Abstract
Introduction
Dent disease is an X-linked recessive disorder associated with low molecular weight proteinuria (LMWP), nephrocalcinosis, kidney stones, and kidney failure in the third to fifth decade of life. It consists of Dent disease 1 (DD1) (60% of patients) because of pathogenic variants in the CLCN5 gene and Dent disease 2 (DD2) with changes in OCRL.
Methods
Retrospective review of 162 patients from 121 different families with genetically confirmed DD1 (82 different pathogenic variants validated using American College of Medical Genetics [ACMG] guidelines). Clinical and genetic factors were compared using observational statistics.
Results
A total of 110 patients had 51 different truncating (nonsense, frameshifting, large deletions, and canonical splicing) variants, whereas 52 patients had 31 different nontruncating (missense, in-frame, noncanonical splicing, and stop-loss) changes. Sixteen newly described pathogenic variants were found in our cohort. Among patients with truncating variants, lifetime stone events positively correlated with chronic kidney disease (CKD) evolution. Patients with truncating changes also experienced stone events earlier in life and manifested a higher albumin excretion rate than the nontruncating group. Nevertheless, neither age of nephrocalcinosis nor CKD progression varied between the truncating versus nontruncating patients. A large majority of nontruncating changes (26/31; 84%) were clustered in the middle exons that encode the voltage ClC domain whereas truncating changes were spread across the protein. Variants associated with kidney failure were restricted to truncating (11/13 cases), plus a single missense variant previously shown to markedly reduce ClC-5 functional activity that was found in the other 2 individuals.
Conclusion
DD1 manifestations, including the risk of kidney stones and progression to kidney failure, may relate to the degree of residual ClC-5 function.
Keywords: CKD, Dent disease, hypercalciuria, nephrocalcinosis, proteinuria
Graphical abstract
See Commentary on Page 1127
Dent disease (DD) is an X-linked recessive proximal renal tubulopathy characterized by LMWP, hypercalciuria, and hypophosphatemia. Nephrocalcinosis, kidney stones, hematuria, and progressive CKD are also common.1,2 DD is caused by pathogenic variants in CLCN5 (DD1; OMIM #300009; ∼60% of cases) and OCRL (DD2; OMIM #300555; ∼ 20% of cases). The remaining ∼25% of cases with an apparent DD phenotype are, to date, genetically unresolved with no specific gene implicated.2, 3, 4, 5, 6 However, characteristic DD features (nephrocalcinosis, LMWP) can be associated with other monogenic causes of kidney stone disease,7 and X-linkage can be difficult to establish if a detailed pedigree is unavailable.7
CLCN5 consists of 15 exons (13 coding exons)8 that encode the voltage-gated chloride transporter ClC-5. This protein is largely expressed in the proximal tubule and α-intercalated cells of the collecting duct, predominantly in early endosomes.9 Therefore, the phenotype of DD1 appears to be limited to the kidney. Although over 100 pathogenic CLCN5 variants have been described to date,10,11 DD1 genotype-phenotype correlations have not been clearly described.11, 12, 13, 14 However, there is great phenotype variability among DD1 patients that could potentially be influenced by the effects of specific mutations on protein processing and folding in the endoplasmic reticulum, failure of endosomal acidification, abnormal subcellular localization of the protein, and/or production of a nonfunctional protein that reaches the plasma membrane but exhibits reduced membrane currents.2,15, 16, 17, 18 Therefore, in the current study we examined DD1 genotype-phenotype correlations among patients in the Rare Kidney Stone Consortium DD registry, focusing on kidney stone events, nephrocalcinosis, urinary calcium and protein excretion, and CKD progression.
Methods
The Study Cohort
The Mayo Clinic Institutional Review Board approved this study. Patients enrolled in the Rare Kidney Stone Consortium DD Registry as of January 1, 2021, were included.19 All patients provided informed written consent at the time of enrollment.
Genetic Analysis
Genetic analysis was performed either by Sanger sequencing of the 2 DD genes or via a kidney stone disease targeted next-generation sequencing panel that included CLCN5.7 All positive results by the targeted next-generation sequencing were confirmed by Sanger sequencing of the implicated CLCN5 exons. Variants within CLCN5 were classified according to the ACMG and American College of Pathologists guidelines.20 All variants were described based on the transcript NM_001127898.4 (mRNA, 9863 base pairs, 15 exons [13 coding], and a 2451 base pairs coding sequence encoding an 816 aa protein),21 as listed in gnomAD version 2.1.1. Exons are numbered from the first coding exon. Frameshifting indels, nonsense, and canonical splicing variants were defined as truncating, whereas missense, in-frame indels, noncanonical splicing variants, and stop-loss variants were defined as nontruncating. Only patients with molecularly confirmed DD1 were included in this study.
Clinical Phenotype
Clinical information was retrieved from the Rare Kidney Stone Consortium DD Registry and/or medical record review. All laboratory measurements were performed in clinical laboratories. Serum creatinine and urine biochemical parameters (random or 24-hour urine collections) were analyzed, including total protein, albumin, α-1 microglobulin, β-2 microglobulin, citrate, and calcium. This cohort consisted of both adults and children, and therefore the estimated glomerular filtration rate was calculated using the Full Age Spectrum equation.22 To pool data from children and adults, all urine excretions for subjects <18 years old at the time of collection were normalized to the average adult body surface area of 1.73 m2 calculated from height and weight using Du Bois and Du Bois formula23
CKD was classified according to the K/DOQI CKD staging system.24 Family history of confirmed DD1, CKD, lifetime stone events, and nephrocalcinosis were also abstracted. All clinical stone events in a subject’s lifetime were ascertained from clinician medical record review (with or without imaging), including symptomatic events (pain, gross hematuria, or infection attributed to stones), spontaneous passage, or a urologic procedure for stone management were combined as lifetime stone events. The age of first stone formation, nephrocalcinosis by imaging, and CKD stage 3 or greater were obtained. Diagnostic urine parameters were extracted at the first time point they were available from the medical record.
