Abstract
Advances in the functional genomics and bioinformatics toolkits for Strongyloides species have positioned these species as genetically tractable model systems for gastrointestinal parasitic nematodes. As community interest in mechanistic studies of Strongyloides species continues to grow, publicly accessible reference genomes and associated genome annotations are critical resources for researchers. Genome annotations for multiple Strongyloides species are broadly available via the WormBase and WormBase ParaSite online repositories. However, a recent phylogenetic analysis of the receptor-type guanylate cyclase (rGC) gene family in two Strongyloides species highlights the potential for errors in a large percentage of current Strongyloides gene models. Here, we present three examples of gene annotation updates within the Strongyloides rGC gene family; each example illustrates a type of error that may occur frequently within the annotation data for Strongyloides genomes. We also extend our analysis to 405 previously curated Strongyloides genes to confirm that gene model errors are found at high rates across gene families. Finally, we introduce a standard manual curation workflow for assessing gene annotation quality and generating corrections, and we discuss how it may be used to facilitate community-driven curation of parasitic nematode biodata.
This article is part of the Theo Murphy meeting issue ‘Strongyloides: omics to worm-free populations’.
Keywords: Strongyloides, nematodes, WormBase, comparative genomics, genome annotation, community curation
1. Introduction
Soil-transmitted gastrointestinal parasitic nematodes in the genus Strongyloides are a major source of neglected disease and economic burden worldwide [1,2]. In humans, Strongyloides stercoralis is the primary causative agent of strongyloidiasis, a potentially fatal disease [1,3,4]. The basic biology of Strongyloides species, like other soil-transmitted parasitic nematodes, is not well understood; the mechanistic basis of parasitism in these species is similarly understudied. This is owing, in part, to the historical lack of functional genomics techniques adapted for use in these species. However, recent efforts have significantly advanced the number of functional genomics pipelines and bioinformatics tools in Strongyloides species, including protocols for transgenesis, CRISPR/Cas9-mediated mutagenesis, RNA interference (RNAi), chemogenetic neuronal silencing, fluorescent biosensor imaging and on-demand analysis of gene expression and codon usage [5–12].
Technical advances in the functional Strongyloides toolkit have been enabled by publicly available descriptive genome biodata, including high-quality reference genomes, RNA-sequencing (RNA-Seq) datasets and automated genome annotations that identify predicted gene models [9,10,13–15]. Genome annotations are a cornerstone of high-throughput and targeted genomics studies: they serve as a scaffold for alignment and quantification of RNA-Seq data; they also enable researchers to more easily identify putative promoter regions for transcriptional reporters, exon sequences for RNAi, target sites for CRISPR/Cas9-mediated mutagenesis and predicted protein sequences for comparative genomics and ectopic expression studies [5,6,8,11,13–18].
For genome annotations to facilitate research efforts equitably and effectively within and across scientific communities, it is critical that the information be both broadly available and highly accurate. Central repositories that provide free access to current genome assemblies and annotations are one method for accomplishing equitable distribution of genome biodata. When responsively maintained, these repositories can also support efforts to continuously improve annotation quality through in-house or community-driven curation pipelines. In the case of Strongyloides species, genome sequences and annotations are publicly distributed through two linked online resources: WormBase and WormBase ParaSite, which together maintain genomic records for multiple free-living and parasitic nematode species despite chronic underfunding and understaffing [19–22]. Currently, WormBase hosts genomic biodata for the rodent-parasitic nematode Strongyloides ratti, while WormBase ParaSite hosts three additional Strongyloides species (S. stercoralis, Strongyloides papillosus and Strongyloides venezuelensis) and mirrors S. ratti data [20,21]. Although some Strongyloides gene models were generated de novo, the majority of the Strongyloides genome annotations were predicted through a semi-automated annotation procedure in which an in-house automatic pipeline created first-pass gene models followed over time by targeted manual curation of selected gene families by the Strongyloides research community [11,13–15,23–31]. Notably, only a few Strongyloides gene families have been manually curated, particularly in comparison to the Caenorhabditis elegans reference annotation, which has been extensively refined via both in-house and community-based improvement pipelines and is regarded as the canonical standard [19]. Although the published reference annotations for Strongyloides genomes are an important baseline, the accuracy of the first-pass gene models, and thus the predicted Strongyloides proteomes, is generally unknown.
Here, we highlight the limitations of relying on first-pass annotations of the Strongyloides genomes and the need for community-driven improvements of gene model accuracy, using examples identified during a recent investigation of the receptor-type guanylate cyclase (rGC) gene family in S. stercoralis and S. ratti [11]. These examples illustrate three common gene model errors as well as potential strategies for developing corrections using both indirect (e.g. comparative genomics between Strongyloides species and C. elegans) and direct (e.g. complementary DNA or transcriptome sequencing) evidence [11]. We extend this analysis to a larger set of 405 previously curated S. stercoralis genes, to demonstrate that high error rates are not restricted to the rGC gene family. Finally, we propose a standard manual curation workflow that we hope will help facilitate community-driven improvements to the Strongyloides genome annotation.
2. Strongyloides gene model errors and curation solutions
The rGC gene family encodes single-pass transmembrane receptors involved in sensory transduction in both free-living and parasitic nematodes [11,32–37]. The rGC gene family is particularly expanded in nematodes compared to mammalian genomes [11,37–39]. A recent phylogenetic comparison between S. stercoralis, S. ratti, and C. elegans revealed that although these species' genomes share similar numbers of rGC genes, the degree of protein similarity between individual Strongyloides and C. elegans genes varies [11]. An accurate phylogenetic analysis of free-living and parasitic rGC genes was possible only following manual curation of the Strongyloides gene family. The gene model updates identified during the curation process demonstrate three gene model errors that are probably common throughout the genome annotations of Strongyloides species: (i) inappropriately merged genes, (ii) genes with intron–exon boundary errors, and (iii) genes with incorrect start/end regions. Below, we use three example rGC genes to illustrate a process for identifying annotation errors and determining an appropriate update to the gene models.
(a) . Example 1: separating merged genes to resolve protein homologies
In C. elegans, the rGC gene daf-11 contributes to a range of physiological processes, including: olfaction and pheromone sensing, phototransduction, dauer formation/recovery, ageing and the oxidative stress response [32,40–44]. In the predatory nematode Pristionchus pacificus, daf-11 links the sensation of environmental stimuli to developmental switch genes that drive the transition between alternative predator and bacterial feeder mouth-forms [45]. To identify the S. ratti daf-11 homologue, we first used a BLASTP search to compare Ce-DAF-11 against S. ratti predicted proteins; this revealed a match to the SRAE_1000175800 coding sequence (figure 1a; electronic supplementary material, S1). A subsequent TBLASTN search to identify regions of the S. ratti genome encoding putative homologues of C. elegans DAF-11 also identified the gene SRAE_1000175800 as the nearest match. To confirm homology, we conducted a reciprocal BLASTP search of the C. elegans proteome using the predicted coding sequence of SRAE_1000175800 retrieved from WormBase. Surprisingly, this BLASTP search identified Ce-LET-418 and Ce-CHD-3 as the nearest homologues to SRAE_1000175800 instead of Ce-DAF-11 (figure 1a; electronic supplementary material, S1). Ce-LET-418 and Ce-CHD-3 both encode subunits of the Mi-2 chromatin-remodelling protein and play essential roles during C. elegans embryonic development [46–48]. Correspondingly, the automatically generated gene description and gene homology record for SRAE_1000175800 on WormBase predicted involvement in chromatin remodelling and homology to Ce-let-418 and Ce-chd-3.
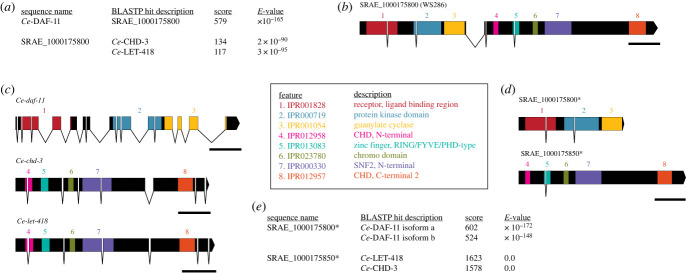
Figure 1.
