Abstract
PURPOSE
GemPred, a transcriptomic signature predictive of the efficacy of adjuvant gemcitabine (GEM), was developed from cell lines and organoids and validated retrospectively. The phase III PRODIGE-24/CCTG PA6 trial has demonstrated the superiority of modified folinic acid, fluorouracil, irinotecan, and oxaliplatin (mFOLFIRINOX) over GEM as adjuvant therapy in patients with resected pancreatic ductal adenocarcinoma at the expense of higher toxicity. We evaluated the potential predictive value of GemPred in this population.
PATIENTS AND METHODS
Routine formalin-fixed paraffin-embedded surgical specimens of 350 patients were retrieved for RNA sequencing and GemPred prediction (167 in the GEM arm and 183 in the mFOLFIRINOX [mFFX] arm). Survival analyses were stratified by resection margins, lymph node status, and cancer antigen 19-9 level.
RESULTS
Eighty-nine patients' tumors (25.5%) were GemPred+ and were thus predicted to be gemcitabine-sensitive. In the GEM arm, GemPred+ patients (n = 50, 30%) had a significantly longer disease-free survival (DFS) than GemPred– patients (n = 117, 70%; median 27.3 v 10.2 months, hazard ratio [HR], 0.43 [95% CI, 0.29 to 0.65]; P < .001) and cancer-specific survival (CSS; median 68.4 v 28.6 months, HR, 0.42 [95% CI, 0.27 to 0.66]; P < .001). GemPred had no prognostic value in the mFFX arm. DFS and CSS were similar in GemPred+ patients who received adjuvant GEM and mFFX (median 27.3 v 24.0 months, and 68.4 v 51.4 months, respectively). The statistical interaction between GEM and GemPred+ status was significant for DFS (P = .008) and CSS (P = .004). GemPred+ patients had significantly more adverse events of grade ≥3 in the mFFX arm (76%) compared with those in the GEM arm (40%; P = .001).
CONCLUSION
This ancillary study of a phase III randomized trial demonstrates that among the quarter of patients with a GemPred-positive transcriptomic signature, survival was comparable with that of mFOLFIRINOX, whereas those receiving adjuvant gemcitabine had fewer adverse events.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive cancer, with only 11% of patients alive 5 years after their diagnosis, all stages considered.1 After surgery, adjuvant gemcitabine- or fluoropyrimidine-based monotherapies have been the standard since two decades.2-4 A modified folinic acid, fluorouracil, irinotecan, and oxaliplatin (mFOLFIRINOX) adjuvant regimen combining fluorouracil, leucovorin, irinotecan, and oxaliplatin has shown in the PRODIGE-24/CCTG PA6 trial to significantly improve the outcome with a disease-free survival (DFS) rate at 5 years of 26% versus 19% and a median DFS of 21.4 months versus 12.8 months with mFOLFIRINOX and gemcitabine, respectively.5,6 This superiority of mFOLFIRINOX was obtained at the cost of greater toxicity (76% of adverse events being grade 3 or 4 in the mFOLFIRINOX group v 53% in the gemcitabine group). The choice of adjuvant chemotherapy is based on general performance status. About a third of patients are unfit to receive postoperative mFOLFIRINOX, in which case gemcitabine-based regimens (gemcitabine with capecitabine or gemcitabine alone in frail patients) are preferred.7
CONTEXT
Key Objective
Can sensitivity to adjuvant gemcitabine be predicted in patients with resected pancreatic ductal adenocarcinoma using GemPred, a transcriptomic signature?
Knowledge Generated
In an ancillary study of the PRODIGE24 trial, the patients in the adjuvant gemcitabine treatment arm predicted to be sensitive to gemcitabine (GemPred+, 25% in total) had a significantly longer disease-free survival (DFS) and cancer-specific survival (CSS) then those predicted as resistant (GemPred–). GemPred+ patients' DFS and CSS were similar while using adjuvant gemcitabine or modified folinic acid, fluorouracil, irinotecan, and oxaliplatin (mFOLFIRINOX), yet significantly less toxicity was observed with gemcitabine. The tumor transcriptome appears to be useful in predicting the sensitivity to adjuvant gemcitabine in patients with resected pancreatic adenocarcinoma using the GemPred signature.
Relevance (A.H. Ko)
-
With further clinical validation and commercialization, this tissue-based transcriptomic signature may become a helpful tool to guide selection of adjuvant chemotherapy—in particular using a gemcitabine-based regimen instead of mFOLFIRINOX—for patients with resected pancreatic cancer.*
*Relevance section written by JCO Associate Editor Andrew H. Ko, MD, FASCO.
While the mFOLFIRINOX regimen became a standard of care for the adjuvant chemotherapy, several biomarker-based stratification approaches have been developed in retrospective studies trying to identify a subgroup of patients who would benefit from adjuvant gemcitabine. A retrospective analysis of the ESPAC-3 trial comparing gemcitabine with 5-fluorouracil/folinic acid in the adjuvant setting showed that the expression of human equilibrative nucleoside transporter 1 (hENT1), a transmembrane glycoprotein transporter that localizes to the plasma membrane of PDAC cells, was predictive of adjuvant gemcitabine efficacy with a median overall survival (OS) of 26.2 months in hENT1-high patients.8 Similar results were found in a retrospective analysis of the RTOG9704 trial comparing gemcitabine with 5-fluorouracil pre- and postchemoradiation.9 Finally, a multicentric retrospective analysis showed that the combined quantification of the expression of hENT1 and deoxycytidine kinase (dCK) may significantly predict the benefit of gemcitabine in the adjuvant setting.10 This predictive combination allowed us to identify a subgroup of 29.6% of patients who achieved an impressive median OS of 61.4 months using adjuvant gemcitabine. However, the studies quantifying the expression of hENT1 were based on unavailable, noncommercialized antibodies and therefore cannot be applied in a routine clinical setting.11
We have previously developed from preclinical models of PDAC a whole-transcriptome signature associated with a strong benefit of adjuvant gemcitabine.12,13 This signature, named GemPred, while having no prognostic value, has been validated on a multicentric retrospective cohort of consecutively operated patients, showing that GemPred+ patients had a great sensitivity to gemcitabine, with a higher OS and DFS compared with patients who did not have adjuvant chemotherapy (91.3 v 31.7 and 42.5 v 13.4 months, respectively). This result prompted us to test the GemPred signature on the surgical specimens of the PRODIGE-24/CCTG PA6 trial (ClinicalTrials.gov identifier: NCT01526135; EudraCT number, 2011-002026-52) to evaluate its predictive value and assess whether gemcitabine could favorably compare with mFOLFIRINOX in the subgroup of GemPred+ patients.
