Abstract
To enhance the understanding of molecular mechanisms and mine previously unidentified biomarkers of pediatric atopic dermatitis, PBMC gene expression profiles were generated by RNA sequencing in infants with atopic dermatitis and age-matched controls. A total of 178 significantly differentially expressed genes (DEGs) (115 upregulations and 63 downregulations) were seen, compared with those in healthy controls. The DEGs identified included IL1β, TNF, TREM1, IL18R1, and IL18RAP. DEGs were validated by real-time RT- qPCR in a larger number of samples from PBMCs of infants with atopic dermatitis aged <12 months. Using the DAVID (Database for Annotation, Visualization and Integrated Discovery) database, functional and pathway enrichment analyses of DEGs were performed. Gene ontology enrichment analysis showed that DEGs were associated with immune responses, inflammatory responses, regulation of immune responses, and platelet activation. Pathway analysis indicated that DEGs were enriched in cytokine‒cytokine receptor interaction, immunoregulatory interactions between lymphoid and nonlymphoid cells, hematopoietic cell lineage, phosphoinositide 3-kinase‒protein kinase B signaling pathway, NK cell‒mediated cytotoxicity, and platelet activation. Furthermore, the protein‒protein interaction network was predicted using the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database and visualized with Cytoscape software. Finally, on the basis of the protein‒protein interaction network, 18 hub genes were selected, and two significant modules were obtained. In conclusion, this study sheds light on the molecular mechanisms of pediatric atopic dermatitis and may provide diagnostic biomarkers and therapeutic targets.
Abbreviations: AD, atopic dermatitis; BP, biological process; CC, cellular component; DEG, differentially expressed gene; GO, gene ontology; MF, molecular function; PIP, protein‒protein interaction; RNA-seq, RNA sequencing
Introduction
Atopic dermatitis (AD) is the most common chronic inflammatory skin disease in early childhood. It affects children with a prevalence of up to 20% and adults with prevalence rates of 7‒10% (Weidinger et al., 2018). AD is a complex multifactorial disease, thought to result from an interplay between environmental factors, an impaired skin barrier, and immune dysfunctions; however, the overall pathologic mechanisms are still not fully understood (Langan et al., 2020). Although much has been learned about the molecular basis of AD, most investigations have focused on adult AD with years of disease activity that is remarkably different from those of early-onset AD in children. Few studies have profiled skin tissue in infants with AD (Brunner et al., 2019, 2018; Cole et al., 2014; Esaki et al., 2016). Over the last decade, RNA sequencing (RNA-seq)-based transcriptome profiles have been implemented in identifying transcripts and pathways in many diseases; however, limited studies using this method were performed on skin tissues in AD, particular in children with AD (Brunner et al., 2019; Cole et al., 2014). Only one study conducted has compared transcriptome profiles of both blood and skin tissue in children with AD at various ages up to age 5 years (Brunner et al., 2019).
Given that AD is an early childhood disease that generates a systemic immunological response and that 60% of all cases of AD begin during the first year of life (Bieber, 2008), we aimed to discover signature biomarkers of AD in infants that might help to identify new diagnostic biomarkers and molecular targets for treatment modalities in pediatric AD.
For this purpose, we performed an integrative study comprising RNA-seq transcriptome profile of peripheral blood cells obtained from infants with AD or healthy infants, quantitative RT-PCR, and systems biology analysis.
Results
Analysis of gene expression by RNA-seq
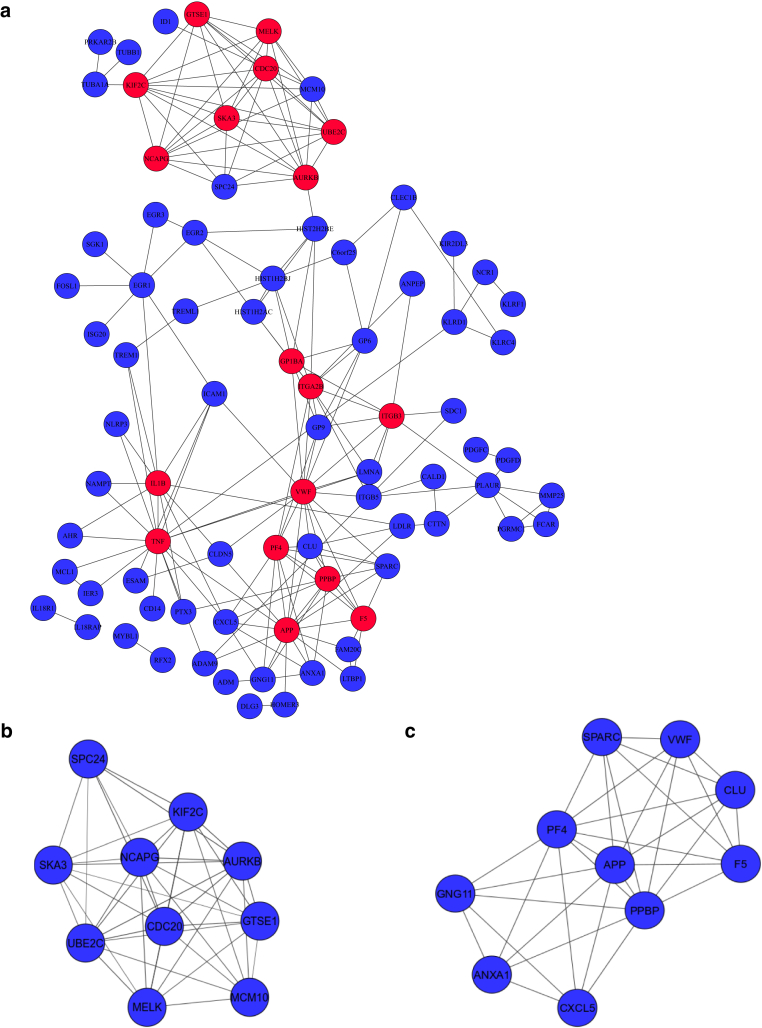
A total of 100 infants with moderate or severe AD in the first year of life and 20 age-matched healthy control infants were initially recruited (McAleer et al., 2019). PBMCs were isolated from 42 patients and 19 controls. RNA samples were extracted, and only 27 samples from patients with AD and 17 controls passed quality control and were used in this study. The use of samples is presented in a schematic flow chart (Figure 1). We performed RNA-seq profiles on PBMCs from randomly selected infants with AD (n = 8) and controls (n = 5) using the Illumina platform. Differential expression analysis was conducted to identify differentially expressed genes (DEGs) between AD and controls on the basis of the following criteria: false discovery rate <0.05 and fold change ≥1.5. We identified a total of 178 significantly DEGs with 115 upregulations and 63 downregulations in AD PBMCs when compared with those in control PBMCs. Among highly upregulated genes, we identified IL1β, previously shown to be upregulated in the serum of adult patients with AD (Thijs et al., 2018); TNF, a proinflammatory cytokine whose role in the pathogenesis of AD is well-known (Jacobi et al., 2005; Sumimoto et al., 1992); and early growth response genes EGR2 and EGR3, known to have a crucial role in the regulation of the immune system (Li et al., 2012). Other upregulated genes included TREM1, previously shown to be elevated in lesional skin and serum in patients with AD (Suarez-Farinas et al., 2015), and CXCL5, an inflammatory chemokine found to be at elevated levels in the blood of patients with AD (Brunner et al., 2017). Among downregulated genes, we identified IL18R1 and IL18RAP found to be associated with AD (Hirota et al., 2012). We summarized the 10 DEGs randomly selected from our study and compared their expression with published data (Table 1). A complete list of DEGs in blood cells is shown in Table 2.
Figure 1.
The overall framework of study design. AD, atopic dermatitis; QC, quality control.
Table 1.
A List of 10 Differentially Expressed Genes Randomly Selected from Our Study and Their Changes Identified in Older Children and Adults with AD on the Basis of the Literature
| DEG | Children Aged 0‒1 y |
Tissue, Down/Up | Older Children | Tissue, Down/Up | Adults | Tissue, Down/Up |
|---|---|---|---|---|---|---|
| IL18RAP | This study | PBMCs; Down |
Brunner et al. (2019) 4 mo, 5 y |
AD LS versus HC skin; Up | Ewald et al. (2015) and Hirota et al. (2012) | Susceptibility loci for AD; AD LS versus NS, Up |
| IL18R1 | This study | PBMCs; Down |
NA | Hirota et al. (2012) | Susceptibility loci for AD | |
| IL1β | This study | PBMCs; Up |
Cole et al. (2014) 6‒16 years |
AD NS versus HC skin; Down |
McAleer et al. (2019) and Thijs et al. (2018) | AD versus HC Serum, Up; AD versus HC SC, Down |
| TNF | This study | PBMCs; Up |
Brunner et al. (2019) Sumimoto et al. (1992) 1‒15 y |
AD LS versus HC skin, Up; AD NS versus HC skin, Up; AD versus HC Plasma, Up |
Suarez-Farinas et al. (2015) and Thijs et al. (2018) | AD LS versus NS, Up AD versus HC Serum, Up |
| TREM1 | This study | PBMCs; Up |
NA | Suarez-Farinas et al. (2015) and Thijs et al. (2018) | AD versus HC Serum, Up AD LS versus NS, Up |
|
| EGR3 | This study | PBMCs; Up |
NA | Suarez-Farinas et al. (2011) | AD LS versus HC skin, Down; AD NS versus HC skin, Down | |
| EGR2 | This study | PBMCs; Up |
Brunner et al. (2019) 4 mo, 5 y |
AD LS versus HC skin, Down; AD NS versus HC skin, Down |
Hirota et al. (2012) | Susceptibility loci for AD |
| EGR1 | This study | PBMCs; Up |
Brunner et al. (2019) 4 mo, 5 y Cole et al. (2014) 6‒16 y |
AD LS versus HC skin, Down; AD NS versus HC skin, Down; AD NS versus HC skin; Up |
Suarez-Farinas et al. (2011) | AD LS versus HC skin, Up; AD NS versus HC skin, Up |
| NLRP3 | This study | PBMCs; Up |
Brunner et al. (2019) | AD LS versus HC skin, Down; AD NS versus HC skin, Down |
Niebuhr et al. (2014) | AD LS versus HC skin, Down |
| FOSL1 | This study | PBMCs; Up |
Brunner et al. (2019) 6‒16 y Cole et al. (2014) 6‒16 y |
AD LS versus HC skin, Up AD NS versus HC skin; Up |
Ewald et al. (2015) and Suarez-Farinas et al. (2011) | AD LS versus HC skin, Up; AD LS versus AD NL skin, Up; AD LS versus NS, Up |
Abbreviations: AD, atopic dermatitis; DEG, differentially expressed gene; Down, downregulated; HC, healthy control; LS, lesional skin; NA, nonavailable; NS, nonlesional skin; SC, stratum corneum; Up, upregulated.
Table 2.
Differentially Expressed Genes in PBMCs from Infants with AD
| Gene | Log2FC | FC | P-Value | FDR |
|---|---|---|---|---|
| GLDC | ‒2.265 | 0.208 | 3.34E−11 | 5.29E−07 |
| HRASLS2 | ‒2.086 | 0.236 | 2.18E−05 | 0.00576 |
| SDC1 | ‒1.809 | 0.285 | 0.00044 | 0.03940 |
| CDC20 | ‒1.749 | 0.297 | 6.14E−05 | 0.01085 |
| IL18RAP | ‒1.702 | 0.307 | 1.56E−07 | 0.00031 |
| INTU | ‒1.664 | 0.316 | 2.70E−05 | 0.00658 |
| IGKV3-11 | ‒1.655 | 0.317 | 0.00031 | 0.03247 |
| KIR2DL3 | ‒1.524 | 0.348 | 6.36E−05 | 0.01095 |
| AL645929.1 | ‒1.497 | 0.354 | 4.41E−05 | 0.00906 |
| IGKV5-2 | ‒1.487 | 0.357 | 1.46E−07 | 0.00031 |
| PLEKHG7 | ‒1.472 | 0.360 | 1.29E−05 | 0.00474 |
| BHLHA15 | ‒1.437 | 0.369 | 0.00021 | 0.02593 |
| IGHV3-21 | ‒1.412 | 0.376 | 0.00038 | 0.03721 |
| GTSE1 | ‒1.402 | 0.378 | 1.17E−05 | 0.00465 |
| AC007278.2 | ‒1.389 | 0.382 | 0.00032 | 0.03341 |
| FASLG | ‒1.367 | 0.388 | 7.97E−05 | 0.01202 |
| MCM10 | ‒1.357 | 0.390 | 4.54E−05 | 0.00923 |
| NCAPG | ‒1.350 | 0.392 | 1.58E−05 | 0.00534 |
| IGJ | ‒1.349 | 0.393 | 0.00015 | 0.02018 |
| SPC24 | ‒1.328 | 0.398 | 2.10E−06 | 0.00210 |
| UBE2C | ‒1.326 | 0.399 | 0.00038 | 0.03721 |
| AL365475.1 | ‒1.321 | 0.400 | 7.85E−05 | 0.01196 |
| CCDC150 | ‒1.276 | 0.413 | 0.00016 | 0.02125 |
| KLRD1 | ‒1.257 | 0.418 | 7.68E−05 | 0.01192 |
| MELK | ‒1.255 | 0.419 | 0.00010 | 0.01551 |
| SKA3 | ‒1.252 | 0.420 | 0.00013 | 0.01832 |
| AC007278.1 | ‒1.226 | 0.428 | 0.00063 | 0.04921 |
| IGLV1-44 | ‒1.225 | 0.428 | 3.86E−05 | 0.00849 |
| NCR1 | ‒1.207 | 0.433 | 0.00056 | 0.04636 |
| IGLV3-19 | ‒1.192 | 0.438 | 2.91E−05 | 0.00687 |
| KLRC4 | ‒1.176 | 0.443 | 0.00020 | 0.02444 |
| RNF165 | ‒1.134 | 0.456 | 0.00046 | 0.04029 |
| IGLV8-61 | ‒1.111 | 0.463 | 7.35E−05 | 0.01176 |
| MYBL1 | ‒1.104 | 0.465 | 2.63E−08 | 0.00011 |
| AC006480.2 | ‒1.093 | 0.469 | 0.00013 | 0.01832 |
| AURKB | ‒1.073 | 0.475 | 0.00026 | 0.02917 |
| SLCO4A1 | ‒1.057 | 0.481 | 7.15E−05 | 0.01167 |
| IGLV2-8 | ‒1.057 | 0.481 | 0.00022 | 0.02608 |
| IGLV2-23 | ‒1.037 | 0.487 | 2.85E−05 | 0.00684 |
| IL18R1 | ‒1.030 | 0.490 | 5.41E−08 | 0.00014 |
| IGLC2 | ‒1.022 | 0.493 | 0.00019 | 0.02387 |
| AL683813.1 | ‒1.020 | 0.493 | 0.00058 | 0.04670 |
| AC010536.1 | ‒1.013 | 0.496 | 0.00056 | 0.04625 |
| STRIP2 | ‒1.002 | 0.499 | 0.00030 | 0.03156 |
| KLRF1 | ‒0.993 | 0.502 | 0.00023 | 0.02686 |
| XYLB | ‒0.971 | 0.510 | 0.00039 | 0.03721 |
| SH2D2A | ‒0.964 | 0.513 | 0.00018 | 0.02284 |
| IGKV1-5 | ‒0.958 | 0.515 | 1.51E−05 | 0.00519 |
| IGKV4-1 | ‒0.958 | 0.515 | 0.00011 | 0.01580 |
| IGLV2-11 | ‒0.947 | 0.519 | 4.07E−05 | 0.00859 |
| PIWIL2 | ‒0.931 | 0.525 | 0.00042 | 0.03910 |
| SLFN13 | ‒0.890 | 0.540 | 8.03E−06 | 0.00340 |
| IGKV1-12 | ‒0.870 | 0.547 | 0.00055 | 0.04604 |
| PDGFD | ‒0.834 | 0.561 | 0.00018 | 0.02275 |
| ABCB9 | ‒0.821 | 0.566 | 0.00043 | 0.03940 |
| ISG20 | ‒0.796 | 0.576 | 0.00025 | 0.02887 |
| KIF2C | ‒0.782 | 0.582 | 0.00037 | 0.03676 |
| IGKV3-15 | ‒0.777 | 0.584 | 0.00057 | 0.04636 |
| TTC22 | ‒0.762 | 0.590 | 0.00044 | 0.03940 |
| C5orf56 | ‒0.705 | 0.613 | 2.15E−05 | 0.00576 |
| DLG3 | ‒0.696 | 0.617 | 0.00054 | 0.04512 |
| COLQ | ‒0.609 | 0.656 | 3.98E−05 | 0.00859 |
| SPATS2 | ‒0.608 | 0.656 | 5.20E−05 | 0.00992 |
| APP | 0.598 | 1.514 | 4.61E−08 | 0.00014 |
| MCL1 | 0.661 | 1.581 | 4.80E−06 | 0.00256 |
| MAML3 | 0.661 | 1.581 | 1.85E−05 | 0.00576 |
| KCNQ1 | 0.732 | 1.661 | 0.00040 | 0.03721 |
| RFX2 | 0.754 | 1.687 | 7.45E−05 | 0.01180 |
| PGRMC1 | 0.779 | 1.716 | 6.33E−05 | 0.01095 |
| PRXL2C | 0.786 | 1.724 | 0.00061 | 0.04852 |
| FRMD4B | 0.842 | 1.793 | 0.00036 | 0.03648 |
| ANXA1 | 0.869 | 1.827 | 5.64E−05 | 0.01051 |
| ESAM | 0.912 | 1.881 | 0.00049 | 0.04315 |
| HIST1H2AC | 0.916 | 1.887 | 0.00045 | 0.04004 |
| RAPH1 | 0.923 | 1.896 | 0.00028 | 0.03031 |
| ADAM9 | 0.940 | 1.918 | 0.00027 | 0.03031 |
| TUBA1A | 1.021 | 2.029 | 0.00060 | 0.04766 |
| LDLR | 1.068 | 2.097 | 1.29E−05 | 0.00474 |
| CPNE2 | 1.098 | 2.140 | 3.93E−06 | 0.00244 |
| ZNF185 | 1.183 | 2.270 | 0.00055 | 0.04604 |
| MYADM | 1.187 | 2.278 | 6.10E−05 | 0.01085 |
| F5 | 1.301 | 2.464 | 2.45E−06 | 0.00210 |
| AHR | 1.343 | 2.536 | 0.00058 | 0.04670 |
| LMNA | 1.382 | 2.606 | 7.10E−05 | 0.01167 |
| HIST2H2BE | 1.467 | 2.765 | 4.13E−06 | 0.00244 |
| GNG11 | 1.491 | 2.810 | 0.00023 | 0.02700 |
| HIST1H2BJ | 1.560 | 2.948 | 0.00033 | 0.03358 |
| FAM129B | 1.570 | 2.968 | 0.00034 | 0.03455 |
| SH3BGRL2 | 1.617 | 3.067 | 0.00028 | 0.03031 |
| PADI4 | 1.625 | 3.085 | 0.00040 | 0.03721 |
| LTBP1 | 1.635 | 3.106 | 0.00057 | 0.04636 |
| ICAM1 | 1.645 | 3.128 | 0.00057 | 0.04636 |
| FAM20C | 1.662 | 3.165 | 0.00049 | 0.04315 |
| C2orf88 | 1.675 | 3.194 | 2.45E−05 | 0.00625 |
| NAMPT | 1.701 | 3.252 | 0.00029 | 0.03156 |
| CD14 | 1.703 | 3.256 | 0.00042 | 0.03898 |
| SCN1B | 1.728 | 3.312 | 5.16E−05 | 0.00992 |
| NRGN | 1.740 | 3.340 | 1.77E−05 | 0.00560 |
| LGALS12 | 1.748 | 3.359 | 0.00011 | 0.01596 |
| GGTA1P | 1.751 | 3.367 | 0.00026 | 0.02917 |
| PPP1R15A | 1.762 | 3.392 | 0.00038 | 0.03721 |
| SEPT5 | 1.765 | 3.399 | 3.99E−06 | 0.00244 |
| TAL1 | 1.767 | 3.402 | 2.23E−06 | 0.00210 |
| PDGFC | 1.769 | 3.408 | 6.82E−05 | 0.01149 |
| PRKAR2B | 1.781 | 3.436 | 1.96E−06 | 0.00210 |
| RAB20 | 1.807 | 3.499 | 0.00039 | 0.03721 |
| PLAUR | 1.810 | 3.505 | 0.00051 | 0.04372 |
| CTTN | 1.813 | 3.513 | 1.97E−05 | 0.00576 |
| GP6 | 1.813 | 3.515 | 1.72E−-05 | 0.00557 |
| GAS2L1 | 1.834 | 3.565 | 0.00016 | 0.02125 |
| CLDN5 | 1.864 | 3.641 | 0.00018 | 0.02321 |
| PEAR1 | 1.867 | 3.649 | 1.71E−05 | 0.00557 |
| KLF4 | 1.871 | 3.657 | 0.00039 | 0.03721 |
| SOWAHC | 1.872 | 3.661 | 0.00051 | 0.04372 |
| TSPAN9 | 1.879 | 3.679 | 4.20E−06 | 0.00244 |
| MMP25 | 1.903 | 3.740 | 7.48E−06 | 0.00329 |
| LGALSL | 1.942 | 3.843 | 4.86E−06 | 0.00256 |
| ANPEP | 1.943 | 3.846 | 7.25E−05 | 0.01171 |
| SPARC | 1.954 | 3.875 | 0.00028 | 0.03031 |
| FAXDC2 | 1.966 | 3.907 | 2.52E−06 | 0.00210 |
| ATP2B1-AS1 | 1.969 | 3.914 | 0.00014 | 0.01999 |
| ITGA2B | 1.979 | 3.942 | 0.00015 | 0.02018 |
| NLRP3 | 1.995 | 3.987 | 4.40E−05 | 0.00906 |
| ITGB3 | 1.999 | 3.998 | 0.00016 | 0.02125 |
| ITGB5 | 2.001 | 4.004 | 5.57E−05 | 0.01051 |
| BEND2 | 2.012 | 4.034 | 0.00051 | 0.04372 |
| TRIB1 | 2.015 | 4.042 | 0.00030 | 0.03156 |
| PTX3 | 2.039 | 4.109 | 0.00024 | 0.02744 |
| VWF | 2.054 | 4.152 | 1.32E−05 | 0.00474 |
| DUSP6 | 2.066 | 4.186 | 0.00014 | 0.01973 |
| ELOVL7 | 2.109 | 4.314 | 2.10E−05 | 0.00576 |
| ALOX12 | 2.134 | 4.389 | 2.96E−05 | 0.00690 |
| HOMER3 | 2.140 | 4.406 | 3.76E−05 | 0.00839 |
| ADM | 2.142 | 4.414 | 0.00045 | 0.04004 |
| PDE5A | 2.151 | 4.442 | 0.00022 | 0.02608 |
| TMEM40 | 2.166 | 4.489 | 0.00039 | 0.03721 |
| MPIG6B | 2.178 | 4.526 | 1.16E−06 | 0.00147 |
| ENKUR | 2.178 | 4.527 | 0.00062 | 0.04886 |
| LRP3 | 2.194 | 4.575 | 5.93E−05 | 0.01085 |
| AC245128.3 | 2.195 | 4.579 | 2.51E−05 | 0.00630 |
| TUBB1 | 2.212 | 4.633 | 1.31E−05 | 0.00474 |
| TREML1 | 2.258 | 4.785 | 3.55E−06 | 0.00244 |
| CAVIN2 | 2.277 | 4.847 | 5.78E−06 | 0.00269 |
| IER3 | 2.290 | 4.891 | 6.17E−05 | 0.01085 |
| WLS | 2.302 | 4.930 | 2.16E−05 | 0.00576 |
| NRIP3 | 2.330 | 5.030 | 0.00021 | 0.02593 |
| EMP1 | 2.340 | 5.062 | 1.21E−06 | 0.00147 |
| GP1BA | 2.342 | 5.069 | 5.15E−06 | 0.00256 |
| PF4 | 2.349 | 5.095 | 2.92E−06 | 0.00231 |
| TNF | 2.352 | 5.107 | 2.47E−06 | 0.00210 |
| PPBP | 2.379 | 5.204 | 9.63E−07 | 0.00139 |
| SGK1 | 2.443 | 5.439 | 5.32E−06 | 0.00256 |
| CLU | 2.464 | 5.517 | 0.00052 | 0.04420 |
| TREM1 | 2.464 | 5.519 | 9.14E−07 | 0.00139 |
| FCAR | 2.472 | 5.549 | 1.91E−05 | 0.00576 |
| LUCAT1 | 2.560 | 5.898 | 0.00050 | 0.04362 |
| GP9 | 2.577 | 5.968 | 4.27E−06 | 0.00244 |
| CMTM5 | 2.580 | 5.981 | 2.02E−05 | 0.00576 |
| AL391903.1 | 2.604 | 6.079 | 4.05E−05 | 0.00859 |
| ABLIM3 | 2.620 | 6.146 | 2.44E−05 | 0.00625 |
| CALD1 | 2.629 | 6.185 | 2.17E−05 | 0.00576 |
| B3GNT5 | 2.681 | 6.415 | 6.12E−06 | 0.00277 |
| AC007032.1 | 2.695 | 6.474 | 0.00062 | 0.04876 |
| SPOCD1 | 2.713 | 6.556 | 5.34E−06 | 0.00256 |
| ZNF503 | 2.843 | 7.177 | 8.15E−06 | 0.00340 |
| CXCL5 | 2.848 | 7.202 | 0.00016 | 0.02125 |
| SEC14L5 | 2.856 | 7.241 | 3.18E−05 | 0.00730 |
| AQP10 | 2.879 | 7.356 | 0.00044 | 0.03940 |
| PDZK1IP1 | 3.014 | 8.080 | 0.00043 | 0.03921 |
| IL1B | 3.127 | 8.736 | 7.60E−05 | 0.01192 |
| SPX | 3.184 | 9.090 | 3.38E−06 | 0.00244 |
| HRAT92 | 3.192 | 9.140 | 1.82E−09 | 1.44E-05 |
| CLEC1B | 3.356 | 10.238 | 8.70E−07 | 0.00139 |
| ID1 | 3.458 | 10.986 | 3.66E−05 | 0.00827 |
| EGR3 | 3.496 | 11.279 | 0.00030 | 0.03156 |
| EGR1 | 3.517 | 11.447 | 0.00033 | 0.03344 |
| FOSL1 | 3.557 | 11.767 | 1.48E−05 | 0.00519 |
| EGR2 | 3.763 | 13.572 | 0.00024 | 0.02800 |
Shown is a list of genes differentially expressed in the blood of children with AD children versus in age-matched healthy control, meeting criteria of FC ≥1.5 and FDR <0.05.
Abbreviations: AD, atopic dermatitis; FC, fold change; FDR, false discovery rate; MMP, matrix metalloproteinase; VWF, Von Willebrand factor.
Validation of RNA-seq data by RT-qPCR
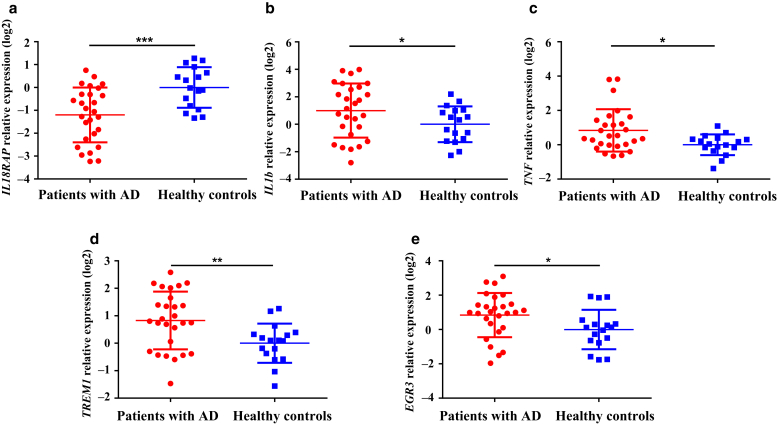
To confirm the results of RNA-seq, real-time RT-qPCR was performed to detect the mRNA expression of five randomly selected DEGs in PBMCs from controls (n = 17) and infants with AD (n = 27). As shown in Figure 2a-e, the mRNA levels of all five genes—IL18RAP, IL1β, TNF, TREM1, and EGR3—had significant differences between the AD and control groups in accordance with RNA-seq results.
Figure 2.
Validation of RNA-sequencing data by RT-qPCR. RT-qPCR analyses for five genes from the top 10 differentially expressed genes identified by high-throughput RNA sequencing: (a) IL18RAP, (b) IL1β, (c) TNF, (d) TREM1, and (e) EGR3 in children with AD (n = 27) and healthy controls (n = 17). Fold change was calculated by 2-ΔΔCT method. The normalized expression data were log2 transformed and shown as the means ± SD. Significant difference among groups was calculated by unpaired t test with Welch’s correction for normal distribution or with Mann‒Whitney rank-sum test for non-normal distribution data. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001
Effect of infant’s age on gene expression in the blood of infants with AD
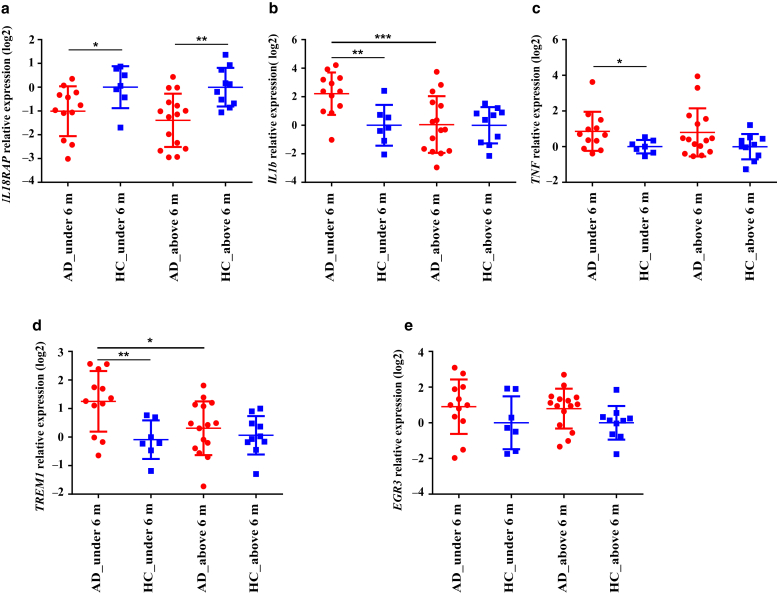
We wondered whether the infant’s age has an effect on gene expression in the blood of patients with AD in the first year of life. Previous studies have shown that differences in skin microbiome depend on infant’s age in healthy infants (Capone et al., 2011) and AD infants (Nakamura et al., 2020). To elucidate the effect of age, we stratified patients with AD and healthy controls accordingly (0‒6 months and 7‒12 months) and performed differential expression analysis on RNA-seq data. Table 3 summarizes the DEGs between four groups: patients with AD aged >6 months (n = 5) versus age-matched healthy controls (n = 2), patients with AD aged <6 months (n = 3) versus age-matched healthy controls (n = 3), patients with AD aged >6 months versus patients with AD aged <6 months, and healthy controls aged >6 months versus healthy controls aged <6 months. Interestingly, four DEGs that have been identified between AD infants and healthy controls and validated by RT-qPCR in this study (IL1B, TNF, TREM1, and EGR3) were differentially expressed in patients with AD aged <6 months compared with those in age-matched healthy controls, suggesting its unique differential expression in the first 6 months of life in patients with AD. IL18RAP has been shown to be differentially expressed in both age groups between patients with AD and healthy controls, suggesting no effect of age stratification on this DEG in patients with AD (Table 4). Data were further validated by RT-qPCR (Figure 3a-e). Four DEGs (IL1B, TNF, TREM1, and IL18RAP) showed significant differential expression affected by age in accordance with RNA-seq results. Expression of EGR3 showed a trend to be affected by age; however, it was not significant as shown by RNA-seq analysis. Altogether, these data indicate that identified DEGs in patients with AD in the first year of life could be affected by age; however, more samples are required to approve this effect.
Table 3.
