ABSTRACT
Xanthomonas citri subsp. citri is the cause of bacterial citrus canker, responsible for major economic losses to the citrus industry. X. citri subspecies and pathovars are responsible for diseases in soybean, common bean, mango, pomegranate, and cashew. X. citri disease has been tracked using several typing methods, but recent studies using genomic sequencing have been key to understanding the evolutionary relationships within the species, including fundamental differences among X. citri subsp. citri pathotypes. Here, we describe a core-genome multilocus sequence typing (cgMLST) scheme for X. citri based on 250 genomes comprising multiple examples of X. citri subsp. citri pathotypes A, A*, and Aw; X. citri subsp. malvacearum; X. citri pv. aurantifolii, pv. fuscans, pv. glycines, pv. mangiferaeindicae, pv. viticola, and pv. vignicola; and single isolates of X. citri pv. dieffenbachiae and pv. punicae. This data set included genomic sequencing of 100 novel X. citri subsp. citri isolates. cgMLST, based on 1,618 core genes across 250 genomes, is implemented at PubMLST (https://pubmlst.org/organisms/xanthomonas-citri/). GrapeTree minimum-spanning tree and Interactive Tree of Life (iTOL) neighbor-joining phylogenies generated from the cgMLST data resolved almost identical groupings of isolates to a core-genome single nucleotide polymorphism (SNP)-based neighbor-joining phylogeny. These resolved identical groupings of X. citri subsp. citri pathotypes and X. citri subspecies and pathovars. X. citri cgMLST should prove to be an increasingly valuable resource for the study of this key species of plant-pathogenic bacteria. Users can submit genomic data and associated metadata for comparison with previously characterized isolates at PubMLST to allow the rapid characterization of the local, national, and global epidemiology of these pathogens and examine evolutionary relationships.
IMPORTANCE Xanthomonas citri is a plant pathogen that causes major economic losses to the citrus industry and sweet orange production in particular. Several subspecies and pathogens are recognized, with host ranges including soybean, common bean, mango, pomegranate, and cashew, among others. Recent genomic studies have shown that host-adapted X. citri subspecies and pathovars and X. citri subsp. citri pathotypes form distinct clades. In this study, we describe a core-genome multilocus sequence typing (cgMLST) scheme for this species that can rapidly and robustly discriminate among these ecologically distinct, host-adapted clades. We have established this scheme and associated databases containing genomic sequences and metadata at PubMLST, which users can interrogate with their own genome sequences to determine X. citri subspecies, pathovars, and pathotypes. X. citri cgMLST should prove to be an invaluable tool for the study of the epidemiology and evolution of this major plant pathogen.
KEYWORDS: citrus canker, MLST, Xanthomonas citri, cgMLST
INTRODUCTION
Bacterial citrus canker has a major economic impact on the production of all commercial citrus crops, including oranges, limes, tangerines, lemons, and grapefruit. Three pathotypes of canker are recognized: A, B, and C. Type A, caused by Xanthomonas citri subsp. citri, is the most widespread and economically damaging, whereas types B and C, caused by X. citri pv. aurantifolii, have much-reduced virulence on sweet orange and have very limited geographical spread (1). The type A (2, 3) pathotype has the broadest host range and infects most economically important citrus plants worldwide, particularly causing a major economic burden on the South American and Californian orange industries (2, 4). Two variants of pathotype A have evolved: A*, which can cause canker on all citrus but with some isolates that can infect only key lime (Citrus aurantifolia), and Aw, which infects only key lime and alemow (Citrus macrophylla) (1).
Xanthomonas citri subspecies and pathovars other than X. citri subsp. citri infect other important crop species, including common bean (X. citri pv. fuscans), Mexican lime (X. citri pv. aurantifolii), mango (X. citri pv. mangiferaeindicae), grape (X. citri pv. viticola), cotton (X. citri subsp. malvacearum), soybean (X. citri pv. glycines), Araceae (X. citri pv. dieffenbachiae), cashew (X. citri pv. anacardii), and pomegranate (X. citri pv. punicae). Previous genome sequencing studies have examined the evolution of X. citri pathovars and subspecies (5) and X. citri subsp. citri pathotypes (4), and these studies have produced robust phylogenies that clearly resolve clades corresponding to individual X. citri pathovars and X. citri subsp. citri pathotypes. Genomic sequencing has also proven useful in investigations of host-pathogen interactions through the identification of host-specific virulence factors (6).
Whole-genome sequencing has greatly advanced the study of the epidemiology and evolution of pathogenic bacteria, greatly improving the discriminatory power and portability of other approaches such as ribotyping or pulsed-field gel electrophoresis (7). Genomic sequencing and analysis tools, developed primarily for the study of human bacterial pathogens to track and investigate outbreaks of disease caused by particularly virulent or antimicrobial-resistant clones, can also be usefully employed for the study of bacterial plant disease epidemiology and evolution.
Whole-genome sequences from isolates of pathogenic bacteria are usually compared using SNP (single nucleotide polymorphism)-based approaches that involve whole-genome alignments. Such SNP-based approaches have been used in recent studies of Xanthomonas citri biology (8, 9); however, they involve identifying genomes of isolates from the literature and downloading their sequences, followed by the computationally intensive alignment of multiple genomes to generate SNP profiles, which are then used to produce phylogenetic trees using methods such as neighbor joining (NJ), maximum parsimony, or maximum likelihood. Core-genome multilocus sequence typing (cgMLST) uses whole-genome sequence data to examine genetic similarities between isolates. It is based on allelic variations at a large number of core-genome loci that are present in all, or nearly all, members of a species (10). It differs from other whole-genome sequencing approaches in that it does not include noncore, accessory genes in comparisons of genomes, and it examines variation in allelic profiles rather than core-genome SNPs. In addition, cgMLST is computationally efficient, scalable, and suited for the representation of very large numbers of genomic comparisons. cgMLST schemes have been established for a diverse range of human pathogens, and some schemes contain many thousands of genomes. For example, the curated, open-source database PubMLST (https://pubmlst.org/) contains genomic data and metadata for 655,340 genomes of >100 bacterial species, and the EnteroBase database (https://enterobase.warwick.ac.uk) contains 379,370 Salmonella and 237,066 Escherichia coli/Shigella genomes and corresponding metadata alone (as of 23 November 2022).
In this study, we describe a cgMLST scheme and website resource that can be used to rapidly and easily identify X. citri subsp. citri variants from genome sequences without the need for computationally intensive and time-consuming core-genome SNP extraction, genome alignment, and phylogenetic comparisons. The X. citri cgMLST database at https://pubmlst.org/organisms/xanthomonas-citri represents an invaluable resource for tracking the spread of pathovars of this devastating pathogen, which should also prove to be a useful, scalable tool in future national and international efforts to control citrus canker and other crop diseases.
RESULTS
Genome sequencing.
Assemblies from each X. citri isolate consisted of between 61 and 161 contigs, with N50 values of between 96,324 and 1,044,915 nucleotides (nt) and an average depth of coverage of 102× (range = 31× to 900×). For all isolates, more than 99% of reads were mapped to the family Xanthomonadaceae using Kraken (11).
rMLST.
Ribosomal MLST (rMLST) confirmed the species designations of the 250 X. citri isolates listed in Table 1 as well as 20 other Xanthomonas spp. and the 4 other species examined and listed in Table S1 in the supplemental material. Figure 1 shows a neighbor-joining tree of all 274 genomes in this study based on the 53 concatenated rRNA gene loci used in the rMLST scheme. It can be clearly seen that all X. citri isolates form a separate and distinct clade whose closest neighbors are genomes of other X. citri pathotypes and subspecies. From this analysis, the Xanthomonas species X. vasicola and X. perforans appear to be the most closely related to X. citri, with the genomes of other xanthomonads such as X. euvesicatoria being separated by greater genetic distances. Example genomes of E. coli, Pseudomonas aeruginosa, Xylella fastidiosa, and Stenotrophomonas maltophilia are separated by even larger genetic distances from the genomes of Xanthomonas spp., including X. citri.
TABLE 1.
