Abstract
The ability of neural stem/progenitor cells (NSPCs) to proliferate and differentiate is required through different stages of neurogenesis. Disturbance in the regulation of neurogenesis causes many neurological diseases, such as intellectual disability, autism, and schizophrenia. However, the intrinsic mechanisms of this regulation in neurogenesis remain poorly understood. Here, we report that Ash2l (Absent, small or homeotic discs-like 2), one core component of a multimeric histone methyltransferase complex, is essential for NSPC fate determination during postnatal neurogenesis. Deletion of Ash2l in NSPCs impairs their capacity for proliferation and differentiation, leading to simplified dendritic arbors in adult-born hippocampal neurons and deficits in cognitive abilities. RNA sequencing data reveal that Ash2l primarily regulates cell fate specification and neuron commitment. Furthermore, we identified Onecut2, a major downstream target of ASH2L characterized by bivalent histone modifications, and demonstrated that constitutive expression of Onecut2 restores defective proliferation and differentiation of NSPCs in adult Ash2l-deficient mice. Importantly, we identified that Onecut2 modulates TGF-β signaling in NSPCs and that treatment with a TGF-β inhibitor rectifies the phenotype of Ash2l-deficient NSPCs. Collectively, our findings reveal the ASH2L-Onecut2-TGF-β signaling axis that mediates postnatal neurogenesis to maintain proper forebrain function.
Subject terms: Cognitive ageing, Neural ageing
Introduction
Neural stem/progenitor cells (NSPCs) are self-renewing, multipotent cells that give rise to the cerebral cortex during fetal development and persist throughout adult life in the forebrain [1, 2]. The process of generating new neurons from NSPCs in neurogenesis is required for normal brain development and function [3]. Sustained neurogenesis throughout life occurs in the subgranular zone (SGZ) of the dentate gyrus in the hippocampus and is hypothesized to be involved in cognitive processes and in neurological diseases [4]. However, there remains much to learn about the regulatory mechanisms that dictate NSPC fate decision and function during postnatal neurogenesis [5]. The maintenance and differentiation of NSPCs in the postnatal stage are tightly controlled by intricate molecular networks [6]. A better understanding of these regulatory mechanisms in postnatal neurogenesis may provide deep clarification of the plasticity of the brain and more clues for realizing the therapeutic potential of NSPCs for treating neurological diseases.
Chromatin structure is influenced by histone modifications, a wide variety of which are associated with active transcription or gene silencing [7]. A set of conserved chromatin regulatory factors, known as trithorax group (TrxG) and Polycomb group (PcG) components, act antagonistically to orchestrate the expression of key genes in cellular and developmental processes [8–10]. PcG-associated H3K27me3 correlates with gene repression, whereas H3K4me3 mediated by TrxG correlates with gene activation [11–13]. Bivalent chromatin, an epigenetic state characterized by the presence of both H3K4me3 and H3K27me3, has emerged as a critical mechanism for lineage determination and developmental gene regulation [14]. However, how bivalent chromatin is established, maintained, and resolved during developmental progression, such as cell fate specification, remains largely unexplored [15].
ASH2L (absent, small, or homeotic 2-like), a critical TrxG protein, together with WDR5, RBBP5, and DPY30 (WRAD complex), is responsible for H3K4 methylation [16], which is crucial to maintain the expression of lineage-specific genes [17, 18] and transcriptional stability [19–21]. ASH2L has been shown to play important roles in maintaining stem cell commitment and differentiation. Early evidence demonstrates that Ash2l is a key regulator of open chromatin in mouse ES cells. RNAi knockdown of Ash2l is sufficient to reduce H3K4 methylation levels and drive mouse ES cells to a silenced chromatin state [22]. Ash2l also forms an enhancer-bound Ash2l/OSN complex that can drive enhancer activation and govern the pluripotency network and stemness circuitry of mouse ES cells [23]. In lineage stem cells, ASH2L is essential for hematopoietic stem and multipotent progenitor cell physiology [24]. In the nervous system, ASH2L is an epigenetic regulator of cortical development in the embryonic stage [25], strongly suggesting an essential role of ASH2L in neuronal formation during cortical development. Moreover, Ash2l mutations may cause intellectual disability in humans [26], suggesting a potential role of ASH2L in postnatal neurogenesis. However, the function and intrinsic mechanism of ASH2L in NSPCs during postnatal neurogenesis is largely unknown.
In the present study, we provide multiple lines of evidence to show that ASH2L is an essential player in forebrain formation by mediating NSPC fate decision at the postnatal stage. First, Ash2l-deficient mice show reduced proliferation and differentiation of NSPCs during postnatal development. Adult-specific deletion of Ash2l in NSPCs results in decreased dendritic growth in adult-born hippocampal neurons. Second, Ash2l deletion in NSPCs is associated with impaired ability of learning and memory in adult mice. Third, by combining transcriptome analysis, we show that the proliferation and differentiation defects of NSPCs caused by Ash2l deletion can be rescued by the overexpression of ONECUT2 (OC2), a key direct functional downstream target gene of ASH2L and characterized by histone bivalent marks. Finally, we found that inhibition of transforming growth factor (TGF)-β signaling, which is present at high levels in Ash2l-deficient NSPCs, restores proliferation and differentiation deficits in postnatal Ash2l-abalated NSPCs. Together, our data support the idea that ASH2L, OC2, and TGF-β signaling form a regulatory network that controls NSPC fate determination in postnatal neurogenesis.
Results
Ash2l is required for postnatal forebrain formation in vivo
To investigate the role of ASH2L in the nervous system, we first generated Ash2l-cKOEmx mice by crossing Ash2lf/f mice with Emx1-Cre transgenic mice (Fig. S1a), a line in which Cre recombinase is expressed under the control of the endogenous Emx1 promoter in the forebrain at E9.5. Littermates with Ash2lf/f were used as wild-type (WT) controls. The expression of ASH2L was abolished in Ash2l-cKOEmx mice, which was confirmed by western blotting and quantitative real-time PCR (qRT-PCR) with forebrain tissues (Fig. S1b–d). Intriguingly, deletion of Ash2l in Ash2l-cKOEmx mice resulted in postnatal lethality, and Ash2l-cKOEmx mice showed significantly reduced body size (Fig. S1e) and aberrant brain morphology (Fig. S1f) at postnatal day 7 (P7) compared to WT mice. We then examined the impact of Ash2l deletion on forebrain formation. Ash2l-cKOEmx mice exhibited a thinner cerebral cortex, larger midbrain, and no identifiable dentate gyrus (DG) regions (Fig. S1g). To understand the cellular mechanisms by which Ash2l regulates cortical lamination, cortical sections were immunostained for neuronal markers of different layers at P7. The quantification results demonstrated that the numbers of different layer-specific cortical neurons of Tbr1+, Ctip2+, Brn2+, Cux1+, and Satb2+ cells were significantly decreased and misplaced in the Ash2l-cKOEmx neocortex (Fig. S1h, i), indicating that ASH2L is essential for neuronal differentiation of NSPCs during nervous system development.
Ablation of Ash2l in NSPCs impairs neurogenesis during postnatal development
We reasoned that Ash2l deletion might disrupt the proliferation and differentiation of NSPCs, leading to decreased neurons and defects in brain morphology in Ash2l-deficient mice. To examine whether Ash2l regulates NSPC homeostasis, we first verified its expression at the mRNA level. qRT-PCR suggested that the expression level of Ash2l was higher in NSPCs than in neurons and astrocytes (Fig. 1a). We then confirmed the expression of ASH2L proteins in NSPCs, as shown by Nestin-Td and SOX2 colocalization in the subgranular zone (SGZ) (Fig. 1b). To determine whether Ash2l is indispensable for neurogenesis during postnatal development, we deleted Ash2l by injection of tamoxifen (TAM) at different time points in vivo or (Z)-4-hydroxytamoxifen (4-OHT) in vitro, followed by assessment of postnatal neurogenesis in Nestin-CreERT2; Ash2lf/f mutant mice (hereafter referred to as Ash2l-iKONes-CreERT2) and control Nestin-CreERT2 mice (hereafter referred to as WT). After the infusion of 4-OHT, we observed depletion of ASH2L, as well as a significant reduction in the core subunits WDR5, RBBP5, and DPY30, in P7 Ash2l-iKONes-CreERT2 NSPCs compared to WT NSPCs by western blot (Figs. 1c and S2a).
