Abstract
Background and Aim:
The high diversity of Aeromonas spp. results in various pathogenicity levels. This group of bacteria causes a serious disease named motile Aeromonas septicemia (MAS) in catfish (Clarias spp.). This study aimed to characterize the species and virulence gene diversity of Aeromonas spp. isolated from diseased catfish.
Materials and Methods:
Nine Aeromonas spp. were isolated from infected catfish cultivated in Java, Indonesia, and they were identified at the phenotypic and molecular levels (16S rDNA). The virulence genes assessed included aer/haem, alt, ast, flaA, lafA, and fstA.
Results:
Phylogenetic analysis identified nine isolates of Aeromonas spp.: Aeromonas hydrophila (11.11%), Aeromonas caviae (11.11%), Aeromonas veronii bv. veronii (44.44%), and Aeromonas dhakensis (33.33%). Virulence genes, such as aer/haem, alt, ast, flaA, lafA, and fstA, were detected in all isolates at frequencies of approximately 100%, 66.67%, 88.89%, 100%, 55.56%, and 66.67%, respectively. This study is the first report on A. dhakensis recovered from an Indonesian catfish culture. Furthermore, our study revealed the presence of A. veronii bv veronii, a biovar that has not been reported before in Indonesia.
Conclusion:
This finding confirms that MAS was caused by multiple species of Aeromonas, notably A. dhakensis and A. veronii bv veronii, within Indonesian fish culture. The presence of these Aeromonas species with multiple virulence genes poses a significant threat to the freshwater aquaculture industry.
Keywords: 16S rDNA, aeromoniasis, motile Aeromonas septicemia, pathogenicity, phenotype, phylogenetic
Introduction
Catfish (Clarias spp.) is a freshwater aquaculture species with global potential. However, the disease caused by Aeromonas spp. is one of the most serious and devastating causes of losses in catfish culture [1–3]. These Gram-negative, oxidase-positive, and facultative anaerobic, bacteria normally live in an aquatic environment [4–6]. Aeromonas spp. causes considerable losses to the aquaculture industry and ubiquitous pathogens. Several Aeromonas spp. can infect marine fish, other organisms [7–10], and humans [11].
Most Aeromonas spp. cause aeromoniasis or motile Aeromonas septicemia (MAS), which features specific clinical signs of erosion and hemorrhage in the mouth and on the body surface, erosion, hyperemia, congestion, abscess, necrosis, dropsy, abdominal dropsy, abdominal ascites, and exophthalmia [12–18]. Infection by Aeromonas spp. also causes histological changes in the organs of the affected fish. Such conditions result in decreased levels of hemocyte aggregation in the digestive system of infected fish, massive aggregation of hemocytes and pyknotic nuclei in the hepatopancreas, and hemocyte aggregation and necrotic cells in gills [19]. In humans, Aeromonas spp. infect skin, soft tissues, and gastrointestinal tract and causes hepatobiliary disease, diarrhea, and bacteremia [11, 20, 21].
Twenty-four species of the Aeromonas complex had been identified as of 2010 [22]. By 2017, this number had grown to 32 [23]. The Aeromonas complex includes 36 species as reported in 2020 [24], which indicates the possibility of further expansion of this genus.
Based on biochemical characterization and 16S rDNA sequences, the main etiologic agents for aeromoniasis in eels in Europe, Japan, and Korea include Aeromonas hydrophila, Aeromonas veronii, Aeromonas sobria, Aeromonas caviae, Aeromonas jandaei, Aeromonas aquariorum, Aeromonas media, and Aeromonas trota [25–27]. These species may also infect other aquaculture species, including catfish. In addition to its high species diversity, most members of the genus Aeromonas include are pathogenic. Their pathogenicity is related to their virulence factors. The complex virulence of Aeromonas features is caused by some factors contributing to the development of the infection process [28]. The virulence is strictly related to the expression of genes recognized in Aeromonas spp. Extracellular products, such as hemolysin, protease and lipase, toxin, and structural components (flagella, fimbriae, lipopolysaccharide, and outer-membrane protein), whether acting together or individually, enable the microorganisms to colonize and infect the host [29]. Several virulence genes have been identified to play a role in the pathogenicity of Aeromonas spp. [30].
The total catfish production in Indonesia reached 1.06 million tons in 2021 and 1.03 million tons in 2020, which indicate a production increment of 2.8% [31]. Disease outbreaks have resulted in a low increment in total fish production in Indonesia in 2020–2021. The previous reports by Mulia et al. [32], Marnis and Iswanto [33], Sarjito et al. [34], Rejeki et al. [35], and Yuliantoro et al. [36] have described Aeromonas spp. as a causative disease agent in catfish. The Aeromonas species identified from fish in Indonesia include A. hydrophila, Aeromonas salmonicida, Aeromonas sobria, A. caviae, A. media, Aeromonas taiwanensis, and A. jandaei [37–39], while those from pond water comprise A. hydrophila and A. veronii [40]. Records of the diverse types of pathogenic Aeromonas are not yet available at the global level or in Indonesia. Therefore, a study on the etiological agents of catfish aeromoniasis is required. In addition, the identification of Aeromonas spp. based on phenotype characteristics along is very difficult and inaccurate. Thus, genotypic identification supported by phenotypic characters and virulence data is required.
This study aimed to characterize the species and virulence gene diversity of Aeromonas spp. isolated from diseased catfish.
Materials and Methods
Ethical approval
The management, conditions, and procedures of the experiment in this study were approved by the Ethical Clearance Commission of Universitas Gadjah Mada (approval # certificate: 00137/04/LPPTI/|/201).
Study period and location
The study was conducted from May 2017 to January 2019. The samples were collected from aquaculture ponds in four provinces in the Special Region of Yogyakarta, Central Java, West Java, and East Java, Indonesia.
Isolation and bacterial culture
Sixty-seven fish with a size of 3.5–30 cm was obtained. Bacteria were isolated from the kidneys of diseased catfish The bacteria were cultured in glutamate starch phenyl (GSP) medium (Merck, Darmstadt, Germany), a selective medium for Aeromonas and Pseudomonas, at 30°C for 24 h. Aeromonas and Pseudomonas grew in yellow and red colonies, respectively. Furthermore, yellow single colonies were recovered in the tryptic soy broth (TSB) medium (Merck). The isolates were stored in TSB medium (Merck) with 20% glycerol at −80°C for further assay.
Phenotypic identification of Aeromonas spp.
Phenotypic identification was performed by observing the colonies, cell morphology, and biochemical properties of the bacteria. The bacteria were grown on tryptic soy agar (TSA) medium (Merck) at 30°C for 24 h. Biochemical characterizations included the oxidative/fermentative (F), Voges-Proskauer (VP) test, citrate utilization, lysine utilization, ornithine utilization, urease production, phenylalanine deamination, nitrate reduction, H2S production, glucose utilization and gas production, adonitol, lactose, arabinose, sorbitol utilization, and aesculin hydrolysis which were conducted using the biochemical identification kit (HiMedia, India).
Bacterial genomic DNA extraction
A total of 1 mL of bacterial culture in TSB medium was incubated at 30°C for 24 h and centrifuged at 13,000× g for 2 min. The bacterial genomic DNA was extracted using a bacterial DNA kit (Promega, Madison, USA) in accordance with the manufacturer’s protocol. The extracted bacterial DNA was stored at −20°C for further assay.
