Abstract
The selective turnover of macromolecules by autophagy provides a critical homeostatic mechanism for recycling cellular constituents and for removing superfluous and damaged organelles, membranes, and proteins. To better understand how autophagy impacts seed maturation and nutrient storage, we studied maize (Zea mays) endosperm in its early and middle developmental stages via an integrated multiomic approach using mutants impacting the core macroautophagy factor AUTOPHAGY (ATG)-12 required for autophagosome assembly. Surprisingly, the mutant endosperm in these developmental windows accumulated normal amounts of starch and Zein storage proteins. However, the tissue acquired a substantially altered metabolome, especially for compounds related to oxidative stress and sulfur metabolism, including increases in cystine, dehydroascorbate, cys-glutathione disulfide, glucarate, and galactarate, and decreases in peroxide and the antioxidant glutathione. While changes in the associated transcriptome were mild, the proteome was strongly altered in the atg12 endosperm, especially for increased levels of mitochondrial proteins without a concomitant increase in mRNA abundances. Although fewer mitochondria were seen cytologically, a heightened number appeared dysfunctional based on the accumulation of dilated cristae, consistent with attenuated mitophagy. Collectively, our results confirm that macroautophagy plays a minor role in the accumulation of starch and storage proteins during maize endosperm development but likely helps protect against oxidative stress and clears unneeded/dysfunctional mitochondria during tissue maturation.
Endosperm, the main storage tissue in maize seeds for both protein and starch, engages autophagy to eliminate dysfunctional mitochondria and dampen oxidative stress during its maturation.
Introduction
Plants employ various catabolic routes for recycling cellular constituents to promote survival and new growth and to remove unwanted/aberrant proteins, membranes, and even entire organelles that could elicit proteotoxic stress if allowed to accumulate. One major recycling route involves autophagy whereby intracellular material is sequestered into specialized vesicles for degradation within vacuoles (Yang and Bassham 2015; Marshall and Vierstra 2018). Numerous studies with several plant species have shown that this clearance is critical to their nutrient economy through its roles in catabolizing proteins, lipids, and nucleic acids, and by helping reuse fixed nitrogen and carbon and various essential elements (Avin-Wittenberg et al. 2015; Masclaux-Daubresse et al. 2017; Marshall and Vierstra 2018; Naumann et al. 2019; Shinozaki et al. 2020). Often autophagic-deficient mutants display stunted growth, accelerated senescence, and reduced fecundity under such limitations. Autophagy mutants are also hypersensitive to pathogens, possibly through a failure to break down the foreign invaders or their constituents (Dagdas et al. 2016; Hafren et al. 2017; Ustun et al. 2018; Leong et al. 2022). Additional roles for autophagy have been seen for hormone signaling (Zhan et al. 2018; Wang et al. 2021), lipid metabolism (Fan et al. 2019; Barros et al. 2021), oxidative stress protection (Sun et al. 2018; Zhou et al. 2018; Ma et al. 2021), and various developmental processes, including anther and seed development and programmed cell death (Sera et al. 2019; Feng et al. 2022). Nonetheless, many strong autophagy mutants are phenotypically normal when grown under nutrient-rich conditions, suggesting that other degradative systems become engaged in its absence.
In plants, as in animals and fungi, two main autophagic routes exist: microautophagy and macroautophagy (Yang and Bassham 2015; Marshall and Vierstra 2018). During microautophagy, the tonoplast deforms locally to directly sequester autophagic cargo into vacuolar vesicles called autophagic bodies. By contrast, macroautophagy assembles a cup-shaped phagophore (or isolation) membrane from the endoplasmic reticulum (ER) that envelops unwanted material in a double membrane-bound autophagosome; these cytosolic vesicles then fuse with the tonoplast to release the internal compartment as an autophagic body. In both cases, autophagic bodies and their cargo are degraded by vacuolar hydrolases with the resulting products then exported back to the cytoplasm for reuse.
The mechanisms underpinning microautophagy are poorly understood at present but appear to be at least partially distinct from those that direct macroautophagy (Reyes et al. 2011; Chanoca et al. 2015; Ding et al. 2022). By contrast, numerous factors have been connected to macroautophagy, designated hereafter as autophagy (Marshall and Vierstra 2018). Key components include (i) an upstream AUTOPHAGY (ATG)-1 kinase complex that responds to nutritional deficits, (ii) an ATG2/9/18 membrane delivery system that promotes phagophore expansion, (iii) a phosphatidylinositol-3-phosphate kinase complex that marks autophagic surfaces with its lipid product, and (iv) an enzymatic cascade that decorates the autophagic membranes with an adduct of ATG8-bearing phosphatidylethanolamine (PE). ATG8-PE conjugation employs a reaction scheme analogous to ubiquitylation, which culminates with an E3 ligase complex containing an ATG12-ATG5 conjugate that links the ethanolamine moiety of PE to the C-terminal glycine of ATG8.
The embedded ATG8-PE not only helps recruit factors needed for autophagosome assembly and tonoplast fusion, but it also provides docking sites for a plethora of autophagic receptors that recruit appropriate cargo (Marshall and Vierstra 2018; Bu et al. 2020; Stephani and Dagdas 2020). A rapidly expanding plant catalog of known ATG8-binding receptors has connected autophagy to the selective clearance of numerous cellular compartments, including mitochondria, peroxisomes, chloroplasts and ER, and the breakdown of protein aggregates and dysfunctional proteasomes and ribosomes. Specific receptors include NEIGHBOR OF BRCA1 (NBR)-1 and REGULATORY PARTICLE NON-ATPase-10 (RPN10) that respectively commit protein aggregates and dysfunctional proteasomes for autophagic clearance by tethering them to lipidated ATG8 (Marshall et al. 2015; Jung et al. 2020).
To better appreciate autophagy and its substrates, we have developed an integrated omic platform that combines maize (Zea mays) atg12 mutants, which attenuate ATG8-PE conjugation and thus autophagosome assembly (Li et al. 2015), with deep metabolomic, ionomic, transcriptomic, and proteomic profiling (McLoughlin et al. 2018, 2020). Consistent with roles for autophagy in nutrient recycling, the mutants are hypersensitive to nitrogen and fixed-carbon deprivation and show reduced nitrogen-use efficiency even when well fertilized. Striking alterations in leaf metabolite, mRNA, and protein profiles were discovered, indicating that autophagy is critical to many physiological processes, ranging from organelle recycling and lipid droplet consumption to primary and secondary metabolism homeostasis (McLoughlin et al. 2018, 2020). Elevated levels of numerous proteins in atg12 backgrounds also identified substrates potentially cleared by autophagy.
Our and others’ studies indicate that autophagy is also active during maize seed development (Li et al. 2015; Ding et al 2022; Arcalís et al. 2023). As the deposition of nutrients into seeds and their subsequent reuse during germination are central aspects of plant reproduction, understanding how autophagy contributes could have important agronomic ramifications. For maize and other cereals, the main seed storage compartment is the endosperm, a specialized triploid tissue created as part of double fertilization of the female gametophyte (Sabelli and Larkins 2009). At maturity, it includes an outermost single-cell aleurone layer and a prominent starchy endosperm which occupies most of the seed and contains dense aggregates of prolamin storage proteins inside the ER (protein bodies) and large granules of starch within plastids (Woo et al. 2001; Reyes et al. 2011). While the starchy endosperm ultimately undergoes programmed cell death, the peripheral aleurone layer remains alive and secretes a cache of hydrolytic enzymes into the starchy endosperm during seed germination whose activities release amino acid and sugar building blocks for the developing embryo.
Understanding how autophagy contributes to maize endosperm development and storage protein accumulation is complicated at present. The major alcohol-soluble, prolamin storage proteins (called Zeins in maize) accumulate in protein bodies within the starchy endosperm cells, but in aleurone cells, similar bodies are delivered to vacuoles for storage via a microautophagy route independent of ATG8 (Reyes et al. 2011; Ding et al. 2022). Nonetheless, these aleurone cells also have an active ATG8-dependent macroautophagic route that promotes ER homeostasis (Zhang et al. 2020).
Here, we further examined the roles of autophagy during maize seed development through an omic analysis of atg12 endosperm harvested at early and middle developmental windows (Chen et al. 2014). We confirmed that ATG8-PE-dependent macroautophagy contributes little to storage protein and starch accumulation at these time points but substantially influences the physiology of the endosperm as it matures. Notably, the mutant endosperm displayed altered redox metabolism and hyperaccumulated mitochondrial proteins and aberrant mitochondria, revealing that autophagy protects against oxidative stress and helps clear dysfunctional/unwanted mitochondria as the endosperm matures.
Results
Maize endosperm at early and later developmental times have markedly altered omic profiles
To assess how autophagy impacts maize endosperm development, we combined two well-characterized atg12 mutants (Li et al. 2015) with a previously described non-targeted omic platform (McLoughlin et al. 2018, 2020) that quantified the metabolome, ionome, transcriptome, and proteome from endosperm collected at 6 and 18 d after pollination (DAP) (Fig. 1A). Six-DAP endosperm was chosen as it would interrogate an early developmental period when this tissue undergoes rapid syncytial divisions followed by cellularization, while 18-DAP endosperm features robust accumulation of starch and Zeins well before programmed cell death of the starchy endosperm and dehydration of the entire seed (Young and Gallie 2000; Chen et al. 2014). Here, we confirmed that the wild-type parent (W22) accumulates little starch and members of the Zein family at 6 DAP but robust storage of both at 18 DAP (Fig. 1, B to E). While separating the starchy endosperm from the embryo and aleurone layer at 18 DAP was relatively easy, it was more problematic at 6 DAP as the aleurone had not yet differentiated, and the embryo was microscopic (Fig. 1A; Chen et al. 2014). However, given their small size, embryos likely contributed little to the analyses.
Figure 1.
Atg12 mutations, while attenuating autophagy, do not impair starch and seed storage protein accumulation in the maize endosperm. A)atg12 seeds develop normally. Photographs of homozygous atg12-1 and atg12-2 cobs and individual whole and dissected seeds next to those from their corresponding wild type (WT) at 8 and 18 DAP. The images were digitally placed on a black background. Endo, endosperm; Emb, embryo. Scale bar = 1 mm. B and C)atg12 seeds accumulate major seed storage proteins normally. Total aqueous extracts B) and ethanol-soluble extracts enriched for Zeins C) from endosperm harvested at 6 and 18 DAP. Equal volumes of extracts were subjected to SDS–PAGE and stained for protein with Coomassie Blue. D) Ethanol-soluble extracts from C) subjected to immunoblot analysis with antibodies against α- and γ-Zeins. E)atg12 mutants accumulate starch normally. Starch levels were quantified spectrophotometrically after hydrolysis to glucose from 5 biological replicates (±Sd) from WT and atg12 endosperm at 6 and 18 DAP. F and G)atg12 seeds have defective autophagy as evidenced by the hyperaccumulation of ATG8 and NBR1. atg12 and WT samples from 6 and 18 DAP endosperm were either subjected to immunoblot analysis with antibodies against Arabidopsis ATG8a or NBR1 F) or quantified for the ATG8d (GRMZM2G134613) and NBR1a (GRMZM2G09447) isoforms in total cell extracts by LC-MS/MS G). Near equal loading in F) was confirmed by immunoblot analysis with anti-histone H3 antibodies. Mean values in G) were determined from the MS1 precursor ion intensities from 5 biological replicates (±Sd) each analyzed by 4 technical replicates. All values were normalized to the mean value for WT endosperm at 6 or 18 DAP. H) Levels of the ATG8d and NBR1a mRNAs in the endosperm are unaffected by the atg12 mutations. polyA+ mRNA isolated from the same endosperm as in G) were subjected to RNA-seq. Each bar represents the mean log2 FC of 3 biological replicates (±Sd). All values were normalized to the mean value for WT endosperm at 6 or 18 DAP. Log2 FC for the ATG8d and NBR1a proteins and mRNAs in G and H) used in the same floating scale dimensions to permit direct comparisons.
