Abstract
Objectives
To assess A-kinase anchor protein 95 (AKAP95), B-Raf, extracellular regulated protein kinases 1/2 (ERK1/2), and Elk-1 expression in colon cancer tissue, and characterize AKAP95 associations with B-Raf, ERK1/2, Elk-1, and colon cancer clinicopathological indices.
Methods
The immunohistochemistry streptavidin-perosidase (SP) method was used to determine protein expression levels in 64 colon cancer and 32 para-carcinoma tissue specimens.
Results
(1) Positive AKAP95 expression rates in colon cancer tissue were higher when compared with para-carcinoma tissue (92.19% vs. 59.38%, P < 0.05). Similar findings were determined for B-Raf (76.56% vs. 25%, P < 0.05), ERK1/2 (90.63% vs. 31.25%, P < 0.05), and Elk-1 levels (92.19% vs. 40.63%, P < 0.05). (2) No significant associations were identified between AKAP95, B-Raf, ERK1/2, and Elk-1 protein expression and degree of differentiation, histological type, and lymph node metastasis in colon cancer samples (P > 0.05); however, in The Cancer Genome Atlas and Gene Expression Omnibus datasets, AKAP95 was closely related to immune infiltration, and highly expressed AKAP95 was negatively associated with overall survival and relapse free survival rates in colon cancer patients. (3) Correlations were observed between AKAP95 and ERK1/2, AKAP95 and Elk-1, B-Raf and ERK1/2, B-Raf and Elk-1, and ERK1/2 and Elk-1 (all P < 0.05), but no correlation was observed between AKAP95 and B-Raf (P > 0.05).
Conclusions
AKAP95 may affect immune infiltration levels in colon cancer by participating in ERK1/2–Elk-1 signal transduction.
1. Background
A-kinase anchor protein 95 (AKAP95) is an anchoring protein for protein kinase A (PKA) and appears to regulate cyclic adenosine monophosphate (cAMP) accumulation by forming a distinct microdomain with PKA and phosphodiesterase (PDE4) in the nucleus [1]. We previously showed that when cAMP was activated by extracellular factors, AKAP95 was elevated and promoted cell proliferation via cyclin D/E and phospho-retinoblastoma (p-Rb) mechanisms [2–4]. Recent studies also showed that AKAP95 participated in tumorigenesis by regulating gene transcription and RNA clipping [5–7]. In addition overexpressed AKAP95 was detected in lung, ovarian, and rectal cancers [8–10]. Hence, AKAP95 is considered a cancer promoting protein; however, its role in tumorigenesis and participation in associated signaling pathways remains unclear.
The mitogen-activated protein kinase/extracellular signal-regulated (MAPK/ERK) pathway plays an important role in colorectal cancer [11]. MAPK/ERK pathway is closely related to cAMP/PKA signaling [12]. cAMP induces the sequential phosphorylation of Raf, MEK, and ERK in MAPK/ERK pathway. B-Raf is a selective target of cAMP in thyroid cells [13] and intercedes in PKA-induced ERK1/2 activation [14]. In addition, cAMP/PKA directly activates ERK [12, 13]. MAPK/ERK pathway plays an important role in transducing cAMP into the nucleus to activate Elk and other substrates [15]. However, it is unclear if AKAP95, like other AKAP family members [16–19], is involved in this signaling pathway.
Both AKAP95 and MAPK/ERK signaling pathways are critically involved in cAMP/PKA regulation [1, 12]. In this study, AKAP95, B-Raf, ERK1/2, and Elk-1 protein levels were examined in cancer and normal tissue samples, and associations between them characterized. This approach provided evidence showing that AKAP95 participated in Raf–MEK–ERK1/2 signaling to promote tumorigenesis or immune cells infiltration in colon cancer.
2. Materials and Methods
2.1. Tumor Sources
Tissue samples from 64 invasive colon cancer cases with definite pathological diagnoses were collected from the Affiliated Zhongshan Hospital of Dalian University, Dalian, China. Patient ages ranged from 51 to 82 years (average age = 70.3 ± 8.1 years) and 39 males and 25 females participated. In total, 62 patients had tubular or papillary adenocarcinoma, and two had mucinous adenocarcinoma. In 32/64 patients, para-carcinoma tissue was obtained from normal colonic tissue at least 3 cm away from cancerous tissue. Pathological examinations were also performed in para-carcinoma tissue to confirm the absence of cancer cells. Study protocols were approved by the Medical Ethics Committee of Affiliated Zhongshan Hospital of Dalian University (Ethics reference number: 2020010).
