antibodies (Abs) against FLAG (F1804, Sigma-Aldrich, 1:2000 dilution) and GAPDH (KC-5G4, Kangcheng Biotech, 1:3000 dilution), horseradish peroxidase (HRP)-conjugated goat anti-mouse/rabbit secondary Abs (Fisher Scientific, 1:3000 dilution), an Alexa Fluor 594-conjugated goat anti-mouse Ab (Molecular Probes, 1:2000 dilution), and 4′-6-diamidino-2-phenylindole (DAPI; Molecular Probes).
Cell Culture
NPEC-Bmi1 cells (N2 and N5) and NP69 cells, immortalized nasopharyngeal epithelial cells induced by Bmi-1 and SV40T, respectively, were grown in keratinocyte serum-free medium (KSFM; Invitrogen). All human NPC cell lines (HONE1, CNE1, CNE2, and HNE1) maintained in our laboratory were cultured in RPMI 1640 medium (Gibco) supplemented with 5% fetal bovine serum (FBS; Gibco). HEK-293FT cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) supplemented with 10% FBS. Cells were grown in a humidified 5% CO2 incubator at 37 °C and passaged using standard cell culture techniques.
RNA-seq
A total of 18 NPC samples were obtained from patients, and all related procedures were performed with the approval of the Internal Review and Ethics Board of our hospital (approval number: B2022-412-01). After extracting RNA from 18 NPC tissues, a library was constructed and sequenced by Igenomics Ltd. The raw sequencing data reported in this paper have been deposited in the Genome Sequence Archive of the BIG Data Center at the Beijing Institute of Genomics, Chinese Academy of Science, under accession number HRA000087 (accessible at http://bigd.big.ac.cn/gsa-human). A human genome (UCSC hg19) was used as the reference genome. Fusion genes were identified using TopHat-Fusion with default parameters.
Identification of the Fusion Gene
Genomic DNA from tumor tissue was extracted with an E.Z.N.A.® Tissue DNA Kit (Omega, D3396-01) according to the manufacturer's instructions, and RNA was extracted with TRIzol and reverse transcribed into cDNA. Then, the 2 extraction products were subjected to PCR followed by agarose gel electrophoresis. Finally, the target band was excised and sent for Sanger sequencing. The primers used for PCR were as follows: BS (genomic level) forward primer, 5′-CAGCGAGAGCCACTCACCACTCTAC-3′; BS (genomic level) reverse primer, 5-GATTTAAAATTGGCTCTACCCCACC-3′; BS (RNA level) forward primer, 5′-ATGGCCTCGCCGGCTGACAGCTGTA-3′; and BS (RNA level) reverse primer, 5′-TCAGGAACTCATGATGGAAGAGGTG-3′.
Reverse Transcription-PCR (RT-PCR)
BCL6 expression levels in multiple NPC cell lines were determined by RT-PCR, and N2, N5, and NP69 were set as normal control. After extracting RNA with TRIzol, the mRNA was reverse transcribed into cDNA with random primers, quantitative PCR (qPCR) was carried out with SYBR Green I to measure the BCL6 expression level. GAPDH mRNA expression was also measured as the internal reference. The primers used were as follows: BCL6 forward primer, 5′-GGAGTCGAGACATCTTGACTGA-3′; BCL6 reverse primer, 5′-ATGAGGACCGTTTTATGGGCT-3′.
Western Blotting (WB)
Cell lysates were prepared in RIPA lysis buffer (Biosharp, BL504A) with protease inhibitors (Roche, Indianapolis, IN, USA). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane. The membrane was blocked with 5% PBS-T (25 mM Tris, 0.8% NaCl, 2.68 mM KCl (pH 7.4), and 0.1% Tween 20) and subsequently incubated with an anti-Flag (Sigma-Aldrich, F1804, 1:2000 dilution), anti-BCL6 (Sigma-Aldrich, HPA004899, 1:500 dilution), anti-SPECC1L (SAB, 29423-1, 1:500 dilution), anti-P21 (CST, 2947S, 1:1000 dilution), and anti-p27 (CST, 3686T, 1:1000 dilution) Abs. The membrane was then incubated with an HRP-conjugated secondary Ab (Pierce, Thermo Fisher Scientific Inc., Rockford, IL, USA).
