Abstract
Spatial transcriptomics (ST) has demonstrated enormous potential for generating intricate molecular maps of cells within tissues. Here we present iStar, a method based on hierarchical image feature extraction that integrates ST data and high-resolution histology images to predict spatial gene expression with super-resolution. Our method enhances gene expression resolution to near-single-cell levels in ST and enables gene expression prediction in tissue sections where only histology images are available.
Keywords: spatial transcriptomics, super-resolution, histology, machine learning
The rapid advancement of ST technologies has made it possible to measure gene expression within the original tissue context1–4, enabling researchers to characterize spatial gene expression patterns5–7, study cell-cell communications8, 9, and resolve the spatiotemporal order of cellular development10. Despite the availability of many ST platforms, none of them provide a comprehensive solution. An ideal ST platform should offer single-cell resolution, cover the entire transcriptome, capture a large tissue area, and be cost-effective. While generating such ST data with existing platforms remains challenging, computational approaches can be employed to reconstruct such data in silico.
Popular experimental methods for ST include in situ sequencing or hybridization-based technologies, such as STARmap11, seqFISH12–14, and MERFISH15, 16, and spatial barcoding followed by next-generation sequencing-based technologies, such as 10x Visium, SLIDE-seqV217, and Stereo-seq18. These platforms differ in their spatial resolution and gene coverage. In situ sequencing or hybridization-based methods typically have a higher spatial resolution and sensitivity but relatively lower multiplexity for genes, whereas sequencing-based methods cover the entire transcriptome but have a lower spatial resolution, which limits their ability in studying detailed gene expression patterns.
Previous studies have shown that gene expression patterns are correlated with histological image features, suggesting the possibility of predicting gene expression from histology19–21. However, these existing methods do not fully utilize the rich cellular information provided by high-resolution histology images. In practice, a pathologist examines a histology image hierarchically. In this process, the first step is to identify a region of interest (ROI) through the examination of high-level image features that capture the global tissue structure. After a ROI is identified, low-level image features that reflect the local cellular structure of the tissue are examined. To mimic this process, we propose to use a hierarchical image feature extraction approach that aims to capture both local and global tissue structures. We further develop a super-resolution gene expression prediction model by leveraging high-resolution tissue information obtained from hierarchically extracted image features. The resulting gene expression enables cell type annotation with a near-single-cell resolution. We have implemented these procedures in iStar (Inferring Super-resolution Tissue ARchitecture).
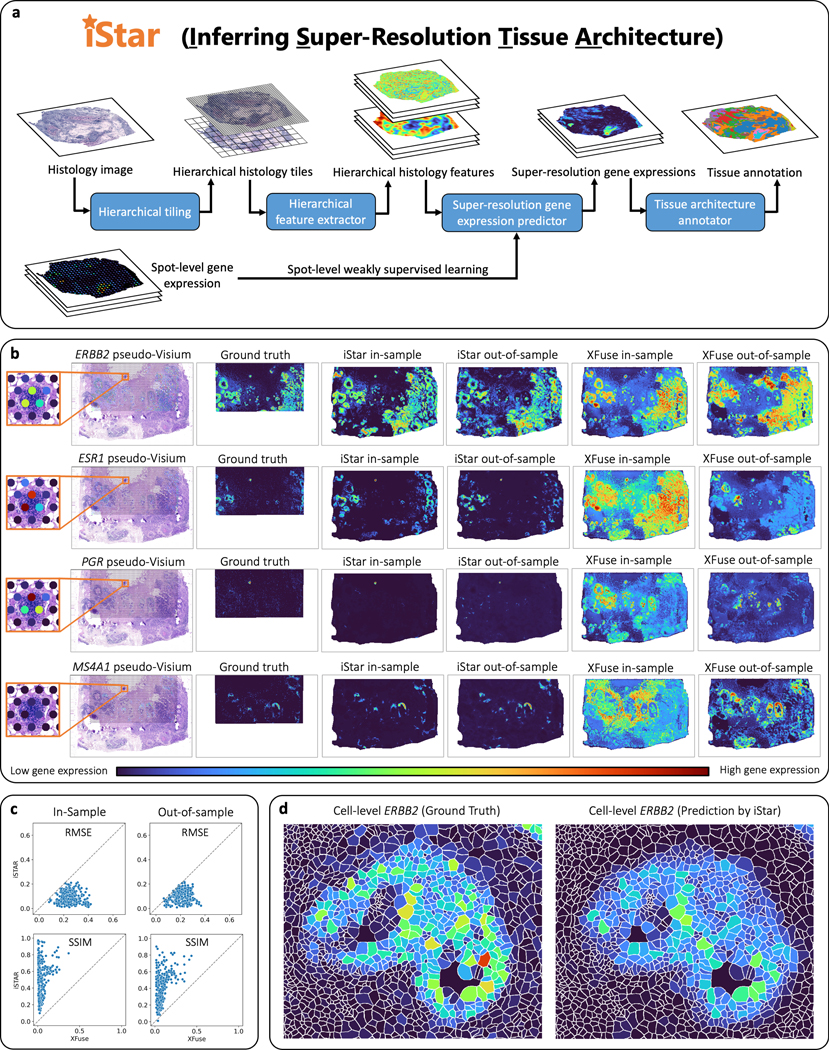
An overview of iStar is shown in Fig. 1a. Our method employs a hierarchical vision transformer22–24 (HViT) that has been pre-trained on publicly available hematoxylin-and-eosin-stained histology image datasets using self-supervised learning25, 26. The HViT initially extracts histology features at a -pixel scale to capture fine-grained tissue characteristics, followed by -pixel scale to capture global tissue structures. Subsequently, a feed-forward neural network, trained through weakly supervised learning, uses these features to predict superpixel-level gene expressions. This model divides the gene expression measurement at a given spot for each gene into multiple values, assigning one to each superpixel, facilitated by the histology features at every superpixel. Additionally, the model predicts superpixel-level gene expressions outside the spots as well as in external tissue sections, as long as histology images are available.