2.2. Reagents and Methods
Specimens were fixed in 10% neutral formaldehyde, paraffin embedded, and sliced into continuous 4 μm sections. The SP-9000 immunohistochemical staining kit (Zhongshan Jinqiao Biotechnology Company, Beijing, China) was used for protein expression analyses according to manufacturer's instructions. The assay involved 3, 3′-diaminobenzidine staining and hematoxylin counterstaining. Mouse anti-human AKAP95, B-Raf, ERK1/2, and Elk-1 monoclonal antibodies were purchased from Santa Cruz (Dallas, TX, USA). Phosphate buffered saline (PBS) (pH 7.20) was used in negative control samples. Rabbit anti-AKAP8 (ab140628) was purchased from Abcam Company (Cambridge, UK); Cy3-affinipure goat anti-rabbit IgG (111-165-003) and 488-affinipure donkey anti-mouse IgG (715-545-150) were purchased from Jackson ImmunoResearch Laboratories Inc. (West Grove, PA, USA); Protein A/G Plus-Agarose (sc-2003) was from Santa Cruz; Cell lysis buffer for Western blot and Immunoprecipitation (IP) (P0013) were purchased from Beyotime Institute of Biotechnology (Haimen, China).
2.3. Co-Immunoprecipitation (Co-IP)
When HCT116 cells cultured to 80% confluence, cells were collected and lysed with the WB/IP lysate buffer. 500 μg proteins were incubated with the antibody for 1 hour on a shaker, then incubated with Protein A/G Plus-Agarose overnight. The precipitant was centrifuged and washed three times in PBS in 4°C, then resuspended by using a sample buffer and identified by western blot assay.
2.4. Western Blot Assay
Proteins were Sodium Dodecyl Sulfate PolyAcrylamide Gel Electrophoresis (SDS-PAGE) separated, electro-transferred to a membrane, and then incubated with primary antibody at 4°C overnight, incubated with secondary antibody at room temperature for 1 hour, Enhanced Chemiluminescence (ECL)-developed, exposed, and imaged using the Tanon-4600SF Imaging System (Shanghai, China).
2.5. Immunofluorescence
HCT116 cells grown on slides were treated with 0.5% Triton-X 100 at room temperature for 30 minutes, blocked in 3% bovine serum albumin (BSA), incubated with primary antibody at 4°C overnight, and labeled by CY-3 and 488 fluorescent antibody (at a dilution of 1 : 300) at 37°C in a dark room. Nuclei were counter-stained by 4′,6-Diamidino-2′-phenylindole (DAPI), and smears were observed under a fluorescence microscope (BX53, Olympus, Tokyo, Japan).
2.6. Criteria Indicating Positive Protein Expression
A brown-yellow stain indicated positive protein expression, whereas its absence indicated no protein expression. Ten different fields/sections were randomly evaluated, with 200 tumor cells counted/field. Positive total cell ratios were used as metrics to assess positive protein expression: “−” indicated that <10% of cancer cells were yellow or brown; “±” indicated that ≥10% and <25% of cancer cells were yellow or brown; “+” indicated that ≥25% and <50% of cancer cells were yellow or brown; “++” indicated that ≥50% and <75% of cancer cells were yellow or brown; and “+++” indicated that >75% of cancer cells were yellow or brown.
For data analyses, “−” and “±” indicated negative expression. In addition “+” indicated low expression, “++” indicated moderate expression, and “+++” indicated high expression levels; thus, all indicated positive expression.
2.7. Bioinformatics and Functional Enrichment Analysis
Genomic data of colon cancer patient were collected from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. We analyzed the relationship between AKAP95 (AKAP8) expression and overall survival (OS) and relapse free survival (RFS) in colon cancer patients among these datasets. The data of relationship between AKAP95 (AKAP8) and tumor infiltrates immune cells were analyzed with methods including CIBERSORT, CIBERSORT-ABS, EPIC, ESTIMATE, MCPCOUNTER, QUANTISEQ, TIMER, and XCELL [20, 21].
2.8. Statistical Analysis
The SPSS20.0 software (SPSS Inc., Chicago, IL, USA) was used for statistical analysis.
Expression ratio comparisons were performed using Chi-square tests, and protein expression correlation analyses were performed using Spearman's rank correlations. A P < 0.05 value indicated statistical significance [22, 23].
