Abstract
KDEL receptor (KDELR) is a key protein that recycles escaped endoplasmic reticulum (ER) resident proteins from the Golgi apparatus back to the ER and maintains a dynamic balance between these two organelles in the early secretory pathway. Studies have shown that this retrograde transport pathway is partly regulated by two KDELR-interacting proteins, acyl-CoA-binding domain-containing 3 (ACBD3), and cyclic AMP-dependent protein kinase A (PKA). However, whether Golgi-localized ACBD3, which was first discovered as a PKA-anchoring protein in mitochondria, directly interacts with PKA at the Golgi and coordinates its signaling in Golgi-to-ER traffic has remained unclear. In this study, we showed that the GOLD domain of ACBD3 directly interacts with the regulatory subunit II (RII) of PKA and effectively recruits PKA holoenzyme to the Golgi. Forward trafficking of proteins from the ER triggers activation of PKA by releasing the catalytic subunit from RII. Furthermore, we determined that depletion of ACBD3 reduces the Golgi fraction of RII, resulting in moderate, but constitutive activation of PKA and KDELR retrograde transport, independent of cargo influx from the ER. Taken together, these data demonstrate that ACBD3 coordinates the protein secretory pathway at the Golgi by facilitating KDELR/PKA-containing protein complex formation.
Keywords: protein kinase A, A-kinase anchoring protein, KDEL receptor, ACBD3, membrane trafficking, Golgi, ER
Protein kinase A (PKA) is a cyclic adenosine monophosphate (cAMP)-dependent protein kinase that plays fundamental roles in regulating a variety of physiological activities by specific phosphorylation. The PKA holoenzyme is a heterotetramer composed of two catalytic (C) subunits and a dimer of two identical regulatory (R) subunits, which bind and restrict C subunits in their inactive state. In mammalian cells, there are three isoforms of C subunits (Cα, Cβ, and Cγ) and four isoforms of R subunits (RIα, RIβ, RIIα, and RIIβ). Association of secondary messenger, cAMP, with the R subunit releases catalytic C subunits (1, 2).
More refined regulation of localized PKA activation is facilitated by the A-kinase anchoring proteins (AKAPs), which contain an anchoring domain for specific subcellular locations, thereby compartmentalizing PKA activity with its substrates, effectors, and/or other signaling proteins. These signaling complexes organized by scaffolding AKAPs ensure the specificity and efficiency of the PKA enzyme and orchestrate the crosstalk between various signaling pathways (3, 4, 5). The PKA signaling is well known to be stably associated with the Golgi membranes on the cytosolic side and regulate its morphology and function (6, 7, 8, 9). A few AKAPs have been identified in the Golgi apparatus to provide docking sites for assembly of kinases, phosphatase, receptors, and phosphodiesters (10, 11, 12, 13).
In this study, we sought to unravel the coordinating function of an AKAP for the retrograde transport from the Golgi to the endoplasmic reticulum (ER) in this PKA-dependent process. In the early steps of the secretory pathway, de novo synthesized proteins from the ER enter the Golgi apparatus for posttranslational modification and subsequent delivery to their destination. During this process, many ER resident proteins slowly escape to the Golgi and need to be returned to the ER by KDEL receptor (KDELR) that recognizes the tetrapeptide KDEL—or similar sequences—present in soluble ER proteins (14, 15). KDELR binding to its substrates initiates a PKA signaling pathway that leads to the phosphorylation of numerous proteins in retrograde trafficking (9, 16). However, the AKAP that orchestrates this transport signaling has not been reported.
Our previous studies have demonstrated that acyl-CoA-binding domain-containing 3 (ACBD3), a Golgi scaffolding protein, directly interacts with KDELR and regulates its trafficking via PKA (17). ACBD3 was originally described as an AKAP-like scaffold platform in mitochondria that mediates hormone induced cholesterol transport and steroidogenesis through its interaction with the R subunit of PKA, the peripheral-type benzodiazepine receptor, the translocator protein in the outer mitochondrial membrane, and the steroidogenic acute regulatory protein (a PKA substrate in mitochondria) (18, 19). Although, in mammalian cells, a major portion of ACBD3 localizes to the Golgi apparatus, its potential role in regulation of Golgi-linked PKA signaling pathways remains incompletely understood.
Our results indicate that ACBD3 interacts specifically with the RII, but not RI subunit. At steady state, ACBD3 appears to orchestrate localization of KDELR, RII, and C subunits to the Golgi and facilitates the formation of an inactive KDELR–PKA holoenzyme complex. Once there is an anterograde traffic wave from the ER to the Golgi, binding of KDELR and soluble ER proteins releases the catalytic subunit of PKA from RII and turns on the kinase activity of PKA. Taken together, these results suggest that ACBD3 is a bona fide AKAP that compartmentalizes KDEL receptor and PKA kinase activity at the Golgi for spatial and temporal control of retrograde transport.
Results
ACBD3 is a RII-binding AKAP at the Golgi
After its initial discovery in the early 2000s as a RIα binding protein in the mitochondria, ACBD3 has been studied extensively as a Golgi scaffold protein with diverse functions. To determine whether ACBD3 could recruit PKA to the Golgi as well, we compared the localization of the R subunits in control and ACBD3 KO HeLa cells. Since RIα and RIIα are more ubiquitously expressed than RIβ and RIIβ, both of which are found mostly in brain (20), we focused on RIα and RIIα in our study.
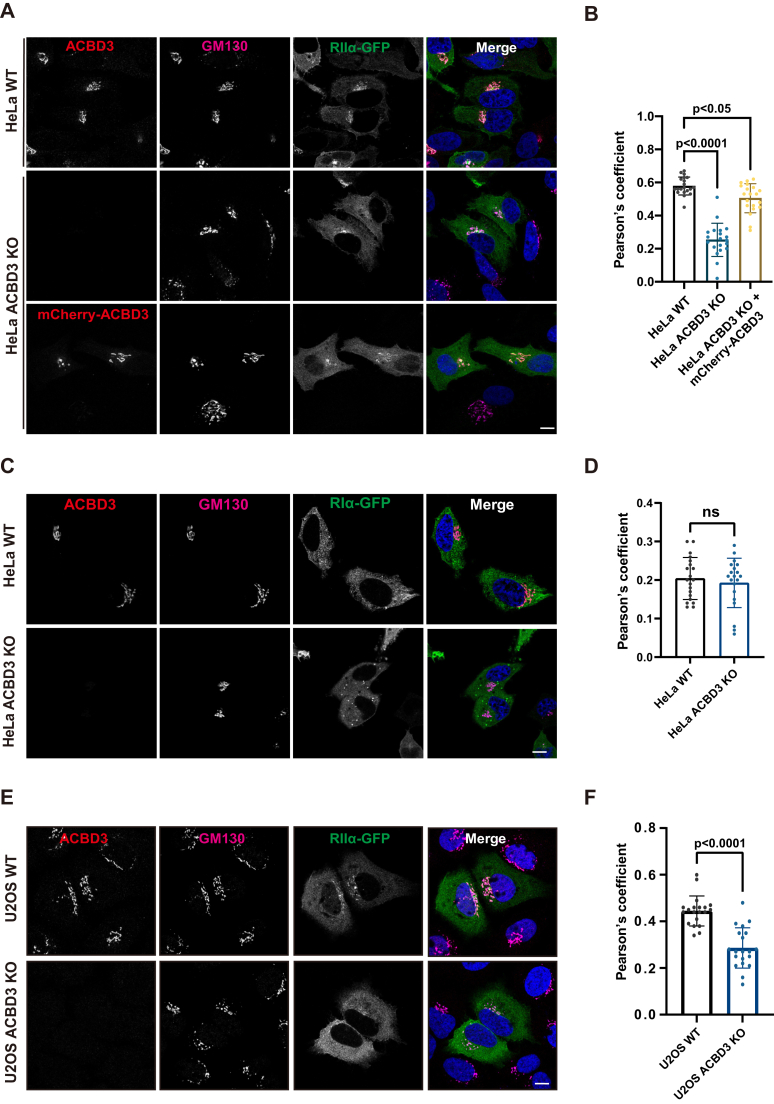
We transiently overexpressed RIα- and RIIα-GFP and utilized a confocal microscope to check whether they were targeted to the Golgi. As shown in Figure 1, A and B a large fraction of RIIα-GFP was indeed colocalized with a Golgi marker, GM130, in HeLa cells. When ACBD3 was depleted, Golgi-targeted RIIα-GFP was significantly decreased, which could be partly restored by overexpression of N terminally mCherry-tagged ACBD3. On the other hand, RIα-GFP showed poor colocalization with GM130 in control and ACBD3 KO HeLa cells, suggesting that ACBD3 does not anchor RIα-GFP to the Golgi (Fig. 1, C and D). Unexpectedly, re-expression of mCherry-tagged ACBD3 was not able to fully restore the Golgi localization of RIIα in ACBD3-depleted cells. This may be due to the high-abundance of endogenous ACBD3 in cells, even compared to transiently overexpressed ACBD3.