Fig. 1 |. Workflow and super-resolution gene expression prediction accuracy of iStar.
a, Model summary of iStar. The histology image is hierarchically divided into tiles, which are then converted into hierarchical histology image features. These features, combined with the spot-level gene expression data, are then utilized to predict super-resolution gene expression. Finally, tissue architecture is inferred based on the super-resolved gene expression prediction. b, c, Evaluation of super-resolution gene expression prediction accuracy using the Xenium breast cancer dataset, which includes two consecutively cut tissue sections. The Xenium data served as the ground truth and were used to simulate spot-level gene expression based on the spot size and layout of Visium. For in-sample prediction, both model training and prediction were performed using section 2’s pseudo-Visium data. For out-of-sample prediction, section 1 was used for model training and section 2 was used for prediction in which only its histology image was used as input. b, Visual comparison between iStar and XFuse. ERBB2, ESR1, and PGR are genes that encode biomarkers for breast cancer prognosis, while MS4A1 is a B cell marker gene. Super-resolution gene expressions are visualized at the scale of 8x resolution enhancement. Visualizations of additional genes at the scale of 8x resolution enhancement are presented in Supplementary Figs. 1–5. Visualizations at other resolutions are available in Supplementary Fig. 6 and at https://upenn.box.com/v/istar-results-benchmark. c, Numerical comparison between iStar and XFuse for 128x resolution enhancement of gene expression. The degree of resolution enhancement is defined as the number of superpixels in the super-resolution prediction divided by the number of spots in the training data. Each dot represents one of the 313 genes. Additional numerical evaluations are reported in Supplementary Fig. 7. d, Predicted single cell-level gene expression, which was computed from the predicted superpixel-level gene expression using the cell segmentation masks provided in the dataset. Additional examples are presented in Supplementary Fig. 9.
To assess the accuracy of iStar in super-resolution gene expression prediction, we applied it to a simulated dataset derived from a Xenium breast cancer dataset generated by 10x Genomics27. The Xenium dataset comprises sub-cellular ST data for 313 genes, measured in two consecutively cut tissue sections from a single patient. To simulate Visium data, we binned the Xenium gene expressions based on Visium’s spot size and layout. We assessed prediction accuracy for both in-sample and out-of-sample predictions. For in-sample prediction, super-resolution gene expression prediction was performed on Section 2’s pseudo-Visium data. For out-of-sample prediction, the pseudo-Visium data from Section 1 was used as the training data, and super-resolution gene expression prediction was performed on Section 2 using only its histology image as the input. We compared the prediction accuracy of iStar to that of the state-of-the-art method XFuse28, and visually, iStar’s predictions more closely match the ground truth as measured by Xenium compared to XFuse (Fig. 1b). To numerically evaluate the performance, we calculated the root mean square error (RMSE) and structural similarity index measure29 (SSIM) between the predicted and ground truth gene expressions for each gene. iStar outperformed XFuse for virtually all genes across all resolutions (Fig. 1c, Extended Data Figs. 1–3 and Supplementary Figs. 1–5). iStar not only enhances the resolution of gene expression within the measured spots but also predicts high-resolution gene expression outside the measured spots, such as tissue gaps between spots and adjacent tissue sections where only the histology image is available. We further assessed iStar’s ability to predict single-cell level gene expression. As shown in Fig. 1d and Extended Data Figs. 4 and 5, iStar predicted gene expression resembles that measured by Xenium.
Next, we assessed iStar’s capability for high-resolution annotation of tissue architecture of multiple tissue sections. In contrast to existing frameworks, which often involve challenging image registration tasks, we show that iStar can bypass the image registration step. To illustrate this capability, we used the breast cancer data in Fig. 1 as an example, assuming pseudo-Visium training data were available for Section 1 but not for Section 2. To perform super-resolution gene expression prediction for the two sections, we concatenated their histology images into one and treated it as a single image in downstream analyses to perform super-resolution gene expression prediction for both sections. We used the second last layer of the feed-forward neural network as features for the k-means algorithm30, and the resulting segmentation highly agreed with the manual annotation and successfully separated invasive cancer (cluster Brown), ductal carcinoma in situ (DCIS) #1 (cluster Grey), and DCIS #2 (cluster Cyan) from the rest of the tissue (Fig. 2a, Supplementary Fig. 6). By contrast, segmentation using super-resolution gene expression predicted by XFuse failed to separate DCIS #2 from invasive cancer or DCIS #2 from DCIS #1. Moreover, iStar was able to annotate tissue regions outside the spot-covered tissue area. Finally, the annotation for Section 2 closely resembled that of Section 1, demonstrating iStar’s consistency across multiple samples.
Fig. 2 |. Tissue annotation using iStar.
a, Comparison of unsupervised tissue segmentation by iStar and XFuse with manual annotation of one of the two consecutively cut tissue sections of a breast cancer patient in the Xenium dataset. The model was trained using the pseudo-Visium spot-level gene expression simulated from Section 1 (in-sample) of the Xenium data. Section 2 was treated as the out-of-sample section, and its super-resolution gene expression was predicted only using its histology image. Super-resolution was performed with 128x resolution enhancement. b, Tissue architecture annotation of a breast cancer tissue in the HER2ST breast cancer dataset (three consecutively cut tissue sections in Subject H). Super-resolution was performed with 128x resolution enhancement. c, iStar assigned biologically meaningful labels to the tissue clusters by performing superpixel-level cell type inference, followed by a cell type enrichment analysis, where depletion, i.e., negative enrichment, was not shown in the heatmap. d, iStar’s unsupervised tissue segmentation revealed intraltumoral heterogeneity that agreed with the pathologist’s manual annotation. e, A small cancer region detected by iStar that was missed in manual annotation provided in the original publication. f, Detection of Tertiary Lymphoid Structures (TLS) in the HER2ST breast cancer dataset (Subject G) by iStar. The TLS score was calculated as the mean of the standardized super-resolution gene expressions of the TLS marker genes shown in Supplementary Table 1.