Statistical Analysis
Continuous variables are reported as median and interquartile range (IQR), and categorical variables as percentages. Fisher’s exact test was used to compare categorical and binominal clinical data between the 2 groups (patients with truncating vs. those with nontruncating variants). Concordance of proteinuria and hypercalciuria was defined by clinical thresholds of 150 mg/d for proteinuria and 300 mg/d for hypercalciuria. Individuals who had a proteinuria level greater than 150 mg/d and a hypercalciuria levels of more than 300 mg/d within families containing 2 or more individuals were considered to have the phenotypes. In addition, stone events were assessed by the presence or absence of kidney stones throughout the patient’s lifetime during the follow-up period. One Proportion Z-test was used to compare low (0%–50%) and high (51%–100%) concordance in families regarding the presence of kidney stones, proteinuria, and hypercalciuria. The variable of “CKD stages 3, 4, and 5” is an ordinal variable that we used a numeric coding scheme to represent (absence of CKD, CKD stage 1 or 2 = 0; CKD stage 3a = 1; CKD stage 3b = 2; CKD stage 4 = 3; and CKD stage 5 = 4). In addition, “lifetime stone events” was a continuous variable with the number of events experienced per each individual. Spearman rank test was used between urine parameters in Table 1 (the entire cohort) and in Table 2 (the cohort divided by genotype category) and the outcomes of CKD and lifetime stone events. Given the skewed data distribution, nonparametric tests, like Mann-Whitney U test, were used to compare continuous values. Kaplan–Meier survival analysis was employed to differentiate kidney survival of the 2 groups for the onset of CKD stage 3 (estimated glomerular filtration rate 30–59 ml/d per 1.73 m2), or CKD stage 5 (estimated glomerular filtration rate <15 ml/d per 1.73 m2), and age of nephrocalcinosis (that is, when first detected by available imaging). Statistical analyses were performed using BlueSky statistics software V. 7.20 (BlueSky statistics LLC, Chicago, IL, USA), and a P-value < 0.05 was accepted as statistically significant.
Table 1.
Correlations between urine biochemistry of the entire cohort and the outcomes of lifetime stone events and chronic kidney disease
| Urine parameter | Chronic kidney disease | Lifetime stone events |
|---|---|---|
| 24-hr urine calcium (mg/d) | n: 104 | n: 26 |
| r: 0.3 | r: 0.5 | |
| P: 0.2 | P: 0.01 | |
| 24-hr total urine protein (gram/d) | n: 51 | n: 20 |
| r: 0.3 | r: 0.1 | |
| P: 0.12 | P: 0.9 | |
| 24-hr Urine Phosphorus (mg/d) | n: 51 | n: 26 |
| r: 0.2 | r: 0.4 | |
| P: 0.08 | P: 0.06 | |
| Urine calcium/creatinine ratio | n: 121 | n: 20 |
| r: 0.1 | r: 0.4 | |
| P: 0.7 | P: 0.08 |
n, number; P,P-value; r, correlation coefficient.
Subjects less than 18 years of age were extrapolated to average adult size (mg/1.73 m2/ d). “CKD stage” is an ordinal variable with values ranging from 0 to 4 and has been treated as such in the analysis. “Lifetime stone events” is a continuous variable representing the actual number of stone events an individual has experienced through last follow-up.
“Chronic kidney disease” is an ordinal variable with values ranging from 0 to 4 and has been treated as such in the analysis. “Lifetime stone events” is a continuous variable representing the actual number of stone events an individual has experienced through last follow-up.
Table 2.
Correlation analysis of predictors and outcomes, comparing patients with truncating and with nontruncating variants
| Variant group | Urine parameter | Lifetime stone events | Chronic kidney disease |
|---|---|---|---|
| Truncating group | 24h urine calcium mg/d | n: 18 | n: 68 |
| n: 110 | r: 0.4 | r: 0.2 | |
| P: 0.02 | P: 0.11 | ||
| 24h urine phosphorus m/d | n: 12 | n: 32 | |
| r: 0.6 | r: 0.2 | ||
| P: 0.04 | P: 0.67 | ||
| Total urine protein gram/d | n: 5 | n: 43 | |
| r: 0.1 | r: 0.3 | ||
| P: 0.85 | P: 0.04 | ||
| Calcium/creatinine ratio | n: 16 | n:79 | |
| r: 0.4 | r: 0.1 | ||
| P: 0.07 | P: 0.6 | ||
| Nontruncating group | 24h urine calcium mg/d | n: 8 | n: 37 |
| n: 52 | r: 0.34 | r: 0.13 | |
| P: 0.46 | P: 0.62 | ||
| 24h urine phosphate mg/d | n: 8 | n: 20 | |
| r: 0.2 | r: −0.2 | ||
| P: 0.63 | P: 0.50 | ||
| Total urine protein gram/d | n: 5 | n: 24 | |
| r: −0.2 | r: 0.2 | ||
| P: 0.71 | P: 0.28 | ||
| Calcium/creatinine ratio | n: 33 | n: 43 | |
| r: 0.1 | r: 0.1 | ||
| P: 0.5 | P: 0.6 |
n, number; P,P-value; r, correlation coefficient.
Subjects less than 18 years of age were extrapolated to average adult size (mg/1.73 m2/ d). “CKD stage” is an ordinal variable with values ranging from 0 to 4 and has been treated as such in the analysis. “Lifetime stone events” is a continuous variable representing the actual number of stone events an individual has experienced through last follow-up.
