Abstract
CRISPR-Cas9 genome engineering is a powerful technology for correcting genetic diseases. However, the targeting range of Cas9 proteins is limited by their requirement for a protospacer adjacent motif (PAM) and in vivo delivery is challenging due to their large size. Here, we use phage-assisted continuous directed evolution (PACE) to broaden the PAM compatibility of CjCas9, the smallest Cas9 orthologue characterized to date. The identified variant, termed evoCjCas9, primarily recognizes N4AH and N5HA PAM sequences, which occur ten-fold more frequently in the genome than the canonical N3VRYAC PAM site. Moreover, evoCjCas9 exhibits higher nuclease activity than wild-type CjCas9 on canonical PAMs, with editing rates comparable to commonly used PAM-relaxed SpCas9 variants. Combined with deaminases or reverse transcriptases, evoCjCas9 enables robust base- and prime editing, with the small size of evoCjCas9 base editors allowing tissue-specific installation of A-to-G or C-to-T transition mutations from single adeno-associated virus (AAV) vector systems.
Introduction
Genome editing with RNA-guided programmable nucleases has been transforming biomedical research and holds great promise for therapeutic application in patients with genetic diseases1–3. Cas9 endonucleases of the type II CRISPR immune defence system are the most widely used RNA-guided nucleases for genome editing4. They are paired with programmable single guide (sg)RNAs, which contain a spacer sequence that targets the complementary protospacer on genomic DNA when a downstream protospacer adjacent motif (PAM) is present. Binding of Cas9 endonucleases to target sites subsequently induces double-strand (ds)DNA breaks, which lead to the formation of insertion/deletion (indel) mutations or precise editing from DNA donor templates via homologous recombination. More recently, the fusion of deaminases or reverse transcriptases (RTs) to catalytically impaired Cas9 led to the establishment of base editors (BEs)5,6 and prime editors (PEs)7. Importantly, these Cas9-based genome editing tools function independently of dsDNA break formation and homologous recombination, enabling precise editing in all cell types, including postmitotic cells8.
Cas9 originating from Streptococcus pyogenes (SpCas9) was the first RNA-programmable endonuclease employed for genome editing1,9,10. Due to its high activity and relatively simple PAM sequence (NGG) it also remains the most widespread Cas9 ortholog used to date. The large size of SpCas9 (1,368 aa), however, presents a challenge for efficient in vivo delivery, particularly when using adeno-associated virus (AAV) vectors. AAVs are among the most promising nucleic acid delivery vehicles for the clinics, as they show low rates of genome integration, minor immunogenicity, and cover a broad range of tissue tropisms11–13. Nevertheless, their limited cargo capacity of ~5 kb (including ITRs)14,15 prohibits the packaging of full-length SpCas9 together with its sgRNA on a single AAV vector. Partly circumventing this issue, the intein system16 has previously been used to split SpCas9 nucleases, SpCas9-BEs or SpCas9-PEs into two parts to express them from two separate AAVs17–19. This approach, however, requires the use of high vector doses to ensure co-infection of cells and to compensate for the low efficiency of generating full-length Cas9 from the intein-split moieties19. A promising alternative to two-vector systems is the replacement of SpCas9 with more compact Cas9 orthologs. Recent studies generated ABEs with Cas9 of Staphylococcus aureus (SaCas9 - 1,053 aa), Neisseria Meningitidis (Nme2Cas9 - 1,082 aa), or Campylobacter jejuni 3 (Cje3Cas9 − 1000 aa) (Supplementary Fig. 1), and packaged them together with their sgRNAs on single AAV vectors. Nevertheless, these constructs were still close to the packaging limit of AAV and require to use of small minimal promoters (e.g. EF-1α or the nuclear RNA (U1a) promoter)20–22.
With 984 aa Campylobacter jejuni (Cj)Cas9 is the smallest known Cas9 ortholog, making it a particularly attractive candidate for in vivo genome editing applications. Similar to other small Cas9 orthologs, CjCas9 recognizes a relatively complex PAM sequence (N3VRYAC), which occurs less frequently in the genome than the NGG motif of SpCas9. This greatly restrains the application of CjCas9, especially for base- and prime editing where Cas9 has to bind to the DNA at a precisely defined distance to the targeted base.
Here, we use phage-assisted continuous directed evolution (PACE) to generate evoCjCas9. This CjCas9 variant primarily recognizes N4AH and N5HA PAM sequences, which occur 10-times more frequently in the genome than the canonical N3VRYAC PAM sequence. Importantly, evoCjCas9 also possesses a higher nuclease activity than wild-type CjCas9 and other small sized Cas nucleases, and enables robust base- and prime editing at canonical and non-canonical PAM sites. Finally, we provide proof-of-concept for tissue-specific in vivo adenine and cytosine base editing with evoCjCas9 using a one-vector AAV system.
Results
Phage-assisted continuous and non-continuous evolution
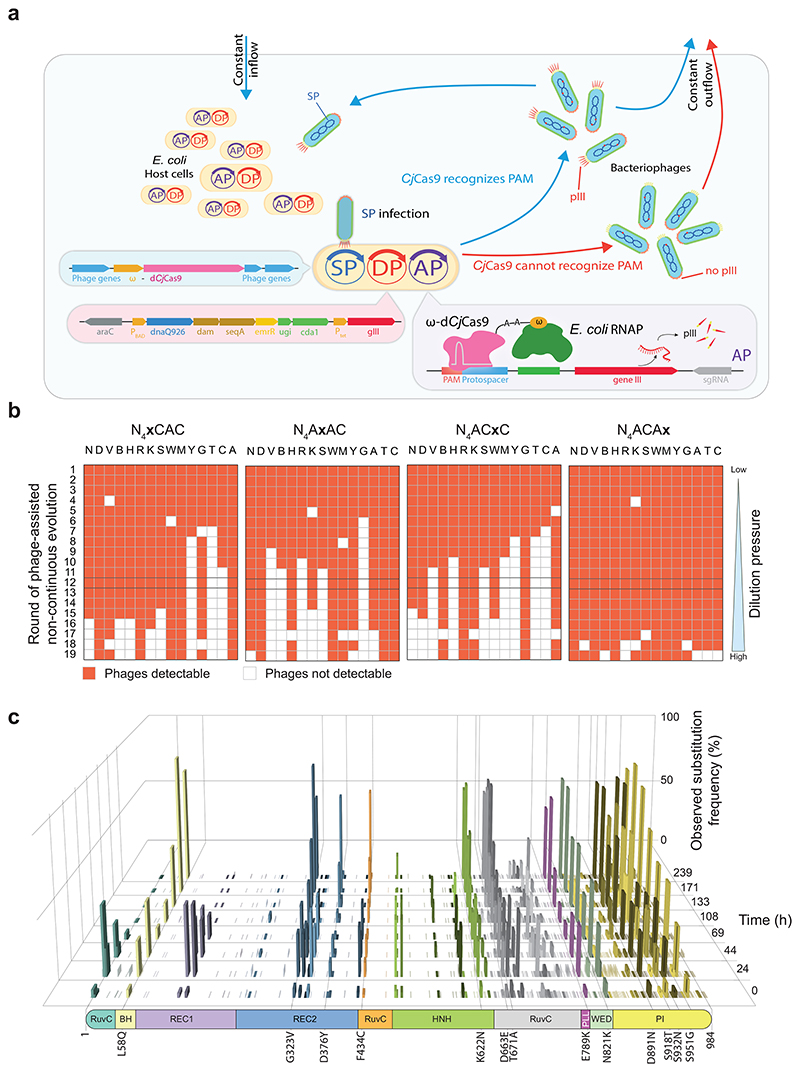
PACE is a technique for directed protein evolution, where the gene of interest is continuously mutated and transferred from bacterial host cell to host cell via a modified M13 filamentous bacteriophage in an activity of interest-dependent manner23–27. In our PACE setup, the essential gene III on the M13 selection phage (SP) is replaced by a catalytically dead CjCas9 (dCjCas9), which is N-terminally fused to the ω subunit of bacterial RNA polymerase. SP infection of E. coli host cells carrying the accessory plasmid (AP) results in the complexation of dCjCas9 with a sgRNA that directs the complex to the promoter region of gene III (Fig. 1a). Recognition of the protospacer and PAM sequence leads to stable binding of the dCjCas9-ω complex, recruitment of the RNA polymerase and subsequent expression of gene III. This completes the life cycle of the phage and enables the assembly of functional and infectious phage particles. The additional presence of the drift plasmid (DP)28 in E. coli host cells enables random mutagenesis of the phage genome during replication while continuous influx and efflux of media with fresh host cells leads to a steady phage dilution, thus setting the evolutionary pressure. By installing a library of non-canonical PAM sequences on the AP, we link phage propagation to dCjCas9 variants that have acquired mutations to recognize these novel PAMs in our experimental setup (Fig. 1a). Since the first four positions of the canonical CjCas9 PAM show only minor nucleotide preferences (NNNV), we focused our efforts on the relaxation of the last 4 PAM positions (RYAC)29,30. Our first attempt to start PACE with a PAM library that is fully randomized at these 4 positions, however, was not successful as the selection was too stringent and phages could not be propagated (determined by RT-qPCR; CT > 30). Therefore, we next tried to pre-select CjCas9 variants with relaxed PAM recognition using phage-assisted non-continuous evolution (PANCE)25. In this setup, we used the same evolution logic but reduced the dilution pressure by propagating phages via serial phage dilution instead of dilution in a continuous flow. However, the moderate dilution rates used in PANCE (Supplementary Table 1) resulted in infrequent gene III activation and loss of phages, as determined by RT-qPCR (CT > 30, Supplementary Fig. 2a). To further reduce stringency, we next performed PANCE in an arrayed fashion where we individually replaced nucleotides at PAM positions 5-8 on the AP in all possible combinations (15 different nucleotide compositions per position, resulting in 56 different combinations plus 4 canonical N4ACAC PAMs - Fig. 1b). In this setup we identified phages to withstand 200-fold dilution steps for PAM combinations close to the canonical PAM, while more unfavourable PAM sequences (e.g. T, G and C at positions 5, 6 and 7, respectively) led to a phage wash-out unless the AP was mixed with a fraction of plasmids with canonical PAM nucleotides (Fig. 1b; Supplementary Fig. 2b). Interestingly, lagoons with all nucleotide variations at position 8 supported phage propagation, while positions 6 and 7 possessed more stringent PAM requirements (Fig. 1b; Supplementary Fig. 2b). After 12 rounds of dilution (a total of 5*1023-fold dilution; Supplementary Table 1), 45 lagoons maintained detectable phage genomes. These were mixed in a 1:1 ratio with wild-type ω-dCjCas9-phages and used to seed a lagoon of a PACE apparatus with a continuous inflow of fresh host cells containing an AP with a randomized PAM library for positions 6-8. The pre-relaxation with PANCE allowed phages to sustain a gradual increase of flow rates from 0.5 to 3 lagoon volumes per hour over 10 days, with phage wash-out starting to occur at higher flow rates. In the initial phase of PACE (first 100 hours), we modulated the selection pressure by fluctuating the dilution rates between 0.5 and 1 lagoon volume per hour (LV/h) in order to facilitate crossing of potential fitness valleys, followed by a more stringent selection with a dilution rate that was increased to 3 LV/h (Supplementary Fig. 3a). To assess the shift of phage genotypes during PACE, we collected phage samples at different time points during the experiment and analysed them by nanopore long read sequencing. Evaluation of the sequencing data revealed a mutation bias of G-C to A-T conversions (51.3%), with A-T to T-A conversions being the least frequently observed mutations (2.0%, Supplementary Fig. 3b; Supplementary Data 1), in line with previous findings characterizing the mutation spectra of DP28. Importantly, amino acid changes occurred most often in the RuvC III domain, phosphate lock loop domain, wedge domain and PAM interacting domain of CjCas9 (Fig. 1c). After 108 hours of PACE, a consensus sequence with the mutations L58Q, G323V, D376Y, F434C, K622N, D663E, T671A, E789K, N821K, D891N, S918T, S932N and S951G started to occur in the lagoon. These mutations were further enriched until termination of the experiment, where this genotype made up the bulk of the phage population (Supplementary Fig. 3c).
