Abstract
Botulinum neurotoxin-producing species of Clostridium are highly diverse. Clostridium botulinum could represent at least four different species of Clostridium. In addition, strains that do not produce botulinum neurotoxin are closely related to toxigenic strains, probably representing the same species. Although reclassification of these organisms has been proposed in the past, their species names have remained unchanged, mainly because of the premise that changing names of medically relevant organisms might cause confusion in the healthcare and scientific community. In this review, we discuss the possible unintended consequences of reclassifying botulinum neurotoxin-producing species of Clostridium, which are of public health, medical, and biodefense interest.
Keywords: Botulinum neurotoxin, Clostridium botulinum, Botulinum neurotoxin-producing clostridia, Taxonomy
1. Introduction
Clostridium botulinum is a Gram-positive, anaerobic, spore-forming bacterium that produces botulinum neurotoxin (BoNT) (Sobel, 2005). BoNT can also be produced by rare strains of Clostridium baratii and Clostridium butyricum (Sobel, 2005). BoNT and BoNT-producing species of Clostridium are of public health and biodefense interest because they present risk of misuse causing mass casualties, thereby posing a severe threat to public health and safety. These organisms are considered Tier 1 Select Agents in the United States, and they are classified as Very High Threat Agents in the European Union (Tian and Zheng, 2014). BoNT blocks the release of acetylcholine at the neuromuscular junction, resulting in paralysis (Sobel, 2005). There are seven serologically distinct types of BoNT: serotypes A through G. Additional serotypes have been proposed (X, En, and H) but have not been accepted by consensus as new BoNT serotypes within the scientific community (Barash and Arnon, 2014; Mansfield et al., 2015; Brunt et al., 2018).
C. botulinum was first described in 1897 by E. van Ermengem, during an investigation of a foodborne botulism outbreak in Belgium. The bacterium was originally named Bacillus botulinus, and it was later assigned to the genus Clostridium (Collins and East, 1998). Similar organisms isolated during subsequent botulism investigations were later designated as C. botulinum. According to the Bergey’s Manual of Systematics of Archaea and Bacteria, all those organisms are classified as C. botulinum despite their physiological characteristics “because of the unique and similar action of the toxins produced by all strains” (Rainey et al., 2015).
C. botulinum is a highly diverse group of organisms. Historically, the species has been divided into four metabolically distinct groups (Holdeman and Brooks, 1970): Group I, formed by C. botulinum type A and proteolytic strains of C. botulinum types B and F are proteolytic, grow optimally at 37 °C, and form spores with high heat resistance; Group II, formed by C. botulinum type E and non-proteolytic strains of C. botulinum types B and F are non-proteolytic, saccharolytic, grow optimally at 30 °C, and form spores with low heat resistance; Group III, formed by C. botulinum types C and D are non-proteolytic, grow optimally at 40 °C, and form spores with intermediate heat resistance; Group IV, formed by C. botulinum type G are proteolytic, grow optimally at 37 °C, and their spore heat resistance is similar to that of Group III. In addition, other non-BoNT-producing species are closely related to these groups. C. botulinum Group I and C. sporogenes are closely related; these two species cannot be distinguished by biochemical methods or 16 S rRNA sequences, and can only be identified by toxin neutralization tests in mice (Rainey et al., 2015). Group III strains and Clostridium novyi type A are closely related; moreover, C. botulinum type C can be cured of type C phage and converted to C. novyi type A following infection by a C. novyi type A phage (Rainey et al., 2015). Also, Group IV strains are closely related to Clostridium subterminale. Suen et al. (1988) proposed renaming C. botulinum Group IV as C. argentinense, a new species which would include nontoxigenic strains of Clostridium subterminale and Clostridium hastifome.
Reclassification of BoNT-producing species of Clostridium has been discussed in the past, as each of the four Groups could represent separate species (Collins and East, 1998), but the name has remained unchanged, mainly because of the premise that changing names of medically relevant organisms can cause confusion in the healthcare and scientific community (Lawson et al., 2016). In 1998, Collins and East (1998) proposed that, because of the clinical and veterinary importance of Clostridium botulinum, any future nomenclature must consider BoNT production, in addition to phenotypic and genotypic information. Recently, in 2018, Smith et al. (Smith et al., 2018) proposed that seven distinct species of Clostridium would be capable of producing BoNT. The authors proposed that the current metabolic group designations should be replaced by the following classification: 1) C. botulinum Group I and related strains of C. sporogenes should be referred to as C. parabotulinum; 2) C. botulinum Group II should be referred to as C. botulinum; 3) C. botulinum Group III and related strains of novyi type A should be referred to as C. novyi sensu lato; and 4) C. argentinense, C. baratii, C. butyricum, and C. sporogenes should retain their current species names (Smith et al., 2018). This new classification has not been formally accepted, as it has not been recorded on the validation lists published by the International Journal of Systematic and Evolutionary Microbiology (https://www.bacterio.net/ and https://lpsn.dsmz.de/). In this review, we discuss possible unintended consequences of reclassifying these organisms.
