Abstract
The establishment of experimental conditions for transcriptional regulator network (TRN) reconstruction in bacteria continues to be impeded by the limited knowledge of activating conditions for transcription factors (TFs). Here, we present a novel genome-scale model-driven workflow for designing experimental conditions, which optimally activate specific TFs. Our model-driven workflow was applied to elucidate transcriptional regulation under nitrogen limitation by Nac and NtrC, in Escherichia coli. We comprehensively predict alternative nitrogen sources, including cytosine and cytidine, which trigger differential activation of Nac using a model-driven workflow. In accordance with the prediction, genome-wide measurements with ChIP-exo and RNA-seq were performed. Integrative data analysis reveals that the Nac and NtrC regulons consist of 97 and 43 genes under alternative nitrogen conditions, respectively. Functional analysis of Nac at the transcriptional level showed that Nac directly down-regulates amino acid biosynthesis and restores expression of tricarboxylic acid (TCA) cycle genes to alleviate nitrogen-limiting stress. We also demonstrate that both TFs coherently modulate α-ketoglutarate accumulation stress due to nitrogen limitation by co-activating amino acid and diamine degradation pathways. A systems-biology approach provided a detailed and quantitative understanding of both TF’s roles and how nitrogen and carbon metabolic networks respond complementarily to nitrogen-limiting stress.
INTRODUCTION
The transcriptional regulatory network (TRN) in bacteria is sophisticatedly composed, responding to diverse environmental stimuli. Comprehensively revealing the TRN is indispensable in understanding metabolic flexibility and robustness in response to various environmental changes (1). Elucidation of the TRN at the systems level starts with the integration of omics datasets, such as transcription factor (TF) binding sites and expression profiling (2–8). The typical approach to designing a TF profiling experiment is determining the relevant growth conditions where a TF of interest is expected to be maximally active. Therefore, the establishment of an appropriate experimental environment is essential in describing and understanding a clear regulatory mechanism of the TRN. However, knowledge about activating conditions for a number of TFs is limited, which hinders the establishment of optimal experimental conditions for these TFs. This remains a challenging biological problem. Therefore, choosing experimental conditions based on the literature could limit exploration of alternative conditions not mentioned in prior studies that might stimulate the activity of TFs. Some of these conditions might be better than previously reported experimental conditions.
Genome-scale metabolic models generated by reconstructions of metabolic networks are a key component of systems biology, providing a mathematical representation and displaying metabolic capabilities. Metabolic network models (M-models) have made significant progress in predicting cellular metabolism in organisms of interest by integrating all metabolic reactions and experimental information (9–13). These metabolic models were used to simulate the maximal growth of a cell with constraints using flux-balance analysis (FBA), resulting in a valuable understanding of optimal metabolic flux states (14,15). Furthermore, macromolecular expression models (ME-models), which include an integrated network of metabolic and gene product expression pathways, were constructed from M-models to compute gene expression and translation levels (16,17). Extended ME-models were used to predict gene expression, resulting in the interpretation of iron metabolism and acid stress responses in Escherichia coli (6,7,18). Recently, the development of M-models (19,20) and ME-models (17,21) for E. coli has enabled in silico exploration of virtually every imaginable experimental condition. Comparisons of different network states computed under different candidate experimental conditions can shed light on various cellular responses to environmental changes, thus identifying regulatory requirements for each condition. In this study, a model-based prediction workflow for TF activation was constructed, which combines a ME-model with existing regulon information from public databases. It can predict transcriptional changes to present a cellular response on the transcriptional level against an environmental stimulus and assess gene enrichment in the experimental conditions in silico using regulon information. Thus, this workflow can predict conditions where a TF could be activated. While the conventional, non-systems approach can suggest experimental conditions from limited information, the extended, model-driven approach is able to generate multiple promising candidate conditions through in silico simulation, which can also render possible differential activation of TFs.
Since nitrogen metabolism is one of the key components in E. coli metabolism, post-translational regulation of the Ntr system has been extensively investigated through a number of studies (22–29). In E. coli, NtrC and Nac are known to play important roles in transcriptional regulation under nitrogen limitation. It is well known that NtrC responds to nitrogen-limitation stress primarily by regulating a small subset of genes involved in nitrogen metabolism and strongly activates Nac to expand transcriptional regulation (30,31). Therefore, in this study, NtrC was used as a standard indicator for measuring nitrogen availability in E. coli. In case of Nac, previous studies have also shown that it is capable of regulating a large number of genes in various metabolic pathways, including nitrogen metabolism (30,32–34). However, limited information on the genome-wide regulatory role of Nac in response to nitrogen limitation is available (30,35,36). Previous studies used binding profiling experiments with low resolution, such as ChIP-chip and ChIP-seq, or different induction methods for TF activation, such as inducible Flag-tagging (30,32)
In this study, a model-based workflow was applied to predict optimal conditions for activating these two TFs, and the differential activation of the TFs under nitrogen-limiting conditions was investigated. Subsequently, ChIP-exo (Chromatin immunoprecipitation with exonuclease treatment) using λ exonucleases was used to identify NtrC and Nac in vivo binding sites with near-base pair resolution (37). In addition, transcription profiling (RNA-seq) on different nitrogen sources was performed with E. coli K-12 MG1655 wild-type and knockout mutants (ΔntrC and Δnac) to combine TF-binding profiles, revealing causal transcriptional regulatory relationships of both TFs. Furthermore, integration of experimentally derived data and model-derived in silico flux calculations elucidated the distinct roles of both regulons on the genome-scale level.
MATERIALS AND METHODS
Prediction of activation conditions for transcription factor
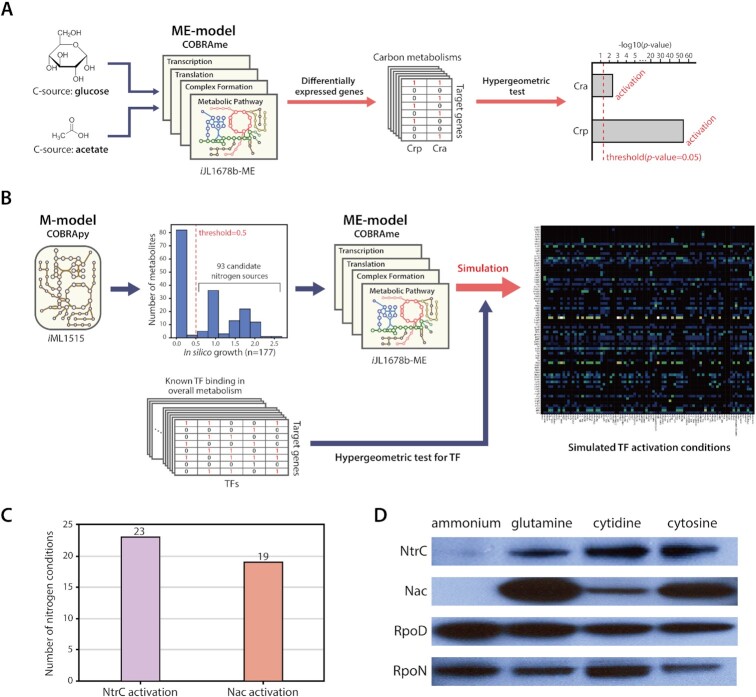
In order to simulate ME-models (17) for in silico E. coli growth under various conditions, candidate nitrogen sources were chosen from simulation with M-models (19) that share the same metabolites with the ME model. There are 1071 cytoplasmic metabolites in iML1515 M-model Among them, 628 cytoplasmic metabolites have more than one nitrogen molecule (Supplementary Figure S1A). From those metabolites, 177 nitrogen-containing cytoplasmic metabolites with exchange reactions in the model were preserved. With glucose as the sole carbon source with a −10 mmol/gDCW/h uptake rate, and each nitrogen-containing metabolites with exchange reactions with a −10 mmol/gDCW/h uptake rate, M-models were used in simulation to decide if the nitrogen-containing molecule supported in silico growth. From the set of in silico growth with 177 nitrogen-containing metabolites, a threshold of biomass objective function (0.5 mmol/gDCW/h) was used to decide growth and non-growth, which resulted in 93 candidate nitrogen sources, which support in silico growth with glucose.
The ME-model was used to simulate expression of genes under 93 candidate nitrogen sources with −10 mmol/gDCW/h and glucose as the carbon source with −10 mmol/gDCW/h (Supplementary Figure S1B). As a result of the ME-model simulation, the feasible solutions were obtained from 90 candidate nitrogen sources. For each simulation under an alternate nitrogen source, expression was compared to simulated growth on ammonium, with a cutoff of a factor of 2 set to identify predicted sets of genes. The hypergeometric enrichment of each known TF regulon taken from Ecocyc (38) in each of these differential gene sets was used to predict the likelihood of transcription factor activity under each condition. For NtrC and Nac, 23 conditions and 19 conditions respectively resulted in predictions of TF activity (Supplementary Figure S1C).
Bacterial strains, media, and growth conditions
All strains used in this study were E. coli K-12 MG1655 and its derivatives, including knock-out strains and a myc tagging strain. For ChIP-exo experiments, E. coli strains harboring NtrC-8myc and Nac-8myc were generated as described previously (39). For expression profiling by RNA-seq, deletion mutants ΔntrC and Δnac were constructed by a λ red-mediated, site-specific recombination system (40). M9 minimal media (41) was used for ammonia and W2 minimal media (42) was used for nitrogen-limiting conditions. For nitrogen-limiting conditions, 0.2% (w/v) glutamine, cytidine, or cytosine was added for alternative nitrogen sources. M9 or W2 minimal media was supplemented with 0.2% glucose (w/v) and 1 ml trace element solution (100X) containing 1 g EDTA, 29 mg ZnSO4.7H2O, 198 mg MnCl2.4H2O, 254 mg CoCl2.6H2O, 13.4 mg CuCl2 and 147 mg CaCl2. Glycerol stocks of E. coli strains were inoculated into M9 or W2 minimal media and cultured overnight at 37°C with constant agitation. Cultures were then diluted 1:100 into 50 mL of fresh minimal media and cultured at 37°C to mid-log phase (OD600 ≈ 0.5 for ammonia, glutamine, and cytidine, OD600 ≈ 0.25 for cytosine) before harvest.
