Abstract
Objective
Classical Cox proportional hazard models tend to overestimate the event probability in a competing risk setup. Due to the lack of quantitative evaluation of competitive risk data for colon cancer (CC), the present study aims to evaluate the probability of CC-specific death and construct a nomogram to quantify survival differences among CC patients.
Methods
Data on patients diagnosed with CC between 2010 and 2015 were collected from the Surveillance, Epidemiology, and End Results Program (SEER) database. Patients were divided into a training dataset for the establishment of the model and a validation dataset to evaluate the performance the model at a ratio of 7:3. To evaluate the ability of multiple variables to predict cause-specific death in CC patients, univariate and multivariate analyses with Fine-Gray models were performed to screen the predictors of cause-specific death, and a nomogram for predicting cause-specific mortality was constructed. Then, the receiver operating characteristic (ROC) curve and the calibration curve were plotted to evaluate the prognostic performance of the nomogram.
Results
The dataset was randomly divided into a training (n = 16,655) dataset and a validation (n = 7,139) dataset at a ratio of 7:3. In the training dataset, variables including pathological subtypes of tumors, pathological grading (degree of differentiation), AJCC staging, T-staging, surgical type, lymph node surgery, chemotherapy, tumor deposits, lymph node metastasis, liver metastasis, and lung metastasis were identified as independent risk factors for cause-specific death of CC patients. Among these factors, the AJCC stage had the strongest predictive ability, and these features were used to construct the final model. In the training dataset, the consistency index (C-index) of the model was 0.848, and the areas under the receiver operating characteristic curve (AUC) at 1, 3, and 5 years was 0.852, 0.861, and 0.856, respectively. In the validation dataset, the C-index of the model was 0.847, and the AUC at 1 year, 3 years, and 5 years was 0.841, 0.862, and 0.852, respectively, indicating that this nomogram had an excellent and robust predictive performance.
Conclusion
This study can help clinical doctors make better clinical decisions and provide better support for patients with CC.
Keywords: CC, Competing risk model, SEER database, Cause-specific mortality
Introduction
Colorectal cancer proves to be one of the most common cancers and leading causes of cancer-related death worldwide [1]. The morbidity and mortality caused by colorectal cancer among men are higher than those among women [2, 3]. It is reported that approximately 20% of colorectal cancer arises in the cecum [4]. Patients with CC are more likely to be diagnosed with a more advanced stage due to the non-specific obscure symptoms at an early stage [5]. Compared with patients with other types of colorectal cancers, the prognosis of patients with CC is often poorer. In addition, epidemiological studies have revealed a declining incidence of left colon cancer (LCC) and an increased incidence of right colon cancer (RCC) [6, 7], in which the incidence of primary colon cancer has the highest increase. RCC is more likely to be exophytic than LCC and lower overall survival is seen in patients with RCC [8–10]. The two most prevalent types of RCC are CC and ACC. Both of them are believed to form in the midgut, but differences may emerge between the adenocarcinomas of them due to different origins and development processes. A recent study demonstrated that the prognosis of CC patients was worse than that of ACC patients, indicating that CC patients have to carry a heavier burden [11]. Therefore, identifying the predictor variables that affect the prognosis of CC patients is significant to help clinicians to formulate more appropriate personalized strategies for the diagnosis and treatment of patients.
It is common practice to use Cox proportional hazards models and the Kaplan–Meier estimator to determine prognostic factors for CC patients [12–14]. Considering the significance of personalized treatment, it is necessary to identify cancer-related and non-cancer-related factors affecting patient mortality, as non-cancer factors such as suicide and traffic accidents other than cancers are often reported to cause death [15, 16]. In studies investigating prognostic factors for cancer patients, non-cancer factors responsible for the mortality of patients are generally considered competitive events in the presence of which multiple endpoints coexist and compete with one another to generate competing risk data [17–19]. Due to the existence of competing risks, survival analysis targeting a single endpoint of interest will yield biased results [20–22]. In terms of traditional approaches to survival analysis such as standard survival analysis using Cox proportional hazard model, the probability of one event over time is estimated, in which the occurrence of one type of death will prevent the occurrence of the death from other factors. However, in our study, CC-specific death and death from other factors are competing events. In this scenario, the use of Cox model to analyze competing event data tends to bias the results, that is, overestimate the mortality in CC patients. However, a competing risk model can be used to investigate the predictive variables that affect the prognosis of patients with CC. What’s more, the comparison of results from competing risk models and those from conventional methods for survival analysis helps to illustrate the actual effects of multiple predictors and presents more accurate estimates of outcome probabilities.
The present study was conducted to construct a nomogram based on clinical data to predict cause-specific mortality for CC patients. Clinical information was obtained from the SEER database. The results of survival analysis to estimate the probability of cause-specific mortality over time using a Cox proportional hazards model vs a competing risk model were compared, and it was found that the Cox model produced inaccurate estimates, that is, underestimated the probability of survival in CC patients. Therefore, a competing risk model was constructed to reduce the likelihood of biased estimates. CC was identified as the clinical outcome, and non-cancer causes of death as competing risks to more accurately predict the cause-specific mortality for patients with CC.
