Abstract

Membrane disruptive α-helical antimicrobial peptides (AMPs) offer an opportunity to address multidrug resistance; however, most AMPs are toxic and unstable in serum. These limitations can be partly overcome by introducing D-residues, which often confers protease resistance and reduces toxicity without affecting antibacterial activity, presumably due to lowered α-helicity. Here, we investigated 31 diastereomers of the α-helical AMP KKLLKLLKLLL. Three diastereomers containing two, three, and four D-residues showed increased antibacterial effects, comparable hemolysis, reduced toxicity against HEK293 cells, and excellent serum stability, while another diastereomer with four D-residues additionally displayed lower hemolysis. X-ray crystallography confirmed that high or low α-helicity as measured by circular dichroism indicated α-helical or disordered structures independently of the number of chirality switched residues. In contrast to previous reports, α-helicity across diastereomers correlated with both antibacterial activity and hemolysis and revealed a complex relationship between stereochemistry, activity, and toxicity, highlighting the potential of diastereomers for property optimization.
Introduction
Membrane disruptive antimicrobial peptides (AMPs), which occur naturally as part of the innate immune system, offer an opportunity to address multidrug-resistant (MDR) bacteria because of their unspecific mechanism of action, against which resistance does not occur easily.1−3 Such AMPs are however unstable in serum and most often toxic owing to their membrane disruptive amphiphilic and usually α-helical structure triggering their antibacterial effect. Their properties can be improved by sequence optimization,4−7 whereby the most versatile approach consists in introducing non-natural structural elements8 such as d-amino acids,9−13 non-natural residues,14 β- or γ-amino acids,15,16 isopeptide bonds,17 or entirely non-peptidic elements such as spermine18 or fatty acids.19,20 A complete redesign of AMPs is also possible in the form of dimers,21 cyclic or bicyclic staples,22−24 small molecules,25 peptoids,26,27 foldamers,28 or dendrimers.29,30
For α-helical AMPs and analogues, the toxicity reduction effect observed upon introducing D-residues or similar perturbations, often measured as lower lysis of red blood cells, is generally attributed to a reduced α-helical folding, which would block pore formation on the membrane surface as a trigger for hemolysis. On the other hand, coating and destabilization of the bacterial membrane, and therefore the antibacterial effect, would still be possible with the modified peptide in the absence of folding.12,31−33 However, very little structural evidence or systematic studies support the hypothesis that reduced α-helical folding should generally preserve antibacterial activity while reducing toxicity.
In our own search for new antibacterial compounds, we have discovered several AMP dendrimers (AMPDs) with very low hemolysis and strong activity against Gram-negative bacteria including MDR clinical isolates.34−37 By investigating stereorandomized sequences, which are obtained by solid-phase synthesis using racemic building blocks and consist of a mixture of all possible diastereomers, we found that stereorandomized (sr-) AMPDs also exhibit strong antibacterial effects and very low hemolysis, suggesting an intrinsically disordered bioactive conformation.38,39 The same effect was observed with the intrinsically disordered AMP indolicidin40 but not with α-helical linear AMPs such as DJK-5,41 which lost their activity when stereorandomized.38
In a separate series of experiments with antimicrobial bicyclic peptides,22,42 we discovered a short membrane disruptive antibacterial but somewhat hemolytic linear undecapeptide, KKLLKLLKLLL (ln65), which did not appear, even as partial sequence, in databases of AMPs,43,44 proteins,45 or ChEMBL (Figure 1).46 The activity of this AMP was preserved upon inverting its four lysine residues to D-enantiomers to form kkLLkLLkLLL (ln69), while its hemolysis was strongly reduced.47 Strikingly, both the all-L sequence ln65 and its diastereomer ln69 were strongly α-helical, as established by circular dichroism (CD) and X-ray crystallography, showing that in this case lowered hemolysis was not related to a reduced α-helical folding. Intrigued by this observation, we set out to prepare and test the stereorandomized version sr-ln65 as well as multiple diastereomers of ln65 in search for analogues with possibly improved activity and/or reduced toxicity. Systematic studies of multiple diastereomers have shown significant activity modulations in the case of short, non-helical arginine–tryptophan containing AMPs.48−50
Figure 1.

(a) Chemical structure of ln65. (b) Helix wheel of ln65 sequence predicted by HeliQuest.51 Blue and yellow indicate, respectively, cationic and hydrophobic residues. The arrow inside the helix wheel indicates the magnitude and direction of the hydrophobic moment. (c) Structure of ln65 (PDB 7NEF, chain I) obtained by X-ray crystallography of a fucosylated analogue in complex with the bacterial lectin LecB. Cationic side chains are colored in blue, and hydrophobic side chains are colored in red.
Results and Discussion
Enantiomeric and Stereorandomized Sequences
We first investigated dln65 and dln69, the enantiomers of ln65 and its diastereomer ln69, to check that they displayed similar activities as expected for enantiomeric membrane disruptive AMPs (Table S1). CD spectra of dln65 and dln69 in aqueous phosphate buffer in the presence of either 5 mM dodecylphosphocholine (DPC), which forms micelles mimicking a membrane environment,52 or 20% trifluoroethanol (TFE) as a folding inducer,53,54 were mirror images from those of the L-enantiomers and confirmed their α-helical folding (Figure 2a,b). The enantiomeric pair ln65/dln65 gave essentially the same minimal inhibitory concentration (MIC) values against the five bacterial species used in this study (Pseudomonas aeruginosa, Klebsiella pneumoniae, Acinetobacter baumannii, Escherichia coli, and methicillin-resistant Staphylococcus aureus), as well as the same minimum hemolytic concentration (MHC) on human red blood cells (hRBCs) indicating significant hemolysis (125 μg/mL, Table 1, Figure S1 and Table S2). In line with these activities, the membrane disruptive effects of both enantiomers on fluorescein-loaded vesicles55 made of the anionic egg yolk phosphatidyl glycerol (EYPG) mimicking bacterial membranes as well as on vesicles made of zwitterionic egg yolk phosphatidyl choline (EYPC) mimicking eukaryotic membranes were comparably strong (Table 1, columns 10 and 11 and Figure S2). A similar behavior of ln65 and its enantiomer dln65 was consistent with membrane disruption as the primary mechanism of action for these α-helical AMPs. On the other hand, despite the mirror image CD-spectra and comparable vesicle leakage activities of ln69 and dln69, dln69 was four-fold more antibacterial and hemolytic than ln69, which might reflect an additional activity of dln69 unrelated to its membrane activity.
Figure 2.

CD-spectra of ln65, dln65, ln69, dln69, sr-ln65, sr-ln65L6, HP5, HP7, and HP9, measured with 0.1 mg/mL peptide in phosphate buffer at pH 7.4 with 10 and 20% v/v 2,2,2-TFE and with 5 mM DPC of (a) ln65 (full lines) and dln65 (dashed lines), (b) ln69 (full lines) and dln69 (dashed lines), (c) sr-ln65 (full lines) and sr-ln65L6 (dashed lines), (d) HP5, (e) HP7, and (f) HP9.
Table 1. Activity of Linear AMPs.
| MIC (μg/mL)c |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cpd. | sequencea | α-helix content (%)b | E. coli W3110 | P. aeruginosa PAO1 | A. baumannii ATCC19606 | K. pneumoniae NCTC418 | S. aureus COL MRSA | MHCd (μg/mL) | EYPG vesicle leakage (%)e | EYPC vesicle leakage (%)e |
| ln65 | KKLLKLLKLLL | 73 | 4 | 2–4 | 2–4 | 4 | 4 | 125 | 79 | 85 |
| dln65 | kkllkllklll | 67 | 2–4 | 2–4 | 4 | 4 | 2–4 | 125 | 82 | 90 |
| ln69 | kkLLkLLkLLL | 61 | 4 | 8 | 2–4 | 8 | 16 | 1000 | 98 | 26 |
| dln69 | KKllKllKlll | 59 | 0.5–1 | 2–4 | 2 | 4 | 2 | 250 | 94 | 24 |
| sr-ln65 | KKLLKLLKLLL | 10 | 4 | 4 | 4 | 16–32 | 8 | 1000 | 90 | 22 |
| sr-ln65L6 | KKLLKLLKLLL | 17 | 4 | 8 | 8 | 16–32 | 16 | 2000 | 91 | 12 |
| Diastereomers of ln65 | ||||||||||
| HP1 | KkLLKLLKLLL | 73 | 2 | 4 | 4 | 4–8 | 2 | <15.6 | 73 | 87 |
| HP2 | kkLLKLLKLLL | 69 | 4 | 4 | 4 | 2–4 | 4 | <15.6 | 70 | 87 |
| HP3 | KkLLkLLKLLL | 69 | 2 | 4 | 2 | 8 | 2–4 | <15.6 | 90 | 55 |
| HP4 | KkLlKLLKLLL | 46 | 2–4 | 2–4 | 2–4 | 2 | 2–4 | <15.6 | 74 | 83 |
| HP5 | kKLLKLLKLLl | 90 | 0.5 | 2 | 0.5 | 2 | 2 | 62.5 | 64 | 39 |
| HP6 | KKLLKllKLLL | 29 | 2 | 8 | 4 | 64 | 16 | 500 | 90 | 9 |
| HP7 | kkLLKLLKLLl | 60 | 0.5 | 2 | 0.5 | 4 | 2 | 125 | 90 | 28 |
| HP8 | KkllKLLKLLL | 37 | 2 | 4 | 2–4 | 4 | 4 | <15.6 | 91 | 63 |
| HP9 | KKLLkllKLLL | 10 | 8 | 64 | >64 | >64 | >64 | 31.3 | 59 | 5 |
| HP10 | kkLLkLLKLLL | 90 | 2 | 4 | 2 | 4 | 2 | <15.6 | 82 | 54 |
| HP11 | KkllKLlKLLL | 52 | 2 | 2–4 | 2 | 4–8 | 2 | 62.5 | 94 | 43 |
| HP12 | KkllKlLKLLL | 17 | 2–4 | 4–8 | 8 | 8–16 | 4 | 250 | 97 | 34 |
| HP13 | KKLLkllkLLL | 9 | 4–8 | 16–32 | 16–32 | >64 | >64 | 1000 | 42 | 8 |
| HP14 | KKllKllKLLL | 13 | 2 | 8 | 4–8 | >64 | 16–32 | 1000 | 95 | 10 |
| HP15 | KKlLkLlKlLL | 6 | 8 | 16 | 32–64 | >64 | >64 | >2000 | 34 | 3 |
| HP16 | KKLlkLLklLL | 8 | 4 | 32 | 32–64 | 8–16 | >64 | 125 | 50 | 2 |
| HP17 | KklLKLLKllL | 16 | 8 | 16 | 32 | 32–64 | 32–64 | 1000 | 20 | 12 |
| HP18 | kkLLKLLKLll | 63 | 4–8 | 8 | 8 | >64 | 16–32 | 1000 | 61 | 11 |
| HP19 | kkLLkLLKLLl | 55 | 2–4 | 8 | 4 | 16 | 8 | 1000 | 95 | 9 |
| HP20 | KKllKLLklLL | 15 | 8 | 16 | 32–64 | >64 | >64 | 1000 | 58 | 12 |
| HP21 | KklLKlLKlLL | 11 | 4 | 8 | 32 | 16–32 | 16–32 | >2000 | 44 | 5 |
| HP22 | KKllKLlKLlL | 23 | 2 | 8 | 8 | >64 | 16 | >2000 | 76 | 5 |
| HP23 | KKLlkLLkLLl | 10 | 2 | 4–8 | >64 | >64 | 32 | >2000 | 51 | 10 |
| HP24 | KkllKllKLLL | 7 | 4–8 | 16 | 8 | >64 | 32 | 250 | 81 | 13 |
| HP25 | KKllKllKlLL | 12 | 8–16 | 8–16 | 32 | >64 | >64 | >2000 | 70 | 4 |
| HP26 | kkLLkLLKLll | 41 | 4 | 4 | 16 | >64 | 32 | >2000 | 68 | 6 |
| HP27 | kkLLkLLkLLl | 23 | 4 | 8 | 32 | >64 | 32 | >2000 | 31 | 5 |
| HP28 | kKLLkllKLLl | 10 | 2–4 | 4 | 64 | >64 | 32 | 1000 | 29 | 3 |
| HP29 | KKLlkllkLLL | 7 | 2 | 4 | 8 | 64 | 32 | 250 | 53 | 6 |
| HP30 | KkLlKlLkLlL | 7 | 8 | 8–16 | 32 | >64 | 64 | >2000 | 11 | 10 |
| HP31 | kKlLkLlKlLl | 5 | 8 | 8 | 32 | >64 | 32 | >2000 | 22 | 9 |
| Lys → Arg and Leu → Ile Analogues of ln65/ln69 and Dimers | ||||||||||
| HP32 | RRLLRLLRLLL | 62 | 4–8 | 8–16 | 4 | 4–8 | 4–8 | 15.6 | 30 | 99 |
| HP33 | rrLLrLLrLLL | 63 | 4–8 | 4–8 | 4 | 4–8 | 2–4 | 125 | 85 | 56 |
| HP34 | KKIIKIIKIII | 68 | 32 | >64 | 8–16 | >64 | >64 | 62.5 | 95 | 10 |
| HP35 | kkIIkIIkIII | 22 | 4 | 16 | 8 | >64 | >64 | 125 | 98 | 13 |
| HP36 | RRIIRIIRIII | 60 | 16 | 64 | 8–16 | 16 | 16–32 | 62.5 | 99 | 39 |
| HP37 | rrIIrIIrIII | 50 | 8 | 4–8 | 8–16 | 32–64 | 8–16 | 250 | 98 | 8 |
| 2ln65 | (KKLLKLLKLLL)2 | 91 | >64 | >64 | >64 | >64 | >64 | <15.6 | 73 | 40 |
| 2ln69 | (kkLLkLLkLLL)2 | 82 | >64 | >64 | >64 | >64 | >64 | <15.6 | 71 | 74 |
One letter for amino acids. d-amino acids are shown in lower case and bold, and stereorandomized residues (ratio 1:1 of L and D) are underlined.