siRNA Transfection
First, we obtained 3 custom siRNAs targeting BCL6 from RiboBio. The sequences of the 2 single siRNA duplexes targeting BCL6 (RIBOBIO, China) were as follows: siBCL6-1, GTCGAGACATCTTGACTGA; siBCL6-2, GAAGAGGCACGAAGTGATA.
siRNAs were transfected into cells using Lipofectamine RNAiMAX (Invitrogen). Twenty-four hours post-transfection, the interference efficiency was verified using qPCR and WB. Transfected cells were used for cell functional assays.
Plasmid Construction, Retroviral Infection, and Establishment of Stable Cell Lines
The coding sequence of BCL6 was amplified from C666-1 cellular cDNA, and the coding sequence of the fusion gene BS was amplified from fusion gene-positive tissue. The coding sequence was subcloned into the pMSCV vector to generate pMSCV-BCL6 and pMSCV-BS. Retrovirus was produced as described previously.9 HNE1 and HONE1 cells were infected with retrovirus carrying BCL6, BS, or pMSCV (empty vector). HNE1 or HONE1 cells expressing BCL6, BS, or empty vector were selected with puromycin (Sigma-Aldrich, catalog no. 101-58-58-2) for 7 days after retroviral infection. After 10 days of selection, the stable cell lines were verified, and the resulting cells were cultured in fresh medium.
MTT Assay
To evaluate the cell proliferation ability, cells were seeded in 5 96-well plates. Beginning the next day, one plate was measured every day after 20 µL of MTT solution was added to each well. After 4 h, the culture medium was removed, and 150 µL of DMSO was added. Ten minutes later, the OD was measured at 490 nm. Finally, the growth curve was plotted with time on the horizontal axis and OD on the vertical axis.
In vivo Experiments
Female BALB/c nude mice (3-4 weeks old) were obtained from the Animal Center of Guangdong Province (Guangzhou, China) and housed under specific pathogen-free conditions. A total of 1 × 106 cells in 50 µL of PBS were injected into the thighs of nude mice. After tumors formed, the tumor size was measured every other day, and a tumor growth curve was drawn. All animal experiments were performed in accordance with a protocol approved by our institutional Animal Care and Use Committee (approval number: L102042015050A). The random animal allocation was performed by computer generation of random number.
Immunofluorescence
First, coverslips were placed in a 24-well plate, and a certain number of cells (HONE1-BCL6 or HONE1-BS) were seeded in each well. After the cells grew to a suitable density, the medium was discarded, and the cells were washed with PBS and fixed with precooled 4% paraformaldehyde for 15 min. After lysis with 0.5% Triton X-100 for 10 min and blocking with 5% BSA for 25 min, the cells were incubated with an anti-FLAG primary Ab (F1804, Sigma-Aldrich, 1:200 dilution) overnight and were then washed with 0.1% PBST 3 times for 5 min each. Then, the cells were protected from light, incubated with a fluorescent secondary Ab for half an hour, and counterstained with DAPI for 5 min. An antifade mounting medium was used to preserve fluorescence, and confocal fluorescence microscopy was used for imaging.
Luciferase Reporter Assay
5X BCL6 reporter plasmid containing the BCL6 binding site was constructed according to a previous study.19 5 µg of BCL6 reporter plasmid plus 5 µg( + ) or 10 µg( + +) of BCL6, BS, or BCL61–513 plasmid were transfected into 293 cells. About 48 h later, luciferase activity was detected using a luciferase assay kit (Luc-Pair Duo-Luciferase HS Assay Kit, iGenobio, LF004) according to the manufacturer's instructions.
