Abstract
Self-incompatibility (SI) is a widespread genetically determined system in flowering plants that prevents self-fertilization to promote gene flow and limit inbreeding. S-RNase–based SI is characterized by the arrest of pollen tube growth through the pistil. Arrested pollen tubes show disrupted polarized growth and swollen tips, but the underlying molecular mechanism is largely unknown. Here, we demonstrate that the swelling at the tips of incompatible pollen tubes in pear (Pyrus bretschneideri [Pbr]) is mediated by the SI-induced acetylation of the soluble inorganic pyrophosphatase (PPA) PbrPPA5. Acetylation at Lys-42 of PbrPPA5 by the acetyltransferase GCN5-related N-acetyltransferase 1 (GNAT1) drives accumulation of PbrPPA5 in the nucleus, where it binds to the transcription factor PbrbZIP77, forming a transcriptional repression complex that inhibits the expression of the pectin methylesterase (PME) gene PbrPME44. The function of PbrPPA5 as a transcriptional repressor does not require its PPA activity. Downregulating PbrPME44 resulted in increased levels of methyl-esterified pectins in growing pollen tubes, leading to swelling at their tips. These observations suggest a mechanism for PbrPPA5-driven swelling at the tips of pollen tubes during the SI response. The targets of PbrPPA5 include genes encoding cell wall–modifying enzymes, which are essential for building a continuous sustainable mechanical structure for pollen tube growth.
In the self-incompatibility response, acetylation of PbrPPA5 enhances the repression of PbrPME44, resulting in the accumulation of methyl-esterified pectins of the pollen tube cell wall and tip swelling.
IN A NUTSHELL.
Background: Self-incompatibility (SI) is a widespread genetic mechanism in flowering plants that prevents self-fertilization and promotes outbreeding. S-RNase–based SI is characterized by the arrest of pollen tube growth through the pistil, resulting in disrupted and swollen pollen tube tips that do not reach the ovule and therefore fail to complete fertilization. However, the underlying molecular mechanism responsible for pollen tube tip swelling is still largely unknown.
Question: Which factors play roles in pollen tube tip swelling during the SI response?
Findings: Here, we report that the swelling at the tips of incompatible pollen tubes in pear is mediated by SI-induced acetylation of the soluble inorganic pyrophosphatase PbrPPA5. Acetylation at Lys-42 of PbrPPA5 by the acetyltransferase PbrGNAT1 caused PbrPPA5 to accumulate in the nucleus. Acetylated PbrPPA5 bound to the transcription factor PbrbZIP77 to form a transcriptional repression complex that inhibited the expression of the pectin methylesterase gene PbrPME44. Downregulating PbrPME44 resulted in increased levels of methyl-esterified pectins in growing pollen tubes, leading to swelling at their tips.
Next steps: In future studies, we aim to further investigate how the acetyltransferase PbrGNAT1 responds to SI signaling and how the soluble inorganic pyrophosphatase PbrPPA5 accumulates in the nucleus.
Introduction
Self-incompatibility (SI) prevents self-fertilization and promotes outbreeding in a wide range of flowering plant species (de Nettancourt 2001; Silva and Goring 2001; Fujii et al. 2016; Zhao et al. 2022). SI is controlled by a single highly polymorphic locus, the S locus, which encodes both male and female S determinants. The SI response occurs when the pollen and stigma have the same allele at the S locus, for example, during self-fertilization. This causes the arrest of pollen that is genetically identical to the stigma (self-pollen) via various mechanisms, ranging from a rapid response that occurs immediately upon pollen germination on the pistil surface to a much slower response that takes place during pollen tube growth through the pistil.
S-RNase–based SI, a major type of self-recognition response that occurs in many flowering plant species, is an example of the slow response in which pollen tube growth through the pistil is arrested (Brugière et al. 2000; Franklin-Tong and Franklin 2003; Liang et al. 2020; Sun et al. 2022). The female S determinants in S-RNase–based SI are pistil-specific glycoproteins that have type T2 ribonuclease activity (Anderson et al. 1986; McClure et al. 1989, 1990). S-RNases from the pistil become internalized by pollen tubes, where they act as cytotoxins that selectively inhibit tube growth in incompatible self-pollen (Luu et al. 2000; Goldraij et al. 2006).
Pear (Pyrus bretschneideri) is a fruit tree species belonging to the Rosaceae family that shows a typical S-RNase–based SI response (Sassa et al. 1992). During SI, S-RNase induces a series of events in the incompatible pollen tube, including depolymerization of the actin cytoskeleton (Liu et al. 2007; Chen et al. 2018), scavenging of tip-localized reactive oxygen species (Wang et al. 2011), swelling at tube tips (Hiratsuka and Tezuka 1980; Liu et al. 2007), growth arrest (Liu et al. 2007; Chen et al. 2018), and ultimately programmed cell death (Wang et al. 2009, 2011). Swollen tips of pollen tubes are also observed in other species that exhibit S-RNase–based SI, such as sweet tobacco (Nicotiana alata) (Newbigin et al. 1993), petunia (Petunia inflata) (Kao and Mccubbin 1996), passion fruit (Passiflora edulis) (Rêgo et al. 2000), and Japanese plum (Prunus salicina) (Jia et al. 2008). Disruption of polarized growth of SI pollen tubes ultimately causes their arrest in the pistil, preventing them from reaching the ovary to fertilize the egg cells. However, the molecular mechanism by which S-RNase leads to the swelling of growing pollen tube tips is unknown.
During the delivery of sperm cells into the ovary, the cell wall of the pollen tube provides structural support and protection to prevent overexpansion (Geitmann and Steer 2006; Parrotta et al. 2019). The cell wall of the pollen tube is composed primarily of cellulose, hemicellulose, and pectin, while the wall at the growing tip mainly consists of pectins. The pectins play a critical role in the delicate regulation of the structural and mechanical properties of polarized pollen tube elongation (Parre and Geitmann 2005; Zhang et al. 2018) and pollen–pistil interactions (Lu et al. 2019; Duan et al. 2020).
Pectin methyltransferases add methyl substituents to carboxylate groups in homogalacturonan (HG) pectins in the Golgi; these pectins are then delivered to the growing pollen tube tip by transport vesicles (Haas et al. 2020). Methyl esters in pectins have low affinity for Ca2+ and weaken crosslinks in the cell wall (Willats et al. 2001), whereas carboxylate anions that form after deesterification can form complexes with Ca2+ and increase cell wall strength (Bosch and Hepler 2005). During the expansion of pollen tube tips, the methyl ester moieties in pectins are hydrolyzed by tip-secreted pectin methylesterases (PME; EC 3.1.1.11). A gradual decrease in the levels of methyl-esterified pectins and an increase in the levels of deesterified pectins due to localized PME activity are observed at the flanks of pollen tubes (Chebli et al. 2012).
PME activity is important for the elongation and integrity of pollen tubes (Bosch et al. 2005; Tian et al. 2006). In Arabidopsis (Arabidopsis thaliana), the PME mutant vanguard1 (vgd1) shows an uprise in pollen tube tip rupture and reduced male fertility (Jiang et al. 2005). AtPPME1, a pollen-expressed PME gene in Arabidopsis, affects the shape and growth of pollen tubes (Tian et al. 2006).
Many plant species contain large PME gene families and express multiple PME isoforms in pollen (Bosch and Hepler 2005; Kim et al. 2020). We recently identified 81 PME genes in pear (PbrPME), 47 of which are expressed in pollen (Tang et al. 2020b). Knocking down PbrPME44, a highly expressed pollen PME gene, significantly inhibited pollen tube growth (Tang et al. 2020b). However, the biological function of PbrPME44 in regulating pollen tube growth and the molecular mechanism of growth arrest, especially during the SI response, are not fully understood. Reduced activities of soluble inorganic pyrophosphatases (sPPases, EC 3.6.1.1), which hydrolyze pyrophosphate (PPi) and prevent it from accumulating to toxic levels, result in inhibition of pollen tube growth during the SI response of common poppy (Papaver rhoeas) (de Graaf et al. 2006) and apple (Malus×domestica) (Li et al. 2018).
In this study, we determined that PbrPPA5, a pollen-specific sPPase of pear, is a transcriptional regulator that regulates pollen tube growth and the SI response independently of its sPPase activity. PbrPPA5 binds to the transcription factor (TF) basic LEUCINE ZIPPER 77 (PbrbZIP77) and functions as a cosuppressor of PbrPME44 expression. Following a treatment that mimics the SI response in pollen tubes, PbrPPA5 was acetylated by GCN5-related N-acetyltransferase 1 (PbrGNAT1), thereby promoting its accumulation in the nucleus. The increase in PbrPPA5 levels in the nucleus reinforced the inhibition of PbrPME44 expression via the PbrPPA5–PbrbZIP77 complex. The reduction in PbrPME44 levels in pollen tubes altered the composition and distribution of pectins in the cell wall, resulting in an imbalance between methyl-esterified pectins and deesterified pectins from the tips to the flanks of SI pollen tubes. Thus, our results show that the SI response in pear dramatically alters the mechanical properties of the cell wall and causes swelling at the tips of pollen tubes.
Results
SI challenge decreases the expression of PbrPME44 in pear pollen tubes
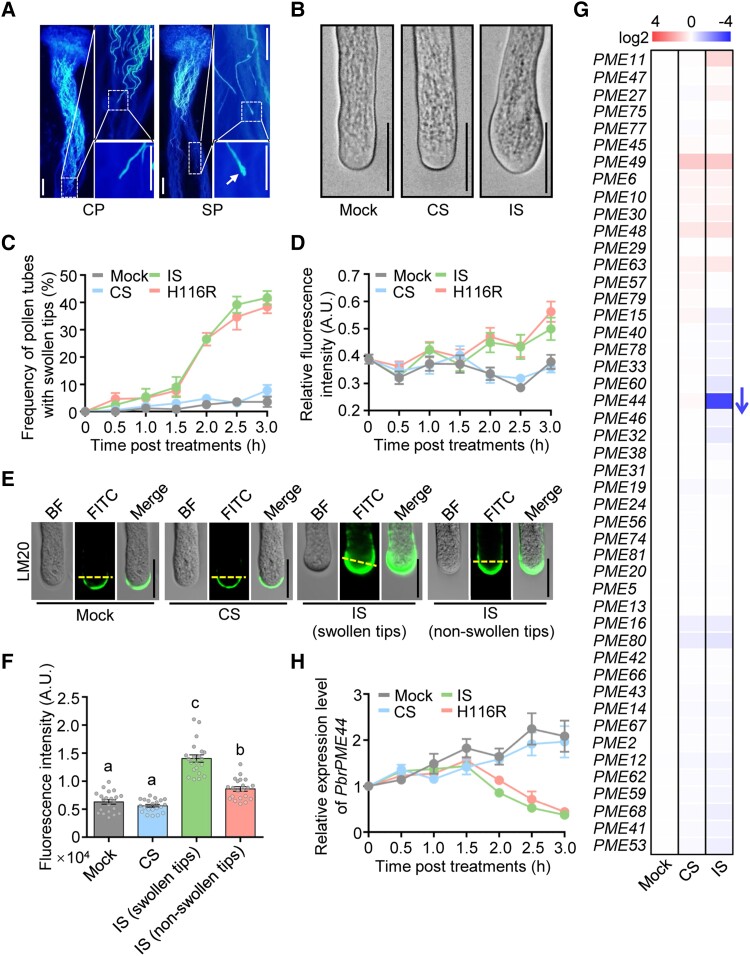
To examine the growth of incompatible pollen tubes in vivo, we cross-pollinated (CP; compatible reaction) or self-pollinated (SP; incompatible reaction) pear pistils. At 48 h after pollination, the growth of SP pollen tubes arrested growth halfway through the pistil and the pollen tubes exhibited swelling at the tip, while CP pollen tubes reached the bottom of the pistil (Fig. 1A).
Figure 1.
PbrS-RNase induces swelling at the tips of pollen tubes and suppresses PbrPME44 expression during the self-incompatibility response. A) Aniline blue staining of P. bretschneideri cv. Dangshansuli pistils 48 h after pollination with P. bretschneideri cv. Huanghua pollen (CP; compatible) and Dangshansuli pollen (SP; incompatible). Insets showing high-magnification views of the boxed areas are shown on the right. The SP tubes in SP had swollen tips (indicated by arrow). Scale bars, 100 μm. B) His-tagged rPbrS-RNases induce the swelling of SI pollen tubes in vitro after a 2.5-h treatment. Mock, liquid germination medium; CS, compatible rPbrS-RNase; IS, incompatible rPbrS-RNase. Scale bars, 20 μm. C) Time course analysis of the frequency of pollen tubes with swollen tips under mock, CS, IS, and H116R mutation treatments. H116R, incompatible rPbrS-RNases (rPbrS7-RNaseH116R + rPbrS34-RNaseH116R). Data are means ± SEM, n ≥ 30 pollen tubes. D) Quantitative analysis of the time course of fluorescence intensity of the LM20 antibody at the apical region (the area within 5 μm from the tip to the shank) of pollen tubes under mock, CS, IS, and H116R mutation treatments. Data are means ± SEM, n = 20 pollen tubes. E) rPbrS-RNases induce an increase in the level of methyl-esterified pectin in pollen tubes 2 h after mock, CS, and IS treatments regardless of whether the tips were swollen. Methyl-esterified pectins in the pollen tube were immunolabeled with LM20. BF, brightfield image; FITC, antirat IgG-FITC antibody. Scale bars, 20 μm. F) Quantitative analysis of the fluorescence intensity of LM20 at the apical regions of pollen tubes 2.0 h after mock, CS, and IS treatments. Data are means ± SEM, n ≥ 20 pollen tubes. G) Relative expression levels of 47 pollen-expressed PbrPME genes (Supplemental Table S2) 2.5 h after mock, CS, and IS treatments using the in vitro pollen culture system. Relative expression levels were normalized to that under mock treatment. The arrow indicates the only gene that was downregulated (fold change > 2). PbrTUB-2 was used as the reference gene. Data are means ± SEM, n = 3 biological replicates. H) Time course analysis of the expression levels of PbrPME44 in pollen tubes under mock, CS, IS, and H116R mutation treatments in vitro. For these treatments, pollen tubes (Dangshansuli; S7 or S34) were precultured in liquid germination medium for 1.5 h. PbrTUB-2 was used as the reference gene for normalization. Data are means ± SEM, n = 7 biological replicates. Statistical analysis results for all figures and supplemental figures are presented in Supplemental Data Set 4.
We recently developed an in vitro pear pollen tube growth system in which adding His-tagged recombinant PbrS-RNases (rPbrS-RNases) to the culture medium mimics the SI response in pollen tubes (Chen et al. 2018), and we refer to this SI-inducing treatment as SI challenge. Following treatment with incompatible rPbrS-RNases (IS), we observed swelling at the tip in pollen tubes (Fig. 1B). The frequency of pollen tubes with swollen tips was significantly higher following treatment with IS than with compatible rPbrS-RNases (CS) or a mock treatment. This occurred in a time-dependent manner that was initiated 2 h after IS treatment (Figs. 1C and S1, A and B). Consistently, pollen tubes were significantly shorter following 2 h of IS treatment compared to mock and CS treatments (Supplemental Fig. S2).
Because methyl-esterified pectins contribute to the swelling at the tips of pollen tubes (Tian et al. 2006; Tang et al. 2020b), we measured the levels of methyl-esterified pectins in the apical regions of incompatible pollen tubes. Total pectin contents did not significantly differ among the mock, CS, and IS treatment groups (Supplemental Fig. S3). However, the Leeds Monoclonal 20 (LM20) antibody, which recognizes methyl-esterified pectins, detected significant increases in fluorescent signals in the apical region of pollen tubes (the area within 5 μm from the tip to the shank) 2 h after IS treatment (Fig. 1D). Fluorescent signals also increased in incompatible pollen tubes without tip swelling, but the increase was lower compared to pollen tubes showing tip swelling (Figs. 1, E and F, and S4).
