Abstract
Background.
Colorectal cancer leads to peritoneal metastases (CRPM) in 10% of cases. Cytoreductive surgery with hyperthermic intraperitoneal chemoperfusion (CRS-HIPEC) improves survival. Primary tumor location and abnormalities in RAS, BRAF, and mismatch repair/microsatellite stability (MMR/MSI) may affect post-CRS-HIPEC survival, but studies have not been consistent. We estimated the effects of primary tumor site and genomic alterations on post-CRS-HIPEC survival.
Methods.
This retrospective cohort study included CRS-HIPEC cases for CRPM at a high-volume center from 2001 to 2020. Next-generation sequencing and microsatellite testing defined the RAS, BRAF, and MMR/MSI genotypes. Adjusted effects of tumor sidedness and genomics on survival were evaluated using a multivariable Cox proportional hazards model. We analyzed these variables’ effects on progression-free survival and the effects of immune checkpoint-inhibitors.
Results.
A total of 250 patients underwent CRS-HIPEC with testing for RAS, BRAF, and MMR/MSI; 50.8% of patients were RAS-mutated, 12.4% were BRAF-mutated, and 6.8% were deficient-MMR/MSI-high (dMMR/MSI-H). Genomic alterations predominated in right-sided cancers. After adjustment for comorbidities and oncological and perioperative variables, rectal origin [hazard ratio (HR) 1.9, p = 0.01], RAS mutation (HR 1.6, p = 0.01), and BRAF mutation (HR 1.7, p = 0.05) were associated with worse survival. RAS mutation was also associated with shorter progression-free survival (HR 1.6, p = 0.01 at 6 months post-operatively), and dMMR/MSI-H status was associated with superior survival (HR 0.3, p = 0.01 at 2 years). dMMR/MSI-H patients receiving immune checkpoint-inhibitors trended toward superior survival.
Conclusions.
Rectal origin, RAS mutations, and BRAF mutations are each associated with poorer survival after CRS-HIPEC for CRPM. Patients with CRPM and dMMR/MSI-H status have superior survival. Further research should evaluate benefits of immune checkpoint-inhibitors in this subgroup.
Cytoreductive surgery with hyperthermic intraperitoneal chemoperfusion (CRS-HIPEC), in combination with systemic chemotherapy, improves the survival of well-selected patients with colorectal peritoneal metastases (CRPM).1–3 Unfortunately, selection criteria for surgery remain incompletely defined, resulting in poor postoperative outcomes in some, and the omission of surgery for others who would have gained a clinically significant survival benefit from the procedure. Optimal CRS-HIPEC involves resection of all visible peritoneal metastases, followed by circulation of heated chemotherapy intraperitoneally to treat microscopic disease.4–6 Major factors associated with survival following CRS-HIPEC include lower peritoneal disease burden, good physical performance status, less aggressive histopathology, responsiveness to systemic chemotherapy, or prolonged disease-free/progression-free interval.7 Despite knowledge of these factors, the patient population with CRPM considered for surgery remains heterogenous and with disparate outcomes. Recent studies suggest an association between distinct CRPM gene expression patterns and survival.8 Therefore, to further improve the precision of patient selection (and thereby optimize survival), knowledge is needed of associations between location/origin of colorectal cancer (CRC), genomic profile of peritoneal metastases, and long-term survival.
Previous work showed that sidedness of the primary tumor affects the prognosis of CRC. Right-sided CRC more commonly presents with aggressive histopathologic features (e.g., poor differentiation, signet ring cells) and a higher proportion of specific genomic alterations, such as Rat sarcoma viral oncogene homologue (RAS) mutations, v-raf murine sarcoma viral oncogene homologue B (BRAF) mutations, and deficient mismatch repair or high microsatellite instability (dMMR/MSI-H) status.9,10 Both in non-metastatic CRC 11 and in patients with colorectal liver metastases (CRLM),12 right-sided tumors lead to worse survival than left-sided tumors. However, the effect of primary tumor site on survival with CRPM is controversial and inconsistent between studies.7,9,13–16 Rates of peritoneal metastasis are higher among right-sided CRC.9 Furthermore, while prognostic differences between colonic and rectal primary cases have been studied in patients with non-metastatic disease17 or with liver metastases,18 the significance of rectal versus colonic primary site remains unknown in patients with peritoneal metastases.14–16
Aside from tumor sidedness/location, genomic alterations such as RAS mutations, BRAF mutations, and dMMR/MSI-H status also independently affect survival in CRC. Most studies of non-metastatic CRC report worse survival secondary to RAS or BRAF mutations,19 while dMMR/MSI-H status confers a survival advantage for stage II–III CRC.20,21 For patients with CRLM, survival is generally worse with RAS mutation,22 BRAF mutation,23–26 and dMMR/MSI-H status.27 However, for patients with CRPM, the effect of dMMR/MSI-H status is less clear, with one study reporting superior survival.28
Finally, primary tumor location and genomic alterations interact to further influence survival. In a multinational study of patients with stage I–III CRC, KRAS mutation independently worsened survival only in patients with left-sided colonic tumors.29 Survival is similarly impaired by KRAS mutation in patients with CRLM whose primary site was the left hemi-colon, but not the right hemi-colon or rectum.30–32 In contrast, for CRPM, the data on interaction between primary tumor site and genetics are limited to date.
Given the uncertainty regarding primary tumor location, genomic effects, their interaction, and long-term survival for CRPM, especially post-CRS-HIPEC, we examined the frequency and distribution of primary tumor location and specific genomic alterations in patients with CRPM undergoing CRS-HIPEC and assessed the oncologic associations of these variables. We hypothesized that the association between tumor location and postoperative survival would be driven by underlying histopathologic features and genomic alterations rather than tumor location itself.
METHODS
Study Cohort
Study approval was obtained from the University of Pittsburgh’s institutional review board (#20080139). A retrospective review of a prospectively maintained institutional database was performed for all consecutive patients with CRPM who underwent CRS-HIPEC between May 2001 and October 2020. Patients were excluded if data were missing for primary tumor location (right sided, left sided, or rectal) or specific genomic variables (RAS/BRAF mutational status and MMR/MSI status), or if they had concomitant CRLM. Patients with transverse colonic primary sites were included only if they could be classified as right- versus left-sided, on the basis of primary tumor location relative to the middle colic artery and using available information in the patients’ medical records. All patients received HIPEC with mitomycin C, 40 mg total, immediately following CRS using a closed-abdomen technique to maintain a target intraperitoneal tissue temperature of 41–42 °C. Patients are followed-up in clinic with routine surveillance cross-sectional imaging every 3–6 months up to 5 years postoperatively, and annually thereafter.
Data Collection and Definitions
Demographic variables included age, gender, race/ethnicity, admission weight, body mass index (BMI), and age-adjusted Charlson Comorbidity Index (AA-CCI). Clinicopathologic variables included histopathologic features [primary tumor location, WHO tumor differentiation/grade, lymph node (LN) stage of the primary tumor, mucinous versus non-mucinous features, presence of signet ring cells], synchronous versus metachronous CRPM presentation, time interval from primary tumor diagnosis until diagnosis of peritoneal metastases, and use/type of neoadjuvant/adjuvant systemic chemotherapy. Perioperative variables included peritoneal carcinomatosis index (PCI), completeness of cytoreduction score (CC score), and 90-days comprehensive complication index (90-days CCI), which was a composite of all complications occurring in hospital and up to 90-days post-discharge following CRS-HIPEC.
We abstracted data on the mutational status of KRAS, HRAS, NRAS, BRAF, PIK3CA, TP53, and SMAD4 by next-generation sequencing (NGS) performed at University of Pittsburgh Medical Center (UPMC) (icolon, oncomine panels using established in-house protocols) or at established outside facilities (FoundationOne) as previously reported.33 To assess MMR/MSI status, immunohistochemistry (IHC) for the mismatch repair proteins MLH1, MSH2, MSH6, and PMS2 proteins was performed if enough tumor tissue was available, and a tumor deficient in any of these proteins was categorized as dMMR/MSI-H. If tissue was insufficient for IHC, polymerase chain reaction (PCR) was utilized to detect instability of the microsatellite markers (BAT-25, BAT-26, D2S123, D5S346, D17S250), and dMMR/MSI-H status was defined as instability of two or more of these markers.
