Abstract
Objective
To assess the diagnostic value and clinical significance of nucleoporin 107 (NUP107) in hepatocellular carcinoma (HCC), and explore the possible mechanisms.
Methods
The transcriptomic and clinical data of HCC patients were retrieved from The Cancer Genome Atlas (TCGA) and GEO databases. Tissue specimens were collected from HCC patients in the Guangxi area. According to the expression levels and prognostic characteristics of NUP107, ROC curves and nomogram models were constructed using the R package.
Results
NUP107 was highly expressed in 26 human cancers including HCC, and was associated with advanced HCC staging and worse prognosis. NUP107 showed satisfactory ability to predict the prognosis of HCC patients (AUC >0.8). Results of gene set enrichment analysis (GSEA) further showed that NUP107 was mainly associated with cell cycle‐related pathways such as the cell cycle, DNA replication, G2M checkpoint, E2F target, and mitotic spindle. In addition, NUP107 was also associated with immune infiltration in HCC and showed significant positive correlation with immune checkpoints (PD‐L1 and TIM‐3).
Keywords: cell cycle, hepatocellular carcinoma, immune cells, immune checkpoints, nucleoporin 107 (NUP107)
NUP107 was highly expressed among 26 human cancers including HCC, and associated with advanced HCC staging and worse prognosis. Results of GESA further showed that NUP107 was mainly associated with cell cycle‐related pathways such as the cell cycle, DNA replication, G2M checkpoint, and E2F target along with mitotic spindle. In addition, NUP107 was also associated with immune infiltration in HCC and showed significant positive correlation with immune checkpoints (PD‐L1 and TIM‐3).

1. INTRODUCTION
In 2020, primary liver cancer ranked the sixth globally in terms of incidence, and third in terms of mortality among all malignancies. 1 , 2 Despite advances in treatment modalities such as radiofrequency ablation, surgery, liver transplantation, targeted therapy, and immunotherapy, the overall survival (OS) of HCC patients is not satisfactory, mainly due to drug resistance as well as the high rate of postoperative recurrence. 3 Furthermore, most patients are diagnosed at the advanced stage of the disease, 4 which precludes the possibility of curative resection. Therefore, novel therapeutic strategies are needed for patients with advanced HCC. The immune microenvironment is critical for tumor progression, invasion, recurrence, and metastasis. 5 , 6 Therefore, understanding the role of immune‐related genes can provide new insights into the mechanisms of tumor progression, and help identify novel prognostic biomarkers and drug targets.
In eukaryotic cells, the nuclear pore complex (NPC) is located on the nuclear envelope and acts as a conduit for transport between the cytoplasm and the nucleus. 7 , 8 NPCs are assembled from approximately 30 different nucleoporins (NUPS), which are arranged in octagonal symmetry around a central transport channel. 9 , 10 , 11 An NPC consists of three subcomplexes: a loop formed by two NUP107/NUP160 complexes, a core scaffold formed by the NUP93 complex and an inner structure formed by the NUP62 complex. 12 The NUP107/160 complex is assembled into a Y‐shaped complex consisting of nine NUPS (NUP160, NUP133, NUP107, NUP96/98, NUP85, NUP43, NUP37, SEH1, and SEC13) 13 , 14 and plays an important role in the synthesis of NPCs. 15 , 16 Studies increasingly show that NPCs are closely related to tumorigenesis. 17 For example, Sakuma et al. showed that inhibiting NPC formation can selectively induce cancer cell death. 18 In addition, NUP88 is a novel cancer biomarker that is closely related to tumor progression and invasion, while NUP107 promotes survival of cervical cancer cells. 19 Furthermore, a recent study showed that NUP160‐SLC43A3 is a recurrent fusion oncogene in patients with angiosarcoma. 20
NUP107, a key component of the NUP107/160 complex, is located in the core scaffold of NPC, and is a key driver of NPC formation 16 , 21 and nucleocytoplasmic molecule trafficking. 22 During mitosis, NUP107 drives NPC assembly 23 , 24 and regulates microtubule polymerization at the kinetochore. 25 Studies have shown that loss of the NUP107 protein in zebrafish resulted in defective pharyngeal skeleton, intestinal degeneration, and defects in other tissues. 26 In addition, a strong association has been reported between childhood steroid‐resistant nephrotic syndrome and NUP107 mutations. 27 , 28 , 29 NUP107 exerts a significant effect on cell cycle arrest during DNA damage‐induced genotoxic stress as well, 30 induces apoptosis, 31 and regulates the fate of senescent cells through growth factor signaling. 32 NUP107 is a novel predictive marker of sensitivity to platinum‐based chemotherapy among patients with ovarian cancer. 33 In addition, NUP107 can also improve the ability of cervical cancer cells to resist oxidative damage. 34 These findings imply that NUP107 is closely related to tumor genesis and development. The objectives of this study were to elucidate the diagnostic value, prognostic significance and signaling pathways of NUP107 in HCC, and its role in immune infiltration.
2. MATERIALS AND METHODS
2.1. Patient datasets
Transcriptome sequencing data of 34 human cancers was retrieved from The Cancer Genome Atlas (TCGA) and GTEx through the UCSC XENA platform (https://xenabrowser.net/datapages/). 35 In addition, the transcriptomic and clinical data of 374 liver cancer tissues and 50 para‐cancerous tissues were obtained from TCGA (https://portal.gdc.cancer.gov/repository). The details are shown in Table 1. The GSE14520 (106 HCC tissues and 106 para‐cancerous tissues), GSE76427 (115 HCC tissues and 52 para‐cancerous tissues), GSE121248 (70 HCC tissues and 37 para‐cancerous tissues), GSE62232 (81 HCC tissues and 11 para‐cancerous tissues), and GSE136247 (39 HCC tissues and 30 para‐cancerous tissues) datasets were downloaded from GEO database (https://www.ncbi.nlm.nih.gov/geo/) for further validation. The details are shown in Table 2. Finally, the amplification and splice mutation data of NUP107 in two HCC datasets (INSERM, Nat Genet 2015, TCGA, Firehose Legacy) were retrieved from cBioPortal for Cancer Genomics website (http://www.cbioportal.org/).
TABLE 1.
