Abstract
Coronavirus disease 2019 (COVID-19) was reported to be associated with severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection, and patients present mostly with respiratory symptoms. There have been an increasing number of reports on oral manifestations, and some of these signs are informative in terms of identifying SARS-CoV-2 infection. The goal of present study was to review and synthesize the clinical characteristics and underlying mechanisms of COVID-19 oral manifestations, as well as to evaluate the factors influencing SARS-CoV-2 infectivity, in order to conduct further in-depth investigations and help clinicians diagnose COVID-19 patients exhibiting oral symptoms.
Keywords: COVID-19, SARS-CoV-2, oral manifestations, ACE2 receptor, TMPRSS2, influential factors
1. Introduction
More than 110.38 million people have been affected by the coronavirus disease 2019 (COVID-19) outbreak, which has now spread to 224 countries and killed more than 2.44 million people (Chan et al., 2020; Harrison et al., 2020). The International Committee on Classification of Viruses has named the virus formally severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) (Gorbalenya et al., 2020). The genome of SARS-CoV-2 isa linear single-stranded sense RNA, containing 14 open reading frames (ORFs), which encode proteins including spike protein (S), envelope protein (E), membrane protein (M), and nucleocapsid protein (N) (Kim et al., 2020; Papageorgiou and Mohsin, 2020; Arya et al., 2021; Yan et al., 2022). S protein is responsible for the viral infectivity and affinity for host cells. It is necessary for receptor binding and encouraging the fusion of the virus and cell membranes during viral invasion of host cells (Jackson et al., 2022).
The most common signs of COVID-19 are fever, coughing, dyspnea, and in severe cases, even death. More cases of COVID-19 extrapulmonary symptoms, including oral signs, are being reported (Chen N. et al., 2020; Rodriguez-Morales et al., 2020; Wang D. et al., 2020; da Rosa Mesquita et al., 2021). According to statistics, two-thirds of COVD-19 patients have at least one oral symptom (El Kady et al., 2021), and roughly one-third of patients have dysgeusia as their initial symptom (Biadsee et al., 2020). Dysgeusia and xerostomia are the most common oral manifestations of COVID-19 patients. The former one refers to patients’ inability to identify the taste of food or drink, and the latter one means that patients cannot smell the odor of food or drink. Additionally, the majority of patients with oral symptoms exhibited anomalies in their oral cavities 3 months after being released from the hospital, indicating that oral symptoms might be one of COVID-19’s aftereffects (Gherlone et al., 2021). The oral symptoms of COVID-19 patients have been the subject of numerous studies, and the appearance of oral symptoms is generally viewed as a reminder of viral infection.
The purpose of the current study was to review the most recent research on SARS-CoV-2 infection in the oral cavity.
2. Methods
The PubMed, Scopus and Web of Science databases were used for literature search to determine the literature related to the oral manifestations and related mechanisms of COVID-19. The keywords used were: “oral manifestations,” “dysgeusia,” “xerostomia,” “oral mucosal lesions,” “central nervous system,” “peripheral nervous system,” “Olfactory dysfunction,” “entry factors,” “ACE2,” “TMPRSS2,” “Furin,” “cathepsin,” “mechanisms,” “influential factors,” “SARS-CoV-2,” “Corona virus disease pandemic,” “COVID-19,” “2019-nCoV.” Studies were limited to those in English language included in PubMed, Scopus and Web of Science databases. Exclusion criteria included non-English language studies and those not included in PubMed, Scopus and Web of Science databases. According to the exclusion and inclusion criteria, all studies were independently screened by two reviewers, first by the title/abstract, and then the full text. Data or the research results extracted from the included studies were used for analysis.
3. Oral manifestations of COVID-19
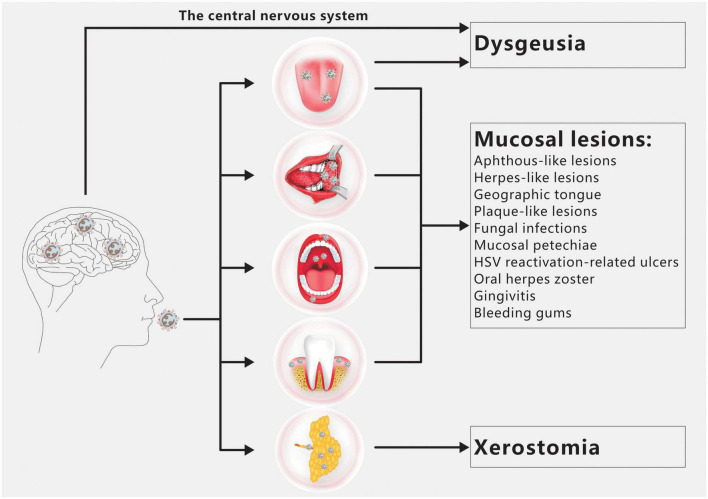
Dysgeusia, xerostomia, and oral mucosal lesions are the three oral symptoms of COVID-19 most frequently observed (Figure 1; Amorim dos Santos et al., 2021a). There are a number of additional oral symptoms, such as facial paralysis, trigeminal neuralgia, Melkersson-Rosenthal syndrome, macroglossia, anomalies of the temporomandibular joint, pain and swelling of the masticatory muscles, etc., although these secondary symptoms have not been widely documented (Amorim dos Santos et al., 2021b; El Kady et al., 2021; Farid et al., 2022; Sharma et al., 2022).
FIGURE 1.
Overview of COVID-19 oral manifestations. Dysgeusia, oral mucosal lesions and xerostomia are the main oral symptoms of COVID-19. SARS-CoV-2 infection of the CNS and/or taste buds is the main cause of dysgeusia. Oral mucosal lesions include aphthous-like lesions, herpes-like lesions, geographic tongue, plaque-like lesions, fungal infections, mucosal petechiae, HSV reactivation-related ulcers, oral herpes zoster, gingivitis and bleeding gums. The primary cause of xerostomia is SARS CoV-2 infection of salivary gland acinar cells and ductal epithelium.
3.1. Dysgeusia
Different sources have reported on the prevalence of dysgeusia, and anosmia frequently coexists with it (Sharma et al., 2022). However, Patients occasionally struggled to discern between dysgeusia and anosmia (El Kady et al., 2021). One of the earliest signs of SARS-CoV-2 infection has been reported as dysgeusia, which was typically seen in female patients with mild to moderate COVID-19 (Amorim dos Santos et al., 2021a; Iranmanesh et al., 2021). However, dysgeusia in COVID-19 patients is not substantially correlated with patients’ age, gender, or employment (El Kady et al., 2021). Dysgeusia can manifest in the majority of patients within 5 days of receiving a COVID-19 diagnosis, and it typically lasts for 2 weeks, or up to 4 weeks in more severe cases (Amorim dos Santos et al., 2021a). It was discovered that the degree of dysgeusia was strongly correlated with the severity of COVID-19, and that severe dysgeusia served as a warning sign (Amorim dos Santos et al., 2021a; Kumar, 2021). It is interesting that among COVID-19 patients, there was no statistically significant difference in the alterations of the tastes of sour, sweet, salty, or spicy (Biadsee et al., 2020).
