Abstract
Anginosus group streptococci (AGS) are opportunistic human pathogens of the oral cavity. The β-hemolytic subgroup of Streptococcus anginosus subsp. anginosus secretes streptolysin S (SLS) and exhibits not only hemolytic activity but also cytotoxicity toward cultured human cell lines. However, the detailed mechanism of action of SLS and the cellular responses of host cells have not yet been fully clarified. To determine the pathogenic potential of SLS-producing β-hemolytic S. anginosus subsp. anginosus, the SLS-dependent response induced in the human oral squamous cell carcinoma HSC-2 cells was investigated to determine the pathogenic potential of SLS-producing β-hemolytic S. anginosus subsp. anginosus. This study revealed that the Ca2+ influx and the expression of immediate early genes (IEGs) encoding transcription factors such as early growth responses (EGRs) and activator protein-1 (AP-1) were greatly increased in HSC-2 cells incubated with the culture supernatant of SLS-producing β-hemolytic S. anginosus subsp. anginosus. Moreover, this SLS-dependent increase in expression was significantly suppressed by Ca2+ chelation, except for jun. These results suggest that SLS caused Ca2+ influx into the cells following greatly enhanced expression of IEG-encoding transcription factors. The results of this study may help in understanding the pathogenicity of SLS-producing AGS.
Subject terms: Cell biology, Microbiology, Diseases, Pathogenesis
Introduction
Streptolysin S (SLS) is a β-hemolysin produced by strains belonging to the pyogenic and anginosus groups of β-hemolytic streptococci1,2. SLS is generated as the product of an operon composed of 9 genes (sagA–sagI) called the “sag operon”3, except for the sag operon of β-hemolytic S. anginosus subsp. anginosus, which is composed of 10 genes (sagA1, sagA2, and sagB–sagI)4,5. The formation of heterocycle (thiazole and oxazole) rings in the precursor peptide is essential for the hemolytic activity of SLS. Thus, SLS is one of the bacteriocins called “thiazole and oxazole modified microcins (TOMMs)”6. The SLS has mainly been investigated in the human pathogenic Streptococcus pyogenes, and its contributions to hemolysis, cytotoxicity, and pathogenicity have been demonstrated both in vitro and in vivo7–12.
We are investigating the β-hemolytic factors produced by human opportunistic anginosus group streptococci (AGS). Among the species belonging to AGS, Streptococcus intermedius produces a cholesterol-dependent cytolysin (CDC) called intermedilysin (ILY) which is the sole β-hemolytic factor of the species with human cell-specific action13,14. In contrast, the β-hemolytic subgroup of other AGS species, such as S. anginosus and S. constellatus, produce the homologs of SLS as the sole β-hemolytic factors4,15. Interestingly, β-hemolytic strains of S. anginosus subsp. anginosus produce two functional mature SLSs with different amino acids4. Moreover, the SLS secreted by β-hemolytic AGS strains shows not only hemolytic activity but also cytotoxicity15,16. Recently, we reported that SLS secreted by hemolytic AGS strains and other SLS-producing streptococci was stabilized in the presence of human serum albumin (HSA) and showed enhanced hemolysis and cytotoxicity17. Based on this information, we suggested that the presence of gingival crevicular fluid (GCF) is suitable for the growth of SLS-producing AGS strains that exhibit SLS-dependent hemolytic activity and cytotoxicity. In lesions such as gingivitis and periodontal disease, the secreted SLS is apparently stabilized in the presence of HSA and may contribute to disorders in the oral cavity as well as in other parts where the β-hemolytic AGS is observed ectopically. However, the exact mechanism of the action of SLS secreted by β-hemolytic AGS on host cells and the cellular responses to SLS remain unclear.
In the present study, to determine the detailed mechanism of action of SLS secreted by β-hemolytic AGS strains, the HSA-stabilized SLS-dependent cellular response in the human oral squamous cell carcinoma cell line HSC-2 was investigated, focusing on the effect on gene expression.
Results
SLS-dependent cytotoxicity against HSC-2 cells
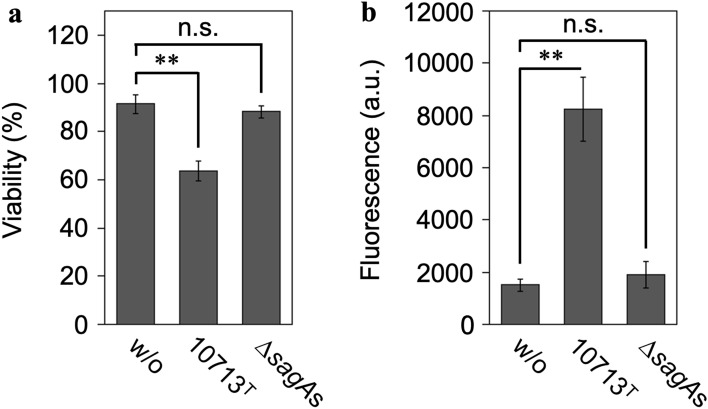
The SLS-dependent cytotoxicity toward HSC-2 cells in the culture supernatant of the tested strains was investigated. The Cell Counting Kit-8 (CCK-8; Dojindo, Masuki, Kumamoto, Japan) assay, which measures the cellular metabolic activity, revealed a partial but significant decrease in the viability of HSC-2 cells incubated with the culture supernatant of the β-hemolytic S. anginosus subsp. anginosus strain NCTC10713T. In contrast, no significant decrease in viability was observed in HSC-2 cells incubated with the culture supernatant of the non-β-hemolytic mutant strain ΔsagAs (Fig. 1a). In addition, the CellTox™ Green Cytotoxicity Assay (Promega, Madison, WI, USA), which reflects the intactness of the cell membrane, showed significant cytotoxicity in HSC-2 cells incubated with the culture supernatant of NCTC10713T but not in those incubated with the culture supernatant of ΔsagAs (Fig. 1b). Therefore, it was confirmed that SLS secreted into the culture medium containing 1.0% (w/v) HSA could induce significant cytotoxicity via cell membrane damage and decrease the viability of HSC-2 cells.
Figure 1.
SLS-dependent cytotoxicity toward the human oral squamous cell carcinoma cell line HSC-2. The viability of HSC-2 cells incubated with the culture supernatant prepared from S. anginosus subsp. anginosus strain NCTC10713T or its sagA-genes (sagA1 and sagA2) deletion mutant (∆sagAs) was evaluated by CCK-8 assay (Dojindo) (a) and by CellTox™ Green assay (Promega) (b). The “w/o” indicates HSC-2 cells incubated with the co-cultivation medium as the control. Triplicate samples were assayed, and the result is presented as the mean ± SD. The significance of the SLS-dependent cytotoxicity was evaluated by Students’ t-test (**p < 0.01, n.s. not significant). a.u. arbitrary unit.