Correlation between nucleoporin 107 (NUP107) expression and clinicopathologic in The Cancer Genome Atlas.
| Variables | Patients (n = 374) | NUP107 expression | p‐value | χ 2 | |
|---|---|---|---|---|---|
| Low | High | ||||
| n | 187 | 187 | |||
| Gender | |||||
| Female | 121 | 60 (16%) | 61 (16.3%) | 1.000 | 0 |
| Male | 253 | 127 (34%) | 126 (33.7%) | ||
| Age | |||||
| ≤60 | 177 | 76 (20.4%) | 101 (27.1%) | 0.011 | 6.44 |
| >60 | 196 | 111 (29.8%) | 85 (22.8%) | ||
| T stage | |||||
| T1 | 183 | 99 (26.7%) | 84 (22.6%) | 0.360 | 3.21 |
| T2 | 95 | 44 (11.9%) | 51 (13.7%) | ||
| T3 | 80 | 36 (9.7%) | 44 (11.9%) | ||
| T4 | 13 | 5 (1.3%) | 8 (2.2%) | ||
| Pathologic stage | |||||
| Stage I | 173 | 93 (26.6%) | 80 (22.9%) | 0.129 | 5.67 |
| Stage II | 87 | 41 (11.7%) | 46 (13.1%) | ||
| Stage III | 85 | 35 (10%) | 50 (14.3%) | ||
| Stage IV | 5 | 4 (1.1%) | 1 (0.3%) | ||
| Histologic grade | |||||
| G1 | 55 | 40 (10.8%) | 15 (4.1%) | < 0.001 | 21.51 |
| G2 | 178 | 94 (25.5%) | 84 (22.8%) | ||
| G3 | 124 | 46 (12.5%) | 78 (21.1%) | ||
| G4 | 12 | 4 (1.1%) | 8 (2.2%) | ||
| AFP (ng/mL) | |||||
| ≤400 | 215 | 127 (45.4%) | 88 (31.4%) | <0.001 | 16.63 |
| >400 | 65 | 19 (6.8%) | 46 (16.4%) | ||
| Vascular invasion | |||||
| No | 208 | 115 (36.2%) | 93 (29.2%) | 0.162 | 1.95 |
| Yes | 110 | 51 (16%) | 59 (18.6%) | ||
TABLE 2.
Correlation between nucleoporin 107 (NUP107) expression and clinicopathological features in GSE14520.
| Variables | Patients (n = 212) | NUP107 expression | p‐value | χ 2 | |
|---|---|---|---|---|---|
| Low | High | ||||
| n | 106 | 106 | |||
| Gender | |||||
| F | 29 | 13 (6.1%) | 16 (7.5%) | 0.689 | 0.16 |
| M | 183 | 93 (43.9%) | 90 (42.5%) | ||
| Age | |||||
| >60 | 37 | 16 (7.5%) | 21 (9.9%) | 0.469 | 0.52 |
| ≤60 | 175 | 90 (42.5%) | 85 (40.1%) | ||
| ALT (>/≤50 U/L) | |||||
| High | 88 | 46 (21.7%) | 42 (19.8%) | 0.676 | 0.17 |
| Low | 124 | 60 (28.3%) | 64 (30.2%) | ||
| Tumor size (>/≤5 cm) | |||||
| >5 cm | 74 | 40 (19%) | 34 (16.1%) | 0.440 | 0.6 |
| ≤5 cm | 137 | 65 (30.8%) | 72 (34.1%) | ||
| Multiple/single lobules | |||||
| Multiple | 45 | 24 (11.3%) | 21 (9.9%) | 0.737 | 0.11 |
| Single | 167 | 82 (38.7%) | 85 (40.1%) | ||
| Cirrhosis | |||||
| No | 17 | 9 (4.2%) | 8 (3.8%) | 1.000 | 0 |
| Yes | 195 | 97 (45.8%) | 98 (46.2%) | ||
| TNM stage | |||||
| I + II | 165 | 77 (36.3%) | 88 (41.5%) | 0.098 | 2.73 |
| III | 47 | 29 (13.7%) | 18 (8.5%) | ||
| BCLC stage | |||||
| 0 | 20 | 11 (5.2%) | 9 (4.2%) | 0.268 | 3.94 |
| A | 143 | 65 (30.7%) | 78 (36.8%) | ||
| B | 22 | 14 (6.6%) | 8 (3.8%) | ||
| C | 27 | 16 (7.5%) | 11 (5.2%) | ||
| AFP (>/≤300 ng/mL) | |||||
| High | 94 | 56 (26.8%) | 38 (18.2%) | 0.030 | 4.74 |
| Low | 115 | 50 (23.9%) | 65 (31.1%) | ||
2.2. Differential expression analysis
The NUP107 expression levels in 34 human cancers, HCC tumor samples, and adjacent non‐tumor samples were analyzed and compared using Student's t‐test. The expression levels of NUP107 in various HCC stages (T stage, pathological stage, histological grade and AFP level) were analyzed with the ggplot2 package using R.
2.3. Survival analysis
Survival analysis was performed by the Kaplan–Meier method along with Cox regression model. The Kaplan–Meier method was used to assess the survival of NUP107high and NUP107low patient groups across various clinical subgroups. Cox regression model was used to analyze the effect of multiple factors (age, gender, T staging, pathological staging, histological grading, AFP, and venous invasion) on the survival of HCC patients.
2.4. Diagnostic efficiency and nomogram
The diagnostic and predictive power of NUP107 was evaluated by plotting ROC curves using the pROC and timeROC R packages. The area under the curve (AUC) value was calculated, and AUC >0.8 is indicative of satisfactory discriminative ability. 36 A nomogram was further developed by adding the scores for each prognostic factor. The OS of HCC patients was predicted using the survival and rms R packages. The predictive accuracy of the nomogram was validated through calibration plots.
2.5. Screening for differentially expressed gene
The differentially expressed genes (DEGs) between the NUP107high and NUP107low expression HCC samples (cutoff value was 50%) in TCGA database were screened using the DESeq2 R package. 37 The top 10 DEGs were visualized by heatmaps.
2.6. Functional enrichment analysis
The ClusterProfiler package in R was adopted to functionally annotate the DEGs and NUP107‐related genes according to Gene Ontology (GO) terms as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. 38
2.7. Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed using the R package ClusterProfiler (3.14.3) to investigate the differences in biological functions and KEGG pathways between the NUP107high and NUP107low HCC groups (cutoff value was 50%). p < 0.05 and false discovery rate (FDR) <0.25 were the criteria for statistical significance. 38
2.8. Immunohistochemistry
Paired tumors and para‐cancerous tissues were collected from the First Affiliated Hospital of Guangxi Medical University from April 2021 to April 2022 from 40 patients with surgically resected and postoperative pathological diagnosis of HCC. The tissue specimens were formaldehyde‐fixed and paraffin‐embedded followed by cutting into sections. According to the instructions of a general two‐step IHC kit (PV‐9000, ZSGB‐Bio, China), the tissue sections were incubated overnight with anti‐NUP107 antibody (1:400, Proteintech) at 4°C, and thereafter with the enhanced enzyme‐labeled goat anti‐mouse/rabbit IgG polymer for 30 min. Finally, the tissue sections were stained with DAB chromogenic kit (ZLI‐9018, ZSGB‐Bio, China) and counterstained with hematoxylin. Two pathologists independently examined and scored the tissues.
2.9. Immune infiltration analysis
TIMER (https://cistrome.shinyapps.io/timer/) is a web server used for analyzing the correlation between immune cells and gene expression levels in the TCGA dataset. 39 , 40 The website TIMER and GSVA package in R were used to evaluate the expression level of NUP107 in different immune cells and its correlation with the degree of immune infiltration. p < 0.05 and correlation coefficient >0.300 were the criteria for statistical significance.
