Significance
Genetic differences between individuals can have a large effect on susceptibility to infectious disease. We have identified a gene called lectin-24A that is important in the immune response that protects fruit flies against one of their main natural enemies—parasitic wasps. However, in nature, many flies carry mutated copies of this gene that are likely to be no longer functional. We found that the high frequency of these loss-of-function mutations can only be explained if they have a selective advantage in some populations. We conclude that this genetic variation in susceptibility is maintained because in some locations, susceptible flies are fitter than resistant flies.
Keywords: loss of function, Leptopilina boulardi, melanization, C-type lectin, cis-regulatory polymorphism
Abstract
Polymorphisms in immunity genes can have large effects on susceptibility to infection. To understand the origins of this variation, we have investigated the genetic basis of resistance to the parasitoid wasp Leptopilina boulardi in Drosophila melanogaster. We found that increased expression of the gene lectin-24A after infection by parasitic wasps was associated with a faster cellular immune response and greatly increased rates of killing the parasite. lectin-24A encodes a protein that is strongly up-regulated in the fat body after infection and localizes to the surface of the parasite egg. In certain susceptible lines, a deletion upstream of the lectin-24A has largely abolished expression. Other mutations predicted to abolish the function of this gene have arisen recurrently in this gene, with multiple loss-of-expression alleles and premature stop codons segregating in natural populations. The frequency of these alleles varies greatly geographically, and in some southern African populations, natural selection has driven them near to fixation. We conclude that natural selection has favored the repeated loss of an important component of the immune system, suggesting that in some populations, a pleiotropic cost to lectin-24A expression outweighs the benefits of resistance.
Parasites can impose strong selection on host populations, driving resistance alleles up in frequency when infection is common (1, 2). Despite the advantages of resistance, genetic variability in susceptibility to infection is abundant in humans (3, 4), plants (5, 6), and insects (7–10). The polymorphisms underlying this variation may be transient as resistant alleles are spread through populations, or they can be maintained by temporal and spatial differences in selection pressures (11, 12) or negative frequency-dependent selection (13).
Variability in susceptibility can be maintained in populations when resistance trades-off with other fitness-related traits. These costs may occur in the absence of infection (14), due to the diversion of resources from growth and reproduction (15, 16), autoimmune damage (17), or when resistance to one pathogen increases susceptibility to a different pathogen (18). However, not all resistance alleles are costly (19), and over time, compensatory mutations that reduce or negate fitness costs may spread (20, 21). Alternatively, fitness costs could be avoided by reverting to susceptibility when the pathogen pressure is low (22).
Early demonstrations of the costs of evolving resistance came from Drosophila and parasitoid wasps, where populations selected for increased resistance had reduced competitive ability (23, 24) and lower feeding rates (25). Female parasitoid wasps lay their eggs inside the larvae of Drosophila, and if the host is unable to mount a successful immune response, the parasitoid larva feeds on the host tissue and eventually kills it. Flies can kill parasitoid wasps through a cellular immune response known as melanotic encapsulation, in which hemocytes (blood cells) are recruited to the parasitoid egg and surround it (26). The capsule is then melanized, killing the parasitoid egg. Despite parasitoids being common in nature, there is considerable variation within and between populations in susceptibility to the parasitoids Asobara tabida (Braconidae) (27) and Leptopilina boulardi (Figitidae) (28). Early work found polymorphisms in two regions of chromosome 2R that are involved in resisting parasitoid infection: one for resistance against L. boulardi and the other against A. tabida (29–31). However, a later study using populations that were artificially selected for resistance to the parasitoid A. tabida identified a 600-Kbp region on chromosome 2R that did not overlap with the previously identified loci (32). This could be the result of differences in the host genetic backgrounds or because intense artificial selection favors different loci to those favored by natural selection in the field (33). In the light of these observations and a lack of concrete evidence for selection acting on specific genes or alleles, we have attempted to identify the genetic basis of resistance against L. boulardi in a natural population of Drosophila melanogaster.
Results
Resistance to Parasitoid Infection Is Associated with a Faster Immune Response.
We selected two inbred lines, DGRP-437 and DGRP-892, that had a marked difference in their ability to survive parasitoid infection to investigate the genetic basis of parasitoid resistance (Fig. 1A; binomial GLMM, z = −7.6, P < 0.001). The antiparasitoid defense response involves the wasp being surrounded by immune cells called hemocytes and melanized. In the resistant DGRP-437, the melanization phenotype becomes apparent at 24 h post-infection (hpi), and the wasp embryo is completely melanized at 26 hpi (Fig. 1B). In contrast, no melanization is seen in the susceptible DGRP-892 in the first 26 hpi (Fig. 1B).
Fig. 1.
Genetic differences in the immune response to parasitoid infection. (A) The proportion of L. boulardi wasp embryos melanized in two inbred lines, DGRP-437 and DGRP-892. Each point is an independent replicate (six/line), bars are 95% CIs, and the number of larvae across all assays is above the bar. (B) Wasp embryo melanization at 18, 24, and 26 h after infection. (C and D) Larvae were injected with oil droplets containing homogenized L. boulardi (C, nDGRP-437 = 853, nDGRP-892 = 816) or A. tabida (D, nDGRP-437 = 128, nDGRP-892 = 128). Bars represent SEs.
This host immune response must be fast to succeed because once the wasp larva emerges from the egg chorion 24 to 48 hpi, it is mobile and better at escaping the cellular capsule (34). Parasitoids can suppress host immunity by injecting venoms along with the egg. To examine the speed of the immune response in the absence of this immune suppression, we triggered the immune response by injecting larvae with droplets of mineral oil containing homogenized parasitoid wasps (Fig. 1C). Both Drosophila lines had melanized the oil droplets by 48 h post-injection. However, the resistant line mounted this immune response faster—89.8% of the oil droplets were melanized at 28 h post-injection compared to 3.4% in the susceptible line (binomial GLM, logistic regression χ2 = 432.1, df = 1, P < 0.001). This difference is not specific to the parasitoid L. boulardi, as we obtained similar results when injecting oil droplets containing A. tabida homogenate (Fig. 1D; binomial GLM, genotype: logistic regression χ2 = 97.3, df = 1, P < 0.001; time post-injection: logistic regression χ2 = 111.5, df = 1, P < 0.001)]
Resistance Results from Epistatic Interactions between Genes on Different Chromosomes.
We next investigated the genetic basis of resistance. We chose to use classical genetic crosses rather than a genome-wide association study based on our experience of identifying genetic polymorphisms affecting susceptibility to infection. Specifically, this approach can identify genetic variants in the presence of epistasis, allelic heterogeneity, and low allele frequencies. This can be challenging using genome-wide association studies in Drosophila, as the available resources mean such analyses frequently use under 200 fly lines. When we crossed the resistant and susceptible lines, the F1 progeny were highly resistant, indicating that resistance was dominant (Fig. 2A). By swapping whether the resistant parent was the mother or father, we generated male offspring that only differed in their X chromosome (Fig. 2A). Neither the sex of progeny (likelihood ratio test, χ2 = 3.46, P = 0.06) nor the X genotype (Tukey’s HSD, z = 0.45, P = 0.65) had a significant impact on susceptibility (Fig. 2A). These results demonstrate that resistance is an autosomal dominant trait.