SLS-dependent Ca2+ influx into HSC-2 cells
SLS-induced intracellular Ca2+ influx was investigated. A significant increase in intracellular Ca2+ was observed in HSC-2 cells incubated with the culture supernatant of NCTC10713T (Fig. 2). In contrast, no such increase was observed in HSC-2 cells incubated with the culture supernatant of ΔsagAs, and the intracellular Ca2+ level was almost the same as that in HSC-2 cells incubated with the co-cultivation medium alone (w/o) (Fig. 2). These results suggest that SLS secreted by NCTC10713T induces significant Ca2+ influx into HSC-2 cells.
Figure 2.

SLS-dependent influx of extracellular Ca2+ into HSC-2 cells. The influx of Ca2+ into HSC-2 cells incubated with the culture supernatant prepared from the tested strains was evaluated using Fura-2 Calcium Kit (Dojindo). HSC-2 cells treated with 0.1 μM ionomycin were used as the Ca2+-permeabilized control for this assay. The “w/o” indicates HSC-2 cells incubated with the co-cultivation medium as the control without cultivating the tested strains. Triplicate samples were assayed, and the result is presented as the mean ± SD. The significance of the SLS-dependent increase of Ca2+ influx was evaluated by Students’ t-test (**p < 0.01, n.s. not significant).
SLS-dependent gene expression in HSC-2 cells
RNA-seq analysis was conducted to investigate SLS-dependent changes in gene expression. Differentially expressed genes (DEG) were analyzed in HSC-2 cells incubated with culture supernatants prepared from NCTC10713T and ΔsagAs. The results are presented as a volcano plot showing the changes in gene expression (Fig. 3). SLS-dependent increases and decreases in expression were observed for 433 and 269 genes, respectively. Interestingly, the expression of the genes encoding transcription factors categorized as “immediate-early genes (IEGs)” and some cytokine-encoding genes was remarkably increased in HSC-2 cells incubated with the culture supernatant containing SLS. Among these, genes encoding early growth response (EGR), Fos family proteins, Jun family proteins, and inflammatory cytokines (IL-6 and CXCL8) were selected for further investigation.
Figure 3.

Volcano plot of the SLS-dependent induction (magenta) and reduction (cyan) of the gene expression in HSC-2 cells incubated with the culture supernatant prepared from S. anginosus subsp. anginosus strain NCTC10713T. The SLS-dependent induction or reduction of the expression of genes in HSC-2 cells was evaluated using the results of RNA-seq analysis outsourced to Macrogen Japan. The genes focused on in this study are indicated with black symbols (IEGs) and green symbols (cytokine genes) with the product name.
SLS-dependent IEG expression is significantly suppressed in HSC-2 cells by Ca2+ chelation
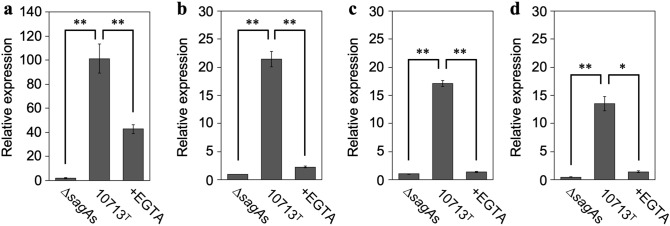
The results of the DEG analysis (Fig. 3) showed increased expression of three egr genes (egr1, egr3, and egr4). As four egr genes (egr1, egr2, egr3, and egr4) have been reported to date, they were further investigated using quantitative real-time polymerase chain reaction (RT-qPCR) to reveal the relationship between SLS-dependent increases in the expression of IEGs and Ca2+ influx. The expression of all egr genes was greatly increased by incubation with culture supernatants containing SLS (Fig. 4a–d, 10713T compared with ∆sagAs). The SLS-dependent increase in expression was significantly decreased by Ca2+ chelation (Fig. 4a–d, + EGTA compared with 10713T). These results showed that the SLS-dependent expression of all egr genes was triggered by Ca2+ influx, according to the SLS-dependent damage to the cellular membrane.
Figure 4.
SLS-dependent and Ca2+-influx-induced expression of the egr genes, egr1 (a), egr2 (b), egr3 (c), and egr4 (d). HSC-2 cells were incubated with the culture supernatant prepared from S. anginosus subsp. anginosus strain NCTC10713T or its sagA-genes (sagA1 and sagA2) deletion mutant (∆sagAs). The contribution of Ca2+ influx to the expression of the genes was also evaluated in the presence of 5 mM EGTA. Triplicate samples were assayed, and the representative result is presented as the mean ± SD. These results were described as the relative expression against the control sample of HSC-2 cells incubated with co-cultivation medium. The significance of the increase of SLS-dependent and Ca2+-influx-dependent expression was evaluated by Welch’s or Students’ t-test (**p < 0.01, *p < 0.05).
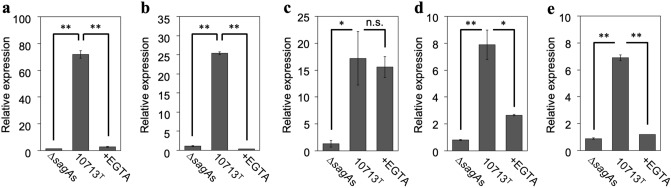
Next, we investigated the SLS-dependent increase in fos gene expression. As shown in Fig. 5a and b, the expression of fos and fosB was greatly increased after incubation with the SLS-containing culture supernatant (10713T compared with ∆sagAs). This SLS-dependent increase in expression was significantly suppressed to the background levels by Ca2+ chelation (Fig. 5a and b, + EGTA compared with 10713T). Thus, these results indicate that the SLS-dependent expression of fos genes was upregulated in a Ca2+-dependent manner, similar to that of egr genes.
Figure 5.
SLS-dependent and Ca2+-influx-induced expression of fos (a), fosB (b), jun (c), junB (d), and junD (e). HSC-2 cells were incubated with the culture supernatant prepared from S. anginosus subsp. anginosus strain NCTC10713T or its sagA-genes (sagA1 and sagA2) deletion mutant (∆sagAs). The contribution of Ca2+ influx to the expression of the genes was also evaluated in the presence of 5 mM EGTA. Triplicate samples were assayed, and the representative result is presented as the mean ± SD. These results were described as the relative expression against the control sample of HSC-2 cells incubated with co-cultivation medium. The significance of the increase of SLS-dependent and Ca2+-influx-dependent expression was evaluated by Welch’s or Students’ t-test (**p < 0.01, *p < 0.05, n.s. not significant).