Figure 1.
ACBD3 interacts with the RII subunit and localizes it to the Golgi.A and B, ACBD3 facilitates the Golgi localization of RIIα in HeLa cells. A, after transfected with RIIα-GFP or RIIα-GFP/mCherry-ACBD3 for 18 h, control and ACBD3 KO HeLa cells were fixed, stained with anti-GM130 antibody, and then examined by confocal microscopy. B, colocalization of RIIα and GM130 was measured using Pearson’s coefficient and subjected to one-way ANOVA analysis with Dunnett’s post-hoc test for multiple comparisons (mean ± SD, n = 20 cells). F(2, 57) = 83.8, p < 0.0001. C and D, ACBD3 has no effect on the Golgi localization of RIα. C, control and ACBD3 KO HeLa cells overexpressing RIα-GFP were analyzed by confocal experiments. D, colocalization of RIα-GFP with GM130 was calculated and evaluated by two-tailed, unpaired t-tests (mean ± SD, n = 20 cells). ns: not significant. E and F, ACBD3 is required for the Golgi localization of RIIα in U2OS cells. E, control and ACBD3 KO U2OS cells were transfected with RIIα-GFP and observed by confocal microscopy. F, colocalization of RIIα-GFP with GM130 was quantified by two-tailed, unpaired t-tests (mean ± SD, n = 20 cells). Scale bars: 10 μm. ACBD3, acyl-CoA-binding domain-containing 3; RII, regulatory subunit II.
To check whether ACBD3 is required for the localization of RII at the Golgi in other cell lines, we selected a commonly used cell line, U2OS, and examined the localization of RII by immunofluorescence assay. In wildtype U2OS cells, a significant subset of overexpressed RIIα-GFP was localized at the Golgi, similar to what was observed in HeLa cells. Upon ACBD3 depletion, the Golgi-localized RIIα-GFP fraction was greatly reduced, resulting in most of RIIα-GFP in the cytoplasm (Fig. 1, E and F).
Taken together, these results suggest that ACBD3 may be a genuine AKAP that recruits PKA holoenzyme to the Golgi with RII specificity. The discrepancy between ACBD3’s preference for the R subunit at different organelles may be explained by the fact that RI and RII are highly enriched at the mitochondria and the Golgi apparatus, respectively (21, 22).
The Golgi dynamics domain of ACBD3 interacts with RIIα
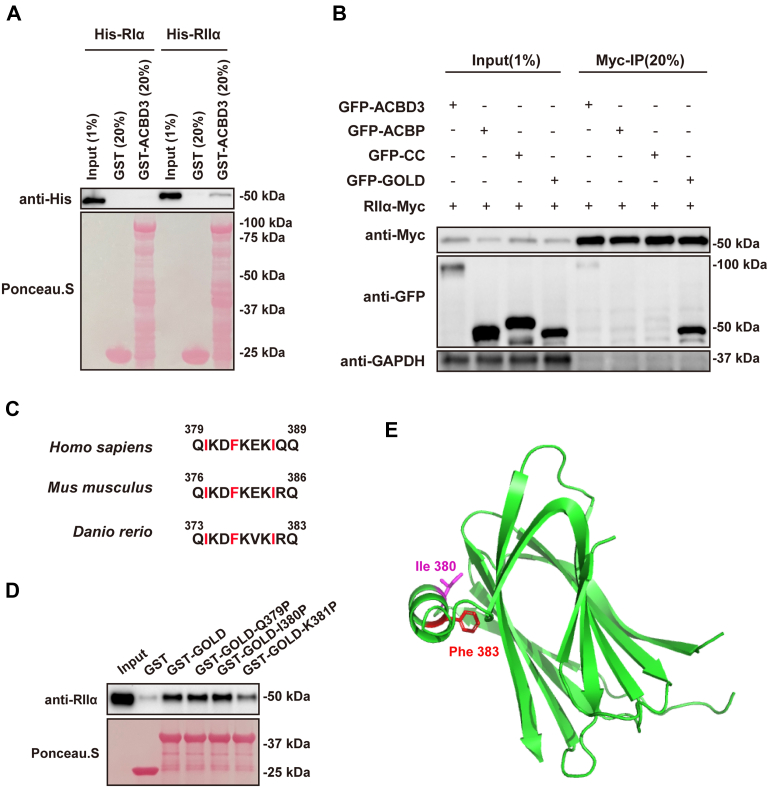
To confirm ACBD3 binding to the RII subunit, a pull-down assay was used to analyze the interaction between recombinant GST-tagged ACBD3 (GST-ACBD3) and His-tagged RIα and RIIα (His-RIα and His-RIIα). As shown in Figure 2A, His-RIIα, but not His-RIα, was readily detected in the pulldown fraction. As a typical dual-specific AKAP interacts with both RI and RII, but with 10 ∼ 100-fold lower affinity for RI than RII (23), it is possible that ACBD3 binds RIα in such low affinity that this binding is below the detection limit in our assay.
Figure 2.
GOLD domain of ACBD3 binds the RIIα subunit directly.A, His-RIIα, but not His-RIα, interacts with GST-ACBD3. GST and GST-ACBD3 immobilized on glutathione beads were incubated with recombinant His-tagged RIα and RIIα. Bound proteins were analyzed by immunoblotting with anti-His antibody (top) or Ponceau S staining (bottom). B, the GOLD domain of ACBD3 binds RIIα. RIIα-myc was cotransfected with GFP-tagged full-length ACBD3, ACBP, CC, or GOLD domains, in HeLa cells for 18 h. Cell lysates were prepared and immunoprecipitated with anti-myc beads, followed by Western blotting analysis. C, a cross-species alignment of the α helix in GOLD domain of ACBD3. Conserved hydrophobic residues are marked in red. D, the α helix in the GOLD domain is responsible for RII binding. Purified GST, GST-GOLD, and GST-GOLD mutants (Q379P, I380P, K381P) were immobilized on glutathione beads and incubated with recombinant His-RIIα. Bound proteins were detected by Western blotting with anti-RIIα antibody (top) or Ponceau S staining (bottom). E, structure of the GOLD domain in ACBD3. Structural illustration is generated by PyMOL using the crystal structure file 5LZ1. The side chains of residue 380 and 383 are shown in magenta and red, respectively. ACBD3, acyl-CoA-binding domain-containing 3; CC, coiled-coil domain; GOLD, C-terminal Golgi dynamics domain; GST-ACBD3, GST-tagged ACBD3; His-RIα, His-tagged RIα; His-RIIα, His-tagged RIIα; KDELR, KDEL receptor; RII, regulatory subunit II.