Materials and methods
Data collection
SEER * Stat software (version 8.4.1) was used to extract the data in the SEER database (https://seer.cancer.gov/) about patients who were diagnosed with CC between 2010 and 2015. The diagnosis of CC is based on the third edition of the International Classification of Diseases Oncology (ICD-O-3). We collected demographic information such as age, marital status, gender, and race of patients, as well as clinical information related to tumor pathology, tumor differentiation level, AJCC staging (7th edition), T staging, N staging, surgical procedures, lymph node dissection, distant metastasis site surgery, radiotherapy, chemotherapy, tumor deposits, lymph node metastasis, bone metastasis, brain metastasis, liver metastasis, lung metastasis, tumor size, and patient survival time and status. Data were excluded if (1) patients had a survival time of less than one month were excluded, (2) patients were younger than 18 or older than 100 years old and information patients were involved in missing information; (3) data involving unknown causes of death (COD). As for continuous variables, patients were split into three groups by age: < 40 years old, 40 to 60 years old, and > 60 years old. They were divided into three groups by tumor size: < 30 mm, 30 to 50 mm, and > 50 mm. In the present study, CC-specific death and death from factors except CC compete with each other to deliver the event of interest.
Statistical analysis
The demographic and clinical characteristics of patients in the training and validation dataset were analyzed. The Chi-square test was utilized for describing distributional differences between two datasets. Multiple variables were screened by univariate and multivariate analysis using the Fine and Gray regression model. Variables with P < 0.05 in the univariate analysis were then included in multivariate analysis, in which factors with P < 0.05 were then used to construct the final competing risk model and nomogram. We calculated the concordance index (C-index) of the final model to evaluate its performance. Calibration plots and receiver operating characteristic (ROC) curves were used to compare predicted and observed probabilities to analyze the performance of the model. We performed statistical analyses using software R version 4.1.2 (https://www.r-project.org/). The tableone package (version 0.13.2) was employed for data description and riskRegression (2021.10.10) for conducting Fine and Gray regression analysis and establishing the competing risk model. Two-tail P value less than 0.05 was the threshold of statistical significance.
Results
Patient characteristics
Data on 23,794 patients were extracted. These patients were split into a training set (N = 16,655; 70%) and a validation set (N = 7,139; 30%). Among them, 10,992 patients died during the follow-up period (7,013 dying from CC and 3,979 dying from other causes). Patients dying from non-cancer-related causes accounted for 36.20% of the total deaths. Among all patients, those over 60 years old accounted for 95.85% of the total (n = 22,807); 13,083 patients (54.98%) were female, and 10,711 (45.02%) were male. Most patients (n = 14,173; 59.57%) had tumors with a size of fewer than three centimeters, and 22.98% (n = 5469), 30.25% (n = 30.25), 32.2% (n = 7662), and 14.57% (n = 3466) have stage I, II, III, and IV tumors, respectively. No significant difference was observed regarding the follow-up data in the training and the validation data set (Table 1).
Table 1.
Basic characteristics of patients
| Factors | Define | Train(N = 16,655) | Test(N = 7139) | All(N = 23,794) |
|---|---|---|---|---|
| Age | < 40 | 37(0.22) | 17(0.24) | 54(0.23) |
| 40 ~ 60 | 656(3.94) | 277(3.88) | 933(3.92) | |
| 60 ~ | 15,962(95.84) | 6845(95.88) | 22,807(95.85) | |
| Marriage | Married | 8862(53.21) | 3777(52.91) | 12,639(53.12) |
| Divorced | 2452(14.72) | 989(13.85) | 3441(14.46) | |
| Single | 3548(21.3) | 1546(21.66) | 5094(21.41) | |