Statistical Analysis
For the functional assays, data from all experiments that were repeated 3 times are presented as the means ± standard deviations (SDs) as determined by Student's t-test (unpaired, 2-tailed). The sample size was chosen based on the need for statistical power. Differences with P < .01 (**) and P < .05 (*), as determined by GraphPad Prism 5 (La Jolla, CA, USA), were considered statistically significant.
Results
Discovery and Verification of the BS Fusion Gene in NPC
Transcriptome sequencing was performed on 18 NPC tissues to identify novel fusion genes in NPC, and the characteristics of the corresponding 18 patients are summarized in Supplemental Table 1. After bioinformatics analysis, a new fusion gene, BS, was identified in NPC tissue 271. The fusion occurred at exon 6 of BCL6 and exon 2 of SPECC1L (Figure 1A). Based on the sequencing results, we designed upstream primers in exon 6 of BCL6 and downstream primers in exon 2 of SPECC1L, performed qPCR on cDNA reverse transcribed from the sample, and found specific bands (Figure 1B, left). The fusion gene was confirmed using Sanger sequencing (Figure 1C), and the fusion site was consistent with that indicated by transcriptome sequencing. Thus, the accuracy of the high-throughput sequencing results was verified by Sanger sequencing in the sample.
Figure 1.
Discovery of the BS fusion gene in NPC. (A) Detection of the BS fusion gene using transcriptome sequencing. The reads are aligned across the junctions of the predicted fusion transcripts. (B) The existence of BS at the transcriptional level (left) and genomic level (right) in NPC tissue 271 was confirmed by RT-PCR. (C) Identification of the fusion breakpoint at the transcriptional level using Sanger sequencing. (D) Identification of the fusion breakpoint at the genomic level using Sanger sequencing. The black bar indicates the 4 bp (TTCC) microhomologous region in the junction. (E) RNA structure (left) and protein structure (right) of the predicted fusion. All experiments were repeated 3 times and representative results are shown.
According to current research, gene fusion may occur at the genomic level due to chromosomal translocation. However, fusion transcripts generated by cis-splicing at the RNA level may be present only at the mRNA level (eg, SLC45A3-ELK4 fusion occurs only at the mRNA level).19 To identify whether the fusion gene was present at the genomic level, a specific primer was used, and PCR of genomic DNA from the sample revealed specific bands (Figure 1B, right). The results of Sanger sequencing are shown in Figure 1D.
The fusion gene BS was also further identified by qPCR in more samples of NPC, as well as in esophageal squamous cell carcinoma (ESCC) and head and neck cancer (HNC). Finally, we found that this fusion gene was present in 5.34% (11/206) of primary NPC biopsies, 2.13% (1/47) of HNC biopsies, and 0.70% (1/143) of ESCC biopsies. The Sanger sequencing results for these fusion events are shown in Supplemental figure 1. Therefore, this fusion gene may be a multivariant, multicancer, and multilevel fusion gene.
Through ORF analysis of the fusion gene, we found that SPECC1L fusion occurred at exon 2 in the 5′UTR of the gene and predicted that the fusion gene can be translated into 21 new amino acid sequences and then produce terminators. Therefore, this sequence is a new protein sequence. The predicted structural variation map is shown in Figure 1E, and the position of the new 21 amino acid sequence is denoted by “?” in the figure. By comparison with the human genome in NCBI, these 21 amino acids were identified as specific amino acid sequences, and there were no homologous sequences.
BCL6 Functions as a Tumor Suppressor Gene in NPC
BCL6 has a total length of 706 amino acids, and a protein containing amino acids 1-513 is expressed after fusion. The fusion gene is speculated to be a truncation of the BCL6 gene body that lacks the complete DNA-binding region—that is, zinc finger domain—of BCL6, thus without the transcription factor function of BCL6. Therefore, we hypothesized that the effect of BS on the development of NPC does not depend on the transcription factor function of BCL6. Furthermore, we hypothesized that BCL6 plays a tumor suppressor role in NPC and that the fusion gene abolishes this tumor suppressor role, thus promoting the occurrence and development of NPC.