Effect of Infant’s Age on Gene Expression in Infants with AD and HCs
| Gene | Log2FC | FC | P-Value | FDR |
|---|---|---|---|---|
| AD aged >6 mo (n = 5) versus HCs (n = 2) | ||||
| MTCO3P12 | ‒8.823 | 0.002 | 5.674E-09 | 9.209E-05 |
| IGKV1-16 | ‒3.584 | 0.083 | 4.496E-06 | 6.483E-03 |
| GLDC | ‒2.244 | 0.211 | 1.331E-06 | 3.086E-03 |
| IL18RAP | ‒1.968 | 0.256 | 1.241E-06 | 3.086E-03 |
| MIF-AS1 | ‒1.916 | 0.265 | 1.954E-05 | 1.885E-02 |
| WASHC1 | ‒1.739 | 0.300 | 1.974E-05 | 1.885E-02 |
| SNHG22 | ‒1.500 | 0.354 | 7.604E-05 | 4.255E-02 |
| ENSG00000273295 | ‒1.294 | 0.408 | 5.821E-07 | 2.362E-03 |
| ENSG00000260404 | ‒1.045 | 0.485 | 4.011E-06 | 6.483E-03 |
| KLRF1 | ‒1.011 | 0.496 | 2.685E-05 | 2.294E-02 |
| ITGB1 | 0.883 | 1.844 | 8.051E-05 | 4.355E-02 |
| H2AC6 | 1.538 | 2.904 | 9.485E-05 | 4.966E-02 |
| PRKAR2B | 2.157 | 4.460 | 5.766E-05 | 3.743E-02 |
| GNG11 | 2.278 | 4.849 | 3.485E-05 | 2.507E-02 |
| PEAR1 | 2.424 | 5.366 | 6.523E-05 | 4.072E-02 |
| MPIG6B | 2.499 | 5.651 | 1.643E-05 | 1.885E-02 |
| PF4 | 2.520 | 5.738 | 7.597E-05 | 4.255E-02 |
| FAXDC2 | 2.540 | 5.816 | 5.347E-05 | 3.616E-02 |
| CAVIN2 | 2.585 | 6.000 | 8.349E-06 | 1.042E-02 |
| PPBP | 2.589 | 6.018 | 3.512E-06 | 6.483E-03 |
| ITGB3 | 2.595 | 6.043 | 3.553E-05 | 2.507E-02 |
| TUBB1 | 2.612 | 6.112 | 3.955E-06 | 6.483E-03 |
| TREML1 | 2.710 | 6.542 | 2.979E-05 | 2.417E-02 |
| FPR1 | 3.011 | 8.060 | 2.393E-05 | 2.158E-02 |
| CLEC4F | 3.292 | 9.798 | 1.938E-07 | 1.048E-03 |
| ABLIM3 | 3.372 | 10.355 | 3.510E-05 | 2.507E-02 |
| FPR2 | 3.438 | 10.836 | 7.061E-05 | 4.244E-02 |
| LIN7A | 4.010 | 16.113 | 4.794E-06 | 6.483E-03 |
| SEC14L5 | 4.348 | 20.367 | 8.718E-07 | 2.830E-03 |
| DDX11L10 | 6.851 | 115.408 | 1.897E-05 | 1.885E-02 |
| IGHV5-10-1 | 9.633 | 793.981 | 2.177E-08 | 1.767E-04 |
| AD aged <6 mo (n = 3) versus HCs (n = 3) | ||||
| ENSG00000271993 | ‒6.451 | 0.011 | 1.429E-04 | 6.675E-03 |
| LOC107987373 | ‒6.102 | 0.015 | 9.330E-04 | 2.534E-02 |
| ENSG00000254851 | ‒4.136 | 0.057 | 7.154E-04 | 2.125E-02 |
| ENSG00000266302 | ‒4.021 | 0.062 | 3.886E-04 | 1.392E-02 |
| GPR82 | ‒3.736 | 0.075 | 3.149E-04 | 1.211E-02 |
| ENSG00000244167 | ‒3.326 | 0.100 | 1.761E-05 | 1.365E-03 |
| GAPDHP1 | ‒3.207 | 0.108 | 7.216E-04 | 2.133E-02 |
| CHL1 | ‒3.046 | 0.121 | 7.483E-04 | 2.173E-02 |
| SDC1 | ‒2.950 | 0.129 | 5.232E-04 | 1.712E-02 |
| RAVER2 | ‒2.943 | 0.130 | 4.456E-05 | 2.817E-03 |
| FCGR3B | ‒2.920 | 0.132 | 6.584E-05 | 3.805E-03 |
| SLC4A10 | ‒2.904 | 0.134 | 2.548E-06 | 2.678E-04 |
| ZBED2 | ‒2.888 | 0.135 | 2.211E-03 | 4.666E-02 |
| SSPN | ‒2.633 | 0.161 | 4.028E-04 | 1.426E-02 |
| LOC730101 | ‒2.581 | 0.167 | 2.129E-03 | 4.537E-02 |
| GLDC | ‒2.464 | 0.181 | 2.327E-05 | 1.709E-03 |
| IGLV2-18 | ‒2.420 | 0.187 | 3.579E-05 | 2.360E-03 |
| IGHV3-13 | ‒2.394 | 0.190 | 5.888E-04 | 1.865E-02 |
| IGHV3-21 | ‒2.390 | 0.191 | 1.176E-12 | 7.064E-10 |
| CHAC2 | ‒2.389 | 0.191 | 2.231E-05 | 1.660E-03 |
| IGKV3D-20 | ‒2.308 | 0.202 | 3.317E-04 | 1.251E-02 |
| TNFRSF17 | ‒2.299 | 0.203 | 1.029E-06 | 1.254E-04 |
| CDC20 | ‒2.240 | 0.212 | 4.534E-09 | 1.230E-06 |
| IGHV3-15 | ‒2.202 | 0.217 | 3.022E-08 | 5.775E-06 |
| MIR3142HG | ‒2.184 | 0.220 | 8.995E-05 | 4.727E-03 |
| LINC01355 | ‒2.181 | 0.220 | 3.300E-04 | 1.249E-02 |
| ENSG00000230521 | ‒2.172 | 0.222 | 1.578E-04 | 7.192E-03 |
| BHLHE41 | ‒2.121 | 0.230 | 1.844E-03 | 4.116E-02 |
| BHLHA15 | ‒2.112 | 0.231 | 8.746E-04 | 2.427E-02 |
| PPP1R17 | ‒2.103 | 0.233 | 2.441E-04 | 1.004E-02 |
| OR2A9P | ‒2.084 | 0.236 | 2.116E-03 | 4.522E-02 |
| ENSG00000275481 | ‒2.066 | 0.239 | 1.494E-04 | 6.923E-03 |
| PARS2 | ‒2.045 | 0.242 | 2.013E-03 | 4.369E-02 |
| EOMES | ‒2.011 | 0.248 | 1.357E-04 | 6.399E-03 |
| IGHGP | ‒1.983 | 0.253 | 8.716E-04 | 2.426E-02 |
| JCHAIN | ‒1.982 | 0.253 | 1.711E-06 | 1.918E-04 |
| IGHV3-72 | ‒1.981 | 0.253 | 7.772E-05 | 4.244E-03 |
| CISH | ‒1.930 | 0.262 | 7.131E-08 | 1.187E-05 |
| CD180 | ‒1.920 | 0.264 | 1.204E-03 | 3.039E-02 |
| MCM10 | ‒1.919 | 0.264 | 6.847E-05 | 3.890E-03 |
| LINC02273 | ‒1.918 | 0.265 | 1.005E-04 | 5.058E-03 |
| KLRC4 | ‒1.911 | 0.266 | 8.811E-08 | 1.398E-05 |
| IGKV1-6 | ‒1.891 | 0.270 | 7.272E-04 | 2.138E-02 |
| TRGV8 | ‒1.860 | 0.276 | 1.861E-03 | 4.134E-02 |
| IGLV3-19 | ‒1.857 | 0.276 | 8.468E-08 | 1.356E-05 |
| SLC23A3 | ‒1.846 | 0.278 | 1.713E-03 | 3.930E-02 |
| C1orf220 | ‒1.844 | 0.279 | 8.631E-04 | 2.407E-02 |
| IGKV1-17 | ‒1.841 | 0.279 | 9.117E-07 | 1.136E-04 |
| IGHG1 | ‒1.835 | 0.280 | 9.853E-04 | 2.625E-02 |
| ENSG00000258810 | ‒1.826 | 0.282 | 1.423E-03 | 3.409E-02 |
| TRGV10 | ‒1.785 | 0.290 | 1.548E-03 | 3.615E-02 |
| IGHV3-11 | ‒1.780 | 0.291 | 2.487E-04 | 1.016E-02 |
| FASLG | ‒1.779 | 0.291 | 1.466E-05 | 1.174E-03 |
| NCAPG | ‒1.772 | 0.293 | 2.036E-04 | 8.800E-03 |
| LINC01560 | ‒1.768 | 0.294 | 6.509E-04 | 1.979E-02 |
| ZNF781 | ‒1.755 | 0.296 | 4.745E-04 | 1.622E-02 |
| IGHV1-18 | ‒1.754 | 0.297 | 3.218E-05 | 2.164E-03 |
| IGHJ3 | ‒1.740 | 0.299 | 5.973E-04 | 1.886E-02 |
| TRAV4 | ‒1.727 | 0.302 | 1.912E-03 | 4.220E-02 |
| IGKV1D-8 | ‒1.722 | 0.303 | 1.633E-04 | 7.322E-03 |
| TRGC2 | ‒1.708 | 0.306 | 7.025E-09 | 1.773E-06 |
| CUTALP | ‒1.700 | 0.308 | 2.636E-04 | 1.060E-02 |
| IGLV1-44 | ‒1.699 | 0.308 | 8.700E-10 | 2.703E-07 |
| AKAP6 | ‒1.688 | 0.310 | 9.866E-04 | 2.625E-02 |
| IGLV1-51 | ‒1.669 | 0.314 | 8.440E-09 | 1.999E-06 |
| ZNF181 | ‒1.667 | 0.315 | 1.396E-05 | 1.123E-03 |
| GIMAP4 | ‒1.663 | 0.316 | 1.074E-06 | 1.281E-04 |
| KLRK1 | ‒1.654 | 0.318 | 1.775E-07 | 2.608E-05 |
| SAMD9 | ‒1.637 | 0.321 | 1.956E-06 | 2.156E-04 |
| MMACHC | ‒1.624 | 0.324 | 7.667E-05 | 4.227E-03 |
| IGKV5-2 | ‒1.617 | 0.326 | 7.368E-04 | 2.155E-02 |
| THNSL1 | ‒1.600 | 0.330 | 1.009E-03 | 2.665E-02 |
| PDE4DIPP6 | ‒1.599 | 0.330 | 3.369E-07 | 4.644E-05 |
| ZNF658 | ‒1.595 | 0.331 | 7.522E-05 | 4.176E-03 |
| GIMAP7 | ‒1.586 | 0.333 | 1.304E-03 | 3.220E-02 |
| ZNF816 | ‒1.571 | 0.337 | 1.156E-07 | 1.817E-05 |
| ZNF772 | ‒1.571 | 0.337 | 7.636E-09 | 1.834E-06 |
| IGLV3-25 | ‒1.563 | 0.338 | 1.939E-03 | 4.269E-02 |
| ZNF501 | ‒1.561 | 0.339 | 3.145E-04 | 1.211E-02 |
| ENSG00000272913 | ‒1.554 | 0.340 | 3.354E-04 | 1.260E-02 |
| MOCS2-DT | ‒1.552 | 0.341 | 2.297E-03 | 4.756E-02 |
| SAMD9L | ‒1.545 | 0.343 | 9.236E-04 | 2.521E-02 |
| ZNF404 | ‒1.543 | 0.343 | 4.938E-04 | 1.657E-02 |
| ZNF470 | ‒1.539 | 0.344 | 8.973E-05 | 4.727E-03 |
| TAS2R4 | ‒1.523 | 0.348 | 8.426E-04 | 2.381E-02 |
| ENSG00000279696 | ‒1.519 | 0.349 | 1.312E-05 | 1.061E-03 |
| PAQR8 | ‒1.516 | 0.350 | 1.029E-06 | 1.254E-04 |
| ZNF594 | ‒1.502 | 0.353 | 5.013E-04 | 1.666E-02 |
| LINC02397 | ‒1.500 | 0.354 | 2.194E-05 | 1.654E-03 |
| IGLV8-61 | ‒1.496 | 0.355 | 1.777E-07 | 2.608E-05 |
| TIGD7 | ‒1.494 | 0.355 | 2.569E-06 | 2.683E-04 |
| IGKV1-39 | ‒1.490 | 0.356 | 8.397E-06 | 7.511E-04 |
| MOXD1 | ‒1.490 | 0.356 | 8.500E-04 | 2.386E-02 |
| CEP19 | ‒1.480 | 0.358 | 2.381E-03 | 4.866E-02 |
| KLRD1 | ‒1.475 | 0.360 | 4.971E-04 | 1.662E-02 |
| FRMPD3 | ‒1.465 | 0.362 | 1.696E-05 | 1.326E-03 |
| IGKV1-9 | ‒1.457 | 0.364 | 1.158E-04 | 5.679E-03 |
| ZNF780A | ‒1.443 | 0.368 | 1.961E-06 | 2.156E-04 |
| RTP4 | ‒1.434 | 0.370 | 7.186E-04 | 2.128E-02 |
| IGLV2-8 | ‒1.433 | 0.370 | 5.396E-05 | 3.288E-03 |
| BTN3A2 | ‒1.431 | 0.371 | 1.402E-06 | 1.626E-04 |
| TMEM60 | ‒1.414 | 0.375 | 7.592E-04 | 2.192E-02 |
| FAM111A-DT | ‒1.389 | 0.382 | 3.412E-04 | 1.272E-02 |
| ENSG00000279267 | ‒1.387 | 0.382 | 2.038E-03 | 4.405E-02 |
| SLAMF7 | ‒1.384 | 0.383 | 2.446E-03 | 4.961E-02 |
| GEMIN6 | ‒1.375 | 0.386 | 1.182E-06 | 1.399E-04 |
| IGKV2-30 | ‒1.374 | 0.386 | 1.834E-04 | 8.071E-03 |
| CASP4LP | ‒1.370 | 0.387 | 2.150E-03 | 4.565E-02 |
| JRKL | ‒1.370 | 0.387 | 4.453E-04 | 1.535E-02 |
| BTLA | ‒1.366 | 0.388 | 3.303E-06 | 3.306E-04 |
| ZNNT1 | ‒1.364 | 0.388 | 1.516E-06 | 1.734E-04 |
| ENSG00000246596 | ‒1.363 | 0.389 | 2.840E-04 | 1.121E-02 |
| UBE2T | ‒1.361 | 0.389 | 1.501E-04 | 6.935E-03 |
| ZBTB32 | ‒1.356 | 0.391 | 2.469E-03 | 4.990E-02 |
| EEF1AKNMT | ‒1.356 | 0.391 | 2.746E-05 | 1.938E-03 |
| IGHV3-33 | ‒1.355 | 0.391 | 1.341E-04 | 6.372E-03 |
| ENSG00000232611 | ‒1.353 | 0.392 | 2.330E-03 | 4.784E-02 |
| CXCR3 | ‒1.350 | 0.392 | 3.158E-04 | 1.211E-02 |
| CD200 | ‒1.349 | 0.393 | 1.689E-04 | 7.535E-03 |
| IGKV4-1 | ‒1.347 | 0.393 | 9.343E-04 | 2.534E-02 |
| DTX3L | ‒1.342 | 0.394 | 5.186E-04 | 1.703E-02 |
| IGHV1-46 | ‒1.339 | 0.395 | 6.659E-05 | 3.835E-03 |
| GCSAM | ‒1.337 | 0.396 | 3.056E-05 | 2.096E-03 |
| ZNF66 | ‒1.325 | 0.399 | 5.140E-04 | 1.691E-02 |
| GIMAP1 | ‒1.324 | 0.399 | 7.225E-09 | 1.773E-06 |
| AURKA | ‒1.324 | 0.400 | 6.992E-04 | 2.096E-02 |
| CENPBD1 | ‒1.322 | 0.400 | 2.913E-04 | 1.144E-02 |
| IL18RAP | ‒1.309 | 0.404 | 1.963E-03 | 4.305E-02 |
| NA | ‒1.304 | 0.405 | 2.527E-09 | 7.081E-07 |
| ZNF737 | ‒1.302 | 0.406 | 5.127E-04 | 1.690E-02 |
| IGLV2-23 | ‒1.300 | 0.406 | 3.615E-04 | 1.324E-02 |
| IGLV3-27 | ‒1.298 | 0.407 | 1.261E-03 | 3.136E-02 |
| PGBD2 | ‒1.296 | 0.407 | 8.355E-06 | 7.511E-04 |
| IGKV1-12 | ‒1.292 | 0.409 | 5.242E-07 | 6.996E-05 |
| IGKV1-16 | ‒1.291 | 0.409 | 1.932E-05 | 1.477E-03 |
| DENND2D | ‒1.283 | 0.411 | 3.314E-05 | 2.202E-03 |
| IGKV3-11 | ‒1.281 | 0.412 | 3.948E-07 | 5.354E-05 |
| NA | ‒1.279 | 0.412 | 9.526E-04 | 2.571E-02 |
| CMTR2 | ‒1.273 | 0.414 | 7.274E-08 | 1.199E-05 |
| BBS10 | ‒1.266 | 0.416 | 1.207E-03 | 3.041E-02 |
| FIGNL1 | ‒1.265 | 0.416 | 1.068E-06 | 1.281E-04 |
| IGLV3-1 | ‒1.264 | 0.416 | 9.491E-04 | 2.566E-02 |
| ARSK | ‒1.263 | 0.417 | 4.091E-05 | 2.626E-03 |
| IGLV1-47 | ‒1.258 | 0.418 | 8.770E-04 | 2.430E-02 |
| ZKSCAN7 | ‒1.242 | 0.423 | 8.287E-04 | 2.354E-02 |
| IGKV3-15 | ‒1.240 | 0.423 | 4.808E-05 | 2.994E-03 |
| HCP5 | ‒1.238 | 0.424 | 2.807E-05 | 1.966E-03 |
| TMED2-DT | ‒1.231 | 0.426 | 1.984E-03 | 4.328E-02 |
| C14orf119 | ‒1.227 | 0.427 | 6.280E-04 | 1.941E-02 |
| ZFP3 | ‒1.218 | 0.430 | 5.653E-05 | 3.383E-03 |
| ZNF583 | ‒1.216 | 0.430 | 5.982E-04 | 1.886E-02 |
| IGLC2 | ‒1.212 | 0.432 | 1.360E-03 | 3.330E-02 |
| TMEM140 | ‒1.207 | 0.433 | 1.390E-03 | 3.373E-02 |
| ZNF613 | ‒1.202 | 0.435 | 1.454E-03 | 3.459E-02 |
| ACKR3 | ‒1.202 | 0.435 | 1.373E-04 | 6.451E-03 |
| ZNF780B | ‒1.201 | 0.435 | 3.300E-04 | 1.249E-02 |
| IGKV3-20 | ‒1.198 | 0.436 | 1.620E-04 | 7.318E-03 |
| ZNF626 | ‒1.191 | 0.438 | 1.015E-03 | 2.674E-02 |
| ZNF607 | ‒1.184 | 0.440 | 1.295E-05 | 1.052E-03 |
| ZNF175 | ‒1.181 | 0.441 | 2.765E-06 | 2.853E-04 |
| LRIF1 | ‒1.178 | 0.442 | 2.817E-04 | 1.117E-02 |
| STAT1 | ‒1.175 | 0.443 | 1.256E-03 | 3.134E-02 |
| TRGC1 | ‒1.175 | 0.443 | 7.599E-05 | 4.203E-03 |
| RBM12B | ‒1.174 | 0.443 | 7.139E-06 | 6.560E-04 |
| IGLV2-11 | ‒1.166 | 0.446 | 1.560E-03 | 3.629E-02 |
| GIMAP6 | ‒1.163 | 0.447 | 2.190E-04 | 9.323E-03 |
| IGKV1-5 | ‒1.159 | 0.448 | 1.131E-05 | 9.558E-04 |
| MRPL35 | ‒1.159 | 0.448 | 1.822E-05 | 1.399E-03 |
| ZNF721 | ‒1.157 | 0.449 | 2.130E-07 | 3.088E-05 |
| ZNF799 | ‒1.155 | 0.449 | 1.154E-03 | 2.937E-02 |
| CARD8-AS1 | ‒1.154 | 0.449 | 6.959E-04 | 2.090E-02 |
| ZNF665 | ‒1.152 | 0.450 | 3.676E-04 | 1.338E-02 |
| ZNF226 | ‒1.150 | 0.450 | 5.285E-09 | 1.411E-06 |
| ZNF616 | ‒1.147 | 0.451 | 1.652E-03 | 3.821E-02 |
| LXN | ‒1.145 | 0.452 | 6.716E-04 | 2.028E-02 |
| IGLV4-69 | ‒1.138 | 0.455 | 1.009E-03 | 2.665E-02 |
| C5orf51 | ‒1.132 | 0.456 | 1.835E-03 | 4.104E-02 |
| FCRL3 | ‒1.127 | 0.458 | 2.841E-04 | 1.121E-02 |
| MYBL1 | ‒1.127 | 0.458 | 9.251E-04 | 2.521E-02 |
| TIGD2 | ‒1.118 | 0.461 | 4.773E-04 | 1.627E-02 |
| IL18R1 | ‒1.117 | 0.461 | 1.725E-03 | 3.947E-02 |
| LBH | ‒1.117 | 0.461 | 4.585E-05 | 2.866E-03 |
| ENSG00000279059 | ‒1.114 | 0.462 | 1.704E-03 | 3.916E-02 |
| ENSG00000259877 | ‒1.113 | 0.462 | 8.156E-04 | 2.324E-02 |
| ZNF189 | ‒1.100 | 0.467 | 6.862E-04 | 2.068E-02 |
| C17orf80 | ‒1.096 | 0.468 | 9.718E-06 | 8.380E-04 |
| PREPL | ‒1.095 | 0.468 | 5.271E-04 | 1.721E-02 |
| ZKSCAN3 | ‒1.094 | 0.468 | 2.583E-04 | 1.047E-02 |
| OGFOD1 | ‒1.094 | 0.468 | 1.696E-04 | 7.547E-03 |
| ZBTB38 | ‒1.090 | 0.470 | 1.106E-03 | 2.849E-02 |
| N6AMT1 | ‒1.087 | 0.471 | 8.993E-04 | 2.471E-02 |
| NAPEPLD | ‒1.084 | 0.472 | 1.109E-03 | 2.853E-02 |
| INTS5 | ‒1.080 | 0.473 | 4.717E-06 | 4.533E-04 |
| IGKC | ‒1.079 | 0.473 | 7.877E-04 | 2.253E-02 |
| GIMAP8 | ‒1.076 | 0.474 | 9.667E-06 | 8.379E-04 |
| ABCB1 | ‒1.073 | 0.475 | 7.390E-04 | 2.158E-02 |
| TRDC | ‒1.073 | 0.475 | 2.020E-03 | 4.378E-02 |
| ZNF230 | ‒1.072 | 0.476 | 9.037E-05 | 4.734E-03 |
| UMPS | ‒1.071 | 0.476 | 2.068E-05 | 1.574E-03 |
| ZNF681 | ‒1.067 | 0.477 | 2.312E-03 | 4.767E-02 |
| SLC25A20 | ‒1.067 | 0.477 | 1.269E-05 | 1.036E-03 |
| RNASEL | ‒1.063 | 0.479 | 1.762E-03 | 4.014E-02 |
| MCM8 | ‒1.062 | 0.479 | 9.754E-05 | 5.016E-03 |
| PCDH9 | ‒1.059 | 0.480 | 5.797E-04 | 1.839E-02 |
| MRPL50 | ‒1.055 | 0.481 | 2.564E-04 | 1.042E-02 |
| HIBCH | ‒1.048 | 0.484 | 6.758E-06 | 6.245E-04 |
| HERPUD2-AS1 | ‒1.043 | 0.485 | 1.263E-03 | 3.137E-02 |
| LOC101927151 | ‒1.040 | 0.486 | 1.764E-03 | 4.014E-02 |
| SLAMF6 | ‒1.038 | 0.487 | 9.397E-06 | 8.187E-04 |
| L3MBTL3 | ‒1.023 | 0.492 | 1.079E-04 | 5.384E-03 |
| SP4 | ‒1.022 | 0.492 | 1.445E-03 | 3.448E-02 |
| ZNF397 | ‒1.019 | 0.494 | 2.382E-07 | 3.409E-05 |
| WEE1 | ‒1.017 | 0.494 | 3.585E-04 | 1.319E-02 |
| PURA | ‒1.015 | 0.495 | 1.806E-05 | 1.393E-03 |
| ENSG00000259820 | ‒1.010 | 0.497 | 1.386E-03 | 3.367E-02 |
| TTC9C | ‒1.006 | 0.498 | 4.920E-05 | 3.031E-03 |
| IGLV2-14 | ‒1.003 | 0.499 | 4.237E-04 | 1.478E-02 |
| SIT1 | ‒1.001 | 0.500 | 6.669E-04 | 2.017E-02 |
| CHURC1 | ‒1.000 | 0.500 | 7.051E-04 | 2.104E-02 |
| ZNF234 | ‒0.996 | 0.501 | 6.351E-04 | 1.956E-02 |
| UTP14C | ‒0.995 | 0.502 | 2.641E-04 | 1.060E-02 |
| GTF2E1 | ‒0.994 | 0.502 | 4.997E-04 | 1.666E-02 |
| LPAR6 | ‒0.983 | 0.506 | 6.115E-04 | 1.909E-02 |
| CEACAM1 | ‒0.981 | 0.507 | 7.600E-04 | 2.192E-02 |
| GSDMB | ‒0.980 | 0.507 | 2.299E-04 | 9.642E-03 |
| PM20D2 | ‒0.974 | 0.509 | 4.898E-05 | 3.028E-03 |
| ALDH5A1 | ‒0.969 | 0.511 | 1.091E-03 | 2.826E-02 |
| CPT1A | ‒0.965 | 0.512 | 4.082E-04 | 1.439E-02 |
| KCTD11 | ‒0.963 | 0.513 | 2.092E-04 | 8.998E-03 |
| DHFR2 | ‒0.962 | 0.513 | 4.011E-04 | 1.423E-02 |
| YIPF4 | ‒0.961 | 0.514 | 5.389E-06 | 5.063E-04 |
| ENSG00000260719 | ‒0.957 | 0.515 | 1.277E-04 | 6.170E-03 |
| C21orf91 | ‒0.954 | 0.516 | 3.112E-04 | 1.211E-02 |
| TRNT1 | ‒0.948 | 0.518 | 2.113E-04 | 9.062E-03 |
| TMEM186 | ‒0.946 | 0.519 | 9.766E-04 | 2.615E-02 |
| SPDL1 | ‒0.939 | 0.522 | 9.564E-04 | 2.577E-02 |
| ZFP82 | ‒0.919 | 0.529 | 8.385E-04 | 2.374E-02 |
| DARS2 | ‒0.918 | 0.529 | 1.614E-03 | 3.744E-02 |
| PAXIP1-AS2 | ‒0.906 | 0.534 | 8.944E-04 | 2.466E-02 |
| MRM1 | ‒0.895 | 0.538 | 5.773E-04 | 1.839E-02 |
| PYGO2 | ‒0.891 | 0.539 | 3.044E-05 | 2.096E-03 |
| IRF4 | ‒0.888 | 0.540 | 2.427E-03 | 4.942E-02 |
| ZNF486 | ‒0.888 | 0.540 | 6.370E-04 | 1.958E-02 |
| PRAG1 | ‒0.888 | 0.540 | 7.519E-04 | 2.176E-02 |
| ZNF320 | ‒0.887 | 0.541 | 9.588E-04 | 2.580E-02 |
| GGPS1 | ‒0.883 | 0.542 | 1.508E-05 | 1.202E-03 |
| TRMT13 | ‒0.875 | 0.545 | 7.919E-05 | 4.282E-03 |
| ZNF420 | ‒0.873 | 0.546 | 1.863E-03 | 4.134E-02 |
| FBXO22 | ‒0.872 | 0.546 | 3.694E-05 | 2.398E-03 |
| ENSG00000268027 | ‒0.866 | 0.549 | 1.188E-03 | 3.010E-02 |
| NUP43 | ‒0.865 | 0.549 | 1.418E-03 | 3.406E-02 |
| TIA1 | ‒0.862 | 0.550 | 1.475E-06 | 1.699E-04 |
| ZNF785 | ‒0.862 | 0.550 | 2.190E-03 | 4.638E-02 |
| ZNF480 | ‒0.861 | 0.550 | 4.853E-05 | 3.011E-03 |
| ZNF260 | ‒0.856 | 0.552 | 2.416E-03 | 4.924E-02 |
| MAT2B | ‒0.853 | 0.554 | 1.065E-05 | 9.093E-04 |
| RAB29 | ‒0.845 | 0.557 | 4.962E-04 | 1.662E-02 |
| LEO1 | ‒0.845 | 0.557 | 5.540E-05 | 3.327E-03 |
| GIMAP2 | ‒0.844 | 0.557 | 1.041E-03 | 2.714E-02 |
| ZNF74 | ‒0.843 | 0.557 | 1.478E-03 | 3.501E-02 |
| ZNF200 | ‒0.836 | 0.560 | 2.650E-05 | 1.896E-03 |
| FGFR1OP2 | ‒0.833 | 0.561 | 3.555E-05 | 2.354E-03 |
| PABIR1 | ‒0.833 | 0.561 | 9.817E-04 | 2.620E-02 |
| KLHL9 | ‒0.829 | 0.563 | 1.518E-03 | 3.569E-02 |
| HSPA8 | ‒0.828 | 0.563 | 3.539E-04 | 1.308E-02 |
| THAP5 | ‒0.828 | 0.563 | 1.208E-03 | 3.041E-02 |
| PPIL1 | ‒0.824 | 0.565 | 2.268E-03 | 4.732E-02 |
| FANCF | ‒0.817 | 0.568 | 1.850E-03 | 4.121E-02 |
| STT3A | ‒0.810 | 0.570 | 2.334E-04 | 9.667E-03 |
| VKORC1L1 | ‒0.800 | 0.574 | 2.362E-03 | 4.832E-02 |
| CTR9 | ‒0.800 | 0.574 | 8.579E-04 | 2.400E-02 |
| UBE4A | ‒0.799 | 0.575 | 1.130E-04 | 5.605E-03 |
| IRF1-AS1 | ‒0.797 | 0.576 | 2.316E-03 | 4.767E-02 |
| RBM4B | ‒0.795 | 0.576 | 5.011E-04 | 1.666E-02 |
| KCTD21 | ‒0.795 | 0.576 | 2.442E-03 | 4.961E-02 |
| RPE | ‒0.793 | 0.577 | 4.809E-04 | 1.630E-02 |
| UBA6-AS1 | ‒0.787 | 0.580 | 1.551E-04 | 7.125E-03 |
| POLH | ‒0.778 | 0.583 | 2.054E-03 | 4.417E-02 |
| TMEM223 | ‒0.775 | 0.584 | 1.512E-03 | 3.561E-02 |
| ZNF561 | ‒0.772 | 0.586 | 1.749E-03 | 3.995E-02 |
| TRIM4 | ‒0.769 | 0.587 | 7.730E-05 | 4.234E-03 |
| ZNF557 | ‒0.765 | 0.589 | 2.316E-03 | 4.767E-02 |
| FCMR | ‒0.762 | 0.590 | 2.407E-06 | 2.546E-04 |
| ZKSCAN1 | ‒0.759 | 0.591 | 4.671E-04 | 1.600E-02 |
| ENSG00000239665 | ‒0.757 | 0.592 | 5.955E-05 | 3.526E-03 |
| EIF2S1 | ‒0.752 | 0.594 | 6.725E-05 | 3.860E-03 |
| C15orf40 | ‒0.751 | 0.594 | 6.441E-04 | 1.966E-02 |
| ZNF154 | ‒0.749 | 0.595 | 6.572E-04 | 1.995E-02 |
| BRD8 | ‒0.748 | 0.595 | 2.083E-05 | 1.578E-03 |
| ARCN1 | ‒0.748 | 0.595 | 1.313E-04 | 6.280E-03 |
| ERGIC2 | ‒0.741 | 0.598 | 6.239E-04 | 1.935E-02 |
| FASTKD1 | ‒0.737 | 0.600 | 7.642E-04 | 2.200E-02 |
| ZNF146 | ‒0.736 | 0.600 | 1.282E-03 | 3.172E-02 |
| ZNF841 | ‒0.736 | 0.601 | 2.956E-04 | 1.159E-02 |
| TRIM27 | ‒0.736 | 0.601 | 1.358E-04 | 6.399E-03 |
| KRBOX4 | ‒0.729 | 0.603 | 3.577E-04 | 1.319E-02 |
| HSPH1 | ‒0.713 | 0.610 | 1.531E-03 | 3.587E-02 |
| NCOR1 | ‒0.713 | 0.610 | 1.901E-03 | 4.202E-02 |
| HNRNPF | ‒0.713 | 0.610 | 1.906E-04 | 8.346E-03 |
| COL19A1 | ‒0.699 | 0.616 | 2.265E-03 | 4.732E-02 |
| MON2 | ‒0.686 | 0.622 | 2.158E-03 | 4.576E-02 |
| MITD1 | ‒0.684 | 0.622 | 3.227E-04 | 1.233E-02 |
| ATAD2B | ‒0.679 | 0.624 | 1.235E-03 | 3.092E-02 |
| TUBGCP4 | ‒0.676 | 0.626 | 5.429E-04 | 1.762E-02 |
| MRPS14 | ‒0.674 | 0.627 | 2.343E-03 | 4.805E-02 |
| GLS | ‒0.673 | 0.627 | 1.256E-03 | 3.134E-02 |
| VEZT | ‒0.668 | 0.629 | 2.260E-03 | 4.732E-02 |
| TARS1 | ‒0.653 | 0.636 | 1.282E-04 | 6.179E-03 |
| PIK3R4 | ‒0.641 | 0.641 | 2.285E-03 | 4.740E-02 |
| OBI1 | ‒0.641 | 0.641 | 2.286E-03 | 4.740E-02 |
| ZNF671 | ‒0.635 | 0.644 | 1.380E-03 | 3.359E-02 |
| LINC00667 | ‒0.635 | 0.644 | 6.179E-04 | 1.924E-02 |
| MCM3 | ‒0.626 | 0.648 | 3.529E-04 | 1.307E-02 |
| PIGF | ‒0.612 | 0.654 | 2.230E-03 | 4.694E-02 |
| THRAP3 | ‒0.604 | 0.658 | 1.783E-04 | 7.869E-03 |
| PLRG1 | ‒0.599 | 0.660 | 2.278E-03 | 4.736E-02 |
| APOL2 | ‒0.596 | 0.662 | 1.351E-03 | 3.318E-02 |
| GART | ‒0.591 | 0.664 | 2.004E-03 | 4.359E-02 |
| NUP42 | ‒0.588 | 0.665 | 1.143E-03 | 2.922E-02 |
| ZNF559 | ‒0.587 | 0.666 | 8.986E-04 | 2.471E-02 |
| ZNF362 | 0.588 | 1.503 | 2.457E-03 | 4.978E-02 |
| WBP11 | 0.591 | 1.506 | 1.572E-03 | 3.652E-02 |
| MAPRE1 | 0.593 | 1.509 | 1.099E-03 | 2.834E-02 |
| INSIG1 | 0.599 | 1.515 | 1.037E-03 | 2.711E-02 |
| CCNY | 0.602 | 1.517 | 7.865E-04 | 2.253E-02 |
| JARID2 | 0.621 | 1.537 | 1.219E-03 | 3.059E-02 |
| FBRS | 0.622 | 1.539 | 3.948E-04 | 1.404E-02 |
| MAPK1IP1L | 0.625 | 1.542 | 1.989E-03 | 4.332E-02 |
| RBM3 | 0.629 | 1.547 | 1.531E-03 | 3.587E-02 |
| CSGALNACT2 | 0.632 | 1.549 | 1.552E-03 | 3.615E-02 |
| INTS1 | 0.646 | 1.565 | 1.496E-03 | 3.529E-02 |
| HNRNPH2 | 0.649 | 1.568 | 3.683E-04 | 1.338E-02 |
| TMEM167B | 0.655 | 1.575 | 3.365E-04 | 1.260E-02 |
| LAPTM5 | 0.655 | 1.575 | 4.076E-04 | 1.439E-02 |
| MLF2 | 0.660 | 1.581 | 1.411E-03 | 3.403E-02 |
| CHCHD2 | 0.660 | 1.581 | 1.233E-03 | 3.090E-02 |
| RREB1 | 0.661 | 1.581 | 9.899E-05 | 5.044E-03 |
| TMCC1 | 0.663 | 1.584 | 2.444E-03 | 4.961E-02 |
| EME2 | 0.664 | 1.584 | 6.241E-04 | 1.935E-02 |
| TRA2B | 0.664 | 1.584 | 8.481E-04 | 2.385E-02 |
| RAB1A | 0.669 | 1.590 | 4.183E-04 | 1.469E-02 |
| UBC | 0.676 | 1.597 | 5.208E-05 | 3.196E-03 |
| NCOR2 | 0.677 | 1.599 | 1.943E-03 | 4.272E-02 |
| DAZAP2 | 0.677 | 1.599 | 5.074E-04 | 1.680E-02 |
| ARF1 | 0.681 | 1.603 | 8.186E-05 | 4.398E-03 |
| ADIPOR1 | 0.685 | 1.608 | 9.716E-05 | 5.012E-03 |
| EIF5A | 0.685 | 1.608 | 1.589E-04 | 7.222E-03 |
| MKNK2 | 0.688 | 1.611 | 1.371E-03 | 3.345E-02 |
| RAPGEF2 | 0.691 | 1.614 | 4.612E-04 | 1.583E-02 |
| HNRNPA0 | 0.691 | 1.614 | 1.675E-03 | 3.854E-02 |
| TET3 | 0.692 | 1.615 | 1.974E-03 | 4.317E-02 |
| HNRNPL | 0.693 | 1.617 | 1.246E-05 | 1.022E-03 |
| MAP1LC3B | 0.694 | 1.618 | 8.722E-05 | 4.627E-03 |
| RPS9 | 0.707 | 1.633 | 2.099E-03 | 4.490E-02 |
| RHBDD2 | 0.711 | 1.637 | 1.781E-03 | 4.030E-02 |
| SLC25A3 | 0.715 | 1.641 | 6.243E-05 | 3.645E-03 |
| SYF2 | 0.719 | 1.646 | 1.787E-03 | 4.033E-02 |
| OTULINL | 0.726 | 1.654 | 8.728E-04 | 2.426E-02 |
| ENSG00000279117 | 0.734 | 1.664 | 1.777E-03 | 4.028E-02 |
| RNF26 | 0.746 | 1.677 | 9.815E-04 | 2.620E-02 |
| PAFAH1B2 | 0.748 | 1.680 | 1.165E-04 | 5.695E-03 |
| HERPUD1 | 0.750 | 1.682 | 1.949E-04 | 8.511E-03 |
| RSL24D1 | 0.752 | 1.684 | 1.096E-03 | 2.830E-02 |
| MCL1 | 0.752 | 1.685 | 1.803E-03 | 4.054E-02 |
| MRFAP1 | 0.755 | 1.688 | 1.623E-04 | 7.318E-03 |
| PNRC1 | 0.762 | 1.696 | 8.879E-04 | 2.452E-02 |
| SUMO3 | 0.765 | 1.700 | 2.090E-04 | 8.998E-03 |
| BUD31 | 0.776 | 1.712 | 1.796E-03 | 4.049E-02 |
| SLC25A33 | 0.790 | 1.729 | 1.775E-03 | 4.028E-02 |
| COX4I1 | 0.792 | 1.732 | 3.267E-04 | 1.246E-02 |
| ANXA1 | 0.793 | 1.732 | 7.262E-04 | 2.138E-02 |
| AP1G2-AS1 | 0.796 | 1.736 | 1.838E-04 | 8.071E-03 |
| SRF | 0.798 | 1.739 | 1.130E-03 | 2.892E-02 |
| MMP24OS | 0.799 | 1.739 | 1.058E-04 | 5.309E-03 |
| CYP4V2 | 0.799 | 1.740 | 1.630E-04 | 7.322E-03 |
| BCL7B | 0.801 | 1.743 | 3.883E-04 | 1.392E-02 |
| PNPLA2 | 0.803 | 1.745 | 6.248E-04 | 1.935E-02 |
| TGIF1 | 0.809 | 1.752 | 7.705E-05 | 4.234E-03 |
| SNN | 0.811 | 1.754 | 2.741E-04 | 1.095E-02 |
| UAP1L1 | 0.819 | 1.764 | 7.464E-04 | 2.172E-02 |
| EEPD1 | 0.831 | 1.778 | 3.783E-04 | 1.368E-02 |
| TRAF4 | 0.847 | 1.798 | 4.790E-04 | 1.627E-02 |
| CARD19 | 0.849 | 1.801 | 5.571E-04 | 1.789E-02 |
| TUBB4B | 0.850 | 1.802 | 1.602E-05 | 1.264E-03 |
| ADGRL1-AS1 | 0.863 | 1.818 | 1.448E-03 | 3.448E-02 |
| CLK1 | 0.869 | 1.826 | 2.140E-03 | 4.555E-02 |
| IRF1 | 0.870 | 1.828 | 9.341E-06 | 8.187E-04 |
| SLC3A2 | 0.876 | 1.835 | 1.523E-04 | 7.017E-03 |
| RNF19B | 0.876 | 1.836 | 1.819E-03 | 4.085E-02 |
| IDI1 | 0.876 | 1.836 | 4.200E-06 | 4.154E-04 |
| KLHL26 | 0.888 | 1.850 | 4.394E-04 | 1.522E-02 |
| VAMP3 | 0.888 | 1.850 | 3.411E-04 | 1.272E-02 |
| ETF1 | 0.889 | 1.852 | 9.769E-07 | 1.208E-04 |
| AKT1S1 | 0.892 | 1.856 | 2.312E-05 | 1.705E-03 |
| MAPK7 | 0.892 | 1.856 | 2.053E-03 | 4.417E-02 |
| CDKN2D | 0.893 | 1.857 | 3.615E-05 | 2.374E-03 |
| EIF1B | 0.894 | 1.859 | 8.468E-04 | 2.385E-02 |
| SNIP1 | 0.895 | 1.859 | 1.426E-04 | 6.675E-03 |
| TMEM201 | 0.896 | 1.861 | 4.790E-04 | 1.627E-02 |
| DDX21 | 0.902 | 1.869 | 3.152E-05 | 2.138E-03 |
| NECAP1 | 0.902 | 1.869 | 1.724E-06 | 1.920E-04 |
| VPS18 | 0.905 | 1.873 | 6.841E-05 | 3.890E-03 |
| RFX2 | 0.910 | 1.879 | 1.151E-03 | 2.932E-02 |
| MEF2D | 0.911 | 1.881 | 8.729E-09 | 2.039E-06 |
