Abstract
How enhancers interpret morphogen gradients to generate gene expression patterns is a central question in developmental biology. Recent studies have proposed that enhancers can dictate whether, when, and at what rate promoters engage in transcription, but the complexity of endogenous enhancers calls for theoretical models with too many free parameters to quantitatively dissect these regulatory strategies. To overcome this limitation, we established a minimal promoter-proximal synthetic enhancer in embryos of Drosophila melanogaster. Here, a gradient of the Dorsal activator is read by a single Dorsal DNA binding site. Using live imaging to quantify transcriptional activity, we found that a single binding site can regulate whether promoters engage in transcription in a concentration-dependent manner. By modulating binding site affinity, we determined that a gene’s decision to transcribe and its transcriptional onset time can be explained by a simple model where the promoter traverses multiple kinetic barriers before transcription can ensue. A record of this paper’s transparent peer review process is included in the supplemental information.
eToc
Developmental enhancers dictate whether a locus engages in transcription and, if so, at what rate it will produce mRNA. Using theory and live imaging in Drosophila embryos, Alamos & Reimer et al. propose a model for how a transcriptional activator binding to an enhancer may dictate these multiple regulatory dynamics.
1. Introduction
The adoption of distinct cellular identities in multicellular organisms relies on the formation of spatial gene expression domains driven, in large part, by transcriptional regulatory programs. The positional information giving rise to these mRNA patterns is typically provided by transcription factor gradients (Fig. 1A) whose concentrations are interpreted by enhancer DNA sequences that, in turn, regulate transcription of developmental genes [1, 2]. A long-standing goal in quantitative developmental biology is to precisely predict gene expression from knowledge of the DNA regulatory sequence and morphogen concentration [3, 4]. Achieving this predictive understanding requires theoretical models that calculate how DNA sequence dictates the functional relation between input morphogen concentration and output transcriptional activity, and calls for testing these predictions by measuring input-output functions [3]. Precise genetic manipulations [5, 6] and powerful imaging technologies [7, 8, 9] have rendered the early embryo of the fruit fly Drosophila melanogaster (Drosophila) a prime model system for quantitatively dissecting these input-output functions in development.
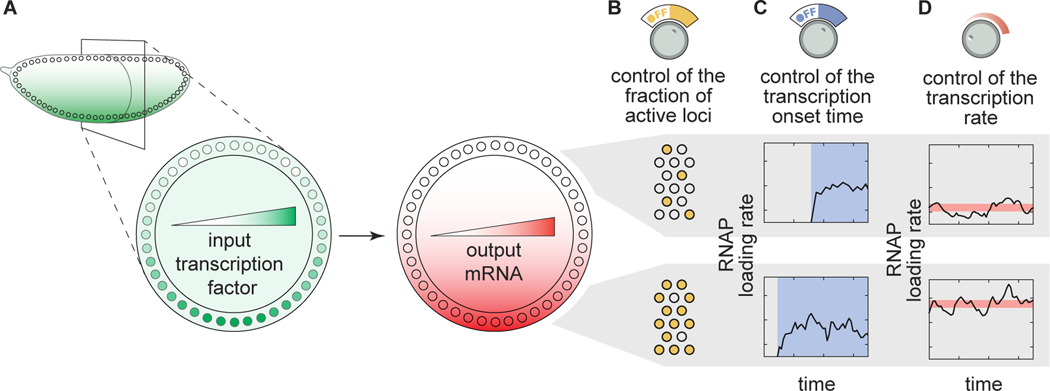
Figure 1. Transcriptional regulatory strategies of enhancers in response to transcription factor concentration gradients.
(A) A Drosophila embryo with a transcription factor gradient along its dorsoventral axis. This input transcription factor dictates the emergence of output gene-expression patterns by controlling a combination of three enhancer regulatory ‘knobs’: (B) the probability of loci becoming transcriptionally active, (C) the transcriptional onset time, and (D) the mean transcription rate of active loci.
In recent years, several studies have reported that Drosophila enhancers can control various, potentially independent aspects of transcriptional dynamics in early embryonic development [10, 8, 11, 12, 13, 14, 15, 16, 17]. First, for a given gene, a fraction of loci remain transcriptionally inactive throughout entire mitotic cycles in development, even when exposed to the same activator concentration as active loci (Fig. 1B)-a behavior usually quantified through the fraction of active nuclei or loci. This stochastic decision for a locus to become active is a ubiquitous and potentially important regulatory feature for shaping gene-expression patterns in the embryo [8, 18, 12, 17]. However, it remains unclear whether this feature constitutes a regulatory ‘knob’ or whether inactive loci are artifacts of experimental detection thresholds. Second, the timing of transcription onset (and cessation, which is not addressed in the present investigation) can also be controlled by input transcription-factor dynamics (Fig. 1C; [19, 20, 18, 14, 12, 21, 17] ). Finally, the rate of transcriptional initiation in active loci is under regulatory control (Fig. 1D) and has been the focus of most studies to date (e.g., [8, 22, 23, 12, 15, 16] ). Thus, multiple regulatory strategies together realize gene-expression patterns in space and time.
Intense theoretical scrutiny [19, 24, 25, 26, 27, 18, 23, 14, 28] has generated a compelling hypothesis: that the regulation of transcriptional dynamics can be separated into two stages. First, a promoter must pass through a series of kinetic barriers consisting of reactions catalyzed by transcription factors in order for loci to engage in transcription. Previous analyses of the mean and distribution in transcriptional onset times have suggested that the number of inactive promoter states can range from one to three [18, 14, 17]. These reactions could be associated with, for example, the stepwise unwrapping of DNA from nucleosomes [19, 18, 14] and/or the sequential recruitment of general transcriptional cofactors [29]. Second, after initial promoter activation, the rate of mRNA production is proportional to the probability of finding RNA polymerase II (RNAP) bound to the promoter. Statistical mechanical (also called thermodynamic) models have been used to calculate this probability of finding RNAP bound to the promoter, and have successfully been used to predict mRNA production rates in bacteria [30]. However, whether they can be applied to the more complex context of eukaryotic transcriptional regulation—let alone to the dynamical processes of cellular decision-making in development—is still an open question [31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 26, 41, 23, 14].
One of the main challenges to systematically testing these models is the complexity of endogenous regulatory regions [24, 42, 25, 18, 23, 14]. Because endogenous enhancers contain multiple binding sites for different transcription factors, accounting for these sites and their interactions leads to a combinatorial explosion of model parameters [43, 3]; determining the values of these parameters from simple experiments constitutes a computational—and conceptual—challenge [4, 43, 3]. To render complex transcriptional regulatory systems tractable to theory, minimal synthetic enhancers have been engineered in bacteria [44, 45, 30, 46], eukaryotic cells [47], and developing organisms [24, 25]. In such experiments, a short, synthetic DNA sequence with only one to a few binding sites for a single transcription factor drives the expression of a reporter gene. As shown in detail in Box 1, measuring the concentration of the transcription-factor input and reporter mRNA output makes it possible to test models of transcriptional regulation and to infer molecular parameters that can be used to predict the behavior of more complex regulatory architectures [46]
Box 1: Bending Nature to Understand It.
The inherent complexity of endogenous enhancers, with their plethora of binding sites for multiple transcription factors and protein-protein interactions, calls for complex theoretical descriptions with a multitude of free parameters. This explosion of free parameters makes it challenging to confront theoretical models against experiments.
An alternative to describing the complex reality of endogenous enhancers using complex theoretical models is to first reach a predictive understanding of simpler, synthetic regulatory architectures. As our predictive understanding increases, so too can the complexity of the regulatory regions assayed in an iterative cycle that, hopefully, will culminate with the understanding of endogenous regulatory regions [43, 3].
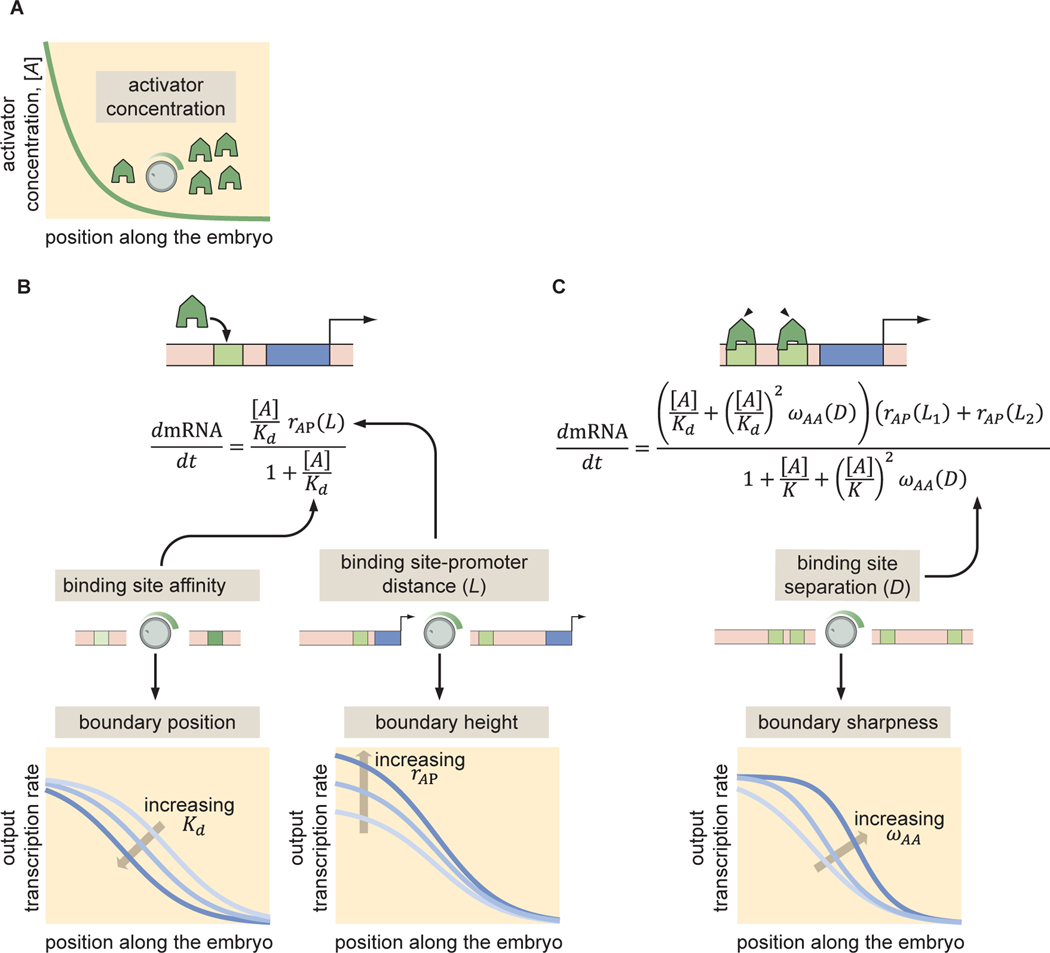
This idea of bending nature to make it simpler is illustrated in Figure 2. Consider, for example, an activator that is distributed in an exponential gradient along one of the axes of the fruit fly embryo (Fig. 2A). Endogenous enhancers might contain multiple binding sites for this activator However, a simpler synthetic enhancer bearing only one binding site for this activator could be created. As illustrated in Figure 2B, a theoretical description of the rate of mRNA production driven by this enhancer-based on thermodynamic models [50, 51, 52, 53, 54, 55, 56] for this particular illustrative example-would only have two free parameters: a dissociation constant between for activator-DNA binding, , and a parameter that captures the efficiency with which the activator increases transcription, . As shown in the figure, each of these free parameters dictate different aspects-boundary position and height -of the transcription profile. As a result, by measuring this profile and fitting to the model, a numerical estimate of each parameter can be obtained.
With a solid understanding of the single-activator enhancer system, the next iteration in this synthetic dissection calls for the addition of a second binding site for the same activator. As shown in Figure 2C, the prediction for the mRNA production rate looks more complicated than for its single-binding site counterpart. However, a closer examination of the expression reveals that it only contains one free parameter: the activator-activator interaction term , which dictates the sharpness of the boundary. As a result, by taking the parameters inferred from the previous iteration, the inference of becomes much simpler.
Once a predictive understanding of this architecture with two binding sites is reached, the complexity can be further increased. Each iteration brings us closer to describing an endogenous enhancer. Of course, these regions do not exist in nature. Yet, we argue that there is little hope of predicting the input-output functions of endogenous enhancers if we cannot accomplish this feat in the much simpler context of the synthetic enhancers presented here.
Here we sought to use synthetic minimal promoter-proximal enhancers to challenge our integrated model of transcriptional control using the dorsoventral patterning system in Drosophila embryos, in which a concentration gradient of the Dorsal transcription factor specifies spatial domains of transcription, as a case study. To test the integrated model of transcriptional dynamics (Fig. 3A, B), we performed simultaneous quantitative live-cell measurements of Dorsal concentration (input) and transcription (output) driven by minimal synthetic Dorsal-dependent promoter-proximal enhancers in single nuclei. By repurposing the parS-ParB DNA labeling technology [48, 49] to quantify transcriptional activity independent of RNA detection, we determined that the inactive loci described by our model constitute a distinct transcriptional state under regulatory control and are not the result of detection artifacts. Further, our theoretical model predicted how, through the Dorsal-mediated catalysis of reactions prior to transcriptional onset, regulatory architecture dictates both the transcriptional onset time and the fraction of active loci. Finally, once promoters turn on, we found that our measurements are compatible with an equilibrium model. Thus, the present investigation provides quantitative evidence supporting a unified model of transcriptional regulation in eukaryotes that accounts for whether loci become transcriptionally active, when this activity ensues, and, once transcription ensues, at what rate nascent RNA molecules are produced. More generally, our work demonstrates the feasibility of using minimal synthetic enhancers to engage in a dialogue between theory and experiment in the context of transcriptional control in development.
