Abstract
Background
DNA damage response (DDR) pathways are essential to sustain genomic stability and play a critical role in cancer development and progression. Here, we investigated the profile of DDR gene mutations in early‐stage non‐small cell lung cancer (NSCLC) and their prognostic values.
Methods
We first examined 74 DDR genes involved in seven DDR pathways and then focused on six specific genes: ATM, BRCA1, BRCA2, CHEK1, BARD1, and BRIP1. A total of 179 stage I and IIIa NSCLC patients who received curative resection in Peking Union Medical College Hospital and their corresponding samples were collected for DNA sequencing, immunohistochemistry and survival analysis.
Results
A total of 167 eligible patients were finally analyzed. Mutation frequencies were 82% and 26.3% for the selected 74 genes and six genes, respectively. Mismatch repair (MMR) and nucleotide excision repair (NER) alterations were observed more frequently in lung squamous cell carcinoma (LUSC) and smokers were more likely to develop the selected six DDR gene mutations than those who never smoked. Deleterious mutations in the six genes were independent prognostic indicators of significantly longer disease‐free survival and overall survival. No association was found between DDR gene status and PD‐L1 expression, CD8 positive lymphocyte and tumor‐associated macrophage infiltration in tumor area. However, numbers of mutations were significantly increased among patients with DDR alterations.
Conclusions
Deleterious mutations of these six genes were common in resected NSCLC and could serve as prognostic biomarkers.
Keywords: DNA damage response, mutation landscape, prognosis, resected NSCLC, tumor microenvironment
No correlation was discovered between the condition of DNA damage response (DDR) genes and the tumor microenvironment.
Harmful mutations in the ATM, BRCA1, BRCA2, CHEK1, BARD1, and BRIP1 genes, which participate in DDR, increased the number of mutations and extended disease‐free survival as well as overall survival in stage I and IIIa non‐small cell lung cancer (NSCLC) patients.

INTRODUCTION
Lung cancer is the most frequent cancer and the leading cause of cancer‐related mortality worldwide, of which non‐small cell lung cancer (NSCLC) accounts for about 85%. 1 With the development of the diagnostic techniques and widespread screening, more patients are being diagnosed with localized NSCLC. 2 Surgery resection is the major treatment for stage I–II NSCLC patients and an alternative treatment for selected stage IIIA patients who are likely to undergo complete resection. However, 5‐year survival rate after surgery is 36%–92% 3 and the 5‐year recurrence rate is 45%–76% 4 according to the pathological stage, which is disappointing. If there are reliable biomarkers that can predict the risk of postoperative recurrence in these patients, stratified management of the population can be performed.
DNA is the carrier of genetic information. Maintaining DNA integrity faithfully is crucial for sustaining life. However, every cell in our body undergoes more than 10 000 DNA damage per day. 5 In order to sense, signal, and repair these DNA lesions, cells have developed complex mechanisms collectively termed as DNA damage response (DDR), which include base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), nonhomologous end joining (NHEJ), homologous recombination (HR) and Fanconi anemia (FA) pathways. Deleterious mutations of genes involved in DDR will lead to genomic instability. Hence, there is no doubt that defects in DDR genes play an important role in tumorigenesis and development. 5 , 6 , 7 , 8 However, on the other hand, emerging studies have found that deleterious DDR gene mutations predicted improved survival in patients who treated with platinum‐based chemotherapy in advanced urothelial carcinoma, triple‐negative breast cancer as well as ovarian cancer and resected pancreatic ductal adenocarcinoma, suggesting that tumors harboring deleterious DDR gene mutations were sensitive to platinum‐based chemotherapy. 9 , 10 , 11 , 12 What is more, several studies have shown that for a variety tumors, including NSCLC, deleterious DDR mutations indicated prolonged survival in patients treated with immunotherapy. 13 , 14 , 15 Furthermore, a previous study demonstrated that among NSCLC patients who received immunoneoadjuvant therapy, those with DDR and HR mutations were more likely to develop a major pathological response (MPR). 16
In this study, we first investigated the mutation profile of DDR in resected NSCLC patients and explored the association of DDR genes status with immune infiltration properties and PD‐L1 expression, to explore why DDR mutations are more effective for immunotherapy. Second, we investigated the association of deleterious DDR mutations and disease‐free survival (DFS) as well as overall survival (OS) in NSCLC patients undergoing radical surgical resection.
METHODS
Patients and samples
Patients who received curative resection at Peking Union Medical College Hospital from January 2009 to December 2012 and were diagnosed with pathological stage I and IIIa NSCLC were retrospectively included in this study. The cohort used in this study has previously been described in a published article, 17 and was based on the seventh edition lung cancer staging system to determine the stage. No stage I patients received adjuvant chemotherapy after surgery. All stage IIIa patients received four cycles of postoperative chemotherapy, typically platinum‐based doublet chemotherapy. DFS was calculated from the date of surgery to the date of confirmed recurrence or death. OS was defined as the interval between diagnosis and death. Clinical information including age, smoking status and stage was acquired by consulting electronic medical records and survival information including DFS and OS was obtained from outpatient clinics or by telephone. The last date of follow‐up was November 15, 2018. In addition, corresponding paraffin‐embedded tumor samples were obtained from the Pathology Department for DNA sequencing and assessment of immune infiltration properties and PD‐L1 expression. DNA sequencing, immunohistochemistry and double immunofluorescence were performed in April 2019. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Peking Union Medical College Hospital (no. S‐K146) and individual consent for this retrospective analysis was waived.