16S rDNA amplification
The 16S rDNA was amplified using the universal oligonucleotide primers 27F and 1492R (Table-1) [31–35]. The total volume of 25 μL contained 12 μL Mytaq HS Red Mix (Bioline, Meridian Life Science, Memphis, UK), 2×polymerase chain reaction [PCR] Master Mix (Bioline, Meridian Life Science, Memphis, UK), 1 μL forward primer, 1 μL reverse primer, 1 μL DNA template (20 ng), and 10 μL nuclease-free water (NFW) [16]. The PCR cycling program was implemented as follows: An initial denaturation at 95°C for 3 min; 30 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 90 s; and a final extension at 72°C for 5 min. The PCR products were separated by electrophoresis on a 1% agarose gel before sequencing (1st BASE Laboratories, Selangor, Malaysia).
Table-1.
Primer sets used in this study.
| Gene | Gene product | Primer sequence (5’- 3’) | Product size (bp) | Annealing temperature (°C) | Reference |
|---|---|---|---|---|---|
| 16 S | 16 S rRNA gene | F: AGA GTT TGA TCM TGG CTC AG R: TAC GGY TAC CTT GTT ACG ACT T | 1500 | 55 | [31] |
| aerA/haem | Aerolysin/Hemolysin | F: CCT ATG GCC TGA GCG AGA AG R: CCA GTT CCA GTC CCA CCA CT | 431 | 56 | [32] |
| Alt | Heat-labile cytotonic enterotoxin | F: TGA CCC AGT CCT GGC ACG GC R: GGT GAT CGA TCA CCA CCA GC | 442 | 56 | [33] |
| ast | Heat-stabile cytotonic enterotoxin | F: TCT CCA ATG CTT CCC TTC ACT R: GTG TAG GGA TTG AAG AAG CCG | 331 | 56 | [33] |
| flaA | Polar flagellum | F: TCC AAC CGT YTG ACC TC R: GMY TGG TTG CGR ATG GT | 608 | 56 | [33] |
| lafA | Lateral flagellum | F: CCA ACT T (T/C) G C (C/T) T C (T/C) (C/A) TGA CC R: TCT TGG TCA T (G/A) T TGG TGC T (C/T) | 736 | 50 | [34] |
| fstA | Ferric siderophore receptor | F: CGC TCG CCC ATC CCC CTC TG R: GCC CCT TGC ACC CCC ACC ATT | 452 | 55 | [35] |
Detection of virulence genes
Virulence genes were amplified by PCR. The total PCR volume of 25 μL contained 12 μL Mytaq HS Red Mix (Bioline), 2× PCR Master Mix (Bioline), 1 μL forward primer, 1 μL reverse primer, 1 μL DNA sample (20 ng), and 10 μL NFW [16]. The virulence genes detected included aerA/haem, alt, ast, flaA, lafA, and fstA (Table-1). The PCR products were subjected to electrophoresis on a 1.5% agarose gel.
Sequencing and phylogenetic analysis
Descriptive analysis was conducted on phenotypic identification and virulence gene detection results. The DNA sequences were edited and assembled using the DNA Baser program (Dacia/P7, Mioveni 115400, Arges, Romania, http://www.dnabaser.com/news/index.html) [41]. The degree of similarity was analyzed using the basic local alignment search tool (BLAST) program (http://www.ncbi.nlm.nih.gov/BLAST). The Clustal W program (Bio.tools organization, http://www.clustal.org/) was used for multiple sequence alignments. Phylogenetic trees with 1000 replications of bootstrap analysis were constructed using the maximum likelihood MEGA 7.0.26 package (The Pennsylvania State University, USA; https://www.megasoftware.net/) [42].
Results
Clinical signs of diseased catfish
Figure-1 shows the clinical signs of diseased catfish. The clinical signs observed included skin depigmentation, necrosis, hemorrhagic, hyperemia, ulcer, abdominal dropsy, abdominal ascites, abscess, and paleness in the kidney.
Figure-1.
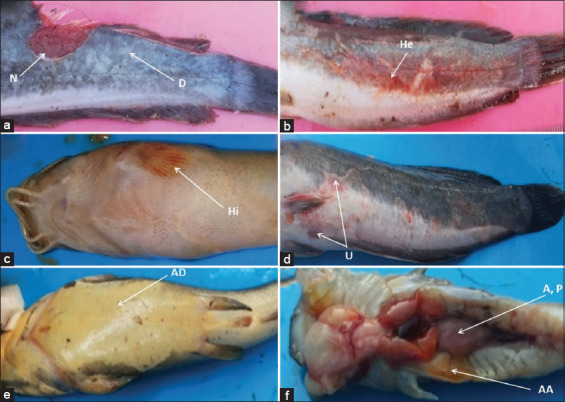
Clinical signs of moribund hybrid catfish diseased. a: Depigmentation (D) of the skin, necrosis (N), b: Hemorrhagic (He), c: Hyperemia (Hi), d: Ulcer (U), e: Abdominal dropsy (AD), abdominal ascites (AA), f: Abscess (S), pale (P) in kidney, abdominal ascites (AA).
Phenotypic characteristics of Aeromonas spp.
Morphological and biochemical characteristics suggested that the isolates observed in this study were members of Aeromonas. These bacteria grew well on GSP and TSA media at 30°C. They were rod-shaped, Gram-negative, motile, and F and can reduce nitrate (100%). These bacteria can also decarboxylate ornithine, deaminate phenylalanine, and utilize adonitol in the range of 11%–89% (v), but cannot produce acetyl methyl carbinol (VP), urease, and H2S. Two isolates of Aeromonas spp. (22.22%) can decarboxylate lysine, and seven isolates (77.78%) can decarboxylate lysine in the range of 11%–89% (v). However, only one isolate can utilize citrate (11.11%). Seven isolates (77.78%) were acid-forming (from glucose), with two isolates (22.22%) forming acids (from glucose) in the range of 11%–89% (v). One isolate (11.11%) produced gas from glucose. Five isolates (55.56%) utilized lactose, and four isolates (44.44%) used lactose in the range of 11%–89% (v). Eight isolates (88.89%) can utilize arabinose, and one isolate (11.11%) can utilize arabinose in the range of 11%–89% (v). Four isolates (44.44%) utilized sorbitol, and one isolate (11.11%) did not. Four isolates (44.44%) used sorbitol in the range of 11%–89% (v). Three isolates of Aeromonas spp. (33.33%) hydrolyzed aesculin (Table-2).
Table-2.
The phenotypic characters of Aeromonas spp. bacteria isolated from diseased catfish.