3. Results
3.1. AKAP95, B-Raf, ERK1/2, and ELK-1 Expression Levels in Colon Cancer Tissue
AKAP95, ERK1/2, ELK-1, and B-Raf expression levels in 64 colon cancer and 32 para-carcinoma samples were assessed (Table 1). AKAP95 positive rates were 92.19% in colon cancer (Table 1; Figures 1(b1), 1(c1), 1(d1), and 1(e1)) and 59.38% in para-carcinoma specimens (Table 1, Figure 1(a1)). B-Raf positive rates were 76.56% in colon cancer (Table 1; Figures 1(b2), 1(c2), 1(d2), and 1(e2)) and 25% in para-carcinoma specimens (Table 1; Figure 1(a2)). ERK1/2 positive rates were 90.63% in colon cancer (Table 1; Figures 1(b3), 1(c3), 1(d3), and 1(e3)) and 31.25% in para-carcinoma specimens (Table 1; Figure 1(a3)). Elk-1 positive rates were 92.19% in colon cancer (Table 1; Figures 1(b4), 1(c4), 1(d4), and 1(e4)) and 40.63% in para-carcinoma specimens (Table 1; Figure 1(a4)). Differences were all statistically significant (Table 1; all P < 0.001). We also observed that AKAP95 (Figures 1(d1) and 1(e1)) and Elk-1 (Figure 1(e4)) were mainly expressed in nuclei in colon carcinoma tissue, whereas B-Raf (Figures 1(c2), 1(d2), and 1(e2)) and ERK1/2 (Figures 1(c3), 1(d3), and 1(e3)) subcellular localization were predominantly cytoplasmic.
Table 1.
AKAP95, ERK1/2, ELK-1, and B-Raf protein levels in colon cancer and normal tissue.
| Protein | Status | Colon cancer | Normal tissue | χ 2 | P-value |
|---|---|---|---|---|---|
| AKAP95 | Positive | 59 | 19 | 15.07 | <0.001 |
| Negative | 5 | 13 | |||
| B-Raf | Positive | 49 | 8 | 23.51 | <0.001 |
| Negative | 15 | 24 | |||
| ERK1/2 | Positive | 58 | 10 | 36.40 | <0.001 |
| Negative | 6 | 22 | |||
| ELK-1 | Positive | 59 | 13 | 30.25 | <0.001 |
| Negative | 5 | 19 |
χ 2: chi-square tests.
Figure 1.
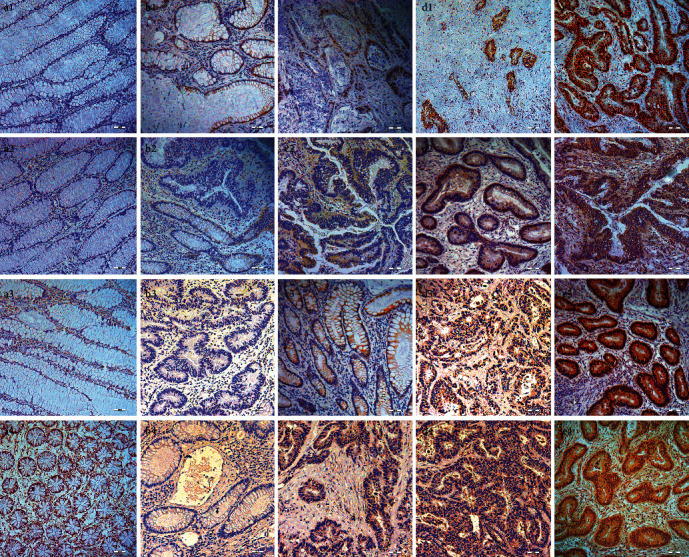
Representative images showing AKAP95, B-Raf, ERK1/2, and ELK-1 expressions in colon cancer and para-carcinoma tissue (magnification = 10×). (a1) No AKAP95 expression in para-carcinoma tissue. (b1) AKAP95 showing some expression (− or ±) in colon cancer tissue. (c1) Lowly expressed AKAP95 (+) in colon cancer tissue. (d1) Moderately expressed AKAP95 (++) in colon cancer tissue. (e1) Highly expressed AKAP95 (+++) in colon cancer tissue. (d1) and (e1) AKAP95 expression not only in the nuclei, but also in the cytoplasm. (a2) No B-Raf expression in para-carcinoma tissue. (b2) B-Raf showing some expression (− or ±) in colon cancer tissue. (c2) Lowly expressed B-Raf (+) in colon cancer tissue. (d2) Moderately expressed B-Raf (++) in colon cancer tissue. (e2) Highly expressed B-Raf (+++) in colon cancer tissue. (c2)–(e2) B-Raf expression in the cytoplasm. (a3) No ERK1/2 expression in para-carcinoma tissue. (b3) ERK1/2 showing some expression (− or ±) in colon cancer tissue. (c3) Lowly expressed ERK1/2 (+) in colon cancer tissue. (d3) Moderately expressed ERK1/2 (++) in colon cancer tissue. (e3) Highly expressed ERK1/2 (+++) in colon cancer tissue. (c3) ERK1/2 mainly expression in the cytoplasm. (d3) and (e3) ERK1/2 expression in the cytoplasm and nucleus. (a4) No ELK-1 expression in para-carcinoma tissue. (b4) ELK-1 showing some expression (− or ±) in colon cancer tissue. (c4) Lowly expressed ELK-1 (+) in colon cancer tissue. (d4) Moderately expressed ELK-1 (++) in colon cancer tissue. (e4) Highly expressed ELK-1 (+++) in colon cancer tissue. (c4) and (d4) ELK mainly expression in the cytoplasm. (e4) ELK-1 expression in the cytoplasm and nucleus.
