Graphical abstract
Keywords: Spontaneous fermentation, Microbial diversity, Correlation analysis, Volatile compounds, Flavor
Highlights
-
•
Fungal diversity was significantly lower in rain-shelter than in the open-field, while bacteria diversity was incoherent.
-
•
PLSR characterized the association between volatile compounds and wine sensory properties.
-
•
OPLS-DA model identified 39 species of differentially variable volatiles under two cultivations.
-
•
RDA and Spearman analysis elucidated the correlations between microbial species and wine metabolites.
-
•
SEM equation revealed that rain-shelter mode strongly affected wine aromas mainly through fungal microbiota.
Abstract
Microbiota succession in spontaneous fermentation of Cabernet Sauvignon cultivated under the rain-shelter was characterized, with open-field cultivation as the control. For both cultivation modes, Saccharomyces, Starmerella, and Mycosphearella were the principal fungi, and Tatumella, Gluconobacter, and Acinetobacter were the prevailing bacteria. Rain-shelter reduced the abundance of Hanseniaspora, Candida, Starmerella, Gluconobacter, and Lactococcus. During fermentation, fungal microbiota diversity in samples from the rain-shelter cultivation decreased more drastically than the control (p < 0.05). In terms of the correlation between microbiota and volatile compounds production, the abundance of Hanseniaspora uvarum, Candida apicola, Starmerella bacillaris, Gluconobacter oxydans, and Lactococcus lactis were positively correlated with the production of esters and higher alcohols. Instead of bacterial microbiota, fungal community succession exhibited a positive correlation with the final wine volatiles under the rain-shelter cultivation. These findings demonstrated rain-shelter cultivation influences the succession pattern of microbial communities and in turn impacts the wine aromas and flavors.
Introduction
Microorganisms largely influence the quality of wine. Grapevine-associated indigenous microbiota has been exploited in winemaking for their capability of improving wine sensory characteristics. Grape-derived metabolites are spontaneously released by the microbes in fermentations, creating wines with unique characteristics representing their geographical identity, which benefit from the regional vineyard microbiota known as microbial terroir (Li et al., 2021, Liu et al., 2021).
Non-Saccharomyces yeasts such as Debaryomyces, Hansenula (Pichia), Candida, and Hanseniaspora possess high β-glucosidase activity and significantly alter wine aroma and flavor by releasing volatile compounds from non-volatile precursors in different grape varieties (Del Fresno et al., 2021, Morrison-Whittle and Goddard, 2018, Wei et al., 2022). Bacteria are also involved in the biochemical process that converts grape must into wine. Acetic acid bacteria (e.g., Acidobacterium, Gluconacetobacter) can spoil wine fermentation and lead to wine faults due to the over-production of acetic acid (Barata et al., 2012, del Carmen Portillo and Mas, 2016). Albeit these microbiota are sensitive to stressors, metabolites produced by indigenous microbes may still affect sensory profiles during fermentation (Barata et al., 2012).
The major wine regions in China have a monsoon continental climate (Wang et al., 2018). The high precipitation during the grape maturation may cause grape disease which decrease the quality and yield of grapes. To avoid the harmful impacts of surplus rainfall, rain-shelter cultivation has been applied as an emerging viticulture practice (Duan et al., 2019). The shelters protect vines from rainfall, which reduces physical damages to fruit, and lowers the chance of microbial diseases such as powdery mildew, downy mildew and botrytis (Duan et al., 2019, Li et al., 2014). In addition, the rain-shelter can also change the microclimate of grape bunches, particularly in temperature, humidity and UV radiation (Holcman et al., 2018, Huang et al., 2022, Li et al., 2014). The altered microclimate will in turn influence the microbial community structure (Huang et al., 2022). At grape harvest, these microbial communities can be transferred to grape must/juice via pressing or destemming (Barata et al., 2012, Morrison-Whittle and Goddard, 2018). However, few studies showed the effects of rain-shelter cultivation on microbiota diversity and their succession in spontaneous fermentation as well as simultaneously quantified the contribution of yeast, bacteria, and fungi to the sensory profile of the final wine.
To understand how microbial variation can be transferred from vineyard to wine fermentation and whether rain-shelter cultivation led to a unique wine profile. In this work, microbial communities were monitored during the spontaneous fermentation of Cabernet Sauvignon harvested from rain-shelter and open-field cultivation. Volatile compounds and sensory characteristics of the resulting wines were evaluated and correlations between microbial communities and volatiles in the wines were elucidated. Furthermore, partial least squares regression (PLSR) analyses revealed the relationship between volatiles linked to multiple layers of microbiota and the sensory characteristics of the wines.
Materials and methods
Grape sampling
Cabernet Sauvignon (Vitis vinifera L. self-rooted) was cultivated under both the rain-shelter and open-field modes at the vineyard in Jingyang, Shaanxi, China (34°40′56′′ N, 108°38′53′′ E). The vineyard setup was described in the previous study (Huang et al., 2022). The chemical parameters of harvested grapes such as titratable acidity, ◦Brix, and pH were measured based on the National Standard of the People’s Republic of China (GB/T 15038-2006) listed in Table S1. Cabernet Sauvignon grapes with a titratable acidity of 5.93 to 6.38 g/L (tartaric acid) and total soluble solids of 16.10 to 16.68 °Brix were harvested for spontaneous fermentations. 50 Kg of each cultivation group was collected based on the 5-point sampling method (Huang et al., 2022). Harvested grapes were immediately placed in a sterile box and transferred to the laboratory within 4 h.
Spontaneous fermentation
Destemmed grape musts (∼17 kg) were transferred into 20 L sterilized tanks and incubated at 23–25 °C in triplicates. When grape mash density was reduced to 1.000, the fermenting wine was separated from the solids for continuous alcoholic fermentation. The wine was racked when 0.992 g/mL density and less than 4 g/L residual sugar were reached. 50 mL samples at four fermentation stages from each tank were collected for high-throughput Illumina sequencing. The four stages were (1) grape juice (GJ, after destemming and crushing), (2) at the beginning of alcoholic fermentation (BF, density = 1.064 ∼ 1.067 g/mL), (3) in the middle of alcoholic fermentation (MF, density = 1.038 ∼ 1.041 g/mL) and (4) at the end of alcoholic fermentation (EF, density = 0.995 ∼ 0.992 g/mL). The resultant wine samples were analyzed for basic physicochemical parameters, such as alcohol and residual sugar, following the protocols reported in OIV-INT-00-2020 (OIV, 2020).
Volatiles analysis
The analysis of volatile compounds by gas-chromatographic mass-spectrometric (GC–MS) was based on Ju and coworkers’ study (Ju et al., 2018). The volatile compounds were identified using the NIST14.L standard library (National Institute of Standards and Technology, USA). The peak area on the total ion chromatogram was used for quantification. Calibration curves for target compounds were constructed by graphing the area ratio of an individual target compound to the internal standard (ITSD, 4-methyl-2-pentanol) and calculating against the ITSD concentration. Agilent ChemStation (USA, version 10.0) was employed to calculate the regression correlation coefficients of the calibration curves. Information of the analytes regarding CAS number, retention time, reference Wang et al. (2016) (retention index; RI), calculated RI, and quantifier/qualifier ions are shown in Table S2.
Sensory analysis
Fourteen trained panelists (seven males, and seven females, aged 20 to 23 years) participated in the sensory evaluation of Cabernet Sauvignon wines. The participants had more than 2 years’ experience in wine evaluation at the College of Enology at Northwest A&F University (China). Wine sensory attributes were evaluated according to the protocol reported by (Wang et al., 2016). Five to six descriptors for evaluating the characteristics of rain-shelter and control treatment wines were carried out by descriptive analysis, using a 5-point scale (1 = weak; 2 = slightly weak; 3 = medium; 4 = slightly intense; 5 = intense). The tasting session was conducted in a tasting room at 20 °C, where 20 mL of each wine sample was served in a XL5 (ISO standard) clear wine glass covered with a plastic lid. The wine samples were evaluated by sniffing the aroma for 5 to 7 s followed by 8 to 10 s shaking before tasting. The calculation was performed by the mixed modified frequency (MF) of detection intensity and frequency, as shown in the below equation:
F% is the detection frequency, and I% is average intensity.
DNA extraction and sequencing analysis
Microbial DNA was extracted using HiPure Soil DNA Extraction Kit (Magen, Guangzhou, China) following the manufacturer’s protocol. DNA extracts as templates were stored at −20 °C for later PCR amplification. PCR mixture preparation, reaction conditions, and further product purification were conducted following the procedures described in Huang et al. (2022). The purified PCR amplicons were sent to Gene Denovo Biotechnology Co. Ltd. (Guangzhou, China) for Illumina HiSeq 2500 sequencing analysis. The sequencing raw data was filtered with operational taxonomic unit (OTU) annotation operations, consistent with our previous study (Huang et al., 2022).