| Characterization | Isolates | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Ah-01 | Ac-01 | Av-01 | Av-02 | Av-03 | Av-04 | Ad-01 | Ad-02 | Ad-03 | |
| Colony morphology | |||||||||
| Form | Circular | Circular | Circular | Circular | Circular | Circular | Circular | Circular | Circular |
| Edge | Even | Even | Even | Even | Even | Even | Even | Even | Even |
| Elevation | Convex | Convex | Convex | Convex | Convex | Convex | Convex | Convex | Convex |
| Color in TSA | White | White | White | White | White | White | White | White | White |
| Color in GSP | Yellow | Yellow | Yellow | Yellow | Yellow | Yellow | Yellow | Yellow | Yellow |
| Bacterial morphology | |||||||||
| Form | Rod | Rod | Rod | Rod | Rod | Rod | Rod | Rod | Rod |
| Gram | - | - | - | - | - | - | - | - | - |
| Motility | + | + | + | + | + | + | + | + | + |
| O/F | F | F | F | F | F | F | F | F | F |
| Voges-Proskauer | - | - | - | - | - | - | - | - | - |
| Citrate utilization | - | - | - | - | - | - | - | + | - |
| Lysine decarboxyzation | v | v | v | v | + | v | v | v | + |
| Ornithine decarboxyzation | v | v | v | v | v | v | v | v | v |
| Urease | - | - | - | - | - | - | - | - | - |
| Phenylalanine deamination | v | v | v | v | v | v | v | v | v |
| Nitrate reduction | + | + | + | + | + | + | + | + | + |
| H2S production | - | - | - | - | - | - | - | - | - |
| Glucose, acid | + | + | v | + | + | + | v | + | + |
| Glucose, gas | - | - | - | - | + | - | - | - | - |
| Adonitol | v | v | v | v | v | v | v | v | v |
| Lactose | + | v | + | v | + | v | v | + | + |
| Arabinose | + | + | + | + | + | v | + | + | + |
| Sorbitol | v | v | v | v | + | - | + | + | + |
| Aesculin hydrolysis | - | + | - | - | - | - | + | - | + |
+, >90% of strains positive; –, >90% of strains negative; v, 11%–89% of strains positive; F=Fermentative, TSA=Tryptic soy agar, GSP=Glutamate starch phenyl, O/F=Oxidative/Fermentative
Molecular identification of Aeromonas spp. based on 16S rDNA
The results of 16S rDNA amplification for nine isolates revealed base lengths of approximately 1500 bp (Figure-2). The 16S rDNA sequences of these isolates were compared with those of 28 isolates from the GenBank database (NCBI, USA). The homology search based on the 16S rDNA sequence (1410–1479 bp) showed that the nine isolates demonstrated similarities to A. hydrophila, A. caviae, A. veronii bv veronii, and Aeromonas dhakensis. Queries reached up to 96%–100% with a similarity range of 97%–99.86% (Table-3).
Figure-2.
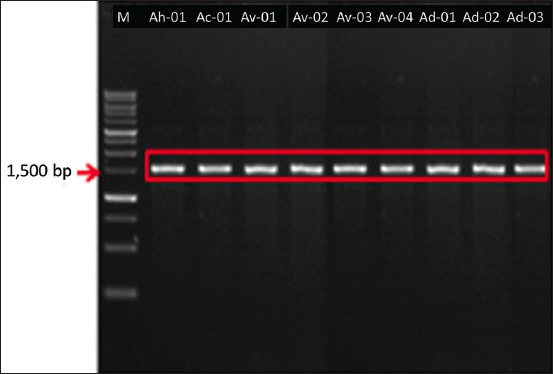
Results of amplification of bacterial DNA isolated with 16S rDNA primers. Arrow: 1500 bp, Marker: 1 kb.
Table-3.
Identity of Aeromonas spp. bacteria isolated from diseased catfish.
| Isolates | Closest comparator | Query (%) | Identity (%) | Accession number |
|---|---|---|---|---|
| Ah-01 | Aeromonas hydrophila ATCC 7966 | 99 | 99.45 | NR118944 |
| Ac-01 | Aeromonas caviae ATCC 15468 | 97 | 99.57 | NR029252 |
| Av-01 | Aeromonas veronii bv. veronii ATCC 35624 | 96 | 99.51 | NR118947 |
| Av-02 | Aeromonas veronii bv. veronii ATCC 35624 | 100 | 99.72 | NR118947 |
| Av-03 | Aeromonas veronii bv. veronii ATCC 35624 | 99 | 98.55 | NR118947 |
| Av-04 | Aeromonas veronii bv. veronii ATCC 35624 | 96 | 99.72 | NR118947 |
| Ad-01 | Aeromonas dhakensis P21 | 99 | 97.91 | NR042155 |
| Ad-02 | Aeromonas dhakensis P21 | 100 | 99.86 | NR042155 |
| Ad-03 | Aeromonas dhakensis P21 | 100 | 99.86 | NR042155 |
Isolate Ah-01 was closest to A. hydrophila ATCC 7966 (NR118944) [43], with a similarity rate of 99.45% and query match of 99%. Isolate Ac-01 was closest to A. caviae ATCC 15468 (NR029252), with a similarity rate of 99.57% and 97% query matching. The Av-01, Av-02, Av-03, and Av-04 isolates were closest to A. veronii bv veronii ATCC 35624 (NR118947) [44]. The levels of similarity of these isolates were in the range of 98.55%–99.72% with 96%–100% query matching. Three isolates (33.33%), namely, Ad-01, Ad-02, and Ad-03 were the most closely related to A. dhakensis strain P21 (NR042155) [45]. Their levels of similarity were 97.91%–99.37% with 99%–100% query matching.
Phylogenetic tree
Phylogenetic analyses showed that Ah-01 and Ac-01 isolates were included in the clade of A. hydrophila ATCC 7966 and A. caviae ATCC 15468, respectively. Av-01, Av-02, Av-03, and Av-04 isolates were grouped into the clade of A. veronii bv veronii ATCC 35624 and Ad-01, Ad-02, and Ad-03 isolates into the clade of A. dhakensis P21 (Figure-3).
Figure-3.
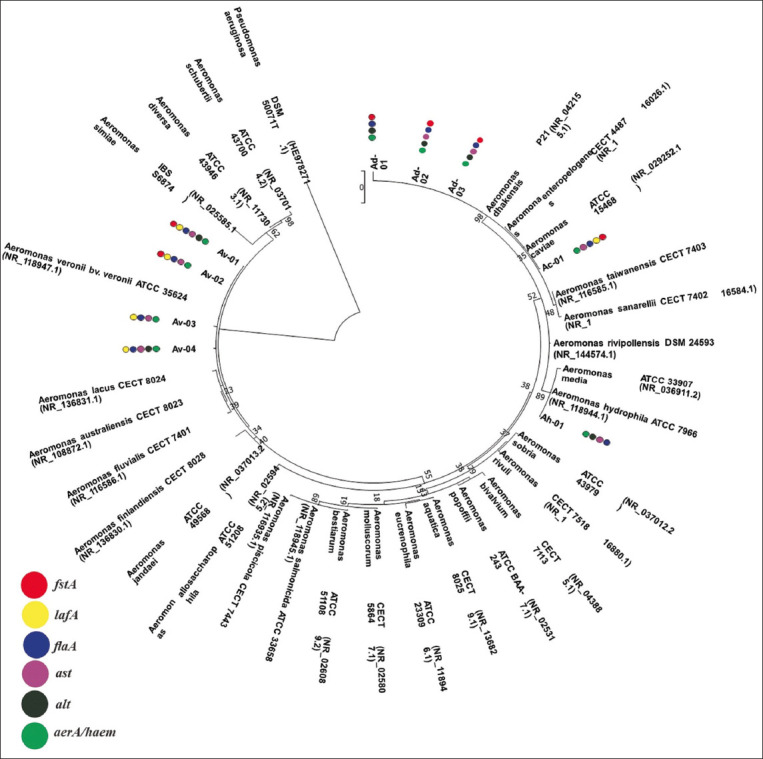
Phylogenetic tree constructed from the 16S rDNA sequences of Aeromonas sp. and other Aeromonas species (class of Gammaproteobacteria) and detected virulence genes. Pseudomonas aeruginosa was used as an outgroup. The topology was obtained by maximum likelihood with bootstraps of 1000 replications. The scale bar signifies 0.02 substitutions per nucleotide position (Knuc). Colored circles indicate the presence of the virulence genes for each isolate,  aer/haem: aerolysin/hemolysin;
aer/haem: aerolysin/hemolysin;  alt: heat-labile cytotonic enterotoxin;
alt: heat-labile cytotonic enterotoxin;  ast: heat-stable cytotonic enterotoxin;
ast: heat-stable cytotonic enterotoxin;  flaA: polar flagellum;
flaA: polar flagellum;  lafA: lateral flagellum;
lafA: lateral flagellum;  fstA: ferric siderophore receptor.
fstA: ferric siderophore receptor.