Updated annotations to separate incorrectly fused genes can reveal hidden protein homologues. (a) WormBase BLASTP results for Ce-DAF-11 searched against the S. ratti proteome as well as the S. ratti SRAE_1000175800 protein sequence searched against the C. elegans proteome. Protein sequences are from WormBase release WS286. Although Ce-DAF-11 appears most similar to SRAE_1000175800, the reciprocal search identifies matches to Ce-CHD-3 and Ce-LET-418, not Ce-DAF-11. (b) Intron–exon diagram and protein motifs of the SRAE_1000175800 gene annotation from WormBase release WS286 (corresponding to WormBase ParaSite version 16). Protein motifs common to receptor-type guanylate cyclases are present in the 5′ end of the gene model (features 1–3); the 3′ end of the gene includes additional protein motifs (features 4–8). Scale bar is 500 bp. (c) Intron–exon diagrams and protein motifs of Ce-daf-11, Ce-chd-3, and Ce-let-418. Scale bar is 500 bp. (d) Intron–exon diagrams and protein motifs of updated SRAE_1000175800 and SRAE_1000175850. Scale bar is 500 bp. (e) WormBase BLASTP results for updated S. ratti protein sequences searched against the C. elegans proteome. The updated SRAE_1000175800 protein sequence is most similar to Ce-DAF-11; the new SRAE_1000175850 protein sequence retains the original match to Ce-CHD-3 and Ce-LET-418. Asterisks indicate updated gene models.
To resolve this discrepancy, we performed an in-depth examination of the SRAE_1000175800 gene model. We found that the SRAE_1000175800 annotation described a 2876 amino acid protein; by contrast, the Ce-DAF-11 isoform A protein is a significantly shorter 1077 amino acids. We next used WormBase to inspect predicted protein motifs in the SRAE_1000175800 protein sequence. An rGC protein is composed of several distinct domains: an N-terminus extracellular domain that may contain a ligand-binding region, a transmembrane domain and a C-terminus intracellular domain that contains a protein kinase domain and a guanylate cyclase domain. Motifs for a ligand-binding region, protein kinase domain and guanylate cyclase domain were all present in the predicted SRAE_1000175800 protein sequence, as well as Ce-DAF-11 (figure 1b,c; electronic supplementary material, S1). However, the SRAE_1000175800 protein sequence also included multiple protein motifs not generally associated with rGCs, including: chromodomain, helicase, DNA-binding (CHD) N- and C- terminals, a plant homeodomain zinc-finger motif, a Chromo domain, and an SNF2 N-terminal domain [46,47]. Notably, these motifs are all commonly associated with Chromo-like domain superfamily proteins, including Ce-CHD-3 and Ce-LET-418 (figure 1c), providing a likely explanation for the results of our S. ratti-to-C. elegans BLASTP search and indicating that the SRAE_1000175800 gene annotation required revision.
The extended protein length and additional protein motifs together suggested that the SRAE_1000175800 gene model represented multiple genes (Sr-daf-11 and Sr-chd-3/let-418) incorrectly annotated as a single gene. The location of the protein motifs within the original SRAE_1000175800 gene model further suggested that the gene fusion involved annotation errors occurring after the guanylate cyclase domain in exon 3, but before the N-terminal CHD domain in exon 5 (figure 1b; electronic supplementary material, S1). To separate the annotations, we first searched the unspliced SRAE_1000175800 DNA sequence for a stop codon that would terminate the gene before the region encoding the erroneous protein motifs. We found a TGA stop codon located 28 base pairs (bp) downstream of the original SRAE_1000175800 exon 3 boundary; we designated this stop codon as the putative termination site for an updated SRAE_1000175800 gene model. InterProScan analysis demonstrated that the updated SRAE_1000175800 gene encodes a 1092 amino acid protein that retains the rGC-associated protein domains (figure 1d; electronic supplementary material, S1). A BLASTP search of the C. elegans proteome with the updated SRAE_1000175800 protein sequence revealed a match to both isoforms of Ce-DAF-11 (figure 1e; electronic supplementary material, S1).
Next, we searched for an open reading frame (ORF) that could encode the 5′ region of a distinct Sr-chd-3/let-418 gene. We found an ATG sequence 8 bp downstream of the original SRAE_1000175800 exon 5 boundary that retains in-frame protein coding. We combined this marginally shortened exon with the original SRAE_1000175800 exon 6 into a separate gene annotation, named SRAE_1000175850 following consultation with WormBase. The new SRAE_1000175850 gene encodes a 1781 amino acid protein; an InterProScan search confirmed the presence of all five protein motifs found in Ce-CHD-3 and Ce-LET-418 (figure 1d; electronic supplementary material, S1). A BLASTP search of the C. elegans proteome using the new SRAE_1000175850 protein sequence identified matches to Ce-CHD-3 and Ce-LET-418 (figure 1e; electronic supplementary material, S1). The proposed modifications thus preserve a putative member of the CHD gene family while clarifying a previously ambiguous protein homology (i.e. the identity of the S. ratti DAF-11 homologue). This error illustrates the potential unreliability of the first-pass predicted proteomes of Strongyloides species; WormBase-facilitated data mining and descriptive databases that rely on proteome predictions (e.g. AlphaFold) should be considered with caution in the absence of previous curation efforts [49].
(b) . Example 2: resolving missing protein domains by adjusting intron–exon boundaries
The rGC gene Ce-gcy-23 encodes a thermoreceptor protein that is selectively expressed in C. elegans AFD neurons and contributes to thermotaxis behaviours, along with two other AFD-specific rGCs (Ce-gcy-8 and Ce-gcy-18) [35,36,50–52]. As part of our efforts to identify the S. ratti thermosensory rGCs, we used reciprocal BLASTP and TBLASTN searches to identify SRAE_2000430600 as a potential homologue of Ce-gcy-23 [11]. Like other rGCs, the Ce-GCY-23 protein contains four key elements: an extracellular domain containing a ligand-binding region, a transmembrane domain, an intracellular protein kinase domain, and an intracellular guanylate cyclase domain. When we examined the SRAE_2000430600 predicted protein sequence, we observed the absence of a transmembrane domain located between the ligand-binding region and the protein kinase domain, and instead the presence of two introns separated by a short 22 bp exon (figure 2a; electronic supplementary material, S1). The transmembrane domain is an essential feature of the rGC gene family; we therefore sought to determine whether adjusting the intron–exon boundaries of SRAE_2000430600 could uncover the missing transmembrane domain.
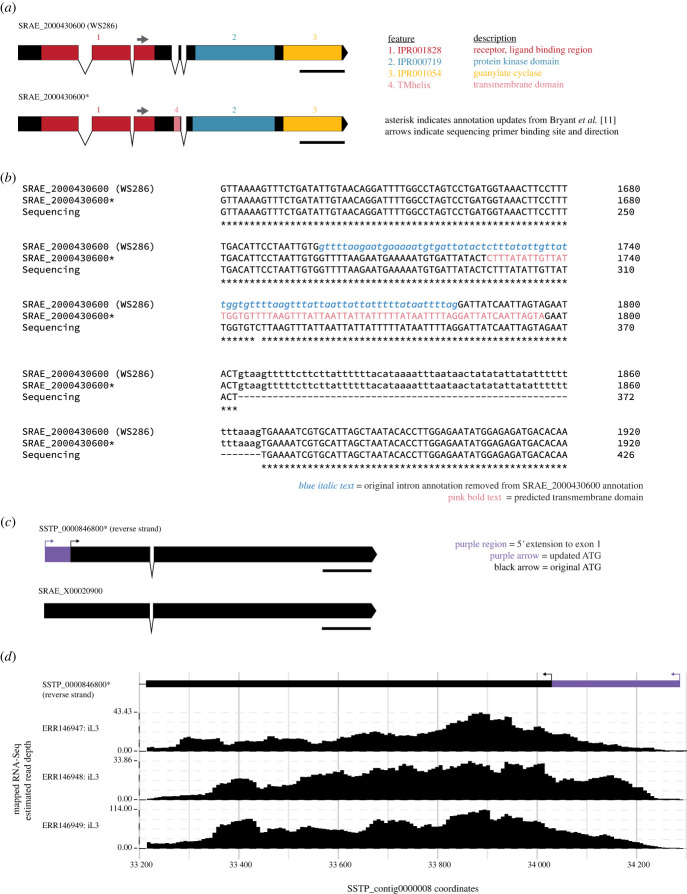
Figure 2.