Comparative omic analyses of wild-type endosperm confirmed the unique maturation states at 6 and 18 DAP that emphasized cell division/enlargement versus storage. Quantitative profiles of 440 metabolites by nontargeted liquid chromatography-mass spectrometry (LC-MS), ∼33,000 transcripts by RNA-seq, and almost 3,300 proteins by LC-MS/MS revealed distinct differences related to their early and middle developmental stages (Supplemental Fig. S1, A to C). For example, metabolomic profile heat maps revealed substantial differences between the 6- and 18-DAP wild-type samples for almost 90% of the compounds and identified increases in the metabolism of fatty acids, nucleotides, and several amino acid classes at 6 DAP but found enhancements in phospholipid accumulation at 18 DAP (Supplemental Fig. S1A).
Gene ontology (GO) analyses of RNAs from 11,341 differentially expressed genes (DEGs) and 3,267 differentially accumulating proteins confirmed this view, with GO term enrichments seen at 6 DAP favoring anion/nucleotide-sugar transport, cell cycle, microtubules, and the plasma membrane, and those at 18 DAP favoring amide and starch biosynthesis, macromolecules metabolism, plastids, and translation, all likely connected to a robust accumulation of storage compounds (Supplemental Fig. S1, D and E). As expected (Chen et al. 2014; Yi et al. 2019; Dai et al. 2021; Ding et al. 2022), transcripts for representative Zein genes (19-kDa α-Zein and 50-kDa γ-Zein), which can become as much as 65% of the endosperm mRNAs, and those related to Zein expression and localization or involved in Suc and starch synthesis were all strongly upregulated in the endosperm samples at 18 versus 6 DAP (Supplemental Fig. S2C). Conversely, the expression of genes related to early endosperm differentiation, cell cycle and cell expansion, and mitosis/cytokinesis (Yi et al. 2019; Dai et al. 2021) were all emphasized at 6 DAP (Supplemental Fig. S2, A and B). Notably, mRNA levels for several autophagic components rose marginally at 18 versus 6 DAP, suggesting that this recycling route is not appreciably upregulated as the endosperm develops (Supplemental Fig. S2D).
Our omic comparisons of atg12 versus wild-type endosperm used two UniformMu mutant alleles (Li et al. 2015) and involved numerous biological replicates, each generated by pooling samples from 30 to 50 individual seeds, to help find robust connections. Previous studies showed that the atg12-1 mutation is a likely null allele, whereas the atg12-2 mutation is a weaker transcript knockdown allele (Li et al. 2015; McLoughlin et al. 2018, 2020). Here, when grown under nutrient-rich conditions, the developing seeds and the endosperms from both atg12 alleles were morphologically similar to their wild-type cohort except for a slightly reduced embryo size (Figs. 1A and S3).
Surprisingly, we saw little difference in the accumulation of starch and Zeins, which eventually comprise the bulk of the mature starchy endosperm (Sabelli and Larkins 2009), in the mutants at either 6 or 18 DAP (Figs. 1, B to E, and S4). By contrast, the atg12-1 allele at both time points had substantially elevated levels of the core autophagy factor ATG8 and autophagic receptor NBR1 as seen either by immunoblotting for the entire family with ATG8a antibodies or by LC-MS/MS quantification of the ATG8d and the NBR1a isoforms specially identified in the MS data sets (Fig. 1, F and G). These elevations were not concomitant with increases in their corresponding mRNAs (Fig. 1H). As ATG8 and NBR1 are both consumed during autophagy (Li et al. 2015; Jung et al. 2020), their increased levels in the atg12 backgrounds especially at 18 DAP confirmed that the mutations substantially dampened endosperm autophagy as seen previously in leaves (McLoughlin et al. 2018, 2020). Furthermore, while NBR1 was stabilized only by the strong atg12-1 allele, ATG8 was stabilized by both the atg12-1 and atg12-2 alleles, implying that it is a more sensitive readout for compromised autophagy.
Effects of autophagy on the endosperm metabolome and ionome
Next, we compared the omic profiles of the atg12 lines to appreciate how autophagy globally impacts maize endosperm physiology. The metabolome and ionome analyses employed 5 biological replicates, the transcriptomes were analyzed using the 3 most representative biological replicates based on the average metabolomic profiles, and the proteome analyses included the same 5 biological replicates each measured in quadruplicate. See Supplemental Data Set S1 to S10 for the raw data associated with each omic profile. Principal component analyses (PCA) showed clear separation of all 3 data sets between the atg12 mutants and wild type at both developmental stages (Fig. 2A), with the atg12-2 samples expectedly having intermediate values (Li et al. 2015). The ionome, as measured by ion-coupled plasma MS, was least affected by genotype, where less separation of the PCA values was observed (Fig. 2A). In agreement, focused quantifications saw little variations in abundance between mutant and wild type for the 20 common inorganic elements with a modest change seen only for calcium at 18 DAP (Supplemental Fig. S5).
Figure 2.
Metabolic responses of maize atg12 endosperm at 6 and 18 DAP. A) PCA of the metabolome, transcriptome, proteome, and ionome data sets for 6- and 18-DAP endosperm collected from atg12-1, atg12-2, and wild-type (WT) seeds. The values were calculated from log2-transformed ion counts for the metabolome and ionome data sets (n = 5 biological replicates), median-normalized log2-transformed transcript counts for the transcriptomic data sets (n = 3 mean biological replicates), and log2-transformed MS1 precursor ion intensities for the proteomic data sets (n = 5 biological replicates, each with 4 technical replicates). The amounts of variation explained by the first 2 components are indicated on the axes. The dashed circles outline the PC coordinates for the biological replicates from each genotype/sample. B) Heat maps comparing the abundances of 440 metabolites measured in atg12-1, atg12-2, and WT endosperm at 6 and 18 DAP. The metabolites were quantified by LC-MS from 5 biological replicates each prepared from 30 to 50 seeds. The relative abundance of each metabolite was calculated by Z-scores after normalization of each mean value generated with WT at each developmental time point. The metabolites were clustered based on chemical type and specific subcategory. C) Scatterplots showing strong correlations for affected metabolites in atg12-1 and atg12-2 endosperm at 6 and 18 DAP. The dashed line shows the correlation within each comparison. Log2 FC values (atg12/WT) for metabolites significantly impacted in both atg12 backgrounds (P ≤ 0.05; q ≤ 0.05; 80 for 6-DAP and 70 for 18-DAP samples) and for the rest of the 440 metabolites are shown in red and gray, respectively. The Pearson correlation coefficient (Corr) and fit (R2) values are indicated. D) Metabolic pathway overrepresentation and topology analysis determined by MetaboAnalyst for metabolites that differed significantly in both atg12 alleles compared to those from WT. P-values generated as in (Chong et al. 2019) reflect overrepresentation of each category, while the pathway impact weighs the importance of the affected metabolites within the pathway. Significantly impacted pathways/clusters are indicated. Yellow box highlights pathways with insignificant impact values (impact <0.15 and −log10P < 2). E) Metabolic enrichment of specific metabolic pathways showed in the heat maps. The enrichment was generated by clusterProfiler package in R and was based on the −log10P-value enrichment comparisons of the metabolite levels within each pathway between WT and atg12 endosperm.
In-depth analysis of the metabolome showed clear differences between wild type and the mutant endosperm, with the abundances of 239 and 247 metabolites (∼55% of total) significantly altered (P ≤ 0.05 and q ≤ 0.10) by the atg12 mutations at 6 and 18 DAP, respectively (Fig. 2B). The strength of the metabolite changes—either up or down—were well correlated between the atg12-1 and atg12-2 backgrounds by Pearson correlation coefficient and fit values, strongly implying that the differences were caused by a block in autophagy (Fig. 2C). When clustered into groups either based on chemical subcategories (Fig. 2B), metabolic impact and pathway enrichment as defined by Kyoto Encyclopedia of Genes and Genomes (KEGG) and MetaboAnalyst (Chong et al. 2019) (Fig. 2, D and E), or organized into networks by Cytoscape (Supplemental Figs. S6 and S7), robust differences were seen with respect to amino acid, primary carbon, phospholipid, and nucleotide metabolism. The strongest metabolite impact for the atg12 mutants, as calculated by MetaboAnalyst, was for the 2 nonproteinogenic sulfur amino acids—taurine and hypotaurine (Figs. 2D, S6, and S7—which have been connected to antioxidant metabolism and stress defense (Song et al. 2019). Additionally, robust differences were seen for various phospholipids (e.g. 1-linoleoyl-GPA [18:2] and 1-steroyl-GPA [18:2]), along with a strong drop (8.6-fold) in the auxin amide conjugate indoleacetylaspartate at 6 DAP, with the latter suggesting an early compromise in auxin homeostasis (Figs. 3B and S6).
Figure 3.
Specific metabolic clusters that are substantially impacted by ATG12 in maize endosperm. A) Cytoscape clustering of metabolites related to glutathione/ascorbate, serine, and glutamate metabolism that showed significant FCs for atg12-1 versus wild-type (WT) endosperm at 18 DAP. The sizes of the circles reflect FC in abundance between the WT and atg12-1; red indicates FC significantly higher in atg12-1, blue indicates FC significantly higher in the WT, and gray indicates no significant difference. FC values ≥5 are indicated. Cytoscape clusterings of all 440 metabolites in 6 and 18 DAP samples are displayed in Supplemental Figs. S6 and S7. NS, not significant. B) Specific metabolites that differentially accumulate in atg12-1 endosperm at 6 and/or 18 DAP. Each bar represents the mean of 5 biological replicates (±Sd). Glucarate and galactarate are the oxidized products of glucose and galactose, respectively, S-methylcysteine is a methylated derivative of cysteine, hydroxyproline is the hydroxylated version of proline, 1-linoleoyl-GPA (18:2) and 1-steroyl-GPA (18:2) are intermediates of phospholipid degradation, N-acetylglutamate is a condensate of glutamate and acetyl-CoA, and indoleacetylaspartate is connected to auxin metabolism.
The metabolite differences from wild type were more evident in the 18-DAP samples, suggesting that the physiological consequences of the autophagy mutants become more acute as the endosperm matures (Supplemental Fig. S7). At this developmental age, robust changes were seen for compounds related to glutathione and ascorbate metabolism, such as an 8.1-fold increase in cystine combined with 22-fold less glutathione and 3.1-fold less ascorbate and 6.5- and 8.1-fold more glucarate (saccharate) and galactarate (mucic acid), which are generated by oxidation of glucose and galactose, respectively (Fig. 3, A and B). Other compounds that hyperaccumulated in the mutants included cys-glutathione disulfide (4.9-fold), numerous γ-glutamyl amino acids (1.2- to 1.7-fold), S-methylcysteine (4.1-fold), N-acetylglutamate (4.2-fold), and hydroxyproline (3.8-fold). Most of the strongly affected compounds have been connected to thiol metabolism and oxidative stress, implying that they protect the endosperm against reactive oxygen species (ROS) that accumulate without autophagy.
Atg12 mutations alter the endosperm transcriptome
Assuming that the metabolic changes in the atg12 endosperm are linked to gene expression and protein composition, we first examined the transcriptome in search for informative loci. By RNA-seq of polyA+ RNA, we identified 33,066 shared transcripts in the wild-type and the atg12-1/atg12-2 lines. Comparisons among the replicates for each genotype/developmental time point showed strong coincidences, confirming the reliability of the data sets (Supplemental Fig. S8A). When the genotypes were compared, we identified 539 DEGs based on a log2 fold change (FC) in abundance ≥0.5 (≥1.4 FC) together with a significant expression difference (false discovery rate (FDR) ≤0.05); 460 among the 6-DAP samples but only 91 among the 18-DAP samples (Fig. 4, A and B). These differences were unexpectedly diminutive (only 1.6% of total transcripts), implying that autophagy only has a mild impact on endosperm gene expression. As with the metabolome, the strength of the transcript differences strongly correlated between the atg12-1 and atg12-2 backgrounds by Pearson correlation coefficient and fit values based on log2-transformed data at 6 and 18 DAP, implying that the DEGs we did find arose due to a block in autophagy (Fig. 4C).
