Summary
Organoids are unique tools to mimic how tumors evolve in a 3D environment. Here, we present a protocol to embed spheroids invading a 3D matrix into a paraffin mold. We describe steps for preparing spheroids, collagen and agarose inclusion, and paraffinization. We then detail procedures for sectioning, staining, and visualization. This protocol allows histological identification of markers expressed in cells escaping the tumor.
For complete details on the use and execution of this protocol, please refer to Guyon et al. (2022).1
Subject areas: Cell Biology, Cell Culture, Cancer
Graphical abstract

Highlights
-
•
Histological analysis of invasive spheroids
-
•
Detailed description of paraffin inclusion and sectioning of invasive spheroids
-
•
Immunostaining and detection of markers in invasive cells
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Organoids are unique tools to mimic how tumors evolve in a 3D environment. Here, we present a protocol to embed spheroids invading a 3D matrix into a paraffin mold. We describe steps for preparing spheroids, collagen and agarose inclusion, and paraffinization. We then detail procedures for sectioning, staining, and visualization. This protocol allows histological identification of markers expressed in cells escaping the tumor.
Before you begin
We hereby present a histological method for obtaining transverse or sagittal sections of organoids or spheroids invading a three-dimensional matrix that are embedded in paraffin.1 Capturing cell adaptations during invasive processes is central to better understanding tumor development. Three-dimensional cell structures such as spheroids, neurospheres, or organoids are unique tools to study invasive process. Immunofluorescence of spheroids invading in a 3D gel is challenging for visualizing subcellular localizations of intracellular proteins or organelles. This detailed protocol overcomes difficulties to include spheroids invading into 3D matrices in a paraffin mold for future sectioning and imaging.
Cell culture and preparation of homogeneous size spheroids
Timing: 3 days
This protocol has been developed using non-adherent glioblastoma cells (P3 glioblastoma stem-like cells) which grow as heterogeneous spheroids in the culture flasks. Cells are cultured in complete neurobasal medium (NBM) without serum. It corresponds to NBM supplemented with B27 1×, heparin (100 U/μL), 20 ng/mL basic FGF, and antibiotics (penicillin-streptomycin 1,000 U/mL) at 37°C in a 5% CO2 incubator. For complete details on the use and execution of this protocol, please refer to Guyon et al.2
-
1.
Collect heterogeneous spheroid in 10 mL tube.
-
2.Spheroid rinsing:
-
a.centrifuge the tube at 1,000 × g for 5 min,
-
b.remove the supernatant,
-
c.wash with PBS.
-
a.
-
3.Spheroid dissociation:
-
a.centrifuge the tube at 1,000 × g for 5 min,
-
b.remove the supernatant,
-
c.dissociate spheroids with 1 mL of Accutase (700 U/mL) for 15 min at 37°C.
-
a.
-
4.
Recover cells with 9 mL of complete NBM.
-
5.
Count the cells and prepare a solution at 100,000 cells per 1 mL with complete NBM and 0.4% of methylcellulose.
-
6.
Add 100 μL (10,000 cells) per well into 96 round bottom well-plate.
Note: Spheroids can be used after 3 days of culture.
