Significance
Color (chromatic) vision is mediated by cone photoreceptors with sensitivity to different regions of the light spectrum, typically green and blue wavelength regions in mammals. However, knowledge is limited of transcriptional controls underlying this cone diversity, because cones are sparse and poorly defined apart from opsin photopigments. Using methodical single-cell analyses in mice, we find that cone subtypes differentially express groups of genes. Furthermore, we demonstrate a key role for a thyroid hormone receptor in determining chromatin accessibility and transcriptional distinctions between cone subtypes. Our findings suggest that receptor-mediated control of a bipotential transcriptional state promotes cone diversity and the full chromatic potential of the species.
Keywords: cone photoreceptor, color vision, thyroid hormone receptor, THRB, retina
Abstract
Cone photoreceptor diversity allows detection of wavelength information in light, the first step in color (chromatic) vision. In most mammals, cones express opsin photopigments for sensitivity to medium/long (M, “green”) or short (S, “blue”) wavelengths and are differentially arrayed over the retina. Cones appear early in retinal neurogenesis but little is understood of the subsequent control of diversity of these postmitotic neurons, because cone populations are sparse and, apart from opsins, poorly defined. It is also a challenge to distinguish potentially subtle differences between cell subtypes within a lineage. Therefore, we derived a Cre driver to isolate individual M and S opsin-enriched cones, which are distributed in counter-gradients over the mouse retina. Fine resolution transcriptome analyses identified expression gradients for groups of genes. The postnatal emergence of gradients indicated divergent differentiation of cone precursors during maturation. Using genetic tagging, we demonstrated a role for thyroid hormone receptor β2 (TRβ2) in control of gradient genes, many of which are enriched for TRβ2 binding sites and TRβ2-regulated open chromatin. Deletion of TRβ2 resulted in poorly distinguished cones regardless of retinal location. We suggest that TRβ2 controls a bipotential transcriptional state to promote cone diversity and the chromatic potential of the species.
Cone photoreceptors are specialized light-sensitive neurons that function in daylight and mediate color vision. Color vision requires cone populations with opsin photopigments for response to different regions of the light spectrum, typically medium/long (M) and short (S) wavelengths in most mammals (1). These cone populations, sometimes termed cone subtypes, are described principally by opsin immunoreactivity but are otherwise poorly defined. Cone distribution patterns over the retina vary between species reflecting adaptations to lifestyle and habitat (2, 3). For example, in humans, a dense concentration of cones resides in the foveal area of high-acuity vision with cone subtypes arrayed in mosaics (4, 5). In mice, cones are dispersed over the retina with subtypes arrayed in counter-gradients: M opsin is enriched in superior (dorsal) areas and S opsin in inferior (ventral) areas (2).
Cones are generated early during retinal neurogenesis (6, 7) then acquire mature morphology and opsin characteristics during a poorly understood process of terminal differentiation. Studies in rats suggested that an M opsin immunopositive identity is gained by switching from a “default S” identity (8). We reported that thyroid hormone receptor β2 (TRβ2) encoded by the Thrb gene (9) is critical for M opsin expression (10). TRβ2-deficient mice lack M opsin and express S opsin in all cones suggesting that both M and S opsin identities arise from an S opsin-positive cell type under the control of TRβ2 (10, 11). Thyroid hormone and the Thrb gene have since been found to promote expression of opsins for sensitivity to medium-long wavelengths in mice (12, 13), rats (14, 15), and fish (16–19) and in human retinal organoids (20). Human THRB mutations are associated with monochromacy and impaired responses to longer wavelengths of light (21–23).
The remarkable ability of TRβ2 to impose counter-gradients of both opsins in mice (10, 11) led us to postulate a wider role in cone diversity. Therefore, to elucidate cone subtype distinctions, we devised a methodical screen of cones from specific regions of mouse retina. To distinguish subtle differences, we employed a high-resolution transcriptome screen allowing in-depth analysis of each individually isolated cone. To investigate the transcriptional role of TRβ2, we exploited genetic tagging to analyze chromatin binding and accessibility in cones. Our results suggest that TRβ2 controls bipotential transcriptional plasticity in cone precursors to promote cone diversity.
Results
Fine Resolution Screen of Cone Transcriptome Diversity.
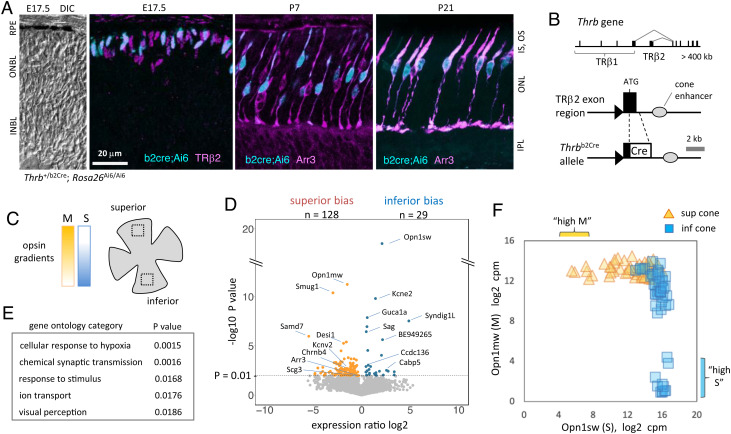
We devised a selective screen to overcome the scarcity of cones (which represent ≤2% of retinal cells in mice) using a cone-specific Cre knockin at the TRβ2 exon of the endogenous Thrb gene. TRβ2 is one of the earliest specific factors induced in cones (10) (Fig. 1 A and B). The Thrbb2cre driver activated cone-specific fluorescent signals (ZsGreen) from an Ai6 reporter (24), as shown using double-staining with embryonic (TRβ2) and postnatal (Arr3) cone markers. Labeled cells displayed characteristic cone morphology. The peak expression of TRβ2 near birth in the newly formed cone population (25) allowed us to use the Thrbb2cre driver to select cones from immature to adult stages. The Thrbb2cre driver was ~80% efficient (% of total cones, defined as ZsGreen+;Arr3+ / Arr3+ cells) and labels cones in all retinal regions, consistent with TRβ2 as a pan-cone marker. The driver was ~98% cone specific with sporadic noncone cells distinguished by morphology, laminar location, and marker stains (SI Appendix, Fig. S1).