Statistical Analysis
Descriptive statistics of demographic, oncological, and perioperative variables were generated for all analyzed patients. Comparisons were performed by primary tumor site and by RAS mutation status. The Kruskal–Wallis test was used for continuous variable comparisons and the Pearson’s chi-squared test for categorical variables. Kaplan–Meier curves were utilized to compare survival differences (if present) by decade between our study sample and all patients with CRPM receiving CRS-HIPEC at our institution to assess study sample representativeness. The log-rank test detected differences between survival curves.
Variables were assessed for missing data. For those with data missing for ≥ 10% of the patients, the missing data were obtained retrospectively via electronic medical record, such that there was < 10% data missing per variable for all variables studied. Loss to follow-up was determined as the percentage of all patients in the study who were not known to be deceased and had not been seen in clinic for > 1 year. Their survival times were censored in the survival analyses, as was done for patients still alive and in active follow-up.
Univariable and multivariable survival analyses were performed using Cox proportional hazards models. For the multivariable analysis, we included all variables assessed in the univariable models, then performed backward elimination with the elimination criterion of p ≥ 0.05. In this algorithm, we successively removed the variable with the largest p-value, that is, ≥ 0.05, but did not eliminate genetic variables, given our primary focus on these variables. The outcome for the main survival analysis was overall survival, calculated from date of CRS-HIPEC. Each variable in the final model was checked for satisfaction of the proportional hazards (PH) assumption (i.e., hazard ratio is constant with time) using the Grambsch–Therneau test. Continuous variables were treated as continuous in the analyses, unless they did not meet the PH assumption, in which case they were binarily categorized (less than versus greater than the sample median) and the PH assumption reverified. For categorical variables not satisfying the PH assumption, we included an interaction term between the variable and survival time to allow for the calculation of a time-varying hazard ratio at multiple prespecified timepoints (1, 3, 6, 12, and 24 months post-CRS-HIPEC).
We performed a progression-free survival (PFS) analysis using a Cox proportional hazards model and the same backward elimination scheme as above. Further exploratory analyses included interactions between genomic alterations and primary tumor site, differences in genetic mutation distributions in patients with both CRLM and CRPM (compared with only CRPM), and the association of immune checkpoint inhibitors with survival in patients with dMMR/MSI-H status. Statistical significance level was set to α = 0.05. All analyses were performed using Stata software version 17 (StataCorp LP, College Station, TX, USA).
RESULTS
Between 2001 and 2020, 487 consecutive patients with CRPM underwent CRS-HIPEC (defined as the starting cohort). After excluding ineligible patients (Fig. 1), we analyzed data for 250 patients, for whom complete data was available for RAS/BRAF mutations and MMR/MSI status (defined as the study cohort). In the study cohort, missingness of patient demographic, clinicopathologic, perioperative, and oncologic outcome data were minimal (1.5%). Notably, routine molecular testing was less common during the time period 2001–2010 compared with 2011–2020 (58% versus 93% patients tested, respectively); therefore, a larger proportion of patients in the earlier time period was excluded from the study cohort. However, the selected study cohort appeared to be representative of the starting cohort given that the frequency of molecular alterations (RAS/BRAF mutations and dMMR/MSI-H status), distribution of primary tumor location (right-sided, left-sided, rectal cancers), and median OS were similar for the two cohorts (data not shown).
FIG. 1.
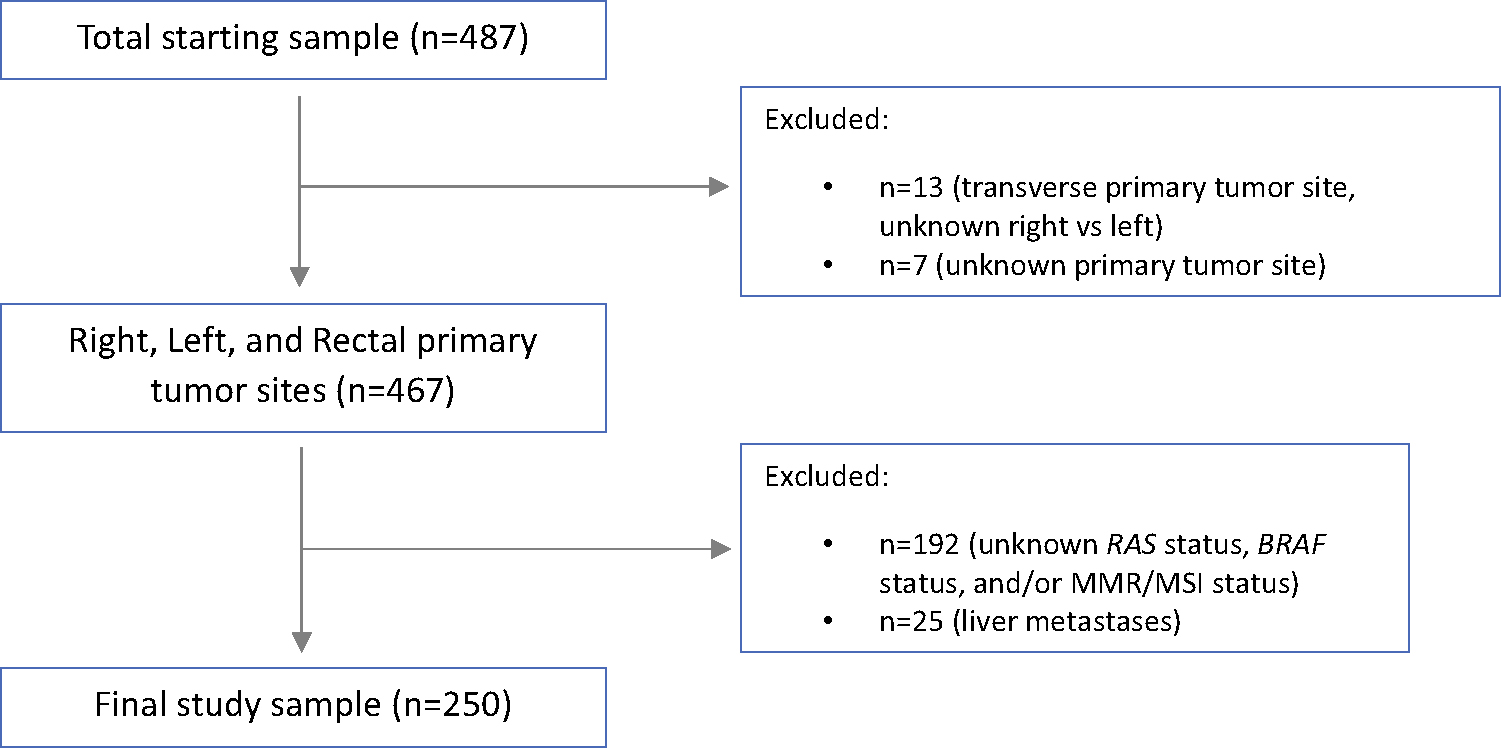
Study enrollment flowchart
The median age at time of CRS-HIPEC in the study cohort was 56.3 years and 48.4% were female. CRS-HIPEC was performed for right-sided cancers in 122 patients (48.8%), left-sided cancers in 91 patients (36.4%), and rectal cancers in 37 patients (14.8%). The median PCI was 12 (IQR 7–18) and CC-0 resection was achieved in 212 patients (84.8%). Most patients had moderately differentiated tumors by World Health Organization (WHO) criteria (well-differentiated/grade 1, 3.3%; moderately differentiated/grade 2, 71.8%; poorly differentiated/grade 3, 24.9%); 41.1% of patients had mucinous adenocarcinomas, and 14% had signet ring cell features/cancers. The incidence of genomic alterations in order of decreasing frequency was TP53 mutation (60.7%), RAS mutations (50.8%), SMAD4 mutation (24.7%), PIK3CA mutation (13.8%), BRAF mutation (12.4%), and dMMR/MSI-H status (6.8%) (Fig. 2).
FIG. 2.