Values are corresponding to data recorded by CD for the condition 5 mM DPC in 7 mM PB buffer (pH 7.4). Percentage of the α-helix content were extracted from using Dichroweb.58 (Contin LL method, set 459).
MICs were determined after incubation in Mueller–Hinton (MH) broth (pH 7.4) for 16–20 h at 37 °C. Values represent two independent duplicates of MIC determinations.
MHC measured on human red blood cells in PBS (pH 7.4) after 4 h incubation at room temperature.
Lipid vesicles made of EYPG or EYPC were suspended in buffer (10 mM TRIS, 107 mM NaCl, pH 7.4). After 45 s, the indicated compound was added at the desired concentration and after 240 s, 30 μL of Triton X-100 1.2% was added for full fluorescein release. The percentage leakage observed with 10 μg/mL of compound is given. See the Supporting Information for full curves.
To further probe if α-helical folding was required for activity, we prepared the fully stereorandomized sequence sr-ln65, a racemic mixture of the 1024 possible diastereomers, as well as sr-ln65L6 with pure l-leucine at position 6 of the sequence, containing all 1024 diastereomers with single chirality at position 6 such as to make a possible folding detectable by CD. Remarkably, both sr-ln65 and sr-ln65L6 were as antibacterial as ln65 but much less hemolytic, an effect comparable to our previous observation with AMPDs and sr-AMDPs, suggesting that the antibacterial bioactive conformation of ln65 might be disordered while the hemolytic bioactive conformation would be α-helical.38,39 However, while CD spectra of sr-ln65 were nearly flat as expected because the stereorandomized sequence is racemic, those of sr-ln65L6 showed approximately 17% α-helix content in 5 mM DPC or with TFE, suggesting that a significant fraction of the 1024 possible diastereomers of ln65 might be α-helical (Figure 2c). Therefore, the activity of sr-ln65 might also be explained by the presence of some highly active and α-helical diastereomers, such as ln69, mixed with inactive and possibly disordered diastereomers.
Diastereomers and Mutants of ln65
In view of these preliminary experiments, we set out to test a series of diastereomers of ln65 for their α-helicity and antibacterial and hemolytic effects. From the 1024 possible diastereomers, 11 (0.1%) sequences are possible with a single inverted chirality residue, 55 (5.4%) with two, 165 (16.1%) with three, 330 (32.2%, including ln69) with four, and 462 (45.1%) with five inverted chirality residues. Balancing our interest to investigate diastereomers with multiple D-residues related to ln69 with the expectation that α-helical folding was more likely to be preserved with only a few inverted chirality residues,56,57 we selected 31 diastereomers HP1–HP31, one (3%) with a single D-residue, five (16%) with two D-residues, four (13%) with three D-residues, 13 (42%) with four D-resides, and eight (26%) with five D-residues, distributing D-residues in groups or scattered, at N- or C-termini, or in the middle of the sequence (Table 1).
Many of these diastereomers showed substantial α-helical folding in their CD spectra recorded in 5 mM DPC (Table 1, Figures 2d–f, S1 and Table S2). The average α-helicity decreased with increasing D-residues from 73% for ln65 and HP1 (zero and one D-residues), to 61 ± 24% for HP2–HP6 (two D-residues), 49 ± 34% for HP7–HP10 (three D-residues), 23 ± 20% for HP11–HP23 (four D-residues), and 14 ± 12% for HP24–HP31 (five D-residues). Assuming that these average α-helicity values were representative of the average across all ln65 diastereomers with the corresponding number of D-residues giving a predicted weighted average α-helicity of 26% for sr-ln65L6, slightly above the measured 17%.
Diastereomers with one, two, or three D-residues (HP1–HP10) generally showed activities comparable to the full L peptide ln65 against the five bacterial strains (MIC = 0.5–8 μg/mL) but were slightly more hemolytic (MHC = 15.6–62.5 μg/mL) than ln65. Notable exceptions were HP6, which was less active than ln65 against K. pneumoniae (MIC = 64 μg/mL) and MRSA (MIC = 16 μg/mL) and less hemolytic (MHC = 500 μg/mL), and HP9, which had much weaker antibacterial effects than ln65 (MIC = 8–>64 μg/mL) but was quite hemolytic (MHC = 31.3 μg/mL). HP6 and HP9 both had a relatively low α-helicity (29% and 10%). On the other hand, HP5 (2 D-residues) and HP7 (3 D-residues) stood out in this series as particularly antibacterial (MIC = 0.5–4 μg/mL) although somewhat hemolytic diastereomers (MHC = 62.5–125 μg/mL). Both peptides completely killed bacteria within 1 h in the time-kill assay as expected for membrane disruptive compounds (Figure S3). Furthermore, EYPG vesicle leakage activities of HP5 and HP7 were strong in line with antibacterial effects. Except for the non-helical but hemolytic HP9, EYPC vesicle leakage activities varied in line with hemolysis, consistent with a membrane disruptive activity.
Diastereomers with four and five D-residues (HP11–HP31) were generally less active against bacteria, especially against A. baumannii, K. pneumoniae, and MRSA, although they all kept significant EYPG vesicle leakage activities, reflecting the fact that vesicle leakage activity is often not sufficient for antibacterial effects to occur due to the much more complex nature of bacteria compared to lipid vesicles. Furthermore, these diastereomers mostly lost their hemolytic activity in proportion to their low EYPC vesicle leakage activities, except for HP16, HP24, and HP29, which, like HP9, showed significant hemolysis despite being non-helical and inactive on EYPC vesicles. The least active peptides were HP13 with four D-residues and HP25 with five D-residues. Both peptides retained some activity against E. coli, P. aeruginosa, and A. baumannii (MIC = 4–32 μg/mL) but were inactive against K. pneumoniae and MRSA, were non-hemolytic, and were not α-helical (7 and 11% in 5 mM DPC). Gratifyingly, one peptide with four D-residues, HP19, was as strongly antibacterial and low hemolytic as the previously identified ln69 with four D-residues. Another peptide with four D-residues, HP11 (MIC = 2–8 μg/mL), was even slightly more antibacterial than ln69, although slightly more hemolytic (MHC = 62.5 μg/mL). HP11 and HP19 were among the most α-helical in this set (52–55% in 5 mM DPC) although not as much as ln69 (61%).
To compare the effects of diastereomeric changes with more classical sequence variations, we performed conservative mutations in ln65 and ln69 by mutating all lysines to arginines, all leucines to isoleucines, or both, preserving their chirality pattern. In this series, the Lys → Arg exchanges (ln65 → HP32 and ln69 → HP33) preserved α-helicity, antibacterial activity, and EYPG vesicle leakage, but increased hemolysis and EYPC vesicle leakage, which might be related to the better cell-penetrating properties of poly-arginines versus poly-lysines attributed to stronger binding to phospholipids.60 On the other hand, Leu → Ile exchanges (ln65 → HP34, ln69 → HP35, HP32 → HP36, HP33 → HP37) led to reduced antibacterial effects and in part lower hemolysis, accompanied by slightly lower α-helicity as expected since Leu stabilizes and Ile destabilizes α-helices.61 Surprisingly, dimerization of ln65 to 2ln65 and ln69 to 2ln69 gave peptides that were strongly α-helical and hemolytic but entirely inactive against bacteria. Vesicle leakage activities were generally high for EYPG vesicles and partially followed hemolysis trends for EYPC.