Fig. 1.
Cone transcriptome diversity. (A) Cones labeled with Thrbb2Cre and Rosa26Ai6 reporter, costained with cone markers (TRβ2, Arr3). DIC, differential interference contrast view. (B) Thrbb2Cre knockin. Cre replaces the TRβ2-specific exon, leaving the rest of the gene and cone enhancer intact. (C) Diagram of opsin gradients beside a retinal flatmount. Boxes, zones from which cones were isolated. (D) Volcano plot identifying gradient genes with superior or inferior expression bias (P < 0.01; Student t test). RNA-seq data for 6,469 genes, cutoff 20 cpm; 53 superior, 50 inferior cones; 2-mo-old mice. (E) Gene ontology categories for 157 gradient genes. (F) Distribution plot of 103 cones showing opsin counter-gradients. Abbreviations: INBL, inner neuroblastic layer; INL, inner nuclear layer; IS/OS, inner/outer segments; ONBL, outer neuroblastic layer; ONL, outer nuclear layer; RPE, pigmented epithelium.
We analyzed cone subtypes with respect to spatial location by isolation of cones individually (based on ZsGreen label and morphology) from dissociated pieces of superior and inferior retina from Thrb+/b2cre; Rosa26 Ai6/Ai6 adult mice (Fig. 1C and SI Appendix, Fig. S1). This methodical approach yielded high-purity, high-efficiency single-cell libraries. RNA-sequencing at substantial depth (~13 million reads/cell) revealed that ~2.4% of expressed genes (n = 157) displayed differential expression, i.e., gradients, between superior and inferior groups (P < 0.01; 103 cells; mean expression cutoff 20 cpm) (Fig. 1D). Many (~46%) of these genes displayed <2.5-fold gradients indicating the subtle, quantitative nature of cone distinctions in mice. Gradient gene ontology categories included synaptic transmission and ion transport suggesting that subtype distinctions reflect maturational functions (Fig. 1E andSI Appendix, Fig. S2).
To validate the screen and to correlate M (Opn1mw) and S (Opn1sw) opsin expression ratios with the sensitivity of cones in different retinal locations to medium and short wavelength stimuli (26), we analyzed a cone distribution plot. About 10% of cells, all from the inferior, had a high S/M ratio (“high-S”) whereas ~6% of cells, all from the superior, had a high M/S ratio (“high-M”) (Fig. 1F). These small percentages align with estimates for so-called “pure” S or M immunoreactive cones in mice (2, 27, 28). Most cones (84%) expressed Opn1mw and Opn1sw in moderate ratios, in accord with the physiological response of most cones in mice reflecting M- or S-dominant rather than M- or S-exclusive sensitivity (26). High-M and high-S cones displayed similar trends for most gradient genes as the larger groups of superior and inferior cones (SI Appendix, Figs. S2A and S3A), suggesting that these subgroups are also subject to controls that impose graded gene expression on cone subtypes in mice.
Gradient Emergence: Bipotential Differentiation.
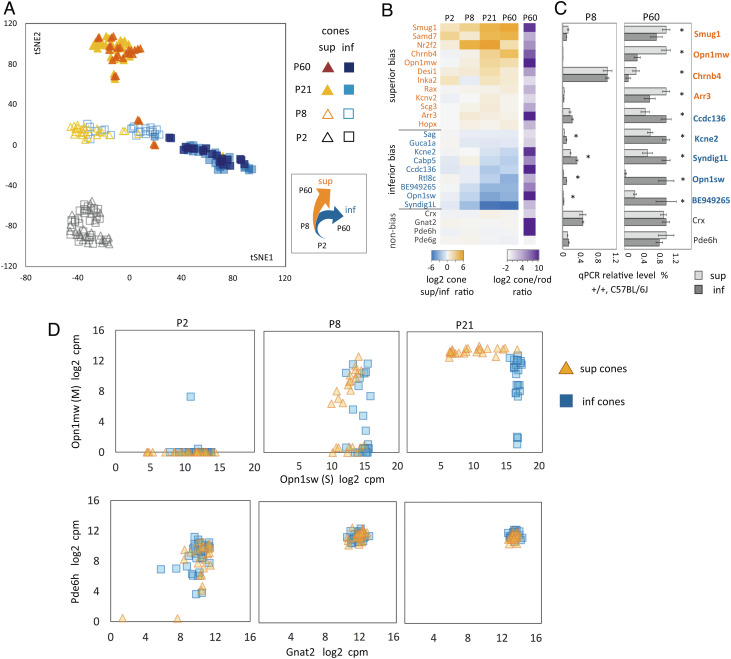
Newly generated, postmitotic cones are histologically indistinct in neonatal mice then mature over the next ~3 wk. To determine when cone subtypes emerge during this period, we analyzed 200 cones spanning the initial appearance of cone morphology (postnatal days P2, P8), early function after eye opening (P21) and mature function in adulthood (P60). In a t-Distributed Stochastic Neighbor Embedding (tSNE) plot, cones clustered together regardless of retinal location at P2 or P8 but diverged into superior and inferior groups by P21 (Fig. 2A). This divergence is shown for example genes in a heatmap of superior/inferior ratios (Fig. 2B). The initial similarity then divergence after ~P8 suggests bipotential differentiation from related cone precursors.