| SIVA1 | 0.915 | 1.886 | 2.287E-05 | 1.694E-03 |
| ATXN1L | 0.915 | 1.886 | 6.338E-06 | 5.888E-04 |
| ANAPC15 | 0.924 | 1.897 | 2.563E-04 | 1.042E-02 |
| CDK2AP1 | 0.930 | 1.905 | 1.783E-03 | 4.030E-02 |
| MED30 | 0.930 | 1.905 | 4.214E-04 | 1.473E-02 |
| PRNP | 0.930 | 1.906 | 1.267E-04 | 6.138E-03 |
| CDKN1B | 0.932 | 1.907 | 1.403E-03 | 3.389E-02 |
| TENT5C | 0.933 | 1.909 | 4.399E-04 | 1.522E-02 |
| PI4K2A | 0.937 | 1.914 | 3.144E-04 | 1.211E-02 |
| ENSG00000280138 | 0.939 | 1.917 | 9.366E-06 | 8.187E-04 |
| TUBA1C | 0.941 | 1.920 | 1.147E-04 | 5.658E-03 |
| UPP1 | 0.945 | 1.925 | 2.448E-04 | 1.004E-02 |
| DENND1A | 0.946 | 1.926 | 2.786E-04 | 1.110E-02 |
| GCNT2 | 0.948 | 1.929 | 1.830E-03 | 4.097E-02 |
| SELENOK | 0.953 | 1.936 | 5.480E-05 | 3.305E-03 |
| PTS | 0.964 | 1.951 | 3.127E-04 | 1.211E-02 |
| RBM7 | 0.974 | 1.964 | 2.460E-03 | 4.979E-02 |
| PRXL2C | 0.980 | 1.973 | 1.258E-03 | 3.134E-02 |
| TWF1 | 0.986 | 1.980 | 1.977E-04 | 8.612E-03 |
| ATF4 | 0.990 | 1.987 | 3.687E-05 | 2.398E-03 |
| ENSG00000269688 | 0.993 | 1.991 | 3.432E-04 | 1.277E-02 |
| WDR74 | 1.000 | 2.000 | 5.483E-05 | 3.305E-03 |
| GPR35 | 1.001 | 2.001 | 1.053E-06 | 1.274E-04 |
| RNF103 | 1.004 | 2.006 | 5.484E-04 | 1.770E-02 |
| EMD | 1.005 | 2.008 | 2.369E-05 | 1.725E-03 |
| MGST3 | 1.014 | 2.020 | 5.682E-04 | 1.816E-02 |
| JMJD6 | 1.015 | 2.021 | 7.918E-05 | 4.282E-03 |
| GRPEL1 | 1.016 | 2.023 | 2.361E-06 | 2.513E-04 |
| CCRL2 | 1.018 | 2.025 | 1.023E-03 | 2.693E-02 |
| CTIF | 1.019 | 2.026 | 8.366E-04 | 2.372E-02 |
| NFKBIB | 1.024 | 2.034 | 3.801E-04 | 1.372E-02 |
| MNT | 1.033 | 2.046 | 7.009E-05 | 3.955E-03 |
| TOM1 | 1.042 | 2.059 | 1.475E-03 | 3.497E-02 |
| TLE3 | 1.044 | 2.062 | 3.236E-06 | 3.258E-04 |
| JOSD1 | 1.044 | 2.062 | 3.044E-04 | 1.190E-02 |
| FCGRT | 1.046 | 2.065 | 1.830E-03 | 4.097E-02 |
| EIF5 | 1.048 | 2.068 | 2.865E-08 | 5.537E-06 |
| NXT1 | 1.050 | 2.071 | 1.717E-05 | 1.336E-03 |
| ARHGDIA | 1.052 | 2.073 | 3.600E-04 | 1.322E-02 |
| BRI3 | 1.054 | 2.076 | 2.099E-03 | 4.490E-02 |
| NDUFV2 | 1.060 | 2.085 | 9.459E-04 | 2.561E-02 |
| CLN8-AS1 | 1.063 | 2.090 | 2.094E-03 | 4.490E-02 |
| FEM1C | 1.065 | 2.092 | 2.259E-04 | 9.522E-03 |
| NR1H2 | 1.067 | 2.095 | 3.494E-04 | 1.297E-02 |
| FASTKD5 | 1.092 | 2.131 | 1.315E-04 | 6.280E-03 |
| YOD1 | 1.093 | 2.134 | 1.716E-03 | 3.931E-02 |
| PTGER4 | 1.096 | 2.138 | 2.812E-06 | 2.884E-04 |
| SMURF1 | 1.097 | 2.139 | 4.288E-06 | 4.168E-04 |
| CCNL1 | 1.102 | 2.147 | 5.947E-09 | 1.562E-06 |
| LDLR | 1.106 | 2.152 | 3.636E-05 | 2.379E-03 |
| SLC22A18 | 1.107 | 2.154 | 2.273E-03 | 4.736E-02 |
| IRGQ | 1.115 | 2.166 | 1.200E-03 | 3.035E-02 |
| CHASERR | 1.116 | 2.167 | 1.599E-04 | 7.247E-03 |
| HMGB3 | 1.118 | 2.170 | 2.043E-03 | 4.409E-02 |
| IER5 | 1.119 | 2.172 | 1.609E-06 | 1.816E-04 |
| EIF2AK3-DT | 1.120 | 2.174 | 2.237E-03 | 4.703E-02 |
| TUBB2A | 1.122 | 2.177 | 7.424E-04 | 2.164E-02 |
| PDE4B | 1.123 | 2.178 | 1.433E-04 | 6.675E-03 |
| ADAM9 | 1.127 | 2.184 | 5.438E-04 | 1.762E-02 |
| GPR183 | 1.127 | 2.184 | 1.572E-06 | 1.786E-04 |
| CKAP4 | 1.128 | 2.185 | 1.157E-04 | 5.679E-03 |
| KLF11 | 1.128 | 2.186 | 3.067E-05 | 2.096E-03 |
| ZMIZ1 | 1.129 | 2.187 | 9.055E-04 | 2.480E-02 |
| RBM34 | 1.132 | 2.192 | 1.973E-03 | 4.317E-02 |
| KCNQ1 | 1.132 | 2.192 | 5.875E-07 | 7.599E-05 |
| GDPD5 | 1.136 | 2.197 | 6.395E-04 | 1.959E-02 |
| C6orf226 | 1.136 | 2.198 | 7.171E-04 | 2.127E-02 |
| MFSD2A | 1.137 | 2.199 | 1.225E-04 | 5.954E-03 |
| BACH1 | 1.140 | 2.204 | 2.817E-04 | 1.117E-02 |
| MYLIP | 1.143 | 2.209 | 9.289E-06 | 8.187E-04 |
| RELL1 | 1.151 | 2.220 | 5.583E-04 | 1.789E-02 |
| EIF1 | 1.159 | 2.234 | 1.296E-07 | 2.017E-05 |
| SLC7A5 | 1.160 | 2.234 | 1.919E-03 | 4.230E-02 |
| ATP6V0C | 1.168 | 2.247 | 3.262E-05 | 2.177E-03 |
| YRDC | 1.169 | 2.249 | 6.586E-08 | 1.119E-05 |
| DDX47 | 1.173 | 2.254 | 1.445E-03 | 3.448E-02 |
| CPNE2 | 1.173 | 2.255 | 2.342E-05 | 1.712E-03 |
| CCDC59 | 1.176 | 2.260 | 2.328E-04 | 9.667E-03 |
| ENSG00000258944 | 1.181 | 2.267 | 2.202E-03 | 4.652E-02 |
| H3-3B | 1.182 | 2.269 | 2.639E-04 | 1.060E-02 |
| GPX1 | 1.182 | 2.270 | 1.148E-03 | 2.930E-02 |
| ZFAND2A | 1.187 | 2.277 | 2.136E-06 | 2.333E-04 |
| RAPH1 | 1.198 | 2.294 | 1.846E-03 | 4.116E-02 |
| CBX4 | 1.204 | 2.304 | 2.524E-10 | 9.430E-08 |
| PRR7 | 1.219 | 2.328 | 2.186E-04 | 9.323E-03 |
| MAFK | 1.221 | 2.332 | 6.495E-08 | 1.119E-05 |
| KIAA1522 | 1.227 | 2.340 | 7.294E-05 | 4.075E-03 |
| FTH1 | 1.227 | 2.341 | 1.395E-03 | 3.380E-02 |
| GABARAPL1 | 1.229 | 2.344 | 6.533E-08 | 1.119E-05 |
| PELI1 | 1.232 | 2.349 | 4.744E-08 | 8.398E-06 |
| IRF2BP2 | 1.236 | 2.356 | 2.639E-11 | 1.082E-08 |
| ENSG00000255847 | 1.243 | 2.367 | 2.218E-05 | 1.658E-03 |
| SULT1A1 | 1.245 | 2.370 | 2.045E-03 | 4.409E-02 |
| KLF6 | 1.245 | 2.371 | 2.311E-04 | 9.642E-03 |
| DUS3L | 1.255 | 2.387 | 1.237E-05 | 1.020E-03 |
| PIGA | 1.265 | 2.404 | 1.580E-05 | 1.253E-03 |
| TUBA1A | 1.269 | 2.410 | 4.847E-04 | 1.640E-02 |
| RNF139 | 1.270 | 2.412 | 9.661E-04 | 2.595E-02 |
| OAF | 1.272 | 2.415 | 1.378E-03 | 3.358E-02 |
| ADCY9 | 1.284 | 2.435 | 3.922E-04 | 1.397E-02 |
| TIPARP | 1.293 | 2.451 | 7.424E-06 | 6.748E-04 |
| CEBPB | 1.296 | 2.456 | 2.245E-03 | 4.711E-02 |
| ENSG00000262766 | 1.307 | 2.474 | 2.312E-03 | 4.767E-02 |
| PTPRE | 1.316 | 2.490 | 6.628E-04 | 2.008E-02 |
| PMAIP1 | 1.317 | 2.492 | 6.762E-07 | 8.680E-05 |
| NRROS | 1.319 | 2.495 | 9.700E-05 | 5.012E-03 |
| CBX6 | 1.319 | 2.495 | 1.784E-07 | 2.608E-05 |
| BCL10 | 1.322 | 2.499 | 1.566E-08 | 3.292E-06 |
| RIPK2 | 1.323 | 2.502 | 1.029E-03 | 2.698E-02 |
| YPEL5 | 1.324 | 2.504 | 2.780E-07 | 3.895E-05 |
| SIAH1 | 1.328 | 2.511 | 2.318E-04 | 9.648E-03 |
| RELT | 1.329 | 2.513 | 1.116E-03 | 2.863E-02 |
| NA | 1.330 | 2.513 | 2.400E-03 | 4.897E-02 |
| MAFG | 1.331 | 2.515 | 1.962E-08 | 3.966E-06 |
| POLR1F | 1.337 | 2.527 | 5.113E-06 | 4.858E-04 |
| ZBTB21 | 1.342 | 2.535 | 5.950E-05 | 3.526E-03 |
| GZF1 | 1.342 | 2.535 | 4.131E-10 | 1.478E-07 |
| MCEMP1 | 1.347 | 2.543 | 1.265E-03 | 3.137E-02 |
| SNHG15 | 1.357 | 2.561 | 3.176E-10 | 1.161E-07 |
| EIF4A1 | 1.357 | 2.561 | 9.866E-09 | 2.242E-06 |
| C5AR1 | 1.374 | 2.592 | 1.495E-03 | 3.529E-02 |
| FBXO33 | 1.377 | 2.598 | 1.552E-03 | 3.615E-02 |
| PLPPR2 | 1.384 | 2.611 | 5.988E-04 | 1.886E-02 |
| ZNF628 | 1.389 | 2.619 | 6.461E-05 | 3.759E-03 |
| OSER1 | 1.392 | 2.625 | 1.015E-08 | 2.277E-06 |
| SIRPA | 1.395 | 2.630 | 1.520E-03 | 3.571E-02 |
| KDM6B | 1.404 | 2.646 | 3.228E-08 | 6.099E-06 |
| SRGN | 1.408 | 2.653 | 7.075E-05 | 3.979E-03 |
| DGCR11 | 1.413 | 2.663 | 2.472E-03 | 4.990E-02 |
| HNRNPD-DT | 1.414 | 2.665 | 8.446E-04 | 2.383E-02 |
| SPATA2 | 1.416 | 2.668 | 7.274E-09 | 1.773E-06 |
| CD69 | 1.416 | 2.669 | 4.169E-04 | 1.466E-02 |
| RBM38 | 1.425 | 2.685 | 1.195E-05 | 9.982E-04 |
| ZNF394 | 1.426 | 2.688 | 3.509E-08 | 6.485E-06 |
| ANKRD9 | 1.427 | 2.690 | 5.212E-04 | 1.708E-02 |
| RBM47 | 1.431 | 2.696 | 3.906E-04 | 1.394E-02 |
| B3GNT7 | 1.433 | 2.700 | 2.744E-05 | 1.938E-03 |
| EAF1 | 1.438 | 2.709 | 3.380E-14 | 2.842E-11 |
| PTP4A1 | 1.442 | 2.717 | 4.180E-07 | 5.624E-05 |
| MXD1 | 1.447 | 2.727 | 3.820E-04 | 1.376E-02 |
| BNIP1 | 1.448 | 2.729 | 5.154E-06 | 4.869E-04 |
| ETS2 | 1.449 | 2.729 | 5.042E-04 | 1.672E-02 |
| HIF1A | 1.457 | 2.745 | 9.755E-09 | 2.242E-06 |
| TOE1 | 1.468 | 2.766 | 4.288E-05 | 2.731E-03 |
| CLP1 | 1.468 | 2.766 | 8.330E-08 | 1.347E-05 |
| ENSG00000260708 | 1.477 | 2.783 | 6.458E-04 | 1.967E-02 |
| DUSP5 | 1.477 | 2.783 | 5.683E-11 | 2.209E-08 |
| C17orf49 | 1.479 | 2.787 | 5.141E-10 | 1.729E-07 |
| MYADM | 1.491 | 2.810 | 3.961E-06 | 3.941E-04 |
| SERTAD1 | 1.491 | 2.811 | 5.429E-06 | 5.072E-04 |
| SERTAD3 | 1.498 | 2.825 | 7.674E-08 | 1.253E-05 |
| TTC34 | 1.499 | 2.826 | 3.359E-04 | 1.260E-02 |
| NFKBID | 1.499 | 2.826 | 1.296E-04 | 6.227E-03 |
| ENSG00000279602 | 1.499 | 2.827 | 4.282E-06 | 4.168E-04 |
| LDLRAD3 | 1.516 | 2.861 | 2.247E-03 | 4.711E-02 |
| CRABP2 | 1.524 | 2.877 | 1.358E-03 | 3.330E-02 |
| ZP3 | 1.525 | 2.878 | 2.169E-04 | 9.279E-03 |
| SMARCD3 | 1.525 | 2.878 | 1.459E-03 | 3.466E-02 |
| MMP14 | 1.527 | 2.882 | 2.038E-03 | 4.405E-02 |
| SDE2 | 1.533 | 2.893 | 4.053E-08 | 7.328E-06 |
| GLT1D1 | 1.536 | 2.900 | 1.483E-03 | 3.506E-02 |
| F5 | 1.539 | 2.907 | 1.117E-03 | 2.863E-02 |
| H2BC21 | 1.547 | 2.921 | 4.282E-04 | 1.491E-02 |
| CEACAM19 | 1.547 | 2.923 | 2.489E-04 | 1.016E-02 |
| ENSG00000277879 | 1.551 | 2.930 | 1.364E-03 | 3.334E-02 |
| DUSP1 | 1.554 | 2.936 | 3.121E-04 | 1.211E-02 |
| ARRDC4 | 1.555 | 2.938 | 1.240E-08 | 2.707E-06 |
| OTUD1 | 1.561 | 2.950 | 9.380E-05 | 4.868E-03 |
| STX11 | 1.567 | 2.963 | 9.889E-05 | 5.044E-03 |
| RNVU1-30 | 1.568 | 2.966 | 8.028E-04 | 2.292E-02 |
| VENTX | 1.576 | 2.981 | 2.028E-04 | 8.796E-03 |
| JDP2 | 1.578 | 2.986 | 5.716E-07 | 7.451E-05 |
| SOX4 | 1.591 | 3.013 | 4.929E-04 | 1.657E-02 |
| RBM38-AS1 | 1.594 | 3.018 | 1.567E-04 | 7.159E-03 |
| HIC1 | 1.595 | 3.020 | 4.478E-06 | 4.328E-04 |
| BTG3 | 1.596 | 3.024 | 7.075E-04 | 2.106E-02 |
| LINC00963 | 1.597 | 3.025 | 1.758E-03 | 4.012E-02 |
| NRGN | 1.599 | 3.029 | 1.192E-04 | 5.809E-03 |
| CXCR4 | 1.600 | 3.032 | 5.429E-08 | 9.511E-06 |
| UBE2FP1 | 1.607 | 3.047 | 8.372E-05 | 4.469E-03 |
| DUSP10 | 1.612 | 3.056 | 5.420E-05 | 3.290E-03 |
| MTSS2 | 1.615 | 3.064 | 8.435E-10 | 2.676E-07 |
| STAB1 | 1.617 | 3.068 | 1.627E-03 | 3.768E-02 |
| LONRF3 | 1.619 | 3.071 | 1.771E-04 | 7.835E-03 |
| IER5L | 1.625 | 3.084 | 4.887E-06 | 4.669E-04 |
| SH3RF1 | 1.630 | 3.096 | 3.304E-04 | 1.249E-02 |
| NIBAN2 | 1.653 | 3.144 | 1.078E-04 | 5.384E-03 |
| SBDS | 1.653 | 3.146 | 5.381E-07 | 7.069E-05 |
| ZC3H12A | 1.654 | 3.148 | 1.620E-13 | 1.048E-10 |
| LOC102723996 | 1.667 | 3.175 | 1.768E-03 | 4.018E-02 |
| TNRC18P1 | 1.675 | 3.194 | 7.768E-04 | 2.229E-02 |
| FAM20C | 1.692 | 3.232 | 3.151E-04 | 1.211E-02 |
| JUND | 1.705 | 3.261 | 4.856E-10 | 1.701E-07 |
| CSF3R | 1.718 | 3.291 | 1.421E-03 | 3.409E-02 |
| BTG2 | 1.727 | 3.310 | 1.556E-04 | 7.131E-03 |
| AHR | 1.735 | 3.328 | 5.842E-05 | 3.483E-03 |
| GGTA1 | 1.740 | 3.341 | 2.319E-03 | 4.767E-02 |
| SKOR1 | 1.742 | 3.346 | 1.901E-03 | 4.202E-02 |
| GADD45A | 1.743 | 3.347 | 1.157E-05 | 9.729E-04 |
| MIR222HG | 1.747 | 3.356 | 9.930E-05 | 5.045E-03 |
| MIDN | 1.749 | 3.361 | 4.288E-05 | 2.731E-03 |
| ZBTB10 | 1.750 | 3.363 | 1.332E-04 | 6.346E-03 |
| KLF10 | 1.753 | 3.371 | 2.696E-04 | 1.079E-02 |
| RAB3A | 1.758 | 3.383 | 7.505E-04 | 2.176E-02 |
| HES1 | 1.759 | 3.384 | 4.456E-04 | 1.535E-02 |
| SNORA75 | 1.760 | 3.388 | 1.416E-03 | 3.406E-02 |
| FSCN1 | 1.760 | 3.388 | 4.913E-04 | 1.656E-02 |
| ENSG00000275056 | 1.761 | 3.390 | 2.029E-04 | 8.796E-03 |
| JAG1 | 1.768 | 3.405 | 8.961E-05 | 4.727E-03 |
| GNA15 | 1.785 | 3.446 | 2.928E-06 | 2.966E-04 |
| ADM | 1.791 | 3.461 | 6.120E-04 | 1.909E-02 |
| MGP | 1.792 | 3.463 | 2.357E-03 | 4.828E-02 |
| JUNB | 1.800 | 3.482 | 1.334E-08 | 2.841E-06 |
| NINJ1 | 1.804 | 3.492 | 3.508E-08 | 6.485E-06 |
| CCR1 | 1.810 | 3.507 | 3.162E-04 | 1.211E-02 |
| COL9A3 | 1.812 | 3.510 | 1.799E-03 | 4.050E-02 |
| ENSG00000280067 | 1.814 | 3.516 | 9.222E-04 | 2.521E-02 |
| RPL32P1 | 1.820 | 3.532 | 8.565E-04 | 2.400E-02 |
| GAS2L1 | 1.832 | 3.560 | 3.706E-04 | 1.343E-02 |
| NFKBIE | 1.850 | 3.604 | 1.032E-08 | 2.283E-06 |
| NOCT | 1.851 | 3.607 | 2.854E-05 | 1.991E-03 |
| SMOX | 1.852 | 3.610 | 5.454E-04 | 1.764E-02 |
| SLC6A8 | 1.859 | 3.628 | 1.896E-03 | 4.200E-02 |
| RGS2 | 1.860 | 3.630 | 1.027E-05 | 8.808E-04 |
| ENSG00000272256 | 1.876 | 3.670 | 8.199E-04 | 2.333E-02 |
| S100A8 | 1.880 | 3.681 | 3.006E-05 | 2.080E-03 |
| LMNA | 1.891 | 3.710 | 4.578E-08 | 8.190E-06 |
| KLF4 | 1.906 | 3.748 | 3.816E-05 | 2.459E-03 |
| JUN | 1.909 | 3.754 | 1.266E-08 | 2.729E-06 |
| PDGFC | 1.912 | 3.764 | 1.164E-03 | 2.952E-02 |
| EREG | 1.914 | 3.769 | 3.635E-04 | 1.328E-02 |
| SLC22A4 | 1.916 | 3.775 | 1.061E-03 | 2.763E-02 |
| ITPRIP | 1.918 | 3.778 | 4.358E-14 | 3.490E-11 |
| BHLHE40 | 1.918 | 3.780 | 1.442E-07 | 2.204E-05 |
| CDKN1A | 1.920 | 3.784 | 2.221E-06 | 2.395E-04 |
| GADD45B | 1.925 | 3.797 | 1.663E-08 | 3.410E-06 |
| IER2 | 1.926 | 3.801 | 1.071E-05 | 9.093E-04 |
| RETN | 1.937 | 3.828 | 4.254E-06 | 4.168E-04 |
| P2RY2 | 1.943 | 3.846 | 1.003E-03 | 2.657E-02 |
| CD14 | 1.953 | 3.872 | 2.367E-04 | 9.780E-03 |
| TNFAIP3 | 1.956 | 3.881 | 4.405E-05 | 2.795E-03 |
| RASGEF1B | 1.958 | 3.885 | 6.554E-05 | 3.801E-03 |
| VEGFA | 1.967 | 3.909 | 2.005E-08 | 3.966E-06 |
| DDIT3 | 1.973 | 3.925 | 3.717E-15 | 4.167E-12 |
| ENSG00000232811 | 1.978 | 3.940 | 2.881E-06 | 2.937E-04 |
| LRG1 | 1.979 | 3.943 | 1.737E-04 | 7.707E-03 |
| ARHGAP22 | 1.979 | 3.943 | 6.293E-04 | 1.942E-02 |
| IGHEP2 | 1.988 | 3.966 | 5.389E-04 | 1.756E-02 |
| ENSG00000279520 | 1.994 | 3.984 | 4.254E-16 | 5.503E-13 |
| RHBDL1 | 2.002 | 4.005 | 7.004E-04 | 2.096E-02 |
| BTBD19 | 2.014 | 4.040 | 2.229E-04 | 9.444E-03 |
| HOMER3 | 2.017 | 4.047 | 9.268E-05 | 4.825E-03 |
| ENC1 | 2.022 | 4.063 | 2.181E-06 | 2.366E-04 |
| ENSG00000273972 | 2.030 | 4.084 | 1.666E-03 | 3.837E-02 |
| RGS17P1 | 2.048 | 4.135 | 1.199E-05 | 9.982E-04 |
| S100A9 | 2.049 | 4.137 | 8.698E-06 | 7.739E-04 |
| FOS | 2.056 | 4.159 | 7.651E-07 | 9.674E-05 |
| ENSG00000270640 | 2.067 | 4.190 | 1.267E-03 | 3.137E-02 |
| SNAI1 | 2.070 | 4.200 | 1.332E-03 | 3.280E-02 |
| LRP3 | 2.087 | 4.249 | 6.814E-05 | 3.890E-03 |
| KCNK7 | 2.089 | 4.253 | 3.744E-05 | 2.421E-03 |
| IFITM10 | 2.095 | 4.273 | 3.236E-05 | 2.168E-03 |
| NECTIN2 | 2.118 | 4.342 | 3.293E-04 | 1.249E-02 |
| SCN1B | 2.129 | 4.374 | 7.524E-05 | 4.176E-03 |
| S100A12 | 2.131 | 4.380 | 7.126E-05 | 3.994E-03 |
| NRARP | 2.152 | 4.443 | 2.014E-03 | 4.369E-02 |
| IRS2 | 2.154 | 4.451 | 6.444E-10 | 2.125E-07 |
| DUSP2 | 2.158 | 4.462 | 9.808E-15 | 9.461E-12 |
| MAFB | 2.167 | 4.491 | 3.263E-07 | 4.535E-05 |
| ZFP36 | 2.168 | 4.493 | 2.525E-09 | 7.081E-07 |
| NFKBIA | 2.170 | 4.500 | 1.211E-11 | 5.817E-09 |
| NR6A1 | 2.176 | 4.518 | 8.261E-05 | 4.424E-03 |
| ENSG00000207525 | 2.182 | 4.539 | 6.065E-04 | 1.906E-02 |
| FOSL2 | 2.187 | 4.552 | 1.416E-11 | 6.582E-09 |
| AREG | 2.189 | 4.561 | 5.583E-04 | 1.789E-02 |
| PPP1R15A | 2.196 | 4.583 | 8.779E-14 | 5.905E-11 |
| ICAM1 | 2.198 | 4.587 | 3.686E-12 | 2.066E-09 |
| SOCS3 | 2.198 | 4.587 | 2.672E-06 | 2.774E-04 |
| MIR616 | 2.208 | 4.619 | 1.664E-03 | 3.837E-02 |
| SLC2A3 | 2.209 | 4.623 | 1.570E-18 | 2.934E-15 |
| PLAU | 2.209 | 4.624 | 2.676E-05 | 1.907E-03 |
| DIP2A-IT1 | 2.209 | 4.625 | 4.566E-05 | 2.865E-03 |
| ENSG00000266993 | 2.215 | 4.642 | 2.278E-03 | 4.736E-02 |
| PPIF | 2.227 | 4.683 | 1.967E-12 | 1.141E-09 |
| NA | 2.248 | 4.749 | 2.364E-08 | 4.622E-06 |
| NAMPT | 2.259 | 4.786 | 4.009E-08 | 7.328E-06 |
| DUSP6 | 2.259 | 4.788 | 1.487E-07 | 2.253E-05 |
| ENSG00000256913 | 2.261 | 4.793 | 1.039E-03 | 2.712E-02 |
| ENSG00000275210 | 2.272 | 4.830 | 9.994E-04 | 2.651E-02 |
| IFI30 | 2.282 | 4.863 | 9.844E-05 | 5.044E-03 |
| PIM3 | 2.285 | 4.874 | 1.380E-25 | 5.800E-22 |
| LGALS12 | 2.286 | 4.876 | 3.171E-05 | 2.141E-03 |
| ASGR2 | 2.290 | 4.890 | 2.306E-04 | 9.642E-03 |
| ANPEP | 2.291 | 4.892 | 1.736E-07 | 2.606E-05 |
| FFAR1 | 2.292 | 4.896 | 1.448E-11 | 6.582E-09 |
| CSRNP1 | 2.299 | 4.923 | 5.836E-14 | 4.267E-11 |
| NFIL3 | 2.304 | 4.939 | 7.050E-08 | 1.185E-05 |
| TCF3P1 | 2.307 | 4.948 | 9.014E-04 | 2.473E-02 |
| HLX | 2.344 | 5.077 | 1.339E-03 | 3.291E-02 |
| PLK3 | 2.346 | 5.085 | 2.505E-25 | 8.425E-22 |
| PDE2A | 2.351 | 5.103 | 2.590E-04 | 1.047E-02 |
| PFKFB3 | 2.356 | 5.121 | 1.178E-17 | 1.981E-14 |
| SOWAHC | 2.363 | 5.143 | 6.411E-04 | 1.960E-02 |
| TNFRSF12A | 2.374 | 5.183 | 6.012E-05 | 3.547E-03 |
| MIR22HG | 2.378 | 5.198 | 2.038E-09 | 5.908E-07 |
| XK | 2.387 | 5.230 | 1.413E-03 | 3.403E-02 |
| BCL3 | 2.393 | 5.254 | 6.803E-12 | 3.690E-09 |
| ENSG00000268734 | 2.402 | 5.284 | 3.668E-07 | 5.015E-05 |
| GADD45G | 2.415 | 5.332 | 1.857E-03 | 4.131E-02 |
| ENSG00000274008 | 2.423 | 5.363 | 6.394E-04 | 1.959E-02 |
| ASTL | 2.435 | 5.409 | 1.479E-04 | 6.872E-03 |
| ELOVL7 | 2.469 | 5.537 | 2.200E-03 | 4.652E-02 |
| MATN1 | 2.469 | 5.537 | 4.546E-04 | 1.563E-02 |
| TRIB1 | 2.476 | 5.562 | 2.206E-05 | 1.656E-03 |
| CHRM4 | 2.507 | 5.683 | 7.352E-04 | 2.154E-02 |
| ENSG00000250138 | 2.514 | 5.711 | 8.650E-05 | 4.603E-03 |
| ATP2B1-AS1 | 2.520 | 5.735 | 2.634E-05 | 1.896E-03 |
| PLAUR | 2.555 | 5.875 | 5.065E-10 | 1.729E-07 |
| TSPEAR-AS1 | 2.582 | 5.989 | 1.314E-03 | 3.239E-02 |
| GP9 | 2.589 | 6.018 | 2.218E-03 | 4.674E-02 |
| AVPI1 | 2.591 | 6.023 | 5.787E-04 | 1.839E-02 |
| RND1 | 2.601 | 6.068 | 8.633E-04 | 2.407E-02 |
| PTX3 | 2.618 | 6.140 | 7.139E-12 | 3.752E-09 |
| NLRP3 | 2.625 | 6.168 | 1.345E-07 | 2.075E-05 |
| MMP25 | 2.630 | 6.191 | 2.027E-14 | 1.794E-11 |
| IER3 | 2.659 | 6.315 | 8.094E-13 | 5.041E-10 |
| FAM238A | 2.669 | 6.359 | 1.596E-11 | 6.884E-09 |
| RBKS | 2.677 | 6.395 | 2.392E-07 | 3.409E-05 |
| EMP1 | 2.694 | 6.471 | 5.779E-11 | 2.209E-08 |
| DUSP8 | 2.700 | 6.500 | 2.755E-05 | 1.938E-03 |
| RRAD | 2.718 | 6.581 | 5.245E-05 | 3.207E-03 |
| MIR23AHG | 2.733 | 6.646 | 6.241E-15 | 6.559E-12 |
| RAB20 | 2.739 | 6.675 | 8.601E-14 | 5.905E-11 |
| NA | 2.741 | 6.684 | 7.517E-17 | 1.149E-13 |
| MAFF | 2.742 | 6.690 | 1.013E-14 | 9.461E-12 |
| TMEM88 | 2.744 | 6.700 | 1.217E-05 | 1.008E-03 |
| MMP9 | 2.766 | 6.802 | 7.058E-04 | 2.104E-02 |
| PHLDA1 | 2.802 | 6.973 | 2.759E-07 | 3.895E-05 |
| CHMP4BP1 | 2.806 | 6.992 | 2.993E-05 | 2.080E-03 |
| NR4A1 | 2.843 | 7.176 | 2.237E-06 | 2.396E-04 |
| IGFBP2 | 2.853 | 7.226 | 1.002E-04 | 5.058E-03 |
| ENSG00000234436 | 2.893 | 7.426 | 5.720E-04 | 1.825E-02 |
| ENSG00000274677 | 2.893 | 7.426 | 1.663E-04 | 7.437E-03 |
| PER1 | 2.898 | 7.452 | 1.029E-25 | 5.765E-22 |
| ENSG00000278022 | 2.940 | 7.677 | 9.969E-05 | 5.050E-03 |
| ATF3 | 2.943 | 7.691 | 4.281E-11 | 1.714E-08 |
| NTSR1 | 2.954 | 7.747 | 7.847E-06 | 7.094E-04 |
| SLC22A16 | 2.984 | 7.913 | 1.026E-03 | 2.696E-02 |
| TP53INP2 | 2.990 | 7.943 | 1.099E-26 | 9.240E-23 |
| TMEM119 | 3.002 | 8.008 | 1.083E-03 | 2.809E-02 |
| CCL2 | 3.020 | 8.110 | 2.218E-04 | 9.420E-03 |
| THNSL2 | 3.021 | 8.116 | 2.647E-05 | 1.896E-03 |
| RNVU1-3 | 3.027 | 8.148 | 7.264E-04 | 2.138E-02 |
| ENSG00000270681 | 3.038 | 8.212 | 7.719E-04 | 2.219E-02 |
| PANX2 | 3.042 | 8.235 | 6.241E-05 | 3.645E-03 |
| TAMALIN | 3.044 | 8.249 | 1.567E-16 | 2.196E-13 |
| TNFSF9 | 3.078 | 8.446 | 1.045E-11 | 5.287E-09 |
| NRIP3 | 3.082 | 8.467 | 4.518E-05 | 2.845E-03 |
| TNF | 3.092 | 8.526 | 8.842E-10 | 2.703E-07 |
| MT1XP1 | 3.114 | 8.655 | 1.089E-04 | 5.419E-03 |
| FCAR | 3.125 | 8.726 | 8.222E-07 | 1.032E-04 |
| ENSG00000274051 | 3.136 | 8.791 | 6.113E-04 | 1.909E-02 |
| PDGFA-DT | 3.137 | 8.797 | 1.983E-03 | 4.328E-02 |
| KRT86 | 3.148 | 8.866 | 1.644E-05 | 1.291E-03 |
| LOC399900 | 3.149 | 8.873 | 3.641E-04 | 1.328E-02 |
| TREM1 | 3.164 | 8.963 | 7.190E-10 | 2.325E-07 |
| MIR4420 | 3.198 | 9.176 | 5.616E-14 | 4.267E-11 |
| ENSG00000224356 | 3.202 | 9.202 | 7.259E-06 | 6.634E-04 |
| FOSB | 3.211 | 9.260 | 1.335E-09 | 4.009E-07 |
| OSM | 3.260 | 9.582 | 1.623E-08 | 3.370E-06 |
| C17orf107 | 3.312 | 9.931 | 8.022E-05 | 4.324E-03 |
| HP | 3.333 | 10.078 | 9.749E-04 | 2.615E-02 |
| RNU5D-1 | 3.342 | 10.140 | 1.076E-03 | 2.797E-02 |
| CD83 | 3.385 | 10.448 | 1.852E-33 | 3.114E-29 |
| CXCL2 | 3.402 | 10.573 | 1.213E-06 | 1.427E-04 |
| HBEGF | 3.505 | 11.350 | 1.069E-11 | 5.287E-09 |
| SGK1 | 3.516 | 11.443 | 1.492E-11 | 6.602E-09 |
| B3GNT5 | 3.536 | 11.600 | 7.350E-07 | 9.363E-05 |
| NR4A3 | 3.586 | 12.011 | 1.649E-11 | 6.932E-09 |
| NR4A2 | 3.622 | 12.313 | 6.717E-23 | 1.883E-19 |
| FAM71A | 3.657 | 12.618 | 5.086E-04 | 1.680E-02 |
| ZNF503 | 3.707 | 13.055 | 6.415E-09 | 1.659E-06 |
| LRRC32 | 3.721 | 13.190 | 7.182E-09 | 1.773E-06 |
| ENSG00000280407 | 3.756 | 13.514 | 2.566E-05 | 1.860E-03 |
| GJB6 | 3.764 | 13.585 | 2.250E-04 | 9.505E-03 |
| PHLDA2 | 3.834 | 14.263 | 5.363E-07 | 7.069E-05 |
| THBS1 | 3.897 | 14.896 | 2.312E-03 | 4.767E-02 |
| IL1B | 3.901 | 14.942 | 2.202E-15 | 2.645E-12 |
| SEMA6B | 3.922 | 15.161 | 1.147E-04 | 5.658E-03 |
| RNVU1-6 | 3.981 | 15.794 | 1.348E-04 | 6.387E-03 |
| RNF152 | 4.010 | 16.115 | 2.091E-03 | 4.490E-02 |
| SPX | 4.028 | 16.315 | 1.538E-03 | 3.597E-02 |
| SNORD3B-2 | 4.092 | 17.048 | 2.855E-04 | 1.124E-02 |
| ENSG00000218809 | 4.104 | 17.192 | 1.987E-08 | 3.966E-06 |
| RN7SL368P | 4.609 | 24.404 | 7.879E-05 | 4.282E-03 |
| TEX45 | 4.751 | 26.919 | 6.112E-04 | 1.909E-02 |
| CCL3L3 | 4.768 | 27.241 | 5.399E-04 | 1.756E-02 |
| EGR1 | 4.840 | 28.632 | 2.278E-04 | 9.579E-03 |
| G0S2 | 4.981 | 31.578 | 5.628E-20 | 1.352E-16 |
| CXCL8 | 4.995 | 31.887 | 9.194E-05 | 4.801E-03 |
| EGR3 | 5.076 | 33.732 | 4.196E-04 | 1.470E-02 |
| ADRA2B | 5.110 | 34.527 | 2.412E-04 | 9.943E-03 |
| LINC01220 | 5.216 | 37.168 | 6.991E-05 | 3.955E-03 |
| SLED1 | 5.246 | 37.958 | 1.372E-06 | 1.603E-04 |
| ID1 | 5.485 | 44.786 | 4.129E-09 | 1.138E-06 |
| ENSG00000224029 | 5.501 | 45.302 | 4.341E-04 | 1.508E-02 |
| EGR2 | 5.788 | 55.266 | 3.139E-05 | 2.137E-03 |
| MMP2-AS1 | 6.120 | 69.545 | 1.158E-03 | 2.943E-02 |
| ENSG00000258413 | 6.192 | 73.113 | 1.445E-03 | 3.448E-02 |
| FOSL1 | 6.357 | 81.941 | 7.520E-19 | 1.581E-15 |
| CXCL1 | 6.439 | 86.742 | 7.316E-04 | 2.147E-02 |
| LERFS | 6.597 | 96.823 | 4.893E-04 | 1.652E-02 |
| ENSG00000261026 | 6.791 | 110.769 | 6.213E-05 | 3.645E-03 |
| CLLU1-AS1 | 8.387 | 334.831 | 1.475E-09 | 4.350E-07 |
| AD aged >6 mo (n = 5) versus AD aged <6 mo (n = 3) | ||||
| WASHC1 | ‒2.328 | 0.199 | 3.255E-10 | 5.279E-06 |
| MYO18B | ‒4.947 | 0.032 | 2.603E-06 | 1.477E-02 |
| IFI27 | ‒3.795 | 0.072 | 2.732E-06 | 1.477E-02 |
| NRIR | ‒2.206 | 0.217 | 8.777E-06 | 3.558E-02 |
| HCs aged >6 mo (n = 2) versus HCs aged <6 mo (n = 3) | ||||
| ENSG00000213058 | ‒7.336 | 0.006 | 8.210E-06 | 3.863E-04 |
| RPS14P1 | ‒6.971 | 0.008 | 4.300E-05 | 1.468E-03 |
| RPL23P3 | ‒6.413 | 0.012 | 5.931E-04 | 1.061E-02 |
| CLEC4F | ‒5.654 | 0.020 | 2.039E-03 | 2.629E-02 |
| FCGR3B | ‒4.753 | 0.037 | 4.584E-03 | 4.626E-02 |
| IFIT3 | ‒4.526 | 0.043 | 6.902E-04 | 1.182E-02 |
| AGAP14P | ‒4.423 | 0.047 | 2.570E-05 | 9.886E-04 |
| NAALAD2 | ‒4.269 | 0.052 | 3.906E-03 | 4.164E-02 |
| IFIT1 | ‒4.029 | 0.061 | 3.432E-03 | 3.804E-02 |
| P2RY12 | ‒4.015 | 0.062 | 4.380E-04 | 8.525E-03 |
| ENSG00000254851 | ‒3.978 | 0.063 | 2.595E-03 | 3.152E-02 |
| EFHB | ‒3.961 | 0.064 | 1.762E-03 | 2.363E-02 |
| PFN1P1 | ‒3.950 | 0.065 | 3.605E-03 | 3.958E-02 |
| CXCL10 | ‒3.923 | 0.066 | 3.740E-07 | 2.960E-05 |
| L1TD1 | ‒3.846 | 0.070 | 3.650E-08 | 3.750E-06 |
| FPR2 | ‒3.678 | 0.078 | 4.660E-13 | 1.850E-10 |
| GPR20 | ‒3.606 | 0.082 | 5.070E-03 | 4.963E-02 |
| LINC02432 | ‒3.506 | 0.088 | 8.400E-07 | 5.800E-05 |
| ENSG00000276758 | ‒3.502 | 0.088 | 2.538E-03 | 3.100E-02 |
| ANKRD22 | ‒3.458 | 0.091 | 6.360E-06 | 3.128E-04 |
| ZBED2 | ‒3.352 | 0.098 | 2.091E-04 | 5.014E-03 |
| P2RY13 | ‒3.293 | 0.102 | 2.920E-10 | 5.790E-08 |
| PPP1R17 | ‒3.272 | 0.104 | 1.290E-08 | 1.500E-06 |
| NAGS | ‒3.192 | 0.109 | 2.293E-03 | 2.863E-02 |
| FAM20A | ‒3.164 | 0.112 | 2.420E-07 | 2.090E-05 |
| FILIP1L | ‒3.149 | 0.113 | 5.340E-04 | 9.756E-03 |
| ASH2LP1 | ‒3.135 | 0.114 | 4.320E-08 | 4.420E-06 |