To further dissect the interaction between ACBD3 and RIIα, we performed co-immunoprecipitation (Co-IP) experiments using GFP-tagged ACBD3 fragments, including the N-terminal acyl-CoA binding domain, the coiled-coil domain, and the C-terminal Golgi dynamics domain (GOLD). The full-length ACBD3 and its fragments were cotransfected with RIIα-myc in HeLa cells followed by immunoprecipitation using agarose beads conjugated with anti-myc antibody. The results of this assay demonstrated that ACBD3 and its GOLD domain were both successfully co-immunoprecipitated with RIIα-myc, indicating that the GOLD domain is the binding site for RII (Fig. 2B).
A classic AKAP contains an amphipathic helix that interacts with a four-helix bundle in the R subunit of PKA (5). The crystal structure of the GOLD domain consists of one α helix and 11 β strands (24, 25). Alignment of the α helix in GOLD domain of ACBD3 from different species showed that residues on the hydrophobic side of the amphipathic helix are well conserved (Fig. 2C). To confirm whether the helical motif in the GOLD domain of ACBD3 is the docking site for RIIα, we generated GST-tagged GOLD mutants (Q379P, I380P, or K381P) for pulldown assays since proline is known to be a potent helix breaker. As indicated by Figure 2D, K381P mutation greatly reduced the interaction between RIIα and the GOLD domain, while Q379P and I380P had almost no effect, suggesting that K381P disrupts the helical structure and the RIIα-binding interface.
As our results indicate that ACBD3 is a RII-preferring AKAP, it is surprising to find a bulky hydrophobic residue, Phe383, in the α helix of GOLD domain, which is usually present in RI-selective AKAPs. To interpret the selectivity of ACBD3 toward RIIα, we examined the crystal structure of the GOLD domain (24). Strikingly, the majority of the hydrophobic surface generated by the side chain of Phe383 is buried inside the GOLD domain, which makes it unlikely to contribute substantially to the binding of R subunit. On the other hand, the side chain of Ile380 is mostly available for protein interactions, likely involved in binding the shallow hydrophobic groove on RII surface (Fig. 2E).
Endogenously tagged RIIα and Cα are enriched at the Golgi by ACBD3
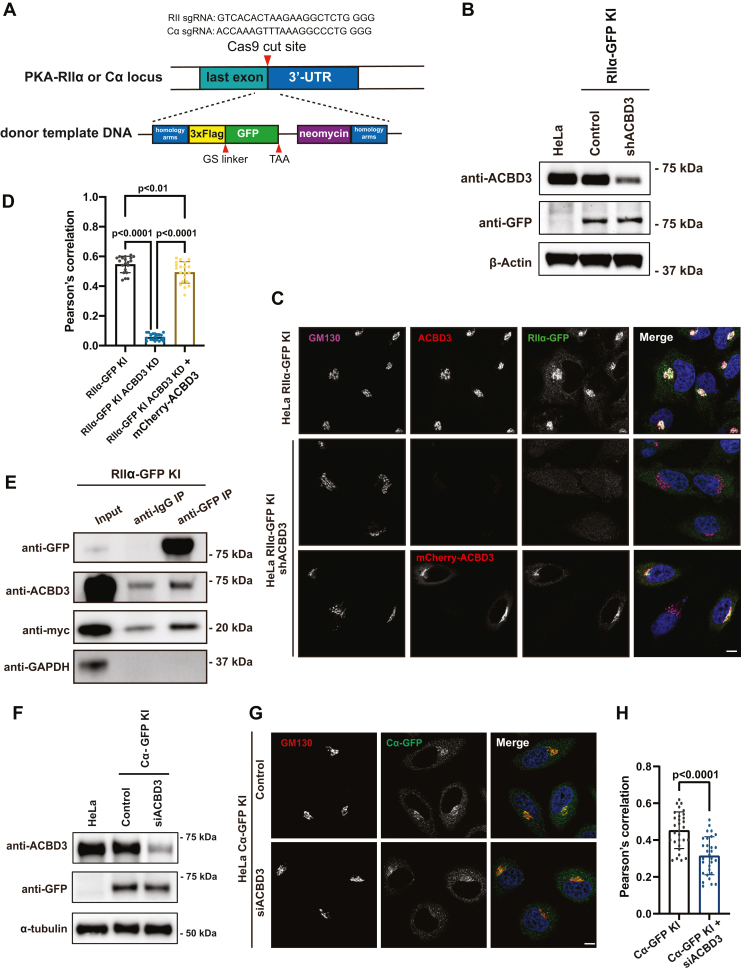
Our results with overexpressed RII subunit clearly showed that ACBD3 is the anchor protein for PKA holoenzyme at the Golgi apparatus. To circumvent possible overexpression artifacts, we studied the localization of endogenous RIIα, tagged with GFP at its C terminus using CRISPR/Cas9 genome engineering technology, due to lack of good commercial antibody for confocal experiments. As illustrated in Figure 3A and the methods, we took advantage of a selection marker, neomycin, and successfully isolated a few monoclonal cell lines verified by genome sequencing and western blots (Fig. 3B).
Figure 3.
ACBD3 is required for Golgi distribution of endogenously tagged RIIα.A, schematic diagram of generating C-terminal 3xFlag-GFP-tagged RIIα or Cα. B, ACBD3 was stably knocked down in RIIα-GFP knock-in HeLa cells (RIIα-GFP KI) using lentivirus expressing ACBD3-shRNA. Western blots analysis shows the expression of endogenously tagged RIIα-GFP and ACBD3 in control and stable knockdown RIIα-GFP KI cells. C, confocal images showing that the localization of RIIα-GFP at the Golgi was depleted by ACBD3 knockdown in RIIα-GFP KI cells. Scale bar: 10 μm. D, colocalization of RIIα and GM130 was represented by bar graphs (n = 20 cells). Statistical analysis was performed using one-way ANOVA with Tukey’s post-hoc test for multiple comparisons (mean ± SD, n = 20 cells). F(2, 57) = 491, p < 0.0001. E, RIIα, ACBD3, and KDELR are in the same protein complex. RIIα-GFP KI cells transfected with KDELR-myc was immunoprecipitated with anti-GFP agarose beads and subjected to immunoblotting using indicated antibodies. F, ACBD3 expression was efficiently depleted in Cα-GFP knock-in HeLa cells (Cα-GFP KI) using siACBD3. The expression of endogenously tagged Cα-GFP and ACBD3 were examined in HeLa, Cα-GFP KI, and ACBD3-depleted Cα-GFP KI cells using Western blots. G, confocal images showing that the localization of Cα-GFP at the Golgi was moderately affected by ACBD3 knockdown in Cα-GFP KI cells. Scale bar: 10 μm. H, colocalization of Cα and GM130 was represented by bar graphs (n = 30 cells). Statistical analysis was performed using two-tailed, unpaired t test. ACBD3, acyl-CoA-binding domain-containing 3; KDELR, KDEL receptor; RII, regulatory subunit II; KI, knockin.
