Abstract
Understanding the biological roles of all genes only through experimental methods is challenging. A computational approach with reliable interpretability is needed to infer the function of genes, particularly for non-coding RNAs. We have analyzed genomic features that are present across both coding and non-coding genes like transcription factor (TF) and cofactor ChIP-seq (823), histone modifications ChIP-seq (n = 621), cap analysis gene expression (CAGE) tags (n = 255), and DNase hypersensitivity profiles (n = 255) to predict ontology-based functions of genes. Our approach for gene function prediction was reliable (>90% balanced accuracy) for 486 gene-sets. PubMed abstract mining and CRISPR screens supported the inferred association of genes with biological functions, for which our method had high accuracy. Further analysis revealed that TF-binding patterns at promoters have high predictive strength for multiple functions. TF-binding patterns at the promoter add an unexplored dimension of explainable regulatory aspects of genes and their functions. Therefore, we performed a comprehensive analysis for the functional-specificity of TF-binding patterns at promoters and used them for clustering functions to reveal many latent groups of gene-sets involved in common major cellular processes. We also showed how our approach could be used to infer the functions of non-coding genes using the CRISPR screens of coding genes, which were validated using a long non-coding RNA CRISPR screen. Thus our results demonstrated the generality of our approach by using gene-sets from CRISPR screens. Overall, our approach opens an avenue for predicting the involvement of non-coding genes in various functions.
Keywords: Functional genomics, Long noncoding RNA (long ncRNA, LncRNA), Gene regulation, General transcription factor (GTF), Epigenetics, Gene function prediction, Coregulation of functions
Graphical Abstract

1. Introduction
A biochemical pathway in a cell includes the role of both coding and non-coding RNA (ncRNA). The functional role of ncRNA is prominently in the trans or cis-regulation of the coding genes whose products are the backbone of biochemical pathways [1]. The ncRNAs have various molecular mechanisms through which they exert their functions in myriad biological and cellular functions at multiple regulatory levels, making it harder to study their functions experimentally [2], [3]. Moreover, unlike protein-coding genes, the sequences of non-coding genes are most often not conserved across species, and finding their homologs is challenging [4]. Therefore, experimental validation of functions in model organisms for ncRNAs does not necessarily reflect their role in human cells. Low homology and low sequence conservation of multiple genes, including ncRNAs, also create hurdles for sequence-based prediction of their function, traditionally done by many scientific groups.
A promising way to dissect the functions of the genes is through computational analysis by leveraging the existing knowledge of gene-function or gene-disease relationships. Gene ontologies represent empirically annotated relationships between disease, functions, and genes. Multiple research groups have previously utilized these ontologies to predict genes' associations with functions and diseases [5]. Here we have used the word “function” to represent ontological gene-sets of molecular functions and biological processes for ease of reading. Predictive models are good at identifying similarities between data points and are extensively used in gene function identification by comparing the features of unknown and known genes. However, using the most relevant biological signals to train a predictive model is still crucial. The features of the functionally related genes must essentially represent the functional classes for robust prediction. A straightforward approach to gene function prediction is comparing the primary nucleotide and amino acid sequences of genes and proteins of known function with the genes of unknown function [5], [6], [7], [8]. However, it has been shown that alternative isoforms can be functionally divergent [9], and primary sequence comparison for non-coding genes would be of limited use because of the lack of reference to non-coding genes whose biological functions are known. Some researchers have used the ontological relationships of genes to identify disease-related ncRNA genes [10], but the number of annotations of ncRNA genes in the ontologies is less and would result in less coverage. Few studies have used gene expression data as features for identifying the functions of non-coding genes based on the co-expression of the coding genes [11]. Liao et al. constructed a co-expression network of coding and non-coding genes to predict the function of long non-coding RNAs (lncRNAs) [12]. However, a vast number of functionally unrelated genes can show co-expression at a given instant, and genes involved in the same pathway may not exhibit any correlation in expression [13]. Thus, current approaches could be ineffective in utilizing the right input genomic features to predict non-coding gene functions.
It is well known that non-coding RNAs (ncRNAs) regulate the transcription of genes involved in the same biological process through interactions with chromatin, RNA, and protein [14]. A few other computational approaches have also been proposed to predict the function of non-coding genes using different combinations of features. Utilizing lncRNA-protein interaction and protein-protein interaction network, Zhang et al. proposed a bi-random walk model, BiRWLGO, to predict the function of long non-coding RNAs [15]. PLAIDOH is a computational method that integrates transcriptome, subcellular localization, enhancer landscape, genome architecture, chromatin interaction, and RNA-binding data and generates statistically defined output scores for each lncRNA to functionally associate them to coding genes in different cancer conditions [16]. However, epigenome and TF-binding patterns at the promoters have not been explored properly for predicting ncRNA function. Epigenomic features, along with the binding of transcription factors (TFs) and cofactors, are present across coding and non-coding genes as they are required to modulate gene expression [17], [18]. Epigenetic marks and chromatin structure work in tandem with the TFs in the modulation of gene expression [19]. In the past, epigenome profiles have been used to predict gene expression [20] and the association between disease and single nucleotide polymorphism (SNP) [21]. At the same time, a few methods and studies that published TF ChIP-seq profiles have tried associating binding patterns of TFs with genes for individual functions [22], [23], [24]. Using TFs as features can also help make insights into the combinatorics (synergy and cooperativity) involved in regulating different functions [25]. However, a comprehensive analysis of combinatorics of binding patterns of large numbers of TFs at promoters and their associations with the function of genes has rarely been done.
Here, we devised an approach to use combinatorics of epigenome, TF-binding, and CAGE-tag patterns at promoters of genes to predict the ontology-based function of genes. Accordingly, to capture all the signatures of elements involved during the modulation events that would occur during the transcription of a gene, we leveraged a large number of publicly available ChIP-seq data of TFs, histone modification marks, and DNase I hypersensitivity sites along with cap analysis gene expression (CAGE) tags to include the expression of genes including non-polyadenylated ones. In order to gain more insight into the reliability of our method, we performed downstream analysis involving top predictive TF and cofactor binding profiles for clustering of functions and associating genes with those clusters of gene-sets. We also made insights into the specificity of simple combinatorics of TFs (i.e., TF-pair) towards functions.
2. Results
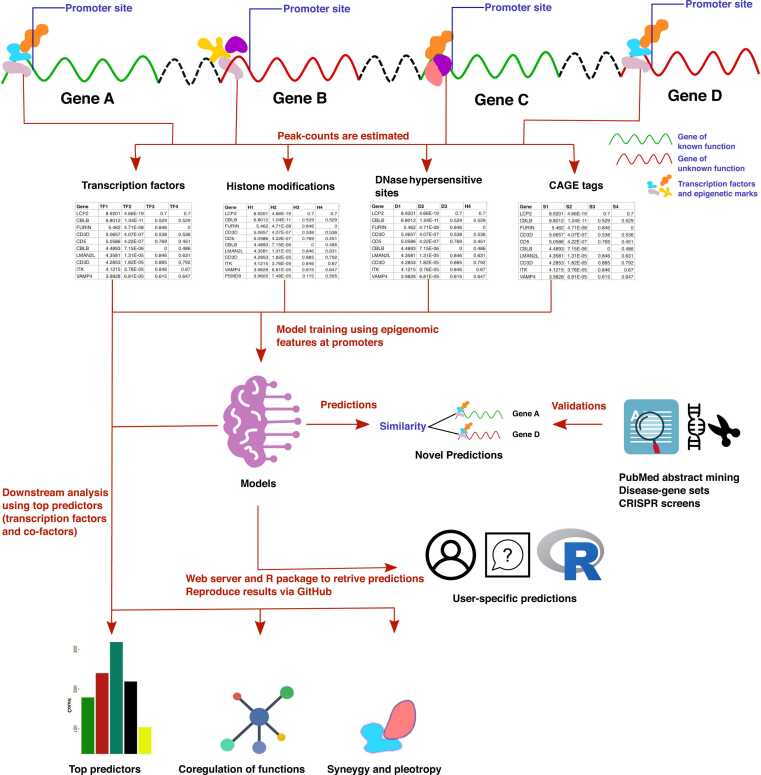
We developed our approach based on the hypothesis that coordinated expression of functionally associated genes is brought about by a few common key regulatory factors that are present across both coding and non-coding genes. We downloaded ChIP-seq profiles of TFs, histone modification marks, and DNase-seq and CAGE-tags from different sources and estimated their read-count within 1 Kbp of transcription start sites (TSS) of genes, in other words, 2Kbp wide region around promoter. The flowchart of our approach (GFPredict) is shown in Fig. 1.
Fig. 1.
Flowchart of our analysis to predict gene functions using epigenome and TF binding profiles.
