Abstract

Highly sensitive detection of microRNAs (miRNAs) is of great significance in early diagnosis of cancers. Here, we develop a palindrome-embedded hairpin structure and its target-catalyzed padlock cyclization for rolling circle amplification, named PHP-RCA for simplicity, which can be applied in label-free ultrasensitive detection of miRNA. PHP-RCA is a facile system that consists of only an oligonucleotide probe with a palindrome-embedded hairpin structure (PHP). The two ends of PHP were extended as overhangs and designed with the complementary sequences of the target. Hence, the phosphorylated PHP can be cyclized by T4 DNA ligase in the presence of the target that serves as the ligation template. This ligation has formed a palindrome-embedded dumbbell-shaped probe (PDP) that allows phi29 polymerase to perform a typical target-primed RCA on PDP by taking miRNA as a primer, resulting in the production of a lengthy tandem repeat. Benefits from the palindromic sequences and hairpin-shaped structure in padlock double-stranded structures can be infinitely produced during the RCA reaction and provide numerous binding sites for SYBR Green I, a double-stranded dye, achieving a sharp response signal for label-free target detection. We have demonstrated that the proposed system exhibits a good linear range from 0.1 fM to 5 nM with a low detection limit of 0.1 fM, and the non-target miRNA can be clearly distinguished. The advantages of high efficiency, label-free signaling, and the use of only one oligonucleotide component make the PHP-RCA suitable for ultrasensitive, economic, and convenient detection of target miRNAs. This simple and powerful system is expected to provide a promising platform for tumor diagnosis, prognosis, and therapy.
1. Introduction
Biomarkers are a kind of small or macromolecular substance that can reflect specific disease information, including but not limited to proteins,1,2 nucleic acids,3 metabolites, and hormones, which is crucial in the early diagnosis, prognosis, and treatment of numerous human diseases such as cancers.4 In the 21st century, with the rapid development of molecular biology, immunology, and genomics, a tumor biomarker (TM) has attracted great attention owing to widespread applications in patient assessment in a variety of clinical settings, such as risk assessment, early diagnosis, drug efficacy assessment, and prognostic diagnosis.5,6
MicroRNAs (miRNAs) are a class of small (∼22 bases) non-coding RNA with the ability to regulate gene expression.7−9 Accumulating experience has revealed that the aberrant expression of certain miRNAs is closely associated with plentiful human diseases, such as Alzheimer’s disease,10 diabetes mellitus,11 and, in particular, highly fatal cancers.12,13 It is intrinsically because of the critical roles of miRNAs across the cellular processes of cancer closely related to initiation and progression including the proliferation, apoptosis, and differentiation.14 The representative case is that the miR-21 has been found to be overexpressed in many cancers, such as chronic lymphocytic leukemia,15 acute myeloid leukemia,16 glioblastoma,17−19 breast cancer,20 and prostate cancer,21 and miR-21 has been designated as a nonspecific biomarker of all maladies.22 Accordingly, miRNAs are considered as important tumor markers to evaluate the initiation, stage, and treatment of tumors, and the miRNA expression assay is of significance for early diagnosis and prognosis of tumors.23,24 However, the miRNA detection remains a great challenge owing to the intrinsic properties of miRNAs, such as a low expression abundance, short length, and sequence homology.23 In general, the conventional methods including quantitative real-time PCR (qRT-PCR), northern blotting, and microarrays have been developed and reached impressive progress for miRNA assays over the past decades; however, inevitable imperfections, such as poor repeatability and sensitivity of the northern blotting, PCR needing a strict and complicated thermal cycler, and requiring trained technicians in microarrays, obviously limits the applications in practical analysis. Therefore, there is an imperative need for developing novel methods with high sensitivity, simplicity, and convenience to reliably detect miRNAs for early diagnosis and prognosis of tumors.
Strand displacement amplification (SDA), for instance, has been reported to be an efficient process that enables rapidly accumulating a mass of nucleic acid sequences under catalysis of various enzymes known as the powerful toolboxes of DNA, and numerous SDA-based methods also have been successfully applied in amplified miRNA detection.25−29 Despite having reached high sensitivity and efficiency, the cost efficiency and stability are compromised due to the fact that SDA-based reactions are always dependent on more than two, even three, kinds of enzymes, such as the Klenow fragment, endonuclease, and Nb.BbvCI, which work together in a precisely combined manner.30,31 Noteworthy, rolling circle amplification (RCA) is a method that enable performs amplification reactions based on a single circular padlock probe and one phi29 enzyme to produce a lengthy tandem repeat sequence.32−34 In spite of the fact that the RCA reaction is powered purely depending on the phi29 enzyme, the impressive amplified capability of RCA has been used to detect targets of interest that are expressed at a trace level, indicating that the RCA-based method is suitable for the development of a sensing platform toward miRNA detection.35,36
The fluorophore- and quencher-modified probes were practicable for signaling in the nucleic acid-based system, and by contrast, a label-free system without any modified groups further decreases the synthesis period of probes and the detection cost. SYBR Green is a kind of luminescent dye that works by inserting into double-stranded DNA while quenched under other circumstances. Theoretically speaking, the efficient accumulation of double-stranded DNA and use of SYBR Green as an indicator are enable us to develop a cost-effective and rapid approach for target detection especially in some unforeseeable emergency circumstances or in resource-limited rural areas.
Consequently, we herein design a palindrome-embedded hairpin structure and its target-catalyzed padlock cyclization for target-primed rolling circle amplification (PHP-RCA), which can be applied in label-free ultrasensitive detection of miRNA. Palindrome fragments are DNA sequences whose base order is the same whether they are read from 5′ to 3′ ends or vice versa and play important roles in biological processes, including gene expression, transcription, chromosome translocation, and so on.37−39 Palindromic sequences have the unique property of hybridizing with each other to form double chains, which provided the potential for exploiting low-cost and rapid sensors.40,41 In this work, a palindrome-embedded hairpin structure (PHP) was designed in the initial state that enables efficiently transforming the single-stranded RCA product into double-stranded DNA and allowing the insertion of SYBR Green dye for label-free signaling. Benefits of a high-efficiency of RCA, a label-free response mechanism, and the use of only one oligonucleotide component, make the PHP-RCA suitable for ultrasensitive, economic, and convenient detection of target miRNAs. This simple and powerful design is expected to develop a promising platform in the ultrasensitive detection of miRNAs to be used for tumor diagnosis, prognosis, and therapy.