“Chronic kidney disease” is an ordinal variable with values ranging from 0 to 4 and has been treated as such in the analysis. “Lifetime stone events” is a continuous variable representing the actual number of stone events an individual has experienced through last follow-up.
Results
Genetic Makeup of the Cohort
All variants were scored according to ACMG guidelines as pathogenic or likely pathogenic (see Supplementary Table S1), except for c.1802G>A (p.Gly601Asp), which was scored as a variant of unknown significance. Thus, this family was excluded from subsequent genotype-phenotype analyses, and the remaining 162 patients with DD1 from 121 kindreds were included in our analysis. Most patients were recruited from the USA (73.3%; 85 pedigrees, 114 patients), including 6 pedigrees commercially genotyped (GeneDx, Inc) and 79 recruited and genetically screened at Mayo Clinic under a research protocol. An additional 17 patients (10.3%; 12 pedigrees) were recruited from the University of Padua, Italy; and 31 patients (22.4%; 26 pedigrees) from the University of Seoul, South Korea.
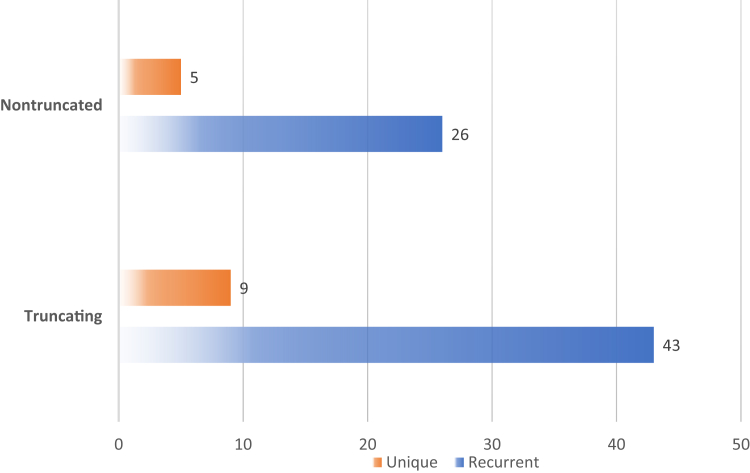
A total of 82 different pathogenic variants were observed in this cohort. Missense variants were the most common (34%), followed by frameshifting (28%) and nonsense (21%) (Figure 1). Truncating pathogenic variants (n = 51) included canonical splicing, nonsense, and frameshifting duplications and deletions, including several multiexon, copy number variant deletions. The 31 nontruncating pathogenic variants included missense, in-frame deletion or duplication, noncanonical splicing, and a stop-loss variant (c.2450A>G; p.Ter817Trpext46Ter). Among the truncating changes, 43 in our cohort were found in more than 1 family and/or described in the prior literature, and 9 were present in only 1 family. The most common truncating variant was p.Arg98Ter, found in 5 families. Most nontruncating pathogenic variants (n = 26) were present in multiple families or previously described, whereas 5 were only present in 1 family, including 2 newly described (Figure 2). Missense variant p.Ser314Leu was the most abundant, found in 9 families. Overall, 17% were unique variants and 83% recurrent pathogenic variants.
Figure 1.
Prevalence of 82 unique pathogenic variant types (a) and mutation groups (b) among the 121 families in the study. Missense mutations (n = 27), frameshift mutations (n = 23) and nonsense mutations (n = 17) were most frequent followed by canonical splice site mutations (n = 6), copy number variants (CNV; n = 5), in-frame dup/ins (n = 2), noncanonical splicing (n = 1), and stop-loss (n = 1).
Figure 2.
Comparison of the number of families with recurrent versus unique pathogenic variants in the truncating and nontruncating groups. Recurrent indicates more than 1 family within our cohort and/or with variants previously described in the literature; Unique indicates the variant present just in 1 family in our cohort.
Truncating mutations were distributed throughout CLCN5 and the protein encoding domains of ClC-5 (Figure 3). However, nontruncating mutations were largely clustered in the middle exons encoding the voltage ClC domain of the protein (Figure 4). Sixteen newly described pathogenic variants were identified, including 3 copy number variant deletions, 4 canonical splicing, 3 nonsense, 3 frameshifting deletions, 2 missense, and 1 in-frame deletion. All variants and their ACMG classification are presented in Supplementary Table S1.
Figure 3.
Location of truncating pathogenic variants (frameshift, nonsense, noncanonical splicing, and CNV) along the entire length of CLCN5 and its known functional domains.
∗Novel variants are in red color
Figure 4.
Location of nontruncating pathogenic variants (missense, in-frame [deletions, insertions], stop-loss, and noncanonical splicing) along ClC-5 and its functional domains. The majority fall within middle exons encoding the voltage ClC domain of the protein.
∗Novel variants are in red color
Clinical Characteristics
Initial data were collected as available at the time of clinical DD diagnosis, or the first available data after that date. Thirteen individuals had data for just one visit. The median (IQR) age at diagnosis was 14 (10–23) years. The median (IQR) follow-up period was 5 (1.5–12) years, and the median (IQR) age at last available follow-up was 17 (11.8–28) years.