Detection of virulence genes
Six virulence genes (aer/haem, alt, ast, flaA, lafA, and fstA) of the Aeromonas spp. were detected (Figure-3 and Table-4). The aer/haem gene was found in all isolates, and the alt gene was present in six isolates. The virulence gene ast was discovered in eight isolates, and flaA was amplified in all isolates. The virulent gene lafA was found in five isolates, and fstA was found in six isolates.
Table-4.
Virulence genes in the Aeromonas spp. isolated from catfish.
| Isolates | Species | The presence of virulence genes | Detection rate (%) | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| aerA/haem | alt | ast | flaA | lafA | fstA | |||
| Ah-01 | Aeromonas hydrophila strain ATCC 7966 | + | + | + | + | - | - | 4 (66.67) |
| Ac-01 | Aeromonas caviae strain ATCC 15468 | + | - | + | + | + | + | 5 (83.33) |
| Av-01 | Aeromonas veronii bv. veronii strain ATCC 35624 | + | + | + | + | + | + | 6 (100.00) |
| Av-02 | Aeromonas veronii bv. veronii strain ATCC 35624 | + | - | + | + | + | + | 5 (83.33) |
| Av-03 | Aeromonas veronii bv. veronii strain ATCC 35624 | + | - | + | + | + | - | 4 (66.67) |
| Av-04 | Aeromonas veronii bv. veronii strain ATCC 35624 | + | + | + | + | + | - | 5 (83.33) |
| Ad-01 | Aeromonas dhakensis strain P21 | + | + | - | + | - | + | 4 (66.67) |
| Ad-02 | Aeromonas dhakensis strain P21 | + | + | + | + | - | + | 5 (83.33) |
| Ad-03 | Aeromonas dhakensis strain P21 | + | + | + | + | - | + | 5 (83.33) |
| Total | 9 (100%) | 6 (66.67%) | 8 (88.89%) | 9 (100%) | 5 (55.55%) | 6 (66.67%) | 10 | |
The four isolates of A. dhakensis harbored aer/haem, alt, flaA, and fstA but not lafA. However, three isolates harbored ast (Table-4 and Figure-3). The aer/haem, ast, flaA, and lafA were detected in all isolates of A. veronii bv veronii. However, alt and fstA were detected in two isolates, whereas only A. caviae isolate had aer/haem, ast, flaA, lafA, and fstA, but not alt. The genes aer/haem, alt, ast, and flaA were detected in A. hydrophila but lafA and fstA were missing.
The genes aer/haem, alt, ast, and flaA were detected in A. hydrophila, but lafA and fstA were absent. A. caviae isolate contained the aer/haem, ast, flaA, lafA, and fstA, but not alt. The aer/haem, ast, flaA, and lafA were detected in all isolates of A. veronii bv veronii. However, alt and fstA were detected in two isolates. The three isolates of A. dhakensis harbored aer/haem, alt, flaA, and fstA but did not harbor the lafA gene. However, two isolates harbored ast (Table-4 and Figure-3).
The PCR assay of all nine isolates detected the aer/haem, alt, ast, flaA, lafA, and fstA at the rates of 100%, 66.67%, 88.89%, 100%, 55.56%, and 66.67%, respectively. The results showed that all isolates had four or more virulence genes, and only one isolate (A. veronii bv veronii Av-01) had all six virulence genes.
Discussion
This study investigated the diversity of Aeromonas spp. causing MAS in Indonesia and infected catfish. Specifically, this study aimed to determine the biodiversity of Aeromonas spp. in diseased catfish cultivated in four provinces on Java Island, Indonesia.
The clinical signs of diseased catfish included depigmentation of the skin, necrosis, hemorrhagic, hyperemia, ulcer, abdominal dropsy, abdominal ascites, abscess, and paleness of the kidney (Figure-1). Similar findings have been reported by Austin and Austin [46] and Dias et al. [47]. Aeromonas spp. can break the blood vessels, which results in ulcerative lesions with a hemorrhagic appearance throughout the tegument [48]. In this study, clinical signs of diseased catfish indicated typical signs of aeromoniasis. This finding suggests that the isolates of Aeromonas spp. considered in this study were pathogenic and infectious with remarkable clinical signs.
Based on phenotypic characteristics, this study identified nine isolates as presumptive Aeromonas species. The isolates were characterized based on their growth on GSP selective agar medium: yellow colonies, rod shape, Gram-negative, motile, and production of acid from glucose (Table-2). These characteristics have also been reported previously by El-Sharaby et al. [5]. In addition, Aeromonas can ferment glucose. A common characteristic of Aeromonas is their ability to produce gases from glucose. However, this study found only one isolate with such a trait, which suggests the inconsistency of this ability among Aeromonas [49]. In the present study, all Aeromonas isolates reduced nitrate, as previously reported by Vega-Sanchez et al. [50] for A. caviae, A. veronii bv veronii, and A. hydrophila. However, Aeromonas cannot produce acetyl methyl carbinol (VP), urease, and H2S. The previous study reported by Khor et al. [51] have also reported that all Aeromonas isolates are unable to produce urease and H2S. The ability of Aeromonas to produce acetyl methyl carbinol (VP) varies. Aeromonas hydrophila and A. veronii bv veronii manufacture this substance [52], but A. caviae does not [53]. The previous study by Khor et al. [51] have also revealed that Aeromonas varies in their ability to use citrate. This research found characteristics that are different from those of previous study by Borty et al. [52], revealing that A. hydrophila cannot utilize lysine; in addition, some species cannot utilize ornithine [51], but A. veronii bv veronii cannot specifically use arabinose [50, 54, 55].
In contrast to the findings of previous studies, A. hydrophila and A. veronii bv veronii cannot utilize sorbitol [50, 52], and hydrolyze aesculin [53], but can hydrolyze aesculin [50, 52, 54, 55]; meanwhile, 50% of A. caviae can hydrolyze aesculin [52]. These results indicate variations in the phenotypic characteristics of Aeromonas. This condition has also been reported previously by Liu [49] and Martin-Carnahan and Joseph [56]. Phenotypic variation is thought to have clinical significance for determining the relationship between biotype and enterotoxin production [53].
The identification of members of Aeromonas spp. at the species level remains a huge challenge given their genetic heterogeneity. Biochemical characteristics are ineffective for the identification of Aeromonas at the species level [57]; thus, species identification should be supported by genotypic analysis. Molecular identification using 16S rDNA is very helpful for accurate species identification [58, 59]. Furthermore, we proved the biodiversity of Aeromonas spp. by 16S rDNA sequencing. We used 16S rDNA sequence rather than gyrB sequence for molecular identification in this study. We performed preliminary screening for Aeromonas spp. using GSP medium based on its morphological characteristics, further confirmed using a series of biochemical tests that highly aligned with Aeromonas characteristics, and finally, 16S rDNA phylogenetical analysis with the inclusion of Aeromonas type strains. Results from our phylogenetic tree also revealed that our isolates formed a tight clade with their corresponding type strains. In addition, the numbers of 16S rRNA gene database in NCBI are much higher available than that of gyrB gene sequence. Based on the availability of the database, the initial molecular identification is likely more appropriate using the 16S rDNA sequence to avoid the mismatch results of similarity analysis.
