Abstract
Biofilms are found in many infections in the forms of surface-adhering aggregates on medical devices, small clumps in tissues, or even in synovial fluid. Although antibiotic resistance genes are studied and monitored in the clinic, the structural and phenotypic changes that take place in biofilms can also lead to significant changes in how bacteria respond to antibiotics. Therefore, it is important to better understand the relationship between biofilm phenotypes and resistance and develop approaches that are compatible with clinical testing. Current methods for studying antimicrobial susceptibility are mostly planktonic or planar biofilm reactors. In this work, we develop a new type of biofilm reactor—three-dimensional (3D) microreactors—to recreate biofilms in a microenvironment that better mimics those in vivo where bacteria tend to form surface-independent biofilms in living tissues. The microreactors are formed on microplates, treated with antibiotics of 1000 times of the corresponding minimal inhibitory concentrations (1000 × MIC), and monitored spectroscopically with a microplate reader in a high-throughput manner. The hydrogels are dissolvable on demand without the need for manual scraping, thus enabling measurements of phenotypic changes. Bacteria inside the biofilm microreactors are found to survive exposure to 1000 × MIC of antibiotics, and subsequent comparison with plating results reveals no antibiotic resistance-associated phenotypes. The presented microreactor offers an attractive platform to study the tolerance and antibiotic resistance of surface-independent biofilms such as those found in tissues.
Keywords: Biofilm microreactors, Hydrogel, Antibiotic susceptibility assays, Biofilm phenotype, Antibiotic resistance
1. Introduction
Persistent bacterial infections are often related to the presence of microbial biofilms, a consortium of microbial communities in a self-derived extracellular matrix (ECM) that interacts with the surrounding environment while adhered to or in the vicinity of biotic or abiotic surfaces [1]. Microbial biofilms protect the embedded microbes against altered pH, topography, osmolarity, mechanical and shear stresses from the external environment [2,3]. In addition, the biofilm matrix allows for microbial survival under harsh chemical conditions, nutrient starvation, and antibiotics, which may provide an ideal environment for the development of antimicrobial resistance [[4], [5], [6]]. Due to the urgent need to address antimicrobial resistance, it is important to develop methods that incorporate native biofilm environments in antimicrobial testing [[7], [8], [9], [10]]. Clinical antimicrobial susceptibility testing (AST) is typically based on planktonic responses [11]. Treatments based on ASTs have only an 8–30% success rate [12]. While cell population size of the bacterial samples being tested is one possible cause [13], another potential confounding variable could be the presence of biofilms in persistent device associated infections. The development of a clinically feasible biofilm AST test format could help narrow the gap between AST results and the clinical success of antimicrobial therapy.
The current gold standards for antimicrobial susceptibility testing, such as agar dilution and broth macrodilution methods, do not incorporate biofilms in testing. Common biofilm models include 96-well microplates [14], modified Robbins device [15], Calgary biofilm device [16], disk reactor [17], and Centers for Disease Control (CDC) rotary biofilm reactor [18]. Biofilms usually form when microbes reach a critical density and produce extracellular polymeric substances through quorum sensing [19,20]. Besides biofilms on hard surfaces such as teeth and medical instruments, in vivo studies have shown that biofilms are also found in small clumps in tissues and often in microcrevices on/around medical devices [[21], [22], [23], [24]]. Biofilms can even form in synovial fluid when cells nucleate due to high viscosity [25]. The biofilms formed in these clinical environments are sparse and rugged, visually distinct from the thick and uniform lawns formed in most current biofilm reactors. Therefore, an antimicrobial susceptibility model incorporating three-dimensional (3D) biofilm microreactors is needed to better recreate these in vivo microenvironmental niches that are different from biofilms grown on surfaces.
Soft materials are ideal for the creation of such a niche and better resemble tissues that bacteria colonize in vivo. Previous research has indicated that biofilms are more robust with increased adhesion and colonization on soft materials resembling living tissues than on abiotic solid surface materials, which enables better outcomes for studies in biofilm function, metabolism, and growth [26]. Several studies have utilized bacteria-encapsulated hydrogels as biofilm models to yield highly reproducible biofilm formation and simulate some key physiochemical characteristics of in vivo biofilms [[27], [28], [29]]. Alginate is widely used in hydrogel-based biomedicine due to biocompatibility and low toxicity. Alginate-based beads/hydrogels have been employed as a facile and highly reproducible in vitro model system enabling bacterial biofilm development in a surface-independent manner [[30], [31], [32], [33]]. Alginate scaffolds with embedded target bacteria have been electrodeposited or flow-assembled in microfluidics as model biofilm to interrogate cell signaling with the convenience of continuous observation [[34], [35], [36]], although the high throughput capability should be further demonstrated with these platforms. Alginate-based beads have also been developed as simple and highly reproducible in vitro biofilm models for Pseudomonas aeruginosa biofilm formation [32]. The formed biofilms possessed small bacterial aggregations and growth characteristics similar to those observed in vivo, although the fabrication was complicated and extra sorting is needed to address the beads individually. An unique advantage of alginate hydrogel is the ability to be dissolved for further testing of biofilm microbes without having to undergo complex removal steps such as scraping and plating. The ability to gently dissolve the matrix for plating may also reduce the possibility for artifacts such as viable but non-culturable organisms.
In this work, we developed a dissolvable alginate hydrogel-based biofilm microreactor assay as schematically summarized in Fig. 1A (details in Methods), which is capable of simultaneously measuring biofilm tolerance and antibiotic resistance. Microscopic characterization of the hydrogel showed large numbers of uniformly distributed bacterial colonies with biofilms throughout the alginate hydrogel in a surface-independent manner. The microreactor could be easily dissolved with chelating agents such as EDTA, citrate, lactate, or phosphate [37], allowing for downstream plating and quantification of bacteria in planktonic form. As shown in Fig. 1B, the pathogens were treated with antibiotics of 1000 times of the corresponding minimal inhibitory concentrations (1000 × MIC), and monitored spectroscopically with microscopy and a microplate reader in a high-throughput manner. Meanwhile, the bacteria in the microreactor biofilms were released and plated either on agar plates to verify the viability of the pathogen (antimicrobial tolerance), or on MIC antibiotic agar plates to determine if mutations were responsible for antibiotic resistance. The study tested the hypothesis that the 3D biofilm microreactors better recreate the well-known antimicrobial tolerance of biofilms while greatly simplifying the retrieval of bacteria for subsequent plating assays by simple dissolution of the medium.
Fig. 1.
Schematic of the dissolvable alginate hydrogel-based biofilm microreactors for antibiotic susceptibility assays. (A) Formation of microreactors: (1) a synthesized bacteria-encapsulated alginate gel was cultured for 24 h to form microreactors; (2) schematic of microreactors within the alginate gel; and (3) chemical structure of cross-linked alginate chains where Ca2+ chelates the G-blocks of alginate chains. (B) The microreactors in alginate gels were exposed to antibiotic assay, followed by downstream analyses: visualized using confocal laser scanning and fluorescence microscopies (left); monitored using microplate reader for fluorescence/luminescence signal (middle); or reversely dissolved to obtain the bacteria in planktonic form for agar plating to assess antibiotic-induced mutation and viability (right).
2. Results and discussion
2.1. Biofilm formation in alginate hydrogel and microscopy
To determine if bacterial growth in the microreactor environment was comparable to liquid culture, we compared growth rates of microbes cultured in tubes to cultures in microreactors. Escherichia coli RP437/pRSH103, P. aeruginosa PAO1/pTdK-GFP, and Staphylococcus aureus AH2547/pCM29 were cultured in tubes or microreactors for 24 h. These strains have previously been reported to form biofilm in the literature [26,[38], [39], [40], [41], [42]]. Subsequently, samples from tube cultures or samples released through hydrogel dissolution of microreactors were plated to measure viability by colony forming units (Log10(CFU/mL)). As shown in Fig. 2, the total number of cells grown in microreactors were slightly more than but within an order of magnitude of those from liquid cultures in tubes. Two-factor ANOVA tests of the Log10(CFU/mL) of cell numbers in tube and alginate gel culture for each strain found that the p values (0.103, 0.101, 0.356 for RP437/pRSH103, PAO1/pTdK-GFP, and AH2547/pCM29, respectively) were higher than the significance level set at p < 0.05. Therefore, it was concluded that there was no significant difference in Log10(CFU/mL) of cell numbers between the tube and alginate gel culture. These results confirm that the alginate hydrogel environment does not interfere with bacterial growth.