Fig. 1. Phage-assisted continuous and non-continuous evolution (PACE) to broaden the PAM compatibility of CjCas9.
a, Schematics of the PACE experiment. Selection phages (SP) infect E. coli host cells and use the hosts machinery to replicate and translate required phage genes. Instead of gene III, SP carries the ω-subunit-dCjCas9 fusion gene. Cas9 recognition of a PAM and protospacer sequence on the accessory plasmid (AP) allows recruitment of endogenous E. coli RNA polymerase via the ω-subunit and subsequent translation of gene III (pIII). Efficient recognition of the PAM and protospacer sequence results in infectious phages that are able to sustain in the lagoon, whereas weak or lack of recognition results in phage wash-out. The mutation rate during phage replication is controlled and elevated via arabinose-inducible expression of mutagenesis genes from a mutagenesis plasmid (DP6). b, Illustrative overview of 19 rounds of PANCE using accessory plasmids containing different PAM sites (according to IUPAC notation). Phages were increasingly diluted from one round to the next. Red indicates presence of phages, white indicates absence. c, Long-read sequencing of individual phages (n = 811) at 8 different time points during PACE. For each time point, the mutation frequency and amino acid position is shown. Amino acid positions substituted in more than 50% of phage genotypes are indicated.
Evolved CjCas9 variant with broad PAM recognition
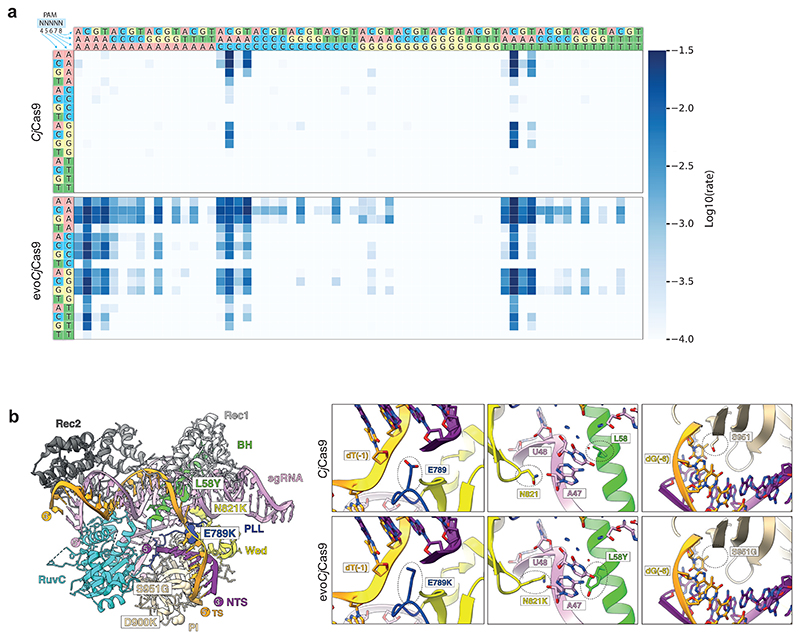
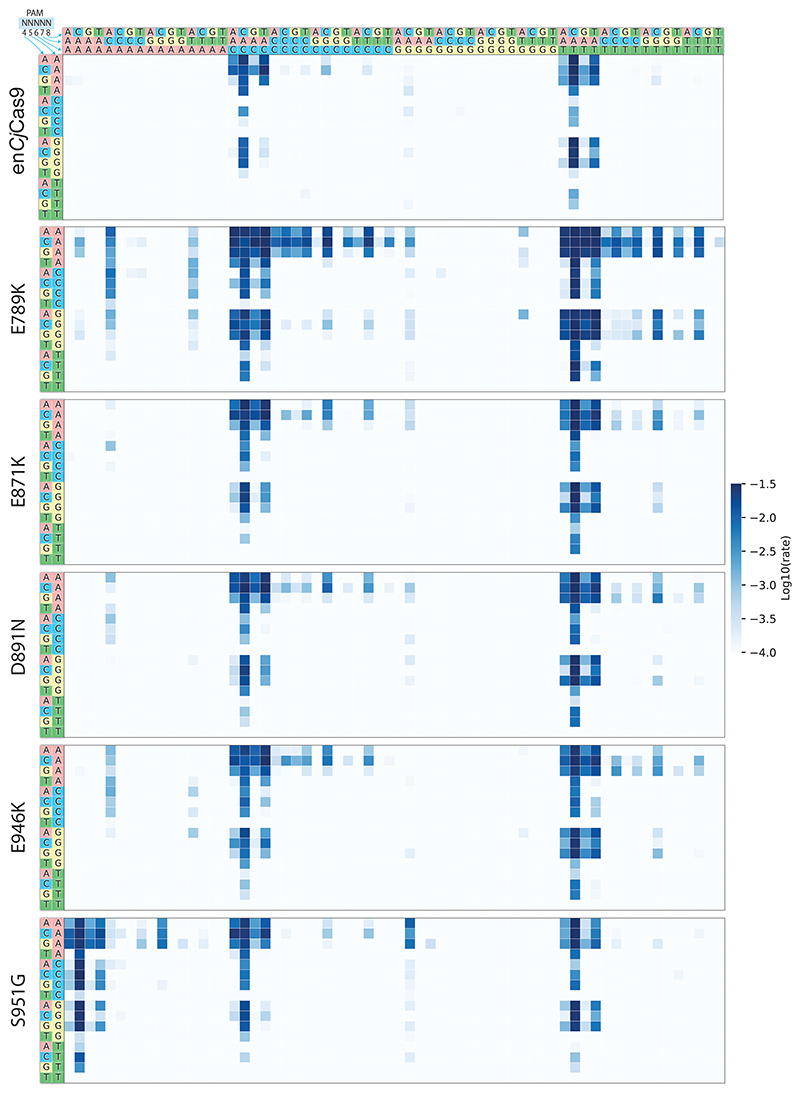
In our PACE and PANCE evolution logic, we selected for PAM recognition via CjCas9 binding, potentially leading to mutations that impair the nuclease activity of Cas931,32. Hence, we first assessed the effect of the mutations in the consensus genotype individually in a kinetic in vitro cleavage assay on a randomized PAM library33 (Supplementary Fig. 4a, b), with the aim of finding a set of mutations that lead to PAM relaxation without impairing the nuclease activity. Of the 13 mutations, E789K, E871K, D891N, and E946K had a strong contribution to PAM relaxation at position 5 and 7 (Fig. 2a; Extended Data Fig. 1; Supplementary Data 2), with all 4 mutations leading to the recognition of non-canonical C and T bases at position 5 and non-canonical C, G and T bases at position 7. Since E789K resulted in a higher cleavage activity at most non-canonical PAMs than the other three variants, and additionally allowed pronounced recognition of N5ACA PAMs (Extended Data Fig. 1), we decided to select this mutation for our evolved (evo)CjCas9 variant. Interestingly, mapping E789K onto the crystal structure of CjCas9 (Protein Data Bank (PDB): 5X2H)30 revealed that it is located in the phosphate lock loop domain and interacts with the phosphate backbone of PAM position 1. The positively charged lysine interaction may therefore increase the non-specific association of CjCas9 with the PAM region, allowing recognition of non-canonical bases at downstream PAM positions (Fig. 2b).
Figure 2. Characterization of the PAM compatibility of evoCjCas9.
a, HT-PAMDA33 characterization of CjCas9 and evoCjCas9 illustrating their PAM preference at PAM positions 4 to 8. The log10 rate constant represents the mean of two replicates against two distinct spacer sequences. b, Protein structure of CjCas9 with evoCjCas9 mutations introduced and highlighted (left, based on PDB: 5X2H). Detailed view illustrating differences between CjCas9 (top panels) and evoCjCas9 (bottom panels) residues interacting with PAM nucleotides of the target strand.
The mutation S951G occurred with the highest frequency of all PAM-relaxing mutations identified in PACE (after 170h, 93.9% of phage genomes contained S951G), and was the only mutation that allowed relaxation of PAM position 6, leading to the recognition of a non-canonical A nucleobase in addition to the canonical C and T (Extended Data Fig. 1). Thus, we also selected this mutation for introduction into evoCjCas9. Notably, S951 is located in the PAM interacting domain, but instead of interacting with nucleobases at position 6 of the PAM it interacts with the nucleobases on the opposing DNA target strand via hydrogen bonding (Fig. 2b)30. The shorter side chain of glycine compared to serine may therefore reduce the interactions with A nucleobases at this position, leading to the detection of non-canonical T at position 6 of the PAM.
Substitutions of residues L58 and N821 were also frequently observed at the end of our PACE experiment (98.6% and 76.7%, respectively), but did not substantially contribute to PAM relaxation of CjCas9 in our PAM detection assay (Supplementary Data 2). Nevertheless, L58Y has previously been described to increase the stability of the CjCas9-sgRNA complex by forming a stacking interaction with U48 in the sgRNA, thereby enhancing the DNA cleavage activity of CjCas9 on canonical PAMs34. Since N821 is in close physical distance and interacts with the same nucleotide of the sgRNA via hydrogen bonding (Fig. 2b), we hypothesize that the positively charged lysine residue at this position could further stabilize the CjCas9-sgRNA complex and enhance cleavage activity, prompting us to introduce L58Y and N821K into evoCjCas9.
Mutations G323V, D376Y, F434C, K622N, D663E, and T671A were also enriched during PACE, but had a detrimental effect on the catalytic activity of CjCas9 nuclease in our cleavage assay (Supplementary Data 2). Thus, while these mutations may pose a fitness advantage regarding target recognition and binding of CjCas9, they would prohibit applications for which cutting or nicking of DNA is required and we did not integrate them into evoCjCas9.
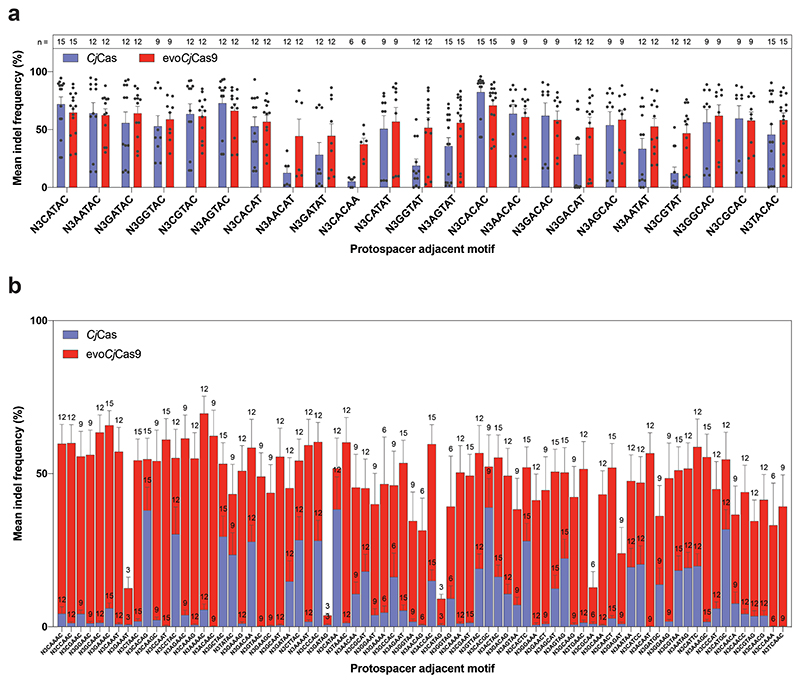
Taken together, we found that the mutations L58Y, E789K, N821K, and S951G increase PAM flexibility and/or cleavage activity. Combined with D900K, a mutation that was not enriched in our PACE experiment but was previously described to marginally enhance CjCas9 cleavage activity34, these mutations were introduced into CjCas9 to generate evoCjCas9. Compared to wild-type CjCas9 and the previously established enhanced (en)CjCas9 variant with the substitutions L58Y and D900K34, evoCjCas9 shows a substantial relaxation in PAM recognition (Fig. 2a; Extended Data Fig. 1); it recognizes all nucleotides at the first three PAM positions (Supplementary Fig. 5) and primarily NAHNN or NNHAN sequences at PAM positions 4-8. This allows evoCjCas9 to detect 10-times more target sequences than wild-type CjCas9, which requires a VRYAC motif at PAM positions 4-8 (198 versus 19 PAM sequences with a kinetic rate constant >10-3, Fig. 2a).
