Abstract
At the core of the CRISPR-Cas9 genome-editing technology, the endonuclease Cas9 introduces site-specific breaks in DNA. However, precise mechanistic information to ameliorating Cas9 function is still missing. Here, multi-microsecond molecular dynamics, free-energy and multiscale simulations are combined with solution NMR and DNA cleavage experiments to resolve the catalytic mechanism of target DNA cleavage. We show that the conformation of an active HNH nuclease is tightly dependent on the catalytic Mg2+, unveiling its cardinal structural role. This activated Mg2+-bound HNH is consistently described through molecular simulations, solution NMR and DNA cleavage assays, revealing also that the protonation state of the catalytic H840 is strongly affected by active site mutations. Finally, ab-initio QM(DFT)/MM simulations and metadynamics establish the catalytic mechanism, showing that the catalysis is activated by H840 and completed by K866, rationalising DNA cleavage experiments. This information is critical to enhance the enzymatic function of CRISPR-Cas9 toward improved genome-editing.
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas9 is a genome-editing tool that has revolutionized basic and applied sciences.1 At the core of this technology, the endonuclease Cas9 associates with a guide RNA structure to recognise and cleave matching sequences of DNA.2 DNA targeting starts with the recognition of a short Protospacer Adjacent Motif, initiating double strand separation, whereby one strand (the target strand) base-pairs with the guide RNA, forming an RNA:DNA heteroduplex, while the other strand non-target is displaced. Then, two catalytic domains, HNH and RuvC, catalyse cleavage of the target and non-target strands of DNA, respectively (Fig. 1A). In this complex biophysical process, the nuclease function of Cas9 is an essential step, yet its characterisation has remained incomplete.
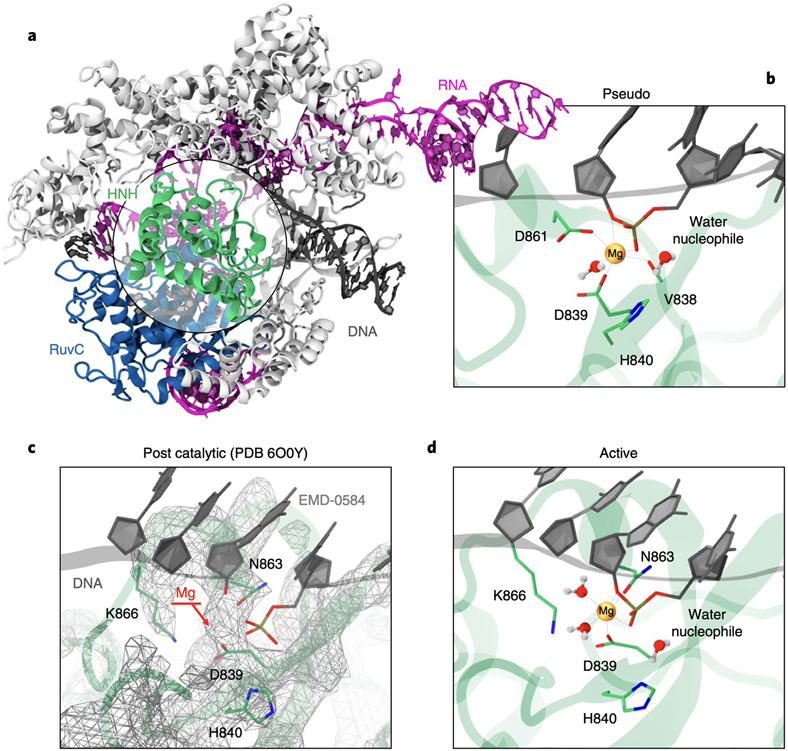
Fig. 1. Overview of the Streptococcus pyogenes CRISPR-Cas9 system.
(A) X-ray structure of the CRISPR-Cas9 system (PDB: 5F9R).12 The Cas9 protein is shown as ribbons, highlighting its catalytic domains HNH (green) and RuvC (blue), in complex with RNA (magenta) and DNA (black). (B) Close-up view on the HNH catalytic site, displaying the D839 and D831 residues coordinating Mg2+ and forming a catalytic triad with H840. This configuration of the catalytic core – referred as pseudo-active – arises from structures capturing HNH in the absence of Mg2+ (e.g., PDB: 5F9R) and from the homology with the T4 endonuclease VII9 (Supplementary Fig. 1). (C) Catalytic core from the cryo-EM structure EMD-0584 (PDB: 6O0Y)14 capturing HNH in the presence of Mg2+ after target strand cleavage, and (D) model of the catalytic site prior DNA cleavage. In this configuration – referred as active – N863 coordinates Mg2+ in place of D861. The atomic coordinates of HNH are shown as cartoon (green), while the electronic density is shown as wireframes (grey). The EMD-0584 map displays a visible density in the position of Mg2+ (C, indicated using an arrow), in agreement with the EMD-24838 map15 (Supplementary Fig. 2), enabling to locate the Mg2+ ion in the catalytic state (D).
Biochemical and computational studies revealed that RuvC cleaves the DNA non-target strand through a two-metal (Mg2+)-dependent catalytic mechanism.3-5 Conversely, the catalysis of target strand cleavage by the HNH nuclease is not understood. Biochemical studies indicated that a single Mg2+ ion catalyses phosphodiester bond cleavage.3 However, HNH displays a complex conformational landscape, which regulates the process of DNA binding and culminates with the docking of HNH at the cleavage site on the target strand.6-8 While this is a precondition for target strand cleavage, two distinct states were reported displaying HNH in close proximity to the cleavage site. Homology with the T4 endonuclease VII (endo VII)9 suggested that D861 and D839 coordinate the catalytic Mg2+ and form a triad with the catalytic H840 (Fig. 1B).6,8,10 This conformation was supported by structures capturing HNH in various non-catalytic states and in the absence of Mg2+ ions (Supplementary Fig. 1).3,4,11,12 A cryo-EM structure of a catalytically dead Cas9 also supported this conformation of HNH.13 This structure (PDB: 5Y36), solved at 5.2 Å resolution, was obtained including Mg2+ ions in the experimental buffer, but the location of Mg2+ was not determined experimentally. The HNH site was reconstructed based on X-ray structures obtained in the absence of Mg2+,4,11 and on the structure of endo VII,9 ultimately suggesting that D839 and D861 coordinate Mg2+.
Recently, the structural determination of the HNH catalytic core, captured right after target strand cleavage and in the presence of Mg2+, displayed a different configuration of the catalytic site (Fig. 1C).14,15 A first cryo-EM structure (PDB: 6O0Y) displayed a visible density in the position of Mg2+ (Fig. 1C), enabling to locate the catalytic ion and to reconstruct the catalytic state. Here, N863 (rather than D861) coordinates Mg2+, forming a catalytic triad with D839 and H840, while D861 points outward. This configuration was confirmed by other recent cryo-EM structures (e.g., PDB: 7S4X), revealing the atomic positions of the catalytic site including the Mg2+ ion (Supplementary Fig. 2).15,16 Biochemical experiments also showed that the D861A substitution retains DNA cleavage activity, while N863A loses gene-editing capability.4,17 These findings suggested that the previously reported state could be an alternative pseudo-active state, advocating also a possible conformational equilibrium between the two states.17 In this scenario, studies of the catalytic mechanism inferred information from endo VII,18 and were based on a cryo-EM structure of the pseudo-active state,19 sustaining the coordination of Mg2+ by both D861 and D839.