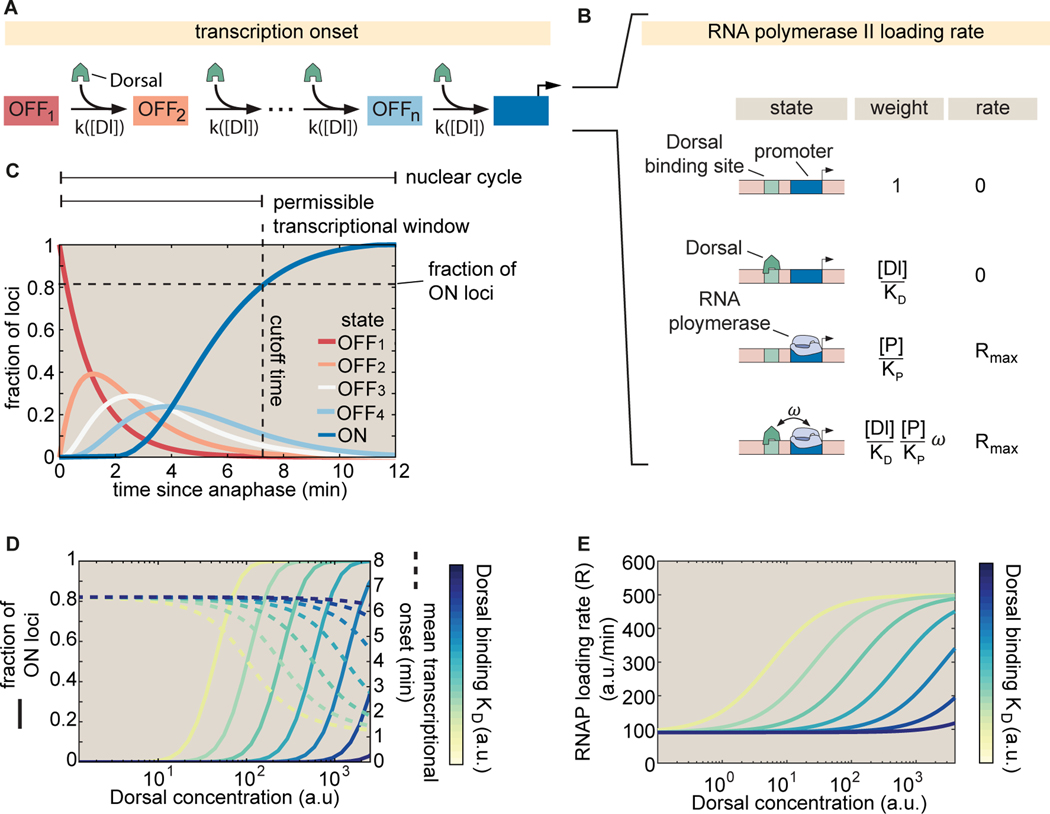
Figure 3. Integrated kinetic and thermodynamic model of simple activation by Dorsal.
(A) The promoter undergoes kinetic transitions from transcriptionally inactive states ( to to an active state with Dorsal accelerating the transition rate, , by a factor proportional to the Dorsal occupancy at the promoter. (B) Thermodynamic states and weights for the simple activator model. The probability of finding RNAP bound to the promoter can be calculated from the statistical weights associated with all possible occupancy states of the proximal enhancer-promoter system. (C) Visualization of a particular solution of the kinetic scheme from (A) showing the probability of finding a given locus in each of the states for an illustrative, representative set of parameters ( a.u., a.u., states, and 7 min nuclear cycle duration). The predicted fraction of active loci (dashed horizontal line) is calculated as the probability of being in the ON state by the end of the permissible time window (dashed vertical line) that is determined by mitotic repression. (D) Predictions for the fraction of active loci (solid lines plotted against the left y-axis) and mean transcriptional onset times (dashed lines plotted against the right -axis) as a function of Dorsal concentration for different, logarithmically-spaced values of the Dorsal dissociation constant in arbitrary units of Dorsal concentration. Note that under some parameter regimes, mean turn on times are similar across Dorsal concentrations. (E) Rate of mRNA production across active loci as a function of Dorsal concentration for different values of based on the model in (B) a.u., Dorsal ranging from 10 a.u. to a.u., .
2. Results
2.1. An integrated model of transcriptional dynamics driven by a single activator binding site
To probe the transcriptional regulatory strategies of a minimal synthetic enhancer (Fig. 1), we posit a theoretical model that predicts the fraction of loci that will become active, their transcriptional onset time, and RNAP loading dynamics once transcription ensues. Specifically, we consider a simplified case in which only one activator is present and can only bind to one site only a few base pairs away from the promoter (Fig. 3).
In order to explain the transcriptional onset dynamics of a locus and the probability of loci becoming active, we invoke recent experiments leading to a ‘kinetic barrier’ model [19, 18, 14] proposing that, after exiting mitosis, all promoters are in an inactive state. In this state, labeled as ‘OFF1’ in Figure 3A, transcription is not possible. Promoters must then traverse a series of distinct inactive states (labeled ‘OFF2’ to ‘OFFn’ in Fig. 3A) before reaching an active state in which transcription proceeds (labeled ON in Fig. 3A).
The temporal evolution of the transcriptional dynamics as it traverses the states shown in Figure 3 A can be simulated by computing the probability that the promoter occupies each state. Here, the transition rate between states, , determines how the states probability spreads from the initial condition where the promoter is in state to the active state as time passes (see Methods S1.1 for details).
We propose that a transcriptional activator such as Dorsal can catalyze the transition between states in an affinity-dependent manner via binding to its cognate site in the enhancer. In this model, we assume that the transition rate is much slower than transcription factor DNA binding, which has been shown to be on the order of a few seconds for transcription factors in the fly embryo [9, 57], and for the mammalian homolog of Dorsal [58]. As a result of this separation of time scales, we posit that Dorsal visits the enhancer multiple times before a transition between OFF states takes place such that the probability of finding Dorsal bound to the enhancer is proportional to its equilibrium occupancy. As a result, the transition rate is given by
| (1) |
where is a rate constant, is the Dorsal concentration at time , and is the Dorsal-DNA dissociation constant.
Because Dorsal concentration varies in time, the model cannot be solved analytically. Thus, we numerically calculated the probability of the promoter being in each state as a function of time using a particular set of model parameters (see details in Methods S1.1). As seen in Figure 3C, since individual loci must traverse a sequence of intermediate states before reaching the ON state, this model introduces a delay in activation.
This kinetic barrier model accounts for loci that never transcribe during the nuclear cycle. Specifically, the model predicts that if nuclear cycles lasted indefinitely, all promoters would eventually reach the ON state as shown in Figure 3C. However, due to the rapid mitotic cycles that characterize early embryonic development in Drosophila, this duration is limited: transcription cannot initiate during mitosis and thus is only permissible during a time window within interphase (Fig. 3C, vertical dashed line; [59, 8, 14]. Consequently, if the time it takes a promoter to reach the ON state is longer than the duration of this window, then this promoter will not initiate transcription at all during the nuclear cycle (Fig. 3C, horizontal dashed line).
The kinetic barrier model can be used to predict two of the three regulatory strategies, fraction of active loci and transcription onset times, that we aim to dissect quantitatively (Fig. 1). First, the model predicts how the fraction of active loci is determined by Dorsal nuclear concentration and binding affinity (Fig. 3D, left y-axis). Second, this same model calculates the mean transcriptional onset time of those loci that turn on as a function of these same Dorsal parameters (Fig. 3D, right -axis).
To model a locus once it is active, we follow [14] and propose a simple thermodynamic model [60, 55] that assumes that the RNAP loading rate, , is proportional to the probability of finding RNAP bound to the promoter , such that
| (2) |
where is a constant coefficient that dictates the maximum possible polymerase loading rate.
Thermodynamic models enable the calculation of by assigning a statistical weight to each possible state in which the regulatory system can be found. In the case of a minimal promoter-proximal enhancer with one activator binding site, the enhancer-promoter DNA can be empty, occupied by Dorsal, occupied by RNAP, or simultaneously bound by Dorsal and RNAP (Fig. 3B). The statistical weight associated with each of these terms is shown in Figure 3B. Here, is the statistical weight associated with finding Dorsal (with concentration and binding dissociation constant ) bound to the promoter alone, while is the weight of finding RNAP (with concentration and binding dissociation constant ) bound to the promoter alone. Note that the weight of having both Dorsal and RNAP bound simultaneously includes an extra glue-like cooperativity coefficient, , that determines how strongly Dorsal recruits RNAP to the promoter. The value of is constrained to be so that higher Dorsal occupancy leads to higher RNAP occupancy. This thermodynamic modeling approach also allows for more indirect forms of RNAP recruitment by Dorsal such as binding mediated by cofactors. As shown in Figure S6 and Figure S7, these more complex models make theoretical predictions that are essentially indistinguishable from those made by the simplest case considered in Figure 3B. As a result, throughout this work, we choose to entertain only the simplest model of direct Dorsal-RNAP recruitment.
To calculate , we divide the sum of the weights featuring a bound RNAP molecule by the sum of all possible weights. Substituting this calculation into Equation 2 yields
| (3) |
which is plotted in Figure 3E. As shown in the figure, increasing shifts the concentration at which the RNAP loading rate reaches half its maximum value toward higher Dorsal concentrations, but does not change the overall shape of the curve. We also note the presence of a non-zero baseline of RNAP loading rate due to the Dorsal-independent term in the numerator of Equation 3. This baseline suggests that it could be possible for a promoter in the ‘ON‘ state to produce low, basal-level transcription in the absence of bound Dorsal.
Together, the kinetic barrier model outlined in Figure 3A and the thermodynamic model’s Equation 3 define a comprehensive quantitative framework that predicts how the fraction of active loci, the transcriptional onset time, and the RNAP loading rate as a function of Dorsal concentration vary as model parameters such as the Dorsal dissociation constant are modulated (Fig. 3D, E). These predictions constitute hypotheses that we experimentally tested throughout the remainder of this work.
2.2. Establishing a minimal synthetic enhancer system to test theoretical predictions
To test our model’s predictions, it is necessary to simultaneously measure transcription factor input and transcriptional output driven by a minimal regulatory system containing a single activator binding site. Thus, we sought to engineer and validate such a system in a developing embryo. To this end, we constructed single binding site promoter-proximal enhancers driven by the Dorsal activator, one of the best characterized transcription factors in Drosophila and a classic example of a morphogen [61, 62]. Dorsal is provided maternally and forms a dorsoventral gradient of nuclear localization (Fig. 4A; [63]), acting as an activator by default [64, 65] and as a repressor in the presence of nearby binding sites for corepressors [66, 67]. Prior to activation of the zygotic genome (up to the 12th mitotic cycle), Dorsal is the only known transcription factor with a nuclear protein gradient across the dorsoventral axis [68, 69]. Thus, the Dorsal nuclear concentration is the sole source of dorsoventral positional information for developmental enhancers at this stage in development. These features, combined, make Dorsal an ideal input transcription factor for activating a minimal synthetic reporter system.
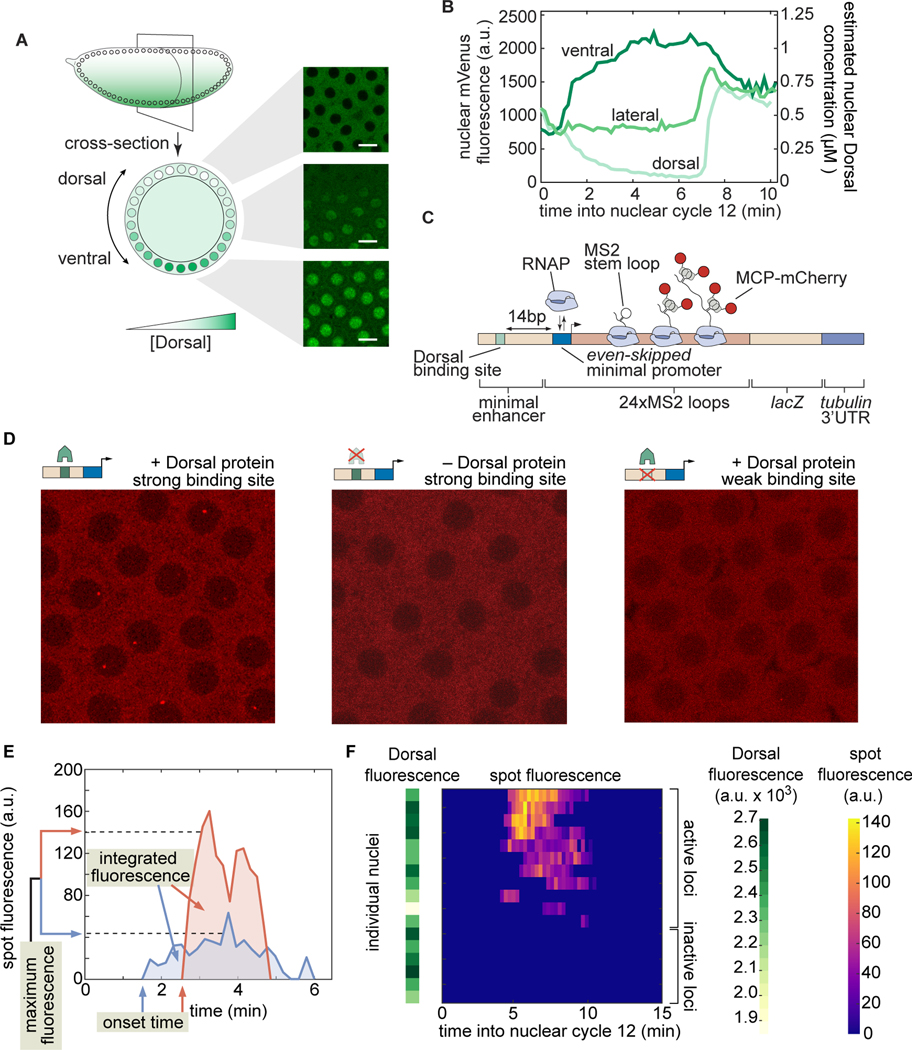
Figure 4. Simultaneously measuring transcription factor protein input and transcriptional output.