Similar to the exogenously transfected RIIα-GFP, the endogenously tagged RIIα mostly localized to the perinuclear region (Fig. 3C). ACBD3 knockdown by shRNA lentivirus resulted in a significant loss of endogenous RIIα-GFP at the Golgi, which could be largely recovered by subsequent overexpression of mCherry-ACBD3 (Fig. 3, C and D), suggesting that ACBD3 is the anchoring protein for endogenous RIIα at the Golgi apparatus.
Next, we tested whether ACBD3, as an AKAP, provides a platform for its interactors to assemble into a protein complex. To this end, we transfected RIIα-GFP knockin (KI) cell line with KDELR-myc and performed Co-IP experiment to validate protein interactions in complex formation. The results showed that both ACBD3 and KDELR-myc were efficiently pulled down by endogenous RIIα-GFP using anti-GFP agarose (Fig. 3E). As our previous data already proved that ACBD3 interacts with KDELR at the Golgi (17), these results indicated RII, ACBD3, and KDELR are indeed in a protein complex.
Since a typical AKAP recruits PKA in its holoenzyme state, we then examined whether the C subunit of PKA is also recruited to the Golgi by ACBD3. Although there are three isoforms of the C subunit, Cα has been studied the most because it has a higher expression level than Cβ and less cell type specificity than Cγ (26, 27). As our previous data demonstrated that ACBD3 was co-immunoprecipitated with Cα, but not Cβ, in HeLa cells (17), we decided to examine the effect of ACBD3 on localization of Cα only. Similar to RIIα-GFP KI cells, we generated endogenously GFP-tagged Cα KI HeLa cells using CRISPR/Cas9 system (Fig. 3, A and F). As expected, GFP-tagged Cα was localized to the Golgi as well, but to a less extent than RIIα-GFP. Depletion of ACBD3 with small-interfering RNA had a statistically significant but modest effect on the distribution of Cα-GFP, compared to RIIα (Fig. 3, G and H).
Traffic from the ER induces activation of PKA complex anchored by ACBD3
Although our results have shown so far that ACBD3 serves as an anchoring protein for PKA and KDELR, these results did not prove whether the ACBD3-mediated complex is physiologically relevant. One major obstacle in studying PKA is to selectively monitor its activity at the location of interest with an imaging sensor that can yield easily detectable signals. To this end, we modified a fluorescence resonance energy transfer (FRET)-based PKA activity reporter by adding a Golgi localization signal at its C terminus (28, 29).
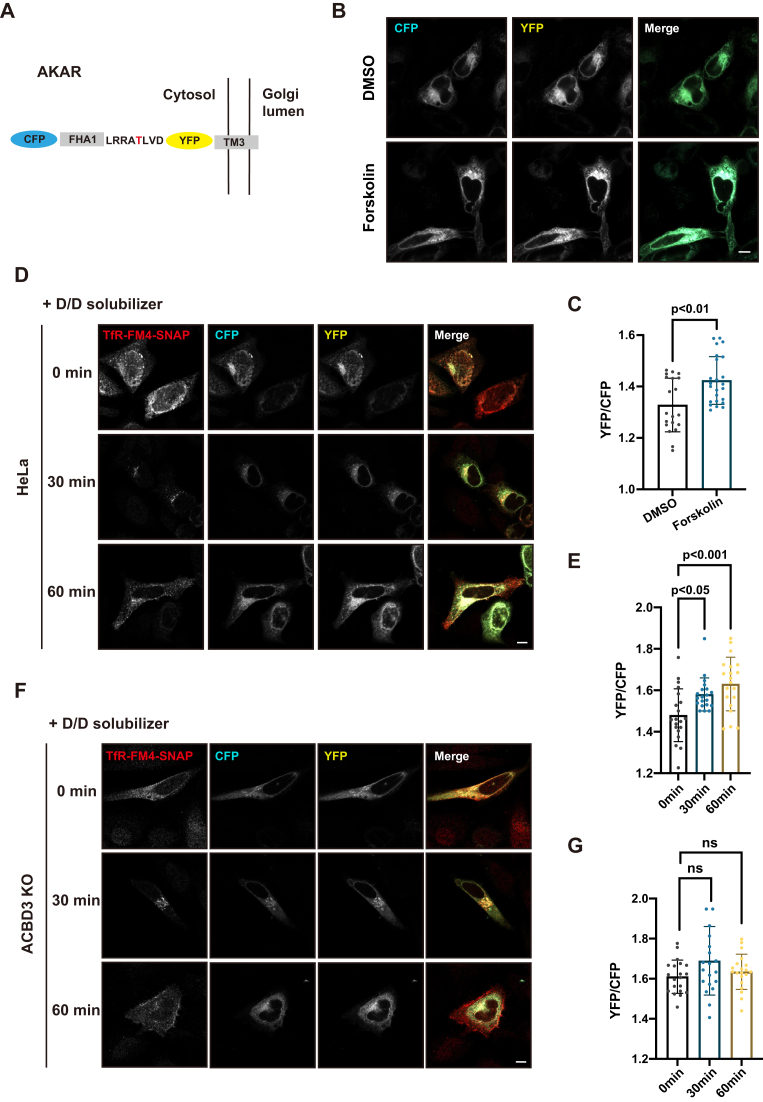
As illustrated in Figure 4A, this sensor construct (A kinase activity reporter, AKAR) contains a cyan fluorescent protein (CFP), a phosphopeptide binding motif, a consensus sequence for PKA phosphorylation, a yellow fluorescent protein (YFP), and a transmembrane domain that localizes the fusion protein to the Golgi. Upon PKA activation, the phosphopeptide binding motif domain binds the phosphorylated substrate sequence, placing CFP and YFP in a close proximity to generate FRET signals. To test whether the FRET sensor, AKAR, generates detectable FRET changes upon PKA activation, HeLa cells were transfected with AKAR and then incubated with DMSO or forskolin, an adenylyl cyclase and PKA activator (Fig. 4B). As expected, forskolin caused a significant increase in the fluorescence intensity ratio of YFP over CFP, compared to DMSO, suggesting that AKAR is a valid sensor for PKA activation (Fig. 4C).
Figure 4.
Cargo wave from the ER activates ACBD3-localized PKA at the Golgi.A, schematic illustration of a FRET-based sensor (AKAR) monitoring PKA activity on the cytoplasmic side of Golgi membrane. The threonine residue in red marks the phosphorylation site by PKA. B and C, forskolin induces FRET signal. HeLa cells were transfected with AKAR for 18 h and incubated with DMSO or 10 μM forskolin at 37 °C for 15 min, prior to confocal microscopy analysis for FRET signals (B). The fluorescence intensity ratio of YFP to CFP is quantified by two-tailed, unpaired t test (C). n = 20 cells. D–G, influx cargos from ER activate ACBD3-anchored PKA at the Golgi. Control (D) and ACBD3 KO (F) cells were transfected with TfR-FM4-SNAP and AKAR for 18 h. Cells were then incubated with D/D solubilizer to dissolve TfR-FM4-SNAP aggregates in the ER and induce synchronized ER-to-Golgi traffic. FRET signals were recorded by confocal microscopy at 0, 30, and 60 min after solubilizer treatment. Quantitative analysis of the fluorescent intensity ratio YFP to CFP in control (E) and ACBD3 KO (G) cells was performed using one-way ANOVA with Dunnett’s post-hoc test for multiple comparisons (mean ± SD, n = 20 cells). F(2, 57) = 8.99, p = 0.00040 (E), F(2, 57) = 2.25, p = 0.11 (G). ns: not significant. Scale bars: 10 μm. AKAR, A kinase activity reporter; CFP, cyan fluorescent protein; ER, endoplasmic reticulum; FRET, fluorescence resonance energy transfer; PKA, protein kinase A; TfR, transferrin receptor; YFP, yellow fluorescent protein.