2. Results and Discussion
2.1. Principle of the PHP-RCA for Label-Free Target Detection
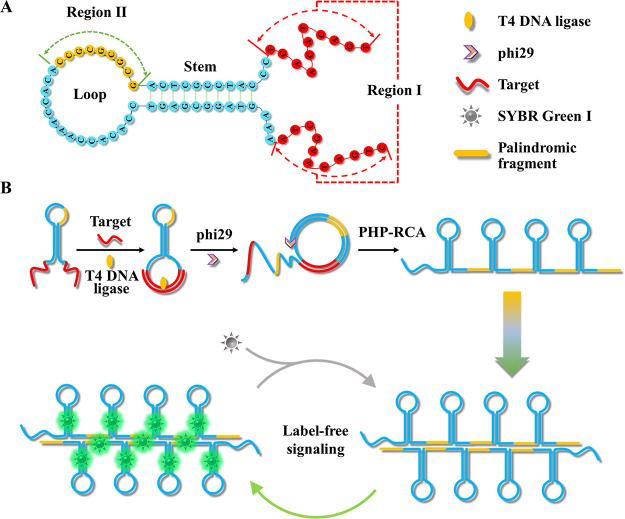
PHP-RCA is a facile system that consists of only an oligonucleotide probe with a palindrome-embedded hairpin structure (PHP). In this work, PHP is structurally designed to a hairpin structure, which contains two functional regions as follows: two overhangs with the complementary sequence of the target at the two ends for target recognition and initiating the reaction (region I) and a palindromic fragment embedded in the loop of PHP for enhancing the self-hybridization of RCA production (region II). The detailed sequence of PHP is listed in Table S1. The molecule structure of PHP as well as the locations of regions I and II are depicted in Scheme 1A. Under natural conditions, the PHP spontaneously folds into a hairpin-shaped structure attached with two overhangs that move freely. Upon being introduced to the target (Scheme 1B), the two overhangs can be pulled adjacently and form another loop-shaped structure for benefiting the subsequent ligation by the specific hybridization between the target and region I. After the ligation by the T4 DNA ligase, PHP was transformed into a dumbbell-shaped probe (PDP) that allows phi29 polymerase to perform a typical target-primed rolling circle amplification by taking the target as a primer, resulting in the production of a lengthy tandem repeat. Benefiting from the initial hairpin-shaped structure in PHP, namely, the stem configuration in PDP, the RCA products folded into a continuous hairpin-shaped secondary structure during the RCA reaction. Additionally, repeated palindromic fragments in RCA products that originate from the design of palindromic sequences embedded in PHP further promote the self-hybridization of RCA production or the hybridization between different molecules. In this case, the massive double-stranded structures provide numerous binding sites for SYBR Green I, a double-stranded dye, to produce a sharp response signal. Considering the high efficiency of RCA, the label-free response mechanism, and the use of only one oligonucleotide component, the PHP-RCA is suitable for ultrasensitive, economic, and convenient detection of target miRNAs.
Scheme 1. (A) Molecule Structure of PHP and Functional Region Analysis and (B) the Principle of PHP-RCA for Label-Free Ultrasensitive Detection of Target miRNA.
2.2. Feasibility of the PHP-RCA Sensing System for Target miR-21 Detection
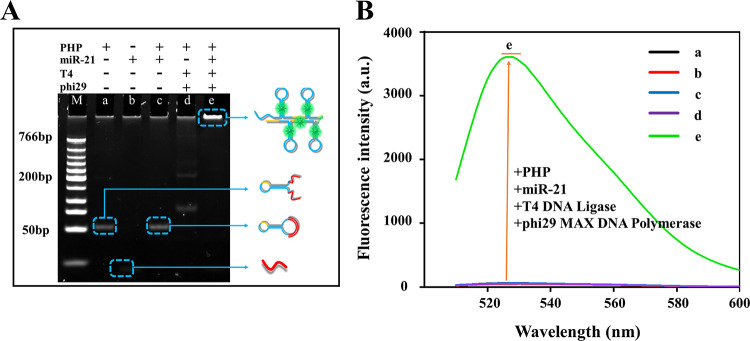
Accumulating evidence has revealed that microRNA-21 (miR-21) is closely associated with various human cancers, inflammation, and cardiovascular diseases, and miR-21 has been considered as a crucial and accepted biomarker for the diagnosis and therapy of different human diseases.42,43 Hence, we chose miR-21 as the target model for exploring the performance of the PHP-RCA system for miRNA detection. The feasibility of the PHP-RCA system for the label-free amplified detection of miR-21 was first investigated by 8% native polyacrylamide electrophoresis (PAGE). Five samples were studied, and the specified components are listed in Figure 1A. In this section, SYBR Green I was used as an indicator to show the DNA structures. A fast-moving and shallow band is observed in sample b, indicating the natural feature of a short (22 bases) and rare secondary structure in miR-21. The hairpin-shaped PHP in sample a displays a slower-moving and brighter band due to the longer sequences (71 bases) and the stem structure that provided more binding sites for SYBR Green I. After hybridizing with target miR-21, no intrinsic difference of the band in sample c can be observed because of the similar structure of the PHP/miR-21 duplex compared with PHP in sample a. The PHP-based RCA reaction indeed occurred, as evidenced by the appearance of a bright band at the top lane (sample e) due to the double-stranded lengthy tandem repeat of RCA production. On the contrary, the RCA production cannot be detected in the absence of the target (sample d). Moreover, the four samples were further characterized by 1% agarose gel electrophoresis in Figure S1 and produced the same results. For quantification analysis, samples a–e in Figure 1A were scanned for fluorescence as shown in Figure 1B. The fluorescence spectra results show that the RCA reaction cannot occur without either the phi29-based polymerase or target miR-21, as supported by the weak and almost negligible signal intensities of curves a–d. As expected, the free ends in PHP were ligated to form the dumbbell structure, thus triggering the rolling circle amplification reaction by phi29 polymerase, and we received a high signal response (sample e) when the reaction mixture coexisted with miR-21 and phi29 enzymes. As shown in Figure S2, a remarkable fluorescence intensity can be observed only when using the integrated PHP structure as the probe, while we obtained a weak signal when using the defective PHP probe that lacks the hairpin and/or palindromic fragment. These results have strongly proved the feasibility of PHP-RCA for label-free detection of miR-21.