High cleavage activity at canonical and non-canonical PAMs
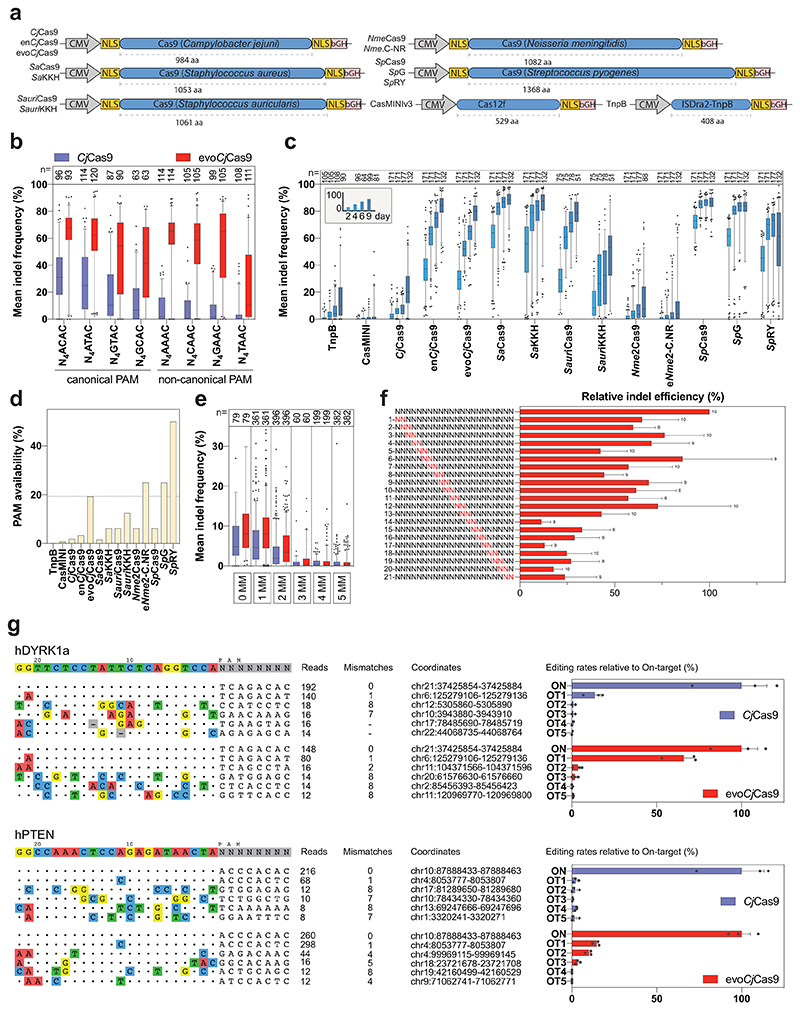
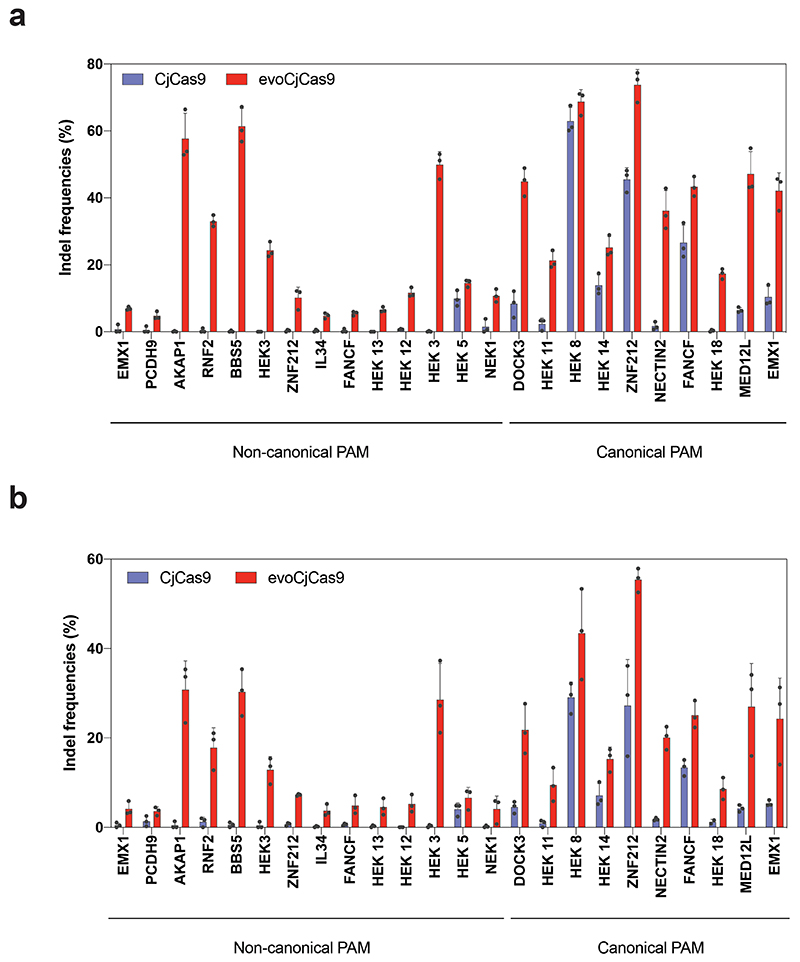
To comprehensively characterize the activity of evoCjCas9, we compared indel frequencies of CjCas9 and evoCjCas9 in mammalian cells at multiple sites with canonical and non-canonical PAM sequences. We generated a lentiviral library containing CjCas9 sgRNA constructs paired with their target sites (self-targeting library), with the different target sites comprising a set of canonical (N4ACAC, N4ATAC, N4GTAC and N4GCAC) and non-canonical (N4AAAC, N4CAAC, N4GAAC and N4TAAC) PAM sites. After integrating the library into the genome of HEK293T cells, plasmids expressing wild-type CjCas9 or evoCjCas9 were transfected (Fig. 3a; Supplementary Data 3), and after selection genomic DNA was isolated and analysed by high throughput sequencing (HTS). Importantly, evoCjCas9 was already 2-fold more efficient than wild-type CjCas9 in generating indels at canonical PAM sites (on average 54.0% versus 24.7%), and reached average editing rates of 48.6% at non-canonical PAM sites at which wild-type CjCas9 did not show substantial editing (Fig. 3b). Confirming these results, evoCjCas9 showed considerably higher activity than wild-type CjCas9 on 24 endogenous target sites containing non-canonical (n=14) and canonical (n=10) PAMs (Extended Data Fig. 2). To provide additional support for the wider targeting range of evoCjCas9, we next generated a self-targeting library containing 5 target sites combined with 100 different PAMs. Except for 8 canonical PAMs ending with RYAC, evoCjCas9 consistently outperformed wild-type CjCas9 in terms of indel formation rates (Extended Data Fig. 3, Supplementary Data 4).
Figure 3. Indel formation rates of evoCjCas9 compared to CjCas9 and other commonly used RNA-guided endonucleases.
a, Illustration and size comparison of the expression vectors encoding for different Cas enzymes. b, Observed indel frequencies for CjCas9 and evoCjCas9 on 8 different PAM sites. c, Observed indel frequencies for different commonly used RNA-guided endonucleases and PAM relaxed Cas variants on different target sites with the most optimal PAM for each nuclease. Colors represent different timepoints. d, PAM frequency of the different RNA-guided endonucleases, illustrated as the probability of encountering a PAM on a randomly selected target site in a random DNA sequence (in %). Calculations are based on TTGAT for TnpB; TTTR for CasMINIv3.1; PAM sequences with a kinetic rate constant >10-3 for CjCas9 variants; N2GRRT for SaCas9; N3RRT for SaKKH; N2GG for SauriCas9; N2RG for SauriKKH; N4CC for Nme2Cas9; N4C for eNme2-C.NR; NGG for SpCas9; NG for SpG and NR for SpRY. e, Mean indel frequencies of CjCas9-variants at on-target sites (0 MM) and potential off-target sites containing up to 5 mismatches (MM). f, Indel frequencies of evoCjCas9 at sites with dinucleotide mismatches normalized to the on-target site. Dinucleotides highlighted in red represent mismatch positions. PAM at position 23-30. Number above bars indicate amount of different target sites. g, Relative indel frequencies evaluated by HTS at off-target sites detected by CHANGE-seq for two sgRNAs. Boxplots in (b, c, e) represent the 25th, 50th, and 75th percentiles. Whiskers indicate 5 and 95 percentiles. Numbers (n) above plots indicate number of biologically independent datapoints. Bars in (f, g) represent mean, with error bars representing standard error of the mean of indicated number of target sites (f) or biologically independent replicates (g, n=3, and n= 2 for OT2, 3, 4, 5 of CjCas9). b, c, e, f, g, Indels refer to insertion, deletions, or substitutions.
Since other PAM relaxed Cas9 variants often come with the trade-off of reduced activity35,36, we next benchmarked indel formation rates of evoCjCas9 with commonly used Cas9 orthologs (CjCas9, enCjCas9, SaCas9, SauriCas9, Nme2Cas9, SpCas9 - Fig. 3a), their PAM-relaxed variants (SaKKH, SauriKKH, eNme2-C.NR, SpRY and SpG). In addition, we benchmarked it to engineered UnCas12f1 (CasMINIv3.1) and transposon-associated ISDra2 TnpB1,3,29,32,36–41. For this purpose, we designed a self-targeting library where each target site contains the optimal protospacer length and PAM sequence for each ortholog (22 bp and N3AACAC for CjCas9, 22 bp and N2GRRT for SaCas9, 22 bp and N2GG for SauriCas9, 19 bp and NGG for SpCas9, 22 bp and N4CC for Nme2Cas9, 20 bp and TTTR for CasMINI, 20 bp and TTGAT for TnpB - Supplementary Data 5). The library was cloned into a lentiviral plasmid and after virus production, integrated into the genome of HEK293T cells. The cell pool was subsequently transfected with plasmids expressing the different RNA-guided endonuclease variants and after selection genomic DNA was extracted for analysis by HTS. Confirming previous studies, we found that PAM relaxed SaCas9-, SauriCas9-, Nme2Cas9- and SpCas9 variants showed lower activity than the wild-type orthologs on canonical PAMs (Fig. 3c)42,43. In contrast, evoCjCas9 showed substantially higher activity than wild-type CjCas9 on the canonical N3AACAC PAM sequence, which could be explained by the activity enhancing mutations that are also present in enCjCas9 (Fig. 3c). Cross-comparison to other Cas9 orthologs further revealed that evoCjCas9 works with similar efficiency as SpG, a frequently used SpCas9 variant detecting NGN PAM sequences that occur with comparable frequencies to that of evoCjCas9 PAM sequences. In addition, we observed that evoCjCas9 outperforms SpRY, the most PAM-relaxed SpCas9 variant that has been established so far, as well as the considerably smaller endonucleases ISDra2 TnpB and CasMINIv3.1 (Fig. 3c, d).