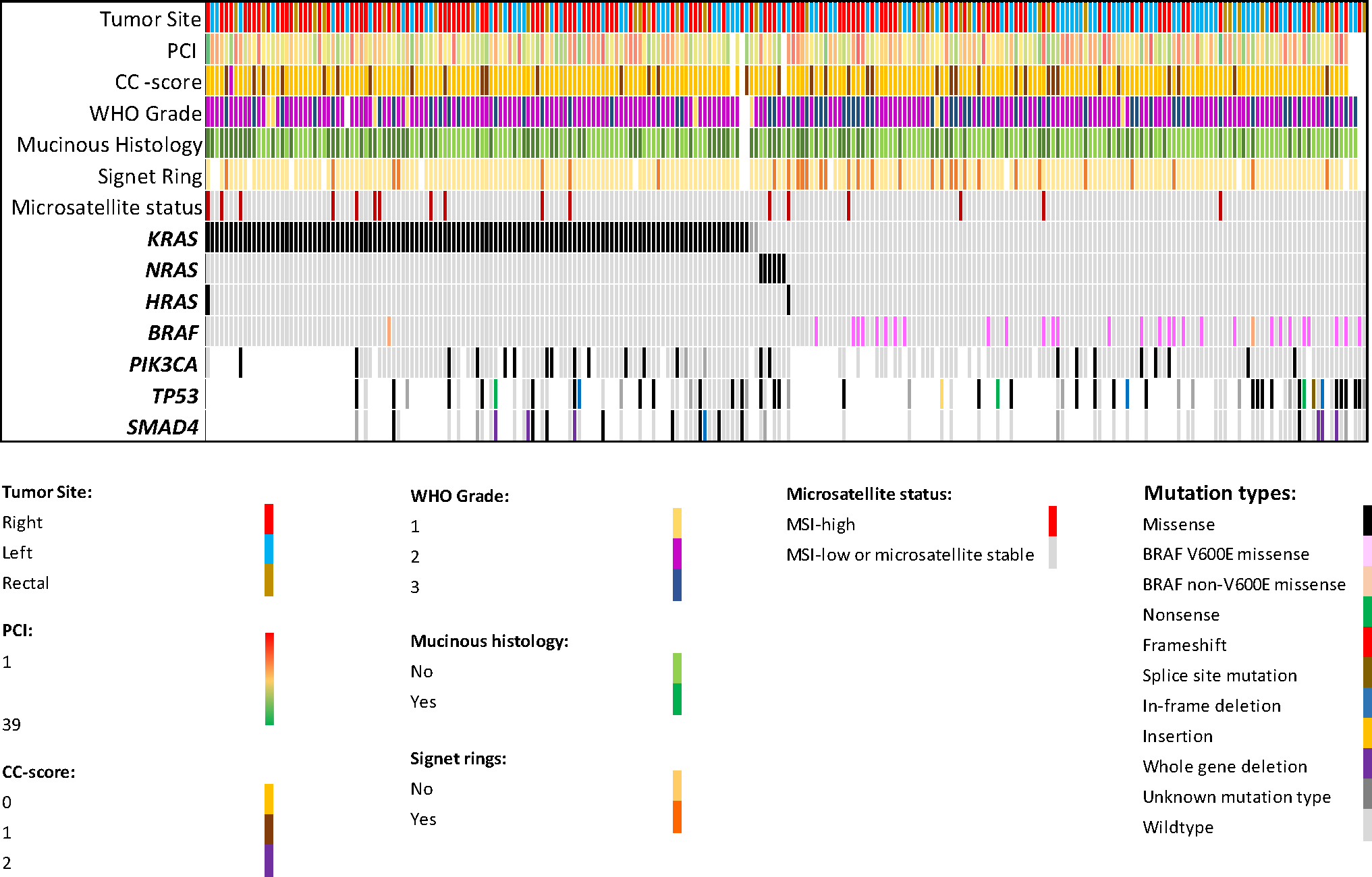
Distribution of key pathologic variables and genomic alterations in patients with CRPM who underwent CRS-HIPEC; (WHO grade: pathologic grade, CC-score: cytoreduction completeness score)
Associations Between Tumor Sidedness and Clinicopathologic Features/Molecular Alterations
Patients with right-sided cancers were significantly more likely to harbor RAS mutations (59% versus 40.7% versus 48.7%; p = 0.03) and BRAF mutations (18% versus 9.9% versus 0%; p = 0.005), compared with those with left-sided and rectal cancers. Notably, none of the patients with rectal primaries had BRAF mutations. There was a trend toward proportionately greater dMMR/MSI-H status and LN involvement in right-sided cancers, compared with left-sided and rectal cancers. There was a trend toward a higher frequency of poorly differentiated tumors, signet ring cell features/cancers, LVI, PNI, and mucinous cancers in right-sided and rectal cancers, compared with left-sided cancers. There was no difference in PCI, CC-score, receipt of neoadjuvant/adjuvant chemotherapy (most often FOLFOX or FOLFIRI ± biological agents), or 90-days CCI when stratified by primary tumor location (Table 1).
TABLE 1.
Baseline, pathologic, oncologic, and perioperative factors in patients with CRPM who underwent CRS-HIPEC, stratified by primary tumor site
| Variable | All patients | Right | Left | Rectal | p Value# |
|---|---|---|---|---|---|
|
| |||||
| Number of patients | 250 | 122 | 91 | 37 | |
| Age at HIPEC, median (IQR) | 56.3 (15.7) | 57.4 (15.4) | 56.0 (14.2) | 52.4 (16.0) | 0.55 |
| Gender, male, n (%) | 129 (51.8) | 67 (55.4) | 41 (45) | 21 (56.8) | 0.27 |
| Race, Caucasian, n (%) | 230 (94.3) | 110 (94) | 87 (95.6) | 33 (91.7) | 0.65 |
| BMI, preop, kg/m2, median (IQR) | 27.8 (7.7) | 28.4 (8.3) | 26.9 (7.9) | 27.5 (5.6) | 0.5 |
| AA-CCI, median (IQR) | 9 (2) | 9 (2) | 8 (2) | 8 (2) | 0.2 |
| Primary tumor grade, poorly differentiated, n (%) | 61 (24.9) | 34 (28.3) | 16 (18) | 11 (30.6) | 0.16 |
| Primary tumor N-stage, n (%) | 0.28 | ||||
| N0 | 62 (25.7) | 24 (20.3) | 26 (29.6) | 12 (34.3) | |
| N1 | 83 (34.4) | 40 (33.9) | 32 (36.4) | 11 (31.4) | |
| N2 | 96 (39.8) | 54 (45.8) | 30 (34.1) | 12 (34.3) | |
| LVI, n (%) | 147 (60.3) | 71 (60.2) | 51 (57.3) | 25 (67.6) | 0.56 |
| PNI, n (%) | 104 (41.8) | 52 (43) | 35 (38.5) | 17 (46) | 0.69 |
| Mucinous, n (%) | 101 (41.1) | 52 (43.3) | 32 (35.6) | 17 (47.2) | 0.38 |
| Signet rings, n (%) | 35 (14) | 20 (16.4) | 7 (7.7) | 8 (21.6) | 0.07 |
| Months between primary tumor and PC, median (IQR) | 0.85 (14.3) | 0.94 (11.9) | 0.53 (15.0) | 3.63 (16.5) | 0.47 |
| Neoadjuvant chemotherapy within 3 months before HIPEC, n (%) | 177 (74.7) | 89 (78.1) | 59 (67.1) | 29 (82.9) | 0.1 |
| PCI, median (IQR) | 12 (11) | 12 (11) | 12 (11.5) | 12 (7.5) | 0.7 |
| Cytoreduction completeness score > 0, n (%) | 38 (15.6) | 20 (16.8) | 14 (15.9) | 4 (11.1) | 0.71 |
| 90-days CCI, median (IQR) | 30.8 (26.2) | 30.8 (28.5) | 30.8 (23.9) | 32.0 (22.6) | 0.47 |
| Adjuvant chemotherapy after HIPEC, n (%) | 99 (40.7) | 49 (41.5) | 34 (38.6) | 16 (43.2) | 0.87 |
| RAS, mutated/lost, n (%) | 127 (50.8) | 72 (59) | 37 (40.7) | 18 (48.7) | 0.03 |
| BRAF, mutated/lost, n (%) | 31 (12.4) | 22 (18) | 9 (9.9) | 0 (0) | 0.005 |
| dMMR/MSI-H, n (%) | 17 (6.8) | 12 (9.8) | 4 (4.4) | 1 (2.7) | 0.26 |
| PIK3CA, mutated/lost, n (%)* | 25 (13.8) | 8 (9.6) | 14 (19.7) | 3 (11.1) | 0.20 |
| TP53, mutated/lost, n (%)* | 51 (60.7) | 21 (65.6) | 23 (59) | 7 (53.9) | 0.73 |
| SMAD4, mutated/lost, n (%)* | 21 (24.7) | 10 (30.3) | 7 (18) | 4 (30.8) | 0.41 |
Bolded variables are statistically significant (p < 0.05)
p Value for comparison of the three primary tumor locations (right, left, and rectal primary tumors)
Data from patients who were tested for the gene (181 patients tested for PIK3CA, 84 for SMAD4, and 85 for TP53)
IQR interquartile range, BMI body mass index, AA-CCI age-adjusted Charlson Comorbidity Index, LVI lymphovascular invasion, PNI perineural invasion, PC peritoneal carcinomatosis, 90-days CCI cumulative complication index during hospital stay and up to 90 days postoperatively, PCI peritoneal carcinomatosis index
Associations Between Molecular Alterations and Clinicopathologic Features
RAS mutations were more common in right-sided cancers than left-sided or rectal cancers (56.7% versus 29.1% versus 14.2%, p = 0.03). RAS-mutant cancers were also more likely to be associated with mucinous histology (48% versus 33.9%; p = 0.02), dMMR/MSI-H status (10.2% versus 3.3%; p = 0.03), PIK3CA mutations (19.5% versus 8.5%; p = 0.03), and TP53 mutations (33.3% versus 15%; p = 0.05), while they were less likely to be associated with poorly differentiated histology (17.7% versus 32.2%; p = 0.01), signet-ring cell features/cancers (7.9% versus 20.3%; p = 0.01), BRAF mutations (0.79% versus 24.4%; p < 0.001), and SMAD4 mutations (48.9% versus 74.4%; p = 0.02), compared with RAS wildtype cancers. Notably, none of the patients with RAS-mutated rectal cancers had dMMR/MSI-H status (Table 2). For RAS-wildtype cancers, 68 of 123 (55%) were on regimens involving anti-EGFR agents (cetuximab or panitumumab) prior to or after CRS-HIPEC.
