Abstract
The natural world is constantly changing, and planetary boundaries are issuing severe warnings about biodiversity and cycles of carbon, nitrogen, and phosphorus. In other views, social problems such as global warming and food shortages are spreading to various fields. These seemingly unrelated issues are closely related, but it can be said that understanding them in an integrated manner is still a step away. However, progress in analytical technologies has been recognized in various fields and, from a microscopic perspective, with the development of instruments including next-generation sequencers (NGS), nuclear magnetic resonance (NMR), gas chromatography-mass spectrometry (GC/MS), and liquid chromatography-mass spectrometry (LC/MS), various forms of molecular information such as genome data, microflora structure, metabolome, proteome, and lipidome can be obtained. The development of new technology has made it possible to obtain molecular information in a variety of forms. From a macroscopic perspective, the development of environmental analytical instruments and environmental measurement facilities such as satellites, drones, observation ships, and semiconductor censors has increased the data availability for various environmental factors. Based on these background, the role of computational science is to provide a mechanism for integrating and understanding these seemingly disparate data sets. This review describes machine learning and the need for structural equations and statistical causal inference of these data to solve these problems. In addition to introducing actual examples of how these technologies can be utilized, we will discuss how to use these technologies to implement environmentally friendly technologies in society.
Keywords: Biodiversity, Environmental analysis, Network, Machine learning, Statistical inference
1. Introduction
Tilman et al. indicated the importance of new incentives and policies to ensure the sustainability of agriculture and ecosystem services [1], [2]. Subsequently, the concrete framework of the planetary boundary was emphasized [3]. In particular, biodiversity conservation and the efficient recycling of carbon, nitrogen, and phosphorus resources are urgent global issues [3], [4], [5]. Along with these problems, global warming [6], [7], food problems [8], [9], and hybrid problems [10], [11] are emerging. For nitrogen control, nitrogen fixation or denitrification balance in soil [12], lakes [13], and oceans [14], [15] is important. Nitrogen fixation will lead to a reduction in the number of chemical fertilizers produced with petroleum [16] and indirectly contribute to the suppression of global warming. In addition, the generation of nitrous oxide, a greenhouse gas, has a direct impact on global warming: it is essential to control denitrification. These controls are expected to involve symbiotic relationships between multiple organisms, including nitrogen-fixing bacteria, anammox bacteria, fungi [17], [18], ticks [19], and insects [20]. Furthermore, management in recycling [21], [22] and livestock management technology [23] will also be affected. Phosphorus is an essential element for organisms, and in agriculture [24], animal husbandry [25], [26], [27], and fisheries [28], the dynamics on the input and output sides is closely watched. In particular, excess phosphorus in soil and water affects eutrophication [29], [30], [31], so a well-balanced management is necessary to reduce the environmental impact. The management of phosphorus from livestock feed is closely related to soil through fecal manure compost [27]. Control of phosphorus and nitrogen by livestock fecal manure of livestock [32] requires management, and research to consider the balance between nitrogen and phosphorus at the global level is emphasized [33]. The control of the balance between nitrogen and phosphorus is closely related to biodiversity. Generally, it is difficult to evaluate the viewpoint of biological diversity, and methods based on DNA classification, mainly targeting microorganisms [34], [35], methods focusing on metabolites [36], [37], or combinations [20], [38] are performed. Land research has advanced in assessing diversity on land and sea, and it has been pointed out that at least the diversity of soil symbionts is extremely important in the animal-plant-human relationship [39]. Many researchers have been conducting research to gain insight into these complex environmental factors from various angles in the fields of agriculture, animal husbandry, fisheries, and environmental purification. This environmental complexity can be elucidated by combining a data science approach with physical environmental monitoring data and biochemical analytical data. Namely, visualization of homeostatic plasticity can show “environmental health (or illness)”, like as an assessment of a healthy human state [40], [41].
We will introduce a multifaceted approach method from the viewpoint of computational science to these research subjects and describe future perspectives.
2. Approach
2.1. Scheme
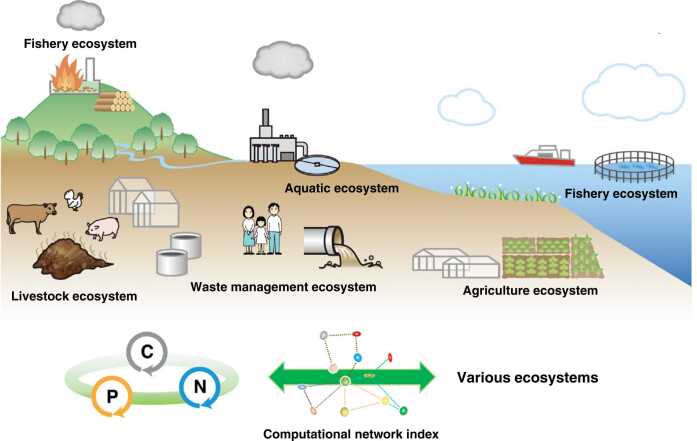
This review will discuss how the various impacts caused by human activities should be captured throughout the ecosystem from the viewpoint of water, soil, forests, agriculture, livestock, the water industry, and recycling (Fig. 1). Each ecosystem is connected in the cycles of carbon, nitrogen, and phosphorus, and there is a network of related factors. The overall balance of the material cycles of carbon, nitrogen, and phosphorus will have a significant impact on the ecosystem (Fig. 2). Therefore, we will introduce case studies in each field on how to rearrange the exhaustive data analyzed with the latest technologies to enable an integrated understanding. Data acquired by various analysis technologies can be analyzed by cluster analysis, correlation analysis, machine learning, structural equations, and causal inference. They can be viewed as a way of seeing ecosystem patterns in computational science. The practice patterns are viewed can also provide hints for preserving ecosystems, not only from a one-sided view of each ecosystem but also from a comprehensive view of the ecosystem as a wholistic way of thinking.
Fig. 1.
Various ecosystems where computational science can be applied. It shows that the multiple impacts that human activities can generate affect each ecosystem and the integrated ecosystem, and we are now at a crossroads. Carbon (C), nitrogen (N), and phosphorus (P) flows are indicated by gray, blue, and orange arrows, respectively. Computational network indicators, which serve as markers of the circulation of carbon, nitrogen, and phosphorus, are connected to various ecosystems.
Fig. 2.
Relationship between carbon, nitrogen, and phosphorus flow balance and ecosystem homeostasis. The flow balance of carbon, nitrogen, and phosphorus on the left side negatively affects ecosystem homeostasis; the flow balance on the right side positively impacts ecosystem homeostasis.
2.2. Ecosystem monitoring and measurements
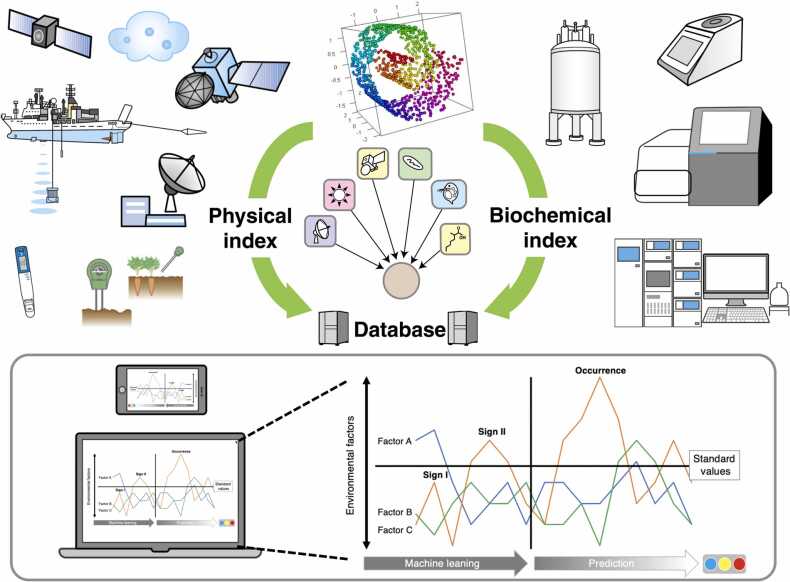
Various instruments and measuring devices have been developed for environmental assessment. From semiconductor instruments for easy measurement to data from satellites and observation ships, there are tools to promote environmental monitoring (Fig. 3). That is, real-time environmental monitoring data in terms of physical factors can be very useful to elucidate time-dependent changes in homeostatic plasticity. In addition, there have been remarkable developments in analytical technologies for laboratory-level instruments [42], such as NGS NMR, GC/MS, and LC/MS. However, the work to organize and integrate these data has not progressed as much as expected. A substance equivalent to the glue that connects these on-site data from macro- and micro-perspectives is needed. The advancement of these efforts will be an urgent task in data science in the future to build a sustainable society. These efforts are also expected to lead to the development of methods that can predict changes in environmental factors and technologies that can develop countermeasures based on such predictions.