(A) Schematic of the Dorsal protein gradient in early Drosophila embryos. Dorsal protein accumulates in ventral nuclei and is progressively excluded from more dorsal nuclei. Example snapshots show Dorsal-mVenus in various positions along the dorsoventral axis. (B) Representative time traces of nuclear Dorsal-mVenus fluorescence in various positions along the dorsoventral axis in Dorsal embryos. The right -axis shows the approximate nuclear Dorsal concentration according to the estimation described in Figure S9. (C) Schematic of minimal synthetic promoter-proximal enhancer system containing a single binding site for Dorsal that drives transcription of a reporter tagged with MS2 loops, which are visualized through the binding of MCP-mCherry. The Dorsal binding site is placed upstream of the even-skipped minimal promoter. (D) Snapshots from embryos containing an optimal binding-site reporter in the presence (left) or absence (middle) of Dorsal, or containing a strongly mutated Dorsal binding site (right). (E) Examples of fluorescence time traces and quantitative metrics of transcriptional activity used throughout this work. (F) Fluorescence of all transcription spots in individual nuclei in the field of view of one embryo as a function of time (heatmap) and their corresponding Dorsal-Venus fluorescence midway through the nuclear cycle (green bars on the left). If a transcription spot was detected within a nucleus at any point during the interphase of nuclear cycle 12, then the locus was considered active; otherwise, the locus was classified as inactive.
In order to relate output transcriptional activity to the time-variant input Dorsal concentration throughout development, we measured the instantaneous Dorsal concentration in live embryos by creating a CRISPR knock-in Dorsal-mVenus fusion allele based on a previous Dorsal fusion [62] that rescues embryonic development [70, 71]; Materials and Methods). Further, in order to increase the dynamic range of Dorsal concentration in our experiments, we combined this CRISPR allele with a Dorsal-mVenus transgene [62], resulting in a line that will hereafter be referred to as Dorsal flies. This fusion made it possible to quantify the concentration dynamics of the Dorsal protein input (Fig. 4A, B) in individual nuclei (Supplementary Video 1, left; Materials and Methods). Dorsal-mVenus nuclear fluorescence time traces quantified over nuclear cycle 12 confirmed the dynamic nature of Dorsal concentration and were quantitatively similar to previous measurements (Fig. 4B; [62]; details of Dorsal-mVenus quantification in Fig. S8A, B). Nuclear cycle 12 nuclei in 4x Dorsal flies experience a Dorsal concentration gradient spanning several orders of magnitude, from less than to more than (Fig. 4B; details of Dorsal-mVenus calibration in Fig. S9).
To visualize the dynamics of Dorsal-dependent transcription, we built a reporter transgene containing a minimal synthetic promoter-proximal enhancer consisting of a single high affinity, consensus Dorsal binding site (Fig. 4C; [72, 73, 74]. Hereafter we refer to this strong site enhancer as DBS_6.23 for Dorsal Binding Site, followed by its binding affinity score according to the Patser algorithm ([75]; Materials and Methods). To quantify the transcriptional activity of this enhancer, we used the MS2-MCP system to fluorescently label nascent RNA molecules in our reporter constructs, which appear as nuclear fluorescent puncta (hereafter “transcription spots”) in laser-scanning confocal microscopy movies (Supplementary Video 1, right; [76, 8, 10] ). We performed image analysis of the MS2 movies using a custom data analysis pipeline in Matlab and Fiji (Materials and Methods; [77, 12]
To validate this minimal synthetic system, we determined that DBS_6.23-MS2 drives quantifiable levels of transcription, and that this transcriptional activity is mainly governed by Dorsal. We compared the transcriptional activity of DBS_6.23-MS2 in embryos laid by 4x Dorsal females with the activity in embryos laid by females homozygous for the dorsal null allele. While transcription spots were clearly present in the Dorsal background (Fig. 4D, left), they were extremely rare in dorsal null embryos (Fig. 4D, middle): not a single transcription spot was detected during nuclear cycle 12 in any of 4 replicates containing nuclei in total. Dorsal is therefore necessary for transcriptional activity in our reporter constructs.
We next sought to determine whether the detected transcriptional activation is solely due to Dorsal interacting with the binding site we explicitly engineered into the construct or whether there are cryptic Dorsal binding sites contributing to gene expression. We generated a second reporter, DBS_4.29-MS2 in which the Dorsal binding site was strongly perturbed using known point mutations [72]. Transcription was rarely detectable in DBS_4.29-MS2 embryos (Fig. 4D, right), with the average transcriptional activity (mean instantaneous fluorescence) per nucleus being less than 10% of the optimal DBS_6.23 enhancer at any Dorsal concentration (Fig. S10). Thus, the Dorsal site placed within the synthetic enhancer is necessary for robust activation and is the main driver of its transcriptional activity.
Next, we asked whether the MS2 signal could be used as a reporter of Dorsal-dependent transcriptional activity that can be directly compared to our model predictions in terms of transcription onset time, transcription rate and fraction of active nuclei. We collected DBS_6.23-MS2 time traces of MCP-mCherry fluorescence from transcription spots during nuclear cycle 12 along with the aforementioned three metrics of transcriptional activity (Fig. 4E, F). First, the transcriptional onset time is defined as the time since the previous mitosis at which a transcription spot is first detected. Second, the maximum spot fluorescence corresponds to the 95th percentile of intensity over time, which is proportional to the transcription rate (Section S1.2). Further, the integrated spot fluorescence corresponds to the time integral of the spot fluorescence and is directly proportional to the amount of mRNA produced by the locus ([8]; Materials and Methods). Finally, as previously observed in other genes in flies [8, 18, 12, 17] and predicted by our model, not all nuclei exposed to the similar nuclear Dorsal concentration exhibited detectable transcription (Fig. 4F). This failure of some nuclei to turn on and engage in transcription throughout the nuclear cycle is consistent with previous results from Dorsal-dependent synthetic enhancers which displayed a ‘salt and pepper’ pattern even at peak Dorsal concentrations [74]. As a result, we quantified the fraction of active loci-regardless of their level of activity or temporal dynamics—by measuring the number of nuclei with observable transcription signal in at least one movie frame throughout nuclear cycle 12, divided by the total number of nuclei. Thus, we have established an experimental platform and quantitative metrics for Dorsal activity that enable us to engage in a dialogue between experiment and theory.
2.3. Transcriptionally active and inactive loci correspond to functionally distinct populations
contrast the predicted fraction of active loci with experimental observations, it is important to ensure that this fraction is the result of Dorsal action and not simply due to false negatives in our experimental setup. Transcriptionally silent loci that remain inactive throughout interphase, such as those revealed by our experiment (Fig. 4F), have been observed using MS2 (and its sister mRNA labeling tool, PP7) in live-imaging experiments in flies [8, 12, 15], plants [78], and mammalian cells [79]. However, so far it has not been possible to determine whether these inactive loci correspond to a separate transcriptional state from active loci, or whether they are an artifact of the fluorescence detection thresholds associated with these microscopy techniques.
To answer this question, it is necessary to quantify MS2 fluorescence at loci undetected by our image analysis pipeline and determine whether they differ from loci not exposed to activators, which do not transcribe (Fig. 4D, middle). However, to date this approach has not been feasible because most MS2 measurements have relied on the presence of an MS2 signal itself to segment transcription spots and quantify their fluorescence. We hypothesized that, if undetected loci correspond to a distinct and weaker, Dorsal-independent state, then detected and undetected spots in embryos carrying wild-type Dorsal would appear as two distinct populations. In this scenario, the mCherry fluorescence of inactive loci in wild-type Dorsal embryos would be similar to that observed in Dorsal null embryos, and clearly distinct from the mCherry fluorescence of active loci in the presence of Dorsal.
To quantify MS2 fluorescence independently of whether a MS2 spot was detected, we implemented the parS-ParB DNA labeling system [48, 49]. Here, fluorescently labeled ParB proteins bind the parS DNA sequence resulting in a fluorescence spot appearing at the locus independently of the transcriptional state of the locus (Fig. 5A). We created flies with and without functional Dorsal expressing ParB2-eGFP (subsequently referred to as ParB-eGFP) and MCP-mCherry. We then crossed these flies to flies containing parS-DBS_6.23-MS2 to generate embryos that have our locus of interest labeled with ParB-eGFP colocalized with the transcriptional signal in the MCP-mCherry channel (Fig. 5A, B; Supplementary Video 2).
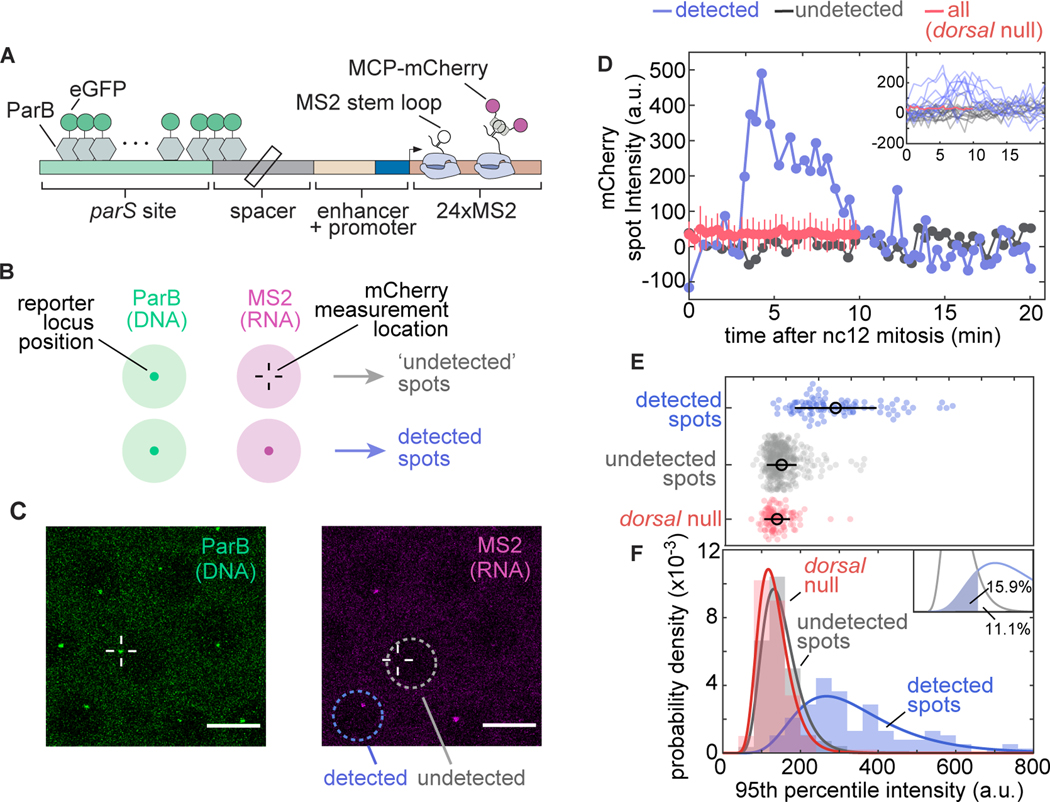
Figure 5. Transcriptionally independent ParB labeling confirms that transcriptionally inactive loci are functionally distinct from active loci.
(A) Schematic of ParB-eGFP construct. ParB-eGFP molecules bind and polymerize out from parS sequences, which are placed bp upstream of the enhancer. The enhancer and promoter together drive transcription of MS2 loops that subsequently bind MCP-mCherry. (B) Schematic of the experiment. Loci are located by detecting a signal in the ParB-eGFP channel; these locations were used to fit a 2D Gaussian to the same area in the MS2-mCherry channel to estimate fluorescence intensity regardless of whether an MS2-mCherry signal was detected (Materials and Methods Section 4.3). (C) Example images of ParB-eGFP (left) and MCP-mCherry (right) channels. Detected and undetected loci are found based solely on the MCP-mCherry signal. (D) Example time traces of MCP-mCherry fluorescence over time at the ParB-eGFP loci in nuclei with (blue) and without (grey) detected MS2-mCherry spots of the DBS_6.23 enhancer showing clear qualitative differences between the two populations. For comparison, the mean mCherry fluorescence at the ParB-eGFP loci in a representative Dorsal null embryo is also shown. Inset, all detected and undetected fluorescence traces obtained in the same embryo along with the mean fluorescence of all traces in a dorsal null embryo. Negative intensity values are due to spot intensities very close to the background fluorescence. (E) Swarm plots of 95th percentile MCP-mCherry fluorescence at loci with detected (blue; nuclei pooled from 20 embryos) and undetected MS2-mCherry transcription (gray; nuclei pooled from 20 embryos) driven by the DBS_6.23 enhancer in wild-type Dorsal embryos. Red (N = 96 nuclei pooled from 6 embryos), maximum fluorescence of all loci in Dorsal null embryos, defined as the 95 th percentile of intensity over time (black circles, mean; bars, standard deviation). Detected spots are significantly different from both null (ANOVA, p) and undetected spots (ANOVA, p) (F) Histograms of the data shown in (E). Solid lines correspond to log-normal fits performed for ease of visualization. Inset, undetected and detected distribution fits and the area used to estimate the false-negative detection rate of and the false-positive detection if .
Guided by the spatial positions reported by ParB-eGFP, we measured the MCP-mCherry signal at all DBS_6.23 reporter loci in embryos carrying wild-type Dorsal (Fig. 5C) or laid by mothers homozygous for the null allele (Dorsal null embryos). We then classified loci from wild-type Dorsal embryos into two categories, detected and undetected, depending on whether they were identified as spots in the MCP-mCherry channel by our analysis pipeline (Fig. 5B, C; Materials and Methods Section 4.3). As shown in the the examples presented in Figure 5D, there are clear qualitative differences between MCP-mCherry fluorescence time traces corresponding to detected and undetected transcriptional spots from wild-type embryos. Thus, our analysis made it possible to quantify MS2 fluorescence in three populations: detected loci and undetected loci in wild-type embryos, and all loci in Dorsal null embryos.
To compare these populations, we computed the 95th percentile value over each locus’ MCP-mCherry fluorescence time trace (Fig. 5E). The distribution of mCherry fluorescence from undetected spots in wild-type Dorsal embryos largely overlapped with that of all spots in Dorsal-null embryos (Fig. 5F), consistent with these two populations corresponding to loci expressing Dorsal-independent levels of activity. Moreover, both distributions were clearly distinct from the distribution of detected spots in wild-type Dorsal embryos (Fig. 5E, F). Thus, our results provide strong evidence that inactive loci are not artifacts of the detection limit of our imaging technique. Rather loci can belong to one of two distinct populations: those that transcribe at a high, Dorsal-dependent level and those that are transcriptionally inactive (or active at a low, undetectable level that is comparable to that of embryos lacking Dorsal). We therefore conclude that the decision to transcribe made by each locus is an additional regulatory strategy controlled by Dorsal.