1.1. C. botulinum Group I and related species
Recent studies using whole genome sequence analyses have confirmed a marked similarity between C. botulinum and C. sporogenes. For instance, Weigand et al. in 2015 (Weigand et al., 2015) analyzed the genomes of 28 strains of C. sporogenes and 9 strains of C. botulinum Group I by core genome phylogeny and variable gene content analysis. The genomes formed two separate clades; 5 of the 9 C. botulinum strains were more closely related to C. sporogenes strains than to other C. botulinum Group I strains, and 2 of the 28 C. sporogenes strains were more closely related to C. botulinum Group I than to other C. sporogenes strains. The Average Nucleotide Identity (ANI) between the two clades was 93%, which is below 95%, the cutoff value frequently used for species demarcation (Kim et al., 2014). Weigand et al. (2015) also suggested that C. sporogenes clade-specific genes could provide a genomic signature for “true” C. sporogenes strains. This approach could be a useful research tool but would require sequencing isolates before classifying them as C. sporogenes or C. botulinum, which could delay botulism case investigations.
Similarly, Williamson et al. (2016) reported in 2016 that four C. sporogenes and five C. botulinum type B strains clustered together by core genome phylogeny. The study also reports two clades within Group I. ANI values were above 95% within each of the two clades, and fell below 95% (minimum, ~92%) when comparing all strains within Group I. The authors proposed that the ability to produce BoNT type B within the C. sporogenes/C. botulinum type B clade appears to be plasmid-mediated. A recent study by Wentz et al., published in 2021 provides data supporting this idea (Wentz et al., 2021).
A study by Cruz-Morales et al. (2019) reported in 2019, analyzed 779 genomes of Clostridium, including 106 genomes of Clostridium botulinum Group I (types A, B, and F) and C. sporogenes, by core genome phylogeny, using 27 conserved proteins among those genomes. Based on those 27 conserved proteins, the study showed that strains C. botulinum types A, B, and F and C. sporogenes clustered together, separated from other C. botulinum strains. Further analysis revealed that this subgroup had an open pangenome, meaning that the number of new gene families continuously increased in this lineage, demonstrating larger genetic diversity. The observations from this study support the presence of distinct lineages among C. botulinum strains.
In a more recent study published in 2020, Brunt et al. (2020a) reported a comparative genomic analysis of 556 strains of C. botulinum Group I and C. sporogenes, using core genome single-nucleotide polymorphism. The study reported that 23 of 452 (5%) strains assigned to a C. botulinum Group I lineage did not possess a bont gene, and that the genome of 20 of 104 (19%) of strains assigned to a C. sporogenes lineage possessed bont gene(s). Brunt et al. also reported observing two main clades within Group I. The study did not report ANI values.
1.2. C. botulinum Group II and related species
Williamson et al. (2016) reported in 2016 the analysis of 15 genomes of C. botulinum Group II by core genome single-nucleotide polymorphism analysis. They found that the 15 genomes were divided into two distinct clades; one clade contained C. botulinum type E only, and the other clade included C. botulinum types B, E and F. ANI values within each clade were greater than 97%, and ANI for all strains fell below the species-delineating threshold of 95% (minimum of ~94%). The study did not include non-toxigenic strains.
Cruz-Morales et al. (2019) reported in 2019 the analysis of 779 genomes of Clostridium, including 20 genomes of C. botulinum type E by core genome phylogeny, and showed that this group clustered separately from other C. botulinum strains. The study also indicates that these strains had an almost closed pangenome, implying loss of genetic diversity, as fewer gene families were being added to the pangenome.
Brunt et al. (2020b), reported in 2020 the analysis of 208 genomes of non-proteolytic C. botulinum strains by core genome single-nucleotide polymorphism. They also reported two major lineages, one with type E-producing strains only, the other one with strains producing BoNT types B, E or F. ANI was not calculated. In addition, the study reported that 31 of the 208 (15%) strains did not harbor a bont gene but still clustered with C. botulinum Group II strains. Both major lineages included non-toxigenic strains. The strains lacking a bont gene were closely related to strains possessing the bont gene on a plasmid or the chromosome. Although no specific name has been used to describe these strains, non-toxigenic organisms that resemble C. botulinum Group II have been reported (Collins and East, 1998). Unfortunately, most of the non-toxigenic strains used in the study by Brunt et al. did not list a known source, country, or year of isolation; therefore, epidemiological inferences of the observed similarity are limited.