3.2. Xerostomia
Saliva secretion is frequently impaired after SARS-CoV-2 infection, and xerostomia is the most common oral symptom in COVID-19 patients (Amorim dos Santos et al., 2021b). Initially, Chen L. et al. (2020) found that 46.3% of patients had xerostomia, with no discernible gender difference but severe cases appeared to be more prone to develop. Patients with xerostomia frequently experienced various symptoms along with their main complaint of dry mouth, such as a burning feeling, dysgeusia, angular stomatitis, and dysphagia (Biadsee et al., 2020; Eghbali Zarch and Hosseinzadeh, 2021; Tsuchiya, 2021). Despite not lethal, xerostomia can have a substantial impact on a patient’s life quality and dental health (Tsuchiya, 2021). Notably, sialadenitis may also be found in patients. In a case described by Fisher et al. (2021) the patient experienced symptoms of both acute bacterial suppurative parotitis and viral parotitis. Lechien et al. (2020) reported 3 cases of COVID-19-related parotitis. All three patients sought care for unilateral ear pain and retromandibular edema, and magnetic resonance imaging (MRI) indicated the occurrence of intracarotid lymphadenitis. Capaccio et al. (2020) demonstrated that the COVID-19-related acute parotitis may be one of the virus’s first symptoms. Amorim dos Santos et al. (2021b) summarized numerous reports of sialadenitis, and discovered that unilateral parotid gland lesions were frequently recorded. Furthermore, the most frequent oral sequelae are xerostomia and dilation of salivary ducts, indicating that there was a significant inflammatory response in the salivary glands of COVID-19 patients (Gherlone et al., 2021).
3.3. Oral mucosal lesions
Less frequently occurring than dysgeusia and xerostomia, oral mucosal lesions were seen in about 20.5% of COVID-19 patients (Amorim dos Santos et al., 2021b). The majority of patients reportedly experienced oral mucosal lesions within 10 days of infection, and they were often treated within 1–3 weeks after receiving photobiomodulation therapy (PBMT) and/or antiviral medication (Amorim dos Santos et al., 2021a; Brandão et al., 2021). Elderly, long-term hospitalized, unhygienic, or diabetic people are more likely to have oral mucosal lesions, and these individuals also tend to have more severe, long-lasting, and wide-ranging oral lesions (Iranmanesh et al., 2021). In addition, it had been stated that aphthous-like lesions, herpes-like lesions, Kawasaki-like disease (geographic tongue), plaque-like lesions, fungal infections (candidiasis and mucormycosis), mucosal petechiae, herpes simplex virus (HSV) reactivation-related ulcers, oral herpes zoster, gingivitis, and bleeding gums are frequently seen (Amorim dos Santos et al., 2021a; Iranmanesh et al., 2021; Orilisi et al., 2021; Sharma et al., 2022). The most frequent lesions, according to a recent study, are aphthous-like lesions, which are distinguished by many, round or irregular shapes, an erythematous halo, a surface coated in a purulent membrane, a white pseudomembrane, etc (Brandão et al., 2021). It is noteworthy that patients with oral lesions resembling Kawasaki are more likely to develop severe COVID-19 or require hospital admission (Erbaş et al., 2022).
4. The mechanisms of oral manifestations
4.1. The expression of entry factors and the entry pathways of SARS-CoV-2 in oral cavity
Severe acute respiratory syndrome coronavirus-2 assaults host cells through interacting with angiotensin-converting enzyme 2 (ACE2) receptors, inducing inflammatory responses in corresponding tissues and organs, which is similar to SARS-CoV. Additionally, the S protein containing S1 and S2 domains can be cleaved by Furin or transmembrane serine protease 2 (TMPRSS2) to accelerate the virus-cell membrane fusion and increase the viral tropism to organs, which may justify why SARS-CoV-2 has a higher infection rate than SARS-CoV. There are also some airway proteases such as TMPRSS4, TMPRSS11A, TMPRSS11E, TMPRSS13, human airway trypsin-like protease (HAT), matriptase, differentially expressed in squamous cell carcinoma 1 (DESC1), secreted neutrophil elastase, etc. appear to contribute to respiratory virus infection (Laporte and Naesens, 2017; Zou et al., 2020; Jackson et al., 2022). Therefore it is necessary to summarize the expression of SARS-CoV-2 entry factors in distinct structures of oral cavity in order to predict the infection of oral cavity by the virus and to reveal the mechanism of oral symptoms in COVID-19 patients (Table 1).
TABLE 1.
The expression and function of SARS-CoV-2 entry factors in varying structures of oral cavity.
| Entry factor | Function | Location | Sample source | Expression of the factor | References |
| ACE2 | SARS-CoV-2 receptor | Tongue (taste bud) | Human | In the type II and III taste cells of the fungiform and circumvallate papillae. | Sakaguchi et al., 2020; Okada et al., 2021; Drozdzik and Drozdzik, 2022 |
| SD rat | In the taste bud cells of the fungiform papillae. | Park et al., 2022 | |||
| Tongue (mucosa) | Human | In the keratinized stratified squamous epithelium and endothelial cells. Oral tongue >buccal and gingival tissues. |
Sakaguchi et al., 2020; Xu H. et al., 2020; Huang et al., 2021; Okada et al., 2021; Sawa et al., 2021; Drozdzik and Drozdzik, 2022 | ||
| SD rat | In the mature keratinocytes in the suprabasal layer of the squamous epithelium. Ventral mucosa >dorsal of the tongue, sporadically in the lamina propria and muscle. |
Park et al., 2022 | |||
| Salivary glands | Human | Similar in men and women. Salivary glands >oral cavity mucosa. Minor salivary glands >parotid >submandibular >sublingual glands. Salivary gland: Ducts, serous, and mucous acini clusters. Parotid gland: Serous cells, ductal epithelium, and adipocytes. Sublingual gland and buccal gland: Serous demilunes, ductal epithelium, and endothelial cells. Submandibular glands: Serous cells, ductal epithelium, and saliva of the ductal cavity. |
Baughn et al., 2020; Sakaguchi et al., 2020; Huang et al., 2021; Matuck et al., 2021; Okada et al., 2021; Drozdzik and Drozdzik, 2022 | ||
| SD rat | Strongly expressed in all salivary gland ductal cells. Strongly expressed in the acinar cell of parotid gland, Sporadically present in the submandibular and minor salivary gland. |
Park et al., 2022 | |||
| Lips | Human | In the non-keratinized stratified squamous epithelia in labial mucosa. In the mucous acini and serous acini in labial gland. |
Sawa et al., 2021; Drozdzik and Drozdzik, 2022 | ||
| Gingiva | Human | In the sulcular epithelium and periodontal pocket epithelium. In the spinous–basal cell layer, but not the epithelial surface and horny layer. Suprabasal >basal cells. |
Sakaguchi et al., 2020; Huang et al., 2021; Drozdzik and Drozdzik, 2022 | ||
| Buccal mucosa | Human | In non-keratinized stratified squamous epithelia. Suprabasal >basal cells. |
Huang et al., 2021; Okada et al., 2021; Sawa et al., 2021; Drozdzik and Drozdzik, 2022 | ||
| SD rat | Intermediate layer >basal and superficial layers. Not expressed in immature keratinocytes or in the basal layers of the squamous epithelia. |
Park et al., 2022 | |||
| Soft palate | Human | Suprabasal >basal cells. | Huang et al., 2021; Drozdzik and Drozdzik, 2022 | ||
| SD rat | Weak expression in a few superficial keratinocytes. Not expressed in immature keratinocytes/basal layers of the epithelia/the taste buds. |
Park et al., 2022 | |||
| Tonsil | Human | In the tonsillar crypt. Suprabasal >basal cells. |
Huang et al., 2021 | ||
| Blood vessel | Human | In arterial/venous endothelial cells and arterial smooth muscle cells in oral cavity. | Hamming et al., 2004; Okada et al., 2021 | ||
| SD rat | Not in venules/arterioles. Strongly in the capillaries of the salivary glands. |
Park et al., 2022 | |||
| TMPRSS2 | Cleave S2’ site of S protein | Tongue (taste bud) | Human | Strongly in the taste bud cells of the fungiform papilla. | Sakaguchi et al., 2020; Okada et al., 2021; Drozdzik and Drozdzik, 2022 |