Figure 4.
ATG12 mildly impacts the maize endosperm transcriptome. A) Heat maps showing the log2 FC in transcript abundance in 6 and 18 DAP endosperm from the atg12-1 and atg12-2 mutants versus wild type (WT). Abundances were calculated using EBSeq and were normalized to the mean values obtained for WT. Of the 33,066 total transcripts detected, 539 were found to be DEGs with either log2 FC ≥ 0.5 up or ≤−0.5 down in both mutants. B) Venn diagram showing the overlap in the numbers of transcripts that were consistently up- or downregulated in both atg12 mutants versus WT in 6- and 18-DAP endosperm. C) Scatterplots of significantly affected transcripts identified in B) that compared the 2 atg12 mutants versus WT. The total number of significantly affected transcripts analyzed is indicated in each plot, along with Pearson correlation coefficient (Corr) and fit (R2) values. The dashed line shows the correlation within each comparison. D) Specific GO terms for DEGs that were significantly enriched or depleted in the atg12 backgrounds versus WT for 6- and 18-DAP endosperm. Log10 fold enrichment/depletion values were based on a singular enrichment of specific GO terms for transcripts consistently altered in abundance for 2 atg12 backgrounds. E) Response of representative endosperm transcripts differentially impacted by development and the atg12 mutations. Shown are the responses of transcripts encoding a putative GDH, glutamine synthetase (GLN)-2, S-adenosylmethionine synthase (SAM)-1, spermine synthetase (SPDS)-1, the DNA replication complex subunit B (RPA70B), and RNR-1. Each bar represents the mean log2 FC of 3 biological replicates (±Sd) from endosperm collected at 6 and 18 DAP from WT, atg12-1, and atg12-2 seeds.
Notably, only 12 DEGs overlapped between 6- and 18-DAP samples (Fig. 4B), in line with the distinct maturation states of the endosperm at these time points (Chen et al. 2014; Fig. 1). Included in this short list were mRNAs encoding ATG12 (direct target of the mutagenesis), the SPIKE1 guanyl-nucleotide exchange factor, a phosphoinositol 3-phosphate kinase, the mitochondrial enzyme aconitase, a vacuolar glucose transporter, and VESICLE TETHERING (VETH)-1 that regulates microtubule dynamics, all of which have been connected to autophagy, membrane trafficking, and/or metabolic flux (Supplemental Data Set 5). These distinct mRNA profiles were also evident when aligning the transcriptome heat maps by transcript (Fig. 4A), which showed that many DEGs at 6 DAP were not impacted at 18 DAP.
When analyzed for GO, we identified a number of transcript groupings differentially influenced by the atg12 mutations despite the low DEG numbers. Particularly at 6 DAP, there were increases in GO terms related to S-adenosylmethionine (SAM), gamma-(γ) aminobutyric acid (GABA), and sulfur compound metabolism but decreases in responses to abiotic stress and peroxide, and protein folding, implying specific compromises in metabolic, redox, and protein homeostasis (Fig. 4D). At 18 DAP, elevated GO terms included DNA replication and chromosomes, suggesting that the mutations delayed the syncytial divisions associated with endosperm maturation (Fig. 4D). Transcriptomic analyses by MapMan (Usadel et al. 2005) also identified other GO terms enriched in the atg12 endosperm (Supplemental Fig. S9), which might be related to the observed changes in glutamate, ammonia, and polyamine metabolites (Fig. 2, B, D, and E).
A focus on specific transcripts revealed informative changes in endosperm expression in the atg12 backgrounds. Included were an upregulation of mRNAs encoding methionine adenosyltransferase (MAT)-1 and spermine synthase (SPMS)-1, which respectively participate in SAM and polyamine biosynthesis, and a downregulation of mRNAs related to nitrogen assimilation and glutamate metabolism, such those encoding glutamine synthetase (GS)-2 and a putative glutamate dehydrogenase (GDH) that likely underlie the observed differences in the glutamate-related compounds (Fig. 4E). Intriguing changes at 18 DAP were for mRNAs encoding ribonucleotide reductase (RNR)-1 and the DNA replication complex 70 subunit B (RPA70B), which again might reflect altered DNA replication and chromosome maintenance (Fig. 4E). As noted above, modest mRNA differences were seen between wild type and the atg12 backgrounds for ATG8d and NBR1a either at 6 or 18 DAP (Fig. 1H), implying that the block in autophagy did not transcriptionally upregulate the process.
Autophagy strongly influences the endosperm proteome
To define how autophagy impacts the endosperm proteome, we analyzed total tissue extracts by shotgun LC-MS/MS followed by label-free quantification based on MS1 precursor ion intensities (McLoughlin et al. 2018, 2020). Only those proteins detected in at least 3 biological replicates were included in the final data sets; strong coincidence among the replicates confirmed their reliability (Supplemental Fig. S8B). We then used 150 proteins whose abundances varied little among the backgrounds/time points to develop a normalization factor (ranging from 0.85 to 1.27) for each data set (McLoughlin et al. 2018, 2020). This approach was validated by the statistically similar histone levels observed among samples after correction (Figs. 5A and S10). In total, ∼3,160 and 2,800 proteins were shared among the data sets for atg12-1, atg12-2, and wild-type endosperm at 6 and 18 DAP, respectively.
Figure 5.
The maize endosperm proteome is substantially altered by atg12 mutations. A) Volcano plots comparing protein abundance in 6- and 18-DAP endosperm from the atg12-1 mutant versus wild type (WT). Protein abundances were quantified by LC-MS/MS based on the MS1 precursor ion intensities, normalized by a collection of 150 stable proteins, and plotted based on their log2 FC in abundance (atg12-1/WT) and their −log10P-value in significance. The dashed yellow boxes outline the collection of proteins whose abundances did not significantly differ in atg12-1 versus WT (FC < 1.5-fold or −log10P > 0.05). The mean value for each protein was determined from 5 biological replicates, each analyzed by 4 technical replicates. The enlargements highlight a section of the volcano plots containing proteins with significant P-values that were at least 1.5-fold and 4-fold more abundant in the atg12-1 mutant at 6 and 18 DAP, respectively. Select significantly affected proteins are indicated. Proteins connected to autophagy, histones, and the collection of normalization proteins are colored in cyan, orange, and red, respectively. Comparable volcano plots for atg12-2 endosperm at 6 and 18 DAP can be found in Supplemental Fig. S10. The total number of proteins analyzed (n) and the numbers in the indicated quadrants are shown. B) Scatterplots comparing significantly affected proteins in the 2 atg12 alleles versus WT. The total number of significantly affected proteins analyzed is indicated in each plot, along with Pearson correlation coefficient (Corr) and fit (R2) values. Dashed lines show the correlation within each comparison. C and D) Specific GO terms for proteins that were significantly enriched or depleted in the atg12-1 endosperm versus WT at 6 DAP C) and 18 DAP D). Log10 fold enrichment/depletion values were based on a singular enrichment of specific GO terms for proteins consistently altered in abundance for the atg12-1 background.
In contrast to the endosperm transcriptome, a much greater influence for the atg12 mutations was observed with respect to the proteome. As seen by volcano plots assessing FC in abundance and P-value of significance, 13.4/13.6% and 29.1/22.2% of the proteins detected showed a significant abundance difference in the mutants versus wild type at 6 and 18 DAP (FC ≤ 0.67 or ≥1.5 and P-value ≤ 0.05) (Figs. 5A and S10). The strength of the protein changes strongly correlated between the atg12-1 and atg12-2 backgrounds by Pearson correlation coefficient and fit values (Fig. 5B), clearly implying that the differences were caused by attenuated autophagy. In contrast to the transcriptome, the proteome differences were greater at 18 DAP than at 6 DAP and were strongly skewed in the volcano plots toward greater abundance in the atg12 mutants (Figs. 5A and S10). For example, the number of proteins whose abundance increased in the atg12-1 endosperm went from 7.4% of total proteins to 23.7% as the endosperm matured (Fig. 5A).
Among the significantly increased proteins in atg12 backgrounds at 18 DAP were the autophagy components ATG8d and NBR1a and various organellar proteins (Figs. 1G, 5A, and S11). GO analysis identified term enrichments for the nucleolus, nuclear body, ribosomes, RNA processing, and translation-related events in atg12-1 endosperm at 6 DAP, and for mitochondria, ribonucleoprotein complexes, proteasomes, rough ER, and metabolic functions associated with these compartments/complexes at 18 DAP (Fig. 5, C and D). The enrichment for mitochondria was particularly intriguing given that they are known autophagy substrates (Li et al. 2014; Ma et al. 2021; Kacprzak and Van Aken 2022).
Elevated protein levels in the atg12 backgrounds could have arisen from increased synthesis, reduced turnover, or both, which can be discriminated in part by directly comparing transcript and protein levels (McLoughlin et al. 2018, 2020). Here, we examined these possibilities by scatter plots comparing FC in abundance for each protein with the FC in abundance for its corresponding mRNA (FC values ≤ 0.5 or ≥2). Of the 3,073 and 2,700 proteins and associated transcripts shared between the atg12-1 and wild-type data sets at 6 and 18 DAP, respectively, a large percentage showed little change in both values (75.7% at 6 DAP and 67.9% at 18 DAP), indicating that most endosperm proteins were not differentially regulated by either transcription or autophagy (central sector; Fig. 6A). Nonetheless, we identified a smaller group whose protein levels were significantly affected by either or both processes. Of particular interest were proteins with elevated levels in the atg12 endosperm that were not paralleled by increased mRNA (blue sector; Fig. 6A), which likely represent autophagy cargo/adaptors/receptors (11.7% of total proteins analyzed at 6 DAP and 21.4% at 18 DAP for atg12-1). (However, we cannot discount the possibilities that changes in translation efficiency or an increase in another recycling route also contributed ([Li et al. 2022a]). NBR1d was notably in this blue sector (Fig. 6A), in agreement with its likely consumption during cargo clearance in wild type (Mcloughlin et al. 2018; Jung et al. 2020).
Figure 6.
Comparisons between transcript and protein abundances for atg12 endosperm. A) Scatterplots showing the relation between log2 FC in protein abundance versus the log2 FC in mRNA abundance in atg12-1 versus wild-type W22 (WT) endosperm at 6 and 18 DAP. The different colored regions indicate sectors that contain proteins and/or mRNAs whose abundances were impacted ≥2-fold by the atg12-1 mutation. Yellow, proteins (but not mRNA) that were more abundant in WT; orange, mRNAs (but not proteins) that were more abundant in atg12-1; blue, proteins (but not mRNAs) that were more abundant in atg12-1; green, proteins and their mRNAs that were both more abundant in atg12-1; and gray, mRNAs (but not proteins) that were less abundant in WT. The total number of proteins/transcripts (n) analyzed and the number of proteins in each sector are shown in parentheses. Proteins connected to autophagy and histones are colored in red and orange, respectively. Specific outlier proteins/mRNAs are labeled. The dashed yellow boxes outline the collection of proteins whose protein and transcript abundances did not significantly differ in atg12-1 versus WT (<2-fold). The white cross shows the mean FC value for all protein detected. B) GO term enrichment for proteins found within the blue and yellow sectors in (A) based on a FC ≥ 2 (blue) or ≤−2 (yellow) in protein abundance but not mRNA abundance in atg12-1 versus WT at each developmental point. C) Scatterplots highlighting the protein/mRNA ratios for proteins associated with mitochondria (blue) and the ribonucleoprotein complex (green). All other proteins are in gray. Pearson correlation coefficient (Corr) and fit (R2) values are given for each selection. The dashed lines show the correlation for the highlighted proteins. The dashed red boxes outline the collection of proteins whose protein and transcript abundances did not significantly differ in atg12-1 versus WT (FC < 2). The yellow crosses show the mean FC value for all detected protein from mitochondria and the ribonucleoprotein complex. The number of proteins (but not mRNA) significantly up or down (FC ≥ 2 or ≤−2) is shown in parentheses. n, total number of selected proteins analyzed.