Figure 1.
Feasibility of the PHP-RCA-based sensing system for the detection of miR-21. (A) Native PAGE (8%) analysis of various reaction mixtures: (a) miR-21, (b) PHP, (c) PHP + miR-21, (d) PHP + T4 DNA ligase + phi29 polymerase, and (e) PHP + miR-21 + T4 DNA ligase + phi29 polymerase. The sensing system was 40 μL, and the concentrations of the target miR-21, PHP, T4 DNA ligase, phi29 polymerase, and dNTPs are 5 nM, 5 nM, 4 U/μL, 0.1 U/μL, and 10 mM, respectively. (B) Fluorescence spectra of the same reaction mixtures from A. SYBR Green I was used as an indicator.
2.3. Optimization of the Reaction System
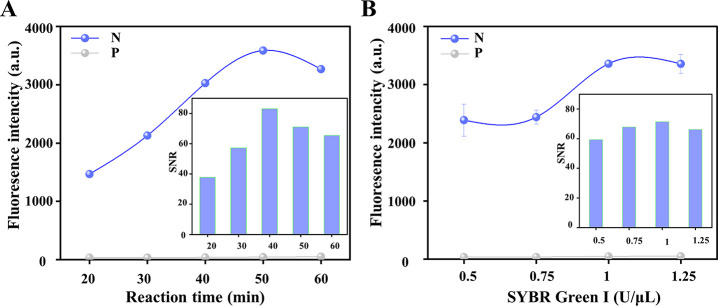
The signal transduction efficiency and accuracy of the sensor platform are greatly affected by experimental conditions. Considering the optimal activity of the phi29 enzyme, the reaction temperature and buffer of the RCA reaction are separately chosen to be 30 °C and phi29 buffer, respectively, which is also consistent with previous reports.44,45 To achieve optimal stimuli–response and economic performances, two related experimental parameters were investigated. The reaction time is an important parameter to ensure the low background and high signal of the entire experiment, and therefore, the reaction time was first optimized. As shown in Figure 2A, the response signal increased with the increase of the reaction time from 20 to 50 min then slightly decreased at 60 min, which may be due to the generated polymer that formed flocs and affected the stability of the fluorescence signal, indicating that the fluorescence intensity has reached a plateau at 50 min. One can notice that the background fluorescence gradually and slightly increased when the reaction time increased from 20 to 60 min. Taken together, the signal-to-noise ratio (SNR), which refers to the ratio of the fluorescence peak generated by the target to the background fluorescence, reached the highest value within 40 min and demonstrated a rapid detection capacity of the system. Therefore, a reaction time of 40 min was adopted in the following experiments. For SYBR Green I, the crucial indicator for signaling, the concentration of this indicator was also optimized. As shown in Figure 2B, the target-initiated signal gradually increased with the concentration of SYBR Green I, increasing from 0.5 to 1.25 U/μL, and reached a plateau at 1 U/μL. Considering the slightly increased background, the SNR reached the peak value at 1 U/μL that was chosen as the optimal concentration.
Figure 2.
Effects of (A) the RCA reaction time and (B) the concentration of SYBR Green I on the performance of the PHP-RCA sensing system. The concentration of target miR-21, PHP, T4 DNA ligase, phi29 polymerase, and dNTPs are 5 nM, 5 nM, 4 U/μL, 0.1 U/μL, and 10 mM, respectively. Error bars represent the standard deviations (SD, n = 3). The signal-to-noise ratio (SNR) refers to the ratio of the fluorescence peak generated by the target to the background fluorescence. “N” and “P” denote negative samples without miR-21 and positive samples with miR-21, respectively.
2.4. Sensitivity of the Assay
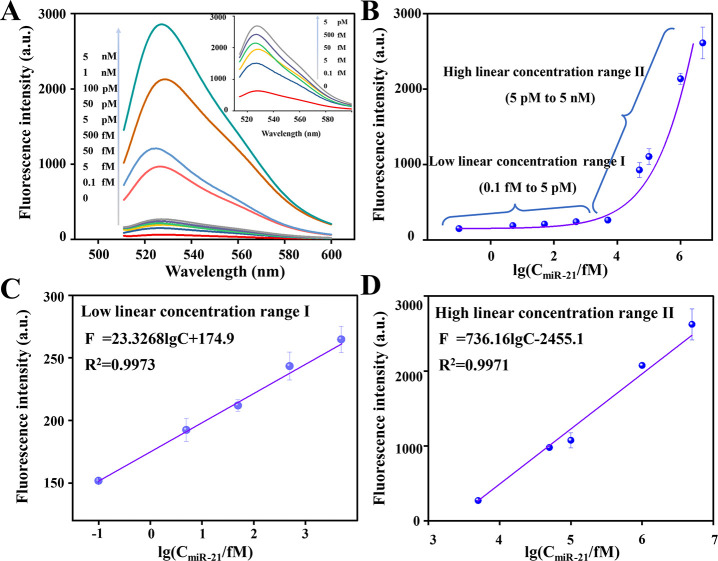
Sensitivity is an important parameter for evaluating the performance of a sensing system.46,47 To evaluate the analytical performance of PHP-RCA for quantitatively detecting miR-21, we measured the fluorescence intensity of the system presented with different concentrations of miR-21 under optimized experimental conditions. Figure 3A plots the typical fluorescence spectra of the assay for miR-21 with concentrations of 0, 1 × 10–16, 5 × 10–15, 5 × 10–14, 5 × 10–13, 5 × 10–12, 5 × 10–11, 1 × 10–10, 1 × 10–9, and 5 × 10–9 M. Inset is the fluorescence spectra of PHP-RCA for detecting miR-21 at low concentrations. Figure 2B depicts two different dose–response curves, namely, the low linear concentration range I and high linear concentration range II, that are further presented in Figure 2C,D, respectively. In the low-concentration range from 0.1 fM to 5 pM, the fluorescence intensity and the logarithm of the miRNA-21 concentration display a linear relationship, and the regression equation is F = 23.3268logC + 174.9 with a correlation coefficient (R2) of 0.9973. Similarly, in the high target concentration range from 5 pM to 5 nM, the linear regression equation between the signal and target concentration can be expressed as F = 736.16logC – 2455.1 with a correlation coefficient R2 of 0.9971. The symbols F and C in the formula represent the fluorescence signal intensity and target concentration, respectively. Noteworthy, as low as 0.1 fM miR-21 is able to produce an observable signal compared with the blank and thus 0.1 fM is defined as the detection limit (LOD). As shown in Table S2, a desirable assay sensitivity is exhibited by further comparison with previous studies.