Curating single gene models can reveal missing protein domains and novel start codons. (a) Intron–exon diagram and protein motifs of the original SRAE_2000430600 (upper, from WormBase version WS286) and the updated SRAE_2000430600 (lower, identified as Sr-gcy-23.1). In the original SRAE_2000430600 gene annotation, three protein motifs common to receptor-type guanylate cyclases are present: (i) a ligand-binding region; (ii) a protein kinase domain; and (iii) a guanylate cyclase domain. The updated version also contains an additional domain: (iv) transmembrane domain, which appears in the site formerly annotated as a third intron. Scale bars are 500 bp. (b) Sequencing of SRAE_2000430600 cDNA confirms the proposed update to the gene model. Nucleotide sequences show the original annotation (top), the updated annotation (middle), and the cDNA sequencing results (bottom). Asterisks indicate agreement between all three sequence sources. Blue italic text indicates the erroneous third intron included in the original gene annotation. Pink bold text indicates nucleotides that are predicted to encode a transmembrane domain. (c) Intron–exon diagrams of the SSTP_0000846800 gene model (upper, identified as Ss-gcy-23.3) and the one-to-one S. ratti homologue, SRAE_X00020900 (lower). For the SSTP_0000846800 gene model, the black region indicates the original gene model (from WormBase ParaSite version 16); the black arrow indicates location of the original start codon. The purple region shows a 5′ extension that shifts the ATG start codon upstream by 258 bp; the purple arrow indicates the new start codon site. The updated SSTP_0000846800 gene model was released in WormBase ParaSite version 17. Scale bars are 500 bp. (d) RNA-Sequencing (RNA-Seq) tracks aligned relative to the SSTP_0000846800 exon 1 gene model, showing abundant RNA-Seq reads aligning to the 5′ extension. RNA-Seq data show transcript abundance from three replicate samples of S. stercoralis third-stage infective larvae (iL3); genome-aligned tracks were exported from the WormBase ParaSite Region in Detail view in the Location widget [14,20]. Note that SSTP_0000846800 is located on the reverse strand, thus the orientation of the gene is flipped relative to panel (c).
We retrieved the unspliced SRAE_2000430600 sequence and examined the region from the end of exon 3, which encodes the C-terminus of the ligand-binding receptor motif, to the start of exon 5, which encodes the protein kinase and guanylate cyclase domains (figure 2a). These exons 3 and 5 were separated by an 84 bp intron, a 22 bp exon 4 and then a 64 bp intron. We found an in-frame ORF that spanned the end of exon 3 through exon 4, terminating in the 64 bp intron. We preliminarily redrew the intron–exon boundaries, removing the 84 bp intron and generating a longer exon 3 that extended through to the predicted start of the 64 bp intron. We analysed the resulting 1129 amino acid protein with InterProScan and found a transmembrane domain located within the extended exon 3 (figure 2a; electronic supplementary material, S1).
To confirm the proposed intron–exon adjustments, we performed RNA extraction from S. ratti third-stage infective larvae, then used reverse transcriptase polymerase chain reaction (RT-PCR) to amplify SRAE_2000430600 complementary DNA (cDNA). We sequenced the cDNA amplicon using a forward primer binding in exon 3; the results matched our proposed exon boundaries. Specifically, we confirmed cDNA sequencing reads that spanned the original 84 bp intron; if an intron existed in that location, those nucleotides would be excluded from the cDNA amplicon (figure 2b). Notably, this strategy for confirming intron–exon identity does not require access to genome-aligned RNA-Seq data and is therefore helpful for cryptic Strongyloides species or other parasitic nematode species for which high-quality RNA-Seq data is not yet available (e.g. Parastrongyloides trichosuri) [53,54].
(c) . Example 3: adjusting a truncated gene model using RNA-Sequencing data
Our investigation of putative AFD-specific rGCs also included the S. stercoralis genome; for these genes we specifically sought to characterize functional properties as part of a broader study of the molecular and cellular basis of temperature-driven host-seeking behaviours of S. stercoralis infective larvae [11]. We again used reciprocal BLASTP and TBLASTN searches, which identified SSTP_0000846800 as a putative homologue of Ce-gcy-23. The SSTP_0000846800 gene model described a 1032 amino acid protein that was notably shorter than other S. ratti and S. stercoralis putative AFD-specific rGCs, as well as Ce-GCY-23, by approximately 40–100 amino acids. An initial multiple sequence alignment with other rGCs suggested that the N-terminus of SSTP_0000846800 was truncated. A comparison of the intron–exon structures of SSTP_0000846800 and a projected one-to-one S. ratti homologue, SRAE_X00020900, supported the need to elongate the SSTP_0000846800 N-terminus (figure 2c). Examination of publicly available genome-aligned S. stercoralis RNA-Seq tracks confirmed the presence of contiguous transcript reads extending into the intergenic region upstream of the original start codon (figure 2d) [13,20,21]. To identify an updated start codon location, we first downloaded the unspliced SSTP_0000846800 sequence plus approximately 500 bp of 5′ untranslated region sequence. We then identified an in-frame ORF that shifted the ATG upstream by 258 bp and added an additional 86 amino acids to the N-terminus of the protein. Finally, we confirmed that the 5′ exon addition overlapped with genome-aligned RNA-Seq track reads from WormBase ParaSite (figure 2c,d). This example illustrates the potential for more subtle errors in Strongyloides gene structures that are most visible as part of a systematic analysis of the gene family across closely related species. Observed in isolation, the truncated N-terminal region of the SSTP_0000846800 protein was initially attributed to the divergence between the C. elegans and Strongyloides genomes [13]. Only when SSTP_0000846800 was viewed in comparison to its one-to-one S. ratti homologue, as well as other S. ratti and S. stercoralis AFD-specific rGCs, was the annotation error revealed. This example also highlights the benefit of high-quality, publicly accessible, genome-aligned RNA-Seq tracks for guiding gene model corrections.
3. A workflow for manual curation of Strongyloides gene models
The above sections present three examples of the most common types of gene annotation errors that we identified while investigating the rGC gene family in S. stercoralis and S. ratti. Of the genes originally present in the S. stercoralis and S. ratti WS283/WBPS16 annotations that we identified as putative rGC genes based on TBLASTN homology to C. elegans rGCs, our analysis proposed corrections to 11 gene models: six for S. ratti and five for S. stercoralis. The S. ratti corrections resolved four cases in which the original gene model represented two genes incorrectly merged (as in Example 1). In two cases, SRAE_0000025200/50 and SRAE_2000417200/50, the merged genes were both putative rGCs; their separation increased the total number of identified S. ratti rGC genes by two and revealed previously obscured one-to-one homology with S. stercoralis genes. In the other two cases, SRAE_1000175800 and SRAE_1000137850, only one of the genes was an rGC and their separation thus significantly improved our phylogenetic comparison of rGC gene families across species. The remaining two S. ratti updates (SRAE_2000024700 and SRAE_2000430600), as well as all five S. stercoralis updates (SSTP_0000937800, SSTP_0000214200, SSTP_0000489700, SSTP_0000846800 and SSTP_0000912800) involved adjusting the structure of the gene model (as in examples 2 and 3). Ultimately, our manual curation pipeline identified 23 fully sequenced rGC genes in the S. ratti genome, and 24 in S. stercoralis, a significant fraction of which required updates (8 out of 23 for S. ratti and 5 out of 24 for S. stercoralis).
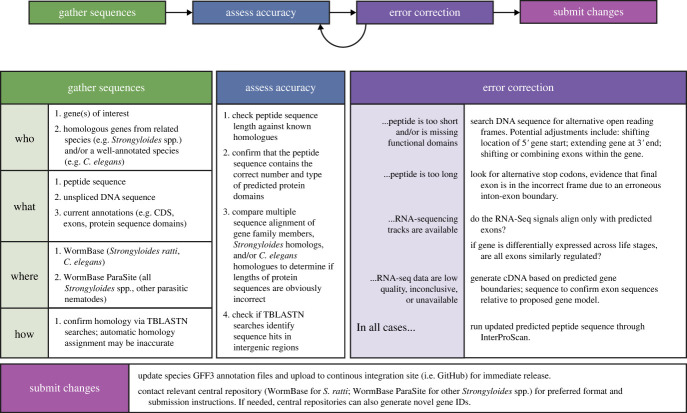
High error rates for in silico gene models are not restricted to the rGC gene family. Manual curation of a set of 405 S. stercoralis genes involved in dauer pathways, G-protein/G-protein-coupled receptor (GPCR) signalling, lipid metabolism, dafachronic acid synthesis, and sex determination identified 125 genes requiring updates to first-pass gene models (approx. 31% error rate; electronic supplementary material, S1 and S2) [14,24,25,30,31]. The high incidence of gene model errors in the S. ratti and S. stercoralis rGC gene families highlights the importance of manual curation efforts by the research community. To date, S. stercoralis and S. ratti have been the most common targets of investigation for researchers seeking to perform genomics studies of Strongyloides biology. The clear need for manual curation in these relatively prominent species highlights the likelihood that similar curation efforts will be essential in less scrutinized Strongyloides species, as well as non-Strongyloides parasitic nematodes with less advanced genomics toolkits [55]. In support of this concern, a recent effort to manually curate software-derived gene models for Caenorhabditis briggsae, another prominent model nematode, identified corrections to a substantial proportion of existing in silico gene models (8000 out of 21 000 total genes, approx. 38% error rate) [56]. To facilitate the annotation curation process within our laboratories, we developed a standard curation workflow that includes four primary steps: assembling sequences of interest, assessing annotation quality, generating annotation updates, and submitting changes to reference databases (figure 3; electronic supplementary material, S3). Below, we provide a brief description and discuss key aspects of each step.