3.2. Relationships between Clinical Pathological Parameters or Immune Cell Infiltration and AKAP95, ERK1/2, Elk-1, and B-Raf Expression
We observed no AKAP95 associations with tumor-node-metastasis stages, degree of differentiation, vascular invasion, lymph node metastasis, and distant metastasis (Table 2; P > 0.05). ERK1/2, Elk-1, and B-Raf also showed similar results (Supplemental Tables S1, S2, and S3). When we analyzed TCGA and GEO datasets, elevated AKAP95 expression was negatively associated with OS and RFS in colon cancer patients (Supplemental Figure S1(a) and (b)).
Table 2.
AKAP95 protein expression associations with clinical–pathological parameters.
| Item | AKAP95 status | χ 2 | P-value | |
|---|---|---|---|---|
| Positive | Negative | |||
| TNM stage | ||||
| T1–T2 | 31 | 1 | 1.95 | 0.16 |
| T3–T4 | 28 | 4 | ||
| Differentiation | ||||
| High | 1 | 0 | 0.76 | 0.68 |
| Moderate | 51 | 5 | ||
| Low | 7 | 0 | ||
| T stage | ||||
| T1–T2 | 3 | 0 | 0.27 | 0.61 |
| T3–T4 | 56 | 5 | ||
| Lymph node | ||||
| Yes | 31 | 2 | 0.29 | 0.59 |
| No | 28 | 3 | ||
| Metastasis | ||||
| Yes | 4 | 0 | 0.93 | 0.54 |
| No | 55 | 5 | ||
| Vascular invasion | ||||
| Yes | 17 | 0 | 1.96 | 0.16 |
| No | 42 | 5 | ||
χ 2: chi-square tests. TNM stage: tumor node metastasis staging classification; differentiation: differentiation degree of colon cancer cells; T stage: local invasion of colon cancer cells; lymph node: colon cancer cells metastasis in regional lymph node; metastasis: colon cancer cells distant metastasis; vascular invasion: colon cancer cells invasion vascular, lymphatic vessel, or neural invasion.
Since tumor infiltration by immune cells is a vital parameter for patient survival, correlations between AKAP95 and immune cells were also analyzed in TCGA and GEO datasets (Supplemental Figure S2(a)). AKAP95 showed positive relationships with CD4 naïve T cells, M0 macrophages, and resting dendritic cells (Supplemental Figures S2(b), (c), (d) and (e)). AKAP95 showed negative relationships with Immune Score and ESTIMATE Score (Supplemental Figures S2(f) and (g)).
3.3. AKAP95, ERK1/2, Elk-1, and B-Raf Associations in Colon Cancer Tissue
We also analyzed associations between AKAP95 and B-Raf, ERK1/2, and Elk-1 in colon cancer tissue. We identified significant correlations between AKAP95 and ERK1/2 (P < 0.05; Table 3) and AKAP95 and ELK-1 (P < 0.05; Table 4). No significant associations were identified between AKAP95 and B-Raf (Table 5; P > 0.05). Additionally, significant correlations were identified between ERK1/2 and B-Raf (Table 6; P < 0.05), ERK1/2 and Elk-1 (Table 7; P < 0.05), and Elk-1 and B-Raf (Table 8; P < 0.05).