Fig. 2.
Gradient emergence during cone maturation. (A) Distribution (tSNE) plot of 200 cones; RNA-seq; 21–30 cells/group. Initially, cones cluster together then diverge by location after ~P8 (interpretative diagram, Right) (B) Heatmap of gradient emergence for representative genes (as sup/inf expression ratios). (C) Selected gradient genes corroborated by qPCR. Levels are relative to the highest expression (assigned as “1”) among the four groups at both ages; three pools of ≥3 mice; *P < 0.05, Student’s t test. (D) Distribution plots (152 cones) showing that all cones express Opn1sw before Opn1mw is induced and counter-gradients appear. Pde6h and Gnat2, control genes lacking gradients.
The development of gradients was corroborated by qPCR analysis of retinal tissue from wild-type C57BL/6J mice at P8 and P60 (Fig. 2C) for genes that were sufficiently detectable in cones relative to highly abundant cell types such as rods. Several gradient genes were consistent with genes indicated previously by northern blot, immunostaining (29, 30), and microarray analysis of retina (31). We found that although the developmental profile of each gene varies, gradients often became pronounced as expression rises, suggesting a coordinated process during maturation (Fig. 2 B and C).
We analyzed opsin gene expression to delineate the timing of cone subtype differentiation and to test the hypothesis suggested in rats that cones have a default S opsin identity before a population gains an M identity (8). Cone distribution plots show that in mice all immature cones regardless of location express Opn1sw before Opn1mw is induced (Fig. 2D). After Opn1mw is induced, both opsin genes acquire counter-gradients between P8 and P21, suggesting an opsin shift. However, in mice, this shift is quantitative rather than absolute, resulting in graded expression of Opn1mw and Opn1sw over the retinal vertical plane.
Cone Gene Expression Determined by TRβ2.
The unique ability of TRβ2 to determine both opsin gradients led us to test a wider role in control of gradient genes by transcriptome analysis of 98 TRβ2-KO (Thrbb2cre/2cre) cones and 103 control (Thrb+/b2cre) cones. A tSNE plot revealed dispersed clusters of cones from superior and inferior regions in control mice but intermingled clusters in KO mice, indicating diminished gradient distinctions (Fig. 3A). Analyses of averaged expression (Fig. 3B) and superior/inferior ratios (Fig. 3C) showed that deletion of TRβ2 disrupted expression of gradient genes in both superior and inferior bias categories resulting in poorly distinguished cones regardless of location. Overall, TRβ2-deficiency disrupted expression of a high proportion of gradient genes (~40%, 63/157 genes) and a comparatively small proportion of other, nongradient genes (6.3%, 398/6,312 genes) (Fig. 3D).
Fig. 3.
Disrupted gene expression in TRβ2-KO cones. (A) tSNE plots showing dispersed clusters of superior and inferior cones in control mice and mixed clusters in TRβ2-KO mice. RNA-seq, adults, 48–53 cones/group. (B) Box plots of normalized expression averaged for gradient gene groups, showing loss of gradients in KO cones. Z score for each gene = (each cone value−mean of both genotypes)/SD. P values, calculated by Student’s t test (and in panel E). (C) Heatmap of superior/inferior gene expression ratios. Superior bias, yellow; inferior bias, blue. Reference gradient genes noted. (D) TRβ2-KO disrupts expression of a high proportion of gradient genes (~40%; 63/157) relative to other genes (6.3%; 398/6,312). (E) Box plots for nongradient, TRβ2-dependent genes.
In summary, TRβ2-KO cones tend to homogeneity and as expected all express Opn1sw but lack Opn1mw, consistent with diversity arising from a default S-cone pathway. However, the lack of overt expression changes for ~60% of gradient genes suggests that TRβ2-KO cones retain remnants of other gradients and are not homogeneous S cones. Furthermore, the disrupted expression of nongradient genes in TRβ2-KO cones (Fig. 3E) suggests that gradient formation is coordinated with a broader, TRβ2-mediated maturational process. Nongradient gene ontology categories include visual perception and response to stimulus (SI Appendix Fig S2. SI Appendix Fig S3 shows nongradient target gene examples, Rd3L and Pgc).
Chromatin Control by TRβ2 at Gradient Genes.
To support a transcriptional basis for control by TRβ2, we investigated TRβ2 binding sites (i.e., putative control sites) in chromatin, using tagged endogenous receptors that are expressed in natural patterns by a knockin ThrbHAB allele (Fig. 4A andSI Appendix, Fig. S2). TRβ2-HAB protein is biotinylated in vivo by BirA ligase expressed by Rosa26 BirA, facilitating high-affinity purification of receptors with bound chromatin despite low numbers of cones in retinal tissue. Binding sites were identified by chromatin affinity purification-sequencing (ChAP-seq) of HAB (ThrbHAB/HAB;Rosa26 BirA/BirA) vs. control BirA (Rosa26 BirA/BirA) tissue, which had low background. ChAP-seq analysis of retina at embryonic day 18, the peak of TRβ2 expression, identified 4,590 binding sites, most of which were distal to the transcription start site (TSS) of genes, indicative of enhancers (Fig. 4B). The top consensus motif at these sites was a direct repeat of AGGTCA-like sequences with four base spacing (DR4) (Fig. 4C), consistent with binding sites for TR identified in vitro (32). TRβ2-HAB binding sites were enriched near gradient genes, including both superior and inferior bias categories, suggesting versatile control of a range of gradient genes (Fig. 4 D and E).
Fig. 4.