To evaluate iStar’s generalizability in super-resolution tissue segmentation and annotation, we applied it to another HER2+ breast cancer dataset31 (denoted as HER2ST) generated using the legacy Spatial Transcriptomics technology32, which has a lower spatial resolution than Visium (Fig. 2b). We considered three consecutively cut tissue sections from Subject H, where manual annotation was provided for only one section in the original publication. To segment all three sections, we carried out multi-sample tissue segmentation using the same approach as in Fig. 2a and found iStar showed strong agreement with the coarse manual annotation while providing increased granularity (Fig. 2b, Supplementary Figs. 7–9). Moreover, the three sections exhibited similar structures, demonstrating the consistency of iStar across samples. After segmenting the tissue, we conducted cell-type annotation at the superpixel level (Fig. 2c) and inferred cell types based on predicted gene expressions of marker genes33. The cell type annotation yielded cell type proportion estimates within each tissue cluster, enabling the evaluation of cell type enrichment. As shown in Fig. 2c, Clusters 9 (cyan), 6 (pink), and 4 (purple), and 3 (red) closely matched with the invasive and in situ cancer regions based on the manual annotation and were enriched with cancer epithelial cells. Furthermore, Clusters 8 (yellow) and 5 (brown) were enriched with B cells and T cells, as expected from the manual annotation. Fig. 2c and Supplementary Fig. 10 visualize the superpixels annotated as B cells, T cells, cancer epithelial cells, and other cell types. The underlying biological relevance of each tissue cluster is also hinted at by the most over-expressed genes in the cluster. For example, FABP4 (encodes fatty acid binding protein) was enriched in Cluster 1 (orange), CD8A (a lineage marker of T cell) was enriched in Cluster 5 (brown), and MS4A1 (a lineage marker of B cell) was enriched in Cluster 8 (olive) (Supplementary Fig. 11).
Further examination of the iStar unsupervised segmentation revealed intratumoral heterogeneity, as shown in Fig. 2d, where the refined annotation was provided by a board-certified pathologist (E.E.F). Overall, superpixel-level cell type annotation provides biologically meaningful interpretations of the automatically detected tissue clusters, closely aligned with manual labels while revealing fine tissue structures. Notably, iStar was even able to detect a positive surgical margin (Fig. 2e) missed in the original manual annotation, and the validity of this cancer region was confirmed by E.E.F, demonstrating that iStar can identify small regions of interest that are neglected during the initial manual annotation. Our findings in this dataset suggest that iStar can accurately annotate tissue architecture even for the legacy Spatial Transcriptomics platform. The identification of biologically relevant genes within tissue clusters further supports the potential utility of iStar in uncovering new insights into tissue biology and diseases.
Next, we show that iStar can be utilized to detect multicellular structures, such as tertiary lymphoid structures (TLSs), which are clusters of highly organized immune cells formed in non-lymphoid tissues, often found at sites of inflammation, including a variety of solid tumors34, 35. The presence of TLSs has been shown to be associated with positive clinical outcomes and responses to immunotherapy36–38. However, the manual detection of TLSs using the spot-resolution Visium data is labor-intensive and imprecise, due to the small size and the fine-grained characteristics of TLSs. To demonstrate the ability of iStar for automatic TLS detection, we analyzed three consecutively cut tissue sections of another patient (Subject G) in the HER2ST dataset. In order to detect TLSs, we curated a list of unique TLS marker genes (Supplementary Table 1) and computed TLS gene signature scores by standardizing and averaging the predicted gene expression of the TLS marker genes (Fig. 2f). We identified multiple TLSs, all of which were confirmed by a board-certified pathologist (E.E.F.), and the TLS marker gene expressions are shown in Supplementary Fig. 12. By contrast, the original HER2ST study31 detected several TLSs, but the analyses were based on low-resolution spot-level gene expression, resulting in a much lower resolution compared to our results (Extended Data Fig. 6).
In addition to the two breast cancer datasets analyzed above, we also analyzed one additional breast cancer dataset generated using Visium by 10x Genomics. As shown in Supplementary Fig. 13, iStar revealed fine-grained tissue structures. Although we have primarily focused on the applications to breast cancer in this study, iStar is a generic tool that can be applied to various diseased or healthy tissue types. To demonstrate iStar’s capability in analyzing healthy tissues, we conducted benchmarking evaluations using a Xenium dataset generated from mouse brain by 10x Genomics. The benchmarking was designed similarly to the experiments for the Xenium-derived pseudo-Visium breast cancer dataset. As shown in Extended Data Figs. 7–8a and Supplementary Figs. 14–17, iStar achieved high accuracy in this evaluation across all resolutions and outperformed XFuse. In addition, our super-resolution gene expression-based segmentation (Extended Data Fig. 8b) revealed a fine-grained tissue structure that matches closely with the Allen Brain Atlas annotation.
Finally, to demonstrate iStar’s broad applicability to diverse cancer and healthy tissue types, we applied it to additional Visium datasets generated from mouse brain (Extended Data Fig. 9), mouse kidney (Extended Data Fig. 10a), prostate cancer (Extended Data Fig. 10b, Supplementary Figs. 18), colorectal cancer (Extended Data Fig. 10c), and kidney cancer (Extended Data Fig. 10d). In all applications, iStar was able to characterize tissue architecture with high resolution. For example, iStar accurately detected TLSs that aligned well with pathologist’s manual annotation in kidney cancer (Extended Data Fig. 10d).
In summary, we have presented iStar, a method for rapid annotation of super-resolution tissue architecture based on ST data generated from platforms that lack single-cell resolution. This holds implications for practical studies, as existing ST platforms lack either single-cell resolution or whole-transcriptome coverage. However, iStar allows us to generate ST data that cover the entire transcriptome with near-single-cell resolution (Supplementary Fig. 19). A key step of iStar is to leverage the high-resolution histology image obtained from the same ST tissue section to reconstruct the unobserved super-resolution gene expression. Through the analysis of several datasets across multiple cancer types and healthy tissues, we have demonstrated that the super-resolution gene expressions predicted by iStar are accurate. These predictions not only preserve the original gene expression at the spot-level (Supplementary Figs. 20 and 21) but also have practical applications in various tissue architecture inference tasks. Moreover, we have shown that iStar can perform out-of-sample prediction for tissue sections where only histology images are available. iStar is computationally efficient, with the end-to-end analysis of the Xenium-derived pseudo-Visium breast cancer data taking only 9 minutes (Supplementary Table 2). By contrast, XFuse took 1,969 minutes (more than 32 hours) to analyze the same data, which was 218 times slower. This advantage in computational efficiency allows iStar to generate virtual ST data from a large number of consecutively cut tissue sections with histology images, enabling a comprehensive characterization of gene expression variations in 3D tissues39.