PATIENTS AND METHODS
Patients and Samples
Surgical samples of the patients included in the PRODIGE-24/CCTG PA6 trial were retrieved and centralized by UNICANCER. All samples were re-examined by the same expert pathologist (J.C.) to assess the sufficient amount of tumor cells. Tumor areas were manually microdissected for RNA extraction. Clinical data from the last update of the PRODIGE-24/CCTG PA6 trial were used.6 This study is compliant with the REMARK criteria. This study was approved by the PRODIGE ethical review board (EudraCT: 2011-002026-52).
Human Investigations
The trial protocol was approved by an independent ethics committee in France (Comité de Protection des Personnes Est III) and by ethics committees at participating centers in Canada. All the patients provided written informed consent. The trial was conducted in accordance with the latest version of the Declaration of Helsinki, the Good Clinical Practice guidelines of the International Conference on Harmonisation, and relevant French, European, and Canadian laws and directives.
Transcriptome Profile Generation and Analysis
Total RNA was extracted for every tumor sample using the QIAGEN AllPrep protocol.14 Overall, 150 ng of total RNA was used to generate RNAseq sequencing library using the Lexogen QuantSeq 3′ FWD kit and sequenced by the Institut du cerveau iGenseq plateform on a NovaSeq 6000 aiming for a minimum of 10 million reads. Raw RNAseq reads were mapped to the human genome (Ensembl GRCH38) and Ensembl's reference transcriptome using STAR.15 Gene counts were obtained using FeatureCount,16 normalized by a Upper Quartile procedure, and logged on a base 2. The improved GemPred signature13 was applied to the normalized counts to dichotomize in a GemPred+ or GemPred– status for each sample.
Statistical Analysis
Qualitative variables were compared by Pearson's chi-square test, and continuous variables were compared with binary variables or discrete variables with more than two modalities using a nonparametric Mann-Whitney or Kruskal-Wallis test, respectively.
Survival analyses were performed to measure DFS, cancer-specific survival (CSS), or postrelapse CSS. DFS was calculated from the date of random assignment until the date of first cancer-related event, second cancer, or death of any cause. CSS was calculated from the date of random assignment until death because of the treated cancer or a treatment-related complication. Postrelapse CSS was calculated from the date of relapse until death because of the treated cancer or a treatment-related complication. Hazard ratio with 95% CI and survival comparison was performed using a Cox proportional hazards model and stratified, when associated with DFS or CSS, by resection margin, lymph node status (pN0 with <12 lymph nodes examined, pN0 with 12 or more lymph nodes examined, and pN1), and cancer antigen 19-9 (CA 19-9) as in the original PRODIGE-24/CCTG PA6 study.5 The proportional hazards assumption was verified by the Schoenfeld residual method.17 Survival rate estimates were compared using a stratified log-rank test. The predictive value of GemPred was evaluated using a stratified (using resection margin, lymph node status, and CA 19-9) Cox proportional hazards model including the effect of the treatment group, the GemPred subgroup, and the interaction of both. The predictive significance of GemPred is evaluated by the estimation of the P value of the interaction term. The difference in proportions of grade ≥3 adverse events is reported as relative risk (RR), and proportion differences between two groups are tested using Fisher's exact test.
RESULTS
Characteristics of the Cohort
Of the 493 patients included in the trial, an informative sample from the surgical specimen was available for 357 patients. Seven were then excluded because of the lack of tumor cells on the retrieved sample. A sample of the surgical specimen for 350 patients was processed and used for whole-transcriptome profiling using an RNA-sequencing approach. In this PRODIGE-24/CCTG PA6 trial subset, 262 relapses occurred with a median DFS of 15.6 months. Two hundred and six cancer-specific deaths had occurred with a median CSS of 45.2 and a median follow-up of 69.8 months. In this subset of 350 patients included in this molecular study, 183 (52%) and 166 (48%) patients had received adjuvant mFOLFIRINOX (mFFX group) and gemcitabine (GEM group), respectively.
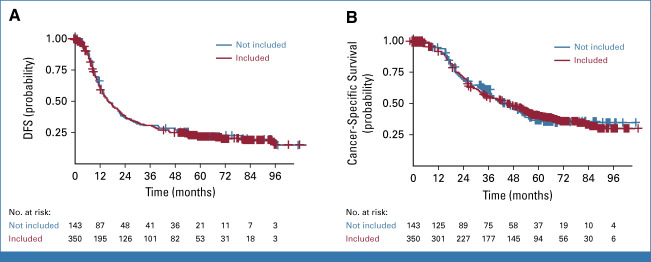
Clinicopathologic characteristics, including treatment group, age, sex, Eastern Cooperative Oncology Group (ECOG) status, CA 19-9 serum level, stage T, lymph node status, histologic grade, resection margins, DFS, and CSS, of the patients for whom no valid tumor sample could be obtained (n = 143) were similar to those whose tumor could be profiled (n = 350; Appendix Table A1 and Fig A1 [online only]). In the latter group, median DFS and CSS were significantly longer in the mFFX group than in the GEM group (21.6 v 12.1 months, stratified HR, 0.62 [95% CI, 0.48 to 0.80]; P < .001, and 56.1 v 34.1 months, stratified HR, 0.62 [95% CI, 0.471 to 0.826]; P < .001).
GemPred Status by Treatment Group
Among the 350 patients included, 89 (26%) and 261 (75%) patients had GemPred+ and GemPred– signatures, respectively. The GemPred status was not significantly associated with any specificities in clinicopathologic characteristics including ECOG, CA 19-9 level, resection margin, lymph node status, age, sex, stage T, and histologic grade (Table 1).
TABLE 1.
