Abstract
Evolution is driven by the accumulation of competing mutations that influence survival. A broad form of genetic variation is the amplification or deletion of DNA (≥50 bp) referred to as copy number variation (CNV). In humans, CNV may be inconsequential, contribute to minor phenotypic differences, or cause conditions such as birth defects, neurodevelopmental disorders, and cancers. To identify mechanisms that drive CNV, we monitored the experimental evolution of Saccharomyces cerevisiae populations grown under sulfate-limiting conditions. Cells with increased copy number of the gene SUL1, which encodes a primary sulfate transporter, exhibit a fitness advantage. Previously, we reported interstitial inverted triplications of SUL1 as the dominant rearrangement in a haploid population. Here, in a diploid population, we find instead that small linear fragments containing SUL1 form and are sustained over several generations. Many of the linear fragments are stabilized by de novo telomere addition within a telomere-like sequence near SUL1 (within the SNF5 gene). Using an assay that monitors telomerase action following an induced chromosome break, we show that this region acts as a hotspot of de novo telomere addition and that required sequences map to a region of <250 base pairs. Consistent with previous work showing that association of the telomere-binding protein Cdc13 with internal sequences stimulates telomerase recruitment, mutation of a four-nucleotide motif predicted to associate with Cdc13 abolishes de novo telomere addition. Our study suggests that internal telomere-like sequences that stimulate de novo telomere addition can contribute to adaptation by promoting genomic plasticity.
Keywords: copy number variation, gene amplification, evolutionary genomics, de novo telomere addition, Cdc13, DNA repair, Saccharomyces cerevisiae
Through experimental evolution, Hoerr et al. show that long-term growth in sulfate-limiting conditions selects for amplification of the primary sulfate transporter gene SUL1 in diploid yeast cells through generation of acentric linear fragments stabilized by acquisition of a telomere within the nearby gene SNF5. This telomere-like sequence from SNF5 acts as a hotspot of de novo telomere addition when integrated at an ectopic location, suggesting that internal hotspots of telomerase recruitment can contribute to adaptation by promoting genomic plasticity.
Introduction
Genomic variation is the basis of evolution. Mutations, including both single-nucleotide polymorphisms and larger genomic rearrangements, can rise in prevalence within a population if they confer a fitness advantage. Copy number variation (CNV) refers to a genomic rearrangement that results in loss or gain of DNA (Alkan et al. 2011; MacDonald et al. 2014). CNV is most likely to have a phenotypic impact when the DNA that is duplicated or lost contains one or more genes that impact cellular fitness. In humans, CNV is associated with a variety of disorders primarily (but not exclusively) related to intellectual disabilities, developmental delays, and predisposition to cancers (reviewed in Zhang et al. 2009; Shaikh 2017; Doran and Pennington 2022; Song, Gao, et al. 2022).
CNV can arise through a variety of chromosomal rearrangements stemming from errors in replication fork progression, recombination, or in DNA damage repair pathways (Hastings et al. 2009). The frequency of CNVs is often increased in cancer cells due to dysregulation of DNA repair or replication pathways (Doran and Pennington 2022). Gene amplification can present in a variety of configurations including both direct and inverted tandem duplication (additional copies at the original location), dispersed duplication (integration of additional copies elsewhere in the genome), or double minute chromosomes that are small circular extrachromosomal fragments of DNA that lack a centromere (Biedler et al. 1983; Chakraborty and Ay 2018; Doran and Pennington 2022).
The origin of CNVs is not well-understood, in part because the initiating event may be replaced over time by more beneficial secondary and tertiary events that confer increased growth advantages (Ilić et al. 2022). For example, double minute chromosomes may in some cases serve as recombinational intermediates in the formation of tandem or dispersed duplications (Turner et al. 2017; Rosswog et al. 2021; Song, Minami, et al. 2022). Since most studies of CNV in humans rely on samples from a single point in time, they provide only a snapshot of the most dominant rearrangement (Chakraborty and Ay 2018). This limitation highlights the need for experimental systems in which the temporal relationship between events can be observed.
The budding yeast Saccharomyces cerevisiae utilizes DNA repair pathways similar to those of human cells but is more easily manipulated and monitored for genomic rearrangements (Krogh and Symington 2004; Wellinger and Zakian 2012). For example, growth under specific environmental conditions selects for amplification of one of several genes (e.g. HXT6/7, CUP1, GAP1, or SUL1) (Brown et al. 1998; Gresham et al. 2008; Kao and Sherlock 2008; Gresham et al. 2010; Zhang et al. 2013). Growth of continuous yeast cultures in a chemostat allows the genetic composition of the population to be monitored over time, providing an attractive system to monitor CNV formation and persistence in a heterogeneous population. Previously reported evolution experiments with S. cerevisiae describe several different types of chromosomal rearrangements that increase gene copy number. Extrachromosomal circular DNAs, similar to the double minute chromosomes described above, have been reported (Gresham et al. 2010). Additionally, persistent linear fragments have also been described (Dorsey et al. 1992; Narayanan et al. 2006). Circular molecules lack DNA ends, but linear fragments must protect their ends from nucleolytic degradation. In most cases, protection of the ends is afforded by the telomere, a repetitive, protein-bound DNA sequence that resists nuclease action and engagement with the DNA repair machinery (Blackburn 1991). Where such linear fragments have been described, they arise from palindromic duplication, resulting in an isochromosomal fragment capped at both ends by copies of the same pre-existing telomere (Dorsey et al. 1992; Narayanan et al. 2006).
In haploid laboratory yeast strains grown in sulfate-limiting chemostats, the fitter variants almost always contain an inverted interstitial triplication of the primary sulfate-transporter gene (SUL1) and variable amounts of the flanking regions in an otherwise unrearranged chromosome (Araya et al. 2010; Payen et al. 2014; Brewer et al. 2015). Here, we report that diploids of the same laboratory strain appear to follow a different path to SUL1 amplification. Instead of chromosomally stabilized amplicons, we find extrachromosomal linear fragments that contain SUL1 and the nearby endogenous telomere at one end and a new telomere added to a sequence centromere proximal to SUL1 at the other end. Similar fragments arose independently multiple times, suggesting that one or more sequences centromere proximal to SUL1 have a propensity to stimulate new telomere formation. Indeed, we show that a sequence within the SNF5 gene is sufficient to stimulate de novo telomere addition when moved to an ectopic location. The ability of this sequence to stimulate de novo telomere addition requires at least two short sequence motifs that are predicted to associate with the telomere-binding protein Cdc13 (Eldridge et al. 2006) and can be functionally replaced by artificial recruitment of Cdc13.
Methods and materials
Strains and plasmids
Eighteen sulfate-limited chemostat experiments (S701-S712, S10101-3, and S10105-7) were performed on diploid strains of the S288c background (FY3 and FY5) (Gresham et al. 2008; Miller et al. 2013) that were isogenic except for their ARS228 status—ARS228/ARS228, ars228/ars228, and ARS228/ars228. Creation of the ars228 mutation has been described (Brewer et al. 2015). All strains for SiRTA analyses were derived from S288c and are listed in Supplementary File 1. Plasmid pAB180 (pRS414-ADHpromoter-GBD-CDC13) and pAB220 (pRS414-ADHpromoter-GBD) were gifts of A. Bianchi. Strains for the HO cleavage assay were generated as described in Ngo et al. (2020). In brief, the HO endonuclease recognition sequence was integrated on chromosome VII by one-step gene replacement using plasmid pHO invC (based on JH2017; gift of J. Haber). The URA3 gene was integrated by one-step gene replacement using pRS306 (Sikorski and Hieter 1989) as template. Putative SiRTAs were inserted on chromosome VII using CRISPR/Cas9 as described (Anand et al. 2017) using gRNA sequence 5′-TGCGGCAAGTTCATCTTCCA. Sequences of different lengths within SNF5 were amplified from genomic yeast DNA with appropriate primers (Supplementary File 1) and additional sequences were added that lie upstream (5′-TTTCTTTGGAAAACGTTGAAAATGAGGTTCTATGATCTAC) and downstream (5′-AGAACATAGAATAAATTTGGTACTGGAACGTTGATTAACT) of the gRNA recognition site. RAD52 was replaced by one-step gene replacement using pFA6a-TRP1 (Longtine et al. 1998) as template.