Considering this knowledge, fundamental questions remain unmet. First, the biological relevance of the pseudo-active state in the presence of the Mg2+ ions is ambiguous. Indeed, structural evidence supporting the pseudo-active site captured HNH far from the cleavage site and/or without Mg2+.3,4,11-13 Moreover, the possible equilibrium between the two states in the presence of the catalytic Mg2+ has not been investigated. The molecular details of this equilibrium are central to complete understanding of the complex conformational landscape of HNH, and to inform the conformational requirements underlying DNA cleavage. Finally, and perhaps more intriguingly, considering the newly reported structural data.14-16 the catalytic mechanism of target strand cleavage and how it proceeds through the aid of a single Mg2+ ion has not been addressed. This knowledge is important to improve the efficiency of the Cas9 enzyme and can help in overcoming unselective DNA cleavages.
Here, multi-μs molecular dynamics (MD) simulations and free energy methods were combined with extensive ab-initio MD and quantum mechanics/molecular mechanics (QM/MM) approaches to establish the molecular details of the equilibrium between active and pseudo-active states and resolve the mechanism of DNA cleavage. Supported by solution NMR and biochemical assays, our findings reveal a critical structural role for Mg2+, determining the chemical mechanism and which of the known conformation of HNH is responsible for target DNA cleavage in Cas9.
Results
Molecular preference of the competent HNH
To determine the conformation of the catalytic HNH domain in the presence of Mg2+, and to characterise the relevance of the pseudo-active state, we performed free energy simulations. We investigated the pseudo-active–to–active transition (and vice versa) using Umbrella Sampling simulations,20 rigorously sampling the populations of the two states and of their intermediates through ~28 μs of MD runs. The transition was sampled along the difference in Root Mean Square Deviation (RMSD) of the heavy atoms’ positions of HNH with respect to both states (used as Reaction Coordinate, RC), in the presence and absence of Mg2+. Our classical model described the metal sites in line with QM/MM simulations, posing the basis for classical and free energy simulations (Supplementary Figs. 3-6).
The simulations show that without Mg2+ (w/o Mg2+), the free energy reaches a minimum at RC ~−2.3 Å (Fig. 2A), corresponding to the pseudo-active conformation. Contrarywise, with Mg2+ (w Mg2+) we detect an opposite trend in the free energy profile, displaying a well-defined minimum at RC ~2.2 Å, consistent with the active state. Two close-up views show the conformation of the pseudo-active and active states at their energetic minima (Fig. 2A). Hence, the conformational state of HNH critically depends on the presence of Mg2+, which favours the formation of the active conformation, while in the absence of Mg2+, HNH mainly populates the pseudo-active state.
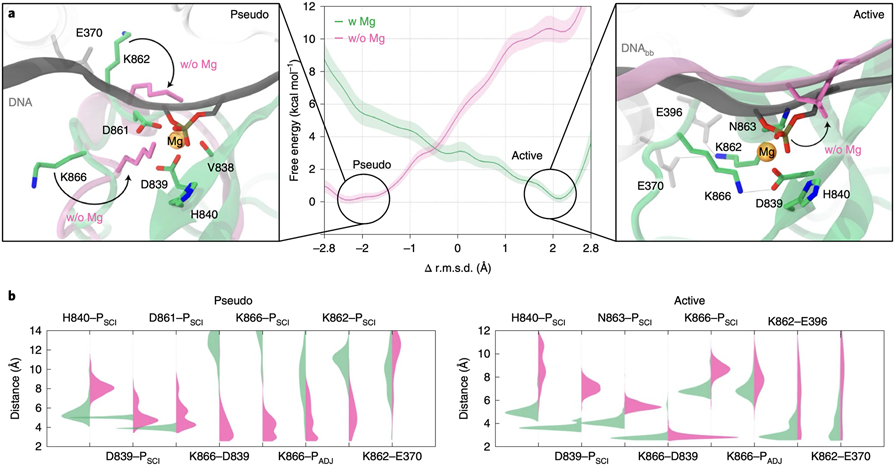
Fig. 2. Transition of the HNH domain from pseudo-active to active states.
(A) Free energy profiles for the HNH conformational transition in the presence of Mg2+ (w Mg, green) and without Mg2+ ions (w/o Mg, magenta). Two close-up views show the conformation of the pseudo-active (i.e., pseudo) and active states at their energetic minima. The HNH catalytic site bound to Mg2+ is shown as in Fig. 1C. Residues in green refer to HNH bound to Mg2+, while residues in magenta show the conformational change occurring in the absence of Mg2+ (also indicated using an arrow). (B) Probability distributions of critical interaction distances in the presence of Mg2+ (green) and absence of Mg2+ (magenta) for the pseudo-active and active states at their energetic minima (i.e., at −2.5 Å ≤ RC ≤ −1.5 Å and 1.5 Å ≤ RC ≤ 2.5 Å, respectively). The Welch’s t-test was used to assess the statistical significance of the differences in the distributions of the interaction distances with and without Mg2+. At the confidence level of 95 %, we rejected the null hypothesis in favour of the alternative with the p value < 0.0001. This observation was true for all cases but for the K866–D839 distance in the active system, where p = 0.437, as arising from overlapping distributions.
To understand this observation, we analysed the conformational ensembles at the free energy minima. The pseudo-active state at its energetic minimum (at −2.5 Å ≤ RC ≤ −1.5 Å) revealed that, in the presence of Mg2+, the K862 and K866 sidechains orient away from the DNA (Fig. 2A, left). Instead, w/o Mg2+, K866 binds the scissile phosphate (PSCI) and its adjacent phosphate (PADJ), stabilised also by the interaction with D839. This is evidenced by the probability distributions of the distances involving K866, K862 and the DNA phosphate groups assuming values < 6 Å (Fig. 2B). This is consistent with the cryo-EM structure of the pseudo-active state, where the K866 side-chain locates in proximity to the DNA.13 Hence, K866 and K862 stabilise the pseudo-active state in the absence of Mg+.
The active state at its energetic minimum (at 1.5 Å ≤ RC ≤ 2.5 Å) showed that, in the presence of Mg2+, H840, D839 and N863 stably maintain their position with respect to PSCI, while D839 binds K866 (Fig. 2). In the absence of Mg2+, the catalytic residues move away from the DNA, destabilising the latter with respect to the catalytic core. With Mg2+, K862 interacts with E396 and E370, while these interactions are lost w/o Mg2+. The active state is thereby stabilised by Mg2+, whose loss leads to unproductive conformations for catalysis.
These findings clarify why structural studies not including Mg2+ supported a pseudo-active configuration.3,4,11-13 Indeed, the absence of Mg2+ leads to interactions that anchor HNH at the DNA, stabilising the pseudo-active conformation. Inversely, with Mg2+, HNH shifts its preference toward an active state, consistent with cryo-EM findings obtained in the presence of Mg2+.14-16
Chemical environment enabling catalysis
To examine the chemical environment enabling catalysis and to determine the protonation state of the catalytic H840, we performed solution NMR, DNA cleavage experiments and extensive MD simulations (reaching ~54 μs of collective sampling).