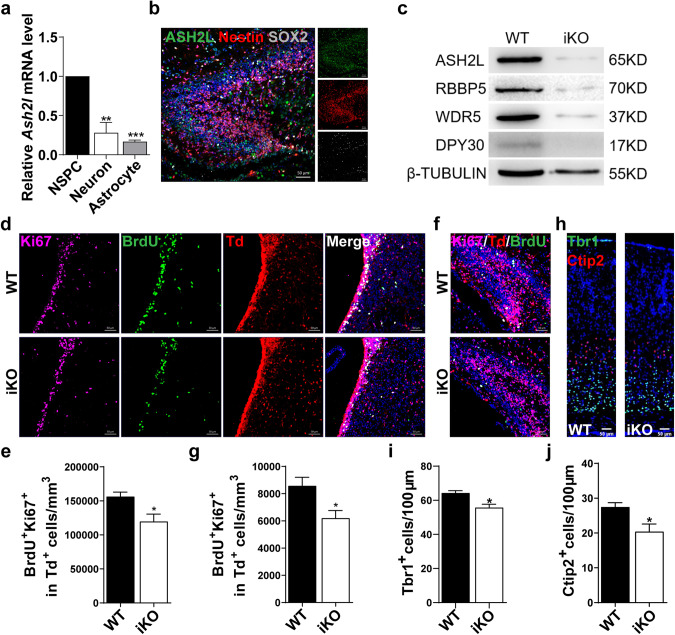
Fig. 1. Ash2l is essential for the maintenance of proliferation and differentiation of NSPCs.
a Expression level of Ash2l in NSPCs, neurons and astrocytes isolated from P0 mice measured by qRT-PCR. (n = 3 mice). **p = 0.0052, ***p < 0.0001. b Representative images of ASH2L and SOX2 immunostaining in the SGZ of P7 Nestin-CreERT2-Td mice. c Expression levels of the indicated WRAD components (ASH2L, WDR5, RBBP5, and DPY30) were quantified by western blot analysis of P7 NSPC protein lysates in WT and Ash2l-iKONes-CreERT2 mice treated with 4-OHT. (n = 3). Tubulin was used as the loading control. d, e Representative image of P7 WT and Ash2l-iKONes-CreERT2-Td brain sections stained with BrdU and Ki67 in the SVZ (d) and quantification of BrdU+ Ki67+ cells in the SVZ (e). (n = 3 mice). *p = 0.0324. f, g Representative images of P7 WT and Ash2l-iKONes-CreERT2-Td brain sections stained with BrdU and Ki67 in the DG (f) and quantification of BrdU+ Ki67+ cells in the DG (g). (n = 3 mice). *p < 0.05. h–j Representative images of P7 WT and Ash2l-iKONes-CreERT2 brain sections stained with Tbr1 and Ctip2 (h) and quantification of Tbr1+ and Ctip2+ neurons in the cortex (i, j). (n = 3 mice). *p < 0.05. Data information: The data shown are the mean ± SEM; Student’s t test.
To assess the function of Ash2l in postnatal neurogenesis, we generated a cohort of WT/Ash2l-iKONes-CreERT2-Td reporter mice by crossing WT/Ash2l-iKONes-CreERT2 with Rosa26 loxP-flanked stop tdTomato reporter mice. We injected TAM at P1 to induce Ash2l deletion and examined mice at P7 to assess whether Ash2l deficiency affects NSPC fate specification during postnatal development (Fig. S2b). We found that the numbers of BrdU+ Ki67+ proliferating Td-positive NSPCs in the SVZ/SGZ were significantly lower in Ash2l-iKONes-CreERT2-Td mice than in WT mice (Fig. 1d–g). Meanwhile, Ash2l-iKONes-CreERT2 mice also exhibited slightly decreased Tbr1+ and Ctip2+ neurons (Fig. 1h–j). These results suggest that Ash2l is indispensable for the proliferation and differentiation of postnatal NSPCs. Next, we examined whether and how Ash2l deletion in NSPCs affects adult neurogenesis (Fig. S2c, d). We observed a similar decrease in BrdU+ Ki67+ proliferating NSPCs in the SVZ/SGZ at P60 and P180 (Fig. 2a–f) upon Ash2l deletion. At both time points, 1 week after BrdU labeling, Ash2l-iKONes-CreERT2 mice exhibited significantly reduced numbers of DCX+ immature neurons in the DG, as well as reduced numbers of BrdU+ DCX+ neurons (Fig. 2g–k). We also observed decreased numbers of NeuN+ neurons at P180 (Fig. S2e, f). To exclude the possibility that these alterations may be due to increased cell death, we analyzed TUNEL+ apoptotic cells throughout the paradigm and observed no difference between WT and Ash2l-iKONes-CreERT2 mice (Fig. S2g, h).
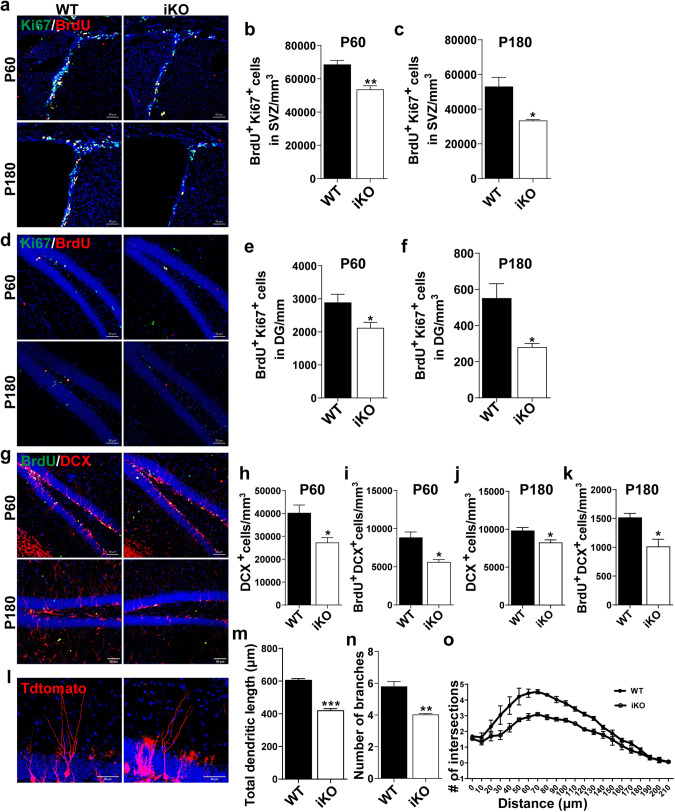
Fig. 2. Ablation of Ash2l in NSPCs impairs adult neurogenesis.
a–c Representative images of P60 (top) and P180 (bottom) WT and Ash2l-iKONes-CreERT2 brain sections stained with BrdU and Ki67 in the SVZ (a) and quantification of BrdU+ Ki67+ cells in the SVZ at P60 (b) and P180 (c). (n > 3 mice). **p = 0.0041, *p = 0.0202. d–f Representative images of P60 (top) and P180 (bottom) WT and Ash2l-iKONes-CreERT2 brain sections stained with BrdU and Ki67 in the DG (d) and quantification of BrdU+ Ki67+ cells in the DG at P60 (e) and P180 (f). (n > 3 mice). *p < 0.05. g, k Representative images of P60 and P180 WT and Ash2l-iKONes-CreERT2 brain sections stained with BrdU and DCX in the DG (g) and quantification of DCX+ immature neurons at P60 (h) and P180 (j) and BrdU+ DCX+ neurons at P60 (i) and P180 (k). (n = 3 mice). *p < 0.05. l Representative images of Td+ newborn neurons in the DG of WT and Ash2l-iKONes-CreERT2-Td reporter mice at 30 dpi after TAM injection. m–o Quantification of total dendritic length (m), dendrites (n) and dendritic complexity (o) of Td+ newborn neurons. (n = 30 per group). ***p < 0.001, **p < 0.01. Data information: The data shown are the mean ± SEM; Student’s t test.
Next, we examined the functional consequences of Ash2l ablation on adult-born neurons during dendritic development. Given that the morphology of dendritic adult-born neurons beyond 4-week-old is reported to be similar to that of 8-week-old neurons [27], we analyzed Td+ cells in the WT/Ash2l-iKONes-CreERT2-Td reporter mice at 30 dpi after TAM injection. In contrast to dendrite elaboration in WT, Ash2l-iKONes-CreERT2-Td reporter mice showed reduced total dendritic length, arborization, and dendritic complexity (Fig. 2l–o), indicating significant impairment of dendritic development of newborn neurons in the DG. Collectively, our findings suggest that Ash2l plays a critical role during postnatal and adult neurogenesis.
Ablation of Ash2l in NSPCs impairs learning and memory in adult mice
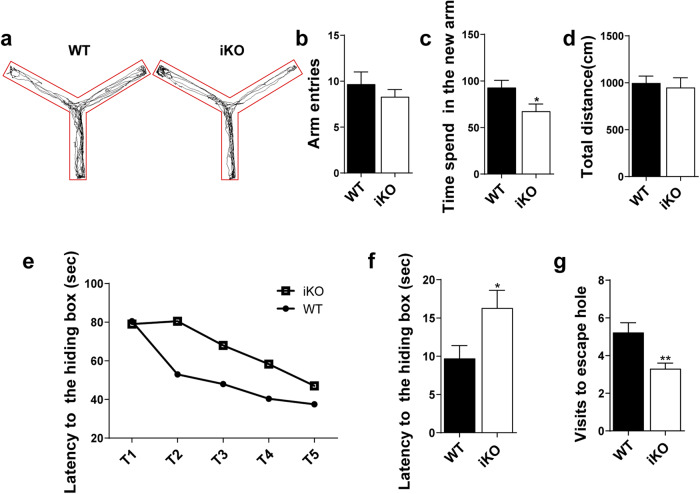
Adult neurogenesis is considered to be involved in human cognitive functions [28]. In particular, Ash2l mutations may lead to intellectual disability in humans [26]. To determine whether Ash2l depletion affects learning and memory, we conducted open field, Y-maze and Barnes maze behavioral tests in adult WT and Ash2l-iKONes-CreERT2 mice. First, the open field test results showed that there was no difference in the total moving distance, entries and time in the center zone, suggesting similar locomotive activities between the WT and Ash2l-iKONes-CreERT2 mice (Fig. S3a–d). In the Y-maze behavioral tests, the Ash2l-iKONes-CreERT2 mice entered the new arm fewer times than the WT mice and stayed in the new arm for a shorter time, but there was no significant difference in the total distance traveled between the two groups (Fig. 3a–d). Furthermore, we found that the depletion of Ash2l in NSPCs significantly impaired mouse performance in the Barnes maze, as they visited the target hole less often and displayed a significant delay in locating the hiding box (Fig. 3e–g). Our results suggested that the impairment of postnatal neurogenesis caused by Ash2l deletion leads to working memory deficits.