Taken together, these experiments showed that diastereomers of ln65 featured new analogues with interesting activity profiles, while other simple modifications such as Lys → Arg, Leu → Ile mutations or dimerization were not as profitable. For further evaluation, we selected the most strongly antibacterial diastereomeric AMPs irrespective of their hemolytic properties, namely, ln65, ln69, dln69, and all diastereomers HP1–HP11 except HP6 and HP9. These AMPs showed good activities (MIC = 2–8 μg/mL) against additional Gram-negative and Gram-positive bacteria including several drug-resistant P. aeruginosa variants,62 although none of them were active against Burkholderia cenocepacia, a Gram-negative bacterium which is naturally resistant to AMPs like colicin (Table 2).63 Furthermore, most diastereomers were much more stable against serum degradation than the full L-sequence ln65 (Figure 3a). Interestingly, inverting the chirality of only the N- and C-termini (ln65 → HP5) was sufficient to entirely stabilize the peptide in line with the non-recognition of d-amino acids by proteases preventing the proteolysis from peptide extremities. On the other hand, dln69 with 7 d-leucine residues was entirely degraded due to proteolytic scission at the N-terminal l-lysine residue presumably from trypsin-like proteases (Figure S4).
Table 2. Antimicrobial Activity of Diastereomeric AMPse.
| Cpd. | PA14b | PA14 4.13 (phoQ)a,b | PA14 4.18 (pmrB)a,b | PA14 2P4 (pmrB)a,b | ZEM-1Aa,c | ZEM9Aa,c | K. pneumoniae Oxa-48a,c | Enterobacter cloacaea,c | Stenotrophomonas maltophiliaa,c | Burkolderia cenocepaciaa,c | Staphylococcus epidermisc,d | S. aureus Newmand |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ln65 | 2–4 | 4 | 32 | 32 | 4 | 4 | 2 | 2 | 2 | >64 | 2 | 2 |
| ln69 | 2 | 4 | 16 | 32 | 2 | 8 | 4 | 4 | 2 | >64 | 4 | 8 |
| dln69 | 1 | 2 | 8 | 16 | 4 | 8 | 8 | 4 | 2 | >64 | 4 | 1 |
| HP1 | 2 | 4 | 32 | 64 | 2 | 4 | 8 | 2 | 4 | >64 | 2 | 2 |
| HP2 | 2 | 4 | 32 | 64 | 2 | 4 | 2 | 2 | 2 | >64 | 2 | 2 |
| HP3 | 4 | 8 | 32 | 32 | 2 | 8 | 4 | 4 | 4 | >64 | 4 | 2 |
| HP4 | 2 | 4 | 32 | 64 | 2 | 4 | 2 | 2 | 2 | >64 | 2 | 2 |
| HP5 | 2 | 4 | 16 | 32 | 2 | 4 | 2 | 2 | 2 | >64 | 2 | 2 |
| HP7 | 2 | 4 | 16 | 16 | 1 | 4 | 4 | 2 | 2 | >64 | 2 | 4 |
| HP8 | 2 | 4 | 16 | 32 | 2 | 8 | 4 | 4 | 2 | >64 | 4 | 4 |
| HP10 | 2 | 4 | 16 | 32 | 2 | 4 | 2 | 2 | 2 | >64 | 2 | 2 |
| HP11 | 1 | 2 | 8 | 16 | 2 | 8 | 4 | 4 | 4 | >64 | 4 | 2 |
| Pol B | <0.125 | 0.25 | 1 | 1 | <0.125 | 2 | 2 | 1 | 0.5 | >16 | ||
| Vancomycin | 0.5 | 0.5 |
Gram-negative strains.
Strains carrying spontaneous mutations in the indicated genes, all leading to polymyxin B resistance.
MDR strains.
Gram-positive strains.
MIC were determined after incubation in MH broth pH 7.4 for 16–20 h at 37 °C. Values represent two independent duplicate MIC determinations.
Figure 3.

(a) Percentage of undegraded peptide after 24 h incubation in 12.5% human serum in TRIS buffer (pH 7.4) at 37 °C. Data are presented in mean ± SD, n = 3. See the Supporting Information for full curves. (b) Toxicity on HEK293 and A549 cells represented as the IC50 measured by Alamar blue assay after 24 h treatments with concentrations of peptide ranging from 0 to 200 μM. Data are presented as the mean ± SD, n = 3. See the Supporting Information for all data and procedure.
While most of these strongly antibacterial diastereomers were equally or more hemolytic than the full L-sequence ln65, they showed reduced cytotoxicity against human embryonic kidney HEK293 cells (Figure 3b). Diastereomer HP7, which was the most active AMP against bacteria, showed the lowest toxicity in the series (IC50 = 128 ± 5 μM). Furthermore, diastereomers were generally toxic against A549 lung cancer cells, with HP11 showing the strongest toxicity (IC50 = 3.6 ± 0.1 μM), in line with the fact that many AMPs are often active against cancer cells (Figures S5 and S6).64,65 The observed differences between diastereomers in hemolysis, toxicity against HEK293 cells or A549 lung cancer cells, are probably caused by diastereomeric interactions with the different membrane components of the different cell types and possibly proteins in the cell culture medium.66
X-ray Crystallography
To establish whether the CD signal observed with diastereomeric AMPs was indeed caused by α-helical folding, we prepared derivatives with their N-termini acylated with an α-C-fucosylacetyl group for crystallization as complexes with lectin LecB,67 an approach which we have successfully used for oligonucleotides,68 cyclic,69 bicyclic,47 and linear peptides,70 as well as for peptide dendrimers.71,72 We considered the nine most potent diastereomers detailed above, the Lys → Arg mutants HP32 and HP33, and the inactive, alternating chirality diastereomers HP30 and HP31. Crystallization screening provided good diffracting LecB crystals with well-resolved ligand electron density for complexes with the fucosylated analogues FHP5, FHP8, FHP30, and FHP31 (Table 3).
Table 3. X-ray Crystallography of Mixed-Chirality AMPs.
| Cpd. | sequencea | conditions | composition | PDB ID |
|---|---|---|---|---|
| FHP5 | (*)kKLLKLLKLLl | crystal screen G7 | 0.1 M HEPES pH 7.5, 20% v/v Jeffamine M-600 | 8AN9 |
| FHP8 | (*)KkllKLLKLLL | index screen D8 | 0.1 M HEPES pH 7.5, 25% w/v polyethylene glycol 3350 | 8ANO |
| FHP30 | (*)KkLlKlLkLlL | index screen H4 | 0.2 M ammonium citrate tribasic pH 7.0, 20% w/v polyethylene glycol 3350 | 8ANR |
| FHP31 | (*)kKlLkLlKlLl | index screen G8 | 0.2 M ammonium acetate, 0.1 M HEPES pH 7.5, 25% w/v polyethylene glycol 3350 | 8AOO |
One letter code for amino acids. * = α-l-fucosyl-acetyl. Many high-quality LecB crystals were also obtained in complex with fucosylated ln65R and ln69r; however, in these two cases, electron density only revealed the l-fucose and the adjacent two arginine residues.
In the X-ray crystal structure of FHP5 in complex with LecB, the undecapeptide was visible in full α-helical conformation in two of the four different fucose-binding sites present in the asymmetric unit, while the other two fucose-binding sites only showed electron density for the fucosyl group, probably due to a disordered conformation (PDB 8AN9, 1.3 Å resolution, Figure 4a, Table S3 and Figure S7). The two α-helices in the well-resolved binding sites are superimposable and interact through intermolecular hydrophobic interactions between leucine side chains (Figure 4b). We observed a similar situation for the structure of FHP8 in complex with LecB (PDB 8ANO, 1.3 Å resolution, Figure 4c,d, Table S4 and Figure S8). Both structures were very similar to the previously reported structure of fucosylated dln69 with LecB.47
Figure 4.
X-ray crystallography of mixed-chirality AMPs. (a) X-ray crystal structure of the FHP5·LecB complex (PDB 8AN9, chain E). Left panel: peptide is represented in stick, Ca2+ atoms in magenta spheres, and LecB in green cartoon. Blue mesh represents electron density (0.5σ level). Right panel: Stick model of the FHP5 crystal structure, lysine side chains shown in blue, and leucine side chains shown in red. (b) Superposition of the two complete non-equivalent peptides in the unit cell of PDB 8AN9. Fucose is omitted for more clarity. (c) Same as (a) for X-ray structure of the FHP8· LecB complex (PDB 8ANO, chain H). Electron density is shown for a 0.7σ level. (d) Same as (b) for the two complete non-equivalent peptides in the unit cell of PDB 8ANO. (e) X-ray structures of the two different asymmetric peptides in the FHP30·LecB complex (PDB 8ANR). Same color code as shown in (a). Electron densities are shown for a 1.0σ level. (f) Left panel: H-bonds between two symmetrical FHP30 chains. Right panel: full bundle of four symmetrical FHP30 chains. Lectin monomers and calcium atoms were omitted for clarity in the right panel. Same color code as shown in (a).
Although the inactive undecapeptides HP30 and HP31 with alternating L- and D-residues in their sequences had almost the same number of L- and D-residues, their flat CD spectra most likely indicated a disordered conformation considering that an excess of just one chiral residue was sufficient to indicate folding with sr-ln65L6. Indeed, the structure containing two asymmetric units of the LecB complex with FHP30 showed two different undefined structures, one forming a four members bundle maintained by H-bonds and hydrophobic interactions between the four symmetric peptides and the other one forming H-bonds with LecB (PDB 8ANR, 1.6 Å resolution, Figure 4e,f, Table S5 and Figure S9). A similar situation was observed in the LecB complex with fucosylated HP31 containing four different asymmetric units. In this case, only two of them were completely resolved and showed unordered conformations interacting with LecB via H-bonds but also with symmetrical peptides (PDB 8AOO, 1.2 Å resolution, Table S6 and Figure S10).
Molecular Dynamics
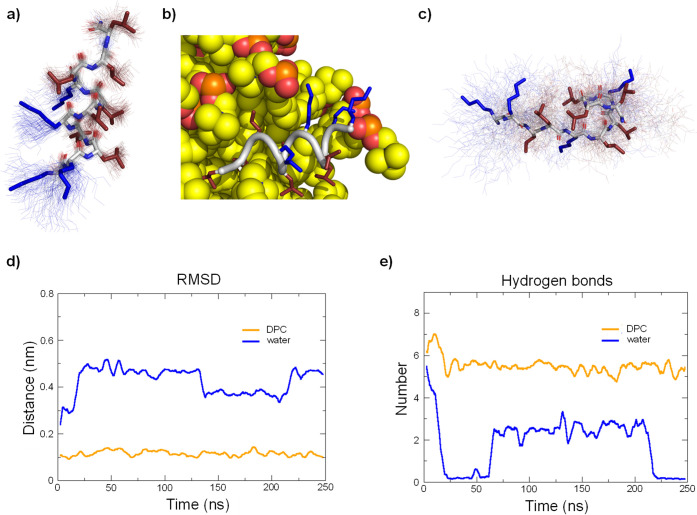
To further investigate the α-helical folding of our diastereomers, we performed molecular dynamics (MD) simulations over 250 ns using GROMACS73 starting from a pre-folded α-helical structure in water with or without a DPC micelle. For active diastereomers such as HP5 in the presence of DPC micelles, the peptide first entered in contact with the micelle surface by salt bridges between lysine side chain ε-ammonium groups and phosphate groups of DPC and later remained in an α-helical conformation at the micelle surface (Figure 5a,b). The peptide did not deviate significantly from the starting α-helical conformation (Figure 5d) and retained the full set of backbone H-bonds (Figure 5e).
