Abstract
Arapaima gigas, known as Pirarucu in Brazil, is one of the largest freshwater fish in the world. Some individuals could reach 3 m in length and weight up to 200 kg. Due to extinction risks and its economic value, the species has been a focus for preservation and reproduction studies. Thyrotropin (TSH) is a glycoprotein hormone formed by 2 subunits α and β whose main activity is related to the synthesis of thyroid hormones (THs)—T3 and T4. In this work, we present a combination of bioinformatics tools to identify Arapaima gigas βTSH (ag-βTSH), modeling its molecular structure and express the recombinant heterodimer form in mammalian cells. Using the combination of computational biology, based on genome-related information, in silico molecular cloning and modeling led to confirm results of the ag-βTSH sequence by reverse transcriptase-polymerase chain reaction (RT-PCR) and transient expression in human embryonic kidney (HEK293F) cells. Molecular cloning of ag-βTSH retrieved 146 amino acids with a signal peptide of 21 amino acid residues and 6 disulfide bonds. The sequence has a similarity to 39 fish species, ranging between 43.1% and 81.6%, whose domains are extremely conserved, such as cystine knot motif and N-glycosylation site. The Arapaima gigas thyrotropin (ag-TSH) model, solved by AlphaFold, was used in molecular dynamics simulations with Scleropages formosus receptor, providing similar values of free energy ΔGbind and ΔGPMF in comparison with Homo sapiens model. The recombinant expression in HEK293F cells reached a yield of 25 mg/L, characterized via chromatographic and physical-chemical techniques. This work shows that other Arapaima gigas proteins could be studied in a similar way, using the combination of these techniques, recovering more information from its genome and improving the reproduction and preservation of this prehistoric fish.
Keywords: Arapaima gigas, AlphaFold, thyrotropin, in silico prediction
Introduction
Arapaima gigas, in Brazil commonly known as Pirarucu, is a teleost fish native to the Amazon basin that reaches up to 3 m in length and weighs more than 200 kg. Its morphological characteristics provoke a commercial appeal within the riverine community and the region. Besides the flavor of its meat, several parts can be used when it is slaughtered. For example, its bony tongue has been commercialized and used as sandpaper or adornments, and its skin can become a luxury item, as a covering for handbags, sneakers, blouses, and so on. There is also a great interest in the fish farming business, as its captive breeding stimulates the local economy. In 2010, Brazilian production reached levels of 10 tons, retrieving a farm-gate of US$130.000.1
A. gigas is a fish of high economic and cultural value, making it the most studied floodplain fish among the existing ones. It is evident in fact that the capture of immature Pirarucu in their natural habitat affects the reproduction of individuals, as it completely alters the biological cycle of the species, inducing unnecessary migrations.2 Furthermore, it is well known that habitat degradation, such as local logging and human settlement, has a detrimental impact on this species. Reinforcing restrictive measures, such as limiting fishing of the species by size, as larger fish is often breeding stock, are crucial factors for the A. gigas conservation.3 Management of the fish reproduction in captivity still faces some issues, eg, the seasonality of reproduction.4 In other words, the spawning only occurs under certain conditions, such as in rainy times (if both sexes are fit at the same time). Attempts to change the environment (temperature, humidity, space, feeding, lighting) have already been made in fish farms and none had a direct success. We believe that one of the possibilities may be an increase at the physiological level of hormones that are directly linked to reproduction.5
Thyroid-stimulating hormone—or thyrotropin—(TSH) is a hormone belonging to the family of “glycoprotein hormones,” which contains oligosaccharides linked to the protein chain in specific glycosylation sites. All 4 pituitary glycoprotein hormones, ie, TSH, chorionic gonadotropin (CG), follicle-stimulating hormone (FSH), and luteinizing hormone (LH)6 are heterodimers (formed by 2 subunits) that share a common alpha subunit and different beta subunit determining the hormone specificity. The alpha subunit from A. gigas (ag-αGTH) was previously characterized and used to produce in mammalian cells other pituitary glycoproteic hormones.7
TSH is synthesized by the anterior portion of the pituitary gland upon stimulation of the thyrotropin-releasing hormone (TRH), whose synthesis and release occur in the hypothalamus. Both hormones (TSH and TRH) have their secretion and production controlled by the negative feedback mechanism (self-regulation): as one hormone reaches a high concentration in the blood circulation and the other hormone regulates its release, and vice versa.8 TSH, in turn, stimulates the thyroid cells to secrete thyroxine (T4) in larger amounts, which, in turn, will undergo the deiodation process—by enzymes known as deiodinases—and keeps high the levels of triiodothyronine (T3) in the bloodstream.9 THs, however, are known for their iodine uptake, acquired through food intake. However, THs participate in intra-cellular metabolism, regulation of gene expression, and even neuroprotective ways. We note that THs participate directly in the control of mechanisms that are linked to reproduction. Some intra-cellular signalization played by THs has direct influence on the development, maturation, and maintenance of organs like liver, brain, and gonads, specially regulating the release of some hormones.10-12
In this research, we propose the application of a combination of bioinformatics techniques focused on protein studies from A. gigas. Using thyrotropin as a model, the coding sequence prediction allowed us to identify the 3’ untranslated region (UTR) of the gene and to synthesize the recombinant protein in mammalian cells with its characterization in high-performance size exclusion chromatography (HPSEC) and reverse-phase high-performance liquid chromatography (RP-HPLC). Furthermore, the TSH structure was modeled using the deep learning algorithm AlphaFold2,13 and the generated structure was employed for molecular dynamics (MD) simulations and free energy calculations to explore its binding to the human and A. gigas TSH receptors (hTSHR and ag-TSHR, respectively).
Material and Methods
In silico prediction of ag-βTSH coding gene sequence
We used the National Center for Biotechnology Information (NCBI) Eukaryotic gene prediction tool “Gnomon” (http://www.ncbi.nlm.nih.gov/genome/guide/gnomon.html) to predict the coding sequence of ag-βTSH. The method is an ab initio homology modeling composed of 5 steps: Blastn/BlastP, Compart, Splign/SplignPro, Chainer, and Gnomon. Briefly, each step is described as follows:
Blastn/BlastP: used to identify the position of ag-βTSH in the A. gigas genome.
Compart: searches for gene duplication on the genome, analyzing Blast hits. In this case, the βTSH coding sequence retrieved 1 unique position.
Splign or SplignPro: constructs the exons and simulates a splicing based on cDNA.
Chainer: evaluates the best position for transcript variants clustering the exons.
Gnomon: extends predictions of missing start or stop codons using HMM-based algorithm.