Sequencing and analysis
Total DNA was extracted from 179 FFPE specimens using the QIAamp DNA FFPE Tissue Kit (Qiagen). DNA libraries were prepared with the KAPA Hyper Prep Kit (Kapa Biosystems) and captured with custom‐designed probes covering 509 cancer related genes (Genetron Health.Inc). High‐throughput sequencing was performed on the Illumina NovaSeq platform (Illumina) with pair‐end 150 bp (PE150) aiming for the average of 500×.
Raw data were trimmed and filtered using Trimmomatic (version 0.33) and then mapped to the reference genome hg19 using BWA (version 0.7.10). Single‐nucleotide variants (SNVs) and insertions and deletions (indels) were detected using Pisces (version 5.1.6.54) without normal control. The effects of variants were annotated using the Variant Effect Predictor (version 83) and Oncotator (version 1.5.1.0). A series of procedures was performed to filter germline variants using public databases (1000 Genomes Project, NHLBI‐ESP, gnomAD) and an in‐house database. All mutations that remained were manually checked using the Integrative Genomics Viewer (version 2.3.34).
Genes selected and pathogenicity assessment
Our subsequent analysis focused on genes which play a major role in the DNA damage repair process. We first analyzed 74 DDR‐related genes and grouped them into HR, BER, NER, MMR, FA, NHEJ and cell cycle checkpoint based on the study by Dai et al. 18 (Table S1). Then, we further analyzed six selected genes, which (1) were involved in the HRR pathway and defined in the PROfound trial (NCT02987543), 19 (2) were listed in the functional category of DNA repair of the Molecular Signatures Database (MSigDB). The gene set included BRCA1, BRCA2, ATM, CHEK1, BARD1 and BRIP1, which were involved in at least two DDR pathways (Table 1).
TABLE 1.
The six genes identified were involved in at least two DDR pathways.
| Gene | GO terms | ||
|---|---|---|---|
| ATM | Homologous DNA pairing and strand exchange | Cell cycle checkpoint | Nonrecombinational repair |
| BRCA1 | Homologous DNA pairing and strand exchange | Cell cycle checkpoint | Nonrecombinational repair |
| BRIP1 | Homologous DNA pairing and strand exchange | Cell cycle checkpoint | Nucleotide excision repair |
| CHEK1 | Homologous DNA pairing and strand exchange | Cell cycle checkpoint | |
| BRCA2 | Homologous DNA pairing and strand exchange | Nucleotide excision repair | |
| BARD1 | Homologous DNA pairing and strand exchange | Nonrecombinational repair |
Abbreviations: DDR, DNA damage response; GO, gene ontology.
Loss‐of‐function mutations including nonsense, splice site and frameshift alterations were identified as deleterious mutations. A missense mutation would also be identified as deleterious if it was reported as pathogenic or likely pathogenic in the ClinVar database or predicted to be deleterious by at least two out of three prediction tools (Polyphen2, SIFT and MutationAssessor). Patients would be excluded if they had only missense mutations which were predicted to be deleterious by only one prediction tool. Patients harboring deleterious mutations were defined as DDR‐mut, otherwise as DDR‐wild.
Immunohistochemistry and double‐immunofluorescence
PD‐L1 expression, tumor‐associated macrophage (TAM) (M1 and M2 macrophage) infiltration and CD8 positive lymphocyte were analyzed as reported previously. 17 Briefly, expression of PD‐L1 was evaluated by immunohistochemistry, and the infiltration of TAM and CD8 positive lymphocyte were analyzed by double‐immunofluorescence. Samples with tumor proportion score (TPS) ≥1% were considered to be positive for PD‐L1 expression (shown in Figure S1). The location of M1 and M2 macrophages was recorded in both tumor and nontumor areas (T&NT) or only in nontumor areas (NT). CD8 positive lymphocyte infiltration was recorded in the tumor area or not.
Comparison of the number of gene mutations and TMB between the DDR‐mut group and the DDR‐wild group
We first calculated the number of gene mutations in each patient and compared them between the DDR‐mut and the DDR‐wild groups. Then we further compared the tumor mutation burden (TMB) of DDR‐mut and DDR‐wild in the Pan‐Lung Cancer dataset (TCGA, Nat Genet 2016) obtained from cBioPortal for Cancer Genomic (cBioPortal, https://www.cbioportal.org/).
Statistical analysis
To determine the differences between groups, Pearson's chi‐square test and independent sample t‐test were used, respectively for categorical variables and continuous variables. The number of gene mutations between the two groups was compared using the Wilcoxon rank sum test. Survival was analyzed using the Kaplan–Meier method, and p‐values were determined using the log‐rank test. Variables with p < 0.15 in the univariate analysis were included in multivariate Cox proportional hazards models. p‐values less than 0.05 were considered statistically significant. All statistical analyses were performed with the IBM SPSS statistical software package version 25.0 (SPSS) and visualizations were performed with R software (version 4.2.0) and GraphPad Prism 9.