Cinicopathologic Characteristics of GemPred– Versus GemPred+ Patients
| Characteristic | GemPred– (n = 261) | GemPred+ (n = 89) | Overall (n = 350) | P |
|---|---|---|---|---|
| Age, years, median (IQR) | 62 (55-67) | 63 (56-68) | 63 (55-67) | .73a |
| Sex, No. (%) | .68b | |||
| Female | 118 (45) | 38 (43) | 156 (45) | |
| Male | 143 (55) | 51 (57) | 194 (55) | |
| ECOG, No. (%) | .90b | |||
| 0 | 131 (50) | 45 (51) | 176 (51) | |
| 1 | 129 (50) | 43 (49) | 172 (49) | |
| Unknown | 1 | 1 | 2 | |
| Postoperative CA 19-9 level, U/mL, No. (%) | .86b | |||
| ≤90 | 242 (93) | 83 (93) | 325 (93) | |
| >90 | 19 (7.3) | 6 (6.7) | 25 (7.1) | |
| Margin, No. (%) | .70b | |||
| R0 | 185 (71) | 65 (73) | 250 (71) | |
| R1 | 76 (29) | 24 (27) | 100 (29) | |
| LN, No. (%) | .88b | |||
| N0l12 | 21 (8.0) | 7 (7.9) | 28 (8.0) | |
| N0m12 | 41 (16) | 16 (18) | 57 (16) | |
| N1 | 199 (76) | 66 (74) | 265 (76) | |
| Grade, No. (%) | .55b | |||
| Moderate | 129 (52) | 42 (54) | 171 (52) | |
| Poor | 41 (17) | 9 (12) | 50 (15) | |
| Well | 78 (31) | 27 (35) | 105 (32) | |
| Unknown | 13 | 11 | 24 | |
| TNM2017.T, No. (%) | .51b | |||
| T1 | 53 (21) | 13 (15) | 66 (19) | |
| T2 | 152 (59) | 55 (63) | 207 (60) | |
| T3 | 52 (20) | 19 (22) | 71 (21) | |
| Unknown | 4 | 2 | 6 | |
| Treatment group, No. (%) | .064b | |||
| GEM | 117 (45) | 50 (56) | 167 (48) | |
| mFFX | 144 (55) | 39 (44) | 183 (52) |
Abbreviations: CA 19-9, cancer antigen 19-9; ECOG, Eastern Cooperative Oncology Group; GEM, gemcitabine; LN, lymph node; mFFX, modified FOLFIRINOX.
Wilcoxon rank-sum test.
Pearson's chi-squared test.
In the GEM group, GemPred+ patients (n = 50, 30%) had significantly longer DFS (median 27.3 v 10.2 months, stratified HR, 0.43 [95% CI, 0.29 to 0.65]; P < .001; Fig 1A) and CSS (median 68.4 v 28.6 months, stratified HR, 0.42 [95% CI, 0.27 to 0.66]; P < .001) than GemPred– patients (n = 117, 70%; Fig 1B).
FIG 1.
Kaplan-Meier estimates of (A) DFS and (B) CSS in each treatment group and according to GemPred status with respective median; (C) 3-year and 5-year survival and comparison of GemPred+ versus GemPred– in each treatment group for (C, left) DFS and (C, right) CSS. Crosses indicate censored data. Values in tables are shown with 95% CIs. CSS, cancer-specific survival; DFS, disease-free survival; sHR, stratified hazard ratio.
At a 3-year follow-up, in the GEM group, GemPred+ patients had DFS and CSS rates of 41% (95% CI, 29 to 57) and 73% (95% CI, 62 to 87), compared with 13% (95% CI, 8.4 to 21) and 38% (95% CI, 30 to 48) in the GemPred– patients, respectively (Fig 1C).
In the mFFX group, the GemPred status (GemPred+ n = 39, 21%; GemPred– n = 144, 79%) did not influence median DFS (24.0 v 21.4 months, P = .85; Fig 1C) or CSS (median 51.4 v 56.5 months, P = .54), confirming the lack of prognostic value of the signature for the mFFX schema (Fig 1D).
GemPred Subgroup and Multivariate Analyses
In the subset of GemPred+ patients (n = 89), patients receiving mFOLFIRINOX (n = 39, 44%) had a similar median DFS and CSS than those who received gemcitabine (n = 50, 56%; DFS 24.0 v 27.3 months; stratified HR, 1.05 [95% CI, 0.60 to 1.83]; P = .87; Fig 2A; CSS 51.4 v 68.4 months; stratified HR, 1.22 [95% CI, 0.65 to 2.3; P = .54), respectively (Fig 2B).
FIG 2.
Forest plot of GemPred subgroup analysis for comparison between mFFX versus GEM in terms of (A) DFS and (B) cancer-specific survival. P value int., P value of interaction; sHR, stratified hazard ratio.
The efficacy of the GemPred status as a predictive tool of adjuvant gemcitabine sensitivity was evaluated by the statistical interaction between the GEM group and the GemPred+ status. We found a significant interaction between treatment and the GemPred subgroup for DFS (interaction test: P = .008; Fig 2A) and CSS (interaction test: P = .004; Fig 2B).
Toxicity by Treatment and Subgroup
From the 342 patients with available toxicity evaluations, 225 (66%) reported an adverse event of grade 3 or higher. When comparing the highest-grade event per patient, and specifically highest-grade events ≥3, patients in the mFFX group had significantly more grade ≥3 adverse events (n = 140, 79%) than those in the GEM group (n = 85, 52%, Fisher's exact test P < .001, RR, 1.81 [95% CI, 1.47 to 2.23]; Fig 3). In the GemPred+ subgroup, 29 of 38 (76.3%) events of grade ≥3 were reported for patients in the mFFX group compared with 20 of 50 (40%) in the GEM group (Fisher's exact test P = .001, RR, 1.88 [95% CI, 1.31 to 2.81]). This was similar to the comparison of all mFFX versus GemPred+/Gem patients (Fisher's exact test: P < .001, RR, 3.58 [95% CI, 2.2 to 5.82]).
FIG 3.

Distribution of highest-grade adverse events. The number of patients with each reported highest-grade adverse event in each group and corresponding percentage (excluding patients with unreported toxicity) are shown. Statistical comparison between groups is shown, as well as comparison of grade 3, 4, and 5 events versus grade 0, 1, and 2 events. Adverse events were not reported for eight patients, six in the mFFX group (one GemPred+, five GemPred–) and two in the Gemcitabine group (both GemPred–). Results of Fisher's exact test are shown.