ChIP-exo experiment
To identify TF and σ-factor binding maps in vivo, we isolated the DNA bound to NtrC, Nac or RpoN and RpoD from formaldehyde cross-linked E. coli cells by chromatin immunoprecipitation (ChIP) with the antibodies that specifically recognize myc tag (9E10, Santa Cruz Biotechnology) or RpoD (2G10, Neoclone), and RpoN (6RN3, Neoclone) subunits of RNA polymerase complex, respectively, and Dynabeads Pan Mouse IgG magnetic beads (Invitrogen) followed by stringent washings as described previously (43). ChIP materials (chromatin-beads) were used to perform on-bead enzymatic reactions detailed in the ChIP-exo method (37) with following modifications. Briefly, the sheared DNA of chromatin-beads was repaired using the NEBNext End Repair Module (New England Biolabs) followed by the addition of a single dA overhang and ligation of the first adaptor (5’-phosphorylated) using the dA-Tailing Module (New England Biolabs) and NEBNext Quick Ligation Module (New England Biolabs), respectively. Nick repair was performed using PreCR Repair Mix (New England Biolabs). Lambda exonuclease- and RecJf exonuclease-treated chromatin was eluted from the beads and the protein–DNA cross-link was reversed by overnight incubation at 65°C. RNA- and proteins-removed DNA samples were used to perform primer extension and second adaptor ligation with the following modifications. The DNA samples incubated for primer extension as described previously (37) were treated with the dA-Tailing Module (New England Biolabs) and NEBNext Quick Ligation Module (New England Biolabs) for second adaptor ligation. The DNA sample purified by GeneRead Size Selection Kit (Qiagen) was enriched by polymerase chain reaction (PCR) using Phusion High-Fidelity DNA Polymerase (New England Biolabs). The amplified DNA samples were purified again by GeneRead Size Selection Kit (Qiagen) and quantified using Qubit dsDNA HS Assay Kit (Life Technologies). Quality of the DNA sample was checked by running Agilent High Sensitivity DNA Kit using Agilent 2100 Bioanalyzer (Agilent) before being sequenced using MiSeq or NextSeq550 (Illumina) in accordance with the manufacturer's instructions. Each modified step was also performed in accordance with the manufacturer's instructions. ChIP-exo experiments were performed in biological duplicate.
RNA-seq expression profiling
Three milliliters of cells from mid-log phase culture were mixed with six ml RNAprotect Bacteria Reagent (Qiagen). Samples were mixed immediately by vortexing for 5 s, incubated for 5 min at room temperature, and then centrifuged at 5000 × g for 10 min. The supernatant was decanted, and any residual supernatant was removed by inverting the tube once onto a paper towel. Total RNA samples were then isolated using RNeasy Plus Mini kit (Qiagen) in accordance with the manufacturer's instructions. Samples were then quantified using a NanoDrop 1000 spectrophotometer (Thermo Scientific) and quality of the isolated RNA was checked by running an RNA 6000 Pico Kit using Agilent 2100 Bioanalyzer (Agilent).
Paired-end, strand-specific RNA-seq was performed using the dUTP method (44) with the following modifications. The ribosomal RNAs were removed from 2 μg of isolated total RNA with Ribo-Zero rRNA Removal Kit (Epicentre) in accordance with the manufacturer's instructions. Subtracted RNA was fragmented for 2.5 min at 70°C with RNA Fragmentation Reagents (Ambion), and then fragmented RNA was recovered with ethanol precipitation. Random primer (3 μg) and fragmented RNA in 4 μl was incubated in 5 μl total volume at 70°C for 10 min, and cDNA or the first strand was synthesized using SuperScript III first-strand synthesis protocol (Invitrogen). The cDNA was recovered by phenol–chloroform extraction followed by ethanol precipitation. The second strand was synthesized from this cDNA with 20 μl of fragmented cDNA:RNA, 4 μl of 5× first strand buffer, 30 μl of 5× second strand buffer, 4 μl of 10 mM dNTP with dUTP instead of dTTP, 2 μl of 100 mM DTT, 4 μl of E. coli DNA polymerase (Invitrogen), 1 μl of E. coli DNA ligase (Invitrogen), 1 μl of E. coli RNase H (Invitrogen) in 150 μl of total volume. This reaction mixture was incubated at 16 °C for 2 h, and fragmented DNA was recovered with PCR clean-up kit (QIAGEN) and eluted in 30 μl of nuclease-free water. The fragmented DNA was end-repaired with End Repair Kit (New England Biolabs), and dA-tailed with dA-Tailing Kit (New England Biolabs), and then ligated with 7.5 μg of DNA adaptor mixture with Quick Ligation Kit (New England Biolabs). The adaptor-ligated DNA was size-selected to remove un-ligated adaptors with GeneRead Size Selection Kit (QIAGEN). It was treated with 1 U of USER enzyme (New England Biolabs) in 30 μl of total volume, and incubated at 37°C for 15 min followed by 5 min at 95°C. The USER-treated DNA was amplified by PCR to generate sequencing libraries for Illumina sequencing. The samples were sequenced using MiSeq or NextSeq550 (Illumina) in accordance with the manufacturer's instructions. All RNA-seq experiments were performed in biological duplicate.
Peak calling for ChIP-exo dataset
Sequence reads generated from ChIP-exo were mapped onto the reference genome (NC_000913.2) using bowtie (45) with default options to generate SAM output files. The MACE program (https://code.google.com/p/chip-exo/) was used to define peak candidates from biological duplicates for each experimental condition with sequence depth normalization. To reduce false-positive peaks, peaks with signal-to-noise (S/N) ratio <1.5 were removed. The noise level was set to the top 5% of signals at genomic positions because the top 5% make a background level in plateau and the top 5% intensities from each ChIP-exo replicate across conditions correlate well with the total number of reads. The calculation of S/N ratio resembles the way ChIP-chip peak intensity is calculated, where IP signal is divided by Mock signal. Then, each peak was assigned to the nearest gene. Genome-scale data were visualized using MetaScope (https://sites.google.com/view/systemskimlab/software).
Classification of regulatory and non-regulatory binding sites
For binding sites of NtrC, Nac, RpoN and RpoD, a binding site was categorized as regulatory if it was located within 300 bp upstream of a target gene, and as non-regulatory if not. Non-regulatory regions cover intragenic regions without downstream genes and intergenic regions without a promoter nearby.
Motif search from ChIP-exo peaks
Sequence motif analysis for TFs and σ-factors was performed using the MEME software suite (46). For NtrC, Nac and RpoN, sequences in binding regions were extracted from the reference sequence (NC_000913.2). For RpoD, sequences were extended by 20 bp away from target genes, because only −10 box was found without that extension. MEME was run for regulatory bindings for all conditions, regulatory bindings for at least one condition, and non-regulatory bindings of NtrC, Nac, RpoN and RpoD.
Calculation of differentially expressed gene
Sequence reads generated from RNA-seq were mapped onto the reference genome (NC_000913.2) using bowtie (45) with the maximum insert size of 1000 bp, and 2 maximum mismatches after trimming 3 bp at 3’ ends. SAM files were generated from bowtie mapping. To compare expression changes according to nitrogen sources or genetic deletion mutants, DESeq2 was used to calculate transcripts per million (TPM) value and differential expression (47). From DESeq2 output, genes with differential expression with log2 fold change ≥1.0 and q-value ≤0.05 were considered differentially expressed genes. Genome-scale data were visualized using MetaScope (https://sites.google.com/view/systemskimlab/software).
Clusters of orthologous groups (COG) analysis
NtrC and Nac regulons were categorized according to their annotated clusters of orthologous groups (COG) category. Functional enrichment of COG categories in NtrC and Nac target genes was determined by performing a hypergeometric test, and a P-value <0.05 was considered significant.
Measuring growth rate and extracellular metabolites on different nitrogen sources
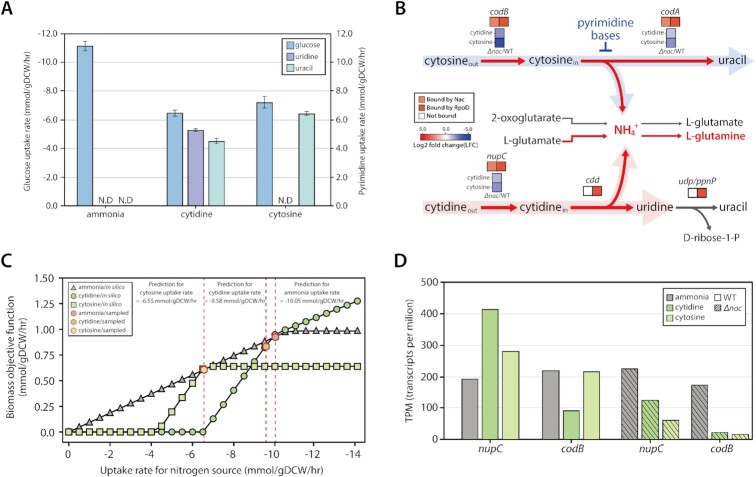
M9 minimal media (41) was used for ammonia and W2 minimal media (42) was used for glutamine, cytidine, and cytosine as nitrogen-limiting conditions. A glycerol stock of E. coli K-12 MG1655 strain was inoculated into fresh M9 or W2 minimal media and cultured overnight at 37°C with constant agitation. Cultures were then diluted 1:200 into 100 ml of fresh minimal media and cultured at 37°C to late-log phase, having been sampled six or seven times during early to mid-log phase. Optical density at OD600 was measured to get growth rates using different nitrogen sources. For each time point, extracellular metabolites were measured by HPLC using a 300 mm × 7.8 mm Aminex HPX-87H (Bio-Rad, USA) column at 35°C with 5.41 mM H2SO4 as the mobile phase. Growth rate and extracellular metabolite measurement was performed in biological triplicates. Measured glucose uptake rates were −11.18, −6.48 and −7.25 mmol/gDCW/hr on ammonia, cytidine and cytosine, respectively. The measured uridine export rate for cytidine was 5.49 mmol/gDCW/hr, and uracil export rate for cytidine and cytosine were 4.94 and 6.44 mmol/gDCW/h, respectively.
FBA analysis and MCMC sampling to calculate the metabolic flux
FBA analysis and MCMC sampling were performed with the iML1515 E. coli metabolic model (19) and COBRApy (48). For a parameter of nitrogen uptake rate, uptake rate for the glucose and export rate for pyrimidine measured by HPLC were used to constrain the model. Glucose uptake rate (−11.18 mmol/gDCW/h) was used to simulate ammonia condition. Glucose uptake rate (−6.48 mmol/gDCW/h), uridine export rate (5.49 mmol/gDCW/h), and uracil export rate (4.94 mmol/gDCW/h) were used for constrains on cytidine condition. In cytosine condition, glucose uptake rate (−7.25 mmol/gDCW/h) and uracil export rate (6.44 mmol/gDCW/h) were used. An unspecified uptake rate with a lower bound of −15 mmol/gDCW/hr was used for ammonia, cytidine and cytosine, because they were all used less after optimization and this parameter calculated close growth rates to measured rates. For cytidine, the uptake rate was set to −9.58 mmol/gDCW/h to match the in vivo growth rate with the measured one, otherwise the model chose to uptake all cytidine available, generating much higher, unrealistic growth rates. This is because cytidine can be utilized as a carbon and energy source. In addition, the distribution of feasible fluxes for each reaction in the iML1515 model was determined through MCMC sampling using experimentally measured and computationally predicted constrains (Table 1) (49). The biomass objective function was provided a lower bound of 95% of the optimal growth rate as computed by FBA. Thus, the sample flux distributions by MCMC sampling method represented sub-optimal flux distributions. MCMC sampling was used to obtain 10 000 feasible flux distributions, and the average of flux samples for each reaction was used.
Table 1.
Measured extracellular metabolites profiling and predicted uptake rates
| Ammonia | Cytidine | Cytosine | |
|---|---|---|---|
| Measured uptake and export rates (mmol/gDCW/h) | |||
| Glucose | −11.18 | −6.48 | −7.25 |
| Uridine | N/A | 5.29 | N/A |
| Uracil | N/A | 4.52 | 6.44 |
| Predicted nitrogen sources uptake rates (mmol/gDCW/h) | |||
| Nitrogen source | −10.05 | −9.58 | −6.55 |
Prediction of three-dimensional structure and interaction of proteins
The three-dimensional structure of each TF and sigma factor were predicted by AlphaFold, which was created by Deepmind (50). The AlphaFold-Multimer pipeline was used to predict the complex structure model of NtrC multimers or TF and sigma factor multimers (51). These amino acid sequences were obtained from EcoCyc. The monomer confidence was determined by the predicted Local Distance Difference Test (pLDDT). The complex structure confidence was calculated with predicted Template Modeling score (pTM score), and interface pTM (ipTM). The open-source molecular visualization program, PyMOL participated in implementation of structure models.
