Abstract
Background
Genome-wide sequencing may extensively identify potential pathogenic variants, which helps to understand mechanisms of tumorigenesis, but such study has not been reported in benzene-induced leukemia (BIL).
Methods
We recruited 10 BIL patients and conducted the whole-exome sequencing on their peripheral blood samples. The obtained sequencing data were screened for potential pathogenic and novel variants, then the variants-located genes were clustered to identify cancer-related pathways. Shared or recurrent variants among the BIL cases were also identified and evaluated for their potential functional impact.
Results
We identified 48,802 variants in exons in total, 97.3% of which were single nucleotide variants. After filtering out variants with minor allele frequency ≥ 1%, we obtained 8667 potentially pathogenic variants, of which 174 were shared by all the BIL cases. The identified variants located in genes that could be significantly enriched into certain cancer-related pathways such as PI3K-AKT signaling pathway and Ras signaling pathway. We also identified 1010 novel variants with no record in the Genome Aggregation Database and in dbSNP, and one of them was shared by 90% cases. The recurrent and novel variant caused a missense mutation in SESN3.
Conclusions
We examined variations of the whole exome in BIL patients for the first time. The commonly shared variants implied a relation with BIL, and the recurrent and novel variant might be specifically related to BIL. The related variants may help unravel the carcinogenic mechanisms of BIL.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-023-01442-w.
Keywords: Benzene, Exome, High-throughput sequencing, Leukemia, Genetic variant
Background
Benzene has been wildly used as chemical intermediate and organic solvent in the chemical, petroleum and drug industries all over the world for about a century, though it was gradually proved to have various adverse health effects, especially to the haematopoietic system [1]. Carcinogenicity of benzene was first established in 1982 based on definite evidence of animal experiments as well as epidemiological studies [1], and benzene-induced leukemia (BIL) is diagnosed in China according to the current national criteria GBZ 94-2017 “Diagnosis of Occupational Tumor” [2]. Apart from exposure to benzene of over 6 mg/m3 for at least 6 months, the diagnosis requires a latent period of 2 or more years [2]. Besides, leukemia developed with a medical history of chronic benzene poisoning can be directly diagnosed as BIL [2]. Large cohort studies estimated that the incidence of leukemia among benzene exposure workers was 13.59 per 100,000, and the relative risk for all hematologic neoplasms combined was 2.6 (95% CI 1.4–4.7) [3, 4].
It was reported that about 80% of BIL were clinically classified as acute myeloid leukemia (AML), and about 20% were acute lymphocytic leukemia (ALL) and chronic myeloid leukemia (CML) combined [5, 6]. We observed more CML in clinics than ALL. Both AML and CML are malignant bone marrow cancers showing the characteristic of abnormal growth of myeloid cells at different maturation stages. Clinically, CML can be classified into chronic phase (CP), accelerated phase (AP), and blast phase (BP) or blast crisis (BC) [7]. Although most CML patients remain in the CP which is relatively benign, their conditions will undesirably progress to the myeloid or lymphoid BP if left untreated. CML in BP is similar to AML, both of them show an increased frequency of blast cells, aggressiveness and poor prognosis [8]. The Philadelphia chromosome (Ph) is a unique hallmark of CML cells, which results from the t(9;22)(q34;q11) chromosomal translocation and encodes a carcinogenic fusion protein BCR-ABL1, a tyrosine kinase with deregulated activity. The tyrosine kinase inhibitors (TKIs) targeting BCR-ABL1 were developed 2 decades ago to effectively treat CML [8]. Comparatively, AML was found to have at least 24 different genetic subtypes, suggesting quite more heterogeneous molecular features than CML [9, 10]. Until this day, cytotoxic chemotherapy is still the same standard treatment for AML as in half a century ago [8].
The key to develop more efficient cancer detection methods and therapeutic approaches is uncovering the mechanisms of tumorigenesis, which is considered primarily driven by genetic mutations [11]. Nowadays genetic studies have reached to the genome-wide scope at a single-base resolution thanks to the next-generation sequencing technologies. AML was among the first malignancies that had been extensively studied by novel high-throughput microarray and sequencing technologies [12]. After first reported in 2008, genomic studies in AML identified numerous novel recurrent somatic alleles, such as mutations in the DNA methyltransferase 3A gene (DNMT3A) and the isocitrate dehydrogenase genes (IDH1 and IDH2), and most of them were of biologic, prognostic, or therapeutic relevance [12]. As for CML, studies focused on detecting mutations in BCR-ABL1 gene, because the fusion gene plays an important role in the pathogenesis of CML, and mutations in it might engender failure of one or more of the currently effective TKIs [13]. Such reported mutations include T315I, Y253F/H, E255K, V299L and L237M, etc. (transcript ENST00000318560.6). [13]. Besides, studies also discovered mutations in ASXL1, DNMT3A, EZH2, RUNX1, TET2 and TP53 genes in the chronic phase of CML, and in CBL, CDKN2A, GATA-2, IDH1, IDH2, IKZF1, KRAS, NPM1, NRAS, RB1, RUNX1, TET2, TP53 and WT1 genes in the advanced phase [13, 14]. Unfortunately, there has been no genome-wide sequencing studies reported on BIL to date regardless of the clinical types.