| LINC01506 | ‒3.090 | 0.117 | 1.024E-03 | 1.591E-02 |
| CCDC121 | ‒3.043 | 0.121 | 1.007E-03 | 1.572E-02 |
| NDN | ‒2.991 | 0.126 | 1.018E-03 | 1.585E-02 |
| CX3CR1 | ‒2.971 | 0.128 | 1.589E-04 | 4.006E-03 |
| SCARA5 | ‒2.957 | 0.129 | 9.880E-04 | 1.550E-02 |
| GPR82 | ‒2.940 | 0.130 | 3.116E-03 | 3.563E-02 |
| RAVER2 | ‒2.931 | 0.131 | 4.369E-03 | 4.490E-02 |
| C3AR1 | ‒2.920 | 0.132 | 7.920E-12 | 2.400E-09 |
| SFTPD | ‒2.851 | 0.139 | 1.863E-03 | 2.459E-02 |
| GATA2 | ‒2.842 | 0.139 | 4.630E-04 | 8.797E-03 |
| CYP4F22 | ‒2.805 | 0.143 | 1.490E-06 | 9.340E-05 |
| PTGDR2 | ‒2.782 | 0.145 | 3.070E-14 | 1.600E-11 |
| NA | ‒2.742 | 0.149 | 2.330E-07 | 2.020E-05 |
| OLFML2B | ‒2.707 | 0.153 | 2.643E-03 | 3.192E-02 |
| OAS1 | ‒2.704 | 0.153 | 9.279E-04 | 1.478E-02 |
| CXCR1 | ‒2.653 | 0.159 | 2.508E-04 | 5.733E-03 |
| TLR8 | ‒2.651 | 0.159 | 1.510E-09 | 2.310E-07 |
| NLRC4 | ‒2.642 | 0.160 | 3.610E-06 | 2.014E-04 |
| CCR2 | ‒2.640 | 0.160 | 9.070E-13 | 3.360E-10 |
| S1PR3 | ‒2.621 | 0.163 | 2.945E-03 | 3.436E-02 |
| CD180 | ‒2.618 | 0.163 | 5.740E-18 | 5.030E-15 |
| ENSG00000261655 | ‒2.597 | 0.165 | 1.793E-04 | 4.432E-03 |
| RET | ‒2.594 | 0.166 | 6.200E-05 | 1.931E-03 |
| TMEM51 | ‒2.583 | 0.167 | 3.675E-03 | 4.002E-02 |
| HGF | ‒2.582 | 0.167 | 6.970E-05 | 2.100E-03 |
| IGSF6 | ‒2.544 | 0.171 | 3.770E-07 | 2.960E-05 |
| XAF1 | ‒2.542 | 0.172 | 4.601E-03 | 4.632E-02 |
| ENSG00000244167 | ‒2.540 | 0.172 | 5.150E-05 | 1.696E-03 |
| CISH | ‒2.536 | 0.172 | 7.870E-16 | 4.860E-13 |
| SIGLEC5 | ‒2.509 | 0.176 | 2.174E-04 | 5.166E-03 |
| C3 | ‒2.503 | 0.176 | 5.271E-04 | 9.661E-03 |
| MOCS2-DT | ‒2.497 | 0.177 | 2.500E-06 | 1.454E-04 |
| TNFSF10 | ‒2.480 | 0.179 | 3.380E-20 | 4.340E-17 |
| CEACAM3 | ‒2.413 | 0.188 | 5.356E-04 | 9.773E-03 |
| AATBC | ‒2.413 | 0.188 | 2.713E-03 | 3.243E-02 |
| KCNE3 | ‒2.388 | 0.191 | 1.140E-05 | 5.160E-04 |
| PRICKLE1 | ‒2.382 | 0.192 | 8.130E-07 | 5.640E-05 |
| HSPA1A | ‒2.329 | 0.199 | 2.020E-11 | 5.360E-09 |
| HECW2 | ‒2.328 | 0.199 | 3.273E-03 | 3.677E-02 |
| PDK4 | ‒2.317 | 0.201 | 2.060E-10 | 4.460E-08 |
| DAZL | ‒2.287 | 0.205 | 3.266E-03 | 3.675E-02 |
| CDKN1C | ‒2.230 | 0.213 | 2.010E-19 | 2.230E-16 |
| CARD6 | ‒2.187 | 0.220 | 2.910E-11 | 7.450E-09 |
| ICAM4 | ‒2.168 | 0.222 | 2.367E-04 | 5.508E-03 |
| MYOF | ‒2.148 | 0.226 | 3.050E-06 | 1.735E-04 |
| FPR1 | ‒2.123 | 0.230 | 2.210E-26 | 5.830E-23 |
| FGL2 | ‒2.122 | 0.230 | 3.672E-03 | 4.001E-02 |
| LILRA6 | ‒2.119 | 0.230 | 1.780E-05 | 7.353E-04 |
| TLR10 | ‒2.117 | 0.231 | 2.050E-11 | 5.360E-09 |
| CCR1 | ‒2.092 | 0.235 | 2.990E-08 | 3.170E-06 |
| RGS18 | ‒2.089 | 0.235 | 4.505E-04 | 8.648E-03 |
| CXCR2 | ‒2.082 | 0.236 | 3.500E-05 | 1.250E-03 |
| GTF2H2 | ‒2.073 | 0.238 | 1.280E-07 | 1.210E-05 |
| FZD1 | ‒2.061 | 0.240 | 4.600E-05 | 1.547E-03 |
| BATF2 | ‒2.057 | 0.240 | 3.069E-03 | 3.531E-02 |
| SOWAHD | ‒2.056 | 0.241 | 1.090E-06 | 7.290E-05 |
| IRAG1 | ‒2.052 | 0.241 | 4.760E-07 | 3.590E-05 |
| MIR5195 | ‒2.035 | 0.244 | 6.010E-06 | 2.991E-04 |
| CDC42EP2 | ‒2.033 | 0.244 | 5.500E-05 | 1.775E-03 |
| ENSG00000227615 | ‒2.030 | 0.245 | 6.680E-10 | 1.140E-07 |
| GPBAR1 | ‒2.010 | 0.248 | 4.630E-09 | 6.270E-07 |
| OAS2 | ‒2.009 | 0.248 | 4.692E-03 | 4.699E-02 |
| LINC01504 | ‒1.998 | 0.250 | 6.430E-05 | 1.983E-03 |
| LRRC25 | ‒1.991 | 0.252 | 1.420E-08 | 1.630E-06 |
| SLC31A2 | ‒1.987 | 0.252 | 3.510E-07 | 2.810E-05 |
| SENCR | ‒1.986 | 0.252 | 3.747E-03 | 4.041E-02 |
| PELATON | ‒1.984 | 0.253 | 1.100E-11 | 3.110E-09 |
| GAPT | ‒1.978 | 0.254 | 2.060E-11 | 5.360E-09 |
| IGHV5-78 | ‒1.977 | 0.254 | 1.280E-08 | 1.500E-06 |
| TMEM60 | ‒1.973 | 0.255 | 1.690E-09 | 2.550E-07 |
| ROR1 | ‒1.962 | 0.257 | 3.031E-03 | 3.500E-02 |
| TLR7 | ‒1.961 | 0.257 | 2.450E-08 | 2.670E-06 |
| FFAR2 | ‒1.938 | 0.261 | 9.490E-05 | 2.687E-03 |
| HSPA6 | ‒1.938 | 0.261 | 3.690E-04 | 7.515E-03 |
| FCGR3A | ‒1.937 | 0.261 | 1.270E-13 | 5.880E-11 |
| NFE2 | ‒1.908 | 0.266 | 1.430E-08 | 1.640E-06 |
| FNDC5 | ‒1.900 | 0.268 | 1.033E-04 | 2.882E-03 |
| SAMD9L | ‒1.876 | 0.273 | 1.471E-03 | 2.076E-02 |
| GEMIN6 | ‒1.873 | 0.273 | 5.100E-07 | 3.780E-05 |
| FMNL2 | ‒1.871 | 0.273 | 1.701E-04 | 4.248E-03 |
| IGLV3-27 | ‒1.864 | 0.275 | 1.210E-05 | 5.409E-04 |
| PRSS30P | ‒1.864 | 0.275 | 1.497E-03 | 2.106E-02 |
| MS4A7 | ‒1.862 | 0.275 | 3.310E-08 | 3.460E-06 |
| GIMAP4 | ‒1.853 | 0.277 | 1.070E-26 | 3.560E-23 |
| SAMD9 | ‒1.848 | 0.278 | 4.260E-10 | 7.880E-08 |
| FZD2 | ‒1.845 | 0.278 | 3.042E-03 | 3.507E-02 |
| ISL2 | ‒1.841 | 0.279 | 4.605E-03 | 4.633E-02 |
| LINC02576 | ‒1.838 | 0.280 | 3.872E-03 | 4.140E-02 |
| ENSG00000257275 | ‒1.817 | 0.284 | 4.550E-07 | 3.460E-05 |
| ERAP2 | ‒1.808 | 0.286 | 7.980E-32 | 6.650E-28 |
| CCDC126 | ‒1.803 | 0.287 | 3.840E-05 | 1.343E-03 |
| TASL | ‒1.801 | 0.287 | 3.530E-14 | 1.780E-11 |
| SOS1-IT1 | ‒1.799 | 0.287 | 5.103E-04 | 9.447E-03 |
| AQP9 | ‒1.797 | 0.288 | 3.120E-05 | 1.147E-03 |
| LOC105377623 | ‒1.790 | 0.289 | 2.519E-03 | 3.082E-02 |
| UGGT2 | ‒1.754 | 0.296 | 1.282E-03 | 1.885E-02 |
| CD24 | ‒1.748 | 0.298 | 8.600E-09 | 1.090E-06 |
| FKBPL | ‒1.747 | 0.298 | 1.998E-04 | 4.838E-03 |
| PLA2G2D | ‒1.745 | 0.298 | 1.614E-03 | 2.228E-02 |
| OLIG1 | ‒1.737 | 0.300 | 9.447E-04 | 1.503E-02 |
| KLK1 | ‒1.735 | 0.300 | 4.544E-03 | 4.608E-02 |
| TCL1A | ‒1.731 | 0.301 | 3.480E-10 | 6.590E-08 |
| PARS2 | ‒1.729 | 0.302 | 7.571E-04 | 1.266E-02 |
| ENSG00000261222 | ‒1.720 | 0.303 | 5.548E-04 | 1.010E-02 |
| MTMR11 | ‒1.714 | 0.305 | 2.341E-04 | 5.470E-03 |
| CALHM2 | ‒1.713 | 0.305 | 1.930E-09 | 2.870E-07 |
| MIR223HG | ‒1.712 | 0.305 | 8.440E-14 | 4.020E-11 |
| MSR1 | ‒1.710 | 0.306 | 4.511E-03 | 4.591E-02 |
| ENSG00000279696 | ‒1.702 | 0.307 | 4.450E-06 | 2.370E-04 |
| MGC16275 | ‒1.696 | 0.309 | 1.449E-03 | 2.056E-02 |
| SMCO4 | ‒1.677 | 0.313 | 1.040E-06 | 6.980E-05 |
| GCNT1 | ‒1.664 | 0.315 | 1.510E-13 | 6.640E-11 |
| GUCY1B1 | ‒1.657 | 0.317 | 2.921E-03 | 3.417E-02 |
| SPTA1 | ‒1.648 | 0.319 | 5.100E-03 | 4.983E-02 |
| CEBPA | ‒1.638 | 0.321 | 1.121E-04 | 3.076E-03 |
| LINC01355 | ‒1.635 | 0.322 | 2.380E-10 | 4.970E-08 |
| CKB | ‒1.632 | 0.323 | 5.940E-05 | 1.885E-03 |
| SLAMF8 | ‒1.618 | 0.326 | 1.039E-03 | 1.606E-02 |
| LPCAT2 | ‒1.614 | 0.327 | 2.918E-03 | 3.417E-02 |
| HS3ST1 | ‒1.594 | 0.331 | 1.160E-07 | 1.110E-05 |
| C9orf64 | ‒1.583 | 0.334 | 5.090E-06 | 2.633E-04 |
| C1orf220 | ‒1.583 | 0.334 | 8.730E-06 | 4.051E-04 |
| TRIM69 | ‒1.581 | 0.334 | 6.100E-07 | 4.400E-05 |
| B3GNT8 | ‒1.580 | 0.334 | 2.682E-03 | 3.222E-02 |
| KCNQ1OT1 | ‒1.575 | 0.336 | 1.056E-04 | 2.932E-03 |
| DTX3L | ‒1.568 | 0.337 | 1.530E-05 | 6.538E-04 |
| FCRLB | ‒1.566 | 0.338 | 1.747E-03 | 2.346E-02 |
| SECTM1 | ‒1.563 | 0.339 | 6.220E-05 | 1.935E-03 |
| LMO2 | ‒1.553 | 0.341 | 6.500E-05 | 1.997E-03 |
| C14orf119 | ‒1.553 | 0.341 | 1.010E-15 | 6.010E-13 |
| ENSG00000279611 | ‒1.546 | 0.343 | 4.701E-03 | 4.702E-02 |
| HMOX1 | ‒1.543 | 0.343 | 1.680E-05 | 7.022E-04 |
| ADPRH | ‒1.538 | 0.344 | 8.100E-06 | 3.835E-04 |
| MIR4645 | ‒1.537 | 0.345 | 2.188E-03 | 2.774E-02 |
| ATP9A | ‒1.537 | 0.345 | 9.535E-04 | 1.514E-02 |
| TNFAIP8L2 | ‒1.533 | 0.346 | 1.380E-13 | 6.210E-11 |
| TNFRSF8 | ‒1.532 | 0.346 | 5.600E-07 | 4.090E-05 |
| CD101 | ‒1.532 | 0.346 | 1.404E-03 | 2.012E-02 |
| LOC100287896 | ‒1.532 | 0.346 | 3.405E-03 | 3.785E-02 |
| MAFB | ‒1.528 | 0.347 | 2.330E-07 | 2.020E-05 |
| IGHJ3 | ‒1.527 | 0.347 | 4.816E-03 | 4.776E-02 |
| DUSP6 | ‒1.524 | 0.348 | 2.180E-09 | 3.180E-07 |
| ZNF404 | ‒1.523 | 0.348 | 6.190E-05 | 1.930E-03 |
| HECW2-AS1 | ‒1.521 | 0.348 | 4.760E-05 | 1.584E-03 |
| CCR5 | ‒1.521 | 0.348 | 4.240E-07 | 3.270E-05 |
| RTP4 | ‒1.518 | 0.349 | 6.874E-04 | 1.179E-02 |
| ZNF594 | ‒1.514 | 0.350 | 1.640E-08 | 1.850E-06 |
| LINC00324 | ‒1.511 | 0.351 | 9.090E-07 | 6.230E-05 |
| ZNF2 | ‒1.507 | 0.352 | 3.583E-04 | 7.393E-03 |
| CPM | ‒1.504 | 0.353 | 2.150E-04 | 5.116E-03 |
| LILRB3 | ‒1.502 | 0.353 | 7.820E-06 | 3.756E-04 |
| LACTB2 | ‒1.501 | 0.353 | 1.611E-04 | 4.048E-03 |
| TREML2 | ‒1.488 | 0.357 | 3.180E-07 | 2.580E-05 |
| HYAL2 | ‒1.475 | 0.360 | 8.192E-04 | 1.342E-02 |
| ABCC3 | ‒1.473 | 0.360 | 1.356E-03 | 1.967E-02 |
| APOBEC3A | ‒1.471 | 0.361 | 9.350E-05 | 2.667E-03 |
| SLC37A2 | ‒1.456 | 0.364 | 4.837E-04 | 9.086E-03 |
| FASLG | ‒1.456 | 0.365 | 5.238E-04 | 9.622E-03 |
| C2orf74-DT | ‒1.454 | 0.365 | 7.110E-04 | 1.209E-02 |
| SIGLEC9 | ‒1.452 | 0.365 | 6.010E-04 | 1.072E-02 |
| METTL7A | ‒1.451 | 0.366 | 4.190E-09 | 5.770E-07 |
| MERTK | ‒1.440 | 0.368 | 5.084E-04 | 9.422E-03 |
| MPEG1 | ‒1.436 | 0.370 | 2.350E-06 | 1.372E-04 |
| ZNF772 | ‒1.432 | 0.371 | 2.930E-08 | 3.130E-06 |
| ENSG00000213976 | ‒1.432 | 0.371 | 2.061E-04 | 4.963E-03 |
| EEF1AKNMT | ‒1.431 | 0.371 | 1.190E-12 | 4.200E-10 |
| FANCL | ‒1.431 | 0.371 | 1.368E-03 | 1.974E-02 |
| PTPN13 | ‒1.429 | 0.371 | 1.868E-03 | 2.465E-02 |
| CARD8-AS1 | ‒1.423 | 0.373 | 9.660E-08 | 9.580E-06 |
| EOMES | ‒1.417 | 0.374 | 2.810E-05 | 1.047E-03 |
| ARHGEF2-AS2 | ‒1.417 | 0.375 | 8.446E-04 | 1.370E-02 |
| RIN2 | ‒1.410 | 0.376 | 6.321E-04 | 1.113E-02 |
| DOK1 | ‒1.409 | 0.376 | 7.160E-10 | 1.200E-07 |
| RPP40 | ‒1.408 | 0.377 | 9.037E-04 | 1.444E-02 |
| IKBIP | ‒1.406 | 0.377 | 9.800E-05 | 2.758E-03 |
| PPIL1 | ‒1.403 | 0.378 | 1.960E-06 | 1.186E-04 |
| IL12RB2 | ‒1.401 | 0.379 | 3.335E-03 | 3.732E-02 |
| ZNF28 | ‒1.395 | 0.380 | 2.710E-10 | 5.540E-08 |
| CALHM6 | ‒1.394 | 0.381 | 2.730E-06 | 1.585E-04 |
| BATF3 | ‒1.392 | 0.381 | 9.785E-04 | 1.544E-02 |
| PLEKHO2 | ‒1.387 | 0.382 | 3.450E-05 | 1.240E-03 |
| CFD | ‒1.386 | 0.383 | 1.290E-05 | 5.688E-04 |
| PAQR8 | ‒1.378 | 0.385 | 1.370E-15 | 7.600E-13 |
| LYL1 | ‒1.377 | 0.385 | 3.210E-09 | 4.490E-07 |
| MME | ‒1.375 | 0.385 | 1.296E-04 | 3.434E-03 |
| ACKR3 | ‒1.374 | 0.386 | 2.048E-03 | 2.637E-02 |
| ERI2 | ‒1.373 | 0.386 | 3.010E-03 | 3.486E-02 |
| MIR3142HG | ‒1.372 | 0.386 | 1.712E-03 | 2.314E-02 |
| LOC100507642 | ‒1.370 | 0.387 | 7.370E-04 | 1.244E-02 |
| PHF23 | ‒1.358 | 0.390 | 9.300E-13 | 3.370E-10 |
| SLC1A5 | ‒1.354 | 0.391 | 1.930E-07 | 1.740E-05 |
| FCGR2A | ‒1.350 | 0.392 | 2.890E-03 | 3.394E-02 |
| TOR4A | ‒1.347 | 0.393 | 5.020E-07 | 3.740E-05 |
| GBP1 | ‒1.347 | 0.393 | 7.940E-07 | 5.560E-05 |
| TRMT5 | ‒1.336 | 0.396 | 3.285E-04 | 6.876E-03 |
| ZNF583 | ‒1.334 | 0.397 | 7.960E-06 | 3.809E-04 |
| HSD17B7P2 | ‒1.331 | 0.397 | 1.180E-06 | 7.710E-05 |
| NEURL1 | ‒1.331 | 0.397 | 2.470E-07 | 2.110E-05 |
| TMEM187 | ‒1.331 | 0.398 | 4.428E-04 | 8.558E-03 |
| IRAK3 | ‒1.329 | 0.398 | 1.075E-03 | 1.652E-02 |
| FCRLA | ‒1.323 | 0.400 | 1.210E-05 | 5.393E-04 |
| UBE2T | ‒1.321 | 0.400 | 1.458E-03 | 2.065E-02 |
| PCTP | ‒1.315 | 0.402 | 5.240E-05 | 1.724E-03 |
| PLP2 | ‒1.314 | 0.402 | 9.830E-04 | 1.544E-02 |
| LILRA2 | ‒1.314 | 0.402 | 2.914E-03 | 3.416E-02 |
| FAM111A-DT | ‒1.309 | 0.404 | 2.550E-05 | 9.866E-04 |
| SCIMP | ‒1.304 | 0.405 | 4.207E-04 | 8.286E-03 |
| HOMEZ | ‒1.295 | 0.408 | 1.380E-07 | 1.290E-05 |
| CEP19 | ‒1.295 | 0.408 | 2.026E-03 | 2.618E-02 |
| ZNF613 | ‒1.289 | 0.409 | 3.699E-04 | 7.515E-03 |
| LPAR6 | ‒1.288 | 0.410 | 1.960E-09 | 2.890E-07 |
| SRD5A3 | ‒1.288 | 0.410 | 7.341E-04 | 1.240E-02 |
| SLAMF7 | ‒1.286 | 0.410 | 1.300E-05 | 5.718E-04 |
| ZNF626 | ‒1.284 | 0.411 | 1.163E-03 | 1.753E-02 |
| DNASE1L3 | ‒1.283 | 0.411 | 3.735E-03 | 4.040E-02 |
| STAT2 | ‒1.263 | 0.417 | 2.690E-05 | 1.015E-03 |
| TBC1D8 | ‒1.261 | 0.417 | 4.760E-05 | 1.584E-03 |
| CPPED1 | ‒1.260 | 0.418 | 2.761E-03 | 3.279E-02 |
| ASCL2 | ‒1.254 | 0.419 | 1.172E-04 | 3.191E-03 |
| LBH | ‒1.253 | 0.420 | 3.350E-11 | 8.220E-09 |
| CENPBD1 | ‒1.251 | 0.420 | 1.053E-03 | 1.624E-02 |
| ANKRD50 | ‒1.250 | 0.420 | 2.015E-03 | 2.610E-02 |
| SLFN12 | ‒1.243 | 0.423 | 2.490E-07 | 2.110E-05 |
| ZNF181 | ‒1.238 | 0.424 | 3.000E-05 | 1.115E-03 |
| STAT1 | ‒1.237 | 0.424 | 2.053E-04 | 4.963E-03 |
| CLEC4C | ‒1.236 | 0.424 | 8.864E-04 | 1.424E-02 |
| S100A11 | ‒1.235 | 0.425 | 2.330E-05 | 9.237E-04 |
| UGDH | ‒1.232 | 0.426 | 2.366E-03 | 2.935E-02 |
| TMEM102 | ‒1.231 | 0.426 | 1.510E-05 | 6.490E-04 |
| TNFSF13B | ‒1.229 | 0.426 | 4.534E-03 | 4.603E-02 |
| NA | ‒1.229 | 0.427 | 3.618E-04 | 7.403E-03 |
| KIAA0040 | ‒1.223 | 0.428 | 3.210E-07 | 2.600E-05 |
| MKKS | ‒1.221 | 0.429 | 1.460E-07 | 1.350E-05 |
| IL18 | ‒1.219 | 0.430 | 4.469E-03 | 4.562E-02 |
| C5AR2 | ‒1.218 | 0.430 | 1.241E-03 | 1.833E-02 |
| IGHV1-46 | ‒1.215 | 0.431 | 3.240E-05 | 1.176E-03 |
| ARL11 | ‒1.214 | 0.431 | 4.450E-04 | 8.582E-03 |
| INTS5 | ‒1.211 | 0.432 | 1.080E-07 | 1.050E-05 |
| FCF1P2 | ‒1.210 | 0.432 | 1.403E-03 | 2.012E-02 |
| APOBEC3G | ‒1.207 | 0.433 | 2.488E-04 | 5.708E-03 |
| ZNNT1 | ‒1.206 | 0.433 | 5.280E-05 | 1.725E-03 |
| RMI2 | ‒1.202 | 0.435 | 1.260E-03 | 1.855E-02 |
| WDCP | ‒1.201 | 0.435 | 6.150E-06 | 3.038E-04 |
| TNFSF13 | ‒1.201 | 0.435 | 2.655E-03 | 3.196E-02 |
| DUSP18 | ‒1.200 | 0.435 | 5.990E-05 | 1.895E-03 |
| HCK | ‒1.199 | 0.435 | 4.173E-04 | 8.248E-03 |
| PILRA | ‒1.198 | 0.436 | 2.697E-03 | 3.232E-02 |
| BBS10 | ‒1.198 | 0.436 | 1.015E-04 | 2.848E-03 |
| ATP6AP1L | ‒1.198 | 0.436 | 1.217E-03 | 1.813E-02 |
| HSD17B1-AS1 | ‒1.196 | 0.436 | 1.510E-07 | 1.390E-05 |
| POLR1G | ‒1.196 | 0.436 | 1.774E-03 | 2.370E-02 |
| LRRN3 | ‒1.195 | 0.437 | 3.750E-05 | 1.317E-03 |
| USP27X | ‒1.192 | 0.438 | 2.640E-04 | 5.947E-03 |
| LOC101927151 | ‒1.191 | 0.438 | 1.292E-04 | 3.433E-03 |
| TNFRSF1A | ‒1.188 | 0.439 | 5.630E-07 | 4.090E-05 |
| DDX60 | ‒1.185 | 0.440 | 3.927E-03 | 4.173E-02 |
| TOB2 | ‒1.183 | 0.440 | 3.710E-07 | 2.950E-05 |
| PECAM1 | ‒1.183 | 0.440 | 5.970E-10 | 1.030E-07 |
| PNP | ‒1.183 | 0.441 | 5.320E-06 | 2.736E-04 |
| RNASEL | ‒1.182 | 0.441 | 2.120E-12 | 6.910E-10 |
| C1orf162 | ‒1.180 | 0.441 | 4.700E-10 | 8.410E-08 |
| ADAMTSL4 | ‒1.180 | 0.441 | 7.991E-04 | 1.318E-02 |
| BTN3A2 | ‒1.180 | 0.441 | 2.363E-04 | 5.506E-03 |
| GIMAP1 | ‒1.178 | 0.442 | 1.960E-10 | 4.290E-08 |
| JRKL | ‒1.176 | 0.443 | 4.600E-05 | 1.547E-03 |
| IL1RN | ‒1.175 | 0.443 | 9.194E-04 | 1.466E-02 |
| ZNF285 | ‒1.174 | 0.443 | 2.190E-05 | 8.742E-04 |
| TPPP3 | ‒1.174 | 0.443 | 9.537E-04 | 1.514E-02 |
| RNASE6 | ‒1.173 | 0.443 | 2.160E-14 | 1.160E-11 |
| ENSG00000272335 | ‒1.173 | 0.443 | 2.948E-03 | 3.437E-02 |
| LAP3 | ‒1.173 | 0.444 | 8.283E-04 | 1.349E-02 |
| CD86 | ‒1.172 | 0.444 | 5.029E-04 | 9.348E-03 |
| CASP1 | ‒1.172 | 0.444 | 3.550E-06 | 1.983E-04 |
| LTBR | ‒1.170 | 0.445 | 2.660E-03 | 3.200E-02 |
| ADAP2 | ‒1.168 | 0.445 | 3.579E-04 | 7.393E-03 |
| KLRD1 | ‒1.162 | 0.447 | 1.602E-03 | 2.224E-02 |
| VMP1 | ‒1.160 | 0.447 | 7.660E-05 | 2.259E-03 |
| PLEK | ‒1.149 | 0.451 | 1.570E-06 | 9.790E-05 |
| TRAFD1 | ‒1.144 | 0.453 | 6.480E-07 | 4.630E-05 |
| ABI3 | ‒1.144 | 0.453 | 1.180E-06 | 7.710E-05 |
| HAL | ‒1.141 | 0.453 | 2.249E-03 | 2.830E-02 |
| KCTD21 | ‒1.137 | 0.455 | 1.600E-05 | 6.781E-04 |
| ENSG00000256448 | ‒1.137 | 0.455 | 1.332E-03 | 1.940E-02 |
| CYBB | ‒1.135 | 0.455 | 3.195E-04 | 6.754E-03 |
| ENSG00000260285 | ‒1.133 | 0.456 | 1.768E-03 | 2.368E-02 |
| FAM241A | ‒1.132 | 0.456 | 5.080E-04 | 9.422E-03 |
| NMI | ‒1.131 | 0.457 | 2.150E-06 | 1.273E-04 |
| SLC45A3 | ‒1.129 | 0.457 | 3.666E-03 | 3.999E-02 |
| PPP1R18 | ‒1.127 | 0.458 | 5.600E-06 | 2.835E-04 |
| HCG11 | ‒1.127 | 0.458 | 2.639E-04 | 5.947E-03 |
| GIMAP8 | ‒1.124 | 0.459 | 3.390E-10 | 6.580E-08 |
| FCRL5 | ‒1.124 | 0.459 | 6.368E-04 | 1.118E-02 |
| FGD2 | ‒1.123 | 0.459 | 5.260E-05 | 1.724E-03 |
| PTPRO | ‒1.123 | 0.459 | 1.100E-08 | 1.330E-06 |
| PGBD2 | ‒1.122 | 0.460 | 1.100E-07 | 1.060E-05 |
| TPTEP1 | ‒1.120 | 0.460 | 4.080E-05 | 1.408E-03 |
| UTP14C | ‒1.116 | 0.461 | 4.080E-06 | 2.210E-04 |
| INIP | ‒1.115 | 0.462 | 2.610E-05 | 1.000E-03 |
| UBE2L6 | ‒1.113 | 0.462 | 2.381E-03 | 2.943E-02 |
| BTLA | ‒1.111 | 0.463 | 9.090E-06 | 4.206E-04 |
| APOBEC3C | ‒1.109 | 0.464 | 3.420E-10 | 6.580E-08 |
| ZKSCAN7 | ‒1.108 | 0.464 | 4.324E-04 | 8.457E-03 |
| RCBTB2 | ‒1.108 | 0.464 | 1.027E-03 | 1.591E-02 |
| ZNF470 | ‒1.106 | 0.465 | 2.219E-04 | 5.237E-03 |
| ZNF681 | ‒1.105 | 0.465 | 7.397E-04 | 1.247E-02 |
| CD79B | ‒1.105 | 0.465 | 5.150E-06 | 2.657E-04 |
| ENSG00000224376 | ‒1.103 | 0.465 | 3.860E-03 | 4.136E-02 |
| FLVCR1-DT | ‒1.103 | 0.466 | 2.981E-03 | 3.461E-02 |
| KLRK1 | ‒1.102 | 0.466 | 2.644E-03 | 3.192E-02 |
| RASSF4 | ‒1.102 | 0.466 | 1.567E-04 | 3.985E-03 |
| CTSS | ‒1.102 | 0.466 | 4.441E-03 | 4.547E-02 |
| DHFR2 | ‒1.100 | 0.467 | 8.841E-04 | 1.422E-02 |
| SPN | ‒1.100 | 0.467 | 1.170E-06 | 7.680E-05 |
| RALB | ‒1.099 | 0.467 | 1.720E-05 | 7.147E-04 |
| UICLM | ‒1.098 | 0.467 | 7.470E-05 | 2.216E-03 |
| LRIF1 | ‒1.096 | 0.468 | 4.036E-04 | 8.043E-03 |
| GLRX | ‒1.095 | 0.468 | 1.990E-07 | 1.780E-05 |
| TUBB6 | ‒1.095 | 0.468 | 1.079E-04 | 2.991E-03 |
| SLC29A1 | ‒1.095 | 0.468 | 6.040E-05 | 1.898E-03 |
| CD200 | ‒1.094 | 0.469 | 1.800E-06 | 1.105E-04 |
| FCER1G | ‒1.091 | 0.469 | 1.248E-03 | 1.840E-02 |
| IRF5 | ‒1.089 | 0.470 | 8.010E-04 | 1.320E-02 |
| GNS | ‒1.087 | 0.471 | 2.959E-03 | 3.441E-02 |
| TCF7L2 | ‒1.084 | 0.472 | 6.690E-06 | 3.258E-04 |
| ZNF189 | ‒1.084 | 0.472 | 1.020E-07 | 1.010E-05 |
| CMTR2 | ‒1.084 | 0.472 | 1.730E-11 | 4.720E-09 |
| FAM214B | ‒1.083 | 0.472 | 3.420E-08 | 3.560E-06 |
| MARCKS | ‒1.082 | 0.472 | 4.835E-03 | 4.787E-02 |
| GIMAP7 | ‒1.082 | 0.473 | 6.790E-05 | 2.063E-03 |
| LINC01013 | ‒1.081 | 0.473 | 2.989E-04 | 6.441E-03 |
| PUS3 | ‒1.078 | 0.474 | 2.080E-05 | 8.385E-04 |
| SLC15A2 | ‒1.078 | 0.474 | 2.390E-05 | 9.377E-04 |
| GCSAM | ‒1.075 | 0.475 | 8.230E-09 | 1.050E-06 |
| TRGC2 | ‒1.074 | 0.475 | 3.750E-10 | 7.020E-08 |
| FAM110A | ‒1.074 | 0.475 | 7.300E-10 | 1.210E-07 |
| LACTB | ‒1.072 | 0.476 | 2.431E-04 | 5.617E-03 |
| GPATCH11 | ‒1.070 | 0.476 | 8.420E-07 | 5.800E-05 |
| WARS1 | ‒1.069 | 0.477 | 1.080E-11 | 3.090E-09 |
| DYNLL1 | ‒1.068 | 0.477 | 2.960E-06 | 1.686E-04 |
| CHRNB1 | ‒1.068 | 0.477 | 2.403E-04 | 5.577E-03 |
| FCRL3 | ‒1.067 | 0.477 | 2.550E-07 | 2.150E-05 |
| ATP6V1D | ‒1.066 | 0.478 | 1.420E-06 | 8.930E-05 |
| CETN3 | ‒1.064 | 0.478 | 3.247E-04 | 6.821E-03 |
| MRPL18 | ‒1.061 | 0.479 | 2.270E-07 | 2.000E-05 |
| TMEM223 | ‒1.059 | 0.480 | 7.130E-06 | 3.462E-04 |
| PYGO2 | ‒1.059 | 0.480 | 1.300E-08 | 1.500E-06 |
| KYNU | ‒1.057 | 0.481 | 3.775E-03 | 4.063E-02 |
| PLSCR1 | ‒1.055 | 0.481 | 4.601E-03 | 4.632E-02 |
| C5AR1 | ‒1.053 | 0.482 | 2.546E-03 | 3.105E-02 |
| LST1 | ‒1.053 | 0.482 | 3.431E-04 | 7.127E-03 |
| RNF144B | ‒1.051 | 0.483 | 1.890E-05 | 7.709E-04 |
| FAM220A | ‒1.045 | 0.485 | 2.789E-04 | 6.162E-03 |
| RIN1 | ‒1.043 | 0.485 | 1.947E-03 | 2.540E-02 |
| CRTAM | ‒1.041 | 0.486 | 1.554E-03 | 2.168E-02 |
| EVI2B | ‒1.040 | 0.486 | 3.140E-05 | 1.148E-03 |
| LILRB1 | ‒1.039 | 0.487 | 1.270E-05 | 5.627E-04 |
| RAB10 | ‒1.038 | 0.487 | 5.030E-07 | 3.740E-05 |
| IL3RA | ‒1.037 | 0.487 | 1.840E-03 | 2.437E-02 |
| FCGR2B | ‒1.035 | 0.488 | 4.392E-04 | 8.539E-03 |
| DCLRE1B | ‒1.033 | 0.489 | 1.202E-03 | 1.800E-02 |
| SPIB | ‒1.033 | 0.489 | 3.184E-03 | 3.606E-02 |
| ZNF429 | ‒1.033 | 0.489 | 2.480E-04 | 5.700E-03 |
| OPA3 | ‒1.031 | 0.489 | 7.460E-05 | 2.215E-03 |
| GPR171 | ‒1.029 | 0.490 | 2.780E-06 | 1.602E-04 |
| CDC42EP3 | ‒1.029 | 0.490 | 4.700E-03 | 4.702E-02 |
| SIGLEC10 | ‒1.029 | 0.490 | 2.730E-10 | 5.540E-08 |
| TIGD7 | ‒1.026 | 0.491 | 2.080E-05 | 8.376E-04 |
| RNF122 | ‒1.024 | 0.492 | 6.710E-07 | 4.760E-05 |
| SLC7A7 | ‒1.024 | 0.492 | 2.823E-03 | 3.322E-02 |
| KIAA0930 | ‒1.023 | 0.492 | 1.210E-12 | 4.200E-10 |
| ERLIN1 | ‒1.021 | 0.493 | 4.880E-06 | 2.555E-04 |
| RUNDC1 | ‒1.021 | 0.493 | 1.790E-05 | 7.398E-04 |
| ZNF616 | ‒1.019 | 0.493 | 8.790E-05 | 2.521E-03 |
| LINC02397 | ‒1.019 | 0.494 | 2.840E-06 | 1.630E-04 |
| FASTKD1 | ‒1.019 | 0.494 | 1.100E-07 | 1.060E-05 |
| MED11 | ‒1.019 | 0.494 | 5.590E-07 | 4.090E-05 |
| VPREB3 | ‒1.017 | 0.494 | 1.564E-03 | 2.180E-02 |
| WSB1 | ‒1.016 | 0.494 | 1.170E-07 | 1.120E-05 |
| SMIM14 | ‒1.015 | 0.495 | 1.700E-06 | 1.049E-04 |
| LOC101929698 | ‒1.012 | 0.496 | 2.827E-04 | 6.213E-03 |
| SAMD4A | ‒1.011 | 0.496 | 6.553E-04 | 1.142E-02 |
| KEAP1 | ‒1.009 | 0.497 | 5.880E-05 | 1.873E-03 |
| MRPL35 | ‒1.007 | 0.498 | 6.320E-05 | 1.958E-03 |
| DHRS4-AS1 | ‒1.006 | 0.498 | 1.310E-05 | 5.726E-04 |
| NAGA | ‒1.006 | 0.498 | 7.165E-04 | 1.217E-02 |
| TMEM69 | ‒1.004 | 0.499 | 8.350E-06 | 3.911E-04 |
| MED20 | ‒1.002 | 0.499 | 9.210E-06 | 4.249E-04 |
| ZNF747-DT | ‒1.000 | 0.500 | 2.951E-03 | 3.438E-02 |
| NAPSB | ‒0.996 | 0.501 | 6.440E-05 | 1.983E-03 |
| FAM200A | ‒0.996 | 0.501 | 9.017E-04 | 1.444E-02 |
| SLFN11 | ‒0.994 | 0.502 | 8.150E-11 | 1.860E-08 |
| SAC3D1 | ‒0.993 | 0.502 | 8.717E-04 | 1.405E-02 |
| GBGT1 | ‒0.993 | 0.502 | 2.652E-04 | 5.947E-03 |
| HERPUD2-AS1 | ‒0.992 | 0.503 | 1.313E-04 | 3.467E-03 |
| DAPP1 | ‒0.991 | 0.503 | 6.790E-11 | 1.620E-08 |
| TRDC | ‒0.991 | 0.503 | 5.847E-04 | 1.052E-02 |
| THNSL1 | ‒0.988 | 0.504 | 2.648E-03 | 3.192E-02 |
| IFI16 | ‒0.986 | 0.505 | 9.310E-06 | 4.287E-04 |
| POU2F2-AS1 | ‒0.985 | 0.505 | 4.342E-03 | 4.469E-02 |
| LILRA5 | ‒0.984 | 0.506 | 4.738E-03 | 4.716E-02 |
| LILRB2 | ‒0.977 | 0.508 | 3.090E-03 | 3.548E-02 |
| LAT2 | ‒0.974 | 0.509 | 5.718E-04 | 1.033E-02 |
| ALDH6A1 | ‒0.974 | 0.509 | 3.542E-03 | 3.898E-02 |
| SLC43A3 | ‒0.974 | 0.509 | 6.830E-05 | 2.074E-03 |
| AIF1 | ‒0.971 | 0.510 | 2.695E-03 | 3.232E-02 |
| YWHAH | ‒0.968 | 0.511 | 6.491E-04 | 1.134E-02 |
| MPZL1 | ‒0.967 | 0.512 | 5.960E-05 | 1.888E-03 |
| HCP5 | ‒0.965 | 0.512 | 1.300E-07 | 1.230E-05 |
| PRCP | ‒0.964 | 0.513 | 4.170E-05 | 1.428E-03 |
| CD300LB | ‒0.963 | 0.513 | 2.151E-03 | 2.744E-02 |
| C6orf47 | ‒0.962 | 0.513 | 1.250E-05 | 5.549E-04 |
| LPAR5 | ‒0.961 | 0.514 | 7.540E-05 | 2.228E-03 |
| GLIPR1 | ‒0.959 | 0.514 | 1.412E-04 | 3.652E-03 |
| TTC9C | ‒0.958 | 0.515 | 2.160E-05 | 8.654E-04 |
| ARV1 | ‒0.958 | 0.515 | 2.602E-04 | 5.899E-03 |
| SELPLG | ‒0.958 | 0.515 | 3.480E-06 | 1.960E-04 |
| LINC00847 | ‒0.956 | 0.516 | 2.380E-05 | 9.366E-04 |
| QRSL1 | ‒0.955 | 0.516 | 1.520E-10 | 3.370E-08 |
| NCF1 | ‒0.953 | 0.516 | 2.739E-04 | 6.060E-03 |
| RRAS | ‒0.946 | 0.519 | 2.787E-03 | 3.295E-02 |
| TIGD2 | ‒0.945 | 0.519 | 4.460E-04 | 8.591E-03 |
| CYSLTR1 | ‒0.943 | 0.520 | 1.079E-03 | 1.656E-02 |
| TNFRSF1B | ‒0.941 | 0.521 | 5.580E-05 | 1.799E-03 |
| IGLV2-14 | ‒0.939 | 0.521 | 6.289E-04 | 1.109E-02 |
| MCM8 | ‒0.938 | 0.522 | 6.061E-04 | 1.079E-02 |
| WEE1 | ‒0.938 | 0.522 | 4.960E-05 | 1.641E-03 |
| BORCS6 | ‒0.938 | 0.522 | 3.530E-05 | 1.258E-03 |
| GNB4 | ‒0.937 | 0.522 | 1.619E-03 | 2.231E-02 |
| HNRNPF | ‒0.936 | 0.523 | 2.560E-08 | 2.770E-06 |
| PWWP2B | ‒0.936 | 0.523 | 3.854E-03 | 4.133E-02 |
| BTK | ‒0.935 | 0.523 | 8.560E-05 | 2.467E-03 |
| CD1C | ‒0.934 | 0.523 | 6.590E-05 | 2.021E-03 |
| NCOA4 | ‒0.933 | 0.524 | 8.071E-04 | 1.328E-02 |
| POLH | ‒0.932 | 0.524 | 2.100E-04 | 5.021E-03 |
| SKAP2 | ‒0.932 | 0.524 | 1.219E-03 | 1.814E-02 |
| ENSG00000246596 | ‒0.932 | 0.524 | 1.175E-03 | 1.768E-02 |
| RXRA | ‒0.931 | 0.524 | 2.741E-03 | 3.266E-02 |
| ZDHHC7 | ‒0.929 | 0.525 | 3.696E-04 | 7.515E-03 |
| LAX1 | ‒0.929 | 0.525 | 2.646E-04 | 5.947E-03 |
| SDHAF1 | ‒0.927 | 0.526 | 1.095E-03 | 1.674E-02 |
| IFITM2 | ‒0.927 | 0.526 | 3.236E-03 | 3.655E-02 |