Figure 5.
MD simulations of HP5 with and without DPC micelle. (a) Average structure (stick model) in the presence of DPC micelle over 100 structures sampled during the last 100 ns (thin lines). Hydrophobic side chains are colored in red, and cationic side chains are colored in blue. DPC micelle was omitted for clarity. (b) Last frame of the 250 ns run with DPC micelle. Peptide backbone is in gray cartoon, cationic side chains are colored in blue, hydrophobic side chains are colored in red, and DPC molecules are represented in spheres. (c) Same as (a) for run in water. (d) Comparison of root-mean-square deviation of the peptide backbone relative to the starting coordinates of the α-helix built in PyMol between run with DPC and run in water. (e) Comparison of the number of intramolecular backbone hydrogen bonds between run with DPC and run in water.
In water by contrast, the α-helix of HP5 completely and irreversibly unfolded to an unordered conformation (Figure 5c). This unordered conformation strongly differed from the starting α-helix (Figure 5a) with complete loss of backbone H-bonds (Figure 5b). Similar results were obtained for the other active compounds (HP1, HP2, HP3, HP4, HP7, HP8, HP10, and HP11, Figures S11–S19). For the inactive, non-helical diastereomers HP16 and HP29 by contrast, the starting α-helical conformation rapidly unfolded to an unordered conformation with complete loss of backbone H-bonds even in the presence of the DPC micelle (Figures S20 and S21).
Statistical Analysis
In view of the structural studies above showing in several cases that the degree of α-helicity of ln65 diastereomers as measured by CD corresponded to an observable α-helix or structural disorder, we assumed that the CD signal could be used as indication of folding across the entire series. Strikingly, increasing α-helicity (% in 5 mM DPC) was linearly correlated with increasing antibacterial activity measured as log2(MIC) against K. pneumoniae (r2 = 0.57), A. baumannii (r2 = 0.59), and MRSA (r2 = 0.62), but to a lesser extent with activity against P. aeruginosa (r2 = 0.41) and with hemolysis (r2 = 0.37), and only quite poorly with activity against E. coli (r2 = 0.29) against which most diastereomers were active (Figures 6a,b and S22).
Figure 6.
Statistical analysis of data set measured on ln65 derivatives. (a) Scatter plot of % helicity in 5 mM DPC against log2(MIC) for A. baumannii ATCC19606. (b) Same as (a) for log2(MHC). (c) Loading analysis of principal components 1 and 2. α = α-helix, β = β-sheet, t = turn, and u = unordered. Visualization of the (PC1 and PC2) plane. Each point represents one compound and is color coded depending on (d) activity on A. baumannii and (e) hemolytic activity.
To gain an overview of the series, we performed principal component analysis of the complete data set of antimicrobial activity, hemolysis, and folding under the different conditions measured. The first principal component PC1 covered 64% of data variance and reflected the variation of antimicrobial activities, hemolysis, and vesicle leakage activities with α-helicity measured in any of the four conditions (Figure 6c). The second principal component PC2 covered another 10% of data variance and reflected a modulation of antimicrobial activities with α-helicity and EYPG vesicle leakage independent of hemolysis and EYPC vesicle leakage. The distribution of the diastereomers on the (PC1 and PC2) plane separated active from inactive compounds from right to left and separated the two most active diastereomers identified, HP5 and HP7, from the majority of tested diastereomers (Figures 6d,e and S23). Both optimized AMPs stood out by their increased antimicrobial activity, which was particularly strong against A. baumannii, while keeping a moderate level of hemolysis, and reduced toxicity against HEK293 cells.
Conclusions
The above experiments with diastereomers of undecapeptide ln65 supported by X-ray crystallography show that this α-helical AMP preserves folding and activity across many of its diastereomers. In contrast to previous studies of diastereomers focused on cases with reduction in toxicity and preservation of antibacterial effects, our study across a broad set of diastereomers shows that introducing D-residues in an α-helical AMP can affect antibacterial effects at least as much as toxicity as measured by hemolysis. Although these activities were correlated, sufficient variability was available to identify two diastereomers with improved properties, HP5 and HP7, as two AMPs with increased antibacterial effects compared to the full L-AMP ln65, and moderate hemolysis and reduced toxicity against HEK293 cells.
In the present study, the preservation of folding and activity across many diastereomers of ln65 was anticipated by characterizing its stereorandomized version sr-ln65L6, which showed only modest reduction in antibacterial effects and significant α-helicity. Identifying active diastereomers might be more difficult for other α-helical AMPs if they lose their activity in the stereorandomized form as reported for DJK-5 (vqwrairvrvir) and SB1 (KYKKALKKLAKLL).38 Testing the stereorandomized sequence might therefore be the first step to address diastereomeric optimization of other AMPs.
Experimental Section
Peptide Synthesis
Materials and Reagents
N,N-dimethylformamide (DMF) was purchased from Thommen-Furler AG. Ethyl cyanohydroxyiminoacetate (Oxyma Pure) was purchased from SENN AG. N,N′-diisopropyl carbodiimide (DIC) was purchased from Iris BIOTECH GMBH. Piperazine, butanol, and 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU) were purchased from Alfa Aesar. Triisopropylsilane and trifluoroacetic acid (TFA) were purchased from Fluorochem Ltd. d-amino acids were purchased from GL Biochem Shanghai Ltd, and l-amino acids were purchased from Shanghai Space Peptides Pharmaceuticals Co., Ltd. Chemicals were used as supplied, and solvents were of technical grade. Amino acids were used as the following derivatives: Fmoc-Leu-OH, Fmoc-(D)-Leu-OH, Fmoc-Lys(Boc)-OH, Fmoc-(D)-Lys(Boc)-OH, Fmoc-Ile-OH, Fmoc-Arg(Pbf)-OH, and Fmoc-D-Arg(Pbf)-OH. Tentagel S RAM resin was purchased from RAPP Polymer. Analytical RP-HPLC–MS was performed with an Ultimate 3000 Rapid Separation LC–MS System (DAD-3000RS diode array detector) using an Acclaim RSLC 120 C18 column (2.2 μm, 120 Å, 3 × 50 mm, flow 1.2 mL/min) from Dionex. The HPLC is directly linked to a Thermo Scientific LCQ-Fleet Ion-trap MS. Data recording and processing were carried out with Dionex Chromeleon Management System Version 6.80 (analytical RP-HPLC) and FreeStyle software. All RP-HPLC were using HPLC-grade acetonitrile and Milli-Q deionized water. The elution solutions were A: MilliQ deionized water containing 0.05% TFA and D: MilliQ deionized water/acetonitrile (10:90, v/v) containing 0.05% TFA. Preparative RP-HPLC was performed with a Waters automatic Prep LC Controller System containing the four following modules: Waters2489 UV/vis detector, Waters2545 pump, Waters Fraction Collector III, and Waters 2707 Autosampler. A Dr. Maisch GmbH Reprospher column (C18-DE, 100 × 30 mm, particle size 5 μm, pore size 100 Å, and flow rate 40 mL/min) was used. Compounds were detected by UV absorption at 214 nm using a Waters 248 Tunable Absorbance Detector. Data recording and processing were performed with Waters ChromScope version 1.40 from Waters Corporation. MS spectra, recorded on a Thermo Scientific LTQ OrbitrapXL, were provided by the MS analytical service of the Department of Chemistry, Biochemistry and Pharmaceutical Sciences at the University of Bern (group of PD Dr. Stefan Schürch).
Solid-Phase Peptide Synthesis
Peptides were synthesized manually using Tentagel S RAM resin (0.22–0.25 mmol/g) and standard Fmoc solid phase peptide synthesis at 60 °C under nitrogen bubbling. The resin was swollen in DMF during 10 min. Double deprotections of the Fmoc group were performed using a solution of 5% w/v piperazine/2% DBU with 10% of butanol in DMF for 1 and 4 min. The resin was washed five times (5 × 8 mL DMF) after deprotection. Double couplings (2 × 8 min) were performed with 3 mL of amino acid (0.2 M), 2 mL of DIC (0.8 M), and 1.5 mL of Oxyma (0.8 M) in DMF. Resin was washed twice (2 × 8 mL DMF) between couplings and three times (3 × 8 mL DMF) after second coupling. The reaction mixture was removed by filtration, and the resin was washed with DMF and MeOH before cleavage.
On-Beads Sugar Coupling and Deprotection for Fucosylated Compounds
Peracetylated α-l-fucosyl-acetic acid (3 equiv), Oxyma (3 equiv), and DIC (3 equiv) were dissolved in 6 mL of DMF. Double coupling (2 × 1 h) was performed at 50 °C under nitrogen bubbling on resin. Deacetylation of sugar was performed directly on-bead using a mixture of MeOH/H2O/NH3 (8:1:1, v/v/v). Reaction was stirred overnight at room temperature. The reaction mixture was removed by filtration, and the resin was washed with DMF and MeOH before cleavage.
Cleavage from the Resin
Cleavage was carried out by treating the resins with 7 mL of a TFA/TIS/H2O (94:5:1, v/v/v) solution for 3 h at room temperature. The peptide solutions were precipitated with 25 mL of cold TBME, centrifuged for 10 min at 3500 rpm, evaporated, and dried with argon.
Purification and Characterization
The dried crude was dissolved in a water/ACN mixture, filtered (pore size 0.22 μm), and purified by preparative RP-HPLC with gradients of 60 min. Fractions were analyzed by analytical LCMS. Peptides were obtained as white foamy solids after lyophilization and analyzed by both LCMS and HRMS. Yields were calculated for the TFA salts. All compounds are >95% pure by HPLC.