Contour-clamped homogeneous electric field (CHEF) gels
Agarose plugs for CHEF gel electrophoresis were generated using the method by Lucas Argueso (described in Kwan et al. 2016). For separation of whole chromosomes, run conditions in the BioRad CHEF-DRII were 1% LE agarose in 0.5XTBE in 2.3 L 0.5% TBE running buffer at 14 °C. Switch times were either 47″ to 170″ at 165 V for 39–62 h or 300″ to 900″ at 100 V for 68 h. Linear fragments were separated from chromosomes by CHEF gels with switch times 1.7″ to 2.6″ at 198 V for 20 h. Restriction analysis of linear fragments was performed by conventional agarose gel electrophoresis in 0.4% ME agarose gels in 1XTBE at 1 V/cm for 19 h at room temperature. Standard protocols for restriction digestion, Southern blotting and hybridization using 32P-labeled PCR probes were used. Linear fragments were concentrated from plug supernatants using Centricon Filter devices (Millipore) and then ethanol precipitated.
Sequence analysis of linear fragments
Whole-genome sequencing of fragments purified from plug supernatants and split read analysis were as described (Payen et al. 2014). Briefly, sequencing libraries were generated using Nextera tagmentation (Illumina). Libraries were sequenced on an Illumina MiSeq instrument with paired end 150 bp reads. The number of reads obtained per sample ranged from 42,619 to 345,290 (exact reads for each sample can be found on each data deposit record, see below). Reads were then mapped onto the S. cerevisiae genome with BLAST and filtered for those mapping to chromosome II. Coordinates on chromosome II with large differences in upstream and downstream read coverage were identified as putative junction points. Reads that mapped to these coordinates were aligned to generate junction sequences. Raw sequencing data are deposited at the NIH Sequence Read Archive (SRA) under BioProject ID PRJNA909857.
HO cleavage assay
The HO cleavage assay was performed as described in Ngo et al. (2020). In brief, cells were grown in SD-Ura medium (+2% raffinose) overnight to an optical density at 600 nm (OD600) of 0.6–0.9. Cells were plated on yeast extract peptone medium with 2% galactose (YEPGal). Simultaneously, cells were serially diluted in water and plated on yeast extract peptone medium with 2% dextrose (YEPD) to determine total cell number. When plasmid selection was required (pRS414, pAB220, and pAB180), cells were plated on synthetic medium lacking tryptophan and containing either 2% glucose or 2% galactose. After 3 days, colonies were counted and at least 100 individual colonies were transferred to plates containing 5-fluoroorotic acid (5-FOA) to determine the frequency of URA3 loss. For each experiment, at least 30 5-FOA-resistant clones were grown in YEPD for DNA extraction by MasterPure Yeast DNA Purification Kit (Lucigen). The relative location of each chromosome truncation event was determined using multiplex PCR with primers that anneal centromere or telomere proximal to the SiRTA (Supplementary File 1). De novo telomere addition events were amplified using CHECKCHR7GUIDE1F and TelomereRev as primers. The purified PCR product was sequenced using Sanger sequencing to determine the site of de novo telomere addition. Full data sets corresponding to Figs. 3 and 4 are found in Supplementary File 2.
Results
Linear fragments containing SUL1 accumulate in cells grown in sulfate-limiting conditions
Under sulfate-limiting conditions, S. cerevisiae cells with increased capacity for sulfate transport exhibit a selective growth advantage (Gresham et al. 2008). The most common genomic rearrangement observed after prolonged competitive growth of haploid lab strains in low-sulfate conditions is an interstitial, inverted triplication of a chromosomal region containing the primary sulfate-transporter gene SUL1 and its adjacent origin of replication ARS228, located ∼25 kb from the right telomere of chromosome II (Payen et al. 2014; Brewer et al. 2015). From the structure of these amplicons (the central copy of the triplication is in inverted orientation), Brewer et al. (2011) proposed a model [Origin Dependent Inverted Repeat Amplification (ODIRA)] involving template switching at pre-existing short interrupted inverted repeats that flank the SUL1 gene and its adjacent origin of replication (ARS228). This model proposes that, following fork stalling and reversal, inverted repeats at the 3′-end of the leading strands anneal to the complementary repeats on the lagging strand template. Extension of the 3′-ends by DNA polymerase and ligation to the nearest Okazaki fragment creates a closed loop that is subsequently displaced from the replicating chromosome structure (Brewer et al. 2011). Replication of this evicted molecule in the next cell cycle forms a dimeric, inverted circular DNA molecule that can recombine at the SUL1 locus to generate the interstitial triplication with the inverted central copy (Brewer et al. 2011). The requirement for an origin of replication within the region that is triplicated is supported by the observation that deletion of ARS228 results in larger amplicons that encompass the next nearest replication origin (Brewer et al. 2015).
A key feature of the ODIRA model is the generation of an extrachromosomal intermediate that later recombines with the chromosomal locus (Brewer et al. 2011). In an attempt to gather evidence for the unique extrachromosomal intermediate, we constructed a diploid heterozygous for the ARS228 deletion (ARS228/ars228Δ; Brewer et al. 2015) and two diploid control strains (ARS228/ARS228 and ars228Δ/ars228Δ) and carried out independent evolution experiments in sulfate-limited chemostats (n = 10, 4, and 4, respectively). We reasoned that if the intermediate arose from the ARS228 chromosome and subsequently recombined with the ars228Δ chromosome, the resulting amplicon would contain copies of both ARS228 and ars228Δ.
Contrary to expectations, these diploids did not produce the same type of inverted interstitial amplicons of SUL1 that has been observed in haploid strains. Array comparative genome hybridization (aCGH) of each of the diploid populations after ∼180 generations (33–36 days) under sulfate-limitation revealed consistent amplification of the SUL1 region (e.g. Fig. 1a; Supplementary Fig. 1). However, in each case, the amplicon was not interstitial but continued through the right telomere. The amplified sequences were limited to this region of chromosome II and the new junctions were narrowly clustered in a small region of chromosome II near the SNF5 gene.
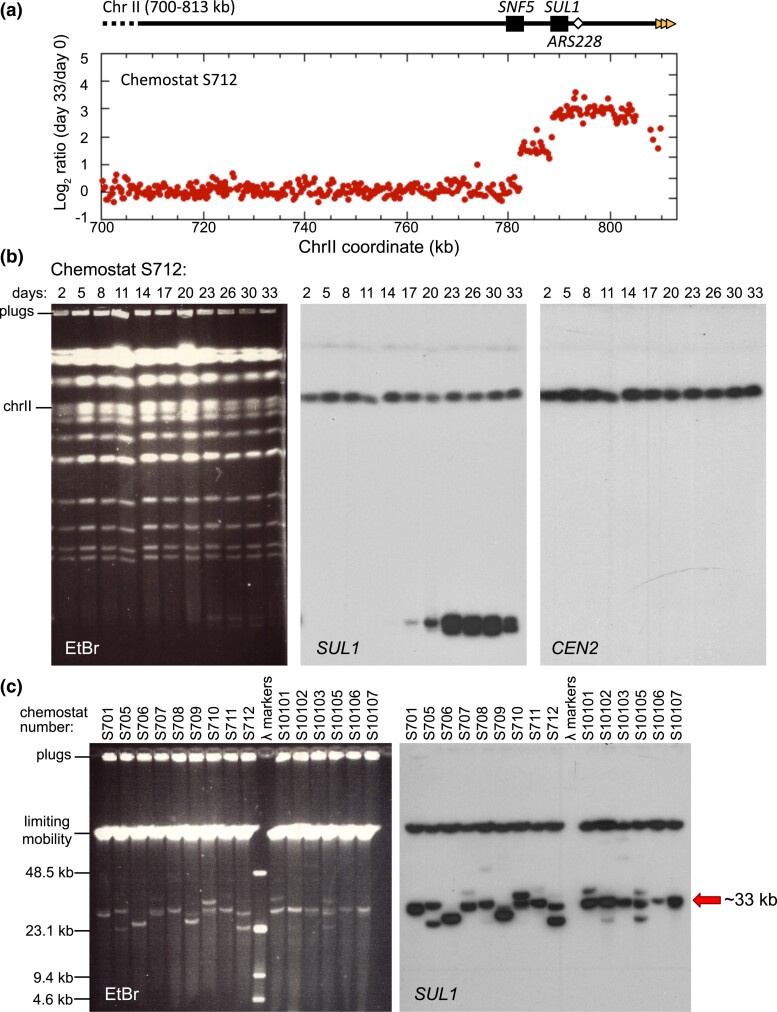
Fig. 1.