Chromatin binding and remodeling by TRβ2. (A) HAB-tagged endogenous TRβ2 is biotinylated by BirA, then affinity purified (AP) from mice with ThrbHAB (HAB) and Rosa26BirA (BirA) alleles. (B) TRβ2-HAB peaks relative to TSS determined by ChAP-seq (Left) and association with TRβ2-regulated ATAC peaks determined by differential analysis of control and TRβ2-KO datasets; cutoff >1.5-fold, FDR q < 0.05 (Right). (C) Top consensus motifs at TRβ2 binding and TRβ2-regulated chromatin sites, identified by Homer analysis. (D) Association of TRβ2-HAB binding sites with gradient genes; heatmap as in Fig. 3C (157 gradient genes, 85 [54%] of which have a binding site within 50 kb). (E) TRβ2-HAB binding and TRβ2-regulated open chromatin (ATAC) associated with gradient genes, total cone-expressed genes (6,469) or total genes in genome (25,672). (F) Inka2 gradient gene illustrating TRβ2 binding (HAB) and TRβ2-regulated open chromatin (ATAC) sites (red arrows; >2.5-fold difference, both ATAC peaks). BirA, Rosa26BirA/BirA control for ChAP-seq. IGV reads scale, numbers on Left.
We also demonstrated an association of TRβ2-regulated open chromatin, an indicator of active enhancers and promoters (33), with gradient genes. We used Thrbb2cre to activate a Rosa26 Sun1Gfp marker (34) for immunopurification of cone nuclei for ATAC-seq (assay for transposase-accessible chromatin-sequencing). TRβ2-regulated chromatin sites were identified by differential analysis of ATAC peaks in control (Thrb+/b2cre) and TRβ2-KO (Thrbb2cre/b2cre) cones, at P30, a relatively mature end point (cutoff >1.5-fold; FDR q < 0.05). TRβ2-regulated ATAC peaks, particularly induced (open chromatin) peaks, were enriched near gradient genes in both superior and inferior bias categories (Fig. 4E). Strikingly, the top motif at TRβ2-induced ATAC peaks was DR4 (Fig. 4C) consistent with receptor binding, which was supported in many cases by alignment with TRβ2-HAB binding sites (Fig. 4B). In contrast, the low frequency of TRβ2-HAB binding (or DR4 motifs) at TRβ2-suppressed ATAC peaks suggests that chromatin closing is often indirect without receptor binding to specific DNA elements. Fig. 4F illustrates regulated chromatin sites at Inka2, a TRβ2-induced gradient gene. In summary, ATAC analysis supports the control by TRβ2 of gradient and nongradient (SI Appendix, Fig. S2E) genes.
Gradient Patterning by TRβ2.
Gradient gene responses to TRβ2-deficiency varied by gene and by cone location on the retina suggesting complex controls of gradient formation. However, common trends emerged indicating that TRβ2 coordinates both activation and repression over the retina to establish gradients (Fig. 5A). TRβ2 deficiency decreased expression of an M opsin-like (superior bias) gene group (e.g., Opn1mw, Arr3) in all retinal regions, implying activation by TRβ2. In contrast, TRβ2 deficiency elevated expression of an S opsin-like (inferior bias) group (e.g., Opn1sw, Ccdc136) particularly in superior regions, implying repression by TRβ2. Thus, for these genes, superior bias is established by activation and inferior bias by repression with a net outcome of opposite gradients that contribute to cone subtype distinctions.
Fig. 5.
TRβ2-mediated gradient formation. (A) Dot plot of representative gradient genes with diminished gradients in KO cones; RNA-seq, 48–53 cones/group. TRβ2-independent gradient examples, below. (B) In situ hybridization of example genes. Sagittal cryosections; outer nuclear layer (ONL) of wild-type and TRβ2-KO mice at P28. (Scale bar applies to all panels.) Arrowheads, cone nuclei location. (C) Opsin gradient genes with TRβ2-regulated open chromatin (ATAC) sites (control vs. KO; FDR q < 0.05; fold changes: Opn1mw >60, Opn1sw = 1.8, Ccdc136 = 1.4). Red arrows; sites with TRβ2 binding; red outline arrow, site lacking detectable TRβ2 binding; IGV reads scale, numbers on Left. (D) TRβ2-dependent gene expression. Single cones ordered from lowest to highest expression; +, mean; **P < 0.01 control vs. KO (Student’s t test).
Representative genes in these gradient groups were further corroborated by in situ hybridization (Fig. 5B), qPCR, and immunostaining analyses (SI Appendix, Fig. S3) and by association with TRβ2 binding and/or regulated open chromatin (Fig. 5 C and D). As examples, Opn1mw displays major opening of chromatin at the promoter consistent with induction by TRβ2. In contrast, Opn1sw and Ccdc136 which reside in a tail-to-tail cluster ~18 kb apart display striking dual negative regulation whereby TRβ2 modestly decreases chromatin opening at upstream binding sites and represses expression of both genes, particularly in superior retinal regions.
Discussion
Cone Diversity Genes in a Mammal with Graded Cone Arrays.
Cone diversity illustrates the importance of subtle cellular heterogeneity in biological systems. Using a methodical, in-depth approach for individual cones, we identified groups of gradient genes in addition to opsins in cone subtypes and revealed that TRβ2 controls many of these genes (Fig. 6). The results highlight the role of fine transcriptional control within a neuronal lineage in generating diversity. Fine control of diversity has also been suggested for retinal amacrine cells, which form varied morphological subtypes (35).
Fig. 6.
TRβ2 and cone diversity. A simplified model in which TRβ2 controls gene expression gradients underlying cone diversity in mice. Gradient genes are also influenced by spatial location on the superior–inferior plane of the retina. Gradient emergence postnatally occurs during a maturation phase for many organs (including eye opening) that is prompted by rising levels of thyroid hormone (L-triodothyronine [T3], active form; thyroxine [T4], precursor of T3).