Statistical analysis
The α-diversity (Shannon, Goods-coverage, and Phylogenetic diversity) was calculated in QIIME (v 1.9.1). Shannon index comparisons among multiple groups were analyzed using Tukey’s HSD test with the R Vegan package (v 2.5.3), as well as Welch’s t-test for comparing the difference of species abundance between the rain-shelter and control groups (Huang et al., 2022). Principal coordinates analysis (PCoA) based on Bray–Curtis similarity distance matrices and the β-distance Permanova (Permutational multivariate analysis of variance) were performed by the Vegan package in R (v 3.4.1) to analyze the microbial community structure (Liu et al., 2021). LEfSe software (v 1.0) (Liu et al., 2021) and random forest package (v 4.6.12) (Wei et al., 2022) were used to conduct and verify the screening of Biomarker species. A Circos map plotted via Circos software (v 0.69-3) was used to describe the dominant microbial species in each treatment. Partial least squares (PLS) regressions were performed on chemical compounds (x-variables) and sensory data (y-variables) using the Unscrambler (v 9.7, Camo, Trondheim, Norway) (Wang et al., 2016). Student’s t-test performed by IBM SPSS Statistics 22.0 software (SPSS Inc., Chicago, IL, USA) was to analyze the significant differences of parameters related to wines from the two treatments. Principal component analysis (PCA) as a multivariate analysis was employed to visualize the contribution of the volatiles of different cultivation treatments via the SIMCA 14.1 software.
Key metabolites were selected based on variable importance in projection (VIP) values > 1 and p < 0.05 in orthogonal partial least squares-discriminant analysis (OPLS-DA) to investigate the association between microbial communities and the production (Lu et al., 2020). Redundancy analysis (RDA) was applied to explore the association of microbial genera with volatiles, using R software (v 2.5.3). Spearman correlation coefficient value ρ ≥ 0.7 showed the correlation between microbial species and volatile compounds was visualized using the Cytoscape 3.5.1 software. Structural equation modeling (SEM) was employed to assess the effects of cultivation patterns on wine aroma. The fungal and bacterial phylogenetic diversity (PD) in grape must under both cultivations were selected as the initial microbiota. Shannon indices of fungal and bacterial diversity during the whole alcoholic fermentation of must (from must to the end of fermentation) were used to characterize the microbiota succession. Z-score normalization of the final wine aromas was applied before the SEM model analysis, which was done with the AMOS software (v20.0, AMOS IBM, NY, USA). The good fit of SEM model was verified by χ2 test (P > 0.05) using the comparative fit index (CFI > 0.9), the goodness of fit index (GFI > 0.8), and Tucker-Lewis index (TLI > 0.9) (Liu et al., 2021).
Results
Microbiota variation in grape juice under two cultivation modes
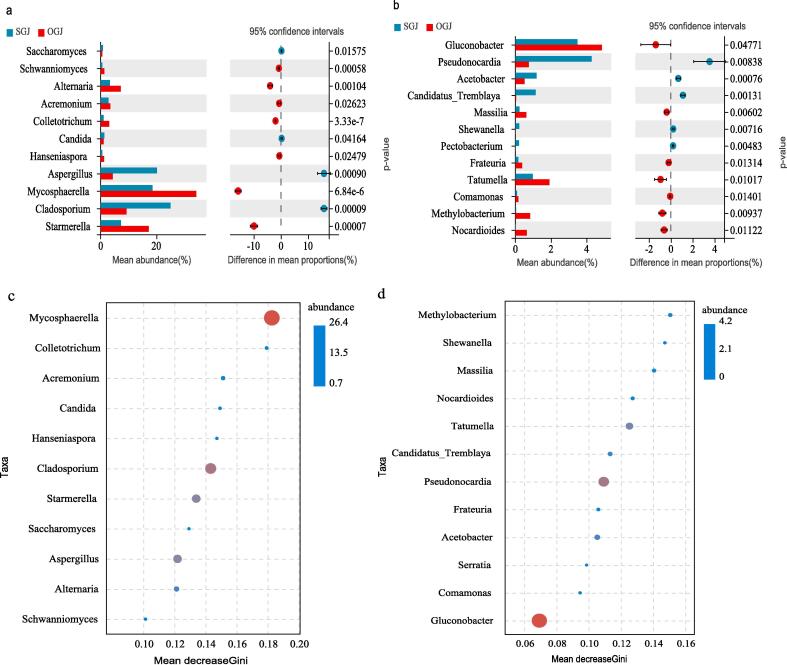
Fermentation samples from four stages (mentioned in methods) were collected for microbial community analyses. A total of 2,799,155 ITS and 2,727,245 16S rRNA high-quality sequences were generated, which were clustered into 9178 fungal and 15,545 bacterial OTUs with a threshold of 97% pairwise identity, respectively (Table S3). The sequencing results demonstrated that the fungal genera with significant differences in abundance from both cultivation modes were mainly Mycosphaerella, Aspergillus, Cladosporium, and Starmerella (Welch’s t-test, p < 0.05; Fig. 1a). To differentiate the fungal taxa of community patterns between the two cultivation modes, random forest analysis was performed. A larger Mean Decrease Gini in the random forest model suggested that the corresponding species were more effective in distinguishing between the rain-shelter and the control. In addition, circles with different sizes were used to reflect species abundance. The random forest model indicated that the fungal microbiota with high abundance in SGJ and OGJ were Mycosphaerella, Cladosporium and Aspergillus (Fig. 1c).
Fig. 1.
Main microbial community compositions in grape juices harvested from two cultivation modes. S: rain-shelter mode; O: open-filed mode. The species abundance of fungal (a) and bacterial (b) communities were characterized at the genus level (Welch’s t-test, p-value ≤ 0.05). Random forest analysis based on Mean Decrease Gini determined significant discriminatory taxa of fungal (c) and bacterial (d) microbiota associated with the cultivation modes.
For bacterial communities, Pseudonocardia and Gluconobacte were the core species that differed significantly in grape juice between the two cultivation modes (Fig. 1b). Random forest analysis revealed that Methylobacterium, Shewanella and Massilia had higher Mean Decrease Gini, indicating that these species may have higher impact on distinguishing the two cultivations despite their low abundance during fermentation (less than 1%) (Fig. 1d).
Fungal microbiota dynamics during spontaneous wine fermentation
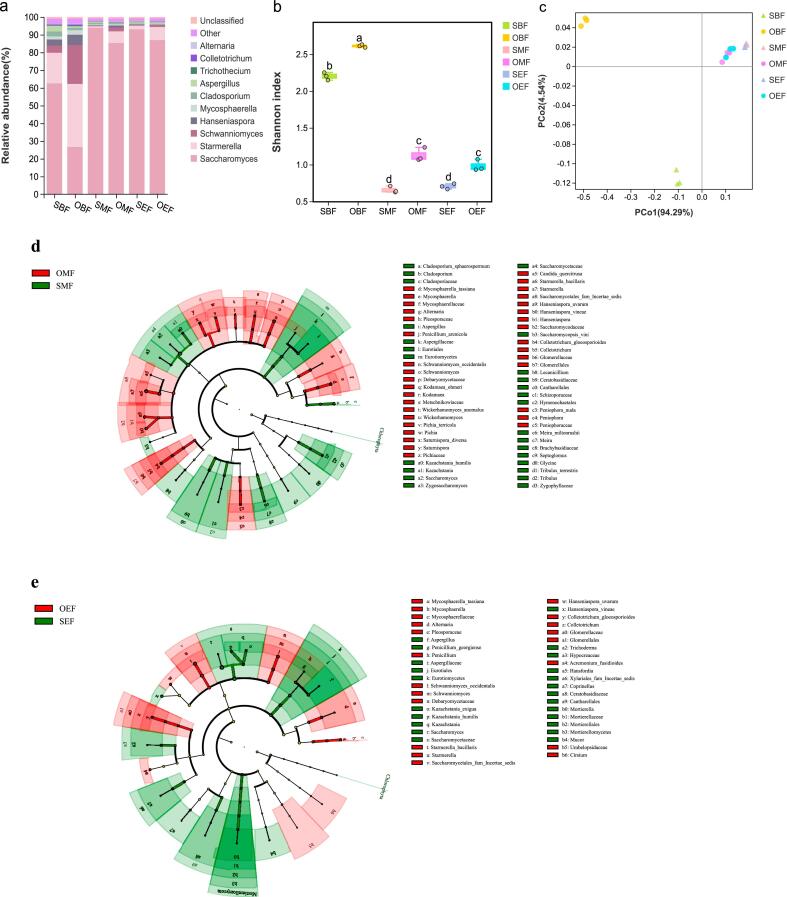
The fungal community succession during spontaneous fermentation was determined for each cultivation method, and the top 10 abundant genera identified throughout the fermentation were listed in Fig. 2a. Microbe with low abundance were categorized as “other”. For rain-shelter cultivation (SBF), Saccharomyces, Starmerella, and Schwanniomyces were the dominant species at the beginning of fermentation with relative abundances of 62.54%, 20.13% and 18.52%, respectively (Fig. 2a). Additionally, higher percentage of Starmerella and Schwanniomyces present in the open-field cultivation at the beginning of fermentation (OBF; 35.56% and 21.94%, respectively). Despite the low initial abundance in grape juice, Saccharomyces in rain-shelter (SBF) was 38.87% higher than that in the control when fermentation just started. In the middle and at the end of the fermentation, the dominant specie was Saccharomyces, with an abundance of at least 85.35%. At the end of fermentation (SEF), the abundance of Starmerella of rain-shelter treatment was lower than 5.37% compared to the control (OEF; 7.39%) (Fig. 2a). Based on the Shannon index, the cultivation method apparently influenced the fungal community at the beginning, in the middle and at the end of fermentation (Fig. 2b). A significant decrease in fungal diversity was observed with the progress of the fermentation, regardless of the cultivation mode (Fig. 2b; Table S3; p < 0.05).
