Abstract
PURPOSE: To explore the relationship between ATM, ATR and CAT polymorphisms and prognosis of lung cancer patients received platinum-based chemotherapy.
METHODS: 404 patients with lung cancer who received platinum-chemotherapy were enrolled and DNA typing was performed. Cox regression analysis and stratification analyses was performed to assess relationships between OS and PFS with SNPs genotypes. The prognosis of lung adenocarcinomaand squamous cell carcinomapatients was analyzed with The Cancer Genome Atlas (TCGA) database according to the grouping of CAT expression.
RESULTS: CAT rs769217 was significantly related to PFS of patients with lung cancer who received platinum-chemotherapy. In the Additive model, rs769217 was associated with PFS (HR = 0.747, 95% CI = 0.581–0.960, p = 0.023). In the Dominant model, CT and TT genotypes led to lung cancer progression 0.738 times more than CC genotype. In stratification analyses of association between CAT rs769217 polymorphisms and PFS, the HR of patients at stage IV in additive model was 0.73, and HR was 0.745 (p = 0.034) in dominant model. For OS analyses, HR was 0.672 in the older lung cancer patients (>55 years old) in additive model. Meanwhile, in the Dominant model, it was found that the older patients with CT and TT genotypes had better prognosis, and the risk of death after receiving platinum-based chemotherapy was 0.692 times that of patients with CC genotype (p = 0.037). TCGA data shows that LUAD patients with high CAT expression have longer OS (p = 0.020).
CONCLUSION: CAT rs769217 is significantly related to PSF of platinum-based chemotherapy in lung cancer patients and may be a biomarker for predicting the prognosis of lung cancer patients with platinum-based chemotherapy.
Keywords: lung cancer, platinum-based chemotherapy, snps, cat, ATM, ATR, precious medicine
1 Introduction
Lung cancer is still the main cause of cancer death in the worldwide. There were an estimated 2,206,771 new cases and 1,796,144 cancer deaths of lung ancer worldwide in 2020 according to GLOBOCAN 2020[1]. Chemotherapeutic drugs have been widely used in the treatment of cancer disease for about 70 years. The development of new treatments has not hindered their use, and oncologists still prescribe them routinely, alone or in combination with other antineoplastic agents[2]. Platinum-based chemotherapy, as a conventional treatment, is usually used in combination with immunotherapy as a first-line treatment for most patients with metastatic non-small-cell lung cancer[3]. Platinum drugs can lead to an intrastrand or interstrand cross-linkage by interacting with DNA, thereby activating cellular processes leading to apoptosis[4]. There are many factors that affect the sensitivity of cancer cells to platinum chemotherapeutic drugs[5–7]. Based on previous studies and publication research, we hypothesized that ATM (ataxia telangiectasia mutant gene), ATR (ataxia telangiectasia and Rad3 related) and CAT (catalase) polymorphisms may be related to the prognosis of lung cancer patients receiving platinum-chemotherapy[8–10].
ATM and ATR kinases were the key mediators of DNA damage response (DDR), which induce cell cycle arrest and facilitate DNA repair via their downstream targets[11–13]. Obviously, here is consequently a strong rationale that ATM and ATR may be potential targets to affect platinum chemosensitivity[14,15]. Research shows that functional loss of ATR leads to abrogation of the DNA damage-induced G2/M cell cycle arrest and sensitization of cells to a variety of DNA damaging chemotherapeutic agents[11]. ATM and ATR SNPs may regulate kinase to enhance the DNA-damaging effect of Pt-based chemotherapy in cancer cells[9,16,17].
Platinum-based chemotherapy causes cancer cells death through inducing oxidative stress to highly toxic level[18]. Emerging data indicate that abnormally high ROS(reactive oxygen species) levels may instigate chemoresistance[19]. ROS can provide metabolic reprograming, promoting PGC-1α expression and mitochondrial mass that are in favor of cisplatin resistance in non-small cell lung cancer[20,21]. Therefore, the genetic polymorphism of oxidative stress related genes is likely to affect the platinum chemosensitivity. CAT are well studied enzymes that play critical roles in protecting cells against the toxic effects of hydrogen peroxide, which is a key enzyme in the metabolism of H2O2, reducing the production of ROS in cells[22]. Studies have shown that CAT rs769218 is related to the prognosis of gastric cancer patients receiving platin and fluorouracil-based adjuvant therapy, but the correlation between the prognosis of lung cancer patients and CAT polymorphism has not been reported[18]. Herein, we investigated the association of potentially functional SNPs in ATM, ATR and CAT with platinum-based chemotherapy outcome of lung cancer patients.
2 Materials and methods
2.1 Study population and data collection
All patients in this study were recruited from Xiangya Hospital of Central South University and Hunan Cancer Hospital (Changsha, China), from 2012 to 2019. All patients were diagnosed with NSCLC by histopathological examination and confirmed the absence of driver genetic alterations that could be targeted. All patients received first-line platinum-based chemotherapy regimens for two to six cycles: cisplatin (75 mg/m2) or carboplatin (AUC 5), which were both administered on Day 1 every 3 weeks, in combination with pemetrexed (500 mg/m2) on Day 1 every 3 weeks, gemcitabine (1,250 mg/m2) on Days 1 and 8 every 3 weeks, paclitaxel (175 mg/m2) on Day 1 every 3 weeks, docetaxel (75 mg/m2) on Day 1 every 3 weeks, or navelbine (25 mg/m2) on Days 1 and 8 every 3 weeks. Patients did not undergo surgery, targeted therapy, radiotherapy, or other anti-tumor therapy before chemotherapy. A physical examination as well as a detailed inquiry into each patient’s medical history was carried out. Patients with serious concomitant diseases that might greatly affect their physical condition were excluded. The study protocol was approved by the Ethics Committee of Institute of Clinical Pharmacology, Central South University (Changsha, China), and all subjects were provided with written informed consents. We applied this study for clinical admission in the Chinese Clinical Trial Register (registration number: ChiCTR-RO-12002873)[23].