To conduct the in silico prediction thyrotropin beta subunit (ag-βTSH) coding gene sequence, the A. gigas genome (NCBI access no. GCA_900497675.1) and the βTSH sequences of other 4 different fish species, ie, Anguilla anguilla (NCBI access no. XM_035387135.1), Danio rerio (NCBI access no. NM_181494), Anguilla japonica (NCBI access no. AB175833), and Scleropages formosus (NCBI access no. XM_018725541), were employed for the identification of related position of βTSH coding sequence on the genome assembly, using Nucleotide and Protein BLAST. After retrieving 1 unique position and no other duplications for the predicted gene (scaffold540), the SplignPro methodology (using NCBI Genome Workbench, available for free at: https://www.ncbi.nlm.nih.gov/tools/gbench/) aligned XM_018725541 with scaffold540 (genome position: 160600-161733) evidencing 2 exons (160600-160869 and 161566-161733). The final result of the prediction tool is a 1 single transcript variant, used as a theoretical sequence (named in silico ag-βTSH) for in vitro confirmation on next steps.
Fish materials and in vitro cloning
A. gigas pituitary glands were gently donated by Embrapa Pesca e Aquicultura (Palmas, Tocantins, TO, Brazil) in 1 mL of RNAlater (Thermo Fisher Scientific, Waltham, MA, USA). Total Ribonucleic Acid (RNA) extraction was performed using the RNeasy kit protocol (Qiagen, Hilden, Germany). For in vitro cloning of 3’ UTR cDNA, the protocol of 3’ Rapid Amplification of complementary DNA (cDNA) Ends (3’RACE, commercialized by Thermo Fisher Scientific) was conducted following the instructions of the company. One single primer “AACACCACCATCTGCATGGG” was used and designed based on the theoretical sequence obtained from the in silico prediction of ag-βTSH coding gene sequence. Analysis of the product was performed in agarose gel electrophoresis followed by the Sanger sequencing methodology. The result was compared as “in silico ag-βTSH vs in vitro ag-βTSH” cloning. To confirm the sequence, the βTSH coding sequences from 37 different fish species were aligned using ClustalΩ.14 Geneious 2022.2.1 free version software provided the images in this article.15 Percentage of identity, coverage, and pairwise identity from alignment were calculated using the same software.
Phylogenetic tree
The phylogenetic tree of the coding sequence corresponding to ag-βTSH was constructed using the maximum likelihood (ML) method. We used 38 amino acid sequences from different fish species (including A. gigas) deposited in a database. Only the coding regions were used. The whole process was performed with the freely accessible software MEGA 11.16 The first step consists of the alignment of all sequences, using ClustalΩ.14 Then, the software determines which substitution model is the best. According to the following values (Supplemental Table S1) for BIC (Bayesian information criterion) and AICc (Akaike information criterion), the best substitution model available, used in this analysis, was the Jones-Taylor-Thornton (JTT; BIC: 10079.19979; AICc: 9596.47366). The methodology is a refinement of automation derived from maximum parsimony (MP) assuming that a select pair of sequences were >85% identical.13
The tree was constructed with 500 bootstraps for each taxon included in the tree. Finally, the construction was carried out with the Subtree-Pruning-Regrafting (SPR) algorithm, which dismembers a portion of the best constructed tree and allocates it into other trees already constructed. The algorithm then searches through all the subtrees until the result does not exceed the best likelihood of the topologies already built. Its search parameters were “search_level = 1,” and the initial trees were obtained through 10 replicates.
Three-dimensional structure modeling
The 3-dimensional (3D) structure corresponding to the coding sequences of the α (NCBI access no. AIG51239.1) and β subunits (ag-βTSH has an NCBI access no. OP125854, related with this work) of A. gigas thyrotropin was independently modeled using the AlphaFold217 implementation at ColabFold.18 The model is available in Model Archive at https://modelarchive.org/doi/10.5452/ma-plm8v. Coding sequences from both subunits excluding signal peptides were inferred with SignalP 6.0.19 An identical approach was followed to model the structure of the extracellular domain of S. formosus thyrotropin receptor (sfTSHR, NCBI access no. XM_018732307.1).
Structures of Arapaima gigas thyrotropin (ag-TSH) in complex with hTSHR and sfTSHR were modeled through structural alignment using the crystal structure of the α and β subunits of human follicle-stimulating hormone (hFSH) bound to its receptor (hFSHR) (PDB: 4AY9). A control system made up of human thyroid stimulating hormone (hTSH) in complex with hTSHR was built in a similar fashion after modeling the structure of the β subunit of hTSH with AlphaFold2 and taking the crystal structures of the α subunit of hTSH and hTSHR from PDBs: 4AY9 and 3G04, respectively. Although hTSH:hTSHR has a recently published model solved by electron microscopy (RCSB PDB ID: 7T9I), our work has been developed with high accuracy methods, evidenced between aligned structures (Supplemental Figure S1) which has the same binding orientation, with a Root Mean Square Deviation (RMSD) of 1.581 angstrom (Å), for 5548 atoms. The documentation of the program with details is described in Supplementary data. For isoelectric point (pI) and electrostatic potential surface calculation, we use PDB2PQR/APBS server20 and other properties described in results with the database.20 For visualization of the protein structure, Maestro 2022.2 (Maestro. Schrödinger Release 2022-3: Maestro, Schrödinger, LLC, New York, NY, 2021) and PyMOL 2.0 (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC) free version software were used.
MD and umbrella sampling
The ligand:receptor complexes (ag-TSH:hTSHR, ag-TSH:sfTSHR, and hTSH:hTSHR) were prepared for MD simulations. First, all the complexes were protonated at pH = 7.4 at the H++ web server (http://newbiophysics.cs.vt.edu/H++/). Then, the complexes were embedded in octahedral boxes, filled with Transferable Intermolecular Potential with 3 Points (TIP3P) water molecules, and sufficient counter-ions using tleap of Amber20. MD simulations were parameterized with the ff14SB force field. All systems were subjected to energy minimization (EM), heating in the Canonical Ensemble (NVT) from 10 to 300 K using a linear temperature gradient, and an Isothermal-Isobaric Ensemble (NPT) equilibration at 300 K, at 1 bar pressure. Finally, 1 μs productive MD simulations at 300 K in the NVT ensemble were carried out.
Representative structures of the complexes obtained through clustering analysis of the 1 μs trajectories were employed for umbrella sampling (US) free energy calculations. The complexes were carefully oriented so that it was possible to separate the TSH molecule from the receptors along the z-axis without steric clashes. Rotational and translational motions of the receptor and rotational motions of TSHs were prevented by attaching these proteins to 3 dummy particles (D1, D2, D3) added to the solvation boxes in a noncollinear fashion and kept tightly fixed by means of stiff harmonic restraints. The distance between D1 and a Cα atom of TSHs was restrained along the z-axis every 1 Å with a harmonic constant of 10 kcal/mol/Å2. A total of 23 windows were created, and the weighted histogram analysis method (WHAM) was employed to obtain the potentials of mean force (PMFs) along the selected axis.21
Expression of recombinant A. gigas thyrotropin
A chemically competent strain of Escherichia coli DH5⍺ was transformed with pcDNA 3.4 ag-βTSH and pcDNA 3.4 ag-αGTH plasmids expression vectors (GenScript Biotech Corp, Piscataway, New Jersey, USA) by the heat shock method.22
To obtain the plasmids for transient transfection, a purification protocol plasmid was conducted following the instructions of the kit (NucleoBond Xtra Midi kit transfection-grade, Macherey-Nagel, Düren, Nordrhein-Westfalen, Germany). The plasmid was quantified in a spectrophotometer and stored at −20°C in deionized nuclease-free water.