Fig. 2.
Log10(CFU/mL) of microbial growth in alginate hydrogels and in tubes, indicating they were within the same order of magnitudes.
The bacteria in microreactors formed spherical clusters of cells that resemble biofilms, and therefore, we consider them as a new type of biofilm reactor, unlike existing 2D models. Fig. 3A shows confocal laser scanning microscopy (CLSM) images of the representative microreactors (populated by E. coli RP437/pRSH103), self-assembled into spherical clusters with an average diameter of 24 ± 5 μm after one day of growth. The microreactors were distributed over the gel with some aggregates in the center of the gel (Fig. 3B). Live/dead staining of E. coli RP437/pRSH103 in the microreactors showed that the vast majority of cells are viable (Fig. 3C). These confocal imaging and staining results also demonstrate robust microbial growth in the alginate hydrogel environment comparable to liquid culture.
Fig. 3.
Distribution of biofilm microreactors in alginate gel. (A) Confocal image of alginate-encapsulated RP437/pRSH103 microreactors grown in vitro for 24 h. (B) Rebuilt 3D image of the microreactors. (C) Live/dead staining of the corresponding gels. Scale bars as indicated.
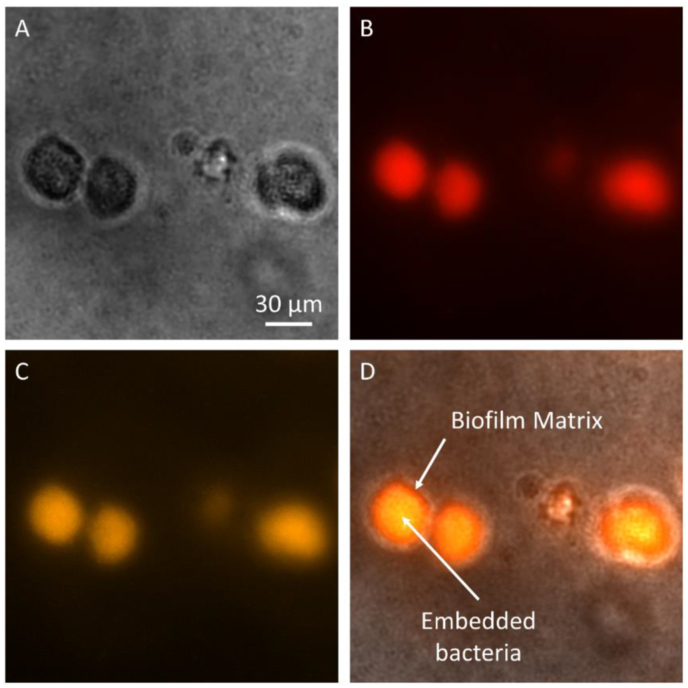
To determine the degree of biofilm formation in the alginate hydrogel, P. aeruginosa clusters as shown in Fig. 4A were stained with dyes specific to either the biofilm matrix (FilmTracer™ SYPRO® Ruby) or the cells inside biofilms (FilmTracer™ FM® 1–43). P. aeruginosa strain PA14 was used since the GFP signal from the PAO1/pTdK-GFP strain would confound measurements of fluorescence signals emitted by the biofilm-specific dyes. Fig. 4B shows the biofilm matrix that was stained in red, while Fig. 4C shows the embedded microbes that were stained in orange. For the control experiment (results not shown), the biofilm dyes were tested in empty alginate hydrogel and showed no fluorescence emission from the empty alginate hydrogel, which demonstrated that the SYPRO® Ruby is specific to the secreted ECM but not the alginate chains in the hydrogel. When the biofilm matrix in red was co-registered with the embedded microbes in orange, Fig. 4D clearly shows that the stained microbes were embedded within their self-derived ECM or biofilm matrix in red: it is orange in the center of the aggregation where the matrix and cells were both abundant; it is mainly red at the peripheral layer where there was less cells than matrix. The microreactors formed inside alginate hydrogel had an approximate size of around 29 ± 8 μm in diameter, similar to those observed under CLSM. Similar results were also observed in other studies of biofilms in alginate hydrogels [32,43].
Fig. 4.
Microscopic images of biofilm matrix and cell aggregate of P. aeruginosa strain PA14. (A) Brightfield image of the microreactors grown in vitro for 24 h. (B) SYPRO® Ruby staining of biofilm matrix in red. (C) FM® 1–43 staining of cells within the microreactors in orange. (D) Co-registered image of two stains showing microbes embedded in biofilm matrix. Scale bar as indicated. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.2. Assessment of biofilm tolerance, cell viability, and antibiotic resistance mutation
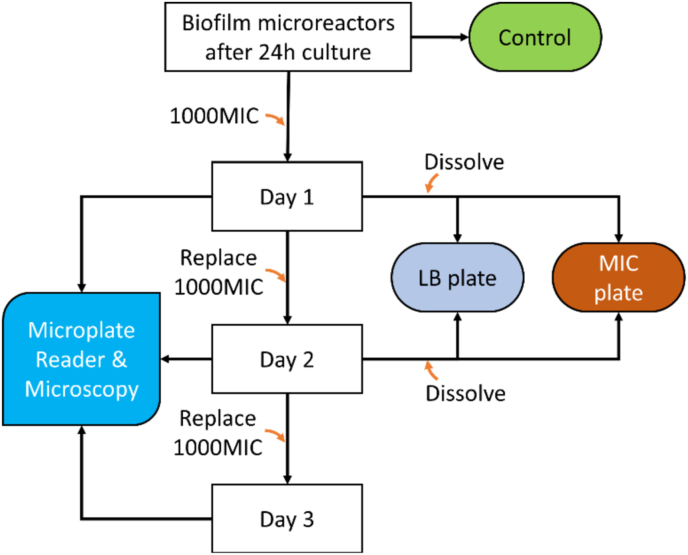
We performed minimal inhibitory concentration (MIC) tests of the target strains in liquid cultures and have confirmed their comparability with Clinical and Laboratory Standards Institute (CLSI) M100 guidelines (29th edition) for standard testing and the literature with details in Table S1 in Supplementary Information. The MIC of planktonic bacteria served as the baseline for the biofilm susceptibility assay, and only effective antibiotics for each susceptible strain were selected, as summarized in Table 1. Next, we assessed the biofilm tolerance in the presence of 1000 × MIC antibiotics using the alginate hydrogel-based microreactors. Among the four selected antibiotics in Table 1, ampicillin [44], ciprofloxacin [45], and vancomycin [46] have often been encapsulated in alginate beads for drug delivery due to their limited interactions with calcium alginate, chloramphenicol has weak interactions with alginate and can release most of the drug content from alginate hydrogel [47]. Antibiotics with strong interaction with alginate hydrogel such as tetracycline [48] were excluded. It has been previously shown that small molecules such as fluorescein, glucose, peptides, and antibiotics can freely diffuse through the alginate hydrogels [34,35,[49], [50], [51]] as the pore size of the alginate hydrogel prepared with 1% w/v should be around tens of nanometers [52,53]. Together, these observations suggest that the alginate hydrogel did not limit the diffusion of the selected antibiotics. On the other hand, it has previously been reported that biofilms can tolerate up to 1000 × MIC that their planktonic forms can tolerate [54]. To study how bacterial biofilm in the microreactor responds to antibiotics, the synthesized alginate biofilm microreactors were treated with 1000 × MIC for three days, 200 μL of antibiotics for each microreactor in a well, as described in the flowchart in Fig. 5.
Table 1.