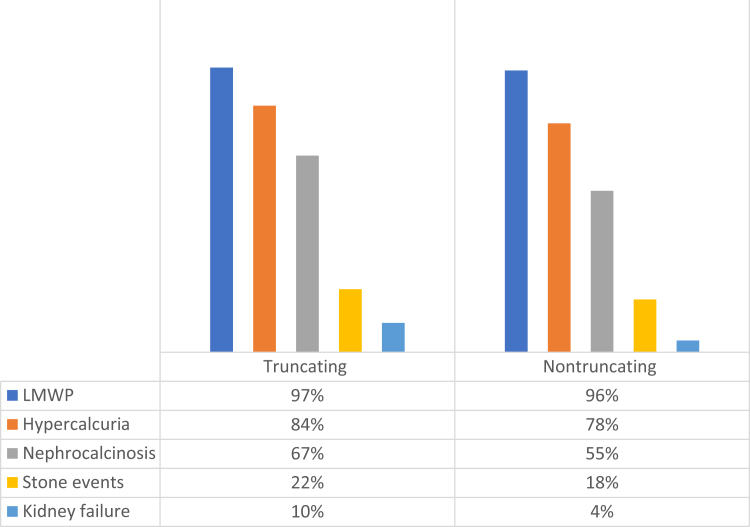
At the last available follow-up visit, nephrocalcinosis was present in 67% of patients with truncating variants and in 55% of patients with nontruncating variants (Table 3). LMWP was documented in 97% of the patients with truncating variants and 96% of those with nontruncating variants. Hypercalciuria was present in 81% and 78% of patients with truncating versus nontruncating, respectively. Kidney stone events by the last follow-up were present in 22% of patients with truncating variants versus 18% of the patients with nontruncating variants (Figure 5).
Table 3.
Clinical endpoints in patients with truncating versus those with nontruncating variant groups
| Group type patients pedigrees |
Truncating n:110 n:82 |
Nontruncating n:52 n:41 |
P-value |
|---|---|---|---|
| Clinical stone events (yes/no) | n: 23 (20%) | n: 9 (18%) | 0.87 |
| Nephrocalcinosis | |||
| At first visit | n: 35/69 (50%) | n: 15/40 (38%) | 0.12 |
| At last, follow-up | n: 46/69 (67%) | n: 22/40 (55%) | 0.11 |
| Hematuria at the time of diagnosis (yes/no) | n: 17 (15%) | n: 18 (37%) | <0.01 |
| Family history of Dent disease-1 (N = pedigrees) | n: 19/82 (23%) | n: 4/41 (9%) | <0.01 |
| Family history of nephropathy (N = pedigrees) | n: 21/82 (25%) | n: 11/41 (26%) | 0.88 |
| Age at 1st stone event (yr) | 14 (10–20) | 27 (21–27) | 0.01 |
| Lifetime stone events/yr | 0.12 (.057–0.321) | 0.38 (0.237–0.667) | 0.06 |
| Urine calcium/creatinine ratio (24h urine) | 0.24 (0.14–0.33) | 0.22 (0.15–0.42) | 0.89 |
| Urine citrate/creatinine ratio (24h urine) | 0.56 (0.317–0.706) | 0.47 (0.367–0.688) | 0.77 |
| Total urine protein (gram/24h urine) | |||
| At first visit | 1.5 (0.7–1.87) | 1.4 (0.7–2.2) | 0.60 |
| At last, follow-up | 1.07 (0.6–1.6) | 1.3 (0.8–2.1) | 0.10 |
| Urine albumin (mg/24h urine) | 180.6 (119.15–279.5) | 106.6 (74.45–176.95) | 0.02 |
| Low molecular weight proteinuria: | |||
| Alpha-1M mg/l | 502.5 (321.5–602) | 157 (71–522) | 0.27 |
| Beta-2 microglobulin mg/l | 48.6 (30.9–62.6) | 50.8 (38.7–61.6) | 0.86 |
For nephrocalcinosis, individuals missing imaging were excluded from the analysis. Comparisons were between all individual subjects except for family history of DD1 and family history of nephropathy which were between pedigrees.
Figure 5.
Prevalence of clinical phenotypes at last follow-up in the truncating and nontruncating groups.
Among the entire cohort, associations between urine parameters and the clinical outcomes of lifetime stone events and CKD progression are summarized in Table 1. Urine calcium excretion (mg/d) positively associated with lifetime stone events (P = 0.01), whereas urine phosphorus excretion (mg/d) did not significantly correlate with lifetime stone events (P = 0.06) or CKD risk (P = 0.08). Total urine protein excretion (g/d) was not associated with CKD progression (P = 0.12) or lifetime stone events (P = 0.9). The urine calcium/creatinine ratio did not correlate with CKD (P = 0.7) or with lifetime stone events (P = 0.08).
Among subjects with truncating mutations, lifetime stone events positively correlated with urine calcium excretion (P = 0.02) and urine phosphorus excretion (P = 0.04). CKD progression positively correlated with total urine protein excretion (P = 0.04). Interestingly, among patients with truncating variants, a greater number of lifetime stone events correlated with progression of CKD stage (r = 0.5, P = 0.02). Among subjects with nontruncating mutations, lifetime stone events and CKD progression did not correlate with urine biochemistry (Table 2).
Genotype/Phenotype Correlations
A total of 110 patients had truncating, and 52 patients had nontruncating pathogenic variants. No difference was observed regarding age at diagnosis between the truncating group and the nontruncating group (14 [8–22.3] vs. 11 [7–17.5] years; P = 0.56) or at last available follow-up visit (17.5 [11–23.8] vs. 17 [12–28.3] years; P = 0.58). Thirteen patients (8%) developed kidney failure at a median age of 35 years (IQR: 32.5–43.75); 11 of these had truncating changes, including 1 family (2 individuals) carrying the variant p.Arg774Ter., and 2 unrelated individuals had the recurrent missense variant c.941C>T (p. Ser314Leu).25 Multiple stone events in more than 1 family member were reported in 4 families with truncating changes (p.Arg774Ter, p. Phe758fs, p. Arg417Ter, and p.Glu55fs) and in 1 family with a nontruncating pathogenic variant (p.Gly330Val). Within DD1 families of 2 or more confirmed cases, concordance of kidney stone events, hypercalciuria (mg/d), and proteinuria (g/d) were assessed. There was significant concordance in families with 50% or more of their individuals with proteinuria (g/d) (P ≤ 0.05), and hypercalciuria (mg/d) (P ≤ 0.05) adjusted for age and body surface area of 1.73 m2. However, familial concordance for the presence of kidney stone events was not statistically significant (P = 0.18; Table 4)
Table 4.