2.2 DNA extraction and genotyping, SNP selection
In the morning, 5 mL blood of lung cancer patients undergoing chemotherapy was collected. After centrifugation at 4000rpm, plasma was separated and stored at −20 °C. DNA was extracted using Genomic DNA Purification Kit Genomic (Promega, Madison, WI, United States) and stored at −20°C according to the instructions. Each DNA sample was genotyped by Sequenom Mass Array Genotype Platform (Sequenom, SanDiego, CA, United States). After polymerase chain reaction (PCR), the product was purified by resin and analyzed by the Mass Array system (Sequenom)[24]. Then we used Plink (version 1.9, http://pngu.mgh.harvard.edu/purcell/plink/)to detect and control the quality of SNPs data, requiring that the MAF (Minor allele frequency) ≥0.05, the call rate≥90%, and conform to Hardy-Weinberg equilibrium. Finally, we selected 7 SNPs of ATR, ATM, and CAT for follow-up (Table 1).
TABLE 1.
Association of comorbidities with CAT genotypes in lung cancer patients.
| Comorbidities | Statue | CAT rs769217 genotype | p-Value | ||
|---|---|---|---|---|---|
| CC | CT | TT | |||
| Hypertension | Yes | 17 (4.3) | 29 (7.34) | 7 (1.77) | 0.389 |
| No | 98 (24.81) | 171 (43.29) | 73 (18.48) | ||
| Diabetes mellitus | Yes | 6 (1.52) | 11 (2.78) | 8 (2.03) | 0.318 |
| No | 109 (27.59) | 189 (47.85) | 72 (18.23) | ||
| COPD | Yes | 17 (4.30) | 32 (8.10) | 17 (4.30) | 0.457 |
| No | 98 (24.81) | 168 (42.53) | 63 (15.95) | ||
| coronary heart disease | Yes | 1 (0.25) | 10 (2.53) | 2 (0.51) | 0.137 |
| No | 114 (28.86) | 190 (48.10) | 78 (19.75) | ||
| pulmonary tuberculosis | Yes | 7 (1.77) | 4 (1.01) | 4 (1.01) | 0.144 |
| No | 108 (27.34) | 196 (49.62) | 76 (19.24) | ||
| Chronic viral hepatitis | Yes | 4 (1.01) | 13 (3.29) | 9 (2.28) | 0.111 |
| No | 111 (28.10) | 187 (47.34) | 71 (17.97) | ||
2.3 Statistical analysis
OS (Overall survival) was defined as the time from treatment initiation until either the date of death, the date of last follow-up or the date of end of analysis. PFS (Progression-free survival) was defined as the time from treatment initiation until disease progression or death, whichever occurred first[25]. Kaplan-Meier analysis was used to calculate the median survival time of lung cancer patients undergoing chemotherapy. COX proportional hazards regression analysis was used to analyze the correlation between genotypes and prognosis in each Genetic Model after adjusting for age, sex, smoking, histology, and stage confounding factors. Stratified analysis was used to analyze the correlation between SNPs in each layer and the prognosis of lung cancer patients with platinum-based chemotherapy, so as to control the influence of covariates on the research results to a certain extent. Hazard ratio (HR) was used to evaluate the degree of effect between genotype and patient prognosis. 95% confidence interval (95% CI) is the range of HR estimated by 95% probability. All the p-values were two-sided, p < 0.05 were supposed to be significant. All the above analyses were performed by The SPSS version 25.0 (SPSS Inc., Chicago, IL, United States). UCSC Xena (https://xena.ucsc.edu/kaplan-survival-analysis/) was used to analyze the prognosis of LUAD and LUSC patients in TCGA database.
3 Results
3.1 Characteristics and prognosis of lung cancer patients
A total of 404 lung cancer patients received platinum-based chemotherapy were included in this research. There were 175 patients with age ≤55 years old, accounting for 43.32%. Most of the patients were male, accounting for 77.48% (313). 40.10% (162) patients have the habit of smoking. 97.52% of the patients (394) were in stage III or IV. 50% of the patients (202) were adenocarcinoma and 42.33% (171) were small cell carcinoma (Table 2).
TABLE 2.