Fig. 3.
Concept of environmental prediction based on the accumulated machine learning database. Data accumulation is critical to better prediction performance by machine learning computations. To do that, environmental monitoring based on physical parameters is quite valuable for accumulating time-dependent data. Furthermore, occasional samplings from environments of interest allow us to elucidate biochemical data measured by NMR, MS, and NGS instruments. Even physical and biochemical data can be formatted with RDF; such a multifactored database can be computed by machine learning. In the future, computational devices, such as quantum innovation, might significantly reduce computational time, as well as device size. Therefore, “field computation (prediction)” may be possible in the fields of agriculture, fishing, livestock, and forest.
2.3. Database accumulation
To accomplish these objectives, it is necessary to organize and integrate databases (Fig. 3). In addition to using the data as raw data, it is required to simplify the data that are affected by environmental conditions. In other words, it is the preprocessing of data in dry analysis. When accumulating environmental data, it is difficult to say for sure whether the data correspond to a Gaussian distribution or a non-Gaussian distribution. Based on the data that do exist, we can only computationally infer the character of distributions. A linear or non-linear perspective is also required when time-series data are included. One way to overcome these problems is to view the data itself as relative values or by binarizing the data so that they can be viewed from a bird's eye view (see elsewhere on how to use mean assignment or regression, etc., for data with missing values). Another critical point is the data management mechanism. Traditionally, data formatting has been based on the extensible markup language (XML) [43], [44]. However, in recent years, it has become necessary to use the Resource Description Framework (RDF) to organize large amounts of data [45]. This paper does not discuss RDF in detail. In any case, it is essential to organize the accumulated data efficiently.
2.4. Machine learning
Aggregated data are the primary subject of data science for classification or regression by machine learning. Machine learning is currently used in many scientific fields. Two types of machine learning are known: unsupervised learning and supervised learning.
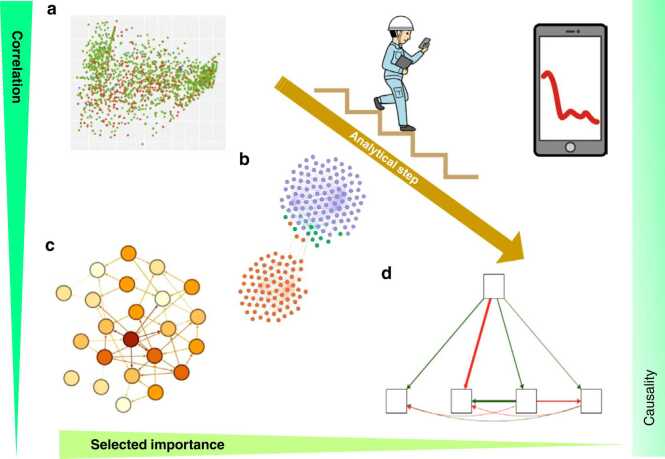
In unsupervised learning, the system learns complex patterns more autonomously and identifies them to summarize, explore, and discover (Fig. 4). For example, PCA, k-means clustering, hierarchical clustering, and self-organizing maps. These have a strong relationship with the correlation strength and can also be represented by a correlation heatmap [40]. A known analysis method that uses raw data is Bayesian network analysis. Association analysis (AA) (market basket analysis) [20] and energy landscape analysis (ELA) [46] are also known as methods that analyze data after binarizing them as a preprocessing step. Although the resolutions of the three analyses do not necessarily coincide and depend on the analysis conditions, it should be noted that AA and ELA, which use raw data that have been binarized by preprocessing, are relatively unaffected by the data characteristics of whether the raw data are Gaussian or non-Gaussian distribution.
Fig. 4.
Analytical step from multivariate and association analysis to machine learning and statistical causal inference (From left-top to right-bottom). (a) represents a diverse set of data. (b) Association analysis (market basket analysis), (c) Bayesian network analysis (HC: hill climbing), and (d) structural equations are shown, respectively. The stair walker is representatively showing how a person performing normal work might be able to check by portable computational device for changes in environmental factors at the field and industrial site. The multifactor environmental database can be computed and selected as casual importance factors. First, a data-driven approach can calculate all the accumulated data sets using multivariate and association analysis. The second computational step is a selection of essential factors by machine learning. Finally, causal relationships can be visualized by SEM, BayesLiNGAM, and other methods (see in the text).
There are probability-aware calculation methods, and one of their bases is Bayes' Theorem [47]. Similarly, the Markov model, a probability-aware concept, and its development, the hidden Markov model (HMM) [48] are now indispensable techniques in gene classification. Along with this method, an important methodology is dynamic programming (DP) [49]. It has a long history and is best known for DP matching using the DP algorithm, which is used for speech recognition [50] and image recognition [51]. It does not rely primarily on probability theory but rather calculates the value of each state from the data itself and evaluates the value based on the Bellman equation (DP equation) under the assumption that the behavior is to transition to the state with the highest value.
Supervised learning, on the other hand, involves training in an annotated data set and defining outputs. The goal is to determine the association between the response and predictor variables (often called covariates) and to make accurate predictions. A classification method of analysis that compares discrete variables (e.g., control and test groups) is discriminant analysis. Linear discriminant analysis (LDA) [52] is a well-known method for comparing microbial communities. It should be noted that Latent dirichlet allocation [53], a machine learning method for natural language processing, is also shortened by the same acronym as LDA used for linear discriminant analysis. Latent dirichlet allocation is used to detect genetic displacement in the biological domain.
Both are forms of machine learning. Partial least squares discriminant analysis (PLS-DA) [40] is a known method used for metabolite analysis. In addition, structured support vector machines [54] are a method that can be applied to complex linear and nonlinear cases. Conventionally, the support vector machine (SVM) [40] is an algorithm that increases the dimension of the target data by maximizing the kernel function and the margin and can divide and interpret non-linear data as linear data. This method further extends its properties and can be applied to various data structures. It allows one to predict the outputs of new data, suggesting that, in addition to SVM, random forest (RF) and artificial neural networks (ANN) may apply to the analysis of various environmental factors [40].
2.5. Statistical structure equations and causal inference
Structural equation modeling (SEM) is used with the same meaning as covariance structure analysis and refers to an analysis in which relationships among various variables are modeled in the form of linear combinations (Fig. 4). It can integrate correlation analysis, regression analysis, and factor analysis, and it can statistically assess complex relationships between factors. The foundation of structural equations was created by Wright S [55], [56], [57]: it is a research method that began as a way to understand genetics, but is now used in a variety of fields, including ecology, economics, and psychology.
SEM can be used to establish and explore hypotheses, although the former is more common. It is a particularly useful analytical method when testing the validity of a hypothesis, and there are several statistical indices available. For example, SEM is evaluated by the chi-square p-value (p > 0.05, not significant), the comparative fit index (cfi/CFI) (>0.95), the root mean square error of the approximation (rsmea/RSMEA) (<0.05), and the standardized mean residual (srmr/SRMR) (<0.08) [58]. When there are many candidate models, the lower the value of the Akaike information criterion (AIC), the better, and the model with the smaller difference between the goodness of fit index (gfi/GFI) and the adjusted goodness of fit index (agfi/AGFI) (>0.95) is selected. The path diagram of the superior model can be visualized.