1.3. C. botulinum Group III and related species
Skarin et al. (2011) reported in 2011 the analysis of whole genome sequences of six strains of C. botulinum Group III and one strain of C. novyi A by average similarity of the conserved core. The study reports that the genome of the C. novyi strain belonged to the same lineage as C. botulinum type C strains. The authors proposed classifying C. botulinum as dual species: a pathospecies C. botulinum, which would include all BoNT-producing strains (types A through G), and a genospecies C. novyi sensu lato which would include C. botulinum Group III, C. novyi and C. haemolyticum (Skarin et al., 2011). This proposed classification has not been formally recorded on the lists published by the International Journal of Systematic and Evolutionary Microbiology (https://www.bacterio.net/ and https://lpsn.dsmz.de/).
Skarin and Segerman (2014) also showed that 24 strains of C. botulinum Group III, C. novyi, and C. haemolyticum were closely related when analyzed by pairwise average BLASTN score similarities. Genomic comparisons of the 24 genomes and 61 plasmids revealed four separate lineages, which did not strictly correlate with the species designations, highlighting the genomic complexity within the C. novyi sensu lato group (Skarin and Segerman, 2014). Lineage I included only C. botulinum group III strains, while lineages II, III, and IV included strains of C. botulinum, C. haemolyticum, and C. novyi.
In 2019, Cruz-Morales et al. (2019) reported the analysis of 779 genomes of Clostridium, including 42 genomes of C. botulinum types C, D, and mosaic CD, C. haemolyticum, and C. novyi, and showed high synteny among these strains. In addition, the study showed that this group has an almost closed pangenome, meaning that a few gene families were added to the pangenome, suggesting loss of genetic diversity.
In a more recent study reported in 2021, Fillo et al. (2021) used overall genomic similarity to analyze the phylogenetic relations among genomes of 60 C. botulinum types C, D, C/D, and D/C (newly sequenced) and the genomes of 36 C. botulinum, 9 C. novyi, and 3 C. haemolyticum (previously sequenced). The same four lineages mentioned above were also identified by Fillo et al., who further divided lineage I into two branches, IA and IB. Analysis of the botulinum neurotoxin gene revealed that the four BoNT serotypes produced by C. botulinum group III (C, D, C/D and D/C) were highly conserved; no new subtypes were identified.
1.4. C. botulinum Group IV and related species
Worldwide, few BoNT type G-producing strains have been identified; these strains have been classified as C. botulinum type G or C. argentinense. Suen et al. (1988) reported in 1988 the use of DNA hybridization studies to characterize 9 strains of C. botulinum type G, 11 strains of C. subterminale, 3 strains of C. hastiforme, and several other strains of C. botulinum types A, B, and F, C. sporogenes, and other related species. The study showed that all nine C. botulinum type G strains, two C. subterminale strains, and one C. hastiforme strain were included in the same hybridization group, sharing 94% intragroup relatedness. The authors proposed renaming C. botulinum Group IV as C. argentinense, a new species that would include non-toxigenic strains of C. subterminale and C. hastiforme (Suen et al., 1988). C. argentinense has since been formally recorded on the lists published by the International Journal of Systematic and Evolutionary Microbiology (https://www.bacterio.net/ and https://lpsn.dsmz.de/). No phylogenetic analyses of C. argentinense have been published recently.
2. Relatedness among C. botulinum and related species by ANI analysis
ANI is used to calculate the relatedness between genome sequences, to determine if they belong to the same or separate species. The proposed and generally accepted species boundary for ANI values is ~95% (Chun et al., 2018). We used Mashtree v.0.37 (Katz et al., 2019) to find nearest neighbors among representatives of metabolic groups I, II, and III (Fig. 1); we also determined ANI for eight clusters within metabolic groups I, II, and III (Table 1) by using publicly available sequences. Unfortunately, whole genome sequences from only two of the C. argentinense strains characterized by Suen et al. (Holdeman and Brooks, 1970) were publicly available; thus, ANI was not calculated for this group. In addition, ANI was not calculated for C. butyricum type E and C. baratii type F as no substantial genomic variability has been shown between toxigenic and non-toxigenic strains within the two species, and the two groups are clearly separated from other BoNT-producing species of Clostridium (Smith et al., 2018).
Fig. 1.

Neighbor-Joining tree drawn using mash distances between whole genome sequences. Tree created using mashtree 0.37 and annotated with the Interactive Tree of Life v. 6.5.4 (Letunic and Bork, 2021).
Table 1.