GO analysis of this blue sector (360 proteins at 6 DAP and 569 proteins at 18 DAP) detected strong enrichment based on P-values and FC for proteins associated with the nucleolus, RNA processing, and ribosomes at 6 DAP and for mitochondria, proteasomes, and various metabolic functions including respiration at 18 DAP (Fig. 6B). The early and later impact of the atg12 mutations on ribosomes and mitochondria, respectively, became more evident when plotting those proteins specifically. As seen in Fig. 6C, a skew of ribosomal proteins toward increased protein levels but not for mRNA was selectively seen at 6 DAP, while a skew of mitochondrial proteins toward increased protein levels but not for mRNA was evident at 18 DAP. Also of interest were proteins whose levels declined in the atg12 backgrounds but without a commensurate drop in mRNA (yellow sector: Fig. 6, A and B). These might represent substrates of other proteolytic routes that activate in the absence of autophagy.
Atg12 endosperm hyperaccumulates mitochondrial proteins and defective mitochondria
Given the possibility that mitochondria selectively undergo autophagy as the endosperm matures by a process referred to as mitophagy (Li et al. 2014; Ma et al. 2021; Nakamura et al. 2021; Kacprzak and Van Aken 2022, 2023; Li et al. 2022b), we more closely analyzed the mitochondrial proteome using GO to identify proteins uniquely assigned to this organelle. When their relative levels were analyzed by volcano plots, a strong skew in abundance toward the atg12-1 background was clearly evident at 18 DAP (90 of 205 proteins or 44% of this collection) and to a lesser extent (6%) at 6 DAP, consistent with a retention of mitochondria in the absence of autophagy. Upon subdividing this collection by functional categories, including mitochondrial membranes, TCA cycle, and the respirasome supermolecular complex, we found significant abundance increases of all 3 in the atg12-1 background at 18 DAP (49%, 66%, and 73% of each subcategory, respectively, based on FC ≥1.5 and P-value ≤ 0.05), implying that the entire mitochondrion was impacted (Fig. 7A).
Figure 7.
Maize endosperm mitochondria and their proteomes are affected by autophagy. A) Volcano plots showing the preferential accumulation of proteins assigned by GO to mitochondria (total) or to specific mitochondrial sub-compartments/complexes in atg12-1 versus wild-type (WT) endosperm at 6 and 18 DAP. Protein abundances were quantified by LC-MS/MS as in Fig. 5 and plotted based on their log2 FC in abundance (atg12-1/WT) and their −log10P-value in significance. The dashed yellow boxes outline those proteins whose abundances did not significantly differ in atg12-1 versus WT (FC < 1.5-fold or P > 0.05). Representative proteins specific for each sub-compartment/complex with significantly altered levels (up or down) are labeled. The total number of detected proteins assigned to mitochondria or each sub-compartment/complex (n) and those that significantly increased or decreased are in parentheses. B) Simplified flow chart of the mitochondrial electron transport chain and TCA cycle describing how levels of the associated metabolites and relevant proteins and their corresponding mRNAs were altered by the atg12-1 mutation in 18-DAP endosperm. FCs as compared to WT of each metabolite and associated protein and mRNA are indicated by the size of the geometric shapes (see legend). NS and ND, not significant and not detected, respectively. C) Examples of specific proteins associated with the mitochondrial respiratory Subcomplexes I, II, III, and V that differentially accumulated in atg12-1 endosperm. Log2 FCs in protein abundances were quantified by LC-MS/MS as in A). Each bar represents the mean of 5 biological replicates each analyzed by 4 technical replicates (±Sd). All values were normalized to the mean value for WT at either 6 or 18 DAP. D) The atg12-1 mutation alters the morphology of starchy endosperm mitochondria. Shown are transmission electron microscopic images from representative normal mitochondria in WT and abnormal mitochondria with dilated/abnormal cristae (red stars) in atg12-1 starchy endosperm cells harvested at 20 DAP. Scale bars = 500 nm. E) Quantification of total, normal, and dilated mitochondria and mitochondrial area in WT and atg12-1 endosperm cells analyzed as in D). Mitochondrial area per cell and numbers of normal/dilated mitochondria per µm2 were quantified from 32 cells from 2 to 3 kernels for each genotype. In each box plot, the center orange box indicates the median, the green/blue box encompasses the upper and lower quartiles, the error bar shows the maximum and minimum of the distribution, and circles indicate individual data points (n = 32). Asterisks indicate significant differences based on the two-tailed Students’ t test.
Similar analysis of several other major organelles/complexes also detected increased protein levels in the atg12-1 endosperm at 18 DAP, suggesting that they are also autophagy substrates; examples included proteins associated with plastids (24% of assigned total), ribonucleoprotein complexes (18%), peroxisomes (53%), and proteasomes (56%) (Supplemental Fig. S11). These increases starkly compared to Golgi and ER proteins, in which only a small fraction of their proteins changed in abundance in the atg12-1 background; a modest spread likely signifying noise (Supplemental Fig. S11). Of all the organelles/complexes examined at 6 DAP, only nucleolar proteins were significantly enriched in the atg12-1 background (28%), possibly reflecting a disruption in ribosome biogenesis or increased turnover (Supplemental Fig. S11).
For a deeper analysis of mitochondria, we generated a simplified diagram of its metabolic, transcriptomic, and proteomic profiles at 18 DAP, with a focus on the TCA cycle and the protein complexes associated with oxidative respiration (I to IV) and ATP synthesis (V). As seen in Fig. 7B, strong abundance increases were observed in the atg12-1 endosperm for proteins associated with the NADH oxidase (I), succinate dehydrogenase (II), cytochrome-c reductase (III), and ATP synthase (V) complexes, which were not paralleled by mRNA increases. Notable examples included the GAMMA CA-2 subunit of Complex I, succinate dehydrogenase (SDH)-2 of Complex II, MITOCHONDRIAL PROTEIN PROCESSING (MPP) subunit A of Complex III, and the ATPB subunit of Complex V (Fig. 7C). A similar but less pronounced response was also seen for TCA cycle enzymes, where increases in protein abundances for some components in the atg12-1 background (e.g. malate dehydrogenase, citrate synthase, and isocitrate dehydrogenase) were not coincident with more mRNA (Fig. 7B). Surprisingly, most of these increases in TCA cycle enzymes were not paralleled by changes in the levels of their precursors/products, suggesting a homeostatic mechanism modulating metabolite flow (Fig. 7B). Taken together, it was clear that the lack of autophagy globally disrupted mitochondrial proteostasis in the endosperm.
We anticipated that the higher levels of mitochondrial proteins seen in atg12-1 endosperm would follow an increased number of mitochondria. To test this, we measured mitochondria numbers and area in endosperm harvested at 20 DAP by transmission electron microscopy. Contrary to expectations, we actually found by cytology less mitochondria by both numbers and area in the atg12-1 endosperm (Fig. 7E). We also found an unusually large number of aberrant mitochondria with dilated/abnormal cristae, seen as electron-translucent areas within their lumens (Fig. 7, D and E). Our most parsimonious conclusion is that the atg12-1 endosperm collects dysfunctional mitochondria that hyperaccumulate resident proteins.
While the mechanism(s) underpinning mitophagy in maize are not understood, several reports have recently showed that the Arabidopsis (Arabidopsis thaliana) protein FRIENDLY helps clear depolarized mitochondria possibly by clustering them for autophagy and thus might serve as a mitophagy receptor (Ma et al. 2021; Kacprzak and Van Aken 2023). Searches of the maize genome identified a putative maize ortholog with 59% amino acid sequence identity, whose mRNA and protein were evident in our endosperm transcriptomic and proteomic data sets. Interestingly, its mRNA level rose in the atg12-1 endosperm compared to wild type at 6 DAP, while its protein level dropped by 3-fold at 18 DAP (Supplemental Fig. S12).
Autophagy influences antioxidant and thiol metabolism
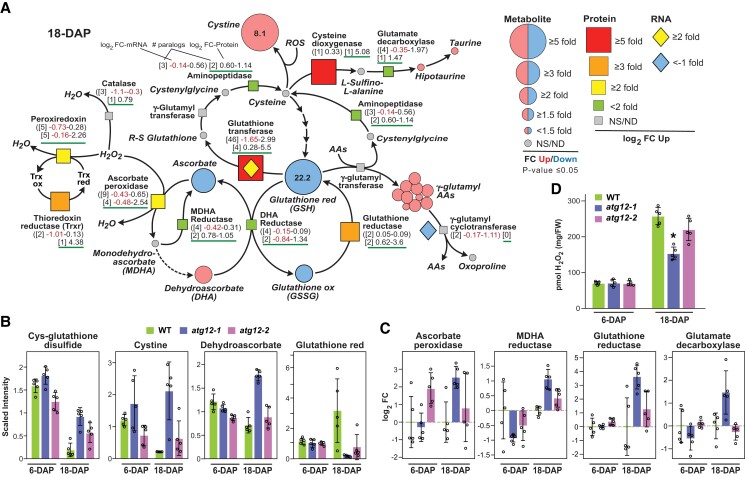
The strong increases in metabolites associated with redox metabolism in atg12-1 backgrounds implied that mutant endosperm also experienced oxidative stress. To further explore this possibility, we comprehensively examined various aspects of redox metabolism with an emphasis on glutathione/ROS-related pathways and cysteine metabolism (Fig. 8A). Particular attention was on the ascorbate/glutathione cycle that uses ascorbate peroxidase (APX) to convert hydrogen peroxide (H2O2) to water, thus yielding monodehydroascorbate (MDHA), which is recycled back to ascorbate by monodehydroascorbate reductase (MDHR) or is spontaneously converted to dehydroascorbate using reduced glutathione (GSH) (Hasanuzzaman et al. 2019). The oxidized form of glutathione (GSSG) is then regenerated back to GSH by glutathione reductase (GR). As shown in Fig. 8, A to C, lower levels of ascorbate together with increases in dehydroascorbate were observed in atg12-1 endosperm, likely through increases in APX whose levels rose 5.8-fold in atg12-1 mutant versus wild type at 18 DAP. Despite high levels of dehydroascorbate, we measured less GSH and GSSG (22.2- and 2.1-fold reduction) in the atg12-1 endosperm, suggesting a dysregulation of glutathione synthesis or increased consumption.
Figure 8.
Atg12 mutations alter redox metabolism in maize endosperm. A) Simplified flow chart of various pathways involved in redox metabolism describing how the associated metabolites are impacted by the atg12 mutations in 18-DAP endosperm and how relevant enzymes changed with respect to their protein and corresponding mRNA levels. FCs as compared to wild type (WT) of each metabolite and associated protein and corresponding mRNA are indicated by the size of the geometric shapes (see legend). NS and ND, not significant and not detected, respectively. B) Examples of key redox metabolites that differentially accumulate in atg12-1 starchy endosperm. Each bar represents the mean of 5 biological replicates (±Sd). C) Examples of key redox enzymes that differentially accumulate in atg12-1 endosperm at 6 and 18 DAP. Log2 FCs in protein abundance were quantified by LC-MS/MS based on the MS1 precursor ion intensities. Each bar represents the mean of 5 biological replicates (±Sd), each analyzed by 4 technical replicates. All values were normalized to the mean value for WT endosperm at 6 and 18 DAP. D) Levels of H2O2 in 6- and 18-DAP endosperm. Each bar represents the mean of 5 biological replicates (±Sd), each assayed in duplicate by the Amplex Red assay. Asterisks indicate a significant difference based on the 2-tailed Students’ t test.
