Abstract
Pancreatic cancer is a highly lethal disease with obesity as one of the risk factors. Oncogenic KRAS mutations are prevalent in pancreatic cancer and can rewire lipid metabolism by altering fatty acid (FA) uptake, FA oxidation (FAO), and lipogenesis. Identification of the underlying mechanisms could lead to improved therapeutic strategies for treating KRAS mutant pancreatic cancer. Here, we observed that KRASG12D upregulated the expression of SLC25A1, a citrate transporter that is a key metabolic switch to mediate FAO, fatty acid synthesis (FAS), glycolysis, and gluconeogenesis. In genetically engineered mouse models and human pancreatic cancer cells, KRASG12D induced SLC25A1 upregulation via GLI1, which directly stimulated SLC25A1 transcription by binding its promoter. The enhanced expression of SLC25A1 increased levels of cytosolic citrate, FAs, and key enzymes in lipid metabolism. In addition, a high-fat diet (HFD) further stimulated the KRASG12D-GLI1-SLC25A1 axis and the associated increase in citrate and FAs. Pharmacological inhibition of SLC25A1 and upstream GLI1 significantly suppressed pancreatic tumorigenesis in KrasG12D/+ mice on a HFD. These results reveal a KRASG12D-GLI1-SLC25A1 regulatory axis with SLC25A1 as an important node that regulates lipid metabolism during pancreatic tumorigenesis, thus indicating an intervention strategy for oncogenic KRAS-driven pancreatic cancer.
Keywords: Mutant KRAS, Pancreatic cancer, SLC25A1, GLI1, Lipid metabolism
Introduction
Pancreatic cancer has a dismal five-year survival rate of 11% with obesity as a known risk factor (1-5). In pancreatic ductal adenocarcinoma (PDAC), 86% of cases have KRAS mutations, which predominantly occur at codon G12 (91%), including about 45% as G12D oncogenic mutation (6). The acquisition of KRAS mutations is an early change in the pancreatic tumorigenesis (6). Corroborating with epidemiologic studies, HFD accelerates tumorigenesis in mouse models of PDAC (7-9) and causes severe fat accumulation in murine pancreas, possibly due to elevated fatty acid synthesis (FAS), although the molecular mechanisms exploited by pancreatic cells to store and utilize fat to promote tumorigenesis are not well defined (4,10-12).
Lipid metabolism is often drastically altered in cells undergoing transformation to the malignancy (13,14). Tumor cells increase de novo lipogenesis, FA uptake, FAO, and lipid accumulation. The canonical rationale for cancer cells to elevate lipid metabolism is the increased lipid demand for plasma membrane synthesis and energy production (15). However, most studies did not address the unique properties of rewired lipid metabolism in the context of obesity, a known pancreatic cancer risk factor with a direct link to altered lipid metabolism (16-20).
SLC25A1 is a key metabolic switcher to mediate pathways related to FAO, FAS, glycolysis, and gluconeogenesis (21). In the pancreas with wild-type KRAS, limited SLC25A1 is incapable of unrestrictedly boosting cytoplasmic and mitochondrial pools of citrate (22). However, it is not known whether pancreatic SLC25A1 and KRASG12D conspire to reprogram metabolism for tumorigenesis, especially under an HFD challenge, and whether targeted inhibition of their interplay provides novel preventive and therapeutic strategies for PDAC.
In this work, we observed an enhanced level of SLC25A1 and altered lipid metabolism in pancreatic acinar cell-specific KRASG12D-expressing mouse models and pancreatic cancer cells. Through increasing cytosolic citrate, SLC25A1 altered lipid metabolism as indicated by the significantly increased levels of ACSL1 (acyl-CoA synthetase long-chain family member 1), CPT1A & 2 (carnitine palmitoyl transferase 1A and 2), and fatty acid synthase (FASN). KRASG12D upregulated SLC25A1 expression via GLI1 to increase cytosolic citrate. HFD promoted the KRASG12D-GLI1-SLC25A1 regulatory axis. Pharmacological inhibition of SLC25A1 and its upstream GLI1 significantly suppressed HFD-accelerated pancreatic tumorigenesis in KrasG12D/+ mice. Thus, KRASG12D-GLI1-SLC25A1 is a novel regulatory axis with SLC25A1 as a critical node to regulate lipid metabolism for accelerated pancreatic tumorigenesis. This study identifies a novel intervention target for oncogenic KRAS-driven PDAC.
Materials and Methods
Mouse strains
CreERT-LoxP recombination was used to control gene expression in pancreatic acinar cells. LSL-KrasG12D/+ mice, which possess conditional knock-in of the endogenous level of mutant KrasG12D/+, were obtained as described (8). fElasCreERT mice, which express tamoxifen (TM)-regulated Cre recombinase under a full-length elastase promoter specifically in pancreatic acinar cells, were described previously (23). LSL-KrasG12D/+ mice and fElasCreERT mice were crossed to generate fElasCreERT/ LSL-KrasG12D/+ double transgenic mice (called KrasG12D/+) (24). All animal experiments were reviewed and approved by the institutional IACUC of Stony Brook University and the University of Texas at El Paso.
Animal treatment
fElasCreERT and KrasG12D/+ mice, including both male and female, were randomly recruited and given TM by intraperitoneal (i.p.) injection for five consecutive days to fully activate Cre recombinase in pancreatic acinar cells at 60 days of age. KrasG12D/+ and fElasCreERT mice were fed with either a normal laboratory-supplied diet or an HFD (60% fat, Test Diet DIO 58Y1 van Heek Series; Lab Supply). After being HFD-fed for six weeks, the mice were given either GANT61 or CTPI-2 at 50 mg/kg (i.p.) twice per week for another six weeks. The same volume of vehicle (90% corn oil and 10% DMSO) was used as a control. The body weight of each mouse was measured at least once a week. At 150 days of age or 192 days of age, mice were euthanized, and the pancreas, liver, and/or fat tissues were harvested for further experiments.
Cells, reagents, and antibodies
BxPC3, PANC1, MIA PaCa-2, AsPC1, SW1990, Su.86.86, and HPDE cells were purchased from American Type Culture Collection (ATCC). GANT61 and CTPI-2 were purchased from MedChemExpress LLC (MCE). The primary antibodies for Western blot and IHC were purchased as follows: SLC25A1 (NBP2-93363, Novus), ACSL1 (NBP1-60016, Novus), GLI1 (NB600-600, Novus; sc515751, Santa Cruz Biotechnology), CPT1A (NBP1-59608, Novus), CPT2 (NBP2-67699, Novus), FASN (3180s, Cell Signaling Technology), Vimentin (5741s, Cell Signaling Technology), α-SMA (19245s, Cell Signaling Technology), phospho-SAPK/JNK (Thr183/Tyr185) (4668s, Cell Signaling Technology), phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (4370s, Cell Signaling Technology), GAPDH (sc-32233, Santa Cruz Biotechnology), α-tubulin (sc-5286, Santa Cruz Biotechnology), β-Actin (sc-47778, Santa Cruz Biotechnology; NB600-501, Novus), CK19 (Ab52625, Abcam), and amylase (sc-46657, Santa Cruz Biotechnology).
Cell culture
BxPC3, PANC1, MIA Paca-2, AsPC1, SW1990, and Su.86.86 cells were cultured in RPMI 1640 or DMEM medium supplemented with 10% FBS, as well as penicillin and streptomycin. HPDE cells were cultured in a serum-free keratinocyte (KSF) medium supplemented by epidermal growth factor (EGF) and bovine pituitary extract (Life Technologies, Inc., Grand Island, NY).
Isolation of cytoplasm and mitochondria
Isolation of cytoplasm and mitochondria was performed using a commercial mitochondria isolation kit (89874, Thermo Scientific) according to the manufacturer’s instructions. Cytoplasm and mitochondria were isolated under sterile conditions at 4°C and the isolated samples were stored at −80°C for citrate assay.
Histology analysis
Mouse tissues were dissected, rinsed with PBS, and fixed in 10% formalin overnight. Formalin-fixed tissues were placed in cassettes, dehydrated in alcohol gradients, and then embedded in paraffin for sectioning. 5-μm-thick tissue sections were dewaxed in xylene, rehydrated through reversed ethanol gradients, washed in PBS thoroughly, and stained with H&E.
Pancreatic fibrosis and Alcian blue staining
To evaluate pancreatic fibrosis, we analyzed the levels of pancreatic collagens using Sirius red staining on pancreatic tissue sections. Briefly, tissue sections were deparaffinized, hydrated in ethanol gradients and distilled water, and then incubated in adequate Picro-Sirius Red Solution (Abcam, MA, USA) for 60 min. After rinsing quickly in two changes of acetic acid solution followed by dehydration with two changes of absolute alcohol, the Sirius Red-stained sections were sealed and photographed under light microscopes. For Alcian blue staining, rehydrated paraffin sections were stained with 1% Alcian blue 8GX in 3% acetic acid (pH 2.5) for 30 min and counterstained with Nuclear Fast Red (ab150662, Abcam). Three digitized pictures of each pancreatic section were photographed using an Olympus iX81 microscope at 200x magnification. The images were analyzed using Fiji Image J software. Levels of pancreatic collagen deposition were presented as the ratio of the mean collagen-stained area to the whole mean area of the section and finally converted to relative collagen contents.