RESULTS
Model-driven prediction showed distinct activating pattern of NtrC and Nac by pyrimidine-based nitrogen sources
A model-driven elucidation workflow was conceived to predict experimental conditions and TF activation based on genome-scale metabolic models. This provides the ability to render optimal conditions that activate target TFs. It is also possible to predict which TFs are activated in a specific environment. By using ME-models, which are an extension of M-models, in a computational approach, gene expression values enable computation in growing E. coli (17). To demonstrate the applicability of ME-model to predict TF activation in E. coli, our previous experimental data for Cra and CRP were used to simulate and investigate its consistency with observable results (5). Based on simulations, when comparing glucose conditions with acetate conditions, differentially expressed genes (DEGs) in silico were significantly enriched in Cra and CRP regulon of the carbon metabolism (hypergeometric test P-value < 0.05). The results of the simulation stated the two TFs were activated in carbon metabolism under acetate conditions, which is consistent with actual experimental data (Figure 1A).
Figure 1.
Workflow of model-driven experimental design. (A) A model-driven workflow including macromolecular expression (ME-model) predicted the activation of Cra and Crp due to different carbon sources in carbon metabolisms. (B) Model-driven experimental design was achieved using genome-scale models of metabolism (M-model) and ME-model of the metabolic network in E. coli K-12 MG1655, with a list of viable nitrogen sources, and known TF binding sites. (C) Results of model-driven TF prediction. NtrC was activated on 23 nitrogen sources and Nac was activated on 19 nitrogen sources. (D) Protein expression of two major nitrogen-responsive TFs, NtrC and Nac, and two σ-factors, RpoD and RpoN, was measured by western blotting. Ammonia was used as a negative control as it is known not to activate NtrC and Nac. Glutamine was used as a positive control to activate the two TFs. Expression of NtrC and Nac increased on alternative nitrogen sources. However, Nac expression on cytidine increased significantly less than on glutamine or cytosine.
To expand this knowledge, a model-driven prediction workflow, including overall metabolism, was constructed to explore nitrogen source candidates that activate NtrC and Nac (Figure 1B). A genome-scale model of metabolism (M-model: iML1515) of E. coli (19) was used to determine viable nitrogen-containing molecules. A total of 93 nitrogen-containing nutrients with known transporters that support in silico growth were chosen and used for further analysis (Supplementary Figure S1A, Supplementary Table S1). Then, a model of metabolism and macromolecular expression (ME-model: iJL1678b-ME) of E. coli (17) was used to simulate growth on glucose with each of the candidates as the sole nitrogen source (Supplementary Figure S1B). From simulated results, predicted gene expression for each alternative nitrogen candidate was compared with predicted gene expression for the control nitrogen source, ammonia, to find a set of predicted differentially expressed genes. Annotated TF binding information from EcoCyc (38) was used to calculate which TFs were enriched in the predicted set of differentially expressed genes for each alternative nitrogen source. Out of 93 nitrogen source candidates, 23 and 19 sources were predicted to significantly enrich the expression of NtrC and Nac regulons, respectively (Figure 1C, Supplementary Table S1).
For instance, glutamine was also predicted to activate both two TFs. This is consistent with the previous studies, which used glutamine as a nitrogen source to cause nitrogen-limiting stress (30,43,52). To increase the reliability of model-driven prediction, nitrogen sources activating only Nac were excluded. This is due to the fact that Nac activation must be preceded by NtrC activation under unfavorable nitrogen sources. Therefore, cytosine was predicted to be the most potent inducer among predicted nitrogen candidates that simultaneously activate both NtrC and Nac (Supplementary Figure S1C). However, cytidine, belonging to pyrimidine-based substances like cytosine, was predicted to activate only NtrC. Considering that the two molecular structures are different due to the additional five carbon, cytosine degradation involves only nitrogen metabolism, whereas cytidine degradation requires both nitrogen and carbon metabolisms (Supplementary Figure S2A). Therefore, this result was expected to be a promising predictive result. Thus, it was predicted that firstly, NtrC would be activated by both pyrimidine-based nitrogen sources and that secondly, Nac would be differentially activated depending on the presence of a five-carbon source.
To ascertain activation of NtrC and Nac, expression profiling on RNA and protein levels, through RNA-seq and western blots, respectively, were performed. E. coli nitrogen source candidates were selected based on analysis of model-driven prediction results. Under these alternative nitrogen conditions, transcription of ntrC and nac was significantly up-regulated, but no significant change in RNAP subunits was observed (Supplementary Figure S2B). Moreover, analysis of environmental differentially expressed genes (DEGs) showed that a total of 371 and 575 DEGs were uncovered under cytidine and cytosine conditions, respectively (Supplementary Figure S2C, Supplementary Table S2). Among these, expression of 247 DEGs changed in both conditions. In agreement with RNA-seq, the protein expression levels of NtrC and Nac increased with cytidine, or cytosine as the sole nitrogen source (Figure 1D). Consistent with the model-driven prediction, the amount of Nac protein on cytidine was much lower than the amount on cytosine. This lower protein expression may explain why the number of DEGs on cytidine was lower than on cytosine. Thus, transcriptional expression changes of the genes encoding two key TFs in response to unfavorable nitrogen sources were significantly up-regulated, which was reflected in an increase in protein abundance of those TFs.
Genome-wide reconstruction of NtrC and Nac regulons associated with σ-factors
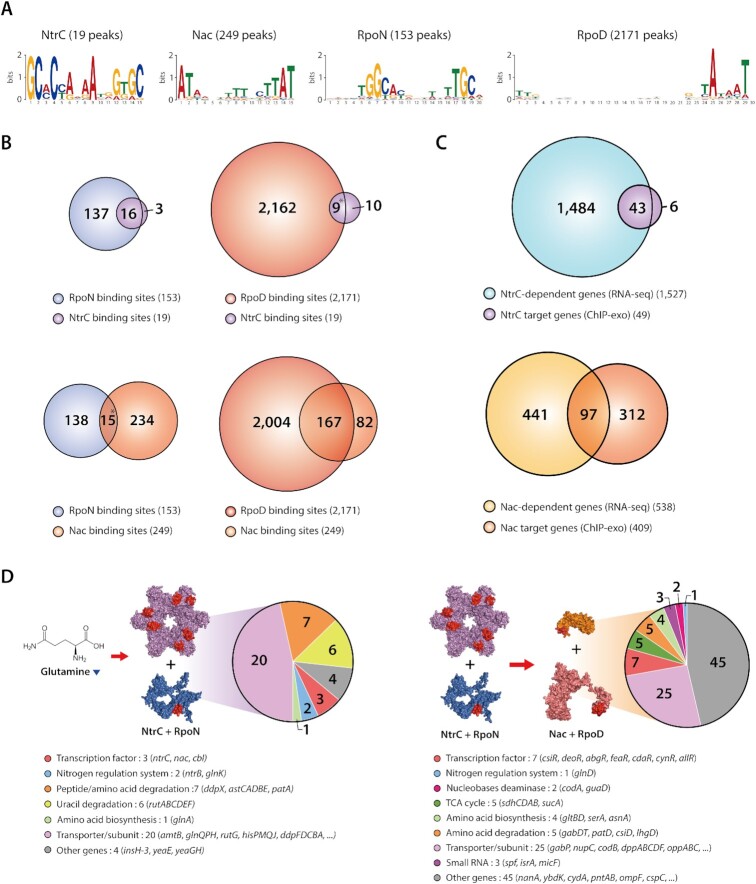
To shed light on transcriptional regulation by NtrC and Nac, genome-wide binding profiles of both TFs and σ-factors were generated under alternative nitrogen sources including cytosine and cytidine (Supplementary Table S3). Across the three-growth conditions, a total of 19 NtrC and 249 Nac binding sites were identified in regulatory regions (Supplementary Figure S3A). 153 and 2171 binding sites were identified for RpoN and RpoD, respectively. The number of binding sites for NtrC increased from 5 to 19, and Nac bindings increased from 15 to over 240 for each condition on alternative nitrogen sources compared to ammonia condition, whereas sigma factors binding sites did not change. The sequence motifs from NtrC, Nac, RpoN and RpoD binding sites were GCaCcaaaAtgGtGC, ATAagnaaaanttAT, tGGcacgattttTGCa, and ttgaca-15bp-gntAtaaT (Figures 2A and S3B). Motifs of TFs and σ-factors were identical to previous results (38,43,53,54). Previous studies demonstrated that NtrC is a member of the RpoN-dependent activator family (55) and interacts with RpoN (56). Nac, activated by NtrC, is postulated to serve as an adaptor between NtrC and RpoD-dependent promoters (30). To identify in vivo association of NtrC with RpoN and Nac with RpoD, binding sites of both TFs and their corresponding σ-factors were investigated. Out of 19 NtrC binding sites, 16 were found to be located near an RpoN binding site upstream of the same gene (Figure 2B). Nine of these were from complex promoters, having both RpoN and RpoD binding sites. The majority of Nac bindings (167, or 67.1%) adjoined RpoD bindings, and 15 binding sites were also found in complex promoters.
Figure 2.
Genome-wide identification of NtrC and Nac binding sites and regulons associated with sigma factors. (A) Motif analysis on ChIP-exo binding sites for NtrC, Nac, RpoN and RpoD resulted in previously known sequence motifs (lower-case characters indicate an information content < 1 bit). (B) Overlaps between NtrC and RpoN or RpoD binding sites under the alternative nitrogen sources. 16 NtrC binding events out of 19 were accompanied by RpoN binding. 9 NtrC bindings were identified near RpoD binding sites. In the case of Nac, 167 Nac binding events out of 249 were accompanied by RpoD binding. 15 Nac bindings were identified near RpoN binding sites. (*9 NtrC and RpoD bindings were identified near RpoN binding sites, *15 Nac and RpoN bindings were also detected near RpoD binding sites. These cases are complicated promoters with both RpoD and RpoN binding sites.) (C) Comparison of ChIP-exo binding results and differentially expressed gene profiles to define direct NtrC and Nac regulons. (D) NtrC associates with RpoN-dependent promoters, and regulates 20 transporter genes, 7 peptide/amino acid degradation genes, 6 uracil degradation genes, 3 TF genes, 2 nitrogen regulation system genes, 1 amino acid biosynthesis genes and 4 other enzymes. Nac associates with RpoD-dependent promoters, and regulates 25 transporter genes, 7 TF genes, 5 TCA cycle genes, 5 amino acid degradation genes, 4 amino acid biosynthesis genes, 3 sRNAs, 2 nucleobases deaminase genes, 1 nitrogen regulation system gene and 45 other enzymes (3D structure of Nac, RpoD, NtrC and RpoN were predicted by AlphaFold).