| Other | 1793(10.77) | 827(11.58) | 2620(11.01) | |
| Race | White | 13,563(81.44) | 5828(81.64) | 19,391(81.5) |
| Black | 2121(12.73) | 901(12.62) | 3022(12.7) | |
| Other | 971(5.83) | 410(5.74) | 1381(5.8) | |
| Sex | Female | 9168(55.05) | 3915(54.84) | 13,083(54.98) |
| Male | 7487(44.95) | 3224(45.16) | 10,711(45.02) | |
| Behav | Behav1 | 10,993(66) | 4676(65.5) | 15,669(65.85) |
| Behav2 | 1880(11.29) | 744(10.42) | 2624(11.03) | |
| Behav3 | 1648(9.89) | 745(10.44) | 2393(10.06) | |
| Behav4 | 928(5.57) | 455(6.37) | 1383(5.81) | |
| Other | 1206(7.24) | 519(7.27) | 1725(7.25) | |
| Grade | I | 1311(7.87) | 532(7.45) | 1843(7.75) |
| II | 11,230(67.43) | 4808(67.35) | 16,038(67.4) | |
| III | 3324(19.96) | 1441(20.18) | 4765(20.03) | |
| IV | 790(4.74) | 358(5.01) | 1148(4.82) | |
| Stage | I | 3879(23.29) | 1590(22.27) | 5469(22.98) |
| II | 5018(30.13) | 2179(30.52) | 7197(30.25) | |
| III | 5342(32.07) | 2320(32.5) | 7662(32.2) | |
| IV | 2416(14.51) | 1050(14.71) | 3466(14.57) | |
| Tstage | T1 | 1874(11.25) | 763(10.69) | 2637(11.08) |
| T2 | 2808(16.86) | 1194(16.73) | 4002(16.82) | |
| T3 | 8313(49.91) | 3618(50.68) | 11,931(50.14) | |
| T4 | 3660(21.98) | 1564(21.91) | 5224(21.96) | |
| Nstage | N0 | 9211(55.3) | 3915(54.84) | 13,126(55.17) |
| N1 | 4012(24.09) | 1772(24.82) | 5784(24.31) | |
| N2 | 3432(20.61) | 1452(20.34) | 4884(20.53) | |
| Surgery | Surg1 | 13,275(79.71) | 5751(80.56) | 19,026(79.96) |
| Surg2 | 2777(16.67) | 1122(15.72) | 3899(16.39) | |
| Surg3 | 191(1.15) | 88(1.23) | 279(1.17) | |
| Surg4 | 412(2.47) | 178(2.49) | 590(2.48) | |
| LNSur | 4 ~ | 15,845(95.14) | 6838(95.78) | 22,683(95.33) |
| 1 ~ 3 | 490(2.94) | 172(2.41) | 662(2.78) | |
| None | 320(1.92) | 129(1.81) | 449(1.89) | |
| SurgOth | None | 15,446(92.74) | 6621(92.74) | 22,067(92.74) |
| Yes | 1209(7.26) | 518(7.26) | 1727(7.26) | |
| Radiation | None | 16,416(98.56) | 7020(98.33) | 23,436(98.5) |
| Yes | 239(1.44) | 119(1.67) | 358(1.5) | |
| Chemotherapy | None | 5657(33.97) | 2386(33.42) | 8043(33.8) |
| Yes | 10,998(66.03) | 4753(66.58) | 15,751(66.2) | |
| Deposits | None | 4339(26.05) | 1892(26.5) | 6231(26.19) |
| Yes | 12,316(73.95) | 5247(73.5) | 17,563(73.81) | |
| LnPositive | No | 9179(55.11) | 3954(55.39) | 13,133(55.19) |
| 1 ~ 3 Positive | 3705(22.25) | 1586(22.22) | 5291(22.24) | |
| 4 ~ 6 Positive | 1556(9.34) | 676(9.47) | 2232(9.38) | |
| > 7 Positive | 2215(13.3) | 923(12.93) | 3138(13.19) | |
| Bone | None | 16,585(99.58) | 7108(99.57) | 23,693(99.58) |
| Yes | 70(0.42) | 31(0.43) | 101(0.42) | |
| Brain | None | 16,636(99.89) | 7133(99.92) | 23,769(99.89) |
| Yes | 19(0.11) | 6(0.08) | 25(0.11) | |
| Liver | None | 15,064(90.45) | 6441(90.22) | 21,505(90.38) |
| Yes | 1591(9.55) | 698(9.78) | 2289(9.62) | |
| Lung | None | 16,314(97.95) | 6973(97.67) | 23,287(97.87) |
| Yes | 341(2.05) | 166(2.33) | 507(2.13) | |
| Size | < 3 cm | 9941(59.69) | 4232(59.28) | 14,173(59.57) |
| 3 ~ 5 cm | 2937(17.63) | 1289(18.06) | 4226(17.76) | |
| > 5 cm | 3777(22.68) | 1618(22.66) | 5395(22.67) |
(1) Disease classification: Behav1-adenocarcinoma, NOS; Behav2-adenocarcinoma in tubulovillous adenoma; Behav3-mucinous adenocarcinoma; Behav4-adenocarcinoma in adenomatous poly; (2) Operation types: Sugery1-subtotal colectomy/hemicolectomy (partial colectomy but less than total colectomy, right or left colectomy (resection of left or right colon and partial transverse colon), or additional resection of other organs); Surgery2-partial colectomy but less than hemicolectomy or additional resection of adjacent organs; Surgery3 -no operation or tumor destruction, Surgery4-other extended operations
Variable selection
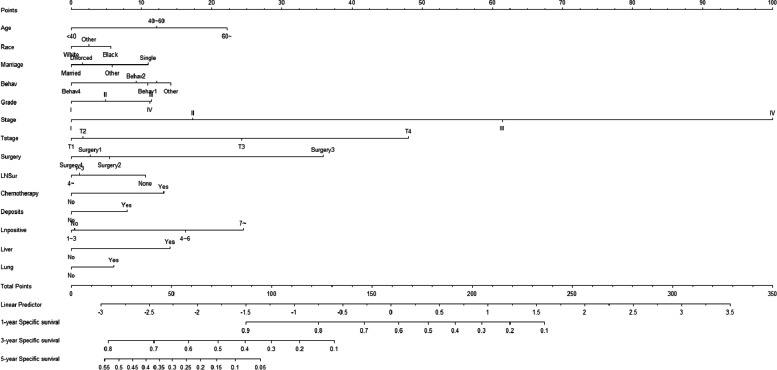