Fig. 2.
A single locus on chromosome II is associated with resistance to parasitoid wasp infection. (A) The mean proportion of wasps melanized in the F1 progeny of crosses between two Drosophila lines. Bars are 95% CIs. (B) The mean proportion of wasps melanized in lines with different chromosome combinations. Bars are SEs, and points are replicates of 20 larvae. Letters show significantly different groups (Tukey’s test, P < 0.05 between groups). Numbers above bars in (A) and (B) indicate the number of larvae that were assayed. (C) QTL on chromosome II associated with the melanization rate measured by dissecting larvae. The black line is interval mapping and the red line composite interval mapping. The blue region is the CI on the QTL location (1.5 LOD drop). X axis ticks are marker locations. (D) Fine-scale QTL in the region containing the gene. Only informative recombinants were genotyped. The χ2 statistic represents the deviation from the expected Mendelian frequency (0.5) of each marker among 152 adults that contained a capsule and tested positive for parasitoid wasp DNA. The shaded region indicates the 95% CI (χ2 drop of 4.6).
To identify the chromosomes affecting susceptibility, we generated lines carrying varying combinations of the X, II, and III chromosomes (Fig. 2B and SI Appendix, Fig. S1). When comparing lines that differ only in their second chromosome, having chromosome II from the resistant parent always resulted in greater melanization (Fig. 2B). When paired with a third chromosome from the resistant parent, swapping chromosome II could convert a fully resistant line into a fully susceptible line (Fig. 2B). However, when paired with a third chromosome from the susceptible parent, chromosome II only had a small effect. This is reflected in a statistical interaction between the two chromosomes (GLM with logit link, Wald test: χ2= 24.7, P < 0.001), indicating that there is a multiplicative epistatic interaction between the second and third chromosomes.
A Major Effect Locus on Chromosome II Affects Resistance.
We used quantitative trait locus (QTL) mapping to locate the region on chromosome II affecting susceptibility to infection. We crossed fly lines that differed only in the second chromosome (X892; II892; III437 and X892; II437; III437) and then backcrossed the F1 progeny to the susceptible parent. The resulting larvae were parasitized, and we genotyped 386 individual larvae using 10 molecular markers spanning chromosome II. We identified a single region where the chromosome II genotype was associated with differences in susceptibility (Fig. 2C, black line). Based on a 1.5 logarithm of the odds (LOD) drop, the QTL encompassed 11 to 25 cM (Fig. 2C, blue box). Composite interval mapping, which searches for additional QTLs while accounting for the main peak, indicated that there is a single locus on chromosome II affecting susceptibility (Fig. 2C, red line).
As this QTL contained many genes, we conducted a second round of genetic mapping using only flies that were recombinant within the QTL. We repeated the genetic cross, parasitized the backcrossed larvae, and selected adults that contained visible capsules in their body and had therefore likely survived infection (susceptible flies were killed). We genotyped these individuals using molecular markers flanking the QTL (3 to 27 cM) to identify recombinants. Out of 1,486 adults, 298 had a recombination breakpoint between 3 cM and 27 cM—a recombination fraction of 0.20. We genotyped 12 molecular markers within this region for 152 individuals where we could amplify wasp DNA to confirm that they had been infected. As we only genotyped resistant flies, we tested whether marker allele frequencies departed from the 50:50 Mendelian expectation. Sixty-one out of 348 uninfected flies had a breakpoint between 3 cM and 27 cM, which is only marginally lower than expected suggesting little segregation distortion. A χ2 drop to identify informative markers was selected by simulating 1,000 datasets based on the observed risk ratio estimated from nonrecombinant flies and the observed recombination fraction. The χ2 drop defined a region that included the gene in 95% of simulations. Using this approach, we identified a single QTL at 10.3 cM on the left arm of chromosome II (Fig. 2D). By simulating 1,000 replicate datasets, we estimated that the 95% CI on the location of the QTL contained 84 protein-coding genes and 23 long noncoding RNAs (10.0 to 11.6 cM, genome v6: 3.43-4.03Mbp, SI Appendix, Table S1). Among the 1,188 nonrecombinant flies, 906 carried the resistant allele and 282 the susceptible allele, indicating a risk ratio of 3.21 (homozygous susceptible versus heterozygotes).
A C-Type Lectin Underlies Resistance.
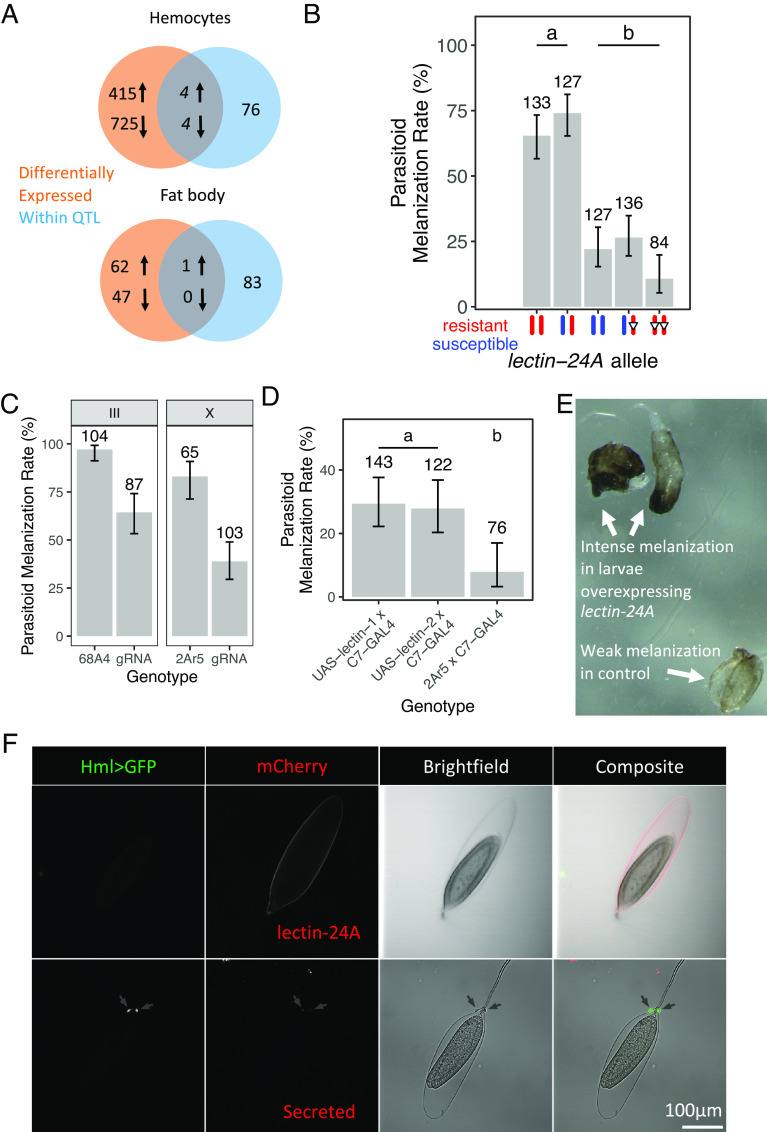
Parasitoid wasp infection induces a large transcriptional response in the two main immune tissues of Drosophila—the fat body and hemocytes. Using our open access RNA sequencing data (35), we searched for genes within the QTL that were up-regulated after antiparasitoid immune induction in these tissues (log2 fold change > 2). We found that eight genes were differentially expressed in hemocytes and one gene, lectin-24A, in the fat body (Fig. 3A). When we measured the expression of these nine genes in the lines we are studying, only lectin-24A showed differential induction following parasitic wasp infection (SI Appendix, Fig. S2). lectin-24A has previously been found to be massively up-regulated following parasitoid wasp infection (36–38). Furthermore, the marker most strongly associated with the melanization rate in the fine-scale QTL analysis is located within lectin-24A. As lectins are important receptors in innate immune systems, we focused on this gene.