RESULTS
A total of 179 patients and corresponding pathological samples were involved in this study. In further analysis, 12 samples were excluded, respectively in the 74‐gene group and six‐gene group because they only had missense mutations of interest genes and were predicted to be deleterious by only one software package. Thus, it was worth noting that although the 74‐gene group and the six‐gene group both included 167 patients at the end, the patients were slightly different due to the different genes of interest.
Patient characteristics and mutation profile
In this study, we first analyzed 74 DDR‐related genes. More than half of the patients were male (109/167, 65.3%), smokers (92/167, 55.1%) and younger (<65 years, 97/167, 58.1%). Patients with lung adenocarcinoma (LUAD) accounted for 50.9% (85/167) and patients with stage I disease accounted for 50.3% (84/167) (Table 2). A total of 137 (82.0%) patients had deleterious DDR mutations and 77 (46.1%) patients had more than two gene mutations. ATM (10.2%), PRKDC (9.6%), POLE (7.8%), BRCA1 (7.2%), PARP4 (6.6%), FANCM (6.6%) and BRCA2 (6.6%) were found to be the genes with the most frequent mutations in this study (Figure 1a). The DDR‐mut and DDR‐wild groups showed no significant differences in terms of age, gender, smoking status or stage (Table 3). However, patients with lung squamous cell carcinoma (LUSC) were more likely to harbor DDR gene mutations (p = 0.056). Specifically, MMR and NER alterations were more frequently observed in LUSC (in comparison with LUAD, 15.9% vs. 4.71%, p = 0.017; 17.1% vs. 5.89%, p = 0.023, respectively) (Figure 1b). In addition, HR alterations were more likely to develop in males and smokers (males vs. females, 42.2% vs. 25.9%, p = 0.039; smokers vs. never smokers, 43.5% vs. 28.0%, p = 0.037), whereas NHEJ alterations were more likely to develop in females (females vs. males, 17.2% vs. 5.5%, p = 0.014) (Figure 1c–e).
TABLE 2.
Clinical characteristics of all patients.
| 74 DDR genes | 6 DDR genes | |
|---|---|---|
| Clinical characteristics | All (n = 167) (%) a | All (n = 167) (%) a |
| Age | ||
| <65 | 97 (58.1) | 95 (56.9) |
| ≥65 | 70 (41.9) | 72 (43.1) |
| Gender | ||
| Female | 58 (34.7) | 57 (34.1) |
| Male | 109 (65.3) | 110 (65.9) |
| Smoking status | ||
| Nonsmoker | 75 (44.9) | 76 (45.5) |
| Smoker | 92 (55.1) | 91 (54.5) |
| Pathology | ||
| LUAD | 85 (50.9) | 82 (49.1) |
| LUSC | 82 (49.1) | 85 (50.9) |
| Stage | ||
| I | 84 (50.3) | 88 (52.7) |
| IIIa | 83 (49.7) | 79 (47.3) |
| Mutation | ||
| No | 30 (18.0) | 123 (73.7) |
| Yes | 137 (82.0) | 44 (26.3) |
Abbreviations: DDR, DNA damage response; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Samples which only had missense mutations of interest genes and were predicted to be deleterious by only one software were excluded. Thus, the patients finally included in the analysis set were slightly different due to the different genes of interest.
FIGURE 1.

Mutation profile of the 74 DDR genes in stage I and IIIa non‐small cell lung cancer (NSCLC) patients. (a) Mutation profile of the 74 DNA damage response (DDR) genes. (b) Distribution of LUAD or LUSC in individual DDR pathway mutate samples. LUSC patients were prone to NER and MMR mutations. (c–e) Males and smokers were more likely to develop HR alterations. Females were more likely to develop NHEJ mutations. LUAD, lung adenocarcinoma. LUSC, lung squamous cell carcinoma. NER, nucleotide excision repair. MMR, mismatch repair. NHEJ, nonhomologous end joining. HR, homologous recombination.
TABLE 3.
Clinical characteristics between the DDR‐wild and DDR‐mut subgroups.