The protocol below describes the specific steps including the invasion experiment in a Transwell insert, fixation, embedment in paraffin, and cutting for immunohistochemical labeling.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Tom20 (1:200 dilution) | Santa Cruz Biotechnology | Sc-11415 |
| Donkey anti-rabbit IgG (1:500 dilution) | Invitrogen | A16028 |
| Chemicals, peptides, and recombinant proteins | ||
| Agarose D5 | Euromedex | D5-E |
| FGF-basic | PeproTech | 100-18B |
| Collagen type I | Corning | 354236 |
| B27 50× | Thermo Fisher Scientific | 17504044 |
| Bovine serum albumin | Euromedex | 04100812C |
| DAPI (1:2,000) | Thermo Fisher Scientific | D1306 |
| DMSO | Sigma-Aldrich | D8418 |
| Ethanol absolute | VWR Chemicals | 20821.310 |
| Ethanol 96% | VWR Chemicals | 20905.320 |
| Donkey serum | Sigma-Aldrich | S30-100ML |
| Heparin | Sigma-Aldrich | H3149 |
| NaOH 1 M | Sigma-Aldrich | 655104 |
| Neurobasal medium | Gibco | 2110349 |
| Methylcellulose | Sigma-Aldrich | M0262 |
| Paraformaldehyde 4% | ChemCruz | Sc-281692 |
| Paraffin | Leica | 39602006 |
| Phalloidin rhodamine (1:100) | Thermo Fisher Scientific | R415 |
| Phosphate-buffered saline | Gibco | 14190-094 |
| ProLong Gold antifade reagent | Thermo Fisher Scientific | P36934 |
| Sterile distilled water (dH2O) | Thermo Fisher Scientific | 10977015 |
| Toluene | VWR | 28676.322P |
| Triton X-100 | Sigma | X100 |
| Experimental models: Cell lines | ||
| P3 glioblastoma stem-like cells (human) | N/A | N/A |
| Other | ||
| Permeable support for 24-well plate with 0.8 μm transparent PET membrane | Corning | 353097 |
| 24-Well plate | Falcon | 353504 |
| Microtome | Leica | RM 2235 |
| Mold | Thermo Fisher Scientific | NC9991740 |
| Cassette | VWR | 720-2232 |
| Slides | Dako | K8020 |
Materials and equipment
Collagen matrix
| Reagent | Final concentration | Amount |
|---|---|---|
| Collagen type I | 1 mg/mL | N/A |
| NaOH 1 M | N/A | 0,023 ∗ Volume of collagen |
| PBS 10× | 1× | N/A |
| dH2O | N/A | Depending on number of wells |
The collagen solution should be prepared at 0°C and kept at 0°C for 30 min before use. It cannot be stored for later use; it must be used quickly after preparation.
Agarose gel 1%
| Reagent | Final concentration | Amount |
|---|---|---|
| Agarose | 1% | 0.5 g |
| dH2O | N/A | 50 mL |
We microwave the solution in a 250 mL Erlenmeyer flask until the agarose is completely dissolved. The solution can be stored 1 month at 24°C. To reuse the agarose solution, re-melt the solidified gel in a microwave.
Citrate buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Citric acid (anhydrous) | 10 mM | 1.92 g |
| dH2O | N/A | 1,000 mL |
Shake to dissolve the solution by using a magnetic stirrer. Adjust pH to 6.0 with 1 M NaOH. This buffer can be stored at 24°C for 3 months or at 4°C for 6 months.
Step-by-step method details
Spheroid inclusion into collagen matrix
Timing: 24 h
This step contains the inclusion of the spheroid into the collagen matrix. This setting generates a microenvironment that permits tumor spheroid to invade three-dimensionally.
-
1.
Collect a spheroid with a cut tip in a 0.5 mL Eppendorf tube.
-
2.
Remove the supernatant and let the spheroid in a small residual volume.
-
3.
Wash carefully the spheroid with 100 μL PBS twice.
-
4.
Remove the PBS and let the spheroid in a small residual volume.
-
5.Include the spheroid in collagen:
-
a.pipette 150 μL of collagen matrix with a cut tip,
-
b.resuspend the spheroid in the collagen matrix,
-
c.aspirate the spheroid and the collagen matrix with the same tip, and,
-
d.place it gently on the permeable support for 24-well plate with a 0.8 μm transparent PET membrane (Figures 1A and 1B).
-
a.
Note: Center the spheroid in the permeable support. Troubleshooting 1
-
6.
Incubate 30 min at 37°C.
-
7.
Add 150 μL of NBM in the well.
-
8.
Incubate 24 h at 37°C (Figure 1C). Troubleshooting 2.
CRITICAL: Before removing the supernatant, wait for the spheroid to pellet in the tube.
CRITICAL: Do not remove the whole liquid in which the spheroid is immersed.