Fig. 3.
lectin-24A is required for parasitoid resistance. (A) Protein-coding genes within the QTL and differentially expressed after immune induction (log2 fold change > 2). The gene in the intersection of the fat body Venn diagram is lectin-24A. (B) Mean parasitoid melanization rate in lines with resistant, susceptible, and mutated (open inverted triangles, Δ129) lectin-24A. Error bars represent 95% CIs. Tukey’s test, P < 0.05 between groups. (C) Mean parasitoid melanization rates in larvae expressing Act-Cas9 and guide RNAs targeting lectin-24A compared to control larvae. Guide RNAs on either the X or the III chromosome were used, and 2Ar5 and 68A4 are the lines in which the construct was microinjected. Each of these lines was crossed to Act-Cas9; II437; III437, and the F1 heterozygotes were assayed. Error bars represent 95% CIs. A Fisher’s exact test was used to identify significant differences for each pair of comparisons. (D) Mean parasitoid melanization rates in larvae overexpressing lectin-24A under the C7-GAL4 driver in the larval fat body compared to control larvae. UAS-lectin-1 and UAS-lectin-2 are independent insertions of a construct expressing guide RNAs targeting lectin-24A, and 2Ar5 is the original line in which the construct is microinjected into. Error bars represent 95% CIs. Tukey’s test, P < 0.05 between groups. Numbers above bars in (B–D) indicate the number of larvae that were assayed. (E) Representative image of partially melanized wasp larva and melanized wasp eggs in Drosophila larvae overexpressing lectin-24A and control larvae. (F) Confocal microscopy image of wasp eggs dissected 2 to 12 h post-infection. The presence of hemocytes (arrows) was checked with HmlΔ-GAL4 UAS-GFP and confirmed in the brightfield (BF) channel. lectin-24A-mCherry can be observed on the egg chorion in regions without hemocytes (top row). The same is not observed in secreted-mCherry alone (bottom row). The scale bar represents 100 μm.
To test whether lectin-24A is necessary for resistance to parasitoid wasps, we created a germline mutation in the resistant lectin-24A allele using CRISPR-Cas9 in the resistant X892; II437; III437 flies. We created a 4-bp insertion that introduced a premature stop codon 129 bp downstream of the start codon, which we named lectin-24AΔ129. The change in reading frame introduces a premature stop codon and abolishes the carbohydrate-binding domain of the protein. This mutation made flies susceptible to infection (Fig. 3B; Tukey’s HSD test: P < 0.001).
To confirm this result, we generated somatic lectin-24A mutants in the F1 progeny of a cross between flies that ubiquitously express Cas9 and fly lines that we created to express guide RNAs targeting the lectin-24A gene (SI Appendix, Fig. S3A). This efficiently generated somatic mutants (SI Appendix, Fig. S3B), and the mutant larvae showed significantly reduced melanization rates when parasitized compared to larvae that express Cas9 but not the gRNAs (Fig. 3C; Fisher’s exact test: P < 0.001). We repeated this experiment using an independently generated fly line that carried the guide RNA on a different chromosome and obtained the same result (Fig. 3C; Fisher’s exact test: P < 0.001).
We next overexpressed lectin-24A in the larval fat body. We generated flies carrying a UAS-driven Flag-tagged lectin-24A construct with the DGRP-437 coding sequence, under the control of the C7-GAL4, which drives expression in the larval fat body. The overexpression of lectin-24A increased the melanization rate in Drosophila larvae against parasitic wasp infection (Fig. 3D; Tukey’s HSD test: P < 0.005 between groups). In larvae overexpressing lectin-24A, we frequently observed partially melanized wasp larvae (Fig. 3E). There was also a striking and consistent increase in the intensity of melanization between the larvae overexpressing lectin-24A and the controls (Fig. 3E).
lectin-24A is a C-type lectin, a family of proteins which frequently act as pattern recognition receptors in the innate immune system due to their specificity in binding ligands (39). To investigate the role of lectin-24A in the immune response, we created transgenic flies that expressed lectin-24A fused to mCherry fluorescent protein under the control of the gene’s native promoter. When larvae from these lines were infected by a parasitoid, the protein localized to the surface of the wasp egg at an early time point (Fig. 3F). To understand at what point in the immune response this occurred, we also visualized hemocytes using both brightfield microscopy and by expressing GFP in plasmatocytes (at this time point lamellocytes have not yet differentiated). This revealed that lectin-24A is found on the parasitoid egg before hemocytes attached to the egg (Fig. 3F). This is consistent with this molecule being an opsonin involved in the initial recognition of the parasite, guiding the subsequent cellular immune response.
C-type lectins are named due to their ability to bind to specific carbohydrates in a calcium-dependent manner (39). By aligning the peptide sequence of the lectin-24A carbohydrate recognition domain with other members of the protein family, we found that residues required for the interaction with the calcium ions have been lost, suggesting that it is not involved in calcium-dependent carbohydrate binding (SI Appendix, Fig. S4A). This is reminiscent of another well-characterized group of C-type lectins that have lost the calcium-binding ability—natural killer cell receptors—which bind ligands including proteins (40). Alongside conserved cysteines involved in forming the Ca2+ binding site, lectin-24A contains an additional cysteine within the carbohydrate domain and four cysteines elsewhere in the protein (SI Appendix, Fig. S4A), suggesting that it may form multimers. By expressing affinity-tagged lectin-24A in Drosophila cells, we confirmed the protein forms tetramers (SI Appendix, Fig. S4B).