While uncertain about glutathione synthesis from cysteine, increased glutathione use in the atg12 mutants was supported by changes in several routes (Fig. 8, A to C). One shunt was the conjugation of glutathione to various γ-glutamyl amino acids through transpeptidation of GSH by γ-glutamyl transferases (Noctor et al. 2012). The levels of these γ-glutamyl conjugates (10 of 12 measured) collectively rose by ∼1.5-fold in the atg12-1 endosperm (Fig. 3A). Another shunt could involve cysteine which is converted to glutathione by a complex conjugation cascade (Fig. 8A). Our omic profiles implied that cysteine levels were impacted by multiple events in the atg12 backgrounds, including its oxidation to cystine whose level rose 8.1-fold in the mutant, and its conversion to taurine- and hypotaurine metabolites through cysteine dioxygenase whose protein level increased 5.1-fold without a concomitant increase in transcript level (Fig. 8A).
Apart from ascorbate and glutathione, plants adopt other mechanisms to detoxify H2O2 and ROS, such as the nitric oxide-driven oxidation of glucose and galactose into glucarate and galactarate, respectively, or by the glutaperoxidase (GPX) and peroxiredoxin (PRX) scavenging pathways operating in conjunction with thioredoxin (Smirnoff and Arnaud 2019). As described above, glucarate and galactarate levels were both strongly elevated in the atg12-1 background (Fig. 3, A and B), whereas, the levels of PRX reductase and thioredoxin reductase increased by 2.3- and 4.4-fold, respectively. H2O2 can also be removed by catalase (Smirnoff and Arnaud 2019), but the similar catalase protein levels measured in wild type and the atg12-1 mutant suggested that its activity cannot account for the redox differences seen in the mutant endosperm (Fig. 8A).
Predicting that changes in redox-thiol metabolism might alter ROS accumulation in atg12 endosperm, we measured H2O2 levels in endosperm extracts using the Amplex Red reagent (Brumbarova et al. 2016) (Fig. 8D). In wild-type endosperm, H2O2 levels rose ∼3-fold from 6 to 18 DAP, implying that oxidative stress increased as the endosperm matured. Surprisingly, while the endosperm from the atg12-1 and atg12-2 mutants had similar H2O2 levels as compared to wild type at 6 DAP, they accumulated less H2O2 at 18 DAP (40% less in atg12-1), suggesting that this oxidant is being consumed by end products such as cystine, glucarate, and galactarate (Figs. 3B and 8, A and B).
Discussion
Numerous studies have connected autophagy to plant cell homeostasis, source sink relationships, and metabolic and proteomic dynamics through its roles in nutrient recycling and the clearance of unwanted or dysfunctional constituents (Avin-Wittenberg et al. 2015; Li et al. 2015, 2022a; Masclaux-Daubresse et al. 2017; McLoughlin et al. 2018, 2020; Naumann et al. 2019; Shinozaki et al. 2020; Zhang et al. 2020; Mugume et al. 2022). Here, we extended this view to the developing maize endosperm through integrated omic profiling that compared tissue dissected from wild type and autophagy-compromised mutants and found that canonical autophagy dependent on ATG8 is also important to this nutritive tissue. The strength of our approach was the analysis depth that simultaneously interrogated 440 metabolites, the 20 common inorganic elements, near 33,000 transcripts, and ∼3,800 proteins representing most, if not all, cellular compartments and essential physiological processes. Consequently, our study provides a comprehensive window into the metabolomic and proteomic changes that occur during endosperm maturation beyond the synthesis of starch and storage proteins that dominate its physiology (Woo et al. 2001; Sabelli and Larkins 2009; Reyes et al. 2011). Notably, endosperm harvested at 6 and 18 DAP had substantially different metabolomes and proteomes, implying that these two time points are developmentally distinct in agreement with prior transcriptomic studies (Chen et al. 2014; Yi et al. 2019; Dai et al. 2021). The collective picture is that autophagy (i.e. macroautophagy) aids maturation of this nutritive tissue that becomes essential to the survival of the germinating embryo.
Autophagic disruption could be directly observed in the atg12 samples by the increased levels of both the core component ATG8 (Chung et al. 2009; Li et al. 2015) and the NBR1 autophagic receptor that directs protein aggregate clearance (Jung et al. 2020). Although we might have predicted that the metabolic, transcriptomic, and proteomic profiles would have simplified as most of the endosperm commits to nutrient storage and eventual programmed cell death, our analysis of 18-DAP endosperm failed to see this constriction, with wide swaths of mRNAs and proteins still detectable beyond those connected to starch and storage protein biosynthesis. The collective outcome is that while autophagy is relatively inconsequential to carbohydrate and protein storage by the endosperm at least during the early and middle stages of development, it does regulate metabolic flow and promotes ROS protection and directs the turnover of several organelles as the endosperm develops. The impacted constituents include mitochondria, plastids (likely amyloplasts), peroxisomes, and possibly the ER, protein complexes such as ribosomes, proteasomes and those involved in mitochondrial respiration, and an array of individual proteins that were more abundant in the atg12 backgrounds.
It is likely that some of this turnover is a prelude to programmed cell death and eventual seed dormancy as the endosperm continues to mature. While many elevated proteins are likely autophagic cargo, it is also possible that some are yet-to-be identified adaptors that promote vesicle dynamics or serve as ATG8-binding autophagic receptors (Marshall and Vierstra 2018; Bu et al. 2020; Stephani and Dagdas 2020). It is interesting to note that the levels of some metabolites/proteins were substantially impacted solely by the strong atg12-1 allele, while others were also impacted by the weaker atg12-2 allele (e.g. glucarate/GAMMA CA-2/NBR1 versus cys-glutathionine disulfide/ATPB/ATG8, respectively). In fact, punctual differences in metabolite and proteins between the 2 maize atg12 alleles were observed previously in other developmental and stress contexts (Li et al. 2015; McLoughlin et al. 2018, 2020). The underpinning reasons for these variations are unclear when considering that the overall influences of the two alleles strongly aligned by Pearson correlation coefficient and fit values. We speculate that mutant strength can differentially impact metabolite/protein levels depending on their positions within metabolism, roles in seed maturation, and/or inherent synthesis/degradation rates.
Our data with atg12 endosperm at 6 and 18 DAP support the view that autophagy driven by the ATG8-PE adduct (i.e. macroautophagy) does not impact the formation of protein bodies and protein storage vacuoles (Li et al 2015; Ding et al. 2022) and starch accumulation (this study) in the maize endosperm at least during these stages of seed development. Just measuring seed weight, we also saw only a modest reduction (∼10%) at 18 DAP. This is in contrast to the 25% drop in mature seed weight at >40 DAP measured previously for atg12 seeds grown under identical well-fertilized field conditions (Li et al. 2015). As we expect that starch and storage proteins represent the bulk of mature seed weight, it is conceivable that the lack of autophagy stalls further development beyond 18 DAP, possibly by compromising mitophagy and/or ROS regulation. We also acknowledge that our conclusions with the maize endosperm might not translate to other plant species. For example, Sera et al. (2019) reported that rice (Oryza sativa) atg7-1 plants blocked in macroautophagy have challenged seed yields caused in part by less endosperm starch and more soluble sugars, traits not seen with the maize atg12 mutants at least at 18 DAP.
Comparisons of our omic data of atg12 versus wild type revealed that autophagy strongly modifies the profile of endosperm metabolites, especially those related to redox metabolism, oxidative stress, and conjugated sulfur metabolism, suggesting that it helps prevent cytotoxic ROS accumulation. Elevated oxidation products in the mutants include cystine, glucarate, and galactarate, whose metabolic functions are not well understood, and increases in γ-glutamyl conjugates of glutathione, which coincide with a robust drop in ROS-protective glutathione. Cystine hyperaccumulation is particularly striking, suggesting a unique function for this enigmatic oxidant of cysteine. Taurine and hypotaurine, which were identified as significantly upregulated by MetaboAnalyst, have also been described as antioxidants (Song et al. 2019). The collective hyperaccumulation of these antioxidants and oxidation products might explain why atg12-1 endosperm actually contained less H2O2 than wild type at 18 DAP. An unanticipated link between these compounds and sulfur metabolism was also implied by GO term enrichments and by elevated levels of S-methylcysteine and Cys-glutathione disulfide, but not sulfate, in atg12 endosperm. With respect to amino acid metabolism, we also note that the atg12-1 endosperm hyperaccumulated the tryptophan precursor—anthranilic acid at 6 DAP (Supplemental Fig. S6), which has been connected to stress and herbivory defense (Kollner et al. 2010). While direct links between some of these metabolites, ROS protection, and autophagy are not yet clear (e.g. cystine, glucarate, galacarate, and taurine), we imagine that associations will eventually emerge as our picture of plant metabolism improves.
Our connections between autophagy to oxidative stress fits with prior studies connecting the two. For example, we previously found that maize atg12 leaves under nitrogen or carbon starvation accumulate oxidized fatty acids and secondary compounds considered to be antioxidants, including syringic acid, rutin, and secoisolariciresinol (McLoughlin et al. 2018;, 2020), while Yoshimoto et al. (2009) reported that autophagy-deficient Arabidopsis leaves hyperaccumulate ROS. Protection against ROS damage induced by intense light has also been connected to the autophagic removal of oxidatively damaged peroxisomes (Oikawa et al. 2022), which might explain the increased peroxisome turnover inferred here. In Arabidopsis, an inhibition of the NADPH oxidase, which can generate ROS, also blocks autophagy (Liu et al. 2009), while in tomato (Lycopersicum esculentum), overexpression of mitochondrial alternative oxidase, which regulates mitochondrial ROS, was found to elevate autophagic flux (Zhu et al. 2018). We presume that heightened autophagy during oxidative stress helps remove damaged proteins/organelles as part of a protective homeostatic mechanism.
The sources for this ROS imbalance in the atg12 backgrounds are currently unclear but could involve a hyperaccumulation of defective mitochondria. A role for ATG8-dependent autophagy in clearing dysfunctional mitochondria agrees with prior studies in Arabidopsis that observed mitophagy triggered by starvation-induced senescence, hypoxia, heat, H2O2, UV-B damage, and mitochondrial depolarization (Li et al. 2014; Ma et al. 2021; Nakamura et al. 2021; Kacprzak and Van Aken 2022). A possible scenario is that nonfunctional mitochondria are continually cleared from the endosperm as part of either a normal housekeeping activity that purges dysfunctional organelles or a larger developmental program that transitions physiologically active starchy endosperm cells into those developmentally restricted to nutrient storage and then death. In the absence of mitophagy, the proportion of dysfunctional mitochondria would increase leading to release of cytotoxic ROS that are eventually converted into a host of oxidized metabolites. In agreement, yeast and mammalian cells lacking autophagy also contain aberrant mitochondria resulting in increased ROS levels (Zhang et al. 2007; Tal et al. 2009). The retention of potentially defective peroxisomes in atg12 endosperm might also enhance ROS accumulation.
Despite the limited knowledge of mitophagy regulators in plants, recent studies have identified the Arabidopsis FRIENDLY and TRAB (TRB) proteins as important factors (Ma et al. 2021; Li et al. 2022b; Kacprzak and Van Aken 2023). While putative TRB1 and TRB2 orthologs were not found in our maize proteomic data sets, we could detect a possible FRIENDLY counterpart in both wild-type and atg12 endosperm, with its levels dropping substantially in atg12-1 samples at 18 DAP. At present, it is unclear whether FRIENDLY acts as a mitophagy receptor capable of binding both mitochondria and ATG8.