Most mammals, like mice, possess a dichromatic cone system based on M and S opsins, suggesting that other subtype genes may be conserved. Data are limited for other mammals, but CCDC136 is reported to be enriched in S cones in macaques (36) and humans (37). The OPN1SW-CCDC136 gene cluster is also conserved raising the possibility of dual control by TRβ2 as in mice. In mice, gradient gene categories include phototransduction (e.g., opsins, arrestins) and membrane functions (e.g, Kcne2, Chrnb4) suggesting contributions of gradient genes to wavelength-specific phototransduction responses or the relay of these responses to interneurons for specific visual pathways. For example, the modest enrichment of arrestins Arr3 and Sag in M- and S-dominant cones, respectively, might influence wavelength-specific phototransduction. SAG is also enriched in macaque S cones (36). However, in mice, the gradients are overlapping rather than exclusive and knockouts indicate that both arrestins quench responses to long or short wavelength stimuli (38). Thus, wavelength-specific functions might be better investigated in species with more segregated cone subtypes than mice. The “high-S” cone subgroup we detect may correlate with “pure-S” cones that connect with a subclass of cone bipolar cells forming a putative pathway for short wavelength information in mice (28). We speculate that conserved subtype genes may give insights into wavelength-specific functions, whereas nonconserved subtype genes might reflect retinal specializations within a species.
Cone diversity in mice reflects quantitative, graded expression differences over the retinal vertical plane. This graded array of cone subtypes differs from the mosaic pattern in other mammals such as humans but may be beneficial in a ground habitat, aiding chromatic (26, 39) and contrast discrimination viewed between the ground and sky (40, 41). The varying role of TRβ2 over this vertical plane suggests an interplay with spatial cues arising from broader retinal gradients, involving for example, retinoids (29), morphogens, transcription factors (42), or local availability of L-triiodothyronine (T3), the active form of thyroid hormone (13). How spatial cues signal to cones is unclear. Regarding T3 availability, variations within retinal regions might arise from complex mechanisms including differential transfer from the vasculature, selective uptake in cones by plasma membrane transporters or the amplification and depletion of T3 by deiodinase enzymes (13, 43). We suggest that the interplay between TRβ2 and spatial cues can adapt to variations in retinal anatomy. In mammals with mosaic cone arrays, equivalent spatial cues may be organized in the cellular community around each cone rather than as retinal-wide gradients.
Control of Cone Diversity by TRβ2.
Our findings suggest that TRβ2 promotes cone diversity from bipotential cone precursors (Fig. 6). Early fate-determining factors (44–46) commit multipotent progenitor cells to the cone lineage, then TRβ2, in conjunction with spatial cues, promotes graded gene expression resulting in M- or S-dominant cone outcomes (10). The transcriptional plasticity of cone precursors may be critical for cone diversity in mammals, as suggested by the reported developmental switch of S to M opsin immunoreactivity in rats (8) and a THRB-dependent switch from S to medium/long opsins in human retinal organoids (20) similar to that in mice (10). The extent of the similarities beyond opsin switches is unknown and awaits detailed analyses of cones in other mammals.
The transcriptional actions of TRβ2 in gradient formation are complex, resulting in graded activation or repression of multiple genes over the retina. We showed previously that ectopic expression of TRβ2 knocked into the rod gene Nrl imposed counter-gradients of both M and S opsins in cone-like, Nrl-deficient cells over the mouse retina (11), suggesting that TRβ2 integrates existing spatial cues to determine gradients. TRβ2-regulated chromatin sites (Fig. 4C and SI Appendix, Fig S4A) include consensus motifs for TRβ2 and CRX, an activator of many cone and rod photoreceptor genes (47, 48). Thus, cooperative enhancers may allow TRβ2 to impose graded expression while CRX or other activators stimulate expression levels. The developmental rise of thyroid hormone, augmented by local amplification of T3 levels by deiodinase enzymes within target tissues, prompts the maturation of many organs (43). The postnatal timing of cone gradient emergence in mice suggests that the generation of cone diversity is part of this wider transitional process that facilitates an independent, mature lifestyle.
A possible limitation of the study is that we derived a reference gradient gene list from a heterozygous Thrb+/b2cre model that might underestimate some gradients because of haploinsufficiency. However, we validated the screen by independent methods for selected genes in wild-type mice (Figs. 2C and 5B and SI Appendix, Fig. S3 B and C). Furthermore, the open chromatin pattern detected in Thrb+/b2cre control mice closely resembles that reported for wild-type mice using a different Cre driver (hRGP-Cre) (34) and differs from the pattern in TRβ2-KO mice (SI Appendix, Fig. S4C). It is possible that Cre itself, especially if overexpressed, influences phenotypes. The Thrbb2Cre driver is a knockin such that Cre expression would not exceed levels driven by the endogenous Thrb gene. As a control, we obtained evidence against a major impact of Cre on cone phenotypes by analysis of another driver line (Rora-ires-Cre, that keeps Rora intact) (49), which retained graded expression of opsins and other gradient genes (SI Appendix, Fig. S4B).
Materials and Methods
Gene Targeting and Genotyping.