TABLE 2.
Baseline, pathologic, oncologic, and perioperative factors in patients with CRPM who underwent CRS-HIPEC, stratified by RAS mutation status
| Variable | All patients | RAS-mutated | RAS-wildtype | p ValueƗ |
|---|---|---|---|---|
|
| ||||
| Number of patients | 250 | 127 | 123 | |
| Age at HIPEC, median (IQR) | 56.3 (15.7) | 55.9 (16.4) | 56.5 (15.2) | 0.56 |
| Gender, male, n (%) | 51.8 | 52.4 | 51.2 | 0.86 |
| Race, Caucasian, n (%) | 94.3 | 92.7 | 95.9 | 0.29 |
| BMI, preop, kg/m2, median (IQR) | 27.8 (7.7) | 27.6 (7.1) | 27.8 (9.4) | 0.56 |
| AA-CCI, median (IQR) | 9 (2) | 9 (2) | 9 (2) | 0.82 |
| Primary tumor site (% of all patients) | 0.03 | |||
| Right | 48.8 | 56.7 | 40.7 | |
| Left | 36.4 | 29.1 | 43.9 | |
| Rectal | 14.8 | 14.2 | 15.5 | |
| Primary tumor grade, poorly differentiated, n (%) | 24.9 | 17.7 | 32.2 | 0.01 |
| Primary tumor N-stage, n (%) | 0.5 | |||
| N0 | 25.7 | 28.7 | 22.7 | |
| N1 | 34.4 | 34.4 | 34.5 | |
| N2 | 39.8 | 36.9 | 42.9 | |
| LVI, n (%) | 60.2 | 58.1 | 62.5 | 0.48 |
| PNI, n (%) | 41.8 | 40.2 | 43.4 | 0.6 |
| Mucinous, n (%) | 41.1 | 48 | 33.9 | 0.02 |
| Signet rings, n (%) | 14 | 7.9 | 20.3 | 0.01 |
| Months between primary tumor and PC, median (IQR) | 0.85 (14.3) | 0.76 (13.2) | 0.89 (15.4) | 0.56 |
| Neoadjuvant chemotherapy within 3 months before HIPEC, n (%) | 74.7 | 73.6 | 75.9 | 0.68 |
| PCI, median (IQR) | 12 (11) | 12.5 (10) | 11 (12) | 0.39 |
| Cytoreduction completeness score > 0, n (%) | 15.6 | 14.5 | 16.8 | 0.62 |
| 90-days CCI, median (IQR) | 30.8 (26.3) | 29.6 (24.6) | 32 (28.5) | 0.23 |
| Adjuvant chemotherapy after HIPEC, n (%) | 40.7 | 38.8 | 42.6 | 0.55 |
| BRAF, mutated/lost, n (%) | 12.4 | 0.79 | 24.4 | < 0.001 |
| dMMR/MSI-H, n (%) | 6.8 | 10.2 | 3.3 | 0.03 |
| PIK3CA, mutated/lost, n (%)* | 13.8 | 19.5 | 8.5 | 0.03 |
| TP53, mutated/lost, n (%)* | 24.7 | 33.3 | 15 | 0.05 |
| SMAD4, mutated/lost, n (%)* | 60.7 | 48.9 | 74.4 | 0.02 |
Bolded variables are statistically significant (p < 0.05)
p-Value for comparison by RAS mutational status (RAS-mutated and RAS-wildtype)
Data from patients who were tested for the gene (181 patients tested for PIK3CA, 84 for SMAD4, and 85 for TP53)
IQR interquartile range, BMI body mass index, AA-CCI age-adjusted Charlson Comorbidity Index, LVI lymphovascular invasion, PNI perineural invasion, PC peritoneal carcinomatosis, 90-days CCI cumulative complication index during hospital stay and up to 90 days postoperatively, PCI peritoneal carcinomatosis index
Most BRAF mutations were of the V600E variant (29 out of 31 patients). One patient with a non-V600E BRAF mutation had a concurrent KRAS mutation. BRAF-mutated tumors were rarely associated with dMMR/MSI-H disease status (one patient with a right-sided cancer). Only 2 of 29 BRAF V600E-mutated cases were treated with BRAF inhibitors.
Compared with pMMR/MSS/MSI-L tumors, tumors with dMMR/MSI-H status were more likely to be right-sided than from other sites (70.6% versus 47.2%; p = 0.06) and demonstrated poorly differentiated histology (52.9% versus 22.8%; p = 0.02), signet ring cell features/cancers (29.4% versus 12.9%; p = 0.07), mucinous histology (70.6% versus 38.9%; p = 0.01), and concurrent RAS mutations (76.5% versus 48.9%; p = 0.03). dMMR/MSI-H tumors were rarely located in the rectum (one patient) and generally lacked BRAF mutations (one patient had a non-V600E mutation). Of the 17 patients with dMMR/MSI-H cancers, 4 were treated with immune checkpoint inhibitors within the first 5 years after CRS-HIPEC. All patients with dMMR/MSI-H cancers received similar neoadjuvant/adjuvant chemotherapy as those with pMMR/MSS/MSI-L cancers (FOLFOX/FOLFIRI-based regimen).
Poorly differentiated tumors were less likely to be RAS-mutated (36% versus 55%; p = 0.009), equally likely to be BRAF-mutated (11.5% versus 12.5%; p = 0.8), and more likely to be dMMR/MSI-H (14.8% versus 4.3%; p = 0.02) compared with well–moderately differentiated tumors. Within both the poorly and well–moderately differentiated groups, > 50% of each of the RAS-mutated, BRAF-mutated, and dMMR/MSI-H subgroups consisted of right-sided cases.
Survival Analysis and Prognostic Significance of Tumor Sidedness and Molecular Alterations
The median reverse Kaplan–Meier estimated follow-up duration and 95% CI for the study cohort was 51.6 months (44.8–55.8), with 11.2% of patients lost to follow-up. The median OS and PFS, calculated from CRS-HIPEC, were 28.9 months (24.9–37.8) and 9.7 months (8.2–11.8), respectively. The 3- and 5-years OS were 43.9% and 18.1%, respectively. The 3- and 5-years PFS were 11.5% and 7.8%, respectively.
In a univariate Cox proportional hazards model, significant predictors for OS included tumor sidedness, LN status, PCI, CC score, adjuvant systemic chemotherapy, 90-days CCI, and MMR/MSI status (Table 3). Notably, RAS or BRAF mutations did not affect OS on a univariate basis. However, further exploratory crude univariate comparisons demonstrated that for the subgroup of patients with left-sided colon cancers, the presence of either RAS or BRAF mutations predicted a worse OS (HR 1.97; p = 0.02); for patients with either RAS or BRAF mutations, dMMR/MSI-H status predicted a better OS (HR 0.3; p = 0.01), and combined mutations of BRAF/PIK3CA predicted worse OS (HR 4.9; p = 0.02) (Fig. 3).
TABLE 3.