From the observations in Figures 5E and F, we estimated our error in classifying loci as inactive. This false-negative detection rate, corresponding to the area under the curve shaded in the inset of Figure 5F, is estimated as . However, this false-negative rate is likely an underestimation. For example, this rate may depend on Dorsal concentration, which cannot be controlled for in this experiment. Additionally, the presence of ParB in the locus may itself affect transcriptional dynamics, impacting the false-negative rate. For these reasons, we do not attempt to correct our measurements of the fraction of active loci using this estimated false-negative rate.
2.4. Dorsal-dependent kinetic barriers explain transcription onset dynamics and modulation of the fraction of active loci
Having established that transcriptionally inactive promoters mostly constitute a separate population from transcriptionally active promoters (Fig. 5), we sought to test whether our theoretical model (Fig. 3A) can quantitatively recapitulate the fraction of active loci and their transcription onset times. Tuning transcription factor-DNA binding affinity has been a powerful tool to test models of transcriptional regulation in the past [80, 46]. Inspired by these previous works, we probed our model by adjusting the Dorsal-DNA interaction energy in our minimal synthetic enhancer.
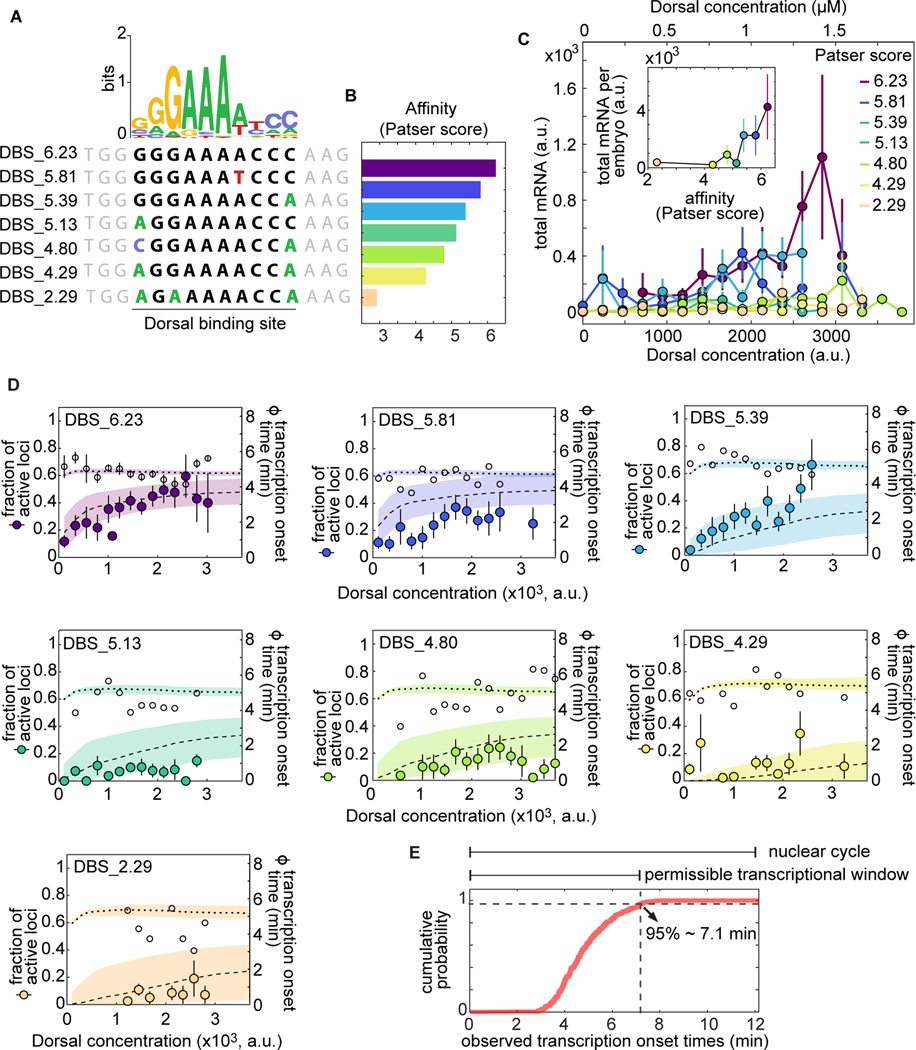
We constructed a series of enhancers containing a single binding site with varying affinities for Dorsal. Building on the optimal DBS_6.23 and the mutated DBS_4.29 sites (Fig. 4D, left vs. right), we created five additional enhancers of varying intermediate strengths by introducing point mutations into the consensus Dorsal binding motif to obtain a range of predicted affinities (Fig. 6A,B; Materials and Methods Section 4.3). As described above, we refer to these enhancers as DBS, followed by their corresponding Patser score.
Figure 6. A multi-step kinetic barrier model predicts the Dorsal-dependent fraction of active loci with constant mean transcriptional onset times.
(A) Top: Dorsal positional weight matrix logo from [100]. Bottom: Sequence of the Dorsal binding sites engineered into our minimal synthetic enhancers. Bold letters, 10 bp Dorsal motif; black letters, consensus bases; colored letters, mutated bases; gray letters, sequence context. (B) Relative affinities of Dorsal binding sites estimated from the Patser algorithm using the Dorsal position weight matrix. (C) Overall transcriptional activity driven by the enhancers containing the binding sites in (A) measured as the total produced mRNA (fluorescence integrated over nuclear cycle 12) as a function of Dorsal concentration. Inset, mean total mRNA produced per embryo integrated across all Dorsal concentrations. Error bars, SEM over embryos containing 3 or more nuclei belonging to that Dorsal fluorescence bin. The to -axis shows the estimated nuclear Dorsal concentration according to the calibration described in Figure S9. (D) Data and model fits for the fraction of active loci (left y-axis) and mean transcription onset time (right y-axis) for each enhancer. Empty black circles, experimentally observed mean transcription onset time; filled circles, experimentally observed mean fraction of active loci. Fitted curves are represented as dashed lines (fraction of active loci) and dotted lines (mean onset times), corresponding to predictions using median parameter values from the joint posterior distribution. Shaded areas, 95% credible interval (see Table S1 for inferred parameter values). Error bars, SEM over embryos containing 3 or more nuclei belonging to that Dorsal fluorescence bin. The total number of embryos per enhancer from lowest to highest Patser score were and 46. (E) Cumulative probability distribution of spot detection over all Dorsal fluorescence bins across all embryos and enhancers ( spots). Vertical dashed line, time at which of spots have turned on corresponding to the end of the permissible transcription time window.
For the purpose of quantifying output transcriptional activity as a function of Dorsal concentration, we assigned a single Dorsal concentration value to each nucleus corresponding to the mVenus fluorescence in the center of that nucleus at a fiducial time point halfway through each nucleus’s lifetime, approximately in the middle of nuclear cycle 12 when Dorsal levels are relatively stable (Fig. S8A, B). We next grouped nuclei into 17 linearly spaced bins that span the dorsoventral axis based on their fiducial Dorsal fluorescence value (Fig. S8B).
We assessed whether these point mutations were sufficient to generate a graded response to Dorsal and to determine the dynamic range of gene expression afforded by these enhancers. To make this possible, we integrated the total mRNA output over nuclear cycle 12 of each enhancer as a function of Dorsal concentration across all nuclei exposed to a given Dorsal concentration. The integrated mRNA output of the four weakest enhancers changed little across the dorsoventral axis (Fig. 6C). However, an appreciable trend in integrated mRNA was observed for the three strongest affinities (Fig. 6C). Further, plotting the total mRNA integrated across the entire dorsoventral axis of the embryo as a function of Patser score revealed that binding-site affinity (as reported by Patser score) is strongly correlated with transcriptional output in our single binding site enhancers (Fig. 6C, inset). In the case of this measure, there was also a threshold affinity: enhancers containing binding sites with affinities below that of DBS_5.13 showed no substantial differences in transcriptional activity among them (Fig. 6C, inset). We note that, while useful to drive qualitative insights about our synthetic constructs, the total mRNA is a quantity that is removed from the transcriptional dynamics that our models aim to predict. As a result, we do not attempt to draw quantitative insights from the analyses shown in Figure 6C.
We used these constructs to measure mean transcriptional onset time as a function of Dorsal concentration and binding affinity, one of the key magnitudes predicted by our model (Fig. 3D). The measured mean onset time was relatively constant at approximately 5 minutes across all Dorsal concentrations and enhancer constructs (Fig. 6D, white circles). This value is consistent with the measured onset times of other early embryonic genes such as the minimal hunchback promoter P2P [8, 10, 14].
We also determined that the fraction of active loci is highly sensitive to Dorsal concentrations and Dorsal binding-site affinity (Fig. 6D, filled circles). The strongest Dorsal binding sites showed a large modulation of the fraction of active loci across Dorsal concentrations, while the weakest drove a relatively constant and low fraction of active loci across all Dorsal concentrations (Fig. 6D).
Our kinetic barrier model assumes that loci which fail to become active during the permissible transcription time window will remain inactive during the rest of the nuclear cycle (Fig. 3C). As a result, to determine whether the kinetic barrier model recapitulates the observations in Figure 6D, it was necessary to assign a value to this time window. We reasoned that the end of this time window determines the time point at which new transcription spots can no longer appear, possibly due to the onset of the next round of mitosis. To estimate the time point when nearly all spots have turned on, we calculated the 95th percentile of the observed spot onset times across all affinities to be approximately 7.1 min after the previous anaphase (Fig. 6E).
Using the measured time window of permissible transcription, we performed a simultaneous fit to the fraction of active loci and mean transcription onset times across all enhancers based on the kinetic barrier model from Section 2.1 (Fig. 6D, Materials and Methods). Consistent with our model, we forced all enhancers to share the same value for the rate constant , and only let the Dorsal dissociation constant , vary for each enhancer separately. By systematically exploring models with different numbers of OFF states (Figs. S11, S12 and S13), we determined that a biochemical cascade with at least 3 to 4 OFF states is capable of capturing the qualitative behavior of our observations: a Dorsal concentration- and binding affinity-dependent fraction of active loci (dashed lines in Fig. 6D) and a mean transcription onset time that is mostly constant across Dorsal concentrations and affinities (dotted lines in Fig. 6D). Alternative functional forms for , such as modeling this transition rate as depending linearly on Dorsal concentration, instead of depending on Dorsal DNA occupancy, resulted in worse fits to the fraction of active loci at saturating concentrations of Dorsal (Section S1.5). Thus, our observations can be explained by a model in which Dorsal, through DNA binding, accelerates the promoter’s transition through a sequence of kinetic barriers to a state of active transcription. We note, however, that this model demanding the sequential transition across inactive states is not the only scenario capable of recapitulating our data. For example, a model in which multiple parallel switches need to be flipped on for transcription to ensue can also lead to a similar behavior as long as their switching rate is accelerated by Dorsal binding (Figs. S14 and S15).
2.5. The experimentally measured RNAP loading rates are compatible with a thermodynamic binding model
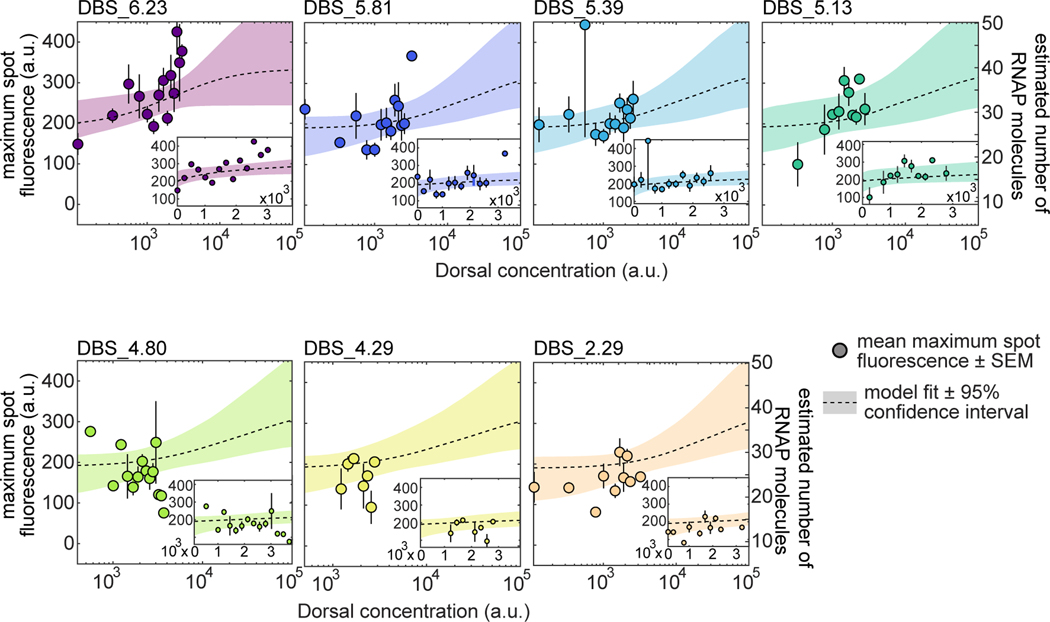
As a next step in our theoretical dissection, we tested the performance of our theoretical model in explaining the rate of transcription after loci become active. Typically, in MS2 experiments, the loading rate is measured from the initial slope of spot fluorescence traces [8, 14, 81]. However, due to the weak expression driven by our enhancers, it was not possible to perform this analysis with confidence (Fig. S16). In lieu of directly measuring the transcription rate, we evaluated a related, more robust and readily observable quantity: the maximum trace fluorescence (Fig. 4E). A theoretical foundation for this approach can be found in Methods S1.2, where we show how we approximately relate the RNAP loading rate predicted by the simple activator model (Equation 3) to the maximum fluorescence using a constant scaling factor, enabling direct comparison between theoretical predictions and experimental data. Examination of previously published live imaging data of transcription driven by the hunchback P2P reporter construct [14] confirms that the maximum fluorescence constitutes a good proxy for the RNAP loading rate (Fig. S2).