According to the results of univariate analysis, risk factors with P < 0.05 included pathological type of tumor, bone metastasis, brain metastasis, liver metastasis, lung metastasis, tumor grade (degree of differentiation), lymph node dissection, T stage, N stage, M stage, lymph nodes-positive, tumor size, AJCC stage, etc. These variables were then included in the multivariate analysis and the results showed that the following factors affected the survival of patients: age (< 40 years as a reference, 40 to 60: HR = 1.638, 95% CI [0.803, 3.342]; > 60: HR = 2.189, 95% CI [1.089, 4.399]), race (White as a reference, Black: HR = 1.162, 95% CI [1.068, 1.264]), marital (being married as a reference; divorced: HR = 1.014, 95% CI [0.93, 1.106]; single: HR = 1.304, 95% CI [1.201, 1.415]), pathological type of tumor (Behav1 as a reference, Behav2: HR = 0.945, 95% CI [0.841, 1.063], Behav3: HR = 1.009, 95% CI [0.921, 1.106]; Behav4: HR = 0.773, 95% CI [0.652, 0.916]), tumor stage (Stage I as a reference, Grade 1 as a reference; Grade 2: HR = 1.094, 95% CI [0.958, 1.249]; Grade 3: HR = 1.338, 95% CI [1.161, 1.542]; Grade 4: HR = 1.283, 95% CI [1.071, 1.537]), AJJC Stage (Stage I as a reference, Stage II: HR = 1.419, 95% CI [1.161, 1.734]; Stage III: HR = 3.68, 95% CI [2.914, 4.64]; Stage IV: HR = 9.704, 95% CI [7.634, 12.335]), T stage (T1 as a reference, T2: HR = 1.14, 95% CI [0.942, 1.378; T3: HR = 1.759, 95% CI [1.445, 2.141]; T4: HR = 3.029, 95% CI [2.48, 3.7]), type of surgeries(Surg1 as a reference, Surg2: HR = 1.047, 95% CI [0.964, 1.136]; Surg3: HR = 2.252, 95% CI [1.56, 3.251]; Surg4: HR = 0.942, 95% CI [0.789, 1.124]), lymph node dissection (> 4 as a reference;1 ~ 3: HR = 0.927, 95% CI [0.705, 1.219]), chemotherapy(None as a reference, Yes:HR = 1.554, 95% CI [1.444, 1.672]), deposits(None as a reference, Yes: HR = 1.179, 95%CI [1.093, 1.272]), lymph nodes-positive (No as a reference, 1 ~ 3: HR = 1.311, 95% CI [1.044, 1.646]; 4 ~ 6:HR = 1.714, 95% CI [1.276, 2.3]; > 7: HR = 2.234, 95% CI [1.681, 2.971]), liver metastasis (None as a reference; Yes: HR = 1.446, 95% CI [1.302, 1.605), lung metastasis (None as a reference; Yes: HR = 1.206, 95%CI [1.064, 1.367]) (Table 2). These variables were used to construct a competing risk model to estimate the probability of cause-specific mortality in CC patients at 1, 3, and 5 years (Fig. 1).
Table 2.
Univariate and multivariate analysis
| Factors | Define | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| HR (95%CI) | Z(P) | HR (95%CI) | Z(P) | ||
| Age | < 40 | Ref | NA | Ref | NA |
| 40 ~ 60 | 1.346(0.648 ~ 2.795) | 0.796(0.43) | 1.638(0.803 ~ 3.342) | 1.356(0.17) | |
| 60 ~ | 1.653(0.808 ~ 1.106) | 1.375(0.17) | 2.189(1.089 ~ 4.399) | 2.2(0.03) | |
| Marriage | Married | Ref | NA | Ref | NA |
| Divorced | 1.133(1.044 ~ 1.230) | 3.004(< 0.01) | 1.014(0.93 ~ 1.106) | 0.323(0.75) | |
| Single | 1.214(1.130 ~ 1.303) | 5.342(< 0.01) | 1.304(1.201 ~ 1.415) | 6.339(< 0.01) | |
| Other | 1.224(1.119 ~ 1.339) | 4.422(< 0.01) | 1.108(1.006 ~ 1.22) | 2.081(0.04) | |
| Race | White | Ref | NA | Ref | NA |
| Black | 1.174(1.084 ~ 1.270) | 3.958(< 0.01) | 1.162(1.068 ~ 1.264) | 3.492(< 0.01) | |
| Other | 0.978(0.865 ~ 1.106) | -0.355(0.72) | 1.017(0.895 ~ 1.156) | 0.262(0.79) | |
| Sex | Female | Ref | NA | Ref | NA |
| Male | 1.007(0.952 ~ 1.064) | 0.231(0.82) | 1.048(0.985 ~ 1.115) | 1.473(0.14) | |
| Behav | Behav1 | Ref | NA | Ref | NA |
| Behav2 | 0.542(0.486 ~ 0.604) | -11.059(< 0.01) | 0.945(0.841 ~ 1.063) | -0.944(0.35) | |
| Behav3 | 1.028(0.938 ~ 1.127) | 0.599(0.55) | 1.009(0.921 ~ 1.106) | 0.201(0.84) | |
| Behav4 | 0.478(0.407 ~ 0.562) | -8.960(< 0.01) | 0.773(0.652 ~ 0.916) | -2.967(< 0.01) | |
| Other | 1.292(1.168 ~ 1.429) | 4.992(< 0.01) | 1.119(0.995 ~ 1.259) | 1.883(0.06) | |
| Grade | I | Ref | NA | Ref | NA |
| II | 1.678(1.464 ~ 1.922) | 7.456(< 0.01) | 1.094(0.958 ~ 1.249) | 1.32(0.19) | |
| III | 3.228(2.802 ~ 3.792) | 16.221(< 0.01) | 1.338(1.161 ~ 1.542) | 4.026(< 0.01) | |
| IV | 3.459(2.917 ~ 4.102) | 14.267(< 0.01) | 1.283(1.071 ~ 1.537) | 2.702(0.01) | |
| Stage | I | Ref | NA | Ref | NA |
| II | 2.508(2.179 ~ 2.889) | 12.795(< 0.01) | 1.419(1.161 ~ 1.734) | 3.415(< 0.01) | |