Changes in the protonation state of the catalytic histidine side chain were monitored through solution NMR using a construct of the HNH nuclease that was shown to properly represent this domain in full-length CRISPR-Cas9, in the presence of Mg2+ and DNA (see Methods, Supplementary Fig. 7).21,22 Two-dimensional 1H-13C correlation spectra depict strong chemical shift perturbations in the catalytic H840 as the pH of the sample is modulated (Fig. 3A, Supplementary Figs. 8-9). We also titrated H799, which does not significantly change upon alanine mutations and is an internal standard. The chemical shift (δH1) range indicates that H799 and H840 are both partially protonated at pH 7.4. The measured pKa was ~7.27 and ~6.83 for H799 and H840, respectively, indicating that, at pH 7.4, H799 is 79% protonated and H840 is 57% protonated (Fig. 3B). To further assess the pKa of H799 and H840 in the full-length CRISPR-Cas9, we performed Constant pH (CpH) MD simulations in explicit solvent, in conjunction with Replica Exchange.23 The computed pKa for H799 and H840 in the active state of HNH resulted ~7.52 and ~6.90, respectively (Fig. 3B), in very good agreement with NMR data. Though the determination of the tautomeric populations of H840 by NMR were complicated by fast relaxation of the histidine sidechain and possible proton exchange with solvent,24,25 in-depth analysis of the simulations revealed that at pH 7.4, H840 is likely to assume the neutral tautomeric form protonated on the ε position (H840-ε >60 % of the simulation, Supplementary Fig. 10), resulting in the δ nitrogen in proximity to the nucleophile for activation and at ~5 Å from PSCI (Fig. 3C). CpH MD simulations of the pseudo-active state revealed a shift of the pKa toward higher values, reducing the fraction of H840-ε at ~50 % at pH 7.4. Classical MD of the two tautomeric forms of H840 also showed that H840-ε leads to a stable catalytic site (Supplementary Fig. 11). Conversely, the tautomeric form of H840 protonated on δ results in the detachment of the activating nitrogen from PSCI at > 6 Å, unlikely for nucleophile activation. Hence, NMR and MD suggest that H840-ε is prone for catalysis, with critical information to study the catalytic mechanism (vide infra).
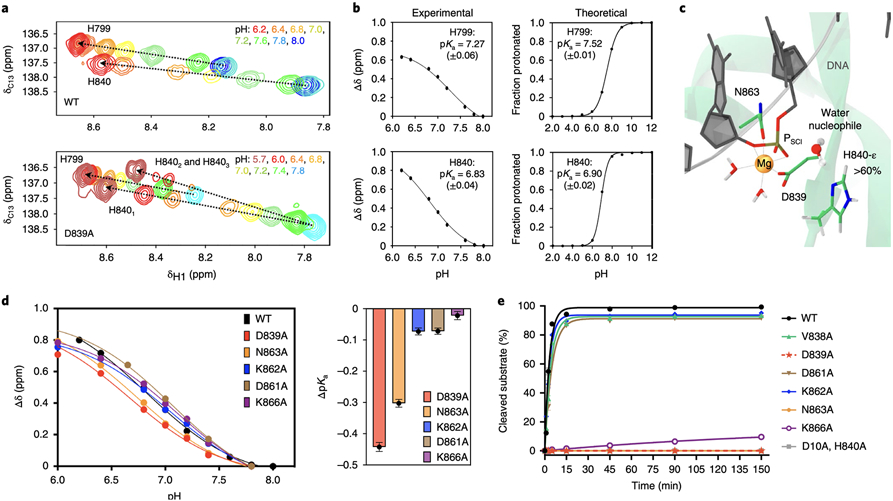
Fig. 3. Chemical environment enabling the catalysis.
(A) Titration of the H799 and H840 side chains in the wild-type HNH (WT, top) and in the D893A mutant (bottom), reporting changes in the 1H-13Cε1 correlation as the histidine nitrogen atoms change protonation state, in the presence of Mg2+ and DNA. Resonances are coloured according to the pH values in the legend. Chemical shifts ~8.5 ppm correspond to fully protonated histidine, while those ~8.2–7.9 ppm refer to partially protonated or deprotonated residues. The trajectories of the chemical shifts are indicated using dashed arrows. In the D893A mutant, H840 shows evidence of two-three conformational states (upper arrow, H8401-3). (B) Fitted titration curves for pKa determination of H799 and H840 in the WT HNH through solution NMR (experimental) and constant pH molecular dynamics (theoretical). A modified version of the Henderson-Hasselbach equation was used for fitting the experimental data. Computational data were obtained by fitting the deprotonated fraction to Equation 3. Each data point represents the ensemble population of the protonated/deprotonated states from constant pH MD simulation (last ~32 ns) for each pH value. pKa values are reported, alongside the error from the fit (see Supplementary Methods). (C) Active state of HNH displaying the tautomeric form of H840 protonated on ε (H840-ε), which occurs for >60 % of CpH MD at pH 7.4 (Supplementary Fig. 11). (D) Titration curves for the catalytic H840 in the WT HNH and its mutants and change in pKa upon mutation (ΔpKa). In plots of the former, error associated with variance in NMR chemical shifts is smaller than the data points depicting Δδ. Values of ΔpKa are depicted as dots with error bars propagated as E(ΔpKa) = E(mutant) – E(wt), where E is the error determined from fits of the individual titration curves in Prism 9.0, centred at the calculated ΔpKa value. Coloured bars are shown as a visual aide to assess the magnitude of ΔpKa. (E) In vitro cleavage kinetics of Cas9 HNH mutants on a double stranded DNA on-target substrate. Line represents a single exponential fit of each individual time course experiment, each data point represents an average of four independent experiments (n = 4) with standard deviation plotted for each data point.
NMR experiments were also performed introducing alanine mutations of D839, D861, N863, K862 and K866. Each of these HNH mutants showed a shift to a lower pKa value (Fig. 3D, Supplementary Fig. 9). D839A most significantly reduced the pKa of the catalytic histidine, suggesting that in this mutant, H840 is a weaker base. This mutant hampers cleavage (Fig. 3E, Supplementary Fig. 12)17 and exhibits the most significant structural perturbations, with altered flexibility throughout the protein and near the catalytic site (Supplementary Fig. 8).
In this mutant, H840 populates at least two (or three) states during pH titrations, which manifests as a series of bifurcated NMR resonances in Fig. 3A. This is consistent with classical MD of the D839A mutant in the active state, where H840 adopts three different conformations and detaches from PSCI (Fig. 4A). Hence, the loss of activity in the D839A Cas9 may be due to the ability of this residue to strongly influence the conformation of the adjacent H840. Titration of the N863A mutant also revealed a reduction of the pKa of H840 (Fig. 3D). This is in line with classical MD of the active state, where N863A results in the detachment of the S860–D868 helix from Mg2+ (Fig. 4B), inducing instability of the site and affecting the location of H840 with respect to PSCI. This is consistent with NMR data showing structural and dynamic changes at the active site and proximal to the S860-D868 helix (Supplementary Figs. 8, 13-14). D861A resulted in a reduced effect on the change in pKa of H840 (Fig. 3D), supporting that D861 resides outside the catalytic site in the active state, at odds with the pseudo-active conformation where it coordinates Mg2+ (Fig. 1C). Finally, K862 and K866 do not significantly affect the pKa of H840, likely due to their distal location with respect to the catalytic residue. K866A, however, reduces the enzymatic activity (Fig. 3E). In this respect, classical MD of K866A in the active HNH reveal that this mutation induces instability of the S860-D868 helix (Fig. 4C) and increases its flexibility with respect to the wild-type (Supplementary Fig. 15), resulting in impeding the K866–D839 interaction that is crucial for the stabilisation of the active state (Fig. 2). Remarkably, mutations that retain catalysis (V838A, D861A and K862A) preserve the stability of the catalytic site in the active HNH, with H840 remaining in the vicinity of PSCI (Supplementary Fig. 16).