| 74 DDR genes | 6 DDR genes | |||||
|---|---|---|---|---|---|---|
| Clinical characteristics | DDR‐mut N = 137 (%) | DDR‐wild N = 30 (%) | p‐value | DDR‐mut N = 44 (%) | DDR‐wild N = 123 (%) | p‐value |
| Age | 0.52 | 0.29 | ||||
| <65 | 78 (56.9) | 19 (63.3) | 28 (63.6) | 67 (54.5) | ||
| ≥65 | 59 (43.1) | 11 (36.7) | 16 (36.4) | 56 (45.5) | ||
| Gender | 0.28 | 0.14 | ||||
| Female | 45 (32.8) | 13 (43.3) | 11 (25.0) | 46 (37.4) | ||
| Male | 92 (67.2) | 17 (56.7) | 33 (75.0) | 77 (62.6) | ||
| Smoking status | 0.15 | 0.03 | ||||
| Smoker | 79 (47.3) | 13 (43.3) | 30 (68.2) | 61 (49.6) | ||
| Nonsmoker | 58 (52.7) | 17 (56.7) | 14 (31.8) | 62 (50.4) | ||
| Pathology | 0.056 | 0.36 | ||||
| LUAD | 65 (47.4) | 20 (66.7) | 19 (43.2) | 63 (51.2) | ||
| LUSC | 72 (52.6) | 10 (33.3) | 25 (56.8) | 60 (48.8) | ||
| Stage | 0.10 | 0.26 | ||||
| I | 73 (53.3) | 11 (36.7) | 20 (45.5) | 68 (55.3) | ||
| IIIA | 64 (46.7) | 19 (63.3) | 24 (54.5) | 55 (44.7) | ||
| PD‐L1 | 0.56 | 0.35 | ||||
| Positive | 93 (67.9) | 22 (73.3) | 33 (75.0) | 83 (67.5) | ||
| Negative | 44 (32.1) | 8 (26.7) | 11 (25.0) | 40 (32.5) |
Abbreviations: DDR, DNA damage response; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Patient characteristics for the six selected genes are listed in Table 2. Approximately 26.3% (44/167) of patients harbored deleterious mutations of these six genes. Most of the patients in DDR‐mut group had one gene mutation, except for three patients who had two gene mutations. ATM (10.2%), BRCA1 (7.2%) and BRCA2 (6.6%) were the most frequently mutated genes, followed by CHEK1 (1.8%), BARD1 (1.2%) and BRIP1 (1.2%) (Figure 2a). Missense mutations were the most common mutation type, accounting for approximately 74.5% (35/47). Furthermore, the mutation frequencies of the tumor suppressor genes TP53, STK11 and driver genes including EGFR, ALK, MET, ROS1, ERBB2, KRAS and BRAF were comparable between the DDR‐wild and DDR‐mut group (Figure 2b). No difference in age, sex, stage or pathological type was observed between the DDR‐mut and DDR‐wild groups. (Table 3) However, it was found that patients with a history of smoking were more likely to develop the six selected DDR gene mutations than those who never smoked (p = 0.034). (Figure 2c) The mutation frequencies of ATM, BRCA1, BRCA2, CHEK1, BARD1 and BRIP1 were comparable between stage I and IIIa. In addition, except for the mutation frequency of BRCA2, which was significantly higher in LUSC than in LUAD (10.6% vs. 2.4%, p = 0.034), there was no statistical difference in the mutation frequency of the other five genes between LUSC and LUAD (Figure 2d).
FIGURE 2.

Mutation profile of the specified six genes in stage I and IIIa non‐small cell lung cancer (NSCLC) patients. (a) Mutation profile of ATM, BRCA1, BRCA2, CHEK1, BARD1 and BRIP1. (b) Comparison of driver mutation frequencies between the six‐gene wild group and the six‐gene mutant group. There was no difference in driver mutation frequency between the six‐gene wild group and the six‐gene mutant group. (c) Smokers were more likely to develop the specified six gene mutations. (d) Distribution of LUAD or LUSC in the specified six genes mutations samples. LUSC were more likely to develop BRCA2 mutations. LUAD, lung adenocarcinoma. LUSC, lung squamous cell carcinoma.
Prognostic significance of DDR gene mutations
For the selected six genes, 45 patients finally died at the end of follow‐up. Among all patients, those with deleterious mutations in DDR genes tended to have longer DFS and OS, although the differences were not statistically significant (p = 0.11 and 0.07, respectively) (Figure 3a) Variables with a p‐value less than 0.15 in the univariate Cox analysis were further included in the multivariate Cox analysis. The results showed that patients below the age of 65, with stage I cancer, and carrying the specified six genes mutations had better DFS (HR = 0.492, 95% CI: 0.303–0.799, p = 0.004; HR = 0.258, 95% CI: 0.154–0.433, p < 0.001; HR = 0.433, 95% CI: 0.240–0.783, p = 0.006, respectively) and OS (HR = 0.423, 95% CI: 0.230–0.777, p = 0.006; HR = 0.238, 95% CI: 0.125–0.455, p < 0.001; HR = 0.359, 95% CI: 0.164–0.785, p = 0.01, respectively) (Figure 3b) To examine whether mutations in DDR genes confer protection in both stage I and IIIa patients, we carried out a subgroup analysis based on clinical stage. The findings illustrated that among stage IIIa patients, the DDR‐mut subgroup displayed a tendency towards improved OS (49.4 months vs. not reached, p = 0.05) and a statistically significant enhancement in DFS (21.3 vs. 100.1 months, p = 0.004) in comparison to the DDR‐wild subgroup. However, no such correlation was identified in patients with stage I. Furthermore, in patients with stage IIIa, we discovered that mutations in DDR extended DFS and OS only in LUSC patients and not in those with LUAD (Figure 3c).
FIGURE 3.

