Summary
A high percentage of patients with brain metastases frequently develop neurocognitive symptoms; however, understanding how brain metastasis co-opts the function of neuronal circuits beyond a tumor mass effect remains unknown. We report a comprehensive multidimensional modeling of brain functional analyses in the context of brain metastasis. By testing different preclinical models of brain metastasis from various primary sources and oncogenic profiles, we dissociated the heterogeneous impact on local field potential oscillatory activity from cortical and hippocampal areas that we detected from the homogeneous inter-model tumor size or glial response. In contrast, we report a potential underlying molecular program responsible for impairing neuronal crosstalk by scoring the transcriptomic and mutational profiles in a model-specific manner. Additionally, measurement of various brain activity readouts matched with machine learning strategies confirmed model-specific alterations that could help predict the presence and subtype of metastasis.
Keywords: cancer neuroscience, brain metastasis, brain circuit impact, biomarkers, electrophysiology, elta oscillations, gamma oscillations, decision trees
Graphical abstract

Highlights
-
•
Brain metastasis experimental models recapitulate neuronal impact heterogeneity
-
•
The underlying mechanism cannot be explained by the tumor mass effect
-
•
A molecular signature is enriched in models imposing high neural impact
-
•
Altered brain activity patterns predict the presence and subtype of metastasis
Patients with brain metastasis experience neurocognitive impairment. Until now, the mass effect of the tumor was the only underlying cause. Sanchez-Aguilera et al. demonstrate that, independently on the size, number, and location, a machine learning approach correctly classifies different models of brain metastasis based on their impact on brain activity.
Introduction
Brain metastases have a dramatic impact in the quality of life (QoL). Apart from the impact in reducing survival,1 44% of patients suffering brain metastases also experience cognitive disabilities.2,3,4 Why some individuals are more affected than others is unknown, since the tumor mass effect has been questioned as the sole cause. In spite of limited research on the crosstalk between metastatic cells and neuronal circuits, a role for N-methyl-D-aspartate receptor has been described in breast cancer brain metastasis.5 However, this crosstalk is described as unidirectional, addressing the benefit of cancer cells by co-opting a neural-like behavior5 while no data exist on the consequences it might have on brain function. Given the neural mimicry of metastatic cells in the brain,6,7,8,9 and the high prevalence of neurocognitive impairment, we decided to couple in vivo electrophysiology and ex vivo calcium imaging of various established brain metastasis models to better understand their cellular, molecular, and electrophysiological profiles.
Here, we unveil an inter-model heterogeneity, which impacts on brain circuits independently of the tumor mass effect. In an effort to characterize this phenotype, we confirm its correlation with specific electrophysiological and molecular signatures suggesting potential molecular mediators. Additionally, to reinforce the model-dependent impact on brain function, we demonstrate that computational analysis of in vivo electrophysiological profiles with machine learning approaches is capable of defining metastasis-dependent brain alterations and subtypes, which could inspire novel strategies for noninvasive diagnosis.
In summary, our findings report a preclinical strategy to address brain functional compromise by metastasis. In addition, our data suggest that, besides tumor size, a molecular crosstalk between metastases and neurons could be exploited to prevent and target brain metastasis-associated impact in QoL.
Results
Effect of brain metastasis in electrophysiological brain activity
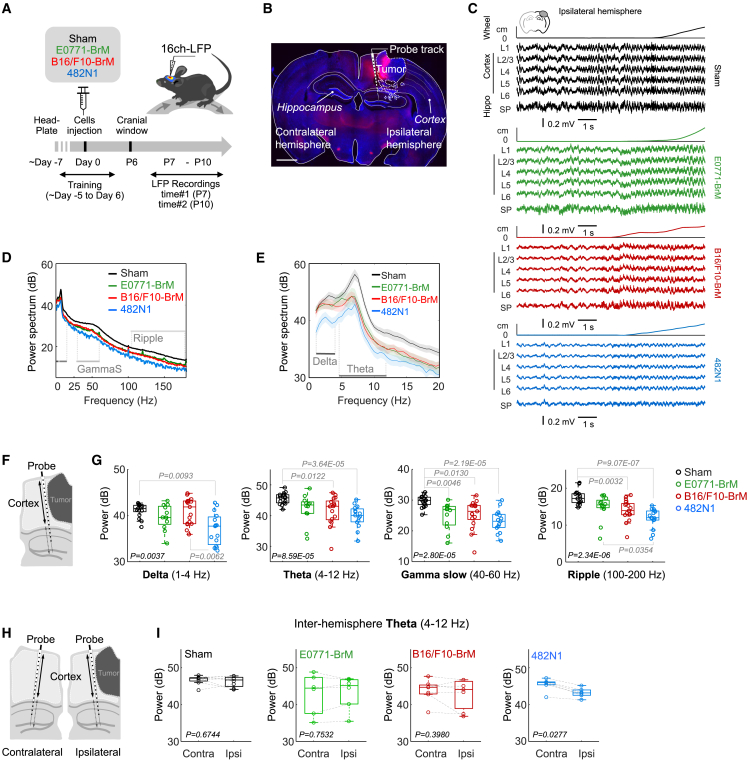
We inoculated three brain organotropic mouse cancer cell lines (482N1,10 E0771-BrM,11 B16/F10-BrM12) directly into the brain to eliminate the variable of metastasis location when systemic inoculation is used,13 which is a crucial aspect for our purpose. We adapted the protocol of opening a cranial window and obtaining longitudinal electrophysiological recordings from head-fixed mice during the survival period of brain metastasis syngeneic models.10,11,12,14 Twenty head-fixed awake mice were trained to rest comfortable (Still) and to move on a wheel (Run) before the experimental session (Figure 1A; sham = 6 mice; B16/F10-BrM = 5; E0771-BrM = 4; 482N1 = 5). Local field potentials (LFPs) where recorded from several penetrations at two time points (7 and 10 days post implantation) in each animal to maximize data acquisition and to compare potential differences along tumor evolution (Figure 1A) within the two weeks survival period of these models.10,11,12 Sixteen-channel linear array silicon probes were inserted in both peritumoral (Right hemisphere) and in the homotopic contralateral tumor-free region (Left hemisphere), as confirmed by posterior histological analysis (Figure 1B).
Figure 1.
Effect of brain metastasis in electrophysiological brain activity
(A) Schema of experimental design. Mice were implanted with head-bars and habituated to stay head-fixed in a wheel. After 7 days, brain metastatic cells from lung cancer (482N1), breast cancer (E0771-BrM), or melanoma (B16/F10-BrM) were inoculated intracranially in the right hemisphere. 7 days later, local field potential (LFP) recordings were obtained during 4 days using a 16-channel linear probe in each hemisphere. On day 10, animals were transcardially perfused with 4% PFA and the brains extracted and processed for histological analysis.
(B) Representative coronal section of a mouse with a 482N1 metastasis. Scale bar: 1500 μm. The silicon probe was stained with Red-DiI to identify the probe track.
(C) LFP signals recorded across cortical and hippocampal layers surrounding the tumor in a representative example from each group. LFP from the ipsilateral side is shown. The position in the wheel is shown at the top of each panel.
(D) Mean power spectrum from ipsilateral cortical layers of the examples shown in C.
(E) Enlarged 1 to 20 Hz band representing mean values of all data.
(F) Schema of experimental design indicating analysis of LFP corresponding to cortical areas.
(G) Differences of cortical LFP power in the delta (1–4 Hz; Chi.2 = 13.4, p = 0.0037), theta (4–12 Hz; Chi.2 = 21.4, p < 0.0001), gamma slow (40–60 Hz, Chi.2 = 23.8; p < 0.0001), and ripple (100–200 Hz; Chi.2 = 28.9, p < 0.0001) bands among mice without or with metastases from three different models, as evaluated during continuous running periods. Values are shown in box-and-whisker plots where every dot represents a different penetration and the line in the box corresponds to the median. The boxes go from the upper to the lower quartiles and the whiskers go 1.5 times the range between percentile 25 and 75 (down from percentile 25, up from percentile 75) (penetration sessions: 17 sham, 12 E0771-BrM, 17 B16/F10-BrM and 15 482N1). p value was calculated using Kruskal-Wallis with a post-hoc Tukey test. Only significant p values are shown.
(H) Schema of experimental design indicating analysis of LFP corresponding to cortical areas at the ipsi- and contralateral sides respect to the location of the metastasis.
(I) Analysis of the interhemispheric as measured by the theta power spectrum. Values are shown in box-and-whisker plots where every dot represents a different penetration and the line in the box corresponds to the median. The boxes go from the upper to the lower quartiles and the whiskers go from the minimum to the maximum. Data from 27 bilateral penetrations (sessions: 8 sham, 6 E0771-BrM, 7 B16/F10-BrM, 6 482N1). See also Figure S1.
Analysis of LFP oscillatory activity from cortical and hippocampal areas showed a general decrease in the power spectrum for cortical penetrations ipsilateral to the tumor (Figure 1C; sham = 17 penetration sessions from 6 mice; B16/F10-BrM = 17 from 4 animals; E0771-BrM = 12 from 5 animals; 482N1 = 15 from 5 animals). Changes affected all frequency bands, from the ripple (100–200 Hz) and slow gamma bands (40–60 Hz,) (Figure 1D), to delta (1–4 Hz) and theta (4–12 Hz) activity (Figure 1E), especially in the 482N1-BrM model (Figures 1F and 1G). Spectral differences between groups could not be explained by the degree of locomotor activity (Figure S1A). Instead, they rather reflect evident differences among brain metastasis models. Remarkably, although limited in prevalence, interictal discharges were detected in all models, while non-convulsive seizures were recorded in the E0771-BrM and the B16/F10-BrM models (Figures S1B and S1C) (8.33% and 5.88% incidence, respectively in all recorded sessions). Similar oscillatory changes were detected from hippocampal penetrations ipsilateral to the tumor (Figures S1D and S1E). To exclude any potential volume conduction effect, we also evaluated the current-source density signals (Figure S1F), which provide a measure of the direct currents flowing through different cortical layers and found comparable results (Figures S1G and S1H). Thus, our findings suggest that individual brain metastasis models might influence neural communication differently both locally (neocortex) and at nearby structures (hippocampus).
To confirm this point, we expanded analysis of LFP recordings to the contralateral tumor-free area in the cortex (Figure 1H) and hippocampus (Figure S1D) to evaluate inter-hemispheric effects (sham = 16 penetrations; B16/F10-BrM = 17; E0771-BrM = 12; 482N1 = 17). We found significant differences only for the 482N1-BrM model (Figures 1I and S1I). Such a heterogeneity in the long-range impact on neural circuits consolidates the metastasis model evaluated as a key variable.
Thus, even though the three different brain metastases models were inoculated in the same cortical area and they share a general negative influence on neural communication, inter-model heterogeneity was evident regarding the qualitative impact influencing brain field potential both locally (peritumoral area) and distally (contralateral tumor-free hemisphere).
Dissociation between altered local field potential and mass effect or inflammation
In order to dissect the underlying cause of the differential impact of metastases on LFP, we interrogated the histology of the brains evaluated with electrophysiology.
In agreement with previous clinical studies,4 histological analysis of the three models at the endpoint of the experiment confirmed no correlation between tumor size and LFP (Figures 2A and 2B). Indeed, no significant differences in tumor size exist among the three models (Figure 2B). If anything, the E0771-BrM model, which tends to generate bigger tumors compared to the other two models (Figure 2B), has electrophysiological patterns that in some of the analyses performed are closer to the control than to the other two models (Figures 1G and S1E). In conclusion, our findings argue against the mass effect of the tumor as the solely contributor to explain its impact in neural circuits.
Figure 2.
Dissociation between altered local field potential and mass effect or inflammation
(A) Representative brain sections affected by metastases from different models (Breast, cell line E0771-BrM; melanoma, B16/F10-BrM; lung, 482N1). Scale bar: 1mm.
(B) Quantification of tumor area. Values are shown in box-and-whisker plots where every dot represents a different brain and the line in the box corresponds to the median. The boxes go from the upper to the lower quartiles and the whiskers go from the minimum to the maximum value (B16/F10-BrM n = 7; E0771-BrM n = 7; 482N1 n = 8 mice per experimental condition). One-way ANOVA (F(2,19) = 1.884, p = 0.1793).
(C) Representative images of immunofluorescence staining labeling GFAP+ cells, Olig2+ cells, Iba1+ cells, and NeuN+ cells in the peritumoral areas (dotted line) of brains from mice inoculated with B16/F10-BrM, E0771-BrM, 482N1, or saline (sham). Blue channel is DAPI. Scale bar, 50μm.
(D) Quantification of the relative GFAP+ area, number of Olig2+ cells, relative Iba1+ area, and number of NeuN+ cells. Values are shown in box-and-whisker plots where every dot represents a different brain and the line in the box corresponds to the median. The boxes go from the upper to the lower quartiles and whiskers go from the minimum to the maximum value (Sham n = 4; B16/F10-BrM n = 5; E0771-BrM n = 4; 482N1 n = 5 mice per experimental condition). One-way ANOVA (relative GFAP+ area: F(3,14) = 1.015, p = 0.4155/number of Olig2+ cells: F(3,14) = 0.9241, p = 0.4547/relative Iba1+ area: F(3,14) = 1.556, p = 0.2442/number of NeuN+ cells: F(3,14) = 0.5621, p = 0.6488).
(E) Bar plot of the relative proportion of astrocytes (left), metastasis-associated macrophages Apoe+ (center), and metastasis-associated macrophages expressing S100a8 (right) against the rest of the cell types identified by CIBERSORTx from different mouse models (B16/F10-BrM n = 6; E0771-BrM n = 6 and 482N1 n = 6 mice). One-way ANOVA (astrocytes: F(2,15) = 0.03133, p = 0.9692). One-way ANOVA (metastasis-associated macrophages Apoe+: F(2,15) = 15,92, p = 0.0002) with a Tukey post hoc test (E0771-BrM Vs. B16/F10-BrM p = 0.0004; B16/F10-BrM Vs. 482N1 p = 0.0007; E0771-BrM Vs. 482N1 p = 0.9499). Kruskal-Wallis test (metastasis-associated macrophages expressing S100a8: H = 10.87, p = 0.0012) with a Dunn's post hoc test (E0771-BrM Vs. B16/F10-BrM p = 0.0044; B16/F10-BrM Vs. 482N1 p = 0.0573; E0771-BrM Vs. 482N1 p > 0.9999). See also Figure S2 and Table S1.