| SD rat | In the taste bud cells of the fungiform papillae. Strongly in the pore cells of the fungiform papillae. |
Park et al., 2022 | |||
| Tongue (mucosa) | Human | In the keratinized stratified squamous epithelia and endothelial cells as well as tongue coating of human (mainly in the stratum granulosum and stratum spinosum). Suprabasal >basal cells. |
Sakaguchi et al., 2020; Huang et al., 2021; Okada et al., 2021; Sawa et al., 2021; Drozdzik and Drozdzik, 2022 | ||
| SD rat | Ventral mucosa >dorsal mucosa. Strongly in the intermediate layer of the squamous epithelia. Rarely in basal cells. Strongly in the muscle layer mast cells. |
Park et al., 2022 | |||
| Salivary glands | Human | Similar in men and women. Salivary glands >oral mucosa. Minor salivary glands >parotid >submandibular >sublingual glands. Salivary gland: Ducts epithelium. Serous acini >mucous acini Parotid gland: Serous cells, ductal epithelium, and adipocytes. Sublingual gland and buccal gland: Serous demilunes, ductal epithelium, and endothelial cells. Submandibular glands: Serous cells, ductal epithelium, and saliva of the ductal cavity. |
Baughn et al., 2020; Sakaguchi et al., 2020; Huang et al., 2021; Matuck et al., 2021; Okada et al., 2021; Drozdzik and Drozdzik, 2022 | ||
| SD rat | Strongly expressed in all salivary gland ductal cells. Strongly expressed in the acinar cell of parotid gland, Sporadically present in the submandibular and minor salivary gland. |
Park et al., 2022 | |||
| Lips | Human | In the non-keratinized stratified squamous epithelia in labial mucosa. In the mucous acini and serous acini in labial gland. Serous acini >mucous acini |
Sawa et al., 2021; Drozdzik and Drozdzik, 2022 | ||
| Gingiva | Human | In the sulcular epithelium and periodontal pocket epithelium. In the spinous cell layer of epithelia, not basal layer. |
Sakaguchi et al., 2020; Huang et al., 2021; Drozdzik and Drozdzik, 2022; Ohnishi et al., 2022 | ||
| Buccal mucosa | Human | In the non-keratinized stratified squamous epithelia. Suprabasal >basal cells. |
Huang et al., 2021; Okada et al., 2021; Sawa et al., 2021; Drozdzik and Drozdzik, 2022 | ||
| SD rat | Intermediate layer >basal and superficial layers. Not expressed in immature keratinocytes or in the basal layers of the squamous epithelia. |
Park et al., 2022 | |||
| Soft palate | Human | Increased suprabasal expression was observed when compared with the basal compartment. | Huang et al., 2021; Drozdzik and Drozdzik, 2022 | ||
| SD rat | Weak expression in a few superficial keratinocytes. Not expressed in immature keratinocytes or in the basal layers of the squamous epithelia. Not expressed in the taste buds of the soft palate mucosa. |
Park et al., 2022 | |||
| Tonsil | Human | Suprabasal >basal | Huang et al., 2021 | ||
| Blood vessel | Human | Vascular endothelial cells. | Okada et al., 2021 | ||
| SD rat | Not in venules/arterioles. Strongly in the capillaries of the salivary glands. |
Park et al., 2022 | |||
| Furin | Cleave a multibasic site (Arg-Arg-Ala-Arg) located at the S1–S2 junction of S protein during biosynthesis and maturation of SARS-CoV-2 in the infected cell. | Tongue (taste bud) | Human | In the lower layers of the taste buds of the fungiform papillae. | Sakaguchi et al., 2020; Okada et al., 2021 |
| Tongue (mucosa) | Human | In the spinous and basal cell layers of the epithelium in a dotted pattern. | Sakaguchi et al., 2020; Okada et al., 2021 | ||
| Salivary glands | Human | Submandibular glands: In the serous cells and saliva of the ductal cavity, not in the ductal epithelium. Parotid gland: In the serous cells and ductal epithelium. Sublingual gland and the buccal gland: in the serous demilunes and ductal epithelium. |
Sakaguchi et al., 2020; Okada et al., 2021 | ||
| Lips | Human | Furin was expressed in lips. | |||
| Gingiva | Human | In the spinous and basal cell layers. | Sakaguchi et al., 2020 | ||
| Buccal mucosa | Human | In the spinous and basal cell layers of the epithelium in a dotted pattern. | Okada et al., 2021 | ||
| Soft palate | Human | Furin was expressed in soft palate. | |||
| Tonsil | NA | NA | |||
| Blood vessel | Human | In vascular endothelial cells. | Okada et al., 2021 | ||
| TMPRSS4 | NA | Tongue (taste bud) | NA | NA | |
| Tongue (mucosa) | Human | Suprabasal >basal | Huang et al., 2021 | ||
| Salivary glands | Human | TMPRSS4 was expressed in salivary glands. | Huang et al., 2021 | ||
| Lips | NA | NA | |||
| Gingiva | Human | TMPRSS4 was expressed in gingival mucosa. details? | Huang et al., 2021 | ||
| Buccal mucosa | Human | Suprabasal >basal | Huang et al., 2021 | ||
| Soft palate | Human | Suprabasal >basal | Huang et al., 2021 | ||
| Tonsil | Human | Suprabasal >basal | Huang et al., 2021 | ||
| Blood vessel | NA | NA | |||
| TMPRSS11D | NA | Tongue (taste bud) | NA | NA | |
| Tongue (mucosa) | Human | Suprabasal >basal | Huang et al., 2021 | ||
| Salivary glands | Human | Enriched in mucosal keratinocytes. | Huang et al., 2021; Drozdzik and Drozdzik, 2022 | ||
| Lips | NA | NA | |||
| Gingiva | Human | Enriched in mucosal keratinocytes. | Huang et al., 2021 | ||
| Buccal mucosa | Human | Suprabasal >basal | Huang et al., 2021 | ||
| Soft palate | Human | Suprabasal >basal | Huang et al., 2021 | ||
| Tonsil | Human | Suprabasal >basal | Huang et al., 2021 | ||
| Blood vessel | NA | NA | |||
| CTSB | Cleave S2’ site of S protein | Tongue (taste bud) | NA | NA | |
| Tongue (mucosa) | NA | NA | |||
| Salivary glands | Human | Abundantly expressed in minor salivary glands. Broadly expressed in the epithelia. |
Huang et al., 2021; Drozdzik and Drozdzik, 2022 | ||
| Lips | NA | NA | |||
| Gingiva | Human | Broadly expressed in the epithelia. | Huang et al., 2021 | ||
| Buccal mucosa | NA | NA | |||
| Soft palate | NA | NA | |||
| Tonsil | NA | NA | |||
| Blood vessel | NA | NA | |||
| CTSL | Cleave S2’ site of S protein | Tongue (taste bud) | NA | NA | |
| Tongue (mucosa) | NA | NA | |||
| Salivary glands | Human | Abundantly expressed in minor salivary glands. Broadly expressed in the epithelia. |
Huang et al., 2021; Drozdzik and Drozdzik, 2022 | ||
| Lips | NA | NA | |||
| Gingiva | Human | Broadly expressed in the epithelia. | Huang et al., 2021; Okada et al., 2021 | ||
| Buccal mucosa | NA | NA | |||
| Soft palate | NA | NA | |||
| Tonsil | NA | NA | |||
| Blood vessel | NA | NA | |||
| TMPRSS3 | NA | Salivary glands | Human | Correlate with the expression of ACE2 in salivary glands. | Song et al., 2020 |
| TMPRSS5 | NA | Salivary glands | Human | Correlate with the expression of ACE2 in salivary glands. | Song et al., 2020 |
| TMPRSS7 | NA | Salivary glands | Human | Correlate with the expression of ACE2 in salivary glands. | Song et al., 2020 |
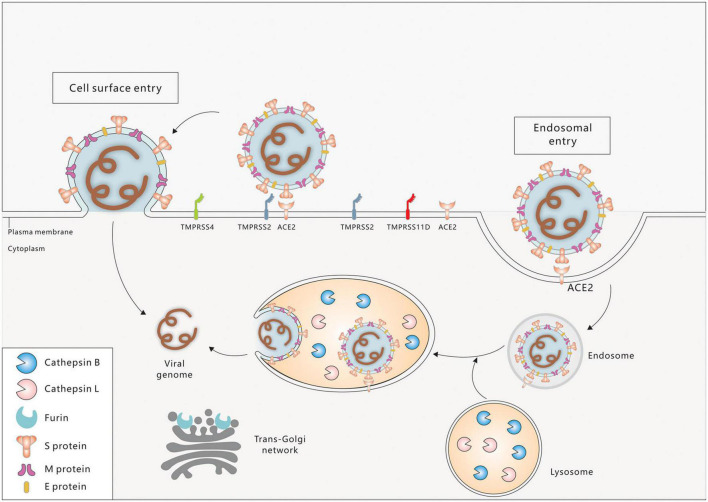
Severe acute respiratory syndrome coronavirus-2 entry factors in the oral cavity include ACE2, TMPRSS2, TMPRSS4, TMPRSS11D, Furin, Cathepsin B (CTSB), Cathepsin L (CTSL), and others. During viral assembly and maturation, the S1/S2 site (multibasic site) of the S protein is recognized and cleaved by Furin, and the S1 and S2 subunits are subsequently stabilized by non-covalent binding. In the process of virus infection of target cells, the S protein binds to the ACE2 receptor on the target cell membrane, inducing a conformational change in the S protein and exposing the S2’ site. If TMPRSS2 is present on the target cell membrane, the S2’ site is cleaved by TMPRSS2 and initiates a membrane fusion process in which the virus fuses directly with the target cell membrane, followed by the release of viral RNA into the cytoplasm. If there are insufficient TMPRSS2 on the target cell membrane or the virus-ACE2 complex does not encounter TMPRSS2, the virus enters into the cell via clathrin-mediated endocytosis and forms an endosome. The S2’ site is then cleaved by cathepsins (CTSL/CTSB) and initiates the membrane fusion process, followed by the release of viral RNA into the cytoplasm (Figure 2; Li et al., 2003; Cai et al., 2020; Hoffmann et al., 2020; Lu et al., 2020; Jackson et al., 2022).