Thrbb2cre mice were derived by homologous recombination in W9.5 embryonic stem (ES) cells using standard methods (10). The construct carried 4.0 kb 5′-homology and 4.4 kb 3′-homology arms, a Cre cassette replacing TRβ2 coding sequences and a self-excising neomycin resistance gene (a loxP-flanked dual cassette tACE-Cre/Neor, abbreviated as ACN) (50). Targeting was confirmed by Southern blot and long template PCR analysis. Founder mice were backcrossed with C57BL/6J mice. Routine genotyping was performed by 3-primer PCR: b2R 5′-GGC CAT GTC CAA GTC AGA GT-3′; b2F 5′-CAT TTT CCC CAG CAA TAA GC-3′; creR 5′-AGG CAA ATT TTG GTG TAC GG-3′, giving band sizes: wt 552 bp, b2cre allele 393 bp. Conditions: 95°C, 2 min; then 36 cycles of 95°C, 30 s; 56°C, 30 s; 72°C, 30 s; end with 72°C, 5 min (SI Appendix, Fig. S1). To isolate cones, Thrb+/b2Cre mice (C57BL/6J × 129/Sv background) were crossed with Rosa26+/Ai6 mice (JAX #007906; 129S6/SvEvTac x C57BL/6NCrl background) (24). Rora-ires-Cre (49) mice were used for a Cre control and NrlpGfp mice for rod isolation (51). For ATAC-seq, Thrb+/b2Cre mice were crossed with Rosa26+/Sun1Gfp mice (34). To biotinylate receptors, Thrb+/HAB mice expressing receptors with a C-terminal HAB peptide tag (52) were crossed with Rosa26+/BirA mice expressing BirA ligase (JAX #010920; 129P2/OlaHSD x C57BL/6J x FVB/N background) (53). The HAB tag includes a hemagglutinin epitope and a biotinylation site. Procedures followed approved protocols at the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) at the National Institutes of Health.
Quantitative Reverse Transcriptase-PCR (qPCR).
Triplicate total RNA samples were prepared using Trizol (Life Technologies) and RNeasy Micro kit (Qiagen). Samples represented 4–6 retinas of ≥3 wild-type mice (male C57BL/6J) (Fig. 2) or wild-type and TRβ2-KO groups (10) (SI Appendix, Fig. S3B). The superscript III Reverse Transcriptase kit (Life Technologies) and oligo-dT primers were used for complementary DNA (cDNA) synthesis and then 1 μg of cDNA was analyzed by qPCR with FastStart Universal SYBR Green Master-Mix (Millipore-Sigma/Roche; Cat #04913914001) and normalization to β-actin levels. SI Appendix, Table S1 shows primer pairs for genes.
In Situ Hybridization and Immunostaining.
Eyes were fixed in 2% paraformaldehyde, and 10 μm cryosections were analyzed as described (11). In situ hybridization with digoxigenin-labeled riboprobes followed described methods (11). Probes were generated by PCR (SI Appendix, Table S1), cloned into pCDNA3.1, and sequenced to confirm identity. Antibodies are listed in SI Appendix, Table S2.
Single-Cell Isolation and RNA-Sequencing.
Fluorescent cells were isolated from ~3-mm2 dissociated pieces of superior or inferior retina of control (Thrb+/b2Cre) and KO (Thrbb2Cre/b2Cre) mice on a Rosa26Ai6/Ai6 background with males and females equally represented. Briefly, following described procedures (54), each cell was collected promptly using a micromanipulator and a Zeiss Axiovert 200M microscope and assessed visually for integrity and cone morphology. Cells were lysed and cDNA prepared immediately using SMARTer UltraLow Input RNA Kit-V4 (Takara #634890). Size and concentration of cDNA were determined using an Agilent kit and 2100 Bioanalyzer (Agilent Technologies, #5067–4626). A water bath bioruptor (Diagenode UCD-200) was used to shear cDNA into 200–500-bp fragments, then ~0.5 µg used to make libraries with a SMARTer ThruPLEX DNA-Seq Kit (Takara # R400676 for adult cones; #R400427 for developmental series). Libraries were quantified using a dsDNA Assay kit and Qubit 2.0 fluorimeter (Invitrogen Life Technologies, #Q32854).
RNA-Seq Analysis.
Control (Thrb+/b2Cre) and TRβ2-KO (Thrbb2Cre/b2Cre) adult groups contained 48–53 cones. Developmental groups (P2–P60) of control (Thrb+/b2Cre) mice contained 21–30 cones. Groups represented ≥3 mice. Rod groups (NrlpGfp adults) contained 29 cells.
Libraries were sequenced on an Illumina HiSeq-2500 apparatus at the NIDDK Genomics Facility. Ten–20 million single-end 50 base reads/per library were collected, converted by bcl2fastq (version 2) into fastq files and aligned on (GRCm38/mm10) with STAR (version 2.7.3a) (55). Output bam files were analyzed as counts per million mapped reads (cpm). Quality control: Libraries represented: adult groups on average ~13 million reads/cell (range 5.6–30.6 million) and ~15,000 genes/cell; developmental groups ~3.3 million reads/cell (range 1.1–6.4 million). Libraries were edited to remove reads for pseudogenes, Y-linked genes, mitochondrial-encoded genes (56) and artifactual reads for Calu and Tex28, which lie on the opposite strands of Opn1sw and Opn1mw, respectively. Calu reads were confirmed as artifactual by in situ hybridization with single-strand riboprobes.
ChAP-Seq and ATAC-Seq Analyses.
For ChAP-seq, pooled retinas from 20–30 ThrbHAB/HAB;Rosa26BirA/BirA (HAB;BirA) and control Rosa26BirA/BirA (BirA) E18 embryos were divided into two replicates for cross-linking and affinity-purification with streptavidin. For ATAC-seq, cone nuclei were immunopurified from two replicates of pooled retinas from ≥5 control (Thrb+/b2Cre) and TRβ2-KO (Thrbb2Cre/b2Cre) mice on a Rosa26Sun1Gfp/Sun1Gfp background, then ~40,000 nuclei analyzed using a Tagment DNA kit (Illumina 20034197) (34).
Dataset Analyses.
RNA-seq.
Gene ontology was analyzed with whole-genome background reference and selected terms with P < 0.05 per category using DAVID (Database for Annotation, Visualization, and Integrated Discovery)(https://david.ncifcrf.gov/home.jsp). To identify TRβ2-dependent genes, the top 6,469 genes in control and KO cones combined (201 cells, including superior and inferior groups) were analyzed using Student's t test (P < 0.01). Most analyses represented genes with average expression >20 cpm. For tSNE plots, the top 3,000 expressed genes in single cones were analyzed (Rtsne: T-SNE, https://github.com/jkrijthe/Rtsne) with cluster analyses using Rtsne (initial dims = 20, iterations = 3,000) (version 0.15) (57).