The brain microenvironment includes glial cells that are a major component reacting to the presence of the metastasis and contributing very significantly to the local progression of the tumor as part of the inflammatory milieu.10,11,12 Consequently, we explored whether different experimental metastases could have a different impact on surrounding glial cells. Neither reactive GFAP+ astrocytes, Iba1+ microglia/macrophages, nor Olig2+ oligodendrocytes were different in numbers respect to the sham or between different brain metastasis models (Figures 2C and 2D), as they are against control non-operated mice (Figures S2A and S2B). These findings discard a correlation between general markers for glial populations, which was confirmed with additional ones in the case of astrocytes (Figures S2C and S2D) and microglia/macrophages (Figures S2E and S2F), and the electrophysiology patterns, as they do in response to the procedure (i.e., sham versus control) (Figures S2A and S2B). In order to go beyond specific markers, we applied CIBERSORTX15,16,17 on bulk RNA sequencing (RNA-seq) from established brain metastases. None of the thirteen cell types identified from the brain metastasis microenvironment18 after deconvolution of the transcriptomic data were differentially enriched in the 482N1 model with respect to B16/F10-BrM and E0771-BrM (Figures 2E, S2G and Table S1).
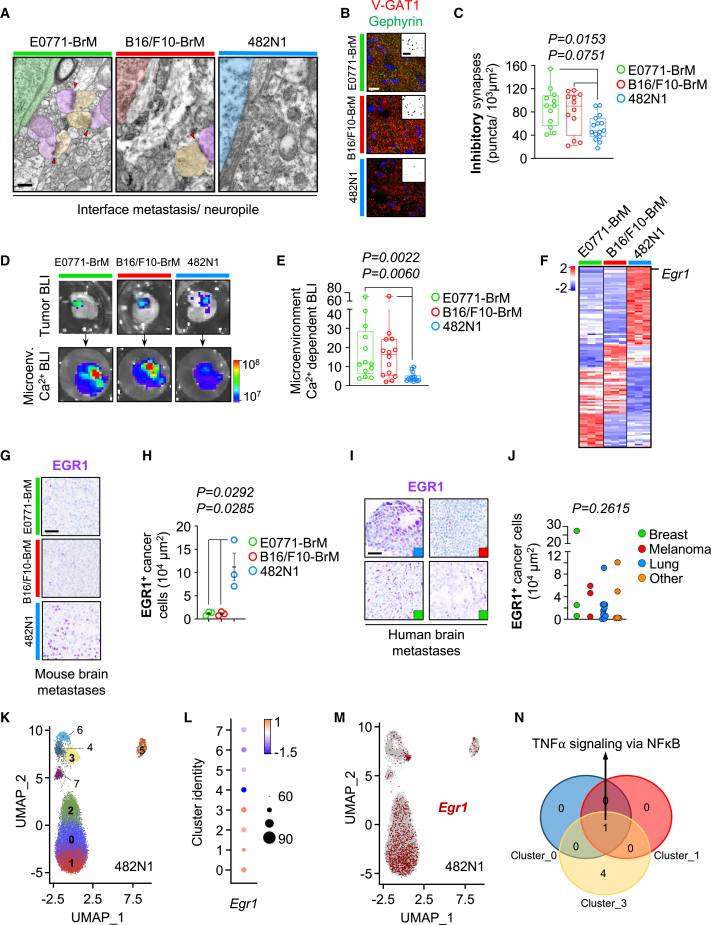
Of note, the total number of peritumoral neurons was not affected either, reflecting a limited impact of the tumor on their cell density (Figures 2C, 2D, S2A and S2B). Indeed, we confirmed with electron microscopy that the immediate vicinity of the tumor was densely populated by well-preserved neuronal dendritic and axonal processes, which might suggest that metastases do not massively destruct the neuropile (Figure 3A), which is a prerequisite to study a potential crosstalk with neural circuits.
Figure 3.
Correlation between electrophysiological impact and the transcriptomic profile of brain metastases
(A) Electron microscopy images at the interface between the metastasis and the neuropile. Some synapses could be observed (red arrowheads). Green/red/blue: tumor cell (E0771-BrM/B16/F10-BrM/482N1, respectively); yellow: pre-synaptic terminal; purple: post-synaptic terminal. Scale bar: 0.4 μm.
(B) Representative images of pre-synaptic (V-GAT1) and post-synaptic (Gephyrin) markers of inhibitory synapses and their colocalization indicative of mature synapsis. Scale bar: 15 μm.
(C) Quantification of mature inhibitory synapses in the peritumoral area associated with different brain metastasis models. Values are shown in box-and-whisker plots where every dot represents a different field of view and the line in the box corresponds to the median. The boxes go from the upper to the lower quartiles and the whiskers go from the minimum to the maximum (E0771-BrM n = 12 field of view from 4 brains; B16/F10-BrM n = 12 field of view from 4 brains; 482N1 n = 15 field of view from 5 brains). One-way ANOVA (F(2,36) = 4.889, p = 0.0132) with a Tukey post hoc test (E0771-BrM Vs. B16/F10-BrM p = 0.7943; B16/F10-BrM Vs. 482N1 p = 0.0751; E0771-BrM Vs. 482N1 p = 0.0153).
(D) Representative images of organotypic cultures established from the genetically engineered mice model LSL-CamBI containing firefly luciferase-expressing brain metastases at the experimental endpoint after being stimulated with appropriate substrates. Color bars show bioluminescence intensity in p s−1 cm−2 sr−1.
(E) Quantification of microenvironment-derived calcium-dependent bioluminescence normalized for metastasis-derived bioluminescence. Values are shown in box-and-whisker plots where every dot represents a different organotypic culture and the line in the box corresponds to the median. The boxes go from the upper to the lower quartiles and the whiskers go from the minimum to the maximum (E0771-BrM n = 13 slices; B16/F10-BrM n = 14; 482N1 n = 14. Slices were obtained from four animals each group, and two independent experiments). Kruskal-Wallis test (H = 14.09, p = 0.0009) with a Dunn's post hoc test (E0771-BrM Vs. B16/F10-BrM p > 0.9999; B16/F10-BrM Vs. 482N1 p = 0.006; E0771-BrM Vs. 482N1 p = 0.0022).
(F) RNA-seq from the three different brain metastasis models where genes previously reported in synaptic signatures significantly upregulated in 482N1 are shown. Scale bar corresponds to Z Score.
(G) Validation of increased levels of EGR1 in 482N1 compared to E0771-BrM and B16/F10-BrM. Scale bar: 50 μm.
(H) Quantification of EGR1 levels in metastatic cells in situ. Every dot represents an individual mouse where 4 fields of view were analyzed. The line represents the mean and the error bars the sem. (n = 3 brains). p value was calculated using unpaired t test.
(I) Representative images of human brain metastases showing EGR1 levels. Selected samples illustrate the heterogeneity among patients as well as the primary tumor source (colored squares in the lower right corner). Blue square: lung cancer brain metastasis; red square: melanoma brain metastasis; green square: breast cancer brain metastasis. Scale bar: 50 μm.
(J) Quantification of EGR1 levels in metastatic cells in situ. Every dot represents an individual patient where 4 fields of view were analyzed. Samples are grouped according to the primary source of the brain metastasis. Other: additional primary tumor types. Kruskal-Wallis test (H = 1.202, p = 0.7526).
(K) Uniform manifold approximation and projection (UMAP) labeling clusters identified at 0.75 resolution (clusters 0–7) for the 482N1 cancer cells. Each cluster is represented by a different color. Clusters labeled in bold correspond to the Egr1 enriched ones.
(L) Dotplot showing Egr1 enriched cells, split by cluster. Average expression is indicated by a colored scale: low in blue, high in red. Dot size represents the percentage of cells with Egr1 expression by cluster.
(M) Projection of Egr1 gene expression of 481N1 cancer cells. Pre-defined Egr1-positive cells are highlighted in red.
(N) Venn diagram of gene hallmarks significantly upregulated in Egr1-positive cells within clusters 0, 1, and 3 identified by GSEA. See also Figure S3 and Table S2.
Thus, we addressed and clarified that the two more evident potential causes of neural circuit impairment (i.e., mass effect of the growing tumor and the induction of an inflammatory peritumoral milieu) are not responsible for the inter-model heterogeneity observed among brain metastasis models under this specific experimental setting. Interestingly, the maintenance of a well-conserved neuropile is highly suggestive on the possibility that metastases could influence neural communication.
Correlation between electrophysiological impact and the transcriptomic profile of brain metastases
A closer look to the peritumoral area on the three experimental models demonstrates intact peritumoral synapses in the immediate vicinity (<1 μm) of the metastasis, thus susceptible to be influenced by the cancer cells (Figures 3A and S3A). In order to evaluate the basic components of neural circuits, we interrogated excitatory and inhibitory synapses. Interestingly, the model with higher degree of alteration in electrophysiology, 482N1, showed a reduced number of inhibitory synapses, which was not matched by a parallel loss of interneurons, in the peritumoral area, (Figures 3B, 3C, S3D and S3E) while no differences were detected with respect to the excitatory ones (Figures S3B and S3C). This phenotype affecting interneurons mimics previous findings in glioma,19 although the underlying mechanism might be different given the specific effect on synapses but not in the number of interneurons.
Additionally, we reinforced the superior brain impact imposed by the 482N1 model by using a genetically engineered mouse model (GEMM) LSL-CamBI, which reports calcium activity through bioluminescence in a Cre-dependent manner.20 Brain organotypic cultures from this GEMM affected with metastases were established ex vivo; D-luciferin for firefly-expressing BrM cells was used to identify bioluminescence from tumor cells (day 0) and discard those without metastasis. Subsequently, selected brain slices were infected with adenoviruses (Ad5-CMV-Cre) to induce Cre-dependent activation of calcium-dependent luciferase in the metastasis-associated microenvironment (Figure S3F). 3 days post Ad5-CMV-Cre infection, hydrofluoromizine was used to activate CamBI-expressing cells from the microenvironment and the bioluminescence from the non-cancer compartment was registered (Figure S3F). Microenvironment-derived calcium-dependent bioluminescence normalized to the cancer cells-derived bioluminescence obtained at day 0, which minimizes any influence of tumor size, indicates that the 482N1 metastasis has a stronger impact also influencing calcium activity in the microenvironment (Figures 3D and 3E).
Given previous findings on specific mutational profiles in glioma and the incidence of epilepsy,21 we performed whole exome sequencing on the three models. We found 35 specific genetic alterations in the 482N1 model (Table S2); however, these remain to be functionally evaluated in the context of their influence on neuronal impact. Complementarily, we obtained the individual transcriptomic profiles of the three tested models and evaluated whether their different impact on brain functional readouts (i.e., LFP, calcium activity) might have a specific correlate on gene expression. First, we focused the analysis on those genes involved in neuronal communication.5,21,22,23,24,25,26,27 Remarkably, 51 genes from these signatures were differentially expressed by 482N1 compared to the other two brain metastasis models (Figures 3F and Table S2). In addition, unbiased analysis of differential gene expression among the three models identified additional molecules and signatures (Figures S3G–S3L and Table S2) specifically enriched in 482N1 that have been also described to have the potential to modulate neuronal communication (i.e., ECM) and to be used by cancer cells to communicate with brain-resident cells (i.e., nanotubes).28 Indeed, the transcription factor Egr1, which was the only gene deregulated both within the analysis of the top 50 differentially expressed genes in the 482N1 model and part of the pre-selected 51 genes from selected signatures, was validated at the protein level in situ (Figures 3G and 3H). Additionally, EGR1 was also expressed in brain metastases from patients independently of the primary tumor source and with high interpatient heterogeneity (Figures 3I, 3J and Table S2), which might be compatible with the various degrees in neurocognitive impact detected in patients. Besides its role in modulating synaptic plasticity,29 EGR1 has been shown to affect angiogenesis,30,31,32,33,34 which might influence neuronal activity as well. This mechanism might be also present in the 482N1 model where we detected a trend toward increased peritumoral vascular density (Figures S3M and S3N). In order to characterize in more detail the Egr1+ cancer cells, we performed single-cell RNA-seq on established metastases from the 482N1 model (Figure 3K). We detected Egr1+ cells distributed in four major clusters (Figures 3L, 3M and Table S2). Gene signature enrichment analysis for each Egr1+ cluster identified the tumor necrosis factor alpha signaling via nuclear factor κB hallmark signature as commonly enriched in three out of the four Egr1+ clusters (Figure 3N and Table S2), suggesting a potential mechanistic link between cytokines and neuronal communication.35
Overall, our data suggest that a molecular program rather than or in addition to the mass effect caused by brain metastasis might influence neural communication. Although the molecular program remains to be functionally evaluated, we wondered whether the impact on neural circuits could be further globally exploited computationally.
A generalized linear model identifies key components defining the diversity of electrophysiological profiles among brain metastases
The data described previously suggest that the molecular profile of cancer lines underlies a bidirectional crosstalk that may specifically affect microcircuit function in colonized brain territories. We thus applied data science strategies to uncover the complex contributions underlying the differential effect of each metastasis model in the LFPs.36 To this purpose, we aggregated all spectral data per session, per mice, per group, resulting into a large dataset (492 samples). We also included categorical variables, such as hemisphere (Left/Right), behavioral state (Run/Still), and the origin of the data according to the electrode location (Cortex/Hippo) (Table S3).