FIGURE 2.
Cell entry mechanisms of SARC-CoV-2. Spike protein (S), envelope protein (E), and membrane protein (M) are three major proteins on the membrane of SARS-CoV-2. Cell surface entry and endosome entry are two distinct SARS-CoV-2 entry mechanisms. ACE2 is the primary receptor for SARS-CoV-2 entrance into the oral cavity. Virus binding to ACE2 triggers the conformational changes of the S1 subunit, exposing the S2’ site, which is then cleaved by the membrane protease TMPRSS2/4/11D (Cell surface entry pathway). In the absence of TMPRSS2, ACE2-virus complex is internalized via endocytosis into endosome/endolysosome where the S2’ site is cleaved by Cathepsin B/L (Endosome entry pathway). The fusion peptide is exposed and fuses with the cell membrane. The viral genome is then released into the cytoplasm of the host cell. In the virus producing cell, Furin cleaves the S1–S2 boundary in the trans-golgi network, contributing to the virus assembly and maturation.
4.2. The possible mechanisms of dysgeusia
Patients with severe acute respiratory syndrome (SARS) and the middle east respiratory syndrome (MERS), beta-coronavirus infections, rarely suffered from dysgeusia (Pellegrino et al., 2020). The causes of dysgeusia in COVID-19 patients have been the subject of numerous investigations and theories.
4.2.1. Dysfunction of taste buds
Severe acute respiratory syndrome coronavirus-2 may infect taste bud cells directly, resulting in dysgeusia (Mahmoud et al., 2021). According to previous studies, taste bud cells co-expressed ACE2 and TMPRSS2, which provided SARS-CoV-2 with receptors and hydrolases for invasion (Sakaguchi et al., 2020; Park et al., 2022). Doyle et al. (2021) found that ACE2 was expressed in human type II taste cells and SARS-CoV-2 could be replicated in type II taste cells through in situ hybridization. Additionally, it was revealed that the patient’s fungiform papillae taste stem cell layer has been damaged for several weeks, which could explain why dysgeusia lasted for a longer time. However, Wang Z. et al. (2020) showed that mice tongue papillae without taste buds had higher levels of ACE2 expression. Thus, SARS-CoV-2 may also potentially infect the squamous epithelial cells of the tongue, resulting in localized inflammation and edema and impairing the normal function of taste buds (Finsterer and Stollberger, 2020).
Additionally, the taste buds of mice had certain renin-angiotensin system (renin, angiotensinogen, and angiotensin-converting enzyme 1), and these constituents can locally create angiotensin II (Ang II), which can influence taste responses and be broken down into Ang 1–7 by ACE2. Accordingly, some studies hypothesized that patients’ taste buds may have less local Ang II degradation, which would lead to Ang II buildup and compromise the function of taste buds (Mariz et al., 2020).
4.2.2. Dysfunction of the nervous system
Dysgeusia could be a complication of SARS-CoV-2 infection of the central nervous system (CNS) or peripheral nervous system (PNS). Dysgeusia occurring in COVID-19 patients was one of the most common neurological symptoms (Garg et al., 2020). ACE2 was expressed in some brain regions, such as motor cortex and posterior cingulate, nigra substance, ventricles, middle temporal gyrus, olfactory bulb, ventrolateral medulla, solitary tract nucleus, and vagus nerve, as well as some cells in CNS, including neurons, microglia, astrocytes, and oligodendrocytes, making the CNS a possible target organ for SARS-CoV-2 (Baig et al., 2020; Generoso et al., 2021). The autopsy examinations on COVID-19 patients revealed varied degrees of brain injury as well as the presence of viral RNA in the brain (Generoso et al., 2021; Maiese et al., 2021). Other studies have found that COVID-19 patients’ cerebrospinal fluid (CSF) contains SARS CoV-2 (Elmakaty et al., 2022). All the above evidences supported that SARS CoV-2 can target CNS. Two primary mechanisms for ACE2 related CNS infection have been found, namely hematogenous pathway and neural pathways. The former one is SARS-CoV-2 crossing the blood-brain barrier (BBB) via infecting the cerebral vascular endothelial cells or leukocytes. The latter refers to the virus traveling through the olfactory, trigeminal nerves (nasal cavity and nasopharynx) and the vagus nerve (lower respiratory tract) (Baig et al., 2020; Generoso et al., 2021). However, further studies are needed to determine whether SARS-CoV-2 infection of the CNS directly contributes to the development of dysgeusia in patients. Additionally, because SARS-CoV-2 is neurotropic, it may directly harm the cranial nerves (CN VII, CN IX, and CN X) responsible for transmitting taste (Lozada-Nur et al., 2020).
Impaired synaptic transmission may also contribute to dysgeusia. The neurotransmitters dopamine and 5-hydroxytryptamine (5-HT), both essential for synaptic transmission (including taste), are produced by the enzyme aromatic L-amino acid (DOPA) decarboxylase. However, SARS-CoV-2 may suppress the expression of dopamine decarboxylase in target cells, resulting in lower levels of dopamine and 5-HT that might impair regular synaptic transmission and cause dysgeusia (Finsterer and Stollberger, 2020).
4.2.3. Olfactory dysfunction
Olfactory dysfunction (OD), one of the most common sensory dysfunction in patients with COVID-19, may also be one of the important causes for dysgeusia in patients (Mehraeen et al., 2021). About 41.5% of patients had both dysgeusia and OD as their primary symptom (Samimi Ardestani et al., 2021). The brain integrates the taste, smell, texture, temperature, appearance or sound of food or drink to form flavor. The insula, caudal orbitofrontal cortex (OFC), and anterior cingulate cortex (ACC) of the brain showed overlapping activation in response to independently presented tastes and scents, indicating that these areas may be crucial in the integration of taste and smell. Interestingly, putative primary gustatory areas occasionally respond to olfactory stimuli, whereas primary olfactory cortex does not seem to respond to gustatory stimuli. The integration of taste and smell in the insula, OFC, and ACC with other brain regions is also influenced by the olfactory delivery patterns and prior exposure to taste/smell combinations (Small et al., 2004; Landis et al., 2005; Small and Prescott, 2005; Hannum et al., 2018; Olofsson and Freiherr, 2019). Therefore, COVID-19 individuals who experience OD may also experience dysgeusia as a result of decreased olfactory delivery without stimulation of the gustatory cortex in the brain or defective integration of smell and taste.