Details of isolates and genomes used in this studya
| MLST ID | Isolate | Alias | Country | Region | Yr of isolation | Source | Plant host species | X. citri pathovar or subspecies | Pathotype | BioProject accession no. | BioSample accession no. | Reference | cgMLST group (≤200 mismatches) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 306 | IBSBF 1594 | Brazil | Paraná | Leaf | Sweet orange | citri | A | PRJNA779375 | NA | 14 | 1 | |
| 2 | FDC102 | Brazil | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028269 | This study | 1 | |||
| 3 | FDC103 | Argentina | Corrientes | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028270 | This study | 1 | ||
| 4 | FDC104 | Paraguay | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028271 | This study | 1 | |||
| 5 | FDC1053 | Brazil | Ilha Solteira, São Paulo | 2004 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028272 | This study | 1 | |
| 6 | FDC107 | Uruguay | Salto | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028273 | This study | 1 | ||
| 7 | FDC1083 | IBSBF 256 | Brazil | Assis, São Paulo | 1980 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028274 | This study | 1 |
| 8 | FDC1085 | IBSBF 314 | Brazil | Araçatuba, São Paulo | 1979 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028275 | This study | 1 |
| 9 | FDC1087 | IBSBF 338 | Brazil | Cândido Mota, São Paulo | 1981 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028276 | This study | 1 |
| 10 | FDC1088 | IBSBF 340 | Brazil | Lins, São Paulo | 1981 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028277 | This study | 1 |
| 11 | FDC1091 | IBSBF 353 | Brazil | São Pedro do Turvo, São Paulo | 1981 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028278 | This study | 1 |
| 12 | FDC1094 | IBSBF 438 | Brazil | Cajobi, São Paulo | 1982 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028279 | This study | 1 |
| 13 | FDC1095 | IBSBF 491 | Brazil | Santa Mercedes, São Paulo | 1983 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028280 | This study | 1 |
| 14 | FDC1098 | IBSBF 947 | Brazil | Presidente Prudente, São Paulo | 1992 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028281 | This study | 1 |
| 15 | FDC1101 | IBSBF 1287 | Brazil | Moji-Mirim, São Paulo | 1996 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028282 | This study | 1 |
| 16 | FDC1102 | IBSBF 1403 | Brazil | São João da Boa Vista, São Paulo | 1998 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028283 | This study | 1 |
| 17 | FDC1104 | IBSBF 1415 | Brazil | Engenheiro Coelho, São Paulo | 1998 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028284 | This study | 1 |
| 18 | FDC1107 | IBSBF 1428 | Brazil | Itirapina, São Paulo | 1999 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028285 | This study | 1 |
| 19 | FDC1115 | IBSBF 1440 | Brazil | General Salgado, São Paulo | 1999 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028286 | This study | 1 |
| 20 | FDC1116 | IBSBF 1449 | Brazil | Presidente Bernardes, São Paulo | 1999 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028287 | This study | 1 |
| 21 | FDC1118 | IBSBF 1453 | Brazil | Botucatu, São Paulo | 1999 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028288 | This study | 1 |
| 22 | FDC1120 | IBSBF 1484 | Brazil | Clementina, São Paulo | 2000 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028289 | This study | 1 |
| 23 | FDC1121 | IBSBF 1485 | Brazil | Luiziânia, São Paulo | 2000 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028290 | This study | 1 |
| 24 | FDC1125 | IBSBF 1491 | Brazil | Sud Mennucci, São Paulo | 2000 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028291 | This study | 1 |
| 25 | FDC1129 | IBSBF 1518 | Brazil | Guzolândia, São Paulo | 2000 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028292 | This study | 1 |
| 26 | FDC1139 | IAPAR 12426 | Brazil | Araraquara, São Paulo | 1999 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028293 | This study | 1 |
| 27 | FDC1142 | Brazil | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028294 | This study | 1 | |||
| 28 | FDC1143 | IAPAR 12778 | Brazil | Avaré, São Paulo | 2000 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028295 | This study | 1 |
| 29 | FDC1144 | IAPAR 12822 | Brazil | Guaimbê, São Paulo | 2000 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028296 | This study | 1 |
| 30 | FDC1145 | IAPAR 12989 | Brazil | Marília, São Paulo | 2000 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028297 | This study | 1 |
| 31 | FDC1148 | IAPAR 12991 | Brazil | Tarumã, São Paulo | 2001 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028298 | This study | 1 |
| 32 | FDC1150 | IAPAR 12001 | Brazil | Salto Grande, São Paulo | 2001 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028299 | This study | 1 |
| 33 | FDC1182 | Brazil | Ourizona, Paraná | 2005 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028300 | This study | 1 | |
| 34 | FDC122 | China | Hong Kong | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028301 | This study | 1 | ||
| 35 | FDC1227 | Brazil | Adolfo, São Paulo | 2005 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028302 | This study | 1 | |
| 36 | FDC124 | Japan | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028303 | This study | 1 | |||
| 37 | FDC1252 | Brazil | Sales, São Paulo | 2005 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028304 | This study | 1 | |
| 38 | FDC126 | Philippines | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028305 | This study | 1 | |||
| 39 | FDC1277 | Brazil | Marinópolis, São Paulo | 2005 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028306 | This study | 1 | |
| 40 | FDC129 | France | Reunion | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028307 | This study | 1 | ||
| 41 | FDC1291 | Brazil | Rubiácea, São Paulo | 2007 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028308 | This study | 1 | |
| 42 | FDC130 | China | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028309 | This study | 1 | |||
| 43 | FDC131 | Thailand | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028310 | This study | 1 | |||
| 44 | FDC133 | Mauritius | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028311 | This study | 1 | |||
| 45 | FDC1387 | Brazil | Urânia, São Paulo | 2007 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028312 | This study | 1 | |
| 46 | FDC1424 | Brazil | Suzanápolis, São Paulo | 2007 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028313 | This study | 1 | |
| 47 | FDC1488 | Brazil | Urânia, São Paulo | 2007 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028314 | This study | 1 | |
| 48 | FDC15 | Argentina | Entre Rios | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028315 | This study | 1 | ||
| 49 | FDC1531 | Brazil | Pereira Barreto, São Paulo | 2007 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028316 | This study | 1 | |
| 50 | FDC1533 | Brazil | Palmeira d’Oeste, São Paulo | 2007 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028317 | This study | 1 | |
| 51 | FDC1539 | Brazil | Ilha Solteira, São Paulo | 2007 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028318 | This study | 1 | |
| 52 | FDC1580 | Brazil | Boa Esperança do Sul, São Paulo | 2007 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028319 | This study | 1 | |