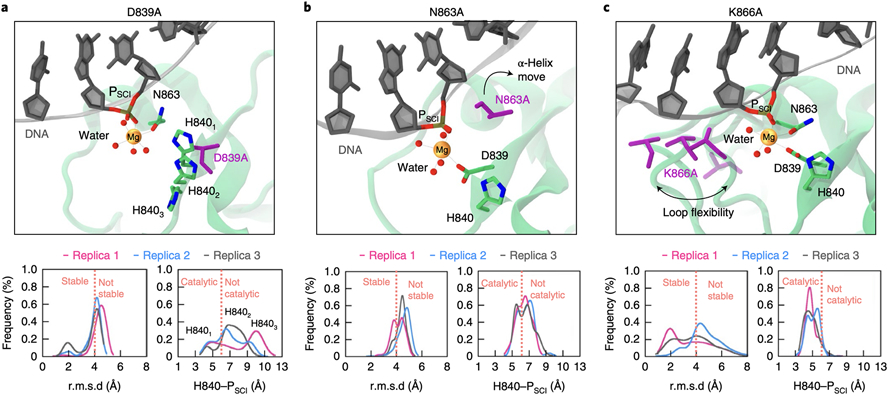
Fig. 4. Effect of alanine mutations on the catalytic site.
Data are shown for the D839A, N863A and K866A mutants in the active state of HNH. Alanine mutations are shown in violet. (A) D839A affects the conformation of H840, resulting in three main conformations (H8401-3). (B) N863A results in the detachment of the S860–K866 α-helix from the catalytic Mg2+, destabilising the catalytic core. (C) K866A destabilises the catalytic core, with flexibility of the S860–D868 loop (four configurations are shown). Bottom graphs: stability of the catalytic site, computed as probability distribution of the heavy atoms’ Root Mean Square Deviation (RMSD) within 8 Å of the catalytic Mg2+; and location of the catalytic H840 with respect to the scissile phosphate (PSCI), computed considering the interatomic distance between H840 (Nδ) and PSCI. Data are reported for three simulation replicas of ~1 μs each. Vertical dashed lines (orange) indicate the cutoffs for the stability of the catalytic site (i.e., RMSD < 4 Å) and for the catalytic function of H840 (i.e., H840–PSCI < 6 Å allows the water nucleophile to position between H840 and PSCI, Fig. 3C, while H840–PSCI > 6 Å results in the detachment of H840 from the catalytic centre).
Overall, the dynamics of the active state agrees with NMR and DNA cleavage experiments. Molecular simulations of the pseudo-active state do not provide a rationale for the experimental evidence. Indeed, only V838A preserves the HNH site, maintaining its backbone interactions, while all other mutants result in the detachment of H840 from PSCI (Supplementary Figs. 17-18).
Catalytic Mechanism of Target Strand Cleavage
To further resolve which of the known HNH conformations is catalytic, and to characterise the cleavage mechanism, we employed ab-initio MD and free energy methods.26 We performed Thermodynamic Integration27 with QM/MM simulations28 of HNH in both the active and pseudo-active states at the DFT-BLYP level of theory.29,30 Phosphodiester bond cleavage was studied along the difference in the distance between the breaking and forming P–O bonds (used as RC, Fig. 5). This approach was used on several nucleases,31-34 including the Cas9 RuvC domain.5 The reactant states were carefully selected from classical MD (Supplementary Fig. 19), and considered the tautomeric form of H840 protonated on ε, as from NMR and computational studies (vide supra).
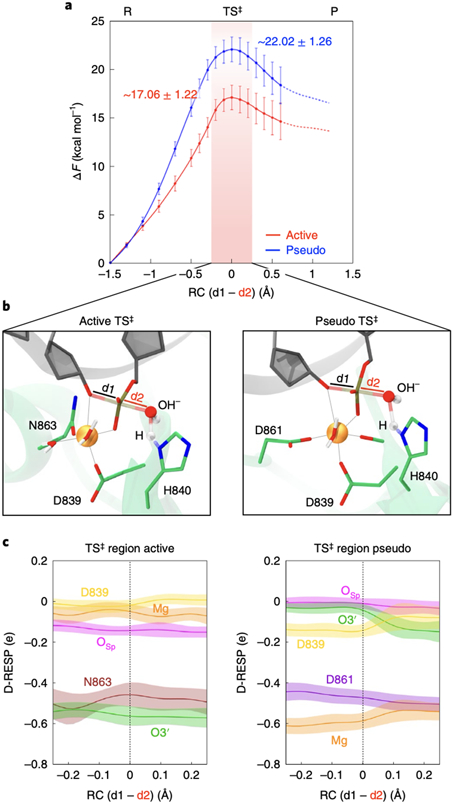
Fig. 5. Free energy profiles for phosphodiester bond cleavage.
(A) Free energy profiles (ΔF, in kcal/mol) for the active (red) and pseudo-active (blue) states of HNH, obtained through QM(BLYP)/MM MD and Thermodynamic Integration. The difference in distance between the breaking and forming P–O bonds is the reaction coordinate (RC = d1–d2, shown in panel B). The chemical step evolves from the Reactants (R) to Products (P) passing through a Transition State (TS‡, region indicated using a red vertical bar). Error bars show standard deviations obtained from error propagation analysis of the primary data set in which each data point represents the mean from the last ~5 ps of converged ab-initio MD. (B) Close-up view of the TS‡ structure in the active (left) and pseudo-active (right) states of HNH. (C) Dynamical RESP (D-RESP)28 charges within the TS‡ region (i.e., −0.2 Å < RC < 0.2 Å) for the active and pseudo-active states. At each value of the RC, data are presented as mean (solid line) and standard deviation (shaded bands), computed over the last ~5 ps of converged ab-initio MD.
For each system, we collected >200 ps of ab-initio MD, obtaining the free energy profiles for the active and pseudo-active states of HNH (Fig. 5A). The chemical step proceeds from the reactants (R) to the products (P), separated by a transition state (TS‡) maximum. The reaction is activated by H840, acting as a general base, and proceeds through an SN2-like mechanism. The activation free energy for the chemical step in the active HNH was 17.06 ± 1.22 kcal/mol (details, cross-validation and error analysis in Supplementary Figs. 20-21). This is consistent with the catalytic rate of 4.3 s−1 (corresponding to ΔG‡ ~16/17 kcal/mol) measured for the HNH catalysis, distinct from the RuvC catalysis and from nucleic acid binding.35
The free energy barrier for the pseudo-active state was ~22.02 ± 1.26 kcal/mol, in line with studies of the catalytic mechanism based on the pseudo-active configuration.18,19 This activation barrier is considerably higher than that for the active state (~5 kcal/mol with no overlapping error bars between the energy peaks). This indicates the catalytic preference of the active conformation, and that the catalysis is unlikely to proceed through the pseudo-active state. To understand the origin of this difference in the free energy barrier, we examined the polarization effects in the catalytic centres. We analysed how the dynamical electrostatic potential derived charges (D-RESP)28 change at the TS‡ (Fig. 5B-C) and along the chemical steps (Supplementary Fig. 22). At the TS‡, the charge of Mg2+ and of the O3’ leaving group display opposite values in the active and pseudo-active conformations of HNH. In the pseudo-active HNH, the charge of Mg2+ is lower than that in the active state, mainly due to the coordination of both D839 and D861 carboxylates. D839 also reduces its charge with respect to the active HNH, with increased polarization on Mg2+. The latter loosely binds O3’ in the pseudo-active HNH (Supplementary Fig. 22), with an increase of the O3’ charge compared to the active state. This charge increase reduces the ability of O3’ to be a good leaving group, resulting in a larger barrier at the TS‡.