Summary of BoNT-producing species of Clostridium, divided into phylogenetic clades.
| Metabolic Group | Cluster | Species included in each clade | Reference |
|---|---|---|---|
| I | 1 | All C. botulinum type A Proteolytic C. botulinum type B Proteolytic C. botulinum type F C. sporogenes |
(Smith et al., 2018; Weigand et al., 2015; Williamson et al., 2016; Brunt et al., 2020a) |
| I | 2 |
C. botulinum type B C. sporogenes |
(Weigand et al., 2015; Williamson et al., 2016; Brunt et al., 2020a) |
| II | 3 |
C. botulinum type E Non-proteolytic, non-toxigenic C. botulinum |
(Williamson et al., 2016; Brunt et al., 2020b) |
| II | 4 |
C. botulinum type E Non-proteolytic C. botulinum type B Non-proteolytic C. botulinum type F Non-proteolytic, non-toxigenic C. botulinum |
(Williamson et al., 2016; Brunt et al., 2020b) |
| III | 5 |
C. botulinum type D C. botulinum type C/D C. botulinum type D/C |
(Skarin and Segerman, 2014; Fillo et al., 2021) |
| III | 6 |
C. botulinum type D C. botulinum type C C. novyi C. haemolyticum |
(Skarin and Segerman, 2014; Fillo et al., 2021) |
| III | 7 |
C. botulinum type C/D C. novyi |
(Skarin and Segerman, 2014; Fillo et al., 2021) |
| III | 8 |
C. botulinum type D/C C. novyi |
(Skarin and Segerman, 2014; Fillo et al., 2021) |
| IV | 9 |
C. argentinense (C. botulinum type G, C. subterminale, and C. hastiforme) |
Suen et al. (1988) |
| N/A | 10 | C. butyricum type E | Collins and East (1998) |
| N/A | 11 | C. baratii type F | Collins and East (1998) |
Clusters 1 and 2 within metabolic group I, which include C. botulinum type A, proteolytic C. botulinum types B and F, and C. sporogenes strains, yielded an average ANI value of 93.1% between the two clusters (maximum 93.9%) (Fig. 2), confirming findings from other authors that the ANI between these two clusters fall outside values corresponding to a single species (Weigand et al., 2015; Williamson et al., 2016).
Fig. 2.

ANI matrix for metabolic Group I. Two-way ANI and coverage values were determined using an in-house script that utilizes MUMmer v.4. All pairwise comparisons within the group resulted in query coverage >70%, and ANI values delineated 2 clusters within the group, each with >95% similarity within the cluster by ANI. All pairwise ANI values in the group are >90%.
Clusters 3 and 4 within metabolic group II, which include C. botulinum type E, non-proteolytic C. botulinum types B and F, and non-toxigenic strains, resulted in an average ANI value of 94% (maximum 94.1%) (Fig. 3), slightly below the species threshold of 95%.