| SLC40A1 | ‒0.926 | 0.526 | 2.075E-03 | 2.663E-02 |
| AK6 | ‒0.925 | 0.527 | 9.934E-04 | 1.556E-02 |
| PREPL | ‒0.925 | 0.527 | 1.990E-05 | 8.112E-04 |
| MON1A | ‒0.924 | 0.527 | 1.628E-03 | 2.235E-02 |
| NA | ‒0.924 | 0.527 | 1.399E-03 | 2.011E-02 |
| UEVLD | ‒0.924 | 0.527 | 1.906E-04 | 4.658E-03 |
| IGHV1-2 | ‒0.924 | 0.527 | 2.565E-03 | 3.122E-02 |
| SPTLC2 | ‒0.923 | 0.528 | 1.290E-06 | 8.310E-05 |
| NCF1B | ‒0.921 | 0.528 | 2.070E-06 | 1.247E-04 |
| GGPS1 | ‒0.920 | 0.528 | 1.520E-06 | 9.540E-05 |
| RBBP8 | ‒0.920 | 0.529 | 4.720E-05 | 1.581E-03 |
| ADRB2 | ‒0.917 | 0.530 | 4.135E-03 | 4.321E-02 |
| RHEX | ‒0.917 | 0.530 | 2.283E-03 | 2.857E-02 |
| FAM98B | ‒0.913 | 0.531 | 6.730E-04 | 1.163E-02 |
| SIT1 | ‒0.910 | 0.532 | 1.312E-03 | 1.916E-02 |
| CHST15 | ‒0.910 | 0.532 | 1.979E-04 | 4.799E-03 |
| C17orf80 | ‒0.910 | 0.532 | 1.520E-07 | 1.390E-05 |
| FGR | ‒0.909 | 0.533 | 3.613E-04 | 7.403E-03 |
| C5orf51 | ‒0.906 | 0.534 | 1.616E-03 | 2.229E-02 |
| KCTD15 | ‒0.905 | 0.534 | 2.207E-03 | 2.792E-02 |
| ANXA4 | ‒0.905 | 0.534 | 6.878E-04 | 1.179E-02 |
| HK3 | ‒0.904 | 0.534 | 1.316E-04 | 3.469E-03 |
| CPNE5 | ‒0.904 | 0.534 | 4.818E-03 | 4.776E-02 |
| RWDD2B | ‒0.903 | 0.535 | 2.958E-03 | 3.441E-02 |
| APOBEC3F | ‒0.902 | 0.535 | 1.834E-03 | 2.433E-02 |
| KMO | ‒0.902 | 0.535 | 2.494E-04 | 5.708E-03 |
| GOLIM4 | ‒0.901 | 0.536 | 2.739E-03 | 3.266E-02 |
| CAMK1 | ‒0.899 | 0.536 | 5.070E-03 | 4.963E-02 |
| TOR1B | ‒0.898 | 0.537 | 1.202E-04 | 3.246E-03 |
| MFAP1 | ‒0.898 | 0.537 | 1.393E-03 | 2.004E-02 |
| TMEM106A | ‒0.897 | 0.537 | 4.068E-04 | 8.080E-03 |
| PLAGL2 | ‒0.897 | 0.537 | 3.079E-04 | 6.603E-03 |
| RHOC | ‒0.895 | 0.538 | 4.344E-04 | 8.465E-03 |
| PFKFB4 | ‒0.894 | 0.538 | 2.956E-03 | 3.441E-02 |
| KCTD11 | ‒0.891 | 0.539 | 6.009E-04 | 1.072E-02 |
| ARCN1 | ‒0.891 | 0.539 | 2.290E-07 | 2.010E-05 |
| HAVCR2 | ‒0.891 | 0.539 | 4.945E-03 | 4.875E-02 |
| PIK3AP1 | ‒0.890 | 0.539 | 1.296E-04 | 3.434E-03 |
| EIF2S1 | ‒0.890 | 0.540 | 3.690E-07 | 2.940E-05 |
| NRM | ‒0.889 | 0.540 | 4.771E-03 | 4.743E-02 |
| B3GALT4 | ‒0.887 | 0.541 | 7.940E-05 | 2.334E-03 |
| DAPK1 | ‒0.887 | 0.541 | 6.595E-04 | 1.146E-02 |
| MYD88 | ‒0.887 | 0.541 | 1.820E-05 | 7.493E-04 |
| CARD16 | ‒0.886 | 0.541 | 4.070E-05 | 1.408E-03 |
| COA3 | ‒0.885 | 0.542 | 1.571E-03 | 2.189E-02 |
| MYCBP | ‒0.885 | 0.542 | 4.448E-04 | 8.582E-03 |
| PABIR1 | ‒0.884 | 0.542 | 2.450E-05 | 9.530E-04 |
| TMEM63C | ‒0.884 | 0.542 | 3.373E-03 | 3.762E-02 |
| TMEM250 | ‒0.883 | 0.542 | 3.470E-05 | 1.243E-03 |
| IGLV1-51 | ‒0.882 | 0.543 | 1.730E-03 | 2.330E-02 |
| TRIM21 | ‒0.882 | 0.543 | 1.918E-03 | 2.514E-02 |
| DCLRE1A | ‒0.880 | 0.543 | 2.415E-03 | 2.979E-02 |
| PLD4 | ‒0.879 | 0.544 | 5.697E-04 | 1.033E-02 |
| GPN3 | ‒0.878 | 0.544 | 5.717E-04 | 1.033E-02 |
| HCCS | ‒0.877 | 0.544 | 8.760E-05 | 2.517E-03 |
| FIGNL1 | ‒0.877 | 0.545 | 6.853E-04 | 1.179E-02 |
| ANKEF1 | ‒0.875 | 0.545 | 4.877E-03 | 4.824E-02 |
| ATP23 | ‒0.874 | 0.546 | 1.076E-03 | 1.653E-02 |
| ZFP3 | ‒0.873 | 0.546 | 1.860E-04 | 4.570E-03 |
| AHCYL1 | ‒0.872 | 0.546 | 7.540E-05 | 2.228E-03 |
| CYB561A3 | ‒0.870 | 0.547 | 3.738E-04 | 7.576E-03 |
| NDUFAF1 | ‒0.868 | 0.548 | 7.415E-04 | 1.248E-02 |
| SP110 | ‒0.865 | 0.549 | 1.025E-03 | 1.591E-02 |
| DCAF12 | ‒0.865 | 0.549 | 3.621E-04 | 7.403E-03 |
| TOR1A | ‒0.864 | 0.549 | 5.340E-05 | 1.737E-03 |
| ZNF816 | ‒0.862 | 0.550 | 2.478E-03 | 3.036E-02 |
| CTSC | ‒0.861 | 0.550 | 4.344E-04 | 8.465E-03 |
| IL7R | ‒0.861 | 0.551 | 1.050E-05 | 4.793E-04 |
| SCRN1 | ‒0.860 | 0.551 | 8.573E-04 | 1.387E-02 |
| SNX11 | ‒0.860 | 0.551 | 3.940E-05 | 1.374E-03 |
| DUSP7 | ‒0.859 | 0.551 | 7.930E-04 | 1.311E-02 |
| TMEM186 | ‒0.858 | 0.552 | 1.522E-03 | 2.131E-02 |
| PARP14 | ‒0.857 | 0.552 | 4.924E-03 | 4.857E-02 |
| GVINP1 | ‒0.857 | 0.552 | 6.777E-04 | 1.169E-02 |
| TXNDC9 | ‒0.854 | 0.553 | 3.630E-03 | 3.971E-02 |
| ALAS1 | ‒0.853 | 0.554 | 1.150E-06 | 7.630E-05 |
| CDK14 | ‒0.850 | 0.555 | 2.801E-03 | 3.307E-02 |
| ZNF737 | ‒0.849 | 0.555 | 1.275E-04 | 3.399E-03 |
| VASP | ‒0.848 | 0.555 | 6.150E-05 | 1.925E-03 |
| TLR6 | ‒0.847 | 0.556 | 4.776E-03 | 4.745E-02 |
| IDH1 | ‒0.847 | 0.556 | 5.150E-04 | 9.514E-03 |
| CAMKK2 | ‒0.846 | 0.556 | 3.040E-05 | 1.121E-03 |
| GTF2E1 | ‒0.846 | 0.556 | 3.155E-04 | 6.722E-03 |
| STT3A | ‒0.846 | 0.556 | 7.780E-07 | 5.470E-05 |
| FBXO45 | ‒0.845 | 0.557 | 4.381E-03 | 4.500E-02 |
| DPYSL2 | ‒0.845 | 0.557 | 9.826E-04 | 1.544E-02 |
| SLC25A19 | ‒0.844 | 0.557 | 2.963E-04 | 6.401E-03 |
| MTF1 | ‒0.844 | 0.557 | 3.516E-04 | 7.278E-03 |
| DYNC1I2 | ‒0.843 | 0.557 | 3.290E-06 | 1.857E-04 |
| MGME1 | ‒0.841 | 0.558 | 3.640E-06 | 2.023E-04 |
| CD300A | ‒0.840 | 0.559 | 4.670E-03 | 4.684E-02 |
| LIG4 | ‒0.838 | 0.559 | 1.942E-03 | 2.536E-02 |
| SNAPC3 | ‒0.837 | 0.560 | 1.417E-04 | 3.661E-03 |
| NMT1 | ‒0.836 | 0.560 | 2.370E-05 | 9.334E-04 |
| ZW10 | ‒0.836 | 0.560 | 8.179E-04 | 1.341E-02 |
| CD244 | ‒0.835 | 0.560 | 2.957E-04 | 6.397E-03 |
| ZC3H10 | ‒0.835 | 0.561 | 2.703E-03 | 3.238E-02 |
| ZNF226 | ‒0.835 | 0.561 | 7.200E-06 | 3.487E-04 |
| CHSY1 | ‒0.834 | 0.561 | 2.531E-04 | 5.770E-03 |
| GBP4 | ‒0.833 | 0.561 | 2.213E-03 | 2.796E-02 |
| CHCHD4 | ‒0.832 | 0.562 | 2.198E-03 | 2.784E-02 |
| SH2B2 | ‒0.832 | 0.562 | 4.452E-03 | 4.550E-02 |
| WDR5B | ‒0.828 | 0.563 | 3.222E-04 | 6.787E-03 |
| SLC25A24 | ‒0.828 | 0.563 | 2.719E-03 | 3.246E-02 |
| TRIM68 | ‒0.828 | 0.563 | 1.426E-03 | 2.036E-02 |
| PLPBP | ‒0.825 | 0.565 | 4.750E-05 | 1.584E-03 |
| GVQW3 | ‒0.824 | 0.565 | 1.770E-03 | 2.368E-02 |
| HSPA8 | ‒0.822 | 0.565 | 2.737E-04 | 6.060E-03 |
| CRYZ | ‒0.818 | 0.567 | 2.996E-04 | 6.450E-03 |
| DNAJA1 | ‒0.818 | 0.567 | 3.882E-04 | 7.840E-03 |
| FASTKD3 | ‒0.816 | 0.568 | 8.419E-04 | 1.367E-02 |
| ZKSCAN3 | ‒0.814 | 0.569 | 2.612E-03 | 3.163E-02 |
| MTPN | ‒0.812 | 0.570 | 2.568E-04 | 5.831E-03 |
| CHMP5 | ‒0.812 | 0.570 | 3.901E-03 | 4.162E-02 |
| CTR9 | ‒0.811 | 0.570 | 6.160E-05 | 1.925E-03 |
| SNAPIN | ‒0.811 | 0.570 | 1.112E-04 | 3.059E-03 |
| SLAMF6 | ‒0.807 | 0.572 | 8.430E-05 | 2.444E-03 |
| NMNAT1 | ‒0.806 | 0.572 | 4.680E-03 | 4.691E-02 |
| WASF1 | ‒0.805 | 0.572 | 3.541E-03 | 3.898E-02 |
| ARMT1 | ‒0.804 | 0.573 | 4.017E-03 | 4.246E-02 |
| CHURC1 | ‒0.800 | 0.574 | 3.104E-03 | 3.559E-02 |
| RBM6 | ‒0.800 | 0.574 | 3.925E-04 | 7.917E-03 |
| DCAF7 | ‒0.799 | 0.575 | 5.390E-07 | 3.970E-05 |
| PRDX3 | ‒0.799 | 0.575 | 2.480E-04 | 5.700E-03 |
| HSCB | ‒0.798 | 0.575 | 3.370E-03 | 3.761E-02 |
| RGS19 | ‒0.795 | 0.576 | 1.740E-05 | 7.220E-04 |
| JPT2 | ‒0.795 | 0.577 | 6.867E-04 | 1.179E-02 |
| PLXNB2 | ‒0.794 | 0.577 | 1.454E-03 | 2.061E-02 |
| SLC35A5 | ‒0.793 | 0.577 | 2.649E-04 | 5.947E-03 |
| CD72 | ‒0.791 | 0.578 | 4.041E-04 | 8.043E-03 |
| SPRYD3 | ‒0.791 | 0.578 | 8.190E-05 | 2.384E-03 |
| DARS2 | ‒0.790 | 0.578 | 3.140E-04 | 6.699E-03 |
| ZNF200 | ‒0.788 | 0.579 | 2.690E-05 | 1.015E-03 |
| TMOD3 | ‒0.787 | 0.579 | 1.359E-03 | 1.970E-02 |
| ARL6IP5 | ‒0.787 | 0.580 | 1.060E-07 | 1.040E-05 |
| DCTPP1 | ‒0.786 | 0.580 | 2.789E-03 | 3.295E-02 |
| CALML4 | ‒0.786 | 0.580 | 4.506E-03 | 4.589E-02 |
| YWHAG | ‒0.786 | 0.580 | 2.025E-03 | 2.618E-02 |
| TMBIM1 | ‒0.782 | 0.581 | 2.160E-06 | 1.273E-04 |
| VKORC1L1 | ‒0.779 | 0.583 | 1.136E-04 | 3.103E-03 |
| CNP | ‒0.778 | 0.583 | 3.691E-03 | 4.011E-02 |
| CAP1 | ‒0.776 | 0.584 | 2.844E-04 | 6.234E-03 |
| ZNF234 | ‒0.775 | 0.584 | 1.437E-04 | 3.706E-03 |
| BASP1 | ‒0.773 | 0.585 | 5.920E-05 | 1.881E-03 |
| TBC1D24 | ‒0.771 | 0.586 | 1.164E-03 | 1.754E-02 |
| AHSA1 | ‒0.771 | 0.586 | 4.257E-04 | 8.354E-03 |
| LAMP2 | ‒0.769 | 0.587 | 1.589E-04 | 4.006E-03 |
| SYK | ‒0.769 | 0.587 | 2.178E-03 | 2.772E-02 |
| MRPS18B | ‒0.769 | 0.587 | 1.626E-03 | 2.235E-02 |
| TARS1 | ‒0.769 | 0.587 | 4.520E-05 | 1.530E-03 |
| MCMBP | ‒0.768 | 0.587 | 6.120E-05 | 1.921E-03 |
| OSTM1 | ‒0.768 | 0.587 | 4.755E-04 | 8.963E-03 |
| CD53 | ‒0.767 | 0.588 | 1.440E-05 | 6.211E-04 |
| EVI2A | ‒0.767 | 0.588 | 4.626E-04 | 8.797E-03 |
| B4GALT5 | ‒0.765 | 0.588 | 1.846E-03 | 2.443E-02 |
| TXLNA | ‒0.763 | 0.589 | 1.096E-04 | 3.023E-03 |
| JAK2 | ‒0.763 | 0.589 | 6.770E-05 | 2.063E-03 |
| TPP1 | ‒0.763 | 0.589 | 3.500E-03 | 3.861E-02 |
| BPNT1 | ‒0.762 | 0.590 | 3.441E-03 | 3.811E-02 |
| NDUFA8 | ‒0.762 | 0.590 | 2.972E-03 | 3.453E-02 |
| HLA-DOA | ‒0.762 | 0.590 | 1.671E-04 | 4.181E-03 |
| TAPBPL | ‒0.762 | 0.590 | 2.182E-03 | 2.772E-02 |
| GIMAP6 | ‒0.759 | 0.591 | 3.270E-05 | 1.186E-03 |
| SLC35C1 | ‒0.759 | 0.591 | 2.860E-03 | 3.360E-02 |
| UBXN2B | ‒0.756 | 0.592 | 2.081E-04 | 5.003E-03 |
| ZNF397 | ‒0.756 | 0.592 | 3.820E-07 | 2.990E-05 |
| CEP89 | ‒0.756 | 0.592 | 3.606E-03 | 3.958E-02 |
| PARP4 | ‒0.755 | 0.593 | 2.203E-04 | 5.215E-03 |
| GSTO1 | ‒0.754 | 0.593 | 8.860E-05 | 2.537E-03 |
| IRF2 | ‒0.754 | 0.593 | 5.590E-06 | 2.835E-04 |
| VPS41 | ‒0.753 | 0.593 | 1.407E-03 | 2.013E-02 |
| ZNF213 | ‒0.753 | 0.593 | 3.088E-03 | 3.548E-02 |
| TRIM27 | ‒0.753 | 0.594 | 1.370E-06 | 8.750E-05 |
| YIPF4 | ‒0.752 | 0.594 | 5.883E-04 | 1.055E-02 |
| ACADM | ‒0.752 | 0.594 | 2.713E-03 | 3.243E-02 |
| CAPZA2 | ‒0.751 | 0.594 | 6.572E-04 | 1.144E-02 |
| ZKSCAN4 | ‒0.749 | 0.595 | 1.423E-03 | 2.033E-02 |
| ZNF780B | ‒0.749 | 0.595 | 7.360E-05 | 2.194E-03 |
| TNFAIP1 | ‒0.748 | 0.595 | 2.610E-04 | 5.901E-03 |
| NFE2L1 | ‒0.748 | 0.596 | 8.379E-04 | 1.362E-02 |
| HLA-DPA1 | ‒0.747 | 0.596 | 5.163E-04 | 9.526E-03 |
| GYG1 | ‒0.746 | 0.596 | 1.608E-03 | 2.225E-02 |
| PURA | ‒0.744 | 0.597 | 2.421E-04 | 5.602E-03 |
| LFNG | ‒0.743 | 0.598 | 1.014E-03 | 1.581E-02 |
| C11orf68 | ‒0.742 | 0.598 | 7.261E-04 | 1.231E-02 |
| C2orf42 | ‒0.740 | 0.599 | 2.705E-03 | 3.238E-02 |
| GID4 | ‒0.739 | 0.599 | 1.028E-04 | 2.875E-03 |
| CCDC117 | ‒0.738 | 0.600 | 1.763E-03 | 2.363E-02 |
| TSPAN31 | ‒0.737 | 0.600 | 1.678E-03 | 2.281E-02 |
| PRUNE1 | ‒0.735 | 0.601 | 4.325E-04 | 8.457E-03 |
| SWAP70 | ‒0.734 | 0.601 | 2.087E-04 | 5.011E-03 |
| THRAP3 | ‒0.731 | 0.602 | 8.680E-08 | 8.770E-06 |
| MOB3C | ‒0.730 | 0.603 | 3.011E-03 | 3.486E-02 |
| RAB8A | ‒0.730 | 0.603 | 1.505E-04 | 3.858E-03 |
| RNGTT | ‒0.729 | 0.603 | 4.728E-03 | 4.716E-02 |
| MRPL15 | ‒0.726 | 0.605 | 4.012E-04 | 8.025E-03 |
| DHRS7 | ‒0.724 | 0.606 | 3.188E-04 | 6.754E-03 |
| C1GALT1C1 | ‒0.723 | 0.606 | 3.879E-03 | 4.146E-02 |
| ZNF146 | ‒0.723 | 0.606 | 2.930E-07 | 2.440E-05 |
| SDF2 | ‒0.723 | 0.606 | 2.940E-04 | 6.372E-03 |
| IFNGR2 | ‒0.722 | 0.606 | 4.709E-04 | 8.916E-03 |
| E2F5 | ‒0.722 | 0.606 | 9.697E-04 | 1.533E-02 |
| HVCN1 | ‒0.721 | 0.606 | 3.394E-04 | 7.077E-03 |
| JKAMP | ‒0.721 | 0.607 | 1.312E-03 | 1.916E-02 |
| PRKCD | ‒0.720 | 0.607 | 7.843E-04 | 1.303E-02 |
| RP2 | ‒0.719 | 0.608 | 3.960E-03 | 4.203E-02 |
| STIP1 | ‒0.717 | 0.608 | 6.692E-04 | 1.159E-02 |
| ORMDL2 | ‒0.717 | 0.608 | 1.371E-03 | 1.977E-02 |
| SHMT1 | ‒0.716 | 0.609 | 1.467E-03 | 2.074E-02 |
| SLC25A20 | ‒0.716 | 0.609 | 5.278E-04 | 9.664E-03 |
| HSBP1 | ‒0.715 | 0.609 | 1.230E-04 | 3.306E-03 |
| CTSH | ‒0.714 | 0.610 | 1.713E-03 | 2.314E-02 |
| ARPC5 | ‒0.713 | 0.610 | 1.708E-04 | 4.253E-03 |
| LARS2 | ‒0.712 | 0.611 | 2.247E-04 | 5.287E-03 |
| DOK3 | ‒0.711 | 0.611 | 2.647E-03 | 3.192E-02 |
| BRD8 | ‒0.710 | 0.611 | 4.840E-07 | 3.630E-05 |
| NEK4 | ‒0.708 | 0.612 | 2.598E-03 | 3.152E-02 |
| RBM4B | ‒0.707 | 0.612 | 6.340E-04 | 1.114E-02 |
| ZBTB7B | ‒0.707 | 0.612 | 7.401E-04 | 1.247E-02 |
| SLC35B3 | ‒0.704 | 0.614 | 3.171E-04 | 6.739E-03 |
| HAT1 | ‒0.704 | 0.614 | 3.129E-03 | 3.569E-02 |
| GIMAP2 | ‒0.704 | 0.614 | 2.470E-07 | 2.110E-05 |
| ATP6V0E1 | ‒0.704 | 0.614 | 1.282E-04 | 3.411E-03 |
| SNX27 | ‒0.703 | 0.614 | 1.127E-03 | 1.711E-02 |
| PDCD7 | ‒0.703 | 0.614 | 2.816E-04 | 6.198E-03 |
| ECHDC1 | ‒0.702 | 0.615 | 4.178E-04 | 8.248E-03 |
| ZNF486 | ‒0.701 | 0.615 | 4.571E-03 | 4.624E-02 |
| MTREX | ‒0.699 | 0.616 | 2.644E-04 | 5.947E-03 |
| SNX19 | ‒0.697 | 0.617 | 2.376E-03 | 2.940E-02 |
| OMA1 | ‒0.697 | 0.617 | 7.030E-05 | 2.113E-03 |
| DLAT | ‒0.696 | 0.617 | 4.279E-03 | 4.420E-02 |
| RPP25L | ‒0.695 | 0.618 | 2.389E-03 | 2.950E-02 |
| VEZT | ‒0.693 | 0.619 | 5.126E-04 | 9.478E-03 |
| OGFOD1 | ‒0.690 | 0.620 | 1.115E-03 | 1.699E-02 |
| TTI2 | ‒0.690 | 0.620 | 4.590E-03 | 4.630E-02 |
| TBL2 | ‒0.689 | 0.620 | 1.505E-04 | 3.858E-03 |
| ZNF608 | ‒0.688 | 0.621 | 2.782E-03 | 3.295E-02 |
| UBE4A | ‒0.687 | 0.621 | 3.200E-05 | 1.165E-03 |
| MCEE | ‒0.687 | 0.621 | 2.028E-03 | 2.620E-02 |
| DAXX | ‒0.686 | 0.622 | 3.421E-04 | 7.124E-03 |
| IFNAR1 | ‒0.685 | 0.622 | 2.224E-04 | 5.241E-03 |
| PAXIP1-AS2 | ‒0.683 | 0.623 | 1.178E-03 | 1.771E-02 |
| TOLLIP | ‒0.683 | 0.623 | 2.260E-05 | 9.004E-04 |
| BUD13 | ‒0.683 | 0.623 | 4.870E-05 | 1.615E-03 |
| TIA1 | ‒0.682 | 0.623 | 5.530E-06 | 2.817E-04 |
| PCMT1 | ‒0.682 | 0.623 | 7.390E-05 | 2.200E-03 |
| ATP6V0D1 | ‒0.682 | 0.624 | 1.010E-04 | 2.837E-03 |
| HSD17B4 | ‒0.680 | 0.624 | 1.400E-05 | 6.077E-04 |
| LAIR1 | ‒0.680 | 0.624 | 4.060E-05 | 1.405E-03 |
| ITGAX | ‒0.679 | 0.624 | 9.025E-04 | 1.444E-02 |
| RITA1 | ‒0.679 | 0.625 | 6.116E-04 | 1.087E-02 |
| SFT2D1 | ‒0.679 | 0.625 | 9.540E-05 | 2.694E-03 |
| UTP6 | ‒0.679 | 0.625 | 1.660E-05 | 6.972E-04 |
| CNIH4 | ‒0.678 | 0.625 | 1.843E-04 | 4.542E-03 |
| LYN | ‒0.677 | 0.625 | 4.031E-04 | 8.043E-03 |
| CTNND1 | ‒0.675 | 0.626 | 1.995E-03 | 2.590E-02 |
| PSMB8-AS1 | ‒0.675 | 0.626 | 1.674E-03 | 2.279E-02 |
| REEP5 | ‒0.675 | 0.627 | 1.024E-03 | 1.591E-02 |
| TLR1 | ‒0.674 | 0.627 | 3.392E-03 | 3.776E-02 |
| MGAT2 | ‒0.673 | 0.627 | 8.250E-04 | 1.346E-02 |
| CRKL | ‒0.670 | 0.628 | 3.963E-04 | 7.965E-03 |
| SLC49A3 | ‒0.670 | 0.629 | 3.924E-03 | 4.173E-02 |
| SEC23IP | ‒0.668 | 0.629 | 1.127E-04 | 3.089E-03 |
| NADK | ‒0.668 | 0.630 | 1.154E-03 | 1.744E-02 |
| HSPH1 | ‒0.666 | 0.630 | 7.160E-05 | 2.136E-03 |
| GPR65 | ‒0.665 | 0.631 | 1.014E-03 | 1.581E-02 |
| APEX2 | ‒0.665 | 0.631 | 4.314E-03 | 4.450E-02 |
| CPLANE1 | ‒0.664 | 0.631 | 3.376E-03 | 3.762E-02 |
| MAP1S | ‒0.663 | 0.631 | 1.552E-03 | 2.168E-02 |
| SNAPC5 | ‒0.663 | 0.632 | 1.127E-03 | 1.711E-02 |
| HLA-DPB1 | ‒0.663 | 0.632 | 3.619E-04 | 7.403E-03 |
| APOBEC3D | ‒0.661 | 0.633 | 6.868E-04 | 1.179E-02 |
| LYSMD2 | ‒0.660 | 0.633 | 1.488E-03 | 2.095E-02 |
| FCMR | ‒0.660 | 0.633 | 5.730E-07 | 4.150E-05 |
| ZBTB38 | ‒0.659 | 0.633 | 1.204E-04 | 3.246E-03 |
| GDI2 | ‒0.659 | 0.633 | 6.120E-04 | 1.087E-02 |
| VTA1 | ‒0.657 | 0.634 | 7.080E-04 | 1.207E-02 |
| MR1 | ‒0.654 | 0.635 | 7.883E-04 | 1.306E-02 |
| ICAM2 | ‒0.654 | 0.635 | 3.458E-03 | 3.826E-02 |
| PHF11 | ‒0.653 | 0.636 | 1.591E-03 | 2.213E-02 |
| ACTR2 | ‒0.652 | 0.636 | 5.789E-04 | 1.044E-02 |
| CRLS1 | ‒0.650 | 0.637 | 4.520E-04 | 8.652E-03 |
| GM2A | ‒0.649 | 0.638 | 9.820E-04 | 1.544E-02 |
| SLC2A6 | ‒0.648 | 0.638 | 1.200E-06 | 7.840E-05 |
| MARCHF7 | ‒0.648 | 0.638 | 2.294E-03 | 2.863E-02 |
| ALOX5AP | ‒0.646 | 0.639 | 3.902E-03 | 4.162E-02 |
| COX15 | ‒0.646 | 0.639 | 1.360E-03 | 1.970E-02 |
| DOCK8 | ‒0.646 | 0.639 | 1.360E-05 | 5.939E-04 |
| TRIM4 | ‒0.644 | 0.640 | 7.500E-06 | 3.611E-04 |
| ILK | ‒0.644 | 0.640 | 1.388E-04 | 3.601E-03 |
| LCP1 | ‒0.643 | 0.640 | 4.403E-03 | 4.518E-02 |
| CLTA | ‒0.642 | 0.641 | 1.195E-04 | 3.237E-03 |
| IL10RB | ‒0.640 | 0.642 | 3.723E-03 | 4.034E-02 |
| TMBIM6 | ‒0.639 | 0.642 | 2.440E-05 | 9.504E-04 |
| TIMMDC1 | ‒0.639 | 0.642 | 1.759E-04 | 4.362E-03 |
| HEIH | ‒0.638 | 0.643 | 3.753E-04 | 7.598E-03 |
| DENND5B | ‒0.638 | 0.643 | 6.972E-04 | 1.191E-02 |
| PPP1R11 | ‒0.638 | 0.643 | 2.980E-05 | 1.109E-03 |
| UBE2V2 | ‒0.637 | 0.643 | 2.890E-04 | 6.312E-03 |
| C11orf24 | ‒0.637 | 0.643 | 2.280E-03 | 2.857E-02 |
| NME6 | ‒0.636 | 0.643 | 3.406E-03 | 3.785E-02 |
| BIRC2 | ‒0.636 | 0.643 | 6.780E-05 | 2.063E-03 |
| NIT1 | ‒0.636 | 0.644 | 1.581E-04 | 3.997E-03 |
| ACTR1A | ‒0.635 | 0.644 | 1.921E-03 | 2.517E-02 |
| ZNF260 | ‒0.635 | 0.644 | 2.911E-03 | 3.415E-02 |
| C21orf91 | ‒0.634 | 0.644 | 2.697E-04 | 5.994E-03 |
| COPB2 | ‒0.633 | 0.645 | 1.357E-04 | 3.539E-03 |
| VCL | ‒0.632 | 0.645 | 4.889E-03 | 4.832E-02 |
| FRAT1 | ‒0.630 | 0.646 | 4.730E-04 | 8.934E-03 |
| S100A4 | ‒0.630 | 0.646 | 3.008E-03 | 3.486E-02 |
| AIMP2 | ‒0.629 | 0.647 | 2.159E-03 | 2.752E-02 |
| CREBL2 | ‒0.629 | 0.647 | 2.557E-03 | 3.114E-02 |
| RNF5 | ‒0.627 | 0.647 | 7.217E-04 | 1.225E-02 |
| AHNAK | ‒0.627 | 0.647 | 4.531E-03 | 4.603E-02 |
| AFTPH | ‒0.626 | 0.648 | 2.755E-03 | 3.276E-02 |
| KRCC1 | ‒0.625 | 0.648 | 1.733E-04 | 4.303E-03 |
| BLNK | ‒0.625 | 0.648 | 4.971E-03 | 4.889E-02 |
| ETFA | ‒0.623 | 0.649 | 1.257E-04 | 3.368E-03 |
| SP1 | ‒0.622 | 0.650 | 1.998E-03 | 2.592E-02 |
| AMMECR1L | ‒0.621 | 0.650 | 2.412E-04 | 5.588E-03 |
| MITD1 | ‒0.621 | 0.650 | 1.673E-03 | 2.279E-02 |
| MRS2 | ‒0.620 | 0.651 | 1.214E-03 | 1.810E-02 |
| GRK3 | ‒0.619 | 0.651 | 4.105E-03 | 4.302E-02 |
| SNX20 | ‒0.618 | 0.651 | 5.810E-05 | 1.862E-03 |
| PTPN6 | ‒0.618 | 0.652 | 3.015E-04 | 6.473E-03 |
| ZKSCAN1 | ‒0.618 | 0.652 | 3.728E-04 | 7.566E-03 |
| NIPA2 | ‒0.617 | 0.652 | 2.034E-03 | 2.625E-02 |
| POC5 | ‒0.616 | 0.652 | 8.359E-04 | 1.360E-02 |
| CCT5 | ‒0.616 | 0.653 | 1.356E-04 | 3.539E-03 |
| ANXA1 | ‒0.616 | 0.653 | 4.253E-03 | 4.404E-02 |
| FGFR1OP2 | ‒0.614 | 0.653 | 3.125E-04 | 6.675E-03 |
| MED12 | ‒0.614 | 0.653 | 3.740E-05 | 1.314E-03 |
| ZNF672 | ‒0.613 | 0.654 | 1.240E-03 | 1.833E-02 |
| RPE | ‒0.611 | 0.655 | 6.204E-04 | 1.096E-02 |
| COPS4 | ‒0.610 | 0.655 | 7.730E-04 | 1.289E-02 |
| FXR2 | ‒0.610 | 0.655 | 2.358E-04 | 5.502E-03 |
| COP1 | ‒0.610 | 0.655 | 1.446E-03 | 2.053E-02 |
| DENND2D | ‒0.609 | 0.656 | 1.316E-03 | 1.921E-02 |
| FUCA1 | ‒0.609 | 0.656 | 9.178E-04 | 1.465E-02 |
| KRBOX4 | ‒0.607 | 0.656 | 1.301E-03 | 1.907E-02 |
| MAT2B | ‒0.603 | 0.658 | 1.130E-05 | 5.131E-04 |
| NUP62 | ‒0.603 | 0.659 | 7.100E-05 | 2.122E-03 |
| NA | ‒0.602 | 0.659 | 3.171E-03 | 3.599E-02 |
| MTHFD2 | ‒0.602 | 0.659 | 4.403E-04 | 8.540E-03 |
| ZNF721 | ‒0.601 | 0.659 | 5.302E-04 | 9.696E-03 |
| HERC2P2 | ‒0.601 | 0.659 | 3.240E-06 | 1.834E-04 |
| MSANTD4 | ‒0.600 | 0.660 | 4.577E-03 | 4.625E-02 |
| DNAJC7 | ‒0.600 | 0.660 | 3.606E-04 | 7.403E-03 |
| ELF4 | ‒0.600 | 0.660 | 2.594E-03 | 3.152E-02 |
| GLT8D1 | ‒0.599 | 0.660 | 6.020E-05 | 1.898E-03 |
| SPIDR | ‒0.599 | 0.660 | 3.138E-03 | 3.577E-02 |
| NLRX1 | ‒0.599 | 0.660 | 4.019E-03 | 4.246E-02 |
| ACTR3 | ‒0.598 | 0.661 | 1.266E-04 | 3.380E-03 |
| ZNF175 | ‒0.597 | 0.661 | 4.158E-03 | 4.334E-02 |
| NDUFB3 | ‒0.596 | 0.662 | 7.103E-04 | 1.209E-02 |
| SH2D3C | ‒0.596 | 0.662 | 3.037E-03 | 3.504E-02 |
| ZNF197 | ‒0.596 | 0.662 | 4.523E-04 | 8.652E-03 |
| UMPS | ‒0.595 | 0.662 | 2.930E-03 | 3.423E-02 |
| SEPHS2 | ‒0.594 | 0.662 | 3.244E-03 | 3.660E-02 |
| ZNF586 | ‒0.594 | 0.662 | 1.264E-04 | 3.380E-03 |
| LEO1 | ‒0.594 | 0.663 | 3.921E-03 | 4.173E-02 |
| RAC2 | ‒0.593 | 0.663 | 3.166E-03 | 3.596E-02 |
| SLC16A3 | ‒0.593 | 0.663 | 2.532E-04 | 5.770E-03 |
| PPP1R8 | ‒0.592 | 0.663 | 3.268E-03 | 3.675E-02 |
| RBL2 | ‒0.592 | 0.664 | 3.084E-04 | 6.605E-03 |
| ABRACL | ‒0.591 | 0.664 | 2.282E-03 | 2.857E-02 |
| ARFIP1 | ‒0.590 | 0.664 | 4.555E-03 | 4.613E-02 |
| KLHL9 | ‒0.590 | 0.664 | 8.259E-04 | 1.346E-02 |
| TBC1D1 | ‒0.589 | 0.665 | 1.603E-03 | 2.224E-02 |
| AGTRAP | ‒0.588 | 0.665 | 1.624E-03 | 2.235E-02 |
| MICB | ‒0.588 | 0.665 | 4.557E-03 | 4.613E-02 |
| ASH2L | ‒0.587 | 0.666 | 3.380E-05 | 1.215E-03 |
| RTCB | ‒0.586 | 0.666 | 1.095E-03 | 1.674E-02 |
| MRM3 | 0.587 | 1.503 | 2.585E-03 | 3.143E-02 |
| HNRNPA1 | 0.588 | 1.503 | 2.640E-05 | 1.007E-03 |
| REXO1 | 0.589 | 1.504 | 4.524E-03 | 4.599E-02 |
| TRA2B | 0.589 | 1.505 | 2.448E-04 | 5.648E-03 |
| EME2 | 0.590 | 1.505 | 3.269E-03 | 3.675E-02 |
| DDX51 | 0.592 | 1.507 | 9.971E-04 | 1.558E-02 |
| CHD1 | 0.592 | 1.508 | 1.627E-03 | 2.235E-02 |
| MED9 | 0.592 | 1.508 | 4.072E-03 | 4.282E-02 |
| ASNS | 0.593 | 1.508 | 2.197E-04 | 5.207E-03 |
| ZNF831 | 0.596 | 1.511 | 3.658E-04 | 7.469E-03 |
| CLDND1 | 0.599 | 1.514 | 3.464E-04 | 7.188E-03 |
| FBXO32 | 0.601 | 1.516 | 2.468E-03 | 3.026E-02 |
| SREBF1 | 0.603 | 1.519 | 2.245E-03 | 2.829E-02 |
| GMFB | 0.604 | 1.519 | 1.149E-03 | 1.739E-02 |
| ATN1 | 0.605 | 1.521 | 1.502E-03 | 2.112E-02 |
| DNAJB14 | 0.606 | 1.522 | 2.373E-03 | 2.940E-02 |
| KANSL2 | 0.606 | 1.522 | 4.408E-04 | 8.540E-03 |
| MLLT3 | 0.609 | 1.525 | 1.763E-04 | 4.365E-03 |
| KLF11 | 0.609 | 1.525 | 4.225E-03 | 4.389E-02 |
| FOXP1 | 0.610 | 1.526 | 5.710E-05 | 1.833E-03 |
| MZT2B | 0.611 | 1.527 | 4.147E-03 | 4.330E-02 |
| NOP53 | 0.612 | 1.528 | 3.736E-03 | 4.040E-02 |
| POU2F1 | 0.612 | 1.528 | 1.041E-04 | 2.900E-03 |
| RFX3 | 0.612 | 1.529 | 7.849E-04 | 1.303E-02 |
| KRR1 | 0.617 | 1.534 | 4.080E-06 | 2.210E-04 |
| SSBP1 | 0.617 | 1.534 | 6.140E-06 | 3.038E-04 |
| MCRIP2 | 0.619 | 1.536 | 1.407E-03 | 2.013E-02 |
| LRP5L | 0.620 | 1.537 | 3.344E-03 | 3.739E-02 |
| LEMD3 | 0.621 | 1.537 | 4.250E-06 | 2.286E-04 |
| MEF2D | 0.621 | 1.538 | 9.140E-07 | 6.240E-05 |
| ENSG00000261490 | 0.621 | 1.538 | 3.129E-03 | 3.569E-02 |
| PAG1 | 0.622 | 1.539 | 7.514E-04 | 1.261E-02 |
| CCDC57 | 0.623 | 1.540 | 4.723E-04 | 8.933E-03 |
| PRDM2 | 0.623 | 1.540 | 2.620E-05 | 1.002E-03 |
| SFXN1 | 0.624 | 1.541 | 1.611E-03 | 2.228E-02 |
| FRS2 | 0.625 | 1.542 | 2.355E-03 | 2.924E-02 |
| SGMS1 | 0.625 | 1.542 | 2.255E-03 | 2.833E-02 |
| H1-10 | 0.626 | 1.544 | 3.870E-06 | 2.106E-04 |
| INTS1 | 0.626 | 1.544 | 1.236E-03 | 1.831E-02 |
| TRERF1 | 0.631 | 1.549 | 1.605E-03 | 2.225E-02 |
| PPP1R15A | 0.632 | 1.549 | 1.870E-05 | 7.664E-04 |
| RHNO1 | 0.635 | 1.553 | 3.557E-03 | 3.912E-02 |
| ANAPC15 | 0.635 | 1.553 | 1.214E-04 | 3.267E-03 |
| SLC5A6 | 0.636 | 1.554 | 4.500E-06 | 2.390E-04 |
| COX4I1 | 0.637 | 1.555 | 8.740E-08 | 8.770E-06 |
| MRPL44 | 0.637 | 1.555 | 3.803E-04 | 7.690E-03 |
| FBXL3 | 0.638 | 1.556 | 4.040E-05 | 1.404E-03 |
| LCOR | 0.639 | 1.557 | 5.050E-06 | 2.628E-04 |
| TGIF1 | 0.641 | 1.560 | 3.228E-03 | 3.649E-02 |
| FKBP11 | 0.642 | 1.560 | 1.951E-03 | 2.542E-02 |
| KDM7A | 0.642 | 1.560 | 2.144E-04 | 5.111E-03 |
| SNHG16 | 0.643 | 1.562 | 2.941E-04 | 6.372E-03 |
| SNRK | 0.644 | 1.562 | 5.018E-04 | 9.348E-03 |
| SLC25A36 | 0.644 | 1.563 | 4.191E-04 | 8.263E-03 |
| SUPV3L1 | 0.644 | 1.563 | 5.874E-04 | 1.055E-02 |
| PNMA1 | 0.644 | 1.563 | 4.122E-03 | 4.314E-02 |
| PAFAH1B2 | 0.644 | 1.563 | 1.967E-04 | 4.783E-03 |
| ATG2A | 0.645 | 1.564 | 7.069E-04 | 1.207E-02 |
| ELF2 | 0.646 | 1.565 | 2.100E-06 | 1.258E-04 |
| FAM229A | 0.646 | 1.565 | 4.983E-03 | 4.899E-02 |
| ATF4 | 0.647 | 1.566 | 3.570E-05 | 1.269E-03 |
| FAM102A | 0.648 | 1.567 | 2.165E-03 | 2.757E-02 |
| EAF1 | 0.649 | 1.569 | 1.721E-03 | 2.321E-02 |
| TMEM120B | 0.650 | 1.570 | 3.680E-03 | 4.004E-02 |
| ARID2 | 0.652 | 1.572 | 3.384E-04 | 7.066E-03 |
| TRIM52-AS1 | 0.654 | 1.573 | 2.059E-03 | 2.645E-02 |
| RPS14 | 0.654 | 1.574 | 2.686E-04 | 5.994E-03 |
| NFKBIE | 0.655 | 1.574 | 5.094E-03 | 4.981E-02 |
| MEGF6 | 0.655 | 1.574 | 1.771E-03 | 2.368E-02 |
| SNRPA1 | 0.658 | 1.578 | 1.883E-04 | 4.620E-03 |
| LMTK2 | 0.658 | 1.578 | 2.750E-05 | 1.027E-03 |
| NSMCE3 | 0.659 | 1.579 | 2.660E-05 | 1.008E-03 |
| EPB41L4A-AS1 | 0.660 | 1.580 | 2.530E-05 | 9.788E-04 |
| POU6F1 | 0.661 | 1.581 | 1.190E-03 | 1.785E-02 |