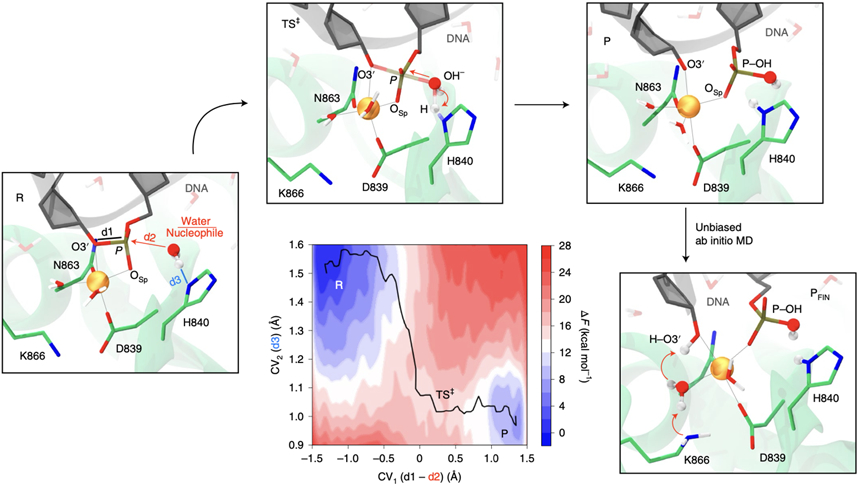
To fully characterise the catalytic mechanism in the active HNH, we performed QM/MM metadynamics simulations.36 This method enabled explicitly describing phosphodiester bond cleavage on one dimension (the first collective variable, CV1), and the deprotonation of the water nucleophile on the other (CV2). Through ~120 ps of sampling, the free energy surface confirmed an SN2-like mechanism (Fig. 6A, Supplementary Figs. 23-24). H840 extracts the water’s proton right before the free energy peak (the TS‡), consistent with in-depth analysis of Thermodynamic Integration (Supplementary Fig. 22), and similar to other His-activated nucleases,31,37 including the RuvC domain of Cas9.5 An overall free energy barrier of ~17.38 ± 0.84 kcal/mol is consistent with our mono-dimensional profile (Fig. 5) and with kinetic studies.35 Interestingly, during unbiased QM/MM simulations of the P state, O3’ gets protonated by one water molecule coordinating Mg2+. By including the neighbouring K866 in the QM part, K866 releases a proton to the water molecule, which simultaneously protonates the O3’ (Fig. 6, Supplementary Fig. 25), reaching a final product (PFIN) that is consistent with cryo-EM structures of the post-cleavage state (Supplementary Fig 26). Indeed, as shown by CpH MD simulations, the pKa of K866 is 10.9 (Supplementary Fig 27), making it a suitable proton donor for the basic alkoxide O3’ (pKa ~16). To further understand the role of K866, we computed the D-RESP charges from ab-initio QM/MM simulations in the presence of K866 and for its alanine mutant (both included in the QM part). We found that the K866A substitution dramatically increases the negative charge on PSCI, making it a worse electrophile (Supplementary Fig. 28). This analysis also reveals that all three K866 hydrogen atoms are positive in the P state and thereby prone to shuttle toward the negatively charged oxygen atoms of water and the O3’. Hence, K866 creates a favourable electronic environment and is critical for leading the catalysis to completion. This clarifies DNA cleavage experiments, showing that K866A remarkably reduces the enzymatic activity (Fig. 3E).
Fig. 6. Catalytic mechanism of DNA cleavage in the HNH domain of CRISPR-Cas9.
Two-dimensional free energy surface for phosphodiester bond cleavage reporting the progress of the chemical step from the reactants (R), transition state (TS‡) and products (P) along two collective variables (CVs, shown on the 3D structure of the R). CV1 denotes the nucleophilic attack on scissile phosphate, while CV2 accounts for the proton transfer from the water nucleophile to H840. The free energy surface was obtained through QM/MM metadynamics. Unbiased QM/MM simulations of the P state reveal that the K866 side chain releases a proton to the water molecule coordinating Mg2+, which protonates the DNA O3’, leading to the final product (PFIN) of DNA cleavage. This clarifies DNA cleavage experiments (Fig. 3E), showing that the K866A substitution remarkably reduces the enzymatic activity.
Discussion
We combined extensive molecular simulations with solution NMR and biochemical experiments to decipher the catalysis of target DNA cleavage in CRISPR-Cas9. To this point, the structure of the catalytic state was ambiguous, with two possible conformations – referred as active and pseudo-active – for which a conformational equilibrium was suggested.17 Free energy simulations were used to investigate this equilibrium, revealing that Mg2+ favours the formation of the active state, while its absence leads to the pseudo-active conformation (Fig. 2). Here, D861 and D839 point toward the catalytic core, while N863 orients in opposite direction, consistent with structures of Cas9 obtained without Mg2+ ions.3,4,11-13 This pseudo-active state is stabilised by two second-shell lysine (K866, K862), anchoring the DNA backbone in place of the ion. Contrarywise, when Mg2+ is bound to the catalytic site, N863 reorients to coordinate the ion and engage in the catalysis. This is in line with multiple sequence alignment of Cas9 orthologs showing that N863 is conserved, whereas D861 is highly variable.4 It also agrees with recent cryo-EM structures of the post-catalytic state, obtained in the presence of Mg2+.14-16 Hence, Mg2+ is important not only for the catalysis, but holds a critical structural role. Accordingly, single-molecule experiments have shown that HNH adopts different states in the presence/absence of Mg2+.38 Another single-molecule study showed that HNH is unlikely to transition to the active state for DNA cleavage without divalent cations.7 Our findings thereby support a model by which Mg2+ shifts the conformational equilibrium of HNH toward the catalytic state.
Solution NMR revealed that at pH 7.4, the pKa for the catalytic H840 is ~6.83. Molecular simulations using a Constant pH MD method23 reported an excellent agreement for the pKa of H840 in the active state (~6.90), and indicated that H840 likely assumes the neutral tautomeric form protonated on ε (Fig. 3C), posing the basis for QM/MM studies of the catalysis. Solution NMR and MD also consistently reported structure and dynamics of the active HNH in the presence of alanine mutations. Indeed, mutations disrupting the catalytic activity (D839A, N863A, K866A) more sensibly reduce the pKa of H840, and alter its location of H840 with respect to PSCI (Figs. 3-4), while mutants preserving the catalysis (D861A, K862A) result in minor conformational effects. The consistency of molecular simulations with the experiments also suggests to harness computations for residual mutations in the active HNH. For instance, R844 (in place of Q844) stably binds PSCI, mimicking the stabilising role of R976 in the RuvC site (Supplementary Fig. 29),15,39 which could be exploited to ameliorate the HNH function.