Methods
The algorithm of iStar
The algorithm of iStar consists of three components: the histology feature extractor, super-resolution gene expression predictor, and tissue architecture annotator.
Histology feature extractor
To facilitate the processing of histology images with different resolutions, we first rescale each image such that the size of one pixel is . This rescaling ensures a -pixel tile corresponds to ., which is about the size of a single cell. To simplify the subsequent tiling procedure, we pad the rescaled image so that its height and width are both divisible by 256.
Next, we partition the whole image into image tiles hierarchically such that the large (high-level) tiles reflect the global tissue structure, whereas the small (low-level) tiles within a large tile reflect the local fine-grained cellular structure of the tissue. Let be the RGB-channel histology image with height and width . We first partition into a -row, -column rectangular grid of -pixel image tiles: , where each . Next, each -pixel image tile is further partitioned into a 16-row, 16-column rectangular grid of -pixel image tiles: , where each .
To extract hierarchical histology features, we use a hierarchical vision transformer architecture22–24 that consists of a local vision transformer (ViT) and a global ViT . First, within each -pixel image tile, the local ViT maps each -pixel sub-tile into a low-level local feature vector of length , that is,
and then maps all the 256 low-level local feature vectors within the -pixel image tile into a high-level local feature vector of length , that is,
Next, to model long-range dependencies of histology features within the whole image, the global ViT maps all the high-level local features within the whole image into high-level global features of the same dimension:
After this hierarchical histology feature extraction procedure, we have
the high-level global feature image , which is an image of size with channles,
the low-level local feature image , which is an image of size with channels, and
the original RGB image, which is an image of size with 3 channels.
To align these feature images, we use bicubic interpolation to resize each image into the desired size and stack the channels of the resized images, which results in a combined histology feature image of size with channels, where each is the histology feature vector at pixel . In our implementation, we set and . For the image size, we varied among , , , and .
To train the histology feature extractor, we optimize the ViTs through self-supervised learning (SSL). Because of the benefits of transfer learning on ViTs40, the model is pretrained on publicly available histology datasets. In this step, since only histology images are needed and no gene expression data or image-level labels are required, many publicly available histology datasets, such as the Cancer Genome Atlas (TCGA), the Genotype-Tissue Expression (GTEx) project, and the kidney biopsies in Holscher et al. (2023) 41, are suitable for pretraining the model. Moreover, for the choice of the SSL algorithm, any SSL method for analyzing image data, such as DINO25 or BEIT26, is suitable for our purpose. In our implementation, we adopted the pretrained model in Chen et al. (2022) 22, which uses DINO to train the ViTs hierarchically on the TCGA data. In our experiments, we found that the pretrained model was able to capture the histology characteristics well and thus decided to skip the fine-tuning step to improve computation efficiency.
Super-resolution gene expression predictor
Once the histology feature images have been extracted, we use them to predict super-resolution gene expression. The histology features at every superpixel contain information on not only its local cellular characteristics but also its global relationships to other regions in the whole image. Thus the gene expression predictor does not need to explicitly model the spatial dependencies, since correlation between superpixels are already reflected in the similarity between their high-level global histology features (i.e. the channels in the combined histology feature image ), even if the superpixels are physically far away from each other. Therefore, when predicting the gene expression at a superpixel, the input of the predictor only includes the histology features at this superpixel, and no convolution, attention, or any other mechanisms with spatial awareness are needed, which substantially reduces the computation cost.
To train the super-resolution gene expression predictor, since the model output is at the superpixel level but the training data are at the spot level, we adopt a weakly supervised learning framework. We model the gene expression observed at each spot as the sum of the superpixels’ gene expression inside that spot. This model design mimics the data collection procedure of sequencing-based ST platforms, which barcodes and combines all the transcripts inside a spot into a sample and sends it for next-generation sequencing. To express the loss function, let be the number of spots in the whole image, be the number of genes to predict, be the gene expression prediction model for gene , be the observed gene expression for gene at spot , be the spot mask of (i.e. the collection of superpixels covered by spot ), and be the histology feature vector at superpixel . Then the weakly supervised loss function is
Superpixels outside the spot masks, including the between-spot gaps and the background image, are excluded during model training. After model training, the predicted gene expression for gene at superpixel is , which gives us the gene expression image . Furthermore, if cell segmentation masks are provided, single cell-level gene expression can be obtained using the predicted superpixel-level gene expressions, where the former is computed as a weighted sum of the latter, with the weight equal to the proportion of the superpixel that overlaps with the cell mask42. In our experiments, we only predicted the union of the top 1000 most highly variable genes in each dataset and the marker genes for the user-defined structures (e.g. TLS), since lowly variables genes had low signal-to-noise ratios and would introduce extra noise to the model training procedure. The only two exceptions are the benchmark experiments using the Xenium breast cancer dataset (313 genes) and the Xenium mouse brain dataset (248 genes), in which case we predicted all the genes due to their small number and the need for method evaluation.
For network architecture of the gene expression prediction model, we use a feed-forward neural network with 4 hidden layers and 256 nodes per hidden layer. The leaky rectified linear unit (ReLU) 43 is used as the activation function for the hidden layers. The output layer is a linear layer with 256 input nodes and output nodes, and the outputs are activated by an exponential linear unit (ELU) 44 to ensure that the predicted gene expressions are non-negative.