The SLS-dependent expression of jun genes was also investigated. As shown in Fig. 5c–e, a significant increase in the expression of the three jun genes was observed after incubation with SLS-containing culture supernatant (10713T compared with ∆sagAs), with a similar trend to that observed for the egr and fos genes. The expression of these genes is associated with Ca2+ influx since the SLS-dependent expression of junB and junD was significantly decreased by Ca2+ chelation (Fig. 5d and e, + EGTA compared with 10713T), as were the egr genes and fos genes. However, only the expression of jun gene under the Ca2+-chelating conditions was different (Fig. 5c, + EGTA). No significant change was observed in the SLS-dependent increase in the expression of jun even under Ca2+-chelating conditions (+ EGTA compared with 10713T). This interesting result suggests that the SLS-dependent pathway that induces jun expression differs from that of junB and junD.
SLS-dependent increased expression of cytokine genes is also dependent on the Ca2+ influx in HSC-2 cells
We also investigated whether the SLS-dependent increase in the expression of the inflammatory cytokine genes encoding IL-6 and CXCL8 (IL-8) depended on Ca2+ influx (Fig. 6). RT-qPCR analysis revealed that the expression of both genes was greatly increased in HSC-2 cells treated with culture supernatants containing SLS (10713T compared with ΔsagAs), and this SLS-dependent expression was significantly decreased by Ca2+ chelation (+ EGTA compared with 10713T). These results strongly suggest that the SLS-dependent increase in the expression of the genes encoding IL-6 and CXCL8 in HSC-2 cells also depend on Ca2+-influx.
Figure 6.
SLS-dependent and Ca2+-influx-induced expression of the genes encoding IL-6 (a) and CXCL8 (b). HSC-2 cells were incubated with the culture supernatant prepared from S. anginosus subsp. anginosus strain NCTC10713T or its sagA-genes (sagA1 and sagA2) deletion mutant (∆sagAs). The contribution of Ca2+ influx to the expression of the genes was also evaluated in the presence of 5 mM EGTA. Triplicate samples were assayed, and the representative result is presented as the mean ± SD. These results were described as the relative expression against the control sample of HSC-2 cells incubated with co-cultivation medium. The significance of the increase of SLS-dependent and Ca2+-influx-dependent expression was evaluated by Welch’s or Students’ t-test (**p < 0.01).
Discussion
S. anginosus is an opportunistic pathogen that generally inhabits the human oral cavity. However, reports on the pathogenicity of this species in humans are increasing. For example, this species has been isolated from parts other than the oral cavity in cases of infection and other disorders18. Therefore, S. anginosus is currently attracting attention as a causative bacterium of ectopic infections in humans. There are only two subspecies of S. anginosus: subsp. anginosus and subsp. whileyi19. S. anginosus subsp. whileyi exhibits a β-hemolysis due to the production of SLS encoded by the sag operon which includes nine genes15. In contrast, both β-hemolytic and non-β-hemolytic strains are present in the S. anginosus subsp. anginosus. The hemolytic factor of β-hemolytic subgroup of S. anginosus subsp. anginosus consists of two SLSs with different amino acid sequences4,5. The amino acid sequence of each of SLS precursor (SagA1 and SagA2) was highly conserved among the strains of β-hemolytic S. anginosus subsp. anginosus (Table 1).
Table 1.
Amino acid sequence identity and similarity of SLS precursor (SagA1 and SagA2) among sequenced strains of S. anginosus subsp. anginosus.
| SAA strain (GenBank ID) | SagA1 | SagA2 | ||
|---|---|---|---|---|
| Identity | Similarity | Identity | Similarity | |
| R87/1657 (JN619422.1) | 54/54 (100%) | 54/54 (100%) | 50/50 (100%) | 50/50 (100%) |
| R84/4972 (JN619421.1) | 54/54 (100%) | 54/54 (100%) | 49/50 (98%) | 50/50 (100%) |
| CCUG27928 (AP018548.1) | 54/54 (100%) | 54/54 (100%) | 50/50 (100%) | 50/50 (100%) |
| K20 (JAASHW010000002.1) | 54/54 (100%) | 54/54 (100%) | 50/50 (100%) | 50/50 (100%) |
| T5 (BASY01000013.1) | 54/54 (100%) | 54/54 (100%) | 50/50 (100%) | 50/50 (100%) |
| 47S1 (CP088916.1) | 54/54 (100%) | 54/54 (100%) | 50/50 (100%) | 50/50 (100%) |
SLS is a peptide hemolysin generally produced by pyogenic group streptococci such as S. pyogenes. In vitro studies have revealed that SLS contributes to epithelial barrier translocation9, programmed cell death and inflammatory signaling10, inflammation and cytotoxicity via NBCn112, and mitochondrial damage and macrophage death by inhibition of GSK-3β degradation20. Moreover, in vivo investigations using mouse and zebrafish models21 have reported the recruitment of neutrophils at an early stage of infection. Based on the results of numerous studies, SLS produced by S. pyogenes is generally recognized as an important pathogenic factor in this species.
Although the cytotoxicity of SLS produced by S. anginosus has been previously reported15,16,22, the detailed mechanism of SLS-dependent cytotoxicity and the contribution of S. anginosus-produced SLS to human pathogenicity remain unclear. Recently, we reported that the cytotoxicity of SLS secreted from the β-hemolytic S. anginosus subsp. anginosus strain NCTC10713T and other SLS-producing streptococci was significantly stabilized in the presence of HSA17. This property shows that β-hemolytic S. anginosus subsp. anginosus can exhibit cytotoxicity in the presence of serum albumin in, for example, the GCF and lesions of gingivitis and periodontal disease. Furthermore, if SLS-producing streptococci translocate into the bloodstream, SLS-dependent cytotoxicity may be induced at the ectopic site of infection. This finding confirms that SLS-producing streptococci can establish ectopic infections, explaining the clinical perspective of ectopic infections by oral streptococci in humans.