Low tolerance for mismatches between spacer and target site
Off-target editing at genomic sites that share sequence similarities to the targeted locus are limiting the clinical application of CRISPR-Cas nucleases. Therefore, we assessed the mismatch tolerance of wild-type and evoCjCas9. To this end, we designed a self-targeting library with 687 members, consisting of 37 target sites and up to 19 target matched off-target sites with 1-5 mismatches (Supplementary Data 6). The library was stably integrated into HEK293T cells using lentiviral vectors, and cells were transfected with plasmids expressing wild-type or evoCjCas9. HTS analysis of the target sites revealed that both variants have a similar mismatch tolerance; the mean off-target (1-5 mismatches) to on-target ratio for CjCas9 and evoCjCas9 was 0.36 and 0.39, respectively, with both variants not tolerating more than 2 mismatches between the spacer of the sgRNA and the protospacer (Fig. 3e). To further investigate how the location of the mismatches within the protospacer influences off-target editing activities, we designed a dual-mismatch permutation library for 10 CjCas9 target sites. As expected from previous observations with SpCas9, mismatches in the seed region of the sgRNA (position 14-22) were more detrimental to target recognition than PAM distal mismatches (Fig. 3f; Supplementary Fig. 6). Despite the similar mismatch tolerance of wild-type and evoCjCas9, we speculated that the higher prevalence of non-canonical PAM sites in the genome could lead to elevated off-target editing with evoCjCas9. Therefore, we also examined the specificity of CjCas9 and evoCjCas9 with two sgRNAs (hDYRK1a, hPTEN) in the human genome using circularization for high-throughput analysis of nuclease genome-wide effects by sequencing (CHANGE-seq)44 (Fig. 3g; Supplementary Fig. 7). In line with our hypothesis, HTS analysis of the top 5 CHANGE-seq identified off-target sites in treated HEK293T cells revealed elevated off-target editing with evoCjCas9 (Fig. 3g); with the hDYRK1a targeting sgRNA we obtained 2 confirmed off-target sites with evoCjCas9 vs. 1 confirmed off-target site with wild-type CjCas9, and with the hPTEN targeting sgRNA we obtained 3 vs. 0 confirmed off-target sites with evoCjCas9. Thus, similar to the observations made with PAM-relaxed SpCas9 variants36, the broader targeting scope of evoCjCas9 comes at the cost of lower specificity.
Efficient base- and prime editing with evoCjCas9
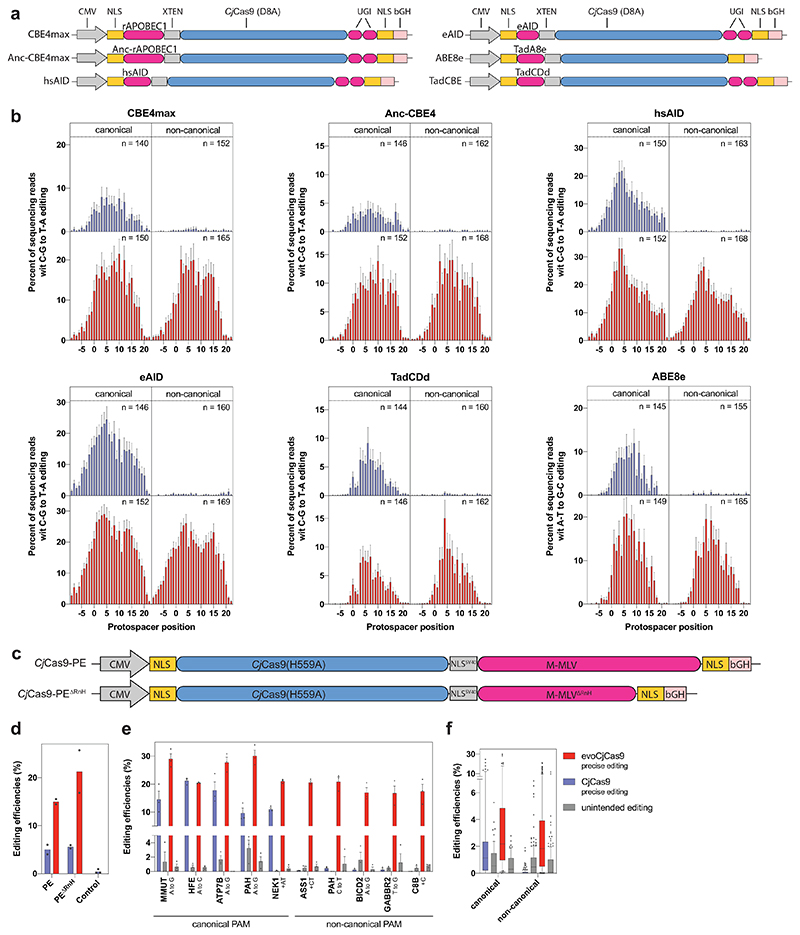
The broad PAM recognition and high activity of evoCjCas9 make it an ideal nuclease for generating size optimized BEs and PEs. To first assess base editing with evoCjCas9, we fused evo- or wild-type CjCas9 variants that nick the target strand (D8A) N-terminally to either an adenosine deaminase (ABE8e45) or to a cytosine deaminase (hsAIDmax6, eAID46, BE4max, AncBE4max47 or TadCBEd48) (Fig. 4a). Cytosine BEs were furthermore C-terminally fused to an uracil glycosylase inhibitor (UGI) to block the conversion of the installed U back to a C. We next designed a self-targeting library consisting of target sites with canonical and non-canonical PAM sites for base editing with CjCas9-BEs. The library was stably integrated into HEK293T cells using lentiviral vectors, which were subsequently transfected with plasmids expressing the different base editors. In line with our results obtained with the evoCjCas9 nuclease, evoCjCas9 BEs showed higher editing efficiency than wild-type CjCas9 BEs at canonical PAMs. In addition, evoCjCas9 BEs allowed base editing at non-canonical PAMs, where wild-type CjCas9 BEs were largely inactive (Fig. 4b; Supplementary Fig. 8). Similar to previously reported BEs established from other compact Cas9 orthologs20, CjCas9 BEs had a broader editing window than the typical editing window of SpCas9 BEs49. We reasoned that this could be caused by a less constrained R-loop formation. Hence, we next replaced the HNH domain of evoCjCas9 with a heterodimer of TadA-TadA8e to constrain the deaminase as close to the R-loop as possible. However, while the editing window of HNHxevoCjCas9 was indeed more constrained, similar to reports with HNHxSpCas9 BEs50, editing rates were substantially lower than for CjCas9 BEs with N-terminally fused TadA8e (Fig. 4b, last panel, and Supplementary Fig. 8). Finally, we also assessed if CjCas9 base editors show preferences for certain trinucleotide motifs (bases flanking the targeted A or C). Consistent with recent reports for SpCas9 BEs49, we found that evoCjCas9 BEs using the rAPOBEC1 deaminase show a preference for editing of C bases that are preceded by T, while evoCjCas9 BEs using AID show a preference for editing of C bases that are preceded by A or G. In addition, we observed that evoCjCas9 BEs using the TadA8e deaminase largely disfavor target bases with a preceding A (Supplementary Fig. 9).
Figure 4. Base- and Prime editing with CjCas9 and evoCjCas9.
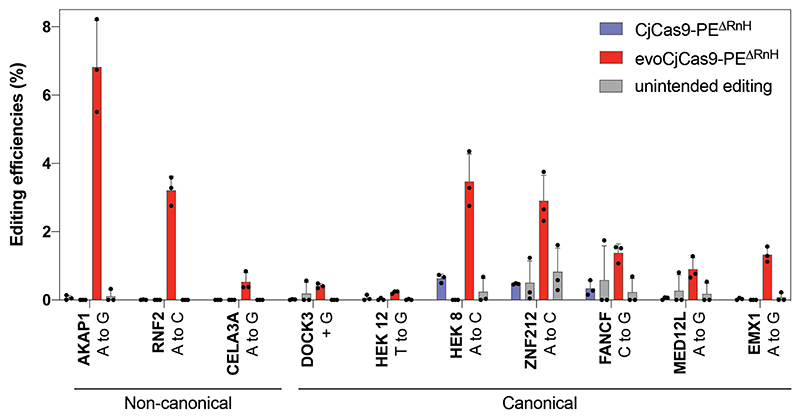
a, Illustration of expression vectors encoding for different CjCas9 BEs. b, Observed nucleotide transitions within the target sites with canonical and non-canonical PAMs. Bars represent mean of three independent biological replicates on N4ACAC, N4ATAC, N4GTAC, and N4GCAC (canonical) or N4AAAC, N4CAAC, N4GAAC and N4TAAC (non-canonical) PAM sites. Number of target sites (n) is indicated within each plot and bars represent mean of three independent biological replicates for each target site. Protospacer positions from -9 to 22 are shown. c, Illustration of expression vectors encoding for CjCas9 prime editors with the full length (top) or RnaseH depleted M-MLV reverse transcriptase (bottom). d, Comparison of CjCas9 or evoCjCas9 PEs installing an A to G transition at the AAVS1 locus. e, Mean editing efficiencies of CjCas9 or evoCjCas9-PEΔRnH on a selection of loci with canonical or non-canonical PAM sites. The loci of the target sites and type of edits are indicated. Bars (d, e) represent mean of n=2 (d) or n=3 (e) independent biological replicates in HEK293T cells, with error bars indicating standard error of the mean. f, Mean prime editing efficiencies and indel rates of biologically independent replicates (n=3) on 64 (canonical) and 172 (non-canonical) integrated target sites. Boxplots represent the 25th, 50th, and 75th percentiles. Whiskers indicate 5 and 95 percentiles. CjCas9 in blue, evoCjCas9 in red (b, d − f).
Prime editing, a more versatile genome editing technology than base editing, enables the introduction of all 12 possible base conversions as well as multi-base replacements and small insertions or deletions. To generate miniature PE systems based on CjCas9, we first introduced the H559A mutation into wild-type CjCas9 and evoCjCas9 to obtain variants that only nick the non-target strand. These variants were then C-terminally fused to either the full-length M-MLV reverse transcriptase (PE) or an M-MLV reverse transcriptase that lacks the RNaseH domain (PEΔRnH)19 (Fig. 4c). When we tested these variants by introducing an A to G mutation into the AAVS1 locus, we observed that wild-type CjCas9 PEs only achieved 6% editing while evoCjCas9 PEs achieved up to 26% editing, with no significant difference between full-length PE and PEΔRnH (Fig. 4d). We next generated a self-targeting library containing 236 prime editing guide (peg)RNAs paired with their target sites. These target sites include human disease loci with canonical- and non-canonical PAMs, which are corrected back to the wild-type sequence by their corresponding pegRNAs. The library was integrated into HEK293T cells using lentiviral vectors, and cells were subsequently transfected with plasmids expressing CjCas9-PEΔRnH or evoCjCas9-PEΔRnH. In line with the results from the AAVS1 locus, evoCjCas9-PEΔRnH achieved higher editing rates (up to 30.1%) than wild-type CjCas9-PEΔRnH (up to 24.6%) at target sites with canonical PAM sequences. In addition, evoCjCas9-PEΔRnH enabled editing at target sites with non-canonical PAMs (up to 22%), whereas wild-type CjCas9-PEΔRnH was unable to edit these sites (Fig. 4e; Supplementary Data 7). The same pattern, albeit with lower overall editing rates, was also observed on endogenous target sites (Extended Data Fig. 4) or when the library was integrated into K562 cells (Supplementary Fig. 10a). Further analysis of CjCas9 prime editing outcomes on the library revealed that single nucleotide replacements were more efficient than insertions or deletions, with A to C and C to T conversions being the most efficient edits (Supplementary Fig. 10b). Notably, within our library we also observed multiple loci with low or no prime editing, suggesting that at these sites our pegRNA design was not optimal (Fig. 4f). Therefore, we next assessed if we could identify pegRNA designs that correlate with editing rates in our library (n=238). We found that the most influential features correlating with CjCas9 prime editing efficiency were the melting temperature of the primer binding sequence (PBS) (R=0.43) and the reverse transcriptase template (RTT) (R=0.26, Supplementary Fig. 10c, d), suggesting that extending the pegRNA length downstream of the scaffold sequence may benefit editing rates.