The suspension Freestyle Human Embryonic Kidney cells 293 (HEK293F) were transfected at the density of 3.0 × 106 cells/mL in 30 mL of Expi293 media, with subunits α and β (1:1 plasmid mass proportion, final mass: 30 μg) vectors for recombinant thyrotropin expression. Expi293 Transient Expression System (Thermo Fisher Scientific) protocol was followed, using ExpiFectamine™ (Thermo Fisher Scientific) as a transfection agent (cationic liposome-mediated transfection). Transfected cells were maintained in a 125-mL Corning® Erlenmeyer cell culture flasks (CLS431143, Merck, Darmstadt, Germany) at 37°C, 8% CO2, and 130 r/min, for 7 days, without changing the media. A total of 1 mL fractions of the third, fifth, and seventh days after transfection were collected and kept at −80°C for sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
Physico-chemical characterization
After SDS-PAGE analysis and confirmation of the best day of expression, RP-HPLC was performed, using as positive control 5 µg of commercial human LH (National Hormone Pituitary Program—NHPP, NIH, Bethesda, MD, USA) at a concentration of 1 µg/µL; as negative control 5 ml of concentrated media from untransfected cells (30 mL initial volume) and 4 mL of concentrated media from the fifth day (30 mL initial volume) after transfection. The collection of the fractions was conducted from 30 to 50 min, 0.5 mL/tube, at a flow rate of 0.5 mL/min, under detection wavelength of 220 nm. A C4 214TP54 Grace Vydac (W.R. Grace and Company, Columbia, MD, USA), 300 Å pore size, 5 μm particle size, internal diameter 4.6 mm, length 250 mm, and Si60 pre-column was run under isocratic conditions.
HPSEC step was used for the characterization from RP-HPLC pools collections, using H. sapiens recombinant TSH (Thyrogen®, Sanofi, Paris, France; 5 µg/5 µL) as reference preparation. Samples were concentrated and dialyzed in a mobile phase buffer for HPSEC (described below) on Amicon® 10 kDa MWCO system (Merck). The total running time was 40 min. Sample flow rate was 1.0 mL/min with reading at 220 nm wavelength, using a TSKGel G2000SW (Tosoh, Tokyo, Japan) column as stationary phase.
Both chromatographic analyses were carried out in a Shimadzu LC 20 Prominence equipment (Shimadzu Corporation, Kyoto, Japan). RP-HPLC and HPSEC conditions are described as follows, distinguished by (1) and (2), respectively:
Mobile phase consisting of 2 buffers A and B: (A) sodium phosphate 50 mM pH 7.0 and (B) acetonitrile. A total of 5 steps for elution were applied. Step one started with 15% of B for 7 min. A second step was a gradient of the B buffer until it reached 60% between 7 and 45 min. The third one maintained 60% up to 50 min. The fourth and fifth steps were a slow decline of B reaching 15% over the end. The flow rate was 0.5 mL/min with a 2-mL loop and column temperature of 30°C. The total running time was 75 min.
The mobile phase consisted of 1 single buffer of sodium phosphate 20 mM pH 7.0 + 150 mM NaCl. The flow rate was 1.0 mL/min with a 0.5-mL loop at room temperature. Total running time was 40 min.
For the qualitative analysis of conditioned medium and RP-HPLC collection, a 15% SDS-PAGE technique with silver staining was used. A total of 0.5 mL of samples were prepared via trichloroacetic acid (TCA) precipitation protocol, and the protein pellet was resuspended in 100 µL of 1× phosphate-buffered saline (PBS) + 100 µL 4× sample buffer.
Results
Predicted coding sequence and in silico confirmation with 3’UTR
A. gigas beta-thyrotropin coding sequence and 3’UTR (5’UTR is under developing) was successfully identified through in silico and reverse transcriptase-polymerase chain reaction (RT-PCR) methodologies, cloning 677 nucleotides (146 amino acids compose the coding sequence) (Figure 1). SignalP 6.019 indicates the cleavage site between Serine-21 and Phenylalanine-22, inferring that the signal peptide is composed of 21 amino acids. There is a prevalence of Guanine-Cytosine (GC) in the coding sequence (58.3%); if we consider only the mature peptide, GC content remains close (57.4%). The 12 cysteines involved in disulfide bridges and N-glycosylation sites (asparagine-45) are clearly identified and conserved. The deduced glycoprotein subunit has a theoretical pI of 6.34 and a molecular mass of 15.7 kilodaltons (kDa). It has a high content of leucine, glycine, and cysteine, each at 10.3%, 10.3%, and 9.6% (molecule%/total amino acids), respectively. The total of positively and negatively charged residues is 9 and 11, respectively. Finally, ag-βTSH has 78.00% of query cover and 47.57% of identity with native H. sapiens TSH structure (RCSB PDB ID: 7T9I).
Figure 1.
ag-βTSH coding sequence (1–402 nt) and 3’ untranslated region (402–677 nt). The first line corresponds to the nucleotides, and the second one corresponds to the translated amino acids.
* indicates stop codon.
The predicted sequence was confirmed by in vitro reaction, using the 3’ RACE system for cloning the complete 3’UTR gene. 3’ UTR polyadenylation signal (631-ATTAAA-636) is identified according to the literature, and the poly-A tail starts after 19 nucleotides (656–677).23 The provided information was used to submit the sequence to the NCBI GenBank database with the access number OP125854. Moreover, Table 1 shows the pairwise sequence identities retrieved by ClustalΩ multiple alignment of 40 species including ag-βTSH, which range from 81.60% (S. formosus) to 40.10% (Mus musculus). Conserved amino acids with 100% sequence identity are shown in Figure 2.
Table 1.
Arapaima gigas thyrotropin identity among species, from the most similar to the less similar.