Based on the ChIP-exo datasets, a total of 49 target genes in 19 TUs of NtrC and 409 target genes in 234 TUs of Nac were identified in their regulatory regions during growth on alternative nitrogen sources. These genome-wide NtrC and Nac binding sites were also compared to binding sites from previous studies (Supplementary Text S1) (30,32,33). Of the 22 reported NtrC binding sites, 16 (72.7%) were identified by ChIP-exo experiments in this study. Of the 234 reported binding sites of Nac, 129 (55.1%) of them were identified, and 120 novel Nac binding sites were discovered. The number of binding sites of Nac was much higher than that of NtrC, suggesting that Nac may play a broader role in various metabolisms in E. coli.
To determine the causal relationships between the binding of TFs and changes in transcription levels of regulon genes, transcription levels of the wild-type and ntrC and nac deletion mutants (ΔntrC and Δnac) were compared under alternative nitrogen conditions. From ΔntrC and Δnac, a total of 1527 and 538 genes were differentially expressed under least one condition, respectively (Supplementary Table S4). Only 43 out of 1527 DEGs were differentially expressed by NtrC, and 97 out of 538 DEGs were found to be directly regulated by Nac under alternative nitrogen sources (Figure 2C, Supplementary Table S5). Notably, NtrC binding was mainly found upstream of the RpoN-dependent promoter. It works predominantly as a transcription activator in many cases (Supplementary Figure S4A). In contrast, Nac works as a dual regulator on the RpoD-dependent promoter. It binds upstream of a promoter when up-regulating the target gene, and it binds downstream of a promoter when down-regulating the target (Supplementary Figure S4B).
Functional analysis for regulons of both TFs was performed to shed more light on distinct functions of NtrC and Nac in response to unfavorable nitrogen sources. NtrC up-regulated 40 genes under both alternative nitrogen sources, and an additional two genes in cytidine conditions alone (Supplementary Figure S5A). Only a single gene, yeaE, was down-regulated by NtrC under both conditions. The NtrC regulon mostly contains 20 transporters, or their subunits, related to nitrogen sources (Figures 2D and S4C). NtrC also up-regulates transcription factors and major nitrogen regulatory proteins, including three TFs, ntrC itself, nac, cbl and two additional nitrogen regulatory proteins (ntrB and glnK). In addition, glutamine synthetase (glnA) is up-regulated by NtrC to maintain intracellular glutamine. Metabolic enzymes that scavenge nitrogen-containing molecules to produce ammonia or glutamate are also activated by NtrC.
While NtrC regulates a smaller set of genes and activates the expression of these target genes, Nac regulates a large number of genes and works as a dual regulator by up-regulating 73 genes and down-regulating 24 genes (Supplementary Figure S4D). Functional analysis of the Nac regulon showed Nac regulated 53 genes under all alternative nitrogen sources, and 44 genes in only one condition (Supplementary Figure S5B). The Nac regulon is comprised of 25 transporters or their subunits relating to the transport of various compounds (nitrogen sources: 12, carbon sources: 4, zinc: 1, multidrug: 2, putative transporter: 6) (Figure 2D). This regulon also includes a number of mostly locally acting TFs, some of which known to be related to carbon metabolism or found in both carbon and nitrogen metabolism (Supplementary Text S2). An interesting property of Nac regulatory mechanisms is that Nac activates nitrogen-containing molecule catabolic enzymes and represses amino acid anabolic enzymes. Moreover, Nac up-regulates succinate dehydrogenase (sdhCDAB) and 2-oxoglutarate dehydrogenase (sucA), which are some key enzymes in the tricarboxylic acid (TCA) cycle. Three sRNAs, nitrogen regulatory protein (glnD), and 48 other enzymes were found in the Nac regulon. Similarly, clusters of orthologous groups (COG) analysis showed most NtrC regulon genes are functionally enriched in amino acid metabolism (E) and signal transduction (T) (Supplementary Figure S5C). Nac regulon genes are only enriched in amino acid metabolism (E). But this regulon is also highly involved in carbohydrate metabolism (G) and energy production and conversion (C).
Differences in response to nitrogen-limiting stress caused by cytidine and cytosine
Under nitrogen limitation conditions, a low concentration of intracellular ammonium ions in E. coli reduces glutamate biosynthesis flux, resulting in the accumulation of intracellular ɑ-ketoglutarate. Studies investigating the effect of accumulated ɑ-ketoglutarate on E. coli metabolisms have been extensively conducted, and the level of accumulation occurs in proportion to nitrogen-limiting stress (57–59). Moreover, a high level of ɑ-ketoglutarate decreases expression of important target genes (e.g. TCA cycle) to insufficient levels, leading to growth deficiency under nitrogen limitation conditions (58).
Through high-performance liquid chromatography (HPLC), supernatants were analyzed to obtain a complete profile of extracellular metabolites for each nitrogen source respectively. For the cytidine condition, extracellular uridine and uracil were detected, but only uracil was found under cytosine condition (Table 1, Figure 3A). 3-hydroxypropionic acid (3-HP) was not detected at all, which is the final molecule of the pyrimidine degradation pathway. This extracellular metabolite profiling provides a better understanding of how cytidine and cytosine are used as the sole nitrogen source (Supplementary Figure S6A). Cytidine and cytosine have three nitrogen atoms but only one, in the form of an ammonium ion, is expected to be harvested during enzymatic reactions in E. coli. In the case of cytidine, it is first converted to uridine and an ammonium ion by cytidine deaminase (cdd), which has a Nac-independent promoter (Figure 3B). Subsequently, a portion of the uridine is broken into uracil and ribose 1-phosphate by uridine phosphorylase (udp) or nucleoside phosphorylase (ppnP). Ultimately, this ribose 1-phosphate is converted to ribose 5-phosphate, which can go into the pentose phosphate pathway (PPP). Similar to cytidine, only one ammonium ion is harvested from cytosine, by cytosine deaminase (codA). However, codA is repressed by the presence of pyrimidine in the medium (60). Thus, cytosine catabolism when using cytosine as the sole nitrogen source seems to have a ‘bug’ that leads to a slowed generation of ammonium ions. Interestingly, it was observed that codA was significantly upregulated by Nac. From this, it was inferred that the ‘bug’ can be debugged by Nac activation, which maintains the expression of codA to supply ammonium ions. These results also indicate that the pyrimidine degradation pathway (rutABCDEF), when activated by NtrC, is not used on both nitrogen sources. Additionally, these suggest that Nac plays a more important role in cytosine conditions than cytidine conditions to maintain growth.
Figure 3.
Different transcriptional regulatory response by Nac under cytidine and cytosine. (A) Glucose uptake rate and pyrimidine export rate measured by HPLC under alternative nitrogen sources. (B) Different metabolic enzymes of cytidine and cytosine regulated by Nac with RpoD or only RpoD. Cytidine and cytosine are transported by nupC and codB, respectively. These genes are activated by Nac with RpoD. Then, through cytosine deaminase (codA), regulated by Nac and RpoD, cytosine is degraded to uracil and ammonium ion. However, cytidine is converted to uridine and ammonium ion by cytidine deaminase (cdd), which is transcribed by only RpoD. (C) Prediction of nitrogen uptake rate for different nitrogen sources. Simulation of flux balance analysis (FBA) used experimentally measured glucose uptake rate and extracellular pyrimidine export rate to calculate nitrogen source uptake rates (ammonia shown as triangles; cytidine shown as circles; cytosine shown as squares). Sample points for flux analysis are also denoted (shown as red-lined hexagons). (D) Changes in mRNA expression of cytidine and cytosine transporters on different nitrogen sources. Cytidine transporter (nupC) is significantly up-regulated on cytidine, but cytosine transporter (codB) expression doesn’t change on cytosine. Both transporters are significantly down-regulated in the nac deletion strain. This indicates that Nac up-regulates the expression of both genes.
Through growth profiles and extracellular metabolite profiles, each nitrogen source uptake rate was calculated by flux balance analysis (FBA) using iML1515 (Figure 3C). On cytidine, the cytidine uptake rate was −9.58 mmol/gDCW/hr, which is not significantly different from the ammonia uptake rate, −10.05 mmol/gDCW/h. However, the cytosine uptake rate was calculated to be −6.55 mmol/gDCW/h. mRNA expression profiling of transporters for both nitrogen sources supported the predictive uptake rates generated by iML1515 (Figure 3D). The expression of cytidine transporter (nupC) is significantly increased with cytidine as the sole nitrogen source, whereas the expression of codB, which transports cytosine, did not change under cytosine conditions. Additionally, these transporter genes were both significantly up-regulated by Nac under nitrogen limitation. Markov Chain Monte Carlo (MCMC) sampling with iML1515 provided insight into nitrogen availability and catabolic pathways of both pyrimidine molecules. CYTD (cdd) had a higher flux value than CSND (codA), suggesting higher nitrogen availability under cytidine conditions (Supplementary Figure S6B). Likewise, flux of glutamate biosynthesis (GLUSy-GLUDy), which is indicative of nitrogen availability, showed the lowest value under the cytosine condition, compared to ammonia or cytidine. This result is also consistent with mRNA expression values (Supplementary Figure S6C). Thus, these results indicate that cytosine is the most stressful condition through induction of nitrogen limitation and accumulation of ɑ-ketoglutarate simultaneously.
Nac has unique regulatory mechanisms to restore intracellular glutamine pools under nitrogen-limiting stress
Intracellular glutamine is the central molecule with which E. coli cells sense nitrogen-limiting conditions (61). The Ntr regulatory cascade is triggered by low levels of glutamine, resulting in recovery of intracellular glutamine levels (Supplementary Figure S7A). On the transcription level of the regulatory cascade, NtrC up-regulated five genes (ntrC, nac, ntrB, glnA and glnK) while Nac up-regulated one gene (glnD) and down-regulated two genes (gltB and gltD) (Supplementary Figure S7B). Thus, NtrC and Nac control a majority of regulatory components in this cascade.
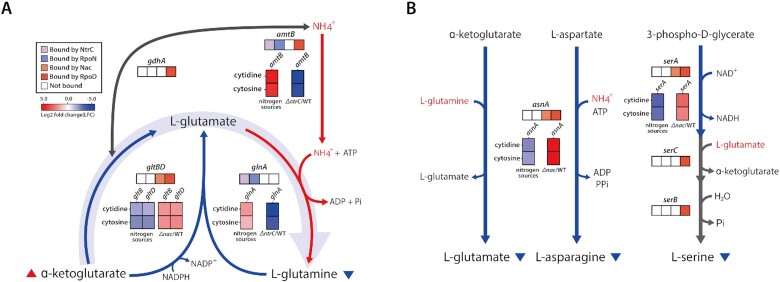
Based on the results of gene expression profiling, the primary regulatory mechanism of NtrC acts by restoring the glutamine concentration decreased by nitrogen-limiting stress. First, ammonium ion transporters (amtB) are up-regulated to increase intracellular ammonium ions used to produce glutamine (Figure 4A) (30). Second, synthesis of cytoplasmic glutamine from glutamate is increased by activating glutamine synthetase (glnA) (30,31). Apart from the NtrC responses, an important mechanism of Nac in responding to nitrogen limitation was also observed in the repression of amino acid biosynthesis to alleviate nitrogen-limiting stress. As amino acid synthesis requires a large amount of nitrogen, genes involved in the process may accelerate nitrogen-limiting stress under nitrogen depletion conditions caused by unfavorable nitrogen sources. To sustain growth and survival, Nac is activated by NtrC and significantly represses enzymes involved in amino acid biosynthesis, including gltBD (glutamate), asnA (asparagine) and serA (serine) (Figure 4B). Glutamate, a precursor of glutamine, can be built up from ɑ-ketoglutarate mediated by two different enzymes. One of the enzymes is glutamate synthase, which consumes glutamine with ɑ-ketoglutarate to generate glutamate. It is encoded by gltBD, an operon with a RpoD-dependent promoter. Nac significantly represses the expression of gltBD to minimize glutamine usage on unfavorable nitrogen sources (log2 fold change > 2) (Figure 4A). The other enzyme is glutamate dehydrogenase (gdhA), which also has a RpoD-dependent promoter for constitutive expression, thus, the expression of gdhA did not change significantly with alternative nitrogen sources.