Principal component (PC) analysis identified 9 major directions explaining 99% of the data variance (Figure 4A). Some components mostly discriminated between categorical variables (e.g., PC2, PC3, PC4), while others better explained changes of spectral features (e.g., PC1, PC5) and their combination (e.g., PC7, PC8, PC9). Projecting data from the different experimental groups into the 9-dimensional PC space suggested segregation between metastasis models (Figure 4B). This was clearly appreciated in two-dimensional representations for categorical variables (Figures 4C and S4A–S4C), spectral features (Figures 4D and S4D–S4F), and experimental groups (Figures 4E and S4G). Group distribution suggested different contribution of spectral features to their separation.
Figure 4.
A generalized linear model identifies key components defining the diversity of electrophysiological profiles among brain metastases
(A) Weights of the different LFP features contributing to the different principal components (PC). The scale bar indicates PC weights.
(B) Variance of data from different experimental groups as projected over PCs. The scale bar indicates PC weights.
(C–E) 2D projections of data over different PC pairs that illustrate distribution of categorical variables (C), oscillations (D), and metastasis models (E).
(F) Results of a generalized linear model (GLM) analysis fitted to distinguish E0771-BrM in data from all sessions (n = 492). Beta values (βi) shown in magenta to blue represent the GLM coefficients. Positive numbers represent a positive correlation and negative numbers represent a negative correlation. P-values represent the significance of factors explaining the GLM output. The discontinuous line represents the threshold at p < 0.05.
(G) Same as in F for B16/F10-BrM. H. Same as in F for 482N1. See also Figure S4 and Table S3.
To understand this point, we built a generalized linear model to test what features had better identified brain metastasis (Figure S4H). We found that slow oscillations in the delta and theta range characterized all metastases, while unique signatures were appreciated for each type separately. For the E0771-BrM breast model, changes of alpha and gamma activity also contributed (Figure 4F), while only alpha activity characterized the B16/F10-BrM melanoma model (Figure 4G). This, together with the ratio between oscillatory bands (Delta/Theta and Gamma slow/fast) helped to identify breast and melanoma from the 482N1 lung cancer model (Figure 4H).
Altogether, this analysis suggests that PC analysis of spectral features should help to discriminate between brain metastasis subtypes.
Machine learning identifies experimental brain metastasis subtypes
Based on our findings suggesting that each brain metastasis model has a different impact on brain microcircuits and their emerging oscillations, we hypothesized that PC analysis operating over spectral features could help to classify across metastasis subtypes.
Five different machine learning classifiers were tested: a Linear Support Vector Classifier, the standard nonlinear Support Vector Classifier, a Decision Tree (DecisionTree), an Extra-Tree Decision Tree (ExtraTree), and XGBoost. These models were based on data projected over the PC space. They were trained using randomly selected subsets of data recorded 9–10 days after cancer cell injections, and tested in the remaining subset (Figure 5A). We trained algorithms independently, and chose the one showing the best accuracy representativeness (Figures S5A and S5B). We found that Decision Tree provided the most accurate and significant performance (Figure 5B; see non-selected models in Figure S5C).
Figure 5.
Machine learning identifies experimental brain metastasis subtypes
(A) Scheme of the training and test approach.
(B) An example of one Decision Tree Classifier trained on data projected in the PC space used to predict the group classes.
(C) Class predictor applied to data generated from sessions 9–10 days post implantation of brain metastatic cells. The model is able to classify the presence of different metastasis subtypes using PC analysis of LFP spectral features with a p value of 0.00001.
(D) Class predictor applied to early data obtained 7 days post implantation of brain metastatic cells. The model is able to identify the presence of a tumor with a p value < 0.00001.
(E) Scheme of the prediction strategy. The model (4 Decision Trees) trained with previous data was used to classify new data from the two additional brain metastasis cell lines from lung cancer 393N1 and 2691N1. Note that new data will be ascribed to one of the four categories resulting from the previous training.
(F) Mean output of all decision trees when data from new lung cancer lines 393N1 (dark blue) and 2691N1 (light blue) were evaluated with the already trained predictors. Most trees (3 out of 4) correctly classified all samples from both 393N1 and 2691N1 lines as 482N1 (lung), while 1 decision tree misclassified samples as B16/F10-BrM (melanoma). Local field potential (LFP) data from 22 ipsilateral and 17 contralateral recordings from 3 mice injected with 393N1 cells; 26 ipsilateral and 24 contralateral sessions from 3 mice injected with 2691N1 cells.
(G) Same as in F for the most-voted output of the 4 decision trees. Here, all 393N1 samples and all 2691N1 samples but 1 were correctly classified as 482N1 (lung). See also Figure S5 and Table S4.
Four Decision Tree models were selected from the pool and used to predict group classes. The prediction successfully identified data coming from the different groups with a mean accuracy of 0.77 ± 0.02 (Figure 5C; p < 0.00001 when tested against shuffled distribution of model labels; Table S4). Interestingly, using a random forest approach on Decision Tree models trained in the later metastatic stages identified the presence of some metastatic tumor as early as 7 days post injection (Figure 5D; mean accuracy 0.73 ± 0.09; p < 0.00001 significantly different from shuffles; Table S4). The ability of machine learning tools to predict the presence of metastasis early in advance was better than relying on standard statistical analysis of individual LFP spectral signatures (Figures S5D and S5E).
One potential caveat could be related with the ability of the predictor to generalize beyond data in which they were trained. To address this issue, we adopted two different strategies. First, we run a leave-one-out cross-validation test using sessions from all mice but one per group (sham, B16/F10-BrM, E0771-BrM, 482N1) to train new Decision Trees. Then, sessions from the mice that were left out were used for testing. We found consistent performance supporting generalizing ability of the prediction approach (Figures S5F and S5G).
Second, we obtained LFP recordings from mice injected with two additional brain metastasis cell lines from lung cancer (393N1 and 2691N1)10 (Figure S5H). Previously, we showed that these lines, which were derived from the same GEMM harboring Kras mutation G12D and Trp53 null alleles,37 exhibited a range of metastatic power similar to the 482N1.10 We detected that both 393N1 and 2691N1, which generate tumors of equivalent size to the other models evaluated (Figure S5I), induced similar electrophysiological properties as the 482N1 model (Figure S5J). Subsequently, we used data from these mice to test performance of Decision Trees trained on the original dataset (Figure 5E). The predictor would thus classify the new data into one of the original categories. We found that data from lines 393N1 and 2691N1 were more likely to be classified as the 482N1 than to any of the other lines (Figure 5F). Importantly, we noticed that one of the Decision Trees misclassified 393N1 and 2691N1 as if they were B16/F10-BrM, and so we tested a most-voted strategy reaching 98% accuracy for the 2691N1 line and 100% for the 393N1 line (Figure 5G).
Discussion
As patients with brain metastasis start to live longer thanks to recent therapeutic breakthroughs,38,39,40 the demand to maintain organ function is expected to increase. Because the association between tumor mass and the degree of organ compromise is not always well correlated, even metastases under proliferative control by an effective therapy might still give rise to deleterious effects in the brain. Thus, our research provides a solid proof of concept to study such phenomena in vivo by exploiting preclinical models with state-of-the-art techniques. The three models used have been previously established10,11,12 and although similar in terms of their aggressiveness of brain colonization, their impact on organ function is different, the 482N1 lung adenocarcinoma model being the most influential one. Our data are not conclusive on the association of different primary tumor sources and the incidence of neurocognitive impact since we have not used several individual models representing each primary tumor entity. However, clinical studies do not support such association41 and we do not find a correlation in human sample between the primary source of the metastasis and the expression of our candidate gene. In addition, another limitation of our study is the need to compare our findings with treatment-induced toxicities that frequently affect the brain.42
The use of electrophysiology to identify brain metastases is not a novel approach;43 however, its impact in clinical practice has been minimal, if any, due to the lack of underlying cellular and molecular mechanisms that preclude any therapeutic translation. However, emerging data suggest that neuronal activity can favor the progression of primary brain tumors such as glioma, which integrates into neuronal circuits24,44 or challenges the interplay among neurons, microglia, and astrocytes.21,45,46 More strikingly, however, is that colonization of the brain by invading metastatic cancer cells is fueled by neuronal activity itself.47 Our results of metastasis-dependent functional alterations of LFP oscillatory activity suggest that communication between different cell entities can uniquely shape the form and function of the underlying brain circuits, which is in agreement with recent data in glioblastoma.44 Consistently, we found that a model trained in LFP signatures surrounding the three different brain metastasis can help to disambiguate between subtypes and to anticipate metastasis. Indeed, our findings might also point out to the possibility that the molecular makeup of the metastasis could be linked to the impact imposed by the tumor on neural circuits. Three models harboring alterations in Kras and Trp53 generate a similar electrophysiology phenotype involving a superior impact on neuronal communication. This suggests there is information that can be integrated across observational levels from molecules to functional readouts48,49 to guide exploration of the underlying mechanisms.
Our approach is aiming to increase the translational nature of such associations by adding the molecular signature associated with neurological impact. This effort has identified Egr1, a transcription factor, that could directly29 and indirectly, through its role in angiogenesis,30,31,32,33,34 modulate synaptic communication. Our current efforts are aimed to dissect the functional implication of the gene signature identified in both preclinical models as well as patients. Such an approach would not only offer the foundation for using electrophysiology with emerging noninvasive multimodal approaches50 in early diagnosis, which will minimize the use of surgery to define and characterize brain metastases, but also the possibility to prevent or minimize impact of metastasis on QoL, since new therapeutic lines could be developed to target the underlying molecular mediators. Importantly, while our data reflect results from reduced laboratory models and a limited sample size, diagnosis of the complexities of human brain metastases still requires an advanced approach. To achieve this goal, it is critical to incorporate systematic neurocognitive assessment with available objective and validated tests51 in the clinical practice. This effort will provide well-curated clinical databases of neurocognitive impairment associated with brain metastases that could be used to validate preclinical findings and to design potential clinical interventions.