| Common name | Scientific name | Similarity (%) | GenBank number |
|---|---|---|---|
| Arapaima | Arapaima gigas | – | OP125854 |
| Asian arowana | Scleropages formosus | 81.60 | XP_018581057 |
| Indo-pacific tarpon | Megalops cyprinoides | 70.40 | XP_036387986.1 |
| Brown grousea | Paramormyrops kingsleyae | 68.80 | XP_023693336.1 |
| Allis shad | Alosa alosa | 64.52 | XP_048111532.1 |
| Denticle herring | Denticeps clupeoides | 64.46 | XP_028854856.1 |
| Japanese eel | Anguilla japonica | 64.29 | Q7ZZV4.1 |
| American shad | Alosa sapidissima | 63.71 | XP_041945432.1 |
| European eel | Anguilla anguilla | 63.49 | XP_035243023.1 |
| Atlantic Salmon | Salmo salar | 63.48 | NP_001117000.1 |
| Atlantic herring | Clupea harengus | 62.90 | XP_041945431.1 |
| Rainbow trout | Oncorhynchus mykiss | 62.90 | XP_021425667.1 |
| Giant grouper | Epinephelus lanceolatus | 59.68 | XP_033482843.1 |
| Greater pipefish | Syngnathus acus | 58.82 | XP_037107826.1 |
| Electric eel | Electrophorus electricus | 58.54 | XP_026875009.1 |
| Common coral trout | Plectropomus leopardus | 58.06 | XP_042353710.1 |
| Milkfish | Chanos chanos | 56.45 | XP_030633387.1 |
| Rhinoceros horn fishb | Sinocyclocheilus rhinocerous | 56.45 | XP_016395895.1 |
| Anshui clove fishc | Sinocyclocheilus anshuiensis | 56.45 | XP_016312618.1 |
| Ninespine stickleback | Pungitius pungitius | 56.10 | XP_037346720.1 |
| Common carp | Cyprinus carpio | 55.65 | XP_018973503.2 |
| Golden-line barbel | Sinocyclocheilus grahami | 55.65 | XP_016143109.1 |
| Tambaqui | Colossoma macropomum | 55.60 | XP_036426973.1 |
| Banded archerfish | Toxotes jaculatrix | 55.60 | XP_040889707.1 |
| Fathead minnow | Pimephales promelas | 54.84 | XP_039548072.1 |
| Goldfish | Carassius auratus | 54.47 | XP_026111649.1 |
| Channel catfish | Ictalurus punctatus | 54.10 | XP_017335429.1 |
| Striped catfish | Pangasianodon hypophthalmus | 54.10 | XP_026777430.1 |
| Striped bass | Morone saxatilis | 53.50 | XP_035518185.1 |
| Ballan wrasse | Labrus bergylta | 53.23 | XP_020486388.1 |
| Burton’s Mouthbrooder | Haplochromis burtoni | 52.10 | XP_014195782.1 |
| Chinese large-mouth catfish | Silurus meridionalis | 51.70 | XP_046710878.1 |
| Zebra fish | Danio rerio | 49.70 | NP_852471.1 |
| Grass carp | Ctenopharyngodon idella | 49.70 | ACF33511.1 |
| Chinese sturgeon | Acipenser sinensis | 49.60 | AHV85521.1 |
| Siberian sturgeon | Acipenser baerii | 48.80 | CAB93505.1 |
| Mummichog | Fundulus heteroclitus | 45.80 | XP_012720582.2 |
| Humphead wrasse | Cheilinus undulatus | 43.10 | XP_041636575.1 |
| Human | Homo sapiens | 40.80 | AAA36782.1 |
| Mouse | Mus musculus | 40.10 | AAA40494.1 |
Translated from Czech dialect: Rypoun hnědý.
Translated from mandarin Chinese dialect: 犀角金線魮.
Translated from Estonian dialect: Anshui sõõrhuul.
Figure 2.
ClustalΩ alignment of 40 species with corresponding coding sequence for βTSH. Amino acids with 100% identity are highlighted.
Number 1 indicates Arapaima gigas.
Phylogenetic tree
The phylogenetic tree was rooted on the species that are within the order Acipenseriformes (Figure 3). The best tree model has obtained a log-likelihood value of −4603.86. The initial tree for heuristic search is obtained by the MP method, with calculation of the evolutionary rates of the species by gamma distribution (G) = 1.0371. In the end, 182 positions were made to compose the tree. Phylogenetic inference on our studies supports the evidence of A. gigas, which is been included in Osteoglossomorpha superorder (S. formosus and A. gigas), is a sister group of Elopomorpha (A. anguilla, A. japonica, Megalops cyprinoides, and Alosa sapidissima).
Figure 3.
Phylogenetic tree for coding sequence of βTSH from 37 fish species.
The numbers represents the node scores.
Three-dimensional structure and MD
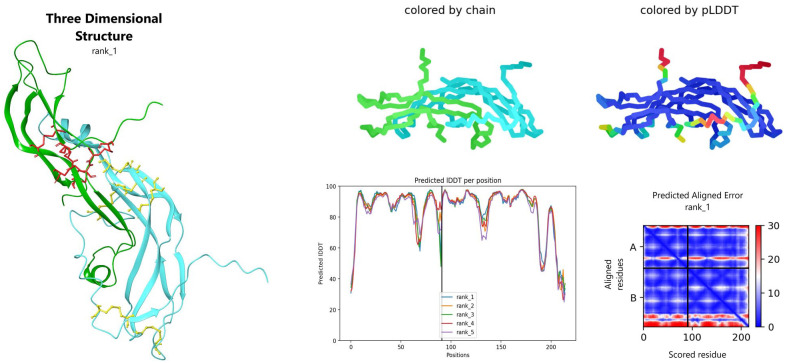
Among the 5 models generated for ag-TSH, the fourth showed the highest pLDDT score: 86.17. The results obtained, as well as the 3D model employed for further analyses, are illustrated (Figure 4). The molecular weight of the hormone is estimated to be ~24 kDa, based on the primary amino acid sequence (post-translational modification as glycosylation is not included in the reported molecular weight). The theoretical pI for the protein heterodimer is 9.40 at 298 K.
Figure 4.
AlphaFold model for ag-TSH, with local Distance Difference Test (IDDT) score per residue and alignment error. Right upper-side: two representations for chain (α: cyan, β: green) and predicted Local Distance Difference Test (pLDDT) colorful representation (high confident values = dark blue, less confident: red). Predicted alignment error and IDDT values per residue also showed. On the left side, best AlphaFold ag-TSH model with disulfide bridges sticks representation (yellow on β chain and red in α chain). The 3-dimensional structure was visualized on Maestro 2022.2.
ag-TSH indicates Arapaima gigas thyrotropin.
Root mean square deviations between ag-TSH (named as model3) and human glycoproteic crystal structures already published in the database were: 1.196 Å (942 atoms) for PDB: 1FL7 (H. sapiens follicle-stimulating hormone, named as model1), 1.252 Å (982 atoms) for PDB: 1HRP (H. sapiens chorionic gonadotropin, named as model2), and 1.388 Å (1928 atoms) for PDB: 7T9I (H. sapiens thyroid-stimulating hormone, named as model4). All protein surfaces images were colored with YRB script24 and cartoon representations created with PyMOL 2.0 (Figure 5). RMSD values between alignment of structures were performed running the following command on PyMOL: \extra_fit model1, model2, model3, model4 \method = align, \cycles = 5, \cutoff = 2.0, \mobile_state = −1, and \target_state = −1.
Figure 5.
Surface and cartoon representation of ag-TSH and 3 human glycoproteic hormones (RCSB PDB ID: 1FL7 for hFSH, RCSB PDB ID: 1HRP for hCG, and RCSB PDB ID:7T9I for hTSH). Aligned structures were performed in PyMOL 2.0. Protein surface representation had transparency set at 0.6.
ag-TSH indicates Arapaima gigas thyrotropin; hFSH, human follicle-stimulating hormone; hTSH, human thyroid-stimulating hormone.