Figure 3.
Capability of the PHP-RCA for target miR-21 detection. (A) Fluorescence spectra of the PHP-RCA system upon addition of different concentrations of target miR-21, ranging from 0 to 5 nM. Inset: fluorescence spectra in the presence of a low target concentration of the miR-21. (B) Dependence of the peak fluorescence response on the concentration of target miR-21 in a range from 0.1 fM to 5 nM. Linear response of the sensing system at (C) the low and (D) high target concentration ranges. Error bars are standard deviations from three repetitive experiments.
2.5. Specificity of the PHP-RCA Sensor
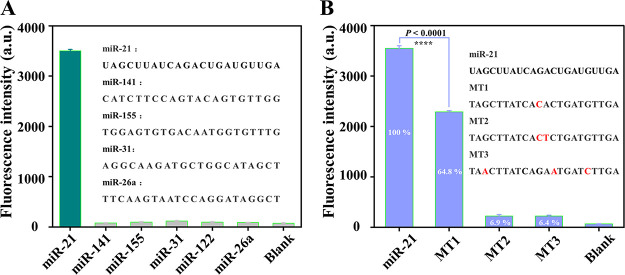
The challenges for miRNA detection are greatly due to the high homology of endogenic miRNA, and the accuracy is important for diagnosis and treatment, especially in the early diagnosis of tumors. Therefore, the specificity is another crucial parameter to evaluate the performance of an assay system for miRNA detection. To investigate the selectivity of the PHP-RCA system for miR-21 detection, the signal response toward other non-target miRNAs, including miR-141, miR-155, miR-31, miR-122, and miR-26a, were also detected under the same conditions. As presented in Figure 4A, non-target miRNAs can hardly induce an observed signal, while the response signal toward miR-21 produces a sharp increasement. The results demonstrated that the PHP-RCA-based sensing system clearly discriminates target miRNA from other non-target miRNAs. For mutation detection, the target sequences with one, two, and three mutation points (named MT1, MT2, and MT3 for simplicity) were chosen as targets to initiate the PHP-RCA-based reaction. As shown in Figure 4B, the fluorescence intensity induced by MT1, MT2, and MT3 are 64.8, 6.9, and 6.4%, respectively, if the fluorescence signal of miR-21 is defined as 100%. The detectable signal of MT1 suggests that this method could not distinguish these closely related analytes in a biological sample where the concentrations are unknown. Nonetheless, the PHP-RCA sensor shows a competitive performance compared with other reported methods48−50 that similarly failed to distinguish such analytes. All of the results above have strongly proven the high specificity of the proposed PHP-RCA method.
Figure 4.
(A) Fluorescence signal induced by other miRNA (miR-141, miR-155, miR-31, miR-122, and miR-26a) and (B) target miR-21 with one, two, and three mutation points (MT1, MT2, and MT3, respectively) against miR-21 at a concentration of 5 nM. The fluorescence signal-to-noise ratio was calculated by F/F0 where F and F0 represent the fluorescence intensity of PHP-RCA in the presence and absence of target miR-21, respectively. The error bar represents the standard deviation (SD) estimated from three replicate measurements. ****P < 0.0001, independent-sample t-test.
2.6. Application of the PHP-RCA Sensor in Real-Sample Analysis
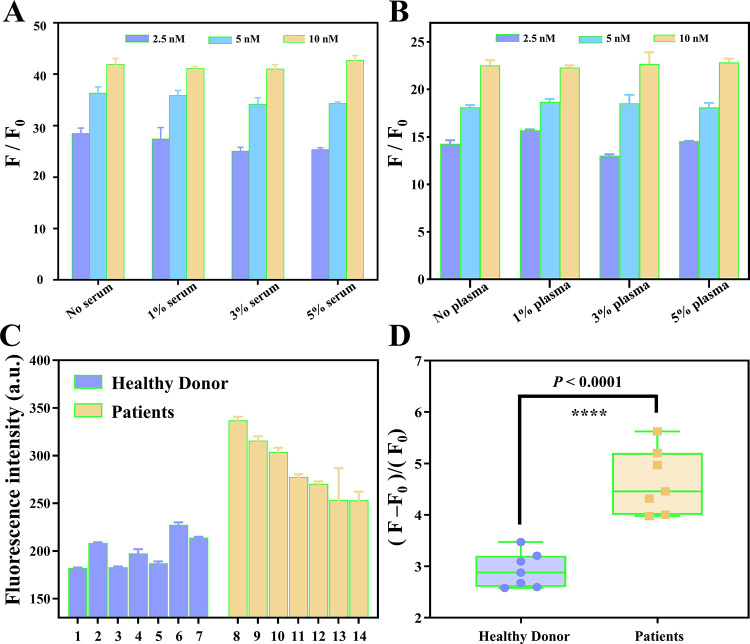
DNA probes have shown great potential in tumor diagnosis, prognosis, and therapy; however, the intrinsic properties of DNA of susceptibility to nuclease degradation have hampered their clinical application. To validate the practical applicability of the PHP-RCA system, miR-21 was detected in human serum and plasma in this work. The fluorescence intensity of the PHP-RCA system presented with three different concentrations (2.5 nM, 5 nM, and 10 nM) of miR-21 was first measured in the reaction buffer without the serum or plasma, and the reaction in the absence of target miR-21 was used as a blank under the same optimized experimental conditions. Then, the practical application of the PHP-RCA was investigated by incubating the same samples with different concentrations of the human serum and plasma. As the result presents in Figure 5A,B, a gradually increased signal-to-noise ratio (SNR) can be observed when the PHP-RCA system presented an increased concentration of miR-21 in the reaction buffer without the serum or plasma. Noteworthy, the human serum and plasma have almost no influence on the performance of the PHP-RCA system as supported by the similar SNRs in the different concentrations of the serum or plasma from 1 to 5%. The results show that the proposed sensing system possesses a satisfactory stability in complex biological environments and potential for clinical application. To directly evaluate the performance of the method for detecting real samples, here, we detected several clinical samples, including seven healthy donors and seven patients. As shown in Figure 5C,D, samples from patients that produced significantly stronger signal intensities than those from healthy donors and the data exhibit a statistically significant difference, demonstrating that the proposed method is reliable for real-sample assay.