| III | 6.391(5.608 ~ 7.282) | 27.831(< 0.01) | 3.68(2.914 ~ 4.648) | 10.942(< 0.01) | |
| IV | 26.820(23.538 ~ 30.560) | 49.387(< 0.01) | 9.704(7.634 ~ 12.335) | 18.566(< 0.01) | |
| Tstage | T1 | Ref | NA | Ref | NA |
| T2 | 1.056(0.875 ~ 1.273) | 0.569((0.57) | 1.14(0.942 ~ 1.378) | 1.345(0.18) | |
| T3 | 3.311(2.841 ~ 3.858) | 15.330(< 0.01) | 1.759(1.445 ~ 2.141) | 5.626(< 0.01) | |
| T4 | 8.809(7.549 ~ 10.278) | 27.639(< 0.01) | 3.029(2.48 ~ 3.7) | 10.866(< 0.01) | |
| Nstage | N0 | Ref | NA | Ref | NA |
| N1 | 3.075(2.854 ~ 3.314) | 29.486(< 0.01) | 0.74(0.58 ~ 0.945) | -2.415(0.02) | |
| N2 | 6.890(6.428 ~ 7.386) | 54.482(< 0.01) | 0.792(0.585 ~ 1.073) | -1.504(0.13) | |
| Surgery | Surg1 | Ref | NA | Ref | NA |
| Surg2 | 0.936(0.866 ~ 1.012) | -1.657(0.098) | 1.047(0.964 ~ 1.136) | 1.090(0.28) | |
| Surg3 | 4.172(3.489 ~ 4.988) | 15.662(< 0.01) | 2.252(1.56 ~ 3.251) | 4.333(< 0.01) | |
| Surg4 | 1.280(1.084 ~ 1.510) | 2.920(0.003) | 0.942(0.789 ~ 1.124) | -0.664(0.51) | |
| LNSur | 4 ~ | Ref | NA | Ref | NA |
| 1 ~ 3 | 1.884(1.637 ~ 2.169) | 8.823(< 0.01) | 0.927(0.705 ~ 1.219) | -0.54(0.59) | |
| None | 1.400(1.164 ~ 1.680) | 3.590(< 0.01) | 1.252(1.021 ~ 1.535) | 2.161(0.03) | |
| SurgOth | None | Ref | NA | Ref | NA |
| Yes | 2.431(2.243 ~ 2.634) | 21.678(< 0.01) | 0.956(0.873 ~ 1.048) | -0.954(0.34) | |
| Radiation | None | Ref | NA | Ref | NA |
| Yes | 2.219(1.890 ~ 2.605) | 9.749(< 0.01) | 1.167(0.968 ~ 1.407) | 1.624(0.10) | |
| Chemotherapy | None | Ref | NA | Ref | NA |
| Yes | 0.455(0.431 ~ 0.482) | -27.591(< 0.01) | 1.554(1.444 ~ 1.672) | 11.774(< 0.01) | |
| Deposits | None | Ref | NA | Ref | NA |
| Yes | 1.586(1.479 ~ 1.702) | 18.878(< 0.01) | 1.179(1.093 ~ 1.272) | 4.253(< 0.01) | |
| LnPositive | No | Ref | NA | Ref | NA |
| 1 ~ 3 | 3.025(2.801 ~ 3.268) | 28.119(< 0.01) | 1.311(1.044 ~ 1.646) | 2.335(0.02) | |
| 4 ~ 6 | 4.451(4.998 ~ 5.945) | 38.318(< 0.01) | 1.714(1.276 ~ 2.3) | 3.584(< 0.01) | |
| > 7 | 8.537(7.909 ~ 9.214) | 55.064(< 0.01) | 2.234(1.681 ~ 2.971) | 5.534(< 0.01) | |
| Bone | None | Ref | NA | Ref | NA |
| Yes | 6.977(5.267 ~ 9.242) | 13.544(< 0.01) | 1.23(0.879 ~ 1.722) | 1.208(0.23) | |
| Brain | None | Ref | NA | Ref | NA |
| Yes | 8.483(5.457 ~ 13.186) | 9.500(< 0.01) | 1.521(0.827 ~ 2.797) | 1.35(0.18) | |
| Liver | None | Ref | NA | Ref | NA |
| Yes | 6.956(6.541 ~ 7.398) | 61.225(< 0.01) | 1.446(1.302 ~ 1.605) | 6.898(< 0.01) | |
| Lung | None | Ref | NA | Ref | NA |
| Yes | 5.844(5.281 ~ 6.468) | 34.124(< 0.01) | 1.206(1.064 ~ 1.367) | 2.924(< 0.01) | |
| Size | < 3 cm | Ref | NA | Ref | NA |
| 3 ~ 5 cm | 1.088(1.001 ~ 1.173) | 2.186(0.03) | 0.979(0.902 ~ 1.062) | -0.514(0.61) | |
| > 5 cm | 1.172(1.096 ~ 1.254) | 4.621(< 0.01) | 0.959(0.893 ~ 1.029) | -1.158(0.25) | |
Fig. 1.
The competing risk nomogram for predicting 1-year, 3-year, 5-year cause-specific survival probability of cecal carcinoma
Validation of the model
The predictive accuracy of the nomogram was evaluated using the C-index and the area under the RCO curve (AUC). The calibration performance of the nomogram was assessed using the calibration curve. In the training set, the C-index of the model was 0.848 (se:0.0009). The AUC of 1-, 3-, and 5-year survival was 0.852 (95% CI [0.842, 0.861], 0.861 (95% CI [0.853, 0.868]), and 0.856 (95% CI [0.848, 0.864]), respectively (Figure 2), indicating that the model performed well regarding risk prediction and the calibration curve showed that the predicted probability was in good agreement with the observed one (Figure 3). In the validation set, the C-index of the model was 0.847 (se:0.0015), and the AUC at 1-, 3-, and 5-year survival was 0.841 (95% CI [0.825, 0.856]), 0.862 (95% CI [0.851, 0.874]), and 0.852 (95CI [0.839, 0.864], respectively (Figure 4), and the calibration curve showed that the predicted probability was in good agreement with the observed one (Figure 5). Taken together, the nomogram performed well regarding prediction and calibration.