Figure 3.
Standard workflow for manual curation of Strongyloides genome annotations. This workflow consists of four partially iterative steps: assembling sequences of interest, assessing annotation quality, generating annotation updates and submitting changes to reference databases.
(a) . Step 1: gather sequences of interest
Most mechanistic studies in Strongyloides have taken a comparative genomics approach to identifying genes of interest, using the well-described genes of C. elegans; typical experiments involve identifying Strongyloides homologues of individual C. elegans genes (e.g. tax-4, gcy-23, age-1, gpa-3, act-2, rps-21, daf-16, etc.) [5,6,11,16,17,26,57]. Alternative data mining approaches, such as searching for specific protein motifs, have promise for unbiased gene identification, as they are theoretically more likely to detect parasite-specific genes without direct homologues in other model systems [58,59]. For homology-based approaches, we strongly recommend the use of TBLASTN searches; as demonstrated in example 1, BLASTP searches are susceptible to errors in predicted protein models. Furthermore, owing largely to substantial variation in GC content across nematode genera, codon usage patterns in Strongyloides species are distinct, particularly in comparison to C. elegans [9,13,60]. As such, we highly discourage the use of nucleotide BLAST searches across nematode genera. WormBase and WormBase ParaSite both provide TBLASTN interfaces that can be used to query current Strongyloides genomes. Alternatively, researchers may wish to perform BLAST searches locally; this can be accomplished by downloading full-genome sequences from WormBase/WormBase ParaSite and searching with the user's software of choice (e.g. Geneious). We have observed some differences between different TBLASTN interfaces, particularly for identifying hits in regions with no previous annotations; this is probably owing to variations in underlying algorithm parameters across interfaces. Thus, for maximum robustness users should record underlying BLAST parameters; users might also consider comparing TBLASTN results across interfaces.
The results of these searches will be a list of potential genes of interest, or regions of the genome not yet associated with a particular gene, that should undergo annotation quality assessment. Some specific recommendations related to TBLASTN search results:
-
(i)
consider whether your ‘search’ gene/protein has multiple isoforms (protein isoforms in C. elegans are particularly well-described). The ‘A’ isoform may be arbitrary; the most rigorous approach is to repeat this process with each isoform;
-
(ii)
sort results by either ‘score’ or ‘E-value’, then copy the gene identities (IDs) of close hits. How many unique gene IDs should you copy? We have found no hard rule, as it very much depends on how many close matches the genome contains. At a bare minimum, researchers should collect all gene IDs with E-values = 0.0, as well as genes with high score values (around more than 300; this is a rough value). We recommend looking for significant changes in E-value; hits prior to a large shift are worth pursuing for manual curation, especially if supported by high score, query coverage and identity values. When investigating an entire gene family, we prefer to err on the side of more genes (first 15–20 genes), as moderate matches are probably additional gene family members or indicate genes that require manual correction to improve homology scores. Note that since E-value is partially a function of gene length, users working with short genes should probably consider all hits;
-
(iii)
when evaluating numerical values associated with BLAST hit metrics, keep in consideration the distance between the ‘search’ species and the species of interest. Greater evolutionary distances and potential lower degrees of conservation can influence how high (or low) scores/values are. For this reason, we also strongly recommend that comparative genomics approaches include comparisons across Strongyloides species, which possess higher gene homology and synteny than is found across nematode genera [11,13,15]. The four sequenced Strongyloides species can be grouped into two phylogenetic subclades: S. stercoralis and S. ratti, and S. venezuelensis and S. papillosus [13,15]. Users seeking to assess gene model accuracy in one of these species are therefore encouraged to also identify and curate the homologous gene in the relevant subclade partner. In particular, users may expect that intron–exon structure will be more conserved between Strongyloides subclade partners; minimal conservation of intron placement and size should be expected from C. elegans [13];
-
(iv)
pay particular attention to low E-value, high complexity hits that are not associated with an overlapping gene, as these can strongly indicate the presence of an incorrect annotation; and
-
(v)
the high-quality reference genomes of Strongyloides species are not yet fully contiguous. Thus, TBLASTN searches may identify partial coding regions located on contig fragments. For example, when characterizing the S. stercoralis rGC gene family, our TBLASTN searches identified three gene fragments located on short contigs: SSTP_0000270000, SSTP_0000334600 and SSTP_0000962200. These fragments each encode at least one guanylate cyclase-associated protein motif; reconstruction of these fragmented gene models would require improvements in genome contiguity (e.g. via de novo assembled short-read RNA-Seq reads or long-read DNA/RNA-sequencing [61]). In some cases, it is possible that the pattern of TBLASTN hits could be used to assemble contig fragments and thus enable full gene sequence reconstruction. Nevertheless, the presence of partial gene fragments will probably complicate phylogenetic comparisons, as contig-truncated genes can be difficult to assess for homology and should be excluded from multiple sequence alignments.
(b) . Step 2: assess annotation accuracy
Once a list of potential genes of interest has been assembled, the next step is to assess the accuracy of the gene annotations archived on WormBase and WormBase ParaSite. When performing quality assessments, users may find online tools such as GeneValidator or WBPS Gene Trees pages helpful for identifying certain errors [21,62]. In the case of the S. ratti rGC gene family, we found that although GeneValidator did successfully identify all cases in which the original gene annotation reflected two inappropriately merged genes, the results included both false negatives and false positions: cases in which manual curation found gene models lacking key protein domains were missed (including SRAE_2000430600 from example 2), and a gene model that was validated by manual curation was incorrectly categorized as being too long (electronic supplementary material, S1). The WBPS Gene Trees page depicts areas of synteny between predicted homologues across nematode species and may be helpful as a method of identifying current homology predictions or as an alternative to users performing their own preliminary multiple sequence alignments. Relying on the WBPS Gene Tree alone is probably insufficient, since the phylogenetic comparisons are based on current homology predictions, which may be inaccurate. For example, the WBPS Gene Trees comparison for Ce-daf-11 does not include an alignment to an S. ratti gene; correctly identifying and updating SRAE_1000175800 required additional targeted BLAST searches (see example 1). However, the WBPS Gene Trees alignments are probably useful for comparing predicted orthologues across many species, and for identifying potential cases where annotation errors are sufficiently severe as to disrupt alignment and homology assessments. Irrespective of the specific tools used, the answers to several questions can help determine if a gene annotation is incorrect:
-
(i)
did TBLASTN searches identify homology in a genomic region not associated with a specific gene? Often these regions will be immediately up/downstream of a BLAST-identified gene, although hits may occur in regions containing no previous gene annotation;
-
(ii)
is the length of the peptide sequence comparable to that of homologous proteins? Is it obviously too short or too long? A predicted sequence that is too long may indicate the presence of inappropriately merged genes that require splitting;
-
(iii)
are the lengths of introns obviously too long or too short? The mean/median intron lengths of Strongyloides species are as follows: 196/51 bp (S. stercoralis), 188/52 bp (S. ratti), 143/48 bp (S. papillosus) and 207/50 bp (S. venezuelensis) [13]. Abnormally large introns may suggest the need to split the existing gene model;
-
(iv)
is the TBLASTN hit to a relatively unique nucleotide sequence or to a low-complexity region? Strongyloides genomes are extremely AT-rich and highly repetitive [13,60,63]. Thus, a TBLASTN hit to a low-complexity region may be erroneous. Alternatively, hits to unique nucleotide sequences are more likely to indicate the presence of a gene and may indicate a previously unannotated gene if they are not in proximity to an existing annotation;
-
(v)
when comparing two or more Strongyloides species, how similar are the intron–exon structures? Note that Strongyloides species have significantly fewer introns compared to C. elegans [13]. Thus, the intron–exon structure of C. elegans genes will be less informative;
-
(vi)
does the transcript contain the correct number of predicted protein domains? Are there domains that are missing or added?