While being clinically heterogeneous, BIL has the same oncogenous origin, that is benzene exposure. Whether there are shared (or recurrent) genetic variants among the BIL patients is still unknown to the best of our knowledge. Furthermore, if the putative variants were specific to BIL and/or had potential impact on biological structures or functions, they would be quite valuable for further studies on carcinogenic mechanisms. Intrigued by the aforementioned hypothesis, we tentatively recruited 10 BIL patients and conducted the whole-exome sequencing (WES) on their peripheral blood samples. The sequencing data were first screened for potential pathogenic variants by using a cutoff value of minor allele frequency (MAF) < 1%, and then screened for novel variants by excluding those recorded in the Genome Aggregation Database (gnomAD) and in Database of Short Genetic Variation (dbSNP). The pathogenic variants- and the novel variants-located genes were separately evaluated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to identify cancer related pathways. Thereafter, commonly shared pathogenic variants and recurrent novel variants among the BIL cases were preliminarily examined for their potential functional impact.
Methods
Cases information and samples
We recruited 10 BIL cases from all 39 leukemia patients hospitalized in Shenzhen Prevention and Treatment Center for Occupational Diseases (SPTCOD) during the year 2016–2020, excluded were non-BIL and patients undergone hematopoietic stem cell transplantation therapy. BIL was strictly diagnosed according to the GBZ 94-2017 criteria [2]. Originally, we planned to select 5 CML and 5 AML from the BIL cases successively by date of admission, but only 4 AML cases met the selection criteria, so we finally included one more CML to get 10 study cases in total. Written informed consent for participating the study was obtained from all the cases or their guardians.
We collected and sorted demographic, occupational and medical information of the cases from their electronic medical records. Peripheral blood samples of the cases were collected after their routine laboratory examination during the hospitalization, and stored at − 80 °C for later use. Unfortunately, paired normal samples were not obtained at the same time. The use of patient information and peripheral blood samples for further studies beyond routine laboratory examination was approved by the Ethics Committee of SPTCOD (No. LL-202036). This study abides by the Helsinki Declaration on ethical principles for medical research involving human subjects.
Whole-exome sequencing
Extracted from 1.0 mL of each blood sample, each genomic DNA sample was quantified by using a spectrofluorometer (Gemini™ XPS Microplate Reader, Molecular Devices, USA), and about 200 ng of each DNA sample were randomly fragmented by Covaris sonication (LE220-plus Focused-ultrasonicator, Covaris, USA). The DNA fragments with the size mainly distributed between 150 and 250 bp were repaired with an "A" base added at the 3′-end of each strand, thereafter adapters were ligated to both ends of the end repaired/dA tailed DNA fragments, which were selected by size, amplified by ligation-mediated PCR (S1000 Thermal Cycler, Bio-Rad, USA), purified [QIAquick PCR Purification Kit, QIAGEN China (Shanghai)], and hybridized to the exome array for enrichment. After washing out non-hybridized fragments, captured products were circularized and the rolling circle amplification was performed to produce DNA nanoballs. Each resulting qualified captured library was then loaded on BGISEQ-500 sequencing platforms (MGI Tech, China) to perform high-throughput sequencing [15, 16]. Sequencing-derived raw image files were processed by BGISEQ-500 base calling software with default parameters to generate the sequence data of each case as paired-end reads.
Data analysis and plotting
The raw sequencing data were first filtered to obtain high-quality clean data by the following methods: (a) removing the adapter sequences from all reads, (b) removing the pair of reads if the percentage of low-quality base in either of the two end reads exceeds 50% or the percentage of N base in either of the two end reads exceeds 10%. All clean data of each sample were then mapped to the human reference genome (GRCh38) using Burrows–Wheeler Aligner (BWA V0.7.15) [17, 18]. Local realignment around small insertions and deletions (InDels) and base quality score recalibration were performed using the Genome Analysis Toolkit (GATK) [19, 20], with duplicate reads removed by Picard tools [21]. The strict data analysis quality control system was built through the whole process to guarantee qualified sequencing data. All genomic variations including single nucleotide variants (SNVs) and InDels were detected by the state-of-the-art software, such as HaplotypeCaller of GATK (v3.7). After that, the hard-filtering method was applied to get high-confident variant calls, and the SnpEff tool was applied to perform a series of annotations for variants [22, 23]. Detailed description of the bioinformatics analysis has been reported before [15, 16].
Advanced data analysis and plotting were performed using R v4.0.4 [24]. We first used MAF < 1% as a cutoff value to filter out potential polymorphic or benign variants, and the obtained variants were examined for their located genes, which were then put into KOBAS v3.0 for pathway enrichment using the KEGG database with P value and corrected P value both < 0.05 [25]. Commonly shared variants among the BIL cases were identified and assessed for their functional impact. Subsequently, we further excluded those recorded in dbSNP and in gnomAD to get novel variants, and the variants-located genes were subjected to a similar pathway analysis. Thereafter, the recurrent novel variants were examined for their potential impact on normal functions of the genes.
Results
Basic information of the cases
The 10 BIL cases had an average age of 44 years old, and 30% of them were male. Only one male patient (10%) had smoking and drinking habits. The mean time of benzene exposure of the cases was 6.47 years, and showed no significant difference between the AML and BML cases (P = 0.459). The BCR-ABL1 gene had been detected in all the 6 CML cases before their TKIs therapy, but the fusion gene was not detected in the AML cases who were therefore treated with the chemotherapy. The cases were all alive at the end of follow-up, and the average of survival time (from date of clinical diagnosis to end of follow-up) was 8.56 years for all cases, and was not significantly different between the AML and BML cases (P = 0.173). The blood cell indices of the samples used for WES, including counts of white blood cell, neutrophil and platelet, and immature granulocyte percentage were all within their clinical references, respectively; the indices were not significantly different between the AML and BML cases (P = 0.091, 0.156, 0.591 and 0.507, respectively). Detailed information of the 10 BIL cases was listed in Table 1.
Table 1.