2.10. Statistical analysis
Statistical analysis was performed using R studio (Version 1.2.5033, R 3.6.2). NUP107 expression between tumor and para‐tumor tissues were compared by Student's t‐test. Survival analysis was performed using the Kaplan–Meier method and Cox regression model. Multivariate COX regression analysis was performed to screen for the prognostic factors. Diagnostic value was assessed by ROC analysis. Spearman's correlation test was performed to analyze the correlation between two groups. p < 0.05 was considered statistically significant.
3. RESULTS
3.1. NUP107 is highly expressed in human cancers and HCC
We analyzed NUP107 mRNA levels across 33 human cancers and the corresponding normal tissues, and found that NUP107 was significantly elevated in 26 cancer types, such as liver, bladder, colorectal, breast, prostate, lung, and thyroid cancers. However, no significant difference was observed in NUP107 expression between the tumor and normal tissues in renal clear cell carcinoma, ovarian serous, pheochromocytoma, paraganglioma, and uterine corpus endometrioid carcinoma (Figure 1A). We also analyzed the genome and copy number of NUP107 in two HCC datasets in the cBioPortal for Cancer Genomics website (INSERM, Nat Genet 2015, TCGA, Firehose Legacy), and found that the frequency of NUP107 gene amplification and splice mutation was 1.3% (Figure 1B). NUP107 expression was also significantly higher in the HCC tissues compared to the paired/unpaired normal liver tissues in TCGA, GSE14520, GSE76427, GSE121248, GSE62232, and GSE136247 datasets (Figure 1C–I).
FIGURE 1.

Nucleoporin 107 (NUP107) is highly expressed in human cancers and hepatocellular carcinoma (HCC). (A) Expression levels of NUP107 in 31 different human cancers, ns: p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001. (B) NUP107 expression profile in the cBioPortal OncoPrint plot. The expression levels of NUP107 in (C) 370 HCC tissues and 50 para‐carcinoma tissues in The Cancer Genome Atlas (TCGA) LIHC dataset, (D) 50 paired HCC tissues and para‐carcinoma tissues in TCGA LIHC dataset, (E) 106 HCC tissues and 106 para‐carcinoma tissues in GSE14520 dataset, (F) 115 HCC tissues and 52 para‐carcinoma tissues in GSE76427 dataset, (G) 70 HCC tissues and 37 para‐carcinoma tissues in GSE121248 dataset, (H) 81 HCC tissues and 11 para‐carcinoma tissues in GSE62232 dataset, (I) 39 HCC tissues and 30 para‐carcinoma tissues in GSE136247 dataset.
3.2. High NUP107 expression portends poor prognosis in patients with HCC
To assess the prognostic relevance of NUP107 in patients with HCC, we analyzed its expression levels across different HCC stages (T‐stage, pathological stage, and histological grading) and AFP levels. NUP107 overexpression was related to more advanced HCC stages (Figure 2A–C), as well as higher AFP levels (Figure 2D). In addition, NUP107 overexpression was associated with worse OS (p = 0.011), disease‐specific survival (DSS) (p = 0.032), and progression‐free interval (p < 0.001) in TCGA (Figure 2E–G). In the GSE14520 dataset, NUP107 overexpression correlated with worse OS (p = 0.003) and relapse‐free survival (RFS) (p = 0.009) (Figure 2H,I). Univariate analysis further showed that NUP107 (high versus low, HR 1.624, p < 0.001), T staging (T1 and T2 versus T3 and T4, HR 2.598, p < 0.001), and pathological staging (stages I and II versus III and IV, HR 2.504, p < 0.001) were significantly associated with poor prognosis (Figure 3A). Multivariate analysis demonstrated that NUP107 overexpression was an independent risk factor for HCC (p < 0.05) (Figure 3B), especially in the male (p < 0.001), female (p = 0.004), T1 and T2 (p = 0.004), T3 and T4 (p < 0.001), stage I/II (p = 0.006), stage III/IV (p = 0.001), vascular invasion (p = 0.023), and no vascular invasion (p = 0.005) subgroups (Figure 3C–J). Taken together, NUP107 overexpression is a risk factor for HCC.
FIGURE 2.

High expression of nucleoporin 107 (NUP107) was associated with more advanced hepatocellular carcinoma staging and poor prognosis. Dot plots show the expression levels of NUP107 in different T‐stages (A), pathological stages (B), histological grades (C), and AFP levels (D). Kaplan–Meier curves showing (E) overall survival (OS), (F) disease‐specific survival and (G) PFS in The Cancer Genome Atlas (TCGA), and (H) OS and (I) relapse‐free survival in GSE14520, * p < 0.05; ** p < 0.01; *** p < 0.001.
FIGURE 3.

Prognostic value of nucleoporin 107 (NUP107) in hepatocellular carcinoma (HCC). (A) Univariate Cox regression model. (B) Multivariate Cox regression model. Kaplan–Meier survival curves of NUP107high and NUP107low patients in (C) male, (D) female, (E) T1 and T2, (F) T3 and T4, (G) stage I and II, (H) stage III and IV, (I) vascular invasion, and (J) non‐vascular invasion subgroups.
3.3. Diagnostic and predictive ability of NUP107 in HCC
The diagnostic and predictive utility of NUP107 for HCC was evaluated by ROC analysis. NUP107 exhibited satisfactory diagnostic ability (AUC >0.8) in several datasets, including TCGA (AUC = 0.914, Figure 4A), GSE14520 (AUC = 0.946, Figure 4B), GSE76427 (AUC = 0.832, Figure 4C), GSE121248 (AUC = 0.855, Figure 4D), in GSE62232 (AUC = 0.889, Figure 4E), and GSE136247 (AUC = 0.879, Figure 4F). In addition, the time‐dependent ROC curves constructed using TCGA data also indicated good predictive ability, with AUC values above 0.6 for 1‐, 3‐, and 5‐year OS (Figure 4H). More patients in the high‐risk score group reached the end event in a shorter duration (Figure 4G). To further predict 1‐, 3‐, and 5‐year survival in patients with HCC, we developed a nomogram on the basis of gender, T‐stage, pathological stage, vascular invasion, and NUP107 expression level (Figure 4I). We obtained the total score by adding the scores for each prognostic factor, and the OS of HCC patients was predicted by determining the probability of the endpoint event by the total score corresponding to the outcome axis. NUP107 expression level was a better predictor of prognosis compared to T‐stage or vascular invasion. The calibration plots of the prediction model suggested that the predicted outcome was less biased (Figure 4J).
FIGURE 4.

Predictive ability of nucleoporin 107 (NUP107) for hepatocellular carcinoma (HCC). (A–F) Diagnostic ROC curves in The Cancer Genome Atlas (TCGA), GSE14520, GSE76427, GSE121248, GSE62232, and GSE136247. (G) Scatter plot of survival status and NUP107 expression heat map corresponding to HCC patients sorted by risk score. (H) Predictive power of NUP107 for 1‐, 3‐, and 5‐year overall survival (OS) in HCC patients by time‐dependent ROC analysis. (I) Prediction of 1‐, 3‐, and 5‐year OS by column line plots. (J) Calibration plots were used to validate the column line graph model.