The level of methyl-esterified pectins is largely determined by PME activity (Bosch and Hepler 2005). Total PME enzyme activity decreased in incompatible pollen tubes beginning at 2 h after IS treatment (Supplemental Fig. S5). Transcriptome analysis of SP pistils revealed that 5 PbrPME genes were downregulated during the SI response (Supplemental Fig. S6 and Table S1). Reverse transcription quantitative PCR (RT-qPCR) analysis of the 47 pollen-expressed PbrPME genes (Tang et al. 2020b) revealed that only PbrPME44 was significantly downregulated (fold change > 2) after IS treatment (Fig. 1G and Supplemental Table S2). These results suggest that pistil-expressed PbrPME genes are also involved in the SI response in pear. Here, we focus on the function of PbrPME44, which is significantly downregulated in incompatible pollen.
The expression of PbrPME44 declined in a time-dependent manner, beginning 2 h after IS treatment (Fig. 1H). To confirm the function of PbrPME44 in the growth of pollen tubes, we performed antisense oligodeoxynucleotide (as-ODN) inhibition to knock down PbrPME44 transcript levels. Upon knockdown of PbrPME44 (Supplemental Fig. S7A), the total PME activity in pollen tubes decreased significantly 2 h after as-ODN treatment (Supplemental Fig. S7B). Also, the frequency of pollen tubes with swollen tips was higher compared to mock-treated pollen tubes (Supplemental Fig. S8, A and B). These results are consistent with our previous observation that knocking down PbrPME44 expression in pear pollen increases the contents of methyl-esterified pectins in the apical region of pollen tubes (Tang et al. 2020b). Together, these results suggest that PbrPME44 affects pollen tube growth in pear.
We tested whether the RNase activity of PbrS-RNase is essential for the repression of PbrPME44 and the tip swelling of incompatible pollen tubes. An H116R mutation in PbrS-RNases substantially reduces their RNase activity (Chen et al. 2018). The H116R mutant and the wild-type His-Tagged rPbrS-RNases had similar effects on the suppression of PbrPME44 expression (Fig. 1H), the accumulation of methyl-esterified pectins (Fig. 1D), and the swelling at the tips of pollen tubes (Fig. 1C). These results suggest that the RNase activity of PbrS-RNase is not required for suppressing PbrPME44 expression in incompatible pollen tubes. In addition, the PbrS-RNaseH116R had similar effects on the inhibition of incompatible pollen tube growth as did the wild-type PbrS-RNase 2 h following the IS treatment (Supplemental Fig. S9), indicating that the RNase activity is likely not required in the early stage of SI response.
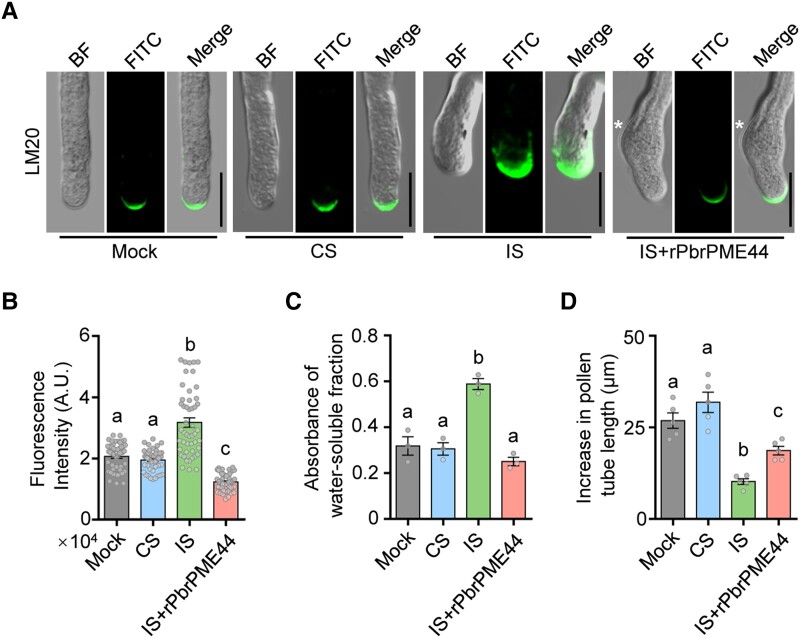
To explore whether and how PbrPME44 contributes to the SI response, we added His-tagged recombinant PbrPME44 (rPbrPME44) to the culture medium to compensate for the reduced endogenous PME activity in the SI challenge (Supplemental Fig. S10). We treated the pollen tubes with rPbrS-RNases for 2 h, followed by the addition of rPbrPME44 and incubation for an additional 1 h. Pollen tubes contained enlarged knots after IS treatment, whereas their diameters returned to normal and they continued to grow after the addition of rPbrPME44 (Fig. 2A). Moreover, the addition of rPbrPME44 reduced the amounts of methyl-esterified pectins in IS-treated pollen tubes to levels similar to those observed in mock control and CS-treated pollen tubes (Fig. 2, A and B).
Figure 2.
PbrPME44 alleviates the PbrS-RNase–induced increase in methyl-esterified pectin levels. A) Methyl-esterified pectins in the pollen tube were immunolabeled with the LM20 antibody 2 h after mock, CS, IS, and IS plus His-tagged rPbrPME44 protein treatments. For PME compensation experiments, pollen tubes were treated with rPbrS-RNases for 2 h, followed by the addition of rPbrPME44 and incubation for 1 h. The asterisks indicate the positions of swelling. Mock, liquid germination medium; CS, compatible rPbrS-RNase; IS, incompatible rPbrS-RNase; BF, brightfield image; FITC, antirat IgG-FITC antibody. Scale bars, 20 μm. B) Quantitative analysis of the fluorescence intensity of LM20 at the apical regions of the pollen tubes from A). Different lowercase letters indicate significant differences, as determined by ANOVA followed by Tukey's multiple comparison test (P < 0.05, n = 49 pollen tubes). Data are means ± SEM. C) Quantification of levels of methyl-esterified pectins (using ELISA) in alcohol-insoluble residues extracted from pollen tubes under the treatments indicated in A). Water was used as the elution buffer. The LM20 monoclonal antibody was used to detect methyl-esterified pectins. Different lowercase letters indicate significant differences as determined by ANOVA followed by Tukey's multiple comparison test (P < 0.05, n = 3 biological replicates). Data are means ± SEM. D) Quantitative analysis of the increase in pollen tube length under the indicated treatments in A). Five biological replicates were performed, with each replicate containing at least 30 pollen tubes. Different lowercase letters indicate significant differences, as determined by ANOVA followed by Tukey's multiple comparison test (P < 0.05). Data are means ± SEM.
Using enzyme-linked immunosorbent assays (ELISAs), we observed that the addition of rPbrPME44 restored the contents of methyl-esterified pectins in pollen tubes (Figs. 2C and S11) and partially alleviated the inhibited growth of pollen tubes induced by SI challenge (Fig. 2D). However, if the pollen tubes were treated with incompatible rPbrS-RNase for more than 3 h, the addition of rPbrPME44 failed to restore pollen tube growth (Supplemental Fig. S12), suggesting that PbrS-RNase activity reaches a point of no return at 3 h after IS treatment. These results suggest that the downregulation of PbrPME44 expression plays an important role in the inhibited growth of incompatible pollen tubes rather than representing an effect of the cessation of pollen tube growth.
The TF PbrbZIP77 suppresses PbrPME44 expression
Using a yeast 1-hybrid (Y1H) assay, we investigated the transcriptional regulation of the PbrPME44 gene using a 1.3-kb promoter sequence upstream of its start codon as a bait to screen a pear cDNA library prepared from total RNA extracted from pollen. We isolated 3 TFs (Supplemental Table S3), including a basic leucine zipper (bZIP) TF (Fig. 3A; Pbr031203.1, named PbrbZIP77; Ma et al. 2021). PbrbZIP77 shares 55.9% amino acid sequence identity with Arabidopsis AtbZIP34 (At2g42380) (Supplemental Fig. S13A). Mutations in AtbZIP34 cause morphological defects in the pollen grain wall (Gibalová et al. 2009). Reverse transcription PCR (RT-PCR) analysis of pear tissues showed that PbrbZIP77 was expressed in pollen tubes (Supplemental Fig. S13B), supporting its role in pollen tube growth.
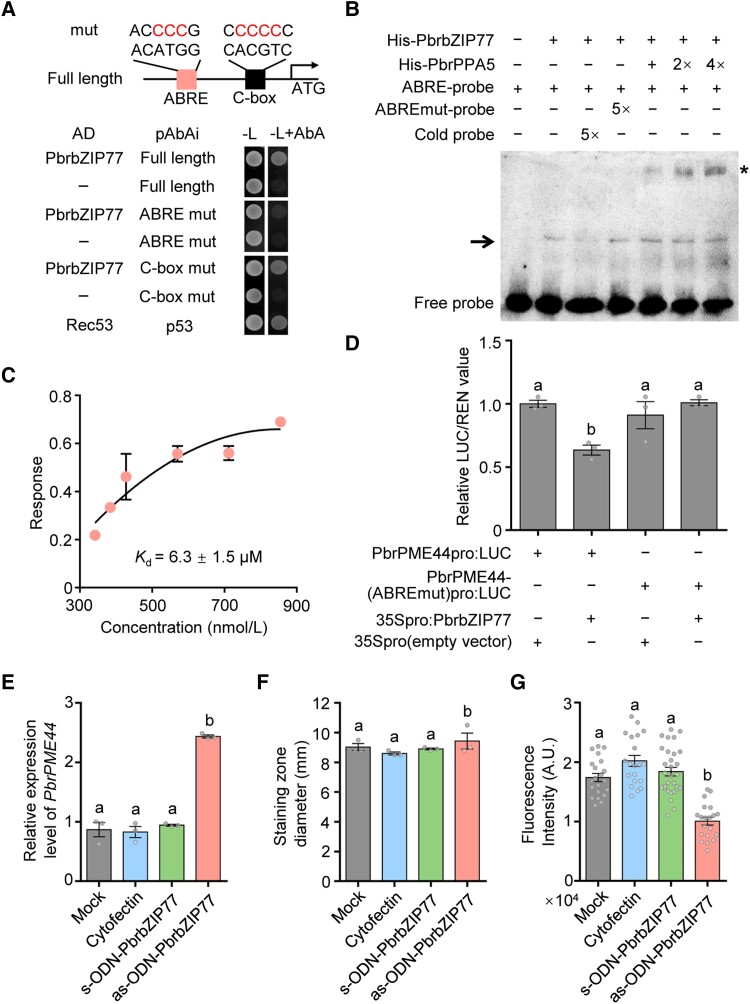
Figure 3.
PbrbZIP77 suppresses PbrPME44 expression by binding to the ABRE in its promoter. A) Y1H assays showing that the ABRE motif is essential for the binding of PbrbZIP77 to the PbrPME44 promoter. The 1,300-bp sequence upstream of the start codon of PbrPME44 was cloned into pAbAi as described in Materials and methods. The left box represents the ABA-responsive element (ACATGG) or its mutated form (ACCCCG). The right box represents the C-box element (CACGTC) or its mutated form (CCCCCC). B) Binding of His-tagged rPbrbZIP77 to the ABRE of the PbrPME44 promoter region and a supershift EMSA of the PbrPPA5–PbrbZIP77 complex. The 30-bp PbrPME44 promoter fragment containing the ABRE was labeled with biotin and used as a probe. − represents absence; + represents presence; the arrow indicates the shifted band; and the asterisk indicates the supershifted band. C) Quantification of the interaction between PbrbZIP77 and the 30-bp PbrPME44 promoter fragment containing the ABRE using a BLI assay. Three technical replicates were performed. D) PbrbZIP77-repressed PbrPME44 promoter activity depends on the ABRE in a dual-LUC assay. The empty vector (35Spro) served as a negative control, and the LUC/REN ratio of 35Spro was set to 1. − represents absence and + represents presence. Different lowercase letters indicate significant differences, as determined by ANOVA followed by Tukey's multiple comparison test (P < 0.05, n = 3 biological replicates). Data represent the means ± SEM. E)PbrPME44 is upregulated in pollen tubes 2 h after treatment with as-ODN-PbrbZIP77. Cytofectin (Lipofectamine 2000) was used as the transfection reagent. The sense oligodeoxynucleotide-PbrbZIP77 (s-ODN-PbrbZIP77) was used as the negative control. Mock, liquid germination medium. Different lowercase letters indicate significant differences, as determined by ANOVA followed by Tukey's multiple comparison test (P < 0.05, n = 3 biological replicates). Data are means ± SEM. F) Analysis of total PME activity by ruthenium red staining under mock, cytofectin, s-ODN-PbrbZIP1, and as-ODN-PbrbZIP1 treatments. The diameters of the stained zones were measured with calipers. Different lowercase letters indicate significant differences, as determined by ANOVA followed by Tukey's multiple comparison test (P < 0.05, n = 3 biological replicates). Data are means ± SEM. G) Quantitative analysis of the fluorescence intensity of LM20 at the apical regions of pollen tubes under mock, cytofectin, s-ODN-PbrbZIP1, and as-ODN-PbrbZIP1 treatments. Different lowercase letters indicate significant differences, as determined by ANOVA followed by Tukey's multiple comparison test (P < 0.05, n ≥ 20 pollen tubes). Data are means ± SEM.
Analysis of the regulatory cis-elements in the PbrPME44 promoter fragment identified 2 potential bZIP binding sites: an abscisic acid–responsive element (ABRE) at position −1,192 to −1,187 and a C-box element at position −139 to −134 (Fig. 3A and Supplemental Data Set 1). To determine whether PbrbZIP77 binds 1 or both of these cis-elements, we introduced mutations at these sites and performed another Y1H analysis. The ACATGG to ACCCCG mutation in the ABRE motif blocked the interaction between the PbrPME44 promoter and PbrbZIP77, whereas the CACGTC to CCCCCC mutation in the putative C-box did not (Fig. 3A).
To further explore the interaction between PbrbZIP77 and a 30-bp promoter fragment containing the putative ABRE binding site, we performed electrophoretic mobility shift assays (EMSAs) and biolayer interferometry (BLI) assays. In the EMSA, recombinant His-PbrbZIP77 specifically bound to the biotin-labeled ABRE fragment, and this specific binding was reduced by competition with cold probe included at a 5-hold higher concentration, but not affected by the ABRE mutated probe (Fig. 3B). Quantification of the interaction using BLI showed that the binding affinity (Kd) between His-PbrbZIP77 and the ABRE fragment was 6.3 ± 1.5 μM (Fig. 3C).
To better understand the function of PbrbZIP77 in PbrPME44 expression, we performed dual-luciferase (LUC) reporter assays. PbrbZIP77 decreased firefly LUC activity derived from the PbrPME44pro:LUC reporter construct, but it had no significant effect on activity from PbrPME44-(ABREmut)pro:LUC (Fig. 3D). In pollen tubes containing a PbrbZIP77-knockdown generated using as-ODN (Supplemental Fig. S14A), we detected a significant increase in PbrPME44 expression (Fig. 3E). This was accompanied by higher total PME activity (Fig. 3F) and lower levels of methyl-esterified pectins (Fig. 3G). In addition, PbrbZIP77-knockdown pollen tubes were significantly shorter than those of the mock control (Supplemental Fig. S14B). These results suggest that PbrbZIP77 in the pollen binds to the ABRE cis-element of the PbrPME44 promoter, resulting in the suppression of PbrPME44 transcription.