In the present study, it was shown that the SLS secreted by the β-hemolytic S. anginosus subsp. anginosus strain NCTC10713T not only exhibited cytotoxicity against HSC-2 cells (Fig. 1) but also greatly induced the expression of IEGs encoding transcription factors and genes encoding inflammatory cytokines in a Ca2+-dependent manner (Figs. 4, 5, 6). The Ca2+-dependent expression of IEGs has been reported previously23, and the influx of extracellular Ca2+ is thought to be an important event that triggers increased expression of IEGs. IEGs are general terms for genes that represent rapid and transient but protein synthesis-independent increase in expression in response to extracellular signals such as growth factors and neurotransmitters24. Generally, although IEGs are mainly investigated in the field of neuroscience25,26, many studies on microbial infections have also been reported, such as infection by human pathogenic viruses27–30 and the pathogenicity of bacteria. Examples of the latter include bacterial pore-forming exotoxins, the relationships of IEGs to alpha-toxin produced by Staphylococcus aureus31, intermedilysin produced by S. intermedius32, and other pore-forming toxin33, have been reported. However, to date, there have been no reports on the relationship between IEGs and the streptococcal peptide hemolysin (SLS), also known as bacteriocin TOMMs34. To the best of our knowledge, this is the first report to demonstrate an SLS-dependent cellular response that induces the expression of IEGs that encode transcription factors. Ionomycin was used as a control to investigate Ca2+ influx into HSC-2 cells (Fig. 2). As both the secreted SLS and ionomycin induced Ca2+ influx into HSC-2 cells, our finding may provide the useful information for elucidating the characteristic SLS-induced cellular responses to compare the cellular responses, such as gene-expression profiles, of the HSC-2 cells treated with secreted SLS and ionomycin.
RNA-seq results of SLS-treated HSC-2 cells showed enhanced expression of 433 genes. Among these genes, we focused on IEGs encoding transcription factors, such as EGR, Fos, and Jun, and genes encoding inflammatory cytokines, such as IL-6 and CXCL8. EGRs are zinc finger transcription factors, and Fos and Jun are components of the heterodimeric transcription factor activator protein 1 (AP-1). These transcription factors regulate the transcription of various downstream genes in the cells, and the expression of these transcriptional factors have been associated with various disorders. For example, the expression of EGR1 has been associated with prostate cancer development35, esophageal tumor tissues36, and human gastric cancer progression37; the expression of EGR2 has been associated with the pathogenesis of fibrosis38 and Guillain-Barré Syndrome39; and the expressions of EGR3 and EGR4 has been associated with prostate cancer40 and small cell lung cancer41, respectively. In the case of AP-1, the c-Fos/JunD heterodimer is the most prevalent complex in human oral cancer tissues42. In addition, IL-1β-enhanced c-Fos/c-Jun heterodimer expression led to dose-dependent transcriptional activation of the collagenase promoter43. According to the relationship between IEG-encoding transcription factors and disorders in humans, the greatly enhanced expression of transcription factors induced by secreted SLS may trigger abnormalities in host cells and tissues. Therefore, human opportunistic AGS, which show SLS-dependent-hemolysis, should be re-evaluated as human pathogens, such as S. pyogenes.
Interestingly, the IEGs investigated this study are also related to disorders of the central nervous system. For example, associations have been found between EGR1 and Alzheimer's disease44, EGR2 and Charcot-Marie-Tooth disease45–47, EGR3 and schizophrenia48–50, and c-Jun and Krox24 (EGR1) and Alzheimer's disease51. In nerve cells, the expression of IEGs encoding transcription factors, such as c-Fos and EGR1, is induced by an increase in intracellular Ca2+ as a result of synaptic activity. This intracellular Ca2+-dependent increase in the expression of IEGs that encode transcription factors was also observed in HSC-2 cells treated with SLS secreted from β-hemolytic S. anginosus subsp. anginosus. HSC-2 cells are thought to regulate the expression of genes downstream of IEGs in response to membrane damage caused by SLS, and HSC-2 cells can quickly respond to infection by SLS-producing streptococci, such as by the induction of inflammatory cytokines. Judging from the results of the RNA-seq analysis, not only the expression of IEGs but also the expression of genes encoding cytokines such as IL-6 and CXCL8 increased in HSC-2 cells after incubation with the culture supernatant containing SLS (Figs. 3 and 6). Therefore, SLS-dependent initiation and/or enhancement of inflammation by SLS-producing streptococci, including β-hemolytic S. anginosus, may occur in vivo. Moreover, an association between increased expression of inflammatory cytokines and diseases such as Alzheimer’s disease and schizophrenia has been reported52–54. Therefore, the findings of this study are important for understanding the pathogenesis of SLS-dependent ectopic infections caused by β-hemolytic AGS. Further investigation is currently underway to elucidate the detailed mechanism of SLS-dependent pathogenicity of human habitual hemolytic oral streptococci in disorders of the host ectopically infected by these streptococci.
Methods
Bacterial strains and culture conditions
In this study, S. anginosus subsp. anginosus strain NCTC10713T showing SLS-dependent β-hemolysis4 and a non-β-hemolytic mutant derived from NCTC10713T without both sagA1 and sagA2 genes (∆sagAs)4 were used. The tested strains were preincubated in BHI broth (Becton Dickinson and Company, Franklin Lakes, NJ, USA) at 37 °C overnight with 5% CO2. The preincubated bacteria were inoculated into the co-cultivation medium [Eagle's minimum essential medium (EMEM, FUJIFILM Wako, Osaka, Osaka, Japan) containing 1.0% (w/v) recombinant human serum albumin (HSA; No. 19597-14, Nacalai Tesque Inc., Kyoto, Kyoto, Japan), 10% (v/v) BHI, 25 mM HEPES (pH 7.4)] to an optical density at 600 nm (OD600) of 0.01 and then incubated for 4 h at 37 °C in 5% CO2. Culture supernatants of the tested strains were obtained by centrifugation (13,000 × g, 5 min, RT). HSC-2 cells were incubated with the prepared bacterial culture supernatants in the presence of antibiotics (200 U/mL penicillin G and 200 U/mL streptomycin; FUJIFILM Wako) to inhibit bacterial growth. The absence of bacterial growth during the incubation of HSC-2 cells with the prepared bacterial culture supernatant was confirmed by microscopic observation.
Human cell line and culture condition
The human oral squamous cell carcinoma cell line HSC-2 (RCB1945; RIKEN BRC, Ibaraki, Tsukuba, Japan) was cultured in EMEM cell culture medium (FUJIFILM Wako) containing 10% (v/v) heat-inactivated fetal bovine serum (FBS) and antibiotics (100 U/mL penicillin G and 100 U/mL streptomycin; FUJIFILM Wako) under cell culture conditions (37 °C, 5% CO2 atmosphere).