Minimal inhibitory concentration (μg/mL) of antibiotics against planktonic bacteria as the baseline for biofilms susceptibility assay.
| Bacterial strains | Antibiotics |
|||||
|---|---|---|---|---|---|---|
| Ampicillin | Ciprofloxacin | Chloramphenicol | Tetracycline | Vancomycin | ||
| E. Coli | RP437/pRSH103 | 4 | 0.035 | 4 | N.A. | N.A. |
| Xen14 | N.A. | 0.035 | 2 | 0.5 | N.A. | |
| P. Aeruginosa | PAO1/pTdK-GFP | N.A. | 1 | N.A. | N.A. | N.A. |
| S. Aureus | AH2547/pCM29 | 0.5 | 0.125 | N.A. | 0.5 | 1 |
N.A.: not applicable, the ineffective antibiotic is not considered for biofilm antibiotic assay.
Fig. 5.
Procedure for the antibiotic assays of biofilm microreactors. Alginate hydrogels were formed in a 96-well microplate followed by 24-h culturing at 37oC to form microreactors. Antibiotic sensitivity was assessed by treating each microreactor in gel with antibiotic dose of 1000 × MIC for three days followed by reading fluorescence/bioluminescence signals daily with a microplate reader.
The microreactors were assessed using a microplate reader for fluorescence or bioluminescence signal over the three days. Meanwhile, a set of microreactors for each bacteria-antibiotic combination was dissolved to assess the viability and antibiotic resistance-induced phenotypic change of the released cells over time. Fluorescence or bioluminescence are common spectroscopic techniques used to monitor the growth of bacteria or biofilm in vitro and in vivo. However, it remains unclear whether the production of fluorescence or bioluminescence is an accurate measure of bacterial viability inside a biofilm. The ability to dissolve the hydrogels after antibiotic treatment to test for antibiotic sensitivity provides information about the accuracy of these signals with respect to cell viability.
The bacterial tolerance testing results are shown in Fig. 6, Fig. 7, Fig. 8. Overall, for the negative controls of bleach-treated bacteria, the fluorescent or luminescent signal never persisted past the first day's measurements. For most bacteria-drug combinations, the fluorescent or luminescent signal was observed to persist for up to three days, suggesting that bacteria in the microreactors might persist despite very high concentrations (1000 × MIC) of common antibiotics in many combinations. Retrieved cells from the dissolved microreactors were plated and cultured overnight on two sets of agar plates. The first was a set of nutrient plates to validate the viability of cells retrieved from the microreactors after 1000 × MIC antibiotic treatments over time. The second was a set of MIC plates to confirm that one MIC antibiotics were effective in killing the retrieved cells and to verify that the microbes did not mutate to develop resistance to the antibiotics. In the following sub-sections, spectroscopic measurements are compared with the plating and culturing results from nutrient plates and MIC plates, and the advantages of the dissolvable alginate hydrogel-based biofilm microreactors are emphasized.
Fig. 6.
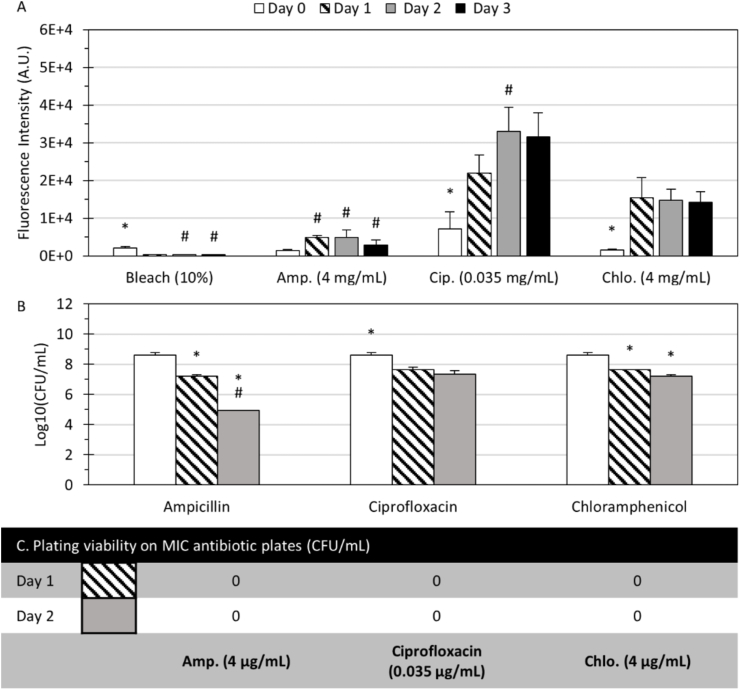
Biofilm tolerance, cell viability, and antibiotic resistance mutation-induced phenotype of E. coli. (A) Fluorescence signals of E. coli RP437/pRSH103 treated with bleach, and 1000 × MIC of antibiotics from Day 0 to Day 3. (B) Log10(CFU/mL) counts on nutrient plates of the retrieved cells from dissolved biofilm microreactors with 1000 × MIC antibiotic treatments for 0, 1 and 2 days. (C) CFU counts on MIC antibiotic plates of the corresponding retrieved cells. Day 0 indicates 24-h microreactors in CMHB before antibiotic treatments. (*) indicates significant differences (p < 0.05) between groups or time points, (#) indicates significant differences (p < 0.05) within the group or time points. All columns and error bars represent mean and ± standard deviation.
Fig. 7.
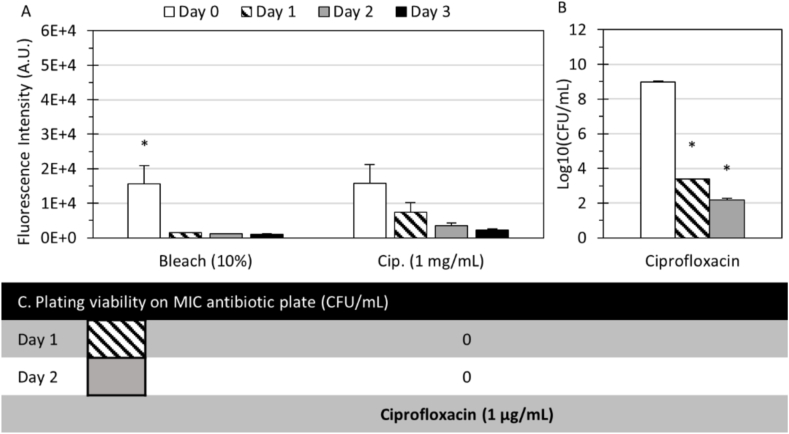
Biofilm tolerance, cell viability, and antibiotic resistance mutations of P. aeruginosa. (A) Fluorescence signals of PAO1/pTdK-GFP treated with bleach and 1000 × MIC ciprofloxacin from Day 0 to Day 3. (B) Log10(CFU/mL) counts on nutrient plates of the retrieved cells from dissolved microreactors with 1000 × MIC ciprofloxacin treatments for 0, 1, and 2 days. (C) CFU counts on MIC ciprofloxacin plates of the corresponding retrieved cells. Day 0 indicates 24-h microreactors in CMHB before antibiotic treatments. (*) indicates significant differences (p < 0.05) between groups or time points, (#) indicates significant differences (p < 0.05) within the group or time points. All columns and error bars represent mean and ± standard deviation.
Fig. 8.
Biofilm tolerance, cell viability, and antibiotic resistance mutations of S. aureus. (A) Fluorescence signals of AH2547/pCM29 treated with bleach and 1000 × MIC effective antibiotics from Day 0 to Day 3. (B) Log10(CFU/mL) counts on nutrient plats of the retrieved cells from dissolved microreactors with 1000 × MIC antibiotic treatments for 0, 1, and 2 days. (C) CFU counts on MIC antibiotic plates of the corresponding retrieved cells. (Day 0) indicates 24-h microreactors in CMHB before antibiotic treatments. (*) indicates significant differences (p < 0.05) between groups or time points, (#) indicates significant differences (p < 0.05) within the group or time points. All columns and error bars represent mean and ± standard deviation.