-
(vii)
does RNA-Seq data archived on WormBase ParaSite indicate the presence or the absence of transcripts not associated with annotated exons? For example, do the mapped RNA-Seq reads start and end where the annotation predicts? Or does the RNA-Seq data map to high-complexity sequences in the surrounding intronic regions? and
-
(viii)
does a multiple protein sequence alignment across members of the gene family reveal obvious misalignments?
(c) . Step 3: correct errors
If the answers to the above questions suggest the presence of an incorrect annotation, the next step is to generate a proposed correction. We recommend the following actions:
-
(i)
to guide adjustments to the gene start site, the gene termination site and/or intron–exon boundaries, first search for potential ORFs. DNA sequence visualization and analysis programs like ApE or Geneious have built-in tools for detecting ORFs [64]. More specialized annotation and curation tools (e.g. Apollo and Artemis) may also be useful for editing feature boundaries, particularly when aligning gene models to RNA-Seq evidence [65–67];
-
(ii)
consult RNA-Seq data associated with the genomic locus. Is the genomic sequence differentially expressed across life stages? If so, check that all predicted exons are similarly regulated; if not, this could be a clue that the gene model includes two distinct genes that need to be split. However, note that life-stage dependent exon regulation could also indicate the presence of alternative splicing events. Abundant RNA-Seq reads aligning with intronic or intergenic regions are helpful for guiding adjustments to exon boundaries, as are reads where one side of an RNA-Seq amplicon maps to one locus and the other part of the read maps to a close-by locus. Conversely, the absence of RNA-Seq reads within an exon can suggest the presence of an unannotated intron, although this interpretation is more ambiguous as low sequencing depth may also be causal;
-
(iii)
if a TBLASTN hit matched to a relatively unique nucleotide sequence that is not adjacent to an existing gene model, search for overlooked ORFs and check RNA-Seq data for mapped reads to that region;
-
(iv)
if the location of introns needs to be adjusted, remember that intron sequences will probably include canonical 5′-GT…AG-3′ splice recognition sites [61,68,69]. The conserved invertebrate exon splice sites (5′-AG^G-3′, 5′-AG^A-3′; the ‘^’ symbol indicates the exact intron insertion site) may flank intron sequences, although their conservation in Strongyloides species has not been systematically evaluated [68,69]. Generally, introns will coincide with genomic sequences containing extraneous stop codons (i.e. not ORFs);
-
(v)
in the absence of clear RNA-Seq data, internal intron–exon boundaries can be confirmed by isolating messenger RNA (mRNA) from the species of interest, then performing RT-PCR amplification of the gene of interest (ideally using primers that bind near the predicted start and stop codons), followed by sequencing of the amplicon. In addition, the start/end of coding sequences can be experimentally characterized using the 5′ or 3′ rapid amplification of cDNA ends (RACE) technique, which pairs a primer binding a defined internal sequence with unknown sequences at either the 5′ or 3′ end of the mRNA [23,70,71]; and
-
(vi)
once a potential updated gene annotation has been produced, determine the updated peptide sequence. Use InterProScan, or a similar program, to compare the updated peptide against a database of protein domain signatures; the updated peptide should contain the appropriate number and type of protein domains.
(d) . Step 4: submit changes to databases
The final step is to release updated annotations to the Strongyloides research community. We propose a tandem approach in which modified gene annotations are both submitted to the relevant curated central repository (i.e. WormBase or WormBase ParaSite) and shared via an open-source platform such as GitHub. Integration of community-submitted updates to the repository-hosted reference annotations can be delayed owing to processing requirements and periodic release schedules. The use of an open-source platform like GitHub is intended as an interim resource to permit continuous integration and release of updated annotations prior to packaged release through the centralized repositories. Below, we discuss a method for efficiently sharing updated gene models with central repository staff and on continuous integration platforms. Laboratories seeking to provide updated gene models may wish to contact central repository staff directly for additional instructions or alternative submission procedures. However, laboratories should be warned that owing to funding and personnel restrictions, repository staff may have limited bandwidth for facilitating curation efforts.
To share updated gene models across platforms, our laboratories modify full-genome annotation files in the standard General File Format with version 3 specifications (GFF3). Producing an updated full-genome annotation file can significantly reduce the infrastructure burden for central repository staff and conceptually streamlines consolidation by version-control-enabled platforms like GitHub. We also generate a running edit log containing the GFF3-formatted data of the subset of genes that have received manual curation. Editing a GFF3 file should be accomplished using a text file reader (e.g. Sublime Text, TextWrangler, etc.). Users should not edit GFF3 files using spreadsheet software programs such as Excel, as GitHub cannot track changes made using these programs. Furthermore, although specialized sequence data analysis software (e.g. Geneious, Apollo, Artemis) can be useful in calculating updated genomic coordinates, researchers are cautioned that the process for exporting annotations may introduce unwelcome formatting changes that will preclude GitHub version control.
When accessed via a text file reader, the Strongyloides GFF3 genome annotation files are organized by gene ID, with each gene feature consisting of a one-line row with nine tab-delimited data columns (electronic supplementary material, S1 and S2). To split a single gene annotation into multiple genes (as in example 1), researchers should add additional gene ID sections. New gene ID numbers can be requested from WormBase (S. ratti) or WormBase ParaSite (other Strongyloides species); in general, novel gene IDs will be assigned to conserve the established accumulative numerical progression along the genome scaffold. To modify the structure of individual gene features (as in examples 2 and 3), laboratories will primarily adjust the feature start and end data (columns 4 and 5). To help track the provenance of updates, the ‘source’ of all gene features in the updated annotation (including individual lines that were not manually adjusted) should be changed to reflect the laboratory responsible for the update. To enable indexing of community curated annotations (CCAs) across laboratories and dates, we encourage users to use the following source name construction: CCA_ < WormBase Laboratory identifier > _year + month + day (e.g. CCA_EAH_230103 for edits proposed by the Hallem Laboratory and shared with the community on 3 January 2023). Researchers should also update feature attributes stored as name-value pairs in column 9, as needed (electronic supplementary material, S1). After editing the full-genome annotation file, researchers should copy all gene features from the updated annotations into a separate GFF3-formatted edit log file. Once updated GFF3 annotation files have been generated, researchers should contact the relevant central repository to submit updates; we urge laboratories to consider the limited bandwidth of repository staff and restrict the frequency of individual submissions as much as possible. Note that future changes to repository resources may alter the ability of repository staff to accommodate submissions from individual laboratories. Finally, we propose a shared GitHub repository for rapid sharing of updated annotation files across laboratories. We have established a public repository (https://github.com/HallemLab/Parasite_Genome_Annotations) for sharing updated annotation files between researchers in our laboratories. The goal of this GitHub repository is to allow individuals to submit potential edits for merging into the most recent central repository-generated GFF3 annotation file possible and to collect community-generated annotations into a running edit log file that that can be uploaded as a WormBase/WormBase ParaSite JBrowse track. To mitigate issues arising from multiple groups proposing conflicting edits to overlapping genes, we have instituted branch protection on the main repository branch such that merges require approval from repository moderators. In addition, we highly encourage individual laboratories/editors to commit edits back to GitHub frequently; we have provided discussion forums in the GitHub repository that can be used by individuals to announce new annotations or discuss potential conflicts. Within the GitHub repository, README files are used to track when repository files are superseded by a central repository release and a species-specific gene history file is used to track changes associated with gene names and identifiers. The gene history file should match formatting in the WormBase Gene Name Sanitizer tool and should be used to record changes associated with the following identifiers: WBGene numbers (for S. ratti), SRAE/SSTP/SPAL/SVE numbers, and gene names (e.g. Sr-daf-11) (electronic supplementary material, S1). To upload new genome annotations, researchers may generate their own repository branch for merging to the main repository or can choose to directly contact the authors of this manuscript for help from repository administrators.
4. Discussion
In recent years, rapid growth in the functional genomics and bioinformatics toolkits in Strongyloides species has positioned members of this genus as highly tractable model systems for soil-transmitted parasitic nematodes. Although large-scale, genome-wide datasets for multiple Strongyloides species are publicly available, improvements to current gene models are needed. As the scientific community continues to expand our knowledge of the molecular and genetic basis of parasitism in Strongyloides species, we urge researchers to assess, and if necessary, update annotations of genes relevant to their scientific research. The workflow presented here relies primarily on manual BLAST-driven annotation assessments and improvements. In the future, accumulation of greater numbers of validated Strongyloides gene models, representing a variety of gene families, will enable the production of more robust training and refinement datasets for gene annotation software tools (e.g. Maker, Maker2) that would enable a more high-throughput approach to improving Strongyloides genome annotations [72–74]. Ultimately, we hope that the workflow presented here, as well as the examples discussed, will help facilitate community-driven improvements to the Strongyloides reference genomes.