Ab-initio QM/MM simulations were used to resolve the catalytic mechanism, revealing the catalytic preference of the active conformation (Fig. 5), and that the catalysis is unlikely in the pseudo-active state. In the active HNH, an activation barrier for phosphodiester bond cleavage of ~17 kcal/mol was in line with the experimental catalytic rate.35 The catalysis proceeds through an SN2 mechanism, activated by H840 and critically aided by K866. The latter fosters an optimal electronic environment and can intervene in the protonation of the O3’, clarifying why the K866A substitution reduces the enzymatic activity (Fig. 3E). Notably, restriction/homing endonuclease enzymes using a single catalytic metal commonly display a lysine/arginine residue in the position of K866, to complement the divalent metal.40 In light of this observation, our findings also offer a mechanistic rationale on the role of the additional positively charged residue in other one-metal dependent enzymes.
Overall, the extensive multiscale approach implemented here resolves the catalytic mechanism and which of the known conformation of HNH is responsible for target DNA cleavage in CRISPR-Cas9. These findings are foundational to ameliorate the function and specificity of CRISPR-Cas9, helping the development of improved genome-editing tools.
Methods
Molecular Dynamics simulations.
Molecular simulations were based on the cryo-EM structures EMD-0584 (PDB: 6O0Y, at 3.37 Å resolution)14 and EMD-23838 (PDB: 7S4X, at 2.76 Å resolution),15 which captured the active state of the HNH domain; and on the X-ray structure PDB: 5F9R (at 3.40 Å resolution),12 which was used to model the pseudo-active configuration of HNH.6 Each systems was embedded in explicit waters, leading to periodic simulation cells of ~180*120*140 Å3 and ~340,000 total atoms. The Amber ff12SB force field was employed, including the ff99bsc0 corrections for DNA41 and the ff99bsc0+XOL3 corrections for RNA.42,43 The Li & Merz model was used for Mg2+,44 describing the metal sites in agreement with QM/MM simulations (Supplementary Fig. 3). An integration time step of 2 fs was used. Temperature control (300 K) was performed via Langevin dynamics, while pressure control was accomplished by coupling the system to a Berendsen barostat.45 The simulation protocol is described in the Supplementary Methods. Production runs were carried out in the NVT ensemble on the active and pseudo-active states of the wild-type (WT) Cas9 and of six mutants (i.e., V838A, D839A, D861A, K862A, N863A, K866A). The WT Cas9 was also simulated considering two tautomeric forms of H840 (protonated on δ and ε). The active HNH was also simulated in the presence of the Q844R mutation. For each system, three MD replicas of ~1 μs each were performed, for a total of ~54 μs of MD runs. The GPU-empowered version of AMBER 2046 was used as MD engine.
Umbrella Sampling Simulations.
The umbrella sampling (US) method was used to compute the free energy profiles associated with the conformational change of the HNH domain from the pseudo-active to active states (and vice versa).20 In this method, a number of simulations (US windows) are run in parallel with additional harmonic bias potential applied to selected Reaction Coordinates (RCs):
| (Eq. 1) |
where V(RC) is the value of the bias potential, k is a bias force constant, RC(t) is the value of RC at given time t and RC* is the reference value of RC. By using different RC* values in each US window, one can sample the biased probability distribution pb(RC) along the whole RC range of interest. The difference in Root Mean Square Deviation (RMSD) of the HNH heavy atoms’ positions with respect to pseudo-active and active conformations was used as a RC (details in the Supplementary Methods). Two independent sets of US simulations were performed: (i) in the presence of Mg2+ ions (i.e., with Mg2+) and (ii) in the absence of Mg2+ ions (i.e., w/o Mg2+). In both cases, the system was simulated in 14 overlapping windows from RC = −3.5 Å to RC = 3.5 Å using a harmonic restrain with a spring constant of 30 kJ mol−1 Å2. The centre of the harmonic bias potential was distributed along the RC in 14 windows separated by 1.0 Å or 0.5 Å, to allow proper overlapping of the probability distributions. Approximately 1 μs-long trajectories were obtained for each US window, reaching ~14 μs of collective sampling per system (a total of ~28 μs). The free energy profiles were computed using the Weighted Histogram Analysis (WHAM) method.20 Analysis of the conformational ensembles has been performed on the reweighted trajectories. Details in the Supplementary Methods.
Constant pH (CpH) Molecular Dynamics.
CpH MD simulations were performed in explicit solvent, in conjunction with a Replica Exchange method to enhance the sampling of the protonation states.23 pH values were sampled from 1 to 14, with exchanges between adjacent replicas every 200 fs, reaching ~40 ns in each replica. The pKa was computed from the distribution of the protonation states using the Hill equation:
| (Eq. 2) |
in which xi is deprotonated fraction of residue i, and n is the Hill Coefficient. The titration curves have been derived by fitting the deprotonated fraction xi to Equation 3, using the Levenberg–Marquardt nonlinear optimization method.
| (Eq. 3) |
Good titration curves display small deviations of each point from the fitted titration curve and Hill coefficients between 0.5 and 1.5, indicating that the protonation states are properly sampled at the simulated pH values.23 By using this method, the pKa of H840 and H799 in the active and pseudo-active states of HNH was computed. The pKa of H840 in the active HNH was also computed in the presence of the Q844R mutant. Furthermore, CpH MD simulations were carried out to compute the pKa of K866 in the active state. For each system, CpH MD built on ~560 ns of MD sampling (i.e., ~40 ns for 14 replicas), resulting in a total of ~3.4 μs of CpH MD simulations. Details in the Supplementary Methods.
Quantum Mechanics/Molecular Mechanics (QM/MM) simulations.
QM/MM simulations were performed on the active and pseudo-active states of HNH. In the active state, the QM part included the Mg2+ ion and its coordinating residues (D839, N863), the catalytic H840, the DNA bases G-3, T-4 and C-5, and nine water molecules. In the pseudo-active state, the QM part comprised the Mg2+ ion and its coordinating V838, D839 and D861, H840, the DNA bases G-3, T-4 and C-5, and eight water molecules. For both systems, capping hydrogens were used to saturate the valence of the terminal QM part, resulting in 123 and 111 QM atoms for the active and pseudo-active systems, respectively. The catalytic H840 was simulated in the neutral tautomeric form protonated in ε, as indicated by classical and CpH MD (Supplementary Figs. 10-11), and NMR data (Fig. 3). The QM part was described at the QM DFT/BLYP29,30 level, and the MM part was treated using the force field above. QM/MM simulations were performed using the CPMD code.47 The wave functions were expanded in a plane wave basis set up to a cutoff of 75 Ry in a QM cell of dimensions ~26*28*26 Å3. A rigorous Hamiltonian treatment of the electrostatic interaction between the QM and MM regions was used.28 The temperatures of the QM and MM subsystems were kept constant at 300 K using a Nosé-Hoover thermostat.48,49 Car-Parrinello QM/MM simulations50 were performed with a time step of 5 au (~0.12 fs) and a fictitious electron mass of 600 au. Upon ~40 ps of unconstrained ab-initio MD, the Mg2+-bound configurations were used to start for free-energy simulations (vide infra).
Thermodynamic Integration.