Basic information of the benzene-induced leukemia cases
| Variables | Total (n = 10) | Clinical diagnosis | |
|---|---|---|---|
| AML (n = 4) | CML (n = 6) | ||
| Age [year, mean (SE)] | 44.00 (1.69) | 44.25 (3.01) | 43.83 (2.21) |
| Gender [male (%)] | 30.00 | 25.00 | 33.33 |
| Race [Han (%)] | 100.00 | 100.00 | 100.00 |
| Smoking [yes (%)] | 10.00 | 25.00 | 0 |
| Drinking [yes (%)] | 10.00 | 25.00 | 0 |
| Benzene exposure duration [year, mean (SE)] | 6.47 (1.13) | 7.93 (1.92) | 5.74 (1.46) |
| White blood cell count [× 109/L, mean (SE)] | 4.77 (0.68) | 6.50 (1.20) | 3.62 (0.41) |
| Neutrophil count [× 109/L, mean (SE)] | 2.76 (0.53) | 3.95 (1.05) | 1.97 (0.28) |
| Platelet count [× 109/L, mean (SE)] | 187.90 (24.95) | 208.25 (52.69) | 174.33 (25.79) |
| Immature granulocyte percentage [%, mean (SE)] | 0.17 (0.05) | 0.12 (0.08) | 0.20 (0.08) |
| BCR-ABL1 fusion gene [positive (%)] | 60.00 | 0 | 100.00 |
| Treatment | |||
| Chemotherapy (%) | 40.00 | 100.00 | 0 |
| TKIs therapy (%) | 60.00 | 0 | 100.00 |
| Survival time [year, mean (SE)] | 8.56 (0.66) | 7.52 (0.62) | 9.24 (0.96) |
AML acute myeloid leukemia, CML chronic myeloid leukemia, SE standard error
Overview of the WES-identified variants
The WES yielded an average of 138,116,902 raw reads per sample. After removing low-quality reads, we obtained averagely 137,989,040 clean reads per sample, thus the mean clean data rate was 99.9%. The clean reads of each sample had high Q20 and Q30, showing high sequencing quality. The average GC content was 49.64%. All WES data production was summarized in Additional file 1.
On average, we captured 60.46 Mb target region, and successfully mapped 99.93% of the clean reads to the human reference genome. After removing the duplicate reads, we obtained a mean of 114,812,614 effective reads. The capture specificity that is the percentage of total effective bases mapped on target regions was 50.55%. We attained a 95.49 fold of mean sequencing depth on target regions. On average, 99.42% of targeted bases per sample were at least sequenced by 1 × coverage and 98.73% by 10 × coverage (see Additional file 2).
As shown in Table 2, we identified averagely 20,213 InDels and 121,407 SNVs per sample. The mean numbers of novel InDels and SNVs presented neither in dbSNP nor in gnomAD were 354 and 195, respectively. Of the InDels on average, 250 were frameshift, 82 were non-frameshift insertion, 120 were non-frameshift deletion, 2 were startloss, and 53 were splice site; of the SNVs on average, 10,467 were synonymous, 10,397 were missense, 40 were stoploss, 113 were stopgain, 33 were startloss, and 153 were splice site.
Table 2.
Statistics of the whole-exome sequencing identified variants
| Variant type | Sample | Total variants | Fraction in dbSNP (%) | Fraction in gnomAD (%) | Novel | Homozygous | Heterozygous | Intronic | 5′-UTR | 3′-UTR | Upstream | Downstream | Intergenic |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| InDel | Case_1 | 20,008 | 94.59 | 87.02 | 332 | 7535 | 12,473 | 15,959 | 307 | 750 | 752 | 568 | 149 |
| Case_2 | 20,168 | 94.98 | 86.47 | 324 | 7666 | 12,502 | 16,072 | 333 | 761 | 792 | 558 | 138 | |
| Case_3 | 19,731 | 95.61 | 87.55 | 261 | 7784 | 11,947 | 15,823 | 311 | 715 | 738 | 581 | 129 | |
| Case_4 | 20,758 | 95.10 | 86.65 | 324 | 7803 | 12,955 | 16,624 | 336 | 770 | 809 | 566 | 132 | |
| Case_5 | 20,193 | 95.40 | 85.43 | 327 | 7915 | 12,278 | 16,149 | 338 | 769 | 782 | 557 | 113 | |
| Case_6 | 20,220 | 95.51 | 81.64 | 397 | 7781 | 12,439 | 16,188 | 330 | 748 | 762 | 564 | 126 | |
| Case_7 | 20,149 | 94.95 | 83.30 | 405 | 7596 | 12,553 | 16,167 | 317 | 750 | 735 | 548 | 133 | |