The microsecond-long MD simulation conducted for the control system, ie, hTSHR in complex with hTSH, led to relatively low RMSDs with respect to the starting structure, which suggests the stability of the complex (Table 2). This result was confirmed by the molecular mechanics/generalized born surface area (MM-GBSA, ΔGeff) and US free energy (ΔGPMF) calculations, which yielded favorable values of −79.9 and −13.3 kcal/mol, respectively. Likewise, similar RMSD values and favorable free energies were obtained for sfTSHR in complex with ag-TSH (ΔGeff = −73.0 kcal/mol and ΔGPMF = −13.1 kcal/mol), which indicates that this complex is also stable. However, the results obtained for ag-TSH in complex with hTSHR suggest that this assembly might be unstable, probably as a consequence of the difference in residue composition between hTSHR and sfTSHR. The graphs from MD simulations were shown in 2 columns (A) for human and (B) fish parameters (Supplemental Figure S2) and binding site amino acid difference composition between hTSH and ag-TSH pointed on (Supplemental Figure S3).
Table 2.
Molecular dynamics (MD) results from different models involving thyrotropin and your receptor.
| Model | ΔGbind (kcal/mol) | RMSD (Å) | ΔGPMF (kcal/mol) |
|---|---|---|---|
| 1. hTSH:hTSHR | −79.9 ± 0.1 | 3.2–4.4 | 13.3 ± 0.4 |
| 2. ag-TSH:sfTSHR | −73.0 | 5.8 | 13.1 ± 0.5 |
| 3. ag-TSH:hTSHR | −38.0 ± 2 | 6.3–12.6 | 5.6 ± 0.6 |
Abbreviations: PMF, potential of mean force; RMSD, root mean square deviation.
Physico-chemical characterization
SDS-PAGE, RP-HPLC, and HPSEC were the techniques used to characterize the protein. After transient expression collection, an initial electrophoresis was performed to look for the best day of expression (Figure 6). The supposed retention time (TR) of the recombinant hormone is between 37 and 46 min. The ag-TSH chromatogram compared with the negative control (Figure 7) helps visualize the background of ag-TSH production in HEK293F cells. The definition of the resulting peaks between 37 and 47 min was crucial for collection and further purification. According to these minutes, 2 pools #P1 (37–40 min) and #P2 (40–45 min) were separated, concentrated and dialyzed in a mobile phase buffer for HPSEC, using the Amicon® 10 kDa MWCO system (Merck). Data were analyzed by SDS-PAGE in parallel with HPSEC (Figure 8).
Figure 6.

Silver-stained SDS-PAGE from transient expression of ag-TSH in HEK293F cells. The samples are described as follow: (I) BlueEye PreStained Protein Ladder (Sigma-Aldrich, St. Louis, MO, USA); (II) Thyrogen® (hTSH) 600 ng; (III) conditioned medium of non-DNA transfected HEK293F cells from fifth day—negative control; (IV) conditioned medium of transfected HEK293F cells from third day; (V) conditioned medium of transfected HEK293F cells from fifth day; (VI) conditioned medium of transfected HEK293F cells from seventh day. Except the ladder, all samples were precipitated with TCA and heated for 3 min at 85°C.
ag-TSH indicates Arapaima gigas thyrotropin; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; TCA, trichloroacetic acid.
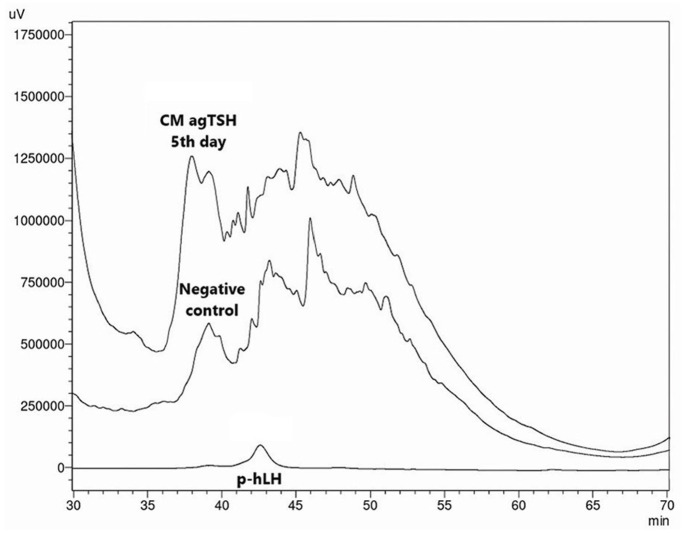
Figure 7.
Super compare the concentrated conditioned medium from the fifth day after transfection, applied in RP-HPLC. Negative control from the fifth day and pituitary human LH (p-hLH) hormone were used for better identification. Retention time of p-hLH was around 42.597 min.
RP-HPLC indicates reverse-phase high-performance liquid chromatography; LH, luteinizing hormone.
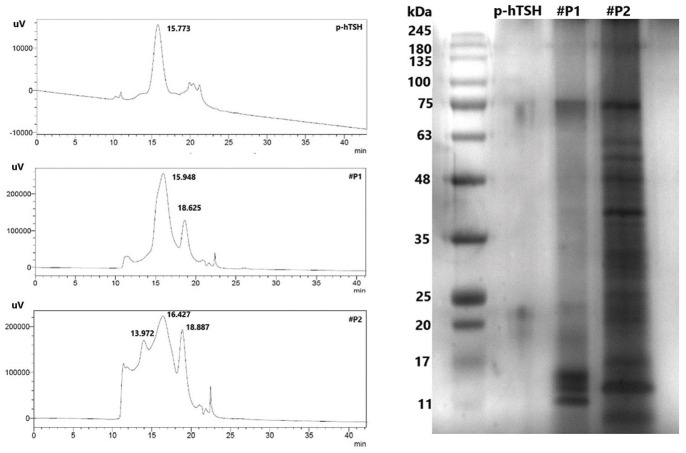
Figure 8.
HPSEC and SDS-PAGE chromatograms of pools #P1 and #P2 collected from the first RP-HPLC characterization step.
HPSEC indicates high-performance size exclusion chromatography; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; RP-HPLC, reverse-phase high-performance liquid chromatography.
In the chromatograms on the left, “p-TSH” represents native hTSH (5 µg/5 µL), with TR = 15.773 and relative area (RA) = 1569; “#P1” pool collected in RP-HPLC (#37–40 min) concentrated to 500 µL and “#P2” pool collected in RP-HPLC (#40–45 min) concentrated to 500 µL. There is a proximity between the TR of the human and A. gigas preparation. The TR differs by only 0.161 min. We were able to visualize the dissociated subunits in #P1 with 17 and 11 kDa.