Fig. 2.
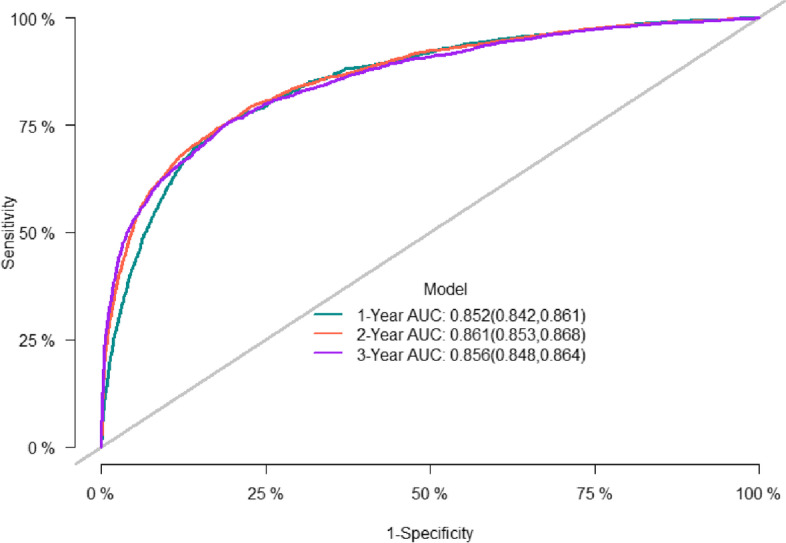
The AUC for OS of 1-, 3- and 5-year of training cohort
Fig. 3.
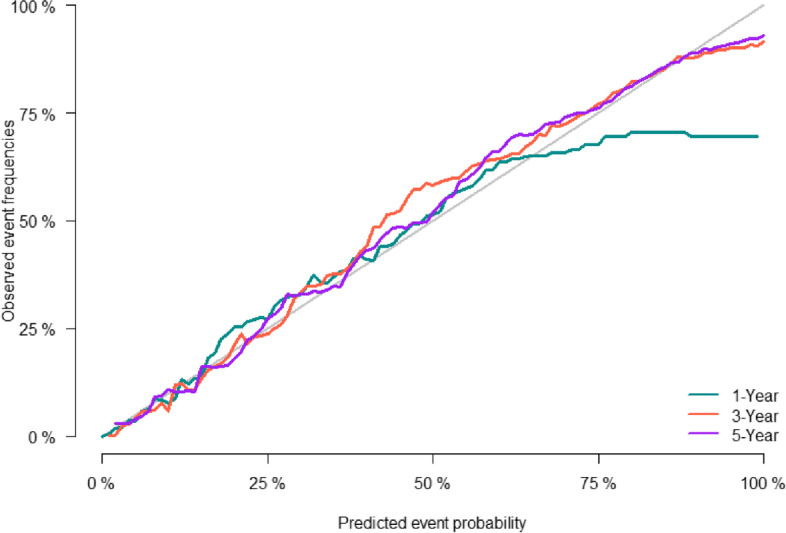
Calibration curves of nomogramfor 1-, 3-, and 5-year CSS in training cohort
Fig. 4.

The AUC for OS of 1-, 3- and 5-year of validation cohort
Fig. 5.
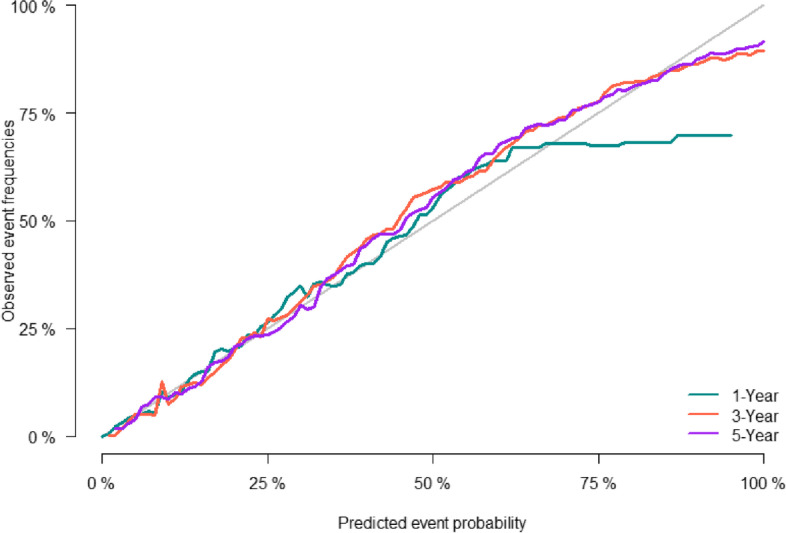
Calibration curves of nomogramfor 1-, 3-, and 5-year CSS in validation cohort
Comparison of prediction with Cox proportional hazard model and competing risk model
Traditional approaches to survival analysis such as Cox proportional hazard model are used to estimate the probability of one event over time, in which the occurrence of one type of death will prevent the occurrence of the death from other factors. However, in our study, CC-specific death and death from other factors are competing events. Patients dying from non-cancer-related causes accounted for 36.20% of the total deaths. In this scenario, the use of Cox model to analyze competing event data tends to overestimate the mortality in CC patients. In the present study, the predicted probability of mortality at 1 year, 3 years, and 5 years among CC patients estimated using the classical Cox model was 12.98%, 28.37%, and 35.06%, respectively, and that using the competing risk model was 10.8%, 23.59%, and 29.03%, respectively, demonstrating a relatively significant difference between these two models in risk prediction. Specifically, the mortality rate estimated by traditional COX survival analysis was higher than that estimated by the competitive risk model (Table3).