Prognostic values of the six gene mutations in patients with stage I and stage IIIa non‐small cell lung cancer (NSCLC). (a) Among all patients, no association between DNA damage response (DDR) gene status and OS or DFS was found. (b) Multivariate Cox proportional hazard regression analysis demonstrated that age younger than 65 years, stage I and harboring the identified six gene mutations were independent prognostic parameters for significantly longer DFS and OS. (c) Among stage IIIa patients, DDR‐mut subgroup showed a trend towards improved OS and a statistically significant improvement in DFS. No similar findings were found in stage I patients. Among stage IIIa patients, LUSC patients with DDR mutations experience extended DFS and OS, whereas no such benefits are observed in LUAD patients. LUAD, lung adenocarcinoma. LUSC, lung squamous cell carcinoma. OS, overall survival. DFS, disease‐free survival.
However, for the 74 genes, there was no association between gene status and OS or DFS in all patients (p = 0.74 and 0.47, respectively) or in stage IIIa (p = 0.53 and 0.41, respectively) or LUSC patients (p = 0.99 and 0.89, respectively). However, among never‐smokers, DDR mutations were associated with longer DFS (43.7 months vs. not reached, p = 0.014), and patients with two or more DDR gene mutations had the longest DFS (p = 0.039). We then analyzed the association of different DDR pathways with survival and found that there was no association between survival and BER, FA, HR, MMR and NER alterations. However, univariate analysis showed that mutations in cell cycle checkpoint‐related genes were associated with significantly longer DFS in LUSC patients (p = 0.04). Nevertheless, the above conclusions lost significance in the multivariate Cox regression analysis.
Association of DDR gene phenotype and tumor microenvironment
No association was observed between DDR gene status and PD‐L1 expression or infiltration of CD8‐positive lymphocytes, M1 and M2 macrophages in the tumor area, regardless of whether 74 or six genes were used (Figure 4a–d). However, further analysis showed that infiltration of CD8‐positive lymphocytes in the tumor area was decreased in samples with BER alterations (p = 0.003), whereas it was increased in samples with HR alterations (p = 0.018) (Figure 4e,f). In addition, infiltration of M1 macrophages in the tumor area was reduced in samples with BER alterations (p = 0.005) and tended to be reduced in samples with NER alterations (p = 0.054), whereas infiltration of M2 macrophages in the tumor area was reduced in samples with FA alterations (p = 0.032) (Figure 4g,h).
FIGURE 4.
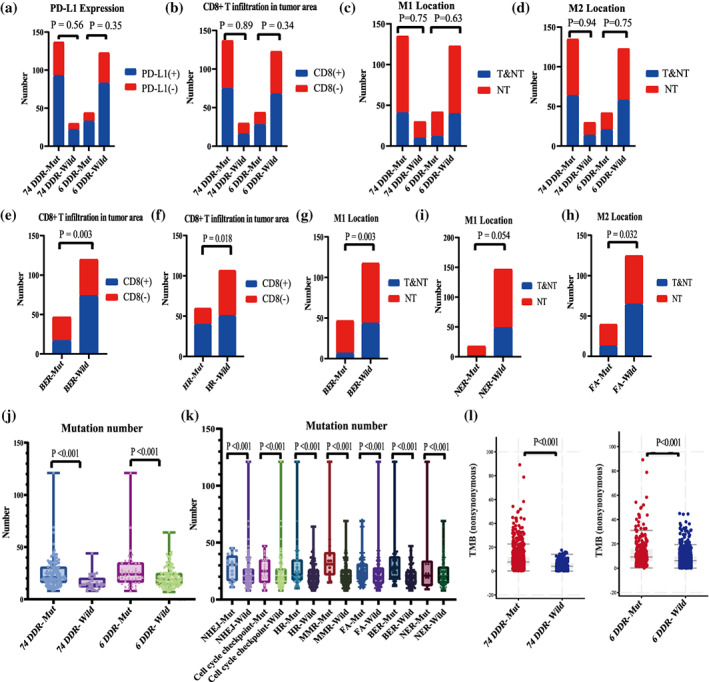
Association of DNA damage response (DDR) gene status with tumor microenvironment. (a–d) DDR gene status did not affect PD‐L1 expression and infiltration of CD8‐positive T lymphocytes, M1 and M2 macrophage in tumor area. (e–h) Different DDR pathway alterations indicate different immune cell infiltration. (i–k) The DDR‐mut group has higher numbers of mutation, whether for 74 genes and six genes. The same conclusion was maintained when the individual DDR pathways were analyzed separately. (l) TMB was significantly higher in the DDR‐mut subgroup in the Pan‐Lung cancer dataset, both for 74 DDR genes and the specified six genes.
DDR alteration indicated increased numbers of gene mutations
The number of gene mutations was significantly higher in the DDR‐mut subgroup, whether for 74 genes and six genes (p < 0.001) (Figure 4j). The same conclusion was maintained when the individual DDR pathways were analyzed separately (Figure 4k). We also validated the relationship between DDR gene mutations and TMB using an external database, and the results showed that TMB was significantly increased in the DDR mutation group, both for 74 genes (median TMB, 7.6 vs. 3.92 mutations/megabase, p < 0.001) and six genes (median TMB, 9.17 vs. 6.22 mutations/megabase, p < 0.001) (Figure 4l).