To test this hypothesis, we first used qPCR and WB to detect BCL6 expression in NPC cell lines and found that BCL6 was significantly downregulated in NPC cell lines (Figure 2A and B). Bioinformatics analysis of the NPC microarray data in the NCBI GEO database also revealed that BCL6 expression was downregulated in NPC samples compared with normal control samples (Figure 2C). These results suggested that BCL6 might play a tumor suppressor role in NPC.
Figure 2.
BCL6 is a tumor suppressor gene in NPC. The expression level of BCL6 in various NPC cell lines was measured by qPCR (2A) and WB (2B). (C) In 2 GEO datasets, BCL6 was downregulated in NPC. MTT assays of HNE1 cells (D) and HONE1 cells (E) seeded 24 h after transfection with siRNA and then cultured for the indicated period; the siRNA interference efficiency is shown on the left. n = 6 wells per group. * refers to differences between NC and siBCL6-1 or siBCL6-2 (* P < .05; ** P < .01; and *** P < .001 (two-way analysis of variance (ANOVA) followed by the Bonferroni post-hoc test)). Growth curve (F) and images (G) of tumors from nude mice inoculated with HONE1 cells stably expressing BCL6 or pMSCV (vector). All in vitro experiments were repeated 3 times and representative results are shown.
Experiments with siRNA-mediated interference with BCL6 expression were carried out to further verify this conclusion. First, 2 siRNAs with knockdown effects were identified by WB (Figure 2D, left; Figure 2E, left), and the MTT assay showed that knockdown of BCL6 promoted the proliferation of the NPC cell lines HNE1 and HONE1 (Figure 2D and E). In contrast, overexpression of BCL6 in HNE1 and HONE1 cells significantly inhibited their proliferation (Figure 3A and B). Then, we carried out in vivo experiments in nude mice with the HONE1 cell line overexpressing BCL6. Overexpression of BCL6 significantly inhibited the tumorigenic ability of NPC cells in vivo; for example, reducing tumor proliferation (Figure 2F) and tumor weight (Figure 2G).
Figure 3.
Effect of the fusion gene on the anticancer effect of BCL6 and the underlying mechanism. MTT assays of HNE1 cells (A) and HONE1 cells (B) stably expressing BCL6 or BS. MTT assays of HNE1 cells (C) and HONE1 cells (D) seeded 24 h after transfection with SPECC1L siRNA and then cultured for the indicated period. n = 6 wells per group. * refers to differences between vector and BCL6 or BS or between NC and siSPECC1L-1 or siSPECC1L-2 (* P < .05; ** P < .01; and *** P < .001 (two-way ANOVA followed by the Bonferroni post hoc test). (C) Immunofluorescence staining was performed to detect the localization of BCL6 and BS in NPC cells. Nuclei were counterstained with DAPI. (D) The transcriptional inhibition ability of BCL6, BS, and BCL61-513 was identified by a luciferase reporter assay. * refers to differences between BCL6 and BS or BCL61-513 (* P < .05; ** P < .01; and *** P < .001 (ANOVA followed by the Bonferroni post-hoc test). All experiments were repeated 3 times and representative results are shown.
In summary, we demonstrated that BCL6 might play a tumor suppressor role in NPC.
Effect of the Fusion Gene on the Anticancer Effect of BCL6 and the Underlying Mechanism
To explore the influence in the tumor suppressor role of BCL6 after its fusion, we overexpressed BCL6 and its fusion gene in the NPC cell lines HONE1 and HNE1 and found that wild-type BCL6 inhibited the proliferation of these cells, but BS had a significantly weaker ability to inhibit proliferation (Figure 3A and B).
The SPECC1L gene encodes a coiled-coil domain-containing protein that may play a critical role in actin cytoskeleton reorganization during facial morphogenesis. Next, we interfered with the expression of SPECC1L via siRNA transfection in the nasopharyngeal cancer cell lines HNE1 and HONE1 and found that cell proliferation was not affected, ruling out the possibility that the fusion gene promotes the proliferation of NPC cells by affecting SPECC1L (Figure 3C and D).