2.1. Gene functions are predictable using the epigenomic and TF-binding signals at the promoter regions
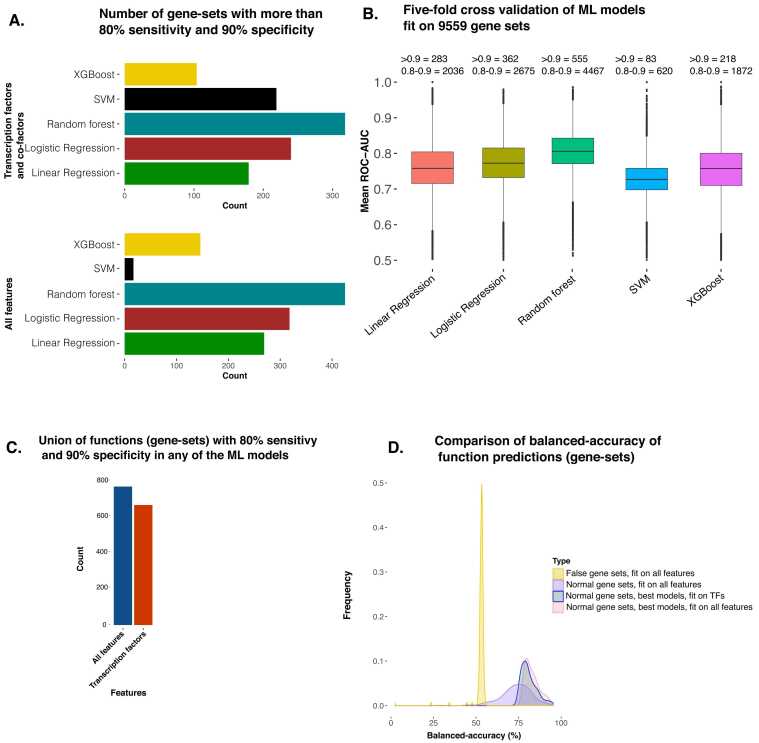
Machine learning (ML) algorithms were trained for each of the biological functions of the ontologies. We trained five different ML models using TFs binding patterns and other features (ChIP-seq data of histone modifications and cofactors, DNase hypersensitivity profile, and CAGE tags) for a total of 9559 function gene-sets downloaded from the MSigDB database [26]. We performed two approaches for predictive modeling. For the first approach, we used ChIP-seq profiles of TF and cofactor (n = 823) and histone modifications ChIP-seq (n = 621) and DNase-seq (n = 255) and CAGE tags (n = 255) from a total of 1954 non-diseased samples. While using this approach, we achieved very good predictions for many functions, such as using random forest; the sensitivity was above 80%, and the minimum specificity was 90% for 425 gene-sets. The other four ML models (Lasso-based linear regression, logistic regression with L2-regularization (ridge), SVM, and XGBoost) showed 100–300 gene-sets with a sensitivity of 80% and specificity of 90% (Fig. 2 A). Further, we found that AUROC (area under the receiver operating characteristics curve) for 555 gene-sets was greater than 0.9 (considered excellent) using the random forest model with a balanced test set (Fig. 2B). Whereas 4467 functions (gene-sets) had good AUC (between 0.8 and 0.9) with the random forest model on the balanced test sets [27].
Fig. 2.
An overview of the predictive power of epigenome profiles, especially transcription factor binding patterns at promoters for predicting gene function. A) Bar plot showing the number of functional gene-sets which had good predictions on the test set (80% sensitivity and 90% specificity) using five different machine learning (ML) models. The upper panel shows the number of functions with the good prediction by ML models using 853 transcription factor (TF) ChIP-seq profiles. The lower panel shows the ML models using five different types of profiles (TF, cofactor, and histone modifications ChIP-seq, DNase-seq, CAGE-tags). B) The box plots of AUC-ROC (area under the curve of receiver operating characteristic) for all gene-sets are shown for five ML models. The AUC values here are an average of five-fold runs for every gene-set. The number of gene-sets with AUC above 0.9 and between 0.8 and 0.9 is mentioned above the boxes. C) The bar plot shows the number of union sets of functions with good predictability (80% sensitivity and 90% specificity) using any of the 5 ML models. D) A plot to show the sanity of our approach. Here the density plot in yellow shows the distribution of balanced accuracy achieved with false gene-sets (gene-sets created by random sampling). Other density plots show the distribution of balanced accuracy achieved using empirically annotated gene-sets. The density plot for some functions with balanced accuracy above the 35 percentile among all the functions is also shown.
For the second approach, we used 823 TF and cofactor ChIP-seq libraries (736 TFs and 87 cofactors) from normal (non-diseased) samples for estimating feature scores. However, with this second approach, the number of functions with similar predictability did not reduce substantially. Using the threshold criteria of 80% sensitivity and 90% specificity, we had 318 functions using random forest. We took the union of functions with very good predictability (sensitivity > 80%, specificity > 90%) from 5 ML models. A total of 670 functions had very good predictability from at least one of the five ML models using only TF and cofactors. However, using all features (TF, Histone modification, CAGE-tags, DNase-seq), in the first approach, we had an increase of only 15% in the number of functions (total number = 773) (Fig. 2 C) with very good predictability (sensitivity > 80%, specificity > 90%) with at least one ML model. When we used the criteria of sensitivity greater than 70%( with specificity > 90%), the number of functions based on union from 5 ML models was above 1300 using the second approach (with TF and co-factor ChIP-seq) as features (see Supplementary Figure 1B in Supplementary File 1). The evaluation metrics for ML model fit on each gene-set, are provided in Supplementary File 2.
2.2. Non-random nature and relevance of high predictability
To ensure that the high predictability achieved using our approach is non-stochastic, we constructed a null model as a control. For this purpose, we checked if modeling on ‘false gene-sets’ is possible apart from the gene-sets annotated empirically. We created 200 false gene-sets by randomly shuffling the genes from existing gene-sets. The best-performing random forest algorithm trained on these false gene-sets showed an overall balanced accuracy of less than 55% on average. In comparison, the balanced accuracy of the models on the empirically annotated gene-sets is 75% on average, as shown in Fig. 2D. Our result indicates that good predictability is possible only for biologically relevant gene-sets, and there is an inherent pattern of regulation exhibited by a set of common regulators at the promoter sites of the genes associated with the same biological function.
3. Inference from clustering of functions
Further, we used a direct approach in studying the function coregulation due to the combinatorics of TFs to get an insight into major functional groups of coding and non-coding RNA. We performed clustering of functions with more than 60% confidence score (1423 gene-sets) using the shared top predictive TFs and cofactors (see Methods). We found 50 (Fig. 3 A) prominent clusters of functions (Supplementary File 3) based on shared top predictors. In addition, we found that in some clusters, the majority of the functions were either involved in similar major cellular activity (Supplementary File 3). Therefore, we manually curated and labeled the clusters with a major cellular process term. For example, one of the large clusters (cluster-47) is related to the cell cycle process and consists of members ‘regulation of cell cycle process’, ‘cytokinesis’, ‘microtubule-organizing center’, ‘nucleolus’, ‘regulation of cellular protein localization’ (Fig. 3 A). Some of the top predictive transcription factors and cofactors shared among the functions of cluster-47 are CTCF, XRN2, BRD4, SMARCA4, and PARP1 (Fig. 3B). The role of CTCF, MYC, PARP-1, and SMARCA4 in cell cycle regulation has been reported by previous studies [28], [29], [30], [31]. One other cluster (cluster-26) shown in Fig. 3 A consisted of early development and morphogenesis-related terms. Some of the shared top predictors for functions in cluster-26 included POU5F1 [32], RNF2, and SMARCB1 [33], [34], SIX1 [33], which are known to regulate genes involved in early development.
Fig. 3.
Clustering functions based on shared predictive TFs and cofactors ChIP-seq profiles reveal their potential overlap for major cellular processes A) tSNE plot and visualization of DBSCAN-based clustering of functions (gene-sets). Here, every dot in the tSNE plot shows a gene-set. The details about the two clusters are displayed as a heatmap showing the similarity in the number of common top predictors (ChIP-seq profiles in top 20 predictors). The two clusters are cluster-47, consisting of functions related to the cell cycle, and cluster-26, which is related to organ development. B) The dot plot shows the value of feature importance of ChIP-seq profiles of TFs and cofactors for functions belonging to cluster-47 (cell cycle-related functions). The feature importance value not lying in the top 20 is shown with a minimum dot size.
In some clusters, the members consist of unrelated ontological function terms but can have a non-discernible role in the overall major cellular process. For example, in cluster-47, the majority of the members of the clusters have an apparent role in the major cellular process– cell cycle activity but a gene-set ‘negative regulation of catabolic process’ (part of cluster 47) might look different. A detailed analysis reveals that, during the cell cycle, there is an increase in anabolic processes to build large molecules (DNA and structural components) [35], [36] needed for proliferation and reduction in the breakdown of protein complexes (catabolic digestion) [37]. Hence, “negative regulation of catabolic processes” could be a part of groups of functions involved in the cell cycle [37]. Such indirect role of functions in major cellular processes like reproduction (cluster-44) and immune system (cluster-7) can be deduced in other clusters (Supplementary File 3). Thus, the emergence of clusters of functions broadens the scope of linking gene-sets to major cellular processes and provides an opportunity to study the specificity of the binding patterns of the regulators (TFs and cofactors) at the systems biology level.