CD Spectroscopy
CD experiments were measured on a Jasco J-715 spectropolarimeter. All the experiments were performed using Hellma Suprasil 110-QS 0.1 cm cuvettes. For each peptide, the measurements were performed in phosphate buffer (PB, pH = 7.4, 7 mM), 10% TFE, 20% TFE, 5 mM DPC. The buffer was degassed for 10 min under high vacuum before each set of experiments. The concentration of the peptides was 0.100 mg/mL, and each sample was measured in one accumulation. The scan rate was 20 nm/min, pitch was 0.5 nm, response was 16 s, and bandwidth was 1.0 nm. The nitrogen flow was kept >8 L/min. After each measurement, the cuvettes were washed successively with milli-Q H2O and PB (pH 7.4). The baseline was recorded under the same conditions and subtracted manually. Primary CD spectra were analyzed using DichroWeb58 and Contin-LL method (set 4).74
Antimicrobial Activity
Antimicrobial activity was determined for all peptides on E. coli W3110, P. aeruginosa PAO1, A. baumannii ATCC19606, K. pneumoniae NCTC418, and methicillin-resistant S. aureus COL and for selected peptides on P. aeruginosa PA14 and the polymyxin B-resistant derivatives PA14 4.13, PA14 4.18, and PA14 2P4 as well as the clinical isolates ZEM-1A and ZEM9A, K. pneumoniae OXA-48, Enterobacter cloacae, Stenotrophomonas maltophilia, B. cenocepacia, and Staphylococus epidermis. To determine the MIC, the broth microdilution method was used.75 A single bacterial colony was grown in LB medium overnight at 37 °C and 180 rpm shaking. The compounds were prepared as stock solutions of 2 mg/mL in sterilized milliQ deionized water, added to the first well of 96-well sterile, polypropylene round-bottom microtiter plates (TPP, untreated, Corning Incorporated, Kennebunk, USA), and diluted serially by 1/2. The concentration range tested was 0.5–64 μg/mL. The bacterial inoculum was prepared by measuring the absorbance of the overnight culture, which was diluted to concentration of the bacteria was quantified by measuring absorbance at 600 nm and diluted to an OD600 of 0.022 in MH medium pH 7.4 (Sigma-Aldrich, Buchs, Switzerland). The sample solutions (150 μL) were mixed with 4 μL of diluted bacterial suspension with a final inoculation of about 5 × 105 CFU. For each test, two columns of the plate were kept for sterility control (MH medium only) and growth control (MH medium with bacterial inoculum, no compound). Positive control was carried out using either polymyxin B for Gram-negative or vancomycin for Gram-positive strains (starting with a concentration of 16 μg/mL) in MH medium. The plates were incubated at 37 °C for 16–20 h under static conditions. 15 μL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)76 (1 mg/mL in sterilized milliQ deionized water) was added to each well, and the plates were incubated at room temperature until MTT staining was completed. The MIC was defined as the lowest concentration of the dendrimer that inhibits the visible growth of the tested bacteria (yellow) with the unaided eye.
Hemolysis Assay
To determine the MHC stock solutions, 8 mg/mL peptide in PBS (pH 7.4) was prepared, and 50 μL was diluted serially by l/2 in 50 μL of PBS (pH 7.4) in a 96-well plate (Costar or Nunc, polystyrene, untreated). The concentration range tested was 15.6–2000 μg/mL. hRBCs were obtained by centrifugation of 1.5 mL of whole blood, from the blood bank of Bern, at 3000 rpm for 15 min at 4 °C. Plasma was discarded, and the pellet was re-suspended in a 15 mL Falcon tube in 5 mL of PBS. The washing was repeated three times, and the remaining pellet was re-suspended in 10 mL of PBS. The hRBC suspension (50 μL) was added to each well, and the plate was incubated at room temperature for 4 h. MHC end points were determined by visual determination of the wells after the incubation period. Controls on each plate included a blank medium control (50 μL PBS + 50 μL of hRBCs suspension) and a hemolytic activity control (mQ-deionized water 50 μL + 50 μL hRBC suspension).
Vesicle Leakage Assay
5(6)-carboxyfluorescein (CF) was purchased from Sigma. EYPC, EYPG, and a Mini-Extruder were purchased from Avanti Polar Lipids. Egg PC or Egg PG thin lipid layers were prepared by evaporating a solution of 100 mg of EYPC or EYPG in 4 mL of MeOH/CHCl3 (1:1) on a rotary evaporator at room temperature and then dried in vacuo overnight. The resulting film was then hydrated with 2 mL of CF buffer (50 mM CF, 10 mM TRIS, 10 mM NaCl, pH 7.4) for 30 min at room temperature under stirring and then subjected to freeze–thaw cycles (7×) and extrusion (15×) through a polycarbonate membrane (pore size 100 nm). Extravesicular components were removed by gel filtration (Sephadex G-50) with 10 mM TRIS, 107 mM NaCl, pH 7.4 buffer. Final conditions: ∼2.5 mM PC or PG; inside: 50 mM CF, 10 mM TRIS, 10 mM NaCl, pH 7.4 buffer; outside: 10 mM TRIS, 107 mM NaCl, pH 7.4. PC or PG stock solutions (37.5 μL) were diluted to 3000 μL with a buffer (10 mM TRIS, 107 mM NaCl, pH 7.4) in a thermostated fluorescence cuvette (25 °C) and gently stirred (final lipid concentration ∼31 μM). The CF efflux was monitored at λem 517 nm (λex 492 nm) as a function of time after addition of the desired volume of peptide from 2 mg/mL stock in mQ water at t = 45 s, 10 and 50 μg/mL were monitored for both EYPC and EYPG. Finally, 30 μL of 1.2% Triton X-100 was added to the cuvette (0.012% final concentration) at t = 240 s to reach the maximum intensity. Fluorescence intensities were then normalized to the maximal emission intensity using I(t) = (It – I0)/(I∞ – I0) where I0 = It at peptide addition, I∞ = It at saturation of lysis.
Time-Killing Kinetic Assay
A single colony of P. aeruginosa PAO1 was picked and grown overnight with shaking (180 rpm) in LB (Sigma-Aldrich, Buchs, Switzerland) medium 5 mL overnight at 37 °C. The overnight bacterial culture was diluted to OD600 0.002 (2 × 106 CFU/mL) in fresh MH medium. Stock solutions of AMPs in sterilized milliQ water were prepared in 1 mg/mL and were diluted to two times more than required concentration in fresh MH (Sigma-Aldrich, Buchs, Switzerland) medium at pH 7.4. 100 μL of prepared bacteria solution in MH and 100 μL of samples in MH were mixed in a 96-well microtiter plate (TPP, untreated, Corning Incorporated, Kennebunk, USA). Untreated bacteria at 1 × 106 CFU/mL were used as a growth control. 96-well microtiter plates were incubated in 37 °C with shaking (180 rpm). Surviving bacteria were quantified at 0, 0.5, 1, 2, 3, 4, 5, and 6 h by plating 10-fold dilutions of the sample in sterilized normal saline on LB agar plates. LB agar plates were incubated at 37 °C for 10 h, and the number of individual colonies was counted at each timepoint. The assay was performed in triplicate in the biosafety level 2 lab.
Serum Stability Assay
Human serum was diluted in 0.1 M filtered TRIS buffer (pH 7.4) (25%, 1:3, v/v). Selected peptides were diluted in 0.1 M filtered TRIS buffer (pH 7.4) to a concentration of 400 μM, and 0.1 mg/mL 4- hydroxybenzoic acid was added as an internal standard. Aliquots of peptide solution (50 μL) were added to aliquots of serum (50 μL) in sterile Eppendorf tubes, to reach a peptide concentration of 200 μM during the assay. Samples were incubated at 37 °C under gentle stirring (350 rpm). Different samples (triplicates) were quenched at different time points (0/1/6/12/24 h) by precipitating serum proteins through the addition of ZnSO4·7 H2O/ACN (1:1) (0.1 M, 100 μL) and cooling in ice bath for 10 min. Protein precipitates were pelleted under centrifugation, and supernatants were then sampled and analyzed by LC–MS. Experiment controls included two references, one known to be degraded and one known to be undegraded. Peaks corresponding to the internal standard and the undegraded peptides were integrated, with the ratio peptide/standard at t = 0 h as 100%.
Cytotoxicity Assay
Cell Culture
The A549 human lung adenocarcinoma cells are derived from a patient and were kindly given to us by Dr. Georgia Konstatinidou (Pharmacology Institute, Bern University). HEK293T cells were obtained from ATCC (CRL-11268). A549 and HEK293 were cultured in an incubator at 37 °C with 5% CO2 in RPMI-1640 (Gibco) and DMEM (Gibco) containing 10% fetal bovine serum (Thermo Fisher), 100 I.U./mL penicillin, and 100 μg/mL streptomycin (Gibco).
Cell-Viability Assay
The viability of the cells was assessed with an AlamarBlue assay (ThermoFisher). Cells were seeded into 96-well plates, 4000 cells/well (HEK293) and 8000 cells/well (A549), the day before the experiment. Cells were then treated with the increasing concentration of the compound and incubated for 24 h at 37 °C in the presence of 5% CO2. The next day, the medium was removed and replaced by a 10% AlamarBlue solution in full growth medium (DMEM or RPMI-1640). The cells were incubated for 3–5 h at 37 °C with 5% CO2 in a humidified atmosphere. The fluorescence was then measured on a Tecan Infinite M1000 Pro plate reader at λex 560 nm and λem 590 nm. The value was normalized according to the untreated cells.
Crystallography Experiment and Data Acquisition
Suitable diffracting crystals were obtained via co-crystallization of the C-fucosylated derivatives with the bacterial lectin LecB. The sitting drop vapor diffusion method was used, screening 192 different conditions per compound. The lyophilized protein was dissolved in milli-Q water (5 mg/mL) in the presence of salts (6 mM CaCl2 and MgCl2). The peptides were added to the protein at a 5:1 molar excess related to the LecB lectin monomer. Crystals were obtained within 1–3 months after mixing 1.5 μL of the LecB ligand complex with 1.5 μL of reservoir solution and incubation at 18 °C. All crystallization conditions were found in Index screens I/II (96 conditions) and Crystal Screen I/II (96 conditions) (Hampton Research, Laguna Niguel, CA, USA). Diffraction data were collected at the Paul Scherrer Institute (Villigen, Switzerland) on beamline X06DA PX-III using a DECTRIS PILATUS 2M-F detector and a multi-axis PRIGo goniometer. The structures were solved and visualized with the help of Phenix,77 ccp4,78 PyMol,79 coot,80 and XDS.81
MD Simulation
MD simulations were performed using GROMACS73 software version 2018.1 and the GROMOS53a6 force field.82 The starting topologies were built from PyMOL. A dodecahedral box was created around the peptide 1.0 nm from the edge of the peptide and filled with extended simple point charge water molecules. Sodium and chloride ions were added to produce an electroneutral solution at a final concentration of 0.15 M NaCl. The energy was minimized using a steepest gradient method to remove any close contacts before the system was subjected to a two-phase position-restrained MD equilibration procedure. The system was first allowed to evolve for 100 ps in a canonical NVT (N is the number of particles, V the system volume, and T the temperature) ensemble at 300 K before pressure coupling was switched on, and the system was equilibrated for an additional 100 ps in the NPT (P is the system pressure) ensemble at 1.0 bar.
MD in the Presence of DPC Micelle
MD simulations in the presence of a DPC (n-DPC) micelle were performed as follows. Parameters and references for the DPC molecule for the GROMOS53a6 forcefield are given in the Supporting Information. Peptides were manually placed at a distance from the pre-equilibrated micelle (of 65 DPC molecules) equal to the diameter of said peptide. Box, solvation, and NVT equilibration procedures were performed as explained previously. For each peptide/micelle system, 10 runs of 50 ns were generated to show the possibility for the peptide to either interact or diffuse away from the micelle. Then, runs of interest were extended up to 250 ns.
Clustering of Stable Structures
To obtain a representative conformer for each run, the last 100 ns (10001 frames) were clustered using an RMSD cutoff adapted to get a good balance between the number of clusters and the size of the main cluster. Many clusters combined with a very large percentage of structures in the top cluster is an indication of the stability of the one main conformer in each case. The PyMol Molecular Graphics System, version 1.8 (Schrödinger, LLC), was used to create structural models.