Basically, the maximum likelihood method is often used, but Pearl proposed non-parametric SEM, which now exists as a command.
There are the following algorithms for causal inference. Statistical causal inference and machine learning are becoming interconnected trends, and here we describe a methodology that falls into the category of statistics. One is the causal mediation analysis (CMA) [59] as a mediation analysis, which allows a statistical evaluation of the direct or indirect causal relationship between three or more factors. There are two indices: the estimated average causal mediation effect (ACME) and the average direct effect (ADE). Next, the causal structure can be estimated assuming that the model is a linear non-Gaussian acyclic model (LiNGAM) [60] and independent factors. BayesLiNGAM [61] is also a Bayesian score-based approach that allows one to estimate the proportion of causal factors within a group that forms a causal structure. Furthermore, Granger causality [62], LiNGAM model with the classic vector autoregressive models (VARLiNGAM) [63], autoregressive integrated moving average (ARIMA) model [64], convergent cross mapping (CCM) [65], and Learning Interactions from Microbial Time Series (LIMITS) [66], microbial dynamical systems inference (MDSINE) [67], and EcohNet [68] have been proposed for time-series evaluation. EcohNet combines a type of recurrent neural network (RNN), called an echo state network (ESN) [68]. The advantages and disadvantages of each analysis have been discussed [38], [69], but the details are omitted here.
These analyses are only computational evaluations based on data. Therefore, a view of the accuracy of the data source is essential. It is important to understand that this may need to be proven by consistent characterization in other literature or by experimental science. Data calculated from measuring instruments and facilities as continuous data have a low rate of internal variation in each data itself. Similarly, for data such as omics analysis data, for which a series of analyses are conducted simultaneously using next-generation sequencers, NMR, GC/MS, LC/MS, etc., the rate of variation does not need to be taken into account as much as for individual analysis data. Therefore, the relationship between other factors within the group being measured can be measured as a single pattern with little variability. However, caution is required when the variability of the data itself needs to be considered for different measurement devices. In such a case, binarization can be a powerful tool that can absorb variability. In the case of different measuring instruments and different standards of data accuracy, the aforementioned binarization process as a preprocessing of data may lead to a suitable analysis in bird's-eye view, as the effect of data errors can be drastically reduced by this process. Viewed another way, the statistical causality can be viewed as a computational marker that combines the influence of environmental conditions, rather than necessarily as a definite causal relationship. If the data are used as a pattern to evaluate the characteristics of the target group from a bird's eye view, they can be used in various ways as a group of marker factors for various environmental factors.
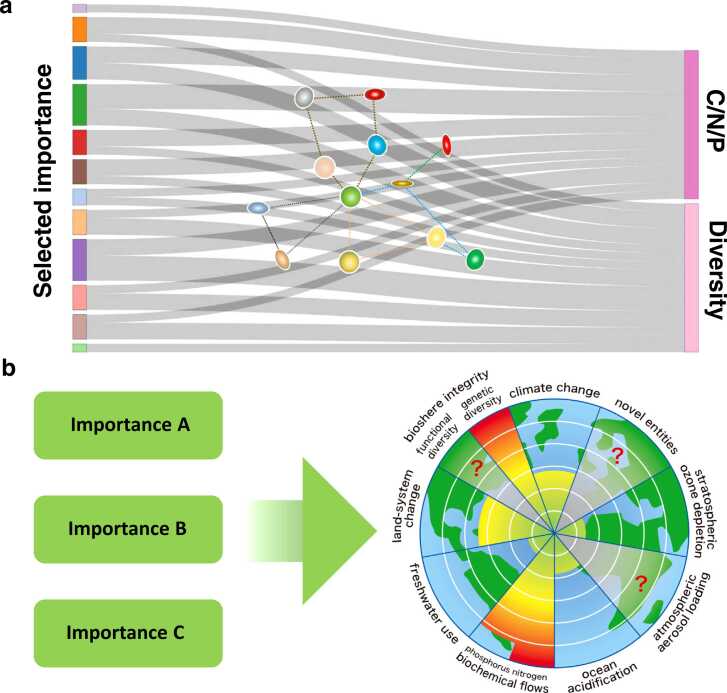
One method of expressing the cause-and-effect relationship of a group of factors by combining the viewpoint of visualizing the flow of material circulation further is the Sankey diagram [20]. It is possible to visualize what kind of outputs are ultimately affected by a group of factors selected based on different criteria; Fig. 5a shows an image visualizing the increase or decrease of carbon (C), nitrogen (N), and phosphorus (P), or the influence of an arbitrary group of factors on biodiversity. This is an effective method for expressing relationships to outputs that are essentially influential, rather than a complex cascade, based on the selection of factors whose relationships are clear through structural equations and causal inference.
Fig. 5.
Impact assessment of technologies that contribute to planetary boundaries. (a) Various conditions and [68] diagrams related to nitrogen, phosphorus, and biodiversity are shown. To make these evaluations, it is necessary to implement integrated understanding using computational science. (b) demonstrate the setting of goals for various technologies and planetary boundaries; Various methodologies are assumed for technologies aimed at environmental conservation and restoration. The current problem is that these distributed technologies are not understood in an integrated manner. In order to realistically implement these fusions, various simulations are also necessary.
In the following, we will discuss the potential of computational science in building a sustainable society by introducing the findings of other research groups as well as the analysis data that the authors have conducted so far, based on these perspectives.
3. Applications to various ecosystems
3.1. Aquatic ecosystem
Water is essential for the sustainability of life and is also involved in various industrial activities and the movement of ecosystems. We introduce examples of the application of machine learning to understanding such water cycles.
A study of Spanish rivers used random forest and regression tree algorithms to predict biological responses to different concentrations of nutrients in river water and to investigate the influence of different regular thresholds on ecological status [70]. It proposes an integrated approach for restoring ecosystem conditions. The Japanese study focuses on estuarine sediments and uses abundant sediment polypeptide and mineral profiles along with microbial structure to demonstrate significant estuarine eutrophication in the Kanto region [71]. In particular, such eutrophicated mad sediment can be evaluated by electronic potentials, such as redox homeostasis: thus, regulation of redox homeostasis can be one of the critical factors for environmental purification [72]. Another report describes a strategy for integrating and understanding the data obtained by omics analysis of surface water samples from 681 sites in Japan, grouped into three clusters, through machine learning, factor mapping, and predictive-error variance decomposition [73]. The results have revealed an overarching understanding of the characteristics of certain microalgae, inorganic ions, and organic acids in some patterns. Some examples of the importance of machine learning include efficient water management, remediation, and transport to conserve water resources [74].
Machine learning can effectively identify and characterize specific contaminants and has comprehensive applications in predicting water quality, mapping groundwater contaminants, classifying water resources, tracking contaminant sources, assessing contaminant toxicity in natural water systems, modeling treatment technologies, supporting characterization analysis, purifying and distributing drinking water, collecting and treating sewage water in engineered water systems; and collecting and treating sewage water in engineered water systems, among other comprehensive applications.
3.2. Forestry ecosystems
In conserving forest ecosystems, it is crucial to understand the symbiotic relationship with insects, which are deeply involved in recycling in the ecosystem. Insects are responsible for the forest's decomposition chain, and an assessment of the diversity and metabolite characteristics of the symbiotic bacterial community (gut symbionts) can lead to an understanding of the regularity of the forest ecosystem. Here, we present a case study on the carbon and nitrogen cycle for termites and beetle larvae.
Termites skillfully exploit the symbiotic relationship between the gut microbial community, certain protists and fungi, and the soil microbial community to utilize plant-derived carbohydrates. Plant-derived carbohydrates are degraded by the soil microbial community and ultimately returned to the soil. To understand these degradation processes, NMR analysis using stable isotope 13C has been promoted as a method that is less sensitive to analyte characteristics by analytical instruments, such as water solubility and fat solubility, and an integrated database has been successfully created [75]. This allows for integrated evaluation through machine learning and statistical inference.