Cytotoxicity assay
The cytotoxicity of the bacterial culture supernatant toward HSC-2 cells was evaluated using CCK-8 (Dojindo) and CellTox™ Green Cytotoxicity Assay (Promega). For the CCK-8 assay, HSC-2 cells were inoculated at 2.0 × 104 cells/0.1 mL/well in a 96-well plate, incubated overnight, and then incubated with the culture supernatant prepared from each strain for an adequate time. After washing the cells with EMEM, 90 µL of fresh culture medium and 10 µL of CCK-8 reagent were added and the cells were incubated for 1 h at 37 °C in 5% CO2. The absorbance at 450 nm was measured using a plate reader (Infinite® 200 PRO M Nano +; TECAN, Männedorf, Zürich, Switzerland) with a reference wavelength of 600 nm. To prepare the background control for this assay, cells were treated with 0.1 N HCl and incubated for a few minutes prior to the assay. For the CellTox™ Green Cytotoxicity Assay, HSC-2 cells were inoculated at 2.0 × 104 cells/0.1 mL/well in a clear-bottom 96-well black plate and incubated overnight. The culture supernatant prepared from each tested strain was added to each well (0.1 mL/well) and incubated for 2 h. The cytotoxicity of the bacterial culture supernatants was evaluated according to the manufacturer’s instructions. The fluorescence of the samples was measured using a plate reader (Infinite® 200 PRO M Nano +) at excitation and emission wavelengths of 490 and 525 nm, respectively.
Measurement of intracellular Ca2+ concentration
Intracellular Ca2+ concentration was measured using a Fura-2 Calcium Kit (Dojindo), according to the manufacturer’s instructions. HSC-2 cells were inoculated at 2.0 × 104 cells/0.1 mL/well in a clear-bottom 96-well black plate and incubated overnight. Briefly, the Fura 2-AM solution prepared using the loading buffer in the kit was added to the cell culture (0.1 mL/well) and incubated for 1 h under cell culture conditions to allow the incorporation of Fura 2 dye into the cells. After washing, culture supernatant prepared from the test strain was added to each well (0.1 mL/well) and incubated for 30 min. HSC-2 cells were also treated with 0.1 µM of ionomycin as the Ca2+-permeabilization positive control of the assay. After incubation, the cell culture supernatant was replaced with the Recording Medium (0.1 mL/well), and fluorescence was measured using a plate reader (Infinite® 200 PRO M Nano +) with an emission wavelength of 510 nm and excitation wavelengths of 340 nm and 380 nm. The fluctuation in intracellular Ca2+ concentration was evaluated using the 340/380 nm ratio calculated from the measurements.
RNA-seq analysis
HSC-2 cells were inoculated at 4.0 × 105 cells/2.0 mL/well in a 6-well plate and incubated overnight. After washing the cells with EMEM, 2.0 mL of the culture supernatant of the tested strain containing antibiotics (200 U/mL penicillin G and 200 U/mL streptomycin) was added to each well and incubated for 3 h. Total RNA was extracted from the cells in each well using a NucleoSpin® RNA Plus kit (Takara Bio Inc., Kusatsu, Shiga, Japan) and further purified using a NucleoSpin® RNA Clean-up kit (Takara Bio Inc.), according to the manufacturer’s protocol. RNA-seq analysis was performed by Macrogen, Japan (Koto-ku, Tokyo, Japan).
Quantitative reverse transcription-polymerase chain reaction (RT-qPCR)
HSC-2 cells were seeded at a density of 4.0 × 105 cells/well in a 6-well plate containing 2.0 mL of cell culture medium per well and incubated overnight. The cells were then treated with 2.0 mL/well of culture supernatant prepared from the tested strains for 2 h. To investigate the contribution of Ca2+ influx to the expression of the target genes, HSC-2 cells were treated with the culture supernatant containing 5 mM EGTA. After washing with EMEM, total RNA was extracted from the cells in each well using the NucleoSpin® RNA Plus kit (Takara Bio Inc.) according to the manufacturer’s protocol. Reverse transcription was performed using a High-Capacity cDNA Reverse Transcription Kit with RNase inhibitor (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. An oligo (dT)15 primer (TaKaRa Bio Inc.) was used for the reaction instead of the random primer used in the kit. Quantitative real-time PCR was conducted on a Thermal Cycler Dice® Real-Time PCR System Lite TP700 (TaKaRa Bio Inc.) using TB Green® Premix Ex Taq® II Tli RNase H Plus (TaKaRa Bio Inc.). The primers used are listed in Table 2. Relative quantification of target gene expression was conducted using the ΔΔCt method, using GAPDH as the internal control gene.
Table 2.
Primers for RT-qPCR used in this study.
| Primer name | Primer sequence (5′–3′) | Size (bp) | Tm (°C) |
|---|---|---|---|
| GAPDH_forward | GTCTTCACCACCATGGAGAAGGCT | 24 | 55.3 |
| GAPDH_reverse | CATGCCAGTGAGCTTCCCGTTCA | 23 | 55.0 |
| EGR1_forward | CCCACCATGGACAACTACCCTAA | 23 | 53.2 |
| EGR1_reverse | GGAAAAGCGGCCAGTATAGGTGA | 23 | 53.2 |
| EGR2_forward | CCCTTTGACCAGATGAACGGA | 21 | 50.5 |
| EGR2_reverse | CTGGATGAGGCTGTGGTTGA | 20 | 50.0 |
| EGR3_forward | AGCAGCGACTCGGTAGTCCA | 20 | 52.0 |
| EGR3_reverse | GAGTCGAAGGCGAACTTTCCCA | 22 | 52.9 |
| EGR4_forward | GGCCACCGGCTACCCTGGAG | 20 | 58.2 |
| EGR4_reverse | CTAAGATGCCCGACATGAGGTTGA | 24 | 53.6 |
| FOS_forward | TACTCCAGGGCTGGCGTTGT | 20 | 52.0 |
| FOS_reverse | TCTCCTTCAGCAGGTTGGCAATC | 23 | 53.2 |
| FOSB_forward | AACTGCTTCTAGAAACTCTGGCTCA | 25 | 52.2 |
| FOSB_reverse | GAGAAAAGACAGAGGGAGAGAGACC | 25 | 55.5 |
| JUN_forward | GGAGGACCGGAGACAAGTGG | 20 | 54.1 |
| JUN_reverse | CGCCGTGGAGAAGCCTAAGA | 20 | 52.0 |
| JUNB_forward | GGAAAAGAAACACGCACTTAGTCTC | 25 | 52.2 |
| JUNB_reverse | AACACACACAAACACAAACACGTC | 24 | 50.2 |
| JUND_forward | CATTCCTGTTTGTAATCCTTGGTTC | 25 | 50.6 |
| JUND_reverse | GGCGTAACGAGACTTTACTGAAAAC | 25 | 52.2 |
| IL-6_forward | ATGAACTCCTTCTCCACAAGCGC | 23 | 56.9 |
| IL-6_reverse | GAAGAGCCCTCAGGCTGGACTG | 22 | 60.3 |
| IL-8_forward | ATGACTTCCAAGCTGGCCGTGCT | 23 | 58.7 |
| IL-8_reverse | TCTCAGCCCTCTTCAAAAACTTCTC | 25 | 55.9 |
Amino acid sequence identity/similarity
As β-hemolytic S. anginosus subsp. anginosus possess two sagA genes (sagA1 and sagA2) in the sag operon, the target sequences for the analysis were picked out by both Nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and Streptococcus anginosus Nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&PROG_DEF=blastn&BLAST_PROG_DEF=megaBlast&BLAST_SPEC=MicrobialGenomes_1328&DB_GROUP=AllMG) using the nucleotide sequence from sagA1 to sagA2 of S. anginosus subsp. anginosus NCTC10713T (GenBank ID: JN619420.1) as the query. Amino acid sequence identity/similarity was determined using the GENETYX-MAC Network Version 20.0.4 software (GENETYX Corp., Shibuya-ku, Tokyo, Japan). When required, the nucleotide sequence was translated into its amino acid sequence and used for the amino acid sequence identity/similarity assays.