Amplification of SUL1 chromosomal fragments in sulfate-limited chemostats. a) Array Comparative Genomic Hybridization (aCGH) of the population of a diploid yeast strain (Chemostat run S712) 33 days after continuous growth in a sulfate-limited chemostat. The locations of SNF5, SUL1, and ARS228 are indicated in the diagram at the top. The Log2 hybridization ratio of day 33/day 0 indicates that the right end of chromosome II, containing the gene for the primary sulfur transporter SUL1, is present in 8 or more copies in excess of the genomic copies. The presence of two discontinuities at 782 and 788 kb suggests two discrete amplicons within the cell population. b) Contour-clamped homogeneous electric field (CHEF) gel electrophoresis of a single population (S712) sampled across the 33 days of continuous growth in sulfate-limited medium. The ethidium bromide-stained gel displays the total yeast karyotype. The CHEF gel was Southern blotted and probed sequentially for SUL1 and CEN2 as indicated. One extrachromosomal band of low molecular weight was detected by the SUL1 probe at day 17 that persists in the population onward. A second, faster migrating band appears at day 23 (more clearly resolved as two bands in c) and persists in the population onward. c) CHEF gel electrophoresis, under conditions to determine the sizes of extrachromosomal amplicons, from specific days of 15 individual chemostat runs. The ethidium bromide-stained gel and Southern blot hybridized with SUL1 reveal multiple small extrachromosomal fragments. The sizes of extrachromosomal amplicons vary from 26 to 40 kb with a recurring event producing a fragment of approximately 33 kb (arrow).
To determine the genomic location of the amplicons, we analyzed samples taken every few days through the 33 days of chemostat growth (each day is approximately six generations) and analyzed the yeast karyotype by contour-clamped homogeneous electric field (CHEF) gels. Southern blotting and hybridization with probes specific to SUL1 detected chromosome II as expected, but also revealed the generation of small (∼30 kilobases) molecules that typically were detected at around days 14–17 and often persisted through the end of the chemostat run (Fig. 1b; Supplementary Figs. 1 and 2). Similar-sized fragments were present in all samples from all three genotypes (e.g. Fig. 1c; Supplementary Figs. 1 and 2). Because they were generated in diploid cells with both copies of ARS228 deleted (Supplementary Fig. 1c), we propose that the adjacent origin of replication ARS229 can support the creation and maintenance of these fragments. The sizes of many of these molecules estimated from their migration on CHEF gels were consistent with the sizes of the amplicon detected by aCGH suggesting that they were acentric linear fragments.
It is notable that the linear fragments often persisted in the population through the end of the experiment. Because the fragments lack a centromere, mitotic partitioning is likely asymmetric. However, the fragments are presumably retained in the population due to the selective growth advantage provided by additional copies of SUL1, similar to the maintenance of an ARS plasmid under selection. In half of the cases, chromosomally integrated SUL1 amplicons were the predominant form by the end of the chemostat run. In many of these cultures, the linear fragments disappeared concurrently with the emergence of new, slower-migrating SUL1-hybridizing bands (Supplementary Figs. 1 and 2), suggesting either a precursor-product relationship between the linear fragments and the chromosomally integrated form or the independent occurrence of a second chromosome amplification event conferring increased fitness. One of the most intriguing aspects of these free linear fragments is their consistent small size—ranging between ∼26 and 42 kb (Fig. 1c). Since all of the linear fragments extend through the right telomere of chromosome II, the newly formed left ends of the linear fragments appear to arise near or within SNF5, a gene centromere proximal to SUL1.
Linear fragments are stabilized by the addition of a new telomere
While selective pressure can partially compensate for the lack of a centromere, persistence of a linear fragment requires protection at both ends from exonucleases. Normal chromosome ends (telomeres) in S. cerevisiae consist of a thymine and guanine (TG)-rich strand that extends 10–15 bases past the complementary adenine and cytosine (AC)-rich strand to create a 3′ overhang (Blackburn 1991). This TG-rich single-stranded overhang interacts with the enzyme telomerase, which utilizes its reverse transcriptase catalytic subunit and intrinsic AC-rich RNA template to add telomeric repeats to the chromosome end (Singer and Gottschling 1994; Lingner et al. 1997). In yeast, the 3′ terminating strand consists of a heterogeneous sequence pattern with a single thymine followed by one to three guanines (abbreviated TG1-3) (Wellinger and Zakian 2012).
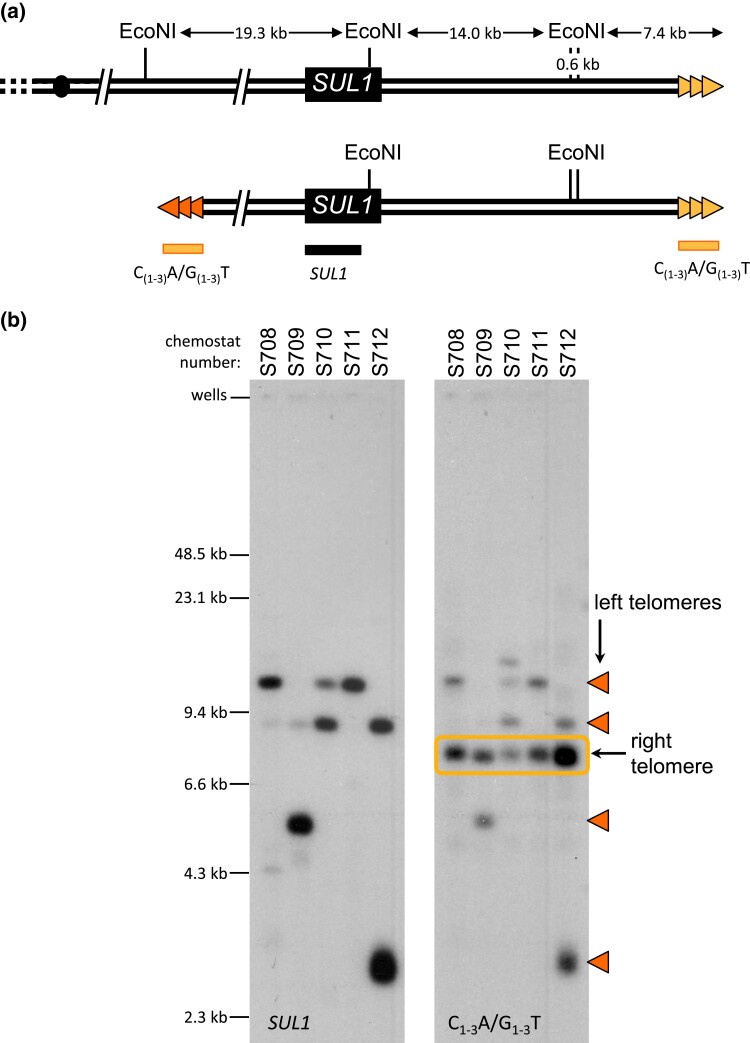
The aCGH analyses (Fig. 1a; Supplementary Fig. 1) suggested that the right end of the linear fragment contained the endogenous right telomere, but the structure of the left end was initially unclear. More detailed analysis of the linear fragments was facilitated by the development of a method to purify these small molecules through passive diffusion from agarose CHEF gel plugs. Over the course of several weeks, the small fragments diffuse out of the agarose plug and into the supernatant where they can be recovered, while the intact chromosomes remain trapped in the plug (Supplementary Fig. 3). Purified linear fragments were digested with EcoNI, which cleaves three times in the terminal region of chromosome II (Fig. 2a). Following separation by CHEF gel, the Southern blotted was probed sequentially with sequences that hybridize to either SUL1 or telomeric DNA (Fig. 2b). EcoNI digestion separated SUL1 from the right telomere and allowed us to query both ends of the linear fragments. The restriction fragments containing the right telomere hybridized to telomeric DNA (C1-3A/G1-3T) and not to SUL1 (Fig. 2b, rectangle). However, fragments of multiple sizes (ranging from ∼3–25 kb) hybridized to both the SUL1 and the telomeric probe (Fig. 2b), suggesting that the left end of each fragment was also capped by telomeric DNA.
Fig. 2.
Linear fragments are capped by new telomeric sequences. a) Top: Positions of the EcoNI restriction sites at the right end of chromosome II (coordinates 770 to 813 kb). Chromosome II-R telomeric sequences are indicated with gold triangles. Bottom: Structure of linear fragments. Proposed telomeric sequences at variable positions at the left margin of the amplicons are indicated by the orange triangles. Positions of two probes [C(1-3)A/G(1-3)T and SUL1 (indicated by the horizontal bars)]. b) Linear fragments purified from the plug supernatant (see Supplementary Fig. 3) for five cultures (S708-S712) were digested with EcoNI and separated on a CHEF gel. Southern blots were sequentially hybridized with a SUL1 probe and a telomere-specific probe (C(1-3)A/G(1-3)T). Cultures S710 and S712 contain two species of extrachromosomal molecules. Triangles indicate the fragments with de novo telomeres. Boxed fragments correspond to the native right telomere.