PbrPPA5 physically interacts with PbrbZIP77
Upon SI challenge, the expression level of PbrbZIP77 was not affected in pear pollen tubes (Supplemental Fig. S15), suggesting that other regulators might be involved in SI. In a yeast 2-hybrid (Y2H) screen, we identified 17 candidate interacting proteins of PbrbZIP77 that are encoded by genes expressed in pollen (Supplemental Table S4). One of these candidates was the soluble inorganic pyrophosphatase (sPPase; Pbr025893.1) PbrPPA5 (Tang et al. 2020a). Five PbrPPA genes are expressed in pear pollen, but only PbrPPA5 is specifically expressed in pollen (Tang et al. 2020a). Y2H assays showed that only PbrPPA5 interacted with PbrbZIP77 (Figs. 4A and S16). We confirmed the interaction between PbrbZIP77 and PbrPPA5 in planta by bimolecular fluorescence complementation (BiFC) and LUC complementation imaging (LCI) assays (Fig. 4, B and C). In addition, microscale thermophoresis (MST) analysis showed that the binding affinity (Kd) between PbrPPA5-His and PbrbZIP77-His was 27.5 ± 4.2 nM (Fig. 4D).
Figure 4.
PbrPPA5 interacts with PbrbZIP77 in vitro and in vivo. A) Y2H assay of the interaction between PbrPPA5 (activation domain, AD) and PbrbZIP77 (binding domain, BD). Coexpression of pGBKT7-P53 and pGADT7-T was used as a positive control and that of pGBKT7-Lam and pGADT7-T as a negative control. Interactions were determined based on growth on synthetic-defined (SD)/–W–L–H–A (–Ade–Leu–Trp–His) medium. B) BiFC assays of the interaction between PbrbZIP77 and PbrPPA5 constructs in N. benthamiana leaves. Vectors YNE and YCE contain the N-terminal and C-terminal fragments of YFP, respectively. Scale bars, 20 μm. C) LCI assay of the interaction between PbrbZIP77 and PbrPPA5 in N. benthamiana leaves. NLuc and Cluc are vectors containing the N-terminal and C-terminal fragments of firefly LUC, respectively. Coinfiltration of 35Spro:NLuc and 35Spro:Cluc was used as a negative control. D) Binding affinity assay showing the interaction kinetics (as indicated by Kd values) between His-tagged rPbrPPA5 and His-PbrbZIP77, as measured by MST analysis.
The pyrophosphatase activity of sPPase catalyzes the hydrolysis of PPi to inorganic phosphate (Pi). To examine whether PbrPPA5 has sPPase activity, we produced and purified His-tagged recombinant PbrPPA5 (rPbrPPA5) from Escherichia coli (Supplemental Fig. S17A). The sPPase activity of rPbrPPA5 (220.80 ± 12.43 μmol Pi mg−1 min−1; Supplemental Fig. 17B) was very similar to that of other plant sPPases, including those of poppy and apple pollen (de Graaf et al. 2006; Li et al. 2018). In addition, PbrPPA5 shares 68.55% amino acid sequence similarity with Prp26.1a/b, the sPPases identified in common poppy (Supplemental Fig. S17C). These results confirm the notion that PbrPPA5 is a classical sPPase.
We asked whether the interaction between PbrbZIP77 and PbrPPA5 requires the intrinsic pyrophosphatase activity of PbrPPA5. The DxDPxDV motif, which is highly conserved in plant PPase enzymes, is part of the Mg2+-binding site and is essential for pyrophosphatase activity (de Graaf et al. 2006; Grzechowiak et al. 2019). Indeed, we identified a DxDPxDV motif in PbrPPA5 (positions 134 to 140, DNDPLDV). We mutated D136 to Asn (N) and P137 to Ala (A) and observed that these mutant sPPases had significantly reduced pyrophosphatase activity (Supplemental Fig. S18A). However, these mutant PbrPPA5s still interacted with PbrbZIP77 in a Y2H assay (Supplemental Fig. S18B). We demonstrated that the segment of PbrPPA5 at positions 214 to 234, which is less conserved among sPPases, is responsible for the interaction with PbrbZIP77 in Y2H assays (Supplemental Fig. S18C). These results suggest that the intrinsic pyrophosphatase activity of PbrPPA5 is not required for its interaction with PbrbZIP77.
PbrPPA5 acts with PbrbZIP77 to cosuppress PbrPME44 expression
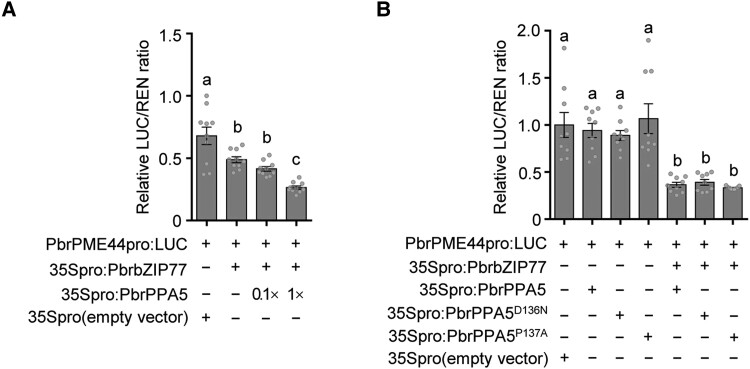
We investigated whether PbrPPA5 is involved in the transcriptional regulation of PbrPME44 in growing pear pollen tubes. Knockdown of PbrPPA5 using as-ODN (Supplemental Fig. S19A) resulted in the significant upregulation of PbrPME44 expression (Supplemental Fig. S19B). Accordingly, relative PME activity was higher in PbrPPA5-knockdown pollen tubes than in the wild-type pollen tubes (Supplemental Fig. S19C), and the levels of methyl-esterified pectins were dramatically reduced at the tips of pollen tubes of these plants (Supplemental Fig. S19, D and E). In dual-LUC assays, PbrPPA5 alone failed to modify the activity of the PbrPME44 promoter; however, when PbrPPA5 and PbrbZIP77 were coexpressed, PbrPME44 expression was significantly reduced (Fig. 5A). In EMSAs, the addition of PbrPPA5 together with His-PbrbZIP77 resulted in the appearance of a supershifted complex on the ABRE site from PbrPME44 promoter. Notably, this complex had a stronger intensity than seen with His-PbrbZIP77 alone, suggesting that the binding of His-PbrbZIP77 to the ABRE was enhanced by adding higher concentrations of His-PbrPPA5 (Fig. 3B). In addition, the suppression of PbrPME44 expression was not affected when the sPPase domain of PbrPPA5 was mutated (D136N or P137A) (Fig. 5B). These results suggest that the presence of PbrPPA5 enhances the suppression of PbrPME44 expression by PbrbZIP77.
Figure 5.
PbrPPA5 enhances PbrbZIP77-mediated suppression of PbrPME44 expression. A) PbrPPA5 promotes the PbrbZIP77-mediated suppression of PbrPME44 expression in a concentration-dependent manner, as measured in a dual-LUC assay. The empty vector (35Spro) served as a negative control, and the LUC/REN ratio of 35Spro was set to 1. The effector constructs consisted of the PbrbZIP77 or PbrPPA5 coding sequences driven by the 35S promoter; PbrPME44pro:LUC was used as the reporter construct. − represents absence, and + represents presence. Different lowercase letters indicate significant differences, as determined by ANOVA followed by Tukey's multiple comparison test (P < 0.05, n = 10 biological replicates). Data are means ± SEM. B) The cosuppressor activity of PbrPPA5 for PbrPME44 expression is independent of its sPPase activity. Dual-LUC assays were performed using N. benthamiana leaves coinfiltrated with the reporter construct PbrPME44pro:LUC and the effector constructs 35Spro:PbrbZIP77, 35Spro:PbrPPA5, 35Spro:PbrPPA5D136N, 35Spro:PbrPPA5P137A, and 35Spro (empty vector), as indicated. − represents absence and + represents presence. Different lowercase letters indicate significant differences, as determined by ANOVA followed by Tukey's multiple comparison test (P < 0.05, n = 9 biological replicates). Data are means ± SEM.
SI-induced acetylation of PbrPPA5 causes it to accumulate in the nucleus
Although a direct interaction between S-RNases and sPPases in SI responses has been reported in apple (Li et al. 2018), we did not observe a direct interaction between PbrS-RNase and PbrPPA5 in our Y2H assays (Supplemental Fig. S20). During the SI response in poppy, the sPPases Prp26.1a/b are rapidly phosphorylated, resulting in a decrease in sPPase activity in pollen (de Graaf et al. 2006; Eaves et al. 2017). However, after 2.5 h of SI challenge, we detected neither a decrease in total sPPase activity nor the phosphorylation of PbrPPA5 in pollen tubes (Supplemental Fig. S21). Notably, we detected acetylation at the Lys-42 (K42) residue (LMASEDQSEEAK) of PbrPPA5 by mass spectrometry of proteins extracted from pollen tubes 1.5 h after SI challenge (Supplemental Fig. S22 and Data Set 2).
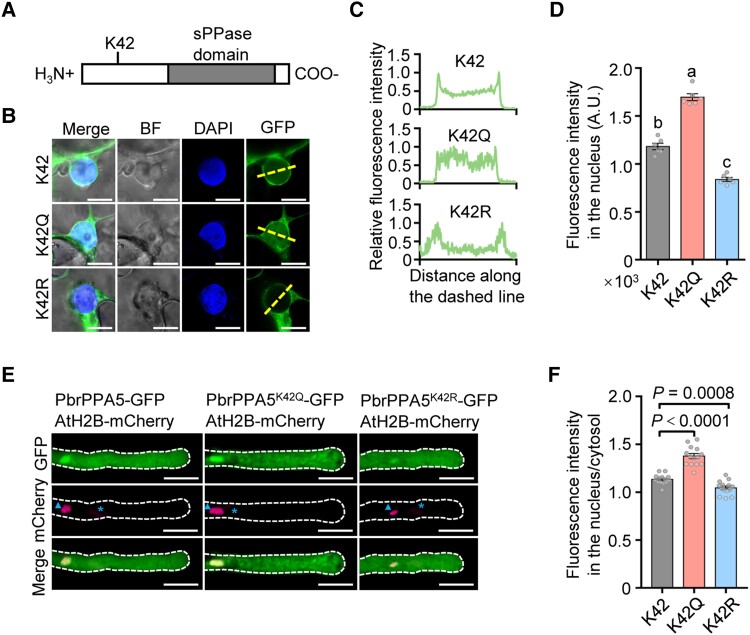
The acetylation of proteins can alter their localizations and functions (Drazic et al. 2016). The substitution of a basic, positively charged Lys residue for an uncharged glutamine (Q) or a charge-conservative arginine (R) can mimic a constitutively acetylated or nonacetylated status, respectively (Kim et al. 2006). We therefore generated PbrPPA5K42Q (constitutively acetylated) and PbrPPA5K42R (nonacetylated) mutant constructs and fused them to green fluorescent protein (GFP) to study the effect of acetylation of the K42 residue on the localization of PbrPPA5. Following transient expression in epidermal leaf cells of Nicotiana benthamiana, the K42Q mutation increased the nuclear localization of PbrPPA5, whereas the K42R mutation reduced its nuclear localization (Fig. 6, A to D). Consistently, when we used particle bombardment to deliver constructs expressing the mutant proteins with K42Q as an acetylation mimic into pollen tubes, the K42Q mutation increased the nuclear localization of PbrPPA5 in the nucleus and led to swelling at the tips of pollen tubes (Figs. 6, E and F, and S23).
Figure 6.
Acetylation of the K42 residue in PbrPPA5 promotes accumulation of PbrPPA5 in the nucleus. A) Structure of PbrPPA5 with the sPPase domain indicated. K42, the Lys residue at position 42 in the N-terminal region of PbrPPA5. B) Acetylation of the K42 residue in PbrPPA5 promotes its accumulation in the nucleus. Substitution of the K42 residue with a charge-conservative (positively charged) Arg residue (R) or an uncharged Glu residue (Q) mimics a nonacetylated or constitutively acetylated state, respectively. PbrPPA5-GFP (K42), PbrPPA5K42Q-GFP (K42Q), and PbrPPA5K42R-GFP (K42R) were transiently expressed in N. benthamiana leaves and observed under a confocal microscope. GFP signals represent the localizaiton of PbrPPA5 isoforms; DAPI was used as the nucleus marker. The dashed lines represent the positions used to measure fluorescence intensity. Scale bars, 10 μm. C) Quantitative analysis of the relative fluorescence intensity of GFP in the last column in B). Fluorescence intensity of PbrPPA5-GFP was measured using ZEN software in the area across the dashed line crossing the nucleus. The values on the x axis represent the distance along the dashed lines. The values on the y axis represent the fluorescence intensity of PbrPPA5-GFP. D) Quantitative analysis of the fluorescence intensity of PbrPPA5-GFP in the nucleus. Different lowercase letters indicate significant differences, as determined by ANOVA followed by Tukey's multiple comparison test (P < 0.05, n = 6 leaf cells). Data are means ± SEM. E) Images of the subcellular localization of PbrPPA5 and its mutant forms in pear pollen tubes. The expression of PbrPPA5-GFP, PbrPPA5K42Q-GFP, PbrPPA5K42R-GFP, and AtH2B-mCherry was driven by the NTP303 promoter. PbrPPA5-GFP, PbrPPA5K42Q-GFP, and PbrPPA5K42R-GFP were transiently coexpressed with AtH2B-mCherry in pear pollen tubes by particle bombardment. The pollen was cultured for 4 h after transformation. The asterisk represents the vegetative nucleus and the triangle represents the generative nucleus. Scale bars, 20 μm. F) Quantitative analysis of the ratio of fluorescence intensity in the nucleus/cytosol for PbrPPA5-GFP, PbrPPA5K42Q-GFP, and PbrPPA5K42R-GFP in the nuclei of pollen tubes from E). P-values were obtained by Student's t test. Data are means ± SEM; n = 10 pollen tubes.
In addition, acetylation of the K42 residue did not affect the pyrophosphatase activity of PbrPPA5 (Supplemental Fig. S24), the interaction between PbrPPA5 and PbrbZIP77 (Supplemental Fig. S25), or the cosuppressor activities of PbrPPA5 for PbrPME44 (Supplemental Fig. S26). We noted that 2 other lysine residues of PbrPPA5, K9 and K98, could be acetylated as well. However, mutating either K9 (Supplemental Fig. S27, A to C) or K98 (Supplemental Fig. S27, D to F) had no visible effect on PbrPPA5 localization, suggesting that these 2 lysine residues are not associated with SI. These results indicate that acetylation of the K42 residue in PbrPPA5 promotes its accumulation in the plant cell nucleus.