Concordance between families with 2 or more individuals confirmed for DD1
| Phenotype | Kidney stones | P-value | Proteinuria | P-value | Hypercalciuria | P-value | |||
|---|---|---|---|---|---|---|---|---|---|
| % Concordance within the family | 0%–50% | 51%–100% | 0%–50% | 51%–100% | 0%–50% | 51%–100% | |||
| N. of families | 8 | 14 | 0.18 | 3 | 12 | < 0.05 | 2 | 10 | < 0.05 |
DD1, Dent disease 1.
Patients with truncating variants had a higher albumin excretion and experienced symptomatic stone passages at a younger age (Table 1). However, patients with nontruncating variants had a greater incidence of hematuria at the time of first presentation. There was no difference between the truncating and nontruncating groups in lifetime stone events, nephrocalcinosis, total urine protein (g/d) either during the first visit or during the follow-up, presence of hypercalciuria, LMWP (β-2 microglobulin or α-1 microglobulin) (mg/d), or family history of CKD. Families with truncating changes had a relatively higher number of siblings and first-degree relatives with a DD1 diagnosis than those with nontruncating mutations.
Time-to-event survival analysis by truncating status suggested no difference in time to reach stage 3 CKD (P = 0.16) or stage 5 CKD (kidney failure) (P = 0.28), nor the age of first recorded nephrocalcinosis (P = 0.45) (Figure 6).
Figure 6.
Kaplan–Meier survival analysis of clinical phenotypes by variant type (truncating vs. nontruncating). Age at CKD stage 3a (eGFR < 60ml/min/1.73m2) (a), kidney failure (eGFR < 15ml/min/1.73 m2) (b), and nephrocalcinosis (c) are shown. eGFR, estimated glomerular filtration rate.
Discussion
This study analyzed a cohort of 162 patients with genetically confirmed DD1, with all pathogenic variants categorized as likely pathogenic or pathogenic using ACMG criteria. We identified truncating (frameshifting deletions, insertions, and duplications, including copy number variants, nonsense, and canonical splicing) pathogenic variants in 62% and nontruncating (missense, noncanonical splicing, in-frame deletions, nonstop) in 38%, consistent with prior reports.11 Clinical outcomes did not vary greatly when patients were compared as truncating versus nontruncating; however, urinary calcium and protein excretion predicted clinical outcomes in the truncating (but not the nontruncating) group. Moreover, an increase in the number of kidney stone events in patients with truncating changes correlated with their risk of progressive loss of kidney function (documented decline in CKD stage).
One of the striking findings in this study was that truncating variants were distributed throughout the protein in no apparent pattern, whereas nontruncating changes were mainly clustered in the voltage ClC domain. Chang et al.25 previously investigated the functional effect of representative mutations in cultured mammalian cells and Xenopus oocytes. The p.Ser314Leu missense variant exhibited a large reduction of ClC-5 current (90%) compared to wildtype, whereas other missense changes were associated with only minimal (p.Arg415Trp; −35%) or no (p.Thr727Ser) change in current amplitude.25,26 The p. Ser314Leu and p. Arg415Trp variants affect the voltage ClC domain, which extends from 220 aa to 620 aa. However, p.Thr727Ser is in the C-terminal cytoplasmic domain that extends from 623 aa to 816 aa. Therefore, the location of p.Thr727Ser probably explains why it does not alter channel function.
In the current study, 13 patients progressed to kidney failure. Among these, 2 unrelated individuals had the missense p.Ser314Leu change whereas the remainder had truncating changes (p.Arg98Ter; p. Arg104Ter, p.Ile106fs; p.Ile159fsTer8; p.Trp189Ter; p.Gly582?; p.Ser756fs4Ter; p.Arg774Ter).11,27 Although there is no published data regarding the functional effects of these truncating changes, it is likely that they do not allow the generation of protein product with any function. Thus, we hypothesize that the risk of kidney failure is related to residual ClC-5 activity, and that missense changes resulting in complete loss of protein function will be associated with a greater CKD risk. Therefore, in vitro assays of the effects of specific variants (especially missense ones) on protein function could be helpful to risk-stratify patients. The lack of a clear differentiation between the truncating versus nontruncating variants and CKD risk may be confounded by a variable mix of residual protein function among the nontruncating variants, and the young age of the cohort, both reducing power for this particular analysis.
The mechanism of hypercalciuria in DD1 is not entirely clear.3,18 However, it does not appear directly related to ClC-5 protein function because this protein is not involved directly or indirectly in calcium reabsorption in the proximal tubule. One potential mechanism is that lack of parathyroid hormone reabsorption in the proximal tubule results in secondary distal effects, notably increased formation of activated 1,25 vitamin-D.3,28 The current study did not show a clear differentiation between urinary calcium excretion in the truncating versus nontruncating groups. However, we observed an association between age of first kidney stone event and truncating status. Therefore, more easily defined clinical phenotypes (e.g., stones) may reflect the underlying biology better than the urinary biochemistries, which can exhibit biologic variability and may not be frequently or repeatedly available. Furthermore, the urinary biochemistry data are not complete in this retrospective observational cohort, perhaps limiting the ability to see genotype correlations with the urinary calcium phenotype.