Association of 7 SNPs polymorphisms and PFS.
| Gene | Polymorphism | Genotype | MPFS (year) | Additive | Dominant | Recessive | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | HR (95%CI) | p-value | Genotype | HR (95%CI) | p-value | Genotype | HR (95%CI) | p-value | ||||
| ATM | rs228589 | AA | 1.855 | AA | REF | 0.409 | AA | REF | TT | REF | ||
| AT | 2.984 | AT | 1.133 (0.837–1.534) | 0.419 | AT + TT | 0.950 (0.716–1.261) | 0.722 | AT + AA | 0.880 (0.694–1.116) | 0.293 | ||
| TT | 4.2199 | TT | 0.96 (0.698–1.320) | 0.800 | ||||||||
| ATR | rs4585 | GG | 4.195 | GG | REF | 0.641 | GG | REF | TT | REF | ||
| GT | 3.014 | GT | 1.116 (0.865–1.440) | 0.397 | GT + TT | 0.924 (0.727–1.174) | 0.516 | GT + GG | 1.062 (0.793–1.421) | 0.687 | ||
| TT | 1.836 | TT | 1.003 (0.723–1.392) | 0.984 | ||||||||
| ATR | rs2227928 | AA | 3.452 | AA | REF | 0.513 | AA | REF | GG | REF | ||
| AG | 3.003 | AG | 1.164 (0.883–1.535) | 0.283 | AG + GG | 0.892 (0.688–1.157) | 0.391 | AG + AA | 1.057 (0.810–1.378) | 0.684 | ||
| GG | 4.381 | GG | 1.043 (0.756–1.438) | 0.799 | ||||||||
| ATR | rs2229032 | CC | 3.164 | CC | REF | 0.533 | CC | REF | TT | REF | ||
| CT | 2.151 | CT | 1.142 (0.829–1.571) | 0.416 | CT + TT | 1.098 (0.802–1.503) | 0.559 | CT + CC | 1.740 (0.427–7.090) | 0.44 | ||
| TT | 5.337 | TT | 0.585 (0.144–2.387) | 0.455 | ||||||||
| CAT | rs564250 | TT | 4.775 | TT | REF | 0.075 | TT | REF | CC | REF | ||
| TC | 3.8 | TC | 1.004 (0.481–2.095) | 0.992 | TC + CC | 1.221 (0.598–2.492) | 0.583 | TC + TT | 0.757 (0.595–0.962) | 0.023 | ||
| CC | 3.003 | CC | 1.326 (0.648–2.714) | 0.44 | ||||||||
| CAT | rs769217 | CC | 2.164 | CC | REF | 0.042 | CC | REF | TT | REF | ||
| CT | 3.8 | CT | 0.747 (0.581–0.960) | 0.023 | CT + TT | 0.738 (0.582–0.936) | 0.012 | CT + CC | 1.159 (0.873–1.539) | 0.308 | ||
| TT | 2.764 | TT | 0.715 (0.518–0.987) | 0.041 | ||||||||
| CAT | rs7943316 | AA | 3.066 | AA | REF | 0.812 | AA | REF | TT | REF | ||
| AT | 3.425 | AT | 1.049 (0.828–1.327) | 0.693 | AT + TT | 1.063 (0.850–1.329) | 0.594 | AT + AA | 0.908 (0.626–1.316) | 0.609 | ||
| TT | 2.307 | TT | 1.126 (0.764–1.661) | 0.547 | ||||||||
3.2 Association of the CAT rs769217 polymorphisms and PFS in lung cancer patients with platinum-based chemotherapy
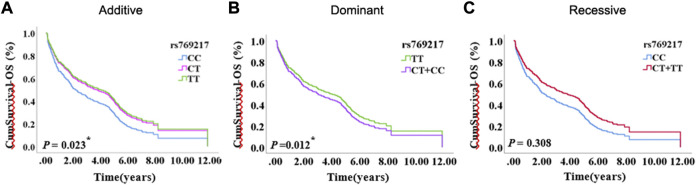
After excluding the effects of age, sex, smoking status, stage, and histology type, we used COX proportional hazards regression analysis to analyze the relationship between SNPs and patient prognosis, and found that CAT rs769217 was significantly related to PFS. In the additive model, rs769217 was associated with PFS, HR = 0.747, 95% CI = 0.581–0.960, p = 0.023. In the dominant model, CT + TT genotypes led to lung cancer progression 0.738 times more than CC genotype. In the recessive model, the p-value > 0.05, which is not significant (Table 3). However, the Kaplan-Meier plot still shows that the PFS of patients with CT and TT genotypes is longer than that of patients with CC genotype (Figure 1). The lung cancer patients’ comorbidities are not associated with CAT genotypes (Supplementary Table S1). The correlation of other SNPs with PFS and OS is shown in Supplementary Table S2, S3.
TABLE 3.
Association of 7 SNPs polymorphisms and OS.