| ENSG00000257354 | 0.662 | 1.582 | 3.623E-03 | 3.971E-02 |
| ZNF408 | 0.662 | 1.583 | 2.807E-03 | 3.310E-02 |
| CHD7 | 0.662 | 1.583 | 1.470E-04 | 3.785E-03 |
| ZNF432 | 0.663 | 1.583 | 1.236E-03 | 1.831E-02 |
| B3GNTL1 | 0.663 | 1.583 | 7.535E-04 | 1.263E-02 |
| NFKBIZ | 0.665 | 1.585 | 3.061E-03 | 3.526E-02 |
| NFKBIB | 0.666 | 1.587 | 6.410E-04 | 1.123E-02 |
| BICDL1 | 0.666 | 1.587 | 4.001E-04 | 8.013E-03 |
| PIK3R1 | 0.668 | 1.589 | 1.102E-03 | 1.682E-02 |
| KAT6B | 0.668 | 1.589 | 2.580E-05 | 9.886E-04 |
| IGF1R | 0.669 | 1.589 | 1.102E-03 | 1.682E-02 |
| BTG2 | 0.669 | 1.590 | 2.494E-04 | 5.708E-03 |
| LEPROTL1 | 0.670 | 1.591 | 1.651E-03 | 2.256E-02 |
| NECAP1 | 0.670 | 1.591 | 1.750E-06 | 1.078E-04 |
| AP1G2-AS1 | 0.672 | 1.593 | 4.120E-05 | 1.420E-03 |
| SVIP | 0.674 | 1.595 | 9.720E-04 | 1.535E-02 |
| ISCA1 | 0.675 | 1.596 | 1.640E-03 | 2.245E-02 |
| PNRC1 | 0.675 | 1.597 | 7.980E-06 | 3.809E-04 |
| PIDD1 | 0.678 | 1.600 | 8.583E-04 | 1.387E-02 |
| VAMP2 | 0.679 | 1.601 | 1.820E-08 | 2.020E-06 |
| ADAMTS10 | 0.680 | 1.602 | 4.540E-03 | 4.607E-02 |
| TCF7 | 0.682 | 1.604 | 1.200E-05 | 5.353E-04 |
| EXD3 | 0.683 | 1.605 | 1.214E-03 | 1.810E-02 |
| TIAM1 | 0.683 | 1.605 | 3.855E-03 | 4.133E-02 |
| LINC-PINT | 0.686 | 1.609 | 2.743E-03 | 3.266E-02 |
| ATXN1L | 0.689 | 1.613 | 8.960E-05 | 2.560E-03 |
| MBIP | 0.691 | 1.615 | 2.098E-03 | 2.688E-02 |
| NOL4L | 0.692 | 1.615 | 4.613E-04 | 8.783E-03 |
| RPL36A | 0.692 | 1.616 | 3.688E-03 | 4.011E-02 |
| VCPKMT | 0.693 | 1.616 | 6.204E-04 | 1.096E-02 |
| CBX6 | 0.693 | 1.617 | 2.331E-04 | 5.460E-03 |
| MRPL41 | 0.695 | 1.618 | 3.350E-03 | 3.744E-02 |
| ENSG00000280135 | 0.695 | 1.618 | 2.653E-03 | 3.196E-02 |
| ADNP2 | 0.695 | 1.619 | 1.100E-05 | 4.997E-04 |
| ZNF800 | 0.701 | 1.626 | 3.030E-05 | 1.118E-03 |
| EPHB4 | 0.704 | 1.629 | 2.807E-03 | 3.310E-02 |
| EIF5 | 0.705 | 1.630 | 1.960E-06 | 1.186E-04 |
| PGM2L1 | 0.705 | 1.630 | 3.167E-04 | 6.738E-03 |
| DYNLT1 | 0.706 | 1.631 | 3.925E-03 | 4.173E-02 |
| CNNM2 | 0.706 | 1.631 | 6.423E-04 | 1.124E-02 |
| PPIL4 | 0.707 | 1.632 | 2.334E-03 | 2.902E-02 |
| TRMO | 0.707 | 1.633 | 1.910E-03 | 2.506E-02 |
| HIVEP1 | 0.707 | 1.633 | 1.326E-04 | 3.484E-03 |
| BEX4 | 0.708 | 1.633 | 2.802E-04 | 6.175E-03 |
| MRPL9 | 0.708 | 1.634 | 1.324E-04 | 3.484E-03 |
| TLE2 | 0.710 | 1.636 | 3.869E-03 | 4.140E-02 |
| EIF1B | 0.712 | 1.638 | 8.150E-05 | 2.379E-03 |
| SNHG1 | 0.712 | 1.638 | 1.213E-03 | 1.810E-02 |
| ZNF814 | 0.714 | 1.640 | 2.058E-04 | 4.963E-03 |
| ENSG00000230551 | 0.715 | 1.642 | 4.259E-03 | 4.407E-02 |
| GOLT1B | 0.716 | 1.642 | 1.880E-03 | 2.473E-02 |
| MFGE8 | 0.717 | 1.644 | 1.714E-03 | 2.314E-02 |
| RICTOR | 0.718 | 1.645 | 1.176E-04 | 3.195E-03 |
| UTP15 | 0.719 | 1.646 | 1.959E-04 | 4.771E-03 |
| SIVA1 | 0.720 | 1.647 | 1.090E-04 | 3.014E-03 |
| SORBS3 | 0.723 | 1.650 | 9.648E-04 | 1.527E-02 |
| EMD | 0.724 | 1.652 | 2.687E-04 | 5.994E-03 |
| TRABD | 0.727 | 1.655 | 6.177E-04 | 1.094E-02 |
| EIF2AK3 | 0.727 | 1.655 | 2.914E-04 | 6.346E-03 |
| ZNF134 | 0.727 | 1.655 | 4.069E-04 | 8.080E-03 |
| PIK3IP1 | 0.728 | 1.656 | 2.966E-04 | 6.401E-03 |
| NCBP2AS2 | 0.728 | 1.656 | 5.660E-05 | 1.821E-03 |
| DDX39A | 0.729 | 1.658 | 1.020E-04 | 2.857E-03 |
| PHF1 | 0.731 | 1.660 | 2.120E-05 | 8.495E-04 |
| ARRDC2 | 0.732 | 1.661 | 1.644E-03 | 2.249E-02 |
| GABARAPL1 | 0.733 | 1.663 | 1.934E-03 | 2.528E-02 |
| SRSF10 | 0.737 | 1.666 | 5.980E-06 | 2.990E-04 |
| CHRM3-AS2 | 0.737 | 1.666 | 1.470E-03 | 2.076E-02 |
| ZNF815P | 0.739 | 1.669 | 4.427E-04 | 8.558E-03 |
| MGST3 | 0.742 | 1.672 | 1.709E-03 | 2.314E-02 |
| AP3M2 | 0.742 | 1.672 | 4.388E-03 | 4.505E-02 |
| JOSD1 | 0.743 | 1.673 | 1.210E-06 | 7.880E-05 |
| ODC1 | 0.743 | 1.674 | 2.740E-05 | 1.027E-03 |
| KPNA5 | 0.749 | 1.680 | 4.024E-04 | 8.039E-03 |
| GOLGA7B | 0.753 | 1.685 | 1.060E-03 | 1.632E-02 |
| MAP3K1 | 0.755 | 1.688 | 3.930E-04 | 7.918E-03 |
| AK5 | 0.756 | 1.689 | 2.327E-04 | 5.460E-03 |
| C5orf24 | 0.756 | 1.689 | 6.860E-07 | 4.840E-05 |
| KLHL24 | 0.758 | 1.691 | 2.270E-05 | 9.013E-04 |
| TAF3 | 0.759 | 1.692 | 1.870E-05 | 7.668E-04 |
| FAM160B1 | 0.760 | 1.693 | 1.333E-04 | 3.498E-03 |
| BICRA | 0.760 | 1.693 | 2.379E-04 | 5.529E-03 |
| AKT1S1 | 0.761 | 1.695 | 8.010E-05 | 2.345E-03 |
| RLIM | 0.763 | 1.697 | 1.340E-06 | 8.610E-05 |
| MAPKAPK5-AS1 | 0.768 | 1.703 | 2.970E-07 | 2.470E-05 |
| NA | 0.772 | 1.707 | 4.109E-03 | 4.303E-02 |
| KLHL15 | 0.772 | 1.707 | 3.060E-05 | 1.127E-03 |
| TIPARP | 0.776 | 1.713 | 1.574E-04 | 3.991E-03 |
| FAM177A1 | 0.776 | 1.713 | 2.670E-04 | 5.980E-03 |
| CIDECP1 | 0.778 | 1.715 | 4.942E-04 | 9.244E-03 |
| MTHFD2L | 0.778 | 1.715 | 2.120E-03 | 2.710E-02 |
| NPIPB5 | 0.780 | 1.717 | 3.472E-03 | 3.836E-02 |
| LMO7 | 0.781 | 1.718 | 5.950E-06 | 2.985E-04 |
| NOXA1 | 0.781 | 1.718 | 2.253E-03 | 2.832E-02 |
| TMEM201 | 0.782 | 1.719 | 6.646E-04 | 1.153E-02 |
| PPP1R32 | 0.782 | 1.720 | 2.050E-03 | 2.637E-02 |
| IL1B | 0.783 | 1.720 | 1.594E-03 | 2.215E-02 |
| MYL5 | 0.783 | 1.720 | 2.121E-03 | 2.710E-02 |
| ENSG00000264112 | 0.787 | 1.726 | 2.120E-06 | 1.266E-04 |
| ENSG00000279838 | 0.789 | 1.728 | 4.723E-03 | 4.716E-02 |
| ZNF805 | 0.789 | 1.728 | 2.189E-04 | 5.196E-03 |
| CLK1 | 0.790 | 1.729 | 4.560E-10 | 8.260E-08 |
| PTS | 0.791 | 1.730 | 3.444E-03 | 3.813E-02 |
| RNF125 | 0.791 | 1.731 | 3.585E-04 | 7.393E-03 |
| PSTK | 0.792 | 1.732 | 3.158E-03 | 3.592E-02 |
| FOXK1 | 0.795 | 1.735 | 8.360E-06 | 3.911E-04 |
| SOX12 | 0.796 | 1.736 | 1.260E-06 | 8.130E-05 |
| ZNF446 | 0.796 | 1.736 | 2.381E-03 | 2.943E-02 |
| MBNL2 | 0.796 | 1.737 | 1.530E-05 | 6.520E-04 |
| SNHG20 | 0.796 | 1.737 | 6.018E-04 | 1.072E-02 |
| RAMAC | 0.798 | 1.738 | 2.212E-03 | 2.796E-02 |
| NA | 0.798 | 1.739 | 1.333E-03 | 1.940E-02 |
| NFKBID | 0.799 | 1.740 | 1.740E-05 | 7.220E-04 |
| HBP1 | 0.800 | 1.741 | 7.925E-04 | 1.311E-02 |
| CHIC1 | 0.805 | 1.747 | 8.110E-05 | 2.370E-03 |
| ISCU | 0.805 | 1.747 | 4.390E-06 | 2.343E-04 |
| SGTB | 0.807 | 1.750 | 7.781E-04 | 1.296E-02 |
| TRAF4 | 0.809 | 1.752 | 3.730E-05 | 1.313E-03 |
| ZFP36 | 0.811 | 1.754 | 1.494E-04 | 3.841E-03 |
| EPHA4 | 0.812 | 1.755 | 1.045E-04 | 2.908E-03 |
| CDK11A | 0.812 | 1.756 | 1.310E-05 | 5.747E-04 |
| ANO9 | 0.813 | 1.757 | 2.292E-03 | 2.863E-02 |
| TYW5 | 0.815 | 1.760 | 4.949E-04 | 9.244E-03 |
| RETREG1 | 0.819 | 1.764 | 5.862E-04 | 1.054E-02 |
| CD28 | 0.822 | 1.768 | 6.437E-04 | 1.125E-02 |
| KDM6B | 0.826 | 1.772 | 4.410E-07 | 3.380E-05 |
| ARL5B | 0.826 | 1.773 | 3.616E-04 | 7.403E-03 |
| SLC2A4RG | 0.826 | 1.773 | 5.712E-04 | 1.033E-02 |
| PET100 | 0.831 | 1.779 | 6.930E-05 | 2.095E-03 |
| ZFYVE28 | 0.835 | 1.784 | 1.633E-03 | 2.237E-02 |
| ZNF587B | 0.837 | 1.787 | 2.430E-05 | 9.504E-04 |
| PLCL1 | 0.847 | 1.799 | 1.886E-03 | 2.478E-02 |
| SMURF1 | 0.850 | 1.802 | 5.870E-05 | 1.873E-03 |
| DDX24 | 0.850 | 1.802 | 2.100E-07 | 1.860E-05 |
| STARD10 | 0.853 | 1.806 | 2.916E-03 | 3.416E-02 |
| POLR1C | 0.853 | 1.806 | 8.020E-06 | 3.816E-04 |
| TIGD1 | 0.855 | 1.809 | 2.612E-03 | 3.163E-02 |
| EIF1 | 0.855 | 1.809 | 4.610E-06 | 2.439E-04 |
| HMGB2 | 0.856 | 1.810 | 7.320E-06 | 3.536E-04 |
| ENSG00000272529 | 0.856 | 1.810 | 5.188E-04 | 9.551E-03 |
| H3-3B | 0.856 | 1.811 | 7.380E-09 | 9.680E-07 |
| ENSG00000235859 | 0.857 | 1.812 | 1.711E-03 | 2.314E-02 |
| CEP85L | 0.858 | 1.812 | 2.426E-03 | 2.982E-02 |
| RBM7 | 0.859 | 1.814 | 5.269E-04 | 9.661E-03 |
| LRRC8B | 0.862 | 1.817 | 6.560E-06 | 3.205E-04 |
| RASGEF1B | 0.862 | 1.818 | 1.402E-04 | 3.632E-03 |
| CNBD2 | 0.863 | 1.819 | 6.823E-04 | 1.176E-02 |
| OTUD1 | 0.863 | 1.819 | 6.160E-06 | 3.038E-04 |
| BCL10 | 0.863 | 1.819 | 1.669E-04 | 4.181E-03 |
| P2RY11 | 0.864 | 1.820 | 1.428E-03 | 2.037E-02 |
| ZNF844 | 0.869 | 1.826 | 7.990E-08 | 8.120E-06 |
| NGDN | 0.871 | 1.829 | 1.550E-08 | 1.760E-06 |
| ENO2 | 0.874 | 1.833 | 2.610E-04 | 5.901E-03 |
| ENSG00000271964 | 0.876 | 1.836 | 1.054E-03 | 1.625E-02 |
| AMMECR1 | 0.877 | 1.837 | 3.840E-06 | 2.096E-04 |
| YPEL5 | 0.880 | 1.840 | 3.000E-07 | 2.470E-05 |
| MGAT4A | 0.881 | 1.841 | 1.650E-05 | 6.972E-04 |
| HSF2 | 0.881 | 1.842 | 7.080E-05 | 2.121E-03 |
| ZXDB | 0.882 | 1.843 | 6.000E-05 | 1.895E-03 |
| ZEB1 | 0.884 | 1.846 | 2.760E-06 | 1.599E-04 |
| HAGHL | 0.886 | 1.847 | 2.784E-03 | 3.295E-02 |
| AEN | 0.886 | 1.847 | 4.430E-07 | 3.390E-05 |
| MYLIP | 0.886 | 1.849 | 3.070E-07 | 2.510E-05 |
| IER2 | 0.888 | 1.851 | 9.330E-08 | 9.310E-06 |
| ENSG00000261526 | 0.889 | 1.852 | 1.907E-04 | 4.658E-03 |
| GZF1 | 0.890 | 1.853 | 9.380E-05 | 2.668E-03 |
| PMAIP1 | 0.890 | 1.854 | 4.041E-03 | 4.256E-02 |
| RHPN1 | 0.894 | 1.859 | 2.180E-03 | 2.772E-02 |
| MAST4 | 0.894 | 1.859 | 4.650E-04 | 8.823E-03 |
| ZFAND2A | 0.899 | 1.865 | 4.370E-06 | 2.341E-04 |
| ZNF628 | 0.900 | 1.866 | 2.684E-03 | 3.222E-02 |
| ASMTL-AS1 | 0.902 | 1.869 | 8.460E-06 | 3.950E-04 |
| ZBTB20 | 0.907 | 1.875 | 1.534E-03 | 2.146E-02 |
| CYP4V2 | 0.908 | 1.877 | 1.304E-04 | 3.449E-03 |
| SETD1B | 0.912 | 1.881 | 3.430E-10 | 6.580E-08 |
| NXT1 | 0.912 | 1.881 | 3.860E-05 | 1.348E-03 |
| IL6ST | 0.912 | 1.882 | 5.010E-05 | 1.654E-03 |
| ENSG00000269688 | 0.912 | 1.882 | 8.641E-04 | 1.395E-02 |
| CTSK | 0.915 | 1.885 | 5.841E-04 | 1.052E-02 |
| GLUD1P3 | 0.915 | 1.886 | 4.271E-03 | 4.417E-02 |
| SCNN1D | 0.916 | 1.887 | 9.470E-05 | 2.687E-03 |
| ENSG00000232545 | 0.917 | 1.888 | 5.076E-03 | 4.966E-02 |
| ZXDA | 0.917 | 1.888 | 1.152E-04 | 3.141E-03 |
| KLHL11 | 0.919 | 1.891 | 1.652E-03 | 2.256E-02 |
| SNIP1 | 0.920 | 1.893 | 3.196E-04 | 6.754E-03 |
| EDAR | 0.922 | 1.895 | 1.294E-03 | 1.899E-02 |
| ABCD2 | 0.926 | 1.900 | 1.365E-04 | 3.547E-03 |
| ZNF256 | 0.927 | 1.901 | 2.885E-04 | 6.312E-03 |
| ANTXRLP1 | 0.927 | 1.902 | 1.811E-03 | 2.410E-02 |
| WDR74 | 0.929 | 1.904 | 9.700E-05 | 2.734E-03 |
| GPRASP1 | 0.930 | 1.905 | 4.769E-04 | 8.968E-03 |
| LINC00852 | 0.930 | 1.905 | 6.850E-05 | 2.074E-03 |
| FASTKD5 | 0.931 | 1.907 | 6.630E-07 | 4.720E-05 |
| MOAP1 | 0.933 | 1.909 | 4.620E-09 | 6.270E-07 |
| TADA1 | 0.936 | 1.913 | 3.640E-05 | 1.289E-03 |
| PPIF | 0.936 | 1.913 | 3.520E-06 | 1.973E-04 |
| MATR3 | 0.938 | 1.916 | 2.239E-03 | 2.826E-02 |
| PDE4B | 0.940 | 1.918 | 2.000E-05 | 8.122E-04 |
| NLRP6 | 0.940 | 1.919 | 5.945E-04 | 1.063E-02 |
| RAB33B-AS1 | 0.945 | 1.925 | 3.465E-03 | 3.830E-02 |
| DBF4 | 0.946 | 1.927 | 2.925E-04 | 6.362E-03 |
| IRF2BP2 | 0.947 | 1.928 | 1.140E-09 | 1.810E-07 |
| CCNT1 | 0.951 | 1.933 | 5.740E-10 | 9.970E-08 |
| SNPH | 0.951 | 1.933 | 4.021E-03 | 4.246E-02 |
| YRDC | 0.953 | 1.936 | 5.730E-06 | 2.887E-04 |
| FEM1C | 0.954 | 1.937 | 2.130E-06 | 1.266E-04 |
| LINC00641 | 0.959 | 1.945 | 2.500E-05 | 9.710E-04 |
| CCNL1 | 0.961 | 1.947 | 1.930E-09 | 2.870E-07 |
| ZNF101 | 0.961 | 1.947 | 1.640E-05 | 6.936E-04 |
| MIRLET7A1HG | 0.963 | 1.949 | 2.058E-04 | 4.963E-03 |
| CDR2 | 0.963 | 1.950 | 1.849E-04 | 4.551E-03 |
| ZNF639 | 0.964 | 1.951 | 8.050E-09 | 1.040E-06 |
| DOC2GP | 0.965 | 1.952 | 1.507E-03 | 2.115E-02 |
| DCTN6 | 0.967 | 1.955 | 1.254E-03 | 1.847E-02 |
| DNAJB9 | 0.968 | 1.957 | 3.380E-05 | 1.215E-03 |
| CREBRF | 0.973 | 1.963 | 6.470E-09 | 8.560E-07 |
| TBKBP1 | 0.978 | 1.970 | 6.293E-04 | 1.109E-02 |
| CCDC85C | 0.980 | 1.972 | 2.530E-03 | 3.092E-02 |
| MAFK | 0.984 | 1.979 | 1.150E-06 | 7.630E-05 |
| SLC22A23 | 0.985 | 1.979 | 1.410E-06 | 8.930E-05 |
| DUSP5 | 0.993 | 1.990 | 6.290E-05 | 1.950E-03 |
| SNHG8 | 1.001 | 2.002 | 6.127E-04 | 1.087E-02 |
| MIR23AHG | 1.002 | 2.003 | 1.224E-03 | 1.820E-02 |
| ENSG00000259623 | 1.006 | 2.008 | 1.670E-05 | 6.989E-04 |
| BCL11B | 1.008 | 2.011 | 3.290E-08 | 3.460E-06 |
| LTBP3 | 1.010 | 2.013 | 2.205E-03 | 2.792E-02 |
| MTFP1 | 1.012 | 2.017 | 3.627E-03 | 3.971E-02 |
| NDUFAF5 | 1.014 | 2.019 | 1.061E-03 | 1.633E-02 |
| RORA | 1.023 | 2.033 | 2.218E-04 | 5.237E-03 |
| TWF1 | 1.027 | 2.037 | 1.270E-08 | 1.500E-06 |
| SYCP2 | 1.029 | 2.040 | 1.876E-03 | 2.471E-02 |
| ENSG00000257497 | 1.029 | 2.041 | 1.512E-03 | 2.118E-02 |
| RNF139 | 1.029 | 2.041 | 3.700E-05 | 1.309E-03 |
| NR4A1 | 1.030 | 2.042 | 1.180E-05 | 5.299E-04 |
| DHRS3 | 1.035 | 2.049 | 2.316E-03 | 2.886E-02 |
| ENSG00000274460 | 1.037 | 2.052 | 2.306E-03 | 2.876E-02 |
| NDST2 | 1.038 | 2.054 | 3.160E-03 | 3.592E-02 |
| SHLD3 | 1.040 | 2.057 | 6.730E-05 | 2.057E-03 |
| SPEG | 1.041 | 2.058 | 2.326E-03 | 2.894E-02 |
| ENSG00000255182 | 1.045 | 2.063 | 1.194E-04 | 3.237E-03 |
| SNHG15 | 1.048 | 2.068 | 7.240E-11 | 1.700E-08 |
| MIR22HG | 1.052 | 2.074 | 2.105E-04 | 5.024E-03 |
| ACTA2-AS1 | 1.054 | 2.076 | 3.290E-03 | 3.689E-02 |
| ENSG00000223511 | 1.063 | 2.089 | 4.530E-05 | 1.530E-03 |
| TSPYL2 | 1.064 | 2.090 | 2.260E-09 | 3.250E-07 |
| SEC14L2 | 1.069 | 2.098 | 1.593E-04 | 4.010E-03 |
| TUBB2A | 1.071 | 2.102 | 5.035E-03 | 4.938E-02 |
| NAF1 | 1.072 | 2.103 | 5.360E-06 | 2.746E-04 |
| NCR3LG1 | 1.074 | 2.105 | 2.084E-03 | 2.673E-02 |
| ENSG00000265625 | 1.079 | 2.113 | 4.492E-03 | 4.577E-02 |
| PGGHG | 1.083 | 2.119 | 5.427E-04 | 9.893E-03 |
| WHAMM | 1.083 | 2.119 | 8.080E-06 | 3.835E-04 |
| PRR7 | 1.084 | 2.120 | 1.139E-03 | 1.725E-02 |
| IKZF5 | 1.084 | 2.120 | 5.300E-13 | 2.050E-10 |
| ENSG00000260404 | 1.085 | 2.122 | 3.780E-06 | 2.072E-04 |
| CD69 | 1.085 | 2.122 | 9.960E-09 | 1.230E-06 |
| ITPRIP | 1.088 | 2.126 | 3.590E-08 | 3.720E-06 |
| IER3 | 1.090 | 2.129 | 3.788E-03 | 4.074E-02 |
| C6orf226 | 1.090 | 2.129 | 7.808E-04 | 1.300E-02 |
| FAM86B3P | 1.095 | 2.135 | 1.933E-03 | 2.527E-02 |
| ADGRB1 | 1.101 | 2.145 | 2.540E-03 | 3.100E-02 |
| NA | 1.104 | 2.149 | 3.240E-04 | 6.815E-03 |
| RBM34 | 1.105 | 2.150 | 3.665E-03 | 3.999E-02 |
| AFAP1 | 1.105 | 2.151 | 3.408E-03 | 3.785E-02 |
| LINC01089 | 1.111 | 2.159 | 5.620E-06 | 2.838E-04 |
| ENSG00000269951 | 1.120 | 2.174 | 4.229E-03 | 4.389E-02 |
| EML5 | 1.123 | 2.177 | 4.451E-03 | 4.550E-02 |
| PASK | 1.127 | 2.183 | 1.010E-05 | 4.609E-04 |
| ARRDC3 | 1.133 | 2.193 | 4.308E-04 | 8.445E-03 |
| TOE1 | 1.133 | 2.194 | 1.010E-11 | 2.960E-09 |
| RBM38 | 1.137 | 2.199 | 6.240E-07 | 4.480E-05 |
| PIGA | 1.144 | 2.210 | 1.900E-06 | 1.160E-04 |
| ENSG00000258944 | 1.146 | 2.213 | 2.697E-04 | 5.994E-03 |
| TRBV30 | 1.147 | 2.214 | 1.401E-03 | 2.012E-02 |
| YOD1 | 1.155 | 2.227 | 7.340E-11 | 1.700E-08 |
| ENSG00000255026 | 1.160 | 2.235 | 7.886E-04 | 1.306E-02 |
| OSER1 | 1.162 | 2.238 | 6.670E-18 | 5.550E-15 |
| IGLV6-57 | 1.163 | 2.239 | 1.796E-03 | 2.396E-02 |
| CCDC59 | 1.164 | 2.240 | 2.610E-08 | 2.800E-06 |
| CHASERR | 1.169 | 2.249 | 5.680E-12 | 1.750E-09 |
| LINC00653 | 1.169 | 2.249 | 5.070E-06 | 2.633E-04 |
| SIAH1 | 1.173 | 2.254 | 3.080E-07 | 2.510E-05 |
| BEX2 | 1.179 | 2.264 | 7.620E-09 | 9.910E-07 |
| CLN8-AS1 | 1.180 | 2.266 | 2.719E-03 | 3.246E-02 |
| ENSG00000273253 | 1.183 | 2.270 | 4.109E-04 | 8.147E-03 |
| LZTS3 | 1.183 | 2.271 | 3.970E-05 | 1.381E-03 |
| DDX47 | 1.184 | 2.272 | 1.040E-09 | 1.690E-07 |
| TENT5C | 1.186 | 2.275 | 2.070E-07 | 1.850E-05 |
| EIF4A1 | 1.186 | 2.275 | 5.590E-10 | 9.810E-08 |
| CACNA1H | 1.187 | 2.276 | 5.840E-05 | 1.867E-03 |
| CLP1 | 1.190 | 2.282 | 3.870E-07 | 3.010E-05 |
| DUSP10 | 1.192 | 2.285 | 6.390E-06 | 3.131E-04 |
| CDKN1B | 1.196 | 2.291 | 2.230E-08 | 2.450E-06 |
| ZBTB21 | 1.204 | 2.303 | 5.340E-09 | 7.110E-07 |
| DUS3L | 1.206 | 2.307 | 1.670E-07 | 1.520E-05 |
| NAP1L5 | 1.211 | 2.315 | 1.250E-04 | 3.353E-03 |
| SLC7A5 | 1.219 | 2.328 | 1.430E-07 | 1.330E-05 |
| SIK1B | 1.222 | 2.333 | 3.780E-06 | 2.072E-04 |
| ERFL | 1.223 | 2.334 | 4.521E-04 | 8.652E-03 |
| SNORD4A | 1.232 | 2.349 | 1.780E-03 | 2.376E-02 |
| CBX4 | 1.232 | 2.349 | 1.650E-12 | 5.500E-10 |
| JUNB | 1.234 | 2.352 | 1.660E-06 | 1.031E-04 |
| C1R | 1.239 | 2.360 | 2.424E-03 | 2.982E-02 |
| ADAMTS7P1 | 1.240 | 2.362 | 1.664E-03 | 2.271E-02 |
| CXCR4 | 1.249 | 2.376 | 2.900E-13 | 1.240E-10 |
| RBM38-AS1 | 1.250 | 2.378 | 1.902E-03 | 2.497E-02 |
| CSRNP1 | 1.250 | 2.379 | 3.480E-12 | 1.090E-09 |
| PTP4A1 | 1.255 | 2.387 | 2.170E-08 | 2.400E-06 |
| SPATA2 | 1.257 | 2.391 | 1.080E-08 | 1.330E-06 |
| ZNF551 | 1.259 | 2.392 | 2.350E-10 | 4.960E-08 |
| ZNF367 | 1.263 | 2.399 | 5.340E-05 | 1.737E-03 |
| HBEGF | 1.268 | 2.408 | 7.978E-04 | 1.317E-02 |
| DNAJB1 | 1.270 | 2.411 | 9.110E-19 | 8.930E-16 |
| ENSG00000280138 | 1.273 | 2.417 | 5.040E-19 | 5.250E-16 |
| ARRDC4 | 1.278 | 2.425 | 1.640E-06 | 1.019E-04 |
| PELI1 | 1.282 | 2.431 | 5.520E-10 | 9.790E-08 |
| CTSF | 1.286 | 2.438 | 2.097E-04 | 5.021E-03 |
| RNF103 | 1.294 | 2.452 | 1.670E-08 | 1.870E-06 |
| BNC2 | 1.298 | 2.460 | 4.575E-03 | 4.625E-02 |
| VEGFA | 1.299 | 2.461 | 1.303E-03 | 1.907E-02 |
| POLR1F | 1.305 | 2.471 | 8.340E-14 | 4.020E-11 |
| SOX4 | 1.306 | 2.472 | 9.710E-09 | 1.210E-06 |
| HIF1A | 1.307 | 2.475 | 4.400E-10 | 8.050E-08 |
| ENSG00000224505 | 1.308 | 2.477 | 1.838E-03 | 2.436E-02 |
| JUND | 1.313 | 2.484 | 9.570E-09 | 1.200E-06 |
| FOSB | 1.313 | 2.485 | 6.710E-11 | 1.620E-08 |
| MIR590 | 1.314 | 2.486 | 1.505E-03 | 2.114E-02 |
| NR3C2 | 1.325 | 2.505 | 4.103E-03 | 4.302E-02 |
| PLK3 | 1.325 | 2.505 | 1.350E-09 | 2.100E-07 |
| IL12A | 1.325 | 2.506 | 3.290E-03 | 3.689E-02 |
| SNHG9 | 1.329 | 2.511 | 8.253E-04 | 1.346E-02 |
| COL18A1 | 1.340 | 2.531 | 1.042E-03 | 1.610E-02 |
| ZNF331 | 1.340 | 2.531 | 1.390E-06 | 8.870E-05 |
| ADAMTS17 | 1.346 | 2.542 | 1.707E-04 | 4.253E-03 |
| RPS6KL1 | 1.348 | 2.546 | 1.230E-03 | 1.824E-02 |
| TAS1R3 | 1.350 | 2.549 | 2.310E-06 | 1.354E-04 |
| IL23A | 1.359 | 2.565 | 3.190E-05 | 1.165E-03 |
| ENSG00000233264 | 1.360 | 2.567 | 7.080E-05 | 2.121E-03 |
| C17orf49 | 1.364 | 2.574 | 8.500E-09 | 1.080E-06 |
| PTCH1 | 1.367 | 2.580 | 4.969E-03 | 4.889E-02 |
| ENSG00000256152 | 1.385 | 2.612 | 2.891E-04 | 6.312E-03 |
| ZNF394 | 1.392 | 2.625 | 4.330E-13 | 1.760E-10 |
| HDGFL3 | 1.397 | 2.634 | 4.760E-05 | 1.584E-03 |
| LINC02390 | 1.404 | 2.646 | 1.660E-05 | 6.972E-04 |
| SBDSP1 | 1.409 | 2.655 | 8.210E-21 | 1.140E-17 |
| SLC2A3 | 1.410 | 2.657 | 2.500E-07 | 2.110E-05 |
| PFKFB3 | 1.412 | 2.661 | 3.230E-11 | 8.030E-09 |
| NTN5 | 1.413 | 2.662 | 1.366E-03 | 1.974E-02 |
| NSG1 | 1.421 | 2.678 | 5.188E-04 | 9.551E-03 |
| SBDS | 1.425 | 2.685 | 6.640E-13 | 2.510E-10 |
| SOCS3 | 1.428 | 2.691 | 4.960E-06 | 2.588E-04 |
| DDIT3 | 1.435 | 2.703 | 3.840E-16 | 2.560E-13 |
| ZNF14 | 1.457 | 2.744 | 7.420E-10 | 1.210E-07 |
| FBXO33 | 1.457 | 2.744 | 5.270E-20 | 6.270E-17 |
| OTUD7A | 1.460 | 2.750 | 1.040E-06 | 6.980E-05 |
| DFFBP1 | 1.464 | 2.758 | 2.560E-05 | 9.866E-04 |
| SDE2 | 1.464 | 2.759 | 1.150E-17 | 8.730E-15 |
| PDE4D | 1.468 | 2.766 | 4.040E-09 | 5.600E-07 |
| CTLA4 | 1.481 | 2.791 | 9.951E-04 | 1.557E-02 |
| RN7SL2 | 1.481 | 2.792 | 1.260E-05 | 5.585E-04 |
| EIF2AK3-DT | 1.482 | 2.793 | 1.190E-08 | 1.430E-06 |
| FOSL2 | 1.487 | 2.804 | 4.440E-17 | 3.220E-14 |
| SNORA75 | 1.491 | 2.812 | 2.518E-03 | 3.082E-02 |
| CITED4 | 1.494 | 2.816 | 2.000E-05 | 8.122E-04 |
| NDUFV2 | 1.496 | 2.820 | 8.150E-12 | 2.420E-09 |
| PIM3 | 1.500 | 2.829 | 9.130E-29 | 5.070E-25 |
| IGLV4-60 | 1.507 | 2.843 | 3.758E-03 | 4.050E-02 |
| GADD45A | 1.519 | 2.866 | 2.780E-10 | 5.590E-08 |
| ENSG00000229299 | 1.519 | 2.867 | 6.000E-06 | 2.991E-04 |
| B3GNT7 | 1.524 | 2.876 | 7.980E-07 | 5.560E-05 |
| EGR1 | 1.538 | 2.905 | 9.542E-04 | 1.514E-02 |
| ENSG00000279520 | 1.540 | 2.907 | 2.340E-09 | 3.340E-07 |
| RAB3A | 1.541 | 2.910 | 1.948E-03 | 2.540E-02 |
| LINC02033 | 1.542 | 2.912 | 4.793E-03 | 4.757E-02 |
| ENSG00000275056 | 1.546 | 2.921 | 1.135E-03 | 1.721E-02 |
| ENSG00000228201 | 1.553 | 2.935 | 5.026E-04 | 9.348E-03 |
| RSPH4A | 1.560 | 2.948 | 4.216E-03 | 4.382E-02 |
| HAR1A | 1.562 | 2.954 | 7.110E-04 | 1.209E-02 |
| NFKBIA | 1.571 | 2.971 | 1.490E-09 | 2.300E-07 |
| FGFR1 | 1.573 | 2.976 | 1.040E-06 | 6.980E-05 |
| WDR86 | 1.578 | 2.986 | 9.610E-06 | 4.400E-04 |
| SCARF2 | 1.580 | 2.989 | 3.076E-03 | 3.537E-02 |
| JUN | 1.589 | 3.008 | 1.260E-21 | 2.090E-18 |
| CEROX1 | 1.593 | 3.016 | 4.580E-07 | 3.470E-05 |
| RPL13P12 | 1.596 | 3.023 | 1.579E-04 | 3.997E-03 |
| ENSG00000260708 | 1.596 | 3.023 | 2.880E-12 | 9.230E-10 |
| BCL3 | 1.613 | 3.060 | 1.080E-09 | 1.720E-07 |
| MMP28 | 1.618 | 3.071 | 7.460E-04 | 1.254E-02 |
| LINC00304 | 1.630 | 3.096 | 3.968E-04 | 7.966E-03 |
| MYRF | 1.634 | 3.103 | 8.451E-04 | 1.370E-02 |
| ENSG00000276517 | 1.640 | 3.117 | 8.122E-04 | 1.333E-02 |
| TNRC18P1 | 1.649 | 3.137 | 1.341E-04 | 3.513E-03 |
| FAM183BP | 1.650 | 3.138 | 1.392E-03 | 2.004E-02 |
| IRS2 | 1.653 | 3.144 | 1.170E-09 | 1.840E-07 |
| TRPM5 | 1.690 | 3.227 | 3.522E-03 | 3.881E-02 |
| ENSG00000276853 | 1.695 | 3.238 | 2.018E-03 | 2.612E-02 |
| MTSS2 | 1.697 | 3.243 | 9.540E-11 | 2.150E-08 |
| RBKS | 1.719 | 3.292 | 1.027E-03 | 1.591E-02 |
| ENSG00000255847 | 1.723 | 3.301 | 3.640E-13 | 1.520E-10 |
| ENSG00000273306 | 1.752 | 3.367 | 1.572E-04 | 3.991E-03 |
| ENC1 | 1.759 | 3.384 | 3.320E-07 | 2.670E-05 |
| ENSG00000279908 | 1.775 | 3.423 | 2.420E-09 | 3.410E-07 |
| FAM153A | 1.799 | 3.480 | 6.660E-05 | 2.041E-03 |
| ENSG00000232891 | 1.804 | 3.492 | 4.911E-04 | 9.204E-03 |
| BTG3 | 1.827 | 3.547 | 1.400E-11 | 3.880E-09 |
| ZBTB10 | 1.829 | 3.552 | 2.450E-26 | 5.830E-23 |
| KCNK7 | 1.835 | 3.568 | 9.350E-06 | 4.291E-04 |
| ZC3H12A | 1.840 | 3.580 | 9.790E-28 | 4.080E-24 |
| EGR2 | 1.848 | 3.599 | 1.150E-05 | 5.160E-04 |
| KLF5 | 1.849 | 3.601 | 1.630E-05 | 6.907E-04 |
| TNFAIP3 | 1.853 | 3.612 | 1.110E-08 | 1.340E-06 |
| ENSG00000279765 | 1.859 | 3.627 | 7.840E-05 | 2.309E-03 |
| CCL3L3 | 1.886 | 3.696 | 3.710E-05 | 1.310E-03 |
| CFL2 | 1.895 | 3.719 | 1.500E-12 | 5.090E-10 |
| TTC34 | 1.896 | 3.721 | 1.450E-05 | 6.245E-04 |
| CRABP2 | 1.915 | 3.771 | 1.857E-03 | 2.455E-02 |
| TP53INP2 | 1.918 | 3.778 | 1.310E-15 | 7.510E-13 |
| UPK3B | 1.928 | 3.805 | 1.020E-06 | 6.930E-05 |
| RGS17P1 | 1.929 | 3.808 | 3.560E-05 | 1.267E-03 |
| LMTK3 | 1.930 | 3.811 | 3.880E-07 | 3.010E-05 |
| KRT18 | 1.933 | 3.820 | 6.919E-04 | 1.184E-02 |
| PSMD10P1 | 1.980 | 3.944 | 2.660E-07 | 2.230E-05 |
| OSM | 1.982 | 3.952 | 3.330E-05 | 1.202E-03 |
| DUSP2 | 2.021 | 4.060 | 3.140E-11 | 7.920E-09 |
| ENSG00000261159 | 2.031 | 4.086 | 1.614E-03 | 2.228E-02 |
| NA | 2.035 | 4.099 | 4.230E-05 | 1.447E-03 |
| MAFF | 2.085 | 4.242 | 1.260E-08 | 1.500E-06 |
| LRRC32 | 2.090 | 4.256 | 7.562E-04 | 1.266E-02 |
| RHBDL1 | 2.123 | 4.357 | 1.977E-04 | 4.799E-03 |
| SNAI1 | 2.148 | 4.432 | 9.033E-04 | 1.444E-02 |
| AREG | 2.166 | 4.489 | 4.989E-04 | 9.308E-03 |
| ENSG00000272256 | 2.171 | 4.504 | 1.440E-05 | 6.211E-04 |
| ENSG00000267655 | 2.203 | 4.603 | 1.972E-03 | 2.567E-02 |
| FAM153B | 2.206 | 4.614 | 3.677E-04 | 7.499E-03 |
| RND1 | 2.213 | 4.635 | 7.490E-04 | 1.258E-02 |
| RPL23AP81 | 2.261 | 4.794 | 4.746E-04 | 8.955E-03 |
| FFAR1 | 2.265 | 4.806 | 3.720E-18 | 3.440E-15 |
| NA | 2.267 | 4.812 | 2.350E-10 | 4.960E-08 |
| ENSG00000245869 | 2.314 | 4.971 | 5.380E-06 | 2.748E-04 |
| DIP2A-IT1 | 2.339 | 5.059 | 1.960E-06 | 1.186E-04 |
| MATN1 | 2.339 | 5.061 | 3.210E-05 | 1.169E-03 |
| ENSG00000231305 | 2.357 | 5.122 | 7.635E-04 | 1.275E-02 |
| ENSG00000256712 | 2.367 | 5.157 | 2.747E-03 | 3.269E-02 |
| GRM2 | 2.392 | 5.248 | 5.290E-09 | 7.110E-07 |
| MIR4420 | 2.399 | 5.274 | 2.860E-06 | 1.639E-04 |
| CXCL8 | 2.400 | 5.279 | 7.380E-10 | 1.210E-07 |