Fig. 3. Ash2l depletion impairs hippocampus-associated behaviors of adult mice.
a Representative motion trail from the Y-maze test. b, c Statistical analysis of entries into the new arm (b) and the time that mice stayed in the new arm (c) between WT and Ash2l-iKONes-CreERT2 mice. n = 8–10 mice, independent replicates. d The total traveling distance was not different between WT and Ash2l-iKONes-CreERT2 mice in the Y-maze test. e Primary latency to escape the hiding box during the training phase in the Barnes maze test. f, g Primary latency (f) and the number of visits to the target hole (g) in the Barnes maze test. n = 8–10 mice, independent replicates. *p < 0.05; **p < 0.01.
Dynamics of gene expression and histone marks are altered by Ash2l deletion
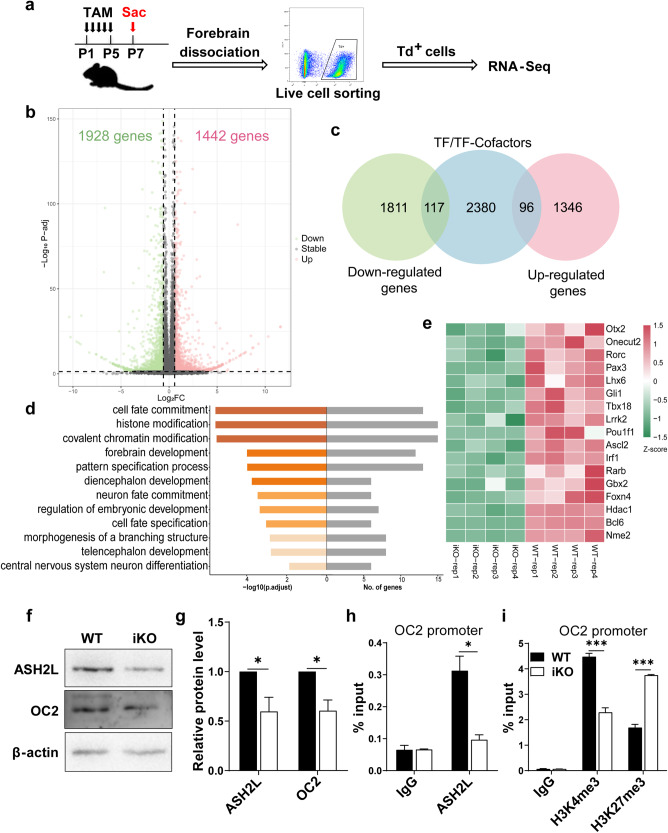
To investigate the molecular mechanisms underlying Ash2l-mediated gene regulation in NSPCs, we performed RNA sequencing (RNA-seq) of Td+ cells by using fluorescence-activated cell sorting (FACS) in P7 WT/Ash2l-iKONes-CreERT2-Td reporter mice to analyze transcriptome-wide changes upon loss of Ash2l (Fig. 4a). We found 1442 differentially expressed genes (DEGs) that were upregulated (p-adjusted < 0.05, fold change > 1.5) and 1928 that were downregulated (p-adjusted < 0.05, fold change < −1.5) upon Ash2l deletion in NSPCs (Fig. 4b). Transcription factors (TFs) and TF cofactors (TcoFs) are crucial for gene transcription in various cellular processes, including cell fate determination, such as NSPC fate specification [29–31]. Therefore, we focused on combined analysis of TFs/TcoFs among DEGs, and we identified 117 downregulated TFs/TcoFs and 96 upregulated TFs/TcoFs upon Ash2l deletion (Fig. 4c). Next, gene ontology (GO) analyses of downregulated TFs/TcoFs revealed enrichment for cell fate commitment, histone modification, forebrain development and neuron fate commitment-related categories (Fig. 4d). These data are consistent with a crucial role for the epigenetic factor Ash2l in NSPC fate determination. We next validated the expression of several key TFs/TcoFs that play pivotal roles in regulating NSPC fate by qRT-PCR. Primary NSPCs isolated from P7 WT/Ash2l-iKONes-CreERT2 mice were treated with 4-OHT to induce Ash2l deletion. Among genes with expression changes in P7 NSPCs after Ash2l deletion (Figs. 4e and S4a), the transcription factor one cut domain family member 2 (Onecut2, OC2) was significantly downregulated in the Ash2l-deficient NSPCs, as verified by western blot and qRT-PCR (Figs. 4f, g and S4a). OC2 has been shown to widely regulate the expression of genes associated with cell proliferation, migration, adhesion, differentiation and cell material metabolism [32]. Notably, OC2 is known to play important roles in cell fate determination, suggesting that OC2 may be involved in Ash2l-mediated NSPC fate specification.
Fig. 4. Global changes in gene expression and histone marks upon Ash2l deletion.
a Experimental scheme for tamoxifen injection and isolated Td+ cells by FACS of P7 WT and Ash2l-iKONes-CreERT2-Td reporter mice. b Volcano plot showing differentially expressed genes in P7 Td+ cells between WT and Ash2l-iKONes-CreERT2-Td reporter mice. (p-adjusted < 0.05, fold change > 1.5 or fold change < −1.5). c Venn diagram between differentially expressed genes in (b) and TFs/TcoFs. d Bar plots depicting the top 12 significantly enriched GO terms (biological processes, BP) of downregulated TFs/TcoFs in (c). e Heatmap of representative downregulated TFs/TcoFs in (d). f, g Western blot analysis showed reduced protein levels of ASH2L and the downstream target OC2 in P7 Ash2l-iKONes-CreERT2 NSPCs compared to WT NSPCs. (n = 3 mice). ***p < 0.001, **p < 0.01. h ChIP-qPCR confirmation of decreased ASH2L enrichment at the promoter of OC2 upon Ash2l deletion in P7 NSPCs. ChIP samples were normalized to input samples and expressed as ‘% Input’. (n = 3 mice). *p < 0.05. i ChIP-qPCR analysis of decreased H3K4me3 and increased H3K27me3 enrichment at the promoter of OC2 upon Ash2l deletion in P7 NSPCs. ChIP samples were normalized to input samples and expressed as ‘% Input’. (n = 3 mice). ***p < 0.001. Data information: The data shown are the mean ± SEM; Student’s t test.
To investigate whether OC2 is directly regulated by Ash2l, we performed ChIP-qPCR analysis and found that ASH2L enrichment was notably decreased at the promoter of the OC2 gene in P7 Ash2l-deleted NSPCs relative to that in WT NSPCs (Fig. 4h). These findings strongly suggest that OC2 is a direct downstream target of Ash2l in NSPCs.
In light of the critical role of Ash2l in regulating the switch of bivalent histone marks, we hypothesized that Ash2l may regulate OC2 expression through modulation of bivalent histone marks. We then analyzed ChIP-seq datasets of NSPCs from our previous study (GEO: GSE172310) [33]. The binding profiles revealed that strong enrichment of H3K4me3 and H3K27me3 occupied the core promoter regions of OC2 in NSPCs (Fig. S4b). Consistent with these findings, our ChIP-qPCR analysis revealed that there was a significant decrease in H3K4me3 enrichment and an increase in H3K27me3 enrichment at the promoter of the OC2 gene in P7 Ash2l-deleted NSPCs compared to WT NSPCs (Fig. 4i). Together, these results suggested that ASH2L regulates OC2 expression by mediating bivalent histone marks at the OC2 promoter.
OC2 is an important regulator for proliferation and differentiation of NSPCs
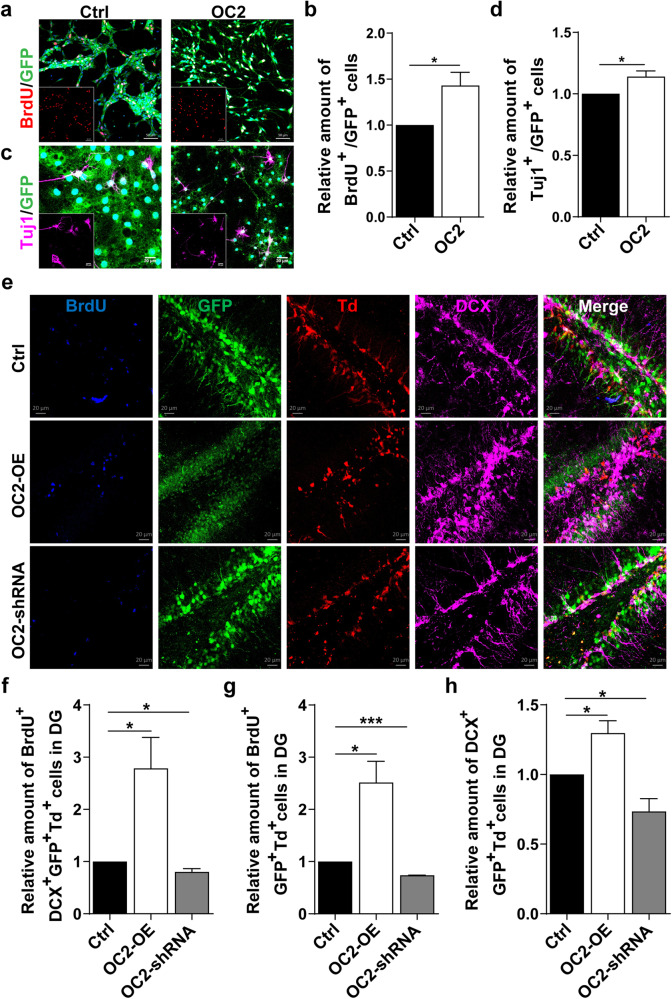
Due to the effect of OC2 on the fate specification of NSPCs has not been examined, we further verified whether modulating OC2 levels impacted NSPC proliferation and differentiation both in vitro and in vivo. We generated a recombinant lentiviral vector coexpressing OC2 and GFP, and western blotting and qRT-PCR confirmed that the expression of OC2 was significantly higher in P7 NSPCs infected with OC2 lentiviruses than in control cells (Fig. S4c–e). To determine the function of OC2, P7 NSPCs isolated from WT mice were infected with the lentiviral OC2 or vehicle control under proliferation cultures. The analysis results showed that OC2 overexpression resulted in a concomitant increase in BrdU+ NSPCs compared to the control (Fig. 5a, b). Next, we cultured P7 NSPCs under differentiation conditions, and the number of differentiated Tuj1+ neurons infected with lentiviral OC2 was higher than that of the control (Fig. 5c, d).