Figure 5.
Performance of PHP-RCA for miR-21 detection in the (A) serum and (B) plasma. (C, D) Performance of PHP-RCA for miR-21 detection in real samples. F and F0 represent the fluorescence intensity of PHP-RCA in the presence and absence of the target, respectively. The error bar represents the standard deviation (SD) estimated from three replicate measurements. ****P < 0.0001, independent-sample t-test.
3. Conclusions
In summary, here, we report a palindrome-embedded hairpin structure and its target-catalyzed padlock cyclization for rolling circle amplification and applied it in label-free ultrasensitive detection of miRNA. The system named PHP-RCA consists of only an oligonucleotide probe with a palindrome-embedded hairpin structure (PHP). Benefits from the design of palindromic sequences and the hairpin-shaped structure in the padlock produced double-stranded structures that provide numerous binding sites for SYBR Green I, a double-stranded dye, to achieve label-free target detection. Advantages of a high-efficiency of the RCA reaction that is powered by one phi29 polymerase, a label-free stimuli–response mechanism, and a single oligonucleotide component make the PHP-RCA suitable for ultrasensitive, economic, and convenient detection of target miRNAs. Specifically, a single probe system integrated the primer, templates, and target recognition region, and there is no need for additional modified fluorescence or quenching groups, thus simplifying the experimental design and steps to a great extent. Second, the reaction does not require a strict temperature control system or heating equipment, leading to a simple and cost-efficient detection. Third, the RCA-based reaction endows the system highly efficient signal amplification. We have demonstrated that the proposed PHP-RCA system exhibits a good linear range from 0.1 fM to 5 nM with a low detection limit of 0.1 fM and the non-target miRNA can be clearly distinguished. Moreover, we also validated the practical applicability of the PHP-RCA by detecting miR-21 in human serum in this work, resulting in negligible influence on the performance of the PHP-RCA system. This simple and powerful platform is expected to provide promising potential in the ultrasensitive detection of miRNAs for tumor diagnosis, prognosis, and therapy.
4. Materials and Methods
4.1. Material and Reagents
All oligonucleotides (Table S1) in this study were HPLC-purified and synthesized by Shanghai Sangon Biological Engineering Technology and Services Co., Ltd. (Shanghai, China). The secondary structures of designed sequences were predicted using online bioinformatic software (NUPACK: Nucleic Acid Package). All DNAs were dissolved in 1× TE buffer (10 mM Tris, 1 mM EDTA, PH 8.0), and the concentration was determined by a NanoDrop 2000 (Thermo Fisher Scientific, USA). Adenosine triphosphate (ATP), T4 DNA ligase, and 10× T4 DNA ligase buffer were provided by Takara Biotechnology Co., Ltd. (Dalian, China). T4 polynucleotide kinase (PNK) and 10× PNK buffer were obtained from Thermo Scientific (MA, USA), and phi29 MAX DNA polymerase and the corresponding reaction buffer (10×) were provided by Vazyme Biotech Co.,Ltd. (Nanjing, China). A low-molecular-weight DNA ladder and deoxyribonucleoside 5′-triphosphate (dNTP) mixture were all supplied by Shanghai Sangon Biological Engineering Technology and Services Co. Ltd. (Shanghai, China). SYBR Green I was purchased from Solarbio Biotechnology Co., Ltd. (Beijing, China). All chemicals were of analytical grade without further purification and dissolved in ultrapure water produced by the MiNiQ-Direct 8 Laboratory Small Pure Water System (electrical resistance of 18.25 MΩ cm at 25 °C) that was purchased from Merck (Merck, GER).
4.2. Instruments
Fluorescence scanning was performed by a Hitachi F-7000 spectrometer and FL fluorescence software. Native polyacrylamide gel electrophoresis and agarose electrophoresis were executed on an electrophoresis apparatus (Bio-Rad, USA) and imaged by a ChemiDoc XRS imaging apparatus (Bio-Rad, USA).
4.3. Gel Electrophoresis Analysis
For gel electrophoresis analysis, 8% native polyacrylamide electrophoresis (PAGE) or 1% agarose gel electrophoresis was freshly prepared and used to verify the rolling circle products. The samples were prepared by mixing 8 μL of the reaction mixture, 2 μL of 6× loading buffer, and 2 μL of 10 × SYBR Green I. After loading into the well, the native PAGE was run under a constant voltage of 80 V in 0.5× TBE buffer (4.5 mM Tris, 4.5 mM boric acid, 0.1 mM EDTA, pH 7.9), while agarose gel was conducted on a constant voltage of 100 V in 0.5× TBE buffer.
4.4. Phosphorylation, Ligation, and PHP-RCA Reaction
The palindrome-embedded hairpin structure (PHP) was first 5′-phosphorylated using PNK. Specifically, 177 μL of PHP was mixed with 20 μL of 10× PNK buffer, 2 μL of ATP (100 mM), and 1 μL of PNK and incubated at 37 °C for 2 h. Subsequently, the mixture was heated at 90 °C for 10 min to terminate the reaction. Finally, the concentration of 5′-phosphorylated PHP (PPHP) was concentrated to 1 μM and stored at 4 °C before usage.