Overall survival analysis of oncologic, perioperative, and genomic variables in patients with CRPM post-HIPEC
| Variable | Univariate |
Multivariate (n = 231 patients) |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p Value | HR | 95% CI | p Value | |
|
| ||||||
| Age, years (each additional year) | 1.01 | 0.99–1.02 | 0.22 | |||
| Male | 1.18 | 0.86–1.62 | 0.3 | |||
| BMI, preop (kg/m2) (each additional point) | 0.98 | 0.95–1.0 | 0.07 | |||
| AA-CCI (each additional point) | 1.04 | 0.95–1.14 | 0.42 | |||
| Primary tumor site | ||||||
| Left | Reference | Reference | ||||
| Right | 1.11 | 0.78–1.58 | 0.58 | 1.01 | 0.68–1.49 | 0.96 |
| Rectal | 1.69 | 1.06–2.71 | 0.03 | 1.93 | 1.17–3.17 | 0.01 |
| Poorly differentiated (grade 3, versus grade 1–2) | 1.21 | 0.84–1.75 | 0.31 | |||
| Primary tumor N-stage | ||||||
| N0 | Reference | Reference | ||||
| N1 | 1.2 | 0.78–1.86 | 0.41 | 1.24 | 0.78–1.97 | 0.37 |
| N2 | 1.83 | 1.21–2.75 | 0.004 | 1.78 | 1.14–2.78 | 0.01 |
| LVI | 1.14 | 0.82–1.58 | 0.42 | |||
| PNI | 1.31 | 0.95–18 | 0.1 | |||
| Mucinous | 0.92 | 0.67–1.28 | 0.64 | |||
| Signet rings | 1.5 | 0.97–2.31 | 0.07 | 1.82 | 1.11–3.0 | 0.02 |
| Months from primary tumor to PC (each additional month) | 1 | 0.99–1.01 | 0.84 | |||
| HIPEC neoadjuvant chemo (within 3 months of HIPEC) | 1.04 | 0.72–1.5 | 0.84 | |||
| PCI (each additional point) | 1.05 | 1.03–1.07 | < 0.001 | 1.03 | 1.003–1.05 | 0.03 |
| Cytoreduction completeness score > 0 | 2.79 | 1.9–4.11 | < 0.001 | 2.06 | 1.32–3.22 | 0.002 |
| HIPEC adjuvant chemo | 0.64 | 0.46–0.90 | 0.009 | 0.58 | 0.41–0.83 | 0.003 |
| 90-days CCI (> median; reference, low) | 2.35 | 1.7–3.24 | < 0.001 | 2.13 | 1.49–3.04 | < 0.001 |
| RAS mutation | 1.04 | 0.76–1.42 | 0.81 | 1.55 | 1.04–2.3 | 0.03 |
| BRAF mutation | 1.21 | 0.76–1.94 | 0.43 | 1.73 | 0.99–2.99 | 0.05 |
| dMMR/MSI-H (overall) | 0.4 | 0.18–0.9 | 0.03 | 0.35 | 0.15–0.82 | 0.02 |
| At specific timepoints: | ||||||
| 1 month | 1.14 | 0.23–5.67 | 0.87 | 1.39 | 0.29–6.62 | 0.68 |
| 3 months | 0.74 | 0.25–2.26 | 0.6 | 0.81 | 0.27–2.45 | 0.7 |
| 6 months | 0.57 | 0.23–1.38 | 0.21 | 0.57 | 0.23–1.43 | 0.23 |
| 12 months | 0.43 | 0.19–0.98 | 0.045 | 0.4 | 0.17–0.95 | 0.04 |
| 24 months | 0.33 | 0.13–0.83 | 0.02 | 0.29 | 0.11–0.74 | 0.01 |
Bolded values indicate statistical significance (p < 0.05)
Median 90-days CCI, 30.82 (in entire study sample). For dMMR/MSI-H, the HR was time varying; an overall HR (without time interaction) is provided in addition to time-specific HRs
HR hazard ratio, CI confidence interval, BMI body mass index, AA-CCI age-adjusted Charlson Comorbidity Index, LVI lymphovascular invasion, PNI perineural invasion, PC peritoneal carcinomatosis, PCI peritoneal cancer index, 90-days CCI cumulative complication index during hospital stay and up to 90 days postoperatively
FIG. 3.
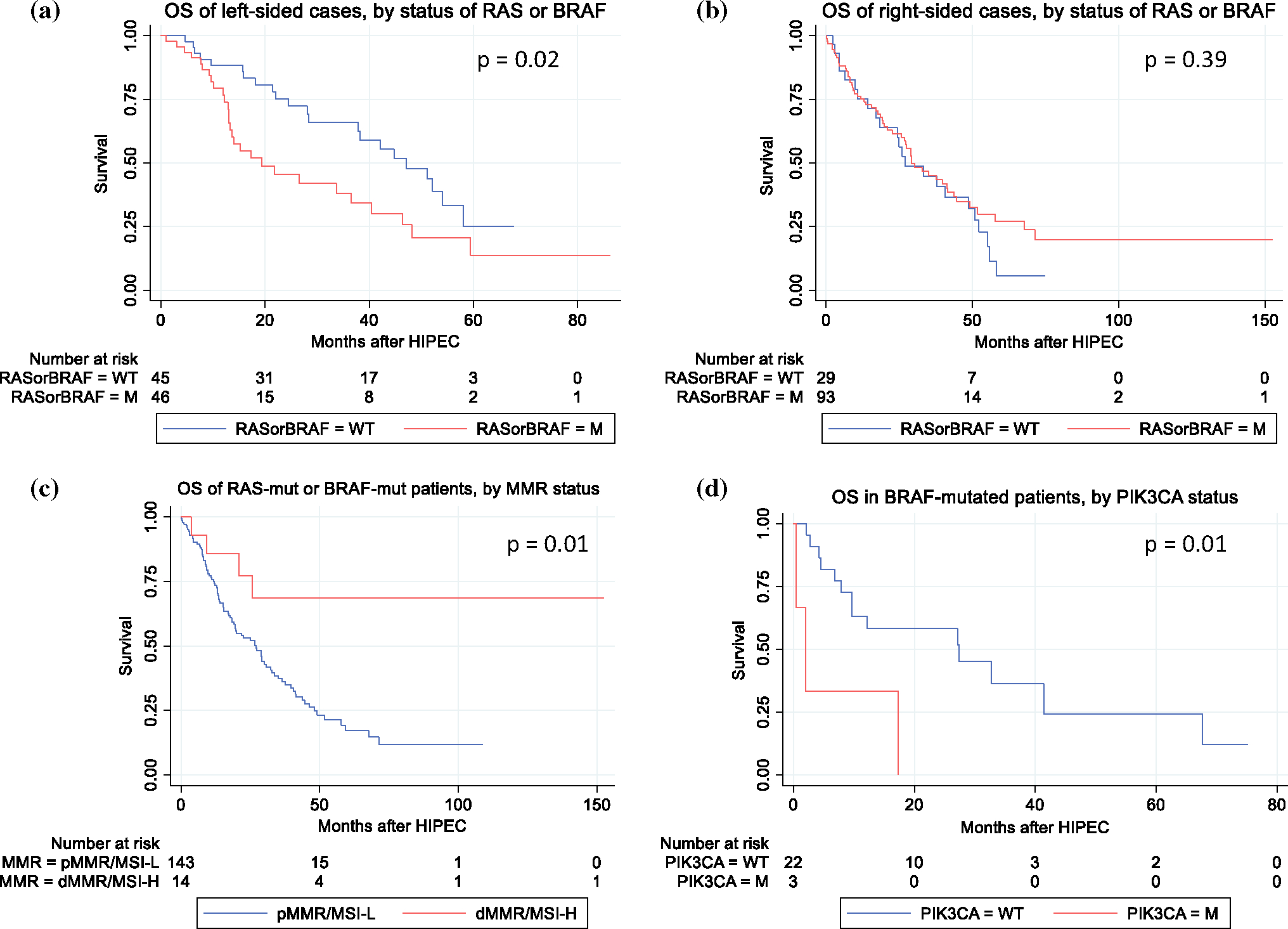
Exploratory associations of primary tumor site, RAS mutation, BRAF mutation, MMR/MSI status, and PIK3CA mutation with survival after CRS-HIPEC; associations of primary tumor site, RAS mutation, and BRAF mutation with survival (a, b). Association of BRAF mutation and MMR/MSI status with survival (c); association of BRAF and PIK3CA mutations with survival (d); p-Value: logrank test p-value; OS overall survival, WT wildtype, M/mut mutated; the mutant category in a, b includes cases with either RAS mutation or BRAF mutation and the wildtype category includes cases that were wildtype for both genes
In a multivariate Cox proportional hazards model, rectal primary (HR 1.9), N2 disease (HR 1.8), signet ring cell features (HR 1.8), higher PCI (HR 1.03 per additional point), non-CC0 resection (HR 2.1), lack of adjuvant systemic chemotherapy (HR 1.7), higher 90-days CCI (HR 2.1), RAS mutation (HR 1.6), and BRAF mutation (HR 1.7) were significant independent prognostic factors for worse OS. In addition, dMMR/MSI-H status had a significant independent time-varying effect, with reduction in mortality risk after 1 year following CRS-HIPEC (HR at 1 year 0.4, HR at 2 years 0.3) (Table 3). In the study cohort, 11 of 17 dMMR/MSI-H cancer patients were still alive at 2 years post-CRS-HIPEC, only 1 of whom had been lost to follow-up. Of the 17 dMMR/MSI-H patients, 4 received immune checkpoint inhibitors within the first 5 years after CRS-HIPEC. There was a trend toward improved overall survival in the dMMR/MSI-H patients receiving immunotherapy (data not shown).