Acknowledgments
This work was supported financially by the European Research Council (grant no. 885076) and the Swiss National Science Foundation (grant no. 200020_178998). We thank Alexandre Lüscher for help in preparing cultures of MDR bacteria and the staff at the Swiss Light Source, Beamline X06DA (PXIII), Villigen, Switzerland for support during data collection.
Glossary
Abbreviations
- ACN
acetonitrile
- AMP
antimicrobial peptides
- AMPD
antimicrobial peptide dendrimer
- ATCC
American-type culture collection
- CD
circular dichroism
- CF
5(6)-carboxyfluorescein
- CFU
colony-forming units
- ChEMBL
Chemical European Molecular Biology Laboratory Database
- DBU
1,8-diazabicyclo[5.4.0]undec-7-ene
- DIC
N,N-diisoprpylcarbodiimide
- DMEM
Dulbecco’s modified Eagle’s medium
- DMF
dimethylformamide
- DPC
dodecylphosphocholine
- EYPC
egg yolk phosphatidyl choline
- EYPG
egg yolk phosphatidyl glycerol
- HEK cells
human embryonic kidney cells
- HEPES
2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid
- HPLC
high-performance liquid chromatography
- hRBC
human red blood cell
- HRMS
high resolution mass spectrometry
- LB
Luria–Bertani
- LCMS
liquid chromatography-mass spectrometry
- MD
molecular dynamics
- MDR
multidrug-resistant
- MIC
minimal inhibitory concentration
- MH
Mueller–Hinton
- MHC
minimum hemolytic concentration
- MRSA
methicillin-resistant Staphylococcus aureus
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NCTC
national collection of type cultures
- Oxyma
ethyl cyanohydroxyiminoacetate
- PB
phosphate buffer
- PBS
phosphate buffer saline
- PCA
principal components analysis
- PDB
protein data bank
- RMSD
root-mean-square deviation
- RP-HPLC
reverse-phase high-performance liquid chromatography
- RPMI
Roswell Park memorial institute medium
- SDS
sodium dodecyl sulfate
- SPPS
solid-phase peptide synthesis
- TBME
terbutylmethyl ether
- TFA
trifluoroacetic acid
- TFE
trifluoroethanol
- TIS
triisopropylsilane
- TRIS
2-amino-2-hydroxymethyl-propane-1,3-diol
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.3c00460.
Peptides yields, MS and analytical data, CD spectra and analysis, vesicle leakage fluorescence curves, time-killing kinetics curves, full serum stability curves, full cytotoxicity data, data collection, refinement statistics, X-ray structures, topology of DPC molecules for MD simulations, MD simulation analyses, additional statistical analysis of ln65 diastereomers, structures, and HPLC–MS chromatograms and HRMS spectra for all compounds (PDF)
SMILES and activity of all tested peptides (CSV)
Accession Codes
Coordinates and structure factors of refined LecB complexed with fucosylated peptides are available from the Protein data Bank with accession codes 8AN9 (FHP5), 8ANO (FHP8), 8ANR (FHP30), and 8AOO (FHP31).
Author Contributions
H.P. designed the project and carried out peptide synthesis, microbiological, hemolysis, serum stability and vesicle leakage assays, CD spectroscopy, MD simulations, and X-ray crystallography and wrote the paper. T.P. performed cytotoxicity assay and wrote the paper. S.F. and S.B. synthesized peptides, performed microbiology and hemolysis assays and CD spectroscopy. T.K. and C.v.D. supervised experiments with clinical and MDR strains. A.S. supervised X-ray crystallography experiment. S.J. supervised MD studies and wrote the paper. J.-L.R. designed and supervised the study and wrote the paper.
The authors declare no competing financial interest.
Notes
Raw data of LC–MS analyses for characterization and serum stability, HRMS, CD measurements, vesicle leakage assay, X-ray crystallography, MD simulations, cytotoxicity assay, and pictures of 96-well plates for microbiological and hemolysis assays are accessible at: 10.5281/zenodo.7824946
Supplementary Material
References
- Lakemeyer M.; Zhao W.; Mandl F. A.; Hammann P.; Sieber S. A. Thinking Outside the Box—Novel Antibacterials To Tackle the Resistance Crisis. Angew. Chem., Int. Ed. Engl. 2018, 57, 14440–14475. 10.1002/anie.201804971. [DOI] [PubMed] [Google Scholar]
- Magana M.; Pushpanathan M.; Santos A. L.; Leanse L.; Fernandez M.; Ioannidis A.; Giulianotti M. A.; Apidianakis Y.; Bradfute S.; Ferguson A. L.; Cherkasov A.; Seleem M. N.; Pinilla C.; de la Fuente-Nunez C.; Lazaridis T.; Dai T.; Houghten R. A.; Hancock R. E. W.; Tegos G. P. The Value of Antimicrobial Peptides in the Age of Resistance. Lancet Infect. Dis. 2020, 20, e216–e230. 10.1016/S1473-3099(20)30327-3. [DOI] [PubMed] [Google Scholar]
- Mookherjee N.; Anderson M. A.; Haagsman H. P.; Davidson D. J. Antimicrobial Host Defence Peptides: Functions and Clinical Potential. Nat. Rev. Drug Discovery 2020, 19, 311–332. 10.1038/s41573-019-0058-8. [DOI] [PubMed] [Google Scholar]
- Lakshmaiah Narayana J.; Mishra B.; Lushnikova T.; Wu Q.; Chhonker Y. S.; Zhang Y.; Zarena D.; Salnikov E. S.; Dang X.; Wang F.; Murphy C.; Foster K. W.; Gorantla S.; Bechinger B.; Murry D. J.; Wang G. Two Distinct Amphipathic Peptide Antibiotics with Systemic Efficacy. Proc. Natl. Acad. Sci. U.S.A. 2020, 117, 19446–19454. 10.1073/pnas.2005540117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X.; Li B.; Yang Y.; Ba Z.; Zhang J.; Zhang T.; Chang L.; Zhang F.; Zhang Y.; Liu H.; Gou S.; Ni J. Improving the Antimicrobial Performance of Amphiphilic Cationic Antimicrobial Peptides Using Glutamic Acid Full-Scan and Positive Charge Compensation Strategies. J. Med. Chem. 2022, 65, 13833–13851. 10.1021/acs.jmedchem.2c01076. [DOI] [PubMed] [Google Scholar]
- Li B.; Ouyang X.; Ba Z.; Yang Y.; Zhang J.; Liu H.; Zhang T.; Zhang F.; Zhang Y.; Gou S.; Ni J. Novel β-Hairpin Antimicrobial Peptides Containing the β-Turn Sequence of -RRRF- Having High Cell Selectivity and Low Incidence of Drug Resistance. J. Med. Chem. 2022, 65, 5625–5641. 10.1021/acs.jmedchem.1c02140. [DOI] [PubMed] [Google Scholar]
- Zhang M.; Ouyang J.; Fu L.; Xu C.; Ge Y.; Sun S.; Li X.; Lai S.; Ke H.; Yuan B.; Yang K.; Yu H.; Gao L.; Wang Y. Hydrophobicity Determines the Bacterial Killing Rate of α-Helical Antimicrobial Peptides and Influences the Bacterial Resistance Development. J. Med. Chem. 2022, 65, 14701–14720. 10.1021/acs.jmedchem.2c01238. [DOI] [PubMed] [Google Scholar]
- Gan B. H.; Gaynord J.; Rowe S. M.; Deingruber T.; Spring D. R. The Multifaceted Nature of Antimicrobial Peptides: Current Synthetic Chemistry Approaches and Future Directions. Chem. Soc. Rev. 2021, 50, 7820–7880. 10.1039/D0CS00729C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai Y.; Oren Z. Diastereomers of Cytolysins, a Novel Class of Potent Antibacterial Peptides (*). J. Biol. Chem. 1996, 271, 7305–7308. 10.1074/jbc.271.13.7305. [DOI] [PubMed] [Google Scholar]
- Hong J.; Oren Z.; Shai Y. Structure and Organization of Hemolytic and Nonhemolytic Diastereomers of Antimicrobial Peptides in Membranes. Biochemistry 1999, 38, 16963–16973. 10.1021/bi991850y. [DOI] [PubMed] [Google Scholar]
- Pratap Verma D.; Ansari M. M.; Verma N. K.; Saroj J.; Akhtar S.; Pant G.; Mitra K.; Singh B. N.; Ghosh J. K. Tandem Repeat of a Short Human Chemerin-Derived Peptide and Its Nontoxic d-Lysine-Containing Enantiomer Display Broad-Spectrum Antimicrobial and Antitubercular Activities. J. Med. Chem. 2021, 64, 15349–15366. 10.1021/acs.jmedchem.1c01352. [DOI] [PubMed] [Google Scholar]
- Ben Hur D.; Kapach G.; Wani N. A.; Kiper E.; Ashkenazi M.; Smollan G.; Keller N.; Efrati O.; Shai Y. Antimicrobial Peptides against Multidrug-Resistant Pseudomonas Aeruginosa Biofilm from Cystic Fibrosis Patients. J. Med. Chem. 2022, 65, 9050–9062. 10.1021/acs.jmedchem.2c00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slingerland C. J.; Kotsogianni I.; Wesseling C. M. J.; Martin N. I. Polymyxin Stereochemistry and Its Role in Antibacterial Activity and Outer Membrane Disruption. ACS Infect. Dis. 2022, 8, 2396–2404. 10.1021/acsinfecdis.2c00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandín D.; Valle J.; Chaves-Arquero B.; Prats-Ejarque G.; Larrosa M. N.; González-López J. J.; Jiménez M. Á.; Boix E.; Andreu D.; Torrent M. Rationally Modified Antimicrobial Peptides from the N-Terminal Domain of Human RNase 3 Show Exceptional Serum Stability. J. Med. Chem. 2021, 64, 11472–11482. 10.1021/acs.jmedchem.1c00795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayouka Z.; Chakraborty S.; Liu R.; Boersma M. D.; Weisblum B.; Gellman S. H. Interplay among Subunit Identity, Subunit Proportion, Chain Length, and Stereochemistry in the Activity Profile of Sequence-Random Peptide Mixtures. J. Am. Chem. Soc. 2013, 135, 11748–11751. 10.1021/ja406231b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X.; Yan T.; Rao J.; An Y.; Yue X.; Miao X.; Wang R.; Sun W.; Cai J.; Xie J. Novel Feleucin-K3-Derived Peptides Modified with Sulfono-γ-AA Building Blocks Targeting Pseudomonas Aeruginosa and Methicillin-Resistant Staphylococcus Aureus Infections. J. Med. Chem. 2023, 66, 1254–1272. 10.1021/acs.jmedchem.2c01396. [DOI] [PubMed] [Google Scholar]