To identify the sites of telomeric repeat addition, we determined the sequence of fragments purified from the plug supernatants by Illumina sequencing. While short read sequencing is limited in its ability to identify large rearrangements—particularly in organisms with genomes rich in repeated sequences—in yeast, it is perfectly suited for identifying split reads that mark rearrangement junctions. The split reads were confined to the junction between the internal chromosome II sequence and a new telomere—no other junctions were detected along the length of the linear fragment. In addition, the Southern blots and aCGH data were consistent with the absence of large structural changes to the linear fragments. The sequencing results revealed that 9 of the 15 fragments contained telomeric repeats clustered within a ∼100 bp region of the gene SNF5, approximately ∼7 kb from SUL1 (Fig. 3). The remaining six events were dispersed across four other regions between SNF5 and SUL1 (Fig. 3).
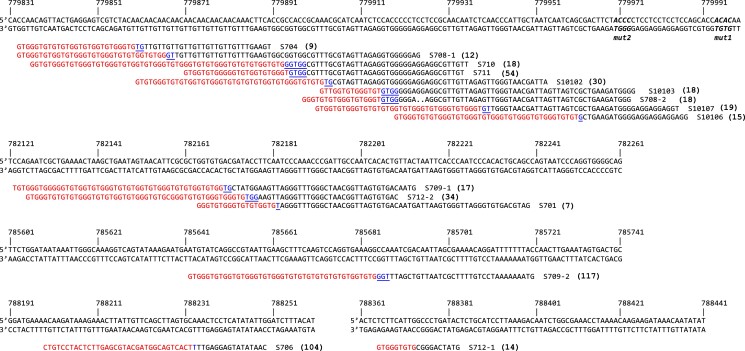
Fig. 3.
Extrachromosomal fragments purified from plug supernatants were prepared for Illumina sequencing. Split reads (red/black sequences) were aligned to chromosome II (top sequence with chromosome II coordinates). The junctions of the left end of the extrachromosomal fragments each contained a new telomeric sequence (red) and potential seed sequences for telomere addition (blue/underline). The number in parentheses to the right of each sequence corresponds to the number of split reads that mapped to that site. One of the 15 sequences (S706) was a junction with an internal portion of a Y’ element. The remaining 14 contained unique junctions with C(1-3)A/G(1-3)T telomere sequences. The four nucleotide 5′-GxGT-3′ motifs (see Fig. 5) are in bold italics in the (top) reference sequence.
Among the 11 experiments for which we had junction data from both population aCGH and sequence of the de novo sites of telomere addition, eight were in perfect agreement. The remaining three identified sequenced junctions that were discordant with the location of the aCGH junction, suggesting an amplicon larger than that predicted by the telomere addition site. These three cases were unique in that the linear fragments had disappeared before the last day of growth—the day on which aCGH was performed. These results are not consistent with the free linear fragments giving rise to the integrated SUL1 fragments. Rather, they support the idea that a second, independent, more stable chromosomal amplification event (that encompassed a larger section of chromosome II) out competed the linear fragments. Among all of the linear fragments that were sequenced, there was only one example where the new left end had acquired subtelomeric sequences (S706). In all of the other examples, the telomeric DNA was appended directly to the chromosome II sequence, consistent with addition of a new telomere to internal sequences by telomerase.
Sequences in the SNF5 gene stimulate unusually high levels of de novo telomere addition
The observation that many of the telomere addition events associated with extrachromosomal SUL1 fragments cluster within a small (∼100 bp) region is reminiscent of sequences in the yeast genome that are hotspots of de novo telomere addition (termed SiRTAs or Sites of Repair-associated Telomere Addition) (Stellwagen et al. 2003; Obodo et al. 2016; Hoerr et al. 2021). SiRTAs contain strand-specific, TG-rich sequence motifs (Obodo et al. 2016; Ngo et al. 2020). These motifs can provide a substrate for the acquisition of telomeric sequences if they become exposed in single-stranded DNA either as a result of 5′-end resection following a double-strand break (DSB) or at nicks in single-stranded gaps. In either case, the resulting TG-rich 3′ overhang can invade a similar sequence and acquire a telomere through break-induced replication or can undergo de novo telomere addition by direct action of telomerase (Putnam and Kolodner 2017). Such events are most easily assayed in cases where the new telomere is added to the centromere-proximal side of the break, causing a terminal truncation. In contrast, the putative SiRTA within SNF5 is oriented in the opposite orientation on the right arm of the chromosome. Rather than stabilizing the centromere-containing region of the chromosome, de novo telomere addition stabilizes a terminal acentric fragment containing SUL1.
Like telomeres, SiRTAs have an asymmetric sequence pattern in which one strand is relatively rich in T and G nucleotides (Obodo et al. 2016; Ngo et al. 2020). For telomerase to act upon a SiRTA, this TG-rich strand must be exposed in single-stranded DNA. SiRTAs have a bipartite structure consisting of one TG-rich region that is most often the site of telomere addition (the “Core”) and a second TG-rich “Stim” sequence located 5′ to the Core on the TG-rich strand (Obodo et al. 2016). The Stim sequence itself is rarely targeted for telomere addition but is required to stimulate de novo telomere addition in the Core (Obodo et al. 2016). Based on this known structure, we speculated that the ∼100 bp region of SNF5 most frequently observed to undergo telomere addition during generation of the linear fragments represents the Core sequence and that an adjacent sequence would provide Stim function. For a telomere to be added to the left end of the acentric fragment of chromosome II, the TG-rich sequence must be on the bottom or 3′ to 5′ strand (Figs. 3 and 4a). Analysis of this strand showed that a 718 nucleotide (nt) region including and extending 5′ to the putative Core sequence (chromosome II coordinates 779784 to 780501) is unusually TG-rich. Sequences of 300 nt from the 3′ to 5′ strand generated as 10 base sliding windows extending across this region contain, on average, 66% T or G nucleotides [(T + G)/300 = 0.66 ± 0.05, n = 42; Supplementary Fig. 4a]. In contrast, 300 nt sequences randomly selected from the yeast genome (n = 493) contain, on average, 50% T or G nucleotides [(T + G)/300 = 0.50 ± 0.04], as expected from an unbiased distribution relative to each other and the two strands (Supplementary Fig. 4a). To ensure that our analysis included any potential Stim sequence, we initially tested this entire 718 nt sequence for its ability to stimulate de novo telomere addition.
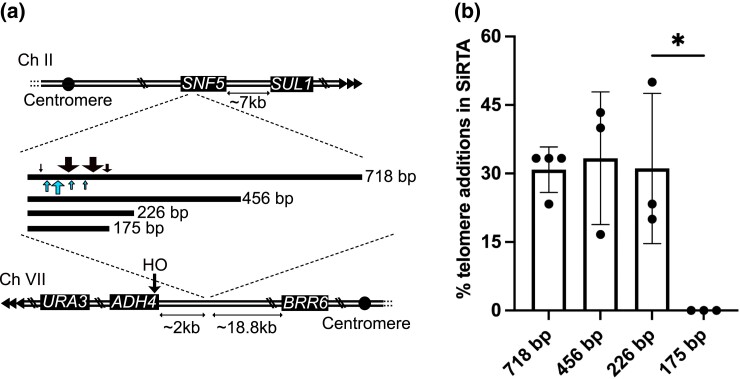
Fig. 4.