| Case_8 | 19,991 | 95.19 | 84.22 | 364 | 7904 | 12,087 | 16,060 | 324 | 720 | 777 | 556 | 122 | |
| Case_9 | 20,822 | 94.60 | 81.88 | 477 | 7596 | 13,226 | 16,712 | 333 | 757 | 804 | 579 | 125 | |
| Case_10 | 20,095 | 94.79 | 86.69 | 329 | 7601 | 12,494 | 16,129 | 319 | 720 | 737 | 548 | 120 | |
| SNV | Case_1 | 120,593 | 99.62 | 97.73 | 165 | 52,586 | 68,007 | 82,056 | 2249 | 3689 | 4236 | 2808 | 475 |
| Case_2 | 120,805 | 99.60 | 97.59 | 198 | 52,520 | 68,285 | 82,454 | 2309 | 3729 | 4280 | 2900 | 431 | |
| Case_3 | 121,785 | 99.62 | 97.66 | 187 | 53,641 | 68,144 | 82,746 | 2308 | 3780 | 4315 | 2860 | 458 | |
| Case_4 | 123,072 | 99.62 | 97.87 | 160 | 52,059 | 71,013 | 84,168 | 2330 | 3805 | 4313 | 2902 | 444 | |
| Case_5 | 122,073 | 99.61 | 97.68 | 178 | 52,677 | 69,396 | 83,687 | 2325 | 3797 | 4238 | 2807 | 418 | |
| Case_6 | 121,331 | 99.62 | 97.57 | 201 | 52,564 | 68,767 | 83,004 | 2323 | 3768 | 4269 | 2797 | 439 | |
| Case_7 | 120,998 | 99.60 | 97.88 | 168 | 51,671 | 69,327 | 82,535 | 2310 | 3752 | 4183 | 2795 | 455 | |
| Case_8 | 120,987 | 99.60 | 97.71 | 182 | 52,956 | 68,031 | 82,752 | 2285 | 3647 | 4210 | 2890 | 461 | |
| Case_9 | 121,267 | 99.58 | 97.61 | 201 | 52,311 | 68,956 | 83,191 | 2265 | 3709 | 4257 | 2828 | 457 | |
| Case_10 | 121,166 | 99.35 | 97.48 | 309 | 52,751 | 68,415 | 82,685 | 2327 | 3720 | 4174 | 2790 | 450 |
dbSNP Database of Short Genetic Variation, gnomAD genome aggregation database, InDel small insertion and deletion, SNV single nucleotide variant
Variants with minor allele frequency < 1%
To filter out potential polymorphic or benign variants in the absence of paired normal samples, we used MAF < 1% as a cutoff value, which is recommended by the Interpretation of Sequence Variants in Somatic Conditions Working Group and also commonly used across many clinical laboratories [26]. In total, we identified 8667 variants with MAF < 1% or not recorded in gnomAD for East Asian. Ninety-three percent of them were SNVs, and 14.8% and 4.5% were recorded in Catalogue of Somatic Mutations in Cancer (COSMIC) and ClinVar databases, respectively. Those variants were listed in Additional file 3.
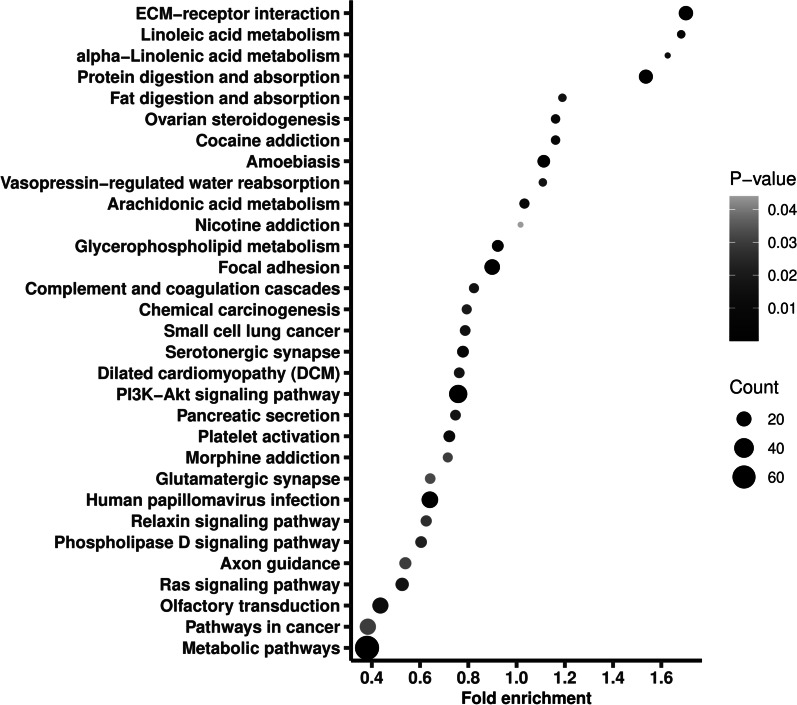
The 8667 variants located in 5665 genes, of which PABPC3 had the largest number of variants, that was 48. Of all the genes, 998 were recorded in COSMIC database, on which we thereafter conducted a pathway enrichment. As shown in Fig. 1, the genes were enriched into 31 pathways, including metabolic pathways, pathways in cancer, PI3K-AKT signaling pathway, and Ras signaling pathway. Notably, metabolic pathways had the largest gene ratio (0.067), and PI3K-AKT signaling pathway had the smallest adjusted P value (9.78 × 10−8).
Fig. 1.
Pathway enrichment of the genes where the identified variants with minor allele frequency < 1% located. Fold enrichment is calculated by dividing gene ratio (the number of genes enriched in the pathway divided by the total number of analyzed genes) with background ratio in KEGG database. P values were adjusted
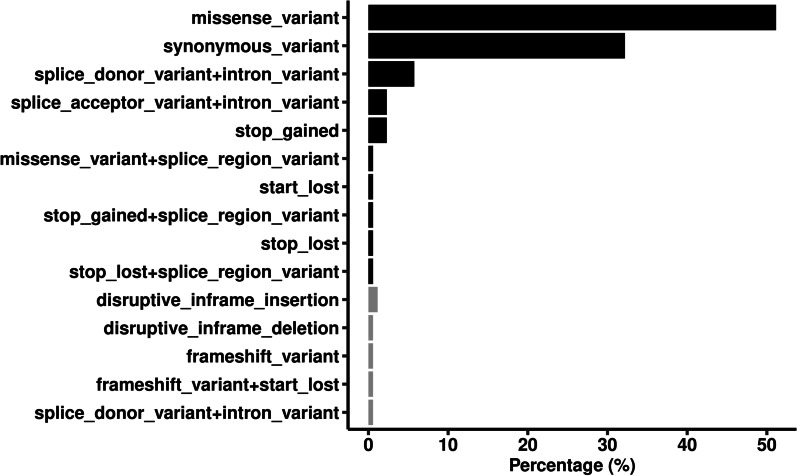
Of the 8667 variants, 174 were shared by all the cases, which were listed in Additional file 4. Ninety-six percent of the shared variants were SNVs, and 14.4% and 2.9% were recorded in COSMIC and ClinVar database, respectively. None of the variants was predicted to be deleterious based on combined annotation-dependent depletion (CADD) score ≥ 10. Those variants located in 123 genes, and their impact on gene function were illustrated in Fig. 2. Over half of the variants (51.1%) were missense mutations caused by SNVs, 1.1% were frameshift mutations caused by InDels, and 67.8% were nonsynonymous in total.