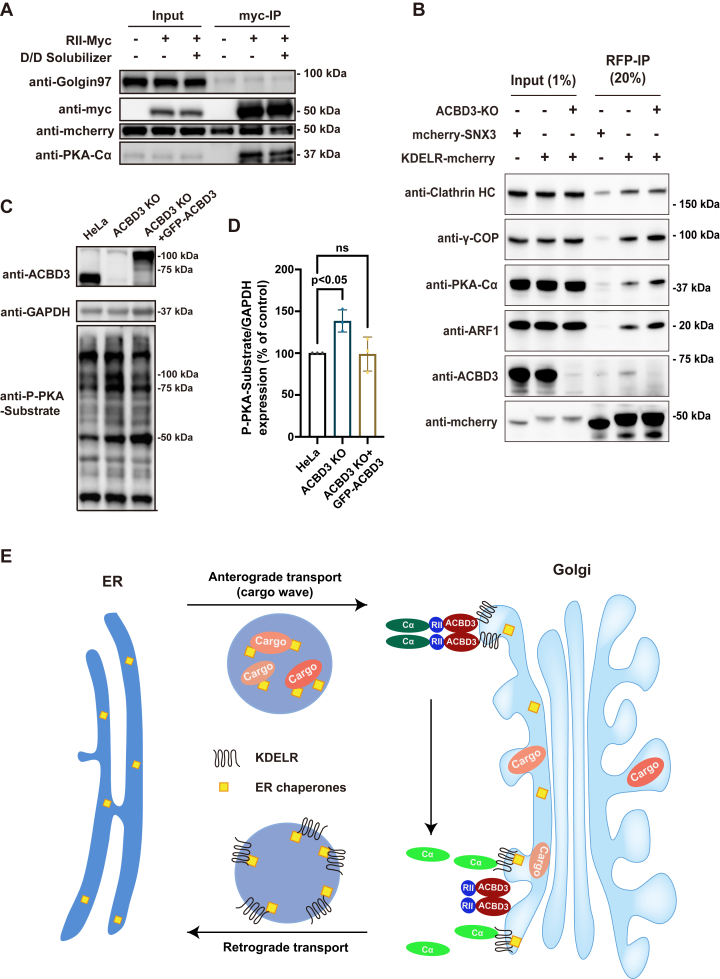
Previous studies have demonstrated that anterograde protein transport from the ER could initiate PKA signaling at the Golgi (16). To explore whether activation of ACBD3-anchored PKA–KDELR complex is triggered by protein traffic, we used a conditional aggregation construct that has been reported before, transferrin receptor (TfR)-FM4-SNAP, to provide controllable protein traffic from the ER and compared PKA activation in control and ACBD3 KO cells (17).
HeLa cells were transfected with TfR-FM4-SNAP and AKAR plasmids for 18 h, followed by addition of D/D solubilizer in the medium to rapidly release TfR-FM4-SNAP from ER. After 30 min, the TfR cargo was already transported to the Golgi and caused enhanced FRET signals, indicating activation of PKA (Fig. 4, D and E). As expected, in ACBD3 KO cells, protein transport from the ER had almost no effect on PKA activation at the Golgi (Fig. 4, F and G), suggesting that ACBD3 is the key protein for PKA signalosome. Interestingly, we noticed that the FRET signal yielded by AKAR before D/D solubilizer treatment appears to be higher in ACBD3 KO cells than control cells. We hypothesized that the KDELR-PKA complex anchored by ACBD3 is inactive until triggered by anterograde transport, whereas KDELR in ACBD3-depleted cells is constantly activated by free C subunit regardless of protein traffic, due to the absence of RII subunit from the Golgi.
PKA activation in ACBD3 KO cells is independent of protein traffic
To test our hypothesis, cells stably overexpressing KDELR-mCherry were transfected with RIIα-myc, prior to Co-IP experiments using anti-myc agarose beads. The amount of both Cα and KDELR pulled down by RIIα-myc was significantly reduced shortly after cargo waves were released from ER, showing that protein transport activates PKA by releasing Cα from RIIα (Fig. 5A). In ACBD3-depleted cells, three key proteins critical for KDELR-dependent retrograde transport, the γ subunit of coat complex, the Cα subunit, and ADP-ribosylation factor 1 (ARF1), had enhanced Co-IP with KDELR-mCherry, compared to the control cells. These results strongly support the notion that lack of ACBD3 as an AKAP results in RII subunit dissociation from the Golgi, leading to activation of KDELR retrograde trafficking regardless of cargo wave (Fig. 5B).
Figure 5.
ER protein influx triggers release of Cα from RIIα subunit anchored by ACBD3.A, cargo wave from ER reduces the binding of Cα and KDELR with RIIα. HeLa cells were transfected with TfR-FM4-SNAP and RII-myc for 18 h, followed by incubation with D/D solubilizer for 30 min. Cell lysates were precipitated with anti-myc beads and analyzed by Western blotting. B, increased pulldown of Cα, γ-COP, and ARF1 by KDELR-mCherry in ACBD3-depleted cells than in wildtype cells. Co-immunoprecipitation experiment was performed in control and ACBD3 KO cells overexpressing KDELR-mCherry or mCherry-sorting nextin 3 (mCherry-SNX3, used as a negative control) using anti-RFP beads. The cell lysate and pulldown fractions were examined by Western blotting. C and D, PKA activity is enhanced in ACBD3 KO cells. C, HeLa WT and ACBD3 KO cells overexpressing control or GFP-ACBD3 plasmids were lysed and subjected to immunoblotting using indicated antibodies. D, statistical analysis of western blots from three independent experiments were performed using one-way ANOVA with Dunnett’s multiple comparison test (mean ± SD, n = 3). F(2, 6) = 7.79, p = 0.022. ns: not significant. E, schematic illustration of the ACBD3–KDELR–PKA signaling complex at the Golgi activated by cargo transport from the ER. ACBD3 anchors KDELR and PKA holoenzyme at the Golgi. Anterograde vesicles carrying ER cargos reach the Golgi and activate Cα subunit of PKA, which then triggers KDELR-mediated retrograde transport of ER chaperones. ARF1, ADP-ribosylation factor 1; ER, endoplasmic reticulum; PKA, protein kinase A; RII, regulatory subunit II; TfR, transferrin receptor.
Thus, we explored whether PKA is constitutively activated in ACBD3 KO cells. Whole cell lysate from control and ACBD3-depleted cells were probed with an antibody that specifically recognizes phosphorylated serine or threonine residues on PKA substrates. The results showed that the phosphorylation of PKA substrates was certainly increased in ACBD3 KO cells, especially around the size of 75 to 100 kD, which could be restored by overexpression of GFP-ACBD3 (Fig. 5, C and D).
Discussion
In this study, we provide evidence that ACBD3 directly interacts with the RII subunit of PKA via its GOLD domain and anchors the PKA holoenzyme to the Golgi, together with KDELR. We also demonstrated that in response to incoming protein traffic from the ER, the C subunit of PKA dissociates from the RII subunit and initiates KDELR-mediated retrieval of ER chaperones in the γ subunit of coat complex–coated vesicles (Fig. 5E).