Consortia
RENACER consortium: Cecilia Sobrino, Nuria Ajenjo, Maria-Jesus Artiga, Eva Ortega-Paino, Virginia García-Calvo, Angel Pérez-Núñez, Pedro González-León, Luis Jiménez-Roldán, Luis Miguel Moreno, Olga Esteban, Juan Manuel Sepúlveda, Oscar Toldos, Aurelio Hernández Laín, Alicia Arenas, Guillermo Blasco, José Fernández Alén, Adolfo de la Lama Zaragoza, Antía Domínguez Núñez, Lourdes Calero, Concepción Fiaño Valverde, Ana González Piñeiro, Pedro David Delgado López, Mar Pascual, Gerard Plans Ahicart, Begoña Escolano Otín.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti-Glial Fibrillary Acidic Protein(GFAP), Unconjugated | Millipore | Cat# AB5541; RRID: AB_177521 |
| Rabbit polyclonal anti-Olig2 | Millipore | Cat# AB9610; RRID: AB_570666 |
| Rabbit polyclonal anti-Iba1 | Wako | Cat# 019-19741; RRID: AB_839504 |
| Rabbit monoclonal anti-NeuN | Abcam | Cat# ab177487; RRID: AB_2532109 |
| Rabbit monoclonal anti-Gephyrin | Synaptic systems | Cat# 147008; RRID: AB_2619834 |
| Guinea pig polyclonal anti-V-GAT1 | Synaptic systems | Cat# 131004, RRID: AB_887873 |
| Guinea pig polyclonal Anti-vesicular Glutamate Transporter 1 (V-GLUT1) Unconjugated | Millipore | Cat# AB5905; RRID: AB_2301751 |
| Rabbit polyclonal anti-Homer1 | Synaptic systems | Cat# 160003; RRID: AB_887730 |
| Chicken polyclonal anti-Vimentin | abcam | Cat# Ab24525; RRID: AB_778824 |
| Rat monoclonal anti mouse CD68 | Biorad | Cat# MCA1957T; RRID: AB_2074849 |
| Guinea pig polyclonal anti-Parvalbumin | Synaptic systems | Cat# 195004; RRID: AB_2156476 |
| Recombinant Rabbit monoclonal anti- CD31 (clone EPR17259) | Abcam | Cat# ab182981; RRID: AB_2920881 |
| Rabbit monoclonal anti- EGR1 (clone 15F7) | Cell Signalling | Cat# 4153; RRID: AB_2097038 |
| Alexa-Fluor goat anti-chicken 488 | Invitrogen | Cat# A11039; RRID: AB_2534096 |
| Alexa-Fluor chicken anti-mouse 488 | Invitrogen | Cat# A21200; RRID: AB_2535786 |
| Alexa-Fluor goat anti-rabbit 555 | Invitrogen | Cat# A21428; RRID: AB_141784 |
| Alexa-Fluor goat Anti-rabbit 488 | Invitrogen | Cat# A32731; RRID: AB_2633280 |
| Alexa-Fluor goat Anti-rabbit 633 | Invitrogen | Cat# A21070; RRID: AB_2535731 |
| Alexa-Fluor goat Anti-Guinea pig 647 | Invitrogen | Cat# A21450; RRID: AB_141882 |
| Alexa-Fluor goat Anti-Guinea pig 555 | Invitrogen | Cat# A21435; RRID: AB_1500610 |
| Alexa-Fluor goat Anti rat 555 | Invitrogen | A21434; RRID: AB_2535855 |
| OmniMap anti-Rabbit HRP | Ventana-Roche | Cat# 760-4311 ; RRID: AB_2811043 |
| Bacterial and virus strains | ||
| Ad5-CMV-Cre | University of Iowa, Gene Transfer Core | Cat# VVC-U of Iowa-5 |
| Biological samples | ||
| Human samples | CNIO Biobank (RENACER) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI-1640 medium | Sigma-Aldrich | Cat# R8758 |
| Penicillin/Streptomycin | Life Technologies | Cat# 15-140-122 |
| DMEM medium | Lonza | N/A |
| Trypsin-EDTA (0.05%) | Gibco | Cat# 25300096 |
| L-Glutamine | Gibco | Cat# 25030-024 |
| Amphotericin B | Fisher Scientific | Cat# 15290018 |
| Fetal bovine serum (FBS) | Life Technologies | Cat# 10500064 |
| Nusieve GTG Agarose | Lonza | Cat# 50080 |
| Hepes | Sigma-Aldrich | Cat# H3375-100G |
| D-Glucose | Sigma-Aldrich | Cat# G7528 |
| CaCl2 | Merck | Cat# 102382 |
| MgCl2 | Sigma-Aldrich | Cat# M2670 |
| NaHCO3 | Sigma-Aldrich | Cat# S6297 |
| 10X HBSS | Gibco | Cat# 4185052 |
| Dulbecco’s Phosphate Buffered Saline (PBS) | Sigma-Aldrich | Cat# D8537-500ML |
| Hydrofluoromizine (HFz) | Su et al.52 | N/A |
| D-Luciferine | Syd Labs | Cat# MB000102-R70170 |
| Isoflurane | Isoflo® | Cat# 571329.8 |
| UltraPure™ BSA | Thermo Fisher Scientific | Cat# AM2616 |
| 20 % Paraformaldehyde | Aname | Cat# 15713 |
| Normal Goat Serum (NGS) | Abcam | Cat# AB7481 |
| Bovine Serum Albumin (BSA) (IGG-Free, protease) | Vitro – Jackson Immunoresearch | Cat# 001-000-162 |
| Triton X-100 | Merck | Cat# T9284-500ML |
| bisBenzimide H 33258 | Sigma-Aldrich | Cat# B2883 |
| Hematoxylin II | Ventana, Roche | Cat# 790-2208 |
| 2.5 % glutaraldehyde | Merck | Cat# 104239.0250 |
| Paraformaldehyde | Sigma-Aldrich | Cat# P6148 |
| Osmium tetroxide | Sigma-Aldrich | Cat# 201030 |
| Durcupan™ ACM | Sigma-Aldrich | Cat# 44611-14 |
| OptiBond™ Universal light-curing glue | Kerr dental | Cat# 36517 |
| Unifast™ LC light-curing acrylic | GC America Inc | Cat# 338006 |
| Kwik-Cast™ silicone sealant | World Precision Instruments | Cat# KWIK-CAST |
| Vybrant™ DiI cell-labeling solution | ThermoFisher Scientific, Waltham | Cat# V22885 |
| Dolethal 200 mg/ml, Sodium pentobarbital | Vetoquinol | Cat# 07400060 |
| Critical commercial assays | ||
| Brain Tumor Dissociation Kit | Miltenyi Biotec | Cat# 130-095-942 |
| SureSelectXT Mouse All Exon kit | Agilent | Cat# G7550 |
| DNAeasy Blood & Tissue Kit | Qiagen | Cat# 69504 |
| Adult Brain Dissociation Kit | Miltenyi Biotec | Cat# 130-107-677 |
| Chromium Single Cell 3′GEM, Library & Gel Bead Kit v3 | 10X Genomics | Cat# PN-1000075 |
| RNAeasy Mini Kit | Qiagen | Cat# 50974104 |
| QIAshredder Kit | Qiagen | Cat# 79656 |
| QuantSeq FWD 3‘ mRNA-Seq Library Prep Kit | Lexogen | Cat# 015 |
| DISCOVERY RUO Purple kit | Ventana, Roche | Cat# No. 760–229 |
| EnVision+/HRP, Rabbit, HRP | Dako | Cat# k400311-2 |
| Target Retrival Solution, High pH | Agilent-Dako | Cat# k8004 |
| EnVision FLEX HRP Magenta Chromogen system (Dako omnis) | Dako/Agilent | N/A |
| NEBNext Ultra II Directional RNA Library Prep Kit for Illumina" (). | New England Biolabs | Cat# NEB #E7760 |
| ChromoMap DAB | Ventana, Roche | Cat# 760-159 |
| Deposited data | ||
| Bulk RNA-Seq data of mice tumours | This paper | GEO: GSE63473 |
| WES data of cancer cell lines | This paper | BioProject: PRJNA975092 |
| Bulk RNA-Seq data of cell lines | This paper | GEO: GSE218071 |
| scRNA-seq data of brain metastases | This paper | GEO: GSE233366 |
| Experimental models: Cell lines | ||
| B16/F10-BrM murine melanoma cell line | Priego et al.12 | N/A |
| 482N1 murine lung cell line | Valiente et al.10 | N/A |
| E0771-BrM murine breast cell line | Monteiro et al.11 | N/A |
| 393N1 murine lung cell line | Valiente et al.10 Winslow et al.37 | N/A |
| 2691N1 murine lung cell line | Valiente et al.10 Winslow et al.37 | N/A |
| Experimental models: Organisms/strains | ||
| Male and Female mice: LSL-CamBI: double hemizygous for CAG-LSL-OrangeCaMBI110 | Su et al.52 | N/A |
| Mouse: Thy1.2-G-CaMP7-DsRed2 | RIKEN Bioresources Center | stock RBRCO6579 |
| Software and algorithms | ||
| Living Image software, v4.5 | Perkin-Elmer | https://www.perkinelmer.com/de/lab-products-and-services/resources/in-vivo-imaging-software-downloads.html#LivingImage |
| Prism, v8 | GraphPad | https://www.graphpad.com/scientificsoftware/prism/ |
| Image J | NIH Image | https://imagej.nih.gov/ij/index.html |
| Synapse Counter plugin for imageJ | Dzyubenko et al.53 | N/A |
| MATLAB 2019a | Mathworks | https://www.mathworks.com |
| Binary decision tree for multiclass clasification (fictree) | MathWorks | https://es.mathworks.com/help/stats/fitctree.html |
| Principal Component Analysis (PCA) | MathWorks | https://es.mathworks.com/help/stats/pca.html |
| R v4.1.1 | R Foundation for Statistical Computing | https://www.R-project.org/ |
| RStudio | PBC | https://www.rstudio.com/ |
| FastQC v0.11.0 and v.0.11.9 | Simon Andrews | https://www.bioinformatics.babraham.ac.uk/projects/fastqc |
| TopHat-2.0.10 | Trapnell et al.54 | https://ccb.jhu.edu/software/tophat/downloads/ |
| Bowtie 1.0.0 | Langmead et al.55 | https://anaconda.org/bioconda/bowtie/files?version=1.0.0 |
| Seurat | Stuart et al.56 | http://satijalab.org/seurat/ |
| Nextpresso | Graña et al.57 | https://github.com/osvaldogc/nextpresso1.9.2 |
| mouse genome (GRCm39) GENCODE release 26 | The Gencode Project (Frankish et al.58) | https://www.gencodegenes.org/mouse/release_M26.html |
| mouse genome (GRCm39) GENCODE release 28 | The Gencode Project | https://www.gencodegenes.org/mouse/release_M28.html |
| Bollito | García-Jimeno et al.59 | https://github.com/cnio-bu/bollito |
| DESeq2 | Love et al.60 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Gene Set Enrichment Analysis (GSEA) | Subramanian et al.61 | https://www.gsea-msigdb.org/gsea/doc/GSEAUserGuideFrame.html |
| varca (v.pre_semver) | GitHub | https://github.com/cnio_bu/varca |
| Ensembl Vep Predictor v.108 | McLaren et al.62 | Conda package |
| SIFT | Ng et al.63 | Conda package |
| COSMIC’s Cancer Gene Census | Sondka et al.64 | https://cancer.sanger.ac.uk/census |
| cluster_rnaseq | GitHub | https://github.com/cnio-bu/cluster_rnaseq |
| bbduk (bbmap v.39.01) | Bushnell et al. | Conda package |
| STAR v2.7.8a | Dobin et al.65 | https://code.google.com/archive/p/rna-star |
| samtools v1.14 and 0.1.19 | Li et al.66 | Conda package |
| featureCounts (subread v.2.0.3) | Liao et al.67 | Conda package |
| CIBERSORTx | Newman et al.15 | https://cibersortx.stanford.edu/ |
| edgeR’s v3.40 | Robinson et al.68 | Conda package |
| Additional code for variant calling of whole exome sequencing reads | Zenodo | https://doi.org/10.5281/zenodo.7957550 |
| Additional code for bulk RNA-Seq deconvolution of cell types | Zenodo | https://doi.org/10.5281/zenodo.7957334 |
| Zen Blue Software | Zeiss | https://www.zeiss.com/microscopy/en/products/software/zeiss-zen.html |
| DigitalMicrograph software package | Gatan | https://www.gatan.com/products/tem-analysis/gatan-microscopy-suite-software |
| Adobe Photoshop CS6 (13.0.1) | Adobe Systems | www.adobe.com |
| Other | ||
| Silver wires | A-M Systems | N/A |
| Implants | Cibertec | N/A |
| Head-fixed apparatus | Cibertec | N/A |
| A1x16-5mm-100-413-A16, 16-channel silicon probes | Neuronexus | N/A |
| USB-ME16-FAI AC amplifier | Multichannel Systems | N/A |
| Digidata 1440 | Molecular Devices | N/A |
| Stereotaxic frame | N/A | N/A |
| Hamilton®TLC syringe | Merck | Cat# HAM7653-01-1EA |
| Needle RN for Hamilton syringe | VWR | Cat# HAMI7803-07 |
| 0.8 mm Whatman Nuclepore TrackEtched Membranes | Sigma-Aldrich | Cat# WHA110409 |
| Automated immunostaining platform (AutostainerLink48 (IHQ) | Dako/Agilent | N/A |
| Automated immunostaining platform (Discovery XT) (IHQ) | Ventana-Roche | N/A |
| Slide scanner (AxioScan Z1) | Zeiss | N/A |
| Sliding microtome | Thermo Fisher Scientific | N/A |
| Leica VT1000 S Vibratome | Leica | Cat# 1404723512 |
| IVIS Xenogen machine Caliper | Life Sciences | N/A |
| BD FACSAria™ Cell Sorter | BD biosciences | N/A |
| FastPrep-24™ 5G lysis system | MPBiomedical | N/A |
| Agilent 2100 Bioanalyzer | Agilent Technologies | N/A |
| NextSeq 550 | Illumina | N/A |
| gentleMACS Octo Dissociator | Miltenyi Biotec | Cat# 130-096-427 |
| gentleMACS™ C Tubes | Miltenyi Biotec | Cat# 130-093-237 |
| Covaris LE220 focused-ultrasonicator | Covaris | N/A |
| TapeStation DNA screentape D1000 | Agilent | N/A |
| NovaSeq platform | Illumina | N/A |
| UltraCut UC7 ultramicrotome | Leica Microsystems | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Manuel Valiente (mvaliente@cnio.es).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Mice
Male and Female adult C57BL6 mice were obtained from the CNIO animal facility. For electrophysiological experiments, male and female adult Thy1.2-G-CaMP7-DFsRed2 mice initially obtained from the RIKEN Bioresources Center (stock RBRCO6579) were maintained at the Instituto Cajal animal faccility. Male and female double hemizygous for CAG-LSL-OrangeCaMBI110 (LSL-CamBI) mice52 (10-12 weeks old) were obtained from the CNIO animal facility after receiving the colony from the original source (M.Z.L.).
All animal experiments were performed in accordance with a protocol approved by the Instituto Cajal CSIC, the CNIO, Instituto de Salud Carlos III and Comunidad de Madrid Institutional Animal Care and Use Committee (PROEX135/19 and PROEX 162/19). All protocols and procedures were performed according to the Spanish legislation (R.D. 1201/2005 and L.32/2007) and the European Communities Council Directive 2003 (2003/65/CE). Experiments were approved by the Ethics Committee of the Instituto Cajal and the Spanish Research Council.
Cell culture
Murine B16/F10-BrM (brain metastatic), 482N1, 393N1 and 2691N1 cells were all cultured in DMEM medium supplemented with 10 % FBS, 100 IU.ml–1 penicillin/streptomycin, and 1 mg.ml–1 amphotericin B. Murine E0771-BrM cells were cultured in RPMI 1640 medium supplemented with 10 % FBS, 100 IU.ml–1 penicillin/streptomycin, 1 mg.ml–1 amphotericin B, and 1 % hepes. All cell lines were tested negative for Mycoplasma. We did not do cell authentification beyond visual morphological and growth rate analyses.
Human samples
All human brain metastases samples were acquired from the CNIO Biobank following protocols approved by the CEI (CEI PI 25_2020 and CEI PI46_2022) and after patients signed the RENACER informed consent.
Method details
Mouse preparation for head-fixed electrophysiological experiments
Wild-type males and females adult mice were implanted with fixation head bars under isofluorane anesthesia (1.5-2 % mixed in oxygen 0.4-0.6 l/min). Two silver wires previously chlorinated were inserted over the cerebellum for reference/ground connections required for electrophysiological experiments. Implant and wires were fixed to skull with light-cured glue (OptibondTM Universal, Kerr dental, Bioggio, Switzerland) and secured with dental cement (UnifastTM LC, GC America Inc, Chicago, IL, USA). Once mice recovered from anesthesia, they were returned to their home cages.
Few days after surgery, mice were habituated to the head-fixed apparatus, consisting on a wheel (20 cm radius) coupled to a stereotactic frame. Habituation sessions (5-7 days, 2 sessions per day) included handling and mounting/dismounting the head for increasing periods of time (from 5-10 min to more than 1 hour). Habituated head-fixed mice typically alternated periods of running and immobility.
Injection of brain metastatic cell lines
Habituated mice were injected intracranially with B16/F10-BrM, 482N1, E0771-BrM, 393N1 and 2691N1 cell lines (40000 cells in 2 μl) in the right hemisphere (1.5 mm lateral and 1 mm caudal from bregma, and to a depth of 1 mm) and with saline (2 μl) in the left hemisphere by using a gas-tight Hamilton syringe and a stereotactic apparatus. Sham animals were injected intracranially with 2 μl of saline in both hemispheres.