Fig. 3.
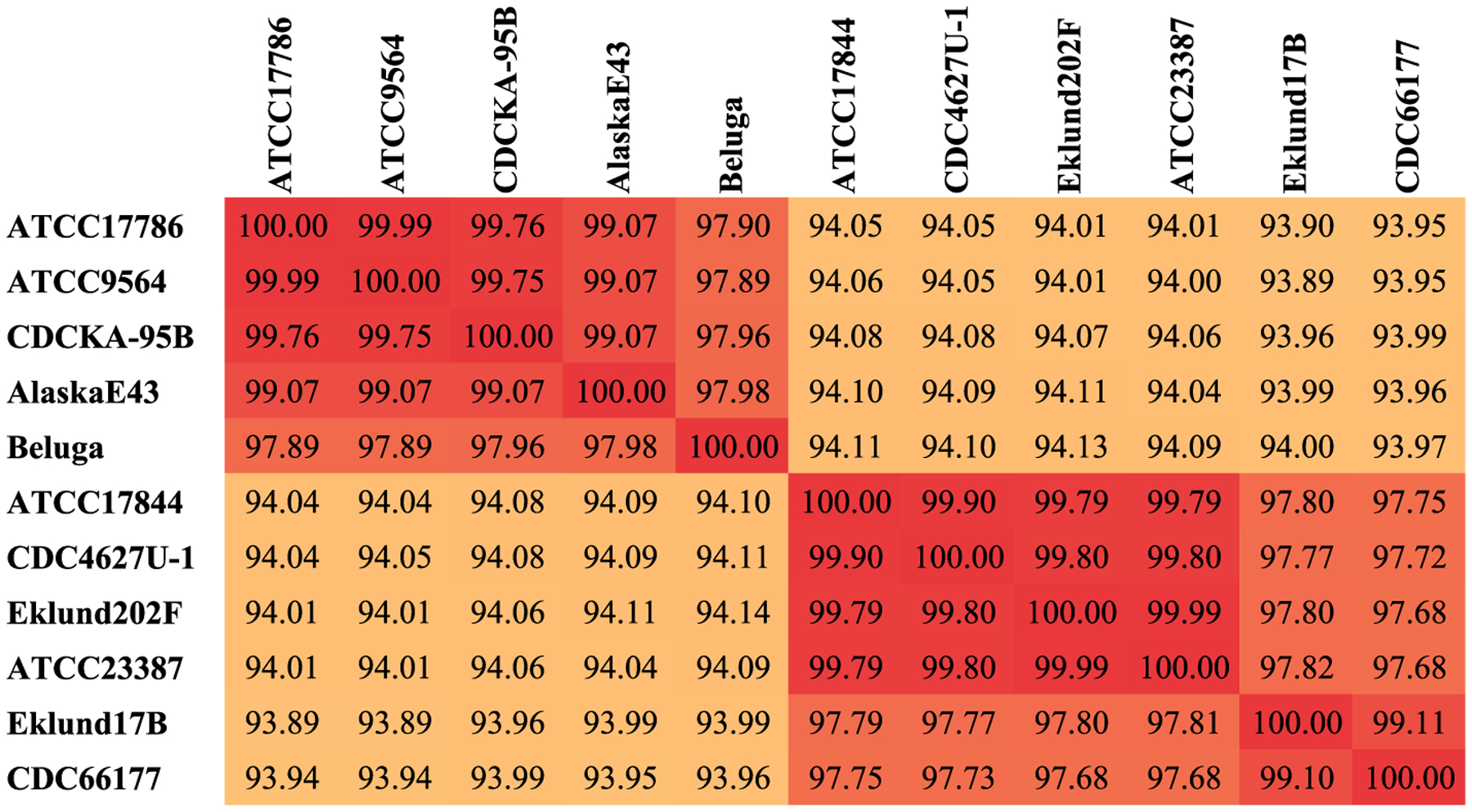
ANI matrix for metabolic group II. Two-way ANI and coverage values were determined using an in-house script that utilizes MUMmer v.4. All pairwise comparisons within the group resulted in query coverage >70%, and ANI values delineated 2 clusters within the group, each with >95% similarity within the cluster by ANI. All pairwise ANI values in the group are >90%.
Clusters 5, 6, 7, and 8 within metabolic group III, which include C. botulinum types C, D, C/D, and D/C, C. novyi, and C. haemolyticum, resulted in the following average ANI values (Fig. 4): 92.3% (maximum 92.8%) between clusters 5 and 6; 86.1% (maximum 87.8%) between clusters 5 and 7; 85.3% (maximum 85.8%) between clusters 5 and 8; 85.9% (maximum 86.9%) between clusters 6 and 7; 85.5% (maximum 85.9%) between clusters 6 and 8; and 90.7% (maximum 91%). These results do not support the proposal of Skarin et al. (2011) to include C. botulinum Group III, C. novyi and C. haemolyticum as part of the genospecies C. novyi sensu lato. As ANI values among these clusters fall below the species threshold of 95%. This discrepancy might be explained by the different methods employed: ANI in this report, and conserved core genome by Skarin et al. (2011).
Fig. 4.
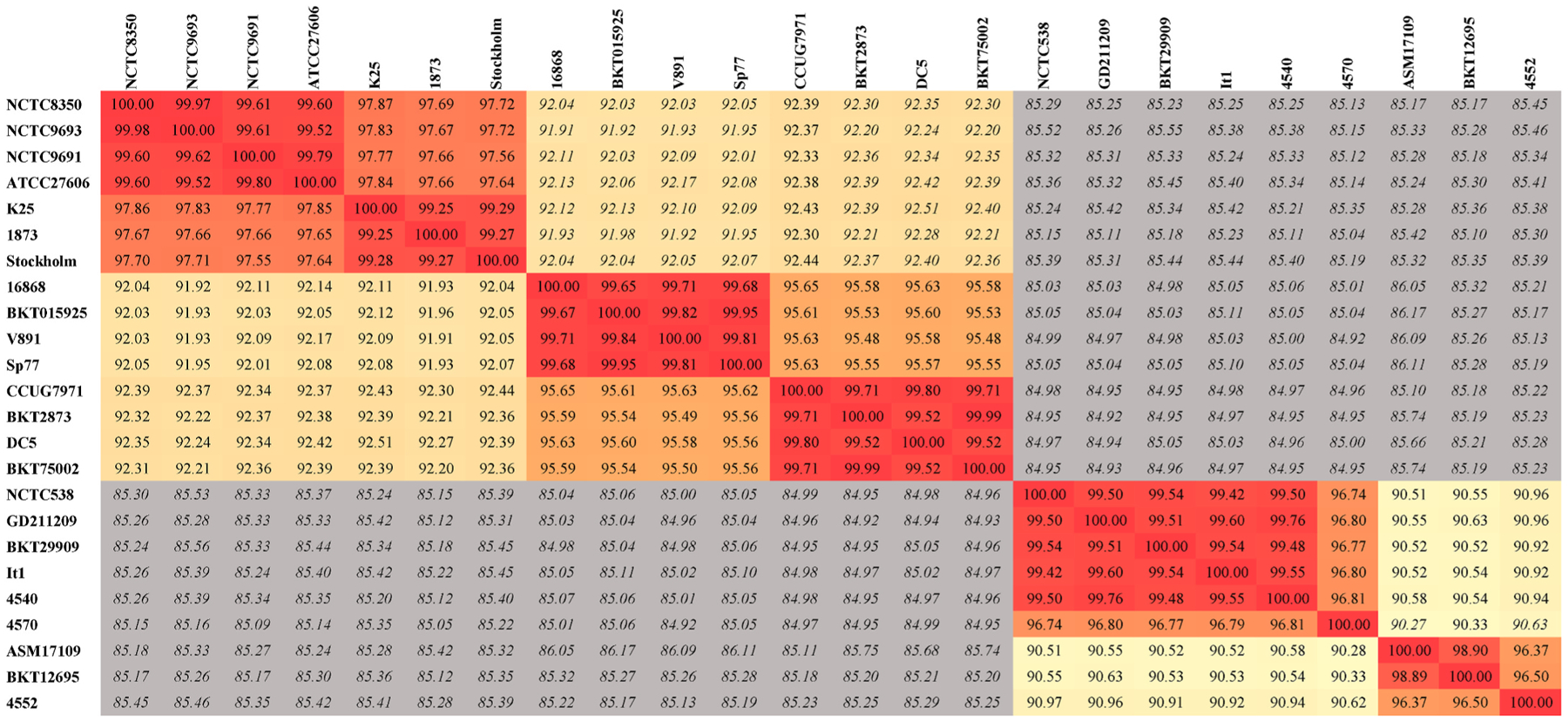
ANI matrix for metabolic group III. Two-way ANI and coverage values were determined using an in-house script that utilizes MUMmer v.4. Values shaded in gray did not meet the query coverage cutoff of 70%, and thus the calculated ANI values are not accurate (values ranged between 30 and 50% coverage). This group breaks into 4 distinct clusters, each with >95% similarity within the cluster.
3. Reclassification of BoNT-producing species of clostridium
Reclassification of Clostridium botulinum and related species has been proposed several times, to separate C. botulinum strains as distinct species and to include non-toxigenic strains as part of those new species. For instance, Bengtson in 1924 (Bengston, 1924) was the first to propose separating proteolytic and non-proteolytic strains of C. botulinum as C. parabotulinum and C. botulinum, respectively. However, Prévot (Pa, 1953) published in 1953 an opinion letter suggesting that all organisms that produce BoNT should be classified as