The results of 16S rDNA amplification of nine isolates showed a base length of approximately 1500 bp (Figure-2). BLAST search of the 16S rDNA Aeromonas spp. revealed a high level of similarity, namely, 97.91%–99.86%, and queries reaching 96%–100% (Table-3). Homology searches conducted on the 16S rDNA sequence indicated that the nine isolates obtained were parallel to A. hydrophila, A. caviae, A. veronii bv veronii, and A. dhakensis. Our results indicate the successful identification of nine isolates of, Aeromonas species, namely, A. hydrophila (11.11%), A. caviae (11.11%), A. veronii bv veronii (44.44%), and A. dhakensis (33.33%). These results were confirmed using phylogenetic relationship analysis (Figure-3). The current findings completed previous results that identified three species, A. hydrophila [33], A. caviae, and A. veronii [34], from catfish in Indonesia during genotypic analysis. Indonesian researchers have previously identified Aeromonas species from catfish by phenotypic identification and collected three Aeromonas species, namely, A. hydrophila, A. caviae, and A. salmonicida [35, 60, 61]. Six species, including A. hydrophila, A. salmonicida, A. veronii, A. media, A. taiwanensis, and A. jandaei were isolated form other freshwater and pond fishes, and successfully identified genotypically [37, 39].
The previous study has also identified Aeromonas spp. from catfish (Clarias gariepinus) in Egypt using genotypic analysis, these bacteria consisted of A. hydrophila (56%) and A. veronii (44%) [62], A. hydrophila (50.4%), and A. caviae (5.6%) according to phenotypic analysis [63]. Other research on catfish in Uganda discovered A. hydrophila (33%) and A. sobria (33%) through genotypic analysis [13]. In the present study, 16S rDNA molecular identification in catfish revealed the high biodiversity of Aeromonas spp., that is, four species, compared with a previous study, which detected one or two Aeromonas species. The high biodiversity of Aeromonas spp. is presumably due to the large sampled locations, which covered three provinces in Java Island: DIY Yogyakarta, Central Java, and West Java. The former surveys only involved West Java of Indonesia [33], center Java of Indonesia [34], and Qena in Egypt [62], as well as the central region and northern region of Uganda [13]. In addition, the differences in type and number of Aeromonas species identified from catfish may be due to the differences in individual fish, cultivation systems, climates, and environmental influences.
In line with this study, A. hydrophila, A. veronii, and A. caviae have frequently been isolated from diseased catfish. Unexpectedly, in this study, the prevalence of A. dhakensis was observed. Although A. dhakensis is rarely found in fish, this species is virulent to Nile tilapia (Oreochromis mossambicus) in Mexico [64] and other fish in Australia [65]. In addition, A. dhakensis was first reported to cause acute hemorrhagic septicemia in experimentally infected pacu fish (Piaractus mesopotamicus) in Brazil [66]. Aeromonas dhakensis was the predominant Aeromonas species isolated from freshwater fish in nine fish farms in Malaysia [67]. Aeromonas dhakensis is a major emerging pathogen of striped catfish in Vietnam [68]. Aeromonas dhakensis is possibly an important pathogen in catfish and other freshwater fishes, either in Indonesia or around the world. In addition, A. veronii bv veronii is the first A. veronii biovar, which has been previously reported as A. veronii, reported in Indonesia [34, 39].
Numerous virulence genes associated with the pathogenicity of Aeromonas spp. act synergistically with each other to exhibit their pathogenicity. In this study, the biodiversity of the Aeromonas complex was due to variations in virulence genes. The pathogenicity of the Aeromonas complex is multifactorial and assumed to involve the products of some genes, such as aerolysin (aer) and hemolysin (haem) (extracellular products), two cytotoxic, heat-labile (alt) and heat-stable (ast) genes, polar flagellum (fla) (structural components), lateral flagellum (laf), and ferric siderophore receptor (fst) [22, 69]. To spread their virulence factors, Aeromonas employs four secretion systems that discharge cellular products into the extracellular environment or directly to the host cell [70].
To recognize the potential virulence of Aeromonas spp., scientists regard the detection of virulence genes as a practical method for screening a large number of Aeromonas isolates with a suitable virulence potential. The previous studies by Mulia et al. [16] and Pessoa et al. [29] have also reported the detection and characterization of virulence factors in Aeromonas spp. In the present study, six virulence genes, that is, aer/haem, alt, ast, flaA, lafA, and fstA, were detected (Table-4). This study also showed that Aeromonas have at least four virulence genes. Similar results have been reported for aer/haem and flaA [30, 62]. Aerolysin is a hemolytic extracellular product coded by the aerolysin gene and plays a significant role in the pathogenicity of Aeromonas spp. It is the most important toxin among potential virulence factors in Aeromonas [71]. Aeromonas has two types of flagella, that is, polar and lateral, which are coded by flaA and lafA genes, respectively [70]. Alt and ast have been detected in some Aeromonas spp. [65]. The present study detected fstA in some isolates of Aeromonas spp. species but it not in A. hydrophila. In contrast, other research on a variety of sources (seawater, water, diseased fish, bivalves, cake, and wound exudate) of Aeromonas detected fstA in A. salmonicida, but not in A. hydrophila, A. caviae, and A. veronii [70].
All findings of this study provide important information regarding the characteristics and species diversity of Aeromonas, virulence genes of Aeromonas spp., and the presence of A. dhakensis in catfish. Aeromonas dhakensis showed virulence potential due to some virulence genes it possesses, such as aer/haem, alt, flaA, and fstA genes (100%), as well as the ast gene (66.67%). Such information may be useful for determining strategies for the control of MAS disease in catfish and other freshwater fishes.
Conclusion
Four species of Aeromonas spp., namely, A. hydrophila, A. caviae, A. veronii bv veronii, and A. dhakensis were identified in the current study. Virulence genes, including aer/haem, alt, ast, flaA, lafA, and fstA were detected at prevalence rates of 100%, 66.67%, 88.89%, 100%, 55.56%, and 66.67%, respectively. This study is the first report on the isolation of A. dhakensis from aquaculture species in Indonesia. Two isolates of A. dhakensis harbored five virulence genes, and one isolate possessed four virulence genes. In addition, the A. veronii bv veronii isolated in this study is the first A. veronii biovar isolated from aquaculture species in Indonesia. The isolates of A. veronii bv veronii possess six, five, or four virulence genes. This study has successfully revealed the diversity of Aeromonas spp. and their virulence genes. The current report contributes considerably to the basic consideration for the development of an Aeromonas vaccine.
Authors’ Contributions
DSM: Conceived and designed, and performed the experiments, analyzed the data, prepared figures and/or tables, authored or revised the manuscript, and approved the final draft. RP and WA: Supervised the experiment and reviewed the manuscript. MA and ISMY: Analyzed and interpreted data and drafted and revised the manuscript. AI: Designed the experiments, proposed the grant proposal, contributed reagents/materials/analysis tools, and reviewed the manuscript. All authors read and approved the final manuscript.
Acknowledgments
The data published in this manuscript are part of the dissertation of DSM under the Doctoral Program, Faculty of Biology, Universitas Gadjah Mada. This publication was supported by the Program of Post-doctoral Batch II, Universitas Gadjah Mada (Grant no. 13602/UN1.P.II/Dit-Lit/PT.01.04/20).