DISCUSSION
Here, we analyzed the mutation profile and prognostic significance of DDR genes in resected NSCLC patients and investigated the association of mutation in DDR genes and the tumor immune microenvironment. Our study found that deleterious mutations of DDR genes were common in resected NSCLC (82.0%) patients and LUSC patients were more prone to DDR mutations, especially for MMR and NER pathway. However, mutations of the 74 DDR genes were not associated with the survival of NSCLC, LUAD or LUSC patients. This is consistent with the report by Dai et al. 18 who found no correlation between DDR mutations and prolonged survival in advanced NSCLC, LUSC and LUAD. We then focused on the selected six genes (ATM, BRCA1, BRCA2, CHEK1, BARD1, BRIP1) and found that their mutations were independent protective factors for prognosis. Previous studies have demonstrated that ALK and KRAS mutations are associated with worse survival in patients with resected NSCLC. 20 , 21 , 22 In this study, the mutation frequencies of KARS and ALK were balanced between the DDR‐wild and DDR‐mut groups. It should be noted that further subgroup analysis revealed that this association only existed in patients with stage IIIa but not in those with stage I. We believe that this may be due to the following two factors. First, the prognosis for patients in stage I who underwent curative surgery is good. In this study, only 30.7% (27/88) of stage I patients experienced relapse at the end of follow‐up. Therefore, the effect of DDR mutation was not significant for stage I. Second, all patients with stage IIIa received postoperative platinum‐based chemotherapy. It has been reported that cancer cells incapable of recovering from replication stress due to defective DNA damage response (DDR) pathways may exhibit increased sensitivity to platinum compounds, 23 whilst tumors with defective HRR were found to be more responsive to chemotherapy. 24 , 25 Consequently, the protective effects of DDR mutations were more pronounced in stage IIIa patients. Notably, our findings suggest that among stage IIIa patients, LUSC patients with DDR mutations experience extended DFS and OS, whereas no such benefits are observed in LUAD patients. These results emphasize the necessity for further research in this area. However, the relatively small sample sizes of each subgroup may limit the reliability of this conclusion.
However, the deleterious mutations of the six genes in this study were 26.3%, which was significantly higher than that in the Pan‐Lung cancer dataset. The reason was that the missense mutations of unknown significance were predicted to be pathogenic by three prediction tools (Polyphen2, SIFT, and Mutation Assessor) in our research, which may have overestimated the frequencies of deleterious gene mutation. However, Ricciuti et al. 26 employed the same method to predict the pathogenicity of missense mutations. They found that in patients with advanced NSCLC who received PD‐(L)1 inhibitor therapy, deleterious DDR gene mutations predicted better prognosis. Importantly, further analysis found that even for missense mutations that were not included in COSMIC and/or ClinVar but were identified as pathogenic by polyphen2, clinical outcomes were also improved when compared to the DDR proficient group. Therefore, it is necessary to further explore the function of DDR mutations of unknown significance in NSCLC. Furthermore, unlike EGFR mutations which were more common in never‐smokers, our findings indicate that deleterious mutations in these six genes were prevalent in former and current smokers. This suggests that smoking can lead to defects in DNA damage repair mechanisms, thereby increasing the frequency of genetic mutations.
Although a few studies have shown that DDR gene mutations can upregulate PD‐L1 expression in gastroesophageal cancer 27 and breast cancer, 28 this study did not find an association between DDR gene status and PD‐L1 expression in early‐stage NSCLC. Similarly, no association was found in advanced NSCLC 26 and small‐cell lung cancer (SCLC). 29 In addition, deleterious mutations in DDR genes did not affect CD8‐positive lymphocytes and TAM infiltration in the tumor. However, it was noted that alterations in the BER, FA and HR pathways could affect immune cell infiltration in the tumor area.
Deleterious DDR mutations have been reported to be associated with high TMB in solid tumors including lung cancer. 10 , 16 , 26 , 29 , 30 , 31 , 32 , 33 , 34 In this study, we found that deleterious alterations in the DDR pathway were associated with a higher number of gene mutations, and we also observed significantly higher TMB in the DDR‐mut subgroup in the pan lung cancer dataset for both the selected 74 DDR genes and six genes. It has been reported that deleterious DDR gene mutations are an independent predictor of better response to immune checkpoint inhibitors in colorectal cancer 15 and NSCLC. 13 , 16 , 26 Therefore, we speculated that deleterious mutations in the six genes may predict better survival with neoadjuvant immunotherapy and immunotherapy, which needs to be validated with a large cohort.
We recognize several limitations of this study. First, it was a retrospective, single‐center study, which may have limited the accuracy and generalizability of the results. However, every effort was made to ascertain the survival status of the patients and to increase confidence in the clinical information. Larger multicenter prospective studies are needed to validate these findings in the future. Second, this study did not include patients who received immune neoadjuvant therapy. The relationship between the DDR gene, in particular the six gene mutations, and the efficacy of neoadjuvant immunotherapy should be further investigated. Third, some PPFE samples have been stored for a long time. This may affect the efficacy of gene mutations. Last but more importantly, this study did not involve stage II patients, which compromises the representativeness of the study's conclusions in early‐stage lung cancer.