In vivo base editing with single AAV constructs
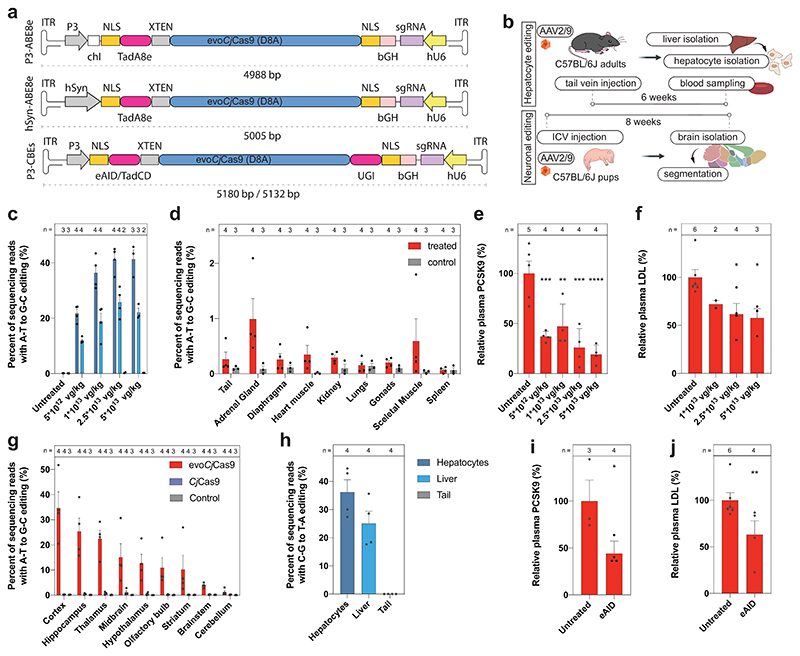
The small size of CjCas9 offers major advantages for in vivo genome editing as it facilitates packaging of BEs together with their sgRNAs on single AAV vectors. To test whether evoCjCas9 BE expression from single AAVs enables efficient editing in vivo in mice, we first cloned an AAV construct that expresses evoCjCas9-ABE8e under the hepatocyte-specific P3 promoter 51,52 and an sgRNA targeting the AG splice acceptor site of the Pcsk9 intron 3 under the hU6 promoter (Fig. 5a). We then packaged the evoCjCas9 BE expression construct spanning 4,988 bp (including ITRs) into hepatotropic AAV9 capsids, which were systemically administered into 5-week-old C57BL/6J mice at different doses between 5 x 1012 or 5 x 1013 vector genomes (vg)/kg body weight (Fig. 5b). 6 weeks post-injection we isolated genomic DNA from liver tissue and hepatocytes to assess editing rates. Since PCSK9 is a secreted protein that functions as a negative regulator of the low-density lipoprotein (LDL) receptor 53,54, we also collected serum from treated mice to analyze the effect of Pcsk9 editing on LDL cholesterol levels. HTS analysis revealed 41% editing in DNA isolated from hepatocytes and 22% editing in DNA isolated from whole liver tissue in mice treated with 5 x 1013 vg/kg (Fig. 5c). Editing remained at similar rates when the dose was reduced down to 1 x 1013 vg/kg, but decreased to 21% in hepatocytes at a dose of 5 x 1012 vg/kg (Fig. 5c). In other tissues editing was below 1%, consistent with the hepatocyte-specific activity of the P3 promoter (Fig. 5d)51,52. Analysis of the serum of treated mice, furthermore, revealed a significant reduction of PCSK9 protein levels and LDL cholesterol levels in animal injected with a dose of 2.5 x 1013 or 5 x 1013 vg/kg (Fig. 5e, f). Next, we assessed the feasibility of adenine base editing from single AAV vectors in the brain and exchanged the P3 promoter with the neuron-specific hSyn promoter55. In addition, we exchanged the PCSK9 sgRNA with an sgRNA targeting a non-canonical PAM site in exon 1 of GPR6, which is a neuronally expressed G protein-coupled receptor (Fig. 5a). Intracerebroventricular injection of AAV9 into newborns led to the installation of the anticipated L128S mutation with an efficiency of 34% in the cortex, and lower but still substantial editing in deeper brain regions (Fig. 5g). As expected, targeting the same locus with wild-type CjCas9-ABE8e, which does not recognize the non-canonical ‘CCGCCAAC’ PAM, did not lead to editing above background (Fig. 5g).
Figure 5. In vivo genome editing with compact evoCjCas9 adenine and cytosine BE.
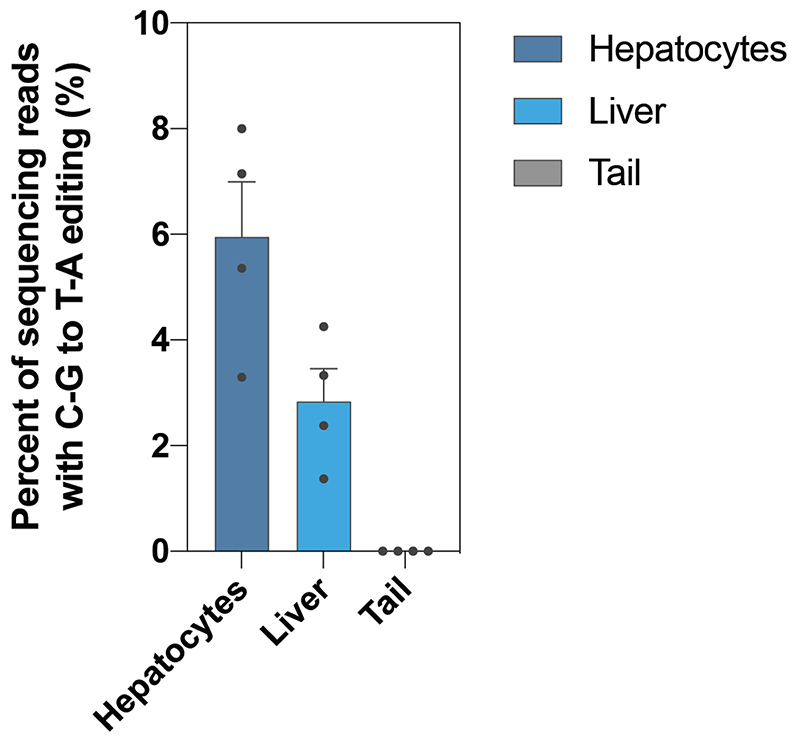
a, Schematic representation of single AAV vectors used for evoCjCas9 base editing. Elements are not illustrated to scale. b, Illustration of the experimental workflow for in vivo adenine or cytosine base editing at the Pcsk9 locus in the liver and of the Gpr6 locus in the brain. c, d, A-to-G editing at the targeted Pcsk9 splice site with different AAV concentrations, analyzed in isolated hepatocytes, whole liver samples, the tail (c, same legend as in h) and other tissues (d). e, Plasma PCSK9 levels relative to untreated control as determined by ELISA. ***P = 0.0006, **P = 0.0029, ***P = 0.0001, ****P < 0.0001 (left to right). f, Plasma LDL cholesterol levels. P = 0.2461, *P = 0.0278, *P = 0.027 (left to right). g, A-to-G editing at the targeted site in the Gpr6 locus (non-canonical CCGCCAAC PAM) in isolated brain tissues for CjCas9 and evoCjCas9. h, C-to-T editing at the targeted Pcsk9 site with evoCjCas9 cytosine base editor (eAID) in isolated hepatocytes, whole liver samples and the tail. i, Plasma PCSK9 levels relative to untreated control as determined by ELISA. *P = 0.0286. j, Plasma LDL cholesterol levels. **P = 0.0095. Means were compared using one-way ANOVA with Dunnett correction (e,f) or one-tailed t-test (Mann-Whitney) (i, j). Bars (c-j) represent mean with error bars representing standard error of the mean. Individual data points as black dots. Numbers (n) above plots indicate number of biologically independent datapoints. vg, vector genomes, chI, chimeric Intron, ICV, intracerebroventricular.
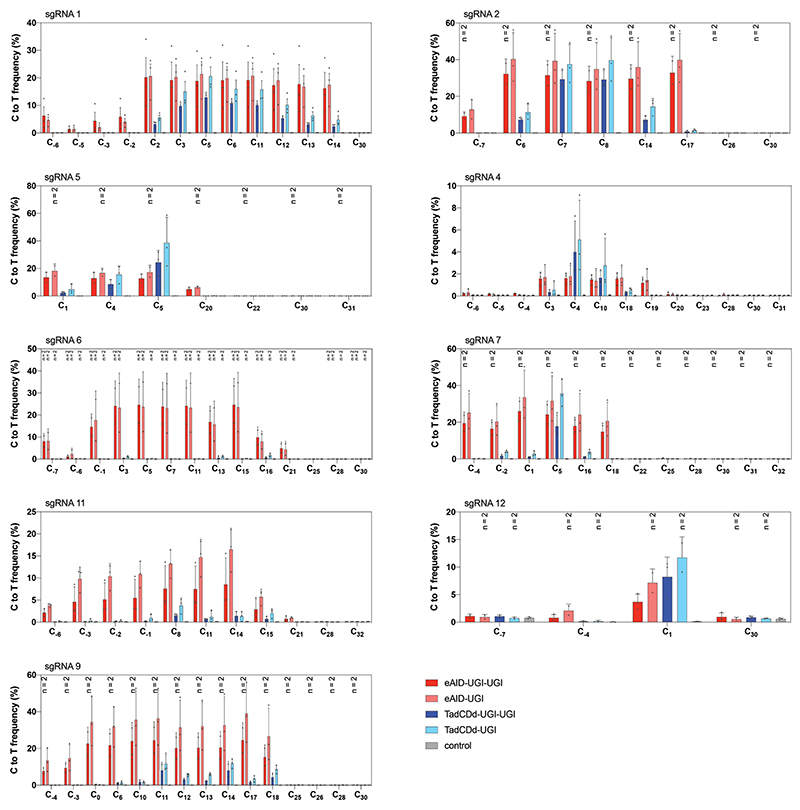
Finally, we assessed the feasibility of in vivo cytosine base editing from single AAV vectors using evoCjCas9. Since the size of APOBEC is quite large, we focused on CBEs utilizing the smaller eAID and TadCDd deaminases. First, we further reduced the size of these CBEs by removing one of the two UGI domains, which did not influence editing rates in Neuro-2a (N2a) cells (Extended Data Fig. 5). Next, we generated AAV vectors expressing the size-optimized evoCjCas9-eAID and evoCjCas9-TadCDd BEs under the P3 promoter, together with an sgRNA that leads to the installation of a CAA to TAA (Q259*) stop mutation in exon 5 of Pcsk9 (Fig. 5a). Systemic delivery of the evoCjCas9-eAID construct spanning 5,180 bp (including ITRs) via tail vein injection at an AAV dose of 2.5 x 1013 resulted in 25% editing in the whole liver and 36% editing in hepatocytes (Fig. 5h), and a significant reduction of plasma PCSK9 and LDL cholesterol levels (Fig. 5i, j). Systemic injection delivery of the TadCBE vector at a dose of 2.5 x 1013 also resulted in the installation of the Q259* mutation, but with lower efficiency (6% in hepatocytes and 2.8% in whole liver samples; Extended Data Fig. 6).Together, these data demonstrate that evoCjCas9 can be employed for efficient and tissue-specific in vivo adenine and cytosine base editing from single AAV vectors at clinically relevant doses56,57.