Our measurements revealed that the maximum spot fluorescence is relatively constant across Dorsal concentration for each of our seven minimal synthetic enhancers-particularly for the weakest of them, DBS_5.13, DBS_4.23, and DBS_2.92 (Fig. 7). However, the sparse and noisy nature of our data makes it challenging to draw confident conclusions from the fits, even for the stronger binding sites (i.e. DBS_6.23, DBS_5.81, and DBS_5.39). In the case of the lower affinity binding sites, the constant maximum fluorescence suggests that the Dorsal concentration level in our embryos is far below the Dorsal dissociation constant , even after increasing the Dorsal dosage by a factor of two with respect to wild-type as in our Dorsal line. The effect of very low Dorsal concentrations relative to their respective values can be clearly seen in Equation 3 and in Figure 3, where, for , the RNAP loading rate, , adopts a basal level given by
| (4) |
Figure 7. Testing RNAP loading rate predictions of the thermodynamic model.
Mean maximum spot fluorescence as a function of Dorsal concentration for minimal synthetic enhancers with different affinities for Dorsal. The right -axis denotes the calibrated number of actively transcribing RNAP molecules. As shown in Equation S15, this calibration depends linearly on the elongation rate which can vary by a factor of two depending on the study [8, 81, 82]. For more details about this calibration, see Section S1.3. Dashed curves correspond to a simultaneous Markov Chain Monte Carlo curve fit to all data using Equation 3. Fits share all parameters except . Shaded areas, 95% prediction intervals. Insets, same data and fits plotted on a linear scale with axis ranges zoomed in on the data. See Table s2 for inferred parameter values. Error bars, SEM across embryos containing 3 or more nuclei in a given fluorescence bin. The total number of embryos per enhancer from lowest to highest Patser score were 19, 27, 18, 26, 16, 35 and 46.
that is independent of Dorsal concentration and binding affinity.
As shown on the right -axes in Figure 7, this basal level corresponds to RNAP molecules actively transcribing the gene of the maximum number of RNAPs that can fit on the gene, as described in Section S1.3). We note that this estimate scales linearly with the magnitude of the RNAP elongation rate (Eq. S15), which can vary by a factor of two depending on the particular experiment [8, 81, 82]. For ease of visual comparison to the thermodynamic model predictions, we also plotted best-fit theoretical curves on top of the data using dashed curves (the insets in Fig. 7 show the same plots but zoomed into the measured data and plotted on a linear scale). These fits further underscore that our data do not explore a wide dynamic range with the precision necessary to determine the magnitude of for each construct and to thoroughly test the thermodynamic model.
3. Discussion
A major obstacle to uncovering the mechanistic and quantitative underpinnings of enhancer action is the inherent complexity of endogenous regulatory sequences. Synthetic minimal enhancers are powerful alternatives to the complex experimental reality faced by modeling efforts in endogenous enhancers (Box 1; [43, 3]). Synthetic minimal enhancers contain binding sites for one or a handful of transcription factors, making them more amenable to theoretical dissection [24, 25, 83] and revealing the complex interplay among activators, repressors, and pioneer factors, as well as their contribution to mRNA transcript accumulation [24, 25, 83]. However, previous synthetic-based efforts to dissect enhancer function always involved fixed-embryo measurements, which cannot reveal the three inherently dynamical features of transcription dictated by enhancer sequences (Fig. 1).
Here we augmented previous synthetic approaches by quantifying the real-time action of minimal enhancers with one binding site for the Dorsal activator in single cells of living, developing Drosophila embryos using the MS2 system. Contrary to theoretical speculations that single binding sites within eukaryotic genomes lack enough information to be recognized by transcription factors in the absence of other nearby binding sites [84], we demonstrated that Dorsal can drive expression when bound to single binding sites (Fig. 4D). Additionally, we demonstrated that the fraction of active loci is a feature under regulatory control in our synthetic system (Fig. 4F; Fig. 5F), confirming the important role of this regulatory strategy in shaping the expression dynamics of endogenous enhancers [8, 18, 12, 17]. Thus, while the signal driven by our minimal synthetic constructs is weak (Fig. 7), it can be quantified and recapitulates biologically relevant dynamic features of transcription that are also at play in endogenous enhancers.
While our minimal enhancer resembles endogenous promoter-proximal enhancers, it likely does not capture all the complexity of long-range distal enhancers where additional regulatory steps such as DNA looping are important. As a first step towards systematically studying the role of enhancer-promoter distance, we generated reporter constructs where we progressively moved the Dorsal binding site upstream up to . These experiments showed that the binding site is fully functional upstream of its original position (Fig. S17). Thus, while our minimal enhancer system could also be used to study the effect of distal activators at short distances in future works, the fact that, after moving the binding site upstream expression is almost completely lost suggests that our minimal regulatory sequence does not capture all the properties of endogenous distal enhancers.
It is important to note that the uncovering of a fraction of inactive loci in many reporter systems by us and others [8, 18, 12, 17] did not necessarily imply that this modulation of transcriptional engagement constitutes a biological control variable. Indeed, because live cell imaging techniques typically lack single-molecule resolution, it was unclear whether undetected loci in our study-and all previous studies-corresponded to a distinct population or were a detection artifact. By simultaneously labeling the locus with the transcription-independent reporter ParB-eGFP and nascent mRNA with MCP-mCherry (Fig. 5A), we demonstrated that a significant number of loci categorized as inactive do not constitute an experimental artifact and instead correspond to a distinct transcriptional state that is comparable to that measured in the absence of Dorsal protein (Fig. 5). In the future, conducting all live transcription measurements with DNA loci labeled by ParB could make it possible to confidently quantify the activity of all loci regardless of their activity.
Our minimal synthetic constructs and our validation of a distinct population of inactive loci enabled us to test an emerging theoretical model of enhancer action in development: a kinetic barrier model of transcriptional engagement (Fig. 3A; [85, 18, 14]. Our model deviated from previous theoretical efforts that assumed that the transition rates between states preceding transcriptional engagement were either constant [18] or depended linearly on activator concentration [14]. Instead, in order to account for the effects of Dorsal binding affinity on transcriptional dynamics, we assumed that this rate was proportional to Dorsal occupancy at its target DNA site. Thus, while the mechanisms underlying several aspects of this model, such as the molecular identity of the various OFF states, remain unknown, this model can generate predictions for how the fraction of active loci and the transcriptional onset time are modulated by the Dorsal concentration and its binding affinity (Fig. 3C–E). Theoretical evidence such as the one presented here can guide the development of new experimental methods to directly test the hypotheses they reveal.
We systematically challenged this model by generating a number of minimal synthetic enhancers spanning a large range of affinities for Dorsal (Fig. 6A). Comparing the fraction of active loci and the transcription onset times of these enhancers revealed that the kinetic barrier model recapitulated our measurements (Fig. 6D).
One interesting feature of our data is the fact that the mean transcriptional onset time is relatively constant as Dorsal concentration and binding affinity are varied. In past studies probing transcription dynamics in the Drosophila embryo [18, 14], the pioneer factor Zelda was found to be largely responsible for ensuring constant mean transcription onset times and for determining the fraction of active loci. We cannot rule out the potential existence of distal or low-affinity Zelda binding sites [86] in our constructs but believe that, just like it has been recently demonstrated for the Bicoid activator [87], Dorsal could also have a pioneering activity. Indeed, the Dorsal homolog NF-B has been recently shown to displace nucleosomes [28]. To test the kinetic barrier model, it would be informative to directly perturb the temporal dynamics of nuclear Dorsal concentration to affect transcriptional engagement. For example, several optogenetics systems have been successfully deployed in the early fly embryo to inactivate transcription factors during discrete time widows [88, 89, 90, 91]. In the future, a version of one of these systems may dissect how the temporal dynamics of Dorsal concentration affect transcriptional activation. To further probe the kinetic barrier model it would be interesting to experimentally extend the nuclear cycle duration. A key prediction of our model is that, given a longer permissible time window, seemingly silent nuclei would eventually engage in transcription. However, this is currently experimentally challenging and thus remains a thought experiment. Existing approaches to extend the duration of nuclear cycles such as lowering the temperature or genetic perturbations have pleiotropic effects on transcriptional dynamics [92, 93]. Further, while it should be possible in principle to image our reporter in the much longer nuclear cycle 14, we observed extremely weak activity even for the strong DBS_6.23 at this stage. This contrasts with endogenous Dorsal targets which are driven by enhancers containing multiple Dorsal sites as well as binding sites for other transcription factors such as Twist [94, 95]
Although the kinetic barrier model predicted the fraction of active loci and onset times (Fig. 6D) relatively well, we were unable to use our data to conclusively test the thermodynamic model’s predictions of the rate of mRNA production (Fig. 7). Such limitation stemmed in part from the fact that only a fraction of loci display detectable transcription that can be used to quantify the mRNA production rate. In addition, for those loci with detectable transcription, the spot fluorescence signal is relatively low and highly variable. Finally although the maximum spot fluorescence is a good proxy for the RNAP loading rate, it is not a perfect one since of the variance in loading rates cannot be predicted from the maximum fluorescence alone (Fig. S2). As a result, our statistics were limited such that it was not possible to perform an unequivocal test of the thermodynamic model
The apparent lack of substantial Dorsal concentration dependence observed in our measurements of RNAP loading rate could be explained in two possible ways. First, it is possible that there is a modulation of this rate in our measurements, but that this modulation is obscured by our ex perimental noise. Second, the Dorsal concentrations accessed by our experiment could be below the of our binding sites. In this scenario, a modulation in the mRNA production rate would become apparent only at Dorsal concentrations higher than those attainable by our experimental system. While our embryos contained double the genetic dosage of Dorsal compared to wild type, perhaps 5–10 times the wild-type Dorsal concentration could be needed to exceed the and modulate the rate of mRNA production. To express this high Dorsal concentration, which is certain to affect normal embryonic development, genetic approaches to increase Dorsal dosage in the embryos similar to those recently applied to flatten the Bicoid gradient might be necessary [87].
It is important to note that, despite not seeing a modulation in the rate of mRNA production, we do see a significant change in the fraction of active loci as Dorsal concentration is varied (Fig. 6). This seeming contradiction could be explained through the presence of two effective dissociation constants in our model (Fig. 3): one dissociation constant for the first part of the model governing the onset of transcription, and a different dissociation constant for the second part of the model dictating the rate of RNAP loading once transcription has ensued. Notably, previous works quantifying transcriptional dynamics of a minimal Bicoid-activated hunchback P2 enhancers also hinted at the existence of these two distinct dissociation constants [8]. These dissociation constants may arise from different binding kinetics depending on the chromatin state of the promoter, represented by the ON and OFF states in our model, which in turn could be modified by a pioneering factor like Zelda, or by the Dorsal activator itself. Further, this model is consistent with our surprising observation of a basal level of transcription in the presence of even extremely weak binding sites (Fig. 7) despite the lack of detected transcription in the absence of Dorsal protein (Fig. 4D, middle). This observation could be explained if Dorsal acted both as a pioneer-like transcription factor triggering the onset of transcription, even at low concentrations relative to its and as an activator of the transcription rate at high concentrations.
Going forward, synthetic minimal enhancers could constitute the foundation for exploring the behavior of more complex regulatory regions. Independently inferring biophysical parameters such as Dorsal-DNA binding and dissociation constants could help constrain models of Dorsal participating in the activation of promoters with additional activators and repressors [24, 25]. For ex ample, multiple Dorsal binding sites might allow for special binding configurations of both Dorsal and co-factors, enabling regulatory modes that are not possible with a single activator binding site While Dorsal is the sole known maternal nuclear-localized input specifying dorsoventral position in Drosophila, it rarely acts alone in endogenous enhancers [96]. For example, the interaction of Dorsal with Twist is a classic example of positive cooperativity in development [74]. Dorsal can also act as a repressor depending on the presence of nearby Capicua binding sites [97].The minimal synthetic enhancers presented here could be used as scaffolds for more complex minimal enhancers incorporating a second binding site for Twist or Capicua. Finally, this minimal system could make it possible to further test the theoretical model beyond the minimal enhancer sequence by probing the effect of modifying the sequence of the minimal basal promoter.
In conclusion, we have developed a minimal synthetic enhancer system that has shed light on fundamental assumptions about transcription in development. By engaging in a dialogue between theory and experiment, we have advanced our understanding of how kinetic processes give rise to important features of transcriptional dynamics in the embryo and made progress toward predictive understanding of how regulatory DNA sequence dictates the functional relation between input transcription factor dynamics and output transcriptional activity in development.
4. STAR METHODS
4.1. LEAD CONTACT AND MATERIALS AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hernan G. Garcia (hggarcia@berkeley.edu).
Materials Availability
Plasmids and fly lines generated in this study are available upon request.
Data and code availability
All data is available upon request.
All code used to analyze confocal imaging files has been previously published [8, 12] and can be found in the Github repository listed in the key resources table.