Association Between GemPred Status and Postrelapse Treatment
Among the 262 patients who had a tumor relapse, postrelapse therapy was unknown for 22 and 186 patients received postrelapse chemotherapy. One was excluded from this analysis because of the missing information on the regimen used, whereas various regimens were used for the remaining 185 patients (Fig 4A). In the mFFX group, 126 patients had a tumor relapse and postrelapse chemotherapy was known for 79 (63%); among them, 52 (66%) patients received a gemcitabine-based regimen (32 gemcitabine single-agent, 17 gemcitabine-nab-paclitaxel combination, and three, other gemcitabine-based combinations). The GemPred status had no significant prognostic value in postrelapse CSS in these patients (Fig 4B). Finally, among the 40 patients with known postrelapse treatment and a GemPred+ status, CSS was not significantly different between the mFFX and GEM group (stratified HR, 1.62 [95% CI, 0.79 to 3.34]; P = .19) despite a difference of 17.9 months in median CSS (Fig 4C).
FIG 4.
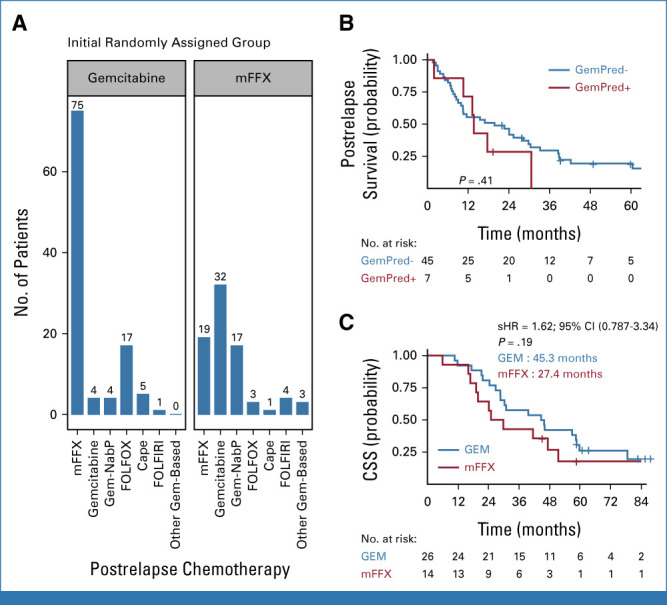
Postrelapse treatment and survival according to GemPred status. (A) Distribution of the number of patients receiving different chemotherapeutic regimens after relapsing in their initial randomized treatment. (B) Postrelapse cancer-specific survival among patients initially randomly assigned in the mFFX group and receiving a gemcitabine-based regimen after relapse. (C) Cancer-specific survival among GemPred+ patients with reported postrelapse chemotherapy. sHR, stratified hazard ratio. Cape, capecitabine; FOLFIRI, fluorouracil, leucovorin, and irinotecan; FOLFOX, fluorouracil, leucovorin, and oxaliplatin; NabP, nab-paclitaxel.
DISCUSSION
This ancillary study of the PRODIGE-24/CCTG PA6 phase III clinical trial confirms that GemPred is a valid predictive signature of gemcitabine efficacy in the adjuvant setting after PDAC resection while it has no prognostic value by itself. Informative RNAseq and GemPred status could be obtained only in a subset of patients (350 of 493, 71%) because of the lack of available material. One limitation of this work is not assessing the impact of BRCA1/2 mutations and homologous repair deficiencies to evaluate whether GemPred+ and HRD tumors should receive adjuvant gemcitabine or mFOLFIRINOX. However, all clinicobiologic characteristics in the subset of the analyzed population were similar to those of the overall population of the trial.
Patients of the GEM arm who had the GemPred+ status had a median DFS of 27.3 months versus 10.2 months in those predicted gemcitabine nonsensitive (GemPred–) patients (HR, 0.43 [95% CI, 0.29 to 0.65]; P < .001). GemPred– patients in the GEM arm had a similar median DFS than those in the gemcitabine arm of CONKO-001 trial (median DFS, 13.4 months).18 Their median OS (29 months) was also similar to that of previous adjuvant gemcitabine arms of the ESPAC-3 (median OS, 23.6 months)2 and the ESPAC-4 trials (median OS, 25.5 months).19 In addition, in our study, GemPred+ patients who had adjuvant gemcitabine achieved a similar DFS to those who received mFOLFIRINOX (27.3 v 24.0 months, respectively). These current results confirm the strong predictive value of this signature for gemcitabine administration and the lack of intrinsic prognostic value for resected PDAC.
Similarly, CSS of GemPred+ patients was similar whether in the mFOLFIRINOX or GEM group (68.4 and 51.4 months, respectively). Even when considering patients with tumor relapse, GemPred+ patients in the GEM arm had a comparable CSS (45.3 months v 27.4 in mFFX arm, P = .19), thus leaving the possibility to receive mFOLFIRINOX after relapse without impaired survival. GemPred+ patients in the GEM arm had significantly fewer grade ≥3 adverse events than any patients in the mFFX arm (P = .001). Given these results, we believe that the subgroup of GemPred+ patients may be eligible for gemcitabine in adjuvant with the same benefit as mFOLFIRINOX yet with less grade ≥3 adverse events. Thus, patients who have slow recovery or are unfit to receive adjuvant mFOLFIRINOX because of an insufficient recovery after a long postoperative delay (>3 months), and more generally patients with poor performance status (ECOG >1), may best benefit from having a tailored adjuvant chemotherapy, that is, gemcitabine for GemPred+ and fluoropyrimidine for GemPred– patients.