As mentioned above, the DNA-binding domain of BCL6 was lost after fusion. By immunofluorescence experiments, we found that wild-type BCL6 was localized mostly in the nucleus, while after fusion, BS was localized mostly in the cytoplasm (Figure 3E). The BS fusion could no longer be localized in the nucleus, indicating that it did not participate in transcriptional inhibition.
To further confirm that the transcriptional repression ability of the fusion gene is affected, we constructed a 5X BCL6 reporter plasmid containing the BCL6 binding site according to a previous study19 and carried out a luciferase reporter assay. The BS fusion, like BCL61-513, which lacks the DNA-binding domain of BCL6, could not inhibit the transcription of pGL3-5X BCL6, whereas wild-type BCL6 could (Figure 3F). In conclusion, the nuclear entry ability and transcriptional inhibition function of BCL6 decreased after fusion. Since BCL6 acts as a transcriptional repressor and a tumor suppressor in NPC, the generation of the fusion gene ultimately leads to the development of NPC.
Anticancer Mechanism of BCL6 in NPC
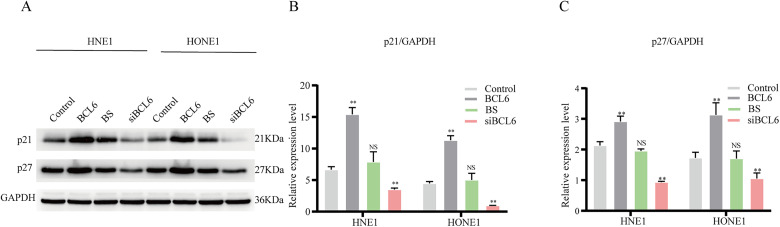
Through cell and animal experiments, we found that BCL6, a gene that plays an important oncogenic role in lymphoma, functions as a tumor suppressor in NPC; thus, identifying its mechanism in NPC was particularly important. Since the previous results demonstrated that BCL6 greatly affected the proliferation of NPC cells, we examined the expression of proteins in cell cycle-related pathways in cells with stable BCL6 or BS overexpression, or knockdown of BCL6 expression. The expression levels of p21 and p27 were significantly increased in BCL6-overexpressing cell lines and significantly decreased after silencing of BCL6 but were not significantly changed after overexpression of the fusion gene BS (Figure 4). These results suggested that BCL6 can block the G1/S transition in NPC cells, thereby inhibiting cell proliferation, but the fusion gene loses this ability.
Figure 4.
Representative Western blot images showing p21, p27, and GAPDH expression in HNE1 and HONE1 cells with BCL6 or BS overexpression, or with knockdown of BCL6 expression. The experiment was repeated 3 times and representative results are shown (A). The blots were quantified using Image Lab software (Bio-Rad, version 6.1), and paired T-test was used for statistical test (B and C).
Discussion
In this study, a novel fusion gene, BS, was identified by transcriptome sequencing. In addition, we found that BCL6 functions as a tumor suppressor gene in NPC and that BS fusion resulted in loss of the DNA-binding domain of BCL6, which changed its cellular localization, thereby abolishing its anticancer function and ultimately promoting the development of NPC. Our results suggest that BS may play an important role in the occurrence and development of NPC.
With the development of high-throughput sequencing technology, an increasing number of fusion genes have been found in various cancers. However, we are the first to discover that BCL6 can fuse with SPECC1L in NPC. We assume that the generation of the fusion gene may be related to the microhomologous sequence, since the sequence denoted by the dotted line in Figure 1D is the sequence shared by intron 6 of BCL6 and intron 1 of SPECC1L. Although microhomologous sequences constitute the basis of DNA repair, they are also a common cause of chromosomal translocation, and several fusion genes have been reported to have similar characteristics.20 Due to the inclusion of microhomologous sequences, we hypothesized that the fusion gene BS might be present in multiple samples. Therefore, we expanded the scope of detection to detect the presence of BS in other NPC tissues, ESCC tissues, and HNC tissues and found that BS was present in 5.34% (11/206) of primary NPC biopsies, 2.13% (1/47) of HNC samples, and 0.70% of ESCC samples. This finding suggested that the fusion gene may exist in various cancers, such as breast cancer and lung cancer, a hypothesis requiring further verification in more relevant samples.
