Abstract
The flowering mechanisms, especially chilling requirement-regulated flowering, in deciduous woody crops remain to be elucidated. Flower buds of northern highbush blueberry cultivar Aurora require approximately 1,000 chilling hours to bloom. Overexpression of a blueberry FLOWERING LOCUS T (VcFT) enabled precocious flowering of transgenic “Aurora” mainly in non-terminated apical buds during flower bud formation, meanwhile, most of the mature flower buds could not break until they received enough chilling hours. In this study, we highlighted two groups of differentially expressed genes (DEGs) in flower buds caused by VcFT overexpression (VcFT-OX) and full chilling. We compared the two groups of DEGs with a focus on flowering pathway genes. We found: 1) In non-chilled flower buds, VcFT-OX drove a high VcFT expression and repressed expression of a major MADS-box gene, blueberry SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (VcSOC1) resulting an increased VcFT/VcSOC1 expression ratio; 2) In fully chilled flower buds that are ready to break, the chilling upregulated VcSOC1 expression in non-transgenic “Aurora” and repressed VcFT expression in VcFT-OX “Aurora”, and each resulted in a decreased ratio of VcFT to VcSOC1; additionally, expression of a blueberry SHORT VEGETATIVE PHASE (VcSVP) was upregulated in chilled flower buds of both transgenic and non-transgenic’ “Aurora”. Together with additional analysis of VcFT and VcSOC1 in the transcriptome data of other genotypes and tissues, we provide evidence to support that VcFT expression plays a significant role in promoting floral initiation and that VcSOC1 expression is a key floral activator. We thus propose a new hypothesis on blueberry flowering mechanism, of which the ratios of VcFT-to-VcSOC1 at transcript levels in the flowering pathways determine flower bud formation and bud breaking. Generally, an increased VcFT/VcSOC1 ratio or increased VcSOC1 in leaf promotes precocious flowering and flower bud formation, and a decreased VcFT/VcSOC1 ratio with increased VcSOC1 in fully chilled flower buds contributes to flower bud breaking.
Keywords: chilling requirement, dormancy release, FLOWERING LOCUS T, flowering mechanism, SHORT VEGETATIVE PHASE, SUPPRESSOR OF OVEREXPRESSION OF CONSTAN 1, transcriptome analysis, Vaccinium corymbosum
1 Introduction
Most of the cultivated deciduous fruit trees and bushes originate and are grown in temperate climates where light and temperature are the key environmental factors that guide flower bud initiation, flowering and fruiting (Saure, 1985). For example, blueberry flower bud initiation and formation usually occur in late summer and fall, and sufficient chilling hour accumulation in winter is the key to enable flower bud breaking in the next spring. The need of chilling hour accumulation is called chilling requirement for woody plants. It is a little different from vernalization in annual plants. Over the past several decades, climate change has caused the onset of the growing season of trees to shift earlier (e.g., 2.3 days/decade in temperate Europe) (Parmesan and Yohe, 2003; Root et al., 2003; Atkinson et al., 2013; Chuine et al., 2016). Generally, reduced winter chill is often associated with insufficient chilling hours. Warm weather sometimes leads to fruit/nut trees flowering out-of-season. And increased temperature fluctuations during plant bloom turns seasonal frost into a greater danger, often causing freezing injuries to flowers and young fruits. Flowering plays a significant role in the life cycle of flowering plants (Angiosperms), and since it is generally a prerequisite for fruiting, many studies have been directed to understand flowering pathways of woody plants to develop genetic solutions for manipulating flowering times to alleviate the negative impact of climate change (Luedeling et al., 2011; Atkinson et al., 2013; Song, 2019). It has been recognized that a complex network of flowering pathway genes controls seasonal flowering (Matsoukas et al., 2012). While flowering mechanisms have been well studied in annual plants, such as Arabidopsis thaliana, rice, and cereals (Cockram et al., 2007; Trevaskis et al., 2007; Greenup et al., 2009; Michaels, 2009; Amasino, 2010; Fornara et al., 2010; Higgins et al., 2010; Lee and Lee, 2010; Pin et al., 2010; Wellmer and Riechmann, 2010; Huijser and Schmid, 2011; Song et al., 2015), chilling-mediated flowering mechanisms in deciduous fruit trees/bushes remain to be revealed (Wilkie et al., 2008; Zhang et al., 2010; Jia et al., 2014; Jameson and Clemens, 2015; Jewaria et al., 2021).
FLOWERING LOCUS T (FT) is a major integrator of signaling that stimulates the transition of meristem tissue into flower buds (Kobayashi et al., 1999). FT is produced in leaves when certain conditions are met and certain pathways are activated (Turck et al., 2008; Fornara et al., 2010; Krzymuski et al., 2015). Constitutive expression of FT induces precocious flowering in many plants, including Arabidopsis (Abe et al., 2005; Wigge et al., 2005), apple (Malus × domestica Borkh.) (Trankner et al., 2010), plum (Prunus domestica) (Srinivasan et al., 2012), eucalyptus (Eucalyptus grandis × E. urophylla) (Klocko et al., 2016), cassava (Manihot esculenta, Crantz) (Adeyemo et al., 2017), petunia (Petunia × hybrid) (Lin et al., 2019a), and blueberry (Gao et al., 2016; Walworth et al., 2016). However, whether constitutive expression of FT induced flowering of all flower buds or just some of them was not clearly reported in these transgenic studies. Recently, we found that while overexpression of a blueberry FLOWERING LOCUS T (VcFT) enabled precocious flowering of transgenic “Aurora” plants during flower bud formation, most of the mature flower buds (i.e., apical and auxiliary floral buds) could not break until they received enough chilling hours (Song et al., 2013b; Gao et al., 2016; Walworth et al., 2016). It appears that FT may not pay a critical role in chilling mediated blueberry flowering. Indeed, in a separate study, chilled and non-chilled flower buds showed no significant difference in VcFT expression in southern highbush blueberry “Legacy”, but several other major flowering pathway genes such as the blueberry LEAFY gene (VcLFY) and MADS-box genes [e.g., SHORT VEGETATIVE PHASE (VcSVP), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (VcSOC1), and APETALA1 (VcAP1)] were differentially expressed and seemed to play significant roles in chilling mediated blueberry flowering (Song and Chen, 2018b). Based on these studies, VcFT appears to be a powerful inducer of flower bud formation (Gao et al., 2016; Walworth et al., 2016; Song et al., 2019), but is not likely a key factor for chilling-mediated dormancy breaking in blueberries (Song and Chen, 2018b).
MADS-box genes play important roles in the vernalization pathway of annual plants (Fornara et al., 2010). FLOWERING LOCUS C (FLC), SVP, and SOC1 are three major MADS-box genes in the vernalization pathway of Arabidopsis (Michaels and Amasino, 1999; Gregis et al., 2006). SVP promotes FLC that represses SOC1 and inhibits flowering prior to plant vernalization (Michaels and Amasino, 1999; Gregis et al., 2006). In woody plants, functional FLC has not been verified. In peach [Prunus persica (L.) Batsch] and other Prunus species, Dormancy-Associated MADS-box (DAM) genes are key regulators of chilling requirement for endodormancy release (Bielenberg et al., 2008; Wells et al., 2015; Zhu et al., 2020a; Yu et al., 2020; Calle et al., 2021) and some of them showed high similarity to FLC, SOC1, and SVP MADS-box genes as defined in Arabidopsis. In peach floral buds the DAM cluster, including the orthologues of SVP, controls dormancy and chilling requirements (Zhu et al., 2020a), but none of these genes have been verified through functional studies due to the difficulty in peach transformation. In other deciduous fruit crops, SOC1 seems to be a significant regulator in chilling-mediated flowering dormancy release. For example,: in kiwifruit (Actinidia delicious), SOC1-like genes may affect the duration of dormancy although they may not have a role in the floral transition (Voogd et al., 2015); in grapevine (Vitis vinifera), chilling accumulation induced expression of its SOC1 (Kamal et al., 2019); and in poplar (Populus tremula × alba), overexpression of a SOC1-like gene promotes bud break and overexpression of a SVP-like repressed flowering (Gómez-Soto et al., 2021; Goralogia et al., 2021). However, in blueberry, no functional blueberry FLC has been identified (Walworth et al., 2016) and ectopic expression of an apple FLC3 did not inhibit, but promoted, blueberry flowering (Zong et al., 2019). Blueberry SOC1 (VcSOC1) showed a high similarity to both peach DAM1 and DAM2 (PmDAM1 and PmDAM2) while VcSVP was similar to PmDAM2. SVP and SVP-like genes in woody fruit crops [e.g., kiwifruit, trifoliate orange (Poncirus trifoliata L. Raf.), apple, and sweet cherry (Prunus avium L.)] suppress budbreak and flowering (Gregis et al., 2006; Li et al., 2010; Wu et al., 2017a; Wu et al., 2017b; Wang et al., 2021), as what was observed in annual species. In general, during vernalization or chilling accumulation, decreased expression of SVP (or SVP-like genes) activates expression of SOC1 (or SOC1-like gene) that promotes budbreak and flowering. However, the roles of SVP expression in flowering may vary among different SVP homologues. For example, in grape, SVP homologues were found to be inconsistent in either promoting or repressing flowering, which contrasts to the negative relationship in Arabidopsis (Diaz-Riquelme et al., 2012; Li-Mallet et al., 2016; Arro et al., 2019; Kamal et al., 2019; Dong et al., 2022). Blueberry SVP (VcSVP) expression in floral buds of southern highbush ‘Legacy’ and its mutant (Mu-Legacy) was upregulated after receiving sufficient chilling hours and downregulated in florescence (Song and Chen, 2018b). It appears that SOC1 is a conserved activator in woody plants, but SVP’s role as a activator or a repressor may depend on bud developmental stage and plant species.
In this study, we identified differentially expressed genes (DEGs) caused by an overexpressed VcFT in leaves and non-chilled flower buds of transgenic blueberry “Aurora”. We investigated the role of FT expression in chilling-dependent floral activation by analyzing transcriptome profiles of chilled and non-chilled flower buds of both non-transgenic and VcFT transgenic blueberry “Aurora”. We found that VcFT and VcSOC1 played critical roles in floral initiation and activation, respectively, and VcSVP acted as a positive regulator in chilling-mediated flowering in blueberry, which contrasts to the SVP roles reported in annual plant species. Based on our current and previous studies, we proposed that the relative ratio of VcFT/VcSOC1 expression at transcript levels is the key factor to determine the flowering developmental process in blueberry.