Discussion
Here, we have implemented a comprehensive approach to successfully cloning the ag-βTSH cDNA sequence. The most recent phylogenetic tree published concerned the genome of A. Gigas,25,26 and bootstrap values greater than 50 on this analysis match with the published papers mentioned, which discuss the evolutionary position of Arapaima. The group has evolutionary roots on Clupeocephala. These results do not diverge from the phylogenetic tree for ag-αGTH.7
The genomic sequence corresponding to the position of thyrotropin beta subunit (βTSH) presents a unique location in chromosomes from different species. For example, in humans (H. sapiens), it was cloned and identified on chromosome 1; in mouse (M. musculus), on chromosome 3, and in zebrafish (D. rerio), on chromosome 6 (https://www.ncbi.nlm.nih.gov/gene). Although the 532 protein coding for βTSH indicates distinct chromosomes among the species, all of them showed a single region in the DNA that is responsible for the expression of βTSH. It helped us obtain the information through the Gnomon gene prediction method, which locates 1 exclusive scaffold on A. gigas genome (NCBI access no. GCA_900497675.1).
The primary amino acid sequence of ag-TSH β-subunit shares specific motifs that have similarity with other species (other than fish). Among these motifs, the most notable for elucidating the sequence and which gave us reassurance for further study was the cystine knot, comprising 12 cysteines forming 6 disulfide bridges that are important in stabilizing the protein.27 The cysteines (C) involved are: 24thC-74thC; 38thC-89thC; 41thC-129thC; 49thC-105thC; 53thC-107thC; 110thC-117thC (the numbering considers the signal peptide). Structural motifs can be found in the Pfam 35.0 cys_knot database (Pfam: 00007) or in the InterPro database “IPR006208.” We can also identify the amino acid sequence “ICMGFCYSRDTNIV” belonging to the dimerization site. Furthermore, the N-glycosylation site (NTT) is present in all the sequences from 40 species that were aligned.
The β-subunit of the glycoprotein hormones exhibits some differences in their charged-residue compositions. The mature chain of ag-TSH has 124 amino acids: 74 polar (48 uncharged and 26 charged) and 50 hydrophobic; Arapaima gigas follicle-stimulating hormone (agFSH) has 109 amino acids: 69 polar (45 uncharged and 24 charged) and 40 hydrophobic amino acids, and finally, Arapaima gigas luteinizing hormone (agLH) has 117 amino acids: 71 polar (47 uncharged and 24 charged) and 46 hydrophobic. The chromatographic analysis via RP-HPLC pointed out that ag-TSH is the glycoprotein that shows the shortest TR, agLH shows an intermediate value, and agFSH shows the longest one, reflecting the lowest hydrophobicity for ag-TSH. These data are in agreement with the results presented in this work and with those published for agFSH.28
One of the regions studied in the β-subunit of TSH is the seat-belt, present between the 10th and 12th cysteines. The amino acids from the beta chain involve the alpha subunit and are linked in the association between the subunits, binding with the receptor and its activation.29 In A. gigas, this is a region with 19 amino acids between 110thC and 129thC. In H. sapiens, however, it corresponds to the region between 88thC and 105thC. The A. gigas seat-belt sequence maintained only 30% amino acids pairwise identity among the 39 aligned species studied in this work. Between C10th-C11th (110thC-117thC), the sequence “DTGSHD” is mostly charged (50%) and among C11th-C12th (117thC-129thC) “THKGGDDNSAQ” shows the same frequency for charged and uncharged polar residues (36.4%). When comparing the same seat-belt region for ag-βLH (GenBank accession no. AJO68014.1) and ag-βFSH (GenBank accession no. AIA09918.1) the charges differ in: (1) C10th-C11th ag-βFSH with higher incidence of uncharged polar residues (66.4%) and C11th-C12th with prevalence of hydrophobic residues (55.6%) and (2) C10th-C11th ag-βLH with uncharged polar residues (50%) and C11th-C12th with hydrophobic residues (44.4%).
The amino acids for the α-chain, R41, T45, M46, L47, and V48 are considered the important “hot spot” residues for binding to the beta subunit. The sequence itself shows 68% similarity with H. sapiens. However, differences in the beta chain sequences of hTSH and ag-TSH such as I/T96 and I/D100 (H. sapiens and A. gigas, respectively) may contribute to the loss of affinity of ag-TSH for hTSHR. In fact, the results obtained via MD simulations of the ag-TSH:hTSHR complex suggest that the hormone from A. gigas is not able to interact with the human TSH receptor.
Moreover, the model resolution and its reliability have 2 major advantages based on the data obtained: the amino acid sequence identity of the hormone when aligned with other species and the extremely novel methodology for building the protein structures. Given this advantages for in silico simulations, we can assume that the recombinant product has potential for biological activity evaluation, as the parameters obtained in ag-TSH:sfTSHR binding show minimal difference when compared with the human model hTSH:hTSHR.
The choice of the plasmid vector pcDNA 3.4 (GenScript Biotech Corp) appeared to be an optimal choice for recombinant protein expression, in which we inserted the coding region obtained in silico without any codon optimization. Such a tool can be tested with the aim of improving expression levels, and may be an option of strategy to be used in future work with these or other proteins from A. gigas.
Regarding the expression efficiency of hTSH, the value obtained in the present work (25 mg/L) was relatively low, but in relation with cultures using adhered cells, the productivity can be high. Concerning some previously works, hTSH expressions yields reach orders of: 1.4,30 0.6,31 3.2 ± 0.64,32 and 17.8 mg/L.33 It is worth mentioning that these values only refer to productions in adherent cells. The study with ag-TSH was done in suspension production, with a final volume of 30 mL.
Therefore, it is still necessary to evaluate the characterization and purification methodologies, as the background of unwanted proteins—visualized in the chromatogram—makes it difficult to identify the heterodimer. A dissociation of the subunits was reported and visualized on SDS-PAGE after RP-HPLC. This was already evidenced34 with hTSH, showing the possibility of the reassociation and biological activity reformed. Another strategy for maintaining the biological activity and integrity of the subunits is adapting the purification methodologies to less aggressive systems, such as ion exchange followed by molecular exclusion. Nevertheless, the methodology was efficient in obtaining the recombinant hormone, with comparable values of 25 mg/L for ag-TSH vs 28 mg/L for agFSH.28
Aiming at the in vivo application of the hormone, a series of assays is necessary before a scale-up in production. These assays are immunological and biological (in vitro determination), such as ELISA (Enzyme-Linked Immunosorbent Assay), western blotting, and even radioimmunoassay (RIE). There are other more recent methodologies that can be employed to measure the biological activity in vitro35 and works that can be referenced in the in vivo evaluation, as already introduced for the recombinant hormone—agFSH—in Oreochromis niloticus.36
With the rapid development of the field of technology, the implementation of artificial intelligence tools and methodologies that expand the information about a given object of study, this work sought to combine bioinformatics techniques to identify a protein sequence, with a high degree of reliability, highlighting the results obtained in vitro. Aiming to improve the reproduction of A. gigas, this work can serve as a basis to explore other proteins, such as growth hormone (ag-GH), prolactin (ag-PRL), and chorionic gonadotrophin (ag-CG), to enhance the strategies that influence the development of the species.