Fig. 2.
Classification of the functional impact of the commonly shared variants with minor allele frequency < 1%. Black bars represent single nucleotide variants, and gray bars represent small insertions and deletions
Newly identified variants
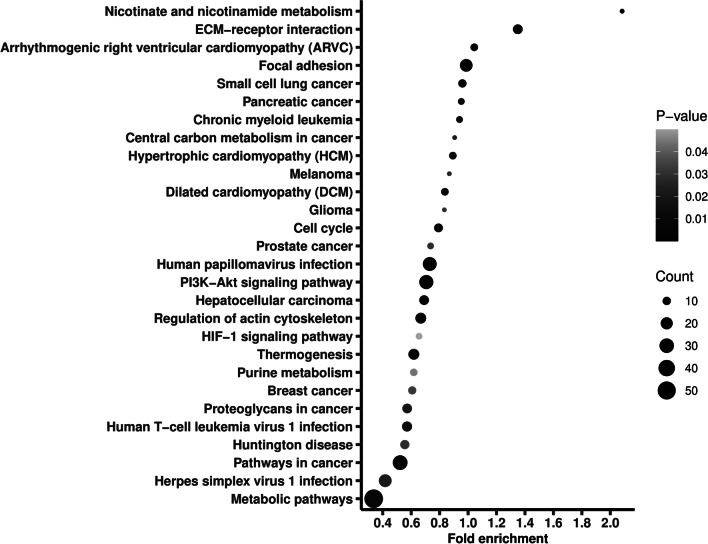
We further excluded variants recorded in gnomAD and in dbSNP, identifying 1010 novel variants (see Additional file 5). Ninety percent of the novel variants were SNVs, and only 2% were recorded in COSMIC database and none of them was found in ClinVar database. The novel variants located in 909 genes, on which we also conducted a pathway enrichment. As shown in Fig. 3, the genes were enriched into 28 pathways, including metabolic pathways, pathways in cancer, PI3K-AKT signaling pathway, and chronic myeloid leukemia.
Fig. 3.
Pathway enrichment of the genes where the newly identified variants located. Fold enrichment is calculated by dividing gene ratio (the number of genes enriched in the pathway divided by the total number of analyzed genes) with background ratio in KEGG database. P values were adjusted
None of the 1010 novel variants was shared by the 10 cases. However, one SNV located in SESN3 gene was found to be shared by 90% cases. It was recorded neither in COSMIC nor in ClinVar database. The SNV (C > G) located in SESN3 (chr11:95230844) was a missense mutation (resulting in Gly > Ala).
Discussion
Despite presenting different clinical types, BIL has been proved to be caused by and is diagnosed primarily on benzene exposure. Recurrent genetic variants may exist among different BIL patients. In order to find variants that may help understand carcinogenic mechanisms of BIL and finally make more effective prevention and treatment measures, we carried out the WES on peripheral blood samples of 10 BIL cases, and identified 48,802 variants in exons in total, 97.3% of which were SNVs. After filtering out variants with MAF ≥ 1%, we obtained 8667 potentially pathogenic variants, of which 174 were shared by all the BIL cases. We also identified 1010 novel variants that might be specifically related to BIL, and one of them was shared by 90% cases.
Because paired normal samples of the cases were not available, we could not accurately differentiate somatic variants from those of germline. By using MAF < 1% as the cutoff value, which is recommended by the Interpretation of Sequence Variants in Somatic Conditions Working Group and also commonly used across many clinical laboratories [26], we obtained variants that were very likely to be somatic and disease-related. However, the number of the variants was much larger than averagely 13 per sample in de novo AML or 8 per sample in newly diagnosed CML as previously reported by the Cancer Genome Atlas Research Network and Togasaki et al. [27, 28] Apart from naturally acquired and accumulated through the long-term course of BIL, some variants might be introduced by the therapies. Unfortunately, we could not exclude such variants in this study due to the fact that BIL is usually diagnosed as occupational cancer long after it is clinically diagnosed and treated in China.