Thermodynamic Integration and the “blue moon ensemble” method27 were used to compute the free-energy profiles for phosphodiester bond cleavage in the active and pseudo-active states of HNH. In this approach, the average converged constraint forces are computed and integrated along a given RC, deriving the associated free energy profile. The difference in distance between the breaking and forming P–O bonds was used as RC, as in previous studies of DNA/RNA processing enzymes.5,31-34 Starting from a RC = −1.5 Å, we sampled in 18 sequential windows along the RC, with a resolution of 0.2 Å (0.1 Å in the region in the vicinity of the TS‡). Each window was simulated for ≥8 ps, reaching convergence of the constraint force, and collecting ~144 ps of ab-initio MD for the active and pseudo-active states, respectively. To estimate the error associated to hysteresis, we also computed the backward free energy pathways, resulting in eight additional sampling windows and ~64 ps of ab-initio MD for each system. The statistical error at each point of free energy profiles (both forward and backward) was computed by error propagation analysis. The overall error on the free energy barrier was estimated as the sum of the statistical error and the error due to hysteresis between the forward and backward pathway. Full details in the Supplementary Methods. Altogether, considering the forward and backward pathways, and unconstrained runs, we collected ~218 ps of ab-initio QM/MM MD for each system.
Metadynamics.
The catalytic mechanism of DNA cleavage in the active CRISPR-Cas9 was characterised through QM/MM simulations and metadynamics.36 This method applies an external history-dependent bias potential to the Hamiltonian of the system as a function of a set of pre-defined degrees of freedom (collective variables, CVs). We used two CVs: (i) the difference in distance between the breaking and forming P–O bonds and (ii) the distance between the H840 δ nitrogen and the water nucleophile’s hydrogen. This enabled mapping the free energy for phosphodiester bond cleavage on one dimension, characterising nucleophile activation on a second dimension. An extended Lagrangian version of the method was used for a proper coupling with the QM/MM simulations. For both CVs, the mass value of the fictitious particles was 16 amu, while the value of the force constant was 0.24 au. The height of the Gaussian terms was 0.5 kcal/mol, which ensures sufficient accuracy for reconstructing the free energy surface. The width of the Gaussian terms was 0.05 Å, in line with the oscillations of the CVs in a free Car-Parrinello QM/MM run. The Gaussian function deposition rate was set to 24 fs. A total of ~5000 Gaussians were deposited, collecting ~120 ps. During this time, the chemical step crossed from the reactants to products multiple times (Supplementary Fig. 23), properly sampling the conformational space. The statistical error associated to the free energy for phosphodiester bond cleavage was estimated by average blocking, i.e., by computing and averaging the standard deviation of the mean free energy in blocks of ~10 ps of the converged metadynamics simulation. Full details in the Supplementary Methods.
NMR Spectroscopy.
The WT HNH domain and the active site alanine mutations (D839A, H840A, D861A, K862A, N863A, K866A) of Cas9 were expressed and purified as previously described.21 Samples of 13C and 15N labelled HNH were expressed in Rosetta(DE3) cells in M9 minimal media containing MEM vitamins, MgSO4, and CaCl2 with 15NH4Cl and U-13C-Dextrose (Cambridge Isotopes) as the sole nitrogen and carbon sources. NMR data were collected in a buffer containing 20 mM HEPES, 80 mM KCl, 1 mM DTT and 7.5% (v/v) D2O at pH 7.4. Samples contained a background of 10 mM Mg2+ and 5 mM DNA (5’-GGACCATAGGATGGTCC-3’). Backbone amide fingerprints and chemical shifts of WT HNH and variants were determined via the sensitivity-enhanced HSQC (hsqcetf3gpsi2 pulse sequence) on a 600 MHz Bruker NEO spectrometer with the 15N dimension centred at 117 ppm. Histidine ε1 sidechain chemical shifts were measured via HMQC (hmqcphpr pulse sequence) using NMR samples that were successively buffer exchanged into NMR buffer at selected pH values between pH 6 – 8. The 13C dimension centred at 130 ppm and the ε1 sidechain chemical shifts of HNH were assigned by mutagenesis of H840 to alanine. H840 pKa values were determined from fitting the 1H chemical shift trajectories to a modified Henderson-Hasselbach equation. Details in the Supplementary Methods.
SpCas9 in vitro cleavage assay.
Single guide sgRNAs and SpCas9 protein used in cleavage assays was produced as described previously.11 Fluorescently labelled target oligonucleotides were synthesised by Integrated DNA Technologies, Inc., with HPLC purification. Cleavage substrates were prepared by mixing complementary oligonucleotides in a 1:2 molar ratio of target strand to non-target strand, heating to 95 °C for 5 minutes and slow cooling to room-temperature. Cas9 protein was first pre-incubated with the sgRNA in a 1:1.25 molar ratio and incubated 10 minutes at room temperature in the reaction buffer containing 20 mM HEPES pH 7.5, 250 mM KCl, 5 mM MgCl2 and 1 mM DTT. The binary complex was rapidly mixed with ATTO-532 labelled dsDNA substrate, to yield final concentrations of 1.67 μM protein and 66.67 nM substrate in a 7.5 μl reaction. Reactions were incubated at 37 °C and time points were harvested at 1, 2.5, 5, 15, 45, 90 and 150 minutes. Substrate cleavage was stopped by addition of 2 μl STOP buffer mix, consisting of 250 mM EDTA, 0.5% SDS and 20 μg of Proteinase K. Reaction products were separated on a 16% denaturing 7M urea PAGE gel and imaged using a Typhoon FLA 9500 gel imager. The average from four independent experiments was fit to a single-exponential function for every time point (Fig. 3E). The sgRNA sequence used in cleavage experiments was GGAAUCCCUUCUGCAGCACCGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUG. The dsDNA cleavage substrate target strand sequence was 5'-[Atto-532]CGACAATACCAGGTGCTGCAGAAGGGATTCCAGATGAGC-3'. The cleavage substrate non-target strand sequence was 5'-GCTCATCTGGAATCCCTTCTGCAGCACCTGGTATTGTCG-3'.
Supplementary Material
Acknowledgement
This material is based upon work supported by the National Institute of Health (Grant No. R01GM141329, to GP) and the National Science Foundation (Grant No. CHE-1905374, to GP). GPL is supported by the National Science Foundation (Grant No. MCB-2143760). MJ acknowledges support from the Swiss National Science Foundation (31003A_182567). MJ is an International Research Scholar of the Howard Hughes Medical Institute and Vallee Scholar of the Bert L & N Kuggie Vallee Foundation. Computer time for MD has been awarded by XSEDE under Grant No. TG-MCB160059 and by NERSC under Grant No. M3807 (to GP).
Footnotes
Competing Interests
The authors declare no competing interests.
Additional Information
Additional information is available as a supplementary information.
Data Availability Statement
Atomic coordinates of the optimised computational models are available in figshare with the identifier DOI: 10.6084/m9.figshare.19158080. NMR resonance assignments for the HNH nuclease are available in the BMRB entry 27949. All other data is available from the authors upon reasonable request.