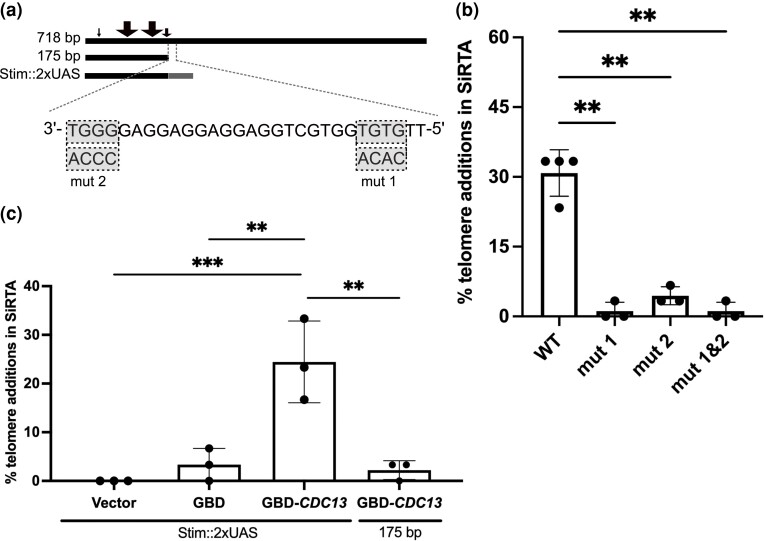
The SNF5 SiRTA is sufficient to stimulate de novo telomere addition at an ectopic location. a) Top schematic: the endogenous right arm of chromosome II; distance between the endogenous SNF5 sequence and SUL1 is shown. Triangles represent the endogenous telomere. Middle schematic: lines representing the relative size of each fragment from SNF5 that was integrated and tested (actual size in bp shown). Black arrows above the 718 bp fragment depict approximate locations of de novo telomere addition events stimulated by HO cleavage. Blue arrows below the 718 bp fragment depict approximate locations of spontaneous de novo telomere addition events from the chemostat experiments. Arrow size is proportional to the number of independent events at that location (see Supplementary Fig. 5 for exact numbers and locations). Bottom schematic: modified left arm of chromosome VII. Distances between the integrated SNF5 sequence and the HO cleavage site or the most distal essential gene (BRR6) on this chromosome arm are shown. b) Chromosome truncation events following HO cleavage preferentially involve de novo telomere addition in the SiRTA. At least 30 clones that survived HO cleavage and lost the URA3 marker were assayed for each experiment. The percentage of clones that incurred telomere addition within the sequence of interest is shown. Bar height is the average of three to four independent experiments; error bars represent standard deviations. A significant difference is indicated (*P < 0.05) by unpaired Student's t-test comparing 226 to 175 bp. See Supplementary File 2 for data used to generate the graph.
To determine the ability of this 718 nt sequence to stimulate telomere addition, we took advantage of an assay in which the sequence of interest is integrated on the left arm of chromosome VII, replacing sequences between chromosome VII coordinates 18086 and 18108 (Ngo et al. 2020). This strain (Lydeard et al. 2010; Obodo et al. 2016) contains a single recognition site for the yeast homothallic switching (HO) endonuclease that allows regulated generation of a DSB approximately 2 kb distal to the sequence being tested (Fig. 4a). Of note, the SNF5 sequence is inserted on chromosome VII in the inverted orientation relative to the centromere compared with its orientation on chromosome II. However, because the endogenous location is on the right arm of chromosome II and the insertion site is on the left arm of chromosome VII, the TG-rich sequence that will stimulate de novo telomere addition is, in both cases, located on the bottom (3′ to 5′) strand. In both configurations, exposure of the TG-rich strand in single-stranded DNA (for example, by resection of a DSB occurring to the left of the sequence of interest) followed by telomerase action on that 3′ overhang will stabilize the chromosome fragment located to the right of the lesion. On chromosome II, de novo telomere addition will create an acentric terminal fragment. On chromosome VII, de novo telomere addition will truncate the chromosome. The site at which the putative SiRTA is integrated is distal to the last essential gene on the chromosome arm (BRR6), ensuring that cells adding a de novo telomere within the sequence of interest remain viable. Because the sequence being tested is present both at its endogenous location and at the integrated site on chromosome VII, we deleted RAD52 to prevent repair by ectopic recombination.
Growth of this strain on media containing galactose induces expression of the HO endonuclease, generating a DSB distal to the putative SiRTA (Lydeard et al. 2010; Obodo et al. 2016). Correct repair restores the cleavage site, leading to a cycle of cleavage and repair that culminates in cell death or mutation/loss of the HO site. Among the cells that survive HO cleavage, those that have lost a URA3 marker distal to the HO site are identified through selection for growth on media containing 5-fluoroorotic acid (5-FOA) and are subsequently screened through a combination of PCR and Sanger sequencing to identify telomere addition events (Obodo et al. 2016). The fraction of 30 analyzed clones that have undergone de novo telomere addition within the sequence of interest represents the “relative” frequency of de novo telomere addition and is used as a measure of SiRTA function (Obodo et al. 2016; Epum et al. 2020). Previously described SiRTAs display relative frequencies of de novo telomere addition ranging from 10 to 35% (Obodo et al. 2016; Ngo et al. 2020).
In this case, analysis of 5-FOA-resistant clones surviving HO cleavage from four independent assays (30 clones each) showed that 30.8% ± 5% had incurred truncation events mapping to the 718 bp sequence (Fig. 4b). Each of the 51 5-FOA-resistant clones was analyzed by PCR using one primer that anneals to the AC-rich telomeric strand and a second primer that anneals centromere-proximal to the 718 bp sequence (Mangahas et al. 2001). In every case, a product of the expected size was produced which, when sequenced, revealed the direct addition of telomeric DNA to the chromosome II sequence (Supplementary File 2). Taken together, these results indicate that this region functions as a SiRTA (subsequently referred to as the “SNF5” SiRTA due to its location within the SNF5 open reading frame). Fifty of the 51 telomere addition events mapped to ∼180 bases at the 3′-end of the 718 base region, relative to the TG-rich strand (Fig. 4a; Supplementary Fig. 5). This ∼180 base region includes the ∼100 base region in which telomere addition was observed in the linear fragments and 69% of the HO-induced events map within three bases of a telomere-addition event observed on the linear molecules. However, six of nine telomere additions in the linear fragments lie 3′ to the vast majority of telomere additions observed after HO induction (Fig. 3; Supplementary Fig. 5). It is unclear why events observed in the linear fragments from the sulfate-limited chemostats are distributed differently than in the HO assay.
De novo telomere addition requires specific motifs associated with binding of Cdc13
To refine the location of sequences required for de novo telomere addition, we tested a series of fragments that included the region within which telomere addition occurred but were progressively truncated at the opposite (5′) end of the TG-rich strand. Fragments of 456 and 226 bp stimulated de novo telomere addition as strongly as the full 718 bp sequence (Fig. 4b). In contrast, no telomere addition events were observed in a fragment of 175 bp, despite retention of virtually all sequences that were targeted by telomerase in the context of the larger fragment (Fig. 4, a and b). We conclude that the SNF5 SiRTA has a structure similar to other characterized SiRTAs, with stimulatory sequences that map to the ∼50 bp region delimited by the 226 and 175 bp fragments. We refer to this 50 bp region as the SNF5 SiRTA Stim. Although the fraction of T and G nucleotides is elevated throughout the 718 bp region (Supplementary Fig. 4b, left axis), the ratio of G to T is notably higher in the centromere proximal 226 nucleotide region that is sufficient for de novo telomere addition than in the remainder of the region (Supplementary Fig. 4b, right axis). Because telomeres conform to a pattern of TG1-3, the higher G to T ratio is consistent with a sequence that has the ability to associate with telomeric proteins and/or the RNA template of telomerase.
In S. cerevisiae, telomerase is primarily recruited to the telomere by the single-stranded binding protein Cdc13 (Pennock et al. 2001; Bianchi et al. 2004). Binding of Cdc13 to TG-rich Stim sequences rendered single-stranded as a result of 5′-end resection following a DSB has been implicated in the high frequency of de novo telomere addition within a SiRTA (Obodo et al. 2016; Epum et al. 2020). Therefore, we hypothesized that the 50 bp SNF5 SiRTA Stim functions through recruitment of Cdc13. The optimal Cdc13 binding site within telomeric repeats has been characterized (5′-GTGTGGGTGTG) (Anderson et al. 2003; Eldridge et al. 2006), but single nucleotide substitutions within most positions are well-tolerated. The largest decrease in Cdc13 binding affinity is observed when G1, G3, or T4 nucleotide positions are substituted (GTGTGGGTGTG), suggesting that a 5′-GxGT-3′ motif may be necessary (but not sufficient) for Cdc13 binding (Anderson et al. 2003; Eldridge et al. 2006). The SNF5 SiRTA Stim sequence defined above contains two 5′-GxGT-3′ motifs on the TG-rich (bottom) strand (Fig. 5a). To determine if these motifs contribute to the high frequency of de novo telomere addition within the SiRTA, we mutated each 5′-GxGT-3′ motif to its complementary CxCA in the context of the 718 bp fragment. Remarkably, mutation of one or both motifs nearly eliminated de novo telomere addition (Fig. 5, a and b), suggesting that both are required for SiRTA function. This result is consistent with previous reports that SiRTA activity likely requires more than one Cdc13 binding site (Strecker et al. 2017). We hypothesize that the SiRTA sequence must reach a threshold of Cdc13 binding to stimulate de novo telomere addition and this threshold is not met in the absence of one or both 5′-GxGT-3′ motifs.
Fig. 5.