Fig. 5. Overexpression of OC2 promotes NSPCs proliferation and differentiation.
a, b Immunostaining with BrdU antibodies in P7 WT NSPCs infected with lentiviruses expressing control GFP or OC2-OE-GFP (a) and quantification of the proportion of BrdU+ cells among GFP+ NSPCs. (n = 3 mice) (b). *p = 0.0423. c, d Immunostaining with Tuj1 antibodies of P7 WT NSPCs induced differentiation for 10 days after infecting NSPCs with lentiviruses (c) and quantification of the proportion of Tuj1+ neurons among GFP+ cells (d). (n = 3 mice). *p = 0.0446. e Lenti-OC2-OE, lenti-OC2-shRNA, or control lenti-NC viruses were grafted into the hippocampus. Representative images of lentivirus-labeled GFP cells costained with BrdU and DCX in the DG of P60 Nes-CreERT2-Td reporter mice. f–h Quantification of BrdU-, DCX-, and BrdU DCX-positive cell numbers among total GFP+ Td+ cells in the DG. (n = 3 mice). *p < 0.05, ***p < 0.001. Data information: The data shown are the mean ± SEM; Student’s t test.
To ensure these changes in NSPC fate, we analyzed the proliferation and differentiation of NSPCs in vivo by stereotactically injecting lentiviruses into the DG of WT (Nes-CreERT2-Td) reporter mice to label NSPCs and euthanized mice 2 weeks after lentiviral injection (Fig. S4f). By evaluating the proportion of BrdU+ GFP+ Td+ proliferating NSPCs, DCX+ GFP+ Td+ immature neurons, and BrdU+ DCX+ GFP+ Td+ neurons, we observed increased proliferation and differentiation of NSPCs with OC2 overexpression compared to the control (Fig. 5e–h). Furthermore, to confirm that OC2 deficiency affects NSPC fate decisions in vivo, we analyzed the proliferation and differentiation of NSPCs by stereotactically injecting OC2-shRNA lentiviruses into the DG. The proportion of BrdU+ GFP+ Td+ proliferating NSPCs, DCX+ GFP+ Td+ immature neurons, and BrdU+ DCX+ GFP+ Td+ neuron differentiation of NSPCs infected with lentiviruses expressing OC2-shRNA was decreased (Fig. 5e–h). These data suggest that OC2 deficiency tips NSPC proliferative capacity and neuronal differentiation both in vivo and in vitro. Altogether, our data indicate that OC2 is an important regulator of the proliferation and differentiation of NSPCs.
OC2 is a main downstream effector of Ash2l to maintain NSPC fate determination
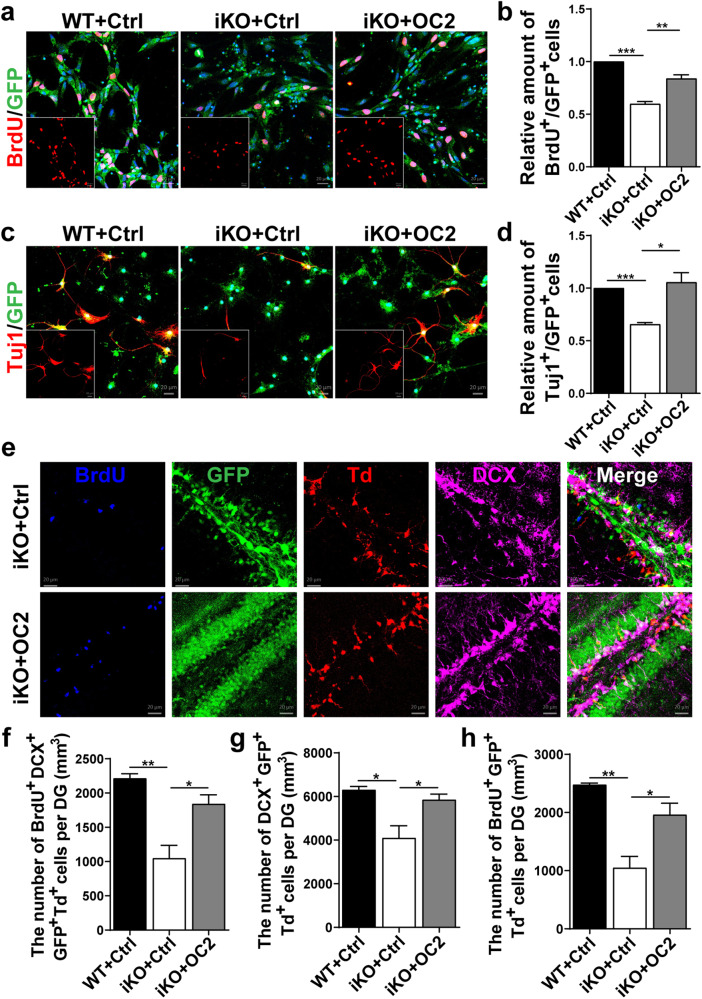
To determine whether Ash2l regulates NSPC fate decision through OC2, we reintroduced OC2 into Ash2l-ablated NSPCs to investigate whether the defects were recovered. We infected Ash2l-deleted NSPCs with lentiviruses that expressed either OC2 and GFP or a control GFP. We found that overexpression of OC2 in Ash2l-depleted NSPCs restored the phenotypes associated with Ash2l deficiency compared to the control (Fig. 6a–d). Next, to consolidate these findings in vivo, we stereotactically injected lentiviruses into the DG of WT/Ash2l-iKONes-CreERT2-Td mice (Fig. S4f). The numbers of BrdU+ GFP+ Td+ proliferating NSPCs, DCX+ GFP+ Td+ immature neurons, and BrdU+ DCX+ GFP+ Td+ neurons were increased in Ash2l-iKONes-CreERT2-Td mice with OC2 overexpression compared to mice with control lentiviral injection (Fig. 6e–h). Together, these results indicate that OC2 is a critical downstream effector for Ash2l-mediated proliferation and differentiation of NSPCs during the postnatal stage.
Fig. 6. OC2 is a main downstream effector of Ash2l and maintains NSPC fate determination.
a, b Immunostaining with BrdU antibodies in P7 WT and Ash2l-iKONes-CreERT2 NSPCs infected with lentivirus-labeled GFP cells (a) and quantification of the proportion of BrdU+ cells among GFP+ NSPCs (b). (n = 3 mice). ***p < 0.001, **p < 0.01. c, d Immunostaining with Tuj1 antibodies of P7 WT and Ash2l-iKONes-CreERT2 NSPCs induced differentiation for 10 days after infecting NSPCs with lentiviruses (c) and quantification of the proportion of Tuj1+ neurons among GFP+ cells (d). (n = 3 mice). ***p < 0.001, *p < 0.05. e Representative images of lentivirus-labeled GFP cells costained with BrdU and DCX in the DG of P60 Ash2l-iKONes-CreERT2-Td reporter mice. f–h Quantification of BrdU+, DCX+, and BrdU+ DCX+ cells among total GFP+ Td+ cells in the DG. (n > 3 mice). **p < 0.01, *p < 0.05. Data information: The data shown are the mean ± SEM; Student’s t test.
Inhibition of TGF-β signaling restores defective neurogenesis in Ash2l-deficient mice
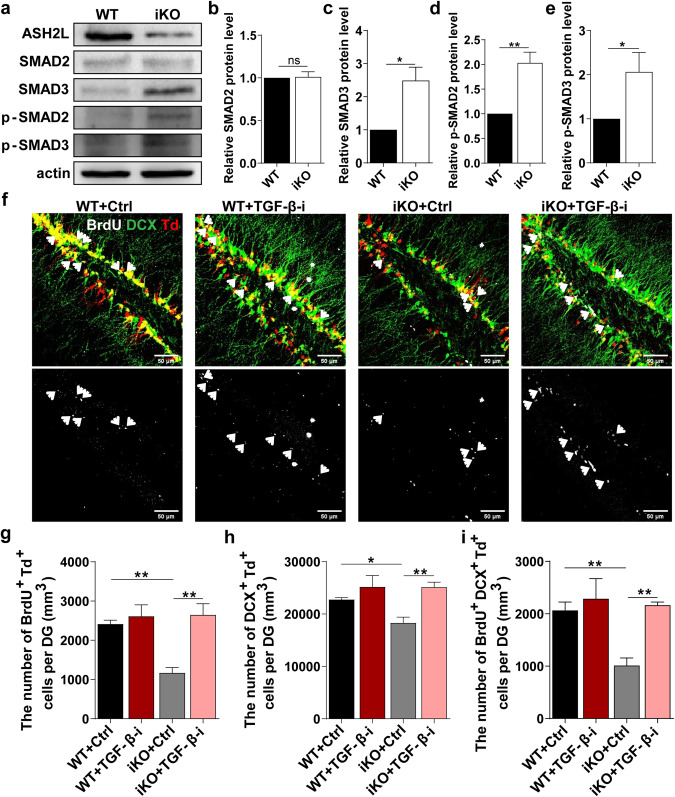
Previous studies have indicated that OC2 inhibits activin/TGF-β signaling in the parenchyma to allow normal hepatocyte differentiation [34]. TGF-β signaling plays an important role in development, patterning, and homeostasis, including cell lineage specification [35–37]. Thus, we hypothesized that Ash2l may control NSPC fate determination through OC2-mediated TGF-β signaling. To test this idea, we first examined the protein levels of SMAD2/3 in P7 NSPCs, the critical components of the TGF-β signaling pathway. We found increased phosphorylation of SMAD2/3 in Ash2l-iKONes-CreERT2 NSPCs (Fig. 7a–e), indicating activation of the TGF-β signaling pathway. These results prompted us to investigate the functional role of the TGF-β pathway in NSPCs by pharmacologically inhibiting TGF-β activation with LY364947 [38]. Quantification of BrdU+ Td+, DCX+ Td+, and BrdU+ DCX+ Td+ cells in the DG showed that LY364947 treatment markedly increased proliferation and differentiation in Ash2l-iKONes-CreERT2-Td reporter mice (Fig. 7f–i). These results indicate that inhibition of TGF-β activation rescues defective proliferation and differentiation in NSPCs caused by loss of Ash2l.