Immunohistochemistry and immunofluorescence
To evaluate the levels of representative protein makers, including α-SMA, SLC25A1, ACSL1, FASN, GLI1, Vimentin, amylase, and CK19, immunohistochemical and immunofluorescent analyses were performed as described (24). Briefly, after deparaffinization in xylene and rehydration in alcohol gradients and rinse with PBS, tissue sections were subjected to heat-mediated antigen retrieval in citrate buffer (pH 6.0), and then treated with 0.5% H2O2 in methanol for 10 min to remove endogenous peroxidase, followed by blocking with normal serum. The treated sections were then incubated with primary antibodies against α-SMA, SLC25A1, ACSL1, FASN, GLI1, vimentin, amylase, and CK19 at 4°C overnight. After washing, the pancreatic tissue sections were further incubated with the corresponding biotinylated secondary antibodies (Vector Laboratories, CA, USA) at room temperature for 1 hour, washed again in PBS, incubated with ABC reagent (Vector Laboratories, CA, USA) for 30 min, and then reacted with diaminobenzidine (DAB, Vector Laboratories, CA, USA) for about 3 min. The resulting tissue sections were then counterstained with hematoxylin until the desired staining intensity developed. Immunofluorescence was performed as described (24). Fiji Image J software was used to obtain data from most IHC images for quantification and statistical analyses, including quantification of the intensity of nuclear IHC staining and subsequent statistical analysis by Prism GraphPad 8, according to our previous publication (24).
Promoter analysis and Chromatin Immunoprecipitation (ChIP)
SLC25A1 promoter (-3000bp from the transcription start site) was found on the NCBI website and manually aligned with the consensus sequence for the GLI1 binding (25,26). PANC1 cells (2x106) were seeded in T75 flasks overnight, treated with or without GANT61 (20 μM) for 4 hours, trypsinized, washed with PBS, and fixed with 1.1% formaldehyde for 10 min at room temperature. Glycine (0.125 M) was added to stop the reaction. The cells were washed with PBS, lysed with a ChIP kit (Ab500, Abcam) according to the manufacturer's instruction, and sonicated (30 sec with 30-sec cooling repeatedly for 15 cycles) to obtain fragmented DNA from 100 bp to 1 kb, which was verified by agarose gel electrophoresis. Lysates were subjected to immunoprecipitation overnight at 4°C with 10 μg of antibodies against GLI1 (Novus), or Histone H3 antibodies as a positive control provided by the Abcam ChIP kit. The complex was subsequently incubated with Dynabeads protein G (Invitrogen) for 2 hours at 4°C on a rotating wheel, washed for 10 min at 4°C with low salt buffer (2x TE, 150mM NaCl, 1% Triton X-100, 0.1% SDS), high salt buffer (2x TE, 500 mM NaCl, 1% Triton X-100, 0.1% SDS), and LiCl buffer (1x TE, 1% NP-40, 0.25M LiCl, 1% deoxycholate) sequentially, washed for 5 min at 4°C twice with TE Buffer (100 mM Tris- HCl, 10 mM EDTA), eluted by heating for 10 min at 70°C with elution buffer, and incubated at 65°C overnight. Purification of DNA was accomplished using a QIAquick PCR purification kit (Qiagen) according to the manufacturer's instructions. qRT-PCR was performed to assess the enrichment of specific proteins along the human SLC25A1 promoter using specific primer pairs: SLC25A1-ChIP-Primer-1-F: TTCGCCAGCCTCCAATGTA, Primer-1-R: TGTGCTGCTGGAGTGACAT. SLC25A1-ChIP-Primer-2-F: TAGCCGACAACAGAGACTG, and Primer-2-R CGGTACCTCCAAACAAGACG were used as a negative control. All qPCR assays were conducted in triplicate. IgG was simultaneously used in the ChIP assay. The results were generated by the enrichment of GLI1 antibody groups normalized with each IgG group.
Protein isolation, nuclear/cytosolic fractionation, and Western blot
To evaluate protein levels of SLC25A1, ACSL1, FASN, GLI1, GAPDH, α-tubulin, and β-Actin, cell and tissue lysates were separated by SDS-PAGE and analyzed by immunodetection as described (24). Briefly, snap-frozen tissues were homogenized in 0.5-1 ml ice-cold lysis buffer (Millipore, MA, USA) containing a protease inhibitor cocktail tablet (Roche, Germany). Tissue homogenates were centrifuged at 15,000 g for 15 minutes at 4°C, and the supernatant was collected. Protein lysates from the tissues were aliquoted to determine protein concentration using a protein assay dye reagent concentrate (Bio-Rad, CA, USA). Nucleus/cytosol fractionation was isolated with NE-PER Nuclear and Cytoplasmic Extraction Kit (78833, Thermo Scientific). Samples were then used for Western blot analysis. The lysates were separated by SDS-PAGE and then transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were rinsed with PBS containing 0.05% Tween-20 (PBST) and probed with antibodies against SLC25A1, ACSL1, FASN, GLI1, GAPDH, α- tubulin, or β-Actin. The membranes were then washed with PBST and probed with respective secondary antibodies conjugated to horseradish peroxidase for one hour at room temperature. Azure Imaging System (LiCor Biosciences, Lincoln, NE) was used to visualize protein bands. Image J software was used to quantify the densitometry of individual bands. The uncropped version of all the Western blots can be found in Supplementary Data.
Gene expression levels of GLI1 and SLC25A1 by qRT-PCR
Total RNA isolation, first-strand cDNA synthesis, and quantitative PCR were performed as described (24). Briefly, tissues or cells were homogenized in 1 mL TRIzol Reagent (Ambion, Life Technologies, Camarillo, CA), and total RNAs were further separated from other cell contents by sequential chloroform and isopropanol precipitation. Aliquots of RNA samples were quantified and examined before reverse transcription by Nanodrop (Thermo Fisher Scientific, Waltham, MA). Gene expression levels of statistically significant alterations were then individually confirmed by qRT-PCR with the primer sets listed below. The gene expression was normalized to the two reference genes and the relative gene expression fold changes were calculated using the ΔΔCT method. Dissociation curves were analyzed and showed for single amplicon products to confirm the specificity of each primer pair, and RT samples were run on an agarose gel to verify the absence of genomic DNA contamination. Data quantification and statistical analysis were performed in Microsoft (Redmond, WA) Excel and GraphPad Prism 8 (San Diego, CA). The primer sequences were as follows: human GLI1 F: CATCTCTCCATTGGCACCAT, R: CTCAGACTTCAGCTGGCAAG; mouse Gli1 F: CGTTTGAAGGCTGTCGGAAG, R: CTTCTCATTGGAGTGGGTCC; human SLC25A1 F: CCTTAGCTCCCTGCTCTACG, R: GAGGTCTGGTCGTGGATGAA; mouse Slc25a1 F: CAGAAGCAGTGGTAGTCGTG, R: TTGAGCCCTGCTTCAGTACA; mouse β-Actin F: CGGTTCCGATGCCCTGAGGCTCTT, R: CGTCACACTTCATGATGGAATTGA; human GAPDH TCAAGGCTGAGAACGGGAAG R: CGCCCCACTTGATTTTGGAG.
Analysis of citrate levels on human pancreatic cells and mouse pancreatic tissues
The cytoplasmic and mitochondrial extracts of HPDE and PANC1 cells were prepared. The pancreatic tissues were obtained from fElasCreERT and KrasG12D/+ mice, respectively. Protein levels of the extracts were determined with BCA assay (Sigma) and citrate levels were measured with a citrate assay kit (MAK057, Millipore/Sigma) according to the manufacturer’s instruction. The citrate level was calculated as nmol citrate per mg protein.
Metabolic assays
Kits from BioAssay Systems were used for measuring total malate (EMAL-100), free fatty acids (EFFA-100), and adipolysis (EAPL-200). In brief, malate assay is based on malate dehydrogenase-catalyzed oxidation of malate in which the formed NADH reduces a formazan (MTT) reagent. The intensity of the product color, measured at 570 nm, is proportional to the malate concentration. The fatty acid assay uses free fatty acids enzymatically converted to acyl CoA and subsequently to H2O2. The resulting H2O2 reacts with a specific dye to form a pink-colored product. The optical density at 570 nm is directly proportional to the free fatty acid concentration in the sample. Adipolysis assay uses a single working reagent that combines glycerol kinase, glycerol phosphate oxidase, and color reactions in one step. The color intensity of the reaction product at 570 nm is directly proportional to glycerol concentration. All these assays follow the manufacturer’s instructions.
Generation of GLI1 knockout in AsPC1 cells using CRISPR-Cas9
Guide RNA sequences for CRISPR-Cas9 were designed at the CRISPR design website (http://crispr.mit.edu/). Insert oligonucleotides for human GLI1 gRNA 5’-GAGTTCGTGTGCCACTGGGGGGG-3’. The GLI1 guide RNA targets exon 8 of the GLI1 gene. The complementary oligonucleotides for guide RNAs (gRNAs) were annealed and cloned into pSpCas9 BB-2A-Puro (PX459) v2.0 vector (GeneScript). The CRISPR-Cas9 plasmid was transfected to AsPC1 cells, and the medium was changed after 12 hours. The transfectants were selected with puromycin (2 μg/ml) for two to four weeks. GLI1 knockout cells were cultured in complete media and then diluted to a final concentration of 50 cells/ml. Cells were resuspended in fresh media, and the cell suspension was transferred to a 100 mm dish. Cells were cultured for 7 days until every single cell grew into a colony. Each colony was removed by trypsin and transferred to a 24-well plate. The cells were allowed to proliferate until there were enough cells for both functional and molecular assays. GLI1 knockout was evaluated by Western blot.
Transwell assay
Pancreatic cancer cell migration was evaluated by a Transwell assay. Cells (1x105) resuspended in 100 μl RPMI 1640 medium with or without GANT61 (30 μM) were placed in the upper chamber of a 24-well chemotaxis plate with 5.5 μm pore diameter (EMD Millipore, Billerica, MA, USA). 500 μl of RPMI-1640 medium containing 10% FBS was added to the lower chamber of the Transwell culture plate. After incubating for 24 h at 37°C under 5% CO2, the number of cells in the lower chamber was counted.