Tissue architecture annotator
Once obtaining the super-resolution gene expression, we segment the tissue by clustering the superpixels using their gene expression information. First, we obtain gene expression embeddings by reducing the dimension of the predicted gene expression vector, where each superpixel is treated as a sample and each gene as a feature. Although any dimension reduction technique (e.g. PCA45 or UMAP46) can obtain gene expression embeddings from the predicted super-resolution gene expression, we recommend treating the intermediate values in the second-last feed-forward layer of the gene expression prediction model as the gene expression embeddings, since they are not only low-dimensional (256 in our setting) but also linearly related to the predicted gene expression vectors, and using these pre-computed values does not incur any additional computational cost. Next, to promote spatial contiguousness in segmentation, we smooth the gene expression embeddings by a Gaussian filter, an approach that is similar in spirit to the sliding-window method for cell neighborhood identification47. Then we treat the smoothed gene expression embedding vector at every superpixel as a sample and cluster all the superpixels with the k-means algorithm30. This procedure partitions the tissue into functionally distinct regions in an unsupervised manner based on their gene expression profiles.
To assign biologically meaningful interpretations to the tissue regions in the segmentation, we perform cell type inference at the superpixel level, where we treat each superpixel as an artificial cell and infer its cell type using its predicted gene expressions along with a marker gene reference panel. Recall that the total number of genes in the model is . Let be the total number of candidate cell types. For each cell type , suppose we have a list of marker gene indices , which is a subset of . For example, in our experiments with breast cancer data, we used the marker gene lists provided by Wu et al. 33. For each marker gene , we standardize its predicted super-resolution gene expression image into the range of and obtain , where . Then for each superpixel , we compute the score for cell type by averaging the standardized gene expressions of all its marker genes: where is the number of genes in . To infer the cell type of superpixel , let be the cell type with the maximal score and be the score of this cell type. Given a predetermined threshold , if , then the cell type of superpixel is predicted to be ; otherwise, the cell type of this superpixel is unclassified. In our experiments, we set and found it effective in most cases. See Supplementary Fig. 22 for a demonstration of the effects of on cell type inference. While this score-based approach was used for cell type inference in our experiments, any cell type annotation tool serves the purpose. For example, when a well-annotated single-cell RNA-seq reference panel is available, the cell types can be annotated by methods such as SingleR48 or ItClust49. Finally, to combine the superpixel-level predicted cell types with the tissue clusters obtained through unsupervised segmentation, an enrichment analysis is applied to every cell type-tissue cluster pair, which elucidates the biological activities inside each cluster by examining the cell types over-represented in the cluster.
In addition to the above-described unsupervised tissue annotation procedure, iStar also allows annotation with user-defined tissue structures. In this procedure, a score image is produced to reflect the intensity of the user-defined structure across the tissue. The computation of the user-defined structure score is similar to the computation of the cell type score described in the previous paragraph. Given a list of user-defined gene indices , which is a subset of , for the structure of interest (e.g. TLS, Supplementary Table 1), for every gene , we first standardize its predicted super-resolution gene expression image into the range of and obtain the standardized image . Then we compute the score image for the user-defined structure by averaging the standardized gene expression images of all the marker genes: where is the number of genes in . The resulting score image reflects the activity of the user-defined structure in the tissue.
Benchmark data generation
To evaluate the super-resolution gene expression prediction accuracy, we generated spot-level pseudo-Visium data using pixel-level Xenium data27. The pixel size of the Xenium gene expression images was , and we rescaled the pixel size to . The gene expression measurements in Xenium were binned into spots based on the spot size, shape and layout of Visium: a hexagonal grid of disc-shaped spots with a spot diameter of and a center-to-center distance of . For the ground truth, we binned the Xenium gene expressions into a rectangular grid of superpixels, where the size of the superpixels varied among , , , and , depending on the experimental settings.
Evaluation criteria for super-resolution gene expression prediction accuracy
To evaluate the accuracy of the predicted super-resolution gene expressions, for each gene, we treated both the ground truth and the predicted gene expression as images, where the image intensity was standardized into the range of . Then the prediction accuracy was measured by the root mean squared error (RMSE) and the structural similarity index measure29 (SSIM). To compute the RMSE, the ground truth and the predicted gene expression images were flattened into vectors, and the RMSE was equal to the Euclidean distance between the two vectors. The RMSE is a straightforward and fast metric for assessing the prediction accuracy of any outcomes that can be vectorized, but for image data, the RMSE ignores the spatial contexts within the images50. Thus, in addition to the RMSE, we also computed the SSIM to evaluate the similarity between the spatial structures of the ground truth and the predicted gene expression images. The SSIM is an image similarity metric that is widely used for super-resolution tasks in computer vision51, 52 and medical imaging53. A higher SSIM indicates a higher degree of similarity between two images. In our context, the SSIM captures both global trends and the fine-grained spatial structures in the super-resolution gene expression images. Our experiments showed that iStar outperformed XFuse as measured by both the RMSE and SSIM.
In addition to RMSE and SSIM, Pearson correlation coefficient (PCC), which is an uncommon metric for super-resolution tasks54, 55, was employed in some previous works28, 56 on spatial transcriptomics as an evaluation criterion for gene expression prediction accuracy. However, these works studied spatial transcriptomics at low resolutions, where the number of spatial units (i.e. superpixels or spots) was no more than 2000, and the size of the spatial units was around . By contrast, the prediction accuracy in our experiments was evaluated at a much higher resolution, where the number of superpixels was as large as and their size was as small as . Due to the high image resolution and high noise magnitude in the ground truth, PCC is sensitive to outlying noisy superpixels, especially for sparsely expressed genes. In our experiments, compared to RMSE and SSIM, PCC had difficulties in differentiating superior and inferior super-resolution predictions when the resolution was high. As the resolution decreased, the noise level in the ground truth also decreased, which led to sharpened contrast between the accuracy of iStar and XFuse as measured by PCC. Furthermore, more spatially variables genes, which were associated with higher signal-to-noise ratio, produced higher PCCs and greater differences in PCC between iStar and XFuse, which again indicates the sensitivity of PCC to the noise level in the ground truth. Overall, PCC had limited power in differentiating super-resolution prediction accuracy for high-resolution, high-noise gene expression images. On the other hand, when the resolution and noise level were low, PCC produced results similar to those by RMSE and SSIM (Supplementary Figs. 23 and 24).
Computational efficiency
Computational efficiency was another area that iStar outperformed XFuse. In the benchmark experiments, iStar was approximately 200 times faster than XFuse. The end-to-end analysis of a typical dataset by iStar usually finishes within ten minutes, while XFuse took about a day. The detailed runtimes for training and prediction are reported in Supplementary Table 2. Experiments were conducted on an NVIDIA GeForce RTX 2080 Ti graphics card.