One might hypothesize that urinary calcium excretion would correlate with kidney stone events and that urinary protein excretion would correlate with CKD risk. Indeed, these correlations were positive in the truncating group, but not observed in the nontruncating group. Although there were significant correlations between the parameters and the outcomes in the truncating group, the correlation coefficient suggested that the associations were not strong. In addition, the underlying reason for these apparent differences is not readily apparent. However, the truncating group may represent a more homogeneous population among whom ClC-5 protein function is largely abolished. In contrast, the nontruncating group is a mixture of patients with absent to poorly functioning protein and those with significant residual activity. Therefore, compensatory mechanisms may be more effective in the nontruncating group and hence reduce correlations of urinary calcium with harder clinical outcomes. Furthermore, this study suggests that strategies to effectively reduce urinary calcium excretion might attenuate the risk of kidney stone events, and possibly nephrocalcinosis, whereas strategies that effectively reduce urinary protein excretion might slow CKD progression, even perhaps in a subset of the nontruncating group.
Recent studies suggest that ClC-5 is expressed in podocytes and that focal global sclerosis is associated with estimated glomerular filtration rate in DD1 patients at the time of kidney biopsy.29,30 This observation suggests the hypothesis that loss of ClC-5 protein activity within podocytes influences DD1 CKD progression. In the current study, urinary albumin excretion was higher in the truncating group. This might reflect more complete loss of protein function and hence greater podocyte dysfunction in this subset. Alternatively, significant albuminuria can result when proximal tubular reabsorption of filtered protein is reduced resulting in a mixture of greater amounts of albumin plus other proteins in the urine. Therefore, the increased albuminuria in the truncating group might also reflect complete loss of ClC-5 activity within the proximal tubule. Indeed, urinary excretion of the low molecular weight protein α-1 microglobulin was also higher, albeit nonsignificantly, in the truncating group.
Our study has several limitations. Clinical data were extracted from clinical records and the breadth and longitudinal nature of data were, in some cases, limited. Moreover, the effect of medications on the urine parameters was not available in sufficient detail in the records of our cohort to account for this potential confounder. Furthermore, in vitro protein function data to assess the nontruncating variants were limited. In addition, the study’s retrospective nature and the differences in the modality and timing of radiologic tests across patients make it difficult to determine the exact time of nephrocalcinosis onset. Nevertheless, we present a detailed natural history of DD in a relatively large number of DD1 patients, and for the first time, certain genotype-phenotype correlations. This study also increases our knowledge regarding the frequency of recurrent variants as well as identifying several novel changes as causes of DD1 and provides new insight regarding the underlying etiology of certain characteristic disease manifestations.
In conclusion, DD1 can be caused by many different truncating and nontruncating CLCN5 variants. Disease-causing truncating changes were localized throughout the protein whereas nontruncating ones were largely confined to the voltage ClC domain of the protein. Variants associated with kidney failure were restricted to truncating (11/13 cases), plus a single missense variant in 2 individuals previously shown to have markedly reduced functional activity. Therefore, genotyping may provide prognostic information in DD1; and in the future functional studies may add prognostic value to genotyping when a missense change is identified. Furthermore, data in this cohort suggest that other disease manifestations including kidney stone risk and progression to kidney failure are related to the degree of residual ClC-5 function.
Disclosure
All the authors declared no competing interests.
Acknowledgments
We thank all the patients and families who have participated in the Rare Kidney Stone Consortium DD registry, as well as the many physicians who collected the detailed clinical records (see below). Further, we thank the study coordinators who collected clinical data and biologic samples. We also thank the Mayo Biospecimens Accessioning and Processing, Genome Analysis, and Bioinformatics Cores for their help with the study.