5. Overview of methods for manual gene curation
Identification and manual curation of rGC genes in S. stercoralis and S. ratti was as previously described [11]. In brief, C. elegans rGC peptide sequences were retrieved from WormBase (versions WS283 and WS286; sequences are not different between these two versions) and used to identify putative Strongyloides rGC-encoding genes. Homology-driven searches (BLASTP and TBLASTN) for Strongyloides rGC genes were performed using WormBase and WormBase ParaSite online BLAST interfaces, as well as locally using Geneious Prime 2022.0.01 (Dotmatics, Boston, MA, USA); results from each interface were compared and used to generate a comprehensive list of putative rGC genes [19–21,75]. DNA, peptide and intron–exon biodata for putative Strongyloides rGC proteins were downloaded from WormBase (versions WS283 and WS286; Strongyloides rGC biodata are not different between these two versions) or WormBase ParaSite (release WBPS16). Protein motif predictions for original protein sequences were accessed via WormBase and WormBase ParaSite. Geneious was used for MUSCLE alignment of protein sequences. Individual gene model adjustments were generated using Geneious or the ApE plasmid editor [64]. Gene-structure diagrams were generated with Exon–Intron Graphic Maker (v4, http://www.wormweb.org). When using RNA-Seq data to guide model updates, RNA-Seq tracks for S. stercoralis and S. ratti were accessed via WormBase ParaSite [13,14,20,21]. To test whether the proposed annotation updates improved the BLASTP and TBLASTN hits of an individual gene/protein, we used both the WormBase and WormBase ParaSite BLAST interfaces [19–21,75]. We used the InterPro web interface, InterProScan to characterize protein motifs in updated sequences [76]. For analysis of original S. ratti rGC gene models with GeneValidator, peptide sequences of putative rGC genes were downloaded from WormBase and analysed using a local terminal installation of GeneValidator (v2.1.12, https://github.com/wurmlab/genevalidator).
To calculate the new genomic coordinates of updated annotation features (i.e. exon start/stop positions), publicly available S. stercoralis and S. ratti GFF3-formatted genome annotation files were downloaded from WormBase ParaSite (release WBPS16) and imported into Geneious along with genomic sequence (FASTA) files. The resulting annotated genomic sequence files were then edited in Geneious to duplicate the proposed gene model edits; the following annotation elements were altered as appropriate: gene, coding sequence, mRNA, exons and Introns. Finally, to generate updated whole-genome GFF3 annotation files, the S. stercoralis and S. ratti GFF3 files were edited directly using Sublime Text, using Geneious-calculated genomic coordinates as a reference. Files were submitted directly to WormBase (S. ratti) and WormBase ParaSite (S. stercoralis) for inclusion in the respective databases and archived for immediate release via GitHub (https://github.com/HallemLab/ Bryant_et_al_2021) [11].
To generate SRAE_2000430600 cDNA, RNA was first extracted from S. stercoralis infective third-stage larvae, then treated with RNase-free DNase to digest contaminating DNA, followed by RNA cleanup and concentration using the Qiagen RNeasy MinElute Cleanup Kit. We then used the Invitrogen SuperScript III One-Step RT-PCR system with Platinum Taq DNA polymerase to amplify SRAE_2000430600 cDNA, using the following primers: ggatccatgattgacaacaaaattttttcattttttatttttattttttac (forward, note the insertion of the underlined 5′ BamH1 recognition site), ggtaccgctcatatcttaccaaattgattttgaagttctactgg (reverse, note the insertion of the underlined 3′ KpnI recognition site). Sequencing of amplified cDNA was performed by Laragen (Culver City, CA, USA), using the sequencing primer tcaattatgaataagacaggtggagatttc (forward).
Manual curation of S. stercoralis genes involved in dauer pathways, G-protein/GPCR signalling, lipid metabolism, dafachronic acid synthesis and sex determination was as previously described [14,24,25,30,31]. The error rate of this dataset was calculated by filtering a full-genome S. stercoralis GFF3 file with UNIX commands (electronic supplementary material, S1 and S2).
Acknowledgements
We thank Dr Michelle Castelletto for advice related to Strongyloides genomics. We also thank Dionysios Grigoriadis, Dr Wen Chen, and Breanna Walsh for advice related to manual curation of parasite gene annotations. We thank Dr Sarah Dyer and Stavros Diamantakis for advice related to WormBase submissions. We thank WormBase and WormBase ParaSite.
Ethics
This work did not require ethical approval from a human subject or animal welfare committee.
Data accessibility
All data for this study are available from the GitHub repository: https://github.com/BryantLabUW/Bryant_etal_2023 [77] and from the Zenodo repository: https://zenodo.org/records/8125688 [78].
Supplementary material is available online [79].
Declaration of AI use
We have not used AI-assisted technologies in creating this article.
Authors' contributions
A.S.B.: conceptualization, funding acquisition, investigation, methodology, project administration, visualization, writing—original draft, writing—review and editing; D.A.: methodology, writing—review and editing; J.D.C.S.: data curation, methodology, resources, writing—review and editing; E.A.H.: conceptualization, funding acquisition, project administration, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
The authors declare no competing interests.
Funding
This work was supported by National Institutes of Health R01AI136976 (E.A.H.), an A.P. Giannini Postdoctoral Fellowship (A.S.B.) and funds provided by the University of Washington School of Medicine (A.S.B.).
References
- 1.Beknazarova M, Whiley H, Ross K. 2016. Strongyloidiasis: a disease of socioeconomic disadvantage. Int. J. Environ. Res. Public Health 13, 517. ( 10.3390/ijerph13050517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotez PJ, Brindley PJ, Bethony JM, King CH, Pearce EJ, Jacobson J. 2008. Helminth infections: the great neglected tropical diseases. J. Clin. Invest. 118, 1311-1321. ( 10.1172/JCI34261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisoffi Z, et al. 2013. Strongyloides stercoralis: a plea for action. PLoS Negl. Trop. Dis. 7, 7-10. ( 10.1371/journal.pntd.0002214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamarozzi F, Martello E, Giorli G, Fittipaldo A, Staffolani S, Montresor A, Bisoffi Z, Buonfrate D. 2019. Morbidity associated with chronic Strongyloides stercoralis infection: a systematic review and meta-analysis. Am. J. Trop. Med. Hyg. 100, 1305-1311. ( 10.4269/ajtmh.18-0895) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gang SS, Castelletto ML, Bryant AS, Yang E, Mancuso N, Lopez JB, Pellegrini M, Hallem EA. 2017. Targeted mutagenesis in a human-parasitic nematode. PLoS Pathog. 13, e1006675. ( 10.1371/journal.ppat.1006675) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lok JB, Shao H, Massey HC, Li X. 2017. Transgenesis in Strongyloides and related parasitic nematodes: historical perspectives, current functional genomic applications and progress towards gene disruption and editing. Parasitology 144, 327-342. ( 10.1017/S0031182016000391) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castelletto ML, Gang SS, Hallem EA. 2020. Recent advances in functional genomics for parasitic nematodes of mammals. J. Exp. Biol. 223, jeb206482. ( 10.1242/jeb.206482) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dulovic A, Streit A. 2019. RNAi-mediated knockdown of daf-12 in the model parasitic nematode Strongyloides ratti. PLoS Pathog. 15, e1007705. ( 10.1371/journal.ppat.1007705) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant AS, Hallem EA. 2021. The Wild Worm Codon Adapter: a web tool for automated codon adaptation of transgenes for expression in non-Caenorhabditis nematodes. G3 (Bethesda) 11, jkab146. ( 10.1093/g3journal/jkab146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryant AS, DeMarco SF, Hallem EA. 2021. Strongyloides RNA-Seq Browser: a web-based software platform for on-demand bioinformatics analyses of Strongyloides species. G3 (Bethesda) 11, jkab104. ( 10.1093/g3journal/jkab104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryant AS, Ruiz F, Lee JH, Hallem EA. 2022. The neural basis of heat seeking in a human-infective parasitic worm. Curr. Biol. 32, 2206-2221. ( 10.1016/j.cub.2022.04.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendez P, Walsh B, Hallem EA. 2022. Using newly optimized genetic tools to probe Strongyloides sensory behaviors. Mol. Biochem. Parasitol. 