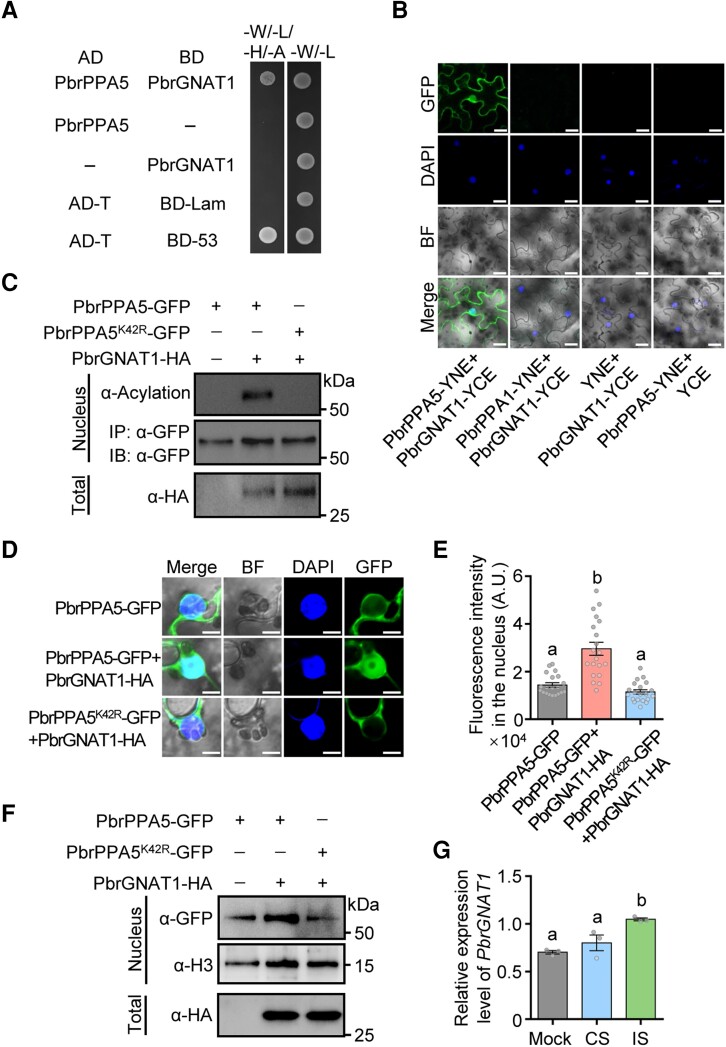
To identify the acetyltransferase responsible for the acetylation of PbrPPA5 during SI, we used a cDNA library generated from pear pollen to perform a Y2H assay to screen for proteins interacting with PbrPPA5. One interacting protein was a GCN5-related N-acetyltransferase (Pbr013581.1, named PbrGNAT1, Fig. 7A). In BiFC assays, PbrGNAT1 interacted with PbrPPA5 in both the nucleus and the cytoplasm (Fig. 7B). In acetylation activity assays, PbrGNAT1 induced the acetylation of PbrPPA5, which was dependent on the K42 residue (Fig. 7C). PbrPPA5 accumulated in the nucleus more frequently when PbrGNAT1 was presented (Fig. 7, D to F). In addition, the PbrGNAT1 transcript level increased at 1 h after IS treatment, supporting the notion that PbrGNAT1 is involved in the SI response in pear (Fig. 7G).
Figure 7.
PbrGNAT1 acetylates PbrPPA5 and promotes its nuclear accumulation. A) Y2H assay of the interactions between PbrPPA5 (activation domain, AD) and PbrGNAT1 (binding domain, BD). Coexpression of pGBKT7-P53 and pGADT7-T was used as a positive control and that of pGBKT7-Lam and pGADT7-T as a negative control. Interactions were determined based on growth on SD/–W–L–H–A (–Ade–Leu–Trp–His) medium. B) BiFC assays of the interaction between PbrPPA5 and PbrGNAT1 in N. benthamiana leaves. Vectors YNE and YCE contain the N-terminal and C-terminal fragments of YFP, respectively. Scale bars, 20 μm. C) PbrGNAT1 acetylates the K42 residue of PbrPPA5 in vivo. Substitution of K42 with a charge-conservative Arg residue (R) mimics a nonacetylated state. PbrGNAT1-HA with PbrPPA5-GFP or PbrPPA5K42R-GFP was transiently coexpressed in N. benthamiana leaves. Nuclear proteins were extracted and immunoprecipitated with magnetic beads containing α-GFP antibody. The protein extracts were immunoblotted with an α-acetylation antibody. The abundance of PbrGNAT1-HA, PbrPPA5-GFP, and PbrPPA5K42R-GFP was verified using an α-HA antibody and an α-GFP antibody, respectively. α-GFP was used as the loading control for nuclear proteins. D) Coexpression of PbrGNAT1 and PbrPPA5 promotes the accumulation of PbrPPA5 in the nucleus. PbrGNAT1-HA with PbrPPA5-GFP or PbrPPA5K42R-GFP was transiently expressed in N. benthamiana leaves. The fluorescence of their encoded proteins was observed under a confocal microscope. GFP signals represent the localizaiton of PbrPPA5 isoforms; DAPI was used as the nucleus marker. Scale bars, 10 μm. E) Quantitative analysis of the fluorescence intensity in the nucleus of PbrPPA5-GFP from D). Different lowercase letters indicate significant differences, as determined by ANOVA followed by Tukey's multiple comparison test (P < 0.05, n = 20 leaf cells). Data represent means ± SEM. F) Immunoblot analysis of PbrPPA5 in the nucleus from transiently infiltrated N. benthamiana leaves with or without coexpression of PbrGNAT1-HA. PbrPPA5-GFP and PbrPPA5K42R-GFP were detected using an α-GFP antibody, and an α-H3 antibody was used as the loading control for nuclear proteins. The expression of PbrGNAT1-HA in total leave protein was detected using an α-HA antibody. G) The expression levels of PbrGNAT1 in pollen tubes under 1 h mock, CS, and IS treatments in vitro. Mock, liquid germination medium; CS, compatible rPbrS-RNase; IS, incompatible rPbrS-RNase. PbrTUB-2 was used as the reference gene for normalization. Different lowercase letters indicate significant differences, as determined by ANOVA followed by Tukey's multiple comparison test (P < 0.05, n = 3 biological replicates). Data are means ± SEM.
Finally, because SI caused the acetylation of the K42 residue of PbrPPA5, we investigated whether PbrPPA5 accumulated in the nuclei of incompatible pollen tubes. PbrPPA5 was significantly more abundant in the nuclei of pollen tubes after IS treatment, as revealed by immunoblot analysis of cellular fractions (Fig. 8A). When PbrPPA5-GFP was transiently expressed in pear pollen tubes via particle bombardment, PbrPPA5-GFP was enriched in the generative nucleus of SI pollen tube 1.5 h after SI challenge (Figs. 8, B and C, and S28). These results suggest that SI induces the nuclear accumulation of PbrPPA5 in pear pollen tubes.
Figure 8.
SI induces PbrPPA5 accumulation in the nucleus. A) Immunoblot analysis showing that 1.5 h of IS treatment induces PbrPPA5 accumulation in the nucleus. Nucleus/cytosolic soluble fractions were isolated from pear pollen. α-H3.3 and α-GAPDH antibodies were used as the loading controls for nuclear and cytoplasmic proteins, respectively. Mock, liquid germination medium; CS, compatible rPbrS-RNase; IS, incompatible rPbrS-RNase. B) Images of the SI-induced PbrPPA5 accumulation in the nuclei of pollen tubes 1.5 h after IS treatment. The expression of PbrPPA5-GFP and AtH2B-mCherry was driven by the NTP303 promoter, and PbrPPA5-GFP and AtH2B-mCherry were transiently coexpressed in pear pollen tubes by particle bombardment. The transformed pear pollen was cultured for 3 h and then treated for 1.5 h with mock, CS, and IS treatments. The asterisks represent the vegetative nuclei and the triangles represent the generative nuclei. Scale bars, 20 μm. C) Quantitative analysis of the ratio of fluorescence intensity of PbrPPA5-GFP in the nucleus/cytosol in the nuclei of pollen tubes from B). Different lowercase letters indicate significant differences, as determined by ANOVA followed by Tukey's multiple comparison test (P < 0.05, n ≥ 12 pollen tubes). Data are means ± SEM.
Discussion
The swelling at the tips of pollen tubes during S-RNase–based SI has been observed for over 40 years (Hiratsuka and Tezuka 1980; Liu et al. 2007), but the underlying molecular mechanisms are still largely unknown. Here, we identified a signaling pathway that led to the repression of PbrPME44 expression and altered the structural composition of pectins in the cell walls of growing pollen tubes during the SI response (Fig. 9). Under SI challenge, PbrGNAT1 was upregulated within 1 h, leading to the acetylation of PbrPPA5 in 1.5 h. In turn, the acetylation of PbrPPA5 promoted its accumulation in the nuclei of pollen tubes, where PbrPPA5 bound to PbrbZIP77 and together cosuppressed PbrPME44 expression. Moreover, the role of PbrPPA5 as a cosuppressor of PbrPME44 expression was independent of its sPPase activity. The reduction in PbrPME44 levels disturbed the balance between methyl-esterified and deesterified pectin at growing pollen tube tips after SI challenge, disrupting polarized growth and leading to swelling at the tips of pollen tubes.
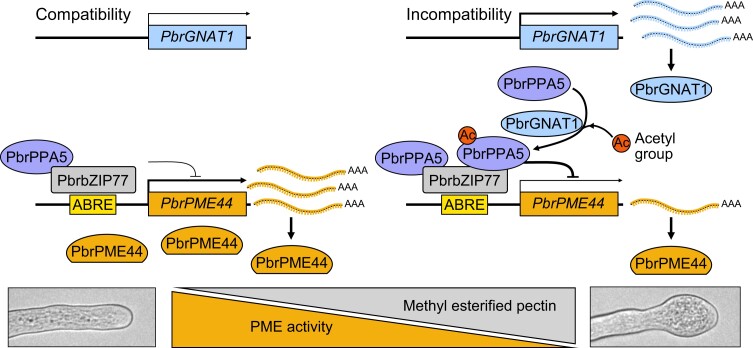
Figure 9.
A working model for the role of PbrPPA5 in tip swelling of pear pollen tubes under SI challenge. In growing pollen tubes, PbrPPA5 interacts with PbrbZIP77, and the PbrPPA5–PbrbZIP77 complex suppresses the expression of PbrPME44 by binding to the ABRE in its promoter to maintain the balance between methyl-esterified and deesterified pectin levels in pollen tubes. During the SI response, the expression level of PbrGNAT1 increases, and PbrGNAT1 acetylates PbrPPA5, inducing the accumulation of PbrPPA5 in the nucleus. PbrPPA5 in the nucleus binds to PbrbZIP77 to enhance the repression of PbrPME44. The downregulation of PbrPME44 leads to an increase in methyl-esterified pectin levels in the pollen tube cell wall, resulting in swelling at the tips of pollen tubes.
PbrPME44 mediates tip swelling of pollen tubes following SI challenge
Maintaining the biomechanical equilibrium and stability of different cell wall components in the pollen tube tip is essential for the polarized growth of pollen tubes (Bosch and Hepler 2005). Methyl-esterified and deesterified pectins are major components of the cell wall whose abundance and ratio play critical roles in determining the shape and integrity of growing pollen tubes (Duan et al. 2020; Zhou et al. 2021). SI-induced tip swelling of pear pollen tubes is not related to changes in total pectin content (Supplemental Fig. S3) but is associated with increased levels of methyl-esterified pectins (Fig. 1). This increase may result in the softening of the pollen tube tip wall (Bosch and Hepler 2005). It should be noted that the tips of most incompatible pollen tubes do not swell, possibly due to insufficient accumulation of methyl-esterified pectin (Fig. 1). Meanwhile, we could not exclude the possibility that some other factors participate in the growth arrest of incompatible pollen tubes in SI response of pear.
Our results uncover the relationship among growth arrest, tip swelling, and methyl esterification of pectin at the apical regions of incompatible pollen tubes in pear. During the early stage following the SI challenge, PbrPME44 restored the level of methyl-esterified pectins and the growth of pollen tubes (Fig. 2). Therefore, downregulating PbrPME44 leads to insufficient PME activity, which affects the methyl esterification of pectin in pollen tubes and contributes to tip swelling.
We propose a holding phase during the SI response in which the pollen tube remains plastic to provide another fertilization opportunity for the arrested pollen tube when the SI conditions weaken for 1 reason or another (Cheung 2022). For example, SI-induced ATP depletion has been suggested to be the holding phase before programmed cell death of pollen tubes during the SI response in common poppy (Wang et al. 2022). Thus, the tip swelling of incompatible pollen tubes could be a holding phase in the SI response of pear. During later stages of the SI challenge, pollen tube growth failed to be restored by PbrPME44 (Supplemental Fig. S12), suggesting that the cessation of growth of incompatible pollen tubes might be determined by other factors, such as actin depolymerization (Chen et al. 2018).
The RNase activity of PbrS-RNase was not required for the downregulation of PbrPME44 (Fig. 1). Combined with results showing S-RNase–mediated actin disruption in apple and pear (Chen et al. 2018; Yang et al. 2018; Zhao et al. 2021), and heat-inactivated S-RNase exerting a more severe inhibition of pollen tubes in N. alata (Gray et al. 1991), we suggest that PbrS-RNase could also function in signaling pathways independent of its enzymatic activity in the SI response.
The PbrbZIP77–PbrPPA5 complex suppresses the expression of PbrPME44
In Arabidopsis, AtbZIP34 regulates PME gene expression and pollen wall development (Gibalová et al. 2009). We demonstrated that PbrbZIP77, a homolog of AtbZIP34, bound to the promoter region of PbrPME44 and inhibited its expression (Fig. 3). We also identified PbrPPA5a as an interacting partner of PbrbZIP77. PbrPPA5 is a classic Mg2+-dependent sPPase; these proteins localize to the nucleus and cytoplasm. Two functions of sPPases in the nucleus have been proposed: (i) sPPases are essential for hydrolysis of PPi, as during nucleic acid biosynthesis in Arabidopsis (Segami et al. 2018), and (ii) sPPases are important for nucleosome remodeling in Drosophila melanogaster (Gdula et al. 1998). Our results propose a role for an sPPase in transcriptional regulation. PbrPPA5 interacted directly with PbrbZIP77 and enhanced PbrbZIP77-mediated transcriptional repression of PbrPME44, whereas PbrPPA5 alone did not inhibit PbrPME44 expression. Moreover, the downregulation of PbrPME44 expression by the PbrPPA5–PbrbZIP77 transcriptional complex was independent of the intrinsic pyrophosphatase activity of PbrPPA5, suggesting that PbrPPA5 functions as a transcriptional regulator in pear pollen (Fig. 5).
Acetylation of PbrPPA5 promotes its accumulation in nuclei during Si
The roles of sPPases in SI signaling have been reported in poppy and apple. During the Ca2+-mediated SI response in poppy, 2 sPPases, Prp26.1a and 1b, become phosphorylated, significantly reducing their sPPase activity (de Graaf et al. 2006; Eaves et al. 2017). In contrast, MdPPa in apple pollen is not phosphorylated during the SI response; rather, the S-RNase responsible for the SI response in apple interacts with MdPPa in pollen and inhibits its enzymatic activity (Li et al. 2018). Unlike in apple and poppy, we established that PbrPPA5 in pear was not phosphorylated and did not interact with PbrS-RNases upon SI challenge. The SI mechanisms for pollen tube growth inhibition are differentially regulated (temporally and spatially) in pear compared to poppy. In pear, the SI response is slow; the female determinants PbrS-RNases enter the growing pollen tubes via endocytosis or an unknown mechanism, after which they exert their toxic effects (Liu et al. 2007; Chen et al. 2018). In poppy, the female determinant PrsS does not enter the cytoplasm of pollen, instead acting as a ligand by interacting with its cognate membrane–localized pollen receptor-like determinant PrpS to induce a series of signaling events in the pollen cytosol, triggering a rapid SI response characterized by inhibited pollen germination and tube growth (Thomas and Franklin-Tong 2004; Bosch and Franklin-Tong 2007; Wheeler et al. 2009; Wang et al. 2019b; Lin et al. 2022).
We determined that PbrPPA5 became acetylated after the SI challenge (Supplemental Fig. S22 and Data Set 2), and posttranslational modifications such as this can alter the localization of a protein (Wang et al. 2010; Baeza et al. 2016; Narita et al. 2019; Zhu et al. 2019). In healthy growing pollen tubes, PbrPPA5 localizes to both the cytoplasm and nuclei. Under SI challenge, the K42 residue of PbrPPA5 was acetylated by PbrGNAT1, leading to the more frequent nuclear accumulation of this modified PbrPPA5 (Fig. 8). The nuclear accumulation of PbrPPA5 could result from an increase in its import rate, a decrease in its export rate, or a sequestration mechanism in which formation of a complex with other nuclear proteins increases its residence time inside the nucleus. Further investigation is needed to determine the underlying mechanism.