4. Independent validations and comparison with other methods
4.1. Pubmed abstract mining of co-occurrence of gene names and function term
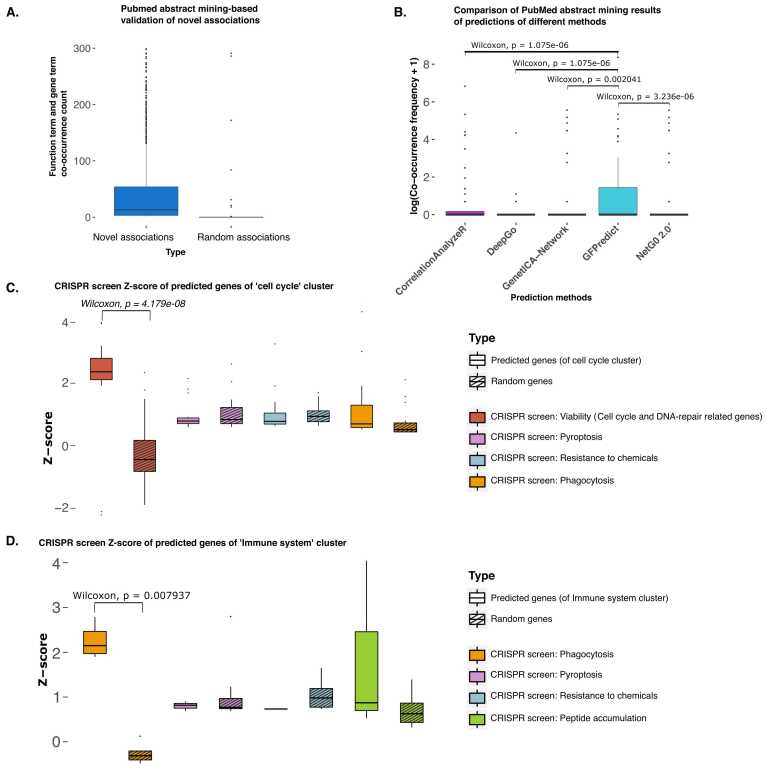
To check if the predicted results are of any biological relevance, the co-occurrence of the predicted gene term and the corresponding biological function term of the ontology was searched in the abstracts of the PubMed articles published from 1990 to 2021. The boxplot in Fig. 4 A shows the total co-occurrence of predicted gene term and function term pairs compared against random gene term and random function term pairs as control. This result adds to the confidence in our predicted results. We also corroborated our prediction with known disease-gene associations (see Supplementary Methods and Supplementary Tables 1 and 2. in Supplementary File 1).
Fig. 4.
Validation of predictions of novel association between function and genes. A) The box plot shows the frequency of co-occurrence of function terms and corresponding gene names in PubMed abstracts. The left box plot shows the frequency of the novel predictions made by GFPredict, while the right one shows random pairs of functions and genes. The novel and random associations between function and genes were not present in the gene-sets we used for training or testing. B) Benchmarking and comparison for five different methods for finding associations between functions and genes. C) Validation using CRISPR screen for ‘Viability’ function for genes predicted to be part of a gene-sets belonging to a cluster associated with a major cellular process, “cell cycle process.” In the corresponding study, authors found that genes with high CRISPR z-score for viability were mostly associated with cell cycle and DNA-repair [38]. The stripped bars indicate the score of random genes, and the non-stripped bars indicate predicted genes' scores. The difference between z-scores for predicted genes (for cell cycle associated cluster) and random genes is not high in other CRISPR screens for ‘pyroptosis,’ ‘resistance to chemicals, and ‘phagocytosis.’ D) Validation using CRISPR screen for the function of ‘Phagocytosis’ for genes predicted to be part of gene-sets of the cluster associated with the ‘immune system.’ Phagocytosis is an important part of immunity [39]. The stripped bars indicate the score of random genes, and the non-stripped bars indicate predicted genes' scores. The difference between z-scores for predicted genes (for the immune system) and random genes is not high in other CRISPR screens for ‘pyroptosis,’ ‘resistance to chemicals, ’ and ‘peptide accumulation.’.
4.2. Comparison of predicted results with other gene function prediction methods
Gene function prediction is one of the classical problems in computational biology. Some of the recent methods to predict the ontology-based functions of genes have utilized different features like primary amino acid sequence (NetGo 2.0, DeepGo), gene expression (correlation AnalyzeR), and network inference using co-functionality of genes obtained through transcriptomic profiles (GenetICA-Network) [40], [41], [42], [43]. We compared the abstract mining results on the predictions of these methods against the novel associations inferred by our approach (Fig. 4B). The co-occurrence of input ontology term and predicted gene term at least once in the PubMed abstracts for randomly selected 20 genesets (see Methods) of our method is significantly more compared to the same input gene terms and their predicted ontology terms by AnalyzeR, DeepGo, NetGo 2.0, and GenetICA-Network [40], [41], [42], [43].
5. CRISPR-based validation of association of genes with major cellular processes of clusters of functions
Our approach of grouping functions based on common top predictors (TFs or cofactors) leads to new ways of finding links (direct and indirect associations) between coding and non-coding genes with few major cellular processes. In order to evaluate the results of discovering such new links between genes and major cellular processes, we analyzed available CRISPR screens. First, we used the CRISPR screen: ‘viability’ in human pluripotent stem cells (hPSC), where the hPSC-enriched essential genes appeared to be mainly encoding transcription factors and proteins related to the cell cycle and DNA-repair [38]. The novel predicted genes in functions belonging to cluster-47 (mainly associated with the cell cycle process) had significantly higher z-scores compared to an equal number of random genes in the same CRISPR screen for the viability of hPSC (Fig. 4 C). However, the novel predicted genes for cluster-47 had comparatively less z-scores in other CRISPR screens: ‘pyroptosis’, ‘resistance to chemicals’, and ‘phagocytosis’ [44], [45], [46].
In another validation, we found a higher difference between the z-score for predicted genes for the gene ontology term ‘immune effector process’ and random genes in the CRISPR for phagocytosis compared to other CRISPR screens (see Supplementary Figure 2.). Further, we validated cluster-7 consisting of functions labeled to be involved in the major cellular process ‘immune system’ (Supplementary File 3). The novel predicted genes of cluster-7 had higher z-scores compared to an equal number of random genes in the same screen for phagocytosis, which is considered a fundamental process of immunity [39]. However, the same novel predicted genes for cluster-7 had comparatively less z-scores in other CRISPR screens (see Fig. 4D) [44], [46], [47]. CRISPR screens' validations assert the associations of novel genes with major cellular processes and link the underlying regulatory factors (top predictors) to those biological processes.
6. Explainability through insight into the association of binding patterns of TF-pairs with functions
The PubMed-based abstract mining result and model’s performance metrics indicate the reliability in the prediction of our approach; however, there could still be a need to study combinatorics of TF-binding for better explainability of the predictions. Hence, we tried to understand the simplest combinatorics of TF-pair binding patterns to gain more interpretability and reliability in our approach. TFs exhibit pleiotropic effects, meaning TFs can have multiple biological functions [48]. As expected, a few TFs had high feature importance scores (from the random forest model) for many functions [49]. To analyze the predictive pleiotropy of TF-pairs, we searched for TF ChIP-seq pairs (from the same cell-types), which emerged as top predictors of different functions. A few TF ChIP-seq pairs were among the top predictors of multiple functions (Fig. 5 A).
Fig. 5.
Insight into the co-occurrence of Transcription factor (TF) pairs among predictors and their synergy. A) The count of functions (pink) and the clusters of functions (green) for which TF ChIP-seq pairs appeared among the top 20 predictors in the same cell type. The panel on the right shows the same counts as a scatter plot. The TF-pairs shown with symbols are C3: E2F4-GATA1, C4: MAZ-GATA1, F3: ZNF366-SPI1, F4: SPI1-STAT1. B) Heatmap showing the significance of overlap of TF ChIP-seq peaks in GM12878 cells at promoters. C) The box plot of values of significance (-log(P-value)) of overlap of promoter-peaks of TF ChIP-seq pairs in GM12878 cells which appeared together as top predictors in one or more functions. On the right is the box plot of the significance of the overlap of promoter peaks for random pairs of TF ChIP-seq profiles in GM12878 cells.
The occurrence of a TF-pair among the top important features across multiple biological functions indicated their pleiotropic predictive power [50] (Fig. 5 A). Further, for every TF pair, we checked for the diversity of functions for which they were top predictors. For diversity estimation, we counted TF-pairs occurrences in the clusters of co-regulated functions (Supplementary File 4). As expected, we found that TF-pairs appeared to have predictive pleiotropy for many functions but had less diversity regarding clusters of co-regulated functions. One of the reasons for such a reduction of diversity of clusters of function for TF-pairs could be that the clustering was done based on common top predictors. However, as described above, the clusters highlighted the coherence of member functions for major cellular processes. Therefore, the diversity of pleiotropic predictive power of TF-pairs using clusters was needed to understand their regulatory effect better. ChIP-seq patterns of BATF and RUNX3 at promoters in B cells (GM12878) appeared together among the top 20 predictors for 11 functional gene-sets. However, these 11 gene-sets belonged to only 2 clusters of functions mainly involved in immune cell activation and differentiation (Supplementary File 5). Similarly, DNA-binding profiles at promoters in adipocytes by CEBPA and E2F4 appeared to be top predictors for 8 gene-sets (functions) belonging to a single cluster and mainly associated with response to the stimulus by peptides (like insulin), monosaccharides, and related metabolic functions. Thus, the utility of our analysis highlighting some TF-pairs with more specificity toward certain clusters of functions is that it can help to confirm the prediction of involvement of coding and non-coding genes for a few major cellular processes. However, a group of TF-pairs was the top predictor of the gene-sets belonging to more diverse clusters of co-regulated functions. Especially pairs involving CTCF showed more diversity in a cluster of co-regulated functions. The TFs, ZCAN5FB, and CTCF appeared as the top predictors for functions belonging to more than 12 different co-regulated clusters. Similarly, TET3 and CTCF appeared as the top predictor of functions from 6 different clusters. CTCF is known to have a more general effect than other TFs in addition to its general role as insulator [51]. Nevertheless, its co-occurrence with certain TFs as the top predictor also highlights another possible role in various cellular processes. Furthermore, the same analysis for pleiotropy and diversity was repeated for TF-pairs ChIP-seq profiles of different cell-types (see Supplementary Figure 3 A in Supplementary File 1).