Oil red O staining
Frozen tissue sections were cut at 7 μm and air dried for 15 min at room temperature, followed by fixation in 10% of formalin for 5 min and washing with running tap water. After washing, tissue sections were rinsed with 60% isopropanol and subsequently stained with freshly prepared Oil Red O working solution (Ab146295, Abcam) for 15 min. The resulting tissue sections were counterstained with hematoxylin until the desired staining intensity developed. The images were acquired under the microscope (DM6000B, Leica) and analyzed using Fiji Image J software.
[U- 13C6]-Glucose tracing for fatty acid synthesis
PANC1 cells were passaged at confluency to nine 10-cm culture plates and grown in complete RPMI 1640 medium per plate to half confluency. Then, 13.3 μl DMSO (0.13% final) was added to the medium of each of the three PANC1 plates, while GANT61 (30 μM) was added to the medium of another three PANC1 plates. Six hours later, the medium was removed completely and changed to 10 ml RPMI 1640 medium without glucose (11879020, Thermo Fisher Scientific) but supplemented with 5% dialyzed FBS (100-108, Gemini Bio Products) containing either 0.13% DMSO or 30 μM GANT61, with 12.5 mM stable isotope D-Glucose-13C6 (CLM-1396-1, Cambridge Isotope). After 24 h of incubation, cell plates were washed with 10 ml ice-cold PBS twice. Then, 5 ml cold PBS containing 1% fatty acid-free bovine serum albumin (FAF-BSA; 126575, Sigma) was added, and cells were scraped by a wide-edge scraper, transferred to 15-ml Fulton tube, and centrifuged at 300 xg for 3 min, at 4°C. After removing the supernatant, 5 ml cold PBS/1% FAF-BSA was added, and cells were washed by centrifugation at 300xg for 3 min, at 4°C. Then, 1.5 ml PBS was added, and cells were transferred to solvent-resistant microtubes (Sorenson BioScience, 12030) and centrifuged again at 300xg for 3 min, at 4°C. Lipids were extracted by organic solvents and then subjected to GC-MS analysis as described (27,28), with modifications as detailed below.
Lipid extraction and analysis of 13C-labeled fatty acids by GC-MS
Otherwise indicated, all reagents used here were MS grade from Thermo Fisher Scientific. All organic solvent extractions and reactions were performed in 22-ml amber glass vials (screw top with solid green Thermoset cap with PTFE liner, cat no. 27004, Sigma-Aldrich). Following [U-13C]-glucose metabolic labeling of fatty acid, mock (DMSO)- and PANC61-treated cell pellet samples were subjected to Bligh and Dyer extraction (29) and analyzed as described (27), with modifications. Briefly, each sample was extracted twice with 10 ml methanol:chloroform:water (38:31:31, v/v/v). The final combined hydrophobic lower phase was washed with 0.15 ml water to remove any contaminating aqueous upper phase, dried under N2 stream, and redissolved in 1.5 ml methanol:toluene (4:1, v/v) plus 0.45 mM butylated hydroxytoluene (BHT), used as an antioxidant. Methyl esterification of free fatty acids was attained by adding 10 μl 2.8 M acetyl chloride (14 mM final). The reaction mixture was heated at 100°C for 1 h. After cooling, 0.56 M aqueous sodium carbonate was added at 2:5 (v/v) to drive the fatty acid methyl esters (FAMEs) into the hydrophobic upper phase. The toluene-rich phase was purified by centrifugation (1,000xg, 5 min, 4°C) and analyzed by gas chromatography-mass spectrometry (GC-MS) in a TSQ 9000 Triple Quadrupole GC-MS, Thermo Fisher, equipped with autosampler Qsert Verex vials (cat. no. ARO-9973-12, Phenomenex). A FAME standard mixture (Supelco 37 Component FAME mix, cat. no. CRM47885, Sigma-Aldrich), diluted at 1:20 in dichloromethane to bring the palmitic acid (C16:0) methyl ester concentration to 30 ng/μl, was used. The FAME mix and experimental samples (1 μl each) were injected into the GC-MS, and data were collected and analyzed using Chromeleon version 7.2.10 (Thermo Fisher Scientific). GC-MS parameters: column: Trace TR-5, 30 mm x 0.25 mm x 0.25 μm (cat. no. 260E113P, Thermo Fisher Scientific); temperature gradient: 40°C for 1 min, ramp to 180°C at 12°C/min rate, 1-min hold, ramp to 260°C at 3°C/min rate, 1-min hold; scan mass: 40-800 m/z; scan time: 0.2 sec; chromatogram filter peak width: 1 sec; carrier gas and flow: helium: 1.5 mL/min, 15 min, then 1.0 ml/min flow until the end of temp gradient (42 min); injection: 1 μL splitless, cold needle injection; ion source temp: 260°C; MS transfer line temp: 260°C. The intensities of the isotopomers of 12C and 13C of methyl-palmitate (Me-C16:0) and methyl-stearate (Me-C18:0), the two major FAMEs selected in the experimental samples (control and GANT61-treated), were calculated using the integrated peak areas from the total-ion chromatogram of the parent ion of Me-C16:0 (m/z 270, M+0) and its 13C-isotopomers (M+2 – M+16, m/z 272-286), and the parent ion of Me-C18:0 (m/z 298, M+0) and its 13C-isotopomers (M+2 – M+18, m/z 300-316).
Statistical analyses
Comparisons between two groups were analyzed by Student t-test unless otherwise indicated. p values of less than 0.05 were considered statistically significant. Results are expressed as group mean ± standard error of the mean or mean ± standard deviation. We used GraphPad Prism 8.0 for statistical analysis.
Data Availability
The data analyzed in this study were obtained from GEPIA database at http://gepia.cancerpku.cn/. All datasets generated for the current study are available from the corresponding author upon request.
Results
Oncogenic KRASG12D upregulates SLC25A1 expression in the pancreas
We induced pancreatic acinar cell-specific expression of fElasCreERT or KrasG12D/+ with TM in 2-month-old male and female mice and analyzed their pancreata when they reached 5 months old. Hematoxylin and eosin (H&E) stains showed that fElasCreERT mice had normal pancreata, while KrasG12D/+ mice developed early-stage PanIN lesions (Fig. 1A). We then performed Alcian blue staining of acidic mucins, a standard method for evaluating PanIN lesions (24) and IHC staining of alpha-smooth muscle actin (α-SMA) for pancreatic stellate cell (PSC) activation. Both Alcian blue and α-SMA were strongly elevated in KrasG12D/+ mice (Fig. 1 A&B), suggesting that KrasG12D/+ mice developed PanIN lesions with active stroma. Concurrently, IHC showed that SLC25A1 levels were significantly increased in KRASG12D-induced PanIN lobules (Fig. 1 A&B), and Western blot revealed that SLC25A1 level was significantly higher in the pancreata of KrasG12D/+ mice compared to fElasCreERT mice (Fig. 1C). qRT-PCR further confirmed that pancreatic Slc25a1 expression is significantly higher in KrasG12D/+ mice compared to fElasCreERT mice (Fig. 1D). Furthermore, SLC25A1 level was increased in KRASG12D-harboring human pancreatic cancer cells, including AsPC1, PANC1, SW1990, and Su.86.86 cells, compared to wild-type KRAS-bearing BxPC3 cells or normal human pancreatic ductal epithelial (HPDE) cells (Fig. 1E), suggesting a close correlation between pancreatic SLC25A1 and KRASG12D. Of note, MIA PaCa-2 cells, which carry mutant KRASG12C, also showed an increase in SLC25A1 protein level compared to cells bearing wild-type KRAS (Fig. 1E), indicating a link between SLC25A1 and mutant KRAS. The less metastatic BxPC3 cells that possess wild-type KRAS have a slightly lower SLC25A1 level compared to HPDE cells (Fig. 1E), indicating that EGF supplemented in the HPDE culture medium could stimulate SLC25A1 expression, as EGF activates GLI1 (30). At the gene expression level, GEPIA analysis of pancreatic adenocarcinoma tumors (PAAD) cancer dataset (http://gepia.cancer-pku.cn) revealed a significant increase in SLC25A1 mRNA level in PAAD (n = 179) compared to matched normal (n = 171) tissues (Supplementary Fig. S1A). Although so far, no available PAAD database differentiates the KRAS status, 91% of KRAS mutations in pancreatic adenocarcinoma occur at G12 (6). All these results imply that SLC25A1 expression is relevant to mutant KRAS. Here, we specifically investigate oncogenic KRASG12D-driven tumorigenesis in the pancreas.
Figure 1. Oncogenic KRAS upregulates SLC25A1 expression in the pancreas.
(A) H&E staining, Alcian blue staining, and IHC staining of α-SMA and SLC25A1 on pancreatic tissue sections of fElasCreERT and KrasG12D/+ mice (n=5) (200x). (B) Quantitation of Alcian blue, α-SMA, and SLC25A1 levels in 1A. (C) Western blot analysis of SLC25A1 in the pancreata of fElasCreERT and KrasG12D/+ mice (n=3). (D) qRT-PCR analysis of Slc25a1 gene expression in the pancreata of fElasCreERT and KrasG12D/+ mice (n=5). (E) Western blot analysis of SLC25A1 in HPDE cells and pancreatic cancer BxPC3, MIA PaCa-2, AsPC1, PANC1, SW1990, and Su.86.86 cells. (F) Citrate levels of pancreatic tissues from fElasCreERT and KrasG12D/+ mice. (G) Cytoplasmic to mitochondrial citrate ratio of pancreatic tissue from fElasCreERT and KrasG12D/+ mice. (H) IHC staining of FASN, ACSL1, CPT1, and CPT2 on pancreatic sections of fElasCreERT and KrasG12D/+ mice (n=5) (200x). (I) Quantitation of FASN, ACSL1, CPT1, and CPT2 levels in 1H. Bars were expressed as means ± SD and Results were statistically evaluated with t-test. **, p<0.01; ***, p<0.001; ****, p<0.0001.