Our observations by transmission electron microscope that the atg12 endosperm has less mitochondria at 18 DAP while containing more mitochondrial proteins as quantified by LC-MS/MS appears contradictory. Insomuch as there was a significant increase of abnormal mitochondria in the mutant endosperm compared to wild type, it is possible that these dysfunctional species in the atg12 backgrounds actually have higher protein densities due to a lack of autophagic recycling. Although less likely, it is also possible that either the aberrant morphology of atg12 mitochondria at 18 DAP complicated microscopic identification given the extensive fusion/fission/condensation/degradation dynamics of this organelle (Ma et al. 2020) or the mitochondrial proteins seen by MS were no longer compartmentalized and either released into the cytoplasm or awaiting turnover inside vacuoles. Clearly, immunocytological localization of mitochondrial proteins will be necessary to resolve this conundrum.
Taken together, our omic profiling of endosperm revealed a complicated developmental history for this nutritive tissue that exploits autophagy to minimize oxidative stress and maintain cellular homeostasis as it eventually transitions to a quiescent storage tissue. As parts of its possible functions are to regulate the levels of mitochondria, peroxisomes, plastids, ER, ribosomes, and proteasomes, it would be informative to study fully mature atg12 seeds to examine how far autophagy goes in managing starchy endosperm biology prior to cell death. Identifying bottlenecks in endosperm development related to autophagy might then reveal opportunities for improving maize grain quality and yield.
Materials and methods
Plant growth
The maize (Z. mays) atg12-1 (mu02975) and atg12-2 (mu02196) mutants in the W22 inbred background were as described (Li et al. 2015). For the omic studies, the plants were field grown on well-fertilized soil (Li et al. 2015). The endosperm from each genotype/developmental age was collected manually from 30 to 50 seeds from self-pollinated cobs, rapidly frozen at liquid nitrogen temperatures, and stored at −80 °C. Unless indicated otherwise, the resulting 5 biological replicates were used for subsequent omic profiling and other quantitative assays. For the seed morphometrics, the plants were greenhouse grown in well-fertilized Metro-Mix 360 (Sun Gro Horticulture) under 16-h light/8-h dark photoperiod at 27 °C/21 °C day/night temperatures (McLoughlin et al. 2018, 2020).
Metabolome and ionome profiling
Endosperm was subjected to unbiased metabolite profiling by Metabolon (www.metabolon.com) using optimized chromatographic platforms in conjunction with a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific) interfaced with a heated electrospray ionization source and an Orbitrap mass analyzer operated at 35,000 mass resolution (Evans et al. 2009; Ohta et al. 2009). The tissues were extracted, dried, and reconstituted in compatible solvents, spiked with MS standards to assess chromatographic consistency, and then separated by 4 chromatography columns designed to interrogate a range of metabolites (see (McLoughlin et al. 2018, 2020) for details). Metabolites (440 total) were identified by automated comparisons with the retention times, ion features, and MS/MS fragmentation patterns of a reference chemical library (DeHaven et al. 2010). Following log2-transformation and imputation of missing values, analysis of variance (ANOVA) identified metabolite abundances that differed significantly among experimental groups (P < 0.05). To correct for multiple testing, FDRs were estimated by calculating q-values to then determine significant differences in metabolite abundance. Compounds were clustered by Metabolon into subpathways and plotted in Cytoscape (Shannon et al. 2003).
Pathway enrichments were calculated in R using clusterProfiler (Wu et al. 2021) based on the mean accumulation pattern of each metabolite. Metabolites were assigned to specific pathways using the KEGG pathway library for A. thaliana (https://www.genome.jp/kegg-bin/show_organism?org=ath) and were tested for overrepresentation by a hypergeometric test. Metabolite heat maps were generated in Perseus (Tyanova et al. 2016), while overrepresentation and the topology of significantly altered metabolites were determined in MetaboAnalyst (Chong et al. 2019).
Elemental ion profiles of the 5 biological replicates above were generated by ICP-MS at the USDA Agricultural Research Service Plant Genetics Facility at the Donald Danforth Plant Science Center as described (McLoughlin et al. 2018). Pulverized samples were lyophilized, digested in HNO3, mixed with 115In (BDH Chemicals) as an internal standard, and heated to 105 °C for 2 h before analysis (Ziegler et al. 2013). ICP-MS employed an ELAN 6000 Dynamic Reaction Cell-e mass spectrometer (PerkinElmer SCIEX) connected to a perfluoroalkoxy MicroFlow nebulizer (Elemental Scientific) and an Apex HF desolvator (Elemental Scientific).
Transcriptome analysis
Total RNA was extracted from 50 to 100 mg of tissue from the 3 mean biological replicates based on the metabolomic profiling using a RNeasy Plant Mini Kit (Qiagen), enriched for mRNA by poly(A) selection, and deep sequenced (20 to 27 million reads per sample) as 100-mers using the Illumina HiSeq-2500 platform (DNA Sequencing Facility, University of Wisconsin Biotechnology Center). FASTQ files were processed with Trimmomatic v0.36 as described (McLoughlin et al. 2018), and the resulting reads were aligned to the B73 reference genome files Zea_mays_AGPv3.31.gff3 and Zea_mays.AGPv3.31.dna.toplevel.fa from ENSEMBL Plants (http://plants.ensembl.org). Transcript abundances were quantified using RSEM v1.2.21 and Bowtie 2 (Langmead and Salzberg 2012). The fragment-length-mean and fragment-length-Sd parameters are summarized in Supplemental Data Set 4. See Supplemental Table S1 for the GRMZM gene identifier numbers for the specific proteins highlighted in this study.
DEGs were identified by EbSeq v1.12.075 (Leng et al. 2013) using pairwise comparisons (McLoughlin et al. 2018). Library sizes were median-normalized before 10 iterations of the EBtest algorithm set to a 5% FDR threshold. Those DEGs (posterior probability of equal expression <0.05 and a log2 FC > 0.5) in both atg12-1 and atg12-2 backgrounds versus wild type for both the 6- and 18-DAP samples were included in the comparisons. For the network analyses, transcripts were grouped by MapMan (Usadel et al. 2005) based on the Zm_GENOME_RELEASE_09 mapping file (https://mapman.gapipd.org). Pathway enrichment tests for significance used the Fisher's exact test and corrected for multiple testing via the Holm–Bonferroni method. Coefficients of variation among the biological replicates for the transcriptome and proteome data sets were determined using Perseus.
Proteome profiling
Proteomic data were collected as described (McLoughlin et al. 2018, 2020). Total endosperm protein (∼300 mg) was isolated from the 5 biological replicates above by extraction in Tris-buffered phenol (pH 8.0) (Invitrogen), mixed with 5 volumes of extraction buffer (50 mM Tris-HCl (pH 7.5), 1 mM Na4EDTA, and 0.9 M sucrose), and then separated by centrifugation at 13,000 × g for 10 min at 4 °C. The top phenol phase was collected, and the aqueous phase was extracted again in 5 volumes of Tris-buffered phenol. Both phenol phases were combined and precipitated at −80 °C for 1 h with 5 volumes of 0.1 M ammonium acetate in methanol. Proteins were collected by centrifugation at 4,500 × g for 10 min at 4 °C, vortexed, and washed twice at −20 °C with 5 volumes of 0.1 M ammonium acetate (w/v) in methanol, and once at −20 °C with 5 volumes of 70% (v/v) methanol, each of which was followed by centrifugal collection.
After the final wash, the pellets were lyophilized to dryness, resuspended in 100 µL of 8 M urea, and reduced and alkylated with iodoacetamide (McLoughlin et al. 2018). The reactions were quenched with dithiothreitol, diluted with 25 mM ammonium bicarbonate to reduce the urea concentrations below 1.5 M, and digested overnight with sequencing-grade modified porcine trypsin (Promega). The resulting peptides were lyophilized to a final volume of 250 µL, acidified with 0.5% (v/v) trifluoroacetic acid (pH < 3.0), and desalted using a 100-µL Bond Elut OMIX C18 pipette tip (Agilent Technologies). Final samples were resuspended in 20 µL of 5% (v/v) acetonitrile and 0.1% (v/v) formic acid.
Nano-scale LC separation of the tryptic peptides was performed using a Dionex Ultimate 3000 Rapid Separation system equipped with a 75 µm × 25 cm Acclaim PepMap RSLC C18 column (Thermo Fisher Scientific), in combination with a 2-h linear 4% to 36% acetonitrile (v/v) gradient in 0.1% (v/v) formic acid and a flow rate of 250 nL/min (McLoughlin et al. 2018). Eluted peptides were analyzed online by a Thermo Fisher Scientific Q-Exactive Plus spectrometer operated in the positive electrospray ionization mode. Data-dependent acquisition of full MS scans was collected using the automatic gain control, intensity threshold, charge states, and dynamic exclusion settings as described (McLoughlin et al. 2018). Each biological replicate was analyzed in quadruplicate, and the first two runs were performed without an exclusion list, while the third and fourth runs were performed with exclusion of the 5,000 most abundant peptides detected in the first two runs to enhance peptide coverage.
The MS/MS data sets were queried by Proteome Discoverer (version 2.0.0.802; Thermo Fisher Scientific) against the maize B73 proteome (Zea-mays.AGPv3.21.pep.all from www.maizegdb.org). Peptides were assigned by SEQUEST HT (Eng et al. 1994), allowing a maximum of 2 missed tryptic cleavages, a minimum peptide length of 6, a precursor mass tolerance of 10 ppm, and fragment mass tolerances of 0.02 Da. Label-free quantification based on MS1 precursor ion intensities was performed in Proteome Discoverer; the “3 Top N” peptides were used for area calculation (Silva et al. 2006).
Abundance comparisons based on the MS1 scans were generated after normalizing the data by sample weight and imputation as described (McLoughlin et al. 2018, 2020); the final values were scaled by the mean intensity of all samples. The intensities were then normalized based on the median values from 150 proteins considered least variable among the samples at each development stage (Sd/average) (McLoughlin et al. 2018). Significant differences in protein abundances were calculated by ANOVA contrasts (P ≤ 0.05) and displayed by volcano plots. Proteins were assigned by GO to specific cellular compartments, protein complexes and/or functions by the AgriGo analysis toolkit (Tian et al. 2017) in combination with the Z. mays AGPv3.21.pep.all database (www.maizegdb.org).
GO and metabolic pathways maps
GO analyses were performed using the maize profile database in g-:Profiler V3.10.1 (Raudvere et al. 2019) as part of the ELIXIR Infrastructure package (http://biit.cs.ut.ee). GO categories were selected based on their P-value of significance, uniqueness, and degree of completeness. Changes in metabolite, protein, and/or transcript abundances were superimposed onto metabolic pathway maps using the maizecyc v.2.2 (pathway.gramene.org) and KEGG databases. Transcripts and proteins were associated with their corresponding metabolic conversions using KEGG.
Storage protein extractions
Zein and non-Zein proteins were enriched from endosperm as described (Wu and Messing 2012). For Zeins, 50 mg of pulverized tissue were vortexed with 400 µL of Zein extraction buffer (70% (v/v) ethanol and 2% (v/v) 2-mercaptoethanol) and incubated at room temperature overnight. After clarification at 16,000 × g for 10 min, 100 µL of the supernatant was mixed with 10 µL of 10% (w/v) SDS, dried, and resuspended in 100 µL of deionized water. For the non-Zein fraction, the pellets obtained as above were washed 3 times with the Zein extraction buffer. The final pellets were resuspended in 400 µL of non-Zein extraction buffer (12.5 mM sodium borate, 5% [w/v] SDS, and 2% [v/v] 2-mercaptoethanol), incubated at 37 °C for 2 h, and clarified at 16,000 × g for 10 min.
Immunoblot analysis
Immunoblot analysis of non-Zein fractions employed antibodies at the indicated dilutions: ATG8 (1:1,000) (Thompson et al. 2005) and NBR1 (1:3,000) (Jung et al. 2020), with the signals generated with antihistone H3 antibodies (1:10,000; Abcam AB1791) used to confirm near equal protein loading. The Zein fractions were diluted at 1:1,000 and probed with anti-α-Zein (1:10,000) and γ-Zein (1:2,000) antibodies (Woo et al. 2001).