The Golgi apparatus has been shown to play an important role in compartmentalizing PKA signaling pathways, which in turn seems essential for regulating Golgi structure and function during protein transport along the secretory pathway. For spatial and temporal control of Golgi-associated PKA activation, local cAMP levels are tightly controlled by the adenyl cyclases and phosphodiesterases (30, 31, 32). For retrograde transport, protein fluxes from the ER to the Golgi activates KDELR, which leads to the activation of the classic PKA signaling cascade of stimulatory G protein, AC9, and PDE7A1 (16). Therefore, KDELR seemingly function as a G-protein coupled receptor–like protein at the Golgi, although it has been shown that the receptor structurally resembles SWEET family of sugar transporters, rather than G-protein coupled receptor family of receptors (16, 33). Our data identify ACBD3 as a basis molecule that ensures a quick and efficient response to traffic signal by integrating KDELR-PKA cascade to designated sites.
It is widely accepted that PKA kinases found in microdomains organized by AKAPs are inactive holoenzymes at steady state until free C subunits dissociate from inhibitory R subunits upon signal activation. Indeed, we observed that the Cα subunits bound to RIIα were greatly reduced once KDELR was activated by protein influxes at the Golgi (Fig. 5A). In ACBD3 knockout cells, Golgi localization of Cα was less than in the control cells, yet with a higher catalytic activity, in agreement with the observation that the RII was greatly decreased from the Golgi upon ACBD3 depletion. These data suggest that a certain fraction of Cα subunits may be always associated with the Golgi membranes. In fact, activated Cα released from the Golgi has been reported to remain in close proximity with this organelle even after prolonged incubation with cAMP (30). Moreover, the C subunit anchoring proteins apart from the R subunit, which could occur as inhibitory or noninhibitory, had been reported to be present in the cytoplasm and the nucleus (34).
Our previous study had shown that KDELR trafficking activated by PKA is regulated by ACBD3 via ARF1-dependent tubular carriers (17). ARF1 activators, brefeldin A-inhibited guanine nucleotide-exchange factors 1 & 2 (ArfGEF 1 & ARFGEF2 or BIG1 & BIG2), contain RI and RII binding motifs as well (10). Activation of PKA anchored by BIG1 & 2 stimulates the transport of exosome-like vesicles and enhances β-catenin activity via ARF1 activation (35, 36). Combined with our data that PKA recruited by ACBD3 has a regulatory role in KDELR transport, it seems likely that multiple PKA microdomains, each anchored by a specific AKAP, may integrate different groups of molecules and respond to various signals by regulating ARF1-dependent tubulovesicular trafficking.
Experimental procedures
Antibodies and reagents
Following antibodies were used: anti-GAPDH (HRP-60004, Proteintech), anti-6∗His (ab18184, Abcam), anti-GFP (11814460001, Roche), anti-Myc (2278S, Cell Signaling Technology), anti-ACBD3 (H00064746-B01P, Abnova), anti-P-PKA-substrate(9621S, Cell Signaling Technology), anti-Golgin97 (13192, Cell Signaling Technology), anti-mCherry (ab167453, Abcam), anti-PKA-Cα (#4782, Cell Signaling Technology), anti-PKA-RIIα (612242, BD), anti-γ-COP (sc-393615, Santa Cruz), anti-GM130 (610822, BD Bioscience), anti-SNAP tag (P9310S, NEB), anti-Clathrin HC (ab21679, Abcam), anti-ARF1 (NB300-505, Novus), anti-β-Actin (3700S, Cell Signaling), and anti-α-Tubulin (3873S, Cell Signaling). Anti-Rabbit Alexa Fluor 488 (A21441), Alexa Fluor 568 (A10042), Alexa Fluor 647 (A21245), and anti-Mouse Alexa Fluor 488 (A21200), Alexa Fluor 568 (A10037), and Alexa Fluor 647 (A21236) for immunofluorescence staining were obtained from ThermoFisher. D/D Solubilizer (635054) was purchased from Clontech. Forskolin (F6886) was purchased from Sigma.
Cell culture, transfection, and lentivirus infection
HeLa (ATCC, CCL-2) and HT-1080 (Stem Cell Bank, Chinese Academy of Sciences) cells were grown in Dulbecco’s modified Eagle medium (Meilunbio) and supplemented with 10% fetal bovine serum (ExCell Bio). Transfection of DNA constructs was performed using Lipofectamine 2000 (11668019, Thermo-Fisher), according to the manufacturer’s instructions. For DNA expression, cells were transfected 18 h before Co-IP and immunofluorescence experiments.
To prepare ACBD3 knockdown and KDELR-mCherry overexpression in RIIα KI and HT1080 cells respectively, lentivirus was used to infect target cells in the presence of polybrene overnight. Two days after infection, the cells were cultured in puromycin (0.3–1 μg/ml, ThermoFisher) for 2 weeks. Lentivirus expressing ACBD3-shRNA (GCTGAAGTTACATGAGCTACA) was packaged and commercially provided by Shanghai GenePharma. To produce KDELR-mCherry lentivirus, a lentiviral vector containing KDELR-mCherry along with packing (psPAX2) and envelop (pMD2.G) vectors were transfected into 293FT cells using Lipofectamine 2000. The supernatants were collected after transfection for 48 h and 72 h, filtrated with 0.45 nm filters, and concentrated using a Lenti-concentration kit (AC04L441, life-ilab).
CRISPR/Cas9 gene editing
We used the CRISPR/Cas9 system to create knock-in cell lines stably expressing C-terminal 3× Flag-EGFP fusion proteins at PKA-RIIα or Cα locus. Design of the guide RNAs was carried out using benchling website (https://www.benchling.com/crispr) to minimize potential off-target effects. The genomic loci of PRKAR2A (Gene ID 5576) and PRKACA (Gene ID 5566) were targeted with the following guide RNA sequences: 5′- GTCACACTAAGAAGGCTCTG GGG -3′ and 5′- ACCAAAGTTTAAAGGCCCTG -3′ for RIIα and Cα respectively. The guide RNA was encoded in bicistronic expression plasmids pX330 (Addgene plasmid #42230). The homologous repair plasmid for genome editing was generated using these four PCR products: the pEGFP-N1 plasmid backbone, the left and right homology arms (∼1000 bp), and the reporter/selection cassette. The left homology arm fusing with 3× Flag-EGFP were synthesized (GenScript) and subcloned into pEGFP-N1 using AseI and NotI. Then, the right homology arm was synthesized and subcloned after the Neor/Kanr resistance cassette to integrate the whole cassette and allow selection of positive recombinants with the drug G418/Geneticin (ThermoFisher Scientific). The protospacer adjacent motif was mutagenized to avoid recutting by the Cas9.
The pX330 plasmid with the PKA-RIIα or Cα guide and the corresponding homologous recombination plasmid were cotransfected in HeLa cells using Lipofectamine 2000. G418 was added to the cells 4 days after transfection. After 2 weeks of selection, cells were subjected to single cell sorting by dilution into 96-well plates. Single clones were expanded and genotyped via Western blot and PCR. Preparation of ACBD3 KO cells was described previously (17).