The reaction mixture for target detection was prepared by mixing PPHP (1 μM, 1 μL), miR-21 (1 μM, 1 μL), 2 μL 10× T4 DNA ligase buffer (660 mM Tris–HCl, 66 mM MgCl2, 100 mM DTT, 1 mM ATP, pH 7.6), and 5 U T4 DNA ligase, and we added ddH2O to a final volume of 20 μL. The ligation reaction was carried out at 16 °C for 2 h to form a palindrome-embedded dumbbell-shaped probe (PDP). After that, 2.5 μL of 10 mM dNTPs, 2.5 U phi29 MAX DNA polymerase, and 4 μL of 10× phi29 MAX DNA polymerase buffer was added and diluted with ddH2O to a final volume of 40 μL then incubated at 30 °C for 40 min. Finally, the mixture was heated at 65 °C for 10 min to inactivate the enzymes and gradually lowered down to room temperature.
4.5. Fluorescence Detection
The sample for fluorescence scanning was prepared by mixing the reaction solution (40 μL) and 40 μL of 10× SYBR Green I for 10 min at room temperature then diluting with 120 μL of 1× phi 29 MAX DNA polymerase buffer to a final volume of 200 μL. Fluorescence measurements were performed using a Hitachi F-7000 fluorescence spectrometer (Hitachi, Ltd., Japan) with a xenon lamp as the excitation light source. We set an excitation wavelength of 492 nm and collected the spectra between 500 and 600 nm at a scan rate of 240 nm/min with the integration time of 0.5 s. The slit widths of both emission and excitation were consistent at 5 nm. The maximum peak of the fluorescence emission at 526 nm was employed to evaluate the response capability for the sensing system.
Acknowledgments
This research was financially supported by the Taizhou Science and Technology Bureau (grant no. 22ywb148).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c06532.
DNA oligonucleotide sequence designed in this work, agarose gel electropherogram on feasibility, fluorescence derived from linear RCA using four different functional structures of MB, and a comparison with previous works reported in the open literature (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Yu Z.; Gong H.; Xu J.; Li Y.; Zeng Y.; Liu X.; Tang D. Exploiting Photoelectric Activities and Piezoelectric Properties of NaNbO3 Semiconductors for Point-of-Care Immunoassay. Anal. Chem. 2022, 94, 3418–3426. 10.1021/acs.analchem.2c00066. [DOI] [PubMed] [Google Scholar]
- Gao Y.; Zeng Y.; Liu X.; Tang D. Liposome-Mediated In Situ Formation of Type-I Heterojunction for Amplified Photoelectrochemical Immunoassay. Anal. Chem. 2022, 94, 4859–4865. 10.1021/acs.analchem.2c00283. [DOI] [PubMed] [Google Scholar]
- Zeng R.; Gong H.; Li Y.; Li Y.; Lin W.; Tang D.; Knopp D. CRISPR-Cas12a-Derived Photoelectrochemical Biosensor for Point-Of-Care Diagnosis of Nucleic Acid. Anal. Chem. 2022, 94, 7442–7448. 10.1021/acs.analchem.2c01373. [DOI] [PubMed] [Google Scholar]
- Hayes J.; Peruzzi P. P.; Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- He B.; Lang J.; Wang B.; Liu X.; Lu Q.; He J.; Gao W.; Bing P.; Tian G.; Yang J. TOOme: A Novel Computational Framework to Infer Cancer Tissue-of-Origin by Integrating Both Gene Mutation and Expression. Front. Bioeng. Biotechnol. 2020, 8, 394. 10.3389/fbioe.2020.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Qiu C.; Wang B.; Bing P.; Tian G.; Zhang X.; Ma J.; He B.; Yang J. Evaluating DNA Methylation, Gene Expression, Somatic Mutation, and Their Combinations in Inferring Tumor Tissue-of-Origin. Front. Cell Dev. Biol. 2021, 9, 619330 10.3389/fcell.2021.619330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorge A. L.; Pereira E. R.; de Oliveira C. S.; Ferreira E. D. S.; Menon E. T. N.; Diniz S. N.; Pezuk J. A. MicroRNAs: understanding their role in gene expression and cancer. Einstein 2021, 19, eRB5996 10.31744/einstein_journal/2021RB5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N.; Aharonov R.; Meiri E.; Rosenwald S.; Spector Y.; Zepeniuk M.; Benjamin H.; Shabes N.; Tabak S.; Levy A.; Lebanony D.; Goren Y.; Silberschein E.; Targan N.; Ben-Ari A.; Gilad S.; Sion-Vardy N.; Tobar A.; Feinmesser M.; Kharenko O.; Nativ O.; Nass D.; Perelman M.; Yosepovich A.; Shalmon B.; Polak-Charcon S.; Fridman E.; Avniel A.; Bentwich I.; Bentwich Z.; Cohen D.; Chajut A.; Barshack I. MicroRNAs accurately identify cancer tissue origin. Nat. Biotechnol. 2008, 26, 462–469. 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- Jeffrey S. S. Cancer biomarker profiling with microRNAs. Nat. Biotechnol. 2008, 26, 400–401. 10.1038/nbt0408-400. [DOI] [PubMed] [Google Scholar]
- Zou H. Y.; Guo L.; Zhang B.; Chen S.; Wu X. R.; Liu X. D.; Xu X. Y.; Li B. Y.; Chen S.; Xu N. J.; Sun S. Aberrant miR-339-5p/neuronatin signaling causes prodromal neuronal calcium dyshomeostasis in mutant presenilin mice. J. Clin. Invest. 2022, 132, e149160 10.1172/JCI149160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R.; Liu C.; He X.; Sun P.; Zhang B.; Yang H.; Shi W.; Ruan Q. MicroRNA-21 promotes pancreatic β cell function through modulating glucose uptake. Nat. Commun. 2022, 13, 3545. 10.1038/s41467-022-31317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Ba Y.; Ma L.; Cai X.; Yin Y.; Wang K.; Guo J.; Zhang Y.; Chen J.; Guo X.; Li Q.; Li X.; Wang W.; Zhang Y.; Wang J.; Jiang X.; Xiang Y.; Xu C.; Zheng P.; Zhang J.; Li R.; Zhang H.; Shang X.; Gong T.; Ning G.; Wang J.; Zen K.; Zhang J.; Zhang C. Y. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Chin L. J.; Slack F. J. A truth serum for cancer--microRNAs have major potential as cancer biomarkers. Cell Res. 2008, 18, 983–984. 10.1038/cr.2008.290. [DOI] [PubMed] [Google Scholar]