BCL6, a transcriptional repressor with a zinc finger domain, is well known as an oncogene in lymphoma and has been reported to be an oncogene in breast cancer and other cancers.21,22 BCL6 was reported to be highly expressed in breast cancer cell lines, which promoted the survival of these cells, while targeting BCL6 with a peptidomimetic inhibitor induced apoptosis in these cells.21 BCL6 induces EMT by enhancing the expression of the transcriptional repressor ZEB1, which binds to the E-cadherin promoter and inhibits E-cadherin transcription, thereby stimulating EMT in breast cancer cells to promote their invasion, migration, and growth.22 However, surprisingly, this study indicated that BCL6 is a tumor suppressor in NPC, and the mechanism is worth exploring. It was initially found that BCL6 could inhibit the G1/S transition but could no longer localize to the nucleus after fusion with SPECC1L, resulting in loss of its transcriptional function. Therefore, we suspected that the gene profiles transcriptionally inhibited by BCL6 vary across cancers and that in NPC, BCL6 can transcriptionally inhibit the expression of certain oncogenes in the nucleus. Sarah R. Walker et al found through ChIP-seq that although some BCL6 binding sites in breast cancer cells overlapped with those identified in lymphoma cells, there might be a unique BCL6 binding pattern in breast cancer.21 However, which genes are specifically regulated by BCL6 in NPC remains to be explored.
The SPECC1L gene encodes a coiled-coil domain-containing protein that may play a critical role in actin cytoskeleton reorganization during facial morphogenesis. A novel fusion gene, RET-SPECC1L, was found in uterine sarcoma tissue from an adult woman, and this fusion enabled the coiled-coil domain of SPECC1L to mediate ligand-independent autophosphorylation of the RET kinase domain, leading to activation of downstream signaling pathways that drive tumor cell proliferation.23 Another novel SPECC1L-MET fusion was detected in circulating tumor DNA in a patient with lung adenocarcinoma following treatment ith erlotinib and osimertinib.24 In addition, SPECC1L was found to be fused with ALK in an NSCLC patient with pretreatment.25 The above results indicate that SPECCL seems to have a tendency to fuse with other genes, but the underlying mechanisms remains to be clarified.
New proteins expressed by fusion genes may also have their own specific functions. For example, the fusion gene BCR-ABL1, which has been suggested to be the causative mutation of chronic myeloid leukemia (CML), has a new function; specifically, it has extremely strong tyrosine kinase activity. This activity leads to constitutive phosphorylation of a series of signaling proteins, affecting cell proliferation, differentiation, and apoptosis, thus promoting CML. However, the fusion gene that we found did not promote the proliferation of NPC cells after overexpression (Figure 3A and B). Thus, this fusion gene might not have an intrinsic cancer-promoting function.
One limitation of our study is that we did not explore the clinical significance of BS. Detecting the existence of BS in more patient tissues and analyzing its relationships with certain clinicopathological characteristics and the survival prognosis of patients will help to identify new diagnostic and therapeutic markers for NPC. This research focus will be completed in the future.
In conclusion, by performing RNA-seq on 18 NPC tissues, we discovered the fusion gene BS for the first time and confirmed its existence in various cancers. Gene fusion causes BCL6 to lose its inhibitory effect on the proliferation of NPC cells, thereby promoting the occurrence and development of NPC.
Conclusion
BCL6 is a tumor suppressor gene in NPC that loses its tumor suppressor function after fusion with SPECCL. BS is expected to become a new diagnostic marker for NPC. This conclusion warrants further study with more clinical samples.