4.3. The possible mechanisms of xerostomia
Salivary gland lesions could be caused by SARS-CoV-2 infection since ACE2 and TMPRSS2 were expressed in the ductal epithelium, serous acini, and mucous acini of the salivary glands. According to the research by Wang C. et al. (2020) SARS-CoV-2 can attack salivary glands via binding to the ACE2 receptor, leading to acute sialadenitis. Subsequently, the salivary glands may be repaired through fibroblast proliferation and fibrous connective tissue formation. However, this will also cause fibrosis of acinar cells and salivary gland ducts, resulting in a decrease in salivary secretion and obstruction of the salivary ducts (Wang C. et al., 2020). This theory offered a potential explanation for how salivary gland lesions manifest in COVID-19 patients. Chronic sialadenitis was the most prevalent histological alteration in infected salivary glands (Huang et al., 2021). They also discovered immune cells in the salivary glands, which suggested that sialadenitis was closely related to T cell responses. Bruno et al., also discovered morphological alterations in the epithelial cells and acinar cells of infected salivary gland (Matuck et al., 2021). The results stated above suggested that SARS-CoV-2 might infect salivary glands and trigger localized inflammation and immunological reactions. Additionally, increased mouth breathing and reduced salivary gland function were brought on by COVID-19 patients’ impaired nasal breathing, which in turn caused secondary symptoms such as xerostomia (Brandão et al., 2021).
4.4. The possible mechanisms of oral mucosal lesions
Since oral epithelial cells have high levels of ACE2, it is possible that SARS-CoV-2 might directly invading oral epithelial cells (Xu H. et al., 2020; Huang et al., 2021). Huang et al. (2021) discovered that SARS-CoV-2 may also infect the basal cells, suprabasal cells, and differentiated cells of the oral mucosal epithelium. Additionally, compared to other tissues, the oral mucosal epithelium has a low risk of contracting SARS-CoV-2 (Sapkota et al., 2022). However, it is unknown if oral mucosal lesions in patients are driven on by a direct infection with SARS-CoV-2 (Erbaş et al., 2022). The causes of oral mucosal lesions may also be related to oral cavity local immune responses, fungus infections, drug side effects, injuries caused by medical devices, vasculitis, microcirculation issues, etc (Viner and Whittaker, 2020; Amorim dos Santos et al., 2021b; La Rosa et al., 2021; Orilisi et al., 2021; Mohseni Afshar et al., 2022). Cell vacuolization, inflammatory cell infiltration, thrombosis, hemorrhage, necrosis, and other pathological abnormalities of the oral mucosa in COVID-19 individuals are additional oral mucosal abnormalities (Silveira et al., 2022). However, HPV infection was a potential source of these pathological manifestations (Hajdu, 2006).
5. Factors affecting SARS-CoV-2 infection
5.1. Periodontal pathogens
Periodontal infections are the common cause of the oral condition known as periodontitis. It has been revealed that 49.4% of COVID-19 patients had severe periodontitis (Anand et al., 2022). It was discovered that periodontal pathogens could impact affect the infectivity of SARS-CoV-2. Fusobacterium nucleatum, a periodontal infection, has been shown to boost ACE2 expression in A549 lung epithelial cells, according to Takahashi et al. (2021a). Despite the fact that this study did not show that Fusobacterium nucleatum could cause an increase in ACE2 expression in oral epithelial cells, it did provide compelling proof that periodontal infections might accelerate SARS-CoV-2 infection. In addition, Sena et al. (2021) reported that Porphyromonas gingivalis lipopolysaccharide (PgLPS) or inflammatory factors/mediators [e.g., interleukin-1β (IL-1β), tumor necrosis factor-α (TNF-α), and PGE2], derived from Porphyromonas gingivalis, could alter the expression levels of ACE2 and TMPRSS2 in human gingival fibroblasts. Some investigations showed that the S protein could be cleaved by the proteases produced by periodontal infections, increasing the infectivity of SARS-CoV-2 (Imai and Tanaka, 2021; Takahashi et al., 2021b).
Marouf et al. (2021) found that patients with periodontal illnesses are more vulnerable to COVID-19-related problems. SARS-CoV-2 was found in gingival crevicular fluid in roughly 63.64% of COVID-19 patients (Gupta et al., 2021). SARS-CoV-2 may spread through the periodontal tissues’ capillaries, promoting systemic infection (Badran et al., 2020; Bao et al., 2020; Drozdzik and Drozdzik, 2022). Furthermore, periodontal bacteria may be found in the bronchoalveolar lavage fluid of COVID-19 patients (Shen et al., 2020), and some research suggested that periodontal pathogens could worsen the symptoms of pneumonia or result in increased levels of systemic inflammatory cytokines (Nagaoka et al., 2014; Benedyk et al., 2016; Takahashi et al., 2021a). However, the precise processes through which periodontal disease affects the severity of COVID-19 are yet unclear.
5.2. Saliva
With sensitivity and specificity of 94.4 and 97.6%, respectively, saliva can be used as one of the dependable samples to diagnose COVID-19 in patients who are still in the early stages of the illness (Vaz et al., 2020). Chen L. et al. (2020) showed that saliva samples from severe patients contained more live viruses. However, a number of studies showed that saliva may offer some protection against the SARS-CoV-2. Immunoglobulin A (IgA), immunoglobulin M (IgM), and immunoglobulin G (IgG) antibodies against the S protein were present in the saliva of the individuals (Isho et al., 2020). IgM and IgG levels in saliva might be used to measure the immune response to SARS-CoV-2. Secretory IgA (SIgA), which can not only cross-react with the S1 subunit of the S protein but also stop the S protein from binding with the ACE2 receptor, was discovered by Tsukinoki et al. (2021) in the saliva of certain uninfected individuals. Lactoferrin, lysozyme, peroxidase, etc., in saliva could operate as general immunological defenses against SARS-CoV-2 infection (Tsukinoki et al., 2021). However, Exfoliated epithelial cells from COVID-19 patients’ saliva were found to be capable of sustaining SARS-CoV-2 infection and replication in a histology research (Huang et al., 2021). However, these exfoliated epithelial cells might have a local protective role against SARS-CoV-2 infection in the mouth (Drozdzik and Drozdzik, 2022).
5.3. Abnormal oral tissues
Sapkota et al. (2022) discovered that the expression level of ACE2 in oral squamous cell carcinoma cells and oral dysplasia tissues did not differ significantly from normal oral tissues whereas Furin expression rose and TMPRSS2 expression considerably decreased. However, it is still unclear how these changes may affect SARS-CoV-2 entry into host cells (Sapkota et al., 2022).
5.4. Oral health management during COVID-19 pandemic
It has been demonstrated that the SARS-CoV-2 spreads through spit droplets produced by talking, sneezing, and breathing (To et al., 2020; Xu R. et al., 2020). According to the research by Herrera et al. (2020) COVID-19 severity and viral excretion may be related to the amount of SARS-CoV-2 in the oral cavity. It has been demonstrated that gargling and tooth cleaning can lessen oral virus load (Mateos-Moreno et al., 2021). Therefore, by lowering the viral load in the mouth, oral healthcare may limit viral transmission. Patients should routinely wash their teeth, gargle, and use surgical masks or N95 masks in order to reduce the spread of SARS-CoV-2. Medical personnel should actively treat patients with periodontal diseases, sanitize the air in the facility to prevent saliva droplets from spreading, and pay attention to reducing the production of aerosols during oral surgeries in addition to wearing masks.