As of today, several biomarkers predictive of gemcitabine-based regimen efficacy have been proposed. Most rely on immunohistochemistry assessment of protein expression levels of gemcitabine-related metabolism enzymes, in particular, hENT1. However, no predictive antibody with reproducible reliability is commercially available for routine use.11 Interestingly, the rate of PDAC identified to be gemcitabine-sensitive was about 40%-50% with hENT1 antibody alone (clone 10D7G2), 30% with the combination of hENT1 and dCK expressions, and 25% with the GemPred signature, suggesting that the latter one can still be improved to more efficiently identify gemcitabine-sensitive tumors. The GemPred signature takes into account the expression of more than 2,000 genes whose combination was shown to correlate with the RNA expression of hENT1 and of the cytidine deaminase gene CDA13 but not dCK. A more comprehensive integration of gemcitabine metabolism genes,10 putative genetic signatures,20 stromal signatures,21 and transcriptome deconvolution22 may be considered to improve the signature. It would also be of interest to combine GemPred with other predictive biomarkers, in particular, BRCA-related alterations, which suggests a benefit from platinum-base regimens,23 especially at the metastatic stage.24 One limitation of this work is to not assess this combination with BRCA1/2 mutations (germline and somatic) or homologous repair deficiencies DNA-based signatures to evaluate whether GemPred+ tumors with a homologous repair deficiency would benefit more from either adjuvant gemcitabine or mFOLFIRINOX, although a gemcitabine plus platinum combination regimen could be considered.25
The GemPred signature is based on the transcriptomic profile of the tumor tissue. RNA sequencing requires several complex processing and analytical steps from the sample process to data analysis. However, the great majority of the process is shared with the now routinely performed DNA molecular profiling, requiring minimal adjustments and additions to add RNAseq to molecular tumor boards and routine mutation identification. In the case of adjuvant chemotherapy, the resected pancreatic specimen provides an abundant amount of tissue. Our group has previously shown that RNAseq can be obtained from endoscopic ultrasound-guided fine needle aspirates, showing that sample quality and quantity are not a prohibitive factors for RNA sequencing in PDAC.26
Neoadjuvant chemotherapy or preoperative radiochemotherapy is now considered in patients with high-risk resectable or borderline-resectable PDAC, and this will certainly modify the approach to resectable PDAC in the next future (NCCN 2022).27 The updated results of the PREOPANC trial and a recent meta-analysis showed that the best course of treatment between neoadjuvant chemotherapy versus up-front surgery for up-front resectable patients (nonborderline) is still an open question.28,29 As up-front surgery of resectable patients remains a relevant course of treatment, the selection of adjuvant chemotherapy still requires strong predictive signatures. A limitation of this work is that it does not evaluate the GemPred signature on gemcitabine used in combination with other drugs, in particular, with capecitabine. It was demonstrated that most combination therapies in oncology lack drug synergy and benefit on a populational level by benefiting different patient subgroups,30 suggesting that the GemPred signature is likely to be associated with the response to gemcitabine-based therapeutic combinations and that development of combinatory signatures will further improve patient selection. A recent study demonstrated that hENT1, a gemcitabine-specific gene, was predictive of response to gemcitabine-nab-paclitaxel in the COMPASS trial, suggesting that GemPred will most likely also predict the response to the dual regimen.31
We also tested the prognostic value of the GemPred+ status in patients treated who had postoperative adjuvant mFOLFIRINOX and who received a gemcitabine-based chemotherapy after relapse. In this comparison, the GemPred+ status obtained on a chemotherapy-naive sample was not predictive of postrelapse CSS for patients retreated using gemcitabine-based therapy after mFOLFIRINOX failure. This suggests either a heavy clonal selection or a strong phenotypic shift during adjuvant chemotherapy and demonstrates that the GemPred signature can currently only be applied before administering an adjuvant gemcitabine-based chemotherapy. Unlike genetic markers (eg, BRCA or KRAS mutations), phenotypic signatures such as GemPred measure the molecular state of a tumor sample at a given point, which may be modified by environmental factors, for instance, by a systemic chemotherapy. The biologic stability of these tumor phenotypes (ie, sensitive to a specific therapy) in time and on treatment is difficult to assess. Therefore, any non–genetic-based markers should be assessed as close as possible to the course of treatment to be selected by predictive signatures, which would require a tissue biopsy or rebiopsy after relapse.
In conclusion, this study shows that the RNAseq GemPred signature may help to select nearly a quarter of patients GemPred+ for choosing gemcitabine as an adjuvant setting after a PDAC resection, with both similar efficacy than mFOLFIRINOX and less toxicity, allowing us to treat patients unfit or who have not enough recovered to be eligible to this more toxic chemotherapy combination. In addition to validating the GemPred signature for the adjuvant setting in a randomized trial, this work paves the way for a prospective validation and requires further validation in other clinical settings including for the selection of neoadjuvant chemotherapy or of first-line treatment in locally advanced and metastatic tumors. Two prospective validations are envisioned: first in an adjuvant setting in which GemPred+ patients would be randomly assigned to receive adjuvant mFOLFIRINOX or gemcitabine to evaluate the noninferiority of gemcitabine in GemPred+ patients and second in the setting of an advanced disease in which patients would receive either mFOLFIRINOX or gemcitabine/nab-paclitaxel conditioned on an additional and appropriate retrospective validation. Finally, further work is also required to uncover signatures predictive of the benefit of mFOLFIRINOX.
APPENDIX
FIG A1.
Kaplan-Meier estimates of (A) DFS and (B) OS stratified by the inclusion of patients in the molecular profiling ancillary study. Crosses indicate censored data. DFS, disease-free survival; OS, overall survival.
TABLE A1.