As an example of analysis from a bird's-eye viewpoint, a case study evaluating humus chains in a forest ecosystem uses computational science with beetle larvae as a target [20]. In this case, after profiling symbiotic bacteria involved in the carbon and nitrogen cycles, we analyzed the relationship between the comprehensive bacterial flora and metabolites based on correlation analysis, association analysis, and structural equation modeling. As a result, we succeeded in predicting the bacterial groups involved in the behavior of carbon and nitrogen and their stable isotopes, respectively. Similarly to these single bacteria, the bacterial groups were consistent in the literature. Furthermore, based on these data, we successfully visualized them by means of a material balance flow model (Sankey diagram).
3.3. Fishery ecosystem
A better understanding of the ecosystems of fish and seagrass is crucial to the conservation of the aquatic environment and the fish industry. Examples of machine learning that focus on these topics will also be presented.
An effort has focused on the recent market development of nuclear magnetic resonance (NMR) systems, not only conventional high-end systems but also popular ones, and have reported the development of peak separation methods from these small NMR systems [76], and classification methods for fish meat quality characteristics dependent upon environmental conditions including symbiotic bacteria [77], [78], [79]. On the other hand, high-end systems can be used to more clearly identify differences in the composition of substances in muscle due to differences in production areas and feeds [77]. Therefore, by introducing advanced machine learning calculations such as deep learning and ensemble deep learning to NMR analysis, we can extract key factors (NMR peaks: metabolites) involved in the identification of the origin of wild fish or essential factors involved in fish growth and predictive model selection methods [80], [81], [82]. These methods can also contribute to important issues in the fisheries industry, such as aquaculture environmental management, including water temperature and oxygen concentration, which affect meat quality, and the identification of important ingredients in food for growth performance [41], [42], [83].
In general, seaweeds have been suggested to play a significant role in terms of blue carbon. Therefore, to understand the relationship between seaweeds characteristics and environmental factors, there is a rare case in which spectral data from various analytical instruments were integrated and used for seaweed classification, providing an example of analysis using various analytical tools at a different angle than previously considered [36]. The paper reveals a close relationship between environmental factors, namely heavy metal elements, and the characteristic components of algae [36]. Next, as an example of an analysis that assess seasonal variations, observations that utilize organic and inorganic chemical data on seaweed components and consider seasonal variations in seaweed composition have been reported [84]. To analyze different target materials with the same criteria, we used the least-squares method to eliminate noise, performed correlation network analysis, and successfully constructed a structural equation model. The concept of these studies is partly being applied to our ongoing bottom sediment characterization study [85] on seagrass growth to evaluate of the symbiotic environment for the seagrass meadow.
The need for machine learning and other network evaluations[4] is being touted in various studies that capture the entire water environment.
3.4. Agriculture ecosystem
A better understanding of soils, crops, and agriculture itself is critical for the conservation of soil and water environments. Case studies focusing on these topics will also be presented.
A case study developed a method to profile tomatoes using inductively coupled plasma atomic emission spectrometry and nuclear magnetic resonance spectrometry to find indicators to assess the effects of pesticides, organic fertilizers, and chemical fertilizers [86]. This profiling has revealed that the use of pesticide affects the correlation between mineral nutrients and metabolite correlations.
In another case study on leafy greens, organic nitrogen increased crop yield [16]. This study utilized multi-omics analysis data on soil metabolites, minerals, and microorganisms to successfully detect changes in plant traits as a collection of multiple network modules.
In addition, there are cases that have used structural equations to typify plant-soil-soil microbial relationships and recommended the evaluation of complex soil ecosystems [87]; evaluated the relationship between microbial networks, including filamentous fungi, and changes in soil properties (total N, C/N, and soil health index) [88]; and evaluated the relationship between soil heavy metals and soil microorganisms that assessed the relationship between soil heavy metals and soil microorganisms [89].
Thus, various attempts are underway to comprehensively evaluate microorganisms (fungi and bacteria), soil metabolites, and plant metabolites in the soil and to find their regularities. Although the observations of each of these studies are essential, what is needed in the future is the integration of these fragmented studies. One of the goals is to understand ecological homeostasis. Along these understandings, each of these case studies needs to be further reintegrated and used for various environmental protection purposes.
3.5. Livestock ecosystem
A better understanding of the livestock industry is also critical to the conservation of agricultural land after application of fecal compost, resulting in effects on crop quality and water environments. Case studies focusing on these issues will also be presented. In particular, the intestinal environment is important to maintain health, and the quality of manure compost is also extremely essential.
First, for broiler health care, we present a case study of a traditional intestinal regulator [37]. The effect of a heat-stable lactic acid bacterium, Weizmannia coagulans, on the intestinal metabolism of broilers was evaluated by correlation network analysis, and particularly the effect of the said lactic acid bacterium on the administration of the bacterium was shown to alter the diversity of metabolites. In a paper showing that the administration of probiotic lactic acid bacteria to sows can reduce the disturbance of the gut microbiota of piglets, the disturbance as a bacterial flora pattern were identified using DP matching technology [90]. Aerobic composting is also commonly used to dispose of livestock manure and is considered one of the most efficient ways to reduce antibiotic resistance genes (ARGs). The process has been estimated using SEM [91]. In addition, other examples are the use of probiotics [92] or antibiotics [93] to manage the intestinal microflora of cattle, which are a source of concern as a source of greenhouse gases. A management method has been suggested to reduce the number of methane-producing bacteria in the stage of calves that have not yet developed a rumen. The results of the correlation analysis of intestinal metabolites and bacterial flora at this stage clearly showed an inverse relationship between the suppression of methane-producing bacteria and growth, depending on the management method. It is hoped that computational evaluation of the calf stage based on such a database will lead to the development of environmentally friendly livestock production technology.
An example of evaluation in relation to disease status is the calculation of the relationship between mastitis in dairy cows, which provides useful information for genetic improvement and management strategies [94]. Another example is the evaluation of the relationship between genetic background and milk quality, meat quality, and other factors [95], [96], [97].
Like agriculture, these studies are highly specialized and segmented. Still, they represent the impact of livestock production on the environment, which is being debated worldwide and needs to be evaluated through an efficient fusion of information. In recent years, issues such as animal welfare have been recognized, but the impact of health and sanitation on the food environment via technology that improves the intestinal environment and the soil environment through appropriate composting has not been considered. A quantitative evaluation would be needed to enable a comprehensive discussion that is not one-sided before decisions could be made. For this purpose, it is necessary to work on an environmental form of estimation that transcends animal species.
3.6. Waste management ecosystems
Wastewater treatment and recycling technologies are essential to control eutrophication in rivers, lakes, and coastal environments and to efficiently implement resource recycling. Therefore, we will introduce some examples of efforts related to this paper and those that will be needed in the future.
For example, in a practical wastewater treatment process, 31P NMR methods were applied to model biofilm phosphorus release performance at different pH [98]. Moreover, 1H NMR and machine learning extreme gradient boosting (XGBoost) has also been used to understand the mechanism of membrane fouling (reduction in membrane permeability), which is a problem in membrane bioreactors (MBRs), an essential part of wastewater treatment operations [99]. Changes in the nature of membrane fouling material as a function of conditions have been investigated using NMR data [100].
Thus, there are examples of analysis based on the need for evaluation from the perspective of wastewater treatment, but there are few examples of the use of machine learning and other methods from the perspective of recycling. It is necessary to discuss the qualitative role of recycled fertilizers [35], [101], [102] and feeds [103], [104], [105], [106], [107], [108], [109], [110], [111], [112] in a recycling-oriented society. Although still in the preparatory stage [113], [114], the environmental impact and its effects on plants and animals are also being investigated through machine learning, structural equation modeling, and causal inference. The results of these studies should provide an essential perspective on the significance of material cycles to the ecosystem as a whole, beyond animal or plant species, and the need for efficient human activities in the context of the ecosystem as a whole.