6. Conclusion
The oral signs, which mostly present as dysgeusia, xerostomia, and oral mucosal lesions that may be directly derived from SARS-CoV-2 or secondary COVID-19 lesions, may be helpful for the early diagnosis of individuals with COVID-19. The precise mechanics, meanwhile, are still not completely understood. Additionally, the infectivity of SARS-CoV-2 may be significantly influenced by factors such periodontal diseases, saliva, and abnormal oral tissues, necessitating the monitoring of patients’ oral health. Further investigation should be made into the diagnosis of COVID-19 using in-depth analysis of patients’ oral symptoms and associated processes.
Author contributions
WL and FG drafted the manuscript. WL, XW, and OS revised the manuscript and prepared the table and figures. NQ and XC contributed to the literature review. KT and CZ participated to the study design. MZ and OS initiated the study and revised the manuscript. All the authors read and approved the final version of the manuscript.
Funding
This study was financially supported by the Fundamental Research Fund of Science and Technology Foundation of Shenzhen City (Key program JCYJ20220818095811026 and general program JCYJ20210324094005015) and Start-up Research Fund of Shenzhen University for Youth Scholars (Grant No. 860-000002112112).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Amorim dos Santos J., Normando A. G. C., Carvalho da Silva R. L., Acevedo A. C., De Luca Canto G., Sugaya N., et al. (2021a). Oral manifestations in patients with COVID-19: A living systematic review. J. Dent. Res. 100 141–154. 10.1177/0022034520957289 [DOI] [PubMed] [Google Scholar]
- Amorim dos Santos J., Normando A. G. C., Carvalho da Silva R. L., Acevedo A. C., De Luca Canto G., Sugaya N., et al. (2021b). Oral manifestations in patients with COVID-19: A 6-month update. J. Dent. Res. 100 1321–1329. 10.1177/00220345211029637 [DOI] [PubMed] [Google Scholar]
- Anand P. S., Jadhav P., Kamath K. P., Kumar S. R., Vijayalaxmi S., Anil S. (2022). A case-control study on the association between periodontitis and coronavirus disease (COVID-19). J. Periodontol. 93 584–590. 10.1002/JPER.21-0272 [DOI] [PubMed] [Google Scholar]
- Arya R., Kumari S., Pandey B., Mistry H., Bihani S. C., Das A., et al. (2021). Structural insights into SARS-CoV-2 proteins. J. Mol. Biol. 433:166725. 10.1016/j.jmb.2020.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badran Z., Gaudin A., Struillou X., Amador G., Soueidan A. (2020). Periodontal pockets: A potential reservoir for SARS-CoV-2? Med. Hypotheses 143 109907–109907. 10.1016/j.mehy.2020.109907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig A. M., Khaleeq A., Ali U., Syeda H. (2020). Evidence of the COVID-19 virus targeting the CNS: Tissue distribution, host-virus interaction, and proposed neurotropic mechanisms. ACS chem. Neuroscience 11 995–998. 10.1021/acschemneuro.0c00122 [DOI] [PubMed] [Google Scholar]
- Bao L., Zhang C., Dong J., Zhao L., Li Y., Sun J. (2020). Oral Microbiome and SARS-CoV-2: Beware of lung co-infection. Front. Microbiol. 11:1840. 10.3389/fmicb.2020.01840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughn L. B., Sharma N., Elhaik E., Sekulic A., Bryce A. H., Fonseca R. (2020). Targeting TMPRSS2 in SARS-CoV-2 infection. Mayo Clin. Proc. 95 1989–1999. 10.1016/j.mayocp.2020.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedyk M., Mydel P. M., Delaleu N., Płaza K., Gawron K., Milewska A., et al. (2016). Gingipains: Critical factors in the development of aspiration pneumonia caused by Porphyromonas gingivalis. J. Innate Immun. 8 185–198. 10.1159/000441724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biadsee A., Biadsee A., Kassem F., Dagan O., Masarwa S., Ormianer Z. (2020). Olfactory and oral manifestations of COVID-19: Sex-related symptoms–a potential pathway to early diagnosis. Otolaryngol. Head Neck Surg. 163 722–728. 10.1177/0194599820934380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão T. B., Gueiros L. A., Melo T. S., Prado-Ribeiro A. C., Nesrallah A. C. F. A., Prado G. V. B., et al. (2021). Oral lesions in patients with SARS-CoV-2 infection: Could the oral cavity be a target organ? Oral Surg.Oral Med. Oral Pathol. Oral Radiol. 131 e45–e51. 10.1016/j.oooo.2020.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S. M., Walsh R. M., et al. (2020). Distinct conformational states of SARS-CoV-2 spike protein. Science 369 1586–1592. 10.1126/science.abd4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaccio P., Pignataro L., Corbellino M., Popescu-Dutruit S., Torretta S. (2020). Acute parotitis: A possible precocious clinical manifestation of SARS-CoV-2 Infection? Otolaryngol. Head Neck Surg. 163 182–183. 10.1177/0194599820926992 [DOI] [PubMed] [Google Scholar]
- Chan J. F.-W., Yuan S., Kok K.-H., To K. K.-W., Chu H., Yang J., et al. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 395 514–523. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zhao J., Peng J., Li X., Deng X., Geng Z., et al. (2020). Detection of SARS-CoV-2 in saliva and characterization of oral symptoms in COVID-19 patients. Cell Prolif. 53:e12923. 10.1111/cpr.12923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 395 507–513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rosa Mesquita R., Francelino Silva Junior L. C., Santos Santana F. M., Farias, de Oliveira T., Campos Alcântara R., et al. (2021). Clinical manifestations of COVID-19 in the general population: Systematic review. Wien. Klin. Wochenschr. 133 377–382. 10.1007/s00508-020-01760-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M. E., Appleton A., Liu Q.-R., Yao Q., Mazucanti C. H., Egan J. M. (2021). Human type II taste cells express angiotensin-converting enzyme 2 and are infected by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Am. J. Pathol. 191 1511–1519. 10.1016/j.ajpath.2021.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdzik A., Drozdzik M. (2022). Oral pathology in COVID-19 and SARS-CoV-2 infection-molecular aspects. Int. J. Mol. Sci. 23:1431. 10.3390/ijms2303143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eghbali Zarch R., Hosseinzadeh P. (2021). COVID-19 from the perspective of dentists: A case report and brief review of more than 170 cases. Dermatol. Ther. 34:e14717. 10.1111/dth.14717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kady D. M., Gomaa E. A., Abdella W. S., Ashraf Hussien R., Abd ElAziz R. H., Khater A. G. A. (2021). Oral manifestations of COVID-19 patients: An online survey of the Egyptian population. Clin. Exp. Dent. Res. 7 852–860. 10.1002/cre2.429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmakaty I., Ferih K., Karen O., Ouda A., Elsabagh A., Amarah A., et al. (2022). Clinical implications of COVID-19 presence in CSF: Systematic review of case reports. Cells 11:3212. 10.3390/cells11203212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbaş G. S., Botsali A., Erden N., Arı C., Taşkın B., Alper S., et al. (2022). COVID-19-related oral mucosa lesions among confirmed SARS-CoV-2 patients: A systematic review. Int. J. Dermatol. 61 20–32. 10.1111/ijd.15889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farid H., Khan M., Jamal S., Ghafoor R. (2022). Oral manifestations of Covid-19-A literature review. Rev. Med. Virol. 32:e2248. 10.1002/rmv.2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterer J., Stollberger C. (2020). Causes of hypogeusia/hyposmia in SARS-CoV2 infected patients. J. Med. Virol. 92 1793–1794. 10.1002/jmv.25903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J., Monette D. L., Patel K. R., Kelley B. P., Kennedy M. (2021). COVID-19 associated parotitis. Am. J. Emerg. Med. 39 254.e251–254.e253. 10.1016/j.ajem.2020.06.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R., Jain R., Sodani A., Chouksey D., Dosi R., Athale S., et al. (2020). Neurological symptoms as initial manifestation of Covid-19 - An observational study. Ann. Indian Acad. Neurol. 23 482–486. 