2.2.1. Biofilm tolerance, cell viability, and antibiotic resistance mutation of E. coli RP437/pRSH103
Biofilm susceptibility assays in terms of biofilm tolerance, cell viability and antibiotic resistance mutation were first performed with 1000 × MIC antibiotics against E. coli RP437/pRSH103 biofilm in microreactors. Fig. 6A shows the fluorescence of E. coli RP437/pRSH103 exposed to bleach and antibiotic concentrations of 1000 × MIC from Day 0 to Day 3. Day 0 indicates 24-h microreactors in cation-adjusted Mueller Hinton Broth 2 (CMHB) before bleach or 1000 × MIC antibiotic treatments. It was observed that cells cultured in CMHB hydrogel outgrew hydrogel space into liquid media within 24 h; thus, all fluorescence readings and CFU counting were compared against the initial signals from microreactors in Day 0 rather than presumed positive controls in CMHB hydrogel to ensure only signals in the hydrogel were counted. The fluorescence signal of bleach (NaOCl) culture as control was expected to be knocked out since hypochlorite ions (ClO⁻) destroyed the cell's outer layer [55], caused leakages of intracellular substances, and oxidized the fluorescence proteins [56].
Fig. 6A shows that significantly increased fluorescence signals were observed in the following days except for the expected minimal signal in the bleach culture, suggesting that the bacteria inside microreactors were persistent in the presence of antibiotics. Specifically, for the microreactors treated with ampicillin, Fig. 6A shows that the fluorescence intensity increased noticeably but was overall lowest compared with those treated with ciprofloxacin and chloramphenicol. Two-factor ANOVA tests found that the fluorescence of E. coli RP437/pRSH103 depended on the type of treatment (F = 59.0, p < 0.001), duration of exposure (F = 26.6, p < 0.001), and the interaction of treatment and duration (F = 5.3, p < 0.001). The CFU counts on nutrient plates for the cells retrieved from the dissolved microreactors in Fig. 6B show a log reduction of 1.4 (96.15%) on Day 1 and 3.7 (99.98%) on Day 2, indicating that ampicillin had inhibitory effects against E. coli RP437/pRSH103 growth in the microreactors. For the microreactors treated with ciprofloxacin, Fig. 6A shows that high fluorescence levels were observed over time as compared to those treated with ampicillin. This is inconsistent with MIC results (Table 1), where ciprofloxacin tended to suppress planktonic bacteria of the same strain more effectively than ampicillin. The plating results for cells retrieved from dissolved microreactors in Fig. 6B also show a log reduction of 0.95 (89.1%) on Day 1 and 1.24 (93.85%) on Day 2, indicating that ciprofloxacin was less effective as compared to ampicillin in inhibiting cell growth in the microreactors. For the microreactors treated with chloramphenicol, the fluorescence signals remained high and stable (Fig. 6A). The plating results for cells retrieved from the dissolved microreactors showed a log reduction of 0.97 (90.13%) on Day 1 and 1.39 (96.16%) on Day 2 (Fig. 6B), indicating that chloramphenicol was also less effective compared to ampicillin in inhibiting cell growth in the microreactors. This might be due to the bacteriostatic mechanism of chloramphenicol that inhibits bacterial protein synthesis or stops bacteria from reproducing [57]. These plating results confirmed that E. coli cells inside the microreactors had significant protection from 1000 × MIC ciprofloxacin and chloramphenicol that were effective against planktonic bacteria, while the protection against ampicillin was moderate. The finding that bacteria embedded within the biofilm microreactors were less susceptible to antibiotics aligns with reports in literature. For example, in a study of E. coli biofilm using the Calgary biofilm device [58], the clinical isolate in biofilm tolerated up to at least 64 × MIC enrofloxacin, a quilonone antibiotic similar to ciprofloxacin, and up to more than 512 × MIC ampicillin (Calculations were based on the study-determined MBEC, minimal biofilm eradication concentration, and MIC).
There are discrepancies between (1) the increasing fluorescence signals from the E. coli microreactors over time during the 1000 × MIC antibiotic treatments and (2) the observed reduction in CFU counts of the retrieved cells from these microreactors. Plating and culturing is the gold standard for quantifying viable cells. Discrepancies in the fluorescence measurements showed that they were not a consistent indicator of cell viability in the microreactors. Bioluminescence in E. coli Xen14 in the microreactor was also tested as an alternative to fluorescence. The test results show that bioluminescence intensity dropped dramatically after one day without losing viability as shown in Fig. S1 in Supplemental Information, similar to a previous report in a static culture [59]. The reduction in luminescence may reflect a change in the number of metabolically active cells, which can produce a luminescent signal by the luciferin-luciferase system [60], and switching to an antibiotic environment may also induce dormancy in bacterial cells without a loss in viability since biofilm cells typically have a lower metabolism and can enter a persister cell state [61]. While spectroscopic measurements of genetically modified cells can be valuable with adequate validation and controls, the discrepancies seen here warrant caution in using them as the sole indication of cell viability for antibiotic susceptibility testing. The dissolvable alginate hydrogel-based biofilm model represents a major advantage in facile retrieval of the microbes over the state-of-the-art biofilm models that require laborious manual scraping or intensive sonication.
Meanwhile, the corresponding retrieved cells from the 1000 × MIC antibiotics-treated microreactors were plated on MIC antibiotic plates. Significantly, Fig. 6C shows that there were no meaningful CFU counts on MIC antibiotic plates. Because these same cells did not show alterations in planktonic resistance to antibiotics, it is unlikely that these changes are due to the development of dominant resistance genes [62,63]. These results suggest that the surviving bacteria after 1000 × MIC antibiotic treatment did not present mutation-induced phenotypes due to the antagonistic environment, but the bacterial tolerance was due to the protection of the biofilm matrix in the hydrogels.
Overall, these studies demonstrated that ampicillin efficacy at 1000 × MIC against E. coli RP437/pRSH103 biofilm in microreactors was higher than chloramphenicol, with ciprofloxacin having the least efficacy among the antibiotics. The surviving bacteria were protected by phenotypic changes associated with biofilm but did not express dominant genotypic mutations.
2.2.2. Biofilm tolerance, cell viability, and antibiotic resistance mutation of P. aeruginosa PAO1/pTdK-GFP
Next, biofilm susceptibility assays were performed with 1000 × MIC ciprofloxacin against P. aeruginosa PAO1/pTdK-GFP. Testing was performed only with ciprofloxacin since other antibiotic-strain combinations showed no effectiveness in MIC testing. Fig. 7A shows that the fluorescence levels of the microreactors treated with 1000 × MIC ciprofloxacin were significantly decreased after 1 day, suggesting that ciprofloxacin was effective in inhibiting P. aeruginosa PAO1/pTdK-GFP biofilm growth. Noticeably, the decrease was gradual throughout the three days and depended on the type of treatment (F = 32.4, p < 0.001) and the interaction of treatment and duration (F = 4.3, p < 0.01, 2-factor ANOVA). Meanwhile, the nutrient plating results in Fig. 7B show that ciprofloxacin effectively suppressed the bacteria with a log reduction of 5.6 (99.992%) on Day 1 and 6.79 (99.999%) on Day 2. In this experiment, the spectroscopic measurements of the biofilm tolerance assay were consistent with the results from the bacterial viability assay. These results demonstrated the efficacy of 1000 × MIC ciprofloxacin against P. aeruginosa PAO1/pTdK-GFP biofilm in the microreactors, and the surviving bacteria were protected by the biofilm matrix and phenotypic changes associated with dormant cells rather than antibiotic resistance mutation. In a study of PAO1 in alginate hydrogel with only qualitative results [32], the efficacy of 100 × MIC tobramycin was demonstrated after 48 h of treatment. In another study of P. aeruginosa biofilm using Calgary biofilm device [58], the clinical isolate in biofilm could only tolerate 4 × MIC enrofloxacin, a quilonone antibiotic similar to ciprofloxacin (Calculations were based on division of the study-determined MBEC and MIC). Finally, Fig. 7C shows that there was zero CFU/mL count on the MIC agar plate of the retrieved cells from 1000 × MIC ciprofloxacin-treated microreactors, confirming that there was no sign of antibiotic-induced mutation.