Cinicopathologic Characteristics of Patients Included in the Ancillary Study Versus Those Whose Tumor Could Not be Profiled
| Characteristic | Not Included (n = 143) | Included (n = 350) | Overall (N = 493) | P |
|---|---|---|---|---|
| Sex, No. (%) | .60a | |||
| Female | 60 (42) | 156 (45) | 216 (44) | |
| Male | 83 (58) | 194 (55) | 277 (56) | |
| Age, years, median (IQR) | 63 (56-68) | 63 (55-67) | 63 (56-67) | .56b |
| ECOG, No. (%) | .76a | |||
| 0 | 74 (52) | 176 (51) | 250 (51) | |
| 1 | 68 (48) | 172 (49) | 240 (49) | |
| Unknown | 1 | 2 | 3 | |
| Postoperative CA 19-9 level, U/mL, No. (%) | .83a | |||
| ≤90 | 132 (92) | 325 (93) | 457 (93) | |
| >90 | 11 (8) | 25 (7) | 36 (7) | |
| Margin, No. (%) | .083a | |||
| R0 | 113 (79) | 250 (71) | 363 (74) | |
| R1 | 30 (21) | 100 (29) | 130 (26) | |
| LN, No. (%) | .50a | |||
| N0l12 | 14 (9.8) | 28 (8.0) | 42 (8.5) | |
| N0m12 | 28 (20) | 57 (16) | 85 (17) | |
| N1 | 101 (71) | 265 (76) | 366 (74) | |
| TNM2017.T, No. (%) | .32a | |||
| T1 | 21 (15) | 66 (19) | 87 (18) | |
| T2 | 95 (67) | 207 (60) | 302 (62) | |
| T3 | 25 (18) | 71 (21) | 96 (20) | |
| Unknown | 2 | 6 | 8 | |
| Grade, No. (%) | .33a | |||
| Moderate | 78 (57) | 171 (52) | 249 (54) | |
| Poor | 14 (10) | 50 (15) | 64 (14) | |
| Well | 44 (32) | 105 (32) | 149 (32) | |
| Unknown | 7 | 24 | 31 | |
| Arm, No. (%) | .13a | |||
| GEM | 79 (55) | 167 (48) | 246 (50) | |
| mFFX | 64 (45) | 183 (52) | 247 (50) |
Abbreviations: CA 19-9, cancer antigen 19-9; ECOG, Eastern Cooperative Oncology Group; GEM, gemcitabine; LN, lymph node; mFFX, modified FOLFIRINOX.
Pearson's chi-squared test.
Wilcoxon rank-sum test.
Jean-Baptiste Bachet
Honoraria: Amgen, Bayer, Merck Serono, Sanofi, Roche, Servier, AstraZeneca, Pierre Fabre, Viatris, MSD Oncology
Consulting or Advisory Role: Amgen, Bayer, Merck Serono, Servier, AstraZeneca, Pierre Fabre, AC BioScience, Acobiom, GlaxoSmithKline, BMS, MSD, Incyte
Travel, Accommodations, Expenses: Merck Serono, Amgen, Roche, Servier
Alexandre Harlé
Honoraria: AstraZeneca, Biocartis, GlaxoSmithKline, MSD Oncology, Pierre Fabre, SOPHiA Genetics
Consulting or Advisory Role: AstraZeneca, GlaxoSmithKline, SOPHiA Genetics, Janssen Oncology
Juan Iovanna
Stock and Other Ownership Interests: PreicitngMed, PanCa therapeutics
Pascal Hammel
Consulting or Advisory Role: VECT-HORUS, ERYTECH Pharma (Inst), AstraZeneca (Inst), Rafael Pharmaceuticals, Mylan, IPSEN (Inst), Servier/Pfizer
Speakers' Bureau: AstraZeneca, Servier, Mylan
Research Funding: ERYTECH Pharma (Inst), AstraZeneca (Inst), Celgene (Inst), Halozyme (Inst)
Travel, Accommodations, Expenses: Ipsen, Halozyme, Shire, Pfizer/EMD Serono, Vect-Horus
Anthony Turpin
Honoraria: SERVIER, Daiichi Sankyo/AstraZeneca, AstraZeneca, BMS, Incyte, MSD Oncology
Meher Ben Abdelghani
Consulting or Advisory Role: Incyte, Pierre Fabre, BMS, Deciphera, Merck Serono
Travel, Accommodations, Expenses: Amgen, Servier, Viatris, Merck Serono
Alice Wei
Honoraria: AstraZeneca Canada
Consulting or Advisory Role: Histosonics, Biosapien
Research Funding: Ipsen (Inst)
Other Relationship: BioNTech SE (Inst)
Emmanuel Mitry
Honoraria: Pierre Fabre, AstraZeneca/Daiichi Sankyo, Servier, Esteve
Anthony Lopez
Honoraria: Amgen
Consulting or Advisory Role: Vifor Pharma, Bayer, Merck, Sanofi, Ipsen, Servier, Pierre Fabre, Viatris
Research Funding: Roche
Travel, Accommodations, Expenses: AbbVie, Amgen, Bayer, MSD, Vifor Pharma, Mundipharma, Ipsen, Novartis
Eric François
Honoraria: Pierre Fabre
Pascal Artru
Honoraria: Servier, Amgen, Roche, Pierre Fabre
Travel, Accommodations, Expenses: Servier
Aurélien Lambert
Consulting or Advisory Role: Janssen, Merck KGaA, Bayer, Servier/Pfizer, Advanced Accelerator Applications, AstraZeneca/MedImmune, GlaxoSmithKline, Ipsen, Merck Serono, Pierre Fabre, Novartis
Travel, Accommodations, Expenses: Merck KGaA, Pfizer
Daniel J. Renouf
Honoraria: Roche, Bayer
Consulting or Advisory Role: Roche, Bayer, Viatris
Research Funding: Bayer, Roche (Inst)
Nelson Dussetti
Stock and Other Ownership Interests: Predicting Med
Patents, Royalties, Other Intellectual Property: PCT/EP2022/065222. Simple transcriptomic signatures to determine chemosensitivity for pancreatic ductal adenocarcinoma. SATT Sud-Est. Inventors: N. Dusetti, N. Fraunhoffer, J. Iovanna, 2021. Licensing to Predicting Med
Jérome Cros
Honoraria: Novartis
Consulting or Advisory Role: HISTALIM
Research Funding: ERYTECH Pharma (Inst)
Travel, Accommodations, Expenses: IPSEN, Novartis
No other potential conflicts of interest were reported.
SUPPORT
PRODIGE 24 was supported by R&D Unicancer, Clinical Research Hospital Program grant (PHRC11-006) from the French Ministry of Health and the Institut National du Cancer, and the French National League against Cancer. The Canadian Cancer Trials Group Pancreatic Adenocarcinoma (CCTG PA.6) part of the trial was supported by a Program Grant (704970) from the Canadian Cancer Society and by grants from 7 Days in May. This ancillary study was supported in part by the translational research program from the Institut National du Cancer (INCa-DGOS_14504, project No. PRT-K19-131) and by the French National League against Cancer.