Extended Data
Extended Data Fig. 1. Correlation between super-resolution gene expression and histology.
Comparison of iStar’s super-resolution gene expression patterns with the paired histology image in the Xenium-derived pseudo-Visium data. Spot boundaries are highlighted.
Extended Data Fig. 2. Resolution enhancement at various scales.
Visualization of predicted super-resolution gene expressions by iStar at various scales of resolution enhancement for three breast cancer-related genes (ESR1, ERBB2, and PGR) in the Xenium-derived pseudo-Visium data.
Extended Data Fig. 3. Numerical evaluation of prediction accuracy.
Prediction accuracy of iStar and XFuse as measured by root mean squared error (RMSE) and structural similarity index measure (SSIM) for all the 313 genes in the Xenium-derived pseudo-Visium data obtained from a breast cancer patient. In each scatter plot, a dot represents a gene. In this analysis, Section 1 was first treated as the training sample in which in-sample prediction was perfromed for Section 1 and then out-of-sample prediction was perfromed for Section 2. We then repeated this analysis by treating Section 2 as the ‘in-sample’ and Section 1 as the ‘out-of-sample’. The evaluation metrics in’Average’ is the average of those in ‘Sections 1’ and ‘Section 2’.
Extended Data Fig. 4. Visualization of single-cell level gene expression prediction.

Visualization of single-cell level gene expression predicted by iStar for the pseudo-Visium breast cancer data derived from Xenium data. Shown on the left is the ground truth single-cell level gene expression directly measured by Xenium, and shown on the right is the single-cell level gene expression predicted by iStar. For each gene, the root mean squared error (RMSE) and Pearson’s correlation coefficient (PCC) between the prediction and the ground truth across the whole tissue and within the shown region are displayed.
Extended Data Fig. 5. Prediction accuracy evaluation of single-cell level gene expression prediction.
Prediction accuracy of iStar and XFuse for single-cell level gene expression prediction as measured by root mean squared error (RMSE) for all the 313 genes in the Xenium-derived pseudo-Visium data obtained from a breast cancer patient. In each scatter plot, a dot represents a gene. In this analysis, Section 1 was first treated as the training sample in which in-sample prediction was performed for Section 1 and then out-of-sample prediction was performed for Section 2. We then repeated this analysis by treating Section 2 as the ‘in-sample’ and Section 1 as the ‘out-of-sample’. The evaluation metrics in’Average’ is the average of those in ‘Sections 1’ and ‘Section 2’. We stratified cells by the quantiles of their cell size.
Extended Data Fig. 6. Super-resolution vs spot-level signature scoring of tertiary lymphoid structures (TLSs).
iStar detected tertiary lymphoid structures (TLSs) in the Anderson et al. (2021) HER2+ breast cancer dataset. Displayed are the predicted TLS scores by iStar and the original publication, along with the pathologist’s manual annotation reported in the original publication. Super-resolution was performed with 128x resolution enhancement.
Extended Data Fig. 7. Super-resolution gene expression prediction in the Xenium-derived pseudo-Visium mouse brain data.
Visualization of the spot-level training data, ground truth gene expression, and predicted super-resolution gene expressions by iStar and XFuse for 24 highly variable genes, whose variances are in the 80%−100% quantiles among all the 248 genes in the Xenium-derived pseudo-Visium data obtained from mouse brain. The variance quantiles of the genes in the order of top-left, top-right, bottom-left, bottom-right are equally spaced from 100% to 80% (in descending order). Super-resolution gene expressions are visualized at the scale of 8x resolution enhancement.
Extended Data Fig. 8. Prediction accuracy evaluation and gene-based segmentation comparison in the Xenium-derived pseudo-Visium mouse brain data.
a. Prediction accuracy of by iStar and XFuse as measured by root mean squared error (RMSE) and structural similarity index measure (SSIM) for all the 248 genes in the Xenium-derived pseudo-Visium data obtained from mouse brain. In each scatter plot, a dot represents one gene. b. Segmentation of the Xenium-derived pseudo-Visium data obtained from mouse brain by iStar and XFuse using all 248 genes available in this dataset. Super-resolution was performed with 128x resolution enhancement.
Extended Data Fig. 9. Gene-based segmentation of mouse brain datasets.
Analyses of a. mouse brain (coronal cut), b. mouse brain posterior (sagittal cut), and c. mouse brain ofactory bulb Visium datasets generated by 10x Genomics. For each dataset, iStar was applied to enhance the resolution of the top 1000 most highly variable genes. Super-resolution was performed with 128x resolution enhancement. Segmentations by iStar identified fine-grained tissue structures in the mouse brain and agreed with the Allen Brain Atlas annotations.
Extended Data Fig. 10. Gene-based tissue segmentation and tertiary lymphoid structure (TLS) signature scoring of cancer datasets and a mouse kidney dataset.
Comparison of gene-based segmentation by iStar with manual tissue annotation in a. mouse kidney and b. prostate cancer Visium datasets generated by 10x Genomics. c. Gene-based segmentation by iStar in a colorectal cancer Visium dataset by 10x Genomics. d. Comparison of tertiary lymphoid structure (TLS) signature score by iStar with manual TLS annotation in a kidney cancer Visium dataset generated by Meylan et al. (2022).
Supplementary Material
Acknowledgments
M.L. was supported by the following NIH grants: R01GM125301, R01EY030192, R01HL150359, and P01AG066597. E.B.L. was supported by NIH grant P01AG066597. L.W. was supported in part by NIH grant R01CA266280, the Cancer Prevention and Research Institute of Texas (CPRIT) award RP200385, the University Cancer Foundation via the Institutional Research Grant Program at the University of Texas MD Anderson Cancer Center, the Andrew Sabin Family Foundation, and the Break Through Cancer Foundation. We thank Drs. Maxime Meylan and Wolf Herman Fridman for sharing the kidney cancer histology image data. We also thank Drs. Erickson, Lamb, and Lundberg for sharing the prostate cancer histology image and clone annotation data.