| ENSG00000207525 | 2.410 | 5.316 | 3.989E-04 | 7.997E-03 |
| RBBP4P1 | 2.416 | 5.336 | 3.657E-03 | 3.995E-02 |
| SULT1A2 | 2.435 | 5.406 | 9.660E-07 | 6.570E-05 |
| TCTE1 | 2.442 | 5.433 | 5.029E-03 | 4.935E-02 |
| AVPI1 | 2.443 | 5.440 | 8.720E-05 | 2.508E-03 |
| HLA-U | 2.444 | 5.440 | 1.685E-03 | 2.288E-02 |
| ENSG00000275927 | 2.458 | 5.494 | 1.932E-03 | 2.527E-02 |
| EREG | 2.501 | 5.660 | 5.270E-05 | 1.724E-03 |
| LOC105378721 | 2.524 | 5.750 | 6.651E-04 | 1.153E-02 |
| TNFSF9 | 2.532 | 5.785 | 8.500E-05 | 2.459E-03 |
| FLT4 | 2.538 | 5.807 | 6.380E-05 | 1.973E-03 |
| CHMP4BP1 | 2.538 | 5.808 | 6.960E-05 | 2.100E-03 |
| DUSP4 | 2.605 | 6.083 | 3.700E-06 | 2.048E-04 |
| ENSG00000274677 | 2.653 | 6.288 | 3.481E-04 | 7.214E-03 |
| ENSG00000278022 | 2.676 | 6.390 | 4.150E-05 | 1.425E-03 |
| ENSG00000256913 | 2.717 | 6.576 | 4.902E-04 | 9.198E-03 |
| NR4A3 | 2.718 | 6.580 | 1.740E-07 | 1.570E-05 |
| PER1 | 2.758 | 6.765 | 6.470E-21 | 9.800E-18 |
| TAMALIN | 2.791 | 6.921 | 3.640E-45 | 6.070E-41 |
| CD83 | 2.850 | 7.210 | 2.750E-24 | 5.100E-21 |
| FAM238A | 2.902 | 7.473 | 2.770E-16 | 1.920E-13 |
| ADRB1 | 2.906 | 7.496 | 2.030E-05 | 8.221E-04 |
| DUSP8 | 2.981 | 7.895 | 8.650E-06 | 4.024E-04 |
| THNSL2 | 3.049 | 8.277 | 3.100E-03 | 3.557E-02 |
| ENSG00000270681 | 3.108 | 8.624 | 3.192E-04 | 6.754E-03 |
| ACTN1-AS1 | 3.126 | 8.732 | 4.670E-06 | 2.452E-04 |
| FOSL1 | 3.150 | 8.877 | 4.726E-03 | 4.716E-02 |
| TPBGL | 3.156 | 8.914 | 4.160E-03 | 4.334E-02 |
| CHRM4 | 3.189 | 9.120 | 2.280E-06 | 1.340E-04 |
| TMEM119 | 3.276 | 9.684 | 1.179E-03 | 1.772E-02 |
| NR4A2 | 3.339 | 10.116 | 7.350E-18 | 5.830E-15 |
| GRK5-IT1 | 3.339 | 10.117 | 1.958E-04 | 4.771E-03 |
| MMP9 | 3.528 | 11.532 | 8.140E-06 | 3.841E-04 |
| GEM | 3.551 | 11.718 | 4.821E-03 | 4.776E-02 |
| G0S2 | 3.560 | 11.793 | 6.010E-16 | 3.850E-13 |
| SAXO2 | 3.803 | 13.956 | 6.210E-26 | 1.290E-22 |
| HAR1B | 3.908 | 15.007 | 5.480E-05 | 1.775E-03 |
| ABHD17AP6 | 3.928 | 15.221 | 3.253E-03 | 3.665E-02 |
| ENSG00000259635 | 4.089 | 17.022 | 4.475E-04 | 8.610E-03 |
| IL13 | 4.111 | 17.283 | 2.698E-04 | 5.994E-03 |
| SLED1 | 4.143 | 17.665 | 3.646E-03 | 3.986E-02 |
| TEX45 | 5.004 | 32.082 | 4.948E-04 | 9.244E-03 |
| YAP1P1 | 5.831 | 56.906 | 3.275E-04 | 6.864E-03 |
| CLLU1-AS1 | 5.963 | 62.401 | 9.653E-04 | 1.527E-02 |
| ENSG00000224029 | 6.334 | 80.649 | 4.380E-05 | 1.492E-03 |
| RPS4XP22 | 6.439 | 86.769 | 2.270E-03 | 2.851E-02 |
| LRRC7 | 6.948 | 123.489 | 1.091E-04 | 3.014E-03 |
| RASD2 | 6.974 | 125.745 | 2.660E-05 | 1.008E-03 |
| TUBB8B | 6.974 | 125.745 | 2.660E-05 | 1.008E-03 |
| MTCO3P12 | 8.403 | 338.535 | 1.370E-05 | 5.963E-04 |
| ZFP57 | 8.785 | 441.159 | 2.190E-09 | 3.180E-07 |
Patients with AD and HCs were stratified by age (0‒6 months and 7‒12 months), and differential expression analysis on RNA-seq data was performed between the four groups: children with AD aged >6 months above versus age-matched HCs, children with AD aged <6 months versus age-matched HCs, patients with AD aged >6 months versus patients with AD aged <6 months, and HCs aged >6 months versus HCs aged <6 months. Meeting criteria were set at FC ≥1.5 and FDR <0.05.
Abbreviations: AD, atopic dermatitis; FC, fold change; FDR, false discovery rate; HC, healthy control; KLK, kallikrein; MMP, matrix metalloproteinase; RNA-seq, RNA sequencing; STAT, signal transducer and activator of transcription; TLR, toll-like receptor.
Table 4.
Effect of Infant’s Age on Validated DEG in Infants with AD
| DEG | Group | LFC | FC | FDR |
|---|---|---|---|---|
| IL18RAP | Patients with AD aged >6 mo versus HCs | ‒1.968 | 0.256 | 0.003 |
| Patients with AD aged <6 mo versus HCs | ‒1.309 | 0.404 | 0.043 | |
| IL1B | Patients with AD aged <6 mo versus HCs | 3.901 | 14.942 | 0.000 |
| TNF | Patients with AD aged <6 mo versus HCs | 3.092 | 8.526 | 0.000 |
| TREM1 | Patients with AD aged <6 mo versus HCs | 3.164 | 8.963 | 0.000 |
| EGR3 | Patients with AD aged <6 mo versus HCs | 5.076 | 33.732 | 0.015 |
Shown is the effect of infant’s age on the expression of selected genes that were differentially expressed in infants with AD and validated by RT-qPCR. Differential expression analysis between patients with AD and HCs was performed using DESeq2 on RNA-seq data after stratifying the dataset by age (0‒6 months and 7‒12 months).
Abbreviations: AD, atopic dermatitis; AIF, apoptosis-inducing factor; DEG, differentially expressed gene; FC, fold change; FDR, false discovery rate; HC, healthy control; LFC, log2 fold change; OSM, oncostatin M; RNA-seq, RNA sequencing; XAF1, XIAP-associated factor 1.
Figure 3.
Effect of infant’s age on the expression of differentially expressed genes in infants with AD. RT-qPCR analyses for five genes from the top 10 differentially expressed genes identified by high-throughput RNA sequencing: (a) IL18RAP, (b) IL1β, (c) TNF, (d) TREM1, and (e) EGR3 in children with AD (n = 27) and healthy controls (n = 17) after stratifying the dataset by age (0‒6 months and 7‒12 months). Fold change was calculated by 2-ΔΔCT method. The normalized expression data were log2 transformed and shown as the means ± SD. Significant difference among groups was calculated by unpaired t test with Welch’s correction for normal distribution or with Mann‒Whitney rank-sum test for non-normal distribution data. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. AD, atopic dermatitis; HC, healthy control.
Gene ontology and pathway analysis of DEGs
To gain insight into the gene ontology (GO) categories of DEGs between the AD group and control group, all DEGs were uploaded to the DAVID (Database for Annotation, Visualization and Integrated Discovery) database. GO analysis contained three categories: biological process (BP), cellular component (CC), and molecular function (MF). In total, 86 significant GO terms were enriched for DEGs identified in the AD group, of which 57 were within the BP category, 21 were within the CC category, and 8 were within the MF category. The enriched BP categories included immune response, inflammatory response, regulation of immune response, leukocyte migration, positive regulation of NF-kB transcription factor activity, cell adhesion, cell surface receptor signaling pathway, and many others. In the category CC, the DEGs were significantly enriched in the plasma membrane, extracellular region, extracellular exosome, cell surface, and cytoplasm. Furthermore, in the category MF, DEGs were mainly enriched in antigen binding, receptor activity, cytokine activity, enzyme binding, and protein binding. The selected pathways significantly enriched in the AD group included hematopoietic cell lineage, NK cell‒mediated cytotoxicity, phosphoinositide 3-kinase‒protein kinase B signaling pathway, cytokine‒cytokine receptor interaction, extracellular matrix‒receptor interaction, and immunoregulatory interactions between a lymphoid and a nonlymphoid cell (Figure 4a and b). Further details of the results of the GO enrichment and pathway analyses are provided in Table 5.
Figure 4.
GO enrichment and pathway analysis of differentially expressed genes. (a) The top 20 enriched GO terms; the x-axis represents gene counts, and the y-axis represents GO terms. (b) Selected KEGG pathways; the x-axis represents gene counts, and the y-axis represents KEGG pathway names. Akt, protein kinase B; ECM, extracellular matrix; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PI3K, phosphoinositide 3-kinase.
Table 5.
GO Enrichment and Pathway Analyses of Differentially Expressed Genes
| GO Term | Gene Count |
P-Value | Genes |
|---|---|---|---|
| Biological process | |||
| Regulation of immune response | 17 | 3.76E-12 |
ICAM1, IGLV1-44, IGKV5-2, NCR1, IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV2-23, IGKV4-1, KLRF1,TREM1, KIR2DL3, TREML1, IGLC2, KLRD1, and IGKV3-15 |
| Receptor-mediated endocytosis | 16 | 8.68E-11 |
LDLR, IGLV1-44, IGKV5-2, JCHAIN, SPARC, IGLV2-11, IGLV2-8, IGLV3-19, CTTN, IGKV1-5, IGLV2-23, IGKV4-1, IGLC2, LRP3, CD14, and IGKV3-15 |
| Complement activation | 11 | 4.37E-09 | IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, CLU, IGKV4-1, IGLC2, and IGKV3-15 |
| Immune response | 20 | 4.97E-09 |
IL18R1, TNF, IL18RAP, CXCL5, IGLV1-44, IGKV5-2, JCHAIN, FASLG, PF4, IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, PPBP, FCAR, IGLV2-23, IGKV4-1, IL1B, KIR2DL3, and IGKV3-15 |
| Complement activation and classical pathway | 11 | 1.56E-08 | IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, CLU, IGKV4-1, IGLC2, and IGKV3-15 |
| Platelet activation | 10 | 7.55E-07 |
VWF, GP6, F5, C6ORF25, PF4, GP1BA, ITGB3, TREML1, CLEC1B, and GP9 |
| Fc-γ receptor signaling pathway involved in phagocytosis | 10 | 1.74E-06 | IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, IGKV4-1, IGLC2, and IGKV3-15 |
| Platelet degranulation | 9 | 3.36E-06 | VWF, APP, F5, PPBP, CLU, PF4, SPARC, ITGB3, and ITGA2B |
| Blood coagulation | 11 | 5.08E-06 |
PRKAR2B, VWF, GP6, F5, C6ORF25, PDGFC, GP1BA, PDGFD, ITGB3, GP9, and PLAUR |
| FCERI signaling pathway | 10 | 2.71E-05 | IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, IGKV4-1, IGLC2, and IGKV3-15 |
| Positive regulation of nitric oxide biosynthetic process | 6 | 3.54E-05 | ICAM1, TNF, CLU, IL1B, PTX3, and KLF4 |
| Proteolysis | 14 | 0.000 |
NRIP3, IGLV1-44, IGKV5-2, ANPEP, MMP25, IGLV2-11, IGKV1-5, IGLV3-19, IGLV2-8, F5, IGLV2-23, IGKV4-1, IGLC2, and IGKV3-15 |
| Inflammatory response | 12 | 0.001 |
SDC1, TNF, IL18RAP, PPBP, CXCL5, ANXA1, IL1B, PF4, PTX3, NLRP3, CD14, and MMP25 |
| Antibacterial humoral response | 5 | 0.001 | APP, ADM, HIST2H2BE, HIST1H2BJ, and JCHAIN |
| Positive regulation of cell division | 5 | 0.001 | TAL1, PPBP, IL1B, PDGFC, and PDGFD |
| Innate immune response | 12 | 0.001 |
APP, CLU, ANXA1, JCHAIN, PADI4, TREM1, PTX3, NLRP3, IGLC2, TREML1, KLRD1, and CD14 |
| ECM organization | 8 | 0.002 | ICAM1, VWF, APP, TNF, ITGB5, SPARC, ITGB3, and ITGA2B |
| Response to glucocorticoid | 5 | 0.003 | SDC1, TNF, ADM, SPARC, and ADAM9 |
| Response to lipopolysaccharide | 7 | 0.003 | PPBP, ADM, CXCL5, FASLG, PF4, SPARC, and TRIB1 |
| Positive regulation of chemokine biosynthetic process | 3 | 0.003 | EGR1, TNF, and IL1B |
| Negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | 4 | 0.004 | TNF, MCL1, IL1B, and PF4 |
| Viral entry into host cell | 5 | 0.005 | ICAM1, LDLR, ITGB5, ANPEP, and ITGB3 |
| Response to yeast | 3 | 0.006 | APP, ADM, and PTX3 |
| Defense response to Gram-positive bacterium | 5 | 0.007 | APP, TNF, ADM, HIST2H2BE, and HIST1H2BJ |
| Negative regulation of gene expression | 6 | 0.007 | TNF, LDLR, GAS2L1, ZNF503, MYADM, and KLF4 |
| Cell adhesion mediated by integrin | 3 | 0.007 | ICAM1, ITGB3, and ADAM9 |
| Positive regulation of membrane protein ectodomain proteolysis | 3 | 0.007 | TNF, IL1B, and ADAM9 |
| Positive regulation of IFN-γ production | 4 | 0.008 | IL18R1, TNF, IL1B, and CD14 |
| Cell surface receptor signaling pathway | 8 | 0.01 |
IL18RAP, ANXA1, GP1BA, TSPAN9, KLRF1, KLRD1, CD14, and CLEC1B |
| Blood coagulation, intrinsic pathway | 3 | 0.011 | VWF, GP1BA, and GP9 |
| Positive regulation of leukocyte chemotaxis | 3 | 0.011 | PPBP, CXCL5, and PF4 |
| Integrin-mediated signaling pathway | 5 | 0.011 | C6ORF25, ITGB5, ITGB3, ADAM9, and ITGA2B |
| Platelet formation | 3 | 0.012 | TAL1, C6ORF25, and CLEC1B |
| Positive regulation of transcription from RNA polymerase II promoter | 17 | 0.012 | EGR1, NAMPT, TNF, EGR2, ABLIM3, PF4, MYBL1, NLRP3, AHR, TAL1, APP, SPX, BHLHA15, IL1B, MAML3, FOSL1, and KLF4 |
| Cellular response to hydrogen peroxide | 4 | 0.014 | IL18RAP, ANXA1, PDGFD, and KLF4 |
| Positive regulation of NF-kB import into the nucleus | 3 | 0.014 | IL18R1, TNF, and IL1B |
| Positive regulation of MAPK activity | 4 | 0.015 | TNF, PDE5A, PDGFC, and PDGFD |
| Positive regulation of smooth muscle cell proliferation | 4 | 0.016 | NAMPT, TNF, PDGFD, and ALOX12 |
| Cellular response to lipopolysaccharide | 5 | 0.017 | ICAM1, TNF, NLRP3, CD14, and ADAM9 |
| Regulation of gastric acid secretion | 2 | 0.017 | SGK1 and KCNQ1 |
| Cell adhesion | 10 | 0.02 | ICAM1, VWF, APP, SCN1B, ITGB5, GP1BA, ITGB3, ADAM9, GP9, and ITGA2B |
| Leukocyte migration | 5 | 0.022 | ICAM1, GP6, ESAM, TREM1, and ITGB3 |
| Regulation of cell proliferation | 6 | 0.024 | TAL1, SGK1, TNF, PPBP, ANXA1, and PF4 |
| Establishment or maintenance of microtubule cytoskeleton polarity | 2 | 0.026 | KIF2C and LMNA |
| positive regulation of calcidiol 1-monooxygenase activity | 2 | 0.026 | TNF and IL1B |
| Positive regulation of phagocytosis | 3 | 0.026 | TNF, IL1B, and PTX3 |
| Positive regulation of gene expression | 7 | 0.028 | TNF, LDLR, ID1, IL1B, PF4, KLF4, and ALOX12 |
| Positive regulation of NF-kB transcription factor activity | 5 | 0.03 | ICAM1, TNF, CLU, IL1B, and NLRP3 |
| Lipopolysaccharide-mediated signaling pathway | 3 | 0.032 | TNF, IL1B, and CD14 |
| Sequestering of triglyceride | 2 | 0.035 | TNF and IL1B |
| IL-1 beta production | 2 | 0.043 | IL1B and NLRP3 |
| Positive regulation of fever generation | 2 | 0.043 | TNF and IL1B |
| Cell matrix adhesion | 4 | 0.044 | ITGB5, ITGB3, ADAM9, and ITGA2B |
| TGFβ receptor signaling pathway | 4 | 0.047 | ID1, CLDN5, ITGB5, and ADAM9 |
| Positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 3 | 0.048 | TNF, NLRP3, and ALOX12 |
| Positive regulation of apoptotic process | 7 | 0.049 | TNF, ADM, CLU, FASLG, FOSL1, MELK, and DUSP6 |
| Platelet aggregation | 3 | 0.049 | GP1BA, ITGB3, and ITGA2B |
| Cellular component | |||
| Plasma membrane | 68 | 4.45E-09 |
SEPT5, IGLV1-44, LDLR, C6ORF25, CLDN5, FASLG, IGKV1-12, TSPAN9, MMP25, GLDC, GP9, ATP2B1, PRKAR2B, CTTN, APP, GP6, HOMER3, IGLV2-23, ZNF185, DLG3, PDGFC, ESAM, FAM129B, KLRD1, KCNQ1, ICAM1, SGK1, IL18RAP, SLCO4A1, WLS, NCR1, MYADM, PLAUR, IGLV2-11, IGKV1-5, SDC1, F5, COLQ, IGKV4-1, TREM1, KIR2DL3, EMP1, MELK, CLEC1B, ITGA2B, TNF, SCN1B, CALD1, IGKV5-2, ITGB5, GNG11, ITGB3, C2ORF88, IGLV2-8, IGLV3-19, GP1BA, KLRF1, IGKV3-15, IL18R1, ANXA1, SPARC, RAPH1, AQP10, FCAR, CPNE2, IGLC2, TREML1, and CD14 |
| Extracellular region | 34 | 1.16E-06 | LTBP1, SCN1B, TNF, IGLV1-44, CXCL5, IGKV5-2, CLU, JCHAIN, FASLG, PF4, IGKV1-12, IGLV2-8, APP, IGLV3-19, IGLV2-23, IL1B, PDGFC, PDGFD, PTX3, IGKV3-15, ANXA1, SPARC, NLRP3, IGLV2-11, VWF, IGKV1-5, F5, ADM, FCAR, PPBP, IGKV4-1, TREM1, IGLC2, and CD14 |
| Extracellular space | 30 | 2.25E-06 | NAMPT, TNF, CXCL5, FAM20C, CLU, JCHAIN, FASLG, PF4, ANPEP, APP, SPX, HIST1H2BJ, IL1B, DLG3, PDGFC, PDGFD, PTX3, ADAM9, ICAM1, ANXA1, SPARC, F5, PPBP, ADM, COLQ, HIST2H2BE, FRMD4B, IGLC2, CD14, and CMTM5 |
| Platelet alpha granule lumen | 7 | 6.55E-06 | VWF, APP, F5, PPBP, CLU, PF4, and SPARC |
| Cell surface | 17 | 1.34E-05 |
ICAM1, TNF, LDLR, CLU, ANXA1, ITGB5, SPARC, and ITGB3, APP, SDC1, GP6, GP1BA, PDGFC, TREML1, INTU, ITGA2B, and ADAM9 |
| Extracellular exosome | 44 | 4.28E-05 |
HIST1H2AC, NAMPT, CLU, FAM20C, CLDN5, JCHAIN, FASLG, ITGB5, ANPEP, ITGB3, ATP2B1, PRKAR2B, CTTN, APP, IGLV3-19, GP6, PGRMC1, IL1B, PDGFC, ESAM, GP1BA, FAM129B, PDGFD, TUBA1A, TUBB1, ADAM9, ICAM1, ANXA1, WLS, MYADM, PLAUR, IGLV2-11, PDZK1IP1, VWF, IGKV1-5, SDC1, HIST2H2BE, SH3BGRL2, CPNE2, IGLC2, CD14, XYLB, ALOX12, and ITGA2B |
| Membrane raft | 9 | 3.55E-04 |
ATP2B1, PRKAR2B, ICAM1, APP, TNF, SDPR, KCNQ1, MYADM, and CD14 |
| External side of the plasma membrane | 9 | 4.43E-04 |
ICAM1, SDC1, TNF, LDLR, FASLG, ANPEP, IGLC2, KLRD1, and ITGA2B |
| Focal adhesion | 11 | 0.002 |
ICAM1, SDC1, CTTN, ZNF185, ANXA1, ITGB5, TSPAN9, ITGB3, ADAM9, PLAUR, and ITGA2B |
| Blood microparticle | 7 | 0.002 |
IGKV1-5, CLU, JCHAIN, IGKV4-1, IGLC2, IGKV3-15, and ITGA2B |
| Platelet alpha granule membrane | 3 | 0.005 | SPARC, ITGB3, and ITGA2B |
| Platelet alpha granule | 3 | 0.006 | VWF, SPARC, and TREML1 |
| Integral component of the plasma membrane | 22 | 0.008 |
ICAM1, TNF, LDLR, SLCO4A1, FASLG, ANPEP, TSPAN9, ITGB3, NCR1, AQP10, PLAUR, GP9, ATP2B1, APP, SDC1, GP6, FCAR, GP1BA, KLRF1, KIR2DL3, CLEC1B, and ITGA2B |
| Phagocytic cup | 3 | 0.011 | TNF, PEAR1, and ANXA1 |
| Clathrin-coated pit | 4 | 0.011 | APP, CTTN, LDLR, and LRP3 |
| Anchored component of the external side of the plasma membrane | 3 | 0.014 | GGTA1P, GP1BA, and CD14 |
| Basolateral plasma membrane | 6 | 0.018 | ATP2B1, LDLR, ANXA1, DLG3, KCNQ1, and ADAM9 |
| Integrin complex | 3 | 0.021 | ITGB5, ITGB3, and ITGA2B |
| Receptor complex | 5 | 0.022 | APP, LDLR, ITGB5, ITGB3, and KLRD1 |
| Dendritic shaft | 3 | 0.029 | PRKAR2B, APP, and DLG3 |
| Cytoplasm | 55 | 0.043 |
NAMPT, MCM10, ISG20, PRKAR2B, CTTN, APP, HOMER3, SDPR, ZNF185, HIST1H2BJ, DLG3, PIWIL2, PDGFC, FAM129B, TUBB1, KCNQ1, EGR1, SGK1, EGR2, PADI4, UBE2C, NLRP3, AHR, SH2D2A, SDC1, ADM, HIST2H2BE, FRMD4B, PPP1R15A, INTU, XYLB, ALOX12, MCL1, ABLIM3, CLU, TRIB1, SPATS2, NCAPG, RNF165, STRIP2, SKA3, GP1BA, HRASLS2, ANXA1, LMNA, CDC20, SPARC, RAPH1, SH3BGRL2, GAS2L1, CPNE2, RFX2, TREML1, KLF4, and DUSP6 |
| Molecular function | |||
| Antigen binding | 12 | 1.30E-09 |
IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, JCHAIN, IGKV4-1, KIR2DL3, IGLC2, and IGKV3-15 |
| Receptor activity | 11 | 1.82E-05 |
ICAM1, IL18R1, GP6, IL18RAP, LDLR, ITGB5, ANPEP, TREM1, ITGB3, KIR2DL3, and PLAUR |
| Serine-type endopeptidase activity | 11 | 7.17E-05 |
IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, F5, IGLV1-44, IGLV2-23, IGKV5-2, IGKV4-1, IGLC2, and IGKV3-15 |
| Virus receptor activity | 5 | 0.003 | ICAM1, LDLR, ITGB5, ANPEP, and ITGB3 |
| Platelet-derived GF receptor binding | 3 | 0.007 | PDGFC, PDGFD, and ITGB3 |
| Collagen binding | 4 | 0.015 | VWF, GP6, SPARC, and ADAM9 |
| ECM binding | 3 | 0.021 | SPARC, ITGB3, and ITGA2B |
| IgA binding | 2 | 0.026 | FCAR and JCHAIN |
| Pathway | |||
| KEGG pathway | |||
| ECM‒receptor interaction | 8 | 4.38E-05 | VWF, SDC1, GP6, ITGB5, GP1BA, ITGB3, GP9, and ITGA2B |
| Hematopoietic cell lineage | 8 | 4.38E-05 | TNF, IL1B, ANPEP, GP1BA, ITGB3, CD14, GP9, and ITGA2B |
| African trypanosomiasis | 4 | 0.006 | ICAM1, TNF, IL1B, and FASLG |
| Pertussis | 5 | 0.009 | TNF, CXCL5, IL1B, NLRP3, and CD14 |
| Hypertrophic cardiomyopathy | 5 | 0.011 | TNF, LMNA, ITGB5, ITGB3, and ITGA2B |
| NK cell‒mediated cytotoxicity | 6 | 0.011 | ICAM1, TNF, FASLG, KIR2DL3, NCR1, and KLRD1 |
| PI3K‒Akt signaling pathway | 10 | 0.013 |
VWF, SGK1, MCL1, ITGB5, FASLG, GNG11, PDGFC, PDGFD, ITGB3, and ITGA2B |
| Dilated cardiomyopathy | 5 | 0.014 | TNF, LMNA, ITGB5, ITGB3, and ITGA2B |
| Platelet activation | 6 | 0.014 | VWF, GP6, GP1BA, ITGB3, GP9, and ITGA2B |
| Malaria | 4 | 0.017 | ICAM1, SDC1, TNF, and IL1B |
| Cytokine‒cytokine receptor interaction | 8 | 0.017 | IL18R1, TNF, IL18RAP, PPBP, CXCL5, IL1B, FASLG, and PF4 |
| Pathogenic E. coli infection | 4 | 0.018 | CTTN, TUBB1, TUBA1A, and CD14 |
| Proteoglycans in cancer | 7 | 0.023 | SDC1, CTTN, TNF, ITGB5, FASLG, ITGB3, and PLAUR |
| Phagosome | 6 | 0.025 | FCAR, ITGB5, ITGB3, TUBB1, TUBA1A, and CD14 |
| Inflammatory bowel disease | 4 | 0.033 | IL18R1, TNF, IL18RAP, and IL1B |
| Arrhythmogenic right ventricular cardiomyopathy | 4 | 0.037 | LMNA, ITGB5, ITGB3, and ITGA2B |
| Reactome pathway | |||
| Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell | 17 | 2.07E-10 |
ICAM1, IGLV1-44, IGKV5-2, NCR1, IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV2-23, IGKV4-1, KLRF1, TREM1, KIR2DL3, TREML1, IGLC2, KLRD1, and IGKV3-15 |
| Scavenging of heme from plasma | 11 | 4.03E-09 |
IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, JCHAIN, IGKV4-1, IGLC2, and IGKV3-15 |
| CD22-mediated B-cell receptor regulation | 10 | 2.14E-08 |
IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, IGKV4-1, IGLC2, and IGKV3-15 |
| FCERI signaling | 10 | 2.14E-08 |
IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, IGKV4-1, IGLC2, and IGKV3-15 |
| Classical antibody-mediated complement activation | 10 | 3.31E-08 |
IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, IGKV4-1, IGLC2, and IGKV3-15 |
| Role of LAT2/NTAL/LAB on calcium mobilization | 10 | 8.38E-08 |
IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, IGKV4-1, IGLC2, and IGKV3-15 |
| FCGR activation | 10 | 7.38E-08 |
IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, IGKV4-1, IGLC2, and IGKV3-15 |
| Initial triggering of complement | 10 | 1.36E-07 |
IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, IGKV4-1, IGLC2, and IGKV3-15 |
| Role of phospholipids in phagocytosis | 10 | 3.31E-07 |
IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, IGKV4-1, IGLC2, and IGKV3-15 |
| FCERI-mediated Ca2+ mobilization | 10 | 5.49E-07 |
IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, IGKV4-1, IGLC2, and IGKV3-15 |
| FCERI-mediated MAPK activation | 10 | 4.97E-07 |
IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, IGKV4-1, IGLC2, and IGKV3-15 |
| Antigen activates B-cell receptor leading to the generation of second messengers | 10 | 8.82E-07 |
IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, IGKV4-1, IGLC2, and IGKV3-15 |
| Regulation of actin dynamics for phagocytic cup formation | 10 | 6.50E-06 |
IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, IGKV4-1, IGLC2, and IGKV3-15 |
| CERI-mediated NF-kB activation | 10 | 2.18E-05 |
IGLV2-11, IGLV2-8, IGLV3-19, IGKV1-5, IGLV1-44, IGLV2-23, IGKV5-2, IGKV4-1, IGLC2, and IGKV3-15 |
| Platelet degranulation | 9 | 1.01E-04 | VWF, APP, F5, PPBP, CLU, PF4, SPARC, ITGB3, and ITGA2B |
| Platelet adhesion to exposed collagen | 4 | 5.69E-04 | VWF, GP6, GP1BA, and GP9 |
| GP1b-IX-V activation signaling | 3 | 0.005 | VWF, GP1BA, and GP9 |
| Mitotic prometaphase | 6 | 0.007 | SPC24, KIF2C, CDC20, AURKB, TUBB1, and TUBA1A |
| ECM proteoglycans | 5 | 0.01 | APP, ITGB5, SPARC, ITGB3, and ITGA2B |
| GRB2:SOS provides linkage to MAPK signaling for Integrins | 3 | 0.012 | SPC24, KIF2C, CDC20, AURKB, TUBB1, and TUBA1A |
| p130Cas linkage to MAPK signaling for integrins | 3 | 0.012 | VWF, ITGB3, and ITGA2B |
| Resolution of Sister Chromatid Cohesion | 6 | 0.012 | VWF, ITGB3, and ITGA2B |
| Integrin cell surface interactions | 5 | 0.016 | ICAM1, VWF, ITGB5, ITGB3, and ITGA2B |
| RHO GTPases Activate Formins | 6 | 0.018 | SPC24, KIF2C, CDC20, AURKB, UBE2C, TUBB1, and TUBA1A |
| Separation of Sister Chromatids | 7 | 0.018 | SPC24, KIF2C, CDC20, AURKB, TUBB1, and TUBA1A |
| Cell surface interactions at the vascular wall | 4 | 0.023 | GP6, ESAM, PF4, and TREM1 |
| Intrinsic pathway of fibrin clot formation | 3 | 0.025 | PRKAR2B, TUBB1, TUBA1A, and NTU |
| Hedgehog 'off' state | 4 | 0.025 | VWF, GP1BA, and GP9 |
| Integrin alpha IIb beta 3 signaling | 3 | 0.027 | VWF, ITGB3, and ITGA2B |
| Syndecan interactions | 3 | 0.037 | SDC1, ITGB5, and ITGB3 |
DEGs with a significant change between children with AD and healthy control children (cutoff FC ≥1.5 and FDR <0.05) were used for GO enrichment and pathway analyses using DAVID database. KEGG and Reactome pathway analyses were used to determine the pathways of DEGs between two groups.