| Gene | Polymorphism | Genotype | Mos (year) | Additive | Dominant | Recessive | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | HR (95%CI) | p-value | Genotype | HR (95%CI) | p-value | Genotype | HR (95%CI) | p-value | ||||
| ATM | rs228589 | AA | 3.186 | AA | REF | 0.959 | AA | REF | TT | REF | ||
| AT | 4.268 | AT | 0.955 (0.689–1.325) | 0.785 | AT + TT | 1.047 (0.768–1.427) | 0.771 | AT + AA | 1.013 (0.789–1.300) | 0.922 | ||
| TT | 4.671 | TT | 0.955 (0.679–1.352) | 0.794 | ||||||||
| ATR | rs4585 | GG | 4.381 | GG | REF | 0.652 | GG | REF | TT | REF | ||
| GT | 4.392 | GT | 0.915 (0.701–1.195) | 0.514 | GT + TT | 1.055 (0.820–1.358) | 0.675 | GT + GG | 0.900 (0.659–1.230) | 0.508 | ||
| TT | 3.186 | TT | 1.053 (0.741–1.495) | 0.775 | ||||||||
| ATR | rs2227928 | AA | 3.83 | AA | REF | 0.428 | AA | REF | GG | REF | ||
| AG | 4.115 | AG | 0.821 (0.610–1.105) | 0.194 | AG + GG | 0.845 (0.640–1.116) | 0.235 | AG + AA | 0.983 (0.742–1.301) | 0.904 | ||
| GG | 4.679 | GG | 0.893 (0.636–1.254) | 0.512 | ||||||||
| ATR | rs2229032 | CC | 4.268 | CC | REF | 0.687 | CC | REF | TT | REF | ||
| CT | 3.449 | CT | 1.15 (0.825–1.604) | 0.409 | CT + TT | 1.154 (0.833–1.598) | 0.389 | CT + CC | 0.826 (0.202–3.369) | 0.789 | ||
| TT | 1.627 | TT | 1.232 (0.302–5.030) | 0.772 | ||||||||
| CAT | rs564250 | TT | 3.066 | TT | REF | 0.178 | TT | REF | CC | REF | ||
| TC | 3.942 | TC | 0.653 (0.313–1.363) | 0.257 | TC + CC | 0.764 (0.375–1.557) | 0.459 | TC + TT | 0.823 (0.639–1.058) | 0.129 | ||
| CC | 4.268 | CC | 0.820 (0.401–1.677) | 0.586 | ||||||||
| CAT | rs769217 | CC | 3.888 | CC | REF | 0.241 | CC | REF | TT | REF | ||
| CT | 4.658 | CT | 0.800 (0.610–1.048) | 0.105 | CT + TT | 0.832 (0.644–1.075) | 0.159 | CT + CC | 0.927 (0.690–1.247) | 0.618 | ||
| TT | 3.205 | TT | 0.931 (0.662–1.310) | 0.683 | ||||||||
| CAT | rs7943316 | AA | 4.392 | AA | REF | 0.806 | AA | REF | TT | REF | ||
| AT | 4.049 | AT | 1.024 (0.800–1.312) | 0.85 | AT + TT | 0.997 (0.788–1.261) | 0.978 | AT + AA | 1.143 (0.754–1.732) | 0.53 | ||
| TT | 4.671 | TT | 0.885 (0.575–1.361) | 0.577 | ||||||||
FIGURE 1.
The Kaplan-Meier plot of COX proportional hazards regression analysis, (A) Additive model, (B) Dominant model, (C) Recessive model.
3.3 Stratification analyses of association between CAT rs769217 polymorphisms and PFS
We used stratification analysis to stratify the clinical data of lung cancer patients receiving platinum-based chemotherapy according to age, sex, smoking status, histology, and stage, and then calculated the association between SNP rs769217 polymorphism and prognosis. In the correlation analysis between SNP polymorphism and PFS, the HR of patients at stage IV in the Additive model was 0.730, which indicates patients with CT genotype are 0.73 times more likely to progress than those with CC genotype, and the prognosis of patients with CT genotype is better (p = 0.031). In the Dominant model, the HR of patients in stage IV was 0.745, p = 0.034 (Figure 2). In the correlation analysis between SNP polymorphism and OS, HR = 0.672, p = 0.031 of the older lung cancer patients (>55 years old) was found in the Additive model. Meanwhile, in the Dominant model, it was found that the older patients with CT and TT genotype had better prognosis, and the risk of death after receiving platinum-based chemotherapy was 0.692 times that of patients with CC genotype (p = 0.037) (Figure 3).
FIGURE 2.
Association of CAT rs769217 and PFS in Stratification analyses, (A) additive model, (B) Dominant model, (C) Recessive model. p* <0.05, p**< 0.01.
FIGURE 3.
Association of CAT rs769217 and OS in Stratification analyses, (A) additive model, (B) Dominant model, (C) Recessive model. p* <0.05, p**< 0.01.
4 Discussion
In 2019, about 2 million people worldwide died from lung cancer, more than any other cancer[26]. Annually, approximately 631,000 deaths were reported because of lung cancer according to Chinese national statistics[27]. Compared with the decline of the incidence in some western countries, the incidence of lung cancer in China is still rising, which is a major public health problem[28]. Chemotherapy and immunotherapy are common therapeutic methods in clinic. Immunotherapy has incomparable advantages over traditional anti-tumor therapy, which can prolong PFS and OS. The current reality, however, is that the majority of patients often cannot benefit from it because of low tumor mutation burden, or other reasons, or terminate the treatment due to serious adverse reactions[29,30]. Therefore, platinum-based chemotherapy, as an effective anti-tumor therapy, is still the first-line drug regimen for lung cancer[23]. After platinum chemotherapeutic drugs enter the tumor cell, a series of chemical reactions occur in the cytoplasm. Platinum binds to DNA by forming intra- and inter-stranded crosslinks, changing the DNA structure, and causing DNA damage, so as to achieve the purpose of anti-tumor[31]. In addition, platinum-based chemotherapy was shown to upregulate tumor cell expression of PD-L1 and has immunostimulatory properties as well, which plays an anti-tumor role in coordination with immunotherapy[32–35]. Genetic abnormalities could influence chemosensitivity, the development of predictive markers to identify patients who will derive significant benefit with minimal toxicity from chemotherapy is a continuing challenge in lung cancer research[36,37]. However, the genetic underpinnings of platinum sensitivity remain poorly understood[38].
DNA is usually considered as the main target of platinum chemotherapeutic drugs, but the cisplatin used in vitro studies lead to acute apoptosis that involves induction of oxidative stress but is largely DNA damage-independent[39]. Cellular exposure to cisplatin causes direct damage to mtDNA resulting in a reduction of mitochondrial protein synthesis, impairment of electron transport chain function, and subsequently, increases in intracellular ROS levels, ultimately promoting cell death[40]. However, due to the double-edged sword role of ROS in cancer as a pro-survival or pro-death mechanism, ROS can result in platinum-chemotherapy resistance[21]. ROS can provide metabolic reprograming, promoting PGC-1α expression and mitochondrial mass that are in favor of cisplatin resistance in non-small cell lung cancer[20].