Fig. 7. Inhibition of TGF-β signaling restores defective adult neurogenesis in Ash2l-deficient mice.
a Western blot analysis of SMAD2, SMAD3, p-SMAD2, and p-SMAD3 levels in P7 WT and Ash2l-iKONes-CreERT2 NSPCs. b–e Quantification of (a). (n > 3 mice). **p < 0.01, *p < 0.05. f Representative images of Td+ cells costained with BrdU and DCX in the DG of P60 WT and Ash2l-iKONes-CreERT2-Td reporter mice injected with DMSO and the TGF-β inhibitor LY364947. g–i Quantification of BrdU+, DCX+, and BrdU+ DCX+ cells among total Td+ cells. (n > 3 mice). **p < 0.01, *p < 0.05. Data information: The data shown are the mean ± SEM; Student’s t test.
Discussion
Ash2l coordinates the proliferation and differentiation capacities of NSPCs
NSPCs, the primary source of neurons and glial cells, exist at various locations and times throughout embryonic and adult development [39]. Understanding the mechanisms underlying the developmental regulation of NSPCs has profound therapeutic implications for neurological diseases [40, 41]. Here, we established novel Ash2l ablation mouse models in which Ash2l was conditionally deleted in NSPCs throughout postnatal neurogenesis. Our work highlights a crucial role for Ash2l in maintaining the ability of NSPCs to proliferate and differentiate during postnatal, young, and adult neurogenesis. Upon Ash2l depletion in NSPCs, the fate determination of NSPCs and dendritic development of newborn neurons in the postnatal hippocampus were perturbed, leading to impaired ability of learning and memory in adult mice. Mechanistically, we demonstrate that Ash2l regulates essential genes involved in neuron lineage commitment and cell fate specification, likely through modulation of bivalent histone marks, and that Ash2l-mediated transcriptional regulation of OC2 is integrated into the TGF-β signaling pathway during neurogenesis. Taken together, our study offers new molecular insights into the developmental regulation of NSPC proliferation and differentiation, and the Ash2l-OC2-TGF-β signaling axis may provide therapeutic potential for neurological diseases.
Ash2l modulates the cell fate determination of NSPCs in a stage-dependent manner
Previous reports showed that conditional deletion of Ash2l at E10.5 using D6-Cre resulted in hydrocephalus and thinning of the cortical plate. Moreover, loss of Ash2l leads to more severe defects in cell cycle progression and cell apoptosis at E16.5 than at E14.5 [25]. In our study, we established a novel Ash2l ablation model in which Ash2l was deleted at E9.5 using Emx1-Cre transgenic mice. We observed markedly decreased and misplaced layer I–VI cortical neurons, malformation of the cortex, and unidentifiable DG regions in the Ash2l-cKOEmx neocortex, thus strongly supporting the notion that Ash2l is essential for proper neurogenesis. Importantly, we further showed that Ash2l coordinates the proliferation and differentiation of NSPCs in postnatal neurogenesis using a Nestin-CreERT2 mouse-crossed Ash2l ablation model. It is worth noting that a previous report found a striking increase in apoptotic cells at E16.5 [25], which was not observed in NSPCs from animals carrying postnatal deletion of Ash2l in our study. Thus, we propose that Ash2l modulates the proliferation, differentiation, and apoptosis of NSPCs in a developmental stage-dependent manner. Further studies are needed to dissect the molecular and cellular mechanisms by which Ash2l regulates NSPC transition throughout embryonic and postnatal development.
Ash2l modulates cognitive functions in mice
In our Ash2l-deleted model, the deficits in learning and memory phenotypes we observed are consistent with previous findings that Ash2l dysfunction is associated with intellectual disability in humans, as identified by rare variant analyses of Mendelian neurologic disease [26]. We propose that the decline in neurogenesis and abnormalities in DG neurons might explain the behavioral phenotypes because DG neurogenesis is necessary for learning and memory [42]. Moreover, newborn neurons have been shown to be essential for hippocampal functions [43, 44]. In particular, we demonstrated that depletion of Ash2l affected the morphology of neurons and reduced the number of NeuN+ neurons in the DG. Neuronal dendrites are an important part of the connections and functional architecture of neurons, and thus, dendritic defects are likely to cause neurodevelopmental disorders [45]. These findings are consistent with the phenotype of neuronal changes we observed in Ash2l-iKONes-CreERT2 mice. We used the Ash2l-iKONes-CreERT2-Td reporter mouse model to conduct neuronal morphology experiments and found that the absence of Ash2l reduced the total dendritic length, arborization, and dendritic complexity of newborn neurons in the DG. In addition, Y-maze and Barnes maze behavioral tests of mice revealed learning and memory deficits after Ash2l deletion. Our experimental results could inspire ideas that the decreased neurogenesis caused by Ash2l depletion may disturb neuronal functions in the hippocampus and cause a series of abnormal cognitive behaviors.
Ash2l regulates TGF-β signaling in NSPCs through OC2
Previous work has implicated Wnt signaling in the early neurodevelopmental defects of Ash2l-deleted brains [25]. However, it remains to be determined what downstream effectors mediate the function of Ash2l in NSPCs. In our study, we demonstrate that the defective postnatal proliferation and differentiation observed in Ash2l-deficient mice was rescued by overexpression of OC2. Moreover, we first demonstrate that OC2 is an important regulator of the proliferation and differentiation of NSPCs, in which overexpression of OC2 promotes cell fate determination. OC2 has been shown to regulate early retinal cell fates during development and to induce neuronal characteristics in fibroblasts [46, 47]. Our observations that OC2 is a direct downstream target gene of Ash2l and that overexpression of OC2 enhanced the capacity of proliferation and differentiation in NSPCs add new molecular insights into the function and regulation of OC2 in neuronal cell fate determination. Moreover, OC2 was reported to control liver cell fate decision through TGF-β signaling [34]. Our work reveals that Ash2l modulates NSPC fate determination by TGF-β signaling in postnatal neurogenesis. The TGF-β family is a key regulator of stem-cell state from the earliest stages of development to homeostasis of the adult organism. The multiple roles of TGF-β signaling are determined by the interactions between transcription factors and signaling receptors that define each cell type in combination with additional signals from their niches [48]. Indeed, we found that Wnt3 was downregulated in P7 Ash2l-deficient NSPCs (Fig. S4g), which suggests a potential role of Wnt signaling in NSPCs. Given that both the Wnt and TGF-β pathways regulate cell fate, proliferation, and differentiation, it comes as no surprise that there is cross-talk at multiple levels between these two pathways [49], which may coordinate with combinatorial activities of other signaling pathways to maintain stem cell homeostasis [50]. For example, in midbrain development, signaling through the canonical TGF-β pathway inhibits Wnt-induced proliferation and expansion of neuroepithelial cells and NSCs of early brain development [48], which suggests that the cross-talk between TGF-β and Wnt pathways plays an essential role in switching on the genetic machinery initiating cell fate in the brain. A better understanding of these mechanisms will open new opportunities for the development of targeted therapeutic approaches against neurological diseases in the future.
Taken together, our studies uncover Ash2l as a crucial regulator for modulating proliferation and differentiation in NSPCs and normal cognitive functions. Our data demonstrate that Ash2l, OC2, and TGF-β form a regulatory network that controls the cell fate determination of NSPCs. These findings represent an important step toward unraveling the molecular mechanisms that underlie NSPC biology and may offer new regenerative potential of stem cell therapy to repair the damaged brain.
Materials and methods
Animals
Mice were kept under standard laboratory conditions. Mice were bred and maintained at a constant temperature with a 12-hour light/dark cycle. All procedures and husbandry were approved by the Animal Committee of the Institute of Zoology, Chinese Academy of Sciences. Generation of Ash2lf/f mice: Mice with a “Knockout-first” allele (REF: A conditional knockout resource for the genome-wide study of mouse gene function) for generating the Ash2l conditional knockouts were generated by CAM-SU Genomic Resource Center of Soochow University. Briefly, ES cells (Clone ID: EPD0127_3_B02) harboring a loxP-flanked exon 4 of Ash2l within a L1L2_Bact_P targeting cassette (frequently used in the EUCOMM/KOMP-CSD resource), which itself is flanked by FRT sites were microinjected into C57BL/6N blastocysts. Male chimeras with germline transmission were used to generate mice with the targeting cassette, which was then removed through mating with FLPeR mice (129S4/SvJaeSor-Gt(ROSA)26Sortm1(FLP1)Dym/J) to obtain the conditional-ready allele (the floxed allele, Ash2lf). Ash2lf/+ mice were interbred to obtain Ash2lf/f and littermate controls. Mice carrying floxed Ash2l alleles with Emx1-Cre transgenic mice, Nestin-CreERT2 mice used for experiments were used to monitor Cre-mediated recombination. The sequences of genotyping primers for Ash2l were: forward, 5′- ACTAGCCAGTACAGCCAATGAGT-3′; reverse, 5′ GCACAGCTCCGAACGACG-3′.