- Lai C.-Y.; Yeh K.-Y.; Lin C.-Y.; Hsieh Y.-W.; Lai H.-H.; Chen J.-R.; Hsu C.-C.; Her G. MicroRNA-21 Plays Multiple Oncometabolic Roles in the Process of NAFLD-Related Hepatocellular Carcinoma via PI3K/AKT, TGF-β, and STAT3 Signaling. Cancers 2021, 13, 940. 10.3390/cancers13050940. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Schmid V. K.; Khadour A.; Ahmed N.; Brandl C.; Nitschke L.; Rajewsky K.; Jumaa H.; Hobeika E. B-cell antigen receptor expression and phosphatidylinositol 3-kinase signaling regulate genesis and maintenance of mouse chronic lymphocytic leukemia. Haematologica 2022, 107, 1796–1814. 10.3324/haematol.2021.279924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussa Agha D.; Rouas R.; Najar M.; Bouhtit F.; Fayyad-Kazan H.; Lagneaux L.; Bron D.; Meuleman N.; Lewalle P.; Merimi M. Impact of Bone Marrow miR-21 Expression on Acute Myeloid Leukemia T Lymphocyte Fragility and Dysfunction. Cell 2020, 9, 2053. 10.3390/cells9092053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Liang C.; Wong K. C.; Jin K.; Zhang Z. Inferring probabilistic miRNA-mRNA interaction signatures in cancers: a role-switch approach. Nucleic Acids Res. 2014, 42, e76 10.1093/nar/gku182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Zheng M.; Jiao M.; Yan C.; Xu S.; Du Q.; Morsch M.; Yin J.; Shi B. Polymeric nanoparticle mediated inhibition of miR-21 with enhanced miR-124 expression for combinatorial glioblastoma therapy. Biomaterials 2021, 276, 121036 10.1016/j.biomaterials.2021.121036. [DOI] [PubMed] [Google Scholar]
- Sintupisut N.; Liu P. L.; Yeang C. H. An integrative characterization of recurrent molecular aberrations in glioblastoma genomes. Nucleic Acids Res. 2013, 41, 8803–8821. 10.1093/nar/gkt656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan T.; Shastri A. A.; Palagani A.; Buraschi S.; Neill T.; Savage J. E.; Kapoor A.; DeAngelis T.; Addya S.; Camphausen K.; Iozzo R. V.; Simone N. L. miR-21 Plays a Dual Role in Tumor Formation and Cytotoxic Response in Breast Tumors. Cancers 2021, 13, 888. 10.3390/cancers13040888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folini M.; Gandellini P.; Longoni N.; Profumo V.; Callari M.; Pennati M.; Colecchia M.; Supino R.; Veneroni S.; Salvioni R.; Valdagni R.; Daidone M. G.; Zaffaroni N. miR-21: an oncomir on strike in prostate cancer. Mol. Cancer 2010, 9, 12. 10.1186/1476-4598-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenike A. E.; Halushka M. K. miR-21: a non-specific biomarker of all maladies. Biomark Res 2021, 9, 18. 10.1186/s40364-021-00272-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zen K.; Zhang C. Y. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med. Res. Rev. 2012, 32, 326–348. 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- Hilton C.; Karpe F. Circulating microRNAs: what is their relevance?. Clin. Chem. 2013, 59, 729–731. 10.1373/clinchem.2013.203166. [DOI] [PubMed] [Google Scholar]
- Zhang B.; Li S.; Guan Y.; Yuan Y. Accurate Detection of Target MicroRNA in Mixed Species of High Sequence Homology Using Target-Protection Rolling Circle Amplification. ACS Omega 2021, 6, 1516–1522. 10.1021/acsomega.0c05279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.; Yu X.; Wang J.; Liu D.; Wang L.; Liu S. Exonuclease III-Powered Self-Propelled DNA Machine for Distinctly Amplified Detection of Nucleic Acid and Protein. Anal. Chem. 2020, 92, 9764–9771. 10.1021/acs.analchem.0c01197. [DOI] [PubMed] [Google Scholar]
- Liu Y. Q.; Zhang M.; Yin B. C.; Ye B. C. Attomolar ultrasensitive microRNA detection by DNA-scaffolded silver-nanocluster probe based on isothermal amplification. Anal. Chem. 2012, 84, 5165–5169. 10.1021/ac300483f. [DOI] [PubMed] [Google Scholar]
- Liang K.; Wang H.; Li P.; Zhu Y.; Liu J.; Tang B. Detection of microRNAs using toehold-initiated rolling circle amplification and fluorescence resonance energy transfer. Talanta 2020, 207, 120285 10.1016/j.talanta.2019.120285. [DOI] [PubMed] [Google Scholar]
- Li H.; Tang Y.; Song D.; Lu B.; Guo L.; Li B. Establishment of Dual Hairpin Ligation-Induced Isothermal Amplification for Universal, Accurate, and Flexible Nucleic Acid Detection. Anal. Chem. 2021, 93, 3315–3323. 10.1021/acs.analchem.1c00007. [DOI] [PubMed] [Google Scholar]
- Goryunova M. S.; Arzhanik V. K.; Zavriev S. K.; Ryazantsev D. Y. Rolling circle amplification with fluorescently labeled dUTP-balancing the yield and degree of labeling. Anal. Bioanal. Chem. 2021, 413, 3737–3748. 10.1007/s00216-021-03322-7. [DOI] [PubMed] [Google Scholar]