All original code written for this paper for post-image processing analyses has been deposited at the Github repository listed in the key resources table and is publicly available as of the date of publication.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Critical commercial assays | ||
| Deposited data | ||
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| yw; ParB2-eGFP; eNosx2-MCP-mCherry; + | ||
| yw; Dorsal-mVenus, pNos-MCP-mCherry; pNos-MCP-mCherry, His2Av-iRFP | ||
| yw; Dorsal-mVenus, pNos-MCP-mCherry; Dorsal-mVenus, pNos-MCP-mCherry, His2Av-iRFP | ||
| yw; dl1, pNos-MCP-mCherry; pNos-MCP-mCherry, His2Av-iRFP | ||
| yw; 1Dg(11) ; + | DBS_6.23 | |
| yw; 1DgS(2) ; + | DBS_5.81 | |
| yw; 1DgW(2) ; + | DBS_5.39 | |
| yw; 1DgAW(3) ; + | DBS_5.13 | |
| yw; 1DgSVW(2) ; + | DBS_4.8 | |
| yw; 1DgVW(1) ; + | DBS_4.73 | |
| yw; 1DgVVW(3) ; + | DBS_4.29 | |
| yw; 2xIntB2–1Dg(4) ; + | ||
| Oligonucleotides | ||
| 5’ ggaacgaaggcagttagttgt | 18.8 | |
| 5’tagttccagtgaaatccaagcattttc | Ori-Seq-F1 | |
| 5’ ccattaaaacggaaccaagaggtgag | OutLHA | |
| 5’ tctaacaatggctcgatttttgcca | OutDlRHA | |
| Recombinant DNA | ||
| pIB-1Dg-evePr-MS2v5-LacZ-Tub3UTR | DBS_6.23 | |
| pIB-1DgS-MS2v5-LacZ-Tub3UTR | DBS_5.81 | |
| pIB-1DgW-MS2v5-LacZ-Tub3UTR | DBS_5.39 | |
| pIB-1DgAW-MS2v5-LacZ-Tub3UTR | DBS_5.13 | |
| pIB-1DgSVW-MS2v5-LacZ-Tub3UTR | DBS_4.8 | |
| pIB-1DgVW-MS2v5-LacZ-Tub3UTR | DBS_4.73 | |
| pIB-1DgVVW-MS2v5-LacZ-Tub3UTR | DBS_4.29 | |
| pIB-4xIntB2-Neutral400–1Dg-MS2v5-LacZ-Tub3UTR | ||
| Dl-mVenus-dsRed | ||
| pU6-DlgRNA1 | ||
| pBPhi-eNosx2-pTrans-NoNLS-MCP-mCherry-tub3’UTR | ||
| pCasper4-His2Av-iRFP | ||
| pCasper4-Pnos-NoNLS-MCP-mCherry-TUB3’UTR | ||
| pCasper-pNos-NoNLS-ParB2-GFP-TUB3’UTR | ||
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Matlab | ||
| Patser | Hertz GZ and Stormo GD, 1999. | |
| Image analysis pipeline | Garcia HG, Tikhonov M, Lin A, Gregor T. Quantitative Imaging of Transcription in Living Drosophila Embryos842 Links Polymerase Activity to Patterning. Current Biology. 2013 Nov; 23(21):2140–2145 Lammers NC, Galstyan V, Reimer A, Medin SA, Wiggins CH, Garcia HG. Multimodal transcriptional control902 of pattern formation in embryonic development. PNAS. 2020 Jan; 117(2):836–847. |
https://github.com/GarciaLab/mRNADynamics |
| Post image processing data analysis pipeline | This paper | https://github.com/GarciaLab/mRNADynamics_dorsal_synthetics/releases/tag/v1.0.0 |
| MCMCSTAT pipeline | H. Haario, M. Laine, A. Mira and E. Saksman, 2006. DRAM: Efficient adaptive MCMC, Statistics and Computing 16, pp. 339–354. doi: 10.1007/s11222-006-9438-0 | https://github.com/mjlaine/mcmcstat |
| Other | ||
4.2. EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila melanogaster (see Key Resources Table)
4.3. METHOD DETAILS
Plasmids and reporter design
To design our minimal construct (Fig. 4), we placed the 10 bp consensus Dorsal binding site [98] upstream of the even-skipped core promoter. This enhancer-promoter construct drives the expression of the MS2v5 sequence containing 24 nonrepetitive MS2 loops [99] followed by the lacZ coding sequence and the tubulin 3’UTR. [8].
In addition to the consensus Dorsal binding site (DBS_6.23), we created six enhancers of varying strength by introducing point mutations to the consensus Dorsal binding motif. Some of these binding sites were taken from known validated Dorsal motifs [98], while others were generated based on mutations known to decrease Dorsal binding [72, 65]. To guide the design of these binding sites, we used an already existing position weight matrix computed with the MEME algorithm [100, 101] using motifs generated by DNAse I footprinting assays [102] and quantified the information content of each base pair using Patser [103].
All plasmid sequences used in this study can be accessed from a public Benchling folder. Injections were carried out by Rainbow inc. or Bestgene inc.
Transgenic Flies
Reporter plasmids were injected into BDSC fly line 27388 containing a landing site in position . Transgene orientation was confirmed by PCR using primers 18.8 (ggaacgaaggcagttagttgt) and Ori-Seq-F1 (tagttccagtgaaatccaagcattttc) binding outside of the 5’ attP site and the even-skipped promoter, respectively. All reporter lines were confirmed to be in the same orientation.
To generate the embryos used in the experiments shown in all figures except for Figure 5, we crossed Dorsal or Dorsal virgins to males carrying synthetic enhancers. The genotype of Dorsal flies is yw;Dl-mVenus (CRISPR), MCP-mCherry; Dorsal-mVenus, MCP-mCherry, His2Av-iRFP. The genotype of Dorsal flies is yw;dl[1], MCP-mCherry; Dorsal-mVenus, MCP-mCherry, His2Av-iRFP. Because there does not seem to be a difference in transcriptional activity between the CRISPR knock-in and the transgene Dorsal-mVenus alleles (Fig. S18), we combined the Dorsal and Dorsal data for some enhancers.
MCP-mCherry and His-iRFP were described before by [81]. The Dorsal-mVenus transgene was developed by [62].
To generate the Dorsal-Venus knock-in allele we used the CRISPR/Cas9 protocol described by [71]. We generated a donor plasmid containing the mVenus sequence followed by a stop codon and a 3XP3-dsRed marker flanked by PiggyBac recombinase sites. This insert was flanked by two homology arms matching surrounding the Dorsal stop codon (plasmid DI-mVenus-dsRed). The Cas9 expressing BDSC line 51324 was injected with the donor plasmid in combination with a plasmid carrying a sgRNA targeting the sequence GTTGTGAAAAAGGTATTACG located in the C-terminus of Dorsal (plasmid pU6-DlgRNA1). Survivors were crossed to and the progeny was screened for dsRed eye fluorescence. Several independent lines were established and tested for rescue. The insertion was confirmed by PCR using primers flanking the homology arms OutLHA (ccattaaaacggaaccaagaggtgag) and OutDIRHA (tctaacaatggctcgatttttgcca). The dsRed eye marker cassette was flipped out of rescuing lines via crossing with a piggyBac recombinase line. The resulting Dorsal-mVenus locus was then resequenced using the same primers. We used the same procedure to generate Dl-mCherry knock-in fusion lines but failed to obtain fertile females.
The data shown in Figure 5 were obtained from embryos laid by yw;ParB2-eGFP, eNosx2-MCP-mCherry;+ (wild-type Dorsal mothers) or yw;ParB2-eGFP, eNosx2-MCP-mCherry, dl[1];+ (Dorsal null mothers).
Microscopy
Fly cages were allowed to lay for 90 to 120 minutes prior to embryo collection. Embryos were then mounted on microscopy slides in Halocarbon 27 oil (Sigma-Aldrich, H8773) in between a coverslip and breathable membrane as described in [8, 104, 105].
Confocal microscopy was performed on a Leica SP8 with HyD detectors and a White Light Laser. We used a oil objective, and scanned bidirectionally with a scan rate of and a magnification of zoom. We did not use line or frame accumulation. Time-lapse -stacks were collected with frame rate and x-y pixel dimensions and separation between z-slices (7 range, 16 slices). resolution was pixels. Pinhole was set to 1.0 Airy units at . mVenus was excited by a laser line calibrated to using the objective and detected in a 520–567 spectral window. mCherry was excited by a laser line calibrated to and detected in a 597–660 nm spectral window. To image His2av-iRFP, the laser line was set to and detected in a spectral window. In all channels, detection was performed using the counting mode of the HyD detectors.
All movies were taken at along the anterior-posterior axis of the embryo.
ParB experiment fly crosses and microscopy
We created flies with and without functional Dorsal expressing ParB2-eGFP maternally driven by the nanos promoter and MCP-mCherry driven by two copies of a minimal nanos enhancer to label our locus DNA and nascent mRNA, respectively. In addition, we added a pars sequence followed by a 400 bp spacer (created with SiteOut, [26]) to our DBS_6.23 enhancer. We then crossed male flies containing parS-DBS_6.23-MS2 to ; ParB2-eGFP; eNosx2-MCP-mCherry; + females to create embryos that have our locus of interest labeled with eGFP colocalized with transcriptional loci in the MCP-mCherry channel (Fig. 5A and B).
After mounting embryos using the protocol described above in Section 4.3, we used the sequential scanning mode on the Leica SP8 confocal microscope to eliminate bleedthrough from eGFP into the mCherry channel, and imaged at approximately 20 s per stack, half the rate used in other imaging experiments in this study.
Image and time-series analysis
Image analysis was performed in Matlab using the custom pipeline described in [8] and [12]. This pipeline is publicly available and can be found in the Github repository listed in the Key Resources Table. Image segmentation was also aided by the Trainable Weka Segmentation plugin in FIII [106, 107]. Further analysis of time-series and other data were likewise performed in Matlab. Movies for publication were made in FIJI [108, 77].
Measuring Dorsal-mVenus concentration
Dorsal-mVenus concentration was calculated as in (Fig. S8). As shown in the figure, we measured the average mVenus fluorescence intensity in a circle of radius at the center of the nucleus in every z-slice of each nucleus. This results in a z-profile of fluorescence values covering the nucleus itself and the cytoplasm below and above it. The reported concentration corresponds to the value at the middle z-plane of each nucleus. To find this plane, we fit a parabola to the fluorescence z-profile. We use as the nuclear concentration the fluorescence value at the plane corresponding to the fitted parabola’s vertex (Fig. S8B). We then plotted this value over time and selected a single time point for each trace corresponding to the middle of each nucleus’s observed trajectory (Fig. S8B). To determine the background fluorescence in the mVenus channel we imaged flies with the same genotype as Dorsal except for the Dorsal-Venus fusions. We calculated the average nuclear fluorescence in the mVenus channel across nuclear cycle 12 and subtracted this value from our Dorsal-Venus measurements.
4.4. QUANTIFICATION AND STATISTICAL ANALYSIS
Curve fitting and parameter inference
Curve fitting and parameter inference were performed in Matlab using the MCMCSTAT Matlab package using the DRAM Markov Chain Monte Carlo algorithm [109]. For simplicity, uniform priors were assumed throughout.
Supplementary Material
VIDEO 1: Confocal microscopy movie taken on the ventral side of a developing fly embryo (yw; MCP-mCherry, DI-mVenus(CRISPR) / DBS_6.23-MS2; MCP-mCherry, DI-mVenus, His-iRFP / +) during nuclear cycle 12. Left: Dorsal-mVenus; Right: MCP-mCherry. The time stamp corresponds to hours:minutes:seconds from the beginning of nuclear cycle 12.
VIDEO 2: Confocal microscopy movie taken on the ventrolateral side of a developing fly embryo (yw; ParB-eGFP, MCP-mCherry / intB2-DBS_6.23-MS2; +) during nuclear cycle 12. Left: MCP-mCherry; Right: ParB-eGFP. The time stamp corresponds to hours:minutes:seconds from the beginning of nuclear cycle 12.
Figure 2. (Box 1) Iterative synthetic dissection of transcriptional control in development.
(A) We consider an activator exponentially distributed along one of the axes of the embryo. (B) A synthetic enhancer containing only one binding site can be described by a thermodynamic model with two parameters, the activator-DNA dissociation constant and the transcription rate enhancement upon activator binding , which control the position and amplitude of the gene expression boundary driven by the enhancer, respectively. (C) Adding one binding site to the synthetic enhancer introduces only one more free parameter describing activator-activator interactions and dictating the sharpness of the developmental boundary. (Adapted from [3].)
Highlights.
A model for whether and when a gene will turn on, and at what rate it will transcribe.
Minimal synthetic enhancers allow a systematic test of model predictions in an embryo.
DNA binding of the Dorsal transcriptional activator catalyzes transcriptional onset.
Once transcription ensues, Dorsal DNA occupancy can dictate the transcription rate.
6. Acknowledgments
We thank Greg Reeves for providing the Dorsal-mVenus and fly lines. We also thank Francois Payre and Philippe Valenti for sharing a ParB2-eGFP plasmid and a 2xIntB2 (aka parS) plasmid. We would like to thank Rob Phillips, Jane Kondev, and members of the Garcia lab for their helpful feedback on the manuscript.
H.G.G was supported by the Burroughs Wellcome Fund Career Award at the Scientific Interface, the Sloan Research Foundation, the Human Frontiers Science Program, the Searle Scholars Program, the Shurl and Kay Curci Foundation, the Hellman Foundation, the NIH Director’s New Innovator Award (DP2 OD024541–01), and an NSF CAREER Award (1652236). AR was supported by NSF GRFP (DGE 1752814).