Mouse neural stem/progenitor cell cultures
Mouse primary NSPCs were isolated as previously described [10]. Mouse brains were harvested, and then sections containing the hippocampus were collected in ice-cold phosphate-buffered saline (PBS, RSBM). The pooled sections were digested with 0.25% trypsin (Gibco) for 10 min at 37 °C. Trypsin was inactivated by incubation with DMEM (Invitrogen) containing 10% fetal bovine serum (Gibco). Tissue was triturated by pipetting up and down and passed through a 40 μm cell strainer to obtain single cells. Cells were suspended and then centrifuged at 400 g for 5 min at room temperature. After washing twice, the cells were resuspended in proliferation medium containing neurobasal medium (Gibco), 1% B27 (Gibco), 1% GlutaMAX (Invitrogen), 1% penicillin–streptomycin (HyClone), 20 ng/mL FGF (PeproTech), and 20 ng/mL EGF (PeproTech) in a 5% CO2 incubator at 37 °C. Half of the medium was changed every 2 days. The differentiation medium was made up of 2% B27, 1% GlutaMAX, 1 mM sodium pyruvate (Sigma-Aldrich), 5% fetal bovine serum, and 1% penicillin–streptomycin in neurobasal medium. For all experiments, neurospheres were trypsinized, and cells were plated as monolayers on either glass coverslips or directly on polystyrene tissue culture plates coated with 10 μg/mL poly-l-ornithine (Sigma-Aldrich) and 5 μg/mL laminin (Invitrogen).
Tamoxifen administration
To induce recombination, 1-month-old mice were intraperitoneally injected with tamoxifen (Sigma-Aldrich) once every 2 days 3 times and then once every week 2 times (180 mg/kg i.p., 30 mg/mL in 10% EtOH/Sesame oil, Sigma-Aldrich). Postnatal mice received a tamoxifen injection from P1 to P5 (90 mg/kg i.p.).
BrdU administration
For analysis of NSPC proliferation, mice were intraperitoneally injected with BrdU (Sigma-Aldrich) at a dosage of 100 mg/kg body weight and analyzed 2 h after injection. For analysis of NSPC differentiation, mice were injected with 50 mg/kg BrdU intraperitoneally 6 times (every twice/day) in 3 days and were analyzed 1 week later to identify BrdU-positive adult-born cells.
Immunostaining
Anesthetized mice were perfused with saline followed by 4% paraformaldehyde (PFA, in 0.1 M phosphate buffer, pH 7.4, Aladdin). Mice brains were removed, postfixed in 4% PFA at 4 °C for 12 h, and then dehydrated in 30% sucrose in PBS at 4 °C. Brains were sectioned into 40–50-μm-thick floating coronal slices using a sliding microtome (Leica SM2010 R). The embryonic and P7 brains were cut into 15 μm sections mounted on a freezing microtome (Leica CM 1950). For immunostaining, the brain slices were washed in PBS, permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) for 15 min, blocked in a buffer containing 3% bull serum albumin (Easybio) and 0.3% Triton X-100 for 1 h and incubated with primary antibodies at 4 °C overnight. The brain slices were washed with PBS and incubated with Alexa Fluor-conjugated secondary antibodies at a concentration of 1:500 for 1.5 h at room temperature. The sections were washed three times and mounted using adhesion antifade medium.
TUNEL assay
The TUNEL assay was performed using the One Step TUNEL Apoptosis Assay Kit (Beyotime) according to the manufacturer’s instructions. Briefly, sections were fixed with fixation solution and permeabilized by TrionX-100. Then, slides were incubated with TUNEL reaction mixture for 1 h at 37 °C. Samples were counterstained with DAPI and analyzed with a fluorescence microscope.
RNA sequencing and bioinformatics analyses
Brains of P7 Nestin-CreERT2; Ash2lf/f; tdTomato and Nestin-CreERT2; tdTomato mice were intracardially perfused with saline and digested following the Papain Dissociation System (Worthington Biochemical Corporation). Single cells were obtained by scattering through repetitive pipetting in PBS and filtered through a 70 μm cell strainer (Falcon) before sorting. tdTomato+ cells were sorted by using MoFlo XDP (Beckman Coulter), centrifuged for 5 min at 500 g and resuspended in TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. The quantity and quality of total RNA were assessed using a Nanodrop spectrophotometer (Thermo Scientific). RNA was amplified by the Smart-Seq2 method. The cDNA production was checked by Qubit 3.0 Fluorometer and Agilent 2100 Bioanalyzer to ensure the expected production, then the cDNA was sheared randomly by ultrasonic waves for Illumina library preparation protocol. After library preparation, PerkinElmer LabChip GX Touch and Step OnePlus Real-Time PCR System were introduced for library quality inspection. Qualified libraries were then loaded on the Illumina HiSeq platform for PE150 sequencing. Global transcriptome sequencing was conducted by Annoroad Gene Technology Co., Ltd. To minimize the effect of library construction bias and sequencing depth, we performed pseudo-replicates of FASTQ files according to a uniform processing pipeline (https://github.com/ENCODE-DCC/atac-seq-pipeline/blob/master/src/encode_task_spr.py) [51, 52]. Briefly, the four biological repetitions were combined together and then randomly divided into four equal parts that called pseudo-replicates [51, 52]. FastQC was used to assess sequencing quality control, and FASTX-Toolkit was used to remove the reads containing adapter sequences. Alignment-based quantification methods were applied to mappings processed with Salmon (v1.0.0, SAF Pattern). The clean reads were mapped to the reference genome via GENCODE vM23 for mouse. Differential gene expression was performed with DESeq2. Significantly differentially expressed genes were identified with p-adjusted < 0.05, fold change > 1.5, or fold change < −1.5. Gene function and pathway enrichment analyses were subjected to gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes. TPM (transcripts per million) was used to estimate the gene expression levels [53].
Chromatin Immunoprecipitation-qPCR
ChIP was performed as described previously [10, 54]. Briefly, NSPCs isolated from P7 WT and Ash2l-iKONes-CreERT2 mice were fixed with 1% formaldehyde (Sigma-Aldrich) for 10 min. After washing with PBS, NSPCs were suspended in cell lysis buffer (5 mM PIPES, pH 8.0, 85 mM KCl, 0.5% NP-40, and 1× proteinase inhibitor) twice, and then nuclei were lysed at room temperature with 500 μl of nuclei lysis buffer (50 mM Tris, pH 8.1, 10 mM EDTA, 1% SDS, and 1× protease inhibitor). DNA was sonicated for 30 min at 4 °C on high power to achieve fragments of sizes ranging from 200–500 bp, and then protein-DNA complexes were immunoprecipitated with 80 μl protein A-bound antibodies (5 μg) at 4 °C overnight. After incubation, chromatin pulled down by the Protein A–bound antibodies were washed sequentially two times each in IP dilution buffer, TSE-500 solution (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8.1, and 500 mM NaCl), LiCl wash solution (100 mM Tris, pH 8.1, 300 mM LiCl, 1% NP-40, and 1% deoxycholic acid), and 1× Tris-EDTA buffer (TE). Protein-DNA complexes were eluted from the protein A-agarose beads twice with IP elution buffer (50 mM NaHCO3 and 1% SDS) for 15 min with rotation. Formaldehyde-induced protein-DNA cross-linking was reversed by incubating protein-DNA complexes at 65 °C overnight. DNA was purified with phenol-chloroform-isoamyl alcohol (25:24:1) isolations and precipitated with two volumes of 100% ethanol and 10 μg linear acrylamide at –20 °C overnight. Purified DNA fragments were resuspended in nuclease-free water. For ChIP-qPCR, the primers used are listed below.
| Onecut2-Forward primer 5′ -3′ | CCGCCCCTTCTATAGCCATT |
| Onecut2-Reverse primer 5′ -3′ | GGAGCAGCCAACCTATCCTC |
qRT-PCR analysis
Total RNA was extracted from collected samples using TRIzol, and cDNA was synthesized using a Reverse Transcription Kit (TransScript One-Step gDNA Removal and cDNA Synthesis Kit, TransGen Biotech) according to the manufacturer’s instructions. cDNA was amplified using SYBR Premix Ex Taq (TliRNaseH Plus, Takara), and quantitative gene expression analysis was performed using the 2−ΔΔCT method. The mRNA expression levels were normalized to those of Actin and GAPDH. The primers used are listed in Supplementary Table 1.
Western blot analysis
Collected samples were lysed in ice-cold RIPA buffer (Beyotime) with protease inhibitors (Beyotime). Protein content was determined using a BCA Protein Assay kit (Beyotime) according to the manufacturer’s instructions. Protein samples were separated on 6–15% SDS-PAGE gels (Bio-Rad) and transferred onto PVDF membranes. Membranes were blocked in 5% milk in TBS-T (0.05% Tween-20) and incubated with primary antibodies overnight at 4 °C. Membranes were then washed and probed with HRP-coupled goat anti-mouse or HRP-coupled goat anti-rabbit (Easybio) for 2 h at room temperature. Membranes were detected with enhanced chemiluminescence reagent (ECL, Pierce). Protein bands were quantified using ImageJ software (NIH, USA). Quantification of protein was normalized using β-Actin or Histone 3. Original western blots for all relevant figures are shown in “Supplementary Material-Original Blots”.