3.4. Validation of NUP107 using patient samples
The results obtained so far were further validated on 40 paired HCC and para‐tumor tissues collected from the First Affiliated Hospital of Guangxi Medical University. Compared to the adjacent tissues, the in situ expression of NUP107 in HCC tissues was stronger (Figure 6A), which corresponded to significantly higher IHC scores in the latter (p < 0.001, Figure 6B). Furthermore, NUP107 showed satisfactory diagnostic performance in the Guangxi cohort (AUC = 0.831, Figure 6C). We also verified that NUP107 overexpression was associated with BCLC staging of more advanced HCC in this cohort (p < 0.01, Figure 6D).
FIGURE 6.

Validation of nucleoporin 107 (NUP107) in liver cancer patients in Guangxi. (A) Representative immunohistochemistry (IHC) images showing in situ expression of NUP107 in hepatocellular carcinoma (HCC) and adjacent liver tissues. (B) Violin chart showing the immunostaining scores of NUP107 in HCC and adjacent liver tissues in the Guangxi cohort. (C) Diagnostic ROC curve of NUP107 for the Guangxi cohort. (D) Expression level of NUP107 in the different BCLC stages.
3.5. DEG identification and functional enrichment analysis
We identified 1260 DEGs between the NUP107high and NUP107low HCC samples in TCGA cohort, including 1080 upregulated and 180 downregulated genes (Figure 5A). The top 5 upregulated/downregulated genes are shown in the heat map in Figure 5B. The DEGs were functionally annotated by GO and KEGG functional enrichment analyses. The significantly enriched biological process (BP) consisted of humoral immune response, acute inflammatory response and digestion, the cellular components (CC) included HDL particles, immunoglobulin complexes and plasma lipoprotein particles, and the significant molecular functions (MF) were passive transmembrane transporter protein activity, channel activity, and hormonal activity (Figure 5C,D). The prominent KEGG pathways associated with the DEGs included neuroactive ligand–receptor interactions, mineral uptake and taste transduction.
FIGURE 5.

Identification and functional annotation of differentially expressed gene (DEGs). (A) Volcano plot showing the DEGs between NUP107high and NUP107low groups. (B) Heat map showing the top five upregulated and downregulated genes. (C) Bubble plot of the significantly enriched Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. (D) Circle diagram showing the GO and KEGG terms corresponding to the DEGs.
3.6. Results of GSEA
To further understand the BPs associated with NUP107 and identify the key signaling pathways involved in HCC, we performed GSEA between the NUP107 expression groups. There were several pathways in the Molecular Signature Database (MSigDB) (C2.cp.v7.2.symbols.gmt and h.all.v7.2.symbols.gmt) that showed significant differences between the NUP107high and NUP107low groups (FDR <0.05, ADJ p < 0.05). We ranked the signaling pathways on the basis of the normalized enrichment score and obtained the signaling pathways that were most significantly enriched (Figure 7A,D). The five KEGG‐annotated pathways positively associated with high NUP107 expression were ECM receptor interaction, axon guidance, neuroactive ligand–receptor interaction, cell cycle and DNA replication (Figure 7B), and those with a positive association with low NUP107 expression were fatty acid metabolism, retinol metabolism, drug metabolism cytochrome P450, metabolism of xenobiotics by cytochrome P450 and peroxisome (Figure 7C). As shown in Figure 7E, the top five HALLMARK‐related annotations associated with high NUP107 expression were G2/M checkpoint, epithelial mesenchymal transition, E2F target, mitotic spindle, and inhibition of KRAS signaling. Furthermore, adipogenesis, fatty acid, bile acid and xenobiotic metabolism, as well as oxidative phosphorylation were the top 5 HALLMARK terms positively correlated to low NUP107 expression (Figure 7F). These results indicate that overexpression of NUP107 may interfere with cell cycle and mitosis in HCC.
FIGURE 7.

Enrichment plots and normalized enrichment score (NES) visualization of gene set enrichment analysis. (A) Bar graph showing the NES values of the top five Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways positively and negatively correlated with nucleoporin 107 (NUP107). Enrichment plots showing the top five (B) positively and (C) negatively correlated KEGG pathways. (D) Bar graphs showing the NES values of the top five HALLMARK pathways positively and negatively correlated with NUP107. Enrichment plots showing the top five (E) positively and (F) negatively associated HALLMARK pathways.
3.7. NUP107 and its related genes are strongly correlated with cell cycle genes in HCC
A protein–protein interaction network of the NUP107‐related genes was constructed through the STRING website. NUP133, NUP85, NUP37, NUP96/98, NUP160, SEC13, SEH1L, NUP43, NUP155, and NUP93 were identified as the 10 genes that interacted most with NUP107 (Figure 9A,B). Eight of these 10 functional partners have also been identified by Lutzmann et al. 14 In addition, NUP133, NUP160, NUP37, NUP107, NUP96/98, NUP43, SEH1, NUP85, and SEC13 are part of the NUP107/160 complex. According to GO analysis, the most significantly enriched BP terms among the NUP107‐related genes were post‐transcriptional gene silencing, viral translocation, tRNA translocation and regulation of tRNA export from the nucleus, the main CC terms included nuclear pore, other organisms, host cells and nuclear pore outer ring, and the MF terms were structural components of the nuclear pore, promoter‐specific chromatin binding, signal sequence binding, and nuclear localization sequence binding. Furthermore, amyotrophic lateral sclerosis, RNA translocation, and mTOR signaling pathway were the KEGG pathways that were most significantly enriched (Figure 9C,D). The above results suggested that NUP107 may influence the progression of HCC via regulation of the cell cycle.
FIGURE 9.