Figure 1.
Collagen invasion in the permeable insert
(A) Upper view of a well containing the permeable insert.
(B) Schematic representation of a well containing a spheroid in a collagen gel.
(C) Bright-field images of spheroid at time 0 and 24 h during the invasion assay in a collagen matrix inside the permeable insert. Scale bar 100 μm.
Sample fixation and partial dehydration
Timing: 5 h
This step includes the fixation of samples in paraformaldehyde and first dehydration stages. Fixation is an important stage in histology, as it stabilizes proteins while preserving tissue structure.
-
9.Fix the sample:
-
a.remove the supernatant by inverting the insert,
-
b.fill the insert with 500 μL paraformaldehyde (PFA) 4%,
-
c.incubate 4 h at 24°C.
-
a.
Note: Due to PFA toxicity, it is mandatory to use a laboratory forceps to handle the permeable insert.
-
10.
Remove PFA 4% by inverting the permeable support.
-
11.
Wash the sample with PBS thrice.
-
12.
Remove PBS by inverting the permeable support.
-
13.
Fill the insert with 500 μL of ethanol 70% and incubate 1 h at 24°C.
-
14.
Remove ethanol 70% by inverting the insert.
-
15.
Fill the insert with 500 μL of ethanol 70% and incubate 12 h at 24°C.
Note: At the end of this step, the sample can be stored in ethanol 70% at 4°C for several days.
Inclusion of samples in an agarose matrix 1%
Timing: 30 min
This step enables samples to be embedded with agarose gel for easier handling.
-
16.
Remove ethanol 70% by inverting the insert.
-
17.Prepare liquid agarose 1%:
-
a.dissolve 1% of agarose in distilled water using a microwave while avoiding boiling,
-
b.allow the gel to cool down to approximately 37°C–40°C,
-
a.
-
18.Include the sample in agarose:
-
a.fill the insert with 500 μL of liquid agarose gel (Figure 2Ai),
-
b.wait for the gel to solidify.
-
a.
-
19.
Gently cut the membrane along the edge of the insert, then remove it from the insert with fine forceps.
-
20.
Flip the insert and carefully remove the gel from it (Figures 2Aii and 2B).
-
21.
Remove a lower part of the agarose gel (Figures 2Aiii and 2C).
-
22.
Place the sample in a mold and fill the mold with non-boiling liquid agarose gel 1% (Figures 2Aiv and 2D).
-
23.
Shape a small parallelepiped and place it in embedding cassettes (Figures 2Av and 2F).
Figure 2.
Embedding of the sample in agarose
(A) Schematic representation of the different steps. (i) The insert is filled with agarose. (ii) The permeable insert is flipped down and the membrane is carefully removed to demold the sample. (iii) A part of the agarose is removed by cutting with a scalpel. (iv) The sample is deposited in a disposable histology mold and is filled a second time with agarose. (v) Cut the agarose into a small cube.
(B) Image of the demolded sample (A ii).
(C) Upper view of the sample (A iii). Scale bar 1 mm.
(D) Image of the sample embedded in agarose inside a disposable histology mold (A vi).
(E) Image of the trimmed sample in a histology cassette (A v).
Total dehydration of the sample and paraffinization
Timing: 24 h
As paraffin is a hydrophobic inclusion medium, the sample must be first completely dehydrated by immersion in alcohol baths of increasing degree, then in toluene baths. The liquid paraffin is then used to infiltrate the sample to protect it and ensure long preservation. The main purpose of paraffin inclusion is to obtain histological sections.
-
24.Immerse the cassette in successive baths of:
-
a.ethanol 96% at 24°C for 1 h, twice.
-
b.ethanol absolute at 24°C 12 h.
-
c.ethanol absolute at 24°C for 1 h.Note: Sample can be stored in ethanol absolute at 4°C for a few days, it will be necessary to repeat an ethanol absolute bath for 1 h before incubating into toluene.
-
d.toluene at 24°C for 45 min, twice. Troubleshooting 3.