Footnotes
Declaration of interest
The authors have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Wolpert L. Positional information and the spatial pattern of cellular differentiation. J Theor Biol, 25(1):1–47, 1969. [DOI] [PubMed] [Google Scholar]
- [2].Briscoe James and Small Stephen. Morphogen rules: design principles of gradient-mediated embryo patterning. Development, 142(23):3996–4009, December 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Garcia HG, Berrocal A, Kim YJ, Martini G, and Zhao J. Lighting up the central dogma for predictive developmental biology. Curr Top Dev Biol, 137:1–35, 2020. [DOI] [PubMed] [Google Scholar]
- [4].Vincent BJ, Estrada J, and DePace AH. The appeasement of doug: a synthetic approach to enhancer biology. Integr Biol (Camb), 8(4):475–84, 2016. [DOI] [PubMed] [Google Scholar]
- [5].Venken Koen J. T. and Bellen Hugo J.. Emerging technologies for gene manipulation in Drosophila melanogaster. Nature Reviews Genetics, 6(3):167–178, March 2005. Number: 3 Publisher: Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- [6].Bier Ethan, Harrison Melissa M., O’Connor-Giles Kate M., and Wildonger Jill. Advances in Engineering the Fly Genome with the CRISPR-Cas System. Genetics, 208(1):1–18, January 2018. Publisher: Genetics Section: FlyBook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gregor T, Bialek W, de RR Ruyter van Steveninck, D. W. Tank, and E. F. Wieschaus. Diffusion and scaling during early embryonic pattern formation. Proc Natl Acad Sci U SA, 102(51):18403–72005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Garcia Hernan G., Tikhonov Mikhail, Lin Albert, and Gregor Thomas. Quantitative Imaging of Transcription in Living Drosophila Embryos Links Polymerase Activity to Patterning. Current Biology, 23(21):2140–2145, November 2013. Publisher: Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mir M, Reimer A, Haines JE, Li XY, Stadler M, Garcia H, Eisen MB, and Darzacq X. Dense bicoid hubs accentuate binding along the morphogen gradient. Genes Dev, 31(17):1784–1794, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lucas T, Ferraro T, Roelens B, De J.Chanes Las Heras, Walczak AM, Coppey M, and Dostatni N. Live imaging of bicoid-dependent transcription in drosophila embryos. Curr Biol, 23(21):2135–9, 2013 [DOI] [PubMed] [Google Scholar]
- [11].Fukaya T, Lim B, and Levine M. Enhancer control of transcriptional bursting. Cell, 166(2):358–368, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lammers Nicholas C., Galstyan Vahe, Reimer Armando, Medin Sean A., Wiggins Chris H., and Garcia Hernan G.. Multimodal transcriptional control of pattern formation in embryonic development. PNAS, 117(2):836–847, January 2020. Publisher: National Academy of Sciences Section: Physical Sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fuqua Timothy, Jordan Jeff, van Breugel Maria Elize, Halavatyi Aliaksandr, Tischer Christian, Polidoro Peter, Abe Namiko, Tsai Albert, Mann Richard S., Stern David L., and Crocker Justin. Dense encoding of developmental regulatory information may constrain evolvability. bioRxiv, page 2020.04.17.046052, April 2020. Publisher: Cold Spring Harbor Laboratory Section: New Results. [Google Scholar]
- [14].Eck Elizabeth, Liu Jonathan, Maryam Kazemzadeh-Atoufi Sydney Ghoreishi, Blythe Shelby A, and Garcia Hernan G. Quantitative dissection of transcription in development yields evi dence for transcription factor-driven chromatin accessibility. elife, 9:e56429, October 2020. Publisher: eLife Sciences Publications, Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Berrocal A, Lammers NC, Garcia HG, and Eisen MB. Kinetic sculpting of the seven stripes of the drosophila even-skipped gene. Elife, 9, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fukaya Takashi. Dynamic regulation of anterior-posterior patterning genes in living Drosophila embryos. Current Biology, 31(10):2227–2236.e6, May 2021. [DOI] [PubMed] [Google Scholar]
- [17].Harden Timothy T., Ben Vincent J, and DePace Angela H.. Defining kinetic roles of transcriptional activators in the early drosophila embryo. bioRxiv, page 2021.02.25.432925, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dufourt Jeremy, Trullo Antonio, Hunter Jennifer, Fernandez Carola, Lazaro Jorge, Dejean Matthieu, Morales Lucas, Saida Nait-Amer Katharine N. Schulz, Harrison Melissa M., Favard Cyril, Radulescu Ovidiu, and Lagha Mounia. Temporal control of gene expression by the pioneer factor Zelda through transient interactions in hubs. Nature Communications, 9(1):5194, December 2018. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Desponds J, Tran H, Ferraro T, Lucas T, Perez Romero C, Guillou A, Fradin C, Coppey M, Dostatni N, and Walczak AM. Precision of readout at the hunchback gene: Analyzing short transcription time traces in living fly embryos. PLoS Comput Biol, 12(12):e1005256, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tran H, Desponds J, Perez Romero CA, Coppey M, Fradin C, Dostatni N, and Walczak AM. Precision in a rush: Trade-offs between reproducibility and steepness of the hunchback expression pattern. PLoS Comput Biol, 14(10):e1006513, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Desponds Jonathan, Vergassola Massimo, and Walczak Aleksandra M. A mechanism for hunchback promoters to readout morphogenetic positional information in less than a minute. eLife, 9:e49758, July 2020. Publisher: eLife Sciences Publications, Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fukaya Takashi, Lim Bomyi, and Levine Michael. Enhancer Control of Transcriptional Bursting. Cell, 166(2):358–368, July 2016. Publisher: Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Park J, Estrada J, Johnson G, Vincent BJ, Ricci-Tam C, Bragdon MD, Shulgina Y, Cha A, Wunderlich Z, Gunawardena J, and DePace AH. Dissecting the sharp response of a canonical developmental enhancer reveals multiple sources of cooperativity. Elife, 8, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fakhouri WD, Ay A, Sayal R, Dresch J, Dayringer E, and Arnosti DN. Deciphering a transcriptional regulatory code: modeling short-range repression in the drosophila embryo. Mol Syst Biol, 6:341, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sayal Rupinder, Jacqueline M Dresch Irina Pushel, Taylor Benjamin R, and Arnosti David N. Quantitative perturbation-based analysis of gene expression predicts enhancer activity in early Drosophila embryo. elife, 5:e08445, May 2016. Publisher: eLife Sciences Publications, Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Estrada J, Ruiz-Herrero T, Scholes C, Wunderlich Z, and DePace AH. Siteout: An online tool to design binding site-free dna sequences. PLoS One, 11(3):e0151740, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Scholes C, DePace AH, and Sanchez A. Combinatorial gene regulation through kinetic control of the transcription cycle. Cell Syst, 4(1):97–108 e9, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cheng Quen J., Ohta Sho, Sheu Katherine M., Spreafico Roberto, Adelaja Adewunmi, Taylor Brooks, and Hoffmann Alexander. NF-B dynamics determine the stimulus specificity of epigenomic reprogramming in macrophages. Science, 372(6548):1349–1353, June 2021. Publisher: American Association for the Advancement of Science Section: Report. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhou Jumin, Zwicker Jörk, Szymanski Paul, Levine Michael, and Tjian Robert. TAFll mutations disrupt Dorsal activation in the Drosophila embryo. PNAS, 95(23):13483–13488, November 1998. Publisher: National Academy of Sciences Section: Biological Sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Razo-Mejia M, Barnes SL, Belliveau NM, Chure G, Einav T, Lewis M, and Phillips R. Tuning transcriptional regulation through signaling: A predictive theory of allosteric induction. Cell Syst, 6(4):456–469 e10, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Polach KJ and Widom J. Mechanism of protein access to specific DNA sequences in chromatin: A dynamic equilibrium model for gene regulation. J Mol Biol, 254(2):130–49, 1995. [DOI] [PubMed] [Google Scholar]
- [32].Schulze Sandra R. and Wallrath Lori L.. Gene Regulation by Chromatin Structure: Paradigms Established in Drosophila melanogaster. Annu. Rev. Entomol, 52(1):171–192, December 2006. Publisher: Annual Reviews. [DOI] [PubMed] [Google Scholar]
- [33].Lam FH, Steger DJ, and O’Shea EK. Chromatin decouples promoter threshold from dynamic range. Nature, 453(7192):246–50, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, Chu HC, Ogawa N, Inwood W, Sementchenko V, Beaton A, Weiszmann R, Celniker SE, Knowles DW, Gingeras T, Speed TP, Eisen MB, and Biggin MD. Transcription factors bind thousands of active and inactive regions in the drosophila blastoderm. PLOS Biol, 6(2):e27, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kim HD and O’Shea EK. A quantitative model of transcription factor-activated gene expression. Nat Struct Mol Biol, 15:1192–1198, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Levine Mike. Transcriptional Enhancers in Animal Development and Evolution. Current Biol ogy, 20(17):R754–R763, September 2010. Publisher: Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Fussner Eden, Ching Reagan W., and Bazett-Jones David P.. Living without 30 nm chromatin fibers. Trends in Biochemical Sciences, 36(1):1–6, January 2011. Publisher: Elsevier. [DOI] [PubMed] [Google Scholar]
- [38].Bai L, Ondracka A, and Cross FR. Multiple sequence-specific factors generate the nucleosome-depleted region on cln2 promoter. Mol Cell, 42(4):465–76, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Li GW, Burkhardt D, Gross C, and Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell, 157(3):624–35, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hansen AS and O’Shea EK. cis determinants of promoter threshold and activation timescale. Cell Rep, 12(8):1226–33, 2015. [DOI] [PubMed] [Google Scholar]
- [41].Li Xiao-Yong and Eisen Michael B.. Zelda potentiates transcription factor binding to zygotic enhancers by increasing local chromatin accessibility during early <em> drosophila melanogaster embryogenesis. bioRxiv, page 380857, 2018. [Google Scholar]
- [42].Foo SM, Sun Y, Lim B, Ziukaite R, O’Brien K, Nien CY, Kirov N, Shvartsman SY, and Rushlow CA. Zelda potentiates morphogen activity by increasing chromatin accessibility. Curr Biol, 24(12):1341–6, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Garcia HG, Brewster RC, and Phillips R. Using synthetic biology to make cells tomorrow’s test tubes. Integr Biol (Camb), 8(4):431–50, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Garcia Hernan G. and Phillips Rob. Quantitative dissection of the simple repression input-output function. PNAS, 108(29):12173–12178, July 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Brewster RC, Weinert FM, Garcia HG, Song D, Rydenfelt M, and Phillips R. The transcription factor titration effect dictates level of gene expression. Cell, 156(6):1312–23, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Phillips R, Belliveau NM, Chure G, Garcia HG, Razo-Mejia M, and Scholes C. Figure 1 theory meets figure 2 experiments in the study of gene expression. Annu Rev Biophys, 48:121–163,2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Achim P Popp Johannes Hettich, Christof J, and Gebhardt M. Transcription factor residence time dominates over concentration in transcription activation. bioRxiv, page 2020.11.26.400069,2020. [Google Scholar]
- [48].Germier Thomas, Kocanova Silvia, Walther Nike, Bancaud Aurélien, Haitham Ahmed Shaban Hafida Sellou, Antonio Zaccaria Politi Jan Ellenberg, Gallardo Franck, and Bystricky Kerstin. Real-Time Imaging of a Single Gene Reveals Transcription-Initiated Local Confinement. Biophys J, 113(7):1383–1394, October 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chen H, Levo M, Barinov L, Fujioka M, Jaynes JB, and Gregor T. Dynamic interplay between enhancer-promoter topology and gene activity. Nat Genet, 50(9):1296–1303, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ackers GK, Johnson AD, and Shea MA. Quantitative model for gene regulation by lambda phage repressor. Proc Natl Acad Sci U S A, 79(4):1129–33, 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Buchler NE, Gerland U, and Hwa T. On schemes of combinatorial transcription logic. Proc Natl Acad Sci U SA, 100(9):5136–41, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Vilar JM and Leibler S. Dna looping and physical constraints on transcription regulation. J Mol Biol, 331(5):981–9, 2003. [DOI] [PubMed] [Google Scholar]
- [53].Bolouri H.and Davidson EH. Transcriptional regulatory cascades in development: initial rates, not steady state, determine network kinetics. Proc Natl Acad Sci U SA, 100(16):9371–6, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, and Phillips R. Transcrip tional regulation by the numbers: models. Curr Opin Genet Dev, 15(2):116–24, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Kuhlman T, and Phillips R. Transcriptional regulation by the numbers: applications. Curr Opin Genet Dev, 15(2):125–35, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Sherman MS and Cohen BA. Thermodynamic state ensemble models of cis-regulation. PLoS Comput Biol, 8(3):e1002407, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Mir M, Stadler MR, Ortiz SA, Hannon CE, Harrison MM, Darzacq X, and Eisen MB. Dynamic multifactor hubs interact transiently with sites of active transcription in drosophila embryos. Elife, 7:e40497, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Callegari Andrea, Sieben Christian, Benke Alexander, Suter David M., Fierz Beat, Mazza Davide, and Manley Suliana. Single-molecule dynamics and genome-wide transcriptomics reveal that NF- κ B (p65)-DNA binding times can be decoupled from transcriptional activation. bioRxiv, pages 1–23, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Shermoen Antony W. and O’Farrell Patrick H.. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell, 67(2):303–310, October 1991. Publisher: Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bintu Lacramioara, Buchler Nicolas E, Garcia Hernan G, Gerland Ulrich, Hwa Terence, Kondev Jané, and Phillips Rob. Transcriptional regulation by the numbers: models. Curr Opin Genet Dev, 15(2):116–124, April 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Roth Siegfried, Stein David, and Nüsslein-Volhard Christiane. A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell, 59(6):1189–1202, 1989. [DOI] [PubMed] [Google Scholar]
- [62].Reeves GT, Trisnadi N, Truong TV, Nahmad M, Katz S, and Stathopoulos A. Dorsal-ventral gene expression in the drosophila embryo reflects the dynamics and precision of the dorsal nuclear gradient. Dev Cell, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Steward Ruth, Zusman Susan B., Huang Leslie H., and Schedl Paul. The dorsal protein is distributed in a gradient in early drosophila embryos. Cell, 55(3):487–495, 1988. [DOI] [PubMed] [Google Scholar]
- [64].C Thisse F Perrin-Schmitt, Stoetzel C, and Thisse B. Sequence-specific transactivation of the Drosophila twist gene by the dorsal gene product. Cell, 65:1191–1201, 1991. [DOI] [PubMed] [Google Scholar]