Footnotes
Code Availability
The iStar algorithm was implemented in Python and is available on GitHub at
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Competing Interests
M.L. receives research funding from Biogen Inc. unrelated to the current manuscript. The other authors declare no competing financial interests.
Data Availability
We analyzed the following publicly available ST datasets: (1) 10x Xenium human breast cancer data (https://www.10xgenomics.com/products/xenium-in-situ/preview-dataset-human-breast); (2) 10x Xenium mouse brain data (https://www.10xgenomics.com/resources/datasets/fresh-frozen-mouse-brain-replicates-1-standard); (3) human HER2-positive breast cancer ST data reported in Anderson et al. (https://github.com/almaan/her2st); (4) 10x Visium human breast cancer data (https://www.10xgenomics.com/resources/datasets/human-breast-cancer-visium-fresh-frozen-whole-transcriptome-1-standard); (5) 10x Visium human colorectal cancer data (https://www.10xgenomics.com/resources/datasets/human-colorectal-cancer-whole-transcriptome-analysis-1-standard-1-2-0); (6) 10x Visium human prostate cancer data (https://www.10xgenomics.com/resources/datasets/human-prostate-cancer-adenocarcinoma-with-invasive-carcinoma-ffpe-1-standard-1-3-0); (7) Human prostate cancer data reported in Erickson et al. (https://doi.org/10.17632/svw96g68dv.1); (8) Human clear cell renal cell carcinoma primary tumors reported in Meylan et al. (GSE175540); (9) 10x Visium mouse kidney data (https://www.10xgenomics.com/resources/datasets/adult-mouse-kidney-ffpe-1-standard-1-3-0); (10) 10x Visium mouse brain coronal cut data (https://www.10xgenomics.com/resources/datasets/mouse-brain-coronal-section-2-ffpe-2-standard); (11) 10x Visium mouse brain sagittal cut posterior data (https://www.10xgenomics.com/resources/datasets/mouse-brain-serial-section-2-sagittal-posterior-1-standard); (12) 10x Visium mouse brain olfactory bulb data (https://www.10xgenomics.com/resources/datasets/adult-mouse-olfactory-bulb-1-standard-1). Details of the datasets analyzed in this paper were described in Supplementary Table 3. Gene expression visualizations for other spatial resolutions in the 10x Xenium breast cancer and mouse brain data are available at https://upenn.box.com/v/istar-results-benchmark.
References
- 1.Burgess DJ Spatial transcriptomics coming of age. Nat. Rev. Genet 20, 317 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Asp M, Bergenstrahle J. & Lundeberg J. Spatially Resolved Transcriptomes-Next Generation Tools for Tissue Exploration. Bioessays 42, e1900221 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Crosetto N, Bienko M. & van Oudenaarden A. Spatially resolved transcriptomics and beyond. Nat. Rev. Genet 16, 57–66 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Moor AE & Itzkovitz S. Spatial transcriptomics: paving the way for tissue-level systems biology. Curr. Opin. Biotechnol 46, 126–133 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Hu J. et al. SpaGCN: Integrating gene expression, spatial location and histology to identify spatial domains and spatially variable genes by graph convolutional network. Nat. Methods 18, 1342–1351 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Sun S, Zhu J. & Zhou X. Statistical analysis of spatial expression patterns for spatially resolved transcriptomic studies. Nat. Methods 17, 193–200 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svensson V, Teichmann SA & Stegle O. SpatialDE: identification of spatially variable genes. Nat. Methods 15, 343–346 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dries R. et al. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 22, 78 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pham D. et al. stLearn: integrating spatial location, tissue morphology and gene expression to find cell types, cell-cell interactions and spatial trajectories within undissociated tissues. Preprint at bioRxiv, 10.1101/2020.05.31.125658 (2020). [DOI] [Google Scholar]
- 10.Asp M. et al. A Spatiotemporal Organ-Wide Gene Expression and Cell Atlas of the Developing Human Heart. Cell 179, 1647–1660 e1619 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Wang X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M. & Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 11, 360–361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah S, Lubeck E, Zhou W. & Cai L. In Situ Transcription Profiling of Single Cells Reveals Spatial Organization of Cells in the Mouse Hippocampus. Neuron 92, 342–357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eng CL et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568, 235–239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moffitt JR et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362, eaau5324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen KH, Boettiger AN, Moffitt JR, Wang S. & Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stickels RR et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol 39, 313–319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792 e1721 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Badea L. & Stanescu E. Identifying transcriptomic correlates of histology using deep learning. PLoS One 15, e0242858 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ash JT, Darnell G, Munro D. & Engelhardt BE Joint analysis of expression levels and histological images identifies genes associated with tissue morphology. Nat. Commun 12, 1609 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmauch B. et al. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat. Commun 11, 3877 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen RJ et al. Scaling vision transformers to gigapixel images via hierarchical self-supervised learning. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 16144–16155 (2022). [Google Scholar]
- 23.Liu Z. et al. Swin transformer: hierarchical vision transformer using shifted windows. Proceedings of the IEEE/CVF international conference on computer vision, 10012–10022 (2021). [Google Scholar]
- 24.Han K. et al. Transformer in transformer. Advances in Neural Information Processing Systems 34, 15908–15919 (2021). [Google Scholar]
- 25.Caron M. et al. Emerging properties in self-supervised vision transformers. Proceedings of the IEEE/CVF international conference on computer vision, 9650–9660 (2021). [Google Scholar]
- 26.Bao H, Dong L, Piao S. & Wei F. Beit: Bert pre-training of image transformers. Preprint at arXiv, 10.48550/arXiv.2106.08254 (2021). [DOI] [Google Scholar]