-
e.liquid paraffin 60°C for 1 h, twice.
-
a.
-
25.
Insert the sample in the desired direction in the inclusion block and embed with paraffin.
Note: Paraffin embedded samples can be stored for a very long time at 24°C.
Blade sectioning
Timing: 24 h
The paraffin block is cut with a microtome to produce thin section for subsequent histological analyses. Sections are collected on glass slides and dried.
-
26.
Section the paraffin block with a thickness of 10 μm (Figures 3A and 3B).
-
27.
On a slide covered with water heated to 37°C, place the paraffin section on the hot water to expand.
-
28.
Remove water.
-
29.
Dry them at 37°C 12–24 h.
Note: Paraffin section can be stored for a very long time at 4°C.
Figure 3.
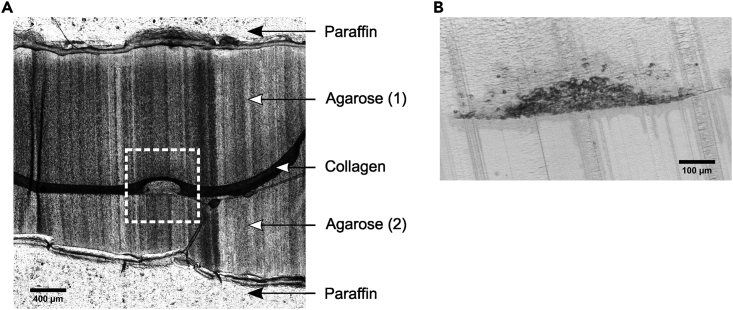
Raw paraffin frontal section of spheroid invading a collagen matrix
(A) Global histological section showed the succession of layers surrounding the spheroid (in the white square), recapitulating the previous steps. Scale bar 400 μm.
(B) Image of the invasive spheroid. Scale bar 100 μm.
Staining procedures and acquisition
Timing: 2 days
Most tissues are transparent, slide staining can be used to identify individual tissue elements. Before immunostaining, fixed paraffin-embedded tissues need to undergo few steps, such as deparaffinization, antigen retrieval and permeabilization.
-
30.Immerse the paraffin-embedded sections in:
-
a.toluene at 24°C for 5 min, twice.
-
b.ethanol at 24°C absolute for 5 min.
-
c.ethanol at 24°C 96% for 5 min.
-
d.ethanol at 24°C 70% for 5 min.
-
e.distilled H2O at 24°C for 5 min.
-
f.citrate buffer at 95°C for 1 h.
-
a.
-
31.Staining.
-
a.Permeabilize the samples with 0.1% Triton X-100 in PBS for 15 min.
-
b.Wash in PBS for 5 min.
-
c.Block with 1% bovine serum albumin and 2% donkey serum in PBS (blocking buffer) for 1 h.
-
d.Delineate the section with hydrophobic pen.
-
e.Incubate with primary antibodies (1:100) in blocking buffer 12–24 h at 4°C.
-
f.Wash in PBS thrice.
-
g.Incubate 1 h at 24°C the samples in blocking buffer containing:
-
i.Secondary antibodies (1:200),
-
ii.DAPI (1:2,000) for staining DNA,
-
iii.Phalloidin rhodamine (1:500) for staining cytoskeleton.
-
i.
-
h.Wash in PBS thrice.
-
i.Mount with ProLong Gold antifade reagent.
-
a.
-
32.
Image with a confocal microscope (Figure 4).
Figure 4.
Deparaffinized, rehydrated, and stained paraffin section
Bright-field and fluorescent images of a spheroid invading a collagen matrix stained for phalloidin (binding to F-actin) in red, DAPI (binding adenine-thymine in DNA) in blue, and TOM20 (binding mitochondrial import receptor subunit) in green. Scale bars 100 μm and 25 μm.
Expected outcomes
This protocol allows a histological analysis of cells organized in spheroid, invading in 3D matrices. Cutting an invasive spheroid on several slides allows the use of many markers on the same sample and reduces the quantity of antibodies used in the study. The information extracted from the images are at the cellular/subcellular level, deciphering cell/cell or cell/matrix interactions (Figure 4).