| 53 | FDC1666 | Brazil | Rondon, Paraná | 2011 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028320 | This study | 1 | |
| 54 | FDC1681 | Brazil | Matão, São Paulo | 2012 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028321 | This study | 1 | |
| 55 | FDC1705 | Brazil | Paranavaí, Paraná | 2013 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028322 | This study | 1 | |
| 56 | FDC1707 | Brazil | Alto Paraná, Paraná | 2013 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028323 | This study | 1 | |
| 57 | FDC1733 | Brazil | Guairaçã, Paraná | 2014 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028324 | This study | 1 | |
| 58 | FDC2 | Brazil | Lins, São Paulo | 2001 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028325 | This study | 1 | |
| 59 | FDC24 | Paraguay | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028326 | This study | 1 | |||
| 60 | FDC4167 | Unknown | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028327 | This study | 1 | |||
| 61 | FDC46 | New Zealand | New Plymouth | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028328 | This study | 1 | ||
| 62 | FDC49 | Japan | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028329 | This study | 1 | |||
| 63 | FDC50 | Fiji | Vanua Levu | Leaf | Sweet orange | citri | A* | PRJNA779375 | SAMN23028330 | This study | 2 | ||
| 64 | FDC502 | New Zealand | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028331 | This study | 1 | |||
| 65 | FDC512 | Brazil | Iacri, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028332 | This study | 1 | |
| 66 | FDC52 | India | New Delhi | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028333 | This study | 1 | ||
| 67 | FDC53 | Iran | Leaf | Sweet orange | citri | A* | PRJNA779375 | SAMN23028334 | This study | 2 | |||
| 68 | FDC54 | Australia | Darwin | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028335 | This study | 1 | ||
| 69 | FDC544 | Brazil | Mira Estrela, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028336 | This study | 1 | |
| 70 | FDC55 | Taiwan | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028337 | This study | 1 | |||
| 71 | FDC550 | Brazil | Rubinéia, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028338 | This study | 1 | |
| 72 | FDC551 | Brazil | Ibitinga, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028339 | This study | 1 | |
| 73 | FDC553 | Brazil | Avaré, São Paulo | 2001 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028340 | This study | 1 | |
| 74 | FDC559 | Brazil | Cafelândia, São Paulo | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028341 | This study | 1 | ||
| 75 | FDC560 | Brazil | Urupês, São Paulo | 2001 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028342 | This study | 1 | |
| 76 | FDC562 | Brazil | Terra Roxa | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028343 | This study | 1 | |
| 77 | FDC565 | Brazil | Barbosa, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028344 | This study | 1 | |
| 78 | FDC575 | Brazil | Barbosa, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028345 | This study | 1 | |
| 79 | FDC601 | IBSBF 1989 | Brazil | Rio Grande do Sul | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028346 | This study | 1 |
| 80 | FDC603 | IBSBF 1990 | Brazil | Mariana Moro, Rio Grande do Sul | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028347 | This study | 1 | |
| 81 | FDC7 | Brazil | Bataguassu, Mato Grosso do Sul | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028348 | This study | 1 | ||
| 82 | FDC704 | Brazil | Nova Canaã, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028349 | This study | 1 | |
| 83 | FDC705 | Brazil | Nova Canaã, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028350 | This study | 1 | |
| 84 | FDC714 | Brazil | Jales, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028351 | This study | 1 | |
| 85 | FDC718 | Brazil | Borborema, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028352 | This study | 1 | |
| 86 | FDC719 | Brazil | Parapuã, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028353 | This study | 1 | |
| 87 | FDC724 | Brazil | Urupês, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028354 | This study | 1 | |
| 88 | FDC748 | Brazil | Osvaldo Cruz, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028355 | This study | 1 | |
| 89 | FDC749 | Brazil | Aparecida do Oeste, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028356 | This study | 1 | |
| 90 | FDC75 | ISBSF 1421 | Brazil | Casa Branca, São Paulo | 1998 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028357 | This study | 1 |
| 91 | FDC755 | Brazil | Caiuá, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028358 | This study | 1 | |
| 92 | FDC764 | Brazil | Sandovalina, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028359 | This study | 1 | |
| 93 | FDC769 | Brazil | Narandiba, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028360 | This study | 1 | |
| 94 | FDC782 | Brazil | Ibitinga, São Paulo | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028361 | This study | 1 | |
| 95 | FDC8 | Brazil | Corbélia, Paraná | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028362 | This study | 1 | ||
| 96 | FDC806 | Brazil | Boa Vista, Roraima | 2002 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028363 | This study | 1 | |
| 97 | FL71 | USA | Florida | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028364 | This study | 1 | ||
| 98 | FL72 | USA | Florida | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028365 | This study | 1 | ||
| 99 | FL75 | USA | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028366 | This study | 1 | |||
| 100 | LM199 | Argentina | 2015 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028367 | This study | 1 | ||
| 101 | FDC4 | Brazil | Cuiabá-Mato Grosso | 2001 | Leaf | Sweet orange | citri | A | PRJNA779375 | SAMN23028368 | This study | 1 | |
| 102 | 12_2 | Thailand | Nakhon Ratchasima | 1991 | Leaf | Soybean | glycines | PRJNA323439 | SAMN05179543 | 3 | |||
| 103 | 1017 | South Korea | Suwon | 1997 | Unknown | glycines | PRJNA556098 | SAMN12340733 | 3 | ||||
| 104 | 1018 | South Korea | Hwaseong | 1997 | Unknown | glycines | PRJNA556099 | SAMN12340735 | 3 | ||||
| 105 | 1045 | South Korea | Hwaseong | 1997 | Unknown | glycines | PRJNA556109 | SAMN12340804 | 3 | ||||
| 106 | 1157 | South Korea | Pocheon | 1997 | Unknown | glycines | PRJNA556102 | SAMN12340780 | 3 | ||||
| 107 | 1566 | Brazil | Unknown | aurantifolii | PRJNA273983 | SAMN03835495 | 4 | ||||||
| 108 | 4834-R | France | Beaucouzé | 1998 | Unknown | Common bean | fuscans | PRJNA176873 | SAMEA3283112 | 25 | 5 | ||
| 109 | 5208 | USA | Florida | 2002 | Unknown | citri | PRJNA255042 | SAMN02911840 | 26 | 1 | |||
| 110 | 12609 | Taiwan | 2015 | Leaf | glycines | PRJNA344018 | SAMN05818161 | 3 | |||||
| 112 | AR81009 | Argentina | 1981 | Unknown | malvacearum | PRJNA396899 | SAMN07447516 | 6 | |||||
| 113 | AS8 | Saudi Arabia | Unknown | citri | A* | PRJNA255042 | SAMN02911853 | 26 | 2 | ||||
| 114 | AS9 | Saudi Arabia | Unknown | citri | A* | PRJNA255042 | SAMN02911854 | 26 | 2 | ||||
| 115 | AS270 | Saudi Arabia | 1988 | Unknown | citri | A* | PRJNA255042 | SAMN02911852 | 26 | 2 | |||
| 116 | AW13 | USA | Florida | 2003 | Unknown | citri | Aw | PRJNA255042 | SAMN02911848 | 26 | 7 | ||
| 117 | AW14 | USA | Florida | 2005 | Unknown | citri | Aw | PRJNA255042 | SAMN02911849 | 26 | 7 | ||