Electron microscopy
Endosperm collected at 20 DAP was high-pressure frozen in an ICE high-pressure freezer (Leica) and freeze-substituted with 2% (w/v) OsO4 in anhydrous acetone at −80 °C overnight followed by slow warming to room temperature (Reyes et al. 2011). Samples were rinsed in acetone, infiltrated in EPON resin (Ted Pella), and stained with 2% (w/v) uranyl acetate in 70% methanol followed by Reynold's lead citrate (2.6% [w/v] lead nitrate and 3.5% [w/v] sodium citrate [pH 12.0]). Mitochondrial numbers, cross-sectional area, and morphology were quantified by ImageJ (http://imagej.nih.gov) from sections imaged by a FEI CM120 electron microscope.
Peroxide quantification
H2O2 content was measured from the 5 biological replicates of field-grown endosperm described above using the Amplex Red H2O2-peroxidase assay kit (Molecular Probes). Thirty mg of pulverized frozen tissue were resuspended in 20 mM K2HPO4 (pH 6.5) and clarified at 16,000 × g. The resulting supernatants were incubated in the dark with 25 µL of 100 mM 10-acetyl-3,7-dihydrophenoxazine and 0.2 U/mL horseradish peroxidase for 30 min at room temperature. The resorufin product was quantified spectrophotometrically at 560 nm using an Infinite M200 Pro plate reader (Tecan).
Starch quantification
Starch was heat extracted from 0.3 mg of endosperm for 3 min in 5 mL of 80% ethanol (v/v) from the 5 field-grown, biological replicates described above and clarified by centrifugation at 5,000 × g for 10 min. The pellets were washed twice with 80% (v/v) ethanol, dried, resuspended in 3 mL water, and boiled for 10 min. The gelatinized starch (500 µL) was converted to glucose by mixing with 500 µL of 200 mM sodium acetate (pH 5.5) and incubating at 37 °C for 4 h either alone or with 6 units of amyloglucosidase (Roche) and 1 unit of α-amylase (Roche). After clarification, glucose was measured spectrophotometrically by the formation of NADH in the presence of hexokinase and glucose-6-phosphate dehydrogenase (Smith and Zeeman 2006).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the GRMZM gene identifier numbers listed in Supplemental Table S1.
Supplementary Material
Acknowledgments
We thank D. Alexander and L. Guo for the metabolome studies; J. Barrett, G. Zeigler, and I. Baxter for help with the ionomics; and M. Dyer for greenhouse assistance.
Contributor Information
Jessica A S Barros, Department of Biology, Washington University in St. Louis, St. Louis, MO 63130, USA.
Elizabeth C Chatt, Department of Biology, Washington University in St. Louis, St. Louis, MO 63130, USA.
Robert C Augustine, Department of Biology, Washington University in St. Louis, St. Louis, MO 63130, USA.
Fionn McLoughlin, Department of Biology, Washington University in St. Louis, St. Louis, MO 63130, USA.
Faqiang Li, Department of Genetics, University of Wisconsin, Madison, WI 53706, USA.
Marisa S Otegui, Department of Botany, University of Wisconsin, Madison, WI 53706, USA; Center for Quantitative Cell Imaging, University of Wisconsin, Madison, WI 53706, USA.
Richard D Vierstra, Department of Biology, Washington University in St. Louis, St. Louis, MO 63130, USA.
Author contributions
J.A.S.B., E.C.C., F.L., and R.D.V. designed the research. J.A.S.B., E.C.C., R.C.A., F.M., and M.S.O. performed the research. J.A.S.B., E.C.C., R.C.A., M.S.O., and R.D.V. analyzed the data. R.D.V. and J.A.S.B wrote the manuscript with input from all authors.
Supplemental data
The following materials are available in the online version of this article.
The following supplemental materials are available.
Supplemental Figure S1. Omic comparisons of wild-type maize endosperm collected early and later in development.
Supplemental Figure S2. Accumulation of specific mRNAs during endosperm development of wild-type maize at either 6 or 18 DAP.
Supplemental Figure S3. Morphological comparisons of atg12 and wild-type maize seeds.
Supplemental Figure S4. Zein storage protein and mRNA levels are unaffected in the atg12 mutant endosperm.
Supplemental Figure S5. Autophagy has minimal influence on the maize endosperm ionome.
Supplemental Figure S6. Cytoscape survey of metabolite changes in atg12-1 versus wild-type endosperm at 6 DAP.
Supplemental Figure S7. Cytoscape survey of metabolite changes in atg12-1 versus wild-type endosperm at 18 DAP.
Supplemental Figure S8. Quality control analysis of the transcriptome and proteome data sets from 6- and 18-DAP endosperm.
Supplemental Figure S9. Specific pathways/processes transcriptionally impacted by autophagy in the maize endosperm.
Supplemental Figure S10. Identification of endosperm proteins whose accumulation is significantly altered by the atg12-2 mutation.
Supplemental Figure S11. The impact of autophagy on protein abundances in specific cellular compartments/complexes in the maize endosperm.
Supplemental Figure S12. FRIENDLY protein but not its mRNA decreases in abundance in the endosperm from the atg12-1 mutant at 18 DAP.
Supplemental Table S1. Maize GRMZM gene identifier numbers for the genes/proteins highlighted in this study.
Supplemental Data Set 1. Metabolome raw data; wild-type endosperm versus atg12 at 6 and 18 DAP.
Supplemental Data Set 2. Analysis of the metabolome data sets.
Supplemental Data Set 3. Ionome raw data; wild-type endosperm versus atg12 at 6 and 18 DAP.
Supplemental Data Set 4. Transcriptomic fragment length and correlation values.
Supplemental Data Set 5. Transcriptome raw data; wild-type endosperm versus atg12 at 6 and 18 DAP.
Supplemental Data Set 6. Analysis of the transcriptome data sets.
Supplemental Data Set 7. Description of the 12 transcripts similarly impacted in the atg12-1 and atg12-2 mutant endosperm versus wild type.
Supplemental Data Set 8. Proteome raw data; wild-type endosperm versus atg12 at 6 and 18 DAP.
Supplemental Data Set 9. Analysis of the proteome data sets at 6 DAP.
Supplemental Data Set 10. Analysis of the proteome data sets at 18 DAP.
Funding
This work was supported by grants from the US National Science Foundation; Plant Genome Research Program (IOS-1339325 and IOS-1840687) to M.S.O and R.D.V., and the US National Institutes of Health; National Institute of General Medical Sciences (R01-GM124452) to R.D.V.
Data availability
The metabolomic, ionomic, transcriptomic, and proteomic data can be found in Supplemental Data Sets S1 to S10. Raw data for the transcriptomics and proteomics are available in the NCBI Sequence Read Archive repository under the BioProject ID PRJNA934885 and the PRIDE repository under accession number PXD022863, respectively.
References
- Arcalís E, Hörmann-Dietrich U, Stöger E. Multiscale imaging reveals the presence of autophagic vacuoles in developing maize endosperm. Front Plant Sci. 2023:13:1082890. 10.3389/fpls.2022.1082890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avin-Wittenberg T, Bajdzienko K, Wittenberg G, Alseekh S, Tohge T, Bock R, Giavalisco P, Fernie AR. Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell 2015:27(2):306–322. 10.1105/tpc.114.134205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros JAS, Magen S, Lapidot-Cohen T, Rosental L, Brotman Y, Araújo WL, Avin-Wittenberg T. Autophagy is required for lipid homeostasis during dark-induced senescence. Plant Physiol. 2021:185(4):1542–1558. 10.1093/plphys/kiaa120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbarova T, Le CTT, Bauer P. Hydrogen peroxide measurement in Arabidopsis root tissue using Amplex Red. Bio-protocols 2016:6:e199. 10.21769/BioProtoc.1999 [DOI] [Google Scholar]
- Bu F, Yang M, Guo X, Huang W, Chen L. Multiple functions of ATG8 family proteins in plant autophagy. Front Cell Dev Biol. 2020:8:466. 10.3389/fcell.2020.00466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanoca A, Kovinich N, Burkel B, Stecha S, Bohorquez-Restrepo A, Ueda T, Eliceiri KW, Grotewold E, Otegui MS. Anthocyanin vacuolar inclusions form by a microautophagy mechanism. Plant Cell 2015:27(9):2545–2559. 10.1105/tpc.15.00589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zeng B, Zhang M, Xie S, Wang G, Hauck A, Lai J. Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 2014:166(1):252–264. 10.1104/pp.114.240689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Wishart DS, Xia J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinformatics. 2019:68(1):e86. 10.1002/cpbi.86 [DOI] [PubMed] [Google Scholar]
- Chung T, Suttangkakul A, Vierstra RD. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 2009:149(1):220–234. 10.1104/pp.108.126714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagdas YF, Belhaj K, Maqbool A, Chaparro-Garcia A, Pandey P, Petre B, Tabassum N, Cruz-Mireles N, Hughes RK, Sklenar J, et al. An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. Elife 2016:5:e10856. 10.7554/eLife.10856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D, Ma Z, Song R. Maize endosperm development. J Integr Plant Biol. 2021:63(4):613–627. 10.1111/jipb.13069 [DOI] [PubMed] [Google Scholar]
- DeHaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform. 2010:2(1):9. 10.1186/1758-2946-2-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Zhang X, Paez-Valencia J, McLoughlin F, Reyes FC, Morohashi K, Grotewold E, Vierstra RD, Otegui MS. Microautophagy mediates vacuolar delivery of storage proteins in maize aleurone cells. Front Plant Sci. 2022:13:833612. 10.3389/fpls.2022.833612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994:5(11):976–989. 10.1016/1044-0305(94)80016-2 [DOI] [PubMed] [Google Scholar]
- Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, non-targeted ultra-high performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009:81(16):6656–6667. 10.1021/ac901536h [DOI] [PubMed] [Google Scholar]
- Fan J, Yu L, Xu C. Dual role for autophagy in lipid metabolism in Arabidopsis. Plant Cell 2019:31(7):1598–1613. 10.1105/tpc.19.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, De Rycke R, Dagdas Y, Nowack MK. Autophagy promotes programmed cell death and corpse clearance in specific cell types of the Arabidopsis root cap. Curr Biol. 2022:32(20):4548. 10.1016/j.cub.2022.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafren A, Macia JL, Love AJ, Milner JJ, Drucker M, Hofius D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc Natl Acad Sci U S A. 2017:114(10):E2026–E2035. 10.1073/pnas.1610687114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasanuzzaman M, Bhuyan M, Anee TI, Parvin K, Nahar K, Mahmud JA, Fujita M. Regulation of ascorbate-glutathione pathway in mitigating oxidative damage in plants under abiotic stress. Antioxidants 2019:8(9):384. 10.3390/antiox8090384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Lee HN, Marshall RS, Lomax AW, Yoon MJ, Kim J, Kim JH, Vierstra RD, Chung T. Arabidopsis cargo receptor NBR1 mediates selective autophagy of defective proteins. J Exp Bot. 2020:71(1):73–89. 10.1093/jxb/erz404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacprzak SM, Van Aken O. Carbon starvation, senescence and specific mitochondrial stresses, but not nitrogen starvation and general stresses, are major triggers for mitophagy in Arabidopsis. Autophagy 2022:18(12):2894–2912. 10.1080/15548627.2022.2054039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacprzak SM, Van Aken O. FRIENDLY is required for efficient dark-induced mitophagy and controlled senescence in Arabidopsis. Free Rad Biol Med. 2023:204:1–7. 10.1016/j.freeradbiomed.2023.04.007 [DOI] [PubMed] [Google Scholar]