- 27.Janesick A. et al. High resolution mapping of the breast cancer tumor microenvironment using integrated single cell, spatial and in situ analysis of FFPE tissue. Preprint at bioRxiv, 10.1101/2022.10.06.510405 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergenstråhle L. et al. Super-resolved spatial transcriptomics by deep data fusion. Nat. biotechnol 40, 476–479 (2022). [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Bovik AC, Sheikh HR & Simoncelli EP Image quality assessment: from error visibility to structural similarity. IEEE transactions on image processing 13, 600–612 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Hamerly G. & Elkan C. Learning the k in k-means. Advances in neural information processing systems 16 (2003). [Google Scholar]
- 31.Andersson A. et al. Spatial deconvolution of HER2-positive breast cancer delineates tumor-associated cell type interactions. Nat. Commun 12, 6012 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stahl PL et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Wu SZ et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. genetics 53, 1334–1347 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sautes-Fridman C, Petitprez F, Calderaro J. & Fridman WH Tertiary lymphoid structures in the era of cancer immunotherapy. Nat. Rev. Cancer 19, 307–325 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Fridman WH et al. B cells and tertiary lymphoid structures as determinants of tumour immune contexture and clinical outcome. Nat. Rev. Clin. Oncol 19, 441–457 (2022). [DOI] [PubMed] [Google Scholar]
- 36.Helmink BA et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 577, 549–555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petitprez F. et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 577, 556–560 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Cabrita R. et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 577, 561–565 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Lin JR et al. Multiplexed 3D atlas of state transitions and immune interaction in colorectal cancer. Cell 186, 363–381 e319 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 40.Steiner A. et al. How to train your vit? data, augmentation, and regularization in vision transformers. Preprint at arXiv, 10.48550/arXiv.2106.10270 (2021). [DOI] [Google Scholar]
- 41.Hölscher DL et al. Next-Generation Morphometry for pathomics-data mining in histopathology. Nat. Commun 14, 470 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rappez L. et al. SpaceM reveals metabolic states of single cells. Nat. Methods 18, 799–805 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu B, Wang N, Chen T. & Li M. Empirical evaluation of rectified activations in convolutional network. Preprint arXiv, 10.48550/arXiv.1505.00853 (2015). [DOI] [Google Scholar]
- 44.Clevert D-A, Unterthiner T. & Hochreiter S. Fast and accurate deep network learning by exponential linear units (ELUs). Preprint at arXiv, 10.48550/arXiv.1511.07289 (2015). [DOI] [Google Scholar]
- 45.Ringnér M. What is principal component analysis? Nat biotech 26, 303–304 (2008). [DOI] [PubMed] [Google Scholar]
- 46.McInnes L, Healy J. & Melville J. Umap: Uniform manifold approximation and projection for dimension reduction. Preprint at arXiv, 10.48550/arXiv.1802.03426 (2018). [DOI] [Google Scholar]
- 47.Schurch CM et al. Coordinated Cellular Neighborhoods Orchestrate Antitumoral Immunity at the Colorectal Cancer Invasive Front. Cell 182, 1341–1359 e1319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aran D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunology 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu J. et al. Iterative transfer learning with neural network for clustering and cell type classification in single-cell RNA-seq analysis. Nat machine intelligence 2, 607–618 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu Y. The level weighted structural similarity loss: A step away from MSE. Proceedings of the AAAI Conference on Artificial Intelligence 33, 9989–9990 (2019). [Google Scholar]
- 51.Lai W-S, Huang J-B, Ahuja N. & Yang M-H Deep laplacian pyramid networks for fast and accurate super-resolution. Proceedings of the IEEE conference on computer vision and pattern recognition, 624–632 (2017). [Google Scholar]
- 52.Dahl R, Norouzi M. & Shlens J. Pixel recursive super resolution. Proceedings of the IEEE international conference on computer vision, 5439–5448 (2017). [Google Scholar]
- 53.Masutani EM, Bahrami N. & Hsiao A. Deep learning single-frame and multiframe super-resolution for cardiac MRI. Radiology 295, 552–561 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anwar S, Khan S. & Barnes N. A deep journey into super-resolution: A survey. ACM Computing Surveys (CSUR) 53, 1–34 (2020). [Google Scholar]
- 55.Wang Z, Chen J. & Hoi SC Deep learning for image super-resolution: A survey. IEEE transactions on pattern analysis and machine intelligence 43, 3365–3387 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Ma Y. & Zhou X. Spatially informed cell-type deconvolution for spatial transcriptomics. Nat. Biotechnol 40, 1349–1359 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We analyzed the following publicly available ST datasets: (1) 10x Xenium human breast cancer data (https://www.10xgenomics.com/products/xenium-in-situ/preview-dataset-human-breast); (2) 10x Xenium mouse brain data (https://www.10xgenomics.com/resources/datasets/fresh-frozen-mouse-brain-replicates-1-standard); (3) human HER2-positive breast cancer ST data reported in Anderson et al. (https://github.com/almaan/her2st); (4) 10x Visium human breast cancer data (https://www.10xgenomics.com/resources/datasets/human-breast-cancer-visium-fresh-frozen-whole-transcriptome-1-standard); (5) 10x Visium human colorectal cancer data (https://www.10xgenomics.com/resources/datasets/human-colorectal-cancer-whole-transcriptome-analysis-1-standard-1-2-0); (6) 10x Visium human prostate cancer data (https://www.10xgenomics.com/resources/datasets/human-prostate-cancer-adenocarcinoma-with-invasive-carcinoma-ffpe-1-standard-1-3-0); (7) Human prostate cancer data reported in Erickson et al. (https://doi.org/10.17632/svw96g68dv.1); (8) Human clear cell renal cell carcinoma primary tumors reported in Meylan et al. (GSE175540); (9) 10x Visium mouse kidney data (https://www.10xgenomics.com/resources/datasets/adult-mouse-kidney-ffpe-1-standard-1-3-0); (10) 10x Visium mouse brain coronal cut data (https://www.10xgenomics.com/resources/datasets/mouse-brain-coronal-section-2-ffpe-2-standard); (11) 10x Visium mouse brain sagittal cut posterior data (https://www.10xgenomics.com/resources/datasets/mouse-brain-serial-section-2-sagittal-posterior-1-standard); (12) 10x Visium mouse brain olfactory bulb data (https://www.10xgenomics.com/resources/datasets/adult-mouse-olfactory-bulb-1-standard-1). Details of the datasets analyzed in this paper were described in Supplementary Table 3. Gene expression visualizations for other spatial resolutions in the 10x Xenium breast cancer and mouse brain data are available at https://upenn.box.com/v/istar-results-benchmark.