In conclusion, DDR gene mutations were common in resected NSCLC. Deleterious mutations of ATM, BRCA1, BRCA2, CEEK1, BARD1 and BRIP1 predicted improved DFS and OS in patients with resected NSCLC. More studies into the prognostic value of these genes in immunotherapy are worthy of investigation in the future.
AUTHOR CONTRIBUTIONS
Haoran Zhang: Writing – original draft, formal analysis. Dongming Zhang: Writing – original draft, software. Jia Liu: Investigation, writing – original draft. Yuequan Shi: Data curation, investigation. Xiaoyan Liu: Investigation, visualization. Jing Zhao: Methodology, formal analysis. Minjiang Chen: Data curation, writing–original draft. Wei Zhong: Writing–review and editing. Yan Xu: Conceptualization, project administration, writing–review and editing. Mengzhao Wang: Conceptualization, writing – review and editing.
CONFLICT OF INTEREST
The authors confirm that there are no conflicts of interest.
ETHICS STATEMENT
The study was conducted in accordance with the Declaration of Helsinki. The study was approved by the ethics committee of Peking Union Medical College Hospital (no. S‐K146) and individual consent for this retrospective analysis was waived.
Supporting information
Figure S1. Typical immunochemical staining pictures of PD‐L1 expression.
Table S1. Seventy‐four DDR genes involved in seven DDR pathways.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of Beijing (to YX) (no: 7194311) and National High Level Hospital Clinical Research Funding (to MW), grant/award no.: 2022‐PUMCH‐B‐106.
The authors appreciate the patients for providing their information for the study and also acknowledge the technical support of Weihua GUO from Genetron Health Technology, Co, Ltd., Beijing, China.
Zhang H, Zhang D, Liu J, Shi Y, Liu X, Chen M, et al. Clinical significance of DNA damage response mutations in stage I and stage IIIa NSCLC . Thorac Cancer. 2023;14(32):3191–3201. 10.1111/1759-7714.15109
Contributor Information
Yan Xu, Email: maraxu@163.com.
Mengzhao Wang, Email: mengzhaowang@sina.com.
REFERENCES
- 1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA‐Cancer J Clin. 2021;71(1):7–33. [DOI] [PubMed] [Google Scholar]
- 2. Lu T, Yang X, Huang Y, Zhao M, Li M, Ma K, et al. Trends in the incidence, treatment, and survival of patients with lung cancer in the last four decades. Cancer Manage Res. 2019;11:943–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goldstraw P, Chansky K, Crowley J, Rami‐Porta R, Asamura H, Eberhardt WEE, et al. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11(1):39–51. [DOI] [PubMed] [Google Scholar]
- 4. Friedlaender A, Addeo A, Russo A, Gregorc V, Cortinovis D, Rolfo CD. Targeted therapies in early stage NSCLC: hype or Hope? Int J Mol Sci. 2020;21(17):6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carusillo A, Mussolino C. DNA damage: from threat to treatment. Cell. 2020;9(7):1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang M, Jia K, Wang L, Li W, Chen B, Liu Y, et al. Alterations of DNA damage response pathway: biomarker and therapeutic strategy for cancer immunotherapy. Acta Pharm Sin B. 2021;11(10):2983–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chabanon RM, Rouanne M, Lord CJ, Soria J‐C, Pasero P, Postel‐Vinay S. Targeting the DNA damage response in immuno‐oncology: developments and opportunities. Nat Rev Cancer. 2021;21(11):701–717. [DOI] [PubMed] [Google Scholar]
- 9. Aktas BY, Guner G, Guven DC, Arslan C, Dizdar O. Exploiting DNA repair defects in breast cancer: from chemotherapy to immunotherapy. Expert Rev Anticancer Ther. 2019;19(7):589–601. [DOI] [PubMed] [Google Scholar]
- 10. Teo MY, Bambury RM, Zabor EC, Jordan E, al‐Ahmadie H, Boyd ME, et al. DNA damage response and repair gene alterations are associated with improved survival in patients with platinum‐treated advanced urothelial carcinoma. Clin Cancer Res. 2017;23(14):3610–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gallagher DJ, Konner JA, Bell‐McGuinn KM, et al. Survival in epithelial ovarian cancer: a multivariate analysis incorporating BRCA mutation status and platinum sensitivity. Ann Oncol. 2011;22(5):1127–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu H, Zhu Y, Pu N, Burkhart RA, Burns W, Laheru D, et al. Association of germline variants in human DNA damage repair genes and response to adjuvant chemotherapy in resected pancreatic ductal adenocarcinoma. J Am Coll Surg. 2020;231(5):527–535.e514. [DOI] [PubMed] [Google Scholar]