A Cis-Regulatory Polymorphism in lectin-24A Is Associated with Resistance.
We used qPCR to examine whether the resistant and susceptible copies of lectin-24A differed in their expression (Fig. 4A). In uninfected larvae, the resistant lectin-24A is expressed at a higher level than the susceptible lectin-24A. After infection, there was a ~2.5-fold upregulation of the resistant lectin-24A, but the susceptible copy was not induced (6 and 18 hpi, Fig. 4A and SI Appendix, Fig. S2; ANOVA, effect of Drosophila line: F = 41.69, df = 1, P < 0.001).
Fig. 4.
Cis-regulatory polymorphisms in lectin-24A. (A) Expression of lectin-24A 6 and 18 h post-infection (hpi) with parasitoid wasp. (B) Allele-specific expression of lectin-24A in heterozygous flies. Read counts (depth) of the resistant (DGRP-437) and susceptible (DGRP-892) allele for complementary DNA (cDNA) and genomic DNA (gDNA). Asterisks indicate a Welch t test, P < 0.0001. (C) Expression of Venus driven by the sequence upstream of the susceptible (LP892) or resistant lectin-24A (LP437) imaged 24 hpi under brightfield (Left) or GFP filter (Right). (D) Reporter constructs with different combinations of variants upstream of lectin-24A in the susceptible (DGRP-892, blue) and resistant (DGRP-437, red) lines and expression of Venus. Each point represents a sample of 15 larvae. Letters are Tukey’s test, P < 0.05 between groups, P > 0.05 within groups. (E) Putative binding site of the NF-κB transcription factors in the lectin-24A promoter regions of DGRP-437 and DGRP-892.
To determine whether lectin-24A expression is controlled in cis or trans, we crossed the two lines, infected them, and Illumina-sequenced the lectin-24A transcript in the heterozygous F1 progeny. The sequence reads were assigned to the resistant and susceptible lectin-24A using SNPs that differ between the two lines, allowing us to measure their relative expression. In these heterozygous flies, we found that the expression of the resistant lectin-24A was 34 times greater than the susceptible lectin-24A (Fig. 4B; Welch t test: t = 135.6, df = 4.7737, P < 0.001). As the two alleles of lectin-24A are present in the same cells, they share the same trans-regulatory environment, these differences in expression are controlled in cis.
Many cis-regulatory elements are found a short distance upstream of the gene they control. The lectin-24A-mCherry transgene described above included 489 bp of sequence upstream of the start codon of the resistant lectin-24A, and this is strongly induced after infection (SI Appendix, Fig. S5). However, when the equivalent transgene was made from the susceptible lectin-24A, there was no detectable expression (SI Appendix, Fig. S5). To confirm this result, we cloned these regulatory sequences in front of GFP to create fluorescent reporters that were inserted into the Drosophila genome. Recapitulating the results from the lectin-24A-mCherry construct, the sequence upstream of resistant lectin-24A drove strong reporter expression in the fat body, but the equivalent sequence upstream of the susceptible lectin-24A did not (Fig. 4C). To accurately quantify expression, we measured fluorescence in proteins extracted from larvae. Confirming the microscopy results (Fig. 4C), we observed a ~30-fold difference in fluorescence between the reporter lines carrying the regulatory sequence of the resistant and susceptible lectin-24A (Fig. 4D, top two constructs). While lectin-24A was never up-regulated after infection in DGRP892 (Fig. 4A), the DGRP-892 promoter can drive GFP induction in our transgenic flies, albeit from a low level (Fig. 4C). The reason for this is unknown, but it suggests that the behavior of this regulatory sequence depends on its genomic location. Together, this demonstrates that this region contains a cis-regulatory polymorphism that differs between the resistant and susceptible lectin-24A.
Comparing the resistant and susceptible lectin-24A, the cis-regulatory sequence used in the reporter constructs differed by three insertion-deletion polymorphisms (indels) and six single nucleotide polymorphisms (SNPs; Fig. 4D, top two rows). To identify which of these causes the differences in expression, we created seven more transgenic fly lines, each carrying a reporter construct that had a different combination of alleles at these sites (Fig. 4D). When we introduced a 21 bp indel (c.-171_-151del) found upstream of the susceptible lectin-24A into the resistant lectin-24A reporter, it greatly reduced the expression levels (Fig. 4D). In contrast, swapping alleles of the other polymorphic sites only resulted in minor but statistically significant changes in expression (~threefold). Therefore, we conclude that the 21 bp indel (c.-171_-151del) is primarily responsible for the differential lectin-24A expression in the resistant and susceptible lines.
To understand why the 21 bp deletion reduces lectin-24A expression, we predicted binding sites of Drosophila immunity-related transcription factors (41–43). A putative binding site of the NF-κB transcription factors Dif and dorsal is lost with the 21 bp deletion (c.-171_-151del) (Fig. 4E). These transcription factors are controlled by the Toll pathway—a major immune signaling pathway—so the loss of this binding site might cause the loss of lectin-24A expression.
In a Population, lectin-24A Expression Is Associated with Susceptibility.
We next investigated genetic variation in lectin-24A expression at the population level. We first sequenced a 557-bp region upstream of the gene in the DGRP panel of inbred lines from North America (44, 45) (SI Appendix, Table S2) and selected 20 lines with different haplotypes at the three indels shown in Fig. 4D. We crossed these to our resistant line (DGRP-437) and Illumina-sequenced the lectin-24A transcript in the F1 progeny to look for evidence of allele-specific expression. This assay produced consistent results across replicates (SI Appendix, Fig. S6 A and B), and when we sequenced genomic DNA, the frequency of the two alleles was close to 0.5, indicating that there is no technical bias toward one allele (Fig. 5A).
Fig. 5.

The frequency of predicted loss-of-function alleles of lectin-24A in natural populations. (A) Twenty-four hours post-infection, lectin-24A complementary DNA (cDNA) and genomic DNA (gDNA) was sequenced in the F1 progeny of a cross between DGRP-437 and 20 different inbred DGRP lines. The lines are grouped by their indel haplotype, which is ordered as c.-439_-433del, c.-334_-333insACATTCAT, and the 21 bp indel (c.-171_-151del), and the deletion “D” or insertion “I” state for each indel is depicted. We estimated the frequency of reads from the test line as opposed to DGRP-437. We had two technical and two biological replicates per cross. Data for DGRP-892 are also presented in Fig. 3. (B) Melanization rates in adult flies from 145 DGRP lines where the three upstream lectin-24A indels were characterized. Lines are grouped by whether they have the upstream indel haplotypes capable of expressing lectin-24A at a high level. The 2 to 10 replicate measurements for melanization rates per DGRP line are shown as individual points. A total of 123 lines have the lectin-24A expressing haplotypes (DII, IDI, III), and 22 lines have the haplotypes that do not express lectin-24A (DDD, DDI). A Welch two-sample t test was used to identify significant differences between the line means of the two groups. (C) The frequency of variants that abolish expression (the 21 bp indel as known as c.-171_-151del) or introduce premature stop codons (other variants). Multiple refers to the co-occurrence of any two loss-of-function variants. Analysis used 672 genome sequences genotyped for >50% of the lectin-24A gene.