2 Materials and methods
2.1 Plant materials
All blueberry plants used in this study were derived from in vitro cultured shoots (Song et al., 2013b). Twelve micropropagated plants for non-transgenic “Aurora” and twelve each of six independent T0 lines of VcFT-OX transgenic “Aurora” were individually grown in 4-gallon pots (top diameter 30 cm, bottom diameter 24 cm, depth 27 cm) in a secured courtyard under natural environmental conditions at Michigan State University, East Lansing, Michigan. All plants were grown healthy, watered as needed, and fertilized once a week using an acidic nutrient solution of 0.2 g/L 21-7-7 (nitrogen-phosphate-potassium). Plants were 3–4 years old when investigated. Mature leaves from the middle of soft-wood shoots and flower buds from three individual plants of non-transgenic “Aurora” (control) and each of the six T0 VcFT-OX transgenic lines (three selected plants per line) were sampled on an individual plant basis. In May, approximately 2 g of mature leaves per plant were harvested when flower buds were visible on the transgenic plants. The mature leaves in this study were selected to differentiate them from the young leaf around shoot tips samples that we analyzed in the previous report (Walworth et al., 2016). A total of 30–50 flower buds per plant were collected in October, November (non-chilled), and December; more were collected in January, February, and March (fully chilled, having received approximately 1,200 chilling hours) of the following year. The bud samples collected in November and March were used for RNA sequencing. All samples were collected into 2-mL cryo-tubes, frozen immediately in liquid nitrogen (LN), and stored at −80°C.
2.2 RNA preparation, sequencing, and transcriptome analysis
Approximately 500 mg of each sample was ground in LN and used for RNA isolation and the excess was archived in 2-mL cryotubes at −80°C. Crude total RNA of each sample was isolated using a CTAB method (Zamboni et al., 2008) and purified using an RNeasy Mini Kit (Qiagen, Valencia, CA, United States). On-Column DNase digestion with the RNase-free DNase Set (Qiagen) was used to remove DNA contaminants. RNA quality was determined using the High Sensitivity RNA ScreenTape system (Agilent technologies, Santa Clara, CA). High quality RNA with an RNA integrity number ≥7.0 for bud and leaf was used for sequencing and reverse transcription (RT) PCR analysis.
Three biological replicates of RNA samples were sequenced for both transgenic and non-transgenic “Aurora” plants. The three biological replicates of non-transgenic “Aurora” were represented by the three individual plants sampled as described earlier. However, for the VcFT-OX transgenic “Aurora”, the three biological replicates were represented by three bulks of RNA samples from VcFT-OX transgenic plants with each bulk having equal amount of RNA from six plants one each from the six transgenic lines. RNA samples were sequenced using the Illumina HiSeq4000 to generate 10–20 million, 150 bp paired-end reads per sample at the Research Technology Support Facility at Michigan State University (East Lansing, Michigan, United States). All newly obtained sequence reads were deposited in GenBank (BioProject: PRJNA900257). FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to assess the quality of sequencing reads for the per-base quality scores. The reads with average scores greater than 30 were obtained and used for transcriptome analysis. The paired reads were aligned to the transcriptome reference [RefTrinity; deposited in GenBank (Accession number: SRX2728,597)] developed in our previous study to estimate and the abundance for each of a single read using Trinity/2.8.5 (Haas et al., 2013; Walworth et al., 2016). The genetic background of different blueberry cultivars varies greatly among different cultivars. Therefore, we used our own transcriptome reference instead of the published blueberry genome sequences for comparative transcriptome comparisons in order to minimize the potential errors caused by the specificity of the cultivars used in this study. The differentially expressed transcripts (DETs) with the false discovery rate (FDR) value below 0.05 were identified using the Trinity command “run_DE_analysis.pl--method edgeR” (Haas et al., 2013). In calculating a VcFT/VcSOC1 ratio, the total number of all isoforms of the gene was used, the data from three biological replicates were used for statistical analysis.
Quantitative RT-PCR (qRT-PCR) using the SYBR Green system (LifeTechnologies, Carlsbad, CA) was conducted to check the selected transcripts on a Roche LightCycler® 480 Instrument II (Roche). Primers for qRT-PCR were designed based on the RNA-seq sequence information (Supplementary Table S1). Transcript levels within samples were normalized to EIF (Eukaryotic translation initiation factor 3 subunit H). Fold changes between samples were calculated using 2−ΔΔCT, where ∆∆Ct = (CtTARGET–CtNOM)transgenic—(CtTARGET–CtNOM)non-transgenic. For relative expression analysis of individual genes, expressions were normalized to the VcACTIN.
2.3 Retrieved datasets for comparing multiple comparisons
DETs from our previously published data were retrieved for conducting the following comparisons in this study: 1) VcFT-OX “Aurora” vs. “Aurora” leaf (Walworth et al., 2016); 2) VcSOC1K-OX “Aurora” vs. “Aurora” leaf “Aurora”; VcSOC1K-OX “Aurora” contains an overexpressed blueberry VcSOC1 K-domain that enabled precocious flowering and more flower bud formation (Song and Chen, 2018a); 3) Legacy_mutant1 vs. Legacy leaf; Legacy_mutant1 has an overexpressed VcDDF1 and a constitutively expressed VcRR2 at the insertion position that drove a reduced chilling requirement and promoted flower bud formation (Song and Walworth, 2018); 4) Legacy_mutant2 vs. Legacy leaf; Legacy_mutant2 is a self-pollinated T1 transgenic line from Legacy_mutant1 and it showed precocious flowering (Lin et al., 2019b); 5) Legacy_on_VcSOC1-OX_Aurora vs. Legacy leaf; Legacy_on_VcSOC1-OX_Aurora refers to non-transgenic shoot/scion grafted on the shoots (the leaves at the basal part were retained) of transgenic VcSOC1-OX “Aurora”, the grafting resulted in a promoted flower bud formation in non-transgenic legacy shoot (Song et al., 2019); 6) Legacy_mutant1 vs. Legacy bud; part of Legacy_mutant1 buds could break under non-chilling conditions while non-chilled “Legacy” buds could not 7) Legacy_mutant1 vs. chilled Legacy_mutant1; chilled Legacy_mutant1 could flower normally (Song and Walworth, 2018); 8) Legacy vs. chilled Legacy bud: chilled “Legacy” buds could break while non-chilled “Legacy” bud could not (Song and Chen, 2018b); 9) Chilled bud vs. Late-pink bud of “Legacy” (Song and Chen, 2018b); and 10) Chilled bud vs. Late-pink bud of “Legacy_mutant1” (Lin et al., 2019b).
2.4 Statistical analysis
Statistical analysis of the VcFT/VcSOC1 ratios was conducted using ANOVA and TukeyHSD in RStudio (Version 3.3.1).
3 Results
3.1 Phenotypic changes induced by VcFT-OX
VcFT-OX plants showed enhanced flower bud formation with multiple flower buds at a single node in a branch (Figure 1A), whereas a single flower bud at each node was observed for non-transgenic “Aurora” (Figure 1B). All VcFT-OX blueberry plants showed earlier flower bud formation than those non-transgenic plants (Figure 1C). Under non-chilling conditions, some of the apical buds in VcFT-OX blueberry plants were able to flower, while none of the buds in non-transgenic plants could (Figure 1D). For mature VcFT-OX transgenic buds, most of them (>90%) were not able to flower without enough chilling hours and even the buds that broke were not flowering normally with only 1-3 flowers per bud in contrast to 5–10 flowers per normally flowering bud (Figure 1D), suggesting that the VcFT overexpression was not sufficient to overcome chilling requirement for these mature buds.
FIGURE 1.
Phenotypic changes in when VcFT-OX transgenic “Aurora” and differentially expressed transcripts detected in four pairs of comparisons. (A) VcFT-OX transgenic “Aurora”. (B) Non-transgenic “Aurora”. (C) Flower bud formation in VcFT-OX transgenic “Aurora” and non-transgenic “Aurora”. Each data point represent an average of data from six plants. (D) Flowering chilled and non-chilled flower buds in transgenic “Aurora” VcFT-OX and non-transgenic “Aurora”. Each data point represents an average of data from three plants. Six transgenic lines (VcFT-OX1 to VcFT-OX6) and three groups of wild type (non-transgenic WT1-WT3) plants were investigated after they reached 2–3 year old. Bars show standard deviation.
3.2 VcFT-OX induced DEGs in leaves and non-chilled floral buds
We compared the RNA-seq profiles of non-transgenic and transgenic blueberry plants for mature leaf and non-chilled bud samples, respectively, and revealed 1,024 unique DEGs in leaves and 3,177 in non-chilled flower buds. The number of DEGs in buds was about three times more than that in leaf samples. This big difference seemed not positively correlated with the relative abundance of the VcFT expression in the two tissues, since VcFT expression in leaves [23.2 reads/million reads (MR)] was much higher than that in non-chilling flower buds (9.2 reads/MR). We identified 387 shared DETs which were annotated to 331 unique genes (Figure 2A). Twelve shared DEGs were in the flowering pathway, including the upregulated VcARP6 (ACT_GOSHI), VcFT (HD3A_ORYSJ), and SEPALLATA 3 (VcSEP3) (AGL9_PETHY) and downregulated VcSVP (SVP_ARATH), VcAPRR5 (APRR5_ARATH), and VcPAF1 (PSA4_SPIOL). Six shared DEGs were not consistently up- or downregulated in both leaf and bud tissues. Noteworthily, VcANR1 (ANR1_ARATH) was upregulated in transgenic VcFT-OX leaves but downregulated in transgenic buds, compared to the non-transgenic control while VcEF4L4 (EF4L4_ARATH) was downregulated in transgenic leaves but upregulated in transgenic buds.
FIGURE 2.