Statistics
Statistical evaluation was conducted using R software for Mac OS X (version 3.6.1; https://cran.r-project.org/bin/macosx/).
Acknowledgements
This work was supported by the JSPS KAKENHI Grant-in-Aid for Scientific Research (C) [grant numbers JP20K09919, JP21K06655, and JP23K09139]. We would like to thank Editage (www.editage.jp) for the English editing.
Author contributions
T.Y., Y.Y. and A.T. wrote the main manuscript and prepared figures and tables. All the authors reviewed the manuscript.
Data availability
All data generated or analyzed during this study are included in this published article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Takuya Yamada and Yugo Yamamori.
References
- 1.Molloy EM, Cotter PD, Hill C, Mitchell DA, Ross RP. Streptolysin S-like virulence factors: The continuing sagA. Nat. Rev. Microbiol. 2011;9:670–681. doi: 10.1038/nrmicro2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tabata A, Nagamune H. Diversity of β-hemolysins produced by the human opportunistic streptococci. Microbiol. Immunol. 2021;65:512–529. doi: 10.1111/1348-0421.12936. [DOI] [PubMed] [Google Scholar]
- 3.Nizet V, Beall B, Bast DJ, Datta V, Kilburn L, Low DE, De Azavedo JC. Genetic locus for streptolysin S production by group A streptococcus. Infect. Immun. 2000;68:4245–4254. doi: 10.1128/IAI.68.7.4245-4254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tabata A, Nakano K, Ohkura K, Tomoyasu T, Kikuchi K, Whiley RA, Nagamune H. Novel twin streptolysin S-like peptides encoded in the sag operon homologue of beta-hemolytic Streptococcus anginosus. J. Bacteriol. 2013;195:1090–1099. doi: 10.1128/JB.01344-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asam D, Mauerer S, Walheim E, Spellerberg B. Identification of β-haemolysin-encoding genes in Streptococcus anginosus. Mol. Oral. Microbiol. 2013;28:302–315. doi: 10.1111/omi.12026. [DOI] [PubMed] [Google Scholar]
- 6.Cox CL, Doroghazi JR, Mitchell DA. The genomic landscape of ribosomal peptides containing thiazole and oxazole heterocycles. BMC Genom. 2015;16:778. doi: 10.1186/s12864-015-2008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betschel SD, Borgia SM, Barg NL, Low DE, De Azavedo JC. Reduced virulence of group A streptococcal Tn916 mutants that do not produce streptolysin S. Infect. Immun. 1998;66:1671–1679. doi: 10.1128/IAI.66.4.1671-1679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyoshi-Akiyama T, Takamatsu D, Koyanagi M, Zhao J, Imanishi K, Uchiyama T. Cytocidal effect of Streptococcus pyogenes on mouse neutrophils in vivo and the critical role of streptolysin S. J. Infect. Dis. 2005;192:107–116. doi: 10.1086/430617. [DOI] [PubMed] [Google Scholar]
- 9.Sumitomo T, Nakata M, Higashino M, Jin Y, Terao Y, Fujinaga Y, Kawabata S. Streptolysin S contributes to group A streptococcal translocation across an epithelial barrier. J. Biol. Chem. 2011;286:2750–2761. doi: 10.1074/jbc.M110.171504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flaherty RA, Puricelli JM, Higashi DL, Park CJ, Lee SW. Streptolysin S promotes programmed cell death and enhances inflammatory signaling in epithelial keratinocytes during group A Streptococcus infection. Infect. Immun. 2015;83:4118–4133. doi: 10.1128/IAI.00611-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higashi DL, Biais N, Donahue DL, Mayfield JA, Tessier CR, Rodriguez K, Ashfeld BL, Luchetti J, Ploplis VA, Castellino FJ, Lee SW. Activation of band 3 mediates group A Streptococcus streptolysin S-based beta-haemolysis. Nat. Microbiol. 2016;1:15004. doi: 10.1038/nmicrobiol.2015.4. [DOI] [PubMed] [Google Scholar]
- 12.Hammers DE, Donahue DL, Tucker ZD, Ashfeld BL, Ploplis VA, Castellino FJ, Lee SW. Streptolysin S targets the sodium-bicarbonate cotransporter NBCn1 to induce inflammation and cytotoxicity in human keratinocytes during Group A Streptococcal infection. Front. Cell Infect. Microbiol. 2022;12:1002230. doi: 10.3389/fcimb.2022.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagamune H, Ohnishi C, Katsuura A, Fushitani K, Whiley RA, Tsuji A, Matsuda Y. Intermedilysin, a novel cytotoxin specific for human cells secreted by Streptococcus intermedius UNS46 isolated from a human liver abscess. Infect. Immun. 1996;64:3093–3100. doi: 10.1128/iai.64.8.3093-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sukeno A, Nagamune H, Whiley RA, Jafar SI, Aduse-Opoku J, Ohkura K, Maeda T, Hirota K, Miyake Y, Kourai H. Intermedilysin is essential for the invasion of hepatoma HepG2 cells by Streptococcus intermedius. Microbiol. Immunol. 2005;49:681–694. doi: 10.1111/j.1348-0421.2005.tb03647.x. [DOI] [PubMed] [Google Scholar]
- 15.Tabata A, Sato Y, Maya K, Nakano K, Kikuchi K, Whiley RA, Ohkura K, Tomoyasu T, Nagamune H. A streptolysin S homologue is essential for β-haemolytic Streptococcus constellatus subsp. constellatus cytotoxicity. Microbiology (Reading) 2014;160:980–991. doi: 10.1099/mic.0.075580-0. [DOI] [PubMed] [Google Scholar]
- 16.Tabata A, Yamada T, Ohtani H, Ohkura K, Tomoyasu T, Nagamune H. β-Hemolytic Streptococcus anginosus subsp. anginosus causes streptolysin S-dependent cytotoxicity to human cell culture lines in vitro. J. Oral Microbiol. 2019;11:1609839. doi: 10.1080/20002297.2019.1609839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokohata S, Ohkura K, Nagamune H, Tomoyasu T, Tabata A. Human serum albumin stabilizes streptolysin S activity secreted in the extracellular milieu by streptolysin S-producing streptococci. Microbiol. Immunol. 2023;67:58–68. doi: 10.1111/1348-0421.13042. [DOI] [PubMed] [Google Scholar]