In order to further make insight into the non-random aspect of the co-occurrence of TF-pairs (Fig. 5B) as top predictors, we investigated the overlap of the peaks of their ChIP-seq profiles in the GM12878 cell line. It was based on the notion that if TF-pair occurrence as top predictors has no relation with corresponding biological processes, then the overlap of their peaks would appear as a random event. For this purpose, we used the R package ‘ChIPpeakAnno’ [52] and analyzed TF ChIP-seq peaks in the GM12878 cell line. We compared the overlap of co-predictor TF-pair ChIP-seq peaks in the same cell-type with random TF-pairs as control. Here, co-predictor TF-pair were defined as pairs of TF ChIP-seq profiles in GM12878 cells, which appeared among the top 20 predictors (co-predictive) for any function (Supplementary Figure 3B in Supplementary File 1). We found that the enrichment of overlap of peaks at promoters in GM12878 for such co-predictive TF-pairs was much more significant than random TF ChIP-seq pairs (Fig. 5 C). Such observations build confidence in our approach and indicate that the top predictors’ analysis offers insights into TF-TF synergy through higher co-binding frequency at promoters of the genes involved in the same biological functions.
7. Broader applicability of GFPredict and its utility for predicting functions of non-coding RNAs
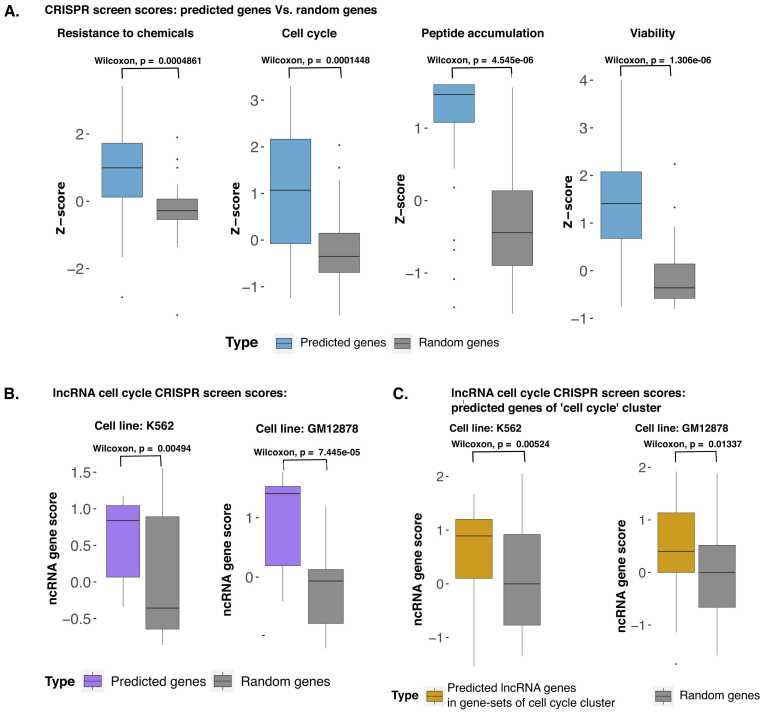
Our results hint at the reliability of our approach for ontology-based function prediction and dependence on binding patterns of TFs. The framework of GFPredict allows for its generalization to apply to the other biologically relevant gene-sets. A vast amount of literature highlights different gene-sets associated with various phenotypes and biological functions. We further focused on a useful application to meet the need for reliable prediction and cost-effective validation of gene-function associations using a smaller set of experiments. We used published CRISPR screen datasets to demonstrate the utility of our approach. We chose the top 50 genes from each CRISPR screen dataset as positives, and the negatives were non-positive random genes in the training data. After training GFPredict, we chose the top predicted 30 genes and validated them using their available CRISPR screen scores. Such as, when we used the top 50 positives from the CRISPR screen for resistance to chemicals (in fibroblasts) [53] to train GFPredict, the top 30 predicted genes for the same function had significantly higher (P-value < 0.004) scores than the random 30 genes in same CRISPR screen [53]. Similarly, in the cell cycle CRISPR screen, the top 30 predicted genes had a significantly higher (P-value < 1e-4) score than the random 30 genes [38]. We have shown results for two more CRISPR screen-based analyses in Fig. 6A. Overall such results show that GFPredict can be used to predict related genes to any of the biologically relevant gene-sets in addition to its utility using traditional ontological functions.
Fig. 6.
Validation of predicted coding and non-coding genes using CRISPR screens. A) CRISPR scores of the top 30 predicted genes from the GFPredict model, which was trained on the top 50 genes of CRISPR screens against CRISPR scores of random genes. The top 30 predicted genes were not part of the training set. B) CRISPR scores of lncRNA genes among the top 30 predicted genes in the lncRNA-CRISPR-screen for cell cycle by GFPredict trained using the top 50 positive coding genes of a different cell-cycle CRISPR screen (Yilmaz et al.). Among the top 30 predicted genes, there were 15 lncRNA genes. C) CRISPR scores of 52 lncRNA genes predicted to be in the cluster with the major cellular process, cell cycle (custer-47 shown in Fig. 3.), compared against the scores of random genes in lncRNA CRISPR screen for cell cycle.
7.1. Application of expanding small CRISPR screens for non-coding genes function prediction
The GFPredict-based analysis enabled the prediction of the association of 1200 long non-coding genes with various biological processes and molecular functions (see Table 1 and Supplementary Table 3 in Supplementary File 1). In order to test the reliability of the function prediction of ncRNAs, we designed a suitable evaluation method. We trained GFPredict on the top 50 genes from the cell cycle CRISPR screen consisting mainly of coding genes [38] and further validated the predictions using a different CRISPR screen built to identify lncRNA genes involved in the cell cycle [54]. The lncRNAs in the top 30 genes predicted to be associated with the cell cycle by GFPredict had significantly higher CRISPR-screen scores than random sets of the same size in two (GM12878, K562) of the cell lines (see Fig. 6B).
Table 1.
List of predicted functions of non-coding RNAs with experimental evidence.
| Ontology | Predicted non-coding gene | Literature evidence |
|---|---|---|
| GO_STEROL_HOMEOSTASIS | LINC02356 | [55] |
| GO_HEART_DEVELOPMENT | AP001528/ENSG00000280339 | [56] |
| GO_EYE_DEVELOPMENT | AC078909.1 | [57] |
| GO_PHOSPHOLIPID_METABOLIC_PROCESS | ENSG00000257023 | [58] |
| GO_SYNAPSE_ORGANIZATION | MIR4281 | [59] |
| GO_NEURON_MATURATION | ENSG00000274367 | [60] |
| GO_NEGATIVE_REGULATION_OF_INTERLEUKIN_6_PRODUCTION | LINC00528 | [61] |
| GO_REGULATION_OF_HOMEOSTATIC_PROCESS | MIR658 | [62] |
| GO_KERATINOCYTE_DIFFERENTIATION | PAUPAR | [63] |
| GO_IN_UTERO_EMBRYONIC_DEVELOPMENT | MIR5001 | [64] |
Our approach for clustering functions based on co-predictive TFs and cofactors helped associate non-coding RNA with a few major cellular processes (Supplementary File 6). We further used ncRNA CRISPR screens to validate the association of ncRNAs to these major cellular processes (see Fig. 3 A). Hence we choose ncRNAs predicted to be part of gene-sets of cluster-47, which is labeled to be associated with the cell cycle (see Supplementary File 3 and Fig. 3). The ncRNA genes predicted to be part of gene-sets in cluster-47 had a substantially higher (p-value < 0.01) CRISPR screen score for the cell cycle in comparison to random genes (see Fig. 6C). Our analysis and validation indicate an impactful application of our approach that our model, trained using a CRISPR screen of coding genes, can be used to predict functions of non-coding genes for which CRISPR screens are rarely available.
8. Discussion
To predict the function of non-coding RNAs, researchers would have to use new assays or genomic features in prediction systems. Here we have shown the feasibility of exploiting TF-binding profiles as features because they are a common set of regulators across coding and non-coding genes involved in the same function, as shown by our results (Fig. 6B) [65].
Using the union of different ML models, we achieved very good predictability (sensitivity > 80% and specificity > 90%) for more than 780 functions with all features and 650 functions using 853 TFs and cofactors ChIP-seq (736 TFs and 87 cofactors). With random forest ML models, for more than 50% of functions (5022 out of 9559 gene-sets), we achieved a minimum of 0.8 AUROC, often considered to represent good prediction [27]. We independently validated our results using different datasets. We also compared predictions of GFPredict to other methods’ predictions that use various other features such as primary amino acid sequence (NetGo 2.0, DeepGo), gene expression (correlation AnalyzeR), and gene-cofunctionality based network inference (GenetICA-Network) [40], [41], [42], [43]. Such comparison with the different features-based models highlights that our approach of using epigenome and TF-binding patterns at promoters can be a very effective feature for predicting the function of genes. Compared to other features, it also provides an additional benefit of predicting the function of non-coding RNA.