The identified variants located in a wide series of genes, some of which have been reported before, such as TP53, TET2, SF3B1 and PTPN11 in AML [27, 29], and ASXL1, TET2, TET3, CENPF, TLE1, PRDM9, TTN, COL7A1 and DLK1 in CML [13, 14, 28]. Those mutated genes have been suggested to play important roles in carcinogenesis. The following pathway analysis showed that some of the mutated genes were significantly enriched into certain popular cancer-related pathways, such as PI3K-AKT signaling pathway and Ras signaling pathway. The PI3K-AKT-mTOR pathway was constantly found activated in a variety of cancers. The PI3K-AKT-mTOR axis consists of many regulators of oncogenic potentials, including the catalytic (p110α) and regulatory (p85α), of AKT, class IA PI3K, mTOR, RHEB, and eIF4E [30]; it may prompt oncogenic transformation by the mechanisms including stimulation of proliferation, metabolic reprogramming, invasion/metastasis, survival, and suppression of autophagy and senescence [30]. The Ras signaling pathway is one of the main pathways to transduce intracellular signals in response to mitogens to control cell growth, survival and anti-apoptotic programs. Three RAS genes encode four main protein products: KRAS4A, KRAS4B, NRAS and HRAS [31]. RAS proteins cycle between the GDP-bound inactive state (RAS-GDP) and the GTP-bound active state (RAS-GTP) [31]. The active RAS-GTP interacts with downstream effector enzymes including RAF, PI3K, and Ral guanine exchange factors (RalGEFs), transducing the signal to regulate biological behavior [32–35]. Ras pathway mutations occur in approximately 19% of all cancers, playing a prominent role in tumorigenesis and tumor progression [36].
We further examined recurrence of the variants and found that some variants were identified in each of the BIL cases, which gives P values no more than 8.8 × 10−14. The result suggested that the shared variants were significantly related to BIL. As previously inferred, some of the shared variants might be involved in the process of carcinogenesis, some might occur through the development of the disease, and others might be induced by the therapies.
By excluding variants recorded in the public databases, we obtained novel variants in BIL. Some of the novel variants might be specifically related to BIL, while others might be previously undiscovered, non-specific variants. Some of the novel variants were also located in genes that can be enriched into cancer-related pathways such as PI3K-AKT signaling pathway, suggesting that they might participate in the carcinogenic mechanisms of BIL.
When examining the recurrence of the novel variants, we found one variant shared by 90% cases. It locates in SESN3 gene. The SESN3 gene encoded protein (Sestrin 3) belongs to a small protein family that has been implicated in multiple biological processes including anti-oxidative stress, anti-aging, cell signaling, and metabolic homeostasis [37]. Sestrin 3 was suggested to play a critical tumor suppressor role through multiple mechanisms, including inhibition of the hedgehog signaling, controlling regeneration of peroxiredoxins to balance reactive oxygen species (ROS) upregulation induced by oncogenic Ras [37–39]. In BCR-ABL expressing cells, SESN3 mediated anti-leukemic responses through inhibition of mTOR signaling cascade [40]. Mutations in this gene might eventually result in oncogenesis.
Conclusions
To sum up, we examined variations of the whole exome in BIL patients for the first time. Although cases of different clinical types were not analyzed respectively due to limited sample size, we identified among all the cases some commonly shared variants that might be related to BIL. We also found novel variants in the exons of BIL cases, and one of them located in a cancer-related gene was shared by most of the cases, suggesting that it might be specifically related to BIL and has potential impact on biological structures or functions of the gene. Our study provided preliminary, genetic information for unraveling carcinogenic mechanisms of BIL.
Supplementary Information
Additional file 1. Statistics of the whole-exome sequencing data of the benzene-induced leukemia cases.
Additional file 2. Statistics of the sequences alignment.
Additional file 3. Whole-exome sequencing identified genetic variants with minor allele frequency < 1% in the gnomAD database for East Asian.
Additional file 4. Whole-exome sequencing identified genetic variants with minor allele frequency < 1% in the gnomAD database for East Asian and shared by all cases.
Additional file 5. Whole-exome sequencing identified genetic variants with no record in the gnomAD database for East Asian and dbSNP.
Acknowledgements
The authors would like to thank colleagues from Medical Laboratory, Shenzhen Prevention and Treatment Center for Occupational Diseases, who had been working so hard to help accomplish this work, and to acknowledge Beijing Genomics Institute (BGI) company for assisting us with the whole-exome sequencing.
Abbreviations
- ALL
Acute lymphocytic leukemia
- AML
Acute myeloid leukemia
- AP
Accelerated phase
- BC
Blast crisis
- BIL
Benzene-induced leukemia
- BP
Blast phase
- CADD
Combined annotation-dependent depletion
- CML
Chronic myeloid leukemia
- COSMIC
Catalogue of Somatic Mutations in Cancer
- CP
Chronic phase
- DNMT3A
DNA methyltransferase 3A
- GATK
The genome analysis toolkit
- IDH
Isocitrate dehydrogenase
- InDel
Small insertion and deletion
- KEGG
The Kyoto encyclopedia of genes and genomes database
- MAF
Minor allele frequency
- ROS
Reactive oxygen species
- SNV
Single nucleotide variant
- SPTCOD
Shenzhen Prevention and Treatment Center for Occupational Diseases
- TKI
Tyrosine kinase inhibitor
- WES
Whole-exome sequencing
Author contributions
DL and NZ conceived and designed the study. PL, DW, ZZ, YZ and LD collected the patients’ blood samples and conducted the whole-exome sequencing. DL, LD and MZ analyzed and interpreted the sequencing data. DL and NZ were major contributors in writing the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Scientific Research Cultivation Project of Shenzhen Prevention and Treatment Center for Occupational Diseases [Grant Number KPIII-202001], Science and Technology Planning Project of Shenzhen Municipality [Grant Numbers JCYJ20180306170306255, KCXFZ20201221173602007], and National Natural Science Foundation of China [Grant Number 81872666].
Availability of data and materials
The whole-exome sequencing datasets generated and analyzed during the current study are available in the National Center for Biotechnology Information SRA database, [https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA830611].
Declarations
Ethics approval and consent to participate
All procedures performed in the study involving human participants were approved by the Ethics Committee of Shenzhen Prevention and Treatment Center for Occupational Diseases (No. LL-202036) and in accordance with the 1975 Declaration of Helsinki and its later amendments or comparable ethical standards. Written informed consent for participating the study was obtained from all the patients or their guardians.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dafeng Lin, Email: david1385@foxmail.com.