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published institutional affiliation.
References
- 1.Alimuddin A, Putri F.M, Wahjuningrum D, Hardiantho D, Sunarma A, Nuryati S. Resistance against Aeromonas hydrophila infection and growth of second generation (F2) African catfish (Clarias gariepinus) using selected molecular markers. Biotropia. 2018;25(2):95–102. [Google Scholar]
- 2.Raji A.A, Junaid Q.O, Oke M.A, Taufek N.H.M, Muin H, Bakar N.H.A, Alias Z, Milow P, Simarani K, Razak S.A. Dietary Spirulina platensis and Chlorella vulgaris effects on survival and haemato-immunological responses of Clarias gariepinus juveniles to Aeromonas hydrophila infection. AACL Bioflux. 2019;12(5):1559–1577. [Google Scholar]
- 3.Mulia D.S, Utomo T, Isnansetyo A. The efficacy of Aeromonas hydrophila GPl-04 feed-based vaccine on African catfish (Clarias gariepinus) Biodiversitas. 2022a;23(3):1505–1510. [Google Scholar]
- 4.Yazdanpanah-Goharrizi L, Rokhbakhsh-Zamin F, Zorriehzahra M.J, Kazemipour N, Kheirkhah B. Isolation, biochemical and molecular detection of Aeromonas hydrophila from cultured Oncorhynchus mykiss. Iran. J. Fish. Sci. 2020;19(5):2422–2436. [Google Scholar]
- 5.El-Sharaby S.M, El-Ballal S, Ali M, El-Hady M. Molecular studies on Aeromonas species isolated from freshwater fishes collected from Delta region, Egypt. J. Curr. Vet. Res. 2021;3(2):8–15. [Google Scholar]
- 6.Mailafia S, Nabilah B, Olabode H.O.K. Phenotypic characterization of Aeromonas hydrophila isolates in fresh water fishes in FCT using Microbact™ GNB 24E identification kit. Open Access Lib. J. 2021;8(1):1–12. [Google Scholar]
- 7.Laviad-Shitrit S, Izhaki I, Arakawa E, Halpern M. Wild waterfowl as potential vectors of Vibrio cholerae and Aeromonas species. Trop. Med. Int. Health. 2018;23(7):758–764. doi: 10.1111/tmi.13069. [DOI] [PubMed] [Google Scholar]
- 8.Luo K, Di J, Han P, Zhang S, Xia L, Tian G, Zhang W, Dun D, Xu Q, Wei Q. Transcriptome analysis of the critically endangered Dabry's sturgeon (Acipenser dabryanus) head kidney response to Aeromonas hydrophila. Fish Shellfish Immunol. 2018;83(12):249–261. doi: 10.1016/j.fsi.2018.09.044. [DOI] [PubMed] [Google Scholar]
- 9.Cunningham F.L, Hanson-Dorr K.C, Ford L, Middleton D.R, Crain A, Durst L, Ware C, Griffin M.J, Mischke C.C, Wan X.F, Hanson L.A. Environmental factor(s) and animal vector(s) associated with atypical Aeromonas hydrophila abundance and dissemination among channel catfish ponds. J. World Aquac. Soc. 2019;51(3):1–13. [Google Scholar]
- 10.Hossain S, De Silva B.C.J, Wimalasena S.H.M.P, Pathirana H.N.K.S, Dahanayake P.S, Heo G.J. Characterization of virulence determinants and multiple antimicrobial resistance profiles in motile Aeromonas spp. isolated from ornamental goldfish (Carassius auratus) J. Exot. Pet Med. 2019;29:51–62. [Google Scholar]
- 11.Yuwono C, Wehrhahn M.C, Liu F, Riordan S.M, Zhang L. The isolation of Aeromonas species and other common enteric bacterial pathogens from patients with gastroenteritis in an Australian population. Microorganisms. 2021;9(7):1440. doi: 10.3390/microorganisms9071440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rozi R, Rahayu K, Daruti D.N, Stella M.S.P. Study on characterization, pathogenicity and histopathology of disease caused by Aeromonas hydrophila in gouramy (Osphronemus gouramy) IOP Conf. Ser. Earth Environ. Sci. 2018;137:012003. [Google Scholar]
- 13.Wamala S.P, Mugimba K.K, Mutoloki S, Evensen O, Mdegela R, Byarugaba D.K, Sørum H. Occurrence and antibiotic susceptibility of fish bacteria isolated from Oreochromis niloticus (Nile tilapia) and Clarias gariepinus (African catfish) in Uganda. Fish. Aquac. Sci. 2018;21(1):6. [Google Scholar]
- 14.El-Son M.A.M, Abdelkhalek N.K.M, El-Ashram A.M.M, Zaki V.H. Phenotypic and biochemical detection of Aeromonas hydrophila isolated from cultured Oreochromis niloticus during disease outbreaks. Int. J. Fish. Aquac. Stud. 2019;7(3):197–202. [Google Scholar]
- 15.Elgohary I, Elatief J.I.A, Fadel N.G, Eissa A.E, Mahmoud M.A. Pathological, bacteriological and seasonal prevalence of Aeromonas hydrophila, Vibrio vulnificus, Proteus vulgaris and Pseudomonas aeruginosa;infecting Oreochromis niloticus in some Egyptian fish farms. Egypt. J. Aquac. Biol. Fish. 2020;24(5):467–482. [Google Scholar]
- 16.Mulia D.S, Isnansetyo A, Pratiwi R, Asmara W. Molecular characterizations of Aeromonas caviae isolated from catfish (Clarias spp.) AACL Bioflux. 2020;13(5):2717–2732. [Google Scholar]
- 17.Mulia D.S, Vauziyah S. The treatment of African catfish, (Clarias gariepinus L.) infected by Aeromonas hydrophila in Banyumas regency using the leaf extract of api-api (Avicennia marina) Sainteks. 2021;18(1):9–24. [Google Scholar]
- 18.Mulia D.S, Karim A, Purbomartono C, Isnansetyo A. Antibacterial activity of mangrove plant (Avicennia marina) to control Aeromonas hydrophila infection in African catfish (Clarias gariepinus) AACL Bioflux. 2022b;15(6):2900–2909. [Google Scholar]
- 19.AlYahya S.A, Ameen F, Al-Niaeem K.S, Al-Sa'adi B.A, Hadi S, Mostafa A.A. Histopathological studies of experimental Aeromonas hydrophila infection in blue tilapia, Oreochromis aureus. Saudi J. Biol. Sci. 2018;25(1):182–185. doi: 10.1016/j.sjbs.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu C.J, Ko W.C, Lee N.Y, Su S.L, Li C.W, Li M.C, Chen Y.W, Su Y.C, Shu C.Y, Lin Y.T. Aeromonas isolates from fish and patients in Tainan city, Taiwan:Genotypic and phenotypic characteristics. Appl. Environ. Microbiol. 2019;85(21):1–9. doi: 10.1128/AEM.01360-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Yu L, Nan Z, Zhang P, Kan B, Yan D, Su J. Taxonomy, virulence genes and antimicrobial resistance of Aeromonas isolated from extra-intestinal and intestinal infections. BMC Infect. Dis. 2019;19(1):158. doi: 10.1186/s12879-019-3766-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janda J.M, Abbott S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010;23(1):35–73. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Figueras M.J, Ballester F, Pujol I, Tena D, Berg K, Hossain M.J, Liles M.R. 'Aeromonas intestinalis'and 'Aeromonas enterica'isolated from human faeces, 'Aeromonas crassostreae'from oyster and 'Aeromonas aquatilis'isolated from lake water represent novel species. New Microb New Infect. 2017;15(C):74–76. doi: 10.1016/j.nmni.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernández-Bravo A, Figueras M.J. An update on the genus Aeromonas:Taxonomy, epidemiology, and pathogenicity. Microorganisms. 2020;8(1):129. doi: 10.3390/microorganisms8010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penders J, Stobberingh E.E. Antibiotic resistance of motile aeromonads in indoor catfish and eel farms in the southern part of The Netherlands. Int. J. Antimicrob. Agents. 2008;31(3):261–265. doi: 10.1016/j.ijantimicag.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Esteve C, Alcaide E. Influence of diseases on the wild eel stock:The case of Albufera lake. Aquaculture. 2009;289(1–2):143–149. [Google Scholar]