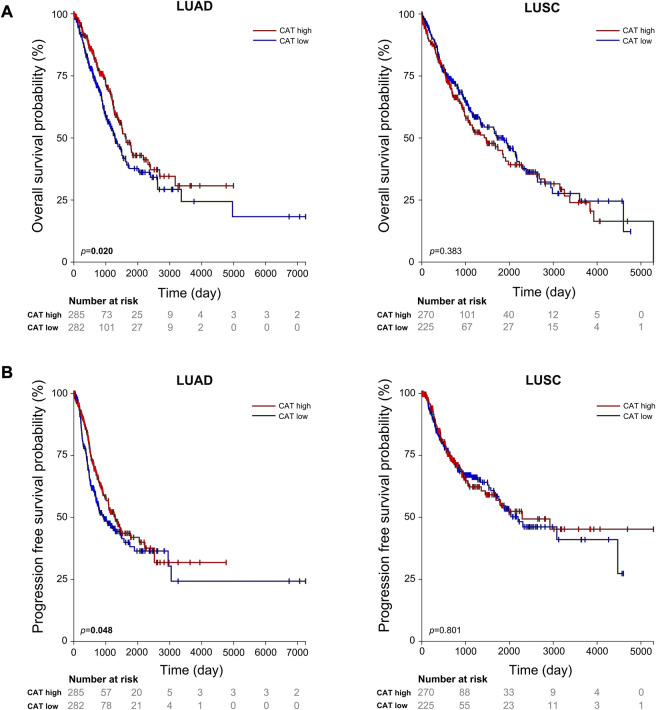
Our results suggest that CAT rs769217 may affect the PFS of lung cancer patients receiving platinum-based chemotherapy (Figure 1). CAT gene encodes catalase, which can regulate reactive oxygen species (ROS), is a key antioxidant enzyme in the bodies defense against oxidative stress[41,42]. Previous studies showed that as compared to normal tissues of the same origin, the expression of CAT in tumors changed[43–45]. Two meta-analyses pointed out a correlation exists between CAT rs1001179 polymorphism and prostate cancer[46,47]. At the same time, the expression of CAT in tumor cells can affect their sensitivity to chemotherapy drugs[48–50]. There are definite experimental results proving that CAT rs769217 can affect the prognosis of patients with biliary tr an act cancer (BTC), that knockdown of CAT induced chemoresistance through elevation of ROS level and activation of Nrf2-ABCG2 pathway in BTC cell lines[51]. But the impact of CAT polymorphism on the prognosis of patients with lung cancer receiving platinum chemotherapy has not been reported. CAT polymorphisms can affect the expression of CAT mRNA in tumor tissues[51], and according to TCGA data analysis, the OS and PFS of LUAD patients with high CAT expression is significantly longer (p = 0.020, p = 0.048 respectively; Figure 4). We speculate that compared with CC genotype, patients with lung cancer who carry TT genotype and receive platinum-chemotherapy have higher CAT expression in tumor cells, thus regulating ROS and making tumor cells sensitive to platinum-chemotherapy, but the specific mechanism needs to be further explored. Therefore, CAT can be used as a potential target to enhance the sensitivity of platinum-chemotherapy, nanocarriers of platinum and CAT enhance the cytotoxicity of drug resistant cancer cells[52]. In addition, we found that compared with LUAD patients, the prognosis of LUSC patients was not associated with CAT expression ( Figure 4). The expression of most DNA repair genes in LUSC tumor cells is upregulated[53]. Lung cancer patients with higher DNA repair capacity had elevated chemoresistance[54]. Therefore, we speculate that the effect of these DNA repair genes on the efficacy of platinum drugs is far greater than that of CAT genes. In conclusion, our study showed that CAT rs769217 is significantly related to PSF of platinum-based chemotherapy in lung cancer patients. CAT rs769217 may be a biomarker for predicting the prognosis of lung cancer patients with platinum-based chemotherapy.
FIGURE 4.
Association of the expression of CAT with lung cancer OS (A) and PFS (B) in TCGA LUAD and LUSC patients.
Funding Statement
National Natural Science Foundation of China (81,874,327,81803640), Key Research and Development Progra m of Hunan Province (2019SK2251), Project Program of National Clinical Research Center for Geriatric Disorders (Xiangya Hospital, 2020LNJJ02), and Science and Technology Program of Changsha (kh2003010), and Sanming Project of Medicine in Shenzhen (No. SZSM201811057).
Data availability statement
The publicly available data sets that supported this study are available from OMIX under accessions OMIX002961.
Ethics statement
The studies involving human participants were reviewed and approved by Ethics Committee of Xiangya Hospital, Central South University. The patients/participants provided their written informed consent to participate in this study.