Electrophysiological recordings from awake head-fixed mice
Six days after cancer cells injection, habituated mice were anesthetized and a cranial window was opened at -2 mm posterior from Bregma and 1.25 mm lateral from midline in each hemisphere. Afterwards, the craniotomy was covered with low toxicity silicone elastomer (Kwik-SilTM, World Precision Instruments, Sarasota, FL, USA). Recordings were performed on days 7, 9 and 10 after the cells/saline injection.
For recordings, we used 16-channel silicon probes consisting in a linear electrode array with 100 μm separation and 413 μm2 electrode area (Neuronexus). Extracellular signals were pre-amplified (4x gain) and recorded with a 16-channel AC amplifier (100x, Multichannel Systems, Reutlingen, Germany), and sampled at 20 kHz/channel (Digidata 1440, Molecular Devices, San Jose, CA, USA). For simultaneous bilateral recordings, we used two different silicon probes and recorded 8 channels per probe with 200 μm separation. Silicon probes were inserted up to 200-300 μm below CA1 stratum pyramidale of the dorsal hippocampus to get simultaneous hippocampal and cortical recordings. The stratum radiatum and stratum pyramidale were identified using characteristic physiological events, including sharp-wave ripples and the reversal of theta oscillations. The position of the animal in the wheel was stored to evaluate periods of running and immobility.
Spectral analysis of LFP activity
Analysis of electrophysiological signals was implemented in MATLAB 2019a (MathWorks). For analysis of the entire frequency band (1–1000 Hz), a Hamming window and the fast Fourier transform (FFT) at 0.5 Hz resolution were used. For spectral analysis, the contributions of 50 Hz and harmonics were filtered out, and data between the filter limits were interpolated. Analysis of the spectral power was estimated from the FFT for delta (1-4 Hz), theta (4-12 Hz), alpha (8-14 Hz), slow gamma (40-60 Hz), fast gamma (70-90 Hz), ripple (100-200 Hz) and HFOs (>200 Hz) bands. For a more standardized comparison, data was transformed to decibels by applying 10·log10(x) (where x is spectral power), and normalizing power spectrum tail to zero. One dimensional current-source density (CSD) signals were calculated from the second spatial derivative of laminar LFPs (100 μm resolution). Smoothing was applied to CSD signals for visualization purposes only. Spectral analysis of the CSD signals was similar to that of the LFP. Multiple probe penetrations were made per animal and hemisphere (typically 2 to 5), resulting in 109 ipsilateral (sham 17; E0771-BrM 12, B16/F10-BrM 17, 482N1 15, 393N1 22 and 2691N1 26) and 103 contralateral penetrations (sham 16, E0771-BrM 12, B16/F10-BrM 17, 482N1 17, 393N1 17 and 2691N1 24) from 26 mice).
After last LFP recordings, the probes were stained with DiI to label the tracks (ThermoFisher Scientific, Waltham, MA USA), and animals transcardially perfused for posterior histological analysis.
RNA-seq extraction and transcriptomic analysis
RNA was obtained from mouse B16/F10-BrM, 482N1 and E0771-BrM cell lines from a confluent well from a 6-well plate. The whole RNA was isolated using RNAeasy Mini Kit (Qiagen). 500ng of total RNA samples were used. Sample RNA Integrity numbers were 9.8 on average (range 9,7-10) when assayed on an Agilent 2100 Bioanalyzer. Sequencing libraries were prepared with the QuantSeq 3‘ mRNA-Seq Library Prep Kit (FWD) for Illumina (Lexogen, Cat.No. 015) by following manufacturer’s instructions. Library generation was initiated by reverse transcription with oligodT priming, and a second strand synthesis was performed from random primers by a DNA polymerase. Primers from both steps contained Illumina-compatible sequences. Libraries were completed by PCR {This kit generates directional libraries stranded in the sense orientation: the read1, the only read in single read format, has the sense orientation (--library-type fr-secondstrand in TopHat, --stranded=yes in HTSeq)}. cDNA libraries were purified, applied to an Illumina flow cell for cluster generation and sequenced on an Illumina NextSeq 550 (with v2.5 reagent kits) by following manufacturer's protocols. Eightysix-base-pair single-end sequenced reads followed adapter and polyA tail removal as indicated by Lexogen. Mouse reads were analysed with the Nextpresso57 pipeline as follows: sequencing quality was checked with FastQC v0.11.0 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were aligned to the mouse genome (GRCm39) with TopHat-2.0.1054 using Bowtie 1.0.055 and Samtools 0.1.19,66 allowing 3 mismatches and 20 multihits. The Gencode vM26 gene annotation for GRCm39 was used. Read counts were obtained with HTSeq.69 Differential expression and normalization were performed with DESeq2,60 filtering out those genes where the normalized count value was lower than 2 in more than 50 % of the samples. From the remaining genes, those that had an adjusted p-value below 0.05 FDR were selected. GSEAPreranked61 was used to perform gene set enrichment analysis for several gene signatures on a pre-ranked gene list, setting 1000 gene set permutations. Only those gene sets with significant enrichment levels (FDR q-value < 0.25) were considered. Access to RNA-seq data is provided from the Gene Expression Omnibus, under the ID GSE231646.
scRNA-seq
Ten days after intracranial injection of different 481N1cell lines, the presence of established brain metastases was confirmed by BLI. Mice were sacrificed and brains were extracted in precooled D-PBS 1x and a pool of three dissected established metastatic lesions from different brains were processed with the Brain Tumor Dissociation Kit (130-095-942, Miltenyi) using gentleMACS C Tubes (130-093-237, Miltenyi) and the gentleMACS Octo Dissociator (130-096-427, Miltenyi). Briefly, dissected lesions were transferred into a gentleMACS C Tub and digested with the enzymatic mixed provided by the kit with the gentleMACS Program 37C_BTDK_01 using the gentleMACS Octo Dissociator with heaters. Resulting cell suspension was filtered with a 70 μm strainer and centrifuged at 300 × g for 10 min at 4°C. For myelin and red blood cell removal, debris removal solution and red blood cell removal solution, included in the Adult Brain Dissociation Kit (130-107-677, Miltenyi), were applied. Single cell suspensión was subjected to fluorescence-activated cell sorting using the BD FACSAria™ Cell Sorter to isolate cancer cells according to GFP expression. Cell suspension was centrifuged at 300 × g for 10 min, and the pellet was resuspended in 0.04% ultrapure BSA (AM2616, Thermo Fisher Scientific) PBS 1x at a concentration of 106 cells/ml. Cells placed on ice, were tested for the optimal viability and free of debris and aggregates. Cell sample was loaded onto a 10x Chromium Single Cell controller chip B (10x Genomics) as described in the manufacturer’s protocol (Chromium Single Cell 3’GEM, Library & Gel Bead Kit v3, ref. PN-1000075). Intended targeted cell recovery of ∼10000 cells. Generation of gel beads in emulsion (GEMs), barcoding, GEM-RT clean-up, cDNA amplification and library construction were all performed as recommended by the manufacturer. scRNA-seq libraries were sequenced with an Illumina NextSeq 550 (using v2.5 reagent kits) in paired-end fashion (28bp + 56bp bases). The bollito59 pipeline was used to perform read analysis, as follows: Sequencing quality was checked with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were aligned to the mouse reference genome (GRCm39, vM28 gene annotation from GENCODE58 with STARsolo (STAR 2.7.8a).65 Each dataset was processed for quality control, normalization, anchoring, integration, dimension reduction, clustering, annotation, and marker identification.
Normalization, integration, and dimension reduction Functions in the Seurat (v3.2.2) package were used for the following analyses.56 Data were log normalized, and the top 2,000 variable features were identified. After scaling the data, linear and non-linear dimension reduction was performed by Principal Component Analysis of variable features and Uniform Manifold Approximation and Progression (UMAP) analysis, respectively, using the top 40 principle components. The number of dimensions used for dimensional reduction analyses was determined based on the inflection point on an Elbow plot. Clustering, annotation, and marker identification Clustering was calculated using the functions FindNeighbors and FindClusters, with a range in resolution between 0.1 and 1. Ultimately, a resolution of 0.75 was used for initial clustering. To identify major sub-states present, the FindAllMarkers function (log2 fold change > 0, using Wilcoxon rank sum test, adjusted p-value < 0.05 using the Bonferroni correction) was used to determine unique and/or highly enriched DEGs in one cluster compared to all other clusters. Data were visualized using Seurat package functions, including DimPlot, FeaturePlot, DotPlot and VlnPlot. For Egr1-positive sorted data, the function Seurat::subset was used (Egr1 > 2). Additionally, DEGs for Egr1-positive sorted were evaluated by pathway analysis in a within cluster mode. In brief, DEG gene IDs were converted to Ensembl IDs (using AnnotationDbi package org.Hs.eg.db::mapIDs) and then to Entrez IDs (using biomaRt::getBM). For each individual cluster in each cell sorted type, Entrez IDs were analyzed using clusterProfiler::enrichGO, and GO terms were identified (adjusted p-values < 0.05 using the Benjamini-Hochberg method, false discovery rate < 0.1).
Whole exome sequencing and somatic variant calling
DNA was obtained from mouse B16/F10-BrM, 482N1 and E0771-BrM cell lines from a confluent well from a 6-well plate. The whole DNA was isolated using DNAeasy Blood & Tissue Kit (Qiagen) and was sent to Macrogen Company in Seoul, South Korea. For the generation of standard exome capture libraries, the Agilent SureSelect Target Enrichment protocol for Illumina paired-end sequencing library (Version C2, December 2018) was used together with 1 μg input gDNA. In all cases, the SureSelectXT Mouse All Exon probe set was used. The quantification of DNA and the DNA quality was measured by PicoGreen and agarose gel electrophoresis. 1 μg of each cell line's genomic DNA diluted in EB Buffer was used and sheared to a target peak size of 150–200 bp using the Covaris LE220 focused-ultrasonicator (Covaris, Woburn, MA) according to the manufacturer's recommendations. 8 microTUBE Strip were loaded into the tube holder of the ultrasonicator and the DNA was sheared using the following settings: mode, frequency sweeping; duty cycle, 10 %; intensity, 5; cycles per burst, 200; duration, 60 sec × 6 cycles; temperature, 4°C–7°C. The fragmented DNA was repaired, an ‘A’ was ligated to the 3′ end, and agilent adapters were then ligated to the fragments. Once ligation had been assessed, the adapter ligated product was PCR amplified. For exome capture, 250 ng of DNA library was mixed with hybridization buffers, blocking mixes, RNase block and 5 μl of SureSelect all exon capture library, according to the standard Agilent SureSelect Target Enrichment protocol. Hybridization to the capture baits was conducted at 65°C using heated thermal cycler lid option at 105 °C for 24 hours on PCR machine. The captured DNA was then washed and amplified. The final purified product was then quantified using qPCR according to the qPCR Quantification Protocol Guide (KAPA Library Quantification kits for Illumina Sequencing platforms) and qualified using the TapeStation DNA screentape D1000 (Agilent). Finally, sequencing was performed using the NovaSeq platform (Illumina, San Diego, USA).
Somatic variant calling was performed using varca (https://github.com/cnio_bu/varca), a snakemake workflow which implements the GATK best-practices guidelines for calling small variants. Due to missing healthy controls and to mitigate the calling of potential germline variants, we included a panel of common single nucleotide variants from the Mouse Genome Project.70 The resulting set of variants was further refined by excluding alterations in intergenic or intronic regions, those at a depth of 50 or less, synonymous variants, those with a low predicted impact by the Ensembl Vep Predictor v.10862 and those annotated as tolerated by SIFT.63 Finally, we summarized each model’s alteration profile as a set of variants spanning known cancer genes present in COSMIC’s Cancer Gene Census.64
Access to WES data is provided from the BioProject database, under the ID PRJNA975092.
Bulk RNA-seq analysis and cell type deconvolution
Ten days after intracranial injection of different cell lines (481N1, B16/F10-BrM, and E0771-BrM), the presence of established brain metastases was confirmed by BLI. Mice were sacrificed and brain tumor and peritumoral area were microdissected and mechanically disgreggated with the FastPrep-24™ 5G lysis system (MPBiomedical) by using zirconium beads at 6.0 m/s for 15 s followed by 10 min incubation on ice. QIAshredder columns (Qiagen,79656) were used to homogenize the preparation and whole RNA was isolated using the RNAeasy Mini Kit (Qiagen).
Total RNA samples [1 μg] were converted into sequencing libraries with the "NEBNext Ultra II Directional RNA Library Prep Kit for Illumina" (NEB #E7760). Briefly, polyA+ fraction is purified and randomly fragmented, converted to double stranded cDNA and processed through subsequent enzymatic treatments of end-repair, dA-tailing, and ligation to adapters. Adapter-ligated library is completed by PCR with Illumina PE primers. The resulting purified cDNA libraries were applied to an Illumina flow cell for cluster generation and sequenced on an Illumina NextSeq 550 (with v2.5 reagent kits) by following manufacturer's protocols.
The raw sequencing reads from the bulk RNAseq samples were processed using cluster_rnaseq (https://github.com/cnio-bu/cluster_rnaseq). Briefly, FastQC (v.0.11.9) was used to generate QC reports of the sequencing reads. Raw reads were then trimmed with bbduk (bbmap v.39.01) to remove the adapter sequences. Trimmed reads were aligned with STAR v2.7.8a65 to the GRCm39 reference and indexed with samtools v1.14.66 Finally, mapped reads were counted and aggregated to a matrix of gene-level counts with featureCounts (subread v.2.0.3).67
Cell type deconvolution from bulk RNAseq samples was performed with CIBERSORTx.15 First, we normalized the gene-level counts matrix using edgeR’s68 TMM normalization and then transformed the normalized matrix to counts per million with the cpm function. Then, we generated a reference expression profile for each cell type. To do so, we downloaded normalized and annotated single cell RNA-Seq data from brain metastases published by Gonzalez H and colleagues.18 CIBERSORTx was used to generate the reference matrix of expression profiles. The parameters used for this job were the following: disabled quantile normalization, kappa=0.999, q-value=0.01, number of barcode genes between 300 and 500 and a minimum of 5 samples. The rest of the settings were left as default.