| 118 | AW15 | USA | Florida | 2005 | Unknown | citri | Aw | PRJNA255042 | SAMN02911850 | 26 | 7 | ||
| 119 | AW16 | USA | Florida | 2005 | Unknown | citri | Aw | PRJNA255042 | SAMN02911851 | 26 | 7 | ||
| 120 | Aw12879 | USA | Florida | Unknown | citri | Aw | PRJNA81931 | SAMN02603165 | 27 | 7 | |||
| 121 | BL18 | USA | Florida | 2011 | Unknown | citri | PRJNA255042 | SAMN02911845 | 26 | 1 | |||
| 122 | CCRMXCV-80 | Brazil | Pernambuco | 2015 | Unknown | viticola | PRJNA407795 | SAMN07664206 | 8 | ||||
| 123 | CFBP6988 | Réunion | 2000 | Unknown | fuscans | PRJNA212252 | SAMN02645882 | 28 | 9 | ||||
| 124 | CFBP1815 | Greece | 1978 | Unknown | Common bean | fuscans | PRJEB23080 | SAMEA104357164 | 28 | 5 | |||
| 125 | CFBP2526 | Sudan | 1956 | Unknown | Soybean | glycines | PRJNA212247 | SAMN02469936 | 29 | 3 | |||
| 126 | CFBP2913 | Brazil | 1974 | Unknown | anacardii | PRJNA232107 | SAMN07766623 | 30 | 10 | ||||
| 127 | CFBP4884 | France | 1998 | Leaf | fuscans | PRJNA254240 | SAMN02902416 | 31 | 5 | ||||
| 128 | CFBP4885 | France | 1998 | Unknown | fuscans | PRJNA384182 | SAMN06829861 | 28 | 5 | ||||
| 129 | CFBP6165 | Canada | 1957 | Unknown | fuscans | PRJNA384145 | SAMN06829306 | 28 | 11 | ||||
| 130 | CFBP6166 | South Africa | 1963 | Unknown | fuscans | PRJNA384183 | SAMN06829862 | 28 | 5 | ||||
| 131 | CFBP6167 | USA | 1954 | Unknown | fuscans | PRJNA384187 | SAMN06829865 | 28 | 5 | ||||
| 132 | CFBP6960 | Réunion | 2000 | Unknown | Common bean | fuscans | PRJEB23080 | SAMEA104357193 | 28 | 5 | |||
| 133 | CFBP6970 | USA | 1990 | Unknown | Common bean | fuscans | PRJEB23080 | SAMEA104357194 | 28 | 5 | |||
| 134 | CFBP6975 | France | 1994 | Unknown | fuscans | PRJNA384188 | SAMN06829867 | 28 | 5 | ||||
| 135 | CFBP6988R | Réunion | 2000 | Unknown | fuscans | PRJNA384160 | SAMN06829542 | 28 | 9 | ||||
| 136 | CFBP6989 | Réunion | 2000 | Unknown | fuscans | PRJNA384161 | SAMN06829585 | 28 | 9 | ||||
| 137 | CFBP6990 | Réunion | 2000 | Unknown | fuscans | PRJNA384163 | SAMN06829587 | 28 | 9 | ||||
| 138 | CFBP6991 | Réunion | 2000 | Unknown | fuscans | PRJNA384178 | SAMN06829858 | 28 | 9 | ||||
| 139 | CFBP6992 | Réunion | 2000 | Unknown | fuscans | PRJNA384177 | SAMN06829854 | 28 | 13 | ||||
| 140 | CFBP6994 | Tanzania | 1990 | Unknown | Common bean | fuscans | PRJEB23080 | SAMEA104357310 | 28 | Singleton | |||
| 141 | CFBP6994R | Tanzania | 1990 | Unknown | fuscans | PRJNA384179 | SAMN06829859 | 28 | 13 | ||||
| 142 | CFBP6996 | Réunion | 2000 | Leaf | Common bean | fuscans | PRJNA212255 | SAMN02645883 | 13 | ||||
| 143 | CFBP6996R | Réunion | 2000 | Unknown | fuscans | PRJNA384180 | SAMN06829860 | 13 | |||||
| 144 | CFBP7111 | USA | Texas | 1942 | Leaf | vignicola | PRJNA390891 | SAMN07252112 | Singleton | ||||
| 145 | CFBP7113 | Sudan | 1966 | Unknown | vignicola | PRJNA390890 | SAMN07251989 | Singleton | |||||
| 146 | CFBP7119 | Brazil | 1981 | Unknown | Soybean | glycines | PRJNA212249 | SAMN02469937 | 29 | 3 | |||
| 147 | CFBP7764 | Brazil | Petrolina | 2012 | Stem | viticola | PRJNA422087 | SAMN08163769 | 8 | ||||
| 148 | CFBP7766 | Cameroon | 2009 | Unknown | Common bean | fuscans | PRJEB23080 | SAMEA104357197 | 28 | 5 | |||
| 149 | CFBP7767 | Cameroon | 2009 | Unknown | Common bean | fuscans | PRJEB23080 | SAMEA104357198 | 28 | 5 | |||
| 150 | EB08 | USA | Central Iowa | 2008 | Unknown | glycines | PRJNA431457 | SAMN08391414 | 32 | 3 | |||
| 151 | FB19 | USA | Florida | 2011 | Unknown | citri | PRJNA255042 | SAMN02911846 | 26 | 1 | |||
| 152 | FDC535 | Brazil | Sao Paulo | 2000 | Unknown | aurantifolii | PRJNA273983 | SAMN03317023 | Singleton | ||||
| 153 | FDC628 | Brazil | Santa Catarina | 2001 | Unknown | citri | PRJNA273983 | SAMN03317028 | 1 | ||||
| 154 | FDC636 | Brazil | Paraná | 1996 | Unknown | citri | PRJNA273983 | SAMN03317019 | 1 | ||||
| 155 | FDC654 | Brazil | Rio Grande do Sul | 1999 | Unknown | citri | PRJNA273983 | SAMN03317029 | 1 | ||||
| 156 | FDC763 | Brazil | São Paulo | 1981 | Unknown | aurantifolii | PRJNA273983 | SAMN03317030 | 17 | ||||
| 157 | FDC828 | Brazil | São Paulo | 1997 | Unknown | citri | PRJNA273983 | SAMN03317018 | 1 | ||||
| 158 | FDC867 | Brazil | São Paulo | 2002 | Unknown | aurantifolii | PRJNA273983 | SAMN03317026 | 17 | ||||
| 159 | FDC1559 | Brazil | São Paulo | 1981 | Unknown | aurantifolii | PRJNA273983 | SAMN03317020 | 17 | ||||
| 160 | FDC1561 | Argentina | 1981 | Unknown | aurantifolii | PRJNA273983 | SAMN03317022 | 4 | |||||
| 161 | FDC1609 | Brazil | Sao Paulo | 2009 | Unknown | aurantifolii | PRJNA273983 | SAMN03317027 | Singleton | ||||
| 162 | FDC1662 | Brazil | Paraná | 2011 | Unknown | citri | PRJNA273983 | SAMN03317017 | 1 | ||||
| 163 | FDC1682 | Oman | 1986 | Unknown | citri | A* | PRJNA273983 | SAMN03317024 | 2 | ||||
| 164 | GD2 | China | Guangdong | 2011 | Unknown | citri | PRJNA255042 | SAMN02911834 | 26 | 1 | |||
| 165 | GD3 | China | Guangdong | 2011 | Unknown | citri | PRJNA255042 | SAMN02911835 | 26 | 1 | |||
| 166 | GSPB1386 | Nicaragua | 1986 | Unknown | malvacearum | PRJNA78127 | SAMN02469610 | 18 | |||||
| 167 | GSPB2388 | Sudan | Unknown | malvacearum | PRJNA79081 | SAMN02469611 | Singleton | ||||||
| 168 | HD-1 | China | Leaf | malvacearum | PRJNA587534 | SAMN13193097 | 6 | ||||||
| 169 | IBSBF2579 | Brazil | 2009 | Unknown | anacardii | PRJNA416784 | SAMN07964563 | 10 | |||||
| 170 | ICPB10535 | Brazil | Unknown | Mexican lime | aurantifolii | PRJNA18835 | SAMN02472096 | 33 | Singleton | ||||
| 171 | ICPB11122 | Argentina | Unknown | aurantifolii | PRJNA18837 | SAMN02472095 | 33 | Singleton | |||||
| 172 | ISO12C3 | Canada | Ontario | Unknown | Common bean | fuscans | PRJNA289080 | SAMN03842216 | 5 | ||||
| 173 | ISO118C1 | Canada | Ontario | Unknown | Common bean | fuscans | PRJNA289080 | SAMN03842217 | 5 | ||||
| 174 | ISO118C5 | Canada | Ontario | Unknown | Common bean | fuscans | PRJNA289080 | SAMN03842218 | 5 | ||||
| 175 | JJ10-1 | Mauritius | Rodrigues Island | 1985 | Unknown | citri | A | PRJEB7180 | SAMEA2844848 | 4 | 1 | ||
| 176 | JK4-1 | China | 1985 | Unknown | citri | A | PRJEB7186 | SAMEA2844844 | 4 | 1 | |||
| 177 | JK48 | Saudi Arabia | 1988 | Unknown | citri | A* | PRJEB7195 | SAMEA2827235 | 4 | Singleton | |||
| 178 | JK143-9 | Thailand | 1990 | Unknown | citri | A* | PRJEB7200 | SAMEA2827230 | 4 | Singleton | |||
| 179 | JK143-11 | Thailand | 1990 | Unknown | citri | A* | PRJEB7184 | SAMEA2844846 | 4 | 2 | |||
| 180 | JM35-2 | Saudi Arabia | 1992 | Unknown | citri | A* | PRJEB7189 | SAMEA2827561 | 4 | 2 | |||
| 181 | JS581 | Iran | 1997 | Unknown | citri | A* | PRJEB7190 | SAMEA2827560 | 4 | 2 | |||
| 182 | JS582 | Iran | 1997 | Unknown | citri | A* | PRJEB7203 | SAMEA2827226 | 4 | Singleton | |||
| 183 | jx-6 | China | Jiangxi | 2014 | Unknown | citri | PRJNA286060 | SAMN03765509 | 1 | ||||
| 184 | JX4 | China | Jiangxi | 2011 | Unknown | citri | PRJNA255042 | SAMN02911836 | 26 | 1 | |||
| 185 | JX5 | China | Jiangxi | 2011 | Unknown | citri | PRJNA255042 | SAMN02911837 | 26 | 1 | |||
| 186 | K2 | South Korea | Danyang | 2017 | Unknown | glycines | PRJNA556107 | SAMN12340803 | 3 | ||||
| 187 | LB100-1 | Seychelles | 2005 | Unknown | citri | A | PRJEB7185 | SAMEA2844845 | 4 | 1 | |||
| 188 | LB302 | USA | Florida | 2002 | Unknown | citri | Aw | PRJEB7197 | SAMEA2827233 | 4 | Singleton | ||
| 189 | LE3-1 | Ethiopia | 2008 | Unknown | citri | A* | PRJEB7194 | SAMEA2827236 | 4 | Singleton | |||