SLC25A1 is the key citrate transporter that promotes the export of citrate from mitochondria to the cytosol to provide substrate for fatty acid synthesis. As expected, both citrate and fatty acid levels were increased in the KrasG12D/+ murine pancreata (Fig. 1F and Supplementary Fig. S1B). Of note, we extracted whole pancreatic lysates, including acinar, stroma, islets, and ducts, therefore, the citrate level of the tissue samples represents the whole level of citrate in the pancreata. Since KRASG12D was induced only in acinar cells, the difference in citrate levels between KrasG12D/+ and fElasCreERT mice should arise from KRASG12D in the pancreatic acinar cells. Meanwhile, in the fatty acid assay (Supplementary Fig. S1B), adipolysis assay (Supplementary Fig. S1C), and malate assay (Supplementary Fig. S1D), the increased FAs, no change of adipolysis, and decreased malate apply to whole cells too. However, the total citrate level could be increased in either mitochondria or cytosol. To evaluate the organelle distribution of citrate in PDAC cells, we separated mitochondria and cytosolic fractions of PANC1 cells that carry oncogenic KRASG12D. A significant increase in the cytosolic to mitochondrial citrate ratio was observed in PANC1 cells compared to that in HPDE control cells (Fig. 1G). There are generally two ways to elevate fatty acid levels: (1) adipolysis, the process of converting fat into free fatty acids; (2) de novo FAS, the generation of fatty acids from acetyl-CoA and NADPH, when citrate is the critical supplier. Our results did not show a significant increase in adipolysis in the pancreata between KRASG12D/+ and fElasCreERT mice (supplementary Fig. S1C). Therefore, increased fatty acid level due to adipolysis was excluded. However, there were increased levels of FASN (Fig. 1H and 1I), indicating a close link to FAS (31). All these are consistent with rewired lipid metabolism by oncogenic KRAS to elevate the cytosolic citrate pool to facilitate FAS. Next, we noticed significantly increased levels of ACSL1 and CPT1A & 2, indicating elevated lipid metabolism in PanIN lobules (Fig. 1H and 1I). Altogether, oncogenic KRAS induces SLC25A1 to regulate lipid metabolism.
KRASG12D increases SLC25A1 expression by activating GLI1
To investigate how oncogenic KRAS regulates SLC25A1, we next analyzed the promoter of SLC25A1 gene and found one putative GLI1 binding site (Fig. 2A), based on the published GLI1 binding sequence (26,32). For promoter analysis, GLI1 promoter sequence (−3000bp from its transcriptional start site) was retrieved from the NCBI website, and manual alignment was performed to find the putative GLI1 binding site. In consistence, the PAAD dataset demonstrated a strong correlation (R>0.6) of GLI1 and SLC25A1 gene expression in PAAD (n = 179) (Fig. 2B). Moreover, GEPIA analysis of the PAAD dataset manifested a significant increase in relative GLI1 mRNA level in PAAD (n = 179) compared to matched normal (n = 171) tissues (Supplementary Fig. S2A). To further confirm the binding of GLI1 to SLC25A1 promoter, chromatin immunoprecipitation (ChIP) was performed by two pairs of primers (Primers-1 and Primers-2). Primers-1 encompassing the predicted GLI1 binding site was designed to detect GLI1 binding, and the binding specificity was confirmed by the absence of signal in control amplification with primers-2 (Fig. 2A). As detected by primers-1, GLI1-binding at SLC25A1 promoter was dramatically increased, suggesting that GLI1 binds to SLC25A1 promoter (Fig. 2C).
Figure 2. KRASG12D increases SLC25A1 expression by activating GLI1.
(A) Consensus GLI1 binding site in SLC25A1 promoter. The reported consensus GLI1 binding site is GACCACCCA. The putative GLI1 binding site was identified in SLC25A1 promoter. P1 primers (red) are designed to flank the putative GLI1 binding sequence, while P2 primers (green) are designed for detecting negative binding as a control. The length of the detected fragment, including the binding site, is 0.2 kb. (B) Correlative expression of GLI1 and SLC25A1 in 179 pancreatic adenocarcinoma tissues based on GEPIA analysis of PAAD dataset. The correlation coefficient was calculated by Spearman's rank for R=0.66. (C) PANC1 cells were used for a ChIP assay after GLI1 immunoprecipitation, with primers that flanked the putative GLI1 binding sequence (P1) and non-binding sites (P2). The enrichment was assayed by qRT-PCR. Also, the RT-PCR products at the end of 40 cycles were analyzed by agarose gel electrophoresis (bottom). (D) IHC staining of GLI1 on pancreatic tissue sections of fElasCreERT and KrasG12D/+ mice (n=5) (400x). (E) Quantitation of GLI1 level in 2D. (F) Western blot analysis of GLI1 in the pancreata of fElasCreERT and KrasG12D/+ mice (n=3). (G) qRT-PCR of GLI1 in the pancreata of fElasCreERT and KrasG12D/+ mice (n=5). (H) Western blot analysis of GLI1 in HPDE cells and pancreatic cancer BxPC3, MIA PaCa-2, AsPC1, PANC1, SW1990, and Su.86.86 cells using the stripped membrane from Figure 1E. β-Actin loading control is the same as in Figure 1E. (I) Western blot analysis of GLI1 in cytosol and nucleus of HPDE, ASPC1, and PANC1 cells. (J) PANC1 cells with or without GANT61 treatment at indicated concentrations for 72 hours and then the cells were harvested to isolate mRNA. The mRNA of SLC25A1 was analyzed by qRT-PCR. (K) PANC1 cells were treated with different concentrations of GANT61 for 72 hours and the cell lysates were subjected to Western blot for SLC25A1, GLI1, FASN, and ACSL1. (L) PANC1 cells were treated with 20μM of GANT61 for 4 hours and the cells were collected for GLI1 ChIP assay with primers that flanked the putative GLI binding sequence. The results were analyzed by qPCR. PCR products were analyzed by agarose gel electrophoresis (bottom). (M) Western blot analysis of the levels of GLI1 and SLC25A1 in WT (wild type) and GLI1 knockout AsPC1 cells. (N) Representative image (left) and quantification (right) of Transwell migration assays in WT and GLI1 knockout AsPC1 cells. (O) Representative image (left) and quantification (right) of Transwell migration assays in AsPC1 cells treated with or without GANT61 (30 μM). Results were expressed as mean ± SD and statistically evaluated with a t-test. ns, not significant; *, p<0.05; **, p<0.01; ****, p<0.0001.
IHC staining of GLI1 in nuclei indicated that GLI1 was significantly increased in KrasG12D/+ mice compared to that in fElasCreERT mice (Fig. 2D&E), reflecting the activation of GLI1 by KRASG12D. Western blot and qRT-PCR assays confirmed that GLI1 had higher pancreatic expression in KrasG12D/+ mice than that in fElasCreERT mice (Fig. 2F&G). To evaluate GLI1 level in mutant KRAS-harboring PDAC cells, we stripped the Western blot membrane used for the evaluation of SLC25A1 in Figure 1E and reprobed it with GLI1 antibody. A significantly increased GLI1 protein level was observed in oncogenic KRAS-harboring human PDAC cells compared to wild-type KRAS-bearing BxPC3 cells and HPDE normal control cells (Fig. 2H, β-actin loading control is the same as in Figure 1E). We also separated the cytosol and nucleus of KRASG12D-harboring AsPC1 and PANC1 cells and found that GLI1 is substantially accumulated in the nuclear portion of these PDAC cells compared to HPDE control cells, suggesting that most GLI1 proteins were in the activated status in PDAC cells, which is consistent with our IHC data observed in KrasG12D/+ mice (Fig. 2I). Meanwhile, inhibition of ERK or JNK, downstream factors of KRAS, decreased GLI1 expression, consistent with the observation that KRASG12D regulated GLI1 expression (Supplementary Fig. S2B). Although ERK/JNK signaling is not unique to KRAS, they are common effectors indicating the KRAS activation (33).
When we treated PANC1 cells with GANT61, a widely used GLI1 inhibitor (34-36), the expression of SLC25A1 was significantly reduced in a dose-dependent manner (Fig. 2J). Western blot data also showed that GANT61 treatment decreased SLC25A1 level in PANC1 cells in a similar dose-dependent manner as it did to GLI1 (Fig. 2K). Moreover, expressions of lipid metabolic enzymes FASN and ACSL1 were reduced, consistent with the decreased SLC25A1 (Fig. 2K). The GLI1-ChIP assay performed with primers-1 showed that GANT61 treatment of PANC1 cells for 4 hours specifically reduced GLI1 binding to the SLC25A1 promoter, although GLI1 protein level was not altered (37,38) (Fig. 2L and Supplemental Fig. S2C). Additionally, we knocked out GLI1 expression using the CRIPSR-CAS9 approach in KRASG12D-harboring AsPC1 cells and Western blot data showed that the ablation of GLI1 substantially decreased SLC25A1 protein level in AsPC1 cells, confirming that SLC25A1 is a downstream target of GLI1 (Fig. 2M). We further performed Transwell assays using GLI1-knockout or GANT61-treated AsPC1 cells to test their migration and invasion ability after inhibition of GLI1. Our data showed that inhibition of GLI1 either by genetic deletion or pharmacological inhibition substantially suppressed the metastasis ability of AsPC-1 cells (Fig. 2N&O). Altogether, our data suggested that oncogenic KRAS upregulates GLI1, which subsequently upregulates SLC25A1 to rewire lipid metabolism for pancreatic tumorigenesis.