10.4103/aian.AIAN_560_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Generoso J. S., Barichello de Quevedo J. L., Cattani M., Lodetti B. F., Sousa L., Collodel A., et al. (2021). Neurobiology of COVID-19: How can the virus affect the brain? Braz. J. Psychiatry 43 650–664. 10.1590/1516-4446-2020-1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherlone E. F., Polizzi E., Tetè G., De Lorenzo R., Magnaghi C., Rovere Querini P., et al. (2021). Frequent and persistent salivary gland ectasia and oral disease After COVID-19. J. Dent. Res. 100 464–471. 10.1177/0022034521997112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Baker S. C., Baric R. S., de Groot R. J., Drosten C., Gulyaeva A. A., et al. (2020). The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5 536–544. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Mohindra R., Chauhan P. K., Singla V., Goyal K., Sahni V., et al. (2021). SARS-CoV-2 detection in gingival crevicular fluid. J. Dent. Res. 100 187–193. 10.1177/0022034520970536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu S. I. (2006). The link between koilocytes and human papillomaviruses. Ann. Clin. Lab. Sci. 36 485–487. [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M., Lely A., Navis G., van Goor H. (2004). Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203 631–637. 10.1002/path.1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum M., Stegman M. A., Fryer J. A., Simons C. T. (2018). Different olfactory percepts evoked by orthonasal and retronasal odorant delivery. Chem. Senses 43 515–521. 10.1093/chemse/bjy043 [DOI] [PubMed] [Google Scholar]
- Harrison A. G., Lin T., Wang P. (2020). Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 41 1100–1115. 10.1016/j.it.2020.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera D., Serrano J., Roldán S., Sanz M. (2020). Is the oral cavity relevant in SARS-CoV-2 pandemic? Clin. Oral Investig. 24 2925–2930. 10.1007/s00784-020-03413-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181 271–280.e278. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N., Pérez P., Kato T., Mikami Y., Okuda K., Gilmore R. C., et al. (2021). SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 27 892–903. 10.1038/s41591-021-01296-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K., Tanaka H. (2021). SARS-CoV-2 infection and significance of oral health management in the Era of “the new normal with COVID-19”. Int. J. Mol. Sci. 22:6527. 10.3390/ijms22126527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranmanesh B., Khalili M., Amiri R., Zartab H., Aflatoonian M. (2021). Oral manifestations of COVID-19 disease: A review article. Dermatol. Ther. 34 e14578–e14578. 10.1111/dth.14578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isho B., Abe K. T., Zuo M., Jamal A. J., Rathod B., Wang J. H., et al. (2020). Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 5:eabe5511. 10.1126/sciimmunol.abe5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C. B., Farzan M., Chen B., Choe H. (2022). Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 23 3–20. 10.1038/s41580-021-00418-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.-Y., Yang J.-S., Kim J. W., Kim V. N., Chang H. (2020). The Architecture of SARS-CoV-2 Transcriptome. Cell 181 914.e–921.e. 10.1016/j.cell.2020.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. (2021). Could the oral cavity be a target organ in SARS-CoV-2 infection? Evid. Based Dent. 22 78–79. 10.1038/s41432-021-0174-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G. R. M., Libra M., De Pasquale R., Ferlito S., Pedullà E. (2021). Association of viral infections with oral cavity lesions: Role of SARS-CoV-2 infection. Front. Med. 7:571214. 10.3389/fmed.2020.571214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis B. N., Frasnelli J., Reden J., Lacroix J. S., Hummel T. (2005). Differences between orthonasal and retronasal olfactory functions in patients with loss of the sense of smell. Arch. Otolaryngol. Head Neck Surg. 131 977–981. 10.1001/archotol.131.11.977 [DOI] [PubMed] [Google Scholar]
- Laporte M., Naesens L. (2017). Airway proteases: An emerging drug target for influenza and other respiratory virus infections. Curr. Opin. Virol. 24 16–24. 10.1016/j.coviro.2017.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechien J. R., Chetrit A., Chekkoury-Idrissi Y., Distinguin L., Circiu M., Saussez S., et al. (2020). Parotitis-like symptoms associated with COVID-19, France, March-April 2020. Emerg. Infect. Dis. 26 2270–2271. 10.3201/eid2609.202059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M. J., Vasilieva N., Sui J., Wong S. K., Berne M. A., et al. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426 450–454. 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozada-Nur F., Chainani-Wu N., Fortuna G., Sroussi H. (2020). Dysgeusia in COVID-19: Possible mechanisms and implications. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 130 344–346. 10.1016/j.oooo.2020.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 395 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud M. M., Abuohashish H. M., Khairy D. A., Bugshan A. S., Khan A. M., Moothedath M. M. (2021). Pathogenesis of dysgeusia in COVID-19 patients: A scoping review. Eur. Rev. Med. Pharmacol. Sci. 25 1114–1134. 10.26355/eurrev_202101_24683 [DOI] [PubMed] [Google Scholar]
- Maiese A., Manetti A. C., Bosetti C., Del Duca F., La Russa R., Frati P., et al. (2021). SARS-CoV-2 and the brain: A review of the current knowledge on neuropathology in COVID-19. Brain Pathol. 31:e13013. 10.1111/bpa.13013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariz B. A. L. A., Brandão T. B., Ribeiro A. C. P., Lopes M. A., Santos-Silva A. R. (2020). New insights for the pathogenesis of COVID-19-related dysgeusia. J. Dent. Res. 99 1206–1206. 10.1177/0022034520936638 [DOI] [PubMed] [Google Scholar]
- Marouf N., Cai W., Said K. N., Daas H., Diab H., Chinta V. R., et al. (2021). Association between periodontitis and severity of COVID-19 infection: A case–control study. J. Clin. Periodontol. 48 483–491. 10.1111/jcpe.13435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Moreno M. V., Mira A., Ausina-Márquez V., Ferrer M. D. (2021). Oral antiseptics against coronavirus: In-vitro and clinical evidence. J. Hosp. Infect. 113 30–43. 10.1016/j.jhin.2021.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuck B. F., Dolhnikoff M., Duarte-Neto A. N., Maia G., Gomes S. C., Sendyk D. I., et al. (2021). Salivary glands are a target for SARS-CoV-2: A source for saliva contamination. J. Pathol. 254 239–243. 10.1002/path.5679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehraeen E., Behnezhad F., Salehi M. A., Noori T., Harandi H., SeyedAlinaghi S. (2021). Olfactory and gustatory dysfunctions due to the coronavirus disease (COVID-19): A review of current evidence. Eur. Arch. Otorhinolaryngol. 278 307–312. 10.1007/s00405-020-06120-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohseni Afshar Z., Barary M., Ebrahimpour S., Janbakhsh A., Afsharian M., Hasanpour A., et al. (2022). Pathophysiology and management of tongue involvement in COVID-19 patients. Indian J. Otolaryngol. Head Neck Surg. 74(Suppl. 2) 3235–3238. 10.1007/s12070-021-03052-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka K., Yanagihara K., Morinaga Y., Nakamura S., Harada T., Hasegawa H., et al. (2014). Prevotella intermedia induces severe bacteremic pneumococcal pneumonia in mice with upregulated platelet-activating factor receptor expression. Infect. Immun. 82 587–593. 10.1128/IAI.00943-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T., Nakamura T., Shima K., Noguchi K., Chiba N., Matsuguchi T. (2022). Periodontitis promotes the expression of gingival transmembrane serine protease 2 (TMPRSS2), a priming protease for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). J. Oral Biosci. 64 229–236. 10.1016/j.job.2022.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Yoshimura K., Toya S., Tsuchimochi M. (2021). Pathogenesis of taste impairment and salivary dysfunction in COVID-19 patients. Jpn. Dent. Sci. Rev. 57 111–122. 10.1016/j.jdsr.2021.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson J. K., Freiherr J. (2019). Neuroimaging of smell and taste. Handb. Clin. Neurol. 164 263–282. 10.1016/b978-0-444-63855-7.00017-4 [DOI] [PubMed] [Google Scholar]