Conclusions
In this work, we have identified through bioinformatic analyses the sequence of A. gigas βTSH coding sequence, in the previously reported genome of the fish. This finding was further confirmed through the RACE method, which allowed us to clone the 3’UTR of the extracted mRNA from A. gigas pituitary gland. Moreover, structural analyses showed that the 3D model of ag-TSH fulfills the main features described of this protein family. MD simulations and free energy calculations suggested the binding of ag-TSH to sfTSHR but not to the human TSHR, thus highlighting that the differences in TSHR sequences from distant species, especially in the interface region, might prevent cross-interaction. Due to the role of TSH in fish reproduction, our work paves the way toward better reproductive management of A. gigas in captivity, in association with other pituitary hormones involved on its development as well.
Supplemental Material
Supplemental material, sj-docx-1-bbi-10.1177_11779322231154148 for Molecular Cloning and AlphaFold Modeling of Thyrotropin (ag-TSH) From the Amazonian Fish Pirarucu (Arapaima gigas) by Renan Passos Freire, Jorge Enrique Hernandez-Gonzalez, Eliana Rosa Lima, Miriam Fussae Suzuki, João Ezequiel de Oliveira, Lucas Simon Torai, Paolo Bartolini and Carlos Roberto Jorge Soares in Bioinformatics and Biology Insights
Supplemental material, sj-png-3-bbi-10.1177_11779322231154148 for Molecular Cloning and AlphaFold Modeling of Thyrotropin (ag-TSH) From the Amazonian Fish Pirarucu (Arapaima gigas) by Renan Passos Freire, Jorge Enrique Hernandez-Gonzalez, Eliana Rosa Lima, Miriam Fussae Suzuki, João Ezequiel de Oliveira, Lucas Simon Torai, Paolo Bartolini and Carlos Roberto Jorge Soares in Bioinformatics and Biology Insights
Supplemental material, sj-tif-2-bbi-10.1177_11779322231154148 for Molecular Cloning and AlphaFold Modeling of Thyrotropin (ag-TSH) From the Amazonian Fish Pirarucu (Arapaima gigas) by Renan Passos Freire, Jorge Enrique Hernandez-Gonzalez, Eliana Rosa Lima, Miriam Fussae Suzuki, João Ezequiel de Oliveira, Lucas Simon Torai, Paolo Bartolini and Carlos Roberto Jorge Soares in Bioinformatics and Biology Insights
Supplemental material, sj-tif-4-bbi-10.1177_11779322231154148 for Molecular Cloning and AlphaFold Modeling of Thyrotropin (ag-TSH) From the Amazonian Fish Pirarucu (Arapaima gigas) by Renan Passos Freire, Jorge Enrique Hernandez-Gonzalez, Eliana Rosa Lima, Miriam Fussae Suzuki, João Ezequiel de Oliveira, Lucas Simon Torai, Paolo Bartolini and Carlos Roberto Jorge Soares in Bioinformatics and Biology Insights
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, process nos: 2020/16549-5 and 2020/10435-8) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasília, Brazil, project no. 305839/2021-7). Research fund for the master’s degree program (R.P.F.) by Coordenação de Aperfeiçoamento Pessoal de Nível Superior (CAPES, no. 88887.506371/2020-00).
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: Methodology: Renan Passos Freire and Jorge Enrique Hernandez-Gonzalez; Validation: Renan Passos Freire, Eliana Rosa Lima, Miriam Fussae Suzuki and João Ezequiel de Oliveira; Investigation: Lucas Simon Torati and Paolo Bartolini; Data curation: Renan Passos Freire, Jorge Enrique Hernandez-Gonzalez and Eliana Rosa Lima; Writing – original draft: Renan Passos Freire, Jorge Enrique Hernandez-Gonzalez and Carlos Roberto Jorge Soares; Writing, editing and review: Renan Passos Freire, Jorge Enrique Hernandez-Gonzalez and Paolo Bartolini; Supervision: Carlos Roberto Jorge Soares; Resources: Paolo Bartolini and Carlos Roberto Jorge Soares; All authors have read for publishing the article. Jorge Enrique Hernandez-Gonzalez acknowledges the National Laboratory for Scientific Computing (LNCC/MCTI, Brazil) for providing HPC resources of the SDumont supercomputer, which have contributed to the research results reported within this work (http://sdumont.lncc.br)
Supplemental Material: Supplemental material for this article is available online.
References
- 1. ©FAO 2012-2022. Cultured Aquatic Species Information Programme. Arapaima gigas. Cultured Aquatic Species Information Programme. Text by Nuñez J. In: FAO Fisheries and Aquaculture Division [https://www.fao.org/fishery/en/culturedspecies/arapaima_gigas/en]. Rome. Accessed September 20, 2022.
- 2. Gurdak DJ, Stewart DJ, Castello L, Arantes CC. Diversity in reproductive traits of arapaima (Arapaima spp., Muller, 1843) in Amazonian varzea floodplains: conservation implications. Aquat Conserv. 2019;29:245-257. [Google Scholar]
- 3. Castello L, Viana JP, Watkins G, Pinedo-Vasquez M, Luzadis VA. Lessons from integrating fishers of arapaima in small-scale fisheries management at the Mamirauá Reserve, Amazon. Environ Manage. 2009;43:197-209. doi: 10.1007/s00267-008-9220-5. [DOI] [PubMed] [Google Scholar]
- 4. Ferreira G, Marcovitch J, Val AL. A systematic review of the production chain of the Arapaima gigas, the giant fish of the Amazon. Manage Environ Qual Int J. 2020;31:349-363. doi: 10.1108/meq-11-2019-0238. [DOI] [Google Scholar]
- 5. Núñez J, Chu-Koo F, Berland M, et al. Reproductive success and fry production of the paiche or pirarucu, Arapaima gigas (Schinz), in the region of Iquitos, Perú. Aquacul Res. 2011;42:815-822. doi: 10.1111/j.1365-2109.2011.02886.x. [DOI] [Google Scholar]
- 6. Cahoreau C, Klett D, Combarnous Y. Structure function relationships of glycoprotein hormones and their subunits’ ancestors. Front Endocrinol (Lausanne). 2015;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Faria MT, Carvalho RF, Sevilhano TCA, et al. Isolation of the pituitary gonadotrophic alpha-subunit hormone of the giant Amazonian fish: pirarucu (Arapaima gigas). Fish Physiol Biochem. 2013;39:683-693. [DOI] [PubMed] [Google Scholar]
- 8. Szkudlinski MW, Fremont V, Ronin C, Weintraub BD. Thyroid-stimulating hormone and thyroid-stimulating hormone receptor structure-function relationships. Physiol Rev. 2002;82:473-502. [DOI] [PubMed] [Google Scholar]
- 9. Luongo C, Dentice M, Salvatore D. Deiodinases and their intricate role in thyroid hormone homeostasis. Nat Rev Endocrinol. 2019;15:479-488. [DOI] [PubMed] [Google Scholar]
- 10. Cyr D, Eales JG. Interrelationships between thyroidal and reproductive endocrine systems in fish. Rev Fish Biol Fish. 1996;6. doi: 10.1007/bf00182342. [DOI] [Google Scholar]