Immunofluorescence
After performing the LFP recordings, at day 10, mice were anesthetized with sodium pentobarbital and perfused with 4 % PFA. Whole brains were dissected and postfixed in the same fixative overnight at 4°C. Slicing of the brain was done by using a sliding microtome (Thermo Fisher Scientific). 80 μm slices were blocked in 10 % NGS, 2 % BSA and 0.25 % Triton X-100 in PBS for 2 h at room temperature (RT). Primary antibodies were incubated overnight at 4°C in the blocking solution and the following day for 30 min at RT. After extensive washing in PBS-Triton 0.25 %, the secondary antibody was added in the blocking solution and incubated for 2h. After extensive washing in PBS-Triton 0.25 %, nuclei were stained with bis-benzamide (1 mg/mL; Sigma-Aldrich) for 7 min at RT. Primary antibodies: GFAP (Millipore, AB5541, dilution 1:700); Olig2 (Millipore, AB9610, dilution 1:500); Iba1 (Wako, 019-19741, dilution 1:500); NeuN (Abcam, ab177487, dilution 1:200)); Gephyrin (Synaptic systems, 147008, dilution 1:1000); V-GAT1 (Synaptic systems, 131004, dilution 1:500); V-GLUT1 (Millipore, AB5905, dilution 1:2000), Homer1 (Synaptic systems, 160003, dilution 1:200); Vimentin (Abcam, ab24525, dilution 1:300); CD68 (Biorad, MCA1957T, dilution 1:1000) and Parvalbumin (Synaptic systems, 195004, dilution 1:500) . Secondary antibodies: Alexa-Fluor goat anti-chicken 488 (Invitrogen, A11039, dilution 1:500); Alexa-Fluor chicken anti-mouse 488 (Invitrogen, A21200, dilution 1:300); Alexa-Fluor goat anti-rabbit 555 (Invitrogen, A21428, dilution 1:300); Alexa-Fluor goat Anti-rabbit 488 (Invitrogen, A32731, dilution 1:300); Alexa-Fluor goat Anti-rabbit 633 (Invitrogen, A21070, dilution 1:300); Alexa-Fluor goat Anti-Guinea pig 647 (Invitrogen, A21450, dilution 1:300); Alexa-Fluor goat Anti-rat 555 (Invitrogen, A-21434, dilution 1:300) and Alexa-Fluor goat Anti-Guinea pig 555 (Invitrogen, A21435, dilution 1:300).
Immunohistochemistry
Whole brains from the brain metastasis models (B16/F10-BrM, E0771-BrM, 482N1, 393N1, 2691N1, H2030-BrM, HCC1954-BrM and MDA231-BrM) were dissected and postfixed in 4 % PFA overnight at 4°C and then stored at PBS. Brains were cut at 3 μm, mounted on SuperFrost Plus® and TOMO® slides and dried overnight. Immunohistochemical reactions were performed on an automated immunostaining platform (AutostainerLink48 (Dako/Agilent and Discovery XT, Ventana-Roche) for CD31 and EGR1 respectively. After antigen retrieval was performed on the PT Link, with target retrieval solution, high pH (k8004, Agilent) for CD31 and with pH buffer (CC1 Ventana, Roche) for EGR1, endogenous peroxidase was blocked (peroxide hydrogen at 3%). The slides were then incubated with the appropriate primary antibody: Rabbit monoclonal anti- CD31 (clone EPR17259, dilution 1/2000, 35 min, Abcam, ab182981) and/or rabbit monoclonal anti- EGR1 (clone 15F7, dilution 1/50, 32 min, Cell Signalling, 4153). After the primary antibody incubation, the slides were incubated with a horseradish peroxidase-conjugated anti-rabbit secondary antibody: i). CD31 with EnVision Rabbit visualisation systems (Dako, k400311-2). The immunohistochemical reaction was developed with EnVision FLEX HRP Magenta Chromogen (Dako/Agilent); ii). ERG1 with visualisation systems (OmniMap anti-Rabbit, 760-4311, Ventana, Roche) and the immunohistochemical reaction was developed with purple (DISCOVERY RUO Purple kit, Cat. No. 760–229, Ventana, Roche).
Finally, nuclei were counterstained with Carazzi’s hematoxylin and slides were dehydrated, rinsed and mounted with permanent mounting medium for microscopic evaluation.
Positive control sections known to be positive for the primary antibody were included in each staining series.
Image acquisition
Immunofluorescence whole slides images were acquired with a Thunder Imaging System (Leica-Microsystems) equipped with a 10X dry objective with a NA of 0.45, led excitation, a DFC9000GTC camera and LAS X v3.5 software. Glial cells and neurons images were acquired with a Leica SP5 up-right confocal microscope equipped with a 20X dry objective with a NA of 0.7 and analysed with ImageJ software.
Excitatory and inhibitory synapses images were acquired with a SP8 up-right confocal microscope equipped with a 63X oil immersion objective with a NA of 1.4. Images were taken as z-stacks (five slices, 0,2 μm intervals) with a scan zoom of 2,78X and an image size of 1024 x 1024 pixels (66,38 x 66,38 μm). Synapses quantification was performed in maximal, intensity projection using Synapse Counter plugin for imageJ.53
Immunohistochemistry whole slides images were acquired with a slide scanner (AxioScan Z1, Zeiss), with a 20X objective and images captured with the Zen Blue Software (Zeiss). An appropriate script was created using Zen Blue Software (additional module for analysis, Zeiss) to analyse each antibody. After training and script optimization, the quantification was run on each image manually, in order to select areas and results exported as excel files with scoring data for each file.
Calcium imaging in organotypic cultures
Adult double hemizygous for CAG-LSL-OrangeCaMBI110 (LSL-CamBI) mice52 were intracranially injected with each BrM cell line (B16/F10-BrM, 482N1 or E0771-BrM). 10 d after intracranial injection of cancer cells, animals were sacrificed and brains with established metastases were processed into organotypic cultures as previously described.10 In brief, brains were dissected in HBSS supplemented with HEPES (pH 7.4, 2.5 mM), D-glucose (30 mM), CaCl2 (1 mM), MgCl2(1 mM), and NaHCO3 (4 mM) and embedded in 4 % low-melting agarose (Lonza) preheated at 42°C. The embedded brains were cut into 300 μm slices using a vibratome (Leica). Brain slices were divided into two halves. Slices were placed with flat spatulas on top of 0.8 μm pore membranes (Sigma-Aldrich) floating on slice culture media (DMEM, supplemented HBSS, FBS 5 %, L-glutamine (1 mM), and 100 IU/ml penicillin/streptomycin).
Brain slices were imaged to identify cancer cells-derived bioluminescence by adding D-Luciferin for firefly-expressing cancer cells using BLI (day 0). Then 20 μl of Ad5-CMV-Cre (103 pfu / μl, University of Iowa) were added to each brain slice to allow CamBI expression.
Brain slices were imaged 3 days after the addition of the AAV-Cre to identify microenvironment derived calcium-dependent bioluminescence by adding Hydrofluoromizine52 using BLI (day 3). Microenvironment derived calcium-dependent bioluminescence (day 3) was then normalized to the cancer cells-derived bioluminescence (day 0) to avoid any variable related to the tumor size. Hydrofluoromizine was synthesized following the synthetic protocols published,52 to afford desired compound as a brown solid. 1H NMR (300 MHz, CDCl3) δ ppm 9.80 (broad s, 1H), 9.43 (br s, 1H), 7.38 (m, 2H), 7.22 (m, 2H), 7.02 (m, 2H), 6.79 (m, 1H), 6.60 (m, 1H), 6.40 (m, 1H), 6.18 (m, 2H), 4.41 (s, 2H), 4.22 (s, 2H). LCMS (ESI, Gemini-NX C18, 100 x 2.0 mm; 5 μm particle size. Eluent A, water with 0.1 % formic acid; eluent B: acetonitrile with 0.1 % formic acid. Gradient 5 % to 100 % of B within 8 min at 50°C, DAD): Rt = 4.667 min, m/z = 398.10 [M+ H]+ (calculated for C24H19N3O3 [M + H]+ m/z 398.1505).
Electron microscopy
Mice bearing brain metastases formed by the intracranial injection of E0771-BrM cells, B16/F10-BrM and 482N1 cells (10 days after the injection) were euthanized by CO2 and perfused with 2.5 % glutaraldehyde and 2 % paraformaldehyde in buffer phosphate 0.1 M (pH 7.2-7.4). Whole brains were dissected and postfixed in the same fixative at room temperature for 6-8 h in slow shaking. Coronal sections (500 μm thick) were obtained with a vibratome (Leica). Slices containing metastases were washed in buffer phosphate 0.1 M (pH7.2-7.4) at room temperature and were then further cut in small pieces of 1 mm x 1.5 mm containing the interface metastasis-neuropil and stored in buffer phosphate 0.1 M (pH7.2-7.4). Samples were then washed in iso-osmolar Sorensen (0.1 M) / Sucrose (4-8 %) buffer and fixed in 1 % (w/v) osmium tetroxide in Sorensen buffer during 2 h in darkness at 4°C. After repeated washes, samples were dehydrated in increasing concentrations of acetone (30, 50, 70, 90, 99 and 100 %) and embedded in epoxi resin in ascending steps of acetone:resin (2:1, 1:1 and 1:2) and then pure resin. Samples were included in a new epoxy resin and were polymerized for 48 h at 55°C. Hardened blocks were cut with a 35° diamond knife (Diatome, USA) using the UltraCut UC7 ultramicrotome (Leica Microsystems, Germany). Ultrathin sections (50-60 nm) were collected on formvar/carbon-coated copper grids and contrast stained with 2 % uranyl acetate and lead citrate. Section were then examined with the JEM 1011 (JEOL, Japan) electron microscope, operating at 80 kV. Micrographs were taken with a camera (Orius 1200A; Gatan, USA) using the DigitalMicrograph software package (Gatan, USA). Electron micrographs were processed using Adobe Photoshop CS6 (13.0.1) (Adobe Systems).
Human sample processing and immunohistochemistry
Human tissue samples were cut at 4 μm, mounted on Thermo Fisher Scientific Gerhard Menzel (Superfrost® Plus) slides and dried overnight. Immunohistochemical reactions were performed on an automated immunostaining platform (Discovery XT, Ventana-Roche). First, antigen retrieval was performed with the appropriate pH buffer (CC1 Ventana, Roche) and endogenous peroxidase was blocked (3 % hydrogen peroxide). Slides were then incubated with the primary antibody: rabbit monoclonal anti-EGR1 antibody (clone: 15F7, dilution 1/50, 32 min, Cell Signaling, 4153). After the primary antibody, slides were incubated with a horse peroxidase-conjugated anti-rabbit secondary antibody with visualization systems (OmniMap anti-rabbit, 760-4311, Ventana, Roche). The immunohistochemical reaction was developed using 3, 30-diaminobenzidine tetrahydrochloride (ChromoMap DAB, 760-159, Ventana, Roche) and nuclei were counterstained with hematoxylin II (modified Mayer's hematoxylin, 790-2208, Ventana, Roche). Finally, the slides were dehydrated, rinsed and mounted with permanent mounting medium for microscopic evaluation. Positive control sections known to be positive for the primary antibody were included in each staining series.
Principal Component Analysis (PCA)
We performed a Principal Component (PC) analysis of an input data matrix that consisted on 492 samples obtained from 123 recording sessions from 20 different animals (control: n=6, breast metastasis: n=4, melanoma metastasis: n=5, lung metastasis: n=5). Each sample data was classified as coming from periods of Run (speed>2 m/s) and Still (speed<2 m/s) and from different recording channels which were separated according to their location (Cortex/Hippo and Ipsi/Contra). Apart from these categorical variables, several spectral features were evaluated for each sample. The mean power (in decibels) for each oscillatory band was computed and reported as: delta (delta_db; 1-4 Hz), theta (theta_db; 4-12 Hz), alfa (alfa_db; 9-14 Hz), slow gamma (gammaS_db; 40-60 Hz), fast gamma (gammaF_db; 70-90 Hz), ripple (ripple_db; 100-200 Hz), high frequency oscillations or HFO (HFO_db; 200-1000 Hz). Ratios between some band were also included, such as delta over theta (delta_over_theta, delta_db/theta_db), and slow over fast gamma (gammaS_over_gammaF; gammaS_db/gammaF_db).
The input data matrix (492 samples x 12 features including categorical variables) was z-scored by column, and PCA was performed over the resulting matrix, using pca MATLAB implementation. We worked with 9 principal components, which were the number of principal components that explained 99% of variance.
Generalized Linear Model (GLM)
Three different Generalized Linear Models (GLMs) were implemented to distinguish a particular brain metastasis from the others. Input data to inform the GLM were the following z-scored variables: delta_db, theta_db, alfa_db, gammaS_db, gammaF_db, ripple_db, HFO_db, delta_over_theta, gammaS_over_gammaF. To compute the GLM, the fitglm function from MATLAB was used with a linear fit, a binomial distribution and logit link.
Comparative between machine learning classifiers
We compared different machine learning classifiers, including several types of decision trees and support vector machines. In particular, standard decision trees (DecisionTreeClassifier from sklearn.tree python package), a random forest that fits several randomized trees (ExtraTreeClassifier from sklearn.tree), XGBoost (xgboost python package), a linear support vector classifier (LinearSVC from sklearn.svm) and a standard non-linear support vector classifier (SVC from sklearn.svm). All methods were tested using default hyperparameters, in python 3.8.5, with sklearn 0.23.2, xgboost 1.6.1. In order to test which classifier was more accurate, we built a pool of 600 different instances of each classifier. Each instance was trained using a different randomized subset of data recorded at late stages (9-10 days after cancer cells injection) projected over the PCA space (60%), and evaluated computing the accuracy over the remaining 40%. Accuracy was defined as:
Models that did not achieved a significant accuracy (tested against 100 shuffled samples, p-value<0.05) for every class (control or breast/melanoma/lung metastasis) were discarded, and only significant models were considered. Accuracies of these instances were compared using the number of significant models, and the mean accuracy of the significant models.