Author contributions
Z-QL and JC contributed to conception and design of the study. X-PL and JC collected the samples. J-SL and J-YL conducted the study and performed the statistical analysis. J-SL wrote the first draft of the manuscript, QX wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2023.1119837/full#supplementary-material
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 71, 209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2. Pagani M, Bavbek S, Alvarez-Cuesta E, Berna Dursun A, Bonadonna P, Castells M, et al. (2022). Hypersensitivity reactions to chemotherapy: an EAACI Position Paper. Allergy 77, 388–403. 10.1111/all.15113 [DOI] [PubMed] [Google Scholar]
- 3. Reck M, Remon J, Hellmann MD. (2022). First-Line Immunotherapy for Non-Small-Cell Lung Cancer. official journal of the American Society of Clinical Oncology 40, 586–97. 10.1200/JCO.21.01497 [DOI] [PubMed] [Google Scholar]
- 4. Yu C, Wang Z, Sun Z, Zhang L, Zhang W, Xu Y, et al. (2020). Platinum-Based Combination Therapy: Molecular Rationale, Current Clinical Uses, and Future Perspectives. Journal of medicinal chemistry 63, 13397–412. 10.1021/acs.jmedchem.0c00950 [DOI] [PubMed] [Google Scholar]
- 5. Ahmad S. (2010). Platinum-DNA interactions and subsequent cellular processes controlling sensitivity to anticancer platinum complexes. CHEMISTRY & BIODIVERSITY 7, 543–66. 10.1002/cbdv.200800340 [DOI] [PubMed] [Google Scholar]
- 6. Carmi YK, Mahmoud H, Khamaisi H, Adawi R, Gopas J, Mahajna J. (2020). Flavonoids Restore Platinum Drug Sensitivity to Ovarian Carcinoma Cells in a Phospho-ERK1/2-Dependent Fashion. INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu EZ, Gareev I, Yuan C, Liang YC, Sun JX, Chen X, et al. (2022). The Mechanisms of Current Platinum Anticancer Drug Resistance in the Glioma. CURRENT PHARMACEUTICAL DESIGN 28, 1863–9. 10.2174/1381612828666220607105746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim H, Kim ST, Yoo KH, Hong JY, Park YS, Lim HY, et al. (2021). ATM Expression as a Prognostic Marker in Patients With Advanced Biliary Tract Cancer Treated With First-line Gemcitabine and Platinum Chemotherapy. IN VIVO 35, 499–505. 10.21873/invivo.12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Teng PN, Bateman NW, Darcy KM, Hamilton CA, Maxwell GL, Bakkenist CJ, et al. (2015). Pharmacologic inhibition of ATR and ATM offers clinically important distinctions to enhancing platinum or radiation response in ovarian, endometrial, and cervical cancer cells. Gynecologic oncology 136, 554–61. 10.1016/j.ygyno.2014.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang HH, Zhao WY, Gu DY, Du ML, Gong WD, Tan YF, et al. (2018). Association of Antioxidative Enzymes Polymorphisms with Efficacy of Platin and Fluorouracil-Based Adjuvant Therapy in Gastric Cancer. CELLULAR PHYSIOLOGY AND BIOCHEMISTRY 48, 2247–57. 10.1159/000492642 [DOI] [PubMed] [Google Scholar]
- 11. Weber AM, Ryan AJ. (2015). ATM and ATR as therapeutic targets in cancer. Pharmacology & therapeutics 149, 124–38. 10.1016/j.pharmthera.2014.12.001 [DOI] [PubMed] [Google Scholar]
- 12. Wagner JM, Kaufmann SH. (2010). Prospects for the Use of ATR Inhibitors to Treat Cancer. Pharmaceuticals (Basel, Switzerland) 3, 1311–34. 10.3390/ph3051311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ueno S, Sudo T, Hirasawa A. ATM. (2022). ATM: Functions of ATM Kinase and Its Relevance to Hereditary Tumors. INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES 23, 523. 10.3390/ijms23010523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Neeb A, Herranz N, Arce-Gallego S, Miranda S, Buroni L, Yuan W, et al. (2021). Advanced Prostate Cancer with ATM Loss: PARP and ATR Inhibitors. European urology 79, 200–11. 10.1016/j.eururo.2020.10.029 [DOI] [PubMed] [Google Scholar]
- 15. Feng WL, Dean DC, Hornicek FJ, Wang JL, Jia YY, Duan ZF, et al. (2020). ATR and p-ATR are emerging prognostic biomarkers and DNA damage response targets in ovarian cancer. THERAPEUTIC ADVANCES IN MEDICAL ONCOLOGY 12, 1758835920982853. 10.1177/1758835920982853 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16. Krajewska M, Fehrmann RSN, de Vries EGE, van Vugt M. (2015). Regulators of homologous recombination repair as novel targets for cancer treatment. FRONTIERS IN GENETICS 6, 96. 10.3389/fgene.2015.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fokas E, Prevo R, Hammond EM, Brunner TB, McKenna WG, Muschel RJ. (2014). Targeting ATR in DNA damage response and cancer therapeutics. Cancer treatment reviews 40, 109–17. 10.1016/j.ctrv.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 18. Zhang H, Zhao W, Gu D, Du M, Gong W, Tan Y, et al. (2018). Association of Antioxidative Enzymes Polymorphisms with Efficacy of Platin and Fluorouracil-Based Adjuvant Therapy in Gastric Cancer. Cellular physiology and biochemistry. international journal of experimental cellular physiology, biochemistry, and pharmacology 48, 2247–57. 10.1159/000492642 [DOI] [PubMed] [Google Scholar]