Table 3.
The relationship between AKAP95 and ERK1/2 protein expression in colon cancer tissue.
| ERK1/2 | AKAP95 | r s | P-value | ||||
|---|---|---|---|---|---|---|---|
| − | ± | + | ++ | +++ | |||
| − | 1 | 1 | 1 | 0 | 1 | 0.265 | 0.034 |
| ± | 0 | 0 | 1 | 0 | 1 | ||
| + | 1 | 2 | 1 | 1 | 6 | ||
| ++ | 0 | 0 | 1 | 1 | 12 | ||
| +++ | 0 | 0 | 0 | 10 | 23 | ||
r s: Spearman's rank correlation's coefficient.
Table 4.
The relationship between AKAP95 and ELK-1 protein expression in colon cancer tissue.
| ELK-1 | AKAP95 | r s | P-value | ||||
|---|---|---|---|---|---|---|---|
| − | ± | + | ++ | +++ | |||
| − | 0 | 1 | 1 | 0 | 2 | 0.252 | 0.045 |
| ± | 1 | 0 | 0 | 0 | 0 | ||
| + | 0 | 2 | 1 | 4 | 10 | ||
| ++ | 1 | 0 | 1 | 2 | 10 | ||
| +++ | 0 | 0 | 1 | 6 | 21 | ||
r s: Spearman's rank correlation's coefficient.
Table 5.
The relationship between AKAP95 and B-Raf protein expression in colon cancer tissue.
| B-Raf | AKAP95 | r s | P-value | ||||
|---|---|---|---|---|---|---|---|
| − | ± | + | ++ | +++ | |||
| − | 0 | 2 | 1 | 0 | 5 | 0.182 | 0.149 |
| ± | 0 | 1 | 1 | 1 | 4 | ||
| + | 1 | 0 | 1 | 2 | 9 | ||
| ++ | 1 | 0 | 1 | 5 | 10 | ||
| +++ | 0 | 0 | 0 | 4 | 15 | ||
r s: Spearman's rank correlation's coefficient.
Table 6.
The relationship between B-Raf and ERK1/2 protein expression in colon cancer tissue.
| B-Raf | ERK1/2 | r s | P-value | ||||
|---|---|---|---|---|---|---|---|
| − | ± | + | ++ | +++ | |||
| − | 1 | 1 | 3 | 3 | 0 | 0.603 | <0.001 |
| ± | 2 | 0 | 3 | 1 | 1 | ||
| + | 0 | 0 | 4 | 2 | 7 | ||
| ++ | 1 | 1 | 1 | 6 | 8 | ||
| +++ | 0 | 0 | 0 | 2 | 17 | ||
r s: Spearman's rank correlation's coefficient.
Table 7.
The relationship between ERK1/2 and ELK-1 protein expression in colon cancer tissue.
| ELK-1 | ERK1/2 | r s | P-value | ||||
|---|---|---|---|---|---|---|---|
| − | ± | + | ++ | +++ | |||
| − | 1 | 0 | 2 | 1 | 0 | 0.421 | 0.001 |
| ± | 1 | 0 | 0 | 0 | 0 | ||
| + | 2 | 1 | 3 | 4 | 7 | ||
| ++ | 0 | 0 | 3 | 5 | 6 | ||
| +++ | 0 | 1 | 3 | 4 | 20 | ||
r s: Spearman's rank correlation's coefficient.
Table 8.
The relationship between B-Raf and ELK-1 protein expression in colon cancer tissue.
| B-Raf | ELK-1 | r s | P-value | ||||
|---|---|---|---|---|---|---|---|
| − | ± | + | ++ | +++ | |||
| − | 2 | 0 | 2 | 2 | 2 | 0.257 | 0.041 |
| ± | 0 | 0 | 4 | 1 | 2 | ||
| + | 2 | 0 | 2 | 4 | 5 | ||
| ++ | 0 | 1 | 5 | 2 | 9 | ||
| +++ | 0 | 0 | 4 | 5 | 10 | ||
r s: Spearman's rank correlation's coefficient.