Decision trees for predictive modeling
Final classification of input data between control or breast/melanoma/lung metastasis classes was performed using decision trees. First, we trained classifiers using four randomly selected subsets of data (60%) recorded at late stages (9-10 days after cancer cells injection) projected over the PCA space, resulting in four different decision trees. Each decision tree model was tested over the remaining subset of late data (40%). Decision trees were trained using fitctree from MATLAB R2019b, choosing MaxNumCategories parameter that better optimized accuracy.
Accuracies were reported as the mean of the four decision trees and tested against shuffled samples (p-value<0.05).
To evaluate the capability to predict the presence of metastasis in advance, we trained a random forest classifier using all late data projected over the PCA space, and test it over data obtained at early stages (7 days after cancer cells injection). The random forest consisted on six decision trees defined using the fitctree MATLAB function with MaxNumCategories = 4, and MaxNumSplits ranging from 2 to 9. Predictions made over the early data were then transformed into control or metastasis (no distinction between metastasis subtypes). The final decision was taken as the most voted of the six decision trees, following a random forest strategy.
Confusion matrices
Predictions were represented using confusion matrices. Each row of the matrix represents the number of predictions of each class, while each column represents the number of instances on the real class. The diagonal therefore contains the number of true positives (or true negatives depending on the visualization), and the off-diagonal elements false positives and false negatives. In order to compensate for imbalances, we divided each column by the total number of instances of the real class.
The confusion matrix is, then:
| T R U E |
|||||
|---|---|---|---|---|---|
| Control (C) | Breast (B) | Melanoma (M) | Lung (L) | ||
| P | C | (#Cp + #Ct) / (#Ct) | (#Cp + #Bt) / (#Bt) | (#Cp + #Mt) / (#Mt) | (#Cp + #Lt) / (#Lt) |
| R | B | (#Bp + #Ct) / (#Ct) | (#Bp + #Bt) / (#Bt) | (#Bp + #Mt) / (#Mt) | (#Bp + #Lt) / (#Lt) |
| E | M | (#Mp + #Ct) / (#Ct) | (#Mp + #Bt) / (#Bt) | (#Mp + #Mt) / (#Mt) | (#Mp + #Lt) / (#Lt) |
| D | L | (#Lp + #Ct) / (#Ct) | (#Lp + #Bt) / (#Bt) | (#Lp + #Mt) / (#Mt) | (#Lp + #Lt) / (#Lt) |
where # means number of, C denotes control, B breast, M melanoma and L lung, t means “true” and p means “predicted”
Testing generalizing capability
We evaluated generalizability of the prediction approach using two strategies. First, a leave-one-out cross validation (LOOCV) test was implemented. To this purpose, we re-trained classifiers using sessions from all mice but one and then tested the predictor in sessions from the remaining mice. We quantified the ability of the predictor to generalize by estimating the mean squared error (MSE). Second, new experiments were obtained using mice from different experimental cancer cell lines to test the generalization capability of the already trained classifiers. New data was z-scored using the mean and SD of the training dataset, and projected into the same PC axes.
Quantification and statistical analysis
Statistical analyzes were run on GraphPad Prism.
Data are represented as the mean or median ± s.e.m. Comparisons between two experimental groups were analysed with unpaired, two-tailed Student’s t-test. Non-parametrical analysis between two experimental groups were performed with Wilcoxon rank sum test and Wilcoxon signed rank test. Analysis between more than two groups with homogenous variances were performed by a one-way ANOVA followed by a post-hoc Tukey test. Non-parametrical multivariate analysis between more than two groups were performed by a Kruskal-Wallis test followed by a post-hoc Tukey test or a Dunn's test. Statistical analysis of machine learning models was implemented in Matlab.
Statistical details of experiments can be found in the figure legends and include the statistical tests used, exact value of n and what n represents.
Acknowledgments
We thank all members of the Brain Metastasis Group, the Prida Lab, and G. Huberfeld and S. Hervey-Jumper for critical discussion of the manuscript; the CNIO Core Facilities and Instituto Cajal Core Facilities for their excellent assistance. We also thank J. Massagué (MSKCC) for some of the BrM cell lines. This study was funded by H2020-FETOPEN-2018-2019-2020-01 (828972) (M.V., L.M.-P.), MICIN/AEI/10.13039/501100011033 by the European Union NextGenerationEU/PRTR (PID2021-124582OB-I00 to M.V., and PID2021-124829NB-I00 to L.M.-P), Fundación Ramón Areces (CIVP20S10662), (E.O.-P.), LAB AECC 2019 (LABAE19002VALI) (M.V.), ERC CoG (864759) (M.V.), NIH grant R21NS122055 (M.Z.L.). M.V. is an EMBO YIP member (4053).
Author contributions
Conceptualization, A.S.-A., M.M.-M., A.N.-O., L.M.P., M.V.; Methodology, A.S.-A., M.M.-M., A.N.-O., L.M.P., M.V., M.L.; Investigation, A.S.-A., M.M.-M., A.N.-O., P.B., C.H.-O., N.P., L.C.-B., L.A.-E., M.L.; Software, C.H.-O., A.N.-O., S.G., F.A.-S.; Formal Analysis, A.S.-A., M.M.-M., A.N.-O., P.B., C.H.-O., N.P., L.C.-B., L.A.-E., M.L., L.M.P., M.V.; Resources, S.M., M.Z.L., C.S., N.A., M.-J.A., E.O.-P., V.G.-C., A.P.-N., P.G.-L., L.J.-R., L.M.-M., O.E., J.M.S., O.T., A.H.L., A.A., G.B., J.F.A., A.L.Z., A.D.N., L.C., C.F.V., A.G.P., P.D.D.L., M.P., G.P.A., B.E.O.; Writing, A.S.-A., M.M.-M., A.N.-O., L.M.P., M.V., M.L.; Visualization, A.S.-A., M.M.-M., A.N.-O., L.M.P., M.V.; Supervision, L.M.P., M.V., F.A.-S.; Project Administration, A.S.-A., M.M.-M., A.N.-O., L.M.P., M.V.; Funding Acquisition, L.M.P., M.V.; Validation, A.S.-A., M.M.-M., A.N.-O.; Data curation, A.N.-O., C.H.-O., S.G., F.A.-S.
Declaration of interests
The authors declare no conflicts of interest.
Published: August 30, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ccell.2023.07.010.
Contributor Information
Liset Menendez de la Prida, Email: lmprida@cajal.csic.es.
Manuel Valiente, Email: mvaliente@cnio.es.
RENACER:
Cecilia Sobrino, Nuria Ajenjo, Maria-Jesus Artiga, Eva Ortega-Paino, Virginia García-Calvo, Angel Pérez-Núñez, Pedro González-León, Luis Jiménez-Roldán, Luis Miguel Moreno, Olga Esteban, Juan Manuel Sepúlveda, Oscar Toldos, Aurelio Hernández Laín, Alicia Arenas, Guillermo Blasco, José Fernández Alén, Adolfo de la Lama Zaragoza, Antía Domínguez Núñez, Lourdes Calero, Concepción Fiaño Valverde, Ana González Piñeiro, Pedro David Delgado López, Mar Pascual, Gerard Plans Ahicart, and Begoña Escolano Otín
Supplemental information
Data and code availability
-
•
Raw sequencing reads from whole exome sequencing data of each of the models described in the paper have been deposited at SRA (BioProject ID: PRJNA975092) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Likewise, bulk RNA-Seq samples from mice models have also been deposited at GEO (GSE231646) and also listed in the key resources table.
-
•
Both of the pipelines used to preprocess whole exome data and RNA-Seq data are available at GitHub: https://github.com/cnio_bu/varca and https://github.com/cnio_bu/cluster_rnaseq Additional DOIs for code used in this study are listed in the key resources table.
-
•
Raw sequencing reads from single-cell RNA-seq data of each of the models described in the paper have been deposited at GEO (GSE233366) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
Original code for scRNA-seq analysis has been deposited at Zenodo. DOI https://doi.org/10.5281/zenodo.7963303.
-
•
Raw sequencing reads from bulk RNA-sequencing data of each cell line described in the paper have been deposited at GEO (GSE218071) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
Pipeline used to preprocess RNA-Seq data is available at GitHub:https://github.com/osvaldogc/nextpresso1.9.2This section does not report original code.
-
•
Data and code from classification algorithms can be found in: https://github.com/PridaLab/metast-tree
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Moss N.S., Beal K., Tabar V. Brain metastasis—a distinct oncologic disease best served by an integrated multidisciplinary team approach. JAMA Oncol. 2022;8:1252–1254. doi: 10.1001/jamaoncol.2022.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerstenecker A., Nabors L.B., Meneses K., Fiveash J.B., Marson D.C., Cutter G., Martin R.C., Meyers C.A., Triebel K.L. Cognition in patients with newly diagnosed brain metastasis: profiles and implications. J. Neuro Oncol. 2014;120:179–185. doi: 10.1007/s11060-014-1543-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson S.W., Damasio H., Tranel D. Neuropsychological impairments associated with lesions caused by tumor or stroke. Arch. Neurol. 1990;47:397–405. doi: 10.1001/archneur.1990.00530040039017. [DOI] [PubMed] [Google Scholar]
- 4.Jena A., Taneja S., Talwar V., Sharma J.B. Magnetic resonance (MR) patterns of brain metastasis in lung cancer patients: correlation of imaging findings with symptom. J. Thorac. Oncol. 2008;3:140–144. doi: 10.1097/JTO.0b013e318161d775. [DOI] [PubMed] [Google Scholar]
- 5.Zeng Q., Michael I.P., Zhang P., Saghafinia S., Knott G., Jiao W., McCabe B.D., Galván J.A., Robinson H.P.C., Zlobec I., et al. Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature. 2019;573:526–531. doi: 10.1038/s41586-019-1576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neman J., Choy C., Kowolik C.M., Anderson A., Duenas V.J., Waliany S., Chen B.T., Chen M.Y., Jandial R. Co-evolution of breast-to-brain metastasis and neural progenitor cells. Clin. Exp. Metastasis. 2013;30:753–768. doi: 10.1007/s10585-013-9576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neman J., Termini J., Wilczynski S., Vaidehi N., Choy C., Kowolik C.M., Li H., Hambrecht A.C., Roberts E., Jandial R. Human breast cancer metastases to the brain display GABAergic properties in the neural niche. Proc. Natl. Acad. Sci. USA. 2014;111:984–989. doi: 10.1073/pnas.1322098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park E.S., Kim S.J., Kim S.W., Yoon S.-L., Leem S.-H., Kim S.-B., Kim S.M., Park Y.-Y., Cheong J.-H., Woo H.G., et al. Cross-species hybridization of microarrays for studying tumor transcriptome of brain metastasis. Proc. Natl. Acad. Sci. USA. 2011;108:17456–17461. doi: 10.1073/pnas.1114210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wingrove E., Liu Z.Z., Patel K.D., Arnal-Estapé A., Cai W.L., Melnick M.-A., Politi K., Monteiro C., Zhu L., Valiente M., et al. Transcriptomic hallmarks of tumor plasticity and stromal interactions in brain metastasis. Cell Rep. 2019;27:1277–1292.e7. doi: 10.1016/j.celrep.2019.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valiente M., Obenauf A.C., Jin X., Chen Q., Zhang X.H.-F., Lee D.J., Chaft J.E., Kris M.G., Huse J.T., Brogi E., et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell. 2014;156:1002–1016. doi: 10.1016/j.cell.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monteiro C., Miarka L., Perea-García M., Priego N., García-Gómez P., Álvaro-Espinosa L., de Pablos-Aragoneses A., Yebra N., Retana D., Baena P., et al. Stratification of radiosensitive brain metastases based on an actionable S100A9/RAGE resistance mechanism. Nat. Med. 2022;28:752–765. doi: 10.1038/s41591-022-01749-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Priego N., Zhu L., Monteiro C., Mulders M., Wasilewski D., Bindeman W., Doglio L., Martínez L., Martínez-Saez E., Ramón Y Cajal S., et al. STAT3 labels a subpopulation of reactive astrocytes required for brain metastasis. Nat. Med. 2018;24:1024–1035. doi: 10.1038/s41591-018-0044-4. [DOI] [PubMed] [Google Scholar]