Differentially expressed flowering pathway genes. (A) VcFT-OX transgenic “Aurora” and non-transgenic “Aurora” were compared in leaves and non-chilled buds, respectively. Non-chilled and chilled flower buds from 3 to 4 year old bushes were collected in late November and February, respectively. Annotated flowering pathway genes (flowering), hormone-related genes (hormone), sugar-related (sugar) genes, and MADS-box (MADS) genes were presented. *The numbers of the shared DEGs were counted according to the shared DETs. (B) Transgenic vs. non-transgenic leaves. (C) Transgenic vs. non-transgenic buds. (D–E) Responses of flowering pathway genes to VcFT-OX in mature leaf (D) and bud (E); the positive or negative regulation sign is based on the information from Arabidopsis, but it may not match the results obtained in this study. White, green, and red boxes indicate no differential expression, upregulated expression, and downregulated expression, respectively. The boxes with “?” indicate inconsistent differences among homologues.
“Aurora” is a heterozygous tetraploid and could have multiple alleles for a given gene. In addition to the DEGs identified from shared DETs, we further identified shared DEGs from non-shared DETs of the two tissues (Figures 2B, C; Supplementary Table S2). Overall, in the comparison of the DEGs identified in transgenic buds and transgenic leaves, the former showed more DEGs of flowering pathway genes (37 DEGs in buds vs.10 in leaves) than those of in transgenic leaves (Figures 2B, C). This suggests that the overexpression of VcFT had a broader impact on the numbers of DEGs in buds than leaves. Of the major flowering pathway genes, MADS-box genes FRUITFUL (VcFUL) (AGL8_ARATH) were shared DEGs in both leaves and buds (Figures 2B, C). FUL is partially redundant to the function of AP1 and CAULIFLOWER (CAL) that promotes floral meristem identity (Ferrandiz et al., 2000). VcAGL8 was upregulated in leaves, which may contribute to early flower bud formation in the VcFT-OX transgenic plants. MADS-box gene VcSOC1 was a shared DEG in both tissues (Figures 2B, C). SOC1 is a positive regulator for plant flowering (Gregis et al., 2009; Lee and Lee, 2010). The repressed expression of SOC1 indicates a potential delay in flowering. Noteworthily: in young VcFT-OX transgenic leaves one SOC1 homolog was upregulated and one was downregulated, the upregulated one showed a higher fold change (Walworth et al., 2016), but in contrast, there was only one downregulated SOC1 homolog in mature VcFT-OX transgenic leaves (Table 1). AGAMOUS-LIKE MADS-BOX PROTEIN AGL9 (VcAGL9) showed upregulation in leaves and downregulation in buds; MADS-BOX TRASCRIPTION FACTOR 6 (VcMADS6) showed upregulation in both leaves and buds; LEAFY (VcLFY) was up-regulated in bud (Figures 2B, C); ENT-KAURENE OXIDASE (KO) gene (VcKO1) was up-regulated in leaves and buds (Figures 2B, C). KO catalyzes a key step in gibberellins (GAs) biosynthesis. The Arabidopsis ga3 mutant, deficient in KO activity, is a gibberellin-responsive dwarf (Helliwell et al., 1998). Increased KO expression suggests a potential increase in GAs, which is associated with the promotion of blueberry flowering driven by VcFT-OX. We conducted qRT-PCR analysis of 9 DEGs to validate the RNA-seq data from leaves and flower buds. These 9 DEGs were selected from flowering, hormones, and sugar pathways. The qRT-PCR results for the selected DEGs were consistent with the RNA-seq data (Supplementary Figure S1).
TABLE 1.
Differentially expressed transcripts of VcFT (HD3A_ORYSJ), VcSOC1 (SOC1_ARATH), and VcSVP (SVP_ARATH) in the 8 pairs of comparisons involving various genetic material in blueberry. LogFC: log2 (fold change) = Log2 (sample 1/sample 2). #N/A: no differential expression. : no annotation. CB: fully chilled bud. NCB: non-chilled flower bud. LPB: Late pink bud.
| Transcript_id | Annotation (sprot_Top_BLASTP_hit) | LogFC: log2 (fold change) = Log2 (sample 1/sample 2) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Aurora, non-transgenic/VcFT-OX transgenic leaf, this study | Aurora, transgenic/non-transgenic NCB, this study | Aurora, CB/NCB, this study | Aurora, transgenic CB/transgenic NCB), this study | Legacy CB/LPB (Song and Chen, 2018b) | Legacy_mutant1, CB/LPB (Lin et al., 2019b) | Legacy_mutant1, CB/NCB (Lin et al., 2019b) | Legacy, CB/NCB (Song and Chen, 2018b) | ||
| c84088_g2_i3 | HD3A_ORYSJ | −10.34 | #N/A | #N/A | #N/A | 4.52 | 4.05 | −1.31 | −0.95 |
| c84088_g2_i5 | HD3A_ORYSJ | −12.34 | −1.52 | #N/A | #N/A | 4.92 | 6.78 | #N/A | 1.11 |
| c84088_g2_i1 | HD3A_ORYSJ | −12.51 | 10.67 | #N/A | −0.98 | 5.06 | 6.60 | −1.13 | #N/A |
| c93787_g3_i1 | SOC1_ARATH | #N/A | #N/A | #N/A | #N/A | −9.35 | −8.72 | #N/A | #N/A |
| c93787_g3_i2 | SOC1_ARATH | #N/A | #N/A | #N/A | #N/A | −10.30 | −9.06 | #N/A | #N/A |
| c86010_g2_i1 | SOC1_ARATH | #N/A | #N/A | #N/A | #N/A | 4.40 | 3.83 | 2.38 | 3.89 |
| c89673_g4_i1 | SOC1_ARATH | #N/A | −2.30 | 2.05 | #N/A | −1.01 | #N/A | 0.89 | #N/A |
| c89673_g4_i2 | SOC1_ARATH | #N/A | #N/A | 1.97 | #N/A | #N/A | #N/A | #N/A | #N/A |
| c94107_g4_i3 | SOC1_ARATH | #N/A | −3.30 | #N/A | #N/A | #N/A | 3.13 | #N/A | #N/A |
| c94107_g4_i4 | SOC1_ARATH | #N/A | −6.10 | #N/A | #N/A | 3.10 | 2.59 | #N/A | #N/A |
| c94107_g4_i2 | SOC1_ARATH | #N/A | −5.38 | #N/A | #N/A | 2.09 | 2.34 | #N/A | #N/A |
| c94107_g4_i5 | SOC1_ARATH | #N/A | −6.26 | #N/A | #N/A | 2.36 | 1.90 | #N/A | #N/A |
| c89673_g3_i1 | SOC1_ARATH | #N/A | #N/A | #N/A | #N/A | 4.04 | #N/A | 3.46 | 2.56 |
| c94107_g4_i1 | SOC1_ARATH | 2.20 | #N/A | #N/A | #N/A | 3.64 | #N/A | #N/A | #N/A |
| c94107_g4_i6 | SOC1_ARATH | #N/A | −2.51 | #N/A | #N/A | 2.95 | 4.02 | #N/A | #N/A |
| c99746_g3_i3 | SOC1_ARATH | #N/A | #N/A | 0.76 | #N/A | #N/A | #N/A | #N/A | #N/A |
| c86010_g1_i3 | SOC1_ARATH | #N/A | #N/A | 1.58 | #N/A | 2.65 | 2.71 | −0.44 | #N/A |
| c86010_g1_i1 | SOC1_ARATH | #N/A | #N/A | 1.08 | #N/A | 2.70 | 2.17 | −0.48 | #N/A |
| c86010_g1_i2 | SOC1_ARATH | #N/A | #N/A | 0.75 | #N/A | 2.06 | 2.05 | −0.82 | #N/A |
| c90289_g1_i4 | SVP_ARATH | #N/A | #N/A | #N/A | #N/A | 3.74 | #N/A | #N/A | #N/A |
| c91377_g1_i9 | SVP_ARATH | 1.83 | #N/A | #N/A | #N/A | #N/A | #N/A | #N/A | #N/A |
| c91377_g1_i7 | SVP_ARATH | 1.30 | −2.38 | 1.05 | #N/A | 1.60 | #N/A | #N/A | 1.01 |
| c90289_g1_i2 | SVP_ARATH | #N/A | #N/A | #N/A | #N/A | 3.92 | #N/A | #N/A | #N/A |
| c90829_g2_i2 | SVP_ARATH | #N/A | #N/A | #N/A | #N/A | #N/A | 1.22 | 0.75 | 1.15 |
| c91377_g1_i14 | SVP_ARATH | 1.32 | −3.21 | 2.55 | #N/A | 1.46 | #N/A | #N/A | #N/A |
| c90289_g1_i1 | SVP_ARATH | #N/A | #N/A | #N/A | #N/A | 6.58 | −1.11 | #N/A | #N/A |
| c90829_g2_i1 | SVP_ARATH | #N/A | #N/A | #N/A | #N/A | 1.20 | 1.90 | 1.02 | 1.19 |
| c91377_g1_i11 | SVP_ARATH | 1.35 | #N/A | #N/A | #N/A | 1.26 | #N/A | #N/A | #N/A |
| c91377_g1_i5 | SVP_ARATH | #N/A | −1.35 | #N/A | 0.80 | #N/A | #N/A | #N/A | #N/A |
3.3 Chilling-indued DEGs in transgenic VcFT-OX and non-transgenic ‘Aurora’ floral buds
The comparison of chilled non-transgenic “Aurora” and non-chilled nontransgenic “Aurora” yielded 3,125 DETs, which were annotated to 1,889 unique genes (Figure 3A). The results of RT-qPCR analysis of 5 selected DETs were consistent with those from RNA-sequencing data (Supplementary Figure S2). The endogenous VcFT showed no differential expression when chilled and non-chilled buds of non-transgenic “Aurora” were compared. The comparison of chilled and non-chilled buds of VcFT-OX transgenic buds yielded 682 DEGs, including a downregulated VcFT expression in transgenic Aurora (Figure 3A).
FIGURE 3.