As expected, we found that the four lines carrying the 21 bp deletion (c.-171_-151del) all had very low expression (Fig. 5A, haplotype DDD). However, four lines that lack the deletion at this site also had strongly reduced expression (Fig. 5A, haplotype DDI). While expression in these lines was low, it was nonetheless 1.7 times higher than lines with the 21 bp deletion (quasibinomial GLM, DDD versus DDI: t = 4.319, P < 0.001). To confirm these results, we also measured lectin-24A expression in a sample of inbred lines by qPCR. These results supported our conclusion that lines with the 21 bp deletion had the lowest expression, but the expression was also reduced in lines with the DDI haplotype (SI Appendix, Fig. S6C). Twenty-five of the 130 fully genotyped lines carried one of the low expression haplotypes (DDD or DDI). After removing lines with the 21 bp deletion, alternate alleles at four sites in the 1,000 bp region upstream of the lectin-24A start codon (3718317, 3718354, 3718388, and 3718452) showed complete association with the loss of capacity to induce lectin-24A expression and are therefore candidate cis-regulatory polymorphisms (SI Appendix, Table S3).
To examine lectin-24A expression in a larger sample at a different life stage, we reanalyzed published RNA-sequencing data from adult DGRP flies (46). We found that the haplotypes that were associated with low expression in infected larvae also had lower expression in uninfected adults (SI Appendix, Fig. S7). This effect in uninfected flies is consistent with the results in uninfected larvae, where alleles of both the gene and reporter construct that carry the 21 bp deletion have reduced expression (Fig. 4 A and D).
To test whether the cis-regulatory polymorphisms in lectin-24A are associated with parasitoid resistance at the population level, we estimated the melanization ability of 194 lines in the DGRP panel. In total, we examined whether 39,696 flies across 1,060 replicate vials melanized wasp larvae. The lines varied greatly in their melanization rate, with most being very susceptible to infection. The DGRP lines with the low expression haplotypes (DDD or DDI) had significantly lower melanization rates compared to lines capable of expressing lectin-24A (Fig. 5B). After accounting for the expression haplotype in an ANOVA, variants in the coding sequence did not have a significant effect on melanization rates (SI Appendix, Table S4). All the lines with the highest melanization rates had high-expression haplotypes. However, most lines with high-expression haplotypes were susceptible to infection, indicating that lectin-24A expression is not sufficient for resistance. This is consistent with our finding above that this gene only provides strong resistance in specific genetic backgrounds. The finding that high expression haplotypes are associated with parasitoid resistance provides confirmation that this gene underlies resistance.
Loss-of-Function Alleles Have Arisen Repeatedly in Natural Populations.
We have evidence for two alleles that cause the loss of lectin-24A expression—the 21 bp deletion and the DDI haplotype (Fig. 5A). We next examined whether loss-of-function variants are segregating in the gene’s protein-coding sequence. It has previously been reported that there are alleles of lectin-24A containing premature stop codons in natural populations (47). As many more genomes have been sequenced since that analysis, we searched 1,039 published genomes from flies collected globally for variants that are likely to result in null alleles of lectin-24A (48). We identified a 165-bp deletion in the protein-coding sequence that resulted in a shift in the reading frame and a premature stop codon (p.Phe217_Glu273del*) and three point mutations that introduced premature stop codons either within or before the lectin-24A carbohydrate recognition domain. Lines containing these premature stop codons were able to up-regulate lectin-24A following parasitoid wasp infection (SI Appendix, Fig. S8), suggesting that these variants cause the loss of gene function independently of the loss-of-expression mutations.
To understand the geographical distribution of putative loss-of-function alleles in lectin-24A, we examined their frequency in 26 populations. Southern African populations have the highest frequency of loss-of-function alleles, with over half of alleles carrying a premature stop codon, and many alleles carrying multiple loss-of-function mutations (Fig. 5C). Outside of southern Africa, premature stop codons are rare, but the 21-bp deletion (c.-171_-151del) that abolished expression is widespread, reaching frequencies over 30% in some locations. These numbers underestimate the true frequency of loss-of-function alleles as we cannot identify the loss-of-expression variant on the DDI haplotype from sequence alone.
Natural Selection Has Driven the Loss of lectin-24A.
Given the importance of lectin-24A in defending flies against parasitoid wasps, the finding that likely nonfunctional alleles are common in nature is unexpected. We therefore explored the evolution of the gene in more detail. First, we examined whether predicted nonfunctional alleles are the ancestral or derived states by aligning the lectin-24A gene region with the homologous region from three closely related species—Drosophila mauritiana, Drosophila simulans, and Drosophila sechellia. All three species contain the 21 bp sequence (AAATAAGGCTATCTGGGATCA; c.-171_-151del; SI Appendix, Fig. S9) that is required for the gene to be induced after infection and do not contain any premature stop codons in the coding sequence. Therefore, in all cases, the loss-of-function allele is the derived state.
The variable frequency of loss-of-function alleles in nature (Fig. 5C) suggests that these alleles may be favored by natural selection in some populations but not others. In line with previous analyses of a smaller dataset (47), multiple SNPs in lectin-24A had very high levels of genetic differentiation in the 1,039 published genomes (48), with several SNPs being below the 0.1% percentile of a null distribution generated using 23,635 variants that occur in neutrally evolving short introns (Fig. 6A). Pairwise comparisons between geographical regions showed that this pattern was driven by the southern African populations being highly differentiated from other regions (Fig. 6B). Across the gene, there are numerous variants that are near fixation across southern Africa but are rare elsewhere in the world (SI Appendix, Fig. S10) due to a divergent haplotype that is common only in southern Africa (SI Appendix, Fig. S11). Two of the three premature stop codons and the coding sequence deletion (p.Phe217_Glu273del*) were at their highest frequency (16.7 to 56.9%) in southern Africa. The other premature stop codon (p.Leu81*) was not found in southern Africa but was segregating at 4 to 5% in North America, Europe, and North Africa and Central Africa.
Fig. 6.

Evidence of local adaptation in lectin-24A. (A) Per site Weir and Cockerham FST (dm5; 2L:3715513–3719050) across 26 global fly populations. FST for the top 1% and 0.1% of 23,635 variants in short introns is indicated by dotted red lines. Red inverted triangles indicate loss-of-function variants. (B) FST for lectin-24A protein coding sequence from pairwise comparisons between seven geographic regions. (C) Population branch statistic (PBS) for 4,433 variants in short introns and three lectin-24A premature stop codons for the three largest geographic regions. The premature stop codons are indicated with arrows. The red dotted line is the 95th percentile. (D) PBS trees for the polymorphic stop codon p.Gln254* and a genome-wide tree based on 4,433 variants in short introns. Both trees are on the same scale.