- 13.Valiente M., Ahluwalia M.S., Boire A., Brastianos P.K., Goldberg S.B., Lee E.Q., Le Rhun E., Preusser M., Winkler F., Soffietti R. The evolving landscape of brain metastasis. Trends Cancer. 2018;4:176–196. doi: 10.1016/j.trecan.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valiente M., Van Swearingen A.E.D., Anders C.K., Bairoch A., Boire A., Bos P.D., Cittelly D.M., Erez N., Ferraro G.B., Fukumura D., et al. Brain metastasis cell lines panel: a public resource of organotropic cell lines. Cancer Res. 2020;80:4314–4323. doi: 10.1158/0008-5472.CAN-20-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., Khodadoust M.S., Esfahani M.S., Luca B.A., Steiner D., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luca B.A., Steen C.B., Matusiak M., Azizi A., Varma S., Zhu C., Przybyl J., Espín-Pérez A., Diehn M., Alizadeh A.A., et al. Atlas of clinically distinct cell states and ecosystems across human solid tumors. Cell. 2021;184:5482–5496.e28. doi: 10.1016/j.cell.2021.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marquez-Galera A., de la Prida L.M., Lopez-Atalaya J.P. A protocol to extract cell-type-specific signatures from differentially expressed genes in bulk-tissue RNA-seq. STAR Protoc. 2022;3:101121. doi: 10.1016/j.xpro.2022.101121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez H., Mei W., Robles I., Hagerling C., Allen B.M., Hauge Okholm T.L., Nanjaraj A., Verbeek T., Kalavacherla S., van Gogh M., et al. Cellular architecture of human brain metastases. Cell. 2022;185:729–745.e20. doi: 10.1016/j.cell.2021.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell S.L., Robel S., Cuddapah V.A., Robert S., Buckingham S.C., Kahle K.T., Sontheimer H. GABAergic disinhibition and impaired KCC2 cotransporter activity underlie tumor-associated epilepsy. Glia. 2015;63:23–36. doi: 10.1002/glia.22730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh Y., Park Y., Cho J.H., Wu H., Paulk N.K., Liu L.X., Kim N., Kay M.A., Wu J.C., Lin M.Z. An orange calcium-modulated bioluminescent indicator for non-invasive activity imaging. Nat. Chem. Biol. 2019;15:433–436. doi: 10.1038/s41589-019-0256-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu K., Lin C.-C.J., Hatcher A., Lozzi B., Kong K., Huang-Hobbs E., Cheng Y.-T., Beechar V.B., Zhu W., Zhang Y., et al. PIK3CA variants selectively initiate brain hyperactivity during gliomagenesis. Nature. 2020;578:166–171. doi: 10.1038/s41586-020-1952-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berto S., Fontenot M., Seger S., Ayhan F., Caglayan E., Kulkarni A., Douglas C., Tamminga C.A., Lega B.C., Konopka G. The genomic underpinnings of oscillatory biomarkers supporting successful memory encoding in humans. bioRxiv. 2019 doi: 10.1101/853531. Preprint at. [DOI] [Google Scholar]
- 23.Cid E., Marquez-Galera A., Valero M., Gal B., Medeiros D.C., Navarron C.M., Ballesteros-Esteban L., Reig-Viader R., Morales A.V., Fernandez-Lamo I., et al. Sublayer- and cell-type-specific neurodegenerative transcriptional trajectories in hippocampal sclerosis. Cell Rep. 2021;35:109229. doi: 10.1016/j.celrep.2021.109229. [DOI] [PubMed] [Google Scholar]
- 24.Venkatesh H.S., Morishita W., Geraghty A.C., Silverbush D., Gillespie S.M., Arzt M., Tam L.T., Espenel C., Ponnuswami A., Ni L., et al. Electrical and synaptic integration of glioma into neural circuits. Nature. 2019;573:539–545. doi: 10.1038/s41586-019-1563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkataramani V., Tanev D.I., Strahle C., Studier-Fischer A., Fankhauser L., Kessler T., Körber C., Kardorff M., Ratliff M., Xie R., et al. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature. 2019;573:532–538. doi: 10.1038/s41586-019-1564-x. [DOI] [PubMed] [Google Scholar]
- 26.Krishna S., Choudhury A., Seo K., Ni L., Kakaizada S., Lee A., Aabedi A., Cao C., Sudharshan R., Egladyous A., et al. Glioblastoma remodeling of neural circuits in the human brain decreases survival. bioRxiv. 2021 doi: 10.1101/2021.02.18.431915. Preprint at. [DOI] [Google Scholar]
- 27.Hai L., Hoffmann D.C., Mandelbaum H., Xie R., Ito J., Jung E., Weil S., Sievers P., Venkataramani V., Azorin D.D., et al. A connectivity signature for glioblastoma. bioRxiv. 2021 doi: 10.1101/2021.11.07.465791. Preprint at. [DOI] [Google Scholar]
- 28.Osswald M., Jung E., Sahm F., Solecki G., Venkataramani V., Blaes J., Weil S., Horstmann H., Wiestler B., Syed M., et al. Brain tumour cells interconnect to a functional and resistant network. Nature. 2015;528:93–98. doi: 10.1038/nature16071. [DOI] [PubMed] [Google Scholar]
- 29.Duclot F., Kabbaj M. The role of early growth response 1 (EGR1) in brain plasticity and neuropsychiatric disorders. Front. Behav. Neurosci. 2017;11:35. doi: 10.3389/fnbeh.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kundumani-Sridharan V., Niu J., Wang D., Van Quyen D., Zhang Q., Singh N.K., Subramani J., Karri S., Rao G.N. 15(S)-hydroxyeicosatetraenoic acid-induced angiogenesis requires Src-mediated Egr-1-dependent rapid induction of FGF-2 expression. Blood. 2010;115:2105–2116. doi: 10.1182/blood-2009-09-241802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheng J., Liu D., Kang X., Chen Y., Jiang K., Zheng W. Egr-1 increases angiogenesis in cartilage via binding Netrin-1 receptor DCC promoter. J. Orthop. Surg. Res. 2018;13:125. doi: 10.1186/s13018-018-0826-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fahmy R.G., Dass C.R., Sun L.-Q., Chesterman C.N., Khachigian L.M. Transcription factor Egr-1 supports FGF-dependent angiogenesis during neovascularization and tumor growth. Nat. Med. 2003;9:1026–1032. doi: 10.1038/nm905. [DOI] [PubMed] [Google Scholar]
- 33.Brown K.C., Lau J.K., Dom A.M., Witte T.R., Luo H., Crabtree C.M., Shah Y.H., Shiflett B.S., Marcelo A.J., Proper N.A., et al. MG624, an α7-nAChR antagonist, inhibits angiogenesis via the Egr-1/FGF2 pathway. Angiogenesis. 2012;15:99–114. doi: 10.1007/s10456-011-9246-9. [DOI] [PubMed] [Google Scholar]
- 34.Shimoyamada H., Yazawa T., Sato H., Okudela K., Ishii J., Sakaeda M., Kashiwagi K., Suzuki T., Mitsui H., Woo T., et al. Early growth response-1 induces and enhances vascular endothelial growth factor-A expression in lung cancer cells. Am. J. Pathol. 2010;177:70–83. doi: 10.2353/ajpath.2010.091164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zipp F., Bittner S., Schafer D.P. Cytokines as emerging regulators of central nervous system synapses. Immunity. 2023;56:914–925. doi: 10.1016/j.immuni.2023.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lotte F., Congedo M., Lécuyer A., Lamarche F., Arnaldi B. A review of classification algorithms for EEG-based brain-computer interfaces. J. Neural. Eng. 2007;4:R1–R13. doi: 10.1088/1741-2560/4/2/R01. [DOI] [PubMed] [Google Scholar]
- 37.Winslow M.M., Dayton T.L., Verhaak R.G.W., Kim-Kiselak C., Snyder E.L., Feldser D.M., Hubbard D.D., DuPage M.J., Whittaker C.A., Hoersch S., et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473:101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters S., Camidge D.R., Shaw A.T., Gadgeel S., Ahn J.S., Kim D.-W., Ou S.-H.I., Pérol M., Dziadziuszko R., Rosell R., et al. Alectinib versus Crizotinib in untreated ALK-positive non-small-cell lung cancer. N. Engl. J. Med. 2017;377:829–838. doi: 10.1056/NEJMoa1704795. [DOI] [PubMed] [Google Scholar]
- 39.Cortés J., Kim S.-B., Chung W.-P., Im S.-A., Park Y.H., Hegg R., Kim M.H., Tseng L.-M., Petry V., Chung C.-F., et al. Trastuzumab deruxtecan versus trastuzumab emtansine for breast cancer. N. Engl. J. Med. 2022;386:1143–1154. doi: 10.1056/NEJMoa2115022. [DOI] [PubMed] [Google Scholar]
- 40.Bartsch R., Berghoff A.S., Furtner J., Marhold M., Bergen E.S., Roider-Schur S., Starzer A.M., Forstner H., Rottenmanner B., Dieckmann K., et al. Trastuzumab deruxtecan in HER2-positive breast cancer with brain metastases: a single-arm, phase 2 trial. Nat. Med. 2022;28:1840–1847. doi: 10.1038/s41591-022-01935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wefel J.S., Parsons M.W., Gondi V., Brown P.D. Neurocognitive aspects of brain metastasis. Handb. Clin. Neurol. 2018;149:155–165. doi: 10.1016/B978-0-12-811161-1.00012-8. [DOI] [PubMed] [Google Scholar]
- 42.Stone J.B., DeAngelis L.M. Cancer-treatment-induced neurotoxicity--focus on newer treatments. Nat. Rev. Clin. Oncol. 2016;13:92–105. doi: 10.1038/nrclinonc.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rowan A.J., Rudolf N. de M., Scott D.F. EEG prediction of brain metastases. A controlled study with neuropathological confirmation. J. Neurol. Neurosurg. Psychiatry. 1974;37:888–893. doi: 10.1136/jnnp.37.8.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishna S., Choudhury A., Keough M.B., Seo K., Ni L., Kakaizada S., Lee A., Aabedi A., Popova G., Lipkin B., et al. Glioblastoma remodelling of human neural circuits decreases survival. Nature. 2023;617:599–607. doi: 10.1038/s41586-023-06036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hatcher A., Yu K., Meyer J., Aiba I., Deneen B., Noebels J.L. Pathogenesis of peritumoral hyperexcitability in an immunocompetent CRISPR-based glioblastoma model. J. Clin. Invest. 2020;130:2286–2300. doi: 10.1172/JCI133316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valiente M., M de la Prida L. Breaking down the wall: the strategic plan of cancer to conquer the brain. Epilepsy Curr. 2020;20:384–386. doi: 10.1177/1535759720949241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venkataramani V., Yang Y., Schubert M.C., Reyhan E., Tetzlaff S.K., Wißmann N., Botz M., Soyka S.J., Beretta C.A., Pramatarov R.L., et al. Glioblastoma hijacks neuronal mechanisms for brain invasion. Cell. 2022;185:2899–2917.e31. doi: 10.1016/j.cell.2022.06.054. [DOI] [PubMed] [Google Scholar]
- 48.Lipkova J., Chen R.J., Chen B., Lu M.Y., Barbieri M., Shao D., Vaidya A.J., Chen C., Zhuang L., Williamson D.F.K., et al. Artificial intelligence for multimodal data integration in oncology. Cancer Cell. 2022;40:1095–1110. doi: 10.1016/j.ccell.2022.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de la Prida L.M., Ascoli G.A. Explorers of the cells: Toward cross-platform knowledge integration to evaluate neuronal function. Neuron. 2021;109:3535–3537. doi: 10.1016/j.neuron.2021.10.025. [DOI] [PubMed] [Google Scholar]
- 50.Pisano F., Masmudi-Martín M., Andriani M.S., Cid E., Pisanello M., Balena A., Collard L., Tantussi F., Grande M., Sileo L., et al. Deep brain cancer metastasis detection with wide-volume Raman spectroscopy through a single tapered fiber. bioRxiv. 2022 doi: 10.1101/2022.06.24.497456. Preprint at. [DOI] [Google Scholar]
- 51.Feenstra H.E., Vermeulen I.E., Murre J.M., Schagen S.B. Online self-administered cognitive testing using the amsterdam cognition scan: establishing psychometric properties and normative data. J. Med. Internet Res. 2018;20:e192. doi: 10.2196/jmir.9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su Y., Walker J.R., Park Y., Smith T.P., Liu L.X., Hall M.P., Labanieh L., Hurst R., Wang D.C., Encell L.P., et al. Novel NanoLuc substrates enable bright two-population bioluminescence imaging in animals. Nat. Methods. 2020;17:852–860. doi: 10.1038/s41592-020-0889-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dzyubenko E., Rozenberg A., Hermann D.M., Faissner A. Colocalization of synapse marker proteins evaluated by STED-microscopy reveals patterns of neuronal synapse distribution in vitro. J. Neurosci. Methods. 2016;273:149–159. doi: 10.1016/j.jneumeth.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 54.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graña O., Rubio-Camarillo M., Fdez-Riverola F., Pisano D.G., Glez-Peña D. Nextpresso: next generation sequencing expression analysis pipeline. Curr. Bioinform. 2018;13:583–591. [Google Scholar]
- 58.Frankish A., Diekhans M., Jungreis I., Lagarde J., Loveland J.E., Mudge J.M., Sisu C., Wright J.C., Armstrong J., Barnes I., et al. GENCODE 2021. Nucleic Acids Res. 2021;49:D916–D923. doi: 10.1093/nar/gkaa1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.García-Jimeno L., Fustero-Torre C., Jiménez-Santos M.J., Gómez-López G., Di Domenico T., Al-Shahrour F. bollito: a flexible pipeline for comprehensive single-cell RNA-seq analyses. Bioinformatics. 2022;38:1155–1156. doi: 10.1093/bioinformatics/btab758. [DOI] [PubMed] [Google Scholar]
- 60.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sondka Z., Bamford S., Cole C.G., Ward S.A., Dunham I., Forbes S.A. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer. 2018;18:696–705. doi: 10.1038/s41568-018-0060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 68.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anders S., Pyl P.T., Huber W. HTSeq — a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adams D.J., Doran A.G., Lilue J., Keane T.M. The Mouse Genomes Project: a repository of inbred laboratory mouse strain genomes. Mamm. Genome. 2015;26:403–412. doi: 10.1007/s00335-015-9579-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw sequencing reads from whole exome sequencing data of each of the models described in the paper have been deposited at SRA (BioProject ID: PRJNA975092) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Likewise, bulk RNA-Seq samples from mice models have also been deposited at GEO (GSE231646) and also listed in the key resources table.
-
•
Both of the pipelines used to preprocess whole exome data and RNA-Seq data are available at GitHub: https://github.com/cnio_bu/varca and https://github.com/cnio_bu/cluster_rnaseq Additional DOIs for code used in this study are listed in the key resources table.
-
•
Raw sequencing reads from single-cell RNA-seq data of each of the models described in the paper have been deposited at GEO (GSE233366) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
Original code for scRNA-seq analysis has been deposited at Zenodo. DOI https://doi.org/10.5281/zenodo.7963303.
-
•
Raw sequencing reads from bulk RNA-sequencing data of each cell line described in the paper have been deposited at GEO (GSE218071) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
Pipeline used to preprocess RNA-Seq data is available at GitHub:https://github.com/osvaldogc/nextpresso1.9.2This section does not report original code.
-
•
Data and code from classification algorithms can be found in: https://github.com/PridaLab/metast-tree
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.