- 27.Yi S.W, You M.J, Cho H.S, Lee C.S, Kwon J.K, Shin G.W. Molecular characterization of Aeromonas species isolated from farmed eels (Anguilla japonica) Vet. Microbiol. 2013;164(1–2):195–200. doi: 10.1016/j.vetmic.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Awan F, Dong Y, Wang N, Liu J, Ma K, Liu Y. The fight for invincibility:Environmental stress response mechanisms and Aeromonas hydrophila. Microb. Pathog. 2018;116:135–145. doi: 10.1016/j.micpath.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 29.Pessoa R.B.G, de Oliveira W.F, Marques D.S.C, Dos Santos Correia M.T, de Carvalho E.V.M.M, Coelho L.C.B.B. The genus Aeromonas:A general approach. Microb. Pathog. 2019;130(1):81–94. doi: 10.1016/j.micpath.2019.02.036. [DOI] [PubMed] [Google Scholar]
- 30.Khor W.C, Puah S.M, Koh T.H, Tan J.A.M.A, Puthucheary S.D, Chua K.H. Comparison of clinical isolates of Aeromonas from Singapore and Malaysia with regard to molecular identification, virulence, and antimicrobial profiles. Microb. Drug Resist. 2018;24(4):469–478. doi: 10.1089/mdr.2017.0083. [DOI] [PubMed] [Google Scholar]
- 31.Marine and Fisheries Ministry. Catfish Production in Indonesia. Jakarta: Marine and Fisheries Ministry; 2022. [Google Scholar]
- 32.Mulia D.S, Maryanto H, Purbomartono C. Isolation, characterization, and identification of bacteria in African catfish infected with disease in Banyumas regency. Sainteks. 2011;7(1):1–15. [Google Scholar]
- 33.Marnis H, Iswanto B. Isolation, Identification and Molecular Characterization of Aeromonas hydrophila Bacteria that Infect Catfish (Clarias gariepinus) Proceedings of the Aquaculture Technology Innovation Forum. 2015:43–50. [Google Scholar]
- 34.Sarjito S, Haditomo A.H.C, Desrina D, Ariyati R.W, Prayitno S.B. The diversity of causative agent associated with bacterial diseases on catfish (Clarias gariepinus) with molecular based from Demak, Indonesia. Omni Akuatika. 2018;14(2):100–106. [Google Scholar]
- 35.Rejeki S, Triyanto T, Murwantoko M. Isolation and identification of Aeromonas spp. from diseased African catfish (Clarias spp.) in Ngawy regency. J. Fish. Gadjah Mada Univ. 2016;18(2):55–60. [Google Scholar]
- 36.Yuliantoro B, Helmizuryani H, Elfachmi E. Diversity of pathogenic bacteria in African catfish (Clarias gariepinus) in several farmers in Palembang city. Fisheries. 2017;6(1):1–6. [Google Scholar]
- 37.Amanu S, Kurniasih K, Indaryulianto S. Identification of Aeromonad disease in freshwater fish culture in Bali. J. Vet. 2014;15(4):474–486. [Google Scholar]
- 38.Syafitrianto I, Aqmal A, Lande M.N.H. Aeromonas variation on eel (Anguilla spp.) which is trafficked through Palu airport. Biogenesis. 2016;4(1):10–15. [Google Scholar]
- 39.Kusumawaty D, Pancoro A, Aryantha I.N.P, Suhandono S. Evaluation of identification techniques for the pathogen, Aeromonas hydrophila from Indonesia. Malays. J. Microbiol. 2016;12(3):191–198. [Google Scholar]
- 40.Manik V.T, Hidayat T, dan Kusumawaty D. Identification and phylogenetics of Aeromonas spp. isolated from fish ponds water in several cities based on 16S rRNA gene sequences [Identifikasi dan filogenetika bakteri Aeromonas spp. isolat air kolam beberapa kota berdasarkan pada sikuen gen 16S rRNA] Formica Online. 2014;1(1):10–19. [Google Scholar]
- 41.Wang D, Dai C, Li Q, Li Y, Liu Z. Complete mitochondrial genome and phylogenic analysis of Rhinogobius cliffordpopei (Perciformes, Gobiidae) Mitochondrial DNA B Resour. 2019;4(2):2473–2474. doi: 10.1080/23802359.2019.1637287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X:Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seshadri R, Joseph S.W, Chopra A.K, Sha J, Shaw J, Graf J, Haft D, Wu M, Ren Q, Rosovitz M.J, Madupu R, Tallon L, Kim M, Jin S, Vuong H, Stine O.C, Ali A, Horneman A.J, Heidelberg J.F. Genome sequence of Aeromonas hydrophila. J. Bacteriol. 2006;188(23):8272–8282. doi: 10.1128/JB.00621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez-Murcia A.J, Benlloch S, Collins M.D. Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing:Lack of congruence with results of DNA-DNA hybridizations. Int. J. Syst. Bacteriol. 1992;42(3):412–421. doi: 10.1099/00207713-42-3-412. [DOI] [PubMed] [Google Scholar]
- 45.Huys G, Kämpfer P, Albert M.J, Kühn I, Denys R, Swings J. Aeromonas hydrophila subsp. Dhakensis subsp. nov., isolated from children with diarrhoea in Bangladesh, and extended description of Aeromonas hydrophila subsp. Hydrophila (Chester 1901) Stanier 1943 (approved lists 1980) Int. J. Syst Evol. Microbiol. 2002;52(2 Pt 3):705–712. doi: 10.1099/00207713-52-3-705. [DOI] [PubMed] [Google Scholar]
- 46.Austin B, Austin D.A. Bacterial Fish Pathogens. 6th ed. Switzerland: Springer International Publishing; 2016. [Google Scholar]
- 47.Dias M.K.R, Sampaio L.S, Proietti-Junior A.A, Yoshioka E.T.O, Rodrigues D.P, Rodriguez A.F.R, Ribeiro R.A, Faria F.S.E.D.V, Ozorio R.O.A, Tavares-Dias M. Lethal dose and clinical signs of Aeromonas hydrophila in Arapaima gigas (Arapaimidae), the giant fish from Amazon. Vet. Microbiol. 2016;188:12–15. doi: 10.1016/j.vetmic.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 48.Van Doan H, Doolgindachbaporn S, Suksri A. The LD50 of Asian catfish (Pangasius bocourti, Sauvage 1870) challenge to pathogen Aeromonas hydrophila FW52 strain. Pensee J. 2013;7(10):287–293. [Google Scholar]