Naixing Zhang, Email: zhangnx@wjw.sz.gov.cn.
References
- 1.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans . Benzene. Lyon: International Agency for Research on Cancer; 2018. [Google Scholar]
- 2.Ministry of Health of the People’s Republic of China. Diagnosis of occupational tumor. National Standards for Occupational Health; 2017.
- 3.Aksoy M. Hematotoxicity and carcinogenicity of benzene. Environ Health Perspect. 1989;82:193–197. doi: 10.1289/ehp.8982193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayes RB, Yin SN, Dosemeci M, Li GL, Wacholder S, Travis LB, Li CY, Rothman N, Hoover RN, Linet MS. Benzene and the dose-related incidence of hematologic neoplasms in China. Chinese Academy of Preventive Medicine-National Cancer Institute Benzene Study Group. J Natl Cancer Inst. 1997;89:1065–1071. doi: 10.1093/jnci/89.14.1065. [DOI] [PubMed] [Google Scholar]
- 5.Wan WG, Zhou HJ. Cases of benzene-related leukemia reported in periodicals in China and analysis of diagnosis. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi. 2010;28:844–847. [PubMed] [Google Scholar]
- 6.Guo JY, Lin QH, Zou WY. Diagnosis analysis on 31 occupational leukemia. Chin J Ind Med. 2018;31:428–431. [Google Scholar]
- 7.Minciacchi VR, Kumar R, Krause DS. Chronic myeloid leukemia: a model disease of the past, present and future. Cells. 2021;10:117. doi: 10.3390/cells10010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vetrie D, Helgason GV, Copland M. The leukaemia stem cell: similarities, differences and clinical prospects in CML and AML. Nat Rev Cancer. 2020;20:158–173. doi: 10.1038/s41568-019-0230-9. [DOI] [PubMed] [Google Scholar]
- 9.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, Gundem G, Van Loo P, Martincorena I, Ganly P, Mudie L, McLaren S, O'Meara S, Raine K, Jones DR, Teague JW, Butler AP, Greaves MF, Ganser A, Döhner K, Schlenk RF, Döhner H, Campbell PJ. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 11.Martínez-Jiménez F, Muiños F, Sentís I, Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, Mularoni L, Pich O, Bonet J, Kranas H, Gonzalez-Perez A, Lopez-Bigas N. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20:555–572. doi: 10.1038/s41568-020-0290-x. [DOI] [PubMed] [Google Scholar]
- 12.Bullinger L, Döhner K, Döhner H. Genomics of acute myeloid leukemia diagnosis and pathways. J Clin Oncol. 2017;35(9):934–946. doi: 10.1200/JCO.2016.71.2208. [DOI] [PubMed] [Google Scholar]
- 13.Soverini S, de Benedittis C, Mancini M, Martinelli G. Mutations in the BCR-ABL1 kinase domain and elsewhere in chronic myeloid leukemia. Clin Lymphoma Myeloma Leuk. 2015;15(Suppl):S120–S128. doi: 10.1016/j.clml.2015.02.035. [DOI] [PubMed] [Google Scholar]
- 14.Branford S, Kim DDH, Apperley JF, Eide CA, Mustjoki S, Ong ST, Nteliopoulos G, Ernst T, Chuah C, Gambacorti-Passerini C, Mauro MJ, Druker BJ, Kim DW, Mahon FX, Cortes J, Radich JP, Hochhaus A, Hughes TP, International CML Foundation Genomics Alliance Laying the foundation for genomically-based risk assessment in chronic myeloid leukemia. Leukemia. 2019;33(8):1835–1850. doi: 10.1038/s41375-019-0512-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, Lin Z, Tang C, Tang Y, Cai Y, Zhong H, Wang X, Zhang W, Xu C, Wang J, Wang J, Yang H, Yang L, Gao Q. A new massively parallel nanoball sequencing platform for whole exome research. BMC Bioinform. 2019;20(1):153. doi: 10.1186/s12859-019-2751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J, Liang X, Xuan Y, Geng C, Li Y, Lu H, Qu S, Mei X, Chen H, Yu T, Sun N, Rao J, Wang J, Zhang W, Chen Y, Liao S, Jiang H, Liu X, Yang Z, Mu F, Gao S. A reference human genome dataset of the BGISEQ-500 sequencer. Gigascience. 2017;6(5):1–9. doi: 10.1093/gigascience/gix024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Picard Tools. http://broadinstitute.github.io/picard/. Accessed 6 Dec 2021.
- 22.Cingolani P, Platts A, le Wang L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, Lu X. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet. 2012;3:35. doi: 10.3389/fgene.2012.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2021. URL https://www.R-project.org/.