2.2.3. Biofilm tolerance, cell viability, and antibiotic resistance mutation of S. aureus AH2547/pCM29
Finally, biofilm susceptibility assays were performed with 1000 × MIC antibiotics against S. aureus AH2547/pCM29 with all three effective antibiotics. Fig. 8A shows that the fluorescence of the S. aureus microreactors treated with 1000 × MIC ampicillin, ciprofloxacin, and vancomycin all increased and remained high over 3 days. In particular, the fluorescence in the ampicillin and ciprofloxacin treated samples remained stable while it increased gradually in the vancomycin treated sample. The fluorescence was affected by the type of treatment (F = 107.9, p < 0.001), duration of exposure (F = 54.7, p < 0.001), and the interaction of treatment and duration (F = 13.9, p < 0.001, 2-factor ANOVA). These fluorescence measurements suggested that the S. aureus biofilm in the microreactors had a high level of tolerance in the presence of ampicillin, ciprofloxacin, and vancomycin. However, the plating results in Fig. 8B show that the Log10(CFU/mL) counts on nutrient plates of the retrieved cells from dissolved microreactors were reduced significantly over time: On Day 1, the log reductions were 0.70 (79.97%), 2.71 (99.80%) and 1.64 log (97.606%) for ampicillin, ciprofloxacin, and vancomycin, respectively; On Day 2, the corresponding log reductions were 2.48 (99.62%), 4.54 (99.996%) and 4.39 log (99.992%). These plating results demonstrated the efficacy of 1000 × MIC ampicillin, ciprofloxacin, and vancomycin against S. aureus AH2547/pCM29 in the microreactors. This discrepancy may have been due to a lag between the time that bacteria died and the degradation of fluorescent molecules, resulting in an artifact signal in the biofilm tolerance assay. Again, the observation that bacteria in the biofilm microreactors were less susceptible to antibiotics is consistent with reports in literature. In a study of S. aureus biofilm using Calgary biofilm device [58], the clinical isolate in biofilm tolerated up to 4 × MIC ampicillin. In other studies of S. aureus biofilm in 96-well plate [64] and on an orthopedic implant surface [65], it was shown an increased tolerance up to 8000 × MIC and 256 × MIC vancomycin, respectively, after 24 h of treatment (Calculations were based on division of the study-determined MBEC and MIC).
When cells were plated to assess for antibiotic resistance mutations (Fig. 8C), S. aureus AH2547/pCM29 was not detectable (0 CFU/mL) on ampicillin, ciprofloxacin, and vancomycin plates. These results highlight the importance of how phenotypic changes in the biofilm community are the primary contributor (within the timeframe of the first few days of treatment) to the “appearance” of resistance to antibiotics, rather than the actual expression of resistance genes.
Overall, these studies demonstrated that 1000 × MIC ampicillin, ciprofloxacin, and vancomycin were all effective against S. aureus AH2547/pCM29 biofilm in microreactors over time. Biofilm matrix and phenotypic changes, but not genotypic mutation, contributed to increased tolerance of S. aureus in the presence of these antibiotics.
3. Conclusions
In conclusion, we have developed an assay that can report antibiotic susceptibility within a surface-independent biofilm environment in a high-throughput manner. The microplate format is readily translatable to studies of clinical samples. The biofilm assay relies on 3D microreactors in the hydrogel, which are able to form robust bacterial biofilms with properties of biofilms found in vivo in terms of morphology and altered susceptibility to antibiotics. A significant advantage of the microreactors for biofilm assay is the ability to dissolve the gel, thereby releasing the embedded bacteria for plating without having to perform extra extraction steps, including scraping, sonicating, vortexing, and validation of extraction efficiency. The viability of bacteria treated with 1000 times the effective concentration of antibiotics (for each strain) were assessed via fluorescence/bioluminescence signals and plating by dissolving the biofilm microreactors. Retrieved cells plated on nutrient plates showed that some microbes in microreactors withstood 1000 times the effective antibiotic concentrations for planktonic cells. While spectroscopic methods allow for high-throughput real-time in-situ interrogation of biofilm, discrepancies were observed between the culturing results (gold standard) and the spectroscopic results. Further research is needed to elucidate these limitations and develop alternative approaches.
4. Methods
4.1. Materials
White flat bottom 96-well microplates were purchased from Thermo Scientific. Sodium alginate and calcium chloride (CaCl2), phosphate buffered saline (PBS), cation-adjusted Mueller Hinton broth 2 (CMHB), powered LB broth, and tryptone soy powder were purchased from Sigma Aldrich (St. Louis, MO). Red fluorescent protein tagged E. coli RP437/pRSH103 were obtained from Dr. Dacheng Ren at Syracuse University (Syracuse, NY) [66]. Green fluorescent protein tagged P. aeruginosa PAO1/pTdK-GFP was obtained from Dr. Joy at The University of Akron (Akron, OH) [67]. Green fluorescent protein tagged S. aureus AH2547/pCM29 were provided by Dr. Alexander Horswill at The University of Iowa (Iowa City, IA) [68]. The bacterial plasmids were encoded with fluorescence proteins. Bioluminescent E. coli Xen14 was purchased from Perkin Elmer. The powder form of antibiotics used in the study, including ampicillin sodium, tetracycline hydrochloride, chloramphenicol, vancomycin hydrochloride, and ciprofloxacin were supplied by Sigma Aldrich. LIVE/DEAD™ BacLight™ Bacterial Viability kit, FilmTracer™ SYPRO® Ruby biofilm matrix stain, and FilmTracer™ FM® 1–43 Green biofilm cell stain was purchased from ThermoFisher Scientific (Waltham, MA). All other chemicals can be purchased from major suppliers.
4.2. Bacterial culture and preparations
A colony of each strain was collected from a tryptone soy agar plate, transferred to a 5 mL CMHB tube, and cultured in a 200-rpm shaking incubator at 37oC for 16–18 h. For E. coli RP437/pRSH103, 30 μg/mL of tetracycline was added to the culture to maintain the red fluorescence expression of pRSH103. For S. aureus AH2547/pCM29 and P. aeruginosa PAO1/pTdK-GFP, 10 μg/mL of chloramphenicol was added to the cultures to maintain the green fluorescence expression of pCM29 and pTdK-GFP. The antibiotic supplement was not needed for the culture of Xen14 since the bacterial chromosome contained a copy of the photorhabdus luminescens luxCDABE operon. The tested E. coli and S. aureus bacterial suspensions were vortexed for 1 min after being taken out of the incubator and 30 s before experiments. P. aeruginosa bacterial suspensions were vortexed for 1 min, filtered with a 0.45 μm filter, sonicated for 5 min, and finally vortexed for another 1 min. Colony forming unit per milliliter (CFU/mL) at an optical density at 600 nm OD600 = 1 was determined by agar plating approximately to be 6.13 × 108 CFU/mL for E. coli Xen14, 6.53 × 108 CFU/mL for P. aeruginosa PAO1/pTdK-GFP, 2.1 × 108 CFU/mL for E. coli RP437/pRSH103, and 4.6 × 108 CFU/mL for S. aureus AH2547/pCM29. Suspension of each bacterial strain with 108 CFU/mL was obtained by dilution for further analysis.
4.3. Antibiotics preparations
The stock solutions for each antibiotic agent used in this study were prepared with sterile deionized (DI) water or solvent and filtered with a 0.22 μm filter. In particular, ampicillin sodium (50 mg/mL) and vancomycin HCl (50 mg/mL) were dissolved in DI water, while tetracycline HCl (12 mg/mL) and chloramphenicol (50 mg/mL) were dissolved in ethanol (EtOH), and ciprofloxacin (30 mg/mL) was dissolved in 0.1 N HCl. For the assessment of bacterial antibiotic tolerance, the stock solutions were diluted in CMHB to the desired concentrations supplemented with 10 mM CaCl2 for maintaining the integrity of alginate hydrogels. For the determination of MIC, antibiotics were not supplemented with 10 mM CaCl2.