We thank the following medical professionals for referring patients who were resolved during the study: Dr. Majid Alfadehel, King Fahad National Guard Hospital, Saudi Arabia; Dr. Amira Al-Uzri, Oregon Health and Science University, Portland, Oregon, US; Dr. Margret Bock, Children’s Hospital Colorado, Aurora, CO, US; Dr. Lorenzo Botto, University of Utah, Salt Lake City, UT, US; Dr. Christine Sethna, Cohen Children's Medical Center at Long Island Jewish Medical Center, New Hyde Park, NY, US; Drs. Cynthia D'Alessandri-Silva and Dr. Samriti Dogra, Connecticut Children's Specialty Group, Hartford, CT, US; Drs. Dean Assimos and Lisa Harvey, University of Alabama, Birmingham, AL, US; Dr. Dharshan Rangaswamy, Sanjay Gandhi Post Graduate Institute, Lucknow, India; Dr. Christy Dunbar, B-L Family Practice, Leesville, SC, US; Dr. Michael Ferguson, Boston Children's Hospital, Boston, MA, US; Dr. Guruprasad Shetty, Jupiter Hospital, Thane, India; Dr. William. E. Haley, Mayo Clinic, Jacksonville, FL, US; Dr. Isa Ashoor, Children's Hospital, New Orleans, LA, US; Dr. J. Bryan Carmody, Children’s Hospital of The King’s Daughters, Norfolk, VA, US; Dr. Justin Kastl, Sanford Children's Hospital, Sioux Falls, SD, US; Dr. Craig B. Langman, Ann & Robert H. Lurie Children's Hospital of Chicago, Chicago, IL, US; Dr. Lawrence Greenbaum, Emory University School of Medicine, Children’s Healthcare of Atlanta, Atlanta, GA, US; Dr. Mangalakumar Veerasamy, Kovai Medical Center and Hospital, Coimbatore, India; Dr. Mini Michael, Texas Medical Center, San Antonio, TX, US; Dr. Sharon Perlman, All Children's Hospital, St. Petersburg, FL, US; Dr. Rasheda Amin, Pediatric Specialists of Virginia, Fairfax, VA, US; Dr. Reem Raafat, Children’s Specialty Group, PLLC, Norfolk, VA, US; Dr. Jeffrey Saland, Mount Sinai Medical Center, NY, NY, US; Dr. Sarah Dugan, Children's Hospital & Clinic of MN, Minneapolis, MN, US; Dr. Christine B. Sethna, Cohen Children's Medical Center-LIJ Health System, New Hyde Park, NY, US; Dr. Nauman Shahid, Vidant Medical Center, Greenville, NC, US; Dr. Sharon Andreoli, Indiana University School Of Medicine, Indianapolis, IN, US; Dr. Danielle Soranno, Children's Hospital, University of Colorado, CO, US; Dr. Troy Zabel, Colorado Kidney Care, Denver, CO, US; Dr. Vasishta Tatapudi, NYU Langone Medical Center, NY, NY, US; Dr. Maria Vaisbich, University of São Paulo School of Medicine, Sao Paulo, Brazil; Dr. Shefali Vyas, Barnabas Health, Children's Kidney Center, Livingston, NJ, US; Dr. Ita Pfeferman, Heilberg, University of Sao Paulo, Sao paulo, Brazil; Dr. Elizabeth Abraham, University of Saint louis, St. louis, MO, US; Dr. Tecile, Andolino, Bethlehm, PA, US; Dr. Gema Ariceta, Hospital materno infantil, Barcelona, Spain; Dr. Balurate H. Jorge, Children’s hospital of Philadelphia, PA, US; Dr. Nicole Beaufort, UNC kidney center, Chapel hill, NC, US; Dr. Yongen Chang, University of California Irvine, Orange, CA, US; Dr. Don Burl, UC Davis medical group, Sacremento, CA, US; Dr. Edvardson Vidar, Landspitali University hospital, Hringbraut, Iceland; David S, Goldfarb, NY Harbor Health care system New York, NY,US; Elaine Ku, University of San Francisco, San Francisco, CA, US; Dr. Lin Jen Jar, Wake Forest University, US; Dr. Robyn Matloff, children’s and women’s physicians of Westchester Maria Farerie children’s hospital New York medical college Valhalla NY, US; Dr. Matsell Douglas, Vancouver, BC; Dr Robert McClellan, CA, US; Dr. Nevins Thomas, University of minnesota medical school MN, US; Dr. Salameh Jamal, Jacksonville, FL, US; Dr. Scheinman, steven, Geisinger commonwealth school of medicine PA, US; Dr. Sharma, Mass general, Boston, MA, US; Dr. Sheena Sharma, Phoenix children’s hospital, Phoenix AZ, US; Dr. Dorella Del Prete, Nephrology, Dialysis and Transplantation Unit, Department of Medicine, University of Padova, Padova, Italy; Dr. Anita Ammenti, Pediatric Institute, University of Parma, Parma, Italy; Dr. Giacomo Colussi, Division of Nephrology, Dialysis and Renal Transplantation, ASST-GOM Niguarda, Milan, Italy; Dr. Fabio Paglialonga, Nephrology Unit, IRCCS Foundation, Ca’ Granda Ospedale Maggiore Policlinico, University of Milan, Milano, Italy.
Funding
Funding for this project was provided by U54-DK083908 and DK133171 from the National Institute of Diabetes and Digestive and Kidney Diseases, National Center for Advancing Translational Sciences, and R21TR003174 from the National Center for Advancing Translational Sciences. JA was supported by the Mayo Clinic Kidney Diseases Training grant (T32 DK07013).
Footnotes
Table S1. Variants in our cohort listed and scored based on the American College of Medical Genetics guidelines categorized into truncating (n = 51) and nontruncating (n = 31). Novel variants are shadowed.
Supplementary Material
Table S1. Variants in our cohort listed and scored based on the American College of Medical Genetics guidelines categorized into truncating (n = 51) and nontruncating (n = 31). Novel variants were shadowed.