Fig. 2.
Fungal community succession during spontaneous fermentation of grapes harvested under two cultivation modes. BF: at the beginning of alcoholic fermentation; MF: in the middle of alcoholic fermentation; EF: at the end of alcoholic fermentation. (a) The dominant fungal species were characterized at the genus level in the fermentation process. (b) Comparison of fungal diversity under two cultivation modes during fermentation (Tukey-HSD test, p < 0.05); different letters indicate significant differences between the two treatments. (c) PCoA analysis (Bray-Curtis distances) of fungal species distribution (OTUs) under two cultivation modes during fermentation. Identification of discriminant fungal taxa in the middle (d) and at the end (e) of fermentation under two cultivation modes by LEfSe (SMF vs. OMF; SEF vs. OEF; Kruskal-Wallis and rank test α < 0.05; LDA score > 2.00). S: rain-shelter mode, green color-coded; O: open-filed mode, red color-coded. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To compare fungal community patterns during alcoholic fermentation of two cultivation modes, principal coordinate analysis (PCoA) was carried out based on the Bray-Curtis distance. Fig. 2c showed that the succession of fungal microbiota differed with cultivation modes and the fermentation stages. The first two principal coordinate axes explained 95.05% of the total variance (Fig. 2c; 95% confidence interval; Fig. S1a; PERMANOVA, R = 0.929, p = 0.001). The fungal community structures of the two cultivation conditions were different at the beginning of fermentation, but was similar in the middle and at the end of the fermentation, indicating that the fungal community structure tended to be similar as the fermentation progressed (Fig. 2c). LEfSe analysis was performed to compare the differences in fungal communities under two cultivations in the middle and at the end of fermentation (Fig. 2d and 2e). In the middle of fermentations, the fungal community under rain-shelter cultivation (SMF) was mainly enriched by Cladosporium (containing Cladosporium sphaerospermum), Aspergillus, Tribulus (e.g., Tribulus terrestris), Kazachstania (specially Kazachstania humilis), and Meira (including Meira miltonrushii), represent 9.21% of the total fungal population. On the other hand, the fungal community in the control group (OMF) mainly consisted of Mycosphaerella (including Mycosphaerella tassiana), Pichia (particularly Pichia terricola), Candida (including Candida quercitrusa), Colletotrichum (especially Colletotrichum gloeosporioides), Schwanniomyces (notably Schwanniomyces occidentalis), Starmerella (notably Starmerella bacillaris) and Hanseniaspora (involving Hanseniaspora uvarum and Hanseniaspora vineae), which accounting for 7.51% of the total fungal group. At the end of fermentation, the abundance of forty-five fungal species exhibit significant differences between two cultivation modes (Fig. 2e). Aspergillus, Trichoderma, Hansfordia (notably Hansfordia pulvinata), Kazachstania (especially K. humilis) and Hanseniaspora vineae were mainly concentrated at the end of fermentation under rain-shelter cultivation (SEF), accounting for 4.32% of the total. The control group (OEF) was principally occupied by Starmerella (including Starm. bacillaris), Hanseniaspora (involving H. uvarum), Mycosphaerella (especially M. tassiana), Penicillium, Colletotrichum (notably Col. gloeosporioides) and Schwanniomyces (including Sch. occidentalis), accounting for 5.36% of the total (Fig. 2e).
Bacterial microbiota dynamics during spontaneous wine fermentation
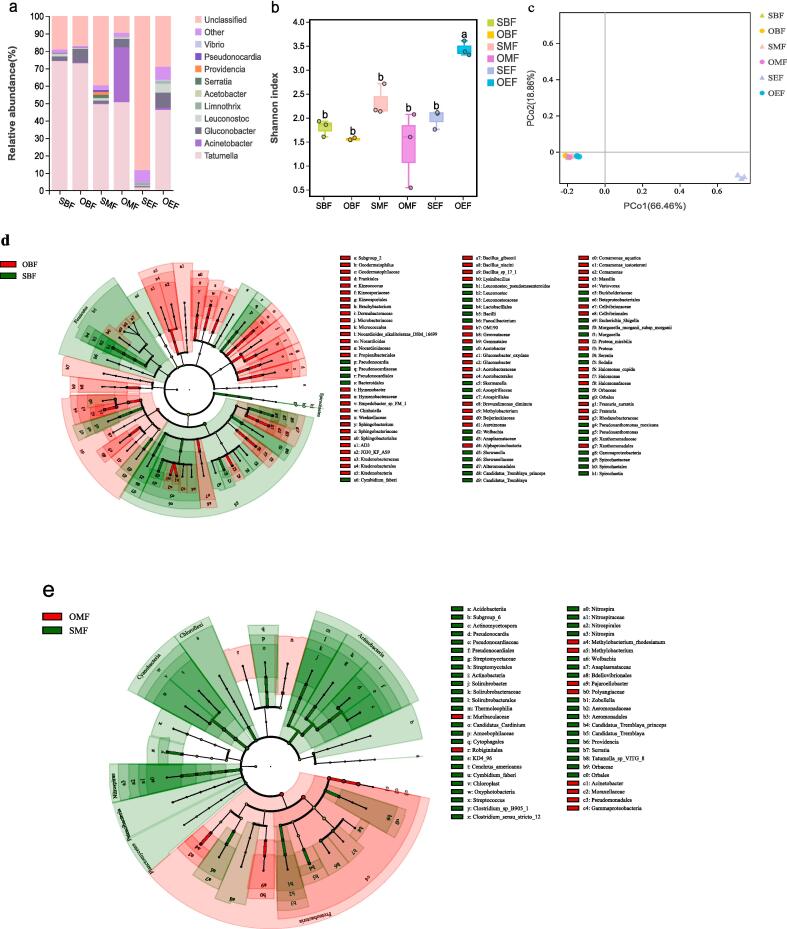
Fig. 3a showed the global features of bacterial community variation during spontaneous fermentation. When fermentation was initiated, Tatumella emerged as the dominant species in the must, accounting for 74.14% and 72.92% of rain-shelter cultivation (SBF) and the control (OBF), respectively (Fig. 3a). As fermentation proceeded, the proportion of Tatumella gradually decreased, but still dominating the bacterial consortia at the middle stage. This was followed by a sharp decrease to ∼1.65% by the end of fermentation for the rain-shelter group whilst the population of Tatumella was marginally reduced for the control. Similarly, for Gluconobacter. The maximum variation in abundance between two cultivation modes was observed at the end of fermentation of 8.27% (OEF; Fig. 3a). Strikingly, a dramatic increase of the Acinetobacter population was observed in the middle of the fermentation (31.41%) for the open-field group. However, the population of Acinetobacter of the rain-shelter group retained at a low level throughout fermentation (OEF; Fig. 3a). Depending on the Shannon indices (Fig. 3b; p < 0.05), the bacterial diversity was comparable between the two cultivation modes at the beginning and in the middle of fermentation.
Fig. 3.