4. Summary and outlook
The fourth industrial revolution that is underway in this century will enable the introduction of digitalization, future forecasting, and feedback control in various fields through the advancement of information technology, especially the IoT (Internet of Things), big data, and AI (artificial intelligence) related technologies. AI-related technologies are also advancing, so too are the technologies that are being used to predict and control the future. However, while AI-related technologies are advancing, it is challenging to introduce omics methods that produce multi-factorial data in the so-called "predictive science" area because the analysis cost to obtain the training data increases when the number of samples is large. Although data accumulation is necessary, it is important to adjust these databases in a form that is easy to use in various fields. In this review, we introduced the flow and examples of data integration and management to avoid these problems. By utilizing machine learning, structural equation modeling, and causal inference in the right places, it may be possible to select and categorize data that are important for ecosystem conservation while shifting to less costly analytical data (Fig. 5). For example, as shown here, it will lead to an understanding of how the selected factors affect the material flow of carbon, nitrogen, and phosphorus, or biodiversity, in terms of their contribution rate (Fig. 5a). Then, it should be important to select factors based on their importance and evaluate their contribution to the planetary boundary in order to assess technological development (Fig. 5b). In the future, large- and small-scale computational devices can be significantly innovated by advancing quantum computing technologies. Therefore, the prediction of multifactored phenomena, such as ecosystem changes accompanied by climate changes, can be well-computed by such innovative computation devices. Then, it will be necessary to integrate research data from many fields and utilize computational science to build a sustainable society.
CRediT authorship contribution statement
Hirokuni Miyamoto and Jun Kikuchi: Writing-original draft, Visualization, review & editing. Jun Kikuchi: Supervision, Conceptualization, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors wish to thank to Ms. Izumi Jouzuka for supporting with the figure design.
References
- 1.Tilman D., Fargione J., Wolff B., D'Antonio C., Dobson A., Howarth R., et al. Forecasting agriculturally driven global environmental change. Science. 2001;292(5515):281–284. doi: 10.1126/science.1057544. [DOI] [PubMed] [Google Scholar]
- 2.Tilman D., Cassman K.G., Matson P.A., Naylor R., Polasky S. Agricultural sustainability and intensive production practices. Nature. 2002;418:671–677. doi: 10.1038/nature01014. [DOI] [PubMed] [Google Scholar]
- 3.RockstrÖm J., Steffen W., Noone K., Persson A., Chapin F.S., Lambin E.F., et al. A safe operating space for humanity. Nature. 2009;461:472–475. doi: 10.1038/461472a. [DOI] [PubMed] [Google Scholar]
- 4.Worden A.Z., Follows M.J., Giovannoni S.J., Wilken S., Zimmerman A.E., Keeling P.J. Environmental science. Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science. 2015;347(6223):1257594. doi: 10.1126/science.1257594. [DOI] [PubMed] [Google Scholar]
- 5.Gruber N., Galloway J.N. An Earth-system perspective of the global nitrogen cycle. Nature. 2008;451(7176):293–296. doi: 10.1038/nature06592. [DOI] [PubMed] [Google Scholar]
- 6.Oreska M.P.J., McGlathery K.J., Aoki L.R., Berger A.C., Berg P., Mullins L. The greenhouse gas offset potential from seagrass restoration. Sci Rep. 2020;10(1):7325. doi: 10.1038/s41598-020-64094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.IPCC . IPCC,; 2014. Climate Change 2014:Synthesis report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. [Google Scholar]
- 8.Barbieri P., Pellerin S., Seufert V., Smith L., Ramankutty N., Nesme T. Global option space for organic agriculture is delimited by nitrogen availability. Nature Food. 2021;2:363–372. doi: 10.1038/s43016-021-00276-y. [DOI] [PubMed] [Google Scholar]
- 9.Costello C., Cao L., Gelcich S., Cisneros-Mata M.A., Free C.M., Froehlich H.E., et al. The future of food from the sea. Nature. 2020;588(7836):95–100. doi: 10.1038/s41586-020-2616-y. [DOI] [PubMed] [Google Scholar]
- 10.Errickson F., Kuruc K., McFadden J. Animal-based foods have high social and climate costs. Nature Food. 2021;2:274–281. doi: 10.1038/s43016-021-00265-1. [DOI] [PubMed] [Google Scholar]
- 11.Gephart J.A., Henriksson P.J.G., Parker R.W.R., Shepon A., Gorospe K.D., Bergman K., et al. Environmental performance of blue foods. Nature. 2021;597(7876):360–365. doi: 10.1038/s41586-021-03889-2. [DOI] [PubMed] [Google Scholar]
- 12.Gouda S., Kerry R.G., Das G., Paramithiotis S., Shin H.S., Patra J.K. Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiol Res. 2018;206:131–140. doi: 10.1016/j.micres.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Pashaei R., Zahedipour-Sheshglani P., Dzingeleviciene R., Abbasi S., Rees R.M. Effects of pharmaceuticals on the nitrogen cycle in water and soil: a review. Environ Monit Assess. 2022;194(2):105. doi: 10.1007/s10661-022-09754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohr W., Lehnen N., Ahmerkamp S., Marchant H.K., Graf J.S., Tschitschko B., et al. Terrestrial-type nitrogen-fixing symbiosis between seagrass and a marine bacterium. Nature. 2021;600(7887):105–109. doi: 10.1038/s41586-021-04063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W.L., Moore J.K., Martiny A.C., Primeau F.W. Convergent estimates of marine nitrogen fixation. Nature. 2019;566(7743):205–211. doi: 10.1038/s41586-019-0911-2. [DOI] [PubMed] [Google Scholar]
- 16.Ichihashi Y., Date Y., Shino A., Shimizu T., Shibata A., Kumaishi K., et al. Multi-omics analysis on an agroecosystem reveals the significant role of organic nitrogen to increase agricultural crop yield. Proc Natl Acad Sci USA. 2020;117(25):14552–14560. doi: 10.1073/pnas.1917259117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su F., Takaya N., Shoun H. Nitrous oxide-forming codenitrification catalyzed by cytochrome P450nor. Biosci Biotechnol Biochem. 2004;68:473–475. doi: 10.1271/bbb.68.473. [DOI] [PubMed] [Google Scholar]
- 18.Shoun H., Fushinobu S., Jiang L., Kim S.W., Wakagi T. Fungal denitrification and nitric oxide reductase cytochrome P450nor. Philos Trans R Soc Lond B Biol Sci. 2012;367(1593):1186–1194. doi: 10.1098/rstb.2011.0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen H., Shiratori Y., Ohta S., Masuda Y., Isobe K., Senoo K. Mitigating N2O emissions from agricultural soils with fungivorous mites. ISME J. 2021;15(8):2427–2439. doi: 10.1038/s41396-021-00948-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyamoto H., Futo Asano F., Ishizawa K., Suda W., Miyamoto H., Tsuji N., et al. A potential network structure of symbiotic bacteria involved in carbon and nitrogen metabolism of wood-utilizing insect larvae. Sci Total Environ. 2022;836 doi: 10.1016/j.scitotenv.2022.155520. [DOI] [PubMed] [Google Scholar]