Supplemental Material
Supplemental material, sj-xls-1-tct-10.1177_15330338221139981 for BCL6-SPECC1L: A Novel Fusion Gene in Nasopharyngeal Carcinoma by Shuo-Gui Fang, Tian-Liang Xia, Jian-Chang Fu, Tong Li, Qian Zhong and Fei Han in Technology in Cancer Research & Treatment
Supplemental material, sj-tif-2-tct-10.1177_15330338221139981 for BCL6-SPECC1L: A Novel Fusion Gene in Nasopharyngeal Carcinoma by Shuo-Gui Fang, Tian-Liang Xia, Jian-Chang Fu, Tong Li, Qian Zhong and Fei Han in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-3-tct-10.1177_15330338221139981 for BCL6-SPECC1L: A Novel Fusion Gene in Nasopharyngeal Carcinoma by Shuo-Gui Fang, Tian-Liang Xia, Jian-Chang Fu, Tong Li, Qian Zhong and Fei Han in Technology in Cancer Research & Treatment
Acknowledgment
We thank Professor Musheng Zeng for his advice and support in this study.
Abbreviations
- PCR
polymerase chain reaction
- BS
BCL6-SPECC1L
- qPCR
quantitative PCR
- HNC
head and neck cancer
- DLCL
diffuse large-cell lymphoma
- Abs
antibodies
- HRP
horseradish peroxidase
- KSFM
keratinocyte serum-free medium
- DMEM
Dulbecco’s modified Eagle’s medium
- WB
Western blotting
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SPF
specific pathogen-free
- SDs
standard deviations
- ESCC
esophageal squamous cell carcinoma
- CML
chronic myeloid leukemia
- ANOVA
analysis of variance
Footnotes
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: All of the tumor tissues were obtained from the patients, and all related procedures were performed with the approval of the Internal Review and Ethics Board of Sun Yat-sen University Cancer Center (approval number: B2022-412-01). All animal experiments were performed in accordance with a protocol approved by Sun Yat-sen University Cancer Center Animal Care and Use Committee (approval number: L102042015050A).
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from Natural Science Foundation of Guang Dong Province (No. 2017A030312003) and the National Natural Science Foundation of China (81772883). The funders had no role in study design, data collection and analysis, the decision to publish, or the preparation of the manuscript.
ORCID iD: Fei Han https://orcid.org/0000-0002-1598-9048
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394(10192):64‐80. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646‐674. [DOI] [PubMed] [Google Scholar]
- 3.Edwards PA. Fusion genes and chromosome translocations in the common epithelial cancers. J Pathol. 2010;220(2):244‐254. [DOI] [PubMed] [Google Scholar]
- 4.Saramäki OR, Harjula AE, Martikainen PM, Vessella RL, Tammela TL, Visakorpi T. TMPRSS2:ERG Fusion identifies a subgroup of prostate cancers with a favorable prognosis. Clin Cancer Res. 2008;14(11):3395‐3400. [DOI] [PubMed] [Google Scholar]
- 5.Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337(6099):1231‐1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561‐566. [DOI] [PubMed] [Google Scholar]
- 7.Robinson DR, Wu YM, Kalyana-Sundaram S, et al. Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat Genet. 2013;45(2):180‐185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung GT, Lung RW, Hui AB, et al. Identification of a recurrent transforming UBR5-ZNF423 fusion gene in EBV-associated nasopharyngeal carcinoma. J Pathol. 2013;231(2):158‐167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong Q, Liu ZH, Lin ZR, et al. The RARS-MAD1L1 fusion gene induces cancer stem cell-like properties and therapeutic resistance in nasopharyngeal carcinoma. Clin Cancer Res. 2018;24(3):659‐673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan L, Liu ZH, Lin ZR, Xu LH, Zhong Q, Zeng MS. Recurrent FGFR3-TACC3 fusion gene in nasopharyngeal carcinoma. Cancer Biol Ther. 2014;15(12):1613‐1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang CC, Ye BH, Chaganti RS, Dalla-Favera R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci U S A. 1996;93(14):6947‐6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye BH, Chaganti S, Chang CC, et al. Chromosomal translocations cause deregulated BCL6 expression by promoter substitution in B cell lymphoma. Embo j. 1995;14(24):6209‐6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCor, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14(14):1810‐1823. [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmad KF, Melnick A, Lax S, et al. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell. 2003;12(6):1551‐1564. [DOI] [PubMed] [Google Scholar]
- 15.Barish GD, Yu RT, Karunasiri MS, et al. The Bcl6-SMRT/NCoR cistrome represses inflammation to attenuate atherosclerosis. Cell Metab. 2012;15(4):554‐562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurtz C, Hatzi K, Cerchietti L, et al. BCL6-mediated Repression of p53 is critical for leukemia stem cell survival in chronic myeloid leukemia. J Exp Med. 2011;208(11):2163‐2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W, Iida S, Louie DC, Dalla-Favera R, Chaganti RS. Heterologous promoters fused to BCL6 by chromosomal translocations affecting band 3q27 cause its deregulated expression during B-cell differentiation. Blood. 1998;91(2):603‐607. [PubMed] [Google Scholar]
- 18.Tiberi L, Bonnefont J, van den Ameele J, et al. A BCL6/BCOR/SIRT1 complex triggers neurogenesis and suppresses medulloblastoma by repressing Sonic Hedgehog signaling. Cancer Cell. 2014;26(6):797‐812. [DOI] [PubMed] [Google Scholar]
- 19.Qin F, Song Z, Babiceanu M, et al. Discovery of CTCF-sensitive Cis-spliced fusion RNAs between adjacent genes in human prostate cells. PLoS Genet. 2015;11(2):e1005001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villarreal DD, Lee K, Deem A, Shim EY, Malkova A, Lee SE. Microhomology directs diverse DNA break repair pathways and chromosomal translocations. PLoS Genet. 2012;8(11):e1003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker SR, Liu S, Xiang M, et al. The transcriptional modulator BCL6 as a molecular target for breast cancer therapy. Oncogene. 2015;34(9):1073‐1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu JM, Sun W, Hua F, et al. BCL6 Induces EMT by promoting the ZEB1-mediated transcription repression of E-cadherin in breast cancer cells. Cancer Lett. 2015;365(2):190‐200. [DOI] [PubMed] [Google Scholar]
- 23.Weisman PS, Altinok M, Carballo EV, et al. Uterine cervical sarcoma with a novel RET-SPECC1L fusion in an adult: A case which expands the homology between RET-rearranged and NTRK-rearranged tumors. Am J Surg Pathol. 2020;44(4):567‐570. [DOI] [PubMed] [Google Scholar]
- 24.Nelson AW, Schrock AB, Pavlick DC, Ali SM, Atkinson EC, Chachoua A. Novel SPECC1L-MET fusion detected in circulating tumor DNA in a patient with lung adenocarcinoma following treatment with erlotinib and osimertinib. J Thorac Oncol. 2019;14(2):e27‐ee9. [DOI] [PubMed] [Google Scholar]
- 25.Ma L, Zhang Q, Dong Y, Li H, Wang J. SPECC1L-ALK: A novel gene fusion response to ALK inhibitors in non-small cell lung cancer. Lung Cancer. 2020;143:97‐100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-xls-1-tct-10.1177_15330338221139981 for BCL6-SPECC1L: A Novel Fusion Gene in Nasopharyngeal Carcinoma by Shuo-Gui Fang, Tian-Liang Xia, Jian-Chang Fu, Tong Li, Qian Zhong and Fei Han in Technology in Cancer Research & Treatment
Supplemental material, sj-tif-2-tct-10.1177_15330338221139981 for BCL6-SPECC1L: A Novel Fusion Gene in Nasopharyngeal Carcinoma by Shuo-Gui Fang, Tian-Liang Xia, Jian-Chang Fu, Tong Li, Qian Zhong and Fei Han in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-3-tct-10.1177_15330338221139981 for BCL6-SPECC1L: A Novel Fusion Gene in Nasopharyngeal Carcinoma by Shuo-Gui Fang, Tian-Liang Xia, Jian-Chang Fu, Tong Li, Qian Zhong and Fei Han in Technology in Cancer Research & Treatment