Figure 4.
Regulatory mechanisms of amino acid metabolism enzymes by Nac and NtrC under unfavorable nitrogen sources. (A) When exposed to unfavorable nitrogen sources, the low ammonium ion concentration decreases nitrogen incorporation from ɑ-ketoglutarate to amino acids. This decreases the concentration of cytoplasmic glutamine and increases the concentration of ɑ-ketoglutarate. Glutamine-producing genes are activated by NtrC, and glutamine consumption genes are repressed by Nac. In E. coli, NtrC and Nac play an important role in maintaining glutamine concentration. (B) Amino acid biosynthesis genes regulated by Nac. Nac directly down-regulates glutamate synthase and asparagine synthetase, which utilize glutamine or ammonium ions. Additionally, enzyme of initial step in serine biosynthesis is repressed by Nac under nitrogen-limiting conditions. (The left stack denotes the relative expression of genes between alternative nitrogen sources and ammonia, right stack indicates the relative expression of genes between TF deletion strain and wild-type strain under alternative nitrogen sources, respectively.)
Asparagine is one of the proteinogenic amino acids containing amide side-chains. Among the two enzymes capable of asparagine synthesis, asnA is known to be the more active asparagine synthetase (62). Interestingly, it was found that asnA was significantly regulated by Nac under nitrogen-limiting conditions. Nac binds upstream of asnA to down-regulate expression, which reduces asparagine biosynthesis, thereby decreasing the consumption of ammonium ions. Therefore, it can be inferred that ammonium ions are preferentially used for glutamine biosynthesis. In addition, phosphoglycerate dehydrogenase (serA), an initial enzyme in the biosynthesis of serine, is down-regulated by Nac. Serine is the precursor of about ∼33% (7/21) of total proteinogenic amino acids (Supplementary Figure S8A), hence Nac can affect amino acid biosynthesis through the strong repression of a single gene, serA. Moreover, the flux state of proteogenic amino acid biosynthesis using MCMC sampling with iML1515 showed that fluxes of reactions repressed by Nac were reduced under unfavorable nitrogen sources (Supplementary Figure S8B). Interestingly, the reduction in the flux of amino acid biosynthesis in the cytosine condition was higher than in the cytidine condition (Supplementary Figure S6B and S8B). Thus, this flux distribution also supports the experimental results of the higher activation of Nac on cytosine conditions.
Taken together, in addition to transcriptional regulation in the cascade, NtrC activates transporters and glutamine biosynthesis genes, and induces gene expression of the other key TF, nac. Furthermore, we expanded precise regulons of Nac under nitrogen-limiting conditions compared to previous studies, which regulate expression by Nac with more than a 2-fold change in expression (30,32) (Supplementary Table S6). Nac plays an important role in transcriptional regulation in amino acid metabolism. To alleviate nitrogen-limiting stress, enzymes responsible for the biosynthesis of three amino acids are repressed by Nac, reducing the usage of ammonium ions and glutamine. In particular, down-regulation of glutamate synthase to maintain the glutamine pool leads to a decrease in the expenditure of ɑ-ketoglutarate, a key molecule in carbon metabolism. The management of production and consumption of ɑ-ketoglutarate in E. coli cells is revealed to be of particular interest under nitrogen-limiting conditions.
Rebalancing carbon flux by Nac on the carbon metabolism
The regulatory network of Nac in E. coli was reconstructed to observe how Nac regulates genes of carbon metabolic processes, including glycolysis and the TCA cycle. 11 of those involved genes were regulated by Nac (Figure 5A). Expression of five TCA cycle genes, sucA and sdhBADC, was significantly up-regulated by Nac. Nac also up-regulated pck, sucB, sucCD and fumC, and down-regulated the glycolysis gene, ppc, but the observed expression fold change was less than two (Supplementary Figure S9A).
Figure 5.
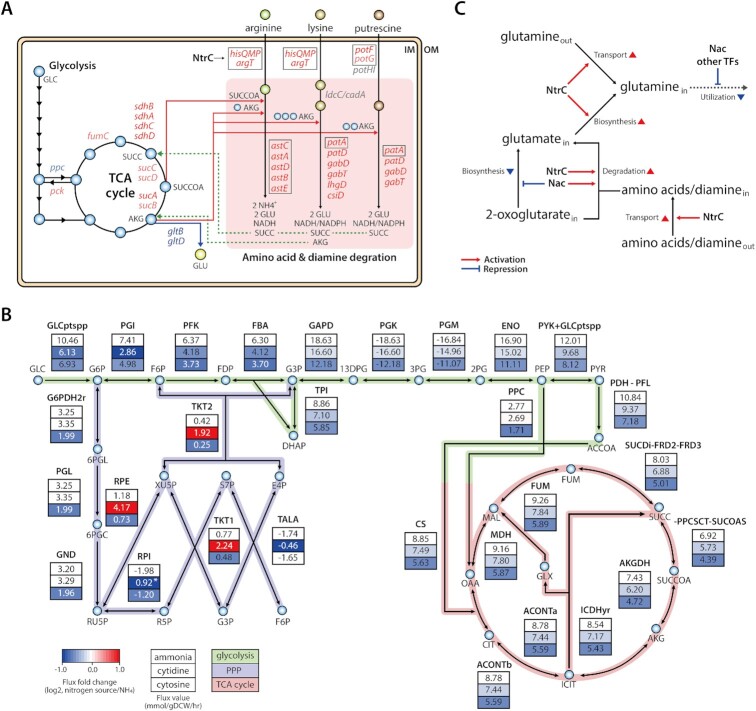
Network-level regulation of carbon metabolism and amino acid and diamine degradation pathways by Nac and NtrC. (A) Carbon metabolism (glycolysis and TCA cycle) and amino acid/diamine degradation pathways are represented. Nac regulates the expression of enzymes in the TCA cycle, including ppc, pck, sdhBADC, sucCD, sucAB and fumC. Transporters or their subunits of amino acids and diamine are up-regulated by NtrC. In amino acid/diamine degradation pathways, the astCADBE operon and patA are activated by NtrC, which encode the enzymes in the arginine and lysine/putrescine degradation pathways, respectively. Moreover, Nac significantly up-regulates lysine/putrescine degradation enzymes that are encoded patD, gabTD, csiD and lhgD. Two lysine degradation genes (ldcC and cadA) and putrescine transporter subunits (potHI) are not regulated by NtrC and Nac. The genes regulated by Nac or NtrC are depicted by bright red (LFC > 1), bright blue (LFC < −1), light red (0.5 < LFC < 1), and light blue (−1 < LFC < −0.5), respectively. And the genes regulated by NtrC are depicted by black boxes. Abbreviation: GLC: glucose, AKG: ɑ-ketoglutarate, SUCCOA: succinyl-CoA, SUCC: succinate, GLU: glutamate, IM, inner membrane; OM, outer membrane. (B) To explain lower activation of Nac on cytidine condition MCMC sampling was performed on ammonia, cytidine, and cytosine with experimentally measured glucose uptake rate and pyrimidine export rates. (There are genes and reactions with minor flux that are omitted for clarity of Figure. *The flux through the reaction was lower than the flux on ammonia; however, the direction of the reaction was reversed.) (C) An overview of the Nac and NtrC regulons under nitrogen limitation. Both TFs induce the conversion of glutamate from ɑ-ketoglutarate and amino acids without using glutamine, to modulate stress response to nitrogen deficiency and accumulated ɑ-ketoglutarate.
An interesting aspect of the expression change in the TCA cycle under nitrogen limitation is that downstream genes of ɑ-ketoglutarate (sucABCD, sdhBADC, lpd) in the pathway were more repressed than upstream genes (icd, acnAB, gltA) (Supplementary Figure S9B). Cytosine was predicted to induce higher nitrogen-limiting stress and ɑ-ketoglutarate accumulation than cytidine. Consistent with this result, the expression levels of sucAB, sucCD and sdhBADC were more strongly down-regulated on cytosine than cytidine. Thus, the regulatory role of Nac found in carbon metabolism is postulated to be regulation of stress caused by the accumulation of ɑ-ketoglutarate. First, Nac represses phosphoenolpyruvate carboxylase (ppc) to reduce carbon flux into the TCA cycle, thereby maintaining the balance of TCA carbons under nitrogen-limiting conditions. Second, in the TCA cycle, ɑ-ketoglutarate downstream genes are activated by Nac, and the expression of TCA cycle genes is maintained by restoring transcription expression caused by high concentration of ɑ-ketoglutarate. These results indicated that need for Nac increases when using more poor nitrogen sources. It was also similar to the model-driven prediction and western blot.
Additionally, Nac and NtrC regulons were mapped to amino acid and diamine degradation pathways, which consume intermediates of the TCA cycle with amino acids to produce glutamate (Figure 5A and Supplementary Figure S10). Under nitrogen-limiting conditions, transporters of amino acids and diamine are activated by NtrC. Arginine is degraded into succinate through AST pathway genes (astCADBE) that are activated by NtrC. This pathway produces two glutamates, one ammonium ion, and one molecule of NADH using one molecule each of succinyl-CoA and ɑ-ketoglutarate. Lysine and putrescine degradation pathways were found to be co-regulated by both TFs, NtrC and Nac. First, NtrC activates putrescine aminotransferase (patA). Subsequently, Nac up-regulates downstream genes of lysine and putrescine degradation pathways, including patD, gabTD. As a result of this co-regulated pathway, lysine and putrescine are degraded into glutarate and succinate, respectively, converting two ɑ-ketoglutarates to two glutamates and generating one molecule each of NADH and NADPH. Additionally, glutarate is degraded by Nac regulon genes, csiD and lhgD, consuming one ɑ-ketoglutarate to convert one molecule of succinate and regenerate one ɑ-ketoglutarate. Thus, these results indicate that NtrC primarily activates amino acid transporter genes to induce uptake of extracellular amino acids, and that both TFs up-regulate amino acid/diamine degradation pathways. It can be inferred that Nac may shift the metabolic flux from glutamate synthesis to amino acid and diamine degradation pathways to convert glutamate from accumulated ɑ-ketoglutarate. This redirection of ɑ-ketoglutarate consumption from glutamate synthesis to amino acid/diamine degradation pathways co-activated by both TFs might relieve stress caused by nitrogen-limitation and ɑ-ketoglutarate accumulation. Furthermore, among the Nac regulon genes in the TCA cycle, succinate dehydrogenase (sdhBADC) was most strongly up-regulated by Nac. This enzyme converts succinate, a final molecule of amino acid/diamine degradation, into fumarate, providing the connection between the amino acid/diamine degradation pathway and the TCA cycle. In addition, two out of these eight Nac regulon genes (gabTD) were known to be directly regulated by Nac under nitrogen limitation in previous study (30) (Supplementary Table S6). These Nac regulon genes were observed to be regulated by Nac with artificial induction of Nac under non-nitrogen-limiting conditions (32). Only sdhBC were shown to overlap with the Nac regulons found in this study. The regulatory role of Nac on the remaining six genes was different. These results demonstrated consistent expansion of the Nac regulon in response to nitrogen-limiting stress.