In conclusion, we present a model describing how the acetylation of PbrPPA5 elicits the swelling of pollen tube tips following SI challenge in pear (Fig. 9). In healthy growing pollen tubes, PbrbZIP77 suppresses the expression of PbrPME44 to maintain the balance between methyl-esterified and deesterified pectin levels at the expanding pollen tube tip. During the SI response, PbrGNAT1 is upregulated, and PbrGNAT1 acetylates PbrPPA5, promoting its nuclear accumulation. Nucleus-localized PbrPPA5 binds to PbrbZIP77, forming a complex that suppresses PbrPME44 expression. Downregulation of PbrPME44 leads to higher levels of methyl-esterified pectin, thus decreasing cell wall rigidity and ultimately resulting in the swelling of pollen tube tips. Overall, our elucidation of the role of PbrPPA5 in SI opens avenues for studying S-RNase–based SI. In addition, our findings shed light on the important role of the PbrPPA5–PbrbZIP77 complex in gene regulation as it fine-tunes the production of 1 plant cell wall–modifying enzyme in order to build a continuous sustainable mechanical structure for plant cell development.
Materials and methods
Plant materials
Pollen from P. bretschneideri cv. Dangshansuli (S7S34) was collected from specimens growing at the Nanjing Agricultural Fruit Experimental Yard, China. After drying for 24 h at room temperature, the mature pollen was stored in silica gel at −20 °C for the subsequent experiments. Roots, stems, leaves, and fruits were collected and frozen immediately in liquid nitrogen and stored at −80 °C. Pistils were isolated and transferred into storage tubes and kept in liquid nitrogen until use.
Aniline blue staining of pollen tubes in pistils
Aniline blue staining was performed as previously described with some modification (Hiratsuka and Tezuka 1980). The preemasculated mature flowers of Dangshansuli (S7S34) were pollinated with either Dangshansuli (S7S34, self-pollination) or Huanghua (S1S2, cross-pollination) pollen. The pistils were collected 48 h after pollination and fixed in ethanol:acetic acid (3:1 v/v) for at least 2 h at room temperature. The fixed pistils were washed with 70%, 50%, and 30% ethanol for 10 min each, then washed with distilled water for 10 min and finally softened overnight in a solution of 2 M NaOH. The softened pistils were washed 3 times with distilled water and stained overnight in aniline blue solution (0.1% w/v aniline blue in 0.1 M K3PO4 buffer, pH 7.3) at 37 °C in the dark. The stained pistils were observed using an IX73 fluorescent microscope (Olympus, Japan).
Pollen culture, measurement, and S-RNase treatments
Pollen was cultured in liquid germination medium (0.5 mM Ca(NO3)2, 1.5 mM H3BO3, 450 mM sucrose, and 25 mM 2-(N-morpholilino)ethanesulfonic acid hydrate (MES), pH 6.0 to 6.5). Pollen tube diameters and lengths were measured under a light microscope using ImageJ software (https://imagej.nih.gov/ij/).
His-tagged rPbrS-RNase proteins were produced according to a previously described method (Chen et al. 2018). For IS treatments, pollen tubes (Dangshansuli; S7 or S34) were precultured in liquid germination medium for 1.5 h and treated with rPbrS7-RNase plus rPbrS34-RNase protein (at a final concentration of 0.15 U) or the same amount of mutant rPbrS7-RNaseH116R plus rPbrS34-RNaseH116R (H116R) protein added to the pollen cultures in vitro. For CS treatments, pollen tubes (Dangshansuli; S7 or S34) were precultured for 1.5 h after which rPbrS1-RNase and rPbrS2-RNase were added. For PME compensation experiments, pollen tubes were precultured for 1.5 h and treated with rPbrS7-RNase plus rPbrS34-RNase proteins or rPbrS7-RNaseH116R plus rPbrS34-RNaseH116R proteins for another 2.0 or 3.0 h, then 0.005 mg mL−1 His-tagged rPbrPME44 proteins were added, respectively, and treated for 1.0 h.
Illumina sequencing and analysis of transcriptome data
At 48 and 72 h after pollination, SP and CP pistils were collected, immediately frozen in liquid nitrogen, and stored at −80 °C. Total RNA was extracted from the frozen tissue mentioned above using a Plant Total RNA Isolation Kit (Foregene, China) according to the manufacturer's instructions. mRNA samples were extracted from total RNA using the VAHTS mRNA-SEQ V2 Library Prep Kit (Vazyme, China) according to the manufacturer's instructions. Double-stranded cDNA was reverse-transcribed and amplified by Illumina gene expression sample preparation kit (Illumina, USA). An Illumina Novaseq 6000 instrument was used to perform the sequencing reactions. Clean reads were obtained by FASTP software (https://github.com/OpenGene/fastp). Hisat2 (http://daehwankimlab.github.io/hisat2/) was used to align clean reads to the reference genome of pear v1.0 (https://doi.org/10.5524/100083). Gene expression levels were calculated based on raw counts and were normalized using DESeq2 (Love et al. 2014). Finally, all differentially expressed genes were annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/).
Immunohistochemical analysis of pollen tubes
The immunohistochemical analysis of methyl-esterified pectins in pollen tubes was performed as previously described (Wang et al. 2018). The primary antibody used in our study was LM20 (University of Leeds, UK; http://www.plantprobes.net/), which labels methyl-esterified HG (1:20 dilution). Antirat IgG-fluorescein isothiocyanate-conjugated (FITC) antibody (1:500 dilution; F1763, Sigma-Aldrich, USA) was used as the secondary antibody. The fluorescence was imaged using a LSM800 confocal microscope with Airyscan (Carl Zeiss, Germany) and quantified using ZEN imaging software (Carl Zeiss, Germany). The excitation wavelength and transmission range for emission were 488 nm/500 to 550 nm for the FITC channel, and the detector gain was 700 V.
Analysis of methyl-esterified HG in the pollen tube cell wall using ELISA
The alcohol-insoluble residue of the pollen tube cell wall was generated as previously described (Silva-Sanzana et al. 2019), and its components were sequentially extracted with 2 mL water and trans-1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid (CDTA) to generate soluble fractions containing HG (Wang et al. 2019a). ELISA was performed as described previously (Willats et al. 1998). A primary HG-directed antibody (LM20) was used as a 10-fold dilution, and the horseradish peroxidase (HRP)–conjugated goat antirabbit IgG antibody that was used as the secondary antibody (Sangon, China) was added at a 1,000-fold dilution. The absorbance of each well at 450 nm was determined using a CYTATION3 microplate reader (BioTek, USA). At least 3 independent experiments were performed.
Measurement of total pectins
After 2.0-h mock, SC, and SI treatments, pollen samples were collected by centrifuging at 13,500 × g and ground in liquid nitrogen. After the addition of 1 mL 95% ethyl alcohol, pollen samples were incubated in a water bath at 80 °C for 30 min. They were then centrifuged at 13,500 × g for 2 min before being sequentially washed with 2 mL 85% ethyl alcohol, 2 mL methyl alcohol:chloroform (1:1), and 2 mL acetone. Samples containing the resulting sediments were left to dry by evaporation at room temperature overnight. Total pectins were measured using a Total Pectin Content Test Kit (Solarbio, China). The absorbance of each well at 530 nm was determined using a CYTATION3 microplate reader (BioTek, USA). At least 3 independent experiments were performed.
RT-PCR analysis
RT-PCR analysis was performed to confirm the presence of PbrbZIP77 transcripts in the pollen tubes, pistils, roots, stems, leaves, and fruits. Total RNA was extracted from the samples mentioned above, then reverse-transcribed using the RevertAid RT Reverse Transcription Kit (Thermo Fisher Scientific, USA), according to the manufacturer's instructions.
The reaction mixture for RT-PCR contained 0.1 μL cDNA, 4 μL 0.1 μM gene-specific primer mixture, 10 μL 2×Taq Plus Master Mix (Vazyme, China), and 5.9 μL double-distilled water in a total volume of 20 μL. PCR was performed with the following program: 3 min at 94 °C; 25 cycles of 30 s at 94 °C, 30 s at 60 °C, and 30 s at 72 °C; and a final 10-min extension at 72 °C. The PbrTUB-2 (XP_009374320.1) was used as the reference gene. PCR products were fractionated in 2% agarose gels. Primers are listed in Supplemental Data Set 3.
RT-qPCR
The expression levels of PbrbZIP77 and 47 PbrPME under mock, SI, and SC treatments were quantified using qPCR. The qPCR reaction mixture contained 0.2 μL cDNA sample, 5 μL 0.5 μM gene-specific primer mixture, 10 μL 2×SYBR Green Master Mix, and 4.8 μL water in a total volume of 20 μL. qPCR was performed using the LightCycler SYBR Green I Master (Roche, Germany). Relative expression levels were calculated using the 2−ΔΔCT method (Livak and Schmittgen 2001), with PbrTUB-2 as the internal standard. A melting curve was performed to verify the specificity of all qPCR primers. At least 3 biological replicates were employed for each experiment. The heat map was generated using TBtools (Chen et al. 2020). The primers are listed in Supplemental Data Set 3.
Point mutants and Y1H assay using the PbrPME44 promoter
The 1.3-kb promoter sequence of PbrPME44 was amplified from pear genomic DNA using PCR and then cloned into the pAbAi vector (Clontech, USA) to use as bait for screening the Dangshansuli pollen cDNA library. Y1H assays were performed using the Matchmaker Gold Yeast One-Hybrid Library Screening System (Clontech, USA).
For the interaction assays, the DNA elements were analyzed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Point mutations (ABRE mut and C-box mut) were introduced using a Fast Mutagenesis System (Transgen, China). Promoters used for the Y1H assay were cloned into pAbAi to generate PbrPME44pro:pAbAi, PbrPME44(ABREmut)pro:pAbAi, and PbrPME44(C-box mut)pro:pAbAi plasmids, which were integrated into the Y1HGold yeast strain as bait. Full-length PbrbZIP77 cDNA was fused into the pGADT7 AD vector (Clontech, USA) to generate PbrbZIP-AD, which was introduced into the 3 bait strains. p53-pAbAi and AD-Rec53 (Clontech, USA) were introduced into the Y1HGold strain as a positive control. The cotransformed yeast cells were cultured on synthetic dropout (SD)/–Leu plates (Clontech, USA) with or without aureobasidin A (AbA) (400 ng mL−1) and incubated at 30 °C for 3 d.
Expression and purification of recombinant proteins
Full-length cDNAs of PbrS1/2/7/34-RNase (without the signal peptide sequence), PbrS7/34-RNaseH116R (without the signal peptide sequence), PbrPPA5, PbrPPA5K42Q, PbrPPA5K42R, PbrPPA5D136N, PbrPPA5P137A, PbrbZIP77, and PbrPME44 (without the N-terminal hydrophobic motif) were amplified by PCR and individually inserted into pCold-TF vectors (Takara, Japan). pCold-TF is a fusion cold shock expression vector that expresses trigger factor chaperone as a soluble tag. Trigger factor is a prokaryotic ribosome-associated chaperone protein (48 kDa) that facilitates cotranslational folding of newly expressed polypeptides. Primers are listed in Supplemental Data Set 3. The rPbrS1/2/7/34-RNase, rPbrS7/34-RNaseH116R, His-tagged rPbrPPA5, His-tagged recombinant PbrPPA5K42Q (rPbrPPA5K42Q), His-tagged recombinant PbrPPA5K42R (rPbrPPA5K42R), His-tagged recombinant PbrPPA5D136N (rPbrPPA5D136N), His-tagged recombinant PbrPPA5P137A (rPbrPPA5P137A), His-tagged recombinant PbrbZIP77 (rPbrbZIP77), and rPbrPME44 were expressed as fusion proteins in E. coli strain BL21 Rosetta (DE3) (Transgen, China). A 10-mL liquid culture of recombinant E. coli protein was incubated overnight at 37 °C in the presence of ampicillin (0.1 mg mL−1) and transferred to fresh Luria-Bertani (LB) medium as a 1:50 dilution (10 mL overnight culture added to 490 mL LB medium). The cultures were grown at 37 °C for another 1 to 4 h. When OD600 of the suspension reached 0.4 to 0.6, the suspension was rapidly cooled to a 15 °C and incubated for 30 min at 15 °C. After this, 0.5 mM IPTG was added and the culture was shaken at 15 °C for 24 h to induce protein expression in the cells, which were then harvested by centrifugation at 13,500 × g and stored at −20 °C until required for protein extraction.
For protein extraction, cells were resuspended in 30 mL lysis buffer (50 mM Tris-HCl, 140 mM NaCl, 2 mM EDTA, and 5 mM imidazole, pH 7.9) supplemented with cOmplete EDTA-free protease inhibitor cocktail (Roche, Germany) and vigorously mixed. The suspension was sonicated 5 times on ice for 7 s each time, with a 20-s rest between each pulse to allow cooling. The lysate was centrifuged (13,500 × g, 15 min) at 4 °C, after which the supernatant was recovered and filtered through a 0.45-μm pore size filter membrane (MilliporeSigma, USA). For rPbrPPA5, rPbrPPA5K42Q, rPbrPPA5K42R, rPbrPPA5D136N, and rPbrPPA5P137A, the supernatant containing the soluble protein was loaded onto a column containing 2 mL NTA-Ni+ His Bind resin in phosphate-buffered saline (PBS) (MilliporeSigma, USA) and washed with 20 mL binding buffer (500 mM NaCl, 5 mM imidazole, and 20 mM Tris-HCl, pH 7.9), then 40 mL wash buffer (500 mM NaCl, 30 mM imidazole, and 20 mM Tris-HCl, pH 7.9) prior to being eluted with 6 mL elution buffer (500 mM NaCl, 50 mM imidazole, and 20 mM Tris-HCl, pH 7.9). For rPbrbZIP77, the wash buffer was changed to 500 mM NaCl, 50 mM imidazole, and 20 mM Tris-HCl (pH 7.9), and the elution buffer was 500 mM NaCl, 90 mM imidazole, and 20 mM Tris-HCl (pH 7.9). For rPbrPME44, the elution buffer was 500 mM NaCl, 60 mM imidazole, and 20 mM Tris-HCl (pH 7.9). The rPbrS-RNases were produced as described previously (Chen et al. 2018). Fractions were desalted and concentrated by centrifugal ultrafiltration (6,000 × g, 5 min) using 30-kDa Amicon Ultra-4 tubes (MilliporeSigma, USA) and equilibrated with 25 mM Tris-HCl (pH 8.0), PBS, or liquid germination medium.
EMSA
An EMSA was performed using the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, USA), according to the manufacturer's protocol. The rPbrbZIP77 and rPbrPPA5 were produced as described above. An oligonucleotide probe (with or without a biotin label) representing the ABRE motif was synthesized by Genewiz (Suzhou, China). The binding activity of rPbrbZIP77 was analyzed using the sequence containing the ABRE motif and its flanking sequence, and additional rPbrPPA5 was added to detect the supershift band. The sequences of the oligonucleotide probes are listed in Supplemental Data Set 3.
BLI assay
The binding affinities of rPbrbZIP77 and the ABRE probe were assessed using an Octet RED96 (FortéBio, USA) equipped with a streptavidin (SA) sensor (Pall, USA). Water was used as a negative control. The experiments were conducted at room temperature in water containing 0.02% Tween-20. The SA sensor tips were immersed into wells containing different concentrations of rPbrbZIP77 (342, 384, 427, 570, 712, and 855 nM). The biotin-labeled oligonucleotide probe of the ABRE motif was synthesized by Genewiz (Suzhou, China). Double-stranded ABRE primers (0.1 μM) were produced by annealing and loaded onto SA biosensors until saturation. Each data set was fitted globally to a 1:1 interaction model (FortéBio, USA) to determine the kinetic parameters, kon and koff. The apparent affinities were then calculated as a ratio (koff/kon) of these rate constants using Data Analysis software v9.0.0.14 (FortéBio, USA). The sequences of the oligonucleotide probes are listed in Supplemental Data Set 3.