Differentially expressed flowering pathway genes. (A) Chilled and non-chilled flower buds in transgenic “Aurora” VcFT-OX and non-transgenic “Aurora”, respectively. Non-chilled and chilled flower buds from 3 to 4 year old bushes were collected in late November and February, respectively. Annotated flowering pathway genes (flowering), hormone-related genes (hormone), sugar-related (sugar) genes, and MADS-box (MADS) genes were presented. *The numbers of the shared DEGs were counted according to the shared DETs. (B) Chilled vs. non-chilled buds of transgenic VcFT-OX “Aurora”. (C) Chilled vs. non-chilled buds of non-transgenic “Aurora”. White, green, and red boxes indicate no differential expression, upregulated expression, and downregulated expression, respectively. The boxes with “?” indicate inconsistent differences among homologues. (D,E) Responses of flowering pathway genes to fully chilled vs. non-chilled buds of non-transgenic plants (D) and VcFT-OX transgenic plants (E). The positive or negative regulation sign is based on the information from Arabidopsis, but it may not match the results obtained in this study.
Interestingly, the VcFT-OX “Aurora” showed a decrease in the number of DEGs between chilled and non-chilled buds when compared to the non-transgenic “Aurora” (Figures 3B, C; Table 1; Supplementary Table S2). The increased number of DEGs (1,889) in the non-transgenic “Aurora” buds (chilled vs. non-chilled) in compared to that of in the VcFT-OX “Aurora” transgenic buds (682 DEGs, chilled vs. non-chilled) suggests the VcFT-OX have already changed some of the chilling-mediated flowering pathway genes prior to their exposure to full chilling.
To identify flowering pathway genes responsive to full chilling in flower buds of both non-transgenic “Aurora” and transgenic VcFT-OX “Aurora”, the transcriptomes of chilled versus non-chilled were compared (Figures 3A–C). In this comparison, the shared DEGs were identified based on annotation (Figures 3B, C). A total of 19 and 47 DEGs in the flowering pathway were identified in chilled buds of transgenic VcFT-OX and non-transgenic “Aurora”, respectively (Figures 3B, C). Of the shared DEGs in the 19 and 47 DEGs, five were downregulated, including VIN3-LIKE 2 (VcVIL2), VcMADS6, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 16 (VcSPL16), PHOSPHATE SYNTHETASE (VcCPS), and GIBBERELLIN 3-OXIDASE 4 (VcGA3OX4); five were upregulated including CONSTITUTIVE PHOTOMORPHOGENIC 1 (VcCOP1), LATE ELONGATED HYPOCOTYL (VcLHY), FLOWERING LOCUS D (VcFLD), VcSVP, and VcSPL8 (Figures 3B, C). These ten shared, consistent up- or downregulated DEGs showed the same behavior in both transgenic and non-transgenic “Aurora” and were most likely the flowering pathway genes responsible for chilling requirement.
3.4 VcFT-induced floral bud formation and VcSOC1-regulated floral bud breaking
The overexpressed VcFT in both leaf and flower bud caused differential expressions of VcSOC1, VcAGL8, VcAGL9, VcLFY, VcMADS6 and VcKO in the flowering pathway (Figures 2D, E). As verified by qRT-PCR analysis, VcFT-OX repressed VcSVP and VcSOC1expression, promoted VcLFY expression, and had no significant effect on AGL8 expression in non-chilled transgenic flower buds (Figure 2E, Figure 3D, Figure 4A and Table 1). Protein FD (FD) is required for FT to promote flowering (Abe et al., 2005; Wigge et al., 2005). VcFT-OX repressed blueberry FD (VcFD) in transgenic bud but did not lead to a significant change in leaf (Figures 2B, C). TERMINAL FLOWER1 (TFL1) [CENTRORADIALIS 1 (CEN1)] has an opposite role of FT. FT competes with TFL1 for FD binding (Hanano and Goto, 2011; Zhu et al., 2020b). Neither VcCEN1 nor VcCEN2 showed differential expression in transgenic leaf and bud (Figures 2B, C).
FIGURE 4.
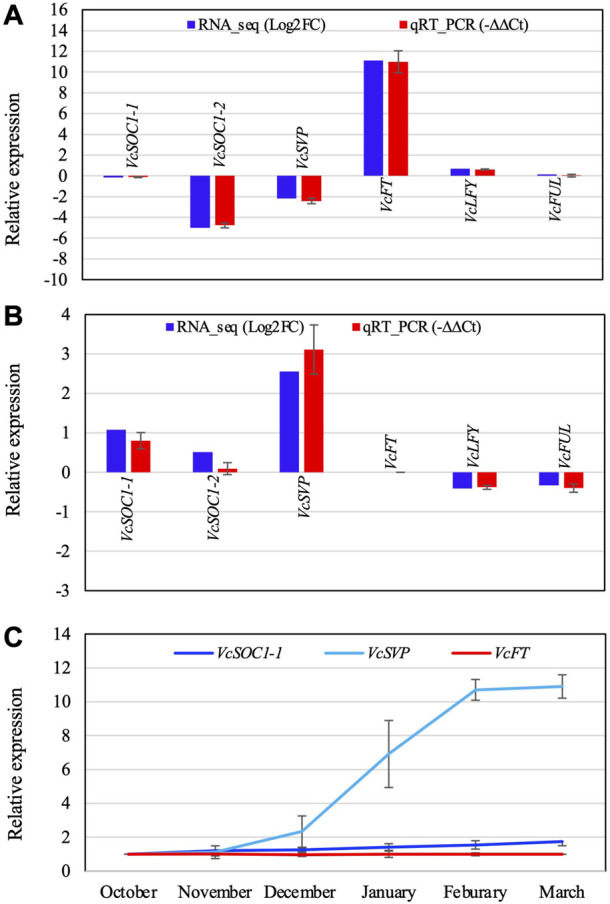
Comparison of the RT-qPCR analysis result and the RNA-seq data of the selected DETs (A,B) and relative expression of three flowering pathway genes in blueberry floral buds during chilling hour accumulation (C). (A) Non-chilled flower buds (VcFT-OX “Aurora” vs. non-transgenic “Aurora” bud). (B) Non-transgenic bud (chilled vs. non-chilled). −∆∆Ct is an average of three biological and three technical replicates for each DET. EUKARYOTIC TRANSLATION INITIATION FACTOR 3 SUBUNIT H was used to normalize the RT-qPCR results. (C) The expression was normalized to VcACTIN. The first normalized data point (October) was arbitrarily set as “1” and then used to normalize the other data points. Each data point is an average of three biological and three technical replicates. The error bars indicate standard deviation. Gene IDs: VcSOC1-1 (SOC1_ARATH), VcSOC1-2 (SOC1_ARATH), VcSVP (SVP_ARATH), VcFT (HD3A_ORYSJ), VcLFY (FLO_ANTMA), and VcFUL (AGL8_SOLTU).
VcSOC1 promoted chilling-mediated flowering in blueberry. In non-transgenic “Aurora”, fully chilled flower buds showed an increased VcSOC1 expression (Figure 3D; Figure 4B). In transgenic VcFT-OX “Aurora”, VcSOC1 was not among the DEGs (Figure 3E). VcMADS6 and VcVIL2 were repressed in chilled buds of both non-transgenic and VcFT-OX transgenic plants (Table 1; Figures 3D, E), suggesting that VcMADS6 and VcVIL2 were negative regulators for flower bud breaking. Importantly, both MADS6 and VIL2 were upregulated by the expression of VcFT in the VcFT-OX transgenic buds (Figure 3C). VcFT-OX in non-chilled flower buds upregulated expression of VcLFY, VcMADS6, and four chilling pathway genes (VcARP6, VcFRI, VcFRL, and VcVIL2), which were, in contrast, all downregulated in full chilled non-transgenic buds (Figures 2, 3). This explains why the VcFT overexpression was insufficient to completely overcome the chilling requirement for mature flower bud breaking. We further checked the VcSOC1 expression using qRT-PCR and found that its expression was increasing over the chilling accumulation period (Figure 4C). The results indicate that increasing VcSOC1 expression occurs during chilling accumulation and is the key to activating budbreak in non-transgenic flower buds.
VcSVP expression was upregulated in chilled flower buds (vs. non-chilled buds) of both “Aurora” and VcFT-OX Aurora (Figures 3D, E; Figure 4B). In addition, similarly to VcSOC1, the VcSVP expression increased with the chilling hours accumulation (Figure 4C), suggesting that the upregulated VcSVP is a positive regulator to promote blueberry bud breaking during chilling accumulation. Interestingly according to the comparisons between fully chilled flower bud and late pink bud during budbreak for both the “Legacy” and the “Legacy” mutant1, VcSVP expression was decreasing comparatively to VcSOC1 expression (Table 1).
While Log2 (fold change) was presented in the transcriptome comparisons using Edge R in Trinity, one problem was that when a gene had both up- and downregulated DETs, it was difficult to determine the overall up- or downregulation of the gene. Therefore, to investigate how VcFT, VcSOC1, and VcSVP1 interact with each other to affect flowering in blueberry, we examined the ratios of VcFT and VcSOC1 expression based on the Fragments Per Kilobase of transcript per Million mapped reads (FPKM) in the transcriptome comparison data from not only this study but also the previous studies for the other blueberry genotypes or tissues (Table 2). The mature leaves of VcFT-OX “Aurora”, in comparison to the non-transgenic “Aurora” leaves, had an increased ratio of VcFT/VcSOC1 associated with an upregulated expression of VcSOC1 (Figures 2B, C; Table 2). This is consistent with the young leaf transcriptome data previously published for VcFT-OX “Aurora” (Table 2). Phenotypically, the increased VcFT/VcSOC1 ratio and VcSOC1 expression were associated with precocious, apical flowering and early flower bud formation (Table 2). Then we re-examined our previous RNA-seq data to investigate specifically the ratios of VcFT and VcSOC1 (Table 2). Interestingly, in the other four cases: 1) in transgenic “Aurora” containing an overexpressed VcSOC1 K domain, VcFT showed no differential expression and VcSOC1 was an upregulated DEG. The VcFT/VcSOC1 ratio increased but not significantly. Phenotypically, the transgenic plants showed precocious, apical flowering and promoted flower bud formation; 2) in non-transgenic “Legacy” grafted on transgenic VcFT-OX “Aurora” flower bud formation was promoted in “Legacy” where there was a non-significant increase in the VcFT/VcSOC1 ratio associated with the upregulated DETs for both VcFT and VcSOC1. In this case, whether or not there was precocious, apical flowering was not tested; 3) in Legacy-mutant1, a transgenic “Legacy” containing an overexpressed blueberry DWARF AND DELAYED FLOWERING 1 (VcDDF1) and a constitutively expressed type-B RESPONSE REGULATOR 2-LIKE gene (VcRR2), there was a significant increase of the VcFT/VcSOC1 ratio was associated with no DEGs for both VcFT and VcSOC1, we found promoted flower bud formation; and 4) in Legacy-mutant2, a derivative from a seedling of the self-pollinated Legacy-mutant1, there was a non-significant change in the VcFT/VcSOC1 ratio associated with upregulated DETs for VcSOC1, precocious flowering was observed. In summary: in leaves, increased VcFT/VcSOC1 ratios (five out the six cases, in which three had significant increases and three had non-significant changes with two increase and one decrease) tended to promote flower bud formation or precocious flowering; and increased VcSOC1 expression was likely associated with precocious flowering (four out of five cases) (Table 2).