The SNF5 SiRTA requires Cdc13 binding to stimulate de novo telomere addition. a) Top: lines representing 718, 175 bp, and SNF5 SiRTA Stim::2XUAS fragments integrated and tested on chromosome VII. Sites of de novo telomere addition are depicted as in Fig. 4. Bottom: Twenty-nine nucleotides of the TG-rich strand of the SNF5 SiRTA Stim sequence are shown from 3′ to 5′. Note that a double-strand break that occurs to the left of this sequence will expose the TG-rich sequence in single-stranded DNA following 5′-end resection. 5′-GxGT-3′ motifs and corresponding mutations are highlighted in gray. b) Percentage of 5-FOA-resistant clones that incurred de novo telomere addition within the 718 bp SNF5 SiRTA in the presence of the indicated mutation. Wild-type (WT) samples are repeated from Fig. 4 for comparison. Samples with statistically significant values by one-way ANOVA with post hoc Dunnett's are indicated (**P < 0.01). c) Percentage of 5-FOA-resistant clones that incurred de novo telomere addition within the 175 bp SNF5 fragment when two copies of the Gal4 UAS are added (Stim::2xUAS) or when the 175 bp fragment is present alone. Cells were transformed with pRS414 (empty vector) or pRS414 expressing either Gal4 DNA binding domain (GBD) or GBD fused to the N-terminus of Cdc13 as indicated. For b and c, at least 30 colonies were assayed for each experiment. Bar height is the average of three to four independent experiments; error bars represent standard deviations. Samples with statistically significant values by one-way ANOVA with post hoc Tukey’s are indicated (*P < 0.01; ***P < 0.001). See Supplementary File 2 for data used to generate the graph.
To further interrogate the requirement of Cdc13 binding to stimulate de novo telomere addition, we replaced the 50 bp region of the SNF5 SiRTA Stim sequence with two copies of the Gal4 upstream activating sequence (Stim::2xUAS) to allow specific recruitment of the Gal4 DNA binding domain (GBD) (Bianchi et al. 2004; Zhang and Durocher 2010; Obodo et al. 2016). We introduced an empty vector, a vector expressing GBD alone, or a vector expressing a fusion of GBD to full-length Cdc13 into cells with the SNF5 SiRTA Stim::2xUAS fragment integrated at the chromosome VII test site. As expected, SNF5 SiRTA Stim::2xUAS did not support high frequency de novo telomere addition with empty vector or GBD alone. Consistent with previous reports that artificial recruitment of Cdc13 to the SiRTA supports de novo telomere addition (Bianchi et al. 2004; Zhang and Durocher 2010; Obodo et al. 2016), expression of the GBD-Cdc13 fusion protein supported de novo telomere addition in 24.4% ± 8.3% of clones tested from three independent assays and stimulation was dependent on the presence of the UAS sequences (Fig. 5c). These results further support our hypothesis that a threshold of Cdc13 binding must be met to stimulate de novo telomere addition and that without the 5′-GxGT-3′ motif-containing Stim sequence, this critical threshold is not satisfied.
Discussion
In this work, we demonstrate that diploid laboratory yeast strains grown in sulfate-limiting conditions generate small, extrachromosomal linear DNA fragments containing the primary sulfate-transporter SUL1 that persist because they provide a selective growth advantage. The linear fragments are capped at one end by sequences from the endogenous right telomere and at the other end by de novo telomere addition events clustered in a region centromere proximal to SUL1. Fragments of similar, but not identical, size arise in multiple independent cultures, suggesting that some sequence feature or features in this region are responsible for the creation and/or stabilization of these linear fragments. These results raise three intertwined questions: (1) What features of the region make it a hotspot for telomere addition? (2) What process generates the substrate for telomere addition in the sulfate-limiting chemostats (in the absence of HO cleavage)? And (3) Why do the diploids produce a different form of SUL1 amplicon than their isogenic haploids?
Telomere addition
We find multiple sequence features within the SNF5 gene that contribute to the stabilization of linear fragments through de novo telomere addition. We demonstrate that a DNA fragment from the SNF5 gene that contains the common sites of telomere addition is sufficient to stimulate de novo telomere addition at an ectopic location, similar to other characterized hotspots of de novo telomere addition (SiRTAs) (Obodo et al. 2016; Ngo et al. 2020). Previous work demonstrated that association of Cdc13 with a SiRTA is critical for the stimulation of telomere addition (Obodo et al. 2016; Epum et al. 2020). However, the number and/or affinity of Cdc13 binding sites sufficient to support de novo telomere formation are unclear. Strecker et al. (2017) placed differing lengths of telomeric repeats adjacent to a DSB to determine a threshold length above which telomere addition is stimulated. Telomere addition is strongly inhibited at sequences below that threshold length (Strecker et al. 2017). Hypomorphic mutations in Cdc13 increase this threshold value, suggesting that the net number and/or affinity of Cdc13 binding sites likely determines the threshold at which a sequence acts to stimulate telomere addition (Strecker et al. 2017). Our results are consistent with this hypothesis. Remarkably, mutation of just four nucleotides corresponding to a predicted binding site for Cdc13 (Anderson et al. 2003; Eldridge et al. 2006) nearly eliminates detectable de novo telomere addition at the SNF5 SiRTA in the ectopic assay. Our results suggest that there are likely two Cdc13 binding sites in the ∼50 bp fragment that together constitute the SNF5 SiRTA Stim. Mutation of either site lowers the total amount of Cdc13 binding below the threshold required for this sequence to function as a hotspot of de novo telomere addition. Consistent with this interpretation, replacement of the 50 bp SNF5 SiRTA Stim sequence with two copies of the Gal4 upstream activating sequence supports de novo telomere addition only when cells express the Gal4 DNA binding domain fused to Cdc13.
Substrate for telomere addition
We showed that SNF5 SiRTA activity is supported by a minimal fragment of 226 bp. This small region comprises approximately ∼1% (226 bp/20.2 kb) of the total region of chromosome VII in which a viable chromosome truncation could occur (between the HO site and the most distal essential gene on this chromosome arm), and yet nearly one third of all truncation events were mapped to the SNF5 SiRTA. This clustering of de novo telomere additions in a small region of SNF5 could result from chromosome fragility in this region; however, observations from the ectopic assay argue against this model. The SNF5 SiRTA undergoes unusually high levels of de novo telomere addition even when the initiating chromosome break is intentionally induced two kilobases distal to the eventual site at which telomerase acts, suggesting that it is a site at which DNA ends are preferentially repaired. We conclude that this region of SNF5 is not necessarily more prone to double-stranded breaks, but rather has the fortuitous combination of features that permit telomere addition.
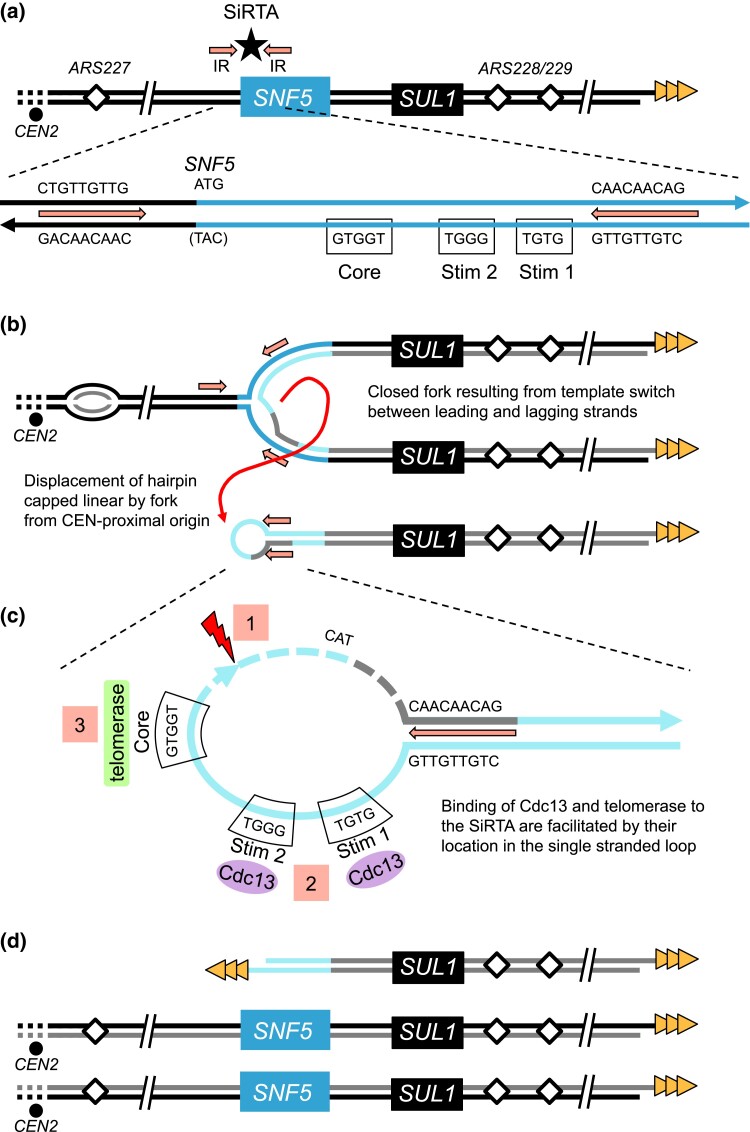
While we have not detected double-stranded breaks in chemostat cultures, we have evidence that replication errors, instead of double-stranded breaks, may generate aberrant intermediates that are subsequently targeted for repair (Brewer et al. 2015). The discontinuous nature of DNA replication forks provides an opportunity for nascent leading strands to anneal to the lagging strand template during fork reversal at sites of interrupted inverted repeats [described as ODIRA in Brewer et al. (2011)]. In a scan of the region at the 5′-end of the SNF5 gene, we find a ∼5-fold increase in the density of short interrupted inverted repeats (Supplementary Fig. 6a) resulting primarily from a sequence in the promoter region (5′-TGTTGTTG-3′) that is complementary to the several stretches of CAA triplet repeats within the N-terminal coding region of SNF5 (Fig. 6a). When compared to >10,000 regions across the genome, the SNF5 region is enriched for short interrupted inverted repeats (Supplementary Fig. 6b). The regression of a replication fork proceeding toward the centromere from ARS228 or another distal origin (e.g. ARS229) could provide multiple opportunities for this sequence on the leading strand to find a match with a complementary sequence in the single-stranded gaps on the lagging strand and thereby create a “closed” fork (Fig. 6b). After completion of replication by a fork established centromere proximal to the SNF5 region, the hairpin-capped linear could be released (Fig. 6b; Brewer et al. 2011). Processing of the single-stranded loop by nucleases provides the appropriate substrate for telomere addition (Fig. 6, c and d).