DATA SHARING STATEMENT
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/JCO.22.02668.
AUTHOR CONTRIBUTIONS
Conception and design: Rémy Nicolle, Jean-Baptiste Bachet, Juan Iovanna, Jérome Cros
Financial support: Rémy Nicolle, Juan Iovanna
Administrative support: Rémy Nicolle, Laure Monard, Jérome Cros
Provision of study materials or patients: Rémy Nicolle, Jean-Baptiste Bachet, Pascal Hammel, Vinciane Rebours, Alice Wei, Anthony Lopez, James Biagi, Eric François, Daniel J. Renouf, Marjorie Mauduit, Thierry Conroy, Jérome Cros
Collection and assembly of data: Rémy Nicolle, Pascal Hammel, Vinciane Rebours, Meher Ben Abdelghani, Emmanuel Mitry, Eric François, Pascal Artru, Daniel J. Renouf, Laure Monard, Marjorie Mauduit, Thierry Conroy, Jérome Cros
Data analysis and interpretation: Rémy Nicolle, Jean-Baptiste Bachet, Alexandre Harlé, Juan Iovanna, Pascal Hammel, Anthony Turpin, Alice Wei, Emmanuel Mitry, Anthony Lopez, James Biagi, Aurélien Lambert, Thierry Conroy, Jérome Cros
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Prediction of Adjuvant Gemcitabine Sensitivity in Resectable Pancreatic Adenocarcinoma Using the GemPred RNA Signature: An Ancillary Study of the PRODIGE-24/CCTG PA6 Clinical Trial
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Jean-Baptiste Bachet
Honoraria: Amgen, Bayer, Merck Serono, Sanofi, Roche, Servier, AstraZeneca, Pierre Fabre, Viatris, MSD Oncology
Consulting or Advisory Role: Amgen, Bayer, Merck Serono, Servier, AstraZeneca, Pierre Fabre, AC BioScience, Acobiom, GlaxoSmithKline, BMS, MSD, Incyte
Travel, Accommodations, Expenses: Merck Serono, Amgen, Roche, Servier
Alexandre Harlé
Honoraria: AstraZeneca, Biocartis, GlaxoSmithKline, MSD Oncology, Pierre Fabre, SOPHiA Genetics
Consulting or Advisory Role: AstraZeneca, GlaxoSmithKline, SOPHiA Genetics, Janssen Oncology
Juan Iovanna
Stock and Other Ownership Interests: PreicitngMed, PanCa therapeutics
Pascal Hammel
Consulting or Advisory Role: VECT-HORUS, ERYTECH Pharma (Inst), AstraZeneca (Inst), Rafael Pharmaceuticals, Mylan, IPSEN (Inst), Servier/Pfizer
Speakers' Bureau: AstraZeneca, Servier, Mylan
Research Funding: ERYTECH Pharma (Inst), AstraZeneca (Inst), Celgene (Inst), Halozyme (Inst)
Travel, Accommodations, Expenses: Ipsen, Halozyme, Shire, Pfizer/EMD Serono, Vect-Horus
Anthony Turpin
Honoraria: SERVIER, Daiichi Sankyo/AstraZeneca, AstraZeneca, BMS, Incyte, MSD Oncology
Meher Ben Abdelghani
Consulting or Advisory Role: Incyte, Pierre Fabre, BMS, Deciphera, Merck Serono
Travel, Accommodations, Expenses: Amgen, Servier, Viatris, Merck Serono
Alice Wei
Honoraria: AstraZeneca Canada
Consulting or Advisory Role: Histosonics, Biosapien
Research Funding: Ipsen (Inst)
Other Relationship: BioNTech SE (Inst)
Emmanuel Mitry
Honoraria: Pierre Fabre, AstraZeneca/Daiichi Sankyo, Servier, Esteve
Anthony Lopez
Honoraria: Amgen
Consulting or Advisory Role: Vifor Pharma, Bayer, Merck, Sanofi, Ipsen, Servier, Pierre Fabre, Viatris
Research Funding: Roche
Travel, Accommodations, Expenses: AbbVie, Amgen, Bayer, MSD, Vifor Pharma, Mundipharma, Ipsen, Novartis
Eric François
Honoraria: Pierre Fabre
Pascal Artru
Honoraria: Servier, Amgen, Roche, Pierre Fabre
Travel, Accommodations, Expenses: Servier
Aurélien Lambert
Consulting or Advisory Role: Janssen, Merck KGaA, Bayer, Servier/Pfizer, Advanced Accelerator Applications, AstraZeneca/MedImmune, GlaxoSmithKline, Ipsen, Merck Serono, Pierre Fabre, Novartis
Travel, Accommodations, Expenses: Merck KGaA, Pfizer
Daniel J. Renouf
Honoraria: Roche, Bayer
Consulting or Advisory Role: Roche, Bayer, Viatris
Research Funding: Bayer, Roche (Inst)
Nelson Dussetti
Stock and Other Ownership Interests: Predicting Med
Patents, Royalties, Other Intellectual Property: PCT/EP2022/065222. Simple transcriptomic signatures to determine chemosensitivity for pancreatic ductal adenocarcinoma. SATT Sud-Est. Inventors: N. Dusetti, N. Fraunhoffer, J. Iovanna, 2021. Licensing to Predicting Med
Jérome Cros
Honoraria: Novartis
Consulting or Advisory Role: HISTALIM
Research Funding: ERYTECH Pharma (Inst)
Travel, Accommodations, Expenses: IPSEN, Novartis
No other potential conflicts of interest were reported.