TABLE 2.
Summary of RNA-seq analysis of VcFT and VcSOC1 expression in different tissues of different genotypes. The ratios of VcFT/VcSOC1 were calculated based on all the transcript reads for each individual gene. There were three biological replicates for each tissue. Statistical analysis was conducted for each tissue in pair comparison, separately. Leaf: developing leaves. Bud: flower bud.
| Tissue | Genotype | VcFT expression | VcSOC1 expression | VcFT/VcSOC1 | p-value | Phenotypic changes | References |
|---|---|---|---|---|---|---|---|
| Leaf | Aurora | 0.0008 | |||||
| Leaf | VcFT-OX Aurora | DET, increased | DET, increased | 1.4294 | 9.41e-05 | 1, 2 | Walworth et al. (2016) |
| Mature Leaf | Aurora | 0.0074 | |||||
| Mature Leaf | VcFT-OX Aurora | DET, increased | DET, decreased | 0.6897 | 0.002 | 1, 2 | This study |
| Leaf | Aurora | 0.0008 | |||||
| Leaf | SOC1K-OX Aurora | non-DET | DET, increased | 0.0011 | 0.721 | 1, 2 | Song and Chen (2018a) |
| Leaf | Legacy | 0 | |||||
| Leaf | Legacy_mutant1 | non-DET | non-DET | 0.0007 | 0.004 | 2 | Song and Walworth (2018) |
| Leaf | Legacy | 0.0035 | |||||
| Leaf | Legacy_mutant2 | non-DET | DET, increased | 0 | 0.091 | 1 | Lin et al. (2019b) |
| Leaf | Legacy | 0.0001 | |||||
| Leaf | Legacy on VcFT-OX Aurora | DET, increased | DET, increased | 0.0004 | 0.592 | 2 | Song et al. (2019) |
| Bud | Aurora | 0.0703 | |||||
| Bud | VcFT-OX Aurora | DET, increased | DET, decreased | 0.1469 | 0.007 | 2 | Unpublished data |
| Bud | Aurora | 0.0443 | This study | ||||
| Bud | VcFT-OX Aurora | DET, increased | DET, decreased | 0.1080 | 0.004 | 2 | |
| Bud | Aurora | 0.3756 | |||||
| Bud | Chilled Aurora | non-DET | DET, increased | 0.1979 | 0.002 | 3 | This study |
| Bud | VcFT-OX Aurora | 0.1080 | |||||
| Bud | Chilled VcFT-OX Aurora | DET, decreased | non-DET | 0.0766 | 0.058 | 3 | This study |
| Bud | Legacy | 0.2429 | |||||
| Bud | Legacy_mutant1 | non-DET | non-DET | 0.3497 | 0.090 | 3 | Song and Walworth (2018) |
| Bud | Legacy | 0.1830 | |||||
| Bud | Chilled Legacy | non-DET | DET, increased | 0.1861 | 0.9195 | 3 | Song and Chen (2018b) |
| Bud | Legacy_mutant1 | 0.1952 | |||||
| Bud | Chilled Legacy_mutant1 | DET, decreased | DET, increased | 0.1142 | 0.003 | 3 | Song and Walworth (2018) |
| Late-pink bud | Chilled Legacy_mutant1 | DET, decreased | DET, decreased | 0.6477 | |||
| Bud | Chilled Legacy_mutant1 | 0.1747 | 0.095 | 4 | Lin et al. (2019b) | ||
| Late-pink bud | Chilled Legacy | DET, decreased | DET, decreased | 0.4895 | |||
| Bud | Chilled Legacy | 0.1326 | 0.001 | 4 | Song and Chen (2018b) |
1: Promoted precocious flowering; 2: Promoted flower bud formation; 3: Promoted flower bud breaking; 4: Flowering.
When the chilled flower buds of four genotypes (non-transgenic “Aurora”, VcFT-OX “Aurora”, “Legacy” and Legacy-mutant1) were compared to non-chilled flower buds after receiving full chilling hours, VcFT showed as a downregulated DEG in two genotypes and as a non-DEG in the other two; upregulated DEGs of VcSOC1 were found in three genotypes and the fourth one was a non-DEG. Three of the four genotypes showed decreased VcFT/VcSOC1 ratios, and only “Legacy” had minimal change in the ratio with the increased VcSOC1 DEG and the non-DE VcFT indicating a decreasing VcFT/VcSOC1 ratio (Table 2). The breaking flower buds at late-pink bud stage for two genotypes tested, compared to the chilled flower buds after full chilling hours, had reduced expression for both VcFT and VcSOC1, of which the more rapidly decreased VcSOC1 contributed to the increased VcFT/VcSOC1 ratio (Table 2).
In non-chilled flower buds, VcFT-OX “Aurora” (vs. non-transgenic “Aurora”) showed an increased VcFT/VcSOC1 ratio with an increased VcFT expression and a decreased VcSOC1 expression (Table 2). This facilitated the formation of endodormant buds, which were able to be broken after sufficient chilling hours repressed the expression of VcFT. Non-chilled flower buds of the Legacy-mutant1 (vs. “Legacy”) exhibited promoted flower bud formation and decreased chilling requirement (Song and Walworth, 2018), however, we did not see increased VcSOC1 expression or a reduced VcFT/VcSOC1 ratio likely due to hormone genes (Table 2) (Lin et al., 2019b).
Taken together, in leaves a high VcFT/VcSOC1 promoted floral initiation and a high VcSOC1 expression could cause precocious flowering. In flower buds, chilled flower buds often had lower VcFT/VcSOC1 ratios due to the increased VcSOC1 expression during the accumulation chilling hours; breaking flower buds had increasing VcFT/VcSOC1 ratios due to a faster decrease in VcSOC1 expression than VcFT. Besides the flowering pathway genes, there exist other pathway genes that can affect floral initiation or floral activation, for example, the altered flowering of the Legacy-mutant1 was not caused by major flowering pathway genes (Song and Walworth, 2018).
4 Discussions
To investigate VcFT roles in controlling flowering in blueberry, we conducted transcriptome analysis of VcFT-OX and its control of non-transgenic “Aurora”. We identified, for the first time, the DETs and DEGs of non-chilled vs. fully chilled buds caused by an overexpressed VcFT gene. RNA sequencing of the non-chilled transgenic flower buds was done twice, including one of a selected representative line from a previous study (not published) and one of pooled samples from six transgenic lines in the current study (Table 1). Results from both studies were similar, providing an assurance of the quality of data presented in this study (Figures 2B, C). The results are invaluable to understand the overall impact and the multifunctional roles of FT expression on blueberry flowering. In this study, we also presented transcriptome data to compare non-chilled and chilled (with full chilling hours) buds in non-transgenic “Aurora” and VcFT-OX (transgenic “Aurora”) to study chilling-mediated flowering (Figures 3B, C). The new data from VcFT-OX “Aurora” allows us to reveal the impact of VcFT-regulated and chilling-mediated flowering simultaneously.
DAM1-6 were identified and named based on a study of the evergrowing locus in peach (Bielenberg et al., 2008). We identified blueberry orthologues of DAM1, DAM2, and DAM5 (E-20 as the cut-off). These orthologues can also be annotated to some specific MADS-box genes based on their annotations in Arabidopsis. For example, DAM2 showed high similarity to the annotated VcSVP, VcSOC1, VcFUL, and VcAP1. To minimize the confusion, we did not use DAMs to refer the blueberry MADS-box genes in this report.
4.1 FT is a main floral inducer
FT is a major integrator in plant flowering pathways (Fornara et al., 2010). It is the top candidate to be florigen (Turck et al., 2008; Turnbull, 2011). There is a long list of reports describing that constitutive expression of FT or its orthologues resulted in precocious flowering and promoted flower bud formation (e.g., apple, poplar, plum, cassava, and blueberry in woody plants) (Zhang et al., 2010; Srinivasan et al., 2012; Song et al., 2013b; Wenzel et al., 2013; Bull et al., 2017; Voogd et al., 2017). However, with the exception of blueberry, it has not been reported that mature flower buds in those precociously flowering woody plants still require chilling to bloom (Walworth et al., 2016). Recent genetic studies have demonstrated that VcFT, VcCOL5 (blueberry CONSTANS-LIKE 5), and VcTFL1 are major flowering regulators in blueberry (Gaire and Wilde, 2018; Omori et al., 2020; Omori et al., 2022). Increased expression of VcFT with decreased VcCOL5 is responsible for the off-season apical flowering of Rabbiteye blueberries (Vaccinium virgatum Aiton) (Omori et al., 2022). A question remains as to why mature flower buds in FT-OX woody plants had a chilling requirement to break their endodormancy. Our transcriptome data analysis of flower buds suggested that decreased VcSOC1 expression in the VcFT-OX flower buds would likely play a major role in forcing mature buds into endodormancy. It sounds contradictory to have precocious flowering occur before the formation of mature buds, but actually, because the observed precocious flowering took place in young buds instead of mature ones, it is not. At the transcriptome level, the developing buds undergoing precocious flowering behaved more like leaves than mature buds. As shown in the transcriptome comparisons (Figure 2; Table 1), VcFT-OX promoted the expression of VcSOC1 in leaves. We believe it was the upregulated VcSOC1 that was responsible for the observed precocious flowering in VcFT-OX. Another piece of evidence to support this is that an overexpressed VcSOC1 K-domain promoted precocious flowering, but had no increase in expression of VcFT (Table 1) (Song and Chen, 2018a) as the overexpressed VcSOC1 K-domain is a truncated VcSOC1 (Song et al., 2013a; Song and Chen, 2018a). Another interesting observation is that VcFT does not respond to chilling (Song and Chen, 2018b). This is deviates from what was reported in kiwifruit in which at least one FT was activated after cold accumulation and dormancy release (Voogd et al., 2017).