Abbreviations: AD, atopic dermatitis; Akt, protein kinase B; Ca2+, calcium ion; DEG, differentially expressed gene; ECM, extracellular matrix; FC, fold change; FCERI, Fc-epsilon receptor; FDR, false discovery rate; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MMP, matrix metalloproteinase; PI3K, phosphoinositide 3-kinase.
Construction of protein‒protein interaction network and module analysis
To explore interactions among the DEG genes, STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) analysis was applied, and the most important modules were then screened and visualized using Cytoscape software. A protein‒protein interaction (PPI) network containing 82 connected nodes (proteins) and 194 interaction edges (interactions of proteins), where the average degree of connectivity (i.e., the average number of neighbors) was 4.732, is presented in Figure 5a. The hub nodes with the greatest number of neighbors (≥8), such as TNF, IL-1β, Von Willebrand factor, and ITGB3, were identified (labeled in red in Figure 5a) and analyzed by GO enrichment and pathway analyses (Table 6). The Kyoto Encyclopedia of Genes and Genomes pathway analysis revealed that the hub genes were involved in the cytokine‒cytokine receptor interaction, hematopoietic cell lineage, extracellular matrix‒receptor interaction, platelet activation, cell division, and other pathways. In addition, two significant modules with 10 nodes were obtained from the PPI network of DEGs using Molecular Complex Detection (Figure 5b and c). Enrichment analysis suggested that the genes in the first significant module (Figure 5b) were mainly associated with functional terms in the category BP, including cell division, cell proliferation, and mitotic nuclear division. In the category CC, the genes in this module were significantly enriched in cytosol and nucleus, and in the category MF, the genes were mainly enriched in protein and adenosine triphosphate binding. The genes in the second module (Figure 5c) were significantly enriched in inflammatory response, chemokine-mediated signaling, platelet degranulation and activation, immune response, and signal transduction in the category BP. In the category CC, the genes were significantly enriched in the extracellular region, extracellular space, and platelet alpha granule lumen, and in the category MF, the genes were mainly enriched in chemokine activity and CXCR chemokine receptor binding. Furthermore, results from the Kyoto Encyclopedia of Genes and Genomes analysis showed that the genes in this significant module were associated with chemokine signaling pathway and cytokine‒cytokine receptor interaction (Table 7).
Figure 5.
PPI networks. (a) PPI network with 82 nodes. In the network, nodes represent proteins, and lines (edges) represent the interactions between proteins. Red nodes represent the hub nodes with a large number of neighbors (≥8). (b) A first significant module with 12 nodes identified by MCODE. (c) A second significant module with 12 nodes identified by MCODE. MCODE, Molecular Complex Detection; PPI, protein‒protein interaction.
Table 6.
GO Enrichment and Pathway Analyses of Hub Genes
| GO Term | Gene Count | P-Value |
|---|---|---|
| Biological process | ||
| Platelet degranulation | 7 | 5.39E-10 |
| Platelet activation | 5 | 4.64E-06 |
| Extracellular matrix organization | 5 | 3.80E-05 |
| Cell division | 5 | 3.56E-04 |
| Negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | 3 | 6.29E-04 |
| Platelet aggregation | 3 | 7.73E-04 |
| Blood coagulation | 4 | 7.86E-04 |
| Cell adhesion | 5 | 9.88E-04 |
| Mitotic nuclear division | 4 | 0.002 |
| Anaphase-promoting complex-dependent catabolic process | 3 | 0.003 |
| Positive regulation of calcidiol 1-monooxygenase activity | 2 | 0.003 |
| Sequestering of triglyceride | 2 | 0.004 |
| Sister chromatid cohesion | 3 | 0.005 |
| Positive regulation of fever generation | 2 | 0.005 |
| Inflammatory response | 4 | 0.006 |
| Regulation of establishment of endothelial barrier | 2 | 0.007 |
| Positive regulation of protein phosphorylation | 3 | 0.007 |
| Cytokine-mediated signaling pathway | 3 | 0.008 |
| Regulation of chromosome segregation | 2 | 0.008 |
| Negative regulation of lipid storage | 2 | 0.008 |
| Immune response | 4 | 0.008 |
| Positive regulation of ubiquitin-protein ligase activity | 2 | 0.009 |
| Positive regulation of chemokine biosynthetic process | 2 | 0.010 |
| Protein ubiquitination involved in ubiquitin-dependent protein catabolic process | 3 | 0.010 |
| Positive regulation of heterotypic cell-cell adhesion | 2 | 0.011 |
| Regulation of cell proliferation | 3 | 0.015 |
| Regulation of I-kB kinase/NF-kB signaling | 2 | 0.015 |
| Positive regulation of membrane protein ectodomain proteolysis | 2 | 0.015 |
| Positive regulation of VEGF receptor signaling pathway | 2 | 0.016 |
| Negative regulation of lipid catabolic process | 2 | 0.016 |
| Cell‒substrate adhesion | 2 | 0.017 |
| Proteasome-mediated ubiquitin-dependent protein catabolic process | 3 | 0.018 |
| Blood coagulation, intrinsic pathway | 2 | 0.018 |
| Positive regulation of leukocyte chemotaxis | 2 | 0.018 |
| Positive regulation of protein export from the nucleus | 2 | 0.019 |
| Positive regulation of NF-kB import into the nucleus | 2 | 0.021 |
| Regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 2 | 0.023 |
| Positive regulation of IL-8 production | 2 | 0.026 |
| Positive regulation of gene expression | 3 | 0.028 |
| Positive regulation of phagocytosis | 2 | 0.029 |
| Lipopolysaccharide-mediated signaling pathway | 2 | 0.032 |
| Protein kinase B signaling | 2 | 0.033 |
| Positive regulation of nitric oxide biosynthetic process | 2 | 0.043 |
| Positive regulation of IL-6 production | 2 | 0.045 |
| Positive regulation of IFN-γ production | 2 | 0.046 |
| Positive regulation of cell division | 2 | 0.047 |
| Cellular component | ||
| Platelet alpha granule lumen | 5 | 1.72E-07 |
| Cell surface | 5 | 0.001 |
| Extracellular region | 7 | 0.002 |
| Kinetochore | 3 | 0.003 |
| Extracellular space | 6 | 0.006 |
| Membrane | 7 | 0.012 |
| Platelet alpha granule membrane | 2 | 0.012 |
| Spindle mid-zone | 2 | 0.018 |
| Anaphase-promoting complex | 2 | 0.021 |
| Cytosol | 8 | 0.023 |
| ER to Golgi transport vesicle | 2 | 0.024 |
| Integrin complex | 2 | 0.025 |
| Integral component of plasma membrane | 5 | 0.038 |
| Spindle microtubule | 2 | 0.040 |
| Cytoplasmic microtubule | 2 | 0.047 |
| Molecular function | ||
| Protein binding | 16 | 0.002 |
| Protease binding | 3 | 0.005 |
| Identical protein binding | 5 | 0.006 |
| Extracellular matrix binding | 2 | 0.026 |
| Enzyme binding | 3 | 0.043 |
| Chemokine activity | 2 | 0.048 |
| KEGG pathway | ||
| Hematopoietic cell lineage | 5 | 1.09E-05 |
| ECM‒receptor interaction | 4 | 3.96E-04 |
| Platelet activation | 4 | 0.001 |
| Cytokine‒cytokine receptor interaction | 4 | 0.008 |
| Hypertrophic cardiomyopathy | 3 | 0.008 |
| Dilated cardiomyopathy | 3 | 0.009 |
| Osteoclast differentiation | 3 | 0.021 |
| Alzheimer's disease | 3 | 0.033 |
| Focal adhesion | 3 | 0.048 |
Hub genes identified in PPI network were used for GO enrichment and pathway analyses using DAVID database.
Abbreviations: ECM, extracellular matrix; ER, endoplasmic reticulum; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein‒protein interaction.
Table 7.
GO Enrichment and Pathway Analyses of Significant Modules
| GO Term | Gene Count | P-Value |
|---|---|---|
| Module 1 | ||
| Biological process | ||
| Cell division | 6 | 4.50E-07 |
| Mitotic nuclear division | 5 | 5.52E-06 |
| Sister chromatid cohesion | 4 | 1.83E-05 |
| Anaphase-promoting complex-dependent catabolic process | 3 | 7.70E-04 |
| Cell proliferation | 4 | 7.82E-04 |
| Protein ubiquitination involved in ubiquitin-dependent protein Catabolic process | 3 | 0.003 |
| Regulation of chromosome segregation | 2 | 0.004 |
| Positive regulation of ubiquitin-protein ligase activity | 2 | 0.005 |
| Proteasome-mediated ubiquitin-dependent protein catabolic process | 3 | 0.005 |
| Regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 2 | 0.012 |
| Negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 2 | 0.037 |
| Positive regulation of ubiquitin-protein ligase activity involved in regulation of mitotic cell cycle transition | 2 | 0.040 |
| Cellular component | ||
| Kinetochore | 3 | 6.88E-04 |
| Cytosol | 7 | 0.002 |
| Anaphase-promoting complex | 2 | 0.011 |
| Spindle microtubule | 2 | 0.022 |
| Nucleus | 7 | 0.024 |
| Cytoplasmic microtubule | 2 | 0.025 |
| Nucleoplasm | 5 | 0.036 |
| Condensed chromosome kinetochore | 2 | 0.042 |
| Molecular Function | ||
| Protein binding | 10 | 0.003 |
| ATP binding | 4 | 0.039 |
| KEGG pathway | ||
| Ubiquitin mediated proteolysis | 2 | 0.039 |
| Module 2 | ||
| Biological process | ||
| Platelet degranulation | 7 | 3.80E-12 |
| Positive regulation of leukocyte chemotaxis | 3 | 3.89E-05 |
| Response to lipopolysaccharide | 4 | 7.36E-05 |
| Chemokine-mediated signaling pathway | 3 | 6.23E-04 |
| Inflammatory response | 4 | 8.66E-04 |
| Platelet activation | 3 | 0.002 |
| Regulation of cell proliferation | 3 | 0.004 |
| Extracellular matrix organization | 3 | 0.005 |
| Response to lead ion | 2 | 0.007 |
| G-protein coupled receptor signaling pathway | 4 | 0.010 |
| Immune response | 3 | 0.020 |
| Signal transduction | 4 | 0.020 |
| Innate immune response | 3 | 0.021 |
| Response to peptide hormone | 2 | 0.023 |
| Positive regulation of TNF production | 2 | 0.025 |
| Negative regulation of angiogenesis | 2 | 0.033 |
| Cellular component | ||
| Platelet alpha granule lumen | 7 | 4.75E-14 |
| Extracellular region | 9 | 3.03E-08 |
| Extracellular space | 8 | 3.74E-07 |
| cell surface | 4 | 0.002 |
| platelet alpha granule | 2 | 0.007 |
| ER to Golgi transport vesicle | 2 | 0.013 |
| mitochondrial membrane | 2 | 0.045 |
| Molecular function | ||
| chemokine activity | 3 | 2.93E-04 |
| CXCR chemokine receptor binding | 2 | 0.005 |
| collagen binding | 2 | 0.032 |
| chaperone binding | 2 | 0.042 |
| KEGG pathway | ||
| Chemokine signaling pathway | 4 | 3.66E-04 |
| Cytokine‒cytokine receptor interaction | 3 | 0.017 |
Two significant modules identified in PPI network were used for GO enrichment and pathway analyses using DAVID database.
Abbreviations: ATP, adenosine triphosphate; ER, endoplasmic reticulum; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein‒protein interaction.
Discussion
AD is a complex disease associated with immunological and epidermal barrier dysfunctions. Most of our knowledge in the field of AD is based on studies performed in adult patients with AD, although remarkable differences between pediatric and adult AD have been shown recently. Therefore, it is of great importance to identify the molecular basis of pediatric AD and elucidate the biomarkers that could help to identify young patients at risk at an earlier stage of life and to explore new therapies in pediatric AD. Given that more than half of all cases of AD begin during the first year of life, we aimed to discover signature biomarkers of AD in infants. Considering that skin biopsies are very difficult to obtain at such a young age, that AD generates a systemic immunological response, and that blood is a noninvasive source of biological tissue, we analyzed blood profiles of pediatric patients with AD in the first year of life.
Using RNA-seq transcriptome profile of peripheral blood cells obtained from infants with AD or healthy infants, we identified 178 genes differentially expressed in pediatric patients with AD: 115 were upregulated, and 63 were downregulated. To further investigate the functions of the DEGs, GO functional annotation and pathway enrichment analysis were used on the basis of the DAVID database. The GO analysis showed that DEGs were associated with immune responses, inflammatory responses, regulation of immune responses, and platelet activation, which are all known to be AD related. The results of the pathway analysis indicated that the DEGs were enriched in immunoregulatory interactions between a lymphoid and a nonlymphoid cell, hematopoietic cell lineage, phosphoinositide 3-kinase‒protein kinase B signaling pathway, cytokine‒cytokine receptor interaction, NK cell‒mediated cytotoxicity, and platelet activation.
Randomly selected DEGs were further validated in a larger number of samples collected from patients with AD in the first year of life using RT-qPCR. Among highly upregulated genes, we identified IL1β, previously shown to be upregulated in the serum of adult AD (Thijs et al., 2018) and stratum corneum of our pediatric AD collection as reported previously (McAleer et al., 2019). It has been shown to be involved in AD development (Bernard et al., 2017). IL-1β is a potent proinflammatory cytokine that can mediate inflammatory responses by supporting T-cell survival, upregulation of the IL-2 receptor on lymphocytes, enhancing antibody production of B cells, and promoting B-cell proliferation and T helper 17 cell differentiation (Lamkanfi et al., 2011). IL-1β activity is regulated at multiple levels, one of which is controlled by inflammasomes (Schroder and Tschopp, 2010). Recent findings suggest that inflammasome-dependent IL-1β activation plays a role in a variety of disorders, including AD. Of note, among upregulated DEGs, we identified NLRP3, one of the important inflammasome proteins.
Another interesting upregulated DEG in our pediatric patients was TREM1. Recently, it has been reported to be highly expressed in lesional skin and serum of adult AD (Suarez-Farinas et al., 2015). It has also been reported to be expressed in psoriasis and has been suggested to be a therapeutic target to modify the effects of inflammatory myeloid dendritic cells in psoriasis (Hyder et al., 2013). TREM1 (CD354) is a cell surface receptor that is expressed on various types of cells: monocytes, neutrophils NK cells, dendritic cells, and B and T cells and has been implicated in innate and adaptive immune responses. Activation of TREM1 was shown to result in the production of a variety of inflammatory cytokines, including TNF, IL6, MCP1, and IL-1β, and amplification of toll-like receptor‒initiated inflammation (Roe et al., 2014). Of interest, TNF was highly expressed in the blood cells of our pediatric AD collection. It has been shown to be involved in inflammatory processes in AD (Jacobi et al., 2005; Sumimoto et al., 1992). Furthermore, TNF together with the T helper 2 cytokines induced AD-like features in a skin model (Danso et al., 2014). In addition, TNF together with TNF-like weak inducer of apoptosis induced keratinocyte apoptosis in AD skin (Zimmermann et al., 2011).
Another interesting group of genes found to be upregulated in the blood of pediatric patients with AD included early growth response genes (EGR1, EGR2, and EGR3), a family of zinc-finger transcription factors. EGR1, an important player in the regulation of cell growth, differentiation, cell survival, and immune responses, has been reported to be upregulated in psoriatic skin lesions (Jeong et al., 2014). EGR2/3 is known to play a crucial role in the regulation of the immune system. They control inflammation, regulate B- and T-cell function in adaptive immune responses, and have been suggested to be involved in preventing the development of autoimmune disease (Morita et al., 2016) and limiting immunopathology during productive adaptive immune responses (Li et al., 2012). Notably, EGR2 is located in a susceptibility locus for AD identified by GWAS in the Japanese population (Hirota et al., 2012).
Among the downregulated genes, we identified IL18R1 and IL18RAP, also previously found to be associated with AD (Hirota et al., 2012). IL18RAP enhances the IL18-binding activity of the IL18 receptor (IL18R1) and plays a role in signaling by IL-18, a pleiotropic immune regulator. IL-18 plays a strong proinflammatory role by inducing IFN-γ. Previous studies have implicated IL-18 in the pathogenesis of AD. It has been shown to contribute to the spontaneous development of AD-like skin lesions in a transgenic mouse model (Konishi et al., 2002). It has been reported to be elevated in skin lesions of adults with AD. In our previous study, we analyzed plasma and stratum corneum biomarkers in this collection of patients and showed that IL-18 was observed in very high levels in the stratum corneum of pediatric patients; however, no difference was observed in IL-18 plasma levels (McAleer et al., 2019). Another study showed that PBMCs from patients with AD have a decreased IL-18 expression and capacity to produce IFN-γ, which is inversely correlated with serum IgE concentrations (Higashi et al., 2001), suggesting an IL-18 role in the skewing of the immune system in patients with AD.
Another gene significantly downregulated in AD infants was GLDC. GLDC, glycine metabolism and the metabolic enzyme glycine decarboxylase, is a key enzyme of the mitochondrial glycine cleavage system (Hiraga and Kikuchi, 1980). GLDC plays important role in many human cancers (Zhang et al., 2012). It has been shown to be differentially expressed in psoriatic skin (Rittie et al., 2016). Interestingly, GLDC has been reported to be differentially expressed in AD-like‒reconstructed human epidermis (Evrard et al., 2021), suggesting its involvement in AD development.
A PPI network among the screened DEGs was predicted. The PPI analysis allowed us to determine significant modules and hub genes. In the resulting PPI network, 18 hub genes with the highest degree of connectivity were selected, which included IL1β, Von Willebrand factor gene VWF, PF4, ITGB3, ITGA2B, APP, F5, AURKB, SKA3, MELK, CDC20, PPBP, NCAPG, GTSE1, KIF2C, GP1BA, UBE2C, and TNF. Pathway analysis revealed that the hub genes were involved in the cytokine‒cytokine receptor interaction, hematopoietic cell lineage, extracellular matrix‒receptor interaction, cell division, platelet activation, and other pathways. In addition, two significant modules were identified. The genes in the first significant module were mainly associated with cell division, cell proliferation, and mitotic nuclear division. The genes in the second module were significantly enriched in inflammatory response, chemokine-mediated signaling, platelet degranulation and activation, immune response, and signal transduction and were associated with chemokine signaling pathway and cytokine‒cytokine receptor interaction.
We wondered whether the hub genes could be linked to AD or other skin inflammatory diseases. The important role of IL-1β and TNF in AD has been shown earlier in this report. Von Willebrand factor, a key player in hemostasis, has been reported in relation to cutaneous inflammation (Hillgruber et al., 2014). Increased expression of PF4 has been proposed to play an important role in the etiology of AD (Watanabe et al., 1999). Increased ITGB3 expression has been reported in T helper 17‒associated skin inflammatory diseases such as psoriasis (Goedkoop et al., 2004) and psoriatic arthritis (Canete et al., 2004). A significant increase in platelet-leukocyte aggregates expressing ITGA2B was found in the blood of mice with chronic hapten-induced allergic dermatitis (Tamagawa-Mineoka et al., 2007). PPBP has been found to be important for regulating excessive inflammation in psoriasis (Oka et al., 2017). Elevated AURKB expression in lesional psoriatic tissues has been suggested to contribute to the development of psoriasis (Liu et al., 2011). Actively proliferating UBE2C+TOP2A+ type 2/type 22 T cells were expanded in lesional AD skin and were either absent or less abundant in nonlesional and healthy samples (He et al., 2020). These findings suggest that identified hub genes could be considered important candidates for prognostic and therapeutic targets of pediatric AD.
Taken together, this study showed that blood gene expression profile identified distinct key genes and pathways of early-onset pediatric AD. Observed dramatic changes in the PBMC transcriptome were predominantly related to immune responses in AD. New data assessed from this study may help to better understand the processes leading to AD in infants and may serve in the development of novel treatment possibilities. However, to decipher the full mechanism involved in pediatric AD pathogenesis, skin RNA profile should be further investigated in infants with AD. Blood profile along with skin profile in infants with AD could provide us with a larger number of potential biomarkers that may contribute to AD prediction, risk of comorbidity development, and responses to AD treatment in infants.
Materials and Methods
Patients
We recruited infants aged <12 months with moderate-to-severe AD who were treatment naive (apart from the use of emollients and 1% hydrocortisone cream or ointment) along with age-matched healthy controls. The study was approved by the Research Ethics Committee of Children's Health Ireland at Crumlin (Dublin, Ireland) and was conducted in compliance with the Helsinki Declaration. Written informed consent was given by parents or legal guardians for all study subjects. The age of onset of AD was recorded. Severity was assessed by the SCORing Atopic Dermatitis index. Clinical and demographic features are summarized in Table 8. Analysis of cytokine and microRNA biomarkers in this collection has previously been reported (McAleer et al., 2019; Nousbeck et al., 2021).
Table 8.
Clinical and Demographical Characteristics of the Study Participants
| Characteristics | Patients with AD | Healthy Controls |
|---|---|---|
| Total | 27 | 17 |
| Sex | ||
| Male | 18 | 11 |
| Female | 9 | 6 |
| Age (mo) | ||
| Average | 6.9 | 7.94 |
| Range | 3-10 | 3-12 |
| Age of AD onset (wk) | ||
| Average | 9 | — |
| Range | 4‒20 | — |
| SCORAD | ||
| Average | 49.4 | — |
| Range | 23.4‒91.3 | — |
Abbreviations: AD, atopic dermatitis; SCORAD, SCORing Atopic Dermatitis.
PBMC preparation and RNA isolation
PBMCs were isolated from whole blood as previously described (Nousbeck et al., 2021) using histopaque double-gradient density centrifugation (Sigma-Aldrich, St. Louis, MO) and cryopreserved for further analysis. Total RNA was isolated from PBMCs according to RNeasy Mini Kit protocol (Qiagen, Hilden, Germany). RNA concentrations, RNA integrity, and quality of RNA were evaluated using Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA) and RNA 6000 Nano Lab Chips on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA samples with optimal RNA integrity number values (≥8) were considered to construct libraries for sequencing.
RNA-seq, data processing, and differential expression analysis
Library preparation (using Illumina TruSeq stranded mRNAseq library kit) and sequencing were conducted by Edinburgh Genomics, The University of Edinburgh (Edinburgh, United Kingdom). The sequencing of the libraries was performed with Illumina NovaSeq 6000 (100 cycles, 50 base pair paired-end sequencing). Sequencing reads showed excellent quality, with the overall Q30 above 94%. After sequencing, reads were trimmed using Cutadapt (Martin, 2011), and clean paired-end reads were mapped to the human reference genome GRCh38 using STAR software (Dobin et al., 2013). The number of reads for each gene was counted using featureCounts (Liao et al., 2014), and the count matrix was used for differential expression analysis. Differential expression was performed using package DESeq2 in R software (version 3.5.2), considering an expression >20 read counts in at least 25% of the samples, a cutoff of at least 1.5-fold change in expression, and a Benjamini‒Hochberg‒corrected false discovery rate < 0.05.
Real-time RT-qPCR
The DEGs were further verified using RT-qPCR. Briefly, total RNA was reverse transcribed using SensiFAST cDNA synthesis kit (Bioline, London, United Kingdom). cDNA PCR amplification was carried out using the SensiFAST SYBR Hi-ROX Kit (Bioline) on 7900HT Fast Real-Time PCR System with gene-specific intron-crossing oligonucleotide pairs. Primers are available in Table 9. Results were normalized to GAPDH mRNA levels. Triplicates of each reaction were performed as the mean ± SD. Relative quantification of target mRNA expression was performed using the 2-ΔΔCT method (Livak and Schmittgen, 2001). The normalized expression data were log2 transformed before data analysis.
Table 9.
Primers Sequences for RT-qPCR
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| IL18RAP | CCAGGGGTGAATAATTCTGGGT | CATTTGTCTGGGGCTTAACTTCT |
| IL1B | TTCGACACATGGGATAACGAGG | TTTTGCTGTGAGTCCCGGAG |
| TNF | CCTCTCTCTAATCAGCCCTCTG | GAGGACCTGGGAGTAGATGAG |
| TREM1 | GAACTCCGAGCTGCAACTAAA | TCTAGCGTGTAGTCACATTTCAC |
| EGR3 | CCAACGACATGGGCTCCATT | GGTCTCCAGAGGGGTAATAGG |
| GAPDH | GAGTCAACGGATTTGGTCGT | GACAAGCTTCCCGTTCTCAGCC |
GO enrichment and pathway analysis
DEGs were submitted to Visualisation and Integrated Discovery analysis (DAVID, version 6.8) (Huang da et al., 2009) for GO term enrichment and pathway analysis using default parameters. Kyoto Encyclopedia of Genes and Genomes and Reactome pathway analyses were used to determine the pathways of DEGs between two groups. Any GO terms and pathways with P < 0.05 were considered significantly enriched.
Construction of PPI network and module analysis
Associations between DEGs were investigated using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (Szklarczyk et al., 2017) (STRING10.5, http://string-db.org/), and a confidence score >0.6 was considered to indicate significance. Cytoscape software (version 3.6.1) was then used to visualize the PPI network (Shannon et al., 2003). In the network, nodes represented proteins, and lines (edges) represented the interactions. In addition, the most significant modules were identified with the plug-in Molecular Complex Detection (version 1.5.1) with the following settings: degree cutoff of 2, node score cutoff of 0.2, k-core of 2, and a maximum depth of 100, and they were identified by the following criteria: Molecular Complex Detection score >5 and number of nodes >5. Finally, the hub genes in the PPI network were determined, defined as those with a degree of connectivity ≥8.
Data availability statement
Datasets related to this article can be found at https://osf.io/hfwyt/?viewonly=ececc20afd5d42e2806b11483edb9d0d, hosted at The Open Science Framework.
ORCIDs
Janna Nousbeck: http://orcid.org/0000-0001-6918-8855
Alan D. Irvine: http://orcid.org/0000-0002-9048-2044
Maeve A. McAleer: http://orcid.org/0000-0001-9958-6504
Conflict of Interest
ADI is a consultant/speaker for Sanofi, Regeneron, Lilly, AbbVie, Pfizer, Benevolent AI, Almirall, LEO, and Arena. The remaining authors state no conflict of interest.
Acknowledgments
Our work was funded by the National Children’s Research Centre through grants to JN and ADI. This work was supported by the National Children’s Research Centre (Dublin, Ireland). We are especially grateful to all members of the family for their participation in this study. We also thank Edinburgh Genomics, The University of Edinburgh (Edinburgh, United Kingdom) that conducted RNA sequencing.
Author Contributions
Conceptualization: ADI, JN; Funding Acquisition: ADI, JN; Investigation: JN, MAM; Methodology: JN, MAM; Resources: JN, MAM; Supervision: ADI, JN; Writing - Original Draft Preparation: JN; Writing - Review and Editing: ADI, JN
accepted manuscript published online XXX; corrected proof published online XXX
Footnotes
Cite this article as: JID Innovations 2022;X:100165
References
- Bernard M., Carrasco C., Laoubi L., Guiraud B., Rozières A., Goujon C., et al. IL-1β induces thymic stromal lymphopoietin and an atopic dermatitis-like phenotype in reconstructed healthy human epidermis. J Pathol. 2017;242:234–245. doi: 10.1002/path.4887. [DOI] [PubMed] [Google Scholar]
- Bieber T. Atopic dermatitis. N Engl J Med. 2008;358:1483–1494. doi: 10.1056/NEJMra074081. [DOI] [PubMed] [Google Scholar]
- Brunner P.M., Israel A., Leonard A., Pavel A.B., Kim H.J., Zhang N., et al. Distinct transcriptomic profiles of early-onset atopic dermatitis in blood and skin of pediatric patients. Ann Allergy Asthma Immunol. 2019;122:318–330.e3. doi: 10.1016/j.anai.2018.11.025. [DOI] [PubMed] [Google Scholar]
- Brunner P.M., Israel A., Zhang N., Leonard A., Wen H.C., Huynh T., et al. Early-onset pediatric atopic dermatitis is characterized by TH2/TH17/TH22-centered inflammation and lipid alterations. J Allergy Clin Immunol. 2018;141:2094–2106. doi: 10.1016/j.jaci.2018.02.040. [DOI] [PubMed] [Google Scholar]
- Brunner P.M., Suárez-Fariñas M., He H., Malik K., Wen H.C., Gonzalez J., et al. The atopic dermatitis blood signature is characterized by increases in inflammatory and cardiovascular risk proteins. Sci Rep. 2017;7:8707. doi: 10.1038/s41598-017-09207-z. [published correction appears in Sci Rep 2018;8:8439] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañete J.D., Pablos J.L., Sanmartí R., Mallofré C., Marsal S., Maymó J., et al. Antiangiogenic effects of Anti-tumor necrosis factor alpha therapy with infliximab in psoriatic arthritis. Arthritis Rheum. 2004;50:1636–1641. doi: 10.1002/art.20181. [DOI] [PubMed] [Google Scholar]
- Capone K.A., Dowd S.E., Stamatas G.N., Nikolovski J. Diversity of the human skin microbiome early in life. J Invest Dermatol. 2011;131:2026–2032. doi: 10.1038/jid.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C., Kroboth K., Schurch N.J., Sandilands A., Sherstnev A., O'Regan G.M., et al. Filaggrin-stratified transcriptomic analysis of pediatric skin identifies mechanistic pathways in patients with atopic dermatitis. J Allergy Clin Immunol. 2014;134:82–91. doi: 10.1016/j.jaci.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danso M.O., van Drongelen V., Mulder A., van Esch J., Scott H., van Smeden J., et al. TNF-α and Th2 cytokines induce atopic dermatitis-like features on epidermal differentiation proteins and stratum corneum lipids in human skin equivalents. J Invest Dermatol. 2014;134:1941–1950. doi: 10.1038/jid.2014.83. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaki H., Brunner P.M., Renert-Yuval Y., Czarnowicki T., Huynh T., Tran G., et al. Early-onset pediatric atopic dermatitis is TH2 but also TH17 polarized in skin. J Allergy Clin Immunol. 2016;138:1639–1651. doi: 10.1016/j.jaci.2016.07.013. [DOI] [PubMed] [Google Scholar]
- Evrard C., Faway E., De Vuyst E., Svensek O., De Glas V., Bergerat D., et al. Deletion of TNFAIP6 gene in human keratinocytes demonstrates a role for TSG-6 to retain hyaluronan inside epidermis. JID Innov. 2021;1:100054. doi: 10.1016/j.xjidi.2021.100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald D.A., Malajian D., Krueger J.G., Workman C.T., Wang T., Tian S., et al. Meta-analysis derived atopic dermatitis (MADAD) transcriptome defines a robust AD signature highlighting the involvement of atherosclerosis and lipid metabolism pathways. BMC Med Genomics. 2015;8:60. doi: 10.1186/s12920-015-0133-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedkoop A.Y., Kraan M.C., Picavet D.I., de Rie M.A., Teunissen M.B., Bos J.D., et al. Deactivation of endothelium and reduction in angiogenesis in psoriatic skin and synovium by low dose infliximab therapy in combination with stable methotrexate therapy: a prospective single-centre study. Arthritis Res Ther. 2004;6:R326–R334. doi: 10.1186/ar1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Suryawanshi H., Morozov P., Gay-Mimbrera J., Del Duca E., Kim H.J., et al. Single-cell transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis. J Allergy Clin Immunol. 2020;145:1615–1628. doi: 10.1016/j.jaci.2020.01.042. [DOI] [PubMed] [Google Scholar]
- Higashi N., Gesser B., Kawana S., Thestrup-Pedersen K. Expression of IL-18 mRNA and secretion of IL-18 are reduced in monocytes from patients with atopic dermatitis. J Allergy Clin Immunol. 2001;108:607–614. doi: 10.1067/mai.2001.118601. [DOI] [PubMed] [Google Scholar]
- Hillgruber C., Steingräber A.K., Pöppelmann B., Denis C.V., Ware J., Vestweber D., et al. Blocking von Willebrand factor for treatment of cutaneous inflammation. J Invest Dermatol. 2014;134:77–86. doi: 10.1038/jid.2013.292. [DOI] [PubMed] [Google Scholar]
- Hiraga K., Kikuchi G. The mitochondrial glycine cleavage system. Functional association of glycine decarboxylase and aminomethyl carrier protein. J Biol Chem. 1980;255:11671–11676. [PubMed] [Google Scholar]
- Hirota T., Takahashi A., Kubo M., Tsunoda T., Tomita K., Sakashita M., et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet. 2012;44:1222–1226. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hyder L.A., Gonzalez J., Harden J.L., Johnson-Huang L.M., Zaba L.C., Pierson K.C., et al. TREM-1 as a potential therapeutic target in psoriasis. J Invest Dermatol. 2013;133:1742–1751. doi: 10.1038/jid.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi A., Antoni C., Manger B., Schuler G., Hertl M. Infliximab in the treatment of moderate to severe atopic dermatitis. J Am Acad Dermatol. 2005;52:522–526. doi: 10.1016/j.jaad.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Jeong S.H., Kim H.J., Jang Y., Ryu W.I., Lee H., Kim J.H., et al. Egr-1 is a key regulator of IL-17A-induced psoriasin upregulation in psoriasis. Exp Dermatol. 2014;23:890–895. doi: 10.1111/exd.12554. [DOI] [PubMed] [Google Scholar]
- Konishi H., Tsutsui H., Murakami T., Yumikura-Futatsugi S., Yamanaka K., Tanaka M., et al. IL-18 contributes to the spontaneous development of atopic dermatitis-like inflammatory skin lesion independently of IgE/stat6 under specific pathogen-free conditions. Proc Natl Acad Sci USA. 2002;99:11340–11345. doi: 10.1073/pnas.152337799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M., Vande Walle L., Kanneganti T.D. Deregulated inflammasome signaling in disease. Immunol Rev. 2011;243:163–173. doi: 10.1111/j.1600-065X.2011.01042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langan S.M., Irvine A.D., Weidinger S. Atopic dermatitis [published correction appears in Lancet 2020;396:758. Lancet. 2020;396:345–360. doi: 10.1016/S0140-6736(20)31286-1. [DOI] [PubMed] [Google Scholar]
- Li S., Miao T., Sebastian M., Bhullar P., Ghaffari E., Liu M., et al. The transcription factors Egr2 and Egr3 are essential for the control of inflammation and antigen-induced proliferation of B and T cells. Immunity. 2012;37:685–696. doi: 10.1016/j.immuni.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu Y., Luo W., Chen S. Comparison of gene expression profiles reveals aberrant expression of FOXO1, Aurora A/B and EZH2 in lesional psoriatic skins. Mol Biol Rep. 2011;38:4219–4224. doi: 10.1007/s11033-010-0544-x. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Martin M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet j. 2011;17 [Google Scholar]
- McAleer M.A., Jakasa I., Hurault G., Sarvari P., McLean W.H.I., Tanaka R.J., et al. Systemic and stratum corneum biomarkers of severity in infant atopic dermatitis include markers of innate and T helper cell-related immunity and angiogenesis. Br J Dermatol. 2019;180:586–596. doi: 10.1111/bjd.17088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Okamura T., Sumitomo S., Iwasaki Y., Fujio K., Yamamoto K. Emerging roles of Egr2 and Egr3 in the control of systemic autoimmunity. Rheumatology (Oxford) 2016;55:ii76–ii81. doi: 10.1093/rheumatology/kew342. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Takahashi H., Takaya A., Inoue Y., Katayama Y., Kusuya Y., et al. Staphylococcus Agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.aay4068. eaay4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebuhr M., Baumert K., Heratizadeh A., Satzger I., Werfel T. Impaired NLRP3 inflammasome expression and function in atopic dermatitis due to Th2 milieu. Allergy. 2014;69:1058–1067. doi: 10.1111/all.12428. [DOI] [PubMed] [Google Scholar]
- Nousbeck J., McAleer M.A., Hurault G., Kenny E., Harte K., Kezic S., et al. MicroRNA analysis of childhood atopic dermatitis reveals a role for miR-451a. Br J Dermatol. 2021;184:514–523. doi: 10.1111/bjd.19254. [DOI] [PubMed] [Google Scholar]
- Oka T., Sugaya M., Takahashi N., Takahashi T., Shibata S., Miyagaki T., et al. CXCL17 attenuates imiquimod-induced psoriasis-like skin inflammation by recruiting myeloid-derived suppressor cells and regulatory T cells. J Immunol. 2017;198:3897–3908. doi: 10.4049/jimmunol.1601607. [DOI] [PubMed] [Google Scholar]
- Rittié L., Tejasvi T., Harms P.W., Xing X., Nair R.P., Gudjonsson J.E., et al. Sebaceous gland atrophy in psoriasis: an explanation for psoriatic alopecia? J Invest Dermatol. 2016;136:1792–1800. doi: 10.1016/j.jid.2016.05.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe K., Gibot S., Verma S. Triggering receptor expressed on myeloid cells-1 (TREM-1): a new player in antiviral immunity? Front Microbiol. 2014;5:627. doi: 10.3389/fmicb.2014.00627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K., Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Fariñas M., Tintle S.J., Shemer A., Chiricozzi A., Nograles K., Cardinale I., et al. Nonlesional atopic dermatitis skin is characterized by broad terminal differentiation defects and variable immune abnormalities. J Allergy Clin Immunol. 2011;127:954–964. doi: 10.1016/j.jaci.2010.12.1124. e1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Fariñas M., Ungar B., Correa da Rosa J., Ewald D.A., Rozenblit M., Gonzalez J., et al. RNA sequencing atopic dermatitis transcriptome profiling provides insights into novel disease mechanisms with potential therapeutic implications. J Allergy Clin Immunol. 2015;135:1218–1227. doi: 10.1016/j.jaci.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Sumimoto S., Kawai M., Kasajima Y., Hamamoto T. Increased plasma tumour necrosis factor-alpha concentration in atopic dermatitis. Arch Dis Child. 1992;67:277–279. doi: 10.1136/adc.67.3.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagawa-Mineoka R., Katoh N., Ueda E., Takenaka H., Kita M., Kishimoto S. The role of platelets in leukocyte recruitment in chronic contact hypersensitivity induced by repeated elicitation. Am J Pathol. 2007;170:2019–2029. doi: 10.2353/ajpath.2007.060881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijs J.L., Strickland I., Bruijnzeel-Koomen C.A.F.M., Nierkens S., Giovannone B., Knol E.F., et al. Serum biomarker profiles suggest that atopic dermatitis is a systemic disease. J Allergy Clin Immunol. 2018;141:1523–1526. doi: 10.1016/j.jaci.2017.12.991. [DOI] [PubMed] [Google Scholar]
- Watanabe O., Natori K., Tamari M., Shiomoto Y., Kubo S., Nakamura Y. Significantly elevated expression of PF4 (platelet factor 4) and eotaxin in the NOA mouse, a model for atopic dermatitis. J Hum Genet. 1999;44:173–176. doi: 10.1007/s100380050136. [DOI] [PubMed] [Google Scholar]
- Weidinger S., Beck L.A., Bieber T., Kabashima K., Irvine A.D. Atopic dermatitis. Nat Rev Dis Primers. 2018;4:1. doi: 10.1038/s41572-018-0001-z. [DOI] [PubMed] [Google Scholar]
- Zhang W.C., Shyh-Chang N., Yang H., Rai A., Umashankar S., Ma S., et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [published correction appears in Cell 2012;148:1066] [DOI] [PubMed] [Google Scholar]
- Zimmermann M., Koreck A., Meyer N., Basinski T., Meiler F., Simone B., et al. TNF-like weak inducer of apoptosis (TWEAK) and TNF-α cooperate in the induction of keratinocyte apoptosis. J Allergy Clin Immunol. 2011;127(200–7):207.e1–207.e10. doi: 10.1016/j.jaci.2010.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Datasets related to this article can be found at https://osf.io/hfwyt/?viewonly=ececc20afd5d42e2806b11483edb9d0d, hosted at The Open Science Framework.