Dual-LUC reporter assay
The full-length cDNAs of PbrPPA5 and PbrbZIP77 were amplified using PCR and cloned into an overexpression vector driven by the 35S promoter to generate 35Spro:PbrPPA5 and 35Spro:PbrbZIP77 (Xie et al. 2014). Point mutations (PbrPPA5K42R, PbrPPA5K42Q, PbrPPA5D136N, and PbrPPA5P137A) were introduced using a Fast Mutagenesis System (Transgen, China). The 1.3-kb promoter sequence of PbrPME44 was amplified by PCR, and the PCR-amplified fragments were then cloned into a pGreenII 0800-LUC vector to generate PbrPME44pro:LUC. Primers are listed in Supplemental Data Set 3. The Renilla luciferase gene (REN) driven by the 35S promoter was used in the pGreenII 0800-LUC as the internal control. The dual-LUC reporter assays in N. benthamiana leaves were performed as described previously (Hellens et al. 2005). Relative LUC activities were quantified using the Dual-Luciferase Reporter Assay System (Promega, USA) with CYTATION3 microplate readers (BioTek, USA). At least 3 independent experiments were performed.
Sequence analysis
The sequence of AtbZIP34 was retrieved from TAIR 10 (http://www.arabidopsis.org/). Sequences of the Prp26.1a/b proteins from poppy were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/). Multiple sequence alignments were performed using DNAMAN (https://www.lynnon.com/qa.html).
Y2H assays
A cDNA library for Y2H assay was constructed by Clontech (Takara, Japan) from the total RNA of Dangshansuli pollen. Full-length cDNAs of PbrbZIP77 and PbrPPA5 were cloned into the pGBKT7 vector (Clontech, USA) to be used as bait for screening the cDNA library using the Matchmaker Gold System (Clontech, USA). The screening was performed on SD/–Trp–Leu–His medium according to the manufacturer's instructions.
For the protein–protein interaction assay, full-length cDNAs of PbrbZIP77, PbrPPA5, PbrS7-RNase, and PbrS34-RNase were cloned into pGADT7 AD (Clontech, USA). PbrPPA1, PbrPPA2, PbrPPA5, PbrPPA9, PbrPPA10, and PbrGNAT1 were cloned into pGBKT7 vectors individually. Point mutations (PbrPPA5D136N and PbrPPA5P137A) were introduced into PbrPPA5 using a Fast Mutagenesis System kit (Transgen, China). After mutation, PbrPPA5D136N and PbrPPA5P137A were inserted into pGBKT7 vectors individually. The coding sequences of different domains in PbrPPA5 (1 to 31, 32 to 54, 55 to 86, 87 to 213, 214 to 234, and 235 to 248 aa) were cloned into pGBKT7 vectors individually. All interactions were detected by cotransforming plasmids into the yeast strain AH109. The interactions were assessed on SD/–Trp–Leu–Ade–His medium. pGBKT7-P53 and pGADT7-T were used as a positive control, and pGBKT-Lam and pGADT7-T were a negative control. All the primers are listed in Supplemental Data Set 3.
LCI assay
PbrPPA5 and PbrbZIP77 were amplified using PCR and cloned into the pCAMBIA1300-CLuc and pCAMBIA1300-NLuc vectors to generate PbrPPA5-CLuc and PbrbZIP77-NLuc, respectively (Zhou et al. 2018). These 2 constructs were transformed into N. benthamiana leaves using Agrobacterium tumefaciens–mediated transformation (Sparkes et al. 2006), and the LUC complementation assay for PbrPPA5 and PbrbZIP77 interaction was performed as described previously (Zhou et al. 2018). Luminescence in N. benthamiana leaves was photographed by a CCD imaging system (PIXIS 1024B, Princeton Instruments, USA).
BiFC assay
PbrPPA5, PbrbZIP77, and PbrGNAT1 were cloned into the pSPYNE-35S and pSPYCE-35S vectors to generate PbrPPA5-YNE, PbrbZIP77-YCE, and PbrGNAT1-YCE, respectively (Walter et al. 2004). These constructs were transformed into N. benthamiana leaf cells using Agrobacterium-mediated transformation (Sparkes et al. 2006). After 3 d of cultivation, tobacco leaves were harvested, stained with DAPI (Thermo Fisher Scientific, USA) as a nucleus marker, and used for imaging. Fluorescence signals were imaged using a LSM800 confocal microscope (Carl Zeiss, Germany). The excitation wavelength and transmission range for emission were 488 nm/500 to 550 nm for the GFP channel, and the detector gain was 700 V.
MST assay
The MST assay was performed using the Monolish NT.115 instrument (Nano Temper Technologies, Germany) according to the manufacturer’s instructions. For protein labeling, the Monolith His-Tag Labeling Kit RED-tris-NTA (Nano Temper Technologies, Germany) was used following the manufacturer's procedure. The purified His-tagged protein, rPbrbZIP77, was concentrated in the labeling buffer (PBS plus 0.05% Tween) by centrifugal ultrafiltration (6,000 × g, 5 min) to a final concentration of 50 nM. For the binding assay, serial 2-fold dilutions of 16 concentrations (initiated from 1,000 nM) of the protein ligands (rPbrPPA5, rPbrPPA5K42Q, or rPbrPPA5K42R) were tested. The labeled rPbrbZIP77 and diluted ligands were mixed into the capillary and analyzed using the Monolith NT.115 instrument. Proteins purified from the empty pCold-TF vector were used as negative controls. MST curves for each capillary were recorded and the Kd of each reaction was calculated using the MO Affinity Analysis software v1.6 (Nano Temper Technologies, Germany). Three biological replicates were employed for each combination.
Measurement of PME activity
Pollen tube samples were collected by centrifugation (13,500 × g, 1 min) and then ground in liquid nitrogen. Crude proteins were extracted using a Plant Protein Extraction Kit (Solarbio, China). Protein concentrations were measured using the Modified Bradford reagent (Sangon, China) with bovine serum albumin as a standard. PME activity was measured as described previously (Downie et al. 1998). In brief, gels were prepared using 0.1% of pectin from citrus (P0024, TCI, Japan), 2% (w/v) agarose (low melting point gel), 0.1 M citric acid, and 0.2 mM Na2HPO4 (pH 7.0). Wells (4-mm diameter) were punched by injector, and 5 μg of each crude protein was loaded into each well. The plates were then incubated at 30 °C for 16 h, and gels were stained with 0.05% (w/v) ruthenium red for 45 min, then washed thoroughly with distilled water. The diameter of the red-stained areas determined by caliper was indicative of the hydrolysis of esterified pectin in the gels. At least 3 biological replicates were employed for each treatment.
Pyrophosphatase activity assay
To determine sPPase activity, 500 ng rPbrPPA5, rPbrPPA5K42Q, rPbrPPA5K42R, rPbrPPA5D136N, or rPbrPPA5P137A was added into 200 μL reaction buffer (1 mM PPi, 5 mM MgCl2, and 100 mM Tris-HCl, pH 7.0) and incubated for 15 min at 37 °C. For pollen samples, 200-mg pollen was collected and extracted using a Plant Protein Extraction Kit (Solarbio, China). The reaction was stopped by adding 5% trichloroacetic acid (TCA) and then assayed as described previously (Visser et al. 1998; de Graaf et al. 2006).
as-ODN treatment
An as-ODN experiment was performed as previously described (Moutinho et al. 2001). Both phosphorothioated as-ODN and phosphorothioated s-ODN were designed using the RNA fold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). For the as-ODN and s-ODN treatments, pollen was precultured in liquid germination medium for 1 h, then 6 μL as-ODNs (30 μM) or s-ODNs was mixed with 12.5 μL germination media and 1.5 μL cytofectin (Lipofectamine 2000, Thermo Fisher Scientific, USA) for 15 min before being added to 180 μL cultivated pollen. Pollen treated with cytofectin and germination medium only was used as a control. The sequences of the phosphorothioated ODNs are listed in Supplemental Data Set 3.
Nuclear fractionation assay of pollen tubes and tobacco leaves
Nuclei were extracted by nuclear fractionation, as described previously (Gao et al. 2015). Pollen tubes were overlaid with nuclei isolation buffer (0.3 M sucrose, 10 mM NaCl, 10 mM MES, 5 mM EDTA, 0.2 mM spermine, 0.5 mM spermidine, 0.2 mM phenylmethylsulfonyl fluoride, 5 mM dithiothreitol, and 0.2% (v/v) Triton X-100, adjusted to pH 7.4 with 2 M NaOH), separated by filtration in a cell strainer I (40 µm pore size) with homogenizing by a glass pestle, and collected in an open Petri dish under a cell strainer II (25 µm pore size). After centrifugation at 200 × g at 4 °C for 3 min, the supernatant was separated and then placed onto a 28% (v/v) Percoll solution (diluted in nuclei isolation buffer) (GE Healthcare, USA). After centrifugation at 3,000 × g for 20 min, the purified nuclei were located at the bottom of the Percoll layer and the upper phase was the cytoplasmic fraction. All the above steps were performed at 4 °C. Protein extracts of tobacco leaf nuclei were obtained using a Nuclear Protein Extraction Kit (Solarbio, China). Crude proteins were extracted using a Plant Protein Extraction Kit (Solarbio, China). All the above steps were performed at 4 °C.
Immunoblotting
Pollen tubes were collected after 1.5-h mock, CS, and IS treatments. Nuclear/cytosolic protein extracts of pear pollen tubes were prepared as described above. Protein extracts were separated using 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred onto an Immobilon-P PVDF membrane (MilliporeSigma, USA). After blocking with 5% solution of nonfat milk powder in Tris-buffered saline containing 0.1% Tween (pH 8.0), the membrane filter was incubated with the anti-PbrPPA5 rabbit polyclonal primary antibody (LPTLQAFEVQKLMASEDQSEEAK; 1:1,000 dilution) overnight, followed by incubation with an HRP-conjugated goat antirabbit IgG antibody (1:5,000 dilution) (Sangon, China) for 2 h at room temperature. The chemiluminescence reagent ECL substrate (Thermo Fisher Scientific, USA) was used for antigen detection, and chemiluminescence was detected using a ChemiDoc MP Imaging System (Bio-Rad, USA). Anti-HA tag mouse monoclonal antibody (1:2,000 dilution; Beyotime, China) was added to detect the presence of PbrGNAT1-HA, and an anti-GFP rabbit polyclonal antibody (1:2,000 dilution; Sangon, China) was added to detect the presence of PbrPPA5-GFP. Antihistone H3 mouse monoclonal antibody (1:2,000 dilution; Beyotime, China), antihistone H3.3 rabbit monoclonal antibody (1:1,000 dilution; Beyotime, China), and anti-GAPDH rabbit polyclonal antibody (1:2,000 dilution; Sangon, China) were used as the loading controls for nuclear and cytoplasmic proteins, respectively.
Subcellular localization
Full-length cDNA of PbrPPA5 lacking the stop codon was amplified using PCR then cloned into pCAMBIA1300-35S:CDS-GFP (Xie et al. 2014). Point mutations (PbrPPA5K9R, PbrPPA5K9Q, PbrPPA5K42R, PbrPPA5K42Q, PbrPPA5K98R, and PbrPPA5K98Q) were introduced using a Fast Mutagenesis System (Transgen, China). Full-length cDNA of PbrGNAT1 lacking the stop codon was cloned into an overexpression vector (pCAMBIA1300-35S:CDS-HA) driven by the 35S promoter to generate the 35Spro:PbrGNAT1-HA vector (Xie et al. 2014). Primers are listed in Supplemental Data Set 3. Constructs were transformed into Agrobacterium strain GV3101 and the transformed cells were infiltrated into N. benthamiana leaf epidermal cells using Agrobacterium-mediated transformation (Sparkes et al. 2006). The blue-fluorescent dye DAPI (Sigma-Aldrich, USA) was used as a nuclear marker. All fluorescence imaging was performed using a LSM800 confocal microscope with Airyscan (Carl Zeiss, Germany) and quantified using ZEN imaging software (Carl Zeiss, Germany). The excitation wavelength and transmission range for emission were 488 nm/500 to 550 nm for the GFP channel and 405 nm/400 to 490 nm for the DAPI channel. The detector gain was 700 V.
Detection of PbrPPA5 modifications during SI using liquid chromatography–tandem mass spectrometry
Pollen was grown in liquid germination medium for 1.5 h at 25 °C and was then subjected to IS and CS treatments for 1.0, 1.5, and 2.0 h. Pollen samples were collected by centrifugation at 6,000 × g for 1 min. After samples were ground in liquid nitrogen, crude proteins were extracted using the Plant Protein Extraction Kit (Solarbio, China). Protein concentrations were measured using a NanoDrop 2000 (Thermo Fisher Scientific, USA). To detect the modification of PbrPPA5 resulting from CS and IS treatments, crude proteins were separated using 12% SDS–PAGE and the bands corresponding to the molecular weight of PbrPPA5 were cut from the gel and sent for mass spectrometric analysis. Mass spectrometry was performed by Shanghai Applied Protein Technology (Shanghai, China) using Q Exactive and Easy-nLC 1000 (Thermo Fisher Scientific, USA). The resulting data were analyzed using Proteome Discoverer1.4 (Thermo Fisher Scientific, USA) and MASCOT (http://www.matrixscience.com).
Particle bombardment
Full-length cDNAs of PbrPPA5, PbrPPA5K42Q, and PbrPPA5K42R were amplified using PCR and individually cloned into an overexpression vector driven by the NTP303 promoter (X69440.1) to generate NTP303pro:PbrPPA5, NTP303pro:PbrPPA5K42Q, and NTP303pro:PbrPPA5K42R. A sequence encoding AtH2B-mCherry was cloned to the same vector and used as a nuclei marker (Motomura et al. 2021). Recombinant plasmids were isolated using a FastPure EndoFree Plasmid Maxi Kit (Vazyme, China) and transiently expressed in pear pollen using a PDS-1000/He particle bombardment device (Bio-Rad, USA) according to a previously described protocol with some modification (Qian et al. 2020). A mixture of 8.5 μL gold particles (Bio-Rad, USA) and 10 μL of 0.15 M spermidine, 2.5 μL of 2 μg/μL recombinant plasmid, and 29 μL of 2 M CaCl2 was used, with constant vortexing for 3 min. The gold microcarriers were centrifuged at 9,500 × g for 5 s and the supernatant was removed. Then, the gold particles were washed with 200 μL of absolute ethanol and centrifuged at 9,500 × g for 5 s. Finally, the gold particles were resuspended in 20 μL of absolute ethanol for transformation. The settings of the PSD-1000/He particle delivery system were as follows: 1,350 psi, 20-mm Hg vacuum, 1-cm gap distance between the rupture disk and macrocarrier, and 9-cm particle flight distance between macrocarrier and pollen samples. The pollen samples were placed on solid germination medium (containing 1.5% agarose) prior to transformation. For DAPI staining, pollen tubes were fixed in 4% paraformaldehyde for 30 min and stained with DAPI. Fluorescence images were performed using a LSM800 confocal microscope (Carl Zeiss, Germany). The excitation wavelength and transmission range for emission were 488 nm/500 to 550 nm for GFP, 405 nm/400 to 490 nm for DAPI, and 561 nm/570 to 700 nm for mCherry. The detector gain was 700 V.