As the loss of lectin-24A has a large effect on susceptibility to infection, we tested whether natural selection has driven the premature stop codons to a high frequency in southern Africa using the population branch statistic (PBS) (Fig. 6C). This used pairwise FST estimates between the three geographical regions with large numbers of published genomes to generate a tree, with longer branches indicative of larger changes in allele frequency along that branch (49). To generate an empirical null distribution, we calculated the PBS for 4,433 variants that short introns (which are regarded as putatively neutral), had a minor allele frequency greater than 5%, and were typed >40% of samples in each region. We found that p.Gln254* had an extremely high PBS in southern Africa when compared to the genome-wide PBS, indicating that this variant is under positive selection in that region (Fig. 6D). We also performed a selection test using Ohana (50) which controls for admixture and historical population structure. We first used genome-wide data to generate a covariance matrix of allele frequencies between ancestry components, and then scanned for local distortions due to positive selection. We used 4,433 variants in short introns to generate an empirical null distribution. We found a strong signature of positive selection on p.Gln254* along the Southern African branch (SI Appendix, Fig. S12; K = 5, log-likelihood ratio statistic = 22.3, P < 0.001). The same pattern was also apparent across a range of different numbers of ancestry components (K; SI Appendix, Fig. S12).
Discussion
We have presented several lines of evidence that polymorphisms on the gene lectin-24A affect susceptibility to parasitoid wasp infection. First, in a QTL analysis, the marker that is most strongly associated with resisting infection falls within this gene. Second, in the resistant line, this gene is strongly up-regulated after infection, resulting in the production of a protein that localizes to the surface of the parasite. In the susceptible line, this gene is not up-regulated due to a mutation in a cis-regulatory sequence. Third, within populations, flies with haplotypes that have low expression are more susceptible to infection. Finally, when we introduce loss-of-function mutations by mutating lectin-24A in the resistant line, the flies become susceptible, and when we overexpress the protein, they become more resistant.
In nature, Leptopilina wasps are capable of infecting and killing 90% of D. melanogaster in some fruits in a single generation (51). Therefore, our finding that numerous loss-of-function alleles are segregating in a gene that protects flies against these parasites is unexpected. Furthermore, our population genetic analysis demonstrated that natural selection has favored null alleles of lectin-24A in southern Africa, suggesting that expressing this gene may sometimes reduce fitness.
Artificial selection experiments in Drosophila have shown that when populations evolve resistance to parasitoid infection, the fitness of uninfected flies is strongly reduced, indicating that alleles that increase parasitoid resistance can pleiotropically reduce other components of fitness (23, 52). This cost has been attributed to an increase in hemocyte numbers in genetically resistant flies (23, 52), and recent experiments have demonstrated that high hemocyte numbers reduce the accumulation of lipids in the fat body that are important during nutrient scarcity (53). Could lectin-24A also contribute to the cost of evolving resistance, explaining why natural selection has favored the loss of this gene? An argument against this idea is that this gene is strongly up-regulated after infection, and inducible expression is thought to avoid the production of costly gene products except in useful contexts (54). However, in the case of lectin-24A, lines with inducible haplotypes also have higher baseline expression, which could give rise to the cost of expression in the absence of infection, perhaps due to autoimmune damage.
Our conclusion that loss-of-function mutations in lectin-24A have a selective advantage in some populations may be an example of the “less is more” process, which postulates that gene loss and pseudogenization can be beneficial, particularly following drastic shifts in environmental conditions (55–57). As natural selection specifically favored loss-of-function alleles of lectin-24A in southern Africa, the balance of these costs and benefits appears to have shifted in different environments, resulting in the susceptible allele having an advantage. Interestingly, studies of Diptericin A, which confers resistance to gram-negative bacteria such as Providencia rettgeri, have also found that loss-of-function alleles segregate at higher frequencies in the south of Africa (58). In both cases, this may reflect differences in the parasite pressure. However, while the L. boulardi group is thought to occur in Southern Africa (59), the prevalence, genotype, and frequency of the parasitoids in that region are unknown. lectin-24A can be triggered by different wasp species (36, 60), and it is possible that this gene does not protect flies against all parasitoid species. Similarly, some genotypes of L. boulardi are highly effective at suppressing the immune response and are rarely melanized. These factors may mean that this immune defense may not provide any protection against infection in some populations, so functional alleles of this gene could reduce fitness due to pleiotropic effects. Alternatively, costs of resistance may only become apparent when food is scarce (23, 52), so geographical differences in selection may be due to differences in harmful pleiotropic effects of the resistant allele on some other trait. Pathogen defense could also employ a different pathway in this geographical region, for instance, via a protective symbiont such as Spiroplasma (61). If this is the case, losing a costly melanization response could be beneficial even in infected flies as the parasite might be killed in another way. If the melanization response against parasitoids is costly, then we might expect other genes involved in this process to be lost when parasitoid pressure is low. This appears to be the case in a species called D. sechellia. Lamellocyte-mediated encapsulation arose recently in the melanogaster subgroup, and this was associated with the appearance of 11 genes that are strongly induced after infection (62). Strikingly, three of these have presumed loss-of-function mutations in D. sechellia (62, 63). This includes two genes—PPO3 (63) and Tep1 (35)— known to have important roles in melanotic encapsulation. Drosophila sechellia is thought to have escaped from parasitoid infection by feeding on a fruit that is toxic to parasitoids (64). Therefore, in this species, the molecular machinery underlying the antiparasitoid immune response appears to have been lost over a short period of evolutionary time once it was no longer required.
lectin-24A appears to be a hotspot of adaptive evolution in the Drosophila immune system (47). It has arisen recently in the common ancestor of D. melanogaster and D. simulans and is one of the most rapidly evolving proteins in the genome (47). In D. simulans, there has been a recent and strong selective sweep (37, 47), while in D. melanogaster, it has exceptionally high geographical variation in allele frequencies (47). These observations suggest that it may be a key player in the coevolution of Drosophila and parasitoids.
Drosophila kills parasitoids using a cellular immune response. However, parasitoid infection also triggers a strong transcriptional response in the fat body, resulting in the secretion of humoral immune factors. The function of these molecules is largely unknown. lectin-24A is massively up-regulated following infection by A. tabida (36, 60) and L. boulardi (37, 38) but not by wounding or bacterial infection (37). We found that it localizes to the surface of the parasitoid egg before the attachment of hemocytes. It may function as an opsonin, binding to the parasite to promote hemocyte attachment. An understanding of lectin-24A’s molecular function may provide insights into why it evolves so fast and why it appears to reduce the fitness of flies in some populations.