- 25.Bu D, Luo H, Huo P, Wang Z, Zhang S, He Z, Wu Y, Zhao L, Liu J, Guo J, Fang S, Cao W, Yi L, Zhao Y, Kong L. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021;49(W1):W317–W325. doi: 10.1093/nar/gkab447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, Tsimberidou AM, Vnencak-Jones CL, Wolff DJ, Younes A, Nikiforova MN. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the association for molecular pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19(1):4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research Network, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley K, Triche TJ Jr, Laird PW, Baty JD, Fulton LL, Fulton R, Heath SE, Kalicki-Veizer J, Kandoth C, Klco JM, Koboldt DC, Kanchi KL, Kulkarni S, Lamprecht TL, Larson DE, Lin L, Lu C, McLellan MD, McMichael JF, Payton J, Schmidt H, Spencer DH, Tomasson MH, Wallis JW, Wartman LD, Watson MA, Welch J, Wendl MC, Ally A, Balasundaram M, Birol I, Butterfield Y, Chiu R, Chu A, Chuah E, Chun HJ, Corbett R, Dhalla N, Guin R, He A, Hirst C, Hirst M, Holt RA, Jones S, Karsan A, Lee D, Li HI, Marra MA, Mayo M, Moore RA, Mungall K, Parker J, Pleasance E, Plettner P, Schein J, Stoll D, Swanson L, Tam A, Thiessen N, Varhol R, Wye N, Zhao Y, Gabriel S, Getz G, Sougnez C, Zou L, Leiserson MD, Vandin F, Wu HT, Applebaum F, Baylin SB, Akbani R, Broom BM, Chen K, Motter TC, Nguyen K, Weinstein JN, Zhang N, Ferguson ML, Adams C, Black A, Bowen J, Gastier-Foster J, Grossman T, Lichtenberg T, Wise L, Davidsen T, Demchok JA, Shaw KR, Sheth M, Sofia HJ, Yang L, Downing JR, Eley G. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–74. 10.1056/NEJMoa1301689. Epub 2013 May 1. Erratum in: N Engl J Med. 2013;369(1):98.
- 28.Togasaki E, Takeda J, Yoshida K, Shiozawa Y, Takeuchi M, Oshima M, Saraya A, Iwama A, Yokote K, Sakaida E, Hirase C, Takeshita A, Imai K, Okumura H, Morishita Y, Usui N, Takahashi N, Fujisawa S, Shiraishi Y, Chiba K, Tanaka H, Kiyoi H, Ohnishi K, Ohtake S, Asou N, Kobayashi Y, Miyazaki Y, Miyano S, Ogawa S, Matsumura I, Nakaseko C, Naoe T. Frequent somatic mutations in epigenetic regulators in newly diagnosed chronic myeloid leukemia. Blood Cancer J. 2017;7(4):e559. doi: 10.1038/bcj.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kishtagari A, Levine RL, Viny AD. Driver mutations in acute myeloid leukemia. Curr Opin Hematol. 2020;27(2):49–57. doi: 10.1097/MOH.0000000000000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aoki M, Fujishita T. Oncogenic roles of the PI3K/AKT/mTOR axis. Curr Top Microbiol Immunol. 2017;407:153–189. doi: 10.1007/82_2017_6. [DOI] [PubMed] [Google Scholar]
- 31.Chen K, Zhang Y, Qian L, Wang P. Emerging strategies to target RAS signaling in human cancer therapy. J Hematol Oncol. 2021;14(1):116. doi: 10.1186/s13045-021-01127-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roskoski R., Jr RAF protein-serine/threonine kinases: structure and regulation. Biochem Biophys Res Commun. 2010;399(3):313–317. doi: 10.1016/j.bbrc.2010.07.092. [DOI] [PubMed] [Google Scholar]
- 33.Krygowska AA, Castellano E. PI3K: a crucial piece in the RAS signaling puzzle. Cold Spring Harb Perspect Med. 2018;8(6):a031450. doi: 10.1101/cshperspect.a031450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hofer F, Fields S, Schneider C, Martin GS. Activated Ras interacts with the Ral guanine nucleotide dissociation stimulator. Proc Natl Acad Sci USA. 1994;91(23):11089–11093. doi: 10.1073/pnas.91.23.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pacold ME, Suire S, Perisic O, Lara-Gonzalez S, Davis CT, Walker EH, Hawkins PT, Stephens L, Eccleston JF, Williams RL. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103(6):931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 36.Prior IA, Hood FE, Hartley JL. The frequency of Ras mutations in cancer. Cancer Res. 2020;80(14):2969–2974. doi: 10.1158/0008-5472.CAN-19-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Kim HG, Dong E, Dong C, Huang M, Liu Y, Liangpunsakul S, Dong XC. Sesn3 deficiency promotes carcinogen-induced hepatocellular carcinoma via regulation of the hedgehog pathway. Biochim Biophys Acta Mol Basis Dis. 2019;1865(10):2685–2693. doi: 10.1016/j.bbadis.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zamkova M, Khromova N, Kopnin BP, Kopnin P. Ras-induced ROS upregulation affecting cell proliferation is connected with cell type-specific alterations of HSF1/SESN3/p21Cip1/WAF1 pathways. Cell Cycle. 2013;12(5):826–836. doi: 10.4161/cc.23723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kopnin PB, Agapova LS, Kopnin BP, Chumakov PM. Repression of sestrin family genes contributes to oncogenic Ras-induced reactive oxygen species up-regulation and genetic instability. Cancer Res. 2007;67(10):4671–4678. doi: 10.1158/0008-5472.CAN-06-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vakana E, Arslan AD, Szilard A, Altman JK, Platanias LC. Regulatory effects of sestrin 3 (SESN3) in BCR-ABL expressing cells. PLoS ONE. 2013;8(11):e78780. doi: 10.1371/journal.pone.0078780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Statistics of the whole-exome sequencing data of the benzene-induced leukemia cases.
Additional file 2. Statistics of the sequences alignment.
Additional file 3. Whole-exome sequencing identified genetic variants with minor allele frequency < 1% in the gnomAD database for East Asian.
Additional file 4. Whole-exome sequencing identified genetic variants with minor allele frequency < 1% in the gnomAD database for East Asian and shared by all cases.
Additional file 5. Whole-exome sequencing identified genetic variants with no record in the gnomAD database for East Asian and dbSNP.
Data Availability Statement
The whole-exome sequencing datasets generated and analyzed during the current study are available in the National Center for Biotechnology Information SRA database, [https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA830611].