ChAP-seq and ATAC-seq.
ChAP-seq peaks and ATAC-seq peaks were called using MACS2 (v2.2.7.1) with false discovery rate (FDR) q < 0.01. Specific TRβ2-HAB peaks were identified by differential analysis of HAB, BirA, and BirA samples using EdgeR (P <0.05; cutoff >twofold). TRβ2-regulated ATAC peaks were identified by differential analysis of control and KO samples using DESeq2 (q < 0.05; fold-change >1.5, except for Ccdc136, >1.4). Motifs were identified using Homer (v4.11) findMotifsGenome.pl function. Peaks were assigned to distal or TSS locations using Homer annotatePeaks.pl function. Libraries were normalized to reads per genomic coverage using deepTools bamCoverage, and peaks visualized on gene maps by Integrative Genomic Viewer (v2.8.12). Peak-to-gene distances were plotted using Bedtools v2.29.2 subcommand and NCBI Refseq genes (~25,000) as control.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Mario Cappechi for pACN plasmid, Colin Stewart for ES cells, Anand Swaroop for NrlpGfp mice, and Nicola Allen and Dennis O'Leary for Rora-Cre mice. We thank Kevin Kelley, Mount Sinai School of Medicine Genetics Core, and Harold Smith, NIDDK Genomics Core, for assistance and Wei Li and Anand Swaroop for discussion. Supported by the intramural research program at NIDDK at the National Institutes of Health.
Author contributions
M.A., X.W., H.L., and D.F. designed research; M.A., X.W., H.L., Y.L., Y.-W.C., M.S., Y.F., and L.N. performed research; M.A., X.W., H.L., Y.L., Y.-W.C., M.S., Y.F., L.N., and D.F. analyzed data; and M.A., H.L., and D.F. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. R.J. is a guest editor invited by the Editorial Board.
Data, Materials, and Software Availability
All data and materials are available as described in the manuscript or SI Appendix. Genomic datasets generated in this work are accessible at GEO, #GSE203481.
Supporting Information
References
- 1.Nathans J., The evolution and physiology of human color vision: Insights from molecular genetic studies of visual pigments. Neuron 24, 299–312 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Hunt D. M., Peichl L., S cones: Evolution, retinal distribution, development, and spectral sensitivity. Vis. Neurosci. 31, 115–138 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Rister J., Desplan C., The retinal mosaics of opsin expression in invertebrates and vertebrates. Dev. Neurobiol. 71, 1212–1226 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roorda A., Williams D. R., The arrangement of the three cone classes in the living human eye. Nature 397, 520–522 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Xiao M., Hendrickson A., Spatial and temporal expression of short, long/medium, or both opsins in human fetal cones. J. Comp. Neurol. 425, 545–559 (2000). [PubMed] [Google Scholar]
- 6.Carter-Dawson L. D., LaVail M. M., Rods and cones in the mouse retina. II. Autoradiographic analysis of cell generation using tritiated thymidine. J. Comp. Neurol. 188, 263–272 (1979). [DOI] [PubMed] [Google Scholar]
- 7.Cepko C. L., Austin C. P., Yang X., Alexiades M., Ezzeddine D., Cell fate determination in the vertebrate retina. Proc. Natl. Acad. Sci. U.S.A. 93, 589–595 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szel A., van Veen T., Rohlich P., Retinal cone differentiation. Nature 370, 336 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Sjoberg M., Vennstrom B., Forrest D., Thyroid hormone receptors in chick retinal development: Differential expression of mRNAs for alpha and N-terminal variant beta receptors. Development 114, 39–47 (1992). [DOI] [PubMed] [Google Scholar]
- 10.Ng L., et al. , A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat. Genet. 27, 94–98 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Ng L., et al. , Two transcription factors can direct three photoreceptor outcomes from rod precursor cells in mouse retinal development. J. Neurosci. 31, 11118–11125 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pessoa C. N., et al. , Thyroid hormone action is required for normal cone opsin expression during mouse retinal development. Invest. Ophthalmol. Vis. Sci. 49, 2039–2045 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Roberts M. R., Srinivas M., Forrest D., Morreale de Escobar G., Reh T. A., Making the gradient: Thyroid hormone regulates cone opsin expression in the developing mouse retina. Proc. Natl. Acad. Sci. U.S.A. 103, 6218–6223 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaschke A., et al. , Thyroid hormone controls cone opsin expression in the retina of adult rodents. J. Neurosci. 31, 4844–4851 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyes W. K., Degn L., George B. J., Gilbert M. E., Moderate perinatal thyroid hormone insufficiency alters visual system function in adult rats. Neurotoxicology 67, 73–83 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki S. C., et al. , Cone photoreceptor types in zebrafish are generated by symmetric terminal divisions of dedicated precursors. Proc. Natl. Acad. Sci. U.S.A. 110, 15109–15114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackin R. D., et al. , Endocrine regulation of multichromatic color vision. Proc. Natl. Acad. Sci. U.S.A. 116, 16882–16891 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deveau C., et al. , Thyroid hormone receptor beta mutations alter photoreceptor development and function in Danio rerio (zebrafish). PLoS Genet. 16, e1008869 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DuVal M. G., Allison W. T., Photoreceptor progenitors depend upon coordination of gdf6a, thrbeta, and tbx2b to generate precise populations of cone photoreceptor subtypes. Invest. Ophthalmol. Vis. Sci. 59, 6089–6101 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Eldred K. C., et al. , Thyroid hormone signaling specifies cone subtypes in human retinal organoids. Science 362, eaau6348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newell F. W., Diddie K. R., Typical monochromacy, congenital deafness, and resistance to intracellular action of thyroid hormone (author’s transl). Klin. Monbl. Augenheilkd. 171, 731–734 (1977). [PubMed] [Google Scholar]