GST-pulldown assays
The DNA coding sequence of RIα and RIIα were inserted in a pET28a plasmid for the expression with a N-terminal 6xHis tag. ACBD3, GOLD (amino acids 376–528), and GOLD-mutants (Q379P, I380P, or K381P) were inserted in pGEX-6p-1 plasmid for the expression of GST fusion proteins. Recombinant proteins expressed in E.coli BL21 (DE3) were purified using Glutathione HiCap Matrix (Qiagen) and Ni-NTA Agarose (Qiagen) for GST and His-tagged proteins, respectively.
For each pulldown assay, 100 μg GST fusion proteins were immobilized by incubating with 30 μl of 50% (vol/vol) Glutathione-Sepharose 4B beads (GE Healthcare) at 4 °C for 1 h. Beads were washed three times and incubated with 0.1 μM prey protein in binding buffer (25 mM Hepes pH 7.4, 150 mM NaOAc, 25 mM Mg(OAc)2, 0.2% Triton, and 1 mM DTT) at 4 °C for 1 h. After pulldown incubation, beads were washed for three times, and 50 μl 2× SDS loading buffer was added. After boiling the samples, 1% input and 20% samples were loaded for Western blot assays.
Immunofluorescence staining and confocal microscopy
HeLa cells grown on glass in 24-well plates coverslips were fixed in 4% paraformaldehyde for 10 min, permeabilized with PBS containing 0.5% Triton X-100 for 10 min, and blocked in blocking buffer (PBS containing 0.05% Triton X-100 and 3% BSA) for 60 min. Then, cells were incubated with primary and secondary antibodies applied in blocking buffer for 1 h. The nucleus was stained with Hoechst-33342 (sc-200908, Santa Cruz Biotechnology). Cells were washed three times with PBS and examined using Zeiss LSM880 with a 63× oil immersion objective.
Förster resonance energy transfer
HeLa WT and ACBD3-KO cells grown on 24-well glass bottom plates were cotransfected with TfR-FM4-SNAP and AKAR for 18 h, followed by D/D solubilizer treatment at a final concentration of 1 μM for 30 and 60 min. Cells were then washed and fixed in paraformaldehyde. Fluorescence images using a 405 nm excitation filter, and two emission filters (487 nm for CFP and 536 nm for YFP) were acquired by Zeiss LSM880 with a 63× oil immersion objective.
Co-immunoprecipitation and immunoblotting
For Co-IP experiments, total lysates were prepared using lysis buffer [20 mM Hepes, pH 7.4, 100 mM NaCl, 2 mM DDM (N-dodecyl-β-D-maltoside, Anatrace), 0.02% CHS (cholesteryl hemisuccinate tris salt, Sigma-Aldrich), and protease inhibitors (Roche)]. Subsequently, the total lysates were passed through a syringe needle (15 times) and then incubated at 4 °C with end-over-end agitation for 1.5 h. The lysates were then cleared by centrifugation at 15,000g for 10 min. The supernatants were incubated with anti-RFP agarose beads (M165-8, MBL Life science) or anti-GFP agarose beads (D153-8, MBL Life science) or myc-trap agarose beads (M20012, Abmart) for 4 h at 4 °C with end-over-end agitation. The beads were washed two times with ice-cold lysis buffer and one time with PBS. Proteins were eluted by boiling in 2× SDS loading buffer and subjected to SDS-PAGE for immunoblotting.
For immunoblotting, proteins were separated by SDS-PAGE (GenScript) and transferred onto nitrocellulose membranes (Amersham). Membranes were probed with specific primary antibodies and then with peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch). The bands were visualized with chemiluminescence (Clarity Western ECL Substrate, Bio-Rad) and imaged by a ChemiDoc Touch imaging system (Bio-Rad). Representative blots are shown from several experiments.
Image processing and statistical analysis
Pearson coefficient was analyzed by Fiji software. Results are displayed as mean ± SD (standard deviation) of results from each experiment or dataset, as indicated in figure legends. Analyses were performed with GraphPad Prism 9.0 software. Statistical analysis was performed using t test or one-way ANOVA with a Tukey's post-hoc test for multiple comparisons. N (number of individual experiments) is noted in the figure legends.
Data availability
All data that support the conclusions in this study are available upon reasonable request.
Conflict of interest
The authors declare no conflict of interest with the contents of this article.
Acknowledgments
Author contributions
I. L. and Y. Q. conceptualization; J. J., S. T., X. Y., S. J., L. Z., C. T., J. G., and Y. D. investigation; Y. Q. and I. L. writing-original draft; J. J., S. T., X. Y., S. J., L. Z., C. T., J. G., Y.D., I. L. and Y. Q. writing-review and editing.
Funding and additional information
Financial support of this study was provided by Shanghai Pujiang Program (16PJ1407 to Y.Q.) and ShanghaiTech University.
Reviewed by members of the JBC Editorial Board. Edited by Roger Colbran
Contributor Information
Intaek Lee, Email: Leeintaek@shanghaitech.edu.cn.
Yi Qian, Email: Qianyi@shanghaitech.edu.cn.
References
- 1.Taylor S.S., Zhang P., Steichen J.M., Keshwani M.M., Kornev A.P. Pka: lessons learned after twenty years. Biochim. Biophys. Acta. 2013;1834:1271–1278. doi: 10.1016/j.bbapap.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor S.S., Ilouz R., Zhang P., Kornev A.P. Assembly of allosteric macromolecular switches: lessons from PKA. Nat. Rev. Mol. Cell Biol. 2012;13:646–658. doi: 10.1038/nrm3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torres-Quesada O., Mayrhofer J.E., Stefan E. The many faces of compartmentalized PKA signalosomes. Cell Signal. 2017;37:1–11. doi: 10.1016/j.cellsig.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Wong W., Scott J.D. AKAP signalling complexes: focal points in space and time. Nat. Rev. Mol. Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 5.Langeberg L.K., Scott J.D. A-kinase-anchoring proteins. J. Cell Sci. 2005;118:3217–3220. doi: 10.1242/jcs.02416. [DOI] [PubMed] [Google Scholar]
- 6.Mayinger P. Signaling at the golgi. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a005314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bejarano E., Cabrera M., Vega L., Hidalgo J., Velasco A. Golgi structural stability and biogenesis depend on associated PKA activity. J. Cell Sci. 2006;119:3764–3775. doi: 10.1242/jcs.03146. [DOI] [PubMed] [Google Scholar]
- 8.Muniz M., Martin M.E., Hidalgo J., Velasco A. Protein kinase a activity is required for the budding of constitutive transport vesicles from the trans-Golgi network. Proc. Natl. Acad. Sci. U. S. A. 1997;94:14461–14466. doi: 10.1073/pnas.94.26.14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabrera M., Muniz M., Hidalgo J., Vega L., Martin M.E., Velasco A. The retrieval function of the KDEL receptor requires PKA phosphorylation of its C-terminus. Mol. Biol. Cell. 2003;14:4114–4125. doi: 10.1091/mbc.E03-04-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H., Adamik R., Pacheco-Rodriguez G., Moss J., Vaughan M. Protein kinase A-anchoring (AKAP) domains in brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BIG2) Proc. Natl. Acad. Sci. U. S. A. 2003;100:1627–1632. doi: 10.1073/pnas.0337678100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi M., Shibata H., Shimakawa M., Miyamoto M., Mukai H., Ono Y. Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the golgi apparatus. J. Biol. Chem. 1999;274:17267–17274. doi: 10.1074/jbc.274.24.17267. [DOI] [PubMed] [Google Scholar]