4.4. Alginate hydrogel biofilm formation
The biofilm microreactors were embedded in alginate hydrogel after 24 h culture. One of the most common approaches to preparing alginate-based hydrogel involves combining aqueous alginate solution with ionic crosslinking agents such as Ca2+ (Fig. 1A) [69]. Ca2+ electrostatically interacts with two negatively charged guluronate residues of alginate (Reaction (1)) to form an “egg-box” junction structure, Fig. 1A(2–3), leading to the gelation of alginate and embedment of target bacteria in the gel [70].
| Ca2+ + 2Alg-COO- ↔ Alg-COO–Ca2+-−OOC-Alg | (1) |
Alginate solution (2% w/v) was prepared by dissolving sodium alginate powder in sterile DI water and stirring overnight. The alginate solution was then autoclaved. CaCl2 powder was dissolved completely in DI water to obtain 1 M CaCl2 solution. Sterilization of stock CaCl2 was performed through a 0.22 μm filter.
The microreactors in alginate hydrogel were formed by mixing the target bacterial strain with alginate solutions. 6 mL of bacterial suspension of each strain (in 2 × CMHB) was thoroughly mixed with 6 mL of 2% w/v alginate solution (ratio 1:1) to make the stock alginate-bacteria solutions (5 × 105 CFU/mL, 1 × CMHB and 1% w/v alginate). Then, 100 μL of alginate-bacteria mixture was added to the wells using a multichannel pipette. Next, 1 M CaCl2 was diluted to the final concentrations of 50 and 250 mM in a fine mist sprayer with a 360-degree rotation atomizer purchased from Nomija through Amazon, USA. First, the 50 mM CaCl2 in one fine mist sprayer was sprayed thoroughly eight inches above the microplate for 30 s. The approximate volume of 50 mM CaCl2 used was 100 μL. The microplate was left at room temperature for 15 min to allow for crosslinking. Next, the 250 mM CaCl2 in another fine mist sprayer was sprayed thoroughly eight inches above the microplate for 30 s. The approximate volume of 250 mM CaCl2 used was 100 μL. Then, 100 μL of 250 mM CaCl2 was introduced to each well using a multichannel pipette. The plate was then left for 1 h at room temperature to further stabilize the gel. Finally, the microplate was washed using a microplate washer (50 TS, Biotek) with sterile DI water. Biofilms in alginate hydrogels were formed in a 96-well microplate (Fig. 1B) with initial 5 × 105 CFU/mL bacteria per hydrogel and cultured in CMHB for 24 h for biofilm formation inside the gel. The hydrogels in microplates were treated with antibiotics followed by assessments through microscopy, high-throughput microplate reading, and dissolution for agar gel plating. For control experiments, a corresponding set of the alginate biofilm gels before antibiotic treatments was dissolved in a solution of 0.05 M Na2CO3 and 0.02 M Nitric acid [71] to obtain the bacteria in planktonic form for agar plating to assess bacterial antibiotic resistance and viability.
The alginate biofilm gel of E. Coli RP437/pRSH103 was assessed using confocal laser scanning microscopy (CLSM, Leica SP8). The E. coli RP437/pRSH103 biofilm was stained with LIVE/DEAD™ BacLight™ Bacterial Viability Kit and observed under microscopy using optimal excitation wavelength/emission wavelength (Ex/Em) of 485⁄498 nm for SYTO®9 and Ex/Em of 535⁄617 nm for propidium iodide. The biofilm staining procedure followed the instruction from the provider. P. aeruginosa PA14 was used to form alginate biofilm gels for biofilm matrix staining. The staining used FilmTracer™ SYPRO® Ruby (Ex/Em of 450/610 nm) to stain the biofilm matrix and FilmTracer™ FM® 1–43 (Ex/Em of 472/580 nm) to stain the cells within the biofilm matrix. The FilmTracer™ FM® 1–43 staining solution was prepared by diluting 10 μL of the stock solution into 990 μL of dimethyl sulfoxide, followed by the 10-time dilution of that mixture with sterile DI water to make the final staining concentration of 1 μg/mL. The staining mixture was prepared by mixing the prepared FM® 1–43 with SYPRO® Ruby at the volume ratio of 1:1. Then, 200 μL of the staining mixture was added to each microwell containing the alginate biofilm gel. The well plate was incubated for 30 min at room temperature in dark conditions. The assessment of biofilm matrix and cell staining was performed under a fluorescence microscope (Zeiss Axio Observer Z1 Inverted Microscope).
4.5. Determination of MIC
The minimum inhibitory concentration, or MIC, is defined as the lowest concentration of antibiotic agents that inhibits the growth of a microorganism. Determination of MICs of the antibiotic agents used in this study against the four proposed strains was carried out on 96-well microplates in compliance with CLSI. Each plate was dedicated to testing one type of bacterium and one row per antibiotic with up to 10 different antibiotic serial dilutions. Briefly, CMHB containing two-fold dilution concentrations of each antibiotic agent were added to the first ten wells (total volume of media and the antibiotic agent was 50 μL). Then, 50 μL of prepared bacterial suspensions (10 [8] CFU/mL) were seeded into each well from the first to the eleventh well to get the final inoculum size of 5 × 105 CFU/mL. Wells at the last two rows were used as a positive control (11th well) which contained 50 of μL bacterial suspension and 50 μL of CMHB; and negative control (12th well), which was filled with 100 μL of CMHB. The plates were incubated at 37oC for 16–20 h. The MIC is recorded as less than or equal to the lowest concentration with no visible turbidity, indicating that no bacterial growth occurred [72].
4.6. Assessment of biofilm tolerance, bacterial antibiotic resistance mutation, and bacterial viability
The assessment of biofilm tolerance, bacterial antibiotic resistance mutation, and bacterial viability was summarized in Fig. 5. At the time of biofilm tolerance measurement, the microplate containing the microreactors was rinsed using a microplate washer (50 TS, Biotek) with sterile DI water. Fluorescence or bioluminescence signals were measured using a microplate reader (Microplate Reader Tecan M1000 PRO) daily for three days. Signals of E. coli RP437/pRSH103 were recorded at Ex/Em of 558/583 nm. Signals of P. aeruginosa PAO1/pTdK-GFP and S. aureus AH2547/pCM29 were obtained at Ex/Em of 500/515 nm and 490/515 nm, respectively. All experiments were conducted in triplicate. Results are reported as mean ± standard deviation. After the measurements, fresh antibiotics and control media were reintroduced to the microplate. The process was repeated for three days.
Bacterial antibiotic resistance mutation was further assessed with the fluorescence strains but not the bioluminescent strain E. coli Xen14. A parallel set of the hydrogels was dissolved each day for two days with a 1 mL solution of 0.05 M Na2CO3 and 0.02 M nitric acid followed by a 5 min sonication, suspended at 4000 RPM for 2 min, and the dissolving medium replaced with PBS to obtain a 1 mL pellet of bacteria for MIC antibiotic agar plating to assess antibiotic resistance mutations. At the same time, a portion from the same dissolved hydrogels was plated on LB agar for viability assessment. For viability control, a triplicate set of dissolved microreactors after 24 h biofilm culturing was plated on LB agar. Cell plating was preceded by serial dilution. The nutrient plates (LB agar) and MIC plates for each strain were cultured at 37oC for 18–20 h, and the cell viability was confirmed by determining Log10(CFU/mL) from an agar plate. Since the solid substrate was dissolved and rinsed with PBS to remove any remaining antimicrobial, there was no need to evaluate the neutralization efficacy and effects of neutralization on the test strains themselves.
4.7. Statistical analysis
All experiments were performed in triplicate unless specified otherwise. Results are reported as mean ± standard deviation. The effects on fluorescence or bioluminescence by type of treatment and duration of exposure were evaluated with 2-factor ANOVA with Tukey's post hoc tests to compare between pairs of groups. All tests were performed using Sigmaplot (version 12.5) with the level of significance set at p < 0.05.
Disclaimer
The findings and conclusions in this paper have not been formally disseminated by the U.S. FDA and should not be constructed to represent any agency determination or policy. The mentions of commercial products, their sources, or their use in connection with material reported herein are not to be construed as either an actual or implied endorsement of such products by the Department of Health and Human Services.