Materials and Methods
We screened the inbred DGRP lines (44, 45) for levels of melanization following parasitoid wasp infection and chose two lines showing a large difference in melanization rates. We mapped the locus responsible for differential melanization ability using QTL mapping. Then, we identified candidate genes by intersecting the genes occurring within the locus governing melanization ability with genes showing differential expression following exposure to parasitoid wasps (35). We confirmed the necessity of our candidate gene (lectin-24A) by knocking it down using CRISPR/Cas9-mediated targeted mutagenesis. We also assessed differential lectin-24A induction in the resistant and susceptible lines following parasitoid infection using qPCR. We investigated whether natural selection had a role in favoring null alleles in lectin-24A in global fly populations. A detailed description of the methods is in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Kurt Drickamer for providing and interpreting the C-type lectin carbohydrate recognition domain alignment. We thank John Pool for providing the SD and SP lines from the Drosophila Genome Nexus. We thank Dagmara Korona for sharing their pCFD5 and a plasmid containing the Flag-StrepII tag with us. We thank Angela Early for providing more detail on the genetic differentiation of lectin-24A in the Global Diversity Lines. We thank Carlos Luque for assisting with Drosophila larval imaging. Natural Environment Research Council grant (NE/P00184X/1) awarded to F.M.J. and A.B.L. European Molecular Biology Organization fellowship (ALT-1556) awarded to A.B.L. Natural Sciences and Engineering Research Council of Canada fellowship (PDF-516634-2018) awarded to R.A. S.O.Z. was supported by the Gates Cambridge Trust. J.P.-G. was supported by an Erasmus+ traineeship, and B.C. was supported by the Balfour-Browne Fund from the University of Cambridge Zoology Department.
Author contributions
R.A., S.O.Z., J.P.D., A.B.L., and F.M.J. designed research; R.A., S.O.Z., J.P.D., S.B., S.P., C.-Y.H., S.O., J.P.-G., B.C., R.J.C., and A.B.L. performed research; R.A., S.O.Z., J.P.D., S.B., S.P., Y.Z., C.-Y.H., S.O., J.P.-G., B.C., R.J.C., A.B.L., and F.M.J. analyzed data; and R.A., S.O.Z., J.P.D., S.B., S.P., A.B.L., and F.M.J. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Preprint Servers: The manuscript was posted on bioRxiv under the “No reuse/adaptation withoutpermission” license.
This article is a PNAS Direct Submission.
Contributor Information
Ramesh Arunkumar, Email: ar885@cam.ac.uk.
Alexandre B. Leitão, Email: alexandre.leitao@research.fchampalimaud.org.
Francis M. Jiggins, Email: fmj1001@cam.ac.uk.
Data, Materials, and Software Availability
Miseq reads for cDNA and gDNA for allele-specific expression of lectin-24A were deposited into the NCBI Sequence Read Archive under Bioproject PRJNA789229 (65). The DGRP-437 lectin-24A coding gene sequence and the germline Cas9 mutant sequence were deposited into GenBank: OM100576–OM100577. Scripts and processed data files are available in Zenodo (66). All other data are included in the manuscript and/or SI Appendix.
Supporting Information
References
- 1.Sabeti P. C., et al. , Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howes R. E., et al. , The global distribution of the Duffy blood group. Nat. Commun. 2, 266 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgner D., Jamieson S. E., Blackwell J. M., Genetic susceptibility to infectious diseases: Big is beautiful, but will bigger be even better? The Lancet Infectious Dis. 6, 653–663 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman S. J., Hill A. V., Human genetic susceptibility to infectious disease. Nat. Rev. Genet. 13, 175–188 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Salvaudon L., Giraud T., Shykoff J. A., Genetic diversity in natural populations: A fundamental component of plant-microbe interactions. Curr. Opin. Plant Biol. 11, 135–143 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Laine A. L., Burdon J. J., Dodds P. N., Thrall P. H., Spatial variation in disease resistance: From molecules to metapopulations. J. Ecol. 99, 96–112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lazzaro B. P., Sceurman B. K., Clark A. G., Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science 303, 1873–1876 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Tinsley M. C., Blanford S., Jiggins F. M., Genetic variation in Drosophila melanogaster pathogen susceptibility. Parasitology 132, 767–773 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obbard D. J., Dudas G., The genetics of host-virus coevolution in invertebrates. Curr. Opin. Virol. 8, 73–78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duxbury E. M., et al. , Host-pathogen coevolution increases genetic variation in susceptibility to infection. Elife 8, e46440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duffy M. A., et al. , Ecological context influences epidemic size and parasite-driven evolution. Science 335, 1636–1638 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Penczykowski R. M., Laine A. L., Koskella B., Understanding the ecology and evolution of host-parasite interactions across scales. Evol. Appl. 9, 37–52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thrall P. H., et al. , Rapid genetic change underpins antagonistic coevolution in a natural host-pathogen metapopulation. Ecol. Lett. 15, 425–435 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westra E. R., et al. , Parasite exposure drives selective evolution of constitutive versus inducible defense. Curr. Biol. 25, 1043–1049 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Boots M., The evolution of resistance to a parasite is determined by resources. Am. Nat. 178, 214–220 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Gwynn D. M., Callaghan A., Gorham J., Walters K. F., Fellowes M. D., Resistance is costly: trade-offs between immunity, fecundity and survival in the pea aphid. Proc. Biol. Sci. 272, 1803–1808 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medzhitov R., Schneider D. S., Soares M. P., Disease tolerance as a defense strategy. Science 335, 936–941 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burmeister A. R., et al. , Pleiotropy complicates a trade-off between phage resistance and antibiotic resistance. Proc. Natl. Acad. Sci. U.S.A. 117, 11207–11216 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rigby M. C., Hechinger R. F., Stevens L., Why should parasite resistance be costly? Trends Parasitol. 18, 116–120 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Levin B. R., Perrot V., Walker N., Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics 154, 985–997 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bjorkman J., Nagaev I., Berg O. G., Hughes D., Andersson D. I., Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287, 1479–1482 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Allen R. C., Engelstadter J., Bonhoeffer S., McDonald B. A., Hall A. R., Reversing resistance: Different routes and common themes across pathogens. Proc. Biol. Sci. 284, 20171619 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kraaijeveld A. R., Godfray H. C., Trade-off between parasitoid resistance and larval competitive ability in Drosophila melanogaster. Nature 389, 278–280 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Fellowes M. D., Kraaijeveld A. R., Godfray H. C., Trade-off associated with selection for increased ability to resist parasitoid attack in Drosophila melanogaster. Proc. Biol. Sci. 265, 1553–1558 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fellowes M. D. E., Kraaijeveld A. R., Godfray H. C. J., Association between feeding rate and parasitoid resistance in Drosophila melanogaster. Evolution 53, 1302–1305 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Lemaitre B., Hoffmann J., The Host Defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Kraaijeveld A. R., Godfray H. C. J., Geographic patterns in the evolution of resistance and virulence in Drosophila and its parasitoids. Am. Nat. 153, S61–S74 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Dupas S., Carton Y., Poirie M., Genetic dimension of the coevolution of virulence-resistance in Drosophila – parasitoid wasp relationships. Heredity (Edinb) 90, 84–89 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Orr H. A., Irving S., The genetics of adaptation: The genetic basis of resistance to wasp parasitism in Drosophila melanogaster. Evolution 51, 1877–1885 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Poirie M., et al. , Drosophila resistance genes to parasitoids: Chromosomal location and linkage analysis. Proc. Biol. Sci. 267, 1417–1421 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hita M., et al. , Mapping candidate genes for Drosophila melanogaster resistance to the parasitoid wasp Leptopilina boulardi. Genet Res. 88, 81–91 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Jalvingh K. M., Chang P. L., Nuzhdin S. V., Wertheim B., Genomic changes under rapid evolution: Selection for parasitoid resistance. Proc. Biol. Sci. 281, 20132303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerritsma S., et al. , Natural and artificial selection for parasitoid resistance in Drosophila melanogaster leave different genetic signatures. Front. Genet. 10, 479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim-Jo C., Gatti J. L., Poirie M., Drosophila cellular immunity against parasitoid wasps: A complex and time-dependent process. Front Physiol. 10, 603 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leitão A. B. Recognition of non-self is necessary to activate Drosophila's immune response against an insect parasite. bioRxiv 2022.06.28.497890 (2022).