C. botulinum. In 1988, Suen (Suen et al., 1988) proposed that C. botulinum type G should be designated as C. argentinense, which also includes a few strains of C. subterminale and C. hastifome. In 1998 Collins and East (1998) proposed that C. botulinum Groups I–IV should constitute separate species; the authors also proposed adding the term “variety” followed by the toxin type to designate toxigenic strains. In 2011, Skarin et al. (2011) proposed classifying C. botulinum Group III as dual species: a pathos-pecies, C. botulinum, which would include all BoNT-producing strains (types A through G), and a genospecies, C. novyi sensu lato, to include C. botulinum Group III, C. novyi, and C. haemolyticum. More recently, in 2018, Smith et al. (2018) proposed a reclassification that would divide C. botulinum into four separate species, and which would add C. sporogenes to the list of BoNT-producing species of Clostridium.
Despite the evidence supporting a reclassification of BoNT-producing species of Clostridium, no formal decision has been made and the scientific community has not yet reached consensus regarding the taxonomic classification of this highly diverse group of organisms. According to Roney et al. (Rainey et al., 2015) “In any other group of organisms, this species would have been divided into four separate species because of the distinct differences in metabolic activity exhibited by strains in the four groups and the lack of DNA homology among groups. However, because of the unique and similar action of the toxins produced by all strains and to facilitate communication between the microbiological and medical professions, they have been retained in one species”. As shown in Table 1, BoNT-producing species of Clostridium could be divided into eleven distinct phylogenetic clades. It would be impractical to give each of those clades a separate species name because all produce BoNT; therefore, they can cause botulism. On the other hand, three of those clades already have a unique species name: C. argentinense, C. baratii, and C. butyricum. The metabolic and microbiological properties of these organisms differ greatly from C. botulinum. Moreover, C. butyricum and C. baratii were recognized as separate species well before the first strains able to produce BoNT were discovered, thus they retained their original species names although they belong to the group of BoNT-producing species of Clostridium.