In order to study the relationship between AKAP95 and ERK1/2 protein, Co-IP and immunofluorescence experiments were performed. The results showed that AKAP95 bound with ERK1/2 (Supplemental Figures S3(a) and (b)), and they were co-localized in HCT116 cells (Supplemental Figure S3(c)).
4. Discussion
AKAP95 promotes cancer cell growth and is highly expressed in lung, rectal, esophageal, ovarian, and breast cancers [4–7]. AKAP95 mechanisms suggest participation in cell cycle regulation by affecting cyclin D/E expression [4, 7]. Moreover, AKAP95 suppresses oncogene-induced senescence by regulating transcription and RNA splicing [10], and which play an important role in tumorigenesis [10]. In our study, the positive AKAP95 protein expression rate was 92.19% (59/64) in colon cancer and 59.38% (19/32) in para-carcinoma specimens, which suggested a functional role for AKAP95 in tumorigenesis. Otherwise, AKAP95 expressed not only in the nuclei but in cytoplasm of colon cancer was also found in present study (Figures 1(d1) and 1(e1)), which was as the same as our previous results [12]. This observation suggested that AKAP95 not only had important roles in the nucleus but was also important in the cytoplasm during tumorigenesis.
AKAP95 participates in cAMP signal transduction by anchoring the RII subunit of PKA [24]. cAMP, which is produced after extracellular receptor stimulation, may regulate cyclin D by ERK1/2 signaling [25, 26]. When cAMP was increased using forskolin, AKPA95 levels were increased and the protein bound to cyclin D/E to promote cell proliferation [3, 4]. In this study, AKAP95 expression was positively correlated with ERK1/2 levels in colon cancer specimens (Table 3). In addition, the results showed that AKAP95 and ERK1/2 bound together in HCT116 cells (Supplemental Figure S3). Therefore, we hypothesize that AKAP95, like its family members, participates in ERK signal transduction. For example, AKAP-Lbc binds to the ERK scaffold protein KSR-1 and forms a growth factor and cAMP reactive signal network to transmit signals from Raf to ERK1/2 via MEK [16, 17]. AKAP79 transmits signals generated by inhibitory GTP-binding protein (Gi) coupled to the beta2-adrenoceptor (β2-AR) receptor and to ERK [18]. mAKAP-β transmits extracellular signals from Angiotensin II (AngII) induced cardiomyocyte hypertrophy through phosphor-extracellular regulated protein kinases p-ERK2 [19]. However, in this current study, AKAP95 was not significantly correlated with B-Raf, but positively correlated with ERK1/2 and Elk-1. Therefore, AKAP95 may be involved in ERK1/2 signal transduction to Elk-1 by binding to ERK1/2 proteins. However, the results need to design detailed experiments to verify in future.
The transcription factor, Elk-1, is a nuclear substrate of ERK. When the ERK/Elk-1/Snail pathway was activated, it promoted epithelial–mesenchymal transition in colon cells and lung tissues [15]. The ERK pathway has also important roles in tumor immune invasion; however, AKAP95, ERK1/2, and Elk-1 expressions were not related to colon cancer invasion and metastasis but were possibly related to our low sample numbers. Fortunately, by using TCGA and GEO datasets, AKAP95 was closely related to tumor immune invasion, and high AKAP95 expression affected OS and tumor free survival rates in the GSE106584 dataset (Supplemental Figures S1 and S2). However, to comprehensively verify these observations, further studies are required.
In conclusion, AKAP95 was closely correlated with Raf–MEK–ERK signaling and was putatively involved in signal transduction from ERK to Elk so as to affect immune cell infiltration, resulting in affecting OS and RFS in colon cancer patients.
Acknowledgments
This study was supported by the Chen Xiao-Ping Foundation for the Development of Science and Technology of Hubei province (CXPJJH11800004-023).
Contributor Information
Feng Lin, Email: linfeng200612@163.com.
Yulong Yang, Email: yyl516@126.com.
Data Availability
Datasets analyzed in this work can be obtained from TCGA (https://portal.gdc.cancer.gov/) and GEO (https://www.ncbi.nlm.nih.gov/geo/).
Ethical Approval
This study was approved by the Ethics Committee of the Affiliated Zhongshan Hospital of Dalian University.
Conflicts of Interest
The author(s) declare(s) that they have no conflicts of interest.