Moreover, the in silico metabolic model, which makes predictions independent of transcriptional regulatory information, provided more insight into the role of Nac. FBA and MCMC sampling on the iML1515 (19,63) were performed to simulate feasible internal flux states during growth on three different nitrogen sources (Figure 5B, Table 3). On ammonia, the glucose uptake rate was −11.18 mmol/gDCW/h, and flux of glucose 6-phosphate from glucose was 10.46 mmol/gDCW/h. About 30% of glucose 6-phosphate went into PPP, leaving about 70% remaining in the glycolysis pathway. Flux distribution through glucose 6-phosphate on cytosine was the same as ammonia, but the glucose uptake rate was lower, at −7.25 mmol/gDCW/h. On cytidine, flux into glucose 6-phosphate was split differently, with more flux towards PPP and less flux into downstream glycolysis. Additionally, the flux of glucose uptake was the lowest among the three nitrogen sources, at −6.48 mmol/gDCW/h. This is expected to be caused by phosphotransferase system (PTS)-independent carbon replenishment with ribose 1-phosphate, which is an additional carbon source generated by cytidine degradation. And most fluxes through PPP increased due to ribose 5-phosphate conversion from ribose 1-phosphate. As a result, flux from fructose 6-phosphate and downstream reactions increased to higher levels compared with the cytosine condition. In particular, flux distribution of the TCA cycle, including sdhCDAB, showed a smaller decrease with cytidine as the sole nitrogen source compared to cytosine. This result showed consistency with experimental data, which found lower repression of TCA cycle genes by ɑ-ketoglutarate accumulation under cytidine conditions. It likely suggests a reduction in the need for expression restored by Nac under cytidine conditions. In addition, flux of PPC, which converts phosphoenolpyruvate into oxaloacetate in glycolysis, does not change with cytidine as the sole nitrogen source, unlike with cytosine. This reaction encodes the ppc gene, repressed by Nac. The unchanged flux of ppc indicates a decrease in the need for repression by Nac. Moreover, growth profiles of the nac deletion mutant (Δnac) indicate growth rate is not as strongly affected by cytidine when comparing cytosine condition (Table 2).
Table 3.
Prediction of the overall metabolic flux of iML1515 on different nitrogen sources
| Reactions | Ammonia | Cytidine | Cytosine |
|---|---|---|---|
| Glycolysis | |||
| GLCptspp | 10.46 | 6.13 | 6.93 |
| PGI | 7.41 | 2.86 | 4.98 |
| PFK | 6.37 | 4.18 | 3.73 |
| FBA | 6.30 | 4.12 | 3.70 |
| TPI | 8.86 | 7.10 | 5.85 |
| GAPD | 18.63 | 16.60 | 12.18 |
| PGK | −18.63 | −16.60 | −12.18 |
| PGM | −16.84 | −14.96 | −11.07 |
| ENO | 16.90 | 15.02 | 11.11 |
| PDH-PFL | 10.84 | 9.37 | 7.18 |
| GLCptspp + PYK | 12.01 | 9.68 | 8.12 |
| Pentose phosphate pathway | |||
| G6PDH2r | 3.25 | 3.35 | 1.99 |
| PGL | 3.25 | 3.35 | 1.99 |
| GND | 3.20 | 3.29 | 1.96 |
| RPI | −1.98 | 0.92 | −1.20 |
| RPE | 1.18 | 4.17 | 0.73 |
| TKT1 | 0.77 | 2.24 | 0.48 |
| TKT2 | 0.42 | 1.92 | 0.25 |
| TALA | −1.74 | −0.46 | −1.65 |
| TCA cycle | |||
| PPC | 2.77 | 2.69 | 1.71 |
| CS | 8.85 | 7.49 | 5.63 |
| ACONTa | 8.78 | 7.44 | 5.59 |
| ACONTb | 8.78 | 7.44 | 5.59 |
| ICDHyr | 8.54 | 7.17 | 5.43 |
| AKGDH | 7.43 | 6.20 | 4.72 |
| -PPCSCT-SUCOAS | 6.92 | 5.73 | 4.39 |
| SUCDi-FRD2-FRD3 | 8.03 | 6.88 | 5.01 |
| FUM | 9.26 | 7.84 | 5.89 |
| MDH | 9.16 | 7.80 | 5.87 |
Table 2.
Growth rate of E. coli K-12 MG1655 wild-type and Δnac strain under alternative nitrogen sources
| Ammonia | Cytidine | Cytosine |
|---|---|---|
| Growth rate of E. coli K-12 MG1655 wild-type (h−1) | ||
| 0.645 ± 0.003 | 0.585 ± 0.012 | 0.433 ± 0.014 |
| Growth rate of E. coli K-12 MG1655 Δnac (h−1) | ||
| 0.601 ± 0.007 | 0.353 ± 0.001 | 0.098 ± 0.001 |
Estimating flux through a metabolic reaction using the E. coli metabolic model under alternative nitrogen sources helped explain the observation of lower activation of Nac on cytidine. To relieve nitrogen limitation and TCA intermediate accumulation stresses, ɑ-ketoglutarate was predicted to be converted to glutamate through amino acid/diamine degradation pathways regulated either by NtrC alone or both NtrC and Nac (Figure 5C). Additionally, down-regulated expression of TCA cycle genes resulted in a decrease in reducing power. To compensate for this, NADH and NADPH are produced through amino acid/diamine degradation pathways. Furthermore, some genes of the TCA cycle are regulated by Nac. Nac restores expression levels of TCA cycle genes that have been reduced by nitrogen-limiting stress, suggesting that Nac may relieve stress caused by TCA intermediate accumulation. And, to balance decreased consumption of TCA carbon, flux through phosphoenolpyruvate to oxaloacetate is repressed by Nac. The overall regulatory consequences indicate Nac is an important dual regulator that modulates core carbon metabolism as well as nitrogen metabolism under nitrogen-limitation conditions.
DISCUSSION
Computation with metabolic models systematizes biochemical, genetic, and genomic knowledge into a mathematical framework that enables a mechanistic description of metabolic physiology. In this study, genome-scale metabolic models were leveraged to improve our understanding of regulation of nitrogen metabolism and its interplay with carbon metabolism. This approach generated an in silico prediction of gene expression of whole the cell, and calculated gene expression was assumed to change in response to TFs. Based on this assumption, experimental conditions were chosen to activate two important, yet relatively uncharacterized, transcription factors involved in responses to nitrogen limitation, NtrC and Nac. Integrative data analysis revealed the important roles of their regulons under nitrogen-limiting conditions. To validate the extension of regulon information for NtrC and Nac, regulon genes of both TFs found in this study were compared to RegulonDB, which is the primary database containing regulon information for E. coli (64). About 95.3% of NtrC regulon genes were found in RegulonDB, while only 16.5% of Nac regulon genes were found (Supplementary Figure S11A). Moreover, iModulonDB, which is a data-driven bacterial TRN knowledgebase using a machine learning algorithm (65), was compared with both experimental regulons as well. In both iModulons, 39 (90.7%) NtrC regulon genes out of 43 were found, and 21 (21.6%) out of 97 Nac regulon genes were found (Supplementary Figure S11B). This result suggests that the TF prediction workflow using the ME-model successfully expanded knowledge of the Nac regulons activated under nitrogen limiting conditions.
Since the advent of the ChIP-exo method used in prokaryotic studies (5,7,66,67) with better resolution than other previously established ChIP methods, ChIP-exo made it possible to measure accurate TF binding events. We also observed unexpected binding of TFs and σ-factors in non-regulatory regions. Given the function of those proteins in gene regulation, they are expected to bind onto regulatory regions mostly near promoters to regulate the gene expression of target genes. It is commonly reported that TFs bind on non-regulatory regions including those inside coding regions. Similarly, a large number of unexpected bindings for NtrC (4, 21%), Nac (281, 113%), RpoN (232, 152%), and RpoD (461, 21%) were observed in non-regulatory regions, respectively. Interestingly, Nac and RpoN showed more bindings on non-regulatory regions than on regulatory regions. The non-regulatory bindings also have identical sequence motifs; however, they have significantly weaker binding intensities (Supplementary Figure S3A, rank sum test P-value < 0.05). The function or consequence of those bindings is still elusive. There are some possible explanations such as traces of evolution (1), regulation of antisense transcripts, or multimeric binding onto genomic DNA as nucleoid-associated proteins (NAPs) like lrp (68). For σ-factors, some binding sites were observed in the coding region associated with weaker transcription on the opposite strand, indicating possible involvement in antisense transcription. Further study is needed to understand the high number of binding sites of Nac on non-regulatory regions. It could simply be evolutionary effect, or Nac may work as an NAP as well.
The role of the Nac regulon was extended by combining ChIP-exo binding profiles of Nac and expression profiles of the nac knock-out strain under nitrogen limitation. 26 out of 32 genes from Zimmer et al. (30) were matched to the Nac regulon found in this study and the role of Nac also coincides (Supplementary Table S6). This network level reconstruction of the Nac regulon increased from 26 to 97 under nitrogen-limiting conditions. Moreover, a comparison of our study with a recent study on the identification of the binding sites using ChIP-seq with artificial induction of Nac showed that the binding sites of 88 out of 97 Nac regulons found in this study were overlapping (32) (Supplementary Table S6). However, only 13 of 34 overlapping regulon genes were regulated by Nac in the same repressive or activating way (Absolute LFC > 1). This result demonstrated that physical induction caused by nitrogen-limiting stress changed the TRN of Nac. Therefore, in this study, we proposed an extended and comprehensively reconstructed TRN of Nac in response to nitrogen limitation.
Furthermore, this study investigated coupling between nitrogen and carbon metabolism. Under nitrogen limitation conditions, the low concentration of intracellular ammonium ions in E. coli reduces glutamate biosynthetic flux. As the reaction consumes ɑ-ketoglutarate and ammonium ions, the decreased consumption of TCA intermediates leads to the accumulation of intracellular ɑ-ketoglutarate. It was reported that accumulated ɑ-ketoglutarate directly blocks glucose uptake by inhibiting the first step of the PTS, resulting in adenylate cyclase (CyaA) deactivation and subsequent intracellular cyclic-AMP (cAMP) depletion (57,69,70) (Supplementary Figure S12). The decreased cAMP level reduces the formation of the CRP-cAMP complex, which downregulates important target genes for cell growth under nitrogen limitation conditions (58). In this study, Nac was found to not only repress glutamine-consuming genes but also play a role in restoring the reduced expression of the TCA cycle. Thus, this study revealed one molecular mechanism through which coupling between nitrogen and carbon metabolism is implemented, mediated by Nac.