| 190 | LE116-1 | Mali | Key lime | 2008 | Unknown | citri | A | PRJEB7201 | SAMEA2827228 | 4 | Singleton | ||
| 191 | LG56-10 | Réunion | 2009 | Unknown | Mango | mangiferaeindicae | PRJNA232105 | SAMN07766891 | 20 | ||||
| 192 | LG81-27 | Réunion | 2009 | Unknown | Mango | mangiferaeindicae | PRJNA232105 | SAMN07766892 | 20 | ||||
| 193 | LG97 | Bangladesh | 2006 | Unknown | citri | A | PRJEB7196 | SAMEA2827234 | 4 | Singleton | |||
| 194 | LG98 | Bangladesh | 2006 | Unknown | citri | A | PRJEB7183 | SAMEA2844847 | 4 | 1 | |||
| 195 | LG102 | Bangladesh | 2006 | Unknown | citri | A | PRJEB7198 | SAMEA2827232 | 4 | Singleton | |||
| 196 | LG115 | India | 2007 | Unknown | citri | Aw | PRJEB7187 | SAMEA2827563 | 4 | 7 | |||
| 197 | LG117 | Bangladesh | 2009 | Unknown | citri | A | PRJEB7188 | SAMEA2827562 | 4 | 1 | |||
| 198 | LH37-1 | Senegal | 2010 | Unknown | Grapefruit | citri | A | PRJEB7192 | SAMEA2827558 | 4 | 21 | ||
| 199 | LH201 | Réunion | 2010 | Leaf | citri | PRJNA344031 | SAMN05823148 | 1 | |||||
| 200 | LH276 | Réunion | 2010 | Leaf | citri | PRJNA344031 | SAMN05823145 | 1 | |||||
| 201 | LJ207-7 | Réunion | 2012 | Leaf | citri | PRJNA344031 | SAMN05823144 | 1 | |||||
| 202 | LL074-4 | Martinique | 2014 | Leaf | citri | PRJNA344031 | SAMN05823143 | 1 | |||||
| 203 | LM180 | Argentina | 2003 | Leaf | citri | PRJNA344031 | SAMN05823141 | 1 | |||||
| 204 | LMG941 | India | 1948 | Unknown | mangiferaeindicae | PRJEA86101 | SAMEA2272013 | 34 | 20 | ||||
| 205 | LMG965 | India | 1969 | Unknown | Grape | viticola | PRJEB5493 | SAMEA3139044 | 35 | 8 | |||
| 206 | LMG712 | Sudan | 1956 | Unknown | glycines | PRJNA298608 | SAMN04145201 | 3 | |||||
| 207 | LMG761 | Sudan | 1958 | Unknown | malvacearum | PRJNA298617 | SAMN04145210 | 6 | |||||
| 208 | LMG826 | Belgium | Merelbeke | 2014 | Unknown | Common bean | fuscans | PRJNA254368 | SAMN02903172 | 13 | Singleton | ||
| 209 | LMG859 | India | 1959 | Unknown | punicae | PRJNA73081 | SAMEA3138416 | 36 | Singleton | ||||
| 210 | LMG7399 | Belgium | Merelbeke | 2014 | Unknown | dieffenbachiae | PRJNA254467 | SAMN02903999 | Singleton | ||||
| 211 | LMG9322 | USA | Florida | 1915 | Unknown | citri | A | PRJNA338819 | SAMN05571463 | Singleton | |||
| 212 | mf20 | USA | Florida | 2011 | Unknown | citri | PRJNA255042 | SAMN02911847 | 26 | 1 | |||
| 213 | MN10 | USA | Florida | 2005 | Unknown | citri | PRJNA255042 | SAMN02911841 | 26 | 1 | |||
| 214 | MN11 | USA | Florida | Unknown | citri | PRJNA255042 | SAMN02911842 | 26 | 1 | ||||
| 215 | MN12 | USA | Florida | 1997 | Unknown | citri | PRJNA255042 | SAMN02911843 | 26 | 1 | |||
| 216 | MS14003 | USA | Wilzone, MS | 2014 | Leaf | malvacearum | PRJNA396899 | SAMN07447517 | 18 | ||||
| 217 | MSCT | USA | Mississippi | 2011 | Leaf | malvacearum | PRJNA299817 | SAMN05595756 | 18 | ||||
| 218 | NCPPB1402 | Uganda | 1962 | Unknown | Common bean | fuscans | PRJNA264810 | SAMN03142810 | 5 | ||||
| 219 | NCPPB381 | Canada | 1957 | Unknown | Common bean | fuscans | PRJNA264772 | SAMN03142398 | 11 | ||||
| 220 | NCPPB670 | Uganda | 1958 | Unknown | Common bean | fuscans | PRJNA263153 | SAMN03097370 | 5 | ||||
| 221 | NCPPB1056 | Ethiopia | 1961 | Unknown | Common bean | fuscans | PRJNA264809 | SAMN03142809 | 5 | ||||
| 222 | NCPPB1058 | Ethiopia | 1961 | Unknown | Common bean | fuscans | PRJNA264815 | SAMN03142815 | 5 | ||||
| 223 | NCPPB1433 | Hungary | 1956 | Unknown | Common bean | fuscans | PRJNA264773 | SAMN03142399 | 5 | ||||
| 224 | NCPPB1654 | South Africa | 1963 | Unknown | Common bean | fuscans | PRJNA264775 | SAMN03142402 | 5 | ||||
| 225 | NCPPB2665 | Italy | 1973 | Unknown | Common bean | fuscans | PRJNA264776 | SAMN03142403 | 5 | ||||
| 226 | NCPPB3607 | India | 1988 | Unknown | citri | A* | PRJEB7191 | SAMEA2827559 | 4 | 2 | |||
| 227 | NCPPB3610 | India | 1988 | Unknown | citri | A | PRJEB7199 | SAMEA2827231 | 4 | Singleton | |||
| 228 | NCPPB3612 | India | Key lime | 1988 | Unknown | citri | A | PRJEB7193 | SAMEA2827237 | 4 | 21 | ||
| 230 | NCPPB3660 | Brazil | 1975 | Unknown | Common bean | fuscans | PRJNA264814 | SAMN03142811 | 13 | 5 | |||
| 231 | NIGEB-88 | Iran | Hashtbandi | 2009 | Unknown | citri | A* | PRJNA283400 | SAMN03649471 | 2 | |||
| 232 | NIGEB-386 | Iran | Nik Shahr | 2009 | Unknown | citri | A* | PRJNA261284 | SAMN03070127 | 2 | |||
| 233 | NT17 | USA | Florida | 2011 | Unknown | citri | PRJNA255042 | SAMN02911844 | 26 | 1 | |||
| 234 | TAQ18 | Brazil | Leaf | anacardii | PRJNA416788 | SAMN07964753 | 24 | ||||||
| 235 | TAQ13 | Brazil | 2009 | Leaf | anacardii | PRJNA416789 | SAMN07964775 | 24 | |||||
| 236 | TX160042 | USA | Rancho Viejo, TX | 2015 | Leaf | citri | Aw | PRJNA381640 | SAMN06685652 | 7 | |||
| 237 | TX160149 | USA | Rancho Viejo, TX | 2015 | Leaf | citri | Aw | PRJNA381640 | SAMN06685654 | Singleton | |||
| 238 | TX160197 | USA | Rancho Viejo, TX | 2015 | Leaf | citri | Aw | PRJNA381640 | SAMN06685696 | 7 | |||
| 239 | UI6 | China | Guangxi | 2011 | Unknown | citri | PRJNA255042 | SAMN02911838 | 26 | 1 | |||
| 240 | UI7 | China | Guangxi | 2011 | Unknown | citri | PRJNA255042 | SAMN02911839 | 26 | 1 | |||
| 241 | WHRI5232 | Sudan | 1959 | Unknown | malvacearum | PRJNA438827 | SAMN08729580 | 37 | 6 | ||||
| 242 | 03-1638-1-1 | Argentina | 2003 | Unknown | citri | PRJNA401937 | SAMN07611881 | 38 | 1 | ||||
| 243 | x8ra | South Korea | Suwon | 1999 | Unknown | glycines | PRJNA556081 | SAMN12340633 | 3 | ||||
| 244 | X18 | Burkina Faso | Unknown | Cotton | malvacearum | PRJNA172044 | SAMN02469929 | 39 | 18 | ||||
| 245 | X20 | Burkina Faso | Unknown | Cotton | malvacearum | PRJNA172045 | SAMN02469930 | 39 | 6 | ||||
| 246 | X621 | South Africa | 1995 | Unknown | Common bean | fuscans | PRJNA272380 | SAMN03281080 | 5 | ||||
| 247 | Xcc29 | China | Jiangxi | 2010 | Unknown | citri | PRJNA407058 | SAMN07665076 | 1 | ||||
| 248 | Xcc49 | China | Chongqing | 2010 | Unknown | citri | PRJNA407058 | SAMN07638001 | 1 | ||||
| 249 | XcmH1005 | USA | Oklahoma | 1968 | Unknown | malvacearum | PRJNA298765 | SAMN04166563 | 6 | ||||
| 250 | XcmN1003 | Burkina Faso | 1967 | Unknown | malvacearum | PRJNA298770 | SAMN04166615 | 6 | |||||
| 251 | XCP631 | Colombia | Quilichao | 2004 | Unknown | Common bean | fuscans | PRJNA272630 | SAMN03284618 | 5 | |||
| 252 | Xff49 | Brazil | Pelotas | 2017 | Unknown | fuscans | PRJNA400313 | SAMN07563171 | Singleton |
NA, not applicable.
FIG 1.

Neighbor-joining tree based on 43 concatenated rRNA gene sequences generated on the PubMLST website. The phylogeny was generated using the iTOL (24) plug-in on the PubMLST website (https://pubmlst.org/). The scale bar represents the genetic distance.
cgMLST.
A total of 1,618 core genes (present in >99% of isolates) were found among 250 X. citri isolate genomes. These genes were numbered XCIT00001 to XCIT01618. Allele calling of the subset of the initial 100 records (from study isolates) resulted in isolates having between 99.4% and 100% of their loci with alleles designated. Core-genome MLST (cgMLST) groupings of the 250 genomes uploaded to the PubMLST website were made based on the number of allelic mismatches. This resulted in 171 groups of genomes with 5 or fewer mismatches (isolates tagged as Xc_cgc_5 on the PubMLST website), 113 with 10 or fewer mismatches (Xc_cgc_10), 53 with 50 or fewer mismatches (Xc_cgc_50), 39 with 100 or fewer mismatches (Xc_cgc_100), and 25 with 200 or fewer mismatches (Xc_cgc_200).
cgMLST groupings.