Discussion
The use of compact Cas9 orthologs is an attractive alternative to the SpCas9 nuclease, which due to the size of 1,368 aa is difficult to be delivered into cells in vivo. With 984 aa, CjCas9 is the smallest known Cas9 ortholog, which in principle fits as a nuclease or a BE fusion protein together with its sgRNA on a single AAV vector. However, like other small-sized Cas9 orthologs, wild-type CjCas9 recognizes a relatively long PAM, reducing the number of targetable sites, and is less efficient than SpCas9. Using phage-assisted evolution (PACE and PANCE), which allows screening of large sequence spaces to evolve proteins in bacterial cells25, we identified a set of amino acid mutations that led to the PAM relaxation of CjCas9.
The novel variant, termed evoCjCas9, can target many − but not all − N4AH and N5HA PAM sequences (see Fig. 2a). These occur 10-fold more frequently in the genome than the canonical N3VRYAC PAM sequences of wild-type CjCas9, and 25- or 6-fold more frequently than the canonical N4CYAT or N4CYA PAM sites of the recently discovered orthologues Cj2Cas9 and Cj3Cas922, respectively. In addition, evoCjCas9 possesses a nuclease activity which is higher than that of wild-type CjCas9 or other small Cas orthologues, and in range with the activity of PAM-relaxed SpCas9 variants such as SpG (Fig. 3c).
While the requirement of a 22 to 24 nucleotide long spacer sequence with a mismatch tolerance of less than 3 bases in principle makes evoCjCas9 a highly precise targetable endonuclease, the higher PAM availability leads to an increase in off-target editing compared to wild-type CjCas9. In addition, previous reports have shown the potential of CjCas9 to bind to and cut RNA58,59, as well as DNA in an sgRNA-independent manner60. Moreover, if a separate tracrRNA is used instead of an sgRNA, endogenous RNAs can act as guiding RNAs for CjCas9, inducing additional DNA damage58. These points warrant further safety investigations prior to clinical use of (evo)CjCas9.
By fusing deaminases or the M-MLV reverse transcriptase to D8A or H559A evoCjCas9 nickase variants we also constructed compact BEs and PEs. Analyzing evoCjCas9 BEs, we observed average editing rates above 20% for various cytosine and adenine BE architectures, albeit with wider editing windows compared to SpCas9 BEs. Similar observations have been reported previously with BEs built on other compact Cas9 proteins, such as SaCas9 and Nme2Cas920,21,45, implying differences in the R-loop accessibility between SpCas9 and smaller Cas9 orthologs. With evoCjCas9 PEs we also achieved >20% editing at specific target sites. Nevertheless, the average editing rates were considerably lower, and it might be necessary to optimize linker architectures or the CjCas9 pegRNA design to achieve robust editing across a broader range of loci.
Due to the small size of evoCjCas9 it can be packaged as an adenine- or cytosine BE on a single AAV vector. While adenine base editing from single AAV vectors has already been demonstrated previously with Cj3Cas922, SaCas920,61 and Nme2Cas921, to our knowledge this study provides the first demonstration for cytosine base editing from a single AAV vector.
Furthermore, the compact size of CjCas9 would also allow for expression of ABEs using tissue-specific promoters other than the demonstrated P3 and hSyn promoters. Examples include the hUPII (bladder; urothelium)62, ProA7 (eye; cone cells)63, Ksp-cadherin (kidney; renal tubules)64, α-MHC (cardiac muscle)65, CK8e or SPc5-12 (skeletal muscle)66,67 promoters.
Taken together, we used PANCE and PACE to develop evoCjCas9, a compact and highly active Cas9 nuclease with a broad targeting range. evoCjCas9 enables efficient base- and prime editing, and owing its small size facilitates in vivo genome editing from single AAV vectors.
Methods
General methods and cloning
PCRs were performed using the Q5 High-Fidelity DNA polymerase (New England Biolabs). All expression vectors were assembled using NEBuilder HiFi DNA assembly (New England Biolabs). Plasmids expressing sgRNAs were cloned using the T4 DNA ligase (New England Biolabs). Plasmids used in mammalian tissue culture were purified using NucleoBond Xtra Midi kits (Macherey-Nagel). Primer sequences (Microsynth AG, Switzerland) used are listed in Supplementary Data 8.
Phage-assisted continuous and non-continuous evolution
PANCE and PACE were performed as recently described23,25. The M13 filamentous selection phage encodes a catalytically dead CjCas9 fused via an Ala-Ala-Linker to the ω subunit of bacterial RNA polymerase. The accessory plasmid harbors the PAM and protospacer sequence in reverse, 114 nt upstream of gene III. To increase the mutagenesis rate, DP6 (Addgene #140446) was used and induced with L-arabinose (10 mM) one hour before selection phage addition. All experiments were performed using the S2060 bacterial strain (Addgene #105064), cultured in Davis Rich Media. PANCE was performed using PAM libraries with all possible nucleobase combinations for either PAM positions 5, 6, 7, or 8. PANCE lagoons were contained in deep well plates at 1 mL volume and selection phages were added once cultures reached the log phase (approx. OD 0.4 − 0.6). Phages were incubated overnight and harvested via centrifugation and sterile filtration (0.2 μm, Sarstedt) before adding a dilution to the next round (Supplementary Table 1). Phage titers were determined using plaque assays or RT-qPCR for higher throughput. qPCR results with CT > 30 were considered to resemble the absence of phages. Similarly, PACE was performed using a pool of PANCE-derived phage variants after round 12, mixed 1:1 with wild-type CjCas9 containing phages. On the accessory plasmid, a PAM library for positions 6-8 was used. S2060 host cells were continuously cultured at OD 0.5 in a custom-built miniature bioreactor. Host cells were pumped through a lagoon and the mutagenesis plasmid DP6 was induced with L-arabinose (10mM) before the cells reached the lagoon. The flow through the lagoon was controlled via a microcontroller and adjusted to increase the selection pressure over time. Phage titers in the bioreactor (no phages detected) and lagoon were monitored using RT-qPCR and phage plaque assays. Regular time points were collected and analysed using long-read Nanopore sequencing (Oxford Nanopore Technologies).
Phylogenetic analysis of directed evolution outcomes
Phage genotypes emerging during PACE were sequenced using a previously established method68. Briefly, from collected phages at different time points ω subunit-CjCas9 variants were PCR amplified using unique molecular identifiers (UMIs) and time point specific barcodes and were subcloned. Per time point, a few hundred colonies were selected and clonally amplified. Plasmids were purified and ω subunit-CjCas9 was extracted using unique restriction sites. Nanopore adapters were ligated, and variants were sequenced using the manufacturer’s protocol. Time points were demultiplexed and consensus reads for each UMI were built using the provided python script.
High-throughput PAM detection assay
HT-PAMDA was performed as recently described33. Cas9-variants were cloned in pCMV-T7-SpCas9-P2A-EGFP (Addgene #139987) and expressed in HEK293T cells for 48 hours. Whole-cell lysates were collected and normalized to a concentration corresponding to 150 nM fluorescein dye. Target-specific sgRNAs were in vitro transcribed using HiScribe T7 High Yield RNA Synthesis Kit. Substrate libraries with different target sites and PAM libraries were cloned into p11-LacY-wtxq (Addgene #69056). gRNA (1.1 μM) and normalized cell lysate (83 nM fluorescein) were complexed for 10 minutes at 37 °C. The RNP mixture (0.5 μM gRNA, 37.5 nM fluorescein lysate) was added to the substrate library (2.5 nM) and the reaction was stopped after different time intervals (1, 8, and 32 minutes). For all time points, substrate libraries were individually PCR amplified using time point-specific barcodes, followed by an amplification using protein variant-specific barcodes. Samples were pooled and sequenced on a NovaSeq 6000 (Illumina). Cas9-variants were characterized on two different target sites, with the PAM library spanning either positions 1 to 5 (Spacer 3) or 3 to 8 (Spacer 1 and 2). Two replicates were performed on different days. Sequencing data were processed using the provided python script. Calculated rate constants from normalized read counts for all characterized CjCas9 variants are listed in Supplementary Data 2.
Target library design using sgRNAs
Self-targeting constructs were designed with guide sequence preceding 5’-G nucleotide (not counted to total guide length) and ordered as single-stranded DNA oligo pools (Twist Bioscience, Supplementary Data 3-6). Oligo pools were amplified according to the manufacturer’s protocols using NEBNext Ultra II polymerase. The amplified pool was cloned into BsmBI-digested Lenti_gRNA-Puro (Addgene #84752) and electroporated into Endura electrocompetent cells (Lucigen). Colonies were harvested (> 2000x coverage) and plasmids were purified.
Target library screen using pegRNAs
A recently developed prime editing prediction tool (PRIDICT)69 was adapted to assist in the design of the pegRNAs for the library (Supplementary Data 7). The library was cloned at > 2000x coverage as described above. Upon harvest, plasmids were digested using BsmBI, purified, and the CjCas9 scaffold sequence was inserted via golden gate assembly. Purified ligation reactions were electroporated, colonies were harvested (> 2000x coverage) and plasmids were purified.
Cell culture and high-throughput sequencing
All cell lines were cultured at 37°C and 5% CO2 in cell culture incubators. HEK293T cells (ATCC CRL-3216) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) plus GlutaMAX (Thermo Fisher Scientific), K562 (ATCC CCL-243) cells were maintained in RPMI-1640 medium (Thermo Fisher Scientific) and Neuro-2a (ATCC CCL-131) cells were maintained in Eagle’s Minimum Essential Medium (EMEM), all supplemented with 10% (v/v) fetal bovine serum (FBS; Sigma-Aldrich) and 1% penicillin/streptomycin (v/v, Thermo Fisher Scientific). Cells were maintained at confluency below 90% and depending on library size seeded on 6-well/48-well cell culture plates or T175 cell culture flasks (Greiner). For base editor target library screens, HEK293T cells were transfected at 70% confluency (day 0) using 10 μl Lipofectamine 2000 (Thermo Fisher Scientific), 3 μg of base editor expression plasmid and 1 μg of Tol2-helper plasmid (Addgene #31828) according to the recommendation of the manufacturer.
Prime editing library screens in HEK293T were performed in T25 flasks by transfection of 2.64 μg of editor plasmid with OptiMEM (total volume of 75.14 μl) and 21.7 μl polyethylenimine (PEI, 1 mg/ml) (Day 0). PE screens in K562 were performed in 48-well plates (16x lipofection per editor or control) by seeding 100,000 K562 cells in 300 μl RPMI++ and transfection of 1000 ng editor plasmid with OptiMEM (total volume of 25 μl) mixed with 1.5 μl of Lipofectamine 2000 (Thermo Fisher Scientific) and 23.5 μl of OptiMEM. Controls for HEK293T and K562 were transfected without editor plasmid. Arrayed editing experiments in HEK293T were performed in 48-well plates by seeding 130,000 cells 6h before transfection. Transfection was performed with 750 ng of editor plasmid and 250 ng of gRNA plasmid with 1 μl of Lipofectamine 2000 as described above for K562 (day 0). On day 1, HEK293T (detached using 1x TrypLE (Gibco)) or K562 cells were transferred into blasticidin (7.5 mg/mL) containing media (no selection for controls) and maintained for 6 (PE) or 9 (BE) more days (unless otherwise noted).
Genomic DNA was isolated by direct lysis and locus-specific primers were used to generate targeted amplicons (NEBNext Ultra II) for deep sequencing, maintaining > 500x coverage. Cells were tested negative for Mycoplasma contamination and were authenticated by the supplier by STR analysis.