Antibodies
The following antibodies were purchased from Cell Signaling Technology: ASH2L (#5019), Nestin (#4760 T), and Doublecortin (#4604 s). BrdU (ab6326), Tbr1 (ab31940), Ctip2 (ab18465), Tuj1 (801202) and Pax6 (901301) were purchased from Biolegend. Antibodies against Brn2 (sc-393324) and SOX2 (sc-17320) were purchased from Santa Cruz.
Plasmid construction and recombinant lentivirus production
The OC2 overexpression lentiviral construct (lenti-OC2-OE) was amplified by PCR from pECMV-Onecut2-m-FLAG of MiaoLingBio and subcloned into the pCD511B-copGFP vector (Youbio company, China) at the NotI/BamHI sites. The primers were as follows: forward, 5′- CGCGGATCCGCCACCATGAAGGCTGCCTACACCGCCTATCGATGC-3′; reverse, 5′ ATTTGCGGCCGCTCATGCTTTGGTACAAGTGCTGGAGGTGGA-3′. pCD511B-copGFP was used as the control for lenti-OC2-OE. For short hairpin RNA (shRNA)-mediated knockdown of OC2, a single-stranded oligonucleotide encoding the gene target and its complement was synthesized as follows (only the sense sequence is shown): 5′- GGAAGAGATCAACACCAAAGA -3′. The oligonucleotide pair was annealed and inserted into the pCD511B-copGFP lentiviral shRNA expression vector.
Lentiviral transfer vector DNA and packaging plasmid DNA (pMDLg/pRRE, pRSV-Rev and pMD2. VSV-G) were co-transfected into 293 T cells using polyethylenimine (PEI). The medium was collected and pooled at 48 and 72 h post-transfection, and filtered through a 0.22 µm filter and then concentrated by ultracentrifugation at 19,000 rpm for 2 h at 4 °C.
Behavior tests
Mice were moved to the testing room 24 hours before testing for acclimation. All experimental areas were cleaned with 75% ethanol between subjects and before the tests. Data were collected using the software Smart V3.3.03 (Harvard Apparatus, Holliston, USA).
For the open field behavior test, the arena was made of a piece of white plywood (72 cm long × 72 cm wide × 36 cm tall). A central zone (18 × 18 cm) was drawn in the middle of the open field chamber. Mice were placed in a corner of the open field chamber and allowed to freely explore for 10 min.
For the Y-maze behavior test, the procedure contained the training phase and the testing phase. During the training phase, each mouse was allowed to explore two arms of the Y-maze with the third arm (the ‘new arm’) blocked for 5 min. In the testing phase, mice were allowed to explore all three arms freely for 5 min during the testing phase [45, 55].
The Barnes maze test was performed in a 120 cm-diameter clear circular platform with 20 equally spaced holes (5 cm in diameter and 2 cm away from the edge) with only one hole leading to a removable hiding box located directly below the platform [56]. The BMT procedure consisted of three phases: habituation (day 1), training (days 2 and 3) and probe (day 5). On day 1, mice were placed in the center of the maze underneath a clear glass beaker while white noise was played. Mice were allowed to enter through the target hole into the hiding box after moving the glass beaker in 3 min. Mice that did not find the hiding box by the end of 3 min were gently guided to the hiding box and stayed for 30 s. In the training session, mice were placed in an opaque box for 15 s, and then mice were allowed to explore the maze for 2 min after moving the opaque box and turning on the buzzer. Three trials on training day 1 and two trials on training day 2 were conducted for each mouse. In the probe phase, the hiding box was removed. Mice were given 2 min to explore the escape hole during which the buzzer was turned on. Animal movements were recorded.
Stereotactic injection of viruses
In vivo lentiviral grating was performed as described [2]. Briefly, mice were anesthetized using tribromoethanol (Avertin, 375 mg/kg), and 1 μl of virus was injected stereotactically into the dentate gyrus using the following coordinates relative to bregma: anteroposterior, −2.0 mm; lateral, ± 1.7 mm; ventral, −1.9 mm (from dura). For each mouse, the control virus was injected into the left DG, and the OC2-OE virus was injected into the right DG.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 6. Student’s t-test was used to determine the statistical significance between different experimental groups, and the levels of significance were *p < 0.05, **p < 0.01, and ***p < 0.001. All values represent the mean ± SEM (standard error of the mean). For dendritic morphology analysis on 50 μm-thick floating brain sections, the tdTomato-labeled neurons were imaged on an LSM 880 confocal microscope (Zeiss) and analyzed by ImageJ software (NIH, Bethesda, Maryland, USA). Data were extracted for Sholl analysis and total dendritic length from each Td tomato-labeled neuron. At least 10 cells per group from at least three different animals were analyzed.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Author contributions
YJX and CML, conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; SKD, CHD, ZHZ, PPL, CL, HZD., and ZQT collection and assembly of data. XKL, SJH, and LL, generation of Ash2l conditional knockout mice.
Funding
This work was supported by grants from the National Key Research and Development Program of China Project (2021YFA1101400), the Informatization Plan of Chinese Academy of Sciences (CAS-WX2021SF-0301), the National Science Foundation of China (82271428), and the Open Project Program of State Key Laboratory of Stem Cell and Reproductive Biology.
Data availability
The RNA‑sequencing data have been deposited in the NCBI GEO database and are available under the accession number GSE234685. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethical approval
Our studies did not include human participants, human data, or human tissue.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhao-Qian Teng, Email: tengzq@ioz.ac.cn.
Chang-Mei Liu, Email: liuchm@ioz.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01189-y.
References
- 1.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu PP, Tang GB, Xu YJ, Zeng YQ, Zhang SF, Du HZ, et al. MiR-203 interplays with polycomb repressive complexes to regulate the proliferation of neural stem/progenitor cells. Stem Cell Rep. 2017;9:190–202. doi: 10.1016/j.stemcr.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Guo Y, Tang C, Han X, Xu M, Sun J, et al. Developmental cytoplasmic-to-nuclear translocation of RNA-binding protein HuR is required for adult neurogenesis. Cell Rep. 2019;29:3101–17.e3107. doi: 10.1016/j.celrep.2019.10.127. [DOI] [PubMed] [Google Scholar]
- 4.Montalban-Loro R, Lassi G, Lozano-Urena A, Perez-Villalba A, Jimenez-Villalba E, Charalambous M, et al. Dlk1 dosage regulates hippocampal neurogenesis and cognition. Proc Natl Acad Sci USA. 2021;118:e2015505118. [DOI] [PMC free article] [PubMed]
- 5.Khacho M, Harris R, Slack RS. Mitochondria as central regulators of neural stem cell fate and cognitive function. Nat Rev Neurosci. 2019;20:34–48. doi: 10.1038/s41583-018-0091-3. [DOI] [PubMed] [Google Scholar]
- 6.Liu PP, Xu YJ, Teng ZQ, Liu CM. Polycomb repressive complex 2: emerging roles in the central nervous system. Neuroscientist. 2018;24:208–20. doi: 10.1177/1073858417747839. [DOI] [PubMed] [Google Scholar]
- 7.Roidl D, Hacker C. Histone methylation during neural development. Cell Tissue Res. 2014;356:539–52. doi: 10.1007/s00441-014-1842-8. [DOI] [PubMed] [Google Scholar]
- 8.Kuroda MI, Kang H, De S, Kassis JA. Dynamic competition of polycomb and trithorax in transcriptional programming. Annu Rev Biochem. 2020;89:235–53. [DOI] [PMC free article] [PubMed]
- 9.Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell. 2017;171:34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Liu PP, Xu YJ, Dai SK, Du HZ, Wang YY, Li XG, et al. Polycomb protein EED regulates neuronal differentiation through targeting SOX11 in hippocampal dentate gyrus. Stem Cell Rep. 2019;13:115–31. doi: 10.1016/j.stemcr.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang H. The complex activities of the SET1/MLL complex core subunits in development and disease. Biochim Biophys Acta Gene Regul Mech. 2020;1863:194560. doi: 10.1016/j.bbagrm.2020.194560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee YT, Ayoub A, Park SH, Sha L, Xu J, Mao F, et al. Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin. Nat Commun. 2021;12:2953. doi: 10.1038/s41467-021-23268-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fossati A, Dolfini D, Donati G, Mantovani R. NF-Y recruits Ash2L to impart H3K4 trimethylation on CCAAT promoters. PLoS ONE. 2011;6:e17220. doi: 10.1371/journal.pone.0017220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harikumar A, Meshorer E. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep. 2015;16:1609–19. doi: 10.15252/embr.201541011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Wu X, Zhang H, Pfeifer GP, Lu Q. Dynamics of RNA polymerase II pausing and bivalent histone H3 methylation during neuronal differentiation in brain development. Cell Rep. 2017;20:1307–18. doi: 10.1016/j.celrep.2017.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bochynska A, Luscher-Firzlaff J, Luscher B. Modes of interaction of KMT2 histone H3 lysine 4 methyltransferase/COMPASS complexes with chromatin. Cells. 2018;7:17. [DOI] [PMC free article] [PubMed]