- 13. Xiong A, Nie W, Zhou Y, Li C, Gu K, Zhang D, et al. Comutations in DDR pathways predict Atezolizumab response in non‐small cell lung cancer patients. Front Immunol. 2021;12:708558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Teo MY, Seier K, Ostrovnaya I, Regazzi AM, Kania BE, Moran MM, et al. Alterations in DNA damage response and repair genes as potential marker of clinical benefit from PD‐1/PD‐L1 blockade in advanced urothelial cancers. J Clin Oncol. 2018;36(17):1685–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Song Y, Huang J, Liang D, Hu Y, Mao B, Li Q, et al. DNA damage repair gene mutations are indicative of a favorable prognosis in colorectal cancer treated with immune checkpoint inhibitors. Front Oncol. 2020;10:549777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou Z, Ding Z, Yuan J, Shen S, Jian H, Tan Q, et al. Homologous recombination deficiency (HRD) can predict the therapeutic outcomes of immuno‐neoadjuvant therapy in NSCLC patients. J Hematol Oncol. 2022;15(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu Y, Li J, Tong B, Chen M, Liu X, Zhong W, et al. Positive tumour CD47 expression is an independent prognostic factor for recurrence in resected non‐small cell lung cancer. ESMO Open. 2020;5(4):e000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dai J, Jiang M, He K, Wang H, Chen P, Guo H, et al. DNA damage response and repair gene alterations increase tumor mutational burden and promote poor prognosis of advanced lung cancer. Front Oncol. 2021;11:708294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Bono J, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Olaparib for metastatic castration‐resistant prostate cancer. N Engl J Med. 2020;382(22):2091–2102. [DOI] [PubMed] [Google Scholar]
- 20. Jianxing HE. Consensus on postoperative recurrence prediction of non‐small cell lung cancer based on molecular markers. Chin J Lung Cancer. 2022;25(10):701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang SM, Zhu QG, Ding XX, Lin S, Zhao J, Guan L, et al. Prognostic value of EGFR and KRAS in resected non‐small cell lung cancer: a systematic review and meta‐analysis. Cancer Manage Res. 2018;10:3393–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tao H, Shi L, Zhou A, Li H, Gai F, Huang Z, et al. Distribution of EML4‐ALK fusion variants and clinical outcomes in patients with resected non‐small cell lung cancer. Lung Cancer. 2020;149:154–161. [DOI] [PubMed] [Google Scholar]
- 23. Kelland L. The resurgence of platinum‐based cancer chemotherapy. Nat Rev Cancer. 2007;7(8):573–584. [DOI] [PubMed] [Google Scholar]
- 24. Helleday T. Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis. 2010;31(6):955–960. [DOI] [PubMed] [Google Scholar]
- 25. Jiang M, Jia K, Wang L, Li W, Chen B, Liu Y, et al. Alterations of DNA damage repair in cancer: from mechanisms to applications. Ann Transl Med. 2020;8(24):1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ricciuti B, Recondo G, Spurr LF, Li YY, Lamberti G, Venkatraman D, et al. Impact of DNA damage response and repair (DDR) gene mutations on efficacy of PD‐(L)1 immune checkpoint inhibition in non‐small cell lung cancer. Clin Cancer Res. 2020;26(15):4135–4142. [DOI] [PubMed] [Google Scholar]
- 27. Cerniglia M, Xiu J, Grothey A, Pishvaian MJ, Hwang JJ, Marshall J, et al. Association of DNA damage response and repair genes (DDR) mutations and microsatellite instability (MSI), PD‐L1 expression, tumor mutational burden (TMB) in gastroesophageal cancers. J Clin Oncol. 2019;37(4):60. [Google Scholar]
- 28. Parkes EE, Walker SM, Taggart LE, McCabe N, Knight LA, Wilkinson R, et al. Activation of STING‐dependent innate immune signaling by S‐phase‐specific DNA damage in breast cancer. J Natl Cancer Inst. 2017;109(1):djw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu S, Zhang Y, Zhang Y, Chen LH, Ouyang HF, Xu X, et al. Mutational landscape of homologous recombination‐related genes in small‐cell lung cancer. Cancer Med. 2022;12:4486–4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang X, Huang Z, Li L, Wang G, Dong L, Li Q, et al. DNA damage repair gene signature model for predicting prognosis and chemotherapy outcomes in lung squamous cell carcinoma. BMC Cancer. 2022;22(1):866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chiu TY, Lin RW, Huang CJ, Yeh DW, Wang YC. DNA damage repair gene set as a potential biomarker for stratifying patients with high tumor mutational burden. Biology. 2021;10(6):528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xiao Y, Lu D, Lei M, Xie W, Chen Y, Zheng Y, et al. Comprehensive analysis of DNA damage repair deficiency in 10,284 pan‐cancer study. Ann Transl Med. 2021;9(22):1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park S, Lee H, Lee B, Lee SH, Sun JM, Park WY, et al. DNA damage response and repair pathway alteration and its association with tumor mutation burden and platinum‐based chemotherapy in SCLC. J Thorac Oncol. 2019;14(9):1640–1650. [DOI] [PubMed] [Google Scholar]
- 34. Chae YK, Davis AA, Raparia K, Agte S, Pan A, Mohindra N, et al. Association of tumor mutational burden with DNA repair mutations and response to anti‐PD‐1/PD‐L1 therapy in non‐small‐cell lung cancer. Clin Lung Cancer. 2019;20(2):88–96.e86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Typical immunochemical staining pictures of PD‐L1 expression.
Table S1. Seventy‐four DDR genes involved in seven DDR pathways.