Authors' Contributions
Xiangyu Kong, Feng Lin and Yulong Yang designed the experiments and wrote the manuscript. Xiangyu Kong and Putian An conducted the experiment of immunohistochemistry and western blotting and so on. Junping Xu performed the statistical analyses. Feng Lin and Wenzhi Liu participated in the coordination of the study. All authors read and approved the final manuscript.
Supplementary Materials
Tables S1, S2, and S3 showed ERK1/2, ELK-1, and B-Raf protein expression associations with TNM stages, degree of differentiation, vascular invasion, lymph node metastasis, and distant metastasis, respectively. Figure S1 showed AKAP95 expression was negatively associated with OS and RFS in colon cancer patients in TCGA and GEO datasets. Figure S2 showed the relationships between AKAP95 expression and immune cells infiltrating the tumor in TCGA and GEO datasets. Figure S3 showed the relationship between AKAP95 and ERK1/2 protein by co-immunoprecipitation and immunofluorescence in HCT116 cells.
References
- 1.Clister T., Greenwald E. C., Baillie G. S., Zhang J. AKAP95 organizes a nuclear microdomain to control local cAMP for regulating nuclear PKA. Cell Chemical Biology . 2019;26(6):885–891.e4. doi: 10.1016/j.chembiol.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kong X., Zhang D., Zhuang W., et al. AKAP95 promotes cell cycle progression via interactions with cyclin E and low molecular weight cyclin E. American Journal of Translational Research . 2016;8(2):811–826. [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X., Kong X., Zhuang W., et al. Dynamic changes in protein interaction between AKAP95 and CX43 during cell cycle progression of A549 cells. Scientific Reports . 2016;6(1):21224–21224. doi: 10.1038/srep21224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen R., Chen Y., Yuan Y., et al. CX43 and AKAP95 regulate G1/S conversion by competitively binding to cyclin E1/E2 in lung cancer cells. Thoracic Cancer . 2020;11(6):1594–1602. doi: 10.1111/1759-7714.13435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang H., Lu X., Shimada M., Dou Y., Tang Z., Roeder R. G. Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95. Nature Structural & Molecular Biology . 2013;20(10):1156–1163. doi: 10.1038/nsmb.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu J., Khodadadi-Jamayran A., Mao M., et al. AKAP95 regulates splicing through scaffolding RNAs and RNA processing factors. Nature Communications . 2016;7(1):13347–13347. doi: 10.1038/ncomms13347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W., Hu J., Shi B., et al. Biophysical properties of AKAP95 protein condensates regulate splicing and tumorigenesis. Nature Cell Biology . 2020;22(8):960–972. doi: 10.1038/s41556-020-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu S., Kong X., Yuan Y., et al. Relationship between AKAP95, cyclin E1, cyclin D1, and clinicopathological parameters in lung cancer tissue. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi . 2013;31(12):890–894. [PubMed] [Google Scholar]
- 9.Liu W., Hua S., Dai Y., et al. Roles of CX43 and AKAP95 in ovarian cancer tissues in G1/S phase. International Journal of Clinical and Experimental Pathology . 2015;8:14315–14324. [PMC free article] [PubMed] [Google Scholar]
- 10.Qi F., Yuan Y., Zhi X., et al. Synergistic effects of AKAP95, cyclin D1, cyclin E1, and CX43 in the development of rectal cancer. International Journal of Clinical and Experimental Pathology . 2015;8:1666–1673. [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan Fang J., Richardson B. C. The MAPK signalling pathways and colorectal cancer. The Lancet Oncology . 2005;6(5):322–327. doi: 10.1016/S1470-2045(05)70168-6. [DOI] [PubMed] [Google Scholar]
- 12.Garza Carbajal A., Ebersberger A., Thiel A., et al. Oncostatin M induces hyperalgesic priming and amplifies signaling of cAMP to ERK by RapGEF2 and PKA. Journal of Neurochemistry . 2021;157(6):1821–1837. doi: 10.1111/jnc.15172. [DOI] [PubMed] [Google Scholar]