Further, in the downstream analysis, the clustering of functions revealed an interesting pattern. Most functions that shared the same top predictors (especially ChIP-seq profile from the same cell type) were related to similar major cellular processes through manual curation and labeling. Thus, despite having a seemingly unrelated biological role, functional gene-sets showed convergence in association with major cellular processes like cell cycle and transport. Such observation is because of shared similarity in patterns of some epigenomic and TF-binding features at promoters of genes. On the same logic, if a few epigenomic and TF-binding features appear to be important common determinants (or predictors) for two known gene-sets, the genes of those gene-sets could likely be involved in the same major function. In other words, our analysis goes beyond the boundary of currently defined gene-sets of function to highlight the effect of TFs. For example, for cluster-47, the top predictors are MYC, PARP-1, CTCF, and SMARCA4, which are involved in the cell cycle [28], [29], [30], [31]. There are few indirect studies on the coregulation of functions by the combinatorics of TF and cofactor binding [66]. However our study could be unique due to analysis of common top predictive TFs that show the interdependence between molecular or biological processes and may also explain the perturbation effect on a key regulator that can potentially affect a myriad of functions. Two major aspects highlight the novelty of our study: i) deciphering combinatorics of TF-binding at promoters for association with functions. ii) grouping known gene-sets using top co-predictors and finding common major cellular process terms for their groups. Such groups of functions with biological and molecular functions have the potential to provide a better explanation in CRISPR screens. CRISPR screens reveal the involvement of coding and non-coding genes in larger cellular contexts like cell cycle and resistance to pathogens [38], [67]. GFPredict’s derivable clusters of ontological functions representing major cellular processes (cell cycle and immune response) can further broaden the link between CRISPR screen genes and those specific ontological molecular and biological functions.
Overall downstream analysis shows the reliability and sensibility of our models, which is directly associated with the prediction of the function of non-coding RNAs. The clustering of functions also highlighted the broader role of a few non-coding RNAs (see Supplementary File 6). For example, the non-coding RNA genes–DLG1-AS1, UBL7-AS1, LINC00441, and LINC01137 were predicted to be associated with at least one of the members of the cluster of functions largely involved in cell cycle activity by our approach. Out of these six non-coding RNAs, DLG1-AS1 [68] and UBL7-AS1 [69] are reportedly involved in proliferation. The other two non-coding RNAs, LINC00441 [70] and LINC01137 [71] are reportedly involved in cancer development. Such inference about the role of non-coding RNAs in a few major cellular processes could help biologists design experiments for validation.
Our approach of combining the TF and cofactor binding pattern as features for gene-function prediction and clustering functions to understand the role of coding and non-coding genes stands out from the typical gene-function prediction methods. There are a few tools and methods for utilizing transcription factor ChIP-seq profiles in different ways, such as the Cistrome BETA suite [72], which predicts transcription factors' repressive or activating behavior. Similarly, Reshef et al. [73] published a method for signed linkage disequilibrium profile regression, which uses a TF-binding profile to identify genome-wide directional effects of functional annotations on diseases. Another tool called MAGIC [74] identifies TFs and cofactors responsible for patterns of gene expression changes between different conditions. However, there is rarely any study on the prediction of the function of coding and non-coding genes using TF-binding patterns at promoters. We could not find any study on the interpretation of the association of combinatorics of the TF-binding pattern at promoters with a cluster of functions, which indicates the uniqueness of our approach and analysis.
We have created a resource for the biologists to corroborate their experimental results and utilize our predictions to design the experiments to understand the molecular and biological roles of non-coding and coding genes. It is to be noted that the current version of GFPredict might not be accurate for all ontological gene-sets. As we have mentioned, out of 9559 gene-sets, our method has a minimum of 80% AUROC for only 5022 gene-sets (52%) using the random forest model. It could be due to three reasons; first, the number of positive genes for some functions could be too low for training the prediction model; second, the number of features (TFs, DNase-seq, CAGE-tags) were insufficient. The third reason could be that additional types of features, in addition to the TF-binding, epigenome, and CAGE-tags patterns, could be needed for many gene-sets. The inclusion of TF-binding signals at promoter-bound enhancers could further improve the prediction of gene function. With the improvement of the consensus list of enhancer-promoter interaction in the context of cell-types in the future, our method with minor modification (by adding enhancers) could become more accurate and useful. Nevertheless, our analysis provides a useful insight into epigenome and TF-binding patterns at the promoters of ncRNA genes, which is indicative and useful for inferring their functions. We hope that with the advent of new technologies of epigenome profiling like FloChIP [75] and multi-CUT&Tag [76], there would be an increase in epigenome profiles that would allow GFPredict to have high accuracy in predictions for more number of functions. Our study advocates using more epigenomic and TF-binding profiles to better understand non-coding RNAs' functions.
9. Methods
9.1. Epigenome and TF-binding features’ score calculation for promoters
Here, we considered each gene ontology as a class and corresponding empirically annotated genes as positive labels, and gene function prediction is treated as a classification problem. We used the read-counts of the epigenome and transcriptome binding profiling assays (DNAse-seq, ChIp-seq, CAGE-tags) as features. All the epigenome, TF, and cofactor ChIP-seq and CAGE-tags profiles were from human cells and tissues and aligned their reads to the hg19 genome version. For the estimation of binding scores at promoters, we counted the number of DNA fragments (read) lying within one kbp of gene transcription start sites (TSS) (see supplementary material for detail). We calculated the number of reads around TSS using ChIP-seq (TF, Histone modifications) and DNAse-seq profile from the ChIP-Atlas database [77] and CAGE-tags from the FANTOM5 database. We used the TSS of non-coding genes from gencode (V30) and RefSeq gene transcripts [78], [79]. For each gene, we allowed multiple transcripts as long as their TSS were at least 500 bp apart from each other. In total, we performed our analysis using 89747 promoter regions.
9.2. Prediction method
For each gene-set in the ontology, we considered genes annotated in them as positive and randomly picked an equal number of genes (not annotated in the same gene-sets) as negatives and gene function prediction is treated as a classification problem. Out of possible 50000 genes, if we assume that the expected number of positive unknown genes belonging to a function is less than 100. Then the background probability of being false negative for one randomly chosen gene would be less than 0.002 (100/500000). We choose an equal number of negatives to positives for each gene-set. Hence if for the same function, we have 50 known positive genes in the training set and 50 randomly chosen genes in the negative set, with a background probability of false negative as 0.002, the probability of one or more positives (false negatives) in a set of 50 randomly chosen genes (as negatives) would be less than 0.005 (using Binomial test). Based on such estimates, we relied on the set of random genes not belonging to a gene-set as negative-set for the corresponding function.
We divided the positives and negatives into a training set (75%) and a test set (25%). The 5 different machine learning models for each gene-sets are Random Forest, XGBoost, SVM (support vector machine), linear regression-based Lasso, and L2-regularization-based logistic regression (Ridge regression). Further, bootstrapping was done to calculate the standard deviation in the balanced accuracy by training the models for five iterations (see Supplementary Figure 4 in Supplementary File 1). We used various criteria to evaluate the test set's prediction: accuracy, balanced accuracy, F1-score and Mathew’s correlation coefficient (MCC), and error rate (Supplementary File 2). The Random Forest models were implemented using the ‘randomForest’ function from the ‘randomForest’ R package. XGBoost models were implemented using the ‘xgb.train’ function from the ‘xgboost’ R package. SVM models were implemented using the “svm” function from the “e1071” R package. Linear regression using ‘cv.glmnet’ (with alpha = 1) (Lasso) from ‘glmnet’ R package and logistic regression with L2-regularization (ridge regression model) was implemented using ‘cv.glmnet’ (with alpha = 0, family = "binomial") from ‘glmnet’ R package.
After evaluating the test set, we used the trained model to make predictions for all promoters (genes) in our list to find novel associations between function and genes. To have a stringent selection of novel/unknown gene-function association predictions, we calculated the confidence score for every function (gene-sets).
9.3. Calculating confidence score for gene-sets
To have robust predictions, we calculated the confidence score of predictions for each function.
For every function (gene-sets), each of their respective trained random forest models were used to predict probability scores (of belongingness to the gene-sets) for all 89747 promoters (genes). The confidence score of a gene-set is the maximum precision, i.e., the maximum value obtained on the ratio of the number of true positive genes to the number of predicted genes (true positive + false positive) by adjusting the threshold to classify the genes as positive or negative, using predicted probabilities of the genes. We have considered gene-sets having more than a 60% confidence score for our downstream analysis.
The prediction model evaluation metrics like accuracy, balanced accuracy, specificity, sensitivity, etc., the balanced datasets (equal number of positives and negatives) were used in training and test sets. Only for the estimation of the confidence score, probabilities of the positives, and all the non-positives (unbalanced datasets) were used.
9.4. Balanced accuracy calculation
Balanced accuracy is a metric used to judge the predictive power of a binary classifier. It is often used when there is an imbalance in the number of positive and negative (imbalanced classes). Balanced accuracy is calculated as the arithmetic mean of sensitivity and specificity:
9.5. Method to make inferences about top regulators
We made inferences about top regulators by estimating feature importance while training random forest models. This approach has also been used by GENIE3, a top performer in gene-network inference in the DREAM 5 challenge [80], [81], [82]. Here, instead of gene expression of transcription factors, we are using binding affinity to promoters as feature scores and predicting the belongingness of a gene to a class. Thus for every function, we chose the top 20 predictors with high feature importance calculated by the random forest-based approach.