- 18.Pilarczyk-Zurek M, Sitkiewicz I, Koziel J. The clinical view on Streptococcus anginosus group—Opportunistic pathogens coming out of hiding. Front. Microbiol. 2022;13:956677. doi: 10.3389/fmicb.2022.956677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen A, Hoshino T, Kilian M. Taxonomy of the Anginosus group of the genus Streptococcus and description of Streptococcus anginosus subsp. whileyi subsp. nov and Streptococcus constellatus subsp. viborgensis subsp. nov. Int. J. Syst. Evol. Microbiol. 2013;63:2506–2519. doi: 10.1099/ijs.0.043232-0. [DOI] [PubMed] [Google Scholar]
- 20.Tsao N, Kuo CF, Cheng MH, Lin WC, Lin CF, Lin YS. Streptolysin S induces mitochondrial damage and macrophage death through inhibiting degradation of glycogen synthase kinase-3β in Streptococcus pyogenes infection. Sci. Rep. 2019;9:5371. doi: 10.1038/s41598-019-41853-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin A, Loughman JA, Zinselmeyer BH, Miller MJ, Caparon MG. Streptolysin S inhibits neutrophil recruitment during the early stages of Streptococcus pyogenes infection. Infect. Immun. 2009;77:5190–5201. doi: 10.1128/IAI.00420-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asam D, Mauerer S, Spellerberg B. Streptolysin S of Streptococcus anginosus exhibits broad-range hemolytic activity. Med. Microbiol. Immunol. 2015;204:227–237. doi: 10.1007/s00430-014-0363-0. [DOI] [PubMed] [Google Scholar]
- 23.Roche E, Prentki M. Calcium regulation of immediate-early response genes. Cell Calcium. 1994;16:331–338. doi: 10.1016/0143-4160(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 24.Abraham WC, Dragunow M, Tate WP. The role of immediate early genes in the stabilization of long-term potentiation. Mol. Neurobiol. 1991;5:297–314. doi: 10.1007/BF02935553. [DOI] [PubMed] [Google Scholar]
- 25.Pérez-Cadahía B, Drobic B, Davie JR. Activation and function of immediate-early genes in the nervous system. Biochem. Cell. Biol. 2011;89:61–73. doi: 10.1139/O10-138. [DOI] [PubMed] [Google Scholar]
- 26.Okuno H. Regulation and function of immediate-early genes in the brain: Beyond neuronal activity markers. Neurosci. Res. 2011;69:175–186. doi: 10.1016/j.neures.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Helin E, Matikainen S, Julkunen I, Heino J, Hyypiä T, Vainionpää R. Measles virus enhances the expression of cellular immediate-early genes and DNA-binding of transcription factor AP-1 in lung epithelial A549 cells. Arch. Virol. 2002;147:1721–1732. doi: 10.1007/s00705-002-0835-1. [DOI] [PubMed] [Google Scholar]
- 28.Chang Y, Lee HH, Chen YT, Lu J, Wu SY, Chen CW, Takada K, Tsai CH. Induction of the early growth response 1 gene by Epstein-Barr virus lytic transactivator Zta. J. Virol. 2006;80:7748–7755. doi: 10.1128/JVI.02608-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buehler J, Carpenter E, Zeltzer S, Igarashi S, Rak M, Mikell I, Nelson JA, Goodrum F. Host signaling and EGR1 transcriptional control of human cytomegalovirus replication and latency. PLoS Pathog. 2019;15:e1008037. doi: 10.1371/journal.ppat.1008037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehman CW, Smith A, Kelly J, Jacobs JL, Dinman JD, Kehn-Hall K. EGR1 upregulation during encephalitic viral infections contributes to inflammation and cell death. Viruses. 2022;14:1210. doi: 10.3390/v14061210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haugwitz U, Bobkiewicz W, Han SR, Beckmann E, Veerachato G, Shaid S, Biehl S, Dersch K, Bhakdi S, Husmann M. Pore-forming Staphylococcus aureus alpha-toxin triggers epidermal growth factor receptor-dependent proliferation. Cell Microbiol. 2006;8:1591–1600. doi: 10.1111/j.1462-5822.2006.00733.x. [DOI] [PubMed] [Google Scholar]
- 32.Susilowati H, Okamura H, Hirota K, Shono M, Yoshida K, Murakami K, Tabata A, Nagamune H, Haneji T, Miyake Y. Intermedilysin induces EGR-1 expression through calcineurin/NFAT pathway in human cholangiocellular carcinoma cells. Biochem. Biophys. Res. Commun. 2011;404:57–61. doi: 10.1016/j.bbrc.2010.11.057. [DOI] [PubMed] [Google Scholar]
- 33.Kao CY, Los FC, Huffman DL, Wachi S, Kloft N, Husmann M, Karabrahimi V, Schwartz JL, Bellier A, Ha C, Sagong Y, Fan H, Ghosh P, Hsieh M, Hsu CS, Chen L, Aroian RV. Global functional analyses of cellular responses to pore-forming toxins. PLoS Pathog. 2011;7:e1001314. doi: 10.1371/journal.ppat.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melby JO, Nard NJ, Mitchell DA. Thiazole/oxazole-modified microcins: Complex natural products from ribosomal templates. Curr. Opin. Chem. Biol. 2011;15:369–378. doi: 10.1016/j.cbpa.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang SZ, Abdulkadir SA. Early growth response gene 1 modulates androgen receptor signaling in prostate carcinoma cells. J. Biol. Chem. 2003;278:39906–39911. doi: 10.1074/jbc.M307250200. [DOI] [PubMed] [Google Scholar]