- 11. Deal CK, Volkoff H. The role of the thyroid axis in fish. Front Endocrinol. 2020;11:596585. doi: 10.3389/fendo.2020.596585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Habibi HR, Nelson ER, Allan ERO. New insights into thyroid hormone function and modulation of reproduction in goldfish. Gen Comp Endocrinol. 2012;175:19-26. doi: 10.1016/j.ygcen.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 13. Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691-699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 14. Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Sys Biol. 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kearse M, Moir R, Wilson A, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647-1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tamura K, Stecher G, Kumar S. MEGA 11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583-589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19:679-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Teufel F, Almagro Armenteros JJ, Johansen AR, et al. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol. 2022;40:1023-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Unni S, Huang Y, Hanson RM, et al. Web servers and services for electrostatics calculations with APBS and PDB2PQR. J Comput Chem. 2011;32:1488-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gismene C, Hernandez Gonzalez JE, Santisteban ARN, et al. Staphylococcus aureus exfoliative toxin E, oligomeric state and flip of P186: implications for its action mechanism. Int J Mol Sci. 2022;23:9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Froger A, Hall JE. Transformation of plasmid DNA into E. coli using the heat shock method. J Vis Exp. 2007;6:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hagemans D, van Belzen IA, Moran Luengo T, Rudiger SG. A script to highlight hydrophobicity and charge on protein surfaces. Front Mol Biosci. 2015;2:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vialle RA, de Souza JES, Lopes KP, et al. Whole genome sequencing of the pirarucu (Arapaima gigas) supports independent emergence of major teleost clades. Genome Biol Evol. 2018;10:2366-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Du K, Wuertz S, Adolfi M, et al. The genome of the arapaima (Arapaima gigas) provides insights into gigantism, fast growth and chromosomal sex determination system. Sci Rep. 2019;9:5293. doi: 10.1038/s41598-019-41457-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun PD, Davies DR. The cystine-knot growth-factor superfamily. Ann Rev Biophys Biomol Struct. 1995;24:269-291. [DOI] [PubMed] [Google Scholar]
- 28. Sevilhano T, Carvalho RF, Oliveira NAJ, et al. Molecular cloning and characterization of pirarucu (Arapaima gigas) follicle-stimulating hormone and luteinizing hormone beta-subunit cDNAs. PLoS ONE. 2017;12:e0183545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grossmann M, Szkudlinski MW, Wong R, Dias JA, Ji TH, Weintraub BD. Substitution of the seat-belt region of the thyroid-stimulating hormone (TSH) beta-subunit with the corresponding regions of choriogonadotropin or follitropin confers luteotropic but not follitropic activity to chimeric TSH. J Biol Chem. 1997;272:15532-15540. [DOI] [PubMed] [Google Scholar]
- 30. Damiani R, Almeida BE, Oliveira JE, Bartolini P, Ribela MT. Enhancement of human thyrotropin synthesis by sodium butyrate addition to serum-free CHO cell culture. Appl Biochem Biotechnol. 2013;171:1658-1672. [DOI] [PubMed] [Google Scholar]
- 31. Oliveira JE, Damiani R, Vorauer-Uhl K, Bartolini P, Ribela MT. Influence of a reduced CO2 environment on the secretion yield, potency and N-glycan structures of recombinant thyrotropin from CHO cells. Mol Biotechnol. 2008;39:159-166. [DOI] [PubMed] [Google Scholar]
- 32. de Mendonca F, de Oliveira JE, Bartolini P, Ribela MT. Two-step chromatographic purification of recombinant human thyrotrophin and its immunological, biological, physico-chemical and mass spectral characterization. J Chromatogr A. 2005;1062:103-112. [DOI] [PubMed] [Google Scholar]
- 33. Peroni CN, Soares CR, Gimbo E, Morganti L, Ribela MT, Bartolini P. High-level expression of human thyroid-stimulating hormone in Chinese hamster ovary cells by co-transfection of dicistronic expression vectors followed by a dual-marker amplification strategy. Biotechnol Appl Biochem. 2002;35:19-26. [DOI] [PubMed] [Google Scholar]
- 34. Carvalho CM, Oliveira JE, Almeida BE, et al. Efficient isolation of the subunits of recombinant and pituitary glycoprotein hormones. J Chromatogr A. 2009;1216:1431-1438. [DOI] [PubMed] [Google Scholar]
- 35. Sendak RA, Wang F, Geagan LB, et al. Comparison of two in vitro methods for the measurement of recombinant human TSH bioactivity. Biologicals. 2002;30:245-254. [DOI] [PubMed] [Google Scholar]
- 36. Aizen J, Kasuto H, Golan M, Zakay H, Levavi-Sivan B. Tilapia follicle-stimulating hormone (FSH): immunochemistry, stimulation by gonadotropin-releasing hormone, and effect of biologically active recombinant FSH on steroid secretion. Biol Reprod. 2007;76:692-700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-bbi-10.1177_11779322231154148 for Molecular Cloning and AlphaFold Modeling of Thyrotropin (ag-TSH) From the Amazonian Fish Pirarucu (Arapaima gigas) by Renan Passos Freire, Jorge Enrique Hernandez-Gonzalez, Eliana Rosa Lima, Miriam Fussae Suzuki, João Ezequiel de Oliveira, Lucas Simon Torai, Paolo Bartolini and Carlos Roberto Jorge Soares in Bioinformatics and Biology Insights
Supplemental material, sj-png-3-bbi-10.1177_11779322231154148 for Molecular Cloning and AlphaFold Modeling of Thyrotropin (ag-TSH) From the Amazonian Fish Pirarucu (Arapaima gigas) by Renan Passos Freire, Jorge Enrique Hernandez-Gonzalez, Eliana Rosa Lima, Miriam Fussae Suzuki, João Ezequiel de Oliveira, Lucas Simon Torai, Paolo Bartolini and Carlos Roberto Jorge Soares in Bioinformatics and Biology Insights
Supplemental material, sj-tif-2-bbi-10.1177_11779322231154148 for Molecular Cloning and AlphaFold Modeling of Thyrotropin (ag-TSH) From the Amazonian Fish Pirarucu (Arapaima gigas) by Renan Passos Freire, Jorge Enrique Hernandez-Gonzalez, Eliana Rosa Lima, Miriam Fussae Suzuki, João Ezequiel de Oliveira, Lucas Simon Torai, Paolo Bartolini and Carlos Roberto Jorge Soares in Bioinformatics and Biology Insights
Supplemental material, sj-tif-4-bbi-10.1177_11779322231154148 for Molecular Cloning and AlphaFold Modeling of Thyrotropin (ag-TSH) From the Amazonian Fish Pirarucu (Arapaima gigas) by Renan Passos Freire, Jorge Enrique Hernandez-Gonzalez, Eliana Rosa Lima, Miriam Fussae Suzuki, João Ezequiel de Oliveira, Lucas Simon Torai, Paolo Bartolini and Carlos Roberto Jorge Soares in Bioinformatics and Biology Insights