REFERENCES
- 1.Siegel RL, Miller KD, Fuchs HE, et al. : Cancer statistics, 2022. CA A Cancer J Clin 72:7-33, 2022 [DOI] [PubMed] [Google Scholar]
- 2.Neoptolemos JP, Stocken DD, Bassi C, et al. : Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: A randomized controlled trial. JAMA 304:1073, 2010 [DOI] [PubMed] [Google Scholar]
- 3.Neoptolemos J, Dunn J, Stocken D, et al. : Adjuvant chemoradiotherapy and chemotherapy in resectable pancreatic cancer: A randomised controlled trial. Lancet 358:1576-1585, 2001 [DOI] [PubMed] [Google Scholar]
- 4.Oettle H, Post S, Neuhaus P, et al. : Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: A randomized controlled trial. JAMA 297:267, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Conroy T, Hammel P, Hebbar M, et al. : FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N Engl J Med 379:2395-2406, 2018 [DOI] [PubMed] [Google Scholar]
- 6.Conroy T, Castan F, Lopez A, et al. : Five-year outcomes of FOLFIRINOX vs gemcitabine as adjuvant therapy for pancreatic cancer: A randomized clinical trial. JAMA Oncol 8:1571-1578, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ducreux M, Cuhna AS, Caramella C, et al. : Cancer of the pancreas: ESMO clinical Practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 26:v56-v68, 2015 [DOI] [PubMed] [Google Scholar]
- 8.Greenhalf W, Ghaneh P, Neoptolemos JP, et al. : Pancreatic cancer hENT1 expression and survival from gemcitabine in patients from the ESPAC-3 trial. J Natl Cancer Inst 106:djt347, 2014 [DOI] [PubMed] [Google Scholar]
- 9.Farrell JJ, Elsaleh H, Garcia M, et al. : Human equilibrative nucleoside transporter 1 levels predict response to gemcitabine in patients with pancreatic cancer. Gastroenterology 136:187-195, 2009 [DOI] [PubMed] [Google Scholar]
- 10.Maréchal R, Bachet J, Mackey JR, et al. : Levels of gemcitabine transport and metabolism proteins predict survival times of patients treated with gemcitabine for pancreatic adenocarcinoma. Gastroenterology 143:664-674.e6, 2012 [DOI] [PubMed] [Google Scholar]
- 11.Raffenne J, Nicolle R, Puleo F, et al. : hENT1 testing in pancreatic ductal adenocarcinoma: Are we ready? A multimodal evaluation of hENT1 status. Cancers 11:1808, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicolle R, Gayet O, Duconseil P, et al. : A transcriptomic signature to predict adjuvant gemcitabine sensitivity in pancreatic adenocarcinoma. Ann Oncol 32:250-260, 2021 [DOI] [PubMed] [Google Scholar]
- 13.Nicolle R, Gayet O, Bigonnet M, et al. : Relevance of biopsy-derived pancreatic organoids in the development of efficient transcriptomic signatures to predict adjuvant chemosensitivity in pancreatic cancer. Transl Oncol 16:101315, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banneau G, Ayadi M, Armenoult L, et al. : Homogenization of cartilage tumors to extract total RNA to microarray and sequencing analysis using Precellys bead-beating technology. BioTechniques 52:196-197, 2012 [Google Scholar]
- 15.Dobin A, Davis CA, Schlesinger F, et al. : STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29:15-21, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao Y, Smyth GK, Shi W: featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923-930, 2014 [DOI] [PubMed] [Google Scholar]
- 17.Grambsch PM, Therneau TM: Proportional hazards tests and diagnostics based on weighted residuals. Biometrika 81:515-526, 1994 [Google Scholar]
- 18.Oettle H, Neuhaus P, Hochhaus A, et al. : Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: The CONKO-001 randomized trial. JAMA 310:1473, 2013 [DOI] [PubMed] [Google Scholar]
- 19.Neoptolemos JP, Palmer DH, Ghaneh P, et al. : Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): A multicentre, open-label, randomised, phase 3 trial. Lancet 389:1011-1024, 2017 [DOI] [PubMed] [Google Scholar]
- 20.Sinn M, Sinn BV, Treue D, et al. : TP53 mutations predict sensitivity to adjuvant gemcitabine in patients with pancreatic ductal adenocarcinoma: Next-generation sequencing results from the CONKO-001 trial. Clin Cancer Res 26:3732-3739, 2020 [DOI] [PubMed] [Google Scholar]
- 21.Neesse A, Michl P, Frese KK, et al. : Stromal biology and therapy in pancreatic cancer. Gut 60:861-868, 2011 [DOI] [PubMed] [Google Scholar]
- 22.Maurer C, Holmstrom SR, He J, et al. : Experimental microdissection enables functional harmonisation of pancreatic cancer subtypes. Gut 68:1034-1043, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blair AB, Groot VP, Gemenetzis G, et al. : BRCA1/BRCA2 germline mutation carriers and sporadic pancreatic ductal adenocarcinoma. J Am Coll Surg 226:630-637e1, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park W, Chen J, Chou JF, et al. : Genomic methods identify homologous recombination deficiency in pancreas adenocarcinoma and optimize treatment selection. Clin Cancer Res 26:3239-3247, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Reilly EM, Lee JW, Zalupski M, et al. : Randomized, multicenter, phase II trial of gemcitabine and cisplatin with or without veliparib in patients with pancreas adenocarcinoma and a germline BRCA/PALB2 mutation. J Clin Oncol 38:1378-1388, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicolle R, Blum Y, Duconseil P, et al. : Establishment of a pancreatic adenocarcinoma molecular gradient (PAMG) that predicts the clinical outcome of pancreatic cancer. EBioMedicine 57:102858, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Comprehensive Cancer Network : NCCN Guidelines Version 1.2022. Pancreatic Adenocarcinoma. http://www.nccn.org
- 28.van Dam JL, Janssen QP, Besselink MG, et al. : Neoadjuvant therapy or upfront surgery for resectable and borderline resectable pancreatic cancer: A meta-analysis of randomised controlled trials. Eur J Cancer 160:140-149, 2022 [DOI] [PubMed] [Google Scholar]
- 29.Versteijne E, van Dam JL, Suker M, et al. : Neoadjuvant chemoradiotherapy versus upfront surgery for resectable and borderline resectable pancreatic cancer: Long-term results of the Dutch randomized PREOPANC trial. J Clin Oncol 40:1220-1230, 2022 [DOI] [PubMed] [Google Scholar]
- 30.Palmer AC, Sorger PK: Combination cancer therapy can confer benefit via patient-to-patient variability without drug additivity or synergy. Cell 171:1678-1691.e13, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perera S, Jang GH, Wang Y, et al. : hENT1 expression predicts response to gemcitabine and nab-paclitaxel in advanced pancreatic ductal adenocarcinoma (PDAC). Clin Cancer Res 28:5115-5120, 2022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/JCO.22.02668.