The groupings of 250 isolates/genomes with fewer than 200 mismatches are shown in Table 1. Group 1 contained 132 X. citri subsp. citri isolates comprising 104 pathotype A isolates (pathotype data are missing for 28 isolates in this group); group 2 contained 12 X. citri subsp. citri genomes, all of which were isolates of pathotype A*; group 3 comprised 12 X. citri pv. glycines isolates; group 4 contained 2 X. citri pv. aurantifolii isolates; group 5 contained 24 X. citri pv. fuscans isolates; group 6 contained 7 X. citri subsp. malvacearum isolates; group 7 contained 8 X. citri pv. citri isolates with pathotype Aw; group 8 contained 3 X. citri pv. viticola isolates; group 9 contained 5 X. citri pv. fuscans isolates; group 10 contained 2 X. citri pv. anacardii isolates; group 11 contained 2 X. citri pv. fuscans isolates; group 13 contained 4 X. citri pv. fuscans isolates; group 17 contained 3 X. citri pv. aurantifolii isolates; group 18 contained 4 X. citri subsp. malvacearum isolates; group 20 contained 3 X. citri pv. mangiferaeindicae isolates; group 21 contained 2 X. citri pv. citri isolates of an unknown pathotype; and group 24 contained 2 X. citri pv. anacardii isolates. Twenty-three isolates had no close matches using any of the allelic mismatch groupings described above, and these are referred to as singleton isolate genomes in Table 1. Other cgMLST groupings (and all other genomic data and metadata) can be found in Table 1.
X. citri phylogeny.
A neighbor-joining tree of concatenated MLST allelic sequences of the 250 X. citri isolates is shown in Fig. 2. This phylogeny was generated using the Interactive Tree of Life (iTOL) plug-in on the PubMLST website. It clearly distinguishes individual X. citri pathovars (colored), with the genomes of isolates belonging to the same pathovar being grouped, although some subgroupings are evident. This is most marked for X. citri pv. fuscans isolate genomes, which are represented by three clades that include isolates from previous studies by Alavi et al. (12) and Aritua et al. (13). These correspond to isolates from three lineages originally named X. citri pv. phaseoli and X. citri pv. phaseoli GL 1 and X. citri pv. phaseoli GL fuscans GL2 and GL3. Genomes from isolates of X. citri pv. aurantifolii resolve as two main clades, with the smaller group showing more genetic similarity to members of the X. citri pv. anacardii group than to other members of X. citri pv. aurantifolii. Overall, this phylogeny displays a high degree of congruence with a neighbor-joining phylogeny based on core-genome SNPs, with the same groupings of genomes and only superficial differences in the tree structure (Fig. S1). A minimum-spanning tree generated using GrapeTree based on cgMLST allele data shows groupings identical to those made using both methods (Fig. 3).
FIG 2.

Neighbor-joining tree based on 250 concatenated core-genome MLST allele sequences of Xanthomonas citri. Isolates are colored according to their original pathovar/subspecies designations and X. citri subsp. citri pathotype. The phylogeny was generated using the iTOL (24) plug-in on the PubMLST website (https://pubmlst.org/). The scale bar represents the genetic distance.
FIG 3.

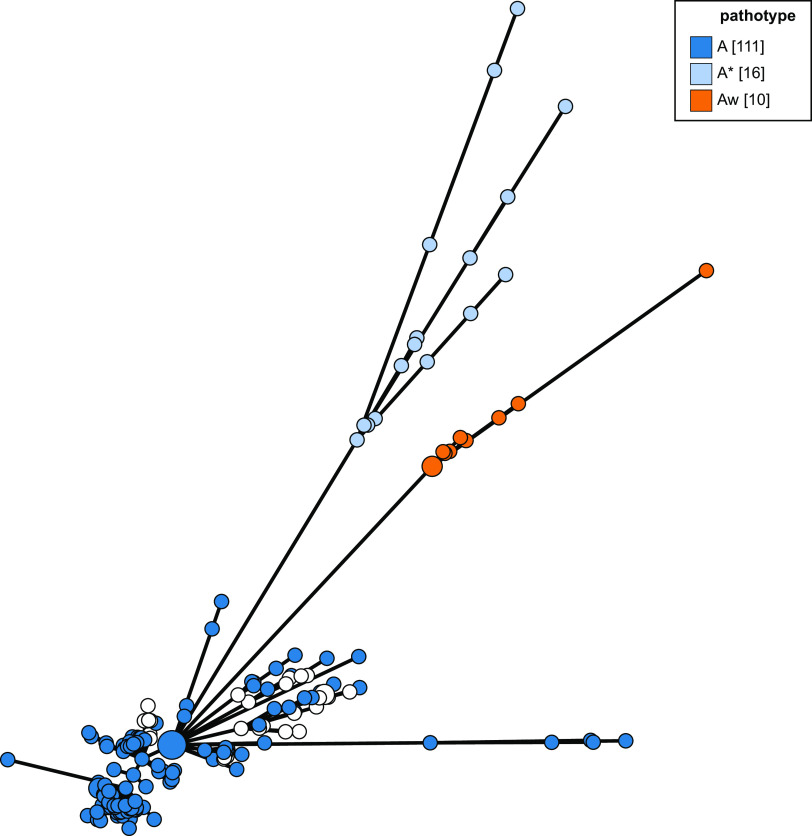
Minimum-spanning tree based on 250 core-genome allelic profiles of Xanthomonas citri. Isolates/groups are colored according to pathovar/subspecies. Pathotype A*, pathotype Aw, and the divergent pathotype A clade containing five genomes, including that of isolate LH37-1, are shown. The phylogeny was generated using the GrapeTree (23) plug-in on the PubMLST website (https://pubmlst.org/).
The times taken to generate phylogenies based on core-genome SNPs using kSNP3 and MEGA 11 (36 h), concatenated allelic sequences using iTOL (15 min), and MLST allele data using GrapeTree (4 min) varied considerably. All tests were run on a 2020 3.6-GHz 10-Core Intel Core i9 iMac with 16 GB RAM.
X. citri subsp. citri pathotype phylogenies.
The genomes of pathotype A strains of X. citri pv. citri represented the largest group in this study, and these isolate genomes resolve as a discrete group in an NJ phylogeny based on concatenated cgMLST allele data (Fig. 2 and 3), with the exception of five isolates whose genomes were listed in the nucleotide submission information as being of pathotype A. These five isolates correspond to a separate lineage of pathotype A isolates examined in a previous study by Gordon et al. (4), which were isolated from grapefruit, key lime, and citrus species. Pathotype A* and Aw isolate genomes also form distinct clades using all three tree-building methods (Fig. 2). This can be seen in Fig. 2 and 3 but is clearer in Fig. 4 and Fig. S1, which include only X. citri subsp. citri isolate genomes.
FIG 4.
Minimum-spanning tree based on 127 Xanthomonas citri pv. citri core-genome allelic profiles, colored according to pathotype. The phylogeny was generated using the GrapeTree (23) plug-in on the PubMLST website (https://pubmlst.org/).
DISCUSSION
We have developed a cgMLST scheme for the study of Xanthomonas citri. Through its implementation on the PubMLST website, this scheme can be used to rapidly and robustly infer the pathovar (or subspecies) and, in the case of X. citri subsp. citri, pathotype designation(s) based on genetic similarity to uploaded and curated genomic sequence data with their associated metadata. The database currently contains 250 isolate genomes and their associated metadata, including 100 novel X. citri subsp. citri isolates sequenced in this study.
A neighbor-joining phylogeny based on concatenated allele sequences generated using the iTOL plug-in on the PubMLST website had a structure very similar to that of a core-genome SNP-based NJ tree but was generated within several minutes, compared to the >36 h required to generate an SNP-based phylogeny that depended on multiple-whole-genome alignment. This is important because as the number of sequenced X. citri genomes deposited in public databases increases, the computational resources required to generate SNP phylogenies de novo will become greater.
We used GrapeTree, implemented at PubMLST, to generate and display minimum-spanning trees of study data, and this plug-in can quickly and clearly display phylogenies colored according to metadata such as country of origin, date, host species, pathovar, and X. citri subsp. citri pathotype. The phylogenies generated using each of the methods used here, SNP-based NJ and iTOL and GrapeTree phylogenies based on concatenated cgMLST locus sequences, were largely congruent, with very similar groupings (see Fig. S1 in the supplemental material). However, GrapeTree, with its ability to easily and quickly display very large genomic data sets such as those present in EnteroBase, is eminently scalable as data sets grow, unlike core-SNP-based methods, which are more computationally intensive and time-consuming.