- Kollner TG, Lenk C, Zhao N, Seidl-Adams I, Gershenzon J, Chen F, Degenhardt J. Herbivore-induced SABATH methyltransferases of maize that methylate anthranilic acid using s-adenosyl-L-methionine. Plant Physiol. 2010:153(4):1795–1807. 10.1104/pp.110.158360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012:9(4):357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, Haag JD, Gould MN, Stewart RM, Kendziorski C. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013:29(8):1035–1043. 10.1093/bioinformatics/btt087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong JX, Raffeiner M, Spinti D, Langin G, Franz-Wachtel M, Guzman AR, Kim JG, Pandey P, Minina AE, Macek B, et al. A bacterial effector counteracts host autophagy by promoting degradation of an autophagy component. EMBO J. 2022:41(13):e110352. 10.15252/embj.2021110352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Chung T, Pennington JG, Federico ML, Kaeppler HF, Kaeppler SM, Otegui MS, Vierstra RD. Autophagic recycling plays a central role in maize nitrogen re-mobilisation. Plant Cell 2015:27(5):1389–1408. 10.1105/tpc.15.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Chung T, Vierstra RD. AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 2014:26(2):788–807. 10.1105/tpc.113.120014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Duckney P, Zhang T, Fu Y, Li X, Kroon J, De Jaeger G, Cheng Y, Hussey PJ, Wang P. TRab family proteins are components of ER-mitochondrial contact sites and regulate ER-mitochondrial interactions and mitophagy. Nat Commun. 2022b:13(1):5658. 10.1038/s41467-022-33402-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lee CP, Ding X, Qin Y, Wijerathna-Yapa A, Broda M, Otegui MS, Millar AH. Defects in autophagy lead to selective in vivo changes in turnover of cytosolic and organelle proteins in Arabidopsis. Plant Cell 2022a:34(10):3936–3960. 10.1093/plcell/koac185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xiong Y, Bassham DC. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 2009:5(7):954–963. 10.4161/auto.5.7.9290 [DOI] [PubMed] [Google Scholar]
- Ma K, Chen G, Li W, Kepp O, Zhu Y, Chen Q. Mitophagy, mitochondrial homeostasis, and cell fate. Front Cell Dev Biol. 2020:8:467. 10.3389/fcell.2020.00467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Liang Z, Zhao J, Wang P, Ma W, Mai KK, Fernandez Andrade JA, Zeng Y, Grujic N, Jiang L, et al. Friendly mediates membrane depolarization-induced mitophagy in Arabidopsis. Curr Biol. 2021:31(9):1931–1944.e4. 10.1016/j.cub.2021.02.034 [DOI] [PubMed] [Google Scholar]
- Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD. Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol Cell. 2015:58(6):1053–1066. 10.1016/j.molcel.2015.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RS, Vierstra RD. Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol. 2018:69(1):173–208. 10.1146/annurev-arplant-042817-040606 [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Chen Q, Have M. Regulation of nutrient recycling via autophagy. Curr Opin Plant Biol. 2017:39:8–17. 10.1016/j.pbi.2017.05.001 [DOI] [PubMed] [Google Scholar]
- McLoughlin F, Augustine RC, Marshall RS, Li F, Kirkpatrick LD, Otegui MS, Vierstra RD. Maize multi-omics reveal roles for autophagic recycling in proteome remodelling and lipid turnover. Nat Plants. 2018:4(12):1056–1070. 10.1038/s41477-018-0299-2 [DOI] [PubMed] [Google Scholar]
- McLoughlin F, Marshall RS, Ding X, Chatt EC, Kirkpatrick LD, Augustine RC, Li F, Otegui MS, Vierstra RD. Autophagy plays prominent roles in amino acid, nucleotide, and carbohydrate metabolism during fixed-carbon starvation in maize. Plant Cell 2020:32(9):2699–2724. 10.1105/tpc.20.00226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugume Y, Ding G, Duenas ME, Liu M, Lee YJ, Nikolau BJ, Bassham DC. Complex changes in membrane lipids associated with the modification of autophagy in Arabidopsis. Metabolites 2022:12(2):190. 10.3390/metabo12020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Hagihara S, Otomo K, Ishida H, Hidema J, Nemoto T, Izumi M. Autophagy contributes to the quality control of leaf mitochondria. Plant Cell Physiol. 2021:62(2):229–247. 10.1093/pcp/pcaa162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann C, Muller J, Sakhonwasee S, Wieghaus A, Hause G, Heisters M, Burstenbinder K, Abel S. The local phosphate deficiency response activates endoplasmic reticulum stress-dependent autophagy. Plant Physiol. 2019:179(2):460–476. 10.1104/pp.18.01379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Chaouch S, Han Y, Neukermans J, Marquez-Garcia B, Queval G, Foyer CH. Glutathione in plants: an integrated overview. Plant Cell Environ. 2012:35(2):454–484. 10.1111/j.1365-3040.2011.02400.x [DOI] [PubMed] [Google Scholar]
- Ohta T, Masutomi N, Tsutsui N, Sakairi T, Mitchell M, Milburn MV, Ryals JA, Beebe KD, Guo L. Untargeted metabolomic profiling as an evaluative tool of fenofibrate-induced toxicology in Fischer 344 male rats. Toxicol Pathol. 2009:37(4):521–535. 10.1177/0192623309336152 [DOI] [PubMed] [Google Scholar]
- Oikawa K, Goto-Yamada S, Hayashi Y, Takahashi D, Kimori Y, Shibata M, Yoshimoto K, Takemiya A, Kondo M, Hikino K, et al. Pexophagy suppresses ROS-induced damage in leaf cells under high-intensity light. Nat Commun. 2022:13(1):7493. 10.1038/s41467-022-35138-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. G:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019:47(W1):W191–W198. 10.1093/nar/gkz369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes FC, Chung T, Holding D, Jung R, Vierstra R, Otegui MS. Delivery of prolamins to the protein storage vacuole in maize aleurone cells. Plant Cell. 2011:23(2):769–784. 10.1105/tpc.110.082156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA. The development of endosperm in grasses. Plant Physiol. 2009:149(1):14–26. 10.1104/pp.108.129437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sera Y, Hanamata S, Sakamoto S, Ono S, Kaneko K, Mitsui Y, Koyano T, Fujita N, Sasou A, Masumura T, et al. Essential roles of autophagy in metabolic regulation in endosperm development during rice seed maturation. Sci Rep. 2019:9(1):18544. 10.1038/s41598-019-54361-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003:13(11):2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki D, Merkulova EA, Naya L, Horie T, Kanno Y, Seo M, Ohsumi Y, Masclaux-Daubresse C, Yoshimoto K. Autophagy increases zinc bioavailability to avoid light-mediated reactive oxygen species production under zinc deficiency. Plant Physiol. 2020:182(3):1284–1296. 10.1104/pp.19.01522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006:5(1):144–156. 10.1074/mcp.M500230-MCP200 [DOI] [PubMed] [Google Scholar]
- Smirnoff N, Arnaud D. Hydrogen peroxide metabolism and functions in plants. New Phytol. 2019:221(3):1197–1214. 10.1111/nph.15488 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC. Quantification of starch in plant tissues. Nat Protoc. 2006:1(3):1342–1345. 10.1038/nprot.2006.232 [DOI] [PubMed] [Google Scholar]
- Song W, Qi N, Liang C, Duan F, Zhao H. Soybean root transcriptome profiling reveals a nonhost resistant response during Heterodera glycines infection. PLoS One 2019:14(5):e0217130. 10.1371/journal.pone.0217130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephani M, Dagdas Y. Plant selective autophagy-still an uncharted territory with a lot of hidden gems. J Mol Biol. 2020:432(1):63–79. 10.1016/j.jmb.2019.06.028 [DOI] [PubMed] [Google Scholar]
- Sun X, Wang P, Jia X, Huo L, Che R, Ma F. Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol J. 2018:16(2):545–557. 10.1111/pbi.12794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A. 2009:106(8):2770–2775. 10.1073/pnas.0807694106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005:138(4):2097–2110. 10.1104/pp.105.060673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017:45(W1):W122–W129. 10.1093/nar/gkx382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016:13(9):731–740. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 2005:138(3):1195–1204. 10.1104/pp.105.060459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustun S, Hafren A, Liu Q, Marshall RS, Minina EA, Bozhkov PV, Vierstra RD, Hofius D. Bacteria exploit autophagy for proteasome degradation and enhanced virulence in plants. Plant Cell 2018:30(3):668–685. 10.1105/tpc.17.00815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Nolan TM, Clark NM, Jiang H, Montes-Serey C, Guo H, Bassham DC, Walley JW, Yin Y. The F-box E3 ubiquitin ligase BAF1 mediates the degradation of the brassinosteroid-activated transcription factor BES1 through selective autophagy in Arabidopsis. Plant Cell 2021:33(11):3532–3554. 10.1093/plcell/koab210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YM, Hu DW, Larkins BA, Jung R. Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. Plant Cell 2001:13(10):2297–2317. 10.1105/tpc.010240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, et al. Clusterprofiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb). 2021:2:100141. 10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Messing J. RNA Interference can rebalance the nitrogen sink of maize seeds without losing hard endosperm. PLoS One 2012:7(2):e32850. 10.1371/journal.pone.0032850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Bassham DC. New insight into the mechanism and function of autophagy in plant cells. Int Rev Cell Mol Biol. 2015:320:1–40. 10.1016/bs.ircmb.2015.07.005 [DOI] [PubMed] [Google Scholar]
- Yi F, Gu W, Chen J, Song N, Gao X, Zhang X, Zhou Y, Ma X, Song W, Zhao H, et al. High temporal-resolution transcriptome landscape of early maize seed development. Plant Cell 2019:31(5):974–992. 10.1105/tpc.18.00961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 2009:21(9):2914–2927. 10.1105/tpc.109.068635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TE, Gallie DR. Programmed cell death during endosperm development. Plant Mol Biol. 2000:44(3):283–301. 10.1023/A:1026588408152 [DOI] [PubMed] [Google Scholar]
- Zhan N, Wang C, Chen L, Yang H, Feng J, Gong X, Ren B, Wu R, Mu J, Li Y, et al. S-nitrosylation targets GSNO reductase for selective autophagy during hypoxia responses in plants. Mol Cell. 2018:71(1):142–154.e6. 10.1016/j.molcel.2018.05.024 [DOI] [PubMed] [Google Scholar]
- Zhang X, Ding X, Marshall RS, Paez-Valencia J, Lacey P, Vierstra RD, Otegui MS. Reticulon proteins modulate autophagy of the endoplasmic reticulum in maize endosperm. Elife 2020:9:e51918. 10.7554/eLife.51918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Qi H, Taylor R, Xu W, Liu LF, Jin S. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy 2007:3(4):337–346. 10.4161/auto.4127 [DOI] [PubMed] [Google Scholar]
- Zhou S, Hong Q, Li Y, Li Q, Wang M. Autophagy contributes to regulate the ROS levels and PCD progress in TMV-infected tomatoes. Plant Sci. 2018:269:12–19. 10.1016/j.plantsci.2017.11.002 [DOI] [PubMed] [Google Scholar]
- Zhu T, Zou L, Li Y, Yao X, Xu F, Deng X, Zhang D, Lin H. Mitochondrial alternative oxidase-dependent autophagy involved in ethylene-mediated drought tolerance in Solanum lycopersicum. Plant Biotechnol J. 2018:16(12):2063–2076. 10.1111/pbi.12939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler G, Terauchi A, Becker A, Armstrong PR, Hudson KA, Baxter I. Ionomic screening of field-grown soybean identifies mutants with altered seed elemental composition. Plant Genome 2013:6(2):2. 10.3835/plantgenome2012.07.0012 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metabolomic, ionomic, transcriptomic, and proteomic data can be found in Supplemental Data Sets S1 to S10. Raw data for the transcriptomics and proteomics are available in the NCBI Sequence Read Archive repository under the BioProject ID PRJNA934885 and the PRIDE repository under accession number PXD022863, respectively.