CRediT authorship contribution statement
Le Hoang Phu Pham: Conceptualization, Methodology, Data curation, Formal analysis, Investigation, Writing – original draft. Khanh Loan Ly: Investigation, Methodology, Writing – original draft. Mariliz Colon-Ascanio: Investigation. Jin Ou: Investigation. Hao Wang: Methodology. Sang Won Lee: Methodology. Yi Wang: Methodology. John S. Choy: Methodology, Funding acquisition, Supervision, Resources, Writing – review & editing. Kenneth Scott Phillips: Conceptualization, Methodology, Data curation, Funding acquisition, Project administration, Supervision, Validation, Visualization, Resources, Writing – review & editing. Xiaolong Luo: Conceptualization, Methodology, Data curation, Funding acquisition, Project administration, Supervision, Validation, Resources, Writing – review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Xiaolong Luo and John S. Choy report financial support was provided by National Institutes of Health. Le Hoang Phu Pham, Xiaolong Luo and Kenneth Scott Phillips have patent pending.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2022.100103.
Contributor Information
John S. Choy, Email: choy@cua.edu.
Kenneth Scott Phillips, Email: Kenneth.Phillips@fda.hhs.gov.
Xiaolong Luo, Email: luox@cua.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Khatoon Z., McTiernan C.D., Suuronen E.J., Mah T.-F., Alarcon E.I. Heliyon. 2018;4 doi: 10.1016/j.heliyon.2018.e01067. e01067-e01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma D., Misba L., Khan A.U. Antimicrob Resist Infect Control. 2019;8:76. doi: 10.1186/s13756-019-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee S.W., Phillips K.S., Gu H., Kazemzadeh-Narbat M., Ren D. Biomaterials. 2021;268 doi: 10.1016/j.biomaterials.2020.120595. [DOI] [PubMed] [Google Scholar]
- 4.A. D. Verderosa, M. Totsika and K. E. Fairfull-Smith, 2019, 7.
- 5.Wilson C., Lukowicz R., Merchant S., Valquier-Flynn H., Caballero J., Sandoval J., Okuom M., Huber C., Brooks T.D., Wilson E., Clement B., Wentworth C.D., Holmes A.E. Res. Rev. J. Eng. Technol. 2017;6 [PMC free article] [PubMed] [Google Scholar]
- 6.Magana M., Sereti C., Ioannidis A., Mitchell C.A., Ball A.R., Magiorkinis E., Chatzipanagiotou S., Hamblin M.R., Hadjifrangiskou M., Tegos G.P. Clin Microbiol Rev. 2018:31. doi: 10.1128/CMR.00084-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee S.W., Gu H., Kilberg J.B., Ren D. Acta Biomater. 2018;81:93–102. doi: 10.1016/j.actbio.2018.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H., Tampio A.J.F., Xu Y., Nicholas B.D., Ren D. ACS Biomater Sci Eng. 2020;6:727–738. doi: 10.1021/acsbiomaterials.9b01199. [DOI] [PubMed] [Google Scholar]
- 9.Gu H., Lee S.W., Carnicelli J., Zhang T., Ren D. Nat Commun. 2020;11:2211. doi: 10.1038/s41467-020-16055-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee S.W., Carnicelli J., Getya D., Gitsov I., Phillips K.S., Ren D. ACS Appl Mater Interfaces. 2021;13:17174–17182. doi: 10.1021/acsami.0c20697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Belkum A., Bachmann T.T., Lüdke G., Lisby J.G., Kahlmeter G., Mohess A., Becker K., Hays J.P., Woodford N., Mitsakakis K., Moran-Gilad J., Vila J., Peter H., Rex J.H., Dunne W.M. Nat Rev Microbiol. 2019;17:51–62. doi: 10.1038/s41579-018-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ersoy S.C., Heithoff D.M., Barnes L., Tripp G.K., House J.K., Marth J.D., Smith J.W., Mahan M.J. EBioMedicine. 2017;20:173–181. doi: 10.1016/j.ebiom.2017.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brukner I., Oughton M. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.01820. 1820-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffey B.M., Anderson G.G. Methods Mol Biol. 2014;1149:631–641. doi: 10.1007/978-1-4939-0473-0_48. [DOI] [PubMed] [Google Scholar]
- 15.L. Hall-Stoodley, J. Rayner, P. Stoodley and H. Lappin-Scott, 2008, vol. 12, pp. 307-318.
- 16.Harrison J., Ceri H., Yerly J., Stremick C., Hu Y., Martinuzzi R., Turner R. Biol Proced Online. 2006;8:194–215. doi: 10.1251/bpo127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banin E., Brady K.M., Greenberg E.P. Appl Environ Microbiol. 2006;72:2064–2069. doi: 10.1128/AEM.72.3.2064-2069.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams D., Bloebaum R. Microsc Microanal : the official journal of Microscopy Society of America, Microbeam Analysis Society, Microscopical Society of Canada. 2010;16:143–152. doi: 10.1017/S143192760999136X. [DOI] [PubMed] [Google Scholar]
- 19.Decho A.W., Gutierrez T. Front Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donlan R.M. Emerg Infect Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phillips K.S., Wang Y. Plast Reconstr Surg. 2017;140:632e–633e. doi: 10.1097/PRS.0000000000003723. [DOI] [PubMed] [Google Scholar]
- 22.Ren Y., Jongsma M.A., Mei L., van der Mei H.C., Busscher H.J. Clin Oral Invest. 2014;18:1711–1718. doi: 10.1007/s00784-014-1240-3. [DOI] [PubMed] [Google Scholar]
- 23.Schwarz E.M., McLaren A.C., Sculco T.P., Brause B., Bostrom M., Kates S.L., Parvizi J., Alt V., Arnold W.V., Carli A., Chen A.F., Choe H., Coraça-Huber D.C., Cross M., Ghert M., Hickok N., Jennings J.A., Joshi M., Metsemakers W.-J., Ninomiya M., Nishitani K., Oh I., Padgett D., Ricciardi B., Saeed K., Sendi P., Springer B., Stoodley P., Wenke J.C., H. f. S. S. B. S. Workgroup J Orthop Res. 2021;39:227–239. doi: 10.1002/jor.24616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong M., Wang Y., Wang H., Marrone A.K., Haugen S.P., Kulkarni K., Basile R., Phillips K.S. Biomed Instrum Technol. 2020;54:397–409. doi: 10.2345/0899-8205-54.6.397. [DOI] [PubMed] [Google Scholar]
- 25.Isguven S., Fitzgerald K., Delaney L., Harwood M., Schaer T., Hickok N. Eur Cell Mater. 2022;43:6–21. doi: 10.22203/eCM.v043a03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., Guan A., Isayeva I., Vorvolakos K., Das S., Li Z., Phillips K.S. Biomaterials. 2016;95:74–85. doi: 10.1016/j.biomaterials.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Pabst B., Pitts B., Lauchnor E., Stewart P.S. Antimicrob Agents Chemother. 2016;60:6294. doi: 10.1128/AAC.01336-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart E.J., Ganesan M., Younger J.G., Solomon M.J. Sci Rep. 2015;5 doi: 10.1038/srep13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kandemir N., Vollmer W., Jakubovics N.S., Chen J. Sci Rep. 2018;8 doi: 10.1038/s41598-018-29269-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bjarnsholt T., Alhede M., Alhede M., Eickhardt-Sorensen S.R., Moser C., Kuhl M., Jensen P.O., Hoiby N. Trends Microbiol. 2013;21:466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Roberts A.E., Kragh K.N., Bjarnsholt T., Diggle S.P. J Mol Biol. 2015;427:3646–3661. doi: 10.1016/j.jmb.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Sonderholm M., Kragh K.N., Koren K., Jakobsen T.H., Darch S.E., Alhede M., Jensen P.O., Whiteley M., Kuhl M., Bjarnsholt T. Appl Environ Microbiol. 2017;83 doi: 10.1128/AEM.00113-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolajeva V., Zommere Ž. Environ. Exp. Biol. 2017;15:105–111. [Google Scholar]