Table 3.
Cumulative specific mortality at different time points generated using survival analysis and competing hazards models
| Time points(months) | Classical Cox proportional hazard model to predict risk of death | Competing risk model | |
|---|---|---|---|
| cause-specific death | death from other factors | ||
| 12 | 12.98 | 10.8 | 3.78 |
| 24 | 22.09 | 18.38 | 6.58 |
| 36 | 28.37 | 23.59 | 9.22 |
| 48 | 32.37 | 26.88 | 11.83 |
| 60 | 35.06 | 29.03 | 14.46 |
| 72 | 37.03 | 30.57 | 17.09 |
| 84 | 38.36 | 31.58 | 19.9 |
| 96 | 39.57 | 32.46 | 22.51 |
| 105 | 40.51 | 33.13 | 24.58 |
Discussion
Colorectal cancer is common worldwide and attracts much attention. An estimated 1.2 million people have diagnosed with colorectal cancer annually and over 0.6 million people die from it every year [23, 24]. Recent research has shown that the increase in LCC and primary colon cancer is the largest. Lower survival is seen in patients with RCC than in those with LCC. Patients with CC have the poorest prognosis. In this sense, it is significant to determine variables to accurately predict the survival and prognosis of CC patients since personalized treatment is laid greater stress nowadays.
Standard methods for survival analysis such as the Kaplan–Meier curve and Cox proportional hazards models evaluate time-to-event probabilities. However, these methods tend to produce inaccurate estimates when competing risks exist. Thus, we constructed a nomogram based on clinical data from the SEER database to predict the cause-specific mortality among CC patients. This nomogram evaluated 14 risk factors including tumor pathological classification, tumor grade (degree of differentiation), AJCC stage, T stage, surgery type, lymph node dissection, chemotherapy, tumor deposits, lymph node metastasis, liver metastasis, lung metastasis, etc. It showed good prediction ability in both training and validation datasets.
In the present study, the AJCC stage was the best predictive variable, followed by the T stage. In previous studies, factors including age, race, tumor grade, tumor size, AJCC stage, and surgical status have been identified as independent risk factors for the prognosis of CC patients [25], and this was confirmed by the present research. Historical studies that extracted clinical information from the SEER database to construct Cox proportional hazards models for survival analysis found that race was an independent risk factor for the prognosis of CC patients, and compared with other races, the White had a higher risk of poorer prognosis [11]. However, the present study based on a competing risk model did not support this finding. This may be explained by the biased estimates attributable to the effects of competing risk events. What’s more, we found that CC patients with larger tumor sizes may have a poorer prognosis. However, a multi-center trial in Iran revealed no relevance between tumor size and prognosis [26]. The deviation between the COX regression model and variable effect estimation is a possible reason. On the other hand, the application of the findings in previous studies to the general population is also limited by sample size.
Our research has several limitations. First, the data from the SEER database for statistical analysis featured a short follow-up duration. Second, the nature of this retrospective study makes it difficult to eliminate selection bias. Third, the prognosis of CC patients in our analysis may be affected by patients’ lifestyle, genotype, and other factors, but the data on these factors cannot be obtained from the SEER database, thus related studies are required for further investigation.
Conclusion
To sum up, this study established a competitive risk model based on clinical data from the SEER database to evaluate the predictor variables for the prognosis of CC patients. Our findings will help clinicians have a better understanding of CC so that they can make appropriate decisions for patients using personalized cancer treatments.
Acknowledgements
Not applicable
Authors’ contributions
All authors contributed to the study conception and design. Qianru Zhou: Conceptualization, Methodology, Software, Writing- Original draft, Data curation, Visualization were performed; Yan Zhan: Investigation, Writing—Original Draft, Writing—Reviewing and Editing were performed; Jipeng Guo: Methodology, Software, Writing- Original draft were performed. All authors read and approved the final manuscript.
Funding
The study did not receive any specific funding from funding agencies in the public, commercial or non-profit sectors.