HFD challenge stimulates the KRASG12D-GLI1 axis to induce SLC25A1 expression
Cancer incidence was remarkably increased in HFD-fed KRASG12D/+ mice (7,24,39), which exhibited more advanced PanIN lobules with extensive fibrosis than the same mice fed with normal diet (ND), as shown by H&E and vimentin staining (Fig. 3A&B). There was a significant increase in nuclear active GLI1 in the pancreas of HFD-fed KrasG12D/+ mice as indicated by IHC (Fig. 3C&D). The nuclear enriched GLI1 after HFD treatment correlated well with an increased level of SLC25A1 (Fig. 3C&D), consistent with a KRAS-GLI1-SLC25A1 regulatory axis. Both Western blot and qRT-PCR data demonstrated that the levels of GLI1 and SLC25A1 were significantly increased in HFD-fed KrasG12D/+ mice compared to ND-fed KrasG12D/+ mice (Fig. 3E&F). Both IHC and Western blot data showed that pancreatic ACSL1 and FASN levels were significantly increased in HFD-fed KrasG12D/+ mice compared to those in fElasCreERT mice (Fig. 3G-I), suggesting that HFD upregulates the KRAS-GLI1-SLC25A1 signaling and lipid metabolism. Remarkably, the levels of both citrate and fatty acids were significantly increased by HFD in the pancreata of KrasG12D/+ mice compared to the same mice fed with ND (Fig. 3J&K), consistent with the upregulated level of the citrate transporter SLC25A.
Figure 3. HFD challenge stimulates the KRAS-GLI1 axis to induce SLC25A1 expression.
(A) H&E staining and IHC staining of vimentin in pancreatic tissue sections of KrasG12D/+ mice fed with ND or high HFD (n=5) (200x). (B) Quantitation of vimentin level in 3A. (C) IHC staining of GLI1 (400X) and SLC25A1 (200X) on pancreatic tissue sections of KrasG12D/+ mice fed with ND or HFD (n=5). (D) Quantitation of GLI1 and SLC25A1 levels in 3C. (E) Western blot analysis for SLC25A1 and GLI1 in the pancreata of KrasG12D/+ mice fed with ND or HFD (n=3). (F) qRT-PCR analysis of Slc25a1 gene expression in the pancreata of KrasG12D/+ mice fed with ND or HFD (n=5). (G) IHC stains of ACSL1 (200X) and FASN (200X) on pancreatic tissue sections of KrasG12D/+ mice fed with ND or HFD (n=5). (H) Quantitation of ACSL1 and FASN levels in 3G. (I) Western blot analysis of FASN and ACSL1 in the pancreata of fElasCreERT and KrasG12D/+ mice fed with ND or HFD. (J&K) Total citrate and fatty acid levels in pancreatic tissues from KrasG12D/+ mice fed with ND or HFD. Results were expressed as mean ± SD and statistically evaluated with a t-test.**, p<0.01; ***, p<0.001; ****, p<0.0001.
Previously, we reported that oncogenic KRAS rewired metabolism by downregulating FGF21 expression, leading to HFD-enhanced inflammation, PanIN lesions, and PDAC (24). Correlated to this observation, we also stained GLI1 and SLC25A1 in pancreatic tissues of HFD-fed KrasG12D/+ mice with or without FGF21 treatment and found significantly decreased levels of GLI1 and SLC25A1 after FGF21 treatment, indicating potential crosstalk between FGF21 and the GLI1-SLC25A1 axis in oncogenic KRAS-driven PDAC (Supplementary Fig. S3A&B).
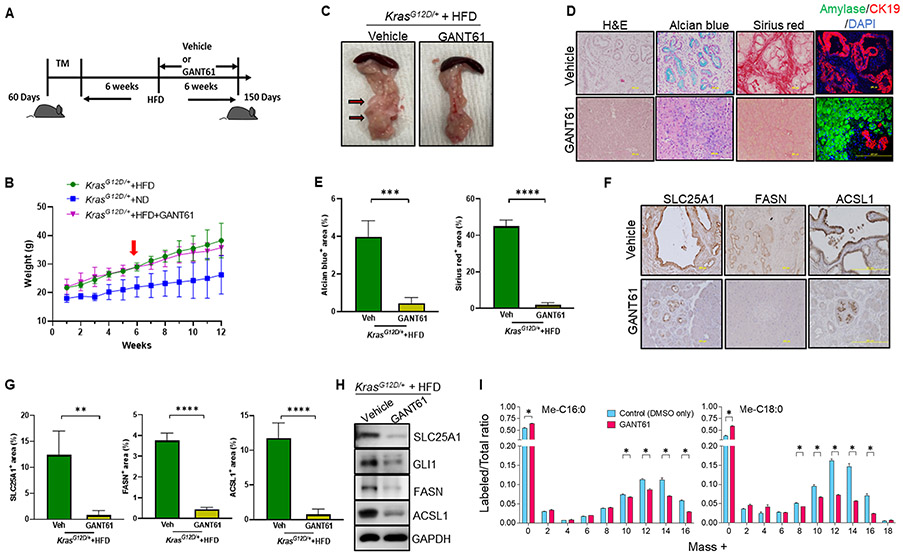
Inhibition of GLI1 alleviates pancreatic precancerous lesions by decreasing SLC25A1 in KrasG12D/+ mice
Although studies confirmed that loss of GLI1 blocks the development of pancreatic preneoplastic lesions in mice carrying pancreas-specific mutant KRAS (38,40), these studies were performed in mice fed with ND, despite that HFD is a strong stimulus in oncogenic KRAS-mediated pancreatic tumorigenesis. Therefore, we investigated whether inhibition of GLI1 was able to suppress pancreatic tumorigenesis in the presence of HFD and whether this inhibition is through the suppression of the KRAS-GLI1-SLC25A1 axis to decrease the lipid metabolism in KrasG12D/+ mice. To test the effects of GLI1 inhibition, KrasG12D/+ mice were first fed with HFD for six weeks, and then randomized to receive a GLI1 inhibitor GANT61 or the vehicle solvent for additional six weeks (Fig. 4A). Body weight curve showed that HFD-fed KrasG12D/+ mice with (n=4) or without GANT61 treatment (n=5) had similar body weights that are significantly higher than the control ND-fed KrasG12D/+ mice (n=5), suggesting that GLI1 inhibition does not have adverse effects on the body weight of the mice (Fig. 4B). However, when assessing the effects of GANT61 treatment on the pancreata, we noticed that HFD-fed KrasG12D/+ mice treated with GANT61 for six weeks had dramatically reduced pancreatic cysts (Fig. 4C). Further histological analysis revealed that GANT61 treatment alleviated formation of pancreatic lesions, and lessened collagen deposition compared to untreated HFD-fed KrasG12D/+ mice, which showed robust signs of PanIN lesions with extensive Alcian blue staining, and collagen deposition (Fig. 4D&E). In addition, compared to vehicle treatment, GANT61 treatment remarkably preserved the acinar cells, as indicated by co-immunofluorescence staining of amylase and keratin 19 (CK19), molecular markers for pancreatic acinar and ductal cells, respectively (Fig. 4D). Moreover, from IHC and Western blot analyses, we observed the significantly decreased levels of SLC25A1, FASN, and ACSL1 after GANT61 treatment, indicating that the decreased pancreatic lesions were closely related to restricted lipid metabolism regulated by the KRAS-GLI1-SLC25A1 axis (Fig. 4F-H).
Figure 4. Inhibition of GLI1 alleviates pancreatic precancerous lesions by decreasing SLC25A1 expression in KrasG12D/+ mice.
(A) Experimental scheme for B-H: 60-day-old male and female ND-fed KrasG12D/+ mice were treated with TM to induce KRASG12D expression in pancreatic acinar cells. These mice were fed HFD for six weeks and then randomly separated into two groups with one group fed HFD with vehicle (n=5) and another group of mice fed with HFD plus GANT61 treatment (i.p. twice per week at 50 mg/kg/time) (n=4) for six weeks. The mice were analyzed at the age of 150 days. (B) Body weight of the KrasG12D/+ mice with ND (n=5), HFD (n=5), or HFD plus GANT61 treatment (n=4). Treatment started at the sixth week of feeding with HFD, as indicated by a red arrow. (C) Gross images for pancreatic tissues of HFD-fed KrasG12D/+ mice with or without GANT61 treatment. The pancreatic cysts were indicated by red arrows with dark edges. (D) Representative H&E stains of the pancreata, Alcian blue staining of acidic mucins, Sirius red staining of collagens on pancreatic tissue sections, and co-immunofluorescence of pancreatic amylase and CK19 in HFD-fed KrasG12D/+ mice with or without GANT61 treatment (n=5) (200X). (E) Quantitation of Alcian blue and Sirius red staining levels in 4D. (F) IHC staining of SLC25A1 (200X), FASN (200X), and ACSL1 (200X) on pancreatic tissue sections of HFD-fed KrasG12D/+ mice with or without GANT61 treatment (n=5). (G) Quantitation of SLC25A1, FASN, and ACSL1 levels in 4F. (H) Western blot analysis of GLI1, SLC25A1, ACSL1, and FASN in the pancreata of HFD-fed KrasG12D/+ mice with or without GANT61 treatment. Results were expressed as mean ± SD and statistically evaluated with a t-test. **, p<0.01; ***, p<0.001; ****, p<0.0001. (I) De novo fatty acid synthesis from glucose is decreased in PANC1 cells following GANT61 treatment. U-13C6-glucose metabolic tracing profile for methyl-palmitate (Me-C16:0) and methyl-stearate (Me-C18:0) chain elongation in PANC1 cells after treatment with either 0.13% DMSO (Control; n=3) or GANT61 (30 μM, also dissolved in 0.13% DMSO; n=2) for 24 h. Free fatty acids were extracted by organic solvents, derivatized as fatty acid methyl esters (FAMEs), and analyzed by GC-MS. The intensity of the endogenous U-12C-Me-C16:0 (M+0) and U-12C-Me-C18:0 (M+0) and the metabolically labeled 13C-isotopomers of Me-C16:0 (M+2 – M+16) and Me-18:0 (M+2 – M+18) are indicated. The ratio of labeled over the total (unlabeled + labeled) was calculated. Results were expressed as mean ± SEM, and statistically analyzed by the multiple unpaired Student t-test with Welch correction. *, p<0.05.