- Li X. Y.; Du Y. C.; Zhang Y. P.; Kong D. M. Dual functional Phi29 DNA polymerase-triggered exponential rolling circle amplification for sequence-specific detection of target DNA embedded in long-stranded genomic DNA. Sci. Rep. 2017, 7, 6263. 10.1038/s41598-017-06594-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng R.; Zhang K.; Wang L.; Ren X.; Sun Y.; Li J. DNA-Sequence-Encoded Rolling Circle Amplicon for Single-Cell RNA Imaging. Chem 2018, 4, 1373–1386. 10.1016/j.chempr.2018.03.003. [DOI] [Google Scholar]
- Deng R.; Zhang K.; Sun Y.; Ren X.; Li J. Highly specific imaging of mRNA in single cells by target RNA-initiated rolling circle amplification. Chem. Sci. 2017, 8, 3668–3675. 10.1039/C7SC00292K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C.; Ouyang P.; Yang Y.; Qing Y.; Han J.; Shang W.; Chen Y.; Du J. MiRNA Detection Using a Rolling Circle Amplification and RNA-Cutting Allosteric Deoxyribozyme Dual Signal Amplification Strategy. Biosensors 2021, 11, 222. 10.3390/bios11070222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng R.; Zhang K.; Li J. Isothermal Amplification for MicroRNA Detection: From the Test Tube to the Cell. Acc. Chem. Res. 2017, 50, 1059–1068. 10.1021/acs.accounts.7b00040. [DOI] [PubMed] [Google Scholar]
- Murakami T.; Sumaoka J.; Komiyama M. Sensitive isothermal detection of nucleic-acid sequence by primer generation-rolling circle amplification. Nucleic Acids Res. 2009, 37, e19 10.1093/nar/gkn1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T.; Kurahashi H.; Emanuel B. S. Chromosomal translocations and palindromic AT-rich repeats. Curr. Opin. Genet. Dev. 2012, 22, 221–228. 10.1016/j.gde.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.; Jia H.; Dröge P.; Li J. The human genome-wide distribution of DNA palindromes. Funct. Integr. Genomics 2007, 7, 221–227. 10.1007/s10142-007-0047-6. [DOI] [PubMed] [Google Scholar]
- Wen J. D.; Gray D. M. Selection of genomic sequences that bind tightly to Ff gene 5 protein: primer-free genomic SELEX. Nucleic Acids Res. 2004, 32, e182 10.1093/nar/gnh179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R.; Luo Z.; Su L.; Zhang L.; Tang D.; Niessner R.; Knopp D. Palindromic Molecular Beacon Based Z-Scheme BiOCl-Au-CdS Photoelectrochemical Biodetection. Anal. Chem. 2019, 91, 2447–2454. 10.1021/acs.analchem.8b05265. [DOI] [PubMed] [Google Scholar]
- Zeng R.; Zhang L.; Luo Z.; Tang D. Palindromic Fragment-Mediated Single-Chain Amplification: An Innovative Mode for Photoelectrochemical Bioassay. Anal. Chem. 2019, 91, 7835–7841. 10.1021/acs.analchem.9b01557. [DOI] [PubMed] [Google Scholar]
- Yang J.; Hui Y.; Zhang Y.; Zhang M.; Ji B.; Tian G.; Guo Y.; Tang M.; Li L.; Guo B.; Ma T. Application of Circulating Tumor DNA as a Biomarker for Non-Small Cell Lung Cancer. Front. Oncol. 2021, 11, 725938 10.3389/fonc.2021.725938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Li Q.; Zhang R.; Dai X.; Chen W.; Xing D. Circulating microRNAs: Biomarkers of disease. Clin. Chim. Acta 2021, 516, 46–54. 10.1016/j.cca.2021.01.008. [DOI] [PubMed] [Google Scholar]
- Cai R.; Zhang S.; Chen L.; Li M.; Zhang Y.; Zhou N. Self-Assembled DNA Nanoflowers Triggered by a DNA Walker for Highly Sensitive Electrochemical Detection of Staphylococcus aureus. ACS Appl. Mater. Interfaces 2021, 13, 4905–4914. 10.1021/acsami.0c22062. [DOI] [PubMed] [Google Scholar]
- Wang L.; Song K.; Qu Y.; Chang Y.; Li Z.; Dong C.; Liu M.; Brennan J. D.; Li Y. Engineering Micrometer-Sized DNA Tracks for High-Speed DNA Synthesis and Biosensing. Angew. Chem. Int. Ed. 2020, 59, 22947–22951. 10.1002/anie.202010693. [DOI] [PubMed] [Google Scholar]
- Gao Y.; Li M.; Zeng Y.; Liu X.; Tang D. Tunable Competitive Absorption-Induced Signal-On Photoelectrochemical Immunoassay for Cardiac Troponin I Based on Z-Scheme Metal-Organic Framework Heterojunctions. Anal. Chem. 2022, 94, 13582–13589. 10.1021/acs.analchem.2c03263. [DOI] [PubMed] [Google Scholar]
- Lv S.; Tang Y.; Zhang K.; Tang D. Wet NH3-Triggered NH2-MIL-125(Ti) Structural Switch for Visible Fluorescence Immunoassay Impregnated on Paper. Anal. Chem. 2018, 90, 14121–14125. 10.1021/acs.analchem.8b04981. [DOI] [PubMed] [Google Scholar]
- Xu H.; Jiang Y.; Liu D.; Liu K.; Zhang Y.; Yu S.; Shen Z.; Wu Z. S. Twin target self-amplification-based DNA machine for highly sensitive detection of cancer-related gene. Anal. Chim. Acta 2018, 1011, 86–93. 10.1016/j.aca.2018.01.022. [DOI] [PubMed] [Google Scholar]
- Xu J.; Dong H.; Shen W.; He S.; Li H.; Lu Y.; Wu Z. S.; Jia L. New molecular beacon for p53 gene point mutation and significant potential in serving as the polymerization primer. Biosens. Bioelectron. 2015, 66, 504–511. 10.1016/j.bios.2014.12.008. [DOI] [PubMed] [Google Scholar]
- Xu J.; Wu Z. S.; Shen W.; Xu H.; Li H.; Jia L. Cascade DNA nanomachine and exponential amplification biosensing. Biosens. Bioelectron. 2015, 73, 19–25. 10.1016/j.bios.2015.05.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.