- 21.Harindintwali J.D., Zhou J., Muhoza B., Wang F., Herzberger A., Yu X. Integrated eco-strategies towards sustainable carbon and nitrogen cycling in agriculture. J Environ Manag. 2021;293 doi: 10.1016/j.jenvman.2021.112856. [DOI] [PubMed] [Google Scholar]
- 22.Wang S., Zeng Y. Ammonia emission mitigation in food waste composting: a review. Bioresour Technol. 2018;248(Pt A):13–19. doi: 10.1016/j.biortech.2017.07.050. [DOI] [PubMed] [Google Scholar]
- 23.Osada T., Nekomoto K., Shiraishi M., Hara M., Hoshiba S., Suzuki K., et al. The concentration of ammonia, methane and nitrous oxide in the barn. J Japan Association on Odor. J Jpn Assoc Odor Environ. 2004;35 [Google Scholar]
- 24.Shen J., Yuan L., Zhang J., Li H., Bai Z., Chen X., et al. Phosphorus dynamics: from soil to plant. Plant Physiol. 2011;156(3):997–1005. doi: 10.1104/pp.111.175232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumari N., Bansal S. Production and characterization of a novel, thermotolerant fungal phytase from agro-industrial byproducts for cattle feed. Biotechnol Lett. 2021;43(4):865–879. doi: 10.1007/s10529-020-03069-8. [DOI] [PubMed] [Google Scholar]
- 26.Humer E., Schwarz C., Schedle K. Phytate in pig and poultry nutrition. J Anim Physiol Anim Nutr. 2015;99(4):605–625. doi: 10.1111/jpn.12258. [DOI] [PubMed] [Google Scholar]
- 27.Zangarini S., Pepe Sciarria T., Tambone F., Adani F. Phosphorus removal from livestock effluents: recent technologies and new perspectives on low-cost strategies. Environ Sci Pollut Res Int. 2020;27(6):5730–5743. doi: 10.1007/s11356-019-07542-4. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y., Ciais P., Goll D.S., Sardans J., Penuelas J., Cresto-Aleina F., et al. The shift of phosphorus transfers in global fisheries and aquaculture. Nat Commun. 2020;11(1):355. doi: 10.1038/s41467-019-14242-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q., Liao Z., Yao D., Yang Z., Wu Y., Tang C. Phosphorus immobilization in water and sediment using iron-based materials: a review. Sci Total Environ. 2021;767 doi: 10.1016/j.scitotenv.2020.144246. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X., Tong C., Taylor W.D., Rudstam L.G., Jeppesen E., Bolotov I., et al. Does differential phosphorus processing by plankton influence the ecological state of shallow lakes? Sci Total Environ. 2021;769 doi: 10.1016/j.scitotenv.2020.144357. [DOI] [PubMed] [Google Scholar]
- 31.Christensen M.L., Keiding K., Nielsen P.H., Jorgensen M.K. Dewatering in biological wastewater treatment: a review. Water Res. 2015;82:14–24. doi: 10.1016/j.watres.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 32.Wan J., Wang X., Yang T., Wei Z., Banerjee S., Friman V.P., et al. Livestock manure type affects microbial community composition and assembly during composting. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.621126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penuelas J., Sardans J. The global nitrogen-phosphorus imbalance. Science. 2022;375(6578):266–267. doi: 10.1126/science.abl4827. [DOI] [PubMed] [Google Scholar]
- 34.Igai K., Itakura M., Nishijima S., Tsurumaru H., Suda W., Tsutaya T., et al. Nitrogen fixation and nifH diversity in human gut microbiota. Sci Rep. 2016;6 doi: 10.1038/srep31942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishikawa K., Ohmori T., Miyamoto H., Ito T., Kumagai Y., Sonoda M., et al. Denitrification in soil amended with thermophile-fermented compost suppresses nitrate accumulation in plants. Appl Microbiol Biotechnol. 2013;97(3):1349–1359. doi: 10.1007/s00253-012-4004-5. [DOI] [PubMed] [Google Scholar]
- 36.Wei F., Ito K., Sakata K., Date Y., Kikuchi J. Pretreatment and integrated analysis of spectral data reveal seaweed similarities based on chemical diversity. Anal Chem. 2015;87(5):2819–2826. doi: 10.1021/ac504211n. [DOI] [PubMed] [Google Scholar]
- 37.Ito K., Miyamoto H., Matsuura M., Ishii C., Tsuboi A., Tsuji N., et al. Noninvasive fecal metabolic profiling for the evaluation of characteristics of thermostable lactic acid bacteria, Weizmannia coagulans SANK70258, for broiler chickens. J Biosci Bioeng. 2022;134(2):105–115. doi: 10.1016/j.jbiosc.2022.05.006. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki K., Abe M.S., Kumakura D., Nakaoka S., Fujiwara F., Miyamoto H., et al. Chemical-mediated microbial interactions can reduce the effectiveness of time-series-based inference of ecological interaction networks. Int J Environ Res Public Health. 2022;19:3. doi: 10.3390/ijerph19031228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wall D.H., Nielsen U.N., Six J. Soil biodiversity and human health. Nature. 2015;528(7580):69–76. doi: 10.1038/nature15744. [DOI] [PubMed] [Google Scholar]
- 40.Kikuchi J., Yamada S. The exposome paradigm to predict environmental health in terms of systemic homeostasis and resource balance based on NMR data science. RSC Adv. 2021;11(48):30426–30447. doi: 10.1039/d1ra03008f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kikuchi J., Yamada S. NMR window of molecular complexity showing homeostasis in superorganisms. Analyst. 2017;142(22):4161–4172. doi: 10.1039/c7an01019b. [DOI] [PubMed] [Google Scholar]
- 42.Kikuchi J. Springer and Humana Press,; USA: 2019. Practical Aspects of the Analysis of Low- and High-field NMR Data from Environmental Complexities. [DOI] [PubMed] [Google Scholar]
- 43.Wang X., Gorlitsky R., Almeida J.S. From XML to RDF: how semantic web technologies will change the design of 'omic' standards. Nat Biotechnol. 2005;23(9):1099–1103. doi: 10.1038/nbt1139. [DOI] [PubMed] [Google Scholar]
- 44.Lyttleton O., Wright A., Treanor D., Lewis P. Using XML to encode TMA DES metadata. J Pathol Inform. 2011;2:40. doi: 10.4103/2153-3539.84233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prud'hommeaux E., Collins J., Booth D., Peterson K.J., Solbrig H.R., Jiang G. Development of a FHIR RDF data transformation and validation framework and its evaluation. J Biomed Inform. 2021;117 doi: 10.1016/j.jbi.2021.103755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki K., Nakaoka S., Fukuda S., Masuya H. Energy landscape analysis elucidates the multistability of ecological communities. Ecol Monogr. 2021;91 [Google Scholar]
- 47.Bayes T.L.I.I. An essay towards solving a problem in the doctrine of chances. By the late Rev. Mr. Bayes, F. R. S. communicated by Mr. Price, in a letter to John Canton, A. M. F. R. S. Philos Trans R Soc Lond. 1763;53:370–418. [Google Scholar]
- 48.Baum L.E., Petrie T. Statistical inference for probabilistic functions of finite state Markov chains. Ann Math Stat. 1966;37(6):1554–1563. [Google Scholar]
- 49.Bellman R. The theory of dynamic programming. Bull Am Math Soc. 1954;60:503–515. [Google Scholar]
- 50.Sakoe H., Chiba S. Dynamic programming algorithm optimization for spoken word recognition. IEEE Trans Acous Speech, Signal Process. 1978:43–49. (ASSP-26) [Google Scholar]
- 51.Furst M.A., Caines P.E. Edge detection with image enhancement via dynamic programming. Comput Vision, Graph Image Process. 1986;33(3):263–279. [Google Scholar]
- 52.Fisher R.A. The use of Multiple Measurements in taxonomic problems Annals of Eugenics;7(2):179–188.