- 19. Conklin KA. (2004). Chemotherapy-associated oxidative stress: impact on chemotherapeutic effectiveness. Integrative cancer therapies 3, 294–300. 10.1177/1534735404270335 [DOI] [PubMed] [Google Scholar]
- 20. Cruz-Bermúdez A, Laza-Briviesca R, Vicente-Blanco RJ, García-Grande A, Coronado MJ, Laine-Menéndez S, et al. (2019). Cisplatin resistance involves a metabolic reprogramming through ROS and PGC-1α in NSCLC which can be overcome by OXPHOS inhibition. Free radical biology & medicine 135, 167–81. 10.1016/j.freeradbiomed.2019.03.009 [DOI] [PubMed] [Google Scholar]
- 21. Mirzaei S, Hushmandi K, Zabolian A, Saleki H, Torabi SMR, Ranjbar A, et al. (2021). Elucidating Role of Reactive Oxygen Species (ROS) in Cisplatin Chemotherapy: A Focus on Molecular Pathways and Possible Therapeutic Strategies. Molecules (Basel, Switzerland) 26, 2382. 10.3390/molecules26082382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goyal MM, Basak A. (2010). Human catalase: looking for complete identity. Protein & cell 1, 888–97. 10.1007/s13238-010-0113-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mao C, Chen J, Zou T, Zhou Y, Liu J, Li X, et al. (2022). Genome-wide analysis identify novel germline genetic variations in ADCY1 influencing platinum-based chemotherapy response in non-small cell lung cancer. Acta pharmaceutica Sinica B 12, 1514–22. 10.1016/j.apsb.2021.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mo JL, Liu JS, Xiao Q, Hong WX, Yin JY, Chen J, et al. (2022). Association of variations in the Fanconi anemia complementation group and prognosis in Non-small cell lung cancer patients with Platinum-based chemotherapy. Gene 825, 146398. 10.1016/j.gene.2022.146398 [DOI] [PubMed] [Google Scholar]
- 25. Kfoury M, Najean M, Lappara A, Voisin AL, Champiat S, Michot JM, et al. (2022). Analysis of the association between prospectively collected immune-related adverse events and survival in patients with solid tumor treated with immune-checkpoint blockers, taking into account immortal-time bias. Cancer treatment reviews 110, 102452. 10.1016/j.ctrv.2022.102452 [DOI] [PubMed] [Google Scholar]
- 26. Brody H. (2020). Lung cancer. Nature 587, S7. 10.1038/d41586-020-03152-0 [DOI] [PubMed] [Google Scholar]
- 27. Zheng RS, Sun KX, Zhang SW, Zeng HM, Zou XN, Chen R, et al. (2019). Report of cancer epidemiology in China, 2015. Zhonghua zhong liu za zhi [Chinese journal of oncology] 41, 19–28. 10.3760/cma.j.issn.0253-3766.2019.01.005 [DOI] [PubMed] [Google Scholar]
- 28. Wu F, Wang L, Zhou C. (2021). Lung cancer in China: current and prospect. Current opinion in oncology 33, 40–6. 10.1097/CCO.0000000000000703 [DOI] [PubMed] [Google Scholar]
- 29. Tan S, Li D, Zhu X. (2020). Cancer immunotherapy: Pros, cons and beyond. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 124, 109821. 10.1016/j.biopha.2020.109821 [DOI] [PubMed] [Google Scholar]
- 30. Yap TA, Parkes EE, Peng W, Moyers JT, Curran MA, Tawbi HA. (2021). Development of Immunotherapy Combination Strategies in Cancer. Cancer discovery 11, 1368–97. 10.1158/2159-8290.CD-20-1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang C, Xu C, Gao X, Yao Q. (2022). Platinum-based drugs for cancer therapy and anti-tumor strategies. Theranostics 12, 2115–32. 10.7150/thno.69424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heinhuis KM, Ros W, Kok M, Steeghs N, Beijnen JH, Schellens JHM. (2019). Enhancing antitumor response by combining immune checkpoint inhibitors with chemotherapy in solid tumors. official journal of the European Society for Medical Oncology 30, 219–35. 10.1093/annonc/mdy551 [DOI] [PubMed] [Google Scholar]
- 33. Tran L, Allen CT, Xiao R, Moore E, Davis R, Park SJ, et al. (2017). Cisplatin Alters Antitumor Immunity and Synergizes with PD-1/PD-L1 Inhibition in Head and Neck Squamous Cell Carcinoma. Cancer immunology research 5, 1141–51. 10.1158/2326-6066.CIR-17-0235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jackaman C, Majewski D, Fox SA, Nowak AK, Nelson DJ. (2012). Chemotherapy broadens the range of tumor antigens seen by cytotoxic CD8(+) T cells in vivo . CII. 61, 2343–56. 10.1007/s00262-012-1307-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nio Y, Hirahara N, Minari Y, Iguchi C, Yamasawa K, Toga T, et al. (2000). Induction of tumor-specific antitumor immunity after chemotherapy with cisplatin in mice bearing MOPC-104E plasmacytoma by modulation of MHC expression on tumor surface. Anticancer research 20, 3293–9. [PubMed] [Google Scholar]
- 36. Wei HB, Hu J, Shang LH, Zhang YY, Lu FF, Wei M, et al. (2012). A meta-analytic review of ERCC1/MDR1 polymorphism and chemosensitivity to platinum in patients with advanced non-small cell lung cancer. Chinese medical journal 125, 2902–7. [PubMed] [Google Scholar]