- 17.Stein AB, Jones TA, Herron TJ, Patel SR, Day SM, Noujaim SF, et al. Loss of H3K4 methylation destabilizes gene expression patterns and physiological functions in adult murine cardiomyocytes. J Clin Invest. 2011;121:2641–50. doi: 10.1172/JCI44641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertero A, Madrigal P, Galli A, Hubner NC, Moreno I, Burks D, et al. Activin/nodal signaling and NANOG orchestrate human embryonic stem cell fate decisions by controlling the H3K4me3 chromatin mark. Genes Dev. 2015;29:702–17. doi: 10.1101/gad.255984.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen K, Chen Z, Wu D, Zhang L, Lin X, Su J, et al. Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat Genet. 2015;47:1149–57. doi: 10.1038/ng.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell SA, McDonald CL, Krentz NAJ, Lynn FC, Hoffman BG. TrxG complex catalytic and non-catalytic activity play distinct roles in pancreas progenitor specification and differentiation. Cell Rep. 2019;28:1830–44.e1836. doi: 10.1016/j.celrep.2019.07.035. [DOI] [PubMed] [Google Scholar]
- 21.Pérez-Lluch S, Blanco E, Tilgner H, Curado J, Ruiz-Romero M, Corominas M, et al. Absence of canonical marks of active chromatin in developmentally regulated genes. Nat Genet. 2015;47:1158–67. doi: 10.1038/ng.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan M, Liang J, Xiong Y, Shi F, Zhang Y, Lu W, et al. The trithorax group protein Ash2l is essential for pluripotency and maintaining open chromatin in embryonic stem cells. J Biol Chem. 2013;288:5039–48. doi: 10.1074/jbc.M112.424515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai PH, Chien Y, Wang ML, Hsu CH, Laurent B, Chou SJ, et al. Ash2l interacts with Oct4-stemness circuitry to promote super-enhancer-driven pluripotency network. Nucleic Acids Res. 2019;47:10115–33. doi: 10.1093/nar/gkz801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luscher-Firzlaff J, Chatain N, Kuo CC, Braunschweig T, Bochynska A, Ullius A, et al. Hematopoietic stem and progenitor cell proliferation and differentiation requires the trithorax protein Ash2l. Sci Rep. 2019;9:8262. doi: 10.1038/s41598-019-44720-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Ruan X, Wen C, Chen P, Liu W, Zhu L, et al. The COMPASS family protein ASH2L mediates corticogenesis via transcriptional regulation of Wnt signaling. Cell Rep. 2019;28:698–711 e695. doi: 10.1016/j.celrep.2019.06.055. [DOI] [PubMed] [Google Scholar]
- 26.Karaca E, Harel T, Pehlivan D, Jhangiani SN, Gambin T, Coban Akdemir Z, et al. Genes that affect brain structure and function identified by rare variant analyses of mendelian neurologic disease. Neuron. 2015;88:499–513. doi: 10.1016/j.neuron.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao C, Teng EM, Summers RG, Jr., Ming GL, Gage FH. Distinct morphological stages of dentate granule neuron maturation in the adult mouse hippocampus. J Neurosci. 2006;26:3–11. doi: 10.1523/JNEUROSCI.3648-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kempermann G, Song H, Gage FH. Neurogenesis in the adult hippocampus. Cold Spring Harb Perspect Biol. 2015;7:a018812. doi: 10.1101/cshperspect.a018812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bond AM, Ming GL, Song H. Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell. 2015;17:385–95. doi: 10.1016/j.stem.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsieh J. Orchestrating transcriptional control of adult neurogenesis. Genes Dev. 2012;26:1010–21. doi: 10.1101/gad.187336.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alam T, Uludag M, Essack M, Salhi A, Ashoor H, Hanks JB, et al. FARNA: knowledgebase of inferred functions of non-coding RNA transcripts. Nucleic Acids Res. 2017;45:2838–48. doi: 10.1093/nar/gkw973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J, Li D, Jiang H. Emerging role of ONECUT2 in tumors. Oncol Lett. 2020;20:328. doi: 10.3892/ol.2020.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai SK, Liu PP, Du HZ, Liu X, Xu YJ, Liu C, et al. Histone crotonylation regulates neural stem cell fate decisions by activating bivalent promoters. EMBO Rep. 2021;22:e52023. [DOI] [PMC free article] [PubMed]
- 34.Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, et al. Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes Dev. 2005;19:1849–54. doi: 10.1101/gad.340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vander Ark A, Cao J, Li X. TGF-beta receptors: In and beyond TGF-beta signaling. Cell Signal. 2018;52:112–20. doi: 10.1016/j.cellsig.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Vogel T, Ahrens S, Buttner N, Krieglstein K. Transforming growth factor beta promotes neuronal cell fate of mouse cortical and hippocampal progenitors in vitro and in vivo: identification of Nedd9 as an essential signaling component. Cereb Cortex. 2010;20:661–71. doi: 10.1093/cercor/bhp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamaguchi M, Muramatsu R, Fujimura H, Mochizuki H, Kataoka H, Yamashita T. Circulating transforming growth factor-beta1 facilitates remyelination in the adult central nervous system. Elife. 2019;8:e41869. [DOI] [PMC free article] [PubMed]
- 38.Mirzamohammadi F, Papaioannou G, Inloes JB, Rankin EB, Xie H, Schipani E, et al. Polycomb repressive complex 2 regulates skeletal growth by suppressing Wnt and TGF-beta signalling. Nat Commun. 2016;7:12047. doi: 10.1038/ncomms12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urban N, Blomfield IM, Guillemot F. Quiescence of adult mammalian neural stem cells: a highly regulated rest. Neuron. 2019;104:834–48. doi: 10.1016/j.neuron.2019.09.026. [DOI] [PubMed] [Google Scholar]
- 40.Yang QQ, Zhai YQ, Wang HF, Cai YC, Ma XY, Yin YQ, et al. Nuclear isoform of FGF13 regulates post-natal neurogenesis in the hippocampus through an epigenomic mechanism. Cell Rep. 2021;35:109127. doi: 10.1016/j.celrep.2021.109127. [DOI] [PubMed] [Google Scholar]
- 41.Hamilton LK, Joppe SE, L MC, Fernandes KJ. Aging and neurogenesis in the adult forebrain: what we have learned and where we should go from here. Eur J Neurosci. 2013;37:1978–86. doi: 10.1111/ejn.12207. [DOI] [PubMed] [Google Scholar]
- 42.Cho KO, Lybrand ZR, Ito N, Brulet R, Tafacory F, Zhang L, et al. Aberrant hippocampal neurogenesis contributes to epilepsy and associated cognitive decline. Nat Commun. 2015;6:6606. doi: 10.1038/ncomms7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anacker C, Hen R. Adult hippocampal neurogenesis and cognitive flexibility - linking memory and mood. Nat Rev Neurosci. 2017;18:335–46. doi: 10.1038/nrn.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luna VM, Anacker C, Burghardt NS, Khandaker H, Andreu V, Millette A, et al. Adult-born hippocampal neurons bidirectionally modulate entorhinal inputs into the dentate gyrus. Science. 2019;364:578–83. doi: 10.1126/science.aat8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma H, Su L, Xia W, Wang W, Tan G, Jiao J. MacroH2A1.2 deficiency leads to neural stem cell differentiation defects and autism-like behaviors. EMBO Rep. 2021;22:e52150. [DOI] [PMC free article] [PubMed]
- 46.Sapkota D, Chintala H, Wu F, Fliesler SJ, Hu Z, Mu X. Onecut1 and Onecut2 redundantly regulate early retinal cell fates during development. Proc Natl Acad Sci USA. 2014;111:E4086–4095. doi: 10.1073/pnas.1405354111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Raadt J, van Gestel SHC, Nadif Kasri N, Albers CA. ONECUT transcription factors induce neuronal characteristics and remodel chromatin accessibility. Nucleic Acids Res. 2019;47:5587–602. doi: 10.1093/nar/gkz273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mullen AC, Wrana JL. TGF-beta family signaling in embryonic and somatic stem-cell renewal and differentiation. Cold Spring Harb Perspect Biol. 2017;9:a022186. [DOI] [PMC free article] [PubMed]
- 49.Kalkman HO. Altered growth factor signaling pathways as the basis of aberrant stem cell maturation in schizophrenia. Pharmacol Ther. 2009;121:115–22. doi: 10.1016/j.pharmthera.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 50.Xu X, Zheng L, Yuan Q, Zhen G, Crane JL, Zhou X, et al. Transforming growth factor-beta in stem cells and tissue homeostasis. Bone Res. 2018;6:2. doi: 10.1038/s41413-017-0005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan P, Liu Z, Song M, Wu Z, Xu W, Li K, et al. Genome-wide R-loop landscapes during cell differentiation and reprogramming. Cell Rep. 2020;32:107870. doi: 10.1016/j.celrep.2020.107870. [DOI] [PubMed] [Google Scholar]
- 52.Fu S, Wang Q, Moore JE, Purcaro MJ, Pratt HE, Fan K, et al. Differential analysis of chromatin accessibility and histone modifications for predicting mouse developmental enhancers. Nucleic Acids Res. 2018;46:11184–201. doi: 10.1093/nar/gky753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu C, Dai SK, Shi RX, He XC, Wang YY, He BD, et al. Transcriptional profiling of microglia in the injured brain reveals distinct molecular features underlying neurodegeneration. Glia. 2021;69:1292–306. doi: 10.1002/glia.23966. [DOI] [PubMed] [Google Scholar]
- 54.Liu C, Teng ZQ, Santistevan NJ, Szulwach KE, Guo W, Jin P, et al. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. 2010;6:433–44. doi: 10.1016/j.stem.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belforte JE, Zsiros V, Sklar ER, Jiang Z, Yu G, Li Y, et al. Postnatal NMDA receptor ablation in corticolimbic interneurons confers schizophrenia-like phenotypes. Nat Neurosci. 2010;13:76–83. doi: 10.1038/nn.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu C, Gao X, Shi RX, Wang YY, He XC, Du HZ, et al. Microglial transglutaminase 2 deficiency causes impaired synaptic remodelling and cognitive deficits in mice. Cell Prolif. 2023: e13439. 10.1111/cpr.13439. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA‑sequencing data have been deposited in the NCBI GEO database and are available under the accession number GSE234685. The data that support the findings of this study are available from the corresponding author upon reasonable request.