Immunoprecipitation and acetylation assays
For immunoprecipitation (IP) assays, Agrobacterium strain GV3101 containing PbrPPA5-GFP, PbrPPA5K42R-GFP, or PbrGNAT1-HA was used as mentioned (Sparkes et al. 2006). Cells transformed with PbrPPA5-GFP or PbrPPA5K42R-GFP were mixed with the strain containing PbrGNAT1-HA at OD600 = 0.6 to 0.8 and infiltrated into N. benthamiana leaves using Agrobacterium-mediated transformation (Sparkes et al. 2006). After 3 d, the total proteins or nuclear proteins were extracted and incubated with GFP nanoantibody–coupled magnetic beads (Nucleotech Scientific, China) at 4 °C for 30 min. After incubation, the antigen–antibody complex was collected using a magnetic separator (Thermo Fisher Scientific, USA; 12321D) and washed 3 times with wash buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, pH 8.0, and 0.5% IGEPAL CA-630), then boiled with 5× SDS sample buffer for immunoblotting. The immunoprecipitates were separated by 12% SDS–PAGE and detected with antiacetylated lysine mouse antibody (1:1,000 dilution; PTM Biolabs, China) which recognized any proteins acetylated on lysine residues. Immunoblotting with an anti-HA tag mouse monoclonal antibody or and anti-GFP rabbit polyclonal antibody was used as the loading control.
Statistical analysis
All experimental data were analyzed by SPSS version 22. Multiple samples were analyzed using 1-way ANOVA followed by Tukey's difference test. The summary of statistical analyses is listed in Supplemental Data Set 4.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: PbrPPA5 (MN610578), PbrGNAT1 (XP_009352002.1), PbrPME44 (XP_009352341.1), PbrbZIP77 (XP_018503142.1), PbrTUB-2 (XP_009374320.1), and NTP303 (X69440.1). The data that support the findings of this study are available from the corresponding authors upon reasonable request. The sequences of the genes and proteins mentioned in this article are available for download at the Pear Genome Project (https://doi.org/10.5524/100083). The raw transcriptome data reported in this paper have been deposited in the Genome Sequence Archive (Chen et al. 2021) at the National Genomics Data Center (CNCB-NGDC Members and Partners 2023), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (PRJCA007552), and are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
Supplementary Material
Acknowledgments
We would like to thank the Bioinformatics Center of Nanjing Agricultural University for support with bioinformatic analysis. We would also like to thank Plant Editors for the English language editing.
Contributor Information
Chao Tang, State Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, Sanya Institute of Nanjing Agricultural University, Nanjing Agricultural University, Nanjing 210095, China; Jiangsu Engineering Research Center for Pear, Nanjing Agricultural University, Nanjing 210014, China.
Peng Wang, State Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, Sanya Institute of Nanjing Agricultural University, Nanjing Agricultural University, Nanjing 210095, China; Jiangsu Engineering Research Center for Pear, Nanjing Agricultural University, Nanjing 210014, China.
Xiaoxuan Zhu, State Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, Sanya Institute of Nanjing Agricultural University, Nanjing Agricultural University, Nanjing 210095, China; Jiangsu Engineering Research Center for Pear, Nanjing Agricultural University, Nanjing 210014, China.
Kaijie Qi, State Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, Sanya Institute of Nanjing Agricultural University, Nanjing Agricultural University, Nanjing 210095, China; Jiangsu Engineering Research Center for Pear, Nanjing Agricultural University, Nanjing 210014, China.
Zhihua Xie, State Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, Sanya Institute of Nanjing Agricultural University, Nanjing Agricultural University, Nanjing 210095, China; Jiangsu Engineering Research Center for Pear, Nanjing Agricultural University, Nanjing 210014, China.
Hao Zhang, State Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, Sanya Institute of Nanjing Agricultural University, Nanjing Agricultural University, Nanjing 210095, China; Jiangsu Engineering Research Center for Pear, Nanjing Agricultural University, Nanjing 210014, China.
Xiaoqiang Li, State Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, Sanya Institute of Nanjing Agricultural University, Nanjing Agricultural University, Nanjing 210095, China; Jiangsu Engineering Research Center for Pear, Nanjing Agricultural University, Nanjing 210014, China.
Hongru Gao, State Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, Sanya Institute of Nanjing Agricultural University, Nanjing Agricultural University, Nanjing 210095, China; Jiangsu Engineering Research Center for Pear, Nanjing Agricultural University, Nanjing 210014, China.
Tingting Gu, State Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, Sanya Institute of Nanjing Agricultural University, Nanjing Agricultural University, Nanjing 210095, China.
Chao Gu, State Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, Sanya Institute of Nanjing Agricultural University, Nanjing Agricultural University, Nanjing 210095, China; Jiangsu Engineering Research Center for Pear, Nanjing Agricultural University, Nanjing 210014, China.
Shan Li, State Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, Sanya Institute of Nanjing Agricultural University, Nanjing Agricultural University, Nanjing 210095, China.
Barend H J de Graaf, School of Biosciences, Cardiff University, Cardiff CF10 3AX, UK.
Shaoling Zhang, State Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, Sanya Institute of Nanjing Agricultural University, Nanjing Agricultural University, Nanjing 210095, China; Jiangsu Engineering Research Center for Pear, Nanjing Agricultural University, Nanjing 210014, China.
Juyou Wu, State Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, Sanya Institute of Nanjing Agricultural University, Nanjing Agricultural University, Nanjing 210095, China; Jiangsu Engineering Research Center for Pear, Nanjing Agricultural University, Nanjing 210014, China; Jiangsu Key Laboratory for Horticultural Crop Genetic Improvement, Jiangsu Academy of Agricultural Sciences, Nanjing 210014, China.
Author contributions
J.W., S.Z., C.T., and P.W. conceived and designed the experiments. C.T., P.W., X.Z., H.Z., X.L., and H.G. performed the experiments. J.W., C.T., and P.W. wrote the manuscript. B.H.J.d.G., T.G., S.L., K.Q., Z.X., C.G., and S.Z. critically read and commented on the manuscript. All authors have read and approved of the manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . The proportion of swollen pollen tubes increases under IS treatment.
Supplemental Figure S2 . rPbrS-RNase treatment inhibits pollen tube growth.
Supplemental Figure S3 . Total pectin content during the IS treatment.
Supplemental Figure S4 . rPbrS-RNases induce an increase in methyl-esterified pectin levels in pear pollen tubes with both normal and swollen tips.
Supplemental Figure S5 . IS treatment results in reduced PME total activity in pollen tubes.
Supplemental Figure S6 . Expression levels of PbrPME genes during the SI response, as determined by transcriptome analysis.
Supplemental Figure S7 . Downregulation of PbrPME44 results in reduced total PME activity in pollen tubes.
Supplemental Figure S8 . Downregulation of PbrPME44 induces swelling at the tips of pollen tubes.
Supplemental Figure S9 . rPbrS-RNaseH116R treatment decreases pollen tube growth.
Supplemental Figure S10 . Purification of His-tagged rPbrPME44.
Supplemental Figure S11 . SI induces an increase in methyl-esterified pectin levels in the pollen tube cell wall.
Supplemental Figure S12 . PbrPME44 fails to alleviate the inhibited growth of incompatible pollen tubes 3.0 h after SI challenge.
Supplemental Figure S13 . Sequence and expression analysis of PbrbZIP77.
Supplemental Figure S14 . Analysis of the expression level of PbrbZIP77 under as-ODN treatment.
Supplemental Figure S15 . The expression of PbrbZIP77 does not change under SI challenge.
Supplemental Figure S16 . PbrbZIP77 does not interact with other pollen-expressed PPA proteins.
Supplemental Figure S17 . Identification of PbrPPA5.
Supplemental Figure S18 . The Mg2+–pyrophosphatase binding site of PbrPPA5 is not required for its interaction with PbrbZIP77 or its cosuppression of PbrPME44 expression.
Supplemental Figure S19 . PbrPPA5 regulates pectin methyl esterification in pollen tubes.
Supplemental Figure S20 . PbrS-RNase does not interact with PbrPPA5.
Supplemental Figure S21 . Analysis of sPPase activity during SI.
Supplemental Figure S22 . Identification of lysine acetylation modifications in PbrPPA5.
Supplemental Figure S23 . K42Q and K42R mutation of the K42 residue in PbrPPA5 promotes its accumulation in the nucleus.
Supplemental Figure S24 . K42Q and K42R mutation of the K42 residue does not affect the pyrophosphatase activity of rPbrPPA5.
Supplemental Figure S25 . Mutations at residue K42 of PbrPPA5 do not alter its interaction with PbrbZIP77.
Supplemental Figure S26 . Mutations at residue K42 of PbrPPA5 do not alter its cosuppressor activity on PbrPME44 expression.
Supplemental Figure S27 . Mutations at residues of K9 or K98 have no effect on nuclear accumulation of PbrPPA5.
Supplemental Figure S28 . SI induces PbrPPA5 accumulation in the nucleus.
Supplemental Table S1 . The 18 differentially expressed PbrPME genes in the transcriptome data for CP and SP.
Supplemental Table S2 . Relative expression levels of all 47 pollen-expressed PbrPME genes under mock, CS, and IS treatments.
Supplemental Table S3 . Positive clones from the Y1H screen of the promoter sequence of PbrPME44.
Supplemental Table S4 . Positive clones from the Y2H screen of PbrbZIP77.
Supplemental Data Set 1 . cis-element analysis of the PbrPME44 promoter.
Supplemental Data Set 2 . The identified peptide sequences of PbrPPA5 in the SI response.
Supplemental Data Set 3 . Primers used in this study.
Supplemental Data Set 4 . Summary of statistical analyses.
Funding
This work was supported by National Key Research and Development Program of China (2022YFD1200503, 2022YFF1003100-02, and 2020YFE0202900), National Natural Science Foundation of China (32172543, 32102358, and 31830081), Jiangsu Agricultural Science and Technology Innovation Fund (CX(22)3044), Fundamental Research Funds for the Central Universities (JCQY202103), Priority Academic Program Development of Jiangsu Higher Education Institutions, Natural Science Foundation of Jiangsu Province (BK20210394), Hainan Yazhou Bay Seed Lab Project (B22E11002), and Earmarked Fund for China Agriculture Research System (CARS-28).
References
- Anderson MA, Cornish EC, Mau S-L, Williams EG, Hoggart R, Atkinson A, Bonig I, Grego B, Simpson R, Roche PJ, et al. Cloning of cDNA for a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata. Nature 1986:321(6065):38–44. 10.1038/321038a0 [DOI] [Google Scholar]
- Baeza J, Smallegan MJ, Denu JM. Mechanisms and dynamics of protein acetylation in mitochondria. Trends Biochem Sci. 2016:41(3):231–244. 10.1016/j.tibs.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Cheung AY, Hepler PK. Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol. 2005:138(3):1334–1346. 10.1104/pp.105.059865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Franklin-Tong VE. Temporal and spatial activation of caspase-like enzymes induced by self-incompatibility in Papaver pollen. Proc Natl Acad Sci U S A. 2007:104(46):18327–18332. 10.1073/pnas.0705826104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Hepler PK. Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell 2005:17(12):3219–3226. 10.1105/tpc.105.037473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugière N, Rothstein SJ, Cui Y. Molecular mechanisms of self-recognition in Brassica self-incompatibility. Trends Plant Sci. 2000:5(10):432–438. 10.1016/S1360-1385(00)01759-3 [DOI] [PubMed] [Google Scholar]
- Chebli Y, Kaneda M, Zerzour R, Geitmann A. The cell wall of the Arabidopsis pollen tube–spatial distribution, recycling, and network formation of polysaccharides. Plant Physiol. 2012:160(4):1940–1955. 10.1104/pp.112.199729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020:13(8):1194–1202. 10.1016/j.molp.2020.06.009 [DOI] [PubMed] [Google Scholar]
- Chen J, Wang P, de Graaf BHJ, Zhang H, Jiao H, Tang C, Zhang S, Wu J. Phosphatidic acid counteracts S-RNase signaling in pollen by stabilizing the actin cytoskeleton. Plant Cell 2018:30(5):1023–1039. 10.1105/tpc.18.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Chen X, Zhang S, Zhu J, Tang B, Wang A, Dong L, Zhang Z, Yu C, Sun Y, et al. The genome sequence archive family: toward explosive data growth and diverse data types. Genom Proteom Bioinf. 2021:19(4):578–583. 10.1016/j.gpb.2021.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY. Self-incompatibility in Papaver rhoeas: a role for ATP. New Phytol. 2022:236(5):1625–1628. 10.1111/nph.18505 [DOI] [PubMed] [Google Scholar]
- CNCB-NGDC Members and Partners . Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2023. Nucleic Acids Res. 2023:51(D1):D18–D28. 10.1093/nar/gkac1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf BH, Rudd JJ, Wheeler MJ, Perry RM, Bell EM, Osman K, Franklin FC, Franklin-Tong VE. Self-incompatibility in Papaver targets soluble inorganic pyrophosphatases in pollen. Nature 2006:444(7118):490–493. 10.1038/nature05311 [DOI] [PubMed] [Google Scholar]
- de Nettancourt D. Incompatibility and incongruity in wild and cultivated plants. Berlin, Heidelberg: Springer; 2001. [Google Scholar]
- Downie B, Dirk LM, Hadfield KA, Wilkins TA, Bennett AB, Bradford KJ. A gel diffusion assay for quantification of pectin methylesterase activity. Anal Biochem. 1998:264(2):149–157. 10.1006/abio.1998.2847 [DOI] [PubMed] [Google Scholar]
- Drazic A, Myklebust LM, Ree R, Arnesen T. The world of protein acetylation. Biochim Biophys Acta. 2016:1864(10):1372–1401. 10.1016/j.bbapap.2016.06.007 [DOI] [PubMed] [Google Scholar]
- Duan Q, Liu MJ, Kita D, Jordan SS, Yeh FJ, Yvon R, Carpenter H, Federico AN, Garcia-Valencia LE, Eyles SJ, et al. FERONIA controls pectin- and nitric oxide-mediated male–female interaction. Nature 2020:579(7800):561–566. 10.1038/s41586-020-2106-2 [DOI] [PubMed] [Google Scholar]
- Eaves DJ, Haque T, Tudor RL, Barron Y, Zampronio CG, Cotton NP, De Graaf BH, White SA, Cooper HJ, Franklin FC, et al. Identification of phosphorylation sites altering pollen soluble inorganic pyrophosphatase activity. Plant Physiol. 2017:173(3):1606–1616. 10.1104/pp.16.01450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin-Tong NV, Franklin FCH. Gametophytic self-incompatibility inhibits pollen tube growth using different mechanisms. Trends Plant Sci. 2003:8(12):598–605. 10.1016/j.tplants.2003.10.008 [DOI] [PubMed] [Google Scholar]
- Fujii S, Kubo K, Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat Plants. 2016:2(9):16130. 10.1038/nplants.2016.130 [DOI] [PubMed] [Google Scholar]
- Gao YB, Wang CL, Wu JY, Wu J, Zhou HS, Zhang SL. A method to isolate male gametic nuclei from pear pollen tubes. J. Hortic. Sci. Biotech. 2015:88(3):313–319. 10.1080/14620316.2013.11512971 [DOI] [Google Scholar]