- 36.Wertheim B., et al. , Genome-wide gene expression in response to parasitoid attack in Drosophila. Genome Biol. 6, R94 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keebaugh E. S., Schlenke T. A., Adaptive evolution of a novel Drosophila lectin induced by parasitic wasp attack. Mol. Biol. Evol. 29, 565–577 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlenke T. A., Morales J., Govind S., Clark A. G., Contrasting infection strategies in generalist and specialist wasp parasitoids of Drosophila melanogaster. PLoS Pathog 3, 1486–1501 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown G. D., Willment J. A., Whitehead L., C-type lectins in immunity and homeostasis. Nat. Rev. Immunol. 18, 374–389 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Natarajan K., Dimasi N., Wang J., Mariuzza R. A., Margulies D. H., Structure and function of natural killer cell receptors: Multiple molecular solutions to self, nonself discrimination. Annu. Rev. Immunol. 20, 853–885 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Hanson M. A., et al. , The Drosophila Baramicin polypeptide gene protects against fungal infection. PLoS Pathog. 17, e1009846 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ko L. J., Engel J. D., DNA-binding specificities of the GATA transcription factor family. Mol. Cell Biol. 13, 4011–4022 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan R., Small S., Desplan C., Dearolf C. R., Darnell J. E. Jr., Identification of a Stat gene that functions in Drosophila development. Cell 84, 421–430 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Huang W., et al. , Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24, 1193–1208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mackay T. F., et al. , The Drosophila melanogaster Genetic Reference Panel. Nature 482, 173–178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Everett L. J., et al. , Gene expression networks in the Drosophila Genetic Reference Panel. Genome Res. 30, 485–496 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Early A. M., et al. , Survey of global genetic diversity within the Drosophila immune system. Genetics 205, 353–366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lack J. B., Lange J. D., Tang A. D., Corbett-Detig R. B., Pool J. E., A thousand fly genomes: An expanded Drosophila genome nexus. Mol. Biol. Evol. 33, 3308–3313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yi X., et al. , Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng J. Y., Stern A. J., Racimo F., Nielsen R., Detecting selection in multiple populations by modeling ancestral admixture components. Mol. Biol. Evol. 39, msab294 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fleury F., et al. , Ecological and genetic interactions in Drosophila-parasitoids communities: A case study with D. melanogaster, D. simulans and their common Leptopilina parasitoids in south-eastern France. Genetica 120, 181–194 (2004). [DOI] [PubMed] [Google Scholar]
- 52.McGonigle J. E., et al. , Parallel and costly changes to cellular immunity underlie the evolution of parasitoid resistance in three Drosophila species. PLoS Pathog. 13, e1006683 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramond E., et al. , The adipokine NimrodB5 regulates peripheral hematopoiesis in Drosophila. FEBS J. 287, 3399–3426 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Karban R., Agrawal A. A., Mangel M., The benefits of induced defenses against herbivores. Ecology 78 (1997). [Google Scholar]
- 55.Olson M. V., When less is more: gene loss as an engine of evolutionary change. Am. J. Hum. Genet 64, 18–23 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Monroe J. G., McKay J. K., Weigel D., Flood P. J., The population genomics of adaptive loss of function. Heredity (Edinb) 126, 383–395 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu Y. C., Guo Y. L., Less is more, natural loss-of-function mutation is a strategy for adaptation. Plant Commun. 1, 100103 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanson M. A., Lemaitre B., Unckless R. L., Dynamic evolution of antimicrobial peptides underscores trade-offs between immunity and ecological fitness. Front Immunol. 10, 2620 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allemand R., et al. , Phylogeny of six African Leptopilina species (Hymenoptera: Cynipoidea, Figitidae), parasitoids of Drosophila, with description of three new species. Ann. de la Société entomologique de France (N.S.) 38, 319–332 (2013). [Google Scholar]
- 60.Salazar-Jaramillo L., et al. , Inter- and intra-species variation in genome-wide gene expression of Drosophila in response to parasitoid wasp attack. BMC Genomics 18, 331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie J., Butler S., Sanchez G., Mateos M., Male killing Spiroplasma protects Drosophila melanogaster against two parasitoid wasps. Heredity (Edinb) 112, 399–408 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salazar-Jaramillo L., et al. , Evolution of a cellular immune response in Drosophila: A phenotypic and genomic comparative analysis. Genome Biol. Evol. 6, 273–289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dudzic J. P., Kondo S., Ueda R., Bergman C. M., Lemaitre B., Drosophila innate immunity: Regional and functional specialization of prophenoloxidases. BMC Biol. 13, 81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salazar-Jaramillo L., Wertheim B., Does Drosophila sechellia escape parasitoid attack by feeding on a toxic resource? PeerJ 9, e10528 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arunkumar R., Jiggins F. M., Allele specific expression for Lectin-24A in hybrid Drosophila Genetic Reference Panel lines. NCBI Sequence Read Archive (SRA). https://www.ncbi.nlm.nih.gov/bioproject/PRJNA789229. Deposited 15 December 2021. [Google Scholar]
- 66.Arunkumar R., Parasitoid-resistance: v3. Zenodo. https://zenodo.org/record/7987828. Deposited 30 May 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Miseq reads for cDNA and gDNA for allele-specific expression of lectin-24A were deposited into the NCBI Sequence Read Archive under Bioproject PRJNA789229 (65). The DGRP-437 lectin-24A coding gene sequence and the germline Cas9 mutant sequence were deposited into GenBank: OM100576–OM100577. Scripts and processed data files are available in Zenodo (66). All other data are included in the manuscript and/or SI Appendix.