- 34.Luo X., Tsao C.-Y., Wu H.-C., Quan D.N., Payne G.F., Rubloff G.W., Bentley W.E. Lab Chip. 2015;15:1842–1851. doi: 10.1039/c5lc00107b. [DOI] [PubMed] [Google Scholar]
- 35.Luo X., Wu H.-C., Tsao C.-Y., Cheng Y., Betz J., Payne G.F., Rubloff G.W., Bentley W.E. Biomaterials. 2012;33:5136–5143. doi: 10.1016/j.biomaterials.2012.03.037. [DOI] [PubMed] [Google Scholar]
- 36.Cheng Y., Tsao C.-Y., Wu H.-C., Luo X., Terrell J.L., Betz J., Payne G.F., Bentley W.E., Rubloff G.W. Adv Funct Mater. 2012;22:519–528. [Google Scholar]
- 37.Mørch Ý.A., Donati I., Strand B.L., Skjåk-Bræk G. Biomacromolecules. 2006;7:1471–1480. doi: 10.1021/bm060010d. [DOI] [PubMed] [Google Scholar]
- 38.Lee S.W., Johnson E.L., Chediak J.A., Shin H., Wang Y., Phillips K.S., Ren D. ACS Appl Bio Mater. 2022;5:3816–3825. doi: 10.1021/acsabm.2c00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y., Borthwell R.M., Hori K., Clarkson S., Blumstein G., Park H., Hart C.M., Hamad C.D., Francis K.P., Bernthal N.M., Phillips K.S. J Biomed Mater Res B Appl Biomater. 2022;110:1932–1941. doi: 10.1002/jbm.b.35050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H., Agrawal A., Wang Y., Crawford D.W., Siler Z.D., Peterson M.L., Woofter R.T., Labib M., Shin H.Y., Baumann A.P., Phillips K.S. Sci Rep. 2021;11:5746. doi: 10.1038/s41598-021-84826-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong M., Wang Y., Wang H., Marrone A.K., Haugen S.P., Kulkarni K., Basile R., Phillips K.S. Biomed Instrum Technol. 2020;54:397–409. doi: 10.2345/0899-8205-54.6.397. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y., Leng V., Patel V., Phillips K.S. Sci Rep. 2017;7 doi: 10.1038/srep45070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sønderholm M., Koren K., Wangpraseurt D., Jensen P.Ø., Kolpen M., Kragh K.N., Bjarnsholt T., Kühl M. NPJ Biofilms and Microbiomes. 2018;4:3. doi: 10.1038/s41522-018-0047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poonguzhali R., Khaleel Basha S., Sugantha Kumari V. Polym Bull. 2018;75:4165–4173. [Google Scholar]
- 45.Blandón L.M., Islan G.A., Castro G.R., Noseda M.D., Thomaz-Soccol V., Soccol C.R. Colloids Surf B Biointerfaces. 2016;145:706–715. doi: 10.1016/j.colsurfb.2016.05.078. [DOI] [PubMed] [Google Scholar]
- 46.Unagolla J.M., Jayasuriya A.C. Eur J Pharmaceut Sci. 2018;114:199–209. doi: 10.1016/j.ejps.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie L., Wei H., Kou L., Ren L., Zhou J. Mater Werkst. 2020;51:850–855. [Google Scholar]
- 48.Zhang X., Lin X., He Y., Chen Y., Luo X., Shang R. Int J Biol Macromol. 2019;124:418–428. doi: 10.1016/j.ijbiomac.2018.11.218. [DOI] [PubMed] [Google Scholar]
- 49.Vo T., Shah S.B., Choy J.S., Luo X. Biomicrofluidics. 2020;14 doi: 10.1063/1.5128739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pham L.H.P., Colon-Ascanio M., Ou J., Ly K., Hu P., Choy J.S., Luo X. Lab Chip. 2022;22:4349–4358. doi: 10.1039/d2lc00728b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu P., Ly K.L., Pham L.P.H., Pottash A.E., Sheridan K., Wu H.-C., Tsao C.-Y., Quan D., Bentley W.E., Rubloff G.W., Sintim H.O., Luo X. Lab Chip. 2022;22:3203–3216. doi: 10.1039/d2lc00481j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gombotz W.R., Wee S. Adv Drug Deliv Rev. 1998;31:267–285. doi: 10.1016/s0169-409x(97)00124-5. [DOI] [PubMed] [Google Scholar]
- 53.Andresen I.-L., Skipnes O., SmidsrØD O., Ostgaard K., Hemmer P.C. Cellulose chemistry and technology. Am Chem Soc. 1977;48:361–381. ch. 24. [Google Scholar]
- 54.Yan J., Bassler B.L. Cell Host Microbe. 2019;26:15–21. doi: 10.1016/j.chom.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.M. Mizozoe, M. Otaki and K. Aikawa, 2019, 11, 2156.
- 56.Starzak K., Matwijczuk A., Creaven B., Matwijczuk A., Wybraniec S., Karcz D. Int J Mol Sci. 2019;20:281. doi: 10.3390/ijms20020281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dinos G.P., Athanassopoulos C.M., Missiri D.A., Giannopoulou P.C., Vlachogiannis I.A., Papadopoulos G.E., Papaioannou D., Kalpaxis D.L. Antibiotics (Basel) 2016;5:20. doi: 10.3390/antibiotics5020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olson M.E., Ceri H., Morck D.W., Buret A.G., Read R.R. Can J Vet Res. 2002;66:86–92. [PMC free article] [PubMed] [Google Scholar]
- 59.Curbelo J., Moulton K., Willard S. Theriogenology. 2010;73:48–55. doi: 10.1016/j.theriogenology.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 60.Gregor C., Gwosch K.C., Sahl S.J., Hell S.W. Proc Natl Acad Sci USA. 2018;115:962. doi: 10.1073/pnas.1715946115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kussell E., Kishony R., Balaban N.Q., Leibler S. Genetics. 2005;169:1807–1814. doi: 10.1534/genetics.104.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ribeiro S.M., Felício M.R., Boas E.V., Gonçalves S., Costa F.F., Samy R.P., Santos N.C., Franco O.L. Pharmacol Therapeut. 2016;160:133–144. doi: 10.1016/j.pharmthera.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 63.Ferrer M.D., Rodriguez J.C., Alvarez L., Artacho A., Royo G., Mira A. J Appl Microbiol. 2017;122:640–650. doi: 10.1111/jam.13368. [DOI] [PubMed] [Google Scholar]
- 64.Castaneda P., McLaren A., Tavaziva G., Overstreet D. Clin Orthop Relat Res. 2016;474:1659–1664. doi: 10.1007/s11999-016-4700-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okae Y., Nishitani K., Sakamoto A., Kawai T., Tomizawa T., Saito M., Kuroda Y., Matsuda S. Front Cell Infect Microbiol. 2022;12 doi: 10.3389/fcimb.2022.896978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hou S., Gu H., Smith C., Ren D. Langmuir. 2011;27:2686–2691. doi: 10.1021/la1046194. [DOI] [PubMed] [Google Scholar]
- 67.Chamsaz E.A., Mankoci S., Barton H.A., Joy A. ACS Appl Mater Interfaces. 2017;9:6704–6711. doi: 10.1021/acsami.6b12610. [DOI] [PubMed] [Google Scholar]
- 68.Pang Y.Y., Schwartz J., Thoendel M., Ackermann L.W., Horswill A.R., Nauseef W.M. J Innate Immunity. 2010;2:546–559. doi: 10.1159/000319855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee K.Y., Mooney D.J. Prog Polym Sci. 2012;37:106–126. doi: 10.1016/j.progpolymsci.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pham P.L.H., Rooholghodos S.A., Choy J.S., Luo X. Advanced Biosystems. 2018;2 [Google Scholar]
- 71.Mater D.D.G., Jean-Noël B., José Edmundo N.S., Nicole T., Daniel T. Biotechnol Tech. 1995;9:747–752. [Google Scholar]
- 72.Wiegand I., Hilpert K., Hancock R.E.W. Nat Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.