Neighbor-joining phylogenies based on concatenated rRNA gene sequences were generated in this study from genomes uploaded to the PubMLST website. This tree delineated the 21 different Xanthomonas species and 4 other more distantly related ones, including E. coli and P. aeruginosa. The rRNA gene is automatically applied to PubMLST genome data and serves as a further check of species designations for uploaded genome data.
The cgMLST scheme implemented on the PubMLST website for X. citri will, we hope, be an increasingly useful tool for the study of the epidemiology and evolution of the major cause of citrus canker, X. citri subsp. citri, but should also be of benefit for the study of other plant-pathogenic X. citri subspecies and pathovars included in this study as well as those not yet included in the database.
MATERIALS AND METHODS
Bacterial isolates.
A total of 101 X. citri subsp. citri isolates were obtained from Fundecitrus, Araraquara, São Paulo, Brazil, an association maintained by citrus growers and juice manufacturers from the State of São Paulo to conduct research, education, and implementation of citrus crop protection. Isolate 306, corresponding to the previously sequenced genome of strain 306 (14), was resequenced as part of this study, resulting in the sequencing of 100 novel isolates. These were sampled from citrus plants from 15 different countries and included 75 isolates from Brazil; 4 from South Korea; 3 each from Argentina and the United States; 2 each from China, New Zealand, and Paraguay; and 1 each from Australia, Fiji, France, India, Iran, Mauritius, Taiwan, Thailand, and Uruguay. One isolate’s country of origin is unknown. Details of the isolates are shown in Table 1. These isolates were all pathotype A isolates from sweet orange, with the exception of two pathotype A* isolates from key lime. Study bacteria were isolated between 1979 and 2015. Data on the year of isolation were not available for 31 of the 100 isolates.
Genomic DNA sequencing.
Genomic sequencing was performed by MicrobesNG (University of Birmingham) from pure culture material stabilized in DNA/RNA Shield buffer (Zymo Research, CA, USA). Genomic DNA libraries were prepared using Nextera XT library prep kits (Illumina, San Diego, CA, USA). Libraries were sequenced using Illumina sequencers (HiSeq), using a 250-bp paired-end protocol. Reads were adapter trimmed using Trimmomatic 0.30 with a sliding window quality cutoff of Q15 (15) and scanned using Kraken (11) to confirm species identity. De novo assembly was performed on samples using SPAdes version 3.7 (16).
A further 150 X. citri genome sequences, downloaded from the European Nucleotide Archive (ENA), were included for analysis, including 65 X. citri subsp. citri isolates (comprising 12 pathotype A, 14 pathotype A*, and 10 pathotype Aw isolates according to their cited literature sources), 9 X. citri pv. aurantifolii isolates, 37 X. citri pv. fuscans isolates, 12 X. citri pv. glycines isolates, 12 X. citri subsp. malvacearum isolates, 3 X. citri pv. mangiferaeindicae isolates, 3 X. citri pv. viticola isolates, 2 X. citri pv. vignicola isolates, and 1 isolate each of X. citri pv. dieffenbachiae and pv. punicae. Details of all X. citri genomes included in this study are shown in Table 1. In addition, 24 genomes representing single examples of 20 different Xanthomonas spp. and single examples of Stenotrophomonas maltophilia, Escherichia coli, Xylella fastidiosa, and Pseudomonas aeruginosa were downloaded from GenBank. Details of these isolates are shown in Table S1 in the supplemental material.
Core-genome MLST.
Complete coding sequences were identified in the finished genome assembly of strain 306 (14) using Prokka (17) with default settings. These were used in Roary (18) to identify 1,618 genes found in all 250 genomes. A BIGSdb database for X. citri was set up on the PubMLST website (19), with loci being defined for each of the identified core genes and named using an XCIT prefix and a five-digit identifier, ranging from XCIT00001 to XCIT01618. The database was seeded with the coding sequence found in strain 306 for each of these loci defined as allele 1. Allelic variants found in the 100-isolate locally sequenced data set were then identified using the BIGSdb allele caller, with thresholds of 98% identity over 98% of the alignment length compared to reference alleles. A further round of allele calling using the same parameters and all previously identified alleles as references was performed, followed by manual scanning to identify more variable alleles containing small indels. The database was then expanded to include all 250 isolates and alleles identified as described above. Start codon positions were adjusted in nine loci as the codon identified in the reference genome was not found consistently across the data set, whereas an alternate consensus start codon was identified nearby. Core-genome sequence types (cgSTs) were defined automatically by BIGSdb for profiles with fewer than 50 missing loci. Single-linkage cluster schemes were set up within the database to identify related isolates using a range of locus mismatch thresholds (200, 100, 50, 25, 10, and 5 locus mismatches).
Ribosomal MLST (rMLST) (20), implemented on the PubMLST website, confirmed the species identity of all 274 study isolates. It was also used to generate concatenated rRNA gene sequences for phylogenetic analysis. As rMLST examines allelic variation at 53 universal rRNA genes, it is ideally suited for the rapid phenotypic analysis of genomes of different species.
Phylogenetic trees.
In common with previous studies of X. citri evolution and epidemiology, we generated phylogenies based on core-genome SNPs using a reference genome. We used the finished genome of X. citri pv. citri strain 306 (14) as a reference and kSNP3 v3.12 (21) to generate fasta nucleotide files of SNPs, which were used in MEGA 11 (22) to generate NJ trees. Genomic DNA sequence data and their associated metadata can be analyzed using a variety of methods implemented on the PubMLST website. Here, we generated minimum-spanning trees from allelic profiles using GrapeTree (23). Neighbor-joining trees based on concatenated nucleotides of cgMLST loci were generated using Interactive Tree of Life (iTOL) (24). Both the GrapeTree and iTOL plug-ins are implemented on the PubMLST website (https://pubmlst.org/). A neighbor-joining tree was constructed for the 250 X. citri study isolates and 24 other species listed in Table S1. This was generated from the 53 concatenated rRNA gene sequences used in the rMLST scheme using the iTOL plug-in as described above.
ACKNOWLEDGMENTS
This work was funded by BBSRC/Newton Fund (https://www.ukri.org/councils/bbsrc/ https://www.newton-gcrf.org/newton-fund/) awards BB/R022720/1 and BB/S018891/1 to M.C.E., CONFAP (https://confap.org.br/) research mobility award 2019/05497-7 to M.C.E., and FAPESP awards 2017/50454-9 and 2018/21164-5 to H.F.
None of the authors have any financial, personal, or professional interests that could be construed to have influenced the work.
Footnotes
Supplemental material is available online only.