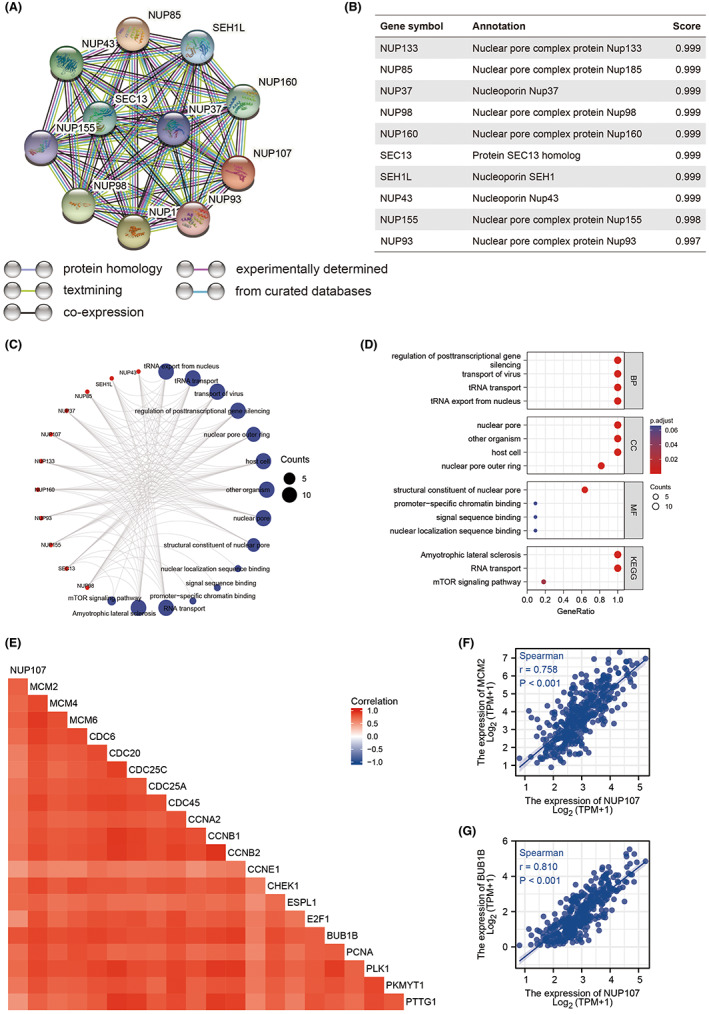
Nucleoporin 107 (NUP107) and its related genes are strongly correlated with cell cycle genes in HCC. (A) PPI network of NUP107‐related genes. (B) Annotation and correlation coefficients of 10 NUP107‐related genes. (C, D) Enriched GO terms and KEGG pathways for the NUP107‐related genes. (E) Correlation matrix showing the correlation between NUP107 and cell cycle regulatory genes including (F) BUB1B and (G) MCM2.
Based on the results of GO and KEGG functional enrichment analysis, we assessed the relationship of NUP107 with cell cycle regulatory genes involved in HCC progression. 41 As shown in Figure 9E, BUB1B, CDC6, CDC20, CDC25A, CDC25C, CDC45, PLK1, MCM6, CCNB1, CCNB2, CHEK1, ESPL1, E2F1, MCM4, PTTG1, PCNA, CCNE1, PKMYT1, MCM2, and CCNA2 were strongly correlated (r > 0.5, p < 0.001) with NUP107, especially BUB1B (r = 0.810, p < 0.001) and MCM2 42 (r = 0.758, p < 0.001) (Figure 9F,G). Thus, NUP107 plays a role in the regulation of the HCC cell cycle.
The expression of these genes in TCGA database was further analyzed, which showed that all 10 genes were significantly upregulated in HCC tissues compared to the para‐tumor tissues (p < 0.001; Figure 8A). Furthermore, NUP160 (p = 0.006), NUP133 (p = 0.042), NUP98 (p = 0.003), NUP85 (p < 0.001) NUP43 (p = 0.004), NUP37 (p < 0.001), SEH1 (p = 0.007), SEC13 (p = 0.002), NUP155 (p = 0.001), and NUP93 (p = 0.026) were significantly associated with the OS of HCC patients (Figure 8B–K). Taken together, NUP107 and other NUP107/160 complex‐related genes are prognostic indicators for HCC.
FIGURE 8.
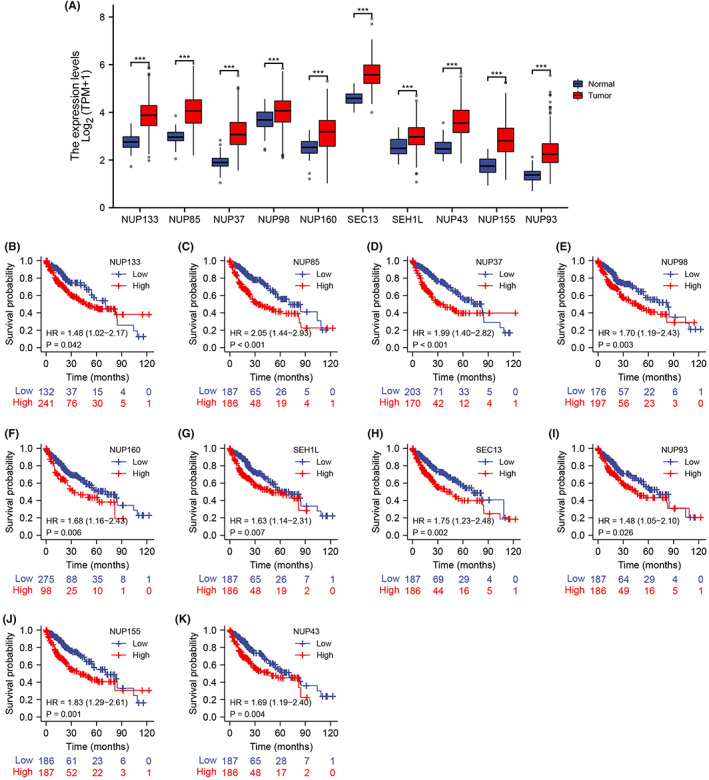
(A) Expression levels of 10 functional partners of nucleoporin 107 in hepatocellular carcinoma (HCC) and para‐carcinoma tissues. Survival curves of HCC patients in The Cancer Genome Atlas cohort demarcated in the basis of (B) NUP133, (C) NUP85, (D) NUP37, (E) NUP98, (F) NUP160, (G) SHE1L, (H) SEC13, (I) NUP93, (J) NUP155, and (K) NUP43 expression. * p < 0.05; ** p < 0.01; *** p < 0.001.
3.8. Role of NUP107 in immune infiltration
HCC is closely related to immune responses during its onset, growth, metastasis, and treatment. 43 The relationship between NUP107 expression and immune infiltration in HCC was explored through GSVA package in R. We observed a higher infiltration of CD8+ T cells, mast cells, cytotoxic cells, dendritic cells (DCs), plasmacytoid dendritic cells (pDCs), neutrophils, NK cells, and γδ T cells in the NUP107low group, whereas NUP107 overexpression was associated with higher infiltration of NK CD56bright cells, NK cells, T helper cells, central memory T cells, follicular helper T cells, and Th2 cells (Figure 10A,B). Furthermore, NUP107 expression correlated positively with the infiltration of Th2 cells (r = 0.482, p < 0.001; Figure 10D), T helper cells (r = 0.410, p < 0.001; Figure 10E), central memory T cells, follicular helper T cells, eosinophils, NK CD56bright cells as well as activated DCs (Figure 10C), but negatively with cytotoxic T cells (r = −0.355, p < 0.001; Figure 10F), pDCs (r = −0.374, p < 0.001; Figure 10G), immature DCs, B cells, T cells, Tregs, NK CD56dim cells, NK cells, mast cells, Th17 cells, CD8 T cells, γδ T cells, neutrophils, and DCs (Figure 10C). The Th1/Th2 ratio is skewed in liver, lung, and breast tumors, 44 , 45 , 46 and the predominance of the Th2 subtype 47 and T helper cells in general are associated with immune escape of tumor cells. 48 Therefore, our findings suggest that NUP107 may promote tumor growth by increasing the infiltration of Th2 cells, central memory T cells, follicular helper T cells, T helper cells and other immunosuppressive populations, and decreasing that of cytotoxic T cells, pDCs, immature DCs, B cells, T cells, Tregs, and NK cells.