- 49.Liu D. Aeromonas. In: Tang Y.W, editor. Molecular Medical Microbiology. Vol. 3. Netherlands: Elsevier Ltd; 2015. pp. 1099–1110. [Google Scholar]
- 50.Vega-Sanchez V, Acosta-Dibarrat J, Vega-Castillo F, Castro-Escarpulli G, Aguilera-Arreola M.G, Soriano-Vargas E. Phenotypical characteristics, genetic identification, and antimicrobial sensitivity of Aeromonas species isolated from farmed rainbow trout (Onchorynchus mykiss) in Mexico. Acta Trop. 2014;130(1):76–79. doi: 10.1016/j.actatropica.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 51.Khor W.C, Puah S.M, Tan J.A.M.A, Puthucheary S.D, Chua K.H. Phenotypic and genetic diversity of Aeromonas species isolated from fresh water lakes in Malaysia. PLoS One. 2015;10(12):e0145933. doi: 10.1371/journal.pone.0145933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borty S.C, Rahman F, Ali Reza A.K.M, Khatun M.S, Kabir M.L, Rahman M.H, Monir M.S. Isolation, molecular identification and antibiotic susceptibility profile of Aeromonas hydrophila from cultured indigenous Koi (Anabus testudineus) of Bangladesh. Asian J. Med. Biol. Res. 2016;2(2):332–340. [Google Scholar]
- 53.Aravena-Roman M, Chang B.J, Riley T.V, Inglis T.J.J. Phenotypic characteristics of human clinical and environmental Aeromonas in Western Australia. Pathology. 2011;43(4):350–356. doi: 10.1097/PAT.0b013e3283463592. [DOI] [PubMed] [Google Scholar]
- 54.Beaz-Hidalgo R, Alperi A, Buján N, Romalde J.L, Figueras M.J. Comparison of phenotypical and genetic identification of Aeromonas strains isolated from diseased fish. Syst. Appl. Microbiol. 2010;33(3):149–153. doi: 10.1016/j.syapm.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 55.Sreedharan K, Philip R, Singh I.S.B. Characterization and virulence potential of phenotypically diverse Aeromonas veronii isolates recovered from moribund freshwater ornamental fishes of Kerala, India. Antonie van Leeuwenhoek. 2013;103(1):53–67. doi: 10.1007/s10482-012-9786-z. [DOI] [PubMed] [Google Scholar]
- 56.Martin-Carnahan A, Joseph S.W. Genus Aeromonas Stainer 1943. In: Brenner D.J, Krieg N.R, Staley J.T, editors. Bergey's Manual of Systematic Bacteriology. The Proteobacteria. 2nd ed. Vol. 2. USA: Springer; 2005. pp. 557–578. [Google Scholar]
- 57.Abu-Elala N, Abdelsalam M, Marouf S, Setta A. Comparative analysis of virulence genes, antibiotic resistance and gyrB-based phylogeny of motile Aeromonas species isolates from Nile tilapia and domestic fowl. Lett. Appl. Microbiol. 2015;61(5):429–436. doi: 10.1111/lam.12484. [DOI] [PubMed] [Google Scholar]
- 58.Zepeda-Velazquez A.P, Vega-Sanchez V, Ortega-Santana C, Rubio-Godoy M, de Oca-Mira D.M, Soriano-Vargas E. Pathogenocity of Mexican isolates of Aeromonas spp. in immersion experimentally infected rainbow trout (Oncorhynchusmykiss, Walbaum 1792) Acta Trop. 2017;169:122–124. doi: 10.1016/j.actatropica.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 59.Nwaiwu O. Data on evolutionary relationships of Aeromonas hydrophila and Serratia proteamaculans that attach to water tanks. Data Brief. 2018;16(C):10–14. doi: 10.1016/j.dib.2017.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sarjito S, Radjasa O.K, Haditomo A.H.C, Prayitno S.B. Causative Agent Motile Aeromonas in the Catfish (Clarias gariepinus) in Central Java Production Center. Indonesian Aquaculture Conference. 2013:146–152. [Google Scholar]
- 61.Wulandari T, Indrawati A, Pasaribu F.H. Isolation and identification of Aeromonas hydrophila on catfish (Clarias gariepinus) Muara Jambi aquaculture, Jambi province. J. Med. Vet. 2019;2(2):89–95. [Google Scholar]
- 62.Emeish W.F.A, Mohamed H.M.A, Elkamel A.A. Aeromonas infections in African sharptooth catfish. J. Aquac. Res. Dev. 2018;9(9):548. [Google Scholar]
- 63.El Tawab A.A.A, Maarouf A.A.A, El Hofy F.I, El Mougy E.E.A. Detection of some virulence genes in A. hydrophila and A. caviae isolated from fresh water fishes at Qalubia governorate. Benha Vet. Med. J. 2017;33(2):489–503. [Google Scholar]
- 64.Soto-Rodriguez S.A, Cabanillas-Ramos J, Alcaraz U, Gomez-Gil B, Romalde J.L. Identification and virulence of Aeromonas dhakensis, Pseudomonas mosselii and Microbacterium paraoxydans isolated from Nile tilapia, Oreochromis niloticus, cultivated in Mexico. J. Appl. Microbiol. 2013;115(3):654–662. doi: 10.1111/jam.12280. [DOI] [PubMed] [Google Scholar]
- 65.Aravena-Roman M, Inglis T.J.J, Riley T.V, Chang B.J. Distribution of 13 virulence genes among clinical and environmental Aeromonas spp. in Western Australia. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33(11):1889–1895. doi: 10.1007/s10096-014-2157-0. [DOI] [PubMed] [Google Scholar]
- 66.Carriero M.M, Maia A.A.M, Sousa R.L.M, Henrique-Silva F. Characterization of a new strain of Aeromonas dhakensis isolated from diseased Pacu fish (Piaractus mesopotamicus) in Brazil. J. Fish Dis. 2016;39(11):1285–1295. doi: 10.1111/jfd.12457. [DOI] [PubMed] [Google Scholar]
- 67.Azzam-Sayuti M, Ina-Salwany M.Y, Zamri-Saad M, Yusof M.T, Annas S, Najihah M.Y, Liles M.R, Monir M.S, Zaidi Z, Amal M.N.A. The prevalence, putative virulence genes and antibiotic resistance profiles of Aeromonas spp. isolated from cultured freshwater fishes in peninsular Malaysia. Aquaculture. 2022;540(6):736719. [Google Scholar]
- 68.Bartie K.L, Ngo T.P.H, Bekaert M, Oanh D.T.H, Hoare R, Adams A, Desbois A.P. Aeromonas hydrophila ST251 and Aeromonas dhakensis are major emerging pathogens of striped catfish in Vietnam. Front. Microb. 2023;13:1067235. doi: 10.3389/fmicb.2022.1067235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tomás J.M. The main Aeromonas pathogenic factors. ISRN Microbiol. 2012;2012:256261. doi: 10.5402/2012/256261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beaz-Hidalgo R, Figueras M.J. Aeromonas spp. whole genomes and virulence factors implicated in fish disease. J. Fish Dis. 2013;36(4):371–388. doi: 10.1111/jfd.12025. [DOI] [PubMed] [Google Scholar]
- 71.Chopra A.K, Xu X, Ribardo D, Gonzalez M, Kuhl K, Peterson J.W, Houston C.W. The cytotoxic enterotoxin of Aeromonas hydrophila induces proinflammatory cytokine production and activates arachidonic acid metabolism in macrophages. Infect. Immun. 2000;68(5):2808–2818. doi: 10.1128/iai.68.5.2808-2818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