References
- 1.Doudna JA & Charpentier E Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Jinek M et al. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinek M et al. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science 343, 1247997 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishimasu H et al. Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell 156, 935–949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casalino L, Nierzwicki Ł, Jinek M & Palermo G Catalytic Mechanism of Non-Target DNA Cleavage in CRISPR-Cas9 Revealed by Ab Initio Molecular Dynamics. ACS Catal. 10, 13596–13605 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palermo G, Miao Y, Walker RC, Jinek M & McCammon JA CRISPR-Cas9 conformational activation as elucidated from enhanced molecular simulations. Proc. Natl. Acad. Sci 114, 7260–7265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dagdas YS, Chen JS, Sternberg SH, Doudna JA & Yildiz A A Conformational Checkpoint between DNA Binding and Cleavage by CRISPR-Cas9. Sci. Adv 3, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sternberg SH, LaFrance B, Kaplan M & Doudna JA Conformational control of DNA target cleavage by CRISPR–Cas9. Nature 527, 110–113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biertümpfel C, Yang W & Suck D Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature 449, 616–620 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Zuo Z & Liu J Structure and Dynamics of Cas9 HNH Domain Catalytic State. Sci. Rep 7, 1–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anders C, Niewoehner O, Duerst A & Jinek M Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang F et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351, 867–871 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huai G et al. Structural Insights into DNA Cleavage Activation of CRISPR-Cas9 System. Nat. Commun 8, 1–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu X et al. Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9. Nat. Struct. Mol. Biol 26, 679–685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bravo JPK et al. Structural basis for mismatch surveillance by CRISPR–Cas9. Nature 603, 343–347 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pacesa M et al. Mechanism of R-loop formation and conformational activation of Cas9. BiorXiv (2021). DOI: 10.1101/2021.09.16.460614. [DOI] [Google Scholar]
- 17.Zuo Z et al. Structural and functional insights into the bona fide catalytic state of Streptococcus pyogenes Cas9 HNH nuclease domain. Elife 8, 1–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon H, Zhao LN & Warshel A Exploring the Catalytic Mechanism of Cas9 Using Information Inferred from Endonuclease VII. ACS Catal. 9, 1329–1336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao LN, Mondal D & Warshel A Exploring alternative catalytic mechanisms of the Cas9 HNH domain. Proteins 88, 260–264 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kästner J Umbrella sampling. Wiley Interdiscip. Rev. Comput. Mol. Sci 1, 932–942 (2011). [Google Scholar]
- 21.East KW et al. Allosteric Motions of the CRISPR–Cas9 HNH Nuclease Probed by NMR and Molecular Dynamics. J. Am. Chem. Soc 142, 1348–1358 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nierzwicki Ł et al. Enhanced Specificity Mutations Perturb Allosteric Signaling in the CRISPR-Cas9 HNH Endonuclease. Elife 10, e73601 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swails JM & Roitberg AE Enhancing conformation and protonation state sampling of hen egg white lysozyme using pH replica exchange molecular dynamics. J. Chem. Theory Comput 8, 4393–4404 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Hansen AL & Kay LE Measurement of histidine pK a values and tautomer populations in invisible protein states. Proc. Natl. Acad. Sci 111, 1705–1712(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimahara H et al. Tautomerism of Histidine 64 Associated with Proton Transfer in Catalysis of Carbonic Anhydrase. J. Biol. Chem 282, 9646–9656 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Brunk E & Rothlisberger U Mixed Quantum Mechanical/Molecular Mechanical Molecular Dynamics Simulations of Biological Systems in Ground and Electronically Excited States. Chem. Rev 115, 6217–6263 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Carter EA, Ciccotti G, Hynes JT & Kapral R Constrained reaction coordinate dynamics for the simulation of rare events. Chem. Phys. Lett 156, 472–477 (1989). [Google Scholar]
- 28.Laio A, VandeVondele J & Rothlisberger U A Hamiltonian electrostatic coupling scheme for hybrid Car–Parrinello molecular dynamics simulations. J. Chem. Phys 116, 6941–6947 (2002). [Google Scholar]
- 29.Becke AD Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A 38, 3098–3100 (1988). [DOI] [PubMed] [Google Scholar]
- 30.Lee C, Yang W & Parr RG Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 37, 785–789 (1988). [DOI] [PubMed] [Google Scholar]
- 31.Dürr SL et al. The Role of Conserved Residues in the DEDDh Motif: the Proton-Transfer Mechanism of HIV-1 RNase H. ACS Catal. 11, 7915–7927 (2021). [Google Scholar]
- 32.Casalino L, Palermo G, Rothlisberger U & Magistrato A Who Activates the Nucleophile in Ribozyme Catalysis? An Answer from the Splicing Mechanism of Group II Introns. J. Am. Chem. Soc 138, 10374–10377 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Borišek J & Magistrato A All-atom Simulations Decrypt the Molecular Terms of RNA Catalysis in the Exon-ligation Step of the Spliceosome ACS Catal. 10, 5328–5334 (2020). [Google Scholar]
- 34.Palermo G et al. Catalytic Metal Ions and Enzymatic Processing of DNA and RNA. Acc. Chem. Res 48, 220–228 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Gong S, Yu HH, Johnson KA & Taylor DW DNA Unwinding Is the Primary Determinant of CRISPR-Cas9 Activity. Cell Rep. 22, 359–371 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laio A & Parrinello M Escaping free-energy minima. Proc. Natl. Acad. Sci. U. S. A 99, 12562–12566 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cisneros GA et al. Reaction Mechanism of the ε Subunit of E. coli DNA Polymerase III: Insights into Active Site Metal Coordination and Catalytically Significant Residues. J. Am. Chem. Soc 131, 1550–1556 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y et al. Real-time observation of cas9 postcatalytic domain motions. Proc. Natl. Acad. Sci. U. S. A 118, 1–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palermo G Structure and Dynamics of the CRISPR–Cas9 Catalytic Complex. J. Chem. Inf. Model 59, 2394–2406 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Galburt EA & Stoddard BL Catalytic Mechanisms of Restriction and Homing Endonucleases. Biochemistry 41, 13851–13860 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Perez A et al. Refinement of the AMBER Force Field for Nucleic Acids: Improving the Description of Alpha/Gamma Conformers. Biophys. J 92, 3817–3829 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banas P et al. Performance of Molecular Mechanics Force Fields for RNA Simulations: Stability of UUCG and GNRA Hairpins. J. Chem. Theor. Comput 6, 3836–3849 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zgarbova M et al. Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J. Chem. Theory Comput 7, 2886–2902 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li P, Roberts BP, Chakravorty DK & Merz KM Rational Design of Particle Mesh Ewald Compatible Lennard-Jones Parameters for +2 Metal Cations in Explicit Solvent. J. Chem. Theory Comput 9, 2733–2748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A & Haak JR Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys 81, 3684–3690 (1984). [Google Scholar]
- 46.Case DA et al. AMBER 2020. Univ. California, San Fr (2020). [Google Scholar]
- 47.Parrinello M, Andreoni W & Curioni A CPMD; IBM. Corporation and Max-Planck Institut: Armonk, New York and. Stuttgart, Germany, 2000. [Google Scholar]
- 48.Hoover WG Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 31, 1695–1697 (1985). [DOI] [PubMed] [Google Scholar]
- 49.Nosé S An extension of the canonical ensemble molecular dynamics method. Mol. Phys 57, 187–191 (1986). [Google Scholar]
- 50.Car R & Parrinello M Unified Approach for Molecular Dynamics and Density-Functional Theory. Phys. Rev. Lett 55, 2471–2474 (1985). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates of the optimised computational models are available in figshare with the identifier DOI: 10.6084/m9.figshare.19158080. NMR resonance assignments for the HNH nuclease are available in the BMRB entry 27949. All other data is available from the authors upon reasonable request.