9.6. Method for clustering functions
To infer clusters of functions (gene-sets), we first estimated similarity scores among functions. The similarity score or closeness among the two functions was defined as the number of the TF and cofactor ChIP-seq (SRX IDs) profiles among the top 20 predictors for both gene-sets in the same directionality with a penalty of the number of common TFs and cofactors (in the top 20) with opposite directionality. The similarity scores (or closeness) were reverted and translated to get distances among functions to apply tSNE-based dimension reduction. Hence the distance between gene-sets A and B was calculated as
| (1) |
Where,
| (2) |
Where,
TFi is one of the common TFs out of total n number of common TFs (TFn) in top 20 features in random forest model. The function is the correlation between the read-count score of TFi at promoters and the association of genes to function A. Thus if is positive, then TF is likely to be more enriched at promoters of genes belonging to function A. Thus we use the directionality of the association of function with the occurrence of top predictive TF to calculate the stringent closeness score. The cohesive index was also calculated for every cluster as the average distance between individual members (Supplementary File 3.).
Dimension reduction was done using the distance matrix, and density-based clustering was performed. For this purpose, the R package ‘Rtsne’ was used with the option ‘is_distance’ equal to TRUE. After low dimensional embedding, DBSCAN was used to find clusters of functions using the 2D embedding coordinates provided by ‘Rtnse’ [83], [84].
9.7. Method for Independent validations using PubMed abstract mining
To gain confidence in novel predictions and compare our approach with other methods, we used PubMed abstract-based validation. Here, the ontology term and corresponding predicted gene term are used as input. In order to have a good match of the ontology term in a potentially relevant abstract, the ontology terms were processed to remove stop words (Supplementary Methods in Supplementary File 1). The ‘Bio.Entrez’ package was used to search for the co-occurrence of the ontology term and its corresponding predicted gene term in the abstracts of the research articles in the PubMed database. As a control to this approach, ontology function terms were paired randomly with gene terms and searched for their co-occurrence with the same parameters.
9.8. Method for comparison of PubMed abstract mining results for predictions of different methods
A list of 50 genes predicted into randomly selected 20 gene-sets out of 1423 gene-sets with more than 60% confidence score (functions) was used as input of other methods–Correlation AnalyzeR, DeepGO, GenetICA-Network, NetGO 2.0 [40], [41], [42], [43]. For the Correlation AnalyzeR method, the R library package ‘correlationAnalyzeR’ was used, the ‘analyzeSingleGenes’ function from the package was used to predict the ontology-based labels for the list of 50 genes, and ontology labels with the highest score were considered as the final predicted label. If the prediction was not available by the method for a gene, its label was left blank.
For DeepGO, GenetICA-Network, and NetGO 2.0 methods, their respective web servers were used to get the predictions on the considered list of genes by feeding the relevant protein sequence FASTA files as input; the top listed isoform was considered from the Uniport database [85]. The prediction label with non-generic terms with the highest score from either Biological Processes or Molecular Functions section was considered the final label. For non-coding genes and coding genes with less than a 50% prediction score, their labels were left blank.
PubMed abstract mining was run on all the predictions of different methods to get the co-occurrence of the predicted ontology term and input gene term using the ‘Bio.Entrez’ package described above. Stop words (Supplementary Methods in Supplementary File 1) were filtered out from the input terms to avoid matching generic terms.
9.9. Methodology for transcription factors synergy and pleiotropy analysis
The occurrence of TFs and cofactors as top 20 predictors for the same and different cell-types across those biological functions with more than 60% confidence score was counted. Similarly, with the set of TFs used as features, a list of TF-pairs was constructed, and the occurrence of each TF-pair among the top 20 predictors across all the biological functions was counted.
9.10. Methodology for evaluation using CRISPR screens
Validation of genes predicted into the clusters ’cell cycle process’ and ‘immune system’ was done using CRISPR screens: viability (cell cycle and DNA-repair related genes) and phagocytosis [38], [45]. The genes which had insignificant p-value (>0.05) were removed. The z-scores of the genes predicted into the cell cycle and immune system clusters were checked against the z-scores of the randomly selected genes. The same procedure was used for other control CRISPR data-set shown in Fig. 4 C-D.
To demonstrate the broader applicability of GFPredict, the function ‘predict_related_genes’ with ‘ml_model = random.forest’, the top 50 genes of different CRISPR screens were used as training sets individually [47], [53], [54], [67], [86]. The models were fine-tuned by changing the n_bootstrap from 3 to 20 to sample more negative points to get the highest balanced accuracy. Among all the predicted genes after training the model using GFPredict, the top 30 predicted genes were selected, and their CRISPR scores were checked and compared against the CRISPR scores of random genes.
To validate the non-coding gene functions, lncRNA CRISPR screens were used by intersecting the non-coding genes predicted in cluster-47 (consisting of cell cycle-related functions) with the CRISPR screen genes, and their corresponding scores were compared against the random genes [54]. The two scripts (lncrna_crispr_validation.R, package_test_crispr.R) used for all the validation are available at https://github.com/reggenlab/GFPredict/tree/main/code.
The lncRNA gene scores, as defined by Liu et al. are as follows: screen score = scale[–log10(adjusted P)] + |scale[log2(sgRNA fold change)]|[54].
9.11. Availability of data and code
Profiles of ChIP-seq of transcription factors, histone marks, and DNase-seq were downloaded from the ChipAtlas database (https://chip-atlas.org/) in bedGraph format, which can be processed by extension of the DFilter tool (https://reggenlab.github.io/DFilter/). The CAGE-tags profiles were downloaded from the FANTOM database (https://fantom.gsc.riken.jp/data/). The read-counts of the epigenome profiles can be obtained using DFilter at https://reggenlab.github.io/DFilter/.
Our method, GFpredict, can predict genes functionally related to a user-provided biologically relevant list of genes. It is available as an R package, ‘GFPredict’.
The code and documentation are provided at https://github.com/reggenlab/GFPredict.
Predictions are available at http://reggen.iiitd.edu.in:1207/gfpredict_server_script/.
Funding and additional contributions
This work was supported by a fellowship provided by the University Grant Commission (UGC) of India to O. C. and computational resources funded by the Department of Biotechnology (DBT) of India under the grant ID. BT/PR40158/BTIS/137/24/2021 to the Department of Computational Biology, IIITD.
CRediT authorship contribution statement
Omkar Chandra: methodology, software, manuscript writing, figure preparation. Madhu Sharma: webserver, literature‐based validation. . Neetesh Pandey: literature-based validation, manuscript writing. Indra Prakash Jha: software testing, methodology. Shreya Mishra: webserver development, software. Say Li Kong: conceptualisation, manuscript revision. Vibhor Kumar: conceptualisation, methodology, manuscript writing, data curation, software.
Declaration of Competing Interest
Author declare no conflict of interest.
Acknowledgments
This work was supported by computing resources obtained from the shared facility of the Department of Computational Biology, IIITD, India. We acknowledge the support of the Department of Biotechnology (DBT), Government of India, under grant No. BT/PR40158/BTIS/137/24/2021.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.07.014.
Appendix A. Supplementary material
Supplementary File 1: Named ‘Supplementary_file1’. It contains Supplementary Methods, Supplementary Figures 1–4, and Supplementary Tables 1–3.
.
Supplementary File 2: Named ‘Supplementary_file2’. Contains the random forest model’s performance metrics for each ontological gene-set.
.
Supplementary File 3: Named ‘Supplementary_file3’. It has three columns, the first column is the cluster members’ names, the second column indicates cluster-ID, and the third column contains major cellular processes.
.
Supplementary File 4: Named ‘Supplementary_file4’. It has ChIP-Seq TF-Pairs from all cell-types appearing as top predictors across ontological gene-sets.
.
Supplementary File 5: Named ‘Supplementary_file5’. ChIP-Seq TF-Pairs from the same cell type appear as top predictors across ontological gene-sets.
.
Supplementary File 6: Named ‘Supplementary_file6’. List of predicted lncRNA genes in the functional gene-sets members of the clusters.
.
Supplementary File 7: Named ‘Supplementary_file7’. It contains metadata of all the features used to train the ML models.
.