- Gdula DA, Sandaltzopoulos R, Tsukiyama T, Ossipow V, Wu C. Inorganic pyrophosphatase is a component of the Drosophila nucleosome remodeling factor complex. Genes Dev. 1998:12(20):3206–3216. 10.1101/gad.12.20.3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geitmann A, Steer M. The architecture and properties of the pollen tube cell wall. In: Malhó R, editors. The pollen tube: a cellular and molecular perspective. Berlin, Heidelberg: Springer Berlin Heidelberg; 2006. p. 177–200. [Google Scholar]
- Gibalová A, Reňák D, Matczuk K, Dupl’Áková N, Cháb D, Twell D, Honys D. AtbZIP34 is required for Arabidopsis pollen wall patterning and the control of several metabolic pathways in developing pollen. Plant Mol Biol. 2009:70(5):581–601. 10.1007/s11103-009-9493-y [DOI] [PubMed] [Google Scholar]
- Goldraij A, Kondo K, Lee CB, Hancock CN, Sivaguru M, Vazquez-Santana S, Kim S, Phillips TE, Cruz-garcia F, McClure BA. Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature 2006:439(7078):805–810. 10.1038/nature04491 [DOI] [PubMed] [Google Scholar]
- Gray JE, McClure BA, Bonig I, Anderson MA, Clarke AE. Action of the style product of the self-incompatibility gene of Nicotiana alata (S-RNase) on in vitro-grown pollen tubes. Plant Cell 1991:3(3):271–283. 10.2307/3869367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzechowiak M, Ruszkowski M, Sliwiak J, Szpotkowski K, Sikorski M, Jaskolski M. Crystal structures of plant inorganic pyrophosphatase, an enzyme with a moonlighting autoproteolytic activity. Biochem J. 2019:476(16):2297–2319. 10.1042/BCJ20190427 [DOI] [PubMed] [Google Scholar]
- Haas KT, Wightman R, Meyerowitz EM, Peaucelle A. Pectin homogalacturonan nanofilament expansion drives morphogenesis in plant epidermal cells. Science 2020:367(6481):1003–1007. 10.1126/science.aaz5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005:1(1):13. 10.1186/1746-4811-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka S, Tezuka T. Changes in proteins in pistils after self- and cross-pollination in Japanese pear. J Japan Soc Hort Sci. 1980:49(1):57–64. 10.2503/jjshs.49.57 [DOI] [Google Scholar]
- Jia HJ, He FJ, Xiong CZ, Zhu FR, Okamoto G. Influences of cross pollination on pollen tube growth and fruit set in Zuili plums (Prunus salicina). J Integr Plant Biol. 2008:50(2):203–209. 10.1111/j.1744-7909.2007.00382.x [DOI] [PubMed] [Google Scholar]
- Jiang L, Yang SL, Xie LF, Puah CS, Zhang XQ, Yang WC, Sundaresan V, Ye D. VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 2005:17(2):584–596. 10.1105/tpc.104.027631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao TH, McCubbin AG. How flowering plants discriminate between self and non-self pollen to prevent inbreeding. Proc Natl Acad Sci U S A. 1996:93(22):12059–12065. 10.1073/pnas.93.22.12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006:23(4):607–618. 10.1016/j.molcel.2006.06.026 [DOI] [PubMed] [Google Scholar]
- Kim YJ, Jeong HY, Kang SY, Silva J, Kim EJ, Park SK, Jung KH, Lee C. Physiological importance of pectin modifying genes during rice pollen development. Int J Mol Sci. 2020:21(14):4840. 10.3390/ijms21144840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Meng D, Gu Z, Yang Q, Yuan H, Li Y, Chen Q, Yu J, Liu C, Li T. Apple S-RNase triggers inhibition of tRNA aminoacylation by interacting with a soluble inorganic pyrophosphatase in growing self-pollen tubes in vitro. New Phytol. 2018:218(2):579–593. 10.1111/nph.15028 [DOI] [PubMed] [Google Scholar]
- Liang M, Cao Z, Zhu A, Liu Y, Tao M, Yang H, Xu Q Jr, Wang S, Liu J, Li Y, et al. Evolution of self-compatibility by a mutant Sm-RNase in citrus. Nat Plants. 2020:6(2):131–142. 10.1038/s41477-020-0597-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Xie F, Triviño M, Zhao T, Coppens F, Sterck L, Bosch M, Franklin-Tong VE, Nowack MK. Self-incompatibility requires GPI anchor remodeling by the poppy PGAP1 ortholog HLD1. Curr Biol. 2022:32(9):1909–1923. 10.1016/j.cub.2022.02.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZQ, Xu GH, Zhang SL. Pyrus pyrifolia stylar S-RNase induces alterations in the actin cytoskeleton in self-pollen and tubes in vitro. Protoplasma 2007:232(1–2):61–67. 10.1007/s00709-007-0269-4 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001:25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014:15(12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Hokin SA, Kermicle JL, Hartwig T, Evans MMS. A pistil-expressed pectin methylesterase confers cross-incompatibility between strains of Zea mays. Nat Commun. 2019:10(1):2304. 10.1038/s41467-019-10259-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu DT, Qin X, Morse D, Cappadocia M. S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature 2000:407(6804):649–651. 10.1038/35036623 [DOI] [PubMed] [Google Scholar]
- Ma M, Chen Q, Dong H, Zhang S, Huang X. Genome-wide identification and expression analysis of the bZIP transcription factors, and functional analysis in response to drought and cold stresses in pear (Pyrus breschneideri). BMC Plant Biol. 2021:21(1):583. 10.1186/s12870-021-03356-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Gray JE, Anderson MA, Clarke AE. Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature 1990:347(6295):757–760. 10.1038/347757a0 [DOI] [Google Scholar]
- McClure BA, Haring V, Ebert PR, Anderson MA, Simpson RJ, Sakiyama F, Clarke AE. Style self-incompatibility gene products of Nicotlana alata are ribonucleases. Nature 1989:342(6252):955–957. 10.1038/342955a0 [DOI] [PubMed] [Google Scholar]
- Motomura K, Takeuchi H, Notaguchi M, Tsuchi H, Takeda A, Kinoshita T, Higashiyama T, Maruyama D. Persistent directional growth capability in Arabidopsis thaliana pollen tubes after nuclear elimination from the apex. Nat Commun. 2021:12(1):2331. 10.1038/s41467-021-22661-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho A, Camacho L, Haley A, Pais MS, Trewavas A, Malhó R. Antisense perturbation of protein function in living pollen tubes. Sex Plant Reprod. 2001:14(1–2):101–104. 10.1007/s004970100086 [DOI] [Google Scholar]
- Narita T, Weinert BT, Choudhary C. Functions and mechanisms of non-histone protein acetylation. Nat Rev Mol Cell Biol. 2019:20(3):156–174. 10.1038/s41580-018-0081-3 [DOI] [PubMed] [Google Scholar]
- Newbigin E, Anderson MA, Clarke AE. Gametophytic self-incompatibility systems. Plant Cell. 1993:5(10):1315–1324. 10.2307/3869784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parre E, Geitmann A. Pectin and the role of the physical properties of the cell wall in pollen tube growth of Solanum chacoense. Planta 2005:220(4):582–592. 10.1007/s00425-004-1368-5 [DOI] [PubMed] [Google Scholar]
- Parrotta L, Faleri C, Guerriero G, Cai G. Cold stress affects cell wall deposition and growth pattern in tobacco pollen tubes. Plant Sci. 2019:283:329–342. 10.1016/j.plantsci.2019.03.010 [DOI] [PubMed] [Google Scholar]
- Qian M, Xu L, Tang C, Zhang H, Gao H, Cao P, Yin H, Wu L, Wu J, Gu C, et al. PbrPOE21 inhibits pear pollen tube growth in vitro by altering apical reactive oxygen species content. Planta 2020:252(3):43. 10.1007/s00425-020-03446-7 [DOI] [PubMed] [Google Scholar]
- Rêgo MM, Rêgo ER, Bruckner CH, Da Silva EAM, Finger FL, Pereira KJC. Pollen tube behavior in yellow passion fruit following compatible and incompatible crosses. Theor Appl Genet. 2000:101(5–6):685–689. 10.1007/s001220051531 [DOI] [Google Scholar]
- Sassa H, Hirano H, Ikehashi H. Self-incompatibility-related RNases in styles of Japanese pear (Pyrus serotina Rehd. Plant Cell Physiol. 1992:33(6):811–814. 10.1093/oxfordjournals.pcp.a078322 [DOI] [Google Scholar]
- Segami S, Tomoyama T, Sakamoto S, Gunji S, Fukuda M, Kinoshita S, Mitsuda N, Ferjani A, Maeshima M. Vacuolar H+-pyrophosphatase and cytosolic soluble pyrophosphatases cooperatively regulate pyrophosphate levels in Arabidopsis thaliana. Plant Cell 2018:30(5):1040–1061. 10.1105/tpc.17.00911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva NF, Goring DR. Mechanisms of self-incompatibility in flowering plants. Cell Mol Life Sci. 2001:58(14):1988–2007. 10.1007/PL00000832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Sanzana C, Celiz-Balboa J, Garzo E, Marcus SE, Parra-Rojas JP, Rojas B, Olmedo P, Rubilar MA, Rios I, Chorbadjian RA, et al. Pectin methylesterases modulate plant gomogalacturonan status in defenses against the aphid Myzus persicae. Plant Cell 2019:31(8):1913–1929. 10.1105/tpc.19.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc. 2006:1(4):2019–2025. 10.1038/nprot.2006.286 [DOI] [PubMed] [Google Scholar]
- Sun L, Cao S, Zheng N, Kao TH. Analyses of Cullin1 homologs reveal functional redundancy in S-RNase-based self-incompatibility and evolutionary relationships in eudicots. Plant Cell 2022:35(2):673–699. 10.1093/plcell/koac357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Qiao X, Zhu X, Khan W, Wu J, Zhang S. Expression and evolutionary analysis of soluble inorganic pyrophosphatase gene family in pear and four other Rosaceae species. Plant Syst Evol. 2020a:306(2):46. 10.1007/s00606-020-01628-0 [DOI] [Google Scholar]
- Tang C, Zhu X, Qiao X, Gao H, Li Q, Wang P, Wu J, Zhang S. Characterization of the pectin methyl-esterase gene family and its function in controlling pollen tube growth in pear (Pyrus bretschneideri). Genomics 2020b:112(3):2467–2477. 10.1016/j.ygeno.2020.01.021 [DOI] [PubMed] [Google Scholar]
- Thomas SG, Franklin-Tong VE. Self-incompatibility triggers programmed cell death in Papaver pollen. Nature 2004:429(6989):305–309. 10.1038/nature02540 [DOI] [PubMed] [Google Scholar]
- Tian GW, Chen MH, Zaltsman A, Citovsky V. Pollen-specific pectin methylesterase involved in pollen tube growth. Dev Biol. 2006:294(1):83–91. 10.1016/j.ydbio.2006.02.026 [DOI] [PubMed] [Google Scholar]
- Visser K, Heimovaara-Dijkstra S, Kijne JW, Wang M. Molecular cloning and characterization of an inorganic pyrophosphatase from barley. Plant Mol Biol. 1998:37(1):131–140. 10.1023/A:1005931003483 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004:40(3):428–438. 10.1111/j.1365-313X.2004.02219.x [DOI] [PubMed] [Google Scholar]
- Wang CL, Xu GH, Jiang XT, Gong C, Wu J, Wu HQ, Zhang SL. S-RNase triggers mitochondrial alteration and DNA degradation in the incompatible pollen tube of Pyrus pyrifolia in vitro. Plant J. 2009:57(2):220–229. 10.1111/j.1365-313X.2008.03681.x [DOI] [PubMed] [Google Scholar]
- Wang CL, Wu J, Xu GH, Gao YB, Chen G, Wu JY, Wu HQ, Zhang SL. S-RNase disrupts tip-localized reactive oxygen species and induces nuclear DNA degradation in incompatible pollen tubes of Pyrus pyrifolia. J Cell Sci. 2011:123(24):4301–4309. 10.1242/jcs.075077 [DOI] [PubMed] [Google Scholar]
- Wang D, Samsulrizal NH, Yan C, Allcock NS, Craigon J, Blanco-Ulate B, Ortega-Salazar I, Marcus SE, Bagheri HM, Perez Fons L, et al. Characterization of CRISPR mutants targeting genes modulating pectin degradation in ripening tomato. Plant Physiol. 2019a:179(2):544–557. 10.1104/pp.18.01187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Holloway MP, Ma L, Cooper ZA, Riolo M, Samkari A, Elenitoba-Johnson KSJ, Chin YE, Altura RA. Acetylation directs survivin nuclear localization to repress STAT3 oncogenic activity. J Biol Chem. 2010:285(46):36129–36137. 10.1074/jbc.M110.152777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lin Z, Carli J, Gladala-Kostarz A, Davies JM, Franklin-Tong VE, Bosch M. Depletion plays a pivotal role in self-incompatibility, revealing a link between cellular energy status, cytosolic acidification and actin remodelling in pollen tubes. New Phytol. 2022:236(5):1691–1707. 10.1111/nph.18350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lin Z, Triviño M, Nowack MK, Franklin-Tong VE, Bosch M. Self-incompatibility in Papaver pollen: programmed cell death in an acidic environment. J Exp Bot. 2019b:70(7):2113–2123. 10.1093/jxb/ery406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang K, Liu X, Liu M, Cao N, Duan Y, Yin G, Gao H, Wang W, Ge W, et al. Pollen-expressed leucine-rich repeat extensins are essential for pollen germination and growth. Plant Physiol. 2018:176(3):1993–2006. 10.1104/pp.17.01241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler MJ, de Graaf BH, Hadjiosif N, Perry RM, Poulter NS, Osman K, Vatovec S, Harper A, Franklin FC, Franklin-tong VE. Identification of the pollen self-incompatibility determinant in Papaver rhoeas. Nature 2009:459(7249):992–995. 10.1038/nature08027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WG, Marcus SE, Knox JP. Generation of a monoclonal antibody specific to (1→5)-α-L-arabinan. Carbohydr Res. 1998:308(1–2):149–152. 10.1016/S0008-6215(98)00070-6 [DOI] [PubMed] [Google Scholar]
- Willats WG, Orfila C, Limberg G, Buchholt HC, van Alebeek GJ, Voragen AG, Marcus SE, Christensen TM, Mikkelsen JD, Murray BS, et al. Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls. Implications for pectin methyl esterase action, matrix properties, and cell adhesion. J Biol Chem. 2001:276(22):19404–19413. 10.1074/jbc.M011242200 [DOI] [PubMed] [Google Scholar]
- Xie Q, Wang P, Liu X, Yuan L, Wang L, Zhang C, Li Y, Xing H, Zhi L, Yue Z, et al. LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 2014:26(7):2843–2857. 10.1105/tpc.114.126573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Meng D, Gu Z, Li W, Chen Q, Li Y, Yuan H, Yu J, Liu C, Li T. Apple S-RNase interacts with an actin-binding protein, MdMVG, to reduce pollen tube growth by inhibiting its actin-severing activity at the early stage of self-pollination induction. Plant J. 2018:95(1):41–56. 10.1111/tpj.13929 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhang B, Chen Z, Zhang D, Zhang H, Wang H, Zhang Y, Cai D, Liu J, Xiao S, et al. A PECTIN METHYLESTERASE gene at the maize Ga1 locus confers male function in unilateral cross-incompatibility. Nat Commun. 2018:9(1):3678. 10.1038/s41467-018-06139-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Song Y, Li J, Zhang Y, Huang H, Li Q, Zhang Y, Xue Y. Primary restriction of S-RNase cytotoxicity by a stepwise ubiquitination and degradation pathway in Petunia hybrida. New Phytol. 2021:231(3):1249–1264. 10.1111/nph.17438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhang Y, Zhang H, Song Y, Zhao F, Zhang Y, Zhu S, Zhang H, Zhou Z, Guo H, et al. Origin, loss and regain of self-incompatibility in angiosperms. Plant Cell 2022:34(1):579–596. 10.1093/plcell/koab266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Lu J, Zhang Y, Guo J, Lin W, Van Norman JM, Qin Y, Zhu X, Yang Z. Membrane receptor-mediated mechano-transduction maintains cell integrity during pollen tube growth within the pistil. Dev Cell. 2021:56(7):1030–1042.e6. 10.1016/j.devcel.2021.02.030 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Bi G, Zhou J. Luciferase complementation assay for protein-protein interactions in plants. Curr Protoc Plant Biol. 2018:3(1):42–50. 10.1002/cppb.20066 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Zou X, Dean AE, Brien JO, Gao Y, Tran EL, Park SH, Liu G, Kieffer MB, Jiang H, et al. Lysine 68 acetylation directs MnSOD as a tetrameric detoxification complex versus a monomeric tumor promoter. Nat Commun. 2019:10(1):2399. 10.1038/s41467-019-10352-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.