To further confirm that GLI1 is critical in regulating lipid metabolism in PDAC cells, we evaluated de novo lipogenesis in PANC1 cells with or without GANT61 treatment by using uniformly 13C-labeled glucose (U-13C6-glucose) and subsequent gas-chromatography-mass spectrometry (GC-MS) analysis. First, we used a FAME mix (with 37 FAMEs) as an external reference and detected Me-C16:0 and Me-C18:0 in the control (DMSO only) and GANT61-treated PANC1 cell samples. To confirm the nature of these two FAMEs, we performed fragmentation analysis in the GC-MS triple-quadrupole collision chamber of the parent ions at m/z 270 and m/z 298, corresponding to the endogenous isotopomers of Me-C16:0 and Me-C18:0, respectively, both containing only 12C (M+0) (Supplementary Fig. S4). Comparing the fragmentation spectra of FAME standards of the same fatty acids (top panel) with those of Me-C16:0 and Me-C18:0 in the control (middle panel) and GANT61-treated PANC1 cell samples (bottom panel), we observe the same pattern of fragment ions, indicating that the two major FAMEs in the experimental samples were indeed Me-C16:0 and Me-C18:0. Next, to confirm that our 13C-tracing de novo lipogenesis was successful, we focused on the m/z regions of the fragmentation spectra where the 13C-isotopomers of Me-C16:0 and Me-C18:0 were observed (dashed open rectangles in Supplementary Fig. S4). We detected the centroid peaks of 13C-isotopomers of Me-C16:0 (M+0), as well as M+2 (m/z 272) to M+16 (m/z 286), and found a considerable reduction of the 13C-isotopomers, especially between M+8 (m/z 278) and M+16 (m/z 286) in GANT61-treated PANC1 cell samples compared to the control (DMSO only) samples (Supplementary Figs. S5A-S5B). A similar trend was observed for the 13C-isotopomers of Me-C18:0 (Supplementary Fig. S5C and S5D) between M+8 (m/z 306) and M+18 (m/z 316). For higher accuracy in the quantification of the 13C-isotopomers of Me-C16:0 and Me-C18:0 in the control (DMSO only) and GANT61-treated PANC1 cell samples, we used the integrated peak areas of the 13C-isotopomers in the original total-ion chromatograms, from where the centroid peaks (Supplementary Fig. S5) were generated. The ion intensities of the integrated areas for the isotopomers (M+0 – M+16 for Me-C16:0, and M+0 – M+18 for Me-C18:0) were converted to labeled fractions (13C-isotopomers) of a total ratio (12C-isotopomer + 13C-isotopomers) (Fig. 4I). We could observe a statistically significant inhibition of de novo synthesized 13C-isotopomers of Me-C16:0 (M+10 – M+16) and Me-C18:0 (M+8 – M+16) following GANT61 treatment in PANC1 cells (Fig. 4I). These results suggest that targeted inhibition of GLI1 is effective in the inhibition of the de novo fatty acid synthesis from glucose in PANC1 cells.
Inhibition of SLC25A1 suppresses HFD-induced acinar cell damage and pancreatic precancerous lesions in KrasG12D/+ mice
To test if SLC25A1 is a major downstream mediator of GLI1, we specifically inhibited SLC25A1 by CTPI-2, the latest SLC25A1 specific inhibitor (Fig. 5A) (41). Unlike GANT61 treatment, CTPI-2 (n=4) treatment led to a significant weight loss of HFD-fed KRasG12D/+ mice (Fig. 5B) that eventually reached a similar body weight as the mice fed with a ND (n=5), although CTPI-2 did not affect feeding and appetite (41). Visual examination of the abdomen of the mice showed that CTPI-2 reduced fat accumulation (Fig. 5C, upper panel). Consistently, Oil Red O staining revealed that CTPI-2 treatment reduced lipid droplet content in pancreatic tissues from the HFD-fed KrasG12D/+ mice (Fig. 5D&E). Remarkably, CTPI-2 treatment significantly alleviated the pancreatic lesions (Fig. 5C, lower panel), and drastically inhibited PanIN lesions and fibrosis compared to the same HFD-fed KrasG12D/+ mice treated with the vehicle, as indicated by Alcian blue, Sirius red, and amylase/CK19 stains (Fig. 5F&G). Moreover, both IHC and Western blot data showed that inhibition of SLC25A1 significantly reduced expression of the key rate-limiting lipid metabolic enzymes FASN and ACSL1 (Fig. 5H&I and Supplementary Fig. S6A). Of note, the tumor-preventive phenotypes elicited by CTPI-2 largely overlapped with those seen in the models of GANT61 treatment, despite that CTPI-2 treatment significantly induced weight loss compared to GANT61 treatment. Thus, SLC25A1 is a preventive target to inhibit pancreatic tumorigenesis in HFD-fed KrasG12D/+ mice.
Figure 5. Inhibition of SLC25A1 suppresses HFD-induced acinar cell damage and pancreatic precancerous lesions in KrasG12D/+ mice.
(A) Experimental scheme for B-G: 60-day-old ND-fed KrasG12D/+ mice were treated with TM to induce KRASG12D expression in pancreatic acinar cells. These male and female mice were fed with HFD for six weeks and then randomly separated into two groups with one group fed HFD with vehicle (n=5) and another group fed with HFD plus CTPI-2 treatment (i.p. twice per week at 50 mg/kg/time) (n=5) for six weeks. The mice were analyzed at the age of 150 days. (B) Body weight of the KrasG12D/+ mice with ND (n=5), HFD (n=5), or HFD+CTPI-2 treatment (n=5). Treatment started after the sixth week of HFD feeding, as indicated by the red arrow. (C) Gross images of HFD-fed KrasG12D/+ mice and their pancreata with or without CTPI-2 treatment. (D) Lipid droplet staining of pancreatic tissue sections isolated from the HFD-fed KrasG12D/+ mice treatment with or without CTPI-2 by using Oil red O staining and Hematoxylin as counterstain. Scale bar, 50 μm. (E) Quantitation of histological sections stained for lipid droplets and depicted in 5D. (F) Representative H&E staining of pancreatic sections with Alcian blue staining of acidic mucins, Sirius red staining of collagens, and co-immunofluorescence of pancreatic amylase and CK19 in HFD-fed KrasG12D/+ mice with or without CTPI-2 treatment (n=5) (200X). (G) Quantitation of Alcian blue and Sirius red stains in 5F. (H) IHC staining of SLC25A1 (200X), FASN (200X), and ACSL1 (200X) on pancreatic tissue sections of HFD-fed KrasG12D/+ mice with or without CTPI-2 treatment (n=5). (I) Quantitation of SLC25A1, FASN, and ACSL1 levels in 5H. Results were expressed as mean ± SD and statistically evaluated with a t-test. **, p<0.01; ***, p<0.001; ****, p<0.0001.
To evaluate the different impacts between GLI1 and SLC25A1 inhibitors on body weight, we compared the liver and pancreas. When we induced KRASG12D expression specifically in pancreatic acinar cells, Gli1 was specifically induced in the pancreas rather than in other organs such as the liver, where KRASG12D is not expressed and GLI1 is limited (Supplementary Fig. S6B). Thus, inhibition of GLI1 mainly alleviates fat accumulation in the targeted pancreas rather than in the liver that is not hit by KRASG12D (Supplementary Fig. S6C). Therefore, the GANT61 treatment did not impact overall body weight. In contrast, SLC25A1, a critical factor in lipid metabolism and energy storage, is highly expressed in white visceral abdominal fat (WAT) and liver (41) (Supplementary Fig. S6D). Thus, after CTPI-2 treatment, a significant reduction of fat accumulation in the liver (Supplementary Fig. S6E) and abdomen (Fig. 5C) was observed. Hence, weight loss was observed. This observation is consistent with previous publications that the weight loss induced by CTPI-2 is due to its pharmacological effect rather than drug toxicity (41,42). Besides, we observed robustly increased adipolysis in HFD-fed KrasG12D/+ mice after CTPI-2 treatment (Supplementary Fig. S6F), while, although significant, only slightly increased adipolysis in HFD-fed KrasG12D/+ mice after GANT61 treatment (Supplemental Fig. S6G). Thus, the different effects induced by GLI and SLC25A1 inhibitors on body weight are due to the varied expression of pharmacological targets among diverse tissues—global organs versus local pancreas.
SLC25A1 inhibitor suppresses PDAC development in KrasG12D/+ mice under a long-term HFD challenge
Our previous studies showed that about 50% of KrasG12D/+ mice developed PDAC after 10 weeks of HFD feeding (24). To test whether inhibition of SLC25A1 has therapeutic potential, TM-treated KrasG12D/+ mice were randomly separated into two groups with one group of mice fed with HFD for twelve weeks and pancreatic tissue samples were collected at 150 days of age (Fig. 6A). Another group of mice was fed with HFD for twelve weeks and then fed with HFD plus CTPI-2 treatment for additional 6 weeks (Fig. 6A). We observed an obvious decrease in abdominal fat accumulation in CTPI-2-treated mice (Fig. 6B). In addition, CTPI-2 treatment suppressed PanIN lesions and fibrosis in HFD-fed KrasG12D/+ mice as indicated by H&E, decreased Alcian blue positive stains, and decreased Sirius red positive stains compared to the pancreata of KrasG12D/+ mice with 12-week HFD treatment (Fig. 6C&D). In conclusion, inhibition of the KRASG12D-GLI1-SLC25A1 axis is a potential preventive and therapeutic strategy for HFD-mediated PDAC development (Fig. 6E).