250, 111491. ( 10.1016/j.molbiopara.2022.111491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunt VL, et al. 2016. The genomic basis of parasitism in the Strongyloides clade of nematodes. Nat. Genet. 48, 299-307. ( 10.1038/ng.3495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoltzfus JD, Minot S, Berriman M, Nolan TJ, Lok JB. 2012. RNAseq analysis of the parasitic nematode Strongyloides stercoralis reveals divergent regulation of canonical dauer pathways. PLoS Negl Trop Dis 6, e1854. ( 10.1371/journal.pntd.0001854) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunt VL, Hino A, Yoshida A, Kikuchi T. 2018. Comparative transcriptomics gives insights into the evolution of parasitism in Strongyloides nematodes at the genus, subclade and species level. Sci. Rep. 8, 5192. ( 10.1038/s41598-018-23514-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoltzfus JD, Massey HC, Nolan TJ, Griffith SD, Lok JB. 2012. Strongyloides stercoralis age-1: a potential regulator of infective larval development in a parasitic nematode. PLoS ONE 7, e38587. ( 10.1371/journal.pone.0038587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Junio AB, Li X, Massey HC, Nolan TJ, Todd Lamitina S, Sundaram MV, Lok JB. 2008. Strongyloides stercoralis: cell- and tissue-specific transgene expression and co-transformation with vector constructs incorporating a common multifunctional 3′ UTR. Exp. Parasitol. 118, 253-265. ( 10.1016/j.exppara.2007.08.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheong MC, Wang Z, Jaleta TG, Li X, Lok JB, Kliewer SA, Mangelsdorf DJ. 2021. Identification of a nuclear receptor/coactivator developmental signaling pathway in the nematode parasite Strongyloides stercoralis. Proc. Natl Acad. Sci. USA 118, e2021864118. ( 10.1073/pnas.2021864118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis P, et al. 2022. WormBase in 2022—data, processes, and tools for analyzing Caenorhabditis elegans. Genetics 220, iyac003. ( 10.1093/genetics/iyac003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howe KL, Bolt BJ, Shafie M, Kersey P, Berriman M. 2017. WormBase ParaSite−a comprehensive resource for helminth genomics. Mol. Biochem. Parasitol. 215, 2-10. ( 10.1016/j.molbiopara.2016.11.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolt BJ, Rodgers FH, Shafie M, Kersey PJ, Berriman M, Howe KL. 2018. Using WormBase ParaSite: an integrated platform for exploring helminth genomic data. Methods Mol. Biol. 1757, 471-491. ( 10.1007/978-1-4939-7737-6_15) [DOI] [PubMed] [Google Scholar]
- 22.Howe KL, et al. 2016. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 44, D774-D780. ( 10.1093/nar/gkv1217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massey HC, Ranjit N, Stoltzfus JD, Lok JB. 2013. Strongyloides stercoralis daf-2 encodes a divergent ortholog of Caenorhabditis elegans DAF-2. Int. J. Parasitol. 43, 515-520. ( 10.1016/j.ijpara.2013.01.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albarqi MMY, Stoltzfus JD, Pilgrim AA, Nolan TJ, Wang Z, Kliewer SA, Mangelsdorf DJ, Lok JB. 2016. Regulation of life cycle checkpoints and developmental activation of infective larvae in Strongyloides stercoralis by dafachronic acid. PLoS Pathog. 12, e1005358. ( 10.1371/journal.ppat.1005358) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoltzfus JD, Bart SM, Lok JB. 2014. cGMP and NHR signaling co-regulate expression of insulin-like peptides and developmental activation of infective larvae in Strongyloides stercoralis. PLoS Pathog. 10, e1004235. ( 10.1371/journal.ppat.1004235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massey HC, Ball CC, Lok JB. 2001. PCR amplification of putative gpa-2 and gpa-3 orthologs from the (A + T)-rich genome of Strongyloides stercoralis. Int. J. Parasitol. 31, 377-383. ( 10.1016/S0020-7519(01)00117-5) [DOI] [PubMed] [Google Scholar]
- 27.Yuan W, et al. 2014. Toward understanding the functional role of Ss-RIOK-1, a RIO protein kinase-encoding gene of Strongyloides stercoralis. PLoS Negl. Trop. Dis. 8, e3062. ( 10.1371/journal.pntd.0003062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan W, Liu Y, Lok JB, Stoltzfus JD, Gasser RB, Lei W, Fang R, Zhao J, Hu M. 2014. Exploring features and function of Ss-riok-3, an enigmatic kinase gene from Strongyloides stercoralis. Parasit. Vectors 7, 561. ( 10.1186/s13071-014-0561-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei W-Q, et al. 2017. Structural and developmental expression of Ss-riok-2, an RIO protein kinase encoding gene of Strongyloides stercoralis. Sci. Rep. 7, 8693. ( 10.1038/s41598-017-07991-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonzalez AD, Dalessandro EJ, Nolan TJ, Stieha CR, Lok JB, Stoltzfus JDC. 2021. Transcriptional profiles in Strongyloides stercoralis males reveal deviations from the Caenorhabditis sex determination model. Sci. Rep. 11, 8254. ( 10.1038/s41598-021-87478-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Stoltzfus J, You Y, Ranjit N, Tang H, Xie Y, Lok JB, Mangelsdorf DJ, Kliewer SA. 2015. The nuclear receptor DAF-12 regulates nutrient metabolism and reproductive growth in nematodes. PLoS Genet. 11, e1005027. ( 10.1371/journal.pgen.1005027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birnby DA, Link EM, Vowels JJ, Tian H, Colacurcio PL, Thomas JH. 2000. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in Caenorhabditis elegans. Genetics 155, 85-104. ( 10.1093/genetics/155.1.85) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.L'Etoile ND, Bargmann CI. 2000. Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron 25, 575-586. ( 10.1016/s0896-6273(00)81061-2) [DOI] [PubMed] [Google Scholar]
- 34.Hallem EA, et al. 2011. Receptor-type guanylate cyclase is required for carbon dioxide sensation by Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 108, 254-259. ( 10.1073/pnas.1017354108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeishi A, Yu YV, Hapiak VM, Bell HW, O'Leary T, Sengupta P. 2016. Receptor-type guanylyl cyclases confer thermosensory responses in C. elegans. Neuron 90, 235-244. ( 10.1016/j.neuron.2016.03.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inada H, Ito H, Satterlee J, Sengupta P, Matsumoto K, Mori I. 2006. Identification of guanylyl cyclases that function in thermosensory neurons of Caenorhabditis elegans. Genetics 172, 2239-2252. ( 10.1534/genetics.105.050013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maruyama IN. 2016. Receptor guanylyl cyclases in sensory processing. Front. Endocrinol. 7, 173. ( 10.3389/fendo.2016.00173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fitzpatrick DA, O'Halloran DM, Burnell AM. 2006. Multiple lineage specific expansions within the guanylyl cyclase gene family. BMC Evol. Biol. 6, 26. ( 10.1186/1471-2148-6-26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ortiz CO, Etchberger JF, Posy SL, Frøkjær-Jensen C, Lockery S, Honig B, Hobert O. 2006. Searching for neuronal left/right asymmetry: genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics 173, 131-149. ( 10.1534/genetics.106.055749) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beckert U, Aw WY, Burhenne H, Försterling L, Kaever V, Timmons L, Seifert R. 2013. The receptor-bound guanylyl cyclase DAF-11 is the mediator of hydrogen peroxide-induced cGMP increase in Caenorhabditis elegans. PLoS ONE 8, e72569. ( 10.1371/journal.pone.0072569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, et al. 2010. C. elegans phototransduction requires a G protein–dependent cGMP pathway and a taste receptor homolog. Nat. Neurosci. 13, 715-722. ( 10.1038/nn.2540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vowels JJ, Thomas JH. 1994. Multiple chemosensory defects in daf-11 and daf-21 mutants of Caenorhabditis elegans. Genetics 138, 303-316. ( 10.1093/genetics/138.2.303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen PAT, Liou W, Hall DH, Leroux MR. 2014. Ciliopathy proteins establish a bipartite signaling compartment in a C. elegans thermosensory neuron. J. Cell Sci. 127, 5317-5330. ( 10.1242/jcs.157610) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murakami M, Koga M, Ohshima Y. 2001. DAF-7/TGF-β expression required for the normal larval development in C. elegans is controlled by a presumed guanylyl cyclase DAF-11. Mech. Dev. 109, 27-35. ( 10.1016/S0925-4773(01)00507-X) [DOI] [PubMed] [Google Scholar]
- 45.Lenuzzi M, Witte H, Riebesell M, Rödelsperger C, Hong RL, Sommer RJ. 2021. Influence of environmental temperature on mouth-form plasticity in Pristionchus pacificus acts through daf-11-dependent cGMP signaling. J. Exp. Zool. B Mol. Dev. Evol. 340, 214-224. ( 10.1002/jez.b.23094) [DOI] [PubMed] [Google Scholar]