4.2 SOC1 is a major floral activator
Another main integrator in the flowering pathway, SOC1 is a downstream gene of FT (Fornara et al., 2010; Lee and Lee, 2010). As shown in this study, VcFT overexpression upregulated VcSOC1 expression in leaves but repressed its expression in mature flower buds (Figure 2). On the other hand, in both “Legacy” and “Aurora”, VcFT expression showed little changes in chilled flower buds when compared to non-chilled buds, and in both cases VcSOC1 expression was upregulated (Table 1). During the process of chilling hour accumulation, VcSOC1 expression gradually increases until the chilled buds begin breaking (Figure 4C). Combined with our previous data showing downregulated VcSOC1 expression in late-pink bud (Song and Chen, 2018b), we believe that VcSOC1 expression is a major floral activator in chilling-mediated blueberry flowering. In annual crops, we recently demonstrated that constitutive expression of a maize SOC1 gene promoted flowering in both maize and soybean (Han et al., 2021; Song et al., 2021). In fruit crops, kiwifruit SOC1-like genes may affect the duration of dormancy although they may not have a role in the floral transition (Voogd et al., 2015). In grapevine, chilling hour accumulation induced expression of its SOC1 (Kamal et al., 2019). In poplar, overexpression of a SOC1-like gene promoted bud break (Gómez-Soto et al., 2021). Taken together, SOC1-like genes are up-regulated during chilling hour accumulation.
4.3 VcFT and VcSOC1 expression ratios in floral initiation and activation in blueberry flowering
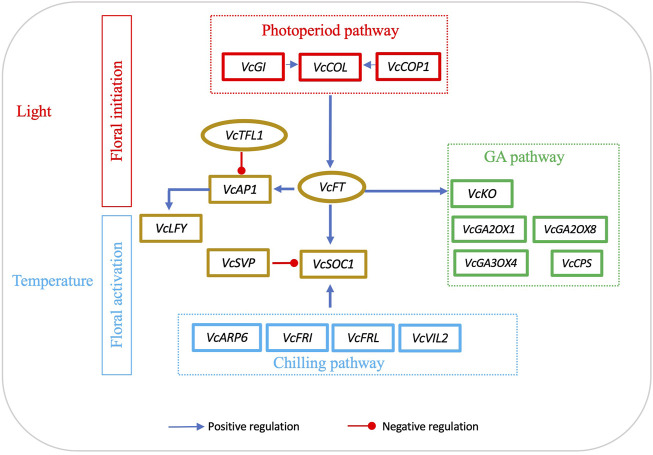
In general, FT expression in leaves is affected by light and SOC1 expression is responsive to temperatures. To date, we have not seen any attempts to use FT/SOC1 expression ratio as a parameter to interpret flowering activities. In blueberry, VcFT has its highest expression level in floral buds, while VcSOC1 and VcSVP have their highest expression in leaves (Walworth et al., 2016). Based on the data presented in this study (Table 1), we believe that using FT/SOC1 ratios could facilitate an understanding of floral initiation and activation. Specifically, we think that, in leaves, an increased FT/SOC1 ratio promotes flower bud formation; however, in mature flower buds, an increased FT/SOC1 ratio makes the buds remain in endodormancy while a decreased FT/SOC1 ratio promotes dormancy release. While this statement is not a perfect explanation of all the data in Table 1, it fits most of them. For additional support, a reduced FT/SOC1 ratio was observed in polar buds during chilling accumulation. This was caused by an increase in SOC1 while FT remained neutral (Gómez-Soto et al., 2021). We believe that this FT/SOC1 ratio can be a determinator of floral initiation in leaves and of floral activation in buds because both genes are conserved integrators in the flowering pathway. As shown in the proposed diagram, light regulates VcCO and VcFT expression in leaves for floral bud initiation; temperature, especially low temperature, regulates VcSOC1 expression in buds for budbreak (Figure 5).
FIGURE 5.
Interactions of FT-mediated floral initiation and SOC1-regulated floral activation in blueberry. Floral initiation signals are mainly produced in leaves. The diagram was drawn based on the data sets in Table 2. The genes listed are the major differentially expressed genes induced by either VcFT overexpression or chilling in blueberry. The positive or negative regulation sign is based on the information from Arabidopsis, but it may not match the results obtained in this study.
While we mainly focused on the roles of VcFT and VcSOC1, there were many other important DEGs in the flowering pathway, inducing ARP6, LFY, and MADS-box genes (e.g., AG1, AGL6, AGL11, J and AGL12) (Figure 5). The roles of these DEGs in blueberry flowering under chilled and non-chilled conditions remain to be analyzed and interpreted.
4.4 Interaction of SOC1, SVP, and other MADS-box genes
SVP_ARATH is a MADS-box gene which controls the identity of the floral meristem by interacting with two other MADS-box genes AGAMOUS-LIKE 24 (AGL24) and APETALA 1 (AP1) (Gregis et al., 2006; 2008; Gregis et al., 2009; Yan et al., 2014). In annual plants, SVP, independent of photoperiod and temperature, inhibits floral transition in the autonomous flowering pathway and promotes EARLY FLOWERING MYB PROTEIN (EFM) expression to suppress flowering (Gregis et al., 2008; Fornara et al., 2010; Yan et al., 2014). In the other words, SVP was considered as a repressor in chilling-promoted flowering (Li et al., 2010; Wu et al., 2017a; Wu et al., 2017b; Arro et al., 2019; Wang et al., 2021; Dong et al., 2022). In woody plants, SVP homologues were found to be either a positive or negative regulator in flowering depending on plant species and homolog (Diaz-Riquelme et al., 2012; Li-Mallet et al., 2016; Wu et al., 2017a; Wu et al., 2017b; Arro et al., 2019; Kamal et al., 2019; Wang et al., 2021; Dong et al., 2022). The results of this study using northern highbush blueberry ‘Aurora’ are consistent with that from previous reports in a southern highbush blueberry “Legacy”, confirming that chilling accumulation promotes expression of VcSVP (Song and Chen, 2018b). Meanwhile, in the flower tissues (e.g., the late pink buds), VcSVP as well as the other major flowering pathway genes (e.g., VcFT, VcSOC1, and VcAP1) were all downregulated (Song and Chen, 2018b). Based on transcriptome comparisons in blueberry, VcSOC1 and VcSVP expressions promoted floral activation through chilling hour accumulation. In the case of the VcFT-OX “Aurora”, an increased VcSVP expression in chilled flower buds seemed to be the key to enable budbreak suggesting that VcSOC1 and VcSVP may function similarly in chilling-mediated floral activation. VcAGL14 (MADS6_ORYSJ) was repressed during chilling accumulation in both non-transgenic “Aurora” and VcFT-OX “Aurora”, but this is contrary to that of in “Legacy” and Mu-Legacy bud (Song and Chen, 2018b).
4.5 Other flowering pathway genes and hormone and sugar pathway genes
In this report, the profiles of DETs and DEGs identified in VcFT-OX and chilled flower buds provide a lot of information that can be used for the interpretation of different pathways, of which we focused mainly on the major flowering pathway genes. Even with that, there remains information on other important DEGs in the flowering pathway to be explained (e.g., ARP6, LFY, and MADS-box genes AGL8, SVP, MADS6, and J).
In addition to the flowering pathway genes, we identified the of DETs and DEGs in hormone pathways and sugar pathways, indicating they were involved, either directly or indirectly, in floral bud initiation or floral activation. Due to the volume of the stacked information, we would interpret these pathways in this report.
5 Conclusion
The transcriptome data generated in this study allowed us to develop new profiles of DETs caused by VcFT overexpression and full chilling, respectively, in blueberry flower buds. These DETs provide invaluable information to reveal the genes associated with flower bud formation and chilling-mediated flower bud breaking in other woody plants. The overall analyses revealed that, in the flowering pathway, VcFT expression in leaves is the major floral initiator and VcSOC1 and VcSVP expression in buds is the key to floral activation. More importantly, the ratio of FT to SOC1 plays a significant role in flower bud formation and chilling-mediated bud breaking. An increased VcFT/VcSOC1 ratio with decreased VcSOC1 expression in buds could induce bud endodormancy. After full chilling, the decreased VcFT/VcSOC1 ratio, due mostly to higher VcSOC1 expression, contributed to an increase in readiness for flower bud breaking. During flower bud breaking, a decreasing VcFT/VcSOC1 ratio occurred because of a more rapid decrease in VcSOC1 expression than VcFT. The results indicate that the proposed FT/SOC1 ratio concept/parameter can facilitate a better understanding of their roles and interactions in floral initiation and activation.