Fig. 6.
A model to explain how an ODIRA event may generate a substrate for de novo telomere addition at the SNF5 SiRTA. a) The endogenous right arm of chromosome II contains interrupted inverted repeats (IR; arrows) flanking the SNF5 SiRTA (star) and the TG-rich strand of the right telomere (triangles). In the expanded schematic the relevant sequences are shown for one possible pair of inverted repeats (arrows), the start codon of SNF5 (ATG), the Core of the SNF5 SiRTA, and the two 5′-GxGT-3′ motifs from Fig. 5 (labeled Stim 1 and 2). b) Ligation of the leading strand to the lagging strand following fork reversal at the leftward moving fork and template switching at the inverted repeats results in a “closed” fork with the rightward moving fork completing replication through the right chromosomal telomere. When a replication fork from ARS227, proceeding toward the telomere, encounters the closed fork structure, a hairpin-capped linear is displaced (curved arrow). c) In the displaced hairpin-capped linear, the inverted repeats form the base of the terminal single-stranded loop that contains the SiRTA Stim and Core sequences. Cleavage of the hairpin to the 3′ side of the SiRTA (1) and resection at the site of the break (dashed lines) exposes a 3′ single-stranded tail containing SiRTA sequences which can be bound by Cdc13 (2) and telomerase (3) to form a de novo telomere. d) In this model, after a single round of replication, the chromosome that experienced the closed fork produces two unaltered sister chromatids and the linear fragment containing SUL1 which is now protected by a de novo telomere addition at the SNF5 SiRTA. (Normal replication of the homolog in this diploid is not shown.) Under sulfate-limiting conditions, the cells containing the linear fragment are retained in the population due to the growth advantage provided by one or more extra copies of SUL1.
Ploidy-specific amplification pathways
In previous experiments with haploid laboratory strains, we have not observed the generation of linear fragments—essentially all events were interstitial inverted triplications on chromosome II (Payen et al. 2014). In some cases, rearrangements that would be lethal in haploids are tolerated in diploids by the presence of a second, unrearranged chromosome (Zhang et al. 2013). It is unlikely that a similar phenomenon is affecting the nature of SUL1 amplification in diploid and haploid cells as there are no essential genes beyond SNF5 on chromosome II, so loss of this terminal fragment should be tolerable in haploids. In point of fact, we do not detect loss of the distal markers on chromosome II in the diploids—both homologs of chromosome II remain intact. The aCGH profiles indicated that there were no regions at the right end of chromosome II that were present at fewer than two copies per diploid genome and CHEF gels did not reveal any truncated variants of chromosome II (Fig. 1; Supplementary Figs. 1 and 2). While these results are consistent with repair of a broken chromosome II by homology directed repair using the homolog or sister chromatid as template (perhaps by break-induced replication), it is also possible that a double-stranded break was not the initiating event. We would like to propose that the same type of ODIRA event that can explain the events that occur in haploid cells is also occurring in the diploids, but that the resolution of the intermediate differs between haploids and diploids. Differences in the regulation of repair pathways between haploid and diploid cells in this laboratory strain may contribute to the different outcomes (Li and Tye 2011). There may be a race between replication of the hairpin intermediate and the processing of the single-stranded loop that is influenced by the levels of repair enzymes. In support of this notion, we have recently observed that in some haploid wild strains with varied genetic backgrounds, linear amplicons similar to those described here, also arising from the vicinity of the SNF5 gene, are the preferred amplicon (manuscript in preparation). Analysis of the transcriptomes and proteomes of these strains could identify co-varying RNA/proteins that influence the amplicon outcome. Regardless of mechanism, our observations in yeast may provide a useful model for the types of rearrangements that occur in humans and other organisms. Finally, results presented here suggest that interstitial TG-rich hotspots of de novo telomere addition that are oppositely oriented relative to the chromosome arm can stabilize an acentric fragment to provide, in some circumstances, a selective advantage. These observations highlight a possible evolutionary consequence of SiRTA sequences regardless of orientation, by increasing adaptation and genome plasticity.
Supplementary Material
Acknowledgments
We thank James Haber and Alessandro Bianchi for generously providing us with strains and plasmids and Natalie Guzman for initial exploratory studies.
Contributor Information
Remington E Hoerr, Department of Biological Sciences, Vanderbilt University, Nashville, TN 37235, USA.
Alex Eng, Department of Genome Sciences, University of Washington, Seattle, WA 98195, USA.
Celia Payen, Department of Genome Sciences, University of Washington, Seattle, WA 98195, USA; IFF, Wilmington, DE 19803, USA.
Sara C Di Rienzi, Department of Genome Sciences, University of Washington, Seattle, WA 98195, USA; Department of Molecular Virology and Microbiology, Baylor College of Medicine, Houston, TX 77030, USA.
M K Raghuraman, Department of Genome Sciences, University of Washington, Seattle, WA 98195, USA.
Maitreya J Dunham, Department of Genome Sciences, University of Washington, Seattle, WA 98195, USA.
Bonita J Brewer, Department of Genome Sciences, University of Washington, Seattle, WA 98195, USA.
Katherine L Friedman, Department of Biological Sciences, Vanderbilt University, Nashville, TN 37235, USA.
Data availability
Strains and plasmids are available upon request. Source data for Figs. 4 and 5 and Supplementary Fig. 5 are provided in Supplementary File 2. Sequencing data are available from the NIH Sequence Read Archive (SRA) under BioProject ID PRJNA909857. aCGH data are available from the NIH Gene Expression Omnibus (GEO) under accession GSE220549.
Supplemental material available at GENETICS online.