- 53.David M.B., Andrew Y.N., Michael I.J. Latent Dirichlet allocation. J Mach Learn Res. 2003;3:993–1022. [Google Scholar]
- 54.Ioannis T., Thorsten J., Thomas H., Yasemin A. Large margin methods for structured and interdependent output variables. J Mach Learn Res. 2005;6:1453–1484. [Google Scholar]
- 55.Wright S. On the nature of size factors. Genetics. 1918;3(4):367–374. doi: 10.1093/genetics/3.4.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wright S. The relative importance of heredity and environment in determining the piebald pattern of guinea-pigs. Proc Natl Acad Sci USA. 1920;6(6):320–332. doi: 10.1073/pnas.6.6.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wright S. Correlation and causation. J Agric Res. 1921;20(7):557–585. [Google Scholar]
- 58.Hooper D., Coughlan J., Mullen M.R. Structural equation modelling: guidelines for determining model fit. Electron J Business Res Methods. 2008;6:53–60. [Google Scholar]
- 59.Tingley D., Yamamoto T., Hirose K., Keele L., Imai K. mediation: R package for causal mediation analysis. J Stat Softw. 2014;59(5):1–38. [Google Scholar]
- 60.Shimizu S., Hoyer P.O., Hyvarinen A., Kerminen A. A linear non-Gaussian acyclic model for causal discovery. J Mach Learn Res. 2006:2003–2030. [Google Scholar]
- 61.Hoyer P.O., Hyttinen A. Bayesian discovery of linear acyclic causal models. arXivorg. 2009 https://doi.org/https://arxiv.org/abs/1205.2641.arXiv:1205.2641. [Google Scholar]
- 62.Stokes P.A., Purdon P.L. A study of problems encountered in Granger causality analysis from a neuroscience perspective. Proc Natl Acad Sci USA. 2017;114(34):E7063–E7072. doi: 10.1073/pnas.1704663114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hyvarinen A., Zhang K., Shimizu S., Hoyer P.O. Estimation of a structural vector autoregression model using non-gaussianity. J Mach Learning Res. 2010;11:1709–1731. [Google Scholar]
- 64.Ives A.R., Dennis B., Cottingham K.L., Carpenter S.R. Estimating community stability and ecological interactions from time‐series data. Ecol Monogr. 2003;73(2):301–330. [Google Scholar]
- 65.Sugihara G., May R., Ye H., Hsieh C.H., Deyle E., Fogarty M., et al. Detecting causality in complex ecosystems. Science. 2012;338(6106):496–500. doi: 10.1126/science.1227079. [DOI] [PubMed] [Google Scholar]
- 66.Fisher C.K., Mehta P. Identifying keystone species in the human gut microbiome from metagenomic timeseries using sparse linear regression. PLOS One. 2014;9(7) doi: 10.1371/journal.pone.0102451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bucci V., Tzen B., Li N., Simmons M., Tanoue T., Bogart E., et al. MDSINE: microbial dynamical systems inference engine for microbiome time-series analyses. Genome Biol. 2016;17(1):121. doi: 10.1186/s13059-016-0980-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki K., Matsuzaki S.S., Masuya H. Decomposing predictability to identify dominant causal drivers in complex ecosystems. Proc Natl Acad Sci USA. 2022;119(42) doi: 10.1073/pnas.2204405119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ridenhour B.J., Brooker S.L., Williams J.E., Van Leuven J.T., Miller A.W., Dearing M.D., et al. Modeling time-series data from microbial communities. ISME J. 2017;11(11):2526–2537. doi: 10.1038/ismej.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Valerio C., De Stefano L., Martinez-Munoz G., Garrido A. A machine learning model to assess the ecosystem response to water policy measures in the Tagus River Basin (Spain) Sci Total Environ. 2021;750 doi: 10.1016/j.scitotenv.2020.141252. [DOI] [PubMed] [Google Scholar]
- 71.Asakura T., Date Y., Kikuchi J. Comparative analysis of chemical and microbial profiles in estuarine sediments sampled from Kanto and Tohoku regions in Japan. Anal Chem. 2014;86(11):5425–5432. doi: 10.1021/ac5005037. [DOI] [PubMed] [Google Scholar]
- 72.Shono N., Ito M., Umezawa A., Sakata K., Li A., Kikuchi J., et al. Tracing and regulating redox homeostasis of model benthic ecosystems for sustainable aquaculture in coastal environments. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.907703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oita A., Tsuboi Y., Date Y., Oshima T., Sakata K., Yokoyama A., et al. Profiling physicochemical and planktonic features from discretely/continuously sampled surface water. Sci Total Environ. 2018;636:12–19. doi: 10.1016/j.scitotenv.2018.04.156. [DOI] [PubMed] [Google Scholar]
- 74.Huang R., Ma C., Ma J., Huangfu X., He Q. Machine learning in natural and engineered water systems. Water Res. 2021;205 doi: 10.1016/j.watres.2021.117666. [DOI] [PubMed] [Google Scholar]
- 75.Tokuda G., Tsuboi Y., Kihara K., Saitou S., Moriya S., Lo N., et al. Metabolomic profiling of C-13-labelled cellulose digestion in a lower termite: insights into gut symbiont function. Proc R Soc B-Biol Sci. 2014;281:1789. doi: 10.1098/rspb.2014.0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hara K., Yamada S., Chikayama E., Kikuchi J. Parameter visualization of benchtop nuclear magnetic resonance spectra toward food process monitoring. Processes. 2022;10(7):1264. [Google Scholar]
- 77.Mekuchi M., Asakura T., Kikuchi J. Springer; 2019. New Aquaculture Technology Based on Host-Symbiotic Co-metabolism. [Google Scholar]
- 78.Shima H., Sato Y., Sakata K., Asakura T., Kikuchi J. Identifying a correlation among qualitative non-numeric parameters in natural fish microbe dataset using machine learning. Appl Sci. 2022;12:12. [Google Scholar]
- 79.Shima H., Murata I., Wei F., Sakata K., Yokoyama D., Kikuchi J. Identification of salmoniformes aquaculture conditions to increase the creatine and anserine levels using multiomics dataset and nonnumerical information. Front Microbiol in press. [DOI] [PMC free article] [PubMed]
- 80.Date Y., Kikuchi J. Application of a deep neural network to metabolomics studies and its performance in determining important variables. Anal Chem. 2018;90(3):1805–1810. doi: 10.1021/acs.analchem.7b03795. [DOI] [PubMed] [Google Scholar]
- 81.Asakura T., Date Y., Kikuchi J. Application of ensemble deep neural network to metabolomics studies. Anal Chim Acta. 2018;1037:230–236. doi: 10.1016/j.aca.2018.02.045. [DOI] [PubMed] [Google Scholar]
- 82.Wei F., Ito K., Sakata K., Asakura T., Date Y., Kikuchi J. Fish ecotyping based on machine learning and inferred network analysis of chemical and physical properties. Sci Rep. 2021;11(1):3766. doi: 10.1038/s41598-021-83194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kikuchi J., Ito K., Date Y. Environmental metabolomics with data science for investigating ecosystem homeostasis. Prog Nucl Magn Reson Spectrosc. 2018;104:56–88. doi: 10.1016/j.pnmrs.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 84.Ito K., Sakata K., Date Y., Kikuchi J. Integrated analysis of seaweed components during seasonal fluctuation by data mining across heterogeneous chemical measurements with network visualization. Anal Chem. 2014;86(2):1098–1105. doi: 10.1021/ac402869b. [DOI] [PubMed] [Google Scholar]
- 85.Miyamoto H., Kawachi N., Kurotani A., Moriya S., Suda W., Suzuki K., et al. Computational estimation of sediment symbiotic bacterial structures of seagrasses overgrowing downstream of onshore aquaculture. Environ Res. 2022 doi: 10.1016/j.envres.2022.115130.115130. [DOI] [PubMed] [Google Scholar]
- 86.Watanabe M., Ohta Y., Sun L.C., Motoyama N., Kikuchi J. Profiling contents of water-soluble metabolites and mineral nutrients to evaluate the effects of pesticides and organic and chemical fertilizers on tomato fruit quality. Food Chem. 2015;169:387–395. doi: 10.1016/j.foodchem.2014.07.155. [DOI] [PubMed] [Google Scholar]
- 87.Mamet S.D., Redlick E., Brabant M., Lamb E.G., Helgason B.L., Stanley K., et al. Structural equation modeling of a winnowed soil microbiome identifies how invasive plants re-structure microbial networks. ISME J. 2019;13(8):1988–1996. doi: 10.1038/s41396-019-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xue R., Wang C., Zhao L., Sun B., Wang B. Agricultural intensification weakens the soil health index and stability of microbial networks. Agric Ecosyst Environ. 2022:339. [Google Scholar]
- 89.Tian Z., Li G., Tang W., Zhu Q., Li X., Du C., et al. Role of Sedum alfredii and soil microbes in the remediation of ultra-high content heavy metals contaminated soil. Agric Ecosyst Environ. 2022:339. [Google Scholar]
- 90.Mori K., Ito T., Miyamoto H., Ozawa M., Wada S., Kumagai Y., et al. Oral administration of multispecies microbial supplements to sows influences the composition of gut microbiota and fecal organic acids in their post-weaned piglets. J Biosci Bioeng. 2011;112(2):145–150. doi: 10.1016/j.jbiosc.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 91.Qiu X., Zhou G., Chen L., Wang H. Additive quality influences the reservoir of antibiotic resistance genes during chicken manure composting. Ecotoxicol Environ Saf. 2021;220 doi: 10.1016/j.ecoenv.2021.112413. [DOI] [PubMed] [Google Scholar]
- 92.Inabu Y., Taguchi Y., Miyamoto H., Etoh T., Shiotsuka Y., Fujino R., et al. Development of a novel feeding method for Japanese black calves with thermophile probiotics at postweaning. J Appl Microbiol. 2022;132(5):3870–3882. doi: 10.1111/jam.15519. [DOI] [PubMed] [Google Scholar]
- 93.Okada S., Inabu Y., Miyamoto H., Suzuki K., Kato T., Kurotani A., et al. Antibiotic-dependent instability of homeostatic plasticity for growth and environmental load. arXiv 2022.preprinted at arxiv.org/abs/2211.14718.