Lentiviral integration of self-targeting libraries
For lentivirus production, HEK293T cells were seeded at 15x106 cells in a T175 flask (Greiner) and transfected at 70% confluency using polyethylenimine (PEI). Briefly, 152 μl PEI (0.1 mg/ml) was mixed with 506 μl Opti-MEM containing 2.6 μg VSV-G (Addgene #12259), 5.2 μg PAX2 (Addgene #12260) and 10.4 μg lentiviral vector plasmid. The mix was incubated for 20 min at room temperature prior to transfection. One day post-transfection, media was replaced and three days post-transfection, the supernatant was harvested and purified by filtration (0.4 μm, Sarstedt). A dilution series of produced lentivirus was used to transduce HEK293T cells. 24 hours post-transduction, cells were selected with 2.5 μg/ml puromycin. Lentiviral concentration with a 50% survival rate 3 days post-transduction was used for subsequent library cell generation. Library cells were generated accordingly, with cell numbers being sufficient for > 2000x coverage (post-selection). 3 days post-transduction, puromycin-containing media was replaced, and 5 days post-transduction library cells were detached and cryopreserved.
Expression and purification of CjCas9 and evoCjCas9
DNA sequences encoding CjCas9 and evoCjCas9 were inserted into the 1S plasmid (Addgene #29659) using ligation-independent cloning. This created a construct containing an N-terminal His6-SUMO tag followed by a tobacco etch virus (TEV) protease cleavage site. The constructs were then expressed in E. coli BL21 Rosetta2 (DE3) cells (Novagen), followed by lysis in 20 mM Tris-HCl pH 8.0, 1 M NaCl, 20 mM imidazole, 1 μg/mL pepstatin, 200 μg/mL 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), 1 mM Tris-(2-Carboxyethyl)phosphine hydrochloride (TCEP) by ultrasonication. Clarified lysate was loaded on a 5 mL Ni-NTA (Sigma-Aldrich) affinity column. The column was washed with 20 mM Tris-HCl pH 8.0, 1 M NaCl, 20 mM imidazole, 1 mM TCEP and bound protein was eluted with 20 mM Tris-HCl pH 8.0, 300 mM NaCl, 300 mM imidazole, 1 mM TCEP. The eluted protein was dialyzed, overnight at 4°C, against a buffer yielding a final imidazole concentration of 40 mM imidazole, in the presence of 1 mM TCEP and TEV protease to remove the His6-SUMO tag. Cleaved protein was passed through a 5 mL Ni-NTA (Sigma-Aldrich) affinity column to remove the cleaved affinity tag and uncleaved protein; the column was washed with 20 mM Tris-HCl pH 8.0, 300 mM NaCl, 40 mM imidazole, 1 mM TCEP, and the flow-through and wash fractions were pooled for subsequent purification using a 5 mL HiTrap HP Heparin column (Cytiva), eluting with a linear gradient up to 1 M NaCl. The elution fractions were pooled, concentrated in the presence of 1 mM Dithiothreitol (DTT) and further purified by size exclusion chromatography using a Superdex 200 (16/600) column (Cytiva) in 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM DTT, yielding pure, monodispersed proteins. Aliquots were flash-frozen in liquid nitrogen and stored at -80 °C.
CHANGE-seq
The sgRNAs were first tested for functionality by digesting a PCR amplicon of the genomic target site. The library was prepared as previously described44. Data (NGS) were processed using the CHANGE-seq analysis pipeline (https://github.com/tsailabSJ/changeseq) with the following parameters: ‘read_threshold: 4, window_size: 3, mapq_threshold: 50, start_threshold: 1, Gap_threshold: 3, mismatch_threshold: 10, search_radius: 30, merged_analysis: True. Identified target sites were deep sequenced to be covered by at least 10,000 reads per site.
Adeno-associated virus production
Vectors (AAV2 serotype 9) were produced by the Viral Vector Facility of the Neuroscience Center Zurich. In brief, the AAV vectors underwent ultracentrifugation and diafiltration. Physical titers (vg/mL) were measured using a Qubit 3.0 fluorometer as previously described.70 Identity of the packaged genomes of each AAV vector were confirmed as size verification of PCR amplicon (gel electrophoresis) and Sanger sequencing.
Animal studies
Mouse experiments were conducted in accordance with protocols approved by the Kantonales Veterinäramt Zürich. Mice were kept in a pathogen-free animal facility at the Institute of Pharmacology and Toxicology at UZH Zurich, maintained under controlled temperature and humidity conditions (21°C, 50% RH) on a 12-h light/dark cycle. Prior to collection of blood from the inferior vena cava and liver perfusion, the mice were fasted for 3−4 h. Each animal was injected with 0.5-1 x 1012 AAV vector genomes per mouse, with injection volumes ranging between 120−150 μl. The experiments were exclusively performed on male C57BL/6J animals at an age of 5-6 weeks.
Primary hepatocyte isolation
Mice were euthanized using CO2 and immediately had Hank’s balanced salt solution (Thermo Fisher Scientific) plus 0.5 mM EDTA perfused through the inferior vena cava, followed by an incision in the portal vein. To obtain whole liver samples for lysate preparation, one liver lobe was excluded from the perfusion by a thread tie. Following liver blanching, the mice underwent perfusion with digestion medium (low-glucose DMEM plus 1x penicillin−streptomycin (Thermo Fisher Scientific), 15 mM HEPES and freshly added Liberase (Roche)) for 5 minutes. Livers were extracted in cold isolation medium (low-glucose DMEM supplemented with 10% (vol/vol) FBS, 1x penicillin−streptomycin (Thermo Fisher Scientific) and GlutaMax (Thermo Fisher Scientific)), and the perfused liver was gently dissociated to create a cell suspension that was strained through a 100-μm filter. The suspension was centrifuged at 50g for 2 min and rinsed with isolation medium 2−3 times until a clear supernatant was achieved. For direct lysis and NGS sample preparation, primary hepatocytes were concentrated via centrifugation.
Clinical chemistry
The levels of PCSK9 in mice were assessed using the Mouse Proprotein Convertase 9/PCSK9 Quantikine ELISA Kit (R&D Systems, cat. no. MPC900) in alignment with the manufacturer’s guidelines. The absorbance was recorded at 450 nm and background at 540 nm; the background was subtracted for the calculation of the final values. Routine parameters (cholesterol, triglyceride and high-density lipoprotein) from all mouse samples were analysed at the Division of Clinical Chemistry and Biochemistry at the University Children’s Hospital Zurich using Alinity ci-series. Low-density lipoprotein levels were determined using the Friedewald equation.
HTS data analysis
Libraries were sequenced on a MiSeq or NovaSeq 6000 (Illumina, 150bp, paired-end). Amplicon sequences were analyzed using custom Python scripts or aligned to their reference sequences using CRIPResso271. In brief, editing rates for BE and PE libraries were determined by calculating the ratio of edited bases (BE) or sequence-stretches (PE; 2bp before nick until 5bp after flap) and subsequently correcting editing rates with background rates from control samples as previously described69.
Statistical analyses
Statistical evaluations were conducted with GraphPad Prism 9.0.0 for macOS or SciPy (1.6.3). Information regarding the sample sizes and the specific statistical tests utilized can be found in the legends of the figures. A p-value less than 0.05 was deemed to indicate statistical significance.
Extended Data
Extended Data Fig. 1.
Extended Data Fig. 2.
Extended Data Fig. 3.
Extended Data Fig. 4.
Extended Data Fig. 5.
Extended Data Fig. 6.
Supplementary Material
Acknowledgments
We thank the Functional Genomics Center Zurich for technical support and access to instruments at the University of Zurich and ETH Zurich, the Genome Engineering and Measurement Lab at the University of Zurich and ETH Zurich and Susanne Kreutzer for preparing CHANGE-seq libraries, the mRNA platform at UZH/USZ and Steve Pascolo, Julia Frei and Conrad Wyss for production and purification of RNAs, the viral vector facility of UZH and Jean-Charles Paterna and Melanie Rauch for production of AAVs, Johannes Häberle and Nicole Rimann for support with blood sample analysis and Sasha Melkonyan for assistance in data visualization, Lukas Villiger and members of the Schwank lab for discussions. We thank Martin Pacesa for comments on the manuscript. This work was supported by the Swiss National Science Foundation (SNSF) grant no. 310030_185293, 310030_214936 (to G.S.) and 31003A_182567 (to M.J.), Novartis Foundation for Medical-Biological Research no. FN20-0000000203 (to D.B.), SNSF Spark fellowship no. 196287 (to D.B.), the URPP Itinerare (to G.S. and to D.B.) and the ETH PhD fellowship (to L.S. and K.M.). M.J. is an International Research Scholar of the Howard Hughes Medical Institute and Vallee Scholar of the Bert L & N Kuggie Vallee Foundation.
Footnotes
Author Contributions
L.S. and G.S. designed the study and wrote the manuscript. L.S., K.F.M., N.M., T.R., C.C., D.B. and J.P.W. performed and analysed in vitro experiments. L.S., T.R., L.K., K.F.M and J.H. performed and analysed in vivo experiments. C.C. and M.J. purified proteins and provided field-specific expertise. L.S., N.M., K.F.M., T.R., D.B. and C.C. prepared figures. All authors reviewed the manuscript.
Competing Interests
L.S. and G.S. have filed a patent application based on evolved CjCas9 variants (European Patent Application No. 23175382.3). The remaining authors declare no competing interests.
Data Availability
Plasmids and sequences of evoCjCas9 constructs are available on Addgene: evoCjCas9 (#194059), evoCjCas9-ABE8e (#194060), evoCjCas9-BE4max (#194061), evoCjCas9-eAID (#194062), evoCjCas9-PEmaxΔRnH (#194063), evoCjCas9-TadCDd (#202558), LentiGuide_CjCas9-Puro (#202564), AAV-P3-evoCjCas9-ABE8e (#202559), AAV-hSyn-evoCjCas9-ABE8e (#202561), AAV-P3-evoCjCas9-eAID (#202562) and AAV-P3-evoCjCas9-TadCDd (#202563). Measured editing rates are provided in Supplementary Data 3-7. DNA-sequencing data is available on NCBI Sequence Read Archive (PRJNA893560).
Code Availability
Custom code used in data analysis is available as Supplementary Code 1 and on GitHub (https://github.com/Schwank-Lab/evoCjCas9).