References
- 1.Rinn J.L., Chang H.Y. Genome Regulation by Long Noncoding RNAs. 2012 [cited 15 Nov 2021]. doi:10.1146/annurev-biochem-051410–092902. [DOI] [PMC free article] [PubMed]
- 2.Kevin C., Wang H.Y.C. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X., Wang W., Zhu W., Dong J., Cheng Y., Yin Z., et al. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci. 2019;20:5573. doi: 10.3390/ijms20225573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noviello T.M.R., Di Liddo A., Ventola G.M., Spagnuolo A., D’Aniello S., Ceccarelli M., et al. Detection of long non–coding RNA homology, a comparative study on alignment and alignment–free metrics. BMC Bioinforma. 2018;19:1–12. doi: 10.1186/s12859-018-2441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y., Wang J., Chen J., Zhang X., Guo M., Yu G. A literature review of gene function prediction by modeling gene ontology. Front Genet. 2020:0. doi: 10.3389/fgene.2020.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H., Hung C.-L., Liu M., Hu X., Lin Y.-Y. NCNet: deep learning network models for predicting function of non-coding DNA. Front Genet. 2019:0. doi: 10.3389/fgene.2019.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulmanov M., Hoehndorf R. DeepGOPlus: improved protein function prediction from sequence. Bioinformatics. 2019;36:422–429. doi: 10.1093/bioinformatics/btz595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou N., Jiang Y., Bergquist T.R., Lee A.J., Kacsoh B.Z., Crocker A.W., et al. The CAFA challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens. Genome Biol. 2019;20:244. doi: 10.1186/s13059-019-1835-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X., Coulombe-Huntington J., Kang S., Sheynkman G.M., Hao T., Richardson A., et al. Widespread expansion of protein interaction capabilities by alternative splicing. Cell. 2016;164:805–817. doi: 10.1016/j.cell.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang P., Li X.-L., Mei J.-P., Kwoh C.-K., Ng S.-K. Positive-unlabeled learning for disease gene identification. Bioinformatics. 2012;28:2640–2647. doi: 10.1093/bioinformatics/bts504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu G., Chen Z., Danilova I.G., Bolkov M.A., Tuzankina I.A., Liu G. Identification of miR-200c and miR141-mediated lncRNA-mRNA crosstalks in muscle-invasive bladder cancer subtypes. Front Genet. 2018:0. doi: 10.3389/fgene.2018.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao Q., Liu C., Yuan X., Kang S., Miao R., Xiao H., et al. Large-scale prediction of long non-coding RNA functions in a coding–non-coding gene co-expression network. Nucleic Acids Res. 2011;39:3864–3878. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uygun S., Peng C., Lehti-Shiu M.D., Last R.L., Shiu S.-H. Utility and limitations of using gene expression data to identify functional associations. PLoS Comput Biol. 2016;12 doi: 10.1371/journal.pcbi.1005244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun X., Wong D. Long non-coding RNA-mediated regulation of glucose homeostasis and diabetes. Am J Cardiovasc Dis. 2016;6:17–25. [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J., Zou S., Deng L. Gene ontology-based function prediction of long non-coding RNAs using bi-random walk. BMC Med Genom. 2018;11:1–10. doi: 10.1186/s12920-018-0414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo X., Gao L., Liao Q., Xiao H., Ma X., Yang X., et al. Long non-coding RNAs function annotation: a global prediction method based on bi-colored networks. Nucleic Acids Res. 2012;41 doi: 10.1093/nar/gks967. e35–e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venters B.J., Pugh B.F. Genomic organization of human transcription initiation complexes. Nature. 2013:502. doi: 10.1038/nature12535. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Yan J., Qiu Y., Ribeiro Dos Santos A.M., Yin Y., Li Y.E., Vinckier N., et al. Systematic analysis of binding of transcription factors to noncoding variants. Nature. 2021;591:147–151. doi: 10.1038/s41586-021-03211-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 20.Kumar V., Muratani M., Rayan N.A., Kraus P., Lufkin T., Ng H.H., et al. Uniform, optimal signal processing of mapped deep-sequencing data. Nat Biotechnol. 2013;31:615–622. doi: 10.1038/nbt.2596. [DOI] [PubMed] [Google Scholar]
- 21.Tak Y.G., Farnham P.J. Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenet. Chromatin. 2015;8:57. doi: 10.1186/s13072-015-0050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roider H.G., Manke T., O’Keeffe S., Vingron M., Haas S.A. PASTAA: identifying transcription factors associated with sets of co-regulated genes. Bioinformatics. 2009;25:435–442. doi: 10.1093/bioinformatics/btn627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed M., Min D.S., Kim D.R. Integrating binding and expression data to predict transcription factors combined function. BMC Genom. 2020;21:610. doi: 10.1186/s12864-020-06977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu W., Zhao X., Wang X., Feng H., Gou M., Jin W., et al. The transcription factor Tox2 drives T follicular helper cell development via regulating chromatin accessibility. Immunity. 2019;51:826–839. doi: 10.1016/j.immuni.2019.10.006. e5. [DOI] [PubMed] [Google Scholar]
- 25.Venkatesh I., Mehra V., Wang Z., Simpson M.T., Eastwood E., Chakraborty A., et al. Co-occupancy identifies transcription factor co-operation for axon growth. Nat Commun. 2021;12:2555. doi: 10.1038/s41467-021-22828-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Interpreting area under the receiver operating characteristic curve. Lancet Digit Health. 2022;4:e853–e855. doi: 10.1016/S2589-7500(22)00188-1. [DOI] [PubMed] [Google Scholar]
- 28.Hyle J., Zhang Y., Wright S., Xu B., Shao Y., Easton J., et al. Acute depletion of CTCF directly affects MYC regulation through loss of enhancer–promoter looping. Nucleic Acids Res. 2019;47:6699–6713. doi: 10.1093/nar/gkz462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H., Lam J., Zhang D., Lan Y., Vermunt M.W., Keller C.A., et al. CTCF and transcription influence chromatin structure re-configuration after mitosis. Nat Commun. 2021;12:1–16. doi: 10.1038/s41467-021-25418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L., Huang K., Li X., Du M., Kang X., Luo X., et al. Identification of Poly(ADP-Ribose) polymerase-1 as a cell cycle regulator through modulating Sp1 mediated transcription in human hepatoma cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0082872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendricks K.B., Shanahan F., Lees E. Role for BRG1 in cell cycle control and tumor suppression. Mol Cell Biol. 2004;24:362–376. doi: 10.1128/MCB.24.1.362-376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakhmet E.I., Tomilin A.N. Key features of the POU transcription factor Oct4 from an evolutionary perspective. Cell Mol Life Sci. 2021;78:7339–7353. doi: 10.1007/s00018-021-03975-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meurer L., Ferdman L., Belcher B., Camarata T. The six family of transcription factors: common themes integrating developmental and cancer biology. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.707854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kenny C., O’Meara E., Ulaş M., Hokamp K., O’Sullivan M.J. Global chromatin changes resulting from single-gene inactivation—the role of SMARCB1 in malignant rhabdoid tumor. Cancers. 2021:2561. doi: 10.3390/cancers13112561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leal-Esteban L.C., Fajas L. Cell cycle regulators in cancer cell metabolism. Biochim Biophys Acta Mol Basis Dis. 2020;1866 doi: 10.1016/j.bbadis.2020.165715. [DOI] [PubMed] [Google Scholar]
- 36.Kaplon J., van Dam L., Peeper D. Two-way communication between the metabolic and cell cycle machineries: the molecular basis. Cell Cycle. 2015;14:2022. doi: 10.1080/15384101.2015.1044172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan S., Pagano M. Linking metabolism and cell cycle progression via the APC/CCdh1 and SCFβTrCP ubiquitin ligases. Proc Natl Acad Sci USA. 2011:20857–20858. doi: 10.1073/pnas.1118443109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yilmaz A., Peretz M., Aharony A., Sagi I., Benvenisty N. Defining essential genes for human pluripotent stem cells by CRISPR-Cas9 screening in haploid cells. Nat Cell Biol. 2018;20:610–619. doi: 10.1038/s41556-018-0088-1. [DOI] [PubMed] [Google Scholar]
- 39.Rosales C., Uribe-Querol E. Phagocytosis: a fundamental process in immunity. Biomed Res Int. 2017:2017. doi: 10.1155/2017/9042851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller H.E., Bishop A.J.R. Correlation analyzeR: functional predictions from gene co-expression correlations. BMC Bioinforma. 2021;22:206. doi: 10.1186/s12859-021-04130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kulmanov M., Khan M.A., Hoehndorf R., Wren J. DeepGO: predicting protein functions from sequence and interactions using a deep ontology-aware classifier. Bioinformatics. 2018;34:660–668. doi: 10.1093/bioinformatics/btx624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urzúa-Traslaviña C.G., Leeuwenburgh V.C., Bhattacharya A., Loipfinger S., et al. Improving gene function predictions using independent transcriptional components. Nat Commun. 2021;12:1464. doi: 10.1038/s41467-021-21671-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao S., You R., Wang S., Xiong Y., Huang X., Zhu S. NetGO 2.0: improving large-scale protein function prediction with massive sequence, text, domain, family and network information. Nucleic Acids Res. 2021;49:W469–W475. doi: 10.1093/nar/gkab398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alimov I., Menon S., Cochran N., Maher R., Wang Q., Alford J., et al. Bile acid analogues are activators of pyrin inflammasome. J Biol Chem. 2019;294:3359–3366. doi: 10.1074/jbc.RA118.005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haney M.S., Bohlen C.J., Morgens D.W., Ousey J.A., Barkal A.A., Tsui C.K., et al. Identification of phagocytosis regulators using magnetic genome-wide CRISPR screens. Nat Genet. 2018:50. doi: 10.1038/s41588-018-0254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krall E.B., Wang B., Munoz D.M., Ilic N., Raghavan S., Niederst M.J., et al. KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. Elife. 