FIGURE 10.

Correlation between nucleoporin 107 (NUP107) expression and immune infiltration in hepatocellular carcinoma. (A, B) Violin plots showing the degree of infiltration of different immune cells in the NUP107high and NUP107low groups. (C) Bubble plots showing the correlation between NUP107 and different immune cells. (D–G) Scatter plots showing the correlation between NUP107 expression levels and (D) Th2 cells, (E) T helper cells, (F) cytotoxic T cells, and (G) plasma DCs. ns: p ≥ 0.05; * p < 0.05; ** p < 0.01; *** p < 0.001.
Chemokines along with their receptors exert a pivotal effect on the anti‐tumor immune response. 49 We found that NUP107 was positively correlated with several chemokines and the specific receptors (Figure S2A,S2D), including CCL28 (r = 0.377, p < 0.001), CXCL8 (r = 0.310, p < 0.001), CCR8 (r = 0.455, p < 0.001), and CXCR4 (r = 0.392, p < 0.001) (FigureS2B, S2C, S2E, S2F). Immune checkpoint inhibitors (ICIs) are able to prolong the survival of patients with cancers, and the response to ICIs relies on the expression levels of immune checkpoint genes. 50 Therefore, we next examined the correlation between NUP107 and HCC‐associated immune checkpoint genes, including PD‐1, PD‐L1, PD‐L2, LAG3, CTLA4, and TIM3 (Figure S1A). NUP107 was correlated (r > 0.3) with PD‐L1 (Figure S1C) and TIM3 (Figure S1D), and the correlation matrix is shown in Figure S1B. To summarize, NUP107 may promote HCC growth by dampening the immune response.
4. DISCUSSION
NUP107 is an important component of the core scaffold of the NUP160/NUP107 complex, which plays a major role in NPC assembly and cell cycle regulation. 32 Several studies have demonstrated the overexpression of NUP107 in cervical cancer, 19 colon cancer, lung cancer, and other tumors, although little is known regarding its role in liver cancer. In the present study, we analyzed NUP107 expression levels and its prognostic relevance in several HCC datasets, and explored the underlying mechanisms using bioinformatics tools. NUP107 was significantly upregulated in most human tumor tissues, including HCC, compared to the corresponding normal tissues, and correlated to more advanced HCC staging and worse prognosis. In addition, NUP107 expression had satisfactory diagnostic efficiency and predicted 1‐, 3‐, and 5‐year survival of HCC patients with high accuracy. The better ROC curves of NUP107 compared to some potential prognostic markers for HCC, such as CDK4 51 and HMGA1, 52 again demonstrate the satisfactory diagnostic value of NUP107. Thus, NUP107 is a prognostic and diagnostic biomarker for HCC.
Tumor cells differ from normal cells in terms of excessive proliferation, abnormal differentiation, and impaired apoptosis. 53 Therefore, we also explored the molecular mechanisms underlying the role of NUP107 in HCC. 54 GO and KEGG analyses indicated that NUP107 is enriched in cell cycle‐related pathways, and the results of GSEA also revealed significant association with the cell cycle, DNA replication, G2M checkpoint, E2F target, and mitotic spindle. The G2/M checkpoint prevents DNA‐damaged cells from entering the mitotic (M) phase 55 and is regulated by the RB‐E2F complex that determines the timing and accuracy of cell cycle replication. 56 NUP107 was closely associated with cell cycle regulatory genes such as BUB1B and MCM2, suggesting that NUP107 may regulate the cell cycle in HCC.
Targeted therapies 57 and immunotherapy 58 have significantly improved the outcomes in HCC patients. Therefore, the exploration of potential therapeutic targets and immune‐related molecules for HCC is crucial for its treatment. Tumor initiation, growth, progression, and metastasis are closely related to the tumor microenvironment, 59 which also includes stromal cells like fibroblasts and infiltrating immune cells such as macrophages. 60 The immune response to tumor cells can be a double‐edged sword, which can activate anti‐tumor pathways but also create an immunosuppressive microenvironment. 61 Studies have shown that tumor infiltrating lymphocytes (TILs) can predict anterior lymph node status and survival in cancer patients. 62 We observed higher infiltration of CD8+ T cells, cytotoxic T cells, DCs, mast cells, neutrophils, NK cells, pDCs, and γδ T cells in the NUP107low group, and that of NK CD56bright cells, NK cells, T helper cells, central memory T cells, follicular helper T cells, and Th2 cells in the NUP107high group. Thus, NUP107 may influence the progression of HCC by regulating the tumor immune microenvironment.
Chemokines control the migration and recruitment of immune cells. 63 , 64 We found that NUP107 was positively correlated with chemokines such as CCL28 and CXCL8 (r = 0.310, p < 0.001), and the chemokine receptors CCR8 and CXCR4 suggesting that NUP107 overexpression may recruit immune cells to the tumor tissues. Activated CCL28 binds to CCR3 and CCR10, and can control the targeted migration of TILs, Tregs, and cancer‐associated stellate cells. 65 Furthermore, CCR8 recruits TAMs and Tregs, and promotes tumor angiogenesis. 66 The CXCR4‐CXCL12 axis attracts Tregs and pDCs to enhance tumor growth, which may be one of the mechanisms through which NUP107 promotes HCC growth. 67
Targeted therapies against immune checkpoints, including programmed cell death‐1 (PD‐1)/programmed apoptosis ligand 1 (PD‐L1) and T‐cell immunoglobulin and mucin structural domain molecule 3 (Tim‐3), have been effective against solid tumors. 68 The Tim‐3 inhibitor cabolimab and the PD‐L1 inhibitor atezumab are promising options for HCC patients. 69 , 70 The expression of immune checkpoints in tumor tissues regulates the degree of immune cell infiltration and also determines the response to immunotherapy. 71 In the present study, we showed that NUP107 is positively correlated with PD‐L1 and Tim‐3, which suggests that the therapeutic effect of ICIs in HCC patients may be enhanced by targeting NUP107.
To summarize, we found that NUP107 is upregulated in HCC and portends poor prognosis, and can predict the survival of HCC patients with reasonable accuracy. NUP107 may exert its oncogenic effects on HCC via the regulation of cell cycle and immune infiltration. Our study has some limitations that ought to be considered. Firstly, the function of NUP107 in HCC cells needs to be validated by in vitro assays. Secondly, the mechanism by which NUP107 controls immune infiltration needs to be elucidated by animal experiments.
5. CONCLUSION
NUP107 correlated with the OS, RFS, and HCC staging in HCC patients and showed diagnostic accuracy. In addition, NUP107 was associated with cell cycle pathways, immune cell infiltration, and immune checkpoints. Thus, NUP107 is a reliable diagnostic and prognostic biomarker for HCC, as well as a potential therapeutic target. Our findings also provide new insights into the mechanisms associated with immune cell infiltration in HCC.