- 13.Vuchak L. A., Tsygankova O. M., Prendergast G. V., Meinkoth J. L. Protein kinase A and B-Raf mediate extracellular signal-regulated kinase activation by thyrotropin. Molecular Pharmacology . 2009;76(5):1123–1129. doi: 10.1124/mol.109.060129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerits N., Kostenko S., Shiryaev A., Johannessen M., Moens U. Relations between the mitogen-activated protein kinase and the cAMP-dependent protein kinase pathways: comradeship and hostility. Cellular Signalling . 2008;20(9):1592–1607. doi: 10.1016/j.cellsig.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J., Ou B., Han D., et al. Tumor-derived CXCL5 promotes human colorectal cancer metastasis through activation of the ERK/Elk-1/snail and AKT/GSK3β/β-catenin pathways. Molecular Cancer . 2017;16(1):p. 70. doi: 10.1186/s12943-017-0629-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith F. D., Langeberg L. K., Scott J. D. Plugging PKA into ERK scaffolds. Cell Cycle . 2011;10(5):731–732. doi: 10.4161/cc.10.5.14902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith F. D., Langeberg L. K., Cellurale C., et al. AKAP-Lbc enhances cyclic AMP control of the ERK1/2 cascade. Nature Cell Biology . 2010;12(12):1242–1249. doi: 10.1038/ncb2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houslay M. D., Baillie G. S. Beta-arrestin-recruited phosphodiesterase-4 desensitizes the AKAP79/PKA-mediated switching of beta2-adrenoceptor signalling to activation of ERK. Biochemical Society Transactions . 2005;33(6):1333–1336. doi: 10.1042/BST0331333. [DOI] [PubMed] [Google Scholar]
- 19.Guo H., Liu B., Hou L., et al. The role of mAKAPβ in the process of cardiomyocyte hypertrophy induced by angiotensin II. International Journal of Molecular Medicine . 2015;35(5):1159–1168. doi: 10.3892/ijmm.2015.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C. Y., Chiao C. C., Phan N. N., et al. Gene signatures and potential therapeutic targets of amino acid metabolism in estrogen receptor-positive breast cancer. American Journal of Cancer Research . 2020;10(1):95–113. [PMC free article] [PubMed] [Google Scholar]
- 21.Folse H. J., Green L. E., Kress A., Allman R., Dinh T. A. Cost-effectiveness of a genetic test for breast cancer risk. Cancer Prevention Research . 2013;6(12):1328–1336. doi: 10.1158/1940-6207.CAPR-13-0056. [DOI] [PubMed] [Google Scholar]
- 22.Kessler A. F., Feldheim J., Schmitt D., et al. Monopolar spindle 1 kinase (MPS1/TTK) mRNA expression is associated with earlier development of clinical symptoms, tumor aggressiveness and survival of glioma patients. Biomedicine . 2020;8(7):p. 192. doi: 10.3390/biomedicines8070192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zubiaur P., Benedicto M. D., Villapalos-García G., et al. SLCO1B1 phenotype and CYP3A5 polymorphism significantly affect atorvastatin bioavailability. Journal of Personalized Medicine . 2021;11(3):p. 204. doi: 10.3390/jpm11030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eide T., Coghlan V., Orstavik S., et al. Molecular cloning, chromosomal localization, and cell cycle-dependent subcellular distribution of the A-kinase anchoring protein, AKAP95. Experimental Cell Research . 1998;238(2):305–316. doi: 10.1006/excr.1997.3855. [DOI] [PubMed] [Google Scholar]
- 25.Densham R. M., Todd D. E., Balmanno K., Cook S. J. ERK1/2 and p38 cooperate to delay progression through G1 by promoting cyclin D1 protein turnover. Cellular Signalling . 2008;20(11):1986–1994. doi: 10.1016/j.cellsig.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Gao F., Li M., Zhou L., Liu W., Zuo H., Li W. Xanthohumol targets the ERK1/2-Fra1 signaling axis to reduce cyclin D1 expression and inhibit non-small cell lung cancer. Oncology Reports . 2020;44(4):1365–1374. doi: 10.3892/or.2020.7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1, S2, and S3 showed ERK1/2, ELK-1, and B-Raf protein expression associations with TNM stages, degree of differentiation, vascular invasion, lymph node metastasis, and distant metastasis, respectively. Figure S1 showed AKAP95 expression was negatively associated with OS and RFS in colon cancer patients in TCGA and GEO datasets. Figure S2 showed the relationships between AKAP95 expression and immune cells infiltrating the tumor in TCGA and GEO datasets. Figure S3 showed the relationship between AKAP95 and ERK1/2 protein by co-immunoprecipitation and immunofluorescence in HCT116 cells.
Data Availability Statement
Datasets analyzed in this work can be obtained from TCGA (https://portal.gdc.cancer.gov/) and GEO (https://www.ncbi.nlm.nih.gov/geo/).