- 22.Campi I., et al. , Retinal photoreceptor functions are compromised in patients with resistance to thyroid hormone syndrome (RTHbeta). J. Clin. Endocrinol. Metab. 102, 2620–2627 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Weiss A. H., Kelly J. P., Bisset D., Deeb S. S., Reduced L- and M- and increased S-cone functions in an infant with thyroid hormone resistance due to mutations in the THRbeta2 gene. Ophthalmic Genet. 33, 187–195 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Madisen L., et al. , A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng L., Ma M., Curran T., Forrest D., Developmental expression of thyroid hormone receptor β2 protein in cone photoreceptors in the mouse. Neuroreport 20, 627–631 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikonov S. S., Kholodenko R., Lem J., Pugh E. N. Jr., Physiological features of the S- and M-cone photoreceptors of wild-type mice from single-cell recordings. J. Gen. Physiol. 127, 359–374 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haverkamp S., et al. , The primordial, blue-cone color system of the mouse retina. J. Neurosci. 25, 5438–5445 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nadal-Nicolas F. M., et al. , True S-cones are concentrated in the ventral mouse retina and wired for color detection in the upper visual field. eLife 9, e56840 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCaffery P., Wagner E., O’Neil J., Petkovich M., Drager U. C., Dorsal and ventral retinal territories defined by retinoic acid synthesis, break-down and nuclear receptor expression. Mech. Dev. 82, 119–130 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Smiley S., et al. , Establishment of a cone photoreceptor transplantation platform based on a novel cone-GFP reporter mouse line. Sci. Rep. 6, 22867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corbo J. C., Myers C. A., Lawrence K. A., Jadhav A. P., Cepko C. L., A typology of photoreceptor gene expression patterns in the mouse. Proc. Natl. Acad. Sci. U.S.A. 104, 12069–12074 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Umesono K., Murakami K. K., Thompson C. C., Evans R. M., Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell 65, 1255–1266 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calo E., Wysocka J., Modification of enhancer chromatin: What, how, and why? Mol. Cell 49, 825–837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mo A., et al. , Epigenomic landscapes of retinal rods and cones. eLife 5, e11613 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng Y. R., et al. , Binary fate choice between closely related interneuronal types is determined by a fezf1-dependent postmitotic transcriptional switch. Neuron 105, 464–474.e466 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng Y. R., et al. , Molecular classification and comparative taxonomics of foveal and peripheral cells in primate retina. Cell 176, 1222–1237.e1222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukowski S. W., et al. , A single-cell transcriptome atlas of the adult human retina. EMBO J. 38, e100811 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikonov S. S., et al. , Mouse cones require an arrestin for normal inactivation of phototransduction. Neuron 59, 462–474 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calderone J. B., Jacobs G. H., Regional variations in the relative sensitivity to UV light in the mouse retina. Vis. Neurosci. 12, 463–468 (1995). [DOI] [PubMed] [Google Scholar]
- 40.Baden T., et al. , A tale of two retinal domains: Near-optimal sampling of achromatic contrasts in natural scenes through asymmetric photoreceptor distribution. Neuron 80, 1206–1217 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Qiu Y., et al. , Natural environment statistics in the upper and lower visual field are reflected in mouse retinal specializations. Curr. Biol. 31, 3233–3247.e3236 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Mui S. H., Kim J. W., Lemke G., Bertuzzi S., Vax genes ventralize the embryonic eye. Genes. Dev. 19, 1249–1259 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hernandez A., Martinez M. E., Ng L., Forrest D., Thyroid hormone deiodinases: Dynamic switches in developmental transitions. Endocrinology 162, bqab091 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishida A., et al. , Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat. Neurosci. 6, 1255–1263 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Sapkota D., et al. , Onecut1 and Onecut2 redundantly regulate early retinal cell fates during development. Proc. Natl. Acad. Sci. U.S.A. 111, E4086–E4095 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brzezinski J. A., Reh T. A., Photoreceptor cell fate specification in vertebrates. Development 142, 3263–3273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen S., et al. , Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 19, 1017–1030 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Furukawa T., Morrow E. M., Li T., Davis F. C., Cepko C. L., Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat. Genet. 23, 466–470 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Chou S. J., et al. , Geniculocortical input drives genetic distinctions between primary and higher-order visual areas. Science 340, 1239–1242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bunting M., Bernstein K. E., Greer J. M., Capecchi M. R., Thomas K. R., Targeting genes for self-excision in the germ line. Genes. Dev. 13, 1524–1528 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akimoto M., et al. , Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc. Natl. Acad. Sci. U.S.A. 103, 3890–3895 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cho Y., Huang C., Liu H., Fu Y., Forrest D., Transcriptional and genomic regulation of pituitary function by thyroid hormone receptor beta. J. Endocr. Soc. 5, A980–A981 (2021). [Google Scholar]
- 53.Driegen S., et al. , A generic tool for biotinylation of tagged proteins in transgenic mice. Transgenic Res. 14, 477–482 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Ng L., Liu Y., Liu H., Forrest D., Cochlear fibrocyte and osteoblast lineages expressing type 2 deiodinase identified with a Dio2CreERt2 Allele. Endocrinology 162, bqab179 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bult C. J., et al. , Mouse genome database (MGD) 2019. Nucleic Acids Res. 47, D801–D806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Maaten L., Hinton G., Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All data and materials are available as described in the manuscript or SI Appendix. Genomic datasets generated in this work are accessible at GEO, #GSE203481.