- 12.Schillace R.V., Andrews S.F., Liberty G.A., Davey M.P., Carr D.W. Identification and characterization of myeloid translocation gene 16b as a novel a kinase anchoring protein in T lymphocytes. J. Immunol. 2002;168:1590–1599. doi: 10.4049/jimmunol.168.4.1590. [DOI] [PubMed] [Google Scholar]
- 13.Salvador L.M., Flynn M.P., Avila J., Reierstad S., Maizels E.T., Alam H., et al. Neuronal microtubule-associated protein 2D is a dual a-kinase anchoring protein expressed in rat ovarian granulosa cells. J. Biol. Chem. 2004;279:27621–27632. doi: 10.1074/jbc.M402980200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelham H.R. Evidence that luminal ER proteins are sorted from secreted proteins in a post-ER compartment. EMBO J. 1988;7:913–918. doi: 10.1002/j.1460-2075.1988.tb02896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis M.J., Sweet D.J., Pelham H.R. The ERD2 gene determines the specificity of the luminal ER protein retention system. Cell. 1990;61:1359–1363. doi: 10.1016/0092-8674(90)90699-f. [DOI] [PubMed] [Google Scholar]
- 16.Cancino J., Capalbo A., Di Campli A., Giannotta M., Rizzo R., Jung J.E., et al. Control systems of membrane transport at the interface between the endoplasmic reticulum and the Golgi. Dev. Cell. 2014;30:280–294. doi: 10.1016/j.devcel.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 17.Yue X., Qian Y., Zhu L., Gim B., Bao M., Jia J., et al. ACBD3 modulates KDEL receptor interaction with PKA for its trafficking via tubulovesicular carrier. BMC Biol. 2021;19:194. doi: 10.1186/s12915-021-01137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J., Rone M.B., Papadopoulos V. Protein-protein interactions mediate mitochondrial cholesterol transport and steroid biosynthesis. J. Biol. Chem. 2006;281:38879–38893. doi: 10.1074/jbc.M608820200. [DOI] [PubMed] [Google Scholar]
- 19.Liu J., Li H., Papadopoulos V. PAP7, a PBR/PKA-RIalpha-associated protein: a new element in the relay of the hormonal induction of steroidogenesis. J. Steroid Biochem. Mol. Biol. 2003;85:275–283. doi: 10.1016/s0960-0760(03)00213-9. [DOI] [PubMed] [Google Scholar]
- 20.Cadd G.G., Uhler M.D., McKnight G.S. Holoenzymes of cAMP-dependent protein kinase containing the neural form of type I regulatory subunit have an increased sensitivity to cyclic nucleotides. J. Biol. Chem. 1990;265:19502–19506. [PubMed] [Google Scholar]
- 21.Ilouz R., Bubis J., Wu J., Yim Y.Y., Deal M.S., Kornev A.P., et al. Localization and quaternary structure of the PKA RIbeta holoenzyme. Proc. Natl. Acad. Sci. U. S. A. 2012;109:12443–12448. doi: 10.1073/pnas.1209538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dimino M.J., Bieszczad R.R., Rowe M.J. Cyclic AMP-dependent protein kinase in mitochondria and cytosol from different-sized follicles and corpora lutea of porcine ovaries. J. Biol. Chem. 1981;256:10876–10882. [PubMed] [Google Scholar]
- 23.Herberg F.W., Maleszka A., Eide T., Vossebein L., Tasken K. Analysis of A-kinase anchoring protein (AKAP) interaction with protein kinase A (PKA) regulatory subunits: PKA isoform specificity in AKAP binding. J Mol Biol. 2000;298:329–339. doi: 10.1006/jmbi.2000.3662. [DOI] [PubMed] [Google Scholar]
- 24.Klima M., Chalupska D., Rozycki B., Humpolickova J., Rezabkova L., Silhan J., et al. Kobuviral non-structural 3A proteins act as molecular harnesses to hijack the host ACBD3 protein. Structure. 2017;25:219–230. doi: 10.1016/j.str.2016.11.021. [DOI] [PubMed] [Google Scholar]
- 25.McPhail J.A., Ottosen E.H., Jenkins M.L., Burke J.E. The molecular basis of Aichi Virus 3A protein activation of phosphatidylinositol 4 kinase IIIbeta, PI4KB, through ACBD3. Structure. 2017;25:121–131. doi: 10.1016/j.str.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 26.Uhler M.D., Chrivia J.C., McKnight G.S. Evidence for a second isoform of the catalytic subunit of cAMP-dependent protein kinase. J. Biol. Chem. 1986;261:15360–15363. [PubMed] [Google Scholar]
- 27.Beebe S.J., Oyen O., Sandberg M., Froysa A., Hansson V., Jahnsen T. Molecular cloning of a tissue-specific protein kinase (C gamma) from human testis--representing a third isoform for the catalytic subunit of cAMP-dependent protein kinase. Mol Endocrinol. 1990;4:465–475. doi: 10.1210/mend-4-3-465. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y., Saulnier J.L., Yellen G., Sabatini B.L. A PKA activity sensor for quantitative analysis of endogenous GPCR signaling via 2-photon FRET-FLIM imaging. Front Pharmacol. 2014;5:56. doi: 10.3389/fphar.2014.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Francis M.J., Jones E.E., Levy E.R., Ponnambalam S., Chelly J., Monaco A.P. A Golgi localization signal identified in the menkes recombinant protein. Hum. Mol. Genet. 1998;7:1245–1252. doi: 10.1093/hmg/7.8.1245. [DOI] [PubMed] [Google Scholar]
- 30.Mavillard F., Hidalgo J., Megias D., Levitsky K.L., Velasco A. PKA-mediated Golgi remodeling during cAMP signal transmission. Traffic. 2010;11:90–109. doi: 10.1111/j.1600-0854.2009.01007.x. [DOI] [PubMed] [Google Scholar]
- 31.Farhan H., Rabouille C. Signalling to and from the secretory pathway. J. Cell Sci. 2011;124:171–180. doi: 10.1242/jcs.076455. [DOI] [PubMed] [Google Scholar]
- 32.Martin M.E., Hidalgo J., Vega F.M., Velasco A. Trimeric G proteins modulate the dynamic interaction of PKAII with the Golgi complex. J. Cell Sci. 1999;112:3869–3878. doi: 10.1242/jcs.112.22.3869. [DOI] [PubMed] [Google Scholar]
- 33.Brauer P., Parker J.L., Gerondopoulos A., Zimmermann I., Seeger M.A., Barr F.A., et al. Structural basis for pH-dependent retrieval of ER proteins from the Golgi by the KDEL receptor. Science. 2019;363:1103–1107. doi: 10.1126/science.aaw2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soberg K., Skalhegg B.S. The molecular basis for specificity at the level of the protein kinase a catalytic subunit. Front. Endocrinol. (Lausanne) 2018;9:538. doi: 10.3389/fendo.2018.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Islam A., Jones H., Hiroi T., Lam J., Zhang J., Moss J., et al. cAMP-dependent protein kinase A (PKA) signaling induces TNFR1 exosome-like vesicle release via anchoring of PKA regulatory subunit RIIbeta to BIG2. J. Biol. Chem. 2008;283:25364–25371. doi: 10.1074/jbc.M804966200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C.C., Le K., Kato J., Moss J., Vaughan M. Enhancement of beta-catenin activity by BIG1 plus BIG2 via Arf activation and cAMP signals. Proc. Natl. Acad. Sci. U. S. A. 2016;113:5946–5951. doi: 10.1073/pnas.1601918113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data that support the conclusions in this study are available upon reasonable request.