Funding Statement
This research was partially supported by AgBioResearch of Michigan State University (http://agbioresearch.msu.edu/programs/info/project_greeen) and by the agreement of Non-Assistance Cooperative Agreement #58-8060-6-009 between the USDA Agricultural Research.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
G-QS conceived and supervised the study; G-QS conducted the experiments; G-QS, BC, and G-YZ analyzed the data; GS and G-YZ wrote the manuscript. All authors read and approved the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1105519/full#supplementary-material
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 (5737), 1052–1056. [pii]. 10.1126/science.1115983 [DOI] [PubMed] [Google Scholar]
- Adeyemo O. S., Chavarriaga P., Tohme J., Fregene M., Davis S. J., Setter T. L. (2017). Overexpression of Arabidopsis FLOWERING LOCUS T (FT) gene improves floral development in cassava (Manihot esculenta, Crantz). PLoS One 12 (7), e0181460. 10.1371/journal.pone.0181460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amasino R. (2010). Seasonal and developmental timing of flowering. Plant J. 61 (6), 1001–1013. TPJ4148 [pii]. 10.1111/j.1365-313X.2010.04148.x [DOI] [PubMed] [Google Scholar]
- Arro J., Yang Y., Song G.-Q., Zhong G.-Y. (2019). RNA-Seq reveals new DELLA targets and regulation in transgenic GA-insensitive grapevines. BMC Plant Biol. 19 (1), 80. 10.1186/s12870-019-1675-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson C. J., Brennan R. M., Jones H. G. (2013). Declining chilling and its impact on temperate perennial crops. Environ. Exp. Bot. 91, 48–62. 10.1016/j.envexpbot.2013.02.004 [DOI] [Google Scholar]
- Bielenberg D. G., Wang Y., Li Z. G., Zhebentyayeva T., Fan S. H., Reighard G. L., et al. (2008). Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet. Genomes 4 (3), 495–507. 10.1007/s11295-007-0126-9 [DOI] [Google Scholar]
- Bull S. E., Alder A., Barsan C., Kohler M., Hennig L., Gruissem W., et al. (2017). FLOWERING LOCUS T triggers early and fertile flowering in glasshouse cassava (manihot esculenta Crantz). Plants (Basel) 6 (2), 22. 10.3390/plants6020022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle A., Grimplet J., Le Dantec L., Wunsch A. (2021). Identification and characterization of DAMs mutations associated with early blooming in sweet cherry, and validation of DNA-based markers for selection. Front. Plant Sci. 12, 621491. 10.3389/fpls.2021.621491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuine I. C., Bonhomme M., Legave J.-M., De Cortázar-atauri I., Charrier G., Lacointe A., et al. (2016). Can phenological models predict tree phenology accurately in the future? The unrevealed hurdle of endodormancy break. Glob. Change Biol. 22, 3444–3460. 10.1111/gcb.13383 [DOI] [PubMed] [Google Scholar]
- Cockram J., Jones H., Leigh F. J., O'Sullivan D., Powell W., Laurie D. A., et al. (2007). Control of flowering time in temperate cereals: Genes, domestication, and sustainable productivity. J. Exp. Bot. 58 (6), 1231–1244. 10.1093/jxb/erm042 [DOI] [PubMed] [Google Scholar]
- Diaz-Riquelme J., Grimplet J., Martinez-Zapater J. M., Carmona M. J. (2012). Transcriptome variation along bud development in grapevine (Vitis vinifera L.). BMC Plant Biol. 12, 181. 10.1186/1471-2229-12-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Khalil-Ur-Rehman M., Liu X., Wang X., Yang L., Tao J., et al. (2022). Functional characterisation of five SVP genes in grape bud dormancy and flowering. Plant Growth Regul. 97 (3), 511–522. 10.1007/s10725-022-00817-w [DOI] [Google Scholar]
- Ferrandiz C., Gu Q., Martienssen R., Yanofsky M. F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127 (4), 725–734. 10.1242/dev.127.4.725 [DOI] [PubMed] [Google Scholar]
- Fornara F., de Montaigu A., Coupland G. (2010). SnapShot: Control of flowering in Arabidopsis. Cell. 141 (3), 550, 550.e1-2. 10.1016/j.cell.2010.04.024 [DOI] [PubMed] [Google Scholar]
- Gaire R., Wilde H. D. (2018). Natural allelic variation in blueberry TERMINAL FLOWER 1. Plant Genet. Resources-Characterization Util. 16 (1), 59–67. 10.1017/s1479262116000435 [DOI] [Google Scholar]
- Gao X., Walworth A. E., Mackie C., Song G. Q. (2016). Overexpression of blueberry FLOWERING LOCUS T is associated with changes in the expression of phytohormone-related genes in blueberry plants. Hortic. Res. 3, 16053. 10.1038/hortres.2016.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Soto D., Ramos-Sánchez J. M., Alique D., Conde D., Triozzi P. M., Perales M., et al. (2021). Overexpression of a SOC1-related gene promotes bud break in ecodormant poplars. Front. Plant Sci. 12, 670497. 10.3389/fpls.2021.670497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goralogia G. S., Howe G. T., Brunner A. M., Helliwell E., Nagle M. F., Ma C., et al. (2021). Overexpression of SHORT VEGETATIVE PHASE-LIKE (SVL) in Populus delays onset and reduces abundance of flowering in field-grown trees. Hortic. Res. 8 (1), 167. 10.1038/s41438-021-00600-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenup A., Peacock W. J., Dennis E. S., Trevaskis B. (2009). The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann. Bot. 103 (8), 1165–1172. mcp063 [pii]. 10.1093/aob/mcp063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V., Sessa A., Colombo L., Kater M. M. (2006). AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. Plant Cell. 18 (6), 1373–1382. 10.1105/tpc.106.041798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V., Sessa A., Colombo L., Kater M. M. (2008). AGAMOUS-LIKE24 and SHORT VEGETATIVE PHASE determine floral meristem identity in Arabidopsis. Plant J. 56 (6), 891–902. 10.1111/j.1365-313X.2008.03648.x [DOI] [PubMed] [Google Scholar]
- Gregis V., Sessa A., Dorca-Fornell C., Kater M. M. (2009). The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 60 (4), 626–637. 10.1111/j.1365-313X.2009.03985.x [DOI] [PubMed] [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8(8), 1494–1512. 10.1038/Nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Wang D. C., Song G. Q. (2021). Expression of a maize SOC1 gene enhances soybean yield potential through modulating plant growth and flowering. Sci. Rep. 11 (1), 12758. ARTN 12758. 10.1038/s41598-021-92215-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano S., Goto K. (2011). Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell. 23 (9), 3172–3184. 10.1105/tpc.111.088641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C. A., Sheldon C. C., Olive M. R., Walker A. R., Zeevaart J. A., Peacock W. J., et al. (1998). Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc. Natl. Acad. Sci. U. S. A. 95 (15), 9019–9024. 10.1073/pnas.95.15.9019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J. A., Bailey P. C., Laurie D. A. (2010). Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS One 5 (4), e10065. 10.1371/journal.pone.0010065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P., Schmid M. (2011). The control of developmental phase transitions in plants. Development 138 (19), 4117–4129. 10.1242/dev.063511 [DOI] [PubMed] [Google Scholar]
- Jameson P. E., Clemens J. (2015). Phase change and flowering in woody plants of the New Zealand flora. J. Exp. Bot. 70, e6488–e6495. 10.1093/jxb/erv472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewaria P. K., Hanninen H., Li X., Bhalerao R. P., Zhang R. (2021). A hundred years after: Endodormancy and the chilling requirement in subtropical trees. New Phytol. 231 (2), 565–570. 10.1111/nph.17382 [DOI] [PubMed] [Google Scholar]
- Jia T. Q., Wei D. F., Meng S., Allan A. C., Zeng L. H. (2014). Identification of regulatory genes implicated in continuous flowering of longan (dimocarpus longan L.). Plos One 9 (12), e114568. ARTN e114568. 10.1371/journal.pone.0114568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal N., Ochßner I., Schwandner A., Viehöver P., Hausmann L., Töpfer R., et al. (2019). Characterization of genes and alleles involved in the control of flowering time in grapevine. PLoS One 14 (7), e0214703. 10.1371/journal.pone.0214703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klocko A. L., Ma C., Robertson S., Esfandiari E., Nilsson O., Strauss S. H. (2016). FT overexpression induces precocious flowering and normal reproductive development in Eucalyptus. Plant Biotechnol. J. 14 (2), 808–819. 10.1111/pbi.12431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 (5446), 1960–1962. 10.1126/science.286.5446.1960 [DOI] [PubMed] [Google Scholar]
- Krzymuski M., Andres F., Cagnola J. I., Jang S., Yanovsky M. J., Coupland G., et al. (2015). The dynamics of FLOWERING LOCUS T expression encodes long-day information. Plant J. 83 (6), 952–961. 10.1111/tpj.12938 [DOI] [PubMed] [Google Scholar]
- Lee J., Lee I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61 (9), 2247–2254. erq098 [pii]. 10.1093/jxb/erq098 [DOI] [PubMed] [Google Scholar]
- Li Z.-M., Zhang J.-Z., Mei L., Deng X.-X., Hu C.-G., Yao J.-L. (2010). PtSVP, an SVP homolog from trifoliate orange (Poncirus trifoliata L. Raf.), shows seasonal periodicity of meristem determination and affects flower development in transgenic Arabidopsis and tobacco plants. Plant Mol. Biol. 74 (1), 129–142. 10.1007/s11103-010-9660-1 [DOI] [PubMed] [Google Scholar]
- Li-Mallet A., Rabot A., Geny L. (2016). Factors controlling inflorescence primordia formation of grapevine: Their role in latent bud fruitfulness? A review. Botany 94 (3), 147–163. 10.1139/cjb-2015-0108 [DOI] [Google Scholar]
- Lin T. Y., Chen Q. X., Wichenheiser R. Z., Song G. Q. (2019a). Constitutive expression of a blueberry FLOWERING LOCUS T gene hastens petunia plant flowering. Sci. Hortic. 253, 376–381. 10.1016/j.scienta.2019.04.051 [DOI] [Google Scholar]
- Lin T. Y., Walworth A., Zong X. J., Danial G. H., Tomaszewski E. M., Callow P., et al. (2019b). VcRR2 regulates chilling-mediated flowering through expression of hormone genes in a transgenic blueberry mutant. Hortic. Res. 6, 96. ARTN 96. 10.1038/s41438-019-0180-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedeling E., Girvetz E. H., Semenov M. A., Brown P. H. (2011). Climate change affects winter chill for temperate fruit and nut trees. PLoS One 6 (5), e20155. 10.1371/journal.pone.0020155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsoukas I. G., Massiah A. J., Thomas B. (2012). Florigenic and antiflorigenic signaling in plants. Plant Cell. Physiol. 53 (11), 1827–1842. 10.1093/pcp/pcs130 [DOI] [PubMed] [Google Scholar]