- 94.Pegolo S., Momen M., Morota G., Rosa G.J.M., Gianola D., Bittante G., et al. Structural equation modeling for investigating multi-trait genetic architecture of udder health in dairy cattle. Sci Rep. 2020;10(1):7751. doi: 10.1038/s41598-020-64575-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Inoue K., Valente B.D., Shoji N., Honda T., Oyama K., Rosa G.J.M. Inferring phenotypic causal structures among meat quality traits and the application of a structural equation model in Japanese Black cattle. J Anim Sci. 2016;94:4133–4142. doi: 10.2527/jas.2016-0554. [DOI] [PubMed] [Google Scholar]
- 96.Pegolo S., Yu H., Morota G., Bisutti V., Rosa G.J.M., Bittante G., et al. Structural equation modeling for unraveling the multivariate genomic architecture of milk proteins in dairy cattle. J Dairy Sci. 2021;104(5):5705–5718. doi: 10.3168/jds.2020-18321. [DOI] [PubMed] [Google Scholar]
- 97.Okamura T., Ishii K., Nishio M., Rosa G.J.M., Satoh M., Sasaki O. Inferring phenotypic causal structure among farrowing and weaning traits in pigs. Anim Sci J. 2020;91(1) doi: 10.1111/asj.13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shi S., Xu G. Identification of phosphorus fractions of biofilm sludge and phosphorus release, transformation and modeling in biofilm sludge treatment related to pH. Chem Eng J. 2019;369:694–704. [Google Scholar]
- 99.Yokoyama D., Suzuki S., Asakura T., Kikuchi J. Chemometric analysis of NMR spectra and machine learning to investigate membrane fouling. ACS Omega. 2022;7(15):12654–12660. doi: 10.1021/acsomega.1c06891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yokoyama D., Suzuki S., Asakura T., Kikuchi J. Microbiome and metabolome analyses in different closed-circulation aquarium systems and their network visualization. ACS Omega. 2022;7(34):30399–30404. doi: 10.1021/acsomega.2c03701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.De Corato U. Agricultural waste recycling in horticultural intensive farming systems by on-farm composting and compost-based tea application improves soil quality and plant health: a review under the perspective of a circular economy. Sci Total Environ. 2020;738 doi: 10.1016/j.scitotenv.2020.139840. [DOI] [PubMed] [Google Scholar]
- 102.Zhao S., Schmidt S., Gao H., Li T., Chen X., Hou Y., et al. A precision compost strategy aligning composts and application methods with target crops and growth environments can increase global food production. Nature Food. 2022;3(9):741–752. doi: 10.1038/s43016-022-00584-x. [DOI] [PubMed] [Google Scholar]
- 103.Miyamoto H., Seta M., Horiuchi S., Iwasawa Y., Naito T., Nishida A., et al. Potential probiotic thermophiles isolated from mice after compost ingestion. J Appl Microbiol. 2013;114(4):1147–1157. doi: 10.1111/jam.12131. [DOI] [PubMed] [Google Scholar]
- 104.Miyamoto H., Shimada E., Satoh T., Tanaka R., Oshima K., Suda W., et al. Thermophile-fermented compost as a possible scavenging feed additive to prevent peroxidation. J Biosci Bioeng. 2013;116(2):203–208. doi: 10.1016/j.jbiosc.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 105.Miyamoto H., Kodama H., Udagawa M., Mori K., Matsumoto J., Oosaki H., et al. The oral administration of thermophile-fermented compost extract and its influence on stillbirths and growth rate of pre-weaning piglets. Res Vet Sci. 2012;93(1):137–142. doi: 10.1016/j.rvsc.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 106.Niisawa C., Oka S., Kodama H., Hirai M., Kumagai Y., Mori K., et al. Microbial analysis of a composted product of marine animal resources and isolation of bacteria antagonistic to a plant pathogen from the compost. J Gen Appl Microbiol. 2008;54(3):149–158. doi: 10.2323/jgam.54.149. [DOI] [PubMed] [Google Scholar]
- 107.Tanaka R., Miyamoto H., Kodama H., Kawachi N., Udagawa M., Miyamoto H., et al. Feed additives with thermophile-fermented compost enhance concentrations of free amino acids in the muscle of the flatfish Paralichthys olivaceus. J Gen Appl Microbiol. 2010;56(1):61–65. doi: 10.2323/jgam.56.61. [DOI] [PubMed] [Google Scholar]
- 108.Tanaka R., Miyamoto H., Inoue S., Shigeta K., Kondo M., Ito T., et al. Thermophile-fermented compost as a fish feed additive modulates lipid peroxidation and free amino acid contents in the muscle of the carp, Cyprinus carpio. J Biosci Bioeng. 2016;121(5):530–535. doi: 10.1016/j.jbiosc.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 109.Satoh T., Nishiuchi T., Naito T., Matsushita T., Kodama H., Miyamoto H., et al. Impact of oral administration of compost extract on gene expression in the rat gastrointestinal tract. J Biosci Bioeng. 2012;114(5):500–505. doi: 10.1016/j.jbiosc.2012.05.026. [DOI] [PubMed] [Google Scholar]
- 110.Ito T., Miyamoto H., Kumagai Y., Udagawa M., Shinmyo T., Mori K., et al. Thermophile-fermented compost extract as a possible feed additive to enhance fecundity in the laying hen and pig: modulation of gut metabolism. J Biosci Bioeng. 2016;121(6):659–664. doi: 10.1016/j.jbiosc.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 111.Miyamoto H., Ohno H. Fundamental grasp of the symbiotic system of intestinal mucosa and microorganisms, and its application to the development of environmental protective livestock technology. J Farm Anim Infect Dis. 2022;11:1–12. [Google Scholar]
- 112.Asano F., Tsuboi A., Moriya S., Kato T., Tsuji N., Nakaguma T., et al. Amendment of a thermophile-fermented compost to humus improves the growth of female larvae of the Hercules beetle Dynastes hercules (Coleoptera: scarabaeidae. J Appl Microbiol. 2022 doi: 10.1093/jambio/lxac006. [DOI] [PubMed] [Google Scholar]
- 113.Miyamoto H., et.al. A novel sustainable role of compost as a universal protective substitute for fish, chicken, pig., and cattle, and its estimation by structure equation modeling. arXiv 2022. https://doi.org/10.48550/arXiv.2201.10895.preprinted at arxiv.org/abs/2201.10895.
- 114.Miyamoto H., et.al. Agricultural quality matrix-based multiomics structural analysis of carrots in soils fertilized with thermophile-fermented compost. arXiv 2022. https://doi.org/10.48550/arXiv.2202.03132.preprinted at arxiv.org/abs/2202.03132.