AUTHOR CONTRIBUTIONS
Ju‐sen Nong: Writing – original draft (equal). Xin Zhou: Writing – original draft (equal). Jun‐qi Liu: Data curation (supporting). Jian‐zhu Luo: Software (supporting). Jia‐mi Huang: Software (supporting). Hai‐xiang Xie: Validation (supporting). Ke‐jian Yang: Validation (supporting). Jing Wang: Resources (supporting). Xinping Ye: Resources (supporting). Tao Peng: Supervision (lead).
FUNDING INFORMATION
Key R&D Plan of Qingxiu District, Nanning (No. 2020056); Innovation Project of Guangxi Graduate Education (YCSM2022227, 02603222064X).
CONFLICT OF INTEREST STATEMENT
The authors have declared that there are no competing interests.
ETHICS STATEMENT
This study had acquired the approval of the Ethics Committee of the first affiliated hospital of Guangxi Medical University before specimen collection. Approval Number: 2022‐KY‐E‐159. Written informed consent was provided by each patient.
Supporting information
Figure S1.
Figure S2.
ACKNOWLEDGMENTS
The authors thank the contributors of GSE14520 and TCGA database for sharing the HCC dataset on open access.
Nong J‐s, Zhou X, Liu J‐q, et al. Nucleoporin 107 is a prognostic biomarker in hepatocellular carcinoma associated with immune infiltration. Cancer Med. 2023;12:10990‐11009. doi: 10.1002/cam4.5807
Ju‐sen Nong and Xin Zhou contribute equally to this work.
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. [DOI] [PubMed] [Google Scholar]
- 3. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589‐604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fitzmaurice C, Allen C, Barber RM, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability‐adjusted life‐years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3(4):524‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bull CJ, Bell JA, Murphy N, et al. Adiposity, metabolites, and colorectal cancer risk: mendelian randomization study. BMC Med. 2020;18(1):396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Llovet JM, Castet F, Heikenwalder M, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19(3):151‐172. [DOI] [PubMed] [Google Scholar]
- 7. Hetzer MW, Walther TC, Mattaj IW. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol. 2005;21:347‐380. [DOI] [PubMed] [Google Scholar]
- 8. Corbett AH, Silver PA. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61(2):193‐211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alber F, Dokudovskaya S, Veenhoff LM, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450(7170):695‐701. [DOI] [PubMed] [Google Scholar]
- 10. Beck M, Förster F, Ecke M, et al. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306(5700):1387‐1390. [DOI] [PubMed] [Google Scholar]
- 11. Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158(5):915‐927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin DH, Stuwe T, Schilbach S, et al. Architecture of the symmetric core of the nuclear pore. Science. 2016;352(6283):aaf1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Belgareh N, Rabut G, Baï SW, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154(6):1147‐1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. Modular self‐assembly of a Y‐shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21(3):387‐397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harel A, Orjalo AV, Vincent T, et al. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11(4):853‐864. [DOI] [PubMed] [Google Scholar]
- 16. Walther TC, Alves A, Pickersgill H, et al. The conserved Nup107‐160 complex is critical for nuclear pore complex assembly. Cell. 2003;113(2):195‐206. [DOI] [PubMed] [Google Scholar]
- 17. de Las Heras JI, Batrakou DG, Schirmer EC. Cancer biology and the nuclear envelope: a convoluted relationship. Semin Cancer Biol. 2013;23(2):125‐137. [DOI] [PubMed] [Google Scholar]
- 18. Sakuma S, Raices M, Borlido J, Guglielmi V, Zhu EYS, D'Angelo MA. Inhibition of nuclear pore complex formation selectively induces cancer cell death. Cancer Discov. 2021;11(1):176‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shi R, Xu L, Huang L, Cheng JX. Nucleoporin 107 promotes the survival of tumor cells in cervical cancers. Gynecol Obstet Invest. 2020;85(1):41‐52. [DOI] [PubMed] [Google Scholar]
- 20. Shimozono N, Jinnin M, Masuzawa M, et al. NUP160‐SLC43A3 is a novel recurrent fusion oncogene in angiosarcoma. Cancer Res. 2015;75(21):4458‐4465. [DOI] [PubMed] [Google Scholar]
- 21. Boehmer T, Enninga J, Dales S, Blobel G, Zhong H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc Natl Acad Sci U S A. 2003;100(3):981‐985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baï SW, Rouquette J, Umeda M, et al. The fission yeast Nup107‐120 complex functionally interacts with the small GTPase ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol Cell Biol. 2004;24(14):6379‐6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zuccolo M, Alves A, Galy V, et al. The human Nup107‐160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26(7):1853‐1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang HJ, Haraguchi T, Hiraoka Y. A nucleoporin that facilitates meiotic kinetochore reorganization. Cell Cycle. 2016;15(3):307‐308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BM, Dasso M. The Nup107‐160 complex and gamma‐TuRC regulate microtubule polymerization at kinetochores. Nat Cell Biol. 2010;12(2):164‐169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng X, Yang S, Han Y, et al. Loss of zygotic NUP107 protein causes missing of pharyngeal skeleton and other tissue defects with impaired nuclear pore function in zebrafish embryos. J Biol Chem. 2012;287(45):38254‐38264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rosti RO, Sotak BN, Bielas SL, et al. Homozygous mutation in NUP107 leads to microcephaly with steroid‐resistant nephrotic condition similar to Galloway‐Mowat syndrome. J Med Genet. 2017;54(6):399‐403. [DOI] [PubMed] [Google Scholar]
- 28. Miyake N, Tsukaguchi H, Koshimizu E, et al. Biallelic mutations in nuclear pore complex subunit NUP107 cause early‐childhood‐onset steroid‐resistant nephrotic syndrome. Am J Hum Genet. 2015;97(4):555‐566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Park E, Ahn YH, Kang HG, Miyake N, Tsukaguchi H, Cheong HI. NUP107 mutations in children with steroid‐resistant nephrotic syndrome. Nephrol Dial Transplant. 2017;32(6):1013‐1017. [DOI] [PubMed] [Google Scholar]
- 30. Jagot‐Lacoussiere L, Faye A, Bruzzoni‐Giovanelli H, Villoutreix BO, Rain JC, Poyet JL. DNA damage‐induced nuclear translocation of Apaf‐1 is mediated by nucleoporin Nup107. Cell Cycle. 2015;14(8):1242‐1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Banerjee HN, Gibbs J, Jordan T, Blackshear M. Depletion of a single nucleoporin, Nup107, induces apoptosis in eukaryotic cells. Mol Cell Biochem. 2010;343(1‐2):21‐25. [DOI] [PubMed] [Google Scholar]
- 32. Kim SY, Kang HT, Choi HR, Park SC. Reduction of Nup107 attenuates the growth factor signaling in the senescent cells. Biochem Biophys Res Commun. 2010;401(1):131‐136. [DOI] [PubMed] [Google Scholar]