- 36.Wang B, Khachigian LM, Esau L, Birrer MJ, Zhao X, Parker MI, Hendricks DT. A key role for early growth response-1 and nuclear factor-kappaB in mediating and maintaining GRO/CXCR2 proliferative signaling in esophageal cancer. Mol. Cancer Res. 2009;7:755–764. doi: 10.1158/1541-7786.MCR-08-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myung E, Park YL, Kim N, Chung CY, Park HB, Park HC, Myung DS, Kim JS, Cho SB, Lee WS, Joo YE. Expression of early growth response-1 in human gastric cancer and its relationship with tumor cell behaviors and prognosis. Pathol. Res. Pract. 2013;209:692–699. doi: 10.1016/j.prp.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 38.Fang F, Ooka K, Bhattacharyya S, Wei J, Wu M, Du P, Lin S, Del Galdo F, Feghali-Bostwick CA, Varga J. The early growth response gene Egr2 (Alias Krox20) is a novel transcriptional target of transforming growth factor-β that is up-regulated in systemic sclerosis and mediates profibrotic responses. Am. J. Pathol. 2011;178:2077–2090. doi: 10.1016/j.ajpath.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doncel-Pérez E, Mateos-Hernández L, Pareja E, García-Forcada Á, Villar M, Tobes R, Romero Ganuza F, Vila Del Sol V, Ramos R, Fernández de Mera IG, de la Fuente J. Expression of early growth response gene-2 and regulated cytokines correlates with recovery from Guillain-Barré Syndrome. J. Immunol. 2016;196:1102–1107. doi: 10.4049/jimmunol.1502100. [DOI] [PubMed] [Google Scholar]
- 40.Pio R, Jia Z, Baron VT, Mercola D, UCI NCI SPECS Consortium of the Strategic Partners for the Evaluation of Cancer Signatures-Prostate Cancer Early growth response 3 (Egr3) is highly over-expressed in non-relapsing prostate cancer but not in relapsing prostate cancer. PLoS One. 2013;8:e54096. doi: 10.1371/journal.pone.0054096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuo T, le Dat T, Komatsu M, Yoshimaru T, Daizumoto K, Sone S, Nishioka Y, Katagiri T. Early growth response 4 is involved in cell proliferation of small cell lung cancer through transcriptional activation of its downstream genes. PLoS One. 2014;9:e113606. doi: 10.1371/journal.pone.0113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mishra A, Bharti AC, Saluja D, Das BC. Transactivation and expression patterns of Jun and Fos/AP-1 super-family proteins in human oral cancer. Int. J. Cancer. 2010;126:819–829. doi: 10.1002/ijc.24807. [DOI] [PubMed] [Google Scholar]
- 43.Hamid QA, Reddy PJ, Tewari M, Uematsu S, Tuncay OC, Tewari DS. Regulation of IL-1-induced gingival collagenase gene expression by activator protein-1 (c-Fos/c-Jun) Cytokine. 2000;12:1609–1619. doi: 10.1006/cyto.2000.0676. [DOI] [PubMed] [Google Scholar]
- 44.Hu YT, Chen XL, Huang SH, Zhu QB, Yu SY, Shen Y, Sluiter A, Verhaagen J, Zhao J, Swaab D, Bao AM. Early growth response-1 regulates acetylcholinesterase and its relation with the course of Alzheimer's disease. Brain Pathol. 2019;29:502–512. doi: 10.1111/bpa.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sevilla T, Sivera R, Martínez-Rubio D, Lupo V, Chumillas MJ, Calpena E, Dopazo J, Vílchez JJ, Palau F, Espinós C. The EGR2 gene is involved in axonal Charcot-Marie-Tooth disease. Eur. J. Neurol. 2015;22:1548–1555. doi: 10.1111/ene.12782. [DOI] [PubMed] [Google Scholar]
- 46.Fusco C, Spagnoli C, Salerno GG, Pavlidis E, Frattini D, Pisani F, Bassi MT. Charcot-Marie-Tooth disease with pyramidal features due to a new mutation of EGR2 gene. Acta Biomed. 2019;90:104–107. doi: 10.23750/abm.v90i1.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grosz BR, Golovchenko NB, Ellis M, Kumar K, Nicholson GA, Antonellis A, Kennerson ML. A de novo EGR2 variant, c.1232A > G p.Asp411Gly, causes severe early-onset Charcot-Marie-Tooth Neuropathy Type 3 (Dejerine-Sottas Neuropathy) Sci. Rep. 2019;9:19336. doi: 10.1038/s41598-019-55875-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamada K, Gerber DJ, Iwayama Y, Ohnishi T, Ohba H, Toyota T, Aruga J, Minabe Y, Tonegawa S, Yoshikawa T. Genetic analysis of the calcineurin pathway identifies members of the EGR gene family, specifically EGR3, as potential susceptibility candidates in schizophrenia. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2815–2820. doi: 10.1073/pnas.0610765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nie F, Zhang Q, Ma J, Wang P, Gu R, Han J, Zhang R. Schizophrenia risk candidate EGR3 is a novel transcriptional regulator of RELN and regulates neurite outgrowth via the Reelin signal pathway in vitro. J. Neurochem. 2021;157:1745–1758. doi: 10.1111/jnc.15225. [DOI] [PubMed] [Google Scholar]
- 50.Marballi KK, Alganem K, Brunwasser SJ, Barkatullah A, Meyers KT, Campbell JM, Ozols AB, Mccullumsmith RE, Gallitano AL. Identification of activity-induced Egr3-dependent genes reveals genes associated with DNA damage response and schizophrenia. Transl. Psychiatry. 2022;12:320. doi: 10.1038/s41398-022-02069-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacGibbon GA, Lawlor PA, Walton M, Sirimanne E, Faull RL, Synek B, Mee E, Connor B, Dragunow M. Expression of Fos, Jun, and Krox family proteins in Alzheimer's disease. Exp. Neurol. 1997;147:316–332. doi: 10.1006/exnr.1997.6600. [DOI] [PubMed] [Google Scholar]
- 52.Sokolova A, Hill MD, Rahimi F, Warden LA, Halliday GM, Shepherd CE. Monocyte chemoattractant protein-1 plays a dominant role in the chronic inflammation observed in Alzheimer's disease. Brain Pathol. 2009;19:392–398. doi: 10.1111/j.1750-3639.2008.00188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boerrigter D, Weickert TW, Lenroot R, O'Donnell M, Galletly C, Liu D, Burgess M, Cadiz R, Jacomb I, Catts VS, Fillman SG, Weickert CS. Using blood cytokine measures to define high inflammatory biotype of schizophrenia and schizoaffective disorder. J. Neuroinflamm. 2017;14:188. doi: 10.1186/s12974-017-0962-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mednova IA, Boiko AS, Kornetova EG, Semke AV, Bokhan NA, Ivanova SA. Cytokines as potential biomarkers of clinical characteristics of schizophrenia. Life (Basel) 2022;12:1972. doi: 10.3390/life12121972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.