Figure 6. SLC25A1 inhibitor suppressed PDAC development in KrasG12D/+ mice under a long-term HFD challenge.
(A) Experimental scheme for B-D: 60-day-old ND-fed KrasG12D/+ mice were treated with TM to induce KRASG12D expression in pancreatic acinar cells. These male and female mice were randomly separated into two groups with one group of mice (n=5) fed with HFD for twelve weeks and pancreatic tissue samples were collected at 150 days of age as indicated by the control group. Another group of mice was fed HFD for twelve weeks and then fed with HFD plus CTPI-2 treatment (i.p. twice per week at 50 mg/kg/time) (n=5) for additional six weeks as indicated by the CTPI-2 group. (B) Gross images of HFD-fed KrasG12D/+ mice with or without CTPI-2 treatment. (C) Representative histology shown by H&E stains, Alcian blue stains, and Sirius red stains on pancreatic tissue sections from the HFD-fed control group and CPTI-2 treatment group (200X). (D) Quantitation of Alcian blue and Sirius red staining levels in 6C. (E) Hypothetical model of the KRAS-GLI1-SLC25A1 axis that regulates lipid metabolism. Oncogenic KRASG12D upregulates SLC25A1 expression by increasing GLI1 transcription. More SLC25A1 exports more citrate from mitochondria to the cytosol for fatty acid synthesis, leading to severe lipid accumulation. Thus, SLC25A1 acts as a nodal transporter to enhance lipid metabolism. Targeted inhibition of the KRASG12D-GLI1-SLC25A1 axis by GANT61 or CTPI-2 could suppress cytosolic citrate and fatty acid accumulation under HFD, therefore hindering pancreatic tumorigenesis. Results were expressed as mean ± SD and statistically evaluated with a t-test. **, p<0.01; ****, p<0.0001.
Discussion
In this study, we first demonstrated that KRASG12D upregulates SLC25A1 via GLI1 to regulate citrate transportation. Slc25a1 expression in pancreatic acinar cells is highly sensitive to KRASG12D, showing remarkable elevation upon the expression of KrasG12D/+ at an endogenous level. Second, we showed that the KRASG12D-GLI1-SLC25A1 axis and its associated citrate and fatty acid are upregulated by HFD. Third, pharmacological inhibition of GLI1 significantly inhibits lipid accumulation and PanIN lesions under an HFD challenge. Finally, pharmacological inhibition of SLC25A1 suppresses fibrosis, PanIN lesions, and pancreatic tumorigenesis even after a long-term HFD challenge.
We observed that KRASG12D upregulated SLC25A1 expression via increasing GLI1 nuclear localization in both KrasG12D/+ mice and human pancreatic cancer cells harboring KRASG12D. Previous studies showed that GLI1 not only functions downstream of KRASG12D signaling to transcribe oncogenes but also plays critical roles in prolonging KRAS activation (37,40,43). Inhibition of GLI1 suppressed pancreatic neoplastic lesions in mice induced by oncogenic KRAS (38). In this study, we identified that GLI1 binds to the promoter of a metabolic transporter SLC25A1 by a ChIP assay. By pharmaceutically blocking GLI1 from binding to the promotor of SLC25A1 with GANT61, both mRNA and protein of SCL25A1 decreased significantly. Moreover, the clinical database indicates a strong correlation between SLC25A1 and GLI1 expressions in pancreatic cancer patients. Furthermore, genetic deletion of GLI1 in AsPC1 cells substantially reduced the SLC25A1 protein level. All these results supported the novel finding that SLC25A1 is a transcriptional target of GLI1 in response to oncogenic KRASG12D.
Besides regulating SLC25A1, GLI1 was reported to regulate I-kappa-B kinase epsilon (IKBKE) to activate NF-κB in pancreatic cancer cells, and NF-κB is a critical downstream mediator for GLI1 in PDAC cell transformation and survival (40,44). Studies showed that NF-kB can induce SLC25A1 (45,46). Given that both GLI1 and NF-kB are transcriptional factors but with different binding sites on target genes, GLI1 might synergistically cooperate with NF-kB to enhance SLC25A1 expression, as online search predicts multiple putative NF-kB binding sites on the first 3 kb promoter of human SLC25A1 (https://alggen.lsi.upc.es).
We previously reported that oncogenic KRAS rewires lipid metabolism by downregulating the expression of FGF21, which in turn creates a vulnerability to obesogenic HFD, leading to extensive inflammation, PanIN lesions, and invasive PDAC (24). In this study, we observed significantly decreased expression of GLI1 and SLC25A1 after FGF21 treatment, indicating that there is a potential crosstalk between FGF21 and the GLI1-SLC25A1 axis in regulating lipid metabolism in KRAS-driven PDAC.
Our data showed that inhibition of GLI1 significantly suppressed KRASG12D-driven pancreatic lesions under an HFD challenge. Of note, the inhibition of SLC25A1 by CTPI-2 and inhibition of GLI1 by GANT61 led to similar outcomes regarding the inhibition of pancreatic tumorigenesis. These two drugs achieved consistent and correlative effects on reducing the expression of SLC25A1, ACSL1, FASN, and the formation of pancreatic lesions promoted by HFD, indicating that restriction of fat metabolism by disrupting the KRAS–GLI1-SLC25A1 regulatory axis inhibits pancreatic tumorigenesis. Altogether, our results demonstrate that inhibition of the KRASG12D-GLI1-SLC25A1 regulatory axis can enable the pancreas to better sustain the HFD challenge for a prolonged period of time without developing significant pancreatic lesions. These are informative for the future development of better intervention strategies for pancreatic cancers.
Although both GANT61 and CTPI-2 alleviated PanIN lesions by disturbing the KRAS-GLI1-SLC25A1 axis, the pharmacological effects of these two compounds on mice still showed some differences. GANT61 treatment did not induce obvious weight loss in HFD-fed KrasG12D/+ mice, whereas CTPI-2 did (Fig. 4B). After CPTI-2 treatment, decreased expression of SLC25A1 was observed (Fig. 5F&G), which is consistent with previous studies that CTPI-2 interacts with SLC25A1 through hydrogen bonds to Lys245, Lys190, Lys50, Lys147, Arg282, and Arg285, leading to a closed conformation and altered citrate binding. CTPI-2 is specific to SLC25A1 (42) and reduces the SLC25A1 protein level (41). CTPI-2 should not directly affect the transcription of SLC25A1, but changes SLC25A1 conformation, and thus could trigger protein degradation because a native structure is altered, and citrate transport is blocked.
Despite the discovery that pancreatic tumorigenesis is significantly suppressed by the inhibition of the KRASG12D–GLI1-SLC25A1 axis to rewire lipid metabolism, there are still limitations. Understanding the metabolic requirements from oncogenic KRAS in tumorigenesis remains challenging due to the dynamic nature of energy conversion, diversity of metabolic substances, and transient response to multiple factors at multiple levels. Therefore, we selected several rate-limiting enzymes, which are closely relevant to SCL25A1 regarding the involvement in lipid metabolism, as the indicators to generally reflect the status of lipid metabolism. However, the current study does not exclude crosstalk with amino acids or glucose metabolism, although HFD was fed as the energy source in our mice model.
In HFD-fed KRASG12D/+ mice, we observed increased levels of GLI1 and SLC25A1, which can pump excessive citrate to the cytosol to stimulate tumorigenesis. With the strategy of feeding KrasG12D/+ mice after 12-week HFD to induce advanced PanIN lesions and PDAC and then giving CTPI-2 with HFD, CTPI-2 suppressed advanced PanIN lesions, indicating that SLC25A1 is a potential therapeutic target for PDAC.
Supplementary Material
Significance:
Upregulation of SLC25A1 induced by KRASG12D-GLI1 signaling rewires lipid metabolism and is exacerbated by high-fat diet to drive the development of pancreatic cancer, representing a targetable metabolic axis to suppress pancreatic tumorigenesis.
Acknowledgments
This work was supported by grants from the Department of Defense W81XWH-20-1-0625 and the NIH 1R01DK123079-01 and 1R01CA240818-01A1 to W. L. Further support came from a STARs Award from the University of Texas System to W. L, as well as a Cancer Prevention and Research Institute of Texas (CPRIT) -Texas Regional Excellence in Cancer (TREC) award (RP210153) to Marc B. Cox. We acknowledge the excellent support by the Research Histology Core Laboratory at Stony Brook University. We thank the Biomolecule Analysis and Omics, Cytometry, Screening, and Imaging, and Genomic Analysis Units at the Border Biomedical Research Center (BBRC), UTEP, supported by the grant # U54MD007592 (to Robert A. Kirken), from the National Institute on Minority Health and Health Disparities (NIMHD), a component of the National Institutes of Health (NIH), for the full access to the GC-MS instrument and other core instruments used in this study. The authors would like to express their gratitude to Dr. Vincent W. Yang (Stony Brook University School of Medicine, Stony Brook, NY) for providing guidance and technical support. The authors would also like to thank Rina D. Koyani for providing kind help with instruments and reagents.
Footnotes
Authors’ Disclosures
No disclosures were reported.