2017:6. doi: 10.7554/eLife.18970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leto D.E., Morgens D.W., Zhang L., Walczak C.P., Elias J.E., Bassik M.C., et al. Genome-wide CRISPR analysis identifies substrate-specific conjugation modules in ER-associated degradation. Mol Cell. 2019;73:377–389. doi: 10.1016/j.molcel.2018.11.015. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chesmore K.N., Bartlett J., Cheng C., Williams S.M. Complex patterns of association between pleiotropy and transcription factor evolution. Genome Biol Evol. 2016;8:3159. doi: 10.1093/gbe/evw228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Breiman L. Random Forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 50.Wang Z., Liao B.-Y., Zhang J. Genomic patterns of pleiotropy and the evolution of complexity. Proc Natl Acad Sci USA. 2010;107:18034–18039. doi: 10.1073/pnas.1004666107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim S., Yu N.-K., Kaang B.-K. CTCF as a multifunctional protein in genome regulation and gene expression. Exp Mol Med. 2015;47 doi: 10.1038/emm.2015.33. e166–e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu L.J., Gazin C., Lawson N.D., Pagès H., Lin S.M., Lapointe D.S., et al. ChIPpeakAnno: a bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinforma. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schinzel R.T., Higuchi-Sanabria R., Shalem O., Moehle E.A., Webster B.M., Joe L., et al. The hyaluronidase, TMEM2, promotes ER homeostasis and longevity independent of the UPR. Cell. 2019;179:1306–1318. doi: 10.1016/j.cell.2019.10.018. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y., Cao Z., Wang Y., Guo Y., Xu P., Yuan P., et al. Genome-wide screening for functional long noncoding RNAs in human cells by Cas9 targeting of splice sites. Nat Biotechnol. 2018 doi: 10.1038/nbt.4283. [DOI] [PubMed] [Google Scholar]
- 55.Raulerson C.K., Ko A., Kidd J.C., Currin K.W., Brotman S.M., Cannon M.E., et al. Adipose tissue gene expression associations reveal hundreds of candidate genes for cardiometabolic traits. Am J Hum Genet. 2019;105:773–787. doi: 10.1016/j.ajhg.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y.-X., Ding J., Zhou W.-E., Zhang X., Sun X.-T., Wang X.-Y., et al. Identification and functional prediction of long non-coding rnas in dilated cardiomyopathy by bioinformatics analysis. Front Genet. 2021;12 doi: 10.3389/fgene.2021.648111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Donato L., Scimone C., Alibrandi S., Rinaldi C., Sidoti A., D’Angelo R. Transcriptome analyses of lncRNAs in A2E-stressed retinal epithelial cells unveil advanced links between metabolic impairments related to oxidative stress and retinitis pigmentosa. Antioxid (Basel) 2020:9. doi: 10.3390/antiox9040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elaine Hardman W., Primerano D.A., Legenza M.T., Morgan J., Fan J., Denvir J. mRNA expression data in breast cancers before and after consumption of walnut by women. Data Brief. 2019;25 doi: 10.1016/j.dib.2019.104050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu P., Pan J., Cai Q.Q., Zhang F., Peng M., Fan X.L., et al. MicroRNA profile as potential molecular signature for attention deficit hyperactivity disorder in children. Biomarkers. 2022:1–10. doi: 10.1080/1354750X.2021.2024600. [DOI] [PubMed] [Google Scholar]
- 60.Li Z., Cai S., Li H., Gu J., Tian Y., Cao J., et al. Developing a lncRNA signature to predict the radiotherapy response of lower-grade gliomas using co-expression and ceRNA network analysis. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.622880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sage A.P., Ng K.W., Marshall E.A., Stewart G.L., Minatel B.C., Enfield K.S.S., et al. Assessment of long non-coding RNA expression reveals novel mediators of the lung tumour immune response. Sci Rep. 2020;10:16945. doi: 10.1038/s41598-020-73787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sánchez-Jiménez C., Carrascoso I., Barrero J., Izquierdo J.M. Identification of a set of miRNAs differentially expressed in transiently TIA-depleted HeLa cells by genome-wide profiling. BMC Mol Biol. 2013;14:4. doi: 10.1186/1471-2199-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen J., Wang Y., Wang C., Hu J.-F., Li W. LncRNA functions as a new emerging epigenetic factor in determining the fate of stem cells. Front Genet. 2020;11:277. doi: 10.3389/fgene.2020.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Whittington C.M., O’Meally D., Laird M.K., Belov K., Thompson M.B., McAllan B.M. Transcriptomic changes in the pre-implantation uterus highlight histotrophic nutrition of the developing marsupial embryo. Sci Rep. 2018;8:2412. doi: 10.1038/s41598-018-20744-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khurana E., Fu Y., Chakravarty D., Demichelis F., Rubin M.A., Gerstein M. Role of non-coding sequence variants in cancer. Nat Rev Genet. 2016;17:93–108. doi: 10.1038/nrg.2015.17. [DOI] [PubMed] [Google Scholar]
- 66.Wu W.-S., Lai F.-J. Detecting cooperativity between transcription factors based on functional coherence and similarity of their target gene sets. PLoS One. 2016;11 doi: 10.1371/journal.pone.0162931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jeng E.E., Bhadkamkar V., Ibe N.U., Gause H., Jiang L., Chan J., et al. Systematic identification of host cell regulators of legionella pneumophila pathogenesis using a genome-wide CRISPR screen. Cell Host Microbe. 2019;26:551–563. doi: 10.1016/j.chom.2019.08.017. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rui X., Xu Y., Huang Y., Ji L., Jiang X. lncRNA DLG1-AS1 promotes cell proliferation by competitively binding with miR-107 and up-regulating ZHX1 expression in cervical cancer. Cell Physiol Biochem. 2018;49:1792–1803. doi: 10.1159/000493625. [DOI] [PubMed] [Google Scholar]
- 69.Cao M., Ma R., Li H., Cui J., Zhang C., Zhao J. Therapy-resistant and -sensitive lncRNAs, SNHG1 and UBL7-AS1 promote glioblastoma cell proliferation. Oxid Med Cell Longev. 2022;2022:2623599. doi: 10.1155/2022/2623599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou J., Shi J., Fu X., Mao B., Wang W., Li W., et al. Linc00441 interacts with DNMT1 to regulate RB1 gene methylation and expression in gastric cancer. Oncotarget. 2018;9:37471–37479. doi: 10.18632/oncotarget.23928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Du Y., Yang H., Li Y., Guo W., Zhang Y., Shen H., et al. Long non-coding RNA LINC01137 contributes to oral squamous cell carcinoma development and is negatively regulated by miR-22-3p. Cell Oncol. 2021;44:595–609. doi: 10.1007/s13402-021-00586-0. [DOI] [PubMed] [Google Scholar]
- 72.Wang S., Sun H., Ma J., Zang C., Wang C., Wang J., et al. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc. 2013:8. doi: 10.1038/nprot.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reshef Y.A., Finucane H.K., Kelley D.R., Gusev A., Kotliar D., Ulirsch J.C., et al. Detecting genome-wide directional effects of transcription factor binding on polygenic disease risk. Nat Genet. 2018;50:1483–1493. doi: 10.1038/s41588-018-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roopra A. MAGIC: a tool for predicting transcription factors and cofactors driving gene sets using ENCODE data. PLoS Comput Biol. 2020:16. doi: 10.1371/journal.pcbi.1007800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dainese R., Gardeux V., Llimos G., Alpern D., Jiang J.Y., Meireles-Filho A.C.A., et al. A parallelized, automated platform enabling individual or sequential ChIP of histone marks and transcription factors. Proc Natl Acad Sci USA. 2020;117:13828–13838. doi: 10.1073/pnas.1913261117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gopalan S., Fazzio T.G. Multi-CUT&Tag to simultaneously profile multiple chromatin factors. STAR protocols. 2022;3(1):101100. doi: 10.1016/j.xpro.2021.101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oki S., Ohta T., Shioi G., Hatanaka H., Ogasawara O., Okuda Y., et al. ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 2018:19. doi: 10.15252/embr.201846255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frankish A., Diekhans M., Jungreis I., Lagarde J., Loveland J.E., Mudge J.M., et al. GENCODE 2021. Nucleic Acids Res. 2021;49:D916–D923. doi: 10.1093/nar/gkaa1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huynh-Thu V.A., Irrthum A., Wehenkel L., Geurts P. Inferring regulatory networks from expression data using tree-based methods. PLoS One. 2010:5. doi: 10.1371/journal.pone.0012776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marbach D., Costello J.C., Küffner R., Vega N.M., Prill R.J., Camacho D.M., et al. Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ester M., Kriegel H.P., Sander J., Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise. KDD. 1996. Available: https://www.aaai.org/Papers/KDD/1996/KDD96–037.pdf?source=post_page.
- 84.Van der Maaten L., Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008:9. https://www.jmlr.org/papers/volume9/vandermaaten08a/vandermaaten08a.pdf?fbclid=IwA (Available) [Google Scholar]
- 85.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu J., Srinivasan S., Li C.-Y., Ho I.-L., Rose J., Shaheen M., et al. Pooled library screening with multiplexed Cpf1 library. Nat Commun. 2019;10:3144. doi: 10.1038/s41467-019-10963-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1: Named ‘Supplementary_file1’. It contains Supplementary Methods, Supplementary Figures 1–4, and Supplementary Tables 1–3.
Supplementary File 2: Named ‘Supplementary_file2’. Contains the random forest model’s performance metrics for each ontological gene-set.
Supplementary File 3: Named ‘Supplementary_file3’. It has three columns, the first column is the cluster members’ names, the second column indicates cluster-ID, and the third column contains major cellular processes.
Supplementary File 4: Named ‘Supplementary_file4’. It has ChIP-Seq TF-Pairs from all cell-types appearing as top predictors across ontological gene-sets.
Supplementary File 5: Named ‘Supplementary_file5’. ChIP-Seq TF-Pairs from the same cell type appear as top predictors across ontological gene-sets.
Supplementary File 6: Named ‘Supplementary_file6’. List of predicted lncRNA genes in the functional gene-sets members of the clusters.
Supplementary File 7: Named ‘Supplementary_file7’. It contains metadata of all the features used to train the ML models.