Bacterial community succession during spontaneous fermentation of grapes harvested under two cultivation modes. S: rain-shelter mode; O: open-filed mode. BF: at the beginning of alcoholic fermentation; MF: at the middle of alcoholic fermentation; EF: at the end of alcoholic fermentation. (a) The dominant bacterial species were characterized at the genus level in the fermentation process. (b) Comparison of bacterial diversity under the two cultivation modes during fermentation (Tukey-HSD test, p < 0.05); different letters indicate significant differences between treatments. (c) PCoA analysis (Bray-Curtis distances) of bacterial species distribution (OTUs) under two cultivation modes during fermentation. Identification of discriminant bacterial taxa in the beginning (d) and middle (e) of fermentation under two cultivation modes by LEfSe (SBF vs. OBF; SMF vs. OMF; Kruskal-Wallis and rank test α < 0.05; LDA score > 2.00). S: rain-shelter mode, green color-coded; O: open-filed mode, red color-coded. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The results of principal coordinate analysis demonstrated that bacterial communities from both cultivation modes were clustered according to the fermentation stages with the first two PC axes explained 69.77% of the total variance (Fig. 3c; Fig. S1b; PERMANOVA, R = 0.789, p = 0.001). The bacterial community structure of BF and MF samples appeared to be similar between the two cultivation modes (Fig. 3c). LEfSe analysis was performed to further identify the key differential bacterial species between the rain-shelter and open-field cultivation patterns associated with the BF and MF stages (Fig. 3d). The major bacterial species at SBF stage were Pseudonocardia, Skermanella, Wolbachia, Acetobacter and Leuconostoc (notably Leuconostoc pseudomesenteroides), accounting for 3.92% (Fig. 3d). On the other hand, in the control (OBF), the main bacterial species were Comamonas (especially Comamonas aquatica), Gluconobacter (particularly Gluconobacter oxydans), Aureimonas, Proteus (e.g., Proteus mirabilis), Aureimonas, Massilia and Methylobacterium, accounting for 4.41% (Fig. 3d). In the middle of fermentation, 58 bacterial taxonomic features with significant differences were observed between SMF and OMF (Fig. 3e). In terms of SMF, Actinomycetospora, Solirubrobacter, Streptococcus, Providencia, Zobellella, Nitrospira, Candidatus_Cardinium (i.e. Candidatus_Tremblaya_princeps) and Pseudonocardia were the main bacteria, accounting for 2.79% of the total. However, for the OMF group, the relatively abundant bacteria were Methylobacterium (majorly Methylobacterium_rhodesianum), Acinetobacter, Robiginitalea, Serratia and Pajaroellobacter, accounting for 2.00% of the total (Fig. 3e).
Chemical composition and organoleptic profiles of wines
After the completion of spontaneous alcoholic fermentation, the resultant wines were subjected to analysis of basic physiochemical parameters, encompassing residual sugar, ethanol, volatile acidity, pH, free SO2, and total SO2 (Table S5). All the physiochemical parameters of the spontaneously fermented wines were within the acceptable ranges referring to the GB/T 15038-2006. Among the tested parameters, only volatile acidity and free SO2 were comparable for the wine samples produced under the two cultivation modes (Table S5). Compared to open-field cultivation, the rain-shelter group displayed a significantly higher ethanol and a lower pH level (Table S5; Student’s t-test analysis, p < 0.05).
Furthermore, the effect of rain-shelter cultivation on wine volatile profiles was evaluated via HS-SPME-GC–MS and a total of 52 volatile compounds were quantified (Table S4). Cultivation methods have significant influences on the production of all the detected volatile compounds except for 1-hexanol, ethyl isobutyrate, and ethyl isovalerate (Table S4). Odor activity values (OAV) of aromas were calculated to investigate the influence of each compound on olfactory characteristics. Volatiles with OAV > 0.1 were considered as main aroma compounds and were subjected to student’s t-test analysis (Table S5). Varietal aromas and fermentative aromas were compared between rain-shelter and open-field cultivation. Among varietal aromas, (E)-3-hexene-1-ol, described as a green grassy aroma, was detected to be reduced by about 2.2-fold by rain-shelter cultivation. Rain-shelter cultivation demonstrated a significant influence on linalool (floral) and β-ionone (violet) compared to open-field cultivation. In terms of fermentative aromas, a total of 14 volatile compounds with OAV > 0.1 were significantly different between the two types of wines. Especially, under rain-shelter cultivation, ethyl 2-methylbutyrate (sweet fruit aroma) was about 2 folds higher than that in open-field group wine. The concentration of octanoic acid (butter), isovaleric acid (cheese), and isoamyl acetate (banana) were about 1.5 folds higher than those of the control. In contrast, ethyl acetate, which contributes to fruity and pineapple aroma, was 2-fold lower than that of the control. Besides, higher alcohols (e.g., isobutyl alcohol, isoamyl alcohol, 1-octen-3-ol) were present in the control at concentrations about 1.2-fold higher than those in the rain-shelter treatment, mainly bringing to the wine with a wide variety of fusel, nail polish and mushroom aromas. Of further note 1-heptanol, characterized as an aromatic plant aroma, was 34 folds above the rain-shelter mode. Phenylacetaldehyde, which presents candy and flower aroma, was reduced by approximately 1.3 folds by the rain-shelter treatment (Table S5).
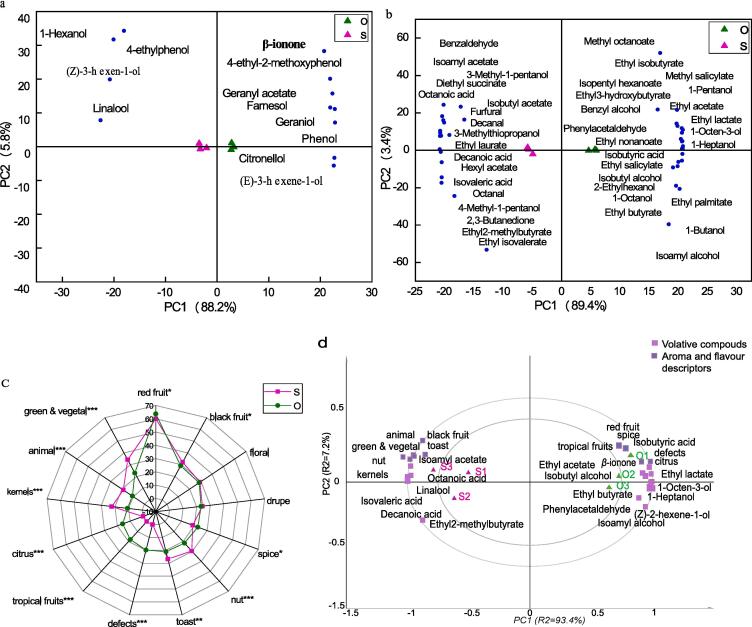
To thoroughly assess the influence of rain-shelter treatment on varietal aroma, PCA analysis was carried out for all 12 volatiles corresponding for varietal aroma. PC1 and PC2 account for 88.2% and 5.8% of the total variance (Fig. 4a). The samples from rain-shelter cultivation (sample S) were located at the negative axis of PC1, whereas the samples from open-field cultivation (sample O) were located on the positive axis of PC1. Specifically, the samples from rain-shelter treatment were grouped for higher production of 1-hexanol, (z)-3-hexen-1-ol and linalool. These compounds could contribute to more intense green grass and floral aromas. Rain-shelter treatment was also positively correlated with 4-ethylphenol, described as an off-flavor from shoe oil, but with OVA values <0.1 in both treatments (Fig. 4a; Table S4). Citronell, geraniol and farnesol, which described as floral, apple, lemon and grass, were the most abundant terpenes in the control treatment (Fig. 4a; Table S4). However, their OAV values were lower than 0.1. Two volatile phenols, 4-ethyl-2-methoxyphenol and phenol were also predominantly related to the control that possibly conferred wine with honey and shoe polish odors (Fig. 4a). However, β-ionone was the only norisoprene volatile identified with OAV value >1, apparently very close to the control, potentially imparting the wine with violet aroma (Fig. 4a; Table S5).
Fig. 4.
Aromatic and organoleptic characteristics of wine samples from two cultivation modes. Cluster analysis of varietal aromas (a) and (b) fermentative aromas. c: MF% of aroma characteristics of wines. (d) PLS regression plots of correlation loadings between chemical and sensory data showing the 75% (inner) and 100% (outer) explained variance limit. S: wine samples from rain-shelter mode; O: wine samples from open-filed mode; *: 0.01 < p ≤ 0.05; **: 0.001 < p ≤ 0.01; ***: p ≤ 0.001.
PCA analysis was also carried out for fermentative aromas, including 10 higher alcohols, 18 esters, 4 volatile fatty acids, 4 carbonyl compounds, 1 sulfur compound, and 3 benzene derivatives. The first two principal components present in the bi-plot in Fig. 4b explained almost 92.8% of the total variance. Sample O was adjacent to 22 volatile compounds located in the positive axis of PC1. On the other hand, sample S is located on the negative axis of PC1with18 fermentative aroma compounds (notably isobutyric acid, octanoic acid, isovaleric acid, furfural, octanal, decanal, 2, 3-butanedione, etc.; Fig. 4b). This suggests that the rain-shelter mode had a greater contribution towards volatile fatty acids and carbonyl compounds, probably endowed the wine with a range of fatty, earthy, caramel and buttery aromas. On the other hand, some compounds contributed to the fruity and floral attributes such as 1-octanol, ethyl acetate, ethyl isobutyrate, ethyl butyrate, ethyl 3-hydroxybutyrate, benzyl alcohol, phenylacetaldehyde, etc. were mainly associated with the control treatment (Fig. 4b). In addition, several compounds with mushroom and plant aroma such as 1-octen-3-ol, 2-ethylhexanol, 1-heptanol and isopentyl hexanoate, were observed closer to the control treatment (Fig. 4b). It further proposed that the open-field treatment seems to promote the production of these compounds (Fig. 4b; Table S4).