The spheroids presented in this protocol were composed of stem-like glioblastoma cells, derived from patient surgical samples and maintained in non-serum based medium. This cell culture type is a good compromise between patient-derived xenografts, mainly passaged in vivo (e.g., in rodents)3 and serum-derived cell lines, which do not recapitulate glioblastoma features.4 We have recently shown that lactate dehydrogenases are differentially expressed in the center of these invading spheroids, spatial localizations which was confirmed in in vivo models derived from these spheroids and patient samples. This confirms the robustness of studying such in vitro cell aggregates for partially replacing in vivo models based on rodent cerebral implantation.1 In a clinical context, better understanding how glioblastoma cells invade surrounding tissues is crucial as current treatments, based on aggressive surgery followed by chemo-radiotherapy, do not eliminate these highly motile cells which are the seeds of recurrent glioblastoma.
Limitations
The main limitations of this protocol are related to inclusion quality of the invasive spheroid into the paraffin. Careful handling of collagen gel with invasive spheroid is the key step for avoiding any destruction of the spheroid polarity.
Troubleshooting
Problem 1
The spheroid in collagen matrix is not centered in the permeable insert (related to major step 1).
Potential solution
As long as the collagen has not polymerized, the insert can be gently tapped to center the spheroid.
Problem 2
The spheroid cells are not invading (related to major step 1).
Potential solution
-
•
If collagen gel has not formed, check that the pH of the collagen matrix is between 6.5 and 7.5.
-
•
If collagen gel has formed, change the density of the collagen matrix, either by softening or rigidifying the gel. Some cells prefer softer or more rigid 3D microenvironment.
Problem 3
Agarose sample shrinks after toluene baths (related to major step 4).
Potential solution
Water was probably not completely removed from the sample during ethanol bath (especially if sample has been stored for some time in the refrigerator in absolute ethanol). To avoid this problem, it may be necessary to incubate the sample for 1 h in a fresh absolute ethanol bath.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr Thomas Daubon (thomas.daubon@u-bordeaux.fr).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This protocol includes all datasets generated or analyzed during this study.
Acknowledgments
This work was supported by the INCa-Canceropole GSO (N2020-EC29), SIRIC BRIO2, Fondation ARC (N216415#1), ARTC (N225119#1), Ligue Contre le Cancer (N228841#1), and PLBIO INCA (N227441#1 and 241284#1).
Author contributions
J.G. and T.D. designed the technique. J.G. developed the detailed technique and performed all experiments. J.G. and T.D. discussed the results and wrote the manuscript. T.D. supervised the work of J.G. Both the authors have read and agreed to the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
References
- 1.Guyon J., Fernandez-Moncada I., Larrieu C.M., Bouchez C.L., Pagano Zottola A.C., Galvis J., Chouleur T., Burban A., Joseph K., Ravi V.M., et al. Lactate dehydrogenases promote glioblastoma growth and invasion via a metabolic symbiosis. EMBO Mol. Med. 2022;14 doi: 10.15252/emmm.202115343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guyon J., Andrique L., Pujol N., Røsland G.V., Recher G., Bikfalvi A., Daubon T. A 3D Spheroid Model for Glioblastoma. J. Vis. Exp. 2020 doi: 10.3791/60998. [DOI] [PubMed] [Google Scholar]
- 3.Golebiewska A., Hau A.-C., Oudin A., Stieber D., Yabo Y.A., Baus V., Barthelemy V., Klein E., Bougnaud S., Keunen O., et al. Patient-derived organoids and orthotopic xenografts of primary and recurrent gliomas represent relevant patient avatars for precision oncology. Acta Neuropathol. 2020;140:919–949. doi: 10.1007/s00401-020-02226-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N.M., Pastorino S., Purow B.W., Christopher N., Zhang W., et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This protocol includes all datasets generated or analyzed during this study.