Another potential consequence of reclassifying BoNT-producing species of Clostridium would be to substantially delay laboratory test results if isolates would have to be sequenced before they could be reported as belonging to a particular species. Moreover, public health laboratories may not have the capacity to characterize BoNT-producing species of Clostridium by whole genome sequencing, further delaying the reporting of results. This is of particular importance in those instances when laboratory confirmation of botulism is achieved by isolation of C. botulinum; e.g., serum specimen is not available for toxin detection testing or it is negative for BoNT, and C. botulinum is isolated from a stool specimen. Knowing the isolate’s specific group (or species) might be valuable for research purposes, but this information would not contribute to clinical management as treatment of botulism does not differ by strain.
BoNT-producing species of Clostridium are listed as Tier 1 Select Agents in the United States. According to current regulations, BoNT-producing species of Clostridium include the following species: C. botulinum, C. baratii, C. butyricum, and C. argentinense. Historically, C. botulinum includes all organisms that produce BoNT and present relevant microbiological characteristics (anaerobic, lipase positive, etc.), and C. sporogenes includes those organisms that microbiologically resemble C. botulinum but do not produce BoNT. Thus, reclassifying C. sporogenes and C. botulinum to include both toxigenic and non-toxigenic strains would require updating the current Select Agents and Toxins regulations. Such changes might not be justified as rule 56a (Brunt et al., 2018) of the International Code of Nomenclature of Prokaryotes (Parker et al., 2019) states that “names whose application are likely to lead to accidents endangering health or life or both or of serious economic consequences” can be rejected.
Moreover, including toxigenic and non-toxigenic in the same species, as the classification noted above proposes, could have adverse consequences. For instance, if a non-toxigenic organism identified as “C. botulinum” by whole genome sequencing was isolated from a commercially-produced food item such as infant formula, it could be misinterpreted as an actionable finding, i.e., safety recall of the product.
4. Conclusions
Since a reclassification of these organisms will not improve clinical or public health measures, we propose that the species name of C. botulinum remain unchanged, to minimize the risk of miscommunication among the public health, medical, and scientific communities. Alternatively, the term “genospecies” could be used when using classifications other than the currently accepted. For instance, “C. botulinum type B, genospecies C. sporogenes” would indicate that the strain produces BoNT type B and belongs to clade 2, with other non-toxigenic C. sporogenes strains.
Acknowledgements
This project was supported by the Center for Preparedness Response, Centers for Disease Control and Prevention. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Credit author statement
Carolina Lúquez: Conceptualization, Investigation, Writing – original draft, review, and editing, Jessica L. Halpin: Methodology, Formal analysis, Writing – review & editing, Janet Dykes: Conceptualization, Writing – review & editing.
Ethical statement
- This material is the authors’ own original work, which has not been previously published elsewhere.
- The paper is not currently being considered for publication elsewhere.
- The paper reflects the authors’ own research and analysis in a truthful and complete manner.
- The paper properly credits the meaningful contributions of co-authors and co-researchers.
- The results are appropriately placed in the context of prior and existing research.
- All sources used are properly disclosed (correct citation). Literally copying of text must be indicated as such by using quotation marks and giving proper reference.
- All authors have been personally and actively involved in substantial work leading to the paper, and will take public responsibility for its content.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
Data will be made available on request.