N2 disease (HR 1.5) and higher PCI (HR 1.04 per additional point) were significant independent prognostic factors for worse PFS on multivariate analysis. In addition, RAS mutation had a significant independent time-varying effect, indicating shorter PFS within the first 6 months following CRS-HIPEC (HR at 6 months 1.6) (Table 4).
TABLE 4.
Multivariate progression-free survival analysis in CRPM patients post-HIPEC (n=237 patients)
| Variable | HR | 95% CI | p |
|---|---|---|---|
|
| |||
| Primary tumor N-statge | |||
| N0 | Reference | ||
| N1 | 0.95 | 0.63–1.41 | 0.79 |
| N2 | 1.46 | 1.0–2.13 | 0.05 |
| PCI (for each additional point) | 1.04 | 1.02–1.06 | < 0.001 |
| RAS mutation (overall) | 1.36 | 0.99–1.87 | 0.06 |
| At specific timepoints: | |||
| 1 month | 3.26 | 1.32–8.05 | 0.01 |
| 3 months | 2.08 | 1.23–3.51 | 0.006 |
| 6 months | 1.56 | 1.1–2.23 | 0.01 |
| 12 months | 1.17 | 0.82–1.68 | 0.38 |
| 24 months | 0.88 | 0.52–1.5 | 0.65 |
| BRAF mutation | 1.03 | 0.62–1.71 | 0.9 |
| dMMR/MSI-H | 1 | 0.55–1.81 | 1 |
For RAS, the HR was time varying; an overall HR (without time interaction) is provided in addition to time-specific HRs. Bolded variables are statistically significant (p < 0.05).
HR hazard ratio, CI confidence interval, PCI peritoneal cancer index
Subgroup Analysis of Patients with Combined Peritoneal and Liver Metastases (CRPM/LM)
We performed an exploratory analysis after adding in the excluded combined CRPM/LM cohort (n = 25) with our CRPM-only study cohort (n = 250). The frequency of molecular alterations (RAS/BRAF mutations, and dMMR/MSI-H status) and the distribution of tumor location (right-sided, left-sided, rectal cancers) were similar for the CRPM/LM and the CRPM-only cohorts (data not shown). The median OS/PFS and 5-years OS/PFS were similar for patients with CRPM-only and CRPM/LM cohorts. We repeated the multivariable Cox proportional hazards analysis on the aggregated CRPM + CRPM/LM cohorts and found that the presence of liver metastases was not an independent prognostic factor for OS (data not shown). Crude univariate comparisons suggested a trend toward worse OS after CRS-HIPEC in patients with concurrent liver metastases and BRAF mutation (data not shown).
DISCUSSION
The aim of this study was to examine unique effects of primary tumor site and genomic alterations on OS in patients who undergo CRS-HIPEC for CRPM. The prevalence of genomic alterations in our post-CRS-HIPEC cohort is consistent with population-level findings in metastatic CRC, specifically that 30–50% are RAS-mutated, 8–20% are BRAF-mutated, and 3–5% are dMMR/MSI-H.10,34,35
We found that RAS mutations are associated with a worse OS and PFS after CRS-HIPEC when adjusted for clinical/pathological factors related to the cancer, the patient’s comorbidities, postoperative complications, and receipt of adjuvant post-CRS-HIPEC chemotherapy. We additionally found that BRAF mutations predict worse OS. dMMR/MSI-H status resulted in a time-varying effect of improved OS over time after CRS-HIPEC. Finally, after adjusting for other variables, colonic right-sidedness did not seem to worsen mortality over left-sided tumors, as suggested in some prior studies, while rectal tumors uniquely had a worse survival. To our knowledge, our study is the first to isolate the independent association of rectal primary site with poorer survival after CRS-HIPEC for CRPM. With only 37 of 250 cases being of rectal origin, future replication of this association is crucial. Nevertheless, our study presents the largest cohort of rectal peritoneal metastases analyzed post-CRS-HIPEC to date. Reasons for correlation with worse survival may include a (albeit non-significantly) higher proportion of cases with signet ring features among rectal cases (21.6%). Other reasons include possible gene expression differences between peritoneal metastases of colonic versus rectal origin, as some studies have shown for non-metastatic disease.36 Of note, the duration between primary tumor diagnosis and peritoneal carcinomatosis, duration between peritoneal carcinomatosis diagnosis and CRS-HIPEC, type of chemotherapy prior to CRS-HIPEC, and response to this chemotherapy (based on imaging, symptoms, and/or serum CEA) did not differ significantly between cases of rectal versus right-sided colonic origin. Our findings related to the rectal cases are in line with prior studies in which stage II rectal cancer was associated with worse prognosis overall than stage II colon cancer,17 and in which liver metastases from rectal cancer are prognostically worse than those from colon cancer.18 In the case of peritoneal metastases, recent studies have yet to elucidate a prognostic difference between rectal and colonic origin due either to exclusion of rectal cases, or to combining the rectal cases with left-sided colonic cases (likely a consequence of the limited number of rectal cases).14–16 The interaction of tumor site and key molecular alterations in CRPM also remains to be explored and was a key aim of our study. Our finding that rectal peritoneal metastases may portend worse survival adds information to the patient selection process and prognostication for CRS-HIPEC, particularly for rectal cases with multiple other negative prognosticators.
Our findings of worsened OS secondary to RAS and BRAF mutations are consistent with a prior study by Schneider et al.,37 in which worse survival is associated with BRAF mutation compared with RAS mutation (higher HR for BRAF mutation). Though the effect of BRAF was only marginally significant in our study, this may be due to the slightly different patient population (our sample had a median PCI of 12, compared with 7 in the study by Schneider et al.), differences in survival measurement (overall survival versus cancer-specific survival), and the relatively small number of BRAF-mutant patients who undergo CRS-HIPEC.37
Our finding of dMMR/MSI-H status being independently associated with improved OS comes amid controversy over the prognostic impact of dMMR/MSI-H status in recent studies of metastatic CRC. Metastatic CRC is uncommonly dMMR/MSI-H (3–5% of cases), thought to be due to the reduced metastatic potential of stage I-III dMMR/MSI-H colorectal tumors.27,35,38 In contrast to the known superior survival in patients with non-metastatic CRC with dMMR/MSI-H tumors,20,21 multiple studies of patients with extraperitoneal metastatic CRC (a majority with hepatic metastases) revealed poorer survival with dMMR/MSI-H status.27,38 An earlier pooled analysis of four large chemotherapeutic trials for patients with metastatic CRC also demonstrated a detrimental survival effect of dMMR/MSI-H status.35 For specifically CRPM, however, the relevance of MMR/MSI status is less clear. A large analysis of the NCDB of colon, rectal, and appendiceal metastatic cancers suggested worse OS with dMMR/MSI-H status (hazard ratio 1.05).39 However, this national cohort differs from ours in that not all received surgical treatment, and those with extraperitoneal metastases were included. A recent study with a more defined cohort from a high-volume Norwegian center found that patients with both dMMR/MSI-H status and BRAF mutation had the best overall survival.40 To our knowledge, our study is the second one to suggest a superior survival association with dMMR/MSI-H status in those undergoing CRS-HIPEC for CRPM. This benefit was seen even though all dMMR/MSI-H patients in the Norwegian study and most (13/17) dMMR/MSI-H patients in our study had not received immune checkpoint inhibitors due to the relatively recent approval of these drugs for this condition (2017 in the USA and 2019 in Norway).40,41 It is less likely that patient age or chemotherapy choice explain the survival difference between dMMR/MSI-H and pMMR/MSS/MSI-L patients in our cohort, given the statistically similar distributions of these two variables in both subgroups. The survival benefit may be due to HIPEC-induced activation of immune cells in the tumor microenvironment in dMMR/MSI-H cancers. This effect is plausible based on recent studies but requires further confirmation.42 Other studies have similarly documented a benefit of dMMR/MSI-H status in the broader cohort of post-surgical metastatic CRC patients, such as a recent pooled analysis of seven adjuvant chemotherapy trials involving patients with resected stage III colon cancer, which showed that among patients who developed recurrent disease, dMMR/MSI-H status was independently associated with superior survival after recurrence.21 In a retrospective study of Japanese patients with metastatic CRC and a previously resected primary tumor, dMMR/MSI-H status was also associated with a survival benefit specifically in the subgroup with peritoneal metastases.27 The findings of these studies and our study are especially relevant in the era of immune checkpoint inhibitors. These drugs have a favorable disease-specific response in patients with metastatic CRC overall,43,44 but it remains to be seen if this survival benefit will be replicated in the post-CRS-HIPEC cohort. Our study suggests that post-CRS-HIPEC patients with CRPM may benefit from utilization of immune checkpoint inhibitors.