- 33. Alanee S, Delfino K, Wilber A, Robinson K, Brard L, Semaan A. Single nucleotide variant in nucleoporin 107 may be predictive of sensitivity to chemotherapy in patients with ovarian cancer. Pharmacogenet Genomics. 2017;27(7):264‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shen Q, Yu M, Jia JK, Li WX, Tian YW, Xue HZ. Possible molecular markers for the diagnosis of pancreatic ductal adenocarcinoma. Med Sci Monit. 2018;24:2368‐2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vivian J, Rao AA, Nothaft FA, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol. 2017;35(4):314‐316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hosmer DW, Lemeshow S, Sturdivant RX. Assessing the fit of the model. In: Applied Logistic Regression. Wiley Series in Probability and Statistics; 2013:153‐225. [Google Scholar]
- 37. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor‐infiltrating immune cells. Cancer Res. 2017;77(21):e108‐e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liping X, Jia L, Qi C, Liang Y, Dongen L, Jianshuai J. Cell cycle genes are potential diagnostic and prognostic biomarkers in hepatocellular carcinoma. Biomed Res Int. 2020;2020:6206157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou X, Luo J, Xie H, et al. MCM2 promotes the stemness and sorafenib resistance of hepatocellular carcinoma cells via hippo signaling. Cell Death Discov. 2022;8(1):418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Makarova‐Rusher OV, Medina‐Echeverz J, Duffy AG, Greten TF. The yin and yang of evasion and immune activation in HCC. J Hepatol. 2015;62(6):1420‐1429. [DOI] [PubMed] [Google Scholar]
- 44. Zhu Y, Yang J, Xu D, et al. Disruption of tumour‐associated macrophage trafficking by the osteopontin‐induced colony‐stimulating factor‐1 signalling sensitises hepatocellular carcinoma to anti‐PD‐L1 blockade. Gut. 2019;68(9):1653‐1666. [DOI] [PubMed] [Google Scholar]
- 45. Anichini A, Perotti VE, Sgambelluri F, Mortarini R. Immune escape mechanisms in non small cell lung cancer. Cancer. 2020;12(12):3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Markosyan N, Chen EP, Evans RA, Ndong V, Vonderheide RH, Smyth EM. Mammary carcinoma cell derived cyclooxygenase 2 suppresses tumor immune surveillance by enhancing intratumoral immune checkpoint activity. Breast Cancer Res. 2013;15(5):R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Basu A, Ramamoorthi G, Albert G, et al. Differentiation and regulation of T(H) cells: a balancing act for cancer immunotherapy. Front Immunol. 2021;12:669474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang D, Chen J, Yang L, et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation‐induced cell death. Nat Immunol. 2018;19(10):1112‐1125. [DOI] [PubMed] [Google Scholar]
- 49. Dyer DP, Medina‐Ruiz L, Bartolini R, et al. Chemokine receptor redundancy and specificity are context dependent. Immunity. 2019;50(2):378‐389.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sharma P, Hu‐Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168(4):707‐723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wei ZL, Zhou X, Lan CL, et al. Clinical implications and molecular mechanisms of cyclin‐dependent kinases 4 for patients with hepatocellular carcinoma. BMC Gastroenterol. 2022;22(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wei YG, Yang CK, Wei ZL, et al. High‐mobility group AT‐hook 1 served as a prognosis biomarker and associated with immune infiltrate in hepatocellular carcinoma. Int J Gen Med. 2022;15:609‐621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Woodruff MF. Cellular heterogeneity in tumours. Br J Cancer. 1983;47(5):589‐594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342‐348. [DOI] [PubMed] [Google Scholar]
- 55. Yasutis KM, Kozminski KG. Cell cycle checkpoint regulators reach a zillion. Cell Cycle. 2013;12(10):1501‐1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kent LN, Leone G. The broken cycle: E2F dysfunction in cancer. Nat Rev Cancer. 2019;19(6):326‐338. [DOI] [PubMed] [Google Scholar]
- 57. Damaskos C, Garmpis N, Dimitroulis D, et al. Targeted therapies for hepatocellular carcinoma treatment: a new era ahead—a systematic review. Int J Mol Sci. 2022;23(22):14117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sperandio RC, Pestana RC, Miyamura BV, Kaseb AO. Hepatocellular carcinoma immunotherapy. Annu Rev Med. 2022;73:267‐278. [DOI] [PubMed] [Google Scholar]
- 59. Wang M, Chang M, Li C, et al. Tumor‐microenvironment‐activated reactive oxygen species amplifier for enzymatic cascade cancer starvation/chemodynamic /immunotherapy. Adv Mater. 2022;34(4):e2106010. [DOI] [PubMed] [Google Scholar]
- 60. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79(18):4557‐4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jiang X, Wang J, Deng X, et al. Role of the tumor microenvironment in PD‐L1/PD‐1‐mediated tumor immune escape. Mol Cancer. 2019;18(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Azimi F, Scolyer RA, Rumcheva P, et al. Tumor‐infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol. 2012;30(21):2678‐2683. [DOI] [PubMed] [Google Scholar]
- 63. Ozga AJ, Chow MT, Luster AD. Chemokines and the immune response to cancer. Immunity. 2021;54(5):859‐874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol. 2014;32:659‐702. [DOI] [PubMed] [Google Scholar]
- 65. Lazarus NH, Kunkel EJ, Johnston B, Wilson E, Youngman KR, Butcher EC. A common mucosal chemokine (mucosae‐associated epithelial chemokine/CCL28) selectively attracts IgA plasmablasts. J Immunol. 2003;170(7):3799‐3805. [DOI] [PubMed] [Google Scholar]
- 66. Korbecki J, Grochans S, Gutowska I, Barczak K, Baranowska‐Bosiacka I. CC chemokines in a tumor: a review of pro‐cancer and anti‐cancer properties of receptors CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 ligands. Int J Mol Sci. 2020;21(20):7619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Susek KH, Karvouni M, Alici E, Lundqvist A. The role of CXC chemokine receptors 1‐4 on immune cells in the tumor microenvironment. Front Immunol. 2018;9:2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 2015;14(8):561‐584. [DOI] [PubMed] [Google Scholar]
- 69. Waidmann O. Recent developments with immunotherapy for hepatocellular carcinoma. Expert Opin Biol Ther. 2018;18(8):905‐910. [DOI] [PubMed] [Google Scholar]
- 70. Lee JH, Lee JH, Lim YS, et al. Sustained efficacy of adjuvant immunotherapy with cytokine‐induced killer cells for hepatocellular carcinoma: an extended 5‐year follow‐up. Cancer Immunol Immunother. 2019;68(1):23‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chae YK, Arya A, Iams W, et al. Current landscape and future of dual anti‐CTLA4 and PD‐1/PD‐L1 blockade immunotherapy in cancer; lessons learned from clinical trials with melanoma and non‐small cell lung cancer (NSCLC). J Immunother Cancer. 2018;6(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.
Figure S2.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