Funding
This work was supported by the National Institutes of Health awards R01GM123292 to KLF, R35GM122497 to BJB, and R01GM094306 to MJD and by the National Science Foundation award 1120425 to MJD. REH was supported by the National Institutes of Health award T32GM137793. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Literature Cited
- Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12(5):363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand R, Memisoglu G, Haber J. Cas9-mediated gene editing in Saccharomyces cerevisiae. Protocol Exchange. 2017. doi: 10.1038/protex.2017.021a. [DOI] [Google Scholar]
- Anderson EM, Halsey WA, Wuttke DS. Site-directed mutagenesis reveals the thermodynamic requirements for single-stranded DNA recognition by the telomere-binding protein Cdc13. Biochemistry. 2003;42(13):3751–3758. doi: 10.1021/bi027047c. [DOI] [PubMed] [Google Scholar]
- Araya CL, Payen C, Dunham MJ, Fields S. Whole-genome sequencing of a laboratory-evolved yeast strain. BMC Genomics. 2010;11(1):88. doi: 10.1186/1471-2164-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Negrini S, Shore D. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol Cell. 2004;16(1):139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Biedler JL, Meyers MB, Spengler BA. Homogeneously staining regions and double minute chromosomes, prevalent cytogenetic abnormalities of human neuroblastoma cells. In: Fedoroff S, Hertz L, editors. Advances in Cellular Neurobiology, Vol. 4. Elsevier; 1983. p. 267–307. [Google Scholar]
- Blackburn EH. Structure and function of telomeres. Nature. 1991;350(6319):569–573. doi: 10.1038/350569A0. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Payen C, Di Rienzi SC, Higgins MM, Ong G, Dunham MJ, Raghuraman MK. Origin-dependent inverted-repeat amplification: tests of a model for inverted DNA amplification. PLoS Genet. 2015;11(12):e1005699. doi: 10.1371/journal.pgen.1005699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Payen C, Raghuraman MK, Dunham MJ. Origin-dependent inverted-repeat amplification: a replication-based model for generating palindromic amplicons. PLoS Genet. 2011;7(3):e1002016. doi: 10.1371/journal.pgen.1002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Todd KM, Rosenzweig RF. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol Biol Evol. 1998;15(8):931–942. doi: 10.1093/OXFORDJOURNALS.MOLBEV.A026009. [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Ay F. Identification of copy number variations and translocations in cancer cells from Hi-C data. Bioinformatics. 2018;34(2):338–345. doi: 10.1093/BIOINFORMATICS/BTX664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran CG, Pennington SR. Copy number alteration signatures as biomarkers in cancer: a review. Biomark Med. 2022;16(5):371–386. doi: 10.2217/BMM-2021-0476. [DOI] [PubMed] [Google Scholar]
- Dorsey M, Peterson C, Bray K, Paquin CE. Spontaneous amplification of the ADH4 gene in Saccharomyces cerevisiae. Genetics. 1992;132(4):943–950. doi: 10.1093/genetics/132.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge AM, Halsey WA, Wuttke DS. Identification of the determinants for the specific recognition of single-strand telomeric DNA by Cdc13. Biochemistry. 2006;45(3):871–879. doi: 10.1021/bi0512703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epum EA, Mohan MJ, Ruppe NP, Friedman KL. Interaction of yeast Rad51 and Rad52 relieves Rad52-mediated inhibition of de novo telomere addition. PLoS Genet. 2020;16(2):e1008608. doi: 10.1371/journal.pgen.1008608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D, Dunham MJ. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4(12):e1000303. doi: 10.1371/JOURNAL.PGEN.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D, Usaite R, Germann SM, Lisby M, Botstein D, Regenberg B. Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus. Proc Natl Acad Sci U S A. 2010;107(43):18551. doi: 10.1073/PNAS.1014023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10(8):551. doi: 10.1038/NRG2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerr RE, Ngo K, Friedman KL. When the ends justify the means: regulation of telomere addition at double-strand breaks in yeast. Front Cell Dev Biol. 2021;9(18):655377. doi: 10.3389/fcell.2021.655377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilić M, Zaalberg IC, Raaijmakers JA, Medema RH. Life of double minutes: generation, maintenance, and elimination. Chromosoma. 2022; 131(3):107–125. doi: 10.1007/S00412-022-00773-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao KC, Sherlock G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat Genet. 2008;40(12):1499–1504. doi: 10.1038/NG.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38(1):233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Kwan EX, Wang XS, Amemiya HM, Brewer BJ, Raghuraman MK. rDNA copy number variants are frequent passenger mutations in Saccharomyces cerevisiae deletion collections and de novo transformants. G3 Genes|Genomes|Genetics. 2016;6(9):2829–2838. doi: 10.1534/g3.116.030296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, Tye BK. Ploidy dictates repair pathway choice under DNA replication stress. Genetics. 2011;187(4):1031. doi: 10.1534/GENETICS.110.125450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR, Hughes TR, Lundblad V. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci U S A. 1997;94(21):11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS , McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR . Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast (Chichester, England). 1998;14(10):953–961. doi:. [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Lipkin-Moore Z, Jain S, Eapen V V, Haber JE. Sgs1 and Exo1 redundantly inhibit break-induced replication and de novo telomere addition at broken chromosome ends. PLoS Genet. 2010;6(5):e1000973. doi: 10.1371/journal.pgen.1000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JR, Ziman R, Yuen RKC, Feuk L, Scherer SW. The database of genomic variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42(D1):D986. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangahas JL, Alexander MK, Sandell LL, Zakian VA. Repair of chromosome ends after telomere loss in Saccharomyces. Mol Biol Cell. 2001;12(12):4078–4089. doi: 10.1091/mbc.12.12.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AW, Befort C, Kerr EO, Dunham MJ. Design and use of multiplexed chemostat arrays. J Vis Exp. 2013;72:1–6. doi: 10.3791/50262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan V, Mieczkowski PA, Kim HM, Petes TD, Lobachev KS. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell. 2006;125(7):1283–1296. doi: 10.1016/J.CELL.2006.04.042. [DOI] [PubMed] [Google Scholar]
- Ngo K, Epum EA, Friedman KL. Emerging non-canonical roles for the Rad51–Rad52 interaction in response to double-strand breaks in yeast. Curr Genet. 2020;66(5):917–926. doi: 10.1007/s00294-020-01081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obodo UC, Epum EA, Platts MH, Seloff J, Dahlson NA, Velkovsky SM, Paul SR, Friedman KL . Endogenous hot spots of de novo telomere addition in the yeast genome contain proximal enhancers that bind Cdc13. Mol Cell Biol. 2016;36(12):1750–1763. doi: 10.1128/mcb.00095-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C, Di Rienzi SC, Ong GT, Pogachar JL, Sanchez JC, Sunshine AB, Raghuraman MK, Brewer BJ, Dunham MJ. The dynamics of diverse segmental amplifications in populations of Saccharomyces cerevisiae adapting to strong selection. G3 (Bethesda). 2014;4(3):399–409. doi: 10.1534/g3.113.009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104(3):387–396. doi: 10.1016/S0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Kolodner RD. Pathways and mechanisms that prevent genome instability in Saccharomyces cerevisiae. Genetics. 2017;206(3):1187–1225. doi: 10.1534/genetics.112.145805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosswog C, Bartenhagen C, Welte A, Kahlert Y, Hemstedt N, Lorenz W, Cartolano M, Ackermann S, Perner S, Vogel W, et al. Chromothripsis followed by circular recombination drives oncogene amplification in human cancer. Nat Genet. 2021;53(12):1673–1685. doi: 10.1038/S41588-021-00951-7. [DOI] [PubMed] [Google Scholar]
- Shaikh TH. Copy number variation disorders. Curr Genet Med Rep. 2017;5(4):183. doi: 10.1007/S40142-017-0129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Source Sci New Ser. 1994;266(5184):404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Song RH, Gao CQ, Zhao J, Zhang JA. An update evolving view of copy number variations in autoimmune diseases. Front Genet. 2022;12:794348. doi: 10.3389/FGENE.2021.794348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Minami JK, Huang A, Dehkordi SR, Lomeli SH, Luebeck J, Goodman MH, Moriceau G, Krijgsman O, Dharanipragada P, et al. Plasticity of extrachromosomal and intrachromosomal BRAF amplifications in overcoming targeted therapy dosage challenges. Cancer Discov. 2022;12(4):1046–1069. doi: 10.1158/2159-8290.CD-20-0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 2003;17(19):2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker J, Stinus S, Pliego Caballero M, Szilard RK, Chang M, Durocher D. A sharp Pif1-dependent threshold separates DNA double-strand breaks from critically short telomeres. Elife. 2017;6:e23783. doi: 10.7554/eLife.23783.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, Li B, Arden K, Ren B, Nathanson DA, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543(7643):122–125. doi: 10.1038/NATURE21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RJ, Zakian VA. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics. 2012;191(4):1073–1105. doi: 10.1534/genetics.111.137851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Durocher D. De novo telomere formation is suppressed by the Mec1-dependent inhibition of Cdc13 accumulation at DNA breaks. Genes Dev. 2010;24(5):502–515. doi: 10.1101/gad.1869110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10(1):451. doi: 10.1146/ANNUREV.GENOM.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zeidler AFB, Song W, Puccia CM, Malc E, Greenwell PW, Mieczkowski PA, Petes TD, Argueso JL. Gene copy-number variation in haploid and diploid strains of the yeast Saccharomyces cerevisiae. Genetics. 2013;193(3):785–801. doi: 10.1534/GENETICS.112.146522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. Source data for Figs. 4 and 5 and Supplementary Fig. 5 are provided in Supplementary File 2. Sequencing data are available from the NIH Sequence Read Archive (SRA) under BioProject ID PRJNA909857. aCGH data are available from the NIH Gene Expression Omnibus (GEO) under accession GSE220549.
Supplemental material available at GENETICS online.