To clarify the effect of rain-shelter cultivation on organoleptic characteristics of wine, sensory analysis was performed on the final wine samples in this study (Fig. 4c). In total, 13 categories of sensory traits were evaluated: red fruit (three descriptors), black fruit (three descriptors), floral (six descriptors), drupe (three descriptors), green and vegetal (five descriptors), spice (six descriptors), nut (three descriptors), toast (four descriptors), tropical fruit (two descriptors), citrus (one descriptor), kernels (one descriptor), animal (two descriptors), and defects (one descriptor; nail polish) (Table S6). Except for the floral and drupe characteristics, the MF% of the remaining aroma characteristics differed significantly between the two cultivations (Fig. 4c). In general, all wine samples possessed red fruit, black fruit, floral, drupe, and spice aromas. The rain-shelter mode enhanced the aromas of kernels (e.g., apple), green-vegetal (e.g., green pepper, mushroom, truffle), toast (e.g., caramel), and nuts (e.g., dried prune). The control wines presented more citrus and tropical fruit odor. In addition to these, defects have been detected in sample O (nail polish odor).
To further reveal the contribution of the core volatiles (OAV ≥ 0.1 and p < 0.05, x-variables; Table S5) to the sensory attributes (p < 0.05, y-variables), PLSR was performed. Correlation coefficients > 0 or < 0 suggested that the perception of each aroma was influenced by a combination of positive and negative contributions of volatiles. The wines from the two cultivation patterns were clustered separately (Fig. 4d). Sample S was grouped by black fruit, green & vegetal and toast characteristics, and were strongly positively correlated with isoamyl acetate, linalool, isovaleric acid, octanoic acid and decanoic acid (Fig. 4d; Table S7). For the control (sample O) sensory attributes, red fruit, and spice aromas were positively correlated with ethyl acetate, ethyl lactate, β-ionone and isobutyl alcohol (Fig. 4d; Table S7). PLSR suggested that ethyl butyrate (−0.042), ethyl2-methylbutyrate (−0.179), and decanoic acid (−0.056) could mask the perception of tropical fruit aromas in under rain-shelter cultivation samples (Table S7).
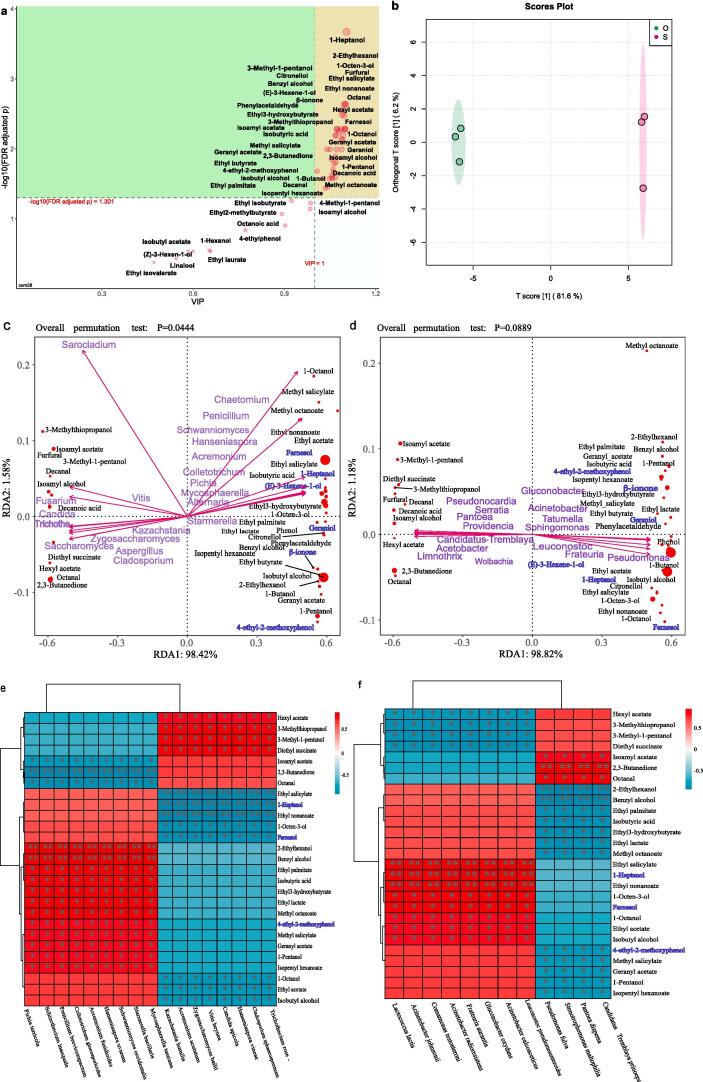
Correlation of microorganisms with major volatile compounds
OPLS-DA analysis was used to screen for differential volatile compounds under two treatments. Differential volatiles were defined as products with a predicted variable importance (VIP) value ≥ 1 and a p-value ≤ 0.05 in the t-test. A list of 39 variable volatiles was screened (Fig. 5a), explaining 81.6% of the variation between the wines under the two cultivation practices (Fig. 5b). To further elucidate the relationship between the microbiota and wine metabolites in the wines under both cultivation modes, species with relative abundance ≥ 0.1% in each sample throughout the fermentation process (from grape juice to EF) were screened against the 39 differential metabolites (Fig. 5c, 5d, 5e, 5f). The correlation was first evaluated at the genera scale via RDA analysis (Fig. 5c, 5d). The fungal genus-differential volatiles, were 98.42% and 1.58% for RDA1 and RDA2, respectively, indicating that the first two axes strongly reflected the relationship between individual fungal genera and volatiles (PERMANOVA, p = 0.0444; Fig. 5c). Particularly, Saccharomyces, Cladosporium, Aspergillus, Trichothecium, Candida and Zygosaccharomyces were mainly distributed on the negative axis of RDA1, with 11 compounds (isoamyl acetate, hexyl acetate, octanal, 3-methyl-1-pentanol, furfural, decanal, 2,3-butanedione, isoamyl alcohol, diethyl succinate, 3-methylthiopropanol, decanoic acid) exhibited a notable positive correlation. Starmerella, Mycosphaerella, Schwanniomyces, Hanseniaspora, Penicillium, Pichia and Chaetomium were positively correlated with the positive axis of RDA1 (Fig. 5c). These fungal genera were positively associated with 24 volatiles (e.g., ethyl acetate, ethyl butyrate, isobutyl alcohol, 1-butanol, 1-pentanol, isopentyl hexanoate).
Fig. 5.
Correlation analysis between microbiota and differential metabolites. OPLS-DA screening of differential metabolites (a) and S-plot distribution of wine samples (b). RDA analysis for predicting fungal (c) and bacterial (d) genera associated with differential metabolites. Spearman correlation heatmap between dominant (e) fungal and (f) bacterial species and volatiles (Spearman correlation coefficient, ρ ≥ 0.7). The blue color indicates a varietal aroma. S: wine samples from rain-shelter mode; O: wine samples from open-filed mode; *: 0.01 < p ≤ 0.05; **: 0.001 < p ≤ 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
For correlations between volatiles and bacterial genera, the RDA analysis reflected eigenvalues of 0.9882 and 0.0118 for the first and second ranking axes, respectively (PERMANOVA, p = 0.0889; Fig. 5d). Isoamyl acetate, hexyl acetate, octanal, 3-methyl-1-pentanol, furfural decanal, 2, 3-butanedione, isoamyl alcohol, diethyl succinate, decanoic acid were located at the negative axis of RDA1 and were positively correlated with Pantoea, Acetobacter, Limnothrix, Providencia Serratia, Candidatus_Tremblaya, Wolbachia. Eight bacterial genera (Tatumella, Acinetobacter, Gluconobacter, Leuconostoc, Pseudonocardia, Pseudomonas, Frateuria, and Sphingomonas) were positively correlated with compounds distributed in the positive half-axis of RDA1 (e.g. ethyl acetate, ethyl butyrate, isobutyl alcohol, 1-butanol, 1-pentanol, ethyl lactate, methyl octanoate, 1-octen-3-ol, isopentyl hexanoate, 2-ethylhexanol) (Fig. 5d).