Contributor Information
M. C. Enright, Email: m.enright@mmu.ac.uk.
Charles M. Dozois, INRS Armand-Frappier Sante Biotechnologie Research Centre
REFERENCES
- 1.Behlau F, Gochez AM, Jones JB. 2020. Diversity and copper resistance of Xanthomonas affecting citrus. Trop Plant Pathol 45:200–212. doi: 10.1007/s40858-020-00340-1. [DOI] [Google Scholar]
- 2.Brunings AM, Gabriel DW. 2003. Xanthomonas citri: breaking the surface. Mol Plant Pathol 4:141–157. doi: 10.1046/j.1364-3703.2003.00163.x. [DOI] [PubMed] [Google Scholar]
- 3.Ference CM, Gochez AM, Behlau F, Wang N, Graham JH, Jones JB. 2018. Recent advances in the understanding of Xanthomonas citri ssp. citri pathogenesis and citrus canker disease management: Xcc pathogenesis and citrus canker management. Mol Plant Pathol 19:1302–1318. doi: 10.1111/mpp.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon JL, Lefeuvre P, Escalon A, Barbe V, Cruveiller S, Gagnevin L, Pruvost O. 2015. Comparative genomics of 43 strains of Xanthomonas citri pv. citri reveals the evolutionary events giving rise to pathotypes with different host ranges. BMC Genomics 16:1098. doi: 10.1186/s12864-015-2310-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Timilsina S, Potnis N, Newberry EA, Liyanapathiranage P, Iruegas-Bocardo F, White FF, Goss EM, Jones JB. 2020. Xanthomonas diversity, virulence and plant-pathogen interactions. Nat Rev Microbiol 18:415–427. doi: 10.1038/s41579-020-0361-8. [DOI] [PubMed] [Google Scholar]
- 6.Buttner D, Bonas U. 2010. Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol Rev 34:107–133. doi: 10.1111/j.1574-6976.2009.00192.x. [DOI] [PubMed] [Google Scholar]
- 7.Armstrong GL, MacCannell DR, Taylor J, Carleton HA, Neuhaus EB, Bradbury RS, Posey JE, Gwinn M. 2019. Pathogen genomics in public health. N Engl J Med 381:2569–2580. doi: 10.1056/NEJMsr1813907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bansal K, Midha S, Kumar S, Patil PB. 2017. Ecological and evolutionary insights into Xanthomonas citri pathovar diversity. Appl Environ Microbiol 83:e02993-16. doi: 10.1128/AEM.02993-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patané JSL, Martins J, Jr, Rangel LT, Belasque J, Digiampietri LA, Facincani AP, Ferreira RM, Jaciani FJ, Zhang Y, Varani AM, Almeida NF, Wang N, Ferro JA, Moreira LM, Setubal JC. 2019. Origin and diversification of Xanthomonas citri subsp. citri pathotypes revealed by inclusive phylogenomic, dating, and biogeographic analyses. BMC Genomics 20:700. doi: 10.1186/s12864-019-6007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maiden MCJ, van Rensburg MJJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alavi SM, Sanjari S, Durand F, Brin C, Manceau C, Poussier S. 2008. Assessment of the genetic diversity of Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans as a basis to identify putative pathogenicity genes and a type III secretion system of the SPI-1 family by multiple suppression subtractive hybridizations. Appl Environ Microbiol 74:3295–3301. doi: 10.1128/AEM.02507-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aritua V, Harrison J, Sapp M, Buruchara R, Smith J, Studholme DJ. 2015. Genome sequencing reveals a new lineage associated with lablab bean and genetic exchange between Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans. Front Microbiol 6:1080. doi: 10.3389/fmicb.2015.01080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.da Silva ACR, Ferro JA, Reinach FC, Farah CS, Furlan LR, Quaggio RB, Monteiro-Vitorello CB, Van Sluys MA, Almeida NF, Alves LMC, do Amaral AM, Bertolini MC, Camargo LEA, Camarotte G, Cannavan F, Cardozo J, Chambergo F, Ciapina LP, Cicarelli RMB, Coutinho LL, Cursino-Santos JR, El-Dorry H, Faria JB, Ferreira AJS, Ferreira RCC, Ferro MIT, Formighieri EF, Franco MC, Greggio CC, Gruber A, Katsuyama AM, Kishi LT, Leite RP, Lemos EGM, Lemos MVF, Locali EC, Machado MA, Madeira AMBN, Martinez-Rossi NM, Martins EC, Meidanis J, Menck CFM, Miyaki CY, Moon DH, Moreira LM, Novo MTM, Okura VK, Oliveira MC, Oliveira VR, Pereira HA, et al. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459–463. doi: 10.1038/417459a. [DOI] [PubMed] [Google Scholar]
- 15.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 18.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jolley KA, Bliss CM, Bennett JS, Bratcher HB, Brehony C, Colles FM, Wimalarathna H, Harrison OB, Sheppard SK, Cody AJ, Maiden MCJ. 2012. Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology (Reading) 158(Part 4):1005–1015. doi: 10.1099/mic.0.055459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardner SN, Slezak T, Hall BG. 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 22.Tamura K, Stecher G, Kumar S. 2021. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol Biol Evol 38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Z, Alikhan N-F, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, Carriço JA, Achtman M. 2018. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res 28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letunic I, Bork P. 2021. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darrasse A, Carrère S, Barbe V, Boureau T, Arrieta-Ortiz ML, Bonneau S, Briand M, Brin C, Cociancich S, Durand K, Fouteau S, Gagnevin L, Guérin F, Guy E, Indiana A, Koebnik R, Lauber E, Munoz A, Noël LD, Pieretti I, Poussier S, Pruvost O, Robène-Soustrade I, Rott P, Royer M, Serres-Giardi L, Szurek B, van Sluys M-A, Verdier V, Vernière C, Arlat M, Manceau C, Jacques M-A. 2013. Genome sequence of Xanthomonas fuscans subsp. fuscans strain 4834-R reveals that flagellar motility is not a general feature of xanthomonads. BMC Genomics 14:761. doi: 10.1186/1471-2164-14-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Jalan N, Zhou X, Goss E, Jones JB, Setubal JC, Deng X, Wang N. 2015. Positive selection is the main driving force for evolution of citrus canker-causing Xanthomonas. ISME J 9:2128–2138. doi: 10.1038/ismej.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jalan N, Kumar D, Yu F, Jones JB, Graham JH, Wang N. 2013. Complete genome sequence of Xanthomonas citri subsp. citri strain Aw12879, a restricted-host-range citrus canker-causing bacterium. Genome Announc 1:e00235-13. doi: 10.1128/genomeA.00235-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruh M, Briand M, Bonneau S, Jacques M-A, Chen NWG. 2017. Xanthomonas adaptation to common bean is associated with horizontal transfers of genes encoding TAL effectors. BMC Genomics 18:670. doi: 10.1186/s12864-017-4087-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darrasse A, Bolot S, Serres-Giardi L, Charbit E, Boureau T, Fisher-Le Saux M, Briand M, Arlat M, Gagnevin L, Koebnik R, Noël LD, Carrère S, Jacques MA. 2013. High-quality draft genome sequences of Xanthomonas axonopodis pv. glycines strains CFBP 2526 and CFBP 7119. Genome Announc 1:e01036-13. doi: 10.1128/genomea.01036-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ah-You N, Gagnevin L, Chiroleu F, Jouen E, Neto JR, Pruvost O. 2007. Pathological variations within Xanthomonas campestris pv. mangiferaeindicae support its separation into three distinct pathovars that can be distinguished by amplified fragment length polymorphism. Phytopathology 97:1568–1577. doi: 10.1094/phyto-97-12-1568. [DOI] [PubMed] [Google Scholar]
- 31.Indiana A, Briand M, Arlat M, Gagnevin L, Koebnik R, Noël LD, Portier P, Darrasse A, Jacques MA. 2014. Draft genome sequence of the flagellated Xanthomonas fuscans subsp. fuscans strain CFBP 4884. Genome Announc 2:e00966-14. doi: 10.1128/genomeA.00966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carpenter SCD, Kladsuwan L, Han S-W, Prathuangwong S, Bogdanove AJ. 2019. Complete genome sequences of Xanthomonas axonopodis pv. glycines isolates from the United States and Thailand reveal conserved transcription activator-like effectors. Genome Biol Evol 11:1380–1384. doi: 10.1093/gbe/evz085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreira LM, Almeida NF, Jr, Potnis N, Digiampietri LA, Adi SS, Bortolossi JC, da Silva AC, da Silva AM, de Moraes FE, de Oliveira JC, de Souza RF, Facincani AP, Ferraz AL, Ferro MI, Furlan LR, Gimenez DF, Jones JB, Kitajima EW, Laia ML, Leite RP, Jr, Nishiyama MY, Rodrigues Neto J, Nociti LA, Norman DJ, Ostroski EH, Pereira HA, Jr, Staskawicz BJ, Tezza RI, Ferro JA, Vinatzer BA, Setubal JC. 2010. Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp. aurantifolii. BMC Genomics 11:238. doi: 10.1186/1471-2164-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Midha S, Ranjan M, Sharma V, Pinnaka AK, Patil PB. 2012. Genome sequence of Xanthomonas citri pv. mangiferaeindicae strain LMG 941. J Bacteriol 194:3031. doi: 10.1128/JB.00433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Midha S, Patil PB. 2014. Genomic insights into the evolutionary origin of Xanthomonas axonopodis pv. citri and its ecological relatives. Appl Environ Microbiol 80:6266–6279. doi: 10.1128/AEM.01654-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma V, Midha S, Ranjan M, Pinnaka AK, Patil PB. 2012. Genome sequence of Xanthomonas axonopodis pv. punicae strain LMG 859. J Bacteriol 194:2395. doi: 10.1128/JB.00181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michalopoulou VA, Vicente JG, Studholme DJ, Sarris PF. 2018. Draft genome sequences of pathotype strains for three pathovars belonging to three Xanthomonas species. Microbiol Resour Announc 7:e00923-18. doi: 10.1128/MRA.00923-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gochez AM, Huguet-Tapia JC, Minsavage GV, Shantaraj D, Jalan N, Strauß A, Lahaye T, Wang N, Canteros BI, Jones JB, Potnis N. 2018. Pacbio sequencing of copper-tolerant Xanthomonas citri reveals presence of a chimeric plasmid structure and provides insights into reassortment and shuffling of transcription activator-like effectors among X. citri strains. BMC Genomics 19:16. doi: 10.1186/s12864-017-4408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cunnac S, Bolot S, Forero Serna N, Ortiz E, Szurek B, Noël LD, Arlat M, Jacques M-A, Gagnevin L, Carrere S, Nicole M, Koebnik R. 2013. High-quality draft genome sequences of two Xanthomonas citri pv. malvacearum strains. Genome Announc 1:e00674-13. doi: 10.1128/genomeA.00674-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.02101-22-s0001.pdf, PDF file, 0.2 MB (244.6KB, pdf)