- Wani N. A.; Stolovicki E.; Hur D. B.; Shai Y. Site-Specific Isopeptide Bond Formation: A Powerful Tool for the Generation of Potent and Nontoxic Antimicrobial Peptides. J. Med. Chem. 2022, 65, 5085–5094. 10.1021/acs.jmedchem.2c00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewangan R. P.; Verma D. P.; Verma N. K.; Gupta A.; Pant G.; Mitra K.; Habib S.; Ghosh J. K. Spermine-Conjugated Short Proline-Rich Lipopeptides as Broad-Spectrum Intracellular Targeting Antibacterial Agents. J. Med. Chem. 2022, 65, 5433–5448. 10.1021/acs.jmedchem.1c01809. [DOI] [PubMed] [Google Scholar]
- Koh J. J.; Lin H.; Caroline V.; Chew Y. S.; Pang L. M.; Aung T. T.; Li J.; Lakshminarayanan R.; Tan D. T.; Verma C.; Tan A. L.; Beuerman R. W.; Liu S. N-Lipidated Peptide Dimers: Effective Antibacterial Agents against Gram-Negative Pathogens through Lipopolysaccharide Permeabilization. J. Med. Chem. 2015, 58, 6533–6548. 10.1021/acs.jmedchem.5b00628. [DOI] [PubMed] [Google Scholar]
- Topman-Rakover S.; Malach E.; Burdman S.; Hayouka Z. Antibacterial Lipo-Random Peptide Mixtures Exhibit High Selectivity and Synergistic Interactions. Chem. Commun. 2020, 56, 12053–12056. 10.1039/D0CC04493H. [DOI] [PubMed] [Google Scholar]
- Li W.; Lin F.; Hung A.; Barlow A.; Sani M.-A.; Paolini R.; Singleton W.; Holden J.; Hossain M. A.; Separovic F.; O’Brien-Simpson N. M.; Wade J. D. Enhancing Proline-Rich Antimicrobial Peptide Action by Homodimerization: Influence of Bifunctional Linker. Chem. Sci. 2022, 13, 2226–2237. 10.1039/D1SC05662J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bonaventura I.; Baeriswyl S.; Capecchi A.; Gan B.-H.; Jin X.; Siriwardena T. N.; He R.; Kohler T.; Pompilio A.; Di Bonaventura G.; van Delden C.; Javor S.; Reymond J.-L. An Antimicrobial Bicyclic Peptide from Chemical Space Against Multidrug Resistant Gram-Negative Bacteria. Chem. Commun. 2018, 54, 5130–5133. 10.1039/c8cc02412j. [DOI] [PubMed] [Google Scholar]
- Mourtada R.; Herce H. D.; Yin D. J.; Moroco J. A.; Wales T. E.; Engen J. R.; Walensky L. D. Design of Stapled Antimicrobial Peptides That Are Stable, Nontoxic and Kill Antibiotic-Resistant Bacteria in Mice. Nat. Biotechnol. 2019, 37, 1186–1197. 10.1038/s41587-019-0222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T.; Xu L.; Hu Y.; Tang X.; Qu R.; Zhao X.; Bai H.; Li L.; Chen W.; Luo G.; Fu G.; Wang W.; Xia X.; Zhang J. Lysine-Tethered Stable Bicyclic Cationic Antimicrobial Peptide Combats Bacterial Infection in Vivo. J. Med. Chem. 2022, 65, 10523–10533. 10.1021/acs.jmedchem.2c00661. [DOI] [PubMed] [Google Scholar]
- Teng P.; Shao H.; Huang B.; Xie J.; Cui S.; Wang K.; Cai J. Small Molecular Mimetics of Antimicrobial Peptides as a Promising Therapy To Combat Bacterial Resistance. J. Med. Chem. 2023, 66, 2211–2234. 10.1021/acs.jmedchem.2c00757. [DOI] [PubMed] [Google Scholar]
- Chongsiriwatana N. P.; Patch J. A.; Czyzewski A. M.; Dohm M. T.; Ivankin A.; Gidalevitz D.; Zuckermann R. N.; Barron A. E. Peptoids That Mimic the Structure, Function, and Mechanism of Helical Antimicrobial Peptides. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 2794–2799. 10.1073/pnas.0708254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J. E.; Alford M. A.; Yung D. B. Y.; Molchanova N.; Fortkort J. A.; Lin J. S.; Diamond G.; Hancock R. E. W.; Jenssen H.; Pletzer D.; Lund R.; Barron A. E. Self-Assembly of Antimicrobial Peptoids Impacts Their Biological Effects on ESKAPE Bacterial Pathogens. ACS Infect. Dis. 2022, 8, 533–545. 10.1021/acsinfecdis.1c00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo H.; Hirano M.; Misawa T.; Demizu Y. Helical Antimicrobial Peptide Foldamers Containing Non-Proteinogenic Amino Acids. ChemMedChem 2021, 16, 1226–1233. 10.1002/cmdc.202000940. [DOI] [PubMed] [Google Scholar]
- Tam J. P.; Lu Y. A.; Yang J. L. Antimicrobial Dendrimeric Peptides. Eur. J. Biochem. 2002, 269, 923–932. 10.1046/j.0014-2956.2001.02728.x. [DOI] [PubMed] [Google Scholar]
- Dhumal D.; Maron B.; Malach E.; Lyu Z.; Ding L.; Marson D.; Laurini E.; Tintaru A.; Ralahy B.; Giorgio S.; Pricl S.; Hayouka Z.; Peng L. Dynamic Self-Assembling Supramolecular Dendrimer Nanosystems as Potent Antibacterial Candidates against Drug-Resistant Bacteria and Biofilms. Nanoscale 2022, 14, 9286–9296. 10.1039/D2NR02305A. [DOI] [PubMed] [Google Scholar]
- Shai Y.; Oren Z. From “Carpet” Mechanism to de-Novo Designed Diastereomeric Cell-Selective Antimicrobial Peptides. Peptides 2001, 22, 1629–1641. 10.1016/S0196-9781(01)00498-3. [DOI] [PubMed] [Google Scholar]
- Di Grazia A.; Cappiello F.; Cohen H.; Casciaro B.; Luca V.; Pini A.; Di Y. P.; Shai Y.; Mangoni M. L. D-Amino Acids Incorporation in the Frog Skin-Derived Peptide Esculentin-1a(1-21)NH2 Is Beneficial for Its Multiple Functions. Amino Acids 2015, 47, 2505–2519. 10.1007/s00726-015-2041-y. [DOI] [PubMed] [Google Scholar]
- Hayouka Z.; Bella A.; Stern T.; Ray S.; Jiang H.; Grovenor C. R. M.; Ryadnov M. G. Binary Encoding of Random Peptide Sequences for Selective and Differential Antimicrobial Mechanisms. Angew. Chem., Int. Ed. Engl. 2017, 56, 8099–8103. 10.1002/anie.201702313. [DOI] [PubMed] [Google Scholar]
- Stach M.; Maillard N.; Kadam R. U.; Kalbermatter D.; Meury M.; Page M. G. P.; Fotiadis D.; Darbre T.; Reymond J.-L. Membrane Disrupting Antimicrobial Peptide Dendrimers with Multiple Amino Termini. MedChemComm 2012, 3, 86–89. 10.1039/c1md00272d. [DOI] [Google Scholar]
- Siriwardena T. N.; Lüscher A.; Köhler T.; van Delden C.; Javor S.; Reymond J.-L. Antimicrobial Peptide Dendrimer Chimera. Helv. Chim. Acta 2019, 102, e1900034 10.1002/hlca.201900034. [DOI] [Google Scholar]
- Gan B.-H.; Siriwardena T. N.; Javor S.; Darbre T.; Reymond J.-L. Fluorescence Imaging of Bacterial Killing by Antimicrobial Peptide Dendrimer G3KL. ACS Infect. Dis. 2019, 5, 2164–2173. 10.1021/acsinfecdis.9b00299. [DOI] [PubMed] [Google Scholar]
- Reymond J.-L. Peptide Dendrimers: From Enzyme Models to Antimicrobials and Transfection Reagents. Chimia 2021, 75, 535–538. 10.2533/chimia.2021.535. [DOI] [PubMed] [Google Scholar]
- Siriwardena T. N.; Gan B.-H.; Köhler T.; van Delden C.; Javor S.; Reymond J.-L. Stereorandomization as a Method to Probe Peptide Bioactivity. ACS Cent. Sci. 2021, 7, 126–134. 10.1021/acscentsci.0c01135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X.; Orsi M.; Capecchi A.; Köhler T.; van Delden C.; Javor S.; Reymond J.-L. An Intrinsically Disordered Antimicrobial Peptide Dendrimer from Stereorandomized Virtual Screening. Cell Rep. Phys. Sci. 2022, 3, 101161. 10.1016/j.xcrp.2022.101161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falla T. J.; Karunaratne D. N.; Hancock R. E. W. Mode of Action of the Antimicrobial Peptide Indolicidin. J. Biol. Chem. 1996, 271, 19298–19303. 10.1074/jbc.271.32.19298. [DOI] [PubMed] [Google Scholar]
- de la Fuente-Núñez C.; Reffuveille F.; Mansour S. C.; Reckseidler-Zenteno S. L.; Hernández D.; Brackman G.; Coenye T.; Hancock R. E. W. D-Enantiomeric Peptides That Eradicate Wild-Type and Multidrug-Resistant Biofilms and Protect against Lethal Pseudomonas Aeruginosa Infections. Chem. Biol. 2015, 22, 196–205. 10.1016/j.chembiol.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bonaventura I.; Jin X.; Visini R.; Probst D.; Javor S.; Gan B.-H.; Michaud G.; Natalello A.; Doglia S. M.; Köhler T.; van Delden C.; Stocker A.; Darbre T.; Reymond J.-L. Chemical Space Guided Discovery of Antimicrobial Bridged Bicyclic Peptides against Pseudomonas Aeruginosa and Its Biofilms. Chem. Sci. 2017, 8, 6784–6798. 10.1039/C7SC01314K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.; Li X.; Wang Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogoladze G.; Grigolava M.; Vishnepolsky B.; Chubinidze M.; Duroux P.; Lefranc M.-P.; Pirtskhalava M. Dbaasp: Database of Antimicrobial Activity and Structure of Peptides. FEMS Microbiol. Lett. 2014, 357, 63–68. 10.1111/1574-6968.12489. [DOI] [PubMed] [Google Scholar]
- Gasteiger E.; Jung E.; Bairoch A. SWISS-PROT: Connecting Biomolecular Knowledge Via a Protein Database. Curr. Issues Mol. Biol. 2001, 3, 47–55. 10.21775/cimb.003.047. [DOI] [PubMed] [Google Scholar]
- Mendez D.; Gaulton A.; Bento A. P.; Chambers J.; De Veij M.; Félix E.; Magariños M. P.; Mosquera J. F.; Mutowo P.; Nowotka M.; Gordillo-Marañón M.; Hunter F.; Junco L.; Mugumbate G.; Rodriguez-Lopez M.; Atkinson F.; Bosc N.; Radoux C. J.; Segura-Cabrera A.; Hersey A.; Leach A. R. ChEMBL: Towards Direct Deposition of Bioassay Data. Nucleic Acids Res. 2019, 47, D930–D940. 10.1093/nar/gky1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeriswyl S.; Personne H.; Di Bonaventura I.; Köhler T.; van Delden C.; Stocker A.; Javor S.; Reymond J.-L. A mixed chirality α-helix in a stapled bicyclic and a linear antimicrobial peptide revealed by X-ray crystallography. RSC Chem. Biol. 2021, 2, 1608–1617. 10.1039/D1CB00124H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strøm M. B.; Haug B. E.; Skar M. L.; Stensen W.; Stiberg T.; Svendsen J. S. The Pharmacophore of Short Cationic Antibacterial Peptides. J. Med. Chem. 2003, 46, 1567–1570. 10.1021/jm0340039. [DOI] [PubMed] [Google Scholar]