In terms of interaction between primary tumor site and genomics, our study uniquely suggests for patients with CRPM that RAS or BRAF mutations are more strongly associated with worse survival for left-sided tumors than for right-sided tumors. This is potentially due to the higher mutational burden of right-sided tumors, in which unassessed genes also influence survival.45 Though it has previously been observed that right-sided tumors have distinct gene expression profiles (e.g., methylation patterns) compared with the rest of the colorectum,46 and that distinct gene expression patterns of CRPM tissues are associated with survival,8 further prospective studies are needed to validate correlations between CRPM gene expression patterns and survival after CRS-HIPEC, and to stratify these correlations by site of the primary tumor, relationships our study begins to probe. We additionally observed trends toward worsened OS for patients with the combination of BRAF and PIK3CA mutations, or with the combination of BRAF mutation and liver metastases. Having either of these combinations may translate to lower candidacy for CRS-HIPEC, especially if other negative prognosticators are present.
Our study possesses several limitations. It is retrospective and susceptible to patient selection biases. However, there were no significant differences in survival, by decade, between patients included in our study and all patients with CRPM who received CRS-HIPEC at our institution. In addition, the distributions of RAS status, BRAF status, MMR/MSI status, and primary tumor site were similar between enrolled patients and all patients operated on at our institution. Finally, 80% of cases performed in 2011–2020 were included in the study, limiting the amount of selection bias. Another limitation is that the sample size was not large enough to investigate survival associations for TP53, PIK3CA, or SMAD4 mutations, which have been associated with higher mortality in less advanced CRC cases.47–50 Future multi-institutional studies are needed to understand the impact of these mutations in patients with CRPM after CRS-HIPEC.
In summary, for patients receiving CRS-HIPEC for CRPM, we demonstrated that mutated RAS or BRAF are associated with poorer postoperative survival, while dMMR/MSI-H status is associated with superior survival. These effects may be more pronounced in cases arising from left-sided tumors. Therefore, our study adds impetus for future prospective assessment of other, less-understood tumor genomic alterations and how they differ between patients who have favorable postoperative survival and those who do not, especially among the right-sided cases that form the majority of CRPM cases. RAS mutations are also associated with shorter PFS. Finally, cases with rectal primary tumors have worse overall survival compared with colonic primary tumors, which may be due to understudied genomic differences between rectal peritoneal metastases and their colonic counterparts. The key novel information highlighted in this study includes the positive prognostic association of dMMR/MSI-H status, the interplay between tumor sidedness and genomic alterations in relation to postoperative survival, and the newly identified worse prognosis related to rectal peritoneal metastases (compared with colonic cases). These findings add molecular and oncologic information useful in the selection/prognostication of patients undergoing CRS-HIPEC for CRPM. Future studies should assess the impact of immunotherapy for dMMR/MSI-H patients; mutations in TP53, PIK3CA, and SMAD4; and high-dimensional gene expression patterns in relation to survival after CRS-HIPEC.
Footnotes
DISCLOSURE None.
REFERENCES
- 1.Franko J, Shi Q, Meyers JP, Maughan TS, Adams RA, Seymour MT, Saltz L, Punt CJA, Koopman M, Tournigand C, Tebbutt NC, Diaz-Rubio E, Souglakos J, et al. Prognosis of patients with peritoneal metastatic colorectal cancer given systemic therapy: an analysis of individual patient data from prospective randomised trials from the analysis and research in cancers of the digestive system (ARCAD) database. Lancet Oncol. 2016;17(12):1709–19. [DOI] [PubMed] [Google Scholar]
- 2.Quenet F, Elias D, Roca L, Goere D, Ghouti L, Pocard M, Facy O, Arvieux C, Lorimier G, Pezet D, Marchal F, Loi V, Meeus P, et al. Cytoreductive surgery plus hyperthermic intraperitoneal chemotherapy versus cytoreductive surgery alone for colorectal peritoneal metastases (PRODIGE 7): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2021;22(2):256–66. [DOI] [PubMed] [Google Scholar]
- 3.Verwaal VJ, van Ruth S, de Bree E, van Sloothen GW, van Tin-teren H, Boot H, Zoetmulder FA. Randomized trial of cytoreduction and hyperthermic intraperitoneal chemotherapy versus systemic chemotherapy and palliative surgery in patients with peritoneal carcinomatosis of colorectal cancer. J Clin Oncol. 2003;21(20):3737–43. [DOI] [PubMed] [Google Scholar]
- 4.Goodman MD, McPartland S, Detelich D, Saif MW. Chemo-therapy for intraperitoneal use: a review of hyperthermic intraperitoneal chemotherapy and early post-operative intraperitoneal chemotherapy. J Gastrointest Oncol. 2016;7(1):45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mistry P, Mohamed F, Dayal S, Cecil TD, Moran BJ. Cytoreductive surgery with intraperitoneal chemotherapy in the management of peritoneal surface malignancy: a pharmacist’s perspective. Eur J Hosp Pharm. 2016;23(4):233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steuperaert M, Debbaut C, Segers P, Ceelen W. Modelling drug transport during intraperitoneal chemotherapy. Pleura Peritoneum. 2017;2(2):73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simkens GA, Wintjens A, Rovers KP, Nienhuijs SW, de Hingh IH. effective strategies to predict survival of colorectal peritoneal metastases patients eligible for cytoreductive surgery and HIPEC. Cancer Manag Res. 2021;13:5239–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lenos KJ, Bach S, Ferreira Moreno L, et al. Molecular characterization of colorectal cancer related peritoneal metastatic disease. Nat Commun. 2022;13:4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong XD. Right sided colon cancer and peritoneal carcinomatosis. Ann Laparosc Endosc Surg. 2019;4:72. [Google Scholar]
- 10.Bylsma LC, Gillezeau C, Garawin TA, et al. Prevalence of RAS and BRAF mutations in metastatic colorectal cancer patients by tumor sidedness: a systematic review and meta-analysis. Cancer Med. 2020;9(3):1044–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baran B, Mert Ozupek N, Yerli Tetik N, Acar E, Bekcioglu O, Baskin Y. Difference between left-sided and right-sided colorectal cancer: a focused review of literature. Gastroenterol Res. 2018;11(4):264–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buisman FE, Galjart B, Buettner S, Groot Koerkamp B, Grünhagen DJ, Verhoef C. Primary tumor location and the prognosis of patients after local treatment of colorectal liver metastases: a systematic review and meta-analysis. HPB. 2020;22(3):351–7. [DOI] [PubMed] [Google Scholar]
- 13.Matsuda K, Tamura K, Iwamoto H, et al. Tumor sidedness is associated with survival in patients with synchronous colorectal peritoneal carcinomatosis. Oncology. 2020;98(4):230–6. [DOI] [PubMed] [Google Scholar]
- 14.Kotha NV, Baumgartner JM, Veerapong J, et al. Primary tumor sidedness is predictive of survival in colon cancer patients treated with cytoreductive surgery with or without hyperthermic intraperitoneal chemotherapy: a US HIPEC collaborative study. Ann Surg Oncol. 2019;26(7):2234–40. [DOI] [PubMed] [Google Scholar]
- 15.Kelly KJ, Alsayadnasser M, Vaida F, et al. Does primary tumor side matter in patients with metastatic colon cancer treated with cytoreductive surgery and hyperthermic intraperitoneal chemotherapy? Ann Surg Oncol. 2019;26(5):1421–7. [DOI] [PubMed] [Google Scholar]