Model-based analysis provided more insight into how Nac-mediated metabolic coupling can respond differently on different nitrogen sources. Genome-scale models have been previously used to analyze cellular responses to environmental stimuli (8,71,72). Therefore, in addition to using modeling to guide experimental design, the iML1515 model was exploited to assess the broader physiological role of the Nac regulon. As nitrogen availability for each condition was provided based on the uptake prediction of nitrogen source, these uptake rates were used to determine quantitative changes in flux through pathways associated with glutamate biosynthesis from model-based simulation in response to alternative nitrogen sources. Lower flux of glutamate biosynthesis under nitrogen limitation conditions indicated the accumulation of ɑ-ketoglutarate might be accelerated. Additionally, it is observed that fluxes through major pathways on carbon metabolism are different depending on which nitrogen sources were used. For example, despite the lowest glucose uptake rate, additional carbon flux from cytidine went the pentose phosphate pathway and increased glycolysis and TCA cycle when using cytidine as the sole nitrogen source than when using cytosine. This provided insight into the lower activity of Nac on cytidine. More broadly, computations with metabolic models predict that other organic nitrogen sources, which contain carbon as well can influence flux through carbon metabolism. This reveals the distinguished importance of the roles of Nac given the interplay of carbon and nitrogen metabolism. Response to nitrogen limitation requires not only response in nitrogen metabolism, essentially regulated by NtrC, but also a response in carbon metabolism to rebalance the flux in the TCA cycle, which is partially regulated by Nac.
Additionally, the three-dimensional structures of the TFs, sigma factors, and complex structures were predicted by AlphaFold pipeline with with high pLDDT average scores (Supplementary Figure S13). However, the protein-protein interactions between TFs and sigma factors (NtrC and RpoN, Nac and RpoD), which were expected to elucidate the mechanistic details of how both TFs recruit or block sigma factors, could not be predicted with an AlphaFold-multimer.
In conclusion, M and ME-model-driven experimental design proved effectiveness in determining activation conditions for regulatory TFs in nitrogen metabolism. Furthermore, the definition of NtrC and Nac regulons with flux prediction from a bacterial metabolic network model deciphered contrasting role of these regulons, emphasizing the unexpected role of Nac in the coupling of nitrogen and carbon metabolism. These results suggest that integration of model-based simulation and genome-wide experimental measurements can be leveraged as a new dimension of systems approaches to transcriptional regulatory network study, allowing better understanding by complementarily providing integrative synergistic effect of each approach.
DATA AVAILABILITY
The whole dataset of ChIP-exo and RNA-seq has been deposited to GEO (https://www.ncbi.nlm.nih.gov/geo/) with the accession number of GSE54905. Scripts for genome-scale metabolic models, which are used to predict nitrogen uptake and to simulate constraint-based models using HPLC for extracellular metabolites concentration, are freely available at the public GitHub repository (https://github.com/SBML-Kimlab/Nitrogen_GSM) and on Zenodo at https://doi.org/10.5281/zenodo.7508655.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Aarash Bordbar for the discussion on interpretation of model-based simulation and Zak King for helping with the visualization of metabolic flux states. We thank Hoa Thi Le for the construction of deletion mutant for E. coli strains. And we also thank Marc Abrams for helpful assistance in writing and editing the manuscript.
Author contributions: J.Y.P., D.K., A.E. and B.O.P. conceived the study. J.Y.P., D.K. and S.W.S. performed experiments. J.Y.P., D.K., A.E. and J.K. performed the computational analysis. B.O.P. supervised the study. J.Y.P., S.M.L., D.K., A.E., Z.K.S.N., J.K., A.S., S.W.S. and B.O.P. wrote the manuscript. All authors helped edit the final manuscript.
Contributor Information
Joon Young Park, School of Energy and Chemical Engineering, Ulsan National Institute of Science and Technology (UNIST), Ulsan 44919, Republic of Korea.
Sang-Mok Lee, School of Energy and Chemical Engineering, Ulsan National Institute of Science and Technology (UNIST), Ulsan 44919, Republic of Korea.
Ali Ebrahim, Department of Bioengineering, University of California, San Diego, La Jolla, CA 92093, USA.
Zoe K Scott-Nevros, School of Energy and Chemical Engineering, Ulsan National Institute of Science and Technology (UNIST), Ulsan 44919, Republic of Korea.
Jaehyung Kim, School of Energy and Chemical Engineering, Ulsan National Institute of Science and Technology (UNIST), Ulsan 44919, Republic of Korea.
Laurence Yang, Department of Chemical Engineering, Queen's University, Kingston, Canada.
Anand Sastry, Department of Bioengineering, University of California, San Diego, La Jolla, CA 92093, USA.
Sang Woo Seo, School of Chemical and Biological Engineering, and Interdisciplinary Program in Bioengineering, and Institute of Chemical Processes, and Bio-MAX Institute, Seoul National University, 1 Gwanak-ro, Gwanak-gu, Seoul 08826, Korea.
Bernhard O Palsson, Department of Bioengineering, University of California, San Diego, La Jolla, CA 92093, USA; Department of Pediatrics, University of California, San Diego, La Jolla, CA 92093, USA; The Novo Nordisk Foundation Center for Biosustainability, Danish Technical University, 6 Kogle Alle, Hørsholm, Denmark.
Donghyuk Kim, School of Energy and Chemical Engineering, Ulsan National Institute of Science and Technology (UNIST), Ulsan 44919, Republic of Korea; Department of Bioengineering, University of California, San Diego, La Jolla, CA 92093, USA; Department of Biomedical Engineering, Ulsan National Institute of Science and Technology (UNIST), Ulsan, Republic of Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (MSIT) [2021M3A9I4024840, 2022M3A9I5018934]; UNIST Center for Waste Plastics Carbon Cycling (UWCC) funded by the Circle Foundation, Republic of Korea; Novo Nordisk Foundation (NNF) Center for Biosustainability at the Danish Technical University; NIH NIGMS (National Institute of General Medical Sciences) [5R01GM057089-19, GM102098].
Conflict of interest statement. None declared.
REFERENCES
- 1. Shimada T., Ishihama A., Busby S.J., Grainger D.C.. The Escherichia coli RutR transcription factor binds at targets within genes as well as intergenic regions. Nucleic Acids Res. 2008; 36:3950–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Myers K.S., Yan H., Ong I.M., Chung D., Liang K., Tran F., Keles S., Landick R., Kiley P.J.. Genome-scale analysis of escherichia coli FNR reveals complex features of transcription factor binding. PLoS Genet. 2013; 9:e1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Park D.M., Akhtar M.S., Ansari A.Z., Landick R., Kiley P.J.. The bacterial response regulator ArcA uses a diverse binding site architecture to regulate carbon oxidation globally. PLos Genet. 2013; 9:e1003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cho B.K., Knight E.M., Palsson B.O.. Transcriptional regulation of the fad regulon genes of Escherichia coli by ArcA. Microbiology. 2006; 152:2207–2219. [DOI] [PubMed] [Google Scholar]
- 5. Kim D., Seo S.W., Gao Y., Nam H., Guzman G.I., Cho B.-K., Palsson B.O.. Systems assessment of transcriptional regulation on central carbon metabolism by Cra and CRP. Nucleic Acids Res. 2018; 46:2901–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seo S.W., Kim D., O’Brien E.J., Szubin R., Palsson B.O.J.N.c.. Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli. Nat. Commun. 2015; 6:7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seo S.W., Kim D., Latif H., O’Brien E.J., Szubin R., Palsson B.O.J.N.c.. Deciphering fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat. Commun. 2014; 5:4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Federowicz S., Kim D., Ebrahim A., Lerman J., Nagarajan H., Cho B.-k., Zengler K., Palsson B.J.P.g.. Determining the control circuitry of redox metabolism at the genome-scale. PLoS Genet. 2014; 10:e1004264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Covert M.W., Knight E.M., Reed J.L., Herrgard M.J., Palsson B.O.J.N.. Integrating high-throughput and computational data elucidates bacterial networks. Nature. 2004; 429:92–96. [DOI] [PubMed] [Google Scholar]
- 10. Feist A.M., Palsson B.Ø.J.N.b.. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat. Biotechnol. 2008; 26:659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bordbar A., Monk J.M., King Z.A., Palsson B.O.J.N.R.G.. Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 2014; 15:107–120. [DOI] [PubMed] [Google Scholar]
- 12. O’Brien E.J., Monk J.M., Palsson B.O.J.C.. Using genome-scale models to predict biological capabilities. Cell. 2015; 161:971–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCloskey D., Palsson B.Ø., Feist A.M.J.M.s.b.. Basic and applied uses of genome-scale metabolic network reconstructions of Escherichia coli. Mol. Syst. Biol. 2013; 9:661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thiele I., Palsson B.Ø.J.N.p.. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010; 5:93–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Orth J.D., Thiele I., Palsson B.Ø.. What is flux balance analysis?. Nat. Biotechnol. 2010; 28:245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O’brien E.J., Lerman J.A., Chang R.L., Hyduke D.R., Palsson B.Ø.J.M.s.b.. Genome-scale models of metabolism and gene expression extend and refine growth phenotype prediction. Mol. Syst. Biol. 2013; 9:693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lloyd C.J., Ebrahim A., Yang L., King Z.A., Catoiu E., O’Brien E.J., Liu J.K., Palsson B.O.. COBRAme: a computational framework for genome-scale models of metabolism and gene expression. PLoS Comput. Biol. 2018; 14:e1006302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Du B., Yang L., Lloyd C.J., Fang X., Palsson B.O.J.P.c.b.. Genome-scale model of metabolism and gene expression provides a multi-scale description of acid stress responses in Escherichia coli. PLoS Comput. Biol. 2019; 15:e1007525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Monk J.M., Lloyd C.J., Brunk E., Mih N., Sastry A., King Z., Takeuchi R., Nomura W., Zhang Z., Mori H.J.N.b.. iML1515, a knowledgebase that computes Escherichia coli traits. Nat. Biotechnol. 2017; 35:904–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Orth J.D., Conrad T.M., Na J., Lerman J.A., Nam H., Feist A.M., Palsson B.Ø.J.M.s.b.. A comprehensive genome-scale reconstruction of Escherichia coli metabolism—2011. Mol. Syst. Biol. 2011; 7:535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dahal S., Zhao J., Yang L.J.B., Engineering B.. Genome-scale modeling of metabolism and macromolecular expression and their applications. Biotechnol. Bioprocess Eng. 2020; 25:931–943. [Google Scholar]
- 22. Garcia E., Rhee S.G.. Cascade control of Escherichia coli glutamine synthetase. Purification and properties of PII uridylyltransferase and uridylyl-removing enzyme. J. Biol. Chem. 1983; 258:2246–2253. [PubMed] [Google Scholar]
- 23. van Heeswijk W.C., Hoving S., Molenaar D., Stegeman B., Kahn D., Westerhoff H.V.. An alternative PII protein in the regulation of glutamine synthetase in Escherichia coli. Mol. Microbiol. 1996; 21:133–146. [DOI] [PubMed] [Google Scholar]