- Albada H. B.; Prochnow P.; Bobersky S.; Langklotz S.; Bandow J. E.; Metzler-Nolte N. Short Antibacterial Peptides with Significantly Reduced Hemolytic Activity Can Be Identified by a Systematic L-to-d Exchange Scan of Their Amino Acid Residues. ACS Comb. Sci. 2013, 15, 585–592. 10.1021/co400072q. [DOI] [PubMed] [Google Scholar]
- Albada H. B.; Prochnow P.; Bobersky S.; Bandow J. E.; Metzler-Nolte N. Highly Active Antibacterial Ferrocenoylated or Ruthenocenoylated Arg-Trp Peptides Can Be Discovered by an L-to-D Substitution Scan. Chem. Sci. 2014, 5, 4453–4459. 10.1039/C4SC01822B. [DOI] [Google Scholar]
- Gautier R.; Douguet D.; Antonny B.; Drin G. HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics 2008, 24, 2101–2102. 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- Townsley L. E.; Tucker W. A.; Sham S.; Hinton J. F. Structures of Gramicidins A, B, and C Incorporated into Sodium Dodecyl Sulfate Micelles. Biochemistry 2001, 40, 11676–11686. 10.1021/bi010942w. [DOI] [PubMed] [Google Scholar]
- Jasanoff A.; Fersht A. R. Quantitative Determination of Helical Propensities from Trifluoroethanol Titration Curves. Biochemistry 1994, 33, 2129–2135. 10.1021/bi00174a020. [DOI] [PubMed] [Google Scholar]
- Arunkumar A. I.; Kumar T. K.; Yu C. Specificity of Helix-Induction by 2,2,2-Trifluoroethanol in Polypeptides. Int. J. Biol. Macromol. 1997, 21, 223–230. 10.1016/s0141-8130(97)00064-0. [DOI] [PubMed] [Google Scholar]
- Hennig A.; Gabriel G. J.; Tew G. N.; Matile S. Stimuli-Responsive Polyguanidino-Oxanorbornene Membrane Transporters as Multicomponent Sensors in Complex Matrices. J. Am. Chem. Soc. 2008, 130, 10338–10344. 10.1021/ja802587j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd N. E.; Hoang H. N.; Abbenante G.; Fairlie D. P. Left- and Right-Handed Alpha-Helical Turns in Homo- and Hetero-Chiral Helical Scaffolds. J. Am. Chem. Soc. 2009, 131, 15877–15886. 10.1021/ja9065283. [DOI] [PubMed] [Google Scholar]
- Nanda V.; DeGrado W. F. Computational Design of Heterochiral Peptides against a Helical Target. J. Am. Chem. Soc. 2006, 128, 809–816. 10.1021/ja054452t. [DOI] [PubMed] [Google Scholar]
- Miles A. J.; Ramalli S. G.; Wallace B. A. DichroWeb, a Website for Calculating Protein Secondary Structure from Circular Dichroism Spectroscopic Data. Protein Sci. 2022, 31, 37–46. 10.1002/pro.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencher S. W.; Gloeckner J. Estimation of Globular Protein Secondary Structure from Circular Dichroism. Biochemistry 1981, 20, 33–37. 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- Takechi Y.; Tanaka H.; Kitayama H.; Yoshii H.; Tanaka M.; Saito H. Comparative Study on the Interaction of Cell-Penetrating Polycationic Polymers with Lipid Membranes. Chem. Phys. Lipids 2012, 165, 51–58. 10.1016/j.chemphyslip.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Lyu P. C.; Sherman J. C.; Chen A.; Kallenbach N. R. Alpha-Helix Stabilization by Natural and Unnatural Amino Acids with Alkyl Side Chains. Proc. Natl. Acad. Sci. U.S.A. 1991, 88, 5317–5320. 10.1073/pnas.88.12.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Jeddou F.; Falconnet L.; Luscher A.; Siriwardena T.; Reymond J.-L.; van Delden C.; Köhler T. Adaptive and Mutational Responses to Peptide Dendrimer Antimicrobials in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2020, 64, 020400. 10.1128/AAC.02040-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutet S. A.; Flannagan R. S.; Kooi C.; Sokol P. A.; Valvano M. A. A Complete Lipopolysaccharide Inner Core Oligosaccharide Is Required for Resistance of Burkholderia Cenocepacia to Antimicrobial Peptides and Bacterial Survival In Vivo. J. Bacteriol. 2006, 188, 2073–2080. 10.1128/JB.188.6.2073-2080.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabernet G.; Müller A. T.; Hiss J. A.; Schneider G. Membranolytic Anticancer Peptides. MedChemComm 2016, 7, 2232–2245. 10.1039/C6MD00376A. [DOI] [Google Scholar]
- Zakharova E.; Orsi M.; Capecchi A.; Reymond J.-L. Machine Learning Guided Discovery of Non-Hemolytic Membrane Disruptive Anticancer Peptides. ChemMedChem 2022, 17, e202200291 10.1002/cmdc.202200291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco I.; Molchanova N.; Holmedal E.; Jenssen H.; Hummel B. D.; Watts J. L.; Håkansson J.; Hansen P. R.; Svenson J. Correlation between Hemolytic Activity, Cytotoxicity and Systemic in Vivo Toxicity of Synthetic Antimicrobial Peptides. Sci. Rep. 2020, 10, 13206. 10.1038/s41598-020-69995-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell E. P.; Sabin C.; Snajdrova L.; Pokorna M.; Perret S.; Gautier C.; Hofr C.; Gilboa-Garber N.; Koca J.; Wimmerova M.; Imberty A. High Affinity Fucose Binding of Pseudomonas Aeruginosa Lectin PA-IIL: 1.0 A Resolution Crystal Structure of the Complex Combined with Thermodynamics and Computational Chemistry Approaches. Proteins 2004, 58, 735–746. 10.1002/prot.20330. [DOI] [PubMed] [Google Scholar]
- Roethlisberger P.; Istrate A.; Marcaida Lopez M. J.; Visini R.; Stocker A.; Reymond J. L.; Leumann C. J. X-Ray Structure of a Lectin-Bound DNA Duplex Containing an Unnatural Phenanthrenyl Pair. Chem. Commun. 2016, 52, 4749–4752. 10.1039/c6cc00374e. [DOI] [PubMed] [Google Scholar]
- He R.; Di Bonaventura I.; Visini R.; Gan B.-H.; Fu Y.; Probst D.; Lüscher A.; Köhler T.; van Delden C.; Stocker A.; Hong W.; Darbre T.; Reymond J.-L. Design, Crystal Structure and Atomic Force Microscopy Study of Thioether Ligated D,L-Cyclic Antimicrobial Peptides against Multidrug Resistant Pseudomonas Aeruginosa. Chem. Sci. 2017, 8, 7464–7475. 10.1039/C7SC01599B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeriswyl S.; Gan B.-H.; Siriwardena T. N.; Visini R.; Robadey M.; Javor S.; Stocker A.; Darbre T.; Reymond J.-L. X-Ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas Aeruginosa Lectin B Complexes. ACS Chem. Biol. 2019, 14, 758–766. 10.1021/acschembio.9b00047. [DOI] [PubMed] [Google Scholar]
- Michaud G.; Visini R.; Bergmann M.; Salerno G.; Bosco R.; Gillon E.; Richichi B.; Nativi C.; Imberty A.; Stocker A.; Darbre T.; Reymond J.-L. Overcoming Antibiotic Resistance in Pseudomonas Aeruginosa Biofilms Using Glycopeptide Dendrimers. Chem. Sci. 2016, 7, 166–182. 10.1039/C5SC03635F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeriswyl S.; Javor S.; Stocker A.; Darbre T.; Reymond J.-L. X-Ray Crystal Structure of a Second-Generation Peptide Dendrimer in Complex with Pseudomonas Aeruginosa Lectin LecB. Helv. Chim. Acta 2019, 102, e1900178 10.1002/hlca.201900178. [DOI] [Google Scholar]
- Abraham M. J.; Murtola T.; Schulz R.; Páll S.; Smith J. C.; Hess B.; Lindahl E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Sreerama N.; Woody R. W. Estimation of Protein Secondary Structure from Circular Dichroism Spectra: Comparison of CONTIN, SELCON, and CDSSTR Methods with an Expanded Reference Set. Anal. Biochem. 2000, 287, 252–260. 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- Wiegand I.; Hilpert K.; Hancock R. E. W. Agar and Broth Dilution Methods to Determine the Minimal Inhibitory Concentration (MIC) of Antimicrobial Substances. Nat. Protoc. 2008, 3, 163–175. 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- Berridge M. V.; Herst P. M.; Tan A. S. Tetrazolium Dyes as Tools in Cell Biology: New Insights into Their Cellular Reduction. Biotechnol. Annu. Rev. 2005, 11, 127–152. 10.1016/S1387-2656(05)11004-7. [DOI] [PubMed] [Google Scholar]
- Liebschner D.; Afonine P. V.; Baker M. L.; Bunkóczi G.; Chen V. B.; Croll T. I.; Hintze B.; Hung L.-W.; Jain S.; McCoy A. J.; Moriarty N. W.; Oeffner R. D.; Poon B. K.; Prisant M. G.; Read R. J.; Richardson J. S.; Richardson D. C.; Sammito M. D.; Sobolev O. V.; Stockwell D. H.; Terwilliger T. C.; Urzhumtsev A. G.; Videau L. L.; Williams C. J.; Adams P. D. Macromolecular Structure Determination Using X-Rays, Neutrons and Electrons: Recent Developments in Phenix. Acta Crystallogr., Sect. D: Struct. Biol. 2019, 75, 861–877. 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M. D.; Ballard C. C.; Cowtan K. D.; Dodson E. J.; Emsley P.; Evans P. R.; Keegan R. M.; Krissinel E. B.; Leslie A. G. W.; McCoy A.; McNicholas S. J.; Murshudov G. N.; Pannu N. S.; Potterton E. A.; Powell H. R.; Read R. J.; Vagin A.; Wilson K. S. Overview of the CCP4 Suite and Current Developments. Acta Crystallogr., Sect. D: Struct. Biol. 2011, 67, 235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
- Emsley P.; Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr., Sect. D: Struct. Biol. 2004, 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr., Sect. D: Struct. Biol. 2010, 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostenbrink C.; Soares T. A.; van der Vegt N. F. A.; van Gunsteren W. F. Validation of the 53A6 GROMOS Force Field. Eur. Biophys. J. 2005, 34, 273–284. 10.1007/s00249-004-0448-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.