References
- 1.Kenner B, Chari ST, Kelsen D, Klimstra DS, Pandol SJ, Rosenthal M, et al. Artificial Intelligence and Early Detection of Pancreatic Cancer: 2020 Summative Review. Pancreas 2021;50:251–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA: a cancer journal for clinicians 2022;72:7–33 [DOI] [PubMed] [Google Scholar]
- 3.Rozeveld CN, Johnson KM, Zhang L, Razidlo GL. KRAS Controls Pancreatic Cancer Cell Lipid Metabolism and Invasive Potential through the Lipase HSL. Cancer research 2020;80:4932–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu M, Jung X, Hines OJ, Eibl G, Chen Y. Obesity and Pancreatic Cancer: Overview of Epidemiology and Potential Prevention by Weight Loss. Pancreas 2018;47:158–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eibl G, Cruz-Monserrate Z, Korc M, Petrov MS, Goodarzi MO, Fisher WE, et al. Diabetes Mellitus and Obesity as Risk Factors for Pancreatic Cancer. Journal of the Academy of Nutrition and Dietetics 2018;118:555–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore AR, Rosenberg SC, McCormick F, Malek S. RAS-targeted therapies: is the undruggable drugged? Nature reviews Drug discovery 2020;19:533–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Philip B, Roland CL, Daniluk J, Liu Y, Chatterjee D, Gomez SB, et al. A high-fat diet activates oncogenic Kras and COX2 to induce development of pancreatic ductal adenocarcinoma in mice. Gastroenterology 2013;145:1449–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang HH, Moro A, Takakura K, Su HY, Mo A, Nakanishi M, et al. Incidence of pancreatic cancer is dramatically increased by a high fat, high calorie diet in KrasG12D mice. PloS one 2017;12:e0184455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu L, Ji J, Ma J, Wang D, Liu M, Du JX, et al. Differential Effects of Dietary Macronutrients on the Development of Oncogenic KRAS-Mediated Pancreatic Ductal Adenocarcinoma. Cancers 2022;14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boden G Obesity and free fatty acids. Endocrinology and metabolism clinics of North America 2008;37:635–46, viii-ix [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baenke F, Peck B, Miess H, Schulze A. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Disease models & mechanisms 2013;6:1353–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das SK, Hoefler G. The role of triglyceride lipases in cancer associated cachexia. Trends in molecular medicine 2013;19:292–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bi J, Ichu TA, Zanca C, Yang H, Zhang W, Gu Y, et al. Oncogene Amplification in Growth Factor Signaling Pathways Renders Cancers Dependent on Membrane Lipid Remodeling. Cell metabolism 2019;30:525–38.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gimple RC, Kidwell RL, Kim LJY, Sun T, Gromovsky AD, Wu Q, et al. Glioma Stem Cell-Specific Superenhancer Promotes Polyunsaturated Fatty-Acid Synthesis to Support EGFR Signaling. Cancer discovery 2019;9:1248–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broadfield LA, Pane AA, Talebi A, Swinnen JV, Fendt SM. Lipid metabolism in cancer: New perspectives and emerging mechanisms. Developmental cell 2021;56:1363–93 [DOI] [PubMed] [Google Scholar]
- 16.Shaw RJ. Glucose metabolism and cancer. Current opinion in cell biology 2006;18:598–608 [DOI] [PubMed] [Google Scholar]
- 17.Sullivan LB, Luengo A, Danai LV, Bush LN, Diehl FF, Hosios AM, et al. Aspartate is an endogenous metabolic limitation for tumour growth. Nature cell biology 2018;20:782–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan MR, Mattaini KR, Dennstedt EA, Nguyen AA, Sivanand S, Reilly MF, et al. Increased Serine Synthesis Provides an Advantage for Tumors Arising in Tissues Where Serine Levels Are Limiting. Cell metabolism 2019;29:1410–21.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagarajan A, Dogra SK, Sun L, Gandotra N, Ho T, Cai G, et al. Paraoxonase 2 Facilitates Pancreatic Cancer Growth and Metastasis by Stimulating GLUT1-Mediated Glucose Transport. Molecular cell 2017;67:685–701.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nature medicine 2017;23:1362–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Icard P, Coquerel A, Wu Z, Gligorov J, Fuks D, Fournel L, et al. Understanding the Central Role of Citrate in the Metabolism of Cancer Cells and Tumors: An Update. International journal of molecular sciences 2021;22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmieri F, Monné M. Discoveries, metabolic roles and diseases of mitochondrial carriers: A review. Biochimica et biophysica acta 2016;1863:2362–78 [DOI] [PubMed] [Google Scholar]
- 23.Ji B, Song J, Tsou L, Bi Y, Gaiser S, Mortensen R, et al. Robust acinar cell transgene expression of CreErT via BAC recombineering. Genesis (New York, NY : 2000) 2008;46:390–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo Y, Yang Y, Liu M, Wang D, Wang F, Bi Y, et al. Oncogenic KRAS Reduces Expression of FGF21 in Acinar Cells to Promote Pancreatic Tumorigenesis in Mice on a High-Fat Diet. Gastroenterology 2019;157:1413–28.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang R, Ma J, Avery JT, Sambandam V, Nguyen TH, Xu B, et al. GLI1 Inhibitor SRI-38832 Attenuates Chemotherapeutic Resistance by Downregulating NBS1 Transcription in BRAF(V600E) Colorectal Cancer. Frontiers in oncology 2020;10:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinzler KW, Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Molecular and cellular biology 1990;10:634–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie H, Tang CH, Song JH, Mancuso A, Del Valle JR, Cao J, et al. IRE1α RNase-dependent lipid homeostasis promotes survival in Myc-transformed cancers. The Journal of clinical investigation 2018;128:1300–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gouw AM, Margulis K, Liu NS, Raman SJ, Mancuso A, Toal GG, et al. The MYC Oncogene Cooperates with Sterol-Regulated Element-Binding Protein to Regulate Lipogenesis Essential for Neoplastic Growth. Cell metabolism 2019;30:556–72.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol 1959;37:911–7 [DOI] [PubMed] [Google Scholar]
- 30.Zhu G, Zhou J, Song W, Wu D, Dang Q, Zhang L, et al. Role of GLI-1 in epidermal growth factor-induced invasiveness of ARCaPE prostate cancer cells. Oncology reports 2013;30:904–10 [DOI] [PubMed] [Google Scholar]
- 31.Fhu CW, Ali A. Fatty Acid Synthase: An Emerging Target in Cancer. Molecules (Basel, Switzerland) 2020;25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winklmayr M, Schmid C, Laner-Plamberger S, Kaser A, Aberger F, Eichberger T, et al. Non-consensus GLI binding sites in Hedgehog target gene regulation. BMC molecular biology 2010;11:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Little AS, Smith PD, Cook SJ. Mechanisms of acquired resistance to ERK1/2 pathway inhibitors. Oncogene 2013;32:1207–15 [DOI] [PubMed] [Google Scholar]
- 34.Gao H, Wang W, Li Q. GANT61 suppresses cell survival, invasion and epithelial-mesenchymal transition through inactivating AKT/mTOR and JAK/STAT3 pathways in anaplastic thyroid carcinoma. Cancer biology & therapy 2022;23:369–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harada K, Ohashi R, Naito K, Kanki K. Hedgehog Signal Inhibitor GANT61 Inhibits the Malignant Behavior of Undifferentiated Hepatocellular Carcinoma Cells by Targeting Non-Canonical GLI Signaling. International journal of molecular sciences 2020;21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ke B, Wang XN, Liu N, Li B, Wang XJ, Zhang RP, et al. Sonic Hedgehog/Gli1 Signaling Pathway Regulates Cell Migration and Invasion via Induction of Epithelial-to-mesenchymal Transition in Gastric Cancer. Journal of Cancer 2020;11:3932–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang R, Wu J, Ferrandon S, Glowacki KJ, Houghton JA. Targeting GLI by GANT61 involves mechanisms dependent on inhibition of both transcription and DNA licensing. Oncotarget 2016;7:80190–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mills LD, Zhang Y, Marler RJ, Herreros-Villanueva M, Zhang L, Almada LL, et al. Loss of the transcription factor GLI1 identifies a signaling network in the tumor microenvironment mediating KRAS oncogene-induced transformation. The Journal of biological chemistry 2013;288:11786–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D, Bi Y, Hu L, Luo Y, Ji J, Mao AZ, et al. Obesogenic high-fat diet heightens aerobic glycolysis through hyperactivation of oncogenic KRAS. Cell communication and signaling : CCS 2019;17:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajurkar M, De Jesus-Monge WE, Driscoll DR, Appleman VA, Huang H, Cotton JL, et al. The activity of Gli transcription factors is essential for Kras-induced pancreatic tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America 2012;109:E1038–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan M, Mosaoa R, Graham GT, Kasprzyk-Pawelec A, Gadre S, Parasido E, et al. Inhibition of the mitochondrial citrate carrier, Slc25a1, reverts steatosis, glucose intolerance, and inflammation in preclinical models of NAFLD/NASH. Cell death and differentiation 2020;27:2143–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandez HR, Gadre SM, Tan M, Graham GT, Mosaoa R, Ongkeko MS, et al. The mitochondrial citrate carrier, SLC25A1, drives stemness and therapy resistance in non-small cell lung cancer. Cell death and differentiation 2018;25:1239–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Avery JT, Zhang R, Boohaker RJ. GLI1: A Therapeutic Target for Cancer. Frontiers in oncology 2021;11:673154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agarwal NK, Kim CH, Kunkalla K, Konno H, Tjendra Y, Kwon D, et al. Active IKKβ promotes the stability of GLI1 oncogene in diffuse large B-cell lymphoma. Blood 2016;127:605–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Infantino V, Iacobazzi V, Menga A, Avantaggiati ML, Palmieri F. A key role of the mitochondrial citrate carrier (SLC25A1) in TNFα- and IFNγ-triggered inflammation. Biochimica et biophysica acta 2014;1839:1217–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santarsiero A, Convertini P, Todisco S, Pierri CL, De Grassi A, Williams NC, et al. ACLY Nuclear Translocation in Human Macrophages Drives Proinflammatory Gene Expression by NF-κB Acetylation. Cells 2021;10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed in this study were obtained from GEPIA database at http://gepia.cancerpku.cn/. All datasets generated for the current study are available from the corresponding author upon request.