Correlations of differential volatiles to fungi and bacteria were further conducted at the species scale (Fig. 5e, 5f). Eight fungal species (C. sphaerospermum, Trichothecium roseum, H. vineae, Candida apicola, Vitis heyneana, Zygosaccharomyces bailii, Acremonium acutatum, Kazachstania humilis) presented significant positive correlations with hexyl-acetate, 3-methyl-1-pentanol, diethyl succinate, 3-methylthiopropanol, while showing remarkable negative correlations with a range of compounds (e.g., ethyl acetate, isobutyl alcohol, 1-pentanol, 1-octen-3-ol, ethyl-nonanoate, 1-octanol, ethyl-salicylate, farnesol). The remaining nine fungal species (Starm. bacillaris, M. tassiana, Schw. occidentalis, H. uvarum, Acremonium fusidioides, Colletotrichum gloeosporioides, Penicillium brevicompactum, Schizothecium inaequale, Pichia terricola) were negatively correlated with 2, 3-butanedione, octanal and isoamyl alcohol, while a noticeable positive association was observed with 12 volatiles (including ethyl butyrate, isoamyl acetate, 1-butanol, ethyl lactate, methyl-octanoate, (E)-3-hexene-1-ol, 1-heptanol, isopentyl hexanoate, furfural, 2-ethylhexanol, decanal, ethyl3-hydroxybutyrate) (Fig. 5e).
For correlations between bacterial species and volatile compounds, Acinetobacter calcoaceticus, Leu. pseudomesenteroides, G. oxydans, F. aurantia, Acinetobacter radioresistens, Comamonas testosteroni, Acinetobacter johnsonii, and L. lactis were found strongly and negatively correlated with hexyl acetate, 3-methylthiopropanol, 3-methyl-1-pentanol and diethyl succinate. These bacteria had a strong correlation with ethyl salicylate, 1-heptanol, ethyl nonanoate, 1-octen-3-ol, farnesol, 1-octanol, ethyl acetate, isobutyl alcohol. Pantoea dispersa, Candidatus_Tremblaya princeps, Stenotrophomonas maltophilia and Pseudomonas fulva positively correlated with isoamyl-acetate, 2, 3-butanedione, octanal, while significantly negatively correlated with 2-ethylhexanol, benzyl alcohol, ethyl-palmitate, isobutyric acid, ethyl3-hydroxybutyrate, ethyl lactate, methyl octanoate, 4-ethyl-2-methoxyphenol, methyl salicylate, geranyl acetate, 1-pentanol, and isopentyl hexanoate (Fig. 5f).
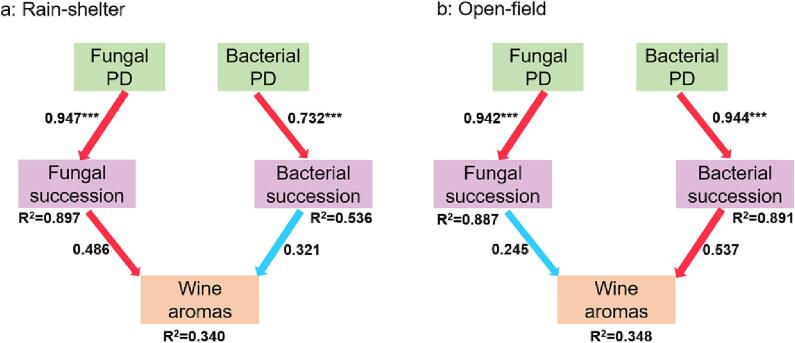
To further disentangle the effect of cultivation patterns on the final wine aroma profile, structural equation modeling (SEM) was used to explore the role of cultivation patterns on the initial microbiota as well as on microbial community succession, transitioning to indirect effects on wine volatiles. The SEM explained 34.0% and 34.8% of the variance in the final contribution of rain-shelter and open-field cultivation to wine aroma, respectively (Fig. 6). The beginning microbiota, represented by phylogenetic diversity (one of the α-diversity indexes), exerted a significant positive effect on microbial succession, regardless of cultivation mode (path coefficients were all significant***; Fig. 6). A positive contribution of fungal communities to wine aromas was predicted under rain-shelter conditions (path coefficient = 0.482; Fig. 6a). The fungal community in the grape juice (path coefficient = 0.947***; Fig. 6a) indirectly drove and regulated wine aroma through its effect on fungal microbiota succession. However, the bacterial community had a lower contribution to wine aroma under rain-shelter mode (path coefficient = −0.321; Fig. 6a). On the contrary, SEM under open-field cultivation suggested that the initial bacterial population played a stronger role on wine volatiles by modulating bacterial community succession (path coefficient = 0.537; Fig. 6b). Compared to bacterial communities, the effect of changes in fungal communities on wine volatiles was low under open-field cultivation (path coefficient = 0.245; Fig. 6b). In summary, rain-shelter cultivation influences wine aromas mainly through the diversity of fungal community, while open-field mode depends mainly on the succession of bacterial communities to influence wine’s volatile characteristics.
Fig. 6.
Direct effects of initial microbiota and microbial community succession in the fermentation on wine aroma profiles. Structural equation model (SEM) fitting of normalized total effects of rain-shelter (a) and open-field (b) cultivations on wine aromas. Measures of overall model fit under rain-shelter cultivation mode are as follows: GFI: 0.881, NFI: 0.976, CFI: 0.974, Pχ2: 0.269. Measures of overall model fit under open-field cultivation mode were as follows: GFI: 0.843, NFI: 0.995, CFI: 0.995, Pχ2: 0.397. PD: phylogenetic diversity of initial microbiota (grape juice). Shannon index of microbiota from grape juice to the end of fermentation as a measure of microbial succession. SEM showed the impact on initial microbial diversity, microbial succession and metabolic diversity (wine aromas). Significance is indicated by *: 0.01 < p ≤ 0.05; **: 0.001 < p ≤ 0.01; ***: p ≤ 0.001. Positive paths were shown in red, but negative paths were indicated in blue. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Discussion
Interactions between fungal diversity and aroma volatiles
Fungal microbiota is ubiquitous in grape juice/must fermentation, with obvious biodiversity (Liu & Howell, 2020). Regardless of the cultivation mode, fungal diversity reduces as alcoholic fermentation progressed (Fig. 2a and b), and the community structure became gradually similar as fermentation progressed to the middle and the end stages (Fig. 2c). This observation is in agreement with recent studies, which described the fungal succession during spontaneous fermentation of Chardonnay and Pinot Noir in the Australian wine region (Liu et al., 2021), and Cabernet Sauvignon wine in China (Li et al., 2021, Wei et al., 2022). Generally, the evolutionary collapse of fungal population diversity in grapes with alcoholic fermentation always follows this trend. It was axiomatic that such a trend in fungal diversity also caused the elimination of other microorganisms (e.g., molds, non-Saccharomyces yeasts), resulting in Saccharomyces with superior succession emerging as the predominant species (Fig. 2a). It was attributed to the low ethanol tolerance of other microbes and the reduction of nutrition (e.g., assimilable nitrogen sources) in the fermentation environment (Liu et al., 2021, Pinto et al., 2015, Wei et al., 2022).
Non-Saccharomyces yeasts can initiate the fermentation (e.g., Starmerella, Hanseniaspora, Candida), but cannot maintain their presence to the end of the fermentation, which is attributed to their intolerance to low oxygen and high ethanol, and weak fermentation metabolism (Bokulich et al., 2016, Liu et al., 2021). In our study, Starmerella, as the dominant fungal genus at the beginning of fermentation, was less abundant in sample S than in sample O (Fig. 2a; Fig. S2a). One of the most important species of this genus is Starm. bacillaris (formerly known as Candida zemplinina) (Englezos et al., 2015). Several studies have shown that strains of Starm. bacillaris produce desirable secondary metabolites (terpenes, lactones) in wine, thus potentially enhancing wine quality (Magyar et al., 2014, Sadoudi et al., 2012). Starm. bacillaris is also able to increase the concentration of higher alcohols, ethyl esters, and short-chain fatty acids in Macabeo wine fermentations (Andorrà et al., 2012). In our study, Starm. bacillaris is positively correlated with the concentration of ethyl butyrate, 1-butanol, and ethyl lactate (Fig. 5e). The concentrations of these three compounds were significantly lower in the rain-shelter wine than in the control (Table S4). Another non-Saccharomyces yeast genera, Hanseniaspora, which also appeared mainly at the beginning of fermentation, composed a higher proportion in the open-field samples (Fig. 2a). H. uvarum exhibits high levels of β-glucosidase and esterase activities, which contributes to the formation of acetate and benzene compounds during wine fermentation (Del Fresno et al., 2021). Our work observed significant positive correlations between H. uvarum and the concentration of esters (e.g. ethyl butyrate, isoamyl acetate, ethyl lactate, ethyl3-hydroxybutyrate) (Fig. 5e). The lower levels of relevant volatiles in wine under rain-shelter conditions may be caused by the lower abundance of this species (Table S4). Under rain-shelter cultivation, the abundance of non-Saccharomyces yeasts (Starmerella, Hanseniaspora) was lower at the beginning of fermentation (Fig. 2a), comparing to the control condition, which may in turn affect the concentration of the corresponding volatile compounds in the final wine. C. apicola secretes proteases which is able to facilitate the production of higher alcohols and acetate esters (Arrizon et al., 2012, Vega-Alvarado et al., 2015). In our work, C. apicola expressed remarkable relationships with the concentration of higher alcohols (isobutanol, 1-pentanol, 1-octen-3-ol) and esters (ethyl acetate, ethyl nonanoate) (Fig. 5e).