- 16.de Boer NL, Rovers K, Burger JWA, et al. A population-based study on the prognostic impact of primary tumor sidedness in patients with peritoneal metastases from colon cancer. Cancer Med. 2020;9(16):5851–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YC, Lee YL, Chuang JP, Lee JC. Differences in survival between colon and rectal cancer from seer data. PLoS One. 2013;8(11):e78709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takamizawa Y, Shida D, Horie T, et al. Prognostic role for primary tumor location in patients with colorectal liver metastases: a comparison of right-sided colon, left-sided colon, and rectum. Dis Colon Rectum. 2023;66(2):233. [DOI] [PubMed] [Google Scholar]
- 19.Gallo G, Sena G, Vescio G, et al. The prognostic value of KRAS and BRAF in stage I-III colorectal cancer: a systematic review. Ann Ital Chir. 2019;90:127–37. [PubMed] [Google Scholar]
- 20.Petrelli F, Ghidini M, Cabiddu M, et al. Microsatellite instability and survival in stage II colorectal cancer: a systematic review and meta-analysis. Anticancer Res. 2019;39(12):6431–41. [DOI] [PubMed] [Google Scholar]
- 21.Taieb J, Shi Q, Pederson L, et al. Prognosis of microsatellite instability and/or mismatch repair deficiency stage III colon cancer patients after disease recurrence following adjuvant treatment: results of an ACCENT pooled analysis of seven studies. Ann Oncol. 2019;30(9):1466–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jácome AA, Vreeland TJ, Johnson B, et al. The prognostic impact of RAS on overall survival following liver resection in early versus late-onset colorectal cancer patients. Br J Cancer. 2021;124(4):797–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margonis GA, Buettner S, Andreatos N, et al. Association of BRAF mutations with survival and recurrence in surgically treated patients with metastatic colorectal liver cancer. JAMA Surg. 2018;153(7):e180996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bachet JB, Moreno-Lopez N, Vigano L, et al. BRAF mutation is not associated with an increased risk of recurrence in patients undergoing resection of colorectal liver metastases. Br J Surg. 2019;106(9):1237–47. [DOI] [PubMed] [Google Scholar]
- 25.Pikoulis E, Margonis GA, Andreatos N, et al. Prognostic role of BRAF mutations in colorectal cancer liver metastases. AR. 2016;36(9):4805–12. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi S, Takahashi S, Kojima M, et al. Clinical impact of BRAF V600E mutations in patients (Pts) with resectable solitary colorectal liver metastases (CRLM). Ann Oncol. 2019;30:v225–6. [Google Scholar]
- 27.Uhlig J, Cecchini M, Sheth A, Stein S, Lacy J, Kim HS. Microsatellite instability and KRAS mutation in stage IV colorectal cancer: prevalence, geographic discrepancies, and outcomes from the national cancer database. J Natl Compr Canc Netw. 2021;19(3):307–18. [DOI] [PubMed] [Google Scholar]
- 28.Fujiyoshi K, Yamamoto G, Takenoya T, et al. Metastatic pattern of stage IV colorectal cancer with high-frequency microsatellite instability as a prognostic factor. Anticancer Res. 2017;37(1):239–47. [DOI] [PubMed] [Google Scholar]
- 29.Kamphues C, Kadowaki S, Amini N, et al. The interplay of KRAS mutational status with tumor laterality in non-metastatic colorectal cancer: an international, multi-institutional study in patients with known KRAS, BRAF, and MSI status. J Surg Oncol. 2021;123(4):1005–14. [DOI] [PubMed] [Google Scholar]
- 30.Amini N, Andreatos N, Margonis GA, et al. Mutant KRAS as a prognostic biomarker after hepatectomy for rectal cancer metastases: does the primary disease site matter? J Hepato Biliary Pancreat. 2022;29(4):417–27. [DOI] [PubMed] [Google Scholar]
- 31.Charlton ME, Kahl AR, Greenbaum AA, et al. KRAS testing, tumor location, and survival in patients with stage IV colorectal cancer: SEER 2010–2013. J Natl Compr Canc Netw. 2017;15(12):1484–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cavallaro P, Bordeianou L, Stafford C, et al. Impact of single-organ metastasis to the liver or lung and genetic mutation status on prognosis in stage IV colorectal cancer. Clin Colorectal Cancer. 2020;19(1):e8–17. [DOI] [PubMed] [Google Scholar]
- 33.Favazza LA, Parseghian CM, Kaya C, et al. KRAS amplification in metastatic colon cancer is associated with a history of inflammatory bowel disease and may confer resistance to anti-EGFR therapy. Mod Pathol. 2020;33(9):1832–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kafatos G, Niepel D, Lowe K, et al. RAS mutation prevalence among patients with metastatic colorectal cancer: a meta-analysis of real-world data. Biomark Med. 2017;11(9):751–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venderbosch S, Nagtegaal ID, Maughan TS, et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: a pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies. Clin Cancer Res. 2014;20(20):5322–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanz-Pamplona R, Cordero D, Berenguer A, et al. Gene expression differences between colon and rectum tumors. Clin Cancer Res. 2011;17(23):7303–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider MA, Eden J, Pache B, et al. Mutations of RAS/RAF proto-oncogenes impair survival after cytoreductive surgery and HIPEC for peritoneal metastasis of colorectal origin. Ann Surg. 2018;268(5):845–53. [DOI] [PubMed] [Google Scholar]
- 38.Aasebø KØ, Dragomir A, Sundström M, et al. Consequences of a high incidence of microsatellite instability and BRAF-mutated tumors: a population-based cohort of metastatic colorectal cancer patients. Cancer Med. 2019;8(7):3623–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherman SK, Schuitevoerder D, Chan CHF, Turaga KK. Metastatic colorectal cancers with mismatch repair deficiency result in worse survival regardless of peritoneal metastases. Ann Surg Oncol. 2020;27(13):5074–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larsen SG, Goscinski MA, Dueland S, et al. Impact of KRAS, BRAF and microsatellite instability status after cytoreductive surgery and HIPEC in a national cohort of colorectal peritoneal metastasis patients. Br J Cancer. 2022;126(5):726–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcus L, Lemery SJ, Keegan P, Pazdur R. FDA approval summary: pembrolizumab for the treatment of microsatellite instability-high solid tumors. Clin Cancer Res. 2019;25(13):3753–8. [DOI] [PubMed] [Google Scholar]
- 42.Fiorentini G, Sarti D, Patriti A, et al. Immune response activation following hyperthermic intraperitoneal chemotherapy for peritoneal metastases: a pilot study. World J Clin Oncol. 2020;11(6):397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Overman MJ, McDermott R, Leach JL, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sahin IH, Akce M, Alese O, et al. Immune checkpoint inhibitors for the treatment of MSI-H/MMR-D colorectal cancer and a perspective on resistance mechanisms. Br J Cancer. 2019;121(10):809–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salem ME, Weinberg BA, Xiu J, et al. Comparative molecular analyses of left-sided colon, right-sided colon, and rectal cancers. Oncotarget. 2017;8(49):86356–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muzny DM, Bainbridge MN, Chang K, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan M, Jiang C, Tse P, et al. TP53 gain-of-function and non–gain-of-function mutations are differentially associated with sidedness-dependent prognosis in metastatic colorectal cancer. JCO. 2022;40(2):171–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li AJ, Li HG, Tang EJ, et al. PIK3CA and TP53 mutations predict overall survival of stage II/III colorectal cancer patients. World J Gastroenterol. 2018;24(5):631–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo Q, Chen D, Fan X, Fu X, Ma T, Chen D. KRAS and PIK3CA bi-mutations predict a poor prognosis in colorectal cancer patients: a single-site report. Transl Oncol. 2020;13(12):100874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fang T, Liang T, Wang Y, et al. Prognostic role and clinicopathological features of SMAD4 gene mutation in colorectal cancer: a systematic review and meta-analysis. BMC Gastroenterol. 2021;21(1):297. [DOI] [PMC free article] [PubMed] [Google Scholar]