- [65].Jiang J, Hoey T, and Levine M. Autoregulation of a segmentation gene in drosophila: combinatorial interaction of the even-skipped homeo box protein with a distal enhancer element. Genes Dev, 5(2):265–77, 1991. [DOI] [PubMed] [Google Scholar]
- [66].Kirov Nikolai, Zhelnin Leonid, Shah Jaymini, and Rushlow Christine. Conversion of a silencer into an enhancer: evidence for a co-repressor in dorsal-mediated repression in Drosophila. 12(8):3193–3199, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Papagianni Aikaterini, Marta Forés Wanqing Shao, He Shuonan, Koenecke Nina, María José Andreu Núria Samper, Ze’ev Paroush Sergio González-Crespo, Zeitlinger Julia, and Jiménez Gerardo. Capicua controls Toll/IL-1 signaling targets independently of RTK regulation. Proceedings of the National Academy of Sciences of the United States of America, 115(8):1807–1812, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sandler JE and Stathopoulos A. Quantitative single-embryo profile of drosophila genome activation and the dorsal-ventral patterning network. Genetics, 202(4):1575–84, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Dufourt Jeremy, Bellec Maelle, Trullo Antonio, Dejean Matthieu, De Rossi Sylvain, and Lagha Mounia. Imaging translation dynamics in live embryos reveals spatial heterogeneities. bioRxiv, page 2020.04.29.058974,2020. [DOI] [PubMed] [Google Scholar]
- [70].Kremers GJ, Goedhart J, van Munster EB, and Gadella TW Jr.. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and fret forster radius. Biochemistry, 45(21):6570–80, 2006. [DOI] [PubMed] [Google Scholar]
- [71].Gratz SJ, Rubinstein CD, Harrison MM, Wildonger J, and O’Connor-Giles KM. Crispr-cas9 genome editing in drosophila. Curr Protoc Mol Biol, 111:31 21–20, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Ip YT, Park RE, Kosman D, Yazdanbakhsh K, and Levine M. dorsal-twist interactions establish snail expression in the presumptive mesoderm of the drosophila embryo. Genes Dev, 6(8):1518–30, 1992. [DOI] [PubMed] [Google Scholar]
- [73].Jiang J.and Levine M. Binding affinities and cooperative interactions with bhlh activators delimit threshold responses to the dorsal gradient morphogen. Cell, 72(5):741–52, 1993. [DOI] [PubMed] [Google Scholar]
- [74].Szymanski P.and Levine M. Multiple modes of dorsal-bhlh transcriptional synergy in the drosophila embryo. EMBO J, 14(10):2229–38, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Stormo and GD 3rd Hartzell, G. W. Identifying protein-binding sites from unaligned dna fragments. Proc Natl Acad Sci U SA, 86(4):1183–7, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, and Long RM. Localization of ash1 mrna particles in living yeast. Mol Cell, 2(4):437–45, 1998. [DOI] [PubMed] [Google Scholar]
- [77].Schindelin Johannes, Ignacio Arganda-Carreras Erwin Frise, Kaynig Verena, Pietzsch Mark Longair Tobias, Preibisch Stephan, Rueden Curtis, Saalfeld Stephan, Schmid Benjamin, Tinevez JeanYves, White Daniel James, Hartenstein Volker, Eliceiri Kevin, Tomancak Pavel, and Cardona Albert. Fiji: an open-source platform for biological-image analysis. Nature Methods, 9(7):676-682, July 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Alamos Simon, Reimer Armando, Niyogi Krishna K., and Garcia Hernan G. Quantitative imaging of rna polymerase ii activity in plants reveals the single-cell basis of tissue-wide transcriptional dynamics. bioRxiv, page 274621, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hafner Antonina, Reyes José, Jacob Stewart-Ornstein Michael Tsabar, Jambhekar Ashwini, and Lahav Galit. Quantifying the Central Dogma in the p53 Pathway in Live Single Cells. Cell Systems, pages 1–11, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, and Yamamoto KR. Dna binding site sequence directs glucocorticoid receptor structure and activity. Science, 324(5925):407–10, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Liu Jonathan, Hansen Donald, Eck Elizabeth, Yang Joon Kim Meghan Turner, Alamos Simon, and Garcia Hernan G.. Real-time single-cell characterization of the eukaryotic transcription cycle reveals correlations between RNA initiation, elongation, and cleavage. PLOS Computational Biology, 17(5):e1008999, May 2021. Publisher: Public Library of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Fukaya T, Lim B, and Levine M. Rapid rates of pol ii elongation in the drosophila embryo. Curr Biol, 27(9):1387–1391, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Crocker J.and Ilsley GR. Using synthetic biology to study gene regulatory evolution. Curr Opin Genet Dev, 47:91–101, 2017. [DOI] [PubMed] [Google Scholar]
- [84].Wunderlich Zeba and Mirny Leonid A.. Different gene regulation strategies revealed by analysis of binding motifs. Trends in Genetics, 25(10):434–440, October 2009. Publisher: Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Fritzsch C, Baumgartner S, Kuban M, Steinshorn D, Reid G, and Legewie S. Estrogen-dependent control and cell-to-cell variability of transcriptional bursting. Mol Syst Biol, 14(2):e7678, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Rushlow CAand Shvartsman SY. Temporal dynamics, spatial range, and transcriptional interpretation of the dorsal morphogen gradient. Curr Opin Genet Dev, 22(6):542–6, 2012. [DOI] [PubMed] [Google Scholar]
- [87].Hannon Colleen E, Blythe Shelby A, and Wieschaus Eric F. Concentration dependent chromatin states induced by the bicoid morphogen gradient. elife, 6:e28275, September 2017. Publisher: eLife Sciences Publications, Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Huang A, Amourda C, Zhang S, Tolwinski NS, and Saunders TE. Decoding temporal interpretation of the morphogen bicoid in the early drosophila embryo. Elife, 6, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].McDaniel SL, Gibson TJ, Schulz KN, Fernandez Garcia M, Nevil M, Jain SU, Lewis PW, Zaret KS, and Harrison MM. Continued activity of the pioneer factor zelda is required to drive zygotic genome activation. Mol Cell, 74(1):185–195 e4, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Irizarry J, McGehee J, Kim G, Stein D, and Stathopoulos A. Twist-dependent ratchet functioning downstream from dorsal revealed using a light-inducible degron. Genes Dev, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Singh Anand P., Wu Ping, Ryabichko Sergey, Raimundo João, Swan Michael, Wieschaus Eric, Gregor Thomas, and Toettcher Jared E.. Optogenetic control of the Bicoid morphogen reveals fast and slow modes of gap gene regulation. Cell Reports, 38(12), 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Isaac JT Strong, Lei Xiaoyun, Chen Fang, Yuan Kai, and O’Farrell Patrick H.. Interphase arrested Drosophila embryos activate zygotic gene expression and initiate mid-blastula transition events at a low nuclear-cytoplasmic ratio. PLoS Biology, 18(10):1–27, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Kuntz SG and Eisen MB. Drosophila embryogenesis scales uniformly across temperature in developmentally diverse species. PLoS Genet, 10(4):e1004293, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hong JW, Hendrix DA, and Levine MS. Shadow enhancers as a source of evolutionary novelty. Science, 321(5894):1314, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Reeves GT and Stathopoulos A. Graded dorsal and differential gene regulation in the drosophila embryo. Cold Spring Harb Perspect Biol, 1(4):a000836, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Hong JW, Hendrix DA, Papatsenko D, and Levine MS. How the dorsal gradient works: insights from postgenome technologies. Proc Natl Acad Sci U S A, 105(51):20072–6, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Dong Hyeon Shinand Joung Woo Hong. Capicua is involved in Dorsal-mediated repression of Zerknüllt expression in Drosophila embryo. BMB Reports, 47(9):518–523, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Markstein Michele, Markstein Peter, Markstein Vicky, and Levine Michael S. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. 99(2):763–768, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Tutucci Evelina, Vera Maria, Biswas Jeetayu, Garcia Jennifer, Parker Roy, and Singer Robert H.. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat Methods, 15(1):81–89, January 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ivan Andra, Halfon Marc S., and Sinha Saurabh. Computational discovery of cis-regulatory modules in Drosophila without prior knowledge of motifs. Genome Biology, 9(1):R22, January 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Bailey Timothy L., Williams Nadya, Misleh Chris, and Li Wilfred W.. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res, 34(suppl_2):W369–W373, July 2006. Publisher: Oxford Academic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Bergman Casey M., Carlson Joseph W., and Celniker Susan E.. Drosophila DNase I footprint database: a systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics, 21(8):1747–1749, April 2005. Publisher: Oxford Academic [DOI] [PubMed] [Google Scholar]
- [103].Hertz GZ and Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics, 15(7):563–577, July 1999. Publisher: Oxford Academic. [DOI] [PubMed] [Google Scholar]
- [104].Bothma JP, Garcia HG, Esposito E, Schlissel G, Gregor T, and Levine M. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living drosophila embryos. Proc Natl Acad Sci U S A, 111(29):10598–10603, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Garcia Hernan G. and Gregor Thomas. Live Imaging of mRNA Synthesis in Drosophila, pages 349–357. Springer New York, New York, NY, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Witten Ian H., Frank Eibe, Hall Mark A., and Pal Christopher J.. Data Mining: Practical Machine Learning Tools and Techniques. Morgan Kaufmann, October 2016. Google-Books-ID: 1SyICgAAQBAJ [Google Scholar]
- [107].Ignacio Arganda-Carreras Verena Kaynig, Rueden Curtis, Eliceiri Kevin W., Schindelin Johannes, Cardona Albert, and H. Sebastian Seung. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics, 33(15):2424–2426, August 2017. [DOI] [PubMed] [Google Scholar]
- [108].Schneider Caroline A, Rasband Wayne S, and Eliceiri Kevin W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods, 9:671, June 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Haario Heikki, Laine Marko, Mira Antonietta, and Saksman Eero. DRAM: Efficient adaptive MCMC. Stat Comput, 16(4):339–354, December 2006. [Google Scholar]
- [110].Eck E, Liu J, Kazemzadeh-Atoufi M, Ghoreishi S, Blythe SA, and Garcia HG. Quantitative dissection of transcription in development yields evidence for transcription factor-driven chromatin accessibility. Elife, 9:e56429, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Talley J. Lambert FP base: a community-editable fluorescent protein database. Nature Meth ods, 16(4):277–278, April 2019. Number: 4 Publisher: Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- [112].Selby Christopher P., Drapkin Ronny, Reinberg Danny, and Sancar Aziz. RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Research, 25(4):787–793, February 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Grimm O.and Wieschaus E. The bicoid gradient is shaped independently of nuclei. Development, 137(17):2857–62, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Gregor Thomas, Tank David W., Wieschaus Eric F., and Bialek William. Probing the Limits to Positional Information. Cell, 130(1):153–164, July 2007. Publisher: Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Liu F, Morrison AH, and Gregor T. Dynamic interpretation of maternal inputs by the drosophila segmentation gene network. Proc Natl Acad Sci U S A, 110(17):6724–9, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
VIDEO 1: Confocal microscopy movie taken on the ventral side of a developing fly embryo (yw; MCP-mCherry, DI-mVenus(CRISPR) / DBS_6.23-MS2; MCP-mCherry, DI-mVenus, His-iRFP / +) during nuclear cycle 12. Left: Dorsal-mVenus; Right: MCP-mCherry. The time stamp corresponds to hours:minutes:seconds from the beginning of nuclear cycle 12.
VIDEO 2: Confocal microscopy movie taken on the ventrolateral side of a developing fly embryo (yw; ParB-eGFP, MCP-mCherry / intB2-DBS_6.23-MS2; +) during nuclear cycle 12. Left: MCP-mCherry; Right: ParB-eGFP. The time stamp corresponds to hours:minutes:seconds from the beginning of nuclear cycle 12.
Data Availability Statement
All data is available upon request.
All code used to analyze confocal imaging files has been previously published [8, 12] and can be found in the Github repository listed in the key resources table.
All original code written for this paper for post-image processing analyses has been deposited at the Github repository listed in the key resources table and is publicly available as of the date of publication.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Critical commercial assays | ||
| Deposited data | ||
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| yw; ParB2-eGFP; eNosx2-MCP-mCherry; + | ||
| yw; Dorsal-mVenus, pNos-MCP-mCherry; pNos-MCP-mCherry, His2Av-iRFP | ||
| yw; Dorsal-mVenus, pNos-MCP-mCherry; Dorsal-mVenus, pNos-MCP-mCherry, His2Av-iRFP | ||
| yw; dl1, pNos-MCP-mCherry; pNos-MCP-mCherry, His2Av-iRFP | ||
| yw; 1Dg(11) ; + | DBS_6.23 | |
| yw; 1DgS(2) ; + | DBS_5.81 | |
| yw; 1DgW(2) ; + | DBS_5.39 | |
| yw; 1DgAW(3) ; + | DBS_5.13 | |
| yw; 1DgSVW(2) ; + | DBS_4.8 | |
| yw; 1DgVW(1) ; + | DBS_4.73 | |
| yw; 1DgVVW(3) ; + | DBS_4.29 | |
| yw; 2xIntB2–1Dg(4) ; + | ||
| Oligonucleotides | ||
| 5’ ggaacgaaggcagttagttgt | 18.8 | |
| 5’tagttccagtgaaatccaagcattttc | Ori-Seq-F1 | |
| 5’ ccattaaaacggaaccaagaggtgag | OutLHA | |
| 5’ tctaacaatggctcgatttttgcca | OutDlRHA | |
| Recombinant DNA | ||
| pIB-1Dg-evePr-MS2v5-LacZ-Tub3UTR | DBS_6.23 | |
| pIB-1DgS-MS2v5-LacZ-Tub3UTR | DBS_5.81 | |
| pIB-1DgW-MS2v5-LacZ-Tub3UTR | DBS_5.39 | |
| pIB-1DgAW-MS2v5-LacZ-Tub3UTR | DBS_5.13 | |
| pIB-1DgSVW-MS2v5-LacZ-Tub3UTR | DBS_4.8 | |
| pIB-1DgVW-MS2v5-LacZ-Tub3UTR | DBS_4.73 | |
| pIB-1DgVVW-MS2v5-LacZ-Tub3UTR | DBS_4.29 | |
| pIB-4xIntB2-Neutral400–1Dg-MS2v5-LacZ-Tub3UTR | ||
| Dl-mVenus-dsRed | ||
| pU6-DlgRNA1 | ||
| pBPhi-eNosx2-pTrans-NoNLS-MCP-mCherry-tub3’UTR | ||
| pCasper4-His2Av-iRFP | ||
| pCasper4-Pnos-NoNLS-MCP-mCherry-TUB3’UTR | ||
| pCasper-pNos-NoNLS-ParB2-GFP-TUB3’UTR | ||
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Matlab | ||
| Patser | Hertz GZ and Stormo GD, 1999. | |
| Image analysis pipeline | Garcia HG, Tikhonov M, Lin A, Gregor T. Quantitative Imaging of Transcription in Living Drosophila Embryos842 Links Polymerase Activity to Patterning. Current Biology. 2013 Nov; 23(21):2140–2145 Lammers NC, Galstyan V, Reimer A, Medin SA, Wiggins CH, Garcia HG. Multimodal transcriptional control902 of pattern formation in embryonic development. PNAS. 2020 Jan; 117(2):836–847. |
https://github.com/GarciaLab/mRNADynamics |
| Post image processing data analysis pipeline | This paper | https://github.com/GarciaLab/mRNADynamics_dorsal_synthetics/releases/tag/v1.0.0 |
| MCMCSTAT pipeline | H. Haario, M. Laine, A. Mira and E. Saksman, 2006. DRAM: Efficient adaptive MCMC, Statistics and Computing 16, pp. 339–354. doi: 10.1007/s11222-006-9438-0 | https://github.com/mjlaine/mcmcstat |
| Other | ||