In summary, based on the joint correlation analysis of RDA and Spearman (Fig. 5c and 5e), fungi act as double-edged sword on the aroma and flavor of the wines. Our study in tend to predict a possible relationship between fungi and volatiles. The specific contribution of these fungi to wine aroma still needs to be further investigated.
Interactions between bacterial diversity and aroma volatiles
In addition to fungi, various bacterial species are also involved in spontaneous wine fermentation. Tatumella was the dominant genera during fermentation (Fig. 3a), which is in line with previous research (Bubeck et al., 2020, Wei et al., 2022). Gluconobacter and Acetobacter were abundant in the early stages of fermentation, but their population gradually decreased with the fermentation progress (Fig. 3a). These species are highly sensitive to ethanol and therefore generally decline during alcoholic fermentation (González et al., 2005, Wei et al., 2022). Acinetobacter presented mainly in the middle and at the end of fermentation (Fig. 3a), and was more predominant in the control grape must (Fig. S2b). Acinetobacter was noted for its ability to secrete esterolytic enzymes (Ahmed et al., 2010), which promote the production of several volatile compounds in fermented foods, especially esters (Gu et al., 2018, Li et al., 2019, Yao et al., 2021). A potential positive correlation between Acinetobacter with higher alcohols (e.g., isobutanol, 1-butanol, 1-pentanol, 1-heptanol) and ethyl esters (ethyl acetate, ethyl butyrate, ethyl lactate) has been unveiled in this study (Fig. 5d). The high abundance of Acinetobacter in the control appeared to result in higher concentrations of these compounds in the control treatment (Table S4). The abundance of Acinetobacter was lowered in the rain-shelter mode, which potentially affected the concentration of higher alcohols in the final wine.
Leuconostoc is indigenous to plant materials and often dominates the initial stage of fermentation (Fig. 3a). In this study, the rain-shelter treatment reduced the abundance of Leuconostoc compared to the control (Fig. S2b). Leuconostoc can secrete various hydrolytic enzymes (e.g., amylase, protease, and lipase), which assist the release of volatile compounds during the fermentation process (He et al., 2019, Kot et al., 2014, Virdis et al., 2021). In our study, the results showed that Leuconostoc was predominantly associated with 1-octen-3-ol, 1-heptanol, isopentyl-hexanoate, and 2-ethylhexanol (Fig. 5d), thus possibly affecting wine characteristics. It can be further speculated that the decreased abundance of Leuconostoc in rain-shelter mode may alter the levels of these volatiles in wine.
L. lactis as lactic acid bacteria affect the aroma matrix in fermented food by producing a wide range of important enzymes, including α-glucosidase and acetyl coenzyme A acyltransferase (Bian et al., 2022, Smid and Kleerebezem, 2014, Virdis et al., 2021). In our work, a noticeable positive correlation has been observed between L. lactis and esters (e.g., ethyl salicylate, ethyl nonanoate, ethyl acetate) as well as alcohols (e.g., 1-heptanol, 1-octen-3-ol, farnesol, 1-octanol, isobutyl alcohol) (Fig. 5f). Rain-shelter mode reduced the abundance of L. lactis (Fig. S2d). This may also lead to a decrease in the concentration of compounds positively associated with L. lactis under rain-shelter conditions (Table S4).
The important influence of fungal and bacterial populations on wine characteristics has been widely investigated (Liu et al., 2021, Lu et al., 2020, Virdis et al., 2021, Wei et al., 2022). In our study, interaction correlation analysis also proved the driving role of microbial communities on wine aroma. To further evaluate the effect of cultivation mode on the wine aroma, based on the SEM equation, it was noticed that for the rain-shelter mode, wine characters were mainly determined by the fungal community succession, while the aroma pattern of wines under open-field cultivation depended more on the bacterial communities (Fig. 6a; 6b). In fact, there are many other factors, such as grape must composition, and interactions between microbial populations may also influence wine aromas (Liu et al., 2017). Our results demonstrated the influence of cultivation modes on wine aroma from the fungal and bacterial succession aspect.
CRediT authorship contribution statement
Rong Huang: Conceptualization, Investigation, Formal analysis, Writing – original draft. Jiao Jiang: Writing – review & editing. Ying Su: Writing – review & editing. Hongfei Yu: Data curation, Software. Lei Shen: Software. Yanlin Liu: Resources. Yi Qin: Methodology, Resources. Yuyang Song: Conceptualization, Funding acquisition, Resources, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the following projects: (1) National Key R&D Program of China (Item no. 2019YFD1002500); (2) National Natural Science Key Foundation of China (U21A20269); (3) Ningxia Hui Autonomous Region Key R&D Project (2020BCF01003); (4) Supported by the earmarked fund for CARS(CARS-29-jg-3).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2023.100618.
Contributor Information
Rong Huang, Email: huangr@nwafu.edu.cn.
Jiao Jiang, Email: jiao.jiang@nwafu.edu.cn.
Ying Su, Email: yingsu@nwafu.edu.cn.
Hongfei Yu, Email: Yuhongfei_xyafu@126.com.
Lei Shen, Email: shenlei@nwafu.edu.cn.
Yanlin Liu, Email: yanlinliu@nwsuaf.edu.cn.
Yi Qin, Email: qinyi@nwsuaf.edu.cn.
Yuyang Song, Email: yuyangsong@nwsuaf.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References:
- Ahmed E.H., Raghavendra T., Madamwar D. An alkaline lipase from organic solvent tolerant Acinetobacter sp. EH28: Application for ethyl caprylate synthesis. Bioresource Technology. 2010;101(10):3628–3634. doi: 10.1016/j.biortech.2009.12.107. [DOI] [PubMed] [Google Scholar]
- Andorrà I., Berradre M., Mas A., Esteve-Zarzoso B., Guillamón J.M. Effect of mixed culture fermentations on yeast populations and aroma profile. LWT. 2012;49(1):8–13. doi: 10.1016/j.lwt.2012.04.008. [DOI] [Google Scholar]
- Arrizon J., Morel S., Gschaedler A., Monsan P. Fructanase and fructosyltransferase activity of non-Saccharomyces yeasts isolated from fermenting musts of Mezcal. Bioresource Technology. 2012;110:560–565. doi: 10.1016/j.biortech.2012.01.112. [DOI] [PubMed] [Google Scholar]
- Barata A., Malfeito-Ferreira M., Loureiro V. The microbial ecology of wine grape berries. International Journal of Food Microbiology. 2012;153(3):243–259. doi: 10.1016/j.ijfoodmicro.2011.11.025. [DOI] [PubMed] [Google Scholar]
- Bian X., Chen J.-R., Fan J., Yang Y., Yu D.-H., Ren L.-K.…Zhang N. Microbial and genes diversity analysis: Relationship between starch conversion and carbohydrate metabolism during Niandoubao fermentation via the glutinous proso millet (GPM) process. Food Control. 2022;140 doi: 10.1016/j.foodcont.2022.109154. [DOI] [Google Scholar]
- Bokulich N.A., Collins T.S., Masarweh C., Allen G., Heymann H., Ebeler S.E., Mills D.A. Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics. MBio. 2016;7(3):e00631–e1616. doi: 10.1128/mBio.00631-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck A.M., Preiss L., Jung A., Dorner E., Podlesny D., Kulis M.…Fricke W.F. Bacterial microbiota diversity and composition in red and white wines correlate with plant-derived DNA contributions and botrytis infection. Scientific Reports. 2020;10(1) doi: 10.1038/s41598-020-70535-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Carmen Portillo M., Mas A. Analysis of microbial diversity and dynamics during wine fermentation of Grenache grape variety by high-throughput barcoding sequencing. LWT-Food Science and Technology. 2016;72:317–321. doi: 10.1016/j.lwt.2016.05.009. [DOI] [Google Scholar]
- Del Fresno J.M., Escott C., Loira I., Carrau F., Cuerda R., Schneider R.…Morata A. The Impact of Hanseniaspora vineae fermentation and ageing on lees on the terpenic aromatic profile of white wines of the Albillo variety. International Journal of Molecular Sciences. 2021;22(4):2195. doi: 10.3390/ijms22042195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan B., Song C., Zhao Y., Jiang Y., Shi P., Meng J., Zhang Z. Dynamic changes in anthocyanin biosynthesis regulation of Cabernet Sauvignon (Vitis vinifera L.) grown during the rainy season under rain-shelter cultivation. Food Chemistry. 2019;283:404–413. doi: 10.1016/j.foodchem.2018.12.131. [DOI] [PubMed] [Google Scholar]
- Englezos V., Rantsiou K., Torchio F., Rolle L., Gerbi V., Cocolin L. Exploitation of the non-Saccharomyces yeast Starmerella bacillaris (synonym Candida zemplinina) in wine fermentation: Physiological and molecular characterizations. International Journal of Food Microbiology. 2015;199:33–40. doi: 10.1016/j.ijfoodmicro.2015.01.009. [DOI] [PubMed] [Google Scholar]
- González Á., Hierro N., Poblet M., Mas A., Guillamón J. Application of molecular methods to demonstrate species and strain evolution of acetic acid bacteria population during wine production. International Journal of Food Microbiology. 2005;102(3):295–304. doi: 10.1016/j.ijfoodmicro.2004.11.020. [DOI] [PubMed] [Google Scholar]
- Gu, J., Liu, T., Sadiq, F. A., Yang, H., Yuan, L., Zhang, G., & He, G. (2018). Biogenic amines content and assessment of bacterial and fungal diversity in stinky tofu – a traditional fermented soy curd. LWT, 88, 26-34. https://doi.org/10.1016/j.lwt.2017.08.085.