References
- Barash JR, Arnon SS, 2014. A novel strain of Clostridium botulinum that produces type B and type H botulinum toxins. J. Infect. Dis 209, 183–191. [DOI] [PubMed] [Google Scholar]
- Bengston IA, 1924. Studies on Organisms Concerned as Causative Factors in Botulism, vol. 136. Hygiene Laboratory Bulletin, pp. 1–101. [Google Scholar]
- Brunt J, Carter AT, Stringer SC, Peck MW, 2018. Identification of a novel botulinum neurotoxin gene cluster in Enterococcus. FEBS Lett. 592, 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunt J, van Vliet AHM, Carter AT, Stringer SC, Amar C, Grant KA, Godbole G, Peck MW, 2020a. Diversity of the genomes and neurotoxins of strains of Clostridium botulinum group I and Clostridium sporogenes associated with foodborne, infant and wound botulism. Toxins 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunt J, van Vliet AHM, Stringer SC, Carter AT, Lindström M, Peck MW, 2020b. Pan-genomic analysis of Clostridium botulinum group II (Non-Proteolytic C. Botulinum) associated with foodborne botulism and isolated from the environment. Toxins 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, De Meyer S, Trujillo ME, 2018. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol 68, 461–466. [DOI] [PubMed] [Google Scholar]
- Collins MD, East AK, 1998. Phylogeny and taxonomy of the food-borne pathogen Clostridium botulinum and its neurotoxins. J. Appl. Microbiol 84, 5–17. [DOI] [PubMed] [Google Scholar]
- Cruz-Morales P, Orellana CA, Moutafis G, Moonen G, Rincon G, Nielsen LK, Marcellin E, 2019. Revisiting the evolution and taxonomy of clostridia, a phylogenomic update. Genome Biol Evol 11, 2035–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillo S, Giordani F, Tonon E, Drigo I, Anselmo A, Fortunato A, Lista F, Bano L, 2021. Extensive genome exploration of Clostridium botulinum group III field strains. Microorganisms 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdeman LV, Brooks J, 1970. Variation Among Strains of Clostridium Botulinum and Related Clostridia, p 278–286. U. S. Government Printing Office, Washington, DC. [Google Scholar]
- Katz LS, Griswold T, Morrison SS, Caravas JA, Zhang S, den Bakker HC, Deng X, Carleton HA A, 2019. Mashtree: a rapid comparison of whole genome sequence files. Journal of Open Source Software 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Oh HS, Park SC, Chun J, 2014. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol 64, 346–351. [DOI] [PubMed] [Google Scholar]
- Lawson PA, Citron DM, Tyrrell KL, Finegold SM, 2016. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) prevot 1938. Anaerobe 40, 95–99. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P, 2021. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield MJ, Adams JB, Doxey AC, 2015. Botulinum neurotoxin homologs in non Clostridium species. FEBS Lett. 589, 342–348. [DOI] [PubMed] [Google Scholar]
- Pa R, 1953. Rapport D’introduction du president du souscomité Clostridium pour L’unification de La nomenclature des types toxinogènes de C. Botulinum. International Bulletin of bacterial Nomenclature 3, 120–123. [Google Scholar]
- Rainey FA,Hollen BJ, Small AM, 2015. Clostridium. In: Whitman WB (Ed.), Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Inc. (in association with Bergey’s Manual Trust). [Google Scholar]
- Skarin H, Segerman B, 2014. Plasmidome interchange between Clostridium botulinum, Clostridium novyi and Clostridium haemolyticum converts strains of independent lineages into distinctly different pathogens. PLoS One 9, e107777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker Charles, T., Tindall BJ, Garrity GM, 2019. International Code of nomenclature of prokaryotes. Int. J. Syst. Evol. Microbiol 69, S1–S111. [DOI] [PubMed] [Google Scholar]
- Skarin H, Håfström T, Westerberg J, Segerman B, 2011. Clostridium botulinum group III: a group with dual identity shaped by plasmids, phages and mobile elements. BMC Genom. 12, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T, Williamson CHD, Hill K, Sahl J, Keim P, 2018. Botulinum neurotoxin-producing Bacteria. Isn’t it time that we called a species a species? mBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel J, 2005. Botulism. Clin. Infect. Dis 41, 1167–1173. [DOI] [PubMed] [Google Scholar]
- Suen JC, Hatheway C, Steigerwalt AG, Brenner DJ, 1988. Clostridium argentinense sp. nov.: a genetically homogeneous group composed of all strains of Clostridium botulinum toxin group G and some nontoxigenic strains previously identified as Clostridium subterminale or Clostridium hastiforme. Int. J. Syst. Bacteriol 375–381. [Google Scholar]
- Tian D, Zheng T, 2014. Comparison and analysis of biological agent category lists based on biosafety and biodefense. PLoS One 9, e101163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigand MR, Pena-Gonzalez A, Shirey TB, Broeker RG, Ishaq MK, Konstantinidis KT, Raphael BH, 2015. Implications of genome-based discrimination between Clostridium botulinum group I and Clostridium sporogenes strains for bacterial taxonomy. Appl. Environ. Microbiol 81, 5420–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentz TG, Tremblay BJM, Bradshaw M, Doxey AC, Sharma SK, Sauer JD, Pellett S, 2021. Endogenous CRISPR-cas systems in group I Clostridium botulinum and Clostridium sporogenes do not directly target the botulinum neurotoxin gene cluster. Front. Microbiol 12, 787726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson CH, Sahl JW, Smith TJ, Xie G, Foley BT, Smith LA, Fernández RA, Lindström M, Korkeala H, Keim P, Foster J, Hill K, 2016. Comparative genomic analyses reveal broad diversity in botulinum-toxin-producing Clostridia. BMC Genom. 17, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.