References
- 1.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zetsche B, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–91. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickar-Oliver A, Gersbach CA. The next generation of CRISPR−Cas technologies and applications. Nat Rev Mol Cell Biol. 2019;20:490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaudelli NMM, et al. Programmable base editing of T to G C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anzalone AV, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeh WH, Chiang H, Rees HA, Edge ASB, Liu DR. In vivo base editing of post-mitotic sensory cells. Nat Commun. 2018;9:1–10. doi: 10.1038/s41467-018-04580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koblan LW, et al. In vivo base editing rescues Hutchinson−Gilford progeria syndrome in mice. Nature. 2021;589:608–614. doi: 10.1038/s41586-020-03086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deverman BE, Ravina BM, Bankiewicz KS, Paul SM, Sah DWY. Gene therapy for neurological disorders: Progress and prospects. Nat Rev Drug Discov. 2018;17:641–659. doi: 10.1038/nrd.2018.110. [DOI] [PubMed] [Google Scholar]
- 13.Mendell JR, et al. Current Clinical Applications of In Vivo Gene Therapy with AAVs. Molecular Therapy. 2021;29:464–488. doi: 10.1016/j.ymthe.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy JM, et al. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat Biomed Eng. 2020;4:97–110. doi: 10.1038/s41551-019-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng C, et al. A flexible split prime editor using truncated reverse transcriptase improves dual-AAV delivery in mouse liver. Molecular Therapy. 2022;30:1343–1351. doi: 10.1016/j.ymthe.2022.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Truong D-JJ, et al. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. 2015;43:6450–6458. doi: 10.1093/nar/gkv601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villiger L, et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med. 2018;24:1519–1525. doi: 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- 18.Rothgangl T, et al. In vivo adenine base editing of PCSK9 in macaques reduces LDL cholesterol levels. Nat Biotechnol. 2021;39:1–9. doi: 10.1038/s41587-021-00933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Böck D, et al. In vivo prime editing of a metabolic liver disease in mice. Sci Transl Med. 2022;14:9238. doi: 10.1126/scitranslmed.abl9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis JR, et al. Efficient in vivo base editing via single adeno-associated viruses with size-optimized genomes encoding compact adenine base editors. Nat Biomed Eng. 2022;6:1–12. doi: 10.1038/s41551-022-00911-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, et al. Adenine Base Editing In Vivo with a Single Adeno-Associated Virus Vector. GEN Biotechnology. 2022;1:285–299. doi: 10.1089/genbio.2022.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S, et al. Compact Cje3Cas9 for Efficient in Vivo Genome Editing and Adenine Base Editing. CRISPR Journal. 2022;5:472–486. doi: 10.1089/crispr.2021.0143. [DOI] [PubMed] [Google Scholar]
- 23.Hu JH, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011;472:499–503. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller SM, Wang T, Liu DR. Phage-assisted continuous and non-continuous evolution. Nat Protoc. 2020;15:4101–4127. doi: 10.1038/s41596-020-00410-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pu J, Disare M, Dickinson BC. Evolution of C-Terminal Modification Tolerance in Full-Length and Split T7 RNA Polymerase Biosensors. ChemBioChem. 2019;20:1547–1553. doi: 10.1002/cbic.201800707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeBenedictis EA, et al. Systematic molecular evolution enables robust biomolecule discovery. Nat Methods. 2022;19:55–64. doi: 10.1038/s41592-021-01348-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Badran AH, Liu DR. Development of potent in vivo mutagenesis plasmids with broad mutational spectra. Nat Commun. 2015;6:8425. doi: 10.1038/ncomms9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim E, et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun. 2017;8:1–12. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamada M, et al. Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems. Mol Cell. 2017;65:1109–1121.:e3. doi: 10.1016/j.molcel.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 31.Miller SM, et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat Biotechnol. 2020;38:471–481. doi: 10.1038/s41587-020-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang TP, et al. High-throughput continuous evolution of compact Cas9 variants targeting single-nucleotide-pyrimidine PAMs. Nat Biotechnol. 2023;41:96–107. doi: 10.1038/s41587-022-01410-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walton RT, Hsu JY, Joung JK, Kleinstiver BP. Scalable characterization of the PAM requirements of CRISPR−Cas enzymes using HT-PAMDA. Nat Protoc. 2021;16:1511–1547. doi: 10.1038/s41596-020-00465-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakagawa R, et al. Engineered Campylobacter jejuni Cas9 variant with enhanced activity and broader targeting range. Commun Biol. 2022;5:1–8. doi: 10.1038/s42003-022-03149-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mir A, Edraki A, Lee J, Sontheimer EJ. Type II-C CRISPR-Cas9 Biology, Mechanism, and Application. ACS Chem Biol. 2018;13:357–365. doi: 10.1021/acschembio.7b00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walton RT, Christie KA, Whittaker MN, Kleinstiver BP. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368:290–296. doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Z, et al. A compact Cas9 ortholog from Staphylococcus Auricularis (SauriCas9) expands the DNA targeting scope. PLoS Biol. 2020;18:e3000686. doi: 10.1371/journal.pbio.3000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu X, et al. Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Mol Cell. 2021;81:4333–4345.:e4. doi: 10.1016/j.molcel.2021.08.008. [DOI] [PubMed] [Google Scholar]
- 40.Karvelis T, et al. Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature. 2021;599:692–696. doi: 10.1038/s41586-021-04058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edraki A, et al. A Compact, High-Accuracy Cas9 with a Dinucleotide PAM for In Vivo Genome Editing. Mol Cell. 2019;73:714–726.:e4. doi: 10.1016/j.molcel.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Legut M, et al. High-Throughput Screens of PAM-Flexible Cas9 Variants for Gene Knockout and Transcriptional Modulation. Cell Rep. 2020;30:2859–2868.:e5. doi: 10.1016/j.celrep.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collias D, Beisel CL. CRISPR technologies and the search for the PAM-free nuclease. Nat Commun. 2021;12:1–12. doi: 10.1038/s41467-020-20633-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lazzarotto CR, et al. CHANGE-seq reveals genetic and epigenetic effects on CRISPR−Cas9 genome-wide activity. Nat Biotechnol. 2020;38:1317–1327. doi: 10.1038/s41587-020-0555-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richter MF, et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat Biotechnol. 2020;38:883–891. doi: 10.1038/s41587-020-0453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Z, et al. Improved base editor for efficient editing in GC contexts in rabbits with an optimized AID-Cas9 fusion. The FASEB Journal. 2019;33:9210–9219. doi: 10.1096/fj.201900476RR. [DOI] [PubMed] [Google Scholar]
- 47.Koblan LW, et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat Biotechnol. 2018;36:843–848. doi: 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neugebauer ME, et al. Evolution of an adenine base editor into a small, efficient cytosine base editor with low off-target activity. Nat Biotechnol. 2022:1–13. doi: 10.1038/s41587-022-01533-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marquart KF, et al. Predicting base editing outcomes with an attention-based deep learning algorithm trained on high-throughput target library screens. Nat Commun. 2021;12:1–9. doi: 10.1038/s41467-021-25375-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villiger L, et al. Replacing the SpCas9 HNH domain by deaminases generates compact base editors with an alternative targeting scope. Mol Ther Nucleic Acids. 2021;26:502–510. doi: 10.1016/j.omtn.2021.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nair N, et al. Computationally designed liver-specific transcriptional modules and hyperactive factor IX improve hepatic gene therapy. Blood. 2014;123:3195–3199. doi: 10.1182/blood-2013-10-534032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grisch-Chan HM, et al. Low-Dose Gene Therapy for Murine PKU Using Episomal Naked DNA Vectors Expressing PAH from Its Endogenous Liver Promoter. Mol Ther Nucleic Acids. 2017;7:339–349. doi: 10.1016/j.omtn.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sahng WP, Moon YA, Horton JD. Post-transcriptional Regulation of Low Density Lipoprotein Receptor Protein by Proprotein Convertase Subtilisin/Kexin Type 9a in Mouse Liver. Journal of Biological Chemistry. 2004;279:50630–50638. doi: 10.1074/jbc.M410077200. [DOI] [PubMed] [Google Scholar]
- 54.Musunuru K, et al. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature. 2021;593:429–434. doi: 10.1038/s41586-021-03534-y. [DOI] [PubMed] [Google Scholar]
- 55.Kügler S, Kilic E, Bähr M. Human synapsin 1 gene promoter confers highly neuron-specific long-term transgene expression from an adenoviral vector in the adult rat brain depending on the transduced area. Gene Ther. 2003;10:337–347. doi: 10.1038/sj.gt.3301905. [DOI] [PubMed] [Google Scholar]
- 56.High-dose AAV gene therapy deaths. Nat Biotechnol. 2020;38:910. doi: 10.1038/s41587-020-0642-9. [DOI] [PubMed] [Google Scholar]
- 57.Morales L, Gambhir Y, Bennett J, Stedman HH. Broader Implications of Progressive Liver Dysfunction and Lethal Sepsis in Two Boys following Systemic High-Dose AAV. Molecular Therapy. 2020;28:1753–1755. doi: 10.1016/j.ymthe.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dugar G, et al. CRISPR RNA-Dependent Binding and Cleavage of Endogenous RNAs by the Campylobacter jejuni Cas9. Mol Cell. 2018;69:893–905.:e7. doi: 10.1016/j.molcel.2018.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiao C, et al. Noncanonical crRNAs derived from host transcripts enable multiplexable RNA detection by Cas9. Science. 2021;372:941–948. doi: 10.1126/science.abe7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saha C, et al. Guide-free Cas9 from pathogenic Campylobacter jejuni bacteria causes severe damage to DNA. Sci Adv. 2020;6:4849–4866. doi: 10.1126/sciadv.aaz4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen Tran MT, et al. Engineering domain-inlaid SaCas9 adenine base editors with reduced RNA off-targets and increased on-target DNA editing. Nat Commun. 2020;11:1–10. doi: 10.1038/s41467-020-18715-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu HJ, et al. Cloning and analysis of human UroplakinII promoter and its application for gene therapy in bladder cancer. Cancer Gene Ther. 2004;11:263–272. doi: 10.1038/sj.cgt.7700672. [DOI] [PubMed] [Google Scholar]
- 63.Jüttner J, et al. Targeting neuronal and glial cell types with synthetic promoter AAVs in mice, non-human primates and humans. Nat Neurosci. 2019;22:1345–1356. doi: 10.1038/s41593-019-0431-2. [DOI] [PubMed] [Google Scholar]
- 64.Bai Y, Pontoglio M, Hiesberger T, Sinclair AM, Igarashi P. Regulation of kidney-specific Ksp-cadherin gene promoter by hepatocyte nuclear factor-1. Am J Physiol Renal Physiol. 2002;283 doi: 10.1152/ajprenal.00128.2002. [DOI] [PubMed] [Google Scholar]
- 65.Pacak CA, Sakai Y, Thattaliyath BD, Mah CS, Byrne BJ. Tissue specific promoters improve specificity of AAV9 mediated transgene expression following intra-vascular gene delivery in neonatal mice. Genet Vaccines Ther. 2008;6:13. doi: 10.1186/1479-0556-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gonçalves MAFV, et al. Transcription Factor Rational Design Improves Directed Differentiation of Human Mesenchymal Stem Cells Into Skeletal Myocytes. Molecular Therapy. 2011;19:1331–1341. doi: 10.1038/mt.2010.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li X, Eastman EM, Schwartz RJ, Draghia-Akli R. Synthetic muscle promoters: Activities exceeding naturally occurring regulatory sequences. Nat Biotechnol. 1999;17:241–245. doi: 10.1038/6981. [DOI] [PubMed] [Google Scholar]
- 68.Zurek PJ, Knyphausen P, Neufeld K, Pushpanath A, Hollfelder F. UMI-linked consensus sequencing enables phylogenetic analysis of directed evolution. Nat Commun. 2020;11:1–10. doi: 10.1038/s41467-020-19687-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mathis N, et al. Predicting prime editing efficiency and product purity by deep learning. Nat Biotechnol. 2023:1–9. doi: 10.1038/s41587-022-01613-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Düring DN, et al. Fast Retrograde Access to Projection Neuron Circuits Underlying Vocal Learning in Songbirds. Cell Rep. 2020;33:108364. doi: 10.1016/j.celrep.2020.108364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clement K, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol. 2019;37:224–226. doi: 10.1038/s41587-019-0032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plasmids and sequences of evoCjCas9 constructs are available on Addgene: evoCjCas9 (#194059), evoCjCas9-ABE8e (#194060), evoCjCas9-BE4max (#194061), evoCjCas9-eAID (#194062), evoCjCas9-PEmaxΔRnH (#194063), evoCjCas9-TadCDd (#202558), LentiGuide_CjCas9-Puro (#202564), AAV-P3-evoCjCas9-ABE8e (#202559), AAV-hSyn-evoCjCas9-ABE8e (#202561), AAV-P3-evoCjCas9-eAID (#202562) and AAV-P3-evoCjCas9-TadCDd (#202563). Measured editing rates are provided in Supplementary Data 3-7. DNA-sequencing data is available on NCBI Sequence Read Archive (PRJNA893560).
Custom code used in data analysis is available as Supplementary Code 1 and on GitHub (https://github.com/Schwank-Lab/evoCjCas9).