Availability of data and materials
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author. (extract the data in the SEER database: https://seer.cancer.gov/).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declared no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Favoriti P, Carbone G, Greco M, Pirozzi F, Pirozzi RE, Corcione F. Worldwide burden of colorectal cancer: a review. Updat Surg. 2016;68(1):7–11. doi: 10.1007/s13304-016-0359-y. [DOI] [PubMed] [Google Scholar]
- 2.Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, Dandona L, et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2017;3(4):524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jemal A, Ward EM, Johnson CJ, Cronin KA, Ma J, Ryerson B, Mariotto A, Lake AJ, Wilson R, Sherman RL, et al. Annual Report to the Nation on the Status of Cancer, 1975–2014, Featuring Survival. J Natl Cancer Inst. 2017;109(9). 10.1093/jnci/djx030. [DOI] [PMC free article] [PubMed]
- 4.Hermann J, Karmelita-Katulska K, Paszkowski J, Drews M, Stajgis M. Diagnosis of a cecal tumour with virtual colonoscopy. Pol J Radiol. 2011;76(2):25–27. [PMC free article] [PubMed] [Google Scholar]
- 5.Ashindoitiang JA. Caecal carcinoma: a review of 3 cases in a private hospital in Lagos. Clinics and practice. 2011;1(2):e42. doi: 10.4081/cp.2011.e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cucino C, Buchner AM, Sonnenberg A. Continued rightward shift of colorectal cancer. Dis Colon Rectum. 2002;45(8):1035–1040. doi: 10.1007/s10350-004-6356-0. [DOI] [PubMed] [Google Scholar]
- 7.Cheng L, Eng C, Nieman LZ, Kapadia AS, Du XL. Trends in colorectal cancer incidence by anatomic site and disease stage in the United States from 1976 to 2005. Am J Clin Oncol. 2011;34(6):573–580. doi: 10.1097/COC.0b013e3181fe41ed. [DOI] [PubMed] [Google Scholar]
- 8.Meguid RA, Slidell MB, Wolfgang CL, Chang DC, Ahuja N. Is there a difference in survival between right- versus left-sided colon cancers? Ann Surg Oncol. 2008;15(9):2388–2394. doi: 10.1245/s10434-008-0015-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss JM, Pfau PR, O’Connor ES, King J, LoConte N, Kennedy G, Smith MA. Mortality by stage for right- versus left-sided colon cancer: analysis of surveillance, epidemiology, and end results–Medicare data. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(33):4401–9. [DOI] [PMC free article] [PubMed]
- 10.Petrelli F, Tomasello G, Borgonovo K, Ghidini M, Turati L, Dallera P, Passalacqua R, Sgroi G, Barni S. Prognostic Survival Associated With Left-Sided vs Right-Sided Colon Cancer: A Systematic Review and Meta-analysis. JAMA Oncol. 2017;3(2):211–219. doi: 10.1001/jamaoncol.2016.4227. [DOI] [PubMed] [Google Scholar]
- 11.Xie X, Zhou Z, Song Y, Wang W, Dang C, Zhang H. Differences between carcinoma of the cecum and ascending colon: Evidence based on clinical and embryological data. Int J Oncol. 2018;53(1):87–98. doi: 10.3892/ijo.2018.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magaji BA, Moy FM, Roslani AC, Law CW. Survival rates and predictors of survival among colorectal cancer patients in a Malaysian tertiary hospital. BMC Cancer. 2017;17(1):339. doi: 10.1186/s12885-017-3336-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassan MR, Suan MA, Soelar SA, Mohammed NS, Ismail I, Ahmad F. Survival Analysis and Prognostic Factors for Colorectal Cancer Patients in Malaysia. Asian Pacific journal of cancer prevention : APJCP. 2016;17(7):3575–81. [PubMed]
- 14.Yuan Y, Li MD, Hu HG, Dong CX, Chen JQ, Li XF, Li JJ, Shen H. Prognostic and survival analysis of 837 Chinese colorectal cancer patients. World J Gastroenterol. 2013;19(17):2650–2659. doi: 10.3748/wjg.v19.i17.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaorsky NG, Zhang Y, Tuanquin L, Bluethmann SM, Park HS, Chinchilli VM. Suicide among cancer patients Nature communications. 2019;10(1):207. doi: 10.1038/s41467-018-08170-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaorsky NG, Churilla TM, Egleston BL, Fisher SG, Ridge JA, Horwitz EM, Meyer JE. Causes of death among cancer patients. Annals of oncology : official journal of the European Society for Medical Oncology. 2017;28(2):400–7. [DOI] [PMC free article] [PubMed]
- 17.Lau B, Cole SR, Gange SJ. Competing risk regression models for epidemiologic data. Am J Epidemiol. 2009;170(2):244–256. doi: 10.1093/aje/kwp107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Austin PC, Lee DS, Fine JP. Introduction to the Analysis of Survival Data in the Presence of Competing Risks. Circulation. 2016;133(6):601–609. doi: 10.1161/CIRCULATIONAHA.115.017719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersen PK, Pohar Perme M. Inference for outcome probabilities in multi-state models. Lifetime Data Anal. 2008;14(4):405–431. doi: 10.1007/s10985-008-9097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varadhan R, Weiss CO, Segal JB, Wu AW, Scharfstein D, Boyd C. Evaluating health outcomes in the presence of competing risks: a review of statistical methods and clinical applications. Med Care. 2010;48(6 Suppl):S96–105. doi: 10.1097/MLR.0b013e3181d99107. [DOI] [PubMed] [Google Scholar]
- 21.Berry SD, Ngo L, Samelson EJ, Kiel DP. Competing risk of death: an important consideration in studies of older adults. J Am Geriatr Soc. 2010;58(4):783–787. doi: 10.1111/j.1532-5415.2010.02767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Putter H, Fiocco M, Geskus RB. Tutorial in biostatistics: competing risks and multi-state models. Stat Med. 2007;26(11):2389–2430. doi: 10.1002/sim.2712. [DOI] [PubMed] [Google Scholar]
- 23.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet (London, England) 2014;383(9927):1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 24.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 25.Wu W, Yang J, Li D, Huang Q, Zhao F, Feng X, Yan H, Lyu J. Competitive Risk Analysis of Prognosis in Patients With Cecum Cancer: A Population-Based Study. Cancer control : journal of the Moffitt Cancer Center. 2021;28:1073274821989316. [DOI] [PMC free article] [PubMed]
- 26.Hosseini S, Bananzadeh AM, Mohammadianpanah M, Salek R, Taghizadeh-Kermani A. Prognostic significance of adjuvant radiation therapy in adenocarcinoma of the cecum. Radiat Oncol J. 2018;36(1):45–53. doi: 10.3857/roj.2017.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author. (extract the data in the SEER database: https://seer.cancer.gov/).