- 24. Blauwkamp T.A., Ninfa A.J.. Physiological role of the GlnK signal transduction protein of Escherichia coli: survival of nitrogen starvation. Mol. Microbiol. 2002; 46:203–214. [DOI] [PubMed] [Google Scholar]
- 25. Vasudevan S.G., Gedye C., Dixon N.E., Cheah E., Carr P.D., Suffolk P.M., Jeffrey P.D., Ollis D.L.. Escherichia coli PII protein: purification, crystallization and oligomeric structure. FEBS Lett. 1994; 337:255–258. [DOI] [PubMed] [Google Scholar]
- 26. Liu J., Magasanik B.. Activation of the dephosphorylation of nitrogen regulator I-phosphate of Escherichia coli. J. Bacteriol. 1995; 177:926–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Atkinson M.R., Blauwkamp T.A., Ninfa A.J.. Context-dependent functions of the PII and GlnK signal transduction proteins in Escherichia coli. J. Bacteriol. 2002; 184:5364–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Heeswijk W.C., Wen D., Clancy P., Jaggi R., Ollis D.L., Westerhoff H.V., Vasudevan S.G.. The Escherichia coli signal transducers PII (GlnB) and GlnK form heterotrimers in vivo: fine tuning the nitrogen signal cascade. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:3942–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Javelle A., Severi E., Thornton J., Merrick M.J.J.o.B.C.. Ammonium sensing in Escherichia coli: role of the ammonium transporter AmtB and AmtB-GlnK complex formation. J. Biol. Chem. 2004; 279:8530–8538. [DOI] [PubMed] [Google Scholar]
- 30. Zimmer D.P., Soupene E., Lee H.L., Wendisch V.F., Khodursky A.B., Peter B.J., Bender R.A., Kustu S.J.P.o.t.N.A.o.S.. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:14674–14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brown D.R., Barton G., Pan Z., Buck M., Wigneshweraraj S.J.N.c.. Nitrogen stress response and stringent response are coupled in Escherichia coli. Nat. Commun. 2014; 5:4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aquino P., Honda B., Jaini S., Lyubetskaya A., Hosur K., Chiu J.G., Ekladious I., Hu D., Jin L., Sayeg M.K.J.B.s.b.. Coordinated regulation of acid resistance in Escherichia coli. 2017; 11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown D.R., Barton G., Pan Z., Buck M., Wigneshweraraj S.. Nitrogen stress response and stringent response are coupled in Escherichia coli. Nat. Commun. 2014; 5:4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rodionova I., Gao Y., Sastry A., Hefner Y., Lim H., Rodionov D., Saier M., Palsson B.. Identification of a transcription factor, PunR, that regulates the purine and purine nucleoside transporter punC in E. coli. Commun. Biol. 2021; 4:991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Camarena L., Poggio S., Garcia N., Osorio A.. Transcriptional repression of gdhA in Escherichia coli is mediated by the Nac protein. FEMS Microbiol. Lett. 1998; 167:51–56. [DOI] [PubMed] [Google Scholar]
- 36. Muse W.B., Bender R.A.. The nac (nitrogen assimilation control) gene from Escherichia coli. J. Bacteriol. 1998; 180:1166–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rhee H.S., Pugh B.F.. ChIP-exo method for identifying genomic location of DNA-binding proteins with near-single-nucleotide accuracy. Curr. Protoc. Mol. Biol. 2012; Chapter 21:Unit 21.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Keseler I.M., Mackie A., Peralta-Gil M., Santos-Zavaleta A., Gama-Castro S., Bonavides-Martínez C., Fulcher C., Huerta A.M., Kothari A., Krummenacker M.J.N.a.r.. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013; 41:D605–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cho B.K., Knight E.M., Palsson B.O.. PCR-based tandem epitope tagging system for Escherichia coli genome engineering. BioTechniques. 2006; 40:67–72. [DOI] [PubMed] [Google Scholar]
- 40. Datta S., Costantino N., Court D.L.J.G.. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006; 379:109–115. [DOI] [PubMed] [Google Scholar]
- 41. Kim D., Hong J.S.-J., Qiu Y., Nagarajan H., Seo J.-H., Cho B.-K., Tsai S.-F., Palsson B.Ø.. Comparative analysis of regulatory elements between Escherichia coli and Klebsiella pneumoniae by genome-wide transcription start site profiling. PLoS Genet. 2012; 8:e1002867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Powell B.S., Court D.L., Inada T., Nakamura Y., Michotey V., Cui X., Reizer A., Saier M.H. Jr, Reizer J. Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli. Enzyme IIANtr affects growth on organic nitrogen and the conditional lethality of an erats mutant. J. Biol. Chem. 1995; 270:4822–4839. [DOI] [PubMed] [Google Scholar]
- 43. Cho B.K., Kim D., Knight E.M., Zengler K., Palsson B.O.. Genome-scale reconstruction of the sigma factor network in Escherichia coli: topology and functional states. BMC Biol. 2014; 12:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Levin J.Z., Yassour M., Adiconis X., Nusbaum C., Thompson D.A., Friedman N., Gnirke A., Regev A.. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods. 2010; 7:709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Love M.I., Huber W., Anders S.J.G.b.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ebrahim A., Lerman J.A., Palsson B.O., Hyduke D.R.J.B.s.b.. COBRApy: constraints-based reconstruction and analysis for Python. BMC Syst. Biol. 2013; 7:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schellenberger J., Palsson B.Ø.J.J.o.b.c.. Use of randomized sampling for analysis of metabolic networks. J. Biol. Chem. 2009; 284:5457–5461. [DOI] [PubMed] [Google Scholar]
- 50. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A.J.N.. Highly accurate protein structure prediction with AlphaFold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Evans R., O’Neill M., Pritzel A., Antropova N., Senior A., Green T., Žídek A., Bates R., Blackwell S., Yim J.et al.. Protein complex prediction with AlphaFold-Multimer. 2022; bioRxiv doi:10 March 2022, preprint: not peer reviewed 10.1101/2021.10.04.463034. [DOI]
- 52. Cho B.-K., Zengler K., Qiu Y., Park Y.S., Knight E.M., Barrett C.L., Gao Y., Palsson B.Ø.J.N.b.. The transcription unit architecture of the Escherichia coli genome. Nat.Biotechnol. 2009; 27:1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pomposiello P.J., Janes B.K., Bender R.A.. Two roles for the DNA recognition site of the Klebsiella aerogenes nitrogen assimilation control protein. J. Bacteriol. 1998; 180:578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Novichkov P.S., Kazakov A.E., Ravcheev D.A., Leyn S.A., Kovaleva G.Y., Sutormin R.A., Kazanov M.D., Riehl W., Arkin A.P., Dubchak I.J.B.g.. RegPrecise 3.0– a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genom. 2013; 14:745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Studholme D.J., Buck M.. The biology of enhancer-dependent transcriptional regulation in bacteria: insights from genome sequences. FEMS Microbiol. Lett. 2000; 186:1–9. [DOI] [PubMed] [Google Scholar]
- 56. Huo Y.X., Nan B.Y., You C.H., Tian Z.X., Kolb A., Wang Y.P.. FIS activates glnAp2 in Escherichia coli: role of a DNA bend centered at -55, upstream of the transcription start site. FEMS Microbiol. Lett. 2006; 257:99–105. [DOI] [PubMed] [Google Scholar]
- 57. Doucette C.D., Schwab D.J., Wingreen N.S., Rabinowitz J.D.J.N.c.b.. α-Ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat. Chem. Biol. 2011; 7:894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bren A., Park J.O., Towbin B.D., Dekel E., Rabinowitz J.D., Alon U.J.S.r.. Glucose becomes one of the worst carbon sources for E.coli on poor nitrogen sources due to suboptimal levels of cAMP. Sci. Rep. 2016; 6:24834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rabinowitz J.D., Silhavy T.J.J.N.. Metabolite turns master regulator. Nature. 2013; 500:283–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Andersen L., Kilstrup M., Neuhard J.J.A.o.m.. Pyrimidine, purine and nitrogen control of cytosine deaminase synthesis in Escherichia coli K12. Involvement of the GlnlG and PurR genes in the regulation of codA expression. Arch. Microbiol. 1989; 152:115–118. [DOI] [PubMed] [Google Scholar]
- 61. Ikeda T.P., Shauger A.E., Kustu S.. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J. Mol. Biol. 1996; 259:589–607. [DOI] [PubMed] [Google Scholar]
- 62. Humbert R., Simoni R.J.J.o.b.. Genetic and biomedical studies demonstrating a second gene coding for asparagine synthetase in Escherichia coli. J.Bacteriol.Res. 1980; 142:212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Orth J.D., Thiele I., Palsson B.Ø.J.N.b.. What is Flux Balance Analysis?. Nat. Biotechnol. 2010; 28:245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Santos-Zavaleta A., Salgado H., Gama-Castro S., Sánchez-Pérez M., Gómez-Romero L., Ledezma-Tejeida D., García-Sotelo J.S., Alquicira-Hernández K., Muñiz-Rascado L.J., Peña-Loredo P.J.N.a.r.. RegulonDB v 10.5: tackling challenges to unify classic and high throughput knowledge of gene regulation in E. coli K-12. Nucleic Acids Res. 2019; 47:D212–D220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rychel K., Decker K., Sastry A.V., Phaneuf P.V., Poudel S., Palsson B.O.J.N.a.r.. iModulonDB: a knowledgebase of microbial transcriptional regulation derived from machine learning. Nucleic Acids Res. 2021; 49:D112–D120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Seo S.W., Kim D., Szubin R., Palsson B.O.J.C.R.. Genome-wide reconstruction of OxyR and SoxRS transcriptional regulatory networks under oxidative stress in Escherichia coli K-12 MG1655. Cell Rep. 2015; 12:1289–1299. [DOI] [PubMed] [Google Scholar]
- 67. Park J.Y., Rimal H., Bang I., Nong L.K., Kim D.J.B., Engineering B.. Genome-wide identification of DNA-protein interaction to reconstruct bacterial transcription regulatory network. Biotechnol. Bioprocess Eng. 2020; 25:944–954. [Google Scholar]
- 68. Dillon S.C., Dorman C.J.. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010; 8:185–195. [DOI] [PubMed] [Google Scholar]
- 69. Bettenbrock K., Sauter T., Jahreis K., Kremling A., Lengeler J.W., Gilles E.-D.J.J.o.b.. Correlation between growth rates, EIIACrr phosphorylation, and intracellular cyclic AMP levels in Escherichia coli K-12. J. Bacteriol. Res. 2007; 189:6891–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. You C., Okano H., Hui S., Zhang Z., Kim M., Gunderson C.W., Wang Y.-P., Lenz P., Yan D., Hwa T.J.N.. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature. 2013; 500:301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. McCloskey D., Gangoiti J.A., King Z.A., Naviaux R.K., Barshop B.A., Palsson B.O., Feist A.M.. A model-driven quantitative metabolomics analysis of aerobic and anaerobic metabolism in E. coli K-12 MG1655 that is biochemically and thermodynamically consistent. Biotechnol. Bioeng. 2014; 111:803–815. [DOI] [PubMed] [Google Scholar]
- 72. Chang R.L., Andrews K., Kim D., Li Z., Godzik A., Palsson B.O.J.S.. Structural systems biology evaluation of metabolic thermotolerance in Escherichia coli. Science. 2013; 340:1220–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole dataset of ChIP-exo and RNA-seq has been deposited to GEO (https://www.ncbi.nlm.nih.gov/geo/) with the accession number of GSE54905. Scripts for genome-scale metabolic models, which are used to predict nitrogen uptake and to simulate constraint-based models using HPLC for extracellular metabolites concentration, are freely available at the public GitHub repository (https://github.com/SBML-Kimlab/Nitrogen_GSM) and on Zenodo at https://doi.org/10.5281/zenodo.7508655.