- Michaels S. D., Amasino R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. PLANT Cell. 11 (5), 949–956. 10.1105/tpc.11.5.949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D. (2009). Flowering time regulation produces much fruit. Curr. Opin. Plant Biol. 12 (1), 75–80. S1369-5266(08)00161-1 [pii]. 10.1016/j.pbi.2008.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori M., Yamane H., Li K.-T., Matsuzaki R., Ebihara S., Li T.-S., et al. (2020). Expressional analysis of FT and CEN genes in a continuously flowering highbush blueberry 'Blue Muffin. Acta Hortic. 1280, 197–202. 10.17660/ActaHortic.2020.1280.27 [DOI] [Google Scholar]
- Omori M., Cheng C.-C., Hsu F.-C., Chen S.-J., Yamane H., Tao R., et al. (2022). Off-season flowering and expression of flowering-related genes during floral bud differentiation of rabbiteye blueberry in a subtropical climate. Sci. Hortic. 306, 111458. 10.1016/j.scienta.2022.111458 [DOI] [Google Scholar]
- Parmesan C., Yohe G. (2003). A globally coherent fingerprint of climate change impacts across natural systems. Nature 421 (6918), 37–42. 10.1038/nature01286 [DOI] [PubMed] [Google Scholar]
- Pin P. A., Benlloch R., Bonnet D., Wremerth-Weich E., Kraft T., Gielen J. J. L., et al. (2010). An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330 (6009), 1397–1400. 10.1126/science.1197004 [DOI] [PubMed] [Google Scholar]
- Root T. L., Price J. T., Hall K. R., Schneider S. H., Rosenzweig C., Pounds J. A. (2003). Fingerprints of global warming on wild animals and plants. Nature 421 (6918), 57–60. 10.1038/nature01333 [DOI] [PubMed] [Google Scholar]
- Saure M. C. (1985). Dormancy release in deciduous fruit trees. Hortic. Rev. 7, 239–300. [Google Scholar]
- Song G.-Q., Chen Q. (2018a). Overexpression of the MADS-box gene K-domain increases the yield potential of blueberry. Plant Sci. 276, 22–31. 10.1016/j.plantsci.2018.07.018 [DOI] [PubMed] [Google Scholar]
- Song G. Q., Chen Q. (2018b). Comparative transcriptome analysis of nonchilled, chilled, and late-pink bud reveals flowering pathway genes involved in chilling-mediated flowering in blueberry. BMC Plant Biol. 18 (1), 98. 10.1186/s12870-018-1311-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G.-Q., Walworth A. (2018). An invaluable transgenic blueberry for studying chilling-induced flowering in woody plants. BMC Plant Biol. 18, 265. 10.1186/s12870-018-1494-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G. Q., Walworth A., Zhao D. Y., Hildebrandt B., Leasia M. (2013a). Constitutive expression of the K-domain of a Vaccinium corymbosum SOC1-like (VcSOC1-K) MADS-box gene is sufficient to promote flowering in tobacco. Plant Cell. Rep. 32(11), 1819–1826. 10.1007/S00299-013-1495-1 [DOI] [PubMed] [Google Scholar]
- Song G. Q., Walworth A., Zhao D. Y., Jiang N., Hancock J. F. (2013b). The vaccinium corymbosum FLOWERING LOCUS T-like gene (VcFT): A flowering activator reverses photoperiodic and chilling requirements in blueberry. Plant Cell. Rep. 32(11), 1759–1769. 10.1007/S00299-013-1489-Z [DOI] [PubMed] [Google Scholar]
- Song Y. H., Shim J. S., Kinmonth-Schultz H. A., Imaizumi T. (2015). Photoperiodic flowering: Time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66, 441–464. 10.1146/annurev-arplant-043014-115555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G. Q., Walworth A., Lin T., Chen Q., Han X., Irina Zaharia L., et al. (2019). VcFT-induced mobile florigenic signals in transgenic and transgrafted blueberries. Hortic. Res. 6, 105. 10.1038/s41438-019-0188-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G. Q., Han X., Ryner J. T., Thompson A., Wang K. (2021). Utilizing MIKC-type MADS-box protein SOC1 for yield potential enhancement in maize. Plant Cell. Rep. 40 (9), 1679–1693. 10.1007/s00299-021-02722-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G.-Q. (2019). “Response of woody plants to low temperature,” in Stress physiology of woody plants. Editor Dai W. (Taylor & Francis Group; ). [Google Scholar]
- Srinivasan C., Dardick C., Callahan A., Scorza R. (2012). Plum (Prunus domestica) trees transformed with poplar FT1 result in altered architecture, dormancy requirement, and continuous flowering. PLoS One 7 (7), e40715. 10.1371/journal.pone.0040715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trankner C., Lehmann S., Hoenicka H., Hanke M. V., Fladung M., Lenhardt D., et al. (2010). Over-expression of an FT-homologous gene of apple induces early flowering in annual and perennial plants. Planta 232 (6), 1309–1324. 10.1007/s00425-010-1254-2 [DOI] [PubMed] [Google Scholar]
- Trevaskis B., Hemming M. N., Dennis E. S., Peacock W. J. (2007). The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 12 (8), 352–357. 10.1016/j.tplants.2007.06.010 [DOI] [PubMed] [Google Scholar]
- Turck F., Fornara F., Coupland G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59 (1), 573–594. 10.1146/annurev.arplant.59.032607.092755 [DOI] [PubMed] [Google Scholar]
- Turnbull C. (2011). Long-distance regulation of flowering time. J. Exp. Bot. 62 (13), 4399–4413. 10.1093/jxb/err191 [DOI] [PubMed] [Google Scholar]
- Voogd C., Wang T., Varkonyi-Gasic E. (2015). Functional and expression analyses of kiwifruit SOC1-like genes suggest that they may not have a role in the transition to flowering but may affect the duration of dormancy. J. Exp. Bot. 66 (15), 4699–4710. 10.1093/jxb/erv234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voogd C., Brian L. A., Wang T., Allan A. C., Varkonyi-Gasic E. (2017). Three FT and multiple CEN and BFT genes regulate maturity, flowering, and vegetative phenology in kiwifruit. J. Exp. Bot. 68 (7), 1539–1553. 10.1093/jxb/erx044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth A. E., Chai B., Song G. Q. (2016). Transcript profile of flowering regulatory genes in VcFT-overexpressing blueberry plants. PLoS One 11 (6), e0156993. 10.1371/journal.pone.0156993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Jiu S., Xu Y., Sabir I. A., Wang L., Ma C., et al. (2021). SVP-like gene PavSVP potentially suppressing flowering with PavSEP, PavAP1, and PavJONITLESS in sweet cherries (Prunus avium L.). Plant Physiol. Biochem. 159, 277–284. 10.1016/j.plaphy.2020.12.013 [DOI] [PubMed] [Google Scholar]
- Wellmer F., Riechmann J. L. (2010). Gene networks controlling the initiation of flower development. Trends Genet. 26 (12), 519–527. 10.1016/j.tig.2010.09.001 [DOI] [PubMed] [Google Scholar]
- Wells C. E., Vendramin E., Jimenez Tarodo S., Verde I., Bielenberg D. G. (2015). A genome-wide analysis of MADS-box genes in peach [Prunus persica (L.) Batsch]. BMC Plant Biol. 15, 41. 10.1186/s12870-015-0436-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel S., Flachowsky H., Hanke M.-V. (2013). The Fast-track breeding approach can be improved by heat-induced expression of the FLOWERING LOCUS T genes from poplar (Populus trichocarpa) in apple (Malus × domestica Borkh.). Plant Cell., Tissue Organ Cult. (PCTOC) 115 (2), 127–137. 10.1007/s11240-013-0346-7 [DOI] [Google Scholar]
- Wigge P. A., Kim M. C., Jaeger K. E., Busch W., Schmid M., Lohmann J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 (5737), 1056–1059. [pii]. 10.1126/science.1114358 [DOI] [PubMed] [Google Scholar]
- Wilkie J. D., Sedgley M., Olesen T. (2008). Regulation of floral initiation in horticultural trees. J. Exp. Bot. 59 (12), 3215–3228. ern188 [pii]. 10.1093/jxb/ern188 [DOI] [PubMed] [Google Scholar]
- Wu R., Tomes S., Karunairetnam S., Tustin S. D., Hellens R. P., Allan A. C., et al. (2017a). SVP-Like MADS box genes control dormancy and budbreak in apple. Front. Plant Sci. 8, 477. 10.3389/fpls.2017.00477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Wang T., Warren B. A. W., Allan A. C., Macknight R. C., Varkonyi-Gasic E. (2017b). Kiwifruit SVP2 gene prevents premature budbreak during dormancy. J. Exp. Bot. 68 (5), 1071–1082. 10.1093/jxb/erx014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y. Y., Shen L. S., Chen Y., Bao S. J., Thong Z. H., Yu H. (2014). A MYB-domain protein EFM mediates flowering responses to environmental cues in Arabidopsis. Dev. Cell. 30 (4), 437–448. 10.1016/j.devcel.2014.07.004 [DOI] [PubMed] [Google Scholar]
- Yu J., Conrad A. O., Decroocq V., Zhebentyayeva T., Williams D. E., Bennett D., et al. (2020). Distinctive gene expression patterns define endodormancy to ecodormancy transition in apricot and peach. Front. Plant Sci. 11, 180. 10.3389/fpls.2020.00180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni A., Pierantoni L., De Franceschi P. (2008). Total RNA extraction from strawberry tree (Arbutus unedo) and several other woodyplants. Iforest-Biogeosciences For. 1, 122–125. 10.3832/Ifor0465-0010122 [DOI] [Google Scholar]
- Zhang H., Harry D. E., Ma C., Yuceer C., Hsu C. Y., Vikram V., et al. (2010). Precocious flowering in trees: The FLOWERING LOCUS T gene as a research and breeding tool in populus. J. Exp. Bot. 61 (10), 2549–2560. 10.1093/jxb/erq092 [DOI] [PubMed] [Google Scholar]
- Zhu H., Chen P. Y., Zhong S., Dardick C., Callahan A., An Y. Q., et al. (2020a). Thermal-responsive genetic and epigenetic regulation of DAM cluster controlling dormancy and chilling requirement in peach floral buds. Hortic. Res. 7 (1), 114. 10.1038/s41438-020-0336-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Klasfeld S., Jeong C. W., Jin R., Goto K., Yamaguchi N., et al. (2020b). TERMINAL FLOWER 1-FD complex target genes and competition with FLOWERING LOCUS T. Nat. Commun. 11 (1), 5118. 10.1038/s41467-020-18782-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong X., Zhang Y., Walworth A., Tomaszewski E. M., Callow P., Zhong G. Y., et al. (2019). Constitutive expression of an apple FLC3-like gene promotes flowering in transgenic blueberry under nonchilling conditions. Int. J. Mol. Sci. 20 (11), 2775. 10.3390/ijms20112775 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.