- 37. Chen X, Zhou C, Sun H, Ren S, Zhang L, Zhou S, et al. (2011). IL-9(+) IL-10(+) T cells link immediate allergic response to late phase reaction. Journal of Clinical Oncology 165, 29–37. 10.1111/j.1365-2249.2011.04394.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yin JY, Shen J, Dong ZZ, Huang Q, Zhong MZ, Feng DY, et al. (2011). Effect of eIF3a on response of lung cancer patients to platinum-based chemotherapy by regulating DNA repair. an official journal of the American Association for Cancer Research 17, 4600–9. 10.1158/1078-0432.CCR-10-2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Berndtsson M, Hägg M, Panaretakis T, Havelka AM, Shoshan MC, Linder S. (2007). Acute apoptosis by cisplatin requires induction of reactive oxygen species but is not associated with damage to nuclear DNA. International journal of cancer 120, 175–80. 10.1002/ijc.22132 [DOI] [PubMed] [Google Scholar]
- 40. Marullo R, Werner E, Degtyareva N, Moore B, Altavilla G, Ramalingam SS, et al. (2013). Cisplatin induces a mitochondrial-ROS response that contributes to cytotoxicity depending on mitochondrial redox status and bioenergetic functions. PloS one 8, e81162. 10.1371/journal.pone.0081162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Glorieux C, Calderon PB. (2017). Catalase, a remarkable enzyme: targeting the oldest antioxidant enzyme to find a new cancer treatment approach. Biological chemistry 398, 1095–108. 10.1515/hsz-2017-0131 [DOI] [PubMed] [Google Scholar]
- 42. Xu D, Wu L, Yao H, Zhao L. (2022). Catalase-Like Nanozymes: Classification, Catalytic Mechanisms, and Their Applications. Small (Weinheim an der Bergstrasse, Germany) 18, e2203400. 10.1002/smll.202203400 [DOI] [PubMed] [Google Scholar]
- 43. Rainis T, Maor I, Lanir A, Shnizer S, Lavy A. (2007). Enhanced oxidative stress and leucocyte activation in neoplastic tissues of the colon. Digestive diseases and sciences 52, 526–30. 10.1007/s10620-006-9177-2 [DOI] [PubMed] [Google Scholar]
- 44. Hwang TS, Choi HK, Han HS. (2007). Differential expression of manganese superoxide dismutase, copper/zinc superoxide dismutase, and catalase in gastric adenocarcinoma and normal gastric mucosa. The journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology 33, 474–9. 10.1016/j.ejso.2006.10.024 [DOI] [PubMed] [Google Scholar]
- 45. Sander CS, Hamm F, Elsner P, Thiele JJ. (2003). Oxidative stress in malignant melanoma and non-melanoma skin cancer. The British journal of dermatology 148, 913–22. 10.1046/j.1365-2133.2003.05303.x [DOI] [PubMed] [Google Scholar]
- 46. Liu K, Liu X, Wang M, Wang X, Kang H, Lin S, et al. (2016). Two common functional catalase gene polymorphisms (rs1001179 and rs794316) and cancer susceptibility: evidence from 14,942 cancer cases and 43,285 controls. Oncotarget 7, 62954–65. 10.18632/oncotarget.10617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang CD, Sun Y, Chen N, Huang L, Huang JW, Zhu M, et al. (2016). The Role of Catalase C262T Gene Polymorphism in the Susceptibility and Survival of Cancers. Scientific reports 6, 26973. 10.1038/srep26973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim HS, Lee TB, Choi CH. (2001). Down-regulation of catalase gene expression in the doxorubicin-resistant AML subline AML-2/DX100. Biochemical and biophysical research communications 281, 109–14. 10.1006/bbrc.2001.4324 [DOI] [PubMed] [Google Scholar]
- 49. Glorieux C, Dejeans N, Sid B, Beck R, Calderon PB, Verrax J. (2011). Catalase overexpression in mammary cancer cells leads to a less aggressive phenotype and an altered response to chemotherapy. Biochemical pharmacology 82, 1384–90. 10.1016/j.bcp.2011.06.007 [DOI] [PubMed] [Google Scholar]
- 50. Zhang J, Yang L, Xiang X, Li Z, Qu K, Li K. (2018). A panel of three oxidative stress-related genes predicts overall survival in ovarian cancer patients received platinum-based chemotherapy. Aging 10, 1366–79. 10.18632/aging.101473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhan M, Wang H, Xu SW, Yang LH, Chen W, Zhao SX, et al. (2019). Variants in oxidative stress-related genes affect the chemosensitivity through Nrf2-mediated signaling pathway in biliary tract cancer. EBIOMEDICINE 48, 143–60. 10.1016/j.ebiom.2019.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen H, He W, Guo Z. (2014). An H₂O₂-responsive nanocarrier for dual-release of platinum anticancer drugs and O₂: controlled release and enhanced cytotoxicity against cisplatin resistant cancer cells. Chemical communications (Cambridge, England) 50, 9714–7. 10.1039/c4cc03385j [DOI] [PubMed] [Google Scholar]
- 53. Wang X, Huang Z, Li L, Wang G, Dong L, Li Q, et al. (2022). DNA damage repair gene signature model for predicting prognosis and chemotherapy outcomes in lung squamous cell carcinoma. BMC cancer 22, 866. 10.1186/s12885-022-09954-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bosken CH, Wei Q, Amos CI, Spitz MR. (2002). An analysis of DNA repair as a determinant of survival in patients with non-small-cell lung cancer. Journal of the National Cancer Institute 94, 1091–9. 10.1093/jnci/94.14.1091 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The publicly available data sets that supported this study are available from OMIX under accessions OMIX002961.