- 46.von Zelewsky T, Palladino F, Brunschwig K, Tobler H, Hajnal A, Müller F. 2000. The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development 127, 5277-5284. ( 10.1242/dev.127.24.5277) [DOI] [PubMed] [Google Scholar]
- 47.Käser-Pébernard S, Pfefferli C, Aschinger C, Wicky C. 2016. Fine-tuning of chromatin composition and Polycomb recruitment by two Mi2 homologues during C. elegans early embryonic development. Epigenetics Chromatin 9, 39. ( 10.1186/s13072-016-0091-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turcotte CA, Sloat SA, Rigothi JA, Rosenkranse E, Northrup AL, Andrews NP, Checchi PM. 2018. Maintenance of genome integrity by Mi2 homologs CHD-3 and LET-418 in Caenorhabditis elegans. Genetics 208, 991-1007. ( 10.1534/genetics.118.300686) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jumper J, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583-589. ( 10.1038/s41586-021-03819-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wasserman SM, Beverly M, Bell HW, Sengupta P. 2011. Regulation of response properties and operating range of the AFD thermosensory neurons by cGMP signaling. Curr. Biol. 21, 353-362. ( 10.1016/j.cub.2011.01.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang D, O'Halloran D, Goodman MB. 2013. GCY-8, PDE-2, and NCS-1 are critical elements of the cGMP-dependent thermotransduction cascade in the AFD neurons responsible for C. elegans thermotaxis. J. Gen. Physiol. 142, 437-449. ( 10.1085/jgp.201310959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramot D, MacInnis BL, Goodman MB. 2008. Bidirectional temperature-sensing by a single thermosensory neuron in C. elegans. Nat. Neurosci. 11, 908-915. ( 10.1038/nn.2157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beknazarova M, Barratt JLN, Bradbury RS, Lane M, Whiley H, Ross K. 2019. Detection of classic and cryptic Strongyloides genotypes by deep amplicon sequencing: a preliminary survey of dog and human specimens collected from remote Australian communities. PLoS Negl. Trop. Dis. 13, e0007241. ( 10.1371/journal.pntd.0007241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frias L, Stark DJ, Lynn MS, Nathan SKSS, Goossens B, Okamoto M, MacIntosh AJJ. 2018. Lurking in the dark: cryptic Strongyloides in a Bornean slow loris. Int. J. Parasitol. Parasites Wildl. 7, 141-146. ( 10.1016/j.ijppaw.2018.03.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doyle SR. 2022. Improving helminth genome resources in the post-genomic era. Trends Parasitol. 38, 831-840. ( 10.1016/j.pt.2022.06.002) [DOI] [PubMed] [Google Scholar]
- 56.Moya ND, et al. 2023. Novel and improved Caenorhabditis briggsae gene models generated by community curation. BMC Genomics 24, 486. ( 10.1186/s12864-023-09582-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bryant AS, Ruiz F, Gang SS, Castelletto ML, Lopez JB, Hallem EA. 2018. A critical role for thermosensation in host seeking by skin-penetrating nematodes. Curr. Biol. 28, 2338-2347. ( 10.1016/j.cub.2018.05.063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wheeler NJ, Heimark ZW, Airs PM, Mann A, Bartholomay LC, Zamanian M. 2020. Genetic and functional diversification of chemosensory pathway receptors in mosquito-borne filarial nematodes. PLoS Biol. 18, e3000723. ( 10.1371/journal.pbio.3000723) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Atkinson LE, et al. 2021. Phylum-spanning neuropeptide GPCR identification and prioritization: shaping drug target discovery pipelines for nematode parasite control. Front. Endocrinol. 12, 718363. ( 10.3389/fendo.2021.718363) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mitreva M, Wendl MC, Martin J, Wylie T, Yin Y, Larson A, Parkinson J, Waterston RH, McCarter JP. 2006. Codon usage patterns in Nematoda: analysis based on over 25 million codons in thirty-two species. Genome Biol. 7, R75. ( 10.1186/gb-2006-7-8-r75) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wheeler NJ, Airs PM, Zamanian M. 2020. Long-read RNA sequencing of human and animal filarial parasites improves gene models and discovers operons. PLoS Negl. Trop. Dis. 14, e0008869. ( 10.1371/journal.pntd.0008869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drăgan M-A, Moghul I, Priyam A, Bustos C, Wurm Y. 2016. GeneValidator: identify problems with protein-coding gene predictions. Bioinformatics 32, 1559-1561. ( 10.1093/bioinformatics/btw015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cutter AD, Wasmuth JD, Blaxter ML. 2006. The evolution of biased codon and amino acid usage in nematode genomes. Mol. Biol. Evol. 23, 2303-2315. ( 10.1093/molbev/msl097) [DOI] [PubMed] [Google Scholar]
- 64.Davis MW, Jorgensen EM. 2022. ApE, a plasmid editor: a freely available DNA manipulation and visualization program. Front. Bioinform. 2, 818619. ( 10.3389/fbinf.2022.818619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, Parkhill J, Rajandream M-A. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24, 2672-2676. ( 10.1093/bioinformatics/btn529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. 2012. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464-469. ( 10.1093/bioinformatics/btr703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee E, et al. 2013. Web Apollo: a web-based genomic annotation editing platform. Genome Biol. 14, R93. ( 10.1186/gb-2013-14-8-r93) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shapiro MB, Senapathy P. 1987. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 15, 7155-7174. ( 10.1093/nar/15.17.7155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blumenthal T, Steward K. 1997. Chapter 6: RNA processing and gene structure. In C. elegans II, 2nd edn (eds Riddle DL, Blumenthal T, Meyer BJ, Priess JR), Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- 70.Scotto-Lavino E, Du G, Frohman MA. 2006. Amplification of 5′ end cDNA with ‘new RACE’. Nat. Protoc. 1, 3056-3061. ( 10.1038/nprot.2006.479) [DOI] [PubMed] [Google Scholar]
- 71.Scotto-Lavino E, Du G, Frohman MA. 2006. 3′ End cDNA amplification using classic RACE. Nat. Protoc. 1, 2742-2745. ( 10.1038/nprot.2006.481) [DOI] [PubMed] [Google Scholar]
- 72.Campbell MS, Holt C, Moore B, Yandell M. 2014. Genome annotation and curation using MAKER and MAKER-P. Curr. Protoc. Bioinform. 48, 4.11.1-4.11.39. ( 10.1002/0471250953.bi0411s48) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Sánchez Alvarado A, Yandell M. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188-196. ( 10.1101/gr.6743907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinf. 12, 491. ( 10.1186/1471-2105-12-491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5-W9. ( 10.1093/nar/gkn201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paysan-Lafosse T, et al. 2023. InterPro in 2022. Nucleic Acids Res. 51, D418-D427. ( 10.1093/nar/gkac993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bryant AS, Akimori D, Stoltzfus JDC, Hallem EA. 2023. Data from: A standard workflow for community-driven manual curation of Strongyloides genome annotations. GitHub repository. (https://github.com/BryantLabUW/Bryant_etal_2023) [DOI] [PubMed]
- 78.Bryant AS, Akimori D, Stoltzfus JDC, Hallem EA. 2023. Code for: A standard workflow for community-driven manual curation of Strongyloides genome annotations. Zenodo. ( 10.5281/zenodo.8125688) [DOI] [PubMed]
- 79.Bryant AS, Akimori D, Stoltzfus JDC, Hallem EA. 2023. A standard workflow for community-driven manual curation of Strongyloides genome annotations. Figshare. ( 10.6084/m9.figshare.c.6888228) [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Bryant AS, Akimori D, Stoltzfus JDC, Hallem EA. 2023. Data from: A standard workflow for community-driven manual curation of Strongyloides genome annotations. GitHub repository. (https://github.com/BryantLabUW/Bryant_etal_2023) [DOI] [PubMed]
- Bryant AS, Akimori D, Stoltzfus JDC, Hallem EA. 2023. Code for: A standard workflow for community-driven manual curation of Strongyloides genome annotations. Zenodo. ( 10.5281/zenodo.8125688) [DOI] [PubMed]
- Bryant AS, Akimori D, Stoltzfus JDC, Hallem EA. 2023. A standard workflow for community-driven manual curation of Strongyloides genome annotations. Figshare. ( 10.6084/m9.figshare.c.6888228) [DOI] [PubMed]
Data Availability Statement
All data for this study are available from the GitHub repository: https://github.com/BryantLabUW/Bryant_etal_2023 [77] and from the Zenodo repository: https://zenodo.org/records/8125688 [78].
Supplementary material is available online [79].