- Orilisi G., Mascitti M., Togni L., Monterubbianesi R., Tosco V., Vitiello F., et al. (2021). Oral manifestations of COVID-19 in hospitalized patients: A systematic review. Int. J. Environ. Res. Public Health 18:12511. 10.3390/ijerph182312511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papageorgiou A. C., Mohsin I. (2020). The SARS-CoV-2 spike glycoprotein as a drug and vaccine target: Structural insights into its complexes with ACE2 and antibodies. Cells 9:2343. 10.3390/cells9112343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G. C., Bang S.-Y., Lee H. W., Choi K. U., Kim J. M., Shin S.-C., et al. (2022). ACE2 and TMPRSS2 immunolocalization and oral manifestations of COVID-19. Oral Dis. 28 (Suppl. 2) 2456–2464. 10.1111/odi.14126 [DOI] [PubMed] [Google Scholar]
- Pellegrino R., Cooper K. W., Di Pizio A., Joseph P. V., Bhutani S., Parma V. (2020). Coronaviruses and the chemical senses: Past, present, and future. chem. Senses 45 415–422. 10.1093/chemse/bjaa031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Morales A. J., Cardona-Ospina J. A., Gutiérrez-Ocampo E., Villamizar-Peña R., Holguin-Rivera Y., Escalera-Antezana J. P., et al. (2020). Clinical, laboratory and imaging features of COVID-19: A systematic review and meta-analysis. Travel Med. Infect. Dis. 34:101623. 10.1016/j.tmaid.2020.101623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi W., Kubota N., Shimizu T., Saruta J., Fuchida S., Kawata A., et al. (2020). Existence of SARS-CoV-2 entry molecules in the oral cavity. Int. J. Mol. Sci. 21:6000. 10.3390/ijms21176000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samimi Ardestani S. H., Mohammadi Ardehali M., Rabbani Anari M., Rahmaty B., Erfanian R., Akbari M., et al. (2021). The coronavirus disease 2019: The prevalence, prognosis, and recovery from olfactory dysfunction (OD). Acta Otolaryngol. 141 171–180. 10.1080/00016489.2020.1836397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota D., Sharma S., Søland T. M., Braz-Silva P. H., Teh M.-T. (2022). Expression profile of SARS-CoV-2 cellular entry proteins in normal oral mucosa and oral squamous cell carcinoma. Clin. Exp. Dent. Res. 8 117–122. 10.1002/cre2.510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa Y., Ibaragi S., Okui T., Yamashita J., Ikebe T., Harada H. (2021). Expression of SARS-CoV-2 entry factors in human oral tissue. J. Anat. 238 1341–1354. 10.1111/joa.13391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena K., Furue K., Setoguchi F., Noguchi K. (2021). Altered expression of SARS-CoV-2 entry and processing genes by Porphyromonas gingivalis-derived lipopolysaccharide, inflammatory cytokines and prostaglandin E(2) in human gingival fibroblasts. Arch. Oral Biol. 129 105201–105201. 10.1016/j.archoralbio.2021.105201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Malik S., Wadhwan V., Gotur Palakshappa S., Singh R. (2022). Prevalence of oral manifestations in COVID-19: A systematic review. Rev. Med. Virol. 32:e2345. 10.1002/rmv.2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L., et al. (2020). Genomic diversity of severe acute respiratory syndrome–coronavirus 2 in patients with coronavirus disease 2019. Clin. Infect. Dis. 71 713–720. 10.1093/cid/ciaa203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira F. M., Mello A. L. R., da Silva Fonseca L., Dos Santos Ferreira L., Kirschnick L. B., Martins M. D., et al. (2022). Morphological and tissue-based molecular characterization of oral lesions in patients with COVID-19: A living systematic review. Arch. Oral Biol. 136 105374–105374. 10.1016/j.archoralbio.2022.105374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small D. M., Prescott J. (2005). Odor/taste integration and the perception of flavor. Exp. Brain Res. 166 345–357. 10.1007/s00221-005-2376-9 [DOI] [PubMed] [Google Scholar]
- Small D. M., Voss J., Mak Y. E., Simmons K. B., Parrish T., Gitelman D. (2004). Experience-dependent neural integration of taste and smell in the human brain. J. Neurophysiol. 92 1892–1903. 10.1152/jn.00050.2004 [DOI] [PubMed] [Google Scholar]
- Song J., Li Y., Huang X., Chen Z., Li Y., Liu C., et al. (2020). Systematic analysis of ACE2 and TMPRSS2 expression in salivary glands reveals underlying transmission mechanism caused by SARS-CoV-2. J. Med. Virol. 92 2556–2566. 10.1002/jmv.26045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Watanabe N., Kamio N., Kobayashi R., Iinuma T., Imai K. (2021a). Aspiration of periodontopathic bacteria due to poor oral hygiene potentially contributes to the aggravation of COVID-19. J. Oral Sci. 63 1–3. 10.2334/josnusd.20-0388 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Watanabe N., Kamio N., Yokoe S., Suzuki R., Sato S., et al. (2021b). Expression of the SARS-CoV-2 Receptor ACE2 and proinflammatory cytokines induced by the periodontopathic bacterium Fusobacterium nucleatum in human respiratory epithelial cells. Int. J. Mol. Sci. 22:1352. 10.3390/ijms22031352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K. K.-W., Tsang O. T.-Y., Yip C. C.-Y., Chan K.-H., Wu T.-C., Chan J. M.-C., et al. (2020). Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 71 841–843. 10.1093/cid/ciaa149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya H. (2021). Characterization and pathogenic speculation of xerostomia associated with COVID-19: A narrative review. Dent. J. 9:130. 10.3390/dj9110130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukinoki K., Yamamoto T., Handa K., Iwamiya M., Saruta J., Ino S., et al. (2021). Detection of cross-reactive immunoglobulin a against the severe acute respiratory syndrome-coronavirus-2 spike 1 subunit in saliva. PLoS One 16:e0249979. 10.1371/journal.pone.0249979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz S. N., Santana D.S.d, Netto E. M., Pedroso C., Wang W.-K., Santos F. D. A., et al. (2020). Saliva is a reliable, non-invasive specimen for SARS-CoV-2 detection. Braz. J. Infect. Dis. 24 422–427. 10.1016/j.bjid.2020.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viner R. M., Whittaker E. (2020). Kawasaki-like disease: Emerging complication during the COVID-19 pandemic. Lancet 395 1741–1743. 10.1016/S0140-6736(20)31129-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wu H., Ding X., Ji H., Du H. (2020). Does infection of 2019 novel coronavirus cause acute and/or chronic sialadenitis? Med. Hypotheses 140:109789. 10.1016/j.mehy.2020.109789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. (2020). Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA 323 1061–1069. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhou J., Marshall B., Rekaya R., Ye K., Liu H.-X. (2020). SARS-CoV-2 Receptor ACE2 is enriched in a subpopulation of mouse tongue epithelial cells in nongustatory papillae but not in taste buds or embryonic oral epithelium. ACS Pharmacol. Transl. Sci. 3 749–758. 10.1021/acsptsci.0c00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zhong L., Deng J., Peng J., Dan H., Zeng X., et al. (2020). High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 12:8. 10.1038/s41368-020-0074-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Cui B., Duan X., Zhang P., Zhou X., Yuan Q. (2020). Saliva: Potential diagnostic value and transmission of 2019-nCoV. Int. J. Oral Sci. 12:11. 10.1038/s41368-020-0080-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Zheng Y., Zeng X., He B., Cheng W. (2022). Structural biology of SARS-CoV-2: Open the door for novel therapies. Signal Transduct. Target. Ther. 7:26. 10.1038/s41392-022-00884-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J., Han P., Hao J., Han Z. (2020). Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 14:185–192. 10.1007/s11684-020-0754-0 [DOI] [PMC free article] [PubMed] [Google Scholar]