References
- 1.Wrong O.M., Norden A.G., Feest T.G. Dent’s disease; a familial proximal renal tubular syndrome with low-molecular-weight proteinuria, hypercalciuria, nephrocalcinosis, metabolic bone disease, progressive renal failure and a marked male predominance. QJM. 1994;87:473–493. [PubMed] [Google Scholar]
- 2.Lieske J.C., Milliner D.S., Beara-Lasic L., Harris P., Cogal A., Abrash E. In: GeneReviews. Adam M.P., Mirzaa G.M., Pagon R.A., et al., editors. University of Washington; 2012; updated 2017; 1993. Dent disease. [Google Scholar]
- 3.Anglani F., Gianesello L., Beara-Lasic L., Lieske J. Dent disease: a window into calcium and phosphate transport. J Cell Mol Med. 2019;23:7132–7142. doi: 10.1111/jcmm.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stolting G., Fischer M., Fahlke C. CLC channel function and dysfunction in health and disease. Front Physiol. 2014;5:378. doi: 10.3389/fphys.2014.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickson F.J., Sayer J.A. Nephrocalcinosis: a review of monogenic causes and insights they provide into this heterogeneous condition. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21010369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claverie-Martin F., Ramos-Trujillo E., Garcia-Nieto V. Dent’s disease: clinical features and molecular basis. Pediatr Nephrol. 2011;26:693–704. doi: 10.1007/s00467-010-1657-0. [DOI] [PubMed] [Google Scholar]
- 7.Cogal A.G., Arroyo J., Shah R.J., et al. Comprehensive genetic analysis reveals complexity of monogenic urinary stone disease. Kidney Int Rep. 2021;6:2862–2884. doi: 10.1016/j.ekir.2021.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheel O., Zdebik A.A., Lourdel S., Jentsch T.J. Voltage-dependent electrogenic chloride/proton exchange by endosomal CLC proteins. Nature. 2005;436:424–427. doi: 10.1038/nature03860. [DOI] [PubMed] [Google Scholar]
- 9.Pusch M., Zifarelli G. ClC-5: physiological role and biophysical mechanisms. Cell Calcium. 2015;58:57–66. doi: 10.1016/j.ceca.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Lourdel S., Grand T., Burgos J., González W., Sepúlveda F.V., Teulon J. ClC-5 mutations associated with Dent’s disease: a major role of the dimer interface. Pflugers Arch. 2012;463:247–256. doi: 10.1007/s00424-011-1052-0. [DOI] [PubMed] [Google Scholar]
- 11.Mansour-Hendili L., Blanchard A., Le Pottier N., et al. Mutation update of the CLCN5 gene responsible for Dent disease 1. Hum Mutat. 2015;36:743–752. doi: 10.1002/humu.22804. [DOI] [PubMed] [Google Scholar]
- 12.Blanchard A., Curis E., Guyon-Roger T., et al. Observations of a large Dent disease cohort. Kidney Int. 2016;90:430–439. doi: 10.1016/j.kint.2016.04.022. [DOI] [PubMed] [Google Scholar]
- 13.Wong W., Poke G., Stack M., Kara T., Prestidge C., Flintoff K. Phenotypic variability of Dent disease in a large New Zealand kindred. Pediatr Nephrol. 2017;32:365–369. doi: 10.1007/s00467-016-3472-8. [DOI] [PubMed] [Google Scholar]
- 14.Scheinman S.J. X-linked hypercalciuric nephrolithiasis: clinical syndromes and chloride channel mutations. Kidney Int. 1998;53:3–17. doi: 10.1046/j.1523-1755.1998.00718.x. [DOI] [PubMed] [Google Scholar]
- 15.Lloyd S.E., Pearce S.H., Fisher S.E., et al. A common molecular basis for three inherited kidney stone diseases. Nature. 1996;379:445–449. doi: 10.1038/379445a0. [DOI] [PubMed] [Google Scholar]
- 16.Smith A.J., Lippiat J.D. Direct endosomal acidification by the outwardly rectifying CLC-5 Cl(−)/H(+) exchanger. J Physiol. 2010;588:2033–2045. doi: 10.1113/jphysiol.2010.188540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorvin C.M., Wilmer M.J., Piret S.E., et al. Receptor-mediated endocytosis and endosomal acidification is impaired in proximal tubule epithelial cells of Dent disease patients. Proc Natl Acad Sci U S A. 2013;110:7014–7019. doi: 10.1073/pnas.1302063110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gianesello L., Del Prete D., Ceol M., Priante G., Calò L.A., Anglani F. From protein uptake to Dent disease: an overview of the CLCN5 gene. Gene. 2020;747 doi: 10.1016/j.gene.2020.144662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lieske J.C., Monico C.G., Holmes W.S., et al. International registry for primary hyperoxaluria. Am J Nephrol. 2005;25:290–296. doi: 10.1159/000086360. [DOI] [PubMed] [Google Scholar]
- 20.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue T., Nagano C., Matsuo M., et al. Functional analysis of suspected splicing variants in CLCN5 gene in Dent disease 1. Clin Exp Nephrol. 2020;24:606–612. doi: 10.1007/s10157-020-01876-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pottel H., Hoste L., Dubourg L., et al. An estimated glomerular filtration rate equation for the full age spectrum. Nephrol Dial Transplant. 2016;31:798–806. doi: 10.1093/ndt/gfv454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du Bois D., Du Bois E.F. A formula to estimate the approximate surface area if height and weight be known 1916. Nutrition. 1989;5:303–311. [PubMed] [Google Scholar]
- 24.National Kidney Foundation K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis. 2002;39(2 suppl 1):S1–S266. [PubMed] [Google Scholar]
- 25.Chang M.H., Brown M.R., Liu Y., et al. Cl(−) and H(+) coupling properties and subcellular localizations of wildtype and disease-associated variants of the voltage-gated Cl(−)/H(+) exchanger ClC-5. J Biol Chem. 2020;295:1464–1473. doi: 10.1074/jbc.RA119.011366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang X., Brown M.R., Cogal A.G., et al. Functional and transport analyses of CLCN5 genetic changes identified in Dent disease patients. Physiol Rep. 2016;4 doi: 10.14814/phy2.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tosetto E., Ghiggeri G.M., Emma F., et al. Phenotypic and genetic heterogeneity in Dent’s disease--the results of an Italian collaborative study. Nephrol Dial Transplant. 2006;21:2452–2463. doi: 10.1093/ndt/gfl274. [DOI] [PubMed] [Google Scholar]
- 28.Silva I.V., Cebotaru V., Wang H., et al. The ClC-5 knockout mouse model of Dent’s disease has renal hypercalciuria and increased bone turnover. J Bone Miner Res. 2003;18:615–623. doi: 10.1359/jbmr.2003.18.4.615. [DOI] [PubMed] [Google Scholar]
- 29.Wang X., Anglani F., Beara-Lasic L., et al. Glomerular pathology in Dent disease and its association with kidney function. Clin J Am Soc Nephrol. 2016;11:2168–2176. doi: 10.2215/CJN.03710416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang L.M., Mao J.H. Glomerular podocyte dysfunction in inherited renal tubular disease. World J Pediatr. 2021;17:227–233. doi: 10.1007/s12519-021-00417-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.