- He G., Huang J., Zhou R., Wu C., Jin Y. Effect of fortified Daqu on the microbial community and flavor in Chinese strong-flavor liquor brewing process. Frontiers in Microbiology. 2019;10 doi: 10.3389/fmicb.2019.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcman E., Sentelhas P.C., Fonseca Conceicao M.A., Zarate Couto H.T. Vineyard microclimate and yield under different plastic covers. International Journal of Biometeorology. 2018;62(6):925–937. doi: 10.1007/s00484-017-1494-y. [DOI] [PubMed] [Google Scholar]
- Huang R., Shen L., Yu H., Jiang J., Qin Y., Liu Y.…Song Y. Evaluation of rain-shelter cultivation mode effects on microbial diversity during Cabernet Sauvignon (Vitis vinifera L.) maturation in Jingyang, Shaanxi, China. Food Research International. 2022;156 doi: 10.1016/j.foodres.2022.111165. [DOI] [PubMed] [Google Scholar]
- Ju Y.-L., Liu M., Tu T.-Y., Zhao X.-F., Yue X.-F., Zhang J.-X.…Meng J.-F. Effect of regulated deficit irrigation on fatty acids and their derived volatiles in 'Cabernet Sauvignon' grapes and wines of Ningxia, China. Food Chemistry. 2018;245:667–675. doi: 10.1016/j.foodchem.2017.10.018. [DOI] [PubMed] [Google Scholar]
- Kot W., Neve H., Heller K.J., Vogensen F.K. Bacteriophages of Leuconostoc, Oenococcus, and Weissella. Frontiers in Microbiology. 2014;5 doi: 10.3389/fmicb.2014.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Li Y., Luo Y., Zhang Y., Chen Y., Lin H.…Liu Z. Shifts in diversity and function of the bacterial community during the manufacture of Fu brick tea. Food Microbiology. 2019;80:70–76. doi: 10.1016/j.fm.2019.01.001. [DOI] [PubMed] [Google Scholar]
- Li R., Lin M., Guo S., Yang S., Han X., Ren M.…Huang W. A fundamental landscape of fungal biogeographical patterns across the main Chinese wine-producing regions and the dominating shaping factors. Food Research International. 2021;150 doi: 10.1016/j.foodres.2021.110736. [DOI] [PubMed] [Google Scholar]
- Li X.X., He F., Wang J., Li Z., Pan Q.H. Simple rain-shelter cultivation prolongs accumulation period of anthocyanins in wine grape berries. Molecules. 2014;19(9):14843–14861. doi: 10.3390/molecules190914843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Howell K. Community succession of the grapevine fungal microbiome in the annual growth cycle. Environmental Microbiology. 2020;23(4):1842–1857. doi: 10.1111/1462-2920.15172. [DOI] [PubMed] [Google Scholar]
- Liu D., Legras J.-L., Zhang P., Chen D., Howell K. Diversity and dynamics of fungi during spontaneous fermentations and association with unique aroma profiles in wine. International Journal of Food Microbiology. 2021;338 doi: 10.1016/j.ijfoodmicro.2020.108983. [DOI] [PubMed] [Google Scholar]
- Liu Y., Rousseaux S., Tourdot-Marechal R., Sadoudi M., Gougeon R., Schmitt-Kopplin P., Alexandre H. Wine microbiome: A dynamic world of microbial interactions. Critical Reviews in Food Science and Nutrition. 2017;57(4):856–873. doi: 10.1080/10408398.2014.983591. [DOI] [PubMed] [Google Scholar]
- Lu Y., Sun F., Wang W., Liu Y., Wang J., Sun J.…Gao Z. Effects of spontaneous fermentation on the microorganisms diversity and volatile compounds during 'Marselan' from grape to wine. LWT-Food Science and Technology. 2020;134 doi: 10.1016/j.lwt.2020.110193. [DOI] [Google Scholar]
- Magyar I., Nyitrai-Sárdy D., Leskó A., Pomázi A., Kállay M. Anaerobic organic acid metabolism of Candida zemplinina in comparison with Saccharomyces wine yeasts. International Journal of Food Microbiology. 2014;178:1–6. doi: 10.1016/j.ijfoodmicro.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Morrison-Whittle P., Goddard M.R. From vineyard to winery: A source map of microbial diversity driving wine fermentation. Environmental Microbiology. 2018;20:75–84. doi: 10.1111/1462-2920.13960. [DOI] [PubMed] [Google Scholar]
- OIV . OIV; Paris, France: 2020. Compendium of international methods of analysis of vines and musts. [Google Scholar]
- Pinto C., Pinho D., Cardoso R., Custodio V., Fernandes J., Sousa S.…Gomes A.C. Wine fermentation microbiome: A landscape from different Portuguese wine appellations. Frontiers in Microbiology. 2015;6 doi: 10.3389/fmicb.2015.00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoudi M., Tourdot-Marechal R., Rousseaux S., Steyer D., Gallardo-Chacon J.J., Ballester J.…Alexandra H. Yeast-yeast interactions revealed by aromatic profile analysis of Sauvignon Blanc wine fermented by single or co-culture of non-Saccharomyces and Saccharomyces yeasts. Food Microbiology. 2012;32(2):243–253. doi: 10.1016/j.fm.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Smid E.J., Kleerebezem M. Production of aroma compounds in lactic fermentations. Annual Review of Food Science and Technology. 2014;5(1):313–326. doi: 10.1146/annurev-food-030713-092339. [DOI] [PubMed] [Google Scholar]
- Vega-Alvarado L., Gómez-Angulo J., Escalante-García Z., Grande R., Gschaedler-Mathis A., Amaya-Delgado L.…Arrizon J. High-quality draft genome sequence of Candida apicola NRRL Y-50540. Genome Announcements. 2015;3(3):e00437–e1415. doi: 10.1128/genomeA.00437-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virdis C., Sumby K., Bartowsky E., Jiranek V. Lactic acid bacteria in wine: Technological advances and evaluation of their functional role. Frontiers in Microbiology. 2021;11 doi: 10.3389/fmicb.2020.612118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Capone D.L., Wilkinson K.L., Jeffery D.W. Chemical and sensory profiles of rose wines from Australia. Food Chemistry. 2016;196:682–693. doi: 10.1016/j.foodchem.2015.09.111. [DOI] [PubMed] [Google Scholar]
- Wang X., Xie X., Chen N., Wang H., Li H. Study on current status and climatic characteristics of wine regions in China. Vitis. 2018;57(1):9–16. doi: 10.5073/vitis.2018.57.9-16. [DOI] [Google Scholar]
- Wei R., Ding Y., Chen N., Wang L., Gao F., Zhang L.…Wang H. Diversity and dynamics of microbial communities during spontaneous fermentation of Cabernet Sauvignon (Vitis vinifera L.) from different regions of China and their relationship with the volatile components in the wine. Food Research International. 2022;156 doi: 10.1016/j.foodres.2022.111372. [DOI] [PubMed] [Google Scholar]
- Yao, D., Xu, L., Wu, M., Wang, X., Zhu, L., & Wang, C. (2021). Effects of microbial community succession on flavor compounds and physicochemical properties during CS sufu fermentation. LWT-Food Science and Technology, 152, 112313. http://doi.org/https://doi.org/10.1016/j.lwt.2021.112313.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.